
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 22 November 2023
Sec. Environmental Informatics and Remote Sensing
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1324932
This article is part of the Research TopicMonitoring, Early Warning and Mitigation of Natural and Engineered Slopes, volume IIIView all 17 articles
Introduction: Eukaryotic plankton plays crucial roles in ecosystem processes, impacting aquatic ecosystem stability. This study focuses on Danjiangkou Reservoir, a canyon lake in central China, that acts as the water source of the Mid-route of the South-to-North Water Diversion Project.
Methods: In this study, high-throughput 18S rDNA gene sequencing was employed to investigate eukaryotic plankton community at four water depths (0.5 m, 5 m, 10 m, and 20 m). The environmental factors including pH, water temperature (WT), nitrate nitrogen (NO3−-N), ammonia nitrogen (NH4+-N), total nitrogen (TN), conductivity (Cond), and dissolved oxygen (DO) in reservoir areas were measured, and their correlations with abundance and diversity of eukaryotic plankton were analyzed.
Results: The results showed the presence of 122 genera of eukaryotic plankton from 38 phyla. Eukaryotic plankton communities were mainly composed of Eurytemora, Thermocyclops, Sinocalanus, Mesocyclops, and Cryptomonas. In particular, significant differences in the diversity of eukaryotic plankton communities were found in vertical distribution. The diversity and abundance of eukaryotic plankton communities in 7 sampling sites decreased with the increase of depth from 0.5 to 10 m, while the diversity and abundance of plankton communities increased at 20 m. RDA analysis indicated that pH, depth, WT, NH4+-N, DO, Cond, and NO3−-N could influence the vertical distribution of the eukaryotic plankton community in the Danjiangkou Reservoir. Among these eukaryotic plankton, Eurytemora, Thermocyclops, and Volvox were negatively correlated with pH and WT and positively correlated with depth.
Discussion: This study revealed a novel perspective on the distribution of the eukaryotic plankton community in Danjiangkou Reservoir, particularly in terms of vertical variation, which will be helpful to comprehensively understand ecological processes and to further ensure the water quality safety in this canyon-style reservoir.
Eukaryotic plankton is important in aquatic communities and plays crucial roles in material cycling and energy flow in aquatic ecosystems (Zubkov and Tarran, 2008; Jiang et al., 2012; Filker et al., 2016). Based on niche theory, environmental variables such as resource availability and abiotic factors could determine species composition in eukaryotic plankton communities. Variations of the eukaryotic plankton community are related to the physicochemical characteristics and biological conditions of the water bodies at space and time scales (Ishida, 2008; MaChado et al., 2019). The study of eukaryotic plankton diversity and community characteristics can indirectly reflect the water quality of the ecological environment. Therefore, studying the distribution characteristics of eukaryotic plankton communities is vital for understanding the characteristics of water ecosystem processes, formulation of scientific and conservation management measures.
In the past decades, studies on the planktonic eukaryotes community mainly depended on microscopy observation. However, this method is not only influenced by sampling conditions and preservation techniques but also there is great variation and disagreement in the identification of these organisms. With the development of biotechniques (Marianne et al., 2015), the diversity of plankton can be assessed by high-throughput sequencing. For eukaryotic plankton high-throughput sequencing of specific PCR products (e.g., eukaryotic 18S rDNA genes) on such platforms can be used to obtain information on eukaryotic community structure, evolutionary relationships, and correlations between eukaryotic plankton and the environment (Sun et al., 2014; Gao et al., 2018; McInnes et al., 2019; Keck et al., 2020). Currently, high-throughput sequencing has been widely used to study plankton community structure in aquatic ecosystems (Zhao et al., 2019; Liu et al., 2020a; Zhong et al., 2021; Wu et al., 2022). Many studies have focused on the community, distribution, and function of phytoplankton, archaea, plant, bacteria, and animal taxa, and their relationship with water quality (Wurzbacher et al., 2010; Nagano and Nagahama, 2012; Sun et al., 2014; Piwosz et al., 2020). High-throughput sequencing technology provides efficient and rapid assessment with specific DNA fragments and provides a more comprehensive profiling of community composition than other methods (Dijk et al., 2014). Its application in water quality monitoring has attracted increasing attention (BoonFei et al., 2015).
Danjiangkou Reservoir, located in Danjiangkou City, Hubei Province and Xichuan County, Henan Province, is the water source of the Mid-route of the South-to-North Water Diversion Project in central China. Benefit from this project, more than 20 cities along the route are supplied with abundant water resources for agriculture, industry, and human consumption. It is important to ensure the safety of water quality in Danjiangkou Reservoir. According to the data provided by the Ecological Environment Department of Henan Province in 2018–2022, the water quality of Danjiangkou Reservoir has reached the standard of class I or II water. Danjiangkou Reservoir is a canyon-style reservoir with a maximum storage capacity of 29.05 billion m3 and a maximum depth of 80 m. As an important water source, many studies have investigated the eukaryotic plankton community structure in surface water and its influencing factors in Danjiangkou Reservoir at temporal and spatial scales (Shen et al., 2011; Tan et al., 2011; Wang et al., 2016; Zheng et al., 2018). Wang et al. (2016) detected a total of 66 phytoplankton species belonging to 7 phyla and 38 species in the reservoir area and found that diatoms were the dominant phyla in spring, autumn, and winter. Furthermore, the results of their study showed that dissolved oxygen, pH, and phosphorus concentration were the main environmental factors affecting the composition of phytoplankton communities. Through six continuous monitoring of periphytic algae in Danjiangkou Reservoir, Zheng et al. (2018) found the community was assigned to 6 phyla and 46 genera, and observed significant spatial and temporal differences of them. He et al. (2021) detected 6 phyla and 57 genera of phytoplankton from the samples collected at 1.5 m, 5 m and 10 m in the reservoir area, and diatoms and green algae were the dominant phyla. Recently, Cui et al. (2023) analyzed the spatial and temporal variations of physicochemical indicators and phytoplankton at seven different water depths in four seasons in Danjiangkou Reservoir to reveal the main factors affecting the vertical distribution of phytoplankton. Additionally, the characteristics of phytoplankton community change in Danjiangkou Reservoir in different seasons and spaces were analyzed, as well as the main environmental factors affecting the distribution of phytoplankton community structure (Zhang et al., 2022; Xiao et al., 2023). However, these studies mainly focused on the eukaryotic plankton community in shallow water column, and the patterns in deep water of this canyon-style reservoir are still limited.
In this study, high-throughput sequencing of 18S rDNA V4 regions was used to investigate eukaryotic plankton community structure in four layers (0.5 m, 5 m, 10 m, and 20 m) of water in Danjiangkou Reservoir. Especially, stratified sampling provided a more comprehensive understanding of eukaryotic plankton distribution in Danjiangkou Reservoir. Our main goals were to analyze the vertical structure of the eukaryotic plankton community in deep water at the Danjiangkou Reservoir, and to reveal the relationship between the eukaryotic plankton community and environmental factors.
According to the overall structural characteristics of the reservoir area, and the basis of previous research methods (Wang et al., 2017; Ferrera et al., 2020; Lee et al., 2018; Song et al., 2019; Liu et al., 2020b), seven sampling sites including Songgang (SG), Tumen (TM), Heijizui (HJZ), Kuxin (KX), Dangzikou (DZK), Wulongquan (WLQ), and Qushou (QS) were set up in the Danjiangkou Reservoir (Figure 1). In June 2021, the samples at 0.5 m, 5 m, 10 m, and 20 m depth of each sampling site were collected respectively for investigating the vertical distribution patterns of the eukaryotic plankton community. A total of 18 L water samples were collected using a Plexiglas water collector, passed through a 0.22 μm filter membrane, and transferred to a 1.5 mL sterile centrifuge tube to be stored in liquid nitrogen for DNA extraction.
The water temperature (WT), pH, dissolved oxygen (DO), and conductivity (Cond) were measured on-site using a portable multifunctional water quality parameter meter. 0.5 L of water samples were taken from each layer and put into polyethylene bottles to be brought back to the laboratory. The physicochemical properties of total nitrogen (TN), ammonia nitrogen (NH4+-N), and nitrate nitrogen (NO3−-N) in the water were determined according to previously described methods (Jin, 1990).
DNA was extracted from the samples using the DNeasy Power Water Kit (Mo Bio/QIAGEN) according to the manufacturer’s instructions. The extracted DNA was analyzed with a fluorescence spectrophotometer (QuantifluorST fluorometer, Promega, E6090; QuantiT PicoGreen dsDNA Assay Kit, Invitrogen, P7589) and 1% agarose gel electrophoresis to determine concentration and purity. V4 hypervariable region of 18S rDNA was amplified using the specific primers, namely 547F (FCCAGCASCYGCGGTAATTCC) and V4R (ACTTTCGTTCTTGATYRA) (Salmaso et al., 2020). Amplification was carried out in 20 μL reactions with 5×reaction buffer 5 μL, 5×GC buffer 5 μL, dNTP (2.5mM) 2 μL, forward primer (10μM) 1 μL, reverse primer (10uM) 1 μL, DNA Template 2 μL, ddH2O 8.75 μL, Q5 DNA Polymerase 0.25 μL as the template. Thermal cycling consisted of an initial of 98°C pre-denaturation for 2 min, 30 cycles (denaturation at 98°C for 15 s, annealing at 55°C for 30 s, extension at 72°C for 30 s), and a final extension at 72°C for 10 min. (PCR instrument: ABI Model 2720) The PCR products were isolated and purified using AxyPrep DNA Gel Extraction Kit, Axygen, AP-GX-500. Paired-end sequencing of the amplicon was performed with the Illumina Novaseq platform. The raw sequences were deposited in NCBI under the Bio Project PRJNA (No.782248).
After quality control, denoising, splicing, and chimera removal with QIIME2 (Quantitative Insights Into Microbial Ecology) software, the best taxonomic unit (Operational Taxonomic Unit) was classified for high-quality sequences at a classification criterion of ≥97% similarity (default is a species-level similarity). The sequenced OTUs results were compared with the Silva 132 rRNA database using the Classify sklearn algorithm of QIIME2 (Chao, 1984; Bokulich et al., 2013) (https://github.com/QIIME2/q2-feature-classifier) for OTUs representative sequence in the QIIME2 software with default parameters, using a pre-trained Naive Bayes classifier for species annotation.
Alpha diversity analysis was performed using QIIME2 software to construct rarefaction curves, the Chao index, Shannon’s index and so on. The Chao (Chao, 1984) and Observed-Species indices were used to characterize richness, the Shannon-Winer (Shannon, 1948) and Simpson (Simpson, 1997) indices to characterize diversity, the Pielou-evenness (Pielou, 1966) index for evenness, and Good’s coverage (Good, 2010) index for cover. Beta diversity was calculated using BrayCurtis and unweighted UniFrac distance matrices. PCoA was used to analyze the similarity at the taxonomic level of the eukaryotic plankton phylum in the vertical direction at each sampling. The relevant data were plotted in Excel and Origin 9.0. Redundancy analysis (RDA) and correlation analysis of the main eukaryotic plankton with environmental factors were performed using genescloud. (https://www.genescloud.cn).
A total of 1,972,914 original sequences were obtained, with an average length of 420 bp. After removal of nontargets and the singletons, a total of 1,645,985 high-quality sequences were obtained in all samples, accounting for 83.4% of original sequences. The rarefaction curve gradually became stable when the sequencing reads reached 25000, which indicated that the number of sequencings was sufficient, and the taxon richness was high enough to cover all taxa for further analysis (Figure 2). All sequences were clustered at the 97% similarity level and 2392 OTUs were obtained from all the samples.
A total of 38 phyla of eukaryotic plankton were identified. The main eukaryotic plankton taxa at the level of phylum in the seven sites were Arthropoda (85.3%), Chordata (4.0%), Chlorophyta (1.6%), while other plankton such as Haptophyceae, Rotifera and Dinophyceae were less than 1% (Figure 3). Obviously, Arthropoda is the dominant species in the reservoir area.
A total of 122 genera of eukaryotic plankton were detected in the 28 samples, all of which contained taxa that could not be identified at the genus level. The eukaryotic plankton community at different depths (0.5 m, 5 m, 10 m, 20 m) comprised 10 higher-level taxon groups (Figure 4). Notably, the relative abundance of major taxa exhibited significant variation across these depth levels. Eurytemora dominated at depths from 0.5 to 10 m, while Thermocyclops was the dominant species at 20 m. Interestingly, the abundance and species richness of eukaryotic plankton were higher at the 20 m depth, while the other depths showed lower abundance and species richness of eukaryotic plankton. Overall, there were distinct differences in the relative abundance and diversity of eukaryotic plankton at various depths.
There was over 99% coverage of sequences at each sample site, fully reflecting the species and structure of the eukaryotic plankton community in the reservoir region. The average number of OTUs (393) at the 20 m depth water was the largest, and the average number of OTUs (265) at the 5 m depth water was the smallest. The eukaryotic plankton α-diversity of the sampling sites with different depths varied greatly (Table 1). The Observed-Species index and Chao index ranged from 86 to 673 and 89 to 708, respectively, the QS4 (Observed-Species 676 and Chao 713) had the highest richness indices, which were much higher compared to other samples. The Simpson index and Shannon-Wiener index have maximum values in SG1, which had the greatest species diversity, and the minimum values of the Simpson index and Shannon-Wiener index were at HJZ2. Pielou-evenness in this study was 0.07–0.68, indicating that the distribution of the number of OUTs varied widely among the samples.
β-diversity analysis showed that different eukaryotic plankton species have unique vertical distribution patterns in Danjiangkou Reservoir (Figure 5). The result showed that the community composition of KX, WLQ and QS at 0.5 m depth water was relatively consistent, and SG, TM, HJZ, DZK each formed a group. At 5 m depth water, HJZ, KX and DZK formed a group with higher similarity, and the remaining four sampling sites each formed a group. At 10 m depth water, KX, DZK and HJZ were more similar as a group, TM and WLQ formed a group, and SG had the same eukaryotic plankton community structure as QS. At 20 m depth water, KX, WLQ, TM and QS formed a group, DZK and HJZ were more similar as a group, and SG formed its own group. The results indicated that the community composition of all sampling sites varied greatly at 0.5–20 m depth water.
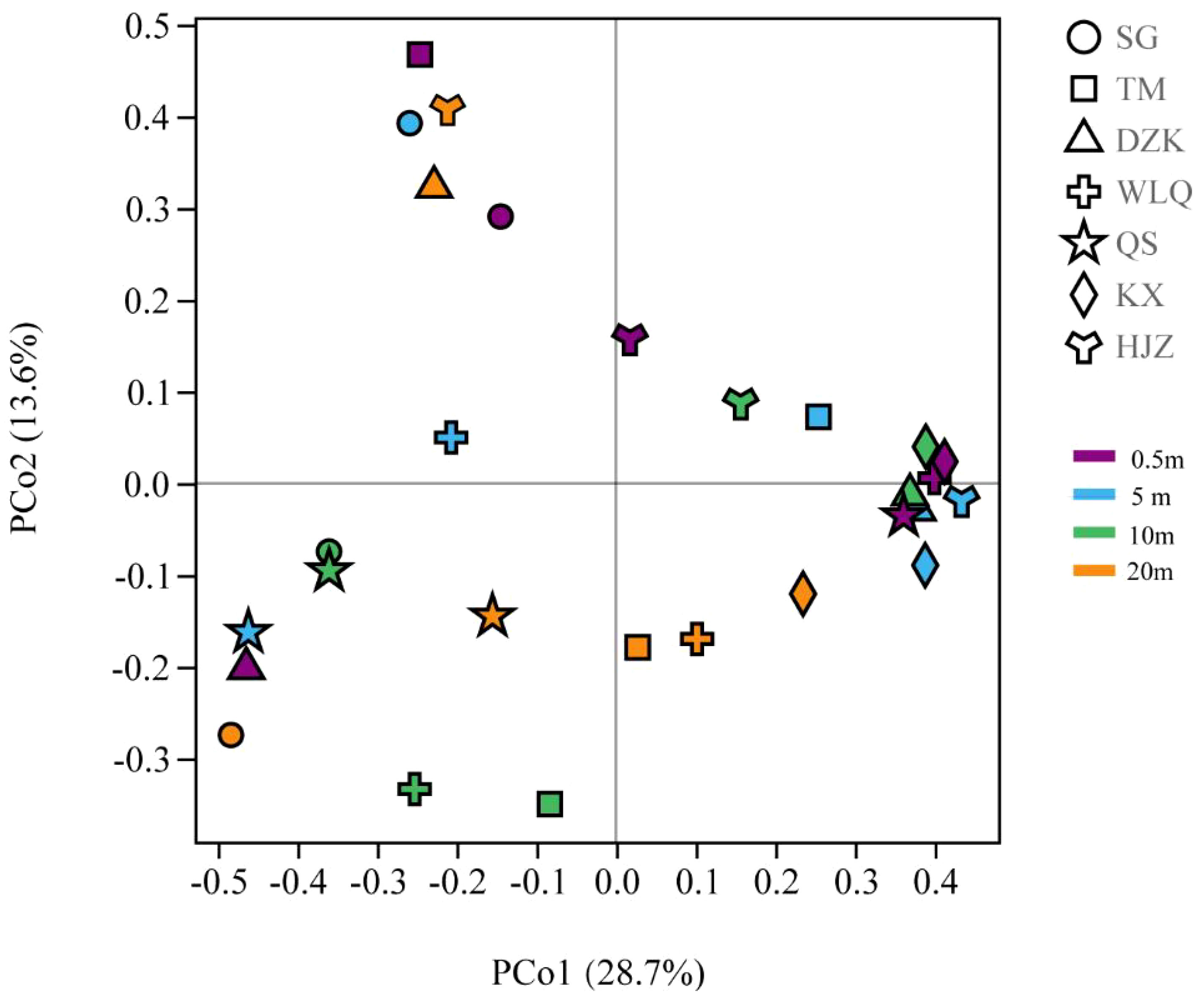
Figure 5 PCoA analysis of eukaryotic plankton community diversity. The percentage in the horizontal and vertical coordinate brackets indicates the proportion of sample difference data that can be explained by the corresponding coordinate axis. The closer the two points are on the coordinate axis, the more similar the community composition of the two samples is.
To further compare differences in species composition between samples, and to achieve a demonstration of trends in species abundance distribution across samples, species composition analysis of all sites could be performed using a heat map (Figure 6). The cross-sectional comparison showed the differences between samples, taking Eurytemora as an example, at the 0.5 m depth water, Eurytemora was the most abundant in DZK, at the 5 m depth water, Eurytemora was the most abundant in QS, at the 10 m depth water, Eurytemora was the most abundant in WLQ, followed by QS and TM, at the 20 m depth water, Eurytemora was the most abundant in SG, followed by QS and TM. The vertical comparison showed the distribution of species within the same sample and the degree of similarity between samples, for example, SG species diversity was the most abundant at 0.5 m depth water, containing Phacotus, Chlamydomonas, Rimostrombidium, Dinobryon, etc.
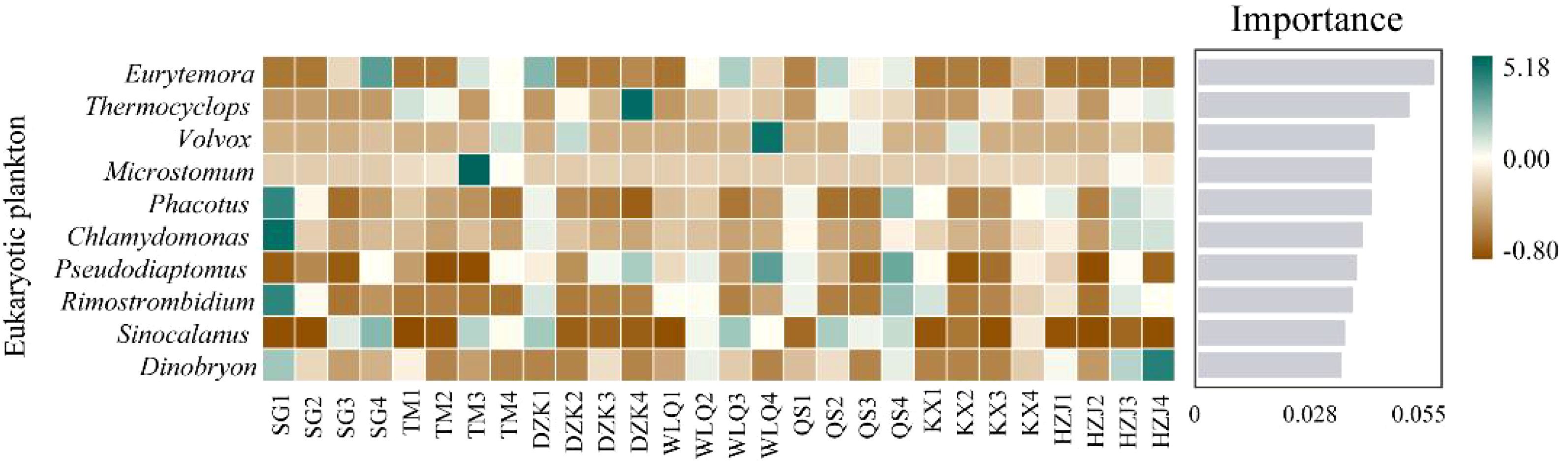
Figure 6 Heatmap of eukaryotic plankton. The figure represents a heatmap of eukaryotic plankton at 0.5 m, 5 m, 10 m, and 20 m depth water. The high and low-abundance species were clustered in blocks, and the color gradient was used to reflect similarities and differences in community composition among the 28 samples at the genus levels. The length of the gray bar indicates the importance of the species to the sample, decreasing from top to bottom.
TN, NH4+-N, DO and pH were measured at the seven sampling sites of Danjiangkou Reservoir (Table 2). The result showed that all sites met the requirement of class I water standards, except for TN (0.68~1.15 mg·L−1). NH4+-N (0.08~0.15 mg·L−1), DO (7.19~8.57 mg·L−1), pH (8.66~8.96) and Cond (274~308 μs·cm−1) had no significant change at all sampling sites. WT at QS (mean value 24.5°C) was higher than other sites (mean value 22.3~24°C). And it was obvious that WT decreased with increasing depth. Remarkably, NO3−-N concentration at depth water was conspicuously higher than those at the surface sample sites. For example, NO3−-N concentration was 0.53 mg·L−1 at SG1, 0.79 mg·L−1 at SG2, 0.93 mg·L−1 at SG3 and 0.98 mg·L−1 at SG4. NO3−-N concentration at TM increased from 0.41 mg·L−1 (0.5 m) to 0.98 mg·L−1 (20 m). Overall, NO3−-N concentration increased with depth at all sampling sites.
The effects of environmental factors in different samples on the eukaryotic plankton communities were analyzed by RDA (Figure 7). The results showed that the percentage of variance explained by the first and second axes were 25.84% and 7.16%, respectively. According to P-values, pH (0.035) was the most important environmental variation factor for the eukaryotic plankton community, followed by depth (0.064), WT (0.097), NH4+-N (0.163), DO (0.195), Cond (0.231), NO3−-N (0.283). Eurytemora, Thermocyclops, and Volvox were negatively correlated with pH and WT and positively correlated with depth.
Microeukaryotes play many roles as primary producers, secondary producers, and decomposers in aquatic ecosystems, affecting the structure of aquatic food webs in terms of taxonomic composition, abundance, biomass, and biodiversity (Tan et al., 2010). However, there are relatively few studies on the composition, distribution characteristics, functions of eukaryotic plankton and their relationship with water quality (Voronin, 2010; Nagano and Nagahama, 2012; Taylor and Cunliffe, 2016). In this study, we used high-throughput sequencing technology to amplify the 18S rDNA V4 region DNA to investigate the community structure of plankton in Danjiangkou Reservoir. As a result, a total of 2392 OTUs belong to 38 phyla and 122 genera of eukaryotic plankton were identified. Based on sequence data, many differences in the diversity of eukaryotic plankton communities were found at a spatial scale, and the environmental factors including pH, depth and WT could influence the vertical distribution of eukaryotic plankton communities. This study would provide a better understanding dynamics of eukaryotic plankton communities in this important water source.
In this study, we found that several taxa including Eurytemora, Thermocyclops, Sinocalanus, Mesocyclops, Cryptomonas, Volvox, Chrysochromulina play an important role in the genus level of eukaryotic plankton in Danjiangkou Reservoir (Figure 4). Among them, Eurytemora, Thermocyclops, Sinocalanus, and Mesocyclops were reported as common species in still-water bodies (Hu et al., 2021). Water quality, water temperature, and hydrological factors affected the phytoplankton community structure (Yan et al., 2021). Some studies have pointed out that Chrysophyta was more suitable for growing in oligotrophic water bodies. Cryptophyta were the dominant species in mesotrophic water bodies, while Cholorophyta were the dominant species in eutrophic water bodies (Jia et al., 2019). In this study, the relative abundance of Cryptomonas, Volvox, Chrysochromulina, and Plagioselmis was ranked in decreasing order. It can be inferred that the degree of eutrophication in the reservoir area was between mesotrophic and oligotrophic water bodies, which was similar to the research conclusions of previous studies on Danjiangkou Reservoir (Wang et al., 2012; Jia et al., 2019). The excellent water quality in the reservoir area allowed Cryptomonas to be one of the dominant species. As an important environmental factor, water temperature could affect the composition of plankton community structure in most water bodies (Yang et al., 2014; Zhang et al., 2019; Wang et al., 2020). For example, Trombetta et al. (2019) reported that the growth temperature of Cholorophyta is higher than that of Cryptophyta. In this study, our results indicated that the reservoir water temperature (Table 2) was favorable for the growth and reproduction of Cryptophyta, which was consistent with the previous study. In addition, Cryptophyta were positively correlated with flow, and Cholorophyta were not significantly correlated with hydrological factors. During the investigation, the water level, flow rate, and flow velocity in the reservoir area were low. The excellent water quality, suitable water temperature, and suitable hydrological conditions probably resulted in a planktonic community composition dominated by Eurytemora, Thermocyclops, Sinocalanus Mesocyclops Cryptomonas Volvox, etc in the reservoir area.
The clustering results of the plankton community structure in each layer of each sampling site in the reservoir area were different (Figure 5). It was found that ecological factors such as season, environmental factors, and sampling sites in the Danjiangkou Reservoir area together explained 39% of the variation in phytoplankton structure, with geographical location accounting for 12% (Yan et al., 2021). Different geographical conditions cause changes in the characteristics of the aquatic environment, leading to spatial differences in the major taxa of plankton. Similar to our results, previous studies reported differences in eukaryotic microbial community structure among sampling sites in Xiamen, Qingdao (Zhang et al., 2018), and Yellowstone Park in the United States (Meadow and Zabinski, 2012). Moreover, there are also quite differences in the diversity of planktonic eukaryotes at different temporal and spatial scales in Danjiangkou Reservoir. KX is located in the center of the Reservoir, with fewer human interference factors and closer to the natural state. QS, KX and WLQ are located near the hilly area in the south of the reservoir, which is upstream of the reservoir area. HJZ is near the downstream tributary. TM is close to the northern dry land. DZK is near the Hanjiang River outlet. SG is closer to the paddy fields and human activities are more frequent. The differences in environmental factors and hydrological conditions were responsible for the differences in eukaryotic plankton community composition among the sampling sites in the reservoir area.
The diversity and abundance of eukaryotic plankton communities in 7 sampling sites decreased with the increase of depth ranging from 0.5 to 10 m, while the diversity and abundance of plankton communities increased at 20 m (Table 1). Notably, our results showed that NO3−-N concentration was positively correlated with the depth of water. As a main nitrogen resource in ecosystems, NO3−-N had a strong effect on the abundance of eukaryotic plankton was reported by investigating eukaryotic plankton communities in the South Yellow Sea (Sun et al., 2021). Relevant studies have found that the nitrate concentration increases with depth due to the sinking of nutrient substances (De Queiroz et al., 2015). Furthermore, the nitrate was absorbed and utilized by primary producers in the surface water (Wei et al., 2018), leading to the low NO3−-N concentration in the surface layer and the high NO3−-N concentration in the deep layer. In deep water, nutrient concentrations change with depth, resulting in a corresponding change in phytoplankton community structure (Tavernini et al., 2005). In this study, the abundance and diversity of eukaryotic plankton communities at 20 m (mean value of OTUs is 393) are higher than those at 0.5–10 m (mean value of OTUs is 265–374) (Table 1). As nitrate increased with depth, the maximum biomass of eukaryotic plankton in deep water was expected (Leal et al., 2009).In deep water bodies, with the gradient of environmental factors such as light, temperature, and nutrient salinity, the peak biomass of the phytoplankton community appeared at different depths (Huovinen, 1999; Ptacnik et al., 2003). In August 2008, it was found that the abundance of phytoplankton in most stations of Danjiangkou Reservoir decreased with the increase of depth, but there was no obvious regularity in other sampling months (Yin et al., 2011). There is no obvious distribution trend of phytoplankton richness and diversity in the range of 0–50 m in the Three Gorges Reservoir. It has been reported that environmental variability explained only 30% of the seasonal succession of microbiota in the eastern English Channel (Logares et al., 2014). The differences in plankton communities at different depths were attributed to the synergistic effects of different environmental factors (Nabout et al., 2009; Zheng et al., 2020). Because of the stratification phenomenon, the upper layers of water had the advantage of nutrient concentrations and temperature. In contrast, the deeper layer of water had extreme environmental conditions such as higher nutrient concentrations, low light, and low temperature, resulting in significant differences in species diversity (Sun et al., 2021). Based on the variation of Chao, Pielou-evenness, Shannon-Wiener, and Simpson index at all sampling sites (Table 1), changes in eukaryotic plankton abundance and diversity in different water samples (Figure 6) indicated that there were large differences in the vertical distribution of eukaryotic plankton communities in Danjiangkou Reservoir. Previous studies have found that there is no significant difference in the abundance of phytoplankton communities in the vertical direction of Danjiangkou Reservoir (Yin et al., 2011). The results of this study are different from the results of previous studies. The first reason is the expansion of the reservoir area. The previous studies were performed time was from 2007 to 2008, and the reservoir area was expanded in 2012. The hydrological conditions and physical and chemical parameters of the reservoir area changed after the expansion, resulting in a corresponding change in the composition of the plankton community in the reservoir area (Hoyer et al., 2009). The second reason is the identification method. The results of plankton characteristics obtained by high-throughput sequencing technology are more comprehensive than those derived from microscopic observations in previous studies (Dijk et al., 2014; Wang et al., 2017).
There is a certain spatial heterogeneity in the ecological environment of the Danjiangkou Reservoir. It is widely accepted that there is a close relationship between plankton composition and environmental factors (Chalar, 2009; Cudowski et al., 2015; Reich et al., 2017). In this study, the RDA showed that pH was the most important environmental factor affecting the community distribution of eukaryotic plankton in the Danjiangkou Reservoir, followed by depth, WT, NH4+-N, DO, Cond and NO3−-N (Figure 7). It can be indicated from RDA that most of the eukaryotic plankton were negatively correlated with pH, which may result from rotifer species increased and decreased in acidic water bodies, in contrast, the opposite situations occurred in alkaline water bodies. Acidic, neutral, and weakly alkaline water bodies are suitable for the survival of Cladocera, and alkaline water bodies are favorable for the survival of Copepods (He et al., 2022). In the water with low pH, the growth of phytoplankton will be limited, and the number of individuals will decrease. The alkaline water with high pH is more conducive to the photosynthesis of phytoplankton to form organic matter, and the number of phytoplankton reproduction will increase (Liu et al., 2010). If the pH value is in the range of 7.5 to 9.0, it is more favorable for the growth of diatoms and cyanobacteria. When the temperature is higher in summer, the suitable pH range is most conducive to the reproduction of algae and the formation of large-scale blooms (Lu, 1987). On the other hand, phytoplankton photosynthesis absorbs carbon dioxide in the water body, changes the pH value of the water body, and the activities of algae themselves will also change the pH value of the water body, thus affecting the pH value of the water environment (Xu et al., 2009). Overall, pH is the main factor affecting the spatial and temporal variation pattern of phytoplankton, and the interaction between water body pH and phytoplankton is bidirectional (Wang et al., 2016).
Water temperature is considered to be an important factor affecting the growth, development, and species composition of plankton, and it is also the main driving factor affecting the seasonal succession of the plankton community (Marques et al., 2006; Chen et al., 2021). Temperature directly affects the growth state of phytoplankton by controlling the intensity of enzymatic reaction during phytoplankton respiration and photoreaction (Blinn, 1993) and is closely related to the relative abundance of phytoplankton (Lu et al., 2013). WT is one of the main environmental factors affecting the distribution of the eukaryotic plankton community in the reservoir area. WT at 0.5m is significantly higher than that at 20 m at each sampling site. Given that positive correlation with WT, Mesocyclops and Cryptomonas were dominant in the surface layer. Correspondingly, Eurytemora and Thermocyclops were negatively correlated with WT, and they were abundant in the deep layer. This study suggested that the change of WT is an important factor leading to the difference in the relative abundance of plankton from 0.5 to 20 m depth in Danjiangkou Reservoir.
The rapid increase in the levels of organic pollutants, suspended solids, and other related oxygen-consuming factors in the water could lead to an imbalance in the production of DO in its water column, which has an impact on zooplankton (He et al., 2022). In addition, oxygen production by phytoplankton photosynthesis is the main source of DO, and phytoplankton respiration and mortality are the largest pathways of DO consumption (Ouyang et al., 2013). It should be noted that the interaction between phytoplankton biomass and the physicochemical indicators of the water column is reciprocal, and changes in phytoplankton biomass can also cause changes in DO concentration, pH, and other indicators in the water column (Wang et al., 2016). As an important index for measuring water quality, DO concentration can reflect the concentration can reflect the degree of water pollution, and the pollution is related to the content of organic compounds (Diaz and Rosenberg, 2008). In this study, Mesocyclops, Cryptomonas, Plagiosemis, and Neodiaptomus are positively correlated with DO, so these taxa can be used as potential biological reference indicators to monitor the water quality in the Danjiangkou Reservoir in the future.
Due to the limited sampling in this study, additional samples from different seasons are needed to provide comprehensive insights into the eukaryotic plankton communities in Danjiangkou Reservoir. It’s worth noting that ArcMap, DEM, and SAR have already found extensive application in ecological environment research (Liu et al., 2022; Qiu et al., 2022; Wang et al., 2022; Ma et al., 2023; Pei et al., 2023). In the future, the integration of sequencing data with ArcMap, DEM, and SAR holds immense promise for portraying the spatial and temporal distribution of eukaryotic plankton within the reservoir area.
Our results provide a novel perspective on the distribution of the eukaryotic plankton community in Danjiangkou Reservoir, particularly in terms of vertical variation. This insight holds significant potential for understanding ecological processes and ensuring the water quality safety of this canyon-style reservoir.
(1) A total of 38 phyla and 122 genera of eukaryotic plankton were identified in Danjiangkou Reservoir. Notably, we observed substantial variations in both the diversity and abundance of the eukaryotic plankton community across different depths. Specifically, diversity and abundance exhibited a decreasing trend from depths of 0.5 to 10 m, while an increase was noted at a depth of 20m.
(2) Environmental factors, including pH, depth, WT, NH4+-N, DO, Cond, and NO3−-N, play crucial roles in influencing the vertical distribution of eukaryotic plankton within the reservoir. It’s important to note that these environmental factors have distinct effects on the eukaryotic plankton community. For instance, Eurytemora, Thermocyclops, and Volvox were found to be negatively correlated with pH and WT, while they showed a positive correlation with depth.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
SM: Conceptualization, Methodology, Writing – original draft. YH: Formal Analysis, Supervision, Writing – original draft. WL: Resources, Supervision, Visualization, Writing – review & editing. TZ: Funding acquisition, Investigation, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (Grant No. U1704241) and the Henan Technological and Innovative Leading Talent Program (Grant No. 194200510010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Blinn D. W. (1993). Diatom community structure along physicochemical gradients in saline lakes. Ecology 74 (4), 1246–1263. doi: 10.2307/1940494
Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods: Techniques Life scientists Chem. 10 (1), 57–59. doi: 10.1038/nmeth.2276
BoonFei T., Charmaine N., Pierre N. J., Leng L. L., G. K. Y. H., T. J. R. (2015). Next-generation sequencing (NGS) for assessment of microbial water quality: current progress, challenges, and future opportunities. Front. Microbiol. 6, 1027. doi: 10.3389/fmicb.2015.01027
Chalar G. (2009). The use of phytoplankton patterns of diversity for algal bloom management. Limnologica 39 (3), 200–208. doi: 10.1016/j.limno.2008.04.001
Chao A. (1984). Nonparametric estimation of the number of classes in a population. Scandinavian J. Stat 11 (4), 265–270. doi: 10.2307/4615964
Chen Y., Peng K., Zhang Q., Cai Y., Zhang Y., Gong Z., et al. (2021). Spatio-temporal distribution characteristics and driving factors of zooplankton in Hongze Lake. Environ. Sci. 42 (8), 3753–3762. doi: 10.13227/j.hjkx.202010195
Cudowski A., Pietryczuk A., Hauschild T. (2015). Aquatic fungi in relation to the physical and chemical parameters of water quality in the Augustow Canal. Fungal Ecol. 13, 193–204. doi: 10.1016/j.funeco.2014.10.002
Cui Z., Gao W., Li Y., Wang W., Wang H., Liu H., et al. (2023). Dissolved oxygen and water temperature drive vertical spatiotemporal variation of phytoplankton community: evidence from the largest diversion water source area. Int. J. Environ. Res. Public Health 20 (5), 4307. doi: 10.3390/ijerph20054307
De Queiroz A. R., Flores Montes M., de Castro Melo P. A. M., da Silva R. A., Koening M. L. (2015). Vertical and horizontal distribution of phytoplankton around an oceanic archipelago of the Equatorial Atlantic. Mar. Biodiversity Records 8, E155. doi: 10.1017/S175526721500130X
Diaz R. J., Rosenberg R. (2008). Spreading dead zones and consequences for marine ecosystems. Science 321 (5891), 926–929. doi: 10.1126/science.1156401
Dijk E. V., Auger H., Jaszczyszyn Y., Thermes C. (2014). Ten years of next-generation sequencing technology. Trends Genet. 30 (9), 418–426. doi: 10.1016/j.tig.2014.07.001
Ferrera I., Rene A., Funosas D., Camp J., Massana R., Gasol J. M., et al. (2020). Assessment of microbial plankton diversity as an ecological indicator in the NW Mediterranean coast. Mar. pollut. Bull. 160, 111691. doi: 10.1016/j.marpolbul.2020.111691
Filker S., Sommaruga R., Vila I., Stoeck T. (2016). Microbial eukaryote plankton communities of high-mountain lakes from three continents exhibit strong biogeographic patterns. Mol. Ecol. 25 (10), 2286–2301. doi: 10.1111/mec.13633
Gao W. L., Chen Z. J., Li Y. Y., Pan Y. D., Zhu J. Y., Guo S. J., et al. (2018). Bioassessment of a drinking water reservoir using plankton: high throughput sequencing vs. Traditional Morphological Method. Water 10 (1), 82. doi: 10.3390/w10010082
Good I. (2010). The population frequency of species and the estimation of the population parameters. Biometrika 40 (1953), 237–246. doi: 10.2307/2333344
He S., Gou J., Yin J., Qiu X., Zhao R. (2022). Community structure of zooplankton and its relationship with water environmental factors in ningxia section of the main stream of the YellowRiver. Water Resour. Power 40 (10), 66–69+18. doi: 10.20040/j.cnki.1000-7709.2022.20212768
He Y., Zheng Y., Li W., Zhang Z., Ma Y., Zhao T., et al. (2021). Characteristics of eukaryotic phytoplankton community structure in early spring and its relationship with environmental factors in Danjiangkou Rservoir. Acta Scientiae Circumstantiae 41 (6), 2912–2200. doi: 10.13671/j.hjkxxb.2020.0550
Hoyer B. A., Blanco M. J., Moreno-Ostos E., Vidal J., Rueda J. F., Palomino T. (2009). The influence of external perturbations on the functional composition of phytoplankton in a Mediterranean reservoir. Hydrobiologia 636, 49–64. doi: 10.1007/s10750-009-9934-2
Hu Y., Peng Y., Li R., Huang J., Zhou Z., Hu S., et al. (2021). Plankton diversity and community charecteristics in Danjiangkou Reservoir based on environmental DNA metabarcoding. J. Lake Sci. 33 (6), 1650–1659. doi: 10.18307/2021.0604
Huovinen (1999). Temporal and vertical dynamics of phytoplankton net growth in Castle Lake, California. J. Plankton Res. 2, 373–385. doi: 10.1093/plankt/21.2.373
Ishida K. N. (2008). Seasonal distribution of photosynthetically active phytoplankton using pulse amplitude modulated fluorometry in the large monomictic Lake Biwa, Japan. J. Plankton Res. 30 (10), 1169–1177. doi: 10.1093/plankt/fbn073
Jia H., Xu J., Lei J. (2019). Relationship of community structure of phytoplankton and environmental factors in Danjiangkou Reservior bay. Yangtze River 50 (5), 52–58. doi: 10.16232/j.cnki.1001-4179.2019.05.011
Jiang Y., Xu H., Zhu M. Z. (2012). Use of planktonic protists for assessing water quality in Jiaozhou Bay, northern China. Protistology 7 (1), 34–41.
Jin X. (1990). Handbook of eutrophication investigation of Lake. 2nd ed (Beijing: China Environmental Sciences Press).
Keck F., Millet L., Debroas D., Etienne D., Galop D., Rius D., et al. (2020). Assessing the response of micro-eukaryotic diversity to the Great Acceleration using lake sedimentary DNA. Nat. Commun. 11 (1), 3831. doi: 10.1038/s41467-020-17682-8
Leal M. C., Sá C., Nordez S., Brotas V., Paula J. (2009). Distribution and vertical dynamics of planktonic communities at Sofala Bank, Mozambique. Estuarine Coast. Shelf Sci. 84 (4), 605–616. doi: 10.1016/j.ecss.2009.07.028
Lee S.-R., Song E. H., Lee T. (2018). Eukaryotic Plankton Species Diversity in the Western Channel of the Korea Strait using 18S rDNA Sequences and its Implications for Water Masses. Ocean Sci. J. 53 (1), 119–132. doi: 10.1007/s12601-018-0005-3
Liu S., Gibson K., Cui Z., Chen Y., Sun X., Chen N. (2020a). Metabarcoding analysis of harmful algal species in Jiaozhou Bay. Harmful Algae 92, 101772. doi: 10.1016/j.hal.2020.101772
Liu Z., Qiu H., Zhu Y., Liu Y., Yang D., Ma S., et al. (2022). Efficient identification and monitoring of landslides by time-series InSAR combining single- and multi-look phases. Remote Sens. 14, 1026. doi: 10.3390/rs14041026
Liu X., Shi X., Qi G., Liu G., Zhao Y., Wu S. (2010). Appliction of freshwater alage in monitoring water quality and sewage purification. J. Biol. 27 (6), 76–78+86. doi: 10.3969/j.issn.1008-9632.2010.06.076
Liu L., Wang S., Ji J., Xie Y., Shi X., Chen J. (2020b). Characteristics of microbial eukaryotic community recovery in eutrophic water by using ecological floating beds. Sci. Total Environ. 711, 134551. doi: 10.1016/j.scitotenv.2019.134551
Logares R., Audic S., Bass D., Bittner L., Boutte C., Christen R., et al. (2014). Patterns of rare and abundant marine microbial eukaryotes. Curr. Biol. 24 (8), 813–821. doi: 10.1016/j.cub.2014.02.050
Lu Q. (1987). The ecological society panel on environmental issues ed. Environmental and biological indicator (water volume) (Beijing: China Environmental Science Press (in Chinese).
Lu Q., Chen H., Shao X., Wang Y., Tao M., He J., et al. (2013). Ecological characteristics of macrobenthic communities and its relationships with environmental factors in Hangzhou Xixi Wetland. Acta Ecologica Sin. 33 (9), 2803–2815. doi: 10.5846/stxb201203080316
Ma S., Qiu H., Zhu Y., Yang D., Tang B., Wang D., et al. (2023). Topographic Changes, Surface Deformation and Movement Process before, during and after a Rotational Landslide. Remote Sens. 15 (3), 662. doi: 10.3390/rs15030662
MaChado K. B., Targueta C. P., Antunes A. M., Soares T. N., d. C. Telles M. P., Logares R., et al. (2019). Diversity patterns of planktonic microeukaryote communities in tropical floodplain lakes based on 18S rDNA gene sequences. J. Plankton Res. 41 (3), 241–256. doi: 10.1093/plankt/fbz019
Marianne S., Purificación L.-G., Philippe D., David M., Gwendal R., Paola B., et al. (2015). Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME J. 9 (9), 1941–1953 doi: 10.1038/ismej.2015.6
Marques S. C., Azeiteiro U. M., Marques J. C., Neto J. M., Pardal M. A. (2006). Zooplankton and ichthyoplankton communities in a temperate estuary: spatial and temporal patterns. J. Plankton Res. 28 (3), 297–312. doi: 10.1093/plankt/fbi126
McInnes A. S., Laczka O. F., Baker K. G., Larsson M. E., Robinson C. M., Clark J. S., et al. (2019). Live cell analysis at sea reveals divergent thermal performance between photosynthetic ocean microbial eukaryote populations. Isme J. 13 (5), 1374–1378. doi: 10.1038/s41396-019-0355-6
Meadow J. F., Zabinski C. A. (2012). Spatial heterogeneity of eukaryotic microbial communities in an unstudied geothermal diatomaceous biological soil crust: Yellowstone National Park, WY, USA. FEMS Microbiol. Ecol. 82 (1), 182–191. doi: 10.1111/j.1574-6941.2012.01416.x
Nabout J. C., Siqueira T., Bini L. M., Nogueira I. (2009). No evidence for environmental and spatial processes in structuring phytoplankton communities. Acta Oecologica 35 (5), 720–726. doi: 10.1016/j.actao.2009.07.002
Nagano Y., Nagahama T. (2012). Fungal diversity in deep-sea extreme environments. Fungal Ecol. 5 (4), 463–471. doi: 10.1016/j.funeco.2012.01.004
Ouyang X., Zhao Q., Wei Y. (2013). A preliminary exploration of dissolved oxygen based on FVCOM in Meiliang Bay, Lake Taihu and its influence mechanism. Lake Sci. 25 (4), 478–488. doi: 10.18307/2013.0404
Pei Y., Qiu H., Zhu Y., Wang J., Yang D., Tang B., et al. (2023). Elevation dependence of landslide activity induced by climate change in the eastern Pamirs. Landslides 20, 1115–1133. doi: 10.1007/s10346-023-02030-w
Pielou E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Piwosz K., Shabarova T., Pernthaler J., Posch T., Simek K., Porcal P., et al. (2020). Bacterial and eukaryotic small-subunit amplicon data do not provide a quantitative picture of microbial communities, but they are reliable in the context of ecological interpretations. mSphere 5, 2, e00052–e00020. doi: 10.1128/mSphere.00052-20
Ptacnik R., Diehl S., Berger S. (2003). Performance of sinking and nonsinking phytoplankton taxa in a gradient of mixing depths. Limnology Oceanography 48 (5), 1903–1912. doi: 10.4319/lo.2003.48.5.1903
Qiu H., Zhu Y., Zhou W., Sun H., He J., Liu Z. (2022). Influence of DEM resolution on landslide simulation performance based on the Scoops3D model, Geomatics. Natural Hazards Risk 13 (1), 1663–1681. doi: 10.1080/19475705.2022.2097451
Reich M., Wichels A., Panzer K., Krause E., Gimenez L., Gerdts G. (2017). Impacts of a reduction of seawater pH mimicking ocean acidification impacts on assemblage, structure and diversity of marine fungal communities. Aquat. Microbial Ecol. 79 (3), 221–233. doi: 10.3354/ame01831
Salmaso N., Boscaini A., Pindo M. J. F. i. M. (2020). Unraveling the diversity of eukaryotic microplankton in a large and deep perialpine lake using a high throughput sequencing approach. Front. Microbiol. 11, 789. doi: 10.3389/fmicb.2020.00789
Shen H., Xu Y., Wang L., Zhang M., Kong L., Cai Q. (2011). Spatial and temporal variations of phytoplankton in Danjiangkou Reservoir and its affecting factor. Plant Sci. J. 29, 683–790. doi: 10.3724/SP.J.1142.2011.60683
Simpson E. H. (1997). Measurement of diversity. J. Cardiothoracic Vasc. Anesth. 11 (6), 812–812. doi: 10.1136/thx.27.2.261
Song L., Wu J., Du J., Li N., Song G., Wang K., et al. (2019). The characteristics and distribution of eukaryotic phytoplankton community in Liaodong bay, China. Ocean Sci. J. 54 (2), 183–203. doi: 10.1007/s12601-019-0007-9
Sun Z., Li G., Wang C., Jing Y., Zhu Y., Zhang S., et al. (2014). Community dynamics of prokaryotic and eukaryotic microbes in an estuary reservoir. Sci. Rep. 4 (1), 6966. doi: 10.1038/srep06966
Sun Y., Liu Y., Wu C., Fu X., Guo C., Li L., et al. (2021). Characteristics of eukaryotic plankton communities in the cold water masses and nearshore waters of the South Yellow Sea. Diversity 13 (1), 21. doi: 10.3390/d13010021
Tan X., Shi X., Liu G., Xu H., Nie P. (2010). An approach to analyzing taxonomic patterns of protozoan communities for monitoring water quality in Songhua River, northeast China. Hydrobiologia 638 (1), 193–201. doi: 10.1007/s10750-009-0040-2
Tan X., Xia X., Zhang Q. (2011). Temporal and spatial pattern of Phytoplankton community and its biodiversity indices in the Danjiangkou Reservoir. Environ. Sci. 32 (10), 2875–2882. doi: 10.13227/j.hjkx.2011.10.008
Tavernini S., Mura G., Rossetti G. (2005). Factors influencing the seasonal phenology and composition of zooplankton communities in mountain temporary pools. Int. Rev. Hydrobiology 90 (4), 358–375. doi: 10.1002/iroh.200510801
Taylor J. D., Cunliffe M. (2016). Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. Isme J. 10 (9), 2118–2128. doi: 10.1038/ismej.2016.24
Trombetta T., Vidussi F., Mas S., Parin D., Simier M., Mostajir B. (2019). Water temperature drives phytoplankton blooms in coastal waters. PLoS One 14 (4), e0214933. doi: 10.1371/journal.pone.0214933
Voronin L. V. (2010). The fungi of small acid lakes. Inland Water Biol. 3 (3), 254–259. doi: 10.1134/S1995082910030089
Wang Y., Chen L., Niu Y., Yu H., Luo M. (2016). Spatio-temporal variation in phytoplankton community and its influencing factors in Danjiangkou Reservoir. J. Lake Sci. 28 (5), 1057–1065. doi: 10.18307/2016.0516
Wang Q., Huang W., Chen K., Yi Q., Li H. (2020). Phytoplankton community structure and trophic status evaluation in Reservior Daxi. Acta Scientiae Circumstantia 40 (4), 1286–1297. doi: 10.13671/j.hjkxxb.2019.0497
Wang L., Qiu H., Zhou W., Zhu Y., Liu Z., Ma S., et al. (2022). The post-failure spatiotemporal deformation of certain translational landslides may follow the pre-failure pattern. Remote Sens. 14, 2333. doi: 10.3390/rs14102333
Wang Z., Tan Y., Li Z., Xiao X. (2012). Assessment on eutrophication in typical bays and tributaries of Danjiangkou Reservoir. Yangtze River 43 (8), 61–64+75. doi: 10.16232/j.cnki.1001-4179.2012.08.016
Wang J., Wang S., Zhang Y., Lin J., Gao X., Zang X., et al. (2017). Community structure characteristics of eukaryotic planktonic algae in Liaohe River through high-throughput Sequencing. Environ. Sci. 38 (4), 1403–1413. doi: 10.13227/j.hjkx.201609133
Wei N., Satheeswaran T., Jenkinson I. R., Xue B., Wei Y., Liu H., et al. (2018). Factors driving the spatiotemporal variability in phytoplankton in the Northern South China Sea. Continental Shelf Res. 162, 48–55. doi: 10.1016/j.csr.2018.04.009
Wu J.-Y., Hua Z.-L., Gu L., Li X.-Q., Gao C., Liu Y.-Y. (2022). Perfluorinated compounds (PFCs) in regional industrial rivers: Interactions between pollution flux and eukaryotic community phylosymbiosis. Environ. Res. 203, 111876. doi: 10.1016/j.envres.2021.111876
Wurzbacher C. M., Baerlocher F., Grossart H.-P. (2010). Fungi in lake ecosystems. Aquat. Microbial Ecol. 59 (2), 125–149. doi: 10.3354/ame01385
Xiao Y., Cheng J., Mo X., Li Y., Liu X., Bi S. (2023). Spatio-temporal variation of phytoplankton community and its relationship with environmental factors in Danjiangkou Reservoir. Lake Sci. 35 (3), 821–832. doi: 10.18307/2023.0306
Xu H., Liu Z., Yuan L., Yang L. (2009). Effect of pH on growth of several freshwater algae. Environ. Sci. Technol. 32 (1), 27–30. doi: 10.3969/j.issn.1003-6504.2009.01.007
Yan X., Zhang Y., Li Y., Jiang Y., Cui Z., Gao X., et al. (2021). Hydrologic and physicochemical factors codrive seasonal changes of phytoplankton during dynamic water diversion processes in the Danjiangkou Reservoir. J. Lake Sci. 33 (5), 1350–1363. doi: 10.18307/2021.0505
Yang L., Yu P., Zhu J., Xu Z., Lü G., Jin C. (2014). Community structure characteristics of phytoplankton and related affecting factors in Heng-shan Reservoir, Zhejiang, China. Chin. J. Appl. Ecol. 25 (2), 569–576. doi: 10.13287/j.1001-9332.2014.0060
Yin D., Zheng L., Song L. (2011). Spatio-temporal distribution of phytoplankton in the Danjiangkou Reservoir, a water source area for the South-to-North Water Diversion Project (Middle Route), China. Chin. J. Oceanology Limnology 29 (3), 531–540. doi: 10.1007/s00343-011-0120-9
Zhang C., He Y., Li W., Guo X., Xiao C., Zhao T. (2022). High-throughput sequencing of diatom community, its spatial and temporal variation and interrelationships with physicochemical factors in Danjiangkou Reservoir, China. Water 14 (10), 1609. doi: 10.3390/w14101609
Zhang W., Pan Y., Yang J., Chen H., Holohan B., Vaudrey J., et al. (2018). The diversity and biogeography of abundant and rare intertidal marine microeukaryotes explained by environment and dispersal limitation. Environ. Microbiol. 20 (2), 462–476. doi: 10.1111/1462-2920.13916
Zhang Q., Song L., Ji D., Fang H., He J., Huo J., et al. (2019). Relationship between water quality of Xiangxi River Reservoir and the algal blooms in non-return area in the Three Gorges Reservoir Area. China Environ. Sci. 39 (7), 3018–3026. doi: 10.19674/j.cnki.issn1000-6923.2019.0356
Zhao Y. J., Sato Y., Inaba T., Aoyagi T., Hori T., Habe H. (2019). Activated sludge microbial communities of a chemical plant wastewater treatment facility with high-strength bromide ions and aromatic substances. J. Gen. Appl. Microbiol. 65 (2), 106–110. doi: 10.2323/jgam.2018.05.002
Zheng B. H., Chen Z. J., Li Y. Y., Fohrer N., Zhang Y., Wu D. Y., et al. (2020). Structural characteristics and driving factors of the planktonic eukaryotic community in the Danjiangkou reservoir, China. Water 12 (12), 3499. doi: 10.3390/w12123499
Zheng B., Zhu J., Xu X., Xin Y., Song J., Shi Y., et al. (2018). Community structure of periphyton algae and water quality in the Danjiangkou Reservoir. J. Henan Normal Univ. (Natural Sci. Edition) 46 (4), 95–101. doi: 10.16366/j.cnki.1000-2367.2018.04.015
Zhong K. X., Cho A., Deeg C. M., Chan A. M., Suttle C. A. (2021). Revealing the composition of the eukaryotic microbiome of oyster spat by CRISPR-Cas Selective Amplicon Sequencing (CCSAS). Microbiome 9 (1), 230. doi: 10.1186/s40168-021-01180-0
Keywords: Danjiangkou Reservoir, eukaryotic plankton, community structure, environmental factor, high-throughput sequencing
Citation: Mai S, He Y, Li W and Zhao T (2023) Effects of environmental factors on vertical distribution of the eukaryotic plankton community in early summer in Danjiangkou Reservoir, China. Front. Ecol. Evol. 11:1324932. doi: 10.3389/fevo.2023.1324932
Received: 20 October 2023; Accepted: 06 November 2023;
Published: 22 November 2023.
Edited by:
Haijun Qiu, Northwest University, ChinaCopyright © 2023 Mai, He, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxiao He, aGV5dXhpYW9AaHB1LmVkdS5jbg==; Weiguo Li, d2dsaUBocHUuZWR1LmNu; Tongqian Zhao, emhhb3RxQGhwdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.