- Department of Ecology and Evolutionary Biology, The University of Arizona, Tucson, AZ, United States
Sexual selection is thought to be a major driver of phenotypic diversity and diversification in animals, but large-scale evolutionary patterns in sexually selected (SS) traits remain largely unknown. Here, we survey and analyze the evolution of these traits across animal phylogeny. We find that female mate choice appears to be the most widespread mechanism of sexual selection, but male-male competition appears to be almost as frequent in chordates and male mate choice is also common in arthropods. Among sensory types, tactile traits appear to be most widespread whereas auditory traits are relatively uncommon. Rather than being ubiquitous or randomly distributed across animals, most of these different types of SS traits are confined to clades in arthropods and chordates, which form “hotspots” for the evolution of these diverse trait types. Thus, different sensory types show accelerated rates of evolution in these clades. Moreover, different types of SS traits are strongly correlated with each other in their evolution across animals. Finally, despite the intensive interest in the role of sexual selection in speciation, we find only limited support for the idea that SS traits drive large-scale patterns of diversification and species richness across all animals.
Introduction
Understanding the evolution of sexually selected (SS) traits is an integral part of evolutionary biology and behavioral ecology. Following a standard definition (Andersson, 1994), we consider SS traits to be those that increase reproductive success, either by influencing mate choice among conspecific individuals or competition for mates among them. These traits are often described as present across animals (Darwin, 1871; Kodric-Brown et al., 2006; Bonduriansky, 2007; Janicke and Morrow, 2018). They include numerous remarkable features of animals, such as frog calls, bird songs, deer antlers, beetle horns, and the conspicuous colors of many spiders, crustaceans, insects, fish, birds, primates, and lizards (reviews in Andersson, 1994; Wiens and Tuschhoff, 2020). These traits are also interesting because many may be involved in speciation (Darwin, 1871; Lande, 1981; West-Eberhard, 1983; Panhuis et al., 2001; Ritchie, 2007; Kraaijeveld et al., 2011; Tsuji and Fukami, 2020). Thus, sexual selection may help drive both phenotypic diversity and species diversity.
Nevertheless, despite intense interest in SS traits, broad-scale patterns in their evolution have gone largely unstudied. Instead, most studies have generally focused on trait evolution within or among populations or closely related species (e.g., within genera and families) or on one specific trait type or mechanism (e.g., male-male competition; Lupold et al., 2014). Analyzing closely related species and populations is an appropriate scale to study sexual selection itself, but many questions about large-scale evolution of SS traits remain unanswered (Wiens and Tuschhoff, 2020). For example, why do some phyla have many SS traits whereas others appear to have few or none? Is a paucity of SS traits associated with certain reproductive systems (e.g., asexual, hermaphroditic) or ecologies that may make mate choice or competition among individuals unlikely or difficult (e.g., internal parasites, marine species with broadcast spawning)? Have SS traits evolved randomly across animal phylogeny, or has there been accelerated evolution in certain clades? Do SS traits associated with different sensory systems (e.g., auditory, visual, olfactory, tactile) evolve independently of each other, or are they correlated in their evolution? If they are correlated, are these correlations negative (i.e., tradeoffs), such that taxa that have one sensory type (e.g., auditory) tend to lack another (e.g., visual), or are they instead positive, such that different types of SS traits tend to accumulate in the same clades or species? Given their potential role in speciation, do SS traits drive large-scale patterns of species diversity across animal phylogeny? For example, do SS traits in arthropods help explain why arthropods encompass ∼80% of all animal species (Jezkova and Wiens, 2017), whereas other phyla have far fewer species? Although there have been some macroevolutionary studies of whether SS traits drive diversification and richness patterns, these studies have generally been within animal phyla (and smaller clades; e.g., Kraaijeveld et al., 2011; Seddon et al., 2013; Huang and Rabosky, 2014; Tsuji and Fukami, 2020; Emberts and Wiens, 2021; Miller et al., 2021) rather than among phyla (but see Janicke et al., 2018 for a possible exception; see section “Discussion”).
Apart from these macroevolutionary patterns, many basic questions about SS traits remain unanswered. For example, which sensory types are most prevalent? Do SS traits evolve most frequently through female mate choice, the primary research emphasis in the field of sexual selection, or through other mechanisms (e.g., male-male competition, male choice)? Do the relative frequencies of these mechanisms vary among groups? Are certain sensory types associated with certain mechanisms? Some of these questions were previously raised in a recent review (Wiens and Tuschhoff, 2020) but they were not explicitly tested nor answered. We address all of these questions here.
We address these questions here by analyzing broad-scale patterns in the evolution of SS traits across the animal Tree of Life. We used time-calibrated phylogenies (Supplementary Dataset 1) that included most animal phyla, in which the tips of the trees were sampled in proportion to the species richness of each phylum. This proportional sampling is crucial for many comparative methods, and for estimating the frequencies of different sensory modalities and mechanisms across animals. We developed a database on the distribution of SS traits for the 1,087 tips in each tree (Supplementary Dataset 2). We categorized each SS trait based on broad types related to different sensory systems (auditory, gustatory, olfactory, tactile, visual; Table 1) and different mechanisms (male-male competition, female choice, female-female competition, male choice, intersexual conflict). We first estimated the frequencies of these sensory types and mechanisms. We then used phylogenetic methods to address the macroevolutionary questions listed above.
Specifically, we tested the following hypotheses. First, we tested whether the absence of most types of SS traits in many phyla was associated with reproductive modes (asexual reproduction and hermaphroditism) or ecologies (internal parasitism, marine habitat) that may not be conducive to mate choice or mate competition. Second, we tested the hypothesis that there are accelerated rates of origin for each sensory type of trait in certain phyla, and that these rates are correlated between types among phyla. We also tested whether the evolution of different types of SS traits was correlated among all taxa across the tree and within arthropods and chordates. Third, we tested the hypothesis that SS traits are positively related to diversification rates across animal phylogeny, and that their relationship to diversification rates helps explain the dramatic differences in richness among phyla. In addition to these three hypotheses, we also evaluate which mechanisms (e.g., female choice, male competition) and sensory types (e.g., auditory, visual) appear to be most prevalent among sampled taxa across animals, and whether these patterns are similar or different among the phyla that are most rich in SS traits (arthropods, chordates).
This research was made possible by continuing advances in generating large-scale, time-calibrated animal phylogenies (and new molecular datasets that make them possible), new macroevolutionary methods, and by decades of work by thousands of scientists, documenting SS traits in hundreds of animal species. We think that it is possible to make progress toward answering these questions even if the distribution of all SS traits among all animal species remains imperfectly known (see section “Discussion”).
Materials and methods
Trait data and trees
We first generated a database on the distribution of SS traits across animals (Supplementary Dataset 2). We surveyed those taxa included in three time-calibrated trees, each containing the same 1,087 terminal taxa (Román-Palacios et al., 2019). These trees differ primarily in the relationships and ages among phyla. They include 28 of the 34 frequently recognized phyla. The 6 other phyla apparently lack documented SS traits (Wiens and Tuschhoff, 2020). In these trees, terminal taxa within phyla were chosen to proportionally represent the number of described species in that phylum (i.e., most animal species belong to Arthropoda, with fewer species in Chordata and Mollusca, and far fewer in other phyla). Overall, there was a strong relationship between the number of tips in the tree for each phylum and its overall species richness (r2 = 0.99, P < 0.0001, n = 28). Most terminal taxa represented families. However, some families were represented by multiple species (e.g., in spiders and mollusks). Sampling within the largest phyla was also roughly proportional to the richness of the major clades within these phyla. For example, in arthropods, the majority of families were from the five largest insect orders, which collectively include the majority of described arthropod species (Bánki et al., 2021). Similarly, in chordates, roughly half of the sampled families were ray-finned fishes, since ∼50% of chordates are actinopterygians (Bánki et al., 2021). Specific tips were selected based on: (i) comprehensive sampling of hexapod families, and (ii) for non-hexapods, their presence in large-scale time-calibrated phylogenies (Román-Palacios et al., 2019). Tips were selected prior to scoring them for SS traits.
We then scored each tip for the presence or absence of SS traits (details in Supplementary Appendix 1). In brief, we searched for papers on sexual selection in each family and phylum. A trait was generally considered to be sexually selected given experimental or observational evidence that it significantly impacted mating success. We then categorized each trait in each taxon based on the sensory systems (auditory, gustatory, olfactory, tactile, visual) and mechanisms (male-male competition, female choice, female-female competition, male choice, intersexual conflict) involved. Only traits that were detectable by another animal using one of these sensory modalities were included. Thus, sperm competition was only included when accompanied by detectable traits such as copulatory plugs. The traits analyzed included both pre-copulatory and post-copulatory traits. In Supplementary Appendix 1, we give definitions and justifications for these trait types and mechanisms and describe how we searched for data and scored each taxon. Note that olfactory and tactile traits were defined very broadly. Again, we address the potential consequences of missing some traits in some taxa in the section “Discussion.”
Overview of statistical analyses
We tested three main hypotheses. First, that the apparent absence of most types of SS traits in certain animal phyla is associated with asexual reproduction, hermaphroditism, parasitism, marine habitat, and/or aquatic habitat. Second, that different sensory types and mechanisms of SS traits have positively correlated rates of evolution, among and within phyla. Third, that the presence of SS traits is associated with increased diversification rates. All analyses were performed on all three trees. Trait correlations were analyzed within Arthropoda and Chordata, and compared to results across animals. These separate analyses were performed because most taxa with SS traits are arthropods, and differing patterns within Chordata may be obscured in the whole-tree analysis. All analyses were performed in R version 4.0.1 (R Core Team, 2018). Data in analysis-ready format are given in Supplementary Dataset 3, and code is in Supplementary Dataset 4.
Distribution of sexually selected traits among phyla
The apparent lack of most types of SS traits in many phyla might be explained by their asexual reproduction, hermaphroditism, parasitism, or a marine or aquatic habitat (Wiens and Tuschhoff, 2020). The estimated proportion of species with any SS trait in each phylum was compared to the estimated proportions of species in each phylum that were asexual, monecious (hermaphroditic), parasitic, marine, and terrestrial (data and definitions from Jezkova and Wiens, 2017). We separately tested for a relationship between each of these traits and SS traits using phylogenetic-generalized-least-squares (PGLS) regression (Martins and Hansen, 1997). PGLS was conducted using the R package caper version 1.0.1 (Orme et al., 2018). Reduced trees among the 28 phyla were used (one tip per phylum; when only one tip per phylum is used in a time-calibrated tree, all tips yields an identical branch length for that phylum in the reduced tree). These reduced phylogenies are in Supplementary Dataset 1.
Rates of trait evolution and relationships between rates among phyla
The rate of origin for each SS trait type and mechanism in each phylum was determined using maximum-likelihood models for discrete characters. We utilized the function “fitDiscrete” in the R package geiger version 2.0.6 (Harmon et al., 2008; Pennell et al., 2014). We first determined the best-fitting model for each SS variable, using the Akaike Information Criterion (AIC; Burnham and Anderson, 2004). We compared the equal-rates model (ER: same rate for gains and losses) and the all-rates-different model (ARD: different rates for gains and losses). ARD generally had the best fit (see Supplementary Table 1). We used the best-fitting model to estimate the rate of trait origin (0-to-1 transition) for each phylum for each variable. However, many phyla apparently lacked most SS traits, and were therefore assigned a rate of zero (no origins). Rates for Rotifera were also set to 0 despite the presence of an olfactory trait and male choice: with only one taxon from Rotifera present in the tree the rate could not be estimated. We assume the rate is low, consistent with this trait and mechanism being documented in only one of ∼3,000 described Rotifera species (Jezkova and Wiens, 2017). A low rate is also consistent with the universal presence of this trait and mechanism among all the species within this phylum (i.e., the presence in all species could be explained by a single origin in the ancestor of the group). However, even if the true rates were high in this phylum, this would impact relatively few rate comparisons (olfactory, male choice). Further, traits in Rotifera were included in our analyses of trait correlations among tips (see below). Finally, we used PGLS to test for relationships between rates of origin of different sensory types among phyla (see above). All type-type combinations and mechanism-mechanism combinations were tested.
Correlated trait evolution
The likelihood approach of Pagel (1994) was also used to test for correlations between evolution of different sensory types of SS traits and between different mechanisms (but not between types and mechanisms). These analyses addressed correlations among tips, not phyla. First, each type/mechanism was checked for co-occurrence with every other type/mechanism (i.e., both present in the same terminal taxon), including all combinations of types and of mechanisms. Combinations that were never present among the sampled tips (i.e., clearly uncorrelated) were not tested. Then, for all other combinations, Pagel’s method was implemented in the R package phytools version 0.6–99 (Revell, 2012). Four models were analyzed. The null model assumed that the two traits evolved independently. The other three models assumed different kinds of dependence: trait 1 depends on trait 2 (Dependent A), trait 2 depends on trait 1 (Dependent B), and both traits depend on each other (Dependent AB). Model fit was assessed using P-values from a Chi-squared test and AIC values. Models were considered different from the null if they had a P-value < 0.05 and ΔAIC > 4 (Burnham and Anderson, 2004). Analyses used the best-fitting model (ARD, see Supplementary Table 1). This approach for testing for trait dependency does not provide correlation coefficients, but it can be assumed that all significant associations are positive (especially since pairs of traits that did not co-occur among taxa were not included).
We note that several studies have suggested that Pagel’s test (and similar methods) can potentially yield significant results when the independent and dependent variable each evolve only once or when one variable evolves once and the other evolves repeatedly (Darwin scenario and unreplicated burst scenario of Maddison and FitzJohn, 2015; see also Uyeda et al., 2018; Gardner and Organ, 2021; Boyko and Beaulieu, 2022). However, our results indicate that most trait types and mechanisms have evolved repeatedly across animals (e.g., in arthropods and chordates; Figure 1), suggesting that neither scenario generally applies here. Furthermore, our results based on rates of trait origins among phyla (see above) provide an independent test that can corroborate or refute the results from Pagel’s test, at least at a broad scale.
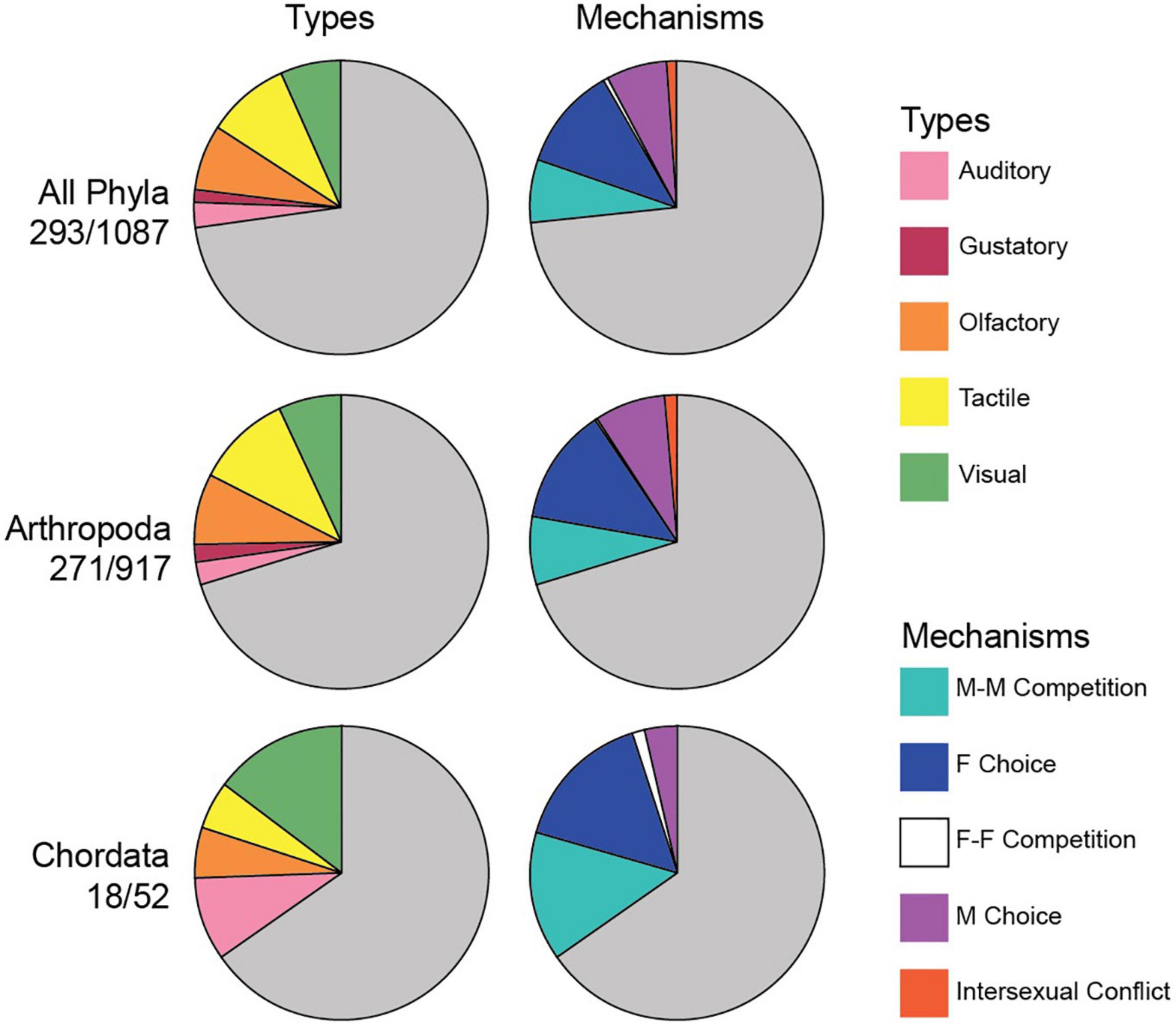
Figure 1. The estimated distribution of sensory types and mechanisms for sexually selected traits across animals. Pie graphs show the proportions of sampled taxa with each type and mechanism, across all phyla, in arthropods, and in chordates. Gray portions represent taxa in which sexually selected traits were not documented. Numbers below each group (all phyla, arthropods, chordates) are the number of taxa with sexually selected traits over the total number of sampled taxa. These estimates are based on the number of terminal taxa (mostly families). Taxa are sampled in proportion to the species richness of phyla, and in proportion to the richness of major clades within arthropods and chordates. When one trait involved multiple types or mechanisms, it was coded as present for each type and mechanism. Percentages for each type and mechanism were adjusted to represent the number of taxa with each trait out of the total number of taxa with any trait (as opposed to the overall total number of taxa) such that taxa with multiple traits did not distort the chart. M-M Comp. (male-male competition), F Choice (female choice), F-F Comp. (female-female competition), M Choice (male choice).
Sexually selected traits and diversification rates
The potential impact of SS traits on diversification rates was assessed using two approaches (full details in Supplementary Appendix 2). First, we used Hidden State Speciation and Extinction (HiSSE) models with the R package HiSSE version 1.8.5 (Beaulieu and O’Meara, 2016; Nakov et al., 2019). For our second approach, we performed PGLS analyses of the relationships between the estimated net diversification rate for each phylum and the proportion of species with each SS trait in that phylum. These rates for phyla were estimated using the method-of-moments estimator (MS estimator) with stem-group ages (Magallón and Sanderson, 2001). Simulations show that this method yields strong relationships between true and estimated diversification rates, even when rates vary strongly within clades over time (Meyer et al., 2018), when rates vary strongly between subclades (Meyer and Wiens, 2018), and when rates are faster in younger clades (Kozak and Wiens, 2016). Thus, this method does not require constant rates within or between clades to be accurate. This overall approach is robust to incomplete sampling of species in the tree, and allowed us to explicitly include multiple traits simultaneously and address how much variance in diversification rates was explained by the SS traits. We address potential criticisms of this method in Supplementary Appendix 2.
We performed analyses including each sensory trait type and each mechanism. We also included analyses in which we treated SS traits as present in general, such that a tip with any type of SS trait was considered to have them. We refer to this variable as the “combined SS” trait.
Multiple comparisons
Many tests were performed that included P-values. However, most conclusions were based on model selection. When P-values were used, a correction for false discovery rates was performed using the method of Benjamini and Hochberg (1995), implemented in the R package stats version 4.0.1 (R Core Team, 2018).
Results
Distribution of traits
Our survey found that SS traits were documented in only 5 of 28 sampled phyla (Table 2; Arthropoda, Chordata, Mollusca, Annelida, Rotifera). Most SS traits were documented in arthropods and chordates, but only in some sampled taxa (arthropods = 30%; chordates = 35%). One trait was reported in one species of Rotifera, two in a family of cephalopod mollusks (1% of sampled mollusks), and two in two annelid families (50% of sampled annelids). Examples of different trait types are given in Table 1.
We tested five potential factors (Supplementary Table 2) that might explain the apparent absence of most SS traits in many animal phyla. Contrary to expectations, phylogenetic regression revealed that asexuality, hermaphroditism, parasitism, occurrence in non-marine habitats, and occurrence in terrestrial habitats were not significantly related to the absence of SS traits (Supplementary Table 2). These factors were also not significantly related to rates of trait origins when rates were averaged or summed across different sensory types (Supplementary Tables 3, 4).
Our proportional sampling allowed us to estimate approximate frequencies of different sensory types and mechanisms across animals (Table 2 and Figure 1). The following trait frequencies represent the percentage of taxa with the given trait out of all sampled taxa in that group with SS traits (where the group is either all animals, arthropods, or chordates). Frequencies can sum to >100% because some tips have multiple traits.
Across animals (Figure 1), the most widespread sensory types among sampled taxa with SS traits were tactile (48% of 293 taxa), olfactory (38%), and visual (35%). Arthropods showed similar patterns with tactile (51% of 271 taxa), olfactory (39%), and visual (33%) being most common, and auditory less common (12%). Among sampled chordates (Figure 1), the most widespread types were visual (61% of 18 taxa with SS traits), auditory (39%), olfactory (22%), and tactile (22%). Across animals, the most frequent mechanisms appeared to be female choice (67%, 196/293 taxa), male-male competition (40%), and male choice (40%). Patterns were similar in arthropods (female choice: 68% of 271 taxa; male choice = 41%; male-male competition = 39%). In chordates, female choice was most common (72% of 18 taxa), followed closely by male-male competition (67%).
Different sensory types could also be associated with different mechanisms at different frequencies (e.g., acoustic predominantly with female choice vs. tactile predominantly with male-male competition). Across animals (Supplementary Dataset 2), female choice was the most common mechanism associated with most sensory types, including auditory, gustatory, olfactory, and visual (51, 61, 44, and 44% of each of the occurrences of these trait types were associated with female choice). This is the pattern expected given that female choice is the most common mechanism overall. Auditory, olfactory, and gustatory traits were also frequently associated with male choice, and visual traits with male-male competition. Tactile traits were more frequently associated with male-male competition (41%) than with female choice (36%), but by a small margin. Among traits associated with female choice, the most frequent sensory type was olfactory (30%). Olfactory was also the most common for male choice (49%).
Rates of trait evolution and correlations among trait types
We estimated the rate of origin for each sensory type and mechanism across animals and in each phylum (Figure 2 and Supplementary Table 1). Most trait types showed higher rates of loss than origins across animals and in arthropods, but rates of gain and loss were often similar in chordates (Supplementary Table 1). Rates of origins were highest for visual and tactile traits and were lowest for auditory and gustatory traits (Supplementary Table 1). All trait types had accelerated rates of origin in arthropods and chordates relative to all other phyla (Figure 2), except annelids (which had very high rates for olfactory and tactile).
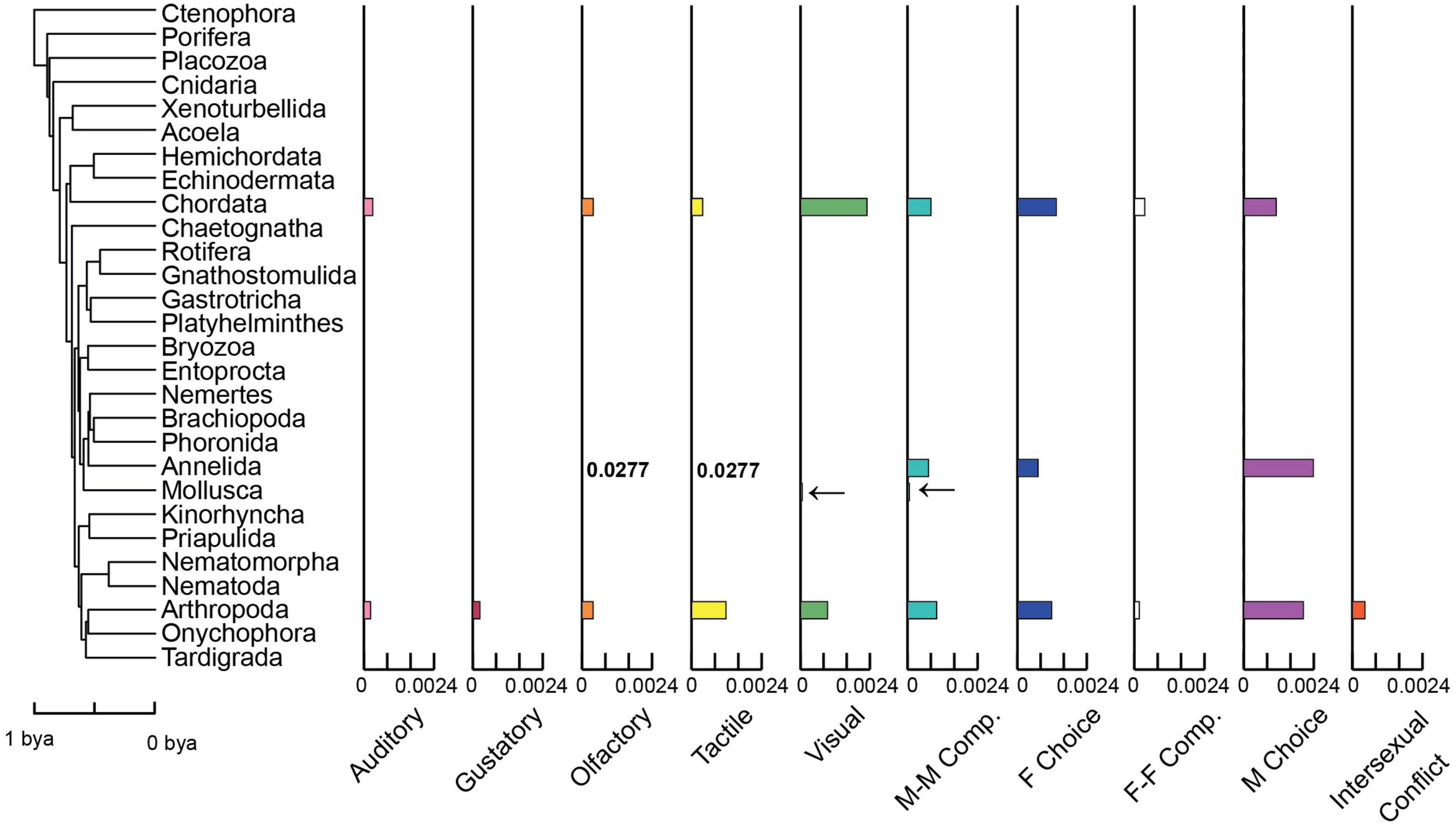
Figure 2. Rates of evolution of different sensory types and mechanisms for sexually selected traits among animal phyla. Bar graphs show the estimated rate of origin for each sensory type and mechanism within each phylum. Bars are aligned with the corresponding phylum on the phylogeny. Rates are shown for Arthropoda, Chordata, Mollusca (indicated by arrows due to low rate), and Annelida (the very high rates for olfactory and tactile traits are shown as numbers instead of bars). Other phyla have no trait origins and a rate of zero (except Rotifera, see section “Materials and methods”). Rates are in origins per million years. Tree 1 is shown along with the rates based on it, but rates are similar using the two alternative trees (Supplementary Table 4). The timescale is in billions of years before present (bya). M-M Comp. (male-male competition), F Choice (female choice), F-F Comp. (female-female competition), M Choice (male choice).
Using phylogenetic regression on trees for the 28 phyla (Supplementary Dataset 1), we found that different sensory types showed significant, positive relationships in their rates of origin, as did different mechanisms (Figure 3 and Supplementary Table 5). The only exception was gustatory with visual. Some pairs were strongly related (auditory-olfactory, auditory-visual, gustatory-tactile, and olfactory-visual, all r2 > 0.80) whereas others were weaker (auditory-gustatory and tactile-visual, r2 = 0.39). All mechanisms also showed significant relationships (Figure 3 and Supplementary Table 5). These relationships are generally explained by rapid rates of trait origins in arthropods and chordates relative to slow rates in most other phyla.
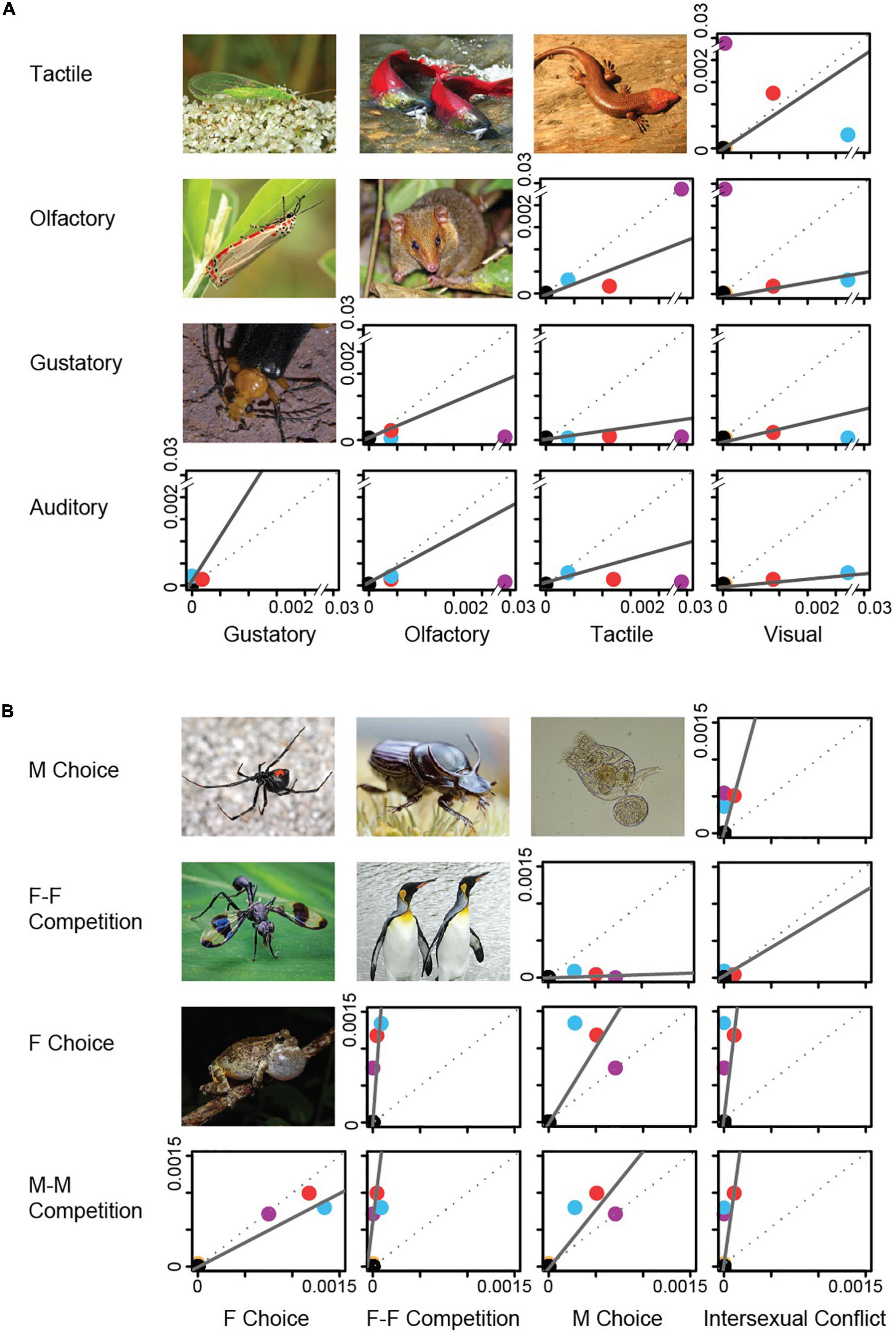
Figure 3. Relationships between rates of trait evolution among phyla. PGLS analyses were performed between pairs of trait types (A) and between pairs of mechanisms (B). For illustrative purposes, results are shown for Tree 1 (full results in Supplementary Table 5). All relationships except gustatory-visual were significant (P < 0.05). Red points indicate the rates of trait origin in Arthropoda, blue points Chordata, purple points Annelida, gold points Mollusca, and black points all other phyla (which lacked sexually selected traits or in the case of Rotifera had rates set to zero, see section “Materials and methods”). Rates are in units of origins per million years. M-M Comp. (male-male competition), F Choice (female choice), F-F Comp. (female-female competition), M Choice (male choice). Photographs show examples of species with the indicated trait combinations. Species shown from top left to bottom right are: (A): Chrysoperla carnea (common green lacewing, Chrysopidae), Oncorhynchus nerka (sockeye salmon, Salmonidae), Plestiodon laticeps (broadhead skink, Scincidae), Utetheisa ornatrix (bella moth, Arctiinae), Monodelphis scallops (long-nosed short-tailed opossum, Didelphidae), Neopyrochroa femoralis (fire-colored beetle, Pyrochroidae); (B): Latrodectus hesperus (western black widow spider, Theridiidae), Onthophagus illyricus (dung beetle, Scarabaeidae), Brachionus plicatilis (Brachionidae), Setellia sp. (Richardiidae), Aptenodytes patagonicus (king penguin, Spheniscidae), Hyla chrysoscelis (gray treefrog, Hylidae). All photographs from Wikimedia Commons. Photograph credits: Ettore Balocchi, Olga Vasik, Ryan Somma, Charles J. Sharp, Lucas Ferreira, Robert Webster, Marshal Hedin, Siga, Sofdrakou, João P. Burini, Andrew Shiva, and Geoff Gallice.
Traits were also generally correlated in their evolution among tips. Using maximum-likelihood tests across the three phylogenies of 1,087 terminal taxa, all sensory types were significantly correlated with each other, as were most mechanisms (Supplementary Table 6). Overall, these two sets of analyses (among phyla and among families) show that SS traits have not evolved randomly across animal phylogeny. Instead their evolution is largely concentrated in particular clades, where a diversity of trait types have evolved together at accelerated rates.
These correlation patterns differed between arthropods and chordates (Supplementary Table 7). Specifically, all types of SS traits (except gustatory-tactile) were significantly correlated in arthropods. However, none were significantly correlated in chordates. Since most animals are arthropods, arthropods drove the overall pattern across animals.
Sexually selected traits and diversification
We used two approaches to test whether SS traits are related to increased diversification rates. First, we used HiSSE models to directly link traits and diversification rates among the 1,087 tips. We compared null models in which SS traits were unrelated to diversification, to models in which only the observed SS traits influenced diversification, to more complex models in which both hidden and observed traits influenced diversification. For all trait types except olfactory, the best-fitting model differed among the three trees (Supplementary Table 8), with a null model (no impact of the observed trait on diversification) being the best supported for at least one tree, and the full HiSSE model supported for another. A model in which SS traits primarily impacted diversification rates was not supported for any trait on any tree. Importantly, when using weighted-mean diversification rates from HiSSE, rates were consistently higher when some widespread trait types were absent (combined SS, olfactory, tactile, and female choice). Results for visual, male-male competition, and male choice differed across trees (Supplementary Table 9). Overall, the HiSSE approach showed little support for a significant, positive effect of SS traits on diversification across animals.
As a second approach, we used phylogenetic regression to test if phyla with higher estimated frequencies of SS traits among species had higher estimated net diversification rates. Diversification rates (Supplementary Table 10) were based on all extant, described species in each clade (not merely those in the tree). Using this approach, some SS traits had positive relationships with diversification rates (Supplementary Table 11), including tactile, visual, and gustatory traits. However, these relationships were relatively weak (r2 = 0.16–0.23) and insignificant after correcting for false-discovery rates. Moreover, the combined SS trait was not significantly related to diversification (Supplementary Table 11). We also included these SS traits (combined, gustatory, tactile, and visual) in phylogenetic regression models with other traits previously shown (Jezkova and Wiens, 2017) to influence large-scale animal diversification (Supplementary Table 12). Adding any SS trait improved model fit somewhat (Supplementary Table 13). The overall best-fitting model included gustatory traits, along with dioecy, parasitism, a skeleton, and vision (Supplementary Table 13). However, the increase in model fit for adding any SS trait was not significant (<4 AIC units), and the additional variance in diversification rates explained was limited (∼5–15%). Overall, SS traits did not have a strong or consistently positive influence on diversification rates among animal phyla.
Discussion
We present here the first analysis of large-scale patterns of evolution in SS traits across animals. We confirmed that most types of SS traits appeared to be absent in most animal phyla (based on a standard definition of sexual selection in which SS traits increase reproductive success through mate choice and/or mate competition; Andersson, 1994). By contrast, most types of SS traits were present in arthropods and chordates, but only in some families. We found that the apparent absence of SS traits in some animal phyla was not significantly linked to asexuality, hermaphroditism, parasitism, or habitat use. We found that in arthropods and chordates, there were accelerated rates of trait evolution in almost all sensory types and mechanisms. These different traits were also generally correlated in their evolution among animal families (but not in chordates). These results together suggest that most types of SS traits are not ubiquitous among animals, nor have they evolved randomly across the tree. Instead there are “hotspots” for their evolution across the animal Tree of Life, in which multiple types and mechanisms evolve in the same group (e.g., insects, lizards, birds, mammals). Across animals, we estimated that most SS traits appear to have evolved through female choice (as expected), but male-male competition was almost as frequent among sampled chordates and male choice was surprisingly prevalent in arthropods (see also Bonduriansky, 2001). We found that SS traits appear to be lost at higher rates than they are gained (see also Wiens, 2001; but see Supplementary Appendix 3). These latter two patterns were implied previously (Bonduriansky, 2001; Wiens, 2001) but not shown quantitatively. We found little support for the idea that SS traits help explain large-scale patterns of diversification and richness across animals, despite intense interest in the role of SS traits in speciation. Overall, these results raise many intriguing questions for future research, which we discuss below. However, we first discuss potential methodological concerns about these conclusions.
Methodological concerns
We acknowledge that the distributions of SS traits across all animals are imperfectly known. Furthermore we estimated the absence of traits based on the absence of documented evidence for their presence within phyla and families (Supplementary Appendix 1). Although the exact frequencies and distributions of traits estimated will doubtless change over time, our overall conclusions need not be overturned.
First, our major conclusions should generally be robust, even if all phyla have one or more undetected SS trait types. For example, if every animal species had olfactory/chemosensory traits and sperm competition, there should still be accelerated rates in the other trait types in arthropods and chordates, and correlated evolution of these other trait pairs among clades. Similarly, if every species had at least one SS trait, this would only reinforce our conclusion that the presence of SS traits does not explain variation in diversification rates among animal clades. In Supplementary Appendix 4, we discuss more fully how our results and conclusions could be impacted by documenting SS traits in additional taxa.
Second, the estimated frequencies of sensory types should not generally be explained by human bias. Although visual and auditory traits are presumably the easiest for humans to detect and olfactory the hardest, we found that olfactory traits were more frequent than visual and auditory traits across animals (Table 2). This is the opposite of the expected pattern if human biases alone explained these frequencies. Furthermore, the estimated frequencies differ strikingly between arthropods and chordates. These frequencies should be similar if they were explained only by human detectability. Similarly, for mechanisms, female choice should not be easier to document than male-male competition (to our knowledge), and yet female choice appeared to be more common (Table 2). Moreover, the frequencies of male-male competition and male choice also differed strikingly between arthropods and chordates (Table 2), which (again) is not expected if human biases alone explained these frequencies. Again, we expect the exact frequencies to change somewhat over time as more information is added across more groups, but the general patterns found need not be overturned. We note that some readers might assume that the reported frequencies of these mechanisms across animals will simply depend on overall trends in research interests over time (e.g., McCullough et al., 2016). But scientists have been documenting diverse mechanisms across taxa for decades. We found no significant difference in the year in which the sampled studies documenting male-male competition were published (mean = 2000, range = 1961–2018, n = 157 studies) relative to studies documenting female choice (mean = 1999, range = 1930–2017, n = 186 studies), based on a t-test in R (P = 0.22, t = 1.22, d.f. = 340.11). Note that some studies included multiple taxa (especially studies of female choice) and some documented both male-male competition and female choice. In many cases, the mechanism of sexual selection involved in a given trait and species may be apparent regardless of overall research trends in the field (e.g., horns used in combat between males, ornaments used in male displays to females).
Third, we acknowledge that our trees of 1,087 taxa represent a small fraction of all animal species. We addressed this in several ways. For phyla that apparently lacked a given type of SS trait, we searched the whole phylum not just the sampled tips (Supplementary Appendix 1). For other phyla, we assessed traits for entire families, not individual species. We may therefore overestimate some trait frequencies within phyla, but sampling species instead could strongly underestimate these frequencies (given that presumably few species per family have been assessed). Many results should be largely insensitive to incomplete taxon sampling. Simulations and empirical analyses suggest that phylogeny-based correlations between traits can be robust to limited sampling, despite reduced statistical power (Ackerly, 2000; Emberts and Wiens, 2022). Similarly, our estimated diversification rates for phyla incorporate all species in each phylum (Supplementary Appendix 2). Simulations also show that relationships between trait frequencies and clade-level diversification rates can be robust to considerable error in estimating trait frequencies (Moen and Wiens, 2017; Emberts and Wiens, 2021). The HiSSE analyses explicitly corrected for incomplete sampling (Supplementary Appendix 2).
Finally, our use of family-level data to score terminal taxa need not be a major source of error. For most analyses (i.e., estimating trait frequencies, rates of origin, and associations between traits), tips are implicitly treated as species. Thus, these analyses should be largely equivalent to randomly sampling 1,087 animal species. We use family-level information to score these tips to help counteract the absence of species-level data for all SS traits across all animal species.
Why are many types of sexually selected traits apparently uncommon?
We found that most types of SS traits were surprisingly uncommon among animal phyla. We tested five potential explanations for this pattern (Wiens and Tuschhoff, 2020). SS traits were hypothesized to be less likely to evolve in taxa that are asexual, lack distinct sexes, are internal parasites, or are marine taxa with broadcast spawning (rarely encountering mates). Yet, these factors were not significantly related to their absence or rates of origin (Supplementary Table 1), although many phyla with these characteristics lack documented SS traits. The problem is that many sexually reproducing, monecious, non-parasitic, and terrestrial taxa also appear to lack most SS traits (thus explaining the lack of a significant relationship). Understanding the causes of the broad-scale distribution of these traits among phyla may require focusing on other ecological traits, such as whether resources or mates are defendable (Emlen and Oring, 1977), operational sex ratios (Emlen and Oring, 1977), and Bateman gradients (Janicke et al., 2016). At the simplest level, sexual selection (following standard definitions; Andersson, 1994) generally involves choice among potential mates or competition among them. However, the aggregations of mating individuals that would facilitate mate choice or mate competition may not be so ubiquitous across animals. An obvious hypothesis is that the presence of such aggregations might help explain the distribution of SS traits among animal phyla.
Correlated trait evolution
Our results show that different types of SS traits have strongly correlated rates of evolution across animals. This pattern arises largely from the repeated origins of each sensory type within arthropods and chordates. For example, most major types have evolved repeatedly in different clades of arthropods (e.g., arachnids, malacostracan crustaceans, dipteran, and coleopteran insects) and chordates (e.g., ray-finned fish, amphibians, lizards, birds, and mammals). Intriguingly, different mechanisms were also correlated, such as mate choice with mate competition.
There are several potential explanations for this pattern of correlated evolution. One is that these “hotspots” simply represent clades with diverse sensory systems (Wiens and Tuschhoff, 2020). Testing this idea will require large-scale analyses of sensory systems across animals, which are presently lacking. Another hypothesis is that once a signal evolves, multiple signals may follow (Møller and Pomiankowski, 1993). Yet, other studies have found trade-offs between different types of sensory signals (Liman and Innan, 2003), especially in chordates (but see Mason et al., 2014). These latter two hypotheses have been studied among relatively closely related taxa, and it remains to be seen whether they apply to broader scales. Our overall results suggest that there are no trade-offs in the evolution of sensory types among animal phyla and possibly not within arthropods (where traits are correlated), but trade-offs might occur within chordates. In chordates, different trait types may be associated with different diel-activity patterns. For example, nocturnal activity seems to drive origins of acoustic communication in tetrapods (Chen and Wiens, 2020), likely because acoustic signals can function in darkness. Conversely, most visual signals may be favored in diurnal lineages, as recently found for conspicuous, sexually dimorphic coloration in tetrapods (Emberts and Wiens, 2022). Overall, further research will be needed to establish the causes of these patterns of correlated evolution across animals and arthropods but not chordates.
Frequencies of types and mechanisms
We also found interesting patterns in the frequencies of different types and mechanisms of SS traits. Female choice has been the most widespread focus of sexual selection research in recent decades (as documented by McCullough et al., 2016). Our systematic survey suggests that the most widespread mechanism across animals (and within arthropods) is indeed female choice (67% of sampled traits), but with male choice (40%) surprisingly common relative to male-male competition (40%). In chordates, female choice again appeared to be most common (72%) but with male-male competition almost as common (67%). Our sampling of chordates was limited, but a similar result was found in a separate, clade-based survey of chordates that included >200 taxa and >200 traits (57% mate choice vs. 43% competition; Supplementary Appendix 4 of Wiens and Tuschhoff, 2020). Our results support the emphasis on female choice in sexual-selection research, but suggest that male competition (in chordates) and male choice (in arthropods) are also widespread.
Sexual selection and diversification
There is strong evidence that SS traits are important in reproductive isolation and speciation in many systems (e.g., Boul et al., 2007; Wagner et al., 2012; Uy et al., 2018). However, the evidence for a strong macroevolutionary effect on speciation and diversification rates has proven elusive, especially at broader scales (Kraaijeveld et al., 2011; Tsuji and Fukami, 2020; Emberts and Wiens, 2021; Miller et al., 2021). Here, we provide evidence that the effect of SS traits on diversification (speciation minus extinction) at the broadest scales is limited. Importantly, our results do not contradict the idea that sexual selection is important for speciation. Instead they suggest that the potential positive effect of these traits on diversification does not scale up to explain large-scale diversity patterns across animals. SS traits might also increase extinction, as suggested theoretically (e.g., Kokko and Brooks, 2003) and found in some organisms (e.g., Martins et al., 2018, 2020). This could negate their positive effects on diversification from increased speciation rates. Indeed, our HiSSE analyses showed lower diversification rates in taxa with SS traits.
We note that a previous study did address the relationship between sexual selection and species richness across animals, and found a positive effect (Janicke et al., 2018). However, that study focused on differences in species richness among families (∼34 to 42 families depending on the analysis) sampled from five phyla, and not richness patterns among phyla. The majority of sampled families were from vertebrates. Diversification rates were not estimated or included. Furthermore, that study related species richness to various proxies for the opportunity for sexual selection (e.g., variance in mating and reproductive success) and for the strength of sexual selection (e.g., the relationship between mating success and reproductive success), and not to the presence of SS traits. Most of these proxies had no significant effect on family-level richness after accounting for phylogeny. In summary, although the results of that important study might appear to contradict ours, there is limited overlap in the scope of these studies. The contrast between that study and ours also supports the idea that the effects of sexual selection on diversification and richness at lower phylogenetic scales (e.g., among families within phyla) do not scale up to drive large-scale richness patterns across animals.
Conclusion
In summary, we analyze large-scale patterns in the evolution of SS traits across animals for the first time. We estimated the distribution of major sensory types and mechanisms across animals, supporting the widespread emphasis on female choice but also showing the prevalence of male-male competition (in chordates) and male mate choice (in arthropods). We found that different types of these traits are not distributed ubiquitously or randomly across animals, but instead show accelerated, correlated evolution of different types in certain clades that are “hotspots” for the evolution of SS traits. We find little effect of these traits on diversification, suggesting that their potential small-scale effects on speciation fail to explain large-scale diversity patterns.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
ET and JW designed the research and wrote the manuscript. ET collected and analyzed the data. Both authors contributed to the article and approved the submitted version.
Funding
JW acknowledges U.S. National Science Foundation grant DEB 1655690.
Acknowledgments
We thank Judie Bronstein, Zachary Emberts, Dan Papaj, and two reviewers for many helpful comments that greatly improved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1042747/full#supplementary-material
References
Ackerly, D. D. (2000). Taxon sampling, correlated evolution, and independent contrasts. Evolution 54, 1480–1492. doi: 10.1111/j.0014-3820.2000.tb00694.x
Andersson, M. B. (1994). Sexual Selection. Princeton, NJ: Princeton University Press. doi: 10.1515/9780691207278
Bánki, O., Roskov, Y., Vandepitte, L., DeWalt, R. E., Remsen, D., Schalk, P., et al. (2021). Catalogue of life checklist (version 2021-08-25). Catalogue of life. Available online at: http://www.catalogueoflife.org (accessed September 4, 2021).
Beaulieu, J. M., and O’Meara, B. C. (2016). Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583–601. doi: 10.1093/sysbio/syw022
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bonduriansky, R. (2001). The evolution of male mate choice in insects: A synthesis of ideas and evidence. Biol. Rev. 76, 305–339. doi: 10.1017/S1464793101005693
Bonduriansky, R. (2007). Sexual selection and allometry: A critical reappraisal of the evidence and ideas. Evolution 61, 838–849. doi: 10.1111/j.1558-5646.2007.00081.x
Boul, K. E., Funk, W. C., Darst, C. R., Cannatella, D. C., and Ryan, M. J. (2007). Sexual selection drives speciation in an Amazonian frog. Proc. R. Soc. Lond. B 274, 399–406. doi: 10.1098/rspb.2006.3736
Boyko, J. D., and Beaulieu, J. M. (2022). Reducing the biases in false correlations between discrete characters. Syst. Biol. syac066. doi: 10.1093/sysbio/syac066
Burnham, K. P., and Anderson, D. R. (2004). Multimodel inference: Understanding AIC and BIC in model selection. Sociol. Methods Res. 33, 261–304. doi: 10.1177/0049124104268644
Chen, Z., and Wiens, J. J. (2020). The origins of acoustic communication in vertebrates. Nat. Commun. 11, 1–8. doi: 10.1038/s41467-020-14356-3
Darwin, C. (1871). The descent of man and selection in relation to sex. London: John Murray. doi: 10.5962/bhl.title.24784
Emberts, Z., and Wiens, J. J. (2021). Do sexually selected weapons drive diversification? Evolution 75, 2411–2424. doi: 10.1111/evo.14212
Emberts, Z., and Wiens, J. J. (2022). Why are animals conspicuously colored? Evolution of sexual versus warning signals in land vertebrates. Evolution 76, 2879–2892. doi: 10.1111/evo.14636
Emlen, S. T., and Oring, L. W. (1977). Ecology, sexual selection, and the evolution of mating systems. Science 197, 215–223. doi: 10.1126/science.327542
Gardner, J. D., and Organ, C. L. (2021). Evolutionary sample size and consilience in phylogenetic comparative analysis. Syst. Biol. 70, 1061–1075. doi: 10.1093/sysbio/syab017
Harmon, L. J., Weir, J. T., Brock, C. D., Glor, R. E., and Challenger, W. (2008). GEIGER: Investigating evolutionary radiations. Bioinformatics 24, 129–131. doi: 10.1093/bioinformatics/btm538
Huang, H., and Rabosky, D. L. (2014). Sexual selection and diversification: Reexamining the correlation between dichromatism and speciation rate in birds. Am. Nat. 184, E101–E114. doi: 10.1086/678054
Janicke, T., and Morrow, E. H. (2018). Operational sex ratio predicts the opportunity and direction of sexual selection across animals. Ecol. Lett. 21, 384–391. doi: 10.1111/ele.12907
Janicke, T., Häderer, I. K., Lajeunesse, M. J., and Anthes, N. (2016). Darwinian sex roles confirmed across the animal kingdom. Sci. Adv. 2:e1500983. doi: 10.1126/sciadv.1500983
Janicke, T., Ritchie, M. G., Morrow, E. H., and Marie-Orleach, L. (2018). Sexual selection predicts species richness across the animal kingdom. Proc. R. Soc. Lond. B 285:20180173. doi: 10.1098/rspb.2018.0173
Jezkova, T., and Wiens, J. J. (2017). What explains patterns of diversification and richness among animal phyla? Am. Nat. 189, 201–212. doi: 10.1086/690194
Kodric-Brown, A., Sibly, R. M., and Brown, J. H. (2006). The allometry of ornaments and weapons. Proc. Natl. Acad. Sci. U.S.A. 103, 8733–8738. doi: 10.1073/pnas.0602994103
Kokko, H., and Brooks, R. (2003). Sexy to die for? Sexual selection and the risk of extinction. Ann. Zool. Fenn. 40, 207–219.
Kozak, K. H., and Wiens, J. J. (2016). Testing the relationships between diversification, species richness, and trait evolution. Syst. Biol. 65, 975–988. doi: 10.1093/sysbio/syw029
Kraaijeveld, K., Kraaijeveld-Smit, F. J. L., and Maan, M. E. (2011). Sexual selection and speciation: The comparative evidence revisited. Biol. Rev. 86, 367–377. doi: 10.1111/j.1469-185X.2010.00150.x
Lande, R. (1981). Models of speciation by sexual selection on polygenic traits. Proc. Natl. Acad. Sci. U.S.A. 78, 3721–3725. doi: 10.1073/pnas.78.6.3721
Liman, E. R., and Innan, H. (2003). Relaxed selective pressure on an essential component of pheromone transduction in primate evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 3328–3332. doi: 10.1073/pnas.0636123100
Lupold, S., Tomkins, J. L., Simmons, L. W., and Fitzpatrick, J. L. (2014). Female monopolization mediates the relationship between pre- and postcopulatory sexual traits. Nat. Commun. 5:3184. doi: 10.1038/ncomms4184
Maddison, W. P., and FitzJohn, R. G. (2015). The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst. Biol. 64, 127–136. doi: 10.1093/sysbio/syu070
Magallón, S., and Sanderson, M. J. (2001). Absolute diversification rates in angiosperm clades. Evolution 55, 1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x
Martins, E. P., and Hansen, T. F. (1997). Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667. doi: 10.1086/286013
Martins, M. J. F., Hunt, G., Thompson, C. M., Lockwood, R., Swaddle, J. P., and Puckett, T. M. (2020). Shifts in sexual dimorphism across a mass extinction in ostracods: Implications for sexual selection as a factor in extinction risk. Proc. R. Soc. Lond. B 287:20200730. doi: 10.1098/rspb.2020.0730
Martins, M. J. F., Puckett, T. M., Lockwood, R., Swaddle, J. P., and Hunt, G. (2018). High male sexual investment as a driver of extinction in fossil ostracods. Nature 556, 366–369. doi: 10.1038/s41586-018-0020-7
Mason, N. A., Shultz, A. J., and Burns, K. J. (2014). Elaborate visual and acoustic signals evolve independently in a large, phenotypically diverse radiation of songbirds. Proc. R. Soc. Lond. B 281:20140967. doi: 10.1098/rspb.2014.0967
McCullough, E. L., Miller, C. W., and Emlen, D. J. (2016). Why sexually selected weapons are not ornaments. Trends Ecol. Evol. 31, 742–751. doi: 10.1016/j.tree.2016.07.004
Meyer, A. L. S., and Wiens, J. J. (2018). Estimating diversification rates for higher taxa: BAMM can give problematic estimates of rates and rate shifts. Evolution 72, 39–53. doi: 10.1111/evo.13378
Meyer, A. L. S., Román-Palacios, C., and Wiens, J. J. (2018). BAMM gives misleading rate estimates in simulated and empirical datasets. Evolution 72, 2257–2266. doi: 10.1111/evo.13574
Miller, E. C., Mesnick, S. L., and Wiens, J. J. (2021). Sexual dichromatism is decoupled from diversification over deep time in fishes. Am. Nat. 198, 232–252. doi: 10.1086/715114
Moen, D. S., and Wiens, J. J. (2017). Microhabitat and climatic niche change explain patterns of diversification among frog families. Am. Nat 190, 29–44. doi: 10.1086/692065
Møller, A. P., and Pomiankowski, A. (1993). Why have birds got multiple sexual ornaments? Behav. Ecol. Sociobiol. 32, 167–176. doi: 10.1007/BF00173774
Nakov, T., Beaulieu, J. M., and Alverson, A. J. (2019). Diatoms diversify and turn over faster in freshwater than marine environments. Evolution 73, 2497–2511. doi: 10.1111/evo.13832
Orme, D., Freckleton, R., Thomas, G., Petzoldt, T., Fritz, S., Isaac, N., et al. (2018). caper: Comparative analyses of phylogenetics and evolution in R. R package version 1.0.1. Available online at: https://cran.r-project.org/package=caper (accessed July, 2019).
Pagel, M. (1994). Detecting correlated evolution on phylogenies: A general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. B 255, 37–45. doi: 10.1098/rspb.1994.0006
Panhuis, T. M., Butlin, R., Zuk, M., and Tregenza, T. (2001). Sexual selection and speciation. Trends Ecol. Evol. 16, 364–371. doi: 10.1016/S0169-5347(01)02160-7
Pennell, M. W., Eastman, J. M., Slater, G. J., Brown, J. W., Uyeda, J. C., FitzJohn, R. G., et al. (2014). Geiger v2.0: An expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218. doi: 10.1093/bioinformatics/btu181
R Core Team (2018). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Ritchie, M. G. (2007). Sexual selection and speciation. Annu. Rev. Ecol. Evol. Syst. 38, 79–102. doi: 10.1146/annurev.ecolsys.38.091206.095733
Román-Palacios, C., Scholl, J. P., and Wiens, J. J. (2019). Evolution of diet across the animal tree of life. Evol. Lett. 3, 339–347. doi: 10.1002/evl3.127
Seddon, N., Botero, C. A., Tobias, J. A., Dunn, P. O., MacGregor, H. E. A., Rubenstein, D. R., et al. (2013). Sexual selection accelerates signal evolution during speciation in birds. Proc. R. Soc. Lond. B 280:20131065. doi: 10.1098/rspb.2013.1065
Tsuji, K., and Fukami, T. (2020). Sexual dimorphism and species diversity: From clades to sites. Trends Ecol. Evol. 35, 105–114. doi: 10.1016/j.tree.2019.09.001
Uy, J. A. C., Irwin, D. E., and Webster, M. S. (2018). Behavioral isolation and incipient speciation in birds. Annu. Rev. Ecol. Syst. 49, 1–24. doi: 10.1146/annurev-ecolsys-110617-062646
Uyeda, J. C., Zenil-Ferguson, R., and Pennell, M. W. (2018). Rethinking phylogenetic comparative methods. Syst. Biol. 67, 1091–1109. doi: 10.1093/sysbio/syy031
Wagner, C. E., Harmon, L. J., and Seehausen, O. (2012). Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487, 366–369. doi: 10.1038/nature11144
West-Eberhard, M. J. (1983). Sexual selection, social competition, and speciation. Q. Rev. Biol. 58, 155–193.
Wiens, J. J. (2001). Widespread loss of sexually selected traits: How the peacock lost its spots. Trends Ecol. Evol. 16, 517–523. doi: 10.1016/S0169-5347(01)02217-0
Keywords: diversification, macroevolution, mate choice, mate competition, sexual selection, phylogeny
Citation: Tuschhoff E and Wiens JJ (2023) Evolution of sexually selected traits across animals. Front. Ecol. Evol. 11:1042747. doi: 10.3389/fevo.2023.1042747
Received: 12 September 2022; Accepted: 04 January 2023;
Published: 19 January 2023.
Edited by:
Alfredo G. Nicieza, University of Oviedo, SpainReviewed by:
Dan Liang, Princeton University, United StatesArild Johnsen, University of Oslo, Norway
Copyright © 2023 Tuschhoff and Wiens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John J. Wiens,  d2llbnNqQGFyaXpvbmEuZWR1
d2llbnNqQGFyaXpvbmEuZWR1
 E. Tuschhoff
E. Tuschhoff John J. Wiens
John J. Wiens