
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 16 March 2023
Sec. Behavioral and Evolutionary Ecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1118748
Understanding the evolution of eusociality in insects has been a long-standing and unsolved challenge in evolutionary biology. For decades, it has been suggested that haplodiploidy plays an important role in the origin of eusociality. However, some researchers have also suggested that eusociality is unrelated to haplodiploidy. Surprisingly, there have been no large-scale phylogenetic tests of this hypothesis (to our knowledge). Here, we test whether haplodiploidy might help explain the origins of eusociality across 874 hexapod families, using three different phylogenetic comparative methods. Two of the methods used support the idea that the evolution of eusociality is significantly associated with haplodiploidy, providing possibly the first phylogenetic support for this decades-old hypothesis across insects. However, some patterns were clearly discordant with this hypothesis, and one phylogenetic test was non-significant. Support for this hypothesis came largely from the repeated origins of eusociality within the haplodiploid hymenopterans (and within thrips). Experimental manipulations of the data show that the non-significant results are primarily explained by the origins of eusociality without haplodiploidy in some groups (i.e., aphids, termites). Overall, our results offer mixed phylogenetic support for the long-standing hypothesis that haplodiploidy helps drive the evolution of eusociality.
Eusociality is often considered the most “advanced” (or complex) form of sociality and is characterized by reproductive division of labor among individuals in a colony, overlap of generations (age polyethism), and cooperative brood care among individuals (Michener, 1969; Andersson, 1984; Crespi and Yanega, 1995). Eusociality occurs in many animals, including bees, wasps, ants, termites, thrips, aphids, and mole rats (Wilson, 1971). However, most species of eusocial animals belong to the insect order Hymenoptera (Andersson, 1984; Kocher and Paxton, 2014).
Why eusociality evolves, and why it evolves more often in some groups than in others, has been an active area of research in evolutionary biology. One of the most well-known explanations is the haplodiploidy hypothesis, which is frequently attributed to Hamilton (1964). Haplodiploidy is a sex-determination system in which haploid males develop from unfertilized eggs and diploid females develop from fertilized eggs (review in Blackmon et al., 2017). In his seminal paper, Hamilton (1964) proposed that the evolution of sociality was especially frequent within the order Hymenoptera because the presence of haplodiploidy in that group results in unusually high relatedness among sisters. This idea was later refined into the explicit haplodiploidy hypothesis: that haplodiploidy was instrumental for the evolution of sociality within Hymenoptera (e.g., West-Eberhard, 1975, 1978; Andersson, 1984; Gadagkar, 1985, 1991). Under the haplodiploidy hypothesis, sisters in haplodiploid species can be more closely related to each other genetically than they are to their own daughters (but depending on the mating system and other factors, see below). This could help explain why workers in many eusocial species sacrifice their own reproduction and take care of the queen’s offspring instead, a key feature of eusociality (Hamilton, 1964).
The haplodiploidy hypothesis has been extensively debated in the literature. Much of the support for the haplodiploidy hypothesis comes from theoretical studies, some of which are summarized in Table 1. On the other hand, there are also several objections to the haplodiploidy hypothesis (Table 1). Haplodiploidy potentially increases the relatedness among sisters (r = 0.75) but decreases relatedness among brothers (r = 0.25). Therefore, given random mating and equal sex ratios, the average relatedness coefficient between a female’s offspring and her siblings is expected to be the same (r = 0.5). Thus, haplodiploidy might not promote evolution of altruistic behavior based on the high relatedness among sisters (Trivers and Hare, 1976). Nevertheless, haplodiploidy could potentially promote evolution of altruism (and eusociality) through mechanisms that allow workers to gain control over sex allocation within the colony (Trivers and Hare, 1976; Gardner et al., 2012). Another related argument is that haplodiploidy may not generate the relatedness among individuals in the colony needed for eusociality to evolve, given the diverse mating systems and mating frequencies observed among eusocial species (Gadagkar, 1991). It has also been argued that the presence of maternal care within Hymenoptera (including nest building, brood feeding, and protection against predators) is more important than haplodiploidy for the evolution of eusociality (Queller and Strassmann, 1998). Another alternative hypothesis is that the relatedness of haplodiploid individuals is a consequence of eusociality rather than a cause of its evolution, rendering the haplodiploidy hypothesis unimportant for the evolution of eusociality (Wilson and Hölldobler, 2005). Some of the studies listed in Table 1 have suggested that haplodiploidy may have contributed to the evolution of eusociality, but not solely through relatedness as Hamilton (1964) proposed. Instead, haplodiploidy may have contributed to the origin of eusociality through other mechanisms, such as through monogamy and haplodiploidy synergistically promoting altruistic genotypes in a colony (Fromhage and Kokko, 2011), through female philopatry in haplodiploid lineages combined with male-based dispersal promoting altruistic behavior (Johnstone et al., 2012), and the ease of sex-ratio adjustment due to female-biased helping (Gardner and Ross, 2013). Other potentially relevant factors linking haplodiploidy and eusociality include lifetime monogamy, sex ratio bias, and overwintering (Quiñones and Pen, 2017), and population growth rate, effective population sex ratio, and a lowered cost of altruism (Rautiala et al., 2019). We refer readers to the original studies for more detailed explanations.
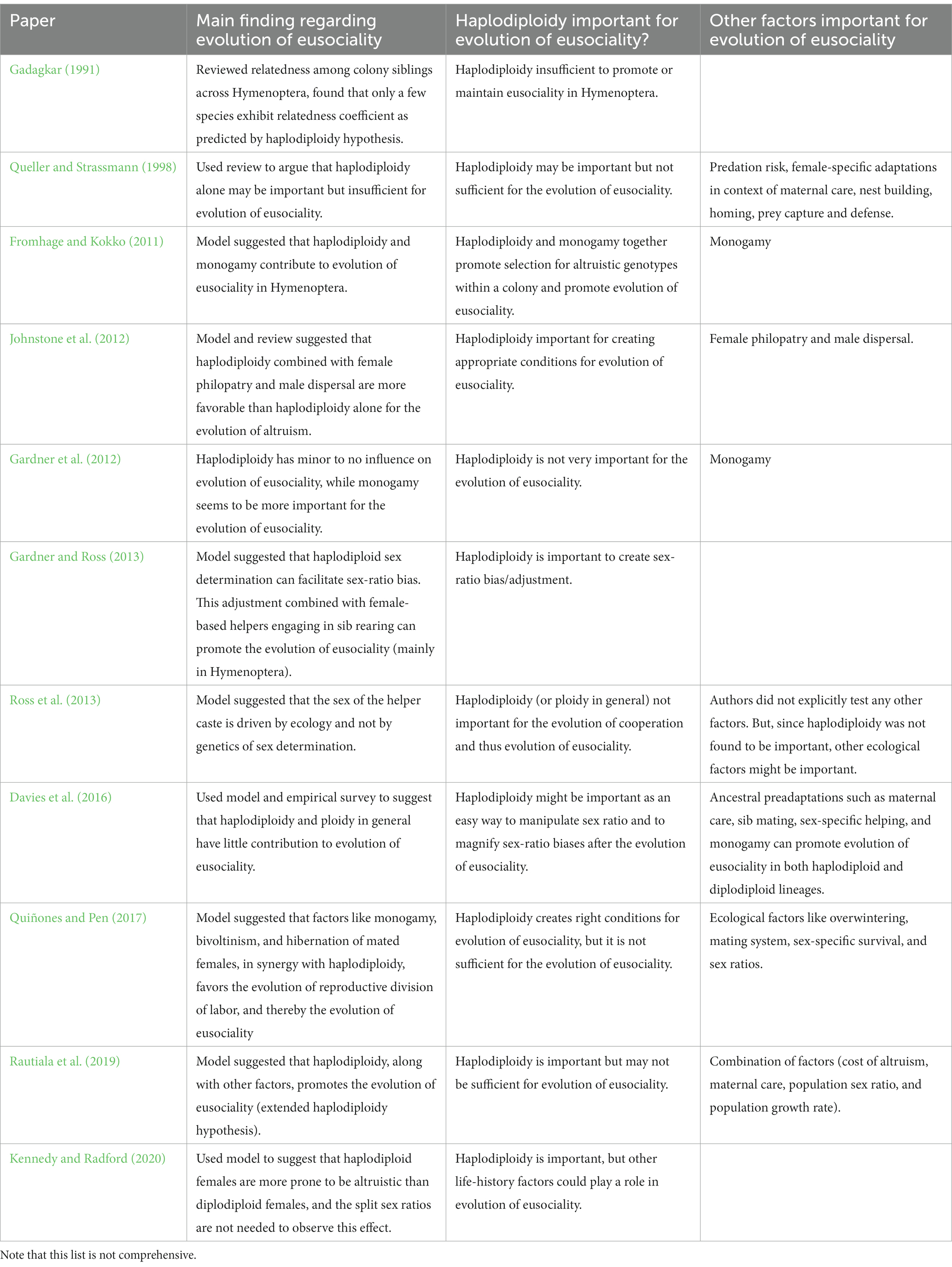
Table 1. Summary of recent reviews and theoretical studies relating haplodiploidy and the evolution of eusociality, listed chronologically.
Despite over 50 years of discussion about the possible role of haplodiploidy in the evolution of eusociality, no studies have offered a statistical phylogenetic test of whether haplodiploidy and eusociality are significantly associated (to our knowledge). We are aware of one formal (but non-phylogenetic) statistical analysis. Crozier (2008) counted the number of occurrences of eusociality among haplodiploid and diplodiploid insect families and concluded that the distribution of eusociality was significantly skewed toward haplodiploid groups, based on Fisher’s exact test. However, this test ignored the phylogenetic non-independence of these data points (Felsenstein, 1985). Crozier (2008) mentioned a similar skew in the number of origins of eusociality, but did not present data or a statistical test. Nowak et al. (2010) stated that there was no statistically significant association between eusociality and haplodiploidy when non-insect groups were included. However, they did not present details of their statistical tests nor of their underlying data. A phylogeny-based maximum likelihood test that could be used to test this hypothesis has been available for >25 years (Pagel, 1994). To our knowledge, this test has not yet been applied to the potential association between haplodiploidy and eusociality across insects, nor have related tests. An important limiting factor for these types of analyses is a detailed time-calibrated phylogeny that spans all or most insects (at least at the family level). Such phylogenies have only become available relatively recently (e.g., Rainford et al., 2014).
In this study, we conduct (to our knowledge) the first phylogenetic test of the haplodiploidy hypothesis in insects. We take advantage of an extensive time-calibrated hexapod phylogeny that includes almost all insect families (Rainford et al., 2014). We generate datasets on the distribution of haplodiploidy and eusociality among these 874 tips. We test whether the evolution of eusociality is associated with haplodiploidy across insect phylogeny using three different phylogeny-based tests of association between traits. Overall, two of the three tests significantly support the hypothesis that eusociality in insects is associated with haplodiploidy, but there is clearly a mixture of concordant and discordant macroevolutionary patterns.
We used the time-calibrated, multi-locus phylogeny of Rainford et al. (2014), which includes almost all families of insects and other hexapods (Supplementary Datafile S1). The tips in this tree are families, with one terminal taxon (tip) per family. However, the analyses we perform are equivalent to treating tips as species and sampling one species per family. Moreover, the number of tips per order is strongly related to the species richness of each order (r2 = 0.804, p < 0.0001; details and data in Supplementary Table S1). Thus, the tree is similar to what would be expected from randomly sampling 874 species across hexapods (in terms of the number of species per order). A time-calibrated, species-level tree spanning all insects is not yet available, to our knowledge.
We selected this tree because it is (as far as we know) the only comprehensive time-calibrated phylogeny available for insects, even if it is only comprehensive at the family level. Furthermore, the strong relationship between the number of tips and the species richness of orders is invaluable for comparative analyses. This tree is similar to those from large-scale phylogenomic analyses (i.e., Misof et al., 2014), both in terms of topology and divergence times among orders (therefore we did not focus on support values or alternative trees). However, the tree of Rainford et al. (2014) has more comprehensive taxon sampling.
To find data on haplodiploidy for each family, we used two main search methods. The first method involved searching for reviews on sex-determination systems in insects as a whole. We conducted searches on Google Scholar using the keywords “sex determination systems,” “determination of sex,” and “insects.” The second method involved searching family by family, for all 874 sampled hexapod families. For these searches, we used “sex determination system,” “haplodiploidy,” and the name of each insect family as keywords. Searches were conducted from February 2020 to September 2020. There are two main types of haplodiploidy: arrhenotoky and parental genome elimination (Blackmon et al., 2017). However, we were primarily interested in the presence (state 1) or absence (state 0) of haplodiploidy, and we did not code them separately in our analysis.
Based on our survey, haplodiploidy is present in 92 families across seven hexapod orders (Figure 1; Coleoptera, Collembola, Diptera, Hemiptera, Hymenoptera, Phthiraptera, Thysanoptera). Several families were variable for the presence of haplodiploidy, in that they included both haplodiploid and non-haplodiploid species (Tree of Sex Consortium, 2014; Blackmon et al., 2017). These included families within Coleoptera (Curculionidae, Micromalthidae), Hemiptera (Aleyrodidae, Coccidae), and Phthiraptera (Pediculidae). In Collembola, Diptera, Hymenoptera, and Thysanoptera, haplodiploidy appeared to be invariant in families in which it is present (Gardner and Ross, 2014; Blackmon et al., 2017). All families with at least one haplodiploid species were coded as having haplodiploidy (state 1).
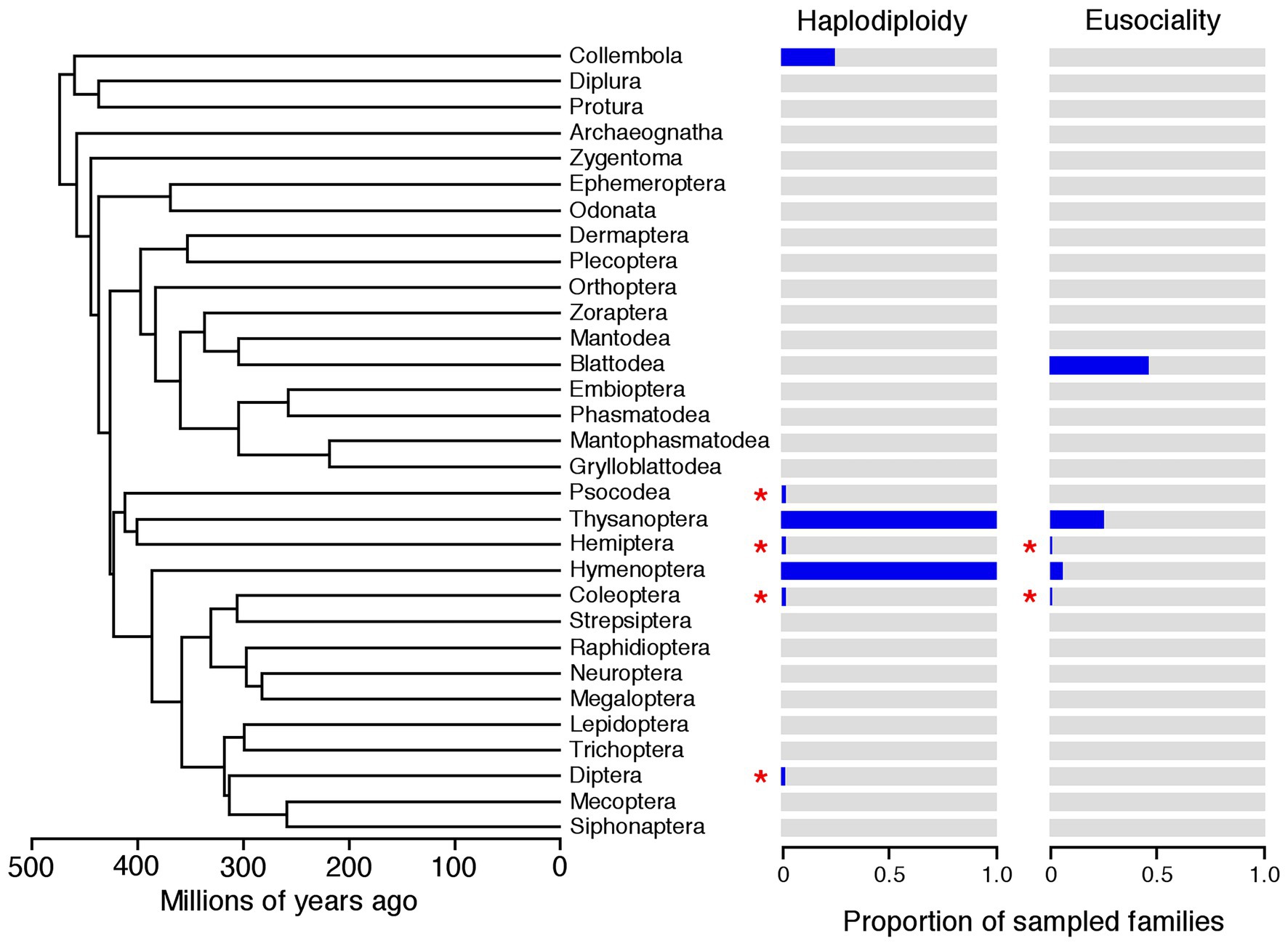
Figure 1. Summary of the distribution of haplodiploidy and eusociality among hexapod orders. For each order, the estimated proportion of sampled families with haplodiploidy and eusociality are shown in dark blue (those without in gray). If haplodiploidy or eusociality were relatively rare in an order, we highlighted that order with a red asterisk to ensure that these traits are visible in these cases. The phylogeny and branch lengths are from Rainford et al. (2014). Note that our analyses used 874 hexapod families from that study as units, not orders (data in Supplementary Datafile S2). Here Blattodea includes Isoptera (termites), and that Psocodea includes Psocoptera and Phthiraptera.
This liberal assignment of families to haplodiploidy could be problematic if some species in a family are haplodiploid and different species are eusocial. We know of only one instance in which this occurred. The beetle family Curculionidae contains one eusocial species (Austroplatypus incompertus) and other species that are haplodiploid, but the eusocial species has diplodiploid sex determination (Smith et al., 2009). We initially assumed that this family contains haplodiploidy. We also conducted analyses treating the family as diplodiploid (state 0) and obtained similar results (see Results). We know of no other cases where this mismatch is problematic. For example, many origins of eusociality are within hymenopterans (see below), in which all species are thought to be haplodiploid. Similarly, the eusocial taxa of Thysanoptera (Kladothrips and Oncothrips) have haplodiploidy as the sex-determination system for all their species (Chapman et al., 2000, 2002). Other eusocial taxa are not haplodiploid. In summary, potential mismatches between eusociality and haplodiploidy within families should not be problematic for our study.
We could not find data on sex-determination systems for seven families in Phthiraptera and four families in Protura. In these cases, we initially assumed that they had the same sex-determination system as the majority of the families in that order (i.e., non-haplodiploid). We also conducted analyses in which these 11 families were excluded and obtained similar results (see Results). Data on haplodiploidy for each family (along with the supporting references) are given in Supplementary Datafile S2.
Various definitions of eusociality have been used (see review of definitions in Boomsma and Gawne, 2018). For this paper, we used the definition proposed by Michener (1969). This definition is relatively explicit and appears to be one of the most well-cited (e.g., Wilson, 1971; Trivers and Hare, 1976; Andersson, 1984; Gadagkar, 1990; Danforth, 2002; Bradley et al., 2009; Kocher and Paxton, 2014; Shell and Rehan, 2018). Following this definition, eusociality is characterized by the presence of three conditions: alloparental care, reproductive division of labor, and overlapping adult generations. We used these three criteria to characterize whether a species was eusocial or not eusocial. Specifically, species that did not satisfy all three criteria were characterized as non-eusocial in our analyses.
We searched for data on eusociality separately for each insect family using Google Scholar. We used the keywords “eusociality” and “eusociality presence” combined with the name of each family. Searches were conducted from February 2020 to September 2020. Within eusociality, many different categories are recognized (e.g., primitive vs. advanced), depending on the extent of division of labor, presence and absence of castes, and other factors (Kocher and Paxton, 2014; Toth and Rehan, 2017). We initially coded families as having (state 1) or lacking (state 0) eusociality, regardless of the type. However, we also performed a set of alternative analyses that distinguished between primitive and advanced eusociality (see below). A family was coded as having eusociality if it occurred in one or more species.
Based on our survey, eusociality is present in 15 families across five hexapod orders (Figure 1). This concurs with other surveys related to eusociality in Hymenoptera and other insect orders (Andersson, 1984; Kent and Simpson, 1992; Thorne, 1997; Chapman et al., 2002; McLeish and Chapman, 2007; Kocher and Paxton, 2014; Abbot, 2015; Toth and Rehan, 2017). Several families have eusocial as well as non-eusocial species, including families within Coleoptera (Curculionidae, one eusocial species), Hymenoptera (Apidae, Crabronidae, Halictidae, Vespidae), Hemiptera (Aphididae), and Thysanoptera (Phlaeothripidae). All species within Formicidae (Hymenoptera) are eusocial, as are all seven families in the order/infraorder Isoptera (Mound, 2005; Bignell et al., 2010; Toth and Rehan, 2017). As for haplodiploidy, we assigned a family as having eusociality (state 1) if it had at least one eusocial species. Data on eusociality for each family (and supporting references) are given in Supplementary Datafile S2.
We also performed alternative analyses in which we distinguished between primitive and advanced eusociality. We considered a family to have advanced eusociality if the species with eusociality have a high degree of morphological variation between the castes (e.g., Michener, 1969; Kocher and Paxton, 2014). Across hexapods, advanced eusociality is present in only 3 families in Hymenoptera and 7 families in Isoptera (Wilson, 1971; Kocher and Paxton, 2014).
We used three different phylogeny-based methods to test for an association between haplodiploidy and eusociality. First, we used Pagel’s (1994) likelihood test to test for a correlation between the evolution of haplodiploidy and eusociality. Four maximum-likelihood models were compared: (i) eusociality and haplodiploidy evolve independently of each other (the null model), (ii) the evolution of eusociality depends on haplodiploidy (the predicted pattern), (iii) the evolution of haplodiploidy depends on eusociality, and (iv) both haplodiploidy and eusociality depend on each other. The test was run in R 4.2.1 (R Core Team, 2019) using the package phytools version 1.0–3 (Revell, 2012) and the function “fitPagel.” The R package treedata.table (Román-Palacios et al., 2021) was used to run the functions from phytools and to shorten the code required to run the models.
The main results of Pagel’s (1994) test are values of AIC (Akaike Information Criterion; Akaike, 1974) for each of the four models, and p-values comparing the fit of each model to the null model (independent evolution of both characters). We also calculated AIC weights (Burnham and Anderson, 2002) for each model, using the function “aic.w” in the base R package (R Core Team, 2019). The preferred model was considered to be that with the highest AIC weight (Burnham and Anderson, 2002). Since there were multiple p-values obtained from Pagel’s test for each dataset, we used the sequential Bonferroni test (Holm, 1979; Rice, 1989) to obtain adjusted p-values. We used the function “p.adjust” within the R package base. Following standard practice, p-values were adjusted within each table, not across all tables in the study.
Prior to running Pagel’s likelihood test, we determined the best-fitting model of character evolution for these two variables. The best-fitting model was then used in the correlation test. We compared the fit of each variable to an equal rates (ER) model (equal rates of gain and loss for each state, where the 0-to-1 rate equals the 1-to-0 rate) and an all-rates-different (ARD) model (rate of gain is different from rate of loss, or the 0-to-1 rate is different from the 1-to-0 rate). Model fit was evaluated using the function “fitMK” in phytools. The fit of different models was evaluated based on their AIC values. If the difference between the AIC values of these models was <4, then both models were considered equivalent for that variable (Burnham and Anderson, 2002), and both were used for Pagel’s (1994) test. If the AIC difference was >4, the model with the lowest AIC was used (Burnham and Anderson, 2002).
Several studies have suggested that Pagel’s test can yield significant results when the independent and dependent variable each evolve only once (“Darwin scenario” of Maddison and FitzJohn, 2015; see also Uyeda et al., 2018; Gardner and Organ, 2021; Boyko and Beaulieu, 2022). This scenario clearly does not apply here, given that haplodiploidy and eusociality have each evolved multiple times across insects (e.g., Figure 1). Another potentially problematic scenario is called the “unreplicated burst” (Maddison and FitzJohn, 2015), in which there is a single origin of the derived state in one character (e.g., the independent variable) but multiple origins of the derived state in the other character. This scenario also does not apply to the characters analyzed here, again given that both eusociality and haplodiploidy evolved multiple times. However, the repeated origins of eusociality within the haplodiploid hymenopterans may yield a somewhat similar scenario, and might be considered phylogenetic pseudoreplication. Therefore, given these (and other) potential issues, we also included two alternative tests, based on very different approaches. We also note that a simple one-to-one association between the evolution of haplodiploidy and the evolution of eusociality is not what is expected based on the previous literature (see Introduction), and that increased origins of eusociality in those parts of the tree with haplodiploidy may be a more reasonable expectation instead.
For the second test of association between haplodiploidy and eusociality, we used phylogenetic logistic regression (Ives and Garland, 2010). Simulations suggest that this general approach may be more conservative under an unreplicated burst scenario (Gardner and Organ, 2021). We ran two phylogenetic logistic regression models: (1) eusociality as a function of haplodiploidy and (2) haplodiploidy as a function of eusociality. The phylogenetic regression was run using the R package phylolm version 2.6.2, using the function “phyloglm.” For each phylogenetic regression, the model which maximizes the penalized likelihood of the logistic regression (MPLE model) was used with 1,000 bootstrap replicates.
We found that the main phylogenetic logistic regression results were not significant (see Results). Therefore, we also performed experimental manipulations of the data to address whether the lack of a significant relationship might be explained by the cases in which eusociality evolves without haplodiploidy (as expected), or by the cases in which haplodiploidy evolves without eusociality. To address the first possibility, we removed cases in which eusociality evolved without haplodiploidy. Specifically, we did the following two manipulations: (1) treated eusociality as absent in Hemiptera (aphids) and (2) treated eusociality as absent in both Hemiptera (aphids) and Isoptera (termites). We then ran the two phylogenetic regression models described above to see if a significant relationship was found (which would suggest that the evolution of eusociality without haplodiploidy explains the non-significant results in the non-manipulated data). We note that we also could have treated eusociality as absent in termites but not in aphids, but since all of these scenarios are entirely hypothetical, it did not seem necessary.
To address the second possibility, we removed cases in which haplodiploidy evolved without eusociality. Specifically, we treated haplodiploidy as absent in the one family of Coleoptera that has haplodiploidy, and in all haplodiploid families in Collembola, Diptera, Hemiptera, and Phthiraptera. We then repeated the phylogenetic logistic regression analyses. A significant result would suggest that these cases in which haplodiploidy evolves without eusociality explain the absence of a significant relationship in the non-manipulated data.
For the third test of association between haplodiploidy and eusociality, we performed posterior predictive D-tests (Huelsenbeck et al., 2003). The D-test is used to measure the difference between the expected and observed association between the states of two characters, where the expected association is based on simulations of independent evolution (using stochastic mapping of character evolution). Positive values of the D-test suggest that the derived character states of the two characters under study are more strongly associated with each other than expected by chance. Here, association is based on shared evolutionary history (i.e., the length of time during which the derived states of the tested two characters are inferred to be present on the same branches of the tree).
To implement this approach, we used the function “Dtest” in phytools. For running the D-test we first created 100 stochastic maps (each) for haplodiploidy and eusociality for both the ER and ARD models. Then we ran the D-test with 100 iterations each on the following combinations of models: (1) ARD stochastic maps (n = 100 each) for both haplodiploidy and eusociality and (2) ARD stochastic maps (n = 100 each) for eusociality and ER stochastic maps for haplodiploidy. We examined these two combinations of models because ARD was the best-fitting model for eusociality, whereas ER and ARD had similar fit for haplodiploidy (Table 2). We also ran D-tests on the dataset that had 11 families removed due to lack of haplodiploidy data and the dataset in which the beetle family Curculionidae was coded as diplodiploid, using the best-fitting transition-rate models in each case.
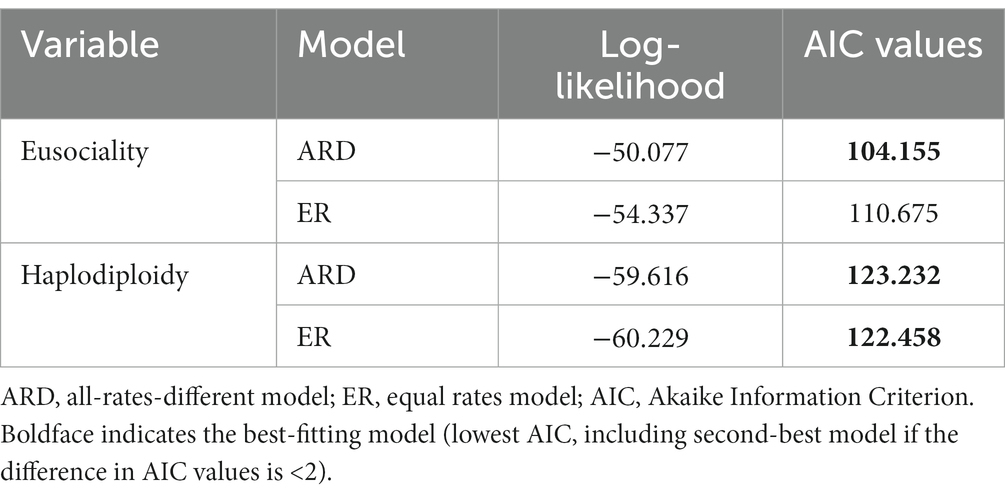
Table 2. Testing the fit of eusociality and haplodiploidy to different models of character evolution across hexapods (all 874 families).
We ran the above three analyses on another dataset to examine the impact of considering only advanced eusociality to be eusociality. Families having only primitive eusociality were treated as lacking eusociality (assigned the state ‘0’). Eusociality was considered present in families having advanced eusociality in at least one species.
The full tree is in Supplementary Datafile S1, and data on eusociality and haplodiploidy for each family are in Supplementary Datafile S2. The reduced tree with 863 taxa is in Supplementary Datafile S3. The analysis-ready csv files are available as Supplementary Datafiles S4–S19, including Supplementary Datafile S4 (main analysis with 874 taxa), Supplementary Datafile S5 (main analysis with 863 taxa), and Supplementary Datafile S6 (with Curculionidae recoded as diplodiploid, not haplodiploid). The various manipulated datasets used in the phylogenetic logistic regression analyses are in Supplementary Datafiles S7–S19. The annotated R code for all analyses is in Supplementary Datafile S20.
The association between haplodiploidy and eusociality was tested across 874 hexapod families. In total, there were 15 families in five orders with eusocial species, including one family in Coleoptera, five families in Hymenoptera, one in Hemiptera (Aphididae), seven in Isoptera/Blattodea, and one in Thysanoptera (Figure 1). There were 92 families in seven orders that had haplodiploid species, including 72 in Hymenoptera. Haplodiploidy and eusociality co-occurred in 7 families in three insect orders (Coleoptera, Hymenoptera, and Thysanoptera). However, haplodiploidy and eusociality did not occur in the same species in the single coleopteran family in which they co-occurred. Eusociality occurred without haplodiploidy in seven families in Isoptera (termites) and one in Hemiptera (aphids).
We used three phylogenetic comparative methods to test for an association between the evolution of eusociality and haplodiploidy. First, using Pagel’s likelihood test of correlation, we found that the model with the evolution of eusociality depending on haplodiploidy had the highest AIC weight, and showed a significant association (Table 3). This result was consistent using both ER and ARD models for both variables. Both models were tested because ARD was the best-fitting model for eusociality but for haplodiploidy the difference in fit between the ER and ARD models was not significant (Table 2). An analysis was also conducted in which 11 families were removed that lacked data on haplodiploidy. This analysis yielded very similar results (Supplementary Tables S2, S3). Another analysis was conducted after recoding the beetle family Curculionidae as diplodiploid (so that there was no association between haplodiploidy and eusociality in this family). This analysis also gave similar results (Supplementary Tables S4, S5), again strongly supporting the idea that haplodiploidy helps drive the evolution of eusociality.
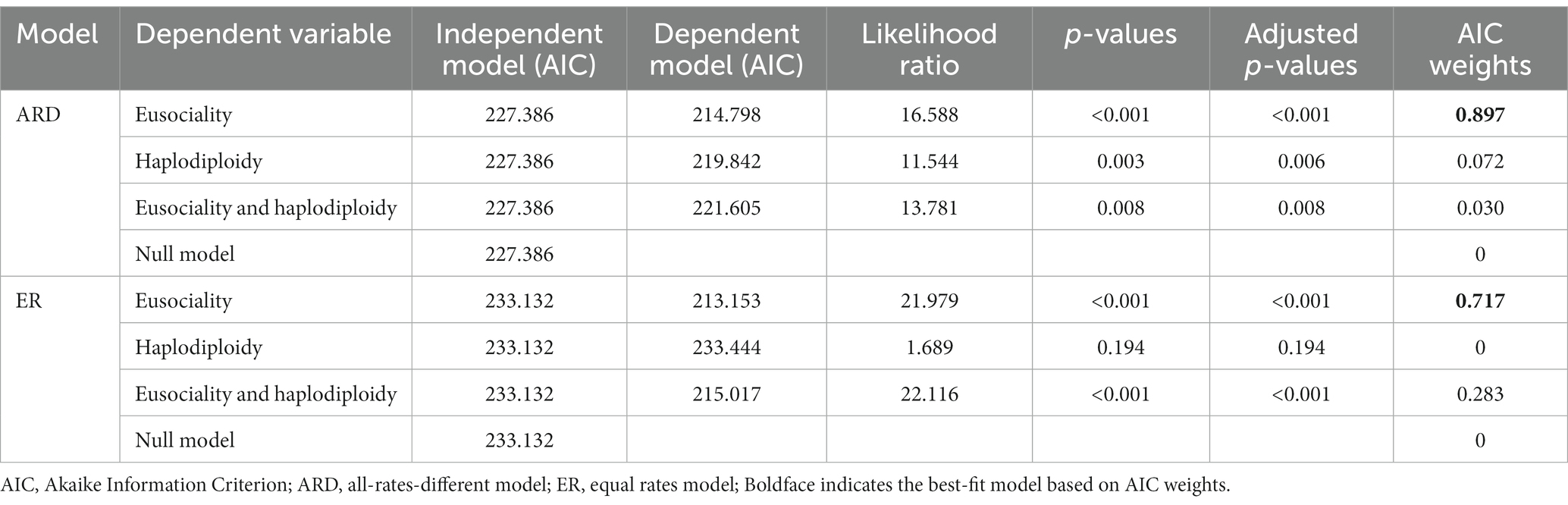
Table 3. Results of Pagel’s correlation test between eusociality and haplodiploidy across hexapods, including all families.
Another analysis was conducted in which only families with advanced eusociality were coded as having eusociality. Both ARD and ER models were tested because there was no significant difference between AIC values for ER and ARD models for eusociality (Supplementary Table S6). A model in which the evolution of eusociality depended on haplodiploidy again had the highest AIC weight (Supplementary Table S7).
In the contrast to the results of Pagel’s test, the phylogenetic logistic regression analyses showed no significant support for the hypothesis that haplodiploidy drives the evolution of eusociality, or vice versa (Table 4). We tested whether this non-significant result occurred because of the evolution of eusociality without haplodiploidy or haplodiploidy without eusociality (Table 4). Coding aphids as non-eusocial also yielded non-significant results (Table 4). However, coding both aphids and termites as non-eusocial showed significant support for the model in which eusociality depended on the evolution of haplodiploidy (Table 4). In a third set of analyses, those haplodiploid lineages that were not eusocial (outside of Hymenoptera) were recoded as diplodiploid (including one family of Coleoptera and all relevant families of Collembola, Diptera, Hemiptera, and Phthiraptera). The results of these analyses were not significant (Table 4). Overall, these manipulations suggest that the phylogenetic regression results in the original data were non-significant because of eusociality evolving without haplodiploidy, not because of haplodiploidy evolving without eusociality. Results were similar using the dataset with 11 families removed because their state for haplodiploidy was unknown (Supplementary Table S8), the dataset in which Curculionidae was considered diplodiploid (Supplementary Table S9), and the dataset in which only families with advanced eusociality were considered to have eusociality (Supplementary Table S10).
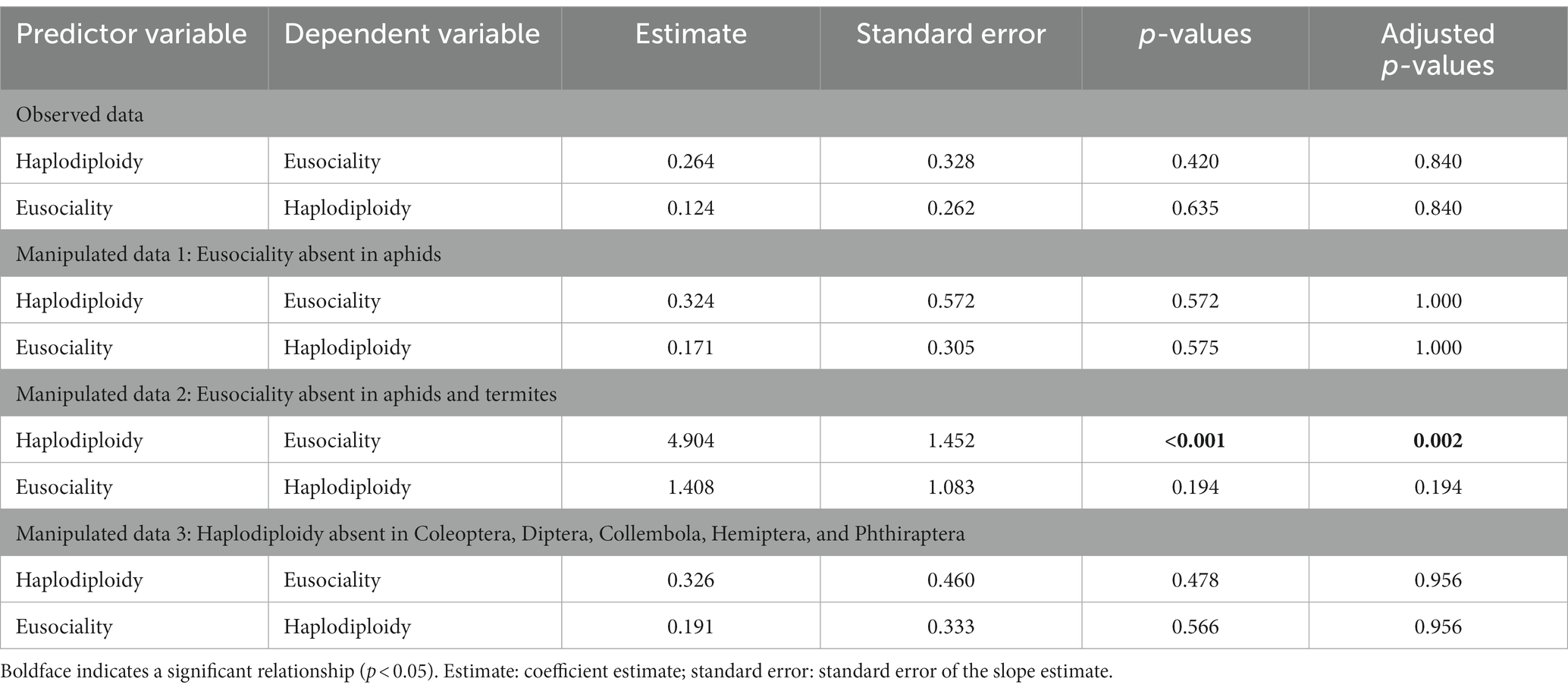
Table 4. Results of phylogenetic logistic regression analyses of haplodiploidy and eusociality across hexapods (including all families).
Using the D-test, we found a significant, positive association between haplodiploidy and eusociality in the main dataset (Table 5) and in all the alternative datasets (11 families removed: Supplementary Table S11; Curculionidae recoded: Supplementary Table S12; recoding for advanced eusociality: Supplementary Table S13). These results show that eusociality and haplodiploidy co-occur more often than expected by chance.
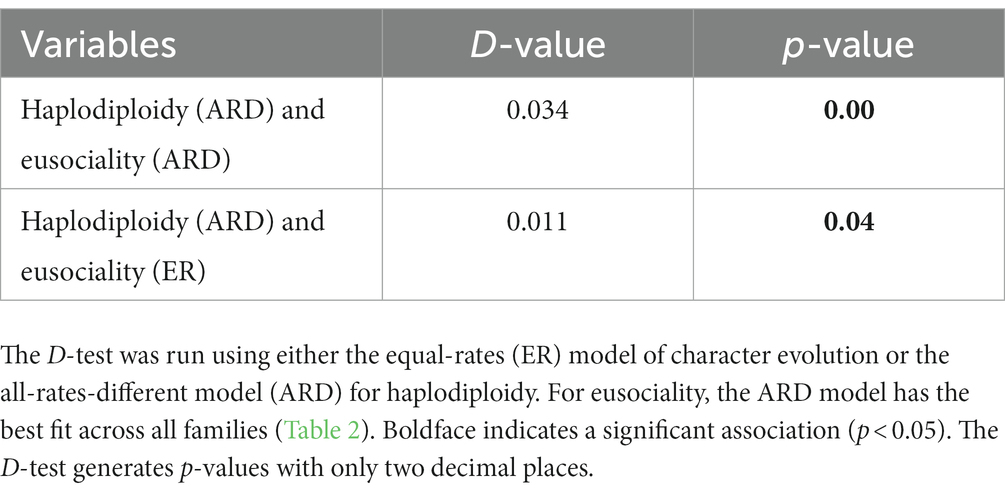
Table 5. Results of the posterior predictive D-test for the original dataset (874 hexapod families).
The evolution of eusociality has puzzled evolutionary biologists for decades. Here we present possibly the first phylogenetic test of the hypothesis that haplodiploidy helped drive the evolution of eusociality across insects (following from Hamilton (1964) and elaborated on by many subsequent authors). We find some support for this hypothesis, but with important caveats. Below, we discuss this support and caveats, the limitations of these analyses, and areas for future research.
Our study shows some phylogenetic support for the long-standing hypothesis that haplodiploidy helps promote the evolution of eusociality. However, that support is mixed. We used three phylogeny-based methods to test this hypothesis. Two of the three methods used significantly supported this hypothesis (Pagel’s test, D-test), whereas a third (phylogenetic logistic regression) gave a non-significant result. We also explored the reasons why the phylogenetic logistic regression results were not significant, by re-running the analyses after manipulating the observed data. These experiments revealed that the origins of eusociality in non-haplodiploid aphids and termites can help explain why eusociality was not significantly linked to haplodiploidy when tested across all insects. Thus, the support for this hypothesis comes primarily from the many repeated origins of eusociality among haplodiploid hymenopterans and the evolution of eusociality among haplodiploid thysanopterans (thrips). Whether the repeated origins of eusociality among haplodiploid taxa (e.g., hymenopterans) outweighed their origins among non-haplodiploid taxa (e.g., aphids, termites) depended on the specific test.
There has been considerable recent discussion of the problem of pseudoreplication in phylogenetic studies of correlation between categorical (discrete) variables (e.g., Maddison and FitzJohn, 2015; Uyeda et al., 2018; Gardner and Organ, 2021; Boyko and Beaulieu, 2022). Much of this discussion has centered around cases where one variable evolves only once, and the other variable either evolves once (Darwin scenario) or repeatedly (unreplicated burst). These scenarios clearly do not apply here, since both haplodiploidy and eusociality evolved repeatedly across insects. On the other hand, much of the evidence for a positive relationship between haplodiploidy and eusociality comes from the repeated origins of eusociality after the single origin of haplodiploidy in Hymenoptera. This might reasonably be considered phylogenetic pseudoreplication. Note that the lack of a significant relationship between haplodiploidy and eusociality from the phylogenetic logistic regression analysis is not necessarily because this method is more conservative with regards to phylogenetic pseudoreplication (e.g., Gardner and Organ, 2021): our experiments show that if eusociality only evolved in Hymenoptera, this method would still yield a significant association with haplodiploidy overall (Table 4). Instead, the non-significant results are explained by the evolution of eusociality without haplodiploidy in termites and aphids (Table 4). An alternative to the pseudoreplication scenario is that there is a one-to-one association between each origin of haplodiploidy and each origin of eusociality. This scenario clearly does not apply here: haplodiploidy alone is not sufficient to explain each origin of eusociality in hymenopterans [nor was this suggested by Hamilton (1964)], since all hymenopterans are haplodiploid and only some are eusocial. Nevertheless, two of our three tests do suggest that haplodiploidy might predispose some lineages toward the evolution of eusociality (as suggested by many authors; Table 1). We also note that other phylogenetic methods could also be applied to test for a relationship between haplodiploidy and eusociality beyond those used here (e.g., Uyeda et al., 2018; Gardner and Organ, 2021; Boyko and Beaulieu, 2022). However, we have already used three diverse methods here, and we think that alternative methods would almost certainly arrive at the same conclusion we found: that the phylogenetic support for the haplodiploidy hypothesis is mixed, rather than being overwhelmingly strong or entirely nonexistent. The results are mixed because most origins of eusociality are among haplodiploid lineages (in hymenopterans and thrips), but with some exceptions (in aphids, beetles, and termites).
We also note some limitations to our analyses. Importantly, our analyses were conducted using a family-level tree. This family-level approach allowed us to include all insects within the same time-calibrated phylogeny (Rainford et al., 2014), with the sampling of tips proportional to the species richness of orders. Furthermore, this analysis is largely equivalent to a species-level analysis (but sampling one species per family). Using family-level sampling, we were able to capture 9 origins of eusociality among five orders (based on the conclusions of Kent and Simpson, 1992; Stern and Foster, 1996; Chapman et al., 2002; Hughes et al., 2008; Bignell et al., 2010; Kocher and Paxton, 2014). These included one in Coleoptera (Curculionidae), one in Hemiptera (Aphididae), five in Hymenoptera (one each in the families Apidae, Crabronidae, Formicidae, Halictidae, Vespidae), one in Isoptera (one origin for seven families), and one in Thysanoptera (Phlaeothripidae).
The main disadvantage of the family-level analysis is that it was not able to capture the origins of eusociality among genera within some families. The origins missed here include three additional origins among genera within Hymenoptera, including one each in Apidae, Halictidae, and Vespidae (Hughes et al., 2008; Kocher and Paxton, 2014). Because hymenopterans are all haplodiploid, including these additional origins of eusociality among haplodiploid hymenopteran genera would strengthen the support for the hypothesis that haplodiploidy helps promote the evolution of eusociality. On the other hand, aphids (Hemiptera) are not haplodiploid and are estimated to have approximately 6 to 9 origins of eusociality (Stern and Foster, 1996; Abbot, 2015). These additional origins were not captured in our analysis and would weaken the correlation between haplodiploidy and eusociality. It is also notable that aphids are unusual in alternating between sexual and asexual generations (Moran, 1992), and eusociality may be favored because many individuals in colonies are clones and therefore closely related to each other. This factor may favor the evolution of eusociality for similar reasons that haplodiploidy favors the evolution of eusociality (Crozier, 2008). Overall, it is clear that the phylogenetic support for the haplodiploidy hypothesis is somewhat mixed among the 874 tips analyzed here, and would continue to be mixed if more tips in hymenopterans and aphids were included. We suggest that future studies should test the haplodiploidy hypothesis using similar approaches to those used here, as more detailed phylogenies become available in insects that span the genus and family levels across orders. However, the overall conclusion seems likely to be similar to what we found here.
Another caveat about our results is that they are based only on hexapods. Some authors (Nowak et al., 2010) have suggested that instances of eusociality among non-insect species that are not haplodiploid show that haplodiploidy does not drive eusociality. Although this is a valid point, this does not prove that haplodiploidy is irrelevant for the evolution of eusociality in insects. Even within insects, eusociality has evolved repeatedly without haplodiploidy (e.g., in aphids, beetles, and termites; Andersson, 1984). Thus, our results support the idea that haplodiploidy may contribute to the evolution of eusociality, but only in some taxa. They do not show that haplodiploidy is the sole explanation for every origin of eusociality in every taxon (not even within hymenopterans).
Most importantly, our results are based on a correlative analysis, and do not prove a causal relationship between haplodiploidy and eusociality. There are likely many factors that led to the evolution of eusociality (review in Table 1). Theoretical studies suggest that haplodiploidy was an important factor in the evolution of eusociality, along with monogamy, overlap of generations in males, population sex ratio, and population growth rate (Fromhage and Kokko, 2011; Quiñones and Pen, 2017; Nonacs, 2019; Rautiala et al., 2019). We suggest that future studies should use broad-scale phylogenetic analyses to test other potential correlates of the evolution of eusociality, as we have done here for haplodiploidy.
A crucial variable to test will be the relatedness coefficient. A haplodiploid sex-determination system results in a haploid male and a diploid female (Blackmon et al., 2017). Therefore, sisters can be more closely related to each other than to their own potential offspring. Under the haplodiploidy hypothesis (Hamilton, 1964), this pattern of relatedness among conspecific individuals is thought to lead to reproductive division of labor, resulting in sterile workers and one or more reproducing queens (i.e., eusociality). However, some eusocial species have a relatedness coefficient < 0.75 among the workers (i.e., less than the crucial value predicted by Hamilton’s equation), due to the presence of polyandry (Queller and Strassmann, 1998; Landi et al., 2003). Therefore, future analyses should use the relatedness coefficient as a predictor variable for the evolution of eusociality (e.g., using phylogenetic logistic regression), to provide a more direct test of Hamilton’s (1964) hypothesis linking haplodiploidy, relatedness, and eusociality. We did not perform such an analysis here because we lacked a large-scale dataset on relatedness coefficients.
In this study, we show mixed phylogenetic support for the decades-old hypothesis that the evolution of eusociality in insects is associated with haplodiploidy, with two methods supporting this hypothesis and one yielding non-significant results. Nevertheless, haplodiploidy is not the sole explanation the origins of eusociality, even within insects. Specifically, eusociality evolves in some lineages without haplodiploidy (e.g., aphids, termites), and haplodiploidy alone cannot explain why some hymenopteran lineages evolve eusociality and others do not. Clearly, our study will not be the last word on what factors drive the evolution of eusociality in insects. Nevertheless, we suggest that future studies on this topic should also include a similar large-scale, statistical phylogenetic approach, if they wish to explain the evolutionary origins of eusociality among clades.
The original contributions presented in the study are included in the Supplementary material, further inquiries can be directed to the corresponding author.
CJ and JW conceptualized the study and wrote the manuscript. CJ performed data collection and analyses. All authors contributed to the article and approved the submitted version.
JW acknowledges U.S. National Science Foundation grant DEB 1655690.
The authors thank Andy Gardner, Jan Oettler, Stephen Trumbo, and William Wcislo for helpful comments that greatly improved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1118748/full#supplementary-material
Abbot, P. (2015). The physiology and genomics of social transitions in aphids. Adv. Insect Phys. 48, 163–188. doi: 10.1016/bs.aiip.2014.12.005
Akaike, H. (1974). A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716–723. doi: 10.1109/TAC.1974.1100705
Andersson, M. (1984). The evolution of eusociality. Annu. Rev. Ecol. Syst. 15, 165–189. doi: 10.1146/annurev.es.15.110184.001121
Bignell, D. E., Roisin, Y., and Lo, N. (eds.) (2010). Biology of Termites: A Modern Synthesis. Dordrecht, Netherlands: Springer Science & Business Media.
Blackmon, H., Ross, L., and Bachtrog, D. (2017). Sex determination, sex chromosomes, and karyotype evolution in insects. J. Hered. 108, 78–93. doi: 10.1093/jhered/esw047
Boomsma, J. J., and Gawne, R. (2018). Superorganismality and caste differentiation as points of no return: how the major evolutionary transitions were lost in translation. Biol. Rev. 93, 28–54. doi: 10.1111/brv.12330
Boyko, J. D., and Beaulieu, J. M. (2022). Reducing the biases in false correlations between discrete characters. Syst. Biol. :syac066. (in press). doi: 10.1093/sysbio/syac066
Bradley, T. J., Briscoe, A. D., Brady, S. G., Contreras, H. L., Danforth, B. N., Dudley, R., et al. (2009). Episodes in insect evolution. Integr. Comp. Biol. 49, 590–606. doi: 10.1093/icb/icp043
Burnham, K. P., and Anderson, D. R. (2002). Model Selection and Multimodel Inference. New York, NY: Springer.
Chapman, T. W., Crespi, B. J., Kranz, B. D., and Schwarz, M. P. (2000). High relatedness and inbreeding at the origin of eusociality in gall-inducing thrips. Proc. Natl. Acad. Sci. U. S. A. 97, 1648–1650. doi: 10.1073/pnas.020510097
Chapman, T. W., Kranz, B. D., Bejah, K. L., Morris, D. C., Schwarz, M. P., and Crespi, B. J. (2002). The evolution of soldier reproduction in social thrips. Behav. Ecol. 13, 519–525. doi: 10.1093/beheco/13.4.519
Crespi, B. J., and Yanega, D. (1995). The definition of eusociality. Behav. Ecol. 6, 109–115. doi: 10.1093/beheco/6.1.109
Crozier, R. H. (2008). Advanced eusociality, kin selection, and male haplodiploidy. Aust. J. Entomol. 47, 2–8. doi: 10.1111/j.1440-6055.2007.00621.x
Danforth, B. N. (2002). Evolution of sociality in a primitively eusocial lineage of bees. Proc. Natl. Acad. Sci. U. S. A. 99, 286–290. doi: 10.1073/pnas.012387999
Davies, N. G., Ross, L., and Gardner, A. (2016). The ecology of sex explains patterns of helping in arthropod societies. Ecol. Lett. 19, 862–872. doi: 10.1111/ele.12621
Felsenstein, J. (1985). Phylogenies and the comparative method. Am. Nat. 125, 1–15. doi: 10.1086/284325
Fromhage, L., and Kokko, H. (2011). Monogamy and haplodiploidy act in synergy to promote the evolution of eusociality. Nat. Commun. 2, 1–6. doi: 10.1038/ncomms1410
Gadagkar, R. (1985). Kin recognition in social insects and other animals—a review of recent findings and a consideration of their relevance for the theory of kin selection. Proc. Anim. Sci. 94, 587–621. doi: 10.1007/BF03191863
Gadagkar, R. (1990). Evolution of eusociality: the advantage of assured fitness returns. Philos. Trans. R. Soc. B 329, 17–25. doi: 10.1098/rstb.1990.0146
Gadagkar, R. (1991). On testing the role of genetic asymmetries created by haplodiploidy in the evolution of eusociality in the hymenoptera. J. Genet. 70, 1–31. doi: 10.1007/BF02923575
Gardner, A., Alpedrinha, J., and West, S. A. (2012). Haplodiploidy and the evolution of eusociality: split sex ratios. Am. Nat. 179, 240–256. doi: 10.1086/663683
Gardner, A., and Ross, L. (2013). Haplodiploidy, sex-ratio adjustment, and eusociality. Am. Nat. 181, E60–E67. doi: 10.1086/669147
Gardner, A., and Ross, L. (2014). Mating ecology explains patterns of genome elimination. Ecol. Lett. 17, 1602–1612. doi: 10.1111/ele.12383
Gardner, J. D., and Organ, C. L. (2021). Evolutionary sample size and consilience in phylogenetic comparative analysis. Syst. Biol. 70, 1061–1075. doi: 10.1093/sysbio/syab017
Hamilton, W. D. (1964). The genetical evolution of social behaviour. II. J. Theor. Biol. 7, 17–52. doi: 10.1016/0022-5193(64)90039-6
Huelsenbeck, J. P., Nielsen, R., and Bollback, J. P. (2003). Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158. doi: 10.1080/10635150390192780
Hughes, W. O., Oldroyd, B. P., Beekman, M., and Ratnieks, F. L. (2008). Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 320, 1213–1216. doi: 10.1126/science.1156108
Ives, A. R., and Garland, T. (2010). Phylogenetic logistic regression for binary dependent variables. Syst. Biol. 59, 9–26. doi: 10.1093/sysbio/syp074
Johnstone, R. A., Cant, M. A., and Field, J. (2012). Sex-biased dispersal, haplodiploidy and the evolution of helping in social insects. Proc. R. Soc. Lond. B 279, 787–793. doi: 10.1098/rspb.2011.1257
Kennedy, P., and Radford, A. N. (2020). Sibling quality and the haplodiploidy hypothesis. Biol. Lett. 16:20190764. doi: 10.1098/rsbl.2019.0764
Kent, D. S., and Simpson, J. A. (1992). Eusociality in the beetle Austroplatypus incompertus (Coleoptera: Curculionidae). Naturwissenschaften 79, 86–87. doi: 10.1007/BF01131810
Kocher, S. D., and Paxton, R. J. (2014). Comparative methods offer powerful insights into social evolution in bees. Apidologie 45, 289–305. doi: 10.1007/s13592-014-0268-3
Landi, M., Queller, D. C., Turillazzi, S., and Strassmann, J. E. (2003). Low relatedness and frequent queen turnover in the stenogastrine wasp Eustenogaster fraterna favor the life insurance over the haplodiploid hypothesis for the origin of eusociality. Insect. Soc. 50, 262–267. doi: 10.1007/s00040-003-0676-2
Maddison, W. P., and FitzJohn, R. G. (2015). The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst. Biol. 64, 127–136. doi: 10.1093/sysbio/syu070
McLeish, M. J., and Chapman, T. W. (2007). The origin of soldiers in the gall-inducing thrips of Australia (Thysanoptera: Phlaeothripidae). Aust. J. Entomol. 46, 300–304. doi: 10.1111/j.1440-6055.2007.00617.x
Michener, C. D. (1969). Comparative social behavior of bees. Annu. Rev. Entomol. 14, 299–342. doi: 10.1146/annurev.en.14.010169.001503
Misof, B. S., Liu, S., Meusemann, K., Peters, R. S., Donath, A., Mayer, C., et al. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. doi: 10.1126/science.1257570
Moran, N. A. (1992). The evolution of aphid life cycles. Annu. Rev. Entomol. 37, 321–348. doi: 10.1146/annurev.en.37.010192.001541
Mound, L. A. (2005). Thysanoptera: diversity and interactions. Annu. Rev. Entomol. 50, 247–269. doi: 10.1146/annurev.ento.49.061802.123318
Nonacs, P. (2019). Hamilton's rule is essential but insufficient for understanding monogamy's role in social evolution. R. Soc. Open Sci. 6:180913. doi: 10.1098/rsos.180913
Nowak, M. A., Tarnita, C. E., and Wilson, E. O. (2010). The evolution of eusociality. Nature 466, 1057–1062. doi: 10.1038/nature09205
Pagel, M. (1994). Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. B 255, 37–45. doi: 10.1098/rspb.1994.0006
Queller, D. C., and Strassmann, J. E. (1998). Kin selection and social insects. Bioscience 48, 165–175. doi: 10.2307/1313262
Quiñones, A. E., and Pen, I. (2017). A unified model of hymenopteran preadaptations that trigger the evolutionary transition to eusociality. Nat. Commun. 8, 1–13. doi: 10.1038/ncomms15920
R Core Team. (2019). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
Rainford, J. L., Hofreiter, M., Nicholson, D. B., and Mayhew, P. J. (2014). Phylogenetic distribution of extant richness suggests metamorphosis is a key innovation driving diversification in insects. PLoS One 9:e109085. doi: 10.1371/journal.pone.0109085
Rautiala, P., Helanterä, H., and Puurtinen, M. (2019). Extended haplodiploidy hypothesis. Evol. Lett. 3, 263–270. doi: 10.1002/evl3.119
Revell, L. J. (2012). Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225. doi: 10.2307/2409177
Román-Palacios, C., Wright, A., and Uyeda, J. (2021). Treedata.Table: a wrapper for data.Table that enables fast manipulation of large phylogenetic trees matched to data. PeerJ 9:12450. doi: 10.7717/peerj.12450
Ross, L., Gardner, A., Hardy, N., and West, S. A. (2013). Ecology, not the genetics of sex determination, determines who helps in eusocial populations. Curr. Biol. 23, 2383–2387. doi: 10.1016/j.cub.2013.10.013
Shell, W. A., and Rehan, S. M. (2018). Behavioral and genetic mechanisms of social evolution: insights from incipiently and facultatively social bees. Apidologie 49, 13–30. doi: 10.1007/s13592-017-0527-1
Smith, S. M., Beattie, A. J., Kent, D. S., and Stow, A. J. (2009). Ploidy of the eusocial beetle Austroplatypus incompertus (Schedl) (Coleoptera, Curculionidae) and implications for the evolution of eusociality. Insect. Soc. 56, 285–288. doi: 10.1007/s00040-009-0022-4
Stern, D. L., and Foster, W. A. (1996). The evolution of soldiers in aphids. Biol. Rev. 71, 27–79. doi: 10.1111/j.1469-185X.1996.tb00741.x
Thorne, B. L. (1997). Evolution of eusociality in termites. Annu. Rev. Ecol. Syst. 28, 27–54. doi: 10.1146/annurev.ecolsys.28.1.27
Toth, A. L., and Rehan, S. M. (2017). Molecular evolution of insect sociality: an eco-evo-devo perspective. Annu. Rev. Entomol. 62, 419–442. doi: 10.1146/annurev-ento-031616-035601
Tree of Sex Consortium (2014). Tree of sex: a database of sexual systems. Sci. Data. 1:140015. doi: 10.1038/sdata.2014.15
Trivers, R. L., and Hare, H. (1976). Haplodiploidy and the evolution of the social insect: the unusual traits of the social insects are uniquely explained by Hamilton's kinship theory. Science 191, 249–263. doi: 10.1126/science.1108197
Uyeda, J. C., Zenil-Ferguson, R., and Pennell, M. W. (2018). Rethinking phylogenetic comparative methods. Syst. Biol. 67, 1091–1109. doi: 10.1093/sysbio/syy031
West-Eberhard, M. J. (1975). The evolution of social behavior by kin selection. Q. Rev. Biol. 50, 1–33. doi: 10.1086/408298
West-Eberhard, M. J. (1978). Polygyny and the evolution of social behavior in wasps. J. Kans. Entomol. Soc. 51, 832–856.
Keywords: eusociality, haplodiploidy, insects, macroevolution, phylogeny
Citation: Joshi CH and Wiens JJ (2023) Does haplodiploidy help drive the evolution of insect eusociality? Front. Ecol. Evol. 11:1118748. doi: 10.3389/fevo.2023.1118748
Received: 07 December 2022; Accepted: 20 February 2023;
Published: 16 March 2023.
Edited by:
Stephen T. Trumbo, University of Connecticut, United StatesReviewed by:
Andy Gardner, University of St Andrews, United KingdomCopyright © 2023 Joshi and Wiens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John J. Wiens, d2llbnNqQGFyaXpvbmEuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.