- 1Department of Botany, St. Petersburg State University, Saint Petersburg, Russia
- 2Laboratory of Biosystematics and Cytology, Komarov Botanical Institute, Russian Academy of Sciences, Saint Petersburg, Russia
- 3Laboratory of Molecular and Ecological Physiology, Komarov Botanical Institute, Russian Academy of Sciences, Saint Petersburg, Russia
- 4Department of Ecology, Environment and Plant Sciences, Stockholm University, Stockholm, Sweden
- 5Department of Artificial Intelligence Technology in Physiology and Medicine, Saint Petersburg Electrotechnical University “LETI”, Saint Petersburg, Russia
Recent advances in plant developmental genetics together with rapid accumulation of transcriptomic data on plants from divergent lineages provide an exciting opportunity to explore the evolution of plant morphology. To understand leaf origin in sporophytes of land plants, we have combined the available molecular and structural data on development of leaves with different morphologies in different plant lineages: clubmosses, spikemosses, leptosporangiate ferns, ophioglossioid ferns, marattioid ferns, whisk ferns, horsetails, and conifers. Specifically, we address the peculiarities of proximo-distal, ad/abaxial, and lateral development; presence/absence of mesophyll differentiation into palisade and spongy parenchyma; and type of leaf vascular bundles (collateral and bicollateral). Furthermore, taxon-specific and morphology-specific features of leaf development are considered in the context of the organization of shoot apical meristems (SAMs)—monoplex, simplex, or duplex—that produce leaf primordia. The data available imply that cellular patterns of leaf initiation correlate strongly with the structure of the SAMs but not with further leaf development or morphology. The later stages of leaf development are neither correlated with SAM structure nor with taxonomy. Occurrence and, if available, patterns of expression of homologs of the angiosperm genes responsible for the development of adaxial (ARP and C3HDZ) and abaxial (YABBY and KANADI) leaf domains, or establishment of the leaf marginal meristem (WOX) are discussed. We show that there is no correlation in the set of homologs of TFs that regulate abaxial and adaxial leaf domain development between leaves containing only spongy and no palisade mesophyll (of spikemosses, clubmosses, whisk ferns, horsetails, and most conifers), and leaves differentiated into palisade and spongy mesophyll (of leptosporangiate ferns, Ginkgo, Gnetum, and angiosperms). Expression of three out of four regulators of leaf development in primordia of both leaves and sporangia—C3HDZ in spikemosses and whisk ferns, YABBY in clubmosses and KANADI in spikemosses and horsetails—indicates that a sporangium developmental program could have been co-opted as a “precursor program” for the origin of microphylls and euphylls. Additionally, expression of leaf development regulators in SAMs of spikemosses (ARP, C3HDZ, and KANADI), clubmosses (YABBY), leptosporangiate ferns (C3HDZ), and horsetails (C3HDZ and KANADI) indicates that at least some mechanisms of SAM regulation were co-opted as well in the pre-program of leaf precursors.
Introduction
Colonization of land by plants in the middle Cambrian—Early Ordovician (Morris et al., 2018) has dramatically changed the ecology of Earth. Although the haploid phase (gametophyte) dominated in the life cycle of the first land plants, it was the transition to the dominance of the diploid phase (sporophyte) in Late Silurian—Early Devonian that led to numerous morphological and reproductive innovations in the plants of the sporophytic evolutionary line (polysporangiophytes; Bowman et al., 2017). One of the most influential morphological innovations was the origin of specialized dorsoventral organs, leaves, that significantly increased the plants’ photosynthetic surface. However, how leaves came into being remains enigmatic (Tomescu, 2009; Harrison and Morris, 2018; Vasco and Ambrose, 2020).
Discussions addressing this question over more than a century resulted in three main scenarios: (1) leaves evolved once in the common ancestor of all tracheophytes (Kaplan, 2001; Schneider et al., 2002); (2) two independent evolutionary events resulted in so-called microphylls in lycophytes and so-called megaphylls in all other vascular plants (Bower, 1935); and (3) leaves evolved more than twice, with the exact number of independent origins subject to debate (Stewart and Rothwell, 1993; Kenrick and Crane, 1997; Boyce, 2010). Three putative mechanisms explaining the origin of leaves are recognized. (1) Gradual elaboration of stem outgrowths of early land plants (e.g., in Drepanophycales) that resulted in lycophyte microphylls (Bower, 1935). (2) A hypothetical series of successive transformations of early land plant axes or telomes (known as Telome Theory): unequal branching (”overtopping”; e.g., in Renalia and Pertica), transition of determinate lateral branching systems from three-dimensional to two-dimensional development (“planation”; e.g., in Eospermatopteris), and the development of lateral outgrowths of photosynthetic tissue filling the space between planar determinate systems of branching axes (”webbing”; e.g., in Rhacophyton and Archaeopteris; Zimmerman, 1952; Kenrick and Crane, 1997; Harrison and Morris, 2018). (3) Sterilization of some lateral branches that originally bore sporangia (e.g., in Adoketophyton; Kenrick and Crane, 1997). As modifications of branching systems were likely independently involved in the evolution of land plant sporophytes, it was assumed that the leaves of ferns and seed plants have independent origins, while the homology of the leaves of horsetails (Equisetidae), whisk ferns (Psilotidae), ophioglossoid ferns (Ophioglossidae), marattioid ferns (Marattiidae), and leptosporangiate ferns (Polypodiidae) remains under debate (Tomescu, 2009). The ambiguous homology of leaves is reflected by the terms “lycophyte leaves,” “fern leaves,” “horsetail leaves,” etc. (Tomescu, 2009; Vasco and Ambrose, 2020), and we will maintain this terminology. Years of research have shown that no morphological or anatomical criteria alone can be used to distinguish unequivocally between a common origin vs. several independent evolutionary origins of leaves.
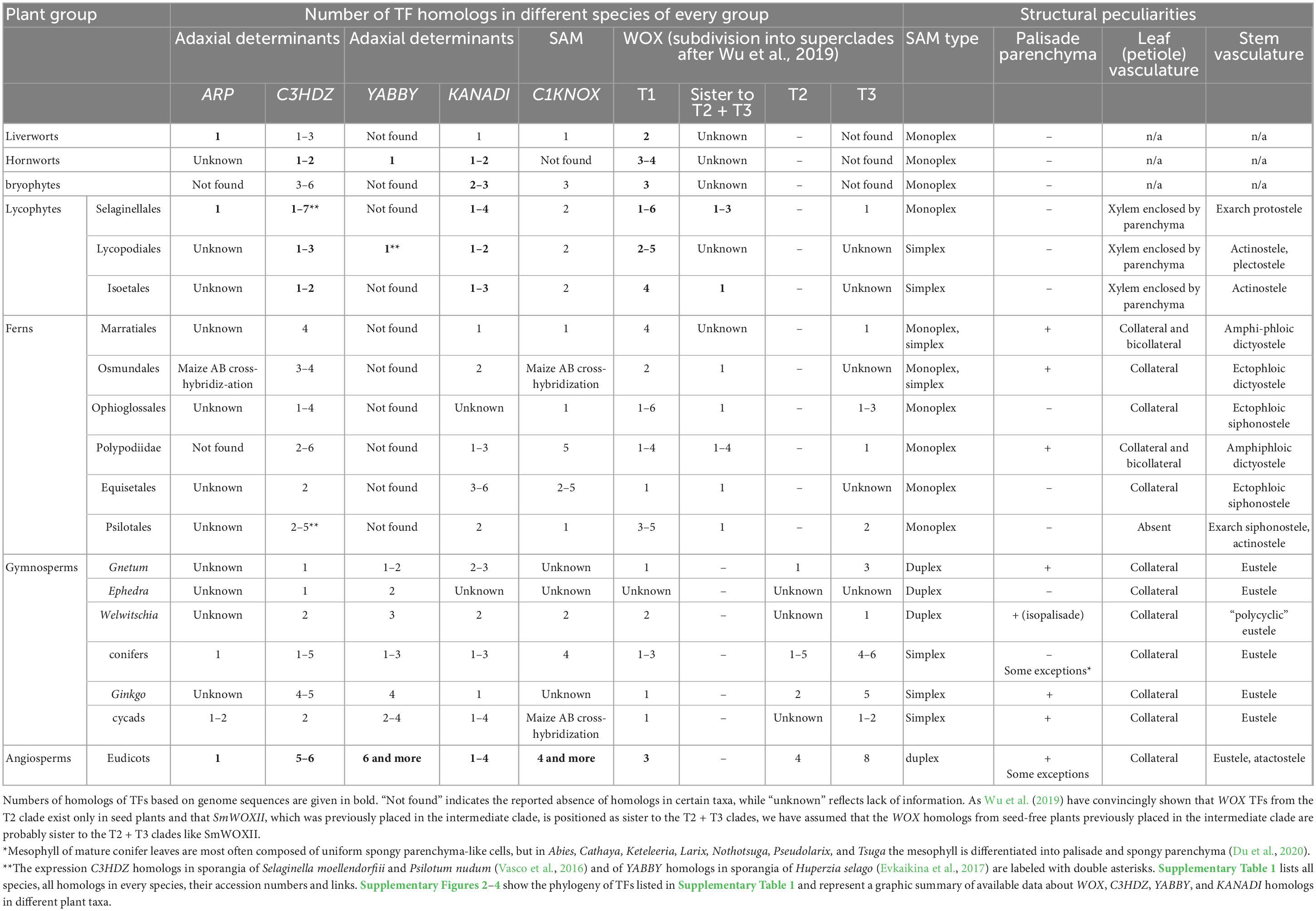
Since all leaves originate from the shoot apical meristem (SAM), the precisely organized pool of undifferentiated proliferating cells on the shoot tips that manages shoot growth and is the origin of lateral organs, it was expected that differences in leaf evolutionary origin would correlate with differences in SAM structure and/or functions. Studies of the diversity of SAMs in land plants resulted in the development of several classifications based on SAM structure (reviewed by Tooke and Battey, 2003). The classification that arose from the studies of Popham (1951), Newman (1965), and Philipson (1990) is best fitted to the evolutionary context. It specifies three structural types of SAMs in land plants according to the number, location and division pattern of the apical initials (AIs; Newman, 1965). Monoplex SAMs contain a single tetrahedral AI located in the surface layer that is the ultimate source of all other cells of the shoot apex (Figure 1A). Simplex SAMs contain several AIs located in the surface layer (Figure 1B). Duplex SAMs contain AIs in several outermost layers of the SAM that belong to two different zones, the tunica specified by anticlinal divisions, and the corpus (Figure 1C). The number of AIs in the SAM is likely correlated with the mechanism of plasmodesmata formation: exclusively during cytokinesis in monoplex SAMs (Figure 1D), and both during and after cytokinesis via local digestion of the cell walls in simplex and duplex SAMs (Figures 1E, F; Imaichi and Hiratsuka, 2007; Evkaikina et al., 2017). However, all three SAM types are likely paraphyletic, and no correlation between SAM structure and the evolutionary origin of leaves produced by a SAM could be found. For instance, monoplex SAMs are found in spikemosses (Selaginellales) while simplex SAMs occur in clubmosses (Lycopodiales) and quillworts (Isoetales), i.e., the leaves of lycophytes are produced by two different SAM types [Gifford and Foster, 1989; Maksimova (Evkaikina) et al., 2021]. On the rather hand, monoplex SAMs give rise to both “microphylls” of spikemosses and “euphylls” of most ferns. However it should be mentioned that some ferns have SAMs with multiple apical initials (Tomescu, 2011, Tomescu and Matsunaga, 2019).
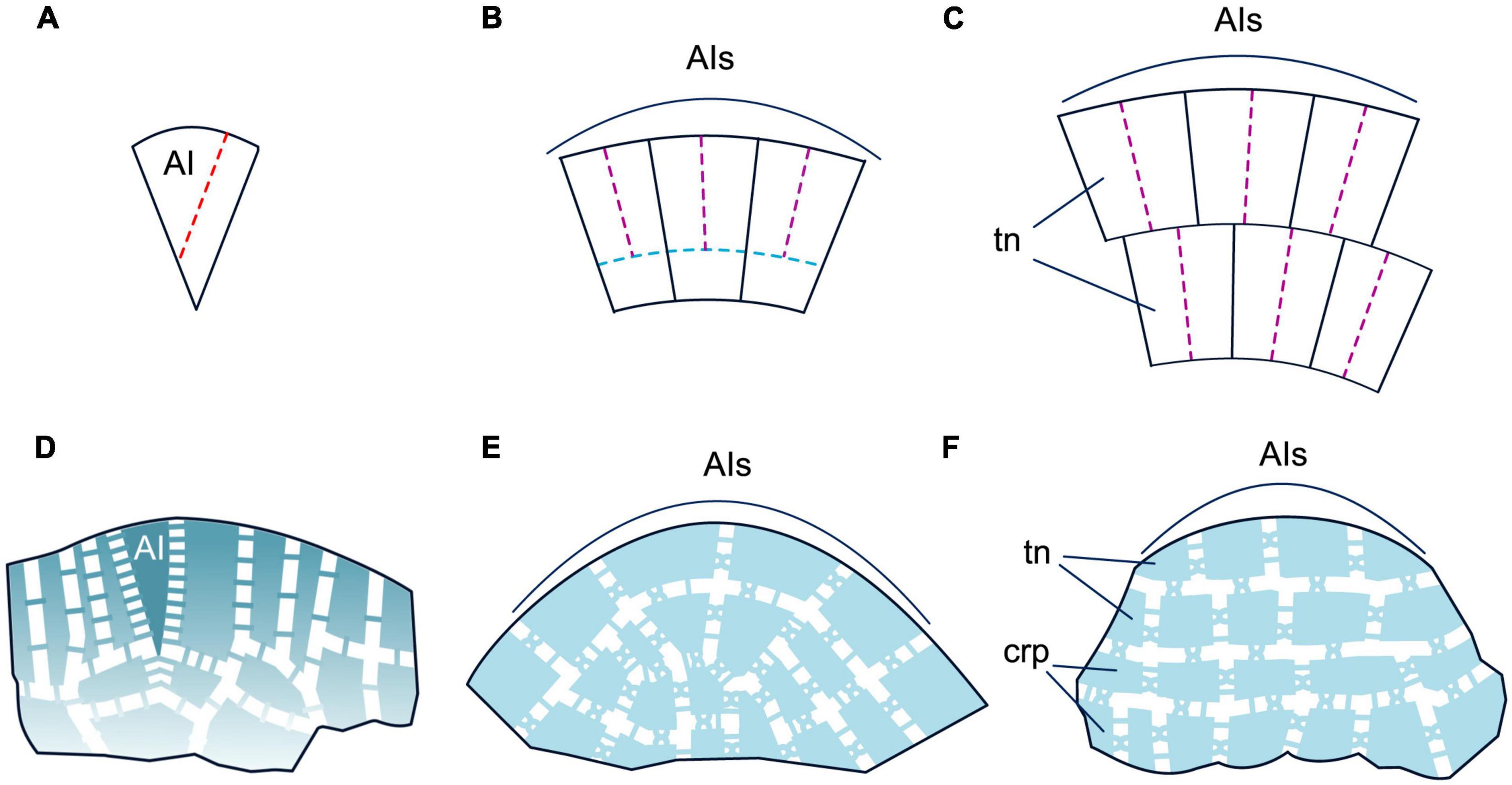
Figure 1. Patterns of cell division and symplastic organization of different types of the shoot apical meristem (SAM) according to Newman (1965), Imaichi and Hiratsuka (2007). (A,D) Monoplex SAM, (B,E) simplex SAM, (C,F) duplex SAM. The colors of dotted lines in panels (A–C) lines denote different planes of cell division: red—oblique anticlinal, purple—anticlinal, blue—periclinal. Plasmodesmata (PDs) formed during cytokinesis are unbranched in outline, while post-cytokinesis PDs are X-shaped in outline. PD density is depicted with a color gradient, where fading color in the peripheral zone (PZ) of the SAM stands for decrease in PD density, and absence of fading for uniform PD density in the central zone (CZ) and PZ.
It is known that leaf development results from cell proliferation localized at certain developmental stages in different parts of growing leaves; these sites of meristematic activity are collectively termed leaf meristems (Esau, 1965; Tsukaya, 2021). Four meristems are usually identified in growing leaves: leaf apical meristem, marginal meristem, intercalary meristem and plate meristem. The term “leaf apical meristem” refers to cell proliferation at the leaf tip that is usually activated at the initial stage of leaf development, while “marginal meristem” refers to the proliferation of cells at the leaf perimeter. Unlike these two meristems, intercalary and plate leaf meristems are rather diffuse; moreover these two terms refer to different planes of division of the same cells. Anticlinal mediolateral divisions of internal cells of the growing leaf blade along the medio-lateral axis (i.e., growth in “width”) are collectively referred to as the plate meristem, while anticlinal proximodistal divisions of these cells that provide growth in “height” are collectively referred to as the intercalary meristem (Esau, 1965; Steeves and Sussex, 1989; Tsukaya, 2021). However, only a few studies have provided structural data about different leaf meristems and their regulation outside angiosperms (e.g., Nardmann and Werr, 2013; Vasco and Ambrose, 2020).
Below we will summarize the cellular and molecular aspects of SAM functions and of leaf development in extant polysporangiophytes. Only genetic regulators that can be discussed in the evolutionary context will be mentioned. For the sake of clarity, for angiosperms we will consider only development of simple leaves of eudicots; the function and interaction of genetic regulators will be described only for Arabidopsis, the most comprehensively studied plant with this leaf morphology. However, much variation in leaf structure, development and regulation exists within angiosperms. For leaf anatomy and development in eudicots and monocots with different morphologies, the reader is referred to Esau (1965) and Steeves and Sussex (1989) and for the diversity of regulation to Tsukaya (2018) and Nakayama et al. (2022). Due to the continuous update of transcriptomics data in public databases, we have added new homologs of the transcription factors under discussion (Supplementary Figures 1–4, Supplementary Table 1, and Supplementary File 1).
Duplex SAMs producing angiosperm leaves
Shoot apical meristems in angiosperms are composed of one to several layers of tunica cells and of underlying corpus cells. The SAMs consist of a central zone (CZ) composed of large, slowly proliferating cells, and a peripheral zone (PZ) consisting of smaller and more quickly proliferating cells (Gifford and Foster, 1989; Figures 2A, E). Molecular genetic studies reveal heterogeneity within the CZ (Figure 2B). Typically, the three outermost layers of the CZ (i.e., two tunica layers and the outermost corpus layer) are marked by the expression of CLAVATA3 (CLV3) and considered true AIs (Fletcher et al., 1999). The subtending CZ cells of the corpus are marked by the expression of the WUSCHEL (WUS) homeobox gene and constitute the organizing center (OC) (Mayer et al., 1998). A negative feedback loop between the CLV3 peptide and the WUS TF, mediated by two more CLV proteins, a receptor-like kinase and a receptor that act together as a heterodimer, maintains a dynamic equilibrium of AIs and OC cells and is essential for maintenance of SAM size (Brand et al., 2000; Schoof et al., 2000; Nimchuk et al., 2011). Another, presumably ancestral, CLE-40 signaling system was shown to have stem-cell-promoting function (Hirakawa, 2022). In both CZ and PZ, the key regulators of the undifferentiated state are transcription factors (TFs) encoded by class I KNOTTED (C1KNOX) genes (Jackson et al., 1994; Figure 2C).
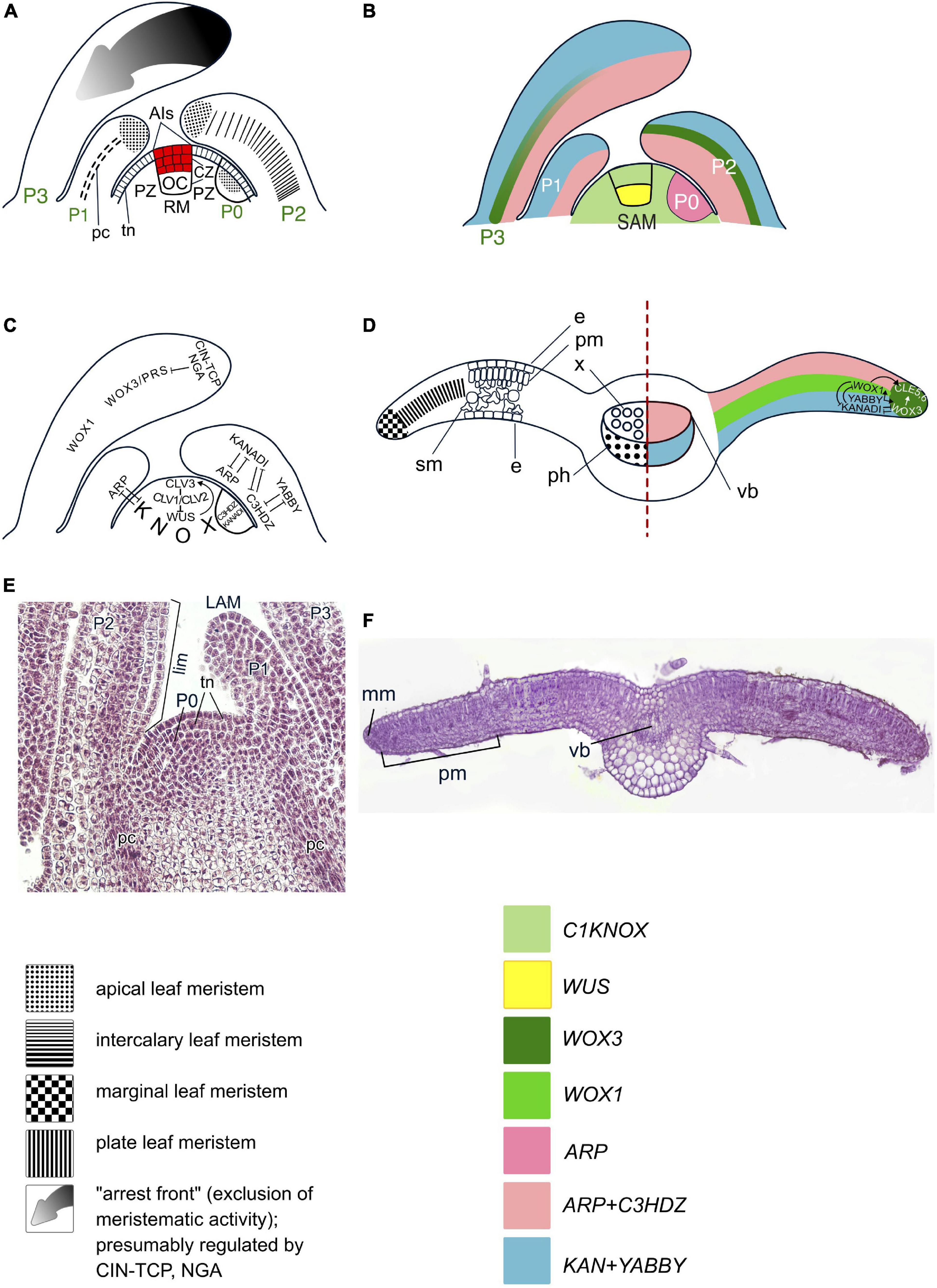
Figure 2. Organogenesis in the duplex SAM of eudicot angiosperms. Line drawings of longitudinal sections through the shoot apex (A–C) and a transversal section of a leaf (D); micrographs of a longitudinal section of the SAM of Syringa vulgaris (E) and a transversal section of a young leaf of Nicotiana tabacum (F). SAM structure, cellular pattern of leaf origin and contribution of different leaf meristems to its development are based on Esau (1965), Steeves and Sussex (1989), Tsukaya (2021); patterns of gene expression are based on in situ RNA–RNA hybridizations reviewed in Nakata et al. (2012), Nardmann and Werr (2013) and Shi and Vernoux (2019). Although procambium strands are mapped only in P1, they are present in all leaf primordia and in young leaves. Apical initials (AIs) are marked in red; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. SAM, shoot apical meristem; AI(s), apical initial(s); OC, organizing center; CZ, central zone; PZ, peripheral zone; P0–P3, successive stages of leaf primordia and leaf development; e, epidermis; pm, palisade mesophyll; sm, spongy mesophyll; vb, vascular bundle; LAM, leaf apical meristem; lim, leaf intercalary meristem; mm, leaf marginal meristem; plm, leaf plate meristem. Putative interactions between the regulators of SAM maintenance and leaf development described in the text are mapped on panels (C,D); the arrows indicate positive regulation, while bars show negative regulation.
Leaves originate in the PZ of the duplex SAM (Esau, 1965; Steeves and Sussex, 1989) after a switch between meristem-specific and leaf-specific programs in a group of initially morphologically indistinguishable cells designated as incipient leaf primordium (LP) (Figures 2A, E). The switch is caused by suppression of C1KNOX expression by an increase in local auxin maxima in the leaf founder cells (Jackson et al., 1994; Long et al., 1996; Reinhardt et al., 2003) followed by upregulation of the expression of the leaf development regulators that in angiosperms comprise “adaxial determinants” and “abaxial determinants” (Yamaguchi et al., 2012; Du et al., 2018). Adaxial determinants regulate the differentiation of palisade parenchyma and upper epidermis in leaves, and of the xylem in the leaf bundles. They are encoded by the ARP genes (ASYMMETRIC LEAVES1, ROUGH SHEATH2 and PHANTASTICA) (Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Guo et al., 2008) and C3HDZ genes (class III HD-Zip) (McConnell et al., 2001; Emery et al., 2003). Abaxial determinants control the differentiation of spongy parenchyma and lower epidermis in leaves and of the phloem in the leaf bundles. They are encoded by the YABBY (Bowman and Smyth, 1999; Sawa et al., 1999; Siegfried et al., 1999) and KANADI (KAN) genes (Eshed et al., 2004; Table 1 and Figures 2B–D). After the onset of the leaf-development program, proliferation of leaf founder cells occurs by anti- and periclinal divisions of the second tunica layer, L2, and of the outermost corpus cells, L3, accompanied by anticlinal divisions of L1, and results in the emergence of a radially symmetric LP (Esau, 1965). The prepatterned expression of the adaxial–abaxial polarity genes C3HDZ and KANADI at the PZ occurs prior to LP formation and persists in incipient LPs (Figure 2C; Caggiano et al., 2017; Yu et al., 2017).
After the onset of LP development, transcription of ARP and C3HDZ genes is confined to the adaxial, whilst that of YABBY and KANADI genes is confined to the abaxial domain of the LP (Yamaguchi et al., 2012; Du et al., 2018). At their juxtaposition along the lateral margins of the LP, YABBY up-regulates the expression of WUSCHEL–RELATED HOMEOBOX (WOX) 1 and WOX3 genes (Figure 2D; Nakata et al., 2012). WOX1 and WOX3 activate two CLAVATA3/ESR–RELATED peptide ligand gene family members, CLE5 and CLE6, and altogether establish a new marginal meristem (DiGennaro et al., 2018). Thus, the meristematic activity in the LP becomes restricted to the marginal regions (Alvarez et al., 2016). While the restriction of the broadly acting leaf meristem to the margins is maintained by WOX1 and WOX3, the cessation of proximodistal lamina growth is mediated by the NGA and CIN-TCP TFs (Figure 2C; Alvarez et al., 2016). During LP transition from radial to dorsoventral symmetry, anticlinal divisions of surface marginal initials produce the leaf epidermis, while both anti- and periclinal divisions of subsurface marginal initials produce the leaf mesophyll. Once the required number of mesophyll layers has been laid down, the activity of the marginal meristem ceases (Esau, 1965).
The further growth of leaf blade along the medio-lateral axis proceeds via anticlinal divisions of the plate meristem (Figures 2D, F; Esau, 1965; Steeves and Sussex, 1989; Tsukaya, 2021). WOX1 is thought to be its key regulator (Nardmann and Werr, 2013; Tsukaya, 2021). Further leaf development is regulated by abaxial and adaxial determinants in concert with auxin and cytokinin (Sarojam et al., 2010; Yamaguchi et al., 2012; Sarvepalli et al., 2019; Satterlee and Scanlon, 2019; Ali et al., 2020). The collateral symmetry of leaf vascular bundles results from the specific pattern of procambium differentiation and activities of the adaxial and abaxial determinants together with the levels of auxin and cytokinin in the precursors of the phloem and the xylem (Yang and Wang, 2016; Heisler and Byrne, 2020).
Leaf growth along the proximodistal axis results from the activity of the intercalary meristem (Figure 2A; Esau, 1965). The limited leaf height growth results from the decline of cell proliferation of its distal-most cells, termed the “arrest front” (Satterlee and Scanlon, 2019; Tsukaya, 2021) where expression of the C1KNOX, WOX1, and WOX3 genes is gradually repressed (Figure 2C; Manuela and Xu, 2020).
Duplex SAMs producing gnetophyte leaves
Structures of gnetophyte SAMs are similar to that of angiosperm SAMs (Gifford and Foster, 1989; Figures 3A, F). The available data suggest that at least some regulatory aspects are similar to those in angiosperms. An immunolocalization study using an antibody raised against KN1 (C1KNOX) from Zea mays yielded a signal in Welwitschia mirabilis SAMs which was absent from LPs (Pham and Sinha, 2003).
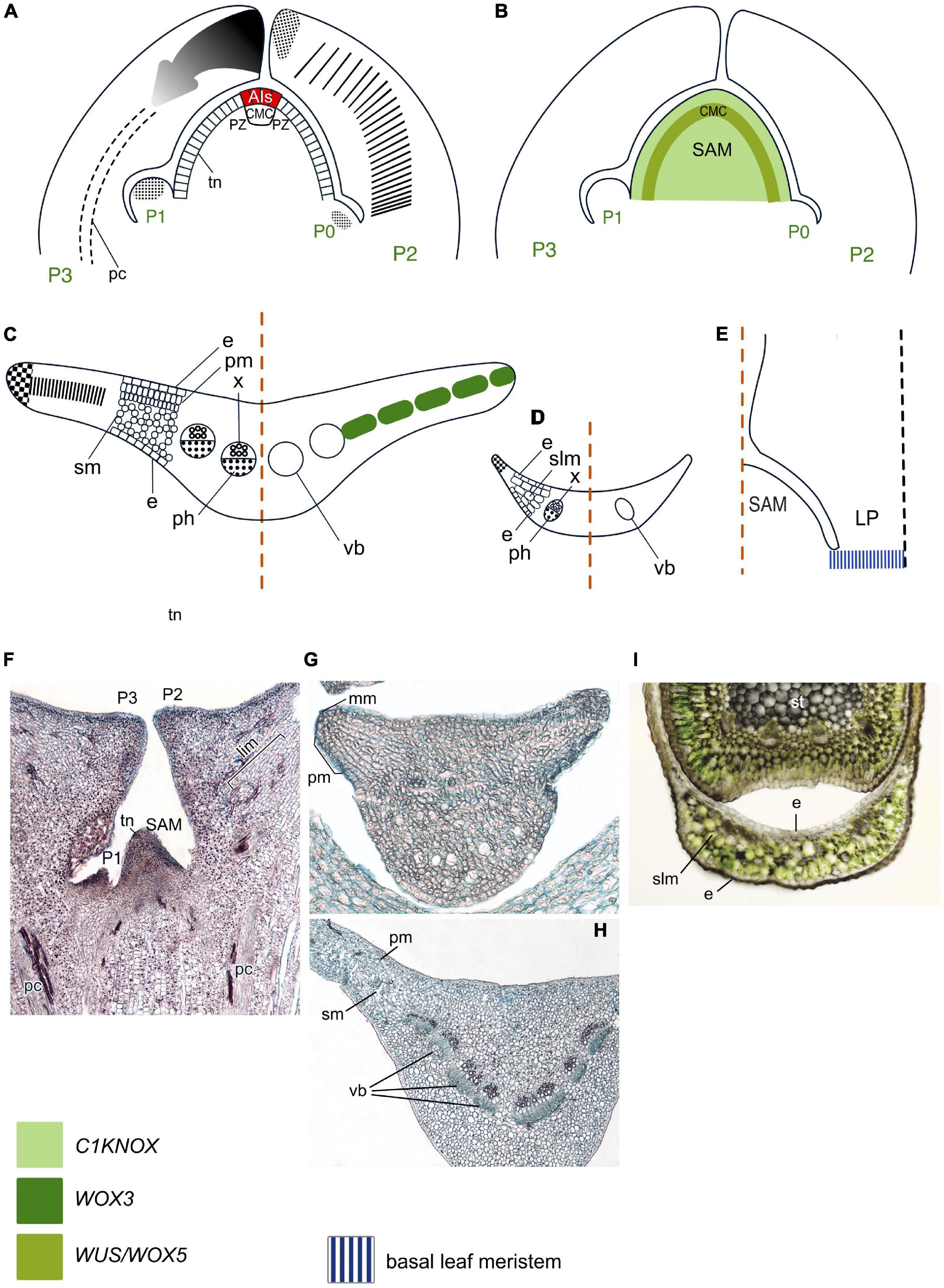
Figure 3. Organogenesis in the duplex SAM of gnetophytes. Line drawings of longitudinal sections through the shoot apex (A,B) and transversal sections of Gnetum (C) and Ephedra (D) leaves and a longitudinal section through the base of a young Welwitschia leaf (E); micrographs of longitudinal section of the SAM of Gnetum gnemon (F) and transversal section of young (G) and mature (H) leaves of G. gnemon and Ephedra sinica (I). SAM structure is based on Gifford and Foster (1989), cellular patterns of leaf origin and contribution of different leaf meristems to its development are inferred from descriptions and illustrations in Rodin (1958a,b, 1967), Nardmann and Werr (2013), Wan et al. (2018) for Gnetum; Dörken (2014), San Martin et al. (2022) for Ephedra; Rodin (1958a,b) for Welwitschia. Patterns of gene expression are based on in situ RNA-RNA hybridizations and immunolocalizations from Nardmann et al. (2009), Nardmann and Werr (2013), Wan et al. (2018), Bueno et al. (2021) for Gnetum; Finet et al. (2016) for Ephedra; Pham and Sinha (2003) for Welwitschia. Apical initials (AIs) are marked in red; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. LP, leaf primordium; CMC, central mother cells; slm, spongy-like mesophyll; other captions and symbols as in Figure 2. Panels (C,G,H) contributed by Ianina Bogdanova.
The major developmental regulators, WOX proteins, form three clades: T1 (previously called the ancient clade) (sensu Hedman et al., 2013; Alvarez et al., 2018; Bueno et al., 2021), T2 (former intermediate clade), and T3 (or WUS/WOX5) clade that were shown to be equally ancient (Wu et al., 2019). A number of WOX proteins from all these clades was found in gnetophytes (Nardmann et al., 2009; Nardmann and Werr, 2013; Bueno et al., 2021; Table 1; Supplementary Figure 1; Supplementary Table 1). A single representative of the T3 clade (GgWUS/WOX5) is expressed in the PZ of the vegetative SAM (Figure 3B) and in the SAMs of male cones of Gnetum gnemon (Nardmann et al., 2009), while in angiosperms, WOX5 is expressed only in the root apical meristem (RAM). Two other genes from the T3 clade of the WOX gene family found in G. gnemon, GgWOXX and GgWOXY, encode proteins lacking homologs in other seed plant genomes, suggesting that they represent ancestral sequences that might be have been lost in other seed plant lineages (Nardmann and Werr, 2013; Wan et al., 2018).
In spite of similar structure of the SAMs in all three genera of gnetophytes, their leaf development differs. Leaves of Gnetum species resemble those of many angiosperms in possessing a broad lamina whose development along the mediolateral axis involves activities of both marginal and plate meristems, while it develops along the proximodistal axis via an intercalary meristem (Figures 3A, C, G, H; Rodin, 1967; Boyce and Knoll, 2002; Tomlinson and Fisher, 2005). No homologs of the marginal and plate meristem regulator WOX1 have been detected in G. gnemon (Nardmann and Werr, 2013; Wan et al., 2018), although expression of its counterpart, a WOX3 homolog, was detected in incipient LPs and along the margins of young leaves (Nardmann et al., 2009). In older LPs, the expression of GgWOX3 marks a stripe between blade margins, resembling the expression pattern of both WOX3 and WOX1 angiosperm homologs and likely regulating both marginal and plate meristems (Wan et al., 2018). Transcriptomic analyses detected C3HDZ, KANADI and YABBY homologs in Gnetum species (Table 1, Supplementary Figures 2–4, and Supplementary Table 1).
Leaves of Ephedra species are reduced to scale-like structures lacking differentiation between palisade and spongy mesophyll (Dörken, 2013, 2014; San Martin et al., 2022; Figures 3D, I). Two YABBY homologs (EdiYABD and EdiYABD) were found in Ephedra distachya, both expressed in the outer layer of the ovule integument in a pattern similar to angiosperm “reproductive” YABBY genes, and in the cone bracts where one of them, EdiYABD, showed a higher expression level on the abaxial side, similar to “vegetative YABBYs” in leaves of angiosperms (Finet et al., 2016). Transcriptomic analyses detected C3HDZ homologs in E. sinica (Table 1, Supplementary Figure 2, and Supplementary Table 1).
Welwitschia mirabilis bears only a single pair of linear isopalisade leaves that develop via a basal meristem (Figure 3E; Rodin, 1958a,b). The fact that immunolocalization with an antibody against Zea mays KN1 (C1KNOX) protein failed to yield a signal in W. mirabilis LPs (Pham and Sinha, 2003) suggests that downregulation of C1KNOX expression is part of the LP initiation program. Obviously, functions of the basal meristem that provides leaf development in Welwitschia are regulated by other, yet unknown proteins. C3HDZ, KANADI, and YABBY homologs were identified in the W. mirabilis transcriptome (Table 1, Supplementary Figures 2–4, and Supplementary Table 1).
No ARP homologs were found in Gnetum, Ephedra, and Welwitschia species (Table 1).
Simplex SAMs producing leaves of conifers, cycads, and Ginkgo
Similar to angiosperms and gnetophytes, the simplex SAMs of conifers, cycads, and Ginkgo biloba can be divided into the CZ and PZ based on differences in cell size and vacuolation. CZ is partitioned into prismatic AIs in the outermost layer and the subtending polygonal central mother cells (Foster, 1938, 1939, 1943). At the periphery and beneath the CZ, respectively, PZ and rib meristem are located (Figures 4A, F).
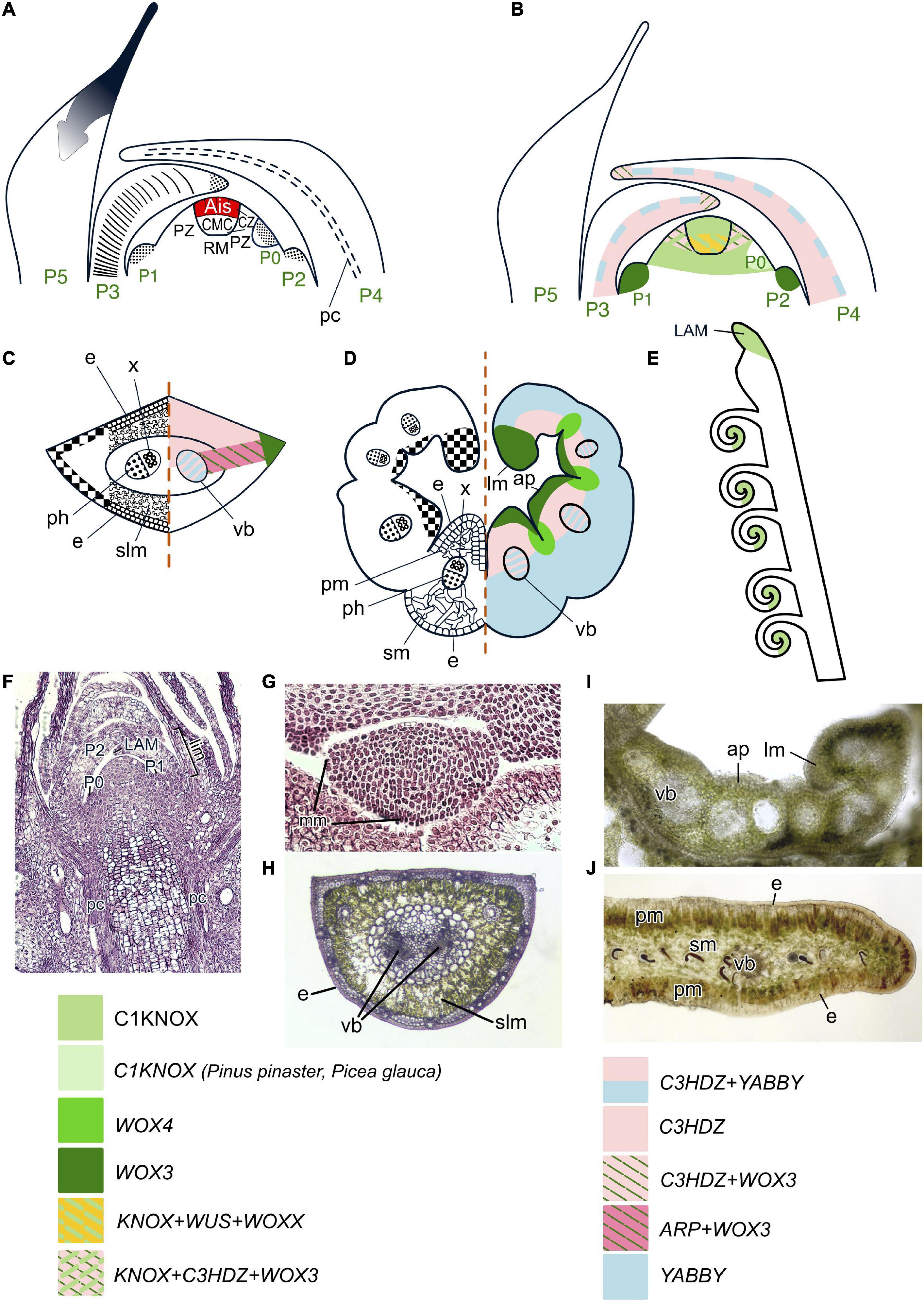
Figure 4. Organogenesis in the simplex SAM of gymnosperms. Line drawings of longitudinal sections through the shoot apex (A,B), transversal sections of leaves of conifers (C) and Ginkgo (D) and schematic of the tip of a young cycad leaf (E); micrographs of longitudinal section of the SAM of Picea abies (F) and transversal sections of young and mature leaves of P. abies (G,H), a young leaf of Ginkgo biloba (I) and a mature leaflet of Cycas revoluta (J). SAM structure is based on Foster (1938, 1939, 1943), cellular pattern of leaf origin and contribution of different leaf meristems to its development are inferred from descriptions and illustrations in Napp-Zinn (1966), Owens (1968), Skupchenko and Ladanova (1984) for conifers, Hara (1980) for Ginkgo; Stevenson (1990, 2020), Boyce and Knoll (2002), Vovides et al. (2020) for cycads. Patterns of gene expression are based on in situ RNA–RNA hybridizations and immunolocalization from Sundås-Larsson et al. (1998), Hjortswang et al. (2002), Floyd et al. (2006), Prigge and Clark (2006), Nardmann and Werr (2013), Alvarez et al. (2015), Finet et al. (2016), Du et al. (2020), Bueno et al. (2021) for conifers; Floyd et al. (2006), Nardmann and Werr (2013), Finet et al. (2016), Zumajo-Cardona et al. (2021) for Ginkgo; Bharathan et al. (2002) for cycads. Apical initials (AIs) are marked in red; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. RM, rib meristem; ap, adaxial protuberance; lm, leaf margin; other captions and symbols as in Figures 2, 3.
Four C1KNOX genes (designated either HBK1–4 or KN1–4) were identified in several spruce and pine species, and one of them, KN3, is phylogenetically very close to AtSTM (Bueno et al., 2020). Two C1KNOX genes are expressed in both CZ and PZ of SAMs of Picea abies (Figure 4B; Sundås-Larsson et al., 1998; Hjortswang et al., 2002) and three in Pinus pinaster (Larsson et al., 2012; Bueno et al., 2020), respectively. An immunolocalization study using an antibody raised against Zea mays KN1 (C1KNOX) protein yielded a signal in the SAM of Zamia integrifolia (Bharathan et al., 2002), suggesting that the key role of C1KNOX TFs in meristem maintenance is shared among simplex SAMs of gymnosperms and duplex SAMs of angiosperms. Several WOX proteins from conifers, cycads and G. biloba belong to the T3 clade (one of them, WOXX, is gymnosperm-specific), or to the T2 and T1 clade (Nardmann and Werr, 2013; Alvarez et al., 2018; Bueno et al., 2021; Table 1; Supplementary Figure 1; Supplementary Table 1). Since conifer WUS and WOXX genes are expressed in cells corresponding to the position of the angiosperm OC (Figure 4B), they are likely to play a similar role in SAM regulation (Bueno et al., 2021). A specific feature of gymnosperm SAMs that is shared with gnetophytes, but different from angiosperms, is its regulatory similarity to root apical meristems (RAMs). Unlike angiosperm C1KNOX genes that are transcribed only in the SAM, all Picea abies C1KNOX homologs are expressed in both SAM and RAM (Hjortswang et al., 2002; Larsson et al., 2012). A number of WUS/WOX5 homologs are as well expressed both in SAM and RAM of Pinus sylvestris, Picea abies, and Ginkgo biloba (Hedman et al., 2013; Alvarez et al., 2018; Bueno et al., 2021).
As in angiosperms and gnetophytes, leaves of non-gnetophyte gymnosperms originate in the PZ of the SAM. LP development starts from enlargement and subsequent periclinal divisions of a group of subsurface and surface cells (Foster, 1938, 1939; Napp-Zinn, 1966; Owens, 1968; Skupchenko and Ladanova, 1984; Skupchenko, 2019). As morphology and development of leaves in conifers, Ginkgo, and cycads differ, the regulation of their leaves development will be discussed separately.
Similar to angiosperms, expression of CIKNOX homologs (HBK2 and HBK4) is downregulated in incipient and developing LPs of P. abies (Larsson et al., 2012); however, they are expressed in leaves of Pinus pinaster and Picea glauca (Bueno et al., 2020). LP development in conifers starts with the proliferation of leaf AIs (Figures 4A, F). Divisions of internal LP cells in the abaxial/adaxial plane sets the number of mesophyll layers, while diffuse proliferation of internal LP cells in the proximodistal plane provides length growth. Then the marginal meristem is activated over the entire circumference of the LP (Figures 4C, G). The activity of the marginal meristem is very short and no plate meristem could be identified (Figure 4H; Owens, 1968; Skupchenko and Ladanova, 1984). The presence of a WOX3 homolog in Pinus sylvestris and P. abies and its expression at leaf margins suggests regulatory similarity between marginal meristems of conifers and angiosperms (Nardmann and Werr, 2013; Alvarez et al., 2015), while the absence of WOX1 expression in P. sylvestris provides molecular support for the absence of a plate meristem (Nardmann and Werr, 2013). On the other hand, WOX3 expression in the PZ of the SAM, in incipient LPs and in the tips of early stage LPs (Nardmann et al., 2009; Hedman et al., 2013; Alvarez et al., 2018; Bueno et al., 2020, 2021) indicates that in conifers it might regulate not only the marginal meristem, but also leaf initiation and leaf apical growth.
A single ARP homolog exists in most of the Pinaceae species (Table 1) and is expressed in young leaves at the juxtaposition between the adaxial and abaxial leaf domains (Figure 4C) in Abies holophylla and Picea smithiana (Du et al., 2020). Thus, ARP TFs seem to be involved in the regulation of leaf development in gymnosperms as well as in angiosperms.
Homologs of adaxial determinants—GymC3HDZ_A to GymC3HDZ_D—were identified in a number of conifers (Floyd et al., 2006; Prigge and Clark, 2006; Du et al., 2020). They are expressed in the PZ of SAMs, in the tips of early stage LPs, adaxial domains of young leaves and the procambium in Pseudotsuga menziesii (Floyd et al., 2006), Abies holophylla and Picea smithiana (Du et al., 2020). Similar expression patterns of C3HDZs in conifers and angiosperms suggest similar functions in regulation of SAM maintenance, leaf development and differentiation of the leaf adaxial domain and of the procambium (Figures 4B, C).
Four clades of YABBY genes (A–D), which encode abaxial determinants, have been identified in conifers (Finet et al., 2016). Unlike in angiosperms and Ginkgoales, where YABBY expression is restricted to the abaxial domain, in conifers with mesophyll only consisting of spongy parenchyma, Pseudotsuga menziesii (Finet et al., 2016) and Picea smithiana, as well as in a species with palisade and spongy mesophyll, Abies holophylla (Du et al., 2020), a YABBY homolog PmeYABC is expressed throughout LPs and in the provascular tissues of young leaves (Figures 4B, C). Homologs of the other abaxial determinant, KANADI, were identified in a number of gymnosperms (Zumajo-Cardona et al., 2019; Table 1; Supplementary Figure 4; Supplementary Table 1), but their expression patterns have not yet been examined.
Ginkgo biloba leaves, similar to angiosperm leaves, possess a broad lamina with the mesophyll differentiated into spongy and palisade parenchyma (see Critchfield, 1970; Hara, 1980 for details). Cells of developing LPs proliferate very unevenly leading to the folding of future leaf blade edges inside and the emergence of adaxial protuberances that split during unfolding of the leaf blade (Figures 4D, I). The discrete sites of meristematic activity are first located along the adaxial surface of leaf margins, but after unfolding of the leaf blade its development in the mediolateral and proximodistal planes proceeds exclusively via proliferation of the ribbon-like marginal meristem. Identification of WOX3 homologs in the genome of G. biloba (Nardmann and Werr, 2013) suggests that regulation of marginal meristems is similar in Ginkgo and seed plants. Two Ginkgo WOX3 homologs are expressed in different parts of the developing leaf lamina: GbWOX3A marks the growing apical part of protuberances, whereas GbWOX3B is expressed at the base of the median furrow or between future protuberances (Nardmann and Werr, 2013). Likely GbWOX3A upregulates cell proliferation, while GbWOX3B suppresses it. Localization of GbWOX3A in the apical part of protuberances supports that most cell proliferation is located at the distal margin of the leaf, contrarily to angiosperms where meristematic activity is first excluded from the distal part (Tsukaya, 2018).
No ARP homologs have been identified in G. biloba thus far (Table 1), but transcriptomics revealed homologs of the other adaxial determinant, C3HDZ (Floyd et al., 2006; Prigge and Clark, 2006). Similar to its counterpart in conifers, GbC3HDZ1 is expressed in the PZ of the SAM, with stronger signal in the incipient LPs, in the tips of young LPs, in the leaf adaxial domain and procambium of the leaf trace (Floyd et al., 2006; Zumajo-Cardona et al., 2021). These data suggest involvement of C3HDZ in regulation of SAM maintenance, leaf development and differentiation of the leaf adaxial domain. However, the similarity of C3HDZ expression patterns in conifer leaves with only spongy mesophyll (Du et al., 2020) and in Ginkgo leaves containing both palisade and spongy mesophyll (Nardmann and Werr, 2013) supports their role in the regulation of ab/adaxial polarity in vascular bundles (which are collateral in both taxa) and not in mesophyll patterning (Table 1 and Figure 4D).
Ginkgo biloba homologs of abaxial determinants, YABBY (GbiYAB_A – GbiYAB_C) are expressed similarly to the “vegetative” YABBYs in angiosperms in abaxial domains of LPs and young leaves (Finet et al., 2016). A KANADI homolog, GibiKAN, is uniformly expressed in all cells of LPs and leaves throughout their development (Zumajo-Cardona et al., 2021; Figure 4D). Notably, both abaxial determinant, GibiKAN (Zumajo-Cardona et al., 2021), and adaxial determinant, GbC3HDZ1 (Floyd et al., 2006), are also expressed in the integument and nucellus of the ovule.
Leaves of Cycads resemble fern leaves by their prolonged apical growth, circinate tips, and leaflets (Stevenson, 1980, 1990, 2020). Marginal position of leaflet veins indicates that leaflets develop exclusively via marginal meristem activity, similar to Ginkgo (Boyce and Knoll, 2002; Boyce, 2005; Figure 4E). Zamia integrifolia is characterized by the presence of C1KNOX TFs at the sites of leaflet initiation in older LPs as suggested by immunolocalization with an antibody against Zea mays KN1 protein (Bharathan et al., 2002; Figure 4E). Thus, C1KNOX TFs in cycads seem responsible for prolonged apical growth of leaves and origin of leaflets (Figure 4J) via establishment of new meristems. ARP, C3HDZ, YABBY, and KANADI homologs were found in transcriptomes of cycad leaves (Table 1, Supplementary File, Supplementary Figures 2–4, and Supplementary Table 1).
Although of all seed-free plants, lycophytes have the lowest similarity to spermatophytes, most of them possess SAMs with several AIs. In order to discuss which molecular differences in similar SAMs could cause differences in leaf development, leaf development in clubmosses and quillworts will be explained next.
Simplex SAMs producing lycophyte leaves
Shoot apical meristems of clubmosses and quillworts resemble those of most gymnosperms, belonging to the simplex type (Newman, 1965; Stevenson, 1976a; Sterling, 1984). AIs and their anticlinal derivatives in simplex SAMs of lycophytes are more elongated than in SAMs of gymnosperms and are termed surface initials (SIs) (Figures 5A, C). AIs differ from other SIs by their larger width and higher vacuolation (Paolillo, 1963; Gola and Jernstedt, 2011; Romanova et al., 2022). The periclinal derivatives of AIs are termed subsurface initials (SSIs) and occupy the position of what in gymnosperms would be the central mother cell zone and the rib meristem (Stevenson, 1976a). Similar to SAMs of angiosperms and gymnosperms, the simplex lycophyte SAMs can be partitioned into the CZ composed of AIs and SSIs and the PZ composed of SIs and their periclinal derivatives (Romanova et al., 2022).
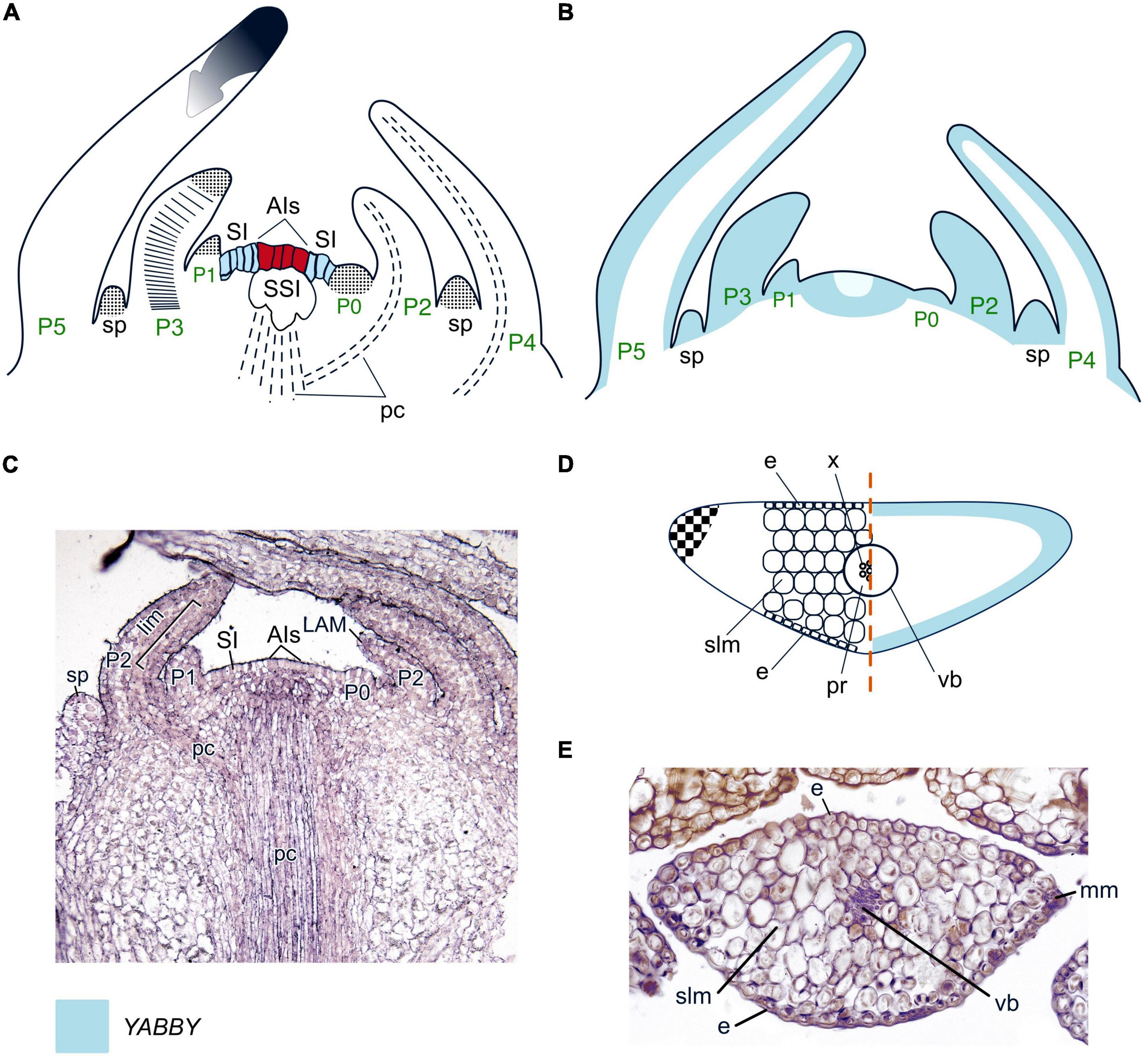
Figure 5. Organogenesis in the simplex SAM of lycophytes. Line drawings (A,B) and a micrograph (C) of longitudinal sections through the shoot apex; line drawing (D) and a micrograph (E) of transversal leaf section of a young leaf of Huperzia selago. SAM structure, cellular pattern of leaf origin and contribution of different leaf meristems to its development are inferred from descriptions and illustrations in Stevenson (1976a), Sterling (1984), Paolillo (1963), Gola and Jernstedt (2011), Evkaikina et al. (2017), and Romanova et al. (2022). Patterns of gene expression are based on in situ RNA–RNA hybridizations from Evkaikina et al. (2017). Apical initials (AIs) are marked in red; surface initials (SI) are marked in blue; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. SI, surface initials; SSI, subsurface initials; pr, parenchymatic cells assumed to represent phloem; cortex cells that contribute to the development of the leaf base are dotted in the same way as the leaf apical meristem; other captions and symbols as in Figures 2–4.
Two C1KNOX homologs have been found in each of the shoot tip transcriptomes of Huperzia selago (Evkaikina et al., 2017) and Isoetes lacustris [Maksimova (Evkaikina) et al., 2021]. Three WOX homologs that are sisters to the T3 clade were recently identified in Isoetes tegetiformans (Wu et al., 2019; Youngstrom et al., 2022). A number of WOX homologs from the T1 clade was found in a Lycopodium annotinum transcriptome, and in the genomes of Diphaziastrum complanatum and Isoetes taiwanensis. The I. taiwanensis genome also contains WOXes that can be mapped as sisters of the T2 + T3 clades (Table 1, Supplementary Figure 1 and Supplementary Table 1). WOX expression patterns in clubmosses and quillworts have not been analyzed yet.
Leaves of clubmosses and quillworts, similar to those of seed plants, initiate in the PZ of the SAM. In Lycopodium species, leaves originate at a considerable distance from AIs. This peculiarity was once interpreted as a fingerprint of their evolutionary origin as outgrowths of the cortex of the axes of rhyniophytes s.l. (Bower, 1935). However, the initiation of leaves in Huperzia and Isoetes species close to the AIs contradicts this interpretation. Leaf initiation in L. lucidulum starts from periclinal divisions of several SIs (Stevenson, 1976a); in Isoetes howellii, I. nuttalii, and I. brainii and Huperzia selago these are accompanied by periclinal divisions of SSIs (Paolillo, 1963; Romanova et al., 2022).
As in seed plants, LPs of clubmosses and quillworts emerge as radial bumps (Figure 5A). Later, LPs acquire an oval shape due to proliferation of cells at the leaf margins that were defined as the marginal meristem (Paolillo, 1963). No cells that would resemble the plate meristem in leaves of seed plants could be identified within the developing leaves of either clubmosses or quillworts. Due to a stronger segmentation of LP cells in the proximodistal than in the mediolateral plane, although this is more pronounced in Isoetes species than in clubmosses (Paolillo, 1963; Stevenson, 1976a), it can be assumed that an intercalary meristem is present in lycophyte leaves. Mature leaves of clubmosses contain homogenous spongy mesophyll (Evkaikina et al., 2017); in quillworts the leaf blade is separated into aerenchyma chambers by narrow trabeculae of rounded mesophyll cells (Liu et al., 2006). The lack of differentiation between palisade and spongy mesophyll is a trait that clubmosses and quillworts share with most conifers (Figures 5D, E).
No ARP homologs were found in the transcriptomes of Huperzia selago (Evkaikina et al., 2017), H. serrata (Yang et al., 2017), indicating that other molecular players are involved in the switch from a meristem-specific to a leaf-specific program in the PZ of lycophyte SAMs. However, homologs of C3HDZ were found in a number of clubmosses and quillworts (Floyd et al., 2014; Vasco et al., 2016; Table 1; Supplementary Figure 2; Supplementary Table 1).
Clubmosses are the only seed-free vascular plants that possess YABBY homologs, expressed in the shoot tips of H. selago (Evkaikina et al., 2017), H. serrata (Yang et al., 2017) and found in the genomes of Phylloglossum drummondii and Diphasiastrum complanatum (Table 1, Supplementary Figure 3, and Supplementary Table 1). Their YABBY proteins form a clade which is sister to the YABBY proteins of seed plants. Expression of HsYABBY in H. selago marks primordia of leaves and sporangia without any abaxial restriction (Evkaikina et al., 2017). HsYABBY is also expressed in the PZ of the SAM, while the expression of its counterparts in angiosperms is always excluded from the SAM. This difference suggests that in lycophytes YABBY TFs might regulate both organogenesis and SAM maintenance. HsYABBY expression in LPs and sporangial primordia supports the hypothesis of lycophyte leaves evolved via co-option and modification of the sporangia development program (Crane and Kenrick, 1997; Vasco et al., 2016; Evkaikina et al., 2017). The uniform expression of HsYABBY in LP cells (Figure 5B) correlates with the absence of leaf differentiation into spongy and palisade mesophyll and suggests that the ancestral YABBYs were not involved in the differentiation of the abaxial domain. KANADI homologs were detected in shoot tip transcriptomes of H. selago (Zumajo-Cardona and Ambrose, 2020), L. annotinum and genomes of Isoetes taiwanensis and Diphasiastrum complanatum (Table 1, Supplementary Figure 4, and Supplementary Table 1). Thus, KANADI TFs evolved before the separation of clubmosses and ferns. Altogether, molecular regulation of simplex SAMs of lycophytes and of gymnosperms shows similar features like C1KNOX-mediated maintenance of the undifferentiated state of the SAM and involvement of the same pool of WOX TFs in maintenance of both SAMs and RAMs.
Monoplex SAMs producing lycophyte leaves
Shoot apical meristems of Selaginella species conform to Newman’s (1965) monoplex structural type. However, analysis of clonal sectors in S. kraussiana revealed a pair of transiently active AIs within the SAM (Harrison et al., 2007; Jones and Drinnan, 2009; Harrison and Langdale, 2010). Oblique anticlinal divisions of AIs produce prismatic cells that together with AIs compose the SI zone (Dengler, 1983; Romanova et al., 2022). Polygonal subsurface cells compose the SSI zone (Figures 6A, C). SIs and SSIs in spikemosses resemble their counterparts in simplex SAMs of clubmosses and quillworts, or cells of CZ in seed plant SAMs. SIs divide anti- and periclinally and retain their prismatic shape, but become more narrow and less vacuolated and, together with their subsurface derivatives, resemble the seed plant PZ. Thus, the monoplex SAM can be partitioned into CZ and PZ similar to simplex and duplex SAMs.
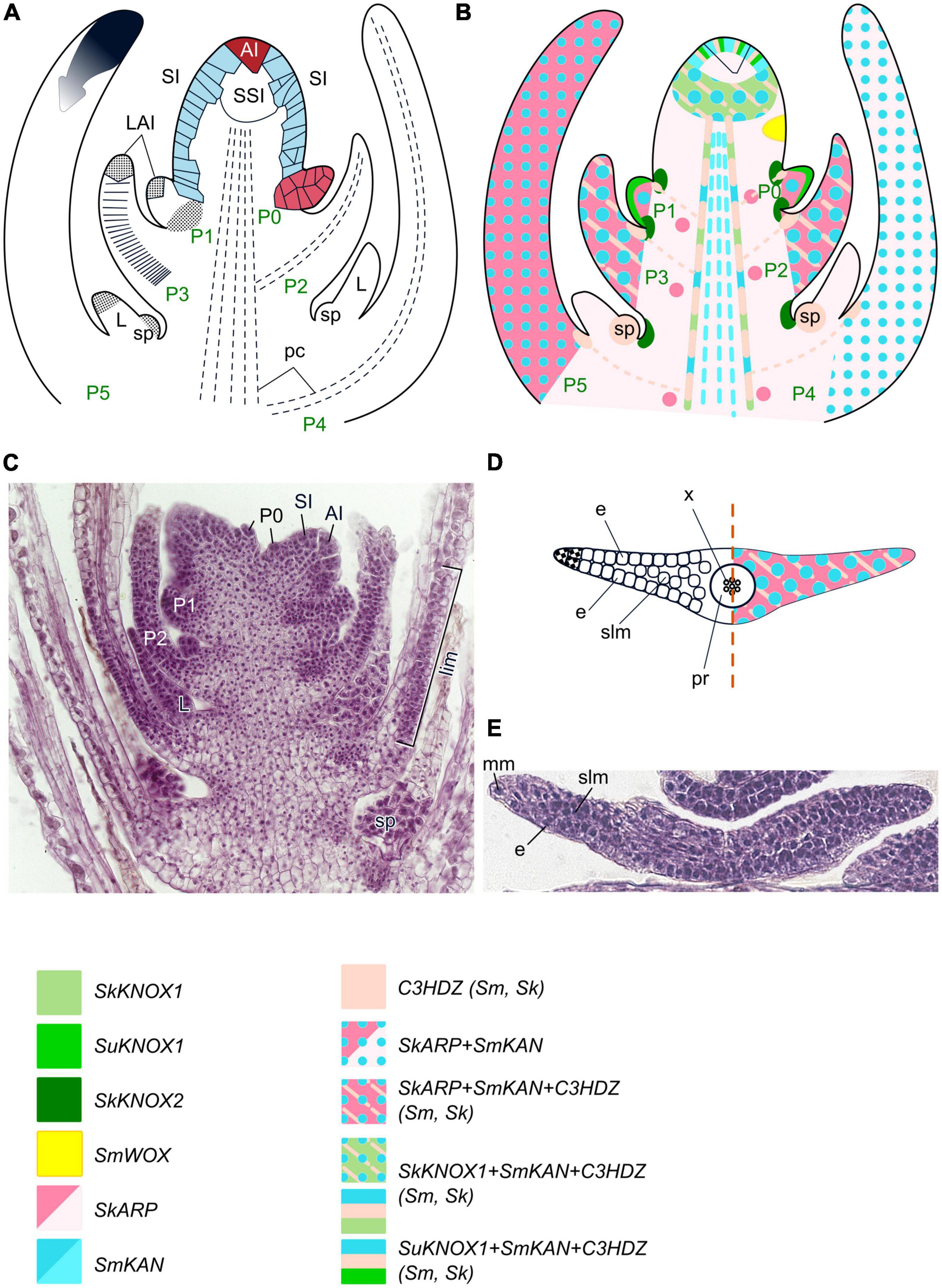
Figure 6. Organogenesis in the monoplex SAM of lycophytes. Line drawings (A,B) and a micrograph (C) of longitudinal sections through shoot apex; line drawing (D) and a micrograph (E) of a transversal section of a young leaf of Selaginella uncinata. SAM structure, cellular pattern of leaf origin and contribution of different leaf meristems to its development are inferred from descriptions and illustrations in Dengler (1983), Harrison et al. (2007), Romanova et al. (2022). Patterns of gene expression are based on Spencer et al. (2021) and Vasco et al. (2016), but expression in different Selaginella species is mapped on the same schematic of the spikemoss apex. For details of gene expression in different Selaginella species see Figure 4 in Spencer et al. (2021). The apical initial (AI) is marked in red; surface initials (SI) are marked in blue; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. LAI, leaf apical initial; L, ligule; sp – sporangium; the cells that contribute to P0 development are labeled in red; other captions and symbols as in Figures 2–5.
Advances in developmental regulation of several Selaginella species have been recently reviewed by Spencer et al. (2021) and will be mentioned only briefly, with some updates. Expression of two CIKNOX genes in Selaginella kraussiana was observed in the SAM, in the peripheral part of the procambium (SkKNOX1) and in the surface cells between the developing leaves (SkKNOX2) (Figure 6B; Harrison et al., 2005). In S. kraussiana, C1KNOX genes are not expressed in the AI and its closest derivatives, while they are expressed in the AI and SIs of S. moellendorffii (Frank et al., 2015) and S. uncinata (Kawai et al., 2010). These findings suggest that multiple C1KNOX genes likely have similar functions in the shoot apex of Selaginella and seed plants, and confirm the functional compartmentalization of monopleõ SAMs into several domains (Frank et al., 2015; Spencer et al., 2021). Some WOX homologs in the spikemosses S. kraussiana and S. moellendorffii have been mapped to the T1 and T2 clades of the WOX genes (Deveaux et al., 2008; van der Graaff et al., 2009; Segatto et al., 2016; Spencer et al., 2021). However, recent studies suggested that SmWOX11 from S. moellendorfii is sister to the T2 and T3 clades, and SkWOX11C is sister to the T3 clade, showing that WUS/WOX TFs were already present in the earliest extant vascular plants, lycophytes (Wu et al., 2019; Youngstrom et al., 2022). Emerging data about expression patterns of WOX genes in Selaginella species suggest that they are functionally different from their angiosperm counterparts. Specifically, RNAseq analysis showed that SmWOX11, a homolog of WOX5 which is expressed root-specifically in Arabidopsis, is ubiquitously expressed in S. moellendorffii (Ge et al., 2016). A WOX homolog from S. moellendorffii (SmWOX) is expressed in peripheral SIs, leaf AIs (LAIs) and other LP cells suggesting that it rather regulates leaf initiation than SAM maintenance (Frank et al., 2015). SkWOX11C in S. kraussiana is expressed only in the phloem of the shoot vasculature while SkWOX11B is expressed throughout shoot apex including in LPs (Youngstrom et al., 2022). Altogether, these data suggest that although genes of the T3 clade emerged already in the last common ancestor of lycophytes and other land plants, they were not recruited to regulation of SAM maintenance at that point but rather involved in the regulation of leaf initiation (Youngstrom et al., 2022).
Leaves of spikemosses originate about a dozen of cells away from the AI. This distance of leaf initiation from the AIs is also found in Lycopodium species, and thus is not correlated with the structural type of the SAM. Development of leaves within the SAM slightly differs between Selaginella species. It was shown for, e.g., S. martensii that leaf initiation starts from anticlinal elongation and subsequent periclinal divisions of several SIs; one of these SIs then divides obliquely, forming a single or a pair of wedge-shaped cells (Dengler, 1983; Figure 6A). However, the clonal analysis of Harrison et al. (2007) revealed that in S. kraussiana, leaves are always initiated from two adjacent epidermal cells. Therefore, the wedge-shaped cell frequently observed at the top of early stage leaves of Selaginella species likely represent only one of these two cells. In S. martensii proliferation of the shoot cortex cells also contributes to the development of the LP base (Dengler, 1983), and this might be considered as a fingerprint of the origin of leaves of Selaginella ancestors as cortex outgrowths. The leaf blade in S. martensii results from divisions of wedge-shaped surface cells along the entire LP edge that act as marginal meristem (Figures 6D, E; Dengler, 1983). As marginal meristem derivatives divide only anticlinally, the leaves in S. martensii are only two cells thick except for the midrib region composed of uniform spongy mesophyll (Dengler, 1983). In S. kraussiana, two adjacent epidermal cells that give rise to the leaves divide to extend leaf development mediolaterally prior to the ad/abaxial divisions that serve proximodistal growth (Harrison et al., 2007).
Expression of CIKNOX homologs is absent from incipient and developing LPs of S. kraussiana like in seed plants (Harrison et al., 2005), but in S. uncinata, SuKNOX1 is expressed in surface cells of the youngest LPs, including LAIs (Kawai et al., 2010; Figures 6B, D). Homologs of the leaf development regulator ARP have been found in S. kraussiana (Harrison et al., 2005), S. viticulosa, S. selaginoides, and S. willdenowii (Hernández-Hernández et al., 2021). Although in seed plants, ARP proteins antagonize the expression of CIKNOX, thus switching the leaf founder cells from a meristematic to a leaf-development program, homologs of both gene groups, SkARP1 and SkKNOX1, are co-expressed in the SAM of S. kraussiana (Harrison et al., 2005). SkARP1 expression also marks LPs and leaf vascular traces (Figures 6B, D). The presence of these genes in S. kraussiana suggests that the C1KNOX/ARP developmental module might have existed in the last common ancestor of lycophytes and other plants, i.e., has a single evolutionary origin in land plants (Harrison et al., 2005). The co-expression of C1KNOX and ARP homologs in the SAM suggests that leaves might have originated via co-option and modification of the dichotomy program of the axes of the first land plants (Harrison et al., 2005). The absence of C1KNOX expression in S. kraussiana leaves allows different interpretations. On the one hand, it can be regarded as a result of ARP-mediated suppression similar to seed plants (Harrison et al., 2005). On the other hand, the absence of C1KNOX expression from S. kraussiana leaves might indicate that C1KNOX genes were never expressed in lycophyte LPs since they originated as cortex outgrowths with limited growth potential (Floyd and Bowman, 2006; Floyd et al., 2006).
A number of C3HDZ homologs are present in Selaginellales (Floyd and Bowman, 2006, 2010; Floyd et al., 2006; Prigge and Clark, 2006; Vasco et al., 2016). C3HDZs of spikemosses are expressed in the SAM similarly to their angiosperm counterparts. SkH3HDZ2 expression marks all SAM cells in S. kraussiana, while SkC3HDZ1 and SmC3HDZ expression in S. moellendorffii is excluded from the AI and its immediate derivatives (Figure 6B; Floyd and Bowman, 2006; Vasco et al., 2016). This expression pattern confirms a multicellular zonal structure of monoplex SAMs and supports the viewpoint of Floyd et al. (2006) that the ancestral role of C3HDZ in land plants was in meristem maintenance. C3HDZ are expressed also in all cells of incipient and early stage LPs (Floyd and Bowman, 2006; Prigge and Clark, 2006; Vasco et al., 2016). In the course of leaf development SkC3HDZ1 and SmC3HDZ expression becomes restricted to the adaxial domain of the leaf base, which later will produce a ligule and a sporangium, and to leaf traces and shoot procambium (Floyd and Bowman, 2006, 2007; Prigge and Clark, 2006).
No homologs of abaxial determinants from the YABBY gene family were identified in spikemosses (Floyd and Bowman, 2007), but KANADI homologs were detected in Selaginella moellendorffii (Zumajo-Cardona et al., 2019). SmKAN1 and SmKAN2 are expressed in the SAM of S. moellendorffii (except in the AI and its closest derivatives) where they co-localize with C3HDZ1, and SmKAN3 is expressed only weakly in the SIs (Zumajo-Cardona et al., 2019). Thus, unlike in seed plants where both KANADI and YABBY antagonistically interact with C1KNOX TFs and are therefore excluded from the SAM, expression of KANADI homologs in S. moellendorffii occurs in the same SAM cells as expression of SkKNOX1 in S. kraussiana. Hence C1KNOX and KANADI TFs are unlikely to act as antagonists in spikemosses. Yet another peculiarity of S. moellendorffii KANADI homologs is their uniform expression in all cells of LPs and young leaves. A possible anatomical consequence of non-polar KANADI expression is the homogenous sponge-like structure of the mesophyll in Selaginella species (Figures 6D, E).
Monoplex SAMs producing leptosporangiate ferns leaves
Shoot apical meristems of most leptosporangiate ferns belong to the monoplex type (Newman, 1965). A single tetrahedral AI produces prismatic derivatives which, in turn, undergo periclinal divisions, producing prismatic SIs and polygonal SSIs (Stevenson, 1976b; Romanova and Jernstedt, 2005; Romanova et al., 2021). Thus, segmentation patterns of AI and SIs in ferns are similar to those in spikemosses (Figures 7A, C). Polygonal cells that enclose SIs and SSIs, are termed the cup-zone and resemble PZ cells in simplex and duplex SAMs (Stevenson, 1976b,1978; Romanova et al., 2022). However, despite this structural similarity, contrarily to PZ cells cup-zone cells are unable to perform organogenesis, and both leaves and branches in ferns are produced by SIs.
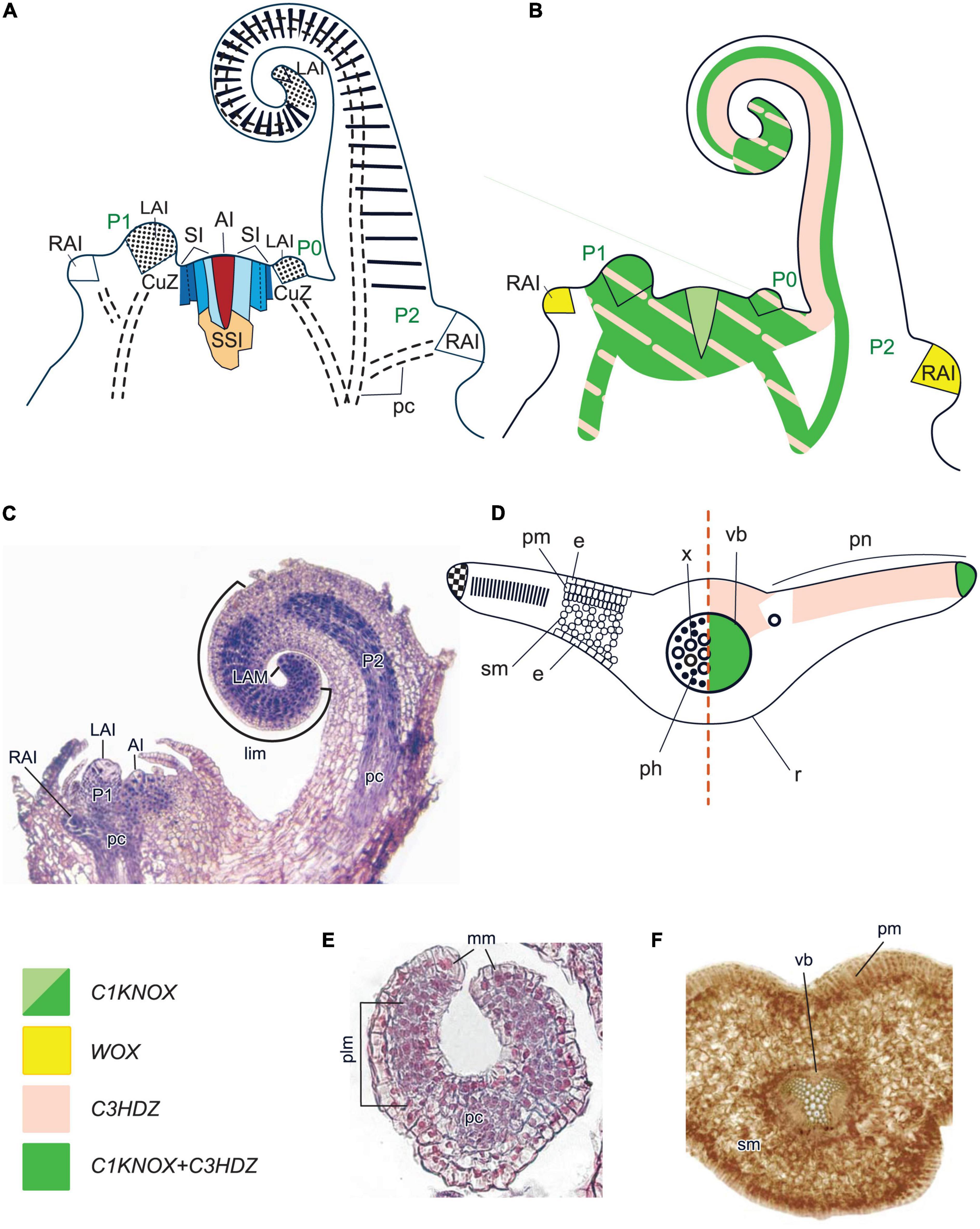
Figure 7. Organogenesis in the monoplex SAM of leptosporangiate ferns. Line drawings (A,B) and a micrograph (C) of longitudinal sections through shoot apex of Gymnocarpium dryopteris; line drawing of a transversal section of a fern leaf (D); micrographs of a transversal section of a young leaf (E) and a fragment of a mature leaflet (F) of Athyrium filix-femina. SAM structure, cellular pattern of leaf origin and contribution of different leaf meristems to its development are inferred from descriptions and illustrations in Imaichi (2008), Romanova and Borisovskaya (2004), Romanova and Jernstedt (2005) and Vasco et al. (2013). Patterns of gene expression are based on in situ RNA–RNA hybridizations and immunolocalizations from Bharathan et al. (2002), Harrison et al. (2005), Sano et al. (2005), Nardmann and Werr (2012), Ambrose and Vasco (2016), Vasco et al. (2016) and Vasco and Ambrose (2020). The apical initial (AI) is marked in red; surface initials (SI) are marked in different shades of blue depending on the number of periclinal divisions they underwent; subsurface initials (SSI) are marked in yellow; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. CuZ, cup-zone; RAI, root apical initial; r, rachis; pn, pinnae; other captions and symbols are as in Figures 2–6.
In a number of leptosporangiate fern species (Ceratopteris richardii, Elaphoglossum lloense), C1KNOX genes are expressed in the cells of the cup-zone (Sano et al., 2005; Ambrose and Vasco, 2016), in E. peltatum they also mark the AI and its closest derivatives (Figure 7B; Vasco and Ambrose, 2020). In Osmunda regalis and Anogramma chaeophylla, CIKNOX was detected in the shoot apex using a heterologous antibody raised against maize CIKNOX (Bharathan et al., 2002; Harrison et al., 2005). These data confirm the partitioning of fern SAMs into functionally different zones and the conservation of KNOX TF function in promoting indeterminacy in all plant lineages. A number of WOX homologs of leptosporangiate and ophioglossoid ferns have been mapped to the T1 and T2 clades (Nardmann and Werr, 2012; Xia et al., 2022). However, since Wu et al. (2019) have shown that the WOX TFs of the T2 clade are only present in seed plants, it can be assumed that the T2 (“intermediate”) clade members are probably sisters to the T2 + T3 clades (Table 1; Supplementary Figure 1; Supplementary Table 1; Wu et al., 2019).
WOX homologs from the T3 clade were discovered in Ceratopteris richardii and Cyathea australis (Nardmann and Werr, 2013). However, unlike its spermatophyte counterparts, the single WUS lineage member of the leptosporangiate fern C. richardii (CrWUL) is expressed only in the procambium, primordia of shoot-borne roots and lateral roots (Nardmann and Werr, 2012). These data suggest that WUS clade genes in ferns already function in the regulation of meristematic tissues such as procambium or RAMs but do not yet regulate SAMs, marginal or plate meristems.
Leaves in leptosporangiate ferns originate via emergence of a lens-shaped LAI from one of SIs (Hou and Hill, 2002; Romanova and Borisovskaya, 2004; Romanova and Jernstedt, 2005; Imaichi, 2008). This LAI produces two files of prismatic cells that constitute the marginal meristem (Figures 7D, E). More active cell proliferation on the abaxial side results in a coiled leaf morphology (Kaplan and Groff, 1995).
Unlike in seed plants and lycophytes, C1KNOX expression in leptosporangiate ferns is not downregulated in LAIs (Figure 7B), with the exception of Elaphoglossum lloense, a fern with simple leaves; nor is it downregulated in LPs, or in the apical part of developing leaves (Bharathan et al., 2002; Harrison et al., 2005; Sano et al., 2005; Ambrose and Vasco, 2016; Vasco and Ambrose, 2020). These data support an indeterminate growth potential of these leaves, similar to that of shoots. In species with simple leaves, marginal meristem cells are marked by continuous C1KNOX transcription, and their uniform proliferation, similar to that in G. biloba leaves, results in the development of the whole leaf blade (Tsukaya, 2018, 2021; Vasco and Ambrose, 2020). Development of pinnae and pinnules in compound leaves proceeds through emergence of new AIs from prismatic surface cells of the marginal meristem (Vasco et al., 2013); accordingly, C1KNOX expression fades in marginal meristem cells except for the initials of pinnae and pinnules (Vasco and Ambrose, 2020).
Thus, leaves of leptosporangiate ferns like those of cycads and unlike those of other plants develop via activities of both the apical and the marginal meristems (Figures 7A, D; Tsukaya, 2021). Apical growth of leaves is nearly indeterminate like shoot growth in some ferns (e.g., in Lygodium venustum a single leaf can reach 30 m in length) (Mueller, 1983). Morphogenetic similarity between fern leaves and shoots was confirmed by extensive experimental studies in the mid-20th century (reviewed in Wardlaw, 1963; White, 1971; Steeves and Sussex, 1989; Vasco et al., 2013).
Leaves of most leptosporangiate ferns are differentiated into adaxial palisade mesophyll and abaxial spongy mesophyll (Gifford and Foster, 1989; Vasco et al., 2013; Figures 7D, F). The leaf veins are collateral in ferns with ectophloic siphonosteles, and are termed amphicribral in ferns with amphiphloic siphonosteles (e.g., Gifford and Foster, 1989). However most if not all phloem elements in the latter are located abaxially and adaxially the xylem, not around it, it would be more accurate to call such veins bicollateral. Similar to SIs of the SAM, marginal meristem cells produce smaller derivatives toward the center that differentiate into leaf mesophyll and vasculature (Cruz et al., 2020). Expression of C1KNOX and the absence of expression of WOX homologs in fern leaf margins indicate a different regulation of the maintenance of marginal meristems compared to seed plants.
Harrison et al. (2005) had detected a putative ARP protein in the SAM and LPs SAM of LPs of Osmunda regalis using antibodies raised against maize ARP. The putative ARP homolog and a C1KNOX homolog occurred in the same cells, unlike mutually exclusive localization of these TFs in angiosperms (Harrison et al., 2005). However, subsequent analyses of Hernández-Hernández et al. (2021) have shown that the genomes of three other leptosporangiate ferns (Azolla filiculoides, Salvinia cucullata and Ceratopteris richardii) do not contain ARP homologs. Thus, the detection of ARP in O. regalis using a heterologous antibody, and the presence of ARP in ferns in general, is questionable.
A number of C3HDZ homologs were discovered in leptosporangiate ferns (Table 1; Supplementary Figure 2; Supplementary Table 1; Aso et al., 1999; Floyd et al., 2006; Plackett et al., 2015; Vasco et al., 2016). In Pilularia globulifera and Elaphoglossum peltatum (Polypodiales), they are expressed in all zones of the SAM, as are their counterparts in seed plants. However, C3HDZ expression was not detected in the SAM of Osmunda regalis (Osmundales) (Vasco et al., 2016). This difference is interpreted as support of a phylogenetic distinction between the two orders. In Polypodiales, C3HDZ homologs are expressed in the entire LPs during the early stages of leaf development. Later, expression becomes restricted to the leaf tip and the adaxial side of the upper circinate leaf domain in both simple and divided leaves, leaf traces and shoot procambium. In O. regalis, a C3HDZ gene is expressed in the adaxial region of developing pinnae (Vasco et al., 2016). Adaxial expression of C3HDZ homologs is similar in leaves of angiosperms and leptosporangiate ferns.
A number of KANADI homologs were detected in leptosporangiate ferns; phylogenetic analyses have shown that they appeared before the diversification of their counterparts in seed plants (Zumajo-Cardona et al., 2019; Table 1; Supplementary Figure 4; Supplementary Table 1). No YABBY homologs could be found (Floyd and Bowman, 2007). To confirm that the specification of the abaxial identity in ferns is regulated solely by KANADI homologs, data on their expression patterns should be obtained.
Monoplex SAMs producing horsetails leaves
Shoot apical meristems in horsetails resemble those in leptosporangiate ferns: the single prominent AI undergoes oblique anticlinal divisions producing prismatic derivatives. Their unequal periclinal divisions produce prismatic SIs and polygonal SSIs (Figures 8A, C). However, unlike SIs of leptosporangiate ferns, SIs of horsetails do not shorten in the course of periclinal divisions; therefore the cup-zone is absent (Gifford and Foster, 1989). SSIs divide predominantly periclinally and form the rib-meristem.
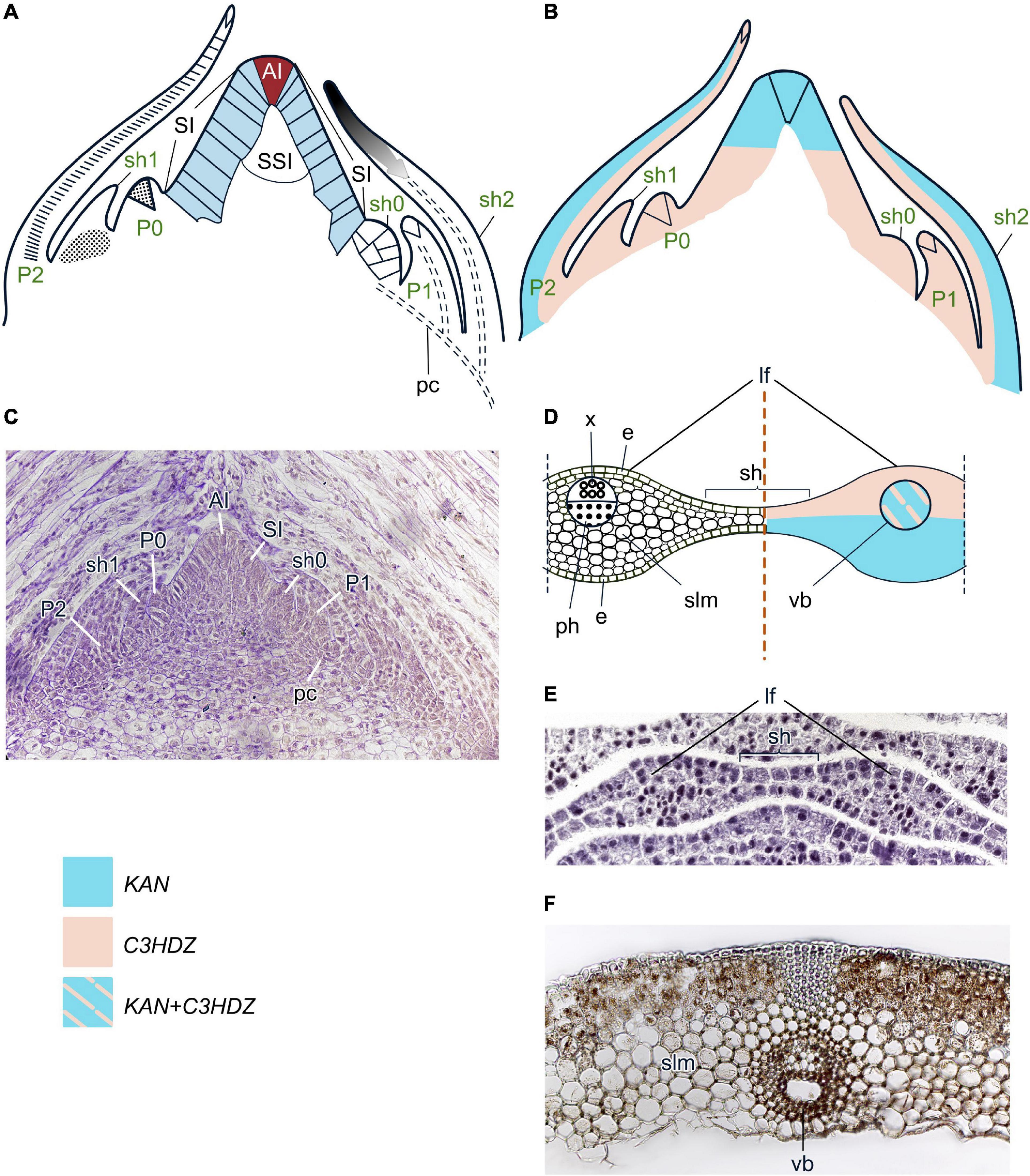
Figure 8. Organogenesis in the monoplex SAM of horsetails. Line drawings (A,B) and a micrograph (C) of longitudinal sections through the shoot apex; line drawing (D) and a micrograph (E) of a transversal section of a fragment of a young whorl that is composed of two leaves and a sheath between them; micrograph of an adult leaf (F) of Equisetum fluviatile. SAM structure, cellular pattern of leaf origin and contribution of different leaf meristems to its development are inferred from descriptions and illustrations in Duval-Jouve (1864), Golub and Wetmore (1948a,b) and Gifford and Foster (1989). Patterns of gene expression are based on in situ RNA-RNA hybridizations from Frank et al. (2015), Vasco et al. (2016) and Zumajo-Cardona et al. (2019). The apical initial (AI) is marked in red; surface initials (SI) are marked in blue; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. Lf, leaf; sh, sheath; other captions and symbols as in Figures 2–7.
Two CIKNOX homologs were discovered in Equisetum diffusum (Vasco and Ambrose, 2020). WOX homologs from the T1 clade were identified in Equisteum giganteum, E. arvense (Xia et al., 2022), E. diffusum (Wu et al., 2019), and E. hyemale (Supplementary Figure 1 and Supplementary Table 1). The WOX members from E. giganteum and E. arvense mapped by Xia et al. (2022) to T2 clade are more likely sisters to the T2 + T3 clades (Table 1), as WOX TFs of the T2 clade are only present in seed plants (Wu et al., 2019). No members of the T3 clade were found in horsetails (Xia et al., 2022).
The leaves of horsetails are highly reduced and connate laterally to form a sheath around the aerial stem, unlike leptosporangiate ferns where leaves are the dominant organs of the plant (Golub and Wetmore, 1948a,b; Tomescu et al., 2017). LPs initiate not one by one, as in most plants, but as a whorl that contains six to more than a dozen LPs. Several detailed anatomical studies (e.g., Duval-Jouve, 1864; Golub and Wetmore, 1948a,b) revealed that leaves from one whorl are produced from three successive segments of the AI. Each LP emerges as result of several periclinal divisions in a group of two to five SIs. Subsequent oblique anticlinal divisions of one of the LP surface cells establishes the LAI which is smaller than the AI in the SAM (Figures 8A, C). In summary, the cellular pattern of LP initiation is similar to that of spikemosses and dissimilar to that of leptosporangiate ferns where LP development starts with the emergence of an LAI.
When LAIs of LPs from the same whorl start their segmentation, SIs between them also undergo periclinal divisions but do not form LAIs. Coordinated proliferation of cells at the base of LPs and between them results in the development of a unitary collar-like structure, the phytomer (Tomescu et al., 2017). The activity of the LAI is very transient. In the fourth or fifth phytomer from the AI, cells in the upper part of LP, including the LAI, significantly elongate, become vacuolated and deteriorate, resulting in abortion of LP development (Golub and Wetmore, 1948a,b).
The entire collar is composed of uniform spongy mesophyll; the part of it located beneath the leaf contains a greater number of mesophyll layers and a single collateral vascular bundle (Figures 8D–F; Golub and Wetmore, 1948a,b). No homologs of the adaxial determinant ARP were identified in the transcriptome of E. diffusum (Hernández-Hernández et al., 2021), similar to leptosporangiate ferns. Homologs of the other adaxial determinant, C3HDZ, were found in E. diffusum (Vasco et al., 2016), E. arvense (Frank et al., 2015) and E. hyemale (Vasco et al., 2016). Accumulation of C3HDZ transcripts was reported for SAMs and leaves of E. arvense (Frank et al., 2015). In E. diffusum, C3HDZ homologs are not expressed in the SAM but are transcribed in the peripheral SIs that are about to originate leaves, in entire early stage LPs, the adaxial protodermal cells and in the procambium of young leaves (Figures 8B, D; Vasco et al., 2016). The absence of C3HDZ expression in horsetail SAMs is similar to the fern O. regalis but different from other leptosporangiate ferns, lycophytes and seed plants. The confinement of C3HDZ expression to the adaxial leaf domain is similar to other ferns and seed plants but different from the leaves of lycophytes where these genes are uniformly expressed over the LPs.
No YABBY homologs were found in horsetails like in most other seed-free plants (Floyd and Bowman, 2007; Romanova et al., 2021). However, three KANADI homologs were found in E. hyemale and in E. diffusum (Zumajo-Cardona et al., 2019; Table 1; Supplementary Figure 4; Supplementary Table 1). EhyKAN1 transcription occurs in all SAM cells including the AI while EhyKAN2 is transcribed in the SAMs of emerging buds. Thus, expression of KANADI homologs in the horsetail shoot apex differs from seed plants and lycophytes, where it is excluded from the SAM (Zumajo-Cardona et al., 2019). EhyKAN1 is transcribed in all cells of the emerging LPs where it co-localizes with C3HDZ transcription. At subsequent developmental stages, expression of all three KANADI homologs (EhyKAN1 – EhyKAN3) becomes confined to the abaxial domain of the sheath (Zumajo-Cardona et al., 2019).
Monoplex SAMs producing whisk fern leaves
Shoot apical meristems of whisk ferns (Psilotales) belong to the monoplex type with a single AI (Figures 9A, C; Takiguchi et al., 1997). SSI and cup-zones are not found (Naumenko and Romanova, 2008).
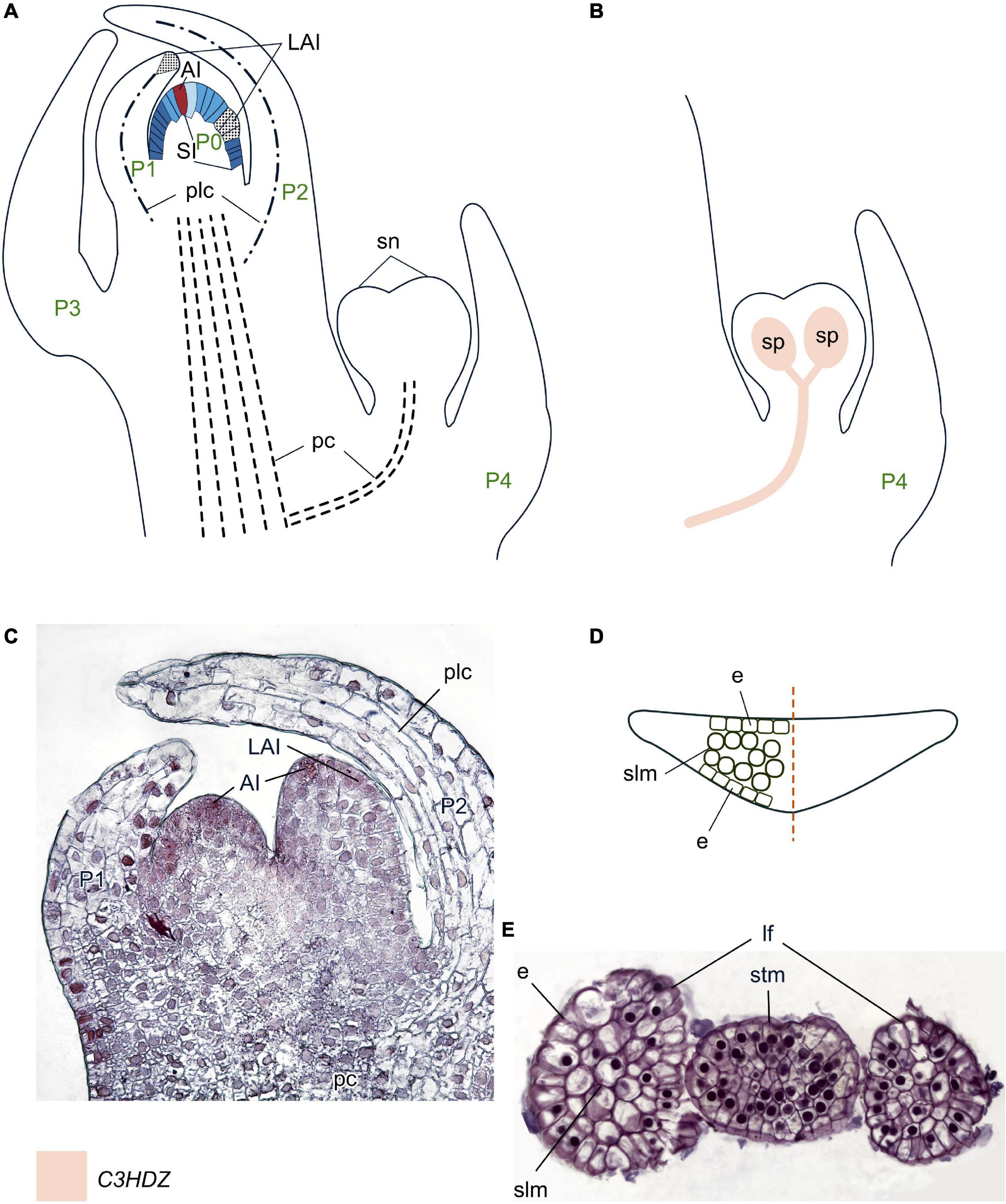
Figure 9. Organogenesis in the monoplex SAM of whisk ferns. Line drawings (A,B) and a micrograph (C) of longitudinal sections through the shoot apex; line drawing of a transversal leaf section (D); a micrograph of a transversal section of a stem tip with two young leaves (E) of Psilotum nudum. SAM structure, cellular pattern of leaf origin and contribution of different leaf meristems to its development are inferred from descriptions and illustrations in Bierhorst (1977), Takiguchi et al. (1997), and Naumenko and Romanova (2008). Patterns of gene expression are based on in situ RNA–RNA hybridizations from Vasco et al. (2016). The apical initial (AI) is marked in red; surface initials (SI) are marked in different shades of blue depending on the number of periclinal divisions they underwent; color codings for gene expression and symbols for different types of leaf meristems and tissues are given. Plc, procambium-like cells that do not further differentiate into vascular tissues; sn, synangium; other captions and symbols as in Figures 2–8.
Leaves of whisk ferns are unique among land plants in lacking vasculature. Their evolutionary homology is debatable and depends on the placement of whisk ferns in plant phylogeny (e.g., Bierhorst, 1968, 1977; Gensel, 1977, Kaplan, 1977, Wagner, 1977; Rothwell, 1999). The inclusion of Psilotales within the ferns as a sister group to Ophioglossales (Rothfels et al., 2015; Qi et al., 2018) suggests that the scale-like appendages in Psilotum nudum are probably best interpreted as highly reduced leaves (Schneider, 2013; Vasco et al., 2013).
Despite their major morphological dissimilarity, scale-like leaves of whisk ferns originate similarly to the leaves of “typical” ferns through the emergence of LAIs via oblique anticlinal divisions of SIs (Bierhorst, 1971; Naumenko and Romanova, 2008). Unlike LAIs of leptosporangiate ferns, and similarly to those of horsetails, LAIs of P. nudum have a very transient meristematic activity and soon become highly vacuolated and crumple. There is no localized meristematic activity within developing LPs. At this stage, an LP looks like LPs of other ferns, as it is tilted toward the SAM, differentiated into parenchymal and prosenchymal cells, although the latter neither proliferate nor differentiate into vascular tissues. Then active proliferation of the SAM results in shoot elongation above the LP (Bierhorst, 1971). Consequently, the LP aborts its morpho- and histogenesis. Therefore, mature scale-like leaves do not show ad/abaxial differentiation of the mesophyll and lack vascular bundles (Figures 9D, E).
Data about the homologs of development regulators in whisk ferns have recently become available (Table 1). Homologs of C1KNOX TFs are found in Psilotum nudum (Huang et al., 2020). WOX TF homologs from the T1 clade are found in P. nudum and Tmesipteris tannensis, and members of the T3 (“modern”) clade are present in P. nudum (Xia et al., 2022). The presumable T2 clade homolog reported for P. nudum (Nardmann and Werr, 2012) might represent a sister to the T2 + T3 clades (Table 1, Supplementary Figure 1, and Supplementary Table 1). Phylogenetic analyses revealed that homologs of both TF groups cluster with their counterparts from ophioglossoid ferns. This topology suggests that the evolution of the KNOX and WOX families took place before the separation of whisk ferns and ophioglossoid ferns (Xia et al., 2022).
No homologs of ARP adaxial determinants were found in whisk ferns so far (Table 1). Several homologs of the other adaxial determinant C3HDZ found in P. nudum form a common clade with their counterparts from horsetails, Ophioglossales and Marattiales (Table 1) (Floyd et al., 2006; Vasco et al., 2016). Similar to their counterparts in horsetails and Osmunda regalis (Vasco et al., 2016) and unlike those in other plant taxa, the C3HDZ homologs are not expressed in the SAMs of P. nudum. Expression of PnC3HDZ2 is confined to the elongated provascular-like cells of the LP and the central shoot procambium (Vasco et al., 2016). Expression of PnC3HDZ2 in provascular-like cells of LPs indicates that they are characterized by initial stages of leaf trace provascular tissue differentiation that is terminated by abortion of LP development. PnC3HDZ2 and PnC3HDZ3 are also expressed throughout sporangium development (Figure 9B), similarly to their counterparts in spikemosses and in the ophioglossoid fern Ophioglossum reticulatum. PnC3HDZ2 marks the sporogenous cells, while PnC3HDZ3 is expressed in the procambium, the basal portion of the synangium and its wall; later its expression becomes confined to the inner synangium wall and the spores (Vasco et al., 2016). These data together with the expression of C3HDZ homologs in both leaves and sporangia of Selaginella moellendorfiii (Vasco et al., 2016) and the expression of YABBY homologs in both leaves and sporangia of Huperzia selago (Evkaikina et al., 2017) might be seen as support of the scenario where the sporangium developmental program was co-opted for leaf development in whisk ferns as well as in ophioglossoid ferns and lycophytes.
Like for other sporophytes of seed-free plants with the exception of clubmosses, no homologs of the adaxial determinant YABBY were found in whisk ferns (Floyd and Bowman, 2007). However, two homologs of another abaxial determinant, KANADI, are present in the transcriptomes of P. nudum (Zumajo-Cardona and Ambrose, 2020) and Tmesipteris parva (Table 1, Supplementary Figures 3, 4, and Supplementary Table 1).
Discussion
All regulators of SAM maintenance and leaf development already existed in bryophytes
Mounting evidence from genomic and transcriptomic studies suggests that most TFs that regulate various developmental programs in polysporangiophytes, SAM functioning and leaf development among them, already existed in bryophytes, the pioneer plants on land (Bowman et al., 2019; Szovenyi et al., 2019). Among them are the homologs of C1KNOX found in the liverwort Marchantia polymorpha (Dierschke et al., 2021) and the moss Physcomitrium (Physcomitrella) patens (Champagne and Ashton, 2001; Sakakibara et al., 2008); C3HDZ homologs in M. polymorpha (Romani et al., 2018), P. patens (Yip et al., 2016) and the hornwort Anthoceros agrestis (Li et al., 2020); YABBY homologs in A. agrestis (Li et al., 2020); KANADI homologs in M. polymorpha (Briginshaw et al., 2022); WOX homologs in P. patens (Sakakibara et al., 2014), A. agrestis (Li et al., 2020) and M. polymorpha (Hirakawa, 2022); ARP homologs that seem to exist in M. polymorpha (Figure 10, Table 1, Supplementary Figures 1–4, Supplementary Table 1, and Supplementary File 1). These TFs function both in sporophytes and gametophytes of bryophytes. C1KNOX promotes cell proliferation in P. patens sporophytes, participates in the regulation of sporangium development (Sakakibara et al., 2008) and promotes the sporophytic program in M. polymorpha (Dierschke et al., 2021); WOX homologs are likely involved in sporophyte development of P. patens (Sakakibara et al., 2014). A C3HDZ homolog regulates leaf development in gametophytes of P. patens (Yip et al., 2016), and a KANADI homolog promotes the development of gametophores in M. polymorpha (Briginshaw et al., 2022). Recently, CLE homologs were found to be involved in stem-cell-promotion and the induction of dichotomy in gametophytes of M. polymorpha (Hirakawa, 2022). How developmental programs regulated by these TFs could have been co-opted and modified at the divergence between bryophytes and polysporangiophytes (protracheophytes + tracheophytes) is discussed elsewhere (e.g., Tomescu et al., 2014; Harrison, 2017; Harrison and Morris, 2018).
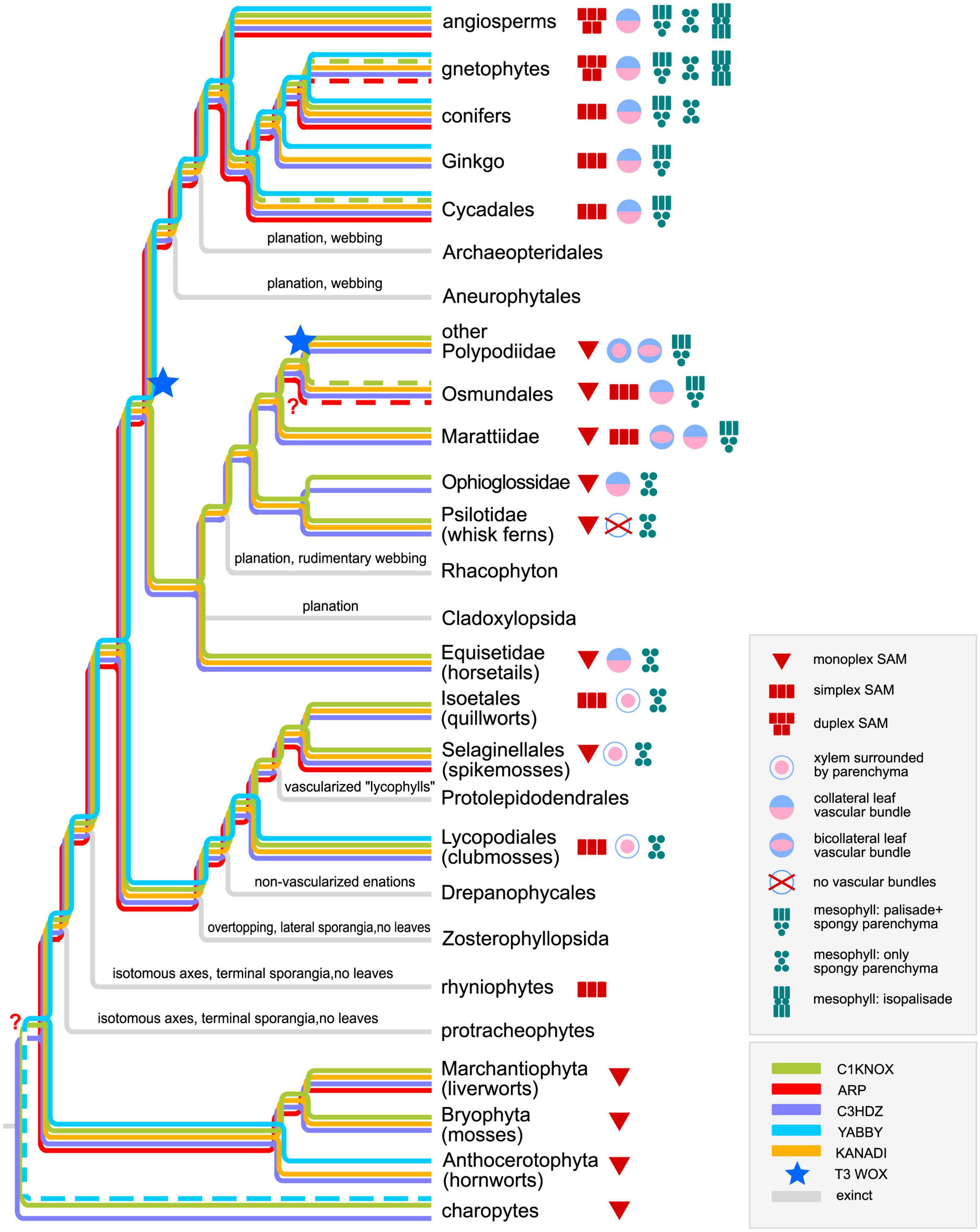
Figure 10. Phylogenetic tree for land plants and their structural and regulatory innovations. From Charophytes to Rhacophyton (with the exception of bryophytes), the tree is based on Harrison and Morris (2018) (Protracheophyta and Rhyniophyta genera are not listed separately); bryophytes are reconstructed according to Harris et al. (2022); ferns are reconstructed according to PPG I (2016); gymnosperms are reconstructed according to Williams (2012). Dashed lines stand for data based on immunolocalization with heterologous antibodies or for ambiguous data for different genera of gnetophytes.
Fingerprints of three hypothetical programs of leaf origin can be found in lycophytes
The complete toolkit of the abovementioned TFs exists in lycophytes. Specifically, C1KNOX homologs were found in the transcriptomes of a number of spikemosses, clubmosses, and quillworts [Harrison et al., 2005; Kawai et al., 2010; Maksimova (Evkaikina) et al., 2021; Supplementary Table 1]; homologs of ARP (Harrison et al., 2005; Hernández-Hernández et al., 2021); C3HDZ (Vasco et al., 2016); and KANADI (Zumajo-Cardona et al., 2019). TFs exist in different Selaginella species; C3HDZ and KANADI homologs were also found in transcriptomes of a number of clubmosses and quillworts as well as in the genomes of Diphasiastrum complanatum and Isoetes taiwanensis (Supplementary Table 1); YABBY homologs were revealed in transcriptomes of the clubmosses Huperzia selago (Evkaikina et al., 2017), H. serrata (Yang et al., 2017), Phylloglossum drummondii and in the genome of Diphasiastrum complanatum (Figure 10, Table 1, Supplementary Figures 1–4, and Supplementary Table 1). These data confirm that SAM and leaf regulators could have been co-opted from bryophytes, suggest that likely all of them existed in leafless polysporangiophytes, and allow us to hypothesize that YABBY homologs may have been lost in spikemosses, and ARP homologs in clubmosses (Figure 10).
Based on the co-expression of genes encoding KNOX and ARP TFs in the SAM of S. kraussiana, Harrison et al. (2005) suggested that these TFs together regulated the dichotomy of leafless axes in the first polysporangiophytes, and that this program was co-opted for leaf development in lycophytes; i.e., lycophyte leaves share deep homology with axes. This hypothesis is supported by the expression of C3HDZ and KANADI in the SAM of Selaginella species (Floyd and Bowman, 2006; Vasco et al., 2016; Zumajo-Cardona et al., 2019; Figure 5) and YABBY in the SAM of H. selago (Evkaikina et al., 2017; Figures 6, 11).
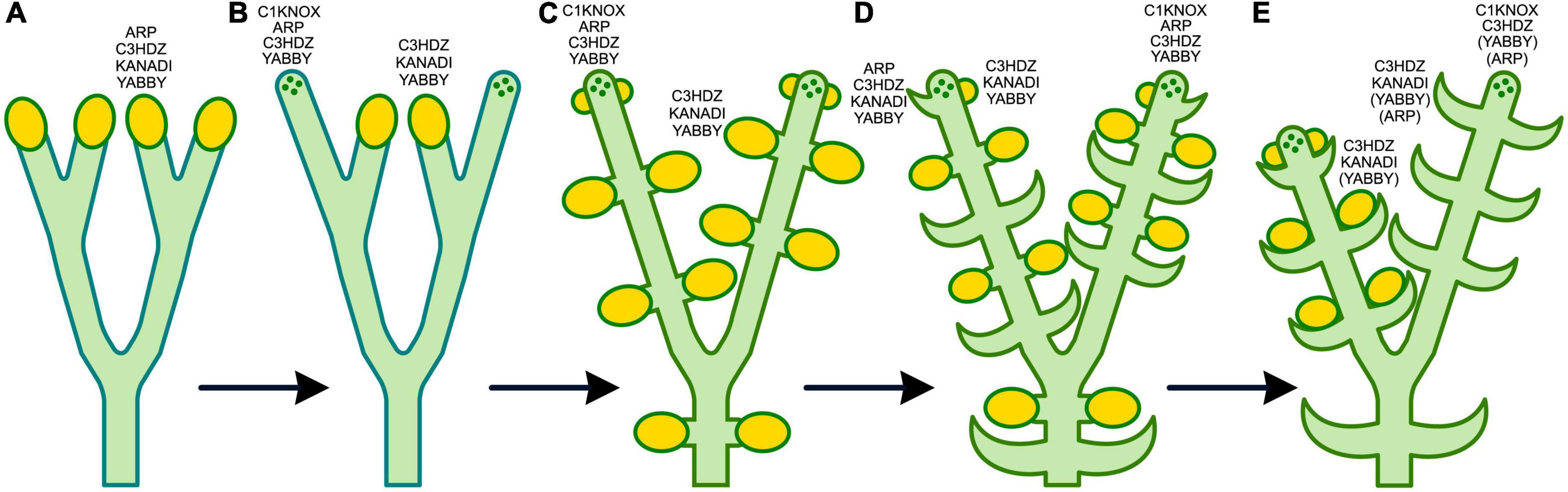
Figure 11. Hypothesized scenario for the evolutionary emergence of leaves in lycophytes. Expression data for shoot apical meristems (SAM), leaf primordia (LP) and sporangia are based on Harrison et al. (2005) for KNOX and ARP in Selaginella kraussiana, Floyd and Bowman (2006), Vasco et al. (2016) for C3HDZ in S. kraussiana and S. moellendorffii, Zumajo-Cardona et al. (2019) for KANADI in S. moellendorffii, Evkaikina et al. (2017) for YABBY in Huperzia selago. The putative regulators of the transition from one stage to the next are indicated near the organs the development of which they regulate; SAMs are dotted. (A) Isotomous axes of protracheophytes with terminal sporangia. The assumption about the putative presence of ARP, C3HDZ, KANADI, and YABBY TFs in sporangia is based on the presence of their homologs both in bryophytes and lycophytes. (B) Gain of indeterminacy by the tips of some axes through sterilization of some terminal sporangia (e.g., in Rhynia). This stage may have involved upregulation of C1KNOX expression in some shoot tips that can be interpreted as the emergence of SAM precursors. ARP, C3HDZ and YABBY TFs also could have been involved in the precursor SAM maintenance because the genes encoding the former two are present in the SAM of spikemosses and those encoding the latter in the SAM of the clubmoss H. selago. C3HDZ, KANADI and YABBY TFs could have retained their role in sporangium development, as C3HDZ and KANADI homologs are expressed in the sporangia of Selaginella, and the YABBY homolog in those of Huperzia. (C) The switch of the sporangium developmental program from dichotomy of the axes to organogenesis in the peripheral zone (PZ) of the newly emerged SAM. This change should have resulted in the displacement of sporangia from terminal to lateral positions (e.g., in Zosterophyllopsida) and could have been caused by the emergence of ARP TFs in the common ancestor of lycophytes and other plants and regulated by KNOX/ARP interaction. C1KNOX, C3HDZ and YABBY TFs could have continued to participate in the regulation of SAM maintenance, while C3HDZ, KANADI and YABBY—in the regulation of sporangium development. (D) Sterilization of some lateral sporangia (e.g., in Adoketophyton). The regulatory prerequisite for this transition is the least clear. However, based on the fact that leaves and sporangia of Selaginella species are characterized by expression of C3HDZ and KANADI homologs, whereas the expression of ARP homologs was reported only in leaves but not in sporangia, it might be hypothesized that through interaction with some yet unknown players, ARP TFs could have suppressed the differentiation of sporogenous tissues in former primordia of sporangia. Consequently the sporangium wall development program that presumably was also regulated by C3HDZ, KANADI, and YABBY TFs in the common ancestor of clubmosses and spikemosses would have been co-opted and modified into the mesophyll differentiation program. However, it should be mentioned that Harrison et al. (2005) have studied only the vegetative shoots of S. kraussiana. (E) A lycophyte with dimorphic vegetative and sporogenous shoots with sporangia on the adaxial side of leaves. C3HDZ and YABBY TFs presumably combine the regulation of the SAM maintenance, leaf and sporangium development, ARP TFs are likely involved in SAM maintenance and leaf development programs, while KANADI TFs control development of both leaf and sporangium. YABBY and ARP are in parenthesis because the former TF is found only in clubmosses, and the latter only in spikemosses.
At the same time, the expression of C3HDZ (Vasco et al., 2016) and KANADI (Zumajo-Cardona et al., 2019) homologs in the primordia of leaves and sporangia of S. moellendorffii, and of YABBY homologs in the primordia of leaves and sporangia of H. selago (Evkaikina et al., 2017) supports the sporangia-sterilization hypothesis of leaf origin in lycophytes (Kenrick and Crane, 1997; Figure 11).
The expression of KANADI homologs in the stem, but not in leaf trace vasculature in S. moellendorffii (Zumajo-Cardona et al., 2019) and the expression of different C3HDZ homologs either in stem or in leaf provascular tissues in S. kraussiana (Floyd and Bowman, 2006) suggest that differentiation of leaf veins in spikemosses differs in regulation from the stem vasculature. Together with the contribution of the stem cortex cells to the development of S. martensii leaves (Dengler, 1983), these facts indicate that the “enation” (axis cortex outgrowth sensu Bower, 1935) program was also involved in the evolutionary origin of leaves in lycophytes.
Despite the presence of the complete toolkit of adaxial and abaxial determinants in different lycophyte species, expression of neither of them is polar in LPs (Harrison et al., 2005; Vasco et al., 2016; Evkaikina et al., 2017; Zumajo-Cardona et al., 2019); the mesophyll is not differentiated into palisade and spongy parenchyma and the vascular bundles are composed from xylem surrounded by the parenchyma cells sometimes interpreted as the phloem (Figures 5, 6 and Table 1). These facts suggest that although ARP, C3HDZ, YABBY, and KANADI TFs control leaf development in lycophytes, they have not gained a role in the regulation of leaf abaxial/adaxial polarity yet.
Although T3 WOX clade members were identified in S. moellendorffii (Wu et al., 2019; Table 1; Supplementary Figure 1), they are closer to RAM-specific WOX5, than to regulators of marginal (WOX3/PRS) and plate (WOX1) meristems and their expression is not leaf-specific (Youngstrom et al., 2022). These facts suggest that WOX-regulated marginal meristems had not yet evolved in lycophytes.
Two programs could be combined during the origin of fern leaves
The last common ancestors of ferns and seed plants (e.g., Trimerophyton, Pertica, and Psilophyton) were leafless and had short dichotomizing lateral axes with terminal sporangia (Stewart and Rothwell, 1993; Harrison and Morris, 2018), while lateral axes of the palaeozoic ferns (e.g., Rhacophyton) have undergone “planation” and “webbing” with photosynthetic tissue between the terminal branchlets (Harrison and Morris, 2018; Figure 10).
The development of leaves in extant ferns also has similarities with the development of shoots: prolonged apical growth via activity of the LAI (Mueller, 1983; Hou and Hill, 2002; Romanova and Borisovskaya, 2004; Imaichi, 2008) and the absence of downregulation of C1KNOX homologs during the initiation and subsequent development of leaves (Harrison et al., 2005; Sano et al., 2005; Ambrose and Vasco, 2016). The expression of C3HDZ homologs both in LPs and the PZ of the SAM in leptosporangiate ferns (Vasco et al., 2016), and of KANADI homologs in LPs and the PZ of the SAM in horsetails (Zumajo-Cardona et al., 2019) also supports the co-option of the shoot development program for the origin of leaves in ferns.
Since C3HDZ homologs are expressed in sporangia of the whisk fern Psilotum nudum and the ophioglossioid fern Ophioglossum reticulatum, as well as in the leaves of a number of leptosporangiate ferns, they are thought to regulate both leaf and sporangium development, with sporangia regulation seen as the ancestral function (Vasco et al., 2016). Thus, the evolutionary origin of fern leaves, like that of lycophyte leaves, might have involved the modification of a sporangium-specific program in fertile axes of Paleozoic ferns (Vasco et al., 2016).
The fact that ferns possess only a single adaxial determinant (C3HDZ; Aso et al., 1999; Floyd et al., 2006; Plackett et al., 2015; Vasco et al., 2016) and a single abaxial determinant (KANADI; Zumajo-Cardona et al., 2019) out of the two present in lycophytes (Figure 10 and Table 1) suggests that the other adaxial and abaxial determinants (ARP and YABBY, respectively) could have been lost in the fern lineage (with the potential exception of Osmunda regalis where an ARP homolog was detected using antibodies raised against maize ARP; Harrison et al., 2005). However, the mesophyll in most leptosporangiate ferns is differentiated into palisade and spongy parenchyma, unlike that of lycophyte leaves, which consists of uniform spongy parenchyma (Gifford and Foster, 1989). Confinement of the expression of C3HDZ homologs to the adaxial domain of developing leaves in the leptosporangiate ferns O. regalis and Elaphoglossum peltatum (Vasco et al., 2016) and abaxial expression of KANADI homologs in leaves of the horsetail Equisetum hyemale (Zumajo-Cardona et al., 2019), despite major morphological differences of the respective leaves, suggests that these two TFs likely gained the function of leaf polarity regulation. However, the presence of bicollateral leaf vascular bundles in many leptosporangiate ferns indicates that the differentiation of ad/abaxial polarity of their mesophyll is not linked to the differentiation of the xylem and phloem of the leaf vein from a regulatory standpoint.
T3 WOX homologs exist in the leptosporangiate fern C. richardii (Nardmann and Werr, 2012), the ophioglossoid ferns O. vulgatum and Botrychium japonicum and the whisk fern P. nudum (Xia et al., 2022). However, T3 WOX (CrWUL) expression is absent from the SAM and leaves of C. richardii (Nardmann and Werr, 2012; Youngstrom et al., 2022). At the same time, C1KNOX expression is upregulated in the cells of the leaf margin that form pinnae and pinnules (Vasco and Ambrose, 2020). These facts suggest that the marginal meristem in fern leaves is likely regulated by C1KNOX, and not by T3 WOX homologs. This regulatory difference might explain the main function of the marginal meristem—initiation of pinnae and pinnulae AIs (Imaichi, 2008; Vasco et al., 2013). It can be speculated that the “partial” ad/abaxiality toolkit is sufficient for dorsoventral leaf polarity, but not for the establishment of a WOX-regulated marginal meristem.
WOX3−regulated marginal meristem appeared in gymnosperms
The modern seed plant lineage arose from progymnosperms (e.g., Aneurophyton and Archaeopteris) that, similarly to paleozoic ferns, had planated lateral axes in some instances “webbed” with laminar tissue (Stewart and Rothwell, 1993; Harrison and Morris, 2018; Figure 10). It was suggested that the independent origin of leaves in ferns and gymnosperms via modification of axes and limited lamina growth was constrained by the absence or underdevelopment of roots and low stomatal density in both lineages and high global temperatures during the Devonian (Osborne et al., 2004).
The morphological similarity between the leaves of ferns and progymnosperms is preserved in the development of leaves in cycads, the oldest extant gymnosperms, that is dominated by the leaf apical meristem. The presence of C1KNOX in the tips of Zamia integrifolia leaves and at the sites of leaflet initiation as detected using a heterologous antibody raised against maize C1KNOX (Bharathan et al., 2002) supports the similarity between fern and cycad leaf development. Interestingly, C1KNOX is not downregulated also during the initiation of needle-like leaves in conifers Pinus pinaster and Picea glauca (Bueno et al., 2020), and C1KNOX homologs are co-expressed with leaf development regulators in LPs of some gymnosperms (Nardmann et al., 2009; Hedman et al., 2013; Finet et al., 2016; Alvarez et al., 2018; Bueno et al., 2020, 2021; Du et al., 2020). These data indicate some common regulation of SAMs and leaves in conifers, like in cycads.
All gymnosperms have collateral vascular bundles in the leaves, but their mesophyll anatomy varies: differentiated into palisade and spongy parenchyma in G. gnemon, G. biloba, cycads and a number of conifers (e.g., Abies, Larix, and Tsuga), isopalisade in Welwitschia mirabilis and composed only of spongy parenchyma in most conifers (Figure 10, Table 1). Gymnosperms possess a bigger set of adaxial and abaxial determinants than ferns: homologs of C3HDZ, YABBY and KANADI TFs are present in all gymnosperms (Figure 10, Table 1, Supplementary Figures 2–4, and Supplementary Table 1), and ARP homologs are found only in cycads and conifers (Du et al., 2020). Correlation between expression of the genes encoding adaxial/abaxial determinants and the leaf anatomy is ambiguous: in leaves of Abies holophylla composed of palisade and spongy mesophyll and in Picea smithiana with only spongy and no palisade mesophyll, an ARP homolog is expressed in a stripe between leaf margins (Du et al., 2020). YABBY expression marks entire young leaves of Pseudotsuga menziesii, A. holophylla, and P. smithiana (Du et al., 2020) despite their anatomical differences, but is confined to the abaxial leaf domain in G. biloba (Finet et al., 2016; D’Apice et al., 2022). Thus ARP, C3HDZ, YABBY and KANADI TFs did not gain the strict role in control of development of the ad/abaxial leaf domains in gymnosperms (Du et al., 2018, 2020). Neither ARP, YABBY nor KANADI are expressed in the gymnosperm SAM, indicating that their function in leaf development is not combined with SAM maintenance, in contrast to lycophytes and ferns.
The important peculiarity of most gymnosperms is the important role of the marginal meristem in leaf development (Nardmann and Werr, 2013). In Gnetum gnemon (Nardmann et al., 2009), conifers Pinus pinaster and Picea abies (Alvarez et al., 2015, 2018) and in Ginkgo biloba (Nardmann and Werr, 2013), the marginal meristem is likely regulated by concerted activities of KNOX and WOX3 homologs. In Gnetum gnemon WOX3 TF seems to control also the plate meristem (Nardmann et al., 2009). However, WOX3−mediated mediolateral leaf growth is not linked to the differentiation of the leaf mesophyll into spongy and palisade parenchyma in conifers.
Diversification in the regulation of leaf meristems is the innovation of angiosperms
Development of simple leaves in angiosperms (recently reviewed by Tsukaya, 2021; Nakayama et al., 2022) starts by downregulation of C1KNOX, upregulation and ad/abaxial polarization of ARP, C3HDZ, YABBY and KANADI expression; the last three of which in turn upregulate WOX3 and WOX1 that stepwise establish the marginal and plate meristems. The regulatory apomorphies of angiosperms (relevant to the scope of this review) are the strictly polar confinement of the expression of adaxial and abaxial determinants and the WOX1-regulated plate meristem. But the innovation in the evolution of angiosperm leaf development is the regulatory diversification of different leaf meristems that resulted in great morphological and anatomical variation. For instance, reactivation of C1KNOX expression can prevent the arrest of the leaf apical meristem and, together with transcriptional peculiarities of the marginal meristem, including C1KNOX and C3HDZ upregulation, results in the production of compound leaves (e.g., in Solanum lycopersicum; Kim et al., 2003). Studies deciphering other putative changes of the basic regulatory network that underlie leaf diversity have started to emerge (reviewed by Nakayama et al., 2022). The existence of broad leaves with only spongy and lacking palisade mesophyll (e.g., in Convallaria majalis) and small leaves with isopalisade mesophyll (e.g., in Prunus scoparia) demonstrates that the expression and/or interaction of abaxial and adaxial determinants, as well as of the marginal and plate meristem regulators, can differ within angiosperms and hopefully will become the subject of future studies.
The simplex SAM may be plesiomorphic in polysporangiophytes
The SAM of one of the three extant lycophyte groups, the spikemosses, is usually classified in the monoplex structural type. However, clonal analyses have shown that no single AI or LAI is present in the SAM or at the leaf tips of S. kraussiana (Harrison et al., 2007). In ferns, in contrast to spikemosses, single AIs (AI of the SAM, LAI, AIs of leaflets, root apical initial) play an exclusive role in morphogenesis (Romanova and Jernstedt, 2005; Imaichi, 2008; Vasco et al., 2013). On the other hand, leaf initiation through divisions of a group of surface and subsurface cells in the PZ of the simplex SAM is similar in clubmosses, quillworts and gymnosperms. The ultrastructure of the monoplex SAM cells of the spikemoss S. kraussiana is more similar to that of the simplex SAM cells of the clubmoss H. selago, than to that of the monoplex SAM of the ferns Athyrium filix-femina, Dryopteris carthusiana and Pteridium aquilinum (Romanova et al., 2022). The composition of the TF toolkit in different groups—spikemosses have ARP, C3HDZ, and KANADI homologs, ferns contain C3HDZ and KANADI homologs; clubmosses possess C3HDZ and YABBY homologs and gymnosperms have ARP, C3HDZ, KANADI, and YABBY homologs—shows that both meristem types can have certain similarities and dissimilarities depending on the group of plants under examination (Figures 1, 10). SAM transcriptomes of the spikemoss S. moellendorffii and the horsetail E. arvense are less similar to each other than either of them is to that of Zea mays (Frank et al., 2015). It is worth mentioning that the SAM of Rhynia contained multiple AIs, i.e., it was simplex (Kidston and Lang, 1920; Edwards, 1994).
These facts suggest that the monoplex SAMs of spikemosses and ferns might have arisen independently, from simplex SAMs, possibly as a result of reversion to the ancestral in embryophytes single apical cell type. This reversion might have resulted from the loss of the ability to form plasmodesmata post-cytokinesis, which may have happened independently in the two groups (Imaichi and Hiratsuka, 2007). Thus, the simplex SAM might be ancestral for polysporangiophytes.
Conclusion
Since lycophytes, ferns and seed plants each had leafless fossil ancestors, leaves in these lineages originated independently (Tomescu, 2009). The cellular pattern of leaf origin within the SAM, contribution of the apical, marginal, plate and intercalary leaf meristems to the leaf development, differentiation of the mesophyll into spongy and palisade parenchyma and the structure of the leaf vascular bundles are likely homoplasic traits. However, molecular fingerprints of a common precursor program that was recruited for the origin of leaves in polysporangiophytes can be hypothesized (Figure 11). In both lycophytes and ferns, similar toolkits are used for the regulation, on the one hand, of SAM and leaves, and on the other hand, of leaves and sporangia. Therefore, leaves in both lineages may have originated via the combination of two processes, the displacement of terminal sporangia to a lateral position through modification of the shoot dichotomy program (Harrison et al., 2005), and the sterilization of some sporangia through modification of their developmental program (Vasco et al., 2016; Evkaikina et al., 2017; Zumajo-Cardona et al., 2019), respectively. The putative presence of C1KNOX proteins in SAM and LPs of Zamia integrifolia (Bharathan et al., 2002), along with the expression of the adaxial determinant GbC3HDZ1 (Floyd et al., 2006) and the abaxial determinant GibiKAN (Zumajo-Cardona et al., 2021) in the integument and nucellus of the Ginkgo biloba ovule, as well as the morphological similarity between the leaves of Paleozoic ferns and gymnosperms, suggests that the same precursor program could have been co-opted for the evolutionary origin of leaves in seed plants. Subsequently, the downstream developmental programs diversified in lycophytes, ferns and spermatophytes in major ways (Vasco et al., 2016), and should be the subjects for future research. The disparate nature of developmental and regulatory data available outside of angiosperms also indicates the need for establishment of true model species for each major plant lineage.
Author contributions
MR, KP, and OV: conceptualization. MR, VD, and AM: writing. KP and OV: review and editing. MR: funding acquisition. All authors reviewed drafts of the manuscript and approved the final draft.
Funding
This work was supported by the grant of Russian Science Foundation, https://rscf.ru/project/22-24-20049/in accordance with the agreement of 25.03.2022 (No. 22–24–20049), and grant of St. Petersburg Science Foundation in accordance with the agreement of 14.04.2022 (No. 36/2022).
Acknowledgments
We are grateful to the editor for the invitation to contribute to the Research Topic Reciprocal Illumination and Consilience in Plant Evolutionary-Development. We thank Peter Romanov (Saint Petersburg) for help with the illustrations, Natalia Ivanova (Department of Botany, Saint Petersburg State University) for the help with hand sections, Yury Ivanenko (Department of Botany, Saint Petersburg State University) for helpful discussions about plant phylogeny, and Ianina Bogdanova (Komarov Botanical Institute, Saint Petersburg) for the kind gift of Gnetum apex sections. We are grateful to the reviewers and AT for the rigorous and constructive review and all the advice that helped to improve the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1097115/full#supplementary-material
Supplementary Figure 1 | Phylogeny of WOX proteins.
Supplementary Figure 2 | Phylogeny of C3HDZ proteins.
Supplementary Figure 3 | Phylogeny of YABBY proteins.
Supplementary Figure 4 | Phylogeny of KANADI proteins.
Supplementary Table 1 | List of homologs of WOX, C3HDZ, YABBY, and KANADI TFs that are referred to in the text and summarized in Table 1. For the angiosperms only model plants for which the developmental genetics data are available are included; for the gymnosperms the homologs were searched in all the genomes available in public databases, transcriptomic data are included only for the groups that lack genomic data; for the seed-free plants most available from public databases genomic and transcriptomic data are included. Homologs found in the genomes are in bold. For every homolog accession numbers and sources are listed. The list of ARP homologs is available from Hernández-Hernández et al. (2021; https://www.journals.uchicago.edu/doi/suppl/10.1086/710579/suppl_file/44514Appendix.pdf), only the homologs absent from this list are included. The list of KNOX homologs is available from Vasco and Ambrose (2020); 10.6084/m9.figshare.1257658.
Supplementary File 1 | Sequences used for bioinformatic search of ARP, WOX, C3HDZ, YABBY, and KANADI TFs, databases used and description of bioinformatics methods.
References
Ali, S., Khan, N., and Xie, L. (2020). Molecular and hormonal regulation of leaf morphogenesis in Arabidopsis. Int. J. Mol. Sci. 2:5132.
Alvarez, J. M., Bueno, N., Cañas, R. A., Avila, C., Cánovas, F. M., and Ordás, R. J. (2018). Analysis of the Wuschel-related homeobox gene family in Pinus pinaster: new insights into the gene family evolution. Plant Physiol. Biochem. 123, 304–318. doi: 10.1016/j.plaphy.2017.12.031
Alvarez, J. M., Sohlberg, J., Engsrom, P., Zhu, T., Englund, M., Moschou, P. N., et al. (2015). The WUSCHEL-related Homeobox3 gene PaWOX3 regulates lateral organ formation in norway spruce. New Phytol. 208, 1078–1088. doi: 10.1111/nph.13536
Alvarez, J. P., Furumizu, C., Efroni, I., Eshed, Y., and Bowman, J. L. (2016). Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 5:e15023. doi: 10.7554/eLife.15023
Ambrose, B. A., and Vasco, A. (2016). Bringing the multicellular fern meristem into focus. New Phytol. 210, 790–793. doi: 10.1111/nph.13825
Aso, K., Kato, M., Banks, J. A., and Hasebe, M. (1999). Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Mol. Biol. Evol. 16, 544–552. doi: 10.1093/oxfordjournals.molbev.a026135
Bharathan, G., Goliber, T. E., Moore, C., Kessler, S., Pham, T., and Sinha, N. R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296, 1858–1860. doi: 10.1126/science.1070343
Bierhorst, D. W. (1968). On the Stromatopteridaceae (fam. nov.) and on the Psilotaceae. Phytomorphol. 16, 232–268.
Bierhorst, D. W. (1977). The systematic position of Psilotum and Tmesipteris. Brittonia 29, 3–13. doi: 10.2307/2805738
Bower, F. O. (1935). Primitive Land Plants-also Known as the Archegoniatae. London: MacMillan and Co.
Bowman, J. L., Briginshaw, L. N., and Florent, S. N. (2019). Evolution and co-option of developmental regulatory networks in early land plants. Curr. Top. Dev. Biol. 131, 35–54.
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha Genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
Bowman, J. L., and Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix- loop-helix domains. Development 126, 2387–2396. doi: 10.1242/dev.126.11.2387
Boyce, C. K. (2005). Patterns of segregation and convergence in the evolution of fern and seed plant leaf morphologies. Paleobiology 31, 117–149.
Boyce, C. K. (2010). The evolution of plant development in a paleontological context. Curr. Opin. Plant Biol. 13, 102–107. doi: 10.1016/j.pbi.2009.10.001
Boyce, C. K., and Knoll, A. H. (2002). Evolution of developmental potential and the multiple independent origins of leaves in Paleozoic vascular plants. Paleobiology 28, 70–100.
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Briginshaw, L. N., Flores-Sandoval, E., Dierschke, T., Alvarez, J. P., and Bowman, J. L. (2022). KANADI promotes thallus differentiation and FR-induced gametangiophore formation in the liverwort Marchantia. New Phytol. 234, 1377–1393. doi: 10.1111/nph.18046
Bueno, N., Alvarez, J. M., and Ordás, R. J. (2020). Characterization of the KNOTTED1-LIKE HOMEOBOX (KNOX) gene family in Pinus pinaster Ait. Plant Sci. 301:110691. doi: 10.1016/j.plantsci.2020.110691
Bueno, N., Cuesta, C., Centeno, M. L., Ordás, R. J., and Alvarez, J. M. (2021). In vitro plant regeneration in conifers: the role of WOX and KNOX gene families. Genes 12:438.
Byrne, M. E., Barley, R., Curtis, M., Arroyo, J. M., Dunham, M., Hudson, A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. doi: 10.1038/35050091
Caggiano, M. P., Yu, X., Bhatia, N., Larsson, A., Ram, H., Ohno, C. K., et al. (2017). Cell type boundaries organize plant development. eLife 6:e27421.
Champagne, C. E. M., and Ashton, N. W. (2001). Ancestry of KNOX genes revealed by bryophyte (Physcomitrella patens) homologs. New Phytol. 150, 23–36. doi: 10.1046/j.1469-8137.2001.00076.x
Crane, P., and Kenrick, P. (1997). Diverted development of reproductive organs: a source of morphological innovation in land plants. Plant Syst. Evol. 206, 161–174.
Cruz, R., Prado, J., and de Albuquerque Melo-de-Pinna, G. F. (2020). Leaf development in some ferns with variable dissection patterns (Dryopteridaceae and Lomariopsidaceae). Flora 270:151658.
D’Apice, G., Moschin, S., Nigris, S., Ciarle, R., Muto, A., Bruno, L., et al. (2022). Identification of key regulatory genes involved in the sporophyte and gametophyte development in Ginkgo biloba ovules revealed by in situ expression analyses. Am. J. Bot. 109, 887–898. doi: 10.1002/ajb2.1862
Dengler, N. G. (1983). The developmental basis of anisophylly in Selaginella martensii. i. initiation and morphology of growth. Am. J. Bot. 70, 181–192.
Deveaux, Y., Toffano-Nioche, C., Claisse, G., Thareau, V., Morin, H., Laufs, P., et al. (2008). Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 8:291. doi: 10.1186/1471-2148-8-291
Dierschke, T., Flores-Sandoval, E., Rast-Somssich, M. I., Althoff, F., Zachgo, S., and Bowman, J. L. (2021). Gamete expression of TALE class HD genes activates the diploid sporophyte program in Marchantia polymorpha. eLife 10:e57088. doi: 10.7554/eLife.57088
DiGennaro, P., Grienenberger, E., Dao, T. Q., Jun, J. H., and Fletcher, J. C. (2018). Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct 2:e00103. doi: 10.1002/pld3.103
Dörken, V. M. (2013). Morphology, anatomy and vasculature in leaves of Ginkgo biloba L. (Ginkgoaceae, Ginkgoales) under functional and evolutionary aspects. Feddes Repert. 124, 80–97.
Dörken, V. M. (2014). Leaf-morphology and leaf-anatomy in Ephedra altissima desf. (Ephedraceae, Gnetales) and their evolutionary relevance. Feddes Repert. 123, 243–255.
Du, F., Guan, C., and Jiao, Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11, 1117–1134.
Du, H., Ran, J., Feng, Y., and Wang, X. (2020). The flattened and needlelike leaves of the pine family (Pinaceae) share a conserved genetic network for adaxial-abaxial polarity but have diverged for photosynthetic adaptation. BMC Evol. Biol. 20:131. doi: 10.1186/s12862-020-01694-5
Edwards, D. (1994). “Towards an understanding of pattern and process in the growth of early land plants,” in Shape and Form in Plants and Fungi, eds D. S. Ingram and A. Hudson (London: Academic Press), 39–59.
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., and Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131, 2997–3006. doi: 10.1242/dev.01186
Evkaikina, A. I., Berke, L., Romanova, M. A., Proux-Wéra, E., Ivanova, A. N., Rydin, C., et al. (2017). The Huperzia selago shoot tip transcriptome sheds new light on the evolution of leaves. Genome Biol. Evol. 9, 2444–2460. doi: 10.1093/gbe/evx169
Finet, C., Floyd, S. K., Conway, S. J., Zhong, B., Scutt, C. P., and Bowman, J. L. (2016). Evolution of the YABBY gene family in seed plants. Evol. Dev. 18, 116–126.
Fletcher, L. C., Brand, U., Running, M. P., Simon, R., and Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Floyd, S. K., and Bowman, J. L. (2006). Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr. Biol. 16, 1911–1917. doi: 10.1016/j.cub.2006.07.067
Floyd, S. K., and Bowman, J. L. (2007). The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 168, 1–35.
Floyd, S. K., and Bowman, J. L. (2010). Gene expression patterns in seed plant shoot meristems and leaves: homoplasy or homology? J. Plant Res. 123, 43–55.
Floyd, S. K., Ryan, J. G., Conway, S. J., Brenner, E., Burris, K. P., Burris, J. N., et al. (2014). Origin of a novel regulatory module by duplication and degeneration of an ancient plant transcription factor. Mol. Phylogenet. Evol. 81, 159–173. doi: 10.1016/j.ympev.2014.06.017
Floyd, S. K., Zalewski, C. S., and Bowman, J. L. (2006). Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173, 373–388. doi: 10.1534/genetics.105.054239
Foster, A. S. (1938). Structure and growth of the shoot apex in Ginkgo biloba. Bull. Torrey Bot. Club 65, 531–556.
Foster, A. S. (1939). Structure and growth of the shoot apex of Cycas revoluta. Am. J. Bot. 26, 372–385.
Foster, A. S. (1943). Zonal structure and growth of the shoot apex in Microcycas calocoma (Miq.) A. DC. Am. J. Bot. 30, 56–73.
Frank, M. H., Edwards, M. B., Schultz, E. R., McKain, M. R., Fei, Z., Sørensen, I., et al. (2015). Dissecting the molecular signatures of apical cell-type shoot meristems from two ancient land plant lineages. New Phytol. 207, 893–904. doi: 10.1111/nph.13407
Ge, Y., Liu, J., Zeng, M., He, J., Qin, P., Huang, H., et al. (2016). Identification of WOX family genes in Selaginella kraussiana for studies on stem cells and regeneration in lycophytes. Front. Plant Sci. 7:93. doi: 10.3389/fpls.2016.00093
Gensel, P. G. (1977). Morphologic and taxonomic relationships of the Psilotaceae relative to evolutionary lines in early land vascular plants. Brittonia 29, 14–29. doi: 10.2307/2805739
Gifford, E. M., and Foster, A. S. (1989). Morphology and Evolution of Vascular Plants. New York: W. H. Freeman.
Gola, E. M., and Jernstedt, J. A. (2011). Impermanency of initial cells in Huperzia lucidula (Huperziaceae) shoot apices. Int. J. Plant. Sci. 172, 847–855. doi: 10.1086/660878
Golub, S. J., and Wetmore, R. H. (1948a). Studies of development in the vegetative shoot of Equisetum arvense L. I. the shoot apex. Am. J. Bot. 35, 755–767. doi: 10.1002/j.1537-2197.1948.tb08146.x
Golub, S. J., and Wetmore, R. H. (1948b). Studies of development in the vegetative shoot of Equisetum arvense L. II. the mature shoot. Am. J. Bot. 35, 767–781. doi: 10.1002/j.1537-2197.1948.tb08147.x
Guo, M., Thomas, J., Collins, G., and Timmermans, M. C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20, 48–58. doi: 10.1105/tpc.107.056127
Hara, N. (1980). Morphological study on early ontogeny of the Ginkgo leaf. Bot. Mag. Tokyo 93, 1–12. doi: 10.1007/BF02489482
Harris, B. J., Clark, J. W., Schrempf, D., Szöllõsi, G. J., Donoghue, P. C. J., Hetherington, A. M., et al. (2022). Divergent evolutionary trajectories of bryophytes and tracheophytes from a complex common ancestor of land plants. Nat. Ecol. Evol. 6, 1634–1643. doi: 10.1038/s41559-022-01885-x
Harrison, C. J. (2017). Development and genetics in the evolution of land plant body plans. Phil. Trans. R. Soc. B. Biol. Sci. 372:20150490. doi: 10.1098/rstb.2015.0490
Harrison, C. J., Corley, S. B., Moylan, E. C., Alexander, D. L., Scotland, R. W., and Langdale, J. A. (2005). Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature 434, 509–514. doi: 10.1038/nature03410
Harrison, C. J., and Langdale, J. A. (2010). Comment: the developmental pattern of shoot apices in Selaginella kraussiana (Kunze) a. Braun. Int. J. Plant Sci. 171, 690–692. doi: 10.1086/653134
Harrison, C. J., and Morris, J. L. (2018). The origin and early evolution of vascular plant shoots and leaves. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20160496. doi: 10.1098/rstb.2016.0496
Harrison, C. J., Rezvani, M., and Langdale, J. A. (2007). Growth from two transient apical initials in the meristem of Selaginella kraussiana. Development 134, 881–889. doi: 10.1242/dev.001008
Hedman, H., Zhu, T., von Arnold, S., and Sohlberg, J. J. (2013). Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in the conifer Picea abies reveals extensive conservation as well as dynamic patterns. BMC Plant Biol. 13:89. doi: 10.1186/1471-2229-13-89
Heisler, M. G., and Byrne, M. E. (2020). Progress in understanding the role of auxin in lateral organ development in plants. Curr. Opin. Plant Biol. 53, 73–79.
Hernández-Hernández, B., Tapia-López, R., Ambrose, B. A., and Vasco, A. (2021). R2R3-MYB gene evolution in plants, incorporating ferns into the story. Int. J. Plant Sci. 182, 1–8.
Hirakawa, Y. (2022). Evolution of meristem zonation by CLE gene duplication in land plants. Nat. Plants 8, 735–740. doi: 10.1038/s41477-022-01199-7
Hjortswang, H. I., Sundås-Larsson, A., Bharathan, G., Bozhkov, P. V., Arnold, S., and Vahala, T. (2002). KNOTTED1-like homeobox genes of a gymnosperm, Norway spruce, expressed during somatic embryogenesis. Plant Physiol. Biochem. 40, 837–843.
Hou, G., and Hill, J. P. (2002). Heteroblastic root development in Ceratopteris richardii (Parkeriaceae). Int. J. Plant Sci. 163, 341–351.
Huang, C. H., Qi, X., Chen, D., Qi, J., and Ma, H. (2020). Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. J. Integrat. Plant Biol. 62, 433–455. doi: 10.1111/jipb.12877
Imaichi, R. (2008). “Meristem organization and organ diversity,” in Biology and Evolution of Ferns and Lycophytes, eds T. A. Ranker and C. H. Haufler (Cambridge: Cambridge University Press), 75–103.
Imaichi, R., and Hiratsuka, R. (2007). Evolution of shoot apical meristem structures in vascular plants with respect to plasmodesmatal network. Am. J. Bot. 94, 1911–1921.
Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413.
Jones, C. S., and Drinnan, A. N. (2009). The developmental pattern of shoot apices in Selaginella kraussiana (Kunze) A. Braun. Int. J. Plant Sci. 170, 1009–1018.
Kaplan, D. R. (1977). Morphological status of the shoot systems of Psilotaceae. Brittonia 29, 30–53.
Kaplan, D. R. (2001). The science of plant morphology: definition, history, and role in modern biology. Am. J. Bot. 88, 1711–1741.
Kaplan, D. R., and Groff, P. A. (1995). “Developmental themes in vascular plants: functional and evolutionary significance”, in Experimental and Molecular Approaches to Plant Biosystematics, eds P. C. Hoch and A. D. Stephenson (Missouri: Missouri Botanical Garden)
Kawai, J., Tanabe, Y., Soma, S., and Ito, M. (2010). Class 1 KNOX gene expression supports the Selaginella rhizophore concept. J. Plant Biol. 53, 268–274.
Kenrick, P., and Crane, P. (1997). The Origin and Early Diversification of Land Plants: A Cladistic Study. Washington and London: Smithsonian Institution Scholarly Press.
Kidston, R., and Lang, W. H. (1920). On old red sandstone plants showing structure, from the Rhynie Chert Bed, Aberdeenshire. Part III. Asteroxylon mackiei, Kidston and Lang. Trans. R. Soc. Edinb. 52, 643–680. doi: 10.1017/S0080456800004506
Kim, M., Pham, T., Hamidi, A., McCormick, S., Kuzoff, R. K., and Sinha, N. (2003). Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130, 4405–4415. doi: 10.1242/dev.00655
Larsson, E., Sitbon, F., and von Arnold, S. (2012). Differential regulation of Knotted1-like genes during establishment of the shoot apical meristem in Norway spruce (Picea abies). Plant Cell. Rep. 31, 1053–1060. doi: 10.1007/s00299-011-1224-6
Li, F. W., Nishiyama, T., Waller, M., et al. (2020). Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants 6, 259–272. doi: 10.1038/s41477-020-0618-2
Liu, H., Wang, Q. F., and Taylor, W. C. (2006). Morphological and anatomical variation in sporophylls of Isoetes sinensis palmer (Isoetaceae), an endangered quillwort in China. Am. Fern J. 96, 67–74.
Long, J. A., Moan, E. I., Medford, J. I., and Barton, M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. doi: 10.1038/379066a0
Maksimova (Evkaikina), A. I., Berke, L., Salgado, M. G., Klimova, E. A., Pawlowski, K., Romanova, M. A., et al. (2021). What can the phylogeny of class I KNOX genes and their expression patterns in land plants tell us about the evolution of shoot development? Bot. J. Linn. Soc. 195, 254–280.
Manuela, D., and Xu, M. (2020). Juvenile leaves or adult leaves: determinants for vegetative phase change in flowering plants. Int. J. Mol. Sci. 21:9753.
Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815.
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713.
Morris, J. L., Puttick, M. N., Clark, J. W., Edwards, D., Kenrick, P., Pressel, S., et al. (2018). The timescale of early land plant evolution. Proc. Natl Acad. Sci. U S A. 115, 2274–2283.
Mueller, R. J. (1983). Indeterminate growth and ramification of the climbing leaves of Lygodium japonicum (Schizaeaceae). Am. J. Bot. 70, 682–690.
Nakata, M., Matsumoto, N., Tsugeki, R., Rikirsch, E., Laux, T., and Okada, K. (2012). Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24, 519–535. doi: 10.1105/tpc.111.092858
Nakayama, H., Leichty, A. R., and Sinha, N. R. (2022). Molecular mechanisms underlying leaf development, morphological diversification, and beyond. Plant Cell 34, 2534–2548. doi: 10.1093/plcell/koac118
Napp-Zinn, K. (1966). Anatomie des Blattes. Blattanatomie der Gymnospermen. Berlin: Gebr. Borntraeger.
Nardmann, J., Reisewitz, P., and Werr, W. (2009). Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol. Biol. Evol. 26, 1745–1755. doi: 10.1093/molbev/msp084
Nardmann, J., and Werr, W. (2012). The invention of WUS-like stem cell-promoting functions in plants predates leptosporangiate ferns. Plant Mol. Biol. 78, 123–134. doi: 10.1007/s11103-011-9851-4
Nardmann, J., and Werr, W. (2013). Symplesiomorphies in the WUSCHEL clade suggest that the last common ancestor of seed plants contained at least four independent stem cell niches. New Phytol. 199, 1081–1092. doi: 10.1111/nph.12343
Naumenko, A. N., and Romanova, M. A. (2008). Apical morphogenesis of Psilotum nudum (Psilotaceae) and Botrychium lunaria (Ophioglossaceae). Vestn. SPb Univ. 3, 15–27.
Newman, I. V. (1965). Pattern in the meristems of vascular plants: III. pursuing the patterns in the apical meristem where no cell is a permanent cell. J. Linn. Soc. Lond. Bot. 59, 185–214.
Nimchuk, Z. L., Tarr, P. T., Ohno, C., Qu, X., and Meyerowitz, E. M. (2011). Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21, 345–352. doi: 10.1016/j.cub.2011.01.039
Osborne, C., Beerling, D., Lomax, B., and Chaloner, W. (2004). Biophysical constraints on the origin of leaves inferred from the fossil record. Proc. Natl Acad. Sci. U S A. 101, 10360–10362. doi: 10.1073/pnas.0402787101
Pham, T., and Sinha, N. (2003). Role of KNOX genes in shoot development of Welwitschia mirabilis. Int. J. Plant Sci. 164, 333–343.
Philipson, W. R. (1990). The significance of apical meristems in the phylogeny of land plants. Plant Syst. Evol. 173, 17–38.
Plackett, A. R. G., Di Stillio, V. S., and Langdale, J. A. (2015). Ferns: the missing link in shoot evolution and development. Front. Plant Sci. 6:5328. doi: 10.3389/fpls.2015.00972
Popham, R. A. (1951). Principal types of vegetative shoot apex organization in vascular plants. Ohio J. Sci. 51, 249–270.
PPG I (2016). A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 54, 563–603. doi: 10.1111/jse.12229
Prigge, M., and Clark, S. (2006). Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8, 350–361. doi: 10.1111/j.1525-142X.2006.00107.x
Qi, X., Kuo, L. Y., Guo, C., Li, H., Li, Z., Qi, J., et al. (2018). A well-resolved fern nuclear phylogeny reveals the evolution history of numerous transcription factor families. Mol. Phylogenet. Evol. 127, 961–977. doi: 10.1016/j.ympev.2018.06.043
Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260.
Rodin, R. J. (1958a). Leaf anatomy of Welwitschia. I. early development of the leaf. Am. J. Bot. 45, 90–95.
Rodin, R. J. (1958b). Leaf anatomy of Welwitschia. II. a study of mature leaves. Am. J. Bot. 45, 96–103.
Romani, F., Reinheimer, R., Florent, S. N., Bowman, J. L., and Moreno, J. E. (2018). Evolutionary history of HOMEODOMAIN LEUCINE ZIPPER transcription factors during plant transition to land. New Phytol. 219, 408–421. doi: 10.1111/nph.15133
Romanova, M., and Jernstedt, J. (2005). Morphogenetic events in the Ceratopteris richardii shoot apex. Fern Gaz. 17:204.
Romanova, M. A., and Borisovskaya, G. M. (2004). Principles of structural organization of the vegetative body in ferns: ontogenetic approach. Bot. Zhurn. 89, 705–717.
Romanova, M. A., Maksimova, A. I., Pawlowski, K., and Voitsekhovskaja, O. V. (2021). YABBY genes in the development and evolution of land plants. Int. J. Mol. Sci. 22:4139. doi: 10.3390/ijms22084139
Romanova, M. A., Yakovleva, O. V., Maksimova, A. I., Ivanova, A. N., and Domashkina, V. V. (2022). Structure of shoot apical meristems and peculiarities of ultrastructure of their cells in lycophytes and ferns. Bot. Zhurn. 107, 65–85.
Rothfels, C. J., Li, F. W., Sigel, E. M., Huiet, L., Larsson, A., Burge, D. O., et al. (2015). The evolutionary history of ferns inferred from 25 low-copy nuclear genes. Am. J. Bot. 102, 1089–1107. doi: 10.3732/ajb.1500089
Rothwell, G. W. (1999). Fossils and in the resolution of land plant phylogeny. Bot. Rev. 65, 188–218.
Sakakibara, K., Nishiyama, T., Deguchi, H., and Hasebe, M. (2008). Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol. Dev. 10, 555–566.
Sakakibara, K., Reisewitz, P., Aoyama, T., Friedrich, T., Ando, S., and Sato, Y. (2014). WOX13-like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss Physcomitrella patens. Development 141, 1660–1670. doi: 10.1242/dev.097444
San Martin, J. A. B., Pozner, R. E., and Di Stilio, V. S. (2022). Heterochrony and repurposing in the evolution of gymnosperm seed dispersal units. Evodevo 13:7. doi: 10.1186/s13227-022-00191-8
Sano, R., Juárez, C. M., Hass, B., Sakakibara, K., Ito, M., Banks, J. A., et al. (2005). KNOX homeobox genes potentially have similar function in both diploid unicellular and multicellular meristems, but not in haploid meristems. Evol. Dev. 7, 69–78. doi: 10.1111/j.1525-142X.2005.05008.x
Sarojam, R., Sappl, P. G., Goldshmidt, A., Efroni, I., Floyd, S. K., Eshed, Y., et al. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22, 2113–2130. doi: 10.1105/tpc.110.075853
Sarvepalli, K., Das Gupta, M., Challa, K. R., and Nath, U. (2019). Molecular cartography of leaf development – role of transcription factors. Curr. Opin. Plant Biol. 47, 22–31. doi: 10.1016/j.pbi.2018.08.002
Satterlee, J. W., and Scanlon, M. J. (2019). Coordination of leaf development across developmental axes. Plants 8:433.
Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E. H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. doi: 10.1101/gad.13.9.1079
Schneider, H. (2013). “Evolutionary morphology of ferns (Monilophytes),” in Annual Plant Reviews, Vol. 45, eds B. A. Ambrose and M. D. Purugganan (New Jersey: John Wiley and Sons).
Schneider, H., Pryer, K. M., Cranfill, R., Smith, A. R., and Wolf, P. G. (2002). “Evolution of vascular plant body plans: a phylogenetic perspective,” in Developmental Genetics and Plant Evolution, eds Q. C. B. Cronk, R. M. Bateman, and J. A. Hawkins (London: Taylor & Francis).
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/s0092-8674(00)80700-x
Segatto, A. L., Thompson, C. E., and Freitas, L. B. (2016). Molecular evolution analysis of WUSCHEL-related homeobox transcription factor family reveals functional divergence among clades in the homeobox region. Dev. Genes Evol. 226, 259–268.
Shi, B., and Vernoux, T. (2019). Patterning at the shoot apical meristem and phyllotaxis. Curr. Top. Dev. Biol. 131, 81–107.
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., and Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. doi: 10.1242/dev.126.18.4117
Skupchenko, V. B. (2019). Cell growth and proliferation in ground tissue of developing terminal shoot in Picea abies (Pinaceae). Rastitelnye Resursy 55, 195–212.
Skupchenko, V. B., and Ladanova, N. V. (1984). The development of Picea obovata (Pinaceae) needles. Bot. Zhurn. 69, 203–206.
Spencer, V., Venza, Z. N., and Harrison, C. J. (2021). What can lycophytes teach us about plant evolution and development? modern perspectives on an ancient lineage. Evol. Dev. 23, 174–196. doi: 10.1111/ede.12350
Sterling, J. (1984). SAM The structure of the apical meristem of Isoetes engelmannii, I. riparia and I. macrospora (Isoetales). Bot. J. Linn. Soc. 89, 77–84.
Stevenson, D. W. (1976a). Observations on phyllotaxis, stelar morphology, the shoot apex and gemmae of Lycopodium lucidulum Michaux (Lycopodiaceae). Bot. J. Linn. Soc. 72, 81–100.
Stevenson, D. W. (1976b). The cytohistological and cytohistochemical zonation of the shoot apex of Botrychium multifidum. Am. J. Bot. 63, 852–856.
Stevenson, D. W. (1978). Observations on shoot apices of eusporangiate ferns. Kew Bull. 33, 279–282.
Stevenson, D. W. (1990). Morphology and systematics of the Cycadales. Mem. N. Y. Bot. Gard. 57, 8–55.
Stevenson, D. W. (2020). Observations on vegetative branching in Cycads. Int. J. Plant Sci. 181, 564–580.
Stewart, W. N., and Rothwell, G. W. (1993). Paleobotany and the Evolution of Plants. Cambridge: Cambridge University Press.
Sundås-Larsson, A., Svenson, M., Liao, H., and Engström, P. (1998). A homeobox gene with potential developmental control function in the meristem of the conifer Picea abies. Proc. Natl. Acad. Sci. U S A. 95, 15118–15122. doi: 10.1073/pnas.95.25.15118
Szovenyi, P., Waller, M., and Kirbis, A. (2019). Evolution of the plant body plan. Curr. Top. Devel. Biol. 131, 1–34.
Takiguchi, Y., Imaichi, R., and Kato, M. (1997). Cell division patterns in the apices of subterranean axis and aerial shoot of Psilotum nudum (Psilotaceae): morphological and phylogenetic implications for the subterranean axis. Am. J. Bot. 84, 588–596.
Timmermans, M. C., Hudson, A., Becraft, P. W., and Nelson, T. (1999). ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284, 151–153. doi: 10.1126/science.284.5411.151
Tomescu, A. M. F. (2009). Megaphylls, microphylls and the evolution of leaf development. Trends Plant Sci. 14, 5–12.
Tomescu, A. M. F. (2011). “The sporophytes of seed-free vascular plants - major vegetative developmental features and molecular genetic pathways,” in Working with Ferns. Issues and Applications, eds H. Fernandez, A. Kumar, and M. A. Revilla (New York, NY: Springer), 67–94.
Tomescu, A. M. F., Escapa, I. H., Rothwell, G. W., Elgorriaga, A., and Cúneo, N. R. (2017). Developmental programmes in the evolution of Equisetum reproductive morphology: a hierarchical modularity hypothesis. Ann. Bot. 119, 489–505. doi: 10.1093/aob/mcw273
Tomescu, A. M. F., and Matsunaga, K. K. S. (2019). “Polar auxin transport and plant sporophyte body plans,” in Reference Module in Life Sciences, ed. B. D. Roitberg (Amsterdam: Elsevier).
Tomescu, A. M. F., Wyatt, S. E., Hasebe, M., and Rothwell, G. W. (2014). Early evolution of the vascular plant body plan – the missing mechanisms. Curr. Opin. Plant Biol. 17, 126–136. doi: 10.1016/j.pbi.2013.11.016
Tomlinson, P. B., and Fisher, J. B. (2005). Development of nonlignified fibers in leaves of Gnetum gnemon (Gnetales). Am. J. Bot. 92, 383–389. doi: 10.3732/ajb.92.3.383
Tsiantis, M., Schneeberger, R., Golz, J. F., Freeling, M., and Langdale, J. A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284, 154–156. doi: 10.1126/science.284.5411.154
Tsukaya, H. (2018). Leaf shape diversity with an emphasis on leaf contour variation, their developmental background, and their adaptive meanings. Semin. Cell Dev. Biol. 79, 48–57. doi: 10.1016/j.semcdb.2017.11.035
Tsukaya, H. (2021). The leaf meristem enigma: the relationship between the plate meristem and the marginal meristem. Plant Cell 33, 3194–3206. doi: 10.1093/plcell/koab190
van der Graaff, E., Laux, T., and Rensing, S. A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10:248.
Vasco, A., and Ambrose, B. A. (2020). Simple and divided leaves in ferns: exploring the genetic basis for leaf morphology differences in the genus Elaphoglossum (Dryopteridaceae). Int. J. Mol. Sci. 21:5180. doi: 10.3390/ijms21155180
Vasco, A., Moran, R. C., and Ambrose, B. A. (2013). The evolution, morphology and development of fern leaves. Front. Plant Sci. 4:345. doi: 10.1111/j.1469-185X.1941.tb01096.x
Vasco, A., Smalls, T. L., Graham, S. W., Cooper, E. D., Wong, G. K., Stevenson, D. W., et al. (2016). Challenging the paradigms of leaf evolution: class III HD-Zips in ferns and lycophytes. New Phytol. 212, 745–758. doi: 10.1111/nph.14075
Vovides, A. P., Pérez-Farrera, M. A., Gutiérrez-Ortega, J. S., Stevenson, D. W., González, D., Avendaño, S., et al. (2020). “Cycad anatomy: Old techniques solving new problems”, in Proceedings of Cycad 2015, the 10th International Conference on Cycad Biology, Chapter: 4, 33–44.
Waites, R., Selvadurai, H. R., Oliver, I. R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. doi: 10.1016/s0092-8674(00)81439-7
Wan, T., Liu, Z. M., Li, L. F., Leitch, A. R., Leitch, I. J., and Lohaus, et al. (2018). A genome for gnetophytes and early evolution of seed plants. Nat. Plants 4, 82–89.
Wardlaw, C. W. (1963). Experimental studies of the sporophytes of ferns. J. Linn. Soc. Lond. Bot. 8, 385–400.
White, R. (1971). Experimental and developmental studies of the fern sporophyte. Bot. Rev. 37, 509–540.
Williams, J. H. (2012). Pollen tube growth rates and the diversification of flowering plant reproductive cycles. Int. J. Plant Sci. 173, 649–661. doi: 10.1073/pnas.0800036105
Wu, C. C., Li, F. W., and Kramer, E. M. (2019). Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS One 14:e0223521. doi: 10.1371/journal.pone.0223521
Xia, Z., Liu, L., Wei, Z., Wang, F., Shen, H., and Yan, Y. (2022). Analysis of comparative transcriptome and positively selected genes reveal adaptive evolution in leaf-less and root-less whisk ferns. Plants 11:1198. doi: 10.3390/plants11091198
Yamaguchi, T., Nukazuka, A., and Tsukaya, H. (2012). Leaf adaxial-abaxial polarity specification and lamina outgrowth: evolution and development. Plant Cell Physiol. 53, 1180–1194. doi: 10.1093/pcp/pcs074
Yang, J. H., and Wang, H. (2016). Molecular mechanisms for vascular development and secondary cell wall formation. Front. Plant Sci. 7:356. doi: 10.3389/fpls.2016.00356
Yang, M., You, W., Wu, S., Fan, Z., Xu, B., Zhu, M., et al. (2017). Global transcriptome analysis of Huperzia serrata and identification of critical genes involved in the biosynthesis of huperzine A. BMC Genom. 18:245. doi: 10.1186/s12864-017-3615-8
Yip, H. K., Floyd, S. K., Sakakibara, K., and Bowman, J. L. (2016). Class III HD-Zip activity coordinates leaf development in Physcomitrella patens. Dev. Biol. 419, 184–197. doi: 10.1016/j.ydbio.2016.01.012
Youngstrom, C. E., Withers, K. A., Irish, E. E., and Cheng, C. (2022). Vascular function of the T3/modern clade WUSCHEL-Related HOMEOBOX transcription factor genes predate apical meristem-maintenance function. BMC Plant Biol. 22:210. doi: 10.1186/s12870-022-03590-0
Yu, T., Guan, C., Wang, J., Sajjad, M., Ma, L., and Jiao, Y. (2017). Dynamic patterns of gene expression during leaf initiation. J. Genet. Genom. 44, 599–601.
Zumajo-Cardona, C., and Ambrose, B. A. (2020). Phylogenetic analyses of key developmental genes provide insight into the complex evolution of seeds. Mol. Phylogenet. Evol. 147:106778. doi: 10.1016/j.ympev.2020.106778
Zumajo-Cardona, C., Little, D. P., Stevenson, D., and Ambrose, B. A. (2021). Expression analyses in Ginkgo biloba provide new insights into the evolution and development of the seed. Sci. Rep. 11:21995. doi: 10.1038/s41598-021-01483-0
Keywords: SAM, leaf evolution, WOX, ARP, C3HDZ, KANADI, YABBY
Citation: Romanova MA, Domashkina VV, Maksimova AI, Pawlowski K and Voitsekhovskaja OV (2023) All together now: Cellular and molecular aspects of leaf development in lycophytes, ferns, and seed plants. Front. Ecol. Evol. 11:1097115. doi: 10.3389/fevo.2023.1097115
Received: 13 November 2022; Accepted: 20 February 2023;
Published: 20 March 2023.
Edited by:
Alexandru Tomescu, Cal Poly Humboldt, United StatesReviewed by:
Jill Harrison, University of Bristol, United KingdomJohn Bowman, Monash University, Australia
Copyright © 2023 Romanova, Domashkina, Maksimova, Pawlowski and Voitsekhovskaja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marina A. Romanova, bS5yb21hbm92YUBzcGJ1LnJ1
†These authors share first authorship
 Marina A. Romanova
Marina A. Romanova Valentina V. Domashkina
Valentina V. Domashkina Anastasiia I. Maksimova3
Anastasiia I. Maksimova3 Katharina Pawlowski
Katharina Pawlowski Olga V. Voitsekhovskaja
Olga V. Voitsekhovskaja