
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 06 April 2023
Sec. Behavioral and Evolutionary Ecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1150083
 Michael R. M. Lawson1*
Michael R. M. Lawson1* Michael G. B. Hayle2
Michael G. B. Hayle2 Asilatu H. Shechonge3
Asilatu H. Shechonge3 Wanja Dorothy Nyingi4
Wanja Dorothy Nyingi4 Antonia G. P. Ford5
Antonia G. P. Ford5 Joseph I. Hoffman6,7
Joseph I. Hoffman6,7 Julia J. Day8
Julia J. Day8 George F. Turner2
George F. Turner2 Kanchon K. Dasmahapatra1
Kanchon K. Dasmahapatra1Characterizing reproductive barriers such as mating preferences within rapid evolutionary radiations is crucial for understanding the early stages of speciation. Cichlid fishes are well-known for their adaptive radiations and capacity for rapid speciation and as such we investigate assortative mating among Alcolapia species; a recent (<10,000 years), small adaptive radiation, endemic to the extreme soda lakes, Magadi (one species) and Natron (three species), in East Africa. In seminatural aquarium conditions, we observed both courtship and mate choice (tested by microsatellite paternity analysis) to be significantly assortative among the three sympatric Natron species in a three-way choice experiment. This was also the case between allopatric species from Natron and Magadi, as found in a two-way choice experiment. However, the proportion of disassortative matings was substantial in both of these experiments, with hybrids comprising 29% of offspring in sympatric species and 11.4% in allopatric species comparisons. Previous work suggests that the Natron/Magadi split might not be much older than the radiation within Natron, so the similar rate of hybridization in the allopatric comparison is surprising and inconsistent with predictions of reinforcement theory, which predicts a faster rate of accumulation of premating isolation in sympatry. The relatively weak assortative mating in sympatry suggests that additional reproductive barriers, such as microhabitat preferences or spatial structuring may contribute to genetic isolation in nature.
Speciation can be best understood by studying the emergence of reproductive barriers within a previously interbreeding population (Coyne and Orr, 2004). As speciation progresses, gene flow between the diverging taxa diminishes as a consequence of the strengthening of existing barriers, and/or the accumulation of other barriers, eventually leading to complete reproductive isolation (Kulmuni et al., 2020). In sexually reproducing organisms, reproductive barriers can be prezygotic or postzygotic. Postzygotic barriers can be extrinsic, and associated with adaptation to divergent environments, or intrinsic and dependent upon genetic incompatibilities that occur irrespective of ecology (Coyne and Orr, 2004). The degree of reproductive isolation changes as speciation proceeds, with the order and appearance of reproductive barriers varying at different stages of the speciation continuum (Drès and Mallet, 2002; Stankowski and Ravinet, 2021). Therefore, characterizing reproductive barriers at different stages along this continuum is important for creating a complete picture of speciation, and mechanisms initiating speciation are best studied by focusing on taxa at the earliest stages along the continuum (Coyne and Orr, 1997; Butlin et al., 2012).
Classical models of speciation focused on the role of geographic barriers in the formation of new species, where populations become physically separated and diverge under the effects of local adaptation and genetic drift (Mayr, 1947; Turelli et al., 2001). Sympatric or parapatric speciation, which occurs in the absence of physical barriers and in the presence of gene flow, generally arises when premating isolation becomes associated with a trait under divergent selection (Coyne and Orr, 2004; Servedio et al., 2011; Smadja and Butlin, 2011). Reproductive character displacement, where there is a greater divergence of reproductive traits in sympatry compared to allopatry, can occur due to reinforcement, whereby selection strengthens premating barriers that reduce hybridization rates (Dobzhansky, 1940; Pfennig and Pfennig, 2009), or via reproductive interference among fully isolated species (Templeton, 1981). However, premating isolation can emerge in allopatry if adaptation to different ecological pressures is associated with reproductive barrier traits (Servedio et al., 2011), as a by-product of sexual selection (Fisher, 1958; Mendelson and Safran, 2021), or through mutation-order effects (Mani and Clarke, 1990). Therefore, to provide evidence of the mechanisms driving speciation under gene flow, it can be informative to include contrasts with recently separated allopatric populations (Coyne and Orr, 1997; Funk, 1998).
Cichlid fishes are well-known for their striking adaptive radiations in the East African Great Lakes, where hundreds of species have evolved from a single or handful of closely related ancestral species often over very short timescales (Turner et al., 2001; Salzburger and Meyer, 2004). Many mechanisms drive speciation in cichlids, including ecologically-mediated processes such as diet or habitat depth (Albertson et al., 2003; Terai et al., 2006) and sexually-mediated processes such as assortative mate choice (e.g., Knight et al., 1998; Seehausen and van Alphen, 1998). Further factors influencing diversification include introgressive hybridization (Salzburger et al., 2002), the reassembly of old genetic variants into new combinations (Meier et al., 2017; Marques et al., 2019) or geographic isolation (Sturmbauer et al., 2001). Additionally, multiple processes may operate together, an example being sensory drive (Seehausen et al., 2008). Between sympatric cichlid species, isolating mechanisms are more often prezygotic, with female choice generally being the ultimate barrier to mating (Kocher, 2004; Henning and Meyer, 2014).
Understanding the emergence of reproductive barriers in larger and/or older systems such as the East African Great Lakes is often difficult due to the complications of historic lake level fluctuations combined with the complex evolutionary history of ancestral lineages, with past periods of gene flow and extensive incomplete lineage sorting making inferences more difficult (Malinsky et al., 2015; Svardal et al., 2021). Therefore, recent and smaller radiations from isolated lakes make for more tractable study systems, where it may be possible to disentangle both the order of emergence and relative contribution of different reproductive barriers (Barluenga et al., 2006; Malinsky et al., 2015; Kautt et al., 2018; Poelstra et al., 2018). Direct tests of the levels of assortative mating in such simple systems have thus far only been reported between Midas cichlids from Nicaraguan crater lakes (Elmer et al., 2009; Machado-Schiaffino et al., 2017).
In this study, we investigate the magnitude of assortative mate choice among the four closely-related Alcolapia species which comprise a young, and isolated adaptive radiation endemic to the East African soda Lakes Magadi, in Kenya, and Natron, in Tanzania (Seegers and Tichy, 1999). Although widely referred to as a separate genus (Kavembe et al., 2014; White et al., 2020), the Alcolapia clade is in fact nested within the genus Oreochromis (Seegers et al., 1999; Ford et al., 2019). Magadi and Natron are volcanic, alkaline lakes dominated by large areas of thick sodium hydrogen carbonate precipitates, with very shallow (<1 m) lagoons, streams, and hot springs interspersed around the lake margins (Kaufman et al., 1990; Seegers and Tichy, 1999). Alcolapia are the only fishes found in these lakes and have evolved several unique adaptations to thrive in extremes of pH, temperature, salinity, UV light and oxygen levels (Trewavas, 1983; Narahara et al., 1996; Walsh et al., 2001; Wood et al., 2012, 2016; White et al., 2020). The much larger and deeper (∼55 m) paleolake Orolonga, which comprised part of rift lake network intermittently connected by rivers, contracted and split to form Natron and Magadi ∼8 Ka. Orolonga itself had more freshwater conditions, and the current highly alkaline and hypersaline conditions are thought to have developed ∼7 Ka (Roberts et al., 1993; Dommain et al., 2022). Therefore, both the adaptive evolution and the speciation of Alcolapia has been extremely rapid.
Three Alcolapia species are described from Lake Natron, Alcolapia alcalica, A. ndalalani, and A. latilabris. A single species, A. grahami, is known from Lakes Magadi and Little Magadi (Coe, 1966; Seegers et al., 2001). The species differ in morphology, size and male nuptial coloration (Seegers and Tichy, 1999; Figure 1). The Natron species have different head and mouth shapes which likely relate to fine-scale niche specialization toward different forms of herbivory (Ford et al., 2016). Alcolapia populations are distributed across the springs, lagoons and small streams around the perimeters of Natron and Magadi. While the distribution of the three species around Lake Natron is uneven, there are sites in the south of the lake where all three species can be found swimming alongside each other at high densities (Ford et al., 2015; Figure 1). Genomic data revealed evidence of ongoing gene flow between all the sympatric Natron species and extremely low genomic differentiation between species (Ford et al., 2015), even when compared to other cichlid radiations (Svardal et al., 2021). Despite this gene flow, these species are genetically distinct (Figure 1).
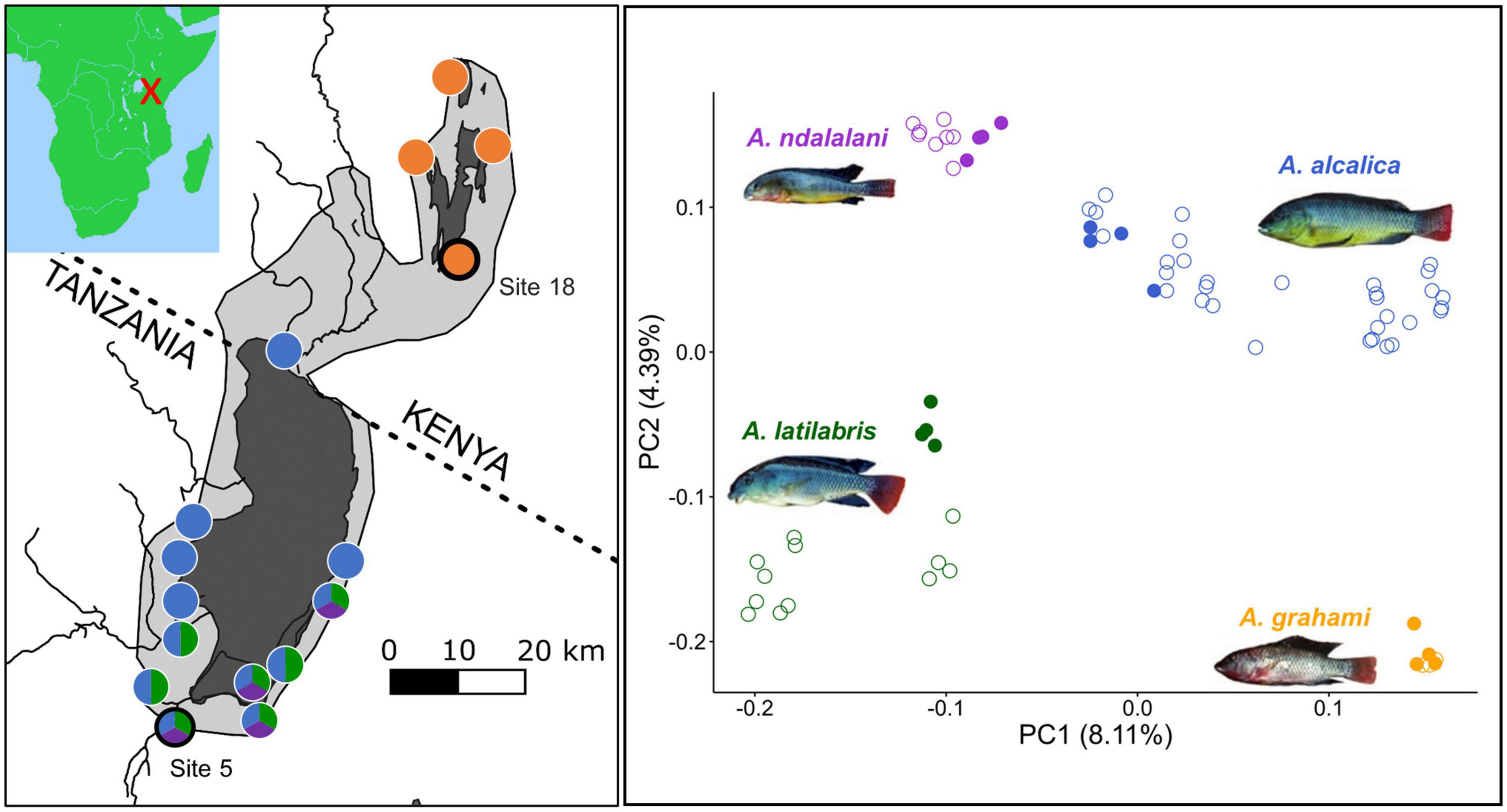
Figure 1. The Alcolapia spp. radiation of lakes Natron and Magadi. (Left panel): Map showing the distribution of the species from sites that have been sampled. The light gray area shows the approximate maximum extent of Palaeolake Orolonga (∼700 Ka) (Williamson et al., 1993). The dark gray area represents the current expanse of the lakes. Colored circles correspond to species present in the right panel with black-ringed circles denoting the populations used in this study. Photos and site information are from Ford et al. (2015). (Right panel): Principal Component Analysis (PCA) of a published Alcolapia RADseq dataset (Ford et al., 2015) demonstrates that the species form distinct genetic clusters even in sympatry. (Left panel): Colors correspond to species. Filled circles represent the populations used in this study (site five for sympatric species and site 18 for A. grahami, the allopatric species); empty circles are individuals from other sites. The covariance matrix was created using PCAngsd (Meisner and Albrechtsen, 2018) using a genotype likelihood file output from ANGSD (Korneliussen et al., 2014).
In Alcolapia, high population density, conspicuous male nuptial coloration, presence of leks and male-biased sex ratios are all predictors of high levels of sexual selection (Seegers and Tichy, 1999; Ford et al., 2016; Maina et al., 2019). Differences in male coloration between the three sympatric Natron species suggest that prezygotic assortative mate choice could be an important factor in their reproductive isolation. Since a comparison between sympatric and allopatric taxa can be valuable in understanding the mechanisms driving speciation with gene flow, we use semi-natural aquarium conditions to quantify the levels of assortative mate choice (male courtship behavior and paternity of embryos) both among sympatric Natron Alcolapia species (A. alcalica, A. latilabris, and A. ndalalani) and also between allopatric species (Natron A. alcalica and Magadi A. grahami). We use this data to test the hypothesis that sympatric Alcolapia exhibit stronger assortative mate compared to the allopatric species A. grahami.
The three Natron species were collected from site 5 in July 2017 (Ford et al., 2015; White et al., 2020), and A. grahami collected from Lake Magadi site 18 (Ford et al., 2015) in March 2019 (Figure 1). Fishes were kept in separate single-sex stock tanks at Bangor University. Appropriate water chemistry was maintained (pH = 9.0, GH = 180 ppm, KH = 180 ppm, specific gravity = 1.005) by the addition of NaHCO3 (0.5 g/L), Na2CO3 (0.07 g/L), MgSO4.7H2O (1.5 g/L) and Instant Ocean® Sea Salt (0.5 g/L). Water conditions were maintained using a continuously recirculating filtration system with the daily addition of a buffer solution. The tanks were kept at 31°C under a 12:12 light:dark cycle. Fish were fed daily with spirulina flake food.
To quantify the degree of assortative mating in Alcolapia and to determine whether this is affected by geographic context, we carried out two separate mate choice experiments in aquarium setups; among species that are found in sympatry, and between species that have allopatric distributions. The strength of assortative mating was assessed through observations of courtship behavior and paternity analysis of resulting offspring.
Behavioral observations and brood collection for the sympatric mate choice experiment involving the three Natron species, A. alcalica, A. latilabris, and A. ndalalani, was carried out between February and March 2019. For the allopatric experiment involving A. alcalica and A. grahami, behavioral observations were carried out between January and February 2020. Broods from the allopatric experiment were collected between March and April 2020. The sympatric mate choice experiment was carried out in a single 8 m (L) × 0.6 m (W) × 0.7 m (D) tank. The allopatric mate choice experiment was carried out in the same tank, but reduced to a length of 6 m, to maintain approximately the same density of fish. An 8 cm layer of silica sand was used as a substrate, with shelter provided by evenly placing five clay pots for every 2 m of tank length.
Fully mature males were selected for both experiments and size-matched with a range in standard length (SL) of no more than 12 mm within species. For the sympatric experiment, fish were all first-generation laboratory-bred and consisted of 10 females and six males of each of the Natron species derived from the wild-caught parents. In the sympatric experiment, males had the following mean SL ± SD: A. alcalica 76.2 ± 4.9 mm, A. latilabris 68.7 ± 4.0 mm, A. ndalalani 65.8 ± 3.0 mm. The larger SL of A. alcalica males relative to the other sympatric species reflects inherent size differences of the species observed in the wild (Seegers and Tichy, 1999). In the allopatric experiment, fish consisted of 12 females and seven males each of first-generation laboratory-bred A. alcalica and wild-caught A. grahami. Alcolapia alcalica was selected as the allopatric Natron species because it is the most widely distributed and forms the basal lineage (Ford et al., 2015). A. alcalica also possesses similar trophic morphology to A. grahami, with both sharing a terminal mouth (Seegers and Tichy, 1999; Ford et al., 2016). Mean male SL was 72.1 ± 3.7 mm for A. alcalica and 71.1 ± 4.0 mm for A. grahami.
For both experiments, males and females were kept in single-sex stock tanks for at least a month before being added simultaneously to the experimental setup. While males of all the Alcolapia species and female Natron species are easily differentiated by their coloration and unique mouth morphology, female A. grahami and A. alcalica are difficult to differentiate. Therefore, the different species of females in the allopatric experiment were made visually distinguishable by caudal fin clips. A. grahami females were fin-clipped along the dorsal section of the caudal fin, whereas A. alcalica were fin-clipped along the ventral section. Every 2 weeks, one species of female was fin-clipped after the fin section had almost re-grown, and the species that was clipped was subsequently alternated.
Mating preferences in Alcolapia and other Oreochromis are primarily displayed by females (Baerends and Baerends-van Roon, 1950; Seegers et al., 2001). While the identification of focal males was possible using reference photos of their unique scale markings along the intersection of the tail and caudal fin, focal observations of females were not possible as we were unable to differentiate between individuals across observation periods. Therefore, behaviors of randomly selected individual males were scored during daily, 5 min focal observations between 10:00 and 13:00 over a period of 15 days. Since individual identification was not possible for females or for male opponents with whom interactions were too rapid, only the species, sex and relative size of the interacting fish was recorded. These behavior measurements provide an assessment of a combination of female and any male courtship preferences.
Since many of the courtship behaviors in Alcolapia were found to be similar to those described for other Oreochromis species (e.g., tilting, circling, and quivering), a reduced ethogram of behaviors was created (Supplementary Table 1) based on the descriptions by Baerends and Baerends-van Roon (1950). Courtship was scored and its duration was recorded if the male performed any courtship behaviors directed at a female within a distance of two SL of the focal male. Courtship directed toward multiple females of different species was recorded, but not assigned a species. For the allopatric experiment, focal observations were video recorded and were carried out blind with respect to the identity of the female species (females were scored as either fin-clipped or non-fin-clipped). Courtship behavior was then scored using Solomon Coder v. 19.08.02 (Péter, 2011) using a custom ethogram.
Alcolapia are maternal mouthbrooders and females were checked visually each day for brooding. The partially developed broods were removed from females, euthanised and counted. Each time a brood was removed from a female, a sample of the female’s DNA was obtained by swabbing the fish along the body using sterile cotton swabs (Breacker et al., 2017). All males were photographed, measured (SL) and swabbed for DNA before being placed into the setup. Swabs and embryos were stored in 95% ethanol at –20°C until required for DNA extraction. In the sympatric experiment, 12–13 separate broods were collected from each species during the same period when courtship measurements were taken. While 15 separate broods were collected per species in the allopatric experiment, these were collected after the period during which the courtship measurements were made.
Swab DNA was extracted following the protocol outlined in Breacker et al. (2017). To assess paternity, DNA was extracted from 2 to 3 mm sections of tissue from up to six randomly selected embryos per brood. For most embryo DNA extractions, a modified version of the swab protocol was used by replacing the swab with dissected tissue. For a few embryos, a DNeasy Blood and Tissue Kit (Qiagen Inc., Hilden, Germany) was used. Extracted DNA was diluted 1:5 using ddH2O for use in PCR.
Dinucleotide microsatellites were detected in the Oreochromis niloticus genome (O_niloticus_UMD_NMBU; Conte et al., 2017) using SciRoKo (Kofler et al., 2007), after which 2–3 primers pairs per chromosome were designed using Primer-BLAST (Ye et al., 2012). Initial screening of 53 dinucleotide-repeat microsatellites for polymorphism was carried out using a panel of eight individuals of the three Natron species using FAM-labeled M13-tailed primers (Supplementary Table 2; Schuelke, 2000). Seven loci exhibiting within and among species polymorphism were selected for use in paternity testing. Microsatellite DNA loci were amplified in two separate multiplexes of fluorescent-tagged primers using the Type-IT Microsatellite PCR Kits (Qiagen Inc.) in 10 μl PCR reaction volumes using the manufacturer’s standard protocol (Supplementary Table 2). The PCRs were run with an initial 95°C denaturation stage for 5 min before conducting 32 cycles: 95°C for 30 s, 60°C for 90 s and 72°C for 30 s, and a final stage of annealing at 60°C for 30 min. PCR products were diluted 1:5 before being analysed on an ABI 3730xl DNA Analyzer. Allele sizes were scored automatically using GeneMarker v. 2.6.2 (SoftGenetics, LLC., State College, PA, USA). All traces were manually inspected and corrected where necessary to ensure high genotype data quality.
Microsatellite genotypes were manually scored using Microsatellite Analysis Software (MSA) (Thermo Fisher). First, to identify which swabbed females had matching genotypes, an identity analysis was carried out using Cervus v.3.0.7 (Kalinowski et al., 2007) using all seven loci (Supplementary Table 2). In addition, sequencing failed for one female (A. ndalalani 10) and therefore to check if this genotype matched with any of the other females, genotype reconstruction was performed in Colony2 using its known offspring and all candidate males (Jones and Wang, 2010).
The clustering of individual males and females to assigned species was visualized using STRUCTURE v. 2.3.4 (Pritchard et al., 2000). All seven loci were used with a default allele frequency parameter (λ = 1). STRUCTURE was run 20 times separately for allopatric and sympatric species using values of k of 2 and 3, respectively with a burn-in of 10,000 and 100,000 iterations.
Parentage analysis was carried out for both the sympatric and allopatric experiments separately using Cervus. Allele frequencies were generated using all the parental genotypes (Flanagan and Jones, 2019) and a simulation of paternity was run with 100,000 offspring using all possible candidate fathers. A threshold minimum of four typed loci was used for paternity assignment. Paternity was assigned based on the LOD score of offspring and parent trios. The trio LOD score is calculated using the genotypes of offspring, known mothers and candidate males while accounting for potential mistyping errors. First, all offspring were assigned a single compatible male if the trio LOD score had assignment confidence of at least 95%. For the remaining offspring, candidate fathers were removed if they had more than one mismatch in either pairwise (offspring-candidate father) or trio comparisons. For the single female with no genotype information, only candidate fathers with a positive pairwise LOD score were kept.
Courtship preference toward different taxa was modeled using Generalized Linear Mixed Models (GLMMs) using two different response metrics: (1) the total amount of time spent courting and (2) courtship frequency, or the total number of courtship behaviors directed toward females. Both response measures were the sum of behaviors carried out over a 5 min focal observation. For both models, fixed effects included the species of male and species of female involved in courtship, whereas individual male ID and date of observation were modeled as random factors, correcting for pseudoreplication. In addition, to account for potential temporal variations in courtship, time of observation was modeled as an additional random effect with times of day split into 5 min intervals. Models with and without the time of day term were selected depending on the Akaike Information Criterion (AIC). Courtship time and courtship frequency behaviors were modeled with a separate model for each experiment. For the courtship time models, the response had a heavily right-skewed distribution with many zeros, therefore models used a zero-inflated gamma distribution with a log link. We allowed zero-inflation to vary within each level of the fixed effects. As courtship frequency consists of count data, these models used a Poisson distribution and log link, but due to overdispersion in the allopatric experiment, a negative binomial distribution (nbinom2) with a log link was used instead.
Assortative mate preferences were also tested using the offspring paternity data. Broods were aggregated for each individual female and offspring were then scored as being either of conspecific or heterospecific paternity. Mating preference toward conspecifics was modeled using a Generalized Linear Model (GLM) with the cbind function and a beta-binomial distribution to account for overdispersion. Due to insufficient data, it was not possible to account for variance in individuals by using a mixed model design. The proportion of conspecific broods was modeled as the response variable and the species of the mother as the independent variable. In addition, differences in total brood size between species were tested using a GLMM with a Poisson distribution and individual female ID modeled as a random effect.
For all models, estimates and post-hoc contrasts were generated using the emmeans package (Lenth et al., 2018). For each species, estimates of their overall courtship propensity towards any species and different species were obtained. The significance (p-values) of differences in courtship between taxa was obtained through pairwise comparisons, with Tukey adjustments to account for multiple contrasts. To obtain estimates of the degree of assortative mating from offspring paternity data, the predicted probabilities of mating with a conspecific male were obtained from the output of the GLM. To test for the significance of assortative mate choice for each taxon, p-values were obtained by testing if the predicted probability of mating with conspecifics was significantly different from the expected proportion of conspecific matings under random mating: 0.5 in the allopatric experiment (two-way choice) and 0.33 in the sympatric experiment (three-way choice). In addition, to test for significant differences in the probability of conspecific mating between species, pairwise tests were carried out using emmeans.
Mating assortativity within and between each experiment was investigated using a network-based approach. Newman’s assortativity coefficients were calculated for each weighted network using the R package assortnet (Farine, 2014). To test whether assortativity was statistically significant, these values were compared to a null distribution of assortativity coefficients, which was generated from 10,000 permutations where species identity was randomly shuffled across nodes without replacement. To test for differences in assortativity between the sympatric and allopatric experiments, the t-statistic was calculated by comparing their assortativity coefficients. The statistical significance of differences in assortativity was obtained by comparing this observed t-statistic with a null distribution of t-statistics generated from the 10,000 permuted assortativity coefficients from each experiment (Heathcote et al., 2016).
All statistical analysis were carried out using R version 4.1.2 (R Core Team, 2013). The packages lme4 (Bates et al., 2014) and glmmTMB (Brooks et al., 2017) were used to generate models, while DHARMa (Hartig, 2020) was used to test model assumptions and the fit of each model. To visualize parental-offspring relationships, a network was created with the R package tidygraph v1.2.1 (Pedersen, 2022a) using additional code maintained by James Ward1 to create a node layout using the Fruchterman-Reingold algorithm. The package ggraph v.2.0.5 (Pedersen, 2022b) was used to plot networks while ggplot2 v3.3.6 was used to generate all other plots (Wickham, 2016).
Males were highly active and performed courtship and territorial behaviors soon after their introduction to the experimental setup. Between courtship and aggressive behaviors, dominant males spent a significant amount of time constructing bowers, simulating the lekking areas found in the wild (Coe, 1969). While individual males often held the same bower for multiple days, bower ownership also changed frequently over the course of the experiments.
Courtship was common, but spawning was observed less frequently. While some females were observed to mate with the same male on multiple occasions, other females were observed to spawn with several males within a single brood. In a small number of cases potential sneak mating was observed, where spawning was interrupted by a rival male as the female released an egg. In sympatric species, brood size ranged from 5 to 31 with a mean ± SD of 20.1 ± 7.9 A. alcalica, 9.8 ± 4.4 A. latilabris and 15.7 ± 6.6 A. ndalalani. A. latilabris had significantly smaller brood sizes than both A. alcalica (GLMM, post-hoc, p < 0.001) and A. ndalalani (p < 0.006). However, comparisons between A. alcalica and A. ndalalani were not significant (GLMM, post-hoc, p < 0.14) (Supplementary Table 4). In the allopatric experiment, brood size ranged from 3 to 51 and A. alcalica had a larger mean (±SD) brood size of 28.2 ± 12.6 compared to 19.1 ± 13.4 in A. grahami, but the difference was marginally non-significant (GLMM, post-hoc, p < 0.064) (Supplementary Table 4).
Male courtship behaviors are based on 1,050 and 1,350 minutes of observation data for the allopatric and sympatric experiments, respectively. In the allopatric experiment, males of both species spent significantly more time courting conspecific than heterospecific females (GLMM post-hoc, A. alcalica: p < 0.001; A. grahami: p < 0.0001, Figure 2A and Supplementary Table 6). Additionally, overall courtship time with any species did not differ between males or females of each species (males: p = 0.54, females: p = 0.65, Supplementary Table 6). Contrastingly, A. alcalica males had a higher courtship frequency (number of courtship attempts) with conspecific females, while A. grahami males did not (A. alcalica: p < 0.001, A. grahami: p = 0.18, Figure 2B and Supplementary Table 7). This was despite there being no differences in overall courtship frequency (courtship toward any species) between males of both species (p = 0.27, Supplementary Table 7). Likewise, A. alcalica females were courted by conspecific males significantly more often, while A. grahami females were not (A. alcalica: p = 0.002, A. grahami p = 0.23, Supplementary Table 7); however, A. alcalica females were courted more often overall compared to A. grahami females (p = 0.015, Supplementary Table 7).
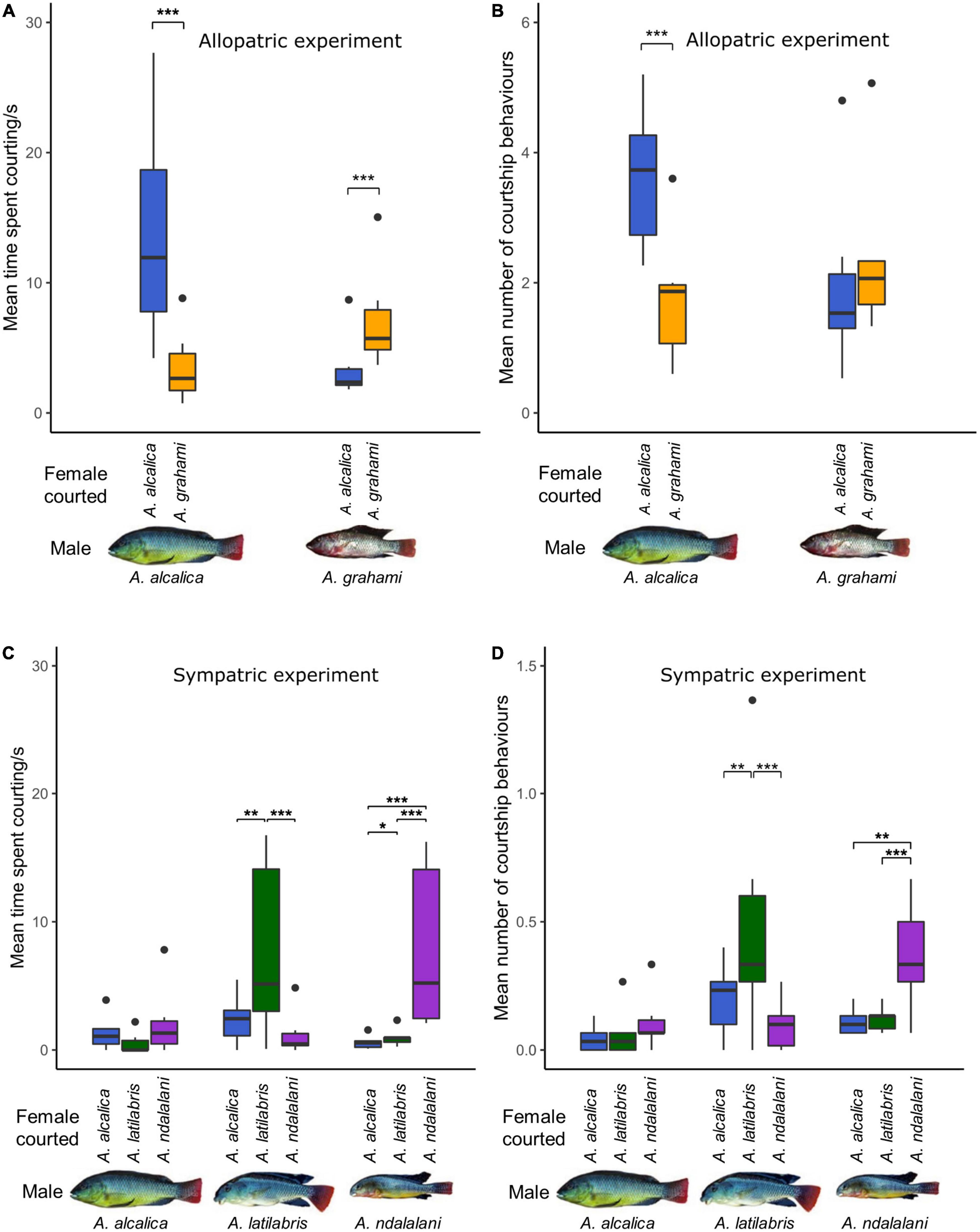
Figure 2. Male courtship with conspecific and heterospecific females in Alcolapia; (A,C) time spent courting and, (B,D) number of courtship attempts for each male during 5 min focal observations. Courtship results are shown for both the allopatric (A,B) and sympatric experiments (C,D). Colors denote the species of female courted with: blue = A. alcalica, green = A. latilabris, purple = A. ndalalani, orange = A. grahami. Large outliers (35.4 and 57.6 s) were removed from panel (C) for clarity (see Supplementary Figure 1 for full figure). Asterisks denote significant levels of pairwise contrasts extracted from GLMMs: *** < 0.001, ** < 0.01, * < 0.05.
In the sympatric experiment, A. latilabris males spent significantly longer courting A. latilabris females than A. alcalica females (p = 0.001) and A. ndalalani females (p < 0.001), but there was no significant difference in time spent courting between both heterospecific species (p = 0.32, Figure 2C and Supplementary Table 9). A. ndalalani males spent significantly longer courting conspecific females compared to A. alcalica females (p < 0.001) and A. latilabris females (p < 0.001), but there was no difference in courtship time in the two heterospecific comparisons (p = 0.21, Figure 2C). In contrast, there was no significant difference in the amount of time spent courting between any of the species by A. alcalica males (A. alcalica–A. latilabris: p = 0.96, A. alcalica–A. ndalalani: p = 0.73, A. ndalalani–A. latilabris: p = 0.52, Figure 2C). There were no significant differences in overall courtship time between males (A. alcalica–A. latilabris: p = 0.99, A. alcalica–A. ndalalani: p = 0.63, A. ndalalani–A. latilabris: p = 0.57); however, courtship was directed toward A. ndalalani females for significantly longer overall compared to A. alcalica females (p = 0.018; Supplementary Table 9).
The number of courtship events largely reflected these results with A. latilabris males courting conspecific females significantly more often than heterospecifics (A. latilabris–A. alcalica: p = 0.003, A. latilabris–A. ndalalani: p < 0.001, A. ndalalani–A. alcalica: p = 0.18, Supplementary Table 11 and Figure 2D). Similarly, A. ndalalani males also spent had a higher courtship frequency with conspecifics compared to heterospecifics (A. ndalalani–A. alcalica: p = 0.002, A. ndalalani–A. latilabris: p = 0.003, A. latilabris–A. alcalica: p = 0.97, Supplementary Table 11 and Figure 2D). There was no significant difference in courtship frequency between any of the species by A. alcalica males (A. alcalica–A. latilabris: p = 0.79, A. alcalica–A. ndalalani: p = 0.24, A. ndalalani–A. latilabris: p = 0.56, Supplementary Table 11 and Figure 2D). There were no significant differences in overall courtship frequency between males of each species (A. alcalica–A. latilabris: p = 0.19, A. alcalica–A. ndalalani: p = 0.16, A. ndalalani–A. latilabris: p = 0.99); however, A. alcalica females were courted more often overall compared to the other species (A. alcalica—A. latilabris: p = 0.01, A. alcalica—A. ndalalani: p = 0.03, A. latilabris—A. ndalalani: 0.91, Supplementary Table 11). Full model outputs and associated test statistics can be found in Supplementary Tables 3–11.
STRUCTURE analysis confirmed that individual males and breeding females predominantly clustered to their respective assigned species (Supplementary Figure 2).
In the allopatric experiment, a total of 140 offspring from 31 broods were successfully genotyped with paternity assigned to an individual male in all cases. The total number of breeding females for each species was 10 for A. alcalica and 11 for A. grahami (Figure 3A). The proportion of conspecific offspring was 88.6% (124/140) for all the offspring in the allopatric experiment. Both A. alcalica (62 of 68 offspring) and A. grahami females (62 of 72 offspring) showed a significant preference toward conspecifics (GLM post-hoc test, A. alcalica: t.ratio = 3.0, df = 18 p = 0.007; A. grahami: t.ratio = 2.8, df = 18, p = 0.012; Figure 3B). There were no significant differences in the probability of conspecific mating between females of each species (GLM, z = 0.98, p = 0.33) and mating was significantly assortative overall (r = 0.77, p < 0.001).
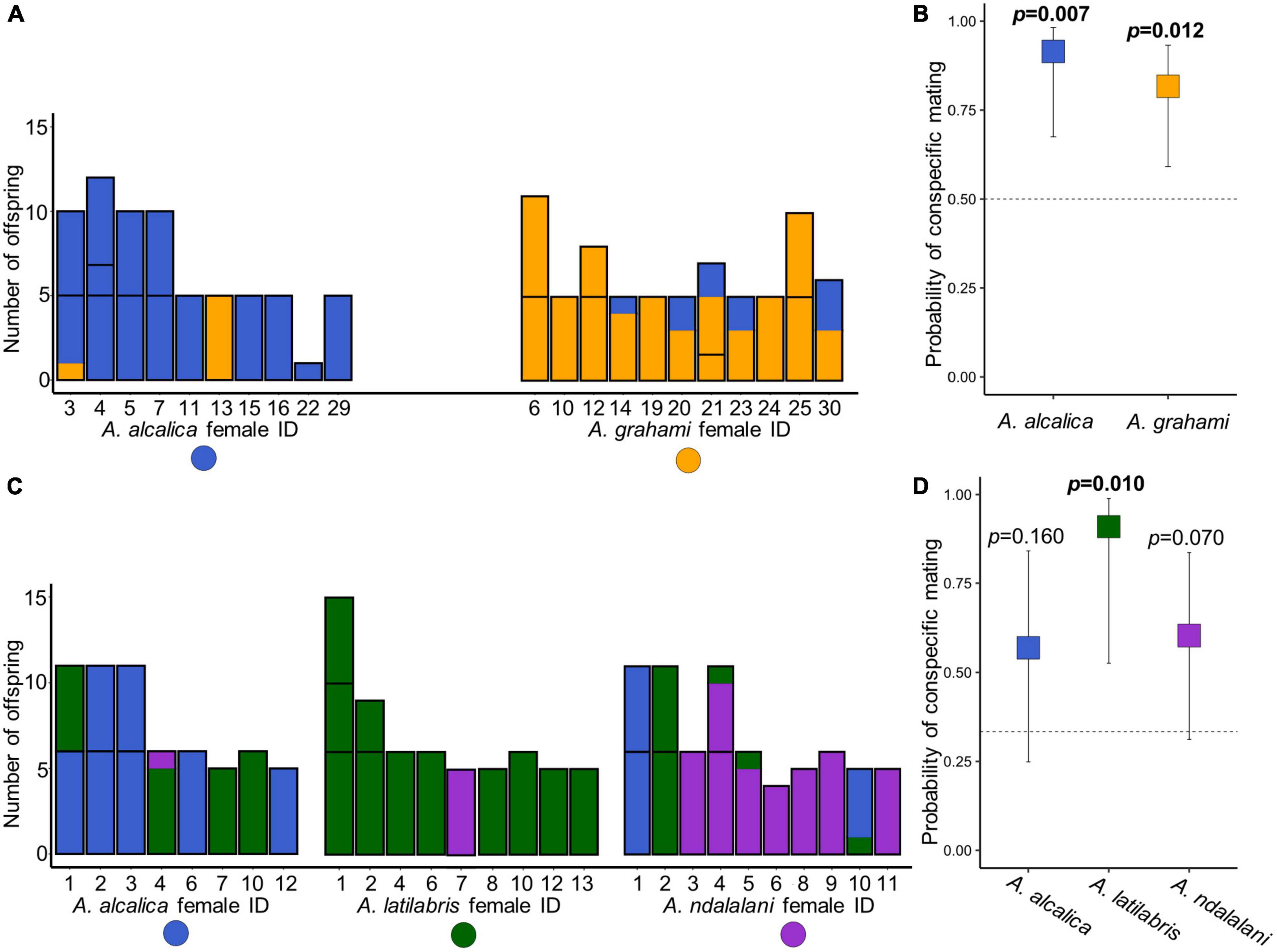
Figure 3. Assortative mate choice in Alcolapia from offspring paternity data. Barplots show paternity of offspring for each spawning female in both allopatric (A) and sympatric (C) experiments. Colors denote the paternal species assigned from the microsatellite data: orange = A. grahami, blue = A. alcalica, green = A. latilabris, purple = A. ndalalani. Each column represents an individual female. Within columns, individual clutches are separated by black bars. (B,D) Boxplots showing the predicted probability of females mating with conspecific species estimated from offspring paternity data, (B) allopatric experiment, (D) sympatric experiment calculated from the model outputs of the GLMs. Error bars denote 95% Wald confidence intervals for the estimates. Dotted lines show the mean expected probability of mating with conspecifics given random mating. In both experiments, mating was assortative overall (p = 0.001). However, in the sympatric experiment, tests for assortative mating were not significant for A. alcalica (p = 0.16) and A. ndalalani (p = 0.07).
In the sympatric experiment, a total of 193 offspring from 36 broods were successfully genotyped with paternity assigned to at least the species level. The total number of breeding females for each species was eight for A. alcalica, nine for A. latilabris, and ten for A. ndalalani (Figure 3C). Of the successfully genotyped offspring, 89.6% (173/193) were assigned to individual males. The proportion of offspring assigned to individual males differed among species; 74.6% (44/59) in A. alcalica, 91.9% (57/62) in A. latilabris and 81.4% (57/70) in A. ndalalani (Figure 4B). The proportion of conspecific offspring was 71.0% (137/193) for all the offspring in the sympatric experiment: 63.9% (39/61) in A. alcalica females, 98.4% (57/62) in A. latilabris females, and 58.6% (41/70) in A. ndalalani females (Figure 3C). Overall, females spawned with conspecific males significantly more than expected given random mating (GLM post-hoc test, t.ratio = 3.65, df = 23, p = 0.001; Supplementary Table 5) and mating was significantly assortative overall (r = 0.56, p < 0.001). While A. latilabris females had a significant preference toward conspecifics (GLM post-hoc test, t.ratio = 2.82, df = 23, p = 0.010; Figure 3D), A. alcalica (t.ratio = 1.45, df = 23, p = 0.160; Figure 3D) and A. ndalalani females did not (t.ratio = 1.90, df = 23, p = 0.070; Figure 3D). There were no significant differences in the probability of conspecific mating between females of each sympatric species [GLM post-hoc test, F.ratio = 1.45, df = (2,23), p = 0.255]. There were no significant differences in mating assortativity between the sympatric and allopatric experiments (t = 2.37, p = 0.25).
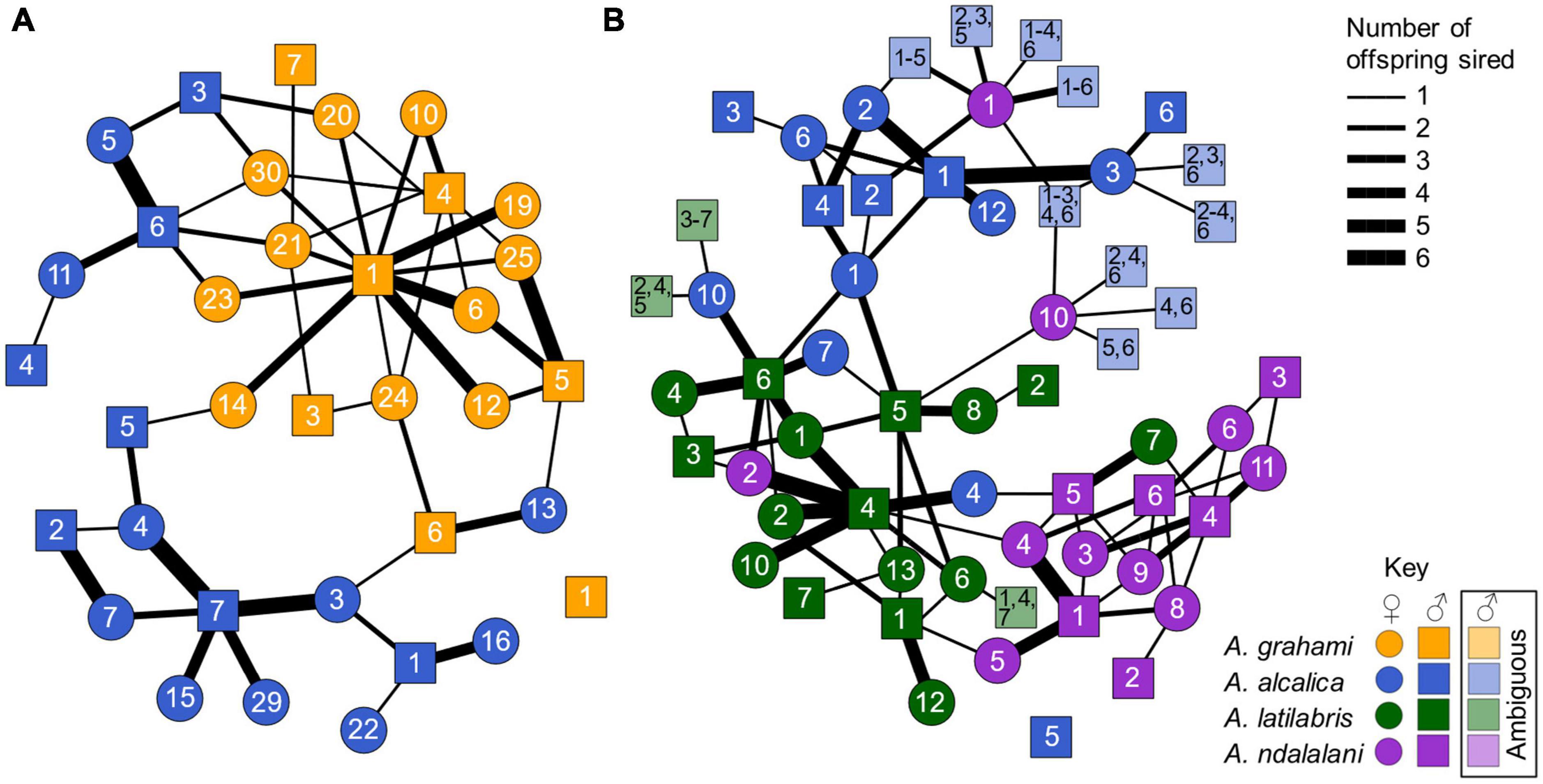
Figure 4. Network showing parent-offspring relationships in the allopatric experiment (A) and sympatric experiment (B) using offspring paternity data. Colors indicate species: blue = A. alcalica, orange = A. grahami, green = A. latilabris, purple = A. ndalalani. Circles = females and squares = males. Numbers correspond to unique male or female ID. Line thickness denotes the number of offspring sired between parents. Lighter colors denote offspring with ambiguous paternity that were not assigned to a single compatible male. Paternity could be assigned unambiguously in 94.0% of offspring. 88.6% (two-way choice) and 71.0% (three-way choice) of offspring in the allopatric and sympatric experiments resulted from conspecific matings. Mating was significantly assortative in both the allopatric (r = 0.77, p < 0.001) and sympatric (r = 0.56, p < 0.001) experiments. There were no significant differences in assortativity between both experiments (t = 2.37, p = 0.25).
Multiple mating was relatively common, with 61.3% (19/31) and 69.4% (25/36) of broods showing multiple paternity in the allopatric and sympatric experiments, respectively (Figure 4). These represent the minimum level of multiple paternity as we only genotyped a maximum of six offspring per brood (brood sizes varied from 3 to 51), and we also could not distinguish between all males in the sympatric experiment. Individual mating success also varied among males. For instance, in the allopatric experiment, one A. grahami male sired at least one offspring with every breeding A. grahami female, but none with any A. alcalica females (Figure 4A). By contrast, a single A. grahami male and A. alcalica male were not assigned parentage to any of the offspring in the allopatric and sympatric experiments, respectively (Figures 4A, B).
We demonstrate that Alcolapia species show evidence of assortative mating in comparisons both between sympatric and allopatric species in a semi-natural aquarium set-up. Hybrid offspring were generated between all species pairs that were tested, indicating that in our experimental setup, reproductive isolation is incomplete. Together with results from previous studies showing differences in trophic morphology between sympatric species (Ford et al., 2016), our findings lend support to the theory that speciation in sympatry is more likely when there is a combination of assortative mating and ecological divergence (Dieckmann and Doebeli, 1999). These findings correspond with other studies of recent and small cichlid adaptive radiations such as Lake Ejagham in Cameroon and the crater lakes of Nicaragua (Martin, 2013; Machado-Schiaffino et al., 2017).
The forces driving speciation may vary depending on the geographic context and amount of gene flow between populations. In general, it is expected that prezygotic barriers will be stronger among sympatric compared to allopatric taxa as there is selection to avoid heterospecific matings in sympatry, but not in allopatry (Butlin, 1987; Coyne and Orr, 1997). Contrary to these expectations, our results cannot detect a difference in the degree of assortative mating between allopatric species and sympatric species.
While reinforcement may be a key driver of premating isolation in some taxa (Yukilevich, 2012), premating barriers are also predicted to increase with genetic distance (Zouros, 1973; Coyne and Orr, 1989). Allopatric A. grahami females showed strong premating isolation, but they are also more genetically distinct compared to the sympatric Natron species (Figure 1; Ford et al., 2015). On the other hand, phenotypic rather than genetic distance is often a better predictor of assortative mate choice (McPeek and Wellborn, 1998). For example, in Pseudocrenilabrus spp. of Lake Mweru and Bangweulu, less closely related but more phenotypically similar species have stronger premating isolation than more distantly related but less phenotypically similar species (Stelkens and Seehausen, 2009). Alcolapia grahami are more phenotypically similar to A. alcalica in terms of trophic morphology (head and mouth shape) compared to the other sympatric Natron species (Ford et al., 2016), but they do differ in male coloration, which is typically a primary cue used by females for mate selection (Seehausen et al., 1997; Selz et al., 2014). A meta-analysis of cichlid premating isolation studies found only slight differences in the levels of assortative mating between sympatric and allopatric species (Rometsch et al., 2020); however, they also found greater variance in premating isolation among allopatric species, and lower levels when comparing allopatric and sympatric populations, suggesting an important role in the duration of the population split. Nevertheless, strong assortative mate choice can evolve among cichlids over relatively short periods of allopatry (Knight and Turner, 2004; Genner et al., 2007), especially if there are divergent ecological pressures resulting from habitat differences (Tyers et al., 2014).
Fossil and geological data date the Magadi-Natron split to approximately 8 Ka (Dommain et al., 2022), and the lakes are currently situated approximately 25 km apart by a topographic sill (Williamson et al., 1993; Figure 1A). Using nuclear genetic data, the A. grahami (Magadi)—Natron Alcolapia species split has been estimated at 0.007–1.55 Ma (95% HPD) (Ford et al., 2019), which suggests that A. grahami populations in Magadi have been separated for at least 7 Ka and may have diverged before the lakes split (Williamson et al., 1993; Dommain et al., 2022). Therefore, our results indicate that strong mate discrimination can either evolve or persist in allopatry without recent (<7 Ka) reinforcement.
The weak species-assortative mating observed in sympatric A. alcalica and A. ndalalani in this experiment differs from other cichlid studies which generally find strong premating isolation among sympatric species (Knight et al., 1998; Plenderleith et al., 2005; Machado-Schiaffino et al., 2017). However, levels of assortative mating can be lower and more variable among more recently separated sympatric cichlid species or populations (Jordan et al., 2003; Selz et al., 2016; Nyalungu and Couldridge, 2020). For instance, Selz et al. (2016) found varying levels of assortative mating between Pundamilia nyererei populations in two-way choice experiments, but mate choice was strongly assortative when females were provided with a choice of closely related P. igneopinnis (96–100%) (Selz et al., 2016). Premating isolation in sympatric Alcolapia may therefore be more comparable to that observed between populations or sub-species in other cichlid systems. Incomplete assortative mating in Alcolapia may be unsurprising given that the radiation is extremely recent (Ford et al., 2015) and that the degree of genetic differentiation is among the lowest measured across all cichlid radiations (Svardal et al., 2021). Moreover, asymmetry in mating preferences or species discrimination may be common among recently diverging cichlids (Nevado et al., 2011; Malinsky et al., 2015; Van Steenberge et al., 2022) and hence it may be unsurprising that some species exhibit weaker assortative mate choice.
The levels of hybridization among Alcolapia species reported in our experiments (Figure 3) are high enough that species would likely hybridize to the point of becoming indistinct within a few generations (Irwin and Schluter, 2022). Yet both genetic (Figure 1) and morphological data indicate that hybrids are comparatively rare in most wild populations (Ford et al., 2015; Ford et al., 2016), suggesting that premating isolation is either stronger in the wild or is accompanied by strong selection against hybrids during early life stages.
Assortative mate choice may be influenced by extrinsic factors such as spatial, temporal and environmental components, some of which may not be present in an aquarium setup. For example, spatially-mediated size-assortative mating has been observed in the cichlid Eretmodus cyanostictus, where larger males dominate high-quality habitats while smaller, subdominant males occur more frequently in low-quality environments (Taborsky et al., 2014). In the wild, Alcolapia may exhibit some spatial separation which could affect encounter rates and influence levels of assortative mating. For instance, Seegers et al. (2001) recorded a higher abundance of A. latilabris in the upper courses of streams. During field collections, we observed that A. latilabris and A. ndalalani mainly occurred in upstream sections and in more rocky habitats, whereas downstream sections with fine-grained substrates were often dominated by A. alcalica. On the other hand, observations of breeding leks in Lake Natron were found to be comprised of multiple different species (Seegers and Tichy, 1999). Furthermore, the occurrence of multiple allopatric sites containing only A. alcalica indicates that its area of sympatry is less extensive compared to A. latilabris and A. ndalalani, which have a significant overlap in their distribution (Figure 1A).
A multitude of ecological and environmental factors may influence premating isolation beyond the primary cues used for mate choice. While previous studies on the sensory cues used by cichlids in assortative mate choice have usually found visual cues such as male coloration to be the primary premating cues (Seehausen et al., 1997; Selz et al., 2014), single cues alone seldom control all premating isolation (Plenderleith et al., 2005; Blais et al., 2009; Rometsch et al., 2020). Instead, sensory cues may be multimodal, with each cue contributing to premating isolation to different degrees (Houck and Verrell, 1993; Rafferty and Boughman, 2006; Keller-Costa et al., 2015; Mérot et al., 2015). Due to the lack of shade from terrestrial vegetation, the shallow depths and the high clarity of the water column, the light environment of Natron and Magadi is extremely bright (Johannsson et al., 2014). This bright visual environment may not be fully replicated in the aquarium setup and subsequently, visual cues involved in mate choice may become less salient (Maan et al., 2010; Wright et al., 2018). In line with this hypothesis, the breakdown of reproductive barriers via the loss of visual cues may already have occurred in Alcolapia, with a high turbidity site on the eastern shore of Natron supporting a potential hybrid population with intermediate morphology (Ford et al., 2015).
Olfactory cues have also been shown to be a component of mate choice in some cichlid species (Plenderleith et al., 2005; Blais et al., 2009). Partitioning of diet among Alcolapia species could potentially promote the differentiation in olfactory cues used in mate choice (Kavembe et al., 2016; Ford et al., 2016). As species were fed an identical diet in these experiments, any diet-related odor discrimination would be eliminated. The combination of ecologically mediated premating cues may have an additive effect, where assortative mating is relatively weak with only primary mate choice cues but strong in the presence of additional factors (Tinghitella et al., 2020). Future studies may investigate the relative contribution that different cues (visual, olfactory etc.) play in Alcolapia reproductive isolation with aquarium experiments (e.g., Knight and Turner, 1999; Selz et al., 2014). Other factors relating to population density and spatial structuring that may also influence mate choice should be considered.
The adaptive radiations of cichlids in the East African great lakes are model systems in speciation research (Kocher, 2004; Seehausen, 2006), but can be challenging when studying early speciation due to their size and complex evolutionary histories. Small and young cichlid radiations are more tractable, but few studies have characterized the reproductive barriers between emerging species. Here, we present evidence of weak to moderate assortative mating both between sympatric and allopatric Alcolapia species. These findings are consistent with most study systems at the early stages of divergence, where premating rather than postmating or postzygotic barriers tend to play a greater role in speciation and reach completion at faster rates (Coyne and Orr, 1997; Grant and Grant, 1997; Seehausen et al., 2014). The similar degree of assortative mate choice in allopatry compared to sympatry observed here suggests that assortative mating can accumulate in allopatry, perhaps through divergent sexual selection. The high rates of hybridization observed between species suggest additional factors such as ecological divergence may also be an important component of their reproductive isolation.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
This animal study was reviewed and approved by Bangor University Animal Welfare and Ethical Review Body (AWERB).
AS, WDN, AF, JD, and GT carried out field collections in Kenya and Tanzania. ML, KD, JD, and GT designed the study. ML, MH, and GT carried out the aquarium experiments. ML and KD carried out laboratory work. JH and KD designed microsatellite markers and assisted with the analysis. ML led the data analysis and wrote the manuscript with input from all authors. All authors contributed to the article and approved the submitted version.
ML was funded by a NERC ACCE DTP Ph.D. studentship. Fieldwork in Kenya was funded by a Fisheries Society of the British Isles research grant to AF.
We thank the National Museums of Kenya and the Tanzania Fisheries Research Institute (TAFIRI) for facilitating the field collections and research permits. We thank Chloe Robinson for carrying out pilot experiments and DNA extractions, Elke Hippauf for screening microsatellite loci, and Lewis White for assisting in field collections. We thank Elva Robinson for offering advice on the network analysis and Roger Butlin for comments on the manuscript. We also thank two reviewers for providing valuable comments that helped improve the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1150083/full#supplementary-material
Albertson, R. C., Streelman, J. T., and Kocher, T. D. (2003). Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc. Natl. Acad. Sci. U.S.A. 100, 5252–5257. doi: 10.1073/pnas.0930235100
Baerends, G. P., and Baerends-van Roon, J. M. (1950). An introduction to the study of the ethology of the cichlid fishes. Behav. Suppl.1:243.
Barluenga, M., Stölting, K. N., Salzburger, W., Muschick, M., and Meyer, A. (2006). Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature 439, 719–723. doi: 10.1038/nature04325
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2014). Fitting linear mixed-effects models using lme4. arXiv [Preprint]. arXiv.1406. 5823. doi: 10.18637/jss.v067.i01
Blais, J., Plenderleith, M., Rico, C., Taylor, M. I., Seehausen, O., van Oosterhout, C., et al. (2009). Assortative mating among Lake Malawi cichlid fish populations is not simply predictable from male nuptial colour. BMC Evol. Biol. 9:53. doi: 10.1186/1471-2148-9-53
Breacker, C., Barber, I., Norton, W. H. J., McDearmid, J. R., and Tilley, C. A. (2017). A low-cost method of skin swabbing for the collection of DNA samples from small laboratory fish. Zebrafish 14, 35–41. doi: 10.1089/zeb.2016.1348
Brooks, M. E., Kristensen, K., Van Benthem, K. J., Magnusson, A., Berg, C. W., Nielsen, A., et al. (2017). glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9, 378–400. doi: 10.32614/RJ-2017-066
Butlin, R. (1987). Speciation by reinforcement. Trends Ecol. Evol. 2, 8–13. doi: 10.1016/0169-5347(87)90193-5
Butlin, R., Debelle, A., Kerth, C., Snook, R. R., Beukeboom, L. W., Cajas, R. C., et al. (2012). What do we need to know about speciation? Trends Ecol. Evol. 27, 27–39. doi: 10.1016/j.tree.2011.09.002
Coe, M. J. (1966). The biology of Tilapia grahami Boulenger in Lake Magadi, Kenya. Acta Trop. 23, 146–177.
Coe, M. J. (1969). Observations on Tilapia alcalica Hilgendorf an endemic cichlid fish from Lake Natron, Tanzania. Rev. Zool. Bot. Afr. 801, 1–9.
Conte, M. A., Gammerdinger, W. J., Bartie, K. L., Penman, D. J., and Kocher, T. D. (2017). A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 18:341. doi: 10.1186/s12864-017-3723-5
Coyne, J. A., and Orr, H. A. (1989). Patterns of speciation in Drosophila. Evolution 43, 362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x
Coyne, J. A., and Orr, H. A. (1997). ‘Patterns of speciation in Drosophila’ revisited. Evolution 51, 295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x
Dieckmann, U., and Doebeli, M. (1999). On the origin of species by sympatric speciation. Nature 400, 354–357. doi: 10.1038/22521
Dobzhansky, T. (1940). Speciation as a stage in evolutionary divergence. Am. Nat. 74, 312–321. doi: 10.1086/280899
Dommain, R., Riedl, S., Olaka, L. A., deMenocal, P., Deino, A. L., Owen, R. B., et al. (2022). Holocene bidirectional river system along the Kenya Rift and its influence on East African faunal exchange and diversity gradients. Proc. Natl. Am. Sci. U.S.A. 119:e2121388119. doi: 10.1073/pnas.2121388119
Drès, M., and Mallet, J. (2002). Host races in plant–feeding insects and their importance in sympatric speciation. Philos. Transact. R. Soc. B Biol. Sci. 357, 471–492. doi: 10.1098/rstb.2002.1059
Elmer, K. R., Lehtonen, T. K., and Meyer, A. (2009). Color assortative mating contributes to sympatric divergence of neotropical cichlid fish. Evolution 63, 2750–2757. doi: 10.1111/j.1558-5646.2009.00736.x
Farine, D. R. (2014). Measuring phenotypic assortment in animal social networks: Weighted associations are more robust than binary edges. Anim. Behav. 89, 141–153. doi: 10.1016/j.anbehav.2014.01.001
Fisher, R. A. (1958). The genetical theory of natural selection. Oxford: Clarendon Press, doi: 10.5962/bhl.title.27468
Flanagan, S. P., and Jones, A. G. (2019). The future of parentage analysis: From microsatellites to SNPs and beyond. Mol. Ecol. 28, 544–567. doi: 10.1111/mec.14988
Ford, A. G. P., Bullen, T. R., Pang, L., Genner, M. J., Bills, R., Flouri, T., et al. (2019). Molecular phylogeny of Oreochromis (Cichlidae: Oreochromini) reveals mito-nuclear discordance and multiple colonisation of adverse aquatic environments. Mol. Phylogenet. Evol. 136, 215–226. doi: 10.1016/j.ympev.2019.04.008
Ford, A. G. P., Dasmahapatra, K. K., Rüber, L., Gharbi, K., Cezard, T., and Day, J. J. (2015). High levels of interspecific gene flow in an endemic cichlid fish adaptive radiation from an extreme lake environment. Mol. Ecol. 24, 3421–3440. doi: 10.1111/mec.13247
Ford, A. G. P., Rüber, L., Newton, J., Dasmahapatra, K. K., Balarin, J. D., Bruun, K., et al. (2016). Niche divergence facilitated by fine-scale ecological partitioning in a recent cichlid fish adaptive radiation. Evolution 70, 2718–2735. doi: 10.1111/evo.13072
Funk, D. J. (1998). Isolating a role for natural selection in speciation: Host adaptation and sexual isolation in Neochlamisus bebbianae leaf beetles. Evolution 52, 1744–1759. doi: 10.2307/2411347
Genner, M. J., Nichols, P., Carvalho, G. R., Robinson, R. L., Shaw, P. W., Smith, A., et al. (2007). Evolution of a cichlid fish in a Lake Malawi satellite lake. Proc. R. Soc. B. 274, 2249–2257. doi: 10.1098/rspb.2007.0619
Grant, P. R., and Grant, B. R. (1997). Genetics and the origin of bird species. Proc. Natl. Am. Sci. U.S.A. 94, 7768–7775.
Hartig, F. (2020). DHARMa: Residual diagnostics for hierarchical (multi-level/mixed) regression models. R package version 0. 3, 3. Available online at: http://florianhartig.github.io/DHARMa/
Heathcote, R. J., While, G. M., MacGregor, H. E., Sciberras, J., Leroy, C., D’Ettorre, P., et al. (2016). Male behaviour drives assortative reproduction during the initial stage of secondary contact. J. Evol. Biol. 29, 1003–1015. doi: 10.1111/jeb.12840
Henning, F., and Meyer, A. (2014). The evolutionary genomics of cichlid fishes: Explosive speciation and adaptation in the postgenomic era. Annu. Rev. Genomics Hum. Genet. 15, 417–441. doi: 10.1146/annurev-genom-090413-025412
Houck, L. D., and Verrell, P. A. (1993). Studies of courtship behavior in plethodontid salamanders: A review. Herpetologica 49, 175–184.
Irwin, D., and Schluter, D. (2022). Hybridization and the coexistence of Species. Am. Nat. 200, E93–E109. doi: 10.1086/720365
Johannsson, O. E., Bergman, H. L., Wood, C. M., Laurent, P., Kavembe, D. G., Bianchini, A., et al. (2014). Air breathing in Magadi tilapia Alcolapia grahami, under normoxic and hyperoxic conditions, and the association with sunlight and reactive oxygen species. J. Fish Biol. 84, 844–863. doi: 10.1111/jfb.12289
Jones, O. R., and Wang, J. (2010). COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 10, 551–555. doi: 10.1111/j.1755-0998.2009.02787.x
Jordan, R., Kellogg, K., Juanes, F., and Stauffer, J. Jr. (2003). Evaluation of female mate choice cues in a group of Lake Malawi mbuna (Cichlidae). Copeia 2003, 181–186. doi: 10.1643/0045-8511(2003)003[0181:EOFMCC]2.0.CO;2
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Kaufman, A., Margaritz, M., Paul, M., Hillaire-Marcel, C., Hollos, G., Boaretto, E., et al. (1990). The 36C1 ages of the brines in the Magadi-Natron basin, East Africa. Geochim. Cosmochim. Acta 54, 2827–2833. doi: 10.1016/0016-7037(90)90017-F
Kautt, A. F., Machado-Schiaffino, G., and Meyer, A. (2018). Lessons from a natural experiment: Allopatric morphological divergence and sympatric diversification in the Midas cichlid species complex are largely influenced by ecology in a deterministic way. Evol. Lett. 2, 323–340. doi: 10.1002/evl3.64
Kavembe, G. D., Kautt, A. F., Machado-Schiaffino, G., and Meyer, A. (2016). Eco-morphological differentiation in Lake Magadi tilapia, an extremophile cichlid fish living in hot, alkaline and hypersaline lakes in East Africa. Mol. Ecol. 25, 1610–1625. doi: 10.1111/mec.13461
Kavembe, G. D., Machado-Schiaffino, G., and Meyer, A. (2014). Pronounced genetic differentiation of small, isolated and fragmented tilapia populations inhabiting the Magadi Soda Lake in Kenya. Hydrobiologia 739, 55–71. doi: 10.1007/s10750-013-1648-9
Keller-Costa, T., Canário, A. V. M., and Hubbard, P. C. (2015). Chemical communication in cichlids: A mini-review. Gen. Comp. Endocrinol. 221, 64–74. doi: 10.1016/j.ygcen.2015.01.001
Knight, M. E., and Turner, G. F. (1999). Reproductive isolation among closely related Lake Malawi cichlids: Can males recognize conspecific females by visual cues? Anim. Behav. 58, 761–768. doi: 10.1006/anbe.1999.1206
Knight, M. E., and Turner, G. F. (2004). Laboratory mating trials indicate incipient speciation by sexual selection among populations of the cichlid fish Pseudotropheus zebra from Lake Malawi. Proc. R. Soc. B. 271, 675–680. doi: 10.1098/rspb.2003.2639
Knight, M. E., Turner, G. F., Rico, C., Van Oppen, M. J. H., and Hewitt, G. M. (1998). Microsatellite paternity analysis on captive Lake Malawi cichlids supports reproductive isolation by direct mate choice. Mol. Ecol. 7, 1605–1610. doi: 10.1046/j.1365-294x.1998.00478.x
Kocher, T. D. (2004). Adaptive evolution and explosive speciation: The cichlid fish model. Nat. Rev. Genet. 5, 288–298. doi: 10.1038/nrg1316
Kofler, R., Schlötterer, C., and Lelley, T. (2007). SciRoKo: A new tool for whole genome microsatellite search and investigation. Bioinformatics 23, 1683–1685. doi: 10.1093/bioinformatics/btm157
Korneliussen, T. S., Albrechtsen, A., and Nielsen, R. (2014). ANGSD: Analysis of next generation sequencing data. BMC Bioinform. 15:356. doi: 10.1186/s12859-014-0356-4
Kulmuni, J., Butlin, R. K., Lucek, K., Savolainen, V., and Westram, A. M. (2020). Towards the completion of speciation: The evolution of reproductive isolation beyond the first barriers. Philos. Transact. R. Soc. B Biol. Sci. 375:20190528. doi: 10.1098/rstb.2019.0528
Lenth, R., Singmann, H., Love, J., Buerkner, P., and Herve, M. (2018). Estimated marginal means, aka least-squares means. R package version, 1(1), 3.
Maan, M. E., Seehausen, O. L. E., and Van Alphen, J. J. M. (2010). Female mating preferences and male coloration covary with water transparency in a Lake Victoria cichlid fish. Biol. J. Linn. Soc. 99, 398–406. doi: 10.1111/j.1095-8312.2009.01368.x
Machado-Schiaffino, G., Kautt, A. F., Torres-Dowdall, J., Baumgarten, L., and Henning, F. (2017). Incipient speciation driven by hypertrophied lips in Midas cichlid fishes? Mol. Ecol. 26, 2348–2362. doi: 10.1111/mec.14029
Maina, J. N., Kavembe, G. D., Papah, M. B., Mashiteng, R., Wood, C. M., Bianchini, A., et al. (2019). Sizes, condition factors and sex ratios of the scattered populations of the small cichlid fish, Alcolapia grahami, that inhabits the lagoons and sites of Lake Magadi (Kenya), one of the most extreme aquatic habitat on Earth. Environ. Biol. Fish. 102, 1265–1280. doi: 10.1007/s10641-019-00905-3
Malinsky, M., Challis, R. J., Tyers, A. M., Schiffels, S., Terai, Y., Ngatunga, B. P., et al. (2015). Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science 350, 1493–1498. doi: 10.1126/science.aac9927
Mani, G. S., and Clarke, B. C. (1990). Mutational order: A major stochastic process in evolution. Proc. R. Soc. B. 240, 29–37. doi: 10.1098/rspb.1990.0025
Marques, D. A., Meier, J. I., and Seehausen, O. (2019). A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34, 531–544. doi: 10.1016/j.tree.2019.02.008
Martin, C. H. (2013). Strong assortative mating by diet, color, size, and morphology but limited progress toward sympatric speciation in a classic example: Cameroon crater lake cichlids. Evolution 67, 2114–2123. doi: 10.1111/evo.12090
McPeek, M. A., and Wellborn, G. A. (1998). Genetic variation and reproductive isolation among phenotypically divergent amphipod populations. Limnol. Oceanogr. 43, 1162–1169. doi: 10.4319/lo.1998.43.6.1162
Meier, J. I., Marques, D. A., Mwaiko, S., Wagner, C. E., Excoffier, L., and Seehausen, O. (2017). Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 1–11. doi: 10.1038/ncomms14363
Meisner, J., and Albrechtsen, A. (2018). Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731. doi: 10.1534/genetics.118.301336
Mendelson, T. C., and Safran, R. J. (2021). Speciation by sexual selection: 20 years of progress. Trends Ecol. Evol. 36, 1153–1163. doi: 10.1016/j.tree.2021.09.004
Mérot, C., Frérot, B., Leppik, E., and Joron, M. (2015). Beyond magic traits: Multimodal mating cues in Heliconius butterflies. Evolution 69, 2891–2904. doi: 10.1111/evo.12789
Narahara, A., Bergman, H. L., Laurent, P., Maina, J. N., Walsh, P. J., and Wood, C. M. (1996). Respiratory physiology of the Lake Magadi tilapia (Oreochromis alcalicus grahami), a fish adapted to a hot, alkaline, and frequently hypoxic environment. Physiol. Zool. 69, 1114–1136. doi: 10.1086/physzool.69.5.30164249
Nevado, B., Fazalova, V., Backeljau, T., Hanssens, M., and Verheyen, E. (2011). Repeated unidirectional introgression of nuclear and mitochondrial DNA between four congeneric Tanganyikan cichlids. Mol. Biol. Evol. 28, 2253–2267. doi: 10.1093/molbev/msr043
Nyalungu, N. P., and Couldridge, V. (2020). Female mate choice and species recognition between two closely related cichlid fish of Lake Malawi, Metriaclima estherae and M. callainos. J. Fish Biol. 97, 75–82. doi: 10.1111/jfb.14327
Pedersen, T. (2022a). tidygraph: A tidy API for graph manipulation. R package version, 1.2.1. Available online at: https://cran.r-project.org/web/packages/tidygraph/
Pedersen, T. (2022b). ggraph: An implementation of grammar of graphics for graphs and networks. Available online at: https://github.com/thomasp85/ggraph (accessed July 29, 2022).
Péter, A. (2011). Solomon coder (version 11.01. 22): A simple solution for behavior coding. Computer programme. Available online at: http://solomoncoder.com (accessed January 5, 2021).
Pfennig, K., and Pfennig, D. (2009). Character displacement: Ecological and reproductive responses to a common evolutionary problem. Q. Rev. Biol. 84, 253–276. doi: 10.1086/605079
Plenderleith, M., van Oosterhout, C., Robinson, R. L., and Turner, G. F. (2005). Female preference for conspecific males based on olfactory cues in a Lake Malawi cichlid fish. Biol. Lett. 1, 411–414. doi: 10.1098/rsbl.2005.0355
Poelstra, J. W., Richards, E. J., and Martin, C. H. (2018). Speciation in sympatry with ongoing secondary gene flow and a potential olfactory trigger in a radiation of Cameroon cichlids. Mol. Ecol. 27, 4270–4288. doi: 10.1111/mec.14784
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
R Core Team (2013). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Rafferty, N. E., and Boughman, J. W. (2006). Olfactory mate recognition in a sympatric species pair of three-spined sticklebacks. Behav. Ecol. 17, 965–970. doi: 10.1093/beheco/arl030
Roberts, N., Taieb, M., Barker, P., Damnati, B., Icole, M., and Williamson, D. (1993). Timing of the younger dryas event in East Africa from lake-level changes. Nature. 366, 146–148. doi: 10.1038/366146a0
Rometsch, S. J., Torres-Dowdall, J., and Meyer, A. (2020). Evolutionary dynamics of pre-and postzygotic reproductive isolation in cichlid fishes. Philos. Trans. R. Soc. B Biol. Sci. 375:20190535. doi: 10.1098/rstb.2019.0535
Salzburger, W., and Meyer, A. (2004). The species flocks of East African cichlid fishes: Recent advances in molecular phylogenetics and population genetics. Sci. Nat. 91, 277–290. doi: 10.1007/s00114-004-0528-6
Salzburger, W., Baric, S., and Sturmbauer, C. (2002). Speciation via introgressive hybridization in East African cichlids? Mol. Ecol. 11, 619–625. doi: 10.1046/j.0962-1083.2001.01438.x
Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18, 233–234. doi: 10.1038/72708
Seegers, L., and Tichy, H. (1999). The Oreochromis alcalicus flock (Teleostei: Cichlidae) from lakes Natron and Magadi, Tanzania and Kenya: A model for the evolution of “new” species flocks in historical times? Ichthyol. Explor. Freshw. 10, 147–174.
Seegers, L., Sonnenberg, R., and Yamamoto, R. (1999). Molecular analysis of the Alcolapia flock from lakes Natron and Magadi, Tanzania and Kenya (Teleostei: Cichlidae), and implications for their systematics and evolution. Ichthyol. Explor. Freshw. 10, 175–199.
Seegers, L., Sonnenberg, R., and Tichy, H. (2001). The Alcolapia group, a remarkable species flock from lakes Natron, Tanzania, and Magadi, Kenya: A further piece of the puzzle of cichlid evolution. J. Aquaric. Aquat. Sci. 9, 335–364.
Seehausen, O. (2006). African cichlid fish: A model system in adaptive radiation research. Proc. R. Soc. B. 273, 1987–1998. doi: 10.1098/rspb.2006.3539
Seehausen, O., and van Alphen, J. J. M. (1998). The effect of male coloration on female mate choice in closely related Lake Victoria cichlids (Haplochromis nyererei complex). Behav. Ecol. Sociobiol. 42, 1–8. doi: 10.1007/s002650050405
Seehausen, O., Butlin, R. K., Keller, I., Wagner, C. E., Boughman, J. W., Hohenlohe, P. A., et al. (2014). Genomics and the origin of species. Nat. Rev. Gen. 15, 176–192. doi: 10.1038/nrg3644
Seehausen, O., Terai, Y., Magalhaes, I. S., Carleton, K. L., Mrosso, H. D. J., Miyagi, R., et al. (2008). Speciation through sensory drive in cichlid fish. Nature 455, 620–626. doi: 10.1038/nature07285
Seehausen, O., van Alphen, J. J. M., and Witte, F. (1997). Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science 277, 1808–1811. doi: 10.1126/science.277.5333.1808
Selz, O. M., Pierotti, M. E. R., Maan, M. E., Schmid, C., and Seehausen, O. (2014). Female preference for male color is necessary and sufficient for assortative mating in 2 cichlid sister species. Behav. Ecol. 25, 612–626. doi: 10.1093/beheco/aru024
Selz, O. M., Thommen, R., Pierotti, M. E. R., Anaya-Rojas, J. M., and Seehausen, O. (2016). Differences in male coloration are predicted by divergent sexual selection between populations of a cichlid fish. Proc. R. Soc. B. 283:20160172. doi: 10.1098/rspb.2016.0172
Servedio, M. R., Van Doorn, G. S., Kopp, M., Frame, A. M., and Nosil, P. (2011). Magic traits in speciation: ‘Magic’ but not rare? Trends Ecol. Evol. 26, 389–397. doi: 10.1016/j.tree.2011.04.005
Smadja, C. M., and Butlin, R. K. (2011). A framework for comparing processes of speciation in the presence of gene flow. Mol. Ecol. 20, 5123–5140. doi: 10.1111/j.1365-294X.2011.05350.x
Stankowski, S., and Ravinet, M. (2021). Defining the speciation continuum. Evolution 75, 1256–1273. doi: 10.1111/evo.14215
Stelkens, R. B., and Seehausen, O. (2009). Phenotypic divergence but not genetic distance predicts assortative mating among species of a cichlid fish radiation. J. Evol. Biol. 22, 1679–1694. doi: 10.1111/j.1420-9101.2009.01777.x
Sturmbauer, C., Baric, S., Salzburger, W., Rüber, L., and Verheyen, E. (2001). Lake level fluctuations synchronize genetic divergences of cichlid fishes in African lakes. Mol. Biol. Evol. 18, 144–154. doi: 10.1093/oxfordjournals.molbev.a003788
Svardal, H., Salzburger, W., and Malinsky, M. (2021). Genetic variation and hybridization in evolutionary radiations of cichlid fishes. Annu. Rev. Anim. Biosci. 9, 55–79. doi: 10.1146/annurev-animal-061220-023129
Taborsky, B., Guyer, L., and Demus, P. (2014). ‘Prudent habitat choice’: A novel mechanism of size-assortative mating. J. Evol. Biol. 27, 1217–1228. doi: 10.1111/jeb.12398
Templeton, A. R. (1981). Mechanisms of speciation–a population genetic approach. Annu. Rev. Ecol. Evol. 12, 23–48. doi: 10.1146/annurev.es.12.110181.000323
Terai, Y., Seehausen, O., Sasaki, T., Takahashi, K., Mizoiri, S., Sugawara, T., et al. (2006). Divergent selection on opsins drives incipient speciation in Lake Victoria cichlids. PLoS Biol. 4:e433. doi: 10.1371/journal.pbio.0040433
Tinghitella, R. M., Lackey, A. C. R., Durso, C., Koop, J. A. H., and Boughman, J. W. (2020). The ecological stage changes benefits of mate choice and drives preference divergence. Philos. Transact. R. Soc. B Biol. Sci. 375:20190546. doi: 10.1098/rstb.2019.0546
Trewavas, E. (1983). Tilapiine fishes of the genera Sarotherodon, Oreochromis and Danakilia. London: Natural History Museum Library. doi: 10.5962/bhl.title.123198
Turelli, M., Barton, N. H., and Coyne, J. A. (2001). Theory and speciation. Trends Ecol. Evol. 16, 330–343. doi: 10.1016/S0169-5347(01)02177-2
Turner, G. F., Seehausen, O., Knight, M. E., Allender, C. J., and Robinson, R. L. (2001). How many species of cichlid fishes are there in African lakes? Mol. Ecol. 10, 793–806. doi: 10.1046/j.1365-294x.2001.01200.x
Tyers, A. M., Bavin, D., Cooke, G. M., Griggs, C., and Turner, G. F. (2014). Peripheral isolate speciation of a Lake Malawi cichlid fish from shallow muddy habitats. Evol. Biol. 41, 439–451. doi: 10.1007/s11692-014-9277-4
Van Steenberge, M., Jublier, N., Kéver, L., Gresham, S., Derycke, S., Snoeks, J., et al. (2022). The initial response of females towards congeneric males matches the propensity to hybridise in Ophthalmotilapia. bioRxiv [Preprint]. doi: 10.26496/bjz.2022.100
Walsh, P. J., Grosell, M., Goss, G. G., Bergman, H. L., Bergman, A. N., Wilson, P., et al. (2001). Physiological and molecular characterization of urea transport by the gills of the Lake Magadi tilapia (Alcolapia grahami). J. Exp. Biol. 204, 509–520. doi: 10.1242/jeb.204.3.509
White, L. J., Sutton, G., Shechonge, A., Day, J. J., Dasmahapatra, K. K., and Pownall, M. E. (2020). Adaptation of the carbamoyl-phosphate synthetase enzyme in an extremophile fish. R. Soc. Open Sci. 7:201200. doi: 10.1098/rsos.201200
Wickham, H. (2016). Data analysis. ggplot2 (189-201). Berlin: Springer. doi: 10.1007/978-3-319-24277-4_9
Williamson, D., Taieb, M., Damnati, B., Icole, M., and Thouveny, N. (1993). Equatorial extension of the younger dryas event: Rock magnetic evidence from Lake Magadi (Kenya). Glob. Planet. Change 7, 235–242. doi: 10.1016/0921-8181(93)90053-Q
Wood, C. M., Bergman, H. L., Bianchini, A., Laurent, P., Maina, J., Johannsson, O. E., et al. (2012). Transepithelial potential in the Magadi tilapia, a fish living in extreme alkalinity. J. Comp. Physiol. 182, 247–258. doi: 10.1007/s00360-011-0614-y
Wood, C. M., Brix, K. V., De Boeck, G., Bergman, H. L., Bianchini, A., Bianchini, L. F., et al. (2016). Mammalian metabolic rates in the hottest fish on earth. Sci. Rep. 6, 1–9. doi: 10.1038/srep26990
Wright, D. S., Rietveld, E., and Maan, M. E. (2018). Developmental effects of environmental light on male nuptial coloration in Lake Victoria cichlid fish. PeerJ 6:e4209. doi: 10.7717/peerj.4209
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13:134. doi: 10.1186/1471-2105-13-S6-S1
Yukilevich, R. (2012). Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution 66, 1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x
Keywords: adaptive radiation, speciation, reproductive isolation, hybridization, behavior
Citation: Lawson MRM, Hayle MGB, Shechonge AH, Nyingi WD, Ford AGP, Hoffman JI, Day JJ, Turner GF and Dasmahapatra KK (2023) Sympatric and allopatric Alcolapia soda lake cichlid species show similar levels of assortative mating. Front. Ecol. Evol. 11:1150083. doi: 10.3389/fevo.2023.1150083
Received: 23 January 2023; Accepted: 21 March 2023;
Published: 06 April 2023.
Edited by:
David Andrew Gray, California State University, Northridge, United StatesReviewed by:
Michael Pauers, Milwaukee Public Museum, United StatesCopyright © 2023 Lawson, Hayle, Shechonge, Nyingi, Ford, Hoffman, Day, Turner and Dasmahapatra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael R. M. Lawson, bXJtbDUwMEB5b3JrLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.