- 1The Key Laboratory of Aquatic Biodiversity and Conservation, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 2College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China
It has been debated whether interspecies niche differences may follow niche conservatism, whether niches are becoming specialists, whether generalists and specialists are maintained in a balance, and how the realized niche may adapt to local or temporal environments. In the present study, by using stable isotope analysis to evaluate the trophic niche of seven Cultrinae species, we investigated niche conservatism among these species from 10 sites of the Yangtze River by testing the Mantel test of trophic niche variables [trophic position (TP), δ13C, and trophic niche breadth]. We also comprehensively investigated the fundamental niche differences as well as spatial and temporal adaptations of the realized niche of these seven Cultrinae species by calculating trophic niche breadths, overlap, and TP. The results showed that TP and fundamental trophic niche breadth exhibited phylogenetic conservatism, while the fundamental trophic niche breadth of the three Hemicultrine species tended to become generalists and four Cultrine species tended to become specialists. Moreover, the realized trophic niche of the same species would exhibit different adaptation at different sites as well as in different seasons. In the wet season, trophic niche breadths of the seven species were significantly larger than that in the dry season with great overlaps between species, while in the dry season, trophic niche separation was found among the seven species due to competitive exclusion of niches. We believe that these findings will contribute to future theoretical and empirical niche investigations.
1 Introduction
The niche concept is often considered as a place in the natural world, particularly a habitat, a microhabitat, or a range of suitable environmental conditions (Grinnell, 1917), or as the role a species plays in the ecosystem defined, in part, by its resources and consumers (Elton, 1927; Ricklefs, 2010). It has been a key concept in the development of ecological and evolutionary theory and holds wider prominence, in areas as diverse as evolutionary theory (Violle et al., 2011; Carscadden et al., 2020). Explorations to niche concept have greatly prompted the advances of ecological science.
Different species should exhibit differences in their ecological niche. It has been debated how species may differentiate in their niches. Niche conservatism proposes that a species tends to retain aspects of its fundamental niche through speciation over space and time. Therefore, closely related species should be most ecologically similar (Harvey and Pagel, 1991; Wiens and Graham, 2005; Losos, 2008; Ricklefs, 2010; Wiens et al., 2010; Crisp and Cook, 2012). However, studies on niche conservatism have received different views. Some studies support niche conservatism (Ackerly, 2003; Hadly et al., 2009; Burns and Strauss, 2011; Pearman et al., 2014), while others do not (Losos et al., 2003; Knouft et al., 2006; Pearman et al., 2014).
For niche divergence (evolving), Loxdale et al. (2011) and Ackermann and Doebeli (2004) suggested evolutionary improbability of “generalism” in nature: over the evolutionary process, species may only utilize limited resources, niche breadth should decrease, and specialists should increase. However, others proposed that during speciation, generalists and specialists coexist and they can be maintained in a balance (Wilson and Yoshimura, 1994; Egas et al., 2004; Abrams, 2006; Dennis et al., 2011). Although these hypotheses have been often discussed, very few studies have focused on experimental validation.
For the ecological niche of a species, Hutchinson further distinguished between the fundamental niche and the realized niche (Hutchinson, 1957). The fundamental niche refers to the ideal conditions in which a given species might be able to survive, grow, and reproduce (Hutchinson, 1957; Roughgarden, 1974; Larose et al., 2018). However, owing to competition or environment constraints, a species actually only occupies a subset of the fundamental niche (realized niche) (Pulliam, 2000; Hurlbert and White, 2007; Colwell and Rangel, 2009; Severtsov, 2013). As such, the realized niche is often narrower than the fundamental niche, and it can change over time as environmental conditions change (Severtsov, 2013). Till now, the adaptability of the fundamental niche of aquatic organisms to temporal and spatial variability has received little attention (Haubrock et al., 2021; Ru et al., 2022).
Trophic niche has been proven more tractable than other niche dimensions. Carbon and nitrogen stable isotope technology as a powerful tool has been widely used to explore trophic structure and niche relationships at the species or community level (Layman et al., 2007; Jackson et al., 2011; Cucherousset and Villéger, 2015). As a proxy of ecological niches, the isotope niche metrics determined not only the space occupied by species in the C-N bi-plot but also relevant descriptors of the overall trophic status and resource use (Newsome et al., 2007), which were used to quantify the trophic diversity and niche overlap of coexisting species (Abrantes et al., 2014; Masese et al., 2018). Furthermore, the community-level metrics and the calculation of the standard ellipse area under the Bayesian framework make it easier to conduct comparative studies on population trophic structure across different temporal and spatial scales (Jackson et al., 2011).
In this study, we selected four fish species (Culter erythropterus, C. mongolicus, C. dabryi, and C. alburnus) as representatives of the Cultrine clade and three fish species (Hemiculter leucisculus, H. bleekeri, and Pseudolaubuca sinensis) as representatives of the Hemicultrine clade to explore the interspecies niche differences and the spatial–temporal adaptations. The seven fish species taxonomically belong to Cypriniformes, Cyprinidae, and Cultrins (Cultrinae) (Cheng et al., 2022), which are also part of the East Asian Endemic group of Cyprinids, and mostly dominant in the local ecosystem. Cultrins (Cultrinae) has been found as a monophyletic group, consisting of two clades, the Cultrine and the Hemicultrine by extensive studies (Cunha et al., 2002; Wang et al., 2004; Saitoh et al., 2006; Mayden et al., 2008; Liu and Chen, 2011; Tang et al., 2013a; Tang et al., 2013b).
In this study, by using stable isotope analysis to evaluate trophic niche of seven Cultrinae species collected from 10 sites of the Yangtze River, we examined phylogenetic conservatism of trophic niche [trophic position (TP), δ13C, and trophic niche breadth]. Moreover, we explored the fundamental niche characteristic of seven Cultrinae species and investigated spatial and temporal variations of the realized niche to comprehensively elucidate the trophic niche adaptation of these seven coexisting species. Our purposes are to elucidate (1) whether fundamental niche may exhibit niche conservatism; (2) whether niche can become more generalist or specialist; and (3) the spatial–temporal adaptations of the realized niche.
2 Materials and methods
2.1 Study areas and sampling sites
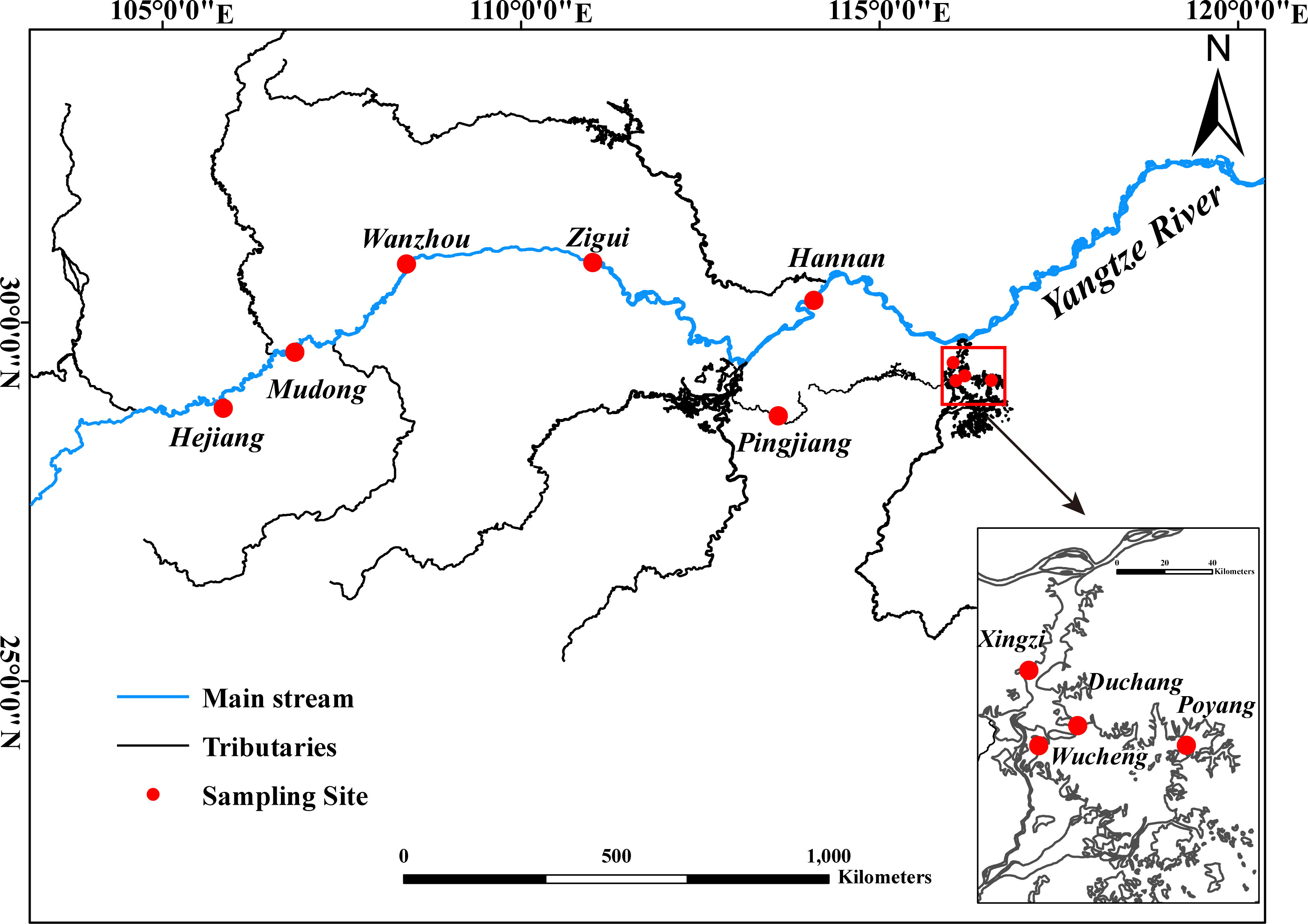
The Yangtze River is the third-longest river in the world, with a length of 6,397 km and a drainage basin of 1.8 × 106 km2 (Gao et al., 2019; Ren et al., 2022). In this study, we collected samples from 10 sites in the Yangtze River (five sites in the main stream and five sites in its tributaries) (Figure 1). All the 10 sampling sites were selected to investigate spatial difference of realized trophic niche, of which, 4 sampling sites of Poyang Lake (Xingzi, Duchang, Wucheng, and Poyang) were used to investigate seasonal variations of realized trophic niche.
2.2 Sample collection and processing
Fish samples for stable isotope analyses were collected using nets in July 2018, and in May, October, and November of 2019. Detailed sampling information is summarized in Supplementary Table 1. The fish were collected by drifting gill nets (50 m long × 1.5 m high, 5 and 10 cm mesh size), trawl nets (8 m net depth; 4.5 m × 1.8 m net opening), and fishhooks. The baseline crustaceans were also caught along with the fish in some sites to correct the differences between sites. Furthermore, the stable isotopes values for the baseline were also referred to the corresponding studies in some sites. Each sampled fish was identified to species level according to Chen (1998). Fishes caught and targeted for further analysis included seven cosmopolitan species of Cultrinae (Culter erythropterus, Culter mongolicus, Culter dabryi, Culter alburnus, Hemiculter leucisculus, Hemiculter bleekeri, and Pseudolaubuca sinensis).
A total of 651 fish belonging to seven Cultrinae species were collected from 10 sites of the Yangzte River and its tributaries. In this study, 1 sample of each fish species was selected for molecular analysis, and 3–16 samples of each fish species and 3 samples of crustacean species were selected for stable isotope analysis. After preserving an appropriate amount of muscle tissue in 95% alcohol for molecular analysis and white dorsal muscle (removing the skin and scales) for stable isotope analysis, the whole body was fixed in 10% formalin and deposited in the Institute of Hydrobiology, Chinese Academy of Sciences. In the laboratory, the white muscle of fish and foot muscle of crustaceans were oven-dried to a constant weight at 60°C for at least 48 h, ground into powder with an automatic grinder, and stored in clean containers until analysis. Fish collection and the experimental protocols were approved by the Institutional Animal Care and Use Committee of the Institute of Hydrobiology, Chinese Academic of Sciences (2018/LL/057).
2.3 Genetic distance and phylogenetic analysis
DNA extraction, PCR amplification, and sequencing of the cytochrome b (Cyt b) were conducted following Tang et al. (2012). Seven sequences of the Cultrinae species were used for the analysis (GenBank accession number: OQ621647, OQ621648, OQ621649, OQ621650, OQ621651, OQ621652, and OQ621653), with three species of Xenocyprininae (Xenocypris argentea MH797098, Xenocypris davidi MH797100, and Pseudobrama simoni MH797105) serving as the outgroup. The DNA sequences were aligned using Clustal X (Thompson et al., 1997) and were manually checked with SEAVIEW (Galtier et al., 1996). The Kimura’s 2-parameter (K2P) genetic distances were calculated using MEGA X (Kumar et al., 2018) for the seven species and were subsequently used for testing their relationships with trophic niche differences. Prior to the phylogenetic analysis, the method of Xia et al. (2003) was used to access substitution saturation of the sequences by comparing the information entropy-based index (ISS) with critical values (ISS.c) in DAMBE 7 (Xia, 2018). If ISS is significantly lower than ISS.c, then sequences have not experienced substitution saturation. The sequences of Cyt b showed no significant substitution saturation (ISS = 0.118< ISS.c = 0.793, p< 0.001), indicating that they can be used for phylogenetic analysis.
Based on the sequences of Cyt b, phylogenetic relationships of seven Cultrinae species sampled in the study areas were reconstructed using Bayesian inference (BI), maximum likelihood (ML), and Neighbor‐joining (NJ) approaches. NJ analysis was performed with MEGA X (Kumar et al., 2018) and bootstrapping with 1,000 pseudo replicates was used to examine the robustness of clades in the resulting tree (Felsenstein, 1985). The best-fit model (GTR+F+I) of nucleotide substitution for the data was identified by ModelFinder (Kalyaanamoorthy et al., 2017) using the Corrected Akaike Information Criterion (AICc) implemented in the PhyloSuite platform (Zhang et al., 2020). BI and ML analyses were performed with MrBayes v3.2.7 (Ronquist et al., 2012) and IQ-TREE 1.5 (Nguyen et al., 2015), respectively, implemented in the PhyloSuite platform (Zhang et al., 2020). In BI, four simultaneous Markov Chain Monte Carlo (MCMC) were run for 6 million generations, sampling every 100 generations and using a burn-in rate of 25%. In ML analysis, the node support was evaluated by 1,000 ultrafast bootstrap replicates (UFBS).
2.4 Stable isotope laboratory analysis
Approximately 200–300 µg of dry mass of muscle tissue were weighted into tin capsules and analyzed for stable isotope values using a Delta V Advantage mass spectrometer coupled to a Flash 2000 HT elemental analyzer (Thermo Fisher Scientific, Waltham, MA, USA). Muscle-tissue samples were not pre-treated with lipid extraction due to the relatively low lipid content of this type of tissue (Passi et al., 2002; Visentainer et al., 2007; Kim et al., 2012; Malek et al., 2016). Stable isotope ratios (part per mil, ‰) were expressed in δ notation as deviations from a standard:
where X is 13C or 15N, and R represents the stable isotope ratio (13C/12C for carbon and 15N/14N for nitrogen). The standard reference materials were Vienna-Pee Dee Belemnite limestone (V-PDB) for 13C/12C and atmospheric nitrogen for 15N/14N ratios (Peterson and Fry, 1987; Fry, 1999; Mendoza-Carranza et al., 2010). In this study, USGS40, USGS41, USGS65, USGS41A, UREA-Thermo, and IAEA-600 were used as working standards and interspersed in the samples. The analytical precision of the measurements was ±0.2‰ for both δ13C and δ15N.
2.5 Stable isotope baseline corrections
As δ13C and δ15N for basal resources might vary considerably between sites (e.g., France, 1995), we corrected for these differences before further analyses (De Santis et al., 2021). δ15N data were converted to trophic position (TP) using the following equation (Olsson et al., 2009; Britton et al., 2022):
where TP and δ15Nfish are the trophic positions and the nitrogen isotope ratios of each individual fish, δ15Nbaseline is the mean δ15N of the putative macroinvertebrate prey resources (Supplementary Table 2), 3.4 is the fractionation factor between adjacent trophic levels, and 2 is the trophic position of these prey resources (as primary consumers) (Post, 2002). δ13C were converted to corrected carbon (δ13Ccorr) using the following equation (Olsson et al., 2009; Britton et al., 2022):
where δ13Cfish is the carbon isotope value of each fish, δ13CmeanMI is the mean δ13C of the macroinvertebrate prey, and CRMI is the carbon range (δ13Cmax − δ13Cmin) of the same macroinvertebrates used for baseline in calculating the trophic position (Olsson et al., 2009; Britton et al., 2022) (Supplementary Table 2).
2.6 Stable isotope statistical analysis
Sampling sites, number of samples, mean stable isotope composition (δ13C and δ15N), and trophic position (TP) of each fish species can be found in Supplementary Table 3. All the data of each species at all sampling sites were pooled to represent the fundamental trophic niche. The δ13Ccorr–TP bi-plot was used to identify the differences in fundamental trophic niche among the seven Cultrinae species (Olsson et al., 2009).
In order to assess and quantify the influence of phylogeny on trophic niche variables, we used the Mantel test, which is a common approach to study niche conservatism (Mantel, 1967; Losos et al., 2003; Warren et al., 2008; Felizola Diniz-Filho et al., 2010; Montaña and Winemiller, 2013; Seger et al., 2013; Wang et al., 2015). The test was performed in the R package “vegan” between the two matrices, trophic niche differences (Euclidean distances) and pairwise genetic distances. The trophic niche differences matrix is then regressed on the genetic distances matrix and the regression is tested for significance. The tests were carried out through the Monte Carlo randomization, whereby the genetic distance matrix is held constant and species in the trait matrix are permutated randomly (procedure with 1,000 random permutations); the null hypothesis was that the two matrices were not related to each other at a significance level of 0.01.
In addition, to describe the trophic niche and estimate the trophic niche breadth, we calculated the corrected standard ellipse areas (SEAc) with the Stable Isotope Bayesian Ellipses (SIBER) package in R (Jackson et al., 2011). The SEAc enclose 40% of the corrected isotope data, thus representing the core trophic niche, indicating typical resource use within a species or population (Jackson et al., 2011; Jackson et al., 2012; Syväranta et al., 2013). We then calculated the fundamental trophic niche breadth and overlapping areas of the seven Cultrinae species to indicate the extent of resource sharing with the “SIBER” and “pheatmap” package in R.
The δ13C-δ15N bi-plots were used to identify trophic niche spatial and seasonal variations among the seven Cultrinae species. We then estimated the realized trophic niche breadths and overlapping areas of the seven Cultrinae species. A box plot with Kruskal–Wallis H Nonparametric Test can be used to test for seasonal variations in trophic niche breadths of the seven Cultrinae species in four sites of Poyang Lake. This analysis was conducted using STATISTICA 18.0, and the significance level was set at 0.05 for all statistical tests.
3 Results
3.1 Fundamental trophic niche of the seven Cultrinae species
For all the samples, the δ13C values ranged from −31.890‰ to −19.502‰ and δ15N values range from 8.041‰ to 19.805‰. The highest average value of δ13C and δ15N was found in H. leucisculus (Mudong) and C. dabryi (Xingzi), and the lowest average value of δ13C and δ15N was found in C. erythropterus (Hannan) and P. sinensis (Hejiang), respectively.
When all the data of each species were pooled together, large degrees of fundamental trophic overlaps were detected among these seven Cultrinae species, indicating similarity in their fundamental trophic niches, of which the highest trophic overlap was found between H. leucisculus and H. bleekeri (3.24‰2) (Figure 2). In addition, comparison of fundamental trophic niche breadth of these seven Cultrinae species showed that H. bleekeri had the greatest breadth and C. erythropterus had the smallest breadth (Figure 2).
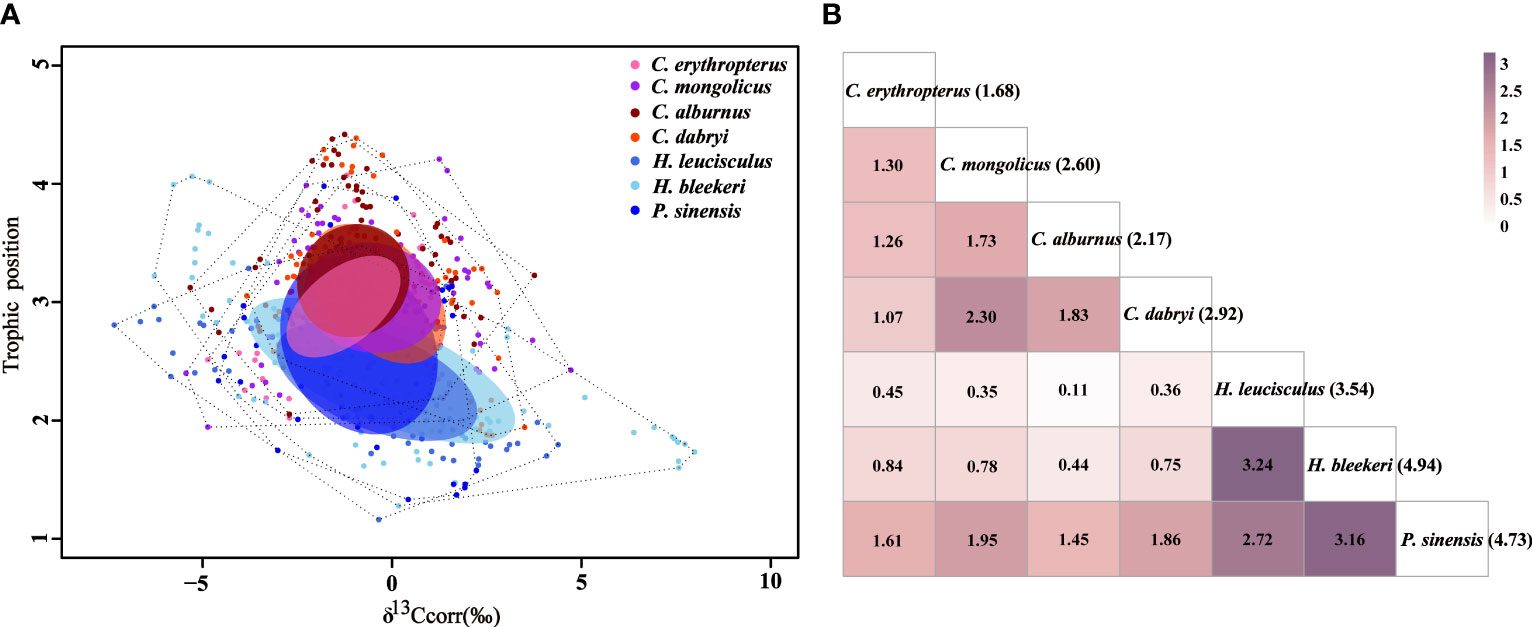
Figure 2 (A) Comparison of stable isotopic values (δ13Ccorr, Trophic position) together with their sample size-corrected standard ellipse area (SEAc) for the seven Cultrinae species from 10 sites in the Yangtze River. The seven species are displayed with different colored circles, respectively. The total area of the convex hull (TA) (dashed black lines) and SEAc represent the total niche space and core niche space occupied by species. (B) The fundamental trophic niche overlap areas (numbers inside boxes) of the corrected standard ellipse area (SEAc) of the seven Cultrinae species estimated using Stable Isotope Bayesian Ellipses (SIBER) in R. Color intensity increases with SEAc overlap. The trophic niche breadth (‰2) of the seven Cultrinae species are given in parentheses.
3.2 Fundamental trophic niche conservatism
Using seven sequences with 1,140 bp from the seven Cultrinae species and three outgroup species, the phylogenetic relationships among them were analyzed, and the genetic distances (Supplementary Table 4) were calculated to test phylogenetic niche conservatism among species (Figure 3). The Mantel test between genetic distances and Euclidean distances showed that phylogenetic relatedness and trophic niche were significantly correlated for TP and fundamental trophic niche breadth (TP: r = 0.642, p = 0.012< 0.05; fundamental trophic niche breadth: r = 0.460, p = 0.033< 0.05), whereas phylogenetic relatedness and trophic niche similarity were not significantly correlated for δ13C (r = −0.148, p = 0.77 > 0.05).
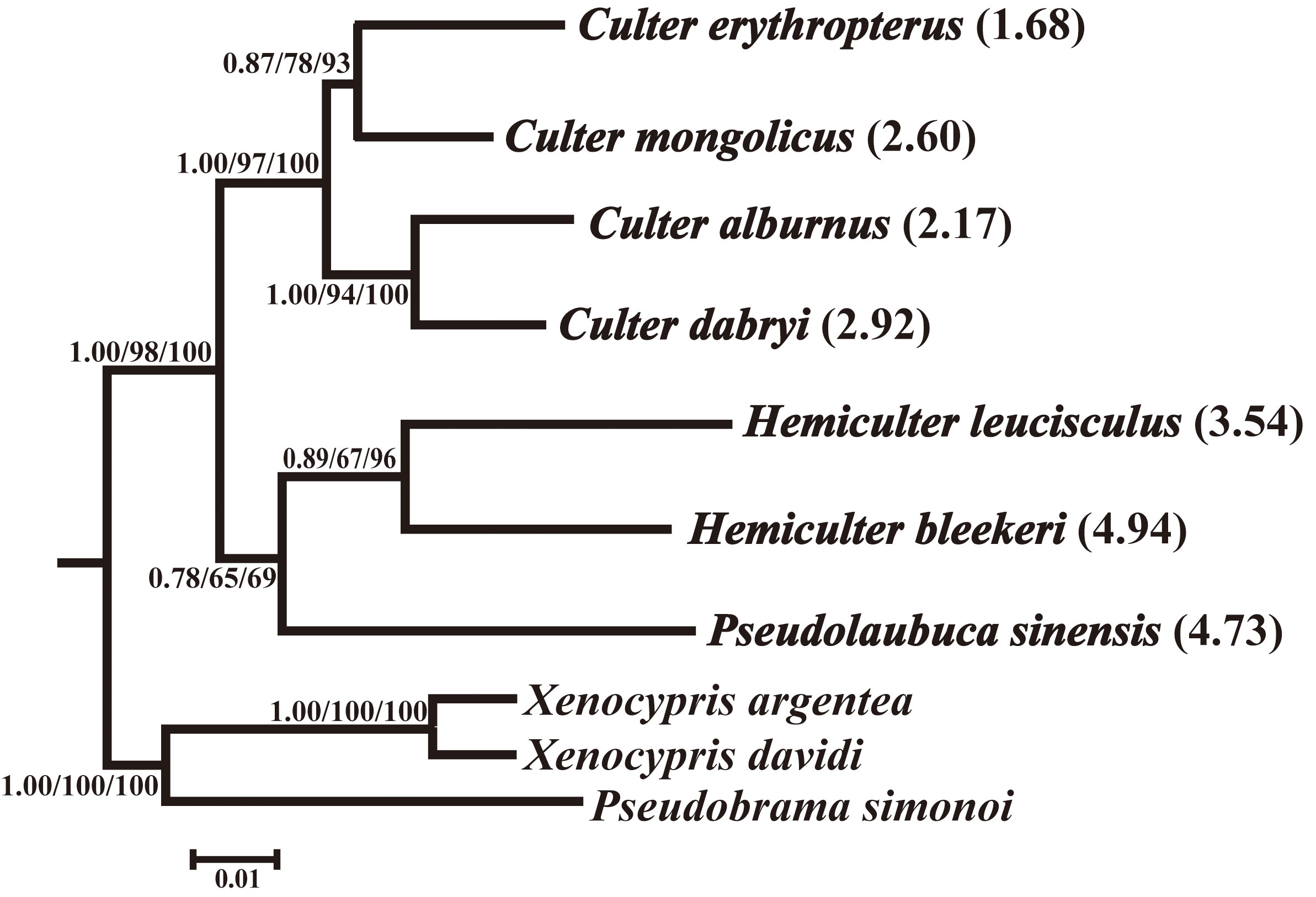
Figure 3 The NJ phylogenetic tree based on Cyt b gene sequences showing the phylogenetic relationship among seven Cultrinae species. Values on nodes correspond to the support values for BI, ML, and NJ methods. The fundamental trophic niche breadth (‰2) of the seven Cultrinae species are given in parentheses.
3.3 Fundamental trophic niche generalism and specialism
We took the average fundamental trophic niche breadth (3.23‰2) of the seven Cultrins species as a reference standard for judging whether a species is a generalist or a specialist: species with trophic niche breadth greater than 3.23‰2 tend to become generalists, whereas species with trophic niche breadth lower than 3.23‰2 tend to become specialists. Comparison of fundamental trophic niche breadth of these seven Cultrinae species showed that four Cultrine species had relatively small fundamental trophic niche breadth, indicating that they tend to become specialists, while three Hemicultrine species had relatively large fundamental trophic niche breadth, indicating that they tend to become generalists (Figure 2).
3.4 Spatial adaptations of realized niche of the seven Cultrinae species
The realized trophic niche of the seven Cultrinae species at different sites showed different results. In some sites, e.g., Mudong and Zigui, evident trophic niche separation among the seven Cultrinae species was detected, with negligible or even no niche overlaps among species, while in other sites, e.g., Hejiang, Wanzhou, Pingjiang, and Hannan, some species showed similarity in their trophic niche, with large degree of overlaps among species (Figure 4).
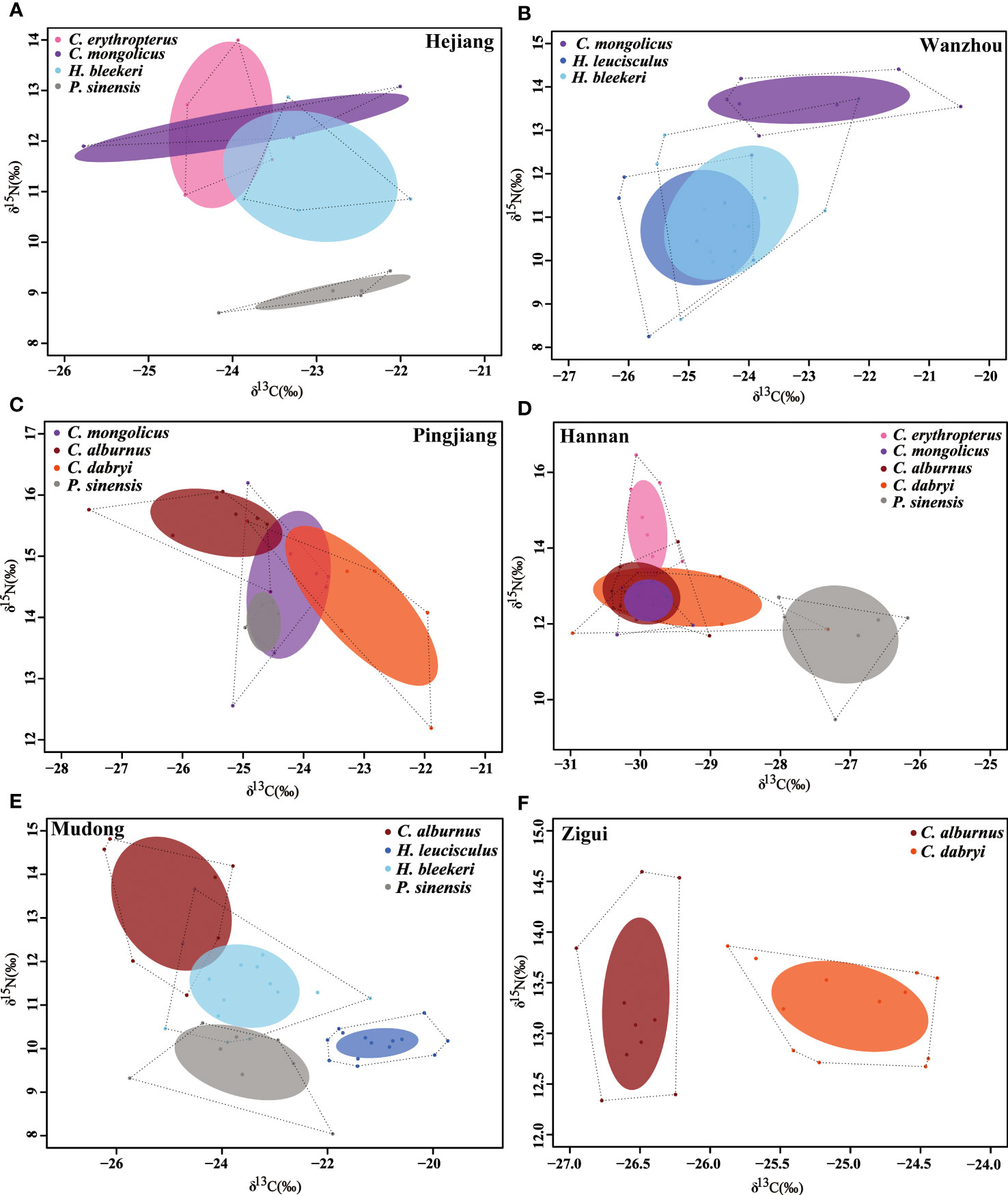
Figure 4 Comparison of stable isotopic values (δ13C, δ15N) together with their sample size-corrected standard ellipse area (SEAc) for the seven Cultrinae species from six sites in the Yangtze River (A–F). Different species are displayed with different colored circles. The total area of the convex hull (TA) (dashed black lines) and corrected standard ellipse area (SEAc) represent the total niche space and core niche space occupied by species.
Trophic niche breadth for the seven Cultrinae species varied over the 10 sites. None of the seven species consistently exhibited the widest or narrowest trophic niche breadth across all 10 sites, even in different seasons of the same sampling site, indicating that no species can always be a generalist or a specialist. Instead, some species exhibited relatively large trophic niche breadth in some sites and tended to become generalist species, while they showed relatively small trophic niche breadth in other sites and were more likely to become specialists. For instance, the relatively large trophic niche breadth of C. alburnus can be found in Mudong and Xingzi (Wet season), indicating that C. alburnus tended to become a generalist in these sites. However, C. alburnus exhibited relatively small trophic niche breadth in Hannan, Pingjiang, and Zigui, suggesting that C. alburnus tended to become a specialist in these sites (Figure 5).
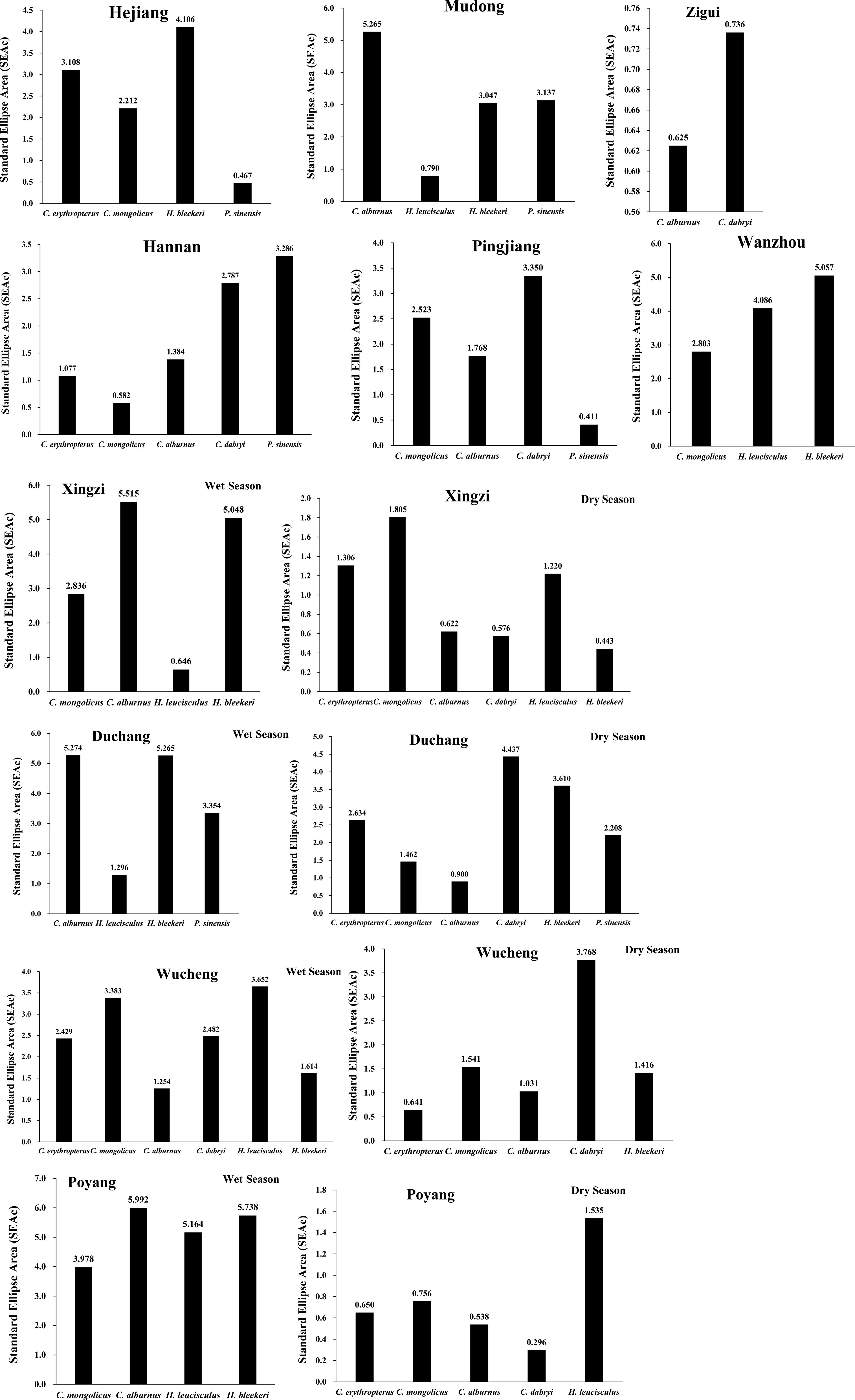
Figure 5 Corrected standard ellipse area (SEAc) representing the trophic niche breadth of the seven Cultrinae species collected from 10 sites of the Yangtze River.
3.5 Seasonal variations of realized niche
The seven Cultrinae species showed similar niche space, with a lot of overlap as well as some differences in four sites of Poyang Lake in the wet season. However, contrary to the wet season, we found trophic niche separation among the seven species, with negligible overlap in the dry season (Figures 6, 7).
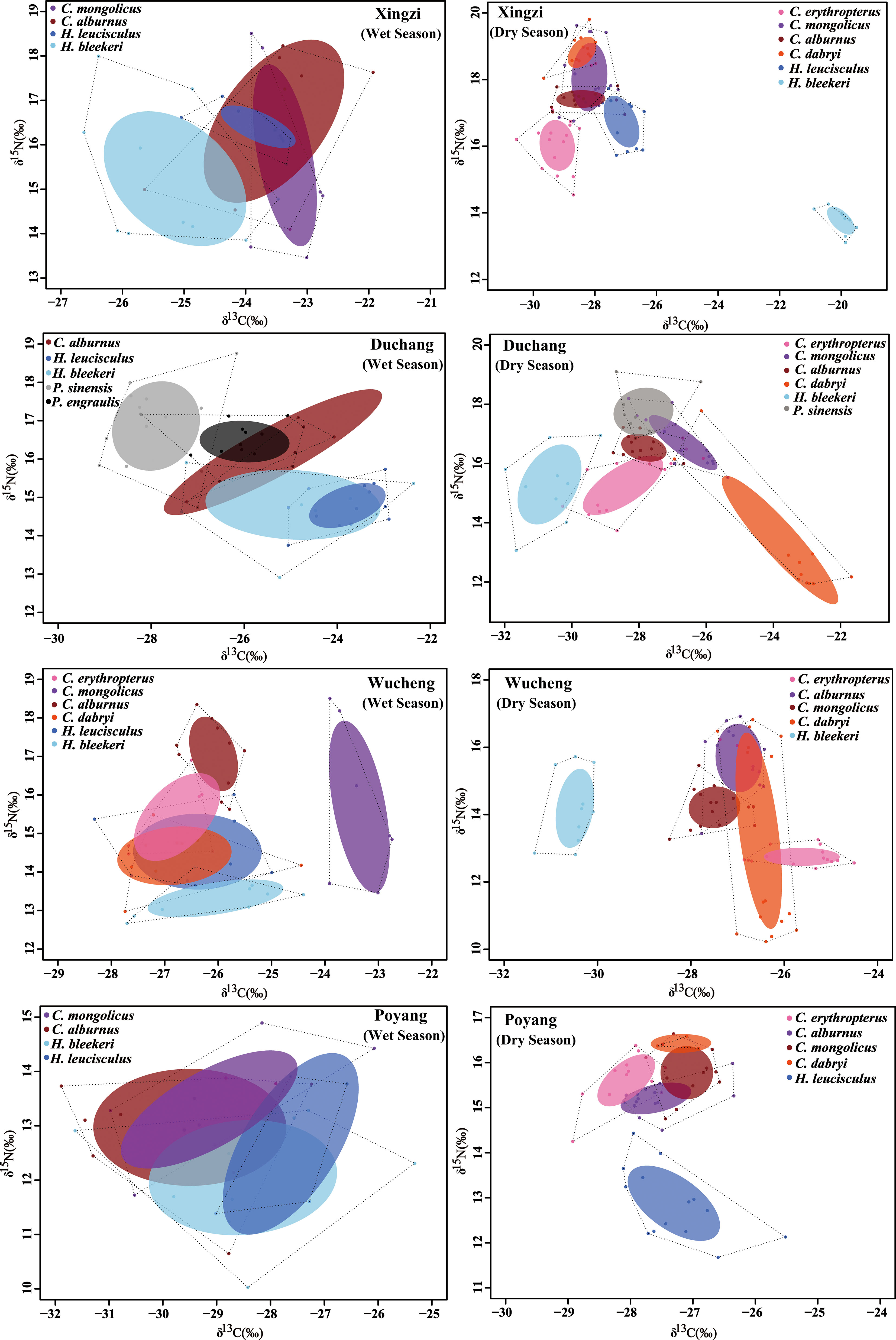
Figure 6 Comparison of stable isotopic values (δ13C, δ15N) together with their sample size-corrected standard ellipse area (SEAc) in the wet and dry season for the seven Cultrinae species from four sites in the Poyang Lake. Different species are displayed with different colored circles. The total area of the convex hull (TA) (dashed black lines) and corrected standard ellipse area (SEAc) represent the total niche space and core niche space occupied by species.
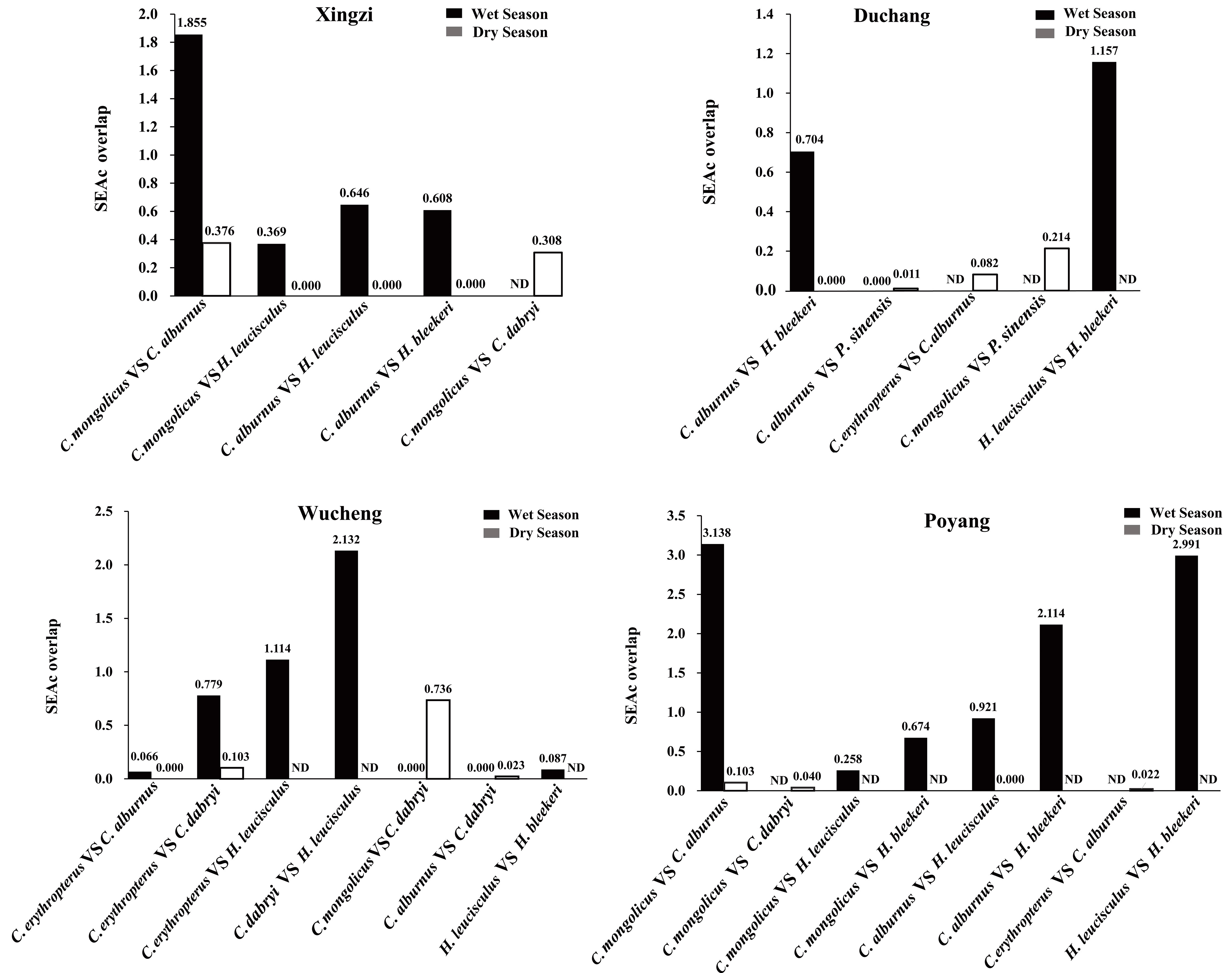
Figure 7 Comparison of sample size-corrected standard ellipse area (SEAc) overlap among the seven Cultrinae species in the wet and dry season in four sites of Poyang Lake. ND represents no data.
While making comparison between the wet season and dry season, we found that the trophic niche breadth of the seven species in the wet season was significantly larger than that in the dry season excluding H. leucisculus (Xingzi) and C. dabryi (Wucheng), indicating abundant food sources in the wet season (Kruskal–Wallis H Nonparametric Test, χ2 = 11.149, df = 1, p = 0.001< 0.01) (Figures 8, 9).
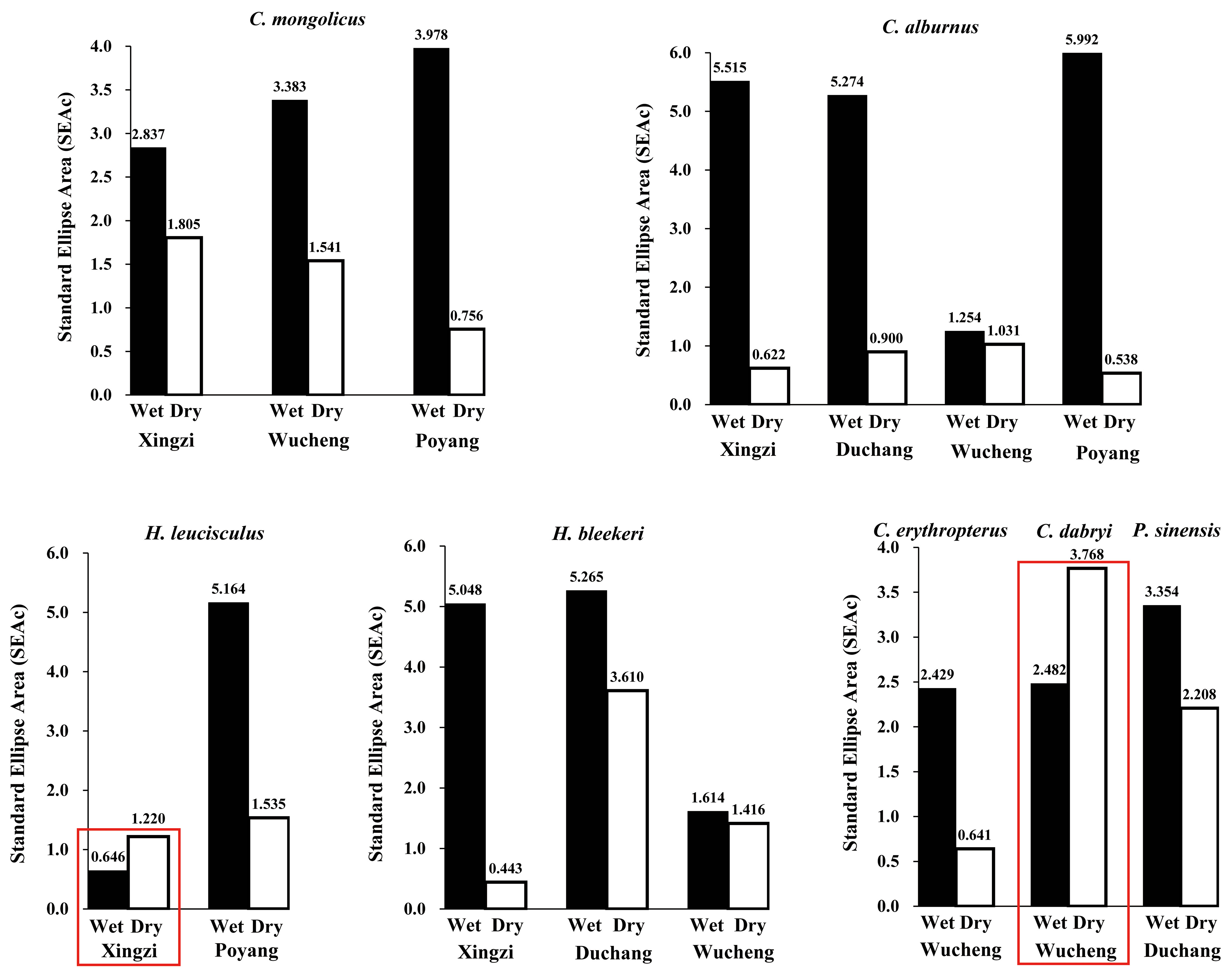
Figure 8 Comparison of corrected standard ellipse area (SEAc) representing the trophic niche breadth of seven Cultrinae species in the wet and dry season in four sites of Poyang Lake.
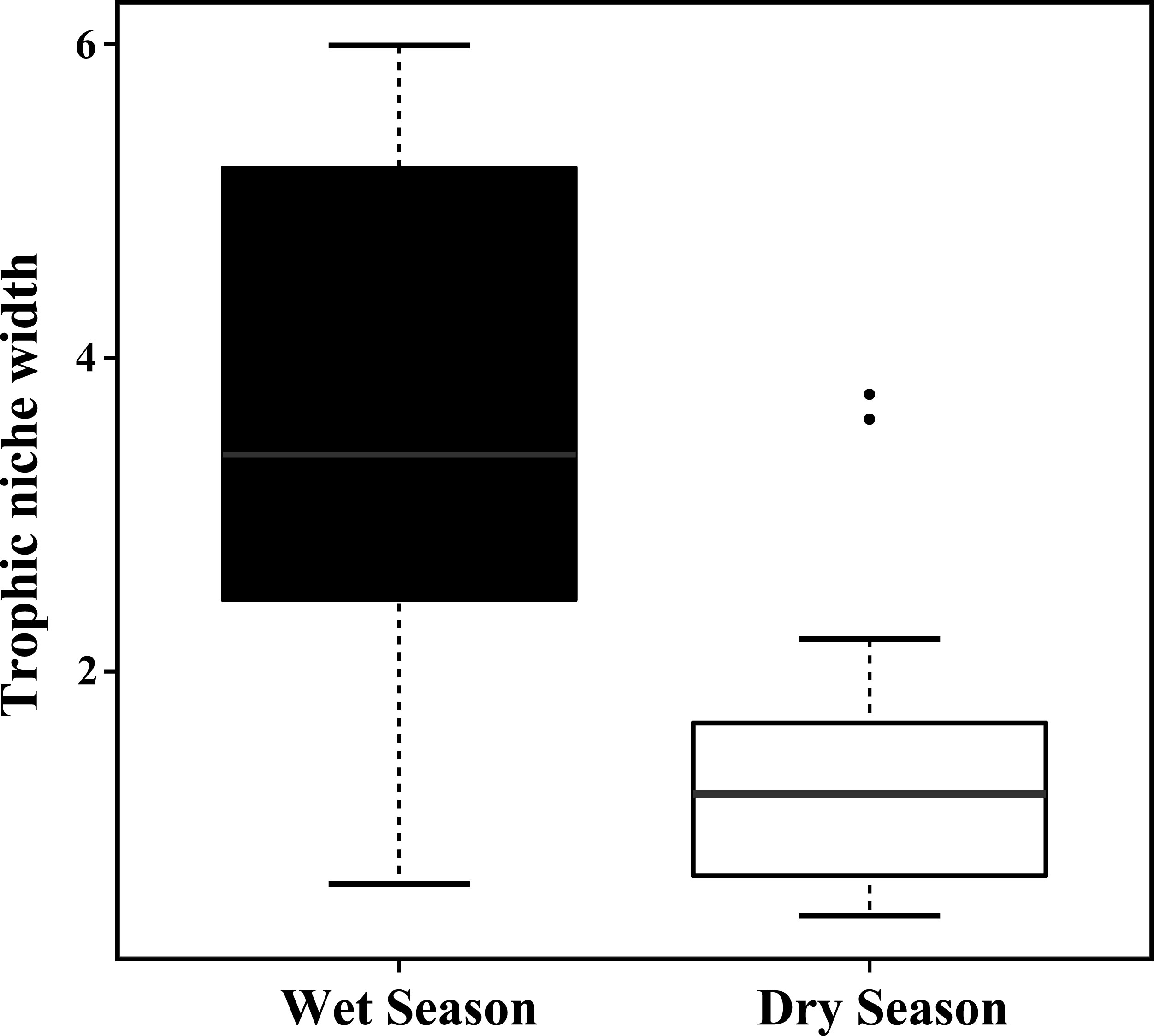
Figure 9 Box-and-whisker plots of trophic niche breadths for the seven Cultrinae species in the wet and dry season from four study sites in the Poyang Lake. Trophic niche breadths for the seven Cultrinae species in the wet season were significantly larger than that in the dry season at an α = 0.01. For the boxplots, lines indicate median, boxes indicate upper and lower quartiles, and whiskers indicate 1.5 quartile ranges. Black dots are outliers.
4 Discussion
4.1 Fundamental trophic niche conservatism
Previous studies have provided empirical support of the niche conservatism (Wiens et al., 2010; Crisp and Cook, 2012; Soberón and Martínez-Gordillo, 2012; Culumber and Tobler, 2016). However, other lines of evidence rather support niche divergence (Herrel et al., 1999; Moreno-Letelier et al., 2013; Pearman et al., 2014). In this study, a species’ trophic niche at all sampling sites were pooled together to represent its fundamental trophic niche. By using the Mantel test between genetic distances and Euclidean distances for the trophic niche variables (TP, δ13C, and trophic niche widths), we found significant phylogenetic effects for TP and fundamental trophic niche breadth, showing niche conservatism. This is consistent with the traditional recognition that Cultrine are carnivorous fish and Hemicultrine are omnivorous fish.
Meanwhile, we also found that different trophic niche variables may exhibit very different patterns, and there was no phylogenetic effect for δ13C. As the results showed that there is a strong phylogenetic effect for TP, close relatives are expected to maintain similar trophic levels, and we guess that even when maintaining a similar trophic level, they could differ in the item they consume (e.g., they feed on a different source of carbon), which leads to no phylogenetic effect in δ13C. Similar results can also be found in Wang et al. (2015) and Schluter (2000). Thus, it is important to remember that the niche is multi-dimensional, and in any clade, some aspects may be conserved while others diversify.
4.2 Fundamental trophic niche generalism and specialism
The evolution of an ecological niche is a process of adaptation in which species with certain traits are favored in a given environment. Along with this process, species in a particular environment will become more specialized in order to better survive and reproduce in that environment (Loxdale et al., 2011). Therefore, specialists are focused on a stable environment, while generalists are favored in an instable environment and cope more easily with environmental stochasticity (Fried et al., 2010; Peers et al., 2012). However, some scenarios of generalist–specialist coexistence have been proposed through recent theoretical work; they argued that during speciation and niche, evolutionary generalist and specialist can be maintained in a balance (Wilson and Yoshimura, 1994; Egas et al., 2004; Abrams, 2006; Dennis et al., 2011).
In this study, comparison of the fundamental trophic niche breadth of seven species showed that the fundamental trophic niche breadths of three omnivorous Hemicultrine species were much greater than that of four carnivorous Cultrine species, indicating that three Hemicultrine species had become generalists in the fundamental trophic niche and four Cultrine species had become specialists. It is important to note that the Mantel test showed that the fundamental trophic niche breadth follows conservatism; the fundamental trophic niches of closely related species (species in Cultrine or in Hemicultrine) were more similar than those of distantly related species (species in Cultrine vs. in Hemicultrine).
4.3 Spatial adaptations of realized trophic niche
This study investigated whether there were variations in the realized trophic niches of the fish species at the 10 sample sites as well as the causes of those variations. Unlike the fundamental niche, the realized niche is a set of ecological interactions between different species. We discovered that when species are distributed in various sites with limited food resources, fish need to balance the utilization of trophic and spatial niche to achieve species coexistence (Pratchett and Berumen, 2008).
The realized niche of the same species varied in different sites. The fish species showed broader trophic niches in some sampling sites, showing to be generalism. In contrast, this fish species exhibited narrower trophic niches in other sampling sites, showing more specialism. We believe that these variations in fish niche breadths at different sites are therefore probably a consequence of abiotic factors and biotic interactions such as food resource availability, water environmental fitness and capacity, predation, or competition (Platell et al., 1997; Guedes et al., 2015; Larose et al., 2018). Therefore, the trophic niches of fish go through different adaptation changes in order to adapt to various aquatic environmental conditions.
4.4 Seasonal variations of realized trophic niche
We identified significant differences in the trophic niche in the wet and dry seasons by comparing the trophic niche breadth and niche overlap of seven species in the Poyang Lake. The trophic niche breadths of all the seven species in the wet season were significantly greater than that in the dry season in four sites of Poyang Lake, except for H. leucisculus (Xingzi) and C. dabryi (Wucheng). The trophic niche overlaps among seven species were larger in the wet season compared to the dry season.
We speculate that this is due to environmental conditions related to seasonal changes in the availability of potential food resources (Wang et al., 2011; Wang et al., 2012). In the dry season, as low-water conditions result in contraction of aquatic habitats, the food resources available to fish are reduced proportionally. When a resource becomes limiting, competing species may specialize according to their species-specific resource preferences, and niche overlap will decrease, which leads to trophic niche differentiation to avoid competitive exclusion (Wiens, 1993; Gabler and Amundsen, 2010; Guedes et al., 2015). However, in the wet season, the water area expansion increased the population size of submerged aquatic macrophyte as well as the organic matter from terrestrial or macrophyte plants, making the food resources substantially more abundant. As a result of adequate food resources, species are no longer competing and competitive exclusion is avoided even when niches broadly overlap (Hastings and Gross, 2012). Our findings are consistent with this viewpoint, that when resources are no longer limiting, niche overlap may have little relationship or even an inverse relationship to competitive intensity: the greater the niche overlap, the less intense is competition (Hastings and Gross, 2012).
5 Conclusions
As one of the most important dimensions of niche, trophic niche plays a crucial role in the coexistence and resource partitioning of fish species in aquatic ecosystems. However, most studies on the trophic niche mainly focused on food chains, food webs, and differences in realized trophic niche among species. The interspecies niche differentiation pattern as well as the spatial and temporal adaptations of the realized niche have received little attention. In this study, by using stable isotope analysis to evaluate the trophic niche of seven Cultrinae species, we investigated interspecies trophic niche differences and spatial–temporal adaptations of the realized niche.
The results showed the following: (1) Significant phylogenetic effects were found for TP and trophic niche breadth except δ13C, indicating the existence of niche conservatism. (2) Comparison of the fundamental trophic niche breadth of seven species showed that three Hemicultrine species tended to become generalists and four Cultrine species tended to become specialists. (3) The trophic niche of the same species would evolve adaptively in different sites, and some species tended to generalize in some sites but specialized in other sites. (4) In the wet season with abundant food resources, trophic niche breadths of the seven species were large and there were great overlaps between species, while in the dry season with limited food resources, there was trophic niche differentiation due to competitive exclusion of niches.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI database with the accession numbers: OQ621647, OQ621648, OQ621649, OQ621650, OQ621651, OQ621652, OQ621653.
Ethics statement
The animal study was reviewed and approved by Fish collection and the experimental protocols were approved by the Institutional Animal Care and Use Committee of the Institute of Hydrobiology, Chinese Academic of Sciences (2018/LL/057).
Author contributions
HL conceived this study. TY wrote the first draft of the manuscript with contributions from DY. TY carried out the experiment with the help of SY, CW, and KS. TY performed data analysis. All authors approved the manuscript for submission.
Funding
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000), the National Key R and D Program of China (2018YFD0900806), the National Natural Science Foundation of China (31872234), and the 2020 consulting project from Hubei Institute of Chinese Engineering Development Strategies (HB2020B19).
Acknowledgments
We sincerely thank Mingzheng Li, Qiaoling Deng, Chen Zhang, Jinghong Li, and other colleagues for their assistance with field sampling. We also thank Ping Yang and Jiaxin Gao for their help with map drawing. Thanks are also given to Qiang Qin for laboratory assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1185413/full#supplementary-material
References
Abrams P. A. (2006). The prerequisites for and likelihood of generalist–specialist coexistence. Am. Nat. 167, 329–342. doi: 10.1086/499382
Abrantes K. G., Barnett A., Bouillon S. (2014). Stable isotope-based community metrics as a tool to identify patterns in food web structure in east African estuaries. Funct. Ecol. 28 (1), 270–282. doi: 10.1111/1365-2435.12155
Ackerly D. D. (2003). Community assembly, niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 164, S165–S184. doi: 10.1086/368401
Ackermann M., Doebeli M. (2004). Evolution of niche width and adaptive diversification. Evolution 58, 2599–2612. doi: 10.1111/j.0014-3820.2004.tb01614.x
Britton J. R., Cusherousset J., Dominguez Almela V. (2022). Novel trophic subsidies from recreational angling transform the trophic ecology of freshwater fishes. J. Appl. Ecol. 59 (9), 2373–2385. doi: 10.1111/1365-2664.14237
Burns J. H., Strauss S. Y. (2011). More closely related species are more ecologically similar in an experimental test. PNAS 108 (13), 5302–5307. doi: 10.1073/pnas.1013003108
Carscadden K. A., Emery N. C., Arnillas C. A., Cadotte M. W., Afkhami M. E., Gravel D., et al. (2020). Niche Breadth: Causes and consequences for ecology, evolution, and conservation. Q. Rev. Biol. 95, 179–214. doi: 10.1086/710388
Cheng P., Yu D., Tang Q., Yang J., Chen Y., Liu H. (2022). Macro-evolutionary patterns of East Asian opsariichthyin-xenocyprinin-cultrin fishes related to the formation of river and river-lake environments under monsoon climate. Water Biol. Secur. 1, 100036. doi: 10.1016/j.watbs.2022.100036
Colwell R. K., Rangel T. F. (2009). Hutchinson’s duality: the once and future niche. PNAS 106, 19651–19658. doi: 10.1073/pnas.0901650106
Crisp M. D., Cook L. G. (2012). Phylogenetic niche conservatism: what are the underlying evolutionary and ecological causes? New Phytol. 196, 681–694. doi: 10.1111/j.1469-8137.2012.04298.x
Cucherousset J., Villéger S. (2015). Quantifying the multiple facets of isotopic diversity: New metrics for stable isotope ecology. Ecol. Indic. 56, 152–160. doi: 10.1016/j.ecolind.2015.03.032
Culumber Z. W., Tobler M. (2016). Ecological divergence and conservatism: spatiotemporal patterns of niche evolution in a genus of livebearing fishes (Poeciliidae: Xiphophorus). BMC. Evol. Biol. 16, 44. doi: 10.1186/s12862-016-0593-4
Cunha C., Mesquita N., Dowling T. E., Gilles A., Coelho M. M. (2002). Phylogenetic relationships of Eurasian and American cyprinids using cytochrome b sequences. J. Fish Biol. 61, 929–944. doi: 10.1111/j.1095-8649.2002.tb01853.x
Dennis R. L., Dapporto L., Fattorini S., Cook. L. M. (2011). The generalism-specialism debate: the role of generalists in the life and death of species. Biol. J. Linn. Soc 104, 725–737. doi: 10.1111/j.1095-8312.2011.01789.x
De Santis V., Quadroni S., Britton R. J., Carosi A., Gutmann Roberts C., Lorenzoni M., et al. (2021). Biological and trophic consequences of genetic introgression between endemic and invasive Barbus fishes. Biol. Invasions 23 (11), 3351–3368. doi: 10.1007/s10530-021-02577-6
Egas M., Dieckmann U., Sabelis M. W. (2004). Evolution restricts the coexistence of specialists and generalists: the role of trade-off structure. Am. Nat. 163, 518–531. doi: 10.1086/382599
Felizola Diniz-Filho J. A., Terribile L. C., Ribeiro da Cruz M. J., Vieira L. C. G. (2010). Hidden patterns of phylogenetic non-stationarity overwhelm comparative analyses of niche conservatism and divergence. Glob. Ecol. Biogeogr. 19, 916–926. doi: 10.1111/j.1466-8238.2010.00562.x
Felsenstein J. (1985). Confidence-limits on phylogenies – an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
France R. L. (1995). Critical examination of stable isotope analysis as a means for tracing carbon pathways in stream ecosystems. Can. J. Fish. Aquat. Sci. 52, 651–656. doi: 10.1139/f95-065
Fried G., Petit S., Reboud X. (2010). A specialist-generalist classification of the arable flora and its response to changes in agricultural practices. BMC Ecol. 10, 20. doi: 10.1186/1472-6785-10-20
Fry B. (1999). Using stable isotopes to monitor watershed influences on aquatic trophodynamics. Can. J. Fish. Aquat. Sci. 56 (11), 2167–2171. doi: 10.1139/f99-152
Gabler H. M., Amundsen P. E. (2010). Feeding strategies, resource utilisation and potential mechanisms for competitive coexistence of Atlantic salmon and alpine bullhead in a sub-Arctic river. Aquat. Ecol. 44, 325–336. doi: 10.1007/s10452-009-9243-x
Galtier N., Gouy M., Gautier C. (1996). SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 12 (6), 543–548. doi: 10.1093/bioinformatics/12.6.543
Gao X., Fujiwara M., Winemiller K. O., Lin P. C., Liu H. Z. (2019). Regime shift in fish assemblage structure in the Yangtze River following construction of the Three Gorges Dam. Sci. Rep. 9 (1), 4212. doi: 10.1038/s41598-019-38993-x
Grinnell J. (1917). The niche-relationships of the california thrasher. Auk. 34, 427–433. doi: 10.2307/4072271
Guedes A. P. P., Araújo F. G., Pessanha A. L. M., Milagre R. R. (2015). Partitioning of the feeding niche along spatial, seasonal and size dimensions by the fish community in a tropical Bay in Southeastern Brazil. Mar. Ecol. 36, 38–56. doi: 10.1111/maec.12115
Hadly E. A., Spaeth P. A., Li C. (2009). Niche conservatism above the species level. PNAS 106 (Suppl 2), 19707–19714. doi: 10.1073/pnas.0901648106
Harvey P. H., Pagel M. D. (1991). The Comparative Method in Evolutionary Biology (Oxford: Oxford University Press).
Hastings A., Gross L. (2012). Encyclopedia of Theoretical Ecology (Berkeley: University of California Press).
Haubrock P. J., Balzani P., Matsuzaki S. S., Tarkan A. S., Kourantidou M., Haase P. (2021). Spatio-temporal niche plasticity of a freshwater invader as a harbinger of impact variability. Sci. Total Environ. 777, 145947. doi: 10.1016/j.scitotenv.2021.145947
Herrel A., Spithoven L., Van Damme R., DE Vree F. (1999). Sexual dimorphism of head size in Gallotia galloti: testing the niche divergence hypothesis by functional analyses. Funct. Ecol. 13, 289–297. doi: 10.1046/j.1365-2435.1999.00305.x
Hurlbert A. H., White E. P. (2007). Ecological correlates of geographical range occupancy in North American birds. Global Ecol. Biogeogr. 16, 764–773. doi: 10.1111/j.1466-8238.2007.00335.x
Hutchinson G. E. (1957). Concluding remarks. Cold Spring Harbor Symp. Quant. Biol. 22, 415–427. doi: 10.1101/SQB.1957.022.01.039
Jackson M. C., Donohue I., Jackson A. L., Britton J. R., Harper D. M., Grey J. (2012). Population-level metrics of trophic structure based on stable isotopes and their application to invasion ecology. PloS One 7 (2), e31757. doi: 10.1371/journal.pone.0031757
Jackson A. L., Inger R., Parnell A. C., Bearhop S. (2011). Comparing isotopic niche widths among and within communities: SIBER - Stable Isotope Bayesian Ellipses in R: Bayesian isotopic niche metrics. J. Anim. Ecol. 80 (3), 595–602. doi: 10.1111/j.1365-2656.2011.01806.x
Kalyaanamoorthy S., Minh B. Q., Wong T., von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi: 10.1038/nmeth.4285
Kim S. L., Casper D. R., Galván-Magaña F., Ochoa-Díaz R., Hernández-Aguilar S. B., Koch P. L. (2012). Carbon and nitrogen discrimination factors for elasmobranch soft tissues based on a long-term controlled feeding study. Environ. Biol. Fishes. 95, 37–52. doi: 10.1007/s10641-011-9919-7
Knouft J. H., Losos J. B., Glor R. E., Kolbe J. J. (2006). Phylogenetic analysis of the evolution of the niche in lizards of the Anolis sagrei group. Ecology 87 (7 Suppl), S29–S38. doi: 10.1890/0012-9658(2006)87[29:paoteo]2.0.co;2
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Larose C., Parker D. J., Schwander T. (2018). Fundamental and realized feeding niche breadths of sexual and asexual stick insects. Proc. R. Soc B. 285 (1892), 20181805. doi: 10.1098/rspb.2018.1805
Layman C. A., Arrington D. A., Montaña C. G., Post D. M. (2007). Can stable isotope ratios provide for community-wide measures of trophic structure? Ecology 88 (1), 42–48. doi: 10.1890/0012-9658(2007)88[42:csirpf]2.0.co;2
Liu H. Z., Chen Y. Y. (2011). Phylogeny of the East Asian cyprinids inferred from sequences of the mitochondrial DNA control region. Can. J. Zool. 81 (12), 1938–1946. doi: 10.1139/z03-169
Losos J. B. (2008). Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11 (10), 995–1003. doi: 10.1111/j.1461-0248.2008.01229.x
Losos J. B., Leal M., Glor R. E., de Queiroz K., Hertz P. E., Schettino L. R., et al. (2003). Niche lability in the evolution of a Caribbean lizard community. Nature 424 (6948), 542–545. doi: 10.1038/nature01814
Loxdale H. D., Lushai G., Harvey J. A. (2011). The evolutionary improbability of “generalism” in nature, with special reference to insects. Biol. J. Linn. Soc 103, 1–18. doi: 10.1111/j.1095-8312.2011.01627.x
Malek A. J., Collie J. S., Taylor D. L. (2016). Trophic structure of a coastal fish community determined with diet and stable isotope analyses. J. Fish Biol. 89 (3), 1513–1536. doi: 10.1111/jfb.13059
Mantel N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27 (2), 209–220.
Masese F. O., Abrantes K. G., Gettel G. M., Irvine K., Bouillon S., McClain M. E. (2018). Trophic structure of an African savanna river and organic matter inputs by large terrestrial herbivores: A stable isotope approach. Freshw. Biol. 63 (11), 1365–1380. doi: 10.1111/fwb.13163
Mayden R. L., Tang K. L., Wood R. M., Chen W. J., Agnew M. K., Conway K. W., et al. (2008). Inferring the Tree of Life of the order Cypriniformes, the earth's most diverse clade of freshwater fishes: Implications of varied taxon and character sampling. J. Syst. Evol. 46 (3), 424–438. doi: 10.3724/SP.J.1002.2008.08062
Mendoza-Carranza M., Hoeinghaus D. J., Garcia A. M., Romero-Rodriguez Á. (2010). Aquatic food webs in mangrove and seagrass habitats of Centla Wetland, a Biosphere Reserve in Southeastern Mexico. Neotrop. Ichthyol. 8 (1), 171–178. doi: 10.1590/S1679-62252010000100020
Montaña C. G., Winemiller K. O. (2013). Evolutionary convergence in perciform fishes. Biol. J. Linn Soc. Lond. 109, 146–164. doi: 10.1111/bij.12021
Moreno-Letelier A., Ortíz-Medrano A., Piñero D. (2013). Niche divergence versus neutral processes: combined environmental and genetic analyses identify contrasting patterns of differentiation in recently diverged pine species. PloS One 8 (10), e78228. doi: 10.1371/journal.pone.0078228
Newsome S. D., del Rio C. M., Bearhop S., Phillips D. L. (2007). A niche for isotopic ecology. Front. Ecol. Environ. 5 (8), 429–436. doi: 10.1890/060150.1
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Olsson K., Stenroth P., Nyström P. E. R., Granéli W. (2009). Invasions and niche width: Does niche width of an introduced crayfish differ from a native crayfish? Freshw. Biol. 54 (8), 1731–1740. doi: 10.1111/j.1365-2427.2009.02221.x
Passi S., Cataudella S., Di Marco P., De Simone F., Rastrelli L. (2002). Fatty acid composition and antioxidant levels in muscle tissue of different Mediterranean marine species of fish and shellfish. J. Agric. Food Chem. 50 (25), 7314–7322. doi: 10.1021/jf020451y
Pearman P. B., Lavergne S., Roquet C., Wüest R., Zimmermann N. E., Thuiller W. (2014). Phylogenetic patterns of climatic, habitat and trophic niches in a European avian assemblage. Global Ecol. Biogeogr. 23 (4), 414–424. doi: 10.1111/geb.12127
Peers M. J., Thornton D. H., Murray D. L. (2012). Reconsidering the specialist-generalist paradigm in niche breadth dynamics: resource gradient selection by Canada Lynx and Bobcat. PloS One 7 (12), e51488. doi: 10.1371/journal.pone.0051488
Peterson B. J., Fry B. (1987). Stable isotope in ecosystem studies. Annu. Rev. Ecol. Syst. 18, 293–320. doi: 10.1146/annurev.es.18.110187.001453
Platell M. E., Sarre G. A., Potter I. C. (1997). The diets of two cooccuring marine teleosts, Parequula melbournensis and Pseudocaranx wright, and their relationships to body size and mouth morphology, and the season and location of capture. Environ. Biol. Fishes. 49, 361–376. doi: 10.1023/A:1007388200920
Post D. M. (2002). Using stable isotopes to estimate trophic position: Models, methods, and assumptions. Ecology 83 (3), 703–718. doi: 10.1111/j.1095-8649.2012.03251.x
Pratchett M. S., Berumen M. L. (2008). Interspecific variation in distributions and diets of coral reef butterflyfishes (Teleostei: Chaetodontidae). J. Fish Biol. 73 (7), 1730–1747. doi: 10.1111/j.1095-8649.2008.02062.x
Pulliam H. R. (2000). On the relationship between niche and distribution. Ecol. Lett. 3, 349–361. doi: 10.1046/j.1461-0248.2000.00143.x
Ren F., Sun Y., Liu J., Chen K., Shi N. (2022). A modified dynamic DEA model to assess the wastewater treatment efficiency: perspective from Yangtze River and Non-Yangtze River Basin. Sci. Rep. 12 (1), 9931. doi: 10.1038/s41598-022-14105-0
Ricklefs R. E. (2010). Evolutionary diversification, coevolution between populations and their antagonists, and the filling of niche space. PNAS 107 (4), 1265–1272. doi: 10.1073/pnas.0913626107
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 (3), 539–542. doi: 10.1093/sysbio/sys029
Roughgarden J. (1974). The fundamental and realized niche of a solitary population. Am. Nat. 108, 232–235. doi: 10.1086/282902
Ru H. J., Zhong L. Q., Wei N., Li Y. F., Sheng Q., Ni Z. H. (2022). Variations of trophic structure and niche space in fish community along a highly regulated subtropical large river. Ecol. Evol. 12, e9424. doi: 10.1002/ece3.9424
Saitoh K., Sado T., Mayden R. L., Hanzawa N., Nakamura K., Nishida M., et al. (2006). Mitogenomic evolution and interrelationships of the cypriniformes (Actinopterygii: ostariophysi): the first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J. Mol. Evol. 63, 826–841. doi: 10.1007/s00239-005-0293-y
Seger G. D. S., Duarte L. D. S., Debastiani V. J., Kindel A., Jarenkow J. A. (2013). Discriminating the effects of phylogenetic hypothesis, tree resolution and clade age estimates on phylogenetic signal measurements. Plant Biol. 15, 858–867. doi: 10.1111/j.1438-8677.2012.00699.x
Severtsov A. S. (2013). Relationship between fundamental and realized ecological niches. Biol. Bull. Rev. 3, 187–195. doi: 10.1134/S2079086413030080
Soberón J., Martínez-Gordillo D. (2012). Occupation of environmental and morphological space: climatic niche and skull shape in Neotoma woodrats. Evol. Ecol. Res. 14, 503–517.
Syväranta J., Lensu A., Marjomäki T. J., Oksanen S., Jones R. I. (2013). An empirical evaluation of the utility of convex hull and standard ellipse areas for assessing population niche widths from stable isotope data. PloS One 8 (2), e56094. doi: 10.1371/journal.pone.0056094
Tang K. L., Agnew M. K., Hirt M. V., Lumbantobing D. N., Sado T., Teoh V., et al. (2013b). Limits and phylogenetic relationships of East Asian fishes in the subfamily Oxygastrinae (Teleostei: Cypriniformes: Cyprinidae). Zootaxa 3681, 101–135. doi: 10.11646/ZOOTAXA.3681.2.1
Tang Q. Y., Liu S. Q., Yu D., Liu H. Z., Danley P. D. (2012). Mitochondrial capture and incomplete lineage sorting in the diversification of balitorine loaches (Cypriniformes, Balitoridae) revealed by mitochondrial and nuclear genes. Zool. Scr. 41 (3), 233–247. doi: 10.1111/j.1463-6409.2011.00530.x
Tang K. L., Lumbantobing D. N., Mayden R. L. (2013a). The phylogenetic placement of oxygaster van hasselt 1823 (Teleostei: cypriniformes: cyprinidae) and the taxonomic status of the family-group name oxygastrinae bleeke. Copeia 2013, 13–22. doi: 10.1643/CG-10-121
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 (24), 4876–4882. doi: 10.1093/nar/25.24.4876
Violle C., Nemergut D. R., Pu Z., Jiang L. (2011). Phylogenetic limiting similarity and competitive exclusion. Ecol. Lett. 14, 782–787. doi: 10.1111/j.1461-0248.2011.01644.x
Visentainer J. V., Noffs M. D. A., de Oliveira Carvalho P., de Almeida V. V., de Oliveira C. C., de Souza N. E. (2007). Lipid content and fatty acid composition of 15 marine fish species from the southeast coast of Brazil. J. Am. Oil Chem. Soc 84, 543–547. doi: 10.1007/s11746-007-1070-4
Wang X. Z., Liu H. Z., He S. P., Chen Y. Y. (2004). Sequence analysis of cytochrome b gene indicates that East Asian group of cyprinid subfamily Leuciscinae (Teleostei: Cyprinidae) evolved independently. Prog. Nat. Sci. 14 (2), 132–137. doi: 10.1080/10020070412331343261
Wang M. R., Liu F., Lin P. C., Yang S. R., Liu H. Z. (2015). Evolutionary dynamics of ecological niche in three Rhinogobio fishes from the upper Yangtze River inferred from morphological traits. Ecol. Evol. 5, 567–577. doi: 10.1002/ece3.1386
Wang Y. Y., Yu X. B., Li W. H., Xu J., Chen Y. W., Fan N. (2011). Potential influence of water level changes on energy flows in a lake food web. Chin. Sci. Bull. 56, 2794–2802. doi: 10.1007/s11434-011-4649-y
Wang Y. Y., Yu X. B., Xu J., Li W. H., Fan N. (2012). Temporal variation of energy sources in a floodplain lake fish community. J. Freshw. Ecol. 27, 295–303. doi: 10.1080/02705060.2012.656714
Warren D. L., Glor R. E., Turelli M. (2008). Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62, 2868–2883. doi: 10.1111/j.1558-5646.2008.00482.x
Wiens J. A. (1993). Fat times, lean times and competition among predators. Trends Ecol. Evol. 8, 348–349. doi: 10.1016/0169-5347(93)90216-C
Wiens J. J., Ackerly D. D., Allen A. P., Anacker B. L., Buckley L. B., Cornell H. V., et al. (2010). Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x
Wiens J. J., Graham C. H. (2005). Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 36 (1), 519–539. doi: 10.1146/annurev.ecolsys.36.102803.095431
Wilson D. S., Yoshimura J. (1994). On the coexistence of specialists and generalists. Am. Nat. 144, 692–707. doi: 10.1086/285702
Xia X. (2018). DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552. doi: 10.1093/molbev/msy073
Xia X., Xie Z., Salemi M., Chen L., Wang Y. (2003). An index of substitution saturation and its application. Mol. Phylogenet. Evol. 26 (1), 1–7. doi: 10.1016/s1055-7903(02)00326-3
Keywords: niche, niche conservatism, specialist, generalist, spatial and temporal adaptations
Citation: Yang T, Yang S, Yu D, Wang C, Shang K and Liu H (2023) Interspecies trophic niche differences and spatial–temporal adaptations found in Cultrinae fishes. Front. Ecol. Evol. 11:1185413. doi: 10.3389/fevo.2023.1185413
Received: 13 March 2023; Accepted: 06 July 2023;
Published: 27 July 2023.
Edited by:
Hung-Du Lin, National Tainan First Senior High School, TaiwanReviewed by:
Cristián E. Hernández, University of Concepcion, ChileJunjie Wang, South China Normal University, China
Copyright © 2023 Yang, Yang, Yu, Wang, Shang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanzhang Liu, hzliu@ihb.ac.cn
 Tingyue Yang
Tingyue Yang Shufan Yang1,2
Shufan Yang1,2 Chunling Wang
Chunling Wang Kunyu Shang
Kunyu Shang Huanzhang Liu
Huanzhang Liu