- 1Human Phenome Institute, Fudan University, Shanghai, China
- 2State Key Laboratory of Genetic Engineering, Department of Life Sciences, School of Life Sciences, Fudan University, Shanghai, China
- 3Institute of Archaeological Science, Fudan University, Shanghai, China
- 4Department of History, Fudan University, Shanghai, China
- 5Criminal Justice College, China University of Political Science and Law, Beijing, China
- 6Ministry of Education Key Laboratory of Contemporary Anthropology and B&R International Joint Laboratory for Eurasian Anthropology, School of Life Sciences, Fudan University, Shanghai, China
- 7Ministry of Education Laboratory for National Development and Intelligent Governance, Fudan University, Shanghai, China
- 8Center for the Belt and Road Archaeology and Ancient Civilizations (BRAAC), Fudan University, Shanghai, China
Abstract: The Kazakhs of Xinjiang province are characterized by their nomadic lifestyle and patrilineal clan system. However, compared to Central Asian Kazakhs, a lack of Y chromosomal high-resolution analysis has hindered our understanding of the paternal history of modern Xinjiang Kazakhs.
Methods: In this study, we present the analysis of 110 Y-SNP data from 209 Altay Kazakhs and 201 Ili Kazakhs in Xinjiang, along with their previously reported 24 Y-STR loci data.
Results and discussion: We found that the Y chromosome haplogroups exhibit greater diversity in Altay Kazakhs compared to Kazakhs in Kazakhstan, Russia, and other regions of China. Y-SNP-based PCA plots reveal that both the Altay and Ili Kazakhs are situated between the Turkic, Mongolia, and Tibeto-Burman clusters. The dominant haplogroup C2a1a3-F1918, which originated in northeast Asia during the Neolithic Age, accounts for nearly half of the Altay and Ili Kazakhs. The Y lineage network of C2a1a3-F1918 contained two subclusters. Approximately 60.6% of the Altay Kazakhs belong to the DYS448-23 subcluster, indicating their Kerey-Abakh ancestry. On the other hand, around three-quarters of the Ili Kazakhs belong to the DYS448-22 subcluster, suggesting their Kerey-Ashmaily heritage. Notably, the TMRCA ages of the DYS448-23 subcluster were calculated to be 289.4 ± 202.65 years, which aligns with the historical immigration of the Kerey clan back to the Altay Mountains after the defeat of the Dzungar by the Qing dynasty in the mid-18th century.
Introduction
Xinjiang, located in the northwest region of China and known as the heart of the ancient Silk Road, has a rich history of population admixture and cultural exchanges (Mair, 2021). The constant influx of western and eastern Eurasian immigrants has contributed to the diverse genetic landscapes of this area (Mair, 2021; Kumar et al., 2022). Among these populations, the Kazakhs, cross-border nomads organized into a hierarchical patrilineal clan system, have complex ancestral sources and ethnic histories. The majority of Xinjiang Kazakhs reside in Altay Prefecture, along the southern Altai Mountains, and in Ili Prefecture along the Ili River Valley. However, there is currently limited genetic information regarding the impact of past population migration on the paternal genetic structure of present-day Xinjiang Kazakhs.
The origins of the Xinjiang Kazakhs can be traced back to the Saka Culture of the Early Iron Age (Di Cosmo, 1996; Chi and Festa, 2020; Gnecchi-Ruscone et al., 2021; Khussainova et al., 2021). During the 3rd-2nd centuries BCE, Kangju and Wusun tribal confederations emerged in this region, contributing to cultural and ethnic continuity (Di Cosmo, 1996; Otarbaeva, 1998; Bi, 2015; Kumar et al., 2022). The Turkic period brought about significant changes in regional ethnic composition, as various steppe nomadic empires like the Xiongnu, Xianbei, Turk, Rouran, Khitan, and others expanded their dominance (Otarbaeva, 1998; Rogers, 2012). The Mongol invasion in the 13th century had a profound influence on the genetic makeup of Xinjiang (Otarbaeva, 1998). In the mid-fifteenth century, the Kazakh Khanate was established as a tribal and political entity, emerging as a new regional power in Xinjiang (Otarbaeva, 1998; Sun, 2022). However, the Kazakhs faced attacks from the Dzungars in the early 18th century, resulting in the loss of their pasturelands (Otarbaeva, 1998).
Genetic investigations have offered new insights into the complex history of Kazakhs, helping us understand how admixture events have shaped the population structure and genetic diversity of modern Kazakhs. Notably, the higher frequencies of C2 haplogroups along the southern and western borders of Kazakhstan align with the historical path of the Mongolian invasion (Khussainova et al., 2021). Moreover, the discovery of the haplogroup C2a1a2-M48 among Western Kazakhs has suggested a founding population from the Middle Ages (Zhabagin et al., 2021). Paternal lineage analysis further supported a common origin of South Uissun clans from Niru’un Mongols, rather than early Wusun peoples (Zhabagin et al., 2020). In China, the paternal origin of Kazakhs residing in Gansu province has been confirmed as Kerey-Abakh (Wen et al., 2020). Similar mtDNA lineage has been observed among Kazakhs from Altai, East Kazakhstan, and Xinjiang (Tarlykov et al., 2013). Moreover, comprehensive genome-wide analyses of Xinjiang Kazakhs have indicated two significant population admixture events: one occurring approximately 120 generations ago and another 20-40 generations ago (Pan et al., 2023).
Our knowledge of the paternal history of the Xinjiang Kazakhs, however, remains limited. Previous studies, using only Y-chromosomal STR data (Shan et al., 2014a; Shan et al., 2014b; Mei et al., 2016; Li et al., 2020; Zhang et al., 2021), have primarily focused on genetic polymorphisms, population comparisons, and forensic applications. Xinjiang Kazakhs have been demonstrated to be genetically close to Eastern Kazakhstan groups but distinct from Northern Kazakhstan groups based on Y-STR loci polymorphism (Ashirbekov et al., 2022; Ashirbekov et al., 2023). In this study, we aimed to gain a deeper understanding of the paternal genetic structure of modern Kazakhs in northwest China by analyzing 110 Y-SNPs and 24 Y-STRs from 410 male Kazakhs in the Altay and Ili Prefecture of Xinjiang. By doing so, our hope was to shed light on how historical events influenced the genetic makeup of this population.
Materials and methods
This study included a total of 410 Kazakh males from the Xinjiang province in China, as previously reported (Mei et al., 2016; Li et al., 2020). Among the participants, 209 were from Altay and 201 were from Ili. Informed consent forms were signed by each participant, and the study was approved by the ethics committee at the School of Life Sciences in Fudan University.
DNA samples were extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Germany). Hierarchical genotyping was performed using the SNaPshot method with the ABI SNaPshot Multiplex Kit, as previously described (Wang et al., 2014; Xu and Li, 2017). Haplogroups were classified according to the ISOGG Y-DNA Haplogroup Tree 2019 (Version: 15.73; Date: 11 July 2020). Y-STR loci data from the 24 analyzed participants had been previously reported (Mei et al., 2016; Li et al., 2020). Haplogroup diversity were calculated using the Arlequin package.
To investigate the genetic relationships between Altay and Ili Kazakhs and other published Eurasian populations, we conducted a Y-SNP-based principal component analysis (PCA) using RStudio software (v. 2021.09.1 + 372). Additionally, we constructed median-joining networks of Y-STR haplotypes C2a1a3-F1918 and C2a1a2-M48 to examine individual-level relationships using the program NETWORK (v. 10.2) and 15 Y-STR data (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635, GATA_H4). We calculated the time to the most recent common ancestor (TMRCA) for each cluster using the average squared distance (ASD) method, with a generation time set to 25 years, as previously described (Wang et al., 2014; Xu and Li, 2017; Zhabagin et al., 2017).
We utilized bcftools mpileup and bcftools call (Li, 2011; Poznik et al., 2013) to analyze the Y-chromosomal SNPs from published ancient DNA data. The haplogroup identification was performed using Yleaf.py script with Yleaf (Ralf et al., 2018). Furthermore, we conducted a thorough verification of the SNPs by visually inspecting them using the IGV software (Helga et al., 2013).
Results
A total of 34 different haplogroups were identified in Altay Kazakhs, and 27 different haplogroups in Ili Kazakhs (Table S1), based on analysis of 110 Y-SNP markers. Altay Kazakhs exhibited the highest haplogroup diversity (HD) among all Kazakh populations in Kazakhstan, Russia, and China, with a value of 0.94 (Figure 1). This finding aligns with a previous study that compared Y-STR diversity among Kazakhs from Xinjiang, Kazakhstan, and Russia (Shan et al., 2014b).
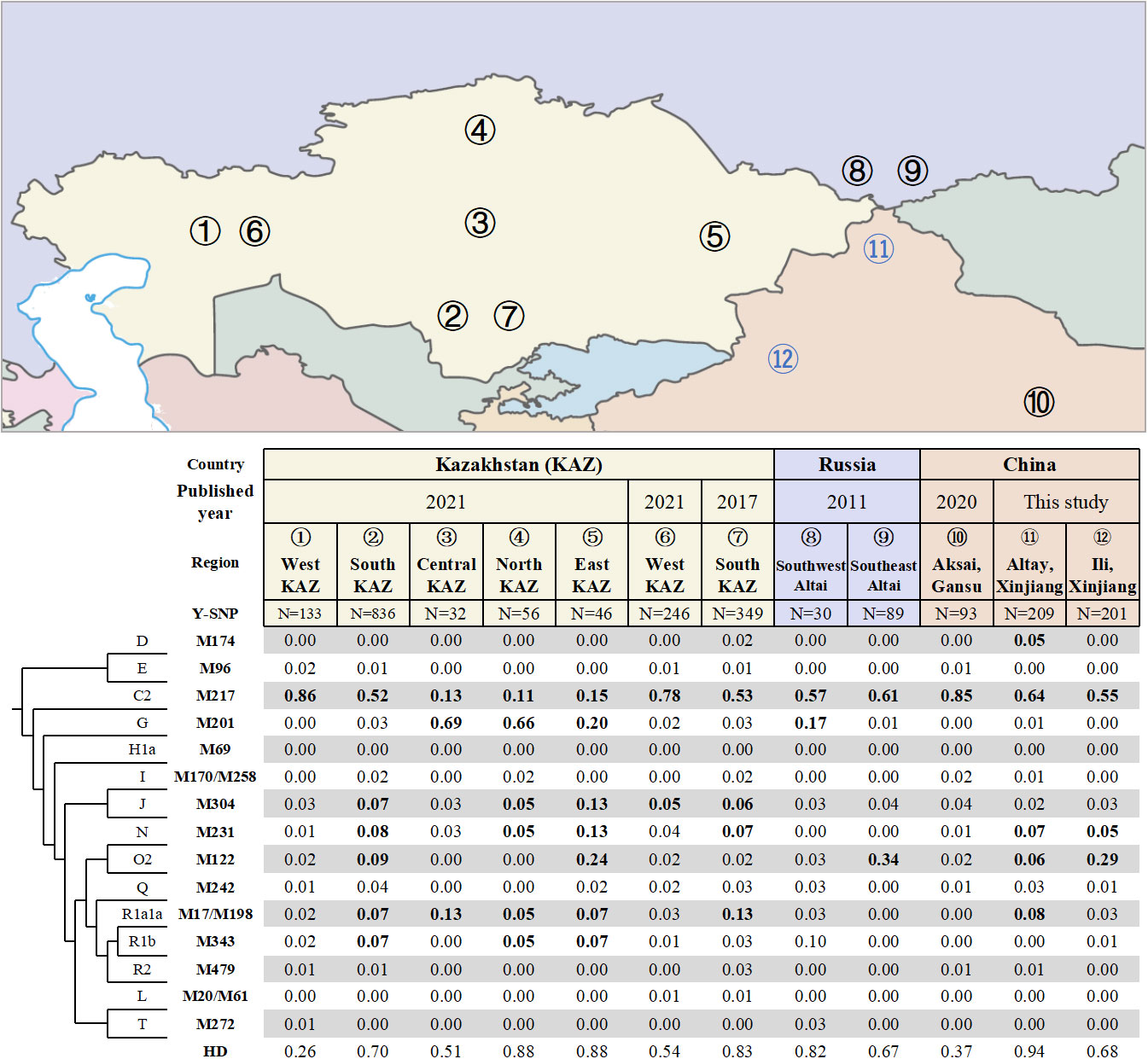
Figure 1 Frequencies of Y-chromosomal haplogroups and geographic locations of different Kazakh populations in Kazakhstan, China and Russia.
The widely distributed haplogroup C2-M217 is prevalent among all Kazakh populations, including Chinese Kazakh populations. In Xinjiang Province, it represents 64.1% among Altay Kazakhs and 55.2% among Ili Kazakhs (Figure 1). In Gansu Province, it accounts for 85% among the Aksai Kazakhs (previously reported) (Wen et al., 2020). Within this haplogroup, there are two major lineages: C2a1a3-F1918, which makes up 49.8% in Altay Kazakhs and 50.7% in Ili Kazakhs, and C2a1a2-M48, which accounts for 9.6% in Altay Kazakhs and 2.5% in Ili Kazakhs (Table S1). Similarly, Gansu Aksai Kazakhs have prominent lineages of C2a1a3a-F3796 (79.6%) and C2a1a2-M48 (0.3%) (Wen et al., 2020).
Haplogroup O2-M122, the most common haplogroup in China, is found at high frequencies in several Kazakh populations. In Ili Kazakhs, it is observed at a frequency of 29.4%, while in Southeast Altai Kazakhs of Russia and East Kazakhstan Kazakhs, it is observed at frequencies of 33.7% and 23.9% respectively. Additionally, Altay Kazakhs and South Kazakhstan Kazakhs have moderate frequencies of 5.7% and 9.4% respectively (Figure 1). The most significant subclade of Haplogroup O2-M122 is O2a2b1a2a-F444, which is present in 25.9% of Ili Kazakhs and 4.8% of Altay Kazakhs (Table S1).
Haplogroup G-M201, primarily found in the Caucasus region (Rootsi et al., 2012), is completely absent in three Chinese Kazakhs. However, it is observed with high frequencies in Central, Northern, and Eastern Kazakhstan Kazakhs (Figure 1).
The PCA was performed by projecting two Xinjiang Kazakh populations onto 156 published modern Eurasian populations (Figure 2, Table S2). The analysis revealed a comprehensive population structure across Eurasia, consisting of nine distinct genetic clusters: Indo-European, Inner Mongolia, Mongolia, Northern Han, Southern Han, Tibeto-Burman, Tungusic, Turkic, and Uralic populations. These clusters aligned well with ethnic and linguistic classifications. Notably, our study found that the Kazakh populations from Altay and Ili Prefecture in Xinjiang province occupied an intermediate position between the Turkic, Mongolia, and Tibeto-Burman clusters, indicating its close genetic affinities with these geographically neighboring populations. Specifically, the Altay Kazakhs shared genetic affinities with the Mongols from Northwest Mongolia, while the Ili Kazakhs showed closer genetic similarities to the Chamdo Tibetans on the eastern Tibetan Plateau.
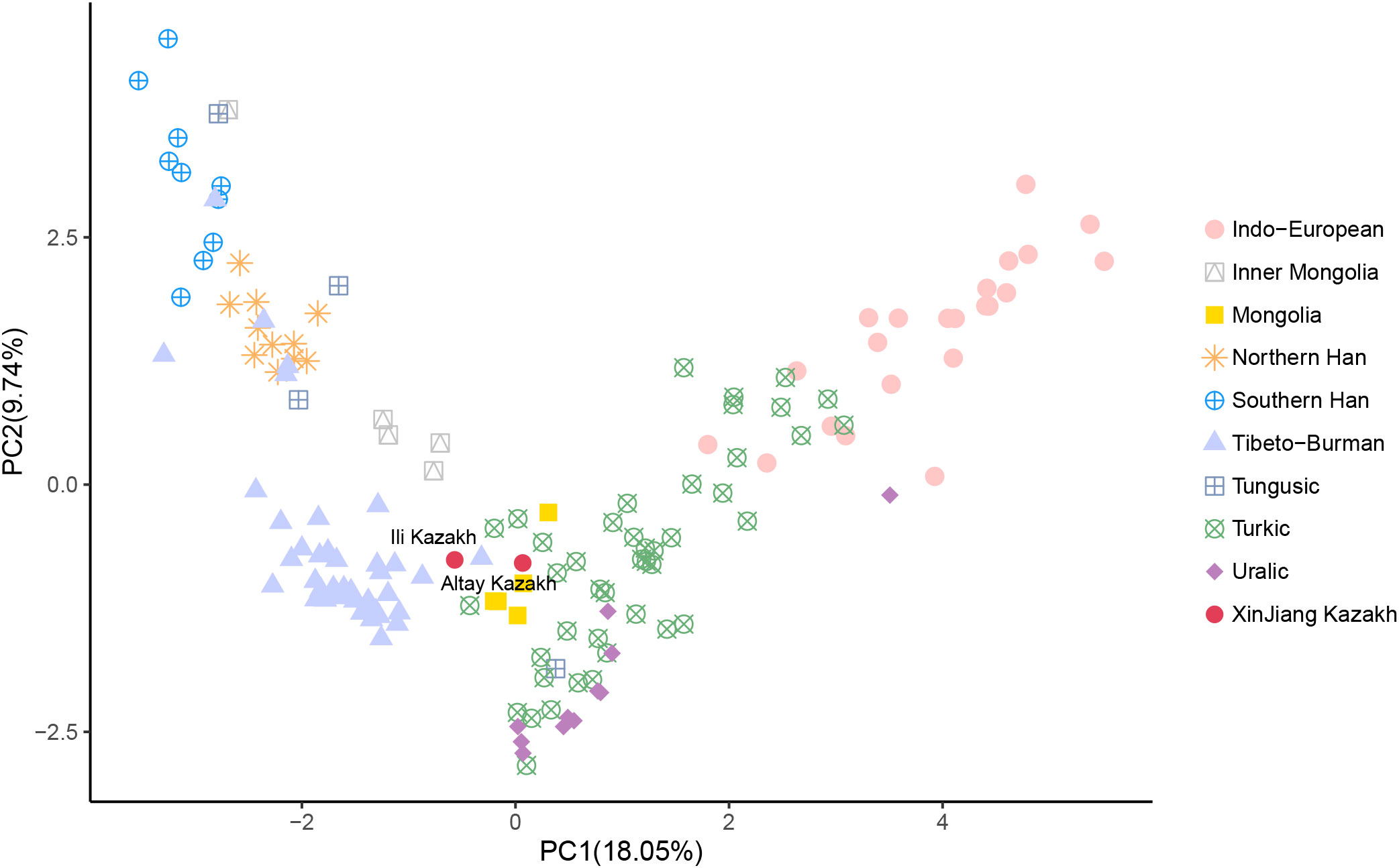
Figure 2 Genetic relationships between Altay Kazakhs and Ili Kazakhs with other reference modern populations from the principal component analysis (PCA) based on Y-chromosome haplogroups.
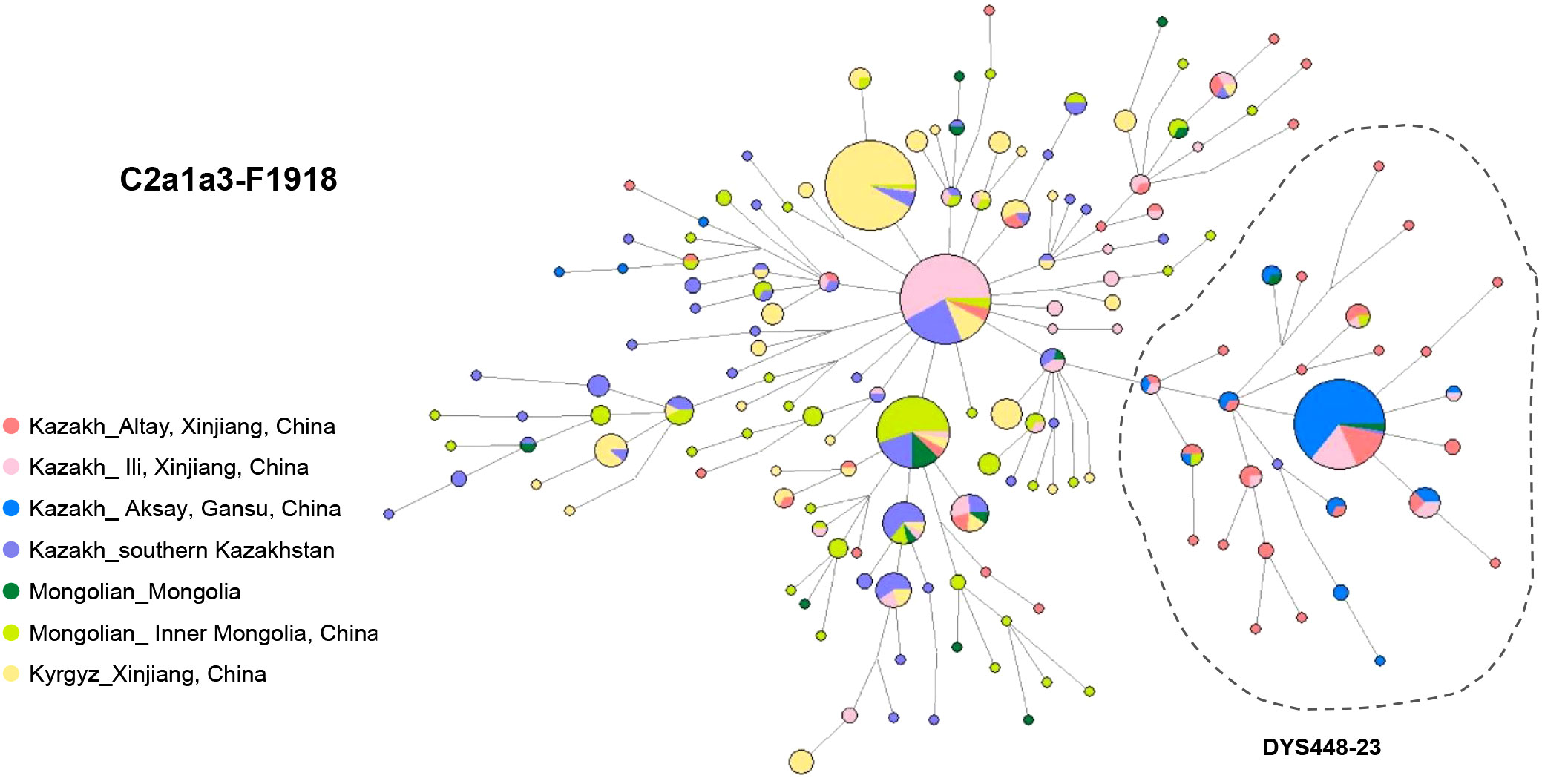
The network of haplogroup C2a1a3-F1918 has been constructed from 15 Y-STR profiles of 614 individuals from 7 populations: Alaty Kazakhs (n=71), Ili Kazakhs (n=102) and Aksay Kazakhs (n=74) from China, Kazakhs from southern Kazakhstan (n=102), Mongolians from Mongolia (n=19) and Inner Mongolia, China (n=89), and Kyrgyz from Xinjiang, China (n=157) (Figure 3; Table S3).
Among them, nearly half of Ili Kazaks (n=52) shared the haplotypes with South Kazakhstan Kazakhs (n=21), Xinjiang Kyrgyz (n=10), Altay Kazakhs (n=4) and Inner Mongolians (n=3), suggesting a potential patrilineal relationship (Table S3). Ili Kazakhs share haplotypes mostly with Kazakhs from southern Kazakhstan and partly with Altay and Aksay Kazakhs from China. This implies a significant influence from southern Kazakhstan on the genetic makeup of Ili Kazakhs.
In the network, a left-side cluster was determined by allele DYS448-23 (Figure 3). This cluster is mainly composed of Altay Kazakhs (34.4%) and Aksay Kazakhs (56.8%) from Northwest China (Table S3). Previous reports have shown that the DYS448-23 subcluster of Kazakhs corresponds to Kereys-Abakh (Abilev et al., 2012). This specific STR haplotype accounts for 20.6% of the Altay Kazakhs’ paternal pool and as much as 76.3% of the Aksay Kazakh pool. These individuals all belong to lineage C2a1a3-F1918* (58.4%) and its downstream lineage C2a1a3a-F3796 (41.6%). A distinct central haplotype suggests that C2a1a3-F1918 may have experienced a series of founder effects or severe bottlenecks in Altay and Aksay. This also reflects the close paternal genetic relationship between these two Kazakh C2a1a3 lineages. TMRCA estimates indicate that this special haplotype cluster is approximately 289.4 ± 202.65 years old, which coincides with historical migration events of Kazakhs after the fall of the Dzungar Kingdom. The Dzungar Kingdom was defeated by the Qing dynasty during the mid-18th century.
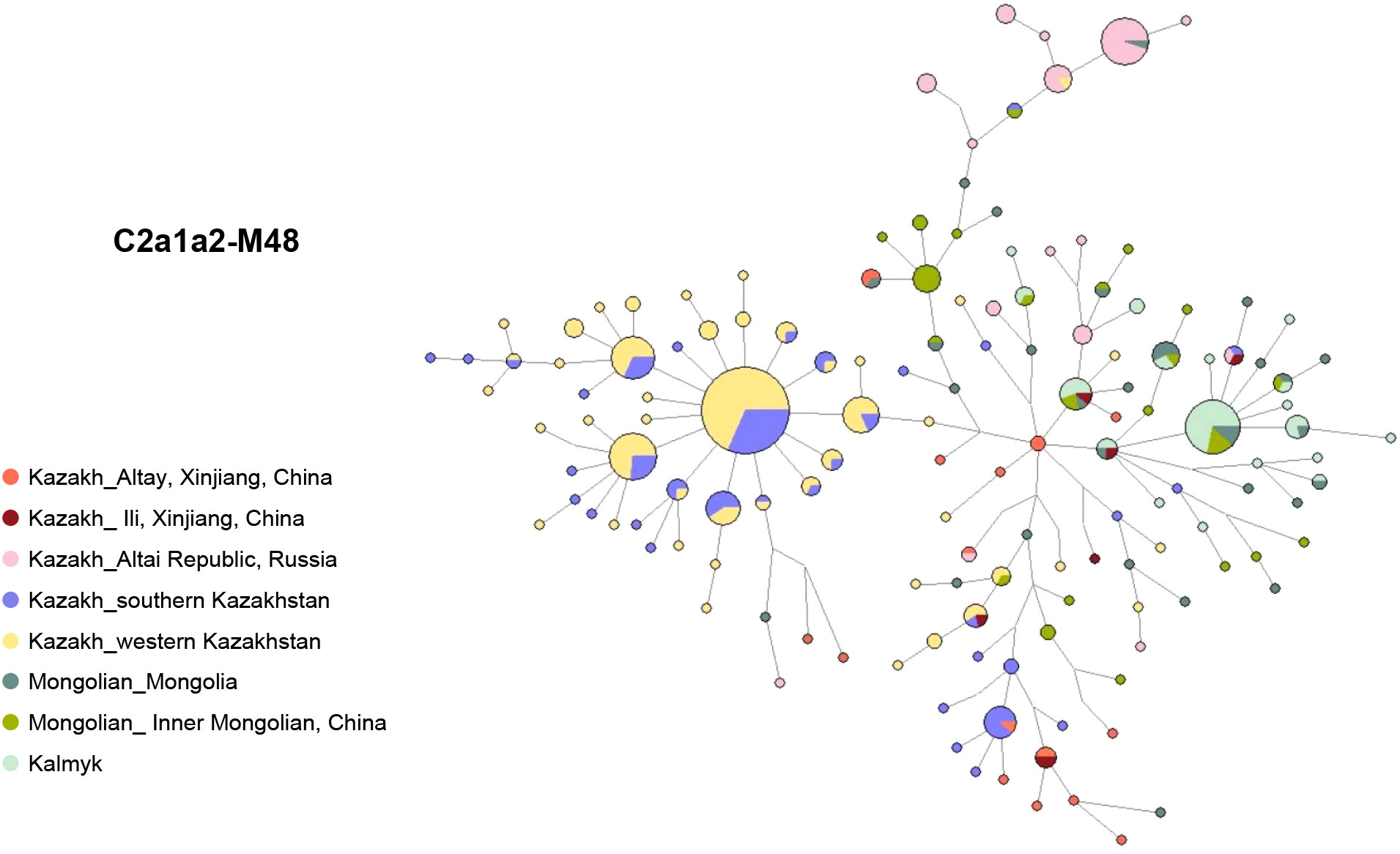
The network of haplogroup C2a1a2-M48 reveals distinct genetic patterns among different populations (Figure 4, Table S4). Specifically, the left star cluster is unique to the south and west Kazakhstan Kazakhs, clearly distinguishing them from the Mongolic-speaking populations on the right. Interestingly, the distribution of the haplogroup C2a1a2-M48 among Ili and Altay Kazakhs is more scattered and does not form a distinct cluster. However, some Altay Kazakhs are centrally located within the network, acting as a bridge between Mongolic-speaking populations and Kazakhstan Kazakhs. This suggests that the Xinjiang Kazakh C2a1a2-M48 lineage has multiple paternal ancestral sources.
Discussion
The ethnogenesis of the Kazakhs can be categorized into three distinct phases: the pre-Turkic, Turkic, and Turko-Mongolian periods. In this study, we utilized 110 Y-SNPs and 15 Y-STRs to investigate the genetic structures and individual relationships within the paternal gene pool of two Kazakh populations in Xinjiang. It is important to highlight that the majority of Y chromosome haplogroups observed in Altay Kazakhs were C2a1a3-F1918 and C2a1a2-M48, while Ili Kazakhs exhibited C2a1a3-F1918 and O2a2b1a2a-F444 as the predominant haplogroups.
The published ancient DNA data revealed the presence of haplogroup C2a1a3-F1918 in East Asia during the middle Neolithic. This haplogroup was primarily found in the Songnen Plain of northeastern China (n=1) (Mao et al., 2021) and the Boisman Culture of the Russian Far East (n=3) (Wang et al., 2021). During the Iron Age, a related lineage was identified in the Xianbei population of China (n=1) (Ning et al., 2020). Subsequently, this lineage was discovered in the Site of Kayalyk (n=1) (Gnecchi-Ruscone et al., 2021) and the Golden Horde (n=1) (Damgaard et al., 2018) in Kazakhstan, as well as in the Mongol empire (n=5) (Jeong et al., 2020) during the Medieval Age. The age estimation of modern east Eurasia Y-chromosome STR data belonging to the C2a1a3-F1918 was determined to be 6327 ± 1736 years (Wei et al., 2018). Based on these findings, we propose that haplogroup C2a1a3-F1918 originated in ancient northeast Asia and expanded to the Mongolia Plateau, later spreading westward to Central Asia and Europe, following the route of its subbranch C2a1a3a-F3796 (Wei et al., 2018). Hence, we speculate that haplogroup C2a1a3-F1918 among Xinjiang Kazakhs may have originated in ancient northeast Asia.
Haplogroup C2a1a2-M48 is widespread in central and northern Asia, with particularly high frequencies in Tungusic and Paleosiberian speaking populations (Zerjal et al., 2002), such as Manchu groups (Xue et al., 2005). It is believed to have originated around 150,000 years ago (Karmin et al., 2015). Ancient DNA data supported the presence of haplogroup C2a1a2-M48 in the Songnen Plain of northeastern China at the end of the last glacial period (n=1) and during the early Neolithic Age (n=1) (Mao et al., 2021). It has also been found in the Boisman Culture of the Russian Far East during the middle Neolithic Age (n=1) (Wang et al., 2021). From the Iron Age onwards, haplogroup C2a1a2-M48 has mainly been identified in Central Asia, including the Xiongnu empire (n=1) and the Mongol empire (n=1) in Mongolia (Jeong et al., 2020), as well as among central steppe nomads (n=1) in the Tian Shan area of Kyrgyzstan (Damgaard et al., 2018). The westernmost ancient individual with haplogroup C2a1a2-M48 was a Hungarian from the Carpathian Basin, dated to the end of the ninth century AD (Fothi et al., 2020). In summary, the spatial and temporal distribution of haplogroup C2a1a2-M48 closely aligned with that of haplogroup C2a1a3-F1918.
Our network analysis and TMRCA estimate of the special branch DYS448-23, a Kerey-Abakh-specific cluster as previously identified (Abilev et al., 2012), indicates a rapid and recent expansion of the Kerey clan in Altay, located at the northern end of Xinjiang province. The Kazakhs had long been under pressure from the Dzungars. In 1757, Ablai Sultan, a Kazakh khan from the Middle zhuz of the Kazakh Khanate, chose to reconcile and submit to the Qing dynasty. As a result, the Kazakhs were able to migrate back to northern Xinjiang and reclaim their pasturelands in the Altay Mountains, which had been taken by the Dzungars (Perdue, 2005).
Conclusion
Our study has revealed the remarkable genetic heterogeneity of Altay Kazakhs, confirming their diverse paternal origins and the population’s genetic admixture in this region. While they possess ancient northeast Asian lineages, our findings also suggested that the present-day Altay Kazakhs and Ili Kazakhs in Xinjiang are primarily descendants of recent waves of migration to the Xinjiang region. Specifically, the Altay Kazakhs are believed to have originated from the Kerey-Abakh subclan, while the Ili Kazakhs trace their ancestry back to the Kerey-Ashmaily subclan.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the ethics committee at the School of Life Sciences in Fudan University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
BW: Conceptualization, Data curation, Formal Analysis, Software, Visualization, Writing – original draft. JL: Investigation, Visualization, Data curation, Validation, Writing – original draft. EA: Investigation, Writing – review & editing, Project administration. XC: Formal Analysis, Data curation, Writing – review & editing. ZJ: Data curation, Software, Formal Analysis, Writing – original draft. YY: Data curation, Formal Analysis, Software, Visualization, Writing – review & editing. MS: Conceptualization, Project administration, Resources, Validation, Writing – review & editing. SW: Conceptualization, Supervision, Writing – review & editing, Project administration, Validation, Writing – original draft.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (32111530227, and 32070576), National Social Science Foundation of China (19VJX074), Qian Duansheng Distinguished Scholars Program of China University of the Political Science and Law (01140065140), Cross disciplinary construction project of evidence investigation (10322308) and European Research Council (ERC) grant (ERC-2019-ADG-883700-TRAM).
Acknowledgments
We are grateful for all sample donors who participated in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1264718/full#supplementary-material
References
Abilev S., Malyarchuk B., Derenko M., Wozniak M., Grzybowski T., Zakharov I. (2012). The Y-chromosome C3* star-cluster attributed to Genghis Khan's descendants is present at high frequency in the Kerey clan from Kazakhstan. Hum. Biol. 84 (1), 79–89. doi: 10.3378/027.084.0106
Ashirbekov Y., Abaildayev A., Neupokoyeva A., Sabitov Z., Zhabagin M. (2022). Genetic polymorphism of 27 Y-STR loci in Kazakh populations from Northern Kazakhstan. Ann. Hum. Biol. 49 (1), 87–89. doi: 10.1080/03014460.2022.2039292
Ashirbekov Y., Nogay A., Abaildayev A., Zhunussova A., Sabitov Z., Zhabagin M. (2023). Genetic polymorphism of 27 Y-STR loci in Kazakh populations from Eastern Kazakhstan. Ann. Hum. Biol. 50 (1), 48–51. doi: 10.1080/03014460.2023.2170465
Bi X. (2015). “Kazak, kirgiz, tajik and tatar,” in Encyclopedia of Chinese nationalities, vol. 19 . Ed. Li D. Z. (Xi’an, China: Xi 'an: World Book Publishing Xi 'an Co. LTD).
Chi Z., Festa M. (2020). Archaeological research in the ili Region: A review. Asian Perspectives-the J. Archaeology Asia Pacific 59 (2), 338–384. doi: 10.1353/asi.2020.0018
Damgaard P. B., Marchi N., Rasmussen S., Peyrot M., Renaud G., Korneliussen T., et al. (2018). 137 ancient human genomes from across the Eurasian steppes. Nature 557 (7705), 369–374. doi: 10.1038/s41586-018-0094-2
Di Cosmo N. (1996). Ancient Xinjiang between Central Asia and China - The Nomadic factor. Anthropology Archeology Eurasia 34 (4), 87–101. doi: 10.2753/AAE1061-1959340487
Fothi E., Gonzalez A., Feher T., Gugora A., Fothi A., Biro O., et al. (2020). Genetic analysis of male Hungarian Conquerors: European and Asian paternal lineages of the conquering Hungarian tribes. Archaeological Anthropological Sci. 12 (1). doi: 10.1007/s12520-019-00996-0
Gnecchi-Ruscone G. A., Khussainova E., Kahbatkyzy N., Musralina L., Spyrou M. A., Bianco R. A., et al. (2021). Ancient genomic time transect from the Central Asian Steppe unravels the history of the Scythians. Sci. Adv. 7 (13), eabe4414. doi: 10.1126/sciadv.abe4414
Helga T., James T. R., Jill P. M. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinf. 14 (2), 178–192. doi: 10.1093/bib/bbs017
Jeong C., Wang K., Wilkin S., Taylor W. T. T., Miller B. K., Bemmann J. H., et al. (2020). A dynamic 6,000-year genetic history of eurasia's eastern steppe. Cell 183 (4), 890–904.e829. doi: 10.1016/j.cell.2020.10.015
Karmin M., Saag L., Vicente M., Wilson Sayres M. A., Järve M., Talas U. G., et al. (2015). A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome. Res. 25 (4), 459–466. doi: 10.1101/gr.186684.114
Khussainova E., Kisselev I., Iksan O., Bekmanov B., Skvortsova L., Garshin A., et al. (2021). Genetic relationship among the kazakh people based on Y-STR markers reveals evidence of genetic variation among tribes and zhuz. Front. Genet. 12. doi: 10.3389/fgene.2021.801295
Kumar V., Wang W., Zhang J., Wang Y., Ruan Q., Yu J., et al. (2022). Bronze and Iron Age population movements underlie Xinjiang population history. Science 376 (6588), 62–69. doi: 10.1126/science.abk1534
Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27 (21), 2987–2993. doi: 10.1093/bioinformatics/btr509
Li X., Zhang J., Li L., Zha L., Shi M., Ding M. (2020). Genetic polymorphism of 24 Y-STR loci in Altay Hui and Kazakh populations from northwest China. Leg. Med. (Tokyo) 47, 101760. doi: 10.1016/j.legalmed.2020.101760
Mair V. H. (2021). The cultures of ancient Xinjiang, Western China: crossroads of the silk roads. Asian Ethnicity 22 (1), 209–211. doi: 10.1080/14631369.2020.1843401
Mao X., Zhang H., Qiao S., Liu Y., Chang F., Xie P., et al. (2021). The deep population history of northern East Asia from the Late Pleistocene to the Holocene. Cell 184 (12), 3256–3266.e3213. doi: 10.1016/j.cell.2021.04.040
Mei T., Zhang L. P., Liu Y. S., Chen J. G., Meng H. T., Yan J. W., et al. (2016). 24 Y-chromosomal STR haplotypic structure for Chinese Kazak ethnic group and its genetic relationships with other groups. Int. J. Legal. Med. 130 (5), 1199–1201. doi: 10.1007/s00414-016-1331-6
Ning C., Li T., Wang K., Zhang F., Li T., Wu X., et al. (2020). Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 11 (1), 2700. doi: 10.1038/s41467-020-16557-2
Otarbaeva B. (1998). A brief history of the kazak people*. Nationalities Papers 26 (3), 421–432. doi: 10.1080/00905999808408575
Pan Y., Wen J., Ning Z., Yuan Y., Liu X., Yang Y., et al. (2023). Comparative genomic and transcriptomic analyses reveal the impacts of genetic admixture in kazaks, uyghurs, and huis. Mol. Biol. Evol. 40 (3), msad054. doi: 10.1093/molbev/msad054
Perdue P. C. (2005). China marches west. The Qing conquest of Central Eurasia (Cambridge: Harvard University Press, Belknap Press).
Poznik G. D., Henn B. M., Yee M. C., Sliwerska E., Euskirchen G. M., Lin A. A., et al. (2013). Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science 341 (6145), 562–565.
Ralf A., Montiel G. D., Zhong K., Kayser M. (2018). Yleaf: Software for human Y-chromosomal haplogroup inference from next-generation sequencing data. Mol. Biol. Evol. 35 (5), 1291–1294. doi: 10.1093/molbev/msy032
Rogers J. (2012). Inner asian states and empires: Theories and synthesis. J. Archaeological Res. 20 (3), 205–256. doi: 10.1007/s10814-011-9053-2
Rootsi S., Myres N. M., Lin A. A., Järve M., King R. J., Kutuev I., et al. (2012). Distinguishing the co-ancestries of haplogroup G Y-chromosomes in the populations of Europe and the Caucasus. Eur. J. Hum. Genet. 20 (12), 1275–1282. doi: 10.1038/ejhg.2012.86
Shan W., Ablimit A., Zhou W., Zhang F., Ma Z., Zheng X. (2014a). Genetic polymorphism of 17 Y chromosomal STRs in Kazakh and Uighur populations from Xinjiang, China. Int. J. Legal. Med. 128 (5), 743–744. doi: 10.1007/s00414-013-0948-y
Shan W., Ren Z., Wu W., Hao H., Abulimiti A., Chen K., et al. (2014b). Maternal and paternal diversity in Xinjiang Kazakh population from China. Genetika 50 (11), 1374–1385. doi: 10.7868/S0016675814110149
Sun Y. (2022). Eurasian crossroads: A history of xinjiang, revised and updated edition. J. Asian Stud. 81 (4), 800–801. doi: 10.1017/S0021911822001541
Tarlykov P. V., Zholdybayeva E. V., Akilzhanova A. R., Nurkina Z. M., Sabitov Z. M., Rakhypbekov T. K., et al. (2013). Mitochondrial and Y-chromosomal profile of the Kazakh population from East Kazakhstan. Croat. Med. J. 54 (1), 17–24. doi: 10.3325/cmj.2013.54.17
Wang C. C., Wang L. X., Shrestha R., Zhang M., Huang X. Y., Hu K., et al. (2014). Genetic structure of Qiangic populations residing in the western Sichuan corridor. PLoS One 9 (8), e103772. doi: 10.1371/journal.pone.0103772
Wang C. C., Yeh H. Y., Popov A. N., Zhang H. Q., Matsumura H., Sirak K., et al. (2021). Genomic insights into the formation of human populations in East Asia. Nature 591 (7850), 413–419. doi: 10.1038/s41586-021-03336-2
Wei L. H., Yan S., Lu Y., Wen S. Q., Huang Y. Z., Wang L. X., et al. (2018). Whole-sequence analysis indicates that the Y chromosome C2*-Star Cluster traces back to ordinary Mongols, rather than Genghis Khan. Eur. J. Hum. Genet. 26 (2), 230–237. doi: 10.1038/s41431-017-0012-3
Wen S. Q., Sun C., Song D. L., Huang Y. Z., Tong X. Z., Meng H. L., et al. (2020). Y-chromosome evidence confirmed the Kerei-Abakh origin of Aksay Kazakhs. J. Hum. Genet. 65 (9), 797–803. doi: 10.1038/s10038-020-0759-1
Xu D., Li H. (2017). Languages and genes in northwestern China and adjacent regions (Singapore: Springer), 107–120.
Xue Y., Zerjal T., Zerjal T., Zhu S., Lim S. K., Shu Q., et al. (2005). Recent spread of a Y-chromosomal lineage in northern China and Mongolia. Am. J. Hum. Genet. 77 (6), 1112–1116. doi: 10.1086/498583
Zerjal T., Wells R. S., Yuldasheva N., Ruzibakiev R., Tyler-Smith C. (2002). A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am. J. Hum. Genet. 71 (3), 466–482. doi: 10.1086/342096
Zhabagin M., Balanovska E., Sabitov Z., Kuznetsova M., Agdzhoyan A., Balaganskaya O., et al. (2017). The connection of the genetic, cultural and geographic landscapes of transoxiana. Sci. Rep. 7 (1), 3085. doi: 10.1038/s41598-017-03176-z
Zhabagin M., Sabitov Z., Tarlykov P., Tazhigulova I., Junissova Z., Yerezhepov D., et al. (2020). The medieval Mongolian roots of Y-chromosomal lineages from South Kazakhstan. BMC. Genet. 21 (Suppl 1), 87. doi: 10.1186/s12863-020-00897-5
Zhabagin M., Sabitov Z., Tazhigulova I., Alborova I., Agdzhoyan A., Wei L. H., et al. (2021). Medieval super-grandfather founder of western kazakh clans from haplogroup C2a1a2-M48. J. Hum. Genet. 66 (7), 707–716. doi: 10.1038/s10038-021-00901-5
Zhang D., Cao G., Xie M., Cui X., Xiao L., Tian C., et al. (2021). RETRACTED ARTICLE: Y Chromosomal STR haplotypes in Chinese Uyghur, Kazakh and Hui ethnic groups and genetic features of DYS448 null allele and DYS19 duplicated allele. Int. J. Legal. Med. 135 (3), 1119. doi: 10.1007/s00414-019-02049-6
Keywords: Altay Kazakhs, Ili Kazakhs, C2a1a3-F1918, ancient northeast Asia, Kerey
Citation: Wang B, Liang J, Allen E, Chang X, Jiang Z, Yu Y, Shi M and Wen S (2023) Y chromosome evidence confirms northeast Asian origin of Xinjiang Kazakhs and genetic influence from 18th century expansion of Kerey clan. Front. Ecol. Evol. 11:1264718. doi: 10.3389/fevo.2023.1264718
Received: 21 July 2023; Accepted: 21 August 2023;
Published: 18 September 2023.
Edited by:
Guanglin He, Sichuan University, ChinaReviewed by:
Wang Zhiyong, Kunming Medical University, ChinaQiyan Wang, Guizhou Medical University, China
Copyright © 2023 Wang, Liang, Allen, Chang, Jiang, Yu, Shi and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meisen Shi, shimeisen2000@163.com; Shaoqing Wen, wenshaoqing@fudan.edu.cn
†These authors have contributed equally to this work and share first authorship
 Bangyan Wang
Bangyan Wang Jiayu Liang2†
Jiayu Liang2† Edward Allen
Edward Allen Xin Chang
Xin Chang Meisen Shi
Meisen Shi Shaoqing Wen
Shaoqing Wen