- 1Faculty of Health and Life Sciences, Applied Sciences, Northumbria University, Newcastle upon Tyne, United Kingdom
- 2Department of Health Sciences, University of Eastern Piedmont, Novara, Italy
- 3Forensic Anthropology Center, Texas State University, San Marcos, TX, United States
- 4Faculty of Humanities, University of Amsterdam, Amsterdam, Netherlands
- 5School of Natural Sciences, University of Central Lancashire, Preston, United Kingdom
Introduction: Human donations are often used in forensic research as they can provide unique insights into post-mortem research that cannot be obtained with animal proxies. This is especially true for forensic microbiome research, as human circumstances such as drug-use or health conditions may influence the post-mortem microbiome. However, it is not always feasible to conduct such research immediately after death. Donors are often stored frozen in human taphonomy facilities, pending the start of any experimental study, yet little is known about how freezing may affect their microbiome.
Methods: We assessed the effects of freezing on the post-mortem human microbiome by analysing the microbial diversity and abundance of seven human donors at the Forensic Anthropology Center at Texas State (FACTS) before and after freezing. Swab samples were taken from five locations on each corpse upon arrival to FACTS and again after they had been frozen in storage for a period ranging between 11 and 40 days and subsequently thawed.
Results: After performing the microbiome analysis of the swabs via 16S rRNA gene metabarcoding, we found changes in the abundance levels of Proteobacteria, Bacteroidota and Firmicutes, as well as the presence of the new phyla Deinococcota and Myxococcota after freezing. However, none of these changes were significant when comparing community diversity before and after freezing.
Discussion: Overall, our results show that the observed changes in the abundance of specific phyla before and after freezing are negligible, that freezing does not significantly alter the human microbiome and that frozen donors are suitable for forensic studies on the human thanatomicrobiome.
Introduction
The use of human bodies in research dates as far back as the 16th century, with the dissection of cadavers for understanding the human anatomy (Varner et al., 2021). Consequently, human corpses have been integrated into teaching for training medical practitioners (Habicht et al., 2018; Holm and Iaizzo, 2019). The study of human cadavers has also been adopted in other disciplines, including forensics – for the purpose of developing and validating new methods to assist forensic experts in death investigations (Gunderman and Wilson, 2005; Shin and Meals, 2005; Turney, 2007; Anyanwu, 2010; Varlet et al., 2020).
Forensic taphonomy, which aims to understand human (or animal, when used as proxies) decomposition over time (Miles et al., 2020; Martin et al., 2021), is a key area of forensic research that more often has utilized human donors for essential research. The importance of such research has been cited in multiple publications (Iqbal et al., 2018; Miles et al., 2020), as this can provide answers to fundamental questions regarding post-mortem interval (PMI) or post-burial intervals, as well as the cause and/or circumstances of death (Miles et al., 2020). Incorporating human donors in forensic taphonomy research can be vital to reaching conclusions and informing standard practice for PMI estimation (Fancher et al., 2017; Matuszewski et al., 2020), discovery of clandestine depositions (Pringle et al., 2020; Doro et al., 2022) and training human remains detection dogs (Komar, 1999; Cablk et al., 2012). This is particularly crucial, as animal replacements may not fully represent true decomposition processes seen in humans (Williams et al., 2019b; Matuszewski et al., 2020). Thus, human taphonomy facilities – where human donors are studied for the purpose of forensic research (Williams et al., 2019a,b) – have been established in different countries for these purposes, among others, and have become centres for multidisciplinary research.
One of such areas of research involves the study of the microbial communities existing on and within human donors, and their related environments during decomposition. Microbiome investigations in forensic human taphonomy studies primarily focus on examining bacterial (and other microbial) succession during human decomposition for predicting PMI or cause of death. However, significant changes to microbial communities and their interaction networks can be driven by even small changes to temperature (Carter et al., 2008; Metcalf, 2019; Burman and Bengtsson-Palme, 2021), setting back our understanding of these succession patterns. Thus, recommendations for sampling in microbiome studies – forensic or otherwise – often suggest the immediate extraction of microbial DNA or the preservation of samples when immediate extraction is not possible (Lauber et al., 2010).
Sample preservation for stabilizing the microbiome often involves storing samples at below freezing temperatures, among other practices. Freezing at −80°C is often considered the gold standard for long-term preservation of microbiome-related samples, particularly fecal matter (Choo et al., 2015; Vandeputte et al., 2017). However, immediate preservation of samples at this temperature is not always feasible, especially at scenes of forensic interest. Hence, some studies have assessed the effect of freezing samples – soil (Bhattacharjee et al., 2021; Ouyang et al., 2021), fecal matter (Choo et al., 2015), saliva (Furuhashi et al., 2022) – at various temperatures and the influence on their associated microbiomes, with conflicting results. Even with such inconsistent findings, it is common practice to freeze these samples, frequently at −20°C, to limit microbial activity or proliferation.
Similarly at human taphonomy facilities, preserving donors by freezing prior to the commencement of decomposition research is relatively common practice, particularly in situations where multiple bodies must be deposited at the same time for decomposition studies involving more than one corpse. Thus, freezing is often used to halt or slow decomposition, and therefore maintain the bodies at a PMI hypothetically close to zero. In meat science studies, it has generally been determined that freezing and thawing do not effectively change the number of viable microbes present in meat (Vieira et al., 2009; Leygonie et al., 2012; da Silva Bernardo et al., 2020). However, it must be noted that these studies primarily assess microbial presence through culture studies. Yet there is information on negative effects of non-optimal freezing due to changes in osmotic pressure (Baust et al., 2009; Li et al., 2018). This can result in the rupture of cells through damaged cell walls which could release sequestered microbes. While the impact of freezing on the microbiome of forensic research samples has been explored, there is no information – to the best of our knowledge – on how freezing whole human donors may impact their microbiome.
Understanding any effects of freezing is vital for forensic human taphonomy research, as shifts to the microbial community composition and abundance prior to the start of a study may influence the results and therefore the interpretation of results. Previously, studies have aimed to understand decomposition related changes to microbial activity in cases where animal carcasses or sections of tissues have been frozen prior to burial. One of such studies reported that freezing plays a significant role in the type of decomposition observed – aerobic decomposition in previously frozen animal cadavers and anaerobic in fresh cadavers (Micozzi, 1986). While it relied on the measurement of carbon dioxide respiration to determine microbial activity, another study concluded that freezing had no significant effect on the microbial activity in surrounding burial soil (Stokes et al., 2009).
Nonetheless, it is essential to also examine any freezing effects immediately after freeze-thaw process and prior to decomposition. This is especially important after the publication of a case study that showed a shift in post-mortem microbial community abundance and structure during the thawing of two pediatric cases (Pechal et al., 2017). While the sample size for the study was limited, their observation called for an in-depth look at the effects of freezing donors and what it might mean for microbiome research in forensic taphonomy.
The purpose of this study therefore was to determine if the storage of donors below freezing at human taphonomy facilities may have any effect on their microbiome. Seven donated bodies at the Forensic Anthropology Center at Texas State (FACTS) were swabbed before they were frozen and after the complete thawing of the bodies. It was also assessed if the freezing of the bodies may have varying effects at different anatomical locations (n = 5), and how the microbial composition at these locations may differ from one another post-mortem.
Materials and methods
Sampling
This research was organized and carried out in collaboration with FACTS as a part of the “Mass Grave Project,” an interdisciplinary taphonomic research project aimed at improving the detection, excavation and documentation of clandestine mass and single graves (Mickleburgh et al., 2022). Whole body donations studied at FACTS are acquired for scientific research purposes, through the expressed and documented willing of the donors and/or their legal next of kin. All human decedents involved in this study were collected through a body donation program which complies with all legal and ethical standards associated with the use of human remains for scientific research in the USA (Gocha et al., 2021). The sampling of tissues from the body donors in the Mass Grave Project was conducted in accordance with the Texas Uniform Anatomical Gift Act.1 Ethical approval for analysis of the samples at Northumbria University was provided by the Northumbria ethics committee (ref: 21514). Nine body donations were received for the Mass Grave Project, however, microbiome analysis for the present study was conducted on only seven of these donors for which both pre- and post-freezing samples were available.
Initial samples were collected upon arrival of the donors at the research facility, either immediately or after a period during which bodies were kept refrigerated (4°C) due to technical reasons (Table 1). Swab samples, collected in triplicates, were taken from five anatomical locations of each corpse. Areas sampled included hand, foot, neck, oral cavity, and rectum.
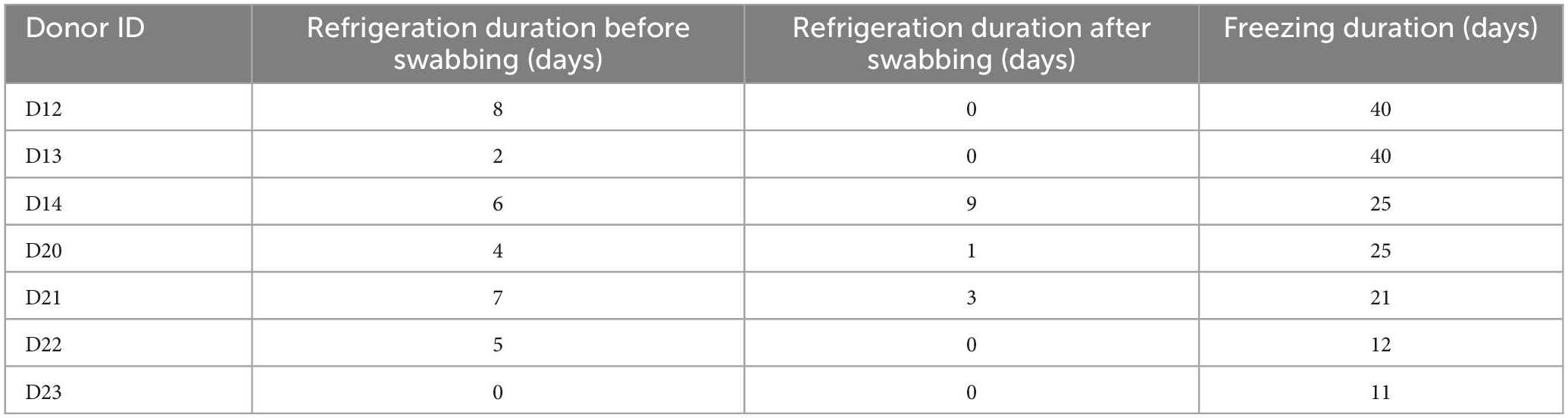
Table 1. Duration of refrigeration (+4°C) between arrival to the facility and swabbing, between swabbing and freezing of the donors, and duration of freezing (−20°C) of the donors.
Hand and foot samples were taken from the dominant limb as identified by the donor information provided. Swabs were collected using sterile swabs by rubbing the swab at the sampling location (i.e., hand, foot, neck, oral cavity, or rectum) for 15 or 30 s, depending on the location. Hand or foot swabs sampled the palm and fingers or sole and toes respectively for 15 s. Neck samples were taken at the back, in the area between the base of the head and the top of the shoulder, by swabbing back-and-forth for 15 s.
For the oral cavity, samples were collected from the left cheek, right cheek, and oral vestibule, including any teeth if available, by swabbing the aforementioned areas using the same swab for 15 s. Finally, rectal samples were obtained by inserting the swab into the anus to about 5 – 10 cm into the rectum, and then rotating for 30 s.
After initial sampling as described, swab samples were stored immediately at −20°C. Bodies were frozen either immediately post-swabbing, or after a short period of time during which they were kept refrigerated (Table 1) due to technical reasons. Due to the differing arrival times of the donors to the facility, the total time spent frozen in storage varied among them. However, all bodies remained in storage at −20°C for a minimum of period of 11 days and a maximum of 40 days (Table 1). For ease of reference, all donors were assigned a reference label – D12, D13, D14, D20, D21, D22, and D23.
After the freezing period, donors were removed from storage and thawed. Donor bodies were frozen and defrosted within a closed and controlled indoor laboratory (21°C) to prevent insect access. Thawing began at about 7:00 pm on the 10th of May 2021, and post-freezing sampling commenced on the 12th of May at about 8:30 am, after complete thawing of the bodies. Swab samples were taken again from each body, and location, as previously described. All swabs were immediately stored frozen at −20°C, along with samples taken prior to freezing. All samples were then shipped to Northumbria University for processing.
DNA extraction
Microbial DNA was isolated from the swab samples using the DNeasy 96 PowerSoil Pro Kit from Qiagen (Qiagen, Hilden, Germany) with a modified protocol. 800 uL of Solution CD4 was added to Eppendorf tubes containing swab samples, vortexed for 30 s and centrifuged at 5000 rpm for 10 min. The supernatant was then transferred to the PowerBead Pro Plate, after which the extraction was performed according to the manufacturer’s protocol.
In addition to swab samples, two extraction negatives – one with a sterile swab, and the other using just kit reagents – were included to identify any potential contamination. A positive swab of a bacterial culture – E. coli Top10 plo3 D55N Hox A – was also used to check the quality of microbial DNA extraction.
Quality check analysis was further analyzed qualitatively using PCR products of extracted microbial DNA via gel electrophoresis. This included the extraction controls (negatives – sterile swab, “N” and only kit reagents, “N1” – and positive, “P”) and a few randomly selected samples from the experiment (Supplementary Figure 1). PCR reaction mixtures for the quality check were set up following Procopio et al. (2021) using 2.0 μL template DNA in a final reaction volume of 25 μL. 1.5% (w/v) agarose gel made with 1× TAE buffer was used to check quality of PCR products obtained prior to library preparation and sequencing.
Sequencing and bioinformatic analysis
Library preparation and sequencing of the 16S rRNA gene (V4 region) was analyzed by NU-OMICS (Northumbria University, Newcastle, UK) based on the Schloss wet-lab MiSeq SOP (Kozich et al., 2013). Briefly, PCR was carried out using 1× Accuprime Pfx Supermix, 0.5 μM each primer and 1 μL of template DNA under the following conditions 95°C 2 min, 30 cycles 95°C 20 s, 55°C 15 s, 72°C 5 min with a final extension 72°C 10 min. One positive and one negative control sample were included in each 96 well plate and carried through to sequencing.
PCR products were normalized using SequalPrep™ Normalization kit (Invitrogen) as described in the manufacturer’s instructions and combined into four pools. Each pool was quantified using fragment size determined by BioAnalyzer (Agilent Technologies) and concentration by Qubit (Invitrogen). Pools were combined in equimolar amounts to create a single library then denatured using 0.2N NaOH for 5 min and diluted to a final concentration of 5 pM, supplemented with 15% PhiX and loaded onto a MiSeq V2 500 cycle cartridge (Illumina Inc., San Diego, CA, USA). Targeting and sequencing of the V4 region of the 16S rRNA for bacterial identification were done following the gold standards suggested by the Human Microbiome Project.
Fastq raw sequencing files generated were imported into QIIME2 software ver. 2021.11 for analysis and quality filtered using DADA2 and trimmed at 240 bp forward and 222 bp reverse. Taxonomy was assigned using the naive Bayes classifier, which was trained on silva-138-99 operational taxonomic units (OTUs). Qiime2 output artifacts were analyzed in R (ver. 4.2.2) using the phyloseq package. Abundance and diversity metrics calculated level were assessed by the aggregation of taxa to phylum or genus level. At genus level, unassigned taxa labeled “NA,” “unassigned,” “metagenome,” “uncultured” were excluded to prevent problems in agglomeration due to multiple taxa being assigned the same name.
Results
A total of 35 pairs of swab samples, taken from seven donors at five anatomical locations – hand, neck, foot, oral and rectum – were analyzed in this study. Each pair was composed by a swab sample taken before freezing (“fresh”), and another taken after freezing the body at −20°C (“thawed”). Qualitative analysis to assess successful extraction and amplification of the 16S rRNA gene using gel electrophoresis (Supplementary Figure 1) showed no amplification signals in both negative controls, a strong band for the positive control, and positive amplification for all samples, although one displayed a weak signal.
Following extraction, amplification and sequencing, the resulting data were standardized to the median sampling depth to account for unequal sequencing depth. This allowed inclusion of all samples to prevent waste of data. The Shannon index – accounting for microbial richness and evenness – was used to determine if there were any differences in the alpha diversity of the bacterial communities within the fresh and thawed samples. No significant differences were observed overall between all fresh and thawed samples (Wilcoxon; p = 0.48). Furthermore, analysis of the alpha diversity, also using the Shannon index, between fresh and thawed samples at each sample source (hand, neck, foot, oral and rectum), showed no significant differences (Figure 1).
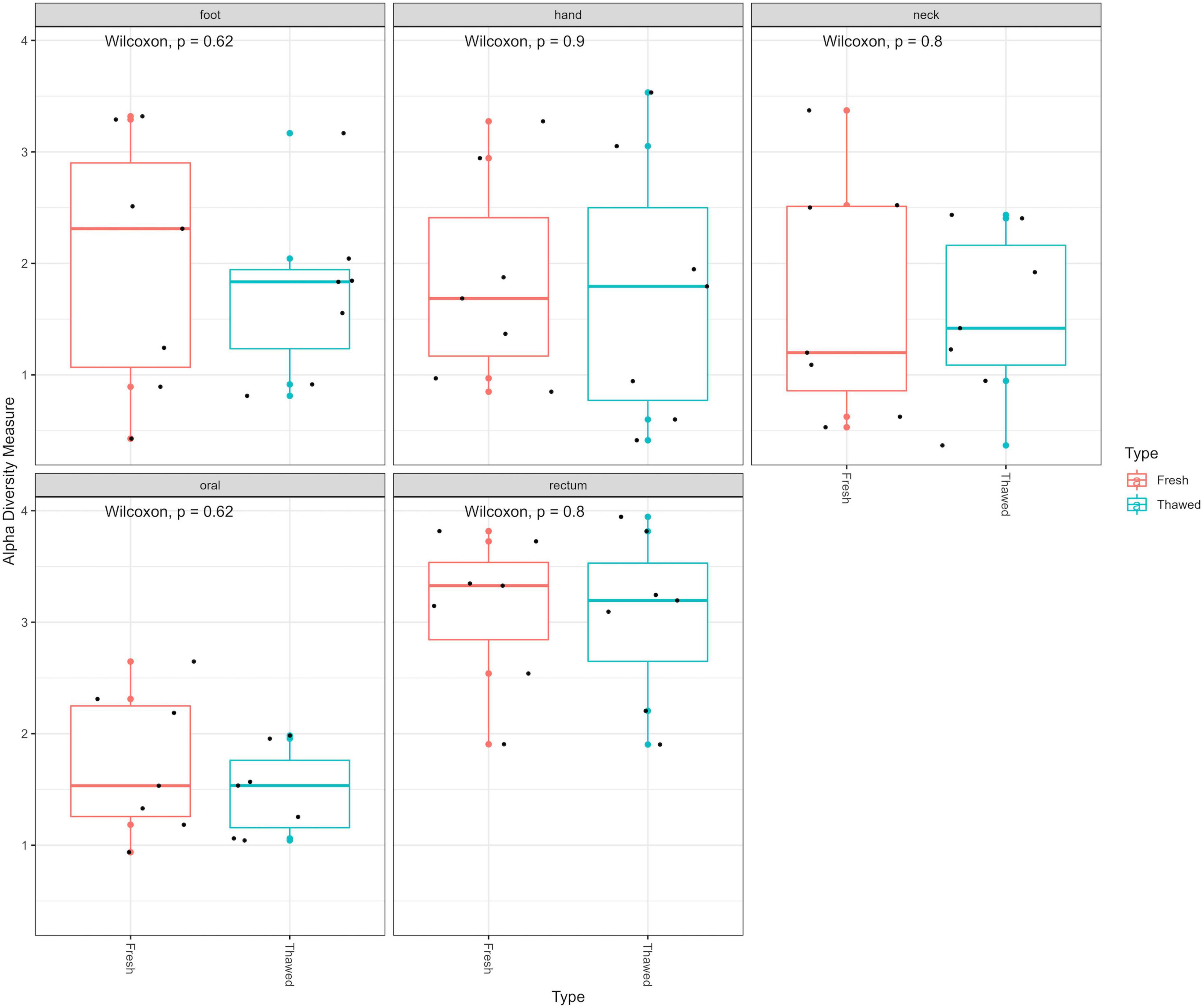
Figure 1. Alpha diversity plot (Shannon diversity) of fresh and thawed samples per anatomical location at ASV level.
The abundance distribution of the bacterial communities in fresh and thawed samples at phylum level was evaluated to detect any microbial compositional changes resulting from the freezing of the donors. Overall, statistical analysis showed that phylum abundances between the two groups were not significantly different. However, Bacteroidota and Firmicutes abundance seemed to decrease in thawed samples, while Proteobacteria appeared to increase after freezing (Figure 2A). At genus level, five taxa Christensenella, Anaerotruncus, Carnobacterium (phylum – Proteobacteria), Acinetobacter, Psychrobacter (phylum – Firmicutes) had significantly different abundances across the two sample groups (Figure 2B). Finally, two phyla, Deinococcota and Myxococcota, were only found in thawed samples, albeit at low abundance levels.
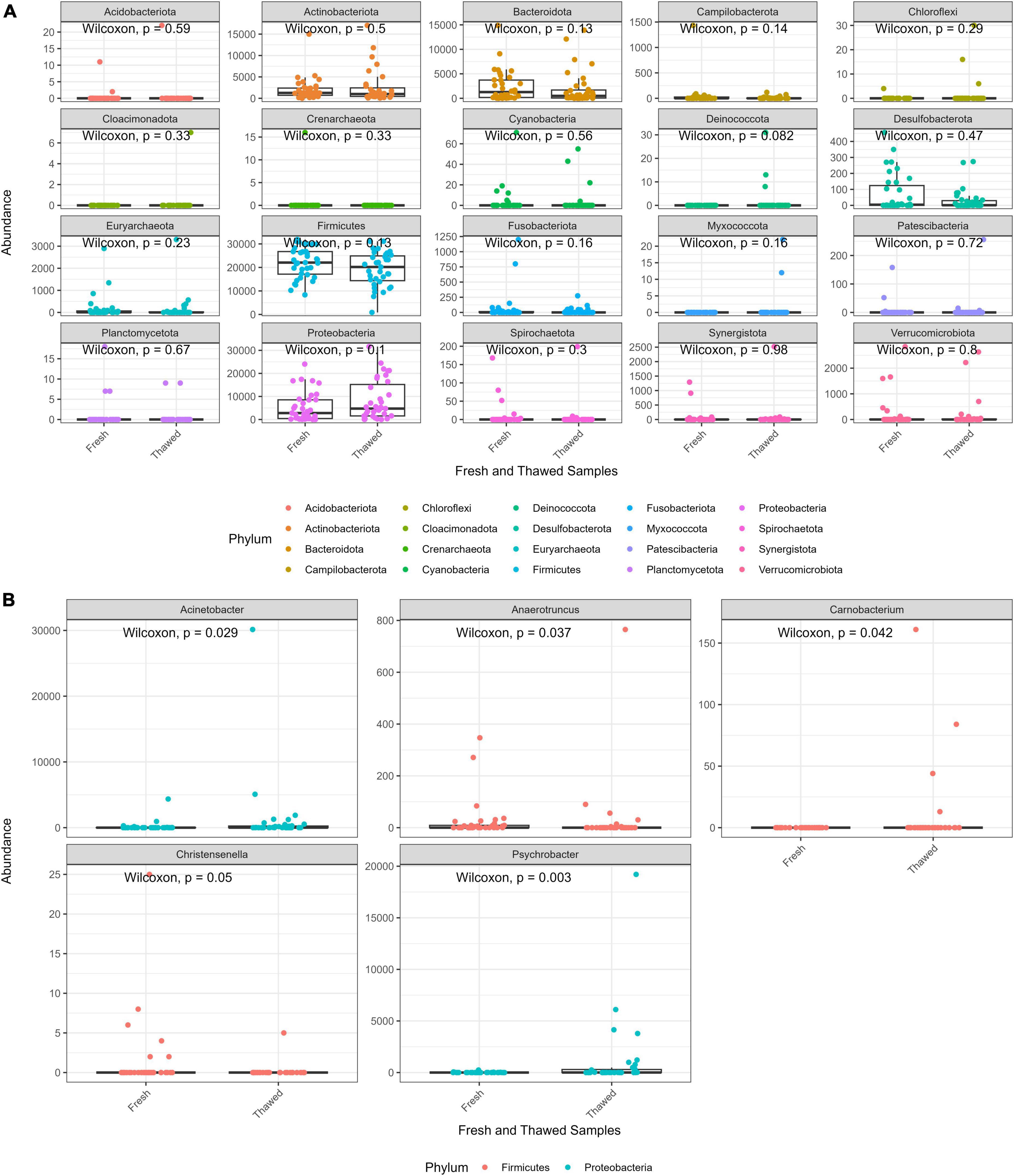
Figure 2. Abundance plot of fresh and thawed samples at phylum level (A) and taxa at genus level (B) with abundance significantly different across fresh and thawed samples.
Shannon diversity of the samples by anatomical location at ASV level, disregarding when they were collected (i.e., before or after freezing), showed that rectum samples were significantly different to other samples taken from any other location (Figure 3). Additionally, we explored sample distribution with Principal Coordinates Analysis (PCoA), using Bray–Curtis distance at ASV level to investigate any inter-individual microbial variation in relation to the sample source or anatomical location and when the samples were collected (Figures 4A, B). There were no observable clusters of fresh or thawed samples, or samples taken from hand, neck, foot, or oral locations. An ADONIS test (with vegan ver. 2.6-2) using Bray–Curtis distance measurement also showed no significant difference at genus level between fresh and thawed samples (p = 0.7439; permutation = 10000; seed = 22).
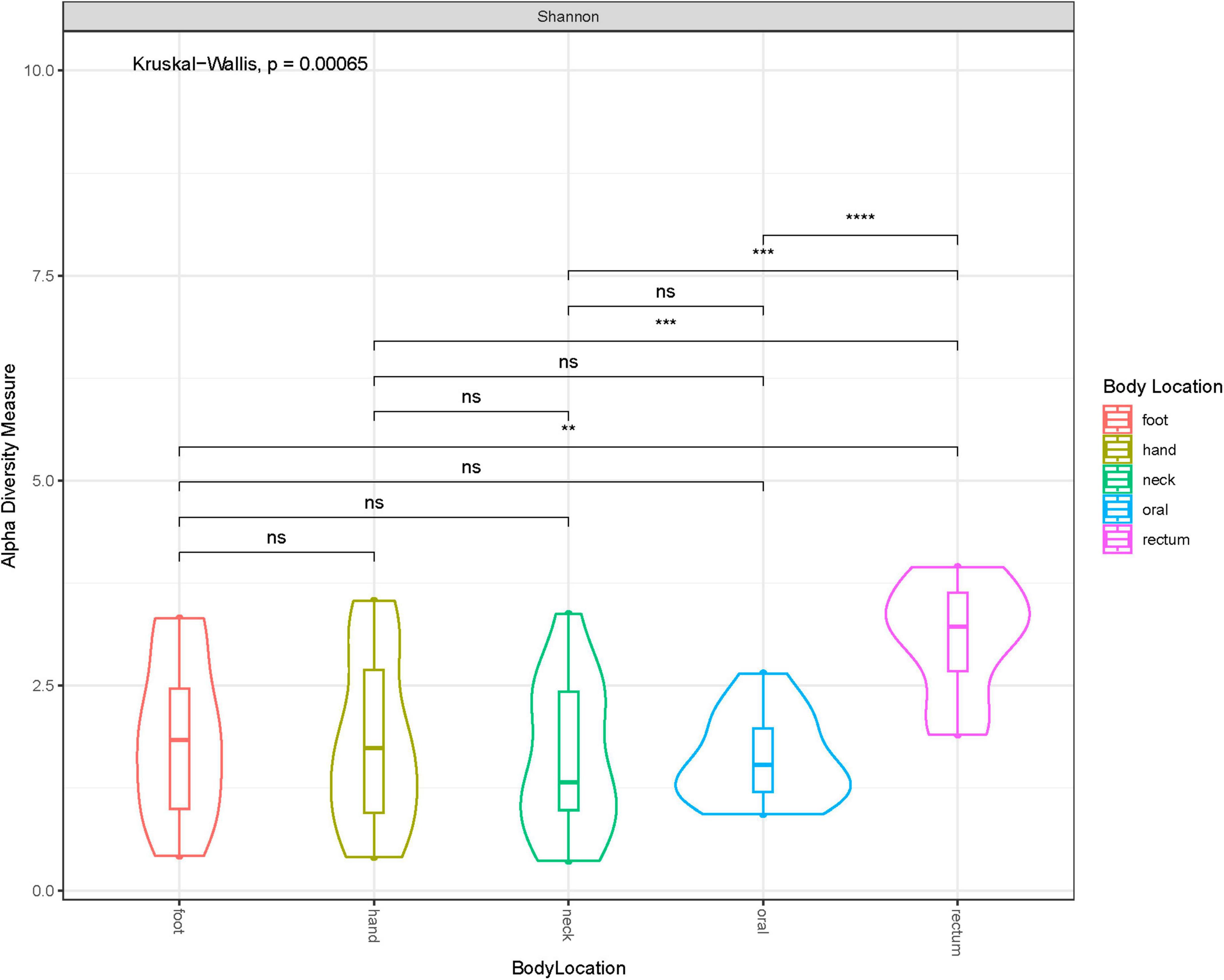
Figure 3. Violin plot showing Shannon diversity at each anatomical location at ASV level. * less than or equal to 0.01; ** less than or equal to 0.001; *** less than or equal to 0.0001.
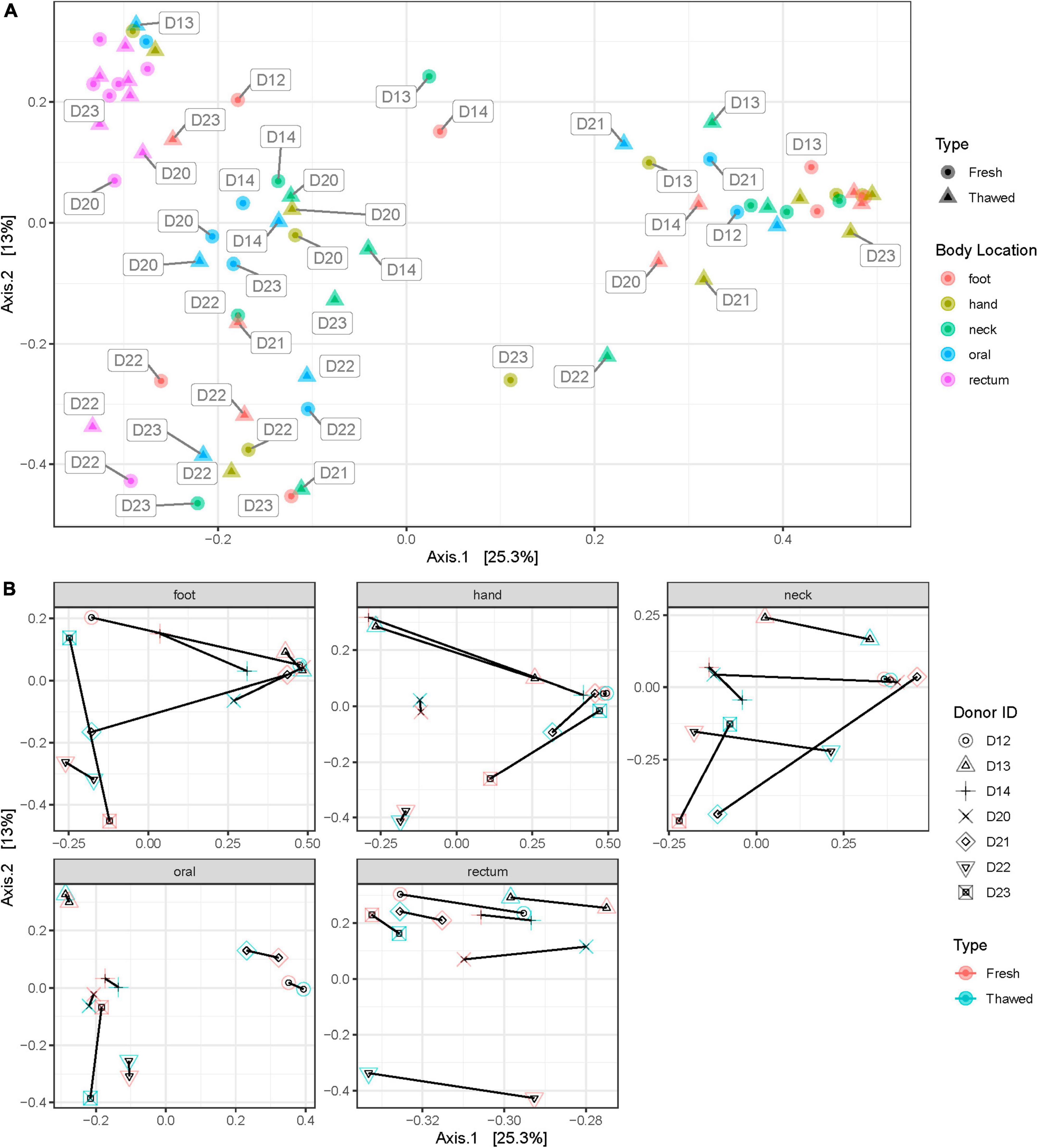
Figure 4. Bray–Curtis distance plot of samples at each anatomical location (A) and plots showing shift in donor microbial community after freezing at each anatomical location (B).
However, there was a clear separation of samples taken from the rectum, reflecting the significant difference in alpha diversity observed in Figure 3. Interestingly, all samples, including rectum samples, collected from one donated donor, D22, were also well separated from all the remaining donors.
Discussion and conclusion
The practice of freezing human bodies as a means of preserving the composition and abundance of associated microbial communities as seen ante-mortem or prior to experimental research is not unusual, especially at human taphonomy facilities (Hyde et al., 2013, 2015). It has been shown that microbial signatures can be obtained from human DNA extracts up to 16 years of being frozen in storage (Sguazzi et al., 2022). However, reports on how freezing human-related samples affects the microbiome has been varied. More specifically, how freezing the entire body affects their microbiome is unknown. Our analysis of seven donors at FACTS facility showed that overall, storing bodies frozen preserves the composition and abundance of their microbiome. Our results also demonstrate that bacterial community composition in the rectum post-mortem is significantly different to other anatomical areas, regardless of freezing.
Observing similar microbiome signatures in the donors before and after freezing is unsurprising as it is generally accepted that for a variety of human-related samples – including fecal matter (Cardona et al., 2012; Pérez-Burillo et al., 2021), skin and saliva (Marotz et al., 2021; Furuhashi et al., 2022), meconium (Xin et al., 2021), and milk (Lyons et al., 2021) – freezing is sufficient to preserve microbial composition. This is supported by the universal knowledge that low temperatures can slow or stop the proliferation of bacteria already present within samples and hinder the establishment of potential contaminants. Hence, the initial baseline composition and abundance of microbial communities in the sample is preserved.
However, although classified as statistically non-significant, our results still show that there were changes to the microbiome after freezing. Pérez-Burillo et al. (2021) have speculated that certain taxa may become impaired due to freezing, allowing competing taxa to thrive in their absence after thawingClick or tap here to enter text.. This change in ecological relations may explain the presence or absence of taxa, such as Deinococcota and Myxococcota phyla, despite the overall stability of the microbiome after freezing. Such observations are important to note, particularly for the interpretation of microbiome research in forensics, as oftentimes “rare” taxa can be presented as trace evidence (Young and Linacre, 2021) or potential biomarkers in human profiling (Hampton-Marcell et al., 2020; Neckovic et al., 2020).
The outcome of this research also provided insights into the post-mortem microbial community at various locations of the human body. Generally, the rectal, oral, and skin microbiomes in life have been reported to differ significantly in their community composition (Costello et al., 2009; Huttenhower et al., 2012). Our results, however, show that apart from the rectum, microbial communities in these habitats are hardly distinguishable after death. While microbial community homogenization occurs after death (Burcham et al., 2019; Liu et al., 2021), particularly with post-mortem transmigration (Burcham et al., 2016), this is unlikely in this case as the donors were stored frozen and we have shown that this storage method does not significantly change the microbiome composition. One thing to note is that all sites sampled – excluding rectum and oral – were all skin sites and this might contribute to the similarity observed. We also propose that this similarity may be caused by the fact that the microbial community differences between oral and skin habitats are not large enough to be detected with our limited sample size. This is supported by another study reporting that communities in various human body habitats are distinct in the post-mortem human microbiome (Pechal et al., 2018). Additionally, the rectal microbiome recorded the smallest shift after freezing and thawing (Figure 4B). This may indicate that the rectal microbiome was the least affected by freezing and is therefore more stable. The sample size included in this study is however not enough to conclusively determine this point.
Furthermore, PCoA grouping of samples (Figure 4A) revealed a deviation in the behavior of rectal samples from one donor, D22, compared to other rectal samples. Closer examination of the plot showed that all samples collected from this donor clustered closely together, regardless of the anatomical origin of the sample. It is unclear why this occurred, although, medical records for this donor were extensive. One thing of note in their record was their gastrostomy status which showed that this donor had a feeding tube antemortem. Studies have shown that enteral feeding tubes can cause alterations in the human microbiome, particularly oral (Takeshita et al., 2011) and gut (Whelan et al., 2009; Fan and Lee, 2021) microbiota. More importantly however, despite the reasons for this alteration, our study shows that complex or deviant microbiomes of donors – even those with complicated medical histories antemortem – can be preserved by freezing, as the microbial shift in D22’s samples prior to and after freezing was not significant.
It is important to note however that this study did not assess the viability of these bacteria as only the genetic material – the microbiome – was evaluated. Research assessing the survival of microorganisms in frozen foods (Speck and Ray, 1977) however, has shown that these microbes can continue to persist, and function. Moreso, another study indicated that lesions in microbial DNA can be repaired at −15°C (Dieser et al., 2013). Therefore, we conclude that freezing human donors at −20°C does not introduce a substantial alteration in their associated microbiome signatures. While we observed small changes to their microbiomes, these changes were not statistically significant to warrant an overhaul in the storage method of donors, particularly at forensic taphonomy facilities when performing thanatomicrobiome research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA925138.
Ethics statement
This study did not involve living human subjects but included only human tissues from deceased individuals from which informed consent was obtained prior to death or from legal next of kin. The study is not subject to institutional review. The ethics code of FACTS was followed and approval for the study was obtained from the FACTS Ethics Review Board as well as from the Ethics Committee at Northumbria University Ref. 21514.
Author contributions
NP, SG, and HM contributed to the conception and design of the study. NP, HM, and TG performed the samplings. NO and AN conducted the laboratory research. NO performed the statistical analysis reviewed by AN. NO wrote the first draft of the manuscript. NP, DW, and HM administered the study. All authors contributed to the manuscript revision, read, and approved the submitted version.
Funding
The mass grave project was supported by the VINNOVA – funded research project “THANATOS” (HM) under grant 2019–05211 and by the UKRI Future Leaders Fellowship (NP) under grant MR/S032878/1.
Acknowledgments
We thank Prof. Darren Smith and Stefano Ghignone for their insightful comments and suggestions regarding statistical analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1151001/full#supplementary-material
Supplementary Figure 1 | Gel electrophoresis of randomly selected samples after DNA extraction and PCR amplification with positive (P) and negative (N and N1) extraction controls. Sample names are listed by donor ID (Dx), anatomical location (H – hand, N – neck, F – foot, O – oral, and R – rectum) and freezing status (F – fresh and T – thawed).
Footnotes
References
Anyanwu, G. E. (2010). Impact of the use of cadaver on student’s ability to pass anatomy examination. Anat. Int. J. Exp. Clin. Anat. 4, 24–38.
Baust, J. G., Gao, D., and Baust, J. M. (2009). Cryopreservation: An emerging paradigm change. Organogenesis 5, 90–6. doi: 10.4161/ORG.5.3.10021
Bhattacharjee, A., Dubey, S., and Sharma, S. (2021). Storage of soil microbiome for application in sustainable agriculture: Prospects and challenges. Environ. Sci. Pollut. Res. 29, 3171–3183. doi: 10.1007/S11356-021-17164-4
Burcham, Z. M., Hood, J. A., Pechal, J. L., Krausz, K. L., Bose, J. L., Schmidt, C. J., et al. (2016). Fluorescently labeled bacteria provide insight on post-mortem microbial transmigration. Forensic Sci. Int. 264, 63–69. doi: 10.1016/J.FORSCIINT.2016.03.019
Burcham, Z. M., Pechal, J. L., Schmidt, C. J., Bose, J. L., Rosch, J. W., Eric Benbow, M., et al. (2019). Bacterial community succession, transmigration, and differential gene transcription in a controlled vertebrate decomposition model. Front. Microbiol. 10:745. doi: 10.3389/FMICB.2019.00745/BIBTEX
Burman, E., and Bengtsson-Palme, J. (2021). Microbial community interactions are sensitive to small changes in temperature. Front. Microbiol. 12:1326. doi: 10.3389/FMICB.2021.672910/BIBTEX
Cablk, M. E., Szelagowski, E. E., and Sagebiel, J. C. (2012). Characterization of the volatile organic compounds present in the headspace of decomposing animal remains, and compared with human remains. Forensic Sci. Int. 220, 118–125. doi: 10.1016/J.FORSCIINT.2012.02.007
Cardona, S., Eck, A., Cassellas, M., Gallart, M., Alastrue, C., Dore, J., et al. (2012). Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 12:158. doi: 10.1186/1471-2180-12-158/FIGURES/3
Carter, D. O., Yellowlees, D., and Tibbett, M. (2008). Temperature affects microbial decomposition of cadavers (Rattus rattus) in contrasting soils. Appl. Soil Ecol. 40, 129–137. doi: 10.1016/J.APSOIL.2008.03.010
Choo, J. M., Leong, L. E. X., and Rogers, G. B. (2015). Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 5:16350. doi: 10.1038/srep16350
Costello, E. K., Lauber, C. L., Hamady, M., Fierer, N., Gordon, J. I., and Knight, R. (2009). Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697. doi: 10.1126/SCIENCE.1177486/SUPPL_FILE/COSTELLO.SOM.PDF
da Silva Bernardo, A. P., da Silva, A. C. M., Francisco, V. C., Ribeiro, F. A., Nassu, R. T., Calkins, C. R., et al. (2020). Effects of freezing and thawing on microbiological and physical-chemical properties of dry-aged beef. Meat Sci. 161:108003. doi: 10.1016/J.MEATSCI.2019.108003
Dieser, M., Battista, J. R., and Christner, B. C. (2013). DNA double-strand break repair at -15°C. Appl. Environ. Microbiol. 79, 7662–7668. doi: 10.1128/AEM.02845-13
Doro, K. O., Kolapkar, A. M., Bank, C. G., Wescott, D. J., and Mickleburgh, H. L. (2022). Geophysical imaging of buried human remains in simulated mass and single graves: Experiment design and results from pre-burial to six months after burial. Forensic Sci. Int. 335:111289. doi: 10.1016/J.FORSCIINT.2022.111289
Fan, L., and Lee, J. H. (2021). Enteral feeding and the microbiome in critically ill children: A narrative review. Transl. Pediatr. 10:2778. doi: 10.21037/TP-20-349
Fancher, J. P., Aitkenhead-Peterson, J. A., Farris, T., Mix, K., Schwab, A. P., Wescott, D. J., et al. (2017). An evaluation of soil chemistry in human cadaver decomposition islands: Potential for estimating postmortem interval (PMI). Forensic Sci. Int. 279, 130–139. doi: 10.1016/J.FORSCIINT.2017.08.002
Furuhashi, H., Takayasu, L., Isshi, K., Hara, Y., Ono, S., Kato, M., et al. (2022). Effect of storage temperature and flash-freezing on salivary microbiota profiles based on 16S rRNA-targeted sequencing. Eur. J. Oral Sci. 130:e12852. doi: 10.1111/EOS.12852
Gocha, T. P., Mavroudas, S. R., and Wescott, D. J. (2021). The Texas state donated skeletal collection at the forensic anthropology center at Texas state. Forensic Sci. 2, 7–19. doi: 10.3390/FORENSICSCI2010002
Gunderman, R. B., and Wilson, P. K. (2005). Viewpoint: Exploring the human interior: The roles of cadaver dissection and radiologic imaging in teaching anatomy. Acad. Med. 80, 745–749. doi: 10.1097/00001888-200508000-00008
Habicht, J. L., Kiessling, C., and Winkelmann, A. (2018). Bodies for anatomy education in medical schools: An overview of the sources of cadavers worldwide. Acad. Med. 93, 1293–1300. doi: 10.1097/ACM.0000000000002227
Hampton-Marcell, J. T., Larsen, P., Anton, T., Cralle, L., Sangwan, N., Lax, S., et al. (2020). Detecting personal microbiota signatures at artificial crime scenes. Forensic Sci. Int. 313:110351. doi: 10.1016/J.FORSCIINT.2020.110351
Holm, M. A., and Iaizzo, P. A. (2019). Importance of human cadaver studies in education and medical device research: Insights derived from various imaging studies and modalities. Eng. Med. Adv. Challenges 2019, 255–280. doi: 10.1016/B978-0-12-813068-1.00010-5
Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., Chinwalla, A. T., et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/NATURE11234
Hyde, E. R., Haarmann, D. P., Lynne, A. M., Bucheli, S. R., and Petrosino, J. F. (2013). The living dead: Bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PLoS One 8:e77733. doi: 10.1371/JOURNAL.PONE.0077733
Hyde, E. R., Haarmann, D. P., Petrosino, J. F., Lynne, A. M., and Bucheli, S. R. (2015). Initial insights into bacterial succession during human decomposition. Int. J. Legal Med. 129, 661–671. doi: 10.1007/S00414-014-1128-4/FIGURES/9
Iqbal, M. A., Ueland, M., and Forbes, S. L. (2018). Recent advances in the estimation of post-mortem interval in forensic taphonomy. Aust. J. Forensic Sci. 52, 107–123. doi: 10.1080/00450618.2018.1459840
Komar, D. (1999). The use of cadaver dogs in locating scattered, scavenged human remains: Preliminary field test results. J. Forensic Sci. 44, 405–8. doi: 10.1520/JFS14474J
Kozich, J., Schloss, P., Baxter, N., Jenior, M., Koumpouras, C., and Bishop, L. (2013). MiSeq_WetLab_SOP. Available online at: https://github.com/SchlossLab/MiSeq_WetLab_SOP/blob/master/MiSeq_WetLab_SOP.md
Lauber, C. L., Zhou, N., Gordon, J. I., Knight, R., and Fierer, N. (2010). Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 307, 80–86. doi: 10.1111/J.1574-6968.2010.01965.X
Leygonie, C., Britz, T. J., and Hoffman, L. C. (2012). Impact of freezing and thawing on the quality of meat: Review. Meat Sci. 91, 93–98. doi: 10.1016/J.MEATSCI.2012.01.013
Li, D., Zhu, Z., and Sun, D. W. (2018). Effects of freezing on cell structure of fresh cellular food materials: A review. Trends Food Sci. Technol. 75, 46–55. doi: 10.1016/J.TIFS.2018.02.019
Liu, R., Wang, Q., Zhang, K., Wu, H., Wang, G., Cai, W., et al. (2021). Analysis of postmortem intestinal microbiota successional patterns with application in postmortem interval estimation. Microb. Ecol. 84, 1087–1102. doi: 10.1007/S00248-021-01923-4/TABLES/1
Lyons, K. E., Fouhy, F., O’ Shea, C. A., Ryan, C. A., Dempsey, E. M., Ross, R. P., et al. (2021). Effect of storage, temperature, and extraction kit on the phylogenetic composition detected in the human milk microbiota. Microbiologyopen 10:e1127. doi: 10.1002/MBO3.1127
Marotz, C., Cavagnero, K. J., Song, S. J., McDonald, D., Wandro, S., Humphrey, G., et al. (2021). Evaluation of the effect of storage methods on fecal, saliva, and skin microbiome composition. mSystems 6, e01329–20. doi: 10.1128/MSYSTEMS.01329-20
Martin, C., Maesen, P., Minchilli, D., Francis, F., and Verheggen, F. (2021). Forensic taphonomy: Characterization of the gravesoil chemistry using a multivariate approach combining chemical and volatile analyses. Forensic Sci. Int. 318:110569. doi: 10.1016/J.FORSCIINT.2020.110569
Matuszewski, S., Hall, M. J. R., Moreau, G., Schoenly, K. G., Tarone, A. M., and Villet, M. H. (2020). Pigs vs people: The use of pigs as analogues for humans in forensic entomology and taphonomy research. Int. J. Legal Med. 134, 793–810. doi: 10.1007/S00414-019-02074-5/TABLES/5
Metcalf, J. L. (2019). Estimating the postmortem interval using microbes: Knowledge gaps and a path to technology adoption. Forensic Sci. Int. Genet. 38, 211–218. doi: 10.1016/J.FSIGEN.2018.11.004
Mickleburgh, H. L., Alexander, M. B., Doro, K. O., Forbes, S. L., Ford, A. L., Gino, S., et al. (2022). “Actualistic experimental replication of a small-sized mass grave. A taphonomic study aimed at developing virtual training tools and advancing methods of human identification, and detection and documentation of mass graves,” in American Academy of Forensic Sciences - Conference Proceeding, (Colorado Springs, CO: American Academy of Forensic Sciences).
Micozzi, M. S. (1986). Experimental study of postmortem change under field conditions: Effects of freezing, thawing, and mechanical injury. J. Forensic Sci. 31:11103. doi: 10.1520/jfs11103j
Miles, K. L., Finaughty, D. A., and Gibbon, V. E. (2020). A review of experimental design in forensic taphonomy: Moving towards forensic realism. Forensic Sci. Res. 5, 249–259. doi: 10.1080/20961790.2020.1792631
Neckovic, A., van Oorschot, R. A. H., Szkuta, B., and Durdle, A. (2020). Challenges in human skin microbial profiling for forensic science: A review. Genes 11:1015. doi: 10.3390/GENES11091015
Ouyang, Y., Chen, D., Fu, Y., Shi, W., Provin, T., Han, A., et al. (2021). Direct cell extraction from fresh and stored soil samples: Impact on microbial viability and community compositions. Soil Biol. Biochem. 155:108178. doi: 10.1016/J.SOILBIO.2021.108178
Pechal, J. L., Schmidt, C. J., Jordan, H. R., and Benbow, M. E. (2017). Frozen: Thawing and its effect on the postmortem microbiome in two pediatric cases. J. Forensic Sci. 62, 1399–1405. doi: 10.1111/1556-4029.13419
Pechal, J. L., Schmidt, C. J., Jordan, H. R., and Benbow, M. E. (2018). A large-scale survey of the postmortem human microbiome, and its potential to provide insight into the living health condition. Sci. Rep. 8:5724. doi: 10.1038/s41598-018-23989-w
Pérez-Burillo, S., Hinojosa-Nogueira, D., Navajas-Porras, B., Blasco, T., Balzerani, F., Lerma-Aguilera, A., et al. (2021). Effect of freezing on gut microbiota composition and functionality for in vitro fermentation experiments. Nutrients 13:2207. doi: 10.3390/NU13072207/S1
Pringle, J. K., Stimpson, I. G., Wisniewski, K. D., Heaton, V., Davenward, B., Mirosch, N., et al. (2020). Geophysical monitoring of simulated homicide burials for forensic investigations. Sci. Rep. 10:7544. doi: 10.1038/s41598-020-64262-3
Procopio, N., Lovisolo, F., Sguazzi, G., Ghignone, S., Voyron, S., Migliario, M., et al. (2021). “Touch microbiome” as a potential tool for forensic investigation: A pilot study. J. Forensic Leg. Med. 82:102223. doi: 10.1016/j.jflm.2021.102223
Sguazzi, G., Mickleburgh, H. L., Ghignone, S., Voyron, S., Renò, F., Migliario, M., et al. (2022). Microbial DNA in human nucleic acid extracts: Recoverability of the microbiome in DNA extracts stored frozen long-term and its potential and ethical implications for forensic investigation. Forensic Sci. Int. Genet. 59:102686. doi: 10.1016/J.FSIGEN.2022.102686
Shin, E. K., and Meals, R. A. (2005). The historical importance of the hand in advancing the study of human anatomy. J. Hand Surg. Am. 30, 209–221. doi: 10.1016/J.JHSA.2004.09.004
Speck, M. L., and Ray, B. (1977). Effects of freezing and storage on microorganisms in frozen foods: A review. J. Food Prot. 40, 333–336. doi: 10.4315/0362-028X-40.5.333
Stokes, K. L., Forbes, S. L., and Tibbett, M. (2009). Freezing skeletal muscle tissue does not affect its decomposition in soil: Evidence from temporal changes in tissue mass, microbial activity and soil chemistry based on excised samples. Forensic Sci. Int. 183, 6–13. doi: 10.1016/J.FORSCIINT.2008.08.013
Takeshita, T., Yasui, M., Tomioka, M., Nakano, Y., Shimazaki, Y., and Yamashita, Y. (2011). Enteral tube feeding alters the oral indigenous microbiota in elderly adults. Appl. Environ. Microbiol. 77, 6739–6745. doi: 10.1128/AEM.00651-11/SUPPL_FILE/TABLES2.DOC
Turney, B. W. (2007). Anatomy in a modern medical curriculum. Ann. R. Coll. Surg. Engl. 89, 104–7. doi: 10.1308/003588407X168244
Vandeputte, D., Tito, R. Y., Vanleeuwen, R., Falony, G., and Raes, J. (2017). Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 41, S154–S167. doi: 10.1093/FEMSRE/FUX027
Varlet, V., Joye, C., Forbes, S. L., and Grabherr, S. (2020). Revolution in death sciences: Body farms and taphonomics blooming. A review investigating the advantages, ethical and legal aspects in a Swiss context. Int. J. Leg. Med. 134, 1875–1895. doi: 10.1007/S00414-020-02272-6
Varner, C., Dixon, L., and Simons, M. C. (2021). The past, present, and future: A discussion of cadaver use in medical and veterinary education. Front. Vet. Sci. 8:720740. doi: 10.3389/FVETS.2021.720740
Vieira, C., Diaz, M. T., Martínez, B., and García-Cachán, M. D. (2009). Effect of frozen storage conditions (temperature and length of storage) on microbiological and sensory quality of rustic crossbred beef at different states of ageing. Meat Sci. 83, 398–404. doi: 10.1016/J.MEATSCI.2009.06.013
Whelan, K., Judd, P. A., Tuohy, K. M., Gibson, G. R., Preedy, V. R., and Taylor, M. A. (2009). Fecal microbiota in patients receiving enteral feeding are highly variable and may be altered in those who develop diarrhea. Am. J. Clin. Nutr. 89, 240–247. doi: 10.3945/AJCN.2008.26219
Williams, A., Rogers, C. J., and Cassella, J. P. (2019b). Why does the UK need a human taphonomy facility? Forensic Sci. Int. 296, 74–79. doi: 10.1016/J.FORSCIINT.2019.01.010
Williams, A., Cassella, J. P., and Pringle, J. K. (2019a). “The ethical considerations for creating a human taphonomy facility in the United Kingdom,” in Ethical approaches to human remains, eds K. Squires, D. Errickson, and N. Márquez-Grant (Berlin: Springer), 367–384. doi: 10.1007/978-3-030-32926-6_16
Xin, Y., Xie, J., Nan, B., Tang, C., Xiao, Y., Wu, Q., et al. (2021). Freeze-thaw pretreatment can improve efficiency of bacterial DNA extraction from meconium. Front. Microbiol. 12:753688. doi: 10.3389/FMICB.2021.753688
Keywords: human microbiome, freezing, thawing, taphonomy, 16S rRNA, metabarcoding, decomposition, forensic science
Citation: Ogbanga N, Nelson A, Gino S, Wescott DJ, Mickleburgh HL, Gocha TP and Procopio N (2023) The impact of freezing on the post-mortem human microbiome. Front. Ecol. Evol. 11:1151001. doi: 10.3389/fevo.2023.1151001
Received: 25 January 2023; Accepted: 08 March 2023;
Published: 30 March 2023.
Edited by:
Gulnaz T. Javan, Alabama State University, United StatesReviewed by:
Tasha M. Santiago-Rodriguez, Diversigen, United StatesAaron Michael Tarone, Texas A&M University, United States
Copyright © 2023 Ogbanga, Nelson, Gino, Wescott, Mickleburgh, Gocha and Procopio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Noemi Procopio, bnByb2NvcGlvQHVjbGFuLmFjLnVr
 Nengi Ogbanga
Nengi Ogbanga Andrew Nelson
Andrew Nelson Sarah Gino
Sarah Gino Daniel J. Wescott
Daniel J. Wescott Hayley L. Mickleburgh
Hayley L. Mickleburgh Timothy P. Gocha
Timothy P. Gocha Noemi Procopio
Noemi Procopio