- 1Department of Pharmaceutical Chemistry, Institute of Pharmaceutical Sciences, University of Graz, Graz, Austria
- 2Department of Pharmacognosy, Institute of Pharmaceutical Sciences, University of Graz, Graz, Austria
- 3Department of Chemistry, Ångström Laboratory, Uppsala University, Uppsala, Sweden
- 4Institute of Biology, University of Graz, Graz, Austria
Millipedes are known to produce various sets of chemical compounds in exocrine defensive glands to protect themselves against predators and microorganisms. Here, we reanalyzed the gland secretion of Polyzonium germanicum, a millipede of the order Polyzoniida, by using a combination of analytical techniques such as GC-MS, LC-HRMS and high field 1D and 2D NMR spectroscopy. Previously only one compound (polyzonimine, 1) had been described, but our approach allowed us to add six compounds to the defensive chemistry of this species. Besides polyzonimine (1), we found nitropolyzonamine (2) and five new compounds: 3 (2,3-dimethyl-7’-nitro-2’,3’,5’,6’,7’,7a’-hexahydrospiro[cyclopent-2-ene-1,1’-pyrrolizine]), 4 (2,3-dimethyl-7’-nitro-2’,3’,5’,6’,7’,7a’-hexahydrospiro[cyclopentane-1,1’-pyrrolizin]-2-ene), 5 ((1Z)-8,9-dimethyl-1-(nitromethylidene)-2-azaspiro[4.4]non-8-en-7-one), and not fully identified compounds A, B. For compounds 3–5 we were able to determine the molecular constitution, for two of them (4, 5) we were able to give relative configurations. Overall, the combination of advanced analytical techniques applied herein allowed detailed insights into the defensive chemistry of P. germanicum with a low number of individuals needed for analysis and without prior compound isolation.
Introduction
Millipedes (Diplopoda) are well known to protect themselves against enemies and microorganisms by defensive secretions (Shear, 2015; Ilić et al., 2018). These fluids are produced by multiple pairs of exocrine glands and show repellent or even toxic properties. Meanwhile an impressive chemical repertoire has been reported from different millipede orders, including various quinones, esters, alcohols, cyanogenic compounds and unusual aromatic alkaloids (Shear, 2015; Bodner et al., 2018). Alkaloids are – with a few exceptions (Bodner et al., 2017) – mainly found in the secretions of two groups of diplopods, Glomerida and Colobognatha. The chemistry of Glomerida appears to be predominated by two unique quinazoline alkaloids: glomerin and homoglomerin (Meinwald et al., 1966; Shear et al., 2011). Most of the published data from Colobognatha refer to the order Polyzoniida, showing that their chemistry comprises monoterpenes such as pinene and limonene as well as heterocyclic alkaloids. Compounds such as polyzonimine (1), nitropolyzonamine (2), buzonamine and various spiropyrrolizidine oximes have been reported (Meinwald et al., 1975; Röper, 1978; Wood et al., 2000; Saporito et al., 2003; Kuwahara et al., 2007). Recently, unique hydroindolizidine alkaloids and deoxybuzonamine isomers were described from the platydesmid colobognathans, Gosodesmus claremontus and Brachycybe lectonii (Hassler et al., 2020; Jones et al., 2022).
The complex chemistry of millipedes prompted researchers to apply increasingly advanced analytical methods for their investigations. The defensive chemistry of European Polyzonium germanicum (Polyzoniida), for instance, was studied in the late 1970’s by Röper (1978), who detected a single compound, polyzonimine (1). At the same time, spirocyclic alkaloids from Polyzoniida were isolated by preparative gas chromatography and further characterized in crystallographic studies (Meinwald et al., 1975; Smolanoff et al., 1975). With the rapid development of analytical techniques, metabolite profiling has been increasingly applied for the analysis of complex natural product extracts, especially in pharmaceutical biology.
Commonly used analytical platforms are either based on NMR or on mass spectrometry (Kellogg et al., 2017). However, in newer publications on millipede chemistry, GC-MS data are already accompanied by NMR data of purified compounds isolated from secretions (Wood et al., 2000; Hassler et al., 2020; Jones et al., 2022). While multi-technique approaches in chemical ecology studies are still rather scarce, and most researchers analyzing gland secretions still focus on GC-MS data which still represent the gold standard when working with thermostable volatiles. As a disadvantage, many other compounds, such as less volatile material and large molecules, may remain inaccessible. NMR spectroscopic evaluations do not underlie these restrictions, but clearly benefit from information by methods of mass spectrometry, e.g., information on exact mass and elementary composition by high resolution mass spectrometry (HRMS). Particularly when dealing with natural compounds, a combination of these methods arises as a most promising strategy to quickly identify novel structures. We here rely on such a multi-technique strategy, expanding the NMR-concept of Schröder and colleagues to analyze crude animal secretions by adding further methods (Taggi et al., 2004; Schroeder et al., 2008). In detail, we re-analyzed the defensive secretion of the millipede species Polyzonium germanicum, combining GC-MS, LC-HRMS and two-dimensional NMR spectroscopy. A high-field NMR spectrometer equipped with a cryo-probe enabled us to record the relatively insensitive HSQC-TOCSY experiment in which the carbon axis resolves signal overlaps in the proton spectrum or proton-proton experiments like COSY and TOCSY. Compared to previous studies, our approach allowed us I) to work with a much lower number of individuals and II) without prior compound isolation. This led to the discovery of novel compounds, and may serve as a model standard for chemical-ecological investigations with arthropods in the future.
Material and methods
Preparation of extracts
Seventeen individuals of both sexes of Polyzonium germanicum were collected by hand on 24th April 2021 in Deutschlandsberg, Styria, Austria (46°48’39.6”N 15°11’48.2”E) under collection permission from the Provincial Government of Styria (department 13; GZ: ABT13-198250/2020-9). Fifteen adult individuals were pooled in 700 μl deutero chloroform (CDCl) which had been deacidified with aluminium oxide before use for about 30 minutes. The extract was then transferred to a 5 mm NMR tube and filled up to 720 μl. The same extract was used for both GC-MS and LC-HRMS analyses.
GC-MS analysis
Aliquots (1 μl) of the deutero chloroform extract were injected to a Trace GC-DSQ I GC-MS system (Thermo Fisher). The GC was equipped with an apolar ZB-5 capillary column (30 m × 0.25 mm × 0.25 μm; Phenomenex). Chromatographic separation and mass spectral settings were the same as previously described (Bodner and Raspotnig, 2012).
LC-HRMS analysis
LC-HRMS experiments were performed on a Dionex Ultimate 3000 HPLC system hyphenated with a QExactive Hybrid Quadrupole Orbitrap MS (Thermo Fisher Scientific, Waltham, MA, USA). The stationary phase consisted of ACQUITY™ Premier HSST3 1.8 µm (2.1 × 100 mm) column and the mobile phase was H2O (A) and CH3CN (B). The following parameters were applied: column temperature: 25°C; flow rate: 0.3 ml/min; gradient: 0–12 min, 5–35% B in A; 12–20 min, 35–100% B in A; 20–22min, 100% B; 22–22.5 min, 100–5% B in A; 22.5–28 min, 5% B in A; MS spectra were acquired in HESI-positive mode; probe heater temperature was set to 300°C, capillary temperature to 250°C, spray voltage to 3 kV, sheath gas flow to 35 arbitrary units, and auxiliary gas flow to 5 arbitrary units. For ddMS2 fragmentation, a stepped NCE (20, 40 70) and an isolation window of 3.0 m/z was applied. Resolution was 70.000 (FWHM) for full MS and 35.000 for ddMS2, stepped NCE was 20, 40, 70. Prior to LC-HRMS analysis, 50 µl of the CDCl3 extract used for NMR-based structure elucidation were evaporated under a stream of nitrogen and redissolved in 50 µl CH3CN. 3 µl of this sample were injected.
NMR spectroscopy
1D (0.5 h) proton and carbon (8 h) spectrum, 2D 1H,1H-COSY (5 h), 1H,1H-NOESY (25 h) with a mixing time of 700 ms, 1H,13C-HSQC (5 h) with multiplicity editing, 1H,13C-HMBC (10 h), and 1H,13C-HSQC-TOCSY (8 h) experiments were recorded with a 700 MHz Bruker Avance III spectrometer equipped with a triple resonance cryoprobe. The 1D proton NMR spectrum was recorded with a 10s relaxation delay to allow proper quantification of the components. The sample temperature was 25°C, TMS was used as internal standard. Processing and analyzing of the NMR data was done in the MestreNova software. The deuteration grade (VWR chemicals, 99.8%) of the used batch of deutero chloroform was confirmed for a 5 ml sample with cyclohexane as internal standard, the residual proton signal (0.21%) in solvent of the same batch was used to quantify the extract components.
Computational methods
Computations were performed using the Gaussian suit of programs (G09 Rev. D01) using a density functional approach. Two functionals with dispersion correction (B3LYP-D3 and M06-2X) and 6-311++G** basis set. Solvent effects have been modelled using a continuum solvation model IEF-PCM for dichloromethane (ϵ = 8.93). All structures are assessed to be local minima by inspection of the first derivative showing no imaginary frequencies.
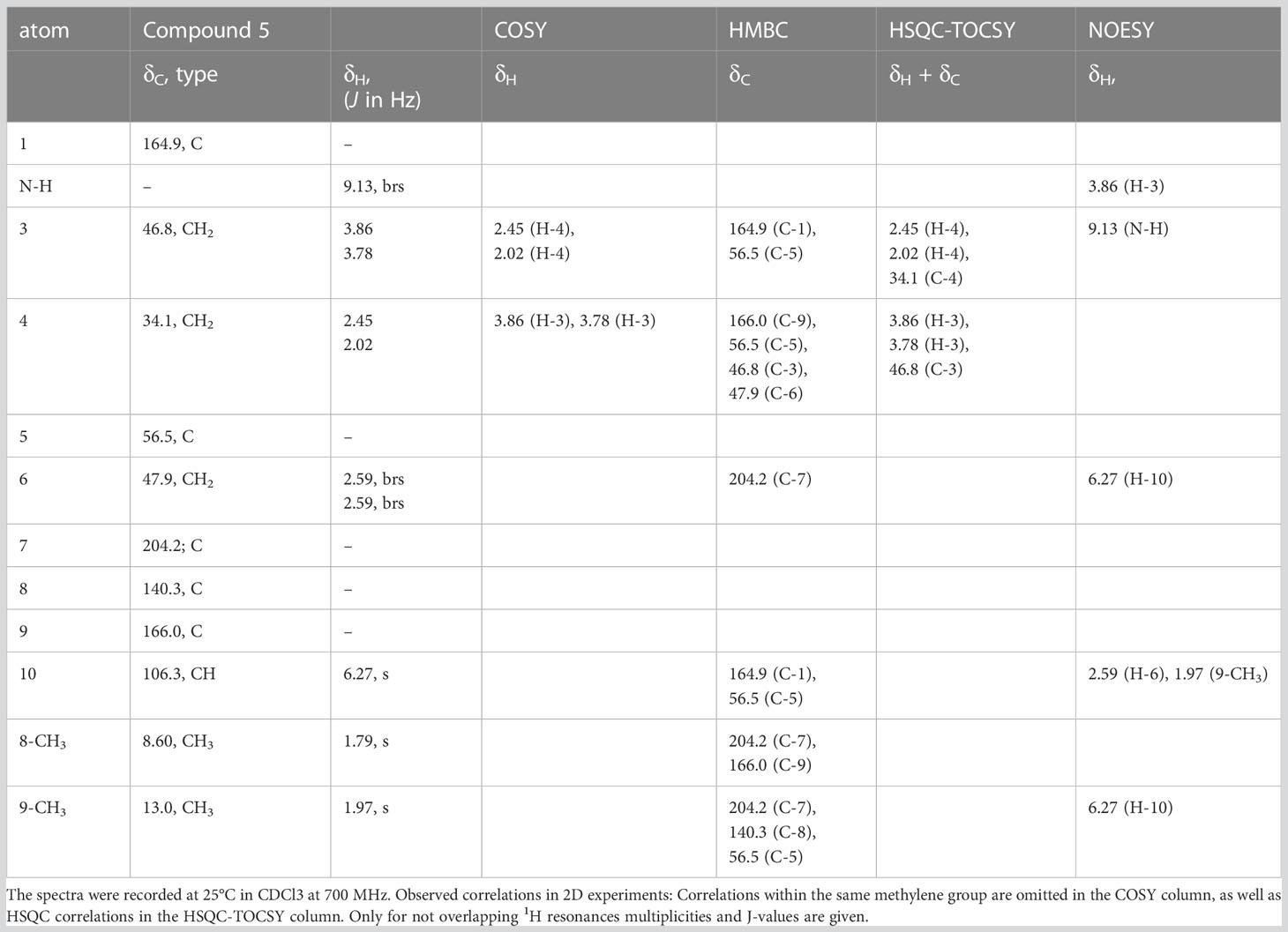
Results
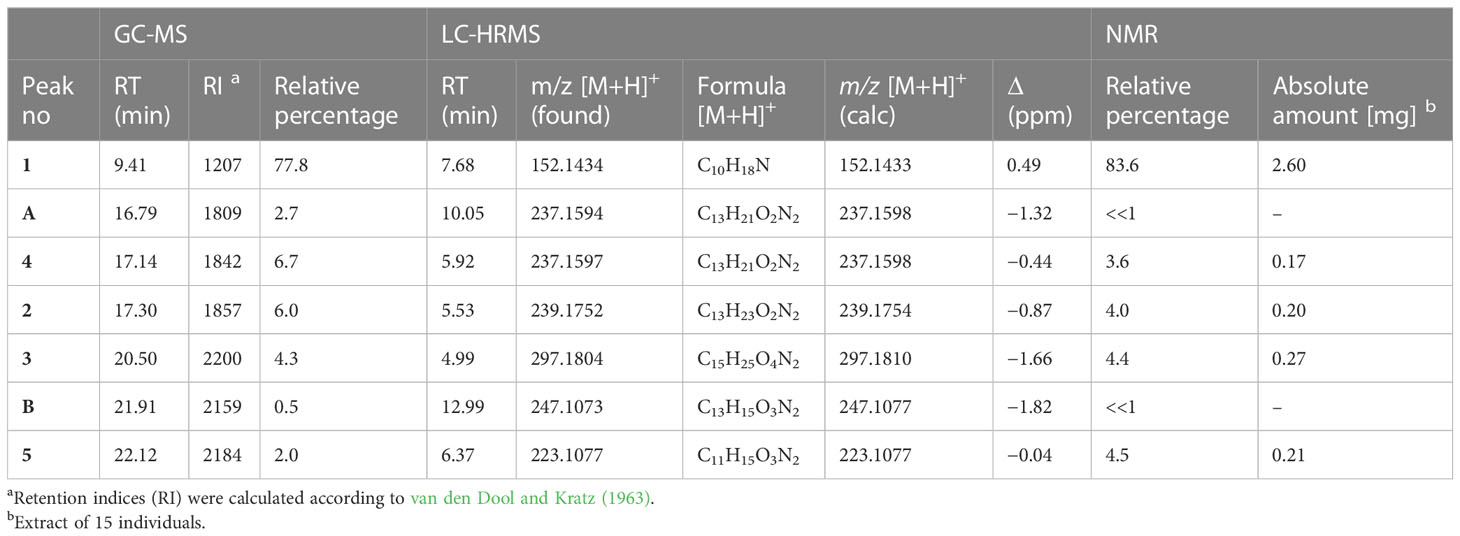
In the pooled extract of Polyzonium germanicum seven compounds were detected by GC-MS analysis (Figure 1, Table 1, SI Figure S1–S7). The structures of compounds 1–5 could be fully elucidated by NMR spectroscopy (Figure 2). Due to their low abundance in the sample, structure elucidation via NMR was not possible for compounds A and B.
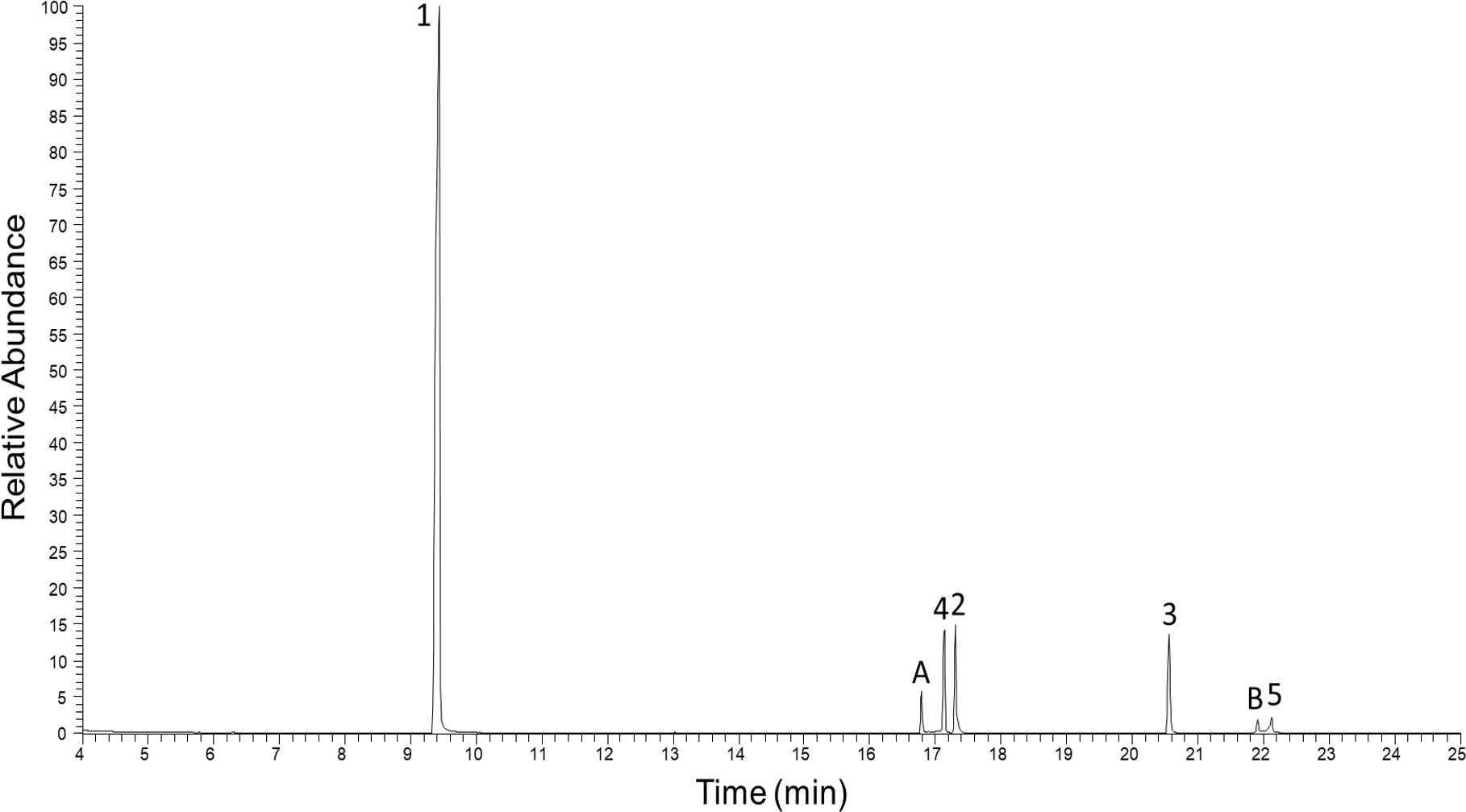
Figure 1 Total ion GC-EIMS chromatogram of full body chloroform extract from Polyzonium germanicum. Peak numbers, and letters refer to given structures (Figure 2, Table 1).
The major compound 1 was the already described polyzonimine (Röper, 1978) (Table 1). The EI-mass spectrum of the compound (RI 1207) showed a base peak at m/z 82 (C5H8N+) and a molecular ion (M+) at m/z (Supplementary Figure S1), corresponding to published data of polyzonimine (Kuwahara et al., 2007). Subsequently, the molecular formula was determined by HESI-HRMS as C10H17N (Table 1). In the proton NMR spectrum, the major component 1 was easily identified as polyzonimine due to its high signal intensity and its characteristic proton and carbon resonance values for methine group C-1. Its 13C NMR data (SI Table S2) are in perfect agreement with NMR data published for synthetic polyzonimine (Mori and Takagi, 2000) indicating that compound 1 has the same relative configuration.
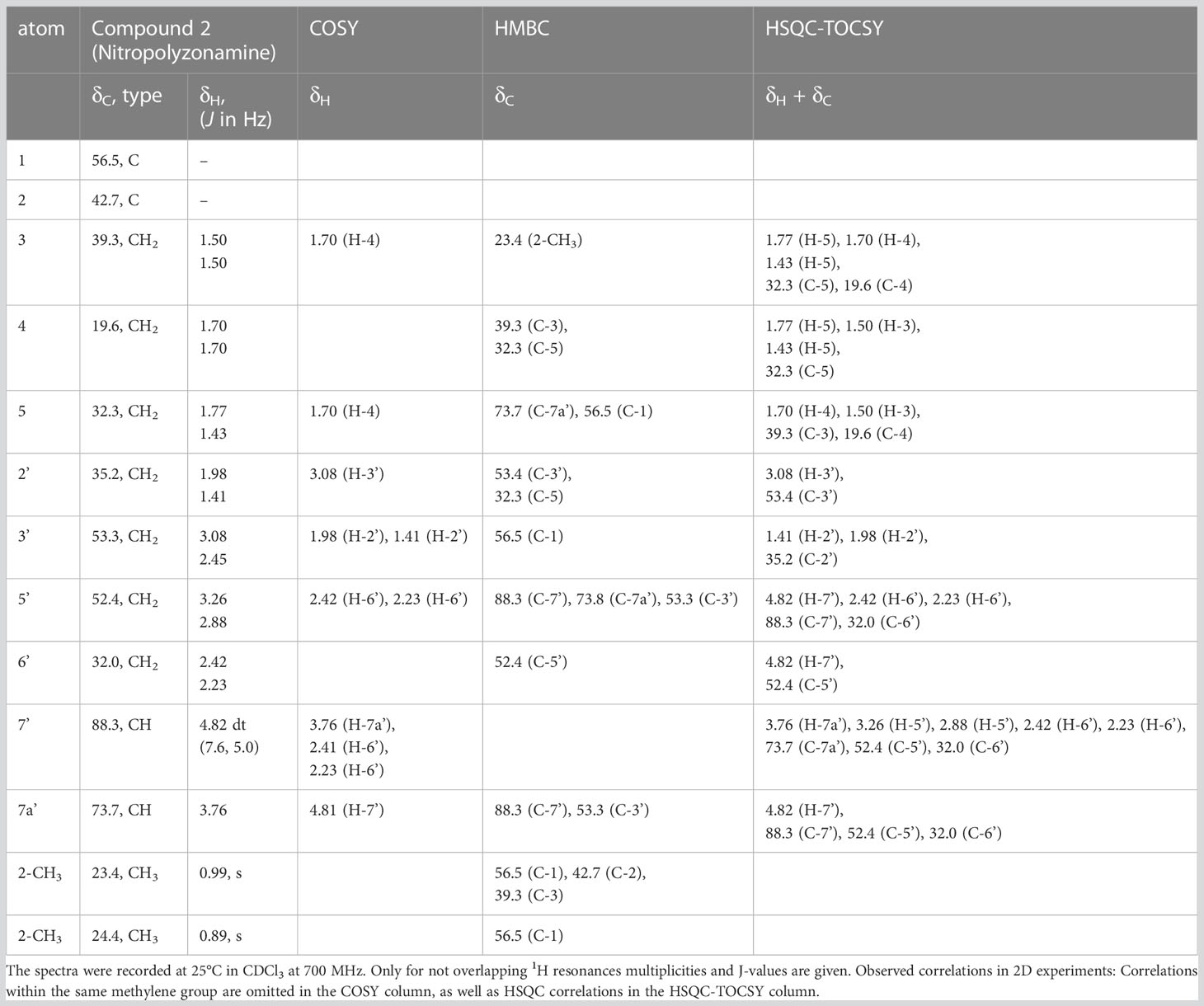
From the correlations observed in the HSQC and HSQC-TOCSY spectra, compounds 2–4 occurring in the extract at medium abundance, were expected to be compounds of the nitropolyzonamine type too. The skeletons of nitropolyzonamine-like molecules comprise three isolated spin systems, the first is C-3, C-4, and C-5, the second C-2’ and C-3’, and the third C-5’, C-6’, C-7’ and C-7a’. The proton and carbon resonances belonging to each of the nine spin systems were identified in the HSQC-TOCSY spectrum (Figure 3) and assigned in the COSY and HSQC spectra.
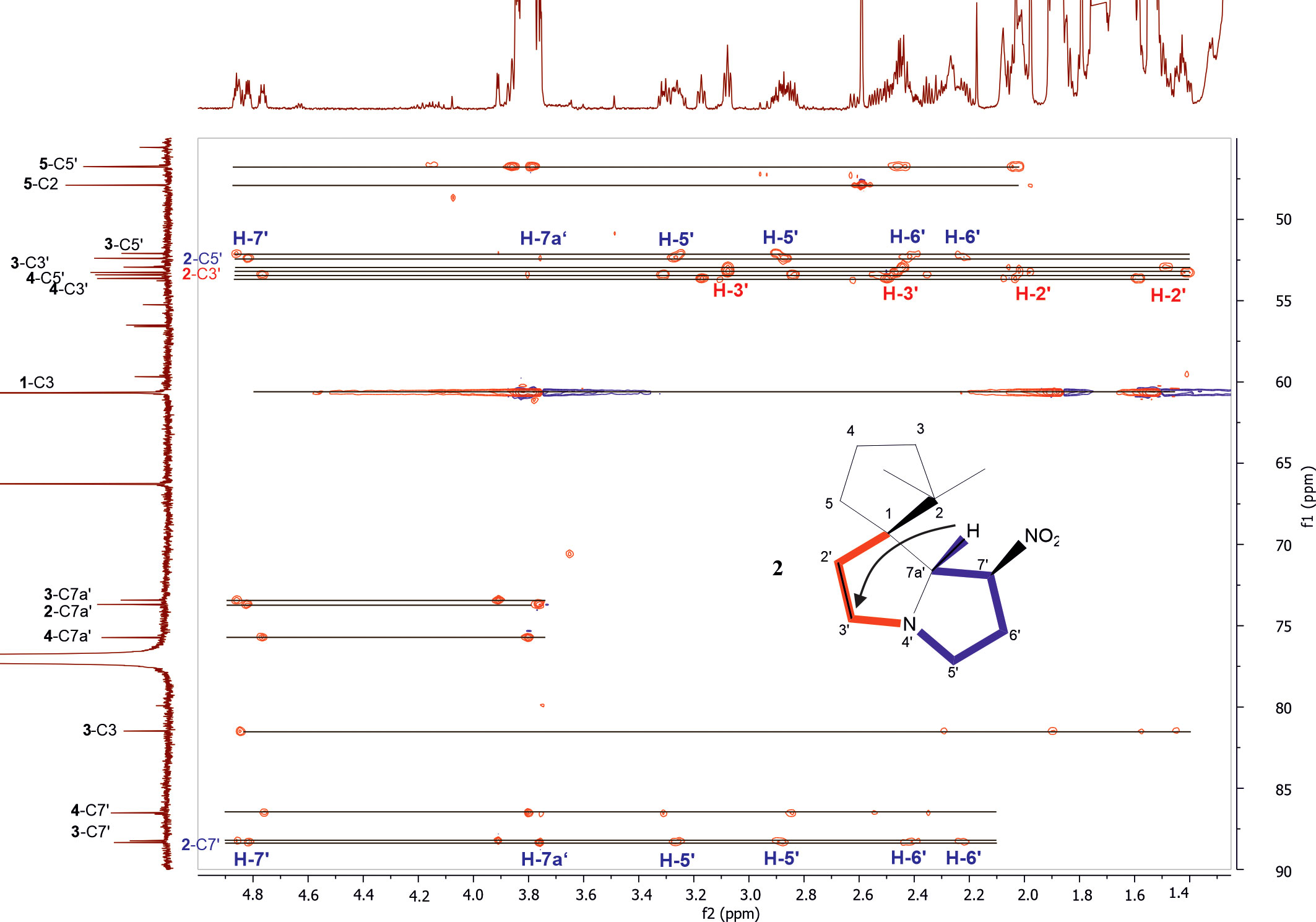
Figure 3 Expansion of HSQC-TOCSY NMR spectrum of the P. germanicum chloroform extract. Numbers in bold indicate compounds, normal print numbers positions within the molecule. Assignments for two isolated spin systems of compound 2 are indicated by proton positions. The arrow in the structure indicates an observed HMBC correlation which connects the isolated spinsystems. Colored spin systems (red/blue) in the chemical structure refer to assignments in the same color in the spectrum.
The EI-MS spectrum of compound 2 (RI 1857) with a weak M+ ion at m/z 238 and a base peak at m/z 82 showed full correspondence with nitropolyzonamine (Meinwald et al., 1975; Saporito et al., 2003; Kuwahara et al., 2007), which has already been described from other polyzoniidan species (Supplementary Figure S2). In general, spiropyrroline polyzonimine and spiropyrrolizidine alkaloids are closely related structures, and EI fragmentation results in a common base peak at m/z 82 (C5H8N+) (Daly et al., 1999). For compound 2, a neutral molecular formula of C13H22O2N2 was deduced from its LC high resolution mass spectrum (Table 1). According to the HSQC and HSQC-TOCSY spectra, the first spin system of this compound consisted of three methylene groups, the second of two methylene groups and the third of two methylene and two methine groups. C-1 and C-3 were identified by HMBC correlations with the two methyl groups attached to C-2 (Figure 4). The proton resonances of the ring C-5’ to C-7a were identified at the carbon resonance C-7’ (88.3 ppm) and are also observed at carbon resonances C-5’ (52.4 ppm), indicating that both carbon resonances belong to the same spin system. The four proton resonances of the methylene groups C-2’ and C-3’ are observed at carbon resonance C-3’ (53.3 ppm) and at the carbon resonance C-2’ (35.2 ppm). The two spin systems were then connected by an HMBC correlation between H-7a’ and C-3’ (black arrow in Figure 3). Then, protons H-2’ and H-3’ were correlated in the HMBC spectrum with the spiro carbon C-1 and carbons C-5 and C-7a’. This indirect approach was necessary because the proton resonance H-7a’ did not show a two-bond HMBC correlation to the spiro carbon C-1 (Table 2). Compound 2 had the same molecular constitution as nitropolyzonamine, and its 13C NMR data were in very good agreement with published primary NMR data for synthesized nitropolyzonamine (Mori and Takagi, 2000). Therefore, it was assigned with the same relative configuration as the synthetic compound.
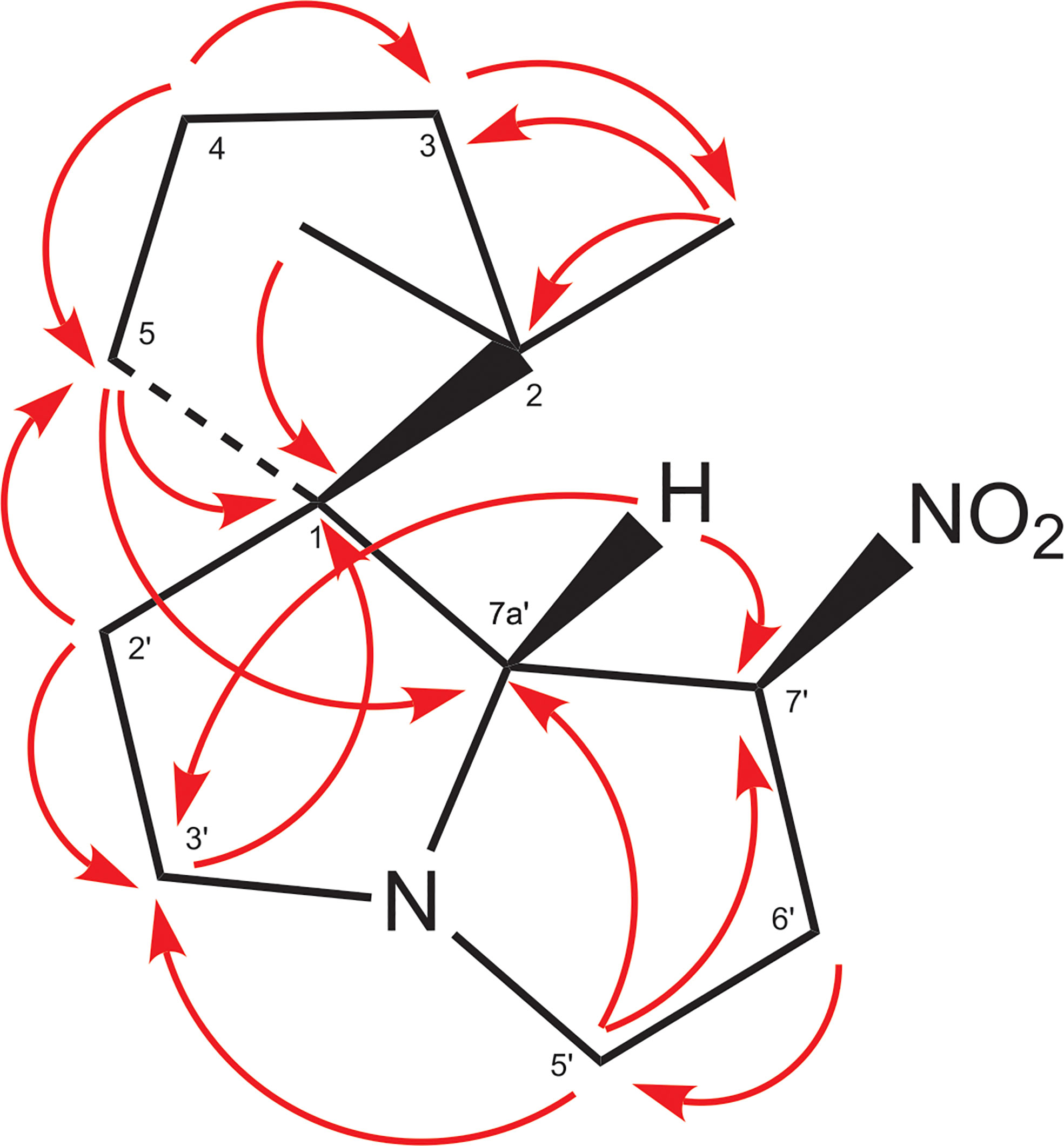
Figure 4 HMBC correlations in compound 2. This is a graphical representation of the HMBC column of Table 2.
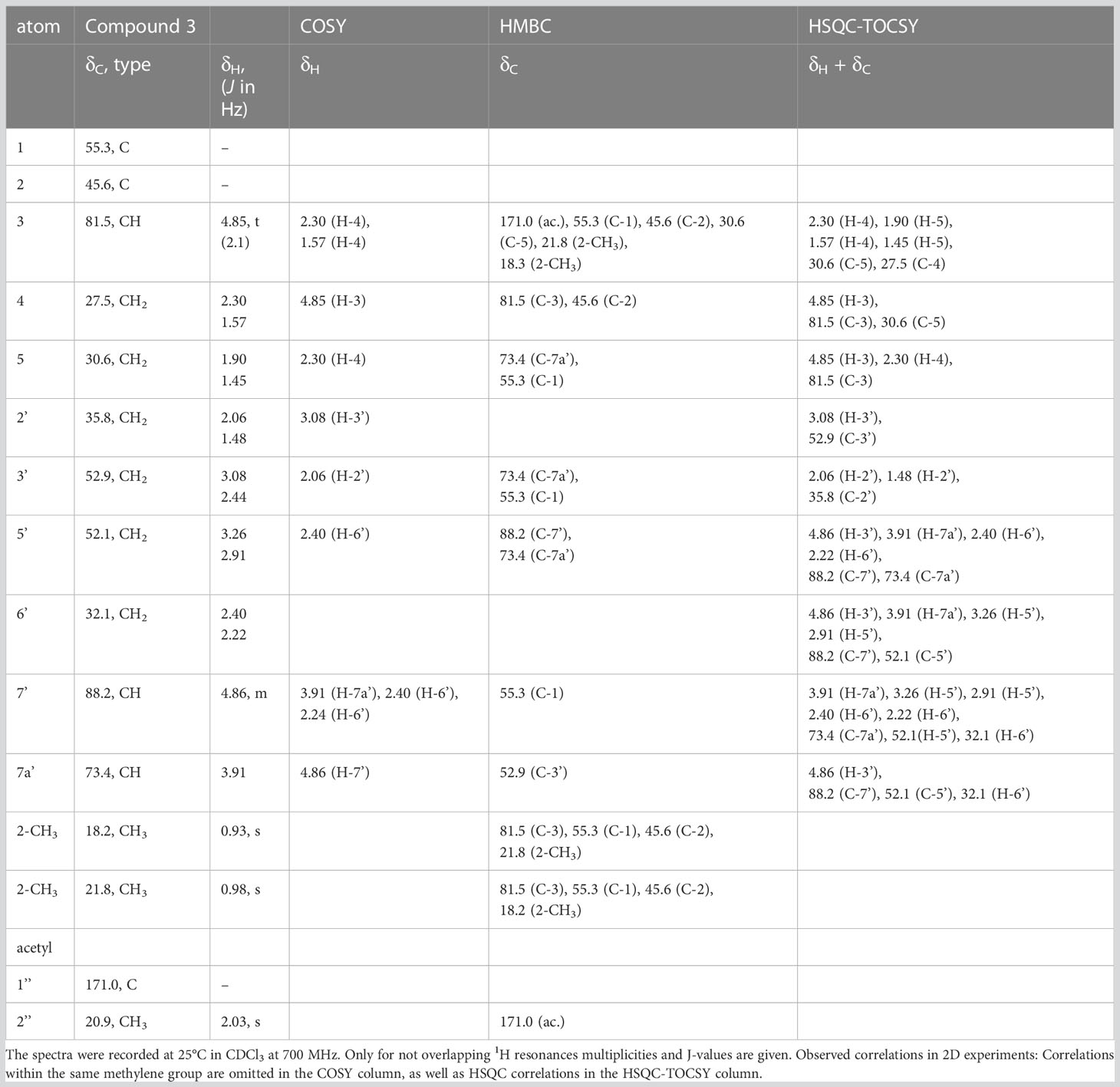
Comparably, compounds 3, 4, and A (RI: 2200, 1842, 1809) provided a base peak at m/z 82 too. They exhibited weak molecular ions in EI-MS at m/z 296, 236 and 236, respectively. For compound 3, which had a neutral molecular formula of C15H24O4N2, the HSQC-TOCSY spectrum indicated that one position in the first spin system comprising C-3, C-4 and C-5 was oxidized. As the two methyl groups at C-2 showed correlations to a methine group with a 13C shift value of 81.5 ppm, it had to be assigned as C-3. In the HMBC spectrum, the proton H-3 was correlated with a carbonyl resonance (δC 171.0 ppm).
The same correlation was also detected for the additionally observed methyl protons of the side chain at 2.03 ppm, indicating an ethanoyloxy moiety (Table 3). The chemical shift values for the other two rings were very similar to compound 2. Therefore, compound 3 was identified as acetylated 3-hydroxynitropolyzonamine. The configuration at C-3 remains unclear as no useful NOEs were observed for the protons in this ring.
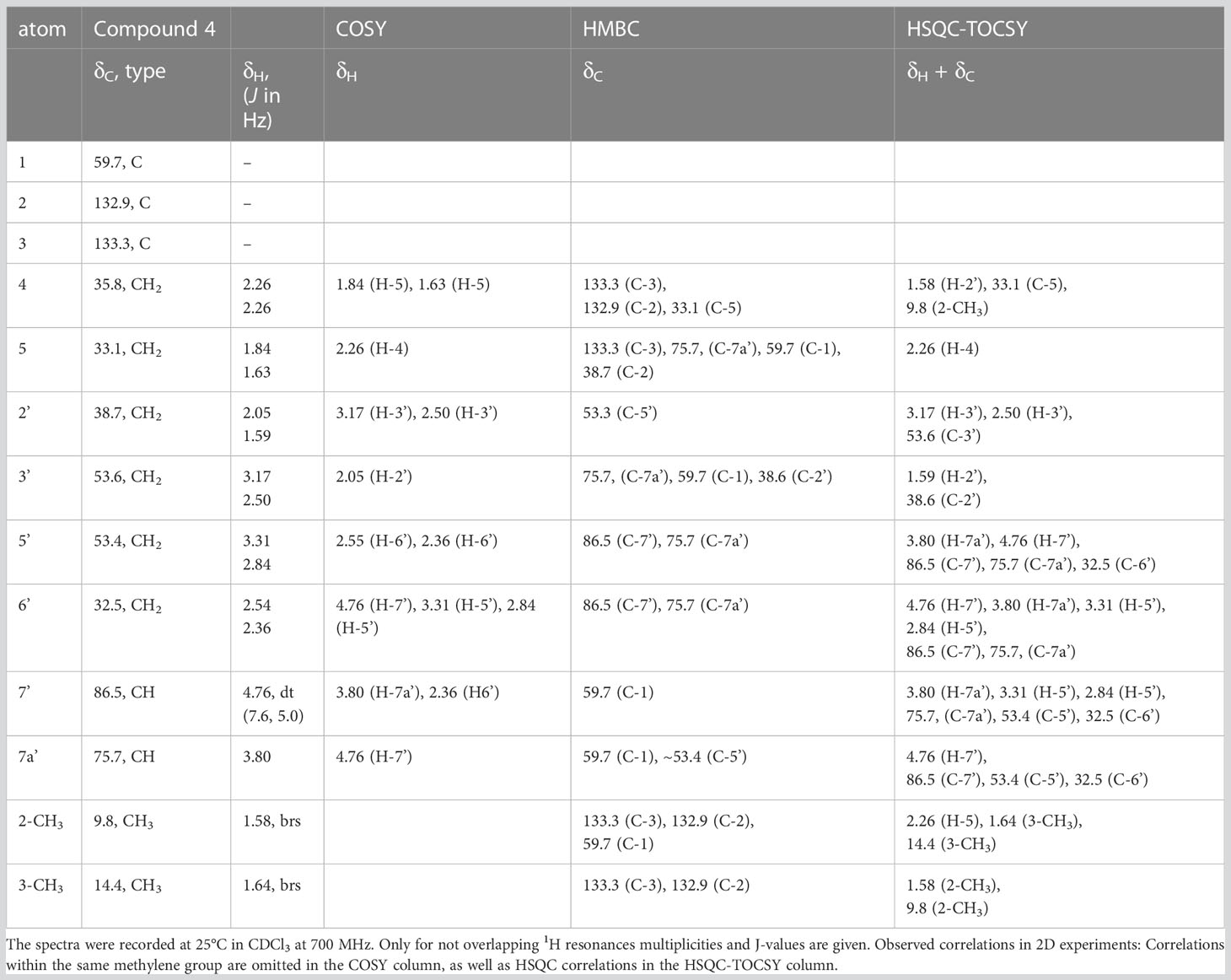
For compound 4, a neutral molecular formula of C13H20O2N2 was calculated. In comparison to nitropolyzonamine (2), very different carbon shift values of C-2 and C-3 suggested a different chemical structure at C-2 and C-3. This was confirmed by analysis of HMBC correlations which indicated a shift of one methyl group from C-2 to C-3 and the presence of a double bond between C-2 and C-3 (Table 4), perfectly explaining the molecular formula of compound 4, that indicated the absence of two hydrogen atoms compared to compound 2. As the 13C chemical shift values values (Tables 2–4) of C-1 to C-7a’ in all three compounds 2–4 were very similar and the stereochemistry of nitropolyzonamine had been established by X-ray crystallography and synthesis (Smolanoff et al., 1975; Mori and Takagi, 2000), the same relative configuration was also assumed at C-7’ and C-7a’ of compounds 3 and 4 in the investigated P. germanicum extract.
The EI-spectra of compounds 5 and B (RI: 2184, 2159) displayed pronounced molecular ions at m/z 222 and m/z 246, respectively. For compound 5, HRMS indicated a neutral molecular formula of C11H14N2O3 (Table 1). The assignments of resonances C-3 to C-9 were straightforward. COSY and HMBC correlations indicated the presence of a spirocyclic terpenoid compound with a keto group at C-7, two methyl groups at olefinic positions C-8 and C-9, and three methylene groups at C-3, C-4 and C-6, respectively. The high carbon resonance values of the methylene group C-3 suggested a nitrogen at position 2. Protons H-6 were correlated with both, 166.0 ppm (C-9) and 164.9 ppm (C-1). The remaining carbon resonance belonged to a methine group (δC 106.3 ppm, δH 6.27 ppm). The proton resonance corresponding to this methine group showed an HMBC correlation with medium intensity to C-1 (164.9 ppm) and one with a very low intensity to C-5 (56.5 ppm). In addition, a labile proton was observed at 9.13 ppm (Table 5). In the HESI-MS/MS spectrum of the compound, the loss of the NO2 group was clearly observable (Supplementary Figure S13). The observed chemical shift values (Table 5) for C-1 and C-10 are consistent with all three tentative structures 5, 5b and 5c depicted in SI Figure S23. However, NOEs observed between H-10 and methylene protons H-6 as well as methyl group at C-9, the NOEs between the labile proton and H-3 (SI Figure S21B), the high proton NMR shift value of the labile proton, and the observed loss of NO2 in the HESI-MS/MS can be explained much more convincingly by structure 5. The NOEs also support the Z-configuration of the exocyclic double bond.
With DFT calculations we aimed to provide additional arguments for the structure of compound 5. We expected a low energy structure. Surprisingly, DFT studies using dispersion corrected hybrid functionals and continuum solvent modelling revealed that 5 – in comparison to the constitutional isomers 5b and 5c (SI Figure S23) – is thermodynamically significantly less favorable by more than 100 kJ/mol. This is confirmed using different density functionals, supporting the notion that this is not a computational artifact but illustrates that the biochemical pathway does not necessarily produce the lowest energy molecule. Other constitutional isomers – not in agreement with the NMR data – that are thermodynamically equally favorable are reported in the supporting information. Nonetheless, the exocyclic nitro group can be confirmed in the depicted cis-conformation being the thermodynamically more favorable (by ca. 35 kJ/mol) conformer showing that only certain aspects of the biogenesis might be thermodynamically controlled. Hence, the structure of 5 was deduced as 8,9-dimethyl-1-(nitromethylidene)-2-azaspiro[4.4]non-8-en-7-one, while structures 5b and 5c were discarded.
Based on data by NMR spectroscopy, it was possible to annotate some of the major fragments observed in HESI-MS/MS spectra. MS/MS fragments provided by HRMS analysis in the HESI positive mode (Supplementary Information Figures S9-S15) confirmed the presence of [C5H8N]+ (m/z 82.066) in compounds 1, 2, 3, 4 and A, corroborating that these compounds possess a similar skeleton. However, m/z 82.066 was the main fragment only in the MS/MS spectra of compounds 2 and 3. In contrast to GC-MS, MS/MS spectra generated by QExactive MS provided a higher range of informative MS/MS fragments (SI TableS1). For example, a fragment indicating the loss of NO2 (Egsgaard and Carlsen, 1996) was detectable in the MS/MS spectra of compounds 2, 3, 4, 5, A and B, suggesting the presence of a NO2 group in these molecules, while it was absent in compound 1. Compounds 2, 3, 4 and B also shared the loss of CH3NO2 (originating from position 7´) and 2, 3 and 4 the loss of C3H5NO2 (originating from positions 7´, 6´and 5´), which is likely due to the degradation of the pyrrolidine ring bearing the NO2 group (SI Table S1). Compound 3 displayed additional major fragment ions at m/z 192.175 [M+H-CH3COONO2]+ and m/z 176.143 [M+H-CH3COOH-CH3NO2]+, indicating the presence of an acetyl function in the molecule. Compound 4 differed from compound 2 by the presence of highly abundant low molecular weight fragment ions in the MS/MS spectrum, e.g. at m/z 107.086, 84.081, and 69.058. This may be due to differences in the spirocyclopentane ring (i.e. the presence of a double bond and different positions of the methyl groups in compound 4). The MS/MS fragmentation pattern of compound 5 strongly deviated from those of the latter compounds. This is likely due to the fact that in this compound, the NO2 group is attached to an exocyclic double bond absent in the skeleton of the latter compounds (Figure 2). The base peak was a fragment at m/z 91.055. An indicative fragment at m/z 205.097 was formed by the loss of H2O. This is likely possible due to migration of the proton at the nitrogen in position 2, which is not present in all the other structures (Egsgaard and Carlsen, 1996). The HRMS data indicated that the neutral molecular formula of A was C13H20O2N2, suggesting that the compound may be an isomer of compound 4. However, despite from showing the presence of a nitro group, the MS/MS fragmentation of A strongly differed from that of 4. The neutral molecular formula of compound B was calculated as C13H14O3N2. Its MS/MS fragmentation pattern was devoid of a fragment at m/z 82.066, but showed the presence of a major fragment at m/z 91.955, indicating potential similarities with compound 5. All carbon NMR resonances with high or medium intensities could be assigned to one of the five described alkaloids, therefore, the compounds A and B observed in GC-MS (Figure 1) and LC-HRMS (Supplementary Figure S8), are presumed to have a much lower concentration than compounds 1–5.
Discussion
We here present a more detailed analysis of extracts from a colobognathan millipedes by a combination of advanced methods, including NMR and different methods of mass spectrometry. Our approach allowed us to directly identify the already known alkaloids polyzonimine (1) and nitropolyzonamine (2) and, in addition, two new compounds (3, 4) as well as a further new alkaloid (5). For two further compounds A and B, at least the molecular formulas could be provided. This analytical strategy feasibly clearly offers the opportunity to identify new chemical compounds, in the best case even at the level of relative configuration. The absolute configuration of the molecules has to be checked via synthesis in future studies.
The complexity of alkaloid chemistry from animal sources has been the topic of several publications, especially regarding the rich repertoire of dendrobatid frogs that sequester toxic alkaloids from their arthropod prey, such as oribatid mites and polyzoniid millipedes, respectively (Saporito et al., 2003; Daly et al., 2005; Takada et al., 2005). The biosynthetic machinery behind alkaloids in arthropods is largely unknown. While our data do not yet allow a detailed discussion of biosynthetic pathways, compound 3 ((2,2-dimethyl-7’-nitro-2’,3’,5’,6’,7’,7a’-hexahydro-spiro[cyclopentane-1,1’-pyrrolizin]-3-yl) acetate) could be a structural link from nitropolyzonamine (2) to compound 4 (2,3-dimethyl-7’-nitro-2’,3’,5’,6’,7’,7a’-hexa-hydrospiro[cyclopentane-1,1’-pyrrolizin]-2-ene) arising via the loss of acetic acid. Furthermore, the exocyclic nitro group of bicyclic compound 5 is in the same position as the second exocyclic nitrogen in common millipede tricyclic pyrrolizidine oximes (Kuwahara et al., 2007) and in nitropolyzonamine 2. Hence, compound 5 ((1Z)-8,9-dimethyl-1-(nitromethyliden)-2-azaspiro[4.4]non-8-en-7-one) could be the structural link between polyzonimine (1) and compounds of the nitropolyzonamine-type (2, 3, 4). However, to get a clearer picture on the formation of these compounds and their biosynthesis, it would be necessary to perform labelling experiments and to analyze additional species.
Already Schröder and colleagues (2008) described a combination of structure elucidation of compounds in mixtures and a more conventional structure elucidation strategy of purified compounds. In this study, structure assignments of venom compounds of spiders mainly relied on data acquired with 500 MHz and 600 MHz NMR spectrometers equipped with room temperature probes. The recorded NMR data sets comprised proton, DQF-COSY, HSQC, and HMBC spectra. We improved this approach by using a 700 MHz NMR spectrometer equipped with a cryoprobe. With our improved hardware we gained a higher spectral resolution and, more importantly, an around 10-fold increase of sensitivity. Therefore, we were not only able to record the aforementioned NMR experiments but also 1D carbon and the 2D HSQC-TOCSY experiments. With the latter, the sampling of resonances for many individual spin systems becomes a much easier task due to an additional carbon axis because ambiguities due to overlapping proton resonances are dramatically reduced. For a mixture of similar compounds, the NMR spectroscopic assignment strategy is in principle the same as used for purified complex natural compounds which consist of several similar building blocks, e.g., sugar units in larger saponins. Proton and carbon resonances of individual spin systems are identified mainly in the 2D HSQC-TOCSY experiment and assigned within the respective spin system by 2D DQF-COSY and HSQC experiments. As a last step, the individual spin systems are combined by HMBC correlations or NOEs to the final structure.
The cryoprobe and the analysis of whole body extracts dramatically reduces the demand for a large sample size of individuals for de novo structure elucidation. Based on this work, it is conceivable that by restricting the NMR measurements to the essential and higher sensitive set of proton-, COSY-, HSQC-, and HMBC-experiments would even allow investigation of more challenging sample quantities, e.g., from smaller arthropods such as oribatid mites (Raspotnig et al., 2023).
Our NMR-spectroscopic data led to the complete assignment of the structure, and in case of mass spectrometry to a tentative annotation of the structures of the detected analytes. This allows so-called dereplication, i.e., the annotation of already known structures without the need for their isolation (Wolfender et al., 2019). NMR spectroscopy has the advantages of simultaneous structural assignment and quantification of the major metabolites in a given mixture, and of being nondestructive to the samples. A useful by-product of the NMR spectroscopic approach is a better quantification of the compounds in the extract. Compared to NMR, MS-based techniques offer a much greater sensitivity, enabling the detection of minor constituents. Mass spectrometers can be easily hyphenated with chromatographic separation techniques like liquid chromatography or gas chromatography, thereby facilitating the in-depth analysis of complex mixtures. However, although mass spectrometric data are highly useful to confirm the presence of known compounds in a mixture or to deduce fragments of a structure, the technique alone does not allow the complete de novo elucidation of unknown compounds. Another limitation of MS-based analysis is the fact that not all analytes are ionized to the same degree. Therefore, the intensity of the signals is not directly proportional to the concentration of the metabolites, and compounds not ionized with the applied technique may remain undetected (Wolfender et al., 2015; Kellogg et al., 2017; Wolfender et al., 2019).
Overall, NMR and MS analysis can be regarded as complementary techniques for high-quality compound identification and quantification in natural product mixtures like in the secretions of P. germanicum.
For future studies on complex mixtures of novel compounds from arthropods, we suggest to rely on a similar approach combining different MS techniques with NMR equipped with a cryo-probe to record 2D experiments (e.g. HSQC-TOCSY). This setup will allow: I) to work with crude extracts from a small sample size, II) to characterize structures in a moderate time, and III) to better quantify the compounds in the extract for further ecological experiments.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because millipedes are not listed in the species conservation law. We applied for a permission to perform collections as stated under “Acknowledgments”.
Author contributions
MB designed the study and performed collection and determination of Polyzonium germanicum. Acquisition and analysis of data was done in case of GC-MS by MB, and in case of HRLC-MS by EP-W. NMR experiments and resonance assignments were done by OK. DFT calculations were done by AO. All authors contributed to the manuscript, read, edited, and approved the final version.
Funding
This work was supported by the Government of Styria, Austria (PN 37), and the Austrian Science Fund (FWF project no. P33840-B). The computations were enabled by resources in project snic2022-5-194 provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX, partially funded by the Swedish Research Council through grant agreement no. 2018-05973.
Acknowledgments
MB is grateful to the Government of Styria for funding and the permission to perform collections of individuals of Polyzonium germanicum. NAWI Graz is thanked for supporting Central Lab Environmental, Plant & Microbial Metabolomics. AO acknowledges the generous support of the Swedish research council (Vetenskapsrådet). The authors thank Robert Weis (University of Graz) for his support with the nomenclature of the new compounds, Jana Kroustkova (Charles University, Czech Republic) for the discussion of the NMR results.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1212452/full#supplementary-material
References
Bodner M., Raspotnig G. (2012). Millipedes that smell like bugs: (E)-alkenals in the defensive secretion of the julid diplopod Allajulus dicentrus. J. Chem. Ecol. 38, 547–556. doi: 10.1007/s10886-012-0127-5
Bodner M., Vagalinski B., Makarov S. E., Raspotnig G. (2017). Methyl N-methylanthranilate: major compound in the defensive secretion of Typhloiulus orpheus (Diplopoda, Julida). Chemoecology 27, 171–175. doi: 10.1007/s00049-017-0242-4
Bodner M., Vagalinski B., Raspotnig G. (2018). Chemotaxonomic potential of exocrine alkyl esters in julid millipedes (Diplopoda: Julidae: Cylindroiulini). Biochem. Systematics Ecol. 81, 1–11. doi: 10.1016/j.bse.2018.08.001
Daly J. W., Martin Garraffo H., Spande T. F. (1999). “Chapter one - alkaloids from amphibian skins,” in Alkaloids: chemical and biological perspectives. Ed. Pelletier S. W. (Pergamon, New York: Pergamon), 1–161.
Daly J. W., Spande T. F., Garraffo H. M. (2005). Alkaloids from amphibian skin: a tabulation of over eight-hundred compounds. J. Natural Products 68, 1556–1575. doi: 10.1021/np0580560
Egsgaard H., Carlsen L. (1996). “Mass spectrometry of nitro and nitroso compounds,” in The chemistry of amino, nitroso and nitro and related groups. Eds. Patai S., Rappoport Z. (Chichester: Wiley Online Library), 249–294.
Hassler M. F., Harrison D. P., Jones T. H., Richart C. H., Saporito R. A. (2020). Gosodesmine, a 7-substituted hexahydroindolizine from the millipede gosodesmus claremontus. J. Natural Products 83, 2764–2768. doi: 10.1021/acs.jnatprod.0c00722
Ilić B., Dimkić I., Unković N., Grbić M. L., Vukojević J., Vujisić L., et al. (2018). Millipedes vs. pathogens: Defensive secretions of some julids (Diplopoda: Julida) as potential antimicrobial agents. J. Appl. Entomol 142, 775–791. doi: 10.1111/jen.12526
Jones T. H., Harrison D. P., Menegatti C., Mevers E., Knott K., Marek P., et al (2022). Deoxybuzonamine isomers from the millipede brachycybe lecontii (Platydesmida: andrognathidae). J.Nat.Prod 85, 1134–1140. doi: 10.1021/acs.jnatprod.2c00077
Kellogg J. J., Graf T. N., Paine M. F., McCune J. S., Kvalheim O. M., Oberlies N. H., et al. (2017). Comparison of metabolomics approaches for evaluating the variability of complex botanical preparations: green tea (Camellia sinensis) as a case study. J. Natural Products 80, 1457–1466. doi: 10.1021/acs.jnatprod.6b01156
Kuwahara Y., Mori N., Tanabe T. (2007). Detection of a neotropical frog alkaloid spiropyrrolizidine 236 from a Japanese polyzoniid millipede Kiusiozonium okai as a major defense component together with polyzonimine and nitropolyzonamine. Jpn. J. Environ. Entomol. Zool. 18, 91–95. doi: 10.11257/jjeez.18.91
Meinwald Y. C., Meinwald J., Eisner T. (1966). 1,2-Dialkyl-4(3H)-quinazolinones in the defensive secretion of a millipede (Glomeris marginata). Science 154, 390–391. doi: 10.1126/science.154.3747.390
Meinwald J., Smolanoff J., McPhail A., Miller A. W., Eisner T., Hicks K. (1975). Nitropolyzonamine: A spirocyclic nitro compound from the defensive glands of a milliped (polizonium rosalbum). Tetrahedron Lett. 28, 2367–2370. doi: 10.1016/0040-4039(75)80013-X
Mori K., Takagi Y. (2000). Enantioselective synthesis of polyzonimine and nitropolyzonamine, spirocyclic compounds from the defensive glands of a millipede, Polyzonium rosalbum. Tetrahedron Lett. 41, 6623–6625. doi: 10.1016/S0040-4039(00)01133-3
Raspotnig G., Bodner M., Fröhlich D., Blesl J., Kunert O. (2023). After chemo-metamorphosis: p-menthane monoterpenoids characterize the oil gland secretion of adult Nothrus palustris. Chemoecology 33, 71–82. doi: 10.1007/s00049-023-00386-y
Röper H. (1978). Ergebnisse chemisch-analytischer Untersuchungen der Wehrsekrete von Spirostreptiden, Spiroboliden und Juliden (Diplopoda), von Peripatopsis (Onychophora) und von Polyzonium (Diplopoda, Colobognatha): Results of chemical-analytical studies of defensive secretions of Spirostrepida, Spirobolida and Iulidae (Diplopoda), of Peripatopsis (Onychophora) and of Polyzonium (Diplopoda, Colobognatha).
Saporito R. A., Donnelly M. A., Hoffman R. L., Garraffo H. M., Daly J. W. (2003). A siphonotid millipede (Rhinotus) as the source of spiropyrrolizidine oximes of dendrobatid frogs. J. Chem. Ecol. 29, 2781–2786. doi: 10.1023/b:joec.0000008065.28364.a0
Schroeder F. C., Taggi A. E., Gronquist M., Malik R. U., Grant J. B., Eisner T., et al. (2008). NMR-spectroscopic screening of spider venom reveals sulfated nucleosides as major components for the brown recluse and related species. Proc. Natl. Acad. Sci. U.S.A. 105, 14283–14287. doi: 10.1073/pnas.0806840105
Shear W. A. (2015). The chemical defenses of millipedes (diplopoda): Biochemistry, physiology and ecology. Biochem. Systematics Ecol. 61, 78–117. doi: 10.1016/j.bse.2015.04.033
Shear W., Jones T., Wesener T. (2011). Glomerin and homoglomerin from the North American pill millipede Onomeris sinuata (Loomis 1943) (Diplopoda, pentazonia, glomeridae). IJM 4, 1–10. doi: 10.3897/ijm.4.1105
Smolanoff J., Kluge A. F., Meinwald J., McPhail A., Miller A. W., Hicks K., et al. (1975). Polyzonimine: A novel terpenoid insect repellent produced by a milliped. Science 188, 734–736. doi: 10.1126/science.1124395
Taggi A. E., Meinwald J., Schroeder F. C. (2004). A new approach to natural products discovery exemplified by the identification of sulfated nucleosides in spider venom. J. Am. Chem. Soc. 126, 10364–10369. doi: 10.1021/ja047416n
Takada W., Sakata T., Shimano S., Enami Y., Mori N., Nishida R., et al. (2005). Scheloribatid mites as the source of pumiliotoxins in dendrobatid frogs. J. Chem. Ecol. 31, 2403–2415. doi: 10.1007/s10886-005-7109-9
van den Dool H., Kratz P. D. (1963). A generalization of the retention index sysstem including linear temperature programmed gas-liquid partition chromatography. J. Chromatogr. 11, 463–471. doi: 10.1016/S0021-9673(01)80947-X
Wolfender J.-L., Litaudon M., Touboul D., Queiroz E. F. (2019). Innovative omics-based approaches for prioritisation and targeted isolation of natural products - new strategies for drug discovery. Nat. Prod Rep. 36, 855–868. doi: 10.1039/c9np00004f
Wolfender J.-L., Marti G., Thomas A., Bertrand S. (2015). Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr A 1382, 136–164. doi: 10.1016/j.chroma.2014.10.091
Keywords: Polyzonium germanicum, alkaloids, defensive secretions, NMR spectroscopy, GC-MS, LC-HRMS
Citation: Kunert O, Pferschy-Wenzig EM, Orthaber A, Raspotnig G and Bodner M (2023) Alkaloids from millipedes: a re-evaluation of defensive exudates from Polyzonium germanicum. Front. Ecol. Evol. 11:1212452. doi: 10.3389/fevo.2023.1212452
Received: 26 April 2023; Accepted: 31 July 2023;
Published: 15 August 2023.
Edited by:
Volker Nehring, University of Freiburg, GermanyReviewed by:
Adrian Brueckner, Inivzyne Technologies Inc, United StatesUte Mettal, University of Giessen, Germany
Christopher S Jeffrey, University of Nevada, Reno, United States
Copyright © 2023 Kunert, Pferschy-Wenzig, Orthaber, Raspotnig and Bodner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michaela Bodner, bWljaGFlbGEuYm9kbmVyQHVuaS1ncmF6LmF0
 Olaf Kunert
Olaf Kunert Eva Maria Pferschy-Wenzig
Eva Maria Pferschy-Wenzig Andreas Orthaber3
Andreas Orthaber3 Günther Raspotnig
Günther Raspotnig Michaela Bodner
Michaela Bodner