- 1College of Plant Protection/Key Laboratory of Green Prevention and Control of Tropical Plant Diseases and Pests, Ministry of Education, Hainan University, Haikou, China
- 2Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 3Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 4Laboratory of Insect Behavior and Evolutionary Ecology, College of Life Science and Technology, Central South University of Forestry and Technology (CSUFT), Changsha, China
Alligator weed Alternanthera philoxeroides is a perennial, worldwide pernicious weed. The beetle Agasicles hygrophila is considered to be a classical biological agent used to control A. philoxeroides. In the insect peripheral olfactory system, the odorant receptor co-receptor (ORco) plays an important function in the perception of odors in insects. However, the function of ORco in the mating and host-finding behaviors of A. hygrophila remains unclear. In this study, we characterized the odorant receptor co-receptor of A. hygrophila (AhygOrco). Real-time quantitative PCR (qRT–PCR) showed that AhygOrco was predominantly expressed in the antennae of both male and female adults, and the difference between male and female antennae was not significant. The RNA interference (RNAi) results showed that compared to the control, the injection of AhygOrco dsRNA strongly reduced the expression of AhygOrco by 90% in male beetles and 89% in female beetles. The mate-seeking and feeding behavior of AhygOrco-silenced beetles were significantly inhibited. Male adults were significantly less successful in finding a mate compared to the control group. Furthermore, host allocation abilities toward A. philoxeroides of both adults were significantly repressed. These results indicated that AhygOrco is associated with A. hygrophila feeding and mate-seeking and that inhibition of AhygOrco expression is one of the causes of reduced host and mate recognition in A. hygrophila. Meanwhile, the study provides support for exploring gene functions based on RNAi.
1. Introduction
Almost all animals respond to pheromones and other chemical signals (Suh et al., 2014) that indicate food, shelter and predators, and they rely heavily on their olfactory system to achieve this process (Wyatt, 2003). The main olfactory structures in Lepidoptera are the antennae and labial palps. Odors are detected in these structures mainly through chemosensory receptors, belonging to the olfactory receptors (ORs) and ionotropic receptors (IRs) of the family of membrane proteins (Larsson et al., 2004; Benton, 2006; Benton et al., 2009). These receptors form ligand-gated ion channels in vivo and can be expressed in different heterologous expression systems in order to study their role (Sato et al., 2008; Wicher et al., 2009; Carey et al., 2010; Wang et al., 2010).
Insect ORs are a large gene family, which is considered as the main family of receptors involved in olfactory function (Zhang and Lofstedt, 2013; Koenig et al., 2015). These genes have relatively low sequence similarity across insect taxa (Krieger et al., 2003; Hansson and Stensmyr, 2011; Zhang and Lofstedt, 2015). One particular class of ORs is called the odorant receptor co-receptors (Orco). Orco genes are highly conserved and have been shown to be homologous in most insect species. Insects use olfactory receptors to distinguish thousands of volatiles or pheromones (Hallem and Carlson, 2006). In the process of olfactory recognition, a specific OR is required together with the universal Orco protein to form the Orco-OR receptor complex as a functional tetramer ligand-gated ion channels to exercise function (Vosshall and Hansson, 2011; Nakagawa et al., 2012; Kumar et al., 2013; Missbach et al., 2014; Turner et al., 2014).
Studies exploring the structure and function of Orco have been uninterrupted, and several recent studies have shown that the structural integrity of Orco influences the channel activity of the insect Orco-OR complex (Nichols and Luetje, 2010; Jones et al., 2011; Nichols et al., 2011; Nakagawa et al., 2012). Abnormal disruptive expression of Orco significantly impairs olfactory function in insects (Zhao et al., 2011; Liu et al., 2017). Orco mutation makes insects lose their sense of smell to various odors (DeGennaro et al., 2013; Koutroumpa et al., 2016; Li et al., 2016; Yan et al., 2017), and the mutation also completely disrupts the feeding behavior of the species (Fandino et al., 2019). In addition, some studies have shown that Orco is often associated to the tissues involved in oviposition, mating, development, resistance and metabolic regulation (Libert et al., 2007; Kato and Touhara, 2009; Fan et al., 2015; Trible et al., 2017; Sun et al., 2020).
Agasicles hygrophila Selma & Vogt (Coleoptera: Chrysomelidae) is native to southern Brazil and northern Argentina and is considered a biological agent for the control of Alternanthera philoxeroides (Mart.) Griseb (Caryophyllales: Amaranthaceae; Lu et al., 2010). After 1964, A. hygrophila was introduced to the United States, Australia, New Zealand and Thailand for the biological control of A. philoxeroides and achieved great success. Chinese scientists introduced the beetle from the United States in 1986, evaluated host specificity and concluded that the insect could be safely utilized in China, followed by extensive rearing and release, biological characterization and evaluation of its control effects. It has an exclusive host recognition ability and can accurately find hosts in the field under complex odorscape (Conchou et al., 2019). However, the molecular mechanism of host recognition in A. hygrophila is still unclear. In the present study, we identified and cloned the Orco gene of A. hygrophila. Afterwards, AhygOrco was silenced by RNAi and mutants were behaviorally tested by host and mate behavioral tests. This study partialy clarified the AhygOrco functions in A. hygrophila and laid the foundation for further study of the olfactory mechanism in A. hygrophila. Because of its specificity and high efficiency, RNAi has attracted widespread attention from biologists and became one of the research hotspots in molecular biology. SiRNA-mediated gene silencing plays a pivotal role in the study of insect gene function.
2. Materials and methods
2.1. Insect rearing
Agasicles hygrophila was obtained from the Institute of Plant Protection, China Academy of Agriculture Sciences. The beetles were reared on A. philoxeroides plants at 26 ± 1°C 85% relative humidity (RH) and a 16:8 h (white light: dark) photoperiod in the laboratory. The flea beetles were cultivated for three generations to eliminate maternal effects before the experiments.
2.2. RNA extraction, cDNA synthesis, and gene cloning
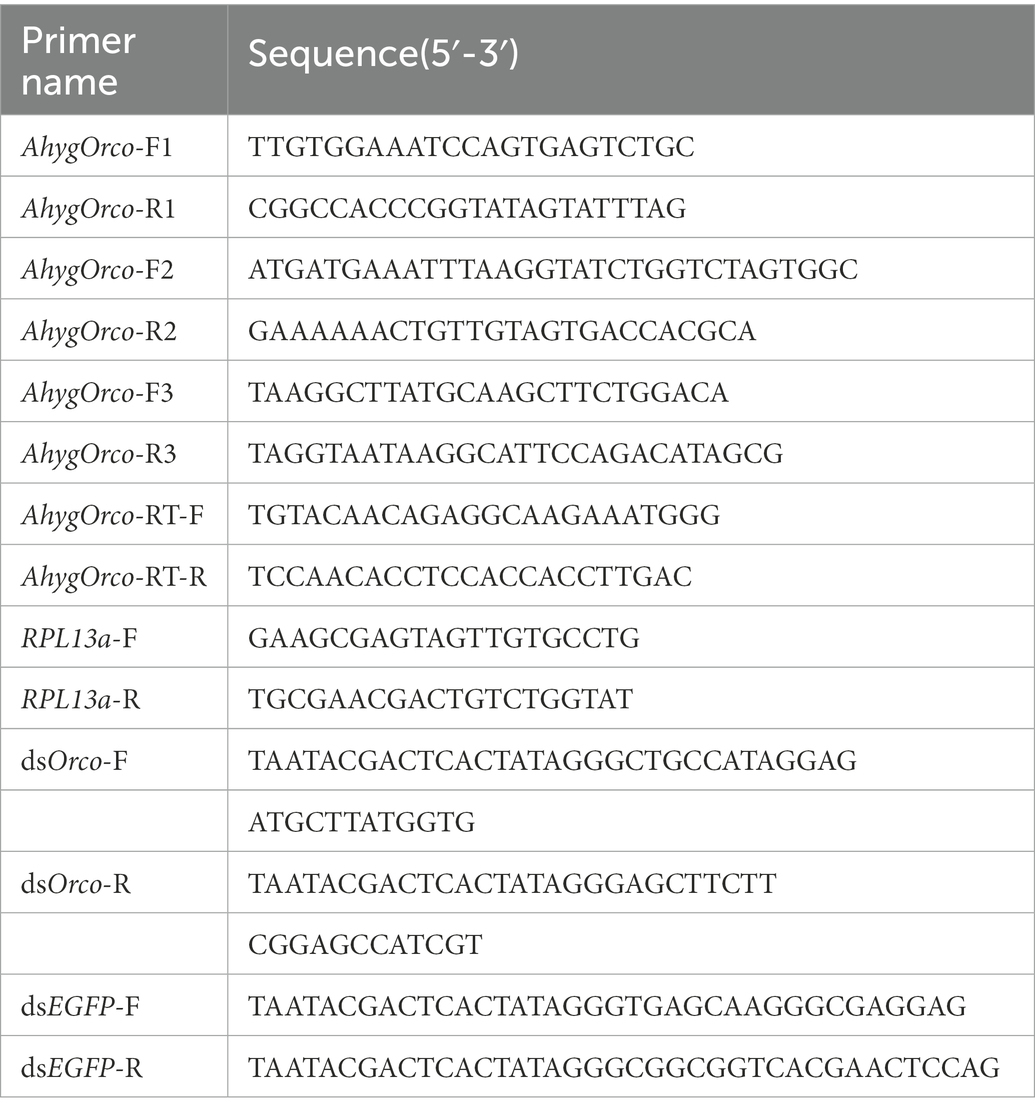
Total RNA was extracted from the dissected antennae of 30 A. hygrophila using a Micro Total RNA Extraction Kit (JianShi Biotech, Beijing, China) following the manufacturer’s protocols. The first strand of the complementary DNA (cDNA) was synthesized from 1 μg of total RNA using a Hifair® III 1st Strand cDNA Synthesis Kit (Yeasen Biotech, Shanghai, China) according to the manufacturer’s protocol. The synthesized cDNAs were stored at −20°C until use. To identify the AhygOrco sequence, three pairs of degenerate primers (AhygOrco-F1: 5′-TTGTGGAAATCCAGTGAGTCTGC-3′, AhygOrco-R1: 5′-CGGCCACCCGGTATAGTATTTAG-3′, AhygOrco-F2: 5′-ATGATGAAATTTAAGGTATCTGGTCTAGTGGC-3′, AhygOrco-R2: 5′-GAAAAAACTGTTGTAGTGACCACGCA-3′, AhygOrco-F3: 5′-TAAGGCTTATGCAAGCTTCTGGACA-3′, AhygOrco-R3: 5′-TAGGTAATAAGGCATTCCAGACATAGCG-3′, Table 1) were designed to amplify the nucleic acid sequence using 2 × Hieff Canace® Gold PCR Master Mix (Yeasen Biotech, Shanghai, China). The amplification was performed under the following thermal program: 98°C for 3 min, 35 cycles at 98°C for 10 s, 55°C for 20 s and 72°C for 45 s, followed by 1 cycle at 72°C for 5 min. The PCR product was purified using a DNA Gel Extraction Kit (Beyotime Biotech, Shanghai, China), cloned into a pClone007 Versatile Simple Vector (Tsingke Biotech, Beijing, China) using a Trelief™ 5α Chemically Competent Cell (Tsingke Biotech, Beijing, China) and sequenced.
2.3. Bioinformatic analysis
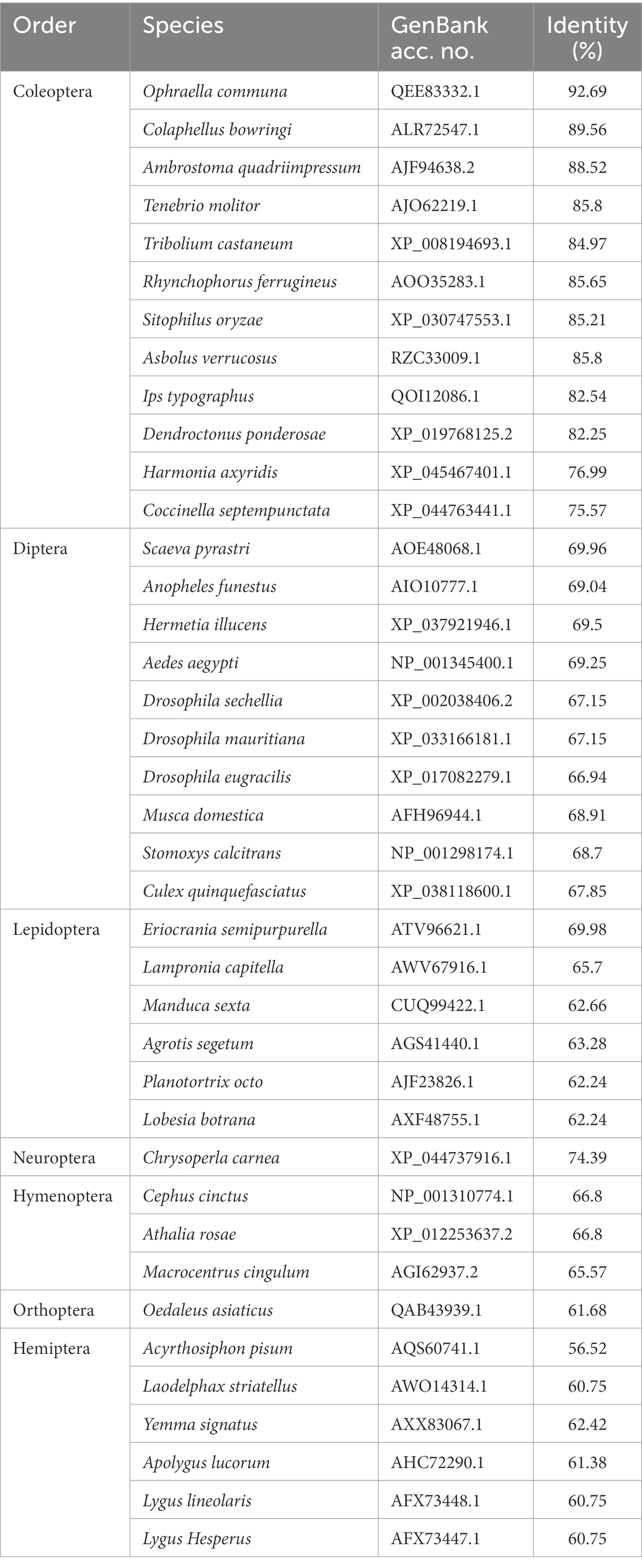
Searching for orthologs of AhygOrco was performed using BLAST.1 Multiple sequence alignment of AhygOrco and orthologs from four other Coleopteran insects was performed with DNAMAN software. The transmembrane domain and topology of the deduced AhygOrco protein were predicted using TMHMM.2 The amino acid sequences of AhygOrco and its orthologs from other insect species were aligned using the ClustalW method implemented in the MEGA X software package. The phylogenetic tree was constructed by the neighbor-joining (NJ) method (Saitou and Nei, 1987) with P-distance modeling (Nei and Kumar, 2000) and partial deletion of gaps using the MEGA X software package (Kumar et al., 2018). The reliability of the tree structure was assessed using a bootstrap procedure based on 1,000 replicates. The Orco amino acid sequences of insects are listed in Appendix B.
2.4. Expression profiling of AhygOrco
The expression profiles of AhygOrco were analyzed using qRT–PCR. Total RNA was isolated from 12 different tissues, including male and female antennae (AN), male and female heads without antennae (H), male and female abdomens (AB), female ovaries (OV), male testes (TE), male and female thoraxes (T), and male and female legs (L), obtained from virgin adult beetles of both sexes within 48 h of adult eclosion. The method of total RNA extraction was the same as described above. The concentration of each RNA sample was standardized to 500 ng/μl, and cDNA was synthesized using a Hifair® III 1st Strand cDNA Synthesis SuperMix for qPCR Kit (Yeasen Biotech, Shanghai, China) according to the manufacturer’s protocol. Based on studies in other Coleoptera (Zhang Y. et al., 2021), ribosomal protein L13a (RPL13a; Jia et al., 2020) was used as an internal control in the present study. Antennal cDNA at 10-fold dilution for qRT–PCR testing. qRT–PCR was performed using ABI StepOne Plus (Thermo Scientific, Waltham, MA, USA) with Hieff® qPCR SYBR Green Master Mix (Yeasen Biotech, Shanghai, China). The reaction program was set as follows: 5 min at 95°C, 40 cycles at 95°C for 10 s and 60°C for 30 s, and 20 s at 72°C. The qRT–PCR primers were designed using Primer Premier 5.0 (PREMIER Biosoft International; AhygOrco -RT-F: 5′- TGTACAACACGAGGCAAGAAATGGG-3′, AhygOrco -RT-R: 5′- TCCAACACCTCCACCACCTTGAC-3′, RPL13a-F: 5′- GAAGCGAGTAGTT-GTGCCTG-3′, RPL13a-R: 5′-TGCGAACGACTGTCTGGTAT-3′, Table 1), and the efficiency of the primers was validated before gene expression analysis. Each qRT–PCR was performed using two technical replicates and three biological replicates. The expression levels of AhygOrco were calculated by the 2 − ΔΔCt method (Livak and Schmittgen, 2001).
2.5. dsRNA synthesis, microinjection, and qRT–PCR validation
Based on the cDNA template, a pair of primers (Saleh et al., 2006; Xiao, 2014; Wang, 2019) containing the T7 RNA polymerase promoter (dsOrco-F: 5′- TAATACGACTCACTATAGGGCTGCCATAGGA-GATGCTTATGGTG-3′, dsOrco-R: 5′- TAATACGACTCACTATAGGGAGCTTCTTCG-GAGCCATCGT-3′, Table 1) was amplified using a 2 × Hieff Canace® Gold PCR Master Mix (Yeasen Biotech, Shanghai, China). The amplification was performed under the following thermal program: 98°C for 3 min, 35 cycles at 98°C for 10 s, 55°C for 20 s and 72°C for 45 s, followed by one cycle at 72°C for 5 min. After PCR amplification using the primers, the obtained targeting fragment was used to synthesize dsOrco with a T7 RNAi Transcription Kit (Vazyme Biotech, Nanjing, China) according to the manufacturer’s instructions. In addition, the dsRNA of the enhanced green fluorescent protein (dsEGFP, the sequence of EGFP is shown in Appendix A, the primers of dsEGFP are shown in Table 1, GenBank acc. No.: MN830806.1) was also synthesized as the control treatment. The concentrations of the synthesized dsOrco and dsEGFP were diluted to 10 μg/μL and stored at −20°C until use. The newly emerged A. hygrophila were separated into male and female beetles and were used for RNAi microinjection. Newly emerged A. hygrophila male adults (within 12 h after eclosion) were placed on a soft plastic plate with their abdomens facing up. Then, 0.1 μl of dsOrco or dsEGFP was injected into the junction site between the first and the second abdominal segment of each individual using a R480 Pico-Injector (RWD Life Science Co. Ltd., Shenzhen, China). After injection, insects were allowed to recover in plastic boxes with A. philoxeroides. After 48 h of injection, the antennae were pulled off with tweezers by grasping at the root of the antennae and were subsequently transferred to a 1.5 ml centrifuge tube. The methods of total RNA extraction and mRNA level validation were performed as described above.
2.6. Behavioral tests after RNAi
Behavioral test on feeding. After 48 h of injection, insects from the dsOrco-injected and dsEGFP-injected groups were individually transferred to one side of a square plastic dish (10 × 10 × 1.7 cm, Figure 1A). Prior to this, a fresh leaf of A. philoxeroides was placed on the opposite side of the dish. The time period from the introduction of an insect to successful leaf contact was measured. If an insect failed to touch the leaf within 2 min, it was judged unable to recognize the feeding (Liu et al., 2016). Insects that had not made a position move within 2 min were abandoned. Behavioral tests were performed on each individual one by one. And the number of preferences for host leaf in a group with 30 individuals of each gender was counted to calculate a success rate (SR), the number (N) of preferences for host leaf as a fraction of the total beetles SR = N/30. The experiment was repeated three times to avoid random error, as a result, a total of 90 individuals of each gender were used. After one completion of the test, the dish was washed with 75% ethanol. The whole bioassays were performed under white lighting at 26 ± 1°C and 85% relative humidity.
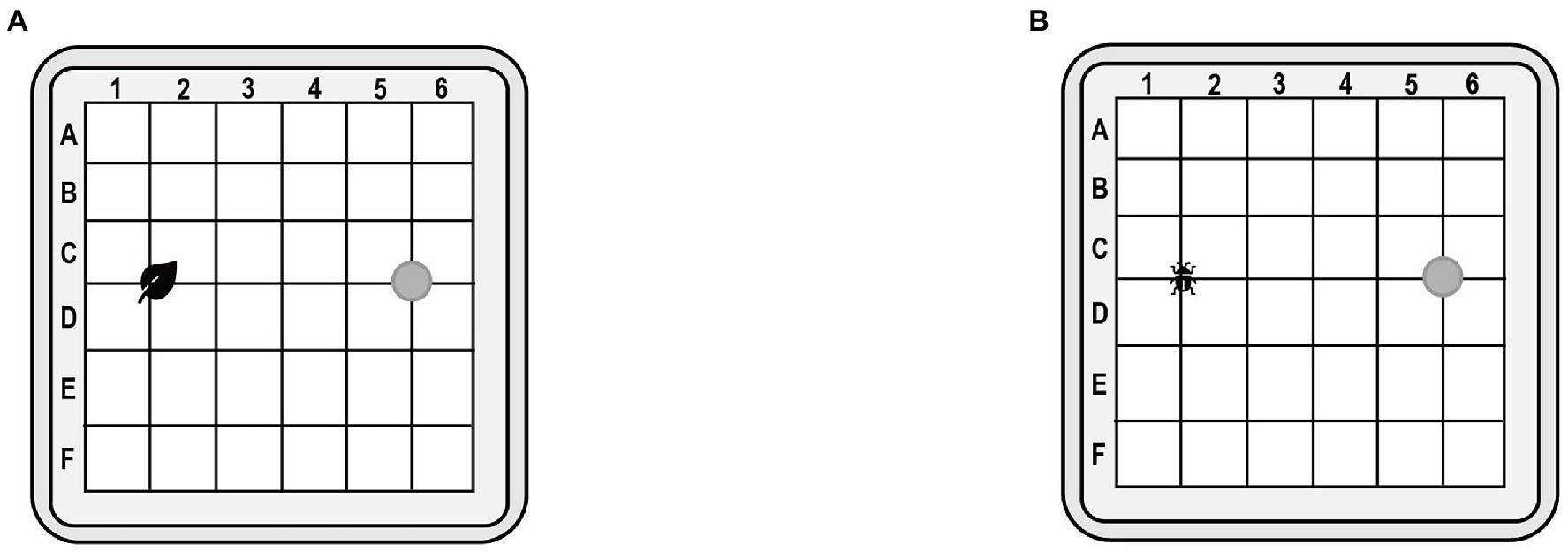
Figure 1. (A) Behavioral test device on feeding. (B) Behavioral test device on mate-finding. Test insects are placed at the gray origin.
Behavioral test on mate-seeking. After 48 h of injection, insects from the dsOrco-injected and dsEGFP-injected groups were individually transferred to one side of a square plastic dish (10 × 10 × 1.7 cm, Figure 1B). Prior to this, an unmated female adult (3 d after eclosion) was fixed on the opposite side of the dish by using double-sided adhesive tape. The time period from the introduction of a male insect to successful contact with the body of the female was measured. If an insect failed to touch the body of the opposite sex within 60 s, it was judged unable to recognize the mate. If an insect failed to touch the body of the female within 60 s, it was judged unable to recognize the mate. Insects that had not made a position move within 60 s were abandoned. Behavioral tests were performed on each pair of A. hygrophila one by one, and the number and time spent by males to successfully found females was counted in a group of 30 pairs of A. hygrophila. In addition, a success rate (SR) was calculated as a fraction of the number of males that successfully found females (N) to the total number of male beetles SR = N/30. To avoid random errors, the experiment was repeated three times. A total of 90 pairs of A. hygrophila were used to behavioral test on mate-seeking. After one completion of the test, the dish was washed with 75% ethanol. The whole bioassays were performed under white lighting at 26 ± 1°C and 85% relative humidity. All of the behavioral tests were performed at 17:00–19:00, when the insects were most active.
2.7. Data analyses
In the behavioral test on feeding, the SR of dsOrco-injected insects was normalized to the SR of dsEGFP-injected females. And in the behavioral test on mate-seeking, the SR of dsOrco-injected insects was normalized to the SR of dsEGFP-injected insects. All data are expressed as the means ± SEMs (standard errors of the mean). Statistical analyses were performed in IBM SPSS Statistics 27. For the expression profiling of AhygOrco in different tissues, the significant differences among different samples (including males and females) were analyzed using one-way analysis of variance (ANOVA) followed by a Tukey’s post hoc test. For the statistical tests of relative expression of AhygOrco and SR of host feeding, as well as mate-seeking between dsOrco-injected insects and dsEGFP-injected insects, we performed the non-parametric tests (Mann–Whitney U test) for males and females, respectively. The level of significance was set at p < 0.05. The figures were constructed using GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA) and Adobe Illustrator CS6 (Adobe, San Jose, CA, USA).
3. Results
3.1. Identification, sequence analysis, and expression profiling of AhygOrco
AhygOrco mRNA was identified from the ovarian transcriptome (unpublished data) of A. hygrophila. Based on the cDNA template and specific primers for PCR, the AhygOrco gene was cloned successfully (the sequence of AhygOrco is shown in Appendix A, GenBank acc. No.: OP972585). The complete open reading frame (ORF) length was 1,440 bp, encoding 479 amino acids. The transmembrane prediction results showed that AhygOrco has seven putative transmembrane regions, with the N-terminus inside the cell membrane and the C-terminus outside, which is similar to a typical insect OR (Figures 2A,B). Orco genes from eight coleopteran insects were used to perform multiple sequence alignment. The identities of AhygOrco with the Orco orthologs of the other insects were in the range of 85%–95% (Table 2). The membrane topology analysis of the AhygOrco protein predicted by TMHMM2.0 indicated that this protein is a seven transmembrane protein with an intracellular N-terminus and an extracellular C-terminus (Figure 2B), which is consistent with the membrane topology of Orco protein demonstrated in L. Hesperus (Hull et al., 2012), D. melanogaster, and A. gambiae (Benton et al., 2006).
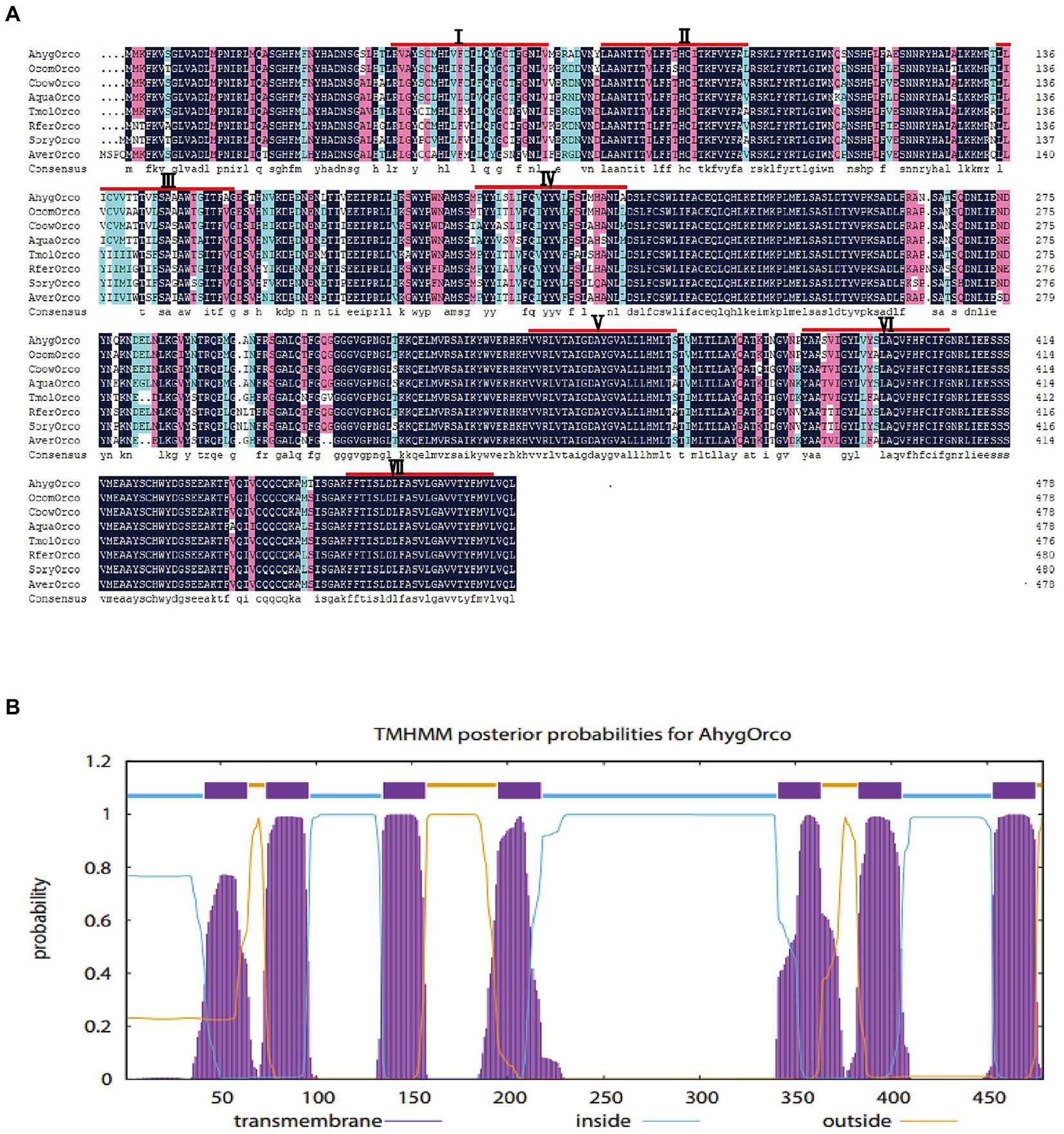
Figure 2. (A) Multiple sequence alignment of the AhygOrco-deduced amino acid sequence with the other Orco from seven Coleoptera insects (Ocom: Ophraella communa, Cbow: Colaphellus bowringi, Aqua: Ambrostoma quadriimpressum, Tmol: Tenebrio molitor, Rfer: Rhynchophorus ferrugineus, Sory: Sitophilus oryzae, Aver: Asbolus verrucosus). The transmembrane domains I–VII are indicated by red lines. (B) Prediction of transmembrane helices in the encoded amino acid sequence.
A phylogenetic tree was constructed by using 40 insect Orco protein sequences. The tree was constructed with MEGA X based on a ClustalW alignment. Bootstrap values are based on 1,000 replicates. The branch lengths are proportional to the percentage of sequence difference (scale bar, 0.05% difference). The results showed a very high level of conservation and a relationship among the Orco subtypes within insect orders (Figure 3). RT–qPCR assays were used to determine the relative expression levels of AhygOrco in different adult tissues. The results showed that AhygOrco was highly expressed in male and female antennae, with transcript levels significantly higher than in other tissues (exceeding at least 200-fold), followed by the abdomen (Figure 3). The remaining tissues had extremely low expression. However, the difference in AhygOrco levels between male and female antennae was not significant (Figure 4).
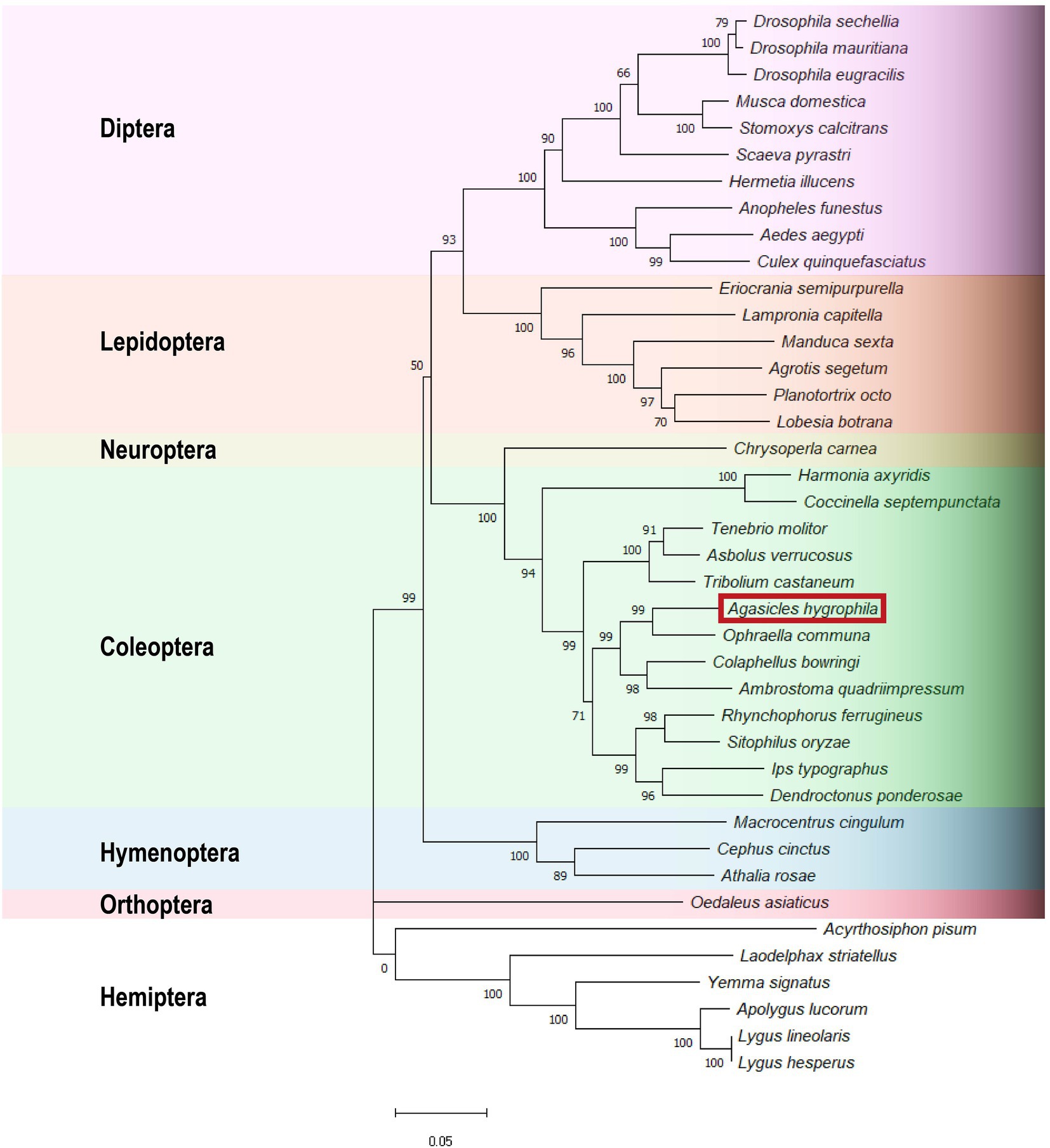
Figure 3. Neighbor-joining tree of Orco orthologs from different insects. Bootstrap support values based on 1,000 replicates are indicated. The branch lengths are proportional to the percentage of sequence difference (scale bar, 0.05% difference). The AhygOrco sequence is shown in the red box. The Orco amino acid sequences of all insects are listed in Appendix B. GenBank accession numbers of the other sequences are listed in Table 2.
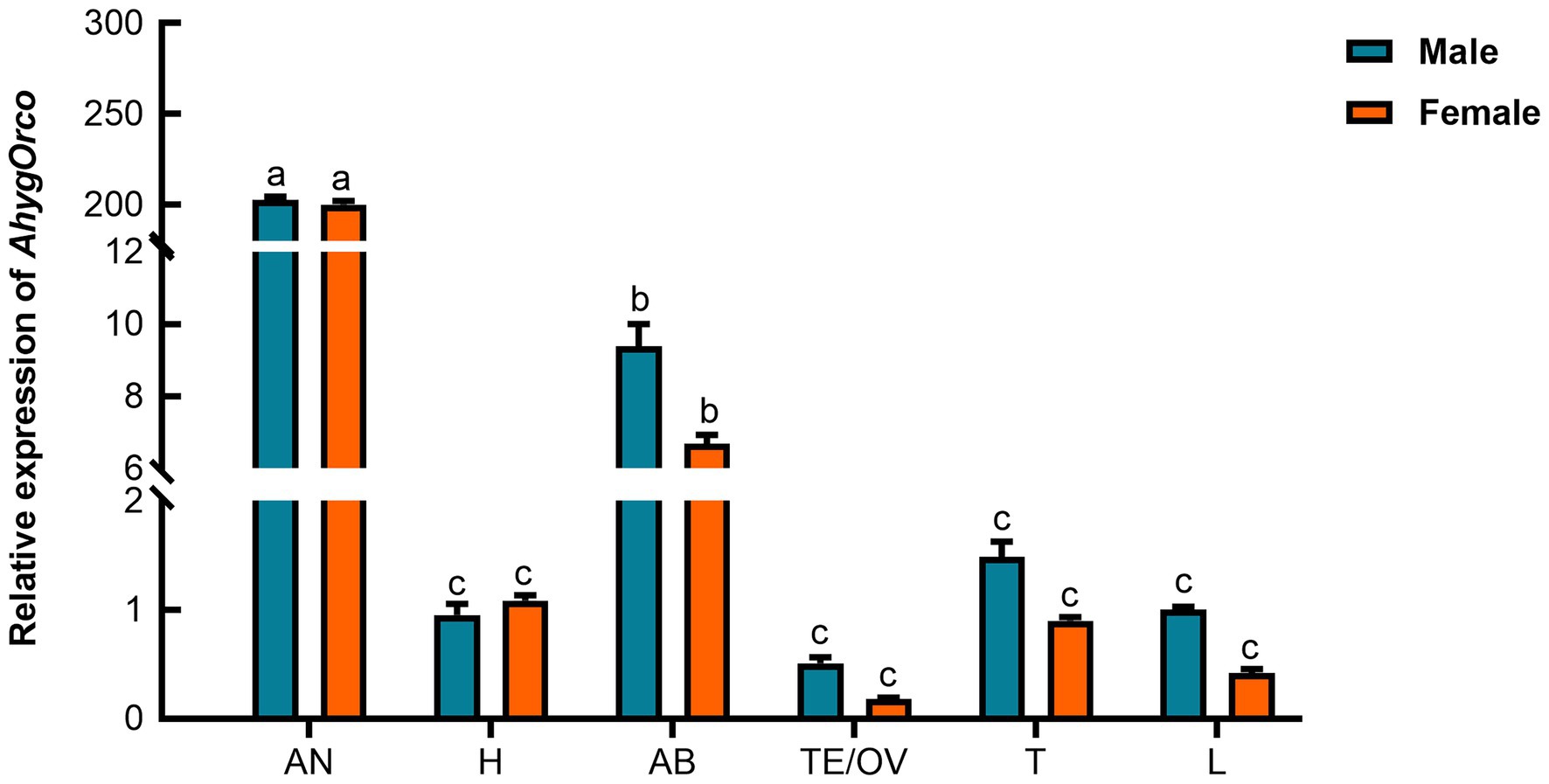
Figure 4. Expression profiling of AhygOrco in different tissues of male and female beetles. All expression fold changes were related to the leg of the male. AN: antennae, H: heads without antennae, AB: abdomens, OV: female ovaries, TE: male testes, T: thoraxes, L: legs. Bars with the same letter are not significantly different from each other at the p < 0.05 level based on Tukey’s test. Each point represents the mean (n = 3) ± SEM. (F 11,24 = 9752.504, p < 0.001).
3.2. Expression analysis of Orco after RNAi
After 48 h (Ma et al., 2020; Tian et al., 2022) of knockdown, we verified the RNAi efficiency of dsOrco by qRT–PCR. The expression level of the AhygOrco gene was significantly lower in dsOrco-injected beetles than in dsEGFP-injected beetles (dsOrco-injected: male, p < 0.0001, female, p < 0.0001), with knockdown levels reaching 90% in males and 89% in females (Figure 5A).
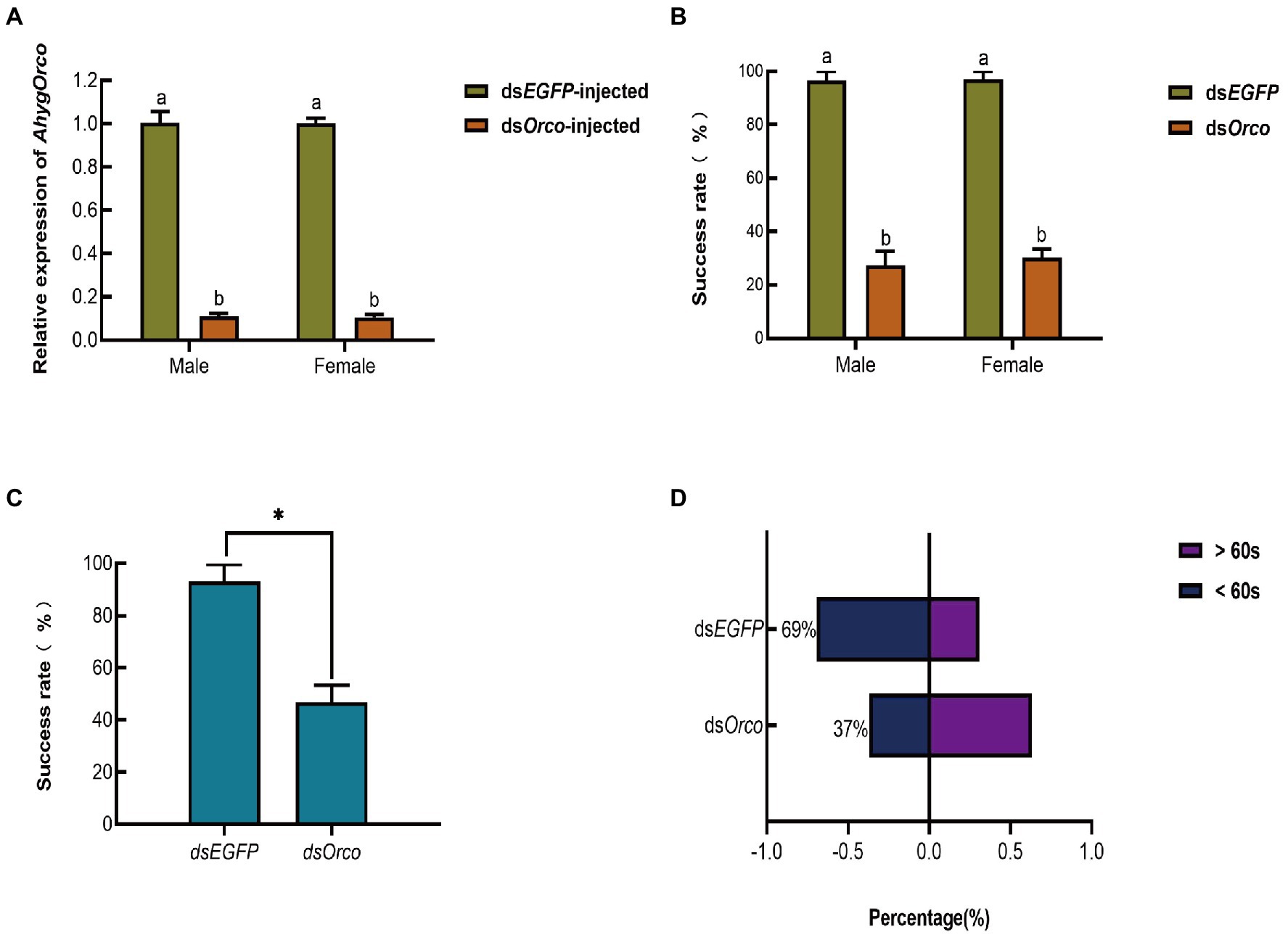
Figure 5. (A) Validation of AhygOrco transcript levels in dsOrco-injected and dsEGFP-injected A. hygrophila. The transcript level of AhygOrco was normalized to the RPL13a control gene. The values marked with different letters are significantly different based on Mann–Whitney U test (Male: Z = −1.964, p < 0.05; Female: Z = −1.964, p < 0.05). Data are presented as the mean (n = 3) ± SEM. (B) The percentage of individuals who successfully found A. philoxeroides within 2 min. (Mann–Whitney U test, Male: Z = −1.993, p < 0.05; Female: Z = −1.993, p < 0.05). (C) The percentage of individuals who successfully found a mate within 60 s. (Mann–Whitney U test, Z = −1.993, p < 0.05) Data are presented as the mean (n = 3) ± SEM. (D) The time these individuals spent searching. dsEGFP, insects injected with dsRNA against EGFP. dsOrco, insects injected with dsRNA against AhygOrco.
3.3. Knockdown of AhygOrco impairs feeding and mate seeking
To study the effect of silent Orco on host and mate recognition, behavioral bioassays were performed. The results showed that the host-seeking behavior of AhygOrco-silenced males and females was significantly inhibited, with only 30% of females and 27% of males finding their hosts within 2 min, which was significantly lower than that of dsEGFP-injected males (Figure 5B). Meanwhile, the host-seeking behavior of AhygOrco-silenced males was significantly inhibited, with only 47% of males finding a mate within 60 s, which was significantly lower than that of insects injected with dsEGFP (Figure 5C). In addition, for those insects that successfully found a host, significantly fewer of the dsOrco-injected group were able to find a host within 60 s compared to the dsEGFP-injected group (dsEGFP: 69%, dsOrco: 37%, Figure 5D).
4. Discussion
In the present study, we characterized the olfactory receptor coreceptor gene AhygOrco of A. hygrophila. AhygOrco has seven TMDs, an Nin-Cout topology and a highly conserved C-terminal structural domain, as predicted by the Orco amino acid sequence comparison and TMHMM program. In detail, the predicted results of the TMHMM procedure show that the topology of AhygOrco is consistent with the topology of Orco of other insects with 7 TMDs (Mombaerts, 1999; Butterwick et al., 2018). It is localized in the plasma membrane, with the N-terminal located inside the cell and the C-terminal located outside the cell. The Nin-Cout topology is used to form the unique Orco-OR complex structure, which is a heteromeric ligand-gated ion channel (Sato et al., 2008; Touhara, 2009; Wicher et al., 2009). Unlike other known families of ion channels, this receptor is not associated with G protein coupling (Smart et al., 2008). This ion channel is unique to insects to cope with the odor environment (Benton, 2006; Touhara and Vosshall, 2009). Through the phylogenetic tree of multiple insect Orco, we found seven insect Orco with high similarity to AhygOrco for amino acid sequence comparison. There are a large number of amino acid sequences in the C-terminal region that are identical to those of other insects. This finding is consistent with previous findings, and it is hypothesized that this high degree of sequence conservation corresponds to a specific function of this region (Krieger et al., 2003; Malpel et al., 2008).
The expression of AhygOrco, in antennae was 200-fold higher than that in other parts of the body, it is not surprising in consideration of its functions in olfaction (Melo et al., 2004; Pitts et al., 2004; Zhang X. F. et al., 2021). In addition, the expression of AhygOrco in non-olfactory tissues suggests that they might be also involved in some non-olfactory chemosensory perception process, such as the Orco genes could mediate the activation of spermatozoa in Aedes aegypti (Pitts et al., 2014) and Cimex lectularius (Hansen et al., 2014).
RNAi technology is increasingly used to explore gene function and has a better knockdown efficiency in Coleoptera (Yan et al., 2017). In this experiment, we adopted the delivery method of injecting dsRNA into the abdomen of adults, and RT–qPCR analysis after treatment showed that the expression level of the AhygOrco gene was reduced by nearly 90% in both males and females (Bettencourt et al., 2002; Tomoyasu and Denell, 2004; Huvenne and Smagghe, 2010). This shows that we successfully and effectively silenced the AhygOrco gene by RNAi.
The results of behavioral tests showed that the experimental group injected with dsOrco significantly reduced its response to A. philoxeroides leaves compared to the control group for both females and males. In addition, dsOrco injection was found to significantly impair the mate detection ability of A. hygrophila. The above results provide evidence that repression of the AhygOrco gene significantly impairs the host and mate detection ability of A. hygrophila. This suggests that AhygOrco plays an extremely important role in the olfactory system, which is consistent with previous studies (Larsson et al., 2004; Jones et al., 2005; Fan et al., 2015; Lin et al., 2015). After silencing Orco, A. hygrophila is still attracted to the host, and we speculate that there are other pathways for A. hygrophila to sense the host, such as the IR pathway. Currently there are two possibilities regarding the relationship between the ORs pathway and the IR pathway. One is as Zhang et al. (2019) discovered that both Orco pathway and IRs pathway are involved in acid sensing. Alternatively, as in Helicoverpa armigera, defects in Orco pathway do not alter the insect’s response to acid (Fan et al., 2022). An interesting question for future research is whether Orco pathway is involved in acid sensing in A. hygrophila along with IRs pathway.
According to the reaction time needed for A. hygrophila to detect its host, the experimental group injected with dsOrco took significantly more time than the control group. For such results, we speculate that the action of A. hygrophila to find a host is performed by multiple senses together. Long range detection relies initially on olfaction (OR and IR pathways) but other cues such as visual ones can be equally involved. Silencing AhygOrco prolongs the time needed for A. hygrophila to detect its host, althought it does not completely disable the ability to find its host. This is consistent with Balkenius’s study (Balkenius et al., 2009; Tateishi et al., 2022). These facts demonstrate that AhygOrco is a key receptor in A. hygrophila olfaction, that the OR pathway is important for insects to be able to carry out normal physiological life activities and that other sensory systems synergistic with olfaction should be further defined in the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XN, CH, and FW conceived and designed the entire research. XQ, JG, and WQ provide experimental resources, XN and CD performed the experiments, XN and CH analyzed the data, XN, CH, JJ, FC, and FW wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work in this paper was supported by the National Key Research and Development Program of China (2021YFD1400100 and 2021YFD1400101), the Shenzhen Science and Technology Program (Grant No. KQTD20180411143628272) and the Agricultural Science and Technology Innovation Program.
Acknowledgments
We sincerely thank Mr. Wu (Qiang Wu) and Ms. Wang (Chenjiaozi Wang) for their scientific support and various types of help with this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1104962/full#supplementary-material
Footnotes
References
Balkenius, A., Bisch-Knaden, S., and Hansson, B. (2009). Interaction of visual and odour cues in the mushroom body of the hawkmoth Manduca sexta. J. Exp. Biol. 212, 535–541. doi: 10.1242/jeb.021220
Benton, R. (2006). On the ORigin of smell: odorant receptors in insects. Cell. Mol. Life Sci. 63, 1579–1585. doi: 10.1007/s00018-006-6130-7
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). Atypical membrane topology and heteromeric function of drosophila odorant receptors in vivo. PLoS Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in drosophila. Cells 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Bettencourt, R., Terenius, O., and Faye, I. (2002). Hemolin gene silencing by ds-RNA injected into Cecropia pupae is lethal to next generation embryos. Insect Mol. Biol. 11, 267–271. doi: 10.1046/j.1365-2583.2002.00334.x
Butterwick, J. A., Del Marmol, J., Kim, K. H., Kahlson, M. A., Rogow, J. A., Walz, T., et al. (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. doi: 10.1038/s41586-018-0420-8
Carey, A. F., Wang, G., Su, C. Y., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi: 10.1038/nature08834
Conchou, L., Lucas, P., Meslin, C., Proffit, M., Staudt, M., and Ronou, M. (2019). Insect Odorscapes: from plant volatiles to natural olfactory scenes. Front. Physiol. 10:972. doi: 10.3389/fphys.2019.00972
Degennaro, M., Mcbride, C. S., Seeholzer, L., Nakagawa, T., Dennis, E. J., Goldman, C., et al. (2013). Orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498, 487–491. doi: 10.1038/nature12206
Fan, X. B., Mo, B. T., Li, G. C., Huang, L. Q., Guo, H., Gong, X. L., et al. (2022). Mutagenesis of the odorant receptor co-receptor (Orco) reveals severe olfactory defects in the crop pest moth Helicoverpa armigera. BMC Biol. 20:214. doi: 10.1186/s12915-022-01411-2
Fan, J., Zhang, Y., Francis, F., Cheng, D., Sun, J., and Chen, J. (2015). Orco mediates olfactory behaviors and winged morph differentiation induced by alarm pheromone in the grain aphid, Sitobion avenae. Insect Biochem. Mol. Biol. 64, 16–24. doi: 10.1016/j.ibmb.2015.07.006
Fandino, R. A., Haverkamp, A., Bisch-Knaden, S., Zhang, J., Bucks, S., Nguyen, T. A. T., et al. (2019). Mutagenesis of odorant coreceptor Orco fully disrupts foraging but not oviposition behaviors in the hawkmoth Manduca sexta. Proc. Natl. Acad. Sci. U. S. A. 116, 15677–15685. doi: 10.1073/pnas.1902089116
Hallem, E. A., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cells 125, 143–160. doi: 10.1016/j.cell.2006.01.050
Hansen, I. A., Rodriguez, S. D., Drake, L. L., Price, D. P., Blakely, B. N., Hammond, J. I., et al. (2014). The odorant receptor co-receptor from the bed bug, Cimex lectularius L. PLoS One 9:e113692. doi: 10.1371/journal.pone.0113692
Hansson, B. S., and Stensmyr, M. C. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
Hull, J. J., Hoffmann, E. J., Perera, O. P., and Snodgrass, G. L. (2012). Identification of the western tarnished plant bug (Lygus hesperus) olfactory co-receptor Orco: expression profile and confirmation of atypical membrane topology. Arch. Insect Biochem. Physiol. 81, 179–198. doi: 10.1002/arch.21042
Huvenne, H., and Smagghe, G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. doi: 10.1016/j.jinsphys.2009.10.004
Jia, D., Ji, Z. Y., and Liu, Y. H. (2020). Screening and validation of internal reference genes in different developmental stages of Agasicles hygrophila. J. Shanxi Agric. Univ. 40, 53–58. doi: 10.13842/j.cnki.issn1671-8151.202001022
Jones, W. D., Nguyen, T. A., Kloss, B., Lee, K. J., and Vosshall, L. B. (2005). Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 15, R119–R121. doi: 10.1016/j.cub.2005.02.007
Jones, P. L., Pask, G. M., Rinker, D. C., and Zwiebel, L. J. (2011). Functional agonism of insect odorant receptor ion channels. Proc. Natl. Acad. Sci. U. S. A. 108, 8821–8825. doi: 10.1073/pnas.1102425108
Kato, A., and Touhara, K. (2009). Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell. Mol. Life Sci. 66, 3743–3753. doi: 10.1007/s00018-009-0111-6
Koenig, C., Hirsh, A., Bucks, S., Klinner, C., Vogel, H., Shukla, A., et al. (2015). A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem. Mol. Biol. 66, 51–63. doi: 10.1016/j.ibmb.2015.09.007
Koutroumpa, F. A., Monsempes, C., Francois, M. C., De Cian, A., Royer, C., Concordet, J. P., et al. (2016). Heritable genome editing with CRISPR/Cas9 induces anosmia in a crop pest moth. Sci. Rep. 6:29620. doi: 10.1038/srep29620
Krieger, J., Klink, O., Mohl, C., Raming, K., and Breer, H. (2003). A candidate olfactory receptor subtype highly conserved across different insect orders. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 189, 519–526. doi: 10.1007/s00359-003-0427-x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, B. N., Taylor, R. W., Pask, G. M., Zwiebel, L. J., Newcomb, R. D., and Christie, D. L. (2013). A conserved aspartic acid is important for agonist (VUAA1) and odorant/tuning receptor-dependent activation of the insect odorant co-receptor (Orco). PLoS One 8:e70218. doi: 10.1371/journal.pone.0070218
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Li, Y., Zhang, J., Chen, D., Yang, P., Jiang, F., Wang, X., et al. (2016). CRISPR/Cas9 in locusts: successful establishment of an olfactory deficiency line by targeting the mutagenesis of an odorant receptor co-receptor (Orco). Insect Biochem. Mol. Biol. 79, 27–35. doi: 10.1016/j.ibmb.2016.10.003
Libert, S., Zwiener, J., Chu, X. W., Vanvoorhies, W., Roman, G., and Pletcher, S. D. (2007). Regulation of drosophila life span by olfaction and food-derived odors. Science 315, 1133–1137. doi: 10.1126/science.1136610
Lin, W., Yu, Y., Zhou, P., Zhang, J., Dou, L., Hao, Q., et al. (2015). Identification and knockdown of the olfactory receptor (OrCo) in gypsy moth, Lymantria dispar. Int. J. Biol. Sci. 11, 772–780. doi: 10.7150/ijbs.11898
Liu, Q., Liu, W., Zeng, B., Wang, G., Hao, D., and Huang, Y. (2017). Deletion of the Bombyx mori odorant receptor co-receptor (BmOrco) impairs olfactory sensitivity in silkworms. Insect Biochem. Mol. Biol. 86, 58–67. doi: 10.1016/j.ibmb.2017.05.007
Liu, X., Zhang, B., Li, S., Rao, X., Wang, D., Hu, X., et al. (2016). Knockdown of the olfactory co-receptor Orco impairs mate recognition in Tenebrio molitor (Coleoptera: Tenebrionidae). J. Asia Pac. Entomol. 19, 503–508. doi: 10.1016/j.aspen.2016.05.005
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J., Zhao, L., Ma, R., Zhang, P., Fan, R., and Zhang, J. (2010). Performance of the biological control agent flea beetle Agasicles hygrophila (Coleoptera: Chrysomelidae), on two plant species Alternanthera philoxeroides (alligatorweed) and A-sessilis (joyweed). Biol. Control 54, 9–13. doi: 10.1016/j.biocontrol.2010.02.012
Ma, C., Cui, S., Bai, Q., Tian, Z., Zhang, Y., Chen, G., et al. (2020). Olfactory co-receptor is involved in host recognition and oviposition in Ophraella communa (Coleoptera: Chrysomelidae). Insect Mol. Biol. 29, 381–390. doi: 10.1111/imb.12643
Malpel, S., Merlin, C., Francois, M. C., and Jacquin-Joly, E. (2008). Molecular identification and characterization of two new Lepidoptera chemoreceptors belonging to the Drosophila melanogaster OR83b family. Insect Mol. Biol. 17, 587–596. doi: 10.1111/j.1365-2583.2008.00830.x
Melo, A. C., Rutzler, M., Pitts, R. J., and Zwiebel, L. J. (2004). Identification of a chemosensory receptor from the yellow fever mosquito, Aedes aegypti, that is highly conserved and expressed in olfactory and gustatory organs. Chem. Senses 29, 403–410. doi: 10.1093/chemse/bjh041
Missbach, C., Dweck, H. K., Vogel, H., Vilcinskas, A., Stensmyr, M. C., Hansson, B. S., et al. (2014). Evolution of insect olfactory receptors. eLife 3:e02115. doi: 10.7554/eLife.02115
Mombaerts, P. (1999). Seven-transmembrane proteins as odorant and chemosensory receptors. Science 286, 707–711. doi: 10.1126/science.286.5440.707
Nakagawa, T., Pellegrino, M., Sato, K., Vosshall, L. B., and Touhara, K. (2012). Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLoS One 7:e32372. doi: 10.1371/journal.pone.0032372
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. Oxford University Press, New York.
Nichols, A. S., Chen, S., and Luetje, C. W. (2011). Subunit contributions to insect olfactory receptor function: channel block and odorant recognition. Chem. Senses 36, 781–790. doi: 10.1093/chemse/bjr053
Nichols, A. S., and Luetje, C. W. (2010). Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J. Biol. Chem. 285, 11854–11862. doi: 10.1074/jbc.M109.058321
Pitts, R. J., Fox, A. N., and Zwiebel, L. J. (2004). A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 101, 5058–5063. doi: 10.1073/pnas.0308146101
Pitts, R. J., Liu, C., Zhou, X. F., Malpartida, J. C., and Zwiebel, L. J. (2014). Odorant receptor-mediated sperm activation in disease vector mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 111, 2566–2571. doi: 10.1073/pnas.1322923111
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Saleh, M. C., van Rij, R. P., Hekele, A., Gillis, A., Foley, E., O'Farrell, P. H., et al. (2006). The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8, 793–802. doi: 10.1038/ncb1439
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Smart, R., Kiely, A., Beale, M., Vargas, E., Carraher, C., Kralicek, A. V., et al. (2008). Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem. Mol. Biol. 38, 770–780. doi: 10.1016/j.ibmb.2008.05.002
Suh, E., Bohbot, J., and Zwiebel, L. J. (2014). Peripheral olfactory signaling in insects. Curr. Opin. Insect. Sci. 6, 86–92. doi: 10.1016/j.cois.2014.10.006
Sun, H. H., Liu, F., Ye, Z., Baker, A., and Zwiebel, L. J. (2020). Mutagenesis of the orco odorant receptor co-receptor impairs olfactory function in the malaria vector anopheles coluzzii. Insect. Biochem. Mol. Biol. 127:103497. doi: 10.1016/j.ibmb.2020.103497
Tateishi, K., Watanabe, T., Nishino, H., Mizunami, M., and Watanabe, H. (2022). Silencing the odorant receptor co-receptor impairs olfactory reception in a sensillum-specific manner in the cockroach. iScience 25:104272. doi: 10.1016/j.isci.2022.104272
Tian, Z., Chen, G., Zhang, Y., Ma, C., Tian, Z., Gao, X., et al. (2022). Rapid evolution of Ophraella communa cold tolerance in new low-temperature environments. J. Pest. Sci. 95, 1233–1244. doi: 10.1007/s10340-021-01461-5
Tomoyasu, Y., and Denell, R. E. (2004). Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 214, 575–578. doi: 10.1007/s00427-004-0434-0
Touhara, K. (2009). Insect olfactory receptor complex functions as a ligand-gated Ionotropic Channel. Ann. N. Y. Acad. Sci. 1170, 177–180. doi: 10.1111/j.1749-6632.2009.03935.x
Touhara, K., and Vosshall, L. B. (2009). Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 71, 307–332. doi: 10.1146/annurev.physiol.010908.163209
Trible, W., Olivos-Cisneros, L., Mckenzie, S. K., Saragosti, J., Chang, N. C., Matthews, B. J., et al. (2017). Orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cells 170, 727–735.e10. doi: 10.1016/j.cell.2017.07.001
Turner, R. M., Derryberry, S. L., Kumar, B. N., Brittain, T., Zwiebel, L. J., Newcomb, R. D., et al. (2014). Mutational analysis of cysteine residues of the insect odorant co-receptor (Orco) from Drosophila melanogaster reveals differential effects on agonist- and odorant-tuning receptor-dependent activation. J. Biol. Chem. 289, 31837–31845. doi: 10.1074/jbc.M114.603993
Vosshall, L. B., and Hansson, B. S. (2011). A unified nomenclature system for the insect olfactory coreceptor. Chem. Senses 36, 497–498. doi: 10.1093/chemse/bjr022
Wang, K. X. (2019). Research on influence factors and the mechanisms of insect RNAi efficiency. Nanjing: Nanjing Agricultural University.
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 107, 4418–4423. doi: 10.1073/pnas.091339210
Wicher, D., Schafer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2009). dOr83b--receptor or ion channel? Ann. N. Y. Acad. Sci. 1170, 164–167. doi: 10.1111/j.1749-6632.2009.04101.x
Wyatt, T. D. (2003). Pheromones and Animal Behaviour: Communication by Smell and Taste. Cambridge University Press: Cambridge, U.K. 566.
Xiao, D. (2014). The mechanism of double-strand RNA up-take and function of related genes in red flour beetle (Tribolium castaneum). Beijing: China Agricultural University.
Yan, H., Opachaloemphan, C., Mancini, G., Yang, H., Gallitto, M., Mlejnek, J., et al. (2017). An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cells 170, 736–747.e9. doi: 10.1016/j.cell.2017.06.051
Zhang, J., Bisch-Knaden, S., Fandino, R. A., Yan, S., Obiero, G. F., Grosse-Wilde, E., et al. (2019). The olfactory coreceptor IR8a governs larval feces-mediated competition avoidance in a hawkmoth. Proc. Natl. Acad. Sci. U. S. A. 116, 21828–21833. doi: 10.1073/pnas.1913485116
Zhang, X. F., Liu, P. J., Qin, Q. J., Li, M., Meng, R. J., and Zhang, T. (2021). Characterizing the role of Orco gene in detecting aggregation pheromone and food resources in Protaetia brevitarsis Leiws (Coleoptera: Scarabaeidae). Front. Physiol. 12:649590. doi: 10.3389/fphys.2021.649590
Zhang, D. D., and Lofstedt, C. (2013). Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PLoS One 8:e77345. doi: 10.1371/journal.pone.0077345
Zhang, D. D., and Lofstedt, C. (2015). Moth pheromone receptors: gene sequences, function, and evolution. Front. Ecol. Evol. 3:105. doi: 10.3389/fevo.2015.00105
Zhang, Y., Yang, J. P., Dai, R. H., Yan, Y., Yang, W. J., and Hu, D. M. (2021). Stability evaluation of candidate reference genes for RT-qPCR normalization in Lasioderma serricorne (F.). J. Stored Prod. Res. 94:101877. doi: 10.1016/j.jspr.2021.101877
Keywords: Agasicles hygrophila, odorant receptor co-receptor, RNA interference, mate recognition, host finding
Citation: Ning X, Huang C, Dong C, Jin J, Qiao X, Guo J, Qian W, Cao F and Wan F (2023) RNAi verifications on olfactory defects of an essential biocontrol agent Agasicles hygrophila (Coleoptera: Chrysomelidae) regarding mating and host allocation. Front. Ecol. Evol. 11:1104962. doi: 10.3389/fevo.2023.1104962
Edited by:
Panagiotis Milonas, Benaki Phytopathological Institute, GreeceReviewed by:
Fotini A. Koutroumpa, Institut National de recherche pour l’agriculture (INRAE), FranceFengqi Li, Guizhou University, China
Copyright © 2023 Ning, Huang, Dong, Jin, Qiao, Guo, Qian, Cao and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengqin Cao, ✉ OTkwOTU3QGhhaW5hbnUuZWR1LmNu; Fanghao Wan, ✉ d2FuZmFuZ2hhb0BjYWFzLmNu
†These authors have contributed equally to this work and share first authorship
 Xiaoyu Ning
Xiaoyu Ning Cong Huang
Cong Huang Changhong Dong4
Changhong Dong4 Jisu Jin
Jisu Jin Xi Qiao
Xi Qiao Wanqiang Qian
Wanqiang Qian Fanghao Wan
Fanghao Wan