
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 14 February 2023
Sec. Biogeography and Macroecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1131408
This article is part of the Research Topic Ecosystem Nutrients Stoichiometry From Local to Global Scales View all 8 articles
Phosphorus (P) application to terrestrial ecosystems affects not only aboveground plants but also soil P forms and phosphatase-associated microbes. The phoD gene is widespread in soil and plays an important role in P transformation. However, it is still unclear how phoD-harboring microbial communities respond to different P application rates, and the relationships between soil properties and phoD-harboring microbial community need to be better understood. In this study, the impacts of seven P application rates [0 (P0), 10 (P10), 20 (P20), 30 (P30), 40 (P40), 50 (P50), and 60 (P60) g⋅m–2⋅a–1] on the soil physicochemical properties, P forms, and phoD-harboring microbial communities were assessed. As the results, inorganic P (i.e., Resin-Pi, NaHCO3-Pi, NaOH-Pi, and HCl-Pi) and Bio-P increased firstly and then decreased with increasing P application rate, with the highest values in the P30 treatment. Soil phoD-harboring microbial community structures in low-P (P0∼P30) treatments were significantly different from that in high-P (P40∼P60) treatments. Soil phoD-harboring microbial Shannon and Simpson diversity increased firstly and then decreased with increasing P application rate, and there was a tipping point at the P application rate of 30 g⋅m–2⋅a–1. The Mantel test and structural equation modeling (SEM) revealed that Bio-P, TC (total carbon), Fe, NaOH-organic P (NaOH-Po), and soil pH were strongly related to the soil phoD-harboring microbial community structure. In conclusion, this study demonstrated that P application affected soil P forms and phoD-harboring microbes in an alpine grassland on the Qinghai-Tibetan Plateau, and there was a P application threshold for optimistic growth of phoD-harboring microbes in an alpine grassland on the Qinghai-Tibetan Plateau.
Phosphorus (P) as the key element of nucleic acid and phosphatide in cells, is a limiting nutrient for plant growth (Hou et al., 2018; Chen et al., 2022; Wang Z. Q. et al., 2022). Soil P is abundant, but a large fraction of P is fixed into organic P or mineral P, which results in very low bioavailability for plants (Liu et al., 2020; Tian et al., 2021). Therefore, the amount of P applied in soil far exceeds the requirement of the plant. In the past 40 years (1980–2020), the P application in China increased by 3.81 × 1012 g, which may lead to soil acidification, P imbalance and even P pollution (Liu et al., 2020; NBS, 2021; Chen et al., 2022). Thus, the appropriate P application rate is vital in achieving high plant productivity and maintaining P balance in terrestrial ecosystems.
Phosphorus application could affect the content of soil P forms (Mahmood et al., 2020; Chen et al., 2022; Hussain et al., 2022) and the P transformation processes (Hu et al., 2018; Wang M. M. et al., 2022; Zeng et al., 2022). In the soils, P forms range from ions in solution to stable compounds, including dissolved P (PO43–, HPO42–, and H2PO4–), absorbed P, mineral-associated P (Fe-P, Al-P, Ca-P, variscite, apatite, xenotime), and organic-associated P (phosphate ester, phosphate and phosphoric acid anhydride) (Cao et al., 2022). P application could boost soluble inorganic P (Pi) supply (Carneiro et al., 2011; Liao et al., 2021) and promote the P assimilation by soil microbes (Hu et al., 2018). The soil microbial cell is a kind of organic P (Po), and the increase in soil microbial biomass might lead to an increase in soil Po (Gu et al., 2017). Additionally, soil microbes, especially phosphatase-associated microorganisms, would decompose Po to orthophosphate with phosphatases, depleting Po and releasing Pi (Fraser et al., 2015a; Zhu et al., 2021). P application might make soil microbes downregulate the expression of the phosphatase genes, given that soil microbes would invest less in the production of phosphatases when the soil available P is sufficient (Wang M. M. et al., 2022). Therefore, soil microbes, especially phosphatase-associated microbes, play a key role in soil P assimilation and mineralization, and it is necessary to understand how the P application rate may affect the soil phosphatase-associated microbes.
PhoA, phoC, phoD, and phoX have been identified as homologous genes in soils relating to phosphatases secretion, which is important in P transformation (Apel et al., 2007; Ragot et al., 2017). PhoD gene has been frequently observed in soils and exclusively derived from soil microbes (Hu et al., 2018). Therefore, PhoD gene is usually employed as a useful molecular marker to investigate the P transformation process in soils (Ragot et al., 2015). P applications could affect soil PhoD-harboring microbial community composition and diversity through P forms or other environmental properties (Luo et al., 2017). The study in paddy soil pointed out that P application could suppress the phoD-harboring microbial diversity, and the content of dissolved organic carbon, citrate-extractable P, HCl-extractable P, and total available P contributed significantly to the variance of phoD-harboring microbial community (Wei et al., 2019). However, the study in a maize field reported an increase in the phoD-harboring microbial diversity in high-P application treatments, and the phoD-harboring microbial diversity was positively related to the soil available P (Chen et al., 2019). Another study in Loess Plateau suggested that phoD-harboring microbial diversity responded insignificantly to increasing P application rate and no environmental factors were significantly correlated with the phoD-harboring microbial community structure (Liu et al., 2020). These different responses of phoD-harboring microbial diversity to P application are puzzling and limit our ability to predict how P application rate will affect phoD-harboring microbial communities in alpine grasslands on the Qinghai-Tibetan Plateau.
Grassland is the largest ecological system in China, which occupies approximately 41.7% of the land area (Ren et al., 2008). Alpine grassland on the Qinghai-Tibetan Plateau is an important livestock husbandry base in China, and soil P in the alpine grassland could be carried out with the output of livestock (dung, milk, or meat) and grass (herb) products. P application is an important anthropogenic activity to maintain the soil P balance in alpine grassland. However, the P application research in alpine grasslands of the Qinghai-Tibetan Plateau mainly focus on the aboveground plants, furthermore, the information about the influence of the P application rates on soil P forms and phoD-harboring microbial communities is very lacking (Sun et al., 2019; Cao et al., 2022; Xiao et al., 2022). In this study, we aim to investigate how P application affects soil P forms and phoD-harboring microbial groups in an alpine grassland on the Qinghai-Tibetan Plateau. We established seven P application rate treatments [0 (P0), 10 (P10), 20 (P20), 30 (P30), 40 (P40),50 (P50), and 60 (P60) g⋅m–2⋅a–1] and determined soil P forms and phoD-harboring microbes in each treatment to evaluate the community composition and diversity of the microorganisms with the potential to produce alkaline phosphatases. We hypothesized that: (1) soil P forms especially bioavailable P would vary significantly between P application treatments; (2) P application would decrease the phoD-harboring microbial diversity; (3) changes in soil phoD-harboring microbial community might be related to the changes in the soil P forms.
The study site is in Hongyuan County (32° 49.823′ N, 102° 35.237′ E), eastern Qinghai-Tibetan Plateau with the typical continental plateau climate. The mean annual temperature (MAT) is 1.1°C and the mean annual precipitation (MAP) ranges from 650 mm to 800 mm. The soil type is Felty soil following the Chinese Soil Classification System. The aboveground plant species mainly include Elymus nutans, Agrostis hugoniana, Kobresia setchwanensis, Kobresia humilis, Saussurea nigrescens etc.
The field experiment was initiated in 2019 and included the following seven treatments with three replicated plots: (1) P0, none fertilization; (2) P10, 10 g⋅m–2⋅a–1 (single superphosphate, containing P2O5 16.0%); (3) P20, 20 g⋅m–2⋅a–1; (4) P30, 30 g⋅m–2⋅a–1; (5) P40, 40 g⋅m–2⋅a–1; (6) P50, 50 g⋅m–2⋅a–1; and (7) P60, 60 g⋅m–2⋅a–1. The P fertilizer was distributed evenly by hand on the soil surface in each plot once a year during natural rain event in early growing season on 6 May 2019 and 4 May 2020. For each replicate, a random quadrat (1 m × 1 m) was assigned as the sampling quadrat, and five scattered soil cores were collected from each quadrat with a 5 cm auger from the topsoil (0∼10 cm) after harvesting on August 15, 2020. All five soil cores from each quadrat were combined into one composite sample and mixed thoroughly to minimize sampling errors. All plant litters and rocks were removed by hand; 50 g of soil were stored at −80°C for DNA extraction; 350 g of soil were stored at 4°C for microbial biomass phosphorus (MBP) analysis within 1 week and the remaining sampled soil was air dried for physicochemical analysis.
Soil moisture content (MC) was determined gravimetrically at 105°C for 24 h and expressed as a ratio of soil water to dry soil weight. Soil pH was measured in the soil-water slurry (soil: water = 1: 2.5) using a pH meter (FE20-FiveEasyTM pH, Mettler Toledo, Germany). The total carbon (TC) and total nitrogen (TN) contents were measured using an elemental analyzer (Vario MAX, Elementar, Germany). Fe, Al, and Ca in acid digested extract were measured by ICP-OES (PerkinElmer Optima 8300, USA). Soil MBP was determined using the chloroform fumigation-extraction method (Wu et al., 2000).
The modified Hedley sequential fractionation method was used to measure soil P forms (Tiessen and Moir, 1993). Air dried soils were milled and sieved to 2 mm. Then 0.5 g soil was extracted sequentially with an anion exchange membrane in deionized water, 30 ml of 0.5 M NaHCO3 at pH 8.5, 30 ml of 0.1 M NaOH and 30 ml of 1 M HCl in a 50 ml screw up centrifuge plastic tube. In each extraction step, samples were shaken continuously for 16 h at 25°C, centrifuged at 3,500 rpm for 15 min at 0°C and thereafter the supernatant was filtered using a 0.45 μm filter paper. The supernatant was stored at 4°C for phosphate determination. Soil residue in the centrifuge plastic tube was then re-suspended using concentrated H2SO4 and 30% H2O2 to extract more chemically stable P (Residual-P). The supernatant was analyzed for dissolved Pi and total P (Pt). Pi was determined directly in the supernatant through the molybdate-ascorbic acid method using a spectrophotometer at 712 nm (Murphy and Riley, 1962), Pt was measured through the same method after digestion with ammonium persulfate and H2SO4 at 121°C in an autoclave. The Po was then calculated as the difference between Pt and Pi. We omitted the addition of hot concentrated HCl from the original methodology. Soil Bio-P was the sum of Resin-Pi and NaHCO3-Pi, which is the biologically most available Pi form for plants (Moro et al., 2015; Tian et al., 2022).
Soil DNA was extracted from the 0.25 g soil using the PowerSoil DNA Isolation Kit (MOBIO Laboratories, Carlsbad, CA, USA) as directed by the manufacturer within 2 weeks after sampling. The quantity and quality of the extracted DNA were measured using NanoDrop ND-2000 (Thermo Fisher Scientific, Waltham, MA, USA). The primer set phoD-F733 (5′-TGGGAYGATCAYGARGT-3′)/phoD-R1083 (5′-CTGSGCSAKSACRTTCCA-3′) (Ragot et al., 2015) was employed to target the microbial phoD gene. The PCR mixture (25 μl) contained 2× Taq MasterMix (TaKaRa, Dalian, China) 12.5 μl, Forward primer 1 μl, Reverse primer 1 μl, ddH2O 9.5 μl, DNA (10 ng⋅μl–1) 1 μl. The PCR amplification program included an initial denaturation at 95°C for 3 min, followed by 35 cycles of 95°C for 40 s, 59°C for 60 s, and 72°C for 60 s, and a final extension at 72°C for 10 min. Triplicate reaction mixtures per sample were pooled and purified using DNA Gel Extraction Kit (Tsingke, Beijing, China). Finally, the amplicons of all samples were pooled at equimolar concentrations, and sequencing was performed using the Illumina MiSeq (Illumina, San Diego, CA, USA) sequencing platform.
The raw sequences of the phoD gene were merged using FLASH with 30 bp overlap (Magoc and Salzberg, 2011), and the joint sequences were processed using the Quantitative Insights Into Microbial Ecology (version 2.0, QIIME2) pipeline (Bolyen et al., 2019). Reads were shorter than 300 bp, reads containing more than two ambiguous bases, and reads with an average quality score lower than 30 were removed. The chimeras were removed and the FrameBot was adopted to correct frameshift mutation sequences in the Ribosomal Database Project (RDA) function gene pipeline (Wang et al., 2013). Using the deblurring algorithm, 10,739 amplicon sequence variants (ASVs) were obtained from the 1,238,300 successfully corrected sequences in 21 samples. The RDP FunGene database was used to assign taxonomic affiliations of phoD sequences (Cole et al., 2014). To eliminate the bias on diversity comparison as caused by uneven sequencing, we rarified sequences to 25,690 for each sample randomly.
One-way ANOVA was performed to examine the effect of P application on soil physicochemical properties, P forms, phoD-harboring microbial diversity, and the relative abundance of the dominant phyla. The comparison of significant means was carried out using Tukey’s Honest Significant Difference (HSD) test. To visualize the community structure of the phoD-harboring microbes among different treatments, the Non-metric multidimensional scaling (NMDS) analysis was constructed based on the Bray--Curtis distance. The significance of the phoD-harboring community structure was tested by Permutational multivariate analysis (PERMANOVA), analyses of similarities (ANOSIM) and Multi Response Permutation Procedure (MRPP) based on 999 permutations with the Vegan package in the R environment.1
Differentially abundant taxa among the seven treatments were identified using the linear discriminant analysis (LDA) effect size (LEfSe) method using online Galaxy,2 and the LEfSe algorithm uses a P-value < 0.05 and an LDA score > 2.0 for the non-parametric factorial Kruskal–Wallis test.
Network analyses were performed to better understand the species relations within the phoD-harboring microbial communities in R as described by Mendes et al. (2014). We calculated all possible Spearman’s rank correlation coefficients, and the high correlations with cutoff at r > 0.8 and statistically significant P-value < 0.01 were kept in the network construction. The networks were constructed using the igraph package. The nodes in the reconstructed network represent ASVs, and the edges represent high and significant correlations between nodes. The network graphs were visualized based on nodes, edges and taxonomic information using the ggraph package. Moreover, the within-module connectivity (Zi) and among-module connectivity (Pi) were calculated using ggClusterNet package. According to Zi = 2.5 and Pi = 0.62, all nodes were divided into four categories: peripherals (Zi < 2.5, Pi < 0.62), connectors (Zi < 2.5, Pi ≥ 0.62), module hubs (Zi ≥ 2.5, Pi < 0.62), and network hubs (Zi ≥ 2.5, Pi ≥ 0.62). From the ecological perspective, connectors, module hubs, and network hubs are the generalists while peripherals are the specialists (Deng et al., 2012; Feng et al., 2022).
The correlations between phoD-harboring microbial community composition and environmental factors were detected by Mantel tests using the linkET package in R. Then, the structural equation modeling (SEM) framework was further applied to test the significance of hypothesized causal relationships between soil properties, P forms and phoD-harboring microbial community structure using AMOS 23.0 (Amos Development, Spring House, Armonk, NY, USA). To simplify the SEM model, the principal component analysis (PCA) was used to create multivariate functional indices for (NMDS1 and NMDS2) phoD-harboring microbial community structure. The first principal component axis (PCA1), explaining 71.24% of the total variance in phoD-harboring microbial community structure, were used in the SEM analysis. The best-fit model was derived using maximum likelihood based on model fit, i.e., chi-square test (χ2), goodness-of-fit index (GFI), and the root mean square error of approximation (RMSEA) (Hooper et al., 2008).
One-way ANOVA analysis showed that soil properties except for soil MC and Ca responded significantly to the P application (P < 0.05) (Table 1). The NaHCO3-Po (Figure 1C) and pH (Figure 1I) declined with increasing P application rate, with the lowest values in the P60 treatment. However, the NaOH-Po (Figure 1E), Al (Figure 1N), Fe (Figure 1O), and MBP (Figure 1P) showed a rough increase pattern with increasing P application rate, with the highest value in the P60 treatment. Soil MC (Figure 1L) and Ca (Figure 1M) varied slightly with increasing P application rate. Soil TN (Figure 1J) and TC (Figure 1K) in the P0 treatment were significantly higher than that in P10∼P40 treatments. Additionally, soil Resin-Pi (Figure 1A), NaHCO3-Pi (Figure 1B), NaOH-Pi (Figure 1D), HCl-Pi (Figure 1F), Residual-P (Figure 1G), and Bio-P (Figure 1H) all peaked at P30 treatment. This indicated that soil Resin-Pi, NaHCO3-Pi, NaOH-Pi, HCl-Pi, Residual-P, and Bio-P showed non-linear relationships with increasing P application amount, instead, there might be a tipping point at the P application rate of 30 g⋅m–2⋅a–1.
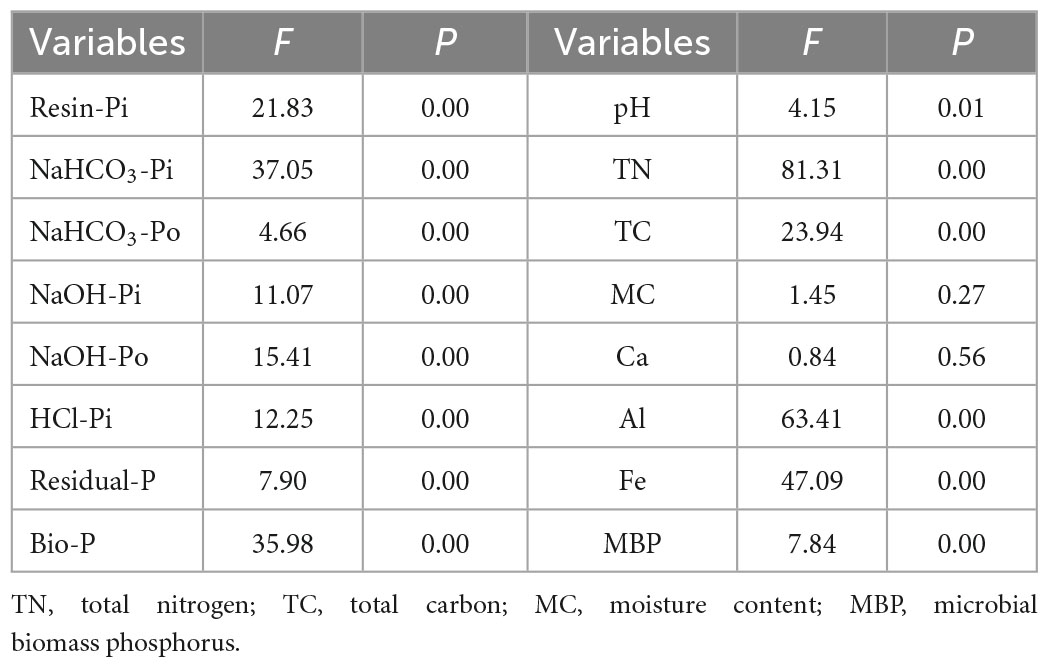
Table 1. The effects of the P application on soil properties and P forms using One-way ANOVA analysis.
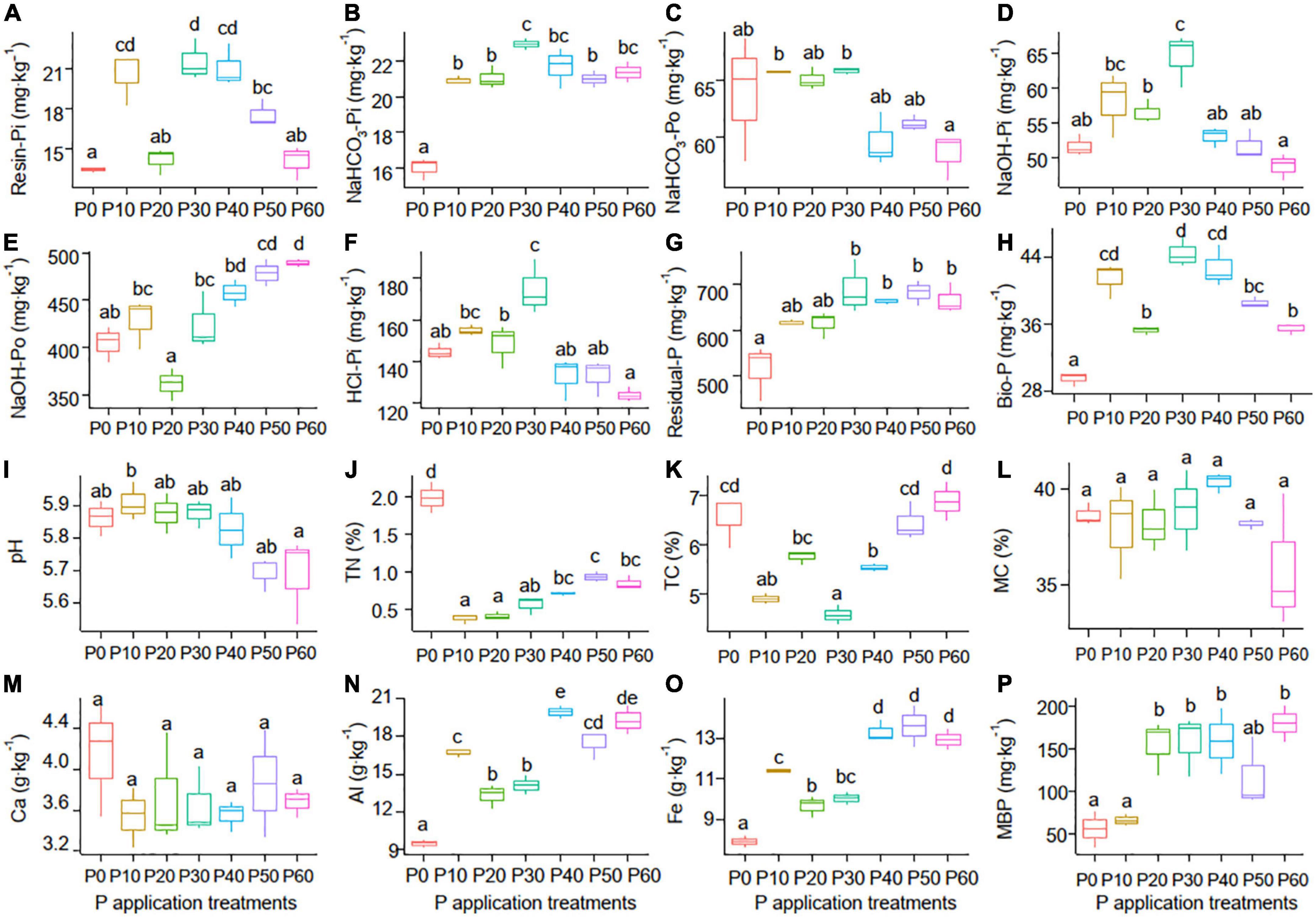
Figure 1. Soil P forms [Resin-Pi (A), NaHCO3-Pi (B), NaHCO3-Po (C), NaOH-Pi (D), NaOH-Po (E), HCl-Pi (F), Residual-P (G), and Bio-P (H)] and physicochemical properties [pH (I), TN (J), TC (K), MC (L), Ca (M), Al (N), Fe (O), and MBP (P)] among the seven P application treatments. P0 (without phosphate fertilizer), P10 (with a rate of 10 g⋅m–2⋅a–1), P20 (with a rate of 20 g⋅m–2⋅a–1), P30 (with a rate of 30 g⋅m–2⋅a–1), P40 (with a rate of 40 g⋅m–2⋅a–1), P50 (with a rate of 50 g⋅m–2⋅a–1), and P60 (with a rate of 60 g⋅m–2⋅a–1). Bio-P, bioavailable P; TN, total nitrogen; TC, total carbon; MC, moisture content; MBP, microbial biomass phosphorus.
A total of 1,238,300 high-quality sequences were obtained from all soil samples. The number of sequences per sample ranged from 25,690 to 76,578, which were clustered into 10,739 ASVs. The number of ASVs in all 21 samples varied from 1,465 to 2,404. The dominant phoD-harboring microbial ASVs (>1%) derived from Proteobacteria (28.16%), Cyanobacteria (5.01%), Gemmatimonadetes (3.68%), and Actinobacteria (3.45%) (Figure 2A). There were significant differences in the relative abundance of Proteobacteria, Deinococcus_Thermus, Firmicutes, and Euryarchaeota among the seven treatments. The relative abundance of Proteobacteria in P60 was higher than that in P0∼P30 (Figure 2I), but the relative abundance of Deinococcus_Thermus in P60 was lower than that in P0∼P30 (Figure 2D). Moreover, both the relative abundance of Firmicutes (Figure 2F) and Euryarchaeota (Figure 2E) peaked at the P30 treatment. The relative abundance of Gemmatimonadetes (Figure 2G) and Planctomycetes (Figure 2H) showed no significant differences between P application treatments.

Figure 2. The dominant phoD-harboring microbial phyla (A) and the variations of those dominant phyla [Actinobacteria (B), Cyanobacteria (C), Deinococcus_Thermus (D), Euryarchaeota (E), Firmicutes (F), Gemmatimonadetes (G), Planctomycetes (H), and Proteobacteria (I)] and P application rates. The asterisk (*) that follows the taxonomic name represents significant differences (P < 0.05) among the seven treatments.
In addition, there were 23 biomarkers with abundance differential, and P30 holds the most biomarkers (Figure 3). Those biomarkers in P30 mainly derived from Actinobacteria (Micromonospora, Jiangellaceae, Jiangella, and Jiangellales) and Cyanobacteria (Tolypothrichaceae and Tolypothrix), but the biomarkers in P60 mainly derived from Cyanobacteria (Acaryochloris, Acaryochloridaceae, and Synechococcales) and Proteobacteria (Rhodospirillaceae and Skermanella). All the five biomarkers in P0, three biomarkers in P10, and four biomarkers in P50 derived from Proteobacteria, Euryarchaeota, and Actinobacteria, respectively.
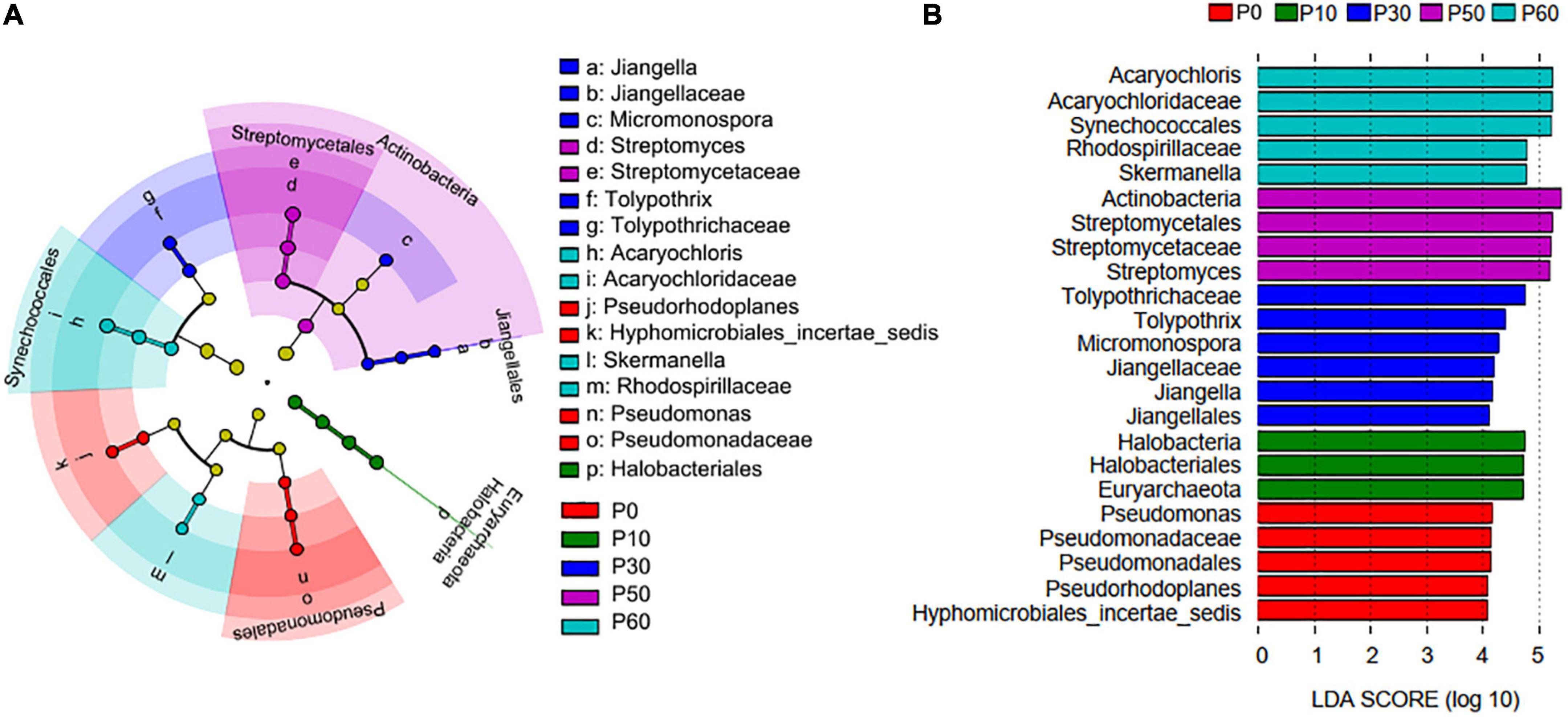
Figure 3. PhoD-harboring microbial biomarkers revealed by Lefse analysis. (A) Linear discriminant analysis (LDA) cladogram between P application treatments. The circles from inside to outside represent phylum, class, order, family, and genus, respectively. (B) The discriminant phoD-harboring microbial taxa based on relative abundance. The color-coded taxa within the cladogram and histogram indicate significantly enriched taxa in a treatment by Kruskal–Wallis test with P < 0.05 and logarithmic LDA > 2.0 (n = 3 for each group).
NMDS analysis indicated that phoD-harboring communities in P0∼P30 treatments differed from that in P40∼P60 treatments (Figure 4A), and the PERMANOVA, ANOSIM, and MRPP (Table 2) results showed that phoD-harboring microbial community structure varied significantly between high-P (P40∼P60) and low-P (P0∼P30) treatments (P < 0.01 in all cases). Both soil Shannon (Figure 4B) and Simpson (Figure 4C) diversity peaked in the P30 treatment, and the soil Shannon and Simpson diversity in P30 were significantly higher than that in P60 (P < 0.05). These results indicated that the phoD-harboring microbial diversity were affected by the P application rate, and 30 g⋅m–2⋅a–1 might be the threshold where microbes-soil interactions diverged.
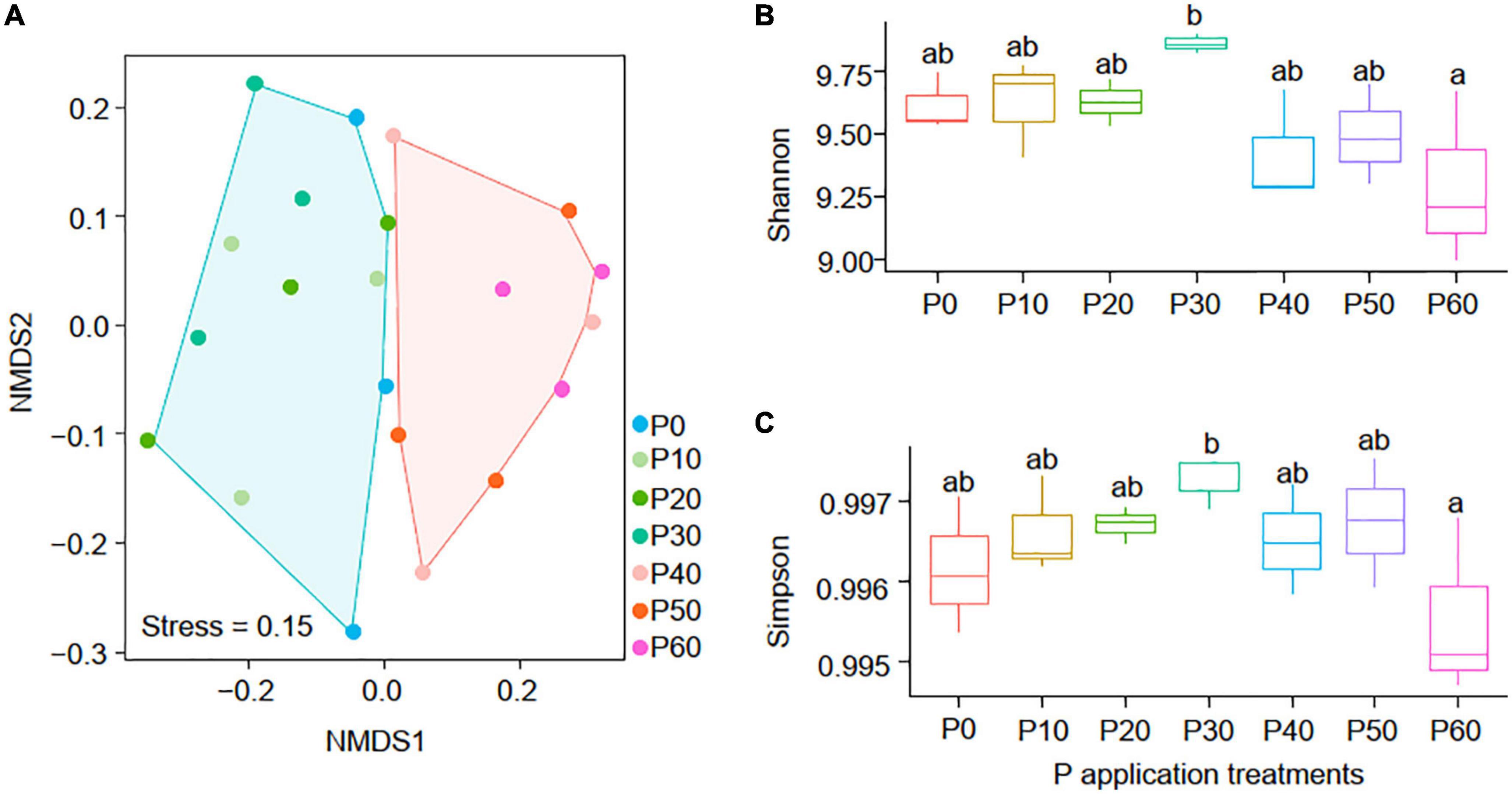
Figure 4. The NMDS of the phoD-harboring microbial composition (A) in all samples and the variations of the phoD-harboring microbial Shannon (B) and Simpson (C) diversity among the seven application treatments.

Table 2. The permutational multivariate analysis (PERMANOVA), analyses of similarities (ANOSIM), and Multi Response Permutation Procedure (MRPP) analyses based on the Bray–Curtis distance of phoD-harboring microbial communities in low-P (P0∼P30) and high-P (P40∼P60) soils.
Microbial co-occurrence networks were reconstructed to explore potential microbial interactions in low-P (P0∼P30) and high-P (P40∼P60) treatments soils. In general, the phoD-harboring microbial network complexity was higher in high-P soils, with the number of nodes, edges, and average degree increased by 20.24, 100.71, and 66.93% in comparison to those in low-P soils (Figures 5A,B). However, 68 edges appeared to be negative in low-P treatments, which was lower than the 98 negative edges in high-P treatments, suggesting high-P fertilization might lead to more conflicting interactions among phoD-harboring microbes. Proteobacteria, Cyanobacteria, and Actinobacteria dominated the nodes of both low-P and high-P networks, accounting for 25.87, 7.73, and 8.32%, respectively, and positive correlations were dominant over negative correlations in both networks. Compared with the low-P fertilization network, the percentage of Proteobacteria, Cyanobacteria, and Actinobacteria affiliated nodes increased by 5.23, 0.56, and 5.43%, respectively in the network of high-P fertilization network. The generalist (Rhodoplanes sp. Z2-YC6860) in low-P network derived from Rhizobiales (Figure 5C), while the generalists Rhizobacter sp. Root29, Pandoraea norimbergensis, Streptomyces sp. WAC00288, and Bradyrhizobium license in the high-P network belonged to Burkholderiales, Burkholderiales, Streptomycetales, and Rhizobiales, respectively (Figure 5D).
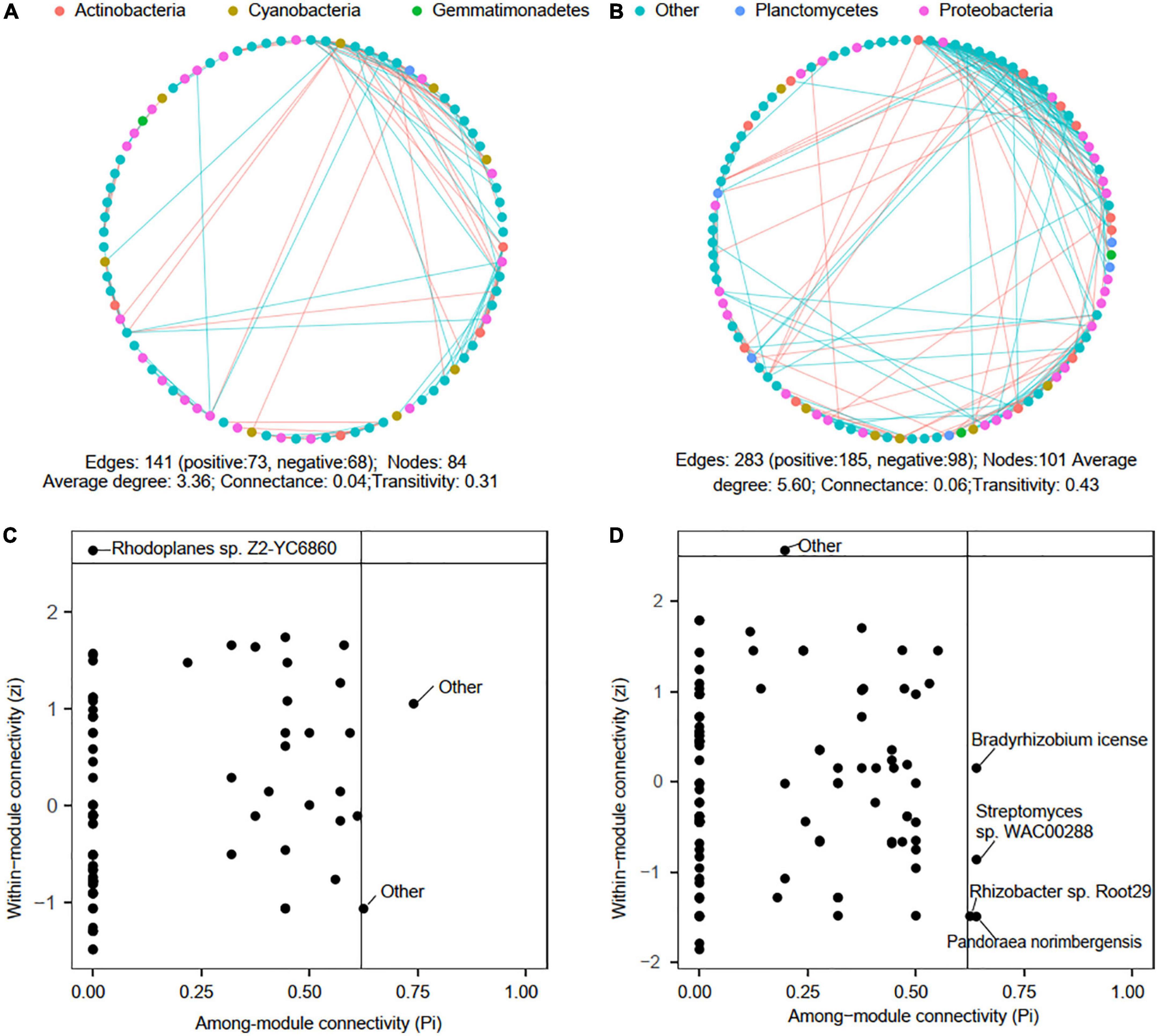
Figure 5. Overview of the reconstructed microbial co-occurrence networks in low-P (P0∼P30) and high-P (P40∼P60) treatments. Panels (A,B) visualized microbial co-occurrence networks in low-P and high-P treatments, respectively (n = 12 for low-P network and n = 9 for high-P network). The node color corresponds to microbial phylum. The edges in cyan represent positive correlation of two ASVs, and edges in red represent negative correlation of two ASVs. Panels (C,D) ZP-plot in the networks of low-P and high-P treatments, respectively. The taxonomy of each module hub (Zi ≥ 2.5, Pi < 0.62) and connector (Zi < 2.5, Pi ≥ 0.62) at species level was presented.
Pearson analysis indicated that soil TC was the most relevant factor affecting soil Resin-Pi (r = −0.77, P < 0.01), NaOH-Pi (r = −0.78, P < 0.01), HCl-Pi (r = −0.74, P < 0.01), and Bio-P (r = −0.74, P < 0.01), while soil TN was the strongest correlate of NaHCO3-Pi (r = −0.82, P < 0.01) (Figure 6). Soil NaOH-Po was negatively correlated with pH (r = −0.57, P < 0.01) but positively correlated with Al (r = 0.69, P < 0.01) and Fe (r = 0.73, P < 0.01). Soil Residual-P was also positively correlated with Al (r = 0.70, P < 0.01) and Fe (r = 0.69, P < 0.01). The links between phoD-harboring microbial community structure and environmental factors were detected by Mantel tests. The phoD-harboring microbial community structure was significantly correlated with NaOH-Po (r = 0.15, P < 0.05), pH (r = 0.18, P < 0.05), and Fe content (r = 0.15, P < 0.05).
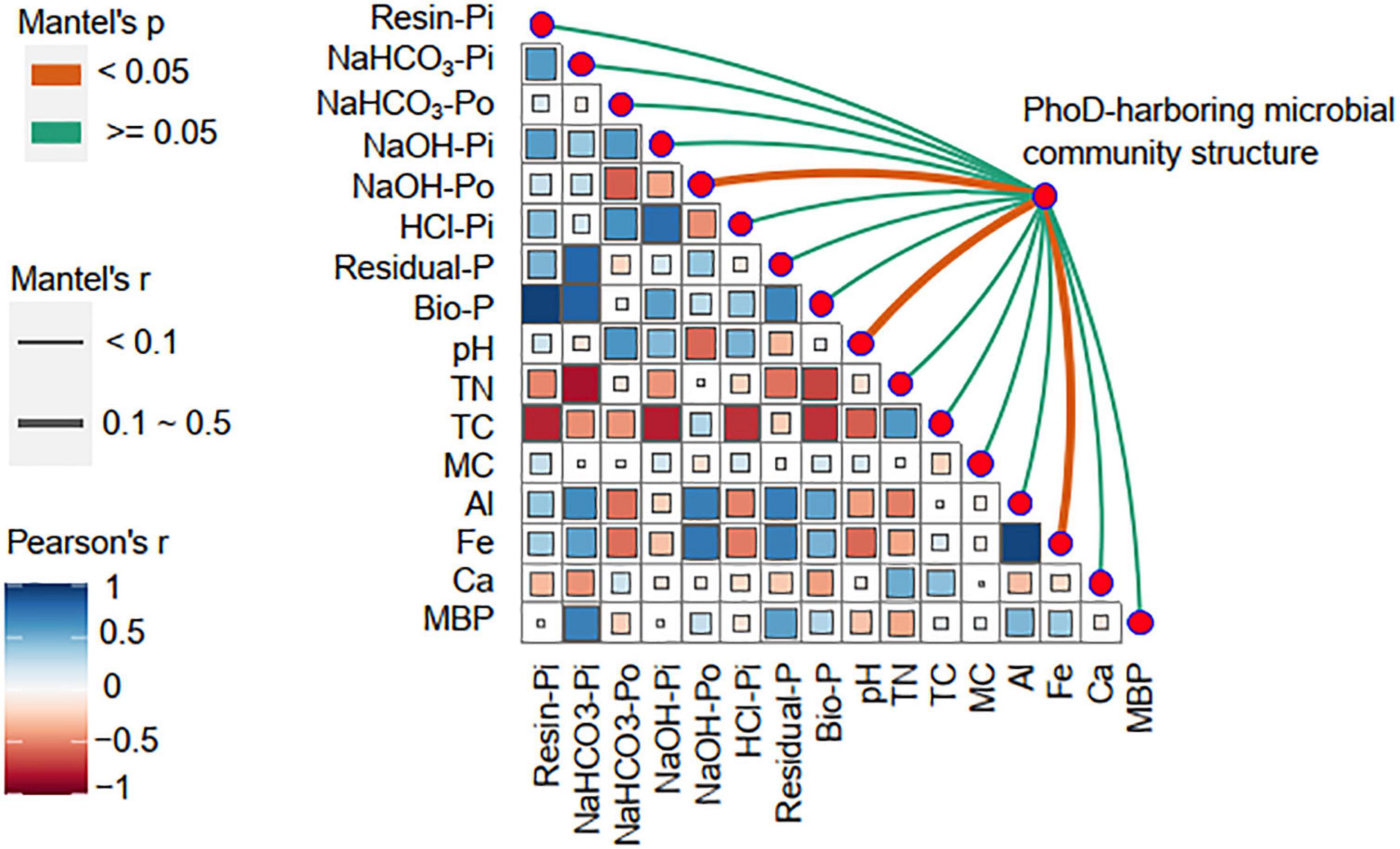
Figure 6. Correlations of environmental factors with phoD-harboring microbial community structure. Pairwise comparisons of environmental factors are shown, with a color gradient denoting Pearson correlation coefficient. The diversity of phoD-harboring microbial community were related to each environmental factor by Mantel tests. Edge width corresponded to the Mantel’s r statistic for the corresponding distance correlations, and edge color denoted the statistical significance based on 999 permutations.
Based on the most relevant factors in Mantel analysis and previous studies (Chen et al., 2019; Xu et al., 2021; Wang M. M. et al., 2022), the NaOH-Po, Fe, pH, TC, and Bio-P were included in the optimal SEM analysis. The model explained 78% of the variance in phoD-harboring microbial community structure (Figure 7A). The Fe (standardized direct effects: −0.61) and NaOH-Po content (standardized direct effects: −0.41) negatively and directly affected phoD-harboring microbial community structure, while Bio-P positively and directly affected phoD-harboring microbial community structure (standardized direct effects: 0.60). Standardized total effects derived from the SEM revealed that Bio-P had the largest positive effect (0.60) on soil phoD-harboring microbial community structure (Figure 7B). However, the standardized total negative effects on phoD-harboring microbial community structure were in the order of P application (−0.65) > TC (−0.55) > Fe (−0.37) > NaOH-Po (−0.31) > pH (−0.03).
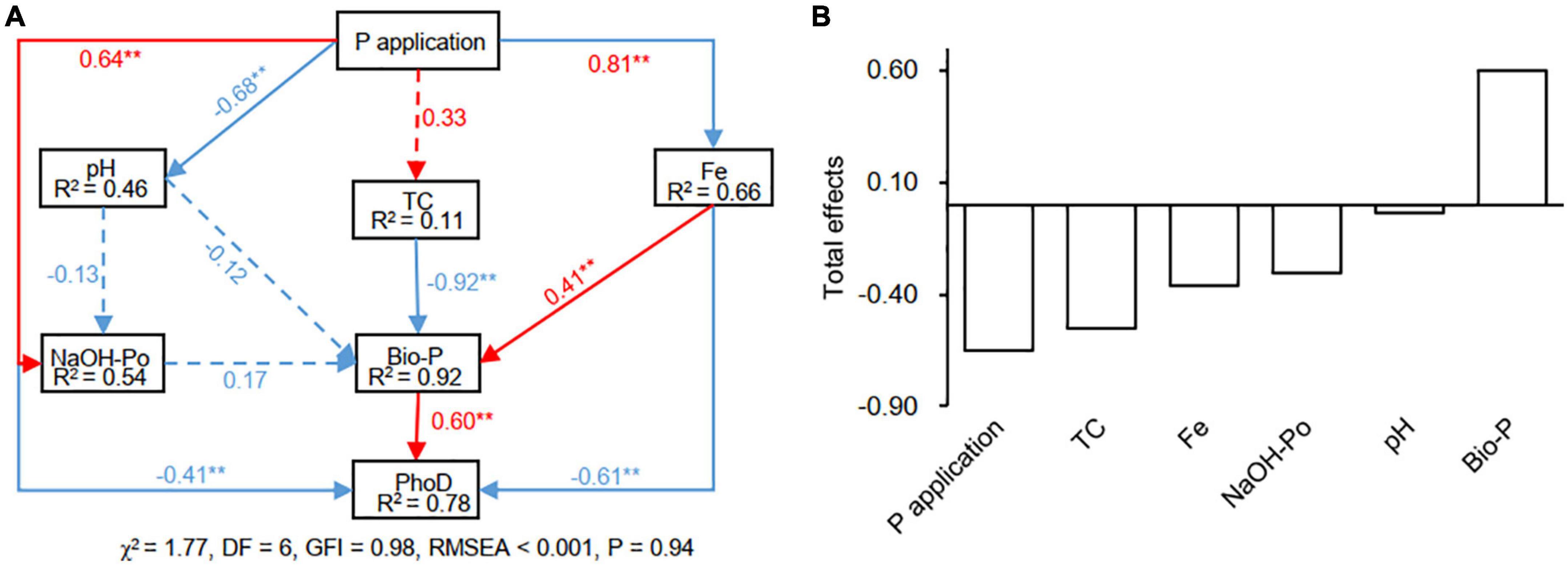
Figure 7. The effects of soil properties on phoD-harboring microbial community structure as estimated using the structural equation model. To simplify the SEM model, the PCA first axis score of each sample was used to represent the phoD-harboring microbial community structure. Panel (A), the numbers in blocks are the variance explained by the model (R2), and the numbers on arrows are standardized direct path coefficients. Red and blue arrows indicate positive and negative effects, respectively. The total effects of environmental factors on phoD-harboring microbial community structure are presented in Panel (B). TC, total carbon; Bio-P, bioavailable P. **P < 0.01.
We investigated the effects of different P application rates on the soil physicochemical properties, P forms and phoD-harboring microbial communities. By comparing the seven P application treatments (0, 10, 20, 30, 40, 50, 60 g⋅m–2⋅a–1), we found that most P forms (Resin-Pi, NaHCO3-Pi, NaOH-Pi, HCl-Pi, and Bio-P) peaked at P30 treatment. Similarly, soil phoD-harboring microbial diversity indices in the P30 were the highest. This indicated that excessive P inputs might suppress the inorganic P accumulation and the growth of phoD-harboring microbes. The observed differences in soil phoD-harboring microbial community structure may be attributed to variations in Bio-P, TC, Fe, NaOH-Po, and pH.
Fertilization is the main way of exogenous P input to soil, and the content of soil P forms is directly affected by P application (Mahmood et al., 2020). Soil Pi (Resin-Pi, NaHCO3-Pi, NaOH-Pi, HCl-Pi, and Bio-P) increased firstly and then decreased with increasing P application rate (0∼9.6 g P2O5⋅m–2), which verified our hypothesis. The study in Loess Plateau pointed out that NaHCO3-Pi, NaOH-Pi, and HCl-Pi increased with P application rate (0∼9.2 g P2O5⋅m–2), which was inconsistent with our result (Mahmood et al., 2020). The decreasing soil pH caused by P application could dissolve Ca-P in calcareous soils of the Loess Plateau, thus resulting in increasing soil Pi. However, most added Pi was absorbed and precipitated with Fe, Al, and Ca in acid soils, and the lower soil pH in high-P treatments would promote the absorption and precipitation (Wang et al., 2014; Maranguit et al., 2017). The increasing trend of NaOH-Po with P application rate may owe to the decrease in soil pH (Figures 1E, 6), because NaOH-Po is often associated with humic and fulvic acids (Wang et al., 2006). Moreover, NaOH-Po also contains the Po absorbed on Al and Fe hydrous oxides (Moro et al., 2015), and the increasing soil NaOH-Po is coupled well with Al and Fe content in current study (Figure 6). Soil Residual-P increased firstly and after that it had an approximate constant with the increase of P application rate, suggesting that soil P coated by Fe/Al hydrous oxides tends to be saturated when P application rate > 4.8 g P2O5⋅m–2.
The hydrolysis of organic P is predominantly mediated by phosphatase, especially acid and alkaline phosphatases (Fraser et al., 2015b). The phoD alkaline phosphatase is solely of microbial origin and widespread in soil (Ragot et al., 2015). Agricultural practices such as fertilization could affect the soil properties and thus influence the phoD-harboring microbial community (Fraser et al., 2015b; Liu et al., 2020). Previous studies have indicated that P fertilization could affect specific taxa related to the phoD-harboring microbial community (Liu et al., 2020; Wan et al., 2020; Chen et al., 2021; Wang M. M. et al., 2022). For example, the study in Loess Plateau found that the relative abundance of Proteobacteria group in high-P treatments (9.2 g P2O5⋅m–2) was higher than that in low-P treatments (0∼6.9 g P2O5⋅m–2) (Liu et al., 2020). Similarly, we found that the relative abundance of Proteobacteria in high-P (9.6 g P2O5⋅m–2) treatments was higher than that in low-P (0∼4.8 g P2O5⋅m–2) treatments (Figure 2I). Proteobacteria was reported to have copiotrophic lifestyle and thrive better under higher nutrient conditions (Veraart et al., 2015; Li et al., 2020), and the higher abundance of Proteobacteria was accordant with the higher soil TC in our study. Though the relative abundance of Actinobacteria varied slightly among the seven treatments (Figure 2B), we found four of six biomarkers in P30 derived from Actinobacteria (Figure 3), and soil TC in P30 was the lowest (Figure 1K). This was consistent with the oligotrophic lifestyle of Actinobacteria (Ho et al., 2017). The Deinococcus_Thermus group could resist the ultraviolet light, and the lower abundance of Deinococcus_Thermus (Figure 1D) in high-P treatments might be owed to the higher plant coverage (Griffiths and Gupta, 2007).
The study sampled from a maize field reported an increasing trend of phoD-harboring microbial diversity with P application rate (Chen et al., 2019). Another study in pasture soils also found PhoD gene diversity increased with P application rate (Tan et al., 2013). In the present study, we found that phoD-harboring microbial diversity also increased with increasing P application rate when P application rate < 4.8 g P2O5⋅m–2 (Figure 4), which disagreed with our second hypothesis. The increasing trend of phoD gene diversity indicated that a small amount of P input could meet the P demand of phoD-harboring microbes and further promote the growth of phoD-harboring microbes (Chen et al., 2019). However, the decreasing trend of phoD-harboring microbial diversity for P application rate > 4.8 g P2O5⋅m–2 (Figure 4) suggested that excessive P inputs might suppress the growth and reproduction of phoD-harboring microbes (Wei et al., 2019). Pimm and Pimm (1982) pointed out that sufficient resource could help to improve the propagation of other competitive species, thus phoD-harboring microbes might lose the advantage to outcompete other competitive microbes after high-P application. The opposite relationship between phoD-harboring microbial diversity and P application amount for below and above P application amount = 4.8 g P2O5⋅m–2 suggested a P fertilization threshold for optimistic growth of phoD-harboring microbes in the alpine grassland on the Qinghai-Tibetan Plateau. Similarly, Liu et al. (2020) conducted a P fertilization experiment (0∼9.2 g P2O5⋅m–2) in Loess Plateau and they also found a P fertilization threshold (6.9 g P2O5⋅m–2) for optimistic growth of soil microbes. Those P fertilization thresholds in the Qinghai-Tibetan Plateau and Loess Plateau further proved the “intermediate disturbance hypothesis” that local species diversity to be maximal at an intermediate level of disturbance (Bongers et al., 2009). Considering the higher percentage of the dominant phyla (e.g., Actinobacteria, Cyanobacteria, and Proteobacteria) in P60 treatment (Figures 2B,C,I), the lower phoD-harboring microbial diversity in P60 treatment likely depended on the growth of dominant taxa, but not rare ones.
The soil phoD-harboring microbes interact with each other and form a connected network to adapt to their habitat. In the current study, the interactions between ASVs (nodes and edges), degrees, connectance, and transitivity were higher in the high-P network (P40, P50, and P60, Figure 5B) than that in low-P network (P0, P10, P20, and P30, Figure 5A), suggesting a larger and more complex network in high-P treatments. The complexity of a network is one of the important traits of ecosystem stability and functioning (Wei et al., 2019). The higher network complexity in the high-P group benefits the stability of the phoD-harboring microbial community, suggesting that phoD-harboring microbes in the high-P group could mediate internal interactions to resist to stresses. Those microbes driving the same biochemical process can compete for similar resources and resulting in conflicting interactions (Le Gac et al., 2012). The more conflicting interactions in the phoD-harboring microbial network under low-P fertilized conditions (percent of the negative edges: 48.23%) than under high-P fertilized conditions (percent of the negative edges: 34.63%) indicated a stronger competition within the phoD-harboring microbes in low-P fertilized condition, and P fertilization could alleviate this competition. Furthermore, the generalists in the phoD-harboring microbial communities in the low-P (Figure 5C) and high-P (Figure 5D) networks were different. In the low-P network, the generalist Rhizobiales is a nitrogen-fixing taxon, but it also can solubilize phosphate (Halder et al., 1990), suggesting that Rhizobiales could increase soil available P in the P deficient condition. Apart from Rhizobiales, Burkholderiales, and Streptomycetales were the generalists in the high-P network. Burkholderiales is well-known as pathogens (Banerjee et al., 2018) while Streptomycetales could excrete antibiotics (Harir et al., 2018). Different functions of the generalists suggested a versatile phoD-harboring microbial community in high-P fertilized conditions.
Phosphorus fertilization can affect the soil phoD-harboring community by changing soil physiochemical properties (Ragot et al., 2016; Hu et al., 2018; Chen et al., 2019; Wei et al., 2019; Liu et al., 2020; Wan et al., 2020). In the present study, P application affected soil phoD-harboring community structure through Bio-P, TC, Fe, NaOH-Po, and pH. The study in a maize field reported that the shift in PhoD gene community structure was related to P availability (Chen et al., 2019). Similarly, soil Bio-P held the largest positive effect on soil phoD-harboring microbial community structure in the present study (Figure 7B). Bio-P is widely known to be a key factor affecting soil phoD-harboring microbes, as P is the essential component of soil phoD-harboring microbial nutrition (Chen et al., 2019; Liu et al., 2020; Wan et al., 2020; Wei et al., 2021; Wang M. M. et al., 2022). The higher Bio-P could stimulate the growth of soil phoD-harboring microorganisms, which could enhance the mineralization of organic P, releasing Pi and supplementing Bio-P in return (Chen et al., 2019). However, the positive feedback could not last forever. Another study reported a negative relationship between P availability and phoD microbial diversity in wheat rhizosphere soil (Xie et al., 2020). This discrepancy may be explained by the different age of fertilization. The P application in the wheat field lasted 7 years, which was longer than that in our study (2 years), and the long-term P application could suppress the reproduction of the phoD microbes (Wang M. M. et al., 2022; Zeng et al., 2022). Previous studies reported the significant effect of soil carbon on phoD-harboring microbial community structure (Ragot et al., 2016; Luo et al., 2017; Hu et al., 2018; Liu et al., 2020; Wang M. M. et al., 2022). Carbon as an energy supply for microbes was demonstrated to promote the propagation of microbial community, and the phoD-harboring microbial respiration could lead to carbon loss (Luo et al., 2017). The lowest soil TC, the highest phoD-harboring microbial diversity and the highest Bio-P in P30 suggested that phoD-harboring microbes likely depleted carbon resource to release Pi. Based on the SEM, the total effect size of Bio-P (0.60) on PhoD gene community structure was higher than that of TC (−0.55), reflecting the more important role of Bio-P. Moreover, the different effects of Bio-P and TC on phoD-harboring microbial community structure suggested that P mineralization is decoupled with C mineralization in the alpine grassland of the Qinghai-Tibetan Plateau.
In addition, the effect of soil Bio-P on soil phoD-harboring community could not be distinguished from that of Fe, given that Fe is commonly correlated with soil available P in acid soils (Wang et al., 2006). Soil Fe associated P acts as a buffer in P supply when the soil P is sufficient, and it could be released into soil solution and transformed into Bio-P when the soil Bio-P is insufficient (Maranguit et al., 2017). Another study in Karst soils also reported that metal ions affected soil phoD microbes through available P (Hu et al., 2018). The negative relationship between NaOH-Po and soil phoD gene (Figure 7) was consistent with the result in subtropical Chinese fir plantations (Wang et al., 2021). NaOH-Po is a kind of organic P that can be slowly mineralized into inorganic P when the soil available P is inadequate (Cao et al., 2022). Conversely, the inorganic P could be transformed into organic P by plant and soil microbial assimilation (Giles et al., 2014). The microbial cell is a kind of NaOH-Po source (Gu et al., 2017), and the high MBP under high-P treatment (Figure 1) was conductive to NaOH-Po accumulation. Moreover, soil phoD-harboring microbes compete with other microbes for resources, the low phoD-harboring microbial diversity and high MBP suggested a strong competition between phoD-harboring microbes and other microbes under high-P conditions. Similarly, P fertilization could promote the growth of the plant (Mahmood et al., 2020), which may enhance the P demand of plants and then intensify the competition between plants and phoD microbes for P. The competitions between soil phoD-harboring microbes, other soil microbes, and plants might be the reason why NaOH-Po negatively correlated with soil phoD microbial community structure (Wang et al., 2021).
Fertilization has been reported to decrease soil pH, and soil pH has also been shown to drive the soil microbial community (Liu et al., 2018; Chen et al., 2019; Xu et al., 2021). Mantel test indicated that soil pH was significantly correlated with phoD-harboring microbial community structure (Figure 6), and this was also confirmed by the negative effect of soil pH on phoD microbial community structure in the SEM model (Figure 7), suggesting that phoD microbial communities are less similar in soils with greater difference in pH. Soil pH has also been found as the important factor affecting phoD microbial communities in soil under grass (Ragot et al., 2015, 2016), Luvisol under maize (Chen et al., 2019), and Karst soil under soybean (Hu et al., 2018). The decreased soil pH enhanced the NaOH-Po content, as NaOH-Po is associated with humic and fulvic acids (Cao et al., 2022), further led to the change in phoD-harboring microbial communities as discussed above.
In conclusion, soil Pi (e.g., Resin-Pi, NaHCO3-Pi, NaOH-Pi, and HCl-Pi), Bio-P and soil phoD-harboring microbial diversity showed a tipping point at the P application rate of 30 g⋅m–2⋅a–1, demonstrating that there might be a P fertilization threshold for optimistic growth of phoD-harboring microbes. The appropriate P application rate for phoD-harboring microbial diversity in the studied alpine grassland on the Qinghai-Tibetan Plateau is about 30 g⋅m–2⋅a–1. In addition, soil Bio-P, TC, Fe, NaOH-Po, and soil pH were the determinant factors for the responses of soil phoD-harboring microbial community structure to P application in the studied alpine grassland on the Qinghai-Tibetan Plateau.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
DL and CW designed the study. XS and YL supported the experimental procedures. DL analyzed the data, prepared the manuscript, acquired the images, and created the figures. All authors read, edited, and approved the manuscript.
This project was supported by the National Natural Science Foundation of China (32101348 and U20A2008), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0302-02), and the “Double First-Class” program of Southwest Minzu University.
We thank Guohong You and Jingwen Bai (Institute of Mountain Hazards and Environment, Chinese Academy of Sciences) for providing experimental procedures and allowing the using of the instruments and reagents. Without their support this study would have not been possible. We also thank Yanhong Wu (Institute of Mountain Hazards and Environment, Chinese Academy of Sciences) and his team for assisting with the access of related materials.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Apel, A. K., Sola-Landa, A., Rodriguez-Garcia, A., and Martin, J. F. (2007). Phosphate control of phoA, phoC and phoD gene expression in Streptomyces coelicolor reveals significant differences in binding of PhoP to their promoter regions. Microbiology 153, 3527–3537. doi: 10.1099/mic.0.2007/007070-0
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bongers, F., Poorter, L., Hawthorne, W. D., and Sheil, D. (2009). The intermediate disturbance hypothesis applies to tropical forests, but disturbance contributes little to tree diversity. Ecol. Lett. 12, 798–805. doi: 10.1111/j.1461-0248.2009.01329.x
Cao, Z., Kühn, P., He, J. S., Bauhus, J., Guan, Z. H., and Scholten, T. (2022). Calibration of near-infrared spectra for phosphorus forms in grassland soils on the Tibetan Plateau. Agronomy 12:783. doi: 10.3390/agronomy12040783
Carneiro, L. F., de Resende, A. V., Neto, A. E. F., Santos, J. Z. L., Curi, N., Reis, T. H. P., et al. (2011). Soil phosphorus fractions in response to phosphate fertilization in an Oxisol under different land uses. Rev. Bras. Cienc. Solo 35, 483–491. doi: 10.1590/S0100-06832011000200017
Chen, S., Cade-Menun, B. J., Bainard, L. D., St Luce, M., Hu, Y. F., and Chen, Q. (2021). The influence of long-term N and P fertilization on soil P forms and cycling in a wheat/fallow cropping system. Geoderma 404:115274. doi: 10.1016/j.geoderma.2021.115274
Chen, X. D., Jiang, N., Condron, L. M., Dunfield, K. E., Chen, Z. H., Wang, J. K., et al. (2019). Impact of long-term phosphorus fertilizer inputs on bacterial phoD gene community in a maize field, Northeast China. Sci. Total Environ. 669, 1011–1018. doi: 10.1016/j.scitotenv.2019.03.172
Chen, X. H., Yan, X. J., Wang, M. K., Cai, Y. Y., Weng, X. F., Su, D., et al. (2022). Long-term excessive phosphorus fertilization alters soil phosphorus forms in the acidic soil of pomelo orchards. Soil Tillage Res. 215:105214. doi: 10.1016/j.still.2021.105214
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113
Feng, K., Peng, X., Zhang, Z., Gu, S., He, Q., Shen, W., et al. (2022). iNAP: An integrated network analysis pipeline for microbiome studies. iMeta 1:e13. doi: 10.1002/imt2.13
Fraser, T. D., Lynch, D. H., Entz, M. H., and Dunfield, K. E. (2015a). Linking alkaline phosphatase activity with bacterial phoD gene abundance in soil from a long-term management trial. Geoderma 257, 115–122. doi: 10.1016/j.geoderma.2014.10.016
Fraser, T. D., Lynch, D. H., Bent, E., Entz, M. H., and Dunfield, K. E. (2015b). Soil bacterial phoD gene abundance and expression in response to applied phosphorus and long-term management. Soil Biol. Biochem. 88, 137–147. doi: 10.1016/j.soilbio.2015.04.014
Giles, C. D., Hsu, P.-C., Richardson, A. E., Hurst, M. R. H., and Hill, J. E. (2014). Plant assimilation of phosphorus from an insoluble organic form is improved by addition of an organic anion producing Pseudomonas sp. Soil Biol. Biochem. 68, 263–269. doi: 10.1016/j.soilbio.2013.09.026
Griffiths, E., and Gupta, R. S. (2007). Identification of signature proteins that are distinctive of the Deinococcus-Thermus phylum. Int. Microbiol. 10, 201–208. doi: 10.2436/20.1501.01.28
Gu, S., Gruau, G., Dupas, R., Rumpel, C., Creme, A., Fovet, O., et al. (2017). Release of dissolved phosphorus from riparian wetlands: Evidence for complex interactions among hydroclimate variability, topography and soil properties. Sci. Total Environ. 598, 421–431. doi: 10.1016/j.scitotenv.2017.04.028
Halder, A. K., Mishra, A. K., Bhattacharyya, P., and Chakrabartty, P. K. (1990). Solubilization of rock phosphate by Rhizobium and Bradyrhizobium. J. Gen. Appl. Microbiol. 36, 81–92. doi: 10.2323/jgam.36.81
Harir, M., Bendif, H., Bellahcene, M., Fortas, Z., and Pogni, R. (2018). “Streptomyces secondary metabolites,” in Basic biology and applications of actinobacteria, ed. S. Enany (London: IntechOpen).
Ho, A., Di Lonardo, D. P., and Bodelier, P. L. E. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93:fix006. doi: 10.1093/femsec/fix006
Hooper, D., Coughlan, J., and Mullen, M. (2008). Structural equation modeling: Guidelines for determining model fit. Electron. J. Bus. Res. Methods 6, 53–60. doi: 10.0000/PMID35188134
Hou, E. Q., Chen, C. R., Luo, Y. Q., Zhou, G. Y., Kuang, Y. W., Zhang, Y. G., et al. (2018). Effects of climate on soil phosphorus cycle and availability in natural terrestrial ecosystems. Glob. Change Biol. 24, 3344–3356. doi: 10.1111/gcb.14093
Hu, Y. J., Xia, Y. H., Sun, Q., Liu, K. P., Chen, X. B., Ge, T. D., et al. (2018). Effects of long-term fertilization on phoD-harboring bacterial community in Karst soils. Sci. Total Environ. 628–629, 53–63. doi: 10.1016/j.scitotenv.2018.01.314
Hussain, A., Chen, L. X., Jamil, M. A., Abid, K., Khan, K., Duan, W. B., et al. (2022). Changes in soil-phosphorus fractions by nitrogen and phosphorus fertilization in Korean pine plantation and its natural forest. Forests 13:527. doi: 10.3390/f13040527
Le Gac, M., Plucain, J., Hindre, T., Lenski, R. E., and Schneider, D. (2012). Ecological and evolutionary dynamics of coexisting lineages during a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, 9487–9492. doi: 10.1073/pnas.1207091109
Li, W. X., Zhang, Y. P., Mao, W., Wang, C. S., and Yin, S. X. (2020). Functional potential differences between Firmicutes and Proteobacteria in response to manure amendment in a reclaimed soil. Can. J. Microbiol. 66, 689–697. doi: 10.1139/cjm-2020-0143
Liao, D., Zhang, C. C., Lambers, H., and Zhang, F. S. (2021). Changes in soil phosphorus forms in response to long-term phosphate fertilization under sole cropping and intercropping of maize and faba bean on a calcareous soil. Plant Soil 463, 589–600. doi: 10.1007/s11104-021-04915-y
Liu, D., Liu, G., Chen, L., Wang, J., and Zhang, L. (2018). Soil pH determines fungal diversity along an elevation gradient in Southwestern China. Sci. China Life Sci. 61, 718–726. doi: 10.1007/s11427-017-9200-1
Liu, J. S., Ma, Q., Hui, X. L., Ran, J. Y., Ma, Q. X., Wang, X. S., et al. (2020). Long-term high-P fertilizer input decreased the total bacterial diversity but not phoD-harboring bacteria in wheat rhizosphere soil with available-P deficiency. Soil Biol. Biochem. 149:107918. doi: 10.1016/j.soilbio.2020.107918
Luo, G., Ling, N., Nannipieri, P., Chen, H., Raza, W., Wang, M., et al. (2017). Long-term fertilisation regimes affect the composition of the alkaline phosphomonoesterase encoding microbial community of a vertisol and its derivative soil forms. Biol. Fertil. Soils 53, 375–388. doi: 10.1007/s00374-017-1183-3
Magoc, T., and Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mahmood, M., Tian, Y., Ma, Q. X., Ahmed, W., Mehmood, S., Hui, X. L., et al. (2020). Changes in phosphorus forms and its availability status in relation to long term P fertilization in Loess Plateau of China. Agronomy 10:1818. doi: 10.3390/agronomy10111818
Maranguit, D., Guillaume, T., and Kuzyakov, Y. (2017). Land-use change affects phosphorus forms in highly weathered tropical soils. CATENA 149, 385–393. doi: 10.1016/j.catena.2016.10.010
Mendes, L. W., Kuramae, E. E., Navarrete, A. A., van Veen, J. A., and Tsai, S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8, 1577–1587. doi: 10.1038/ismej.2014.17
Moro, H., Kunito, T., and Sato, T. (2015). Assessment of phosphorus bioavailability in cultivated Andisols from a long-term fertilization field experiment using chemical extractions and soil enzyme activities. Arch. Agron. Soil Sci. 61, 1107–1123. doi: 10.1080/03650340.2014.984697
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Pimm, S. L., and Pimm, J. W. (1982). Resource use, competition, and resource availability in Hawaiian Honeycreepers. Ecology 63, 1468–1480. doi: 10.2307/1938873
Ragot, S. A., Huguenin-Elie, O., Kertesz, M. A., Frossard, E., and Bunemann, E. K. (2016). Total and active microbial communities and phoD as affected by phosphate depletion and pH in soil. Plant Soil 408, 15–30. doi: 10.1007/s11104-016-2902-5
Ragot, S. A., Kertesz, M. A., and Bunemann, E. K. (2015). Diversity of the phoD alkaline phosphatase gene in soil. Appl. Environ. Microbiol. 81, 7281–7289. doi: 10.1128/AEM.01823-15
Ragot, S. A., Kertesz, M. A., Meszaros, E., Frossard, E., and Bunemann, E. K. (2017). Soil phoD and phoX alkaline phosphatase gene diversity responds to multiple environmental factors. FEMS Microbiol. Ecol. 93:fiw212. doi: 10.1093/femsec/fiw212
Ren, J. Z., Hu, Z. Z., Zhao, J., Zhang, D. G., Hou, F. J., Lin, H. L., et al. (2008). A grassland classification system and its application in China. Rangeland J. 30, 199–209. doi: 10.1071/RJ08002
Sun, X. L., Li, M., Wang, G. X., Drosos, M., Liu, F. L., and Hu, Z. Y. (2019). Response of phosphorus fractions to land-use change followed by long-term fertilization in a sub-alpine humid soil of Qinghai-Tibet plateau. J. Soils Sediments 19, 1109–1119. doi: 10.1007/s11368-018-2132-y
Tan, H., Barret, M., Mooij, M. J., Rice, O., Morrissey, J. P., Dobson, A., et al. (2013). Long-term phosphorus fertilisation increased the diversity of the total bacterial community and the phoD phosphorus mineraliser group in pasture soils. Biol. Fertil. Soils 49, 661–672. doi: 10.1007/s00374-012-0755-5
Tian, J. H., Kuang, X. Z., Tang, M. T., Chen, X. D., Huang, F., Cai, Y. X., et al. (2021). Biochar application under low phosphorus input promotes soil organic phosphorus mineralization by shifting bacterial phoD gene community composition. Sci. Total Environ. 779:146556. doi: 10.1016/j.scitotenv.2021.146556
Tian, X., Bing, H. J., Wu, Y. H., Zhu, H., Zhao, W., He, Q. Q., et al. (2022). Farmland abandonment decreases soil bioavailable phosphorus but increases organic phosphorus in the mid-hills of Nepal. Catena 211:106000. doi: 10.1016/j.catena.2021.106000
Tiessen, H., and Moir, J. O. (1993). Characterization of available P by sequential extraction. Boca Raton, FL: Lewis Publishers.
Veraart, A. J., Steenbergh, A. K., Ho, A., Kim, S. Y., and Bodelier, P. L. E. (2015). Beyond nitrogen: The importance of phosphorus for CH4 oxidation in soils and sediments. Geoderma 259, 337–346. doi: 10.1016/j.geoderma.2015.03.025
Wan, W., Li, X., Han, S., Wang, L., Luo, X., Chen, W., et al. (2020). Soil aggregate fractionation and phosphorus fraction driven by long-term fertilization regimes affect the abundance and composition of P-cycling-related bacteria. Soil Tillage Res. 196:104475. doi: 10.1016/j.still.2019.104475
Wang, C. Q., Xue, L., and Jiao, R. Z. (2021). Soil phosphorus forms, phosphatase activity, and the abundance of phoC and phoD genes vary with planting density in subtropical Chinese fir plantations. Soil Tillage Res. 209:104946. doi: 10.1016/j.still.2021.104946
Wang, G. P., Liu, J. S., Wang, J. D., and Yu, J. B. (2006). Soil phosphorus forms and their variations in depressional and riparian freshwater wetlands (Sanjiang Plain, Northeast China). Geoderma 132, 59–74. doi: 10.1016/j.geoderma.2005.04.021
Wang, M. M., Wu, Y. C., Zhao, J. Y., Liu, Y., Chen, Z., Tang, Z. Y., et al. (2022). Long-term fertilization lowers the alkaline phosphatase activity by impacting the phoD-harboring bacterial community in rice-winter wheat rotation system. Sci. Total Environ. 821:153406. doi: 10.1016/j.scitotenv.2022.153406
Wang, Q., Quensen, J. F. III, Fish, J. A., Lee, T. K., Sun, Y., Tiedje, J. M., et al. (2013). Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio 4:e00592-13. doi: 10.1128/mBio.00592-13
Wang, Y., Tang, J., Zhang, H., Schroder, J. L., and He, Y. (2014). Phosphorus availability and sorption as affected by long-term fertilization. Agronomy 106, 1583–1592. doi: 10.2134/agronj14.0059
Wang, Z. Q., Zhao, M. Y., Yan, Z. B., Yang, Y. H., Niklas, K. J., Huang, H., et al. (2022). Global patterns and predictors of soil microbial biomass carbon, nitrogen, and phosphorus in terrestrial ecosystem. CATENA 211:e106037. doi: 10.1016/j.catena.2022.106037
Wei, X. M., Hu, Y. J., Cai, G., Yao, H. Y., Ye, J., Sun, Q., et al. (2021). Organic phosphorus availability shapes the diversity of phoD-harboring bacteria in agricultural soil. Soil Biol. Biochem. 161:108364. doi: 10.1016/j.soilbio.2021.108364
Wei, X. M., Hu, Y. J., Razavi, B. S., Zhou, J., Shen, J. L., Nannipieri, P., et al. (2019). Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol. Biochem. 131, 62–70. doi: 10.1016/j.soilbio.2018.12.025
Wu, J., He, Z. L., Wei, W. X., O’Donnell, A. G., and Syers, J. K. (2000). Quantifying microbial biomass phosphorus in acid soils. Biol. Fertil. Soils 32, 500–507. doi: 10.1007/s003740000284
Xiao, J. N., Dong, S. K., Shen, H., Li, S., Zhi, Y. L., Mu, Z. Y., et al. (2022). Phosphorus addition promotes Nitrogen retention in alpine grassland plants while increasing N deposition. CATENA 210:105887.
Xie, Y. Y., Wang, F. H., Wang, K., Yue, H. Z., and Lan, X. F. (2020). Responses of bacterial phoD gene abundance and diversity to crop rotation and feedbacks to phosphorus uptake in wheat. Appl. Soil Ecol. 154:103604. doi: 10.1016/j.apsoil.2020.103604
Xu, L., He, N., Li, X., Cao, H., Li, C., Wang, R., et al. (2021). Local community assembly processes shape β-diversity of soil phoD-harbouring communities in the Northern Hemisphere steppes. Glob. Ecol. Biogeogr. 30, 2273–2285. doi: 10.1111/geb.13385
Zeng, Q. C., Mei, T. Y., Delgado-Baquerizo, M., Wang, M. X., and Tan, W. F. (2022). Suppressed phosphorus-mineralizing bacteria after three decades of fertilization. Agric. Ecosyst. Environ. 323:107679. doi: 10.1016/j.agee.2021.107679
Keywords: P application, P forms, phoD gene, microbial diversity, alpine grassland
Citation: Liu D, Song X, Liu Y and Wang C (2023) Effects of phosphorus application on soil phosphorus forms and phoD-harboring microbial communities in an alpine grassland on the Qinghai-Tibetan Plateau. Front. Ecol. Evol. 11:1131408. doi: 10.3389/fevo.2023.1131408
Received: 25 December 2022; Accepted: 30 January 2023;
Published: 14 February 2023.
Edited by:
Heng Huang, University of California, Berkeley, United StatesReviewed by:
Zhanfeng Liu, South China Botanical Garden (CAS), ChinaCopyright © 2023 Liu, Song, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changting Wang,  d2FuZ2N0QHN3dW4uZWR1LmNu,
d2FuZ2N0QHN3dW4uZWR1LmNu,  d2FuZ2N0NkAxNjMuY29t
d2FuZ2N0NkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.