- Stazione Zoologica Anton Dohrn, Naples, Italy
Morphological and molecular characterization of cell types, organs and individual organisms is essential for understanding the origins of morphogenesis. The increased implementation of high throughput methods as a means to address cell type evolution, during the last decade, created the need for an efficient way to assess cell type morphology. Here in order to create a new tool to study cell type morphology, we optimized a fast and easy-to-use whole animal freeze-fracture scanning electron microscopy (WAFFSEM) method. This method was applied on marine experimental systems (echinoderms, mollusks, tunicates, and cephalochordates), that have been widely used to assess environmental, developmental, and evolutionary questions. Our protocol does not require any specialized equipment and the processed specimens are compatible with scanning electron microscopy. This protocol was able to successfully expose the internal cell types of all specimens in which it was tested and to reveal their cellular and subcellular characteristics. We strongly believe that the combination of our protocol with other methods (e.g., light microscopy and single cell transcriptomics) will be beneficial to further improve the way to classify and describe cell types.
1. Introduction
Cells are the building blocks of life, and their development, specification, differentiation, and function are tightly orchestrated by the genetic program operating within them and by how this is properly executed. Multicellular organisms consist of various cell types that can either be found dispersed within the body fluids or organized in distinct units such as tissues and organs. Each specific cell type consists of cells with similar morphological features that are the result of specific gene regulatory programs and epigenetic mechanisms dictating their identity and function (Davidson and Erwin, 2006). Nowadays, high throughput technologies have been developed to assess cell type complexity in an unprecedented resolution. For instance, advances in transcriptomics have allowed the capture of the molecular signature of a given organism, tissue, or organ at a single cell resolution, resolving the identification of putative cell types in an unbiased way. These technologies have been applied on plenty of animals across the evolutionary tree of life including representatives from both invertebrates and vertebrates. Specifically, single cell transcriptomics has been successfully applied on a variety of organisms spanning from sponges, planarians, mollusks, crustaceans, cnidarians and echinoderms to tunicates, cephalochordates, and mammals (Plass et al., 2018; Cao et al., 2019; Tabula Muris, 2020; Chari et al., 2021; Massri et al., 2021; Musser et al., 2021; Paganos et al., 2021; Satoh et al., 2021; Almazan et al., 2022; Murat et al., 2023). The implementation of such approaches has contributed to the reconstruction of distinct cell type atlases as well as the revaluation of embryonic development and cell type evolution. However, in order to fully define a given cell type apart from the identification of the expressed genes within it, it is imperative to understand how this genetic information is translated into phenotypic features. In other words, the assessment of the molecular signature of a given cell type should always go hand in hand with its morphological assessment.
Traditionally, light microscopy has been used to describe the morphology of animals’ cell types and has been heavily exploited in cases where the specimens are optically transparent. On the other hand, for specimens that lack this feature, clearing methods have been developed to allow a better visualization of tissues that otherwise would remain hidden due to the presence of extensive lipid membranes, protective organs, or pigments.
In order to facilitate the study of cell type morphology we optimized a simple, efficient, and low-cost whole animal freeze-fracture scanning electron microscopy (WAFFSEM) protocol. Marine organisms as experimental systems are ideal to approach Evo-Devo research questions. They offer many advantages, such as a high number of transparent embryos/larvae, as well as a well resolved evolutionary history and distances that allow meaningful comparisons (Stracke and Hejnol, 2023). We, therefore, applied the WAFFSEM protocol on a variety of extensively used marine organisms including mollusks, echinoderms, tunicates, and cephalochordates. This freeze fracture protocol combined with subsequent scanning electron microscopy observation allowed us to spatially recognize well-known cell types and to further describe their phenotypic features. Moreover, focusing on echinoderms we also used correlative approaches as proof of concept of the complementarity of our method with other techniques, always in respect to cell type characterization.
2. Materials and methods
2.1. Animal collection, in vitro fertilization and rearing of embryos and larvae
2.1.1. Sea urchin
Adult Paracentrotus lividus individuals were collected from the Gulf of Naples (Italy), while the Pacific Ocean sea urchin Strongylocentrotus purpuratus individuals were collected from the Gulf of Santa Catalina (CA, United States) and distributed by Patrick Leahy (Kerckhoff Marine Laboratory, California Institute of Technology, Pasadena, CA, United States). Gametes were obtained by vigorous shaking of the adult individuals. Fertilization of eggs was performed by using approximately 1:1,000 dry sperm diluted in filtered sea water (FSW) and larvae were let to develop according to their species salinity and temperature biological needs. P. lividus larvae were cultured at 18°C in Mediterranean FSW and S. purpuratus at 15°C in Mediterranean FSW diluted 9:1 with deionized water.
2.1.2. Sea star
Adult Patiria miniata individuals were collected from the Gulf of Santa Catalina (CA, United States) and distributed by Patrick Leahy (Kerckhoff Marine Laboratory, California Institute of Technology, Pasadena, CA, United States). Gametes were collected by performing a 4 mm diameter incision on one side of each animal’s arm. A piece of each gonad was extracted and placed in calcium and magnesium-free artificial sea water (CMF-ASW). In the case of female gametes, immature eggs were treated with 10 μM 1-Methyladenine in Mediterranean FSW for approximately 1 h until the germinal vesicle (GV) disappears, indicating their maturity. Mature eggs were then fertilized with approximately 1:1,000 dry sperm diluted in FSW, and zygotes were let to develop, until they reached the desired developmental stage at 15°C in Mediterranean FSW diluted 9:1 with deionized water.
2.1.3. Sea squirt
Adult Ciona robusta individuals were collected from the Gulf of Taranto (Italy). Gametes from adult individuals were collected for in vitro fertilization and chemical dechorionation was performed prior to fertilization as previously described (D'aniello, 2009). Embryos were allowed to grow until the stage of interest at 18°C in Mediterranean FSW.
2.1.4. Mussel
Adult individuals of Mytilus galloprovincialis were purchased from Irsvem Srl (Naples, Italy) and were used immediately for spawning. Spawning of male and female individuals was induced through mechanical stimulation. Approximately 20–30 mussels were placed in a tank with Mediterranean FSW at 18°C and spread to easily monitor the spawning. When spawning started, each individual was washed and then transferred into a Becker containing 200 mL of Mediterranean FSW to isolate males and females. Mature eggs were fertilized with an eggs/sperm ratio 1:15 in a volume of 50 mL. The resulting zygotes were let to develop at 18°C in Mediterranean FSW until the developmental stage of interest.
2.1.5. Amphioxus
Branchiostoma lanceolatum embryos and larvae were reared from animals collected in Argelès-sur-mer (France) and spawned at Observatoire Océanologique de Banyuls-sur-Mer (France). Gametes were obtained during late spring/early summer period and the spawning was induced through thermal shock (Fuentes et al., 2007). Mature eggs were then fertilized with approximately 1:1,000 dry sperm diluted in FSW. Embryos were let to develop until they reached the desired developmental stage at 18°C in Mediterranean FSW.
2.2. Fluorescent in situ hybridization, immunohistochemistry and F-actin detection
2.2.1. Fluorescent in situ hybridization
Strongylocentrotus purpuratus and P. lividus larvae, intended to be used for FISH, were fixed as previously described in Paganos et al.(2022a). Synthesis of antisense mRNA probes against the genes of interest and FISH were performed as described by Perillo et al. (2021) and Paganos et al. (2022a). Briefly, genes of interest were isolated from a pool of cDNAs, and the amplified products were sequenced. Probes were generated through in vitro transcription using Digoxigenin RNA labeling mix solutions (Roche) and signal was developed through cyanine-based signal amplification (Akoya Biosciences). DAPI was added (1 μg/mL) and specimens were imaged using a Zeiss LSM 700 confocal microscope.
2.2.2. Immunohistochemistry and F-Actin detection
Mytillus galloprovinicialis, P. miniata, P. lividus and S. purpuratus embryos and larvae, intended to be used for IHC and F-Actin detection, were fixed as described in Perillo et al. (2021). In brief, specimens were fixed in 4% PFA in FSW for 15 min at RT. After fixation, samples were washed several times with MOPS Buffer and stored at 4°C. IHC was carried out as described in Perillo et al. (2021) with minor modifications. Specimens were washed several times with MOPS buffer, then they were placed in a blocking solution containing 1 mg/mL Bovine Serum Albumin (BSA) and 4% sheep serum in MOPS buffer for 1 h at room temperature (RT). Primary antibodies were added in the appropriate dilution and incubated for 1 h and 30 min at 37°C. Anti-Endo1 (gift from Dr. David McClay) was used undiluted to mark the sea urchin mid and hindgut domains; anti-Sp-Brn1/2/4 (gift from Dr. Robert Burke) was used instead to label sea urchin neurons and the esophageal regions and used diluted 1:200 in blocking solution; anti-Msp130 labeling the sea urchin skeletal spicules (gift from Dr. David McClay) was used with a dilution factor 1:10 in blocking solution; anti-Acetylated tubulin (Sigma) was chosen to label cilia and was used diluted 1:200 in blocking solution. Specimens were washed with MOPS Buffer (5 times) and incubated for 1 h with the appropriate secondary antibody (AlexaFluor) diluted 1:1,000 in MOPS buffer. F-actin staining was performed using phalloidin solution (Invitrogen) diluted 1:20 in MOPS buffer. Samples were let to incubate for 1 h at RT. Next, specimens were washed 5 times with MOPS buffer and DAPI was added (1 μg/mL). Specimens were imaged using a Zeiss LSM 700 confocal microscope.
2.3. Catecholamine staining (FaGlu)
Paracentrotus lividus early pluteus larvae were collected and placed in a solution containing 4% formaldehyde and 0.5% glutaraldehyde in FSW (FaGlu). Specimens were incubated for 1 h at RT in the dark. Next, a drop of specimens was transferred on a glass slide and FaGlu was used as mounting medium. A coverslip was added and samples were imaged using a Zeiss LSM 700 confocal microscope. Dopaminergic neurons were detected using the 475–480 nm emission peak.
2.4. Whole animal freeze-fracture scanning electron microscopy protocol
• Whole embryos at the developmental stage of interest were fixed in 2.5% glutaraldehyde in FSW and placed at 4°C overnight. The day after, the samples were washed five times with FSW and were let to settle. The duration of each wash was 15 min at RT.
• FSW was then removed and replaced with 25% DMSO in double distilled water (ddH20) in which specimens were incubated for 30 min at RT. This step was repeated twice.
• The 25% DMSO solution was removed and replaced with 50% DMSO in ddH20 and embryos or larvae were incubated for 30 min at RT. This step was repeated twice.
• During the last 30 min of incubation a metal key, a pair of forceps, an aluminum tray and a razor blade were placed in a polyurethane ice bucket and covered with liquid nitrogen to pre-cool.
• Pre-chilled forceps were used to remove the key from the liquid nitrogen bath and placed on the bench. This step is necessary to avoid the immediate freezing of the sample, while being transferred onto the pre-chilled key. Approximately 100 μL of specimens were quickly transferred with a P200 pipette inside the groove of the key and spread throughout the groove surface. The key was quickly returned inside the pre-chilled aluminum tray.
• Using the same pair of forceps, the pre-chilled razor blade was collected and placed in parallel and on top in respect to the key’s groove. Using a hammer, the razor blade was stricken multiple times in a downward motion until complete fracturing of the pellet.
• The key was transferred on the bench and the fractured pellet was let to thaw at RT. Then, using a P200 pipette, the solution was placed in clean Eppendorf tubes.
• Once the fractured specimens settled down, DMSO solution was removed, and specimens were gradually dehydrated by passing them in 30, 50 and 70%, 80, 90% molecular grade ethanol diluted in ddH20. The incubation time for each dehydration step was 15 min.
• To ensure the complete dehydration, samples were incubated 3 times (15 min each) in absolute ethanol.
• Samples were then subjected to the standard processing for SEM including critical point drying and sputtering and were analyzed using a Jeol field emission scanning electron microscope (JSM 6700-F).
3. Results
The accurate characterization of cell type morphology is essential to better understand which are the key features responsible for cell type diversification and to gain insight into their function and interconnectivity with other cells, tissues, or organs. In this study we developed an easy-to-use WAFFSEM protocol that we tested on a variety of marine embryos and larvae. A schematic and detailed representation of this protocol is depicted in Figure 1.
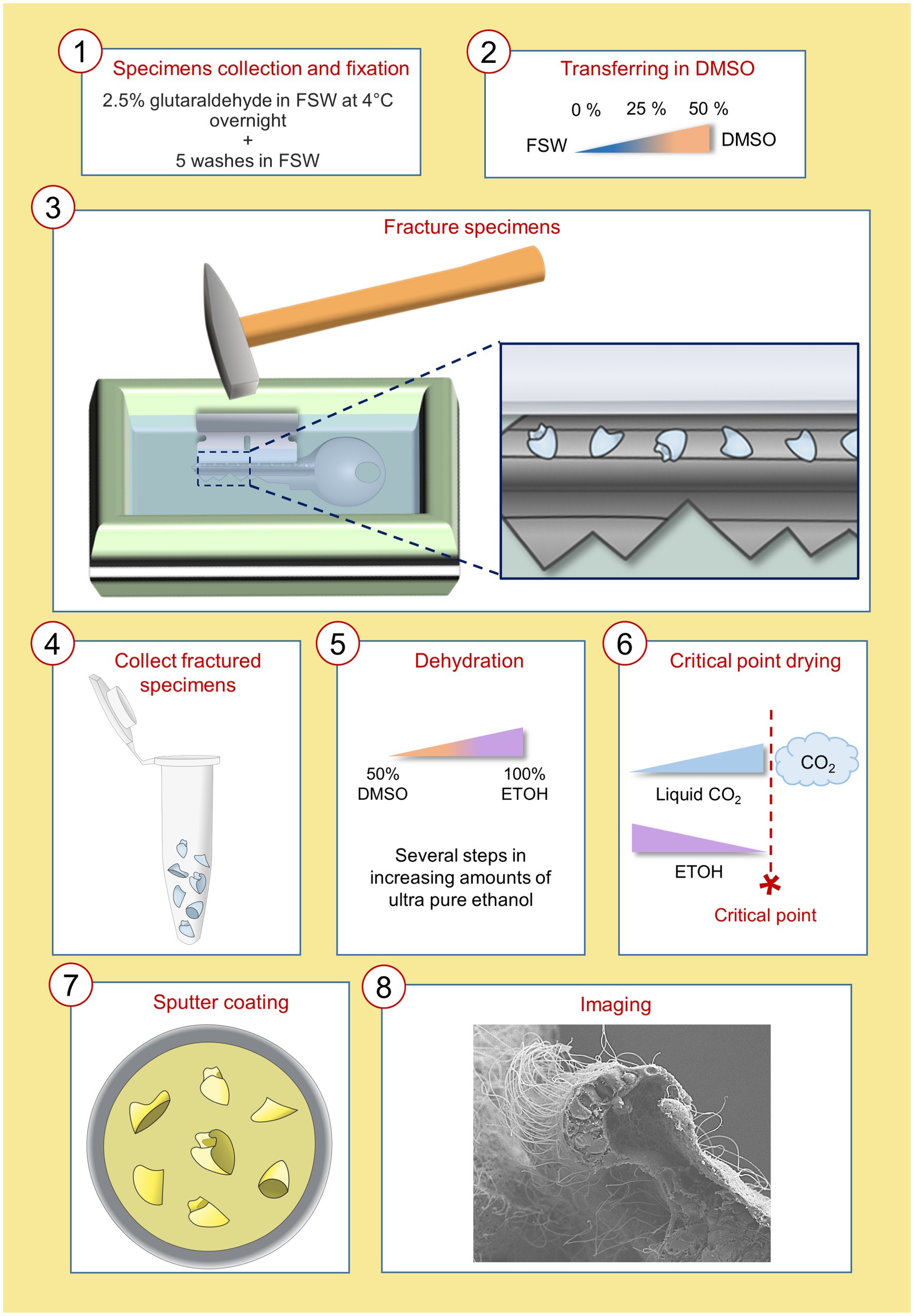
Figure 1. Schematic representation of the WAFFSEM protocol steps from specimen fixation to imaging. Details on the precise steps of the protocol can be found in materials and methods, section 2.4.
The idea for developing this protocol is based on a previous study led by MacDonald et al. (2017), in which the authors developed a method for freeze-fracturing and subsequent observation with scanning electron microscopy of isolated murine mitochondria. Our method is a simplified version of that protocol that has been adapted to be suitable for whole organisms. Here we present WAFFSEM examples for several marine organisms including representatives of both protostomes and deuterostomes. In detail, this protocol was successful to visualize phenotypic features for the tunicate C. robusta, the cephalochordate B. lanceolatum, the mollusk M. galloprovincialis, the sea star P. miniata and for the two sea urchin species P. lividus and S. purpuratus.
3.1. Tunicates, cephalochordates, and mollusks
The WAFFSEM protocol applied on C. robusta and B. lanceolatum embryos and larvae was shown to have high efficiency. An advantage for using those species as test subjects were the availability of great numbers of starting material from each fertilization and their relatively big size (C. robusta mid-tailbud stage ~ 450 μm; T0 stage B. lanceolatum larva ~ 350 μm; L0 stage B. lanceolatum larva ~ 750 μm) that increases the likelihood of specimens being successfully fractured. Taking advantage of the available information about cell types in C. robusta and B. lanceolatum, we were able to recognize key features. In the case of the C. robusta embryo at the mid tailbud stage (Figure 2A) WAFFSEM was able to successfully reveal the morphology of cell type derivatives from all germ layers (Figures 2A–I). In detail, we were able to visualize putative epidermal, mesenchymal, and endodermal cells (Figures 2C,D,F). Furthermore, details on the spatial position of neuronal cell types, such as the ones constituting the nerve cord, are shown and especially in respect to supportive cell types such as muscles and the notochord (Figures 2F–I). Our WAFFSEM protocol was also effective on the cephalochordate B. lanceolatum (Figures 2J–O). We recovered a T0 stage larva approximately longitudinally fractured at the pharynx level, and we were able to recognize morphological characteristics of cell types related to the neural tube, the notochord, longitudinal muscles, and the gut (Figure 2K). Moreover, at the anterior portion of this specimen, we observed a structure potentially related to the primordium of the club-shaped gland (Figure 2K). Immediately posterior to the presumptive club-shaped gland domain, a thickening of the endoderm on the right side of the pharyngeal region is visible and this can be identified as the primordium of the first gill slit (Figure 2K). Concerning the L0 stage larva, depicted in Figures 2L–O and fractured transversally at the posterior part of the body, we can clearly see the lumen of the gut and the cell types around it. Moving from the dorsal to the ventral part of the specimen we observed the anatomy and cell morphology of the neural tube and especially of the cells comprising its wall and the central hollow tube (Figures 2M,N). Immediately below the neural tube we see the notochord and the gut (Figures 2N,O). Moreover, we can clearly see the longitudinal muscles positioned bilaterally to the notochord (Figure 2N).Taken together, we concluded that the WAFFSEM protocol is applicable on marine organisms and is able to reveal the morphological features of various tunicate and cephalochordate cell types.
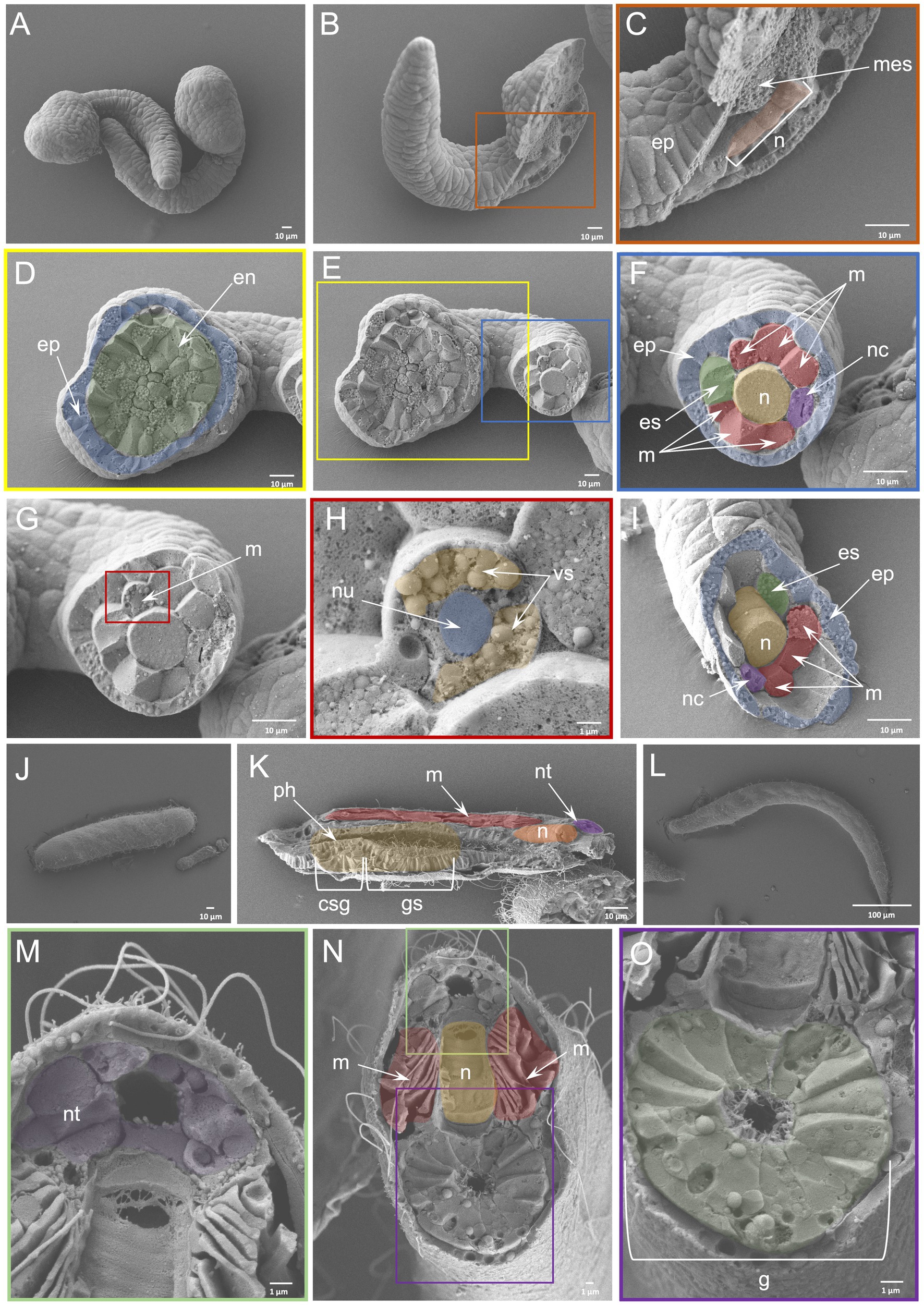
Figure 2. Efficiency of WAFFSEM protocol on tunicate and cephalochordate embryos and larvae. (A) Intact C. robusta mid-tailbud stage embryos. (B) Fractured C. robusta embryo in lateral view. (C) Inset of panel B focused on the fractured region depicting mesenchymal cells and the notochord. (D) Inset showing the head region of the fractured specimen shown in (E), displaying endodermal and epidermal cells. (E) Fractured C. robusta embryo in ventral view. (F) Inset focused on the tail region of the fractured specimen shown in E. A variety of cell types including muscles, notochord, nerve cord, epidermis, and endodermal strand are visible. (F, G) SEM image depicting a C. robusta fractured tail. (H) Inset showing a fractured muscle cell and its ultrastructure. The nucleus and vesicle-like structures are visible. (I) SEM image of fractured C. robusta mid-tailbud stage embryo focused on the tail region and containing the same cell types as labeled in F. (J) SEM image of intact B. lanceolatum T0 stage larva. (K) SEM image of fractured B. lanceolatum T0 stage larva. Several cell types including the primordium of the club-shaped gland and gill slit as well as muscles, notochord, neural tube and pharynx are visible. (L) SEM image of intact B. lanceolatum L0 stage larva. (M–O) SEM images of fractured B. lanceolatum L0 stage larva. (M) Inset showing details of the dorsal region of the specimen depicted in N. (O) Inset depicting the digestive tract of the specimen shown in N. csg, club-shaped gland primordium; en, endoderm, ep, epidermis; es, endodermal strand; (G), gut; gs, gill slit primordium; mes, mesenchyme; (M), muscles; (N), notochord; nc, nerve cord; nt, neural tube; nu, nucleus; ph, pharynx; vs., vesicle. Pseudo-coloring was applied to enhance the visualization of the cell types of interest.
In order to test whether our protocol gives similar results on smaller specimens we also applied it on the D-Larva of the bivalve mollusk M. galloprovincialis (Figures 3A–F). Furthermore, using a specimen that has a calcium-based shell offers the possibility to test whether such structures interfere with the efficiency of our method. Surprisingly, the efficiency of our WAFFSEM protocol did not seem to be altered either by the smaller size of the specimen or the presence of the shell and the results of this are shown in Figures 3C,F. In this case, we also implemented the use of light microscopy in order to recognize distinct cell types and to show the correlative potential of using WAFFASEM combined with other imaging techniques (Figures 3B,E). In Figure 3 panels C, F fractured M. galloprovincialis D-larvae are depicted in oral view and we can see various cell types such as muscles, branchial gills and ciliated cells as well as parts of the gut luminary epithelium. Noteworthy, the combination of our WAFFSEM protocol with light microscopy allowed us to map on the SEM images cell types such as muscles and ciliated cells (Figures 3B,C,E,F). In conclusion, our WAFFSEM protocol can be used as a tool to assess cell type morphology of the M. galloprovincialis larva at a cellular resolution, information that when combined with other imaging approaches can be used for further cell type characterizations.
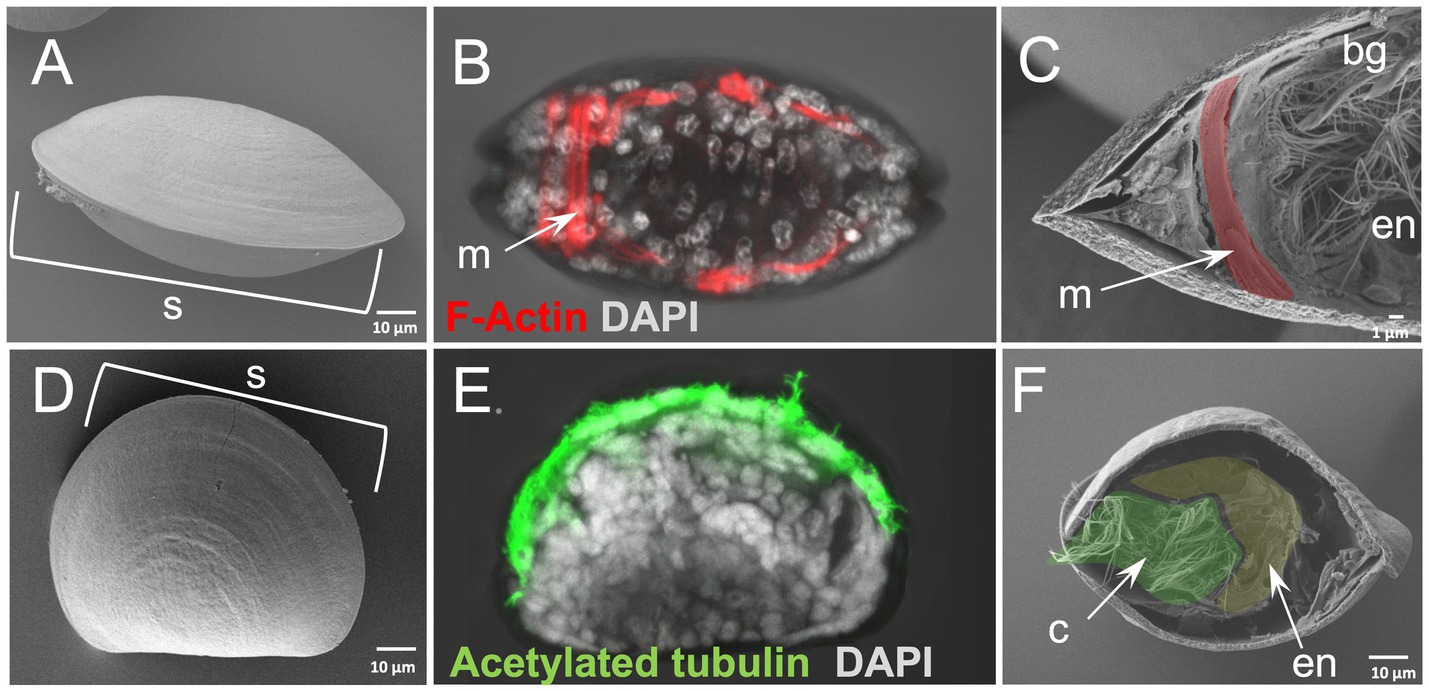
Figure 3. Whole animal freeze-fracture scanning electron microscopy (WAFFSEM) protocol applied on mollusk larvae. (A) SEM image of intact Mytilus galloprovincialis 3 dpf D-larva stage in oral view. (B) Phalloidin staining showing the F-Actin localization in muscle cells seen in oral view. (C) SEM image of a fractured M. galloprovincialis 3 dpf D-larva stage oriented in oral view. Muscle fibers, endodermal structures and the brachial gills are visible. (D) SEM image of intact M. galloprovincialis 3 dpf D-larva stage in lateral view. (E) Immunohistochemical detection of acetylated tubulin enriched cilia in a M. galloprovincialis 3 dpf D-larva stage positioned in lateral view. (F) SEM image of fractured M. galloprovincialis 3 dpf D-larva stage seen in oral view. Cilia and endodermal regions are visible. Nuclei in panels B and E are labeled with DAPI (in gray). bg, branchial gills; c, cilia; en, endoderm; m, muscles; s, shell; Pseudo-coloring was applied to enhance the visualization of the cell types of interest.
3.2. Echinoderms
Echinoderms are non-chordate deuterostomes and represent an ideal model for comparative Evo-Devo approaches at a cell-type resolution level. Historically, echinoderms have been extensively used to describe cell type specific morphological features (Ernst, 2011). Moreover, several molecular biology tools such as ATAC-seq and single cell transcriptomics have been successfully implemented to address chromatin dynamics, cell type composition and evolution during echinoderm embryonic and larval development. Therefore, echinoderms were the perfect candidates to apply our WAFFSEM protocol in a correlative perspective.
To this end we performed the WAFFSEM protocol on early bipinnaria larvae of the sea star P. miniata, which enabled us to assess the cell type morphology of distinct larval cell types (Figure 4). In Figure 4 panel B, a larva in lateral view is depicted in which the foregut and midgut domains have been successfully fractured. In this specimen, we report the presence of distinct muscle fibers and ciliated cells in the esophageal region (Figure 4C), while the midgut domain is enriched in ciliated cells projecting their cilia within the lumen region (Figures 4C,F). In this case the direction of the fracture allowed us to also observe several subcellular components of the midgut cells including nuclei, round structures that are indicative of vesicles as well as small protrusions, corresponding to microvilli (Figure 4C). In the same figure, panels D and E, a larva in oral view is shown and we report fractured cells corresponding to the oral hood and apical organ domains of the larva known to be enriched in neuronal cell types (Zheng et al., 2022). Interestingly, on the same specimen, but focusing on another fractured region, spanning from the lower foregut to the midgut domains, we were able to observe the morphology of the cells forming the constriction between the foregut and midgut domains, known as the cardiac sphincter (Figure 4H). Moreover, the great resolution of these images allowed us also to clearly distinguish two hollow tubes corresponding to the left and right coeloms of the animal (Figures 4D,G,H). In addition to the previous structures observed from this view, we can also see the cell consistency of the posterior enterocoel, a structure that arises from the dorsal side of the digestive tract and later buds off the left endodermal epithelium (Figures 4I,J). This structure has been associated with the metamorphosis process in this species and has been also hypothesized to be a stem cell reservoir (Wessel et al., 2014). Furthermore, we were able to detect many vesicle-like structures and filaments expanding towards the epidermal cells of the larva and the connective tissue holding this structure in place. Ultimately, we report the presence of an extensive extracellular matrix (ECM) network that can be seen in all panels presented in Figure 4.
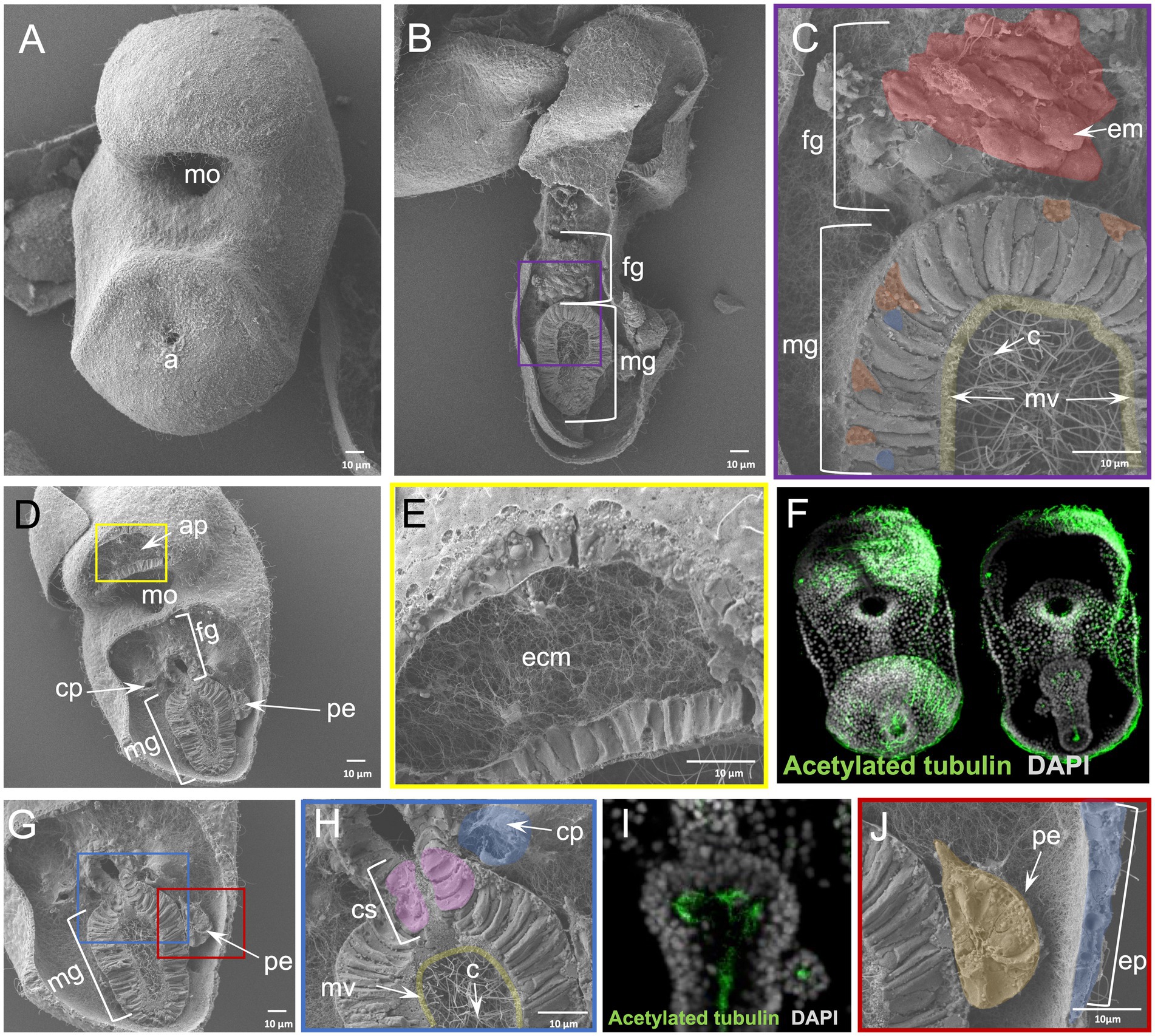
Figure 4. Results of WAFFSEM protocol applied on sea star larvae. (A) SEM image of an intact Patiria miniata ~ 3 dpf bipinnaria larva. (B) SEM image of a fractured P. miniata ~ 3 dpf bipinnaria larva placed in lateral view. (C) Inset of panel B showing in higher magnification the foregut and midgut regions. Muscles patterning the foregut region as well as cilia and microvilli inside the midgut area are highlighted. (D) SEM image of a fractured P. miniata ~ 3 dpf bipinnaria larva placed in oral view. (E) Inset of panel D showing the extensive extracellular matrix (ECM) network in the apical organ/oral hood region of the specimen. (F) Immunohistochemical detection of cilia using anti-acetylated tubulin. (G) SEM image of a fractured P. miniata ~ 3 dpf bipinnaria larva placed in lateral view focused on the digestive tract region. (H) Inset of panel G showing the lower foregut and upper midgut regions of the larva. The cardiac sphincter, left coelom as well as cilia and microvilli of the midgut are evident. (I) Immunohistochemical detection of cilia within the digestive tract, in dorsal view, using anti-acetylated tubulin. (J) Inset of panel G focusing on the posterior enterocoel and the epidermis of the ~ 3 dpf larva. Nuclei in panels F and I are labeled with DAPI (in gray). ap, apical plate; c, cilia; cp, coelomic pouches; cs, cardiac sphincter; e, epidermis; em, esophageal muscle; ecm, extracellular matrix; fg, foregut; mg, midgut; mo, mouth; mv, microvilli; pe, posterior enterocoel. Pseudo-coloring was applied to enhance the visualization of the cell types of interest.
In the case of the sea urchin P. lividus early pluteus larva, a stage for which plenty of molecular information is available (Figure 5A) WAFFSEM was able to reveal distinct features of various larval cell types. In Figure 5 panels B,C, a larva in abanal view is shown and we can easily observe the midgut domain of the digestive tract and the morphology of the cells constituting it, including small vesicle-like structures, the nuclei, microvilli, and cilia. Moreover, we can observe a cell that, based on its spatial location, we hypothesize it to be corresponding to an immune cell that is in contact with the midgut region, but not embedded with it. Interestingly, in Figures 5 panels B, D, we also observed cells that based on their relative position could correspond to the post oral dopaminergic neurons known to be present at sea urchin early pluteus stage as approximately 1–2 cell clusters positioned bilaterally to the midgut region. The distribution of dopaminergic neurons in P. lividus early larvae was visualized using catecholamine staining (FaGlu) and can be seen in Figure 5 panel E. Taking into account the shared morphological features of the dopaminergic neuron shown in Figure 5 panel E and the cell shown in Figure 5 panel D, such as the presence of two axonal-like projections and the spatial position in respect to the post oral arms of the larva, we hypothesize that the cells shown in B and D corresponds to post oral neurons. Furthermore, we can see epidermal cells of various shapes in the regions that correspond to the ciliary band and specifically to the post oral arms of the larva (Figures 5B,D).
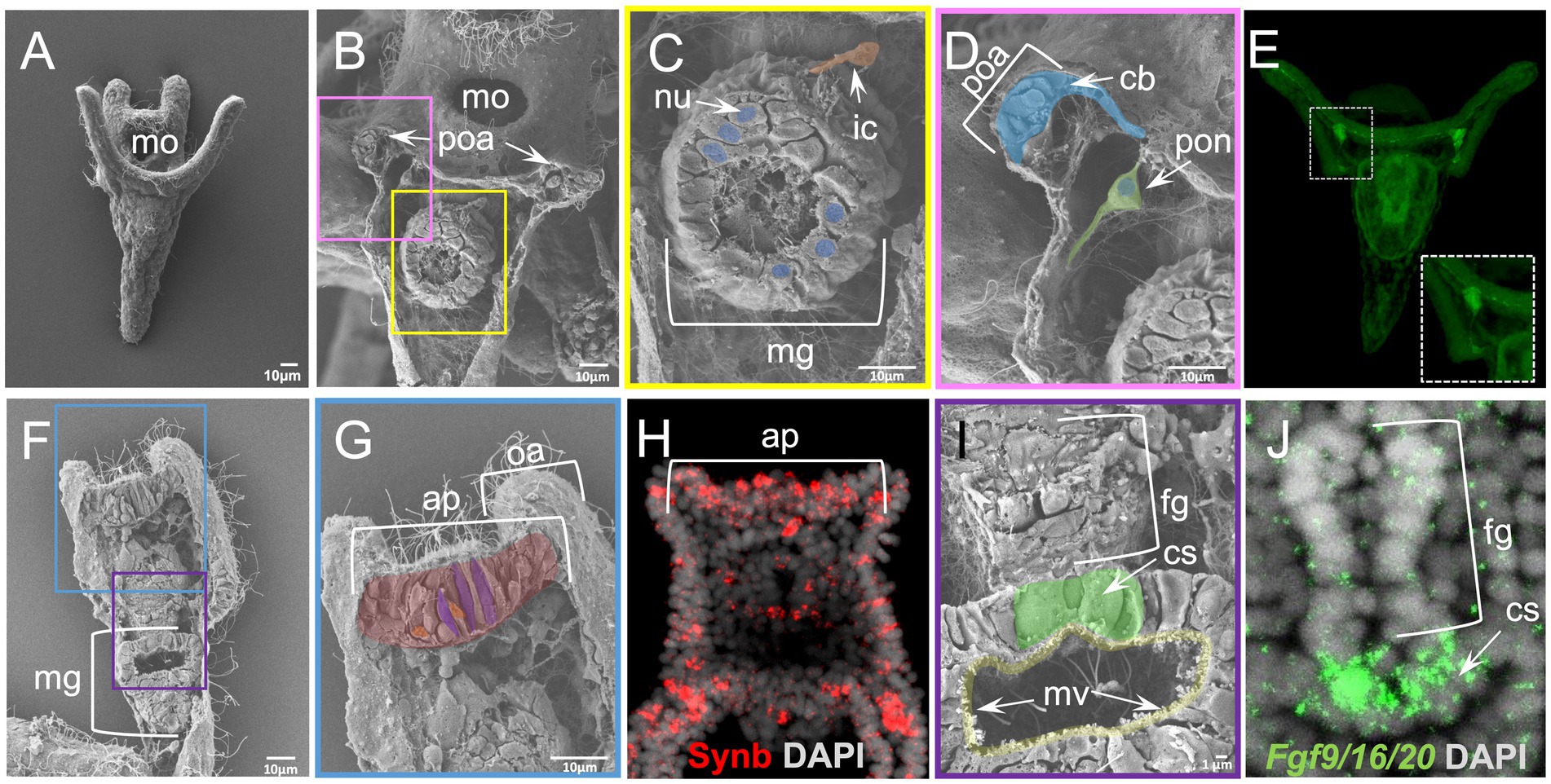
Figure 5. Results of WAFFSEM protocol applied on Paracentrotus lividus early larvae. (A) SEM image of an intact P. lividus 4 arm pluteus larva. (B) SEM image of a fractured P. lividus 4 arm larva seen in abanal view. The post oral arms and midgut regions are visible. (C) Inset of panel B showing in higher magnification the midgut region. An immune cell that is in contact with the midgut region as well as nuclei of midgut cells are visible. (D) Inset of panel B showing in higher magnification the post oral arm cell consistency and the post oral neuron. (E) Fluorescent detection of dopamine through FaGlu staining in a P. lividus larva. The dopaminergic post oral neurons are organized in bilaterally symmetric cell clusters in respect to the digestive tract. Axonal projections are found across the post oral arms and ciliary band. (F) SEM image of a fractured P. lividus 4 arm larva placed in dorsal view. (G) Inset of panel F showing the apical plate region of the specimen. (H) FISH using antisense probe against the sea urchin paneuronal marker Synb. (I) Inset of panel F showing the lower foregut and upper midgut regions of the larva in higher magnification. The cardiac sphincter and midgut microvilli are visible. (J) FISH using antisense probe for Fgf9/16/20 marking the cardiac sphincter region. Nuclei in panels H and J are labeled with DAPI (in gray). ap, apical plate; cb, ciliary band; cs, cardiac sphincter; fg, foregut; ic, immune cell; mg, midgut; mo, mouth; mv, microvilli; nu, nucleus; oa, oral arm; ps, pyloric sphincter; poa, post oral arm; pon, post oral neuron. Pseudo-coloring was applied to enhance the visualization of the cell types of interest.
On another specimen, seen from a dorsal view and fractured longitudinally, we report the presence of cells in various shapes in the apical plate region (Figures 5F,G). Cross-referencing the SEM images showing the exposed apical plate region (Figures 5F,G) with FISH against the paneuronal sea urchin marker Synb (Figure 5H) carried out at the same developmental stage, we speculate that most of the cells shown in panel G correspond to neurons. In support of this, several apical plate cells seem to contain vesicle-like structures and to have a flask-like cell body shape (Figures 5F,G), a known feature of sea urchin apical organ serotonergic neurons. Moreover, in the same specimen we can clearly see the ciliated epithelium of the same region as well as an extensive ECM network. Moving on to a different region of the same specimen, we can clearly see features of the digestive tract and the midgut domain, including cilia, microvilli, and vesicle-like structures (Figures 5G,H). One of the easily observed regions in the fractured specimen in panel I is the constriction corresponding to the cardiac sphincter that is separating the midgut from the foregut domain. Noteworthy, the architecture of the sphincter in the fractured specimen was confirmed when we performed in situ hybridization and compared to the confocal image of Fgf9/16/20, a marker labeling larval sphincters, further showing the power of the combination of these two approaches (Figures 5I,J).
Finally, we applied the WAFFSEM protocol on the 3 dpf S. purpuratus pluteus larva (Figure 6A), a species and developmental stage, for which we recently generated a detailed cell type atlas allowing us to determine its specific cell type composition (Paganos et al., 2021). Briefly, we previously demonstrated that this stage consists of 21 cell type families that we mapped into distinct domains of the larva (Figures 6B,C). Moreover, we recently showed that the implementation of single cell transcriptomics analyses and electron microscopy is sufficient to assess cell type homology (Paganos et al., 2022b).
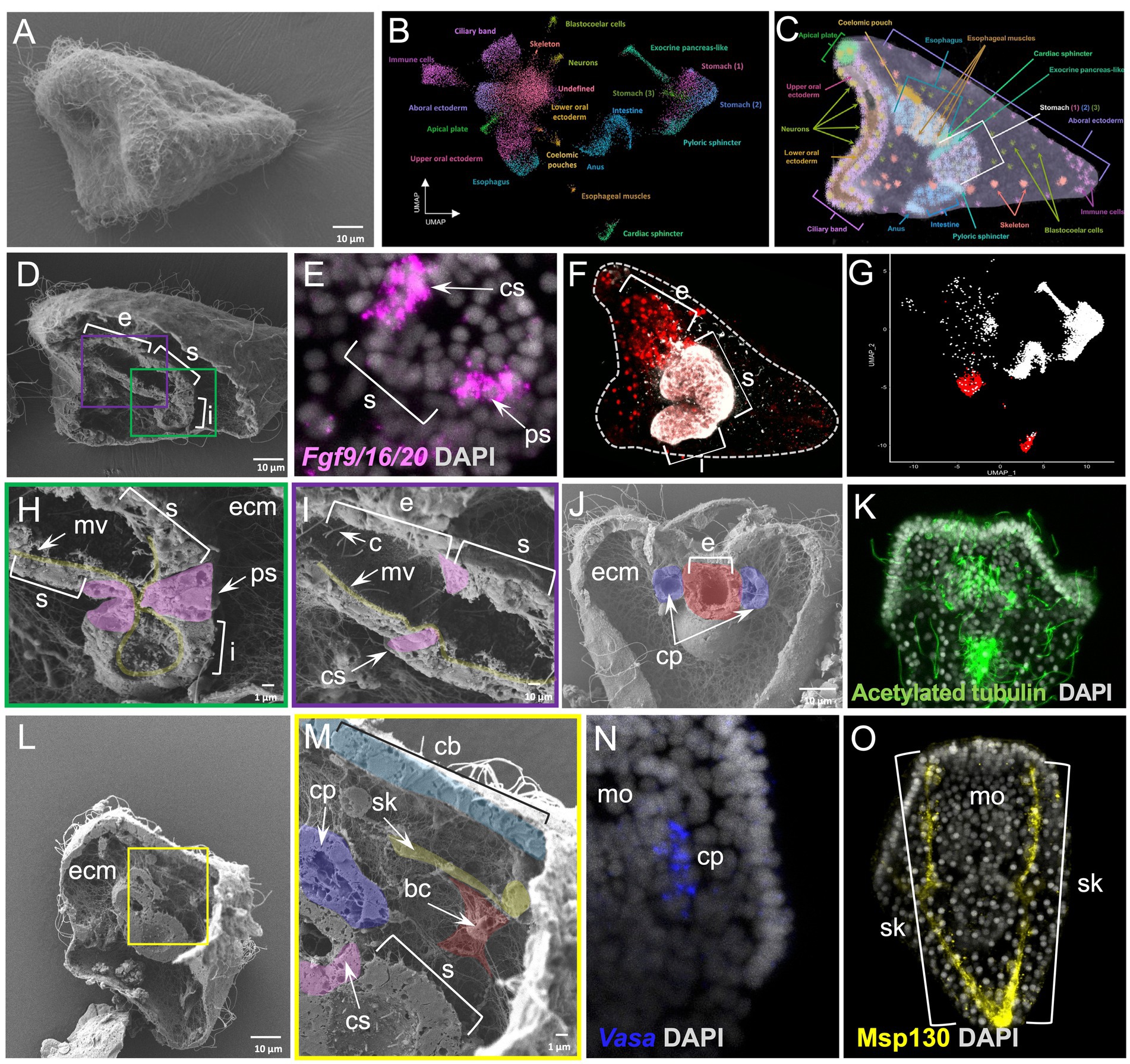
Figure 6. Whole animal freeze-fracture scanning electron microscopy protocol applied on Strongylocentrotus purpuratus 3 dpf larvae combined with FISH, IHC and scRNA-seq. (A) SEM image of an intact S. purpuratus 3 dpf pluteus larva. (B) UMAP representing the cell type atlas of the 3 dpf pluteus larva [adapted from Paganos et al., 2022b]. (C) X-ray microtomography (MicroCT) of the 3 dpf S. purpuratus pluteus larva, placed in lateral view, in which the different cell type families are labeled with pseudo-coloring [adapted from Paganos et al. (2022b)]. (D) SEM images of a fractured S. purpuratus 3 dpf larva seen in lateral view. The three partitions of the digestive tract (esophagus, stomach, and intestine) are visible. (E) FISH using antisense probe for Fgf9/16/20 marking the cardiac and pyloric sphincter regions. (F) Immunohistochemical staining using the Sp-Brn1/2/4 (red) and 5c7 (white) antibodies labeling the esophagus and the rest of the digestive tract regions, respectively, seen in lateral view. (G) Feature plot showing the esophageal (red) and the rest of the digestive tract regions (white) at a single cell resolution. (H) Inset of panel D showing the midgut and posterior gut regions of the larva’s digestive tract in higher magnification. The morphology of the pyloric sphincter cells as well as microvilli are evident. (I) Inset of panel D focused on the esophageal and stomach domains of the digestive tract. The cardiac sphincter, cilia and microvilli are visible. (J) SEM image of a fractured S. purpuratus 3 dpf larva seen in dorsal view. Extensive ECM networks and fractured pieces of the coelomic pouches and esophagus are evident. (K) Immunohistochemical detection of cilia using the antibody against acetylated tubulin. The larva is seen in dorsal view. (L) SEM image of a fractured S. purpuratus 3 dpf larva seen in lateral view. (M) Inset of panel L showing a variety of cell types including the stomach, cardiac sphincter, ciliary band, left coelomic pouch, blastocoelar cells and skeleton. (N) FISH using antisense probe against Vasa mRNA marking the coelomic pouch of the larva. (O) Immunohistochemical detection of the larval skeleton using the antibody against Msp130. Larvae in N and O are seen in ventral view. Nuclei in panels E, K, N, and O are labeled with DAPI (in gray). bc, blastocoelar cell; c, cilia; cp, coelomic pouch; cs, cardiac sphincter; e, esophagus; ecm, extracellular matrix; i, intestine; mo, mouth; mv, microvilli; ps, pyloric sphincter; s, stomach; sk, skeleton. Pseudo-coloring was applied to enhance the visualization of the cell types of interest.
The WAFFSEM protocol applied on 3 dpf pluteus larvae allowed us to correlate the molecular signatures identified with single cell transcriptomics to distinct cell type features. Focusing on the digestive tract of the larva, only WAFFSEM was able to depict morphological diversity of the cell types composing it, while FISH and IHC, as well as single-cell transcriptomics, are able to reveal distinct molecular signatures of the sphincter, esophageal, stomach and intestinal domains (Figures 6D–I). For example, this is the case of the larval pyloric sphincter, in which we can clearly see the constriction formed by the endodermally derived muscles that are morphologically diversified in comparison to the rest of the posterior gut cells (Figure 6H). Specific features of these cells are the elongated cell shape and the presence of vesicle-like structures. Similar cell characteristics can be appreciated in the cardiac sphincter domain, suggesting that the two sphincters share morphological features (Figure 6I). Once more we can clearly see that the blastocoel is full of ECM as well as a complex network of blastocoelar cells that are interconnecting most of the cell types of the larva (Figure 6H).
Moving on to a fractured larva seen from a dorsal view, we can see the two coelomic pouches of the larva at a cell resolution and their relative position and connectivity to the foregut region of the animal (Figure 6J). Furthermore, we also report the presence of distinct ciliated cells patterning those domains, a feature that was also evident by anti-acetylated tubulin immunohistochemistry, although lacking defined cell resolution (Figures 6J,K). Last but not least, a specimen that is placed in lateral view and fractured along the longitudinal axis allowed us to observe the cell morphology of the cells constituting the ciliary band, stomach and left coelomic pouch domains (Figures 6L,M). Especially for the coelomic pouch region, our approach allows us to speculate that the well-known Vasa positive cells, shown by FISH are potentially ciliated as revealed by WAFFSEM (Figures 6M,N). Moreover, on the same specimen, we observed the morphology of the skeletal spicules forming the larval skeleton that we highlighted as Msp130 immunopositive structures (Figure 6O). We identified, as well, the presence of a cell containing filopodia-like structures that appears to be in contact with the skeletal rod and potentially corresponds to a blastocoelar/immune cell (Figure 6M). This hypothesis is based on the previously shown evidence that sea urchin immune cells use the skeletal structures as a scaffold to migrate to different regions inside the larva (Allen et al., 2022). Taken together, our data set an example on how our WAFFSEM protocol combined with gene expression visualization methods, as well as transcriptomics, is able to provide complementary and useful information on assessment of cell type morphology and identity.
4. Discussion
Traditionally the classification of cell types was based on microscopy observation of the phenotypic characteristics of isolated cells, tissues, and organs. Nowadays, technologies have enabled the characterization and re-evaluation of cell types at an unprecedented resolution. However, most of these technologies focus on the identification of cell types at a molecular level and are not able to address their morphological features. On the other hand, electron microscopy-based methods such as Serial Block-face Electron Microscopy (SBEM) and Focused Ion Beam Scanning Electron Microscopy (FIBSEM) are able to successfully address cell type morphology, but currently due to their high price and sample size limitations are not applicable to a wide range of specimens. In this study, we optimized a low-cost method that is easy to use and is sufficient to reveal the cellular and subcellular characteristics of the specimens.
Our WAFFSEM method was tested on different developmental stages of six widely used marine experimental systems allowing the visualization of distinct cell type characteristics that were otherwise undetectable or detectable at a lower resolution with light microscopy. The efficiency of our method does not seem to bear any size or specimen shape restrictions; however, a high number of embryos or larvae is strongly recommended to ensure a high ratio of fractured versus not fractured specimens. In conclusion, we believe that the WAFFSEM protocol we optimized, is a useful and easy-to-use tool to characterize cell types. Nowadays, no matter how powerful, there is no sole technique that is able to solve every biological mystery. Therefore, correlative approaches are needed to thoroughly address complex scientific questions. Along this line we hope that our WAFFSEM protocol combined with other technologies and methods, will promote the advancement of the Evo-Devo research field by improving our understanding of animal cell types from both a morphological and a molecular fingerprint point of view.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
PP, MIA, and GB: conceptualization. PP, FC, and GB: methodology. PP, FC, MC, ED’A, MIA, and GB: validation and writing—review and editing. PP: writing—original draft preparation. PP, FC, MC, and ED’A: visualization. GB: supervision. All authors contributed to the article and approved the submitted version.
Funding
PP was supported by Marie Curie ITN EvoCELL (H2020 grant number: 766053 to MIA) and Human Frontiers Science Program grant number (RGP0002/2019 to MIA). FC was supported by MUR CIR_0029 Human Capital and by Assemble Plus project (BA010618 and 360BA0619) to visit Observatoire Océanologique de Banyuls-sur-Mer (France).
Acknowledgments
The authors would like to thank Davide Caramiello for taking care of the adult sea urchins at Stazione Zoologica Anton Dohrn and Erica Riccio for her assistance in spawning the animals. We are also grateful to the microscopy facility at Stazione Zoologica Anton Dohrn for their assistance in sample preparation for SEM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allen, R. L., George, A. N., Miranda, E., Phillips, T. M., Crawford, J. M., Kiehart, D. P., et al. (2022). Wound repair in sea urchin larvae involves pigment cells and blastocoelar cells. Dev. Biol. 491, 56–65. doi: 10.1016/j.ydbio.2022.08.005
Almazan, A., Cevrim, C., Musser, J. M., Averof, M., and Paris, M. (2022). Crustacean leg regeneration restores complex microanatomy and cell diversity. Sci. Adv. 8:eabn9823. doi: 10.1126/sciadv.abn9823
Cao, C., Lemaire, L. A., Wang, W., Yoon, P. H., Choi, Y. A., Parsons, L. R., et al. (2019). Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349–354. doi: 10.1038/s41586-019-1385-y
Chari, T., Weissbourd, B., Gehring, J., Ferraioli, A., Leclere, L., Herl, M., et al. (2021). Whole-animal multiplexed single-cell Rna-seq reveals transcriptional shifts across Clytia medusa cell types. Sci. Adv. 7:eabh1683. doi: 10.1126/sciadv.abh1683
D'aniello, E. (2009). Studies on Rx Gene Function and Its Involvement in Ciona intestinalis ocellus Differentiation. PhD thesis The Open University.
Davidson, E. H., and Erwin, D. H. (2006). Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800. doi: 10.1126/science.1113832
Ernst, S. G. (2011). Offerings from an urchin. Dev. Biol. 358, 285–294. doi: 10.1016/j.ydbio.2011.06.021
Fuentes, M., Benito, E., Bertrand, S., Paris, M., Mignardot, A., Godoy, L., et al. (2007). Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum).. J. Exp. Zool. B. Mol. Dev. Evol. 308, 484–493. doi: 10.1002/jez.b.21179
Macdonald, J. A., Fowle, W. H., and Woods, D. C. (2017). New insights on mitochondrial heterogeneity observed in prepared mitochondrial samples following a method for freeze-fracture and scanning electron microscopy. Micron 101, 25–31. doi: 10.1016/j.micron.2017.05.002
Massri, A. J., Greenstreet, L., Afanassiev, A., Berrio, A., Wray, G. A., Schiebinger, G., et al. (2021). Developmental single-cell transcriptomics in the Lytechinus variegatus sea urchin embryo. Development 148:dev198614. doi: 10.1242/dev.198614
Murat, F., Mbengue, N., Winge, S. B., Trefzer, T., Leushkin, E., Sepp, M., et al. (2023). The molecular evolution of spermatogenesis across mammals. Nature 613, 308–316. doi: 10.1038/s41586-022-05547-7
Musser, J. M., Schippers, K. J., Nickel, M., Mizzon, G., Kohn, A. B., Pape, C., et al. (2021). Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science 374, 717–723. doi: 10.1126/science.abj2949
Paganos, P., Caccavale, F., La Vecchia, C., D'aniello, E., D'aniello, S., and Arnone, M. I. (2022a). Fish for all: a fast and efficient fluorescent in situ hybridization (Fish) protocol for marine embryos and larvae. Front. Physiol. 13:878062. doi: 10.3389/fphys.2022.878062
Paganos, P., Ronchi, P., Carl, J., Mizzon, G., Martinez, P., Benvenuto, G., et al. (2022b). Integrating single cell transcriptomics and volume electron microscopy confirms the presence of pancreatic acinar-like cells in sea urchins. Front. Cell Dev. Biol. 10:991664. doi: 10.3389/fcell.2022.991664
Paganos, P., Voronov, D., Musser, J. M., Arendt, D., and Arnone, M. I. (2021). Single-cell RNA sequencing of the Strongylocentrotus purpuratus larva reveals the blueprint of major cell types and nervous system of a non-chordate deuterostome. elife 10:e70416. doi: 10.7554/eLife.70416
Perillo, M., Paganos, P., Spurrell, M., Arnone, M. I., and Wessel, G. M. (2021). Methodology for whole mount and fluorescent Rna in situ hybridization in echinoderms: single, double, and beyond. Methods Mol. Biol. 2219, 195–216. doi: 10.1007/978-1-0716-0974-3_12
Plass, M., Solana, J., Wolf, F. A., Ayoub, S., Misios, A., Glazar, P., et al. (2018). Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360:eaaq1723. doi: 10.1126/science.aaq1723
Satoh, N., Tominaga, H., Kiyomoto, M., Hisata, K., Inoue, J., and Nishitsuji, K. (2021). A preliminary single-cell Rna-Seq analysis of embryonic cells that express Brachyury in the Amphioxus, Branchiostoma japonicum. Front. Cell Dev. Biol. 9:696875. doi: 10.3389/fcell.2021.696875
Stracke, K., and Hejnol, A. (2023). Marine animal evolutionary developmental biology–advances through technology development. Evol. Appl. 16, 580–588. doi: 10.1111/eva.13456, PMCID: PMC9923486
Tabula Muris, C. (2020). A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595. doi: 10.1038/s41586-020-2496-1
Wessel, G. M., Fresques, T., Kiyomoto, M., Yajima, M., and Zazueta, V. (2014). Origin and development of the germ line in sea stars. Genesis 52, 367–377. doi: 10.1002/dvg.22772
Keywords: echinoderms, tunicates, mollusks, cephalochordates, morphology, cell types, SEM, freeze fracture
Citation: Paganos P, Caccavale F, Cocurullo M, D’Aniello E, Arnone MI and Benvenuto G (2023) Whole animal freeze-fracture scanning electron microscopy: an easy-to-use method to investigate cell type morphology of marine embryos and larvae. Front. Ecol. Evol. 11:1146749. doi: 10.3389/fevo.2023.1146749
Edited by:
Pedro Martinez, University of Barcelona, SpainReviewed by:
Olga Zueva, Marine Biological Laboratory, United StatesNoriyuki Satoh, Okinawa Institute of Science and Technology Graduate University, Japan
Copyright © 2023 Paganos, Caccavale, Cocurullo, D’Aniello, Arnone and Benvenuto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanna Benvenuto, Z2lvdmFubmEuYmVudmVudXRvQHN6bi5pdA==
 Periklis Paganos
Periklis Paganos Filomena Caccavale
Filomena Caccavale Maria Cocurullo
Maria Cocurullo Enrico D’Aniello
Enrico D’Aniello Maria Ina Arnone
Maria Ina Arnone Giovanna Benvenuto
Giovanna Benvenuto