- 1Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Department of Ocean Science, The Hong Kong University of Science and Technology, Hong Kong, Hong Kong SAR, China
- 4Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 5X-STAR, Japan Agency for Marine-Earth Science and Technology (JAMSTEC), Yokosuka, Japan
- 6Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China
- 7British Antarctic Survey, Cambridge, United Kingdom
- 8Department of Infectious Diseases and Public Health, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
Gastropoda is the most speciose class in Mollusca, the second largest animal phylum. The internal relationships of major gastropod groups remain largely unsettled, partly due to the insufficient data from key deep-water endemic lineages such as the subclass Neomphaliones. Neomphaliones currently includes two orders: Cocculinida, best known from sunken wood habitats, and Neomphalida, best known from hydrothermal vents and often referred to as the “hot vent clade.” Phylogenetic controversy has also been observed in this subclass across different studies, requesting additional investigations. Here, we assembled nine new mitogenomes from two Cocculinida and seven Neomphalida species and analyzed them with published gastropod mitogenomes, with a particular focus on Neomphaliones. The phylogenetic reconstruction of Gastropoda based on 13 mitochondrial protein-coding genes resulted in a topology largely congruent with previous reconstructions based on morphological characters. Furthermore, we recovered characteristic mitochondrial gene order arrangements of Cocculinida and Neomphalida compared to the hypothetical ancestral gastropod gene order, at a level similar to other subclass-level clades. Divergence time estimation showed that Cocculinida and Neomphalida diverged approximately 322.68 million years ago. In addition to characteristic gene order arrangements for the clade, Cocculinida mitogenomes also exhibit some minor rearrangements even among congeners. Within Neomphalida, our tree adds support to monophyletic Peltospiridae and Neomphalidae, with unique gene arrangement recovered for each family. Our results offer new insights into the rearrangement of mitogenomes in Gastropoda, providing another clue to the evolutionary history of gastropods.
Introduction
Mollusca is the second largest animal phylum consisting of eight living classes, among which Gastropoda (snails, limpets, slugs, etc.) is the most speciose, containing approximately 80% of the known species. Gastropoda is currently divided into six major subclass-level clades: Caenogastropoda, Heterobranchia, Neritimorpha, Patellogastropoda, Vetigastropoda, and Neomphaliones (Bouchet et al., 2017). The high species diversity of Gastropoda is linked to its colonization of a wide range of habitats from terrestrial ecosystems to the deepest parts of the ocean, including “extreme” environments such as hydrothermal vents and underground caves. Studying the speciation process is very helpful for our understanding of the biological adaptation of gastropods to “extreme” deep-water environments, for which phylogenetic analyses provides important insights.
Neomphaliones is a key deep-water clade of gastropods, whose taxonomy and systematics is being debated. Some have treated it as a part of Vetigastropoda (Ponder et al., 2020; Uribe et al., 2022a) while others consider the two orders within Neomphaliones—Cocculinida, and Neomphalida—should each be treated at the subclass-level to make a total of seven subclasses (Cunha and Giribet, 2019; Uribe et al., 2019). Even more controversy is present in the phylogenetic relationships among these subclasses, though intensive efforts based on morphology data, nuclear genes, and mitochondrial genes have been carried out, the topologies of phylogenetic reconstructions based on different data have been largely inconsistent (Kocot et al., 2011; Zapata et al., 2014; Cunha and Giribet, 2019; Uribe et al., 2022a). For example, in many morphological analyses, Patellogastropoda has been suggested to be the earliest-diverging group sister to the rest of Gastropoda (Orthogastropoda) (Ponder and Lindberg, 2008). However, Patellogastropoda has been recovered as the sister group to Vetigastropoda based on nuclear genes (Zapata et al., 2014) and sister to Heterobranchia based on some mitochondrial gene phylogenies (Arquez et al., 2014; Osca et al., 2014; Uribe et al., 2016b), although a recent mitogenome phylogeny has recovered Heterobranchia sister to Caenogastropoda, which agrees with morphology (Uribe et al., 2019).
This inconsistency is largely caused by the paucity of sampling in certain key lineages (Sigwart et al., 2021), particularly the deep-water subclass Neomphaliones, including Cocculinida best known from biogenic subtracts and the “hot vent” clade Neomphalida restricted to chemosynthetic habitats. Further molecular data from these deep-water groups is key to convincingly resolve the relationships among the major gastropod lineages. Even within Neomphaliones, the phylogenetic relationships among genera are far from resolved. For example, in Neomphalida, phylogenetic constructions have often used the partial COI gene, which often failed to recover a monophyly of Peltospiridae. The Scaly-foot Snail Chrysomallon squamiferum is often recovered closer to Neomphalidae than Peltospiridae in COI trees, but was placed within Peltospiridae in a five-gene phylogeny (Chen et al., 2015, 2017). A recent study using the published mitogenome of the neomphalid Lamellomphalus manusensis also indicated Peltospiridae to be non-monophyletic (Zhang and Zhang, 2022). However, these issues could be generated by poor taxon sampling or inadequate methodology (only using a single tree and without gene order analysis) in their analyses.
The animal mitochondrial genome is a circular double-stranded DNA molecule, which typically encodes for 13 protein-coding genes, 22 tRNAs and 2 rRNAs. The mitochondrial genomes of gastropods exhibit a high diversity in gene order rearrangements (Kurabayashi and Ueshima, 2000), which providing a suitable approach for exploring the pattern of mitochondrial genome rearrangement mechanisms and the phylogenetic value of the gene arrangement patterns. The need for more complete mitogenomes is thus of great importance in expanding our knowledge on the phylogeny of this group.
To widen the coverage of Neomphaliones in order to better resolve relationships of major clades in Gastropoda as well as groups within Neomphaliones, we assembled nine new mitochondrial genomes from this subclass. This includes two members of Cocculinida (currently including approximately 60 species), Cocculina enigmadonta and Cocculina tenuitesta, as well as seven members of Neomphalida (currently including approximately 50 species) (Sigwart et al., 2021). We also re-annotated some existing gastropod mitogenomes, including Bathysciadiidae indet. (sensu Lee et al., 2019), Cocculina subcompressa and Coccocrater sp. sensu in Cocculinida (Lee et al., 2019), as well as Peltospira smaragdina in Neomphalida (Collins et al., 2020). The evolution of mitochondrial gene order arrangements within Gastropoda was analyzed in order to gain insights into rates and mechanisms of mitogenome rearrangement in this group. To determine rearrangement events and evaluate the phylogenetic utility of mitochondrial gene sequence comparisons, the gene order of gastropod mitogenomes was further mapped to the reconstructed phylogeny.
Materials and methods
Sampling, sequencing, and data curation
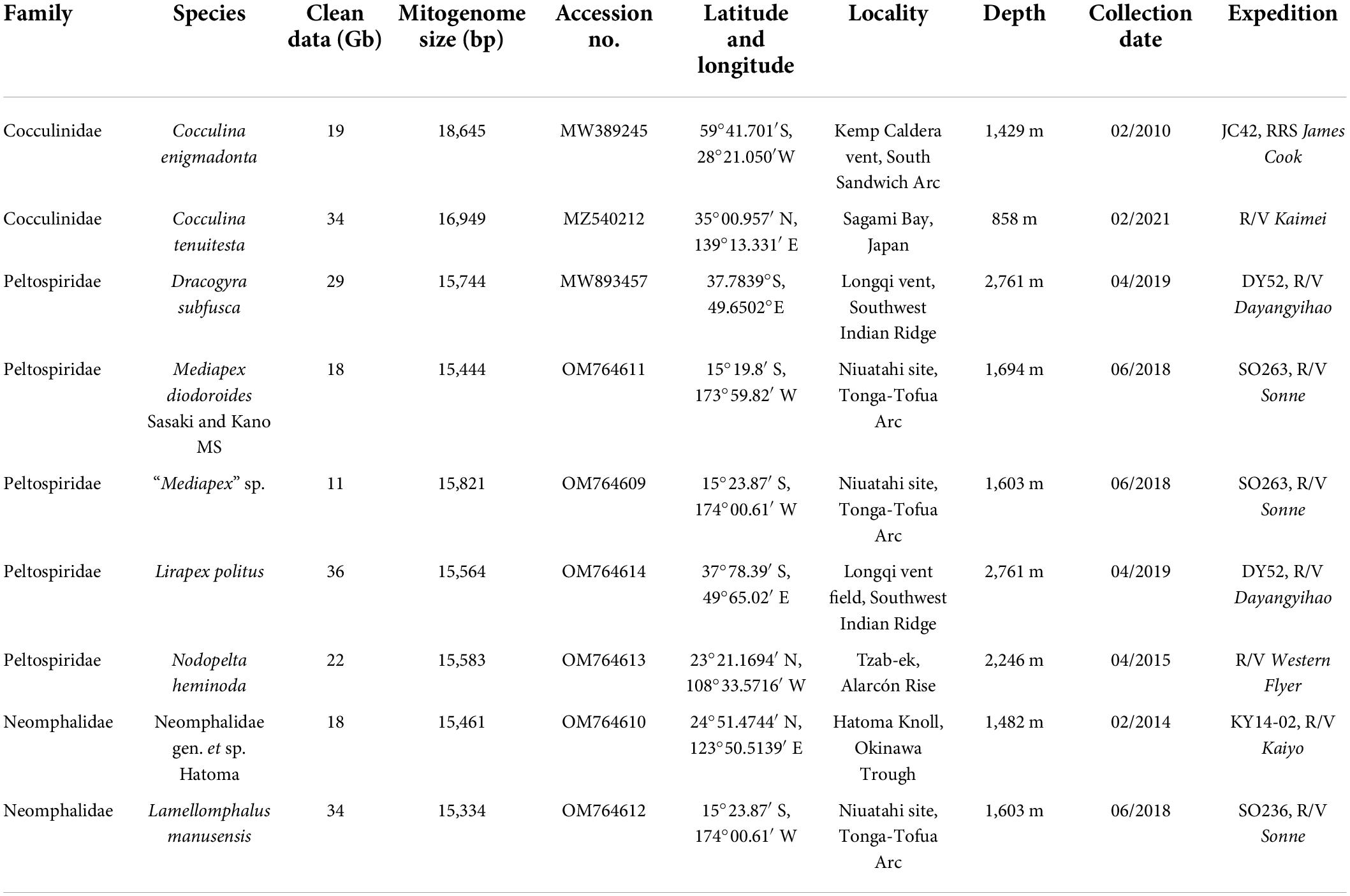
Cocculina enigmadonta was collected from the Kemp Caldera vent site in the Weddell Sea during the expedition JC42 of RRS James Cook and preserved in 96% ethanol (Chen and Linse, 2020), and Cocculina tenuitesta was collected from a piece of plant debris off Hatsushima Sagami Bay, Japan. Mediapex diodoroides Sasaki and Kano MS (sensu Heß et al., 2008; note that the genus Mediapex as well as this species remain undescribed to date and are nomen nodum) was collected from Niuatahi vent site, Tonga-Tofua Arc and a congener, “Mediapex” (sensu Heß et al., 2008) sp. was also collected from the same site. Lirapex politus was collected from Longqi hydrothermal vent field, Southwest Indian Ridge. Nodopelta heminoda was collected from Tzab-ek vent on the Alarcon Rise. Neomphalidae gen. et sp. Hatoma was collected from Hatoma Knoll, Okinawa Trough. Lamellomphalus manusensis was collected from Niuatahi vent site, Tonga-Tofua Arc. The sample information and the corresponding NCBI accession numbers are listed in Table 1.
Genomic DNA of these eight species was extracted using DNeasy Blood and Tissue Kit (Qiagen, Germany) following the manufacturer’s instructions. Genomic DNA was shotgun fragmented, and the library was prepared with NEBNext® Ultra™ DNA Library Prep Kit for Illumina (NEB) and paired-end sequenced on an Illumina Novaseq platform with the read length of 150 bp.
For Dracogyra subfusca, the published genome assembly (Lan et al., 2021) using a specimen collected in Longqi vent field (“Tiamat” chimney), Southwest Indian Ridge did not contain the complete mitogenome. Therefore, the raw sequencing reads of D. subfusca was downloaded from the NCBI SRA database in order to assemble its mitogenome anew.
Assembly and mitogenome annotation
Potential prokaryotic contamination was removed by aligning the Illumina reads with a curated prokaryotic dataset (MiniKraken2 v1) by Kraken2 version 2.0.8. After that, adapters and low-quality reads were removed by Trimmomatic version 0.39 with settings of “LEADING: 10 TRAILING: 10 SLIDINGWINDOW: 4:15, and MINLEN: 40.” The clean reads were used for mitogenome assembly by SPAdes version 3.14.1 (Bankevich et al., 2012) with default settings. To check the assembly accuracy, particularly in the regions with duplicated tRNA or missing coding gene, additional assembly were also generated by one of the three following assemblers: NOVOPlasty version 4.3.31 with the cox1 gene sequence generated from the former SPAdes assembly severing as the “seed” and “genome range” set as 12,000-22,000 (Dierckxsens et al., 2017), MaSuRCA version 4.0.3 (settings: “USE_LINKING_MATES = 1 FLYE_ASSEMBLY = 1”) (Zimin et al., 2013), and ABySS version 2.3.0 (settings: “abyss-pe k=31”) (Simpson et al., 2009).
Mitochondrial genomes were annotated by MITOS version 2.0.6 (Bernt et al., 2013). By comparing the published mitochondrial gene sequence of Gastropoda, the positions of 13 protein-coding genes (PCGs) and two rRNA genes were corrected manually. Then, tRNA gene sequences were confirmed by tRNAscan-SE 2.0 (Lowe and Eddy, 1997). Moreover, we re-annotated other relevant gastropod mitogenomes from NCBI that lacked certain gene order information, including Bathysciadiidae indet. (sensu Lee et al., 2019), Cocculina subcompressa, and Coccocrater sp. (sensu Lee et al., 2019) in Cocculinida, P. smaragdina in Neomphalida (Collins et al., 2020), as well as Cellana radiata (Uribe et al., 2019) in Patellogastropoda using the same strategy (for details see Supplementary Tables 1–6).
Phylogenetic analysis
We used 29 gastropods and two outgroup non-gastropod mollusks (the polyplacophoran Cryptochiton stelleri and the cephalopod Octopus bimaculatus) in our phylogenetic analyses. Nucleotide sequences of the 13 PCGs were translated into amino acid sequences using the code “invertebrate mitochondria.” The amino acid sequences of the 13 PCGs were aligned separately by MUSCLE v3.8.31. Poor alignments were clipped using trimAl v1.4.1 with the setting of “automated1,” optimized for maximum likelihood (ML) tree analysis. Afterward, the resulting alignments were concatenated for downstream ML and Bayesian inference (BI) phylogenetic analyses.
We applied both site-homogenous and site-heterogenous methods for phylogenetic interference. Specifically, ML analysis was conducted by IQ-TREE version 2.0.6 (Nguyen et al., 2014) with the setting of “-m MFP” which would select the optimal models for each partition. The model of mtZOA + Γ4 was selected for cox2, atp6, atp8, nad1, nad4l, and nad6 genes, mtZOA + Γ3 for cox1 gene, mtZOA + I + Γ4 for cox3, cob and nad3 genes, mtInv + Γ4 for nad2 gene, and mtART + I + Γ4 for nad4 and nad5 genes.
Site-heterogenous analysis was suggested to be effective in terms of overcoming the Long Branching Attraction (LBA) issue, defined as the clustering of rapidly evolving taxa in a tree regardless of their true phylogenetic positions, suggested to be common in the gastropod mitogenome phylogeny (Uribe et al., 2019). In the site-heterogenous analysis, ML tree was constructed by IQ-TREE version 2.0.6 with the setting of “–mset mtZOA + Γ4 + C10, mtZOA + Γ4 + C20, mtZOA + Γ4 + C30, mtZOA + Γ4 + C40, mtZOA + Γ4 + C50, and mtZOA + Γ4 + C60” to select the optimal models on each partition, since the mtZOA + Γ4 model was suggested to be the best-fitting model for the most of the partitions in the former analysis.
Another round of ML reconstruction was performed by RAxML version 8.2.12 with the model of CAT-GTR on each partition. The BI analysis was carried out by PhyloBayes-MPI version 1.8 with the model of CAT-GTR on each partition. The subclass Heterobranchia is known to suffer particularly from LBA in mitogenome phylogenies (Uribe et al., 2019), resulting in systematic errors in phylogenetic reconstruction (Bergsten, 2005). After an initial reconstruction (Supplementary Figure 1), we found a poorly supported position for Heterobranchia, which is probably a result of LBA regardless of our efforts to overcome it (as discussed in Uribe et al., 2019). Therefore, we only used the other five subclasses (Bouchet et al., 2017) for downstream phylogenetic reconstruction and analyses.
Divergence times of the major gastropod clades were predicted using MCMCTREE (Reis and Yang, 2011) implemented in PAML version 4.8. The burn-in, sample frequency, and number of samples were set to 10,000, 1,000, and 10,000,000, respectively (Sun et al., 2021). The fossil records and geological events used for time calibration were as follows: (1) the split between Cellana and Nacella at a hard minimum age of 38 Ma (Nakano and Ozawa, 2004); (2) the divergence between Caenogastropoda and other subclasses at a hard minimum age of 390 Ma (Jörger et al., 2010); (3) the first appearance of mollusks between a hard minimum age of 532 Ma and a hard maximum age of 549 Ma (Benton et al., 2015); and (4) the split between Cocculinida and Neomphalida at a hard minimum age of 145 Ma, since the first Neomphalione fossil is known from the Late Jurassic (Kaim et al., 2014); (5) the split between Peltospiridae and Neomphalidae at a hard minimum age of 132.9 Ma, as the earliest good confidence fossil record of Neomphalidae dates back to at least the Early Cretaceous (Valanginian) (Kiel and Campbell, 2005; Campbell et al., 2008).
Gene order analysis and pairwise genetic distance analyses
Considering reversals, reverse transpositions, transpositions and tandem-duplication-random-loss which would shape the mitogenome gene orders, CREx (Common interval Rearrangement EXplorer) (Bernt et al., 2007) was used to analyze each Neomphalione mitochondrial genome vs. the ancestral gastropod mitogenome gene order. In CREx, higher CREx intervals indicate more similar gene orders. Pairwise genetic distance was calculated between Neomphaliones and other subclasses using MEGA X (Kumar et al., 2018). Pearson correlation coefficient between genetic distance and corresponding mitochondrial gene rearrangement was analyzed in R v. 4.0.3.
Amino acid usage analysis
Nucleotide sequences of the 13 PCGs from the 29 gastropods, where stop codons were excluded, were translated into protein sequences using MEGA X (Kumar et al., 2018). Afterward, the proportion of 20 amino acids in each gastropod was counted in MEGA X, arcsine transformed, and Student’s t-test was used to calculate the bias in amino acid usage between Neomphaliones and the other subclasses.
Results
Assembly and characterization of the newly sequenced mitogenome
Approximately 24 Gb of reads were sequenced for Cocculina enigmadonta, 36 Gb reads for Cocculina tenuitesta, 53 Gb reads for Dracogyra subfusca were downloaded from the NCBI SRA database, 20 Gb reads for Mediapex diodoroides Sasaki and Kano MS, 16 Gb reads for “Mediapex” sp., 40 Gb reads for Lirapex politus, 26 Gb reads for N. heminoda, 20 Gb reads for Neomphalidae gen. et sp. Hatoma, and 38 Gb reads for Lamellomphalus manusensis, with 80, 94, 55, 90, 69, 90, 85, 90, and 89% of reads being retained for C. enigmadonta, C. tenuitesta, D. subfusca, M. diodoroides Sasaki and Kano MS, “Mediapex” sp., Lirapex politus, N. heminoda, Neomphalidae gen. et sp. Hatoma, and Lamellomphalus manusensis after removing potential microbe contamination, low-quality, and adapter-contaminated reads. Taking an assumption that the genome size is similar to Gigantopelta aegis (1.27 Gb; Lan et al., 2021), a relatively large genome within Neomphaliones, the sequencing coverage of the genome is at least 12.5X.
The above eight mitogenomes (except Lirapex politus) contained 2 rRNA genes (rrnS and rrnL) and 13 protein-coding genes (cox1, cox2, cox3, cob, nad1, nad2, nad3, nad4, nad4l, nad5, nad6, atp6, and atp8). The mitogenome Lirapex politus. possessed only 12 protein-coding genes with a missing cox2, which were further validated by assembly generated by NOVOPlasty version 4.3.31. Furthermore, there were the 22 standard tRNA genes in D. subfusca, M. diodoroides Sasaki and Kano MS, “Mediapex” sp., Lirapex politus, N. heminoda, Neomphalidae gen. et sp. Hatoma, and Lamellomphalus manusensis. The mitogenome of C. enigmadonta also possessed 22 tRNA genes but with a missing of trnQ and a duplication of trnG, which were further validated by the results generated from another assembly pipeline MaSuRCA version 4.0.3. The 23 tRNA genes for C. tenuitesta, which included two tandem duplicates of trnA, were supported by the results yielding from ABySS version 2.3.0. There were 16 genes located on the heavy strand and 21 genes on the light strand in C. enigmadonta, 15 genes were on the heavy strand and 21 genes on the light strand in C. tenuitesta, 17 genes located on the heavy strand and 20 genes on the light strand in D. subfusca, M. diodoroides Sasaki and Kano MS, “Mediapex” sp., N. heminoda, Neomphalidae gen. et sp. Hatoma, and Lamellomphalus manusensis, and 17 genes located on the heavy strand and 19 genes on the light strand in Lirapex politus. For the 13 PCGs, the start codons were ATG, ATT, ATA, ATC, TTG, and GTG for C. enigmadonta; ATG, ATT, TTG, and GTG for C. tenuitesta; ATA, ATG, TTG, and GTG for D. subfusca; ATG, ATT, and ATC for M. diodoroides Sasaki and Kano MS; ATG and ATT for “Mediapex” sp.; ATG, ATA, TTG, and GTG for Lirapex politus; ATG, ATT, ATA, GTG for N. heminoda; ATG, ATT, and ATA for Neomphalidae gen. et sp. Hatoma and Lamellomphalus manusensis. Similarly, the stop codons were TAG and TAA for C. enigmadonta, M. diodoroides Sasaki and Kano MS, Neomphalidae gen. et sp. Hatoma and Lamellomphalus manusensis; while they were TAG, TAA, and T(AA) for C. tenuitesta, D. subfusca, “Mediapex” sp., Lirapex politus, and N. heminoda.
Phylogenetic relationships of Gastropoda based on the 13 protein-coding genes
Phylogenetic relationships of gastropod species in the five subclass-level clades were reconstructed using the 13 PCGs (Figure 1) with a total of 2,409 aligned amino acids (lacking nad4 and nad4l for Coccocrater sp. sensu Lee et al., 2019). Phylogenetic trees constructed using the four different methods are shown in Supplementary Figure 2. The gastropod species included in the analyses were clustered in agreement with their known systematic and taxonomic affinities at order or subclass-levels. Specifically, monophyly of all five subclasses were fully supported across the four distinct methods (IQ-TREE site-homogenous method, IQ-TREE site-heterogenous method, RAxML, and PhyloBayes). In the phylogenetic tree (Figure 1), Patellogastropoda was recovered sister to the other four subclasses, being the earliest-diverging major clade (not considering Heterobranchia, which was removed from the analysis). Caenogastropoda was supported as the most closely related clade to Neritimorpha (79/95/-/0.81 in four analyses) among the major clades included herein. Within Neomphaliones, a monophyletic Cocculinida was recovered as sister group to the also monophyletic Neomphalida, both with maximum bootstrap support across the four methods. Neomphaliones was in turn recovered sister to Vetigastropoda, although only with moderate support (-/80/99/1).

Figure 1. Time-calibrated phylogeny of major gastropod clades using maximum-likelihood (ML) methods based on 13 mitochondrial protein-coding genes. Age estimates of major divergence events among gastropods (on the top, error bars indicate 95% confidence levels) and node values indicate bootstrap supported by IQ-TREE with the model of MFP and the setting of “–mset mtZOA + Γ4 + C10, mtZOA + Γ4 + C20, mtZOA + Γ4 + C30, mtZOA + Γ4 + C40, mtZOA + Γ4 + C50, mtZOA + Γ4 + C60,” RAxML with the model of CAT-GTR, and posterior probability in PhyloBayes with the model of CAT-GTR, respectively (on the bottom, in bold). Brown dots indicate nodes where all four analyses had full support; Stars indicate fossil calibration points; “−,” unsupported by the corresponding method; Ma, million years ago.
Within Cocculinida, Coccocrater sp. (sensu Lee et al., 2019) was found to be nested within three Cocculina species with good support (100/100/100/0.77) in our analyses. Within Neomphalida, two neomphalids, Lamellomphalus manusensis and Neomphalidae gen. et sp. Hatoma, clustered together to form a clade sister to the eight peltospirids, which formed a monophyletic clade with moderate support (68/52/74/-), interpreted to be Peltospiridae. The Scaly-foot Snail C. squamiferum was recovered sister to all other included species within Peltospiridae. The phylogenetic relationship within Peltospiridae of {[(D. subfusca, G. aegis), (N. heminoda, P. smaragdina)], Lirapex politus} was the same as in a former analysis using partial COI sequence (Chen et al., 2017).
Divergence time analysis
Divergent time estimates (Figure 1) showed that Patellogastropoda diverged at approximately 531.60 million years ago (Ma) (95% confidence level: 548.49–504.58) from the rest of Gastropoda. Vetigastropoda-Neomphaliones diverged from Caenogastropoda-Neritimorpha around 492.91 Ma (95% confidence level: 520.96–460.45); Cocculinida and Neomphalida diverged at 322.68 Ma (95% confidence level: 394.24–251.33). Within Neomphalida, Peltospiridae diverged from Neomphalidae around middle Jurassic at 174.43 Ma (95% confidence level: 232.62–136.29).
Rearrangements of mitochondrial genes
The mitochondrial gene order arrangement of the five subclasses (Figure 2) were compared to the hypothetical ancestral gene order of gastropods (Stöger and Schrödl, 2013; Uribe et al., 2019). Most species in Neritimorpha, Cocculinida, Patellogastropoda, and Vetigastropoda shared an identical gene block of nad5-trnH-nad4-nad4l-trnT-trnS-cob-nad1-trnL-trnL-rnaL-trnV-rnaS, while the majority of species in Neritimorpha, Cocculinida, Caenogastropoda, Neomphalida, and Vetigastropoda shared an identical gene block consisting of cox3-trnK-trnA-trnR-trnN-trnl-nad3-trnS-nad2. The gene order of Neritimorpha was found to be identical to the hypothesized ancestral gastropod mitochondrial genome. The Vetigastropoda gene order closely resembles the ancestral order and only differed by the position of trnE and trnG. Within Patellogastropoda, gene order rearrangements resulted from transpositions of trnM, trnY, trnC, trnW, trnQ, and trnF, as well as the reversal of the trnG-trnE block. The gene order between Caenogastropoda and the hypothesized ancestral gastropod differed by a transposition in the block of trnM-trnY-trnC-trnW-trnQ-trnG-trnE and a reversion in the block of rnaS-trnV-rnaL-trnL-trnL-nad1-trnP-nad6-cob-trnS-trnT-nad4l-nad4-trnH-nad5-trnF.

Figure 2. Comparison of mitochondrial gene rearrangements among main gastropod lineages. Genes for tRNAs are designated by a single letter for the corresponding amino acid. Transposition and inversion are indicated with blue and red box, respectively.
The gene order of Cocculinida differed from the ancestral gastropod order by a transposition of the trnC-trnW block and the reversal of the trnF-atp6-atp8-trnD-cox2 block. However, within Cocculinida, the mitogenome gene order of each species included was found to be unique, with the transposition and even duplication of trnG as well as the transposition trnA in Cocculina tenuitesta. Neomphalida possessed a unique gene order rearrangement with four reversions of trnF, trnG, the two blocks atp6-atp8-trnD-cox2, and trnQ-trnW-trnC-trnY, as well as two transpositions of trnT and trnE. Within Neomphalida, species of Peltospiridae and Neomphalidae shared the same intra-family level mitogenome gene order, but the two families differed by a single transposition of trnT.
Correlation between gene order and pairwise genetic distance and amino acid usage
Pairwise genetic distance and mitochondrial gene rearrangement distance, as revealed by the CREx score, were calculated with the 29 mitogenomes used in the former analysis, and a significant negative correlation was discovered (R = −0.839, P < 0.01). Species in Cocculinida exhibited a general elevated evolutionary rate as well as more extensive gene rearrangements than species in Neomphalida (Figures 3A,B). We also calculated the amino acid usage in the subclass Neomphaliones vs. the other subclasses, and found species in Neomphaliones tend to use more phenylalanine (F), methionine (M), and cystine (C) but less threonine (T), alanine (A), tyrosine (Y), and glutamic acid (E) (Table 2; FDR < 0.05, Student’s t-test).
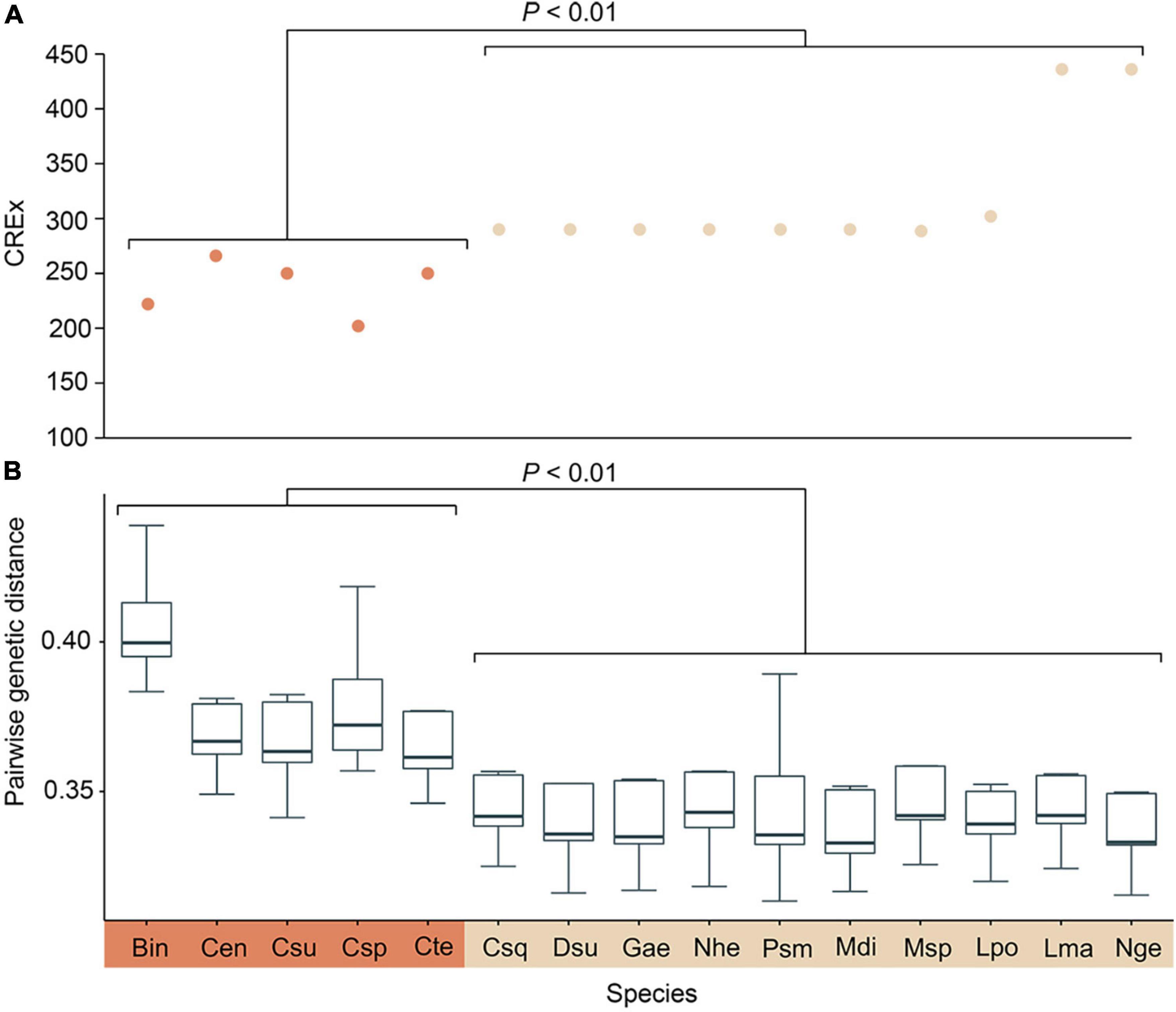
Figure 3. Scatterplot showing the CREx (Common interval Rearrangement EXplorer) score of each Neomphalione mitochondrial genome vs. the ancestral gastropod mitogenome gene order (A) and pairwise genetic distance between Neomphaliones and other subclasses. (B) Statistical differences between these two orders were calculated with Student’s t-test. Bin, Bathysciadiidae indet. (sensu Lee et al., 2019); Cen, Cocculina enigmadonta; Csu, Cocculina subcompressa; Csp, Coccocrater sp. (sensu Lee et al., 2019); Cte, Cocculina tenuitesta; Csq, Chrysomallon squamiferum; Dsu, Dracogyra subfusca; Gae, Gigantopelta aegis; Nhe, Nodopelta heminoda; Psm, Peltospira smaragdina; Mdi, Mediapex diodoroides Sasaki and Kano MS; Msp, “Mediapex” sp.; Lpo, Lirapex politus; Lma, Lamellomphalus manusensis; Nge, Neomphalidae gen. et sp. Hatoma.
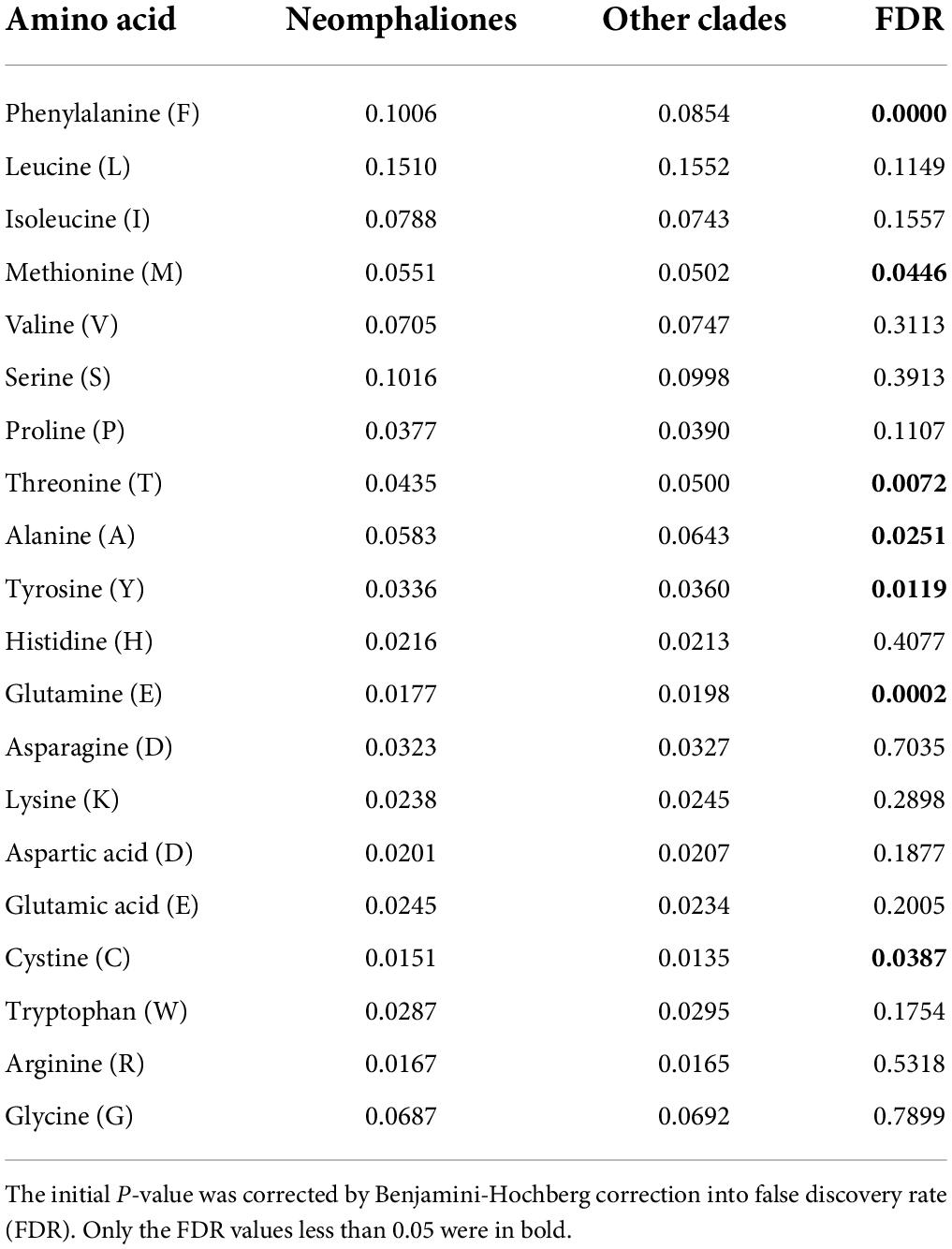
Table 2. Comparisons of average mitochondrial amino acid usage in proportion between Neomphaliones and the other four subclasses included herein.
Discussion
The phylogenetic relationship among the major gastropod lineages has always been conflicting (Cunha and Giribet, 2019; Uribe et al., 2019), with a possible cause being the paucity of sampling efforts in certain deep-water lineages, particularly those within Neomphaliones, for which only three mitogenome studies have been published. We sequenced two complete mitochondrial genomes in Cocculinida, six mitogenomes in Neomphalida, and reassembled the mitogenome of Dracogyra subfusca (also in Neomphalida). Using these data, plus four Neomphaliones mitogenomes from published studies (Lee et al., 2019; Collins et al., 2020), we were able to determine the key mitogenome architecture of the two orders in this subclass (Lee et al., 2019).
It is noted that all five subclasses in Gastropoda included in the analyses herein were found to be monophyletic with maximum support in all four methods, indicating that the assignment of species to each subclass can be done confidently by mitogenome level data. However, the support values of the divergences among these subclasses indicated only moderate support, and additional genes from genome-level studies generated from a broad taxon sampling are required to address phylogenetic issues at the inter-subclass-level (Sun et al., 2020; Uribe et al., 2022a). Even so, the resulting topology from our phylogenetic reconstructions was generally consistent with a previous mitogenome phylogeny (Uribe et al., 2019), a recent genome phylogeny (Uribe et al., 2022a), and data from morphology (Ponder and Lindberg, 2008), lending further support for Patellogastropoda as the earliest-diverging lineage in Gastropoda (Ponder and Lindberg, 2008; Kocot et al., 2011).
We found that Cocculinida and Neomphalida, currently treated as orders within subclass Neomphaliones, were well-separated, with the divergent time going back to late Carboniferous, though with rather sizeable upper and lower confidence intervals. The two families included herein, i.e., Peltospiridae and Neomphalidae, in Neomphalida are suggested to be monophyletic. These results can be further supported by the mitogenome gene order analysis, since these two families have their own unique gene orders but with an inter-family level difference. The monophyly of Peltospiridae, which could not be resolved by partial COI sequencing (Chen et al., 2017) or mitogenome analyses with a limited number of taxa (Zhang and Zhang, 2022), is confirmed in our topology. Within Peltospiridae, the Scaly-foot Snail C. squamiferum was sister to all other peltospirid species included, with its divergent time from other peltospirids dating back to Late Jurassic. The distinctive morphological features of hundreds of chitinous sclerites (Isobe et al., 2022) covering the dorsal foot in Scaly-foot Snail may be linked to its unique phylogeny position. Our mitogenome level molecular phylogeny also lends support to the convergent evolution scenario of endosymbiosis (Chen et al., 2015) in two divergent genera within Peltospiridae, i.e., Chrysomallon and Gigantopelta (Lan et al., 2021), since they are not recovered as sister genera and the other peltospirid snails are asymbiotic (Chen et al., 2015). Within Cocculinida, Coccocrater sp. (sensu Lee et al., 2019) was found to be nested within three Cocculina species, which could indicate either this species is not a Coccocrater (and instead of a Cocculina) or that Cocculina is not monophyletic, as indicated in recent phylogenies of cocculinids (Lee et al., 2022). As no information on the morphology of this species was presented in Lee et al. (2019), this cannot be resolved at present until the mitogenome of another, definitively identified, Coccocrater species is sequenced and analyzed.
Gastropods exhibit a great variety of mitochondrial gene rearrangements among the major lineages, though the organization is thought to be relatively conserved within each lineage (Cunha et al., 2009; Arquez et al., 2014). With broader sampling, the analysis of the correlation between genetic distance and mitochondrial gene rearrangement showed that the evolutionary rate was positively correlated with mitochondrial gene rearrangement, and that the faster the evolutionary rate was, the greater the extent of rearrangement (Figure 3). Lineage-specific mitogenome rearrangements in Gastropoda have also been reported in Lepetellidae and Fissurellidae within Vetigastropoda (Uribe et al., 2022b), Lottidae and Patellidae within Patellogastropoda (Xu et al., 2022), Hydrocenoidea within Neritimorpha (Uribe et al., 2016a), and a number of clades within Caenogastropoda (Osca et al., 2014).
By including additional species, we show that the mitogenome gene orders within Cocculinida, although slightly variable among species, is in general consistent within the clade but different from Neomphalida. The clusters trnC-trnW and trnF-atp6-atp8-trnD-cox2 were unique at the order level in Cocculinida when compared to the hypothetical ancestral gene order of Gastropoda, this unique gene order can therefore be proposed as a molecular synapomorphy. However, no Cocculinida species exhibited identical gene orders, with even the three Cocculina species included having heterogeneous gene orders. Likewise, the evolutionary rate within Cocculinida was inflated compared to Neomphalida, with all Neomphalida species included exhibiting almost identical gene orders with only one transposition of trnT between the two families. The differences in mitogenome organization seen between Cocculinida and Neomphalida is significant, and even comparable to the differences among other subclass-level clades in Gastropoda. However, we should keep in mind that dramatic lineage-specific mitogenome rearrangements are sporadically found in major clades of Gastropoda (Uribe et al., 2022b; Xu et al., 2022).
The mitochondrial gene order of Neritimorpha found herein agrees with previous studies (Castro and Colgan, 2010; Arquez et al., 2014), which is also the same as the hypothetical ancestral gene order of Gastropoda. The representative species of Caenogastropoda used in this study shared the same mitochondrial gene rearrangement (Grande et al., 2008), and the two blocks of trnM-trnY-trnC-trnW-trnQ-trnG-trnE and rnaS-trnV-rnaL-trnL-trnL-nad1-trnP-nad6-cob-trnS-trnT-nad4l-nad4-trnH-nad5-trnF were specific to this subclass. The mitogenome organization of Vetigastropoda is very similar to the ancestral gastropod gene order, as previously acknowledged (Grande et al., 2008). Mitochondrial gene rearrangements of Patellogastropoda include the characteristic trnQ and blocks of trnY-trnM-trnF-trnW-trnC-trnG-trnE, as well as trnR-trnN-nad3-trnA-trnK-trnI. These results collectively indicate that each major clade in Gastropoda, including Cocculinida and Neomphalida, possesses lineage-specific characteristics in their mitogenome (Figure 2). Based on these findings, we consider that the systematic affinity of any particular gastropod species, at order or subclass-level, can be determined with good confidence based on mitogenome organization patterns. Our results offer new insights into the rearrangement of mitogenomes of Gastropoda, as well as provide another clue to untangling the evolutionary history of this diverse group.
Data availability statement
The data presented in this study are deposited in the NCBI repository, accession numbers: MW389245, MZ540212, MW893457, OM764609, OM764610, OM764611, OM764612, OM764613, and OM764614.
Author contributions
JS: study conception and design. CC, JS, YZ, and KL: sample collection. YL: DNA extraction. ZZ, YL, CC, RL, and JS: data analysis and interpretation. ZZ, CC, JS, YL, and KL: manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the National Natural Science Foundation of China (No. 42176110), the Fundamental Research Funds for the Central Universities (Nos. 202172002 and 202241002), the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2022QNLM050102-4), the Young Taishan Scholars Program of Shandong Province (No. tsqn202103036), and the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. SMSEGL20SC02). The ChEsSO consortium and its expedition JC42 were funded by the UK NERC Consortium Grant NE/DO1249X/1.
Acknowledgments
We thank the master and crew of RRS James Cook as well as the pilots and technical teams of ROV Isis on-board the expedition JC42 (cruise PI: Dr. Alex D. Rogers) for their professionalism and support. Fieldwork in the East Scotia Sea during JC42 was carried out under the permit S3-3/2009 issued by the Foreign and Commonwealth Office, London under Section 3 of the Antarctic Act 1994. We are also grateful for the ROV KM-ROV team on-board R/V Kaimei for supporting the sample collection in Sagami Bay. We thank Shannon B. Johnson and Robert C. Vrijenhoek (Monterey Bay Aquarium Research Institute) for providing the specimen of Nodopelta heminoda collected on a cruise funded by the David and Lucile Packard Foundation, and Verena Tunnicliffe (University of Victoria) for providing specimens of “Mediapex” species collected during the R/V Sonne cruise SO263 with ROV Quest “TONGARIFT” (Chief Scientist: Karsten Haase, Friedrich-Alexander University, Germany). We also thank operation team of ROV Sea Dragon III and the crew members on R/V Dayangyihao during the cruise of DY52. Bioinformatics computing was conducted by the high-performance cluster IEMB-1 at Institute of Evolution and Marine Biodiversity. Constructive comments from the three reviewers helped improve an earlier version of this manuscript, for which we are thankful.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LK declared a shared affiliation with the authors ZZ and JS to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.973485/full#supplementary-material
References
Arquez, M., Colgan, D., and Castro, L. R. (2014). Sequence and comparison of mitochondrial genomes in the genus Nerita (Gastropoda: Neritimorpha: Neritidae) and phylogenetic considerations among gastropods. Mar. Genomics 15, 45–54. doi: 10.1016/j.margen.2014.04.007
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Benton, M. J., Donoghue, P. C., Asher, R. J., Friedman, M., Near, T. J., and Vinther, J. (2015). Constraints on the timescale of animal evolutionary history. Palaeontol. Electron. 18, 1–106. doi: 10.26879/424
Bergsten, J. (2005). A review of long-branch attraction. Cladistics 21, 163–193. doi: 10.1111/j.1096-0031.2005.00059.x
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Bernt, M., Merkle, D., Ramsch, K., Fritzsch, G., Perseke, M., Bernhard, D., et al. (2007). CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 23, 2957–2958. doi: 10.1093/bioinformatics/btm468
Bouchet, P., Rocroi, J. P., Hausdorf, B., Kaim, A., Kano, Y., Nützel, A., et al. (2017). Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia 61, 1–526. doi: 10.4002/040.061.0201
Campbell, K. A., Peterson, D. E., and Alfaro, A. C. (2008). Two new species of Retiskenea? (Gastropoda: Neomphalidae) from Lower Cretaceous hydrocarbon-seep carbonates of northern California. J. Paleontol. 82, 140–153. doi: 10.1666/05-025.1
Castro, L. R., and Colgan, D. J. (2010). The phylogenetic position of Neritimorpha based on the mitochondrial genome of Nerita melanotragus (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 57, 918–923. doi: 10.1016/j.ympev.2010.08.030
Chen, C., and Linse, K. (2020). From wood to vent: First cocculinid limpet associated with hydrothermal activity discovered in the Weddell Sea. Antarct. Sci. 32, 354–366. doi: 10.1017/S095410202000022X
Chen, C., Linse, K., Roterman, C. N., Copley, J. T., and Rogers, A. (2015). A new genus of large hydrothermal vent-endemic gastropod (Neomphalina: Peltospiridae). Zool. J. Linn. Soc. 175, 319–335. doi: 10.1111/zoj.12279
Chen, C., Zhou, Y., Wang, C., and Copley, J. T. (2017). Two new hot-vent peltospirid snails (Gastropoda: Neomphalina) from longqi hydrothermal field, southwest Indian Ridge. Front. Mar. Sci. 4:392. doi: 10.3389/fmars.2017.00392
Collins, P. C., Hunter, W. R., Carlsson, J., and Carlsson, J. (2020). Fortuitous insights into the ecology of a recently charted deepsea hydrothermal vent, using snails’ feet. Deep Sea Res. I Oceanogr. Res. Pap. 163:103358. doi: 10.1016/j.dsr.2020.103358
Cunha, R. L., Grande, C., and Zardoya, R. (2009). Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol. Biol. 9:210. doi: 10.1186/1471-2148-9-210
Cunha, T., and Giribet, G. (2019). A congruent topology for deep gastropod relationships. Proc. R. Soc. B Biol. Sci. 286:20182776. doi: 10.1098/rspb.2018.2776
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. doi: 10.1093/nar/gkw955
Grande, C., Templado, J., and Zardoya, R. (2008). Evolution of gastropod mitochondrial genome arrangements. BMC Evol. Biol. 8:61. doi: 10.1186/1471-2148-8-61
Heß, M., Beck, F., Gensler, H., Kano, Y., Kiel, S., and Haszprunar, G. (2008). Microanatomy, shell structre and molecular phylogeny of Leptogyra, Xyleptogyra and Leptogyropsis (Gastropoda: Neomphalida: Melanodrymiidae) from sunken wood. J. Molluscan Stud. 74, 383–401. doi: 10.1093/mollus/eyn030
Isobe, N., Chen, C., Daicho, K., Saito, T., Bissessur, D., Takai, K., et al. (2022). Uniaxial orientation of β-chitin nanofibres used as an organic framework in the scales of a hot vent snail. J. R. Soc. Interface 19:20220120. doi: 10.1098/rsif.2022.0120
Jörger, K. M., Stöger, I., Kano, Y., Fukuda, H., Knebelsberger, T., and Schrödl, M. (2010). On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evol. Biol. 10:323. doi: 10.1186/1471-2148-10-323
Kaim, A., Jenkins, R., Tanabe, K., and Kiel, S. (2014). Mollusks from late Mesozoic seep deposits, chiefly in California. Zootaxa 3861, 401–440. doi: 10.11646/zootaxa.3861.5.1
Kiel, S., and Campbell, K. A. (2005). Lithomphalus enderlini gen. et sp nov from cold-seep carbonates in California–a Cretaceous neomphalid gastropod? Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 232–241.
Kocot, K. M., Cannon, J. T., Todt, C., Citarella, M. R., Kohn, A. B., Meyer, A., et al. (2011). Phylogenomics reveals deep molluscan relationships. Nature 477, 452–456. doi: 10.1038/nature10382
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurabayashi, A., and Ueshima, R. (2000). Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: Systematic implication of the genome organization. Mol. Biol. Evol. 17, 266–277. doi: 10.1093/oxfordjournals.molbev.a026306
Lan, Y., Sun, J., Chen, C., Sun, Y., Zhou, Y., Yang, Y., et al. (2021). Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail Gigantopelta aegis. Nat. Commun. 12:1165. doi: 10.1038/s41467-021-21450-7
Lee, H., Chen, W. J., Puillandre, N., Aznar-Cormano, L., Tsai, M. H., and Samadi, S. (2019). Incorporation of deep-sea and small-sized species provides new insights into gastropods phylogeny. Mol. Phylogenet. Evol. 135, 136–147. doi: 10.1016/j.ympev.2019.03.003
Lee, H., Puillandre, N., Kano, Y., Chen, W.-J., and Samadi, S. (2022). Biodiversity and phylogeny of Cocculinidae (Gastropoda: Cocculinida) in the Indo-West Pacific. Zool. J. Linn. Soc. doi: 10.1093/zoolinnean/zlac023
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Nakano, T., and Ozawa, T. (2004). Phylogeny and historical biogeography of limpets of the order Patellogastropoda based on mitochondrial DNA sequences. J. Molluscan Stud. 70, 31–41. doi: 10.1093/mollus/70.1.31
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Osca, D., Templado, J., and Zardoya, R. (2014). The mitochondrial genome of Ifremeria nautilei and the phylogenetic position of the enigmatic deep-sea Abyssochrysoidea (Mollusca: Gastropoda). Gene 547, 257–266. doi: 10.1016/j.gene.2014.06.040
Ponder, W. F., and Lindberg, D. R. (2008). Towards a phylogeny of gastropod molluscs: An analysis using morphological characters. Zool. J. Linn. Soc. 119, 83–265. doi: 10.1111/j.1096-3642.1997.tb00137.x
Ponder, W. F., Lindberg, D. R., and Ponder, J. M. (2020). Biology and evolution of the mollusca (1st ed.). Boca Raton, FL: CRC Press. doi: 10.1201/9781351115254-1
Reis, M. D., and Yang, Z. (2011). Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol. Biol. Evol. 28, 2161–2172. doi: 10.1093/molbev/msr045
Sigwart, J., Lindberg, D., Chen, C., and Sun, J. (2021). Molluscan phylogenomics requires strategically selected genomes. Philos. Trans. R. Soc. B Biol. Sci. 376:20200161. doi: 10.1098/rstb.2020.0161
Simpson, J., Wong, K., Jackman, S. D., Schein, J., Jones, S., and Birol, I. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123. doi: 10.1101/gr.089532.108
Stöger, I., and Schrödl, M. (2013). Mitogenomics does not resolve deep molluscan relationships (yet?). Mol. Phylogenet. Evol. 69, 376–392. doi: 10.1016/j.ympev.2012.11.017
Sun, J., Chen, C., Miyamoto, N., Li, R., Sigwart, J. D., Xu, T., et al. (2020). The Scaly-foot Snail genome and implications for the origins of biomineralised armour. Nat. Commun. 11:1657. doi: 10.1038/s41467-020-15522-3
Sun, J., Li, R., Chen, C., Sigwart, J. D., and Kocot, K. M. (2021). Benchmarking Oxford Nanopore read assemblers for high-quality molluscan genomes. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 376:20200160. doi: 10.1098/rstb.2020.0160
Uribe, J. E., Colgan, D., Castro, L. R., Kano, Y., and Zardoya, R. (2016a). Phylogenetic relationships among superfamilies of Neritimorpha (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 104, 21–31. doi: 10.1016/j.ympev.2016.07.021
Uribe, J. E., Kano, Y., Templado, J., and Zardoya, R. (2016b). Mitogenomics of Vetigastropoda: insights into the evolution of pallial symmetry. Zool. Scripta 45, 145–159. doi: 10.1111/zsc.12146
Uribe, J. E., González, V. L., Irisarri, I., Kano, Y., Herbert, D. G., Strong, E. E., et al. (2022a). A phylogenomic backbone for gastropod molluscs. Syst. Biol. [Online ahead of print]. doi: 10.1093/sysbio/syac045
Uribe, J. E., Sei, M., and Harasewych, M. G. (2022b). The mitogenome of the sunken wood limpet Notocrater youngi: Insights into mitogenome evolution in Lepetellida (Gastropoda: Vetigastropoda). J. Molluscan Stud. 88:eyac009. doi: 10.1093/mollus/eyac009
Uribe, J. E., Irisarri, I., Templado, J., and Zardoya, R. (2019). New patellogastropod mitogenomes help counteracting long-branch attraction in the deep phylogeny of gastropod mollusks. Mol. Phylogenet. Evol. 133, 12–23. doi: 10.1016/j.ympev.2018.12.019
Xu, T., Qi, L., Kong, L., and Li, Q. (2022). Mitogenomics reveals phylogenetic relationships of Patellogastropoda (Mollusca, Gastropoda) and dynamic gene rearrangements. Zool. Scripta 51, 147–160. doi: 10.1111/zsc.12524
Zapata, F., Wilson, N. G., Howison, M., Andrade, S. C. S., Jörger, K. M., Schrödl, M., et al. (2014). Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc. R. Soc. B Biol. Sci. 281:20141739. doi: 10.1098/rspb.2014.1739
Zhang, S., and Zhang, S. (2022). The mitochondrial genome of Lamellomphalus manusensis Zhang & Zhang, 2017 (Gastropoda: Neomphalida) from deep-sea hydrothermal vent. Mitochondrial DNA 7, 825–827. doi: 10.1080/23802359.2022.2073843
Keywords: Gastropoda, deep sea, mitogenome, gene order, phylogenetic relationship
Citation: Zhong Z, Lan Y, Chen C, Zhou Y, Linse K, Li R and Sun J (2022) New mitogenomes in deep-water endemic Cocculinida and Neomphalida shed light on lineage-specific gene orders in major gastropod clades. Front. Ecol. Evol. 10:973485. doi: 10.3389/fevo.2022.973485
Received: 20 June 2022; Accepted: 16 August 2022;
Published: 18 October 2022.
Edited by:
Joong-Ki Park, Ewha Womans University, South KoreaReviewed by:
Juan E. Uribe, Smithsonian National Museum of Natural History (SI), United StatesLingfeng Kong, Ocean University of China, China
Pawel Mackiewicz, University of Wrocław, Poland
Copyright © 2022 Zhong, Lan, Chen, Zhou, Linse, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Sun, amluX3N1bkBvdWMuZWR1LmNu
 Zhaoyan Zhong
Zhaoyan Zhong Yi Lan
Yi Lan Chong Chen
Chong Chen Yadong Zhou
Yadong Zhou Katrin Linse
Katrin Linse Runsheng Li4,8
Runsheng Li4,8 Jin Sun
Jin Sun