- 1CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, The Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan, China
- 2University of Chinese Academy of Sciences, Beijing, China
DICER-likes (DCLs) proteins are the core component for non-coding RNA (ncRNA) biogenesis, playing essential roles in some biological processes. The DCL family has been characterized in model plants, such as Arabidopsis, rice, and poplar. However, the evolutionary aspect and the expression mechanism under drought stress were scarce and have never been reported and characterized in one of the most important worldwide cultivated fruit trees, peach (Prunus persica). Eight DCLs genes in the Prunus persica genome were detected, in addition to 51 DCLs in the other seven Rosaceae genomes. The phylogenetic analysis with Arabidopsis thaliana and RTL1 gene as outgroups suggested that DCL members are divided into four clades: DCL1, DCL2, DCL3, and DCL4 with several gene gain/loss events of DCL gene copies through the evolutionary tract of the Rosacea family. The number of homologous DCL copies within each clade, along with the chromosomal location indicated gene duplication event of the DCL2 gene occurred once for the subfamily Amygdaloideae and twice for Pyrus communis and Prunus dulics and trice for the P. persica on Chromosome number 7 genes. Another duplication event was found for the DCL3 gene that occurred once for all the eight Rosaceae species with no match in A. thaliana. The DCL genetic similarity and activity was evaluated using BLASTp and previously published RNA-seq data among different tissues and over different time points of peach trees exposed to drought conditions. Finally, the expression pattern of PrupeDCLs in response to drought stress was identified, and two of these members, Prupe.7G047900 and Prupe.6G363600, were found as main candidate genes for response to drought stress. Our data presented here provide useful information for a better understanding of the molecular evolution of DCL genes in Rosaceae genomes, and the function of DCLs in P. persica.
Introduction
Plants are continuously exposed to several biotic and abiotic stresses during their life cycle. Among others, drought is one important abiotic stress that cause a severe loss in the agriculture sector. Over the course of evolution, plant have acquired different drought-tolerance mechanisms that lead to the emergence of new genotypes and varieties with different drought tolerance capabilities. The duration of water deficit in the soil influence the drought stress severity which initiate with water loss from the cells that leads to cellular dehydration, osmotic stress, and reactive oxygen species production (Khan et al., 2015a; Hasanuzzaman et al., 2018). Then activates various molecular pathway cascades of large signaling network and increasing the activities of antioxidant enzymes (Liu et al., 2014; Haider et al., 2018).
Peach (Prunus persica) belongs to the Rosaceae family, which is an important worldwide cultivated fruit tree (Verde et al., 2013; Cao et al., 2014). Because of its short reproductive cycle, nutritional and economic importance, and small genome size (Alves et al., 2016; Zhou et al., 2019; Zhou et al., 2021), P. persica has become an emerging model tree species for genetics, molecular biology, and plant physiology research. Under drought stress, fruit dry matter content and firmness due to the reduction of water in fruits increases, resulting in smaller fruits (Haider et al., 2018; Priya et al., 2019; Singh et al., 2019). Consequently, the fruit surface conductance and its transpiration decrease causing a significant effect on the concentrations of non-structural compounds either through the decrease in dilution and/or modifications of their metabolism (Rahmati et al., 2014). Also, drought has a critical effect on fruit growth and quality properties. Trees yield would be sharply reduced at least 25–50% in yield total weight (Rahmati et al., 2014; Khan et al., 2015b; Belal et al., 2017; Haider et al., 2018).
The small RNA (sRNA) biogenesis and regulation are controlled by three RNAi proteins-encoded gene families, known as Dicer-like (DCL), Argonaute (AGO), and RNA-dependent RNA polymerase (RDR) (Vaucheret, 2006; Chapman and Carrington, 2007; Kapoor et al., 2008; Qu et al., 2008; Cao et al., 2020b). Among them, Dicer and Dicer like (DCLs) genes are an important components of sRNA biogenesis in plants (Xie et al., 2005; Margis et al., 2006; Kapoor et al., 2008; Vaucheret, 2008; Liu et al., 2009b; Mukherjee et al., 2013). In the same context, Acquisition of DCL functions associated with distinct RNA interference branches in plants occurred during, not before, evolution of the plant lineage (Xie et al., 2005). The DCL gene family usually contains inconsistent number of genes copies that vary among plant species (Kapoor et al., 2008; Liu et al., 2009a; Qian et al., 2011; Cao et al., 2020b). For example, the number of recorded DCL copies was four in Arabidopsis (Arabidopsis thaliana), five in poplar (Populus trichocarpa), willow (Salix suchowensis) and maize (Zea mays), seven in tomato (Solanum lycopersicum), and eight in rice (Oryza sativa) (Finnegan and Matzke, 2003; Kapoor et al., 2008; Qian et al., 2011; Qin et al., 2018; Cao et al., 2020b). Based on previous researches, the responses of DCLs to drought, cold, and salt showed to be quite different, indicating that plants might have specialized regulatory mechanism in response to different abiotic stresses (Liu et al., 2009a). In A. thaliana all the DCL genes were fully characterizes, and their role in drought tolerance in addition to the sRNA biogenesis was confirmed (Xie et al., 2005; Deleris et al., 2006). However, the number and function of DCL genes in other plant species (e.g., Roseacea) are poorly studied (Kapoor et al., 2008; Song et al., 2012; Qin et al., 2017).
The current study aimed to characterize the DCL gene family at genomic and transcriptomic levels under drought conditions in Peach (Prunus persica) (International Peach Genome et al., 2013; Verde et al., 2013; Verde et al., 2017) in comparison with additional seven other Rosaceae species. Strawberry (Fragaria vesca) (Li et al., 2019b), China rose (Rosa chinensis) (Hibrand Saint-Oyant et al., 2018; Raymond et al., 2018; Li et al., 2019a; Lin et al., 2019), and Black Raspberry (Rubus occidentalis) (VanBuren et al., 2018) that represent the Rosoideae subfamily. Two species from genus Maleae, apple (Malus × domestica) (Bianco et al., 2014), and pear (Pyrus communis) (Linsmith et al., 2019), in addition to species from genus Prunus, Almond (Prunus dulcis) (Alioto et al., 2020), and Armenian plum (Prunus armeniaca) (Jiang et al., 2019), all represent the Amygdaloideae subfamily.
Materials and Methods
Extraction and Motif Analysis of DCL Genes
The genome sequences of eight Rosaceae species (Supplementary Table 1) were downloaded from the GDR database.1 The DCLs of A. thaliana (AtDCL) were downloaded from TAIR database2 as described by Zhang et al. (2012), Verde et al. (2013), Wu et al. (2013), Daccord et al. (2017), Edger et al. (2018), and Cao et al. (2020b). The HMM model of DCLs was generated based on AtDCL copies by TBtools software (Chen et al., 2020), and then it was used to identify the DCLs members by HMMER3 software (Mistry et al., 2013). All putative genes were validated by using NCBI database3 to confirm and acquire DCL hits from Viridiplantae genomes. To avoid sequence redundancy; we only considered primary transcripts in this study. The primary transcript names, chromosomal locations, and protein lengths of DCLs were obtained from the genome annotation files of these eight Rosaceae genomes. At protein level, Prunus persica DCL copies were investigated for conserved motifs using MEME suite online tool.4 At coding DNA sequence (CDS) level, Similarity levels among different DCLs genes were conducted by using ‘‘Circos’’ plot design software5 that basically visualized by Krzywinski et al. (2009) and described by Zhang et al. (2013) as similar genome regions.
Phylogenetic Analysis and Evolutionary Calibration
The MAFFT software was used to conduct three multiple alignments, one includes all the P. persica DCL copies, the other includes all BLAST top 10 hits of the from NCBI database (Tophits-set) and another includes all Rosaceae-related DCL proteins (Rosa-set). The second and third alignments were performed along with the AthDCL copies (Katoh et al., 2005). The selected Rosaceae species relationship dendrogram was drawn based on Taxonomy database (NCBI) and visualized using iTOL webtool6 and supported with estimated separation time obtained from7 The Fasttree V2 software (Price et al., 2010) was used to construct the phylogenetic tree including retrotransposon-like 1 (RTL1) genes from the same selected Rosaceae species as outgroup using Jones-Taylor-Thorton model at rate categories of sites equal 20. The DCL-based phylogenetic tree was evolutionary calibrated by relative-time maximum likelihood method using MEGA-X software (Kumar et al., 2018).
RNA-Seq Expression Analyses
Two published RNAseq data were used to validate the expression of the detected DCL copies from previous studies on P. persica, one sampled different tissue from high and low altitudes (Leaf, Phloem, Flower, Fruit, Seed, and Root; Bioproject: PRJNA694331) and the other sampled the fruit flesh under drought stress over different time points (0 h, 3 h, 6 h, 12 h, 24 h, 3 days, 6 days, and 12 days; Bioproject: PRJNA694007). For each data set Trimmomatic was used to remove adapter sequences and low-quality sequencing reads with default parameters (Bolger et al., 2014; Cao et al., 2020b). The HISAT2 was used to align the clean reads to the masked genome (Kim et al., 2019), and the Stringtie was used to calculate the FPKM values (Fragments Per Kilobase of transcript per Million mapped reads) of DCLs in the P. persica reference genome (Pertea et al., 2016). The expression of DCL among different tissue was shown as a heatmap drawn by TBtools software (Chen et al., 2020). While the expression of DCL over time was modelized in expression clusters and visualized as parallel coordinate plot using xlstat software.8
Results and Discussion
Identification of DCL Genes in Prunus persica and Other Rosaceae Species
Total of eight DCL copies were detected in P. persica by BLAST search (Supplementary Table 2). Five motifs were defined, all identified as ribonuclease III domain according to pfam database. The five motifs were distributed differently among the eight copies, causing different phylogenetic and clustering signals (Figure 1A). All the four DCL copies as previously reported from A. thaliana and other species were detected and phylogenetically clustered accordingly (Mukherjee et al., 2013; Cao et al., 2020b). The top 10 BLAST hits of the eight P. persica DCL copies were equally clustered into four major clusters, where DCL1 and DCL4 were monophyletic, while DCL2 and DCL3 were paraphyletic, as shown in Figure 1B.
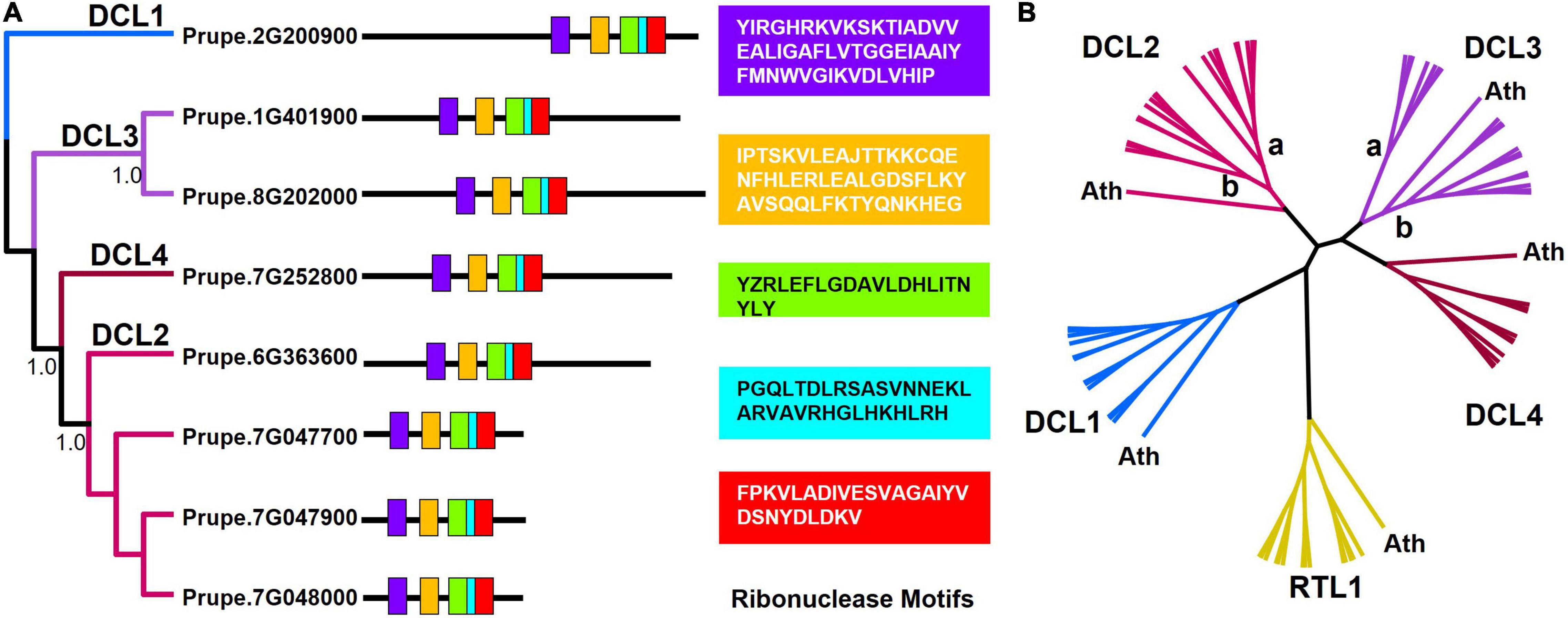
Figure 1. Conserved motifs of Prunus persica DCLs genes. (A) Gene structure and domain compositions of P. persica DCLs based on the presence of conserved Ribonuclease motifs (detailed in colored boxes) were arranged and clustered in four groups. (B) Unrooted maximum likelihood DCL phylogenetic tree shows the RTL1 gene outgroup, and A. thaliana species outgroup along the NCBI BLAST top 10 hits of P. persica DCL copies. A unique cluster is observed for DCL1, and DCL4, while both DCL2 and DCL3 were separated into two subclades.
In Rosaceae, 405 DCL-similar transcripts were detected from the seven selected Rosaceae genomes, including all isoforms. By excluding, the five DCL copies of A. thaliana (AT), and the eight P. persica (Prupe), a total of 51 non-redundant DCLs were retained for further analysis, five F. vesca (Frv), five R. chinensis (Rch), six P. armeniaca (PAR), seven P. dulcis (Prudul), seven R. occidentalis (Roc) (Xie et al., 2005), nine Pyrus communis (pycom), and 12 Malus x domestica (MDP) as displayed in Figure 2A and Supplementary Table 2. Compared with other families, DCL gene family contains relatively few members, which is consistent with the previous studies reported in P. trichocarpa (5) (Cao et al., 2020b), S. suchowensis (5) (Cao et al., 2020b), O. sativa (8) (Kapoor et al., 2008), S. esculentum (7) (Bai et al., 2012) and A. thaliana (4) (Deleris et al., 2006). It does not appear to be a direct correlation between the number of DCL genes and genome size. For example, it has been observed that the numbers of DCL genes differed between P. armeniaca (6) and P. dulcis (8), but their genome size is not significantly differed (P. armeniaca: 206.1 Mb, and P. dulcis: 208.9 Mb). However, F. vesca and P. armeniaca had equal number of DCL copies, although they have different genome size: F. vesca (219.29 Mb) and P. armeniaca (206.10 Mb). Rosaceae species share an ancient whole-genome duplication (WGD) event (Wu et al., 2013; Cao et al., 2020a), while experienced a more recent WGD event (Wu et al., 2013; Daccord et al., 2017), which might explain why M. x domestica contains the highest number of DCL gene copies among the studies Rosaceae species.
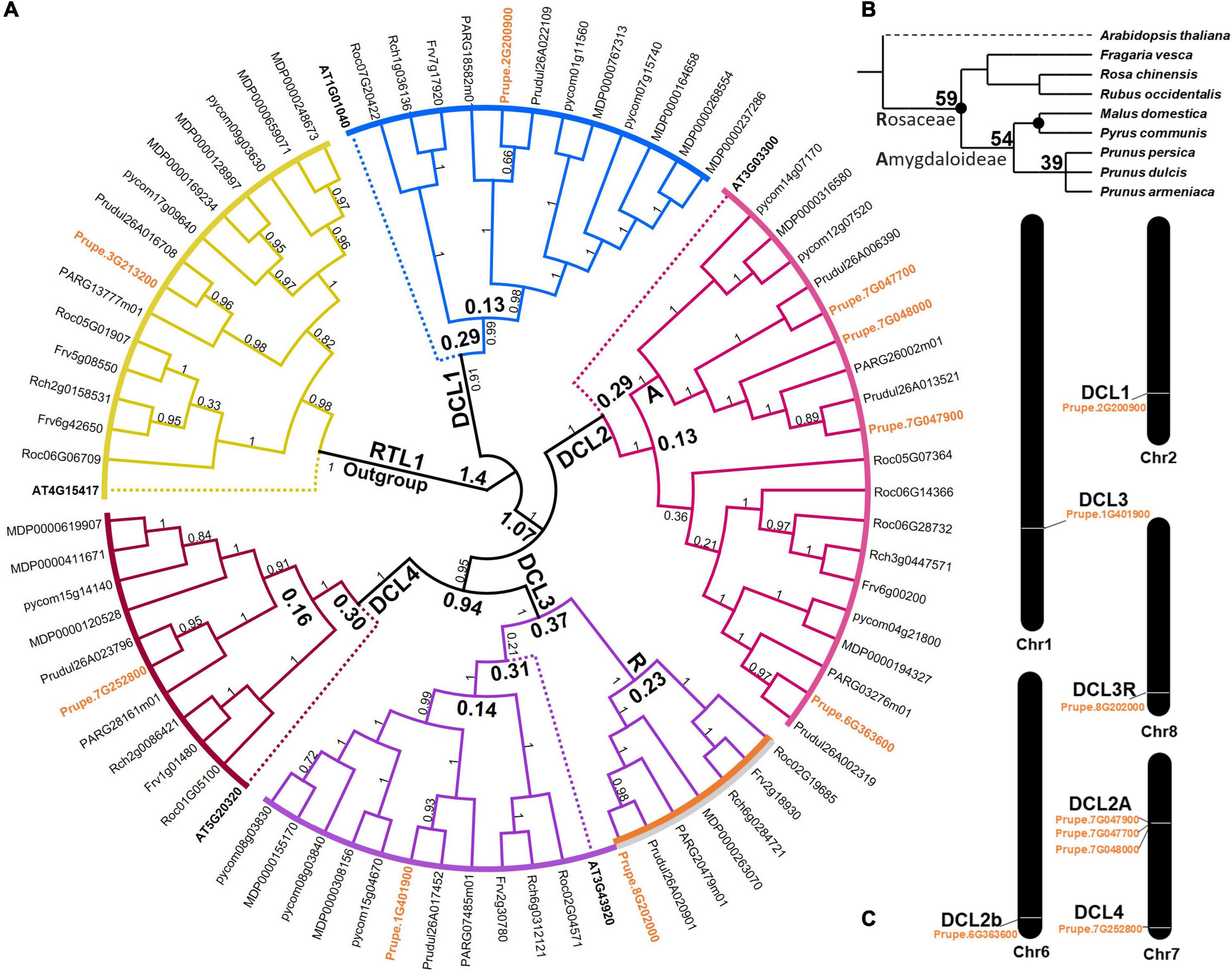
Figure 2. The time-calibrated maximum liklihood phylogenetic tree of DCLs genes of eight Rosacea species. (A) The circular dendogram with colored clusters, each represent one DCL and the RTL1 outgroup. The Prunus persica DCL genes are colored with orange while Arabidopsis DCL genes are marked by dot-lines. The Rosaceae specific DCL3 copy cluster was outlined by orange-gray bolded line and named by letter (R). The Amygdaloideae specific DCL2 copy cluster is named by letter (A). (B) Taxonomy-based relationship of the eight Rosaceae species timely calibrated using data obtained from www.timetree.org/. Black ovals suggest whole-genome duplication (WGD) occurrences. (C) Distributions of DCLs genes on Prunus persica chromosomes. Eight DCLs genes are mapped on five out eight chromosomes. The Amygdaloideae specific DCL2 triple copies are adjacent on chromosome 8.
Phylogenetics and Evolutionary Analysis
All DCL genes were clustered into four clades: DCL1, DCL2, DCL3, and DCL4. This spread on DCLs genes indicating that they play critical roles in plant antiviral response (Necira et al., 2021), as well as contribute to the phased secondary siRNAs production (Gasciolli et al., 2005). clade DCL1 included one PrupeDCL1, one FrvDCL1, four MDPDCL1, two pycomDCL1, one PrudulDCL1, one RocDCL1, one RchDCL1, one PARDCL1, and one AthDCL1 was set as outgroup in the DCL1 clade with the processing of 21 nucleotides (nt) miRNA and contributing to the derivation of siRNAs (Kurihara and Watanabe, 2004; Kurihara et al., 2006), as shown in Figure 2A. Members of the DCL2 clade contained four PrupeDCL2, one FrvDCL2, two MDPDCL2, three pycomDCL2, four PrudulDCL2, three RocDCL2, two PARDCL2 and one RchDCL2 in addition to one AtDCL2 as out group. DCL2A showed tandem duplication between both genes (Prupe.7G047700 and Prupe.7G048000). The members of the DCL3 clade included two PrupeDCL3, two FrvDCL3, three MDPDCL3, three pycomDCL3, two PrudulDCL3, two RocDCL3, two RchDCL3, two PARDCL3 in addition to one AtDCL3 as out group. Our results of phylogenetic analysis showed an important occurrence in the DCLs development of Rosaceae. Clade 3 witnessed a specific split for Rosaceae copies which separated before the Arabidopsis speciation event. That was unlike the usual occurrences in the stages of DCLs duplication of Rosaceae family (0.37 part of the separation time). This event indicates the important occurrence in the evolution of these species and may offer more insights in the coming evolution researches. These copies in clade DCL3 cluster was outlined by orange-gray bolded line and named by letter (R) in Figure 2A. The chronological convergence of the Arabidopsis separation event to the Rosaceae family was noticeable in the phyletic analysis as relative fragments [DCL1 and DCL2(±29); DCL3(±31); DCL4(±30)] which indicates to the normal development and synchronize speciation in different evolution events. Phylogenetic analysis for species was created and the taxonomy-based relationship of the eight Rosaceae species timely calibrated to show the evolutionary relationship between species. The occurrence of the speciation for sub-family Amygdaloideae from Roseaceae family into two branches was demonstrated from the Rosaceae within the estimation study of the relative time 54 million years as time part of the speciation events, as noticed in Figure 2B. The values in the phyletic phases refer to the relative. Black ovals correspond to inferred locations of whole-genome duplication (WGD) events and the values in the head of clades refers to the approximately time of duplications per million years. The distributions of DCLs genes on Prunus persica chromosomes showd eight DCLs genes mapped on five out eight chromosomes as shown in Figure 2C. The Amygdaloideae specific DCL2 triple copies are adjacent on chromosome 7 which include the genes Prupe.7G047900, Prupe.7G047700, and Prupe.7G048000 cluster in a clade DCL2A, indicate that these genes might play different functions in siRNAs production. Previously, the functional differentiation of DCLs in one clade was found in O. sativa DCL3 (Song et al., 2012).
DCLs Genes Similarity
Rosaceae DCLs genes copies were mapped using protein sequences of BLASTP (Altschup et al., 1990) to compare every DCLs annotated with proteome and designated the best hit as a homolog blast data on the different species which were anchored to the PrupeDCLs, As shown in Figure 3. DCL1 copies includes one PrupeDCLs copy (Prupe.2G200900) which scored high similarity with its duplicates while MDPDCLs genes copies showed less similarity than others duplicates. DCL2 has four duplicated copies of PrupeDCLs which have different similarity score with other DCLs. Prupe.6G363600 showed the highest similarity score with other Rosaceae copies in DCL2 compare with the other three PrupeDCLs copies revealed which showed low score. In DCL3, (Prupe.7G252800 and Prupe.8G202000) showed similarity with other DCLs copies of PARDCLs, and PrudulDCLs copies while pycomDCL (pycom08g03830) showed asymmetry with PrupeDCL2 copies. DCL4 has one copy of P. Persica (Prupe.7G252800) which anchored to other copies of species and highly similarity is shown. There is a highly similarity between the DCLs copies hit with PrupeDCLs except MDPDCLs copies which have a noticeable asymmetry score ≤ 0.25. The results clarified that most of PrupeDCLs showed high similarity for their duplicates where this confirms the evolution path that these genes went during the species development for resistance drought.
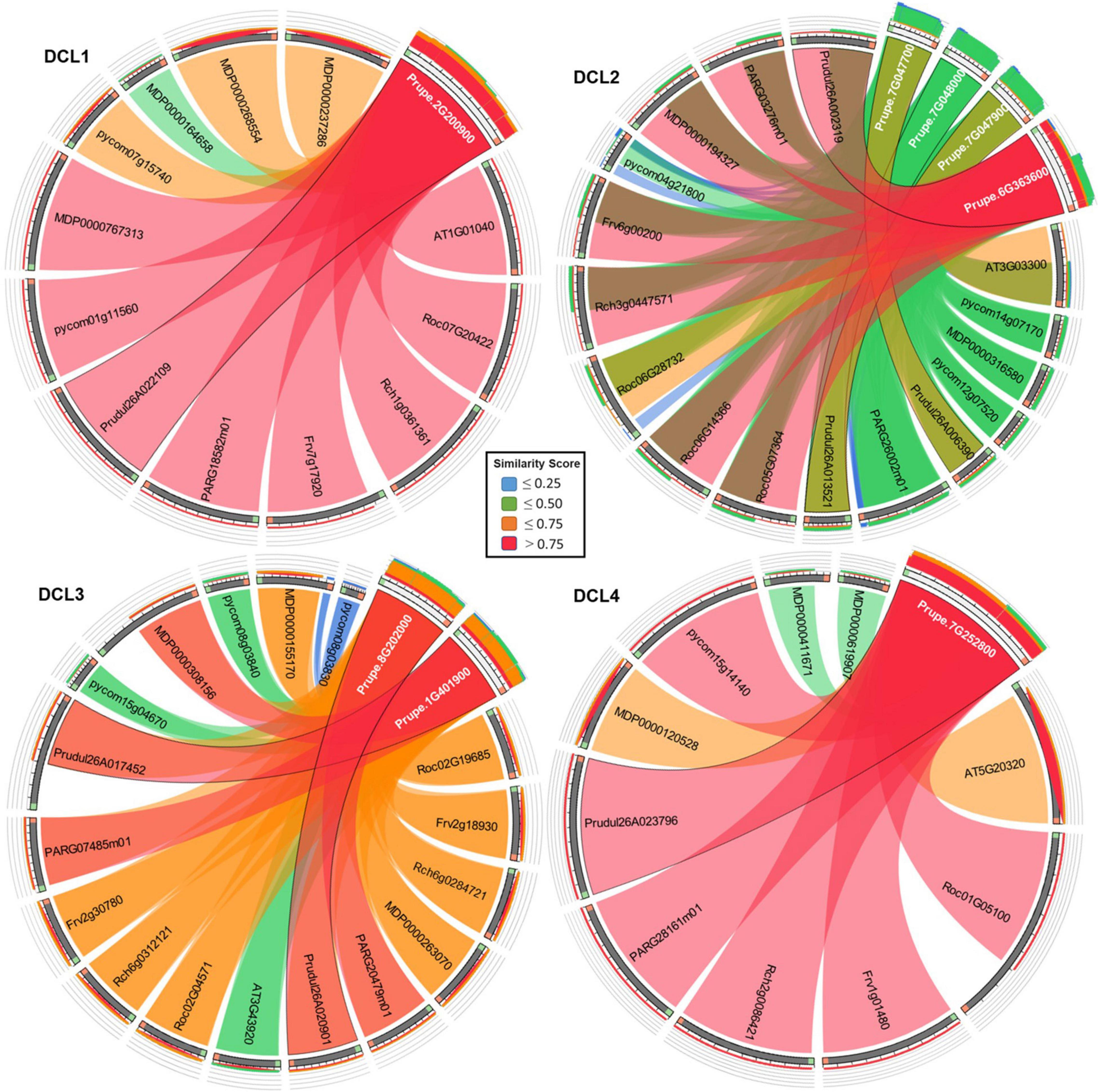
Figure 3. Circos plot of protein similarity among the identified DCLs genes copies. Rosaceae DCLs genes copies were mapped using BLASTP data on the different species which were anchored to the PrupeDCLs. Similarity score is indicated on the inner box and the highlighted arcs in the center of the circos link homologous DCLs genes. P-value<0.05.
Expression Patterns of PrupeDCL Genes in Various Tissues
As the master parts of sRNA biogenesis; DCL genes play important roles in the growth and development of plants (Fang and Spector, 2007; Song et al., 2007). The transcript levels of DCLs may be closely related to their physiological functions. To primarily clarify the functions of PrupeDCLs, the expression patterns of PrupeDCLs were analyzed in six tissues: root, fruit, seed, flower, phloem, and leaf. As shown in Figure 4, Our results found that all PrupeDCLs have expressed in the analyzed P. persica tissues, although they showed different expression patterns that provided a clear vision about how did they work. These results are consistent with the key roles of DCL genes in the miRNAs biogenesis that are involved in plant stress responses and development. Our results exhibited lowest expression showed by PrupeDCL1 (Prupe.2G200900) and PrupeDCL2 (Prupe.7G047700) in the different tissues. Other two PrupeDCL2 genes (Prupe.7G047900, and Prupe.7G048000) showed same expression pattern degree in root tissue. It was observed that PrupeDCL2 (Prupe.6G363600) exclusively expressed in both root tissue and seeds while PrupeDCL4 almost identically expressed in all tissues.
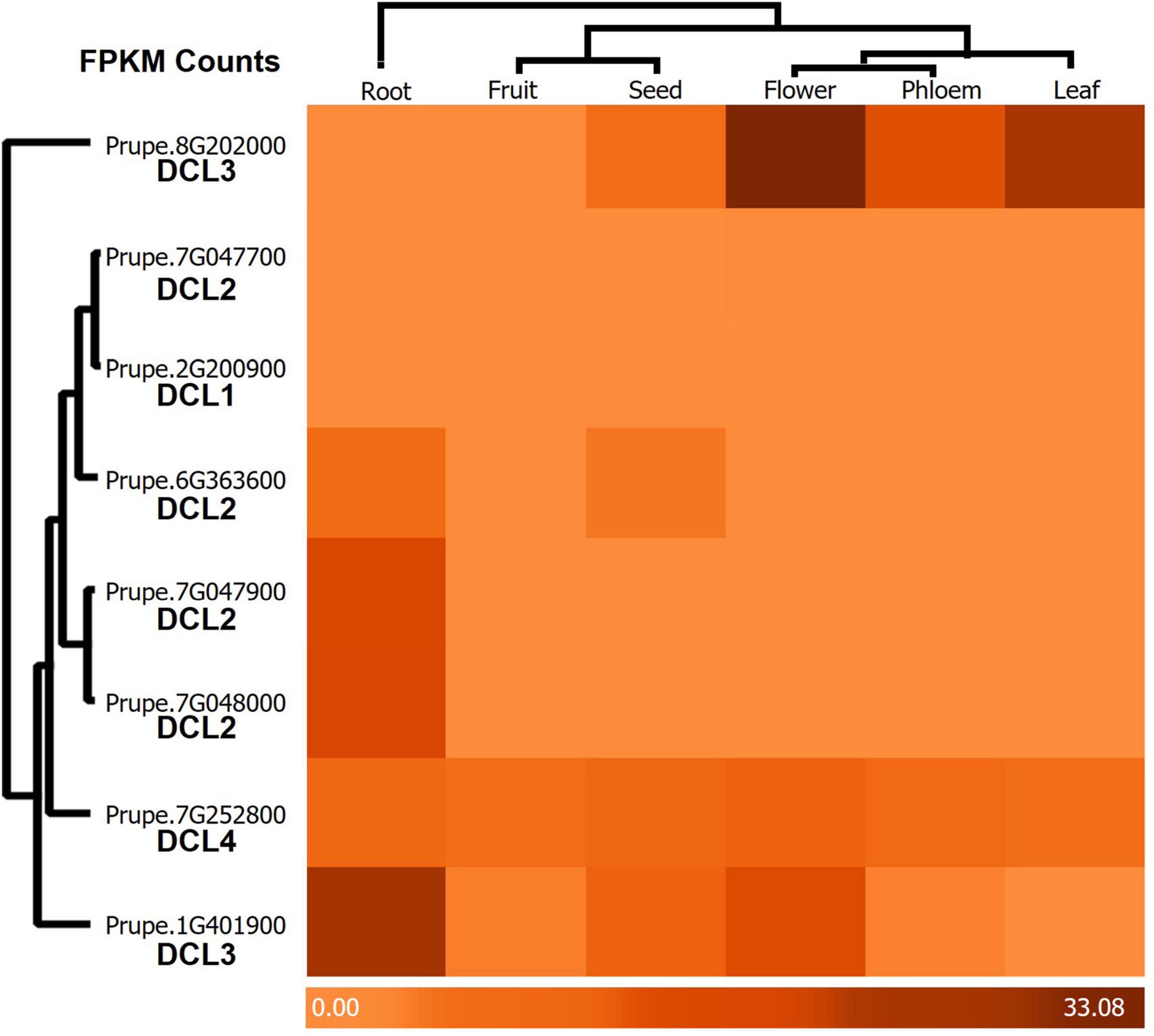
Figure 4. Heatmap of tissue expression pattern of PrupeDCLs blast data in Root, Fruit, Seed, Flower, Phloem, and Leaf by FPKM counts. The color bar showed the expression degree. The phyletic clades were designed within FPKM values counts to identify the most relative items for displaying.
Consistently, we also identified two PrupeDCL3; (Prupe.8G202000 and Prupe.1G401900). The results showed that Prupe.1G401900 was highly expressed in almost all tissues especially in root, and Prupe.1G401900 showed a high level of expression in flower, leaf, phloem and seeds respectively, but recorded low expression in both root and fruit.
Expression Patterns of PrupeDCLs Under Drought Stress
Environmental stress may influence plant growth and health by affecting the regulation of crucial plant genes (Chinnusamy and Zhu, 2009). Many stress-related genes are induced to express under external environmental stresses to help plants cope with adverse conditions (Chinnusamy and Zhu, 2009; Priya et al., 2019). For example, DCLs from P. trichocarpa, S. suchowensis, Saccharum spontaneum, and Solanum tuberosum contribute to plants resist abiotic stress (Esposito et al., 2018; Cao et al., 2020b; Cui et al., 2020). The P. persica is generally widely grown in irrigated semi-arid and arid regions. However, the aggravation of drought caused by the global greenhouse effect has greatly restricted the growth and development of P. persica. To determine the effect of PrupeDCL genes on drought stress, we investigated the expression pattern of PrupeDCL genes in P. persica within the relative expression time. As regards drought stress, the results cleared that all PrupeDCLs were differentially expressed. The expression level of PrupeDCLs have different expression in both of brief and chronical exposure for drought conditions, as shown in Figure 5. DCL1 (Prupe.2G200900) showed high and stable expression in the short-term treatment (time 0:3 days) but it dramatically dropped at the chronic period (3 days: 12 days) while PrupeDCL2 copies (Prupe.7G047900 and Prupe.6G363600) recorded various expression in the short-term treatment but achieved the highest expression in the chronic time of drought treatment as appeared in expression cluster EC1. On the other side, PrupeDCL3 (Prupe.1G401900) and PrupeDCL4 (Prupe.7G252800) have low expression in the relative time of drought treatment in EC3. That could be explained that they worked as chronic co-expression genes to resume resistance drought conditions. PrupeDCL2 (Prupe.7G047700) positively expressed in the brief time (0–12 h) and then continued as moderate expression gene as shown in EC2 was increased at 0–24 h. The transcript levels of PrupeDCLs2 (Prupe.7G04790 and Prupe.6G363600) were significantly upregulated by drought stress for 3–12 day, whereas the two genes from the same clade PrupeDCL2 were unchanged, suggesting that these genes from the same ancestor were functionally redundant or neo-functionalization during evolution.
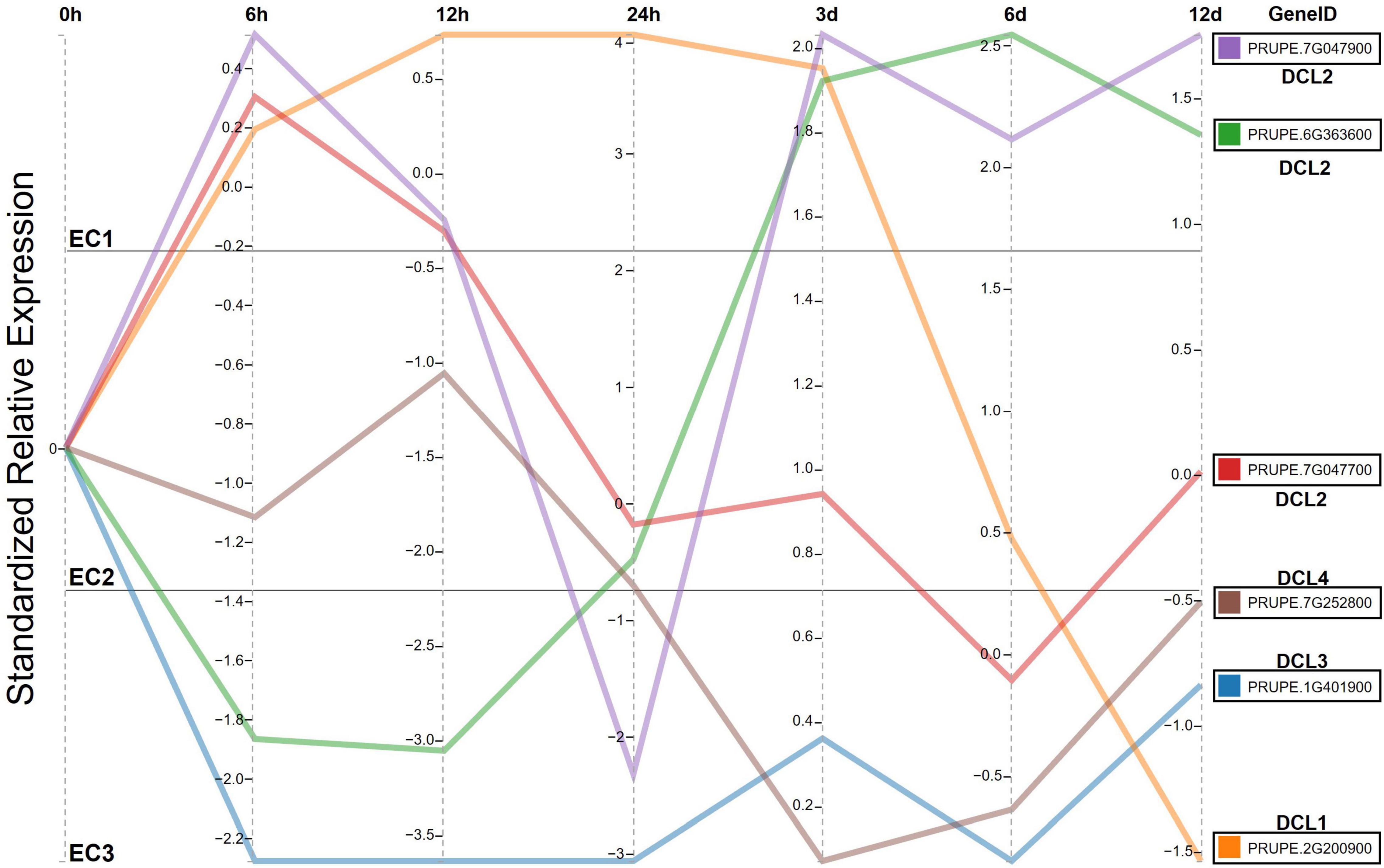
Figure 5. Temporal trend for DCL genes expressed in the fruit flesh of P. persica exposed to drought stress over time periods: 0 h, 6 h, 12 h, 24 h, 3 days, 6 days, and 12 days stratified by 0 timing as a control. The timelines were aligned by the average of DCLs genes expression overtime by standardize relative expression. The expression area is divided into three expression clusters EC1, EC2 and EC3 to evaluate and categorize genes expressions. RNA data were used to measure the expression level of DCLs genes.
Our results provided insights into the molecular evolution of PrupeDCL genes in Rosaceae, and the probable roles in response to drought stresses in P. persica.
Conclusion
Although DICER-like (DCL) genes have been studied because of their critical roles in plant resistance to the abiotic stresses, the focus remains on their role in plants of the Rosaseae species, especially peach (Prunus persica), which needs in-depth studies to clarify their roles, and evolution in Rosaceae. In our study, it was identified about 59 DCLs in eight Rosaceae genomes, 8 of these genes were from P. persica. Based on the phylogenetic analysis, and comparison with homologs from A. thaliana, the DCL family members were divided into four clades: DCL1, DCL2, DCL3, and DCL4. RNA-seq data indicated that the recent WGD might have driven the expansion of DCLs in Prunus Persica and revealed that the identified PrupeDCLs play a pivotal role in different tissues development and drought stress. Overall, our data constitute a foundation for further studies examining the complexity and functioning of Rosaceae DCL genes in the future.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YH and YC planned and designed the experiments. MB, YZ, ZX, and ME performed the experiments and the sequence analysis. YC and MB wrote the manuscript. YH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by funds received from the China Agriculture Research System (grant no. CARS-30).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.923166/full#supplementary-material
Footnotes
- ^ http://www.rosaceae.org/
- ^ https://www.Arabidopsis.org/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://meme-suite.org/meme/
- ^ http://circos.ca/
- ^ https://itol.embl.de/
- ^ www.timetree.org
- ^ https://www.xlstat.com/
References
Alioto, T., Alexiou, K. G., Bardil, A., Barteri, F., Castanera, R., Cruz, F., et al. (2020). Transposons played a major role in the diversification between the closely related almond and peach genomes: results from the almond genome sequence. Plant J. 101, 455–472. doi: 10.1111/tpj.14538
Altschup, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. J. J. M. B. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Alves, P. C., Hartmann, D. O., Núñez, O., Martins, I., Gomes, T. L., Garcia, H., et al. (2016). Transcriptomic and metabolomic profiling of ionic liquid stimuli unveils enhanced secondary metabolism in Aspergillus nidulans. BMC genomics 17:284. doi: 10.1186/s12864-016-2577-6
Bai, M., Yang, G.-S., Chen, W.-T., Mao, Z.-C., Kang, H.-X., Chen, G.-H., et al. (2012). Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 501, 52–62. doi: 10.1016/j.gene.2012.02.009
Belal, M. A., El-Alakmy, H. A., Abdelhameed, A. A. M. M., and Sourour. (2017). influence of reducing irrigation rate and addition of super absorbent polymer on peach trees growth under north sinai conditions. 6, 249–258.
Bianco, L., Cestaro, A., Sargent, D. J., Banchi, E., Derdak, S., Di guardo, M., et al. (2014). Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus x domestica Borkh). PLoS One 9:e110377. doi: 10.1371/journal.pone.0110377
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120.
Cao, K., Zheng, Z., Wang, L., Liu, X., Zhu, G., Fang, W., et al. (2014). Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 15:415. doi: 10.1186/s13059-014-0415-1
Cao, Y., Xu, X., and Jiang, L. (2020b). Integrative analysis of the RNA interference toolbox in two Salicaceae willow species, and their roles in stress response in poplar (Populus trichocarpa Torr. & Gray). Int. J. Biol. Macromol. 162, 1127–1139. doi: 10.1016/j.ijbiomac.2020.06.235
Cao, Y., Meng, D., Li, X., Wang, L., Cai, Y., and Jiang, L. (2020a). A Chinese white pear (Pyrus bretschneideri) BZR gene PbBZR1 act as a transcriptional repressor of lignin biosynthetic genes in fruits. Front. Plant Sci. 11:1087. doi: 10.3389/fpls.2020.01087
Chapman, E. J., and Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Gene. 8, 884–896.
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chinnusamy, V., and Zhu, J.-K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139.
Cui, D.-L., Meng, J.-Y., Ren, X.-Y., Yue, J.-J., Fu, H.-Y., Huang, M.-T., et al. (2020). Genome-wide identification and characterization of DCL, AGO and RDR gene families in Saccharum spontaneum. Sci. Rep. 10:13202. doi: 10.1038/s41598-020-70061-7
Daccord, N., Celton, J.-M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Gene. 49, 1099–1106. doi: 10.1038/ng.3886
Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K. D., Carrington, J. C., and Voinnet, O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71.
Edger, P. P., Vanburen, R., Colle, M., Poorten, T. J., Wai, C. M., Niederhuth, C. E., et al. (2018). Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7:gix124. doi: 10.1093/gigascience/gix124
Esposito, S., Aversano, R., D’amelia, V., Villano, C., Alioto, D., Mirouze, M., et al. (2018). Dicer-like and RNA-dependent RNA polymerase gene family identification and annotation in the cultivated Solanum tuberosum and its wild relative S. commersonii. Planta 248, 729–743. doi: 10.1007/s00425-018-2937-3
Fang, Y., and Spector, D. L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17, 818–823. doi: 10.1016/j.cub.2007.04.005
Gasciolli, V., Mallory, A. C., Bartel, D. P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15, 1494–1500. doi: 10.1016/j.cub.2005.07.024
Haider, M. S., Kurjogi, M. M., Khalil-Ur-Rehman, M., Pervez, T., Songtao, J., Fiaz, M., et al. (2018). Drought stress revealed physiological, biochemical and gene-expressional variations in ‘Yoshihime’ peach (Prunus Persica L) cultivar. J. Plant Interact. 13, 83–90.
Hasanuzzaman, M., Nahar, K., Anee, T. I., Khan, M. I. R., and Fujita, M. (2018). Silicon-mediated regulation of antioxidant defense and glyoxalase systems confers drought stress tolerance in Brassica napus L. South Afr. J. Bot. 115, 50–57.
Hibrand Saint-Oyant, L., Ruttink, T., Hamama, L., Kirov, I., Lakhwani, D., Zhou, N.-N., et al. (2018). A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. 4, 473–484. doi: 10.1038/s41477-018-0166-1
International Peach Genome, I., Verde, I., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–494. doi: 10.1038/ng.2586
Jiang, F., Zhang, J., Wang, S., Yang, L., Luo, Y., Gao, S., et al. (2019). The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic. Res. 6:128. doi: 10.1038/s41438-019-0215-6
Kapoor, M., Arora, R., Lama, T., Nijhawan, A., Khurana, J. P., Tyagi, A. K., et al. (2008). Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 9:451. doi: 10.1186/1471-2164-9-451
Katoh, K., Kuma, K.-I., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518.
Khan, M. I., Fatma, M., Per, T. S., Anjum, N. A., and Khan, N. A. (2015a). Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6:462. doi: 10.3389/fpls.2015.00462
Khan, M. I. R., Asgher, M., Fatma, M., Per, T. S., Khan, N. A. J. C. C., and Sustainability, E. (2015b). Drought stress vis a vis plant functions in the era of climate change. 3, 13–25.
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Krzywinski, M. I., Schein, J. E., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–45 doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. 101, 12753–12758.
Kurihara, Y., Takashi, Y., and Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212. doi: 10.1261/rna.2146906
Li, Y., Pi, M., Gao, Q., Liu, Z., and Kang, C. (2019b). Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic Res. 6:61.
Li, S., Qu, X., Zhong, M., Jiang, X., Dong, X., Yi, T., et al. (2019a). Characterization of the complete chloroplast genome of Rosa chinensis ‘Old Blush’ (Rosaceae), an important cultivated Chinese rose. Acta Horticult. 1232, 119–124.
Lin, W., Huang, J., Xue, M., Wang, X., and Wang, C. J. M. D. P. B. (2019). Characterization of the complete chloroplast genome of Chinese rose. Rosa Chinensis 4, 2984–2985.
Linsmith, G., Rombauts, S., Montanari, S., Deng, C. H., Celton, J. M., Guerif, P., et al. (2019). Pseudo-chromosome-length genome assembly of a double haploid “Bartlett” pear (Pyrus communis L.). Gigascience 8:giz138 doi: 10.1093/gigascience/giz138
Liu, J., Chen, N., Chen, F., Cai, B., Santo, S. D., Tornielli, G. B., et al. (2014). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera).pdf>. Sci. Rep. 6:22783 doi: 10.1186/1471-2164-15-281
Liu, Q., Feng, Y., and Zhu, Z. (2009b). Dicer-like (DCL) proteins in plants. Funct. integr. Genomics 9, 277–286.
Liu, Q., Feng, Y., and Zhu, Z. (2009a). Dicer-like (DCL) proteins in plants. Funct. Integr. Genomics 9, 277–286.
Margis, R., Fusaro, A. F., Smith, N. A., Curtin, S. J., Watson, J. M., Finnegan, E. J., et al. (2006). The evolution and diversification of Dicers in plants. FEBS Lett. 580, 2442–2450.
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121–e121. doi: 10.1093/nar/gkt263
Mukherjee, K., Campos, H., and Kolaczkowski, B. (2013). Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 30, 627–641. doi: 10.1093/molbev/mss263
Necira, K., Makki, M., Sanz-García, E., Canto, T., Djilani-Khouadja, F., and Tenllado, F. (2021). Topical application of Escherichia coli-encapsulated dsRNA induces resistance in Nicotiana benthamiana to potato viruses and involves RDR6 and combined activities of DCL2 and DCL4. Plants 10:644. doi: 10.3390/plants10040644
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT. StringTie and Ballgown. Nat. Prot. 11:1650. doi: 10.1038/nprot.2016.095
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Priya, M., Dhanker, O. P., Siddique, K. H. M., Hanumantharao, B., Nair, R. M., Pandey, S., et al. (2019). Drought and heat stress-related proteins: an update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Gene. 132, 1607–1638. doi: 10.1007/s00122-019-03331-2
Qian, Y., Cheng, Y., Cheng, X., Jiang, H., Zhu, S., and Cheng, B. (2011). Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep. 30, 1347–1363. doi: 10.1007/s00299-011-1046-6
Qin, C., Li, B., Fan, Y., Zhang, X., Yu, Z., Ryabov, E., et al. (2017). Roles of Dicer-Like Proteins 2 and 4 in Intra- and Intercellular Antiviral Silencing. Plant Physiol. 174, 1067–1081. doi: 10.1104/pp.17.00475
Qin, L., Mo, N., Muhammad, T., and Liang, Y. (2018). Genome-wide analysis of DCL, AGO, and RDR gene families in pepper (Capsicum Annuum L.). Int. J. Mol. Sci. 19:1038. doi: 10.3390/ijms19041038
Qu, F., Ye, X., and Morris, T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci.U.S.A. 105, 14732–14737. doi: 10.1073/pnas.0805760105
Rahmati, M., Vercambre, G., Davarynejad, G., Bannayan, M., Azizi, M., and Genard, M. (2014). Water scarcity conditions affect peach fruit size and polyphenol contents more severely than other fruit quality traits. J. Sci. Food Agric. 95, 1055–1065. doi: 10.1002/jsfa.6797
Raymond, O., Gouzy, J., Just, J., Badouin, H., Verdenaud, M., Lemainque, A., et al. (2018). The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50, 772–777. doi: 10.1038/s41588-018-0110-3
Singh, P. K., Srivastava, D., Tiwari, P., Tiwari, M., Verma, G., and Chakrabarty, D. (2019). Drought Tolerance in Plants. Plant Signal. Mol. 105–123.
Song, L., Han, M.-H., Lesicka, J., and Fedoroff, N. (2007). Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. 104, 5437–5442. doi: 10.1073/pnas.0701061104
Song, X., Li, P., Zhai, J., Zhou, M., Ma, L., Liu, B., et al. (2012). Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69, 462–474. doi: 10.1111/j.1365-313X.2011.04805.x
VanBuren, R., Wai, C. M., Colle, M., Wang, J., Sullivan, S., Bushakra, J. M., et al. (2018). A near complete, chromosome-scale assembly of the black raspberry (Rubus occidentalis) genome. Gigascience 7:giy094 doi: 10.1093/gigascience/giy094
Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771.
Verde, I., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S., Marroni, F., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Gene. 45, 487–494.
Verde, I., Jenkins, J., Dondini, L., Micali, S., Pagliarani, G., Vendramin, E., et al. (2017). The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 18:225. doi: 10.1186/s12864-017-3606-9
Wu, J., Wang, Z., Shi, Z., Zhang, S., Ming, R., Zhu, S., et al. (2013). The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23, 396–408.
Xie, Z., Allen, E., Wilken, A., and Carrington, J. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 102, 12984–12989. doi: 10.1073/pnas.0506426102
Zhang, H., Meltzer, P., and Davis, S. J. B. B. (2013). RCircos: an R package for Circos 2D track plots. BMC Bioinformatics 14:244. doi: 10.1186/1471-2105-14-244
Zhang, Q., Chen, W., Sun, L., Zhao, F., Huang, B., Yang, W., et al. (2012). The genome of Prunus mume. Nat. Commun. 3:1318.
Zhou, H., Lin-Wang, K., Wang, F., Espley, R. V., Ren, F., Zhao, J., et al. (2019). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 221, 1919–1934. doi: 10.1111/nph.15486
Keywords: peach, DCL genes, evolution, expression profile, duplication
Citation: Belal MA, Ezzat M, Zhang YQ, Xu Z, Cao YP and Han YP (2022) Integrative Analysis of the DICER-like (DCL) Genes From Peach (Prunus persica): A Critical Role in Response to Drought Stress. Front. Ecol. Evol. 10:923166. doi: 10.3389/fevo.2022.923166
Received: 19 April 2022; Accepted: 14 June 2022;
Published: 11 July 2022.
Edited by:
Lin Zhang, Hubei University of Chinese Medicine, ChinaReviewed by:
Yuanyuan Jiang, South China Agricultural University, ChinaYan Wang, Anhui Agricultural University, China
Mahmoud Magdy, Ain Shams University, Egypt
Copyright © 2022 Belal, Ezzat, Zhang, Xu, Cao and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunpeng Cao, eXBoYW5Ad2JnY2FzLmNu; Yuepeng Han, Y2FveXVucGVuZ0B3YmdjYXMuY24=
 Mohammad A. Belal
Mohammad A. Belal Mohamed Ezzat
Mohamed Ezzat Yuanqiang Zhang1,2
Yuanqiang Zhang1,2 Yunpeng Cao
Yunpeng Cao Yuepeng Han
Yuepeng Han