
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 07 November 2022
Sec. Paleoecology
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.980753
This article is part of the Research TopicIntegrating Conservation Biology and Paleobiology to Manage Biodiversity and Ecosystems in a Changing WorldView all 22 articles
To integrate paleoecological data with the “whole fauna” data used in biological monitoring, analyses usually must focus on the subset of taxa that are inherently preservable, for example by virtue of biomineralized hardparts, and those skeletal remains must also be identifiable in fragmentary or otherwise imperfect condition, thus perhaps coarsening analytical resolution to the genus or family level. Here we evaluate the ability of readily preserved bivalves to reflect patterns of compositional variation from the entire infaunal macroinvertebrate fauna as typically sampled by agencies in ocean monitoring, using data from ten long-established subtidal stations in Puget Sound, Washington State. Similarity in compositional variation among these stations was assessed for five taxonomic subsets (the whole fauna, polychaetes, malacostracans, living bivalves, dead bivalves) at four levels of taxonomic resolution (species, genera, families, orders) evaluated under four numerical transformations of the original count data (proportional abundance, square root- and fourth root-transformation, presence-absence). Using the original matrix of species-level proportional abundances of the whole fauna as a benchmark of “compositional variation,” we find that living and dead bivalves had nearly identical potential to serve as surrogates of the whole fauna; they were further offset from the whole fauna than was the polychaete subset (which dominates the whole fauna), but were far superior as surrogates than malacostracans. Genus- and family-level data were consistently strong surrogates of species-level data for most taxonomic subsets, and correlations declined for all subsets with increasing severity of data transformation, although this effect lessened for subsets with high community evenness. The strong congruence of death assemblages with living bivalves, which are themselves effective surrogates of compositional variation in the whole fauna, is encouraging for using bivalve dead-shell assemblages to complement conventional monitoring data, notwithstanding strong natural environmental gradients with potential to bias shell preservation.
Normalizing the application of paleoecological and other geohistorical data to the direct observations of living systems central to conservation biology is an important, oft-cited objective of conservation paleobiology (Dietl and Flessa, 2011; Kidwell and Tomašových, 2013). Biological monitoring programs are an important component of those observations and require consistent sampling methods, expert taxonomic training, and rigorous quality assurance procedures, especially where regulated by government agencies (e.g., Dutch et al., 2018). The standards for monitoring benthic communities typically include a wide taxonomic breadth (e.g., macroinvertebrate infauna), species-level taxonomic information, and precise numerical estimates of animal density (e.g., raw counts per taxon standardized to benthic area or sediment volume).
Paleoecological data are typically limited in comparability to such monitoring data in several ways. First, death assemblages – i.e., dead and discarded remains encountered during sampling for living individuals – and fully-buried fossil assemblages are almost always restricted to taxa that possess durable hardparts, such as calcifying mollusks, arthropods, and bryozoans. Under oxygenated waters, soft-bodied clades have low preservation potential, an effect that has rarely been fully quantified [but see the seminal analyses by Schopf (1978), Staff et al. (1986)]. Second, owing to fragmentation and other damage accrued during accumulation in the surface mixed layer, not all dead shell specimens are identifiable to the species level, requiring analytic coarsening to, for example, genus or family level (e.g., Kowalewski et al., 2003; Lloyd et al., 2012; Albano, 2014). Finally, some museum and sedimentary archives might only yield presence-absence (occurrence) data for species, in contrast to the numerical counts generated by bulk sampling, spurring efforts by paleontologists to upgrade data post hoc (e.g., Harnik, 2009; Close et al., 2018). Successful integration of paleoecological data with modern biological data thus requires confidence that assemblages of skeletal remains alone can serve as reliable proxies of the broader fauna targeted for study – usually the entire macrobenthos or at least its infaunal portion – and yield reliable versions of the biological metric of interest (e.g., richness, compositional variation, trophic structure).
Fortunately, the challenges that these three limitations place on the reliability of paleoecological data are directly analogous to well-studied issues of “surrogacy” in the biomonitoring literature (Moreno et al., 2007). This term encompasses an array of techniques aimed to reduce the cost (time, labor) and/or expertise required to process biological samples while still attaining adequate biological insights. That is, referring to our challenges above, acquiring an accurate or sufficient picture when only a subset of clades can be sampled, when species-level resolution might not be possible, and/or when numerical abundance is not necessarily trustworthy. Surrogacy falls broadly into two categories (Table 1). “Sufficiency” refers to coarsening the detail of information collected, either via “taxonomic sufficiency” sensu (Ellis, 1985; aggregating species-level data into higher taxonomic ranks, functional guilds, etc.) or via “numerical sufficiency” (simplifying organismal counts into ranked abundances, categorical abundances, or simply presence-absence data). “Subsetting” on the other hand refers to narrowing the scope of information, for example by focusing on a “taxonomic subset” (a singular clade, functional guild, or other group of interest) or on a “numerical subset” of the whole fauna (e.g., the first 100 individuals picked from the sample, as is common in micropaleontology). Other forms of surrogacy include “cross-taxon” approaches in which one group of taxa serves as a proxy for another, completely independent group (Mellin et al., 2011; Gladstone et al., 2020). A death assemblage, which is not part of the living fauna, could be considered a cross-taxon surrogate that is also a temporally coarser sample than the corresponding living assemblage. Surrogacy methods have been the subject of many tests for macroinvertebrate communities from marine (Warwick, 1988; Ferraro and Cole, 1992; Włodarska-Kowalczuk and Kêdra, 2007; Bevilacqua et al., 2009), freshwater (Jones, 2008; Mueller et al., 2013; Heino, 2014), and terrestrial settings (Pik et al., 1999; Timms et al., 2013; Souza et al., 2016). Several studies have examined the sensitivity of surrogacy techniques on compositional variation in the fossil record (e.g., Pandolfi, 2001; Forcino et al., 2012; Zuschin et al., 2017), on the faithfulness of dead-shell assemblages to their living counterparts (e.g., Albano et al., 2016), and on the preservation of broader macrofaunal patterns by the preservable subset (Tyler and Kowalewski, 2017).
Here, we evaluate the individual and interacting effects of three surrogacy methods – taxonomic sufficiency, taxonomic subsetting, and numerical sufficiency (Table 1) – as well as the effects of data transformation, using an infaunal macrobenthic dataset from ten sediment monitoring stations in Puget Sound, Washington State. Spanning 25 classes and 15 phyla, the numerical, species-level data were generated as part of a regional, state-mandated monitoring program of sediments, conducted under strict standards for processing and taxonomic consistency (Dutch et al., 2018). Bivalve death assemblages were rescued from the same sediment grab samples processed for living assemblages, permitting us to directly test the comparability of the preservable subset of the fauna with the whole fauna, including the live-dead agreement of the bivalve subset of that fauna. Long-term biomonitoring has demonstrated that these ten subtidal sites have been remarkably stable in community composition on this regional scale: scatter in ordination space produced by temporal variation over the last three decades is small relative to the separation among stations, despite other evidence of overall deteriorating conditions across Puget Sound (Partridge et al., 2018). This tension of compositional stability, and the possibility of using death assemblages for insights into community states before the onset of monitoring, motivated this collaboration: the test of shelly fauna as surrogates of the whole fauna is an important first step both in assessing the reliability of fossil data for local use and developing a protocol for wider application.
Puget Sound is a fjordic estuarine system in Washington State that, along with the Strait of Georgia and Strait of Juan de Fuca, makes up the Salish Sea (Figure 1), and was formed by the southernmost lobe of the Cordilleran Ice Sheet during the Fraser and Vashon glaciations (∼20–15 ka; Booth, 1994). Puget Sound is the second-largest estuary in the United States by area (2,600 km2) after Chesapeake Bay (11,600 km2), but has considerably greater mean and maximum water depths (140 and 280 m, respectively) than Chesapeake Bay (7 and 53 m) and accommodates ∼250% more water by volume. Macrotidal flushing from the Sound through the Strait of Juan de Fuca to the Pacific Ocean facilitates the import of cold bottom-waters that circulate into major basins of the Sound, with the result that sediment-dwelling macrobenthos encounter fully saline, oceanic waters notwithstanding freshwater-influenced surface lenses (Moore et al., 2008). This flushing is complicated, however, by a series of narrow, silled chokepoints between basins that slow the circulation of water considerably toward the terminal ends of the Sound (Khangaonkar et al., 2011; but see MacCready et al., 2021). The numerous coastal inlets and deep basins within the greater Puget Sound area (US waters of the Salish Sea, henceforth “Puget Sound” for brevity) thus provide a complex array of intertidal, shallow subtidal, and deep subtidal habitats in a hydrographic estuary, with diverse sediment facies and naturally variable hydrologic conditions (e.g., dissolved oxygen, salinity). The Sound also reflects a complex pattern of development in adjacent watersheds, which range from rural to suburban, industrial (historically ore, forestry, military), and densely urbanized cities such as Seattle and Tacoma (Figure 1).
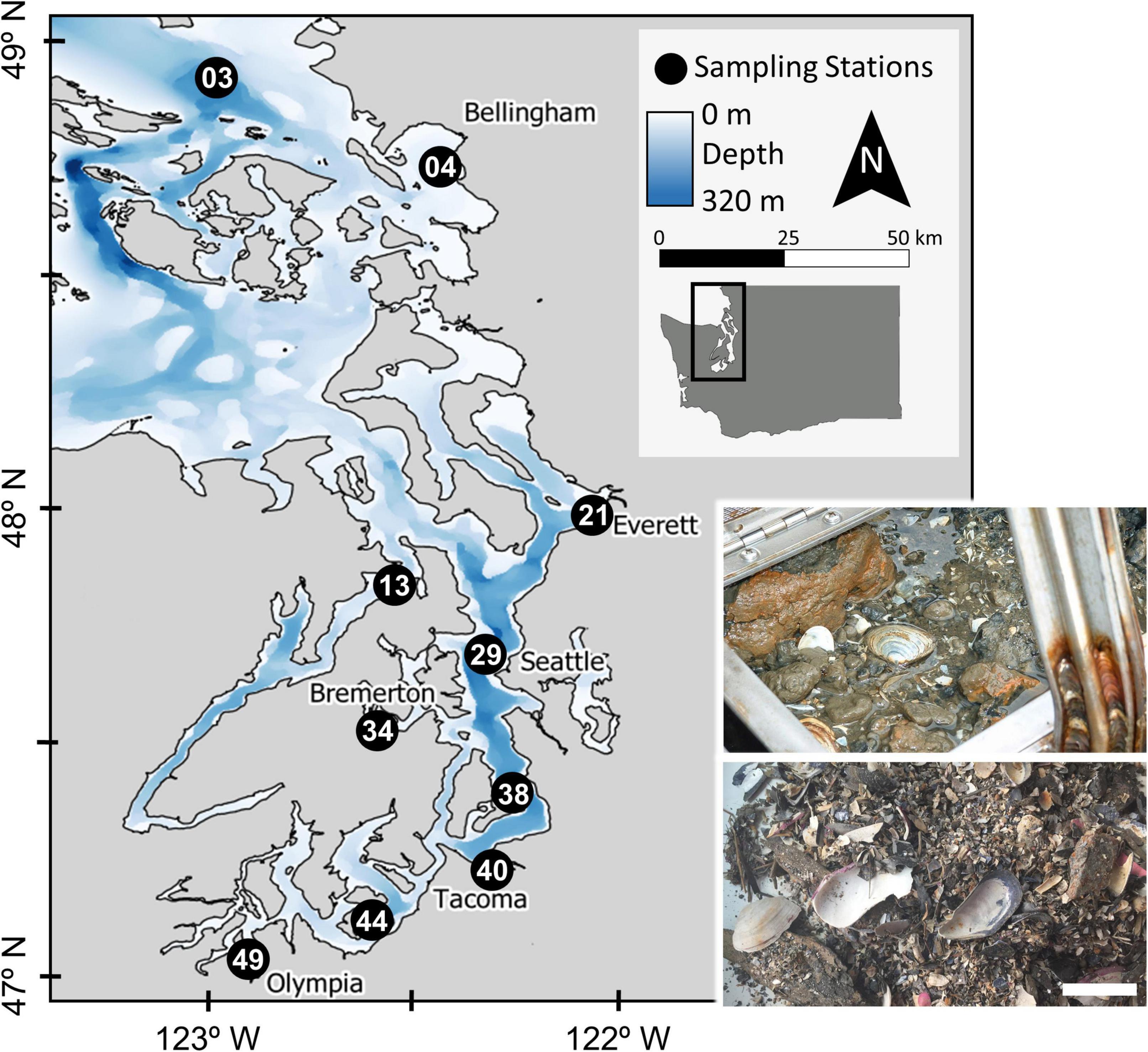
Figure 1. Map of the ten sampling station locations in the greater Puget Sound area, Washington State. Station identification numbers generally increase southwards. Inset images: a van Veen grab with sampled sediment and living organisms (top) and processed residue containing dead bivalve shells (bottom). Photos by ECY and B.S. Kokesh, respectively. Scale bar for dead shell photo = 5 cm.
Benthic macroinvertebrate communities in Puget Sound have been relatively well-documented since the 1960s by a combination of academic (Lie and Evans, 1973), federal (Nichols, 2003), and, over the last 30 years, state agencies (Dutch et al., 2018). Since 1989, ten “sentinel” stations located across Puget Sound have been monitored by the Marine Sediment Monitoring Team at the Washington State Department of Ecology (ECY), focusing on benthic communities, sedimentology, and chemical contaminants as part of the Puget Sound Ecosystem Monitoring Program (PSEMP; Table 2). These ten sentinel stations were selected from a larger set of stations sampled until 1995 because they represent distinct habitats from which distinct benthic communities were observed. PSEMP data reveal long-term declines in benthic condition based on a “sediment quality triad index” that uses community composition (whole fauna), sediment toxicity (using echinoid fertility and amphipod mortality tests), and a suite of chemical contaminants (Long and Chapman, 1985). However, causal mechanisms for this decline have been difficult to identify (Partridge et al., 2018; Weakland et al., 2018). The PSEMP protocol has thus recently been revised to assess a broader suite of variables including macroinvertebrate biomass, nutrients, and stable isotopes to investigate additional possible stressors (Dutch et al., 2018). The overall decline in Puget Sound benthic quality detected by PSEMP since 1989 involved increased toxicity despite no change in contaminant levels, and significant declines in richness and total invertebrate abundance in several areas within the large network of sites (Partridge et al., 2018). However, the annually-sampled set of ten sentinel sites used here exhibited relative stability over this period, despite strong fluctuations in different taxa at different sites: habitat-level differences in sediment grain size and water depth are the strongest correlates of community composition, with date of sampling a secondary and much smaller effect (Figure 7 in Partridge et al., 2018).
The dataset of living assemblage composition used here comes from ECY samples acquired in the years 2017 and 2018, aggregated to achieve adequate sample sizes, at each of ten, long-established monitoring stations in Puget Sound (Figure 1), using the entire living macroinvertebrate community (henceforth “whole fauna”). Sampling followed the Puget Sound Estuary Protocol (Puget Sound Estuary Program [PSEP], 1987), which is standard among many agencies along the US Pacific Coast. Samples were collected using a 0.1-m2 van Veen grab with a minimum sediment penetration of 5 cm (Puget Sound Estuary Program [PSEP], 1987). Sediments were sieved through 1-mm mesh on deck with seawater, fixed in 10% formalin, and then exchanged into 70% ethanol within 3–7 days. ECY taxonomists sorted and identified all invertebrates to the species level, when possible, with reference to a library of voucher specimens; coordination with taxonomists at other agencies in the Pacific US (SCAMIT, 2021) has also contributed to taxonomic consistency.
Dead bivalves were sorted from the dried sediment residue of two years of benthic samples at the same sites collected in the years 2018 and 2019: the death assemblage had not been saved from the 2017 samples and the living whole fauna from 2019 was not yet available. This sediment residue is typically discarded by agencies after living macroinvertebrates have been removed. All bivalve shells with an identifiable hinge were counted as individuals, whether the valve itself was whole or fragmented, and were identified taxonomically to the finest resolution possible. Each fully disarticulated valve or hinge fragment was considered as coming from a different living individual, following the reasoning of Gilinsky and Bennington (1994) and standard protocol for live-dead analysis of mollusks of Kidwell (2009): no correction for the number of skeletal units per living individual (e.g., Kowalewski and Hoffmeister, 2003) was needed because only bivalves were considered. However, to avoid any implicit equating of the population-density of living bivalves with time-averaged abundance data, we used proportional abundances rather than raw counts in all analyses unless otherwise indicated.
From the original whole-fauna dataset, new species-level abundance tables were extracted analytically for polychaetes, malacostracans, and bivalves. A fifth table was produced using species-level abundance data from the rescued dead-shell assemblages. Species from each of these five tables were then coarsened to create abundance tables for genera, families, and orders. Each abundance table was then subjected to four different data transformations that successively reduced the influence of dominant taxa and increased the influence of rare taxa: non-transformed proportional abundances, square-root transformed, fourth-root transformed (the preferred transformation adopted by ECY and widely used in surrogacy studies; Partridge et al., 2018), and binary presence-absence. While the first three of these transformations preserve the abundance structure of the data, presence-absence represents a true test of numerical sufficiency as it does not rely on knowledge of the underlying count data. We note that other transformations commonly applied to ecological data, such as log and arcsine, could also have been tested. Our focus on the series x1, x1/2, x1/4, and x0 instead allowed us to measure the effects of progressively severe transformations – increasing deviation from the original abundance structure – using the same mathematical operator. A log transformation should moreover not be judged a priori as more or less severe than an n-root transformation (although log(x + 1) has been argued to have equivalent effects on the similarities between samples as x1/4; Clarke and Warwick, 2001), whereas we can be confident that fourth-root transformed data will always be less correlated to non-transformed data than will be square-root transformed data. Additional transformations that would qualify as methods of numerical sufficiency, such as semi-quantitative ordinal abundances (e.g., common, rare), were not assessed because the number of ordinal categories and their abundance ranges are largely at the discretion of the investigator.
Distance matrices for each of these surrogate abundance tables were generated using Bray–Curtis dissimilarities (Jaccard in the case of presence-absence) among the ten monitoring stations. This work produced 80 distance matrices (5 subsets × 4 resolutions × 4 transformations), with 79 of these serving as “surrogates” of the original benchmark matrix (proportional abundances at the species level for the whole fauna).
Richness (the raw count of taxa), abundance (raw number of individuals), and evenness (abundance distribution among taxa) for each surrogate dataset were calculated both at the station scale (i.e., at each of the 10 stations) and at the regional scale, that is, after aggregating station-level abundances together. Richness values were converted into the ratio of higher taxa over the number of species (T/S) for genera (G/S), families (F/S), and orders (O/S). T/S values range from >0 to 1, with 1 indicating that all members of the higher taxon are monospecific. Abundance values were converted into the ratio of the abundance of individuals in the subset over the abundance in the whole fauna (N/W). N/W values range from >0 to 1; a value of 0.5 would indicate that the subset represents 50% of all individuals in the whole fauna. We expressed evenness as the probability of interspecific encounter (PIE; Hurlbert, 1971), which is relatively insensitive to sample size and richness > ∼5 (Olszewski, 2004). PIE values have a potential range from 0 to 1, with 1 indicating a perfectly even community (same number of individuals per taxon) in which individuals of any species are equally likely to be randomly sampled. T/S, N/W, and PIE were compared among surrogate datasets using the regional-scale values and the medians and interquartile ranges (IQR) of station-scale values.
Spearman rank correlation coefficients (ρ) between all pairwise combinations of Bray–Curtis distance matrices were calculated and tested for significance using Mantel tests with 999 permutations (Mantel, 1967). Coefficients can range from -1 to +1. A pairwise correlation of + 1 would indicate that the Bray–Curtis distances among stations for two surrogate datasets are in perfect rank order: put another way, the two surrogates would produce identical non-metric multidimensional scaling ordinations (NMDS). We assessed the effects of each surrogacy method (see Table 1) on the perception of compositional variation among sites by evaluating ρ values varied by treatments among the full set of pairwise comparisons. First, taxonomic sufficiency was evaluated using the ρ values generated by comparing matrices of species-level data with those for higher taxa while holding both the subset and data transformation level constant. Second, taxonomic subsetting was evaluated by using the ρ values generated by comparing the whole fauna and each surrogate subset while holding both the taxonomic resolution and data transformation level constant. Third, the effects of data transformations (and numerical sufficiency in the case of presence-absence data) were evaluated using the ρ values generated by comparing matrices based on proportional abundances with those based on more severe data transformations while holding both taxonomic resolution and subset constant. Finally, the composite effect of all three surrogacy methods was evaluated using ρ values generated by comparing the original dataset (species-level whole fauna using proportional abundances) to all surrogate matrices.
In a procedure known as a second-stage analysis introduced by Somerfield and Clarke (1995), pairwise ρ values were themselves compiled into distance matrices by rescaling ρ such that Dρ = (1 – ρ)/2, where Dρ is the distance between two surrogates (see Figure 2 for schematic diagram and Supplementary material for example code). A coefficient of perfect correlation (ρ = +1) thus corresponds to a Dρ = 0. The new distance matrices were then used to generate “second-stage” NMDS. Each point in this ordination represents an entire surrogate’s distance matrix, not the positions of individual samples that are plotted by first-stage, “conventional” ordinations. The more closely that two points plot to each other, the higher their pairwise ρ value relative to that with other surrogates. Second-stage NMDS was performed separately for each of the five taxonomic subsets (16 matrices each subset, i.e., 4 resolutions × 4 data transformations) and then re-run to combine all 80 surrogates simultaneously. Differences between the multivariate means of all surrogates grouped by taxonomic resolution, subset, and data transformation were tested using 1-way analysis of similarity (ANOSIM; Clarke, 1993) based on the ranked distances of the matrix used to generate the combined second-stage NMDS. To test whether patterns observed from the combined second-stage NMDS were consistent across the 30-year time series of available community data, we repeated analyses for each 2-year aggregation of the living community calculating the correlation of each of those resulting second-stage distance matrices to the same, most-recent living community (2017–2018).
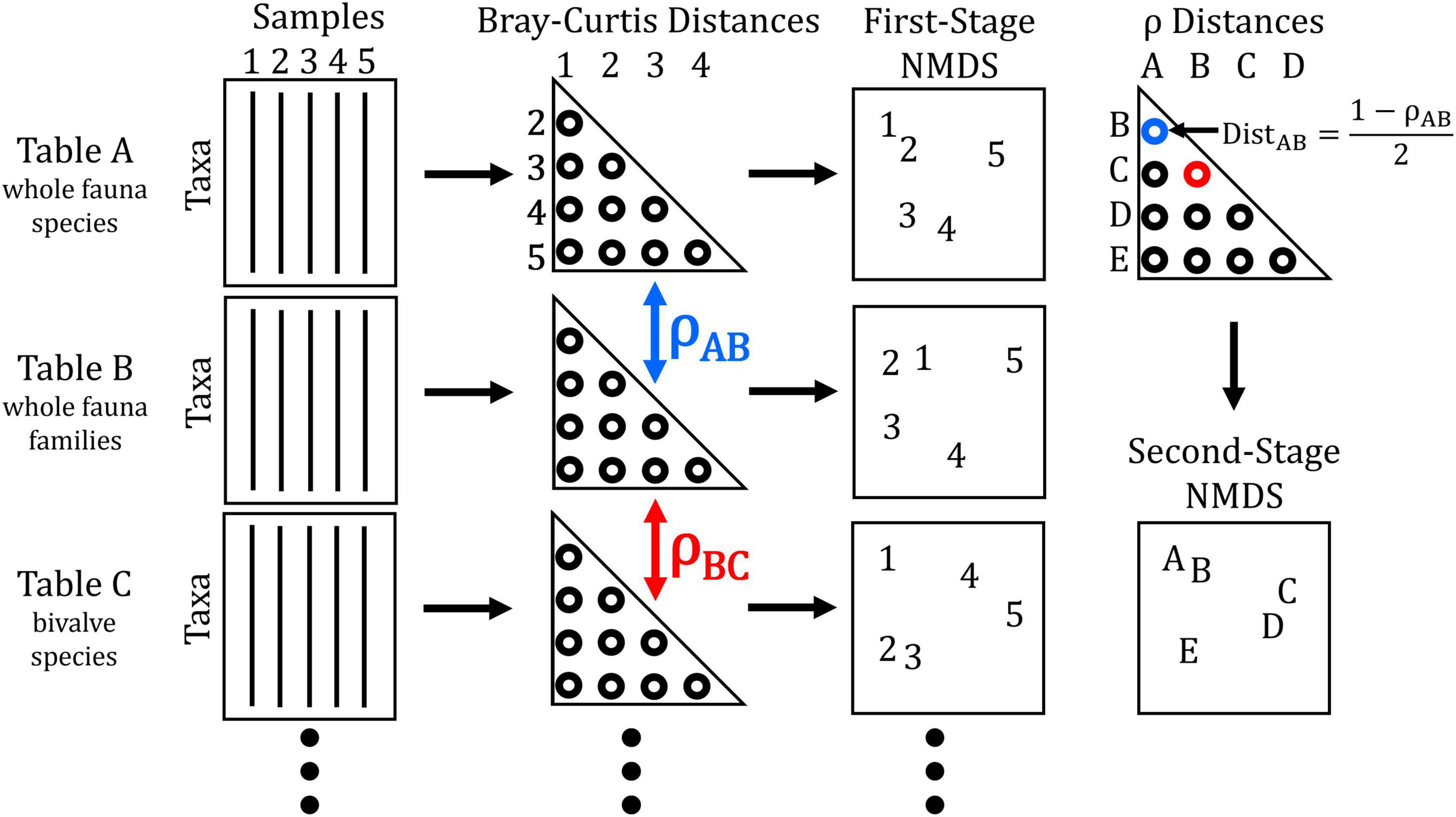
Figure 2. Schematic diagram of the procedure used to quantify inter-matrix correlations and produce second-stage NMDS among surrogate matrices. Starting with data tables for the same set of samples based on different combinations of taxa and numerical transformation, Bray–Curtis distance matrices are generated. These matrices can then be used to directly compute first-order ordinations (where points represent each sample, site, etc.) as typically performed. Correlation coefficients (ρ) calculated between each pair of distance matrices are computed into a distance between matrices (e.g., ρAB in blue is converted to DistAB). These distances are then assembled into a matrix used to compute second-stage ordination (where points represent different surrogate matrices). In the hypothetical example above, the locations of samples 1–5 in the first-stage NMDS plots for data tables A and B are more similar to each other than in C. Thus, the location of matrix C is strongly separated from matrices A and B in the second-stage NMDS plot. After Somerfield and Clarke (1995).
All analyses were conducted in the statistical environment R version 4.2.0 (R Core Team, 2022).
The whole macrobenthic fauna acquired during the two annual samplings of 2017 and 2018 comprised 22,713 individual organisms drawn from 319 species, 240 genera, 146 families, 74 orders, 25 classes, and 15 phyla (Table 3). Polychaetes were the most abundant (4,817 individuals, 43% of the whole fauna) and diverse class (157 species, 49% of the whole fauna). Malacostracans were second in diversity (61 species, 19% of the whole fauna), but third in abundance (433 individuals, 4% of the whole fauna). Bivalves were third in diversity (33 species, 10% of the whole fauna), but second in abundance (4,367 individuals, 39% of the whole fauna). Dead bivalves acquired from annual samples in 2018 and 2019 comprised 5,337 individuals and 43 species.
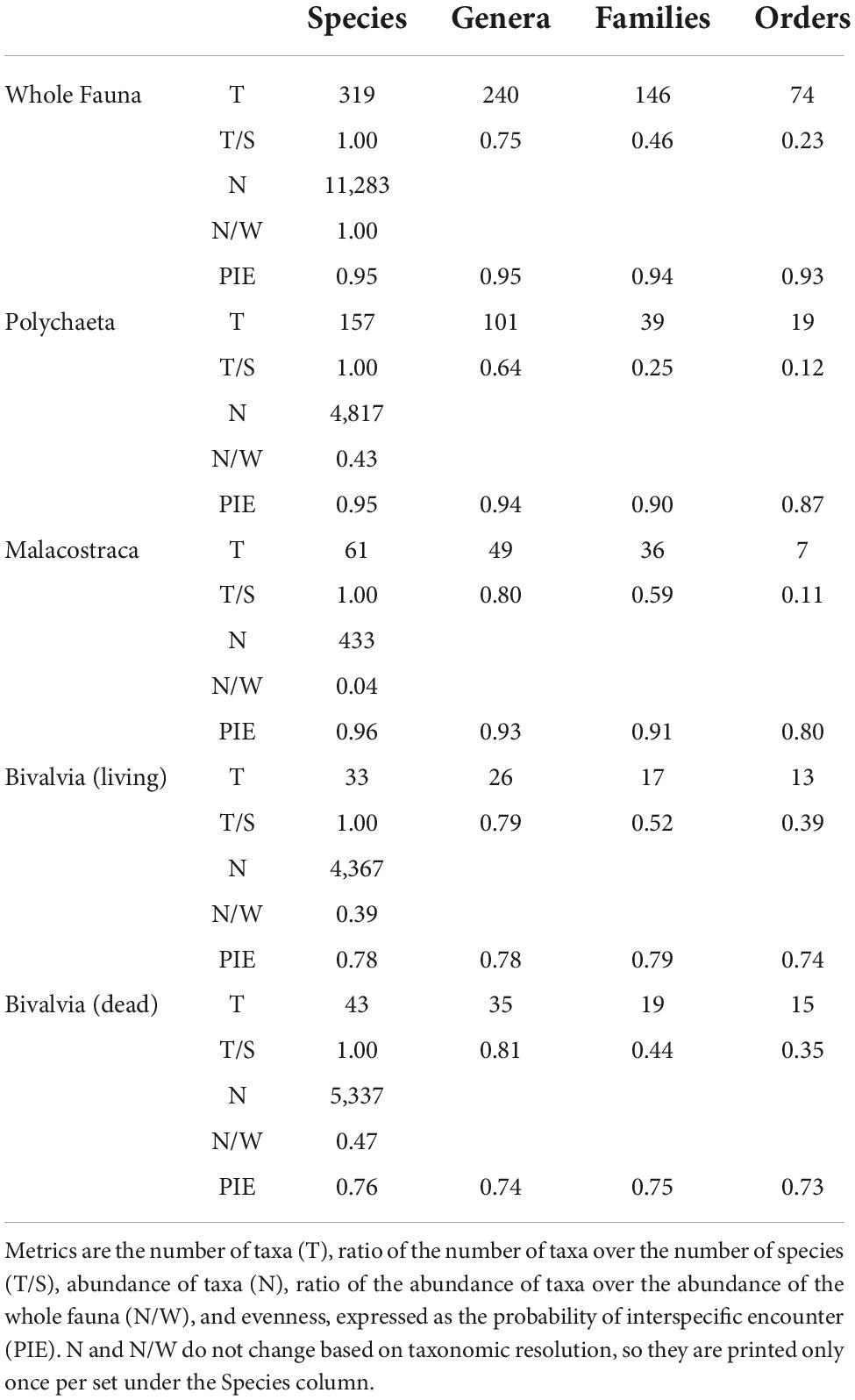
Table 3. Summary of regional-scale macrobenthic community measures divided by taxonomic sets and resolutions.
T/S ratios predictably and significantly declined from genera to orders based on visual separation of IQRs (top row, Figure 3). Trends in T/S for the whole fauna and polychaetes steadily declined to ratios of 0.23 and 0.12, respectively. Malacostracans had a notable decline between families (0.59) and orders (0.11). Living and dead bivalves had similar station- and regional-scale patterns and declined less strongly than did other subsets and the whole fauna. Polychaetes, living bivalves, and dead bivalves had similar regional-scale N/W ratios (middle row, Figure 3). Note that N/W ratios do not change at different taxonomic resolutions, because the same number of individuals are used regardless of the resolution assessed. PIE values at the station- and regional-scale were higher and less variable for the whole fauna and polychaetes compared to malacostracans and bivalves and exhibited scant differences among taxonomic resolutions (bottom row, Figure 3). Regional-scale PIE values for malacostracans were high from the species to family levels (>0.9), but declined for orders (0.8). Living and dead bivalves had the lowest PIE values at the regional-scale (<0.8), and exhibited no significant differences among taxonomic resolutions.
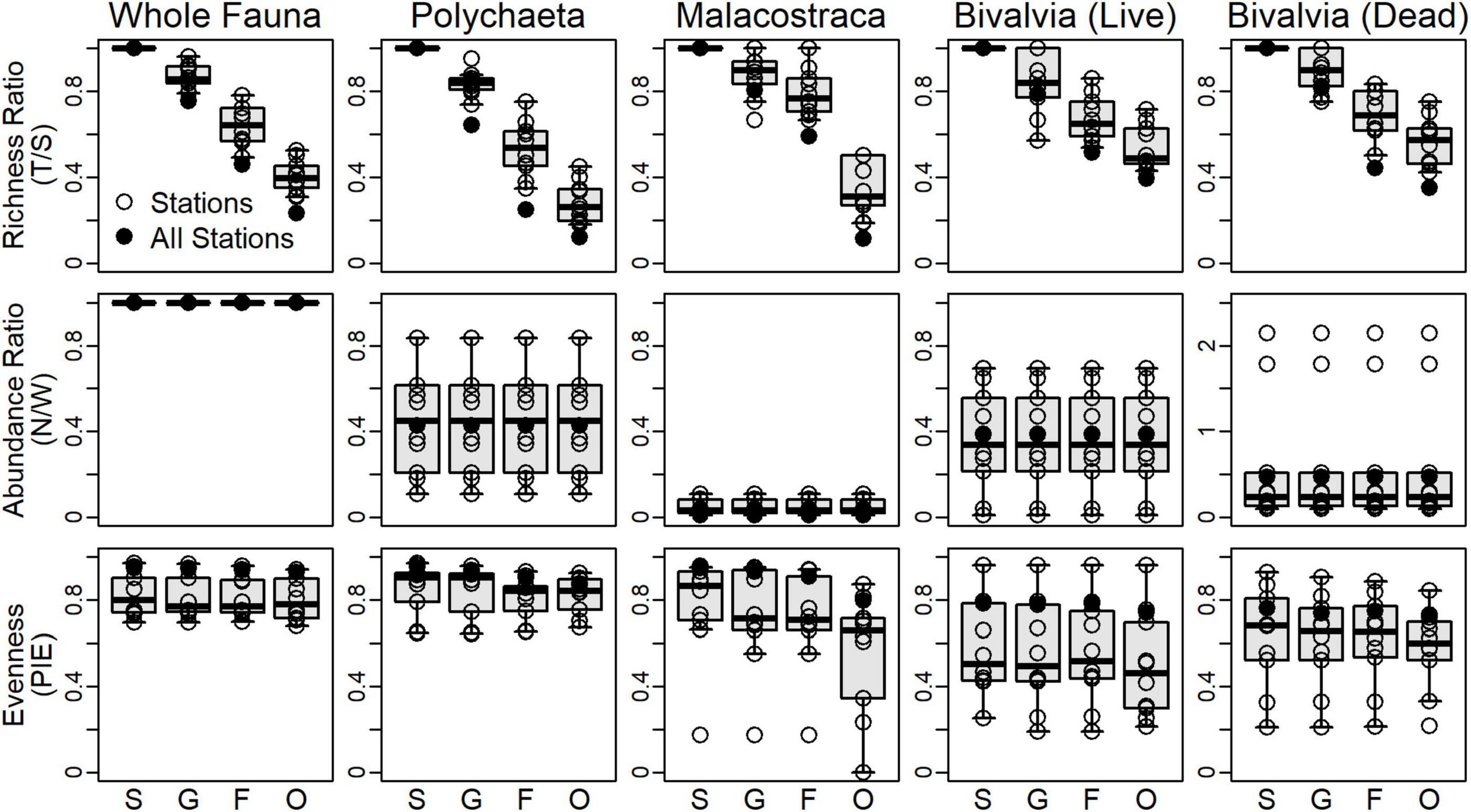
Figure 3. Univariate community metrics measured for surrogate assemblages at each station. Top row: richness ratios as the number of supraspecific taxa to the number of species (T/S). Middle row: abundance ratios as the number of individuals for each set over the number of individuals from the living whole fauna (N/W). Note the different vertical scale for dead bivalves. Bottom row: evenness as the probability of interspecific encounter (PIE) using proportional abundance data. Black circles are values calculated when data from all stations are aggregated. Boxplot whiskers extend to 5th and 95th percentiles. S, species; G, genera; F, families; O, orders.
Focusing on the effect of taxonomic sufficiency (see Table 1): Spearman ρ values between a distance matrix based on species-level data and those based on higher taxonomic resolutions varied by subset but were minimally affected by data transformation (top row, Figure 4). All surrogate matrices of the whole-fauna had a strong correlation (>0.8) with their species-level version. For polychaetes, genera had strong correlation to species, but ρ values dropped for families and orders. Correlations also declined strongly with taxonomic resolution for malacostracans, with ρ values for orders ranging from 0.25 to 0.6, depending on the severity of data transformation. Living bivalves had strong correlations up to the family level, but then declined slightly for orders. Finally, dead bivalves had strong correlations with their species-level version at all taxonomic resolutions, although ρ values notably declined at the genus level using proportional abundance data. All correlations were significantly positive based on Mantel permutation tests (p < 0.05).
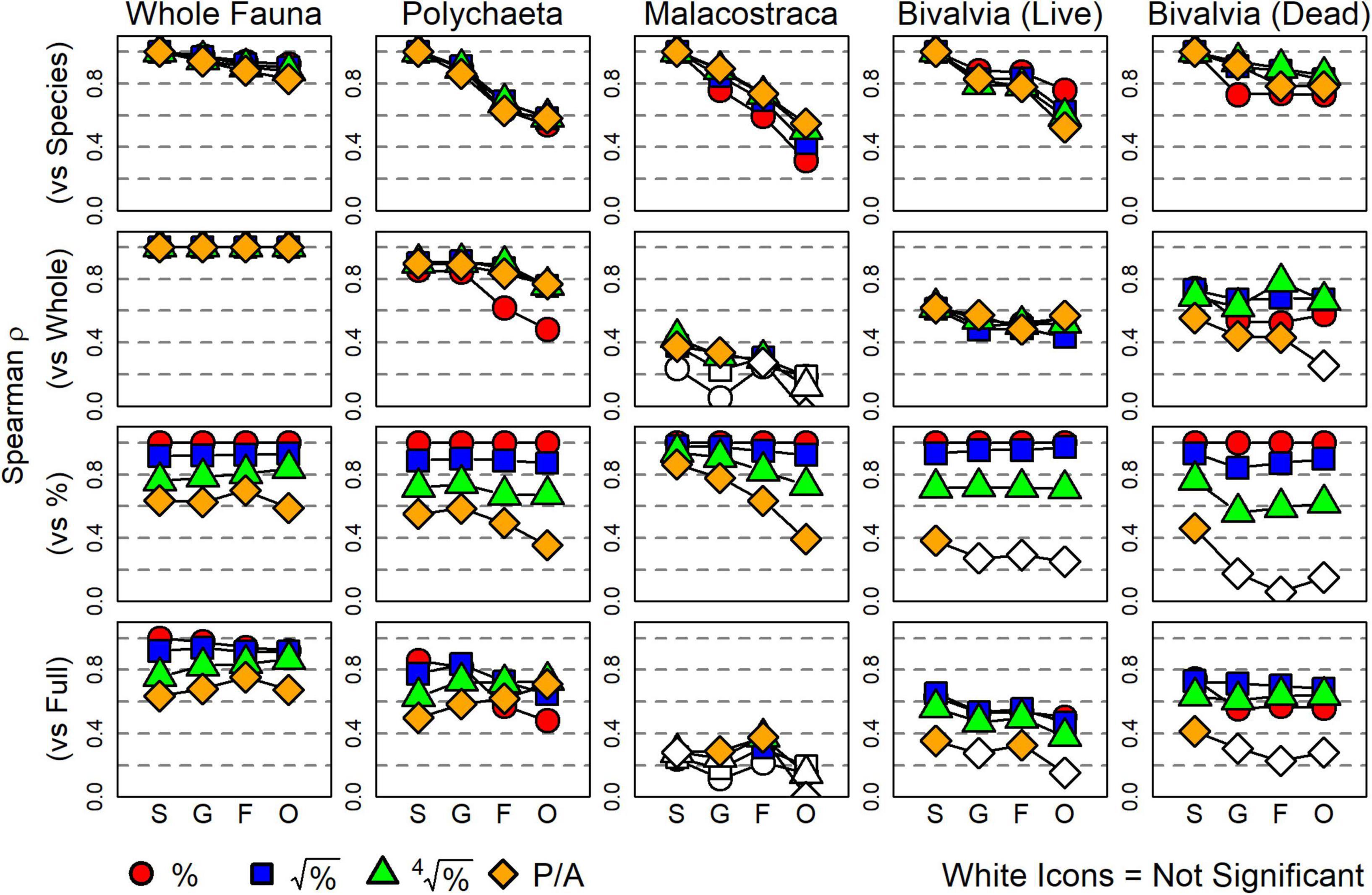
Figure 4. Spearman coefficients (ρ) for inter-matrix correlations representing the effect of each surrogacy method defined in this study. White-filled icons indicate correlations that were not significantly different from zero (p > 0.05). Top row: each matrix is compared to its species-level equivalent to test for taxonomic sufficiency. Correlations decline as the surrogate’s resolution decreases. Second row: each matrix is compared to its whole-fauna equivalent to test for taxonomic subsetting. Correlations are strongest for polychaetes, moderate for bivalves, and poor for malacostracans. Third row: each matrix is compared to its proportional abundance (minimally-transformed) equivalent to test for numerical sufficiency. Correlation strength consistently decreases with increased severity of data transformation. Bottom row: each matrix is compared to the species-level whole-fauna dataset using proportional abundances (the “full” original dataset) to test for the combined effect of all three surrogacy methods. S, species; G, genera; F, families; O, orders.
Concerning the effect of taxonomic subsetting, Spearman ρ values between the whole fauna and the four taxonomic subsets demonstrated that polychaetes were the strongest surrogate for most levels of taxonomic resolution and data transformation (second row, Figure 4). Living and dead bivalves exhibited moderate ρ values to the whole fauna, and malacostracans were notably weak surrogates for all resolutions and data transformations (all ρ < 0.4). Most surrogate matrices for malacostracans were not significantly correlated to their whole-fauna counterparts.
Concerning the effect of numerical sufficiency, Spearman ρ values between a matrix of proportional abundance data and all other transformations of that same subset and resolution demonstrated consistent weakening of correlation with increased severity of data transformation (third row, Figure 4). Generally, square-root-transformed data were strongly correlated to proportional abundance data, whereas fourth-root-transformed data and presence-absence data each decreased correlations detectably. Correlations using presence-absence data for living and dead bivalves were particularly low, with several species- through order-level comparisons not significantly different from null.
For the composite effect of all three surrogacy methods, i.e., Spearman ρ values for comparisons with the original dataset, all three methods had detectable effects (bottom row, Figure 4). Surrogates using polychaetes had strong to moderate ρ values for all resolutions and data transformations. Surrogates using malacostracans exhibited the weakest correlations, most of which were not significantly different from zero. Surrogates using living and dead bivalves were moderately correlated with the original dataset except when using presence-absence data.
For each taxonomic subset, second-stage NMDS plots of inter-matrix correlations – where points separate according to the distinctness of surrogate datasets, not samples within a surrogate dataset – all produced ordinations in which the axes were primarily reflections of the effects of taxonomic and numerical sufficiency (top row, Figure 5). The first NMDS axis for the whole fauna, living bivalves, and dead bivalves was ordered by the severity of data transformation (proportional abundances to presence-absence) from left to right. The second NMDS axis for these three sets generally defined a gradient of taxonomic resolution, with species and genera at the negative end of the axis (bottom of plots) and families and orders at the positive end (top). Polychaetes and malacostracans exhibited a “fanning-out” pattern such that species-level matrices were positioned in the lower right quadrant of each plot and higher taxa of different data transformations progressively spread out toward the left.
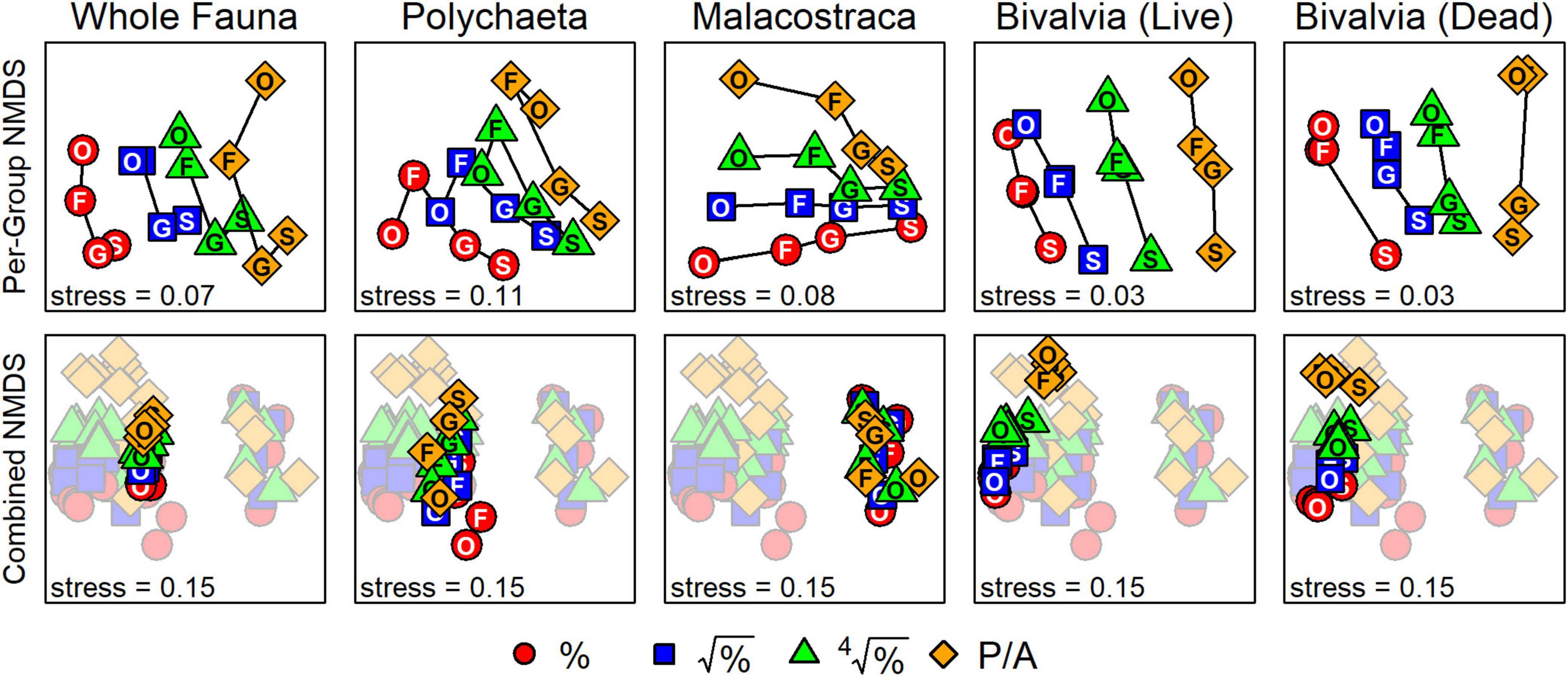
Figure 5. “Second-stage” NMDS ordinations of inter-matrix correlations based on Bray–Curtis distances. Top row: ordinations for each taxonomic set, demonstrating how matrices based on different taxonomic resolutions (species to orders) and data transformations (proportional abundances, %, to presence-absence, P/A) plot along two ordination axes. Bottom row: combined second-stage NMDS with surrogate matrices from all taxonomic sets. Matrices using the whole fauna cluster near those using polychaetes, with living and dead bivalves adjacent. Matrices using malacostracans strongly separate from the others. Data transformation severity also follows the second NMDS axis for all sets except malacostracans. The effect of taxonomic resolution on defining ordination space is weak, as matrices from the same set and based on the same transformation cluster tightly. S, species; G, genera; F, families; O, orders.
For the combined second-stage NMDS – where all points (surrogate datasets) are displayed in a shared ordination space – the first NMDS axis was reflected by clusters of taxonomic sets (bottom row, Figure 5). From left to right, living bivalves, dead bivalves, polychaetes, and the whole fauna were only slightly separated from each other at the negative end of the axis, whereas malacostracans plotted on the positive end of the axis, far from all other subsets. The second NMDS axis reflected a gradient of data transformation severity, with matrices based on proportional abundances plotting at the negative end and those based on presence-absence plotting on the positive end. This gradient along the second NMDS axis was apparent for all sets except malacostracans: their matrices as differentiated by taxonomic resolution (i.e., same set and data transformation) formed tight clusters with no apparent directionality. ANOSIM tests of inter-matrix differences in correlations (Table 4) corroborated the visual patterns from second-stage NMDS. Differences among matrices were significant based on taxonomic subset (R = 0.74, p < 0.001) and much less strongly so for data transformation (R = 0.08, p = 0.003). Differences among matrices based on taxonomic resolution were not significant (Table 4).
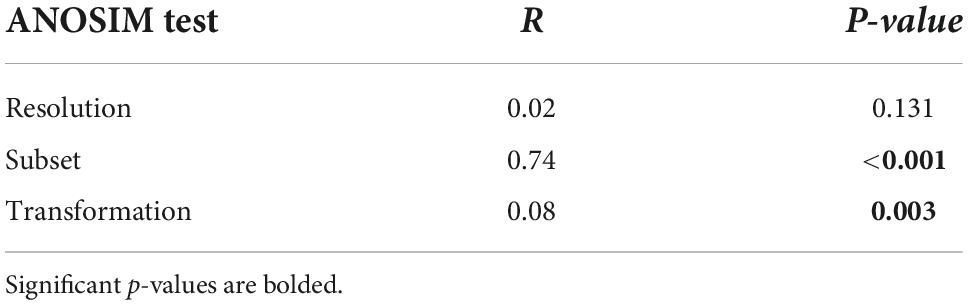
Table 4. Summary of analysis of similarity results testing for the relative effect of different surrogacy methods on the variation of inter-matrix correlations among surrogates.
Finally, rerunning the second-stage NMDS analyses for all two-year sets of the living assemblages drawn from the full 30-year time series produced the same overarching pattern as described in detail above for the 2017–2018 set of living assemblages. Polychaetes were always the strongest surrogate subset of the whole fauna, followed by bivalves, and trailed by poorly-performing malacostracans. Importantly, in each of these replicate tests, the distance matrices produced by bivalve death assemblages collected in 2018–2019 were consistently congruent with those of living bivalves. The consistency of results across the 30-year time series is summarized succinctly by high correlation coefficients (ρ ≥ 0.8) between second-stage distance matrices produced using older sets of living assemblages and that produced using the 2017–2018 set (Figure 6).
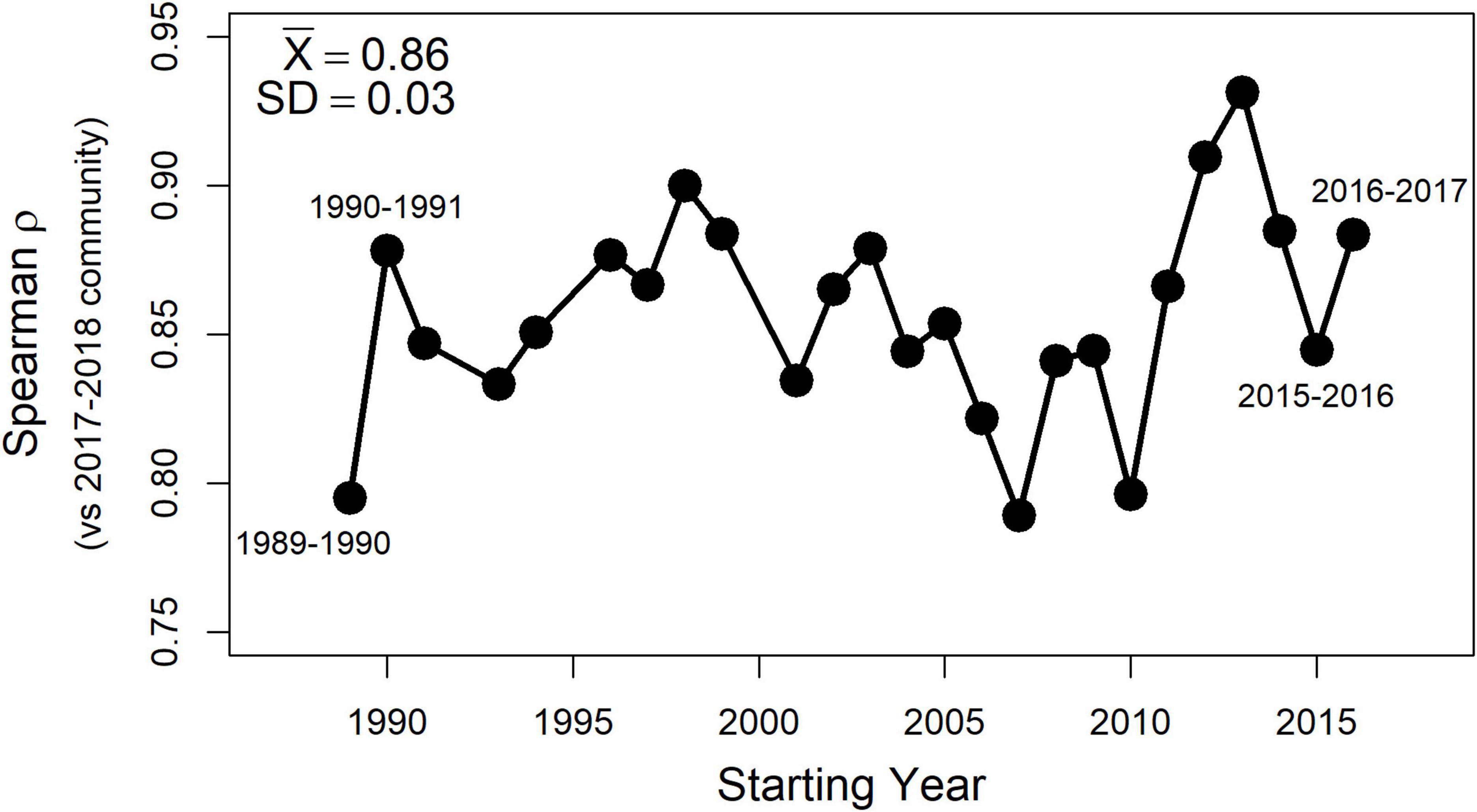
Figure 6. Spearman coefficients (ρ) for correlations between second-stage distance matrices using the 2017–2018 set of living assemblages and those using available 2-year sets from earlier in the 30-year community time series. Mean and standard deviation for the entire set of 2-year comparisons are in the upper left corner. Spearman ρ values are high (≥∼0.8) across the entire time series, indicating that patterns generated the 2017–2018 dataset described in greater detail above have been consistent for the Puget Sound benthos over at least the past 30 years.
The aim of this study was to assess the ability of three surrogacy methods – all commonly adopted to streamline sample processing – to predict the original inter-station compositional distances exhibited by the whole fauna. Are the compositional distances among stations observed using species-level data for the whole fauna retained when data are coarsened to genus or higher resolution, and/or transformed toward presence-absence, and/or subsetted to focus on a single class? In addition, how closely does the local dead-shell assemblage (and its surrogates) mirror the counterpart dead bivalve subset of the whole fauna, relevant to using this usually-discarded part of benthic samples for insights into ecological history? Methods of surrogacy were evaluated for both their individual effects and their effects in various combinations, and they were applied to the class Bivalvia using both living and dead-shell-based data. Importantly, our analyses assess the ability of surrogates to preserve compositional distances among stations over a regional scale: we do not assess the ability of surrogates to detect a known spatial or environmental gradient (i.e., contrary to Tyler and Kowalewski, 2017, and Kokesh et al., 2022) nor their power when formulated as benthic quality metrics (e.g., Tweedley et al., 2014; Dietl et al., 2016; Pruden et al., 2021).
Our analyses could be improved by including additional monitoring stations despite the wide range of depths and substrates the dataset encompasses within Puget Sound (Table 2). However, counterpart analyses of the ability of surrogates to capture regional patterns are definitely needed in other study systems, the better to assess the power of surrogates on local scales, including the generality of the results here. Our discussion focuses on directions needing additional research and some ways to accelerate that progress, such as focusing analyses on gradients in clearly important biological factors, such as the number of species per higher taxon, relative abundances of subset clades, and community evenness.
Variation in compositional distances among stations was reliably preserved despite the coarsening of taxonomic resolution using most taxonomic subsets – i.e., all correlations were significant – even though ρ values did decline with increased coarsening (first row, Figure 4). These results corroborate the findings of numerous earlier studies that inter-matrix correlations do decline with increased taxonomic coarsening (Olsgard and Somerfield, 2000; Włodarska-Kowalczuk and Kêdra, 2007; Bertasi et al., 2009; Bevilacqua et al., 2009) and that family-level identification generally provides a suitable approach to taxonomic surrogacy for the whole fauna (Jones, 2008; Heino, 2010; Pitacco et al., 2019). We found that family-level data were comparable to genus-level data for both living and dead bivalves, even though the level of comparability was weaker than genus-level data for polychaetes and malacostracans. The observed variation among subsets in their robustness to taxonomic coarsening suggests that one should test their retention of power prior to enacting this surrogacy method into practice. In addition, taxonomic sufficiency is a relatively conservative surrogacy method compared to taxonomic subsetting (see next section), as demonstrated here via second-stage NMDS (Figure 5) and discovered earlier by Mellin et al. (2011) using meta-analysis.
The declining power of surrogates with taxonomic coarsening observed here is likely related to the packing of species into higher taxa (e.g., Heino and Soininen, 2007; Bevilacqua et al., 2012; De Oliveira et al., 2020). We found that the decline of ρ values with coarsening resolution (top row, Figure 4) was commensurate with declines in the T/S ratio (top row, Figure 3). Others have postulated that a T/S ratio ≥0.4 (also reported as a S/T ratio ≤2.5) signals that the higher taxon will be able to sufficiently mirror patterns among samples detected using species-level data (Timms et al., 2013). That approximate value has also been identified as a threshold in other systems (Albano et al., 2016; Kokesh et al., 2022). This study thus adds support for the growing consensus that there exists a minimum T/S ratio above which higher taxa are likely to be effective surrogates for species-level patterns, providing a rapid means of predicting success, although this threshold might vary among biological systems (Heino, 2014).
Relevant to the paleoecological potential of taxonomic sufficiency, our finding of particularly strong correlations of species to higher taxa of dead bivalves at multiple resolutions supports the common practice of evaluating fossil assemblages at the resolution of genera or families: such coarsening produces compositional patterns similar to those of species-level information, as also found by others using exclusively fossil data (Forcino et al., 2012; Zuschin et al., 2017). The persistence of live-dead patterns in Puget Sound despite this small magnitude of taxonomic coarsening on live-dead patterns has also been recognized elsewhere (e.g., Albano et al., 2016). We suspect, however, that the power of taxonomic sufficiency will vary over space and time in both modern and ancient faunas, including a likely latitudinal gradient. For example, S/G ratios of living bivalves are particularly high in the tropics and low in boreal settings (Krug et al., 2008), both regions where relatively few sufficiency tests have been conducted compared to mid-latitude settings.
Subset clades exhibited disparate strength as surrogates for the whole fauna, but polychaetes were the most powerful (second row, Figure 4), as has been found in numerous tests of class-level subsets in marine benthos (Olsgard et al., 2003; Włodarska-Kowalczuk and Kêdra, 2007; Kokesh et al., 2022). This high fidelity of polychaetes is usually attributed to their comprising the majority – sometimes the overwhelming majority – of individuals and species richness in soft-sedimentary seafloors. However, polychaetes were closely followed in Puget Sound by bivalves, both alive and dead, in total abundance and were very distantly trailed by malacostracans despite this group being second-ranked in richness in the Puget Sound regional dataset (second row, Figure 4). In general, taxonomic subsetting has been generally found to be less effective, reliable, and/or consistent as a method of surrogacy than has taxonomic sufficiency (coarsening), based on other studies using the same clades as examined here (e.g., Włodarska-Kowalczuk and Kêdra, 2007; Bevilacqua et al., 2009; Mellin et al., 2011; Gladstone et al., 2020; Kokesh et al., 2022). Nonetheless, given the observed variation among subsets in their robustness to taxonomic coarsening, we would suggest that the ability of a subset to preserve whole-fauna patterns be assessed prior to putting this (or any other) surrogacy method into practice. Thus, although the results here are very positive for bivalves in particular, both alive and dead, their reliability should be tested locally prior to formalizing surrogacy in a protocol. Such tests could repeat the same methods described here but use a limited number of stations or a single historic survey to produce a benchmark whole-fauna matrix for assessing the reliability of subsets.
Additional research is also needed to identify the biological properties of taxonomic subsets that convey reliable surrogacy power: these determinants are underexplored compared to those that allow a subset to retain power despite coarser resolution (previous section). One promising attribute is the proportional abundance of the subset within the whole fauna (e.g., Dietl et al., 2016). For example, we found that taxonomic subsets declined in ρ values (second row, Figure 4) congruent with declines in their median station-scale abundance within the whole fauna (N/W; middle row, Figure 3): polychaetes (highest N/W) were a more powerful surrogate subset of the whole fauna than were either living or dead bivalves, which were both more reliable than malacostracans (lowest N/W). Malacostracans are represented by more species than bivalves in this dataset, but the potential leverage of that diversity apparently cannot compensate for their low station- and regional-scale abundances. Thus, the ability of taxonomic subsets to serve as effective surrogates appears to be, at least partially, a function of their relative abundance rather than their absolute or proportional richness in the whole fauna. It would be worthwhile to explore whether some minimum threshold N/W exists that determines the utility of taxonomic subsets as surrogates, even though this threshold might vary among biological systems (especially as a function of community evenness or S/G ratios).
From this single test in Puget Sound, it is difficult to determine the importance of the (quite large) sample size of dead bivalves (N = 5,337) on their performance as a surrogate. Dead bivalves are not technically a subset of the living fauna but rather a cross-assemblage surrogate of much coarser temporal scale. Moreover, their raw abundance might be genuinely biased upward relative to the raw abundance of living bivalves due to our counting disarticulated valves, although that effect on outcome is minimized by our using proportional abundance data. Nevertheless, the high compositional congruence between living and dead bivalves in Puget Sound, as judged by their overlapping clusters in ordination space (bottom row, Figure 5), indicates that bivalve death assemblages have a power similar to that of living bivalve subsets for recognizing compositional variation in the whole fauna, that is, for serving as surrogates of the whole fauna (see section “Bivalve live-dead agreement and taphonomic caveats” below).
We found that the severity of data transformation had a much larger effect on the strength of different surrogates than did the coarsening of taxonomic resolution (third row, Figure 4). Increasing the severity of data transformation resulted in sequential declines in correlation with the original proportional abundances regardless of taxonomic (sub)set or resolution, and downgrading abundance to presence-absence data had a particularly strong impact for both living and dead bivalves, resulting in correlations that lacked statistical significance. Although matrices based on different data transformations were notably separated in second-stage NMDS space (Figure 5), these effects were small compared to differences created by taxonomic subsetting (Table 4).
Ecologists commonly transform data from their original, raw counts in order to better meet the assumptions of univariate tests (usually an assumption of normality; St-Pierre et al., 2018). All such transformations (other than proportional abundance) diminish the influence of dominant taxa on compositional patterns and specifically amplify the role of rare taxa. Effectively, this operation artificially inflates the evenness of a community. Like the predictive power of T/S for taxonomic sufficiency discussed above, the evenness of the raw count or proportional abundance data matrix of a surrogate will determine the extent to which data transformation will alter its composition. For example, the relatively low PIE values (low evenness) observed for bivalve living and (especially) death assemblages here (bottom row, Figure 3) indicate that data transformation should have a particularly large effect, leveling the initially uneven effect of dominant and rare species on composition until, with the extreme of presence-absence data, all species have equally-abundant taxa (PIE = 1). For bivalves in Figure 4 (right two columns), data transformation in fact does have a strong effect, accompanied by large decreases in ρ. In contrast, the whole fauna and polychaetes both had high PIE values relative to bivalves (and malacostracans), and their correlation (resemblance) to the original fauna were much less affected by data transformation, although not immune to it (Figure 4). The particularly large decline in ρ values for bivalves (and for order-level malacostracans; Figure 3) appears to correspond to an outsized response to data transformation by assemblages having relatively low evenness (PIE < ∼0.6). Thus, PIE values for a surrogate’s raw count data can likely serve as an easily-acquired first-order predictor of the magnitude of effect that data transformation will have on community patterns.
Although square-root and fourth-root transformations are commonly employed for biological data analyses (e.g., Thorne et al., 1999; Clarke and Warwick, 2001; Dethier and Schoch, 2006; Domínguez-Castanedo et al., 2007; Vijapure and Sukumaran, 2019), these particular transformations do not themselves constitute true surrogates: they still require that someone generated the underlying original abundance data. In contrast, ranked-abundance (stepwise integers), ordinal abundance data (e.g., “rare,” “common,” “very common,” etc.), and presence-absence data do represent methods that reduce effort during the data collection, and thus are all potential surrogacy techniques (e.g., Carneiro et al., 2010; Landeiro et al., 2012). It should be noted that using numerical sufficiency as a surrogacy method comes at the cost of reducing the range of possible analyses. For example, richness and binary distance metrics like Jaccard similarities can be assessed accurately, but abundance-based metrics such as rarefaction, similarity percentages (SIMPER; Clarke, 1993), and evenness and dominance metrics are no longer viable. The objectives of a study or monitoring program, the pre-determined scope of analytical methods, and the results from preliminary tests of consistency among data transformations thus should all be considered in order to determine whether the implementation of numerical sufficiency is acceptable.
Compositional variation in bivalve death assemblages among the ten Puget Sound stations, where the whole fauna is documented to have been relatively stable since 1989 (Partridge et al., 2018), was congruent with that of living bivalves based on all taxonomic resolutions and data transformations assessed in this study (Figure 5). The ability of dead-shell assemblages to mirror regional variation of bivalve living assemblages in Puget Sound is thus very high, and moreover, is robust to taxonomic coarsening and to multiple levels of data transformation. Bivalve death assemblages have been shown to preserve spatial and environmental gradients in bivalve living assemblages in other systems of relatively pristine or otherwise steady-state conditions (e.g., Warme, 1969; Tomašových and Kidwell, 2009; Martinelli et al., 2016; Hyman et al., 2019). Conversely, poor live-dead agreement in subtidal settings, commonly assumed to reflect postmortem information loss, is now appreciated as being strongly correlated with a recent history of anthropogenic disturbance sufficient to have offset the composition of the living assemblage from its long-term baseline state, of which the surficial time-averaged death assemblage retains a memory (Kidwell, 2007). Stressors with such power include eutrophication (Kidwell, 2007; Korpanty and Kelley, 2014; Leshno et al., 2015; Gilad et al., 2018; Tweitmann and Dietl, 2018), biological invasion (Yanes, 2012; Chiba and Sato, 2013; Steger et al., 2022), bottom trawling (Kidwell, 2009), various combinations of these and other stressors (Haselmair et al., 2021), and warming-associated environmental changes (Powell et al., 2017; Meadows et al., 2019). We note that while live-dead congruence as found here (i.e., living and dead bivalves producing similarly-structured distance matrices of among-station variation) is not the same as live-dead agreement as conventionally determined (i.e., the living and dead bivalves having similar faunal composition), it nevertheless supports the observational data for a stable community organization over the last 30 years at this regional scale (Partridge et al., 2018). Indeed, the consistent results for all two-year sets of the community time series and high correlation to the 2017–2018 dataset (Figure 6) suggests that patterns of taxonomic surrogacy and live-dead congruence among bivalves have both been stable attributes of the Puget Sound benthos for at least the last 30 years.
Based on annual monitoring, overall benthic conditions in Puget Sound are known to have deteriorated over the last 30 years, especially in terminal inlets with poor water exchange and especially using measures of sediment toxicity (Partridge et al., 2018; Weakland et al., 2018). However, changes in benthic communities at the ten sentinel sites used here have been modest and non-directional using compositional measures: most variation is among sites as a function of habitat type, with only a small secondary effect of the date of sampling (Partridge et al., 2018). That said, the ten sites have each exhibited variability including some strong fluctuations in abundance over the 1989–2015 interval: some sites have shown an increase (or a decrease) in total abundance or richness, and some have experienced an increase (or decrease) in a particular taxon (but usually different taxa at different sites) and none of these faunal changes are correlated strongly with tracked contaminants (Partridge et al., 2018). This variability and the uncertain causes stimulated a change in ECY’s monitoring program to include additional environmental measures and increase the number of sites sampled annually (Dutch et al., 2018). It was also a motivation for our collaboration, bringing death assemblages into the evaluation, and thus for this initial investigation of surrogacy.
In light of this known but highly complex and spatially disparate history of communities at these ten sites, which have nonetheless remained compositionally distinct, the high live-dead congruence observed among bivalves suggests that the death assemblages have experienced little net postmortem bias from processes such as lateral transportation and differential preservation of shells. It also suggests that the time averaging of skeletal materials has been either (1) relatively short, i.e., encompassing too few generations of shell input to detect any past shifts in community composition that might have occurred before the onset of monitoring, or (2) fairly long, but compositional changes within that interval of death assemblage accumulation did not exceed the range of variation observed within the past 30 + years (i.e., the distance matrices of community composition have been stable for more than 30 years).
We do not yet have shell-age data to test these alternatives directly. Such data on the window of time averaging (that is, the age of the oldest shells) and the temporal resolution (IQR of the frequency distribution of cohorts) of a death assemblage is typically quantified by radiocarbon or other direct age-dating of shells (for review, see Kidwell, 2013). The per-sample cost and subsample size have declined over the last decade, but shell-age analysis still requires funds and collaborations outside those generally on hand. It thus is still undertaken only to address specific issues relevant to management that emerge from an initial live-dead analysis, such as the date of disappearance of an abundant but dead-only taxon (e.g., Kowalewski et al., 2000; Albano et al., 2016; Tomašových and Kidwell, 2017; LeClaire et al., 2022) and the persistence of an important habitat (e.g., Waldbusser et al., 2013; Casebolt and Kowalewski, 2018; Hyman et al., 2019; Haselmair et al., 2021). The precise scale of time averaging is not critical to the question here, nor have local shell-age data accompanied surrogacy tests by others using live and dead bivalves (e.g., Schopf, 1978; Tyler and Kowalewski, 2017), including surrogacy tests that suppose that dead-shells should serve as good guides to gradients because the living mollusk subset does (Dietl et al., 2016; Kokesh et al., 2022). Age data is in fact lacking in many valuable and almost certainly correct inferences of benthic change based on live-dead discordance (e.g., Tweitmann and Dietl, 2018, building on Kidwell, 2007). Thus, for an initial analysis of the local value of death assemblages, it can suffice to appreciate that dead shells reflect the “past” to some degree, notwithstanding uncertainties in the magnitude of time averaging, contrasting with the “now” represented by living assemblages. This qualitative strategy of analogizing live/dead (or live/Quaternary fossil, or core-top/core-bottom) with now/then underlies much of conservation paleobiologic research and achievements, allowing us to acquire first-order insights before fully knowing the geologic age and age-structure of the local shelly record.
Shell-age analysis is only now getting underway for Puget Sound bivalve assemblages. However, geological inference suggests that time averaging might be quite limited in cold-temperate Puget Sound, perhaps reflecting shell input largely from only the last few decades – that is, encompassing the duration of biomonitoring but perhaps not much more. One argument for such short time averaging is that the ratio of dead-to-live bivalve abundances is only 1.2–1 (Table 3), which is much lower than the global median of 8:1 for tropical and warm-temperate settings (Kidwell, 2009). This low ratio suggests relatively little accrual of past generations in the surface mixed layer, which can have several non-mutually-exclusive explanations. One is that postmortem rates of aragonitic shell disintegration are high, which we would suspect for relatively cold waters like those of Puget Sound. A second explanation for low dead-to-live ratios can be a high rate of sedimentation, which removes shells from the surface mixed layer via burial. Sedimentation rates in Puget Sound are, in fact, relatively high at ∼0.5–2 cm/yr based on 210Pb dating of sedimentary cores (Brandenberger et al., 2011).
Data from other settings with high sedimentation provide the best basis for predicting the scale of time averaging of Puget Sound bivalve death assemblages, pending direct dating. For example, in subtropical siliciclastics of Texas coastal lagoons, with a sedimentation rate of ∼0.3 cm/yr, nearly all dead bivalve shells from the upper 0–10 cm of sediment were dated as younger than ∼10 years (Olszewski and Kaufman, 2015). In tropical lagoonal deposits of the Great Barrier Reef with a sedimentation rate of ∼0.6 cm/yr, the 95% confidence interval of median shell ages from the uppermost 15 cm of sediment ranged from 2 to 167 years with a maximum shell age of ∼200 years old (Kosnik et al., 2015). Given that these are much warmer-water settings with comparable sedimentation rates, these values probably define the upper limit for time-averaging expected in the cold-temperate Puget Sound. Our best expectation for Puget Sound is thus IQRs on the order of a few decades, with few old shells more than ∼100 years old.
Given that variation in living bivalves (a true subset of the whole fauna) demonstrated strong correlation to the whole fauna, it follows that bivalve death assemblages, which map so closely onto living bivalves, possess a similar capacity to that of living bivalves to approximate the among-station compositional distances of the whole living macrobenthic fauna in this system. Polychaetes are the strongest surrogate subset here (as they also are in many other systems, see references above), but they and species-rich malacostracans lack the preservation potential of bivalves under normoxic conditions and thus have little practical paleoecological potential. Albano et al. (2016) found species-level patterns of live-dead agreement to be consistent up to resolutions of families, corroborated further with regard to the effects of data transformation in Miocene assemblages by Zuschin et al. (2017). Using a different methodological approach, namely quantifying compositional variability among environmentally-categorized groups of stations, Tyler and Kowalewski (2017) found that molluscan death assemblages preserve the extent of variation among environments as reflected by the whole living fauna. This study introduces, for the first time, methods already in wide use by applied ecologists for examining surrogacy (particularly second-stage NMDS) and expands to include both living and death assemblages. Our results corroborate those of previous fossil-focused investigations.
Our finding of the reliability of bivalve death assemblages supports their use for retrospectively evaluating human impacts on ecological systems. Conservation biologists typically have to rely on long-term monitoring data to quantify ecological responses to historic human impacts, watching changes (decline, recovery) play out in real time (e.g., Borja et al., 2006; Moritz et al., 2021). Even under the pressure of impending development, investigators still typically employ a Before-After-Control-Impact (BACI) or Beyond-BACI study design (Underwood, 1991, 1994) to establish changes occurring over time, even if observations are limited to only a few seasons. However, human stressors such as wastewater, dredging, and harvesting often predate the onset of even dedicated monitoring efforts (Todd et al., 2019). Demonstrating that shelly death assemblages are congruent with whole fauna, especially their pattern of compositional variation across a region as tested here, encourages their use as tools for estimating pre-monitoring (and potentially pre-impact) biological patterns. It should, of course, be remembered that sampling death assemblages inflicts the same cost on living communities as if the living were the target. The advantages of including death assemblages in monitoring efforts are becoming increasingly clear, however, namely in increasing the sample size of an important part of the benthic community, providing historical perspective on conditions during past decades and centuries, and as undertaken here, establishing the conditions (species:genus ratios, clade representation, evenness, etc.) under which biologists in frontier areas might streamline monitoring to shelly clades alone.
Government-mandated, contracted, and non-profit monitoring programs might thus elect to integrate the retention and processing of death assemblages as part of their routine analytical procedures or, as exemplified here by cooperation between The University of Chicago and ECY, form an academic-agency partnership to explore an environmental issue of particular concern. ECY now saves rather than discards their sediment residue, allowing scientists at Chicago to develop counterpart ecological information from the death assemblage to tackle questions of mutual interest, such as this report. It also permits other applied and academic analyses, now underway, such as establishing scales of time averaging and uncovering environmental and species-level patterns of shell damage and loss, especially those relevant to ocean acidification. The University currently archives the growing library of dead samples from Puget Sound, as it does for counterpart material from cooperative agreements with municipal agencies in southern California (e.g., Bizjack et al., 2017; Tomašových and Kidwell, 2017; Leonard-Pingel et al., 2019). However, although agencies are typically required to retain live-collected material for a set number of years, they cannot accommodate the much larger volume of dead sieve residues, nor necessarily acquire funds for the labor of dead-shell analysis, at least until a formal protocol is established with clear benefits. Relationships with museums and other academic institutions are thus an important step. We strongly encourage academics to be proactive in proposing such collaborative efforts: agency personnel already have the necessary sampling operations in place and high-quality datasets on biological communities and environmental parameters, sometimes over impressive spatial and/or temporal scales as here and in southern California. Like all scientists, they like to see new value from existing data, especially from samples they had been discarding, and educational benefits are always important for public institutions. Additional mutual benefits – already experienced from this ECY-University of Chicago partnership – include (1) projects and connections for undergraduate and graduate students, who are also introduced to careers in applied science, (2) publicity of conservation paleobiology and novel academic-agency partnerships to the public and stakeholders served by agencies, and (3) participation and authorship by agency scientists on publications and deliverables that appeal to a readership of both academic and applied scientists. Eventually, the ECY hopes that this cooperative effort, which extends beyond this first report on surrogacy, will reveal new insights that better inform their monitoring efforts, including the detection of human stressors that predate the PSEMP program, and encourage the implementation of new field methods (e.g., core sampling).
Although our focus here is on supporting modern-day efforts in conservation and environmental management, these results are also encouraging for paleoecological inference in older, pre-Holocene fossil records. The shelly component of a fauna cannot be used to directly reconstruct the composition of the unpreserved soft-bodied fauna, but tests like this support recognizing compositional variation of benthic communities more broadly, especially in the Cenozoic and late Mesozoic (i.e., the realm of the mollusk-rich Modern Evolutionary Fauna; Sepkoski, 1981; Rojas et al., 2021). By extension of death assemblages serving as a congruent proxy for the Modern whole fauna, fossil assemblages might reliably reflect patterns of compositional variation of the unpreserved fauna that was surely present. Surrogates of many types already have a long history of use in paleontology, for example using genus- rather than species-level data and focusing community analysis on a single class (as is also often done in community ecology, e.g., the dynamics of small mammals). Quantifying the confidence of such surrogates for application in the fossil record is thus an important direction for future research, to better understand paleoecological variation among assemblages at basinal and larger regional scales.
This cooperative study between academic paleontologists and applied agency ecologists demonstrates that bivalves, both living and dead, are reliable for recognizing regional-scale compositional variation of the whole macrobenthic fauna of Puget Sound, a cold-temperate estuary with strong natural environmental gradients and potentially challenging taphonomic conditions. The success of surrogacy methods here, especially the suitability of family-level taxonomy and of using proportionally abundant (not necessarily rich) subsets of taxa, should promote the expansion of monitoring to new areas, where the costs and requisite taxonomic expertise needed for whole-fauna analysis might otherwise be discouraging. Such expansion can be an issue even in long-standing monitoring programs. We emphasize the importance of testing for the sensitivity of different surrogates locally prior to or at the onset of new monitoring programs, given variability in our results on numerical sufficiency (coarsening of taxonomic count data) and in local motivations (e.g., surrogates to mirror the whole fauna versus identifying environmental gradients, or comparison with paleoecological evidence from death assemblages and cores). The power of bivalve death assemblages to detect regional compositional variation evident in the living whole fauna, despite potential taphonomic filters on composition and taxonomic identification, are especially encouraging, given the advantages of integrating paleoecological information with modern monitoring programs, a major thread in conservation paleobiology. Given that shelly death assemblages are already encountered by the sampling gear used for living macrobenthic infauna, they are a particularly inexpensive complement to long-term time-series data as well as a means of getting a baseline estimate where long-term data are lacking.
All data and R scripts used in this study are accessible from the online Supplementary material. All further inquiries can be directed to the corresponding author.
BK conceived the study, processed dead shell materials, analyzed the data, and wrote the original manuscript draft. DB, VP, and SW provided the living community data and performed the field sampling. SK advised the project, assisted with writing, and provided funding. All authors contributed editorial feedback and approved the final manuscript version for submission.
This project was funded by a New Directions grant from the Petroleum Research Fund of the American Chemical Society (#58795-ND8).
We thank M. Dutch for her unwavering support of this agency-academic collaboration, as well as A. Eagleston and other scientists at the Washington State Department of Ecology. We thank M. Foote, D. Jablonski, and S. Lidgard for stimulating discussions and comments during the development of this project. We also thank MK and SD for their very helpful comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.980753/full#supplementary-material
Albano, P. G. (2014). Comparison between death and living land mollusk assemblages in six forested habitats in northern Italy. Palaios 29, 338–347. doi: 10.2110/palo.2014.020
Albano, P. G., Tomašových, A., Stachowitsch, M., and Zuschin, M. (2016). Taxonomic sufficiency in a live-dead agreement study in a tropical setting. Palaeogeogr. Palaeoclimatol. Palaeoecol. 499, 341–348. doi: 10.1016/j.palaeo.2016.02.031
Bertasi, F., Colangelo, M. A., Colosio, F., Gregorio, G., Abbiati, M., and Ceccherelli, V. U. (2009). Comparing efficacy of different taxonomic resolutions and surrogates in detecting changes in soft bottom assemblages due to coastal defence structures. Mar. Pollut. Bull. 58, 686–694. doi: 10.1016/j.marpolbul.2009.01.003
Bevilacqua, S., Fraschetti, S., Musco, L., and Terlizzi, A. (2009). Taxonomic sufficiency in the detection of natural and human-induced changes in marine assemblages: A comparison of habitats and taxonomic groups. Mar. Pollut. Bull. 58, 1850–1859. doi: 10.1016/j.marpolbul.2009.07.018
Bevilacqua, S., Terlizzi, A., Claudet, J., Fraschetti, S., and Boreo, F. (2012). Taxonomic relatedness does not matter for species surrogacy in the assessment of community responses to environmental drivers. J. Appl. Ecol. 49, 357–366. doi: 10.1111/j.1365-2664.2011.02096.x
Bizjack, M. T., Kidwell, S. M., Velarde, R. G., Leonard-Pingel, J., and Tomašových, A. (2017). Detecting, sourcing, and age-dating dredged sediments on the open shelf, southern California, using dead mollusk shells. Mar. Pollut. Bull. 114, 448–465. doi: 10.1016/j.marpolbul.2016.10.010
Booth, D. B. (1994). Glaciofluvial infilling and scour of the Puget Lowland, Washington, during ice-sheet glaciation. Geology 22, 695–698. doi: 10.1130/0091-76131994022<0695:GIASOT<2.3.CO;2
Borja, Á, Muxika, I., and Franco, J. (2006). Long-term recovery of soft-bottom benthos following urban and industrial sewage treatment in the Nervión estuary (southern Bay of Biscay). Mar. Ecol. Prog. Ser. 313, 43–55. doi: 10.3354/meps313043
Brandenberger, J. M., Louchouarn, P., and Crecelius, E. A. (2011). Natural and post-urbanization signatures of hypoxia in two basins of Puget Sound: Historical reconstruction of redox sensitive metals and organic matter inputs. Aquat. Geochem. 17, 645–670. doi: 10.1007/s10498-011-9129-0
Carneiro, F. M., Bini, L. M., and Rodrigues, L. C. (2010). Influence of taxonomic and numerical resolution on the analysis of temporal changes in phytoplankton communities. Ecol. Indic. 10, 249–255. doi: 10.1016/j.ecolind.2009.05.004
Casebolt, S., and Kowalewski, M. (2018). Mollusk shell assemblages as archives of spatial structuring of benthic communities around subtropical Islands. Estuar. Coast. Shelf Sci. 215, 132–143. doi: 10.1016/j.ecss.2018.09.023
Chiba, T., and Sato, S. I. (2013). Invasion of Laguncula pulchella (Gastropoda: Naticidae) and predator-prey interactions with bivalves on the tona coast, Miyagi prefecture, northern Japan. Biol. Invasions 15, 587–598. doi: 10.1007/s10530-012-0310-1
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Austral. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Clarke, K. R., and Warwick, R. M. (2001). Changes in marine communities: An approach to statistical analysis and interpretation, 2nd Edn. Plymouth: PRIMER-E.
Close, R. A., Evers, S. W., Alroy, J., and Butler, R. J. (2018). How should we estimate diversity in the fossil record? Testing richness estimators using sampling-standardised discovery curves. Methods Ecol. Evol. 9, 1386–1400. doi: 10.1111/2041-210X.12987
De Oliveira, S. S. Jr., Ortega, J. C., dos Santos, Ribas, L. G., Lopes, V. G., and Bini, L. M. (2020). Higher taxa are sufficient to represent biodiversity patterns. Ecol. Indic. 111:105994. doi: 10.1016/j.ecolind.2019.105994
Dethier, M. N., and Schoch, G. C. (2006). Taxonomic sufficiency in distinguishing natural spatial patterns on an estuarine shoreline. Mar. Ecol. Prog. Ser. 306, 41–49. doi: 10.3354/meps306041
Dietl, G. P., Durham, S. R., Smith, J. A., and Tweitmann, A. (2016). Mollusk assemblages as records of past and present ecological status. Front. Mar. Sci. 3:169. doi: 10.3389/fmars.2016.00169
Dietl, G. P., and Flessa, K. W. (2011). Conservation paleobiology: Putting the dead to work. Trends Ecol. Evol. 26, 30–37. doi: 10.1016/j.tree.2010.09.010
Domínguez-Castanedo, N., Rojas-López, R., Solís-Weiss, V., Hernández-Alcántara, P., and Granados-Barba, A. (2007). The use of higher taxa to assess the benthic conditions in the southern Gulf of Mexico. Mar. Ecol. 28, 161–168. doi: 10.1111/j.1439-0485.2007.00178.x
Dutch, M., Partridge, V., Weakland, S., Burgess, D., and Eagleston, A. (2018). Quality assurance monitoring plan: The puget sound sediment monitoring program. Lacey, WA: Washington State Department of Ecology Publication.
Ferraro, S. P., and Cole, F. A. (1992). Taxonomic level sufficient for assessing a moderate impact on macrobenthic communities in Puget Sound, Washington, USA. Can. J. Fish. Aquat. Sci. 49, 1184–1188. doi: 10.1139/f92-133
Forcino, F. L., Stafford, E. S., and Leighton, L. R. (2012). Perception of paleocommunities at different taxonomic levels: How low must you go? Palaeogeogr. Palaeoclimatol. Palaeoecol. 365–366, 48–56. doi: 10.1016/j.palaeo.2012.09.011
Gilad, E., Kidwell, S. M., Benayuha, Y., and Edelman-Furstenberg, Y. (2018). Unrecognized loss of seagrass communities based on molluscan death assemblages: Historic baseline shift in tropical Gulf of Aqaba, Red Sea. Mar. Ecol. Prog. Ser. 589, 73–83. doi: 10.3354/meps12492
Gilinsky, N. L., and Bennington, J. B. (1994). Estimating numbers of whole individuals from collections of body parts: A taphonomic limitation of the paleontological record. Paleobiology 20, 245–258. doi: 10.1017/S0094837300012719
Gladstone, W., Murray, B. R., and Hutchings, P. (2020). Promising yet variable performance of cross-taxon biodiversity surrogates: A test in two marine habitats at multiple times. Biodivers. Conserv. 29, 3067–3089. doi: 10.1007/s10531-020-02015-4
Harnik, P. G. (2009). Unveiling rare diversity by integrating museum, literature, and field data. Paleobiology 35, 190–208. doi: 10.1666/07062.1
Haselmair, A., Gallmetzer, I., Tomašových, A., Wiester, A. M., Übelhör, A., and Zuschin, M. (2021). Basin-wide infaunalisation of benthic soft-bottom communities driven by anthropogenic habitat degradation in the northwestern Adriatic Sea. Mar. Ecol. Prog. Ser. 671, 45–65. doi: 10.3354/meps13759
Heino, J. (2010). Are indicator groups and cross-taxon congruence useful for predicting biodiversity in aquatic ecosystems? Ecol. Indic. 10, 112–117. doi: 10.1016/j.ecolind.2009.04.013
Heino, J. (2014). Taxonomic surrogacy, numerical resolution and responses of stream macroinvertebrate communities to ecological gradients: Are the inferences transferable among regions? Ecol. Indic. 36, 186–194. doi: 10.1016/j.ecolind.2013.07.022
Heino, J., and Soininen, J. (2007). Are higher taxa adequate surrogates for species-level assemblage patterns and species richness in stream organisms? Biol. Conserv. 137, 78–89. doi: 10.1016/j.biocon.2007.01.017
Hurlbert, S. H. (1971). The nonconcept of species diversity: A critique and alternative parameters. Ecology 52, 557–586. doi: 10.2307/1934145
Hyman, A. C., Frazer, T. K., Jacoby, C. A., Frost, J. R., and Kowalewski, M. (2019). Long-term persistence of structured habitats: Seagrass meadows as enduring hotspots of biodiversity and faunal stability. Proc. R. Soc. B Biol. Sci. 286:20191861. doi: 10.1098/rspb.2019.1861
Jones, F. C. (2008). Taxonomic sufficiency: The influence of taxonomic resolution on freshwater bioassessments using benthic macroinvertebrates. Environ. Rev. 16, 45–69. doi: 10.1139/A07-010
Khangaonkar, T., Yang, Z., Kim, T., and Roberts, M. (2011). Tidally averaged circulation in Puget Sound sub-basins: Comparison of historical data, analytical model, and numerical model. Estuar. Coast. Shelf Sci. 93, 305–319. doi: 10.1016/j.ecss.2011.04.016
Kidwell, S. M. (2007). Discordance between living and death assemblages as evidence for anthropogenic ecological change. Proc. Natl. Acad. Sci. U.S.A. 104, 17701–17706. doi: 10.1073/pnas.070719410
Kidwell, S. M. (2009). “Evaluating human modification of shallow marine ecosystems: Mismatch in composition of molluscan living and time-averaged death assemblages,” in Conservation paleobiology: Using the past to manage for the future, Vol. 15, eds G. P. Dietl and K. W. Flessa (Portland, OR: Paleontological Society), 113–139.
Kidwell, S. M. (2013). Time-averaging and fidelity of modern death assemblages: Building a taphonomic foundation for conservation palaeobiology. Palaeontology 56, 487–522. doi: 10.1111/pala.12042
Kidwell, S. M., and Tomašových, A. (2013). Implications of time-averaged death assemblages for ecology and conservation paleobiology. Annu. Rev. Ecol. Evol. Syst. 44, 539–563. doi: 10.1146/annurev-ecolsys-110512-135838
Kokesh, B. S., Kidwell, S. M., Tomašových, A., and Walther, S. M. (2022). Detecting strong spatial and temporal variation in macrobenthic composition on an urban shelf using taxonomic surrogates. Mar. Ecol. Prog. Ser. 682, 13–30. doi: 10.3354/meps13932
Korpanty, C. A., and Kelley, P. H. (2014). Molluscan live-dead agreement in anthropogenically stressed seagrass habitats: Siliciclastic versus carbonate environments. Palaeogeogr. Palaeoclimat. Palaeoecol. 410, 113–125. doi: 10.1016/j.palaeo.2014.05.014
Kosnik, M. A., Hua, Q., Kaufman, D. S., and Zawadzki, A. (2015). Sediment accumulation, stratigraphic order, and the extent of time-averaging in lagoonal sediments: A comparison of 210Pb and 14C/amino acid racemization chronologies. Coral Reefs 34, 215–229. doi: 10.1007/s00338-014-1234-2
Kowalewski, M., Serrano, G. E. A., Flessa, K. W., and Goodfriend, G. A. (2000). Dead delta’s former productivity: Two trillion shells at the mouth of the Colorado River. Geology 28, 1059–1062. doi: 10.1130/0091-7613200028<1059:DDFPTT<2.0.CO;2
Kowalewski, M., Carroll, M., Casazza, L., Gupta, N. S., Hannisdal, B., Hendy, A., et al. (2003). Quantitative fidelity of brachiopod-mollusk assemblages from modern subtidal environments of San Juan Islands, USA. J. Taphonomy 1, 43–66.
Kowalewski, M., and Hoffmeister, A. P. (2003). Sieves and fossils: Effects of mesh size on paleontological patterns. Palaios 18, 460–469. doi: 10.1669/0883-13512003018<0460:SAFEOM<2.0.CO;2
Krug, A. Z., Jablonski, D., and Valentine, J. W. (2008). Species-genus ratios reflect a global history of diversification and range expansion in marine bivalves. Proc. R. Soc. B Biol. Sci. 275, 1117–1123. doi: 10.1098/rspb.2007.1729
Landeiro, V. L., Bini, L. M., Costa, F. R. C., Franklin, E., Nogueira, A., de Souza, J. L. P., et al. (2012). How far can we go in simplifying biomonitoring assessments? An integrated analysis of taxonomic surrogacy, taxonomic sufficiency and numerical resolution in a megadiverse region. Ecol. Indic. 23, 366–373. doi: 10.1016/j.ecolind.2012.04.023
LeClaire, A. M., Powell, E. N., Mann, R., Hemeon, K. M., Pace, S. M., Sower, J. R., et al. (2022). Historical biogeographic range shifts and the influence of climate change on ocean quahogs (Arctica islandica) on the Mid-Atlantic Bight. Holocene 32, 964–976. doi: 10.1177/09596836221101275
Leonard-Pingel, J. S., Kidwell, S. M., Tomašových, A., Alexander, C. R., and Cadien, D. B. (2019). Gauging benthic recovery from 20th-century pollution on the southern California continental shelf using bivalves from sediment cores. Mar. Ecol. Prog. Ser. 615, 101–119. doi: 10.3354/meps12918
Leshno, Y., Edelman-Furstenberg, Y., Mienis, H., and Benjamini, C. (2015). Molluscan live and dead assemblages in an anthropogenically stressed shallow-shelf: Levantine margin of Israel. Palaeogeogr. Palaeoclimatol. Palaeoecol. 433, 49–59. doi: 10.1016/j.palaeo.2015.05.008
Lie, U., and Evans, R. A. (1973). Long-term variability in the structure of subtidal benthic communities in Puget Sound, Washington, USA. Mar. Biol. 21, 122–126. doi: 10.1007/BF00354608
Lloyd, G. T., Young, J. R., and Smith, A. B. (2012). Taxonomic structure of the fossil record is shaped by sampling bias. Syst. Biol. 61, 80–89. doi: 10.1093/sysbio/syr076
Long, E. R., and Chapman, P. M. (1985). A sediment quality triad: Measures of sediment contamination, toxicity and infaunal community composition in Puget Sound. Mar. Pollut. Bull. 16, 405–415. doi: 10.1016/0025-326X(85)90290-5
MacCready, P., McCabe, R. M., Siedlecki, S. A., Lorenz, M., Giddings, S. N., Bos, J., et al. (2021). Estuarine circulation, mixing, and residence times in the Salish Sea. J. Geophys. Res. Oceans 126:e2020JC016738. doi: 10.1029/2020JC016738
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220.
Martinelli, J. C., Madin, J. S., and Kosnik, M. A. (2016). Dead shell assemblages faithfully record living molluscan assemblages at One Tree Reef. Palaeogeogr. Palaeoclimatol. Palaeoecol. 457, 158–169. doi: 10.1016/j.palaeo.2016.06.002
Meadows, C. A., Grebmeier, J. M., and Kidwell, S. M. (2019). High-latitude benthic bivalve biomass and recent climate change: Testing the power of live-dead discordance in the Pacific Arctic. Deep Sea Res. Part II Top. Stud. Oceanogr. 162, 152–163. doi: 10.1016/j.dsr2.2019.04.005
Mellin, C., Delean, S., Caley, J., Edgar, G., Meekan, M., Pitcher, R., et al. (2011). Effectiveness of biological surrogates for predicting patterns of marine biodiversity: A global meta-analysis. PLoS One 6:e20141. doi: 10.1371/journal.pone.0020141
Moore, S. K., Mantua, N. J., Newton, J. A., Kawase, M., Warner, M. J., and Kellogg, J. P. (2008). A descriptive analysis of temporal and spatial patterns of variability in Puget Sound oceanographic properties. Estuar. Coast. Shelf Sci. 80, 545–554. doi: 10.1016/j.ecss.2008.09.016
Moreno, C. E., Sánchez-Rojas, G., Pineda, E., and Escobar, F. (2007). Shortcuts for biodiversity evaluation: A review of terminology and recommendations for the use of target groups, bioindicators and surrogates. Int. J. Environ. Health 1, 71–86. doi: 10.1504/IJENVH.2007.012225
Moritz, C., Brandl, S. J., Rouzé, H., Vii, J., Pérez-Rosales, G., Bosserelle, P., et al. (2021). Long-term monitoring of benthic communities reveals spatial determinants of disturbance and recovery dynamics on coral reefs. Mar. Ecol. Prog. Ser. 672, 141–152. doi: 10.3354/meps13807
Mueller, M., Pander, J., and Geist, J. (2013). Taxonomic sufficiency in freshwater ecosystems: Effects of taxonomic resolution, functional traits, and data transformation. Freshw. Sci. 32, 762–778. doi: 10.1899/12-212.1
Nichols, F. H. (2003). Interdecadal change in the deep Puget Sound benthos. Hydrobiologia 493, 95–114. doi: 10.1023/A:1025453700512
Olsgard, F., Brattegard, T., and Holthe, T. (2003). Polychaetes as surrogates for marine biodiversity: Lower taxonomic resolution and indicator groups. Biodivers. Conserv. 12, 1033–1049. doi: 10.1023/A:1022800405253
Olsgard, F., and Somerfield, P. J. (2000). Surrogates in marine benthic investigations – Which taxonomic unit to target? J. Aquat. Ecosyst. Sress Recovery 7, 25–42. doi: 10.1023/A:1009967313147
Olszewski, T. D. (2004). A unified mathematical framework for the measurement of richness and evenness within and among multiple communities. Oikos 104, 377–387. doi: 10.1111/j.0030-1299.2004.12519.x
Olszewski, T. D., and Kaufman, D. S. (2015). Tracing burial history and sediment recycling in a shallow estuarine setting (Copano Bay, Texas) using postmortem ages of the bivalve Mulinia lateralis. Palaios 30, 224–237. doi: 10.2110/palo.2014.063
Pandolfi, J. M. (2001). Numerical and taxonomic scale of analysis in paleoecological data sets: Examples from neo-tropical pleistocene reef coral communities. J. Paleontol. 75, 546–563. doi: 10.1666/0022-336020010752.0.CO;2
Partridge, V., Weakland, S., Dutch, M., Burgess, D., and Eagleston, A. (2018). Sediment quality in Puget Sound: Changes in chemical contaminants and invertebrate communities at 10 sentinel stations, 1989-2015. Lacey, WA: Washington State Department of Ecology Publication.
Pik, A. J., Oliver, I., and Beattie, A. J. (1999). Taxonomic sufficiency in ecological studies of terrestrial invertebrates. Austral Ecol. 24, 555–562. doi: 10.1046/j.1442-9993.1999.01003.x
Pitacco, V., Mistri, M., Aleffi, I. F., Lardicci, C., Prato, S., Tagliapietra, D., et al. (2019). The efficiency of taxonomic sufficiency for identification of spatial patterns at different scales in transitional waters. Mar. Environ. Res. 144, 84–91. doi: 10.1016/j.marenvres.2019.01.001
Powell, E. N., Kuykendall, K. M., and Moreno, P. (2017). The death assemblage as a marker for habitat and an indicator of climate change: Georges Bank, surfclams and open quahogs. Cont. Shelf Res. 142, 14–31. doi: 10.1016/j.csr.2017.05.008
Pruden, M. J., Dietl, G. P., Handley, J. C., and Smith, J. A. (2021). Using molluscs to assess ecological quality status of soft-bottom habitats along the Atlantic coastline of the United States. Ecol. Indic. 129:107910. doi: 10.1016/j.ecolind.2021.107910
Puget Sound Estuary Program [PSEP] (1987). Recommended protocols for sampling and analyzing subtidal benthic macroinvertebrate assemblages in Puget Sound. Bellevue, WA: Tetra Tech, Inc.
R Core Team (2022). R: A language and environment for statistical computing. R foundation for statistical computing. Vienna: R Core Team.
Rojas, A., Calatayud, J., Kowalewski, M., Neuman, M., and Rosvall, M. (2021). A multiscale view of the phanerozoic fossil record reveals the three major biotic transitions. Commun. Biol. 4, 309. doi: 10.1038/s42003-021-01805-y
SCAMIT (2021). A taxonomic listing of benthic macro- and megainvertebrates from infauanal and epifaunal monitoring and research programs in the Southern California Bight, 13th Edn, eds D. B. Cadien, L. L. Lovell, K. L. Barwick, and B. M. Haggin (San Diego, CA: Southern California Association of Marine Invertebrate Taxonomists).
Schopf, T. J. (1978). Fossilization potential of an intertidal fauna: Friday Harbor, Washington. Paleobiology 4, 261–270. doi: 10.1017/S0094837300005996
Sepkoski, J. J. (1981). A factor analytic description of the phanerozoic marine fossil record. Paleobiology 7, 36–53. doi: 10.1017/S0094837300003778
Somerfield, P. J., and Clarke, K. R. (1995). Taxonomic levels, in marine community studies, revisited. Mar. Ecol. Prog. Ser. 127, 113–119. doi: 10.3354/meps127113
Souza, J. L. P., Baccaro, F. B., Landeiro, V. L., Franklin, E., Magnusson, W. E., Pequeno, P. A. C. L., et al. (2016). Taxonomic sufficiency and indicator taxa reduce sampling costs and increase monitoring effectiveness for ants. Divers. Distib. 22, 111–122. doi: 10.1111/ddi.12371
St-Pierre, A. P., Shikon, V., and Schneider, D. C. (2018). Count data in biology – Data transformation or model reformation? Ecol. Evol. 8, 3077–3085. doi: 10.1002/ece3.3807
Staff, G. M., Stanton, R. J. Jr., Powel, E. N., and Cummins, H. (1986). Time-averaging, taphonomy, and their impact on paleocommunity reconstruction: Death assemblages in Texas Bays. Geol. Soc. Am. Bull. 97, 428–443. doi: 10.1130/0016-7606198697<428:TTATIO<2.0.CO;2
Steger, J., Bosnjak, M., Belmaker, J., Galil, B. S., Zuschin, M., and Albano, P. G. (2022). Non-indigenous molluscs in the Eastern Mediterranean have distinct traits and cannot replace historic ecosystem functioning. Glob. Ecol. Biogeogr. 31, 89–102. doi: 10.1111/geb.13415
Thorne, R. S. J., Williams, W. P., and Cao, Y. (1999). The influence of data transformations on biological monitoring studies using macroinvertebrates. Water Res. 33, 343–350. doi: 10.1016/S0043-1354(98)00247-4
Timms, L. L., Bowden, J. J., Summersville, K. S., and Buddle, C. M. (2013). Does species-level resolution matter? Taxonomic sufficiency in terrestrial arthropod studies. Insect Conserv. Divers. 6, 453–462. doi: 10.1111/icad.12004
Todd, P. A., Heery, E. C., Loke, L. H., Thurstan, R. H., Kotze, D. J., and Swan, C. (2019). Towards an urban marine ecology: Characterizing the drivers, patterns and processes of marine ecosystems in coastal cities. Oikos 128, 1215–1242. doi: 10.1111/oik.05946
Tomašových, A., and Kidwell, S. M. (2009). Preservation of spatial and environmental gradients by death assemblages. Paleobiology 35, 119–145. doi: 10.1666/07081.1
Tomašových, A., and Kidwell, S. M. (2017). Nineteenth-century collapse of a benthic marine ecosystem on the open continental shelf. Proc. R. Soc. B Biol. Sci. 284:20170328. doi: 10.1098/rspb.2017.0328
Tweedley, J. R., Warwick, R. M., Clarke, K. R., and Potter, I. C. (2014). Family-level AMBI is valid for use in the north-eastern Atlantic but not for assessing the health of microtidal Australian estuaries. Estuar. Coast. Shelf Sci. 141, 85–96. doi: 10.1016/j.ecss.2014.03.002
Tweitmann, A., and Dietl, G. P. (2018). Live-dead mismatch of molluscan assemblages indicates disturbance from anthropogenic eutrophication in the Barnegat Bay-Little Egg Harbor estuary. J. Shellfish Res. 37, 615–624. doi: 10.2983/035.037.0314
Tyler, C. L., and Kowalewski, M. (2017). Surrogate taxa and fossils as reliable proxies of spatial biodiversity patterns in marine benthic communities. Proc. R. Soc. B Biol. Sci. 284:20162839. doi: 10.1098/rspb.2016.2839
Underwood, A. J. (1991). Beyond BACI: Experimental designs for detecting human environmental impacts on temporal variations in natural populations. Mar. Freshw. Res. 42, 569–587. doi: 10.1071/MF9910569
Underwood, A. J. (1994). On beyond BACI: Sampling designs that might reliably detect environmental disturbances. Ecol. Appl. 4, 3–15. doi: 10.2307/1942110
Vijapure, T., and Sukumaran, S. (2019). Optimization of the taxonomic resolution of an indicator taxon for cost-effective ecological monitoring: Perspectives from a heterogeneous tropical coastline. J. Environ. Manage. 247, 474–483. doi: 10.1016/j.jenvman.2019.05.154
Waldbusser, G. G., Powell, E. N., and Mann, R. (2013). Ecosystem effects of shell aggregations and cycling in coastal waters: An example of Chesapeake Bay oyster reefs. Ecology 94, 895–903. doi: 10.1890/12-1179.1
Warwick, R. M. (1988). The level of taxonomic discrimination required to detect pollution effects on marine benthic communities. Mar. Pollut. Bull. 19, 259–268. doi: 10.1016/0025-326X(88)90596-6
Weakland, S., Partridge, V., and Dutch, M. (2018). Sediment quality in Puget Sound: Changes in chemistry, toxicity, and benthic invertebrates at multiple geographic scales, 1989-2015. Lacey, WA: Washington State Department of Ecology Publication.
Włodarska-Kowalczuk, M., and Kêdra, M. (2007). Surrogacy in natural patterns of benthic distribution and diversity: Selected taxa versus lower taxonomic resolution. Mar. Ecol. Prog. Ser. 351, 53–63. doi: 10.3354/meps07127
Yanes, Y. (2012). Anthropogenic effect recorded in the live-dead compositional fidelity of land snail assemblages from San Salvador Island, Bahamas. Biodivers. Conserv. 21, 3445–3466. doi: 10.1007/s10531-012-0373-4
Keywords: taxonomic sufficiency, taxonomic subsets, data transformation, death assemblage, marine, paleoecology, Puget Sound
Citation: Kokesh BS, Burgess D, Partridge V, Weakland S and Kidwell SM (2022) Living and dead bivalves are congruent surrogates for whole benthic macroinvertebrate communities in Puget Sound. Front. Ecol. Evol. 10:980753. doi: 10.3389/fevo.2022.980753
Received: 28 June 2022; Accepted: 27 September 2022;
Published: 07 November 2022.
Edited by:
Gregory P. Dietl, Cornell University, United StatesReviewed by:
Michal Kowalewski, University of Florida, United StatesCopyright © 2022 Kokesh, Burgess, Partridge, Weakland and Kidwell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Broc S. Kokesh, Ymtva2VzaEB1Y2hpY2Fnby5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.