- 1College of Nuclear Technology and Automation Engineering, Chengdu University of Technology, Chengdu, China
- 2Applied Nuclear Techniques in Geosciences Key Laboratory of Sichuan, Chengdu University of Technology, Chengdu, China
- 3College of Life and Environmental Sciences, Minzu University of China, Beijing, China
Studies investigating the diversity and structure of soil microbial systems in response to ionizing radiation are scarce. In particular, effects of long-term low-dose radiation is rarely studied because of its unique conditions. In this study, an area in Chengdu, China, which has been irradiated by the radionuclide thorium-232 for more than 10 years was investigated. Four groups of samples with absorbed dose rates ranging from 192.906 ± 5.05 to 910.964 ± 41.09 nGy/h were collected to analyze the compositional and functional changes of the soil microbial systems in the region. The diversity and structure of the soil microbial systems were determined using high-throughput sequencing. Our results showed that long-term low-dose ionizing radiation had no significant effect on soil bacterial diversity, but had a great impact on fungal diversity. Long-term ionizing radiation strongly affected soil microbial community structure. Long-term low-dose ionizing radiation was shown to have a promoting effect on iron-oxidizing bacteria and ectomycorrhizal fungi and have an inhibiting effect on predatory or parasitic fungi, further affecting the soil C/N ratio. This study is of great reference significance for future research on the impact of long-term low-dose ionizing radiation on soil ecosystems.
1. Introduction
Ecological environment pollution has become a hot topic in recent years. Especially the environmental ionizing radiation problem that affects human life and health, such as lung cancer induced by radon emanation (Mariona et al., 2022), increasing the risk of lens opacity (Gao et al., 2021) and accelerating vascular aging (Su et al., 2016). With the development of science and technology, many projects with large-scale radiation effects have been established, and the surrounding ecosystems have been continuously affected by ionizing radiation. Certain doses of ionizing radiation result in pollution and damage to the environment, thereby affecting the ecological cycle, including soil ecosystems (Andrei et al., 2014). However, most studies on the effects of nuclear radiation on soil microbial communities have focused on long-term exposure to high doses. One study systematically studied the community distribution and composition of fungi in the area around the Chernobyl nuclear power plant leakage and found that the area was rich in fungal diversity (Zhdanova et al., 2000). Distribution characteristics, namely composition and abundance, of the fungal community were significantly influenced by the radiation dose. The proportion of melanized fungi is larger in soil under long-term low-level ionizing radiation (Dighton et al., 2008). Most fungi exhibit amazing adaptability to radiation (Dadachova and Casadeval, 2008). Other studies have shown that radiation is a selective pressure that results in certain bacteria surviving in specific environments (Hoyos-Hernandez et al., 2019). Nevertheless, large-scale nuclear accidents involving short-term high-dose radiation are rare.
Short-term low-dose radiation generated by radioactive substances result in microorganisms reducing radiation damage by rapidly repairing damaged DNA fragments, forming spores to slow down their metabolic rate, generating superoxide dismutase to remove free radicals, and changing the carbon source utilized. These changes are made to adapt to the living conditions caused by nuclear radiation (Yang et al., 2021). Microorganisms develop resistance to radiation through adaptive reactions and can tolerate higher intensities of ionizing radiation over time (Fornalski et al., 2022). However, short-term and high-dose radiation has a sustained and irreversible impact on the composition, distribution, and metabolic pathways of microbial communities (Mohammad et al., 2019). The composition and functional integrity of biofilms are affected as the radiation causes single-strand or double-strand DNA breakages, denaturation of related proteins, and the reduction or inactivation of some enzymes (Romano et al., 2022). Few studies have investigated the effects of low-dose radiation on microorganisms. One study found that the main damage caused by irradiation of Salmonella typhimurium, Escherichia coli, and Listeria monocytogenes was intracellular enzyme inactivation and DNA damage (Cho and Ha, 2019). Another study showed that low-dose radiation increases the diversity of soil microbial communities and changes the metabolic capacity of carbon and nitrogen sources (Senatore et al., 2018). However, it still remains unclear that whether the composition and function of soil microbial systems are significantly affected in long-term low-dose radiation environment.
The exploitation of radioactive minerals, construction of fission reactor nuclear power plants, and research and development of nuclear fusion have all led to long-term radioactivity in the soil of some regions, causing the microorganisms in the soil to endure long-term radiation effects. Microorganisms play an important role in soil ecosystems (Zhang and Li, 2020). Therefore, if long-term low-dose radiation can significantly affect the composition and function of soil microbial systems, it will affect the material cycling processes of the entire ecosystem. To determine whether there is a quantifiable relationship between long-term low-dose ionizing radiation and soil microorganisms, this study analyzed the composition and functional changes of soil microbial systems in an area that has been irradiated by radionuclide thorium−232 for more than 10 years. The results may provide a new reference and basis for solving problems arising from long-term low-dose radiation of soil microbial systems.
2. Materials and methods
2.1. Experimental set-up and sampling
Samples were collected from a grassland in the Chengdu University of Technology campus in Chengdu, China (30°40′29″ North and 108°8′11″ East). A block (75 × 75 × 75 cm cube) of thorium mineral, an isotope of radioactive nuclide thorium−232, was present on the sample site, exposing the surrounding soil to radiation for more than 10 years. The mineral is a technical mineral, and it is exposed in the soil surface. The radioactivity of the thorium mineral is about 2.93 × 106 Bq. The thorium ore is placed in a relatively closed and independent ecological environment, and the Tradescantia fluminensis grows around the ore. After years of generational alternation, the Tradescantia fluminensis is the is the absolute constructive species. The soil type in the sampling location is the dark brown soil. No significant differences are found in other environmental factors such as soil moisture, pH (about 7.5), light and other environmental factors in the plots with different radiation intensity samples. Owing to the rapid propagation of microorganisms in soil, soil microorganisms in this region have formed stable communities that have adapted to the nuclear radiation present.
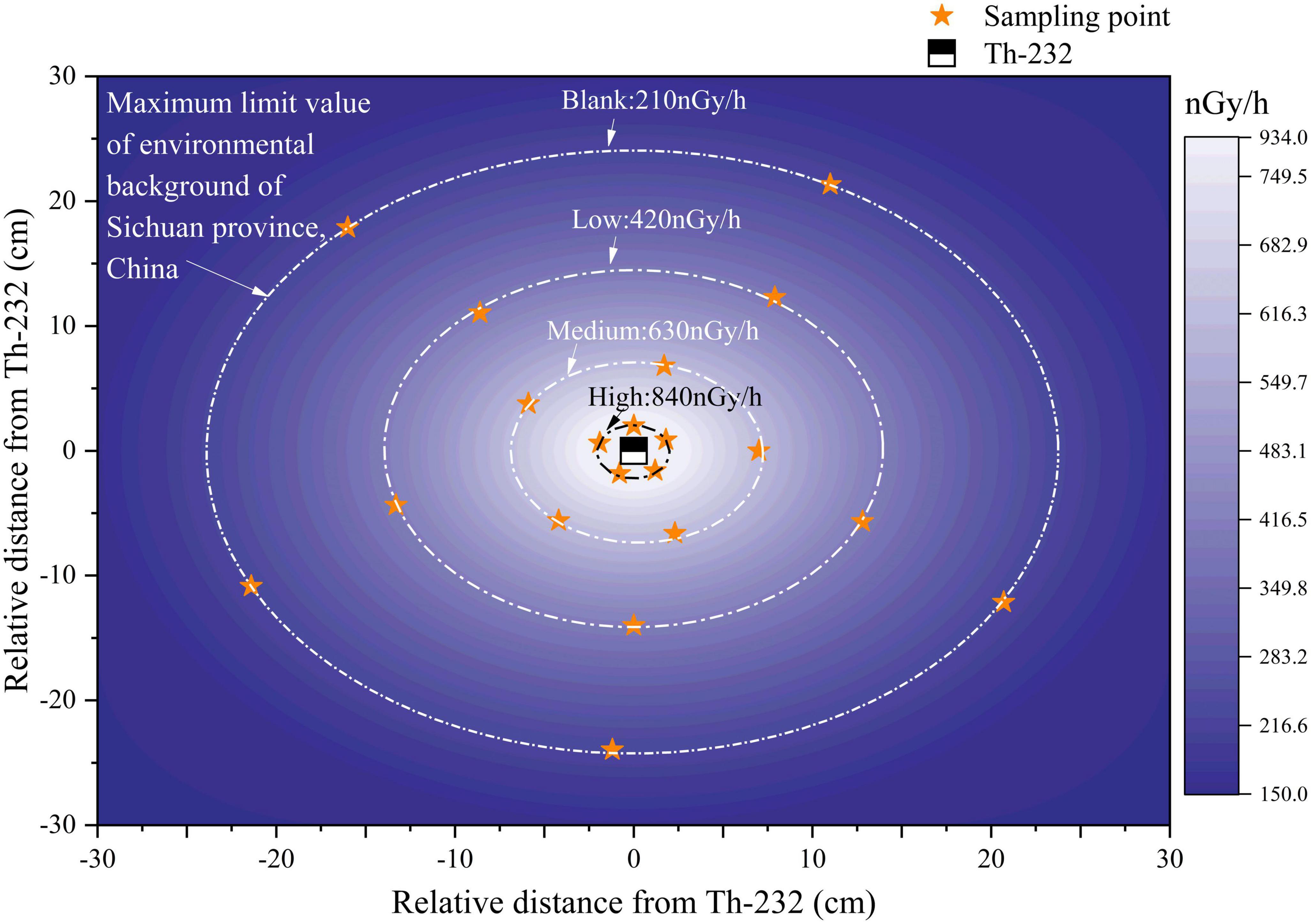
A plane coordinate system was established with the thorium mineral as the coordinate origin. A HPGe portable gamma-ray spectrometer trans-SPEC-DX-100T (AMETEK, San Diego, CA, United states) was used to measure the absorbed dose rate of thorium minerals at different distances. The relative measurement efficiency of this spectrometer was 40%, with the energy resolution at 122 and 233 keV being 1.6 and 2.3 keV, respectively. Sampling around the radioactive source is divided into 3 gradients based on the decay of the absorbed dose distance (2 ± 0.5, 7 ± 0.5, and 14 ± 0.5 cm). Their corresponding values were approximately four, three, and two times the maximum background-environmental radiation limit in Sichuan Province, and these will be referred to as High, Medium, and Low group, respectively (Figure 1). The highest absorbed dose rate was 910.964 ± 41.09 nGy/h (source) and the lowest was 192.906 ± 5.05 nGy/h (at 25 ± 0.5 cm away from the source). In addition, samples were collected at the maximum background-environmental radiation limit, referred to as the Blank group (Figure 1).
In soil sampling, each radiation dose gradient was used as the index, around the source of radiation and the soil core was obtained by 5-point sampling method and 20 samples in total are collected. 0−20 cm topsoil was excavated with a bamboo shovel. After removing stones, plant roots and other sundry materials, the soil was mixed, bagged, sealed and labeled, and stored in a 4°C ice box at low temperature, transported to the laboratory and stored in −20°C prior to analyses.
2.2. DNA extraction and PCR amplification
Soil microbiome DNA extraction was performed using the PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, United states) following the manufacturer’s instructions. Each sample was weighed 0.25 g (fresh weight) soil sample for extraction. The quality and concentration of the DNA extracted was detected by 1% agarose gel electrophoresis and spectrophotometry. The mean of DNA concentration was 212.5 ng/μl with standard deviation of 42.33. Samples were stored at −20°C for subsequent experiments.
The bacterial and fungal species were identified by PCR amplification and sequencing. The v3-v4 region of the bacterial 16 S rRNA gene was amplified using the primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). The ITS1 region of the fungal ITS gene was amplified using the primers ITS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-TGCGTTCTTCATCGATGC-3′). Barcode sequences 8 bp in length were added to the 5′ ends of the upstream and downstream primers to differentiate the different samples. The PCR reaction mixtures were comprised of: 12.5 μL 2xTaq Plus Master Mix, 3 μL BSA (2 ng/μL), 1 μL forward primer (5 μM), 1 μL reverse primer (5 μM), 2 μL DNA (30 ng), and 5.5 μL ddH2O to equal a final volume of 25 μL. The thermocycling conditions of the 16 S rRNA reaction were as follows: pre-denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 60 s; and final extension at 72°C for 7 min. The thermocycling conditions of the ITS reaction were as follows: pre-denaturation at 94°C for 5 min; 34 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s; and final extension at 72°C for 7 min. The PCR products were visualized by 1% agarose gel electrophoresis and purified using an Agencourt AMPure XP nucleic acid purification kit (Beckman Coulter, Brea, CA, United States) following the manufacturer’s instructions.
2.3. Sequencing and data analysis
The PCR products were used to construct a microbial diversity sequencing library, and the Illumina MiSeq PE300 high-throughput sequencing platform (Illumina, Santiago, CA, United States) was used for paired-end sequencing. The original sequences were uploaded to the Sequence Read Archive database of the National Center for Biotechnology Information. The raw data were split using the QIIME 2™ software (Bolyen et al., 2019) according to the barcode sequences, and the data were filtered and spliced using PEAR (v1.1) software. Sequences with scores lower than 20 and those containing non-standard bases and primer mismatch sequences were removed. The minimum overlap when splicing was set to 10 bp and the mismatch rate to 0.1. After splicing, sequences shorter than 230 bp were removed using VSEARCH (v2.8.1) (Edgar, 2010), and chimera sequences were removed using the UCHIME method according to the Gold Database downloaded from http://drive5.com/uchime/uchime_download.html. The VSEARCH (v2.8.1) software program was used to cluster high-quality sequences with operational taxonomic units (OTUs), with the sequence similarity threshold at 97% (Edgar, 2013). This was compared with the RDP classifier algorithm, which Silva and Unite databases use, set at a 70% confidence threshold and used to obtain the species classification data corresponding to each OTU. R (v4.0.2) software was used for data analysis of the species annotations and relative abundance results.
Venn diagrams were drawn using the R package, Venn Diagram (Fouts et al., 2012). The QIIME 2™ software was used for α diversity index analysis (Bolyen et al., 2019). The β diversity distance matrix was calculated using QIIME 2™ and cluster analysis was performed using the UPGMA method within R (v4.0.2), based on the unweighted UniFrac distance. A principal component analysis (PCA) which was previously described was used (Wang et al., 2012). Histogram analysis of species composition was performed, and a correlation heat map was drawn using the R packages psych and pheatmap. Functional guilds of bacteria and fungi were divided using FAPROTAX and FungalTraits (Louca et al., 2016; Põlme et al., 2020). Structural equation models (SEM) were constructed using the R-packages lavaan and piecewiseSEM to investigate the relationships between nuclear radiation, C and N nutrition, microbial groups, and diversity (Lefcheck and Freckleton, 2016). The goodness of fit of the model was evaluated using Fisher’s C statistic method (Bowd et al., 2021).
3. Results
3.1. Soil microbial diversity
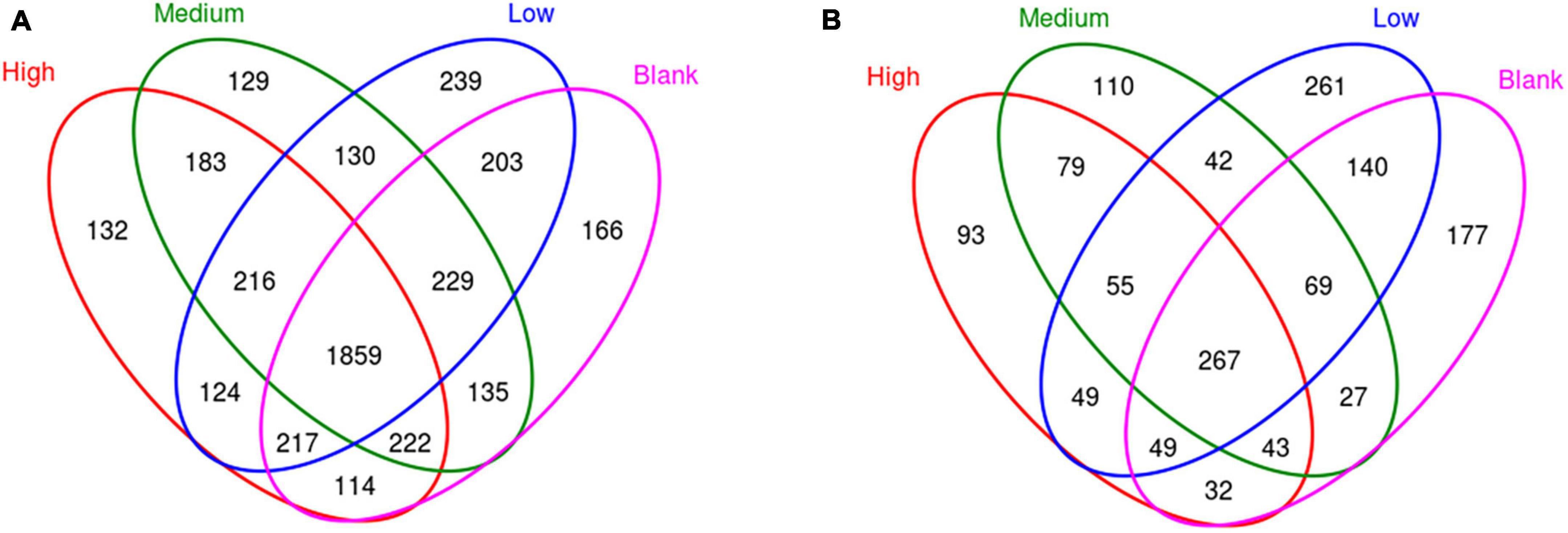
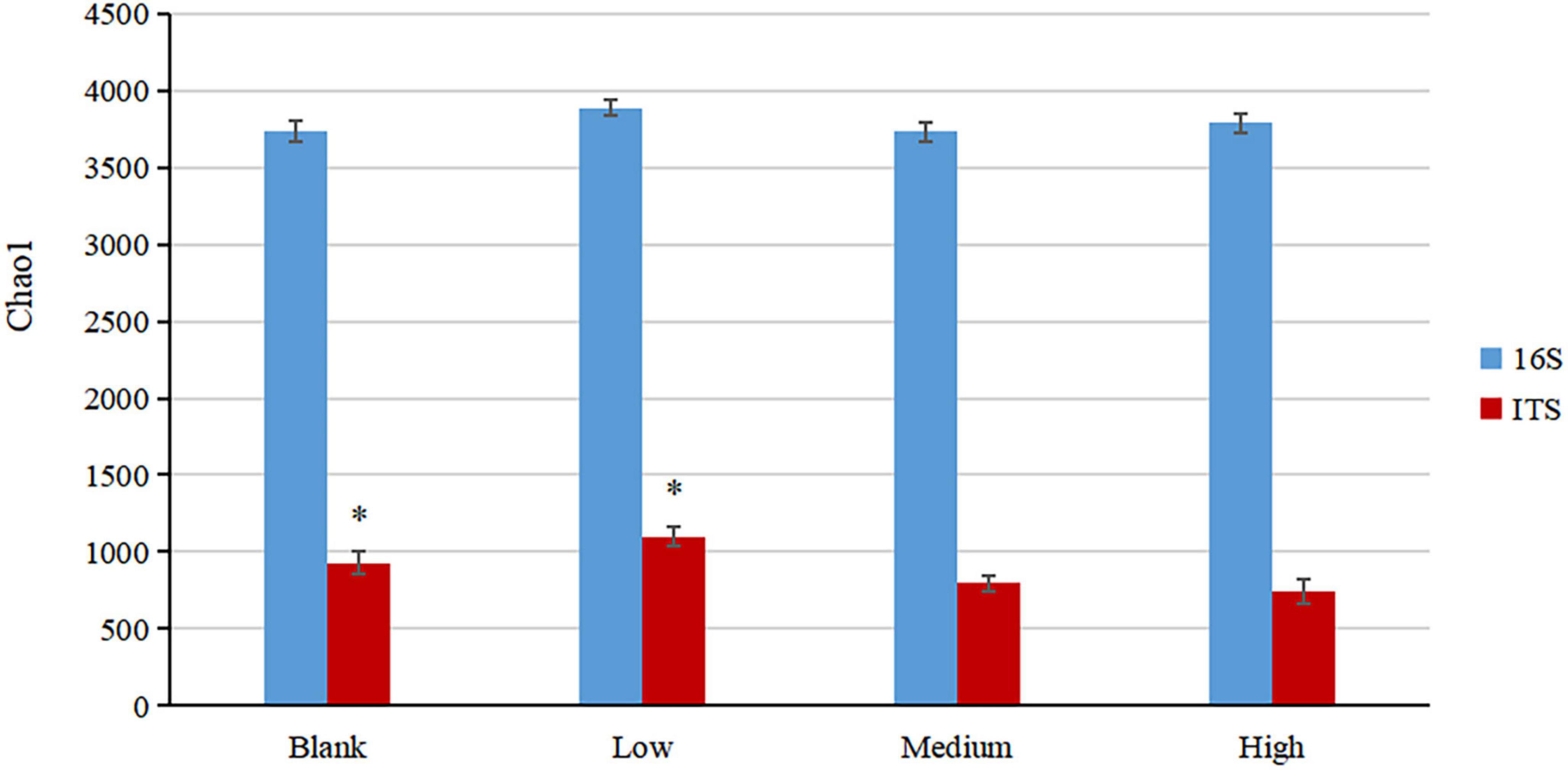
A total of 4,298 bacterial and 1,493 fungal OTUs were identified from the soil samples, of which 1,859 and 267 OTUs were common in each group (Figure 2). The numbers of bacterial OTUs specific to the High, Medium, Low, and Blank groups were 132, 129, 239, and 166, respectively (Figure 2 and Supplementary Table 1), and the respective number of fungal OTUs was 93, 110, 261, 177 (Figure 2 and Supplementary Table 2). There was no significant difference in the bacterial α diversity among the groups, but the fungal α diversity in the Low and Blank groups was significantly higher than that in the High and Medium (Figure 3).
The PCA analysis showed that the bacterial β diversity in each group differed. However, the fungi from the High and Medium groups clustered together whereas those from the Low and Blank groups were dispersed (Figure 4). The unweighted UniFrac distance of the bacterial community in the Medium clustered separately from that of the bacterial community in other groups, whereas the fungal communities in the High and Medium groups clustered together, and Low and Blank groups clustered together (Figure 5).
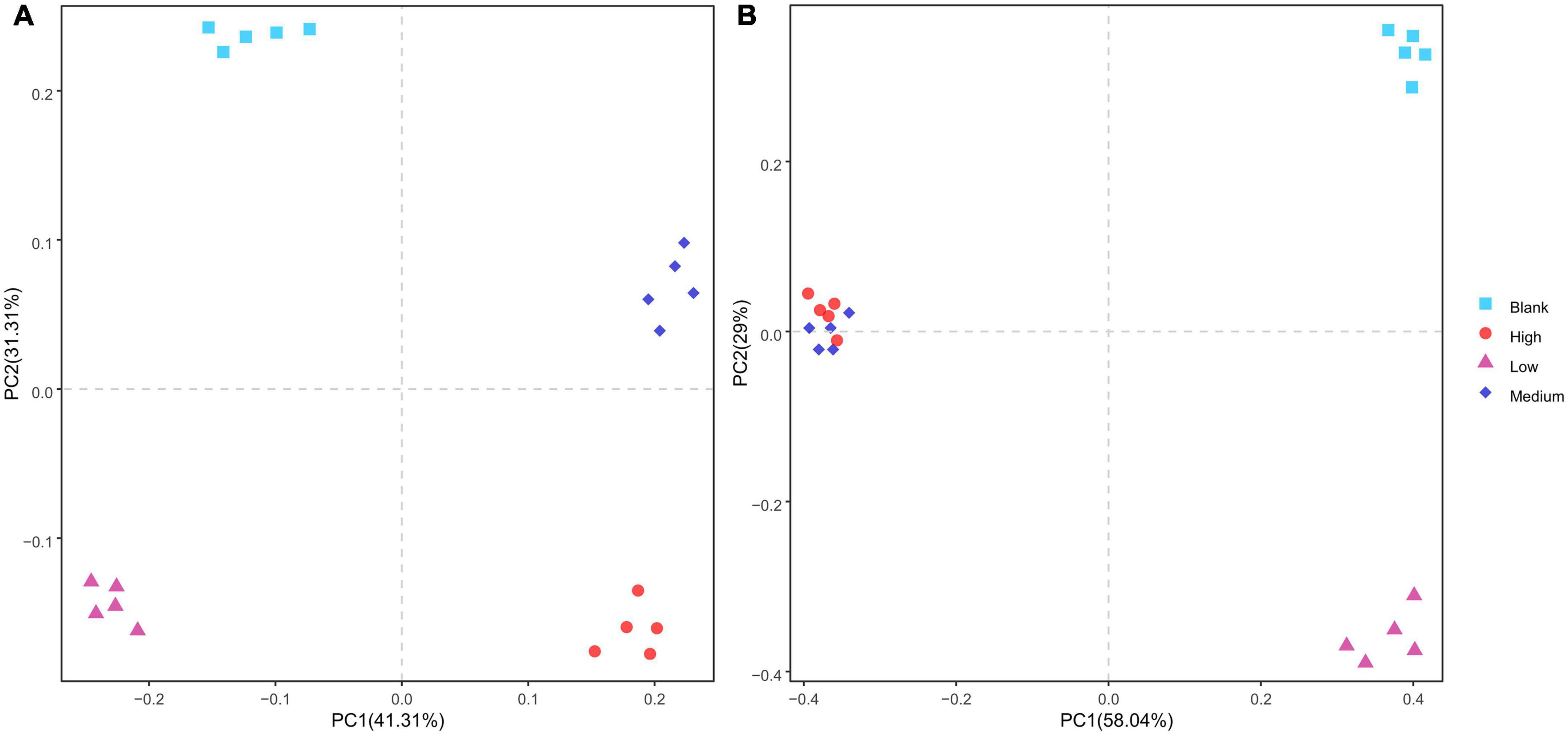
Figure 4. Principal components analysis of the β diversity of panel (A) bacteria and (B) fungi under different radiation intensities.
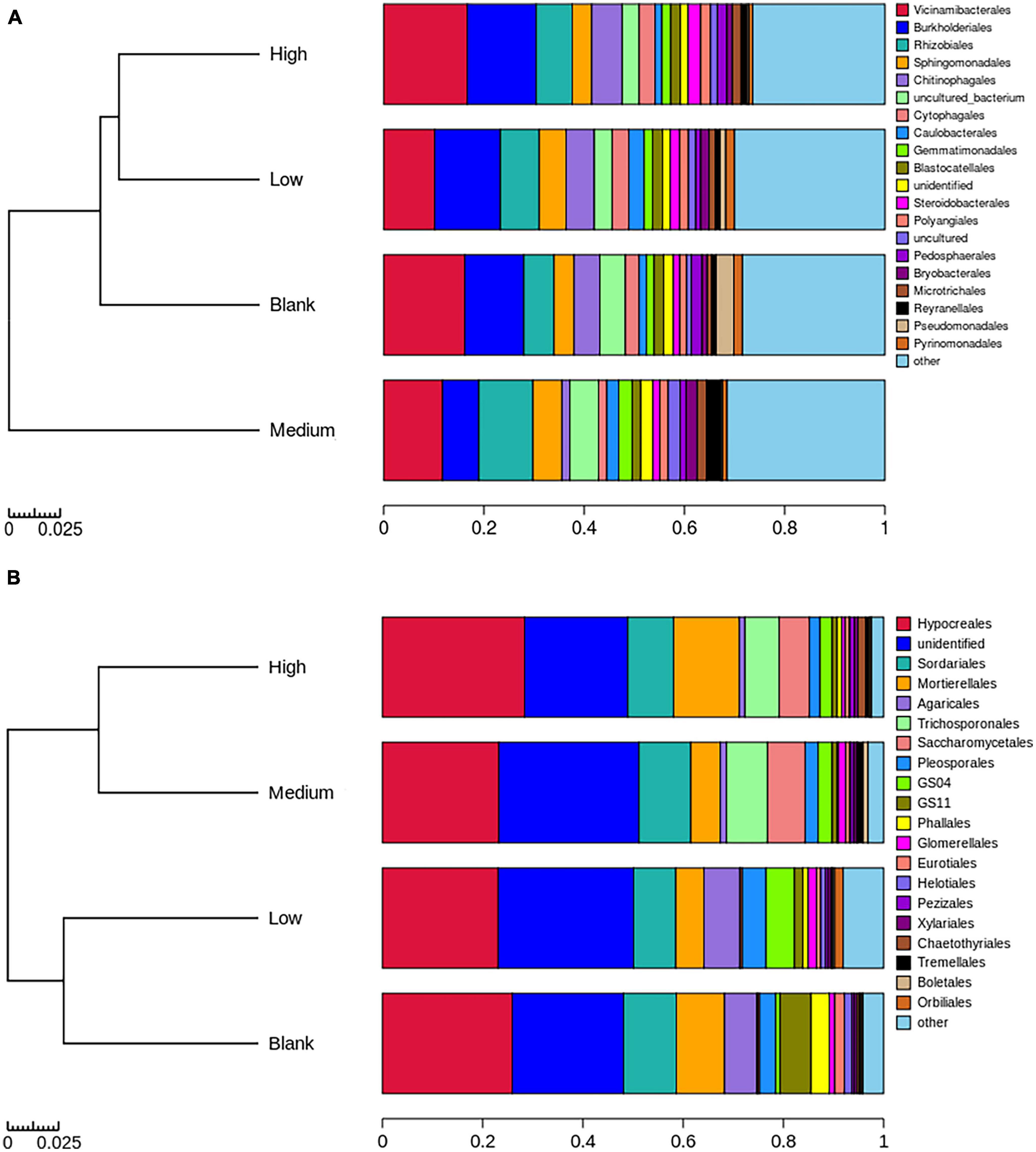
Figure 5. Relative species abundance and unweighted UniFrac distance of panel (A) bacterial and (B) fungal orders under different radiation intensities.
3.2. Composition structure of soil microorganisms
Vicinamibacterales, Rhizobiales, and Burkholderiales were the dominant bacterial orders present in the High, Medium, Low, and Blank groups (Figure 5). The dominant fungal order present in the High, Medium, Low, and Blank groups was Hypocreales (Figure 5). The dominant phyla of endemic bacteria in the High, Medium, Low, and Blank groups were Proteobacteria; Proteobacteria and Bdellovibrionota; Bacteroidota and Proteobacteria; and Verrucomicrobiota and Patescibacteria, respectively (Supplementary Table 1). The dominant phyla of endemic fungi in the High, Medium, Low, and Blank groups were Basidiomycota; Chytridiomycota, Basidiomycota, and Ascomycota; Ascomycota; and Ascomycota, respectively (Supplementary Table 2).
Among the highly abundant bacterial orders present, the relative abundance of Polyangiales was significantly positively correlated with radiation intensity whereas that of Pseudomonadales was significantly negatively correlated with radiation intensity (Figure 6). Among the highly abundant fungal orders present, the relative abundance of Pezizales was significantly positively correlated with radiation intensity and carbon content whereas that of Helotiales was significantly negatively correlated with radiation intensity and C/N ratio (Figure 7). In bacterial functional groups, the relative abundance of iron-oxidizing bacteria was positively correlated with radiation intensity and C/N ratio whereas that of predatory or exoparasitic bacteria was negatively correlated with radiation intensity (Figure 8). In fungal functional groups, the relative abundance of ectomycorrhizal fungi was significantly positively correlated with radiation intensity, carbon content, and C/N ratio (Figure 9). The SEM showed that iron-oxidizing bacteria, predatory or exoparasitic bacteria, and ectomycorrhizal fungi had negative effects on fungal α diversity and microbial community β diversity, but the C/N ratio had positive effects on fungal α diversity and microbial β diversity. The SEM explained 52% of fungal α diversity and 41% of microorganisms β diversity (Figure 10).
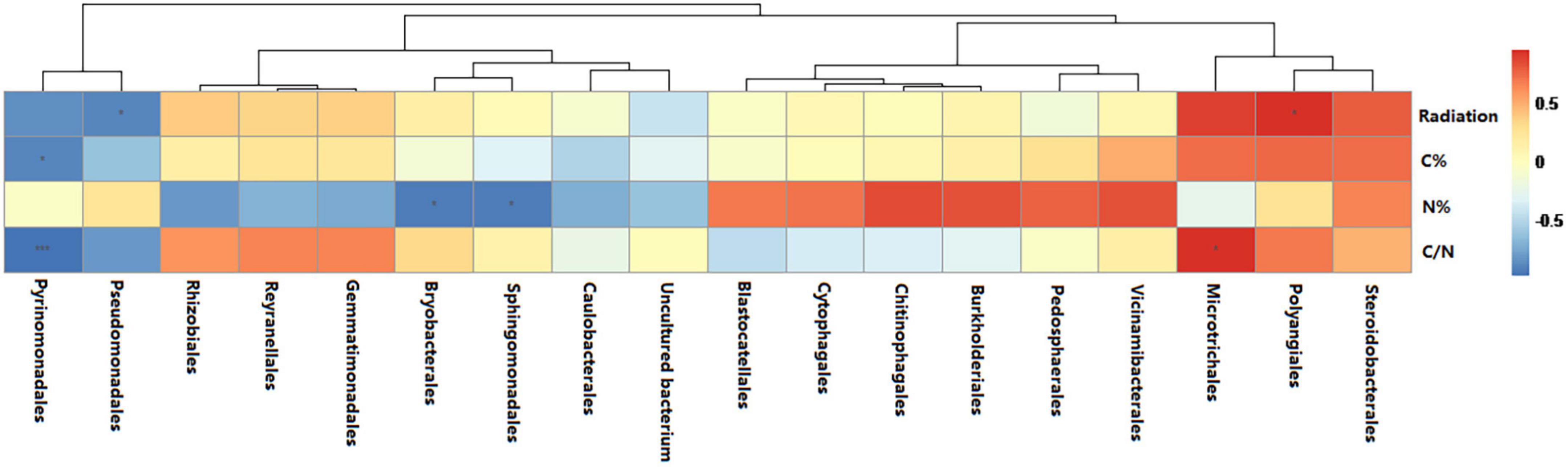
Figure 6. Relationships between the dominant species (at order level) that exhibited higher relative abundance and the radiation intensity, carbon content, nitrogen content, and C/N ratio. The lines represent the relationships from cluster analysis of species/OTUs. Color depth represents the level of correlation. Significance: * < 0.05; *** < 0.001. Distance algorithm: Bray-Curtis, clustering method: Pearson.
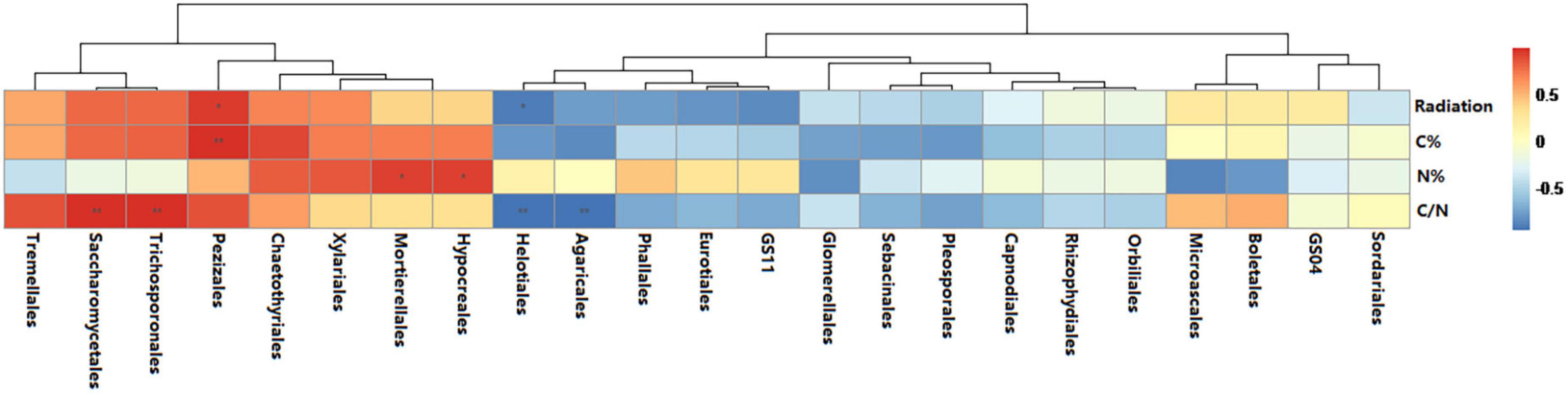
Figure 7. Relationships between the dominant species (at order level) that exhibited higher relative abundance and the radiation intensity, carbon content, nitrogen content, and C/N ratio. The lines represent the relationships from cluster analysis of species/OTUs. Color depth represents the level of correlation. Significance: * < 0.05; ** < 0.01. Distance algorithm: Bray-Curtis, clustering method: Pearson.
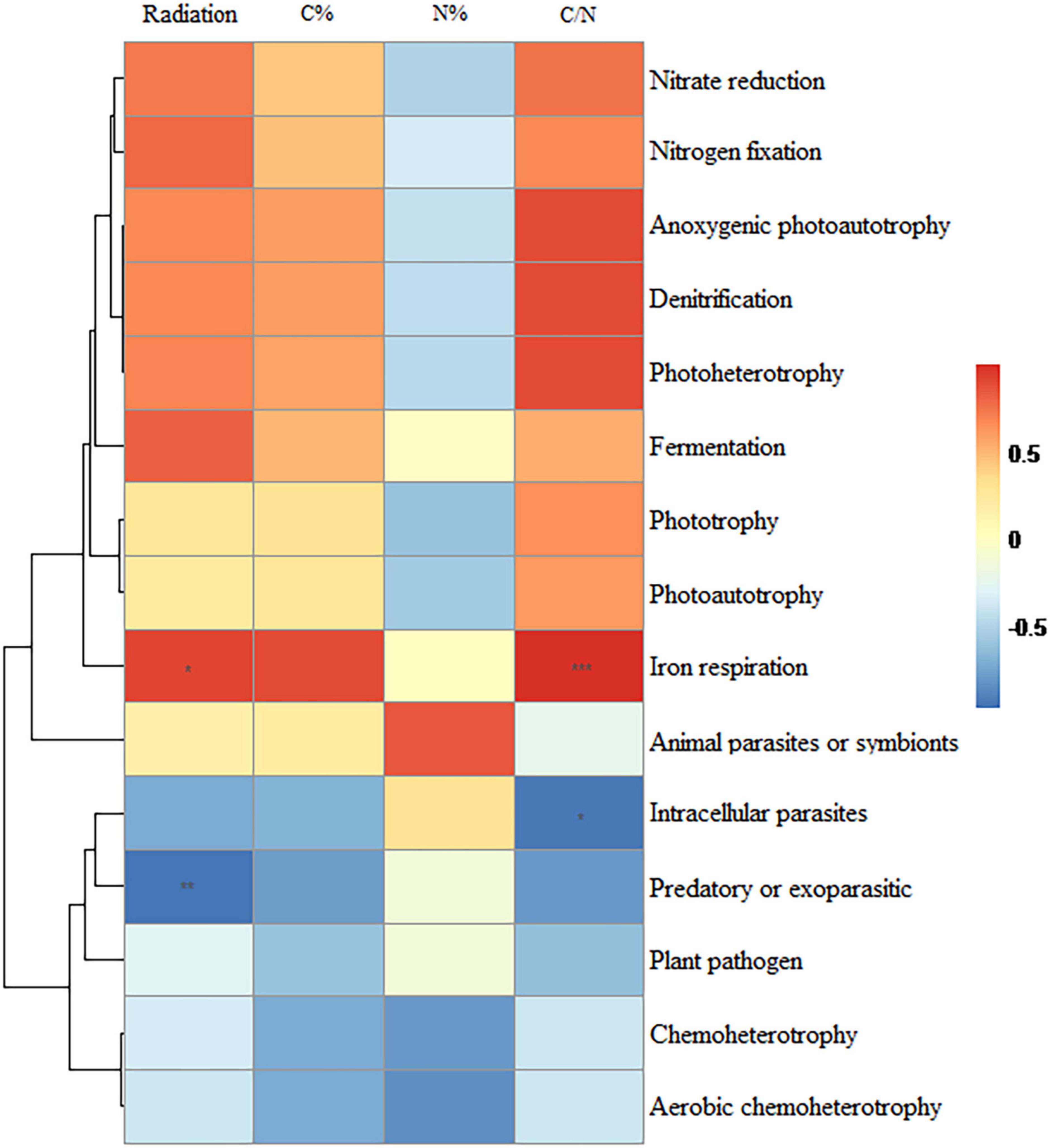
Figure 8. Relationships between bacterial functional taxa and radiation intensity, carbon content, nitrogen content, and C/N ratio. The lines represent the relationships from cluster analysis of species/OTUs. Color depth represents the level of correlation. Significance: * < 0.05; ** < 0.01; *** < 0.001. Distance algorithm: Bray-Curtis, clustering method: Pearson.
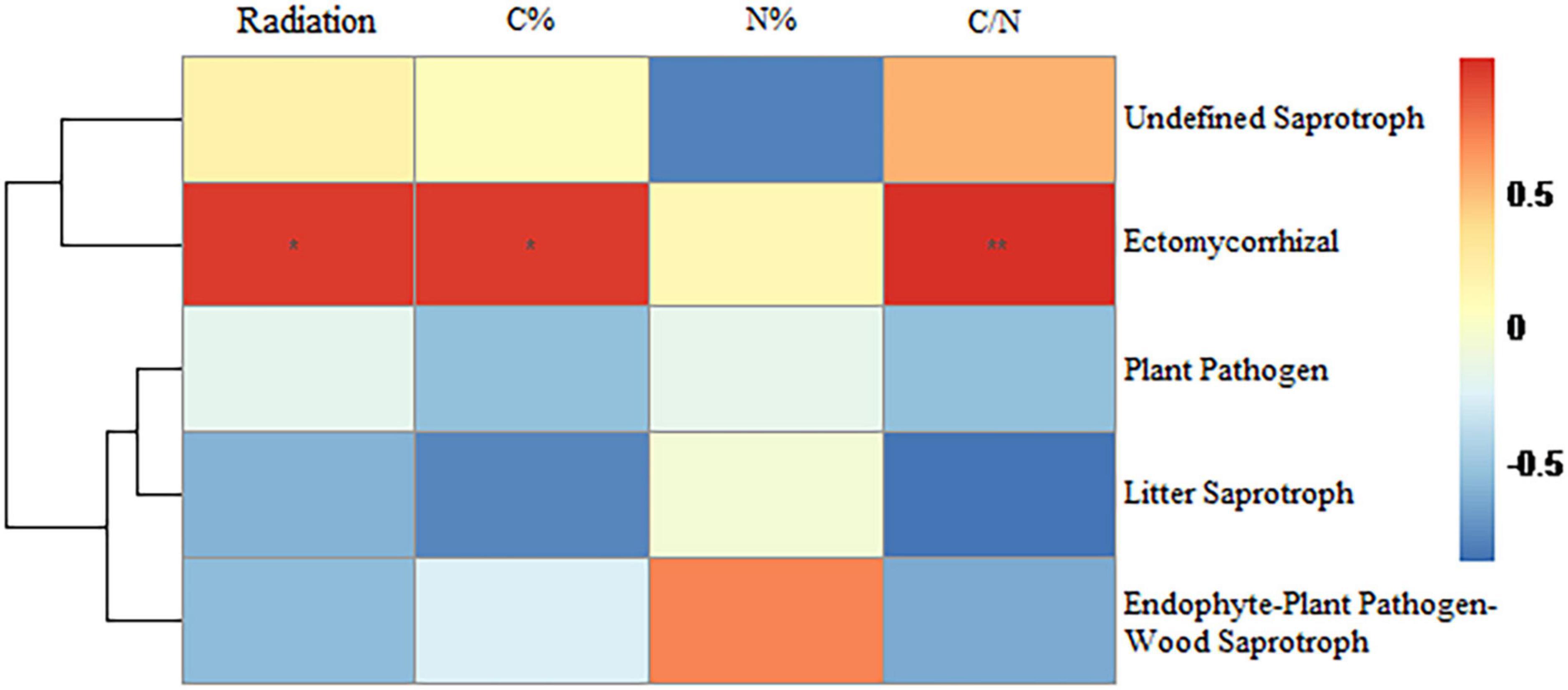
Figure 9. Relationships between bacterial functional taxa and radiation intensity, carbon content, nitrogen content, and C/N ratio. The lines represent the relationships from cluster analysis of species/OTUs. Color depth represents the level of correlation. Significance: * < 0.05; ** < 0.01. Distance algorithm: Bray-Curtis, clustering method: Pearson.
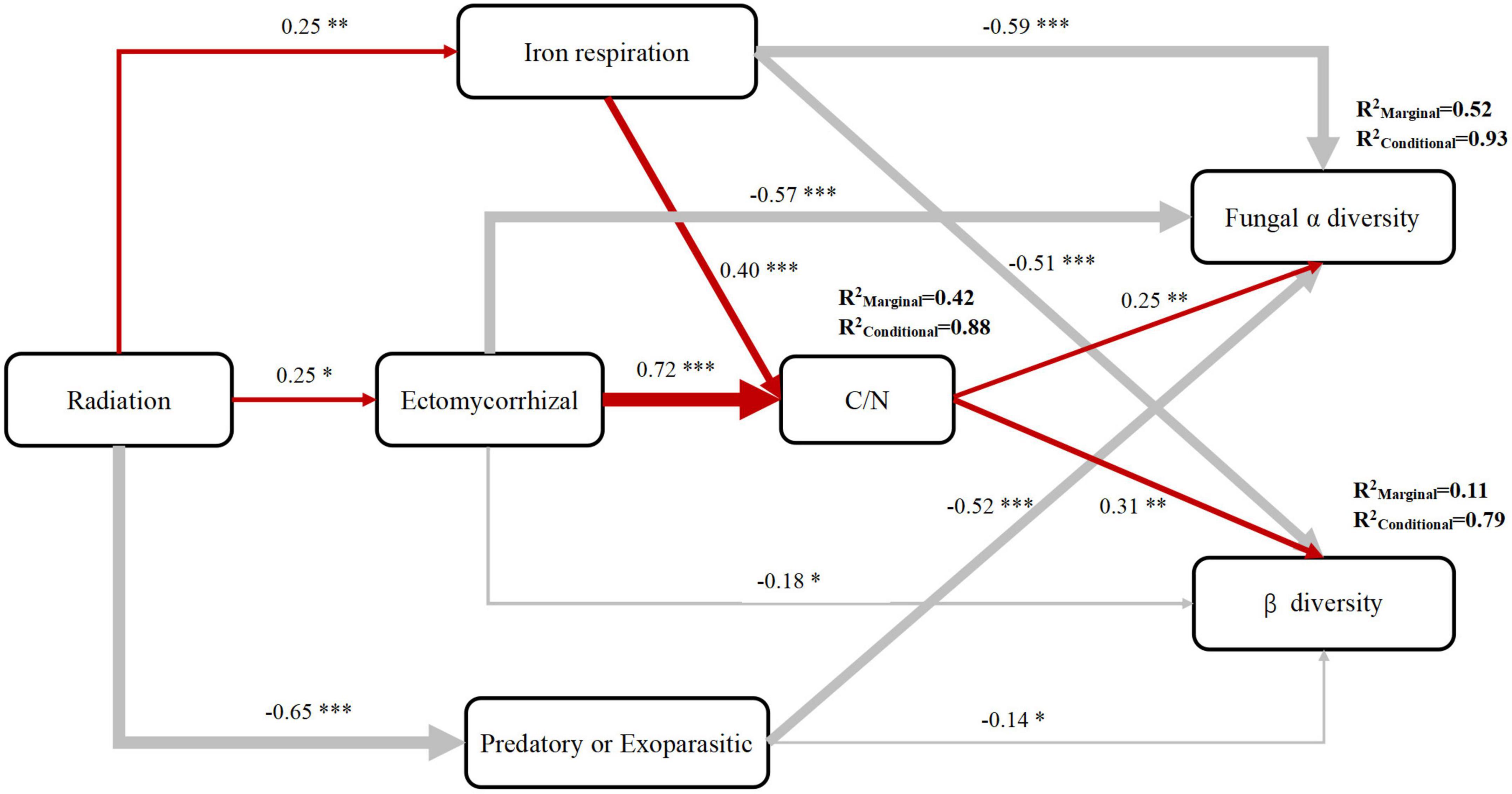
Figure 10. Structural equation model explaining the effect of gamma radiation on the soil microbial community (Fisher’s C = 27.23, P = 0.257, DF = 20, AIC = 73.230). Red and gray arrows indicate positive and negative relationships, respectively. Numbers next to the arrows indicate the normalized path coefficients. *Represents its significance: * < 0.05; ** < 0.01; *** < 0.001. The SEM explained 56% of C/N ratio, 52% of fungi α diversity, and 41% of microorganisms β diversity.
4. Discussion
4.1. Effect of gamma radiation on soil microbial diversity and structure
There was no significant difference in bacterial α diversity among the groups; however, the fungal α diversity in the Low and Blank groups was significantly higher than that in the High and Medium groups. The PCA analysis revealed that fungi from the High and Medium groups clustered together, away from the Low and Blank groups, indicating that the threshold value was reached when the radiation dose was approximately 480 nGy/h, the effect of gamma radiation on fungal diversity was greater than that on bacterial diversity. Under natural conditions, fungal communities are relatively less efficient in recovering from harsh environmental conditions than bacterial communities (Gao et al., 2022). Therefore, it is difficult to recover species lost in environments due to strong radiation (Franciska et al., 2018; Song et al., 2020; Coban et al., 2022). This results in a significant decrease in fungal α diversity in the High and Medium groups and the significant changes in community structure of the Low and Blank groups.
Although there was no significant difference in the α diversity of bacteria in each group, PCA and unweighted UniFrac analysis showed that gamma radiation had a significant effect on bacterial community structure. The regulatory capacity of bacterial communities is usually stronger than that of fungal communities (Liu et al., 2021). Therefore, under radiation conditions, although some bacteria are eliminated because of their inadaptability to the environment, new bacterial groups maintain the stability of the whole community’s diversity, making the bacterial community structure of each group different; however, the biodiversity is not significantly different (Wang et al., 2016). In addition, owing to the existence of a moderate interference effect, the unweighted UniFrac distance of the fungal community in the medium group differs significantly from that of the other groups (Ma and Geng, 2012; Zhang et al., 2018).
4.2. Effect of gamma radiation on soil microbial community
The radiation intensity in this study was not sufficient to change the dominant soil microbial groups, but it was sufficient to change the community composition of these groups. Compared with the control group, the microbial groups in the high, middle and low groups all died out, in the mean time other microbial groups also filled the niche. Proteobacteria, which exhibit strong adaptability, was the predominant group which filled the niche vacated by the extinction of microorganisms not adapted to radiation environment; therefore, the bacterial community remained stable and its diversity was not changed (Souza and Procópio, 2021). The main phyla of endemic fungi in the irradiated areas, namely Basidiomycota, was shown to gradually replace Ascomycota in these areas. Ionizing radiation also inhibited the pathogenic and parasitic bacteria, namely Pseudomonadales and Helotiales bacteria, which indirectly promoted the growth of other fungi such as Polyangiales and Pezizales. This promoted the stability of ecosystem productivity and affected the change in soil nutrients, especially the C/N ratio (Ning et al., 2021). The change in the C/N ratio further affects the biodiversity of the microbial community by the adaptability of microorganisms to soil nutrient changes (Zhou et al., 2017).
It is worth noting that radiation intensity was positively correlated with the relative abundance of iron-oxidizing bacteria, which has not been previously reported. Iron-oxidizing bacteria are involved with the circulation of nutrient elements, such as nitrogen and phosphorus, in the soil. These bacteria utilize large amounts of nitrogen and phosphorus and reduce the loss of organic carbon, thereby increasing the C/N ratio (Weber et al., 2006; Giesler et al., 2012; Cai et al., 2022). Studies have shown that iron-oxidizing bacteria can promote pollutant degradation, protect steel from erosion, and improve ecological resistance (Herrera and Videla, 2009; Qin and Shi, 2017; Zhang et al., 2021). However, there is no clear answer as to whether iron-oxidizing bacteria can improve the resistance of ecosystems to ionizing radiation. This issue needs to be addressed in future research.
In addition, the relative abundance of ectomycorrhizal fungi was significantly positively correlated with radiation intensity, which may be because the hyphae of ectomycorrhizal fungi wrap around the root cells to form a biomembrane. This biofilm not only protects plants from pathogens and chemical pollutants (Martino and Perotto, 2010; Clasen et al., 2018) but also from ionizing radiation. Ectomycorrhizal fungi may function as anti-radiation agents. Plants in high-radiation environments are more inclined to choose ectomycorrhizal fungi as partners, which increases the abundance of ectomycorrhizal fungi (mainly Basidiomycota) significantly. This was clearly observed in the Blank group where the abundance of ectomycorrhizal fungi was zero. Additionally, ectomycorrhizal fungi can help plants absorb soil nutrients, such as nitrogen and phosphorus (Liu et al., 2022), and are beneficial for carbon fixation (Zak et al., 2019). Thus, ectomycorrhizal fungi improve the nutritional status of plants damaged by radiation (Li et al., 2022), increase the soil C/N ratio, and change the soil microbial diversity and community composition (Zheng and Song, 2022). However, more research is needed to prove that ectomycorrhizal fungi have the function of resisting radiation.
5. Conclusion
Long-term ionizing radiation had no significant effect on soil bacterial diversity but had a significant effect on fungal community diversity. High and medium radiation significantly reduced the soil fungal diversity and significantly affected the fungal community structure. Long-term ionizing radiation had a significant promoting effect on iron-oxidizing bacteria and ectomycorrhizal fungi and a significant inhibitory effect on predatory and parasitic fungi. The relative abundances of iron-oxidizing bacteria and ectomycorrhizal fungi were positively correlated with the soil C/N ratio, and the increase in the soil C/N ratio promoted the α diversity of soil fungi and β diversity of microorganisms. The increase in the relative abundance of iron-oxidizing bacteria, ectomycorrhizal fungi, and predatory and parasitic bacteria had a negative effect on soil fungal α diversity and microbial β diversity. The SEM can explain the 56% of C/N ratio, 52% of fungal α diversity, and 41% of microorganisms β diversity. The results of this study thus provide direction for future research on the effects of long-term ionizing radiation on soil ecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA950186.
Author contributions
FL and WS provided the conceptualization and framed the methodology. XH wrote and prepared the original draft. XH and QQ did the review and editing. FC and ZJC provided the validation. All authors made significant contributions to this study and contributed to the article and approved the submitted version.
Funding
This research was supported by the Sichuan Science and Technology Program (Nos. 2020YJ0334 and 2021YJ0522).
Acknowledgments
We thank the assistance of Sichuan Provincial Key Lab of Applied Nuclear Techniques in Geosciences. We also thank the CDUT Team 203 for the English language review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1184582/full#supplementary-material
Supplementary Table 1 | Endemic 16 S OTUs and their relative abundance in each group.
Supplementary Table 2 | Endemic ITS OTUs and their relative abundance in each group.
References
Andrei, S. Z., Konstantin, B. G., Taizo, N., and Nobuhiro, K. (2014). Ionizing radiation effects on soil biota: application of lessons learned from chernobyl accident for radioecological monitoring. Pedobiologia 57, 5–14. doi: 10.1016/j.pedobi.2013.09.005
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bowd, E. J., Banks, S. C., Bissett, A., May, T. W., and Lindenmayer, D. B. (2021). Direct and indirect disturbance impacts in forests. Ecol. Lett. 24, 1225–1236. doi: 10.1111/ele.13741
Cai, Y., Ma, T., Wang, Y. Y., Jia, J., Jia, Y. F., Liang, C., et al. (2022). Assessing the accumulation efficiency of various microbial carbon components in soils of different minerals. Geoderma 407:115562. doi: 10.1016/j.geoderma.2021.115562
Cho, G., and Ha, J. (2019). Application of X-ray for inactivation of foodborne pathogens in ready-to-eat sliced ham and mechanism of the bactericidal action. Food Control 96, 343–350. doi: 10.1016/j.foodcont.2018.09.034
Clasen, B. E., Silveira, A. D., Baldoni, D. B., Montagner, D. F., Jacques, R. J., and Antoniolli, Z. I. (2018). Characterization of ectomycorrhizal species through molecular biology tools and morphotyping. Sci. Agr. 75, 246–254. doi: 10.1590/1678-992X-2016-0419
Coban, O., De, D. G. B., and Van, D. P. M. (2022). Soil microbiota as game-changers in restoration of degraded lands. Science 375:abe0725. doi: 10.1126/science.abe0725
Dadachova, E., and Casadeval, A. (2008). Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Curr. Opin. Microbiol. 11, 525–531. doi: 10.1016/j.mib.2008.09.013
Dighton, J., Tugay, T., and Zhdanova, N. (2008). Fungi and ionizing radiationfrom radionuclides. FEMS Microbiol. Lett. 281, 109–120. doi: 10.1111/j.1574-6968.2008.01076.x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fornalski, K. W., Adamowski, Ł, Dobrzyński, L., Jarmakiewicz, R., Powojska, A., and Reszczyńska, J. (2022). The radiation adaptive response and priming dose influence: the quantification of the raper-yonezawa effect and its three-parameter model for postradiation DNA lesions and mutations. Radiat. Environ. Bioph. 61, 221–239. doi: 10.1007/s00411-022-00963-9
Fouts, D. E., Szpakowski, S., Purushe, J., Torralba, M., Waterman, R. C., MacNeil, M. D., et al. (2012). Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One 7:e48289. doi: 10.1371/journal.pone.0048289
Franciska, T. D. V., Griffiths, R. I., Mark, B., Hayley, C., Mariangela, G., Hyum, S. G., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:3033. doi: 10.1038/s41467-018-05516-7
Gao, C., Xu, L., Montoya, L., Madera, M., Hollingsworth, J., Chen, L., et al. (2022). Co-occurrence networks reveal more complexity than community composition in resistance and resilience of microbial communities. Nat. Commun. 13:3867. doi: 10.1038/s41467-022-31343-y
Gao, Y., Su, Y. P., Li, X. L., Cui, S. Y., Zhang, S. F., Tan, G. X., et al. (2021). Investigation and analysis of lens opacity in residents of high background area in Yangjiang, China. China Occupat. Med. 48, 510–514. doi: 10.1093/jrr/rraa073
Giesler, R., Esberg, C., Lagerström, A., and Graae, B. J. (2012). Phosphorus availability and microbial respiration across different tundra vegetation types. Biogeochemistry 108, 429–445. doi: 10.1007/s10533-011-9609-8
Herrera, L. K., and Videla, H. A. (2009). Role of iron-reducing bacteria in corrosion and protection of carbon steel. Int. Biodeter. Biodegr. 63, 891–895. doi: 10.1016/j.ibiod.2009.06.003
Hoyos-Hernandez, C., Courbert, C., Simonucci, C., David, S., Vogel, T. M., and Larose, C. (2019). Community structure and functional genes in radionuclide contaminated soils in chernobyl and fukushima. Fems Microbiol. Lett. 366, 1–10. doi: 10.1093/femsle/fnz180
Lefcheck, J. S., and Freckleton, R. (2016). piecewiseSEM: piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol. Evol. 7, 573–579. doi: 10.1111/2041-210X.12512
Li, F., Deng, X. T., Chen, H. J., Lu, H., Guo, W. T., Song, W. C., et al. (2022). Long-term gamma radiation effect on functional traits of tradescantia flumnensis L. Pol. J. Environ. Stud. 31, 1153–1160. doi: 10.15244/pjoes/142144
Liu, S. G., García-Palacios, P., Tedersoo, L., Guirado, E., Heijden, M. G. A., Wagg, C., et al. (2022). Phylotype diversity within soil fungal functional groups drives ecosystem stability. Nat. Ecol. Evol. 6, 900–909. doi: 10.1038/s41559-022-01756-5
Liu, W. X., Liu, L. L., Yang, X., Deng, M. F., Wang, Z., Wang, P. D., et al. (2021). Long-term nitrogen input alters plant and soil bacterial, but not fungal beta diversity in a semiarid grassland. Global. Change Biol. 27, 3939–3950. doi: 10.1111/gcb.15681
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Ma, Z. S., and Geng, J. W. (2012). A bird’s eye view of microbial community dynamics, in: microbial ecology theory: current perspectives. Wymondham: Horizon Scientific Press.
Mariona, R., Marta, G. D. H., Benjamin, B., and Laura, M. (2022). Radon and lung cancer: current trends and future perspectives. Cancers 14:3142. doi: 10.3390/cancers14133142
Martino, E., and Perotto, S. (2010). Mineral transformations by mycorrhizal fungi. Geomicrobiol. J. 27, 609–623. doi: 10.1080/01490451003702958
Mohammad, A. A., Al-Najjar, A., and Albokari, M. M. (2019). Shifts in microbial community composition in tannery-contaminated soil in response to increased gamma radiation. Ann. Microbiol. 69, 1567–1577. doi: 10.1007/s13213-019-01541-z
Ning, Q., Chen, L., Zhang, C. Z., Ma, D. H., Li, D., Han, X. R., et al. (2021). Saprotrophic fungal communities in arable soils are strongly associated with soil fertility and stoichiometry. Appl. Soil Ecol. 159:103843. doi: 10.1016/j.apsoil.2020.103843
Põlme, S., Abarenkov, K., Henrik, N. R., Lindahl, B. D., Clemmensen, K. E., Kauserud, H., et al. (2020). FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Div. 105, 1–16. doi: 10.1007/s13225-021-00470-0
Qin, X., and Shi, L. (2017). Electrical interplay between microorganisms and iron-bearing minerals. Acta Chim. Sinica 75, 583–593. doi: 10.6023/A17010021
Romano, I., Camerlingo, C., Vaccari, L., Birarda, G., and Poil, A. (2022). Effects of ionizing radiation and long-term storage on hydrated vs. dried cell samples of extremophilic microorganisms. Microorganisms 10:190. doi: 10.3390/microorganisms10010190
Senatore, G., Mastroleo, F., Leys, N., and Mauriello, G. (2018). Effect of microgravity and space radiation on microbes. Future Microbiol. 13, 831–847. doi: 10.2217/fmb-2017-0251
Song, W. C., Tong, X. J., Liu, Y. H., and Li, W. K. (2020). Microbial community, newly sequestered soil organic carbon, and δ15N variations driven by tree roots. Front. Microbiol. 11:314. doi: 10.3389/fmicb.2020.00314
Souza, D. L. C., and Procópio, L. (2021). The adaptations of the microbial communities of the savanna soil over a period of wildfire, after the first rains, and during the rainy season. Environ. Sci. Pollut. R. 29, 14070–14082. doi: 10.1007/s11356-021-16731-z
Su, Y. P., Tan, G. X., Lei, S. J., Zou, J. M., Zhang, S. F., Liu, J. X., et al. (2016). Preliminary study on the relationship between low-dose radiation exposure and carotid intima thickness in yangjiang female residents with high background. Chin. J. Radiol. Med. Protect. 36, 682–687.
Wang, J. C., Xue, C., Song, Y., Wang, L., Huang, Q. W., and Shen, Q. R. (2016). Wheat and rice growth stages and fertilization regimes alter soil bacterial community structure, but not diversity. Front. Microbiol. 7:1207. doi: 10.3389/fmicb.2016.01207
Wang, Y., Sheng, H. F., He, Y., Wu, J. Y., Jiang, Y. X., Tam, N. F., et al. (2012). Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microb. 78, 8264–8271. doi: 10.1128/AEM.01821-12
Weber, K. A., Urrutia, M. M., Churchill, P. F., Kukkadapu, R. K., and Roden, E. E. (2006). Anaerobic redox cycling of iron by freshwater sediment microorganisms. Environ. Microbiol. 8, 100–113. doi: 10.1111/j.1462-2920.2005.00873.x
Yang, X. B., Song, G. Y., Liu, H., and Hu, D. W. (2021). Microbial diversity formation and maintenance due to temporal niche differentiation caused by low-dose ionizing radiation in oligotrophic environments. Life Sci. Space Res. 31, 92–100. doi: 10.1016/j.lssr.2021.08.003
Zak, D. R., Pellitier, P. T., Argiroff, W., Castillo, B., James, T. Y., Nave, L. E., et al. (2019). Exploring the role of ectomycorrhizal fungi in soil carbon dynamics. New Phytol. 223, 33–39. doi: 10.1111/nph.15679
Zhang, J., and Li, P. Z. (2020). Using soil microorganism to construct a new index of soil quality evaluation. IOP Confer. Seri. Earth Environ. Sci. 440:052023. doi: 10.1088/1755-1315/440/5/052023
Zhang, X. M., Johnston, E. R., Barberán, A., Ren, Y., Wang, Z. P., and Han, X. G. (2018). Effect of intermediate disturbance on soil microbial functional diversity depends on the amount of effective resources. Environ. Microbiol. 20, 3862–3875. doi: 10.1111/1462-2920.14407
Zhang, X. Q., Yuan, Z. G., and Hu, S. H. (2021). Anaerobic oxidation of methane mediated by microbial extracellular respiration. Env. Microbiol. Rep. 13, 790–804. doi: 10.1111/1758-2229.13008
Zhdanova, N. N., Zakharchenko, V. A., Vember, V. V., and Nakonechnaya, L. T. (2000). Fungi from chernobyl: mycobiota of the inner regions of the containment structures of the damaged nuclear reactor. Mycol. Res. 104, 1421–1426. doi: 10.1017/S0953756200002756
Zheng, L. M., and Song, W. C. (2022). Phosphorus limitation of trees influences forest soil fungal diversity in China. Forests 13:223. doi: 10.3390/f13020223
Keywords: long-term gamma radiation, low-dose gamma radiation, soil microorganisms, soil microbial ecosystem, ionizing radiation pollution
Citation: Cheng F, Huang X, Qin Q, Chen Z, Li F and Song W (2023) The effect of aboveground long-term low-dose ionizing radiation on soil microbial diversity and structure. Front. Ecol. Evol. 11:1184582. doi: 10.3389/fevo.2023.1184582
Received: 12 March 2023; Accepted: 04 April 2023;
Published: 18 April 2023.
Edited by:
Jie Gao, Peking University, ChinaReviewed by:
Jianming Wang, Beijing Forestry University, ChinaTimothy A. Mousseau, University of South Carolina, United States
Copyright © 2023 Cheng, Huang, Qin, Chen, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Li, bGlmZWkxN0BjZHV0LmVkdS5jbg==; Wenchen Song, c29uZ3dAbXVjLmVkdS5jbg==
 Feng Cheng1,2
Feng Cheng1,2 Xiaofei Huang
Xiaofei Huang Wenchen Song
Wenchen Song