- 1College of Animal Science, Shanxi Agricultural University, Jinzhong, Shanxi, China
- 2Shandong Vocational College of Light Industry, Zibo, Shandong, China
- 3College of Horticulture, Shanxi Agricultural University, Taiyuan, Shanxi, China
High temperature affects behavior, physiology, survival, and the expression of related genes in adult honeybees. Apis mellifera is the common pollinator in greenhouse and is susceptible to high temperature stress. To further explore the molecular basis related to heat stress, we compared the transcriptome profiles of adult worker bees at 25 and 45°C, and detected the expression patterns of some differentially expressed genes (DEGs) in different tissues by q RT-PCR. Differential expression analysis showed that 277 DEGs were identified, including 167 genes upregulated and 110 genes downregulated after heat stress exposure in adult worker bees. In GO enrichment analysis, DEGs were mostly enriched for protein folding, unfold protein binding, and heme binding terms. Protein processing in endoplasmic reticulum and longevity regulating pathway-multiple species were significantly enriched in KEGG. The expression levels of 16 DEGs were consistent with the transcriptome results. The expression patterns of 9 DEGs in different tissues revealed high levels in the thorax, which was supposed that the thorax may be the most important part in the response to heat stress. This study provided valuable data for exploring the function of heat resistance-related genes.
1. Introduction
As the most valuable pollinators for modern agriculture and ecological diversity (The Honeybee Genome Sequencing Consortium, 2006; Klein et al., 2007; Belsky and Joshi, 2019), the development of honeybee populations is influenced by many factors (Potts et al., 2010; Belsky and Joshi, 2019; Denlinger and Yocum, 2020), including environmental temperature (Abou-Shaara et al., 2012, 2017). Environmental temperature can affect the internal and external activities of honeybee colonies (Tautz et al., 2003; Abou-Shaara et al., 2012, 2017), such as development (Tautz et al., 2003; Groh et al., 2004; Chuda-Mickiewicz and Samborski, 2015; Medina et al., 2020), phenology (Langowska et al., 2017), learnability (Tautz et al., 2003), behavioral performance (Tautz et al., 2003; Al-Qarni, 2006; Blazyte-Cereskiene et al., 2010), and susceptibility to disease (Groh et al., 2004; Conte and Navajas, 2008; Dalmon et al., 2019). And temperature can affect the biochemical and physiological processes of honeybees, including survival, tolerance, loss of body water, and the activity of antioxidant and detoxification enzymes (Dalmon et al., 2019; Li X. Y. et al., 2019). At the same time, Honeybees have evolved a series of measures to cope with temperature stress, such as behavioral avoidance (Nicolson, 2009; Ohashi et al., 2009; Joshi and Joshi, 2010), morphological changes (Langowska et al., 2017), life histories, adjusting metabolism and nutrient supply, adjusting protein synthesis or decomposition, and increasing the content of protective substances, so as to slow down or repair the damage caused by temperature stress (Ahamed et al., 2010; Li X. Y. et al., 2019; Xu et al., 2022).
The foraging behavior of honeybees usually occurs within the temperature range of 10–40°C and is inhibited by high temperatures (Joshi and Joshi, 2010). 20∼25°C is the optimal temperature range for the flight of forager bees (Zeng, 2007; Rameshwor and Resham Bahadur, 2014; Cui et al., 2022). When the external temperature exceeds 40°C, worker bees stop foraging (Joshi and Joshi, 2010) and low the hive temperature by collecting water and rapidly fanning their wings (Nicolson, 2009; Ohashi et al., 2009). Further, heat stress triggers the expression of heat-related genes, which are involved in a series of biological processes such as cell metabolism, protein folding, and degradation (Koo et al., 2015). At present, among the many thermal-resistance genes, the heat shock proteins (HSPs) gene family plays an extremely important role (Jerbi-Elayed et al., 2015; Bhattacharyya et al., 2019; Shih et al., 2021). In addition to HSPs, there are zinc finger proteins (ZFPs), serine/threonine protein kinases (STKs) (Ma et al., 2019), antimicrobial peptide genes and endocrine regulatory genes (Li et al., 2022a,b).
Ambient temperature has a negative effect on foraging behavior (Al-Qarni, 2006; Blazyte-Cereskiene et al., 2010), further affecting pollinator decisions and crop pollination effects. The internal environment of facility agriculture is different from the natural environment, and there will be some problems such as large temperature differences and high temperatures (Su et al., 2017). In the greenhouse, honeybee colonies are prone to adult dysplasia and colony decline, which is unfavorable to reproducing honeybee colonies (Nguyen et al., 2013; Ma et al., 2020). Transcriptome technology has played an increasingly important role in various aspects of honeybee research, including studies of behavioral classification, oxidative stress (Southwick and Heldmaier, 1987), immune responses (Mao et al., 2013), and growth and development (Santos et al., 2021). Therefore, we compared the transcriptome of Apis mellifera workers in control groups (25°C) and in high temperature groups (45°C) were examined using RNA-seq technology and detected the expression patterns of some differentially expressed genes (DEGs) in different tissues by q RT-PCR. Our results provided basic data to better understand the regulatory molecular mechanisms of honeybee adaptation to high temperature.
2. Materials and methods
2.1. Experimental insects
Three healthy colonies (A. mellifera) were obtained from the experimental apiary of Shanxi Agricultural University (Taigu, China). Two capped brood combs were selected from each colony (two combs × three colonies), then put in an incubator at 34 ± 0.5°C and 75 ± 5% relative humidity (RH) to hatch. The hatched bees were marked on their backs with a non-toxic dye, recorded as 1 day old (1 d), and released back to their original colony. The marked bees were collected as samples at 20 d. The marked bees were collected as samples at 20 days old (20 d). The 20 d old worker bees were used as foragers.
According to the temperature data reached over 40°C from the greenhouse in summer and previous experiments on temperature gradients of 25, 30, 35, 40, and 45°C (Li X. Y. et al., 2019). Two treatments were designed: normal temperature, as control (CK: 25°C, 30% RH), and high temperature (HT: 45°C, 30% RH). All samples were put in a 15 mL diameter container and were maintained at 30% RH for 2 h at different temperatures (25 and 45°C). After treatment, 60 bees (10 × two temperature-treatment × three replicates) were randomly selected for RNA-seq, 540 bees (80 × two temperature-treatment × three replicates) for expression patterns of DEGs in different tissues, and then these samples were immediately frozen in liquid nitrogen and stored at −80°C. The head, thorax, abdomen, legs, and wings were dissected (all operations were performed on ice).
2.2. RNA-seq experiment
2.2.1. Library construction and sequencing
Total RNA was extracted using Trizol Reagent according to the manufacturer’s instructions (Ambion, Foster City, CA, USA). The concentration of total RNA was assessed using an RNA NanoDrop ND-2000 spectrophotometer (NanoDrop Inc., Wilmington, DE, USA). Non-denaturing agarose gel electrophoresis and the Agilent 2100 LabChip GX (Agilent, Santa Clara, CA, USA) were used to determine the integrity of extracted nucleic acids for quality control.
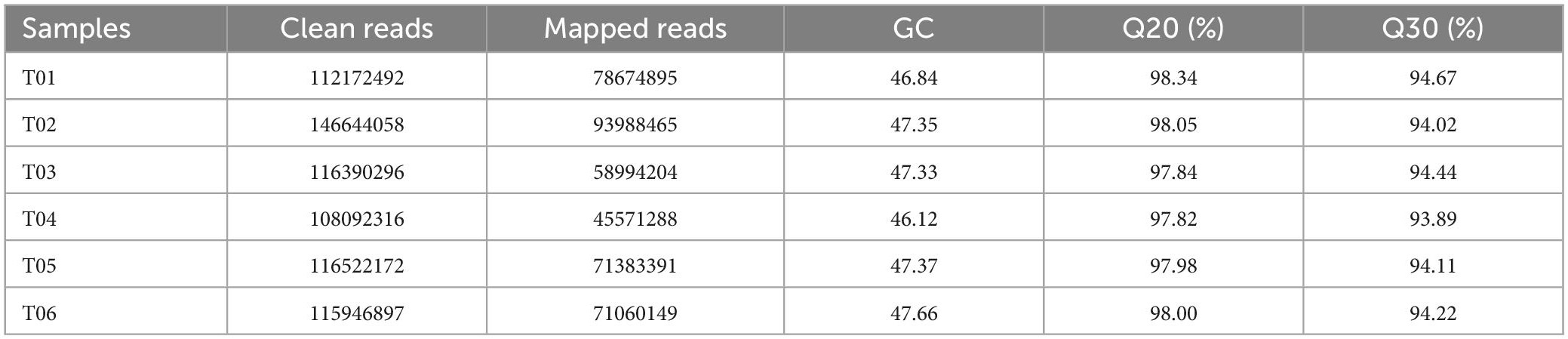
Three individuals were used for each biological replicate for transcriptomic analysis. Six individual cDNA libraries from the 25°C (T01, T02, and T03) and 45°C (T04, T05, and T06) groups were constructed. The mRNA was fragmented and randomized hexamer primers were used to copy the full-length mRNA; reverse transcriptase was used as a template for first strand cDNA synthesis. After purification, the cDNA was connected with adenine and sequencing splitter at the 3′-ends and amplified using PCR to establish a cDNA library. The Illumina NovaSeq 4000 platform (Illumina, San Diego, CA, USA) was used for sequencing. Raw data were stored as FASTQ files and all raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under Submission ID: SUB12053792, BioProject ID: PRJNA880001.
2.2.2. Mapping and transcriptome annotation
Reads with ploy-N (with a ratio of “N” > 10%) and low-quality (with quality scores < 20) were removed to get the clean reads. The clean reads were mapped to the reference genome of A. mellifera using HISAT2 (Li et al., 2014). Based on the selected reference genome sequence, the mapped reads were spliced using StringTie (Kim et al., 2015), and the potential novel transcripts were predicted using Cufflinks (Cole et al., 2012). BLAST was used to compare the discovered new genes with NR (Deng et al., 2006), Swiss-Prot (Rolf et al., 2017), Gene Ontology (GO) (Ashburner et al., 2000), Cluster of Orthologous Groups of proteins (COG) (Tatusov et al., 2000), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2004) databases to obtain annotation information of the new genes.
2.2.3. Differentially expressed gene analysis
Fragments per kilobase of exon model per million (FPKM) were estimated as a measure of transcript levels or gene expression (Love et al., 2014). The density distribution plot and box-whisper plot of FPKM was presented with R software. Differentially expressed genes (DEGs) were identified by DESeq2 (Love et al., 2014). The FDR is the corrected P-value. Genes with | fold change| > 2 and FDR < 0.05 were considered as DEGs. According to the fold change in relative expression levels between the two groups, the DEGs could be classified as upregulated or downregulated. A volcano plot was generated to visualize the results with R software. A hierarchical clustering analysis of the DEGs was performed (Yu et al., 2012), and genes with the same or similar expression patterns were clustered.
2.2.4. Functional enrichment analysis
To determine the functional category, the DEGs were mapped to the GO database by DAVID (Dennis et al., 2003). The annotation was refined and enriched by using the top-GO (v2.40.0) package in R software (Yu et al., 2012). The biological process (BP), molecular function (MF), and cellular components (CC) were particularly upregulated or downregulated in DEGs and were extracted and visualized via DAVID, which is generally referred to as GO analysis. The KEGG pathway was analyzed by using the KOBAS (2.0). P-value < 0.05 was the threshold value for significant enrichment results.
2.2.5. Protein-protein interaction (PPI) analysis
A protein-protein interaction (PPI) network of the DEGs was constructed by STRING (Franceschini et al., 2013). The constructed protein interaction network was imported into Cytoscape (Shannon et al., 2013) for visualization. The PPI consists of nodes and lines. Network nodes represent proteins, and the lines between proteins represent interactions. There are seven protein-protein interactions including neighborhood, co-occurrence, fusion, co-expression, experiments, databases, and text mining.
2.3. Quantitative real-time PCR verification
Quantitative real-time PCR (q RT-PCR) was used to verify the accuracy of RNA-seq (Livak and Schmittgen, 2001). Sixteen genes were randomly selected for verification. Nine DEGs verified were further quantified in different tissues (i.e., head, thorax, abdomen, legs, and wings). According to the CDS region of selected genes, specific primers were designed using Primer 5 (Supplementary Table 1). The primers were synthesized by Shanghai Sangon Co., Ltd (Shanghai, China). First-strand cDNA was synthesized from total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Kusatsu, Japan), according to the manufacturer protocol. The 15 μL reaction consisted of 100 ng/μL diluted cDNA (6.3 μL of double distilled water dilution), 7.5 μL of SYBR® Premix Ex Taq II (TaKaRa, Japan), and 0.6 μL of each primer. The q RT-PCR thermal cycle procedure was: 95°C predefined for 30 s, 45 cycles of denaturation at 95°C for 5 s, annealing at 60°C, and extension for 30 s. The fold changes for RNA-seq were estimated as base 2 logarithms of the FPKM ratio, and the fold changes for q RT-PCR were based on ΔΔCq.
2.4. Expression patterns of DEGs
Nine DEGs verified were further quantified in different tissues (i.e., head, thorax, abdomen, legs, and wings) by q RT-PCR. The reaction system was consistent with the q RT-PCR verification. The mRNA quantification was conducted following the 2–ΔΔCq method (Livak and Schmittgen, 2001). The one-way Anova test and Duncan’s multiple range test (SPSS 17.0) were used for significance analysis. Data were shown mean ± SEM. The results were plotted using GraphPad Prism 8.0.
3. Results
3.1. RNA-sequencing analysis
3.1.1. Reference genome alignment results
The sequencing data of the RNA-seq samples were shown in Table 1. Among six samples, 109.52 Gb of clean data in total was obtained, 15.89 Gb per sample with Q30 values of 93.89% and above. Filtering to remove low-quality reads, there were an average of 134,316,160 and 113,520,461 clean reads in CK and HT (Table 1). The clean reads for each sample were compared with the specified reference genome. The length distribution of single genes was also determined and most of the genes were > 3000 bp (Supplementary Figure 2). A total of 1,743 new genes were discovered, 148 of which were functionally annotated (Supplementary Table 2).
3.1.2. Differential expression gene analysis
Fragments per kilobase of exon model per million values spanned six orders of magnitude from 10–2 to 103 (Figure 1A). The distribution of the six samples was basically the same (Figure 1B). 277 DEGs were identified between CK and HT using DESeq (| log2 FC| ≤ 2, FDR ≤ 0.05). Of these, there were 167 upregulated DEGs and 110 downregulated DEGs (Figure 2). Cluster analysis showed the expression profiles of CK and HT (Figure 3). In the upregulated DEGs, the most enriched and expressive DEGs were the HSPs (HSC70-4, HSC70-5, HSP70-Ab, HSP70-Cb, HSP90, HSP40, and L (2) efl), fatty acid synthase (FAS), BAG domain-containing protein Samui (BAG), as well as several transcripts of unknown function. In the downregulated DEGs, the most enriched DEGs were E3 ubiquitin-protein ligase RNF8 (UbE3), and insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1). Identification and annotation information for all DEGs were provided in the Supplementary Table 3.
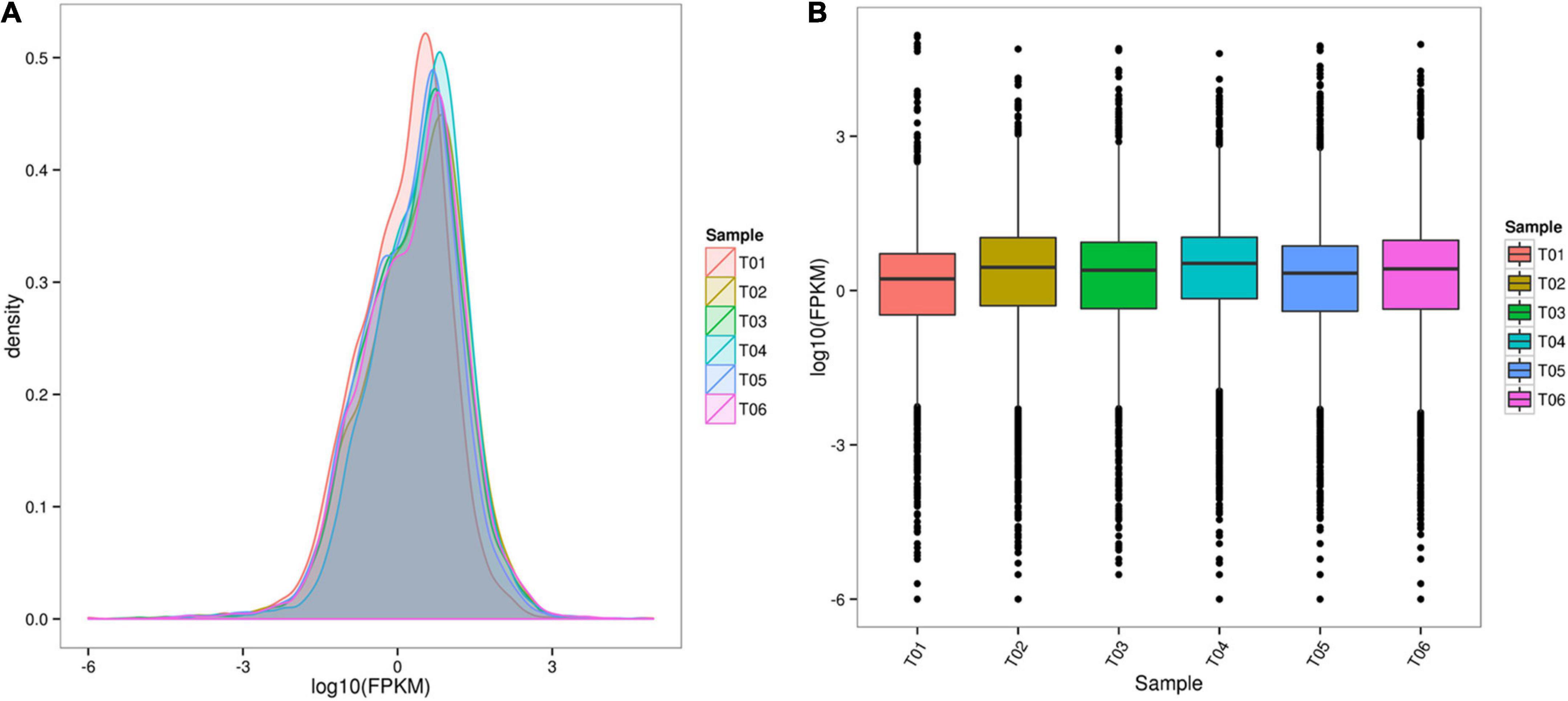
Figure 1. RNA-seq data analysis. (A) Comparison of the fragments per kilobase of exon model per million (FPKM) density distribution for all samples. (B) Box-whisker plot of FPKM values for all samples.
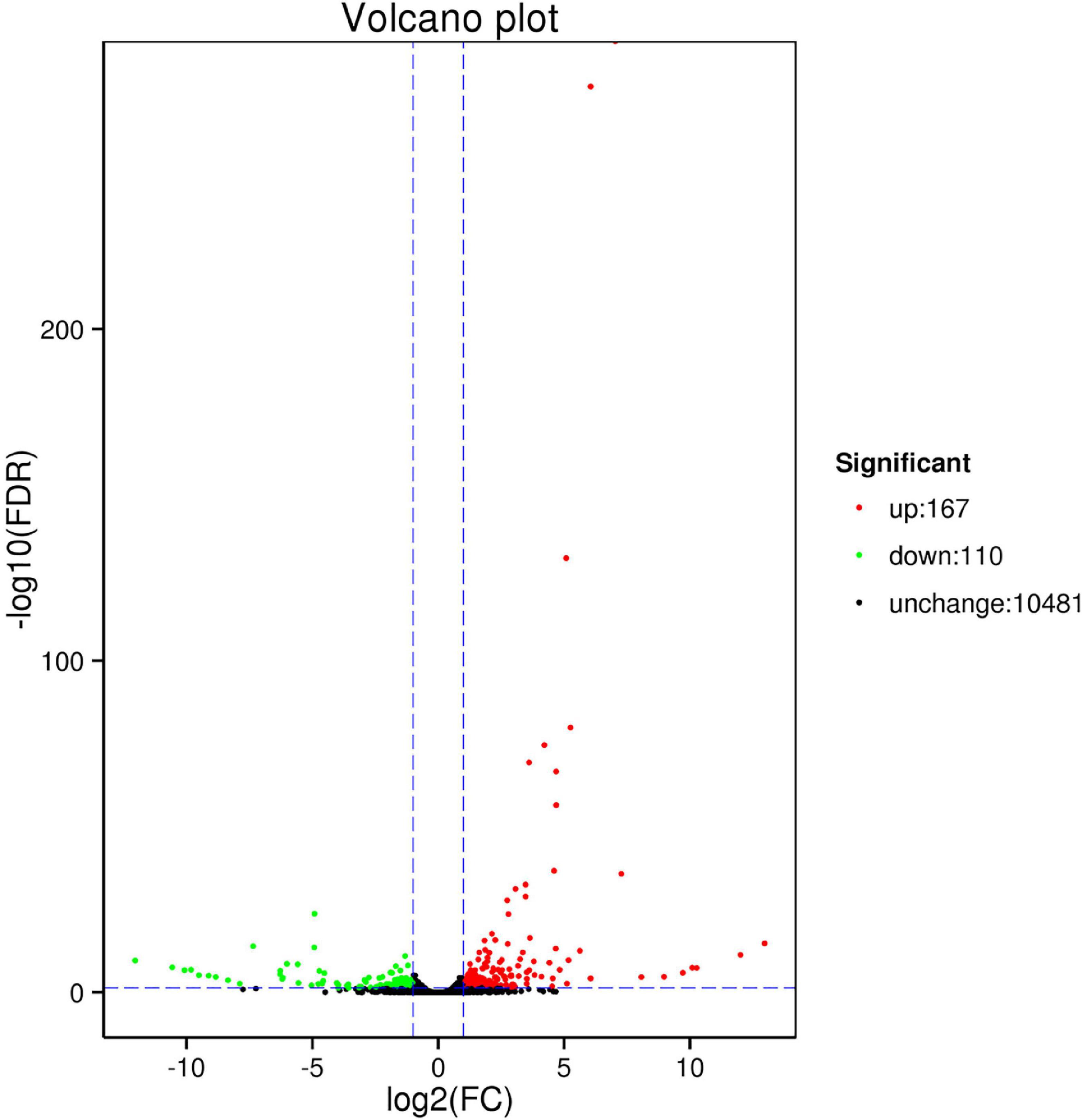
Figure 2. Volcano plot of differentially expressed genes (DEGs) between control (CK) and high temperature (HT).
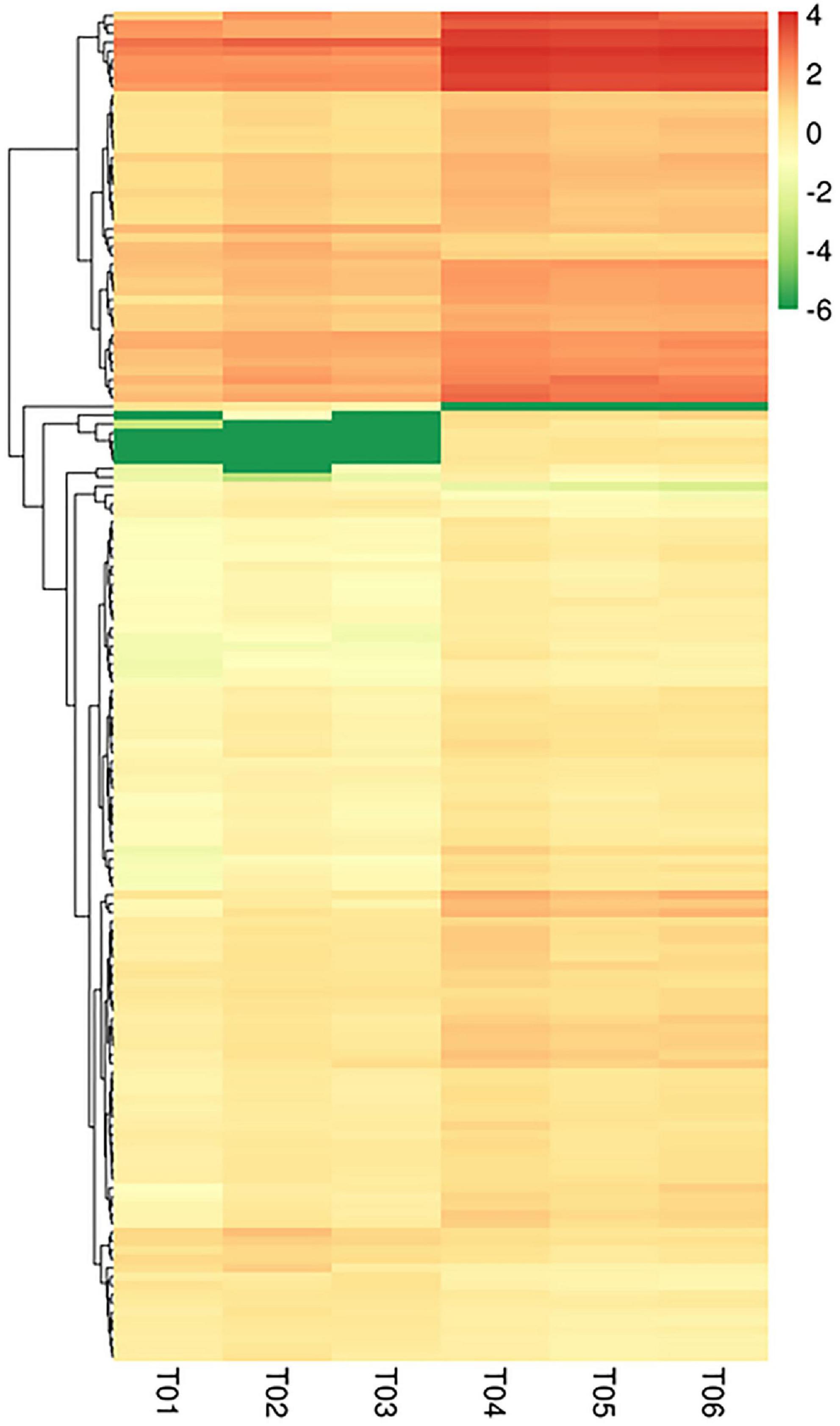
Figure 3. Cluster analysis of DEGs. Columns represent samples and rows represent different genes. The color represents the expression level of the gene in the sample determined by log10 (FPKM + 0.000001).
3.1.3. Enrichment analysis
We performed GO and KEGG enrichment analysis of DEGs (Figures 4, 5). In a GO enrichment analysis of 277 DEGs, 143 genes were annotated, including 25 process categories at P < 0.05 (Table 2). In response to heat stress, DEGs were enriched for various terms in the BP, CC, and MF categories (Figure 4). DEGs were classified into three GO categories: biological processes (33.5%), cellular components (24.1%), and molecular functions (41.15%). DEGs were mostly enriched for protein folding, unfold protein binding, and heme binding terms. The DEGs coregulated under heat stress according to GO categorization were biological process [signal transduction, response to stress, and metabolic process (of cellular amino acid, lipid, and protein)], cellular process (nucleus, cell membrane, and transcription factor complex), and molecular function (ATP binding, unfolding binding, activities of gate channels, and kinases). DEGs were mainly concentrated in the nucleus and cell membrane and were involved in protein folding, response to stress, and some signaling pathways.
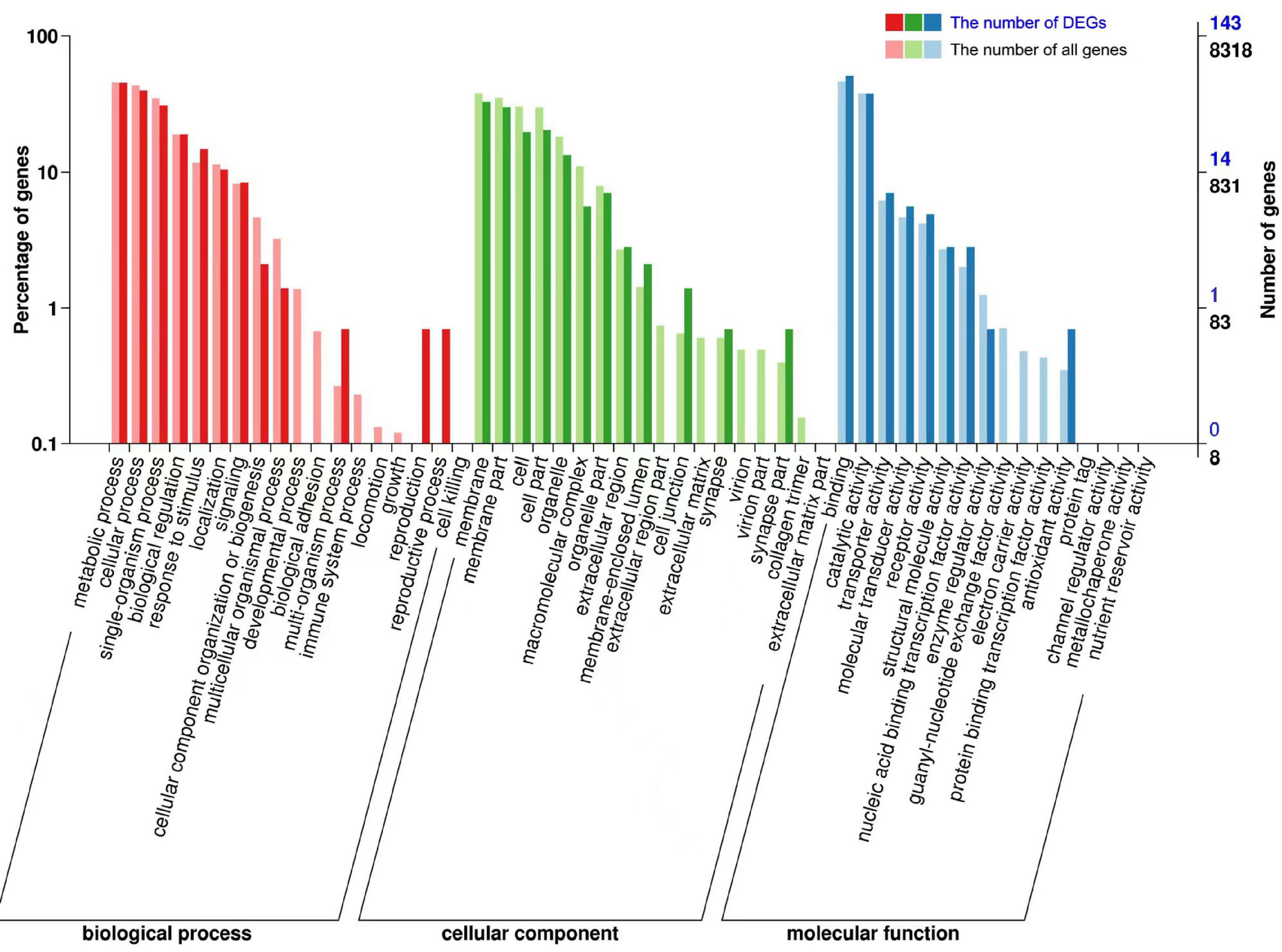
Figure 4. Gene Ontology (GO) enrichment analysis of DEGs. The X-axis indicates the GO classification, the left side of the Y-axis indicates the percentage of genes, and the right side shows the number of genes. Bold colors indicate the number of DEGs, and light colors indicate the number of all genes.
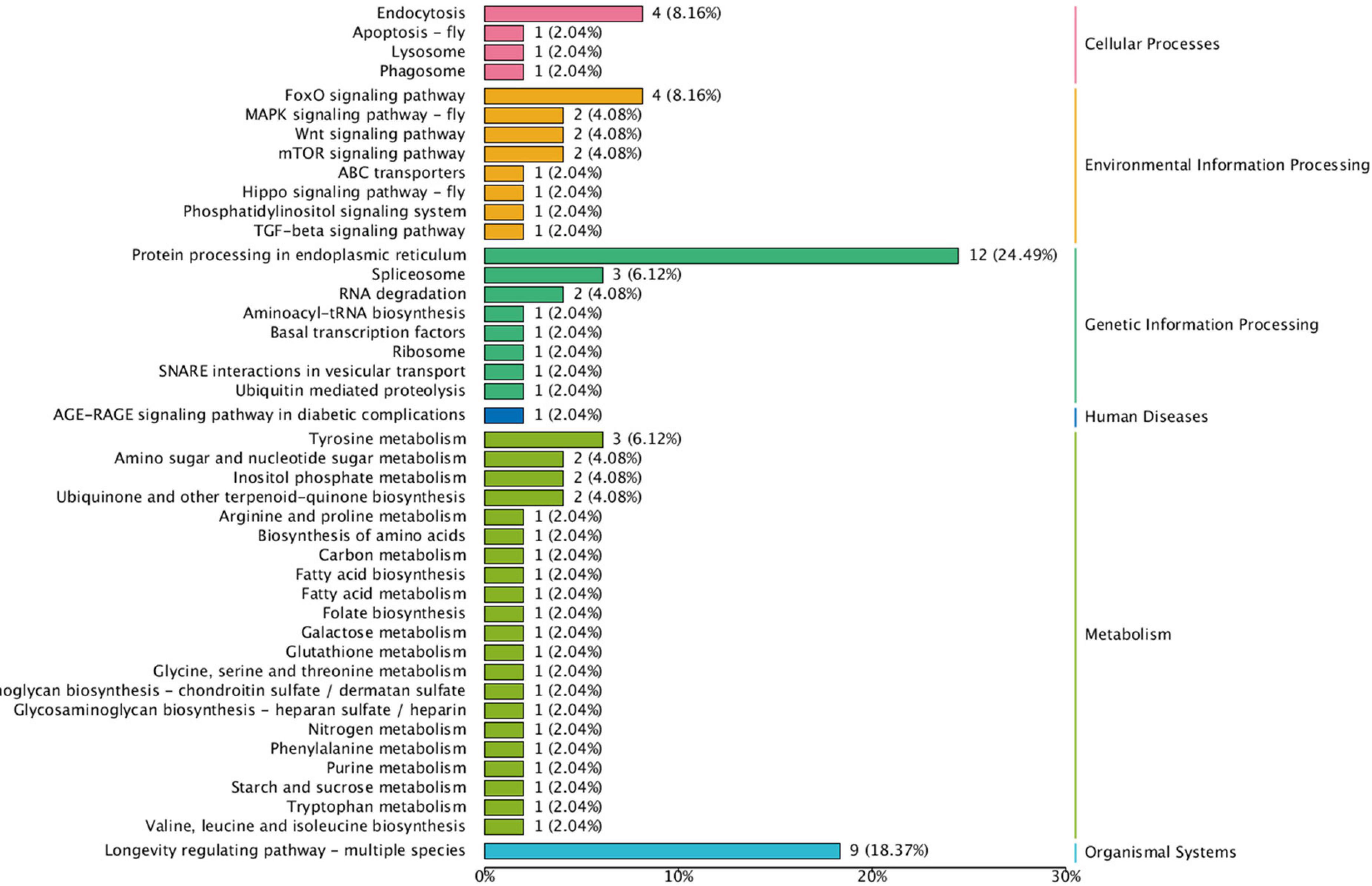
Figure 5. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs. The Y-axis indicates the pathway and the X-axis indicates the number of annotated genes.
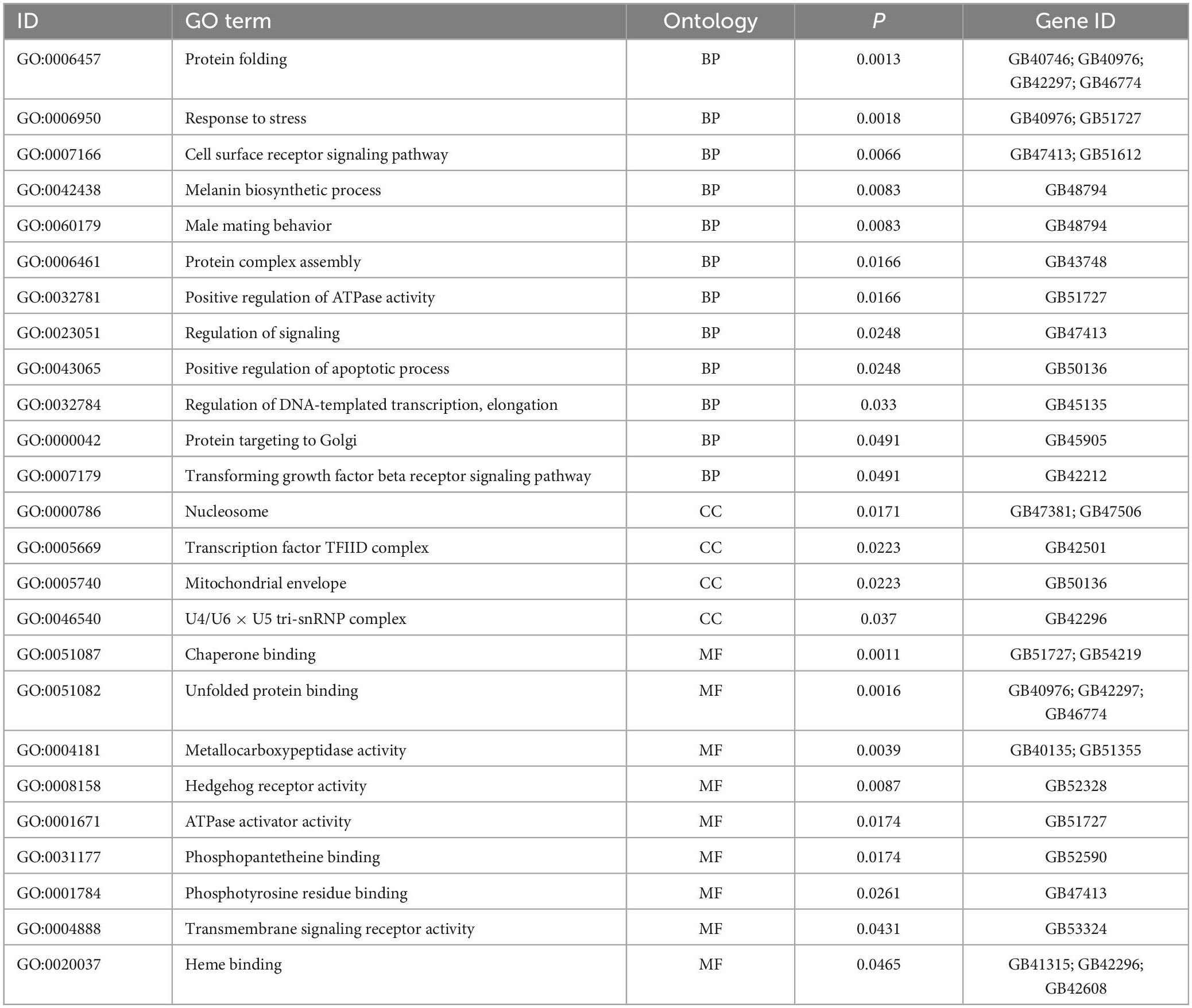
Table 2. Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) enriched in BP, CC, and MF.
The KEGG enrichment analysis of these DEGs was shown in Figure 5. In a KEGG enrichment analysis of 277 DEGs, 79 genes were annotated. KEGG results revealed the main cellular processes (endocytosis and apoptosis), environmental information processes (Fox O signaling pathway, MAPK signaling pathway, Wnt signaling pathway, and mTOR signaling pathway), genetic information processing (protein processing in endoplasmic reticulum, spliceosome, and Ubiquitin mediated proteolysis), metabolism (tyrosine, amino sugar and nucleotide, fatty acid, and amino acid), and organismal systems (longevity regulating pathway) enriched in A. mellifera under heat stress. KEGG enrichment analysis showed that DEGs were significantly enriched in genetic information processing and metabolism, among which protein processing in endoplasmic reticulum and longevity regulating pathway were significantly enriched pathways.
3.1.4. PPI analysis
The PPI analysis was conducted by using String database and visualized using Cytoscape. The result revealed that 18 proteins were involved in heat stress in PPI analysis (Figure 6). 60S Ribosomal protein L7 (ENSBTAG00000020139), Zinc finger proteins (ENSBTAG00000032003, ENSBTAG 00000025246, ENSBTAG00000039523), Heat shock 70 kDa protein (ENSBTAG 00000025441), Ankyrin repeat domain-containing protein (ENSBTAG00000004912), and serine/threonine-protein kinase (ENSBT AG00000004514) had strong interactions. Moreover, the PPI network showed that HSPs form a complex molecular network with other proteins, which promoted protein folding and protected cellular proteins from heat damage.
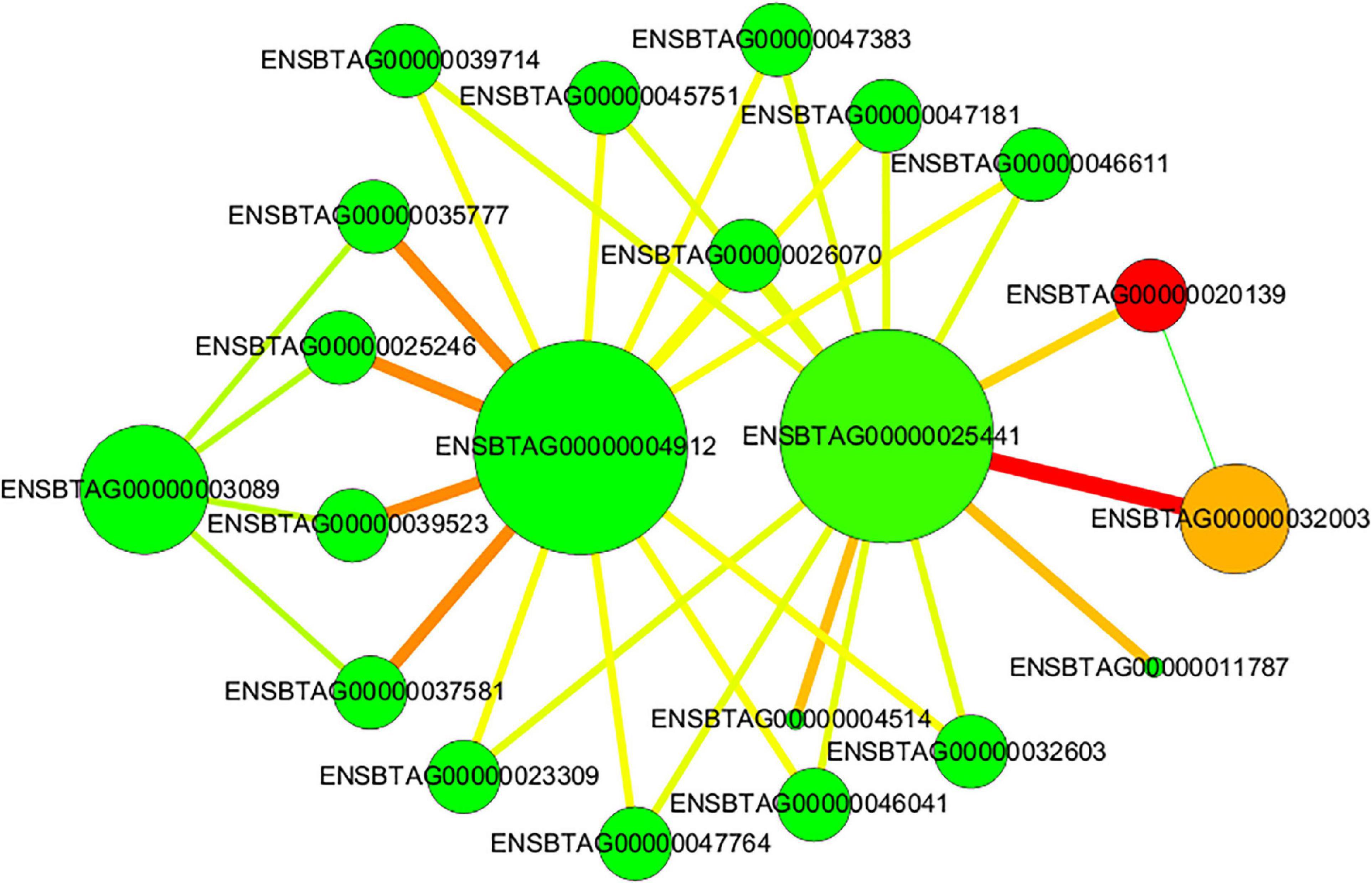
Figure 6. Protein-protein interaction (PPI) networks of DEGs. The nodes represent proteins and the lines represent interactions. The size of node represents the degree. The color of node represents the cluster coefficient, values vary from low to high with the color gradient from green to red. The green line indicates conserved neighborhood evidence, the red line indicates fusion evidence, and the yellow-green line indicates text mining.
3.2. Validation of RNA-seq results
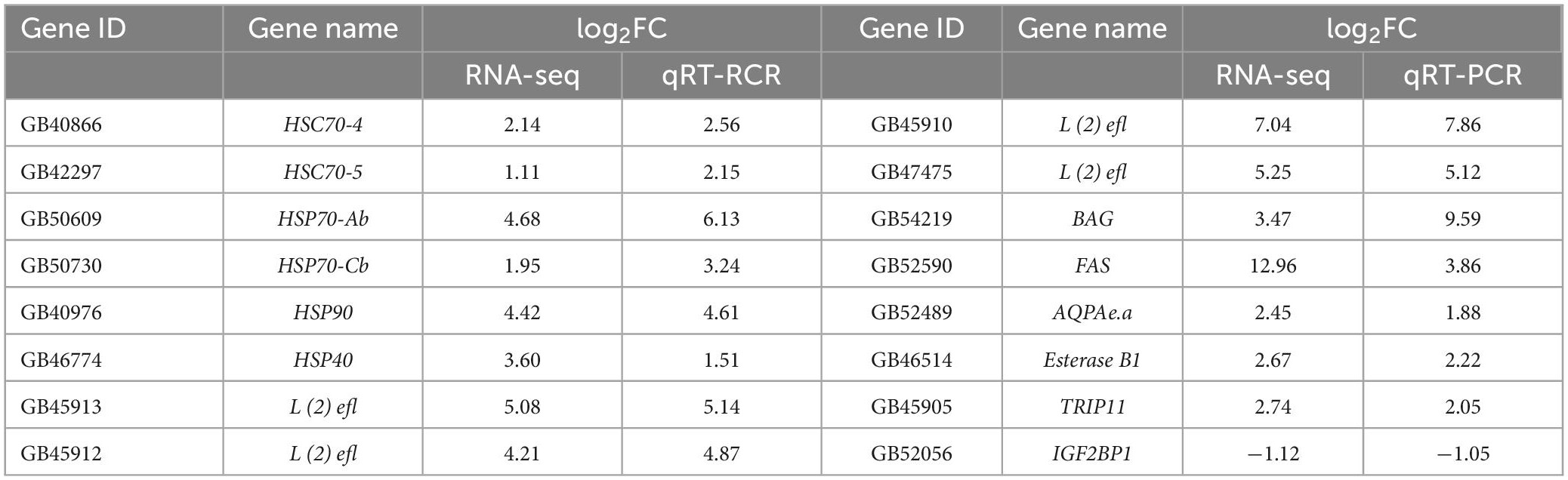
The results of the RNA-seq data showed that hundreds of genes were differentially expressed after high temperature treatment. We randomly selected 16 DEGs for q RT-PCR analysis to verify the results of RNA-seq identification. The results showed that the expression trend detected by q RT-PCR was consistent with the RNA-seq (Table 3). This ensured that the RNA-Seq results were quite reliable for the identification of DEGs.
3.3. Expression patterns of DEGs
The expression levels of 9 heat-related genes in different tissues of A. mellifera were evaluated (Figures 7A–I). The 9 DEGs were L (2) elf genes (GB45910, GB45912, GB45913, GB47475), HSP genes (GB46774, GB50609, GB42297, GB40976), and BAG gene (GB54219). The expression of the DEGs was detected in the head, thorax, abdomen, legs, and wings, and the expression levels of most genes differed among tissues (P < 0.05). The four L (2) efl genes were similarly expressed in different tissues, all being expressed in the thorax (P < 0.05), but with no significant expression in the remaining tissues (P > 0.05) (Figures 7A–D). The four HSP genes were most highly expressed in the thorax and had low expression in other tissues (Figures 7E–H). The BAG gene was most highly expressed in the thorax, followed by the legs, with the lowest expression in the wings. All genes were highly expressed in the thorax with similar expression patterns (P < 0.05) (Figure 7I).
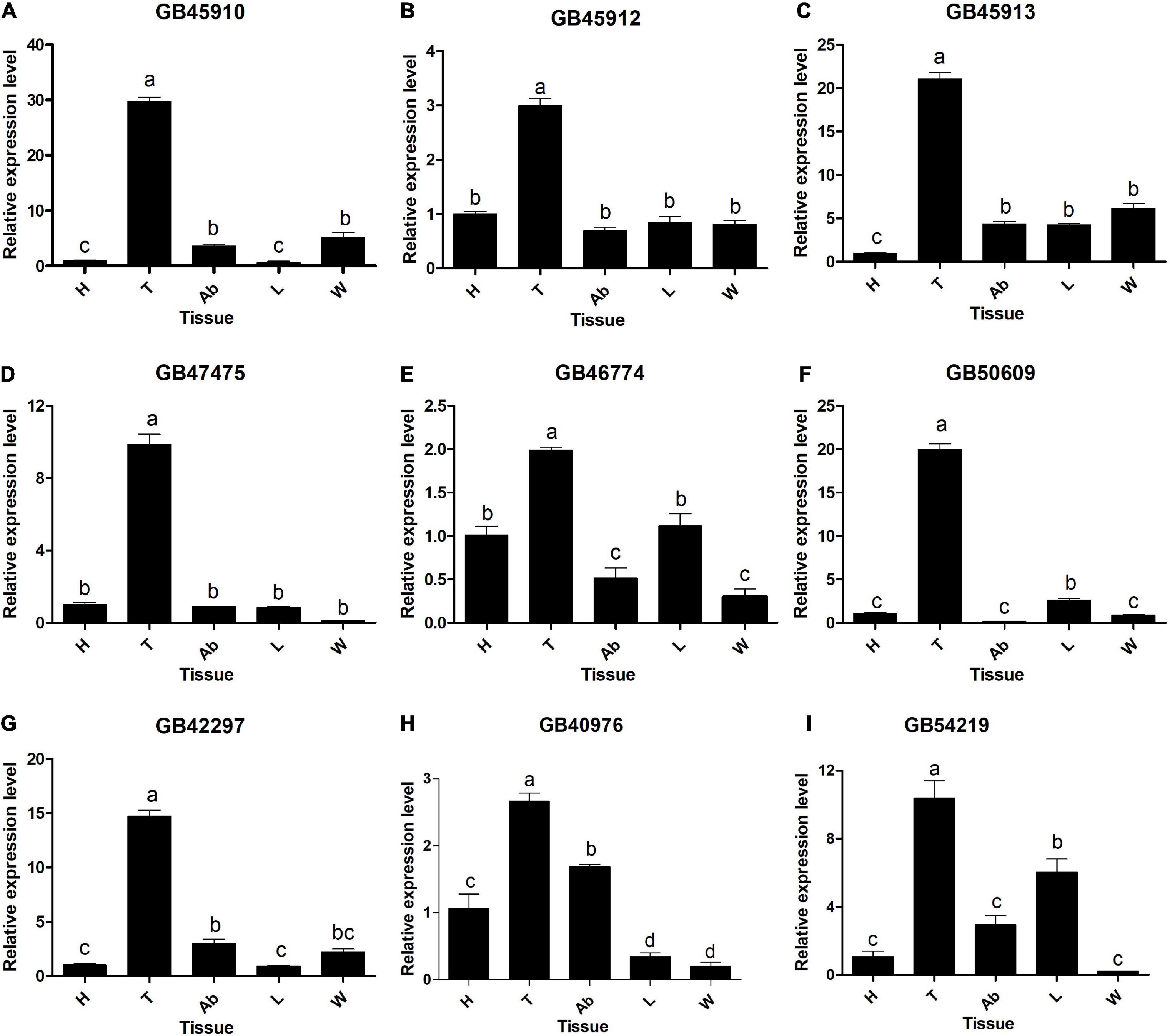
Figure 7. Relative expression levels of DEGs in different tissues. (A) GB45910. (B) GB45912. (C) GB45913. (D) GB47475. (E) GB46774. (F) GB50609. (G) GB42297. (H) GB40976. (I) GB54219. H, head without antenna; Ab, abdomen; T, thorax without wings; L, legs; W, wings. Different capital letters indicate significant differences between tissues at the 0.05 level. Data are presented as mean ± SE.
4. Discussion
Global warming and the pollination of greenhouse crops result in honeybee exposure to heat stress. Honeybees face many adverse effects of heat stress (Medina et al., 2020; Zhao et al., 2021). Ambient temperature can greatly affect foraging activity (Al-Qarni, 2006; Blazyte-Cereskiene et al., 2010), then affect crop pollination. To further understand the molecular mechanism of honeybees exposed to heat stress, a comparative analysis of gene expression among different temperatures was performed using RNA-seq, and the expression patterns of some DEGs were detected in different tissues. This study provided a set of important pathways and DEGs associated with high temperatures.
In our study, 277 DEGs were identified between CK and HT. In all, there were significantly more upregulated DEGs (167) than downregulated DEGs (110). Of these upregulated DEGs, there were 15 HSPs, including HSP40, HSP70, HSP90, L (2) efl, and so on (Supplementary Figure 2). HSPs, as highly conserved molecular chaperones, can influence the folding and functional conformation of proteins (Purschke et al., 2017; Alqarni, 2020). Both GO and KEGG enrichment analysis were associated with the protein processes, which confirmed that heat stress could induce a rapid response by the organism to synthesize resistance proteins and protect against damage (Scheffer et al., 2022). This also provided evidence for the hypothesis that elevated temperature accelerates protein unfolding and initiates molecular mates (Day et al., 2002). Compared with previous studies, the HSP family, such as sHSP, HSP40, HSP70, and HSP90, also participated in honeybee heat stress responses (Liu et al., 2012; Alqarni et al., 2019). We could not find any sHSPs, however, we found 6 L (2) efl genes. Under heat stress conditions, the intracellular environment is destroyed, which leads to cell apoptosis. The apoptosis process is regulated by caspase protease, and the upregulation of HSP90, HSP70, and other stress-related genes could inhibit the activity of caspase protease (Hitomi et al., 2004). We identified increased expression of HSP genes involved in multiple processes, but a striking finding was that L (2) efl genes were dominant, indicating that the genes contributed the most to 20 d old worker bees in 45°C. The expression of HSPs may be a potential target related to the survival and performance of honeybees in harsh climatic conditions.
In addition, these DEGs were found to be associated with the zinc finger protein family (ZFPs, 5 genes), BAGs family (3 genes), and ubiquitin family (5 genes). ZFPs can bind to biological macromolecules and participate in the regulation of resistance mechanisms in response to various biological and abiotic stress factors at the transcriptional level (Andrew et al., 2004; Giri et al., 2011; Droll et al., 2013). BAG proteins may serve as bridging molecules that recruit HSP70/HSC70 to specific target proteins (Takayama et al., 1995; Rhoads and Friedberg, 1997). Thus, the BAG family functionally regulated multiple cellular pathways, such as programmed cell death (PCD) and stress responses (Kang et al., 2006). The association between the ubiquitin family and HSP90 chaperone complex has been reported (Ehrlich et al., 2009). Ubiquitin-protein activity inhibited unfolded proteins in stress-induced cell death (Chen et al., 2015), and ubiquitin mediated proteolysis was one of the recorded KEGG enriched pathways that was exposed at high temperatures in our findings. The results suggested that HSPs, BAGs, and ubiquitin family together form a complex molecular network with ZFPs. More interestingly, HSPs were also associated with immune responses (Parcellier et al., 2001; Lanneau et al., 2010; Zhang et al., 2022). Of the upregulated DEGs, toll-like receptor protein was associated with immune responses. The KEGG pathway showed enrichment of the endocytosis pathway, which was also involved in cellular immunity (Zhu et al., 2020). The endocytosis pathway may be related to the transport of proteins, thus exerting an immune function for cellular protection. Previous transcriptomic studies have indicated the relationship between heat exposure and immunity (Li H. et al., 2019). In addition, 5 genes related to antioxidants were expressed at heat temperatures, suggesting that heat stress can trigger oxidative stress in bees. A. mellifera may promote the body’s defense against external high temperatures through immune and antioxidant responses.
We also discovered 40 DEGs related to metabolism. There were both 26 upregulated and 14 downregulated genes. The KEGG results revealed that metabolic processes were the main pathways (Figure 5). Lipids and amino acids provide energy for organisms via the tricarboxylic acid cycle. Metabolism of energy was essential to maintain homeostasis and growth (Zhu et al., 2020). Heat stress could impair the energy reserve and metabolism of insects (Slimen et al., 2016). This result was consistent with the result of physiological experiments (Yang et al., 2009; Neal, 2015), showing that high temperatures could lead to disruption in metabolism in honeybees. Transcriptome profiles also suggested that high temperatures could cause disruption at metabolic levels (Ma et al., 2019; Shen et al., 2021). In addition, DEGs were found to be associated with stress resistance and protein kinases. A total of 8 protein kinase-related genes were identified, of which 5 were upregulated and 3 were downregulated. Protein kinases can phosphorylate proteins by transferring phosphate groups to specific substrate proteins, usually ATP or GTP as donors and serine, threonine, and tyrosine as receptors (Krupa et al., 2004; Faucher et al., 2008). The regulation of fatty acid synthase and serine protease is part of the diapause program (Robich and Denlinger, 2005; Vatanparast and Park, 2021). The heat stress caused metabolic disorders which cause diapause in honeybees. Simultaneously, adult honeybees had an ability to adapt to heat stress (Stabentheiner et al., 2010). It may be related to the higher content of metabolic enzymes.
Signal transduction pathways could link environmental stress to the organism, thus inducing most of the above-mentioned defense responses (Kawasaki et al., 2002; Ludwig et al., 2005). When organisms were subjected to heat stress, various signal transduction pathways could be activated, such as Notch, MAPK, Fox O, mTOR, Hippo, and Wnt (Badr et al., 2018). In the current experiment, some signal transduction pathways were involved, such as the Fox O, MAPK-fly, mTOR, Wnt, and TGF-beta. In the Wnt signaling pathway, downregulated of the t-cell factor/lymphoid enhancing factor (TCF/LEF) in the nucleus potentially contribute to cell cycle arrest (Kiuchi et al., 2008), and upregulated of the siah-1 interacting protein (SIP) might lead to proteolysis (Supplementary Figure 3). In the TGF-bate singling pathway, upregulated of the Samd2/3 potentially contribute to G1 arrest and lead to angiogenesis extracellular matrix neogenesis, immunosuppression, and apoptosis induction (Supplementary Figure 4). Studies have consistently shown that both Fox O (Martins and Link, 2016) and TGF-beta (Shaw et al., 2007) signaling pathways were involved in life-cycle regulation. Together with Hippo, the Wnt, TGF-beta, and Fox O signaling pathways affected body segment formation, pigmentation, appendage development, and the development of wings in insects (Barry and Camargo, 2013), which may be a key factor in diapause. Thus, the signaling transduction pathways might be the central convergence point in the A. mellifera response to heat stress.
In this experiment, all nine heat-related genes were higher expressed in the thorax of A. mellifera than in other tissues. The thorax temperature is regulated at different levels depending on food quality and demand in the colony (Stabentheiner and Kovac, 2014, 2016). The regulation of the thorax temperature plays a crucial role in honeybee flight ability (Souza-Junior et al., 2020). The thorax temperature can exceed 40°C during flight (Kovac et al., 2018). In A. mellifera, the main mechanisms for the prevention of thorax overheating during the flight are reduced metabolic and increased evaporative when the temperatures rise from 33 to 45°C (Kovac et al., 2018). This may be related to the high expression of heat-responsive genes in the thorax. Therefore, it was supposed that the thorax may be the most important part of the response to heat stress.
5. Conclusion
High temperatures can influence the physiological metabolism in honeybees, and the expression profiles of stress-related genes can also be induced in response to extreme environmental temperatures. Overall, 1,743 new genes were obtained, and 277 DEGs (167 upregulated DEGs and 110 downregulated DEGs) were identified in A. mellifera under heat stress by RNA-seq. Both GO and KEGG pathway enrichment analysis showed that these DEGs were involved in protein folding, binding, response to stress, and signal transduction pathways. DEGs related to metabolism, protein processing, immune response, and signal transduction may work together to protect against heat stress in A. mellifera. This study provided a set of important pathways and DEGs associated with high temperatures. Nine DEGs were highly expressed in the thorax of A. mellifera than in other tissues, which was supposed that the thorax may be the most important part in the response to heat stress. These results will be helpful to the function research of heat resistance-related genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
Author contributions
WM and XL: conceptualization. WM and BZ: methodology and revision. BZ: software, data analysis, and writing the manuscript. YJ and JL: resources. BZ and JZ: visualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the earmarked fund for the Agriculture Research System of China (CARS-44-KXJ22).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1099015/full#supplementary-material
References
Abou-Shaara, H. F., Al-Ghamdi, A. A., and Mohamed, A. A. (2012). Tolerance of two honey bee races to various temperature and relative humidity gradients. Environ. Exp. Biol. 10, 133–138.
Abou-Shaara, H. F., Owayss, A. A., Ibrahim, Y. Y., and Basuny, N. K. (2017). A review of impacts of temperature and relative humidity on various activities of honey bees. Insectes Sociaux 64, 455–463. doi: 10.1007/s00040-017-0573-8
Ahamed, M., Posgai, R., Gorey, T. J., Nielsen, M., Hussain, S. M., and Rowe, J. J. (2010). Silver nanoparticles induced heat shock protein 70, oxidative stress and apoptosis in Drosophila melanogaster. Toxicol. Appl. Pharmacol. 242, 263–269. doi: 10.1016/J.Taap.2009.10.016
Al-Qarni, A. S. (2006). Tolerance of summer temperature in imported and indigenous honeybee (Apis mellifera L.) races in central Saudi Arabia. Saudi J. Biol. Sci. 13, 123–127.
Alqarni, A. S. (2020). Differential foraging of indigenous and exotic honeybee (Apis mellifera L.) Races on nectar-rich flow in a subtropical ecosystem. Insects 11:254. doi: 10.3390/Insects11040254
Alqarni, A. S., Hussain, A. B., Javaid, I., Ayman, A. O., and Brian, H. S. (2019). Expression of heat shock proteins in adult honey bee (Apis mellifera L.) Workers under hot-arid subtropical ecosystems. Saudi J. Biol. Sci. 26, 1372–1376. doi: 10.1016/J.Sjbs.2019.08.017
Andrew, C. J., Thomas, J. C., and Bo Guan Hong Xiao Pamela, M. C. (2004). Controlling gene expression in Drosophila using engineered zinc finger protein transcription factors. Biochem. Biophys. Res. Commun. 348, 873–879.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Badr, G., Ramadan, N. K., Abdel-Tawab, H., Ahmed, S. F., and Mahmoud, M. H. (2018). Camel whey protein protects lymphocytes from apoptosis via the PI3K-AKT, NF-K appaB, ATF-3, and HSP-70 signaling pathways in heat-stressed male mice. Biochem. Cell Biol. 96, 407–416. doi: 10.1139/bcb-2017-0217
Barry, E. R., and Camargo, F. D. (2013). The Hippo superhighway: signaling crossroads converging on the hippo/yap pathway in stem cells and development. Curr. Opin. Cell Biol. 25, 247–253. doi: 10.1016/j.ceb.2012.12.006
Belsky, J., and Joshi, N. K. (2019). Impact of biotic and abiotic stressors on managed and feral bees. Insects 10:233. doi: 10.3390/insects10080233
Bhattacharyya, M., Chakraborty, S. K., and Acharya, S. K. (2019). Proboscis extension reflex in Apis florea (Hymenoptera: Apidae) in response to temperature. J. Entomol. Sci. 54, 238–249.
Blazyte-Cereskiene, L., Vaitkeviciene, G., Venskutonyte, S., and Buda, V. (2010). Honey bee foraging in spring oilseed rape crops under high ambient temperature conditions. Zemdirbyste 97, 61–70.
Chen, L. G., Du, Y. Q., Hu, J. L., Jiao, D. X., Li, J., and Zhou, Z. (2015). Proteomic analysis of ubiquitinated proteins from deltamethrin resistant and susceptible strains of the diamondback moth, Plutella xylostella L. Arch. Insect Biochem. Physiol. 90, 70–88. doi: 10.1002/arch.21245
Chuda-Mickiewicz, B., and Samborski, J. (2015). The quality of honey bee queens from queen cells incubated at different temperatures. Acta Scientiarum Polonorum Zootechnica 14, 25–32.
Cole, T., Adam, R., Loyal, G., Geo, P., Deahwan, K., David, R., et al. (2012). Differential gene and transcript expression analysis of RNA-Seq experiments with Tophat and Cufflinks. Nat. Protoc. 7, 562–578.
Conte, Y. L., and Navajas, M. (2008). Climate change: impact on honey bee populations and diseases. Revue Scientifique et Technique-office International Des Epizooties 27, 499–510.
Cui, J. X., Wang, Y. W., Li, W. H., Yang, J. T., and Zachary, Y. H. (2022). Impact of air temperature on flight capacity of Apis mellifera ligustica forager. Apiculture China 73, 56–59.
Dalmon, A., Peruzzi, M., Conte, Y. L., Alaux, C., and Pioz, M. (2019). Temperature-driven changes in viral loads in the honey bee Apis mellifera. J. Invertebr. Pathol. 160, 87–94. doi: 10.1016/j.jip.2018.12.005
Day, R., Bennion, B. J., Ham, S., and Daggett, V. (2002). Increasing temperature accelerates protein unfolding without changing the pathway of unfolding. J. Mol. Biol. 322, 189–203.
Deng, Y., Li, J. Q., Wu, S. F., Zhu, Y., Chen, Y., and He, F. C. (2006). Integrated nr database in protein annotation system and its localization. Comput. Eng. 32, 71–72.
Denlinger, D. L., and Yocum, G. D. (2020). Physiology of Heat Sensitivity in Temperature Sensitivity in Insects and Application in Integrated Pest Management. Bocan Raton, FL: Chemical Rubber Company Press.
Dennis, G. J., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., et al. (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:R60.
Droll, D., Minia, I., Faddl, A., Singh, A., Stewart, M., Queiroz, R., et al. (2013). Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathogens 9:e1003286. doi: 10.1371/Journal.Ppat.1003286
Ehrlich, E. S., Wang, T., Luo, K., Xiao, Z., Niewiadomska, A. M., Martinez, T., et al. (2009). Regulation of Hsp90 client proteins by a cullin5-ring E3 ubiquitin ligase. Proc. Natl. Acad. Sci. 106, 20330–20335. doi: 10.1073/pnas.0810571106
Faucher, S. P., Viau, C., Gros, P. P., Daigle, F., and Le-Moual, H. (2008). The prpZ gene cluster encoding eukaryotic-type Ser/Thr protein kinases and phosphatases is repressed by oxidative stress and involved in salmonella enterica serovar Typhi survival in human macrophages. FEMS Microbiol. Lett. 281, 160–166. doi: 10.1111/j.1574-6968.2008.01094.x
Franceschini, A., Simonovic, M., Roth, A., Mering, C. V., Szklarczyk, D., Pletscherfrankild, S., et al. (2013). STRING V9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, 808–815. doi: 10.1093/nar/gks1094
Giri, V., Dansana, P. K., and Tyagi, A. K. (2011). Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (Osrlck253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 191, 721–732. doi: 10.1111/j.1469-8137.2011.03740.x
Groh, C., Tautz, J., and Rossler, W. (2004). Synaptic organization in the adult honey bee brain is influenced by brood temperature control during pupal development. Proc. Natl. Acad. Sci. U S A. 101, 4268–4273. doi: 10.1073/pnas.0400773101
Hitomi, J., Katayama, T., Eguchi, Y., Kudo, T., Taniguchi, M., Koyama, Y., et al. (2004). Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J. Cell Biol. 165, 347–356. doi: 10.1083/jcb.200310015
Jerbi-Elayed, M., Lebdi-Grissa, K., and Foray, V. (2015). Using multiple traits to estimate the effects of heat shock on the fitness of Aphidius colemani. Entomol. Exp. Appl. 155, 18–27.
Joshi, N. C., and Joshi, P. C. (2010). Foraging behavior of Apis spp. on apple flowers in a subtropical environment. N. Y. Sci. J. 3, 71–76.
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280.
Kang, C. H., Jung, W. Y., Kang, Y. H., Kim, J. Y., Kim, D. G., Jeong, J. C., et al. (2006). AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 13, 84–95. doi: 10.1038/sj.cdd.4401712
Kawasaki, L., Olivia, S., Shiozaki, K., and Aguirre, J. (2002). Saka MAP Kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45, 1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kiuchi, T., Aoki, F., and Nagata, M. (2008). Effects of high temperature on the hemocyte cell cycle in silkworm larvae. J. Insect Physiol. 54, 454–461. doi: 10.1016/j.jinsphys.2007.11.004
Klein, A. M., Vaissiere, B. E., Cane, J. H., Stean-Dewenter, I., Cunningham, S. A., Kremen, C., et al. (2007). Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B Biol. Sci. 274, 303–313.
Koo, J., Son, T. G., Kim, S. K., and Lee, K. Y. (2015). Differential responses of Apis mellifera heat shock protein genes to heat shock, flower-thinning formulations, and imidacloprid. J. Asia-pacific Entomol. 18, 583–589.
Kovac, H., Kaefer, H., and Stabentheiner, A. (2018). The energetics and thermoregulation of water collecting honeybees. J. Comp. Physiol. 204, 783–790. doi: 10.1007/s00359-018-1278-9
Krupa, A., Preethi, G., and Srinivasan, N. (2004). Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J. Mol. Biol. 339, 1025–1039. doi: 10.1016/j.jmb.2004.04.043
Langowska, A., Zawilak, M., Sparks, T. H., Glazaczow, A., Tomkins, P. W., and Tryjanowski, P. (2017). Long-term effect of temperature on honey yield and honeybee phenology. Int. J. Biometeorol. 61, 11251–11132. doi: 10.1007/s00484-016-1293-x
Lanneau, D., Brunet, M., Frisan, E., Solary, E., Fontenay, M., and Garrido, C. (2010). Heat shock proteins: essential proteins for apoptosis regulation. J. Cell. Mol. Med. 12, 743–761.
Li, H., Zhao, X., Qiao, H., He, X., Tan, J., and Hao, D. (2019). Comparative transcriptome analysis of the heat stress response in Monochamus alternatus hope (Coleoptera: Cerambycidae). Front. Physiol. 10:1568. doi: 10.3389/fphys.2019.01568
Li, J. W., Ma, W., Zeng, P., Wang, J. Y., Geng, B., Yang, J. C., et al. (2014). LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 16, 806–812.
Li, X. Y., Ma, W. H., and Jiang, Y. S. (2022a). Heat stress affects the expression of antimicrobial peptide genes in adult honeybee (Apis cerana and Apis mellifera). Int. J. Trop. Insect Sci. 42, 2465–2471. doi: 10.1007/s42690-022-00774-x
Li, X. Y., Ma, W. H., Du, Y. L., Xu, K., and Jiang, Y. S. (2022b). Differential expression of endocrine regulatory genes in Apis cerana and Apis mellifera (Hymenoptera: Apidae) at high temperature. J. Entomol. Sci. 57, 411–424.
Li, X. Y., Ma, W. H., Shen, J. S., Long, D. L., Feng, Y. J., Su, W. T., et al. (2019). Tolerance and response of two honeybee species Apis cerana and Apis mellifera to high temperature and relative humidity. PLoS One 14:e0217921. doi: 10.1371/journal.pone.0217921
Liu, Z., Xi, D., Kang, M., Guo, X., and Xu, B. (2012). Molecular cloning and characterization of HSP27.6: the first reported small heat shock protein from Apis Cerana Cerana. Cell Stress Chaperones 17, 539–551. doi: 10.1007/S12192-012-0330-X
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 25, 402–408.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with Deseq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ludwig, A. A., Saitoh, H., Felix, G., Freymark, G., Miersch, O., Wasternack, C., et al. (2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. U S A. 102, 10736–10741. doi: 10.1073/pnas.0502954102
Ma, W. H., Li, X. Y., Shen, J. S., Du, Y. L., Xu, K., and Jiang, Y. S. (2019). Transcriptomic analysis reveals Apis mellifera adaptations to high temperature and high humidity. Ecotoxicol. Environ. Saf. 184:109599. doi: 10.1016/j.ecoenv.2019.109599
Ma, W. H., Li, X. Y., Shen, J. S., Li, L. X., and Jiang, Y. S. (2020). The effect of the greenhouse environment on the antioxidant and detoxification enzyme systems of honeybee colonies. Chin. J. Appl. Entomol. 57, 1104–1110. doi: 10.7679/j.issn.2095-1353.2020.111
Mao, W., Schuler, M. A., and Berenbaum, M. R. (2013). Honey constituents up-regulate detoxification and immunity genes in the western honey bee Apis mellifera. Proc. Natl. Acad. Sci. U S A. 110, 8842–8846. doi: 10.1073/pnas.1303884110
Martins, R. L. G., and Link, W. (2016). Long live Fox O: unraveling the role of Fox O proteins in aging and longevity. Aging Cell 15, 196–207. doi: 10.1111/acel.12427
Medina, R. G., Paxton, R. J., HernÁNdez-Sotomayor, S. M. T., Pech-Jimenez, C., Medina-Medina, L. A., and Quezada-Euan, J. J. G. (2020). Heat stress during development affects immunocompetence in workers, queens and drones of Africanized honey bees (Apis Mellifera L.) (Hymenoptera: Apidae). J. Thermal Biol. 89:102541. doi: 10.1016/j.jtherbio.2020.102541
Neal, S. J. (2015). A Functional and Comparative Genomic Analysis of Heat-Induced Gene Expression in Drosophila. Master’s Thesis. Canada: University of Toronto.
Nguyen, T. M., Bressac, C., and Chevrier, C. (2013). Heat stress affects male reproduction in a parasitoid wasp. J. Insect Physiol. 59, 248–254. doi: 10.1016/j.jinsphys.2012.12.001
Nicolson, S. W. (2009). Water homeostasis in bees, with the emphasis on sociality. J. Exp. Biol. 212, 429–434. doi: 10.1242/jeb.022343
Ohashi, M., Okada, R., Kimura, T., and Ikeno, H. (2009). Observation system for the control of the hive environment by the honeybee (Apis Mellifera). Behav. Res. Methods 41, 782–786. doi: 10.3758/BRM.41.3.782
Parcellier, A., Jaattela, M., Larmonier, N., Martin, F., Bruey, J. M., Garrido, C., et al. (2001). Selective depletion of inducible Hsp70 enhances immunogenicity of rat colon cancer cells. Oncogene 20, 7478–7748. doi: 10.1038/sj.onc.1204948
Potts, S. G., Biesmeijer, J. C., Kremen, C., Neumann, P., Schweiger, O., and Kunin, W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353.
Purschke, B., Meinlschmidt, P., Horn, C., Rieder, O., and Henry, J. (2017). Improvement of techno-functional properties of edible insect protein from migratory locust by enzymatic hydrolysis. Eur. Food Res. Technol. 244, 999–1013.
Rameshwor, P., and Resham Bahadur, T. (2014). Foraging behavior of different honeybee species under natural condition in Chitwan. Eur. J. Acad. Essays 1, 39–41.
Rhoads, A. R., and Friedberg, F. (1997). Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340.
Robich, R. M., and Denlinger, D. L. (2005). Diapause in the mosquito Culex pipiens evokes a metabolic switch from blood feeding to sugar gluttony. Proc. Natl. Acad. Sci. U S A. 102, 15912–15917. doi: 10.1073/pnas.0507958102
Rolf, A., Amos, B., Wu, C. H., Barker, W. C., Brigitte, B., Serenella, F., et al. (2017). Uniprot: the universal protein knowledgebase. Nucleic Acids Res. 45, 158–169.
Santos, P. K. F., Natalia, D. S. A., Elaine, F., Rizoo, Z. A., and Cristina, A. M. (2021). Diapause in a tropical oil-collecting bee: molecular basis unveiled by RNA-Seq. BMC Genom. 19:305. doi: 10.1186/s12864-018-4694-x
Scheffer, H., Coate, J. E., Ho, E. K. H., and Schaack, S. (2022). Thermal stress and mutation accumulation increase heat shock protein expression in Daphnia. Evol. Ecol. 36, 829–844. doi: 10.1007/s10682-022-10209-1
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2013). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shaw, W. M., Lou, S., Landis, J., Ashraf, J., and Murphy, C. T. (2007). The C. elegans TGF-Beta dauer pathway regulates longevity via insulin signaling. Curr. Biol. 17, 1635–1645. doi: 10.1016/j.cub.2007.08.058
Shen, H., He, H., Lu, C., Liang, Y., Wu, H., Zheng, L., et al. (2021). Comparative transcriptome analysis of two populations of two populations of Dastarcus helophoroides (Fairmaire) under high temperature stress. Forests 13:13.
Shih, S. R., Bach, D. M., Rondeau, N. C., Sam, J., Lovinger, N. L., Lopatkin, A. J., et al. (2021). Honey bee sHsp are responsive to diverse proteostatic stresses and potentially promising biomarkers of honey bee stress. Sci. Rep. 11:22087. doi: 10.1038/s41598-021-01547-1
Slimen, I. B., Najar, T., Ghram, A., and Abdrrabba, M. (2016). Heat stress effects on livestock: molecular, cellular and metabolic aspects, a review. J. Anim. Physiol. Anim. Nutr. 100, 401–412.
Southwick, E. E., and Heldmaier, G. (1987). Temperature control in honey bee colonies. Bioscience 37, 395–399.
Souza-Junior, J. B. F., Oliveira-Souza, A., Oliveira, P. F., Queiroz, J., and Hrncir, M. (2020). Increasing thermal stress with flight distance in stingless bees (Melipona Subnitida) in the Brazilian tropical dry forest: implications for constraint on foraging range. J. Insect Physiol. 123:104056. doi: 10.1016/j.jinsphys.2020.104056
Stabentheiner, A., and Kovac, H. (2014). Energetic optimisation of foraging honeybees: flexible change of strategies in response to environmental challenges. PLoS One 9:e105432. doi: 10.1371/journal.pone.0105432
Stabentheiner, A., and Kovac, H. (2016). Honeybee economics: optimisation of foraging in a variable world. Sci. Rep. 6:28339. doi: 10.1038/srep28339
Stabentheiner, A., Kovac, H., and Brodschneider, R. (2010). Honeybee colony thermoregulation regulatory mechanisms and contribution of individuals in dependence on age, location and thermal stress. PLoS One 5:e8967. doi: 10.1371/journal.pone.0008967
Su, X. L., Hua, Q. Y., Chen, Y. F., Jiang, X. S., Xi, W. J., Zhao, D. X., et al. (2017). Behavior of Apis cerana cerana and Apis mellifera ligustica as pollinator for long-season cultivated watermelon in tunnel greenhouse under summer high temperature condition. J. Environ. Entomol. 39, 104–110.
Takayama, S., Sato, T., Krajewski, S., Kochel, K., Irie, S., Millan, J. A., et al. (1995). Cloning and functional analysis of BAG-1: a novel BCL-2-binding protein with anti-cell death activity. Cell 80, 279–284. doi: 10.1016/0092-8674(95)90410-7
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/Nar/28.1.33
Tautz, J., Maier, S., Groh, C., Rössler, W., and Brockmann, A. (2003). Behavioral performance in adult honey bees is influenced by the temperature experienced during their pupal development. Proc. Natl. Acad. Sci. 100, 7343–7347.
The Honeybee Genome Sequencing Consortium (2006). Insights into social insects from the genome of the honeybee. Nature 443, 931–949. doi: 10.1038/nature05260
Vatanparast, M., and Park, Y. (2021). Comparative RNA-Seq analyses of Solenopsis japonica (Hymenoptera: Formicidae) reveal gene in response to cold stress. Genes 12:1610. doi: 10.3390/genes12101610
Xu, H., Miao, X., Wang, W., Wang, G., and Li, Y. (2022). Transcriptome analysis reveals the early resistance of zebrafish larvae to oxidative stress. Fish Physiol. Biochem. 48, 1075–1089. doi: 10.1007/S10695-022-01100-5
Yang, L., Tan, G. Y., Fu, Y. Q., Feng, J. H., and Zhang, M. H. (2009). Effects of acute heat stress and subsequent stress removal on function of hepatic mitochondrial respiration, ROS production and lipid peroxidation in broiler chickens. Comp. Biochem. Physiol. 151, 204–208. doi: 10.1016/j.cbpc.2009.10.010
Yu, G., Wang, L. G., Han, Y., and He, Q. (2012). Cluster profiler: an R package for comparing biological themes among gene clusters. J. Integ. Biol. 16, 284–287. doi: 10.1089/Omi.2011.0118
Zhang, X., Duan, Y., Ma, J., Liu, Z., Ding, Y., Xie, H., et al. (2022). Trichloroethylene induces immune renal tubular injury through SIRT 1/HSP 70/TLR 4 Pathway in BALBc mice. Int. Immunopharmacol. 112:109203. doi: 10.1016/j.intimp.2022.109203
Zhao, H., Li, G., Guo, D., Li, H., Liu, Q. X., Xu, B. H., et al. (2021). Response mechanisms to heat stress in bees. Apidologie 52, 388–399.
Keywords: Apis mellifera, temperature, transcriptomics, RNA-seq, gene expression
Citation: Zhang B, Li X, Jiang Y, Liu J, Zhang J and Ma W (2023) Comparative transcriptome analysis of adult worker bees under short-term heat stress. Front. Ecol. Evol. 11:1099015. doi: 10.3389/fevo.2023.1099015
Received: 15 November 2022; Accepted: 06 February 2023;
Published: 22 February 2023.
Edited by:
Xiaobo Wu, Jiangxi Agricultural University, ChinaReviewed by:
Xiangjie Zhu, Fujian Agriculture and Forestry University, ChinaZheguang Lin, Yangzhou University, China
Copyright © 2023 Zhang, Li, Jiang, Liu, Zhang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weihua Ma,  bWF3aDE5OTdAMTYzLmNvbQ==
bWF3aDE5OTdAMTYzLmNvbQ==
 Bing Zhang
Bing Zhang Xinyu Li
Xinyu Li Yusuo Jiang
Yusuo Jiang Jinjia Liu
Jinjia Liu Jiangchao Zhang
Jiangchao Zhang Weihua Ma
Weihua Ma