- 1Northwestern University, Evanston, IL, United States
- 2Negaunee Institute for Plant Conservation Science and Action, Chicago Botanic Garden, Glencoe, IL, United States
- 3Environmental Horticulture Department, University of Florida, Gainesville, FL, United States
- 4Department of Biology, Illinois College, Jacksonville, IL, United States
Orchids grow in diverse habitats worldwide with most (approximately 69%) growing on trees as epiphytes. Although orchid mycorrhizal fungi have been identified as potential drivers for terrestrial orchid distribution, the influence of these fungi on the fine-scale distribution of epiphytic orchids is poorly understood. In this study, we investigated the mycorrhizal fungal community and fine-scale distribution of Dendrophylax lindenii, a rare and endangered epiphytic orchid that is leafless when mature. We used amplicon sequencing to investigate the composition of orchid mycorrhizal fungi in the roots of 39 D. lindenii individuals in their natural habitat, the swamps of Florida. We compared the orchid mycorrhizal fungi of D. lindenii to those of co-occurring epiphytic orchids, as well as to the orchid mycorrhizal fungal communities of bark from potential host trees, with and without D. lindenii. Our results show that D. lindenii has a high specificity for a single Ceratobasidium species, which is widely distributed on phorophytes and detected in both wet and dry periods in the orchid’s habitat. This Ceratobasidium species was mostly absent or only recorded in low frequency in the roots of co-occurring epiphytic orchids. Phylogenetic analysis documented that this Ceratobasidium was conspecific with the strain that is used to germinate D. lindenii ex-situ. However, our findings suggest that laboratory germinated adult D. lindenii transplanted into the field had lower read abundances of this Ceratobasidium compared to naturally occurring plants. These findings suggest that this orchid mycorrhizal fungus may play a significant role in the fine-scale distribution of naturally occurring D. lindenii.
Introduction
Mycorrhizal fungi are well known mutualists that are essential for their plant partners’ abundance and spatial distribution (Smith and Read, 2010; McCormick and Jacquemyn, 2014). While ca. 69% of orchid species are tropical epiphytes (Zotz, 2016), little is known about the orchid mycorrhizal fungi (OMF) they associate with compared to temperate terrestrial orchids. Epiphytic orchids, like their terrestrial counterparts, enlist OMF to facilitate seed germination and seedling development, but it remains unclear to what degree epiphytes continue to utilize OMF into maturity (Dearnaley et al., 2012; Rasmussen et al., 2015; Selosse et al., 2022). Stable isotope work by Gebauer et al. (2016) revealed that a greater number than previously thought of orchids are likely reliant on OMF, and are functioning as myco-heterotrophs even though they are photosynthetic as adults. This finding of likely orchid dependence on OMF as adults, especially epiphytic orchids, raises the question of the potential role that OMF play in driving their fine-scale spatial distribution.
The drivers of fine scale epiphyte spatial distribution and host tree (phorophyte) specificity have been debated within the literature for over a century since the writings of Schimper (1888) see review by Wagner et al. (2015). Debate has focused on the role of various abiotic factors (e.g., microclimate and host bark characteristics) and biotic factors (e.g., symbiotic fungi and co-occurrence with moss). Research by McCormick et al. (2018) has demonstrated that while OMF may restrict terrestrial orchid distributions at local scales, at broad geographic scales terrestrial orchids are not constrained by OMF. Most of these findings were established for terrestrial orchids, with investigations of epiphytic orchids still pending (Li et al., 2021). Recently, studies have investigated fungal communities in the bark of phorophytes of epiphytic orchids, which providing insights into phorophyte specificity and spatial distribution of epiphytic orchids (Izuddin et al., 2019; Eskov et al., 2020; Pecoraro et al., 2021; Petrolli et al., 2021, 2022). Eskov et al. (2020) further explored OMF and revealed that fungi colonizing epiphytic orchid roots were significantly different from the phorophytes’ branches. Pecoraro et al. (2021) studied the phorophyte specificity of two epiphytic orchid species, as well as the environmental factors influencing the relationship between the orchids and their phorophytes. They concluded that the orchid phorophyte associations were influenced by the phorophyte bark’s OMF communities and potentially its pH and water holding capacity. Recent studies have also revealed examples of a strong fungal specificity of epiphytic orchids associated with a single OMF species, Ceratobasidium or Tulasnellaceae species (Rammitsu et al., 2019, 2020) despite inconclusive early studies (Gowland et al., 2013; Wang et al., 2017).
Amplicon sequencing, a type of environmental sequencing, is a cost-effective advancement for investigating fungal communities compared to traditional culture-based methods (McCormick and Jacquemyn, 2014; McCormick et al., 2018). Ectomycorrhizal (ECM) fungi as well as some OMF are known to be recalcitrant to being cultured and recent studies utilizing amplicon sequencing have detected a diversity of ECM fungi in the roots of both epiphytic and terrestrial orchids (Selosse et al., 2022). Additionally, amplicon sequencing can increase the detection of potential OMF in epiphytic orchid roots compared to Sanger Sequencing as Sanger Sequencing is often limited by the need for first culturing the fungi (Waud et al., 2016; Jacquemyn et al., 2017; Novotná et al., 2018; Johnson et al., 2021).
We chose the rare leafless epiphytic orchid Dendrophylax lindenii (Lindl.) Bentham ex Rolfe (Figure 1) as our study taxon to further document OMF communities of rare tropical epiphytic orchids and to examine the potential role of OMF as drivers of their phorophyte specificity. In addition to sampling the roots of D. lindenii we sampled co-occurring epiphytic orchids and the bark of potential phorophytes with and without D. lindenii to uncover evidence of OMF specificity during two periods, flooded and not flooded in the area of its natural distribution in the United States (Supplementary Figures S1, S2).
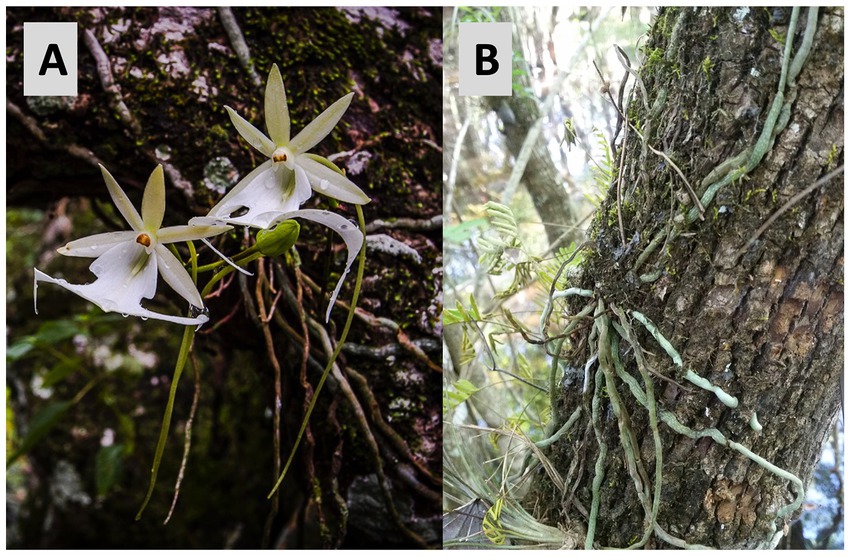
Figure 1. (A) Flowers of Dendrophylax lindenii (photo by Larry W. Richardson). (B) Dendrophylax lindenii roots growing on the tree trunk of a phorophyte, Fraxinus caroliniana.
Dendrophylax lindenii, also known as the Ghost Orchid, is restricted to southwestern Florida and the western tip of Cuba (Brown, 2002) where it remains vulnerable to poaching and environmental changes (Mújica et al., 2018, 2021). In Florida, less than 1,500 individuals are thought to remain (Haaland et al., 2022), and in Cuba, the number is even fewer [<500; (Mújica et al., 2018)]. In the Florida Panther National Wildlife Refuge where about 1/3rd of the state’s Ghost Orchids are found, Mújica et al. (2018) calculated that D. lindenii numbers will decline by 20% during the next decade. Consequently, the species is now a candidate for U.S. Federal protection under the Endangered Species Act (Haaland et al., 2022). The Florida habitats of D. lindenii consist of cypress domes and strand swamps in the Big Cypress Basin. According to a study by Mújica et al. (2018) in 2015, 69% of the growth of D. lindenii in Florida is found on the trunks of Fraxinus caroliniana Mill., while occurring less frequently (36%) on Annona glabra L. These trees are typically located in the lower canopy under Taxodium distichum (L.) Rich. (Brown, 2002; Stewart and Richardson, 2008). Although D. lindenii grows in a moist habitat, it experiences dry periods during the region’s dry season which lacks any standing water (Mújica et al., 2018).
Like all orchids, D. lindenii requires OMF for germination (Hoang et al., 2017). Early seedling stages of D. lindenii have a rudimentary ephemeral leaf. As an adult, the orchid lacks leaves and shoots and photosynthesizes predominantly via its roots (Benzing and Ott, 1981; Benzing et al., 1983; Hoang et al., 2017). Benzing and Ott (1981), have shown that the mature roots of D. lindenii utilizes CAM photosynthesis, and Chomicki et al. (2014) using microscopy hypothesized that it forms a mutualism with an OMF (Ceratobasidiaceae) to obtain carbon to supplement its photosynthesis. Furthermore, seed germination experiments by Hoang et al. (2017) and Mújica et al. (2018) have confirmed that D. lindenii associates with a Ceratobasidium and that this fungus is present in mature roots.
Our primary aim for this study was to identify the OMF associated with D. lindenii and to investigate the potential role of OMF in influencing its fine-scale distribution within naturally occurring populations (i.e., why it was found on some potential phorophytes and not on others). We tested two hypotheses: (1) D. lindenii has a specific community of OMF compared to co-occurring epiphytic orchids; and (2) the OMF colonizing D. lindenii are found in the bark of D. lindenii phorophytes in higher abundances than in the bark of trees without D. lindenii. Given that D. lindenii is currently state-listed as endangered, we primarily restricted our sampling to root tips to minimize damage to the plant. To investigate if additional OMF were missed with this sampling method we also investigated the fungal community of four whole roots.
Materials and methods
Study sites, tree bark and orchid root sampling
During 2016 and 2018 we collected >100 root and bark samples from five sites at the Florida Panther National Wildlife Refuge (FPNWR) a 10,684 ha area (Supplementary Table S1). Four of the sites were natural habitats for D. lindenii. The fifth site lacked naturally occurring plants but had D. lindenii explants that were micropropagated under axenic conditions in the lab and subsequently transplanted (attached) on appropriate species of trees. The site with explants we identified as Site 4 in our study. Most of the sites were dominated (over 90%) by F. caroliniana as the main phorophyte. Sites were either sloughs or strand swamps and were separated by about 1 km from each other. When we collected samples in 2016 (March), FPNWR sites all had standing water in sloughs and swamps (Supplementary Figure S1), but all sites were dry (not flooded) when we sampled in 2018 (April) (Supplementary Figure S2). This sampling period in 2018 was unusually dry. The precise sites at the FPNWR are not disclosed herein because D. lindenii and several co-occurring orchids are state-listed as endangered and remain highly vulnerable to poaching. For each site, Special Use collecting permits were obtained (USFWS, OMB Control # 1018–0102), and permission to access and sample D. lindenii populations was subsequently granted.
In March 2016, root samples were collected from four sites at the FPNWR Sites 1–4 (Supplementary Table S1). Root samples were collected from the leafless epiphytic orchid species D. lindenii (n = 9) and several co-occurring epiphytic orchids: Campylocentrum pachyrrhizum (Rchb.f.) Rolfe (n = 3), Dendrophylax porrectus (Rchb.f.) Carlsward & Whitten (n = 6), Epidendrum amphistomum A. Rich. (n = 4), Epidendrum nocturnum Jacq. (n = 1) and Prosthechea cochleata (L.) W. E. Higgins (n = 3). Simultaneous with the collection of root samples, bark samples were collected from phorophytes adjacent of all epiphytic orchids (Supplementary Table S1).
In April 2018, sampling of the roots of an additional 27 D. lindenii plants was carried out at the original four sites plus one additional site (Site 5). Concurrent with the root tissue collection, bark samples (n = 57) were collected from phorophytes of D. lindenii and trees without D. lindenii (Supplementary Table S1). Five trees with and five trees without D. lindenii individuals were sampled at each of the five sites. The sampling design considered the position of D. lindenii on the tree and bark samples were collected from (1) the base of the tree trunk, (2) above D. lindenii, (3) the side of roots of D. lindenii root; and (4) the opposite side of the tree trunk (Supplementary Figure S3). In instances where D. lindenii was not present bark samples were collected from the base of the tree and three additional samples were taken at a height of at breast height (1.5 M) from base, where D. lindenii would typically grow.
Additionally, we conducted a pilot study to assess the success of amplicon sequencing of root tips for revealing the OMF community of D. lindenii. We obtained 50 mm root samples collected from three mature individuals of D. lindenii at the FPNWR. A root of a D. lindenii that was home cultivated from Redlands, Florida was also sampled. For this pilot study, roots were cut into 5 mm long segments starting from the tip and labelled alphabetically (i.e., A, B, C, etc. see Supplementary Figure S4).
DNA extraction, PCR amplification, and amplicon sequencing
Approximately 5 mm of root tip and bark tissue was collected and stored in cetyltrimethylammonium bromide (CTAB) buffer. Root and bark samples were surface sterilized with 70% ethanol, and 50% Clorox® (2.6% sodium hypochlorite) using the method outlined in Bayman et al. (1997). Next, genomic DNA was extracted from root samples using the Qiagen DNeasy extraction kits (Qiagen, Valencia, CA, United States) following the manufacturer’s instructions. DNA from bark samples was extracted with the modified CTAB method of Murray and Thompson (1980), and for difficult to extract samples the MOBIO Power Soil DNA Extraction kit (MOBIO Laboratories, Carlsbad, CA, United States) was used following the manufacturer’s instruction.
The extracted genomic DNA from the 2016 root samples was amplified using the primers: ITS86f (5′- GTGAATCATCGAATCTTTGAA-3′; Turenne et al., 1999) and ITS4 (5′- TCCTCCGCTTATTGATATGC-3′; White et al., 1990). These fungal primers (ITS86F/ITS4) amplify the internal transcribed region ITS, the standard fungal barcode, for ITS subregion 2 which is shown to be effective for delimiting OMF such as those in the Cantharellales.
Next, amplicons from the PCR products were produced using a three step PCR sequencing protocol (see Johnson et al., 2021 materials and methods). This included PCR steps that used modified primers with indices from the Nextera XT kit for 96 indices to sequence 2 × 250 bp. The final amplicon libraries generated for root and bark samples were quantified using a Qubit dsDNA HS kit (Invitrogen) and a Bioanalyzer-Agilent 2100 (Agilent Technologies, Santa Clara, CA, United States). Final amplicon libraries for root and bark samples were pooled together in equimolar concentrations and the final pool was then sequenced on an Illumina MiSeq at the Pritzker Lab at the Field Museum (Chicago, IL).
The root sections from the pilot study, root tips, and bark samples from 2018 were PCR amplified using modified fungal primers ITS86F/ITS4 with barcodes supplied from Novogene Bioinformatics Institute (Beijing, China) following the protocol applied in 2016. The generated final amplicon libraries were pooled to equimolar concentrations then shipped to Novogene and sequenced on an Illumina HiSeq. Sequences generated from this study were submitted to NCBI’s Sequence Read Archive under the BioProject PRJNA948888.
Bioinformatics and statistical analyses
Initially, bioinformatics analyses were performed on roots and bark collected in 2016 separately. Subsequently, the sequences obtained from the bark, root, and root sections of the 2018 dataset were integrated, and bioinformatics analyses were conducted on these samples collectively to determine patterns of similar Operational Taxonomic Units (OTUs) between sample types.
To conduct bioinformatic analyses, the sequences were first quality filtered, followed by OTU clustering utilizing the PIPITS pipeline (version 1.4.0) default settings as described by Gweon et al. (2015). Briefly, PIPITS joined reads and quality filtered short reads (<50 bp), extracted non ITS fungal reads with the script ITSx (Bengtsson-Palme et al., 2013), then clustered OTUs at 95% sequence similarity. Additional PIPITS scripts assigned taxonomy to OTUs with the Ribosomal Database Project Classifier [a Naïve Bayesian Classifier (Wang et al., 2007)] and the UNITE database (Nilsson et al., 2019). Sequences for the HiSeq dataset was analyzed separately from the 2016 MiSeq dataset. The single difference between analysis of the MiSeq data analyses and HiSeq data analyses was omitting the ITSx step for the HiSeq data.
To further investigate differences between fungal communities we filtered rare OTUs that were less than 1,000 sequences, and the raw read abundances were then normalized with Cumulative Sum Scaling in the R package metagenomeSeq (Paulson et al., 2013). All statistical analyses were conducted within R (R Core Team, 2022). The visualization of abundance of sequences was first accomplished using Krona charts, which were generated using Krona-2.8.1 within R (Ondov et al., 2011). Bar graphs showing relative and read abundances were produced with the R package ggplot2. To better visualize differences between read abundances, the y-axis was truncated using the R package ggbreak (Xu et al., 2021).
Principal coordinate analysis (PCoA) was generated using Bray-Curtis distances with the R package vegan and visualized with ggplot2 (Wickham, 2016). Significance between fungal communities of D. lindenii and epiphytic orchid roots, phorophyte and the bark of trees without D. lindenii present, and location of sites were determined with “permutational manova” (Anderson, 2001) in R package vegan (adonis2 function) by first permuting the raw data with 9,999 permutations (Oksanen et al., 2022). Prior to executing adonis2 for the permutational multivariate analysis of variance (PERMANOVA) we also investigated the dispersion for groups using another vegan function betadisper. In addition, pairwise comparisons were completed for the PERMANOVA using the pairwiseAdonis R package with Bonferroni corrections (Martinez Arbizu, 2017). p-values that are < 0.05 were considered significant.
Phylogenetic analyses were undertaken to investigate relationships among the community of recovered Ceratobasidiaceae sequences from 2016 and 2018 root and bark fungal samples. The phylogenetic tree incorporated Ceratobasidium sequences from NCBI GenBank. A sequence of Tulasnella from the UNITE fungal database was used as an outgroup. We used MUSCLE (Edgar, 2004) in AliView version 1.27 (Larsson, 2014) for multiple sequence alignments and also used AliView to generate a Maximum Likelihood tree using the default settings of the program FastTree version 2.1.10 (Price et al., 2009). The final tree was rooted and visualized using FigTree version 1.4.4.1
Results
Sequence analyses of root and bark samples
Fungal sequence data were obtained from roots of D. lindenii and other co-occurring epiphytic orchids collected in the field in 2016 and 2018. Root samples in 2016 and 2018 yielded 537,371 (n = 26) and 1,691,086 reads (n = 30), respectively, resulting in the identification of 526 and 1,077 Operational Taxonomic Units (OTUs) at the 95% sequence identity level. Sequences generated from sections of whole roots yielded 3,205,959 reads (n = 37 root section samples) and resulted in the identification of 1,372 OTUs. In 2016, we collected 30 bark samples and sequencing yielded 693,482 reads with a total of 550 OTUs. Most phorophytes sampled in 2016 were from F. caroliniana (over 90%) with a small proportion of A. glabra also being sampled (Supplementary Table S1). Additionally, a bark sample was collected from a Taxodium distichum that had an explant affixed to it (Supplementary Table S1). In 2018 we successfully sequenced 57 bark samples mostly from F. caroliniana, from trees with D. lindenii (n = 43) and trees without D. lindenii (n = 14), yielding 7,245,995 reads with a total of 1,141 OTUs resolved.
The increase in read and OTU counts in 2018 can be attributed to the use of the HiSeq platform instead of the MiSeq platform, as well as the greater sampling intensity of root samples. The average OTU richness observed in root and bark samples for 2016 was 67 and 55 respectively, whereas the OTU richness for 2018 samples for roots was 233 and 304 for bark (Supplementary Table S1). Unfortunately, no amplicon libraries were generated for root samples collected at Site 2 in 2016 as the library preps were unsuccessful resulting in sequence data that was unsuitable for data analysis. The analysis of the pilot study examining potential differences in fungal communities in different sections of entire roots of both cultivated and wild collected D. lindenii documented that fungal communities were similar across all sections but were different between cultivated vs. wild collected plants (Supplementary Figure S5).
Ceratobasidiaceae is the dominant OMF associated with Dendrophylax lindenii
The fungal communities of naturally occurring D. lindenii roots across all sites were observed to be similar and dominated by several Ceratobasidiaceae, even during the flooded (2016) (Figure 2; Supplementary Figure S6A) and not flooded (2018) periods (Supplementary Figure S6B). The dominant Ceratobasidiaceae OTUs associated with D. lindenii were OTU 11 (recovered from bark samples), OTU 14 (recovered from 2016 root samples); and OTU 76 (recovered from both 2018 root and bark samples). Phylogenetic analysis resolved each of these dominant Ceratobasidium OTUs as part of a well-supported monophyletic clade, Clade 2, and are considered conspecific (Figure 3). A visual analysis of sequence alignments further supports this finding, as Clade 2 OTUs exhibit a sequence similarity of more than 98%. Ceratobasidium Clade 2 includes other individuals that were previously recovered from mature roots of D. lindenii. Included in Clade 2 is Dlin-394, which was derived from cultures isolated from mature roots of D. lindenii and has been used to germinate seeds of D. lindenii (Hoang et al., 2017).
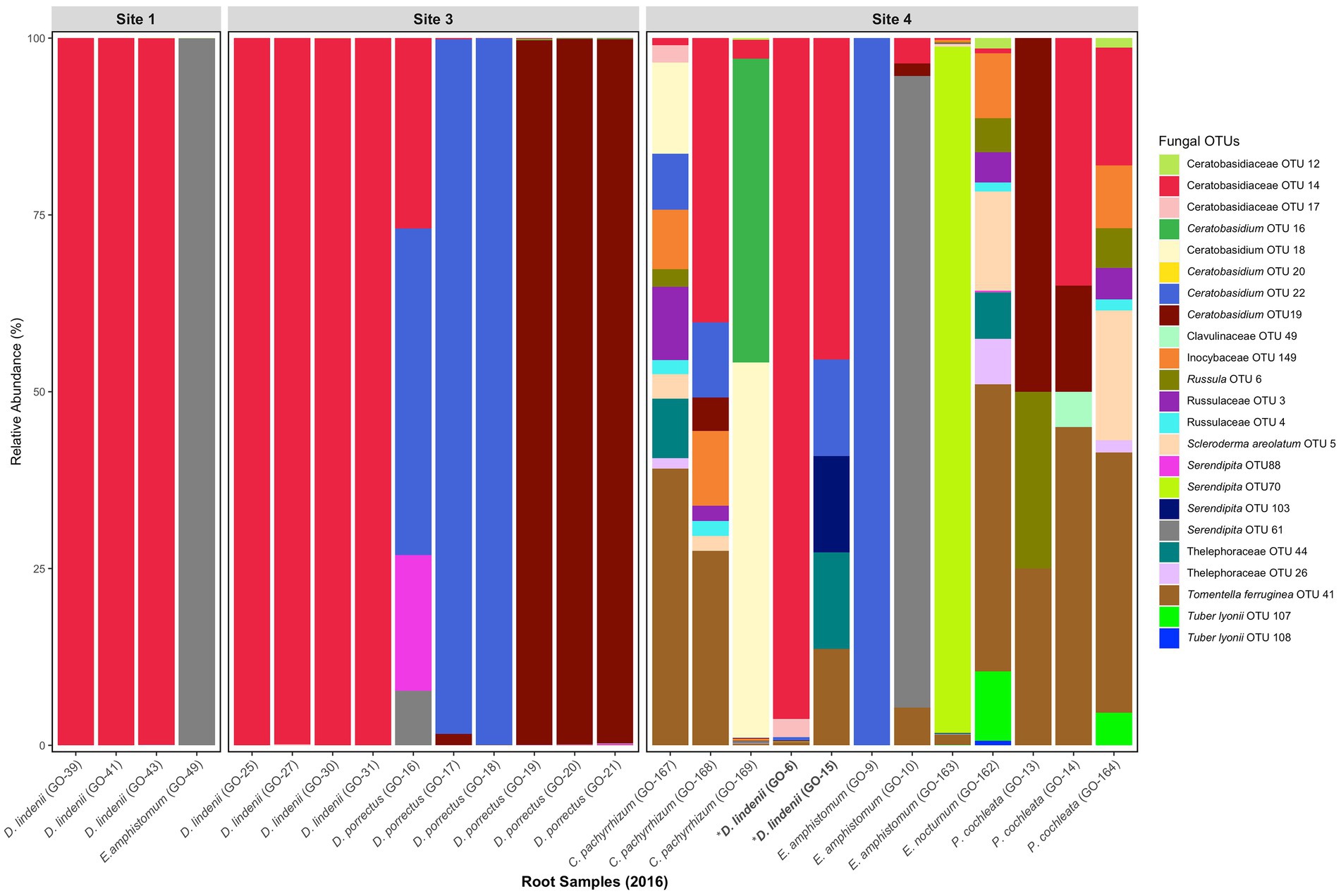
Figure 2. Relative abundance of fungal OTUs (putative OMF) obtained from root samples of D. lindenii and co-occurring epiphytic orchids in 2016. Dendrophylax lindenii explants at site 4 are bolded and are labeled with an asterisk.
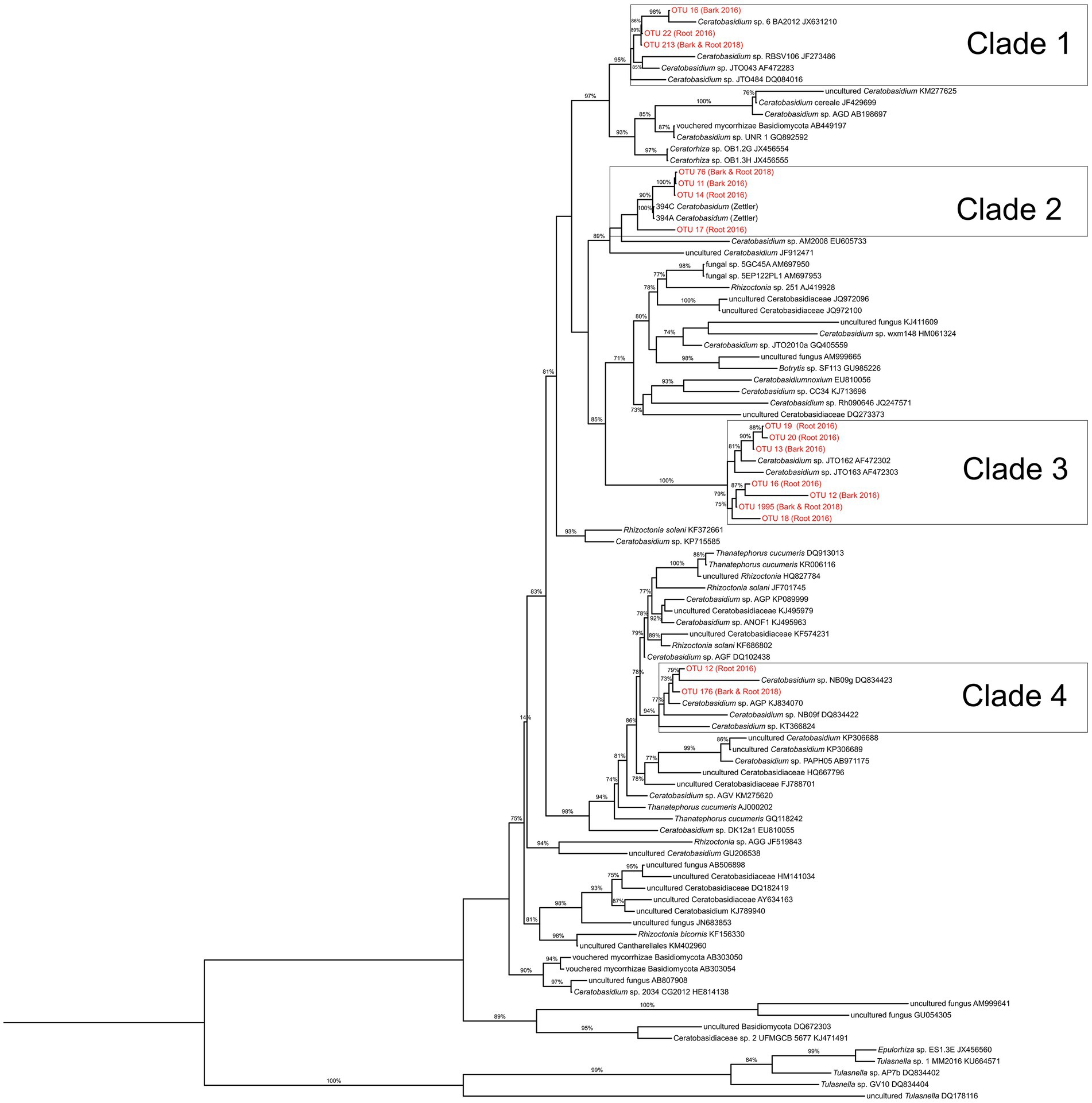
Figure 3. Maximum likelihood phylogeny of putative Ceratobasidiaceae species based on ITS data set of 95 taxa using FastTree constructed with default parameters. A cultured Tulasnella was used as an outgroup taxon. Bootstrap support values above 70% are reported. Sequences generated during this study are indicated in red.
The pilot study undertaken to assess if the OMF community recovered from root tips of D. lindenii provided was reflective of the full root OMF community provided further evidence of the dominance of Ceratobasidium Clade 2 in naturally occurring D. lindenii. The majority of root sections from naturally occurring D. lindenii, including the root tips, were dominated by Ceratobasidium Clade 2 (Supplementary Figure S5). Of note, root section samples of the home-cultivated D. lindenii lacked Ceratobasidiaceae OTUs. Instead, an abundance of Ascomycota OTU reads (Lasiodiplodia OTU 1174 and Diaporthales OTU 627) were recovered (Supplementary Figure S5).
Ceratobasidium Clade 2 was present in most samples of D. lindenii (Supplementary Figure S6C); however, it was absent or only had very low read numbers from the roots of co-occurring epiphytic orchids (Figure 2). Other taxa of Ceratobasidiaceae were recovered from these orchids, e.g., Ceratobasidiaceae OTUs 19 and 22 were abundant in root samples of D. porrectus (Figure 2). These OTUs belonged to different clades (Clade 1 and 3, Figure 3). Other OMF taxa that were recovered from co-occurring epiphytic orchid roots collected in 2016 (Figure 2; Supplementary Figures S6A,B) included taxa of putative OMF Serendipitaceae and ECM fungi such as Inocybaceae, Russulaceae, Scleroderma, Thelephoraceae, Tomentella, and Tuber species. While present in lower proportions (<1%) we did not detect a widespread presence of Tulasnellaceae OTUs, a traditional OMF.
Diversity of OMF in bark of Dendrophylax lindenii phorophytes and trees without Dendrophylax lindenii
Unlike the root fungal community, the bark fungal community was not dominated by Ceratobasidium Clade 2 or other Ceratobasidiaceae OTUs. Ceratobasidiaceae OTUs accounted for less than 5% of the total reads (Supplementary Figures S7A,B). Nonetheless, Ceratobasidium Clade 2 was present in most bark samples (Figure 4). Other putative OMF detected in bark samples were also rare and included a few Serendipitaceae and Tulasnella OTUs. Additional rare OTUs also included ECM fungi including Mycena, Russula, Thelephoraceae, and Tomentella.
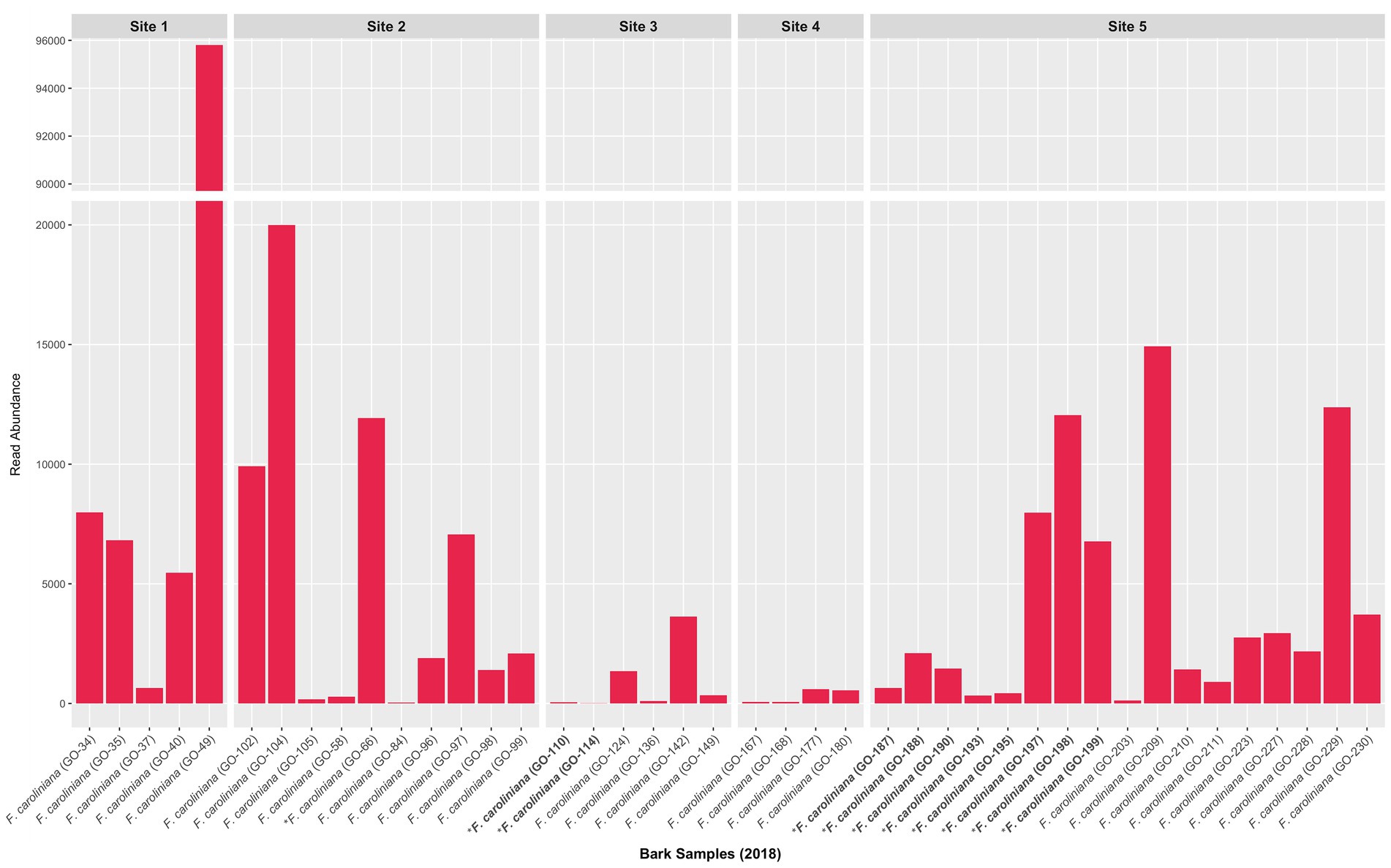
Figure 4. Read abundance of Ceratobasidium clade 2 obtained from bark collected in 2018 from trees with and without naturally occurring D. lindenii. Bar graph is condensed to better visualize the sample with the highest read abundance. Trees without naturally occurring D. lindenii are represented in bold and have an asterisk. All bark samples shown in graph are from F. caroliniana obtained at various positions on the trunk except the base (see Supplementary Table S1 for bark sample positions).
The principal coordinate analysis (PCoA) and subsequent PERMANOVA tests on root samples collected in both 2016 and 2018 showed significant differences between sites and orchid species (Supplementary Figures S8A,B). Specifically, the 2016 root samples reveal potentially significant differences in both orchid species (PERMANOVA: F5, 25 = 2.11, R2 = 0.29, p < 0.05, betadisper: F = 2.08, p = 0.11) and location (PERMANOVA: F2, 25 = 3.20, R2 = 0.18, p < 0.05, betadisper: F = 5.09, p = 0.01). Furthermore, pairwise comparisons showed significant differences between Site 4 (site with only introduced D. lindenii lab grown explants) and the two other sites, Site 1 (adjusted p = 0.009) and Site 3 (adjusted p = 0.003). In addition, Site 4 also differed from Site 3 (adjusted p = 0.03). Similarly, the D. lindenii root samples collected in 2018 revealed significant differences by site (PERMANOVA: F4,24 = 1.26, R2 = 0.20, p = 0.036, betadisper: F = 1.24, p > 0.5). However, pairwise comparisons revealed no significant differences between sites when adjusted p values were generated.
PCoA of bark data collected for 2016 (Supplementary Figure S9A) revealed significant differences for both location (PERMANOVA: F4, 34 = 1.82, R2 = 0.16, p < 0.05, betadisper: F = 1.47, p > 0.05) and the presence of D. lindenii (PERMANOVA: F4, 34 = 1.50, R2 = 0.03, p = 0.028, betadisper: F = 0.69, p = 0.4). Although sites were not different during the flooded period of 2016, pairwise comparisons of the 2018 bark data (corresponding PCoA is Supplementary Figure S9B) revealed differences between Site 1 and Site 3 (adjusted p = 0.01); differences between Site 2 and Site 5 (adjusted p = 0.03); and differences between Site 2 and Site 4 (adjusted p = 0.05).
Discussion
Our study provides strong evidence that D. lindenii may have a high specificity for a single Ceratobasidiaceae OTU (Ceratobasidium Clade 2) in its natural habitat at the Florida Panther National Wildlife Refuge (FPNWR). This OTU was found to be abundant in D. lindenii roots, and rare (<1% of total reads) in other co-occurring epiphytic orchids at the FPNWR. Ceratobasidium Clade 2 was also widespread at all sites in the bark of phorophytes with D. lindenii and potential phorophyte trees without D. lindenii during both flooded (2016) and not flooded periods (2018).
This apparent extreme fungal specificity for one OMF, Ceratobasidium Clade 2, is similar to that reported for mycoheterotrophic orchids (McKendrick et al., 2002; Selosse et al., 2002), terrestrial orchids (Thixton et al., 2020), and some epiphytic orchids (Otero et al., 2002, 2004; Graham and Dearnaley, 2012; Rammitsu et al., 2019, 2021a,b). Our findings of potential high specificity with Ceratobasidium Clade 2 aligns with previous studies demonstrating the importance of Ceratobasidium taxa supporting healthy populations of other epiphytic orchids. For instance, Qin et al. (2021) and Rammitsu et al. (2019) reported on other leafless epiphytic orchids that have a high specificity for single Ceratobasidium species. Furthermore, Ceratobasidium Clade 2 is conspecific (>99% similar) with Ceratobasidium (Dlin-394) that was isolated and brought into culture from roots of D. lindenii that was used to germinate D. lindenii seeds (Hoang et al., 2017).
We also observed evidence of possible specificity in some of the other co-occurring epiphytic orchids, but the sample size was small for many of these epiphytic orchids and clear hypotheses could not be tested. Nevertheless, these orchids associated with different Ceratobasidium. For example, Ceratobasidium OTU 19 and 22 were detected primarily in D. porrectus, another leafless epiphytic orchid. We hypothesize, with a caveat of small sample size, that mature roots of leafless epiphytic orchids are dominated by a single OMF unique to that species.
In addition to traditional OMF, we detected low read abundances of ECM fungi in the roots of the epiphytic orchids examined. This is in contrast to aerial roots of V. planifolia which were heavily colonized by ECM fungi (Johnson et al., 2021). Vanilla planifolia is a hemiepiphytic orchid and it is possible that the ECM fungi in the aerial roots are from systemic colonization emanating from the terrestrial roots. ECM fungi have been commonly reported from terrestrial orchids, but except for those detected by Johnson et al. (2021) an abundance of ECM fungi has not been reported colonizing arial/epiphytic orchid roots.
Foliar orchids exhibited lower read abundances relative to D. lindenii and other leafless epiphytic orchids (data not shown) was observed in our study. We hypothesize that the greater photosynthetic capacity of foliar orchids provided by their leaves reduces their dependence on OMF for supplemental fungal carbon.
Amplicon sequencing enabled us to document the presence of ECM fungi that are resistant to culturing in the orchid root communities. However, we were not successful in recovering species of Tulasnellaceae. Whether this was actually due to very low abundance of these species is not clear. Some primer bias of the primer pair ITS86F/ITS4 for Tulasnella species has been reported and this primer pair is likely poor for detecting Tulasnella spp. (Tedersoo et al., 2015; Vogt-Schilb et al., 2020; Johnson et al., 2021; Rammitsu et al., 2021a). Thus, future work using primers that are not biased towards Tulasnellaceae is needed.
While bark is not a carbon source for orchids (Eskov et al., 2020), it is the likely source of the OMF that epiphytic orchids need for establishment including seed germination and seedling growth (Rasmussen et al., 2015). Pellitier et al. (2019) documented that tree bark can serve as an environmental filter for the fungal communities available to epiphytic orchids. Thus, the distribution of OMF in tree bark throughout an orchid’s range could influence its fine-scale distribution. Ceratobasidium OTU Clade 2 was recovered from all trees with D. lindenii, the fungus was also recovered in low abundance from many potential phorophytes without the orchid, indicating that additional studies are necessary to comprehend the factors that contribute to the fine-scale distribution of D. lindenii beyond the presence of the required OMF. Although several A. glabra trees, the other phorophyte of D. lindenii in Florida, were sampled, we were not successful in obtaining sequences from those samples. Thus, attempts should be made to sample sufficient numbers of A. glabra to better understand the situation in Florida. Additionally, D. lindenii in Cuba is found on several phorophyte species in comparison to the two primary phorophyte species associated with D. lindenii in Florida. Therefore, a fuller understanding of factors influencing the fine-scale distribution of D. lindenii needs to include an analysis of Cuban phorophytes.
When present, Ceratobasidium Clade 2 in bark was recovered at low read abundances, i.e., <5% relative abundance even from bark samples collected adjacent to actively growing root tips of D. lindenii. If Ceratobasidium Clade 2 is functioning as a saprobe in bark, then it is likely an inefficient saprobe and being outcompeted by more efficient saprotrophic fungi in the bark fungal community.
Although some F. caroliniana in Site 4 had Ceratobasidium Clade 2 it may be below the threshold of abundance to facilitate establishment and support the growth of naturally occurring plants (McCormick et al., 2016). Understanding site differences in terms of the presence/abundance of Ceratobasidium Clade 2 and other factors influencing establishment is crucial to sustaining populations of D. lindenii and preventing ‘senile’ populations’, an ageing orchid population that lacks seedling recruitment (Rasmussen et al., 2015).
The findings of this study indicate that D. lindenii, has high specificity for a specific taxon of Ceratobasidium, Clade 2. While this study provides data that suggest that the presence/absence (or very low abundance) of the required OMF influences which tree D. lindenii is likely to establish and persist, the fungus is probably not the sole factor driving fine-scale distribution. Future studies should focus on the role of abiotic factors, such as bark characteristics like pH and phenolics, on Ceratobasidium growth, as well as the ability of the orchid to establish on the tree surface. Pellitier et al. (2019) demonstrated that fungal communities are likely affected by pH and total phenolic content, therefore experiments to test this hypothesis should consider other abiotic factors.
This study establishes the usefulness of amplicon sequencing as a method to examine fungal communities in the roots of endangered orchids such as D. lindenii. Sampling the actively growing root tips provides a non-destructive sampling method for future studies of this and other threatened and endangered orchids. Naturally occurring D. lindenii appears to partner with a specific undescribed species of Ceratobasidium. However, lab-grown explants of D. lindenii have low abundance so long-term survival and successful reintroduction to natural habitats should account for potential phorophytes with abundant Ceratobasidium present for a viable conservation method. Additionally, while the presence of the required fungus is necessary for establishment of the orchid on a particular tree, it is likely that other factors which impact its fine-scale distribution, are also involved. Understanding how the difference in OMF abundance between naturally occurring plants and explants and the factors influencing successful establishment on phorophytes are needed to enhance the success of efforts to augment the population of the Ghost Orchid and refine conservation actions.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found at: NCBI; PRJNA948888.
Author contributions
LJ and MC contributed to the study conception, performed sample preparation, and data collection. LJ and GM wrote the first draft of the manuscript with an initial review by LZ. All authors contributed with comments on the later versions of the manuscript and approved of the final manuscript.
Funding
Financial support for this research was secured by LJ with a Fred Case Grant from the Native Orchid Conference. Additional funds were provided by the Plant Biology and Conservation program, a joint graduate degree training program between Northwestern University and the Chicago Botanic Garden.
Acknowledgments
We gratefully acknowledge Mark Danaher (USFWS) and Larry Richardson (Richardson Nature) for their assistance with field work in the Florida Panther NWR, and other Refuge staff including Mitchell Barazowski, Kevin Godsea, and Ben Nottingham. Thanks are extended to Ernesto Mújica (Orquideo Soroa, Cuba) for providing helpful insight and suggestions, and Adam R. Herdman (Southern Illinois University-Edwardsville). We also extend thanks to Renata Șerban for assisting with molecular methods for the laboratory work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1057940/full#supplementary-material
Footnotes
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Bayman, P., Lebron, L. L., Tremblay, R. L., and Lodge, D. J. (1997). Variation in endophytic fungi from roots and leaves of Lepanthes (Orchidaceae). New Phytol. 135, 143–149. doi: 10.1046/j.1469-8137.1997.00618.x
Bengtsson-Palme, J., Ryberg, M., Hartmann, M., Branco, S., Wang, Z., Godhe, A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919. doi: 10.1111/2041-210X.12073
Benzing, D. H., Friedman, W. E., Peterson, G., and Renfrow, A. (1983). Shootlessness, velamentous roots, and the pre-eminence of Orchidaceae in the epiphytic biotope. Am. J. Bot. 70, 121–133. doi: 10.2307/2443212
Benzing, D. H., and Ott, D. W. (1981). Vegetative reduction in epiphytic Bromeliaceae and Orchidaceae: its origin and significance. Biotropica 13:131. doi: 10.2307/2387715
Brown, P. M. (2002). Wild Orchids of Florida, with References to the Atlantic and Gulf Coastal Plains. Gainsville, Florida: University Press of Florida.
Chomicki, G., Bidel, L. P. R., and Jay-Allemand, C. (2014). Exodermis structure controls fungal invasion in the leafless epiphytic orchid Dendrophylax lindenii (Lindl.) Benth. Ex Rolfe. Flora-Morphol. Distrib. Funct. Ecol. Plants 209, 88–94. doi: 10.1016/j.flora.2014.01.001
Dearnaley, J. D. W., Martos, F., and Selosse, M.-A. (2012). “12 orchid Mycorrhizas: molecular ecology, physiology, evolution and conservation aspects” in Fungal Associations. ed. B. Hock (Berlin, Heidelberg: Springer Berlin Heidelberg), 207–230.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eskov, A. K., Voronina, E. Y., Tedersoo, L., Tiunov, A. V., Manh, V., Prilepsky, N. G., et al. (2020). Orchid epiphytes do not receive organic substances from living trees through fungi. Mycorrhiza 30, 697–704. doi: 10.1007/s00572-020-00980-w
Gebauer, G., Preiss, K., and Gebauer, A. C. (2016). Partial mycoheterotrophy is more widespread among orchids than previously assumed. New Phytol. 211, 11–15. doi: 10.1111/nph.13865
Gowland, K. M., van der Merwe, M. M., Linde, C. C., Clements, M. A., and Nicotra, A. B. (2013). The host bias of three epiphytic Aeridinae orchid species is reflected, but not explained, by mycorrhizal fungal associations. Am. J. Bot. 100, 764–777. doi: 10.3732/ajb.1200411
Graham, R. R., and Dearnaley, J. D. W. (2012). The rare Australian epiphytic orchid Sarcochilus weinthalii associates with a single species of Ceratobasidium. Fungal Divers. 54, 31–37. doi: 10.1007/s13225-011-0106-0
Gweon, H. S., Oliver, A., Taylor, J., Booth, T., Gibbs, M., Read, D. S., et al. (2015). PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 6, 973–980. doi: 10.1111/2041-210X.12399
Haaland, D., Estenoz, S., Williams, M., Director, P. D., and Williams, L. (2022). Petition to List the Ghost Orchid (Dendrophylax lindenii), as Threatened or Endangered Under the Endangered Species Act, and to Designate Critical Habitat.
Hoang, N. H., Kane, M. E., Radcliffe, E. N., Zettler, L. W., and Richardson, L. W. (2017). Comparative seed germination and seedling development of the ghost orchid, Dendrophylax lindenii (Orchidaceae), and molecular identification of its mycorrhizal fungus from South Florida. Ann. Bot. 119, 379–393. doi: 10.1093/aob/mcw220
Izuddin, M., Srivathsan, A., Lee, A. L., Yam, T. W., and Webb, E. L. (2019). Availability of orchid mycorrhizal fungi on roadside trees in a tropical urban landscape. Sci. Rep. 9:19528. doi: 10.1038/s41598-019-56049-y
Jacquemyn, H., Waud, M., Brys, R., Lallemand, F., Courty, P. E., Robionek, A., et al. (2017). Mycorrhizal associations and trophic modes in coexisting orchids: an ecological continuum between auto-and mixotrophy. Front. Plant Sci. 8:1497. doi: 10.3389/fpls.2017.01497
Johnson, L. J. A. N., del Gónzalez-Chávez, M., Carrillo-González, R., Porras-Alfaro, A., and Mueller, G. M. (2021). Vanilla aerial and terrestrial roots host rich communities of orchid mycorrhizal and ectomycorrhizal fungi. Plants People Planet 3, 541–552. doi: 10.1002/ppp3.10171
Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278. doi: 10.1093/bioinformatics/btu531
Li, T., Wu, S., Yang, W., Selosse, M.-A., and Gao, J. (2021). How mycorrhizal associations influence orchid distribution and population dynamics. Front. Plant Sci. 12:647114. doi: 10.3389/fpls.2021.647114
Martinez Arbizu, P. (2017). PairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.4.1.
McCormick, M. K., and Jacquemyn, H. (2014). What constrains the distribution of orchid populations? New Phytol. 202, 392–400. doi: 10.1111/nph.12639
McCormick, M. K., Taylor, D. L., Whigham, D. F., and Burnett, R. K. (2016). Germination patterns in three terrestrial orchids relate to abundance of mycorrhizal fungi. J. Ecol. 104, 744–754. doi: 10.1111/1365-2745.12556
McCormick, M. K., Whigham, D. F., and Canchani-Viruet, A. (2018). Mycorrhizal fungi affect orchid distribution and population dynamics. New Phytol. 219, 1207–1215. doi: 10.1111/nph.15223
McKendrick, S. L., Leake, J. R., Lee Taylor, D., and Read, D. J. (2002). Symbiotic germination and development of the myco-heterotrophic orchid Neottia nidus-avis in nature and its requirement for locally distributed Sebacina spp. New Phytol. 154, 233–247. doi: 10.1046/j.1469-8137.2002.00372.x
Mújica, E. B., Herdman, A. R., Danaher, M. W., González, E. H., and Zettler, L. W. (2021). Projected status of the ghost orchid (Dendrophylax lindenii) in Florida during the next decade based on temporal dynamic studies spanning six years. Plants. 10:1579. doi: 10.3390/plants10081579
Mújica, E. B., Mably, J. J., Skarha, S. M., Corey, L. L., Richardson, L. W., Danaher, M. W., et al. (2018). A comparision of ghost orchid (Dendrophylax lindenii) habitats in Florida and Cuba, with particular reference to seedling recruitment and mycorrhizal fungi. Bot. J. Linn. Soc. 186, 572–586. doi: 10.1093/botlinnean/box106
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Novotná, A., Benítez, Á., Herrera, P., Cruz, D., Filipczyková, E., and Suárez, J. P. (2018). High diversity of root-associated fungi isolated from three epiphytic orchids in southern Ecuador. Mycoscience 59, 24–32. doi: 10.1016/j.myc.2017.07.007
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., et al. (2022). Vegan: Community Ecology Package. R Package Version 2.6-4.
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a web browser. BMC Bioinformatics 12:385. doi: 10.1186/1471-2105-12-385
Otero, J. T., Ackerman, J. D., and Bayman, P. (2002). Diversity and host specificity of endophytic Rhizoctonia-like fungi from tropical orchids. Am. J. Bot. 89, 1852–1858. doi: 10.3732/ajb.89.11.1852
Otero, J. T., Ackerman, J. D., and Bayman, P. (2004). Differences in mycorrhizal preferences between two tropical orchids. Mol. Ecol. 13, 2393–2404. doi: 10.1111/j.1365-294x.2004.02223.x
Paulson, J. N., Colin Stine, O., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. doi: 10.1038/nmeth.2658
Pecoraro, L., Rasmussen, H. N., Gomes, S. I. F., Wang, X., Merckx, V. S. F. T., Cai, L., et al. (2021). Fungal diversity driven by bark features affects phorophyte preference in epiphytic orchids from southern China. Sci. Rep. 11:11287. doi: 10.1038/s41598-021-90877-1
Pellitier, P. T., Zak, D. R., and Salley, S. O. (2019). Environmental filtering structures fungal endophyte communities in tree bark. Mol. Ecol. 28, 5188–5198. doi: 10.1111/mec.15237
Petrolli, R., Augusto Vieira, C., Jakalski, M., Bocayuva, M. F., Vallé, C., Cruz, E. D. S., et al. (2021). A fine-scale spatial analysis of fungal communities on tropical tree bark unveils the epiphytic rhizosphere in orchids. New Phytol. 231, 2002–2014. doi: 10.1111/nph.17459
Petrolli, R., Zinger, L., Perez-Lamarque, B., Collobert, G., Griveau, C., Pailler, T., et al. (2022). Spatial turnover of fungi and partner choice shape mycorrhizal networks in epiphytic orchids. J. Ecol. 110, 2568–2584. doi: 10.1111/1365-2745.13986
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Qin, J., Zhang, W., Feng, J.-Q., and Zhang, S.-B. (2021). Leafless epiphytic orchids share Ceratobasidiaceae mycorrhizal fungi. Mycorrhiza 31, 625–635. doi: 10.1007/s00572-021-01043-4
R Core Team (2022). R: A Language and Environment for Statistical Computing. Available at: https://www.R-project.org/.
Rammitsu, K., Abe, S., Abe, T., Kotaka, N., Kudaka, M., Kudaka, N., et al. (2021a). The endangered epiphytic orchid Dendrobium okinawense has a highly specific mycorrhizal association with a single Tulasnellaceae fungus. J. For. Res. 26, 215–221. doi: 10.1080/13416979.2021.1876587
Rammitsu, K., Kajita, T., Imai, R., and Ogura-Tsujita, Y. (2021b). Strong primer bias for Tulasnellaceae fungi in metabarcoding: specific primers improve the characterization of the mycorrhizal communities of epiphytic orchids. Mycoscience 62, 356–363. doi: 10.47371/mycosci.2021.06.005
Rammitsu, K., Yagame, T., Yamashita, Y., Yukawa, T., Isshiki, S., and Ogura-Tsujita, Y. (2019). A leafless epiphytic orchid, Taeniophyllum glandulosum Blume (Orchidaceae), is specifically associated with the Ceratobasidiaceae family of basidiomycetous fungi. Mycorrhiza 29, 159–166. doi: 10.1007/s00572-019-00881-7
Rammitsu, K., Yukawa, T., Yamashita, Y., Isshiki, S., and Ogura-Tsujita, Y. (2020). The mycorrhizal community of the epiphytic orchid Thrixspermum japonicum is strongly biased toward a single Ceratobasidiaceae fungus, despite a wide range of fungal partners. Am. J. Bot. 107, 1654–1662. doi: 10.1002/ajb2.1575
Rasmussen, H. N., Dixon, K. W., Jersáková, J., and Těšitelová, T. (2015). Germination and seedling establishment in orchids: a complex of requirements. Ann. Bot. 116, 391–402. doi: 10.1093/aob/mcv087
Schimper, A. F. W. (1888). Die Epiphytische Vegetation Amerikas. Jena, Germany: Gustav Fischer Verlag.
Selosse, M.-A., Petrolli, R., Mujica, M. I., Laurent, L., Perez-Lamarque, B., Figura, T., et al. (2022). The waiting room hypothesis revisited by orchids: were orchid mycorrhizal fungi recruited among root endophytes? Ann. Bot. 129, 259–270. doi: 10.1093/aob/mcab134
Selosse, M.-A., WEIss, M., Jany, J. L., and Tillier, A. (2002). Communities and populations of sebacinoid basidiomycetes associated with the achlorophyllous orchid Neottia nidus-avis (L.) L.C.M. rich. And neighbouring tree ectomycorrhizae. Mol. Ecol. 11, 1831–1844. doi: 10.1046/j.1365-294x.2002.01553.x
Stewart, S., and Richardson, L. (2008). Orchid flora of the Florida Panther National Wildlife Refuge. North Am. Native Orchid J. 14, 70–104.
Tedersoo, L., Anslan, S., Bahram, M., Põlme, S., Riit, T., Liiv, I., et al. (2015). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 1–43. doi: 10.3897/mycokeys.10.4852
Thixton, H. L., Esselman, E. J., Corey, L. L., and Zettler, L. W. (2020). Further evidence of Ceratobasidium D.P. Rogers (Basidiomycota) serving as the ubiquitous fungal associate of Platanthera leucophaea (Orchidaceae) in the north American tallgrass prairie. Bot. Stud. 61:12. doi: 10.1186/s40529-020-00289-z
Turenne, C. Y., Sanche, S. E., Hoban, D. J., Karlowsky, J. A., and Kabani, A. M. (1999). Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37, 1846–1851. doi: 10.1128/JCM.37.6.1846-1851.1999
Vogt-Schilb, H., Těšitelová, T., Kotilínek, M., Sucháček, P., Kohout, P., and Jersáková, J. (2020). Altered rhizoctonia assemblages in grasslands on ex-arable land support germination of mycorrhizal generalist, not specialist orchids. New Phytol. 227, 1200–1212. doi: 10.1111/nph.16604
Wagner, K., Mendieta-Leiva, G., and Zotz, G. (2015). Host specificity in vascular epiphytes: a review of methodology, empirical evidence and potential mechanisms. AoB Plants 7:plu092. doi: 10.1093/aobpla/plu092
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., Li, Y., Song, X., Meng, Q., Zhu, J., Zhao, Y., et al. (2017). Influence of host tree species on isolation and communities of mycorrhizal and endophytic fungi from roots of a tropical epiphytic orchid, Dendrobium sinense (Orchidaceae). Mycorrhiza 27, 709–718. doi: 10.1007/s00572-017-0787-7
Waud, M., Busschaert, P., Lievens, B., and Jacquemyn, H. (2016). Specificity and localised distribution of mycorrhizal fungi in the soil may contribute to co-existence of orchid species. Fungal Ecol. 20, 155–165. doi: 10.1016/j.funeco.2015.12.008
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York: Academic Press, Inc), 315–322.
Xu, S., Chen, M., Feng, T., Zhan, L., Zhou, L., and Yu, G. (2021). Use ggbreak to effectively utilize plotting space to deal with large datasets and outliers. Front. Genet. 12:774846. doi: 10.3389/fgene.2021.774846
Keywords: conservation, Ceratobasidium, host tree specificity, amplicon sequencing, ghost orchid
Citation: Johnson LJAN, Kane ME, Zettler LW and Mueller GM (2023) Diversity and specificity of orchid mycorrhizal fungi in a leafless epiphytic orchid, Dendrophylax lindenii and the potential role of fungi in shaping its fine-scale distribution. Front. Ecol. Evol. 11:1057940. doi: 10.3389/fevo.2023.1057940
Edited by:
Tiiu Kull, Estonian University of Life Sciences, EstoniaReviewed by:
Raymond L. Tremblay, University of Puerto Rico, Puerto RicoMaría Isabel Mujica, Pontificia Universidad Católica de Chile, Chile
Copyright © 2023 Johnson, Kane, Zettler and Mueller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynnaun J. A. N. Johnson, THlubmF1bmpvaG5zb24yMDE4QHUubm9ydGh3ZXN0ZXJuLmVkdQ==
 Lynnaun J. A. N. Johnson
Lynnaun J. A. N. Johnson Michael E. Kane3
Michael E. Kane3 Lawrence W. Zettler
Lawrence W. Zettler Gregory M. Mueller
Gregory M. Mueller