- 1Cape Research Centre, Scientific Services, South African National Parks, Cape Town, South Africa
- 2Department of Botany and Zoology, Centre for Invasion Biology, Stellenbosch University, Stellenbosch, South Africa
- 3Threatened Species Unit, South African National Biodiversity Institute, Cape Town, South Africa
- 4Pearson Chair of Botany, University of Cape Town, Cape Town, South Africa
- 5Savanna Research Unit, Scientific Services, South African National Parks, Skukuza, South Africa
Plant and animal checklists, with conservation status information, are fundamental for conservation management. Historical field data, more recent data of digital origin and data-sharing platforms provide useful sources for collating species locality data. However, different biodiversity datasets have different formats and inconsistent naming systems. Additionally, most digital data sources do not provide an easy option for download by protected area. Further, data-entry-ready software is not readily available for conservation organization staff with limited technical skills to collate these heterogeneous data and create distribution maps and checklists for protected areas. The insights presented here are the outcome of conceptualizing a biodiversity information system for South African National Parks. We recognize that a fundamental requirement for achieving better standardization, sharing and use of biodiversity data for conservation is capacity building, internet connectivity, national institutional data management support and collaboration. We focus on some of the issues that need to be considered for capacity building, data standardization and data support. We outline the need for using taxonomic backbones and standardizing biodiversity data and the utility of data from the Global Biodiversity Information Facility and other available sources in this process. Additionally, we make recommendations for the fields needed in relational databases for collating species data that can be used to inform conservation decisions and outline steps that can be taken to enable easier collation of biodiversity data, using South Africa as a case study.
1. Introduction
1.1. The need for collated, standardized species data
Protecting biodiversity requires knowing what plants and animals occur in and around protected areas. As such, the Convention on Biological Diversity (CBD) requires biodiversity monitoring and maintenance of biodiversity information (United Nations, 1992). Despite the inherent value of readily available biodiversity data, biodiversity data management is often overlooked in conservation organizations. Relevant biodiversity data is often inconsistent, incomplete, inaccessible and unusable without relevant metadata (Stephenson et al., 2017). This is not for a lack of available systems, protocols and best practices (see Wilkinson et al., 2016; Hackett et al., 2019). Biodiversity data standards have been produced, such as Darwin Core, which enables comparable data sharing through standardizing data fields and requires certain ancillary data (Wieczorek et al., 2012). Additionally, the Global Biodiversity Information Facility (GBIF) provides a platform for sharing and accessing biodiversity data shared by others (Gaiji et al., 2013). Yet, conservation organizations fall short in their data management. The insights presented here are the outcome of conceptualizing a biodiversity information system for South African National Parks and provide considerations for building capacity to enhance conservation data management more generally, with the Supplementary materials detailing particular processes and tools that can be used for collating biodiversity occurrence data.
1.2. Challenges of making biodiversity data accessible
Biodiversity status and trend data are needed for making conservation decisions (Jimenez-Valverde et al., 2010). However, accessing and sharing biodiversity data is a key challenge of implementing the CBD (Chandra and Idrisova, 2011), and even though data collected using public funds should be publically available (Costello, 2009; Chavan and Penev, 2011; Thessen and Patterson, 2011), it often is not. In Africa, capacity and skills to collect, process and curate data is limited in many institutions (Stephenson et al., 2017), with data sharing not being a priority. This is not surprising: biodiversity scientists globally resist sharing biodiversity data (Mandeville et al., 2021), partly because of the time required to make their data sharable (Enke et al., 2012). Despite the multitude of conservation benefits of publishing biodiversity data (Tulloch et al., 2018), historically, there has not been a data-sharing culture among biodiversity researchers (Huang et al., 2012; Costello et al., 2013), with many researchers being reluctant to share data before publishing (Huang et al., 2012) and not knowing how to share their data (Enke et al., 2012). Funders and journals now require data to be made available (see Costello, 2009), and this approach could be extended to institutions including data sharing as criteria for assessing job performance. However, shared data also needs to be standardized and usable (see Costello et al., 2013).
Disparate data are collected by different people (Alves et al., 2018) using different approaches (Berkley et al., 2001; Heidorn, 2008) to answer different questions, and limited human resources are dedicated to curating conservation data (Heidorn, 2008). In South Africa, one limiting area is data cleaning (Coetzer and Hamer, 2019), which is the correction or removal of inaccurate data and standardization of formatting to enable data to be more useful. However, some data management support and capacity building is being provided by GBIF nodes (Parker-Allie et al., 2021). In conservation organizations, there is often limited post-field processing and availability of expertise to guide this, and even where expertise exists, staff turnover and insufficient hand-overs can lead to substantial data loss (see Wiser et al., 2001; Sato et al., 2019). Therefore, although conservation organizations collect large quantities of biodiversity data, e.g., through rangers, these data need to be digitized, standardized according to protocols, checked for consistency through quality control procedures and collated so that they can contribute to decision making in conservation organizations (see Supplementary material 1). A further complication is obtaining accurate locality data for sensitive species. Locality records for sensitive species are necessarily obscured on data sharing platforms to protect these species from poaching, and obtaining this locality data can be challenging due to its conditional use and the limited data processing capacity of data holders and collators, such as the South African National Biodiversity Institute (SANBI).
1.3. The wealth of biodiversity data sources
Systematic long-term monitoring programmes are fundamental for assessing population trends (see Kamp et al., 2016), but they are resource intensive. Notably, there are unstructured sources of biodiversity data that can be integrated into monitoring programmes (Kühl et al., 2020; Stephenson and Stengel, 2020), including herbarium and museum specimen records and citizen science data (see Supplementary material 2). Most of these data are already collated by the GBIF, which enables access to data stored outside its country of origin, which is useful, as a substantial amount of biodiversity collections and data from the global south is in the global north (Tydecks et al., 2018), and research published in journals may be inaccessible to staff of conservation organizations (see Veríssimo et al., 2020). Additionally, there are increasing opportunities for volunteers to curate, identify and categorize data to assist with data analysis (see Supplementary material 2). However, uploading image and video files to online platforms requires having sufficient bandwidth, and many conservation organizations have slow internet.
1.4. Tools for managing biodiversity data
Although global biodiversity data systems are advancing (Farley et al., 2018) and many tools are available (see Gadelha et al., 2021; Figure 1; Supplementary material 2), awareness of and capacity to use these resources is limited, and there are limited data-entry-ready software options available to collate species data across the multiple available data sources, match names to accepted taxonomies and develop species checklists for protected areas. Database software, e.g., BRAHMS and Specify, has been developed for specimen data, but it does not have the flexibility to incorporate other data types, such as iNaturalist and CyberTracker (see Ansell and Koenig, 2011) observation data, which can be collected and curated much faster than traditional specimen-based data (see Kays et al., 2020).
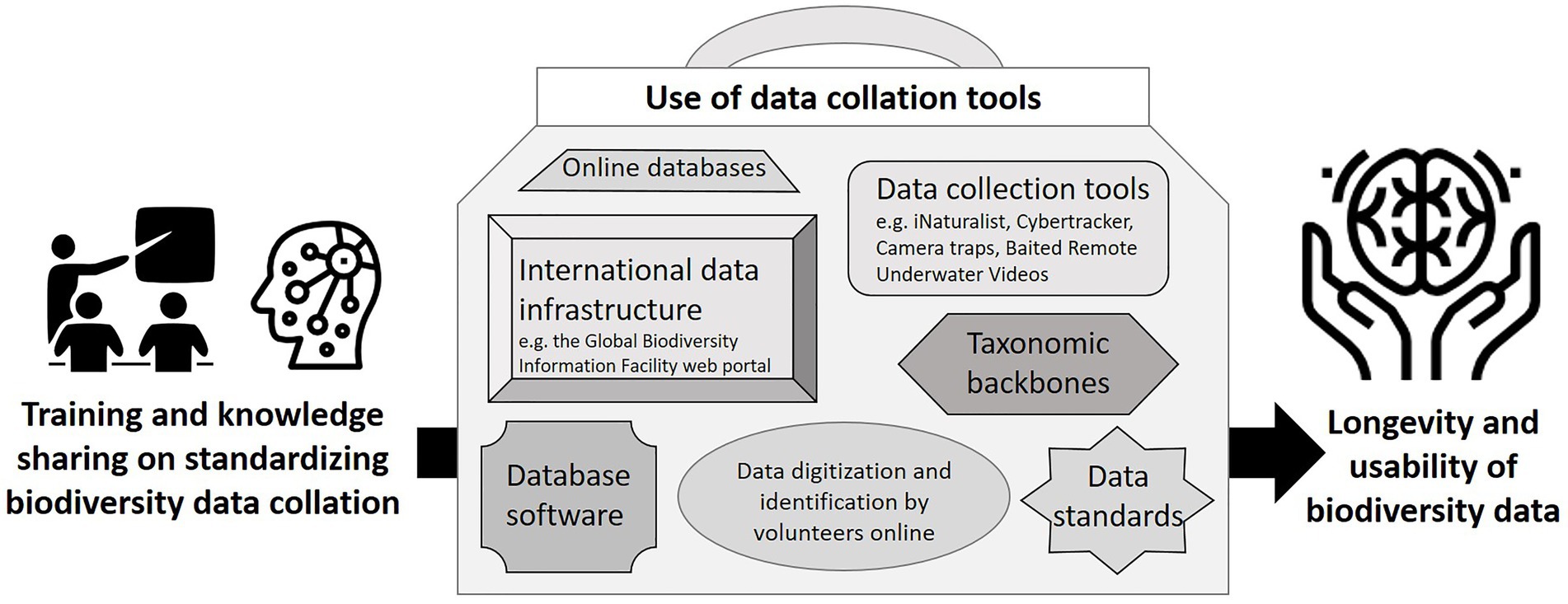
Figure 1. Tools to improve the collation of standardized biodiversity data for conservation. Further examples of resources for accessing biodiversity occurrence data to inform conservation decisions are provided in Supplementary material 2.
Making species data available to inform protected area management requires standardizing existing biodiversity data (see Supplementary material 1), incorporating data from global sources, e.g., GBIF, applying a consistent approach to naming species using taxonomic backbones, using a relational database with relevant fields and formats (Supplementary material 3), implementing data management systems and best practice quality control (Michener et al., 2011; Veiga et al., 2017; Ball-Damerow et al., 2019), and making these data available on a user-friendly platform.
1.5. Taxonomic backbones
Taxonomic backbones are essential foundations of biodiversity information systems (Thomson et al., 2021) and require regular updates. They are exhaustive taxon- and area-specific checklists of names that include unique identifiers for each name, taxonomic information (family, order, phylum, kingdom etc.), taxonomic status (accepted, synonym, inclusive, misapplied), and the unique identifier and accepted name for each synonym. All species names in a database or checklist should be checked against an authoritative taxonomic backbone (Costello and Wieczorek, 2014). There are many online taxonomic backbones that enable checking for accepted names (Grenié et al., 2022). The Catalogue of Life (CoL; Hobern et al., 2021), which is the primary source of names for the GBIF, incorporates many taxonomic databases. Keeping taxonomies up to date is challenging because of continuous changes in nomenclature requiring historical lists to be updated. These updates are complex, as species can change names, swop names with another species, be combined with another species, or a name could apply to several species that were split since an identification was made (Godfray, 2002). Additionally, the subspecies or variety, which may be of conservation interest, is often not specified or ‘aff.’ (similar potentially new species) or ‘cf.’ (uncertain identification) is associated with a listed name.
In South Africa, SANBI is mandated to maintain national species checklists and has made efforts toward compiling checklists of accepted species names in the country. The most comprehensive of these lists is the annually-updated South African National Plant Checklist (SANPC; SANBI, 2022a), which is part of the taxonomic backbone of the Botanical Database of Southern Africa (BODATSA). Updates to this checklist are guided by a policy that requires that only published name changes are included in the checklist, and updates to the checklist have to be checked by taxonomic experts and approved by a committee, which includes three SANBI taxonomists and six external taxonomists (Victor et al., 2013). The BODATSA is maintained in BRAHMS software, and the published version of the checklist includes the necessary information required to form the basis of a taxonomic backbone for South African plants in a new species database.
One challenge with using the SANPC, and likely many other national checklists, is that there is no accessible species matching tool. This is problematic for the staff of conservation organizations, who often have outdated species lists without authorities and lack the technical skills to automate name matching. Matching to online lists, such as World Flora Online, is relatively easy with the use of available fuzzy matching tools, e.g., the Taxonomic Name Resolution Service (Boyle et al., 2013) and an R package called WorldFlora (Kindt, 2020). Additionally, GBIF names are easily matched using GBIF’s species lookup tool1, which would be even more useful if the accepted names of synonyms could be included as part of downloads for matched species names. More manually-intensive methods are available in the absence of a matching tool, including the use of functions in MS Excel (see Supplementary material 4) and R. Another challenge with the SANPC is that it does not include all alien species that are found in the country: it only includes those species that are considered naturalized, and some nationally-regulated alien species are missing from the list. This necessitates having a standardized way of capturing the scientific names of non-naturalized alien species found on protected area species lists. While GBIF or the Global Register of Introduced and Invasive Species (see Pagad et al., 2018), which is available on the GBIF platform, could be used, ideally, the SANPC should be updated. It would be useful for SANPC names to be used across organizations in South Africa, and not just herbaria, to standardize national name usage annually and inform more accurate species assessments and conservation prioritization. Using a national checklist can help keep names relevant and useable, and users can submit relevant published updates, changes and errors.
In contrast to the SANPC, there is no comprehensive animal checklist that has been produced by SANBI. The animal names used by SANBI are also overseen by taxonomic experts and a committee. These taxonomists consult specific databases for different groups, e.g., the Amphibian Species of the World database hosted by the American Museum of Natural History for amphibians (Frost, 2021) and The Reptile Database for reptiles (Uetz et al., 2022). There is limited capacity to develop the animal list, particularly given the large variety of species groups, which have been focused on in isolation, and to date, only some vertebrate and freshwater invertebrate names have been included on SANBI’s national animal checklist (see SANBI, 2022b). In the absence of a comprehensive national list of animals, one interim solution is to use an easily available source of animal names, such as iNaturalist or GBIF, which incorporates the CoL.
1.6. The use of GBIF and iNaturalist to inform conservation
Extensive data sources are easily accessed through the online GBIF data platform (Gaiji et al., 2013). However, locality data specific to protected areas are not easily downloaded (see Supplementary material 5 for how to access species locality data for a protected area using Geographical Information System software). An option to download locality data by protected area administrative boundaries would be a useful addition to the GBIF data platform, as conservation organization staff often do not know how to access biodiversity data for protected areas. While GBIF includes functionality to filter data by IUCN Red List threat status, it would also be useful to be able to filter by occurrence (indigenous or alien) status per country.
iNaturalist data are available directly from the iNaturalist website and research grade data, which are observations for which two-thirds or more than two iNaturalist identifications concur, are available from the GBIF. iNaturalist provides a wealth of data, which can be used to inform conservation (Dobson et al., 2020), particularly when iNaturalist is used in bioblitzes and by specialist groups.
iNaturalist data are often organized into projects and places on the iNaturalist website, making it easy to access and curate locality data for protected areas. However, similar to the case for museum and herbarium specimen data, a key caveat is that not all identifications are accurate. The accuracy of iNaturalist records can be assessed by considering who made the identification, as all iNaturalist users are not equal. Specialists can be asked to verify identifications that are not made by taxon experts. Experts can be identified by looking at the ‘top identifiers’ tab and asking one of the reliable ‘top identifiers’ to verify the species identification. Unlike Cybertracker or Cmore2 data, the images that accompany iNaturalist data make identifications easier to verify. iNaturalist users should also be encouraged to submit diagnostic pictures, such as flowers, seeds and male and female specimens. Further, data users should be aware that because of obscured locality data records may appear at locations where species do not occur. iNaturalist also has a powerful curation tool to assist in rapid and efficient identification of large volumes of records.
2. Discussion
Although species, and lower taxonomic rank, data are a vital aspect of conservation management, these data are not easily available and regularly updated for many protected area networks. Given the parameters that need to be considered for collating, storing and sharing species and locality data for protected areas there are some minimum requirements that need to be considered for plant and animal locality databases, which should form part of organizational data management plans (see Donnelly et al., 2010; Strasser et al., 2011; Michener and Jones, 2012; Hampton et al., 2013). Additionally, capacity building, such as the training provided by GBIF’s biodiversity data mobilization course3 and training on using OpenRefine and Wikibase software, is vital but under resourced.
2.1. Recommendations for collating biodiversity occurrence data
Species databases should include the wide range of heterogeneous species occurrence data (Kühl et al., 2020, Stephenson and Stengel, 2020; see Supplementary material 2), and a relational database should be used with some compulsory fields, such as unique taxon numbers (see Anderson et al., 2020), to allow updates to taxon names and conservation statuses in the taxon table and link to the rest of the database. Although more sophisticated software is available, such a database can be set up in MS Excel with a taxon table in one spreadsheet and occurrence records in another spreadsheet and the use of functions or Power Pivot to link the data between the tables. Additionally, a protocol is required for updating databases regularly to incorporate new data from data sources that are constantly being updated, e.g., searching GBIF for particular time periods and accession dates.
There are several standardized fields that are required to develop a database that will be useful for informing conservation (see Supplementary material 3). Consistent, standardized and accepted naming systems are needed, and while these should ideally be driven by taxonomists, available taxonomic backbones, such as the GBIF backbone taxonomy, provide a work around where resources are limited. Incorrect taxonomic identifications and inaccurate coordinates are well-known issues. Ideally, detail is needed about the accuracy of the locality information (GBIF, 2010; Faith et al., 2013) and the source and reliability of the identification to determine the validity of the identification (Anderson, 2012; Costello and Wieczorek, 2014). For data generated in a conservation organization this would include noting who made the observations and identifications of species. Occurrence status (endemic, indigenous, extralimital, alien) for the protected area and conservation classifications are also needed as these are relevant to conservation management.
2.2. Enabling easier collation of biodiversity occurrence data
The accessibility of biodiversity data for informing conservation in South Africa could be improved through enhanced institutional data management support, inter-organization collaboration and capacity building to enable the use of standardized electronic data capture and data sharing protocols, templates and tools and the use of standardized names for all taxa for species reporting, listing and conservation status assessment.
Increased data collation and sharing by researchers, conservation staff and specimen collectors is possible through the use of iNaturalist, which provides a useful platform for uniform data sharing and access. Additionally, prerequisites and available support for researchers to upload biodiversity occurrence data to GBIF would improve data availability and reduce the data management burden on conservation authorities. An agreed set of backbones for taxa names, e.g., GBIF, that can be used by organizations nationally and national species lookup tools for looking up scientific names would improve consistent name usage nationally. For example, it would be useful if the SANPC could include all alien plants in South Africa to assist with managing and reporting on alien species in a standardized way at the national level. Name matching to the SANPC could also be made easier through an Application Programming Interface being made available for linking the SANPC to existing species matching tools.
Having the functionality to download species occurrence data from global and national platforms using protected area boundaries and the incorporation of GBIF data into local and national biodiversity information systems would improve the accessibility of data to the staff of conservation organizations. The inclusion of a term, such as protectedAreaName, in Darwin Core, through engagement with the TDWG (Biodiversity Information Standards), would also be useful. Further, it would be useful for conservation organizations to have easy ways to securely access occurrence data for sensitive species from SANBI to enable effective monitoring of these species, which is currently constrained by limited access to data as a consequence of human resource constraints.
To conclude, biodiversity data needs an overhaul, with a focus on data sharing, to improve data availability and standardization for biodiversity data to become more useful for informing conservation decisions. Incentives, institutional support and capacity building are needed to enhance the sharing of biodiversity occurrence data to data-sharing platforms, such as GBIF, and enable conservation organizations to access this data.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
DS conceptualized, wrote and revised the manuscript. NvW, AR, and JB conceptualized and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
DS is funded by the JRS Biodiversity Foundation, grant 60916.
Acknowledgments
Our colleagues at SANBI are thanked for explaining their biodiversity data management systems. Three reviewers are thanked for their constructive comments, which improved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1037282/full#supplementary-material
Footnotes
References
Alves, C., Castro, J. A., Ribeiro, C., Honrado, J. P., and Lomba, Â. (2018). “Research data management in the field of ecology: an overview,” in International Conference on Dublin Core and Metadata Applications. 87–94).
Anderson, R. P. (2012). Harnessing the world's biodiversity data: promise and peril in ecological niche modeling of species distributions. Ann. N. Y. Acad. Sci. 1260, 66–80. doi: 10.1111/j.1749-6632.2011.06440.x
Anderson, R. P., Araújo, M. B., Guisan, A., Lobo, J. M., Martínez-Meyer, E., Peterson, A. T., et al. (2020). Optimizing biodiversity informatics to improve information flow, data quality, and utility for science and society. Front. Biogeogr. 12:e47839. doi: 10.21425/F5FBG47839
Ansell, S., and Koenig, J. (2011). CyberTracker: an integral management tool used by rangers in the Djelk indigenous protected area, Central Arnhem Land, Australia. Ecol. Manag. Restor. 12, 13–25. doi: 10.1111/j.1442-8903.2011.00575.x
Ball-Damerow, J. E., Brenskelle, L., Barve, N., Soltis, P. S., Sierwald, P., Bieler, R., et al. (2019). Research applications of primary biodiversity databases in the digital age. PLoS One 14:e0215794. doi: 10.1371/journal.pone.0215794
Berkley, C., Jones, M., Bojilova, J., and Higgins, D. (2001). “Metacat: a schema-independent XML database system,” in Proceedings thirteenth international conference on scientific and statistical database management. SSDBM 2001. IEEE. 171–179.
Boyle, B., Hopkins, N., Lu, Z., Raygoza Garay, J. A., Mozzherin, D., Rees, T., et al. (2013). The taxonomic name resolution service: an online tool for automated standardization of plant names. BMC Bioinf. 14, 1–15. doi: 10.1186/1471-2105-14-16
Chandra, A., and Idrisova, A. (2011). Convention on biological diversity: a review of national challenges and opportunities for implementation. Biodivers. Conserv. 20, 3295–3316. doi: 10.1007/s10531-011-0141-x
Chavan, V., and Penev, L. (2011). The data paper: a mechanism to incentivize data publishing in biodiversity science. BMC Bioinf. 12, 1–12. doi: 10.1186/1471-2105-12-S15-S2
Coetzer, W., and Hamer, M. (2019). Managing south African biodiversity research data: meeting the challenges of rapidly developing information technology. S. Afr. J. Sci. 115, 1–5. doi: 10.17159/sajs.2019/5482
Costello, M. J. (2009). Motivating online publication of data. Bioscience 59, 418–427. doi: 10.1525/bio.2009.59.5.9
Costello, M. J., Michener, W. K., Gahegan, M., Zhang, Z. Q., and Bourne, P. E. (2013). Biodiversity data should be published, cited, and peer reviewed. Trends Ecol. Evol. 28, 454–461. doi: 10.1016/j.tree.2013.05.002
Costello, M. J., and Wieczorek, J. (2014). Best practice for biodiversity data management and publication. Biol. Conserv. 173, 68–73. doi: 10.1016/j.biocon.2013.10.018
Dobson, A. D., Milner-Gulland, E. J., Aebischer, N. J., Beale, C. M., Brozovic, R., Coals, P., et al. (2020). Making messy data work for conservation. One Earth 2, 455–465. doi: 10.1016/j.oneear.2020.04.012
Donnelly, M., Jones, S., and Pattenden-Fail, J. W. (2010). DMP online: the digital curation Centre’s web-based tool for creating, maintaining and exporting data management plans. Int. J. Digit. Curation 5, 187–193. doi: 10.2218/ijdc.v5i1.152
Enke, N., Thessen, A., Bach, K., Bendix, J., Seeger, B., and Gemeinholzer, B. (2012). The user's view on biodiversity data sharing—investigating facts of acceptance and requirements to realize a sustainable use of research data. Ecol. Inform. 11, 25–33. doi: 10.1016/j.ecoinf.2012.03.004
Faith, D., Collen, B., Ariño, A., Koleff, P. K. P., Guinotte, J., Kerr, J., et al. (2013). Bridging the biodiversity data gaps: recommendations to meet users’ data needs. Biodivers. Inform. 8, 41–58. doi: 10.17161/bi.v8i2.4126
Farley, S. S., Dawson, A., Goring, S. J., and Williams, J. W. (2018). Situating ecology as a big-data science: current advances, challenges, and solutions. Bioscience 68, 563–576. doi: 10.1093/biosci/biy068
Frost, D. R. (2021). Amphibian species of the world: An online reference. Version 6.1 (Date of access). (New York, USA: American Museum of Natural History). Available at: https://amphibiansoftheworld.amnh.org/index.php
Gadelha, L. M. Jr., de Siracusa, P. C., Dalcin, E. C., da Silva, L. A. E., Augusto, D. A., Krempser, E., et al. (2021). A survey of biodiversity informatics: concepts, practices, and challenges. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 11:e1394. doi: 10.1002/widm.1394
Gaiji, S., Chavan, V., Ariño, A. H., Otegui, J., Hobern, D., Sood, R., et al. (2013). Content assessment of the primary biodiversity data published through GBIF network: status, challenges and potentials. Biodivers. Inform. 8, 94–172. doi: 10.17161/bi.v8i2.4124
GBIF. (2010). GBIF position paper on future directions and recommendations for enhancing fitness-for-use across the GBIF network, version 1.0. A. W. Hill, J. Otegui, A. H. Ariño, and R. P. Guralnick. Copenhagen: Global Biodiversity Information Facility, 25. Available at: http://www.gbif.org
Godfray, H. C. J. (2002). Challenges for taxonomy. The discipline will have to reinvent itself if it is to survive and flourish. Nature 417, 17–19. doi: 10.1038/417017a
Grenié, M., Berti, E., Carvajal-Quintero, J., Dädlow, G. M. L., Sagouis, A., and Winter, M. (2022). Harmonizing taxon names in biodiversity data: a review of tools, databases and best practices. Methods Ecol. Evol. 14, 12–25. doi: 10.1111/2041-210X.13802
Hackett, R. A., Belitz, M. W., Gilbert, E. E., and Monfils, A. K. (2019). A data management workflow of biodiversity data from the field to data users. Appl. Plant Sci. 7:e11310. doi: 10.1002/aps3.11310
Hampton, S. E., Strasser, C. A., Tewksbury, J. J., Gram, W. K., Budden, A. E., Batcheller, A. L., et al. (2013). Big data and the future of ecology. Front. Ecol. Environ. 11, 156–162. doi: 10.1890/120103
Heidorn, P. B. (2008). Shedding light on the dark data in the long tail of science. Libr. Trends 57, 280–299. doi: 10.1353/lib.0.0036
Hobern, D., Barik, S. K., Christidis, L., T. Garnett, S., Kirk, P., Orrell, T. M., et al. (2021). Towards a global list of accepted species VI: the catalogue of life checklist. Org. Divers. Evol. 21, 677–690. doi: 10.1007/s13127-021-00516-w
Huang, X., Hawkins, B. A., Lei, F., Miller, G. L., Favret, C., Zhang, R., et al. (2012). Willing or unwilling to share primary biodiversity data: results and implications of an international survey. Conserv. Lett. 5, 399–406. doi: 10.1111/j.1755-263X.2012.00259.x
Jimenez-Valverde, A., Lira-Noriega, A., Peterson, A. T., and Soberon, J. (2010). Marshalling existing biodiversity data to evaluate biodiversity status and trends in planning exercises. Ecol. Res. 25, 947–957. doi: 10.1007/s11284-010-0753-8
Kamp, J., Oppel, S., Heldbjerg, H., Nyegaard, T., and Donald, P. F. (2016). Unstructured citizen science data fail to detect long-term population declines of common birds in Denmark. Divers. Distrib. 22, 1024–1035. doi: 10.1111/ddi.12463
Kays, R., McShea, W. J., and Wikelski, M. (2020). Born-digital biodiversity data: millions and billions. Divers. Distrib. 26, 644–648. doi: 10.1111/ddi.12993
Kindt, R. (2020). WorldFlora: an R package for exact and fuzzy matching of plant names against the world Flora online taxonomic backbone data. Appl. Plant Sci. 8:e11388. doi: 10.1002/aps3.11388
Kühl, H. S., Bowler, D. E., Bösch, L., Bruelheide, H., Dauber, J., Eichenberg, D., et al. (2020). Effective biodiversity monitoring needs a culture of integration. One Earth 3, 462–474. doi: 10.1016/j.oneear.2020.09.010
Mandeville, C. P., Koch, W., Nilsen, E. B., and Finstad, A. G. (2021). Open data practices among users of primary biodiversity data. Bioscience 71, 1128–1147. doi: 10.1093/biosci/biab072
Michener, W. K., and Jones, M. B. (2012). Ecoinformatics: supporting ecology as a data-intensive science. Trends Ecol. Evol. 27, 85–93. doi: 10.1016/j.tree.2011.11.016
Michener, W. K., Porter, J., Servilla, M., and Vanderbilt, K. (2011). Long term ecological research and information management. Ecol. Inform. 6, 13–24. doi: 10.1016/j.ecoinf.2010.11.005
Pagad, S., Genovesi, P., Carnevali, L., Schigel, D., and McGeoch, M. A. (2018). Introducing the global register of introduced and invasive species. Sci. Data 5, 1–12. doi: 10.1038/sdata.2017.202
Parker-Allie, F., Pando, F., Telenius, A., Ganglo, J. C., Vélez, D., Gibbons, M. J., et al. (2021). Towards a post-graduate level curriculum for biodiversity informatics. Perspectives from the global biodiversity information facility (GBIF) community. Biodivers. Data J. 9:e68010. doi: 10.3897/BDJ.9.e68010SANBI
SANBI. (2022a). Official yearly release of South African National Plant Checklist for 2022. South African National Biodiversity Institute. Available at: https://hdl.handle.net/20.500.12143/6880 (Accessed June 7, 2022).
SANBI. (2022b). South African animal checklist for reptiles, birds, frogs, freshwater fish, dobsonflies, caddisflies, Mollusca, long-tongue flies, stoneflies, decapods, amphipods and mayflies. South African National Biodiversity Institute. Available at: https://hdl.handle.net/20.500.12143/8511 (Accessed June 7, 2022).
Sato, C., Westgate, M. J., Barton, P., Foster, C., O'Loughlin, L. S., Pierson, J. C., et al. (2019). The use and utility of surrogates in biodiversity monitoring programmes. J. Appl. Ecol. 56, 1304–1310. doi: 10.1111/1365-2664.13366
Stephenson, P. J., Bowles-Newark, N., Regan, E., Stanwell-Smith, D., Diagana, M., Höft, R., et al. (2017). Unblocking the flow of biodiversity data for decision-making in Africa. Biol. Conserv. 213, 335–340. doi: 10.1016/j.biocon.2016.09.003
Stephenson, P. J., and Stengel, C. (2020). An inventory of biodiversity data sources for conservation monitoring. PLoS One 15:e0242923. doi: 10.1371/journal.pone.0242923
Strasser, C., Cook, R., Michener, W., Budden, A., and Koskela, R. (2011). “Promoting data stewardship through best practices,” in Proceedings of the environmental information management conference. 126–131.
Thessen, A. E., and Patterson, D. J. (2011). Data issues in the life sciences. ZooKeys 150, 15–51. doi: 10.3897/zookeys.150.1766
Thomson, S. A., Thiele, K., Conix, S., Christidis, L., Costello, M. J., Hobern, D., et al. (2021). Towards a global list of accepted species II. Consequences of inadequate taxonomic list governance. Org. Divers. Evol. 21, 623–630. doi: 10.1007/s13127-021-00518-8
Tulloch, A. I., Auerbach, N., Avery-Gomm, S., Bayraktarov, E., Butt, N., Dickman, C. R., et al. (2018). A decision tree for assessing the risks and benefits of publishing biodiversity data. Nat. Ecol. Evol. 2, 1209–1217. doi: 10.1038/s41559-018-0608-1
Tydecks, L., Jeschke, J. M., Wolf, M., Singer, G., and Tockner, K. (2018). Spatial and topical imbalances in biodiversity research. PLoS One 13:e0199327. doi: 10.1371/journal.pone.0199327
Uetz, P., Freed, P., Aguilar, R., and Hošek, J. (eds.) (2022). The reptile database. Available at: http://www.reptile-database.org
Veiga, A. K., Saraiva, A. M., Chapman, A. D., Morris, P. J., Gendreau, C., Schigel, D., et al. (2017). A conceptual framework for quality assessment and management of biodiversity data. PLoS One 12:e0178731. doi: 10.1371/journal.pone.0178731
Veríssimo, D., Pienkowski, T., Arias, M., Cugnière, L., Doughty, H., Hazenbosch, M., et al. (2020). Ethical publishing in biodiversity conservation science. Conserv. Soc. 18, 220–225. doi: 10.4103/cs.cs_19_56
Victor, J., Klopper, R. R., Winter, P. J. D., and Hamer, M. (2013). South African National Plant Checklist Policy. Pretoria: South African National Biodiversity Institute. Available at: http://hdl.handle.net/20.500.12143/6880 (Accessed March, 2013).
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., et al. (2012). Darwin Core: an evolving community-developed biodiversity data standard. PLoS One 7:e29715. doi: 10.1371/journal.pone.0029715
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR guiding principles for scientific data management and stewardship. Sci. data 3, 160018–160019. doi: 10.1038/sdata.2016.18
Wiser, S. K., Bellingham, P. J., and Burrows, L. E. (2001). Managing biodiversity information: development of New Zealand’s National Vegetation Survey databank. New Zeal. J. Ecol. 25, 1–17. Available at: http://www.jstor.org/stable/24055293
Keywords: biodiversity data, GBIF, iNaturalist, taxonomic backbones, species checklists, conservation, data management
Citation: Spear D, van Wilgen NJ, Rebelo AG and Botha JM (2023) Collating biodiversity occurrence data for conservation. Front. Ecol. Evol. 11:1037282. doi: 10.3389/fevo.2023.1037282
Edited by:
Vernon Visser, University of Cape Town, South AfricaReviewed by:
Cossi Jean Ganglo, University of Abomey-Calavi, BeninMaarten Trekels, Botanic Garden Meise, Belgium
Copyright © 2023 Spear, van Wilgen, Rebelo and Botha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dian Spear, ZGlhbi5zcGVhckBzYW5wYXJrcy5vcmc=
 Dian Spear
Dian Spear Nicola J. van Wilgen1,2
Nicola J. van Wilgen1,2 Anthony G. Rebelo
Anthony G. Rebelo Judith M. Botha
Judith M. Botha