
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 14 November 2023
Sec. Population, Community, and Ecosystem Dynamics
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1251039
This article is part of the Research Topic Untangling the Complexity: Recent Advances in Understanding Interactions Between Plants, Pathogens and Herbivorous Insects View all 5 articles
Many plant pathogens are transmitted by generalist insects that move between host species, and vector preference and performance on different hosts affects pathogen transmission. Most studies of host–vector–pathogen interactions focus on a single host, however, and it is relatively unknown if viruses affect the host breadth of vectors. For example, although pea aphids (Acyrthosiphon pisum) often form tight associations with specific hosts, viruses transmitted by aphids can break down host defenses, potentially weakening these associations and increasing the host breadth of their vectors. Here, we assessed vector host preference and performance in an experiment with five pea aphid colonies with distinct host associations, five host species, and two viruses (Pea enation mosaic virus [PEMV] and Bean leafroll virus [BLRV]). Our prediction was that pea aphids would have higher performance on their inferior hosts and be less discriminating among hosts when those are infected, compared with noninfected hosts. Our results show that aphid performance, measured as the rate of increase on hosts, differed among hosts based on their host association, and relative performance among hosts was indeed altered by virus infection. Aphid preferences, measured using a choice bioassay in which aphids could settle on any of the five host species, also differed among aphid colonies but was not affected by virus infection status. We also did not find that infection status of hosts altered the performance of aphids on their preferred host. Furthermore, virus infection did not disrupt preference performance relationships for the aphids. Overall, our study suggests that host preference and performance are deeply ingrained in pea aphids, and that host associations may be robust to the presence of host-manipulating plant pathogens.
Many plant viruses and other vector-borne plant pathogens can affect host plant traits as well as vector preferences and performance in ways that facilitate transmission (Ingwell et al., 2012; Mauck et al., 2014; Eigenbrode et al., 2018). However, while many plant viruses have multiple hosts and are transmitted by generalist arthropod vectors (Power and Flecker, 2003), most of the theory and empirical work on host–vector–pathogen interactions has been built upon studies involving a single host species (Eigenbrode et al., 2018). The ability of vectors to move between host species can affect virus prevalence by facilitating interspecific transmission between transient hosts, or by ensuring the maintenance of high vector populations over time even when hosts are scarce (Duffus, 1971; Swei et al., 2011; Ashby et al., 2014; McLeish et al., 2018). Yet, relatively few studies have assessed how viruses may directly or indirectly affect vector preferences or performance across multiple host species.
Many insect herbivores, including vector species, develop specialized races to specific hosts due to natural selection and genetic drift (Drès and Mallet, 2002; Loxdale et al., 2011). Specialized insect populations exhibit higher performance on their associated hosts and prefer these hosts to others in their potential host range. Similarly, adaptation of plant viruses to specific host immune systems often leads to specialization via antagonistic pleiotropies that limit viruses’ ability to overcome host defenses across plant taxa (García-Arenal and Fraile, 2013). On one hand, plant pathogens and vectors experience selection pressure towards host specialization, but specialization can become maladaptive when hosts are transient or pathogens are restricted to a small number of insect vectors.
The pea aphid, Acyrthosiphon pisum Harris (Hemiptera: Aphididae), is a generalist vector that feeds on hundreds of legume species (Fabaceae) (Eastop, 1971; Peccoud and Simon, 2010). However, populations of pea aphids often associate with specific hosts (Via et al., 2000; Peccoud et al., 2008; Eigenbrode et al., 2016). The pea aphid is also a vector for over 30 viruses such as Pea enation mosaic virus (PEMV) and Bean leafroll virus (BLRV), pathogens that infect several legumes (Rashed et al., 2018). Studies show that PEMV and BLRV affect host attractiveness and vector performance (Hodge and Powell, 2010; Wu et al., 2014; Davis et al., 2017), but it is not known whether these viruses affect performance or preferences of pea aphids with ingrained host associations. We predict that if viruses require interspecific transmission for persistence, vector host preference and performance across hosts may be altered by viruses (Chesnais et al., 2019).
Here we tested the idea that host preferences and performance of pea aphids from colonies with distinct host associations will be altered when hosts are infected with viruses. While host preferences and performance seem to be deeply imprinted in pea aphids (Via, 1991; Eigenbrode et al., 2016), whether pathogens persist in transient host populations may cause an evolutionary conflict with consequences for the evolution of host–virus–vector pathosystems. In the Pacific Northwest US (PNW), pea aphids often associate with either alfalfa or pea crops (Eigenbrode et al., 2016), but perennial legumes such as hairy vetch, red clover, and alfalfa can sustain early pea aphid infestations in spring and are reservoirs for PEMV or BLRV (Rashed et al., 2018; Clark et al., 2023). We hypothesize that plants infected with PEMV or BLRV alter pea aphid performance and preference in such a way that interspecific transmission is favored. Here, we used a series of experiments to test whether aphid colonies with specific host associations had altered host preferences and performance when hosts were infected with viruses as compared to uninfected. Overall, our study is among the first to test the notion that viruses manipulate host preferences and performance for multiple hosts in ways that promote interspecific transmission.
We established five clonal colonies of pea aphid, Acyrthosiphon pisum, from aphids that were collected from pea, vetch, clover, or alfalfa fields near Moscow, ID, USA or McMinnville OR, USA in 2017. Two of these colonies (AL-PK-ID, AL-GN-OR) exhibited better performance on alfalfa (“alfalfa colonies”); the other three colonies, two collected from pea (PA-GN-ID, PA-GN-OR) and one from clover (CL-PK-OR), performed better on pea (“pea colonies”). Prior field surveys suggest pea aphids in the PNW feed on species of vetch (Vicia) and clover (Trifolium) in addition to cultivated legumes (Clark et al., 2023). We maintained aphid colonies in greenhouses at the University of Idaho (Moscow, ID, USA) in 60 × 60 × 60cm mesh tents (BugDorm 2120F; BioQuip, Rancho Dominguez, CA). We reared colonies at 20 ± 2°C, with a photoperiod of 18:6 h (L:D), and 50% relative humidity. To remove any effects of the maternal host on pea aphids’ host preference or performance (Slater et al., 2019), the colonies were reared on potted fava bean (Vicia faba L.), a universal host for pea aphid (Peccoud et al., 2014), for at least 20 generations before experiments. We confirmed colonies were genetically separate using 12 autosomal microsatellite loci (described in Eigenbrode et al., 2016).
Our study used six hosts: (i) pea (Pisum sativum L.), (ii) red clover (Trifolilium pratense L.), (iii) common vetch (Vicia sativa L.), (iv) alfalfa (Medicago sativa L.), (v) lentil (Lens culinaris Medik.), and (vi) fava bean (Vicia faba). We grew these host plants for experiments in conditions identical to those for maintaining aphid colonies. For experiments comparing virus-infected to uninfected plants, we used infectious aphids to inoculate plants with either PEMV or BLRV. We collected infectious colonies of pea aphid from infected V. faba, where both viruses are maintained under controlled conditions. We originally collected virus isolates from commercial fields of alfalfa (BLRV) or pea (PEMV) near Moscow, ID, USA, and both insects and plants within the infectious colonies are tested approximately every two weeks for the presence of BLRV or PEMV as described by Vemulapati et al. (2014), to make sure infection levels are >95%.
For bioassays, we grew plants in 15-cm diameter pots filled with commercial potting mix (Sunshine Mix #1; SunGro Horticulture, Bellevue, WA, USA). Fifteen days after emergence (stage 2, BBCH general scale), we inoculated plants by placing five aphids, from an infectious colony of either BLRV or PEMV, in a clip cage (5 cm diameter) onto a leaf from the top node of the test plant. Since alfalfa is not a host for PEMV, alfalfa plants were only exposed to aphids from the BLRV-infectious colony. To ensure virus transmission, we allowed a 3-day inoculation access period, after which aphids were removed using a soft bristled paintbrush. Transmission rates of both BLRV and PEMV by pea aphids range from 90–100% and from 80–100%, for acquisition and inoculation access periods of 2 d and 4 h, and up to 4 h and 4 d, respectively (Fargette et al., 1982; Skaf and Makkouk, 1988). We maintained these plants to be aphid-free until they were used in experiments. To control for the effects of aphid feeding on the test plants, we performed a “sham” inoculation with non-infectious aphids using the same timings and clip cages as virus treatments.
We quantified aphid performance in a no-choice bioassay. We placed eight pea aphids on individual plants infected with PEMV, BLRV, or sham-inoculated among each isolated aphid colony. We counted the total number of aphids present on each plant after 7 d. In this assay, we completed two replicates for each combination of aphid colony × plant species × virus infection status (PEMV, BLRV, sham) across two blocks. In a third block, three additional replicates were completed across all plants, with only two additional replicates for alfalfa and lentil due to low germination rates. In all, six fully factorial replicates of each treatment were conducted. Figure S3 is a photograph of the bioassay set up.
To evaluate the behavioral responses of pea aphids to different host plants relative to virus infection (host preference), we quantified settling behavior of individual aphids in a multiple host species test. We constructed a circular arena 30 cm in diameter with a flat foam core floor and enclosed with a clear plastic wall coated with Fluon (PTFE-30). To hold the stems of test plants, we cut evenly spaced notches around the perimeter. To provide some control over difference in plant sizes, we germinated hosts at different times based on known growth rates in greenhouse conditions. For each trial, we released 50 apterous aphids into the center of the arena. After 24 h, we counted the number of aphids on each test plant, and the remaining aphids that either died or never reached a test plant were discarded. Mortality was only 1.6% for aphids tested across all trials. We conducted five replicate trials for each aphid colony × virus treatment (PEMV, BLRV, sham) followed by an additional five replicate trials in a second time block, and the location of the treatments within the circular arena was assigned randomly each trial to exclude the effect of orientation behaviors triggered by stimulus other than the treatments. Figure S4 is a photograph of the bioassay set up.
All analyses were conducted in R version 4.2.2 (R Development Core Team, 2022). Three models, in which aphid colonies were pooled by their host affinity to increase statistical power, were used to evaluate the outcomes of bioassays. First, we modeled the effects of aphid colony, host species, and virus status on the total number of aphids counted on a host plant at the end of each assay (“performance”). All models fitted to these data followed a negative binomial distribution using the glm.nb function in the MASS package in R (Venables and Ripley, 2002). Second, we modeled effects of aphid colony, host species, and viral status on the number of aphids that moved towards a host plant during each bioassay (“preference”); this model used a normal distribution. Third, we used a linear model to assess correlations between preference and performance with an interaction term of virus status using pooled results from both bioassays. We ran this model to test the hypothesis that virus status modifies the known correlation between preference and performance among aphid colonies. In this third model, the predictor variable aphid counts were log-transformed to meet model assumptions.
For the first two models, we employed stepwise model selection. We started with a fully specified generalized linear model including the three-way interaction between aphid colony, host species, and virus status, as well as all second and first-order terms. Given the relatively small size of the dataset to this large model, we sought to reduce model complexity and avoid overfitting via stepwise regression. After running the full models, the stepAIC function in the MASS package was applied to each model; this function sequentially reduces the number of parameters to produce a model with the lowest AIC (Venables and Ripley, 2002). Both full and reduced models had significance tests completed using the car package in R (Fox and Weisberg, 2018). Post-hoc tests via Tukey HSD were calculated using the emmeans package (Lenth, 2023). Unless otherwise indicated, bar plots present mean and standard error of the means.
Aphid counts on each host plant were significantly different among the host species and followed patterns expected from prior research on aphid performance (Eigenbrode et al., 2016; Clark et al., 2023). The effect of host species was highly significant (χ2 = 23.8, df = 5, P < 0.001, Table S1), with aphid performance being lowest on red clover and highest on fava bean and lentil (Figure 1). There was also a significant effect of virus status on aphid performance (χ2 = 6.66, df = 2, P = 0.040, Figure S1; Table S1). Examination of this virus effect, when averaging across all host species, showed a slight increase in overall aphid abundance among PEMV infected plants compared to BLRV and sham plants. Virus status of hosts also significantly impacted aphid performance, with non-additive affects based on host species (plant:virus interaction: χ2 = 32.2, df = 0, P < 0.001, Table S1). As an example, aphids performed 69.47% better on alfalfa infected with BLRV compared to a sham (Figure 2, Tukey HSD). Conversely, we observed a 80.53% and 73.35% reduction in aphid performance on vetch exposed to BLRV compared to PEMV and sham plants, respectively (Figure 2, Tukey HSD). Remaining host species did not have significant differences in aphid performances based on virus status (Figure 2, Tukey HSD).
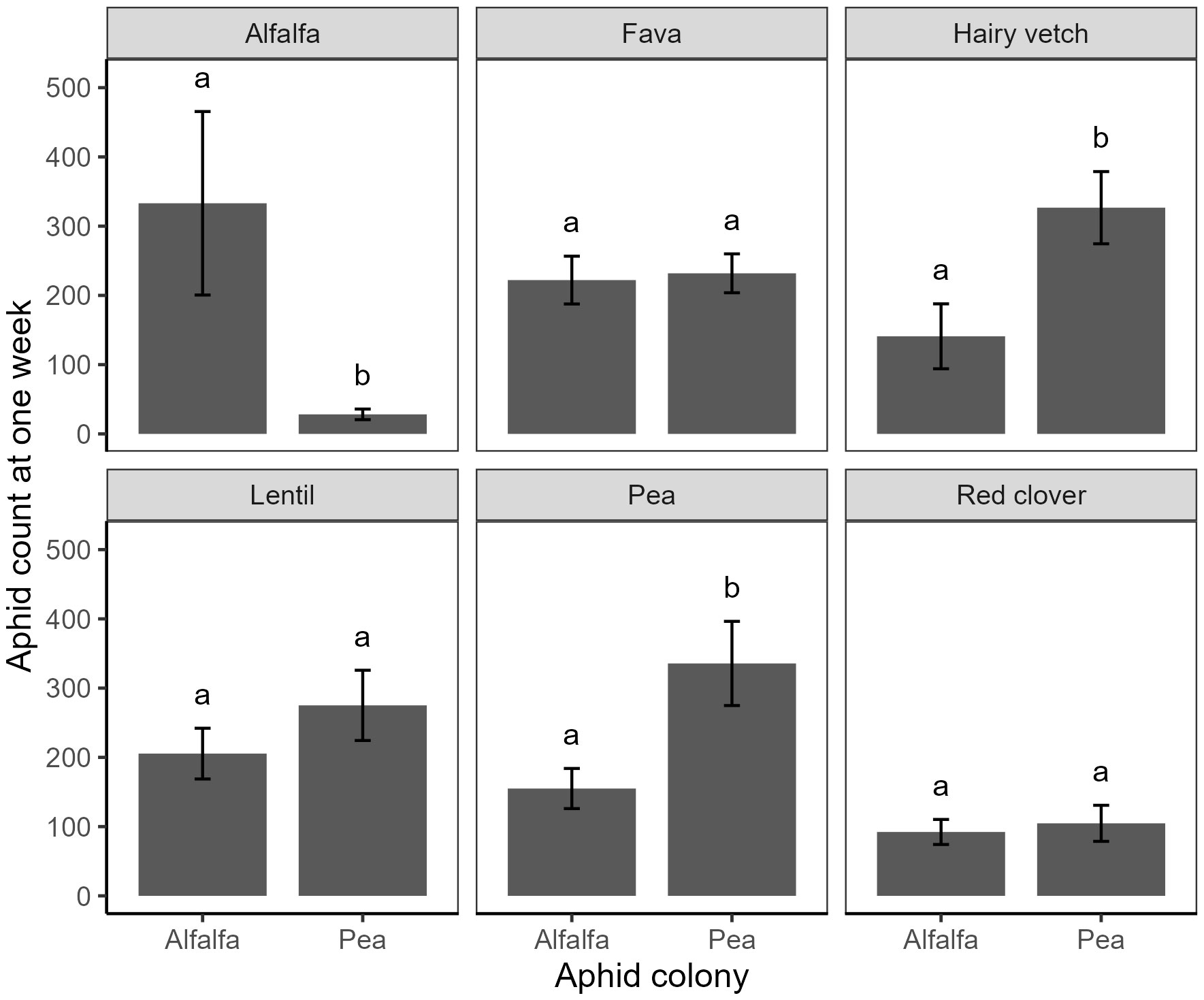
Figure 1 Performance, measured as the number of aphids after one week, of pea aphids from five colonies (two associated with alfalfa and three with pea), in six host species (panels). Means within a panel capped with different letters were significantly different (Tukey HSD: P < 0.05).
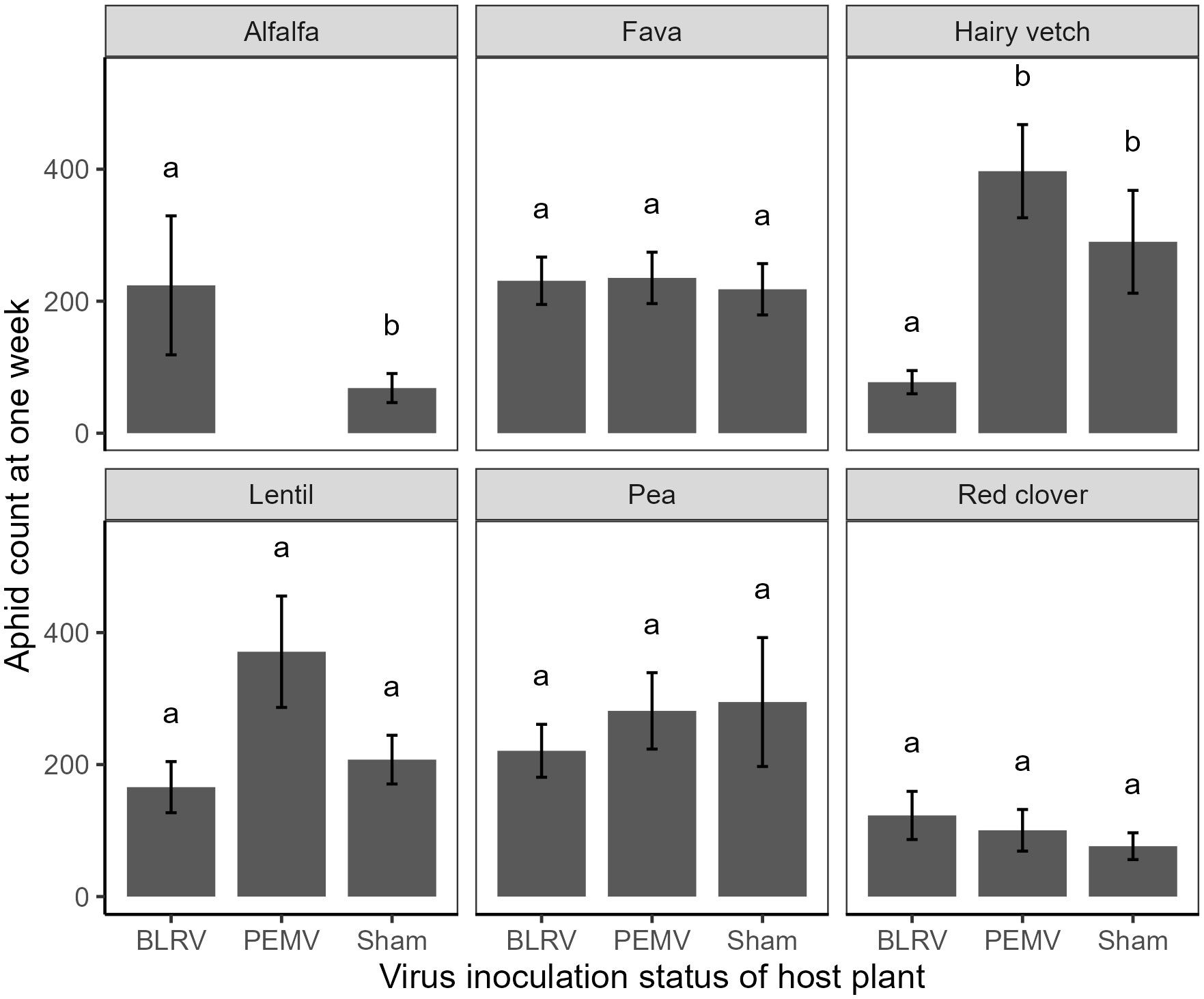
Figure 2 Performance, measured as the number of aphids after one week, pooled across five pea aphid colonies in six host species (panels) previously inoculated with Pea enation mosaic virus (PEMV), Bean leafroll virus (BLRV), or sham inoculated. Means within a panel capped with different letters were significantly different (Tukey HSD: P < 0.05).
Aphid performance also differed based on host species (aphid colony:plant interaction: χ2 = 57.3, df = 5, P < 0.001, Table S1). As predicted, the alfalfa colonies performed better on alfalfa compared to pea colonies (Figure 1, Tukey HSD), while pea colonies performed better on vetch and pea compared to the alfalfa colonies (Figure 1, Tukey HSD). We observed no differences in aphid performance among the remaining host species (Figure 1. Tukey HSD). Given that analysis of deviance tables were calculated using Type II Wald-χ2 tests (Fox and Weisberg, 2018), results suggest the aphid colony:plant interaction term accounted for the effect of the aphid colony on aphid performance (χ2 = 0.45, df = 1, P = 0.50; Table S1).
Aphid preferences differed significantly among host species (χ2 = 111.8, df = 5, P < 0.001, Table S2), and this preference varied according to aphid colony (χ2 = 51.8, df = 5, P < 0.001, Table S2). Matching results from aphid performance, the alfalfa colonies settled more on alfalfa hosts compared to the pea colonies, and the pea colonies settled more on vetch and clover than the alfalfa colonies (Figure 3, Tukey HSD). Paradoxically, we did not see greater settling on pea plants by the pea colonies compared to the alfalfa colonies (Figure 3, Tukey HSD).
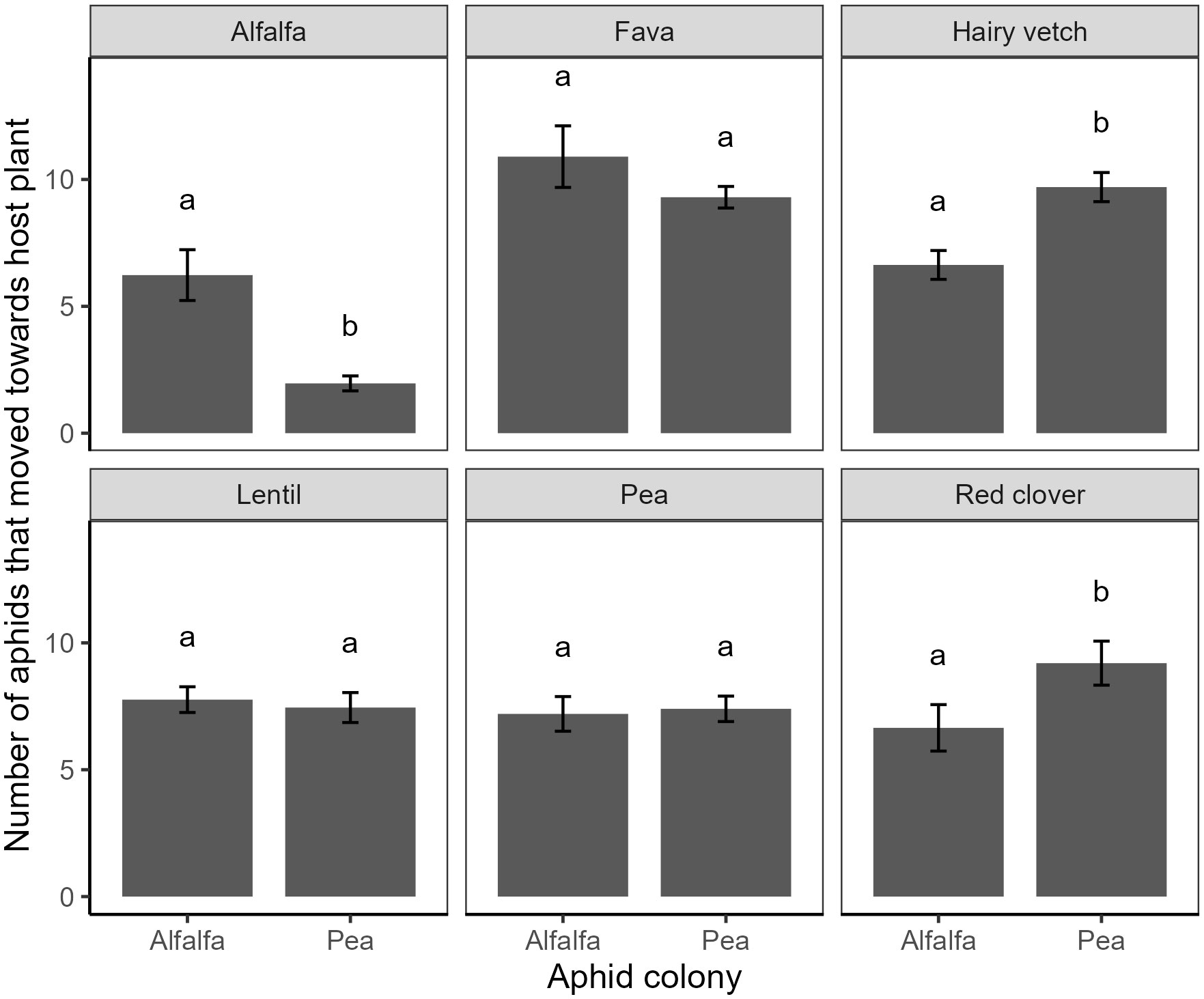
Figure 3 Preference of pea aphids collected from colonies associated with either alfalfa or pea for six host species (panels) measured as the number of aphids that moved towards the host species in a choice bioassay. Means within a panel capped with different letters are significantly different (Tukey HSD: P < 0.05).
All terms in the generalized linear model for settling preference, including virus status, were dropped from final model using our stepwise AIC approach. Consequently, there was no statistical evidence for virus status of the host species altering aphid preference for either the alfalfa or pea colonies (Table S2), nor did we observe any evidence that virus status influenced the effects of aphid colony origin or plant species identity (Table S2).
We observed a positive relationship between aphid host preference and performance, as aphids preferred host plants on which they had higher performance (χ2 = 21.7, df =1, P < 0.001, Table S3; Figure 4). However, this relationship did not appear to be modified for plants infected with BLRV or PEMV (aphid:virus interaction, χ2 = 1.13, df = 2, P = 0.57, Table S3).
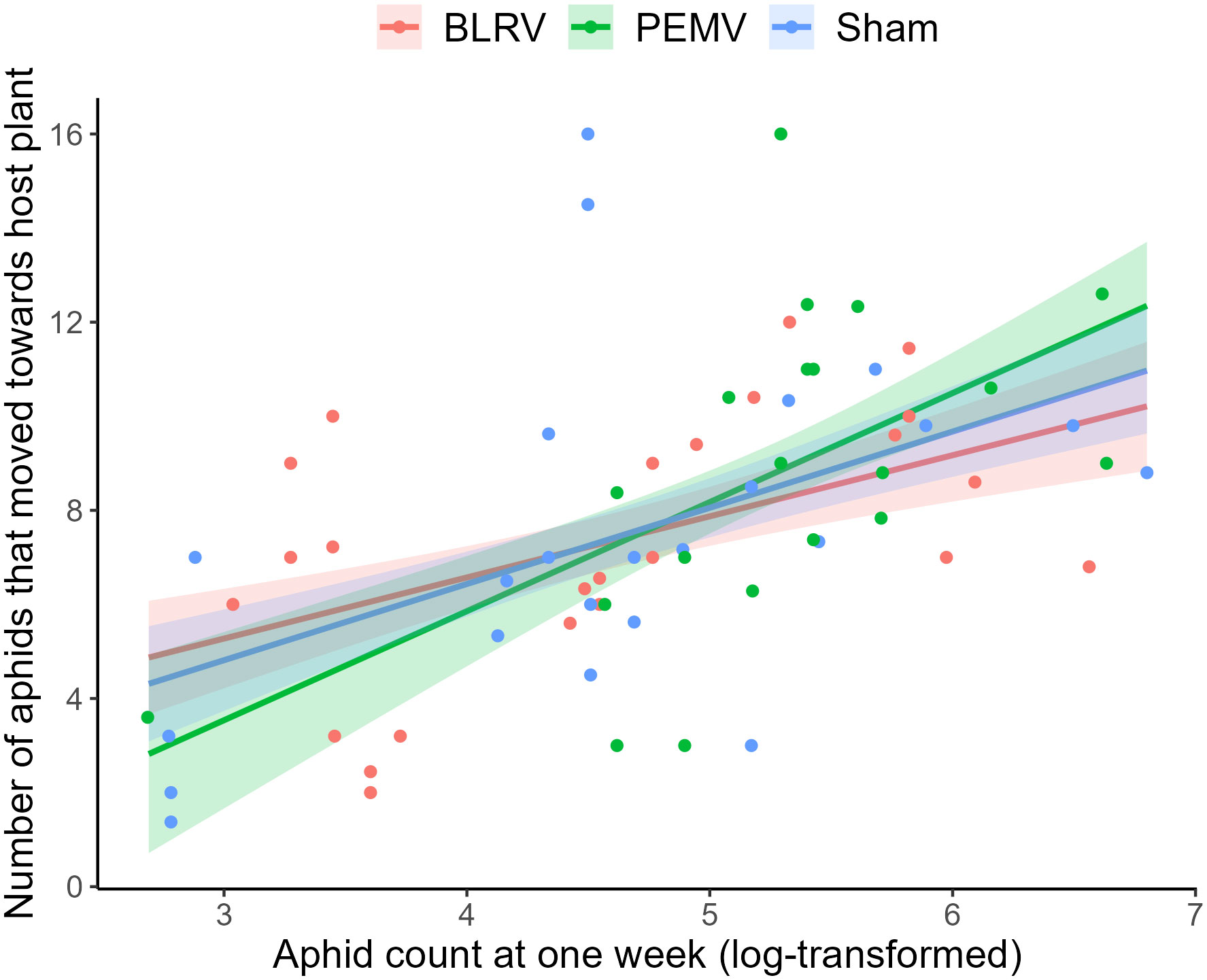
Figure 4 Performance-preference correlation of pea aphids collected from five colonies in six host species previously inoculated with Pea enation mosaic virus (PEMV), Bean leafroll virus (BLRV), or sham inoculated. Performance was measured as the rate of increase of aphids after one week, and preference as the number of aphids that moved towards the host species in a choice bioassay. The lines represent linear models for the performance-preference correlation of pea aphids in plants previously inoculated with the viruses or sham inoculated, and the corresponding bands are the 95% confidence intervals of the fitted lines.
Plant virus infection of hosts can alter performance and preference of pea aphid, consistent with manipulation (Wu et al., 2014; Davis et al., 2017). In this study, we predicted that pea aphid host preferences and performance would depend on virus infection status across host species. We found that the effect of virus infection on aphid performance was not consistent across all tested host species. Plants infected with BLRV affected aphid performance, with higher aphid abundance on BLRV-infected alfalfa and lower on BLRV-infected vetch. While we did not find any changes in aphid performance among PEMV-infected hosts, across all host species, aphids were slightly more abundant on PEMV-infected plants. However, no changes to aphid preferences among infected or sham plants were found, supporting the assertion that viruses may indirectly affect vectors by altering their fitness similarly across host species (Mauck et al., 2018). Without concomitant behavioral changes, we only have partial support for effects of viruses on vector host preferences, although evidence suggests aphids tend to prefer plants where they perform better, regardless of virus-infection status.
Although most studies of vector-borne plant virus transmission focus on a single host plant species, in field settings many plant viruses often rely on transmission among multiple hosts, especially when some hosts are annual or transient (Wilke et al., 2006; Elena et al., 2009). This phenomenon is important for annual crop systems (such as pea, lentil, and garbanzo bean) where vectors and pathogens must use perennial hosts when crop plants are not present. Consequently, it may be adaptive for viruses to facilitate interspecific transmission by affecting vector host preferences and performance. For example, we predicted that host plants infected with PEMV would trigger different aphid host preferences compared to sham-inoculated plants; specifically, we predicted that host associations would be weaker when viruses were present if viruses break down specific plant defenses. However, we did not find evidence that host preferences among aphids differed when presented with PEMV-, BLRV- or sham-infected plants.
It is possible that transient virus infections affect aphid performance, but longer-term virus infection might be required to alter host preferences. Enzyme linked immunosorbent assay (ELISA) tests carried out before the experiments confirmed the presence of PEMV and BLRV in 41.55% and 45.45% of the test plants that were inoculated with infectious aphids, respectively. Although the virus detection did not reach the expected levels, we rely on the high transmission efficiency of both viruses by pea aphids (Fargette et al., 1982; Skaf and Makkouk, 1988) to ensure that inoculated plants had a progressing infection. In fact, the occurrence of false negatives in plant virus detection is common when the disease has not progressed sufficiently due to the irregular distribution of plant virus titers within infected hosts (Hipper et al., 2013; Lacroix et al., 2016). Regardless of the virus detection rate, while pea aphids can detect a wide range of plant volatiles, facilitating host recognition (Robertson et al., 2019; Shih et al., 2023), these cues and others from infected plants may not be altered sufficiently to modify aphid preference until later stages of disease progression than we tested here. We carried out the whole data analysis only using the plants that tested positive for the inoculated viruses and the results were similar, except for the reduced statistical power due to the reduced sample size (Tables S4, S5; Figure S2). Finally, our study examined plant-mediated mechanisms rather than comparing behavioral differences in viruliferous compared to non-viruliferous vectors (Chesnais et al., 2020). For example, beet leafhoppers carrying Beet curly top virus have increased probing behavior (Lee et al., 2022), while bird-cherry oat aphids carrying Barley yellow dwarf virus prefer perennial grasses relative to other hosts. Additional work in pea aphids should consider comparisons of host preferences and performance for viruliferous and non-viruliferous aphids (e.g., Chesnais et al., 2020).
Many generalist insect herbivores exhibit higher performance on single species of hosts, and these often are host species they could have fed on for multiple generations (Garrido et al., 2012). Such mechanisms have long been predicted to drive evolution of dietary specialization, generated by selective pressures to adapt to host resources present in specific environments (Van Zandt and Mopper, 1998). Pea aphids are a globally-distributed species, but their hosts vary worldwide, and unique populations form clonal associations with both crop and non-crop hosts, with some populations exhibiting local adaptation (Caillaud and Via, 2000). Host specialization implies that there will be performance trade-offs that often match host preferences and reduce the diversity of plants that herbivores can exploit (Berlocher and Feder, 2002). For example, our study supports prior work that shows “pea colonies” have dramatically reduced performance on alfalfa compared to competing “alfalfa colonies” (Eigenbrode et al., 2016); however, we did not find evidence for a similar trade-off for “alfalfa colonies” aphids with respect to pea.
We also predicted that virus-infection status of host plants would alter host associations in aphids. For example, our hypothesis was that if viruses increased performance on alternative hosts, that vector host associations might weaken when viruses were present. In other words, if viruses facilitate greater feeding breadth by aphids by increasing performance across hosts, aphid preferences for particular hosts should diminish. However, neither of our factorially-designed bioassays show a disruptive effect of virus-infection status on host preferences by pea aphids, suggesting that indirect effects of viruses on vector host preferences and performance may indeed be operating independently. To resolve this issue, further work could explore how plant responses to infection, and resulting effects on vectors, operate over longer time periods.
Correlations between aphid host preferences, performance, and reproductive isolation have been established as a function of local adaptations or recent co-evolution (Via et al., 2000; Hawthorne and Via, 2001; Queller and Strassmann, 2018). By facilitating higher reproductive output on specific host species, strong host associations can exacerbate outbreaks of insect herbivores in agroecosystems or increase the likelihood of outbreaks occurring in the future. Locally adapted herbivore populations may be more capable of breaking down the defenses of specific hosts that are abundant in the local environment, but not those of hosts that are rare (Scriber, 2002; Garrido et al., 2012). We did indeed observe some additive effects between aphid colonies associated with alfalfa and viral infection. BLRV-infected alfalfa plants had higher pea aphid abundance, and “alfalfa colonies” performed better on alfalfa hosts. While only isolated to a single host species in our bioassays, the pattern suggests synergy between local adaptation and vector-borne pathogens which could exacerbate economic damage from pea aphids. However, host manipulation by viruses may slow viral epidemics within crops by encouraging viruliferous vectors to move into non-crop plants (Shoemaker et al., 2019) or by causing vectors to orient towards otherwise inferior host plants (Mauck, 2016). Overall, our study provides evidence that gauging risk of outbreaks of vectors and associated plant pathogens requires an integrative approach that considers how viruses may alter vector performance and host preferences across a range of locally-relevant host species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: OSF, 10.17605/OSF.IO/KNYTA.
The manuscript presents research on animals that do not require ethical approval for their study.
SE originated the ideas. RC and SE completed bioassays. RC performed analyses under advisement by DC and SE. All authors contributed to the article and approved the submitted version.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by USDA-NIFA awards 2017-67013-26537 and 2002-67013-36422.
We thank students who aided the project including J. Wallis, W. Troutman, and L. Dean.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1251039/full#supplementary-material
Ashby B., Gupta S., Buckling A. (2014). Spatial structure mitigates fitness costs in host-parasite coevolution. Am. Nat. 183, E64–E74. doi: 10.1086/674826
Berlocher S. H., Feder J. L. (2002). Sympatric speciation in phytophagous insects: moving beyond controversy? Annu. Rev. Entomol. 47, 773–815. doi: 10.1146/annurev.ento.47.091201.145312
Caillaud M. C., Via S. (2000). Specialized feeding behavior influences both ecological specialization and assortative mating in sympatric host races of pea aphids. Am. Nat. 156, 606–621. doi: 10.1086/316991
Chesnais Q., Caballero Vidal G., Coquelle R., Yvon M., Mauck K., Brault V., et al. (2020). Post-acquisition effects of viruses on vector behavior are important components of manipulation strategies. Oecologia 194, 429–440. doi: 10.1007/s00442-020-04763-0
Chesnais Q., Mauck K. E., Bogaert F., Bamière A., Catterou M., Spicher F., et al. (2019). Virus effects on plant quality and vector behavior are species specific and do not depend on host physiological phenotype. J. Pest Sci. 92, 791–804. doi: 10.1007/s10340-019-01082-z
Clark R. E., Basu S., Eigenbrode S. D., Oeller L. C., Crowder D. W. (2023). Risk assessment for non-crop hosts of pea enation mosaic virus and the aphid vector Acyrthosiphon pisum. Agric. For. Entomol. 25(3), 427–434. doi: 10.1111/afe.12564
Davis T. S., Wu Y., Eigenbrode S. D. (2017). The effects of Bean leafroll virus on life history traits and host selection behavior of specialized pea aphid (Acyrthosiphon pisum, Hemiptera: Aphididae) genotypes. Environ. Entomol. 46, 68–74. doi: 10.1093/ee/nvw150
Drès M., Mallet J. (2002). Host races in plant–feeding insects and their importance in sympatric speciation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 357, 471–492. doi: 10.1098/rstb.2002.1059
Duffus J. E. (1971). Role of weeds in the incidence of virus diseases. Annu. Rev. Phytopathol. 9, 319–340. doi: 10.1146/annurev.py.09.090171.001535
Eastop V. F. (1971). Keys for the identification of Acyrthosiphon (Hemiptera: Aphididae). Bull. Br. Mus. Entomol. 26, 1–115.
Eigenbrode S. D., Bosque-Pérez N. A., Davis T. S. (2018). Insect-borne plant pathogens and their vectors: ecology, evolution, and complex interactions. Annu. Rev. Entomol. 63, 169–191. doi: 10.1146/annurev-ento-020117-043119
Eigenbrode S. D., Davis T. S., Adams J. R., Husebye D. S., Waits L. P., Hawthorne D. (2016). Host-adapted aphid populations differ in their migratory patterns and capacity to colonize crops. J. Appl. Ecol. 53, 1382–1390. doi: 10.1111/1365-2664.12693
Elena S. F., Agudelo-Romero P., Lalić J. (2009). The evolution of viruses in multi-host fitness landscapes. Open Virol. J. 3, 1–6. doi: 10.2174/1874357900903010001
Fargette D., Jenniskens M.-J., Peters D. (1982). Acquisition and transmission of pea enation mosaic virus by the individual pea aphid. Phytopathology 72 (11), 1386–1390.
Fox J., Weisberg S. (2018). Visualizing fit and lack of fit in complex regression models with predictor effect plots and partial residuals. J. Stat. Softw. 87, 1–27. doi: 10.18637/jss.v087.i09
García-Arenal F., Fraile A. (2013). Trade-offs in host range evolution of plant viruses. Plant Pathol. 62, 2–9. doi: 10.1111/ppa.12104
Garrido E., Andraca-Gómez G., Fornoni J. (2012). Local adaptation: simultaneously considering herbivores and their host plants. New Phytol. 193, 445–453. doi: 10.1111/j.1469-8137.2011.03923.x
Hawthorne D. J., Via S. (2001). Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412, 904–907. doi: 10.1038/35091062
Hipper C., Brault V., Ziegler-Graff V., Revers F. (2013). Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00154
Hodge S., Powell G. (2010). Conditional facilitation of an aphid vector, Acyrthosiphon pisum, by the plant pathogen, Pea enation mosaic virus. J. Insect Sci. 10, 155. doi: 10.1673/031.010.14115
Ingwell L. L., Eigenbrode S. D., Bosque-Pérez N. A. (2012). Plant viruses alter insect behavior to enhance their spread. Sci. Rep. 2, 578. doi: 10.1038/srep00578
Lacroix C., Renner K., Cole E., Seabloom E. W., Borer E. T., Malmstrom C. M. (2016). Methodological guidelines for accurate detection of viruses in wild plant species. Appl. Environ. Microbiol. 82 (6), 1966–1975. doi: 10.1128/AEM.03538-15
Lee H., Stephanus A. P., Fowles T. M., Wintermantel W. M., Trumble J. T., Gilbertson R. L., et al. (2022). Insect vector manipulation by a plant virus and simulation modeling of its potential impact on crop infection. Sci. Rep. 12, 8429. doi: 10.1038/s41598-022-12618-2
Lenth R. (2023) emmeans: Estimated marginal means, aka least-squares means. R package version 1.8.5. Available at: https://CRAN.R-project.org/package=emmeans.
Loxdale H. D., Lushai G., Harvey J. A. (2011). The evolutionary improbability of “generalism” in nature, with special reference to insects. Biol. J. Linn. Soc. Lond. 103, 1–18. doi: 10.1111/j.1095-8312.2011.01627.x
Mauck K. E. (2016). Variation in virus effects on host plant phenotypes and insect vector behavior: what can it teach us about virus evolution? Curr. Opin. Virol. 21, 114–123. doi: 10.1016/j.coviro.2016.09.002
Mauck K. E., Chesnais Q., Shapiro L. R. (2018). “Chapter 7 - Evolutionary determinants of host and vector manipulation by plant viruses,” in Advances in Virus Research. Ed. Malmstrom C. M. (Amsterdam, The Netherlands: Academic Press), 189–250.
Mauck K. E., De Moraes C. M., Mescher M. C. (2014). Evidence of local adaptation in plant virus effects on host–vector interactions. Integr. Comp. Biol. 54, 193–209. doi: 10.1093/icb/icu012
McLeish M. J., Fraile A., García-Arenal F. (2018). “Chapter 9 - Ecological complexity in plant virus host range evolution,” in Advances in Virus Research. Ed. Malmstrom C. M. (Amsterdam, The Netherlands: Academic Press), 293–339.
Peccoud J., de la Huerta M., Bonhomme J., Laurence C., Outreman Y., Smadja C. M., et al. (2014). Widespread host-dependent hybrid unfitness in the pea aphid species complex. Evolution 68, 2983–2995. doi: 10.1111/evo.12478
Peccoud J., Figueroa C. C., Silva A. X., Ramirez C. C., Mieuzet L., Bonhomme J., et al. (2008). Host range expansion of an introduced insect pest through multiple colonizations of specialized clones. Mol. Ecol. 17, 4608–4618. doi: 10.1111/j.1365-294X.2008.03949.x
Peccoud J., Simon J.-C. (2010). The pea aphid complex as a model of ecological speciation. Ecol. Entomol. 35, 119–130. doi: 10.1111/j.1365-2311.2009.01147.x
Power A. G., Flecker A. S. (2003). “Virus specificity in disease systems: are species redundant?,” in The Importance of Species. Eds. Peter K., Simon A. L. (Princeton, USA: Princeton University Press), 330–346.
Queller D. C., Strassmann J. E. (2018). Evolutionary conflict. Annu. Rev. Ecol. Evol. Syst. 49, 73–93. doi: 10.1146/annurev-ecolsys-110617-062527
Rashed A., Feng X., Prager S. M., Porter L. D., Knodel J. J., Karasev A., et al. (2018). Vector-borne viruses of pulse crops, with a particular emphasis on north american cropping system. Ann. Entomol. Soc. Am. 111, 205–227. doi: 10.1093/aesa/say014
R Development Core Team (2022). R: A Language and environment for statistical computing. R version 4.2.2 (Vienna, Austria: The R Foundation for Statistical Computing). Available at: http://www.R-project.org/.
Robertson H. M., Robertson E. C. N., Walden K. K. O., Enders L. S., Miller N. J. (2019). The chemoreceptors and odorant binding proteins of the soybean and pea aphids. Insect Biochem. Mol. Biol. 105, 69–78. doi: 10.1016/j.ibmb.2019.01.005
Scriber J. M. (2002). Evolution of insect-plant relationships: chemical constraints, coadaptation, and concordance of insect/plant traits. Entomol. Exp. Appl. 104, 217–235. doi: 10.1046/j.1570-7458.2002.01009.x
Shih P.-Y., Sugio A., Simon J.-C. (2023). Molecular mechanisms underlying host plant specificity in aphids. Annu. Rev. Entomol. 68, 431–450. doi: 10.1146/annurev-ento-120220-020526
Shoemaker L. G., Hayhurst E., Weiss-Lehman C. P., Strauss A. T., Porath-Krause A., Borer E. T., et al. (2019). Pathogens manipulate the preference of vectors, slowing disease spread in a multi-host system. Ecol. Lett. 22, 1115–1125. doi: 10.1111/ele.13268
Skaf I., Makkouk K. (1988). Aphid transmission of bean yellow mosaic and bean leaf roll viruses in Syria. Phytopathol. Mediterr. 2 (3), 133–137.
Slater J. M., Gilbert L., Johnson D., Karley A. J. (2019). Limited effects of the maternal rearing environment on the behaviour and fitness of an insect herbivore and its natural enemy. PloS One 14 (1), e0209965. doi: 10.1371/journal.pone.0209965
Swei A., Ostfeld R. S., Lane R. S., Briggs C. J. (2011). Impact of the experimental removal of lizards on Lyme disease risk. Proc. R. Soc. Lond. B. Biol. Sci. 278, 2970–2978. doi: 10.1098/rspb.2010.2402
Van Zandt P. A., Mopper S. (1998). A meta-analysis of adaptive deme formation in phytophagous insect populations. Am. Nat. 152, 595–604. doi: 10.1086/286192
Vemulapati B., Druffel K. L., Husebye D., Eigenbrode S. D., Pappu H. R. (2014). Development and application of ELISA assays for the detection of two members of the family Luteoviridae infecting legumes: Pea enation mosaic virus (genus Enamovirus) and Bean leafroll virus (genus Luteovirus). Ann. Appl. Biol. 165 (1), 130–136. doi: 10.1111/aab.12126
Via S. (1991). The genetic structure of host plant adaptation in a spatial patchwork: demographic variability among reciprocally transplanted pea aphid clones. Evolution 45, 827–852. doi: 10.2307/2409692
Via S., Bouck A. C., Skillman S. (2000). Reproductive isolation between divergent races of pea aphids on two hosts. II. Selection against migrants and hybrids in the parental environments. Evolution 54, 1626–1637. doi: 10.1111/j.0014-3820.2000.tb00707.x
Wilke C. O., Forster R., Novella I. S. (2006). “Quasispecies in time-dependent environments,” in Quasispecies: Concept and Implications for Virology. Ed. Domingo E. (Berlin, Heidelberg: Springer Berlin Heidelberg), 33–50.
Keywords: host manipulation, inter-specific transmission, Bean leafroll virus, pea aphid, Pea enation mosaic virus, virus reservoir
Citation: Clark RE, Rincon DF, Wu Y, Crowder DW and Eigenbrode SD (2023) Experimental evidence reveals that vector host preference and performance across host plants is not altered by vector-borne plant viruses. Front. Ecol. Evol. 11:1251039. doi: 10.3389/fevo.2023.1251039
Received: 30 June 2023; Accepted: 25 October 2023;
Published: 14 November 2023.
Edited by:
Li-Long Pan, Zhejiang University, ChinaCopyright © 2023 Clark, Rincon, Wu, Crowder and Eigenbrode. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanford D. Eigenbrode, c2FuZm9yZGVAdWlkYWhvLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.