- 1School of Environment, Faculty of Science, University of Auckland, Auckland, New Zealand
- 2Manaaki Whenua – Landcare Research, Lincoln, Canterbury, New Zealand
- 3School of Biological Sciences, Faculty of Science, Engineering and Technology, University of Adelaide, Adelaide, SA, Australia
Threatened animal taxa are often absent from most of their original habitats, meaning their ecological niche cannot be fully captured by contemporary data alone. Although DNA metabarcoding of scats and coprolites (palaeofaeces) can identify the past and present species interactions of their depositors, the usefulness of coprolites in conservation biology is untested as few endangered taxa have known coprolite records. Here, we perform multilocus metabarcoding sequencing and palynological analysis of dietary plants of >100 coprolites (estimated to date from c. 400–1900 A.D.) and > 100 frozen scats (dating c. 1950 A.D. to present) of the critically endangered, flightless, herbivorous kākāpō (Strigops habroptilus), a species that disappeared from its natural range in Aotearoa-New Zealand (NZ) after the 13th C. A.D. We identify 24 orders, 56 families and 67 native plant genera unrecorded in modern kākāpō diets (increases of 69, 108 and 75% respectively). We found that southern beeches (Nothofagaceae), which are important canopy-forming trees and not an important kākāpō food today, dominated kākāpō diets in upland (c. >900 m elevation) habitats. We also found that kākāpō frequently consumed hemiparasitic mistletoes (Loranthaceae) and the holoparasitic wood rose (Dactylanthus taylorii), taxa which are nutrient rich, and now threatened by mammalian herbivory and a paucity of dispersers and pollinators. No single dataset or gene identified all taxa in our dataset, demonstrating the value of multiproxy or multigene datasets in studies of animal diets. Our results highlight how contemporary data may considerably underestimate the full dietary breadth of threatened species and demonstrate the potential value of coprolite analysis in conservation biology.
1. Introduction
Accurate data that comprehensively reflects the diets and interspecific interactions of endangered animals are often required for supporting decision making around their conservation management. For example, the translocation of an animal species to a new locality may fail, if the new locality lacks the animal species’ favoured food species (Griffith et al., 1989). Animal populations may not recover if important diet species are declining or endangered (e.g., Fritz and Hinckley, 2005). Further, species with important ecological functions, such as pollinators, seed dispersers or keystone predators, make especially ideal targets for conservation (e.g., Dimmerstein and Wemmer, 1988). However, threatened animals are often elusive and/or occupy isolated habitats, presenting challenges for dietary studies (e.g., Shehzad et al., 2012). Furthermore, research efforts may disrupt natural behaviors and stress wild animals, meaning non-invasive research techniques are frequently desirable (Putman, 1995; Romero, 2004). Analyses of scats provide a useful approach to indirectly analyse animal diets and species interactions (Kohn and Wayne, 1997), with DNA metabarcoding methods able to identify even heavily digested or microscopic taxa (de Barba et al., 2014; Kartzinel et al., 2015; Srivathsan et al., 2015; O’Rourke et al., 2020). Due to these advantages, scat metabarcoding is increasingly being used as tool in wildlife management (e.g., Young et al., 2020; Huang et al., 2021; Quéméré et al., 2021; Querejeta et al., 2022).
Contemporary data alone can, however, underestimate a species’ diet or niche space if the study species was formerly more widespread (Monsarrat et al., 2019; Kerley et al., 2020). For example, many relict species are absent from most of their original habitats, and now only exist in areas undesirable or inaccessible to humans, pathogens or invasive predators (Van Riper III et al., 1986; Beauchamp and Worthy, 1988; Kerley et al., 2012, 2020). Coprolites (palaeofaeces), record past diets and interspecific interactions of the depositor, with Late Quaternary coprolites (50 ka to present in age) often retaining amplifiable ancient DNA (aDNA). Due to the degradation of endogenous DNA and the high risk of contaminant DNA in ancient samples, DNA analysis of coprolites can be methodologically challenging. Nonetheless, coprolite metabarcoding has successfully been applied to Aotearoa-New Zealand’s (NZ’s) extinct megaherbivore ratite moa (Dinornithiformes) (Boast et al., 2018; Wood et al., 2021), kurī (the extinct NZ Polynesian dog, Canis lupus familiaris) (Wood et al., 2016), Pleistocene arctic megafauna (Willerslev et al., 2014) and the extinct caprine Myotragus balearicus (Welker et al., 2014), as well as Atacama rodent middens (which comprise coprolite materials) (Wood et al., 2018). As with modern scat metabarcoding, coprolite metabarcoding has expanded the known diets of the depositor taxa. For example, coprolite metabarcoding performed by Boast et al. (2018), resulted in the identification of plant-moa interactions that were not detected through DNA cloning or fossil analyses of the same coprolite specimens (Wood et al., 2008, 2012a, 2013a). However, the potential use of metabarcoding analyses of coprolites in conservation palaeobiology is untested as few threatened animal species are known or suspected to have left extensive coprolite records.
The critically endangered, flightless, nocturnal kākāpō (Strigops habroptilus) endemic to NZ and the world’s heaviest parrot (some individuals exceeding 4 kg) may have one of the most extensive coprolite records of any endangered species. Hundreds to thousands of coprolites believed to originate from kākāpō have been discovered in at least 13 caves and rockshelters in NZ’s South Island (Wood and Wilmshurst, 2014), probably as they were kākāpō roosting or nesting sites (Worthy, 1997) (sites reviewed in Appendix). Fossils show that kākāpō were regionally abundant and occurred across NZ’s mainland (North and South Islands, as well as the smaller Stewart Island/Rakiura) forests and subalpine shrublands adjacent to forest at the time of human arrival in the 13th C. A.D. (Worthy and Holdaway, 2002; Wilmshurst et al., 2008; Boast, 2021; Figure 1). However, habitat loss, predation by introduced mammals and human harvesting reduced the kākāpō population to fewer than 20 aging males in high elevation areas of the Milford Sound catchment of Fiordland (far south-west South Island) observed between the 1950s and 1970s (Johnson, 1976; Atkinson and Merton, 2006; Butler, 2006) and a population of <200 individuals in the Tin Ranges of Southern Stewart Island observed in the 1970s–1990s (Best, 1984; Powlesland et al., 1992; Wilson et al., 2006; Figure 1). Kākāpō are now extinct on the NZ mainland, comprising an intensively managed population of 249 individuals (at the time of writing) on three small predator-free islands, to which they were translocated in the late 20th C. Two of these island refuges occur in the far south of NZ (Whenua Hou/Codfish Island, Pukenui/Anchor Island) and one in the far north of NZ (Te Hauturu-o-toi/Little Barrier Island) (Figure 1). As there is no evidence that they naturally occurred on these islands prior to their translocation, kākāpō are thus considered to be extinct in their natural range (Boast, 2021).
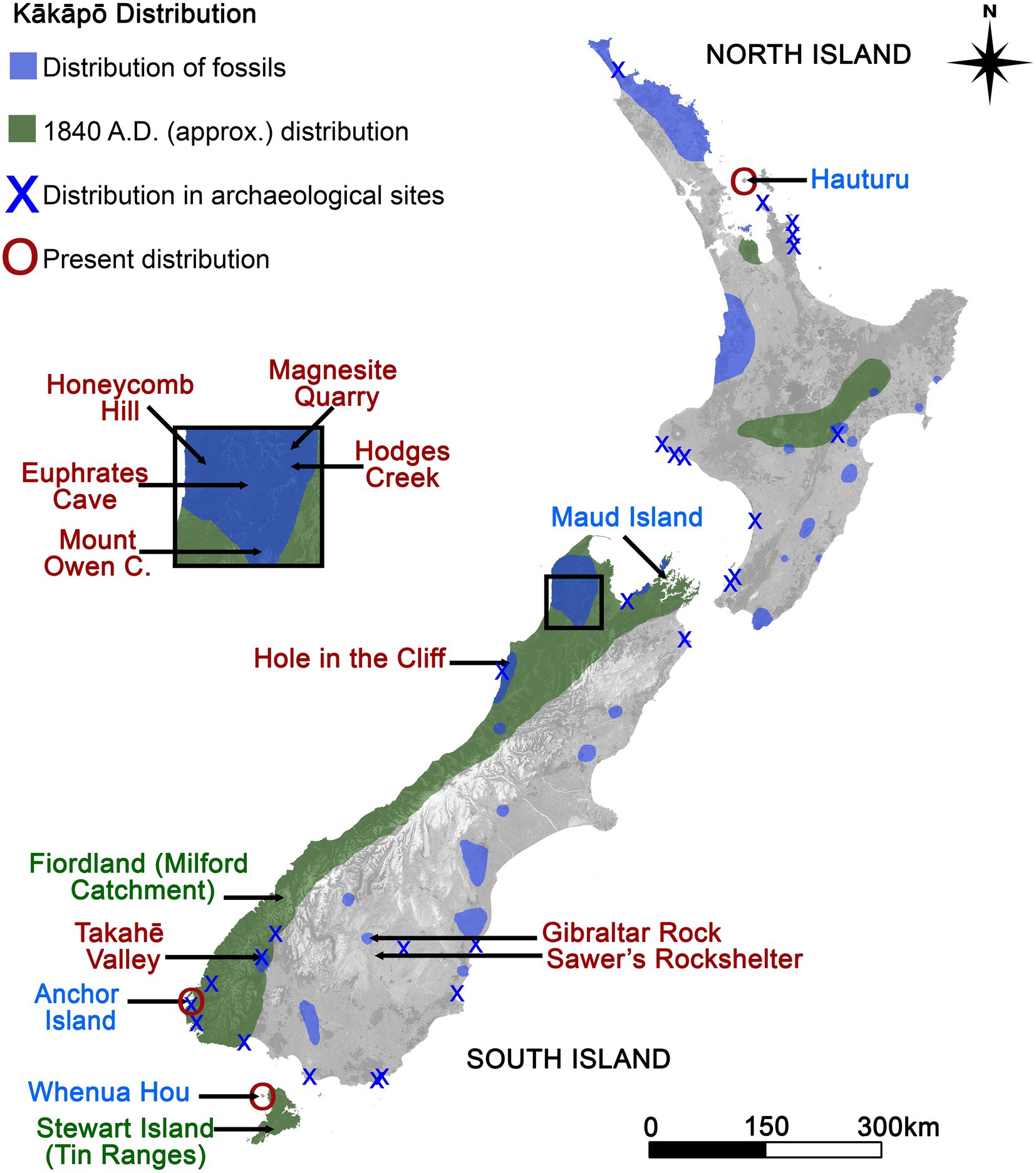
Figure 1. Contemporary and former kākāpō distribution, adapted from Boast (2021). Collection sites are plotted, coloured by age group: (Red) ancient, (Green) historic, (Blue) modern.
Kākāpō are herbivores, consuming leaves, flowers, fruit, nectar, roots, twigs and seeds (Powlesland et al., 2006). Kākāpō diet is mostly of low nutritional value, which may explain why kākāpō have a low basal metabolism (Bryant, 2006) and breed non-annually (Powlesland et al., 1992; Wilson et al., 2006; von Hurst et al., 2015). Recent studies of kākāpō diet have mainly been on Whenua Hou. Observations of kākāpō on their only northerly refuge (Hauturu) are challenging due to the island’s steep and densely forested terrain (Trewick, 1996; Stone et al., 2017). Additionally, few historical observational studies of kākāpō are available for the NZ mainland (Johnson, 1976; Best, 1984; Powlesland et al., 1992; Atkinson and Merton, 2006; Butler, 2006; Wilson et al., 2006). The ecology of kākāpō is thus incompletely understood and enigmatic. For example, the only known reliable trigger for kākāpō breeding are masts (irregular mass-seed falls) of podocarps (Podocarpaceae), specifically rimu (Dacrydium cupressinum), with rimu seeds and “fruit” (fleshy bract scales) comprising most of the food fed to chicks (Powlesland et al., 2006). However, fossil remains confirm that kākāpō were common in habitats where rimu was absent, suggesting other plant taxa must have triggered kākāpō reproduction.
Detailed studies of kākāpō coprolites have focussed on plant microfossil data (Horrocks et al., 2008; Wood et al., 2012b), although kākāpō aDNA has been amplified from several specimens (Wood et al., 2012b, Boast et al., 2018). The study by Wood et al. (2012b) was particularly notable for finding evidence, from one coprolite, that kākāpō pollinated the endangered, parasitic wood rose (Dactylanthus taylorii) in prehistory. As, D. taylorii were thought to be exclusively pollinated by short-tailed bats Mystacina tuberculata (Ecroyd, 1996), and kākāpō and D. taylorii are no longer sympatric (except one reserve where kākāpō were recently translocated), the observation by Wood et al. was of key interest to both kākāpō and D. taylorii conservation. A metabarcoding study using 18S rRNA as a barcode region (Boast et al., 2018) identified parasitic apicomplexans and possible dietary fungi from kākāpō coprolites, but no plants (possibly due to amplification biases). Overall, although kākāpō coprolites have been confirmed to reveal previously unrecorded dietary species or interactions, plants from kākāpō coprolites have yet to be studied by aDNA data.
Here, we perform multilocus metabarcoding sequencing and palynological analysis of dietary plants from coprolites from across NZ’s South Island, frozen scats collected from their last wild populations on the NZ mainland, and recently collected scats from translocated populations on island reserves. We use these data to (a) expand the number of taxa known to be consumed by kākāpō, (b) expand the known habitat range of kākāpō, (c) identify lost ecological interactions between kākāpō and native plants, (d) identify potential breeding triggers in kākāpō and (e) address the potential of coprolite metabarcoding as a resource for conservation biology.
2. Methods
2.1. Materials, field sites and sampling design
We analysed 125 putative kākāpō coprolite samples (Figure 2, hereafter also referred to as “ancient” samples) from ten sites (Figures 1, 2; Table 1; Supplementary Figure S1; Dataset 1; Supplementary Material), two from lowland mixed beech-podocarp forest (Hole in the Cliff Cave, Honeycomb Hill Cave), five in upland or subalpine southern beech forest (Mt. Owen Cave, Euphrates Cave, Hodges Creek, Magnesite Quarry and Takahē Valley), one unidentified site (samples labelled as “Mt. Cook,” an area with no known coprolite deposits) and two sites from semi-arid woodland/shrubland (Gibraltar Rock, Sawer’s Rockshelter).
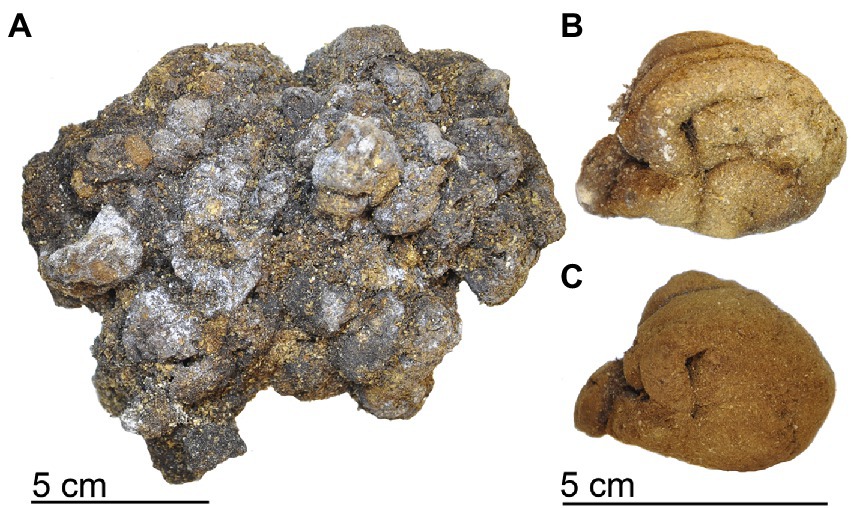
Figure 2. Examples of kākāpō coprolites. (A) Aggregate of coprolites from Mount Owen Cave, (B) Coprolite from Euphrates Cave system (sample X17/14/03). (C) Coprolite sample (X17/14/03) after exterior removal prior to subsampling.
In addition to coprolites, we sampled kākāpō scats collected and frozen by New Zealand’s Department of Conservation (DoC) and former NZ Wildlife Service (Figure 1; Table 1; Dataset 1; Supplementary Material). These originated from two “historic” (c. 1950–1990) extinct, kākāpō populations on the NZ mainland (Fiordland, Stewart Island) (36 samples) and four “modern” (c. 1990-present) translocated kākāpō populations on offshore islands (Anchor Island, Hauturu, Maud Island, Whenua Hou) (81 samples) (Table 1; Dataset 1; Supplementary Material). We hereafter collectively refer to “historic” and “modern” kākāpō scats as “recent,” to contrast them with “ancient” samples (coprolites). Most Whenua Hou and Anchor Island samples were collected during the 2017–2018 breeding season, when birds and chicks were consuming rimu. Maud Island samples came from “Flossie” and her chicks (from early 1998), who were consuming needles and immature cones of radiata pine (Pinus radiata). Most Hauturu samples were collected between 2014 and 2017, although we included specimens from a 1991 nest analysed by Trewick (1996). Overall, modern, historic, and ancient sites covered a suite of different habitats, with some ecosystem types being entirely or only represented by a single age class (Table 1).
2.2. Subsampling
Coprolites were subsampled one at a time, in a clean still-air Perspex box in an isolated laboratory purpose-built for palaeoecological samples, adapting the protocol of Wood and Wilmshurst (2016). The Perspex box was irradiated by UV light (UV-C, peak 253.7 nm wavelength) for >1 h prior to subsampling and was cleaned with 10% Decon and 10% bleach solutions between different coprolites and the surface-removal step (see below). Coprolites were first irradiated by UV light (same parameters as above) for >15 min on each side, and approximately 1 mm thickness of the coprolite exteriors were removed with a sterile scalpel. The exposed surfaces were irradiated a second time, and the coprolite was bisected using a fresh sterile scalpel. Subsamples for DNA extractions (c. 0.5–1 ml), palynomorphs (c. 5 ml), and for (selected samples only) Accelerator Mass Spectrometry (AMS) radiocarbon dating (c. 0.5–1 ml, Table 2), were taken from one of the coprolite halves using a fresh sterile scalpel. The unsampled coprolite half was retained for voucher purposes and stored at the Long-Term Ecology Lab (LTEL), Manaaki Whenua Landcare Research (MWLR), Lincoln, NZ. Frozen recent kākāpō scats were subsampled following the procedure above (omitting UV irradiation steps), in a separate, and physically distant, laboratory from the coprolite subsampling.
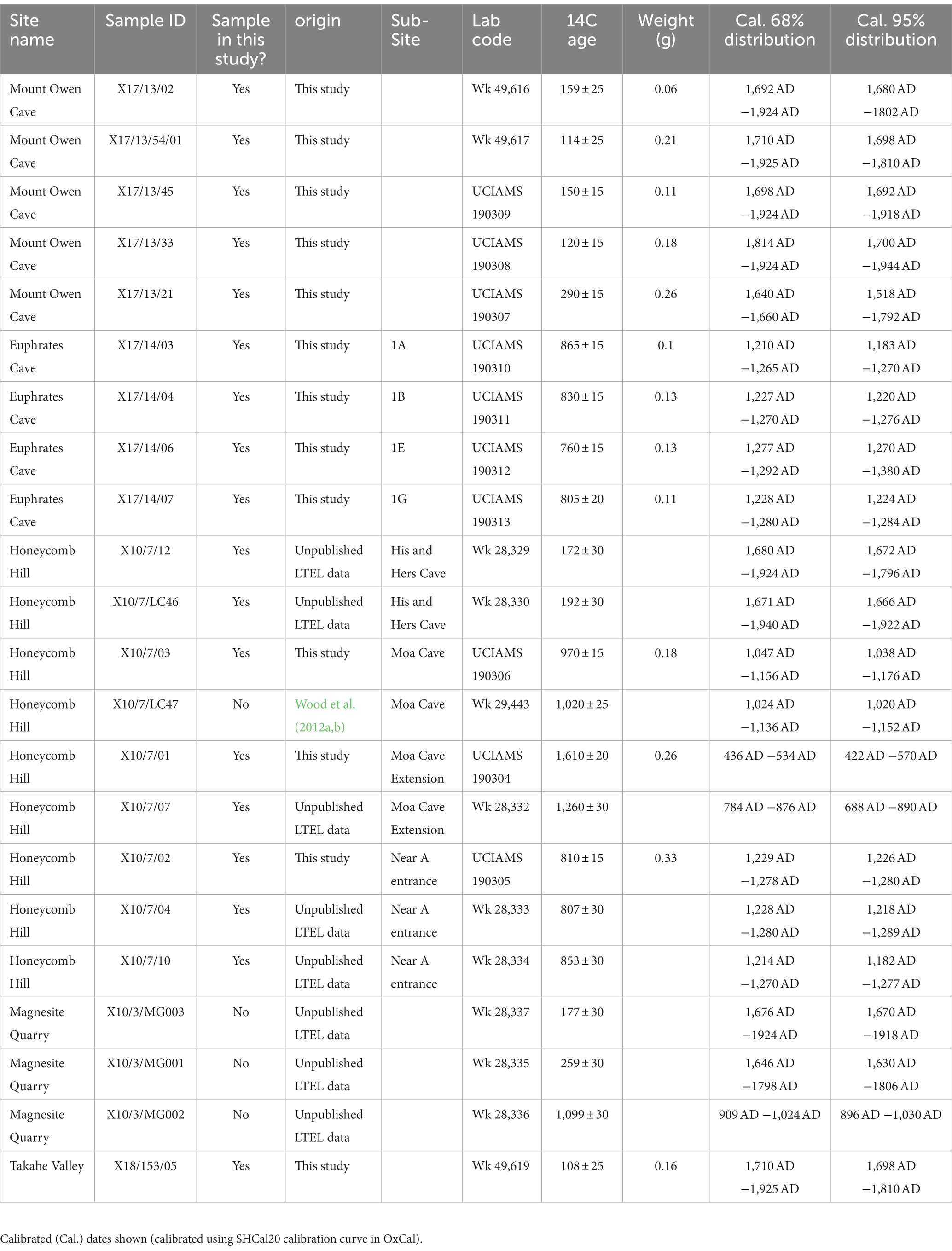
Table 2. List of 14C dates of kākāpō coprolites obtained by the Long-Term Ecology Lab (LTEL), including published and unpublished dates, as well as dates obtained specifically for this study.
2.3. Radiocarbon dating
Coprolite subsamples were radiocarbon dated using Accelerator Mass Spectrometry (AMS) at the Waikato Radiocarbon Dating Laboratory, University of Waikato (WK), and the Keck Carbon Cycle Accelerator Mass Spectrometer, University of California Irvine (UCIAMS) (Table 2). In addition to the 13 dates reported in this study, we included eight earlier dates from putative or confirmed kākāpō coprolites that lacked remnant material for subsampling: three from Magnesite Quarry, and one from Honeycomb Hill cave (Wood et al., 2012b). Radiocarbon dates were calibrated to calendar years before present (1950 A.D.) using the SHCal20 calibration curve (Hogg et al., 2020) in OxCal (version 4.4) (Ramsey, 2009).
2.4. DNA extraction, amplification, and sequencing
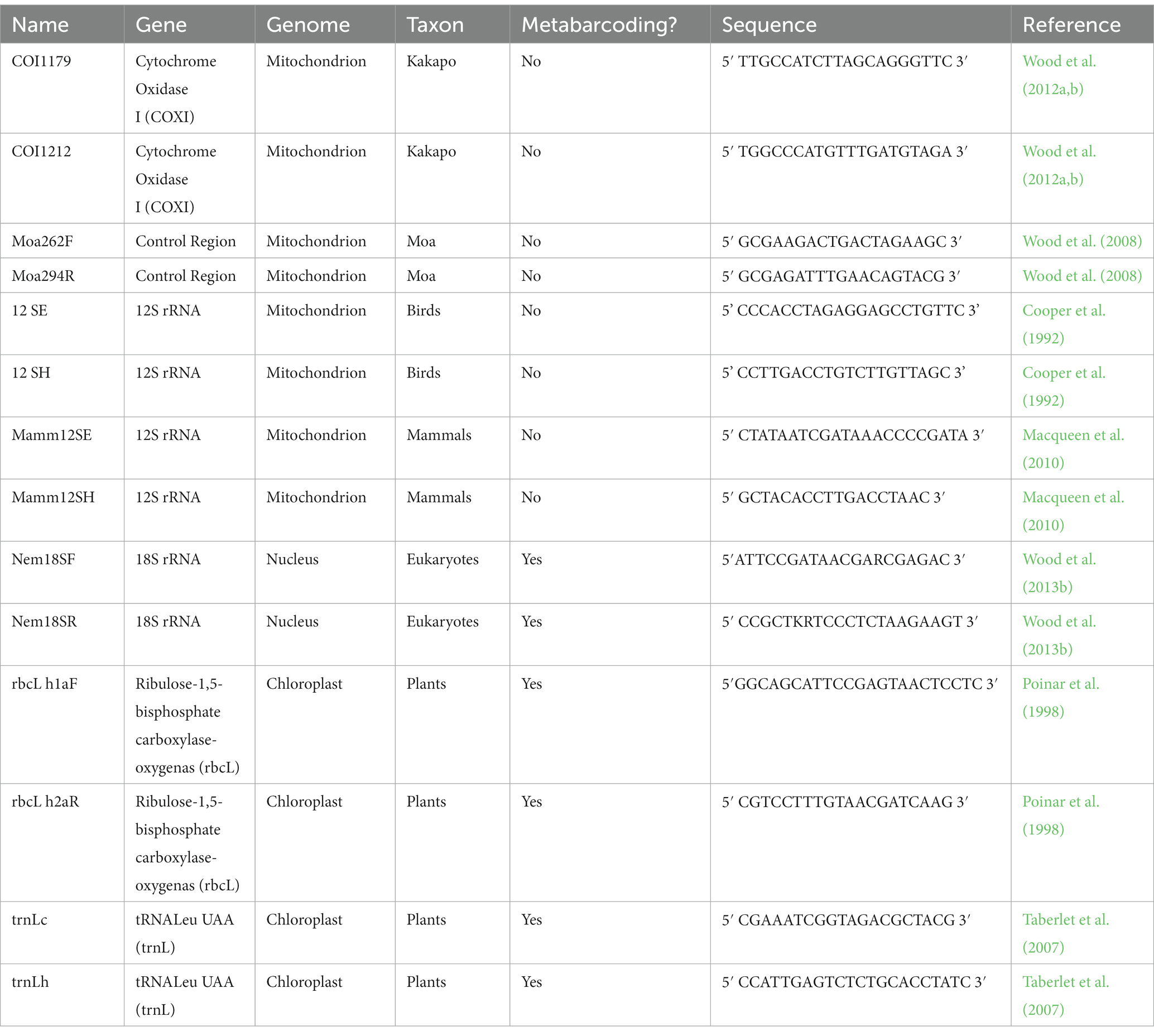
DNA was extracted from coprolites in an isolated, purpose-built ancient DNA laboratory (Long-Term Ecology Lab, Manaaki Whenua Landcare Research, Lincoln, NZ), and from recent samples in a separate multipurpose laboratory. Coprolite subsamples were rehydrated in twice their volume of double-distilled, sterile laboratory grade H2O and left on a rotary mixer at low speed at room temperature for at least 12 h. Subsamples were extracted from recent and ancient samples using the Dneasy powersoil kit (QIAGEN) following manufacturer’s instructions, with each set of extractions containing at least one extraction blank control (EBC) (a reaction processed without subsample material). Coprolite depositors were identified using a range of primers specific to kākāpō, moa, birds (universal) and mammals (universal) (Table 3). Amplified products were purified using EXOSAP-IT (Thermo Fisher, Waltham, MA, United States) and sequenced at the Landcare Research Ecological Genetics Laboratory, Auckland. Sequences were confirmed as kākāpō by using the BLASTn algorithm (Camacho et al., 2009) against all available DNA sequences on GenBank.1
All DNA extractions had fragments of the nuclear 18S rRNA gene (hereafter referred to as 18S), and the chloroplast trnL and rbcL genes, amplified using appropriate primers (Table 3). PCR reactions (12.5 μl) included 1.25 μl BSA (20 mg/Ml), 0.5 μl MgSO4 (50 Mm), 1.25 μl 10x buffer, 0.1 μl dNTPs (25 Mm each), 0.125 μl Platinum HiFi polymerase (Invitrogen), 0.25 μl each primer, 7.775 μl H2O and 1 μl DNA extract. Amplification was performed using a denaturation step of 2 min at 94°C, and 55 cycles of 30 s at 94°C, 30 s at 55°C and 40 s at 68°C, with an extension of 10 min at 72°C. These primers were selected as they targeted a range of amplicon lengths (c. 50–200 bp), controlled for sequencing bias, and amplified both plastid and nuclear DNA. All primers comprised a 5′ linker region for the addition of Illumina indexes and sequencing adapters. Coprolite amplifications were performed in triplicate, as ancient samples were considered to retain a lower portion of endogenous DNA than recent samples and pooled in equal proportions prior to indexing (modern samples were amplified singly). A second round of PCR attached dual indexes to amplicons, with reactions (20 μl) using iTaq DNA polymerase (Bio-Rad) following the manufacturer’s specified reaction mix. Indexing PCR was performed using 8 cycles of 30 s at 95°C, 30 s at 55°C and 40 s at 68°C, with an extension of 10 min at 72°C. Indexed amplicons were purified by SPRI-select magnetic beads (Beckman Coulter, CA, United States), and DNA concentrations were estimated by Qubit fluorometric quantification (Thermo Fisher, MA, United States), following manufacturer’s instructions. Indexed amplicons were pooled in equal concentrations into DNA sequencing libraries and sequenced across three MiSeq runs (two of 250 bp paired end read length for rbcL and trnL, and one run of 150 bp paired end read length for 18S), with all NGS sequencing and demultiplexing performed by Macrogen Inc. (South Korea).
2.5. Raw DNA processing and taxon identifications
Demultiplexed sequencing reads were merged, quality filtered, trimmed of primer sequences and dereplicated using a custom pipeline built around USEARCH v. 11 using default parameters (Edgar, 2010), resulting in single amplicon sequence variants (ASV’s). Read counts of each ASV per sample was retained as metadata. “Global” reference databases were developed for each gene, by downloading all available 18S sequences of eukaryotes, and all available rbcL and trnL sequences of cyanobacteria and chloroplast-bearing eukaryotes, from Genbank, and removing all sequences not identified to family level. We also built secondary “NZ-only” databases, which retained only those genera known to occur in NZ (including native and exotic taxa) based on the NZ Plants database.2
All ASVs were analysed using BLAST+ v. 2.7.1 (Camacho et al., 2009) with the BLASTn algorithm against their respective databases (parameters -max_target_seqs 1,000 -word_size 11 -reward 2 -penalty −3 -gapopen 5 -gapextend 2 -dust no -soft_masking false, rest as default). Raw BLASTn outputs were processed in MEGAN v. 6 (Huson et al., 2016), filtering sequences with <90% query cover and <80% pairwise match and assigning taxonomic identities using the top 1% (for rbcL/trnL) or 5% (18S) of hits, and bitscore cut-offs of 50, 150 and 200 for 18S, rbcL and trnL, respectively. Furthermore, we used the inbuilt “16S percent identity filter” function, constraining the highest taxonomic identifier by pairwise match to the reference (>99% Species, >97% genus, >95% family, >90% order, >85% Class, >80% Phylum). Maximum taxonomic rank was set to the genus level, as very few genera had all their respective species represented in our datasets.
To increase taxonomic resolution, we compared identifications from the “global” and “NZ-plants” databases, selecting the “NZ-plants” identification if the following criteria were followed: (A) the matches differed, (B) the “global” match occurred at a lower resolution than the “NZ-plants” match, (C) both matches were consistent to the highest taxonomic rank obtained by the “global” identifier, and (D) the “global” identifier was resolved at least to order level. If at least one of the above criteria were not met, the “global” match was selected and restricted to family level.
Inferred contaminant sequences were then filtered from all datasets. ASVs which occurred in an EBC (extraction blank control) or were identified at least to family level and shared the same I (IOTU details below) as an ASV in any EBC, were typically filtered from all samples. For example, a rbcL observation would be filtered, if the same taxon was observed in an 18S EBC. However, observations of an ASV or operational taxonomic unit (OTU) were not filtered from a sample if it occurred at ≥10 × the maximum proportion of reads observed in an EBC (e.g., if one OTU or ASV comprised up to 1% of reads in EBCs, only instances where it comprised <10% of reads in samples were filtered). Modern samples used all EBCs as a reference, whereas ancient samples only used ancient EBCs as a reference. To account for potential sequencing error or chimeric reads, all ASVs with fewer than five reads were filtered, as were all samples with <1,000 reads (before and after the filtering steps described above). Finally, all reads not identified as green plants (Viridiplantae), were filtered from subsequent analyses. We justify the final filtering step, as all non-plant sequences for trnL and rbcL likely represent contaminants or bacterial amplicons (as these primers are chloroplast and plant-specific), and non-plant 18S rRNA amplicons (e.g., fungal or parasite sequences) either fell out of the scope of this study or were considered lacking sufficient resolution to be informative.
2.6. Palynological analyses
We processed subsamples for microfossils following similar coprolite studies (Wood et al., 2012a,b), in which samples were boiled in 10% potassium hydroxide (KOH), sieved through a 150 μm mesh sieve, and acetolysed. A known number of exotic Lycopodium clavatum marker spores were added to each sample (University of Lund, batch no. 1031, 20,848 spores per tablet, on average). Samples were stained using fuchsin and mounted on glass slides. Palynomorphs and L. clavatum spores were counted in vertical transects following standard methods (Moore et al., 1991) at 400× magnification. Known weights of the microfossil subsamples, compared with L. clavatum counts, allowed for the mass of counted sample to be estimated. Counts continued until at least 250 palynomorphs were counted following Faegri et al. (1989). If pollen concentrations were insufficiently high, then either an estimated 2.5 mg of sample was counted, or all palynomorphs present on two slides were counted (if available). We used morphological features to identify palynomorphs, using guides by Large and Braggins (1991), Moar (1993), and Moar et al. (2011) and the New Zealand palynology reference slide collection at MWLR, Lincoln, New Zealand.
2.7. Operational taxonomic units and novel taxa
To assemble baseline data on known kākāpō diet, and hence whether any taxa identified in the coprolites were “novel” (hereafter used to refer to taxa previously unrecorded in kākāpō diets), we collated published records of kākāpō food plants on Fiordland, Stewart Island and modern populations (Dataset 2). To directly compare all our datasets and data from previous publications, all available data (including microfossil data, DNA data and observational data from previous studies) were ascribed an OTU comprised of four taxonomic ranks. The primary taxonomic rank comprised either the plant Division (for non-vascular plants), plant Class for (non-Angiosperm Tracheophytes), order for “basal angiosperms” (e.g., “Chloranthales Clade”), or Magnoliidae, eudicotyledons or monocotyledons (for crown-group angiosperms). Sequences not assignable to primary rank were omitted from most analyses Subsequent OTU ranks equated to order, family and genus (blank if unknown). OTUs were considered synonymous with higher resolved, but otherwise taxonomically identical OTUs, to identify unique or previously unrecorded taxa (for example, the unidentified Myrtaceae OTU was not considered novel, as the Myrtaceae genus Leptospermum was recorded in contemporary observations).
2.8. Statistical analyses
We compared representative datasets of each age group (ancient, historic, modern) consisting of presence/absences of all OTUs across all DNA datasets (omitting microfossils, which we only analysed from coprolite samples), using software packages implemented through R v 4.0.1 (R Core Team, 2013. We created a distance-matrix (Bray–Curtis dissimilarity), between datasets, which was ordinated with row totals as weights. We then used this matrix in classical multidimensional scaling (MDS)/principal coordinates analyses (PCoA) (Gower, 1966) to ordinate the data, using vegan v 2.5–6 (Oksanen et al., 2019), correcting for negative eigenvalues using Lingoes method (Legendre and Anderson, 1999). We used permutational multivariate ANOVA (PERMANOVA) (estimating pairwise differences using the pairwise Adonis package v 0.01), and multivariate homogeneity of variance tests, to test if the age groups differed in their location and scatter (significance assessed via permutation n = 999). Finally, “indicator” taxa for each age group (identifying taxa which were common and unique to an age group) were identified using the indicator species approach described by Cáceres and Legendre (2009) using indicSpecies v 1.7.8 (R Core Team, 2013).
3. Results
3.1. Radiocarbon dating
All coprolites (including the Wood et al., 2012b sample) were deposited within the last 2,000 years (calibrated dates presented here with 95% confidence ranges, unless otherwise stated) (Table 2). Date ranges reaching into the early 20th C. were obtained from Honeycomb Hill Cave, Mount Owen Cave, Takahē Valley and Magnesite Quarry, suggesting kākāpō may have persisted near these sites until recently. Non-overlapping age ranges were found in several sites, confirming separate deposition events by different animals including the small deposit of Euphrates Cave (as few as <10 coprolites) (four dates between 1,183 and 1,380 A.D, minimum two depositions). Wider age ranges were identified for the larger sites of Mount Owen Cave (four dates between 1,518 and 1994 A.D., minimum three depositions) and Magnesite Quarry (two dates between 896 and 1918 A.D., minimum two depositions). Most Honeycomb Hill coprolites were dated (nine out of 15, including the Wood et al., 2012b sample), and suggested coprolites sampled from this site represent at least seven depositions (if sample sub-site is also considered). The only Honeycomb Hill coprolite that failed to yield DNA in this study (sample X10/7/03), shared a similar age estimate and the same sub-site as the Wood et al. (2012b) coprolite, strongly suggesting these samples represent the same deposition. As the Wood et al. (2012b) sample yielded kākāpō DNA, we considered sample X10/7/03 to be a kākāpō coprolite and retained it for pollen (but not DNA) analyses.
3.2. Sequencing results
Kākāpō DNA was amplified from most coprolites (112 of 125, 89.6%) (Table 1; Dataset 3); however, a sample from each of Hodges Creek and Takahē Valley were identified as upland moa (Megalapteryx didinus) and so were omitted from this study. All remaining coprolites without a confirmed depositor (except sample X10/7/03), were also omitted from further DNA analysis, including all from Central Otago (Gibraltar Rock and Sawer’s Rockshelter). Several previous attempts to amplify DNA from Central Otago coprolites have failed (Boast, 2016), probably due to the region’s extreme seasonal temperatures affecting DNA preservation (Tait et al., 2001). All metabarcoding loci were successfully amplified and sequenced from most coprolites and modern samples, comprising 223 (18S), 194 (rbcL) and 183 (trnL) samples, although just 154 (18S), 185 (rbcL) and 148 (trnL) samples were retained after contaminant filtering. Post-filtering, the number of reads per sample reached 14,576, 36,556 and 38,956 (Dataset 1), for 18S, rbcL and trnL, respectively. The total number of ASVs that were identified as plants, and retained read counts post-filtering, comprised 91, 364 and 368ASVs, from 18S, rbcL and trnL, respectively (Dataset 3). Gene resolution was similar between rbcL and trnL, although 18S was typically unable to resolve taxa beyond the family level (Figures 3, 4; Supplementary Figures S2, S3).
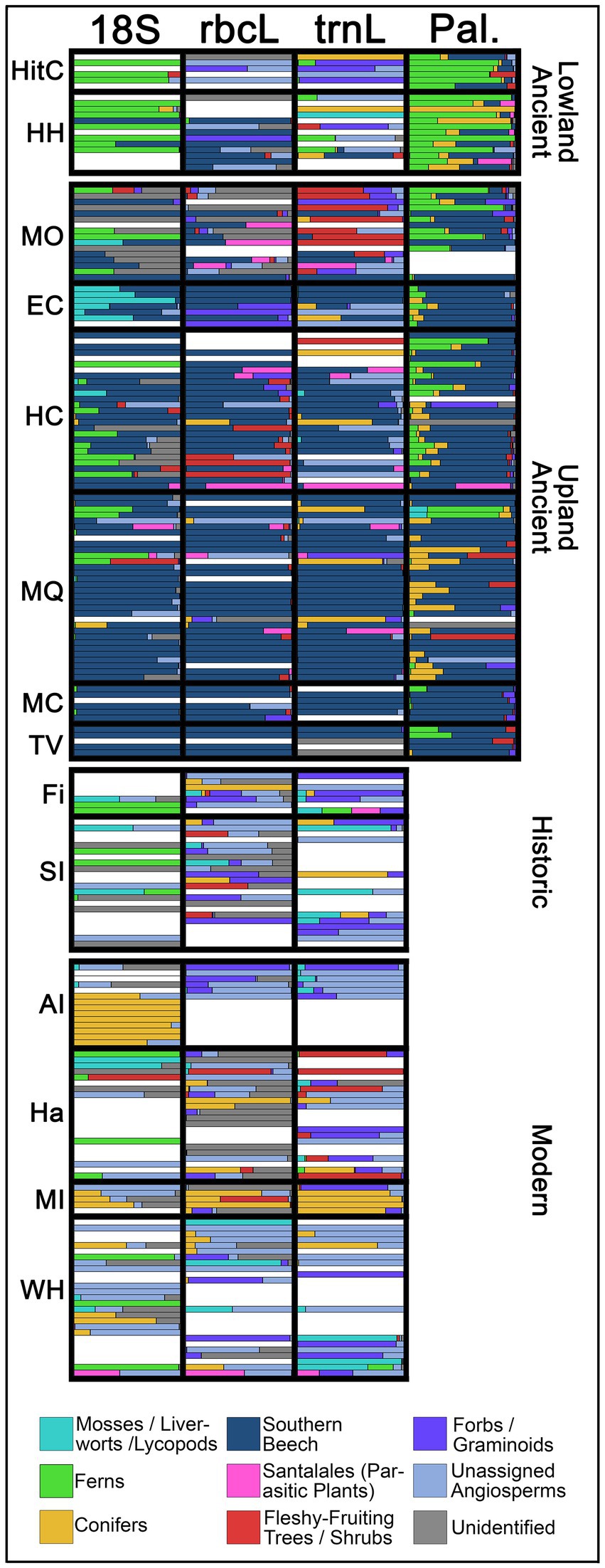
Figure 3. Proportion of key plant taxonomic/ecological groupings, separated by gene (for DNA data), or palynological data (counted for ancient samples only). Sites are further subdivided by age group, and ecology (for ancient samples). White spaces refer to samples that could not be sequenced for the target gene. Refer to Table 1 for site codes.
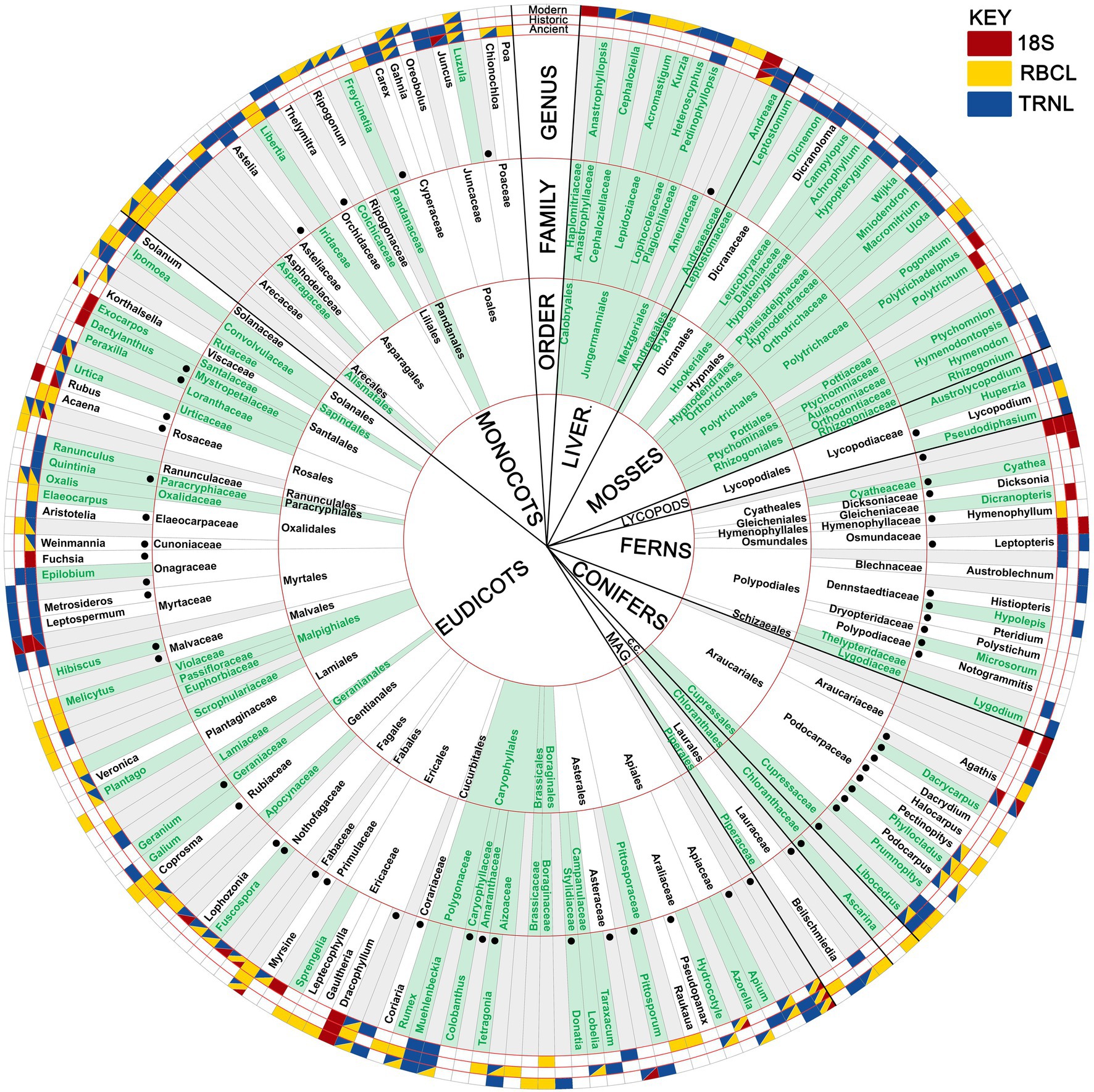
Figure 4. Chart showing all native taxa found in this study. Novel taxa (taxa not recovered from observation-based studies of kakapo post c. 1950), are coloured green. Outer cells (representing presence/absence) are subdivided by age group and coloured by gene. Black circles denote the presence of palynomorphs (ancient samples only). Grey cells represent sequences/palynomorphs not identified to the designating rank. Main ranks not shown in full: MAG (Magnoliidae), C.C. (Chloranthales Clade).
3.3. Taxon composition of DNA data
Overall, we identified 52 Orders, 86 Families and 92 genera from our samples (not including exotic or filtered taxa). Novel taxa (i.e., taxa not identified in past kākāpō diet studies) discussed here are accompanied by an asterisk at first mention (*) (Figures 3–5; Supplementary Figure S3; Datasets 3, 4). Exotic taxa typically occurred as low read counts (excluding Pinus, an observed kākāpō food on Maud Island), and were excluded from subsequent analyses as likely contaminants.
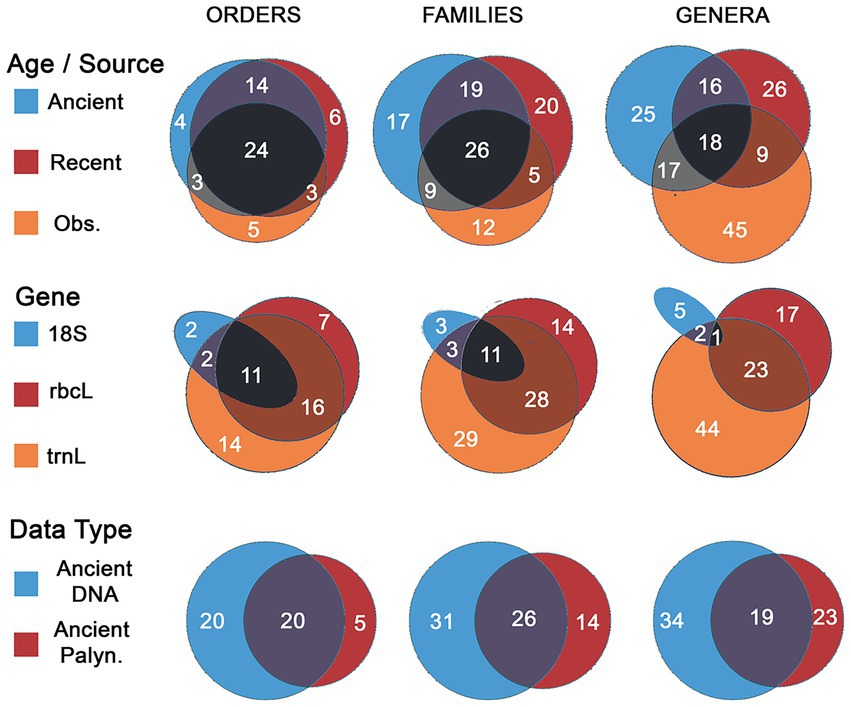
Figure 5. Venn diagram comparing (row 1) counts of plants taxa observed (Obs.) in previous studies of kākāpō (since 1950), and data in our study from the two main age groups (ancient and recent); (row 2) comparing counts of plants taxa observed between the three different genes used in this study (no taxa were observed found only in 18S and rbcL); (row 3) comparing counts of plants taxa observed between DNA (ancient samples only) and palynomorphs (only counted for ancient samples).
Most of the DNA reads (irrespective of gene) in five upland coprolite sites (Euphrates Cave, Hodges Creek, Magnesite Quarry, Takahē Valley and “Mount Cook”), were from southern beech (Fuscospora* and Lophozonia). Although the specific beech species was not identified, Lophozonia certainly represented silver beech (as L. menziesii is the only member of its genus in NZ and often a dominant tree in high elevations). The species of Fuscospora can be estimated for each collection site, as NZ’s four Fuscospora species have elevational and habitat preferences, and still grow near each collection site (Table 1), and DNA reads likely represented either hard beech (F. truncata, low elevations), red beech (F. fusca low to mid elevations), or mountain beech (F. cliffortioides mid to high elevations), with black beech (F. solandri) being less likely (F. solandri is more prevalent in low-rainfall, lowland habitats not represented by this study) (Wardle, 1984). Beech was identified by DNA in most upland coprolites, and comprised on average, 40.8–99% of reads within each site (depending on the gene barcode used). Beech reads were relatively less frequent in Mt. Owen Cave compared to other upland sites (averaging 23.3–40.8% of reads, varying by gene). Most or all beech reads in Euphrates Cave, Magnesite Quarry and Takahē Valley were from Fuscospora* (Supplementary Figure S3). However, only Lophozonia was detected from Mount Owen Cave (Supplementary Figure S3). This result is anomalous because silver beech dominates contemporary forests near Takahē Valley and Euphrates Cave as well as the Mt. Owen Cave site (Supplementary Material). This discrepancy may be explained by kākāpō at Euphrates Cave and Takahē Valley foraging on red beech at lower elevations and using the caves as roosting sites.
The mistletoe family Loranthaceae* (likely to be one of the beech mistletoe species, Alepis flavida, Peraxilla colensoi or P. tetrapetala) often co-occurred with beech in upland coprolites, even dominating reads in some samples from Hodges Creek (up to 80.1% of trnL and 94.3% of rbcL), Magnesite Quarry (up to 26.4% of trnL and 53.3% of rbcL) and Mount Owen Cave (up to 62.1% of trnL and 55% of rbcL). A high proportion of 18S reads were from ferns, especially in the lowland sites of Hole in the Cliff Cave (averaging 35.5% of reads) and Honeycomb Hill Cave (averaging 50.3% of reads). However, ferns comprised a low proportion of trnL or rbcL reads, probably due to sequencing biases or consumption of non-photosynthetic plant tissues such as fern rhizomes by the kākāpō. Although most 18S fern sequences could not be further resolved, the few trnL and rbcL fern reads from lowland coprolites were mostly Osmundaceae (specifically the ground-fern genus Leptopteris), unidentified Polypodiales, and the filmy-fern genus Hymenophyllum*. Bryophytes (mosses) were common in some samples, particularly from 18S reads of Euphrates Cave (averaging 31.6% of reads).
Rimu (Dacrydium cupressinum) DNA was not observed in coprolites (despite being locally common as a tree near Honeycomb Hill and Hole in the Cliff). Conifer taxa included NZ cedars (Libocedrus*, likely pahautea L. bidwillii) in one Hodges Creek (41.4% of rbcL and 70.3% of trnL reads) and three Magnesite Quarry coprolites (up to 7.7% of rbcL and 9.5% of trnL), miro (Pectinopitys ferruginea, 100% trnL) from a single Honeycomb Hill coprolite, and Phyllocladus* (likely mountain toatoa P. alpinus, 79.4% trnL) from a single Magnesite Quarry coprolite. Other taxa, observed in >40% of reads in any sample included Rubiaceae (identified as the fleshy-fruiting tree/shrub genus Coprosma by rbcL) in Hodges Creek; the wineberry genus Aristotelia* in Mount Owen Cave and Hodges Creek; Rosaceae (identified by rbcL as the herb genus Acaena) in Euphrates Cave and Hodges Creek; Onagraceae (possibly Fuchsia, which had high pollen counts in the same samples) in Honeycomb Hill and Hole in the Cliff Cave; the wood-rush genus Luzula* in Mount Owen Cave; and the fleshy-fruiting shrub/liana genus Muehlenbeckia* in Mt. Owen Cave, Hole in the Cliff Cave, Honeycomb Hill Cave and Magnesite Quarry. In addition, low counts of 18S reads were identified as the parasitic wood rose (Dactylanthus taylorii*) from a Honeycomb Hill coprolite (0.8% of reads) and the hemi-parasitic shrub Exocarpos* (presumably E. bidwillii, which is the only NZ member of this genus) from a Magnesite Quarry coprolite (0.3% of reads).
Recent (modern and historic) samples showed key taxonomic differences from ancient samples. For example, recent samples largely lacked beech DNA (Figures 3, 4; Supplementary Figure S3). Our data also corroborated observational data. For example, we identified many of the same kākāpō foods from Stewart Island as past observational studies, including mosses, ferns, lycopods, Myrtaceae, Ericaceae, Asteraceae and the sedge genus Carex (Cyperaceae) (Best, 1984; Powlesland et al., 1992; Wilson et al., 2006). Likewise, conifers (identified by trnL/rbcL as rimu in Whenua Hou and Anchor Island, and the pine genus Pinus in Maud Island) dominated reads from breeding birds, closely matching observation data. However, novel taxa were identified from recent samples, and taxa that had at least 10% of reads included Aizoaceae* (possibly the succulent herb Disphyma) in Stewart Island and Fiordland, the wood-rush genus Luzula* in Stewart Island, the dandelion genus Taraxacum* in Stewart Island, Muehlenbeckia* in Stewart Island and Hauturu, the herb genus Oxalis* in Fiordland, the herb genus Azorella* in Whenua Hou, and the dock genus Rumex* in Maud Island. Taxa with >10% read counts in Hauturu but not found in other sites reflected the sites’ northerly latitude, specifically palms, (Arecaceae, presumably nikau Rhopalostylis sapida), Freycinetia* (presumably kiekie F. banksii), Agathis (likely kauri A. australis), and Lauraceae (likely either taraire Beilschmiedia taraire, tawa B. tawa, or mangeao Litsea calicaris). In Whenua Hou, other taxa found with high read counts included orchids (Orchidaceae) from a single sample (99% of trnL reads), and the dwarf mistletoe genus Korthalsella from two samples (up to 55.1% of rbcL).
3.4. Microfossil data
Plant microfossils (palynomorphs) from coprolites had a wide range of estimated densities (376 grains/g to >5.6 × 107 grains/g) and were generally consistent with local contemporary vegetation, and DNA evidence from the same samples (Figure 3; Supplementary Figure S3; Dataset 4). In upland sites, palynomorph densities were usually low (median 21,201 grains/g), low-richness, and compositionally similar - being dominated by beech (Lophozonia and Fuscospora), with fern spores (especially Hymenophyllum and unidentified ground-fern “monolete” spores) also being common. Upland coprolite palynomorphs included typical alpine taxa, such as Lycopodium varium, Phyllocladus, Apiaceae, Asteraceae, Cyperaceae, Ericaceae and Poaceae. Pollen of tall forest podocarps typical of lowland forests (e.g., rimu) were common in upland coprolites, possibly reflecting windblown grains from forest at lower elevations being present on consumed plants. Lowland coprolites had richer and denser palynomorph counts (median 241,099 grains/g) than upland coprolites, presumably due to the more diverse plant communities found at lower elevations. Lowland coprolite palynomorphs were mostly fern spores, (especially the tree-fern genus Cyathea, the filmy-fern genus Hymenophyllum and the ground-fern genus Leptopteris), which is consistent with DNA data from these sites. Pollen of broadleaf shrubs and trees, such as Weinmannia (likely to be kāmahi, W. racemosa, a dominant tree in these habitats), Coprosma and Fuchsia, were also common in lowland coprolites. As expected, tall podocarp pollen was more common in lowland coprolites than in upland coprolites (tall podocarps are common near both lowland sites).
Many palynomorphs occurred at low densities and may have been ingested passively via food plant surfaces or from water sources. However, some palynomorphs probably reflect feeding behaviours. For example, the pollen of the endangered wood rose Dactylanthus taylorii occurred in five of the 14 Honeycomb Hill coprolites we sampled (up to 31.6% of palynomorphs and 18,376 grains/g, the highest pollen counts occurring in the same sample with D. taylorii DNA), including sample X10/7/03. As Wood et al. (2012b) discussed, D. taylorii pollen is unsuited for wind transport and its presence in coprolites almost certainly reflects direct foraging behaviour. Further, if the Wood et al. (2012b) sample and X10/7/03, are included, D. taylorii was observed in three of the seven minimum deposition events of analysed Honeycomb Hill samples, suggesting that kākāpō-D. taylorii interactions were locally common. Additionally, D. taylorii pollen always occurred in coprolites with probable or possible pre-human settlement ages (prior to. 1,280 A.D.; Wilmshurst et al., 2008), being absent in the one sample with a post-human age (His and Her Cave). Two other taxa, previously unrecorded from kākāpō diets were notable, as they occurred at high pollen densities and dominated DNA reads in the same specimens, specifically Phyllocladus from a single Magnesite Quarry coprolite (25,396 grains/g, 41.9 and 79.4% of palynomorphs and trnL, respectively), and Loranthaceae (c. Peraxilla; 34,873 grains/g, comprising 51.8, 80.9 and 94.3% of palynomorphs, rbcL and trnL, respectively).
Further, unusually high palynomorph densities were observed (>1 × 105 grains/g of a single type) suggesting mature flowers/pollen cones and/or associated foliage, fruits or cones were consumed, including one observation of Cyperaceae (identified as Carex by DNA) from Hodges Creek (>1 × 106 grains/g), one observation of miro from Honeycomb Hill (>2.8 × 106 grains/g), one observation of Fuchsia from Hole in the Cliff (>7.7 × 105 grains/g), five observations of Fuscospora (max. >8.7 × 106 grains/g) from upland coprolites and nine observations of Lophozonia with grains/g (max. >3 × 107 grains/g) from upland coprolites. Fern spores also occasionally reached high densities, including Leptopteris in one Honeycomb Hill coprolite (>2.5 × 107 grains/g) and three observations in Hole in the Cliff (ranging between >2–4 × 105 grains/g). These high fern spore counts may suggest either fern folivory or consumption of subsurface materials such as roots or rhizomes as (fern spores are known to congregate on soil surfaces). In all cases where >1 × 105 grains/g were observed, DNA of the respective palynomorph taxon was identified in the same specimens.
3.5. Novel observations and combined palynomorph-DNA observations
Plant DNA and palynomorphs comprised 191 OTUs (excluding exotic taxa), of which 20, 54, and 111 were identified to order, family, or genus level, respectively. These OTUs represent 54 orders, 96 families, and 111 genera, of which 24 orders, 56 families and 67 genera have not been recorded in modern kākāpō diets, respectively (Figures 3–5). Four orders, 17 families and 25 genera were recovered only from coprolites (7.4, 17.7 and 22.5% of the total found in this study, respectively) (Figure 5). Each gene appeared to be biased towards or against different taxa (Figure 5; Supplementary Figure S2); for example, trnL consistently outperformed rbcL regarding non-spermatophyte taxa (mosses, liverworts, lycopods, ferns). Likewise, combined gene or palynomorph data together contained more taxa than did any single group (Figure 5; Supplementary Figure S2). A high percentage of identified moss (81.8%, 93.3 and 94.4% of orders, families, and genera, respectively), liverwort (100% of orders, families, and genera) and lycopod (75% of genera) taxa were previously unrecorded, possibly as these taxa are difficult to identify to species in the field or from physical remains present in scats (Figure 4).
3.6. Statistical analyses
PERMANOVA and multivariate homogeneity of variance results show differences in location (F2,201 = 8.284, p = 0.001) and dispersion (F2,201 = 12.469, p = 0.001) across age classes. There were significant (p = 0.01) differences in location and dispersion between ancient and modern samples, and ancient and historic samples, but not between modern and historic samples. The most important indicator taxa (at p = 0.05; Supplementary Table S1) for ancient samples were identified as southern beech, Rubiaceae, Araliaceae, unidentified ferns, and Loranthaceae. Key indicator taxa for modern / historic samples included Aizoaceae, Amaranthaceae, unidentified Podocarpaceae, Lauraceae and Fabaceae. Overall, historic and modern samples overlapped in ordination space, although a much higher proportion of recent sample variation occurred within historic sample variation, than the reverse (Figure 6). Further, ancient samples showed a much wider spread than historic or modern samples. These data indicate that modern kākāpō populations are consuming a relatively lower diversity of plants than ancient kākāpō populations, presumably as result of their decreased habitat range and/or the decline of food plant species.
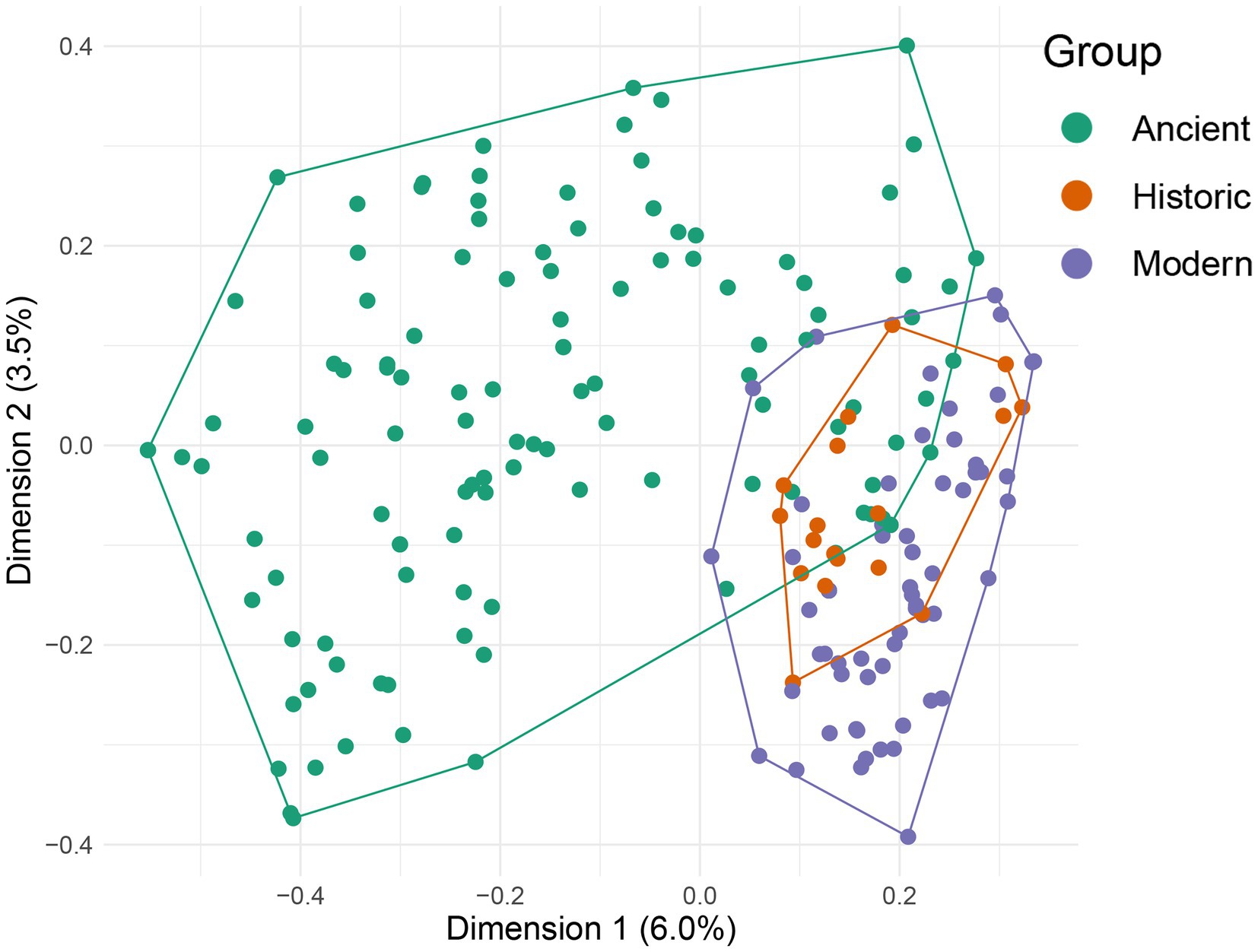
Figure 6. Classical MDA (PcOA) ordination of all observations for all genes (excluding palynomorph data, presence absence only) based on Bray-Curtis distance.
4. Discussion
4.1. Identifying novel taxa
Multilocus metabarcoding and palynomorph data of modern kākāpō scats and coprolites has increased the number of known kākāpō food plants from 35 to 59 orders (+68.6%), 52–108 Families (+107.7%) and 89 to 156 genera (+75.3%), a likely underestimate as many sequences or palynomorphs could not be identified beyond family level, or higher. While our data shows considerable overlap between modern and ancient kākāpō diets, a considerable portion of the ancient dietary range is not represented in modern or historic samples (Figure 6). Further, we recovered taxa from recent samples not recorded from previous studies of the same populations. These data highlight the value of DNA metabarcoding and palaeoecological data for expanding our understanding the past and present diet of threatened animal species. These data also illustrate the extent to which modern observational studies may underestimate the diets and ecological niches of threatened animal species, without the inclusion of palaeoecological evidence.
Some of the novel dietary taxa observed appear to have been key foods (e.g., southern beech, mistletoes), are themselves threatened (e.g., mistletoes, or the wood rose D. taylorii), or may have influenced kākāpō reproduction (southern beech, the native cedar genus Libocedrus, or the podocarp genus Phyllocladus). Other novel dietary taxa observed are widespread and common in contemporary NZ ecosystems, including trees/shrubs (Pittosporum, Quintinia), fleshy-fruiting taxa (Aristotelia, Freycinetia, Elaeocarpus, Muehlenbeckia, Melicytus), forbs (Azorella, Epilobium, Oxalis, Ranunculus, Rumex) or graminoids (Luzula). Some of these data are corroborated by other evidence. For example, Aristotelia was noted as a key component of montane Fiordland shrublands where kākāpō were frequently seen feeding in the late 19th C., referred to as “kākāpō gardens” by naturalist Richard Henry (1903). Further, 19th C. Recorded Māori mātauranga (knowledge) states that Elaeocarpus and Freycinetia were favoured kākāpō foods in the North Island (Williams, 1956; Westerskov, 1981). In addition, most of the moss, liverwort and lycopod taxa we identify are previously unrecorded, presumably because they are difficult to identify in the field, or post-digestion in scats. For example, unidentified mosses have been recorded as a kākāpō food in contemporary (e.g., Trewick, 1996; Butler, 2006; Wilson et al., 2006), and pre-19th C. (Haast, 1864) accounts. Overall, our data is strongly congruent with historic and contemporary observations, reinforcing our results, as well as corroborating the importance of historic (e.g., pre-19th C.) observational data in conservation biology.
4.2. Value of multiproxy data
Our multilocus and multiproxy dataset allowed us to identify a greater richness of taxa than if we had used only a single dataset (Figures 4, 5). For example, although trnL and rbcL provided high taxonomic resolutions for most taxa, their associated primers rarely amplified ferns (Figure 3). Conversely, 18S rRNA universally provided lower resolutions than trnL or rbcL, but the 18S primer set we used consistently amplified a wide taxonomic range. Microfossil data also provided additional resolution, especially for taxa that were predominantly amplified by 18S rRNA (Figure 4) (e.g., ferns), or generally were unresolved across the different markers (e.g., conifers). Further, different taxa were identified between rbcL and trnL, inferring that these gene regions and their associated primers had resolution or amplification biases that affected specific plant families or genera (Supplementary Figures S2, S3). Additionally, as we used nuclear (18S rRNA) and chloroplast (trnL, rbcL) markers, taxa without chloroplasts (e.g., wood rose, Dactylanthus taylorii), were detectable from metabarcoding data.
Our diverse dataset also allowed for inferences on feeding behaviour. For example, ferns dominated palynomorphs and 18S reads in many lowland coprolites. However, ferns were absent or uncommon in rbcL and trnL reads from lowland coprolites, respectively (Figure 3). Although this discrepancy may be explained by amplification biases, it is also possible that kākāpō were feeding on fern rhizomes which are a known kākāpō food and unlikely to contain much chloroplast DNA (Atkinson and Merton, 2006; Wilson et al., 2006). Nonetheless, the higher taxonomic resolution provided by trnL suggested that the unidentified ferns amplified by 18S in lowland coprolites were Leptopteris (Supplementary Figure S3). Congruently, high concentrations of Leptopteris spores were observed from the same specimens (Supplementary Figure S3). Although high fern spore concentrations might suggest reproductive fern fronds were being consumed, rhizome foraging would also result in high spore consumption, as fern spores accumulate on forest floors near to their parent plants and can remain viable for months or even years (Penrod and McCormick, 1996). Many samples had synchronous observations of the same taxon comprising a high proportion of palynomorphs and DNA reads (including chloroplast and nuclear markers), as well as reaching very high pollen densities. Since these observations included chloroplast data, this pattern strongly suggested foraging on reproductively active tissues, specifically, of mistletoes (possibly the genus Peraxilla) silver beech (Lophozonia menziesii), the beech genus Fuscospora, the sedge genus Carex, Onagraceae (strongly indicated to be Fuchsia by pollen data), the podocarp miro (Pectinopitys ferruginea) and the podocarp genus Phyllocladus (c. mountain toatoa P. alpinus).
Overall, our results strongly support the importance of using multilocus or multiproxy data in animal dietary studies, a result congruent with other multilocus metabarcoding studies on animal scats (Mallott et al., 2018), and combined microfossil-DNA studies in coprolites (Wood et al., 2012a, 2013a). For example, the P6 loop of the trnL intron, amplified using primer sets described in Taberlet et al. (2007) (used in our study), are commonplace in plant metabarcoding studies (e.g., de Barba et al., 2014; Srivathsan et al., 2015). However, had we restricted our data to trnL only, we would have failed to detect several key results and resulted in likely incorrect conclusions on kākāpō diet as a result. We therefore recommend that future metabarcoding studies, not just on animal diets, but also on plant communities in general, should ensure that multi-gene or multiproxy datasets in used in their analyses when feasible.
4.3. Importance of southern beech as a kākāpō food
NZ’s five species of southern beech (Nothofagaceae) are major canopy-forming trees; approximately 2/3 and 1/3 of NZ’s surviving forests, contain beech or are predominantly comprised of beech, respectively (Wardle, 1984; Ogden et al., 1996). Kākāpō co-occur with beech on Anchor Island (abundant red beech Fuscospora fusca) and Hauturu (some localised hard beech F. truncata) (Worthy, 1993; Crouchley et al., 2011), and most observations of kākāpō prior to c. 1970 A.D. were in or adjacent to montane beech forest (presumably mountain beech F. cliffortioides and silver beech Lophozonia menziesii) (Williams, 1956). However, the only record of kākāpō feeding on beech are records in the 1970s from Fiordland, which included limited feeding signs on silver beech leaves, and leaf cuticles in scats tentatively identified as beech (Butler, 2006). Beech was also uncommon in modern or lowland coprolite samples. Conversely, beech comprised a high proportion of DNA reads, and reached high palynomorph concentrations, in upland coprolites, suggesting that beech tissues (including leaves, buds, flowers and possibly seeds) comprised the majority of kākāpō diet in some upland forests. These observations concur with Horrocks et al. (2008), who identified that most phytoliths in Hodges Creek coprolites were identical to structures found in Fuscospora leaves. This inference is further supported by several other NZ birds regularly consuming beech foliage, including kererū (a native pigeon, Hemiphaga novaeseelandiae), and three other parrot species: kākā (Nestor meridionalis), kea (N. notabilis) and yellow-fronted parakeets (Cyanoramphus auriceps) (McEwan and McEwan, 1978; O’Donnell and Dilks, 1994; Greer et al., 2015).
The relative absence of beech DNA in lowland coprolites, recent samples, some upland sites, or in most observational studies warrants further explanation. The lowland forests that represent modern kākāpō refuges, and lowland coprolite sites, are biodiverse and structurally complex, with a tall (often >30 m) multi-species canopy comprising conifers and beech (e.g., Wardle, 1984). However, beech forests typically become increasingly low-diversity and structurally simple with increased elevation. For example, forest near to the treeline (c. 1,200 m) may comprise few plants other than dwarfed (e.g., <3 m) mountain or silver beech (Wardle, 1984; Ogden et al., 1996). Although kākāpō are capable climbers (Powlesland et al., 2006), they may have foraged on the nutritionally poor beech leaves only if they were easily accessible and/or if other food sources were absent. For example, kererū were observed to consume beech leaves predominantly during winter when fruits were unavailable (McEwan and McEwan, 1978). Similarly, at high elevations kākāpō would have been able to access subalpine plant communities (which include many fruiting or starch-rich species) during summer months. For example, DNA reads in Mt. Owen Cave samples are dominated by taxa found above the treeline (Aristotelia, Coprosma, Muehlenbeckia, Apiaceae, Ericaceae and graminoids), which is congruent with the sites’ high elevation and proximity to subalpine herbfields. Further, Fiordland kākāpō were mostly observed by researchers above the treeline outside of winter months when the samples we analyse were presumably collected (Atkinson and Merton, 2006; Butler, 2006). Overall, these observations would suggest that in upland forests, beech may have been avoided in favour of fruiting seasonal shrublands when or if these were available (e.g., near forest margins during summer months).
4.4. Lost ecological interactions between kākāpō and parasitic plants
Four of the five families of Santalales (mistletoes and relatives) native to NZ (Loranthaceae, Mystropetalaceae, Santalaceae and Viscaceae, excluding only Nanodeaceae) were identified in our data across all age groups, and comprised a high proportion of DNA reads or palynomorphs in many samples (Figures 3, 4; Supplementary Figure S3). Excluding dwarf mistletoes (Korthalsella, Viscaceae), all Santalales taxa that we observed are unrecorded from recent kākāpō diets (Wilson et al., 2006). Further, most NZ Santalales are no bigger than small shrubs (excluding the small tree Mida salicifolia, Nanodeaceae), suggesting a strong selective bias by kākāpō. These observations are notable because all NZ’s Santalales taxa are photosynthetic hemiparasites, except for Dactylanthus taylorii in the Mystropetalaceae, which is a non-photosynthetic holoparasite.
Studies have shown that hemiparasitic plant tissues have much higher concentrations of some minerals (e.g., phosphorus, potassium and sodium) in their tissues than their hosts, including members of the Loranthaceae (including NZ taxa) (Bannister et al., 2002; Lo Gullo et al., 2012; Gebauer et al., 2018), Santalaceae (including the genus Exocarpos, which we identify) (Patykowski et al., 2018), and Viscaceae (Panvini and Eickmeier, 1993; Türe et al., 2010). Further, in NZ, mistletoes are known to be highly palatable to introduced brushtail possums (Trichosurus vulpecula) and red deer (Cervus elaphus) compared with other plants such as beech (Sweetapple, 2008; Crouchley et al., 2011). Further, palaeoecological data has shown that moa also regularly fed on mistletoe tissues (Wood et al., 2020, 2021). It is therefore possible that kākāpō and other NZ species preferentially forage on Santalales taxa due to their nutritional content. This inference is concerning, as out of NZ’ 12 endemic Santalales taxa, one is already extinct, and eight are classified as declining or threatened; an apparent result of prior forest clearance, browsing by invasive mammals, and a paucity of native dispersers or pollinators (Ecroyd, 1996; Ladley and Kelly, 1996; de Lange et al., 2018). For example, NZ’s beech mistletoes (Peraxilla tetrapetala, P. colensoi or Alepis flavida) are supported by historic and palaeoecological evidence to have been considerably more abundant, and to have occurred closer to the ground, in the past (Wood et al., 2020). As kākāpō are thought to have been abundant and widespread in NZ’s presettlement forests (Boast, 2021), our data suggests that they may have been a key pollinator or dispersers of these Santalales taxa prior to their near extinction.
Kākāpō appear to have consumed hemiparasitic Santalales throughout their range. For example, our data, and previous studies, have shown that kākāpō still feed on stems of the leafless, dwarf mistletoe K. salicornioides in Whenua Hou (and historically also did so on Stewart Island) (Wilson et al., 2006). We also identify low read counts of the genus Exocarpos from a single Magnesite Quarry coprolite. Exocarpos is only represented in NZ by the subalpine shrub E. bidwillii, which could have substituted for mistletoe in some high elevation shrublands. However, most notably, Loranthaceae comprised a high frequency of DNA reads and/or pollen counts in several upland coprolites (e.g., 50% or higher). It is likely that the species of Loranthaceae identified in upland coprolites are beech mistletoes, which the birds would have encountered if feeding on beech foliage. NZ’s beech mistletoes produce fleshy fruits and nectar-rich flowers, both of which are essential energy sources for birds in some forests (Ladley and Kelly, 1996; Murphy and Kelly, 2003). While kākāpō were presumably feeding on mistletoe fruits and flowers (supported by high densities of beech mistletoe pollen in some coprolites), it is highly probable that kākāpō were also consuming nutrient-rich beech leaves and stems year-round. Overall, in low-diversity beech forests, mistletoes may have provided an essential source of nutrients for kākāpō and other endemic species. Beech mistletoes are thus not only threatened by herbivory, and a lack of pollinators or dispersers, but their reduced biomass relative to pre-human levels, also now means that beech forests might not be able to support the communities of kākāpō and other birds (e.g., other frugivorous, herbivorous or nectivorous species such as kererū, kaka, kōkako Callaeas spp. or bellbird Anthornis melanura) as they did in prehistory.
The holoparasitic, threatened wood rose D. taylorii, is today reduced to scattered relict populations in the North Island and appears to be pollinated only by endangered short-tailed bats (Ecroyd, 1996). However, we observed D. taylorii pollen in kākāpō coprolites from Honeycomb Hill, and only in coprolites with confirmed or likely pre-human ages (pre-1,280 A.D., Table 2). These data suggest D. taylorii was either patchily distributed and/or did not occur further south than Honeycomb Hill in the late Holocene and may also show that this species’ range quickly contracted after the arrival of humans and commensal pacific rats (Rattus exulans) in NZ (Ecroyd, 1996). D. taylori was identified in six of 14 (42.5%) Honeycomb Hill coprolites we analysed, where it comprised up to 31.7% of total palynomorphs. Furthermore, D. taylorii was observed in three of the seven (42.9%) minimum Honeycomb Hill deposition events we identify by 14C dating, occurring in samples that differ in age by centuries (Table 2). D. taylorii only appears above ground to flower, and do not disperse pollen widely (Moar et al., 2011), thus all D. taylorii observations in our data must represent direct flower foraging events (Ecroyd, 1996). Although Wood et al. (2012b) identified D. taylorii in a single kākāpō coprolite, these important new records suggest for the first time that kākāpō and D. taylorii flower interactions occurred at a high frequency where the two species co-occurred. There is thus a high potential that the formerly widespread kākāpō was an important pollinator of D. taylorii in prehistory, along with other nectivorous birds and bats. The apparent dependence of D. taylorii on short-tailed bat pollination today is a therefore likely to be a result of extinctions or range contractions of other pollinators, such as kākāpō.
4.5. Potential breeding triggers
Kākāpō appear to depend on an as yet-unknown hormonal “trigger” to reproduce, such as phytoestrogens in unripe fruit or cones, or compounds prevalent in coniferous tissues such as terpenes (Walsh et al., 2006; Fidler et al., 2008; Davis, 2013; Stone et al., 2017). Attempts to increase kākāpō breeding frequency with supplementary feeding have been unsuccessful (Harper et al., 2006), and only rimu masts are confirmed to consistently trigger kākāpō breeding (Powlesland et al., 2006). However, kākāpō have recently bred in the absence of rimu, with females observed feeding their chicks kauri leaves, immature kahikatea (Dacrycarpus dacrydioides) cones and fronds of the ground-fern genus Austroblechnum on Hauturu (Trewick, 1996), and exotic pine needles and immature cones on Maud Island (Walsh et al., 2006). Pink pine (Halocarpus biformis), which co-masts with rimu on Stewart Island, may have helped to stimulate kākāpō breeding (Powlesland et al., 1992), and kākāpō breeding on Hauturu may be linked with large kauri seed falls (Stone et al., 2017). Although all tree species linked to recent kākāpō breeding are conifers, it is possible angiosperms may also trigger kākāpō reproduction. For example, Māori mātauranga recorded in the 19th C. states that kākāpō in the North Island bred when kiekie (Freycinetia banksii) were fruiting (Westerskov, 1981) (supported by our observation of kiekie in Hauturu samples). Southern beech masts have also been proposed as a kākāpō breeding trigger (Harper et al., 2006).
Our data may provide some insights into kākāpō reproduction. For example, two upland sites we sampled (Hodges Creek, Magnesite Quarry) contain kākāpō chick bones and possible eggshell (Worthy, 1997, JRW, personal observation), confirming that the associated coprolites are from breeding populations despite the local absence of rimu in the nearby beech forest (see Supplementary Material). We also show that throughout their habitat range, kākāpō fed on conifers, with miro identified in a lowland coprolite and Libocedrus (c. pāhautea L. bidwillii) and Phyllocladus (c. mountain toatoa P. alpinus) in upland coprolites. NZ’s Phyllocladus and Libocedrus species occur in areas without rimu and thus could have played the role of a coniferous breeding trigger for kākāpō in some habitats (although both genera rarely grow large and are non-masting).
In NZ, beech mast every four to six years (Wardle, 1984), with trees usually producing roughly c. >2,000 nuts m−2, but occasionally >10,000 nuts m−2 (Allen and Platt, 1990; Burrows and Allen, 1991). The total mass of seed during beech forest masts can therefore be considerable despite their small size (e.g., average 368 nuts g−1 for mountain beech F. cliffortioides) (Ledgard and Cath, 1983), and sufficient to support massive irruptions of invasive seed-feeding rodents (Ruscoe et al., 2004). Beech seed has nutritional qualities similar to rimu cones and seed (Murphy, 1992; Beggs, 1999; Ruscoe et al., 2004; von Hurst et al., 2015). Further, while kākā (one of the closest living relatives of kākāpō), breed in response to podocarp masts (including of rimu), they also breed in response to beech masts (Powlesland et al., 2009). Our data demonstrate that beech tissues were the main food source of kākāpō in upland beech forests. As a result, kākāpō would have consumed developing beech seed along with beech foliage, if they were not also deliberately foraging for beech seeds on the forest floor. Therefore, it seems highly plausible that this influx of nutrient-rich food in their diets would have triggered kākāpō breeding. Beech masts are not known to trigger kākāpō breeding in contemporary ecosystems (for example, Anchor Island). This discrepancy may suggest that beech fail to trigger kākāpō breeding if other masting species such as rimu are present, or if kākāpō are not consuming developing beech seed with their regular foods of beech foliage.
5. Conclusion
The kākāpō parrot is a challenging species for conservation, especially due to its vulnerability to mammalian predation, low fecundity and slow maturation rate. Further, masting podocarps (especially rimu) are still the only known reliable breeding stimulus for kākāpō. Despite these difficulties, kākāpō conservation efforts have successfully increased the kākāpō population from 51 birds in the early 1990s, to nearly 250 individuals today. Nonetheless, breeding kākāpō occur on just three small island reserves, meaning that their current carrying capacity is limited and the species remains highly vulnerable to catastrophic events (e.g., accidental mammal introductions, disease or natural disasters). Translocating kākāpō to new habitats is therefore paramount to the success of their conservation, which in turn requires a close understanding of their habitat requirements and dietary range.
Our dataset demonstrates that contrary to modern evidence, kākāpō are a highly versatile species and exploited a much greater number of plant species in prehistory than they do at present. We find no support that kākāpō were solely dependent on rimu masts for reproduction, and from our data we infer that numerous different plant species also triggered and supported kākāpō breeding. For example, our data and other palaeoecological evidence such as fossil distributions (e.g., Boast, 2021), strongly indicate that beech forests are ideal kākāpō habitats, with the beech trees themselves capable of sustaining most of the kākāpō diet and even possibly triggering kākāpō breeding during their non-annual mast events. Predator-free reserves with a high composition of beech or rimu may therefore be ideal focal localities for future kākāpō translocations, especially if these forests also contain a high proportion of nutrient rich species such as mistletoes. These inferences greatly expand the possible range of future kākāpō translocation localities, especially when refuges on the NZ mainland become a possibility. Overall, we strongly support the reintroduction of kākāpō into a broad range of (predator-proof) habitats to ensure population growth and their recovery as a species.
At least one-fifth of terrestrial vertebrate species globally are threatened with extinction and have suffered range contractions due to human activity (Hoffmann et al., 2010). There is an increasing necessity to comprehensively understand the ecological niches, species-interactions, and diets of animal species to guide conservation management. Our study confirms that where available, coprolites can provide new and highly detailed insights into threatened species’ ecology, that may otherwise be inaccessible. Coprolites therefore provide an invaluable tool for informing conservation management plans for threatened taxa with attributed coprolite records such as Eurasian lynx (Lynx pardinus) or European bison (Bison bonasus) (Sanz et al., 2016). However, due to the paucity of extant species with known coprolite deposits, we acknowledge that the application of coprolites in conservation palaeobiology will be limited. Despite these limitations, our coprolite-based study vividly demonstrates how much contemporary observational data may underestimate a threatened species’ ecological niche or dietary range, and how palaeoecological data or historic data of any source, should always be sought, when feasible.
Data availability statement
The data presented in the study are deposited in the Manaaki Whenua Landcare Research datastore repository https://doi.org/10.7931/jddg-tg97.
Ethics statement
Ethical review and approval was not required for the animal study because this research was conducted on faecal samples and no live animals were used.
Author contributions
AB, JWo, GP, and JWi designed and supervised the research project. AB conducted field work. AB, JWo, and NB carried out laboratory work. AB, JWo, and JWi performed microfossil counts. AB and GP conducted data analyses. AB, JWo, GP, and JWi wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by University of Auckland Postgraduate Research Student Support (PReSS), and grants awarded by the University of Auckland Centre for Biodiversity and Biosecurity (CBB), Hauturu Supporters Trust, Ornithological Society of New Zealand (Birds NZ Research Fund), the Royal Society of New Zealand (Hutton Fund), and the Linnean Society of New South Wales (The Joyce W. Vickery Scientific Research Fund). JWi and JWo were supported by funding from the Strategic Science Investment Fund of the New Zealand Ministry of Business, Innovation and Employment’s Science and Innovation Group.
Acknowledgments
Kākāpō are under conservation by the Kākāpō Recovery Programme of the New Zealand Department of Conservation (DoC) in close partnership with Ngāi Tahu Māori, who are key kaitiaki (guardians) of kākāpō and whose rohe (cultural area) includes the last areas kākāpō survived on the NZ mainland and their most important island refugia today. We would like to thank Canterbury Museum and Central Stories Museum and Art Gallery (Alexandra) for loaning specimens, and museum curators Paul Scofield, Maurice Watson, and Alan Tennyson for allowing access to collections; Andrew Digby and other members of the Kākāpō Recovery Programme for research support and for providing specimens; and Trevor Worthy for advising on collection sites. We would also like to give special thanks to our cave guides (John Patterson and Neil Silverwood), field assistants (Michal Kuchar, Lea de Nascimiento, Javier Mendez, Nia-Joelle Weinzweig), and Corey Mosen and the nationally famous, late “kea dog” Ajax who collected samples from near Mount Owen for this project. We also wish to give thanks to Kieren Mitchell for providing edits, and to our reviewers and editor Chris Schneider for their time on this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1058130/full#supplementary-material
SUPPLEMENTARY DATA SHEET 1 | Appendix.
SUPPLEMENTARY DATA SHEET 2 | Supplementary figures and tables.
SUPPLEMENTARY DATA SHEET 3 | Dataset 1.
SUPPLEMENTARY DATA SHEET 4 | Dataset 2.
SUPPLEMENTARY DATA SHEET 5 | Dataset 3.
SUPPLEMENTARY DATA SHEET 6 | Dataset 4.
Footnotes
References
Allen, R. B., and Platt, K. (1990). Annual seedfall variation in Nothofagus solandri (Fagaceae), Canterbury, New Zealand. Oikos 57, 199–206. doi: 10.2307/3565940
Atkinson, I. A., and Merton, D. V. (2006). Habitat and diet of kakapo (Strigops habroptilus) in the Esperance Valley, Fiordland, New Zealand. Notornis 53:37.
Bannister, P., Strong, G. L., and Andrew, I. (2002). Differential accumulation of nutrient elements in some New Zealand mistletoes and their hosts. Funct. Plant Biol. 29, 1309–1318. doi: 10.1071/FP02005
Beauchamp, A. J., and Worthy, T. H. (1988). Decline in distribution of the takahe Porphyrio (= Notornis) mantelli: a re-examination. J. R. Soc. N. Z. 18, 103–118. doi: 10.1080/03036758.1988.10421698
Beggs, J. R. (1999). Comparison of the quality of red and silver beech (Nothofagus) seeds in Nelson Lakes National Park, New Zealand. N. Z. J. Bot. 37, 495–501. doi: 10.1080/0028825X.1999.9512648
Best, H. A. (1984). The foods of kakapo on Stewart Island as determined from their feeding sign. N. Z. J. Ecol. 7, 71–83.
Boast, A. P. (2016). Ancient DNA of New Zealand’s Extinct Avifauna. Unpublished MPhil Thesis, University of Adelaide.
Boast, A. P. (2021). Ancient DNA and Palaeoecology of the Kakapo (Strigops habroptilus). Unpublished Ph.D. Thesis. University of Auckland.
Boast, A. P., Weyrich, L. S., Wood, J. R., Metcalf, J. L., Knight, R., and Cooper, A. (2018). Coprolites reveal ecological interactions lost with the extinction of New Zealand birds. Proc. Natl. Acad. Sci. 115, 1546–1551. doi: 10.1073/pnas.1712337115
Burrows, L., and Allen, R. B. (1991). Silver beech (Nothofagus menziesii (Hook. F.) Oerst.) seedfall patterns in the Takitimu range, South Island, New Zealand. N. Z. J. Bot. 29, 361–365. doi: 10.1080/0028825X.1991.10415489
Butler, D. (2006). The habitat, food and feeding ecology of kakapo in Fiordland: a synopsis from the unpublished MSc thesis of Richard gray. Notornis 53:55.
Cáceres, M. D., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Cooper, A., Mourer-Chauviré, C., Chambers, G. K., von Haeseler, A., Wilson, A. C., and Pääbo, S. (1992). Independent origins of New Zealand moas and kiwis. Proc. Natl. Acad. Sci. 89, 8741–8744. doi: 10.1073/pnas.89.18.8741
Crouchley, D., Nugent, G., and Edge, K. (2011). “Removal of red deer (Cervus elaphus) from Anchor and Secretary Islands, Fiordland, New Zealand,” in Island invasives: eradication and management: proceedings of the International Conference on Island Invasives. eds. C. R. Veitch, M. N. Clout and D.R. Towns (Auckland: IUCN), 422–425.
Davis, C. E. J. (2013). Kākāpō Reproduction: Identification of Steroid Receptors and Oestrogenic Activity in Native Flora. Untitled PhD Thesis. Victoria University of Wellington.
de Barba, M., Miquel, C., Boyer, F., Mercier, C., Rioux, D., Coissac, E., et al. (2014). DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol. Ecol. Resour. 14, 306–323. doi: 10.1111/1755-0998.12188
de Lange, P. J., Rolfe, J. R., Barkla, J. W., Courtney, S. P., Champion, P. D., Perrie, L. R., et al. (2018). “Conservation status of New Zealand indigenous vascular plants, 2017” in New Zealand Threat Classification Series 22. (Wellington: Department of Conservation).
Dimmerstein, E., and Wemmer, C. (1988). Fruits rhinoceros eat: dispersal of Trewia nudiflora (Euphorbiaceae) in lowland Nepal. Ecology 69, 1768–1774. doi: 10.2307/1941155
Ecroyd, C. E. (1996). The ecology of Dactylanthus taylorii and threats to its survival. N. Z. J. Ecol. 30, 81–100.
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Faegri, K., Kaland, P. E., and Krzywinski, K. (1989). Textbook of Pollen Analysis. Chichester: John Wiley & Sons Ltd.
Fidler, A. E., Lawrence, S. B., and McNatty, K. P. (2008). An hypothesis to explain the linkage between kakapo (Strigops habroptilus) breeding and the mast fruiting of their food trees. Wildl. Res. 35, 1–7. doi: 10.1071/WR07148
Fritz, L. W., and Hinckley, S. (2005). A critical review of the regime shift - “junk food” - nutritional stress hypothesis for the decline of the western stock of Steller Sea lion. Mar. Mamm. Sci. 21, 476–518. doi: 10.1111/j.1748-7692.2005.tb01245.x
Gebauer, R., Volařík, D., and Urban, J. (2018). Seasonal variations of Sulphur, phosphorus and magnesium in the leaves and current-year twigs of hemiparasitic mistletoe Loranthus europaeus Jacq. And its host Quercus pubescens Willd. J. For. Sci. 64, 66–73. doi: 10.17221/144/2017-JFS
Gower, J. C. (1966). Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53, 325–338. doi: 10.1093/biomet/53.3-4.325
Greer, A. L., Gajdon, G. K., and Nelson, X. J. (2015). Intraspecific variation in the foraging ecology of kea, the world’s only mountain-and rainforest-dwelling parrot. N. Z. J. Ecol. 39, 254–261.
Griffith, B., Scott, J. M., Carpenter, J. W., and Reed, C. (1989). Translocation as a species conservation tool: status and strategy. Science 245, 477–480. doi: 10.1126/science.245.4917.477
Haast, J. (1864). Notes on the ground-parrot of New Zealand (Strigops habroptilus). Ibis 6, 340–346. doi: 10.1111/j.1474-919X.1864.tb07874.x
Harper, G. A., Elliott, G. P., Eason, D. K., and Moorhouse, R. J. (2006). What triggers nesting of kakapo (Strigops habroptilus)? Notornis 53:160.
Henry, R. T. (1903). The Habits of the Flightless Birds of New Zealand: With Notes on Other New Zealand Birds. Wellington: Government Printer.
Hoffmann, M., Hilton-Taylor, C., Angulo, A., Böhm, M., Brooks, T. M., Butchart, S. H., et al. (2010). The impact of conservation on the status of the world’s vertebrates. Science 330, 1503–1509. doi: 10.1126/science.1194442
Hogg, A. G., Heaton, T. J., Hua, Q., Palmer, J. G., Turney, C. S., Southon, J., et al. (2020). SHCal20 southern hemisphere calibration, 0–55,000 years cal BP. Radiocarbon 62, 759–778. doi: 10.1017/RDC.2020.59
Horrocks, M., D’Costa, D., Wallace, R., Gardner, R., and Kondo, R. (2004). Plant remains in coprolites: diet of a subalpine moa (Dinornithiformes) from southern New Zealand. Emu 104, 149–156. doi: 10.1071/MU03019
Horrocks, M., Salter, J., Braggins, J., Nichol, S., Moorhouse, R., and Elliott, G. (2008). Plant microfossil analysis of coprolites of the critically endangered kakapo (Strigops habroptilus) parrot from New Zealand. Rev. Palaeobot. Palynol. 149, 229–245. doi: 10.1016/j.revpalbo.2007.12.009
Huang, P.-Y., Poon, E. S. K., Wong, A. T. C., So, I. W. Y., Sung, Y.-H., and Sin, S. Y. W. (2021). DNA metabarcoding reveals the dietary composition in the endangered black-faced spoonbill. Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-97337-w
Huson, D. H., Beier, S., Flade, I., Górska, A., El-Hadidi, M., Mitra, S., et al. (2016). MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12:e1004957. doi: 10.1371/journal.pcbi.1004957
Johnson, P. (1976). Vegetation associated with kakapo (Strigops habroptilus gray) in Sinbad gully, Fiordland, New Zealand. N. Z. J. Bot. 14, 151–159. doi: 10.1080/0028825X.1976.10428889
Kartzinel, T. R., Chen, P. A., Coverdale, T. C., Erickson, D. L., Kress, W. J., Kuzmina, M. L., et al. (2015). DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Natl. Acad. Sci. 112, 8019–8024. doi: 10.1073/pnas.1503283112
Kerley, G. I., Kowalczyk, R., and Cromsigt, J. P. (2012). Conservation implications of the refugee species concept and the European bison: king of the forest or refugee in a marginal habitat? Ecography 35, 519–529. doi: 10.1111/j.1600-0587.2011.07146.x
Kerley, G. I., te Beest, M., Cromsigt, J. P., Pauly, D., and Shultz, S. (2020). The protected area paradox and refugee species: the giant panda and baselines shifted towards conserving species in marginal habitats. Conserv. Sci. Pract. 2:e203. doi: 10.1111/csp2.203
Kohn, M. H., and Wayne, R. K. (1997). Facts from feces revisited. Trends Ecol. Evol. 12, 223–227. doi: 10.1016/S0169-5347(97)01050-1
Ladley, J. J., and Kelly, D. (1996). Dispersal, germination and survival of New Zealand mistletoes (Loranthaceae): dependence on birds. N. Z. J. Ecol. 20, 69–79.
Large, M., and Braggins, J. (1991). Spore atlas of New Zealand ferns and ferns allied. New Zeal J Bot 29, 1–68.
Ledgard, N., and Cath, P. (1983). Seed of New Zealand Nothofagus Species: Studies of Seed Weight, Viability, Shape and the Effect of Varying Stratification Periods. Christchurch: New Zealand Forest Service.
Legendre, P., and Anderson, M. J. (1999). Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol. Monogr. 69, 1–24. doi: 10.1890/0012-9615(1999)069[0001:DBRATM]2.0.CO;2
Lo Gullo, M., Glatzel, G., Devkota, M., Raimondo, F., Trifilò, P., and Richter, H. (2012). Mistletoes and mutant albino shoots on woody plants as mineral nutrient traps. Ann. Bot. 109, 1101–1109. doi: 10.1093/aob/mcs033
Macqueen, P., Seddon, J. M., Austin, J. J., Hamilton, S., and Goldizen, A. W. (2010). Phylogenetics of the pademelons (Macropodidae: Thylogale) and historical biogeography of the Australo-Papuan region. Mol. Phylogenet. Evol. 57, 1134–1148. doi: 10.1016/j.ympev.2010.08.010
Mallott, E. K., Garber, P. A., and Malhi, R. S. (2018). trnL outperforms rbcL as a DNA metabarcoding marker when compared with the observed plant component of the diet of wild white-faced capuchins (Cebus capucinus, primates). PLoS One 13:e0199556. doi: 10.1371/journal.pone.0199556
McEwan, W. M., and McEwan, W. (1978). The food of the New Zealand pigeon (Hemiphaga novaeseelandiae novaeseelandiae). N. Z. J. Ecol., 99–108.
Moar, N. T. (1993). Pollen grains of New Zealand dicotyledonous plants. Lincoln: Manaaki Whenua Press.
Moar, N., Wilmshurst, J., and McGlone, M. (2011). Standardizing names applied to pollen and spores in New Zealand quaternary palynology. N. Z. J. Bot. 49, 201–229. doi: 10.1080/0028825X.2010.526617
Monsarrat, S., Novellie, P., Rushworth, I., and Kerley, G. (2019). Shifted distribution baselines: neglecting long-term biodiversity records risks overlooking potentially suitable habitat for conservation management. Philos. Trans. R. Soc. B 374:20190215. doi: 10.1098/rstb.2019.0215
Moore, P. D., Webb, J. A., and Collison, M. E. (1991). Pollen Analysis. Oxford: Blackwell Scientific Publications.
Murphy, E. C. (1992). The effects of a natural increase in food supply on a wild population of house mice. N. Z. J. Ecol. 16, 33–33.
Murphy, D. J., and Kelly, D. (2003). Seasonal variation in the honeydew, invertebrate, fruit and nectar resource for bellbirds in a New Zealand mountain beech forest. N. Z. J. Ecol. 27, 11–23.
O’Donnell, C., and Dilks, P. J. (1994). Foods and foraging of forest birds in temperate rainforest, South Westland, New Zealand. N. Z. J. Ecol. 18, 87–107.
O’Rourke, D. R., Bokulich, N. A., MacManes, M. D., and Foster, J. T. (2020). A total crapshoot? Evaluating bioinformatic decisions in animal diet metabarcoding analyses. Ecol. Evol. 10, 9721–9739. doi: 10.1002/ece3.6594
Ogden, J., Stewart, G., and Allen, R. (1996). “Ecology of New Zealand Nothofagus forests” in The Ecology and Biogeography of Nothofagus Forests. eds. T. T. Veblen, R. S. Hill, and J. Read (New Haven: Yale University Press).
Oksanen, J., Blanchet, F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). Vegan: Community Ecology Package. R package Version 2.5-6. Available at: https://CRAN.R-project.org/package=vegan.
Panvini, A. D., and Eickmeier, W. G. (1993). Nutrient and water relations of the mistletoe Phoradendron leucarpum (Viscaceae): how tightly are they integrated? Am. J. Bot. 80, 872–878. doi: 10.1002/j.1537-2197.1993.tb15307.x
Patykowski, J., Dell, M., Wevill, T., and Gibson, M. (2018). Rarity and nutrient acquisition relationships before and after prescribed burning in an Australian box-ironbark forest. AoB Plants 10:ply032. doi: 10.1093/aobpla/ply032
Penrod, K. A., and McCormick, L. H. (1996). Abundance of viable hay-scented fern spores germinated from hardwood forest soils at various distances from a source. Am. Fern J. 86, 69–79. doi: 10.2307/1547640
Poinar, H. N., Hofreiter, M., Spaulding, W. G., Martin, P. S., Stankiewicz, B. A., Bland, H., et al. (1998). Molecular Coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 281, 402–406. doi: 10.1126/science.281.5375.402
Powlesland, R. G., Greene, T. C., Dilks, P. J., Moorhouse, R., Moran, L., Taylor, G., et al. (2009). Breeding biology of the New Zealand kaka (Nestor meridionalis) (Psittacidae, Nestorinae). Notornis 56, 11–33.
Powlesland, R., Lloyd, B., Best, H., and Merton, D. (1992). Breeding biology of the kakapo Strigops habroptilus on Stewart Island, New Zealand. Ibis 134, 361–373. doi: 10.1111/j.1474-919X.1992.tb08016.x
Powlesland, R., Merton, D. V., and Cockrem, J. F. (2006). A parrot apart: the natural history of the kakapo (Strigops habroptilus), and the context of its conservation management. Notornis 53:3.
Putman, R. (1995). Ethical considerations and animal welfare in ecological field studies. Biodivers. Conserv. 4, 903–915. doi: 10.1007/BF00056197
Quéméré, E., Aucourd, M., Troispoux, V., Brosse, S., Murienne, J., Covain, R., et al. (2021). Unraveling the dietary diversity of Neotropical top predators using scat DNA metabarcoding: a case study on the elusive Giant otter. Environ. DNA 3, 889–900. doi: 10.1002/edn3.195
Querejeta, M., Lefort, M.-C., Bretagnolle, V., and Boyer, S. (2022). Metabarcoding faecal samples to investigate spatiotemporal variation in the diet of the endangered Westland petrel (Procellaria westlandica). Peer Community Ecol. doi: 10.1101/2020.10.30.360289 [E-pub ahead of print].
R Core Team (2013). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Ramsey, C. B. (2009). Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360. doi: 10.1017/S0033822200033865
Romero, L. M. (2004). Physiological stress in ecology: lessons from biomedical research. Trends Ecol. Evol. 19, 249–255. doi: 10.1016/j.tree.2004.03.008
Rowe, P., Millar, I., and Worthy, T. (1994). Exploration on Garibaldi ridge–Euphrates cave, Kahurangi National Park. N. Z. Speleol. Bull. 9, 271–290.
Ruscoe, W. A., Wilson, D., McElrea, L., McElrea, G., and Richardson, S. J. (2004). A house mouse (Mus musculus) population eruption in response to rimu (Dacrydium cupressinum) seedfall in southern New Zealand. N. Z. J. Ecol. 28, 259–265.
Sanz, M., Daura, J., Égüez, N., and Brugal, J.-P. (2016). Not only hyenids: a multi-scale analysis of upper Pleistocene carnivore coprolites in Cova del Coll Verdaguer (NE Iberian Peninsula). Palaeogeogr. Palaeoclimatol. Palaeoecol. 443, 249–262. doi: 10.1016/j.palaeo.2015.11.047
Shehzad, W., McCarthy, T. M., Pompanon, F., Purevjav, L., Coissac, E., Riaz, T., et al. (2012). Prey preference of snow leopard (Panthera uncia) in south Gobi, Mongolia. PLoS One 7:e32104. doi: 10.1371/journal.pone.0032104
Srivathsan, A., Sha, J. C. M., Vogler, A. P., and Meier, R. (2015). Comparing the effectiveness of metagenomics and metabarcoding for diet analysis of a leaf-feeding monkey (Pygathrix nemaeus). Mol. Ecol. Resour. 15, 250–261. doi: 10.1111/1755-0998.12302
Stone, Z. L., Burns, B., Moorhouse, R., and Clout, M. N. (2017). Kakapo habitat selection on Hauturu-o-toi in relation to plant phenology. N. Z. J. Ecol. 41, 207–217. doi: 10.20417/nzjecol.41.32
Sweetapple, P. J. (2008). Spatial variation in impacts of brushtail possums on two Loranthaceous mistletoe species. N. Z. J. Ecol. 32, 177–185.
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., et al. (2007). Power and limitations of the chloroplast trn L (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14–e14. doi: 10.1093/nar/gkl938
Tait, A., Basher, R., Thompson, C., Burgess, S., McKenzie, R., Mullan, B., et al. (2001). The Climate of Otago: Patterns of Variation and Change. Dunedin: Otago Regional Council.
Trewick, S. (1996). The diet of kakapo (Strigops habroptilus), takahe (Porphyrio mantelli) and pukeko (P. porphyrio melanotus) studied by faecal analysis. Notornis 43, 79–112.
Türe, C., Böcük, H., and Aşan, Z. (2010). Nutritional relationships between hemi-parasitic mistletoe and some of its deciduous hosts in different habitats. Biologia (Bratisl.) 65, 859–867. doi: 10.2478/s11756-010-0088-5
Van Riper, C. III, Van Riper, S. G., Goff, M. L., and Laird, M. (1986). The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol. Monogr. 56, 327–344. doi: 10.2307/1942550
von Hurst, P., Moorhouse, R., and Raubenheimer, D. (2015). Preferred natural food of breeding kakapo is a high value source of calcium and vitamin D. J. Steroid Biochem. Mol. Biol. 164, 177–179. doi: 10.1016/j.jsbmb.2015.10.017
Walsh, J., Wilson, K.-J., and Elliott, G. P. (2006). Seasonal changes in home range size and habitat selection by kakapo (Strigops habroptilus) on Maud Island. Notornis 53, 143–149.
Wardle, J. (1984). The New Zealand Beeches: Ecology, Utilisation and Management. Wellington: New Zealand Forest Service.
Welker, F., Duijm, E., van der Gaag, K. J., van Geel, B., de Knijff, P., van Leeuwen, J., et al. (2014). Analysis of coprolites from the extinct mountain goat Myotragus balearicus. Quat. Res. 81, 106–116. doi: 10.1016/j.yqres.2013.10.006
Westerskov, K. (1981). Reischek’s 1890 paper on the kakapo (Strigops habroptilus) in the wild and captivity. Notornis 28, 263–280.
Willerslev, E., Davison, J., Moora, M., Zobel, M., Coissac, E., Edwards, M. E., et al. (2014). Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47–51. doi: 10.1038/nature12921
Williams, G. R. (1956). The kakapo (Strigops Habrotilus, gray): a review and reappraisal of a near-extinct species. Notornis 7, 29–56.
Wilmshurst, J. M., Anderson, A. J., Higham, T. F. G., and Worthy, T. H. (2008). Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc. Natl. Acad. Sci. 105, 7676–7680. doi: 10.1073/pnas.0801507105
Wilson, D. J., Grant, A., and Parker, N. (2006). Diet of kakapo in breeding and non-breeding years on Codfish Island (whenua Hou) and Stewart Island. Notornis 53:80.
Wood, J. R. (2006). Subfossil kakapo (Strigops habroptilus) remains from near Gibraltar rock, Cromwell gorge, Central Otago, New Zealand. Notornis 53:191.
Wood, J. R. (2010). Report on coprolite sampling, Honeycomb Hill and Euphrates Cave systems, March 2010. Unpublished Internal Report. Lincoln: Manaaki Whenua Landcare Research.
Wood, J. R., Crown, A., Cole, T. L., and Wilmshurst, J. M. (2016). Microscopic and ancient DNA profiling of Polynesian dog (kurī) coprolites from northern New Zealand. J. Archaeol. Sci. Rep. 6, 496–505. doi: 10.1016/j.jasrep.2016.03.020
Wood, J. R., Díaz, F. P., Latorre, C., Wilmshurst, J. M., Burge, O. R., and Gutiérrez, R. A. (2018). Plant pathogen responses to late Pleistocene and Holocene climate change in the Central Atacama Desert, Chile. Sci. Rep. 8, 1–8. doi: 10.1038/s41598-018-35299-2
Wood, J. R., Rawlence, N. J., Rogers, G. M., Austin, J. J., Worthy, T. H., and Cooper, A. (2008). Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes). Ice age Refug. Quat. Extinctions Issue Quat. Evol. Palaeoecol. 27, 2593–2602. doi: 10.1016/j.quascirev.2008.09.019
Wood, J. R., Richardson, S. J., McGlone, M. S., and Wilmshurst, J. M. (2020). The diets of moa (Aves: Dinornithiformes). N. Z. J. Ecol. 44, 1–21. doi: 10.20417/nzjecol.44.3
Wood, J. R., Vermeulen, M. J., Bolstridge, N., Briden, S., Cole, T. L., Rivera-Perez, J., et al. (2021). Mid-Holocene coprolites from southern New Zealand provide new insights into the diet and ecology of the extinct little bush moa (Anomalopteryx didiformis). Quat. Sci. Rev. 263:106992. doi: 10.1016/j.quascirev.2021.106992
Wood, J. R., and Wilmshurst, J. M. (2014). Late quaternary terrestrial vertebrate coprolites from New Zealand. Quat. Sci. Rev. 98, 33–44. doi: 10.1016/j.quascirev.2014.05.020
Wood, J. R., and Wilmshurst, J. M. (2016). A protocol for subsampling late quaternary coprolites for multi-proxy analysis. Quat. Sci. Rev. 138, 1–5. doi: 10.1016/j.quascirev.2016.02.018
Wood, J. R., Wilmshurst, J. M., Rawlence, N. J., Bonner, K. I., Worthy, T. H., Kinsella, J. M., et al. (2013b). A Megafauna’s microfauna: gastrointestinal parasites of New Zealand’s extinct moa (Aves: Dinornithiformes). PLoS One 8:e57315. doi: 10.1371/journal.pone.0057315
Wood, J. R., Wilmshurst, J. M., Richardson, S. J., Rawlence, N. J., Wagstaff, S. J., Worthy, T. H., et al. (2013a). Resolving lost herbivore community structure using coprolites of four sympatric moa species (Aves: Dinornithiformes). Proc. Natl. Acad. Sci. 110, 16910–16915. doi: 10.1073/pnas.1307700110
Wood, J. R., Wilmshurst, J. M., Wagstaff, S. J., Worthy, T. H., Rawlence, N. J., and Cooper, A. (2012a). High-resolution Coproecology: using coprolites to reconstruct the habits and habitats of New Zealand’s extinct upland moa (Megalapteryx didinus). PLoS One 7:e40025. doi: 10.1371/journal.pone.0040025
Wood, J. R., Wilmshurst, J. M., Worthy, T. H., Holzapfel, A. S., and Cooper, A. (2012b). A lost link between a flightless parrot and a parasitic plant and the potential role of coprolites in conservation Paleobiology. Conserv. Biol. 26, 1091–1099. doi: 10.1111/j.1523-1739.2012.01931.x
Worthy, T. H. (1997). Fossil deposits in the Hodges Creek cave system, on the northern foothills of Mt Arthur, Nelson. South Island, New Zealand. Notornis 44, 111–108.
Worthy, T. H., and Holdaway, R. N. (2002). The Lost World of the Moa: Prehistoric Life of New Zealand. Christchurch: Canterbury University Press.
Keywords: ancient DNA, conservation palaeobiology, coprolites, metabarcoding, New Zealand, palaeoecology
Citation: Boast AP, Wood JR, Bolstridge N, Perry GLW and Wilmshurst JM (2023) Ancient and modern scats record broken ecological interactions and a decline in dietary breadth of the critically endangered kākāpō parrot (Strigops habroptilus). Front. Ecol. Evol. 11:1058130. doi: 10.3389/fevo.2023.1058130
Edited by:
Chris Schneider, University of Alberta, CanadaReviewed by:
Alan Cooper, BlueSky Genetics, Ashton, AustraliaPiotr Bajdek, Independent Researcher, Częstochowa,Poland
Copyright © 2023 Boast, Wood, Bolstridge, Perry and Wilmshurst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander P. Boast, Ym9hc3RhQGxhbmRjYXJlcmVzZWFyY2guY28ubno=
 Alexander P. Boast
Alexander P. Boast Jamie R. Wood
Jamie R. Wood Nicola Bolstridge2
Nicola Bolstridge2 Janet M. Wilmshurst
Janet M. Wilmshurst