
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 11 January 2023
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.988504
 Wen Wan†
Wen Wan† Zheng Ren†
Zheng Ren† Hongling Zhang
Hongling Zhang Qiyan Wang
Qiyan Wang Ting Wang
Ting Wang Yunteng Yang
Yunteng Yang Jiangtao You
Jiangtao You Kun He
Kun He Jiang Huang*
Jiang Huang* Xiaoye Jin*
Xiaoye Jin*Insertion/deletion polymorphisms (InDels) show great application values in forensic research because they own superiorities of short tandem repeats (STRs) and single nucleotide polymorphisms (SNPs). Whereas, InDels commonly display low genetic diversities in comparison to STRs. Therefore, they may provide limited genetic information in forensic kinship testing. Here, we evaluated forensic application efficiency of a novel multiplex amplification system including two STRs, 59 InDels, and three sex-determination loci in the Guizhou Dong group. In addition, we explored the genetic background of the Guizhou Dong group in comparison to other reported populations based on 59 InDels. We found that 59 InDels displayed relatively high genetic diversities in the Guizhou Dong group. Moreover, the cumulative forensic efficiency of two STRs and 59 InDels could meet the requirement of individual identification and paternity testing in the Guizhou Dong group. For these 59 InDels, we observed that some loci exhibited relatively high genetic differentiations among different continental populations, especially for African and Non-African populations, which could be viewed as candidate ancestry informative markers in the future. Genetic structure results indicated that the Dong group had close genetic relationships with East Asian and some Southern Chinese Han populations. To sum up, we stated that the 64-plex panel could be performed for forensic application of the Guizhou Dong group.
In forensic research, forensic geneticists commonly need to solve two dominant tasks: personal identification and kinship testing. STRs are viewed as the mainstream genetic markers for forensic researchers because of their high genetic diversities and discrimination power (Fan et al., 2021; Harrel et al., 2021; Qu et al., 2021). Nonetheless, there are still some defects for STRs in forensic practice. For example, the relatively high mutation rate of STRs may make incorrect conclusions in complex paternity analyses (Liu et al., 2021; Chandra et al., 2022); the large amplicon of STRs is not conducive to assessing outdated and degraded biological samples (Boelens et al., 2021; Song et al., 2022). To avoid these issues of STRs in practical application, SNPs are usually employed as supplemental genetic markers for forensic research because they have widespread in the human genome, show small amplicon, and possess low mutation rates. Accordingly, a set of panels of SNPs have been constructed (Li et al., 2018; Avent et al., 2019; Hwa et al., 2019; Zhao et al., 2021). In spite of this, SNPs belong to sequence variants, which are commonly involved with complex detection methods and high costs for SNP typing (Wei et al., 2014). Therefore, it is unfavorable for us to promote and popularize SNPs in forensic grassroots laboratories.
InDels are genetic markers that display the insertion or deletion of random DNA fragments in the genome (Weber et al., 2002). InDels have some advantageous features that resemble SNPs: small amplicon and low mutation rate (LaRue et al., 2012; Wei et al., 2014). Intriguingly, alleles of InDels exhibit length differences that are analogous to STRs, which could be compatible with the extant capillary electrophoresis (CE) technology. A set of multiplex amplification systems of InDels for forensic various purposes have been developed: Jin et al. (2019) constructed a panel of 35 InDels for forensic personal identification via the CE; Zhang et al. (2021) developed a multiplex amplification panel of 39 ancestral informative InDels for forensic ancestral origin analyses of different continental populations; Tao et al. (2019) developed a panel of 27 autosomal InDels, 16 X chromosomal InDels, and two Y chromosomal InDels for complicated paternity relationship testing. Even so, these available InDel panels could be applicable for forensic paternity testing as additional tools since InDels possess relatively low genetic diversities. Moreover, these InDels are also unfavorable for dissecting mixed samples consisting of more than two contributors because they exhibit two allelic variations at best. To this end, genetic markers that show multiple allelic variations could be integrated into the extant panels to improve their performance in forensic paternity testing and mixture deconvolution.
Previously, a novel multiplex amplification system (64-plex) that includes two STRs, 59 InDels, and three sex-determination loci has been developed based on the CE (Liu et al., 2022a). In the following study of developmental validation, Liu et al. (2022b) found that the novel panel displayed greater application value in forensic individual identification and kinship testing than the extant InDel and miniSTR panels. In addition, the novel panel showed good tolerance to common inhibitors, strong species specificity, and good applicability for case-type samples. In a nutshell, they proposed that the panel could be performed for forensic research in Chinese populations well. Even so, the application efficiency of the system in Chinese other populations needs to be further evaluated before it will be put into practice.
Dong group, also known as Kam, is one of 55 minority groups in China and mainly lives in Guizhou, Guangxi, and Hunan provinces. Dongs do not have their own traditional script and their language belongs to Tai-Kadai. Regarding the ancestral origins of Dongs, some scholars thought that they were descendants of ancient Liaos (Geary, 2003). Furthermore, Dongs stated that their ancestors might be immigrants from Eastern China and Southern China regions (Skutsch, 2013). At all events, controversy about the ancestral origins of the Dong group still exists. Therefore, ancestral components of the Dong group should be explored by multiple genetic markers to understand its genetic background better.
Currently, we firstly investigated allelic distributions and forensic parameters of the novel panel in the Guizhou Dong group. Besides, population genetic analyses of Guizhou Dong and other previously reported populations (1000 Genomes Project Consortium et al., 2015; Liu et al., 2022a,b) were performed by multiple methods to dissect the genetic structure of the Guizhou Dong group based on 59 InDels.
Bloodstains of 142 Guizhou Dong individuals (86 males and 56 females) who have lived in the Guizhou province for at least three generations were gathered. All participant provided their written informed consent. Furthermore, these Dongs were unrelated healthy individuals. The study was conducted in line with the guideline of the Ethics Committee of Guizhou Medical University and was warranted by the Ethics Committee of Guizhou Medical University (Approval number: 2021-224).
Based on the same 59 InDels, the genetic structure of the Guizhou Dong group was dissected in comparison to the previously reported 29 populations. The general information of populations used in this study was listed in Supplementary Table 1.
1 cm2 card was punched from the bloodstain and added to the PCR mixture. The mixture comprised 2 μL Master Mix (HEALTH Gene Technologies, Ningbo, China), 2 μL Primer Mix, and 6 μL deionized water. Next, the PCR cocktail was amplified on the GeneAmp PCR System 9,700 equipment (Thermo Fisher Scientific, Foster City, CA, United States) according to the recommended parameters (Liu et al., 2022b). Thirdly, we extracted 1 μL amplified product and added it into the cocktail including 0.5 μL SIZE-500 (HEALTH Gene Technologies) and 8.5 μL Hi-Di deionized formamide. And then we utilized the ABI 3500xL Genetic Analyzer (Thermo Fisher Scientific, Foster City, CA, United States) to separate and detect the cocktail. Finally, allelic typing of two STRs, 59 InDels, and three Y-chromosomal loci for all samples were generated by the GeneMapper ID-X Software v1.5 (Thermo Fisher Scientific, Foster City, CA, United States) in comparison to the allelic ladder.
We estimated allelic frequencies and forensic parameters that included polymorphic information content (PIC), matching probability (MP), discrimination power (DP), observed heterozygosity (Ho), expected heterozygosity (He), probability of exclusion (PE), and typical paternity index (TPI) of two STRs and 59 InDels by the STRAF software v1.0.5 (Gouy and Zieger, 2017). In addition, Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) testing of two STRs and 59 InDels were also conducted by the STRAF. Insertion allelic frequency heatmap of 59 InDels in the Guizhou Dong and 29 compared populations was conducted by the Sangerbox online tool v3.0.1 Informativeness-for-assignment metric (In) values of 59 InDels were computed by the infocalc program v1.1 (Rosenberg et al., 2003). Principle component analysis (PCA) of the Guizhou Dong and other compared populations at the population level was conducted by the factorextra package v1.0.7.999 of R software v4.1.0. Besides, PCA of these populations at the individual level was performed and plotted by the STRAF software and ggplot2 package v4.1.3 of R software, respectively. Genetic distances (DA) of the Guizhou Dong and other compared populations were computed by the dispan software2 based on allelic frequencies of 59 InDels. An unweighted pair-group method with the arithmetic means-based tree of these populations was constructed by the MEGA software (Kumar et al., 2018) based on pairwise DA values. Pairwise fixation index (FST) of Guizhou Dong and other reference populations were estimated and plotted by the Arlequin software v3.5 (Excoffier and Lischer, 2010) and corrplot package v0.92 of R software, respectively. Population genetic structure of these populations was performed by the ADMIXTURE software v1.3 (Alexander et al., 2009) at K = 2–7 according to the default parameters. Ancestral components of these populations were further displayed by the AncestryPainter program (Feng et al., 2018) at K = 4.
Results of HWE testing for two STRs and 59 InDels in the Guizhou Dong population were listed in Supplementary Table 2. We found that the p values of rs5877451, rs4024564, rs35453727, rs10649306, and rs148501393 loci were less than 0.05. Even so, these loci did not show statistically significant after Bonferroni’s correction (p = 0.05/61 = 0.00082). Accordingly, we stated that these 61 loci conformed to HWE in the Guizhou Dong group.
p-values of LD analyses for pairwise loci were listed in Supplementary Table 3. Equally, p-values of pairwise loci did not display statistically significant after Bonferroni’s correction (k = 1.83 × 103, p < 0.000027). Therefore, we stated that these 61 loci could be considered independent loci from each other in the Guizhou Dong group.
Allelic frequencies of two STRs and 59 InDels in the Guizhou Dong were given in Supplementary Table 2. There were 136 alleles observed in the Guizhou Dong group for these 61 loci. As expected, D1S1656 and D3S1358 loci exhibited the most number of alleles in the Guizhou Dong group and the number of alleles of these two loci was 12 and 6, respectively. Allelic frequencies of two STRs were distributed from 0.0035 to 0.3451. For the remaining 59 InDels, we found that the majority of loci showed relatively even frequency distributions with the values distributing from 0.3345 to 0.6655.
Forensic parameters of two STRs and 59 InDels in the Guizhou Dong group were presented in Supplementary Table 2. For two STRs, PIC, Ho, He, MP, DP, PE, and TPI values of D1S1656 locus were 0.8171, 0.8239, 0.8391, 0.0489, 0.9511, 0.6442, and 2.8400, respectively; and they were 0.6853, 0.6901, 0.7351, 0.1161, 0.8839, 0.4132, and 1.6136 for D3S1358 locus, respectively. For InDels, the average PIC, Ho, He, MP, DP, PE, and TPI values of 59 InDels were 0.3694, 0.4816, 0.4909, 0.3800, 0.6200, 0.1746, and 0.9712, respectively. Cumulative MP, DP, and PE of these 61 loci in the Guizhou Dong group were 8.34 × 10−28, ~1.00, and 0.999997602, respectively. In addition, we also estimated gene diversities (GD) of two Y-InDels (rs759551978 and rs199815934) in the Guizhou Dong group. GD values of these two loci (rs759551978 and rs199815934) were 0.2079 and 0.2758, respectively.
Insertion allelic frequencies of 59 InDels in the studied Dong and other reference populations were shown in Figure 1. We observed that some loci displayed relatively high allelic frequency divergences among these populations. For instance, rs6481 and rs5833522 loci exhibited relatively low frequencies in African and European populations, respectively; rs3083268, rs35173752, rs144378883, rs10579944, and rs1611025 loci displayed relatively high allelic frequencies in African populations. In can measure genetic differentiations of genetic markers in different populations (Phillips, 2015). Therefore, we also estimated In metrics of these 59 InDels among different continental populations (Figure 1). Obtained results revealed that In values of these loci distributed from 0.0009 (rs10535391) to 0.1198 (rs35173752). Besides, we also estimated In values of these loci between pairwise populations, as given in Supplementary Table 4. Results revealed that there were 13 loci showing relatively high In values between African and other continental populations. For example, In values of rs10579944 locus were greater than 0.1 between African and Non-African populations.

Figure 1. Insertion allelic frequency heatmap and In values of 59 InDels in the Guizhou Dong and other compared populations.
Based on allelic frequencies of 59 InDels, we conducted the PCA of the Guizhou Dong and other reported populations, as shown in Figure 2A. The first two principle components can explain more than 76% variances of the original data set. At PC1, we observed that these populations could be classified into two clusters: seven African populations located in the right part; the remaining populations located in the left part. At PC2, East Asian, African, European, and South Asian populations could be separated from each other. Besides, we found that four American populations were distributed between East Asian and South Asian populations. Next, we further assessed the contributions of 59 InDels to differentiating these populations, as shown in Figure 2B. We found that rs5833522, rs35173752, rs6481, and rs10563906 loci possessed high contributions to PC1; rs5833522, rs35173752, rs71698233, and rs35453727 loci showed high contributions to PC2, implying that these loci could be viewed as candidate ancestry informative markers for forensic ancestral analyses of these continental populations. Thirdly, we performed the PCA of these populations at individual levels (Figure 2C). We found that most Africans could be differentiated from other continental populations at PC1. Furthermore, most East Asians could be separated from European, South Asian, and American populations at PC2. For the studied Dongs, we observed that most Dongs were overlapped with East Asian individuals.
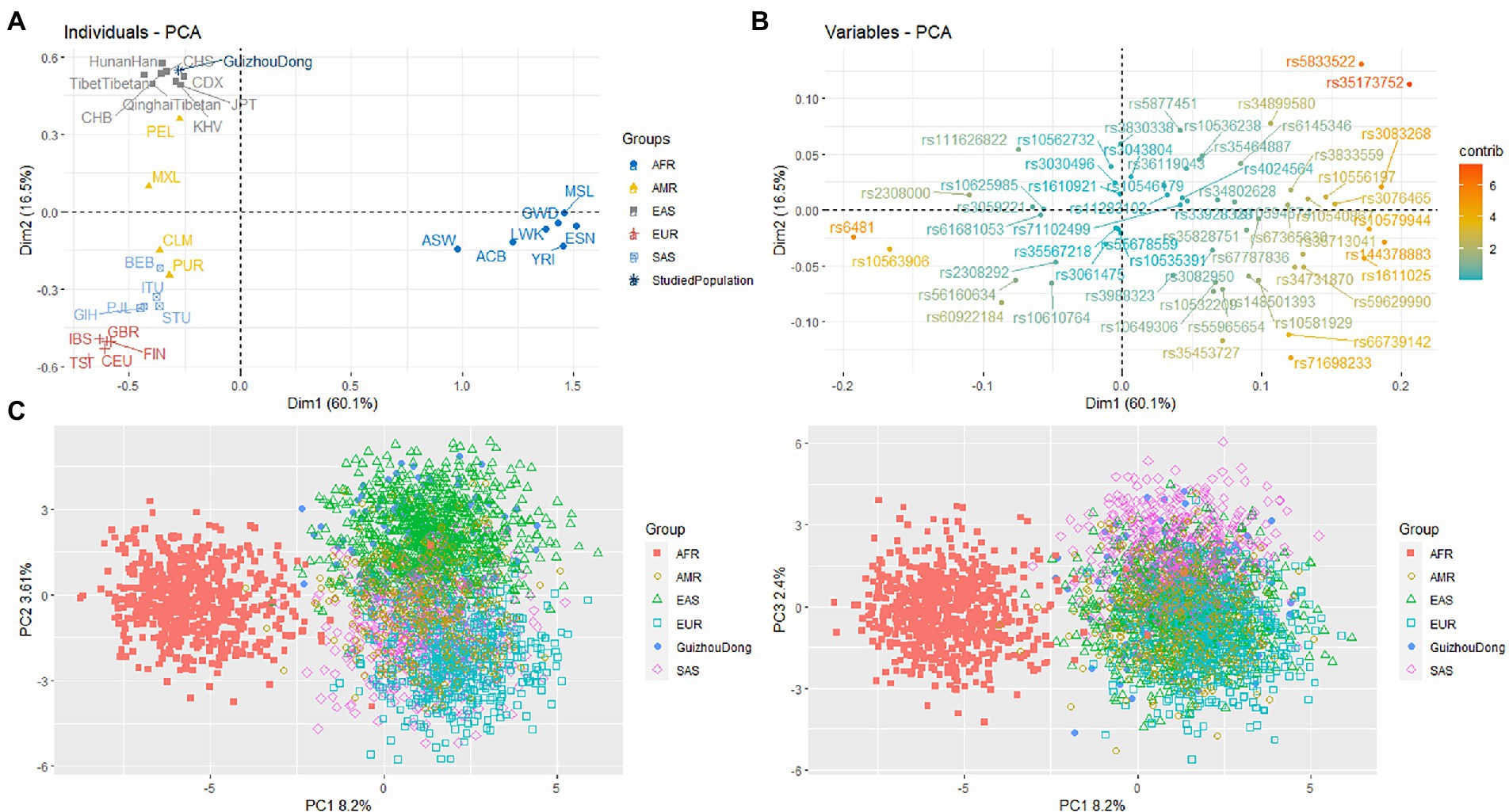
Figure 2. Principle component analysis of the Guizhou Dong and other compared populations. (A) Principle component analysis at population levels. (B) Contributions of each InDel locus to explaining the variability of the data set. (C) Principle component analysis at individual levels.
A phylogenetic tree of the Guizhou Dong and other reference populations was shown in Figure 3A. Five branches were discerned from the tree: seven African populations formed a branch; Guizhou Dong and other East Asian populations clustered in the same branch; five South Asian populations formed a branch; five European and three American populations clustered in the same branch; PEL population formed a branch. In addition, DA genetic distances of the Guizhou Dong and other compared populations were also displayed in Figure 3A; Supplementary Table 5. We found that the studied Dong population had the smallest DA value with CDX, followed by KHV, Hunan Han, CHS, and CHB populations. Instead, the studied Dong group showed high DA values with seven African populations. We also estimated FST values of Guizhou Dong and other reported populations, as shown in Figure 3B; Supplementary Table 6. Likewise, the Guizhou Dong had the lowest FST value with CDX, followed by KHV, CHS, Hunan Han, and CHB populations; whereas, it possessed higher FST values with other continental populations.
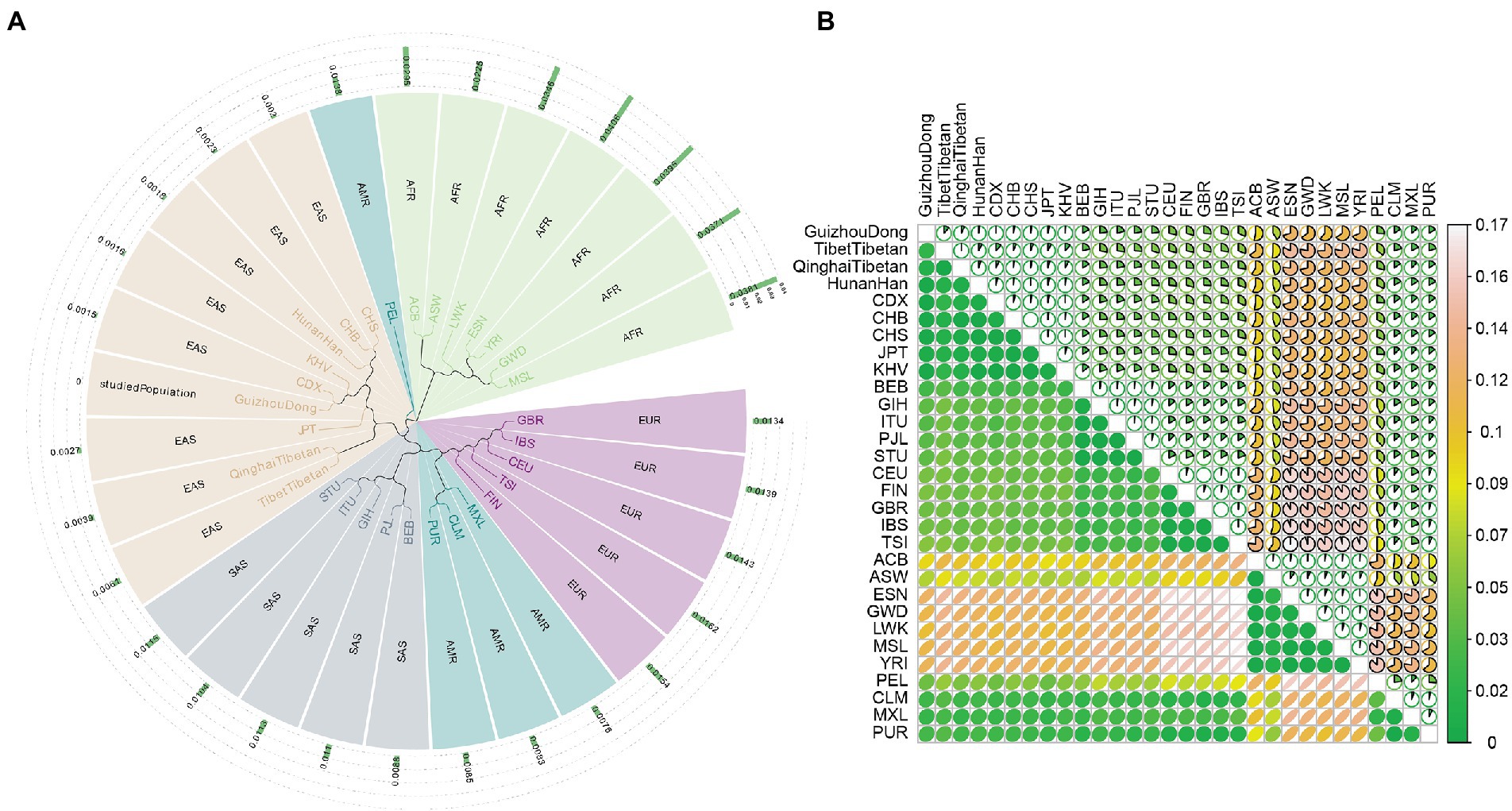
Figure 3. The Unweighted pair-group method with arithmetic means-based tree (A) and FST (B) of the Guizhou Dong and other compared populations.
Population genetic structure analyses of 30 populations at K = 2–7 were conducted, as given in Supplementary Figure 1. Besides, we also calculated the cross-validated error of each K, as shown in Supplementary Figure 2. We observed that the smallest cross-validated error was seen at K = 2, indicating K = 2 was the best K for the data set used in this study. At K = 2, seven African populations exhibited similar ancestral components that could be discriminated from the resting populations. When K increased to 3, nine East Asian populations could be further differentiated from other populations. When K become 4, East Asian, European, South Asian, and African populations showed different ancestral component distributions and could be discriminated from each other, which was also displayed in Figure 4A. However, no further genetic structure could be discerned from these populations at larger K values. Next, we assessed ancestral components of Guizhou Dongs at K = 4, as shown in Figure 4B. We found that these Dongs showed higher ancestral proportions of East Asian populations than other continental populations.
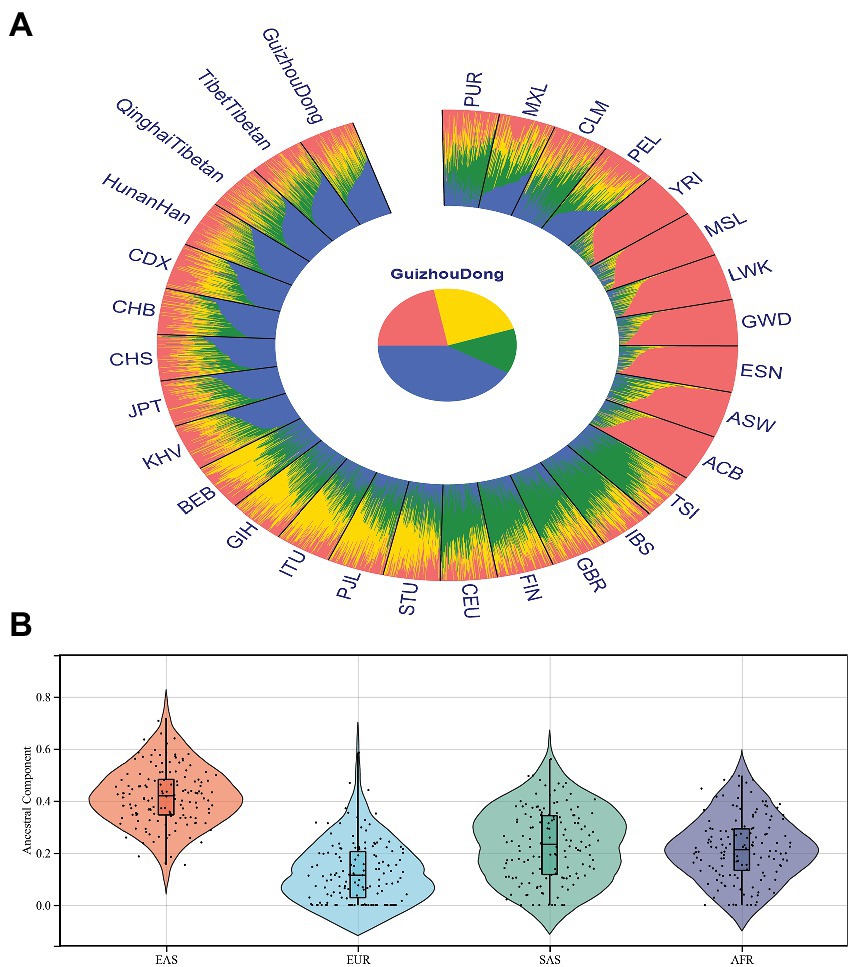
Figure 4. Ancestral component analyses of the Guizhou Dong group in comparison to other reference populations. (A) Population genetic structure of the Guizhou Dong group and other reference populations at K = 4; (B) Ancestral component proportions of the Guizhou Dong group from different continental populations.
Recently, forensic geneticists developed a set of multiplex amplification panels of InDels for forensic personal identification and paternity analyses (Li et al., 2011; Chen et al., 2019; Huang et al., 2020; Jin et al., 2021; Fan et al., 2022). However, InDels of these panels showed lower genetic diversities than commonly used STRs, which could provide limited genetic information in kinship analyses. The combination of InDels and STRs can be developed into a high-efficient tool for forensic research since it could integrate their advantageous features. Hereto, we evaluated the forensic application values of a novel multiplex system comprising two STRs, 59 InDel, and three sex determination loci in the Guizhou Dong group. We found that 59 InDels showed relatively high genetic heterozygosities (He >0.4000) in the Guizhou Dong group. Besides, these InDels also possessed relatively high PIC values (>0.3400), implying that these loci could provide reasonable genetic information content in forensic research. The forensic performance comparisons of different kits were provided in Table 1. We found that the system presented in this study owned higher forensic application values, especially for kinship testing in comparison to previously developed InDel panels (Liu et al., 2020; Jin et al., 2021; Liu J. et al., 2022). In addition, we observed that the panel presented in this study also showed better performance for forensic individual identification than commonly used STRs (Zhang, 2015; Guo, 2017). In conclusion, we proposed that the novel multiplex panel could be suitable for forensic individual identification and paternity testing in the Guizhou Dong group as an independent system.
For 59 InDels, different frequency distribution patterns of some loci were observed among these continental populations. Accordingly, we further assessed In values of these loci among these continental populations. Thirteen loci including rs10540867, rs10563906, rs10579944, rs144378883, rs1611025, rs3076465, rs3083268, rs34731870, rs35173752, rs5833522, rs6481, rs66739142, and rs71698233 displayed relatively high In values between African and Non-African populations. In addition, we also found that these loci provided relatively high contributions to PC1, implying that these loci could be viewed as candidate ancestry informative markers of African populations since PC1 could discriminate Africans and non-Africans (Figure 2). Nonetheless, we found that these 59 InDel loci were hard to differentiate European, East Asian, South Asian, and American populations at individual levels, which might be related to similar frequency distributions of most loci in these populations. Besides, we observed that minor allelic frequencies of these 59 loci in nine East Asian populations were larger than 0.2000, indicating these loci could also be utilized as valuable loci for forensic individual identification in East Asian populations.
For population genetic analyses of the Guizhou Dong group, we found that the studied Dong group showed low DA and FST values with East Asian populations. More importantly, the Guizhou Dong group showed similar ancestral proportions with East Asian populations, implying that they possessed close genetic affinities in comparison to other reference populations. In addition, we observed that the Guizhou Dong group displayed the closest genetic relationships with CDX, KHV, and some southern Han populations, which was similar to results obtained by 30 InDels (Liu et al., 2020). Previous studies of Dong group based on X-STRs and the mitochondrial DNA control region also pointed out that Guizhou Dong group showed relatively intimate genetic affinities with Tai-Kadai-speaking, Hmong-Mien-speaking, and Han populations (Yang et al., 2021; Ren et al., 2022). Dong and CDX groups belong to the same language family, Tai-Kadai, which may result in more interaction between these two populations. Besides, population genetic analyses of the Kinh population also revealed that the Kinh population possessed close genetic affinities with the Dai population based on a large number of SNPs (Huang et al., 2018), which might be related to their similar genetic structure. From the above results, we stated that the studied Dong group had close genetic relationships with Southern Chinese minority groups and Han populations. As more data on these loci in Southern Chinese populations is being reported, we can explore phylogenetic relationships of the Guizhou Dong and its surrounding populations better.
To sum up, the 64-plex panel showed relatively high genetic diversity and could be utilized as the independent system for forensic individual identification and paternity testing in the Guizhou Dong group. In addition, some loci exhibiting high genetic differentiations among different continental populations could be viewed as candidate ancestry informative markers in the future. Population genetic analyses of the Guizhou Dong and other reference populations demonstrated that the studied Dong group displayed intimate genetic affinities with CDX, KHV, and some Southern Chinese Han populations.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The study was conducted in line with the guideline of the Ethics Committee of Guizhou Medical University and was warranted by the Ethics Committee of Guizhou Medical University (Approval number: 2021-224). The patients/participants provided their written informed consent to participate in this study.
ZR and WW wrote the main text. HZ and QW performed experiment and collected samples. TW, YY, JY, and KH conducted statistical analyses. WW revised the manuscript for important intellectual content. JH and XJ provided the conception and designed this research. All authors contributed to the article and approved the submitted version.
This study was supported by the Guizhou Provincial Science and Technology Projects (ZK[2022] General 355); Guizhou Education Department Young Scientific and Technical Talents Project, Qian Education KY NO. [2022]215; Guizhou Scientific Support Project, Qian Science Support [2021] General 448; Shanghai Key Lab of Forensic Medicine, Key Lab of Forensic Science, Ministry of Justice, China (Academy of Forensic Science), Open Project, KF202207; Guizhou Province Education Department, Characteristic Region Project, Qian Education KY No. [2021]065; Guizhou “Hundred” High-level Innovative Talent Project, Qian Science Platform Talents [2020]6012; Guizhou Scientific Support Project, Qian Science Support [2020] 4Y057; Guizhou Science Project, Qian Science Foundation [2020] 1Y353; Guizhou Scientific Support Project, Qian Science Support [2019] 2825; Guizhou Scientific Cultivation Project, Qian Science Platform Talent [2018] 5779-X; Guizhou Engineering Technology Research Center Project, Qian High-Tech of Development and Reform Commission no. [2016] 1345; Guizhou Innovation Training Program for College Students [2019]5200926; National Natural Science Foundation of China (no. 82160324).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.988504/full#supplementary-material
1. ^http://vip.sangerbox.com/login.html
2. ^https://mybiosoftware.com/dispan-genetic-distance-phylogenetic-analysis.html
1000 Genomes Project ConsortiumAuton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi: 10.1038/nature15393
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Avent, I., Kinnane, A. G., Jones, N., Petermann, I., Daniel, R., Gahan, M. E., et al. (2019). The QIAGEN 140-locus single-nucleotide polymorphism (SNP) panel for forensic identification using massively parallel sequencing (MPS): an evaluation and a direct-to-PCR trial. Int. J. Legal Med. 133, 677–688. doi: 10.1007/s00414-018-1975-5
Boelens, D., Fogliatto Mariot, R., Ghemrawi, M., Kloosterman, A. D., and McCord, B. R. (2021). The development of miniSTRs as a method for high-speed direct PCR. Electrophoresis 42, 1352–1361. doi: 10.1002/elps.202100066
Chandra, D., Mishra, V. C., Raina, A., and Raina, V. (2022). Mutation rate evaluation at 21 autosomal STR loci: paternity testing experience. Legal Med. 58:102080. doi: 10.1016/j.legalmed.2022.102080
Chen, L., Du, W., Wu, W., Yu, A., Pan, X., Feng, P., et al. (2019). Developmental validation of a novel six-dye typing system with 47 A-InDels and 2 Y-InDels. Forensic Sci. Int. Genet. 40, 64–73. doi: 10.1016/j.fsigen.2019.02.009
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fan, H., He, Y., Li, S., Xie, Q., Wang, F., Du, Z., et al. (2022). Systematic evaluation of a novel 6-dye direct and multiplex PCR-CE-based InDel typing system for forensic purposes. Front. Genet. 12:744645. doi: 10.3389/fgene.2021.744645
Fan, G. Y., Wang, D. P., Song, D. L., Zheng, X. K., Zhu, J., and Long, B. (2021). Developmental validation study of a 32-plex STR direct amplification system for forensic reference samples. Forensic Sci. Int. 327:110977. doi: 10.1016/j.forsciint.2021.110977
Feng, Q., Lu, D., and Xu, S. (2018). AncestryPainter: a graphic program for displaying ancestry composition of populations and individuals. Genom. Proteom. Bioinform. 16, 382–385. doi: 10.1016/j.gpb.2018.05.002
Gouy, A., and Zieger, M. (2017). STRAF—A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci. Int. Genet. 30, 148–151. doi: 10.1016/j.fsigen.2017.07.007
Guo, F. (2017). Genetic variation of 17 autosomal STR loci in the Dong ethnic minority from Guangxi Zhuang autonomous region, South China. Int. J. Legal Med. 131, 1537–1538. doi: 10.1007/s00414-017-1576-8
Harrel, M., Mayes, C., Houston, R., Holmes, A. S., Gutierrez, R., and Hughes, S. (2021). The performance of quality controls in the investigator® Quantiplex® pro RGQ and investigator® 24plex STR kits with a variety of forensic samples. Forensic Sci. Int. Genet. 55:102586. doi: 10.1016/j.fsigen.2021.102586
Huang, Y., Liu, C., Xiao, C., Chen, X., Yi, S., and Huang, D. (2020). Development of a new 32-plex InDels panel for forensic purpose. Forensic Sci. Int. Genet. 44:102171. doi: 10.1016/j.fsigen.2019.102171
Huang, X., Zhou, Q., Bin, X., Lai, S., Lin, C., Hu, R., et al. (2018). The genetic assimilation in language borrowing inferred from Jing people. Am. J. Phys. Anthropol. 166, 638–648. doi: 10.1002/ajpa.23449
Hwa, H. L., Wu, M. Y., Lin, C. P., Hsieh, W. H., Yin, H. I., Lee, T. T., et al. (2019). A single nucleotide polymorphism panel for individual identification and ancestry assignment in Caucasians and four east and southeast Asian populations using a machine learning classifier. Forensic Sci. Med. Pathol. 15, 67–74. doi: 10.1007/s12024-018-0071-y
Jin, R., Cui, W., Fang, Y., Jin, X., Wang, H., Lan, Q., et al. (2021). A novel panel of 43 insertion/deletion loci for human identifications of forensic degraded DNA samples: development and validation. Front. Genet. 12:610540. doi: 10.3389/fgene.2021.610540
Jin, X. Y., Wei, Y. Y., Cui, W., Chen, C., Guo, Y. X., Zhang, W. Q., et al. (2019). Development of a novel multiplex polymerase chain reaction system for forensic individual identification using insertion/deletion polymorphisms. Electrophoresis 40, 1691–1698. doi: 10.1002/elps.201800412
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
LaRue, B. L., Ge, J., King, J. L., and Budowle, B. (2012). A validation study of the Qiagen investigator DIPplex® kit; an INDEL-based assay for human identification. Int. J. Legal Med. 126, 533–540. doi: 10.1007/s00414-012-0667-9
Li, Y. N., Li, M., Jiang, L., Luan, X. H., Liang, N., Xu, Q. N., et al. (2018). Establishment of 43-plex SNP typing system and its forensic application. J. Forensic Med. 34, 126–131. doi: 10.3969/j.issn.1004-5619.2018.02.004
Li, C., Zhao, S., Zhang, S., Li, L., Liu, Y., Chen, J., et al. (2011). Genetic polymorphism of 29 highly informative InDel markers for forensic use in the Chinese Han population. Forensic Sci. Int. Genet. 5, e27–e30. doi: 10.1016/j.fsigen.2010.03.004
Liu, Y., Cui, W., Jin, X., Wang, K., Mei, S., Zheng, X., et al. (2022a). Forensic efficiency estimation of a homemade six-color fluorescence multiplex panel and in-depth anatomy of the population genetic architecture in two Tibetan groups. Front. Genet. 13, 1–11. doi: 10.3389/fgene.2022.880346
Liu, J., Du, W., Jiang, L., Liu, C., Chen, L., Zheng, Y., et al. (2022). Development and validation of a forensic multiplex InDel assay: the AGCU InDel 60 kit. Electrophoresis 43, 1871–1881. doi: 10.1002/elps.202100376
Liu, Y., Mei, S., Jin, X., Zhao, M., and Zhu, B. (2022b). Independent development and validation of a novel six-color fluorescence multiplex panel including 61 diallelic DIPs and 2 miniSTRs for forensic degradation sample. Electrophoresis 43, 1423–1437. doi: 10.1002/elps.202100225
Liu, Z. Y., Ren, H., Chen, C., Zhang, J. J., Zhang, X. M., Shi, Y., et al. (2021). Actual mutational research of 19 autosomal STRs based on restricted mutation model and big data. Yi Chuan. 43, 949–961. doi: 10.16288/j.yczz.21-197
Liu, Y., Zhang, H., He, G., Ren, Z., Zhang, H., Wang, Q., et al. (2020). Forensic features and population genetic structure of Dong, Yi, Han, and Chuanqing human populations in Southwest China inferred from insertion/deletion markers. Front. Genet. 11:360. doi: 10.3389/fgene.2020.00360
Phillips, C. (2015). Forensic genetic analysis of bio-geographical ancestry. Forensic Sci. Int. Genet. 18, 49–65. doi: 10.1016/j.fsigen.2015.05.012
Qu, Y., Tao, R. Y., Yu, H., Yang, Q., Wang, Z., Tan, R., et al. (2021). Development and validation of a forensic six-dye multiplex assay with 29 STR loci. Electrophoresis 42, 1419–1430. doi: 10.1002/elps.202100019
Ren, Z., Feng, Y., Zhang, H., Wang, Q., Yang, M., Liu, Y., et al. (2022). Genetic analysis of the mitochondrial DNA control region in tai-Kadai-speaking Dong population in Southwest China. Ann. Hum. Biol. 1–7. doi: 10.1080/03014460.2022.2131334
Rosenberg, N. A., Li, L. M., Ward, R., and Pritchard, J. K. (2003). Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 73, 1402–1422. doi: 10.1086/380416
Song, W., Xiao, N., Zhou, S., Yu, W., Wang, N., Shao, L., et al. (2022). Non-invasive prenatal paternity testing by analysis of Y-chromosome mini-STR haplotype using next-generation sequencing. PLoS One 17:e0266332. doi: 10.1371/journal.pone.0266332
Tao, R., Zhang, J., Sheng, X., Zhang, J., Yang, Z., Chen, C., et al. (2019). Development and validation of a multiplex insertion/deletion marker panel, SifaInDel 45plex system. Forensic Sci. Int. Genet. 41, 128–136. doi: 10.1016/j.fsigen.2019.04.008
Weber, J. L., David, D., Heil, J., Fan, Y., Zhao, C., and Marth, G. (2002). Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 71, 854–862. doi: 10.1086/342727
Wei, Y. L., Qin, C. J., Dong, H., Jia, J., and Li, C. X. (2014). A validation study of a multiplex INDEL assay for forensic use in four Chinese populations. Forensic Sci. Int. Genet. 9, e22–e25. doi: 10.1016/j.fsigen.2013.09.002
Yang, M., Jin, X., Ren, Z., Wang, Q., Zhang, H., Zhang, H., et al. (2021). X-chromosomal STRs for genetic composition analysis of Guizhou Dong group and its phylogenetic relationships with other reference populations. Ann. Hum. Biol. 48, 621–626. doi: 10.1080/03014460.2021.2008001
Zhang, L. (2015). Population data for 15 autosomal STR loci in the Dong ethnic minority from Guizhou Province, Southwest China. Forensic Sci. Int. Genet. 16, 237–238. doi: 10.1016/j.fsigen.2015.02.005
Zhang, X., Shen, C., Jin, X., Guo, Y., Xie, T., and Zhu, B. (2021). Developmental validations of a self-developed 39 AIM-InDel panel and its forensic efficiency evaluations in the Shaanxi Han population. Int. J. Legal Med. 135, 1359–1367. doi: 10.1007/s00414-021-02600-4
Keywords: InDels, Dong, STR, forensic application, population genetic background
Citation: Wan W, Ren Z, Zhang H, Wang Q, Wang T, Yang Y, You J, He K, Huang J and Jin X (2023) Insight into forensic efficiency and genetic structure of the Guizhou Dong group via a 64-plex panel. Front. Ecol. Evol. 10:988504. doi: 10.3389/fevo.2022.988504
Received: 07 July 2022; Accepted: 21 December 2022;
Published: 11 January 2023.
Edited by:
Alison G. Nazareno, Federal University of Minas Gerais, BrazilCopyright © 2023 Wan, Ren, Zhang, Wang, Wang, Yang, You, He, Huang and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Huang,  bW1tX2hqQDEyNi5jb20=; Xiaoye Jin,
bW1tX2hqQDEyNi5jb20=; Xiaoye Jin,  MTExNTI1OTgyNUBxcS5jb20=
MTExNTI1OTgyNUBxcS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.