- 1Hebei Key Laboratory of Animal Physiology, Biochemistry and Molecular Biology, College of Life Sciences, Hebei Normal University, Shijiazhuang, China
- 2College of Life Sciences, Capital Normal University, Beijing, China
- 3College of Life Sciences, Shanxi Agricultural University, Jinzhong, China
Cytochrome P450 enzymes (P450s, CYPs) are a superfamily of heme–thiolate proteins involved in the metabolism of endogenous and exogenous substances in insects. In this study, the identification of putative P450 proteins was done and the elimination of the repeated sequences resulted in 57 proteins from Gastropacha populifolia, 63 proteins from Dendrolimus punctatus, and 53 proteins from Dendrolimus tabulaeformis. The putative P450 proteins were aligned together with seven other insect species based on five conserved domains. A total of ten co-orthologous groups were identified. Interestingly, one co-orthologous gene, CYP4g15 in CYP4 clan, was identified and its 3D structure analysis showed that the highly conserved sites of the predicted motifs were close to the active sites of P450. Furthermore, this study revealed that insect CYP4g15 and two bacteria cytochrome P450 were monophyletic. This suggests that insects CYP4g15 are not only functionally conserved but also an ancient gene originating from different bacteria species.
Introduction
Cytochrome P450 is a family of heme-containing enzymes widely existing in all living organisms, such as bacteria, fungi, plants, and animals. These enzymes play important roles in detoxification of xenobiotics (plant toxins, dugs, mutagens, drugs, and other foreign compounds), synthesis and metabolism of juvenile hormone, molting hormone, and fatty acids (Scott et al., 1998; Feyereisen, 1999, 2012). Four clades of insect CYP genes were mitochondrial CYP (Mito.CYP), CYP2, CYP3, and CYP4 clades (Feyereisen, 2006).
It has been reported that CYP306A1 (CYP2 clade), CYP302A1, CYP 315A1, and CYP314A1 genes (Mito.CYP clade), the terminal hydroxylases, are single orthologs of Lepidoptera, Coleoptera, Hymenoptera, and Diptera. CYP302, CYP306, CYP315, and CYP314 (CYP2clade) (Dermauw et al., 2020), and at least five P450 genes of Mito.CYP and CYP2 are involved in different steps of the molting hormone synthesis (Rewitz et al., 2007). CYP15 (CYP2 clade) plays an important function in the biosynthesis of juvenile hormone (Helvig et al., 2004). Therefore, Mito.CYP and CYP2 genes are conserved and involved in physiological functions. CYP3 and CYP4 are species-specific, and some of them function in xenobiotic metabolism, insecticide resistance, and production of natural products (Feyereisen, 2005). P450 genes in these two (CYP3 and CYP4) clades are more numerous among insects (Feyereisen, 2006). CYP9a of Spodoptera litura larvae were were induced by imidacloprid, xanthotoxin or ricin treatments; CYP337a1 and a2, CYP321b1, CYP6ae9 and CYP6b29 of CYP3 clan were also inducible by treatment with toxin (Cheng et al., 2017). CYP6B P450s are critical in furanocoumarin degradation, and this allows papilio to adapt to furanocoumarin-containing host plants (Scott et al., 1998). CYP9A40 in S. litura functions in detoxification of plant allelochemicals and insecticides (Wang et al., 2015). CYP3, CYP5, CYP6, and CYP12 play important roles in pyrethroid resistance among the pyrethroid-resistant populations in S. litura (Xu et al., 2020).
CYP proteins are distributed in two cellular locations, the mitochondria (Mito.CYP) and the endoplasmic reticulum (microsomal CYP containing CYP2, CYP3, and CYP4 clades). Microsomal CYPs are reduced by an integral enzyme NADPH P450 reductase (Paine et al., 2005), while Mito.CYPs are first reduced by adrenodoxin reductase and they receive electrons from an iron–sulfur protein similar to bacterial systems. This implies that Mito.CYPs may be derived from a bacterial ancestor which may differ from microsomal CYPs (Nelson and Strobel, 1987). The function of CYP proteins is related to the earlier biosynthesis of cholesterol prior to the divergence of prokaryotes from eukaryotes (Bloch, 1983). Nelson and Strobel (1987) proposed a hypothesis that all P450 genes evolved from a common ancestor prior to the divergence into eukaryotes and prokaryotes.
The most important function of P450 is the development and in the metabolism of endogenous and exogenous substances in insect. At present, second-generation sequencing platform is the most widely used among sequencing platforms (Feng et al., 2022; Zhang et al., 2021). The study of P450 genes has been carried out in insects through genome or transcriptome analysis (Wan et al., 2013; Raman and Andy, 2018; Antony et al., 2019). There are a number of related studies in lepidoptera (Lin et al., 2013; Wang et al., 2014; Chen and Palli, 2022). However, limited research has been done on P450 in insects that feed on gymnosperms, and comparison to bacteria is also limited.
Majority of the Lepidoptera are agricultural pests, and the use of insecticide results in huge selection pressure (Moulton et al., 2000; Ahmad et al., 2008). P450 plays an important role in insect detoxification and metabolism and help insects to survive in environments exposed to increased use of insecticides. The identification of P450s in target species may help in the understanding of survival strategies used by different species’ living in harsh living conditions and the control of their occurrence.
In this study, transcriptome sequencing of three species were conducted, including Dendrolimus. punctatus, Dendrolimus tabulaeformis, and Gastropacha populifolia (Lepidoptera: Lasiocampidae). G. populifolia feed on angiosperms of Salicaceae and Rosaceae. However, the larva has a wide range of feeding preference, for example, in general, they feed on plant leaves. They also experience two–three generations every year and are mainly distributed in China and South Korea (Kim and Jeong, 2010). D. punctate and D. tabulaeformis (Lepidoptera: Lasiocampidae) are a pair of closely related species of dendrolimus and only feed on pinaceae, D. punctate, and D. tabulaeformis mainly distributed in southern China and northern China, respectively. They both have four life stages: egg, larva, pupa, and adult. The larva stage of this two species moth feeds on the leaves of conifers, they are collectively known as pine moth, and both are serious pest of coniferous trees: D. punctate more likely to feed on Pinus massoniana and D. tabulaeformis more likely to feed on Pinus tabulaeformis (Hou, 1987). P450s are one of the three detoxification enzymes which play an important role in disintegrating plant secondary substances. Therefore, in this study, identification and annotation of P450 genes based on the transcriptome data were done in the three moth species.
Materials and methods
Collection and rearing of insects
Female adults of G. populifolia were collected from Labagoumen County, Beijing, in 2016, and pupa of D. punctatus was collected from Dawu County, Xiaogan City, Hubei Province, in 2016. The adults and pupa were maintained in our laboratory at 25°C ± 1 and 14–16°C, respectively, for 16:8 h light: dark cycle. After both insects had laid eggs, the eggs were reared in an incubator (25°C ± 1, 16:8 h light: dark cycle), after the eggs hatched into larvae, they were raised separately in different culture flasks, the larvae offspring of G. populifolia and D. punctatus were fed on Salix matsudana and P. massoniana, respectively, and after molted for three times, the fourth instar larvae were used for further research. Larvae of D. tabulaeformis were collected from Miyun County, Beijing, on July 2017.
Ribonucleic acid extraction and illumina sequencing
Fourth instar D. punctatus and G. populifolia were taken from the leaves and starved for 24 h to empty their gut content. Before dissecting the larval gut, the larvae were washed using 70% ethanol for 30–60 s until they could no longer move and then washed using sterile water for 10 s. The larva guts were gently dissected in sterilized Petri dishes on ice, and the content put in 1.5 ml tubes and kept in liquid nitrogen to reduce RNA degradation. The experiments were conducted in triplicates for each of the species and stored at −80° until needed for RNA extraction. Larvae of D. tabulaeformis were directly kept in the liquid nitrogen and stored at −80° until RNA isolation (D. tabulaeformis was used to add the transcriptome data of insects feeding on gymnosperms). Qiagen RNeasy Mini Kit (Qiagen) was used for gut RNA extraction, and the entire process was performed on an ultra-clean icebox which was RNase-free following the manufacturer’s instructions. The quality of the extracted RNA was analyzed by 1% agarose gel electrophoresis. NanoDrop2000 (Thermo Scientific) was used to determine the concentration, while Agilent (Agilent 2100 biological analyzer) measured purity. After the construction of RNAseq libraries, they were sequenced using Illumina HiSeq2500 and the sequencing quantity of the raw data was 10G for each sample. The BioProject accession numbers were PRJNA578660.1
P450 protein assembly and prediction
FastQC detected the quality of the raw data. Low-quality reads were filtered by fastx-toolkit software to remove the adapter and reads with unknown “N” content over 5% and reads with low-quality base (q-value < 10) over 20% (Schmieder and Edwards, 2011). Clean reads obtained through quality control were used for subsequent data analysis. Trinity software was used for transcriptome de novo assembly of clean reads (Haas et al., 2013). Left.fq file and right.fq file (three repeated mix assembled) were assembled in Linux system with parameters set to default values and min_kmer_cov set to 2. Trinity.fasta provided a summary of all sequence files after assembly while TGICL software (Pertea et al., 2003) and cd-hit software (Fu et al., 2012) were used for redundant Trinity.fasta file. All parameters were based on the default parameters of the software, and the de-redundant gene is called unigene. TransDecoder2 was used to predict the ORF of the Unigene. P450 family selection starts with the building of a local reference database. The Pfam PF00067 domain is the P450 feature domain used, and P450 protein family was predicted by HMMER (E-value < 1e-5). The proteins predicted by HMMER were annotated by BLASTp (reference databases are shown in Supplementary Table 2), and the unannotated sequences were also filtered in this step. To further confirm the accuracy of the results, the selected P450 protein family was further searched in GenBank NR library. The length of the obtained P450 protein family was statistically analyzed by in-house R script, and P450 proteins that were >300 amino acids in length were retained for further analysis.
Phylogenetic analysis
Lepidoptera P450 phylogenetic tree was constructed using one model insect Bombyx mori (Ai et al., 2011) and the three species (G. populifolia, D. punctatus, and D. tabulaeformis) used in this study. Lepidoptera P450 proteins data of the above four species in combination with two insects from Homoptera [Dendroctonus ponderosae, Tribolium castaneum, P450 downloaded from InsectBase (Yin et al., 2016)], two insects from Coleoptera [Acyrthosiphon pisum, P450 downloaded from InsectBase, Cinara pinitabulaeformis (Wu et al., 2017)], and several bacteria species were used to reconstruct P450 phylogenetic tree. The bacteria species included Macrococcus caseolyticus bacteria (P450 proteins were downloaded from the GenBank) and two human bacterial pathogen species Staphylococcus aureus and Bacillus anthracis which are evolutionarily related species of M. caseolyticus (Baba et al., 2009).
The protein data of CYP4 sequences from nine insect species (eight insects mentioned above and Danaus plexippus) were download from InsectBase. Two M. caseolyticus P450 protein sequences and CYP4g15 protein sequences from Rhopalosiphum maidis, Blattella germanica, Zootermopsis nevadensis, and Lygus hesperus were subjected to phylogenetic analysis.
The reconstruction of NADPH–cytochrome P450 reductase phylogenetic tree was done using ten species: eight from insects including the three species used in this study and B. mori, D. ponderosae, T. castaneum, A. pisum, and C. pinitabulaeformis and two from bacteria including S. aureus and B. anthracis.
Protein sequences were aligned using MAFFT sequence alignment software, and P450 sequence below 300 amino acids was discarded except for WP_206198163_Shimia_sediminis and Cpin_c32126_g1|_NADH_CYP (NADH-CYP Reductase of C. pinitabulaeformis) and CYP4 tree which aligned well. ProtTest was used to test the best model (LG + I + G + F, the lowest BIC score) of amino acid replacement (Darriba et al., 2011). Maximum-likelihood phylogenetic trees were reconstructed by RAxML (Stamatakis, 2014) with 1,000 bootstrap replications. The trees were visualized and colored using iTOL (Letunic and Bork, 2016).
Prediction of motif and 3D structure and GC content analysis
MEME Suite 5.0.1 was used to analyze the predictions of Mito.CYP, CYP2, CYP3, and CYP4 motifs of D. punctatus and G. populifolia (Bailey et al., 2009). TBtools (Chen et al., 2018) were used to redraw the motifs predicted by MEME, and the Motif prediction of CYP4g15 was analyzed in a similar manner.
The secondary structure of CYP4g15 was predicted by SWISS-MODEL3 (Arnold et al., 2006) and ESPript (Robert and Gouet, 2014) (Robert and Gouet, 2014).4 EasyModeller 4.0 was used for 3D structure prediction. Query sequence and PDB file (5t6q.pdb, 6c93.pdb and 6c94.pdb) were imported into EasyModeller 4.0 for multi-template modeling, while the 3D structure of protein PDB files was visualized using the PyMol software.
GC content of M. caseolyticus, CYP4g15, and CYP6AE was analyzed using an in-house Perl script.
Results
Assemblies and de-redundancy
Triplicate data of the gut of D. punctatus and G. populifolia were separately assembled together. In total, 52,230, 86,251, and 87,679 transcripts of D. punctatus, G. populifolia, and D. tabulaeformis, respectively, were obtained. The average sequence length was 1,316, 1,134, and 754 bp with N50 value of 2,073, 1,923, and 1365 bp, respectively (details in Supplementary Table 1). The clean sequence reads data were deposited in Sequence Read Archive at NCBI, and the BioProject accession number was PRJNA578660.
P450 protein gene screening
A total of 103 D. tabulaeformis, 116 D. punctatus, and 143 G. populifolia CYP transcripts were obtained in our study based on domain features. Using BLASTp (aligned to the GenBank protein database with a cutoff), 45 D. tabulaeformis, 55 D. punctatus, and 65 G. populifolia complete CYP transcripts were identified with initial and termination codon. Other sequences were truncated at N-terminal or C-terminal or missed both ends, and the number of the P450 family of the three species is listed in Table 1. The predicted proteins were 58 for D. tabulaeformis, 75 for D. punctatus, and 76 for G. populifolia having greater than 300 amino acids; however, some CYPs protein sequences were same, because their nucleotide sequences are different, whereas the amino acid sequences of the proteins are the same. After removal of the repeat CYPs protein sequences, there were 53 CYPs for D. tabulaeformis, 63 for D. punctatus, and 57 for G. populifolia. The CYPs without repeat sequences were used for further studies.
Except for mitochondrial P450s (Mito.CYP) and endoplasmic reticulum P450s (CYP2, CYP3, and CYP4), this study also identified NADPH–cytochrome P450 reductase in the three species. There is only one CPR (cytochrome P450 reductase) for each insect species except A. pisum (2 CPRs). Our study identified only one CPR for D. punctatus and G. populifolia and two CPR for D. tabulaeformis (Table 2). It indicates that our data are reliable.
Lepidoptera P450 phylogenetic tree
P450 protein family of the three species used in this study and B. mori were clustered into four major divisions in the phylogenetic tree. There were four distinct subfamilies: Mito.CYP, CYP2, CYP3, and CYP4. The bootstrap of each of the four clades branch was above 99. Table 2 shows that there were seven D. punctatus, nine G. populifolia, and six D. tabulaeformis for Mito.CYP, seven D. punctatus, four G. populifolia, and six D. tabulaeformis for CYP2, 30 D. punctatus, 28 G. populifolia, and 25 D. tabulaeformis for CYP3, and 19 D. punctatus, 16 G. populifolia, and 15 D. tabulaeformis for CYP4. The number of each clade was similar to that in B. mori. Notably, CYP3 and CYP4 were more than CYP2 and Mito.CYP, the protein numbers of CYP3 in eight insects were from 25 to 65, and the number of CYP4 was from 15 to 40, The number of CYP2 and Mito.CYP in seven insects except C. pinitabulaeformis was from 4 to 10 (Table 2). There were seven gene clusters in Mito.CYP, and only CYP333 and CYP12 showed a mixed gene cluster in Mito.CYP clade. There were five gene clusters in CYP2 (Figure 1A).
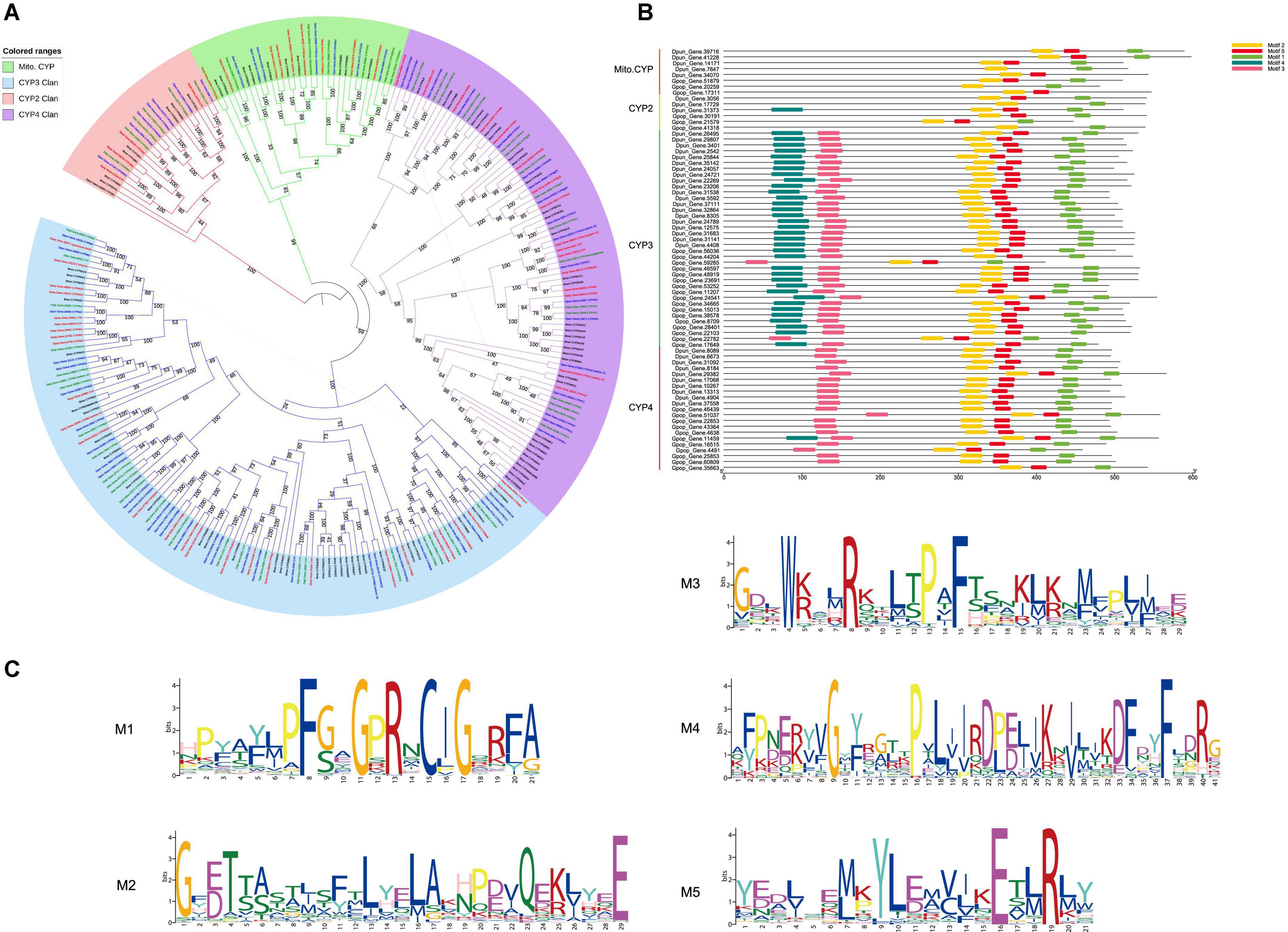
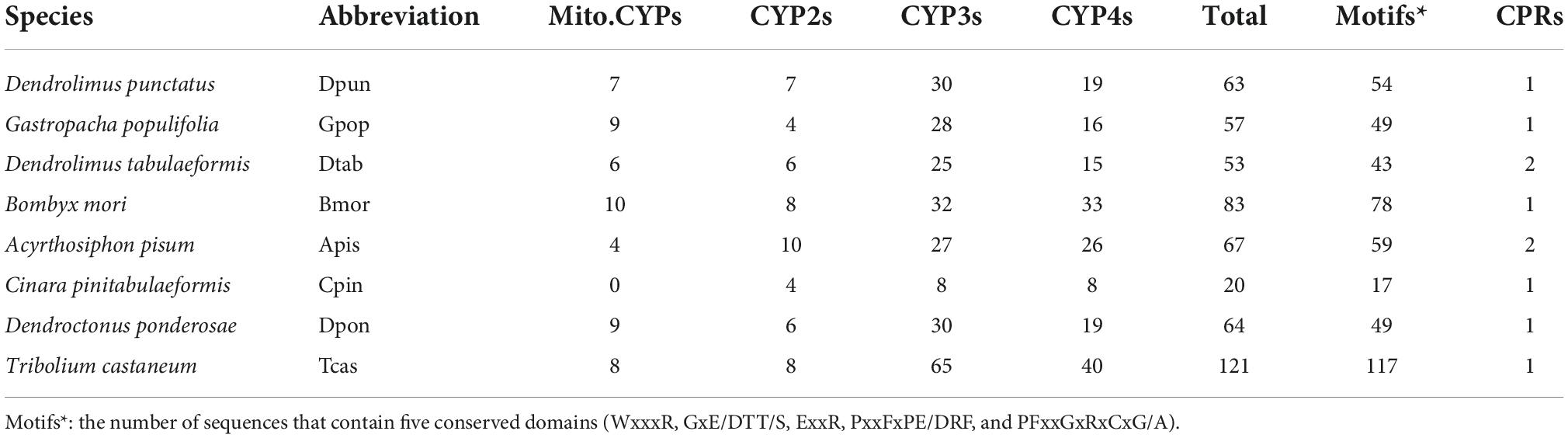
Figure 1. (A) Maximum-likelihood phylogram of cytochrome P450. Sequences from Dendrolimus punctatus (blue), Gastropacha populifolia (red), Dendrolimus tabulaeformis (green), and the genome data of Bombyx mori (Violet). Highlighted clades: Mito.CYP clan is marked in green, CYP2 clan is marked in red, CYP3 clan is marked in blue, and CYP4 clan is marked in purple. (B) Predicted motif of completed P450 sequence of D. punctatus, G. populifolia, and D. tabulaeformis. Five motifs were predicted for CYP3 clan, four motifs were predicted for CYP4 clan, and two motifs were predicted for Mito.CYP and CYP2 clan. (C) The sequence logos of four consensus domains of CYP clan, the larger the letter, the more conservative the amino acid site is.
P450 phylogenetic tree results showed that CYP3 is mainly divided into five groups: CYP9 (including CYP9e, CYP9A, CYP9f, and CYP9G), CYP6 (including CYP6a, CYP6W, CYP6B, CYP6AB, CYP6AE, and CYP6C), CYP332A1, CYP324A1, and CYP337. Mixed gene cluster I (Gpop-Gene.30544 CYP6a14, Gpop-Gene.24541 CYP6B1, and Bmor-CYP365A1; Figure 1A) only contained B. mori and G. populifolia which feeds on angiosperms but mixed gene cluster II (Dpun-Gene.24057-CYP6CV6, Dpun-Gene.8305-CYP6CV1, Dpun-Gene.37111-CYP6CV1, and Dtab–Gene.28411-CYP6B7) only contained D. punctatus and D. tabulaeformis which feeds on gymnosperm. CYP9A22 was only found in B. mori and G. populifolia but not in D. punctatus and D. tabulaeformis. In gene cluster CYP 324A1, there was an obvious expansion in D. punctatus and D. tabulaeformis which feeds on gymnosperm. In CYP4, the species sequenced in this study were mainly divided into two gene clusters CYP4 (CYP4d, CYP4M, CYP4S, CYP4C, CYP4g, and CYP4V) and CYP341. CYP341 gene cluster was only found in B. mori and G. populifolia (Figure 1A).
The complete P450 proteins of D. punctatus and G. populifolia were used for motif prediction by MEME. A total of five motifs were predicted with motif 1, motif 2, and motif 5 found in all the sequences, motif 3 was found in CYP3 and CYP4, and motif 4 was found in most CYP3 except Gpop-Gene.59036 (CYP3) and Gpop-Gene.22782 (CYP3). Motif 4 was also found in Gpop-Gene.31373 (CYP2) and Gpop-Gene.11459 (CYP4; Figure 1B). The characteristics of the five motifs were P(NSC)FxxGxRxCI(NSC)G (motif 1, NSC means it is not a strictly conservative site), GxE/DT and LxxLAxxxxxQxxxxxE (motif 2, the entire site was not strictly conservative), WxxxRxxxxP(NSC)xF (motif 3), GxYF(NSC)xxxxP(NSC) and D(NSC)F(NSC)xxFxxR(NSC) (motif 4), and M/L(NSC)xY(NSC)L(NSC) and ExxR (motif 5; Figure 1C).
CYP4 phylogenetic tree and CYP4g15 second and 3D structure
In the CYP4 tree (Figure 2A), CYP341 and CYP4L6 genes were only found in Lepidoptera. Other genes (such as CYP4d and CYP4C) were not well clustered, and there were mixed branches except for CYP4g15. CYP4g15 clustered in one branch even with additional genes from other insect orders. CYP4g15 exists in all insects (CYP4G22, CYP4G23, and CYP4G24 were the co-orthologous in B. mori) and clusters well with a bootstrap value of 100. P450 proteins from bacteria Macrococcus caseolyticus and Shimia sediminis sp. were found to have high homology with CYP4g15. S. sediminis sp. P450 protein (WP_206198163_Shimia, GenBank accession: WP_206198163) (Zhu et al., 2021) clustered together with Coleoptera, and M. caseolyticus P450 protein (WP_RKO10043 _CYP_Mcase, GenBank accession: WP_RKO10043) clustered together with Lepidoptera. Only M. caseolyticus P450 protein was used for second structure analysis because the S. sediminis sp. P450 protein was too short (< 250AA). The length of M. caseolyticus P450 protein was shorter than insect CYP4g15 as the membrane-binding sequence at the front and a conserved domain (WxxR) were missing (Supplementary Figure 1).
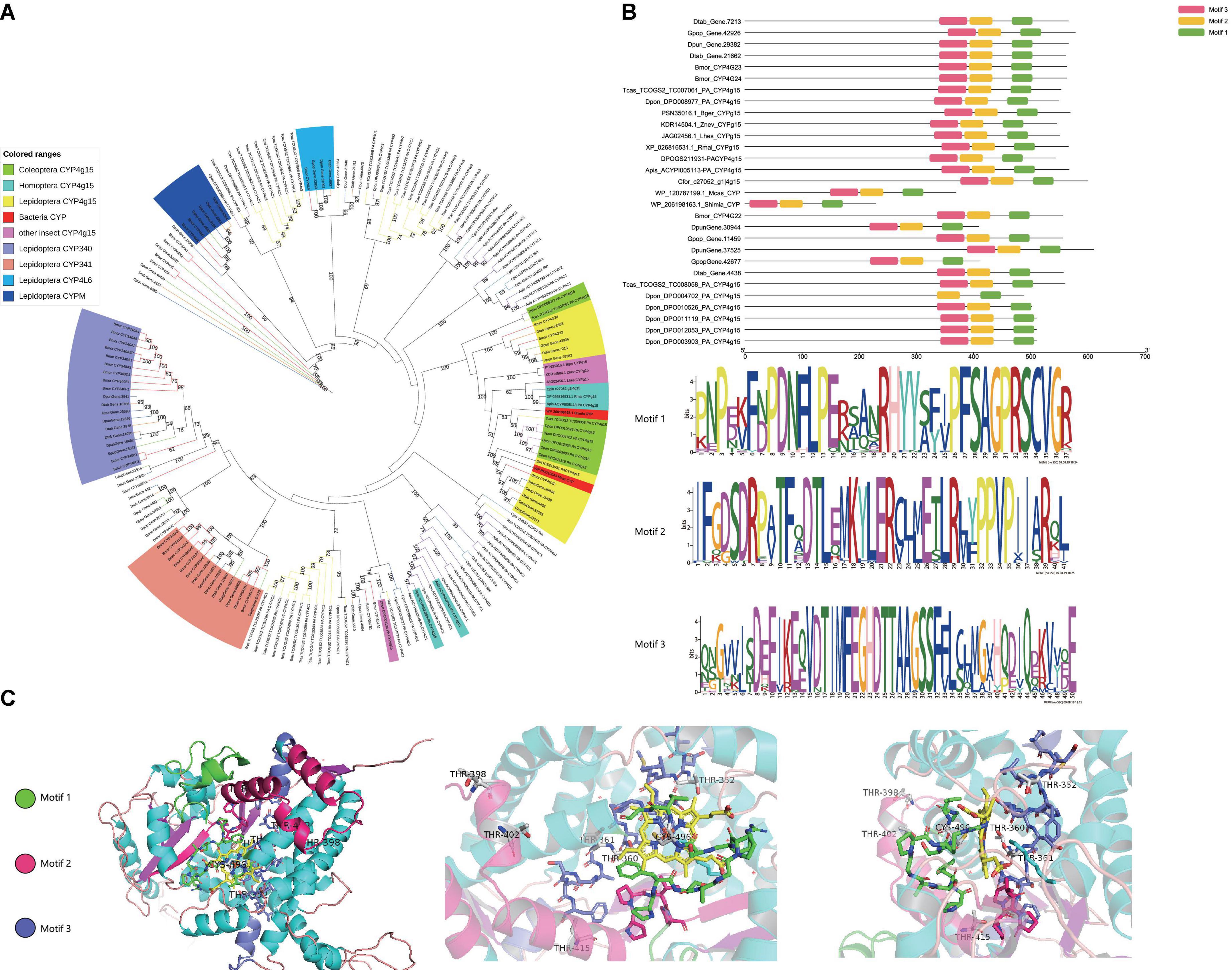
Figure 2. (A) Maximum-likelihood phylogram of CYP4 subfamily. Gastropacha populifolia (blue branch), Dendrolimus punctatus (green branch), Dendrolimus tabulaeformis (orange branch), Bombyx mori (red branch), Acyrthosiphon pisum (purple branch), Cinara pinitabulaeformis (light blue branch), Tribolium castaneum (yellow branch), Dendroctonus ponderosae (pink branch), and other insects are marked with black branches. (B) Motif prediction of D. punctatus and G. populifolia CYP4g15 by MEME, and three motifs were predicted in CYP4g15. (C) 5t6q.pdb, 6c93.pdb, and 6c94.pdb crystals were used as templates to predict the 3D model of CYP4g15. The overall structure of CYP4g15 is shown on the left, the front diagram of HEM in the middle, and the profile diagram of HEM on the right, and the highly conserved sites of amino acids in the middle and right graphs are represented by stick mode.
MEME motif and 3D prediction of CYP4g15 were performed. Three motifs were predicted from all the insects CYP4g15 gene (Figure 2B), and the three predicted motifs contained all the conserved domains of P450 except WxxR (Helix-C). Motif 1 contained PxxFxPE/DRF (PERF motif) and PFxxGxRxCxG/A (heme-binding domain), motif 2 contained ExxR (Helix-K), and motif 3 contained GxE/DTT/S(Helix-I). Compared to insects CYP4g15, about 200 amino acid P450 of M. caseolyticus (GenBank accession: WP_RKO10043) was found to be missing at the N-terminal (Supplementary Figure 1). The N-terminal is the membrane-binding sequence, and this was associated with the fact that free P450 in bacteria and membrane-bound P450 in eukaryotes. Using Gpop-Gene.11459 as an example, the three motifs were colored with different colors to reveal the 3D structure. The conserved continuous sites were indicated by a stick, PFSAGPRSCVG in the right-terminal of motif 1, PPVP in the right-terminal of motif 2, and TIMFEGHDTTAAG/ASSF in the middle of motif 3 (the gene of Dpon_DPO004702_PA_CYP4g15 missing this segment). The conserved sites were all found to be close to the active sites (Heme) in space (Figure 2), an indication that CYP4g15 functions were conserved in insects. Both ends of motif 3 and left terminal of motifs 1 and 2 were far from the activity center of heme and not strictly conserved. However, the diversity of amino acids was lower and this indicated that the function of CYP4g15 is relatively conservative.
P450 phylogenetic tree by conservative structure domain
Five conservative domains [WxxR (Helix-C), PxxFxPE/DRF (PERF motif), PFxxGxRxCxG/A (heme-binding domain), ExxR (Helix-K), and GxE/DTT/S(Helix-I)] were used as the alignment sequences to reconstruct the P450 tree since it is not good to separate the four subfamily clusters (Mito.CYP, CYP2, CYP3, and CYP4). P450 of S. aureus and B. anthracis was clustered together. There were ten co-orthologous genes found in a single clade between eight insects (Supplementary Table 3). In total, there were three for CYP2 (CYP306a, CYP304a1, and CYP307a1), six for Mito.CYP (CYP315A1 CYP301a1, CYP428, CYP314, CYP339A1, and CYP302), and one for CYP4 (CYP4g15), they were based on the five conservative domains, and the bootstrap values of co-orthologous groups were more than 50 (Figure 3). These co-orthologous genes clustered into one branch based on the conservative domains an indication that these co-orthologous genes were highly conserved. P450 proteins of the two bacteria species S. sediminis sp. and M. caseolyticus were also clustered together with CYP4g15 of insects.
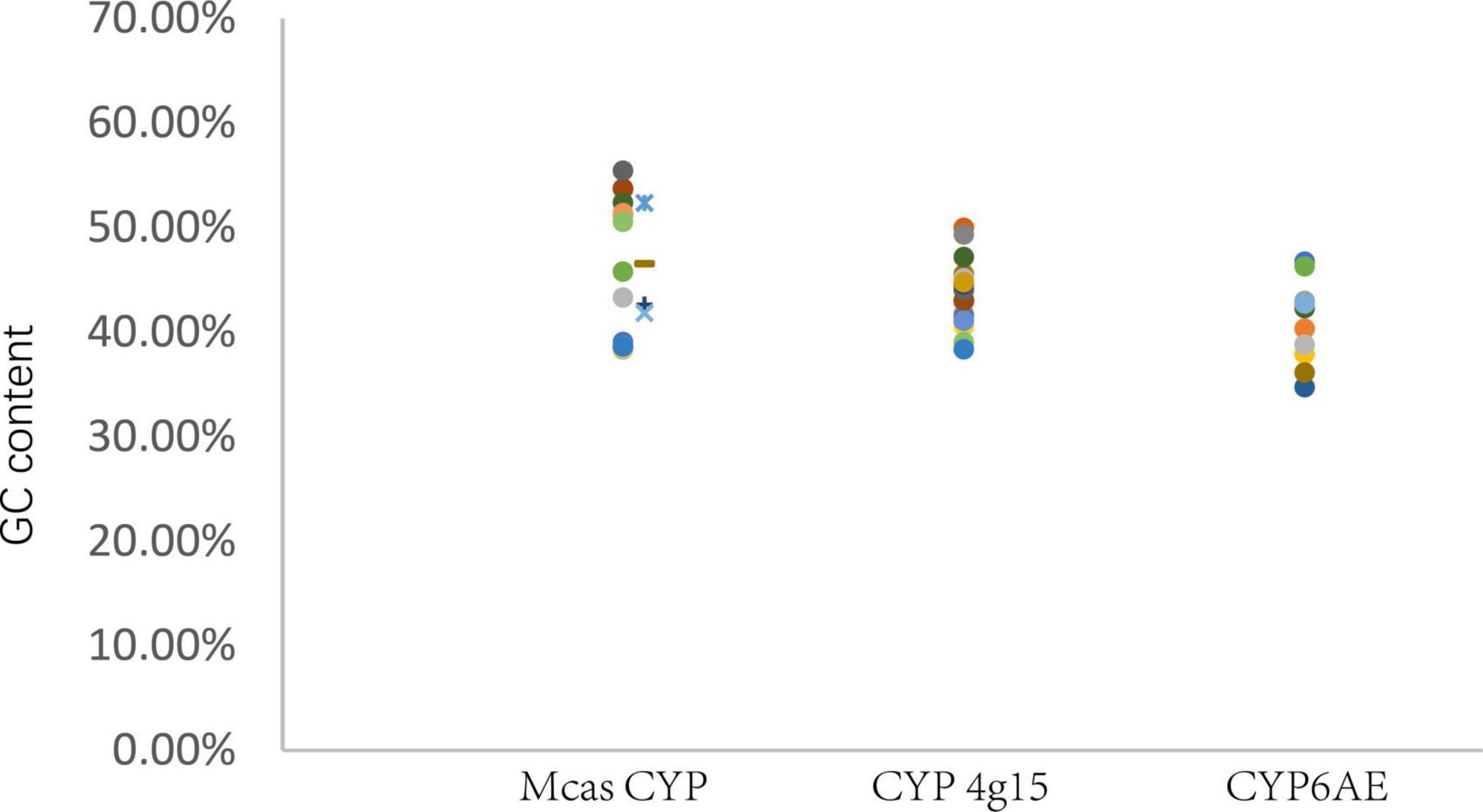
Figure 3. GC content of P450 gene. Mcas CYP: P450 of Macrococcus caseolyticus. CYP4g15 and CYP6AE: P450 of insects. The sequences of Mcas RKO10043.1 and Mcas RKO10043.1 showed high similarity with CYP 4g15, and GC content of Mcas RKO10043 was higher than the maximum GC content of CYP4g15 of insects; the sequences of Mcas RKO09869.1 and Mcas RKO10218.1 showed high similarity with CYP 6AE.
In the phylogenetic P450 tree reconstructed using five conserved structural domains, CYP306A1 was found in six insects (except C. pinitabulaeformis and D. ponderosae), and CYP307a1 was found in five insect species (except in three species) and they were clustered into monophyletic groups identified as co-orthologous, with bootstrap values of 59 and 84, respectively. Also, CYP306 and CYP307 were presumed 1:1 orthologues in six species (Diptera: Drosophila melanogaster, Anopheles gambiae, and Aedes aegypti, Lepidoptera: B. mori, Hymenoptera: Apis mellifera, Coleoptera: T. castaneum) (Feyereisen, 2006). CYP304a1 found in three insect species (D. tabulaeformis, D. punctatus, and T. castaneum) was clustered into a monophyletic group (see Figure 4). These three proteins were clustered into a monophyletic group by only the five conserved domains, indicating that there were an independent group in early CYP2 evolution and they were involved in important physiological functions and thus evolved slowly.
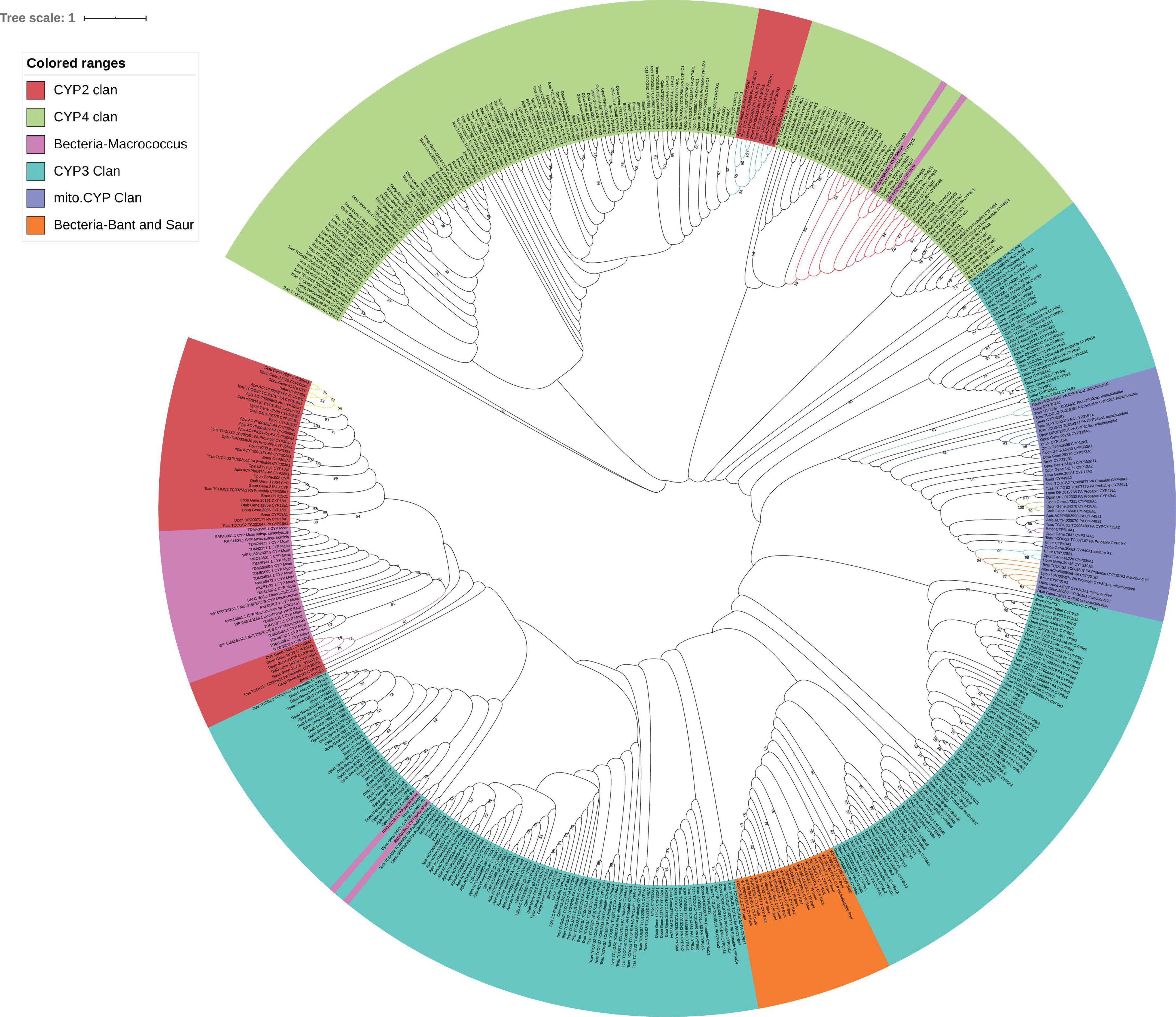
Figure 4. Maximum-likelihood phylogram of cytochrome P450 based on five conserved domains in eight insects (Gastropacha populifolia, Dendrolimus punctatus, Dendrolimus tabulaeformis, Bombyx mori, Acyrthosiphon pisum, Cinara pinitabulaeformis, Tribolium castaneum, and Dendroctonus ponderosae) and bacteria (Macrococcus caseolyticus, Staphylococcus aureus, and Bacillus anthracis). The bootstrap values above 50 (1,000 replicates) are shown. Ten co-orthologous groups are marked with different line colors.
NADPH-cytochrome P450 reductase
The characteristics of CPR were different in the four P450s subfamilies, and there were three conserved domains: FMN-, FAD- and nicotinamide adenine dinucleotide phosphate (NDPH)-binding domains (Wang et al., 1997). Most FAD-binding amino acid sites and NADPH-binding amino acid sites were conserved in both bacteria and insects (Supplementary Figure 2). These amino acid sites were 326H, 461R-464S, 479T, 495G, 496V, and 682W for FAD- and 305R/K,483V, 451G, 452T, 603S, 604R, 609K, 611Y, 639D, 643M, and 646D/E for NDPH-binding domains. The phylogenetic tree of CPR showed that they were closely related in the two species (Figure 5).
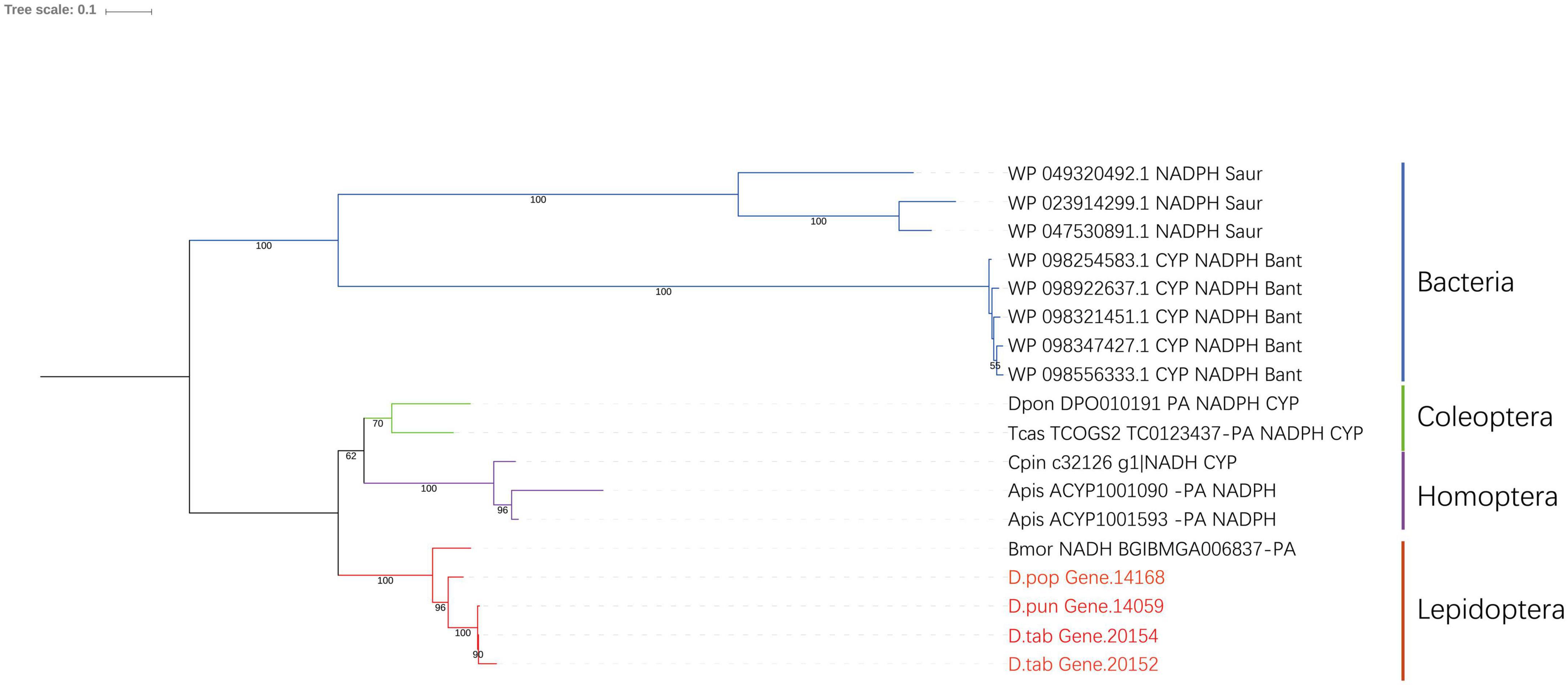
Figure 5. Phylogenic analysis of CPR. Eight insect species and two bacteria species were included in the picture, and Gastropacha populifolia, Dendrolimus punctatus, and Dendrolimus tabulaeformis are marked in red.
GC content analysis
GC content (The ratio of guanine G and cytosine C to all bases in nucleic acids sequence) of M. caseolyticus (Mcas) P450s ranged from 38.34 to 55.40%, the GC content of the eight insects CYP4g15 ranged from 38.32 to 49.91%, and the GC content of the eight insects CYP6AE ranged from 34.69 to 46.69%. The GC content of CYP4g15 was more similar to that of M. caseolyticus and not CYP6AE (Figure 3). The two bacteria species P450 were clustered together with CYP4g15 in CYP4 phylogenetic tree and the P450s tree constructed by the five conservative structural domains (Figures 2, 4). The GC content was 52.34% in M. caseolyticus (GenBank accession: WP_RKO10043) and 45.49% in S. sediminis sp. (GenBank accession: WP_206198163). The GC content of the two bacteria species was compared with insect CYP4g15 and was relatively high. The protein sequence of three M. caseolyticus P450s was similar to the CYP6AE subfamily, and the sequence of M. caseolyticus P450 (GenBank accession: RKO09869) showed the highest similarity to Dpun_Gene.2542 with 61.43% identity. M. caseolyticus P450 (GenBank accession: RKO10724) showed the highest similarity to Gpop_Gene.44204 with 64.13% identity, and M. caseolyticus P450 (GenBank accession: RKO10218) showed the highest similarity to Gpop_Gene.44204 with 62.23% identity. The GC content of the three sequences was 46.47, 42.58, and 41.81%, respectively, and upon comparing them to the insect CYP6AE, the GC content of the three M. caseolyticus P450 sequences was relatively higher (Figure 3).
Discussion
Mito.CYP clade
Mitochondrial P450s are only found in animals and not expressed in plants or fungi (Feyereisen, 2006). It is reported that Mito.CYP may have evolved from microsomal CYPs (Omura, 1993). Here, we found seven monophyletic groups in Mito.CYP clade: CYP428 group, CYP301 group, CYP49 group, CYP339 group, CYP314 group, CYP315 group, and CYP12/CYP333 group (Figure 1). Studies have shown that the genes of CYP12 in Drosophila and CYP333 in Bombyx mori. are homologous pairs, and the two genes may have similar functions, particularly, in exogenous metabolism (Ai et al., 2011). In B. mori, G. populifolia, D. punctatus, and D. tabulaeformis, CYP314A1 and CYP301a1 were found to be 1:1 orthologues. CYP314A1 is a gene for C-20-hydroxylase which converts α-ecdysone into β-ecdysone (Maeda et al., 2008). CYP301a1 plays a role in the formation of insect cuticules (Sztal et al., 2012).
In a phylogenetic tree reconstructed using five conservative domains, six CYP proteins of different insects formed a cluster. Five groups of them were 1:1 orthologues in different insects; they were CYP302a1, CYP315a1, CYP428A1, CYP314A1, and CYP301a1 (Figure 4). This indicates that these proteins are relatively conserved and involved in physiological functions. In this study, CYP428A1 only found in the three species, suggesting that it may be species-specific, and may not be involved in key physiological processes. Other genes such as CYP12/CYP333 were found to be taxon-specific and showed rapid evolution. The CYP12/CYP333 group was clustered into a monophyletic group in the Lepidoptera P450 phylogenetic tree (Figure 1); however, when aligned with other orders insects by conservative domains, they clustered into several groups (Figure 4) and with more members of each species (three for D. tabulaeformis, two for B. mori, two for G. populifolia, and three for B. mori in Figure 1). This implies that the five conservative domains of CYP12/CYP333 protein are not conserved and rather rapidly evolved. CYP12 not only plays a role in metabolism of many xenobiotics but also mediates insecticide resistance (Feyereisen, 2006). Overall, genes associated with environmental adaptation tend to evolve faster and many Mito.CYP are involved in physiological functions.
CYP2 clade
Most genes of CYP2 clan are mainly involved in insect growth and development, and the number and types of genes are relatively conserved. CYP15C1, CYP18a1, and CYP306A1 were found in G. populifolia, D. punctatus, and D. tabulaeformis. CYP15C1, CYP18a1, and CYP306A1 were involved in juvenile hormone biosynthesis and play a major role in the molting process of insects (Helvig et al., 2004; Feyereisen, 2006). CYP305B1 was not found in G. populifolia and so was CYP304a1 in G. populifolia and B. mori. We speculate that they may not be required for the growth and development of insects. It has been reported that CYP304F4 (belonging to CYP304 group) is sex-specific being only expressed in male adults but not in larvae (Pottier et al., 2012). However, CYP304a1 was found to be expressed in the larvae of D. tabulaeformis and D. punctatus in this study. We presume that although CYP304F4 and CYP304a1 belong to CYP304 group, they have different functions. Three Lasiocampidae species larvae lacked CYP307A1 gene (Figure 1). This is consistent with reports that Drosophila CYP307a1 is expressed in adult ovaries and early embryos and may be involved in the synthesis of insect ecdysone (Namiki et al., 2005). Given that the larvae used in our study were forth instar larvae, this may explain why CYP307a1 gene was not detected. These suggests the existence of another pathway for the synthesis of ecdysone at different developmental larval stages (Namiki et al., 2005).
CYP3 clade
Among insect P450 genes, CYP3 clade has the highest number of genes. Among the 13 gene groups, only CYP324A1, CYP332A1, and CYPAE were clustered into monophyletic groups (Figure 1). Most CYP3 members found to be involved in exogenous metabolism and insecticide resistance (Feyereisen, 2005). Genes that encodes CYP324A1 were identified in D. punctatus and D. tabulaeformis which feeds on gymnosperms. It has been reported that CYP324A2 plays a role in plant recognition in Spodoptera littoralis (Pottier et al., 2012). Therefore, we infer that it is likely CYP324A1 participates in the recognition of gymnosperms. Three sequences of P450 in Macrococcus (GenBank accession: RKO09869, RKO10724, and RKO10218) showed high similarity with CYP6AEs sequence of Dpun_Gene.2542, Gpop_Gene.44204, and Gpop_Gene.44204. Among the three species, we predicted a total of six CYP6AEs, two CYP6AE2 in D. tabulaeformis, one CYP6AE2 and CYP6AE22 in D. punctatus, and CYP6AE2 and CYP6AE15v2 in G. populifolia. Members of CYP6AEs subfamily are sensitive to pyrethroids and plant toxins, and nine of CYP6AEs in H. armigera (except CYP6AE20) convert esfenvalerate to 4’-hydroxyesfenvalerate (Shi et al., 2018). Feeding on a diet with 0.16% gossypol upregulates CYP6AE11, CYP6AE12, CYP6AE14, and CYP6AE17 in H. armigera larvae (Celorio-Mancera et al., 2011). Therefore, we speculate that the evolution of CYP6AEs, which regulate plant detoxification, may be a result of horizontal transfer of bacterial genes.
CYP4 clade
Six groups were identified in CYP4 clade of G. populifolia, D. punctatus, and D. tabulaeformis, the most common member of CYP4 group was CYP4C, and the least was the CYP4L6; others members were CYP340, CYP341, CYP4G, and CYP4M. It appeared that CYP340, CYP341, CYP4M, and CYP4L were Lepidoptera-specific, and these four groups were monophyletic (Figure 2). In the CYP4 tree of eight insects, CYP4C was not well clustered, generally the same order clustered into a group. CYP4 clade had numerous P450s, indicating that CYP4s may be involved in the metabolism of foreign substances. For example, exposure of B. mori to pesticide upregulated CYP4M5 (Yamamoto et al., 2010), and CYP341A2 (a degrading enzyme) was expressed in chemosensory tissues in Papilio xuthus suggesting that it is involved in plant recognition (Ono et al., 2005).
Most CYP4Gs were clustered into a monophyletic group (Figure 2). Sequence conservatism indicated that CYP4Gs could be involved in physiological functions (Feyereisen, 2005). Later studies showed that silencing of CYP4G1 in Drosophila melanogaster by RNAi reduced hydrocarbon production (Qiu et al., 2012). CYP4G is a subfamily of cytochromes P450 which converts decarbonylate aldehydes to hydrocarbon. It is involved in the cuticular hydrocarbon biosynthesis and pheromone production (MacLean et al., 2018). In our study, P450 of S. sediminis sp. was clustered with CYP4g15 of Coleoptera. In addition, P450 of M. caseolyticus was clustered together with CYP4g15 of Lepidoptera. In the common predicted motif of eight insects and two bacteria, the highly conserved sequences were all near the active centers (Heme), indicating that CYP4Gs (especially CYP4g15) are highly functionally conserved, and we speculate that the existence of mixed branches of bacterial p450 and insect CYP4g15 may be a result of horizontal transfer of genes or may indicate that insect CYP4g15 was inherited from bacteria. It has been reported that CYPGs may be insect-specific, catalyzing the final step in hydrocarbon production in all insects (Feyereisen, 2012), but the exact function of CYP4g15 requires further exploration.
Cytochrome P450 reductase
CPR shuttles electrons from pyridine nucleotide to heme via the conserved domains FMN and FAD. CPR has been associated with the resistance of Aphis citricidus to abamectin, and silencing of the CPR increased the susceptibility to abamectin indirectly by reducing P450 activity (Jing et al., 2018). CPRs of yeast and animal have high sequence homology, indicating that CPRs play a very important role in the process of biological evolution (Shen and Kasper, 1993). In our study, the CPR of insects shows high sequence homology to CPRs of bacteria S. aureus and B. anthracis (Supplementary Figure 2). The aligned sequence was mainly FMN and FAD, indicating that the functions of bacterial CPR and insect CPR are similar. CPR catalyzes electron transfer from NADPH to all known microsomal CYPs (Wang et al., 1997), pointing to the possibility that P450s may have evolved from bacteria.
Conclusion
In summary, P450s participate in the detoxification and metabolism of endogenous and exogenous substances in insects and therefore play an important role in insect adaptation to host plants and environment. Here, more than 90 P450s of G. populifolia, D. punctatus, and D. tabulaeformis species were investigated. A phylogenetic tree of CYP4s including insect species and bacteria shows that CYP4g15 may exist in all insects and exhibits slow evolution. Insect CYP4g15 and CYP6AE may have evolved from bacteria. The GC content of nucleic acid of these bacteria is higher compared to that of insects. CYP2 and Mito. CYP in eight insects showed high co-orthology than CYP3 and CYP4. The genes of CYP3 and CYP4 were more species-specific in terms of function and environmental adaptation. Yet, the specific functions of some genes require further examination. Further approaches are required to determine the full length of the truncated P450s identified based on transcriptome.
Data availability statement
The data presented in this study are deposited in the GenBank repository, BioProject accession number PRJNA578660.
Author contributions
ABZ and XMZ designed the study. XMZ wrote the manuscript and performed the experiments. XMZ, ZYS, CQY, and JL analyzed data. JZL and ABZ revised the thesis writing. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Nos. 32101282 and 32170421) and a postdoctoral research grant from Hebei Normal University.
Acknowledgments
We thank Sufang Zhang in Research Institute of Forest Ecology, Environment and Protection, and Chinese Academy of Forestry for providing the larvae of Dendrolimus punctatus.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.948043/full#supplementary-material
Supplementary Figure 1 | Second structure of CYP4g15 (Gastropacha populifolia, Dendrolimus punctatus, Dendrolimus tabulaeformis, Bombyx mori, Acyrthosiphon pisum, Cinara pinitabulaeformis, Tribolium castaneum, and Dendrolimus ponderosae). The secondary structure of 6c94.1.A was used as template for the secondary structure prediction, including alpha helix and beta folding and beta corner, the five conserved domains were WxxxR(Helix-C), GxE/DTT/S(Helix-I), ExxR(Helix-K), PxxFxPE/DRF (PERF motif), and PFxxGxRxCxG/A (heme-binding domain), and the three motifs predicted in the Figure 3 are marked with different line colors. All red areas indicate identical amino acids.
Supplementary Figure 2 | Alignment of seven insects (Gastropacha populifolia, Dendrolimus punctatus, Dendrolimus tabulaeformis, Bombyx mori, Acyrthosiphon pisum, Tribolium castaneum, and Dendrolimus ponderosae) and two bacteria (Staphylococcus aureus and Bacillus anthracis) CPRs. The conservative locus of FAD-binding domains is shown in the red box, and the conservative locus of NADP-binding domains is shown in the green box.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA578660
- ^ http://transdecoder.sf.net
- ^ https://swissmodel.expasy.org/interactive
- ^ http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi
References
Ahmad, M., Sayyed, A. H., Saleem, M. A., and Ahmad, M. (2008). Evidence for field evolved resistance to newer insecticides in Spodoptera litura (Lepidoptera: Noctuidae) from Pakistan. Crop Prot. 27, 1367–1372. doi: 10.1016/j.cropro.2008.05.003
Ai, J. W., Zhu, Y., Duan, J., Yu, Q. Y., Zhang, G. J., Wan, F., et al. (2011). Genome-wide analysis of cytochrome P450 monooxygenase genes in the silkworm, Bombyx mori. Gene 480, 42–50. doi: 10.1016/j.gene.2011.03.002
Antony, B., Johny, J., Abdelazim, M. M., Jakše, J., Al-Saleh, M. A., and Pain, A. (2019). Global transcriptome profiling and functional analysis reveal that tissue-specific constitutive overexpression of cytochrome p450s confers tolerance to imidacloprid in palm weevils in date palm fields. BMC Genom. 20:440. doi: 10.1186/s12864-019-5837-4
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: A web-based environmentfor protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Baba, T., Kuwahara-Arai, K., Uchiyama, I., Takeuchi, F., Ito, T., and Hiramatsu, K. (2009). Complete genome sequence of Macrococcus caseolyticus strain jscs5402, reflecting the ancestral genome of the human-pathogenic Staphylococci. J. Bacteriol. 191, 1180–1190. doi: 10.1128/JB.01058-08
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208. doi: 10.1093/nar/gkp335
Bloch, K. H. (1983). Sterol structure and membrane function. CRC Crit. Rev. Biochem. 14, 47–92. doi: 10.3109/10409238309102790
Celorio-Mancera, M. D. L. P., Ahn, S. J., Vogel, H., and Heckel, D. G. (2011). Transcriptional responses underlying the hormetic and detrimental effects of the plant secondary metabolite gossypol on the generalist herbivore Helicoverpa armigera. BMC Genom. 12:575. doi: 10.1186/1471-2164-12-575
Chen, C. C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv [Preprint]. doi: 10.1101/289660
Chen, X., and Palli, S. R. (2022). Midgut-specific expression of CYP321A8 P450 gene increases deltamethrin tolerance in the fall armyworm Spodoptera frugiperda. J. Pest Sci. doi: 10.1007/s10340-022-01483-7
Cheng, T., Wu, J., Wu, Y., Chilukuri, R. V., Huang, L., Yamamoto, K., et al. (2017). Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 1, 1747–1756. doi: 10.1038/s41559-017-0314-4
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Dermauw, W., Van Leeuwen, T., and Feyereisen, R. (2020). Diversity and evolution of the P450 family in arthropods. Insect Biochem. Mol. Biol. 127:103490. doi: 10.1016/j.ibmb.2020.103490
Feng, S., Bai, M., Rivas-Gonzalez, I., Li, C., Liu, S., Tong, Y., et al. (2022). Incomplete lineage sorting and phenotypic evolution in Marsupials. Cell 185, 1646–1660.e18. doi: 10.1016/j.cell.2022.03.034
Feyereisen, R. (1999). Insect P450 enzymes. Annu. Rev. Entomol. 44, 507–533. doi: 10.1146/annurev.ento.44.1.507
Feyereisen, R. (2005). “Insect cytochrome P450,” in Comprehensive Molecular Insect Science, eds L. I. Gilbert, K. Iatrou, and S. Gill (Oxford: Elsevier), 1–77.
Feyereisen, R. (2006). Evolution of insect P450. Biochem. Soc. Trans. 34, 1252–1255. doi: 10.1042/BST0341252
Feyereisen, R. (2012). “Insect CYP genes and P450 enzymes,” in Insect Molecular Biology and Biochemistry, ed. L. Gilbert (London: Elsevier), 236–316.
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., and Bowden, J. (2013). De novo transcript sequence reconstruction from RNA-Seq: Reference generation and analysis with trinity. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Helvig, C., Koener, J. F., Unnithan, G. C., and Feyereisen, R. (2004). Cyp15a1, the cytochrome p450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in cockroach corpora allata. Proc. Natl. Acad. Sci. U.S.A. 101, 4024–4029. doi: 10.1073/pnas.0306980101
Jing, T. X., Tan, Y., Ding, B. Y., Dou, W., Wei, D. D., and Wang, J. J. (2018). NADPH–cytochrome p450 reductase mediates the resistance of aphis (Toxoptera) citricidus) (Kirkaldy) to abamectin. Front. Physiol. 9:986. doi: 10.3389/fphys.2018.00986
Kim, D., and Jeong, M. S. (2010). Summer lepidopterous insect fauna of Mt. Bokju (prov. gangwon-do) in Korea. J. Korean Nat. 3, 223–234. doi: 10.1016/s1976-8648(14)60027-x
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44:W242–W245. doi: 10.1093/nar/gkw290
Lin, Q., Jin, F., Hu, Z., Chen, H., Yin, F., Li, Z., et al. (2013). Transcriptome analysis of chlorantraniliprole resistance development in the diamondback moth Plutella xylostella. PLoS One 8:e72314. doi: 10.1371/journal.pone.0072314
MacLean, M., Nadeau, J., Gurnea, T., Tittiger, C., and Blomquist, G. J. (2018). Mountain pine beetle (D. Ponderosae) CYP4Gs convert long and short chain alcohols and aldehydes to hydrocarbons. Insect Biochem. Mol. Biol. 102, 11–20. doi: 10.1016/j.ibmb.2018.09.005
Maeda, S., Nakashima, A., Yamada, R., Hara, N., Fujimoto, Y., Ito, Y., et al. (2008). Molecular cloning of ecdysone 20-hydroxylase and expression pattern of the enzyme during embryonic development of silkworm Bombyx mori. Comp. Biochem. Physiol. Part B 149, 507–516. doi: 10.1016/j.cbpb.2007.11.015
Moulton, J. K., Pepper, D. A., and Dennehy, T. J. (2000). Beet armyworm (Spodoptera exigua) resistance to spinosad. Pest Manag. Sci. 56, 842–848. doi: 10.1002/1526-4998(200010)56:10<842::AID-PS212>3.0.CO;2-H
Namiki, T., Niwa, R., Sakudoh, T., Shirai, K., Takeuchi, H., and Kataoka, H. (2005). Cytochrome P450 CYP307A1/spook: A regulator for ecdysone synthesis in insects. Biochem. Biophys. Res. Commun. 337, 367–374. doi: 10.1016/j.bbrc.2005.09.043
Nelson, D. R., and Strobel, H. W. (1987). Evolution of cytochrome P-450 proteins. Mol. Biol. Evol. 4, 572–593. doi: 10.1093/oxfordjournals.molbev.a040471
Omura, T. (1993). “Localization of cytochrome P450 in membranes: Mitochondria,” in Cytochrome P450, eds J. B. Schenkman and H. Greim (Berlin: Springer), 61–69.
Ono, H., Ozaki, K., and Yoshikawa, H. (2005). Identification of cytochrome p450 and glutathione-s-transferase genes preferentially expressed in chemosensory organs of the Swallowtail butterfly, Papilio xuthus L. Insect Biochem. Mol. Biol. 35, 837–846. doi: 10.1016/j.ibmb.2005.03.013
Paine, M. J. I., Scrutton, N. S., Munro, A. W., Gutierrez, A., Roberts, G. C. K., and Wolf, C. R. (2005). Electron Transfer Partners of Cytochrome. New York, NY: Springer, 450.
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Pottier, M. A., Bozzolan, F., Chertemps, T., Jacquin-Joly, E., Lalouette, L., and Siaussat, D. (2012). Cytochrome P450s and cytochrome P450 reductase in the olfactory organ of the cotton leafworm Spodoptera littoralis. Insect Mol. Biol. 21, 568–580. doi: 10.1111/j.1365-2583.2012.01160.x
Qiu, Y., Tittiger, C., Wicker-Thomas, C., Le Goff, G., Young, S., Wajnberg, E., et al. (2012). An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 109, 14858–14863. doi: 10.1073/pnas.1208650109
Raman, B., and Andy, M. (2018). Expansion of cytochrome p450 and cathepsin genes in the generalist herbivore brown marmorated stink bug. BMC Genom. 19:60. doi: 10.1186/s12864-017-4281-6
Rewitz, K. F., O’Connor, M. B., and Gilbert, L. I. (2007). Molecular evolution of the insect halloween family of cytochrome p450s: Phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 37, 741–753. doi: 10.1016/j.ibmb.2007.02.012
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42:W320–W324. doi: 10.1093/nar/gku316
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Scott, J. G., Liu, N., and Wen, Z. (1998). Insect cytochromes p450: Diversity, insecticide resistance and tolerance to plant toxins. Comp. Biochem. Physiol. Part C 121, 147–155. doi: 10.1016/S0742-8413(98)10035-X
Shen, A. L., and Kasper, C. B. (1993). “Mechanistic studies on the reductive half-reaction of NADPH-cytochrome P450 oxidoreductase,” in Handbook of experimental pharmacology, eds J. B. Schenkman and H. Greim (New York, NY: Springer), 35–59.
Shi, Y., Wang, H., Liu, Z., Wu, S., Yang, Y., Feyereisen, R., et al. (2018). Phylogenetic and functional characterization of ten P450 genes from the CYP6AE subfamily of Helicoverpa armigera involved in xenobiotic metabolism. Insect Biochem. Mol. Biol. 93, 79–91. doi: 10.1016/j.ibmb.2017.12.006
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sztal, T., Chung, H., Berger, S., Currie, P. D., Batterham, P., and Daborn, P. J. (2012). A cytochrome P450 conserved in insects is involved in cuticle formation. PLoS One 7:e36544. doi: 10.1371/journal.pone.0036544
Wan, P. J., Shi, X. Q., Kong, Y., Zhou, L. T., Guo, W. C., Ahmat, T., et al. (2013). Identification of cytochrome p450 monooxygenase genes and their expression profiles in cyhalothrin-treated Colorado potato beetle, Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 107, 360–368. doi: 10.1016/j.pestbp.2013.10.004
Wang, B., Shahzad, M. F., Zhang, Z., Sun, H., Han, P., Li, F., et al. (2014). Genome-wide analysis reveals the expansion of cytochrome P450 genes associated with xenobiotic metabolism in rice striped stem borer, Chilo suppressalis. Biochem. Biophys. Res. Commun. 443, 756–760. doi: 10.1016/j.bbrc.2013.12.045
Wang, M., Roberts, D. L., Paschke, R., Shea, T. M., Masters, B. S., and Kim, J. J. (1997). Three-dimensional structure of NADPH-cytochrome P450 reductase: Prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. U.S.A. 94, 8411–8416. doi: 10.1073/pnas.94.16.8411
Wang, R. L., Staehelin, C., Xia, Q. Q., Su, Y. J., and Zeng, R. S. (2015). Identification and characterization of CYP9A40 from the tobacco cutworm moth (Spodoptera litura), a cytochrome p450 gene induced by plant allelochemicals and insecticides. Int. J. Mol. Sci. 16, 22606–22620. doi: 10.3390/ijms160922606
Wu, S., Huang, Z., Rebeca, C. L., Zhu, X., Guo, Y., Lin, Q., et al. (2017). De novo characterization of the pine aphid cinara Pinitabulaeformis zhang et zhang transcriptome and analysis of genes relevant to pesticides. PLoS One 12:e0178496. doi: 10.1371/journal.pone.0178496
Xu, L., Mei, Y., Liu, R., Chen, X., Li, D., and Wang, C. (2020). Transcriptome analysis of Spodoptera litura reveals the molecular mechanism to pyrethroids resistance. Pestic. Biochem. Physiol. 169:104649. doi: 10.1016/j.pestbp.2020.104649
Yamamoto, K., Ichinose, H., Aso, Y., and Fujii, H. (2010). Expression analysis of cytochrome P450s in the silkmoth, Bombyx mori. Pestic. Biochem. Physiol. 97, 1–6. doi: 10.1016/j.pestbp.2009.11.006
Yin, C., Shen, G., Guo, D., Wang, S., Ma, X., Xiao, H., et al. (2016). InsectBase: A resource for insect genomes and transcriptomes. Nucleic Acids Res. 44:D801–D807. doi: 10.1093/nar/gkv1204
Zhang, X. M., Shi, Z. Y., Zhang, S. Q., Zhang, P., Wilson, J.-J., Shih, C., et al. (2021). Plant-herbivorous insect networks: Who is eating what revealed by long barcodes using high-throughput sequencing and Trinity assembly. Insect Sci. 28, 127–143. doi: 10.1111/1744-7917.12749
Keywords: cytochrome P450, cytochrome P450 reductase, lepidoptera, evolution, herbivorous insect, bacteria
Citation: Zhang X, Shi Z, Yang C-q, Li J, Liu J and Zhang A-b (2022) Gut transcriptome analysis of P450 genes and cytochrome P450 reductase in three moth species feeding on gymnosperms or angiosperms. Front. Ecol. Evol. 10:948043. doi: 10.3389/fevo.2022.948043
Received: 19 May 2022; Accepted: 24 October 2022;
Published: 17 November 2022.
Edited by:
Sufang Zhang, Chinese Academy of Forestry, ChinaReviewed by:
Rui Tang, Institute of Zoology, Guangdong Academy of Sciences (CAS), ChinaChengxu Wu, Guizhou University, China
Copyright © 2022 Zhang, Shi, Yang, Li, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ai-bing Zhang, emhhbmdhYjIwMDhAY251LmVkdS5jbg==
 Xiaoman Zhang
Xiaoman Zhang Zhiyong Shi
Zhiyong Shi Cai-qing Yang2
Cai-qing Yang2 Jing Li
Jing Li Jingze Liu
Jingze Liu Ai-bing Zhang
Ai-bing Zhang