
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 20 February 2023
Sec. Coevolution
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1126243
 Yanqiong Chen1,2,3
Yanqiong Chen1,2,3 Ying Lai1,2,3
Ying Lai1,2,3 Jingjin Zheng1,2,3
Jingjin Zheng1,2,3 Zheng Liu1,2,3
Zheng Liu1,2,3 Dengpan Nong4
Dengpan Nong4 Jipeng Liang4
Jipeng Liang4 Youbang Li1,2,3
Youbang Li1,2,3 Zhonghao Huang1,2,3*
Zhonghao Huang1,2,3*Investigating gut microbiota is important for understanding the physiological adaptation of animals to food availability changes in fragmented habitats and consequently providing new ideas for the conservation of endangered wild animals. In this study, we explored the gut microbiota of the endangered white-headed black langur (Trachypithecus leucocephalus), which is endemic to the limestone forests of Southwest Guangxi, China, to understand its adaptation strategies to seasonal changes in habitat using 16S rRNA sequencing. Our results revealed significant seasonal variations in the gut microbiota of white-headed black langurs. In particular, the alpha diversity was higher in the rainy season than in the dry season, and the beta diversity was significantly different between the two seasons. At the phylum level, the relative abundance of Firmicutes, Actinobacteriota, and Proteobacteria was higher in the dry season than that in the rainy season, whereas that of Bacteroidetes, Spirochaetota, and Cyanobacteria was significantly higher in the rainy season than that in the dry season. At the family level, Oscillospiraceae and Eggerthellaceae were more abundant in the dry season than in the rainy season, whereas Lachnospiraceae, Ruminococcaceae, and Monoglobaceae were more abundant in the rainy season than in the dry season. These results could have been obtained due to seasonal changes in the diet of langurs in response to food plant phenology. In addition, the neutral community model revealed that the gut microbiota assembly of these langurs was dominated by deterministic processes and was more significantly affected by ecological factors in the dry season than in the rainy season, which could be linked to the higher dependence of these langurs on mature leaves in the dry season. We concluded that the seasonal variations in the gut microbiota of white-headed black langurs occurred in response to food plant phenology in their habitat, highlighting the importance of microbiota in responding to fluctuating ecological factors and adapting to seasonal dietary changes.
The gut microbiota play an important role in host food digestion (Nicholson et al., 2012; Yatsunenko et al., 2012; Froidurot and Julliand, 2022). They directly interact with the digestive system to digest hard-to-degrade dietary components (Zhu et al., 2011; Froidurot and Julliand, 2022). For instance, the gut microbiota break down dietary fibers into short-chain fatty acids (SCFAs) to maintain energy reserves and fight against inflammation (Machado et al., 2021; Sun et al., 2022). In particular, the Firmicutes is a typical taxon that degrade cellulose and hemicellulose in the guts of animals that largely depend on plants, such as the giant panda (Ailuropoda melanoleuca; Zhu et al., 2011; Jin et al., 2021) and Udzungwa red colobus (Procolobus gordonorum; Barelli et al., 2015). In addition, the gut microbiota play an essential role in carbohydrate and lipid metabolism (Flint et al., 2012; Nicholson et al., 2012; Schoeler and Caesar, 2019). For example, a previous study revealed that the proportion of Prevotella in the gut microbiota increased with the provision of high-energy food in captive golden snub-nosed monkeys (Rhinopithecus roxellana; Wang et al., 2021). Thus, disorder in the gut microbiota are disastrous for the host and can even cause diseases (Nicholson et al., 2012); therefore, exploration of gut microbiota is important for monitoring the health of wild animals.
Although host genetics is a commonly considered determinant in structuring host gut microbiota (Amato et al., 2019), the influence of external factors, including habitat (Jin et al., 2021), behavior (Clarke et al., 2014; Barton et al., 2018), and dietary niche (Filippo et al., 2010; Losasso et al., 2018; Rothschild et al., 2018), on gut microbiota is significant. Previous studies have indicated that host diet has a more profound impact on the diversity of gut microbiota when compared to host genetics (Rothschild et al., 2018), which is mainly reflected by the provision of specific substrates for microbiota growth (Ley et al., 2008; Losasso et al., 2018). For example, compared with European children, the microbiota of African children have three unique genera, which are associated with high-fiber diets (Filippo et al., 2010). Moreover, dietary richness is strongly associated with gut microbiota diversity (Lin et al., 2021). An example being when black howler monkeys (Alouatta pigra) live in disturbed habitats, they tend to exhibit lower gut microbiota diversity when compared to others living in undisturbed habitats (Amato et al., 2013). Similarly, captivity commonly results in reduced gut microbiota diversity in animals, mostly due to the simple diets of captive individuals (Clayton et al., 2016; Frankel et al., 2019).
Wild animals adjust their feeding strategies in response to the fluctuations in food resources across seasons (Zhou et al., 2018; Matsuda et al., 2020). This in turn alters the selection pressure acting on the gut microbiota (David et al., 2014; Amato et al., 2015; Li et al., 2021) and results in their dynamic balancing in response to specific diets (Amato et al., 2015; Hicks et al., 2018; Xia et al., 2021). Nonhuman primates (NHPs) tend to have more varied diets in seasons with more low-quality food, which is usually accompanied by higher gut microbiota diversity (Sun et al., 2016; Sawada et al., 2022), as reported in Japanese macaques (Macaca fuscata; Sawada et al., 2022). In addition, the responses of the gut microbiota to food availability changes are reflected in the form of fluctuations in relative abundance (Springer et al., 2017; Hicks et al., 2018). To be specific, the proportion of Bacteroidetes in the gut microbiota of rhesus macaques (Macaca mulatta) and Tibetan macaques (Macaca thibetana) increases when they consume more young leaves and fruits (Sun et al., 2016; Li et al., 2021; Xia et al., 2021). However, the Firmicutes colonizing the guts of these macaques increases when they consume more high-fiber foods (Sun et al., 2016; Li et al., 2021; Xia et al., 2021). Such flexibility in the gut microbiota serves as a buffer, allowing hosts to adapt to varied diets (Alberdi et al., 2016). Similar seasonal variations in gut microbiota have been demonstrated in white-faced capuchin monkeys (Cebus capucinus imitator; Orkin et al., 2019), Verreaux’s sifakas (Propithecus verreauxi; Springer et al., 2017), and Ethiopian geladas (Theropithecus gelada; Baniel et al., 2021).
Although food availability changes are understood to impact the gut microbiota of NHPs, we can utilize neutral community model (NCM; Sloan et al., 2006) to gain a more in depth understanding of how different community assembly processes influence gut microbiota. There are two processes that are thought to influence microbiota assembly: stochastic processes (such as mortality, dispersal, and migration; Chave, 2004) and deterministic processes (such as environmental filtering and biological interactions; Fargione et al., 2003). The relative importance of these processes can be assessed using NCM (Sloan et al., 2006). The white-lipped deer (Cervus albirostris) has been found to enhance the contribution of deterministic processes in the gut microbiota assembly due to changes in diet in captivity (Li B. et al., 2022). In addition, in the four groups of cotton bollworm (Helicoverpa armigera), although the gut microbial community assembly is mainly dominated by stochastic process, the proportion seem to be affected by the maize varieties and locations (Li S. L. et al., 2022). Exploration of complex processes in the gut microbiota assembly in endangered animals could help determine whether survival environment of host interferes with these processes and may contribute to a deeper understanding of microbiota–host interactions (Zhou and Ning, 2017; Heys et al., 2020).
White-headed black langurs (Trachypithecus leucocephalus) are rare and endangered animals included in the IUCN Red List (Bleisch and Long, 2020). Like François’ langurs (Trachypithecus francoisi; Chen et al., 2020), these langurs are highly folivorous (Huang, 2002; Li and Elizabeth Rogers, 2006; Zhou et al., 2013). These langurs have the typical digestive structure of colobine primates, such as sharp molars and an expanded rumen, which are considered adaptation to their high-fiber diet (Huang, 2002). Moreover, white-headed black langurs are exclusively found in the limestone forests of Southwest Guangxi, China, and occupy habitats characterized by little surface water availability, strong seasonality in climatic and plant phenology, and high susceptibility to human disturbance (Guangxi Forestry Department, 1993; Huang, 2002; Tang et al., 2016). These langurs adapt to highly fragmented habitats through behavioral adjustments (Li et al., 2016; Huang et al., 2017; Zhang et al., 2021). Previous studies focusing on behavioral adaptations of these langurs revealed that they heavily rely on mature leaves for their diet, particularly in the dry season when food availability is low (Li and Elizabeth Rogers, 2006; Zhou et al., 2013; Lu et al., 2021). However, when behavioral responses are insufficient to meet the hosts’ need, the animals use physiological adaptations, such as hormonal regulations (Tecot et al., 2019) and gut microbiota adjustments (Amato et al., 2015; Wong and Candolin, 2015; Bertram et al., 2022). Investigating gut microbiota is important to understand the physiological adaptation of white-headed black langurs to fragmented habitats. Previously, the limited studies on the gut microbiota of white-headed black langurs revealed that the gut microbiota of these langurs comprised Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria (Que et al., 2022; Lai et al., 2023). Therefore, it is critical that we need to conduct further studies on the gut microbiota of the white-headed black langurs so that we can comprehend how gut microbiota responds to habitat shifts and propose new strategies for conservation biology.
Gut microbiota is a suitable tool for understanding the physiology of hosts (Amato et al., 2016; Trevelline et al., 2019), which co-evolves with the host and is significantly influenced by host diet (Rothschild et al., 2018). Changes in plant phenology in animals’ habitats would alter their diet, consequently leading to seasonal variations in their gut microbiota (David et al., 2014; Amato et al., 2015; Li et al., 2021). At present, there are limited studies on the gut microbiota of white-headed black langurs, and the data on the seasonality of their gut microbiota have not been available yet. Thus, study on seasonal changes in these langurs’ gut microbiota should be required, which could profoundly reflect the adaptation of these langurs to the habitat shifts and undoubtedly provide new ideas for animal population conservation (Trevelline et al., 2019). In this study, we used 16S rRNA high-throughput sequencing to explore the composition of the gut microbiota of white-headed black langurs in the Guangxi Chongzuo White-headed Langur National Nature Reserve. We first describe the composition and diversity of the gut microbiota and then analyze the community assembly of gut microbiota using a NCM (Sloan et al., 2006). Finally, we discuss the seasonal variations in the gut microbiota of these langurs by testing following predictions:
(1) Dietary diversity is strongly associated with gut microbiota diversity (Lin et al., 2021). High rainfall in the rainy season improves the availability of seasonal foods, such as mature fruits and young leaves, whereas low rainfall in the dry season leads to the lack of food, forcing white-headed black langurs to consume more mature leaves, which is a less varied diet (Huang, 2002; Huang et al., 2015; Tang et al., 2016). We predict that white-headed black langurs will have higher gut microbiota diversity in the rainy season than in the dry season.
(2) Changes in food availability can lead to alterations in the structure of the gut microbiota (Amato et al., 2015; Hicks et al., 2018; Xia et al., 2021). During the dry season, when food availability is reduced, white-headed black langurs consume more mature leaves rich in fibers, such as cellulose (Zhou et al., 2013; Li et al., 2016). Whereas during the rainy season, langurs consume more fruits and young leaves lean in cellulose (Li and Elizabeth Rogers, 2006; Zhou et al., 2013; Li et al., 2016). We predict that white-headed black langurs will have an increased abundance of microbiota taxa associated with cellulose digesting ability in the dry season and will colonize more taxa associated with carbohydrate digestion in the rainy season.
(3) High habitat fragmentation limits the food resources of white-headed black langurs and could increase the importance of environmental factors in community assembly (Burns et al., 2016; Ning et al., 2019; Tang et al., 2021; Li B. et al., 2022). We predict that the value of R2 based on NCM will be less than 0.5, indicating that the gut microbiota assembly of the langurs will be dominated by deterministic processes.
This research was conducted in the Guangxi Chongzuo White-headed Langur National Nature Reserve, China (107°16′53″-107°59′46″E, 22°10′43″-22°36′55″N). This reserve shows marked seasonality in temperature, rainfall, and plant resource availability (Huang, 2002). In this reserve, extreme rainfall usually occurs with high temperatures (Huang, 2002), and the year can be divided into a dry season (September to February) and a rainy season (March to August) based on rainfall patterns (Guangxi Forestry Department, 1993; Li et al., 2021; Zhang et al., 2021). The reserve is covered by unique karst forests, which are vulnerable to disturbance by human activities (farming and/or grazing) and are severely fragmented (Guangxi Forestry Department, 1993). We focused on three groups of white-headed black langurs, namely, the Botanical Garden group (25 individuals), Nongshui group (14 individuals), and Sankeshu group (14 individuals; Supplementary Table S1).
White-headed black langurs use caves on the cliffs as sleeping sites and defecate before leaving in the morning (Huang and Li, 2005). Before sampling, we observed langurs to determine their sleeping site and collected fecal samples under the cliff in the morning of the next day. Fresh feces were collected in the morning under the cliffs with a > 2-m interval between each sampling site to avoid collecting duplicate samples for any given individual. The middle portion of the feces, which was not in contact with the outside environment, was collected by wearing sterile gloves and using germ-free tools, such as a bamboo skewer. Each sample weighed 3–5 g and was placed in a labeled sterile collection tube. The samples transferred to an ultralow temperature refrigerator set at −80°C for storage. The study was conducted in December 2020 and January, June, and July 2021, during which 155 fecal samples were collected (Supplementary Table S1).
All fecal samples were collected with permission from the Administration Center of Guangxi Chongzuo White-headed Langur National Nature Reserve. Collection of animal tissues that could cause injury or fright to the animals was not performed. We collected fecal samples after the langurs left their sleeping sites to avoid causing any stress reactions.
DNA extraction from fecal samples was performed using the E.Z.N.A.® Soil DNA Kit (Omega Bio-Tek, America). The extracted DNA was subjected to electrophoresis in a 1% agarose gel to check its quality, while its purity and concentration were analyzed using a NanoDrop 2000 (Thermo Fisher Scientific, America) micro-ultraviolet–visible spectrophotometer. The V3–V4 region of the 16S rRNA gene of the bacteria was amplified via polymerase chain reaction (PCR) (ABI GeneAmp® 9700, ABI, America), using primers (338F: 5′-ACTCCTACGGGAGGCAGCAG-3′; 806R: 5′-GGACTACHVGGGTWTCTAAT-3′; Mori et al., 2013). The PCR system consisted of 5× FastPfu Buffer (4 μl), 2.5 mM dNTPs (2 μl), 5 μM forward primer (0.8 μl), 5 μM reverse primer (0.8 μl), FastPfu polymerase (0.4 μl) (TransGen, China), BSA (0.2 μl), template DNA (10 ng), and ddH2O. Each sample was amplified thrice, and each amplification consisted of the following three stages: initial denaturation (at 95°C for 3 min), followed by 28 thermal cycles (denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 45 s), and final extension (at 72°C for 10 min and then kept at 10°C until halted by user). The PCR products of three repeats were mixed and extracted from a 2% agarose gel and then purified and quantified using the AxyPrep DNA Gel Extraction Kit (Axygen, America) and Quantus™ Fluorometer (Promega, America). The fluorescently quantified products were used to build a library using the NEXTFLEX Rapid DNA-Seq Kit (Bioo Scientific, America) and sequenced using the Illumina Miseq PE300 platform (Illumina, America).
Trimmomatic software was used to filter and screen the raw sequences by setting a window of 50 bp, truncating reads with average quality values below 20, and filtering reads below 50 bp or containing N bases after quality control. The FLASH 1.2.111 software (Magoč and Salzberg, 2011) was used for splicing. The sequences were matched exactly to the primers (up to two nucleotide mismatches were allowed), and sequences that overlapped by more than 10 bp were merged after discarding those that appeared to be incorrect. Operational taxonomic unit (OTU) clustering with >97% similarity was performed in the software Uparse 7.0.10902 (Edgar, 2013), which resulted in the generation of an OTU table. The OTU sequences were compared in the 16S rRNA SILVA 138 bacteria database3 using RDP classifier 2.134 with a confidence threshold of 0.8 to obtain taxonomic information. Finally, secondary drawdown (by minimum sequence number) was performed, and subsequent analyzes were based on it.
A rank–abundance curve was plotted using the software R to assess the reasonableness of the sequencing data and the adequacy of the data volume. The community abundance of taxa was expressed as mean ± standard deviation (SD), as well as in histogram form (R v.3.3.1) to visualize the composition of the taxa. Alpha diversity analysis was conducted using Mothur software v.1.30.25 and included the ACE index, Chao index, Shannon index, and invSimpson index, which are community richness and diversity indices. The alpha diversity indices were converted to the form of log10 (x) (Warton and Hui, 2011) and visualized by R (v.4.1.2) as box plots. A generalized linear mixed model (GLMM) was constructed in R (v.4.1.2) and used to detect whether there were significant differences in those indices between the two seasons (Zheng et al., 2021; Lai et al., 2023). Wilcoxon rank-sum test was carried out by using R (v.3.3.1) to test the difference in the abundance of each taxon between dry and rainy seasons at the phylum and family levels. The p-value multiple correction was performed using the false discovery rate (FDR), and the results provided corrected p-values. Principal coordinate analysis (PCoA) was performed in R (v.3.3.1) at the OTU level based on weighted unifrac and unweighted unifrac distances, with Adonis test for analyzing differences between the two seasons. All analyzes listed above were performed on the Majorbio Cloud Platform and the significance level was set at p < 0.05.
A random forest (RF) algorithm was built in R (v.4.1.2) using the randomForest package (Breiman, 2001). The test was set up with a training set and a test set, representing 70% and 30% of all samples, respectively. The purpose of the algorithm was to identify representative taxa and obtain the Mean Decrease Gini values, where larger values indicate that the taxa had a larger contribution in variations between the two seasons. In addition, we performed five ten-fold cross-validations to minimize errors. Finally, AUC values from the ROC curves were calculated using the pROC package (Robin et al., 2011). AUC values equivalent to 0.5 indicated the absence of categorical significance, whereas values closer to 1 indicated a stronger predictive power of the model.
To investigate the construction of the community in the gut microbiota of white-headed black langurs, NCM (Sloan et al., 2006) was constructed in R (v.4.1.3) based on the OTU level to quantify the importance of stochastic and deterministic processes in community assembly, and the species migration rate (m) and model fit (0 < R2 < 1) were estimated. R2 represented the relative importance of stochastic processes, 1 − R2 represented the relative importance of deterministic processes.
A total of 7,118,780 original OTU valid sequences were obtained after high-throughput sequencing, with an average of 45,927.61 ± 14,616.09 sequences for each sample (Supplementary Table S2). After rarefying all samples by the minimum number of sample sequence, the sequence number of each sample was 22,886, and clustering yielded 2,706 OTUs, 33 phyla, and 349 families. The rarefaction curves of the Sobs index and Shannon index tended to be asymptote (Figure 1), and the Good’s coverage value of all samples ranged from 98.49% to 99.70% (Supplementary Table S3), indicating that the sequencing data were reasonable and accurately reflected most of the microbial information.
At the phylum level, the gut microbiota of white-headed black langurs were dominated by Firmicutes (87.31% ± 10.85%), Actinobacteriota (5.13% ± 4.36%), and Verrucomicrobiota (2.72% ± 3.72%) in the dry season, whereas they were dominated by Firmicutes (85.32% ± 6.53%), Bacteroidetes (7.30% ± 4.82%), and Actinobacteriota (2.95% ± 2.49%) in the rainy season (Figure 2A; Supplementary Table S4). At the family level, the dominant taxa were norank_o__Clostridia_UCG-014, Oscillospiraceae, and Christensenellaceae. The proportion of those families were 18.97% ± 7.36%, 19.11% ± 6.34%, and 16.48% ± 4.94% in the dry season and 16.45% ± 5.89%, 14.71% ± 3.57%, and 14.93% ± 4.09% in the rainy season, respectively (Figure 2B; Supplementary Table S5).
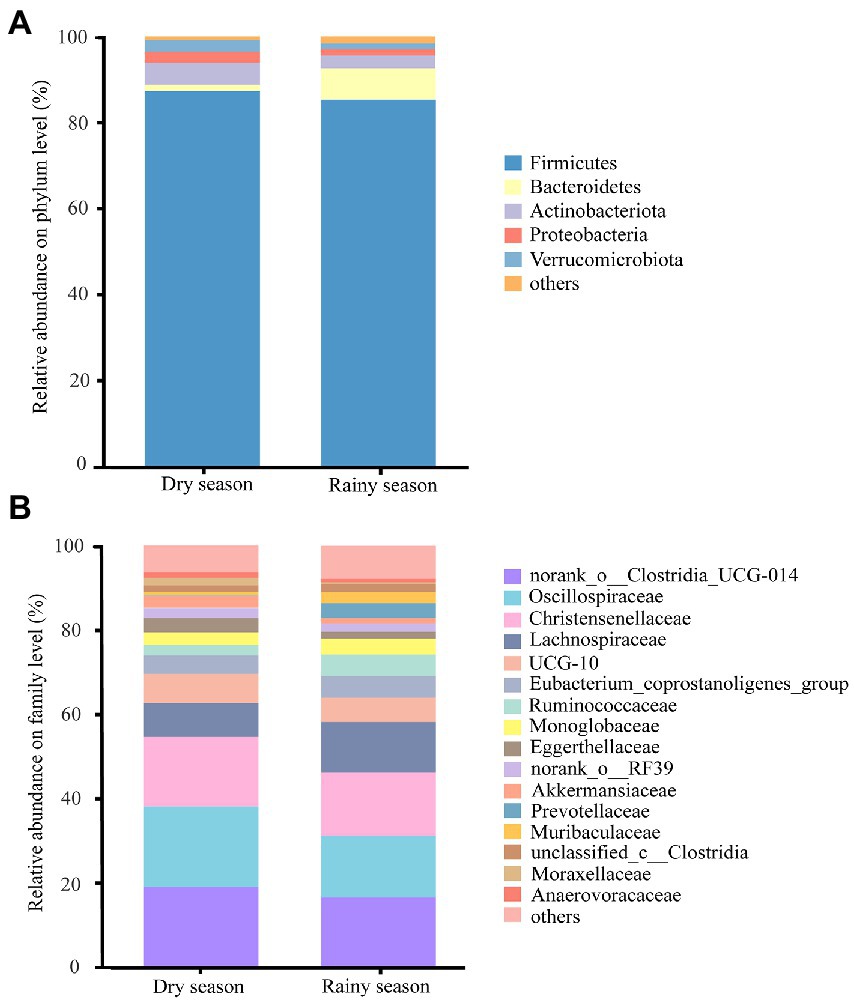
Figure 2. Compositions of the gut microbiota of white-headed black langurs at the phylum level (A) and family level (B). “Others” includes all taxa whose relative abundance is less than 1%.
The Wilcoxon rank-sum test revealed that the relative abundance of Firmicutes, Actinobacteriota, and Proteobacteria at the phylum level was significantly higher in the dry season than in the rainy season. In contrast, the relative abundance of nine phyla, including Bacteroidetes, Spirochaetota, and Cyanobacteria, was significantly higher in the rainy season than in the dry season (Figure 3A; Supplementary Table S4). At the family level, Oscillospiraceae and Eggerthellaceae showed higher abundance in the dry season than in the rainy season. Moreover, the proportion of Lachnospiraceae, Ruminococcaceae, Monoglobaceae, Prevotellaceae, Muribaculaceae, and unclassified_c__Clostridia was higher in the rainy season than in the dry season (Figure 3B; Supplementary Table S5). Furthermore, the results of the RF revealed that among the taxa that differed between the dry and rainy seasons, the greatest contribution was made by Bacteroidetes at the phylum level (AUC = 0.995) and Prevotellaceae at the family level (AUC = 1.000; Figure 4).
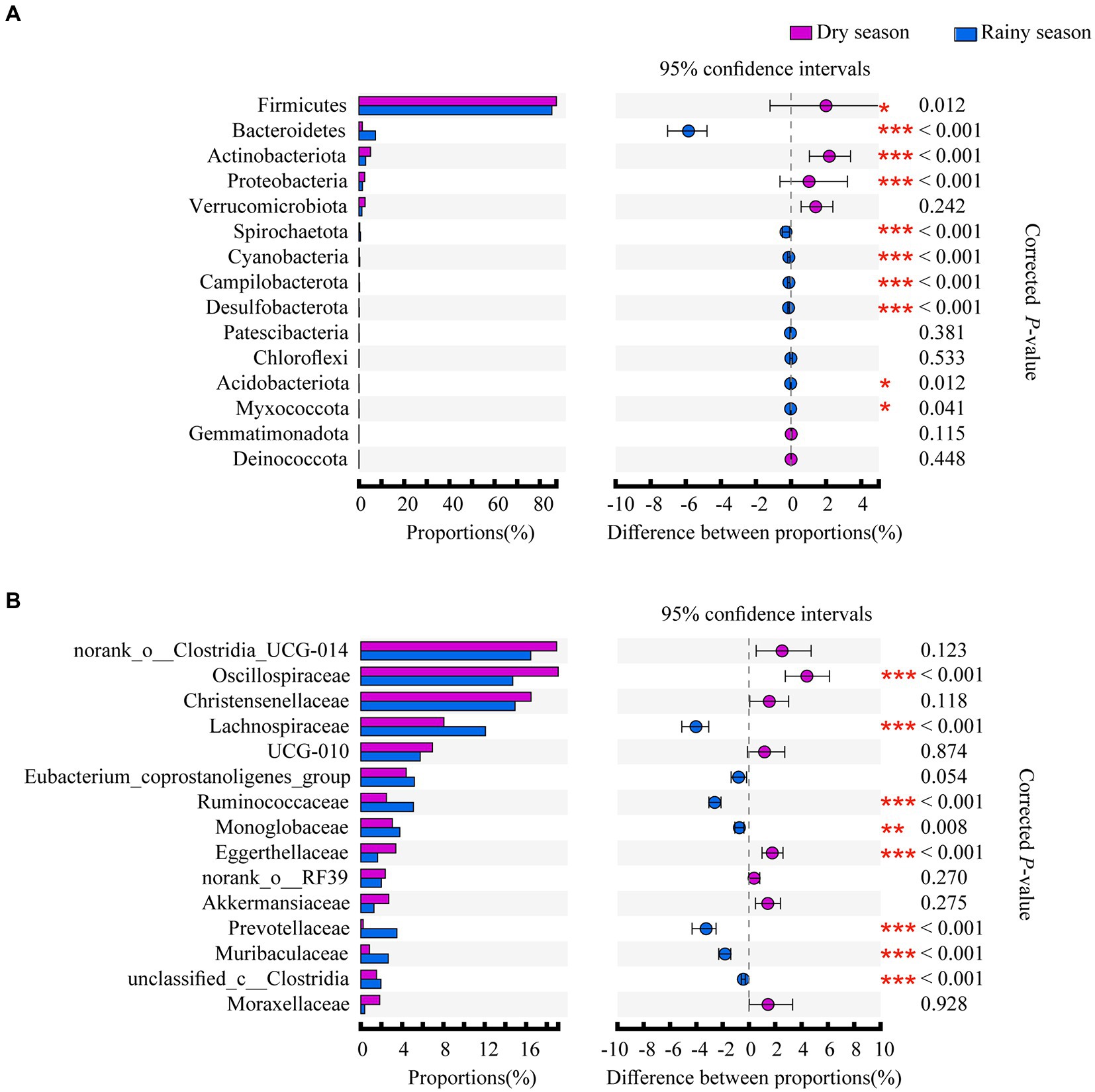
Figure 3. Differences in the composition of the white-headed black langurs gut microbiota at the phylum level (A) and family level (B), with asterisks showing the degree of difference, * for p < 0.05, ** for p < 0.01, and *** for p < 0.001. Only the top 15 taxa are shown in the figure.
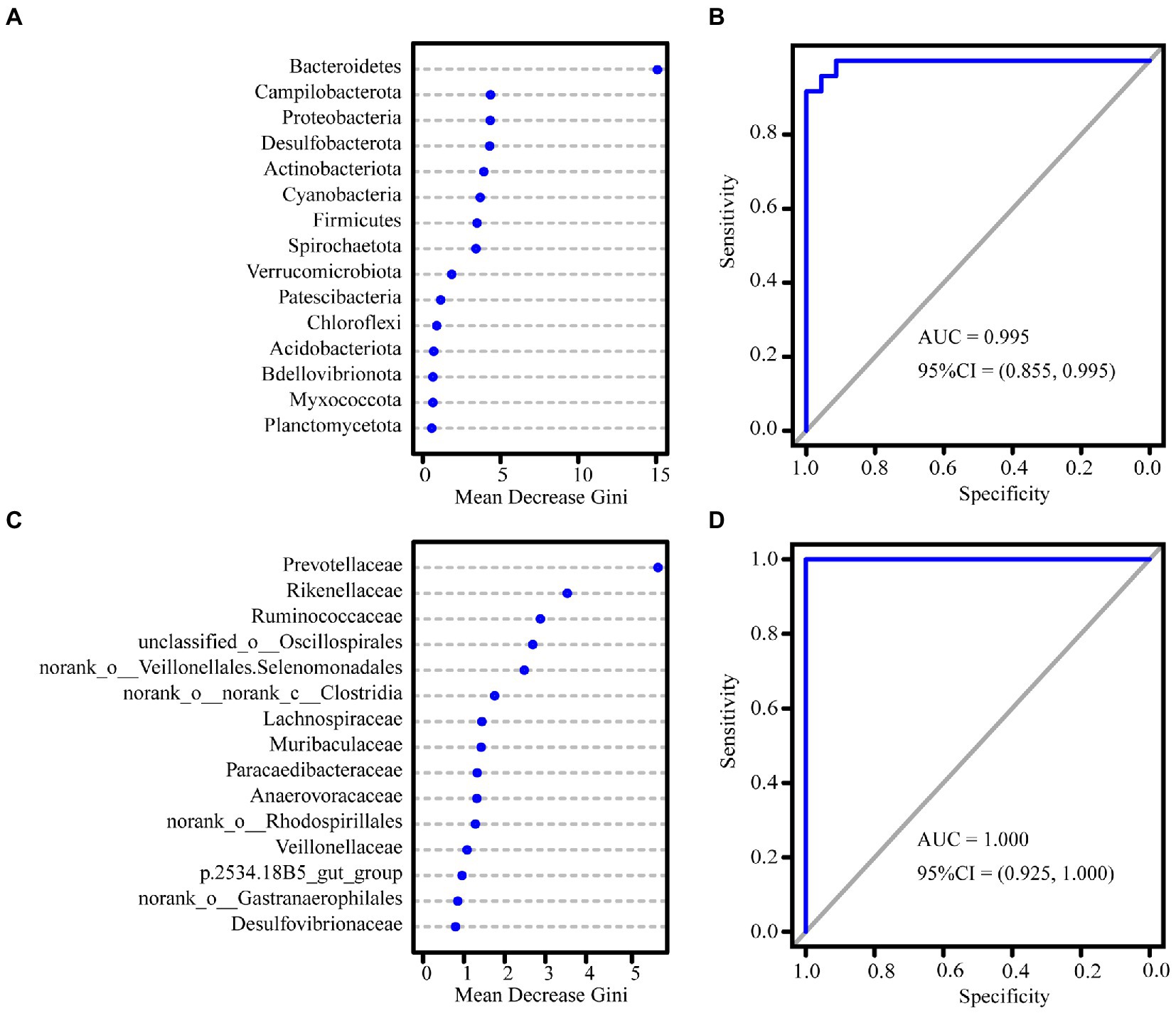
Figure 4. Plots of results for the importance of random forest variables at the phylum level (A), and family level (C), sorted by Mean Decrease Gini values, with higher values indicating greater contribution of the taxon in causing seasonal variations. The ROC curves at the phylum level (B) and at the family level (D).
Alpha diversity analysis based on OTUs showed that the Shannon index (χ2 = 58.384, df = 1, p < 0.001), invSimpson index (χ2 = 80.112, df = 1, p < 0.001), ACE index (χ2 = 6.779, df = 1, p = 0.009), and Chao index (χ2 = 4.724, df = 1, p = 0.030) were noticeably higher in the rainy season than in the dry season (Figure 5; Supplementary Tables S3, S6). Moreover, PCoA indicated significant differences in the community structure of the gut microbiota of white-headed black langurs between the two seasons based on weighted unifrac (R2 = 0.200, p = 0.001) and unweighted unifrac distances (R2 = 0.087, p = 0.001; Figure 6).
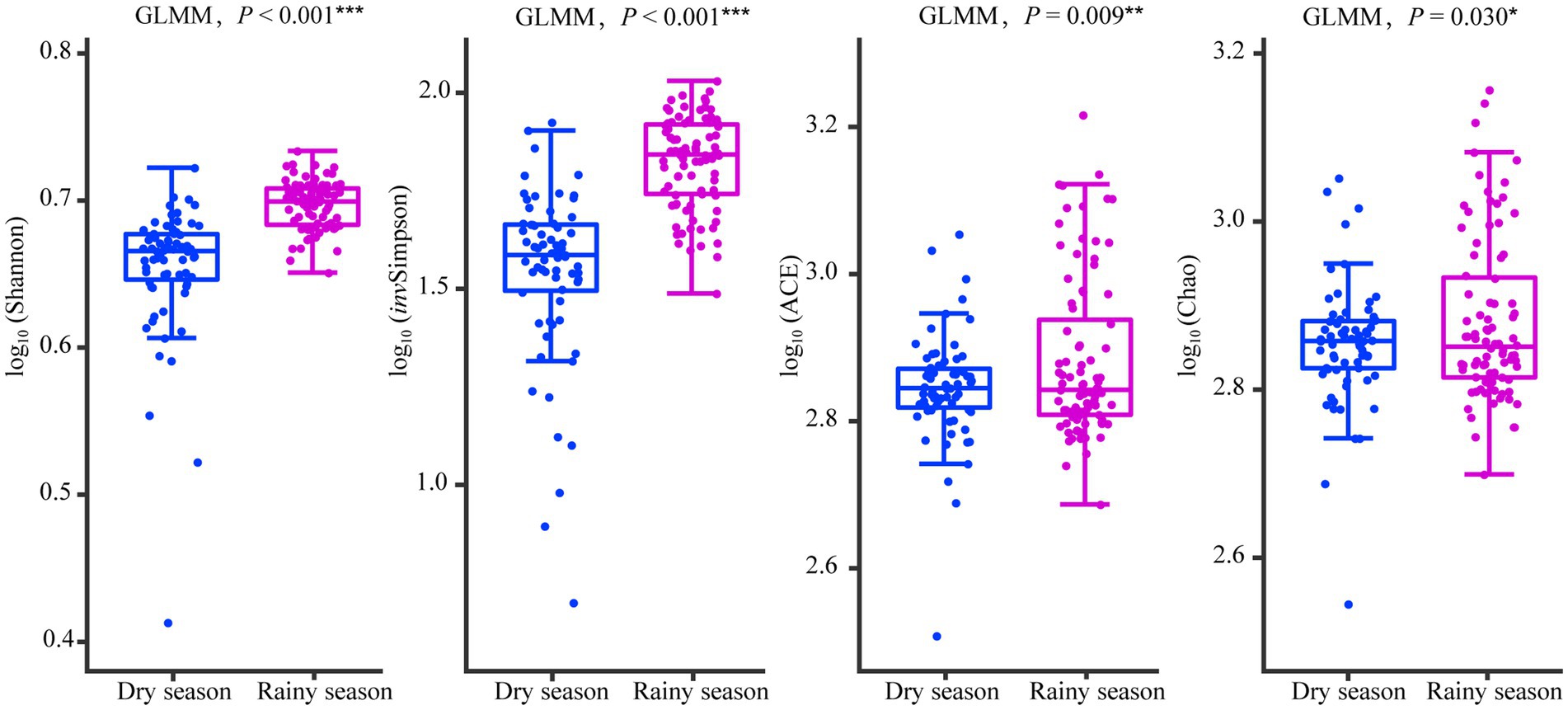
Figure 5. Comparison of alpha diversity in the gut microbiota between the dry season and the rainy season, with asterisks showing the degree of difference, * for p < 0.05, ** for p < 0.01, and *** for p < 0.001.
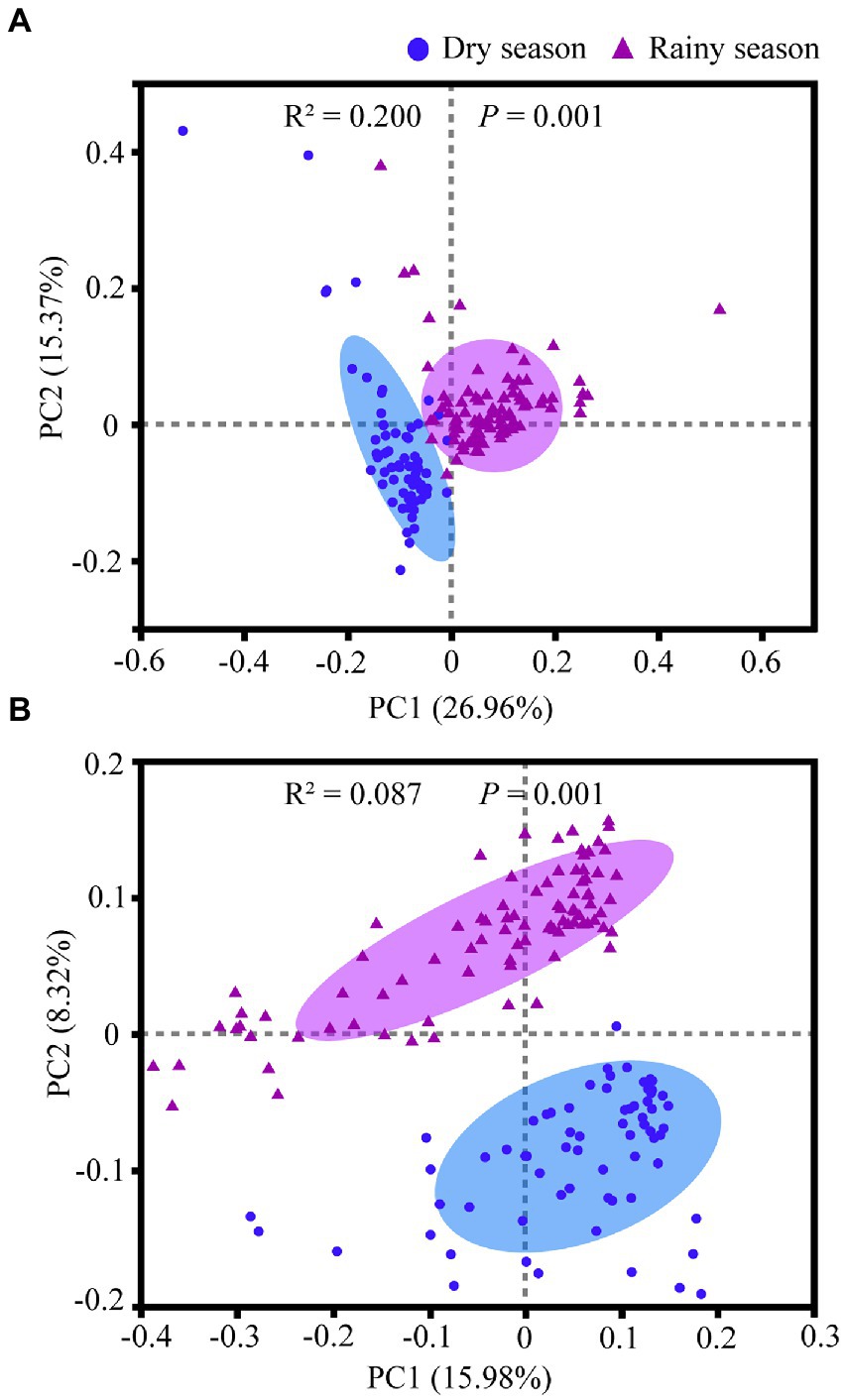
Figure 6. Comparison of beta diversity in the gut microbiota between the dry season and the rainy season based on weighted Unifrac (A) and unweighted Unifrac (B) distances.
Results of the NCM revealed that the explanation of the stochastic process of gut community assembly in white-headed black langurs was 47.5% and the microbial species mobility was 0.125. In this model, 62.7% of the microbiota were within the 95% confidence interval of the theoretical prediction, 25.7% of the microbiota were above the interval, and 11.6% of the microbiota were below the interval (Figure 7A). In addition, differences of gut microbiota assembly between the dry (Figure 7B) and rainy seasons were observed (Figure 7C). This model showed that the microbial species mobility of the gut microbiota assembly in white-headed black langurs was higher in the dry season than in the rainy season (m = 0.188 vs. m = 0.173), and the proportion that could be explained by stochastic processes was lower in the dry season (47.6%) than in the rainy season (49.4%). Moreover, the number of microbial species in the 95% confidence interval was higher in the dry season (75.3%) than in the rainy season (69.3%).
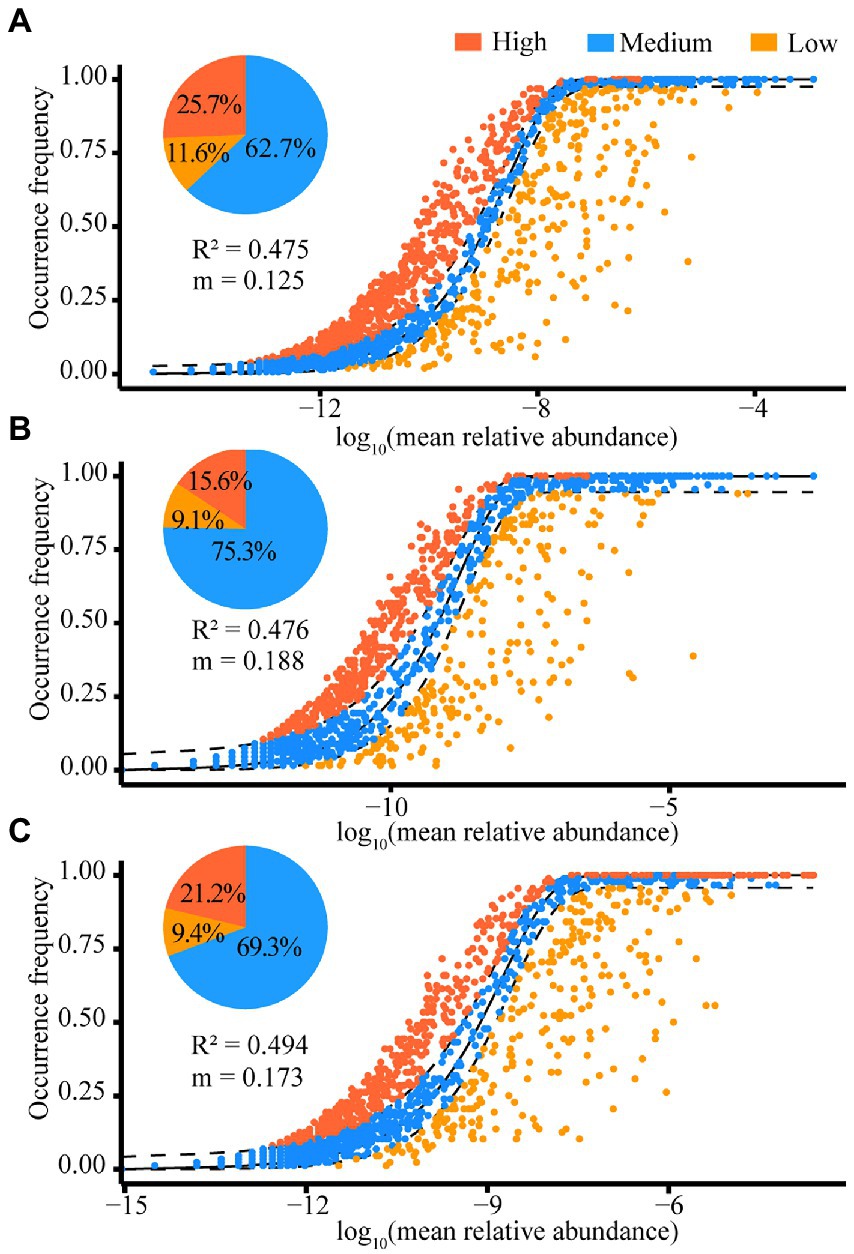
Figure 7. Neutral community model results in all samples (A), the dry season (B) and rainy season (C). The solid black line indicates the best fit and the 95% confidence interval between the two dashed lines, with OTUs above, below, or in the confidence interval marked by different colors. “R2” is the model fit and “m” is the migration rate.
The gut microbiota of white-headed black langurs are characterized by a high proportion of Firmicutes, similar to most colobine primates, including wild golden snub-nosed monkeys and the Udzungwa red colobus. Maintaining high abundance of Firmicutes is commonly considered a digestive strategy that has evolved in active response to highly folivorous diets (Barelli et al., 2015, 2020; Wang et al., 2021). Leaves, especially mature leaves, are rich in lignins and cellulose, both of which are difficult to digest (Que et al., 2022). Firmicutes exhibit genes that encode polysaccharide lyases that degrade dietary fiber into small molecules, such as butyrate, to allow absorption by the host (Sun et al., 2022). For example, there are a large number of genes from glycoside hydrolase families in Ruminococcaceae and Lachnospiraceae, which encode for cellulases and xylanases that degrade dietary fibers into SCFAs to provide energy (Biddle et al., 2013). At the family level, dominant taxa, such as norank_o__Clostridia_UCG-014, Oscillospiraceae, and Christensenellaceae, belong to Firmicutes. This in combination with the expanded rumen is an adaptation response to a large intake of fiber (Sun et al., 2016; Li et al., 2021; Xia et al., 2021).
Bacteroidetes are also a dominant phylum in the gut microbiota of white-headed black langurs and are associated with the digestion of carbohydrates and proteins that are richly available in fruits and young leaves (Wu et al., 2011; Sun et al., 2016; Hicks et al., 2018). A similar pattern of the gut microbiota was identified in sympatric rhesus macaques, in which Prevotella contributed to fructose degradation (Chen et al., 2020). The other dominant phyla occurring in the gut microbiota of the langurs included Actinobacteriota, Verrucomicrobiota, and Proteobacteria. The abundance of Verrucomicrobiota in the gut of the langurs is mainly contributed by Akkermansiaceae, which is associated with the digestion of mucin and might be involved in protecting intestinal mucus layer integrity and maintaining host health (Geerlings et al., 2018). The presence of Akkermansiaceae has also been reported in François’ langurs (Chen et al., 2020). Bifidobacteria, belonging to Actinobacteriota, encode multiple glycosyl hydrolases that can utilize complex carbohydrates from plants (Ventura et al., 2007). Proteobacteria, although reportedly potentially pathogenic, include Succinivibrio, which consumes cellulose and hemicellulose to produce SCFAs (Baniel et al., 2021). These dominant phyla perform major digestive and metabolic functions in the gut, and the taxa with a low proportion maintain the ecological balance of the community and high functional redundancy (Cho and Blaser, 2012; Liu et al., 2022).
The F/B (Firmicutes/Bacteroidetes) ratio has been considered an appropriate proxy for energy balance (Turnbaugh et al., 2006; Springer et al., 2017). A high F/B ratio is efficient at calorie absorption from food (Turnbaugh et al., 2006; Koliada et al., 2017). Compared with previous studies, the F/B ratio of white-headed black langurs (18.07) was higher than that of frugivorous primates [such as red-fronted lemur (Eulemur rufifrons), F/B = 0.98 (Murillo et al., 2022)] and omnivorous primates [such as yellow baboon (Papio cynocephalus), F/B = 2.60 (Barelli et al., 2020) and Tibetan macaques, F/B = 2.65 (Xia et al., 2021)]. The F/B ratio of white-headed black langurs was even higher than that of other folivorous primates [such as ring-tailed lemurs (Lemur catta), F/B = 3.26 (Bennett et al., 2016) and Udzungwa red colobus, F/B = 2.17 (Barelli et al., 2020)]. Higher F/B ratios may be a digestive strategy for white-headed black langurs to adapt to low-quality food resources available in fragmented habitats. Fragmentation has resulted in a reduced foraging range and limited food resources for these karst-dwelling animals (Huang, 2002; Huang et al., 2017). Similar to white-headed black langurs, the sympatric François’ langurs have high F/B ratios, likely enhancing their ability to obtain energy from low-quality foods (Chen et al., 2020). Therefore, high F/B ratios in white-headed black langurs might be an effective strategy to positively respond to low-quality foods, especially in the dry season when mature leaves are intensively consumed (Li et al., 2016; Lu et al., 2021).
Our result indicated that the alpha diversity of the gut microbiota of white-headed black langurs was higher in the rainy season than in the dry season, strongly supporting prediction one. This result could be correlated to higher dietary diversity in the rainy season. A diverse diet provides varied substrates to support the development of specific bacteria, which facilitates high gut microbial diversity (Lin et al., 2021). In the limestone forest of Southwest Guangxi, China, the availability of young leaves and fruits is positively corelated to rainfall (Huang et al., 2015; Tang et al., 2016), which provides the langurs with abundant food resources and high food selectivity in the rainy season (Li and Elizabeth Rogers, 2006; Zhou et al., 2013; Zhang et al., 2021). Several plant species consumed by the langurs, such as Broussonetia papyrifera, show strong seasonality in phenology changes and even lose all their leaves in the dry season (low temperature and rainfall; Huang, 2002; Li and Elizabeth Rogers, 2006). These plants are eaten by the langurs exclusively in the rainy season, resulting in greater dietary and gut microbiota diversities (Zhou et al., 2013). Similar findings have been reported for rhesus macaques (Liu et al., 2022).
Beta diversity analysis indicated significant differences in the gut microbiota of white-headed black langurs across seasons. At the phylum level, the proportion of Firmicutes in the dry season was significantly higher, whereas the proportion of Bacteroidetes was higher in the rainy season; these findings supported prediction two. Enrichment of Firmicutes in the dry season might be associated with the increased utilization of fallback food (mature leaves) by white-headed black langurs (Zhou et al., 2013; Li et al., 2016). The availability of fruits and young leaves is reduced in the dry season and the langurs depend more on mature leaves (Zhou et al., 2013; Li et al., 2016). Firmicutes are powerful in coping with difficult-to-degrade dietary components, compensating for the energy income of animals during the dry season with high-fiber intakes (Zhu et al., 2011; Amato et al., 2015; Sun et al., 2022). Bacteroidetes were found in a higher proportion in the rainy season, probably owing to a rich intake of mature fruits and young leaves (Li et al., 2016; Lu et al., 2021). These food items contain more simple carbohydrates and proteins than mature leaves and can be used effectively by Bacteroidetes, especially Prevotella and Bacteroides (Sun et al., 2016; Hicks et al., 2018; Xia et al., 2021). Research on white-faced capuchin monkeys also indicated a positive correlation between Bacteroidetes and fruit consumption (Orkin et al., 2019). In the present study, the F/B ratio of the langurs was higher in the dry season (59.80) than in the rainy season (11.69), which suggested an adaptation to the large intake of mature leaves in the dry season to cope with the cold weather (Ley et al., 2006; Turnbaugh et al., 2006).
Furthermore, the increase of Proteobacteria in the dry season could be linked to the low temperature. Previous study on mice gut microbiota revealed that exposure to cold conditions significantly increased the number of Proteobacteria, which help the hosts to withstand low temperatures by improving energy accumulation (Chevalier et al., 2015). Limestone forests have lower temperatures in the dry season than in the rainy season (Huang, 2002), and higher proportion of Proteobacteria in white-headed black langurs gut microbiota may be a mechanism to withstand cold. The greater abundance of Actinobacteriota in the dry season was mainly contributed by Eggerthellaceae, which is associated with dietary phenolics metabolism in the human gut (Hervert-Hernández and Goñi, 2011). Dietary phenolics are usually transported to the colon as cellulose carriers to be metabolized by the gut microbiota into bioactive molecules that promote the growth of beneficial bacteria and inhibit the development of harmful bacteria (Jakobek and Matić, 2019). In the current research, the increased intake of fiber in the dry season may result in more dietary polyphenols being transported to the colon, thus providing more metabolic substrates for Eggerthellaceae (Hervert-Hernández and Goñi, 2011; Jakobek and Matić, 2019).
At the family level, Prevotellaceae showed a higher prevalence in the rainy season, which could be linked to the high intake of fruits and young leaves. Prevotellaceae can encode carbohydrate-active enzymes that degrade xylan and pectin (Filippo et al., 2010; Flint et al., 2012). In primate guts, Prevotellaceae is usually considered to respond positively to increased intake of fruits and young leaves (Sun et al., 2016; Springer et al., 2017; Orkin et al., 2019; Liu et al., 2022). Tibetan macaques, for example, have an increased abundance of Prevotellaceae during the transition from the food-lean winter to the fruit-rich spring (Sun et al., 2016). Oscillospiraceae more commonly colonizes the gut of white-headed black langurs in the dry season and might be linked to the digestion of cellulose (Gophna et al., 2017). Some of its members ferment glycans in the host gut to butyrate, which has anti-inflammatory effects, and may be linked to the digestion of leaves (Gophna et al., 2017). It is also noteworthy that the two common taxa of Firmicutes, Ruminococcaceae and Lachnospiraceae, show a significantly higher percentage in the rainy season. This result differs from the findings on most NHPs’ gut microbiota (Amato et al., 2015; Li et al., 2021; Xia et al., 2021). Ruminococcaceae and Lachnospiraceae are commonly associated with high-fiber diets (Zhu et al., 2011; Biddle et al., 2013; Amato et al., 2015). To be specific, the proportion of both taxa in the animals’ gut increases during the leaf-consuming period and decreases during the fruit-consuming period (Amato et al., 2015; Li et al., 2021; Xia et al., 2021). However, the results of this study may be related to the higher gut microbiota diversity in the rainy season. In fact, the diversity of gut microbiota is positively correlated with functional diversity (Lozupone et al., 2012), and the higher percentage of Ruminococcaceae and Lachnospiraceae may be the result of maintaining high functional diversity.
The fit of the NCM indicates that deterministic processes play a predominant role in the assembly of the gut microbiota of white-headed black langurs, which is consistent with prediction three. This result may be linked to fragmented habitats. Changes in environmental factors probably enhance the role of deterministic processes (Burns et al., 2016; Li B. et al., 2022). For example, after a major dietary and living environment adjustment in adult zebrafish, the community assembly shifted from dominance of neutral to deterministic processes (Burns et al., 2016). In this research, the more pronounced effect of deterministic processes could be caused by the limited food resources and low dietary quality in the fragmented habitat. The habitats in which the langurs live suffer from high fragmentation (Huang et al., 2008), characterized by high vegetation diversity but low biomass (Huang, 2002). The limited food resources might increase competition for foods among langur groups (Sun et al., 2016), forcing a greater shift in their diet to mature leaves (Li et al., 2016). The result is a large accumulation of fibers in the gut, which adjusts selection pressure, and causes a specific change in the structure of the microbiota (Amato et al., 2015; Hicks et al., 2018; Li et al., 2021; Xia et al., 2021). Furthermore, leaves have lower nutrients and more secondary metabolites than sugar-rich fruits (Que et al., 2022). Previous studies have demonstrated that reduced nutritional levels intensify competition among bacterial taxa, leading to an intensification of deterministic processes (Ning et al., 2019; Tang et al., 2021). For instance, the gut microbial assembly in the jejunum of Duroc × Bamei pigs shifted to deterministic process dominance due to the addition of dietary fiber in the diet (Tang et al., 2021). Thus, the food with low nutritional level in the habitat may have heightened the importance of deterministic processes in the gut microbiota assembly of white-headed black langurs. According to the NCM, the dry season was affected more by deterministic processes than the rainy season, which might be due to the fact that these langurs heavily depend on mature leaves in the dry season (Zhou et al., 2013; Li et al., 2016; Lu et al., 2021). Such greater addition of dietary fiber alters the selection pressure (David et al., 2014; Amato et al., 2015; Li et al., 2021), emphasizing the contribution of deterministic processes.
In addition, the NCM results showed that migration limitations in the dry season are lower than that in the rainy season, likely owing to close social distance between individuals in the dry season. Generally, social interactions result in similar gut microbiota (Moeller et al., 2016). The white-headed black langurs increase the frequency of sunning and the time spent on resting and grooming behaviors when temperatures are low (Huang, 2002; Zhang et al., 2021). These behaviors bring members closer to each other and reduce the social distance (Zhang et al., 2021), creating opportunities for the spreading of the gut microbiota. In addition, there are fewer food options for the langurs during the dry season when food resources are scarce (Li and Elizabeth Rogers, 2006; Zhou et al., 2013; Lu et al., 2021). The similar food composition among individuals may also facilitate the spread of bacteria. Random taxa in the 95% confidence interval also showed similar seasonal differences. Furthermore, taxa higher than the prediction interval are more likely to colonize the host gut, possibly owing to their easier adaptation to the gut environment (Burns et al., 2016). Besides, the taxa below the prediction interval that were rejected by the host gut may be a protective mechanism, which is associated with resistance to invasion by pathogenic bacteria (Burns et al., 2016).
In summary, changes in the habitat’s plant phenology alters the diet of white-headed black langurs (Huang, 2002; Li and Elizabeth Rogers, 2006; Zhou et al., 2013; Li et al., 2016), further causing seasonal changes in their gut microbiota. Firmicutes with a high percentage of colonization in the dry season could be associated with increased consumption of fiber-rich food such as mature leaves. The abundance of food resources during the rainy season elevated the abundance of the Bacteroidetes as well as gut microbiota diversity. The results demonstrate that dynamic changes in gut microbiota are a strategy for white-headed black langurs to adapt to seasonal changes in food availability, which is amplified by habitat fragmentation. Our results highlight the importance of seasonal variations in the gut microbiota on the adaptation of the white-headed black langurs to the unique environment of limestone forests, providing additional insights into understanding the physiological adaptation strategy used by langurs in response to seasonality in the karst forest.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA904107.
The animal study was reviewed and approved by all fecal samples were collected with permission from the Administration Center of Guangxi Chongzuo White-headed Langur National Nature Reserve.
ZH designed the study. YC analyzed the data and wrote the manuscript. YiL analyzed the data. JZ, ZL, DN, and JL collected samples. YoL and ZH revised the manuscript. All authors read and approved the submitted manuscript.
This work was supported by the National Natural Science Foundation of China (32170488, 31960106, and 31960104).
This study was supported by the substantial helps given by Zhizhang Liang from the Guangxi Chongzuo White-headed Langur National Nature Reserve. We thank Gang Hu from Nanning Normal University, Chuangbin Tang and Shoufu Feng from Guangxi Normal University for Nationalities for their helps in fecal samples collecting and conserving.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1126243/full#supplementary-material
Alberdi, A., Aizpurua, O., Bohmann, K., Zepeda-Mendoza, M. L., and Gilbert, M. T. P. (2016). Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699. doi: 10.1016/j.tree.2016.06.008
Amato, K. R., Leigh, S. R., Kent, A., Mackie, R. I., Yeoman, C. J., Stumpf, R. M., et al. (2015). The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb. Ecol. 69, 434–443. doi: 10.1007/s00248-014-0554-7
Amato, K. R., Metcalf, J. L., Song, S. J., Hale, V. L., Clayton, J., Ackermann, G., et al. (2016). Using the gut microbiota as a novel tool for examining colobine primate GI health. Glob. Ecol. Conserv. 7, 225–237. doi: 10.1016/j.gecco.2016.06.004
Amato, K. R., Sanders, J. G., Song, S. J., Nute, M., Metcalf, J. L., Thompson, L. R., et al. (2019). Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 13, 576–587. doi: 10.1038/s41396-018-0175-0
Amato, K. R., Yeoman, C. J., Kent, A., Righini, N., Carbonero, F., Estrada, A., et al. (2013). Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344–1353. doi: 10.1038/ismej.2013.16
Baniel, A., Amato, K. R., Beehner, J. C., Bergman, T. J., Mercer, A., Perlman, R. F., et al. (2021). Seasonal shifts in the gut microbiome indicate plastic responses to diet in wild geladas. Microbiome 9:26. doi: 10.1186/s40168-020-00977-9
Barelli, C., Albanese, D., Donati, C., Pindo, M., Dallago, C., Rovero, F., et al. (2015). Habitat fragmentation is associated to gut microbiota diversity of an endangered primate: implications for conservation. Sci. Rep. 5:14862. doi: 10.1038/srep14862
Barelli, C., Albanese, D., Stumpf, R. M., Asangba, A., Donati, C., Rovero, F., et al. (2020). The gut microbiota communities of wild arboreal and ground-feeding tropical primates are affected differently by habitat disturbance. mSystems 5:e00061–20. doi: 10.1128/mSystems.00061-20
Barton, W., Penney, N. C., Cronin, O., Garcia-Perez, I., Molloy, M. G., Holmes, E., et al. (2018). The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 67, 625–633. doi: 10.1136/gutjnl-2016-313627
Bennett, G., Malone, M., Sauther, M. L., Cuozzo, F. P., White, B., Nelson, K. E., et al. (2016). Host age, social group, and habitat type influence the gut microbiota of wild ring-tailed lemurs (Lemur catta). Am. J. Primatol. 78, 883–892. doi: 10.1002/ajp.22555
Bertram, M. G., Martin, J. M., McCallum, E. S., Alton, L. A., Brand, J. A., Brooks, B. W., et al. (2022). Frontiers in quantifying wildlife behavioural responses to chemical pollution. Biol. Rev. 97, 1346–1364. doi: 10.1111/brv.12844
Biddle, A., Stewart, L., Blanchard, J., and Leschine, S. (2013). Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5, 627–640. doi: 10.3390/d5030627
Bleisch, B., and Long, Y. (2020). Trachypithecus leucocephalus, the IUCN Red List of Threatened Species 2020. https://dx.doi.org/10.2305/IUCN.UK.2020-2.RLTS.T39872A17988378.en [Accessed October 12,2022].
Burns, A. R., Stephens, W. Z., Stagaman, K., Wong, S., Rawls, J. F., Guillemin, K., et al. (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10, 655–664. doi: 10.1038/ismej.2015.142
Chave, J. (2004). Neutral theory and community ecology. Ecol. Lett. 7, 241–253. doi: 10.1111/j.1461-0248.2003.00566.x
Chen, T., Li, Y. H., Liang, J. P., Li, Y. B., and Huang, Z. H. (2020). Variations in the gut microbiota of sympatric François’ langurs and rhesus macaques living in limestone forests in Southwest Guangxi, China. Glob. Ecol. Conserv. 22:e00929. doi: 10.1016/j.gecco.2020.e00929
Chevalier, C., Stojanović, O., Colin, D. J., Suarez-Zamorano, N., Tarallo, V., Veyrat-Durebex, C., et al. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cells 163, 1360–1374. doi: 10.1016/j.cell.2015.11.004
Cho, I., and Blaser, M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. doi: 10.1038/nrg3182
Clarke, S. F., Murphy, E. F., O’Sullivan, O., Lucey, A. J., Humphreys, M., Hogan, A., et al. (2014). Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63, 1913–1920. doi: 10.1136/gutjnl-2013-306541
Clayton, J. B., Vangay, P., Huang, H., Ward, T., Hillmann, B. M., Al-Ghalith, G. A., et al. (2016). Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. U. S. A. 113, 10376–10381. doi: 10.1073/pnas.1521835113
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fargione, J., Brown, C. S., and Tilman, D. (2003). Community assembly and invasion: an experimental test of neutral versus niche processes. Proc. Natl. Acad. Sci. U. S. A. 100, 8916–8920. doi: 10.1073/pnas.1033107100
Filippo, C. D., Cavalieri, D., Paola, M. D., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A. 107, 14691–14696. doi: 10.1073/pnas.1005963107
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P., and Forano, E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897
Frankel, J. S., Mallott, E. K., Hopper, L. M., Ross, S. R., and Amato, K. R. (2019). The effect of captivity on the primate gut microbiome varies with host dietary niche. Am. J. Primatol. 81:e23061. doi: 10.1002/ajp.23061
Froidurot, A., and Julliand, V. (2022). Cellulolytic bacteria in the large intestine of mammals. Gut Microbes 14:2031694. doi: 10.1080/19490976.2022.2031694
Geerlings, S. Y., Kostopoulos, I., De Vos, W. M., and Belzer, C. (2018). Akkermansia muciniphila in the human gastrointestinal tract: when, where, and how? Microorganisms 6:75. doi: 10.3390/microorganisms6030075
Gophna, U., Konikoff, T., and Nielsen, H. B. (2017). Oscillospira and related bacteria – from metagenomic species to metabolic features. Environ. Microbiol. 19, 835–841. doi: 10.1111/1462-2920.13658
Guangxi Forestry Department. (1993). Nature Reserves in Guangxi. Beijing: China Forestry Publishing House.
Hervert-Hernández, D., and Goñi, I. (2011). Dietary polyphenols and human gut microbiota: a review. Food Rev. Int. 27, 154–169. doi: 10.1080/87559129.2010.535233
Heys, C., Cheaib, B., Busetti, A., Kazlauskaite, R., Maier, L., Sloan, W. T., et al. (2020). Neutral processes dominate microbial community assembly in Atlantic Salmon, Salmo salar. Appl. Environ. Microbiol. 86:e02283–19. doi: 10.1128/AEM.02283-19
Hicks, A. L., Lee, K. J., Couto-Rodriguez, M., Patel, J., Sinha, R., Guo, C., et al. (2018). Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 9:1786. doi: 10.1038/s41467-018-04204-w
Huang, Z. H., Huang, C. M., Tang, C. B., Huang, L. B., Tang, H. X., Ma, G. Z., et al. (2015). Dietary adaptations of assamese macaques (Macaca assamensis) in limestone forests in Southwest China. Am. J. Primatol. 77, 171–185. doi: 10.1002/ajp.22320
Huang, C. M., and Li, Y. B. (2005). How does the white-headed langur (Trachypithecus leucocephalus) adapt locomotor behavior to its unique limestone hill habitat? Primates 46, 261–267. doi: 10.1007/s10329-005-0130-3
Huang, C. M., Li, Y. B., Zhou, Q. H., Feng, Y. X., Chen, Z., Yu, H., et al. (2008). Karst habitat fragmentation and the conservation of the white-headed langur (Trachypithecus leucocephalus) in China. Primate Conserv. 23, 133–139. doi: 10.1896/052.023.0116
Huang, Z. H., Yuan, P. S., Huang, H. L., Tang, X. P., Xu, W. J., Huang, C. M., et al. (2017). Effect of habitat fragmentation on ranging behavior of white-headed langurs in limestone forests in Southwest China. Primates 58, 423–434. doi: 10.1007/s10329-017-0600-4
Jakobek, L., and Matić, P. (2019). Non-covalent dietary fiber - polyphenol interactions and their influence on polyphenol bioaccessibility. Trends Food Sci. Technol. 83, 235–247. doi: 10.1016/j.tifs.2018.11.024
Jin, L., Huang, Y., Yang, S. Z., Wu, D. F., Li, C. W., Deng, W. W., et al. (2021). Diet, habitat environment and lifestyle conversion affect the gut microbiomes of giant pandas. Sci. Total Environ. 770:145316. doi: 10.1016/j.scitotenv.2021.145316
Koliada, A., Syzenko, G., Moseiko, V., Budovska, L., Puchkov, K., Perederiy, V., et al. (2017). Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 17:120. doi: 10.1186/s12866-017-1027-1
Lai, Y., Chen, Y. Q., Zheng, J. J., Liu, Z., Nong, D. P., Liang, J. P., et al. (2023). Gut microbiota of white-headed black langurs (Trachypithecus leucocephalus) in responses to habitat fragmentation. Front. Microbiol. 14:1126257. doi: 10.3389/fmicb.2023.1126257
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, Y. H., Chen, T., Liang, J. P., Li, Y. B., and Huang, Z. H. (2021). Seasonal variation in the gut microbiota of rhesus macaques inhabiting limestone forests of Southwest Guangxi, China. Arch. Microbiol. 203, 787–798. doi: 10.1007/s00203-020-02069-6
Li, Z. Y., and Elizabeth Rogers, M. (2006). Food items consumed by white-headed langurs in Fusui, China. Int. J. Primatol. 27, 1551–1567. doi: 10.1007/s10764-006-9090-8
Li, B., Gao, H. M., Song, P. F., Liang, C. B., Jiang, F., Xu, B., et al. (2022). Captivity shifts gut microbiota mommunities in white-lipped deer (Cervus albirostris). Animals 12:431. doi: 10.3390/ani12040431
Li, S. L., Tang, R., Yi, H., Cao, Z. C., Sun, S. L., Liu, T.-X., et al. (2022). Neutral processes provide an insight into the structure and function of gut microbiota in the cotton bollworm. Front. Microbiol. 13:849637. doi: 10.3389/fmicb.2022.849637
Li, D. Y., Yuan, P. S., Krzton, A., Huang, C. M., and Zhou, Q. H. (2016). Dietary adaptation of white-headed langurs in a fragmented limestone habitat. Mammalia 80, 153–162. doi: 10.1515/mammalia-2014-0152
Lin, R., Huang, X. R., Gao, Y. F., Hu, Q. T., and Chen, W. R. (2021). Dietary variety relates to gut microbiota diversity and abundance. Curr. Dev. Nutr. 5:1166. doi: 10.1093/cdn/nzab054_021
Liu, H. Y., Chen, T., Li, Y. H., Zheng, J. J., Liu, Z., Li, Y. B., et al. (2022). Seasonal variations in gut microbiota of semiprovisioned rhesus macaques (Macaca mulatta) living in a limestone forest of Guangxi, China. Front. Microbiol. 13:951507. doi: 10.3389/fmicb.2022.951507
Losasso, C., Eckert, E. M., Mastrorilli, E., Villiger, J., Mancin, M., Patuzzi, I., et al. (2018). Assessing the influence of vegan, vegetarian and omnivore oriented westernized dietary styles on human gut microbiota: a cross sectional study. Front. Microbiol. 9:317. doi: 10.3389/fmicb.2018.00317
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Lu, S. Y., Chen, T., Huang, Z. H., Li, Y. B., and Lu, C. H. (2021). Interannual variation in food choice of white-headed langur inhabiting limestone forests in Fusui, Southwest Guangxi, China. Ecol. Evol. 11, 9349–9360. doi: 10.1002/ece3.7726
Machado, M. G., Sencio, V., and Trottein, F. (2021). Short-chain fatty acids as a potential treatment for infections: a closer look at the lungs. Infect. Immun. 89:e0018821. doi: 10.1128/IAI.00188-21
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Matsuda, I., Ihobe, H., Tashiro, Y., Yumoto, T., Baranga, D., and Hashimoto, C. (2020). The diet and feeding behavior of the black-and-white colobus (Colobus guereza) in the Kalinzu Forest, Uganda. Primates 61, 473–484. doi: 10.1007/s10329-020-00794-6
Moeller, A. H., Foerster, S., Wilson, M. L., Pusey, A. E., Hahn, B. H., and Ochman, H. (2016). Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2:e1500997. doi: 10.1126/sciadv.1500997
Mori, H., Maruyama, F., Kato, H., Toyoda, A., Dozono, A., Ohtsubo, Y., et al. (2013). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 21, 217–227. doi: 10.1093/dnares/dst052
Murillo, T., Schneider, D., Fichtel, C., and Daniel, R. (2022). Dietary shifts and social interactions drive temporal fluctuations of the gut microbiome from wild redfronted lemurs. ISME Commun. 2:3. doi: 10.1038/s43705-021-00086-0
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Ning, D. L., Deng, Y., Tiedje, J. M., and Zhou, J. Z. (2019). A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. U. S. A. 116, 16892–16898. doi: 10.1073/pnas.1904623116
Orkin, J. D., Campos, F. A., Myers, M. S., Hernandez, S. E. C., Guadamuz, A., and Melin, A. D. (2019). Seasonality of the gut microbiota of free-ranging white-faced capuchins in a tropical dry forest. ISME J. 13, 183–196. doi: 10.1038/s41396-018-0256-0
Que, T. C., Pang, X. W., Huang, H. L., Chen, P. Y., Wei, Y. F., Hua, Y. M., et al. (2022). Comparative gut microbiome in Trachypithecus leucocephalus and other primates in Guangxi, China, based on metagenome sequencing. Front. Cell. Infect. Microbiol. 12:872841. doi: 10.3389/fcimb.2022.872841
Robin, X., Turck, N., Hainard, A., Tiberti, N., Lisacek, F., Sanchez, J. C., et al. (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. doi: 10.1186/1471-2105-12-77
Rothschild, D., Weissbrod, O., Barkan, E., Kurilshikov, A., Korem, T., Zeevi, D., et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. doi: 10.1038/nature25973
Sawada, A., Hayakawa, T., Kurihara, Y., Lee, W., and Hanya, G. (2022). Seasonal responses and host uniqueness of gut microbiome of Japanese macaques in lowland Yakushima. Anim. Microbiome 4:54. doi: 10.1186/s42523-022-00205-9
Schoeler, M., and Caesar, R. (2019). Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Dis. 20, 461–472. doi: 10.1007/s11154-019-09512-0
Sloan, W. T., Lunn, M., Woodcock, S., Head, I. M., Nee, S., and Curtis, T. P. (2006). Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 8, 732–740. doi: 10.1111/j.1462-2920.2005.00956.x
Springer, A., Fichtel, C., Al-Ghalith, G. A., Koch, F., Amato, K. R., Clayton, J. B., et al. (2017). Patterns of seasonality and group membership characterize the gut microbiota in a longitudinal study of wild Verreaux’s sifakas (Propithecus verreauxi). Ecol. Evol. 7, 5732–5745. doi: 10.1002/ece3.3148
Sun, B. H., Wang, X., Bernstein, S., Huffman, M. A., Xia, D. P., Gu, Z. Y., et al. (2016). Marked variation between winter and spring gut microbiota in free-ranging Tibetan macaques (Macaca thibetana). Sci. Rep. 6:26035. doi: 10.1038/srep26035
Sun, Y. G., Zhang, S. S., Nie, Q. X., He, H. J., Tan, H. Z., Geng, F., et al. (2022). Gut firmicutes: relationship with dietary fiber and role in host homeostasis. Crit. Rev. Food Sci. Nutr. 12, 1–16. doi: 10.1080/10408398.2022.2098249
Tang, C. B., Huang, L. B., Huang, Z. H., Krzton, A., Lu, C. H., and Zhou, Q. H. (2016). Forest seasonality shapes diet of limestone-living rhesus macaques at Nonggang, China. Primates 57, 83–92. doi: 10.1007/s10329-015-0498-7
Tang, X. J., Zhang, L. Z., Fan, C., Wang, L., Fu, H. B., Ren, S. E., et al. (2021). Dietary fiber influences bacterial community assembly processes in the gut microbiota of Durco × Bamei crossbred pig. Front. Microbiol. 12:688554. doi: 10.3389/fmicb.2021.688554
Tecot, S. R., Irwin, M. T., and Raharison, J.-L. (2019). Faecal glucocorticoid metabolite profiles in diademed sifakas increase during seasonal fruit scarcity with interactive effects of age/sex class and habitat degradation. Cons. Physiol. 7:coz001. doi: 10.1093/conphys/coz001
Trevelline, B. K., Fontaine, S. S., Hartup, B. K., and Kohl, K. D. (2019). Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B 286:20182448. doi: 10.1098/rspb.2018.2448
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G. F., Chater, K. F., et al. (2007). Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi: 10.1128/MMBR.00005-07
Wang, X. C., Wang, Z. M., Pan, H. J., Qi, J. W., Li, D. Y., Zhang, L. Y., et al. (2021). Captivity influences the gut microbiome of Rhinopithecus roxellana. Front. Microbiol. 12:763022. doi: 10.3389/fmicb.2021.763022
Warton, D. I., and Hui, F. K. C. (2011). The arcsine is asinine: the analysis of proportions in ecology. Ecology 92, 3–10. doi: 10.1890/10-0340.1
Wong, B. B. M., and Candolin, U. (2015). Behavioral responses to changing environments. Behav. Ecol. 26, 665–673. doi: 10.1093/beheco/aru183
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Xia, T. R., Yao, Y. F., Wang, C., Dong, M. M., Wu, Y. H., Li, D. Y., et al. (2021). Seasonal dynamics of gut microbiota in a cohort of wild Tibetan macaques (Macaca thibetana) in western China. Glob. Ecol. Conserv. 25:e01409. doi: 10.1016/j.gecco.2020.e01409
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Zhang, K. C., Zhou, Q. H., Xu, H. L., and Huang, Z. H. (2021). Diet, food availability, and climatic factors drive ranging behavior in white-headed langurs in the limestone forests of Guangxi, Southwest China. Zool. Res. 42, 406–411. doi: 10.24272/j.issn.2095-8137.2020.292
Zheng, J. J., Zhang, K. C., Liang, J. P., Li, Y. B., and Huang, Z. H. (2021). Food availability, temperature, and day length drive seasonal variations in the positional behavior of white-headed langurs in the limestone forests of Southwest Guangxi, China. Ecol. Evol. 11, 14857–14872. doi: 10.1002/ece3.8171
Zhou, Q. H., Huang, Z. H., Wei, H., and Huang, C. M. (2018). Variations in diet composition of sympatric Trachypithecus francoisi and Macaca assamensis in the limestone habitats of Nonggang, China. Zool. Res. 39, 284–290. doi: 10.24272/j.issn.2095-8137.2018.046
Zhou, J. Z., and Ning, D. L. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Bio. Rev. 81:e00002–17. doi: 10.1128/MMBR.00002-17
Zhou, Q. H., Tang, Z., Li, Y. B., and Huang, C. M. (2013). Food diversity and choice of white-headed langur in fragmented limestone hill habitat in Guangxi, China. Acta Ecol. Sin. 33, 109–113. doi: 10.1016/j.chnaes.2013.01.007
Keywords: seasonal variations, gut microbiota, white-headed black langur, limestone forest, Trachypithecus leucocephalus
Citation: Chen Y, Lai Y, Zheng J, Liu Z, Nong D, Liang J, Li Y and Huang Z (2023) Seasonal variations in the gut microbiota of white-headed black langur (Trachypithecus leucocephalus) in a limestone forest in Southwest Guangxi, China. Front. Ecol. Evol. 11:1126243. doi: 10.3389/fevo.2023.1126243
Received: 17 December 2022; Accepted: 31 January 2023;
Published: 20 February 2023.
Edited by:
Timothy J. Colston, University of Puerto Rico at Mayagüez, Puerto RicoReviewed by:
Sierra N. Smith, University of Oklahoma, United StatesCopyright © 2023 Chen, Lai, Zheng, Liu, Nong, Liang, Li and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghao Huang, ✉ aHpoNzczQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.