
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 18 April 2023
Sec. Behavioral and Evolutionary Ecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1099481
This article is part of the Research TopicAdvances in Soft Matter Biological AdhesivesView all 6 articles
 Nadia A. Ayoub1*
Nadia A. Ayoub1* Lucas DuMez2
Lucas DuMez2 Cooper Lazo1
Cooper Lazo1 Maria Luzaran1
Maria Luzaran1 Jamal Magoti1
Jamal Magoti1 Sarah A. Morris2
Sarah A. Morris2 Richard H. Baker3
Richard H. Baker3 Thomas Clarke1
Thomas Clarke1 Sandra M. Correa-Garhwal3
Sandra M. Correa-Garhwal3 Cheryl Y. Hayashi3
Cheryl Y. Hayashi3 Kyle Friend4
Kyle Friend4 Brent D. Opell2*
Brent D. Opell2*Introduction: Orb web and cobweb weaving spiders in the superfamily Araneoidea are distinguished by their ability to make a chemically sticky aqueous glue in specialized aggregate silk glands. Aggregate glue is an environmentally responsive material that has evolved to perform optimally around the humidity at which a spider forages. Protein components and their post-translational modifications confer stickiness to the glue, but the identities of these proteins have not been described for orb web weavers.
Methods: Using biomechanics, gene expression data, and proteomics, we characterized the glue’s physical properties and molecular components in two congeners that live in different environments, Argiope argentata (dry southwest US) and Argiope trifasciata (humid southeast US).
Results: The droplets of A. argentata are less hygroscopic than those of A. trifasciata and have proportionately smaller viscoelastic protein cores, which incorporate a smaller percentage of absorbed water as humidity increases. Argiope argentata protein cores were many times stiffer and tougher than A. trifasciata protein cores. Each species’ glue included ~30 aggregate-expressed proteins, most of which were homologous between the two species, with high sequence identity. However, the relative contribution and number of gene family members of each homologous group differed. For instance, the aggregate spidroins (AgSp1 and AgSp2) accounted for nearly half of the detected glue composition in A. argentata, but only 38% in A. trifasciata. Additionally, AgSp1, which has highly negatively charged regions, was ~2X as abundant as the positively charged AgSp2 in A. argentata, but ~3X as abundant in A. trifasciata. As another example, A. argentata glue included 11 members of a newly discovered cysteine-rich gene family, versus 7 members in A. trifasciata.
Discussion: Cysteines form disulfide bonds that, combined with the higher potential for electrostatic interactions between AgSp1 and AgSp2, could contribute to the greater stiffness of A. argentata glue. The ability to selectively express different glue protein genes and/or to extrude their products at different rates provides a faster mechanism to evolve material properties than sequence evolution alone.
Spiders rank in the top five of the most species rich animal orders and much of their evolutionary success has been attributed to evolutionary innovations in silk types and their use (Bond and Opell, 1998; Bond et al., 2014; Fernández et al., 2018; Coddington et al., 2019). Nowhere is silk innovation more evident than in the hyperdiverse superfamily, Araneoidea, which represents ~25% of all described spider species (Jäger, 2012; World Spider Catalog, 2022). This clade contains orb weaving spiders and other web building spiders derived from orb weaving ancestors. The threads that form an orb web are biomechanically integrated at multiple levels of organization to optimize prey capture. A web’s stiff frame and radial lines absorb most of the force of prey impact, allowing its more elastic, adhesive capture thread spiral to retain prey until a spider can subdue them (Kelly et al., 2011; Eberhard, 2020). In turn, properties of a capture thread’s components are scaled with each other; across species, the stiffness of the pair of supporting flagelliform fibers is correlated with the stiffness of the adhesive protein cores of its glue droplets (Figure 1; Opell et al., 2021; Kelly et al., 2022). Amorphous proteins in a droplet’s outer aqueous layer (Amarpuri et al., 2015a) adhere to a surface and, as force is applied, these outer layer proteins transfer force to the droplet’s viscoelastic protein core (Sahni et al., 2010), in a manner that is not well understood, causing the core to extend and transfer force to the flagelliform fibers (Opell et al., 2022). This causes the thread to bow and sum the adhesion of multiple glue droplets in suspension bridge fashion (Opell and Hendricks, 2009; Guo et al., 2018, 2019).
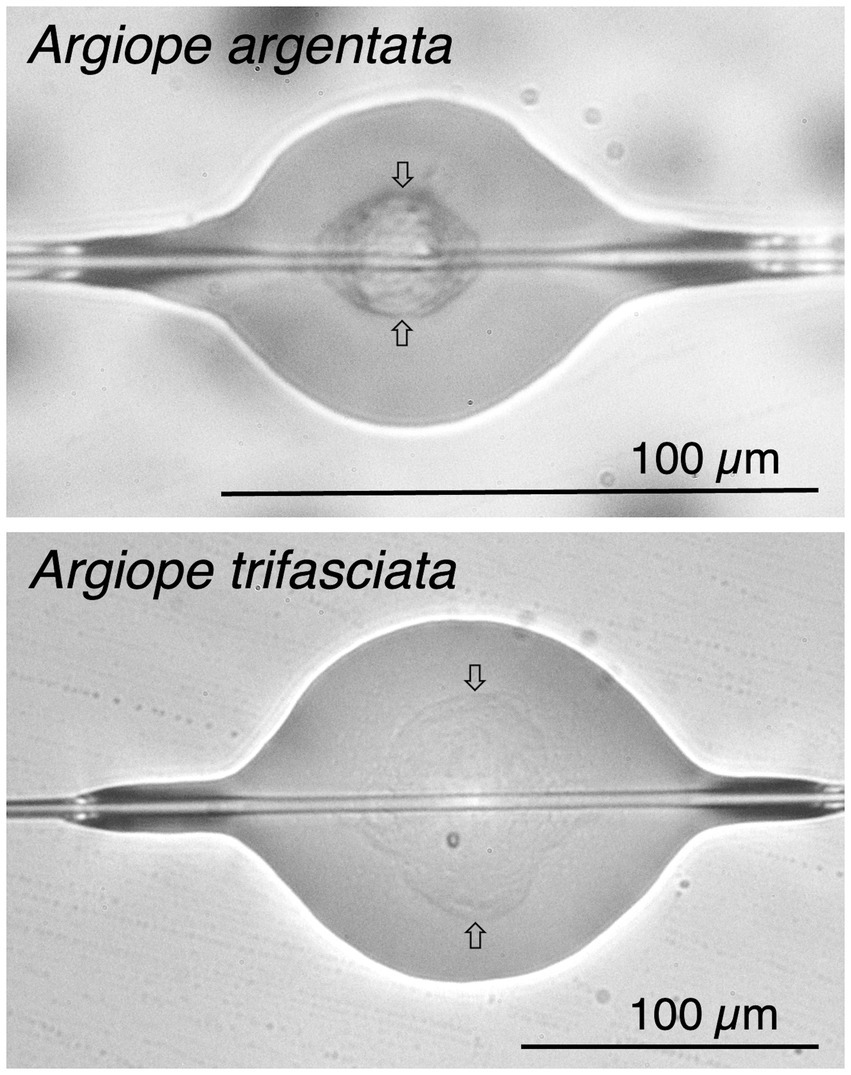
Figure 1. Glue droplets of Argiope argentata and Argiope trifasciata flattened at 72% relative humidity, showing each droplet’s outer aqueous layer, protein core (identified by arrows), and supporting flagelliform fiber pair.
As polymeric substances made largely of protein, spider silks are excellent examples of soft matter. This is especially true of the chemically sticky aqueous glue of orb weavers synthesized in aggregate glands, which open on spigots on each of a spider’s posterior lateral spinnerets. These spigots flank a flagelliform spigot resulting in each of the flagelliform fibers being coated with aggregate material before the two fibers merge to form a larger cylinder. Immediately after extrusion, Rayleigh instability reconfigures the aggregate liquid, which coalesces into sticky droplets evenly distributed along the capture spiral thread (Edmonds and Vollrath, 1992; Jain et al., 2018). As this happens, some of the protein condenses into a droplet’s visible core and some remains as amorphous protein that is not visible by light microscopy (Figure 1; Amarpuri et al., 2015a). The remaining aggregate material forms an aqueous layer that covers droplet cores and flagelliform fibers, both within and between glue droplets (Figure 1).
Aggregate secretions are a composite of proteins, salts, and low molecular mass compounds (LMCCs) (Townley and Tillinghast, 2013). Most of a droplet’s hygroscopicity has been attributed to its LMMCs, although the protein core may also contribute (Jain et al., 2018; Opell et al., 2018b). As humidity increases, the droplets absorb water, become larger, more pliable, and, when flattened on a surface, have an increased contact area for adhesion. Some absorbed water is incorporated into the droplet’s core, increasing the spacing between protein bonds, which causes protein cohesion to decrease and droplet extensibility to increase (Sahni et al., 2011). When water absorption is excessive, protein cores become overlubricated and adhesive force decreases because droplets more easily pull from a surface (Sahni et al., 2011; Opell et al., 2019). Consequently, maximum thread adhesion is achieved at a humidity where droplet adhesive contact and protein core cohesion are optimized, ensuring that the adhesive forces of multiple droplets are summed (Amarpuri et al., 2015b; Opell et al., 2022). Maximum adhesive forces are generally associated with the humidity at which a species forages (Amarpuri et al., 2015b; Opell et al., 2018b, 2021, 2022). These optimal humidities are also correlated with highest retention times of prey (Opell et al., 2017, 2019).
The protein cores of orb web glue droplets are largely responsible for droplet adhesion, contributing an order of magnitude greater adhesive force than the capillary force of the droplet’s surrounding aqueous layer (Sahni et al., 2011). The visible protein cores (Figure 1) of glue droplets were initially termed glycoproteins due to the addition of sugars (Vollrath and Tillinghast, 1991), but aggregate glue proteins are also phosphorylated (Tillinghast et al., 1992; Ayoub et al., 2021). Therefore, we refer to them simply as core proteins. Glycosylation and phosphorylation are common post-translational modifications (PTMs) to proteinaceous biological adhesives (Wagner et al., 1992; Stewart et al., 2011a,b; Tarakhovskaya, 2014; Waite, 2017). The branching groups of sugars likely insinuate into prey surfaces, promoting adhesion; phosphate groups increase ionic interactions, which contributes to the glue’s ability to resist breaking (Stewart et al., 2011a).
Until recently, the identities of aggregate proteins were unknown. Leading candidates were members of the spidroin gene family expressed in aggregate glands, AgSp1 and AgSp2 (Collin et al., 2016; Stellwagen and Renberg, 2019). We confirmed the presence of these two spidroins in aggregate glue droplets of the araneoid cobweb weaving family Theridiidae (Ayoub et al., 2021). Spidroins (spider fibrous proteins) are large, highly repetitive proteins, that have gland-specific expression. Another example besides aggregate spidroins, is flagelleliform spidroin (Flag), the primary component of the capture spiral’s axial fiber, which is highly expressed in flagelliform glands (Hayashi and Lewis, 1998; Chaw et al., 2021). Aggregate spidroins have only been described from araneoid spiders, consistent with the phylogenetic restriction of aggregate glands to this superfamily (Arakawa et al., 2022). Cobweb glue droplets contain many additional non-spidroin proteins (Ayoub et al., 2021). Thus far, however, no protein components have been confirmed from the aggregate glues placed on capture spirals of araneoid orb webs (Araneidae and Tetragnathidae).
Here, we describe the protein components and post-translational modifications (PTMs) of two closely related orb web weaving spider species, Argiope argentata (Fabricius, 1775) and A. trifasciata (Forsskål, 1775) (Agnarsson et al., 2016). We also document the adhesive properties of their glue droplets at five different relative humidities. These species have partially overlapping geographic ranges, but A. argentata is located primarily in dry regions of the southwestern United States and central and south America, while A. trifasciata inhabits more humid environments on multiple continents. Owing to habitat differences in North American populations of these two Argiope species, we hypothesized that the capture thread glue droplets of A. argentata would maximize adhesion at lower humidity than those of A. trifasciata. This assumes that the main difference between the two species’ droplets will be that A. argentata droplets are more hygroscopic than those of A. trifasciata. However, orb web adhesives are complex and other aspects of web architecture and prey capture may lead to additional differences between the material properties and composition of these species’ glue droplet core proteins (Sensenig et al., 2012, 2013; Opell et al., 2021). We relate differences in material properties of these two species and between Argiope and distantly related cobweb weavers to variation in spidroin and other glue protein components and their PTMs. Doing so provides a clearer picture of how spider glue proteins have evolved to contribute optimally to prey retention in the context of a species’ habitat.
Web samples of 14 adult female A. trifasciata were collected near Blacksburg, Virginia in September 2015 during the early morning shortly after they were spun. Aluminum frames whose rims were covered with double-sided tape were pressed against the web and threads pressed into the tape before extending threads were cut or broken. We transported these web samples to the laboratory in closed boxes to prevent contamination. Twelve A. argentata females were collected on Coronado Beach, Coronado, California in late August 2021 and maintained in the laboratory. They were housed in 61 × 61 × 9 cm wooden frames with 4.8 mm diameter wooden rods suspended 14 mm from all sides and maintained on a 12 h light, 12 h dark cycle. To maintain humidity, each frame was covered with Reynolds 916 polyvinyl chloride Foodservice Film® and contained two Humidi Wicks® for room humidifiers that were immersed in distilled water. We completed all tests of droplets from each individual A. trifasciata by late afternoon on the day that threads were collected. Two days were sometimes required to complete all tests of droplets from each individual A. argentata.
Most orb web weaving spiders are generalist predators (Eberhard, 2020). Our personal observations suggest that the A. trifasciata from whose webs we collected threads had a continual supply of diverse, mostly insect, prey in the field. We often found larger orthopterans in the webs and being actively consumed by A. trifasciata at our collecting site. Our observations agree with a study of this species in Indiana and Iowa (USA) that showed acridid grasshoppers made up an average 64% of the A. trifasciata diet (Brown, 1981). A similar diet was found for A. argentata that were studied in Panama, where orthopterans comprised 55% of the mass of prey caught by adult females (Robinson and Robinson, 1970). Consistent with a high proportion of orthopterans in the natural diets of Argiope, we fed an adult Acheta domesticus cricket to A. argentata housed in the laboratory on alternate days. Crickets came from Flukers Farms (Port Allen, LA, USA). We provided these crickets continual access to water and a diet of 50% corn meal and 50% crushed aquatic turtle pellet food (35% protein, 5% fat, and 5% fiber).
Studies that deprived orb web weavers of food, fed them protein-rich and protein-poor liquid diets, and restricted the kind of prey they were fed found that diet affected glue droplet features such as droplet volume, core protein size, and adhesiveness as well as the composition of LMMCs in a droplet’s aqueous layer and the diameters of capture thread flagelliform fibers (Townley et al., 2006; Blamires et al., 2014, 2015, 2016, 2017a,b, 2018a,b). However, given the importance of orthopterans in the natural diets of both A. argentata and A. trifasciata, it is likely that the laboratory diet of A. argentata in our study was both appropriate and like that of A. trifasciata.
Wind has also been shown to affect orb web glue droplets (Wu et al., 2013). However, we can rule out the effect of wind. As noted in the next paragraph, we found no differences in the droplets of the same A. trifasciata individuals from webs that were constructed in the field and in the laboratory. Furthermore, laboratory webs of both A. trifasciata and A. argentata were constructed in wooden frames that were completely covered by plastic food wrap, which blocked all air movement.
The relative humidity at which webs are spun is known to affect the size of an orb web’s glue droplets (Edmonds and Vollrath, 1992). Therefore, to assess the impact of lab conditions on A. argentata glue droplets, in 2022 we compared the droplet volumes of 12 adult female A. trifasciata capture threads that were spun in the field with webs these same individuals produced in the laboratory when housed as described above for A. argentata. We made this comparison during the September – October period that A. argentata were maintained in the laboratory to account for any changes in laboratory humidity that occurred as building conditions were switching from air conditioning to heating. Two droplets from an individual’s field-spun web and two droplets from this individual’s lab-spun web were photographed at 55% RH (as described below) and these droplets’ lengths and widths measured with Image J, allowing droplet volume to be computed (Liao et al., 2015). A comparison of the means of these individual’s field-spun droplet volumes with their lab-spun droplet volumes showed that these volumes did not differ (Matched pairs test two-tailed p = 0.6392). Mean field droplet volume was 37,564 ± SE 5,455 μm3 and mean lab droplet volume was 41,852 ± SE 11,942 μm3. A regression showed that lab-spun droplet volume (LV) was directly related to field-spun droplet volume (FV) (p = 0.0089, LV = FV × 1.566–16,984 μm3). In seven individuals field droplet volume was greater than lab droplet volume and in five individual’s lab droplet volume exceeded field droplet volume. Mean lab droplet volume was 111% of field droplet volume, but decreased to 101% when the individual with the largest mean lab-spun droplet volume was excluded. The core protein volumes of droplets from field- and laboratory-spun capture threads (21,362 ± 4,416 μm3 and 27,753 ± 6,274 μm3, respectively) did not differ (Wilcoxon p = 0.2727). When field and laboratory values are combined, core protein volume and droplet volume are correlated (core volume = 0.55 droplet volume + 2,575 μm3; p < 0.0001, R = 0.93). A residual analysis does not distinguish the deviation of field- and laboratory-produced droplets from this common regression line (p = 0.1939), indicating that core protein makes up a similar proportion of each set of droplets.
The procedures for characterizing capture thread glue droplets have been described and technical considerations related to these methods discussed previously (Opell et al., 2021, 2022). Therefore, we review these only briefly here. To profile glue droplet properties as they responded to changes in a spider’s habitat, we characterized these features at 20%, 37%, 55%, 72%, and 90% relative humidity (RH). This was accomplished by photographing and extending droplets within a glass covered chamber that rested on the mechanical stage of a microscope. We monitored humidity with a digital hygrometer whose probe tip extended into the chamber and used silica gel to lower humidity within the chamber and distilled water saturated Kimwipes® to raise humidity. Small adjustments to humidity were made by either drawing room air into the chamber through a tube connected to the chamber or gently blowing into this tube to increase humidity. A thermostat-controlled Peltier thermocouple array attached to the wall of the chamber maintained a temperature of 23° C. We used forceps whose tips were blocked open and covered with double sided tape to transfer thread strands from collecting frames or rings to supports glued to a microscope slide at 4.8 mm intervals and covered with double-sided tape.
To determined droplet protein core volume, we photographed a suspended droplet, dropped a cover slide on this droplet, flattening it as it might be when contacting an insect surface and revealing its protein core, and then photographed the flattened droplet. Using Image J (2006), we measured a suspended droplet’s length and determined its volume (Liao et al., 2015). Next, we measured a flattened droplet’s area and the area of its protein core. Dividing a droplet’s volume by its surface area yielded droplet thickness and multiplying this thickness by the surface area of its protein core yielded protein core volume. Subtracting protein core volume from droplet volume yielded the volume of the droplet’s aqueous layer. These values allowed us to determine the percent of a droplet’s volume occupied by its protein core, to calculate the volume of water absorbed as humidity increased, and to determine the allocation of this absorbed water to aqueous material and to protein core.
In the case of an A. trifasciata individual, we photographed three suspended droplets, flattened these droplets, and then determined the mean percent droplet volume occupied by protein core. We used this individual- and humidity-specific index to infer the protein core volumes of two additional suspended droplets whose extension was filmed, as described below. We characterized the droplet properties of A. argentata using a simplified procedure developed to characterize the properties of theridiid gumfoot glue droplets, which are less uniformly sized than those of orb web droplets (Ayoub et al., 2021). This involved photographing the suspended droplet that was to be extended, extending this droplet, and then flattening and photographing this droplet to determine its aqueous and protein core volumes.
We prepared for extension by sliding droplets away from a focal droplet at the center of the 4.8 mm suspended thread strand. This ensured that the 413 μm tip of a probe contacted only a single droplet. After the probe’s tip was pressed against a droplet by advancing the microscope stage’s X axis 500 μm into the thread, the thread strand was withdrawn by a stepping motor attached to the stage’s X-axis manipulator at a velocity of 69 μm s−1 while a movie was recorded. We used iMovie® to view these movies and an Onde® screen caliper and protractor to measure the length of an extending droplet filament at the initiation of extension and at 20% intervals until the droplet either pulled from the probe tip or transitioned from phase 1 to phase 2 extension (Opell et al., 2018a). As a thread is pulled from a surface it assumes a suspension bridge configuration with outer droplets extending more than droplets near the center of the thread strand (Opell and Hendricks, 2009). During this time, these droplets exhibit phase 1 behavior, which is characterized by their extending core proteins remaining surrounded by aqueous material. However, in some species whose glue droplets are very hygroscopic, such as the two Argiope species we studied, droplets enter phase 2 extension, which is characterized by tiny droplets of aqueous solution forming along the extending protein core, exposing the core protein. During phase 2 the glue protein filament stiffens, probably due to load-stiffening at greater tensions. At our rate of droplet extension, evaporative loss of water incorporated into the protein may also contribute to protein stiffening. In both A. argentata and A. trifasciata, phase 2 extension was common at 72 and 90% RH.
We believe that more force is necessary to extend a droplet’s denser protein core than its less viscous aqueous layer. Therefore, we consider the stiffness of a droplet’s protein core to be the limiting factor of droplet toughness. If capillary force of the aqueous material contributes to droplet stiffness, just as it contributes to droplet adhesion (Sahni et al., 2010), we believe that this contribution would, likewise, be an order of magnitude less than that of core protein. A possibility that we cannot eliminate is that some of the amorphous proteins in the aqueous layer (Amarpuri et al., 2015a) may polymerize as a droplet extends and contribute to the measured toughness.
We determined the force on an extending glue droplet by measuring the deflection angle of its support line and incorporating the species-specific diameters and elastic modulus value of this line’s paired flagelliform fibers in calculations. For A. trifasciata these values, as taken from the literature (Sensenig et al., 2010), were: diameter of each flagelliform fiber 2.9 ± 0.23 μm standard error and elastic modulus 8 ± 5 MPa. The values for A. argentata were determined at the American Museum of Natural History using the same model of Nano Bionix instrument. Four threads from each of seven spiders were measured and tested, with each individual’s values being averaged before this species’ grand mean values were determined. Threads of 12.7 mm gauge lengths were extended at 10 μm per second for a constant rate of 1% strain/s until failure. We determined fiber diameter by placing two or three 8 mm strands of capture thread on a microscope slide, adding a drop of microscope immersion oil, and placing a glass cover slip on this preparation. For each individual, we photographed three thread regions where flagelliform fibers were contiguous, measured the width of each pair, and then computed this individual’s grand mean single fiber diameter. Argiope argentata mean flagelliform fiber diameter was 3.08 ± 0.23 μm and mean elastic modulus was 1.81 ± 0.19 MPa.
Elastic (Young’s) modulus is computed as the slope of the stress on a material during the linear phase of its extension. Engineering stress, which is used for stiffer materials, is determined by dividing the force on the material by its initial cross-sectional area. True stress, which is used for very extensible materials, is determined by dividing force by the instantaneous cross-sectional area, which decreases to a greater degree as the material extends. A study of A. trifasciata major ampullate threads thoroughly evaluated issues related to the determination of their elastic modulus according to these two indices (Guinea et al., 2006). This study showed that the engineering strain e on these naturally spun threads did not exceed 0.4, which equates to a 140% extension. Even at 55% RH, the core protein of A. argentata, the stiffer of the two species’, had extended by an average of 3,819% at the end of phase 1. The core protein cross sectional area at this extension, which we calculated by dividing core protein volume by the droplet’s extended length, was only 3.5% of the core protein’s initial cross-sectional area. Therefore, the use of true stress in calculations of core protein elastic modulus is justified.
Support line deflection was used to determine the extension of the support line’s flagelliform fibers, which, when combined with material properties, determined the force on either side of an extending droplet. We again used support line angle to resolve these force vectors and compute the force on an extending droplet at the initiation of extension and at five intervals during its extension. The cross-sectional area of a droplet’s protein core at each of these points was determined by dividing protein core volume by droplet filament length. Dividing force by protein core cross-sectional area yielded true stress on a droplet. Plotting true stress against a droplet’s true strain, computed as: natural log (change in droplet length / initial droplet length), produced a true stress – true strain curve, from which the elastic modulus and toughness of each species’ protein core was determined at each test humidity. We determined elastic modulus from the slopes of the linear, elastic region of each species’ humidity-specific true stress – true strain curves. Toughness was determined as the area under each curve minus the area of a thin rectangle defined by the stress on droplet at the initiation of extension and the strain on the droplet at the end of phase 1 extension.
The elastic modulus of flagelliform fibers was determined at laboratory humidity in range of 50% RH while they remained surrounded by aqueous material, which covers them both within and between droplets. The effect of changes in ambient humidity on flagelliform fiber elastic modulus has not been documented. Here we followed a previous study, which progressively increased (stiffened) these values at humidities lower than 55% RH and progressively decreased these values at higher humidities using droplet extension length per protein core volume as an index for this adjustment (Opell et al., 2021). We adjusted the force on a droplet at the end of phase 1 extension by increasing the measured elastic modulus of each species’ flagelliform fibers by 5% at 37% RH and 10% at 20% RH and reduced their elastic modulus by 5% at 72% RH and 10% at 90% RH (Opell et al., 2022).
Mature female A. argentata were collected from Coronado Beach, Coronado, California (USA) in August 2018. Mature female A. trifasciata were collected in Blacksburg, Virginia (USA) in September 2018 and 2019. Individual spiders were housed in large boxes either made of wood frames and plexiglass fronts and backs (81 × 81 × 18 cm, 43 × 61 × 6 cm, or 43 × 46 × 9 cm) or metal frames with wire mesh, wrapped in plastic wrap (60 × 92 × 61 cm or 60 × 61 × 30 cm). Spiders were maintained in Lexington, VA (Washington and Lee campus) in a room kept at approximately 26.5°C with a light cycle of 12 h lights on to 12 h lights off, but the room also received dim natural daylight. Spider webs were sprayed with deionized water daily and wet sponges were kept in the boxes. Each spider was offered a 1–2 cm long cricket 3 days/week.
As discussed above, spider diet can affect material properties of glue droplets and thus potentially also protein composition. Geographic variation in chemical composition has also been documented for Argiope keyserlingi glue droplets from four Australian sites separated by a maximum distance of over 900 km (Henneken et al., 2022). Differences in prey availability at these sites was considered the primary mechanism of chemical composition variation across the A. keyserlingi geographic range. As both A. argentata and A. trifasciata have broad ranges, geographic variability in glue droplet properties is also likely to occur in these species. However, our study was not designed to characterize geographic variation. The A. trifasciata individuals used in all phases of this study were collected on the edges of fields within an area of about 50 hectares. Argiope argentata were collected from low vegetation in similarly sized areas. Furthermore, by housing spiders in constant conditions in the lab and feeding both species the same diet, we should have largely removed the effect of environment on protein composition differences between the species.
Intact capture spirals were collected from the same individual’s spider webs onto stainless steel forceps over two consecutive days for A. argentata and 1–3 consecutive days for A. trifasciata to obtain sufficient silk for protein processing. During collection, radial lines and stabilimenta (rarely made in the lab) were avoided to the greatest extent possible. These silk samples were stored at −80°C as soon as sufficient material had been collected, until processing. Silks were solubilized using hexafluoroisopropanol, resuspended in guanidine hydrochloride, denatured, alkylated, and subjected to tryptic digests following methods described in (Ayoub et al., 2021). Digested silk proteins were sent to the University of Arizona Proteomics Core for LC–MS/MS on a Q Exactive Plus mass spectrometer with chromatography, flow rates, and data collection mirroring methods described in (Ayoub et al., 2021).
The initial protein databases provided for searches with Thermo Proteome Discoverer (ThermoFisher Scientific) were derived from de novo assembled transcriptomes (TRINITY, Grabherr et al., 2011) from 8 silk gland type specific RNA-Seq libraries for A. argentata (3 replicates each gland type except for pyriform, which only had 2 replicates) and 2 pooled silk gland (3 replicates of aggregate and 3 replicates of non-aggregate) RNA-Seq libraries for A. trifasciata. The transcriptomes were annotated using a published pipeline (TrTAP1) that retains a single transcript per Trinity-identified gene based on BLASTx alignments to custom arachnid databases or length of open reading frames in the absence of a BLAST alignment (Correa-Garhwal et al., 2022). The transcriptomes were further reduced by removing lowly expressed transcripts and subjecting predicted proteins to BLASTCLUST with 95% amino acid identity and 95% overlap. From these clusters, the protein with the best BLASTx alignment to a database protein, or the longest protein, were retained for initial mass spec searches. Proteins identified in capture spiral samples from this initial search were then used in future searches with MaxQuant (Cox and Mann, 2008).
In addition to proteins identified from the Proteome Discoverer searches, we added spidroins based on genome sequencing (Stellwagen and Renberg, 2019; Diaz et al., 2022). Because full-length spidroins are highly repetitive, including the full-length sequence in MaxQuant searches would either not complete or would result in very few peptide identifications due to repetitive sequences biasing the FDR calculations, since FDRs are based on scrambled peptides (Cox and Mann, 2008). We thus identified all the potential peptides in each spidroin sequence and determined if they were found uniquely or repeated more than twice. We reduced each spidroin sequence to reflect regions of unique peptides, being careful to include at least one iteration of repeat units to maximize identification of peptides and PTMs. We added these reduced spidroins to the database of proteins identified by Proteome Discoverer for further searches.
We completed label-free quantitation (LFQ) with MaxQuant using default settings. We searched for Post-Translational Modifications (PTMs: both Ser/Thr/Asn glycosylation and Ser/Thr/Tyr phosphorylation), searching for variable modifications to allow detection of both unmodified and modified peptides.
RNA-seq reads are available at NCBI’s Short Read Archive (SRR21593470-SRR21593492 for A. argentata and SRR21591021-2 and SRR21590965-8 for A. trifasciata). Mass spectra are available through ProteomeXchange (PXD037612).
Transcript abundance was determined by mapping raw RNA-Seq reads to the annotated transcriptomes with bowtie2 (Langmead and Salzberg, 2012) using only the reads with an unambiguous match and at most 2 mismatches as previously described (Correa-Garhwal et al., 2022). Transcripts encoding fragments of spidroins were removed based on BLASTn alignments (e-value < e-05, percent identity >95%) to the genome-based full length spidroin sequences. For each full-length spidroin, the conserved non-repetitive C-terminal encoding nucleotide sequence was added to the transcriptome prior to read mapping. Aggregate gland-specific transcripts were identified in two ways. First, transcripts significantly more abundant in aggregate than non-aggregate glands were identified with DESeq2 (Love et al., 2014) by comparing those two library types in the case of A. trifasciata or the anterior and posterior aggregate gland libraries to all other libraries for A. argentata. We used a false discovery rate < 0.05 (Benjamini and Hochberg, 1995) as threshold for significance. Second, for A. argentata the tissue-specific metric, tau (Yanai et al., 2005), was calculated using the RSEM (Li and Dewey, 2011) mean TPM for each gland type as input.
Images of droplets (Figure 1) show that A. argentata droplets are smaller than those of A. trifasciata and have denser and proportionately smaller protein cores, differences that are supported by the following analyses of their material properties. When suspended, A. trifasciata droplets have a similar shape but are slightly wider than A. argentata droplets at all humidities (Wilcoxon p = 0.0017–0.0047), having grand mean width / length ratios of 0.737 ± 0.003, and 0.699 ± 0.004, respectively. However, A. trifasciata droplets are much larger (Figure 2A), a difference that is significant at each humidity (Wilcoxon p = 0.0007–0.0350).
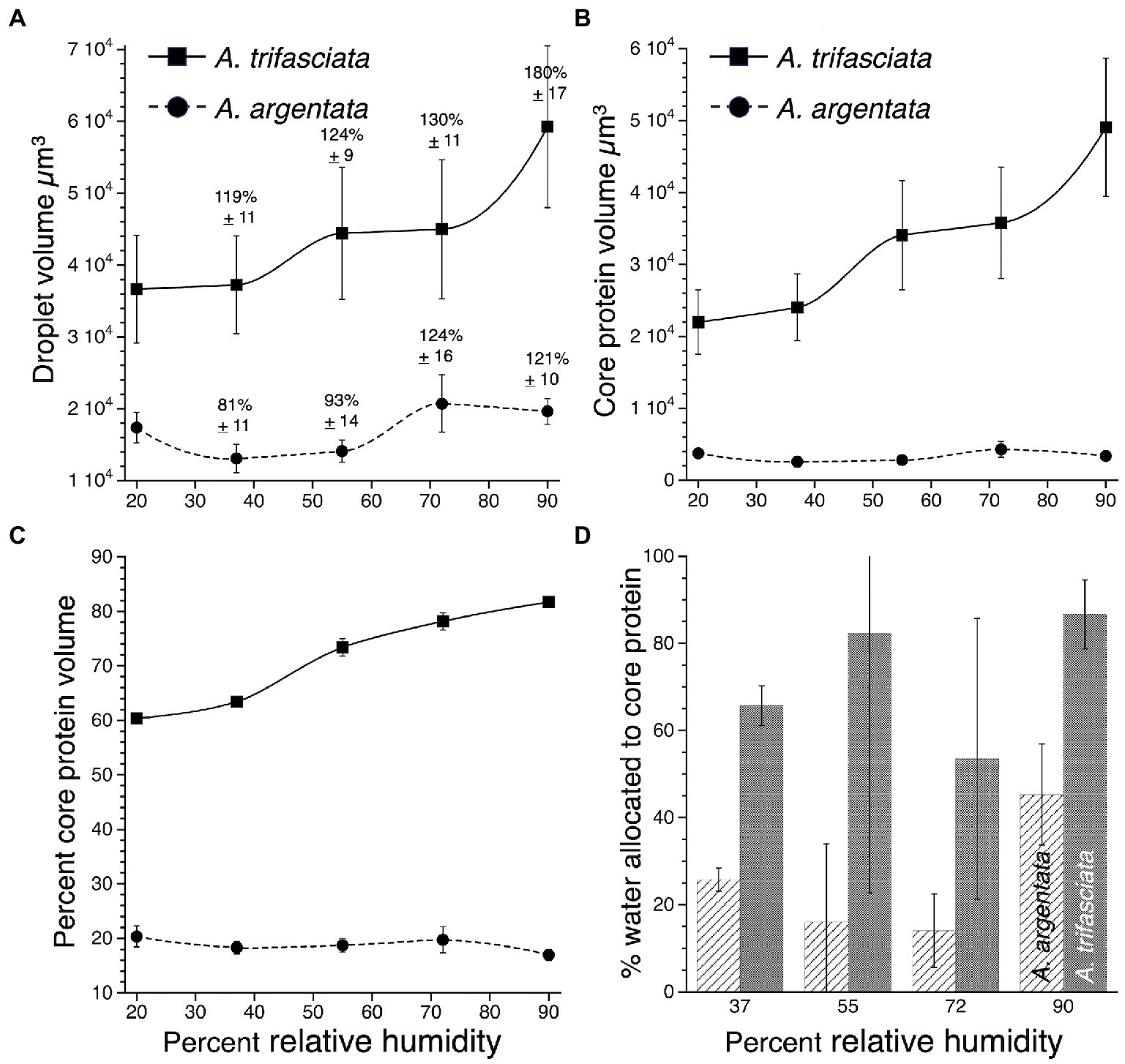
Figure 2. Comparison of A. argentata and A. trifasciata droplet features at five humidities: (A). Droplet volume showing percent changes in volume compared with volume at 20% RH, (B). Protein core volume, (C). Percent of droplet volume occupied by the protein core, and (D). Percent of absorbed atmospheric water allocated to a droplet’s protein core. Values are mean ± standard error.
Argiope trifasciata droplets absorb more water as humidity increases than do A. argentata droplets (Figure 2A). When increased droplet volume is expressed as a percent of droplet volume measured at 20% RH, the percent volume increases of A. trifasciata are greater than those of A. argentata at all humidities except 72% RH (Wilcoxon/Kruskal-Wallis 2-sample test, normal approximation p values: 37% RH 0.0253, 55% RH 0.0253, 72% RH 0.7381, 90% RH 0.0051). Argiope trifasciata droplets have larger protein cores (Figure 2B) and protein cores that comprise a larger portion of the droplet’s volume (Figure 2C), differences that are significant at each humidity (Wilcoxon p ≤ 0.0001). Argiope argentata cores average 24% of droplet volume and A. trifasciata cores average 80% of droplet volume. Argiope argentata protein cores are less hygroscopic than those of A. trifasciata, as a greater percent of atmospheric water absorbed at 37, 55, 72, and 90% RH was incorporated into the protein cores of A. trifasciata droplets (Wilcoxon p ≤ 0.0001) (Figure 2D).
Humidity-specific stress–strain curves constructed from mean true-stress and true-strain values at 20% droplet extension intervals show that A. argentata protein core is stiffer than that of A. trifasciata at all humidities (Figure 3). In both A. argentata and A. trifasciata protein core elastic modulus decreased as humidity increased (p = 0.0143 and 0.0079, respectively) (Figure 3). In A. argentata protein core toughness decreased as humidity increased (p = 0.0002), although in A. trifasciata this decrease did not quite reach the level of significance (p = 0.0514). The variance of A. argentata protein core elastic modulus values exceeded those of A. trifasciata at all test humidities (Bartletts tests of homogeneity p < 0.0001) and the variance of A. argentata toughness exceeded that of A. trifasciata at 72 and 90% RH (p < 0.0001). Argiope argentata protein core elastic modulus exceeded that of A. trifasciata at 20 and 37% RH (Wilcoxon p = 0.0002 and 0.0010, respectively), but did not differ at 55% RH (p = 0.6807). Argiope trifasciata protein core elastic modulus was greater than that of A. argentata at 72 and 90% RH (p = 0.0014 and 0.0024, respectively). The toughness of A. argentata protein exceeded that of A. trifasciata at 20 and 37% RH (Wilcoxon p = 0.0017 and 0.0003, respectively), did not differ from that of A. trifasciata at 55 and 72% RH (p = 0.0803 and 0.0506, respectively), and exceeded A. trifasciata toughness at 90% RH (p = 0.0236). The negative mean elastic modulus values at 72 and 90% RH reflect the limitations of our techniques for proteins that have very low elastic modulus values. Had we removed the negative, outlying values, means would not have been negative, although they would have been very small. However, all values were included in this and all other analyses.
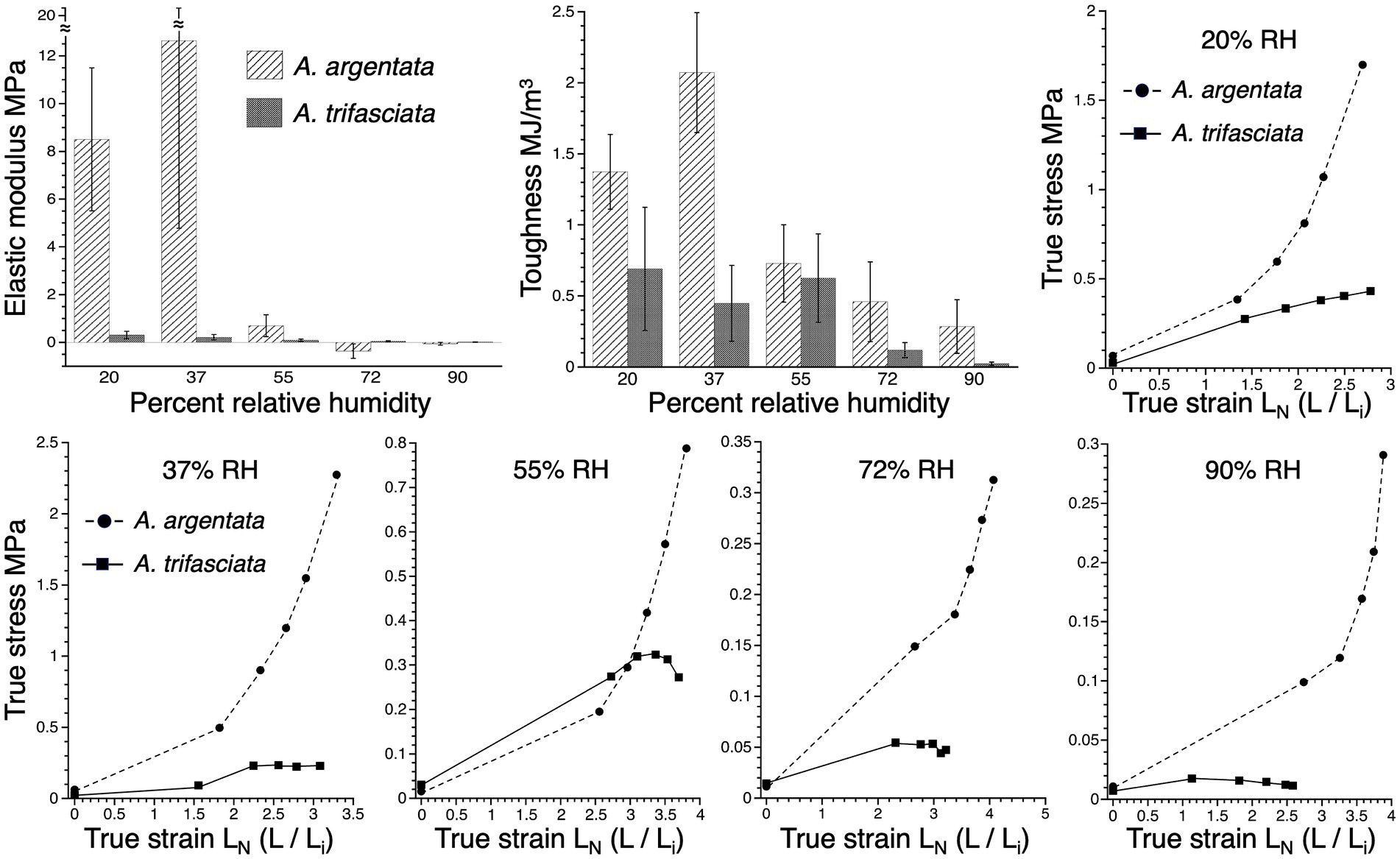
Figure 3. Material properties of A. argentata and A. trifasciata protein cores: elastic modulus and toughness at five humidities and the humidity-specific true stress—true strain curves from these species, based on mean stress and strain values. Material property values are mean ± standard error.
Glue droplet biomechanical behavior has been characterized by examining relationships among droplet contact area, cohesion or elastic modulus, and the force on a thread or droplet at pull-off, or, in the case of Argiope species, at the end of phase 1 droplet extension (Amarpuri et al., 2015b; Opell et al., 2022). Typically, the humidity at which the increase in surface area crosses the decrease in elastic modulus corresponds to the humidity of maximum adhesive force. The shape of A. argentata and A. trifasciata plots are remarkably similar (Figure 4), although, at all humidities, A. argentata droplets have greater protein core elastic modulus values and smaller surface areas due to their smaller droplet volumes. The adhesive force of both species’ droplets decreases with humidity in a similar manner, but, at all humidities, A. argentata droplets exhibited greater force. Field recordings show that the lowest humidity A. trifasciata experiences is around 37% RH (Opell et al., 2021). Therefore, we conclude that under natural conditions the droplets of both species maximize adhesion between 37 and 45% RH.
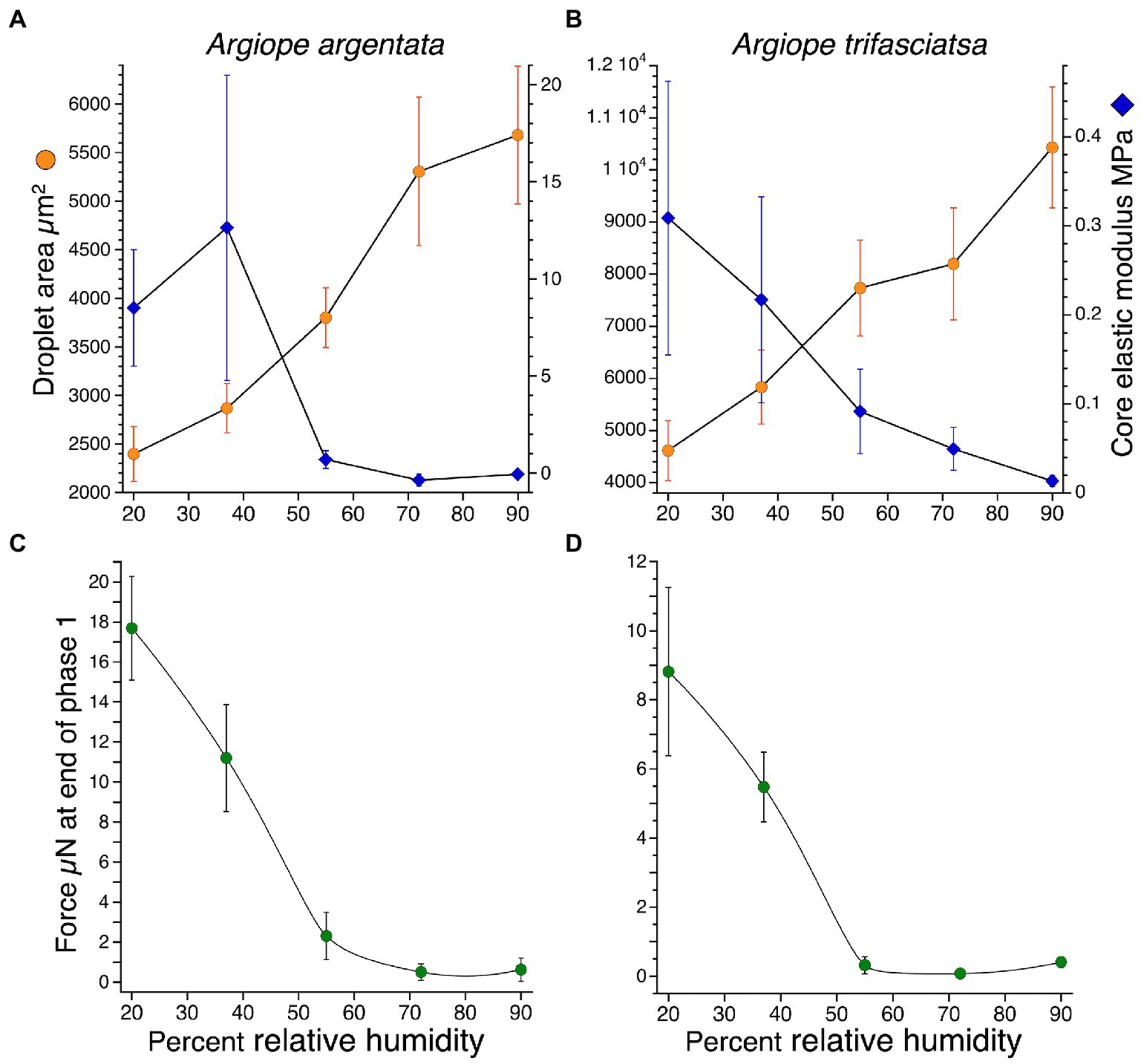
Figure 4. Paired plots of Argiope argentata and A. trifasciata flattened droplet areas and protein core elastic modulus at five humidities (A,B) and the associated forces on droplets at the end of phase one extension (C,D). Values are mean ± standard error.
We identified 47 and 40 proteins of A. argentata (Supplementary Table S1) and A. trifasciata, respectively (Supplementary Table S2), consistently present in capture spirals (MaxQuant score ≥ 50 and all 3 samples with detectable Label Free Quantitation, LFQ > 0). In both species, 30 non-spidroin proteins were identified. The spidroins represented 6 different gland types based on expression, including those expected from the glue forming aggregate glands (AgSp1, AgSp2, Sp8175) and the axial fiber forming flagelliform glands (two copies of Flag in both species and Sp5803). Surprisingly, we found the pyriform spidroin, PySp, in high abundance in both species (4th highest LFQ in A. argentata, 8th highest in A. trifasciata), which is thought to form attachment cement for anchoring radial lines to substrates and bridging lines in the orb web (Blasingame et al., 2009; Chaw et al., 2017). We additionally found aciniform spidroins [stabilimenta and prey wrapping (Foelix, 2011)] and major ampullate spidroins [dragline, radial lines, bridging lines (Foelix, 2011)] in both species, albeit with lower LFQs than aggregate, flagelliform, and pyriform spidroins. We found minor ampullate spidroins (MiSp1 and MiSp2) in A. argentata capture spirals, but not A. trifasciata.
The other 30 proteins are almost all likely aggregate glue proteins based on their expression patterns (Figure 5). In both species, most of these 30 were significantly more abundant in aggregate glands than non-aggregate glands (26 in A. argentata and 22 in A. trifasciata). In A. argentata, only one capture spiral protein was significantly higher in non-aggregate glands; it had highest expression in flagelliform glands with a tau = 0.999, strongly suggesting it is an axial fiber component. In A. trifasciata, three non-spidroin proteins had significantly higher expression in non-aggregate glands (Figure 5).
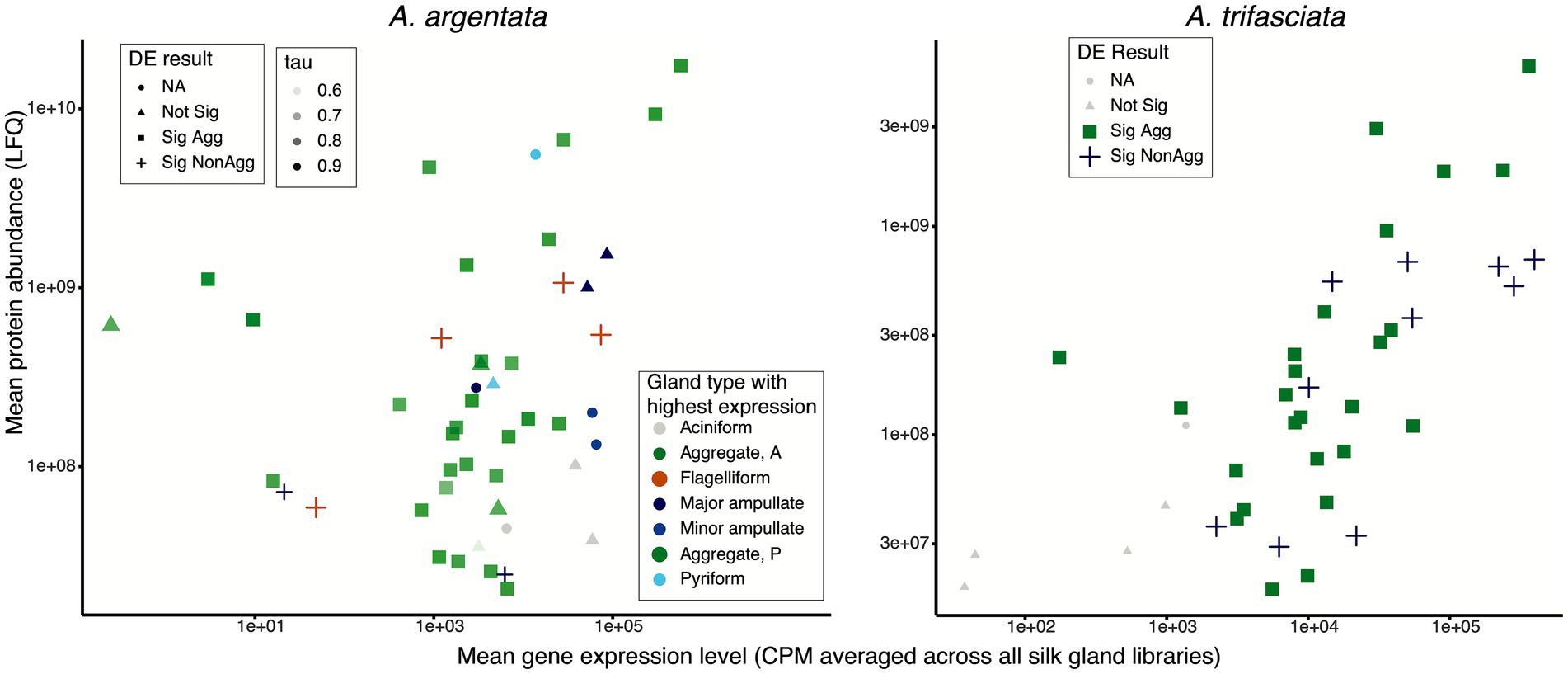
Figure 5. Capture spiral protein abundance (LFQ) and their gene expression levels in silk glands (average CPM) for Argiope argentata and A. trifasciata. Capture spirals are a composite of an axial fiber made in flagelliform glands and an aqueous glue synthesized in aggregate glands. Mean expression level (mean Counts Per Million mapped reads, CPM) was determined by DESeq2. DE result indicates outcome of differential expression analyses (Sig Agg = significantly higher expression in aggregate glands; Sig NonAgg = significantly higher expression in non-aggregate silk glands). For A. argentata, the gland type with highest expression is also indicated with all but two proteins found in capture spirals having highly biased expression to a single silk gland type (45 with tau >0.8, Yanai et al., 2005). The green proteins in this figure are likely aggregate glue proteins.
The non-spidroin proteins found in Argiope capture spirals have a variety of functions, many of which are also found in the gumfoot glues of cobweb weavers (Figure 6). These include macroglobulins, proteins that include a thyroglobulin type-1 repeat, proteases, and protease inhibitors. Some of the proteins found in gumfoot glues overlap with proteins found in only one species of Argiope, including a CRP (Pham et al., 2014), acetylcholinesterase, and CRISP/Allergen in A. trifasciata (Supplementary Table S2), and GMC oxidoreductase in A. argentata (Supplementary Table S1). Only two functions overlapped between the two Argiope species and not with gumfoot glues: calcineurin-like phosphoesterase and phosphatidylethanolamine-binding protein/ OV-16 antigen.
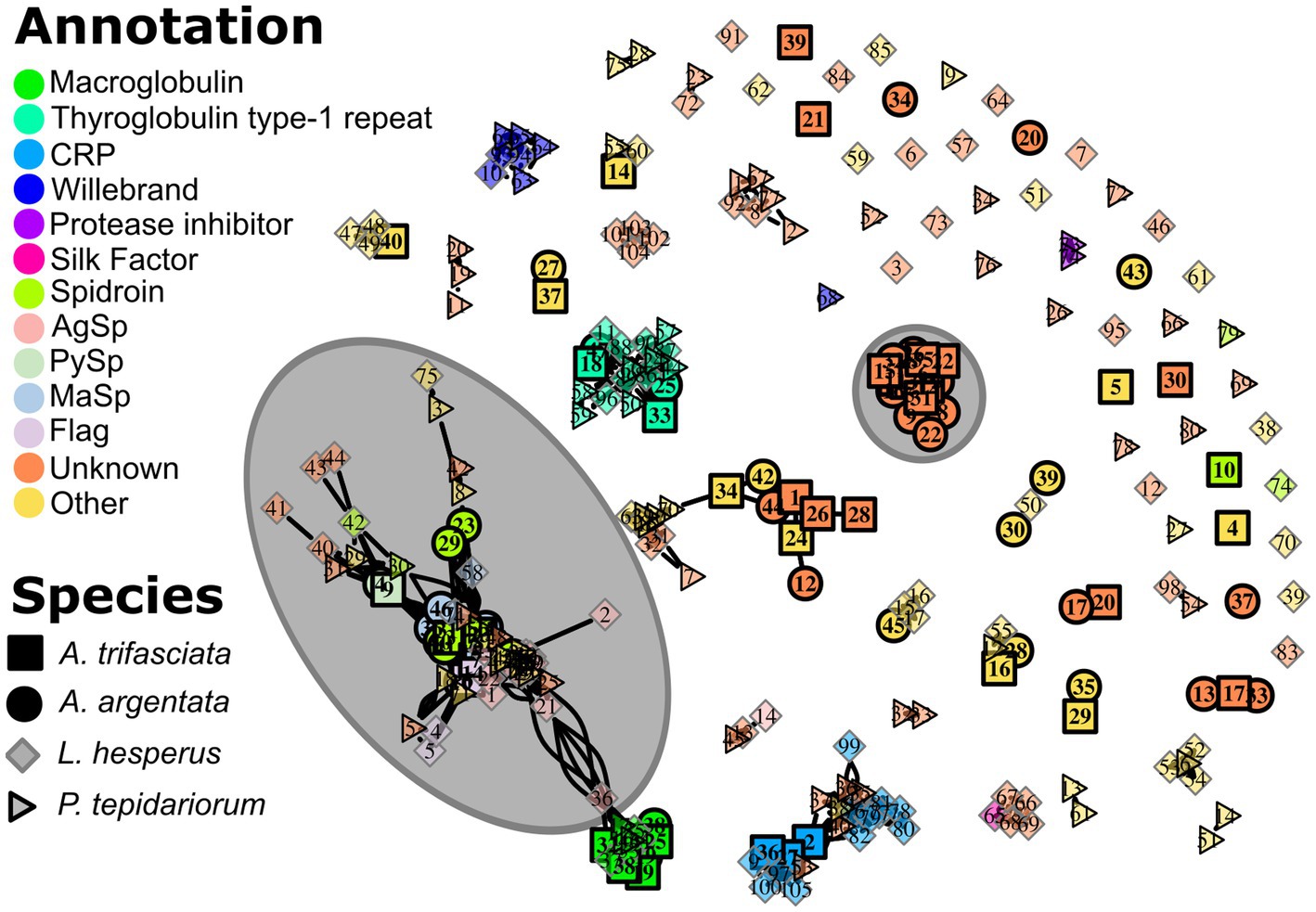
Figure 6. The relationships among capture spiral (A. argentata and A. trifasciata) and gumfoot proteins (Latrodectus hesperus and Parasteatoda tepidariorum, Ayoub et al., 2021) shown as a network graph. A connecting line indicates a significant BLASTP alignment. Interior color indicates the predicted protein annotation and the number is a species-specific protein identifier (see Supplementary Tables S1, S2, and Ayoub et al., 2021). The large grey circle encompasses all the predicted spidroins. Also encircled is a large cluster of Argiope proteins with no gumfoot homolog and no prior annotation (“Unknown”) that are highly abundant in the capture spirals (Figure 7) and have many conserved Cysteine residues (Supplementary Figure S1).
In addition to proteins with predicted functions, we found a large cluster of proteins with no BLAST alignment to gumfoot proteins or to an annotated protein in UniProt or PFAM (Figure 6). When aligned, these proteins have many conserved cysteine residues (Supplementary Figure S1). These proteins, which we refer to as “Cys-rich Unknown,” are reminiscent of the large cluster of cysteine-containing proteins in the gumfoot glues of Latrodectus hesperus and Parasteatoda tepidariorum (Ayoub et al., 2021) despite the lack of a BLAST alignment. The “Cys-rich Unknowns” are highly abundant in capture spirals, accounting for ~30% of the total LFQ in both species (Figure 7). We found 11 members in A. argentata, 5 of which are in the top 10 highest LFQ in capture spirals. We found 7 members in A. trifasciata, with 2 in the top 10 highest LFQ (Supplementary Figure S1).
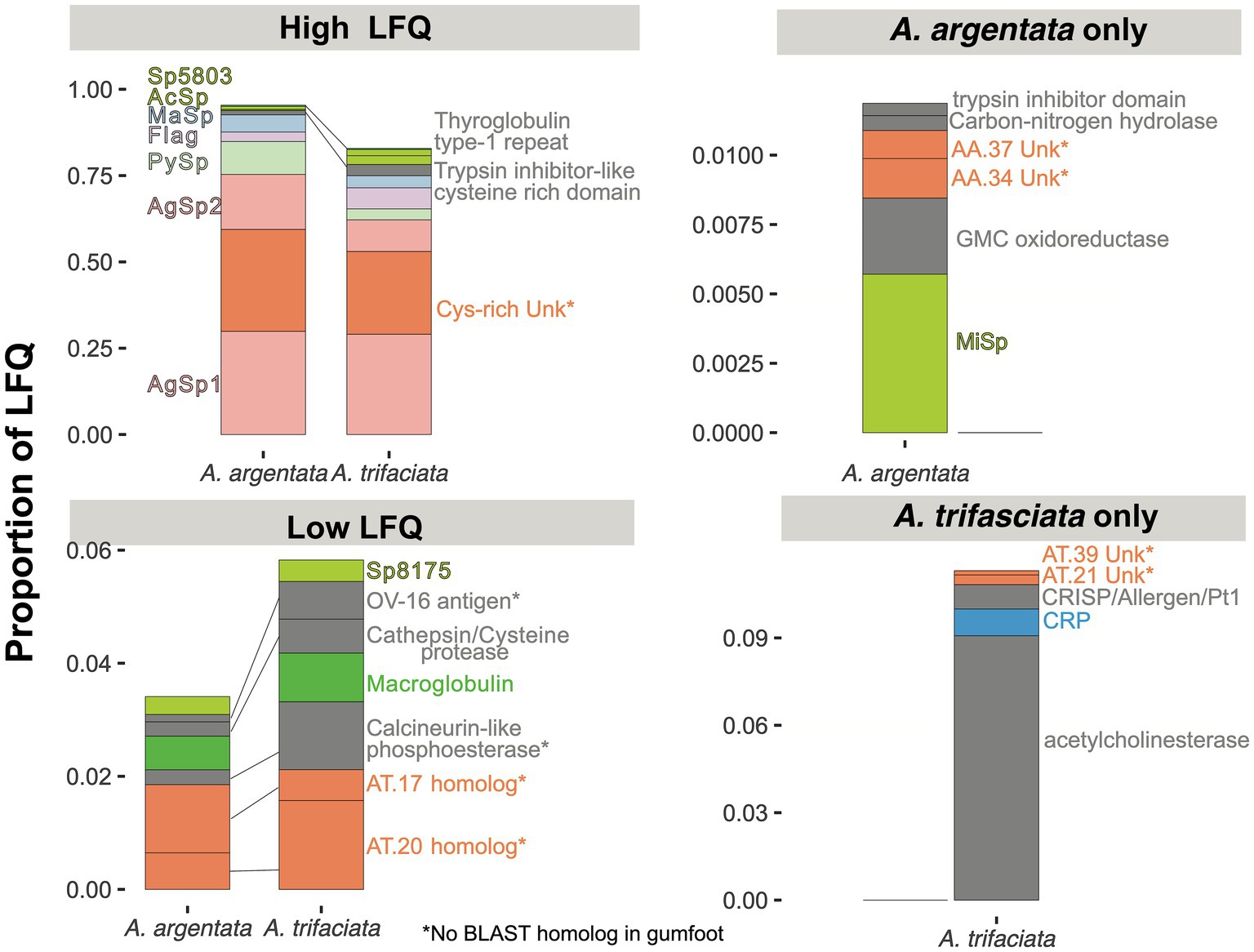
Figure 7. Relative abundance of proteins found in the capture spirals of two Argiope species. Colors for spidroins, proteins with no BLAST alignment (e-value < e-05) to a UniProt or PFAM protein (“Unk,” short for unknown), CRP, and macroglobulins are coded as in Figure 6. Proteins with other functions are in grey. Species abbreviation and numeric identifiers match Figure 6. Some homologous groups shown here include multiple proteins (e.g., 11 Cys-rich Unk in A. argentata and 7 in A. trifasciata, see Supplementary Tables S1, S2). Asterisk (*) indicates that we did not find a significant BLAST alignment between the Argiope protein(s) and a gumfoot protein in L. hesperus or P. tepidariorum (Figure 6).
A few proteins were found to be glycosylated and/or phosphorylated in at least one of the samples of each Argiope species. Only AgSp1 and AgSp2 were modified in all three samples in both species (Supplementary Figure S2). For AgSp1, glycosylated sites are largely conserved across repeat units and between the two species (Supplementary Figure S3). Phosphorylated sites were often conserved between species, but not as clearly maintained across repeat units. For AgSp2, glycosylation far outweighed phosphorylation; we identified only 1 position of phosphorylation in A. argentata and 3 in A. trifasciata compared to 45 and 16 glycosylated positions in AgSp2 of A. argentata and A. trifasciata, respectively (Supplementary Figure S2). The glutamine-rich region of AgSp2 did not have as many positions of modification identified as within the repeat regions, and we did not find conservation of those positions between the two species (Supplementary Figure S4). Many positions of glycosylation within repeat units of AgSp2 were conserved between the two species and across repeats.
In both Argiope species, we found the capture spiral proteins to conform to a weakly bimodal distribution of isoelectric points (Supplementary Figure S5). Capture spiral protein isoelectric point distribution is significantly different (less bimodal and more neutral) than the remainder of the proteome (A. argentata: W = 3,806,377, p = 0.007552; A. trifasciata: W = 1,394,964, p = 0.0001928). Despite a weak bimodal distribution, the opportunity for electrostatic interactions between positively and negatively charged regions of proteins is still present. AgSp1 terminal domains have low isoelectric points (IP ~5 and 4 for N-terminal and C-terminal domains, respectively) and the aspartic acid-rich region adjacent to the N-terminal domain has an exceptionally low isoelectric point (IP < 2.5, Supplementary Figure S3). Furthermore, the repeats found between the N-terminal aspartic acid-rich region and the homogenized repeat region of AgSp1 have considerable phosphorylation, enough to lower the isoelectric point from ~9 to ~6.5 (Supplementary Figure S3). In contrast, the remainder of the AgSp1 repetitive region and most of AgSp2 have an isoelectric point >9. These results suggest the potential for considerable electrostatic interactions between AgSp1 polypeptides or between AgSp1 and AgSp2.
Capture spiral proteins are also slightly more hydrophilic than the remainder of the proteome (Supplementary Figure S5). Wilcoxon rank sum tests indicate significant differences for both species (A. argentata: W = 7,289,166, p < 0.0001; A. trifasciata: W = 3,047,942, p < 0.0001).
Argiope species are large, colorful, conspicuous orb web weaving spiders distributed across the globe (Jäger, 2012; World Spider Catalog, 2022). We collected A. argentata from dry southwestern United States and A. trifasciata from humid southeastern United States. We thus expected these species to differ in the humidity at which their aggregate glue droplets would maximize adhesion, predicting that A. argentata droplets would be more hygroscopic and be more adhesive at a lower humidity than A. trifasciata. Instead, we found that A. trifasciata droplets are more hygroscopic than those of A. argentata and that for both species, droplets maximize adhesion at low relative humidity (RH), ~37% (Figure 4). These species both have very large geographic distributions, with A. argentata found in the Caribbean, most of South America, and the southern portion of North America (with northern limit in southwestern United States, https://scan-bugs.org/ accessed 15 Oct 2022). Argiope trifasciata is even more broadly distributed across most of North and South America (northern limit in southern Canada), northern and southern Africa and even Australia (https://scan-bugs.org/ accessed 15 Oct 2022). Furthermore, both species are daytime foragers in relatively open habitats (Brown, 1981; Ogunsheye, 2016; Velásquez Escalante et al., 2016). Thus, our results are consistent with both species having evolved to maximize glue adhesion at the low end of the wide range of humidities where they forage during much of the day.
Despite the unexpected similarity of the humidity at which glue droplets of both species maximize adhesion, the droplets from these species contrast in important ways. This serves as a reminder that orb web capture threads are complex and highly integrated adhesives whose components can be formed and configured differently. Argiope argentata droplets are significantly smaller than A. trifasciata droplets at all humidities and, as humidity increases, these droplets absorb less water, and their protein cores incorporate proportionately less of the absorbed water (Figure 2). The stiffness of A. argentata glue droplets is many times greater at 55% RH, and toughness is significantly higher at all conditions except 55% RH. The stiffer droplets of A. argentata achieve greater adhesive forces (Figure 4), but we do not suggest that the A. argentata capture thread is necessarily better adapted to capture prey than the thread of A. trifasciata. Instead, these differences, along with those in flagelliform fiber stiffness and the number of glue droplets per mm thread length, probably result in significantly different thread biomechanics. At 37 and 55% RH mean A. argentata phase 1 extensions are only half those of A. trifasciata (458 vs. 935 μm and 778 vs. 1,531 μm, respectively). The diameters of A. argentata and A. trifasciata flagelliform fibers are similar (3.08 μm and 2.9 μm, respectively), but A. trifasciata fibers are much stiffer (8 MPa vs. 1.81 MPa in A. argentata). Argiope argentata threads have an average of 14.8 droplets per mm, whereas A. trifasciata threads average 9.8 droplets per mm (Kelly et al., 2022).
The stiffer adhesive proteins, shorter phase 1 extensions, and more closely spaced glue droplets of A. argentata suggest that the suspension bridges of this species’ adhering threads may be less bowed and incorporate more droplets than the bridges of A. trifasciata threads. However, to complement this set of A. argentata features, one might expect that A. argentata flagelliform fibers would be stiffer than those of A. trifasciata, but the reverse is true. The capture thread of an orb weaving spider whose web has evolved to capture moths had glue of extremely low viscosity, allowing it to flow through scales and contact an insect’s epicuticle (Diaz et al., 2022). The larger droplets and core proteins, greater flattened droplet areas, and more pliable glue protein of A. trifasciata threads may confer an advantage in retaining insects with scaley or very setose surfaces. However, there is no evidence that either species is other than a generalist predator.
The striking differences in water absorption and stiffness between the two Argiope species suggest that chemical interactions among glue proteins differ greatly. Many aspects of aggregate glue proteins could potentially contribute to the observed differences in droplet material properties. Overall, the protein components are largely conserved with most proteins found in the capture spiral of one species having at least one homologous protein in the capture spirals of the other species (Figures 6, 7, Supplementary Tables S1, S2). The homologous proteins vary in the level of conservation (30–90% BLASTP sequence identity), although the less conserved proteins may not be direct orthologs. Furthermore, AgSp1, AgSp2 and the newly discovered aggregate-expressed cysteine-rich family of proteins (“Cys-rich Unk”) make up the bulk of total protein abundance (Figure 7). Sequences of AgSp1 and AgSp2 are highly conserved between the two species for most of the proteins (85–92% identity for terminal domains and repetitive regions), although the Tyrosine-rich region following the N-terminal domain of AgSp1 and the Glutamine-rich regions of AgSp2 are less conserved (60–70% identity, Supplementary Figures S4, S5). In addition, A. trifasciata AgSp1 has more homogenized repeats than A. argentata (Supplementary Figures S2, S3). If the number of repeats is inversely related to elastic modulus, then these differences could explain the lower stiffness of A. trifasciata droplets.
Perhaps more importantly, the relative abundance of protein components differs between species. For instance, the ratio of AgSp1 to AgSp2 is ~3:1 for A. trifasciata compared to ~2:1 for A. argentata. Portions of AgSp1 are highly negatively charged while most of AgSp2 is highly positively charged, and thus there is a possibility of more electrostatic interactions for A. argentata, and potentially other types of associations between these two proteins. Certainly, diluting proteins with water should diminish electrostatic interactions and provides a simple mechanism for the decreased stiffness of glue proteins with increasing water uptake. The higher total abundance and more even distribution of AgSp1 and AgSp2 in A. argentata could mean proportionately more proteins interacting, making it more difficult for water to pry these proteins apart, consistent with the denser protein cores of A. argentata and their lower ability to absorb water as humidity increases. It is possible, however, that AgSp1, AgSp2, and other adhesive proteins are not limited to the core area. Proteins are distributed throughout the droplet that likely contribute to total droplet adhesion (Amarpuri et al., 2015a; Opell et al., 2022).
Another aspect of protein composition that could explain differences in stiffness is that A. argentata includes more members of the cysteine-rich family (11 versus 7) and more of these proteins rank in the top 10 most abundant proteins than in A. trifasciata. Five A. trifasciata cysteine-rich proteins have a direct ortholog with A. argentata, with high sequence identity, but the LFQ ranking can be quite different for orthologs (Supplementary Figure S1). One potentially important difference in sequence is that the most abundant cysteine rich protein in A. argentata has 10 cysteines, while the most abundant one in A. trifasciata only has 5 cysteines. These differences in cysteine-rich protein abundance, composition, and sequence mean there is higher potential for disulfide bond formation in A. argentata, consistent with the higher stiffness of core proteins in A. argentata than A. trifasciata.
Another standout difference in protein composition is the high abundance of acetylcholinesterase in A. trifasciata capture spirals and its absence in A. argentata (Figure 7). This enzyme hydrolyzes acetylcholine into acetic acid and choline, typically to stop the signaling of acetylcholine, but has non-cholinergic functions as well (Appleyard, 1994). Choline is an abundant LMMC in A. aurantia and other araneids (Jain et al., 2018; Opell et al., 2018b), thus the expression of acetylcholinesterase in aggregate glands is unsurprising. Indeed, we found an ortholog of the A. trifasciata acetylcholinesterase expressed at very high levels in A. argentata aggregate and flagelliform glands (TPM > 300) and not in other silk glands (TPM < 11). It is unclear why this enzyme is retained in the glue of A. trifasciata, but not A. argenentata, or how it might contribute to material properties.
Compared to cobweb weaver gumfoot glue droplets, the only other spiders with known glue protein components, the glue droplets of both Argiope species are less stiff, more extensible, and more responsive to changes in humidity (Ayoub et al., 2021). Nevertheless, many protein components are conserved between the Argiope glue droplets and the cobweb gumfoot glues (Figures 6, 7). Aggregate spidroins are abundant in the glue of both groups, suggesting an important role of these proteins in aggregate glue function. Argiope AgSp1 repeat motifs are longer and have more opportunities for glycosylation than cobweb weaver AgSp1 repeat motifs (Stellwagen and Renberg, 2019), although we found abundant glycosylation in both groups. AgSp2 is highly phosphorylated in the cobweb weaving house spider (Ayoub et al., 2021), while it is barely phosphorylated in Argiope. Furthermore, the cobweb weaving black widow has a highly diminished AgSp2 sequence but abundant, and highly phosphorylated Aggregate Silk Factors, a protein family that has not been discovered outside Theridiidae. Both cobweb weavers and the Argiope species also have multiple small cysteine-rich proteins present in glue droplets, although these proteins do not appear to be homologous between the two types of spiders. Cobweb weavers have more members of protein families associated with protein aggregation (e.g., von Willebrand domain-containing; thyroglobulin type-1 repeat domain). Taken together, these differences in protein sequence, modification, and composition may collectively lead to higher cohesion and thus greater stiffness of cobweb gumfoot glues compared to Argiope capture spiral glue droplets.
Argiope argentata and A. trifasciata are both large orb web weavers that build webs on exposed vegetation. However, the properties of their capture threads have diverged significantly during the estimated 17 million years of evolutionary time that separates these species (Agnarsson et al., 2016). Argiope argentata threads have smaller, more closely spaced glue droplets [14.8 vs. 9.8 per mm length in A. trifasciata; (Kelly et al., 2022)]. Also, the protein cores of A. argentata threads are proportionately smaller than those of A. trifasciata, absorb proportionately less water as humidity increases, are stiffer and tougher, and achieve greater adhesive force than do A. trifasciata protein cores. Comparing glue proteins of these congeneric species allows us to better understand the evolutionary plasticity of orb web capture threads and the determinants of droplet adhesion. We find that A. argentata glue is comprised of a more even ratio of AgSp1 and AgSp2 proteins than A. trifasciata glue and includes the products of 11 newly discovered cysteine-rich genes, only seven of which were present in A. trifasciata glue. The disulfide bonds associated with these cysteine-rich proteins could contribute to the greater stiffness of A. argentata glue and, along with the more balanced ratio of AgSp1 and AgSp2, may restrict the amount of water that can be incorporated into the protein core of A. argentata glue droplets as humidity increases. More broadly, our comparison shows that glue gene sequence changes may not fully account for the evolution of orb web glue. Divergence in material properties likely also involves selective gene expression, post translational protein modifications, and differences in the rates of extrusion of constituent glue proteins.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
NA, KF, CH, RB, and BO conceived the study. NA, CL, ML, JM, LD, SM, RB, SC-G, CH, and BO generated data for the study. All authors contributed to the article and approved the submitted version.
This work was supported by Washington and Lee University through Summer Lenfest Research Grants to NA and KF and Summer Research Scholarships to CL, ML, and JM; and the National Science Foundation (Award numbers IOS-1755142 to NA and KF, IOS-1754979 to CH and RB, and IOS-1755028 to BO).
The authors thank Eman Muamar with assistance identifying unique peptides in aggregate spidroins.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MG declared a past co-authorship with CH to the handling editor.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1099481/full#supplementary-material
Agnarsson, I., LeQuier, S. M., Kuntner, M., Cheng, R.-C., Coddington, J. A., and Binford, G. (2016). Phylogeography of a good Caribbean disperser: Argiope argentata (Araneae, Araneidae) and a new ‘cryptic’ species from Cuba. ZooKeys 625, 25–44. doi: 10.3897/zookeys.625.8729
Amarpuri, G., Chaurasia, V., Jain, D., Blackledge, T. A., and Dhinojwala, A. (2015a). Ubiquitous distribution of salts and proteins in spider glue enhances spider silk adhesion. Sci. Rep. 5:9030. doi: 10.1038/srep09030
Amarpuri, G., Zhang, C., Diaz, C., Opell, B. D., Blackledge, T. A., and Dhinojwala, A. (2015b). Spiders tune glue viscosity to maximize adhesion. ACS Nano 9, 11472–11478. doi: 10.1021/acsnano.5b05658
Appleyard, M. E. (1994). Non-cholinergic functions of acetylcholinesterase. Biochem. Soc. Trans. 22, 749–755. doi: 10.1042/bst0220749
Arakawa, K., Kono, N., Malay, A. D., Tateishi, A., Ifuku, N., Masunaga, H., et al. (2022). 1000 spider silkomes: linking sequences to silk physical properties. Sci. Adv. 8:eabo6043. doi: 10.1126/sciadv.abo6043
Ayoub, N. A., Friend, K., Clarke, T., Baker, R., Correa-Garhwal, S., Crean, A., et al. (2021). Protein composition and associated material properties of cobweb spiders’ gumfoot glue droplets. Integr. Comp. Biol. 61, 1459–1480. doi: 10.1093/icb/icab086
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodology 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Blamires, S. J., Blackledge, T. A., and Tso, I.-M. (2017a). Physicochemical property variation in spider silk: ecology, evolution, and synthetic production. Annu. Rev. Entomol. 62, 443–460. doi: 10.1146/annurev-ento-031616-035615
Blamires, S. J., Hasemore, M., Martens, P. J., and Kasumovic, M. M. (2017b). Diet-induced co-variation between architectural and physicochemical plasticity in an extended phenotype. J. Exp. Biol. 220, 876–884. doi: 10.1242/jeb.150029
Blamires, S. J., Liao, C.-P., Chang, C.-K., Chuang, Y.-C., Wu, C.-L., Blackledge, T. A., et al. (2015). Mechanical performance of spider silk is robust to nutrient-mediated changes in protein composition. Biomacromolecules 16, 1218–1225. doi: 10.1021/acs.biomac.5b00006
Blamires, S. J., Martens, P. J., and Kasumovic, M. M. (2018a). Fitness consequences of plasticity in an extended phenotype. J. Exp. Biol. 221:jeb167288. doi: 10.1242/jeb.167288
Blamires, S. J., Nobbs, M., Martens, P. J., Tso, I.-M., Chuang, W.-T., Chang, C.-K., et al. (2018b). Multiscale mechanisms of nutritionally induced property variation in spider silks. PLoS One 13:e0192005. doi: 10.1371/journal.pone.0192005
Blamires, S. J., Sahni, V., Dhinojwala, A., Blackledge, T. A., and Tso, I.-M. (2014). Nutrient deprivation induces property variations in spider gluey silk. PLoS One 9:e88487. doi: 10.1371/journal.pone.0088487
Blamires, S. J., Tseng, Y.-H., Wu, C.-L., Toft, S., Raubenheimer, D., and Tso, I.-M. (2016). Spider web and silk performance landscapes across nutrient space. Sci. Rep. 6:26383. doi: 10.1038/srep26383
Blasingame, E., Tuton-Blasingame, T., Larkin, L., Falick, A. M., Zhao, L., Fong, J., et al. (2009). Pyriform spidroin 1, a novel member of the silk gene family that anchors dragline silk fibers in attachment discs of the black widow spider, Latrodectus hesperus. J. Biol. Chem. 284, 29097–29108. doi: 10.1074/jbc.M109.021378
Bond, J. E., Garrison, N. L., Hamilton, C. A., Godwin, R. L., Hedin, M., and Agnarsson, I. (2014). Phylogenomics resolves a spider backbone phylogeny and rejects a prevailing paradigm for orb web evolution. Curr. Biol. 24, 1765–1771. doi: 10.1016/j.cub.2014.06.034
Bond, J. E., and Opell, B. D. (1998). Testing adaptive radiation and key innovation hypotheses in spiders. Evolution 52, 403–414. doi: 10.2307/2411077
Brown, K. M. (1981). Foraging ecology and niche partitioning in orb-weaving spiders. Oecologia 50, 380–385. doi: 10.1007/BF00344980
Chaw, R. C., Clarke, T. H., Arensburger, P., Ayoub, N. A., and Hayashi, C. Y. (2021). Gene expression profiling reveals candidate genes for defining spider silk gland types. Insect Biochem. Mol. Biol. 135:103594. doi: 10.1016/j.ibmb.2021.103594
Chaw, R. C., Saski, C. A., and Hayashi, C. Y. (2017). Complete gene sequence of spider attachment silk protein (PySp1) reveals novel linker regions and extreme repeat homogenization. Insect Biochem. Mol. Biol. 81, 80–90. doi: 10.1016/j.ibmb.2017.01.002
Coddington, J. A., Agnarsson, I., Hamilton, C. A., and Bond, J. E. (2019). Spiders did not repeatedly gain, but repeatedly lost, foraging webs. PeerJ 7:e6703. doi: 10.7717/peerj.6703
Collin, M. A., Clarke, T. H., Ayoub, N. A., and Hayashi, C. Y. (2016). Evidence from multiple species that spider silk glue component ASG2 is a spidroin. Sci. Rep. 6:21589. doi: 10.1038/srep21589
Correa-Garhwal, S. M., Baker, R. H., Clarke, T. H., Ayoub, N. A., and Hayashi, C. Y. (2022). The evolutionary history of cribellate orb-weaver capture thread spidroins. BMC Ecol. Evol. 22:89. doi: 10.1186/s12862-022-02042-5
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Diaz, C. Jr., Baker, R. H., Long, J. H. Jr., and Hayashi, C. Y. (2022). Connecting materials, performance and evolution: a case study of the glue of moth-catching spiders (Cyrtarachninae). J. Exp. Biol. 225:jeb243271. doi: 10.1242/jeb.243271
Eberhard, W. G. (2020). Spider webs: behavior, function, and evolution. University of Chicago Press, Chicago.
Edmonds, D. T., and Vollrath, F. (1992). The contribution of atmospheric water vapour to the formation and efficiency of a spider’s capture web. Proc. Biol. Sci. 248, 145–148. doi: 10.1098/rspb.1992.0055
Fernández, R., Kallal, R. J., Dimitrov, D., Ballesteros, J. A., Arnedo, M. A., Giribet, G., et al. (2018). Phylogenomics, diversification dynamics, and comparative transcriptomics across the spider tree of life. Curr. Biol. 28, 1489–1497.e5. doi: 10.1016/j.cub.2018.03.064
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guinea, G. V., Pérez-Rigueiro, J., Plaza, G. R., and Elices, M. (2006). Volume constancy during stretching of spider silk. Biomacromolecules 7, 2173–2177. doi: 10.1021/bm060138v
Guo, Y., Chang, Z., Guo, H.-Y., Fang, W., Li, Q., Zhao, H.-P., et al. (2018). Synergistic adhesion mechanisms of spider capture silk. J. R. Soc. Interface 15:20170894. doi: 10.1098/rsif.2017.0894
Guo, Y., Zhao, H.-P., Feng, X.-Q., and Gao, H. (2019). On the robustness of spider capture silk’s adhesion. Extreme Mech. Lett. 29:100477. doi: 10.1016/j.eml.2019.100477
Hayashi, C. Y., and Lewis, R. V. (1998). Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. J. Mol. Biol. 275, 773–784. doi: 10.1006/jmbi.1997.1478
Henneken, J., Blamires, S. J., Goodger, J. Q. D., Jones, T. M., and Elgar, M. A. (2022). Population level variation in silk chemistry but not web architecture in a widely distributed orb web spider. Biol. J. Linn. Soc. 137, 350–358. doi: 10.1093/biolinnean/blac089
Jäger, P. (2012). A review on the spider genus Argiope Audouin 1826 with special emphasis on broken emboli in female epigynes (Araneae: Araneidae: Argiopinae). Beitr. Zur Araneol. 7, 272–331.
Jain, D., Amarpuri, G., Fitch, J., Blackledge, T. A., and Dhinojwala, A. (2018). Role of hygroscopic low molecular mass compounds in humidity responsive adhesion of spider’s capture silk. Biomacromolecules 19, 3048–3057. doi: 10.1021/acs.biomac.8b00602
Kelly, S. D., Opell, B. D., and Correa-Garhwal, S. M. (2022). Correlated evolution between orb weaver glue droplets and supporting fibres maintains their distinct biomechanical roles in adhesion. J. Evol. Biol. 35, 879–890. doi: 10.1111/jeb.14025
Kelly, S. P., Sensenig, A., Lorentz, K. A., and Blackledge, T. A. (2011). Damping capacity is evolutionarily conserved in the radial silk of orb-weaving spiders. Zoology 114, 233–238. doi: 10.1016/j.zool.2011.02.001
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Liao, C.-P., Blamires, S. J., Hendricks, M. L., and Opell, B. D. (2015). A re-evaluation of the formula to estimate the volume of orb web glue droplets. J. Arachnol. 43, 97–100. doi: 10.1636/M14-50.1
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ogunsheye, Z. (2016). Argiope argentata, (silver Argiope spider). Pp. 1–5 in online guide to the animals of Trinidad and Tobago. Available at: https://sta.uwi.edu/fst/lifesciences/sites/default/files/lifesciences/images/Argiope%20argentata%20-%20Silver%20Argiope%20Spider.pdf.
Opell, B. D., Buccella, K. E., Godwin, M. K., Rivas, M. X., and Hendricks, M. L. (2017). Humidity-mediated changes in an orb spider’s glycoprotein adhesive impact prey retention time. J. Exp. Biol. 220, 1313–1321. doi: 10.1242/jeb.148080
Opell, B. D., Burba, C. M., Deva, P. D., Kin, M. H. Y., Rivas, M. X., Elmore, H. M., et al. (2019). Linking properties of an orb-weaving spider’s capture thread glycoprotein adhesive and flagelliform fiber components to prey retention time. Ecol. Evol. 9, 9841–9854. doi: 10.1002/ece3.5525
Opell, B. D., Clouse, M. E., and Andrews, S. F. (2018a). Elastic modulus and toughness of orb spider glycoprotein glue. PLoS One 13:e0196972. doi: 10.1371/journal.pone.0196972
Opell, B. D., Elmore, H. M., and Hendricks, M. L. (2022). Adhesive contact and protein elastic modulus tune orb weaving spider glue droplet biomechanics to habitat humidity. Acta Biomater. 151, 468–479. doi: 10.1016/j.actbio.2022.08.018
Opell, B. D., Elmore, H. M., and Hendricks, M. L. (2021). Humidity mediated performance and material properties of orb weaving spider adhesive droplets. Acta Biomater. 131, 440–451. doi: 10.1016/j.actbio.2021.06.017
Opell, B. D., and Hendricks, M. L. (2009). The adhesive delivery system of viscous capture threads spun by orb-weaving spiders. J. Exp. Biol. 212, 3026–3034. doi: 10.1242/jeb.030064
Opell, B. D., Jain, D., Dhinojwala, A., and Blackledge, T. A. (2018b). Tuning orb spider glycoprotein glue performance to habitat humidity. J. Exp. Biol. 221:jeb161539. doi: 10.1242/jeb.161539
Pham, T., Chuang, T., Lin, A., Joo, H., Tsai, J., Crawford, T., et al. (2014). Dragline silk: a fiber assembled with low-molecular-weight cysteine-rich proteins. Biomacromolecules 15, 4073–4081. doi: 10.1021/bm5011239
Robinson, M. H., and Robinson, B. (1970). Prey caught by a sample population of the spider Argiope argentata (Araneae: Araneidae) in Panama: a year’s census data. Zool. J. Linnean Soc. 49, 345–358. doi: 10.1111/j.1096-3642.1970.tb00746.x
Sahni, V., Blackledge, T. A., and Dhinojwala, A. (2011). Changes in the adhesive properties of spider aggregate glue during the evolution of cobwebs. Sci. Rep. 1:41. doi: 10.1038/srep00041
Sahni, V., Blackledge, T. A., and Dhinojwala, A. (2010). Viscoelastic solids explain spider web stickiness. Nat. Commun. 1:19. doi: 10.1038/ncomms1019
Sensenig, A., Agnarsson, I., and Blackledge, T. A. (2010). Behavioural and biomaterial coevolution in spider orb webs. J. Evol. Biol. 23, 1839–1856. doi: 10.1111/j.1420-9101.2010.02048.x
Sensenig, A. T., Kelly, S. P., Lorentz, K. A., Lesher, B., and Blackledge, T. A. (2013). Mechanical performance of spider orb webs is tuned for high-speed prey. J. Exp. Biol. 216, 3388–3394. doi: 10.1242/jeb.085571
Sensenig, A. T., Lorentz, K. A., Kelly, S. P., and Blackledge, T. A. (2012). Spider orb webs rely on radial threads to absorb prey kinetic energy. J. R. Soc. Interface 9, 1880–1891. doi: 10.1098/rsif.2011.0851
Stellwagen, S. D., and Renberg, R. L. (2019). Toward spider glue: long read scaffolding for extreme length and repetitious silk family genes AgSp1 and AgSp2 with insights into functional adaptation. G3 GenesGenomesGenetics 9, 1909–1919. doi: 10.1534/g3.119.400065
Stewart, R. J., Ransom, T. C., and Hlady, V. (2011a). Natural underwater adhesives. J. Polym. Sci. Part B Polym. Phys. 49, 757–771. doi: 10.1002/polb.22256
Stewart, R. J., Wang, C. S., and Shao, H. (2011b). Complex coacervates as a foundation for synthetic underwater adhesives. Adv. Colloid Interf. Sci. 167, 85–93. doi: 10.1016/j.cis.2010.10.009
Tarakhovskaya, E. R. (2014). Mechanisms of bioadhesion of macrophytic algae. Russ. J. Plant Physiol. 61, 19–25. doi: 10.1134/S1021443714010154
Tillinghast, E. K., Townley, M. A., Wight, T. N., Uhlenbruck, G., and Janssen, E. (1992). The adhesive glycoprotein of the orb web of Argiope aurantia (Araneae, Araneidae). Mater. Resesarch Soc. Symp. Proc. 292, 9–23. doi: 10.1557/PROC-292-9
Townley, M. A., and Tillinghast, E. K. (2013). “Aggregate silk gland secretions of araneoid spiders” in Spider ecophysiology. ed. W. Nentwig (Berlin Heidelberg: Springer), 283–302.
Townley, M. A., Tillinghast, E. K., and Neefus, C. D. (2006). Changes in composition of spider orb web sticky droplets with starvation and web removal, and synthesis of sticky droplet compounds. J. Exp. Biol. 209, 1463–1486. doi: 10.1242/jeb.02147
Velásquez Escalante, R., Cornejo-Escobar, P., and Saenz, R. (2016). Biología y ecología de la araña plateada Argiope argentata (Fabricius, 1775) (Araneidae) en un sector xerófilo del noreste de la Península de Araya, Venezuela. Saber 28, 471–479.
Vollrath, F., and Tillinghast, E. K. (1991). Glycoprotein glue beneath a spider web’s aqueous coat. Naturwissenschaften 78, 557–559. doi: 10.1007/BF01134447
Wagner, V. T., Brian, L., and Quatrano, R. S. (1992). Role of a vitronectin-like molecule in embryo adhesion of the brown alga Fucus. Proc. Natl. Acad. Sci. U. S. A. 89, 3644–3648. doi: 10.1073/pnas.89.8.3644
Waite, J. H. (2017). Mussel adhesion – essential footwork. J. Exp. Biol. 220, 517–530. doi: 10.1242/jeb.134056
Wu, C. C., Blamires, S. J., Wu, C. L., and Tso, I. M. (2013). Wind induces variations in spider web geometry and sticky spiral droplet volume. Journal of Experimental Biology 216, 3342–3349. doi: 10.1242/jeb.083618
Keywords: biomechanics, bioadhesives, Araneoidea, aggregate glands, Argiope, proteomics, spider silk
Citation: Ayoub NA, DuMez L, Lazo C, Luzaran M, Magoti J, Morris SA, Baker RH, Clarke T, Correa-Garhwal SM, Hayashi CY, Friend K and Opell BD (2023) Orb weaver aggregate glue protein composition as a mechanism for rapid evolution of material properties. Front. Ecol. Evol. 11:1099481. doi: 10.3389/fevo.2023.1099481
Received: 15 November 2022; Accepted: 01 March 2023;
Published: 18 April 2023.
Edited by:
Sean Blamires, University of New South Wales, AustraliaReviewed by:
Gabriele Greco, University of Trento, ItalyCopyright © 2023 Ayoub, DuMez, Lazo, Luzaran, Magoti, Morris, Baker, Clarke, Correa-Garhwal, Hayashi, Friend and Opell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nadia A. Ayoub, YXlvdWJuQHdsdS5lZHU=; Brent D. Opell, Ym9wZWxsQHZ0LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.