- 1Laboratorio De Química Ecológica, Departamento De Ciencias Químicas y Recursos Naturales, Facultad De Ingeniería y Ciencias, Universidad De La Frontera, Temuco, Chile
- 2Carrera De Bioquímica, Departamento De Ciencias Químicas y Recursos Naturales, Facultad De Ingeniería y Ciencias, Universidad De La Frontera, Temuco, Chile
- 3Agronomy Department, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia
- 4Chair of Date Palm Research, Department of Plant Protection, College of Food and Agricultural Sciences, King Saud University, Riyadh, Saudi Arabia
- 5Bioscience program, Biological and Environmental Science and Engineering (BESE) Division, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia
Red palm weevil (RPW), Rhynchophorus ferrugineus (Coleoptera: Curculionidae), is rapidly infesting palm trees (Arecaceae) in several countries, threatening coconut, date, and oil cultivations. The male-produced aggregation pheromone in palm weevils has been reported to be secreted through the mouth to the rostrum, a snout-like projection key for pheromone emission and dispersion. The olfactory mechanisms that underlie peripheral odorant detection in RPW have been addressed at the antennal level. However, the rostrum remains unexplored. Through RNA-seq, 27 odorant-binding proteins (OBPs), 6 chemosensory proteins (CSPs), 4 sensory neuron membrane proteins (SNMPs), 21 gustatory receptors (GRs), 25 odorant receptors (ORs) (including one odorant receptor coreceptor, Orco) and 10 ionotropic receptors (IRs), were identified. We reported 27 novel rostrum-specific olfactory proteins (4 IRs, 11 GRs, 2 CSPs, 3 OBPs, and 7 ORs) in R. ferrugineus (Rfer). The OBPs (RferSOBPs) [Rfer with “S” indicating “snout” (rostrum)] were the most abundant transcripts compared with the rest of the olfactory proteins. We identified several rostrum OBPs, which predominately emerged through gene duplication, and were found expressed in both rostrum and antennae. Noticeably, we found R. ferrugineus pheromone-binding protein (RferOBP1768) paralog in the rostrum (RferOBP14) and mapped it in the same scaffold at a different position on the RPW genome as a recent duplicate. We found that an OR (RferSOR1) was the most abundant for both field-collected and lab-reared RPWs, in the rostrum and antennae. Likewise, up-regulated olfactory-related proteins were established in field conditions compared with those from laboratory-reared. We found a rostrum-specific, highly expressing RferSIR1 in IR93a-clade related to hygrosensation. The role of these olfactory proteins as targets for identifying more specific and powerful semiochemicals is discussed in the context of pest management.
Introduction
Olfaction plays a key role during the life cycle of insects, allowing the location of food sources, oviposition sites, mate finding, and avoidance of toxic substances (Leal, 2013). Volatile chemicals mediate this process as odorants, which are recognized by a well-tuned olfactory system in insect antennae (Leal, 2005; Zhou, 2010; Leal, 2013). The insect olfactory system comprises a sophisticated group of proteins that function around olfactory receptor neurons (ORNs) and can discriminate hundreds to thousands of environmental odorants (Bisch-Knaden et al., 2022). Thus, the first filters are odorant-binding proteins (OBPs) and chemosensory proteins (CSPs), which are known as small soluble proteins that transport odorants through the lymph of hair-like structures called sensilla, in which ORNs are present (Vogt and Riddiford, 1981; Klein, 1987; Maida et al., 1993; Jacquin-Joly et al., 2001; Zhou et al., 2006). Furthermore, it is well-studied that odorant receptors (ORs) in complex with an odorant receptor coreceptor (Orco) are responsible for recognizing odorants as semiochemicals (i.e., chemicals that mediate the interaction between organisms) (Larsson et al., 2004; Sato et al., 2008; Wicher et al., 2008; Wicher, 2018). More recently, these seven-transmembrane proteins have been related to sensory neuron membrane proteins (SNMPs), which, at least in Lepidopterans, might function during pheromone detection in complex with ORs and Orco (Cassau and Krieger, 2021). Other receptors, such as gustatory receptors (GRs), are mainly related to contact chemical cues (e.g., sugar, salt, bitter, and nutrients). In contrast, ionotropic receptors (IRs) have been related primarily to acid and amines and thermal or water sensation (Benton et al., 2009; Croset et al., 2010; Zhang et al., 2011; Jiang et al., 2015; Knecht et al., 2016; Ni et al., 2016; Liu et al., 2019; Liu et al., 2020; Hou et al., 2022).
In recent years, these chemosensory proteins have been well-documented in different insect orders, such as Diptera, Hemiptera, Lepidoptera, and Coleoptera (Venthur and Zhou, 2018). For the latter, antennal transcriptomes have revealed hundreds of proteins that have served, for instance, in evolutionary studies such as ORs (Mitchell et al., 2020). Other examples are coniferous forest pests Ips typographus and Dendroctonus ponderosae, for which a repertoire of 80 and 111 chemosensory proteins was identified, respectively (Andersson et al., 2013). Likewise, a repertoire of 149 chemosensory proteins has been reported for the South American palm weevil Rhynchophorus palmarum (Gonzalez et al., 2021), whereas a larger repertoire of 158 chemosensory proteins has been published for the red palm weevil (RPW) R. ferrugineus (Antony et al., 2016).
The RPW highlights as a severe pest of palm trees with a wide geographical distribution, such as Africa, South Asia, the Middle East, and the Mediterranean. It has been described that infestation is led by the attack of RPWs using an aggregation pheromone (4-methyl-4-nonanol and 4-methyl-5-nonanone), which functions as a signal for a coordinated mass attack that often produces the death of palm trees (Faleiro, 2006). Thus, considering the threat that R. ferrugineus represents at a global scale, understanding the mechanisms that underpin its chemical ecology has become of great importance. In that sense, efforts have been focused on chemoreception mechanisms in the antennae of R. ferrugineus, where, as mentioned above, olfactory proteins have been identified, among which OR1 (RferOR1) is demonstrated as pheromone receptor (PR) (Antony et al., 2021). Furthermore, OBPs have also been studied, reporting antennal- and male-biased expression from OBP1 to OBP10 (Yan et al., 2016). More recently, the silencing of R. ferrugineus OBP1768 (RferOBP1768) successfully disrupted the transport of pheromone 4-methyl-4-nonanol (i.e., ferrugineol) (Antony et al., 2018). On the other hand, GRs have also been identified from genomic data, reporting an expanded profile of 65 GR-encoding genes (Engsontia and Satasook, 2021). Although these studies have contributed significantly to the advance in understanding R. ferrugineus chemosensation, they have been focused on detection (i.e., antennae) rather than secretory mechanisms. Whereas, several studies have identified chemosensory proteins in pheromone glands of other insect species, such as the moth Mamestra brassicae, Agrotis ipsilon, Helicoverpa armigera, Ephestia cautella, and H. assulta as well as defensive glands of beetles Tribolium castaneum and Blaps rhynchopetera (Jacquin-Joly et al., 2001; Gu et al., 2013; Li et al., 2013; Antony et al., 2015; Li et al., 2015; Ding et al., 2023)
It is known that R. palmarum and R. ferrugineus can release the male adults produced aggregation pheromone through the mouth in the form of a droplet towards a depression in the dorsal part of the rostrum, which is filled by hairs (Figure 1), helping disperse the chemicals (Sánchez et al., 1996). Besides, pheromone-secreting prothoracic glands (modified salivary gland) occur only in male R. palmarum (Sánchez et al., 1996). Since Rhynchophorus species have similar morphological characteristics (Davis, 2017; Rozziansha et al., 2021), we hypothesize that R. ferrugineus counts with a repertoire of olfactory proteins that function in detection and secretory processes of aggregation pheromone. Therefore, this study aimed to explore chemoreceptor and chemosensory proteins present in the R. ferrugineus rostrum transcriptome of male and female adults. The overexpression of chemoreceptor and chemosensory proteins in field conditions compared to lab conditions may likely pinpoint key olfactory proteins involved in pheromone or plant-volatile detection. Studies revealed higher expression of pheromone receptor (Antony et al., 2021) in field-collected R. ferrugineus, and chemosensory proteins in the field-collected Colorado potato beetle, Leptinotarsa decemlineata (Schoville et al., 2018). Besides, male-biased or did not differ or female-biased expression of chemosensory proteins in antennae, heads, thoraxes, abdomens, snout, eyes, legs, wings, pheromone glands, and reproductive organs reported earlier (Gu et al., 2013; Li et al., 2013; Zhou et al., 2013; Zhu et al., 2016; Wang et al., 2017; Gonzalez et al., 2021). Our study aimed to trace out the differently expressed and overexpressed chemosensory and chemoreceptor proteins in male and female R. ferrugineus in both lab and field conditions, thereby helping to analyze and select future targets for functional studies.

Figure 1 Red palm weevil, Rhynchophorus ferrugineus male and female. The dorsal side of the male rostrum possesses a group of hairs that help disperse pheromones. R, rostrum; A, antenna; H, hairs.
Materials and methods
Insect rearing and rostrum collection
The RPWs were maintained in our laboratory on sugarcane stems at 28–30°C with a photoperiod of 18 h:6 h (light: dark) since 2009, as described previously (Antony et al., 2016). Lab-reared RPW culture was considered pure-line as there was no mix of other populations. Two weeks after pupation, the cocoons were harvested from the sugarcane stems and individually incubated in round 70 mm × 90 mm plastic jars with perforated screw caps and were checked daily for adult emergence. Ten-day-old adult male and female RPW were selected (male and female separately), and the rostrum was carefully dissected (male and female separately) under a light microscope after insects were anesthetized using CO2 for 1–2 min. Immediately after collection, the rostrum was dissected at its base (male and female separately), transferred in RNA later solution, and then stored at −80°C until total RNA extraction. The RPW male and female adults from the field were captured alive from the infested and removed date palm tree materials at Al Qassim (25.8275° N, 42.8638° E) in Saudi Arabia.
Total RNA extraction, cDNA library construction, and sequencing
Total RNA was extracted from ten pairs of RPW male and female rostrum separately using PureLink RNA Mini Kit (Thermo Fisher, MA, USA). A NanoDrop spectrophotometer (Thermo Fisher) was used to quantify and check the quality of the extracted RNA and synthesized cDNA. The quantity and quality of the total RNA were validated using a Qubit 2.0 Fluorometer (Invitrogen, Life Technologies), and RNA integrity was confirmed using a 2100 Bioanalyzer (Agilent Technologies). After ensuring the quality and the characteristic “hidden break” in the 28S RNA profile using 2100 Bioanalyzer, we proceeded to paired-end cDNA library preparation using TruSeq Stranded mRNA preparation Kit (Illumina Inc.) following manufacturer’s protocols, which include the following steps: purification and fragmentation of total RNA, first and second-strand cDNA synthesis, 3′ end adenylation, adapter ligation, and purification. Finally, the purified and PCR-enriched products were used for cDNA library preparation. The cDNA libraries were validated and quantified by Qubit 2.0 Fluorometer. The HiSeq Illumina sequencing was performed at the core sequencing facility of the King Abdullah University of Science and Technology (KAUST), Jeddah, Saudi Arabia. Image deconvolution and quality value calculations were performed using Illumina GAPipeline1.3. Illumina adaptors were removed, and low-quality bases were trimmed off with “Trim Reads” tool of CLC Genomics Workbench/Server suite. Filtered paired-end reads were validated through a “QC for Sequencing Reads” visualization of the same suite. A reference de novo transcriptome assembly was constructed with the “De Novo Assembly” tool of CLC Genomics Workbench/Server with default parameters. Map reads to contigs function on, using paired read files. The resulting contigs files were functionally annotated by following the method described previously (Antony et al., 2016; Gonzalez et al., 2021). Rostrum transcriptome (field and lab RPWs) was uploaded to NCBI under SRA and TSA accession numbers.
Rostrum transcriptome assembly, gene annotation, and differential expression analysis
Data processing, transcriptome assembly, and annotation were carried out following the method described in (Antony et al., 2016; Gonzalez et al., 2021). Contigs were identified and annotated based on a local BLAST search using R. ferrugineus olfactory protein sequences using Geneious v7.1.5 (http://www.geneious.com). Male and female R. ferrugineus (Rfer) rostrum olfactory proteins were named using the format RferS-[Rfer with “S” indicating “snout” (rostrum)]-(olfactory protein)-transcript number. Blast2Go analyses were performed on rostrum transcripts using the BLAST2GO command line tool (v1.5) of the CLC. The top blast hit transcript clusters were extracted from the male and female assembled transcriptomes with an in-house command-line script. Relevant sequences were translated and manually selected based on an e-value score below 1.0E−5, sequence identity to other chemosensory proteins, and an ORF with at least 50% of the average length normally observed for each gene family reported in (Gonzalez et al., 2021). Accordingly, predicted sequences were numbered in relation to their estimated transcript abundance in RPKM values (Mortazavi et al., 2008). Moreover, an abbreviation “S” was used to denote their “snout” (rostrum) biased identification, including those transcripts identical to previously identified proteins from antennae (Antony et al., 2016). The identical olfactory transcripts found in antennae and rostrum with notable nonsynonymous changes in the amino acid level were denoted in this study as paralogs or duplicates. Gene and transcript level quantification was performed, and the transcripts per kilobase per million mapped reads (TPM) value of each gene was calculated manually based on the consensus length of each gene and total read counts. Furthermore, RPKMs and TPM were tabulated and converted to heatmaps using R and R Studio software. To assess transcriptome completeness, an Arthropoda BUSCO database, consisting of 1066 core genes that are highly conserved single-copy orthologues, was used to query the assembled Fasta files. For this process, the gVolante web server (https://gvolante.riken.jp/) was utilized with the following parameters: min_length_of_seq_stats: 1, assembly_type: trans, Program: BUSCO_v2/v3, selected reference_gene_set: Arthropoda.
We generated the RferSOBP expression profile in the male and female rostrum (lab and field). We compared them to male and female antennae (lab and field) transcriptomes. The R. ferrugineus antennae raw reads were downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA SUB12148378). The assembled and cleaned RferOR2 reads in the antennae transcriptomes were visualized in the CLC, with marked gene positions on the R. ferrugineus genome (GenBank: GCA_014462685). We visualize the expression level of the RferSOBP14 in comparison with RferOBP1768 (Antony et al., 2018), with other two OBPs in the same clade, viz.; RferOR1689 and RferOBP19755 in the CLC Genomics Server.
Phylogenetic analysis
Phylogenetic analyses were performed to elucidate evolutionary relationships between antennal- and rostrum-related chemosensory and olfactory proteins from other coleopterans. Antennal-related chemosensory protein sequences of RPW were retrieved from the literature (Antony et al., 2016). Curated nucleotide databases containing chemosensory and chemoreceptor genes (ORs, GRs, IRs, SNMPs, OBPs, and CSPs) of rostrum transcriptome were further annotated and checked for duplications and open reading frame (ORF) identification using the NCBI BLASTx homology search and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The ORF protein sequences were used for phylogenetic tree construction along with selected OBP protein sequences retrieved from NCBI and Protein Data Bank. Likewise, amino acid sequences from coleopterans, such as Anomala corpulenta (Chen et al., 2014), Anoplophora glabripennis (Hu et al., 2016), Tenebrio molitor (Liu et al., 2015), Tomicus yunnanesis (Liu et al., 2018), Tribolium castaneum (Richards et al., 2008), R. palmarum (Gonzalez et al., 2021), Nicrophorus vespilloides (Mitchell et al., 2020), Holotrichia oblita (Li et al., 2017), I. typographus (Yuvaraj et al., 2021), Megacyllene caryae (Mitchell et al., 2012) and D. ponderosae (Andersson et al., 2013; Andersson et al., 2019), Cyrtotrachelus buqueti, Galeruca daurica, Cylas formicarus (Bin et al., 2017) and Agrilus planipennis, were retrieved from GenBank database or literature. More specifically, CSPs and SNMPS from the silk moth Bombyx mori and ORs from the fruit fly Drosophila melanogaster, were included. Homologous full-length amino acid sequences and predicted sequences were aligned using MAFFT (Katoh et al., 2019) with the auto, (FFT-NS-1, FFT-NS-2, FFT-NS-i, or L-INS-i; depending on data size) strategy and default parameters. The auto algorithm and BLOSUM62 were used as the scoring matrix. Poorly aligned and unrelated segments were trimmed using trimal version 1.2 (Capella-Gutiérrez et al., 2009). The final multiple sequence alignments of GR, IR, SNMP, OBP, and CSP proteins were used to construct trees. The OBP, CSP and SNMP, OR, GR, and IR multiple sequence alignments contained the amino-acid sites 805 (206 sequences), 421 (84 sequences), 1497 (40 sequences), 1288 (313 sequences), 1314 (120 sequences) and 2862 (244 sequences) respectively. The automatic model search was performed using ModelFInder (Kalyaanamoorthy et al., 2017), and the LG+F+G4 for OR and GR, WAG+F+G4 for IR, LG+I+G4 for CSP, SNMP, and OBP substitution models were determined as the best-fit model according to Bayesian Information Criterion (BIC). The maximum likelihood analysis was performed using default settings, and ultrafast bootstrap support (Hoang et al., 2018) with 1000 replicates using IQ-tree (Trifinopoulos et al., 2016). FigTree software (http://tree.bio.ed.ac.uk/software/figtree/) was used to highlight clades, specific taxa, and functional evidence. Finally, phylogenetic trees were edited using an image editor Inkscape 0.48 software.
Results
Red palm weevil rostrum transcriptome assembly
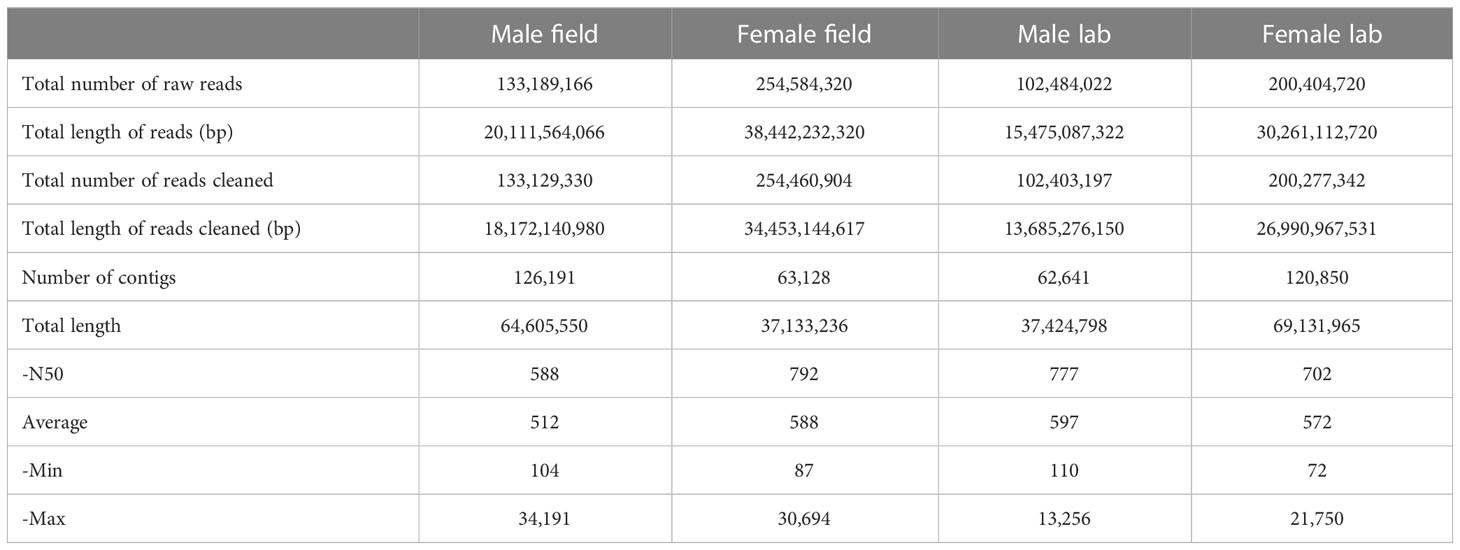
De novo transcriptomes were assembled for each male and female rostrum collected from both field and laboratory conditions, resulting in 4 assembled transcriptomes (male and female, both lab and field conditions). The raw data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database with the accession numbers SRR17732029, SRR17732028, SRR17732027, and SRR17732026 for samples codified as snout_female_field, snout_female_lab, snout_male_field, and snout_male_lab, respectively. The transcriptome Shotgun Assembly (TSA) project was deposited at DDBJ/EMBL/GenBank under the accession PRJNA275430, PRJNA275431, PRJNA275432, and PRJNA275433, BioSamples SAMN25236742, SAMN25236743, SAMN25236744, and SAMN25236745, respectively. A total number of raw reads was generated for each assembled transcriptome, being 133,189,166 for field-collected males, 254,584,320 for field-collected females, 102,484,022 for laboratory-collected males, and 200,404,720 for laboratory-collected females. The total number of clean reads was 133,129,330 for males collected from the field, which yielded 126,191 contigs with an average length of 512 bp and an N50 length of 512 bp. Likewise, 102,403,197 cleaned reads were generated for males collected from the laboratory, which yielded 62,641 contigs with an average length of 597 bp and an N50 length of 777 bp. For females collected from the field, a total number of 254,460,904 clean reads were generated, with 63,128 contigs, an N50 length of 792 bp, and an average length of 588 bp. Finally, 200,277,342 clean reads were generated for females collected from the laboratory, yielding 120,850 contigs, an N50 length of 702 bp, and an average length of 572 bp (Table 1).
GO analysis and transcript abundance
All assembled transcriptomes of RPW (i.e., females and males collected from field and laboratory) were used as BLASTx queries against the non-redundant NCBI protein database and were subjected to InterProScan analyses. For most of the transcripts from all assembled transcriptomes, the greatest number of significant blast hits were to sequences of R. ferrugineus followed by S. oryzae (Figures S1A–D). All assembled transcriptomes (females and males collected from both field and laboratory) showed a consistent GO term distribution, with “cellular process” followed by “metabolic process” the most abundantly assigned Biological Process (BP) (Figures S1A–D). Noteworthy, “localization” was the third most abundantly assigned term for females from the laboratory as well as males from the field (Figures S1A, D). Likewise, “binding,” “catalytic activity,” and “transporter activity” were the most abundant Molecular Functions (MF) assignments for all rostrum transcriptomes. In the Cellular Components (CC) category, “cellular, the anatomical entity” was the most abundant assignment.
For each rostrum transcriptome, namely male field, male laboratory, female field, and female laboratory, Blast2Go (B2G) and InterProScan analyses were performed on 126,195, 62,512, 63,109, and 120,977 transcripts, respectively. All identified transcripts with BLAST hits, 54,327, 28,338, 20,962, and 39,450 transcripts yielded GO annotations in transcriptomes for the male, male, female, and female laboratories, respectively (Figure S1A). Blast hits were identified for 65,815 transcripts in the male field transcriptome and 36,703 transcripts in the male laboratory transcriptome. Likewise, 31,399 and 24,521 transcripts had Blast hits in the female field and laboratory transcriptomes, respectively.
The top 10 most abundant transcripts based on RPKM revealed several uncharacterized proteins in three of the four assembled transcriptomes (Table S1). Interestingly, α-l1 nicotinic acetylcholine receptor with 48,883 RPKM value was identified for female rostrum transcriptome collected from the field. Likewise, a regulator of rDNA transcription protein 15 (19722 of RPKM) is present in the female rostrum collected from the laboratory. Something similar was found in the male rostrum transcriptome collected from the laboratory. Cytoplasmic antigen 1 appeared in the male rostrum (field collected). It is worth noting that, among chemosensory proteins, OBPs were the most abundant according to RPKM (female-lab_contig_145, snout-male-field_contig_137, snout-male-lab_contig_176 and snout-male-lab_contig_194 with 7,792, 12,447, 13,734 and 8,554 of RPKM values) except for the female rostrum field sample (Table S1).
Odorant-binding proteins in red palm weevil rostrum
The RPW rostrum transcriptome analysis revealed 27 full-length candidate sequences related to OBPs (i.e., RferSOBPs hereafter) (Table S2). The average sequence length was 464 bp, and all OBPs presented signal peptides (Figure S2). Three OBPs appeared as rostrum specific (RferSOBP7, 21 and 23), fourteen OBPs were paralogous (duplicated) (RferSOBP6, 8–11, 14, 15, 17–20, 22, 25 and 26) and the remaining ten were identical between the rostrum and antennae (RferSOBP1, 2–5, 12, 13, 16, 24 and 27) (Table S2). Two OBPs (RferSOBP26 and 27) were not present in female rostrum transcriptome, neither field nor laboratory conditions which share high bootstrap (>95%) with RferOBP12511 and RferOBP29, respectively (Table S2). Similarly, RferSOBP16 and 25 were absent in male rostrum transcriptome and shared high bootstrap (>95%) with RferOBP107 and RferOBP17793, respectively. All RferSOBPs were named accordingly to the relative transcript abundance (i.e., RPKM) from the highest to the lowest RPKM value (Table S2). Overall, RferSOBPs were predominantly identified in RPWs collected from the field rather than those reared at laboratory conditions. Nevertheless, RferSOBP1 showed higher transcript abundance in RPWs from the laboratory than in the field. In that sense, RferSOBP1, RferSOBP2, and RferSOBP3 were the most highly abundant transcripts across sex and conditions. We identified several RferSOBPs predominately emerged through gene duplication events (e.g., three paralogs of RferOBP16551 were RferSOBP8, 19, and 20, and three paralogs of RferOBP 1768 were RferSOBP14, RferOBP19755, and RferOBP1689) (Table S2).
Phylogenetic analysis showed the presence of 2 main clades, namely Minus-C and Classic/ABP-II clades (Figure 2). The latter responds to the presence of Classic OBPs (i.e., RferSOBP8, 16, 19, 20, 22, 24, and 27) that follow a conserved cysteine (C) motif (C1-X25-30-C2-X3-C3-X36-42-C4-X8-14-C5-X8-C6) (Figure S2). Furthermore, the RferSOBP22, 24, and 27 that were identical with respect to antennal OBPs, RferOBP23, 3213, and 29, respectively, appeared clustered in the so-called antennal-binding proteins II (ABP-II) subfamily and highlighted within a both antennal- and pheromone-related clade as reported for RferOBP23 and 3213 (Antony et al., 2016; Gonzalez et al., 2021). We retrieved RferOBP23 (Antony et al., 2018) paralog in the rostrum (named RferSOBP22) as it shares 99% amino acid identity. Similarly, RferOBP14511 (Antony et al., 2016) was found expressed in the rostrum (named RferSOBP10), which comes under the Plus-C OBP clade and shares high bootstrap support with D. ponderosae (Andersson et al., 2019). Fifteen of the 27 predicted RferSOBPs were classified as Minus-C, where C2 and C5 are missing (Figure S2). Consequently, these OBPs clustered together in the Minus-C clade. This group’s RferSOBP14 appeared clustered with its ortholog RferOBP1768 (with 99% bootstrap support), reported as an aggregation pheromone carrier (Antony et al., 2018), which shares 65% amino acid identity. Finally, from the identified RferSOBPs, only RferSOBP13 grouped in the CRLBP subfamily, along with its antennal counterpart, RferOBP14025 (Figure 2).
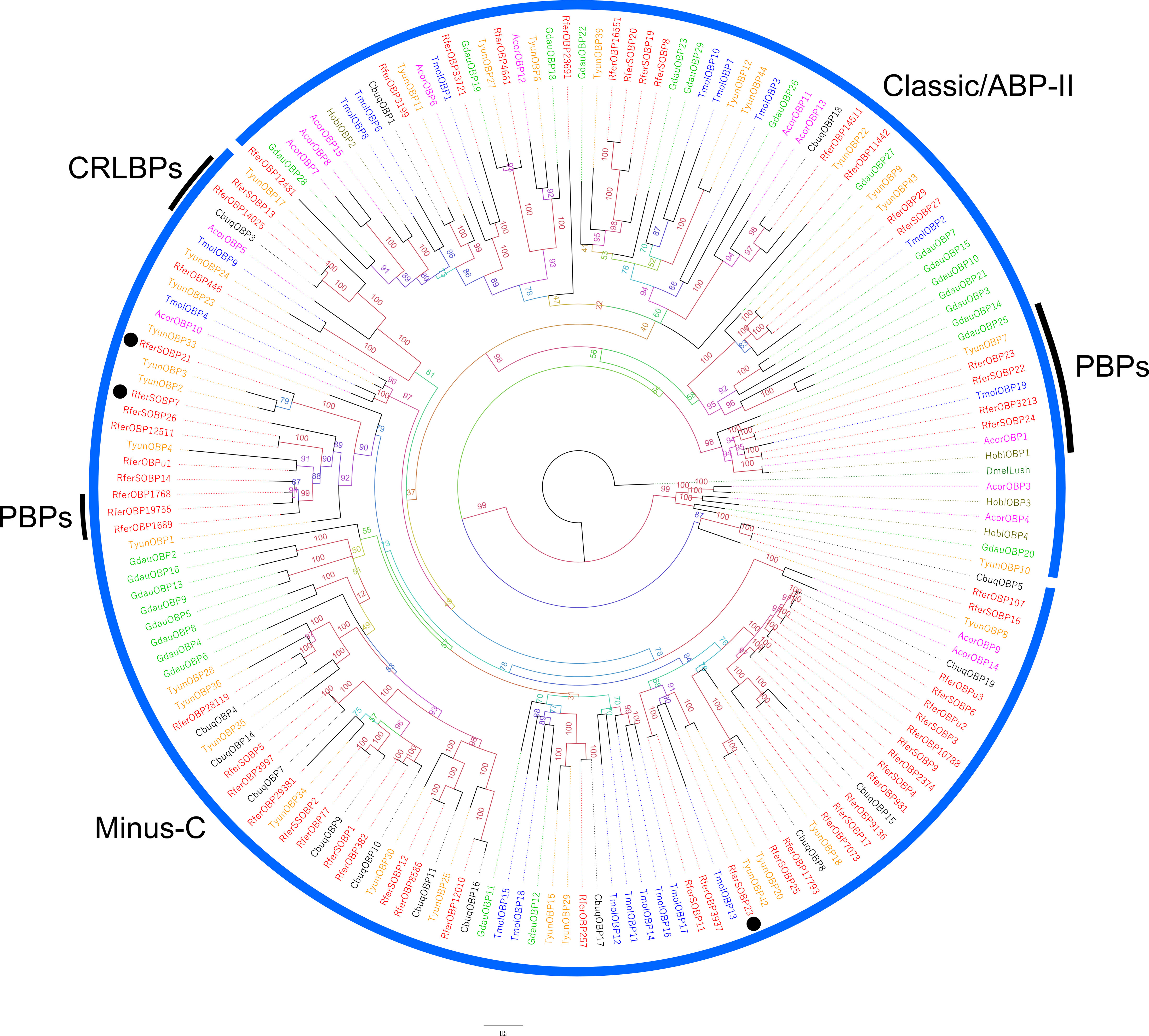
Figure 2 Maximum likelihood rooted tree of R. ferrugineus OBPs identified from rostrum transcriptome. The tree was built from the MAFFT alignment of OBP amino acid sequences of R. ferrugineus, Rfer (red), H. oblita, Hobl (brown), D. melanogaster, Dmel (grey), C. buqueti, Cbuq (black), T. molitor, Tmol (blue), A. corpulenta, Acor (pink), G. daurica, Gdau (green) and T. yunnanesis, Tyun (orange). The branch containing Drosophila OBP LUSH (DmelLush PDB: 2GTE) was used as an outgroup to root the tree and visualized as additive. The major OBP clades are indicated with blue arcs. OBP subfamilies Minus-C, Plus-C, ABPII, CRLBP (black), and classic/GOBPs/PBPs are labeled. RferSOBP7, 21, and 23 showed rostrum-specific expression, denoted in the black dots. Numbers on the branches indicate bootstrap values (UFBoot n = 1000). Scale = 0.5 amino acid substitutions per site.
Chemosensory proteins in red palm weevil rostrum
Six CSPs were identified from RPW rostrum transcriptome (RferSCSPs hereafter), being 5 of them with full-length ORFs (Table S3). An average length of 413 bp was obtained for RferSCSPs. These proteins appeared present in both male and female rostrum transcriptome with the exception of RferSCSP6 (shares 88% bootstrap support with B. mori CSP12, BmoriCSP12), which was found in female transcriptome assemblies neither in field nor laboratory conditions. Two CSPs appeared as rostrum specific (RferSCSP4 and 6), two CSPs (RferSCSP2 and 3) were paralogous (duplicated) and the remaining two were identical (RferSCSP1 and 5) between the rostrum and antennae (Table S3). Interestingly, RferSCSP1, 2, 3, and 5 were present in field conditions for females, whereas RferSCSP4 was only identified in female transcriptome from laboratory conditions. Similarly, RferSCSP1, 3, 4, 5, and 6 were identified in male transcriptome from field conditions rather than laboratory, where RferSCSP2 and 5 were present in the latter. All RferSCSPs were named accordingly to the relative transcript abundance (i.e., RPKM), showing RferSCSP1 and 2 as the most highly abundant.
Five of the 6 predicted RferSCSPs followed the conserved cysteine pattern (C1-X6-C2-X18-C3-X2-C4) proposed for insect CSPs (Lartigue et al., 2002) (Figure S3). Likewise, RferSCSPs were distributed in 5 different clades, where RferSCSP1, 4 and c304 highlight for being present in a clade where functionally studied CSPs, Holotrichia oblita CSP1 and 2 (HoblCSP1 and HoblCSP2), have been reported as carriers of general odorants, namely β-ionone (Sun et al., 2014) (Figure 3). Interestingly, RferSCSP1, 2, 3, and 5 were found to be identical or duplicated to antennae-related RferCSP-c304, c213, c2617, and c19560, respectively (Figure 3).
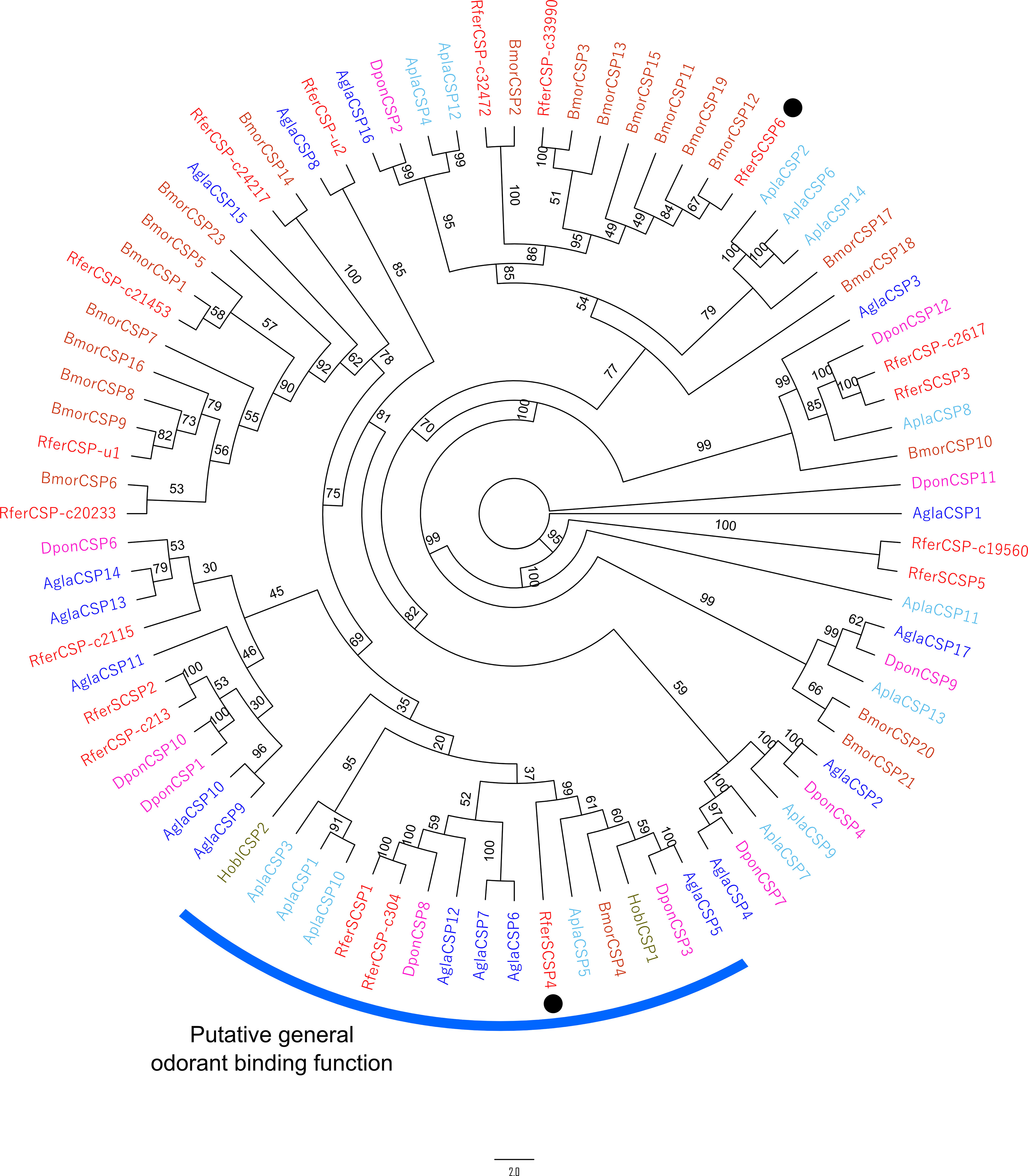
Figure 3 Maximum likelihood unrooted consensus tree of the chemosensory proteins (CSPs) predicted from the rostrum transcriptome of R. ferrugineus. The tree was built from the MAFFT alignment of CSP amino acid sequences of R. ferrugineus, Rfer (red), A. planipennis, Agla (blue), D. ponderosae, Dpon (pink), A. glabripennis, Agla (black), H. oblita (brown) (Sun et al., 2014) and B. mori, Bmor (grey). A clade with putative odorant binding function reported is indicated in a blue arc. RferSCSP4 and 6 showed rostrum-specific expression, denoted in the black dots. The tree is visualized as cladogram. Numbers on the branches indicate bootstrap values (UFBoot n = 1000). Scale = 2.0 amino acid substitutions per site.
Sensory neuron membrane proteins (SNMPs) in red palm weevil rostrum
Four SNMPs were identified from the assembled rostrum transcriptomes (RferS-SNMPs here on), being all full-length with an average of 1872 bp. None of SNMPs appeared as rostrum specific, whereas, two SNMPs (RferSSNMP1b, RferSSNMP2b and RferSSNMP1a) were paralogous (duplicated) and the remaining one was identical (RferSSNM2a) between the rostrum and antennae (Table S4).The 4 RferS-SNMPs have also been reported in antennae (Antony et al., 2016), sharing a 24.2% of sequence identity between RferS-SNMP1a and 2a and 30.1% between RferS-SNMP1b and 2b. High conservation is observed at the N-terminal section of each RferS-SNMP, whereas sequence divergence is evidenced from approximately amino acid 340 (Figure S4). One RferS-SNMP (RferS-SNMP1b) showed 5-fold greater transcript abundance (cumulative RPKM: 124.52) in comparison with the remaining 3. Phylogenetic analysis revealed the presence of 2 main clades (SNMP1 and SNMP2) (Figure 4). As expected, all identified RferS-SNMPs appeared with their antennal counterpart (i.e., Rferc18799, 17112, SNMP_U2, and SNMP_U1 with 100, 99.7, 96.1, and 99.4% of sequence identity, respectively) (Figure S4).
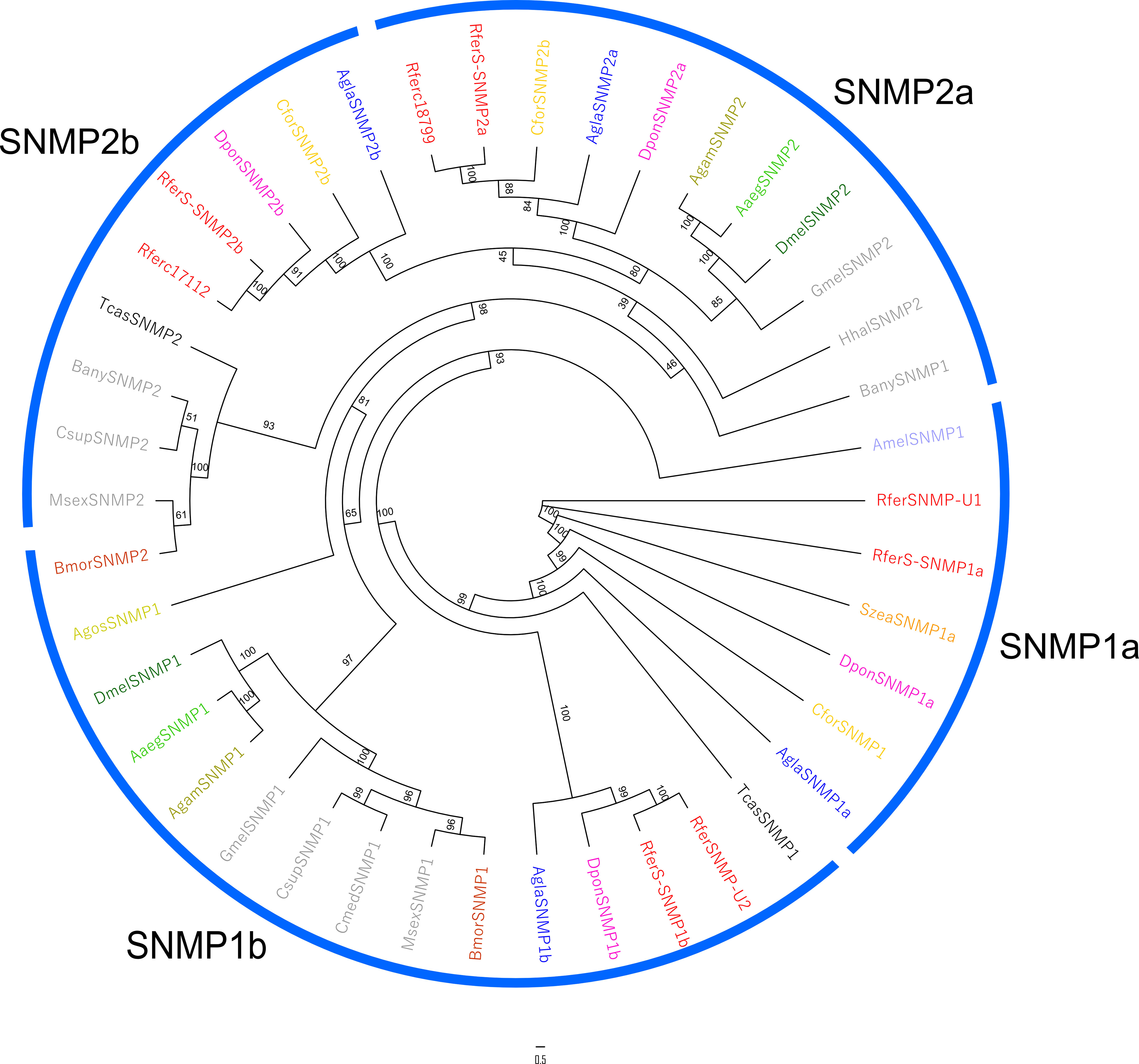
Figure 4 Maximum likelihood unrooted consensus tree of the sensory neuron membrane proteins (SNMPs) annotated from the rostrum transcriptome of R. ferrugineus. The tree was built from the MAFFT alignment of SNMP amino acid sequences of R. ferrugineus, Rfer (red), A. glabripennis, Agla (blue), B. mori, Bmor (grey), D. ponderosae, Dpon (pink), C. formicarus Cfor (green), and T. castaneum, Tcas (black). SNMP subfamilies are indicated in blue arcs. The tree is visualized as cladogram. Numbers on the branches indicate bootstrap values (UFBoot n = 1000). Scale = 0.5 amino acid substitutions per site.
Gustatory receptors in red palm weevil rostrum
Twenty-one GRs were identified from the assembled RPW rostrum transcriptome (hereafter, RferSGRs), which showed an average of 856.9 bp in length (Table S5). Eleven GRs appeared as rostrum specific (RferSGR3, 9, 10, 12, 13, 15 and 17–21), four GRs (RferSGR5, 7, 8 and 11) were paralogous (duplicated) and the remaining six GRs were identical (RferSGR1, 2, 4, 6 14 and 16) between the rostrum and antennae (Table S5). Most of the RferSGRs were present in either male or female transcriptome in field conditions, especially for females, except for RferSGR3, 12, 14, 16, 17, 18, and 19, which were not found in female rostrum transcriptome assemblies. Similarly, RferSGR20 and 21 were not found in the male transcriptome. The C-terminal motif of insect GRs (TY-X5-QF) was present in 10 of the 21 RferSGRs (Figure S5) (Robertson et al., 2003).
Phylogenetic analysis revealed most of the RferSGRs in 3 main clades identified, such as sugar, CO2, and bitter-related clades (Figure 5). In that sense, 9 RferSGRs were also grouped together with their identical antennal-related transcript (Table S5). Besides that, 11 RferSGRs (RferSGR3, 9, 10, 12, 13, 15, 17, 18, 19, 20 and 21) were identified as likely specific to the rostrum and related to a function together with other paralogs. For instance, RferSGR21 was present in a CO2-related clade, whereas RferSGR16 appeared in a sugar-related clade. Likewise, RferSGR9, as paralog of D. ponderosae GR47 (DponGR47), appeared in a bitter-related clade.
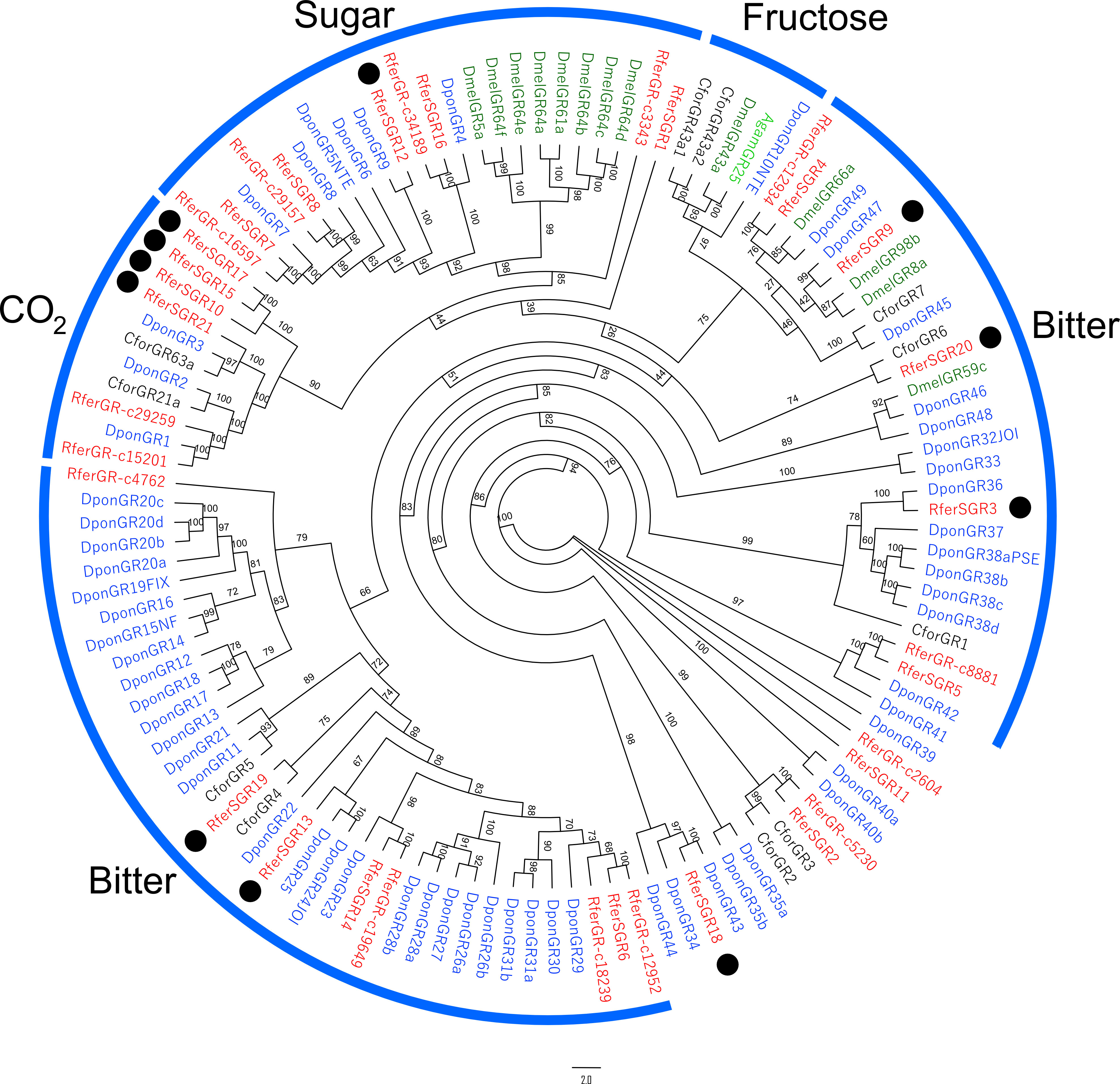
Figure 5 Maximum likelihood rooted tree of R. ferrugineus GRs identified from rostrum. The tree was built from the MAFFT alignment of GR amino acid sequences of R. ferrugineus, Rfer (red), D. ponderosae, Dpon (blue), C. formicarus, Cfor (black), Anopheles gambiae, Agam (green) and D. melanogaster, Dmel (dark green). The branch containing conserved lineage of putative sugar receptors was used as an outgroup to root the tree. Well studied GR clades with high bootstrap support with R. ferrugineus (Antony et al., 2016) and D. ponderosae (Andersson et al., 2019) are indicated by thick blue arcs. RferSGR3, 9, 10, 12, 13, 15, and 17–21 showed rostrum-specific expression, denoted in the black dots. The tree is visualized as additive. Numbers on the branches indicate bootstrap values (UFBoot n = 1000). Scale = 2.0 amino acid substitutions per site.
Odorant receptors and coreceptor in red palm weevil rostrum
Twenty-five ORs, including Orco, were identified from RPW rostrum transcriptome (RferSORs hereafter), which appeared distributed between both sexes and conditions (i.e., field and laboratory) (Table S6). The RferSORs showed an average length of 1001 bp. RferSORs were named accordingly to the relative transcript abundance (i.e., RPKM). Thus, 18 of the 25 RferSORs were identified with their full-length sequence (Table S6). From the highest expressing OR in the RPW rostrum (RferSOR1), we identified its identical OR in antennae (RferOR21) (Figure 6). Seven ORs appeared as rostrum specific (RferSOR3, 4, 8, 14, 15, 16 and 25), six ORs (RferSOR2, 7, 9, 17, 20 and 22) were paralogous (duplicated) and the remaining thirteen ORs were identical (RferOrco, RferSOR1, 5, 6, 10–13, 18, 19, 21, 23 and 24) between the rostrum and antennae (Table S6). Most of the RferSOR transcripts were identified in both female and male rostrum transcriptomes. Exceptions were RferSOR20, 21, 23, 24, and 25, found only in the female rostrum, whereas RferSOR8, 14, 17, 19, and 22 were found in the male rostrum. The remaining RferSOR transcripts were found in the male transcriptome in both field and laboratory conditions, except for RferSOR5, 12, 14, 15, 17, 18, and 19, which were present in one condition.
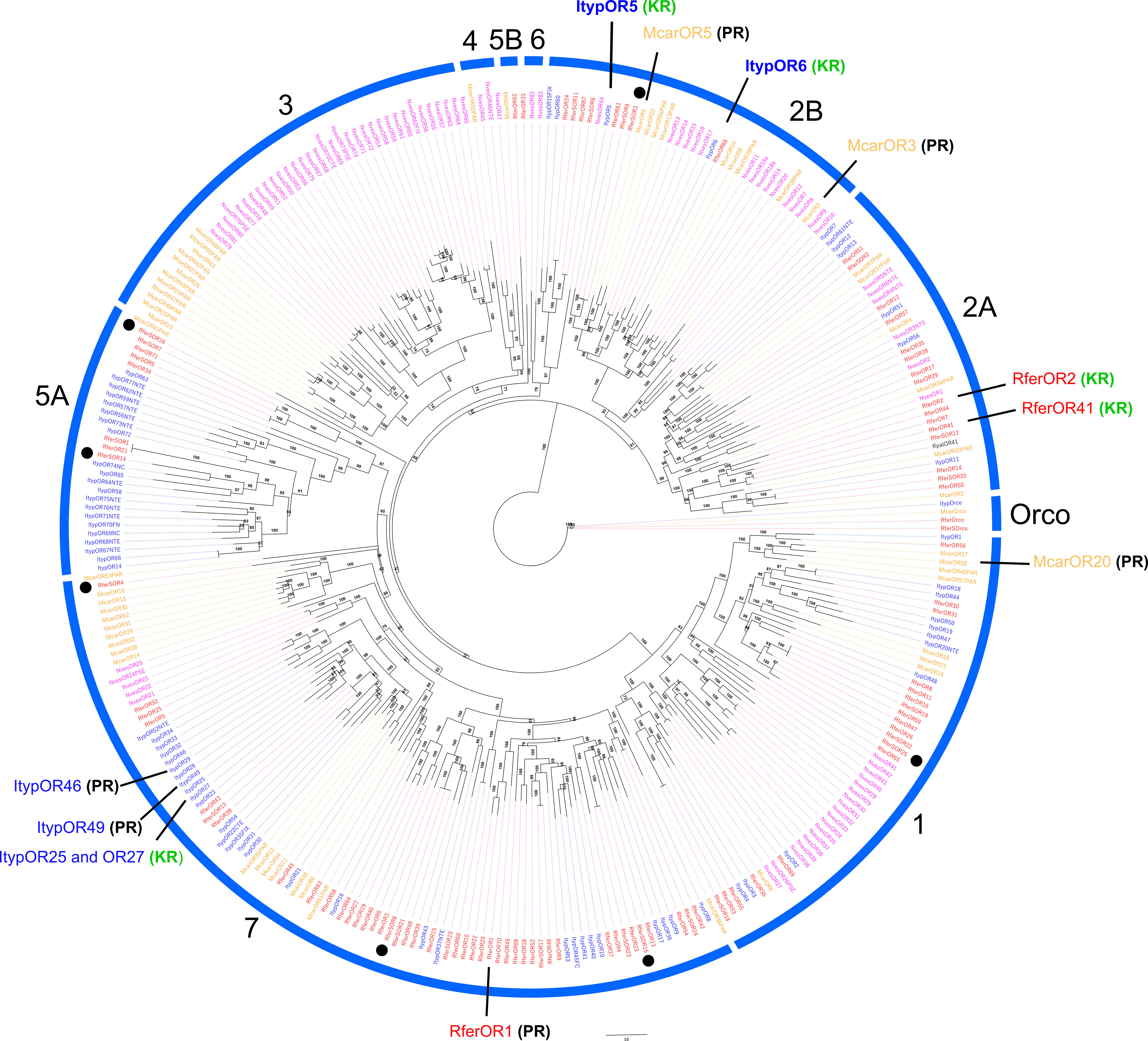
Figure 6 Maximum likelihood rooted tree of R. ferrugineus odorant receptors (ORs) identified from rostrum transcriptome. The tree was built from the MAFFT alignment of OR amino acid sequences of R. ferrugineus, Rfer (red), I. typographus, Ityp (blue), N. vespilloides, Nves (pink), M. caryae, Mcar (orange), and R. palmarum, Rpal (black). The major OR subfamilies are indicated in blue arcs and numbers (1, 2A, 2B, 3, 4, 5A, 5B, 6 and 7). The Orco clade was used as an outgroup to root the tree. The tree is visualized as additive. Clades containing functionally characterized ORs are highlighted as pheromone receptor (PR in black bold letters) or kairomone receptor (KR in green bold letters) based on revised literature (Mitchell et al., 2012; Andersson et al., 2019; Antony et al., 2021; Ji et al., 2021; Yuvaraj et al., 2021; Antony et al., 2023). RferSOR3, 4, 8, 14, 15, 16, and 25 showed rostrum-specific expression, denoted in the black dots. Numbers on the branches indicate bootstrap values (UFBoot n = 1000). Scale = 0.8 amino acid substitutions per site.
The phylogenetic analysis identified nine major coleopteran OR subfamilies, viz., 1, 2A/2B, 3, 4, 5A, 5B, 6, and 7, besides the conserved clade for Orco (Figure 6). We identified RferSOrco sequence that shares 100% amino acid identity with the previously reported RferOrco (Soffan et al., 2016). Either rostrum or antennal RferORs were distributed across 6 of the 9 clades. From these, 4, 3, 4, 4, and 9 RferSORs are present in clades 1, 2A, 2B, 5A, and 7, respectively. Noteworthy, RferSOR10 and 18 were the only rostrum-related ORs in clade 7 found close to the pheromone receptor, RferOR1 (Antony et al., 2021). However, RferSOR10 and 18 shares 24.06% and 31.54% protein identity with RferOR1. RferSOR12 was identical to RferOR41, an R. ferrugineus OR tuned to non-host plant volatile and antagonist, α-pinene (Ji et al., 2021) was located in the same clade of RferOR2, an R. ferrugineus OR tuned to palm-derived volatiles (Antony et al., 2023). Interestingly, RferSOR13 appeared near ItypOR46 and 49, reported as PRs in clade 7 (Andersson et al., 2013; Yuvaraj et al., 2021). Although other functionally studied ORs were included in the phylogeny, no RferSORs were present as orthologs of these. It is worth mentioning that 19 of the 25 RferSORs have also been reported for antennae sharing the same node in the phylogenetic tree (Figure 6) (Antony et al., 2016).
Ionotropic receptors in red palm weevil rostrum
Ten IRs were identified from RPW rostrum transcriptome (RferSIRs hereafter) with an average length of 1840 bp, from which 9 were full-length (Table S7). Four IRs appeared as rostrum specific (RferSIR1, 5, 8 and 10), two IRs (RferSIR6 and 7) were paralogous (duplicated) and the remaining four IRs were identical (RferSIR2, 3, 4 and 9) between the rostrum and antennae (Table S7). Most of the RferSIRs were identified in assembled transcriptomes for field conditions in females and males. The RferSIR5 and 9 were not found in female-assembled transcriptomes (Table S7). Likewise, RferSIR7 was not found in male-assembled transcriptomes. RferSIR1 was the most highly expressed IR (the total RPKM 403) in males and females with rostrum-specific expression (Table S7).
Phylogenetic analysis of RferSIR transcripts along with IRs from D. ponderosae, A. glabripennis, and D. melanogaster revealed five main clades, where RferSIRs were present in 3 of them (Figure 7). The RferSIR4 and 10 were identified in clade coreceptor (IR25a/IR8a), together with RferIR2728 and RferIR_u1. The RferSIR2, 3, 5, 6 and 9 were identified in the antennal IR clade, and RferIR1, 8 and 7 were grouped into divergent IR clade. Furthermore, RferSIR5 and 9 were present in a small clade along with DmelIR41a, and 76a reported with olfactory functions (Benton et al., 2009; Abuin et al., 2011). Finally, the rostrum-specific, highly expressing RferSIR1 appeared in IR93a-clade related to humidity sensation or hygrosensation (Benton et al., 2009; Frank et al., 2017; Knecht et al., 2017; Ni, 2021).
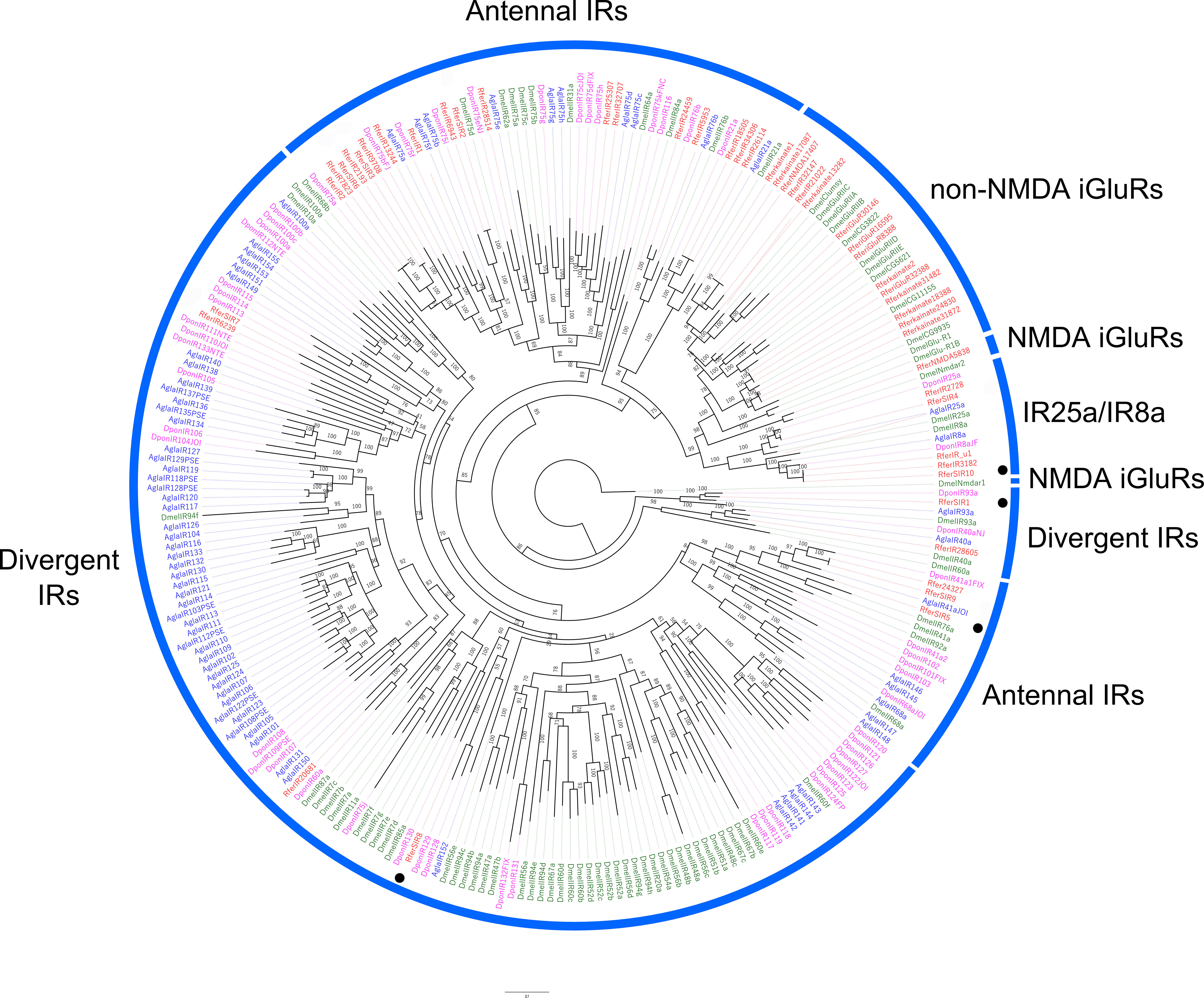
Figure 7 Maximum likelihood rooted tree of R. ferrugineus ionotropic receptors (IRs) identified from rostrum transcriptome. The tree was built from the MAFFT alignment of IR amino acid sequences of R. ferrugineus, Rfer (red), D. ponderosae, Dpon (pink), A. glabripennis, Agla (blue), and D. melanogaster, Dmel (grey). Conserved IR clades, based on the high bootstrap support with R. ferrugineus (Antony et al., 2016) and D. ponderosae (Andersson et al., 2019) are indicated with blue arcs. The branch containing the NMDA-subfamily of IRs was used as an outgroup (Croset et al., 2010; Eyun et al., 2017) to root the tree and visualized as additive. RferSIR1, 5, 8, and 10 showed rostrum-specific expression denoted in the black dots. Numbers on the branches indicate bootstrap values (UFBoot n = 1000). Scale = 3.0 amino acid substitutions per site.
Differential gene expression analysis
Differential gene expression (DEG) analysis revealed differences in the expression of olfactory proteins between field- and laboratory-related RPWs (Figure 8). In terms of OBPs, RferSOBP1 showed the highest expression according to TPM values, with 4438.38 and 12934.94 for females and males reared in the laboratory, respectively. On the other hand, 1102.25 and 1.71 TPM values for females and males collected from the field were obtained for the same OBP. Similarly, RferSOBP2 resulted in 3309.50, and 8056.02 TPM values for females and males reared in the laboratory, respectively, whereas 462.05 for females collected from the field and not found in males from the field. Particularly, RferSOBP1 is identical in sequence identity (i.e., 100%) to antennal RferOBP382 and shows 79.26% of sequence identity with ortholog PBP10 of Cyrtotrachelus buqueti (CbuqPBP10) (APG79371.1). Likewise, RferSOBP2 has 100% of sequence identity to antennal RferOBP77 and 85.71% with CbuqPBP9 (APG79370.1). In general, cumulative RPKM values suggest RferSOBP1 and 2 as the most abundant proteins among rostrum OBPs (Table S2).
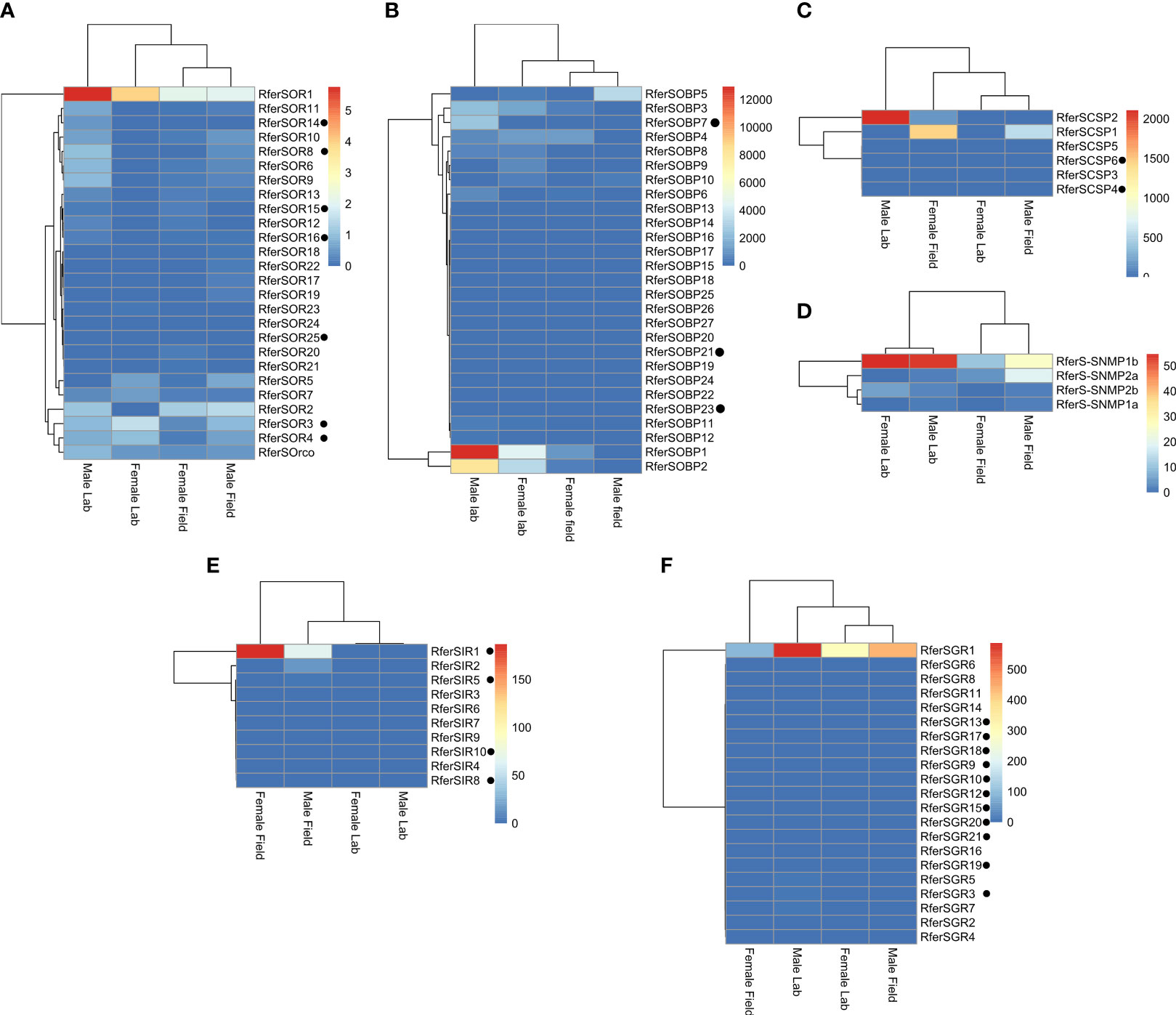
Figure 8 Heatmap showing changes in chemosensory and chemoreceptor expression levels in Rhynchophorus ferrugineus rostrum for lab-reared and field-collected male and female adults. The heatmap colors represent transcript abundance in transcript per million (TPM) from highest (red) to lowest (blue) expression levels. The data represented as log-transformed TPM values tabulated and converted to heatmaps using R and R Studio software. (A) ORs; (B) OBPs; (C) CSPs; (D) SNMPs; (E) IRs; (F) GRs. Those olfactory proteins that exhibited rostrum-specific expression are denoted in the black dots.
For RferSCSPs, RferSCSP1 was more abundant in RPWs collected from the field, with 1438.10 and 556.83 TPM values for females and males, respectively, whereas not found in RPWs reared in the laboratory. Likewise, RferSCSP2 appeared highly abundant in laboratory-collected males (2106.89 of TPM), but with no particular trend between laboratory or field conditions (Figure 8). The identified RferSCSP1 from the assembled rostrum transcriptomes resulted in identical (i.e., 100% of sequence identity) to antennal RferCSP-c304 (Table S3) and with 91.96% of sequence identity to CSP8 of rice water weevil Lissorhoptrus oryzophilus (LoryCSP8) (AHE13803.1). On the other hand, 98.5% of sequence identity was found between RferSCSP2 and RferCSP-c213. Likewise, it shows 64.39% of sequence identity to LoryCSP9 (AHE13804.1).
Regarding SNMPs, RferS-SNMP1b showed higher transcript abundance in RPWs reared in the laboratory (both females and males) than those collected from the field. Thus, RferS-SNMP1b showed TPM values of 54.60 and 53.40 for females and males reared in the laboratory, respectively, and 10.79 and 25.88 for females and males collected from the field. This SNMP has 96.1% sequence identity to the previously identified RferSMP_U2 from antennae. All remaining RferS-SNMPs (RferS-SNMP1a, 2b, and 2a) resulted in TPM values lower than 5.63 in RPWs reared in the laboratory. Similarly, the same RferS-SNMPs showed TPM values lower than 18.84 in RPWs collected from the field.
Although among RferSGRs, no clear differentiation between RPWs from field or laboratory conditions were identified, RferSGR1 highlighted by showing an important difference in abundance compared with the rest of GRs with TPM values of 88.05 and 438.46 for females and males collected from the field as well as 298.45 and 586.18 for females and males reared in the laboratory. Thus, transcript RferSGR1 has 100% sequence identity to the antennal GR, RferGR-c_4762. Likewise, the RferSGR1 showed an identity of 37.98% with GR1 of beetle Colaphellus bowringi (CbowGR1) (ALR72527.1).
For ORs, RferSOR1 showed the highest abundance at laboratory conditions (both females and males) compared with RPWs collected from the field. Thus, females and males reared in the laboratory showed TPM values of 3.95 and 5.75, respectively. On the other hand, females and males collected from the field showed TPM values of 2.12 and 2.05, respectively. The RferSOR1 has 100% sequence identity to antennal RferOR21. It is worth noting that BLAST suggests that the best homologue with annotation is the OR1 of Dastarcus helophoroides (DhelOR1) (AIX97079.1), with 34.43% of sequence identity.
In terms of RferSIRs, RferSIR1 appeared more abundant in field conditions compared with the laboratory. This IR showed TPM values of 186.66 and 65.63 for females and males of RPW collected from the field, respectively. On the contrary, TPM values of 0.14 for females in laboratory conditions were obtained, and no expression was found in males reared in the laboratory.
Overall, except for RferSIR1, none of the highly abundant transcripts was rostrum specific, having its identical sequence identified from RPW antennae (Antony et al., 2016). Besides the already-named transcripts, most R. ferrugineus chemosensory transcripts showed no change in abundance. Generally speaking, RPW lab samples exhibit more abundant olfactory protein expression than field-collected RPWs (Figure 8).
The RPKM value of all olfactory proteins in R. ferrugineus rostrum is provided in the supporting Table S2–S7.
Discussion
In the last few decades, the RPW R. ferrugineus has emerged as an invasive quarantine pest that has spread globally in palm tree-growing countries from South Asia to the Middle East, Europe, Africa, and China (Hoddle et al., 2024). Consequently, sustainable pest control strategies against RPWs have arisen with a basis on their semiochemistry, in which an aggregation pheromone ferrugineol (4-methyl-4-nonanol) and ferrugineone (4-methyl-5-nonanone) functions as a signal for a coordinated mass-attack that often produces the death of palm trees (Faleiro, 2006; Hoddle et al., 2024). Thus, this pheromone blend and kairomones (e.g., food baits and ethyl acetate) have allowed the application of mass trapping methods (Faleiro, 2006). Considering olfaction’s key role in RPW chemical ecology, it is important to understand how these insects recognize the plethora of volatile organic compounds (VOCs) they face during their life cycle. In that sense, insect olfactory systems have been comprehensively studied in the palm weevils (Antony et al., 2016; Gonzalez et al., 2021). Their studies have been made to elucidate chemosensory proteins in RPW in the context of pest management, focusing on antennae as a main olfactory organ. However, no research has been reported on other key chemosensory organs for RPW or other palm weevils, namely the rostrum, despite the palm weevil rostrum plays an important role in releasing and dispersing the male-produced aggregation pheromone (Sánchez et al., 1996). In this study, we annotated a total of 27 OBPs, 6 CSPs, 4 SNMPs, 21 GRs, 25 ORs (including Orco), and 10 IRs were identified from 4 assembled rostrum transcriptomes of the RPW. Several of these chemosensory proteins have also been identified in antennae (Antony et al., 2016). However, here 27 rostrum-specific proteins (4 IRs, 11 GRs, 2 CSPs, 3 OBPs, and 7 ORs) are reported for the first time. Compared with the antennal transcriptome of RPW, the rostrum transcriptomes present a more significant number of reads, though lower N50 values (Antony et al., 2016) (Table 1).
The assembled transcriptomes of both sexes collected from field and laboratory, resulted in 4 datasets, where RferSOBPs were the most abundant transcripts compared with the rest of the olfactory proteins identified in this study. Ten OBPs were identified, and although olfactory proteins have poorly been studied in rostrum of weevils, there is a report where sympatric species Eucryptorrhynchus scrobiculatus and E. brandti, showed 7 and 6 OBPs out of 30 and 28, respectively, with significant expression in rostrum compared with other tissues, such as antennae, legs and heads (Wen et al., 2018). This is also consistent with our hypothesis in terms of the presence of chemosensory proteins in a secretory structure like rostrum. Particularly, RferSOBP1 and 2 showed greater abundance for females and males collected from the field with respect to females or males collected from the laboratory. Both RferSOBPs were classified as Minus-C as identical to RferOBP382 and 77. Both RferOBP382 and 77 are found to be highly expressed in the antennae; however, interestingly, RferOBP77 exhibits male-biased antennal expression (Antony et al., 2018). In contrast, we mined RferSOBP2 transcripts from male and female rostrums in the current study. We found that RferSOBP14 appeared phylogenetically close to RferOBP1768, reported as a pheromone-binding protein (PBP) with binding affinity to pheromone components, such as 4-methyl-5-nonanol (Antony et al., 2018). This clade also contains RferOBP19755 and RferOBP1689 (Figure 2) found to be highly expressed in the antennae (Antony et al., 2018). We manually annotated RferSOBP14, RferOBP1768, RferOBP1689, and RferOBP19755 in the RPW genome (Dias et al., 2021). We visualize the expression level of the RferSOBP14 compared with RferOBP1768 (Antony et al., 2018), with other two OBPs in the same clade, viz.; RferOR1689 and RferOBP19755 in the CLC Genomics Server (Figure 9). Scaffold 22 contains four OBPs including the pheromone binding protein, RferOBP1768. Thus, genome-wide analysis revealed these three OBPs were found to cluster in the same scaffold 22 (GenBank accession JAACXV010000021.1), representing potentially recent duplications (Figure 9). As shown in Figure 9, all three sequences hit different genomic positions, meaning they could be three independent genes. OBP gene duplications are of major interest as insect OBPs are known to evolve rapidly through gene gain-and-loss, allowing them to detect many volatiles (Venthur and Zhou, 2018). Recently, the RferOBP1768 function has already been reported as involved in transporting RPW aggregation pheromone molecules to receptor neurons (Antony et al., 2018). Elucidating the role of the other three OBPs (RferSOBP14, RferOBP1689, and RferOBP19755) would possibly tell us if duplicates retained the similar function of evolved different odorant binding and transport. Thus, RferSOBP14 would represent a potential candidate for further functional studies around the sexual communication of the RPW. On the contrary, RferSOBP22 appeared identical to the reported PBP of RPW, RferOBP23, suggesting that this transcript may likely be related to pheromone transport functions (Antony et al., 2018). However, although RferOBP23 was reported to be one of the highly expressed OBP in the antennae, RferSOBP22 found meager expression in the rostrum (Table S2). Similarly, we identified three duplicates of RferOBP16551, viz., RferSOBP8, 19, and 20 (Table S2). Taken together, we hypothesize that R. ferrugineus OBPs reported in our study as paralogs that may likely function in the detection and secretory processes of aggregation pheromone. However, the meaning of many duplicated OBPs in the R. ferrugineus rostrum remains elusive due to the lack of the functional characterization of any of these genes.In terms of CSPs, 6 RferSCSPs were identified in this study, less than the 12 CSPs reported for the RPW or the 10 CSPs of the American palm weevil, R. palmarum (Antony et al., 2016; Gonzalez et al., 2021). Among these, two CSP transcripts mined from the rostrum transcriptome were not reported in the antennae (Table S4). Interestingly, the RferSCSP1, found identical to RferCSP-c304 (Antony et al., 2016), and RpalCSP4 (Gonzalez et al., 2021) are the most highly expressing CSPs in both rostrum and antennae. Unlike OBPs, CSPs have scant evidence of binding pheromones (Pelosi et al., 2018). Therefore, it is difficult to establish potential functions based solely on tissue-biased expression, transcript abundance, or evolutionary relationships. In that sense, RferSCSPs are not an exception. Nevertheless, it is interesting to notice the presence of RferSCSP1 and 4 phylogenetically close to HoblCSPs that have been reported as carriers of floral compounds, such as β-ionone (Sun et al., 2014). Likewise, RferSCSPs are fewer than RferSOBPs, both as soluble transport proteins. Thus, RferSCSPs might have other non-olfactory functions, such as embryonic development or leg regeneration, as reported for other insects (Maleszka et al., 2007).
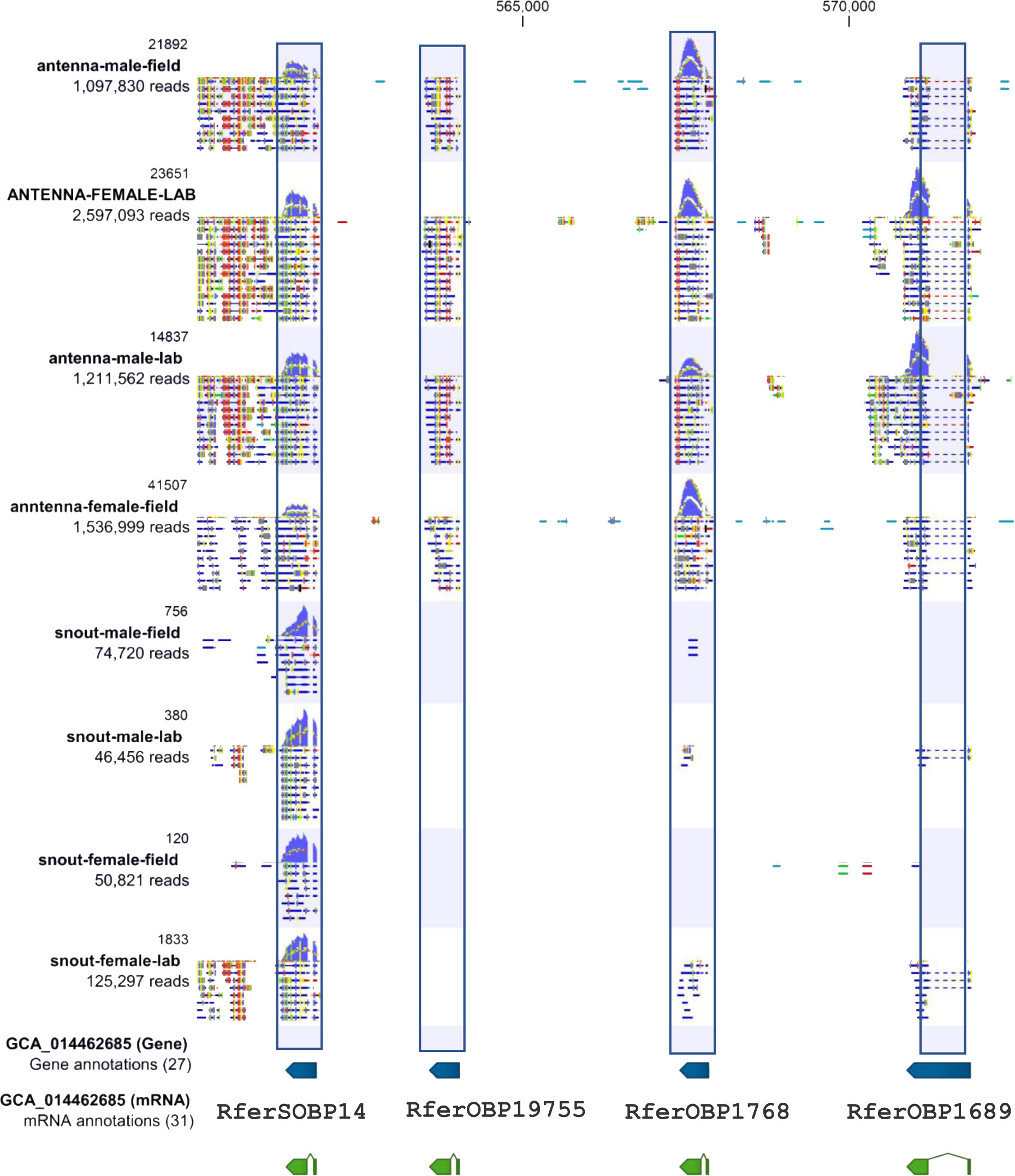
Figure 9 The genome-wide analysis of RferSOBP14, RferOBP1768, RferOR1689, and RferOBP19755 in scaffold 22 (GenBank accession JAACXV010000021.1) (Dias et al., 2021). The OBP gene expression profile in the male and female antennae (lab and field) and rostrum (lab and field) transcriptomes with marked gene position in scaffold 22 of the R. ferrugineus genome (GenBank: GCA_014462685) was generated in the CLC Genomics Server. The RferSOBP14 clade gene sequences hit different genomic positions in the same scaffold. The RferSOBP14 expresses both antennae and rostrum. The other three OBPs (RferOBP1768, RferOR1689, and RferOBP19755) are antennal-specific expressions.
The transcriptome analysis revealed four SNMPs for the RPW, the same number reported for R. palmarum (Gonzalez et al., 2021), although less than the 6 SNMPs identified from antennae of the RPW (Antony et al., 2016). All four rostrum SNMPs were the same expressed in the R. ferrugineus antennae (Table S4). Despite these transmembrane proteins receiving less attention compared with other chemosensory proteins, SNMPs appear to have a key role in signal transduction, probably forming a heteromeric complex of OR/Orco heteromeric complex recently proposed (Zhang et al., 2020). Whether this function is also present in coleopteran SNMPs is still unknown, as no functional studies have been reported so far.
In terms of GRs, our transcriptome analysis revealed more GRs (21 RferSGRs) compared with the reported for RPW antennae (15 GRs) (Antony et al., 2016), where 11 are likely rostrum specific (Table S5). These findings are consistent with the proposal that the number of transcripts might be limited by the analyzed tissue (i.e., antennae, rostrum, legs, etc.). More recently, genome-wide analyses have revealed more GR-encoding genes for R. ferrugineus with 65 candidate sequences (Engsontia and Satasook, 2021). Interestingly, a rostrum-related GR (RferSGR21), identified in female transcriptome collected from the field, appeared in the CO2 clade (Figure 5). It is likely that the RferSGR21, along with antennal orthologs, RferGR-c_29259, and c_15201, could be involved in the selection and oviposition of females of RPW during fermentation of palm trees (Salem et al., 2016). Oviposition preference has also been reported for females of RPW in date palms with higher content of sugars (Al-Ayedh, 2008), where RferSGRs may likely play a key role. In this study, 4 RferSGRs were identified in a sugar clade, RferSGR12 likely rostrum specific and paralog of D. ponderosae GR9 (DponGR9) (Figure 5).
Regarding ORs, RferSORs appeared phylogenetically related to all previously reported RferORs from the RPW antennae. This is particularly true for RferOrco (or RferSOrco in this study), which was grouped in the highly conserved clade for Orco (Soffan et al., 2016). The olfactory role of the RferOrco and its antennal-specific expression have been reported through tissue-specific expression studies and knock-down through RNAi, its encoding gene, plus electroantennographic and behavioral assays (Soffan et al., 2016). In that sense, the identification of RferSOrco in this study supports the chemosensory role of the RPW rostrum. Even though reported an exclusive antennal-specific expression of RferOrco (Soffan et al., 2016), we could successfully mine RferOrco transcripts from the R. ferrugineus rostrum (RferSOrco) from both male and female transcriptome. Nevertheless, in palm weevil antennal OR annotation (Antony et al., 2016; Gonzalez et al., 2021), Orco was the most highly expressing candidate among all ORs, followed by the PR (Antony et al., 2021). However, in R. ferrugineus rostrum, meager Orco expression we found in both males and females (1.49 and 1.25, respectively).
Compared to the expression of the RPW ORs in the antenna, the rostrum OR expression was relatively low, as indicated by the highest RPKM value of 5 and 6 (female and male) for RferSOR1 (identical to RferOR21, and RPKM was 38). In contrast, in the antennae, the highest RPKM was 68 (RferOR1, pheromone receptor) (Antony et al., 2021). RferOR21 (RferSOR1) exhibits ubiquitous expression and is the third highly expressed OR in R. ferrugineus (Antony et al., 2021). Hence, it is assumed that an important odorant detection may likely be vital for palm weevil fitness and survival, suggesting an essential role of RferSOR1. The RferSOR10 and 17 (identical to RferOR60 and RferOR46) were the only rostrum-related transcripts close to the PR clade of RPW, where RferOR1 is present (Figure 6). A manual annotation of RferSOR10 and RferSOR17 along with RferOR1 in the RPW genome revealed these three ORs clustered in the different scaffolds, viz.; 66430, 66071, and 63 (Genbank: JAACXV010014611.1, JAACXV010014285.1, and JAACXV010000062.1 respectively), representing potentially divergent functions. Similarly, we mapped another functionally characterized R. ferrugineus OR, RferOR41 (identical to RferSOR12), tuned to non-host plant volatile, α-pinene (Ji et al., 2021) and found a cluster in the scaffold 66403 (GenBank: JAACXV010014584.1). It is worth mentioning that the most recently reported R. ferrugineus OR (RferOR2) that is tuned to natural palm-emitted odors (Antony et al., 2023) was reported to be an antennae-specific found that comes under this clade (Figure 6). Interestingly, RferSOR13 appeared located near ItypOR46 and 49 (subfamily 7), reported as receptors for enantiomers of pheromones ipsenol (2-methyl-6-methylen-7-octen-4-ol) and ipsdienol (2-methyl-6-methylen2,7-octadien-4-ol) (Yuvaraj et al., 2021). Similarly, RferSOR13 is near ItypOR23, 25, 27, and 29, also present in the subfamily 7, and reported to respond to (+)-trans-(1R,4S)-4-thujanol, (+)-3-carene, p-cymene and (+)-isopinocamphone, respectively (Hou et al., 2021). Furthermore, the synthesis and release of the aggregation pheromone through the rostrum has been well described in R. palmarum, being phylogenetically close species to the RPW (Wattanapongsiri, 1965; Sánchez et al., 1996), which appears to happen after weevils detect ethyl acetate. Thus, it is likely that the identified RferSORs might be broadly tuned to several classes of plant volatiles and pheromone components. Recent evidence suggests that ORs might also participate in host plant volatile detection as reported for H. assulta OR31 (HassOR31), which is present and functionally active in ovipositor (Li et al., 2020).
The annotation of our transcriptome data revealed 10 RferSIRs, four likely rostrum specific, and the remaining six IRs also reported from the antennal transcriptome of RPW (Antony et al., 2016). RferSIR4 and 10 (identical to RferIR2728 and RferIR_u1) appeared as orthologs of RpalIR25a and RpalIR8a, and D. melanogaster IR DmelIR25a and 8a, respectively, reported as coreceptors (Benton et al., 2009; Gonzalez et al., 2021; Ni, 2021). Hence, these IRs might function as coreceptors for R. ferrugineus even though its expression in the rostrum was significantly lower than in antennae (Antony et al., 2016; Gonzalez et al., 2021). Interestingly, it was shown that RferSIR1 expression was the highest (RPKM 333 and 75 for female and male) while scanty expression in the antennae of R. palmarum (RPKM: 0.78 and 0.87 for female and male) was found (Gonzalez et al., 2021). Particularly, phylogeny suggests that RferSIR10 may result from gene duplication events, similar to reported adaptations in the cave beetle Speonomus longicornis (Balart-García et al., 2021). Moreover, RferSIRs have likely suffered contraction compared to expansions of D. ponderosae and D. melanogaster IRs (Divergent IRs). Therefore, no RferSIRs were found with potential hygrosensation nor thermosensation, suggesting that rostrum-related IRs could have chemoreceptive functions, such as RferSIR5 and 9.
Our findings suggest substantial olfactory protein transcript abundance differences between field-collected and lab-reared insects. With sex effect aside, transcripts with the highest abundance were present in lab-reared RPWs except for RferSIRs. Besides that, it is clear that laboratory conditions impact olfaction plasticity, which could be influenced by age, feeding, and circadian rhythm states (Sørensen et al., 2012; Gadenne et al., 2016). An example is a difference in odor composition between field- and laboratory-collected desert locusts, Schistocerca gregaria (Torto et al., 2021). This study report that volatiles emitted by S. gregaria depend on sex and developmental stage, making the profile of volatiles from field-collected locusts richer than those collected from the laboratory.
This comprehensive transcriptome analysis represents the first for rostrum structure, providing a repertoire of candidate chemosensory proteins that could participate in secretory processes of aggregation pheromone, namely RferSOR1, RferSOBP1, 2, 3, and 4, among others. Importantly, R. ferrugineus PBPs (RferOBP1768) splice variant or recent duplicate identified from the rostrum (RferOBP14) needs special attention for further functional studies as this OBP may likely have an essential role in pheromone secretion and dispersion. Likewise, this study has expanded the profile of chemosensory proteins for R. ferrugineus, serving as an entry dataset for genome-wide analysis that could reveal the entire profile of olfactory proteins in this palm pest. Finally, further functional studies on these proteins will significantly contribute to our understanding of the rostrum’s role during RPW infestation, allowing improved integrated pest management strategies.
Conclusion
The red palm weevil, R. ferrugineus, is a harmful pest for palm trees, where olfaction through antennal organs plays a major role in host finding. However, the rostrum, an important sensory organ in weevils, has not been studied. Ninety-three proteins related to olfaction were identified from rostrum transcriptomes, supporting an olfactory role of the rostrum. Among olfactory-related proteins, those for odorant transport appeared highly abundant, with one sharing the same evolutionary origin as a known pheromone transport protein. Differentially expressed genes between females and males were found. We identified up-regulated and male/female-biased transcripts in the weevils reared in laboratory conditions compared with those collected from the field was established. Further functional studies in the context of pest control are discussed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The red palm weevil collections were made with the direct permission of a cooperating land owner [Al-Kharj region (24.1500° N, 47.3000° E), Al Qassim (25.8275° N, 42.8638° E) of Saudi Arabia] and weevil culture was maintained in our laboratory as mentioned in the materials and methods.
Author contributions
BA, MA, and AP conceived the study and acquired the grant. BA, HA, and MA participated in the palm weevil collection and rearing. BA and AP were involved in high throughput sequencing. JJ and BA performed the bioinformatic analysis. HV, BA, IA, and PL were involved in data interpretation, writing—original draft preparation, review, and editing. All authors have read and agreed to the submitted version of the manuscript.
Funding
The authors extend their appreciation to the Deanship of Scientific Research, King Saud University, for funding through the Vice Deanship of Scientific Research Chairs, Chair of Date Palm Research. This work was financially supported through research grants from King Abdullah University of Science and Technology (KAUST) in Saudi Arabia (KAUST-OSR-2018-RPW-3816) to BA and AP, and the faculty baseline fund (BAS/1/1020-01-01) to AP. This work was supported by funds received from Slovenian Research Agency (P4-0077) to JJ.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research, King Saud University, for funding through the Vice Deanship of Scientific Research Chairs, Chair of Date Palm Research. We thank KAUST Bioscience Core laboratory personnel for their support in producing the raw sequence data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1159142/full#supplementary-material
References
Abuin L., Bargeton B., Ulbrich M. H., Isacoff E. Y., Kellenberger S., Benton R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. doi: 10.1016/j.neuron.2010.11.042
Al-Ayedh H. (2008). Evaluation of date palm cultivars for rearing the red date palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Florida Entomologist 91, 353–358. doi: 10.1653/0015-4040(2008)91[353:EODPCF]2.0.CO;2
Andersson M. N., Grosse-Wilde E., Keeling C. I., Bengtsson J. M., Yuen M., Li M., et al. (2013). Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics 14, 1–16. doi: 10.1186/1471-2164-14-198
Andersson M. N., Keeling C. I., Mitchell R. F. (2019). Genomic content of chemosensory genes correlates with host range in wood-boring beetles (Dendroctonus ponderosae, Agrilus planipennis, and Anoplophora glabripennis). BMC Genomics 20, 1–18. doi: 10.1186/s12864-019-6054-x
Antony B., Johny J., Aldosari S. A. (2018). Silencing the odorant binding protein RferOBP1768 reduces the strong preference of palm weevil for the major aggregation pheromone compound ferrugineol. Front. Physiol. 9, 252. doi: 10.3389/fphys.2018.00252
Antony B., Johny J., Montagné N., Jacquin-Joly E., Capoduro R., Cali K., et al. (2021). Pheromone receptor of the globally invasive quarantine pest of the palm tree, the red palm weevil (Rhynchophorus ferrugineus). Mol. Ecol. 30, 2025–2039. doi: 10.1111/mec.15874
Antony B., Montagne N., Comte A., Mfarrej S., Jakse J., Capoduro R., et al. (2023). Reverse chemical ecology approach for sustainable palm tree protection against invasive palm weevils. bioRxiv, 2023.01.13.523742. doi: 10.1101/2023.01.13.523742
Antony B., Soffan A., Jakše J., Abdelazim M. M., Aldosari S. A., Aldawood A. S., et al. (2016). Identification of the genes involved in odorant reception and detection in the palm weevil Rhynchophorus ferrugineus, an important quarantine pest, by antennal transcriptome analysis. BMC Genomics 17, 1–22. doi: 10.1186/s12864-016-2362-6
Antony B., Soffan A., Jakše J., Alfaifi S., Sutanto K. D., Aldosari S. A., et al. (2015). Genes involved in sex pheromone biosynthesis of Ephestia cautella, an important food storage pest, are determined by transcriptome sequencing. BMC Genomics 16, 1–27. doi: 10.1186/s12864-015-1710-2
Balart-García P., Cieslak A., Escuer P., Rozas J., Ribera I., Fernández R. (2021). Smelling in the dark: phylogenomic insights into the chemosensory system of a subterranean beetle. Mol. Ecol. 30, 2573–2590. doi: 10.1111/mec.15921
Benton R., Vannice K. S., Gomez-Diaz C., Vosshall L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Bin S.-Y., Qu M.-Q., Pu X.-H., Wu Z.-Z., Lin J.-T. (2017). Antennal transcriptome and expression analyses of olfactory genes in the sweetpotato weevil Cylas formicarius. Sci. Rep. 7, 11073. doi: 10.1038/s41598-017-11456-x
Bisch-Knaden S., Rafter M. A., Knaden M., Hansson B. S. (2022). Unique neural coding of crucial versus irrelevant plant odors in a hawkmoth. eLife 11, e77429. doi: 10.7554/eLife.77429.sa2
Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cassau S., Krieger J. (2021). The role of SNMPs in insect olfaction. Cell Tissue Res. 383, 21–33. doi: 10.1007/s00441-020-03336-0
Chen H., Lin L., Xie M., Zhang G., Su W. (2014). De novo sequencing, assembly and characterization of antennal transcriptome of Anomala corpulenta Motschulsky (Coleoptera: Rutelidae). PloS One 9, e114238. doi: 10.1371/journal.pone.0114238
Croset V., Rytz R., Cummins S. F., Budd A., Brawand D., Kaessmann H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PloS Genet. 6, e1001064. doi: 10.1371/journal.pgen.1001064
Davis S. R. (2017). The weevil rostrum (Coleoptera: Curculionoidea): internal structure and evolutionary trends. Bull. Am. Mus. Nat. Hist. 2017, 1–76. doi: 10.1206/0003-0090-416.1.1
Dias G. B., Altammami M. A., El-Shafie H. A., Alhoshani F. M., Al-Fageeh M. B., Bergman C. M., et al. (2021). Haplotype-resolved genome assembly enables gene discovery in the red palm weevil Rhynchophorus ferrugineus. Sci. Rep. 11, 9987. doi: 10.1038/s41598-021-89091-w
Ding W. F., Wang C. Y., Zhong J., Zhang L. M., He Z., Sun L., et al. (2023). RNA Sequencing and transcriptome analyses reveal differentially expressed genes in the defensive glands of the medicinal beetle Blaps rhynchopetera. Entomol. Res. 53, 12–28. doi: 10.1111/1748-5967.12629
Engsontia P., Satasook C. (2021). Genome-wide identification of the gustatory receptor gene family of the invasive pest, red palm weevil, Rhynchophorus ferrugineus (Olivier 1790). Insects 12, 611. doi: 10.3390/insects12070611
Eyun S.-I., Soh H. Y., Posavi M., Munro J. B., Hughes D. S., Murali S. C., et al. (2017). Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol. Biol. Evol. 34, 1838–1862. doi: 10.1093/molbev/msx147
Faleiro J. (2006). A review of the issues and management of the red palm weevil Rhynchophorus ferrugineus (Coleoptera: Rhynchophoridae) in coconut and date palm during the last one hundred years. Int. J. Trop. Insect Sci. 26, 135–154. doi: 10.1079/IJT2006113
Frank D. D., Enjin A., Jouandet G. C., Zaharieva E. E., Para A., Stensmyr M. C., et al. (2017). Early integration of temperature and humidity stimuli in the Drosophila brain. Curr. Biol. 27, 2381–2388. doi: 10.1016/j.cub.2017.06.077
Gadenne C., Barrozo R. B., Anton S. (2016). Plasticity in insect olfaction: to smell or not to smell? Annu. Rev. Entomol. 61, 317–333. doi: 10.1146/annurev-ento-010715-023523
Gonzalez F., Johny J., Walker Iii W. B., Guan Q., Mfarrej S., Jakše J., et al. (2021). Antennal transcriptome sequencing and identification of candidate chemoreceptor proteins from an invasive pest, the American palm weevil, Rhynchophorus palmarum. Sci. Rep. 11, 8334. doi: 10.1038/s41598-021-87348-y
Gu S.-H., Wu K.-M., Guo Y.-Y., Pickett J. A., Field L. M., Zhou J.-J., et al. (2013). Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics 14, 1–22. doi: 10.1186/1471-2164-14-636
Hoang D. T., Chernomor O., Von Haeseler A., Minh B. Q., Vinh L. S., Evolution (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. 35, 518–522. doi: 10.1093/molbev/msx281
Hoddle M. S., Antony B., El-Shafie H. F., Chamorro M. L., Milosavljević I., Löhr B., et al. (2024). Taxonomy, biology, symbionts, omics, and management of Rhynchophorus palm weevils (Coleoptera: Curculionidae: Pryophthorinae). Annu. Rev. Entomol. 69. (In press). doi: 10.1146/annurev-ento-013023-121139
Hou X.-Q., Yuvaraj J. K., Roberts R. E., Zhang D.-D., Unelius C. R., Löfstedt C., et al. (2021). Functional evolution of a bark beetle odorant receptor clade detecting monoterpenoids of different ecological origins. Mol. Biol. Evol. 38, 4934–4947. doi: 10.1093/molbev/msab218
Hou X.-Q., Zhang D.-D., Powell D., Wang H.-L., Andersson M. N., Löfstedt C. (2022). Ionotropic receptors in the turnip moth Agrotis segetum respond to repellent medium-chain fatty acids. BMC Biol. 20, 1–19. doi: 10.1186/s12915-022-01235-0
Hu P., Wang J., Cui M., Tao J., Luo Y. (2016). Antennal transcriptome analysis of the Asian longhorned beetle Anoplophora glabripennis. Sci. Rep. 6, 1–12. doi: 10.1038/srep26652
Jacquin-Joly E., Vogt R. G., François M.-C., Nagnan-Le Meillour P. (2001). Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Sens. 26, 833–844. doi: 10.1093/chemse/26.7.833
Ji T., Xu Z., Jia Q., Wang G., Hou Y. (2021). Non-palm plant volatile α-pinene is detected by antenna-biased expressed odorant receptor 6 in the Rhynchophorus ferrugineus (Olivier)(Coleoptera: Curculionidae). Front. Physiol. 12, 701545. doi: 10.3389/fphys.2021.701545
Jiang X.-J., Ning C., Guo H., Jia Y.-Y., Huang L.-Q., Qu M.-J., et al. (2015). A gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect Biochem. Mol. Biol. 60, 39–46. doi: 10.1016/j.ibmb.2015.03.002
Kalyaanamoorthy S., Minh B. Q., Wong T. K., Von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 20, 1160–1166. doi: 10.1093/bib/bbx108
Klein U. (1987). Sensillum-lymph proteins from antennal olfactory hairs of the moth Antheraea polyphemus (Saturniidae). Insect Biochem. Mol. Biol. 17, 1193–1204. doi: 10.1016/0020-1790(87)90093-X
Knecht Z. A., Silbering A. F., Cruz J., Yang L., Croset V., Benton R., et al. (2017). Ionotropic receptor-dependent moist and dry cells control hygrosensation in Drosophila. elife 6, e26654. doi: 10.7554/eLife.26654.013
Knecht Z. A., Silbering A. F., Ni L., Klein M., Budelli G., Bell R., et al. (2016). Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in drosophila. elife 5, e17879. doi: 10.7554/eLife.17879.011
Larsson M. C., Domingos A. I., Jones W. D., Chiappe M. E., Amrein H., Vosshall L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Lartigue A., Campanacci V., Roussel A., Larsson A. M., Jones T. A., Tegoni M., et al. (2002). X-Ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 277, 32094–32098. doi: 10.1074/jbc.M204371200
Leal W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Li R.-T., Huang L.-Q., Dong J.-F., Wang C.-Z. (2020). A moth odorant receptor highly expressed in the ovipositor is involved in detecting host-plant volatiles. elife 9. doi: 10.7554/eLife.53706
Li J., Lehmann S., Weissbecker B., Ojeda Naharros I., Schütz S., Joop G., et al. (2013). Odoriferous defensive stink gland transcriptome to identify novel genes necessary for quinone synthesis in the red flour beetle, Tribolium castaneum. PloS Genet. 9, e1003596. doi: 10.1371/journal.pgen.1003596
Li K., Wei H., Shu C., Zhang S., Cao Y., Luo C., et al. (2017). Identification and comparison of candidate odorant receptor genes in the olfactory and non-olfactory organs of Holotrichia oblita faldermann by transcriptome analysis. Comp. Biochem. Physiol. Part D: Genomics Proteomics 24, 1–11. doi: 10.1016/j.cbd.2017.07.001
Li Z.-Q., Zhang S., Luo J.-Y., Wang C.-Y., Lv L.-M., Dong S.-L., et al. (2015). Transcriptome comparison of the sex pheromone glands from two sibling Helicoverpa species with opposite sex pheromone components. Sci. Rep. 5, 1–11. doi: 10.1038/srep09324
Liu N.-Y., Li Z.-B., Zhao N., Song Q.-S., Zhu J.-Y., Yang B. (2018). Identification and characterization of chemosensory gene families in the bark beetle, Tomicus yunnanensis. Comp. Biochem. Physiol. Part D: Genomics Proteomics 25, 73–85. doi: 10.1016/j.cbd.2017.11.003
Liu S., Rao X.-J., Li M.-Y., Feng M.-F., He M.-Z., Li S.-G. (2015). Identification of candidate chemosensory genes in the antennal transcriptome of Tenebrio molitor (Coleoptera: Tenebrionidae). Comp. Biochem. Physiol. Part D: Genomics Proteomics 13, 44–51. doi: 10.1016/j.cbd.2015.01.004
Liu X.-L., Sun S.-J., Hou W., Zhang J., Yan Q., Dong S.-L. (2020). Functional characterization of two spliced variants of fructose gustatory receptor in the diamondback moth, Plutella xylostella. Pesticide Biochem. Physiol. 164, 7–13. doi: 10.1016/j.pestbp.2019.12.002
Liu X.-L., Yan Q., Yang Y.-L., Hou W., Miao C.-L., Peng Y.-C., et al. (2019). A gustatory receptor GR8 tunes specifically to d-fructose in the common cutworm Spodoptera litura. Insects 10, 272. doi: 10.3390/insects10090272
Maida R., Steinbrecht A., Ziegelberger G., Pelosi P. (1993). The pheromone binding protein of bombyx mori: purification, characterization and immunocytochemical localization. Insect Biochem. Mol. Biol. 23, 243–253. doi: 10.1016/0965-1748(93)90005-D
Maleszka J., Forêt S., Saint R., Maleszka R. (2007). RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 217, 189–196. doi: 10.1007/s00427-006-0127-y
Mitchell R. F., Hughes D. T., Luetje C. W., Millar J. G., Soriano-Agatón F., Hanks L. M., et al. (2012). Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 42, 499–505. doi: 10.1016/j.ibmb.2012.03.007
Mitchell R., Schneider T., Schwartz A., Andersson M. N., Mckenna D. (2020). The diversity and evolution of odorant receptors in beetles (Coleoptera). Insect Mol. Biol. 29, 77–91. doi: 10.1111/imb.12611
Mortazavi A., Williams B. A., Mccue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Ni L. (2021). The structure and function of ionotropic receptors in Drosophila. Front. Mol. Neurosci. 13, 638839. doi: 10.3389/fnmol.2020.638839
Ni L., Klein M., Svec K. V., Budelli G., Chang E. C., Ferrer A. J., et al. (2016). The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. elife 5, e13254. doi: 10.7554/eLife.13254.015
Pelosi P., Iovinella I., Zhu J., Wang G., Dani F. R. (2018). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 93, 184–200. doi: 10.1111/brv.12339
Richards S., Gibbs R. A., Weinstock G. M., Brown S. J., Denell R., Beeman R. W., et al. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955. doi: 10.1038/nature06784
Robertson H. M., Warr C. G., Carlson J. R. (2003). Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. 100, 14537–14542. doi: 10.1073/pnas.2335847100
Rozziansha T., Hidayat P., Harahap I. (2021). “Morphological characters of Rhynchophorus spp.(Coleoptera: Curculionidae) associated with sago, coconut, and oil palm in Indonesia,” in IOP Conference Series: Earth and Environmental Science. (Bogor, Indonesia: IOP Publishing), 694, 012051. Available at: https://iopscience.iop.org/journal/1755-1315.
Salem S., Abd El-Salam A., El-Kholy M. (2016). Field evaluation of red palm weevil Rhynchophorus ferrugineus Oliv.(Coleoptera: Curculionidae) responses to its fermenting date tree volatiles. Int. J. ChemTech Res. 9, 12–17.
Sánchez P., Cerda H., Cabrera A., Caetano F., Materán M., Sánchez F., et al. (1996). Secretory mechanisms for the male produced aggregation pheromone of the palm weevil Rhynchophorus palmarum L. (Coleoptera: Curculionidae). J. Insect Physiol. 42, 1113–1119. doi: 10.1016/S0022-1910(96)00039-X
Sato K., Pellegrino M., Nakagawa T., Nakagawa T., Vosshall L. B., Touhara K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Schoville S. D., Chen Y. H., Andersson M. N., Benoit J. B., Bhandari A., Bowsher J. H., et al. (2018). A model species for agricultural pest genomics: the genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 8, 1931. doi: 10.1038/s41598-018-20154-1
Soffan A., Antony B., Abdelazim M., Shukla P., Witjaksono W., Aldosari S. A., et al. (2016). Silencing the olfactory co-receptor RferOrco reduces the response to pheromones in the red palm weevil, Rhynchophorus ferrugineus. PloS One 11, e0162203. doi: 10.1371/journal.pone.0162203
Sørensen J. G., Addison M. F., Terblanche J. S. (2012). Mass-rearing of insects for pest management: challenges, synergies and advances from evolutionary physiology. Crop Prot. 38, 87–94. doi: 10.1016/j.cropro.2012.03.023
Sun H., Guan L., Feng H., Yin J., Cao Y., Xi J., et al. (2014). Functional characterization of chemosensory proteins in the scarab beetle, Holotrichia oblita Faldermann (Coleoptera: Scarabaeida). PLoS One 9. doi: 10.1371/journal.pone.0107059
Torto B., Kirwa H., Kihika R., Niassy S. (2021). Odor composition of field versus laboratory desert locust populations. J. Insect Physiol. 134, 104296. doi: 10.1016/j.jinsphys.2021.104296
Trifinopoulos J., Nguyen L.-T., Von Haeseler A., Minh B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Venthur H., Zhou J.-J. (2018). Odorant receptors and odorant-binding proteins as insect pest control targets: a comparative analysis. Front. Physiol. 9, 1163. doi: 10.3389/fphys.2018.01163
Vogt R. G., Riddiford L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi: 10.1038/293161a0
Wang R., Li F., Zhang W., Zhang X., Qu C., Tetreau G., et al. (2017). Identification and expression profile analysis of odorant binding protein and chemosensory protein genes in Bemisia tabaci MED by head transcriptome. PloS One 12, e0171739. doi: 10.1371/journal.pone.0171739
Wattanapongsiri A. (1965). A revision to the Genera rhynchophorus and Dynamis (Coleoptera: Curculionidae) (Oregon, USA: Oregon State University).
Wen X., Wang Q., Gao P., Wen J. (2018). Identification and comparison of chemosensory genes in the antennal transcriptomes of Eucryptorrhynchus scrobiculatus and E. brandti fed on Ailanthus altissima. Front. Physiol. 9, 1652. doi: 10.3389/fphys.2018.01652
Wicher D. (2018). Tuning insect odorant receptors. Front. Cell. Neurosci. 12, 94. doi: 10.3389/fncel.2018.00094
Wicher D., Schäfer R., Bauernfeind R., Stensmyr M. C., Heller R., Heinemann S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Yan W., Liu L., Qin W., Luo Y., Ma X., Haider N., et al. (2016). Identification and tissue expression profiling of odorant binding protein genes in the red palm weevil, Rhynchophorus ferrugineus. SpringerPlus 5, 1–8. doi: 10.1186/s40064-016-3198-x
Yuvaraj J. K., Roberts R. E., Sonntag Y., Hou X.-Q., Grosse-Wilde E., Machara A., et al. (2021). Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 19, 1–21. doi: 10.1186/s12915-020-00946-6
Zhang H.-J., Anderson A. R., Trowell S. C., Luo A.-R., Xiang Z.-H., Xia Q.-Y. (2011). Topological and functional characterization of an insect gustatory receptor. PloS One 6, e24111. doi: 10.1371/journal.pone.0024111
Zhang H.-J., Xu W., Chen Q.-M., Sun L.-N., Anderson A., Xia Q.-Y., et al. (2020). A phylogenomics approach to characterizing sensory neuron membrane proteins (SNMPs) in Lepidoptera. Insect Biochem. Mol. Biol. 118, 103313. doi: 10.1016/j.ibmb.2020.103313
Zhou J.-J. (2010). Odorant-binding proteins in insects. Vitamins Hormones 83, 241–272. doi: 10.1016/S0083-6729(10)83010-9
Zhou X.-H., Ban L.-P., Iovinella I., Zhao L.-J., Gao Q., Felicioli A., et al. (2013). Diversity, abundance, and sex-specific expression of chemosensory proteins in the reproductive organs of the locust Locusta migratoria manilensis. Biol. Chem. 394, 43–54. doi: 10.1515/hsz-2012-0114
Zhou J.-J., Kan Y., Antoniw J., Pickett J. A., Field L. M. (2006). Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem. Sens. 31, 453–465. doi: 10.1093/chemse/bjj050
Keywords: Curculionidae, palm weevil, snout transcriptome, aggregation pheromone, olfaction, chemosensory proteins
Citation: Venthur H, Arias I, Lizana P, Jakše J, Alharbi HA, Alsaleh MA, Pain A and Antony B (2023) Red palm weevil olfactory proteins annotated from the rostrum provide insights into the essential role in chemosensation and chemoreception. Front. Ecol. Evol. 11:1159142. doi: 10.3389/fevo.2023.1159142
Received: 05 February 2023; Accepted: 03 July 2023;
Published: 25 July 2023.
Edited by:
Alberto Maria Cattaneo, Swedish University of Agricultural Sciences, SwedenReviewed by:
Patrizia Falabella, University of Basilicata, ItalyDan-Dan Zhang, Lund University, Sweden
Copyright © 2023 Venthur, Arias, Lizana, Jakše, Alharbi, Alsaleh, Pain and Antony. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binu Antony, YmFudG9ueUBrc3UuZWR1LnNh
†ORCID: Binu Antony, orcid.org/0000-0002-6292-620X
 Herbert Venthur
Herbert Venthur Ignacio Arias
Ignacio Arias Paula Lizana
Paula Lizana Jernej Jakše3
Jernej Jakše3 Hatten A. Alharbi
Hatten A. Alharbi Arnab Pain
Arnab Pain Binu Antony
Binu Antony