
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 16 March 2023
Sec. Evolutionary and Population Genetics
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1107034
HOX genes play a central role in the development and regulation of limb patterns. For mammals in the order Carnivora, limbs have evolved in different forms, and there are interesting cases of phenotypic convergence, such as the pseudothumb of the giant and red pandas, and the flippers or specialized limbs of the pinnipeds and sea otter. However, the molecular bases of limb development remain largely unclear. Here, we studied the molecular evolution of the HOX9 ~ 13 genes of 14 representative species in Carnivora and explored the molecular evolution of other HOX genes. We found that only one limb development gene, HOXC10, underwent convergent evolution between giant and red pandas and was thus an important candidate gene related to the development of pseudothumbs. No signals of amino acid convergence and natural selection were found in HOX9 ~ 13 genes between pinnipeds and sea otter, but there was evidence of positive selection and rapid evolution in four pinniped species. Overall, few HOX genes evolve via natural selection or convergent evolution, and these could be important candidate genes for further functional validation. Our findings provide insights into potential molecular mechanisms of the development of specialized pseudothumbs and flippers (or specialized limbs).
The order Carnivora plays important roles not only in wildlife conservation but also as an important model for the adaptive evolution of species (Shubin, 2002; Van Valkenburgh and Wayne, 2010). Morphological modifications of the axial skeleton and limbs in mammals have been broadly recognized as adaptive responses to changes in lifestyle and habitat (Martín-Serra et al., 2015). There are two very interesting changes in limb morphology in Carnivora: the specialized pseudothumbs in giant pandas and red pandas and the flippers or specialized limbs of marine species, including sea lions, seals, walruses, and sea otters.
Belonging to two different families, both the giant panda (Ailuropoda melanoleuca) and the red panda (Ailurus fulgens) possess ‘pseudothumbs,’ i.e., enlarged radial sesamoid bones, which contribute to the ability to grasp bamboo and facilitate foraging (Antón et al., 2006; Arnason et al., 2006). Marine Carnivora species are divided into two categories: pinnipeds, which include species in the Odobenidae, Otariidae, and Phocidae families (Arnason et al., 2006), and sea otters, which are in the Mustelidae family and are more closely related to red pandas. Although sea otters and pinnipeds are relatively distantly related to each other, they all evolved flippers or specialized limbs to adapt to aquatic life (Reidenberg, 2007). Sea lions and fur seals (Otariidae) can walk on land by rotating their hind flippers forward under their body (Uhen, 2007). True seals (Phocidae) crawl on land because their front flippers are small and their hind flippers cannot rotate forward (Fish and Lauder, 2017). Walruses (Odobenidae) can rotate their hind flippers and walk on land (Berta et al., 2018). Sea otters (Enhydra lutris) are the only marine member of Mustelidae and the smallest marine mammals. In the specialized hindfeet of the sea otter, as in pinnipeds, the fovea capitis is absent from the femur, marking the absence of the teres ligament, and the biceps femoris muscle inserts onto the middle of the tibia and maintains the leg in a posterior position (Mori et al., 2015).
HOX genes belong to a large gene family that is responsible for the formation of animal body organs, tissues, bones and body segments. They are also responsible for the correct placement of animal body segments and determine the basic structure and orientation of animal forms (Roelen et al., 2002). HOX genes clearly contribute to the development of secondary axes, particularly the regulation of limb patterns (Roelen et al., 2002; Casaca et al., 2014).
In the mammalian genome, there are 39 HOX genes that are subdivided into 13 paralogous groups (PGs) and are closely linked in four clusters: HOX A, B, C, and D (Ruddle et al., 1994). These HOX genes are evolutionarily conserved (Goodman, 2002). A small mutation in a HOX gene could lead to serious disease in humans (Quinonez and Innis, 2014). Previous studies have provided numerous examples. For example, hand–foot–genital syndrome is caused by polyalanine expansions or point mutations in HOXA13 and rare heterozygous deletions that affect this locus. The progressive reduction of gene expression in HOXA13 and HOXD11-HOXD13 in the Gli3-null background results in progressively more severe polydactyly, with thinner and more densely packed digits (Sheth et al., 2012). HOXA11 mutant mice exhibited abnormal sesamoid bone development in the forelimbs and enlarged sesamoid development in the hindlimbs (Small and Potter, 1993). HOXD11 mutant mice showed the presence of an aberrant sesamoid bone between the radius and ulna and a reduction in the size of a sesamoid bone located next to the tibiale mediale (Davis and Capecchi, 1994).
However, despite their functional importance, the genetic mechanisms controlling limb and digit morphology remain poorly understood. Using a comparative genomics strategy, Hu et al. (2017) identified the adaptively convergent genes DYNC2H1 and PCNT as candidate genes responsible for pseudothumb development in giant and red pandas. However, they did not find any signatures of HOX genes. One possible cause for this was that they analyzed only 16 HOX genes, and potentially adaptively convergent HOX genes may be missing from their analysis. In this study, we focused on the natural selection (positive selection, rapid evolution, and negative selection) and convergent evolution of the HOX9 ~ 13 genes between giant and red pandas and between pinnipeds and sea otter within Carnivora. The aims were to identify the possible candidate HOX genes responsible for the development of pseudothumbs and flippers. In addition, we explored the molecular evolution of other HOX genes. These findings provide insights into HOX gene evolution and the potential relationship with specialized limb development in Carnivora.
Fourteen Carnivora species were chosen for evolutionary analysis based on the completeness of genome data, with humans as the outgroup (Figure 1). Initially, we obtained the full-length coding sequences (CDSs) of 39 HOX genes of 12 species, except for the red panda and northern fur seal, from the Orthologous Mammalian Markers database1 (Douzery et al., 2014). Then, we examined the completeness of the start and stop codons, excluded potential pseudogenes, and eliminated sequences with any deletions or ambiguous sequences.
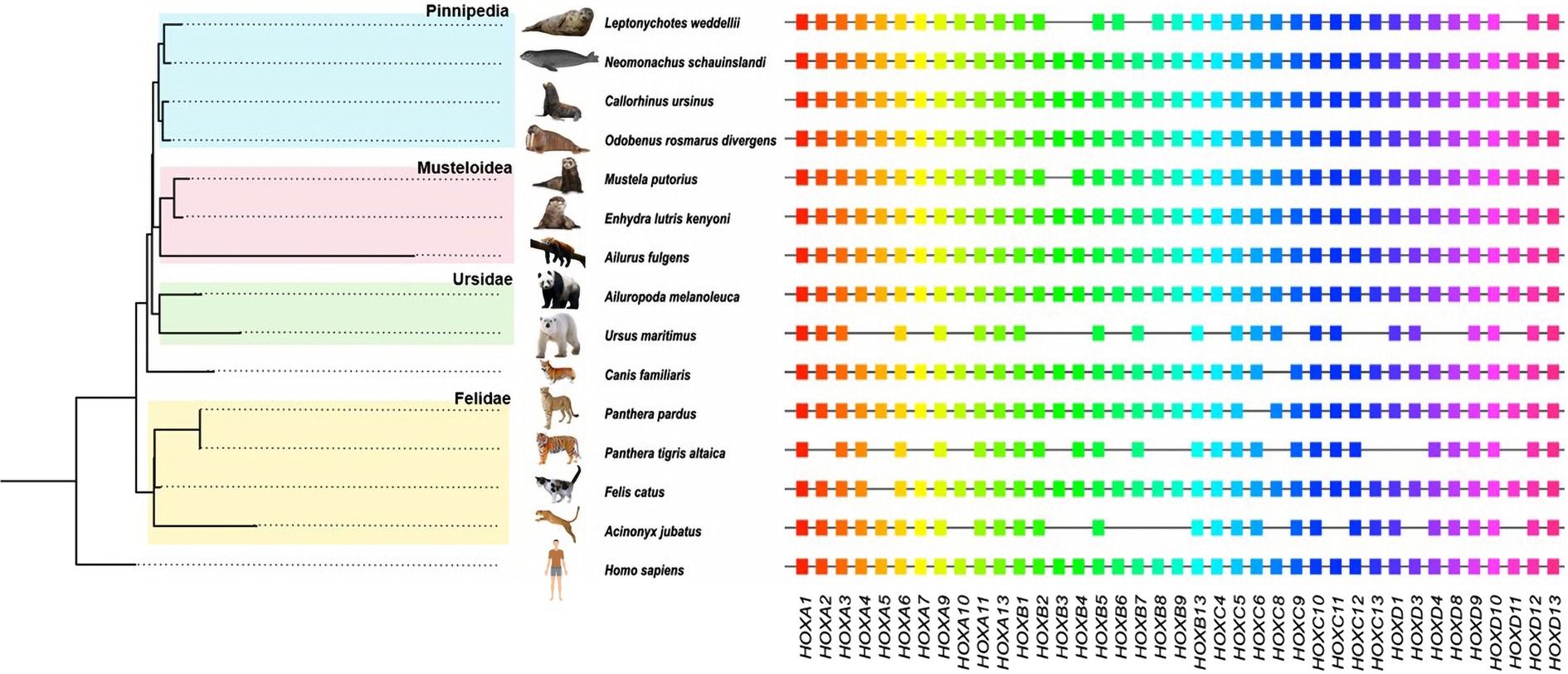
Figure 1. Phylogenetic analyses of Carnivora species based on the concatenated HOX gene family. The phylogenetic tree topology is the same as that of other large-data studies at the family level (Nyakatura and Bininda-Emonds, 2012; Upham et al., 2019). All 39 HOX genes collected from 15 species are shown on the right side.
We obtained the hidden Markov model (HMM) corresponding to the HOX gene family (PF00046) (Finn et al., 2014) and used HMMER (version 3.1b2) (Prakash et al., 2017) for sequence alignment analysis of CDSs of the red panda. To identify homologous pairs of HOX genes, sequence alignment was performed using BLASTN (NCBI-blast-2.6.0+) (Chen et al., 2015) and BLASTP (NCBI-blast-2.6.0+) (Jacob et al., 2008). The database and query alignment included four rounds of mutual alignment. Walrus was used as the reference to identify the HOX genes of the northern fur seal. Similarly, to predict red panda HOX genes that were not annotated in the previous genome version, we performed homology prediction. We extracted the human HOX proteins from Ensembl (release 79) (Zerbino et al., 2018) and aligned them to the red panda genome using TBLASTN (version 2.2.23) (Gertz et al., 2006). Then, we extended the alignment regions by 10 kb at both ends and predicted the gene structure using GeneWise (version 2.2.0) (Birney et al., 2004). The optimal result was taken as the final HOX gene set of the red panda (Figure 2). Sequence analysis was carried out to check the integrity and lengths of all the predicted HOX genes.
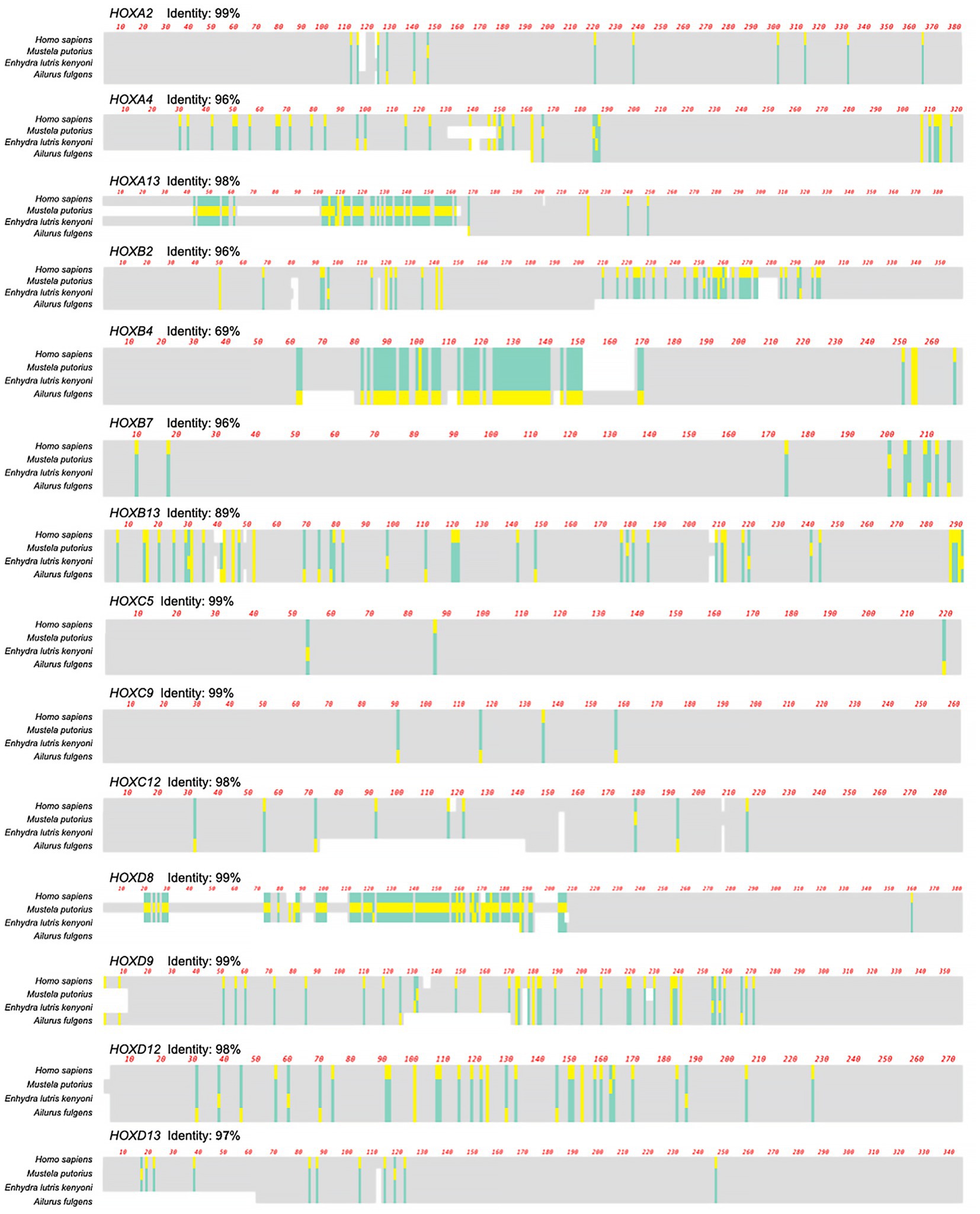
Figure 2. Multiple sequence alignment of 14 reannotated HOX genes in Ailurus fulgens. Three species (Homo sapiens, Mustela putorius, and Enhydra lutris kenyoni) were aligned to show the consistency of HOX gene reannotation. The sequence identity (%) between Homo sapiens and Ailurus fulgens is shown in each panel. The identical amino acid residues are shown in gray, in contrast to the different residues among species, which are colored green and yellow.
Multiple alignments of the DNA and amino acid sequences of the HOX gene set were performed with two methods, MAFFT (version 7) (Katoh and Standley, 2013) and PRANK (Löytynoja, 2014) plus PAL2NAL (Suyama et al., 2006), to form the final gene set alignment. We defined the fourfold degenerate (4D) sites in the alignment sequence using custom scripts. The conserved sequence blocks were extracted from the outputs of multiple sequence alignment by Gblocks (version 0.91) (Castresana, 2000). The selection of the best substitution model for alignment was performed by ProtTest (version 3.4) (Darriba et al., 2011). The phylogenetic tree of the concatenated genes was generated in PhyML (Guindon et al., 2010). The amino acid substitution model was specified as GAMMA + JTTF. The obtained phylogenetic tree was visualized using FigTree viewer v1.4.1.2
To identify positively selected genes (PSGs), we conducted selective pressure analyses using CodeML from PAML 4.8 (Yang, 2007), with branch-site models employed (Yang and dos Reis, 2011). We divided our dataset into two large groups (G1 and G2): G1 included 10 species after removing four pinniped species and the sea otter and focused on the evolutionary analysis of HOX genes related to limb development of bamboo-eating giant and red pandas; G2 targeted the evolution of HOX genes associated with limb development in pinnipeds and sea otter and included 13 species after removing the giant and red pandas.
Furthermore, selective pressure analyses were performed using nine small groups under the same conditions: G1-a: setting the giant panda as the foreground branch; G1-b: the red panda; G1-c: both pandas; G2-a: the Weddell seal; G2-b: the Hawaiian monk seal; G2-c: the walrus; G2-d: the northern fur seal; G2-e: the Weddell seal, Hawaiian monk seal, walrus and northern fur seal; and G2-f: the sea otter. Two categories were used to set different foreground branches in pinnipeds (Figure 3): G2-e1, setting the most recent common ancestor of pinnipeds and the four pinniped species as the foreground branch; and G2-e2, setting only the four pinniped species as the foreground branch.
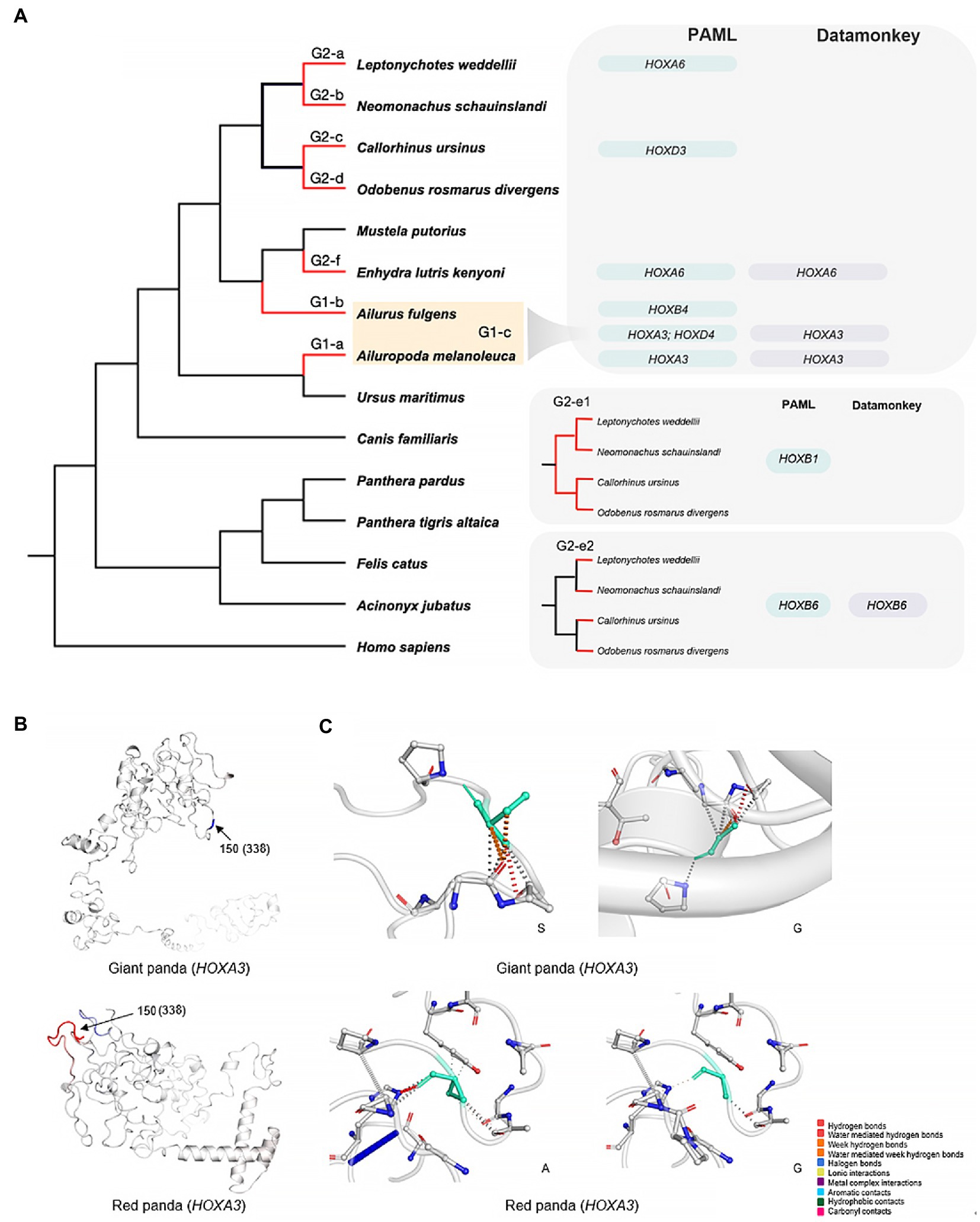
Figure 3. Positively selected genes in 15 species. (A) The red lines (left) represent the foreground branches used in this analysis. Two categories with different foreground branch setting are shown. (B) The 3D structures of HOXA3 for the giant panda and red panda built using a deep-learning-based method, trRosetta. The amino acids are colored according to the vibrational entropy change upon mutation. Blue represents rigidification of the structure, and red represents a gain in flexibility. 150 (338), 150 represents the 150th codon when performing the PAML analysis, and 338 indicates the 338th amino acid using the human gene sequence as the reference (C) The results of interatomic interaction prediction. Wild-type and mutant residues are colored light green and are also represented as sticks alongside the surrounding residues that are involved in any type of interaction.
To confirm the positively selected genes identified in PAML analysis, we further used four models implemented in HyPhy from Datamonkey (Weaver et al., 2018) to examine the signatures of positive selection at three levels (site, gene and branch levels) based on the dN/dS ratio (ω). The analyses at the site level include three methods, which were used to identify individual sites subject to positive selection along subsets of phylogenetic tree branches: (1) FEL (Pond et al., 2005), (2) FUBAR (Murrell et al., 2013), and (3) MEME (Murrell et al., 2012). At the gene level, BUSTED analysis (Murrell et al., 2015) was used to identify genes for episodic diversification. At the branch level, the aBSREL method (Murrell et al., 2015) was used to identify positive selection on individual branches. Finally, to reduce false-positive results, only PSGs detected by both PAML and any one of the Datamonkey methods were considered.
We identified convergent amino acid substitutions between giant and red pandas and among marine mammals with the following rules: (i) the amino acid residues of both extant lineages were identical, and (ii) amino acid changes were inferred to have occurred between the extant lineage and its most recent ancestor lineage (Hu et al., 2017). In this study, calculation of frequencies and rates for the categories and reconstruction of ancestral protein sequences were performed by the CodeML program in PAML 4.8. The relative evolutionary rates of all sites within the gene followed the gamma distribution of sitewise rate variation and the frequency of all amino acids in each site of the gene. We also counted the convergent substitution events and calculated the convergence probability for each branch pair. To filter out noise resulting from chance amino acid substitutions, we performed a Poisson test to verify whether the observed number of convergent substitutions of each gene was significantly greater than the expected number caused by random substitution under the JTT-f gene and JTT-f site amino acid substitution models. We reconstructed the substitutions on all branches in a mammalian phylogeny containing the 15 mammals studied. We then tallied all convergent amino acid substitutions on the giant and red panda branches for all HOX genes, including all possible strictly convergent changes. To further check the strictness of the convergent amino acids, we took advantage of the 100 prealigned vertebrate genomes in the University of California Santa Cruz genome browser.3
Finally, we detected the common amino acid substitutions that considered only the terminal branches rather than the ancestral branches. Specifically, giant and red pandas have the same amino acids that differ from those of other species, and the Weddell seal, Hawaiian monk seal, walrus, northern fur seal and sea otter have the same amino acids that differ from those of other species.
To evaluate the signatures of rapid evolution on each HOX gene, we employed a branch model implemented in the CodeML module of PAML 4.8. Two different tests were conducted to identify lineage-specific effects in the evolution of clades: a two-ratio model versus a one-ratio model and a two-ratio model versus a free-ratio model. The overlap of significant genes from the two tests was thought to be under rapid evolution.
Negative selection plays an important role in maintaining the long-term stability of biological structure and function by purging deleterious mutations. We identified negatively selected genes according to the following criteria: dN/dS of the selected foreground branch is lower than dN/dS of all branches or the background branch, and the p value must be lower than 0.05.
To explore the structural effects of candidate sites, the predicted 3D structure of candidate HOX genes was generated using the trRosetta algorithm (Yang et al., 2020). To evaluate the impacts of amino acid changes on the overall protein structure, we calculated the predicted changes in folding free energy (ΔΔG) between ‘wild-type’ and ‘mutant-type’ amino acids using DynaMut (Yang et al., 2020). We predicted the effects of mutations on protein stability and flexibility. We obtained the PDB files from trRosetta and applied DynaMut on the online website.4 All of the protein 3D structure figures were generated using PyMOL.5
To characterize the functional impact of the mutant, we initially used Protein Variation Effect Analyzer (PROVEAN) (Choi and Chan, 2015) to predict the potential effect of an amino acid substitution. Second, we also obtained the 3D structure of particular proteins. The 3D structure of the wild-type protein was built in silico.
After a series of strict screenings and integrations, we obtained the HOX gene set from 14 Carnivora species with humans as the outgroup. All HOX gene sequences from the above species were intact, without premature stop codons or frame-shift mutations, indicating the presence of functional HOX proteins. The number of HOX genes varied among the 15 species (Supplementary Table S1), and not all 15 species included all members of the HOX gene family (Figure 1). These genes belong to four HOX gene clusters, each of which contains different numbers of HOX genes. Specific information on HOX genes, such as start codons, stop codons and gene lengths, is available in Supplementary Table S1.
For the red panda, only 25 genes could be downloaded from the previously published genome (Hu et al., 2017; Supplementary Table S1). We annotated an additional 14 HOX genes from the genome of the red panda. The results of sequence alignment showed high identities (average > 90%) to corresponding genes in humans (Figure 2). In total, we obtained 39 HOX genes of the red panda for subsequent evolutionary analysis.
Based on the concatenated CDSs of HOX genes of 15 species, we constructed an ML phylogenetic tree to decipher the evolutionary relationships of Carnivora species. We constructed a neighbor-joining phylogenetic tree using 4Dtv sites (Figure 1), which was consistent with the phylogenetic tree constructed based on nuclear and mitochondrial genes (Flynn et al., 2005) or on large-scale integrated analyses (Nyakatura and Bininda-Emonds, 2012; Upham et al., 2019).
By manually checking the multiple sequence alignment, we removed potential false-positive selection sites detected by PAML. When the giant panda, red panda, or both pandas were used as the foreground branch (G1-a; G1-b; G1-c), positive selection was detected for HOXA3 with one positively selected site (176:G:0.811, ω = 280.90, p < 0.001), HOXB4 (174:S:0.55, ω = 999, p < 0.001), HOXA3 (150:G:0.976*, ω = 97.02, p < 0.001) and HOXD4 (38,G:0.822, ω = 178.24, p < 0.001) (Figure 3A; Table 1).
For marine Carnivora species, when the sea otter was the foreground branch, only HOXA6 was detected to be under positive selection with one positively selected site (102:S:0.902, ω = 999, p < 0.001) (Figure 3A; Table 1). When the Weddell seal, Hawaiian monk seal, walrus or northern fur seal were used as the foreground branch separately, no significant PSGs were detected. However, we found several potential PSGs that were marginally significant, including HOXA6 (102:S:0.871; 211:D:0.813, p = 0.061) and HOXD12 (154:L:0.703, p = 0.067) when setting the Weddell seal as the foreground branch and HOXD3 (111:Q:0.833, p = 0.059) when the northern fur seal was used as the foreground branch (Figure 3A; Table 1).
Interestingly, the results of the two categories for the evolution of pinniped species were disparate (Figure 3A). Two HOX genes were identified under positive selection, with no common genes in the two categories. In category 1 (G2-e1), HOXB1 was identified as a PSG with one positively selected site (270:P:0.844, ω = 289.12, p < 0.05) during the origin and evolution of the pinnipeds. In category 2 (G2-e2), HOXB6 was identified as a PSG, with two positively selected sites (92:G:0.795; 140:S:0.998**, ω = 110.09, p < 0.001) (Figure 3A, Table 1).
We further analyzed the data using an additional method implemented in HyPhy of Datamonkey (Supplementary Figure S1 and Supplementary Table S2). When the giant panda was set as the foreground branch, three, seven and nine genes were detected at the gene, branch, and site levels, respectively. After integration, five genes, including eight sites, were detected under positive selection (Supplementary Tables S2–S4). In particular, HOXB3 was detected at all three levels. When the red panda was set as the foreground branch, we detected only one PSG (HOXB3), including two sites, at the branch and site levels (Supplementary Table S3). When both the giant and red pandas were set as the foreground branch, the integrated results showed that four PSGs were detected, including HOXA3, HOXB3, HOXD11, and HOXD12 (Supplementary Table S2). Within these genes, 7 sites were detected by at least two site-based methods (Table 1 and Supplementary Table S4). Codon 150 of HOXA3 was also detected in the PAML analysis.
When the sea otter was set as the foreground branch, HOXA6 was detected as a PSG by gene-level and branch-level HyPhy analyses (Supplementary Table S2) and in the PAML analysis (Table 1). When the Weddell seal was used as the foreground branch, two PSGs, HOXB6 and HOXC13, were detected by integrated methods (Supplementary Table S2). No PSGs were found when the Hawaiian monk seal, northern fur seal or walrus was set as the foreground branch.
When combining the pinniped species together, we detected common PSGs (HOXA6, HOXB6 and HOXC13) in either category 1 or category 2 (Supplementary Table S2). Additionally, codon 270 of HOXB1 was detected both in our site-based results and in PAML, and codon 140 of HOXB6 was found in the integrated analysis and in PAML (Table 1; Supplementary Tables S2–S4). All the above results suggested that a small number of HOX genes and sites were under positive selection in Carnivora.
To determine whether there was convergent evolution of HOX genes between the giant and red pandas and between the pinnipeds and sea otter, we identified the signatures of amino acid convergence in the above pairs. For the giant and red pandas, only one convergent amino acid change was detected for HOXC10. The observed number of convergence events (1) was significantly higher than the number of convergence events expected based on the phylogenetic distance (0.2771; p = 0.0307). The amino acid sequence alignment identified an amino acid substitution of Lys236Gln, with a codon change from AAA to CAA in the giant panda and from AAA to CAG in the red panda (Figure 4A). This substitution was also found to be unique to both pandas when not considering the amino acids of ancestral branches (Supplementary Table S5). In addition, in the gene alignment of 56 mammals, the amino acid at the 236 position is Gln only in the giant and red pandas, whereas this position is Lys in other species, highlighting the potential functional role (Figure 4A).
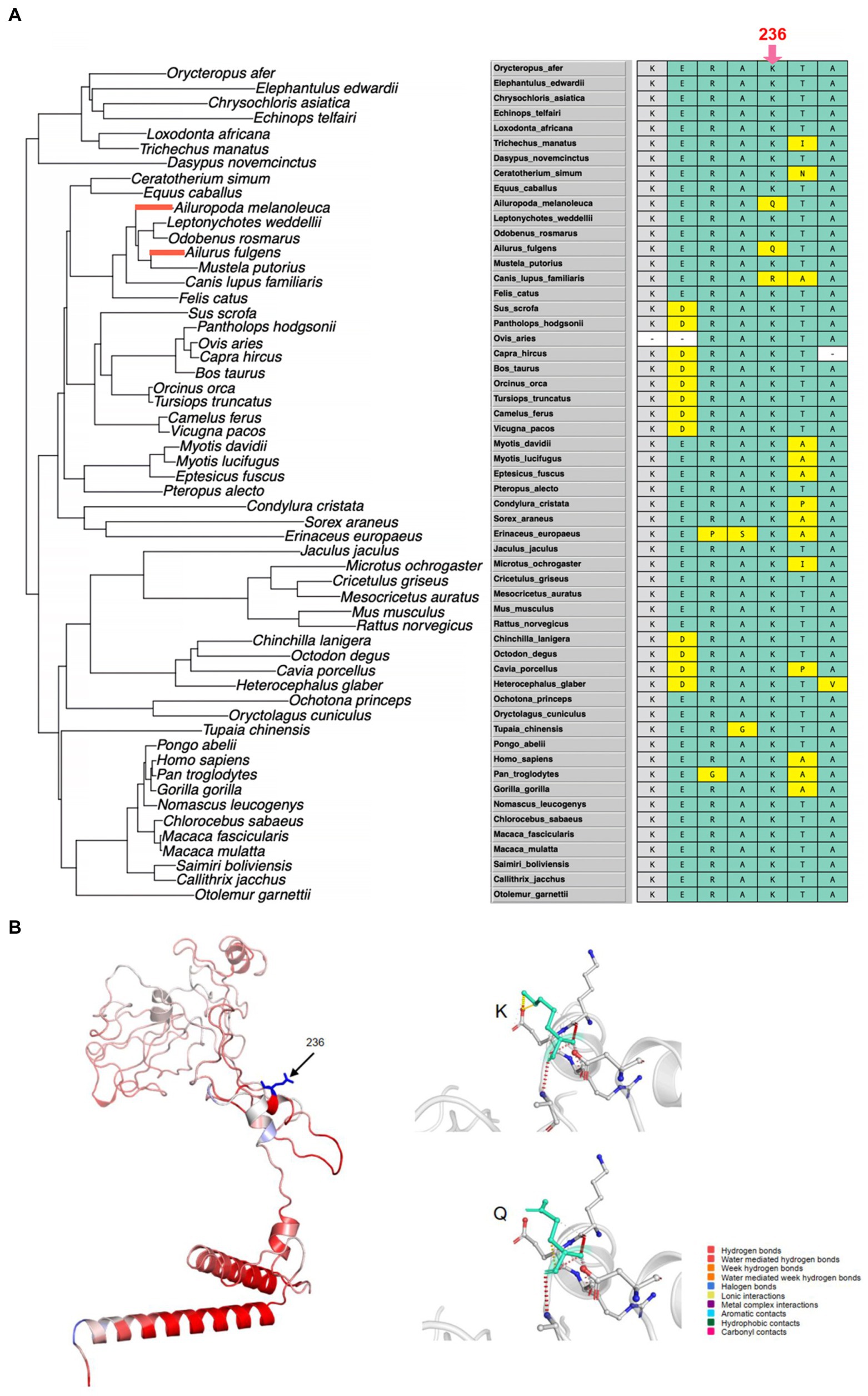
Figure 4. The convergent evolution of HOXC10 in the giant panda and red panda. (A) Left: The phylogenetic tree with the giant and red pandas highlighted by red branches. Right: Multiple sequence alignment of HOXC10 and the convergent amino acid change (Lys236Gln) between the two pandas indicated by the red arrow. (B) Left: The 3D structure of HOXC10 built by using a deep-learning-based method, trRosetta. The amino acids are colored according to the vibrational entropy change upon mutation. Blue represents rigidification of the structure, and red represents a gain in flexibility. Right: The results of interatomic interaction prediction. Wild-type and mutant residues are colored light green and are also represented as sticks alongside the surrounding residues that are involved in any type of interaction.
For the pinnipeds and sea otter, we did not detect convergent amino acid changes in any HOX gene. They did not have any common amino acids when only the extant lineages were compared. However, the four pinniped species had 13 common amino acids from 10 genes that distinguished them from other species, including the limb development-related genes HOXA10, HOXA13, HOXB9, HOXB13, HOXC10, and HOXD12 (Supplementary Table S5).
We detected rapid evolution and negative selection of HOX genes (Table 2; Supplementary Tables S6–S9). With the giant panda as the foreground branch, we identified two rapidly evolving genes, HOXA3 and HOXD4. With the red panda as the foreground branch, HOXD4 was shown to be under rapid evolution. When both giant and red pandas were used as the foreground branch, HOXA3 was detected to be rapidly evolving. When any one of four pinnipeds (Weddell seal, Hawaiian monk seal, walrus, northern fur seal) or sea otter was set as the foreground branch, no rapidly evolving genes were detected. When four pinniped species were used as the foreground branch simultaneously, HOXC13 was detected to be under rapid evolution (Supplementary Tables S6, S7).
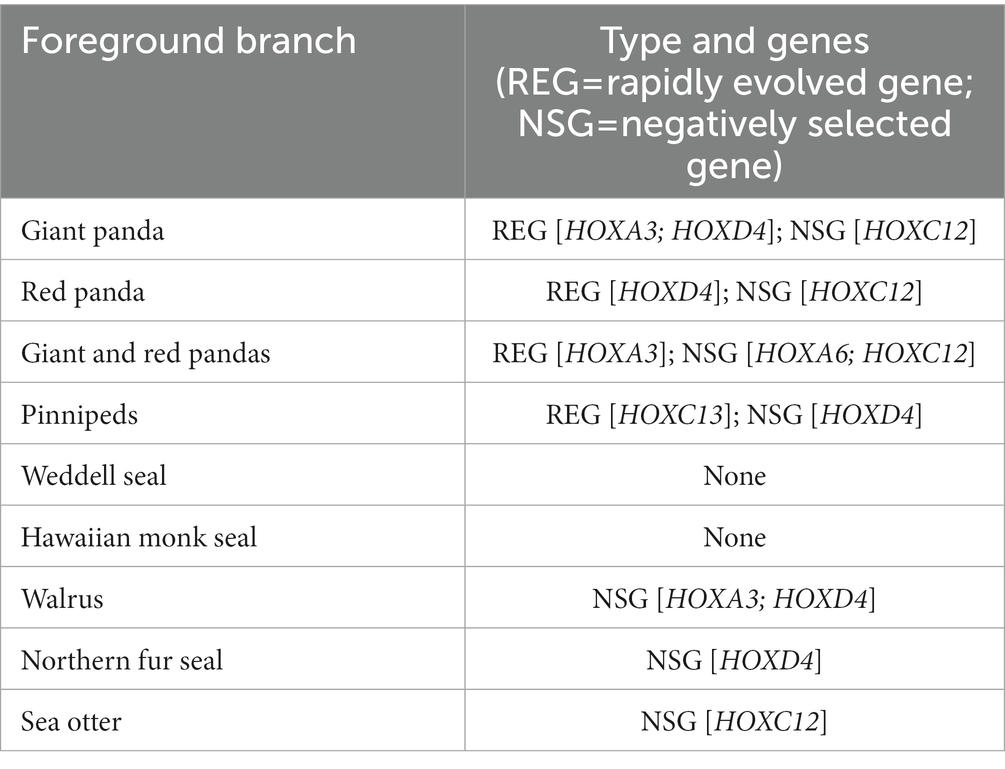
Table 2. Rapid evolution and negative selection based on the test results of both the two-ratio vs. one-ratio model and the two-ratio vs. free-ratio model.
We identified four genes under negative selection (Table 2; Supplementary Tables S8, S9), including HOXC12 for the giant panda, the red panda, both pandas, and the sea otter as the foreground branch; HOXA6 for both giant and red pandas; HOXD4 for four pinniped species, the walrus, and northern fur seal, respectively; and HOXA3 for the walrus.
For the positively selected gene HOXA3, the effect of the amino acid mutation was predicted to be neutral using either the giant panda or red panda protein sequence. The ‘neutral’ outcome was also predicted for HOXC10, a gene convergently evolved between giant and red pandas (Table 3). According to the Pfam database, the convergent amino acid substitution is not located in the Homeodomain. Furthermore, we constructed the 3D structure of HOXA3 and HOXC10 proteins using trRosetta modeling, a deep-learning method, and predicted the dynamic effects of the mutations. For HOXA3, the giant panda G338S mutation destabilized the HOXA3 protein by increasing the molecular flexibility (0.319 kcal/mol/k) and shifting the Gibbs free energy (ΔΔG) value to the negative range (−0.374 kcal/mol) (Figure 2B; Table 3). When using the red panda protein sequence, the G338A mutation decreased molecular flexibility (−0.547 kcal/mol/k) and stabilized the protein. For HOXC10, the K236Q mutation destabilized the protein (−0.085 kcal/mol) but decreased the molecular flexibility (−0.159 kcal/mol/k) (Figure 4B; Table 3).
HOX genes regulate many aspects of embryonic body plan development and patterning in vertebrates (Casaca et al., 2014). They not only ensure the individual developmental program but also regulate and change small developmental traits to adapt individuals to their living environment (Akam, 1998). Our analysis detected rare natural selection in HOX genes in Carnivora, which reflected the strong conservation of HOX genes across a number of species. The findings also provide insights into the common and divergent molecular evolutionary features of HOX gene evolution in Carnivora.
HOXC10, a DNA-binding transcription activator, plays a key role in anterior/posterior pattern specification, proximal/distal pattern formation, embryonic limb morphogenesis, and skeletal system development. Similar to other HOX genes, the expression of HOXC10 has a very broad pattern in terms of temporal and spatial extent at different stages of embryonic development depending on the cell environment and internal state (Wellik and Capecchi, 2003). HOX9 and HOX10 function together to pattern forelimb stylopods, and HOX10 also affects certain phenotypes in zeugopods (Wellik and Capecchi, 2003). Based on Bgee (Bastian et al., 2021), HOXC10 is highly expressed in the triceps brachii, biceps brachii, cartilage tissue, trabecular bone tissue, and upper arm skin in humans.
The two pandas are not closely related, and their sharing of adaptive traits reflects convergent evolution. Sesamoid bones are small auxiliary bones that form near joints and contribute to their stability and function (Antón et al., 2006). Thus far, providing a comprehensive developmental model or classification system for this highly diverse group of bones has been challenging. Based on the latest research, sesamoid bones are regulated by both TGFβ and BMP signaling pathways, and both genetic and mechanical regulation are involved in facilitating developmental diversity (Eyal et al., 2019). All types of sesamoid bones originate from SOX9+/SCX+ progenitors under the regulation of TGFβ and are independent of mechanical stimuli from muscles (Eyal et al., 2019). Similarly, HOXC10 and SOX9 were found to be enriched in the knee at some overlapping time points (Pazin et al., 2012), and HOXC10 is involved in the TGFβ/BMP and Wnt signaling pathways. In addition, PITX1 also strongly associates with many functionally verified limb enhancers that exhibit similar levels of activity in the embryonic mesenchyme of forelimbs and hindlimbs (Park et al., 2014). PITX1 can induce the expression of TBX4, HOXC10 and HOXC11 in chick forelimbs and the expression of TBX4 and HOXC10 in mouse forelimbs (Logan and Tabin, 1999). TBX4, TBX5, and HOX cluster genes are crucial for forelimb development, and mutations in these genes are responsible for congenital limb defects (Jain et al., 2018). TBX4 and HOXC10 interact directly in limbs and synergistically activate transcription via a T-box–HOX composite DNA sequence, and the transcriptional activities of TBX4 and HOXC10 depend on their DNA-binding sites (Jain et al., 2018). This suggests that HOXC10 might play a role in the development of pseudothumbs. In addition, pinnipeds and manatees (belonging to the family Trichechidae) underwent parallel evolution of HOXC10 (Li et al., 2018), which implies a potential important function.
Furthermore, no direct studies have shown that HOXC10 interacts with PCNT or DYNC2H1, two genes that were identified as possibly related to pseudothumb development in a previous convergent evolution study of giant and red pandas (Hu et al., 2017). Thus, HOXC10 could be another candidate gene for pseudothumb development for future functional verification.
Generally, the rates of nonsynonymous and synonymous substitutions in HOX genes are relatively low due to the strong conservation of the HOX gene family. In this study, we identified only one gene, HOXA3, under positive selection in pandas. HOXA3 mutations may be related to parathyroid gland organogenesis and pharyngeal organ development (Gordon, 2018). Furthermore, rapid evolution analysis found that giant and red pandas had a common rapidly evolving gene, HOXD4. Previous studies suggested that HOXD4 acts in parallel to regulate the expression of target genes directing skeletogenesis (Folberg et al., 1999). HOXD4-transgenes are specifically activated in chondrocytes, and mutations in this gene cause severe cartilage defects due to delays in cartilage maturation (Kruger and Kappen, 2010).
For the negatively selected gene HOXC12, there have been few relevant studies. HOXC12 has undergone strong purifying selection, which suggests that mutations in this gene may be harmful to organisms. In addition, HOXA6 was detected to be under purifying selection when both pandas were considered as the foreground branch. Previous studies reported that HOXA6 is expressed at a high level in several types of malignant tumors (Dickson et al., 2013). Whether these genes are involved in body or limb development needs further study.
We found no signatures of positive selection or convergent amino acid substitutions of HOX genes between the sea otter and pinnipeds. This suggests that the phenotypic convergence of marine Carnivora species may be achieved through gene expression or regulatory region variations of HOX genes or the evolution of other relevant genes. Similarly, few signatures of common positive selection on HOX genes were detected across three marine mammalian lineages (pinnipeds, cetaceans, and sirenians), and convergence occurred at a functional level of HOX genes (Nery et al., 2016).
However, focusing only on the pinnipeds, HOXB1 was identified to be under positive selection during the origin and evolution of pinnipeds. This gene is part of a developmental regulatory system that provides cells with specific positional identities on the anterior–posterior axis (from the UniProt database). HOXB6 was identified as a positively selected gene when each pinniped species was set as the foreground branch. Regarding purifying selection, HOXC13 was detected as a negatively selected gene in the ancestral linage of four pinniped species. Studies have shown that HOXC13 has high expression in integument development, and its mutations are associated with skin and appendage development (Wu et al., 2013). Interestingly, this gene showed a rapid evolution signature in the order Sirenia (Wang et al., 2009). A cetacean study showed that the evolution of the cetacean forelimb may be associated with the positive selection or selective relaxation of HOXD12 and HOXD13 (Li et al., 2018).
Although selection pressure appears to have varied among different lineages, HOXD4 was under strong purifying selection in the northern fur seal and walrus lineages, even in the ancestral linage of the four pinniped species, suggesting that HOXD4 may have been under purifying selection over a long evolutionary time. HOXA3 was detected as a negatively selected gene for the northern fur seal. HOXC12 was subject to negative selection in the sea otter lineage and was also a negatively selected gene for both pandas, suggesting that its functional relaxation could be more strictly constrained.
In summary, our study explored the molecular evolution of HOX genes in Carnivora and focused on the potential relationship between HOX9 ~ 13 genes and limb development, providing insights into the potential molecular evolutionary mechanisms of Carnivora limb development. Overall, a few HOX genes undergo positive selection or convergent evolution, most likely because of the functional importance and evolutionary conservativeness of HOX genes. In our study, the identified PSGs and convergently evolved genes among HOX9 ~ 13 genes could be important candidate targets for further functional verification. A combination of evolutionary analyses and functional verification would illuminate the mechanisms of evolutionary developmental biology for specialized limbs or appendages (Hu et al., 2023).
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethical review and approval was not required for the animal study because We just analyzed the genome data of vertebrate animals.
YH designed the study. WF and SM analyzed the data. WF, KL, FW, and YH discussed and interpreted the data. WF, YH, and KL wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (31821001), the Key Project of Science and Technology Department of Qinghai Province, and the Youth Innovation Promotion Association, CAS (Y202026).
We thank Qi Wu for his well-organized UCSC Database and the suggestions about convergence analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1107034/full#supplementary-material
1. ^http://www.orthomam.univ-montp2.fr/orthomam/html/
2. ^http://tree.bio.ed.ac.uk/software/figtree/
3. ^https://hgdownload.soe.ucsc.edu/goldenPath/hg19/multiz100way/
Akam, M. (1998). HOX genes: from master genes to micromanagers. Curr. Biol. 8, R676–R678. doi: 10.1016/S0960-9822(98)70433-6
Antón, M., Salesa, M. J., Pastor, J. F., Peigné, S., and Morales, J. (2006). Implications of the functional anatomy of the hand and forearm of Ailurus fulgens (Carnivora, Ailuridae) for the evolution of the ‘false-thumb’ in pandas. J. Anat. 209, 757–764. doi: 10.1111/j.1469-7580.2006.00649.x
Arnason, U., Gullberg, A., Janke, A., Kullberg, M., Lehman, N., Petrov, E. A., et al. (2006). Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol. Phylogenet. Evol. 41, 345–354. doi: 10.1016/j.ympev.2006.05.022
Bastian, F. B., Roux, J., Niknejad, A., Comte, A., Fonseca Costa, S. S., De Farias, T. M., et al. (2021). The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals. Nucleic Acids Res. 49, D831–D847. doi: 10.1093/nar/gkaa793
Berta, A., Churchill, M., and Boessenecker, R. W. (2018). The origin and evolutionary biology of pinnipeds: seals, sea lions, and walruses. Annu. Rev. Earth Planet. Sci. 46, 203–228. doi: 10.1146/annurev-earth-082517-010009
Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and Genomewise. Genome Res. 14, 988–995. doi: 10.1101/gr.1865504
Casaca, A., Santos, A. C., and Mallo, M. (2014). Controlling HOX gene expression and activity to build the vertebrate axial skeleton. Dev. Dyn. 243, 24–36. doi: 10.1002/dvdy.24007
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chen, Y., Ye, W., Zhang, Y., and Xu, Y. (2015). High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 43, 7762–7768. doi: 10.1093/nar/gkv784
Choi, Y., and Chan, A. P. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. doi: 10.1093/bioinformatics/btv195
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Davis, A. P., and Capecchi, M. R. (1994). Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development 120, 2187–2198. doi: 10.1242/dev.120.8.2187
Dickson, G. J., Liberante, F. G., Kettyle, L. M., O’Hagan, K. A., Finnegan, D. P., Bullinger, L., et al. (2013). HOXA/PBX3 knockdown impairs growth and sensitizes cytogenetically normal acute myeloid leukemia cells to chemotherapy. Haematologica 98, 1216–1225. doi: 10.3324/haematol.2012.079012
Douzery, E. J., Scornavacca, C., Romiguier, J., Belkhir, K., Galtier, N., Delsuc, F., et al. (2014). OrthoMaM v8: a database of orthologous exons and coding sequences for comparative genomics in mammals. Mol. Biol. Evol. 31, 1923–1928. doi: 10.1093/molbev/msu132
Eyal, S., Rubin, S., Krief, S., Levin, L., and Zelzer, E. (2019). Common cellular origin and diverging developmental programs for different sesamoid bones. Development 146:dev167452. doi: 10.1242/dev.167452
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fish, F. E., and Lauder, G. V. (2017). Control surfaces of aquatic vertebrates: active and passive design and function. J. Exp. Biol. 220, 4351–4363. doi: 10.1242/jeb.149617
Flynn, J. J., Finarelli, J. A., Zehr, S., Hsu, J., and Nedbal, M. A. (2005). Molecular phylogeny of the Carnivora (Mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst. Biol. 54, 317–337. doi: 10.1080/10635150590923326
Folberg, A., Nagy Kovács, E., Luo, J., Giguère, V., and Featherstone, M. S. (1999). RARβ mediates the response of HOXD4 and HOXB4 to exogenous retinoic acid. Dev. Dyn. 215, 96–107. doi: 10.1002/(SICI)1097-0177(199906)215:2<96::AID-DVDY2>3.0.CO;2-T
Gertz, E. M., Yu, Y. K., Agarwala, R., Schäffer, A. A., and Altschul, S. F. (2006). Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4:41. doi: 10.1186/1741-7007-4-41
Goodman, F. R. (2002). Limb malformations and the human HOX genes. Am. J. Med. Genet. 112, 256–265. doi: 10.1002/ajmg.10776
Gordon, J. (2018). HOX genes in the pharyngeal region: how HOXA3 controls early embryonic development of the pharyngeal organs. Int. J. Dev. Biol. 62, 775–783. doi: 10.1387/ijdb.180284jg
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hu, Y., Wang, X., Xu, Y., Yang, H., Tong, Z., Tian, R., et al. (2023). Molecular mechanisms of adaptive evolution in wild animals and plants. Sci. China Life Sci. 66. doi: 10.1007/s11427-022-2233-x
Hu, Y., Wu, Q., Ma, S., Ma, T., Shan, L., Wang, X., et al. (2017). Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. U. S. A. 114, 1081–1086. doi: 10.1073/pnas.1613870114
Jacob, A., Lancaster, J., Buhler, J., Harris, B., and Chamberlain, R. D. (2008). Mercury BLASTP: accelerating protein sequence alignment. ACM Trans. Reconfigurable Technol. Syst. 1, 9–44. doi: 10.1145/1371579.1371581
Jain, D., Nemec, S., Luxey, M., Gauthier, Y., Bemmo, A., Balsalobre, A., et al. (2018). Regulatory integration of HOX factor activity with T-box factors in limb development. Development 145:dev159830. doi: 10.1242/dev.159830
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kruger, C., and Kappen, C. (2010). Microarray analysis of defective cartilage in HOXC8- and HOXD4-transgenic mice. Cartilage 1, 217–232. doi: 10.1177/1947603510363005
Li, K., Sun, X., Chen, M., Sun, Y., Tian, R., Wang, Z., et al. (2018). Evolutionary changes of HOX genes and relevant regulatory factors provide novel insights into mammalian morphological modifications. Integr. Zool. 13, 21–35. doi: 10.1111/1749-4877.12271
Logan, M., and Tabin, C. J. (1999). Role of PITX1 upstream of TBX4 in specification of hindlimb identity. Science 283, 1736–1739. doi: 10.1126/science.283.5408.1736
Löytynoja, A. (2014). Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170. doi: 10.1007/978-1-62703-646-7_10
Martín-Serra, A., Figueirido, B., Pérez-Claros, J. A., and Palmqvist, P. (2015). Patterns of morphological integration in the appendicular skeleton of mammalian carnivores. Evolution 69, 321–340. doi: 10.1111/evo.12566
Mori, K., Suzuki, S., Koyabu, D., Kimura, J., Han, S. Y., and Endo, H. (2015). Comparative functional anatomy of hindlimb muscles and bones with reference to aquatic adaptation of the sea otter. J. Vet. Med. Sci. 77, 571–578. doi: 10.1292/jvms.14-0534
Murrell, B., Moola, S., Mabona, A., Weighill, T., Sheward, D., Kosakovsky Pond, S. L., et al. (2013). FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol. Biol. Evol. 30, 1196–1205. doi: 10.1093/molbev/mst030
Murrell, B., Weaver, S., Smith, M. D., Wertheim, J. O., Murrell, S., Aylward, A., et al. (2015). Gene-wide identification of episodic selection. Mol. Biol. Evol. 32, 1365–1371. doi: 10.1093/molbev/msv035
Murrell, B., Wertheim, J. O., Moola, S., Weighill, T., Scheffler, K., and Kosakovsky Pond, S. L. (2012). Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8:e1002764. doi: 10.1371/journal.pgen.1002764
Nery, M. F., Borges, B., Dragalzew, A. C., and Kohlsdorf, T. (2016). Selection on different genes with equivalent functions: the convergence story told by HOX genes along the evolution of aquatic mammalian lineages. BMC Evol. Biol. 16:113. doi: 10.1186/s12862-016-0682-4
Nyakatura, K., and Bininda-Emonds, O. R. (2012). Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biol. 10:12. doi: 10.1186/1741-7007-10-12
Park, S., Infante, C. R., Rivera-Davila, L. C., and Menke, D. B. (2014). Conserved regulation of HOXC11 by PITX1 in Anolis lizards. J. Exp. Zool. B Mol. Dev. Evol. 322, 156–165. doi: 10.1002/jez.b.22554
Pazin, D. E., Gamer, L. W., Cox, K. A., and Rosen, V. (2012). Molecular profiling of synovial joints: use of microarray analysis to identify factors that direct the development of the knee and elbow. Dev. Dyn. 241, 1816–1826. doi: 10.1002/dvdy.23861
Pond, S. L., Frost, S. D., and Muse, S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. doi: 10.1093/bioinformatics/bti079
Prakash, A., Jeffryes, M., Bateman, A., and Finn, R. D. (2017). The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinformatics 60:3.15.1–3.15.23. doi: 10.1002/cpbi.40
Quinonez, S. C., and Innis, J. W. (2014) Human HOX gene disorders. Mol. Genet. Metab., 111(1), 4–15, doi: 10.1016/j.ymgme.2013.10.012
Reidenberg, J. S. (2007). Anatomical adaptations of aquatic mammals. Anat. Rec. 290, 507–513. doi: 10.1002/ar.20541
Roelen, B. A., de Graaff, W., Forlani, S., and Deschamps, J. (2002). HOX cluster polarity in early transcriptional availability: a high order regulatory level of clustered HOX genes in the mouse. Mech. Dev. 119, 81–90. doi: 10.1016/S0925-4773(02)00329-5
Ruddle, F. H., Bartels, J. L., Bentley, K. L., Kappen, C., Murtha, M. T., and Pendleton, J. W. (1994). Evolution of HOX genes. Annu. Rev. Genet. 28, 423–442. doi: 10.1146/annurev.ge.28.120194.002231
Sheth, R., Marcon, L., Bastida, M. F., Junco, M., Quintana, L., Dahn, R., et al. (2012). HOX genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science 338, 1476–1480. doi: 10.1126/science.1226804
Shubin, N. H. (2002). Origin of evolutionary novelty: examples from limbs. J. Morphol. 252, 15–28. doi: 10.1002/jmor.10017
Small, K. M., and Potter, S. S. (1993). Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Dev. 7, 2318–2328. doi: 10.1101/gad.7.12a.2318
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315
Uhen, M. D. (2007). Evolution of marine mammals: back to the sea after 300 million years. Anat. Rec. 290, 514–522. doi: 10.1002/ar.20545
Upham, N. S., Esselstyn, J. A., and Jetz, W. (2019). Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17:e3000494. doi: 10.1371/journal.pbio.3000494
Van Valkenburgh, B., and Wayne, R. K. (2010). Carnivores. Curr. Biol. 20, R915–R919. doi: 10.1016/j.cub.2010.09.013
Wang, Z., Yuan, L., Rossiter, S. J., Zuo, X., Ru, B., Zhong, H., et al. (2009). Adaptive evolution of 5′ HOXD genes in the origin and diversification of the cetacean flipper. Mol. Biol. Evol. 26, 613–622. doi: 10.1093/molbev/msn282
Weaver, S., Shank, S. D., Spielman, S. J., Li, M., Muse, S. V., and Kosakovsky Pond, S. L. (2018). Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 35, 773–777. doi: 10.1093/molbev/msx335
Wellik, D. M., and Capecchi, M. R. (2003). HOX10 and HOX11 genes are required to globally pattern the mammalian skeleton. Science 301, 363–367. doi: 10.1126/science.1085672
Wu, J., Husile, S. H., Wang, F., Li, Y., Zhao, C., and Zhang, W. (2013). Adaptive evolution of HOXC13 genes in the origin and diversification of the vertebrate integument. J. Exp. Zool. B Mol. Dev. Evol. 320, 412–419. doi: 10.1002/jez.b.22504
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, J., Anishchenko, I., Park, H., Peng, Z., Ovchinnikov, S., and Baker, D. (2020). Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U. S. A. 117, 1496–1503. doi: 10.1073/pnas.1914677117
Yang, Z., and dos Reis, M. (2011). Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 28, 1217–1228. doi: 10.1093/molbev/msq303
Keywords: HOX, Carnivora, pseudothumb, flipper, adaptive evolution
Citation: Fang W, Li K, Ma S, Wei F and Hu Y (2023) Natural selection and convergent evolution of the HOX gene family in Carnivora. Front. Ecol. Evol. 11:1107034. doi: 10.3389/fevo.2023.1107034
Received: 24 November 2022; Accepted: 24 February 2023;
Published: 16 March 2023.
Edited by:
Hector Escriva, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
David Ellard Keith Ferrier, University of St Andrews, United KingdomCopyright © 2023 Fang, Li, Ma, Wei and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yibo Hu, eWJodUBpb3ouYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.