
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Ecol. Evol., 22 June 2023
Sec. Paleontology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1100503
This article is part of the Research TopicNew Perspectives on Living FossilsView all 9 articles
The recently proposed concept of ´endangered living fossils’ (ELFs) integrates high-endangered status and evolutionary singularity for any species. In this review, I gathered monotypic genera (single-species genera) that satisfy the three ELF criteria: (i) scarcity and narrow distribution of populations, i.e., considering every species categorized ‘critically endangered’ or contemporary ´extinct´ by IUCN criteria; (ii) evolutionary singularity, i.e., both morphological and phylogenetic singularities of a single-species lineage as a result of a null net diversification rate; and (iii) ancient divergence, i.e., split from the closest extant relatives predating a particular geological epoch. A total of 3,706 monotypic genera of vertebrates and angiosperms were analyzed. I found 109 critically endangered and contemporary extinct genera of which 57 were ELFs. The emergent patterns are: (1) taxonomy (generic level) is a reliable first approach to identifying ELFs; (2) ´morphological singularity´ displayed by monotypic genera does not always help identify ELFs on islands; (3) species of monotypic genera tend to be more threatened than average species; (4) extinction appears to be biased against some animal and plant groups; (5) contemporary extinct genera are strongly associated with distribution on islands, particularly for flightless birds vulnerable to human prosecution; and (6) the ELF approach is a relatively quick method to identify the species of floras and faunas most urgently in need of protection in the world. This approach is complementary to any method searching for phylogenetic diversity (e. g. EDGE), which is also discussed. I argue that ELFs should be prioritized in conservation because they are the most threatened lineages representing an exceptional evolutionary heritage in the world.
Shortly after proposing the concept of ´endangered living fossils’ (ELFs) (Vargas et al., 2020), we are observing its application by zoologists (Bond et al., 2020), botanists (Jiménez-Mejías et al., 2021; Miguez et al., 2022) and science journalists (Davenport, 2021). The reason for this may be related to the benefit of narrowing down the complex interpretations of the living fossil concept that biologists are still debating (Lidgard and Love, 2018; Turner, 2019; Lidgard and Love, 2021). Indeed, any researcher can test whether candidate species are considered ELFs based on three complementary criteria: (1) scarcity and narrow distribution of endangered species recognized by international red lists; (2) ´evolutionary singularity´ in phylogenetic reconstructions; and (3) ancient divergence regarding early geological epochs.
While species abundance and distribution are based on thorough search for populations in the field and locations in museum collections, phylogenies rely on sufficiently representative sampling and sufficiently resolved topologies. In any case, the majority of well-resolved phylogenies are asymmetrical, i.e., some clades are rich in species whereas some others are rather poor, including early diverging clades that contain a low number of species (Ronquist, 2014). The result is a ladderized or asymmetrical topology in which basal-most clades can even show as few species as a single one. Causes for this pattern are typically two: decline of a former species-rich clade (evolutionary extinction) or low diversification over long periods of time (evolutionary stasis) (Eldredge et al., 2005). Phylogenies of living organisms do not help at this point, which make fossils come into play. However, paleontological evidence is missing for the majority of organism groups of the tree of life. In any case, the use of fossils at different calibration points makes time-calibrated phylogenies the most comprehensive tool to test ELFs (Vargas et al., 2020).
The budding speciation pattern found in many phylogenies at low taxonomic levels (species) (see Otero et al., 2022) is congruent with an asymmetrical pattern at deeper levels (classes, phyla, orders) (Rabosky et al., 2012). Indeed, the tree of life is profoundly asymmetrical in such a way that clades currently rich in species show a derived position nested within a group with older clades (Vargas and Zardoya, 2014). In other words, a general phylogenetic pattern shows pervasive paraphyly that affects any taxonomic treatments. Lack of reciprocal monophyly for many group pairs compromises recognition of the same taxonomic rank, as accepted in the past for two groups taxonomically at the same level that turned out to be paraphyletic. In particular, we can observe multiple cases where one of the two groups is included in the other group: monocotyledons in former dicotyledons (angiosperms), bilaterians in former radiolarians (eumetazoans), insects in former crustaceans (panarthropods), birds in former reptiles (sauropsids), tetrapods in former fishes (sarcopterigians), among others.
Since the last century, some studies have acknowledged the importance of phylogenetic singularity in conservation. One of the first approaches evaluating evolutionary trees in conservation used cladistic classifications and a measure of taxonomic distinctness (taxic diversity) (Vane-Wright et al., 1991). Shortly after, molecular (mtDNA) phylogenies were used to prevent from pervasive homoplasy in cladograms based on a few morphological characters (Faith, 1992). The accurate measurement of branch lengths and divergences using DNA sequence-based phylogenies, together with lower impact of convergence and increasingly lower costs of DNA sequencing, made molecular phylogenetics a basic tool in conservation. Currently, the most followed method is Evolutionarily Distinct and Globally Endangered (EDGE), which relies on phylogenetic diversity and distinctiveness (Isaac et al., 2007; Isaac and Pearse, 2018). However, this approach considers taxonomy only for sampling, while evolutionary singularity needs to consider ´morphological singularity´ as well to fit into the concept of ´living fossils´ (see Vargas et al., 2020).
In this paper, the concept of ELF is explored across representative branches of the tree of life (flowering plants and vertebrates) to infer distinctive lineages formed by a single species that is on the brink of extinction. As a working hypothesis, early-divergent lineages of singular animals and plants are investigated, which may predate geological boundaries in the global chronostratigraphic scale (see Vargas et al., 2020). To this aim, single-species groups (monotypic genera) considered IUCN ´critically endangered´ and contemporary ´extinct´ single-species groups are evaluated, followed by searches for reliable publications in academic databases to obtain time-calibrated phylogenies that include those monotypic genera. Emerging patterns related to ancient divergence and survival through geological epochs are further discussed. In more practical terms, any species tested for ELF criteria will be recommended to be further prioritized for conservation based on evolutionary singularity, i.e., including morphological singularity and early divergence from living species.
A spatio-temporal hypothesis is herein considered to identify ELFs that combine both narrow geographic distributions of the species and isolated evolutionary history of the lineage (Vargas et al., 2020). This approach will be performed in plants (flowering plants) and vertebrates (tetrapods) because they are two groups that gather a great deal of detailed information (taxonomy, geographic distributions, population abundance, robust time-calibrated phylogenies), which is essential to evaluate ELFs. In particular, I analyzed monotypic genera of five groups of organisms: eudicots, amphibians, non-avian reptiles, avian reptiles (hereafter birds) and mammals.
For the spatial approach, the ´endangered´ component of ELF includes limited geographic distributions once intensive searches in the field and specimen collections have been performed. In a first approach performed in this study, only single-species genera (hereafter monotypic genera) with few and scarce populations that hold a ‘critically endangered’ category using IUCN (2017) criteria will be considered. In addition, contemporary disappearance (IUCN ´extinct´) is also analyzed to interpret causes of extinction.
For the temporal approach, the ´living fossil´ component is considered when a unique morphology is associated with a lineage illustrating long-term isolation. The evolutionary origin (evolutionary distinctiveness) is also based on the concept of phylogenetic singularity (Isaac et al., 2007; Mishler et al., 2014; Rosauer and Jetz, 2015). Time-calibrated phylogenies using DNA sequences are preferred in the search for early divergence times considering geological boundaries in a global chronostratigraphic scale.1 The Miocene–Pliocene boundary is chosen based on two main reasons: (1) dramatic climatic events have been described worldwide during and after the Miocene, which are responsible for global extinction of numerous plant and animal lineages (Steinthorsdottir et al., 2021); and (2) a considerable time window of over 5 million years has been demonstrated to provide enough time for divergence and speciation. Indeed, active speciation (evolutionary radiations) has even been described in the Pliocene-Pleistocene for angiosperms (Valente et al., 2010; Soltis et al., 2019), lizards (Reaney et al., 2018) and marsupials (Couzens and Prideaux, 2018), among others. The taxonomic rank of ‘genus’ is used because it often reflects considerably morphological differentiation and old divergence, and thus a higher probability of finding evolutionary singularity (Vargas et al., 2020). Besides, the genus rank has historically been used as the taxonomic entity that connects paleontology and neontology (Campbell, 1944). It has also been successfully used as a proxy for pre-Pliocene divergence using time-calibrated phylogenies to investigate the origin of species-rich and species-poor groups in the Mediterranean Floristic Region (Vargas et al., 2018). Lastly, an approach at the genus level fits into the taxonomic standards established by the IUCN Red List of Threatened Species (IUCN, 2022).
A three-step procedure is herein performed to search for ELFs of plants (eudicots) and animals (amphibians, non-avian reptiles, birds and mammals) of the world:
(1) Assessing narrow distribution and population scarcity of endangered species taxonomically considered monotypic genera. For a first search, two web pages that comprehensively collect over 3,000 monotypic genera were used for plants (Plantdrew, JarrahTree, Notwith, 2018a) and vertebrates (Plantdrew, JarrahTree, Notwith, 2018b). Indeed, the use of Wikipedia pages has been successfully used in biodiversity and conservation studies (see Mittermeier et al., 2021). Searches in the IUCN database2 were additionally performed for two purposes: (i) cross-validation on the taxonomy of monotypic genera; and (ii) withdrawing genera with the two highest conservation categories (CR and EX). In addition, specialized databases where used to make final taxonomic and distributional decisions: plants,3 amphibians,4 non-avian reptiles,5 birds6 and mammals (Wilson and Reeder, 2005).
(2) Searching for ´evolutionary singularity´ (phylogenetic and morphological singularities), which includes null net diversification rate as a result of (i) speciation followed by pervasive extinction (high extinction rates); (ii) no speciation at all (evolutionary stasis); or (iii) anagenetic evolution. All available phylogenies found in publications withdrawn using Google Scholar were analyzed. The aim of this phylogenetic search is also to investigate the sister group of every CR and EX monotypic genus retrieved at step (1). Useful phylogenies were considered when including reliable sample size and robust DNA-based phylogenetic resolution.
(3) Identifying ancient divergence regarding a chronostratigraphic scale based on time-calibrated phylogenies. A phylogenetic search for scientific publications including singular lineages of every CR and EX monotypic genus was performed using Google Scholar. Divergence times for the node between the monotypic genus and its siter groups (stem node) were obtained straight from the publication. In a few cases of non-dated phylogenies, the computational tool of TimeTree (Kumar et al., 2017) was used to calculate divergence times. Only phylogenetic divergence that predates the Pliocene was considered for ELFs.
A total of 3,706 monotypic genera were ELF tested (Tables 1, 2). Cross-validation of monotypic genera of plants and animals were checked with the IUCN database, which resulted in high taxonomic congruence. To control for reliable information before the analysis (species numbers, distributions, ´critically endangered´ or ´extinct´ status), I further used specialized websites that were also congruent to a great extent. Adjustments on taxonomy, distributions and updated IUCN categories were performed in a few cases.
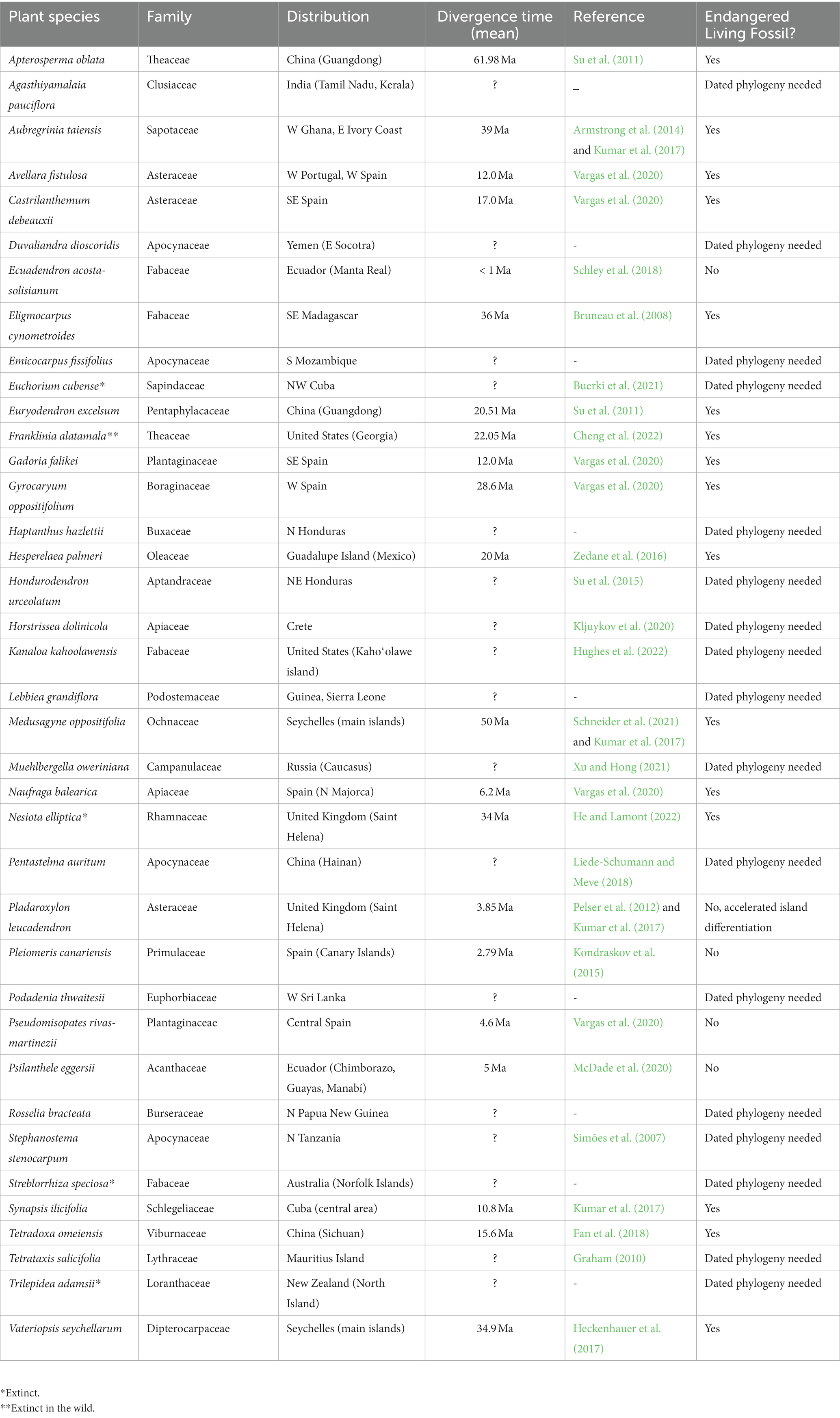
Table 1. List of 38 monotypic genera of plants (Eudicotyledoneae) considered CR or EX in the IUCN website, including species names, family names, distributions, divergence times of the stem group (mean value as provided by every publication), bibliographic references for the phylogenetic studies and consideration of endangered living fossils.
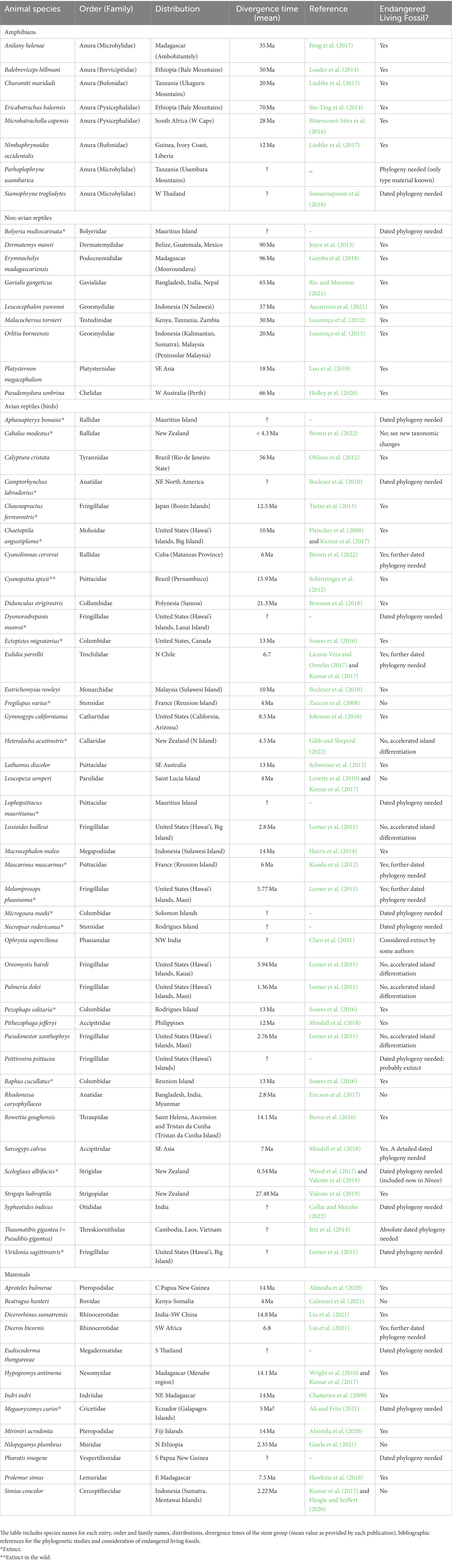
Table 2. List of 71 monotypic genera of animals (amphibians, non-avian reptiles, avian reptiles (birds), mammals) considered CR or EX in the IUCN website.
Searches for monotypic genera resulted in a great congruence among databases. However, I found some taxonomic disagreements. On the one hand, a considerable number resulted in non-monotypic genera, i.e., they showed a higher number (typically two) of living species in the IUCN database for angiosperms (6), non-avian reptiles (4), amphibians (6), birds (47) and mammals (11) (see Supplementary Tables S1, S2). These genera were accordingly not considered. On the other hand, some monotypic genera such as the angiosperms Horstrissea (Crete) and Pleiomeris (Canary Islands) were only found in the IUCN database, and consequently analyzed. The final lists of CR and EX genera analyzed for ELFs are shown in Tables 1, 2.
The IUCN conservation categories herein considered need to be updated for some plants and animals (see Cazalis et al., 2022). Some CR monotypic genera have larger distributions and population abundance than previously considered (see Supplementary Table S2). For instance, the plant Nardostachys jatamansi showed a large distribution to have a CR status: “Bhutan; China; India (Uttaranchal, Sikkim, Himachal Pradesh, Arunachal Pradesh); Myanmar; Nepal” (IUCN, 2022). In addition, insufficient distribution data are observed in some birds (IUCN, 2022), which indicate the need of further field work in many countries as indicated in Supplementary Table S2 for Heliopais personatus, Houbaropsis bengalensis, Necrosyrtes monachus, Rhinoplax vigil. The same is true for the turtle Eretmochelys imbricata. Therefore, these genera were not included in the analysis because they do not meet the requirement of scarce populations in narrow distributions (CR category).
Sister group relationships and divergence times were found in most CR and EX monotypic genera. However, I failed to find 17 of 38 eudicots, 2 of 8 amphibians, 2 of 10 non-avian reptiles, 12 of 41 birds and 3 of 13 mammals (Tables 1, 2). This result indicates that the plant and animal groups chosen for the analysis have complete information for ELF testing in the majority of the cases. We encourage researchers to investigate the remaining monotypic genera to obtain time-calibrated estimates in the near future. Most of the divergence times fall into the Miocene (9 plants, 29 animals), followed by Pleistocene (1 plant, 25 animals), Pliocene (4 plants, 9 animals), Eocene (5 plants, 2 animals), Oligocene (1 plant, 4 animals), Cretaceous (0 plants, 5 animals) and Paleocene (1 plant, 1 animal).
Eudicotyledoneae (eudicots) genera were chosen because they contain the majority (71.1%) of angiosperm diversity in terms of species numbers. In particular, a total of 2,087 monotypic genera of about 10,400 eudicot genera were analyzed, of which 38 are categorized either CR (34) or EX (4). The proportion of CR plus EX genera over the number of total monotypic genera of eudicots is 1.8%, a figure lower than in any animal groups (Table 3). Among the 21 time-calibrated phylogenies, 16 genera meet the three criteria to be considered ELFs. Two families showed the highest number of ELFs (four each), in agreement with high numbers of total genera: Apocynaceae (366) and Fabaceae (765) (Table 1). In contrast, two other small families displayed a higher proportion of ELF genera/total family genera: Theaceae (2/9) and Plantaginaceae (2/90) (Figure 1).
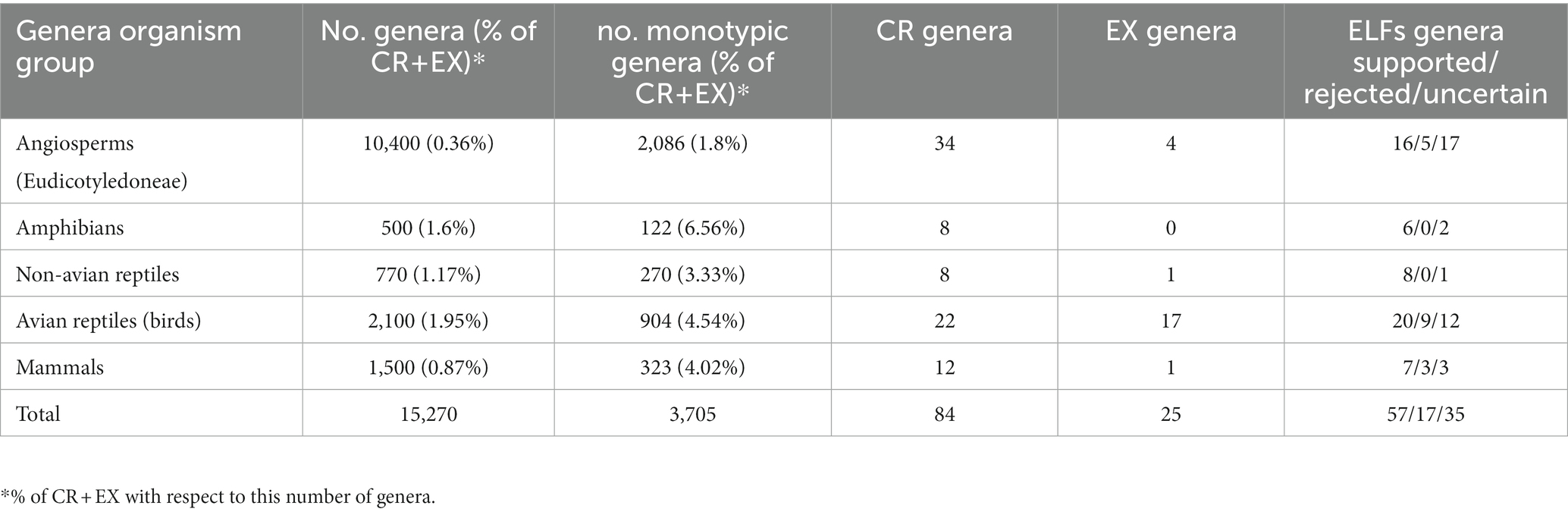
Table 3. Summary figures of total number of genera (data taken from Vargas and Zardoya, 2014), monotypic genera (see the text for databases used), CR genera [data taken from IUCN, 2022], EX genera [data taken from IUCN (2022)] and ELFs genera supported, rejected and uncertain.
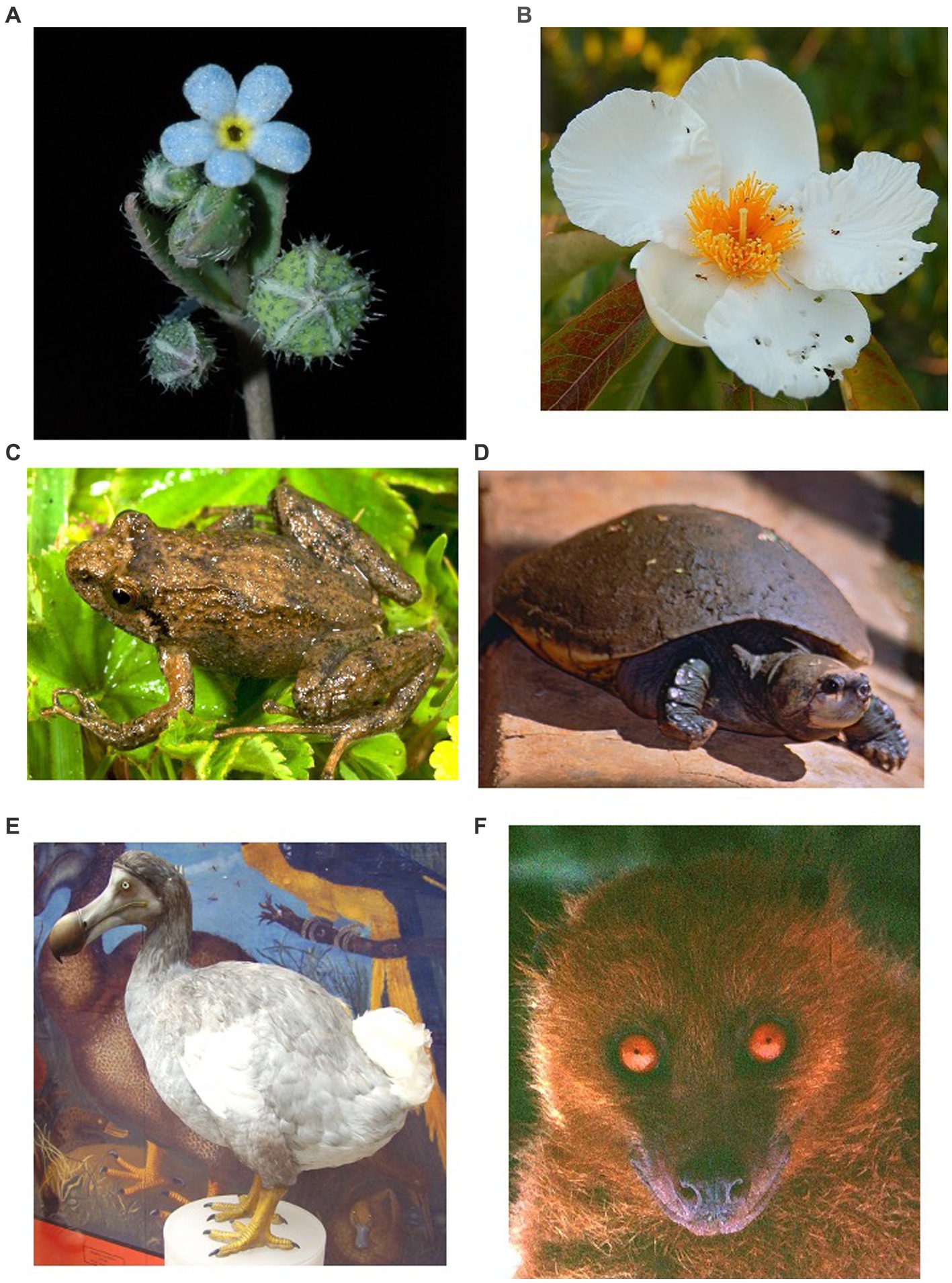
Figure 1. Representatives of ´endangered living fossils´ (ELFs) of angiosperms (A,B), amphibians (C), non-avian reptiles (D), avian-reptiles (birds) (E) and mammals (F) that meet the three criteria to be considered ELFs. (A) Gyrocaryum oppositifolium (CR eudicot, Boraginaceae) from Spain (Madrid and León mountains); (B) Franklinia alatamala (eudicot extinct in the wild, Theaceae) from United States (Georgia); (C) Ericabatrachus baleensis (CR frog, Pyxicephalidae) from Ethiopia (Bale Mountains); (D) Erymnochelys madagascariensis (CR turtle, Podocnemididae) from Madagascar (Mouroundava); (E) Raphus cucullatus (extinct bird, Columbidae) from France (Reunion Island); and (F) Mirimiri acrodonta (CR mammal, Pteropodidae) from Fiji Islands. All photos were taken from Wikipedia (Wikimedia Commons), except for “A” (author: Pablo Vargas).
The four groups of vertebrates (tetrapods) rendered relatively similar proportions of CR (plus EX) genera related to monotypic genera: amphibians (6.56%), non-avian reptiles (3.33%), birds (4.54%) and mammals (4.02%). Likewise, the number of CR (plus EX) genera and the total number of genera of each animal group have comparable proportions (Table 3).
Amphibians. A total of 122 monotypic genera of 500 amphibian genera were investigated, of which eight have an IUCN conservation status of CR (Table 2). There are dated phylogenies only for six of the eight genera, all of which can be considered ELFs. Divergence times from their six living sister groups range between 70 and 12 Ma, which fall into de Cretaceous-Miocene. The family with a higher proportion of ELFs/total family genera is the Brevicipitidae (1/5), followed by the Pyxicephalidae (2/12) and Microhylidae (2/57) (Figure 1).
Non-avian reptiles. A total of 270 monotypic genera of the 770 non-avian reptile genera were analyzed, of which 9 are categorized either CR (8) or EX (1) (Table 2). Time-calibrated phylogenies reveal that at least eight of these 9 genera meet the three criteria to be considered ELFs. The families with a higher proportion of ELFs/total family genera are the Platysternidae (1/1) and Dermatemydidae (1/1), followed by Gavialidae (1/2), Podocnemididae (1/3) and Geoemydidae (2/19) (Figure 1).
Birds. A total of 904 monotypic genera of 2,100 bird genera were investigated, of which 41 have an IUCN conservation status of either CR (22) or EX (19) (Table 2). The majority of them (20 of 29 reliably tested) fit into definition of ELF (Table 3). Numerous monotypic genera of birds from islands are extinct (17), while a considerable number (nine) show divergent times postdating the Miocene, which prevented from being considered ELFs. The bird family with the highest proportion of ELFs/total family genera is the Strigopidae (1/2) and Mohoidae (1/2), followed by Fringillidae (8/49), Megapodiidae (1/7), Psittacidae (4/37), Columbidae (5/51), Otididae (1/9) and Rallidae (3/38). The families with a higher number of extinct genera are: Fringillidae (4), Columbidae (4), Psittacidae (2), Sturnidae (2) and Rallidae (2) (Figure 1).
Mammals. A total of 323 monotypic genera of the 1,500 mammal genera were analyzed, of which 13 are categorized either CR (12) or EX (1) (Table 2). Time-calibrated phylogenies revealed that 7 of these 13 genera meet the criteria for ELF, while three do not. Two of the four genera of Rhinocerotidae are ELFs, while Pteropodidae has also two ELFs of 49 family genera (Figure 1).
The ‘endangered living fossil’ (ELF) concept integrates results from three biological sciences: taxonomy, conservation and phylogenetics. The use of monotypic genera of flowering plants and vertebrates benefits from numerous scientific publications and specialized websites of biodiversity that use taxonomic results (Giles, 2005; Mittermeier et al., 2021; IUCN, 2022). The approach presented herein using 3,706 monotypic genera detected a considerable number of ELFs that require the highest priority in urgent conservation programs (Lean, 2017).
Taxonomy successfully helped the finding of 57 ELFs among the total number of monotypic genera (109) with a conservation status of CR (84) or EX (25), while only 17 of them were ELF rejected. I failed to find reliable information for the rest of them (35). This overall result is congruent with that found for plants of a small area such as the Iberian Peninsula (Vargas et al., 2020). Though the ELF concept has a strong spatial component, it turns out to be useful at both local areas (Iberian Peninsula) and large ones (the world). Similarly, the IUCN conservation categories are closely related to narrow distributions and low number of populations, even though distributions of some plants and animals need to be revised (Supplementary Table S1; see Cazalis et al., 2022). I found also broad utility of this approach across unrelated branches of the tree of life. Indeed, taxonomy at the genus level was a reliable proxy irrespective of the organisms studied (angiosperms, amphibians, non-avian reptiles, birds, mammals).
As expected, a clear pattern of ladderized topologies within each branch of the tree of life was found (Vargas and Zardoya, 2014). The most common phylogenetic relationship found in the phylogenies (109) with CR and EX genera was the monotypic genus sister to a group of multiple species (polytypic genus), followed by a pattern of a monotypic genus sister to a poorly differentiated genus. Furthermore, lineage singularity is observed in both cases because of the lineage in question underwent ancient divergence (ancient relicts; Vargas, 2007) plus no divergence into multiple living species (no changes in diversification rate; Ricklefs, 2007). In absence of a fossil record, it is difficult to distinguish between the two main causes of extant, endangered monotypic genera: decline of a former species-rich clade (evolutionary extinction) or no morphological differentiation over long periods of time (evolutionary stasis) (Eldredge et al., 2005). Indeed, the fossil record is needed to determine causes that account for a ´living fossil´. A textbook example of evolutionary extinction is the ginkgo (Ginkgo biloba) that has a characteristic leaf shape that helps identification of numerous ginkgo species from Cenozoic and late Cretaceous sediments of Eurasia and North America (Royer et al., 2003). When there is lack of macrofossils of an endangered species, the concept of ´endangered living fossils´ gives an alternative perspective.
Phylogenetic methods have been used to perform spatio-temporal analyzes for conservation purposes since the last century (Vane-Wright et al., 1991). On the one hand, geographic areas can be prioritized where accumulated lineages of animals (Rosauer and Jetz, 2015) and plants (Mishler et al., 2014) are located. On the other hand, the temporal component basically relies on early divergent lineages by using relative branching patterns (Isaac et al., 2007). Lineage length based on DNA sequence variation has been largely used to propose multiple metrics for phylogenetic diversity (PD) (see review in Isaac and Pearse, 2018). However, those metrics search for phylogenetic distinctiveness rather than evolutionary distinctiveness because phenotype variation is not considered. Indeed, molecular variation is not always closely related to morphological variation (see Isaac et al., 2007). Needless to say that phenotype variation directly links species and populations with the environment, which is an important component in any studies of the evolutionary change by natural selection.
The ELF approach makes a step further not only by integrating conservation criteria (by choosing critically endangered) and PD (using branch length in time-calibrated phylogenies) but also by integrating the morphological change (by considering genus-level or higher taxonomic ranks). In brief, the integration of three biological sciences in the ELF approach provides a complementary method to PD metrics such as EDGE (and related approaches) because: (1) ELF relies on morphology-based taxonomy, whereas EDGE is not affected by morphology, i.e., taxonomic treatments (Vargas et al., 2020). The lack of the use of taxonomy by EDGE decouples elementary PD with taxonomic ranks, which is the main tenet in IUCN conservation and international legislation. (2) ELF identifies divergence of a particular endangered lineage with respect to its sister group, while EDGE measures accurate scores within large organism groups (e.g., mammals, birds, etc.) (Isaac and Pearse, 2018). (3) Time-calibrated phylogenies against the chronostratigraphic scale (absolute time) help determine association of ELFs with particular geological events (e.g., dramatic climate changes, connection of land masses, emergence of new islands, etc.), while EDGE measures recent-to-early phylogenetic divergence and consequently a branching pattern order (relative time). In other words, ELF is designed to analyze ancient divergence by choosing any time limits of interest, which allow application of thresholds in hypothesis testing (see Vargas et al., 2014; Martín-Hernanz et al., 2023). (4) The use of absolute divergence times together with taxonomy by ELF directly connects neontology and paleontology in the exploration of causes of extinction (Wright et al., 2022). In any case, the two approaches use phylogenetic singularity, which make them partially sharing the same phylogenetic method and thus offering complementary perspectives.
Extension of the ELF approach to some other species-rich organisms such as ferns, gymnosperms, ray-finned fishes (Actinopterygii), butterflies or some other arthropods will be rapidly applied once detailed species distributions, IUCN categories and time-calibrated phylogenies are available. In particular, some gymnosperms have historically been considered living fossils. For instance, the genus Wollemia (W. nobilis) has a high EDGE score (Forest et al., 2018), which agrees with consideration of ELF for Wollemia because of evolutionary distinctiveness in terms of morphological (monotypic genus) and phylogenetic singularity (Lu et al., 2014). However, Araucaria angustifolia and A. araucana also rendered high EDGE scores but do not show significant morphological singularity (same genus), and thus no ELF consideration. In any case, the EDGE method is also based on phylogenetic distinctiveness and extinction risk, which helps propose strong ELF candidates in most of the cases.
In a historical context, it is interesting to note that the use of monotypic genera (and higher taxonomic ranks) in ELF evaluation per se entails considerable morphological differentiation provided by taxonomy (Lean, 2017). This is of paramount importance giving that ELF takes into account ´morphological singularity´, which is expected in any living fossil evaluation (“anomalous forms” in Darwin, 1859).
In our study, absolute divergence times were successfully estimated using time-calibrated phylogenies. The time threshold of 5.33 million years (pre-Pliocene) used in this study helped determine whether diversification times of a CR monotypic genus and its sister group fall into the Miocene epoch or earlier. Our data support a common pattern of pre-Pliocene divergence in the majority of the cases because most of ELF divergence times were found between the Miocene and Cretaceous (see Tables 1, 2).
Though identification of a higher number of ELFs is expected in the near future, the considerable number of plants and animals (3,706 monotypic genera) surveyed in this review helps propose some emergent patterns.
1. Successful implementation of taxonomy (generic level) as a first approach to obtain a list of potential ELF candidates. However, taxonomic ranks vary quite significantly across organism groups of the tree of life. Contrary to Hennig (1966) expectations, dated phylogenies are currently not used to consider taxonomic ranks by temporal banding across the tree of life (Ronquist, 2014). On the contrary, differences in taxonomic treatments render temporal inconsistencies at the genus level among angiosperms, amphibians, non-avian reptiles, birds and mammals.
2. Considerable ´morphological singularity´ claimed for living fossils can be misleading in some cases. An extreme example of rapid morphological differentiation is found in monotypic genera endemic to oceanic islands, where divergent times at the genus level are similar to those at the species level in the continents (see references in Tables 1, 2; see Vargas, 2014). In other words, there is a clear potential for acceleration in rates of morphological change in animals and plants evolving on oceanic islands (Patiño et al., 2017).
3. Species of monotypic genera tend to be more threatened than average species of mammals and birds (Purvis et al., 2000). Indeed, our results additionally suggests that amphibians offer even a higher figure (6.56%) (see Bielby et al., 2006), while plants a lower percentage (1.8%) (Table 3).
4. Mass extinctions have dramatically affected some branches of the tree of life along their evolutionary history. In particular, disappearance of vertebrates appears to be taxonomically selective, rather than random (Purvis, 2008). Similarly, monotypic genera including CR and EX species do not seem to be equally distributed across animal families. The same is true for plants. An extreme case is the family Orchidaceae (c. 28,000 species, 736 genera, 144 monotypic genera; Chase et al., 2015) with no CR or EX monotypic genera, while the small family Aptandraceae (ca. 40 species, 8 genera) has three monotypic genera of which one is CR (Table 1).
5. Extinction of monotypic genera is particularly associated with recent population decrease and species extirpation of insular species. Among the 25 extinct monotypic genera studied in the present study, 22 occurred on islands, which indicate massive extinctions as similarly estimated at the species level (at least 800 insular species of animals and plants in the past 500 years, Fernández-Palacios et al., 2021). In particular, humans have dramatically exterminated flightless animals on remote islands (Fromm and Meiri, 2021; Maderspacher, 2022). These ´extinct´ species can be seen as bellwethers indicators of patterns or trends that can help us shape the most urgent conservation actions (Matthews et al., 2022).
6. The ELF approach is a relatively quick method to identify the species of floras and faunas most urgently in need of protection in the world. Integrating biodiversity and evolutionary results help prioritize resources and conservation efforts in already long lists of endangered species (Forest et al., 2018), particularly in developing countries where vulnerable tropical habitats and remote islands harbor a high number of threatened species with extinction.
Admittedly, we are far from a comprehensive analysis of organisms across the tree of life to identify a significant number of ELFs because of three main reasons: (i) the available lists of monotypic genera overlook some genera; (ii) deficient data, particularly IUCN status and time-calibrated phylogenies of small organisms, prevent from having enough information to test the three ELF criteria for many monotypic genera; and (iii) large genera of hundreds of species will have more likelihood of including ancient lineages, although they may fail in including significant morphological differentiation given that they are not considered different genera. In any case, the sample of five well-known organism groups of two main branches of the tree of life (eudicots and vertebrates) made possible testing the concept of ELF worldwide, which is equally applicable to any other organism groups. Indeed, the monotypic genus choice has already encouraged the search for more study cases in arthropods (Bond et al., 2020) and other angiosperms (Braz et al., 2021).
In this review, a considerable number of ELFs (57) from 3,706 monotypic genera of angiosperms, amphibians, non-avian reptiles, birds and mammals have been identified. The monotypic genera choice provides ideal candidate species in any region of the world, even though only a few genera meet the three ELF criteria (see Vargas et al., 2020; this paper). All these results highlight the necessity of using taxonomic, conservation and evolutionary approaches to provide a quick, albeit comprehensive method to integrate evolutionary information in conservation science (Lean, 2017). It is expected that the relatively small number of 57 ELFs identified so far will increase in the near future thanks to larger sample sizes, new conservation projects and more accurate methods in time-calibrated phylogenies. The test of ELFs can be considered a basic prioritization tool not only to stimulate research on morphologically isolated groups but also to quickly propose the list of urgent species in conservation programs worldwide. Once any species meets all the requirements to be considered an ELF, prioritization among ELFs should be performed by using evolutionary singularity, which has to consider both ancient lineage divergence (lineage singularity) and unique morphological differentiation (morphological singularity) into “anomalous forms” (Darwin, 1859). In sum, the ELF approach will help preserve the most exceptional evolutionary heritage before it is too late.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The author confirms being the sole contributor of this work and has approved it for publication.
The author appreciate the help provided by Ruben Heleno, Mario Fernández-Mazuecos, Pedro Jiménez-Mejías, and Virginia Valcárcel during the discussion of many points when the ELF concept was just a preliminary idea about 10 years ago. Constructive comments and advice given by Alan Love, Derek Turner, Scott Lidgard greatly help improve the quality of the study.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at:
https://www.frontiersin.org/articles/10.3389/fevo.2023.1100503/full#supplementary-material
3. ^powo.science.kew.org; mobot.org
4. ^ amphibiansoftheworld.amnh.org
6. ^avibase.bsc-eoc.org; birdsoftheworld.org; ebird.org; worldbirdnames.org
Ali, J. R., and Fritz, U. (2021). Origins of Galápagos’ land-locked vertebrates: what, whence, when, how? Biol J Linn Soc 134, 261–284. doi: 10.1093/biolinnean/blab085
Almeida, F. C., Simmons, N. B., and Giannini, N. P. (2020). A species-level phylogeny of old world fruit bats with a new higher-level classification of the family pteropodidae. Am. Mus. Novit. 2020, 1–24. doi: 10.1206/3950.1
Armstrong, K. E., Stone, G. N., Nicholls, J. A., Valderrama, E., Anderberg, A. A., Smedmark, J., et al. (2014). Patterns of diversification amongst tropical regions compared: a case study in Sapotaceae. Front. Genet. 5:362. doi: 10.3389/fgene.2014.00362
Ascarrunz, E., Claude, J., and Joyce, W. G. (2021). The phylogenetic relationships of geoemydid turtles from the Eocene Messel Pit quarry: a first assessment using methods for continuous and discrete characters. PeerJ 9:e11805. doi: 10.7717/peerj.11805
Bielby, J., Cunningham, A. A., and Purvis, A. (2006). Taxonomic selectivity in amphibians: ignorance, geography or biology? Anim. Conserv. 9, 135–143. doi: 10.1111/j.1469-1795.2005.00013.x
Bittencourt-Silva, G. B., Conradie, W., Siu-Ting, K., Tolley, K. A., Channing, A., Cunningham, M., et al. (2016). The phylogenetic position and diversity of the enigmatic mongrel frog Nothophryne Poynton, 1963 (Amphibia, Anura). Mol. Phylogenet. Evol. 99, 89–102. doi: 10.1016/j.ympev.2016.03.021
Bond, J. E., Hamilton, C. A., Godwin, R. L., Ledford, J. M., and Starrett, J. (2020). Phylogeny, evolution, and biogeography of the north American trapdoor spider family Euctenizidae (Araneae: Mygalomorphae) and the discovery of a new ‘endangered living fossil’ along California’s central coast. Insect. Syst. Divers. 4:2. doi: 10.1093/isd/ixaa010
Braz, D. M., Daniel, T. F., Kiel, C., Gao, A., Jawadi, S., and Monteiro, R. (2021). Aymoreana (Nelsonioideae, Acanthaceae), a new genus endemic to Brazil. Syst. Bot. 46, 211–217. doi: 10.1002/tax.12600
Brown, A. F., Lawrie, Y., Shannon, T. J., Collinson, J. M., Kirwan, G. M., Kirkconnell, A., et al. (2022). First genetic data for the critically endangered Cuban endemic Zapata Rail Cyanolimnas cerverai, and the taxonomic implications. J. Ornithol. 163, 945–952. doi: 10.1007/s10336-022-02004-0
Bruneau, A., Mercure, M., Lewis, G. P., and Herendeen, P. S. (2008). Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany 86, 697–718. doi: 10.1139/B08-058
Bruxaux, J., Gabrielli, M., Ashari, H., Prŷs-Jones, R., Joseph, L., Milá, B., et al. (2018). Recovering the evolutionary history of crowned pigeons (Columbidae: Goura): Implications for the biogeography and conservation of New Guinean lowland birds. Mol. Phylogenet. Evol. 120, 248–258.
Buckner, J. C., Ellingson, R., Gold, D. A., Jones, T. L., and Jacobs, D. K. (2018). Mitogenomics supports an unexpected taxonomic relationship for the extinct diving duck Chendytes lawi and definitively places the extinct Labrador Duck. Mol. Phylogenet. Evol. 122, 102–109. doi: 10.1016/j.ympev.2017.12.008
Buerki, S., Callmander, M. W., Acevedo-Rodriguez, P., Lowry, P. P., Munzinger, J., Bailey, P., et al. (2021). An updated infra-familial classification of Sapindaceae based on targeted enrichment data. Am. J. Bot. 108, 1234–1251. doi: 10.1002/ajb2.1693
Burns, K. J., Unitt, P., and Mason, N. A. (2016). A genus-level classification of the family Thraupidae (Class Aves: Order Passeriformes). Zootaxa 4088, 329–354. doi: 10.11646/zootaxa.4088.3.2
Calamari, Z. T. (2021). Total evidence phylogenetic analysis supports new morphological synapomorphies for Bovidae (Mammalia, Artiodactyla). Am. Mus. Novit. 2021, 1–38.
Cazalis, V., Di Marco, M., Butchart, S. H., Akçakaya, H. R., González-Suárez, M., Meyer, C., et al. (2022). Bridging the research-implementation gap in IUCN red list assessments. Trends Ecol. Evol. 37, 359–370. doi: 10.1016/j.tree.2021.12.002
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., Van den Berg, C., et al. (2015). An updated classification of Orchidaceae. Bot. J. Linn. Soc. 177, 151–174. doi: 10.1111/boj.12234
Chatterjee, H.J., Ho, S. Y., Barnes, I, and Groves, C. (2009). Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol. Biol. 9:1–19.
Chen, D., Hosner, P. A., Dittmann, D. L., O’Neill, J. P., Birks, S. M., Braun, E. L., et al. (2021). Divergence time estimation of Galliformes based on the best gene shopping scheme of ultraconserved elements. BMC Ecol. Evol. 21:209. doi: 10.1186/s12862-021-01935-1
Cheng, L., Li, M., Han, Q., Qiao, Z., Hao, Y., Balbuena, T. S., et al. (2022). Phylogenomics resolves the phylogeny of Theaceae by using low-copy and multi-copy nuclear gene makers and uncovers a fast radiation event contributing to tea plants diversity. Biology 11:1007. doi: 10.3390/biology11071007
Collar, N. J., and Morales, M. B. (2022). “The little bustard and its family: an overview of relationships” in Little bustard: ecology and conservation. eds. V. Bretagnolle, J. Traba, and M. B. Morales (Berlin: Springer), 9–27.
Couzens, A. M., and Prideaux, G. J. (2018). Rapid Pliocene adaptive radiation of modern kangaroos. Science 362, 72–75. doi: 10.1126/science.aas8788
Darwin, C. (1859). On the origin of the species by means of natural selection, 1st Edn. John Murray, London.
Davenport, E.. (2021). Living fossils are dead! Long live living fossils. Available at: https://nerdfighteria.info/v/XI7_HE3Bkk4/.
Eldredge, N., Thompson, J. N., Brakefield, P. M., Gavrilets, S., Jablonski, D., Jackson, J. B., et al. (2005). The dynamics of evolutionary stasis. Paleobiology 31, 133–145. doi: 10.1666/0094-8373(2005)031[0133:TDOES]2.0.CO;2
Ericson, P. G., Qu, Y., Blom, M. P., Johansson, U. S., Irestedt, M., et al. (2017). A genomic perspective of the pink-headed duck Rhodonessa caryophyllacea suggests a long history of low effective population size. Sci. Rep 7, 1–6. doi: 10.1038/s41598-017-16975-1
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Fan, W. B., Wu, Y., Yang, J., Shahzad, K., and Li, Z. H. (2018). Comparative chloroplast genomics of Dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Front. Plant Sci. 9:689. doi: 10.3389/fpls.2018.00689
Feng, Y. J., Blackburn, D. C., Liang, D., Hillis, D. M., Wake, D. B., Cannatella, D. C., et al. (2017). Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous–Paleogene boundary. Proc. Natl. Acad. Sci. U. S. A. 114, E5864–E5870. doi: 10.1073/pnas.1704632114
Fernández-Palacios, J. M., Kreft, H., Irl, S. D., Norder, S., Ah-Peng, C., Borges, P. A., et al. (2021). Scientists’ warning–the outstanding biodiversity of islands is in peril. Glob. Ecol. Conserv. 31:e01847. doi: 10.1016/j.gecco.2021.e01847
Fleagle, J. G., and Seiffert, E. R. (2020). “The phylogeny of primates” in Evolutionary neuroscience. eds. J. H. Kaas and L. Krubitzer (Cambridge, MA: Academic Press), 483–518.
Fleischer, R. C., James, H. F., and Olson, S. L. (2008). Convergent evolution of Hawaiian and Australo-Pacific honeyeaters from distant songbird ancestors. Curr. Biol. 18, 1927–1931. doi: 10.1016/j.cub.2008.10.051
Forest, F., Moat, J., Baloch, E., Brummitt, N. A., Bachman, S. P., Ickert-Bond, S., et al. (2018). Gymnosperms on the EDGE. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-24365-4
Fromm, A., and Meiri, S. (2021). Big, flightless, insular and dead: Characterising the extinct birds of the quaternary. J. Biogeogr. 48, 2350–2359. doi: 10.1111/jbi.14206
Giarla, T. C., Demos, T. C., Monadjem, A., Hutterer, R., Dalton, D., Mamba, M. L., et al. (2021). Integrative taxonomy and phylogeography of Colomys and Nilopegamys (Rodentia: Murinae), semi-aquatic mice of Africa, with descriptions of two new species. Zool. J. Linnean Soc. 192, 206–235. doi: 10.1093/zoolinnean/zlaa108
Gibb, G. C., and Shepherd, L. D. (2022). Recent evolution of extreme sexual dimorphism in the huia (Heteralocha acutirostris; Callaeidae). Mol. Phylogenet. Evol. 175:107575. doi: 10.1016/j.ympev.2022.107575
Giles, J. (2005). Special report internet encyclopaedias go head to head. Nature 438, 900–901. doi: 10.1038/438900a
Graham, S. (2010). Revision of the Caribbean Genus Ginoria (Lythraceae), Including Haitia From Hispaniola1. Ann. Missouri Bot. Gard 97:34–90. doi: 10.3417/2007028
Gumbs, R., Gray, C. L., Wearn, O. R., and Owen, N. R. (2018). Tetrapods on the EDGE: overcoming data limitations to identify phylogenetic conservation priorities. PLoS One 13:e0194680.
Harris, R. B., Birks, S. M., and Leaché, A. D. (2014). Incubator birds: biogeographical origins and evolution of underground nesting in megapodes (Galliformes: Megapodiidae). J. Biogeogr. 41, 2045–2056. doi: 10.1111/jbi.12357
Hawkins, M. T., Culligan, R. R., Frasier, C. L., Dikow, R. B., Hagenson, R., Lei, R., et al. (2018). Genome sequence and population declines in the critically endangered greater bamboo lemur (Prolemur simus) and implications for conservation. BMC Genomics 19, 1–15.
He, T., and Lamont, B. B. (2022). Ancient Rhamnaceae flowers impute an origin for flowering plants exceeding 250-million-years ago. iScience 25:104642. doi: 10.1016/j.isci.2022.104642
Heckenhauer, J., Samuel, R., Ashton, P. S., Turner, B., Barfusso, M. H., Jang, T.S., and Chase, M. W. (2017). Phylogenetic analyses of plastid DNA suggest a different interpretation of morphological evolution than those used as the basis for previous classifications of Dipterocarpaceae (Malvales). Bot. J. Linn. Soc. 185, 1–26. doi: 10.1093/botlinnean/box044
Hughes, C. E., Ringelberg, J. J., Luckow, M., and Jiménez, J. L. (2022). Mezcala –a new segregate genus of mimosoid legume (Leguminosae, Caesalpinioideae, mimosoid clade) narrowly endemic to the Balsas Depression in Mexico. PhytoKeys 205, 191–201. doi: 10.3897/phytokeys.205.78297
Holley, J. A., Sterli, J., and Basso, N. G. (2020). Dating the origin and diversification of Pan-Chelidae (Testudines, Pleurodira) under multiple molecular clock approaches. Contrib. Zool. 89, 146–174. doi: 10.1163/18759866-20191419
Isaac, N. J. B., and Pearse, W. D. (2018). “The use of EDGE (evolutionary distinct globally endangered) and EDGE-like metrics to evaluate taxa for conservation” in Phylogenetic diversity. eds. R. Scherson and D. Faith (Basel: Springer)
Isaac, N. J. B., Turvey, S. T., Collen, B., Waterman, C., and Baillie, J. E. M. (2007). Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS One 2:e296. doi: 10.1371/journal.pone.0000296
IUCN. (2017) The IUCN red list terms and conditions of use (version 3, May 2017*). Gland, Switzerland: International Union for Conservation of Nature.
IUCN. (2022). The IUCN red list of threatened species (version 2022–1). Gland, Switzerland: International Union for Conservation of Nature.
Jetz, W., Thomas, G. H., Joy, J. B., Redding, D. W., Hartmann, K., and Mooers, A. O. (2014). Global distribution and conservation of evolutionary distinctness in birds. Curr. Biol. 24, 919–930. doi: 10.1016/j.cub.2014.03.011
Jiménez-Mejías, P., Saldivia, P., Gebauer, S., and Martín-Bravo, S. (2021). A new remarkable dwarf sedge (Carex phylloscirpoides, Cyperaceae) from northern Chile, with insights on the evolution of austral Carex section Racemosae. Syst. Bot. 46, 34–47. doi: 10.1600/036364421X16128061189567
Johnson, J. A., Brown, J. W., Fuchs, J., and Mindell, D. P. (2016). Multi-locus phylogenetic inference among New World vultures (Aves: Cathartidae). Mol. Phylogent. Evol. 105, 193–199.
Joyce, W. G., Parham, J. F., Lyson, T. R., Warnock, R. C., and Donoghue, P. C. (2013). A divergence dating analysis of turtles using fossil calibrations: an example of best practices. J. Paleontol. 87, 612–634. doi: 10.1666/12-149
Kondraskov, P., Schütz, N., Schüler, C., Menezes de Sequeira, M., Santos Guerra, A., Caujape-Castells, J., et al. (2015). Biogeography of Mediterranean hotspot biodiversity: Re-Evaluating the ‘Tertiary relict’ hypothesis of Macaronesian laurel forests. PLoS One 10:e0132091. doi: 10.1371/journal.pone.0132091
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Kundu, S., Jones, C. G., Prys-Jones, R. P., and Groombridge, J. J. (2012). The evolution of the Indian Ocean parrots (Psittaciformes): extinction, adaptive radiation and eustacy. Mol. Phylogenet. Evol. 62, 296–305. doi: 10.1016/j.ympev.2011.09.025
Kljuykov, E. V., Petrova, S. E., Degtjareva, G. V., Zakharova, E. A., Samigullin, T. H., and Tilney, P. M. (2020). A taxonomic survey of monocotylar Apiaceae and the implications of their morphological diversity for their systematics and evolution. Bot. J. Linn. Soc. 192, 449–473. doi: 10.1093/botlinnean/boz095
Lean, C. H. (2017). Biodiversity realism: preserving the tree of life. Biol. Philos. 32, 1083–1103. doi: 10.1007/s10539-017-9592-0
Lerner, H. R., Meyer, M., James, H. F., Hofreiter, M., and Fleischer, R. C. (2011). Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Curr. Biol. 21, 1838–1844.
Licona-Vera, Y., and Ornelas, J. F. (2017). The conquering of North America: dated phylogenetic and biogeographic inference of migratory behavior in bee hummingbirds. BMC Evol. Biol 17, 1–17. doi: 10.1186/s12862-017-0980-5
Lidgard, S., and Love, A. C. (2018). Rethinking living fossils. Bioscience 68, 760–770. doi: 10.1093/biosci/biy084
Lidgard, S., and Love, A. C. (2021). The living fossil concept: reply to Turner. Biol. Philos. 36, 1–16. doi: 10.1007/s10539-021-09789-z
Liede-Schumann, S., and Meve, U. (2018). Vincetoxicum (Apocynaceae—Asclepiadoideae) expanded to include Tylophora and allies. Phytotaxa 369, 129–184. doi: 10.11646/phytotaxa.369.3.1
Liedtke, H. C., Müller, H., Hafner, J., Penner, J., Gower, D. J., Mazuch, T., et al. (2017). Terrestrial reproduction as an adaptation to steep terrain in African toads. Proc. Biol. Sci. 284:20162598. doi: 10.1098/rspb.2016.2598
Liu, S., Westbury, M. V., Dussex, N., Mitchell, K. J., Sinding, M. H. S., Heintzman, P. D., et al. (2021). Ancient and modern genomes unravel the evolutionary history of the rhinoceros family. Cells 184, 4874–4885. doi: 10.1016/j.cell.2021.07.032
Loader, S. P., Sara Ceccarelli, F., Menegon, M., Howell, K. M., Kassahun, R., Mengistu, A. A., et al. (2014). Persistence and stability of Eastern Afromontane forests: evidence from brevicipitid frogs. J. Biogeogr. 41, 1781–1792. doi: 10.1111/jbi.12331
Lourenço, J. M., Claude, J., Galtier, N., and Chiari, Y. (2012). Dating cryptodiran nodes: origin and diversification of the turtle superfamily Testudinoidea. Mol. Phylogenet. Evol. 62, 496–507. doi: 10.1016/j.ympev.2011.10.022
Lourenço, J. M., Glémin, S., Chiari, Y., and Galtier, N. (2013). The determinants of the molecular substitution process in turtles. J. Evol. Biol. 26, 38–50. doi: 10.1111/jeb.12031
Lovette, I. J., Pérez-Emán, J. L., Sullivan, J. P., Banks, R. C., Fiorentino, I., Córdoba-Córdobao, S., and Bermingham, E. (2010). A comprehensive multilocus phylogeny for the wood-warblers and a revised classification of the Parulidae (Aves). Mol. Phylogenet. Mol. Phylogenet. Evol 57, 753–770. doi: 10.1016/j.ympev.2010.07.018
Lu, Y., Ran, J. H., Guo, D. M., Yang, Z. Y., and Wang, X. Q. (2014). Phylogeny and divergence times of gymnosperms inferred from single-copy nuclear genes. PLoS One 9:e107679. doi: 10.1371/journal.pone.0107679
Luo, H., Li, H., Huang, A., Ni, Q., Yao, Y., Xu, H., et al. (2019). The complete mitochondrial genome of Platysternon megacephalum peguense and molecular phylogenetic analysis. Genes 10:487. doi: 10.3390/genes10070487
Maderspacher, F. (2022). Flightless birds. Curr. Biol. 32, R1155–R1162. doi: 10.1016/j.cub.2022.09.039
Martín-Hernanz, S., Nogales, M., Valente, L., Fernández-Mazuecos, M., Pomeda-Gutiérrez, F., Cano, E., et al. (2023). Time-calibrated phylogenies reveal mediterranean and pre-mediterranean origin of the thermophilous vegetation of the Canary Islands. Ann. Bot. 131, 667–684. doi: 10.1093/aob/mcac160
Matthews, T. J., Wayman, J. P., Cardoso, P., Sayol, F., Hume, J. P., Ulrich, W., et al. (2022). Threatened and extinct island endemic birds of the world: distribution, threats and functional diversity. J. Biogeogr. 49, 1920–1940. doi: 10.1111/jbi.14474
Miguez, M., Bartolucci, F., Jiménez-Mejías, P., and Martín-Bravo, S. (2022). Re-evaluating the presence of Carex microcarpa (Cyperaceae) in Italy based on herbarium material and DNA barcoding. Plant Biosyst. 156, 628–634. doi: 10.1080/11263504.2021.1897703
McDade, L. A., Kiel, C. A., Daniel, T. F., and Darbyshire, I. (2022). Justicieae II: Resolved placement of many genera and recognition of a new lineage sister to Isoglossinae.. Aliso 38, 1–31. doi: 10.5642/aliso.20213801.02
Mindell, D. P., Fuchs, J., and Johnson, J. A. (2018). “Phylogeny, taxonomy, and geographic diversity of diurnal raptors: Falconiformes, Accipitriformes, and Cathartiformes” in Birds of prey (Cham: Springer), 3–32.
Mishler, B. D., Knerr, N. J., González-Orozco, C. E., Thornhill, A. H., Laffan, S. W., and Miller, J. T. (2014). Phylogenetic measures of biodiversity and neo- and paleo-endemism in Australian acacia. Nat. Commun. 5:4473. doi: 10.1038/ncomms5473
Mittermeier, J. C., Correia, R., Grenyer, R., Toivonen, T., and Roll, U. (2021). Using Wikipedia to measure public interest in biodiversity and conservation. Conserv. Biol. 35, 412–423. doi: 10.1111/cobi.13702
Ohlson, J. I., Irestedt, M., Fjeldså, J., and Ericson, P. G. P. (2012). Nuclear DNA from a 180-year-old study skin reveals the phylogenetic position of the Kinglet Calyptura cristata (Passeriformes: Tyrannides). Ibis 154, 533–541. doi: 10.1111/j.1474-919X.2012.01243.x
Otero, A., Vargas, P., Fernández-Mazuecos, M., Jiménez-Mejías, P., Valcárcel, V., Villa-Machío, I., et al. (2022). A snapshot of progenitor-derivative speciation in Iberodes (Boraginaceae). Mol. Ecol. 31, 3192–3209. doi: 10.1111/mec.16459
Patiño, J., Whittaker, R. J., Borges, P. A., Fernández-Palacios, J. M., Ah-Peng, C., Araújo, M. B., et al. (2017). A roadmap for island biology: 50 fundamental questions after 50 years of the theory of island biogeography. J. Biogeogr. 44, 963–983. doi: 10.1111/jbi.12986
Pelser, P. B., Abbott, R. J., Comes, H. P., Milton, J. J., Moeller, M., Looseley, M. E., et al. (2012). The genetic ghost of an invasion past: colonization and extinction revealed by historical hybridization in Senecio. Mol. Ecol. 21, 369–387. doi: 10.1111/j.1365-294X.2011.05399.x
Plantdrew, JarrahTree, Notwith. (2018a). Category:Monotypic_angiosperm_genera. Available at: https://en.wikipedia.org/wiki (Accessed November 5, 2022).
Plantdrew, JarrahTree, Notwith. (2018b). Category:Monotypic_vertebrate_genera. Available at: https://en.wikipedia.org/wiki (Accessed November 5, 2022).
Purvis, A. (2008). Phylogenetic approaches to the study of extinction. Ann. Rev. Ecol. Evol. Syst. 39, 301–319. doi: 10.1146/annurev.ecolsys.063008.102010
Purvis, A., Gittleman, J. L., Cowlishaw, G., and Mace, G. M. (2000). Predicting extinction risk in declining species. Proceedings Royal Soc. London. Series B 267, 1947–1952. doi: 10.1098/rspb.2000.1234
Rabosky, D. L., Slater, G. J., and Alfaro, M. E. (2012). Clade age and species richness are decoupled across the eukaryotic tree of life. PLoS Biol. 10:e1001381. doi: 10.1371/journal.pbio.1001381
Reaney, A. M., Saldarriaga-Córdoba, M., and Pincheira-Donoso, D. (2018). Macroevolutionary diversification with limited niche disparity in a species-rich lineage of cold-climate lizards. BMC Evol. Biol. 18, 1–12. doi: 10.1186/s12862-018-1133-1
Ricklefs, R. E. (2007). Estimating diversification rates from phylogenetic information. Trends Ecol. Evol. 22, 601–610. doi: 10.1016/j.tree.2007.06.013
Rio, J. P., and Mannion, P. D. (2021). Phylogenetic analysis of a new morphological dataset elucidates the evolutionary history of Crocodylia and resolves the long-standing gharial problem. PeerJ 9:e12094. doi: 10.7717/peerj.12094
Ronquist, F. (2014). “Systematics: charting the tree of life” in The tree of life: Evolution and classification of living organisms. eds. P. Vargas and R. Zardoya (Massachusetts: Sinauer Ass)
Rosauer, D. F., and Jetz, W. (2015). Phylogenetic endemism in terrestrial mammals. Glob. Ecol. Biog. 24, 168–179. doi: 10.1111/geb.12237
Royer, D. L., Hickey, L. J., and Wing, S. L. (2003). Ecological conservatism in the “living fossil” ginkgo. Paleobiology 29, 84–104. doi: 10.1666/0094-8373(2003)029<0084:ECITLF>2.0.CO;2
Schirtzinger, E. E., Tavares, E. S., Gonzales, L. A., Eberhard, J. R., Miyaki, C. Y., Sanchez, J. J., et al. (2012). Multiple independent origins of mitochondrial control region duplications in the order Psittaciformes. Mol. Phylogenet. Evol. 64, 342–356. doi: 10.1016/j.ympev.2012.04.009
Schley, R. J., de la Estrella, M., Pérez-Escobar, O. A., Bruneau, A., Barraclough, T., Forest, F., et al. (2018). Is Amazonia a ‘museum’ for Neotropical trees? The evolution of the Brownea clade (Detarioideae, Leguminosae). Mol. Phylogenet. Evol. 126, 279–292. doi: 10.1016/j.ympev.2018.04.029
Schneider, J. V., Jungcurt, T., Cardoso, D., Amorim, A. M., Töpel, M., Andermann, T., et al. (2021). Phylogenomics of the tropical plant family Ochnaceae using targeted enrichment of nuclear genes and 250+ taxa. Taxon 70, 48–71. doi: 10.1002/tax.12421
Schweizer, M., Guentert, M., and Hertwig, S. T. (2013). Out of the Bassian province: historical biogeography of the Australasian platycercine parrots (Aves, Psittaciformes). Zool. Scr. 42, 13–27. doi: 10.1111/j.1463-6409.2012.00561.x
Simões, A. O., Livshultz, T., Conti, E., and Endress, M.E. (2007). Phylogeny And Systematics Of The Rauvolfioideae (Apocynaceae) Based On Molecular And Morphological Evidence. Ann. Missouri Bot. Gard. 94, 268–297. doi: 10.3417/0026-6493(2007)94[268:PASOTR]2.0.CO;2
Siu-Ting, K., Gower, D. J., Pisani, D., Kassahun, R., Gebresenbet, F., Menegon, M., et al. (2014). Evolutionary relationships of the critically endangered frog Ericabatrachus baleensis Largen, 1991 with notes on incorporating previously unsampled taxa into large-scale phylogenetic analyses. BMC Evol. Biol. 14:44. doi: 10.1186/1471-2148-14-44
Soares, A. E., Novak, B. J., Haile, J., Heupink, T. H., Fjeldså, J., Gilbert, M. T. P., et al. (2016). Complete mitochondrial genomes of living and extinct pigeons revise the timing of the columbiform radiation. BMC Evol. Biol. 16:230. doi: 10.1186/s12862-016-0800-3
Soltis, P. S., Folk, R. A., and Soltis, D. E. (2019). Darwin review: angiosperm phylogeny and evolutionary radiations. Proc. R. Soc. B 286:20190099. doi: 10.1098/rspb.2019.0099
Steinthorsdottir, M., Coxall, H. K., De Boer, A. M., Huber, M., Barbolini, N., Bradshaw, C. D., et al. (2021). The Miocene: the future of the past. Paleoceanogr. Paleoclimatol. 36:4037. doi: 10.1029/2020PA004037
Su, H. J., Hu, J. M., Anderson, F. E., Der, J. P., and Nickrent, D. L. (2015). Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae. Taxon 64, 491–506. doi: 10.12705/643.2
Su, Y., Liao, W., Wang, T., Sun, Y., Wei, Q., and Chang, H. (2011). Phylogeny and evolutionary divergence times in Apterosperma and Euryodendron: evidence of a Tertiary origin in south China. Biochem. Syst. Ecol. 39, 769–777. doi: 10.1016/j.bse.2011.07.004
Suwannapoom, C., Sumontha, M., Tunprasert, J., Ruangsuwan, T., Pawangkhanant, P., Korost, D. V., et al. (2018). A striking new genus and species of cave-dwelling frog (Amphibia: Anura: Microhylidae: Asterophryinae) from Thailand. Peer J. 6:e4422. doi: 10.7717/peerj.4422
Tietze, D. T., Päckert, M., Martens, J., Lehmann, H., and Sun, Y. H. (2013). Complete phylogeny and historical biogeography of true rosefinches (Aves: Carpodacus). Zool. J. Linnean Soc. 169, 215–234. doi: 10.1111/zoj.12057
Turner, D. D. (2019). In defense of living fossils. Biol. Philos. 34, 1–22. doi: 10.1007/s10539-019-9678-y
Valente, L., Etienne, R. S., and Garcia-R, J. C. (2019). Deep macroevolutionary impact of humans on New Zealand’s unique avifauna. Curr. Biol. 29, 2563–2569. doi: 10.1016/j.cub.2019.06.058
Valente, L. M., Savolainen, V., and Vargas, P. (2010). Unparalleled rates of species diversification in Europe. Proc. R. Soc. B Biol. Sci. 277, 1489–1496. doi: 10.1098/rspb.2009.2163
Vane-Wright, R. I., Humphries, C. J., and Williams, P. H. (1991). What to protect?—systematics and the agony of choice. Biol. Conserv. 55, 235–254. doi: 10.1016/0006-3207(91)90030-D
Vargas, P. (2007). “Are Macaronesian island refugia of relict plant lineages?: a molecular survey” in Phylogeography in southern European Refugia: Evolutionary perspectives on the origins and conservation of European biodiversity. eds. S. J. Weiss and N. Ferrand (Dordrecht: Kluwer), 297–314.
Vargas, P. (2014). “Evolution on islands” in The tree of life: Evolution and classification of living organisms. eds. P. Vargas and R. Zardoya (Massachusetts: Sinauer Ass)
Vargas, P., Fernández-Mazuecos, M., and Heleno, R. (2018). Phylogenetic evidence for a Miocene origin of Mediterranean lineages: species diversity, reproductive traits and geographical isolation. Plant Biol. 20, 157–165. doi: 10.1111/plb.12626
Vargas, P., Jimenez-Mejias, P., and Fernandez-Mazuecos, M. (2020). ‘Endangered living fossils’(ELFs): long-term survivors through periods of dramatic climate change. Environ. Exp. Bot. 170:103892. doi: 10.1016/j.envexpbot.2019.103892
Vargas, P., Valente, L. M., Blanco-Pastor, J. L., Liberal, I., Guzmán, B., Cano, E., et al. (2014). Testing the biogeographical congruence of palaeofloras using molecular phylogenetics: snapdragons and the Madrean–Tethyan flora. J. Biogeogr. 41, 932–943. doi: 10.1111/jbi.12253
Vargas, P., and Zardoya, R. (2014). The tree of life: Evolution and classification of living organisms. Massachusetts: Sinauer Ass.
Wilson, D. E., and Reeder, M. R.. (2005). Mammal species of the world. A taxonomic and geographic reference. Baltimore, MA: Johns Hopkins University Press.
Wright, P. C. (2010). Mammals in Madagascar: biogeography, natural history, and new species. J. Mamm. Evol. 17, 65–67.
Wright, A. M., Bapst, D. W., Barido-Sottani, J., and Warnock, R. C. (2022). Integrating fossil observations into phylogenetics using the fossilized birth–death model. Ann. Rev. Ecol., Evol. Syst 53, 251–273. doi: 10.1146/annurev-ecolsys-102220-030855
Wood, J. R., Mitchell, K. J., Scofieldni, R. P., De Pietri, V. L., Rawlence, N. J., and Cooper, A. (2017). Phylogenetic relationships and terrestrial adaptations of the extinct laughing owl, Sceloglaux albifacies (Aves: Strigidae). Zool. J. Linn. Soc. 179, 907–918. doi: 10.1111/zoj.12483
Xu, C., and Hong, D. Y. (2021). Phylogenetic analyses confirm polyphyly of the genus Campanula (Campanulaceae s. str.), leading to a proposal for generic reappraisal. J. Syst. Evol. 59, 475–489. doi: 10.1111/jse.12586
Zedane, L., Hong-Wa, C., Murienne, J., Jeziorski, C., Baldwin, B. G., and Besnard, G. (2016). Museomics illuminate the history of an extinct, paleoendemic plant lineage (Hesperelaea, Oleaceae) known from an 1875 collection from Guadalupe Island, Mexico. Biol. J. Linn. Soc. 117, 44–57. doi: 10.1111/bij.12509
Keywords: angiosperms, EDGE, endangered genera, IUCN, phylogenies, Pleistocene, vertebrates
Citation: Vargas P (2023) Exploring ‘endangered living fossils’ (ELFs) among monotypic genera of plants and animals of the world. Front. Ecol. Evol. 11:1100503. doi: 10.3389/fevo.2023.1100503
Received: 16 November 2022; Accepted: 31 March 2023;
Published: 22 June 2023.
Edited by:
Scott Lidgard, Field Museum, United StatesReviewed by:
Derek Turner, Connecticut College, United StatesCopyright © 2023 Vargas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pablo Vargas, dmFyZ2FzQHJqYi5jc2ljLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.