
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 27 June 2022
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.901295
This article is part of the Research TopicGenetic History of Human Populations Along the Ancient Silk RoadView all 18 articles
 Jianxue Xiong1,2,3†
Jianxue Xiong1,2,3† Yichen Tao1†
Yichen Tao1† Minxi Ben1†
Minxi Ben1† Yishi Yang4
Yishi Yang4 Panxin Du1,5
Panxin Du1,5 Edward Allen2
Edward Allen2 Hui Wang2,3,6
Hui Wang2,3,6 Yiran Xu3,6
Yiran Xu3,6 Yao Yu2
Yao Yu2 Hailiang Meng1
Hailiang Meng1 Haoquan Bao1
Haoquan Bao1 Boyan Zhou7
Boyan Zhou7 Guoke Chen4*
Guoke Chen4* Hui Li1*
Hui Li1* Shaoqing Wen2,3,6*
Shaoqing Wen2,3,6*The relationship between archeological culture and ethnicity is invariably complex. This is especially the case for periods of national division and rapid inter-ethnic exchange, such as China’s Sixteen Kingdoms (304–439 CE) and Northern and Southern Dynasties (420–589 CE). Going by tomb shape and grave goods, the Foyemiaowan cemetery at Dunhuang exhibits a typical third–tenth century Han style. Despite this, the ethnic makeup of the Foyemiaowan population has remained unclear. We therefore analyzed 485 Y-chromosomal SNPs and entire mitochondrial genomes of 34 Foyemiaowan samples. Our study yielded the following discoveries: (1) principal component analysis revealed that the Foyemiaowan population was closely clustered with Tibeto-Burman populations on the paternal side and close to Mongolic-speaking populations on the maternal side; (2) lineage comparisons at the individual level showed that the Foyemiaowan population consisted of primarily Tibeto-Burman and Han Chinese related lineages (Oα-M117, 25%;Oβ-F46, 18.75%), partially Altaic speaking North Eurasian lineages (N-F1206, 18.75%) and a slight admixture of southern East Asian lineages (O1b1a2-Page59, 6.25%; O1b1a1-PK4, 3.13%). Similarly, the maternal gene pool of Foyemiaowan contained northern East Asian (A, 4.17%; CZ, 16.67%; D, 20.83%; G, 4.17%; M9, 4.17%), southern East Asian (B, 12.51%; F, 20.83%) and western Eurasian (H, 4.17%; J, 4.17%) related lineages; (3) we discovered a relatively high genetic diversity among the Foyemiaowan population (0.891) in our ancient reference populations, indicating a complex history of population admixture. Archeological findings, stable isotope analysis and historical documents further corroborated our results. Although in this period China’s central government had relinquished control of the Hexi Corridor and regional non-Han regimes became the dominant regional power, Foyemiaowan’s inhabitants remained strongly influenced by Han culture.
The relationship between archeological culture and ethnic group has been examined extensively and deeply and remains a central issue within archeological research. Some studies suggest a connection between ethnicity and archeological finds, such as diagnostic pottery, tools, metallic objects like brooches, or in some cases residential areas. Yet any approach that simply equates the two has been thoroughly criticized in recent years (Upton, 1996; Jones, 1997; Renfrew and Bahn, 2007).
Cultural and ethnic homogeneity and heterogeneity are especially relevant for archeological contexts involving large-scale migration. For example, the Eurasian Bronze Age (circa. 3000–1000 BC) is widely acknowledged as a period of major cultural change—change intertwined with large-scale population migration (Allentoft et al., 2015). Multiple lines of evidence show that male-driven migration introduced Steppe ancestry to almost all Corded Ware populations in Central Europe (Caramelli et al., 2021), precipitating major shifts in burial practice (Anthony, 2007; Furholt, 2019). In a separate context, archeological analysis has demonstrated that the Quarto Cappello del Prete necropolis, in the heart of the globalized Roman Empire, contained individuals with highly heterogeneous geographical origins. Genomic data also supported these results and proved the applicability of genomic study for drawing out the ethnic contours of such multi-ancestral societies (De Angelis et al., 2021). However, ethnic groups rarely reflect the sum total of similarities and differences in “objective” cultural traits (Jones, 1997). Archeological culture and ethnic background may be distinct, as in the case of Taojiazhai (∼1700–1900 BP). This site was Han Chinese in style, though DNA evidence indicated a strong degree of Tibeto-Burman ancestry, identifying the Taojiazhai population as descendants of Di-Qiang groups (Zhao et al., 2011).
The Hexi Corridor of northwestern China was an important channel for cultural exchange and human migration between the ancient Central Plains and Western Regions (centered around modern Xinjiang). The region may also be considered an important component of the “Northwest National Corridor,” one of three national corridors put forward by Fei Xiaotong in the early 1980s (Fei, 1982). During the Warring States period (476–221 BCE), the Hexi Corridor was inhabited by various nomadic peoples, known historically as the Yuezhi, Wusun and Xiongnu (Si, 2002a). The migration of population as a means of strengthening control over newly-acquired territory was a critical strategy of China’s historical conquests during this and the subsequent Han period. For example, in the “Biography of Chulizi” in Sima Qian’s Historical Records (史记.樗里子传), we learn of Quwo (Lingbao city, Henan, China) occupied by the Qin army in 330 BC, its locals evicted from their hometown and replaced by Qin migrants (Si, 2002b). The Han dynasty government continued this gradual population-based domination of the Hexi Corridor. A series of policies was adopted to isolate Qiang people located in the Hehuang valley and Xiongnu in the Mongolian Steppe, as well as establish contact with the Western Regions. Counties were set up at Wuwei County (武威郡), Zhangye County (张掖郡), Jiuquan County (酒泉郡) and Dunhuang County (敦煌郡) running from east to west along the Hexi Corridor. According to historical documents and unearthed bamboo slips, a considerable population was relocated the Middle and Lower Yellow River watershed (i.e., Hongnong county, He’nei county, Runan county, Julu county, Yingchuan county, among others) (Liu, 2012). A recent study of the middle Hexi Corridor population at Heishuiguo has provided corroborating evidence of this transformation through Y chromosome and mtDNA analysis of individuals from that site, suggesting most Heishuiguo males had migrated from Middle and Lower Yellow River regions, while females were largely natives (Xiong et al., 2022). Meanwhile, localized archeological cultural traits also suggest that the Heishuiguo population inherited a Central Plains tradition. Mass migration of individuals not only impacted the genetic structure of the Hexi population, but also resulted in changes in local subsistence strategy (Chen et al., 2019).
Following the collapse of the Han dynasty in the third century CE, ancient China entered the Wei, Jin and Northern and Southern Dynasties period. Governments occupying the traditional heart of state power in the Central Plains largely abandoned the Hexi corridor; burgeoning regional political forces became the dominant driver of regional developments. The region experienced several dynastic transitions, from the Former Liang (314–376 CE), to Western Qin (385–400 CE and 409–431 CE), Latter Liang (386–403 CE), Northern Liang (397–439 CE), Southern Liang (397–414 CE) and Western Liang (400–421 CE) dynasties. Minority groups also established regimes under the Western Qin, Southern Liang, founded by Xianbei peoples, and the Latter Liang and Northern Liang established by Di peoples and Lushuihu (卢水胡), respectively. Archeological and historical materials, however, both suggest Han Chinese from the Central Plains had become the dominant Hexi Corridor population by the Jin Dynasty—acquiring this position through the continuing exertions of central government, causing Han and Hu (胡, general term for northern non-Han groups) to coexist in local communities (Lv, 2017). Regime changes and warfare were commonplace following the collapse of the Jin Dynasty, particularly as Chinese dynasties and nomadic polities frequently clashed. As such, the region continued to experience recurrent population inflow, through both political or military immigration, with incomers kidnapped by regional political regimes, or refugees fleeing wars and natural disasters (Lv, 2017).
Dunhuang County, at the west end of the Hexi Corridor, lies adjacent to the Western Regions. In the wake of the Han Dynasty, Dunhuang retained its position as an important military town and frontier for central government administration over the Western Regions, successively controlled by the Latter Liang and Western Liang polities. With the population exposed to migration from the Western Regions, Mongolian Steppe, Qinghai-Tibetan Plateau and Central Plain, complex population interaction occurred here and in the entire west of the Hexi Corridor over these centuries. At Foyemiaowan, our first hypothesis anticipated a similarly diverse population.
However, study of the archeological culture at Foyemiaowan supports the Han Chinese origin hypothesis. The Foyemiaowan cemetery, in Wudong county in Dunhuang (Figure 1A), lies north of the Mogao Grottoes, a World Heritage Site renowned for its Buddhist mural paintings. The cemetery dates back as early as the Cao-Wei period (220–280 CE), and remained in use for approximately 600 years. This affords Foyemiaowan almost perfect coverage of the Cao-Wei to Sui/Tang periods. Tomb shape and grave goods suggest a cultural core at Foyemiaowan site that continued to adhere to a Han Chinese tradition (Chen et al., 2022). For example, numerous examples of daily-use potteries (Figure 1D) and bronze mirrors (see Figures 1B,C) in typical Han Chinese style were found in Foyemiaowan burials (Chen et al., 2022).

Figure 1. (A) The geographical location of the Foyemiaowan site. (B) IIIM11:1 Bronze Mirror. (C) IIIM20:1 Bronze mirror. (D) IVM24: Daily-use potteries of Foyemiaowan cemetery.
Were the inhabitants of Foyemiaowan Han or diverse origin? This would have been unanswerable previously, but recent years have seen the widespread study of uniparentally inherited markers in an effort to understand the population history, origin, and migration of human populations (Pamjav et al., 2017). This work offers a suite of methods that can be applied to similar questions at Foyemiaowan. This study aims to update our knowledge of the genetic history of Hexi Corridor populations based on the 485 SNPs Y-chromosome and whole mitochondrial genomes from Foyemiaowan. Such data will allow us to test the Han and diverse origin hypothesis, explore the relationship between archeological culture and ethnic group, as well consider what factors could have affected the genetic profile at Foyemiaowan.
Foyemiaowan site contained thousands of tombs, which were excavated by the Institute of Cultural Relics and Archaeology of Gansu Province in 2015. The cemetery was divided into eight natural areas. The excavators of Foyemiaowan have argued for a four-phase division of the cemetery: Phase 1: Cao-Wei period (220–265 CE); Phase 2: Western Jin Dynasty (265–316 CE); Phase 3: Former Liang to Northern Liang Dynasty (314–439 CE); Phase 4: Sui Dynasty (581–618 CE) and Tang Dynasty (618–907 CE) (Chen et al., 2022). In this study, most samples can be assigned to the Sixteen Kingdoms period (Phase 3). Few samples were preserved from the remaining phases. Sixteen Kingdoms individuals thus form the bedrock of this study and belong to a unique period of chronic dynastic transition. Our survey population was made of up 34 ancient samples. Sex was determined by pelvic (Klales et al., 2012) and skull morphology (Buikstra, 1994), including 32 males and 2 females.
For our 34 samples, we extracted DNA from the temporal bones, teeth and limb bones (Table 1 and Supplementary Table 1A), using a dedicated aDNA facility in Fudan University, Shanghai. Molecular methods used for aDNA extraction and construction of Illumina libraries have been described previously (Zhu et al., 2021; Xiong et al., 2022). One extraction (no sample powder used) and one PCR blank (extract supplemented by water) were set up as negative controls for each batch of samples. Libraries were sequenced on Illumina HiSeq X10 instrument at the Annoroad Company (Beijing, China) using the 150 bp paired-end sequencing design.
MtDNA enrichment was carried out on 13 samples that yielded a lower endogenous rate in the initial shotgun screening. Target enrichment of the mitogenome was performed using a MyGenostics Human Mitochondria Capture Kit (MyGenostics Company, Beijing, China) (Sun et al., 2021). Post-enrichment product was then quantified via qPCR and sequencing was performed using a NovaSeq 6000 platform at Mingma Technologies Company (Shanghai, China). 150 bp paired-end reads were generated according to the manufacturer’s instructions.
We opted for multiplex PCR targeting enrichment with short amplicons based on the NGS (Next Generation Sequencing) platform. In this study, a sensitive short amplifier primer system including 485 Y-SNPs (Wen et al., 2019; Xiong et al., 2022) was conducted to test Y-lineages for each male sample from the Foyemiaowan site. The system covered the common East Asian lineages.
For shotgun and mtDNA captured data, we clipped sequencing adapters and merged these using sequences by ClipAndMerge v1.7.8 (Peltzer et al., 2016), then mapped merged reads to the human reference genome (hs37d5; GRCh37 with decoy sequences) using BWA v0.7.17 (Li and Durbin, 2010). We used Dedup v0.12.3 (Peltzer et al., 2016) to remove PCR duplicates. Utilizing trimBam implemented in BamUtil v1.0.14,1 we clipped four bases from both ends of each read to avoid an excess of remaining C- > T and G- > A transitions at the ends of DNA sequences.
The authenticity of the ancient genome sequence was mainly determined by the combination of two observations of the same specimen. Firstly, we checked DNA damage pattern (Supplementary Figure 1) and estimated the 5′ C > T and 3′ G > A misincorporation rate using mapDamage v 2.0.61 (Jónsson et al., 2013). We then used the Schmutzi program to test mitochondrial contamination rates for all individuals (Renaud et al., 2015).
Y chromosome haplogroups were examined by aligning a set of positions in the ISOGG (International Society of Genetic Genealogy)2 and Y-full3 databases. Haplogroup determination was performed with the script Yleaf.py in Yleaf software (Ralf et al., 2018), which provides outputs for allele counts of ancestral and derived SNPs along a path of branches of the Y-chromosome tree (Supplementary Table 1B). Finally, we re-checked the SNPs by visual inspection with IGV software (Helga et al., 2013).
In order to call mtDNA consensus sequences, we employed a log2fasta program implemented in Schmutzi (Renaud et al., 2015). Mutations that appeared when checked against rCRS were also re-checked in BAM (Binary Alignment Map) files through visual inspection using the IGV software (Helga et al., 2013). Lastly, we used HaploGrep 2 (Weissensteiner et al., 2016) to assign the haplogroups (Supplementary Table 1C).
Haplogroup diversity (H) was calculated using Nei’s formula (Nei and Tajima, 1981). All the above analyses were performed in R 3.6.3. Reference populations are listed in Supplementary Table 2A. Principal component analysis (PCA) was performed using a prcomp () function of R. Visualization of PCA results were conducted using the “ggplot2” package, as well as a pie chart. Reference populations of principal component analysis are listed in Supplementary Table 3. The Map of China was drawn using “mapchina” and “sf” packages.
For comparison with the newly generated 17 ancient mitogenomes from Foyemiaowan Site, previously published data was assembled from the Mitomap database4 (Supplementary Table 4). In total, 417 mitogenomes were employed to construct the maximum-parsimony (MP) phylogenetic trees for each haplogroup (Supplementary Table 5), using mtPhyl v5.003 software.5
Coalescence time estimates were also computed using Bayesian MCMC approach implemented in the BEAST v2.4.7 software package (Drummond et al., 2012). We constructed Bayesian trees using both modern and ancient samples, with the latter dates used as tip dates for molecular clock calibration (Fu et al., 2013; Rieux et al., 2014).
For mitochondrial time estimation, we first aligned all sequences to the rCRS using the MAFFT (Katoh et al., 2017) version 7 program using the iterative refinement method: E-INS-I. Then, we partitioned the alignments into four parts with a scheme adapted in accordance with a published molecular clock calibration for human mtDNA (Rieux et al., 2014): (1) First and second nucleotides in codons of protein coding genes (PC1 + PC2), (2) third nucleotides in codons of protein coding genes (PC3), (3) rRNAs + tRNAs (RNA region) and (4) HVRI + HVRII (control region). Indels were removed from the alignments to avoid potential biases due to possible misalignments and incorrect consensus calling around these regions. Partitioning was performed using the Python script written Margaryan et al. (2017), available through Github account.6 After partitioning, we tested the best-supported substitution models (Supplementary Table 6) using jModelTest v.2.1.10 (Darriba et al., 2012) with the Bayesian Information Criterion (BIC). When running BEAST, we used unlinked strict clock rates and substitution models for the four partitions, but linked the coalescent Bayesian skyline tree model. Each partition was assigned an independent mutation rate prior according to Rieux et al. (2014). We ran each MCMC for 10E8 states, sampling every 10E4 states, and designating the first 10% as burn-in. Three independent runs were combined using LogCombiner v1.8.0 (Drummond et al., 2012). Convergence to the stationary distribution and sufficient sampling and mixing were checked by inspection of posterior samples (effective sample size > 200). Parameter estimation was based on samples combined from different chains. The best supported tree was estimated from the combined samples using the maximum clade credibility (MCC) method implemented in TreeAnnotator v1.8.0 (Drummond et al., 2012).
A total of 20 different Y chromosome sub-haplogroups in 32 Foyemiaowan individuals were determined according to the ISOGG’s Y-DNA Haplogroup Tree 2019 (Table 1, Figure 2, and Supplementary Figure 2). The distribution of paternal haplogroups was revealed as: Oα-M117 (25%), Oβ-F46 (18.75%), N-F1206 (18.75%), O-M188 (6.25%), O-F444 +, F46- (6.25%), O1b1a2-page59 (6.25%), Q-M120 (6.25%), C-F1144 (6.25%), N-F2930 (3.13%) and O1b1a1-PK4 (3.13%). Haplogroup Oα-M117 exhibits high frequency in Tibeto-Burman populations (Xue et al., 2006; Gayden et al., 2007), including Deng (63.33%), Shannan Tibetan (42.31%), Luoba (30.77) and Danba Qiang (22.22%) (Kang et al., 2012; Qi et al., 2013; Wang and Li, 2013; Wang et al., 2014) and present-day Han Chinese (Yan et al., 2011) such as Shandong (24.2%), Beijing (18.5%) and Heilongjiang (16.6%) (Lang et al., 2019). Oβ-F46 is observed at high frequency with ∼11% in Han Chinese (Yan et al., 2011) and moderate to low frequency for Tibeto-Burman (Xinlong Tibetan, 6.52%; Yajing Tibetan, 6.38%; Danba Qiang, 5.56%; Nyingchi Tibetan, 4.27%) groups. However, the haplogroup Oγ-IMS-JST002611 occupying about 14% in Han Chinese, was not detected in the Foyemiaowan population, which suggests that the Foyemiaowan paternal gene pool was somewhat different from Han Chinese. N-F1206, achieving the second high frequency, is widely distributed across Northern Eurasia (Hu et al., 2015) and can be further divided into N-TAT and N-F710. N-TAT exhibited a very high frequency in Altaic (Yakuts, 70.8–91.5%; Evenks, 50.9%; Buryats, 41.4%), Uralic (Udmurts, 66.7%; Finns, 53.8%) and Indo-European (Latvians, 43.0%; Lithuanians, 40.5%) speaking populations (Ilumäe et al., 2016). N-F710 is considered as migrating from Northeast Asia to the Yellow River basin no later than 2.7 kya (Ma et al., 2021). Moreover, the haplogroups O1b1a2-Page59 (6.25%) and O1b1a1-PK4 (3.13%) reflect a minor degree of southern East Asian origins (Yan et al., 2011; Xia et al., 2019).
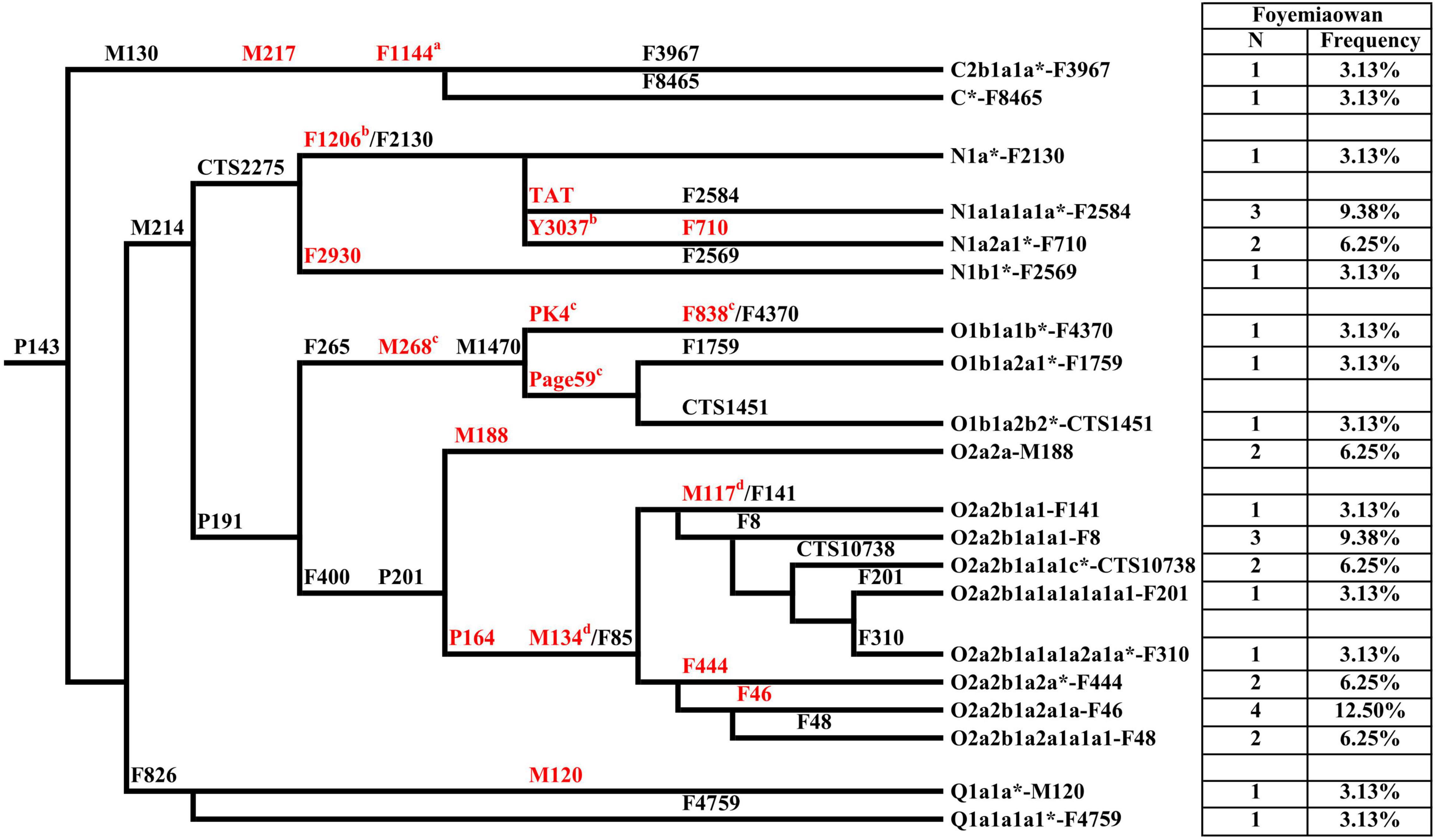
Figure 2. Phylogenetic relationship of Y-chromosome haplogroups in this study, with frequencies are on the right side of the figure. Marker names are shown along the branches and haplogroup names are shown to the right. All Y-chromosome haplogroups are based on the ISOGG-2019. Potentially paraphyletic undefined subgroups are distinguished from recognized haplogroups by the asterisk symbol. Markers in red are key Y-SNPs. a–d: these markers are not designed in the Y-SNP panel but are very common in Y phylogenetic trees.
Library construction of 10 samples failed because of poor sample quality. As a result, we only obtained 24 valid mitochondrial gene data from 34 Foyemiaowan samples. A total of 22 different mtDNA sub-haplogroups were found in 24 individuals (Table 1 and Supplementary Table 1C). Haplogroup frequencies for Cao Wei to Western Jin, Sixteen Kingdoms, Sui to Tang and the entire Foyemiaowan mtDNA data are presented in Supplementary Figure 3 and Figure 3. The entire population of Foyemiaowan is mainly characterized by the most typical eastern Eurasian mtDNA haplogroups (A, B, CZ, D, F, G and M9). Here, Foyemiaowan shows pronounced frequencies (>5%) of haplogroups D (20.83%), F (20.83%), CZ (16.67%), and B (12.5%) and low frequencies (< 5%) of haplogroups A (4.17%), G (4.17%), and M9 (4.17%). Notably, the haplogroup D4 occurs with the highest frequency (16.67%) in the Foyemiaowan population and could be further divided into sub-clades D4*, D4b2b* and D4h1*. D4 haplogroup is observed with high frequency in populations from the Baikal Region (30.9%), Far East (26.6%), Eastern Asia (22.5%), Altai Region (17%), northern Asia (16.7%) and western Siberia (16.4%) (Derenko et al., 2010). Additionally, the haplogroup D4 reaches a high frequency in Han populations from northern China, including Inner Mongolia (22.73%), Liaoning (21.83%) and Heilongjiang (21.51%) (Li et al., 2019). Haplogroup Z (accounting for 12.5% in Foyemiaowan) is most frequent in Northeastern Asia and Siberia populations (Schurr et al., 1999) as well as the Volga-Ural region, such as Udmurts-7.1%, Maris-2.7%, Komis-1.8% (Tambets et al., 2004; Ingman and Gyllensten, 2007). As for haplogroups B and F, these are relatively common in southern East Asia (Wen et al., 2005; Ko et al., 2014; Kutanan et al., 2017; Li et al., 2019). In the meantime, a small number of western Eurasian origin haplogroups H (4.17%) and J (4.17%) were detected. Of these, haplogroup H displays a broadly southeast-northwest distribution pattern in western Eurasia (Pereira et al., 2005) and dominates the mtDNA gene pool of Europeans (∼40–45% on average), decreasing in the Near East and the Caucasus region (∼20–30%). Haplogroup J shows a moderate frequency in Europe (6–15%) (Pliss et al., 2006; Vidrová et al., 2008), and among Norwegians (12.6%), Estonians (10.3%), Germans (8.4%), and Russians (8.0%) (Pliss et al., 2006). In summary, the dominant D4, Z, B, and F mtDNA haplogroups in Foyemiaowan are more frequent in northern and southern East Asian populations, respectively, while haplogroups J and H reveal genetic flow from western Eurasia.
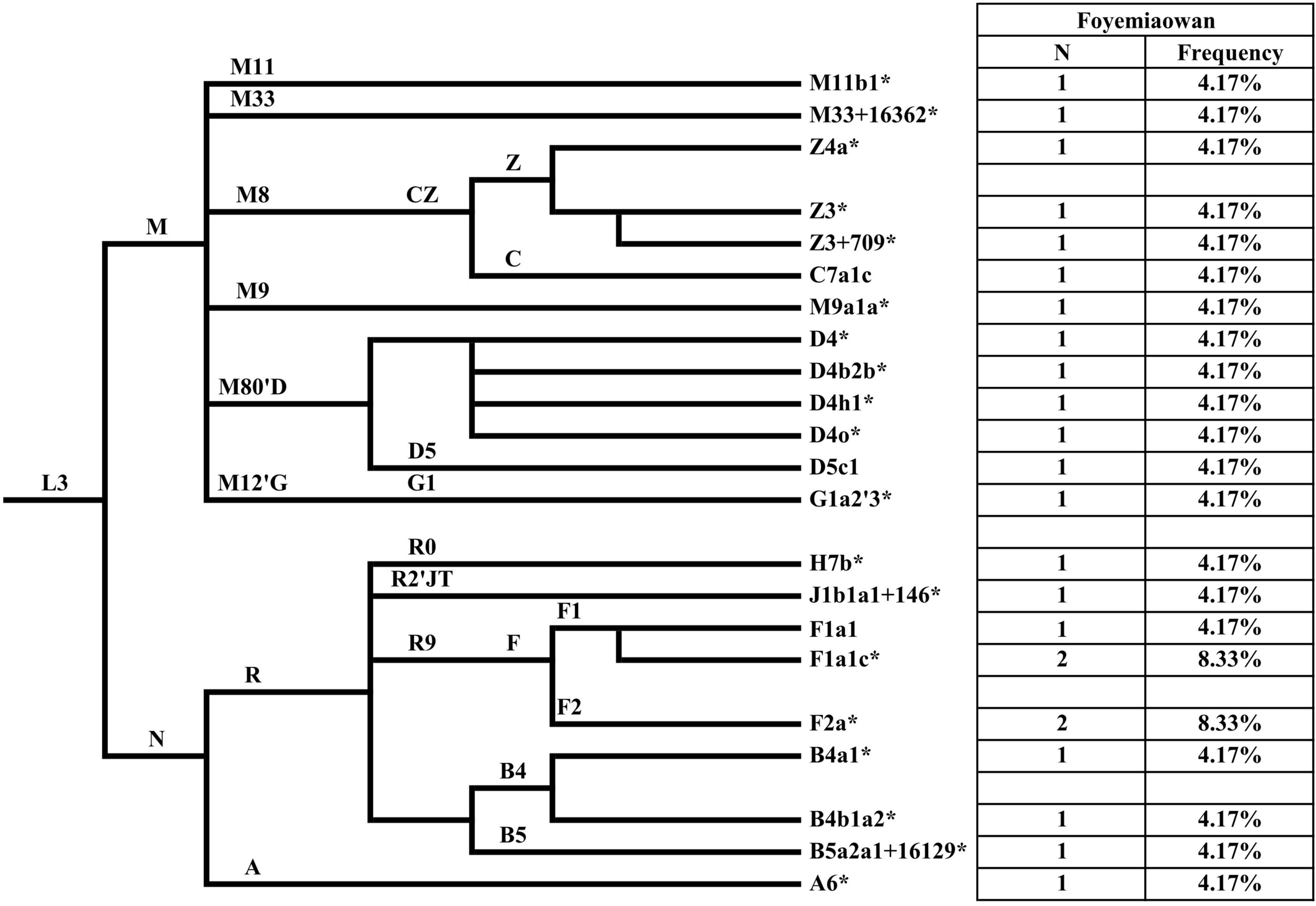
Figure 3. The phylogenetic relationship of mtDNA haplogroups surveyed in this study and their haplogroup-based frequencies in the sampled populations, with haplogroup names shown on the right. Asterisks distinguish potentially paraphyletic undefined subgroups from recognized haplogroups.
In order to compare mtDNA haplogroup diversity and infer possible origins of the Foyemiaowan population across a broad geographical context, we collated 564 individuals from 28 ancient populations, ranging from circa 2000 BCE to 600 CE, and covering Northern China, Mongolia, southern Siberia and eastern Kazakhstan (Supplementary Table 2A). As an initial step, only those ancient populations with a sample size of at least eight and timescales falling mostly within 300 years were selected for further analysis. Secondly, mtDNA haplogroups were classified into 18 macro-haplogroups, including haplogroups A, B, CZ, D, F, G, M7, M8a, M9, N9, U, W, HV, JT, N1, M*, N*, and R. Thirdly, going by distinct geographic origins, these haplogroups were further divided into four groups: northern East Asian (i.e., A, CZ, D, G, M8a, M9, and N9), southern East Asian (i.e., B, F, and M7), western Eurasian (i.e., U, W, HV, JT, and N1) and Undetermined (i.e., M*, N*, and R). Finally, haplogroup diversity in these ancient populations was computed at the level of the 18 macro-haplogroups mentioned above.
The distribution of mtDNA macro-haplogroups (Figure 4) revealed a differential contribution of haplogroups with three distinct geographic origins. As expected, northern East Asian (NEA) haplogroups, with a notable incidence of D, C, G and A, are more frequent in mid-eastern Mongolia and eastern Inner Mongolia regions. These haplogroups reach the highest proportion (∼100%) in Ulaanzuukh Culture and late Xiongnu Culture (central east Mongolia) populations. In the west, the occurrence of western Eurasian (WEu) haplogroups is higher and tends to decrease to the east, occurring with the highest frequency (∼100%) in western Xinjiang, eastern Kazakhstan and Southern Siberia. In the south, the presence of southern East Asian (SEA) haplogroups is notable, especially in Yellow River Valley populations such as Taojiazhai (37.2%) and Foyemiaowan (33.3%). Here, the Foyemiaowan population consisted of 50% NEA, 33.3% SEA, 8.3% WEu and 8.4% Undetermined haplogroups (Supplementary Table 2B), indicating the possibility of three main sources of ancestry in the above-mentioned geographical areas. Finally, examining haplogroup diversity value (Table 2), our highest figure (0.879–0.911) was observed at areas lying at the geographic crossroads between Western and Eastern Steppe, including central west Mongolia and Xinjiang, centers of population migration and mixture over the past four to five millennia (Allentoft et al., 2015; Unterländer et al., 2017; Damgaard et al., 2018). Foyemiaowan exhibited the third highest (0.891) level of diversity, revealing the multi-ancestral admixture history associated with its strategic position along the Silk Road.
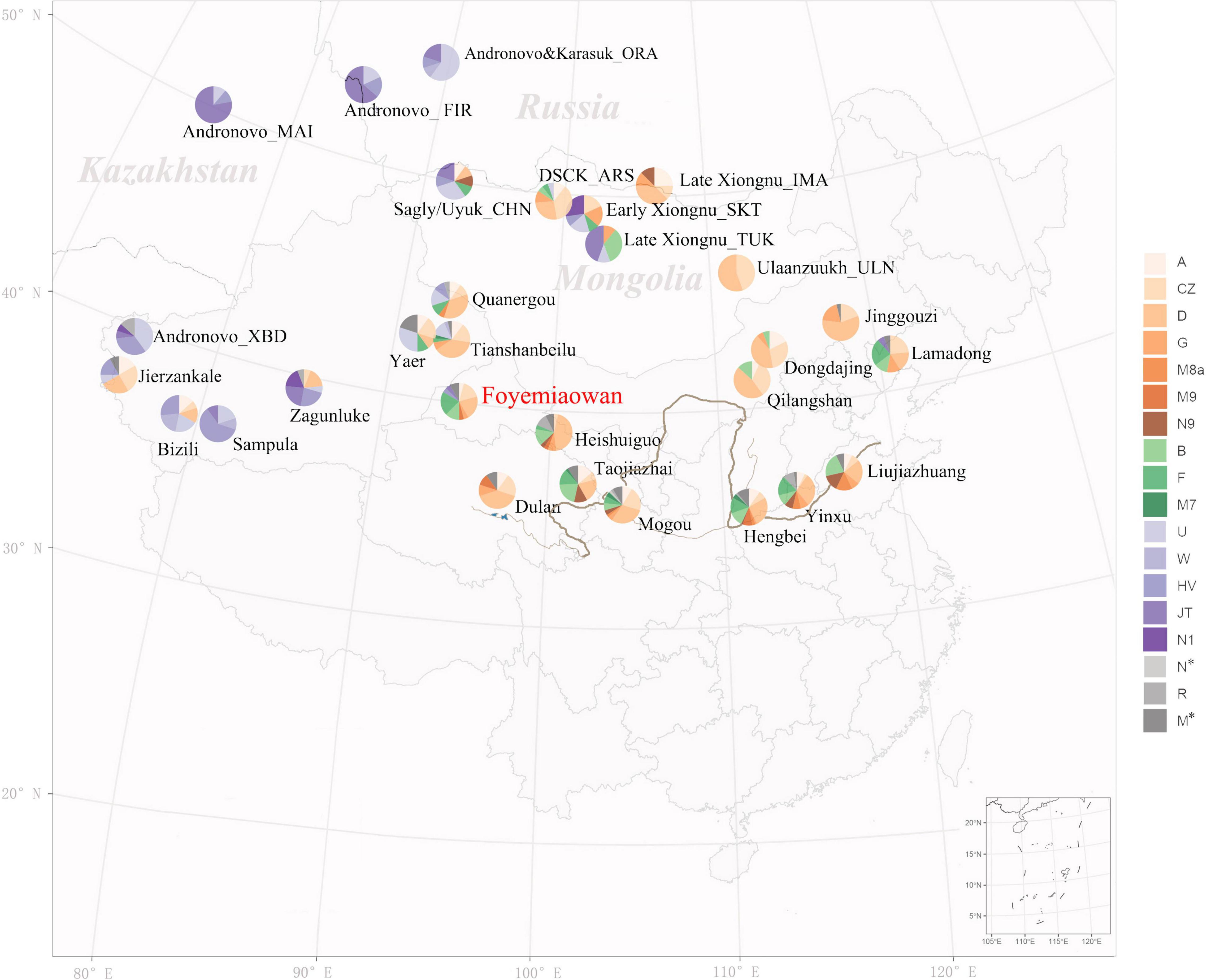
Figure 4. Pie charts of the distribution of mtDNA macro-haplogroups in Foyemiaowan and surrounding populations from 2000BCE to 600CE.
To visualize the relationships between the Foyemiaowan population and reference populations, a PCA plot was constructed according to haplogroup frequencies (Figure 5 and Supplementary Figure 4). Firstly, the Foyemiaowan population was organized into two main temporal groups: a “Sixteen Kingdoms” and an “Overall” group. From the Y-chromosome perspective, the reference populations included 11,940 samples from 172 populations (Supplementary Table 3A). When plotted (Figure 5A), the overall Foyemiaowan group was projected on the Northern Han and Tibeto-Burman cline and clustered closer around Tibeto-Burman populations, especially with Qiangic people from Daofu County, Sichuan and Ü-Tsang Tibetans from Shigatse on the Tibetan Plateau. Specifically, when compared with the “Overall” Foyemiaowan group, the “Sixteen Kingdoms” group was more closely related to Tibetan populations, particularly Ü-Tsang Tibetans from Shigatse and Nyingchi. Traditionally, Ü-Tsang is perceived as the cultural heartland of the Tibetan people, comprising Nagqu, Lhasa, Shannan and Shigatse. A greater degree of genetic flow from Tibetans from the core Tibet region toward Foyemiaowan may have occurred during the Sixteen Kingdoms period. As for the mtDNA perspective, we collected 613 samples from 30 ancient populations (Supplementary Table 3C) and 32,777 samples from 139 modern populations (Supplementary Table 3B). From the mtDNA PCA plot (Figure 5B), we were able to observe both “Overall” and “Sixteen Kingdoms” Foyemiaowan populations clustering closely with each-other as well as Mongolic-speaking populations, especially Dongxiang and Baoan from Linxia in Gansu, Mongolia from Baotou of Inner Mongolia, Daur from northeastern China, Mongolia from Khovd aimag in Mongolia and Mongolia from Dornogovi aimag in Mongolia. Thus, the Foyemiaowan population shows close genetic ties with Mongolic-speaking populations in the north and east of modern Gansu.
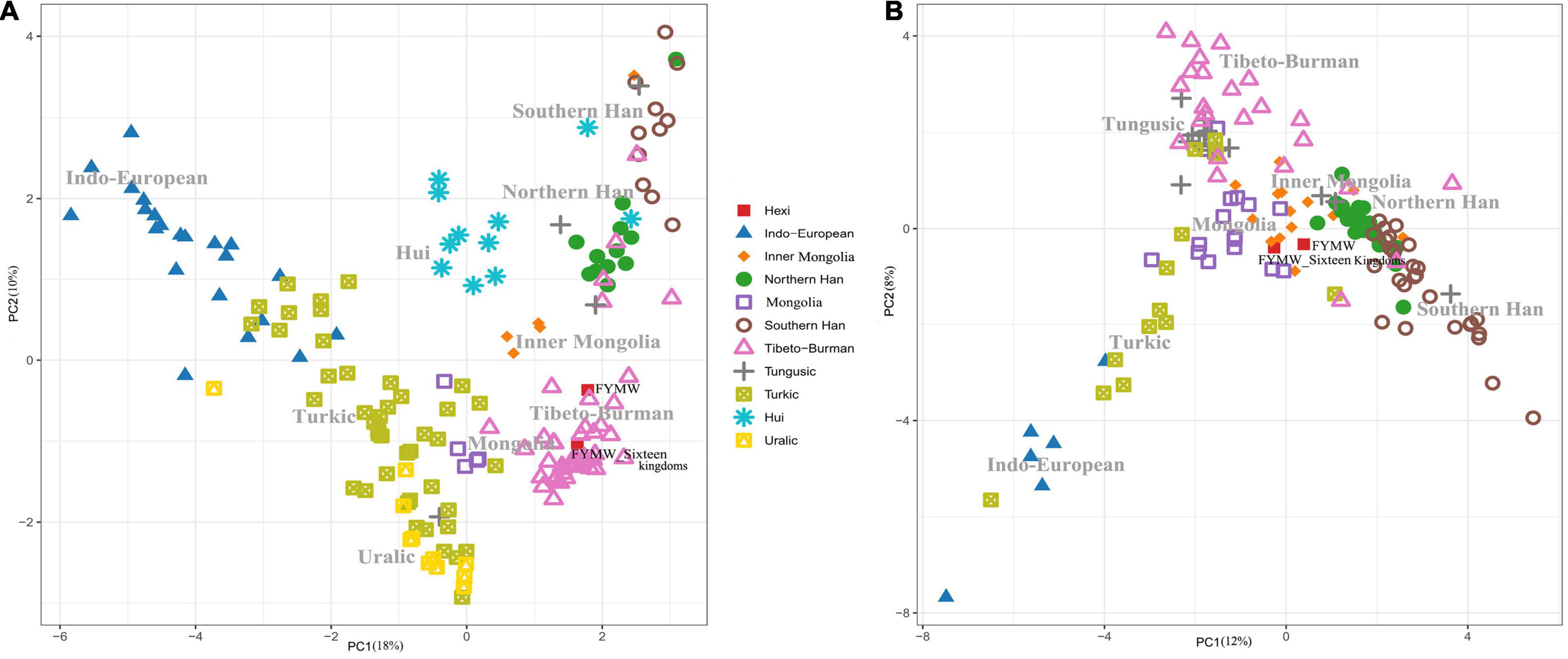
Figure 5. (A) PCA plot based on Y-haplogroups frequency of Foyemiaowan population and reference populations. (B) PCA plot based on mtDNA haplogroups frequency of Foyemiaowan population and reference populations.
In summary, interpopulation comparison shows close affinity between the Foyemiaowan population and Tibeto-Burman populations in terms of paternal structure and Mongolic populations in terms of maternal structure. This might be a consequence of long-term population migration and mixture along the ancient Silk Road.
To discern the fine structure of this Foyemiaowan population, we performed phylogenetic analyses for 14 mtDNA haplogroups (Supplementary Table 5 and Supplementary Figures 5–17) using published mitogenome data, including A6, D4*, D4b2b*, D4h1c, D4o, G1a2’3, M9a1a*, Z3*, Z3 + 709, Z4a, F1a1c, F2a, H7b*, and J1b1a1 + 146. Based on the geographic origins and phylogenetic trees (Supplementary Table 5), we were able to group these 14 haplogroups into three groups: northern East Asian (i.e., A6, D4*, D4b2b*, D4h1c, D4o, G1a2’3, M9a1a*, Z3*, Z3 + 709, and Z4a), southern East Asian (i.e., F1a1c and F2a) and western Eurasian (i.e., H7b* and J1b1a1 + 146). Following this, three representative haplogroups (i.e., D4o, F1a1c, and H7b*) were selected from these three groups for further study.
The phylogeny of D4o sequences is illustrated in Figure 6. The sequence divergence of the 55 D4o complete genomes corresponds to a coalescence time estimate of 14.19 (8.79–20.65) kya (Supplementary Table 7). The D4o tree shows an initial split into two sister subclades, D4o1 and D4o2, encompassing predominantly Turkic, Tungusic, Mongolic and Japonic northern Asian language groups. In particular, three Teleut from the Altai region of southern Siberia, one Buryat from southern Siberia, one Uyghur from the Turpan region of eastern Xinjiang and one from Foyemiaowan clustered into a new branch, harboring the diagnostic motif 146 (back mutation)-183-9196-11809-12612, which implies tight genetic ties between southern Siberian and Foyemiaowan populations. The phylogenetic tree of F1a1c, with the coalescence time estimated as 12.84 (9.01–18.14) kya, could be further subdivided into F1a1c1, F1a1c2, and F1a1c3. F1a1c mostly manifests in Chinese and Austronesian speaking populations in southern China and Southeast Asia. Other subclades included three southern Han from south of the Yangtze River region in China, one southern Han from Zhejiang in southern China, one Naxi from southwestern China, one Japanese and one from Foyemiaowan, falling into one distinct subclade characterized by one coding region mutation at np 15629. As for the basal branch H7b*, it was sporadically found in Indo-European speaking populations in northeastern Europe and central Asia, which shows a relatively younger coalescence time at 4.48 (2.52–7.56) kya. This haplogroup tree contained one Danish, one Russian from Vladimir near to Moscow, one individual with undetermined geographic origin and one from Foyemiaowan, forming a specific branch with one mutation at np 152 within control region. This may be indicative of western Euraisa influx into gene pool of populations in Hexi Corridor along the ancient Silk Road.
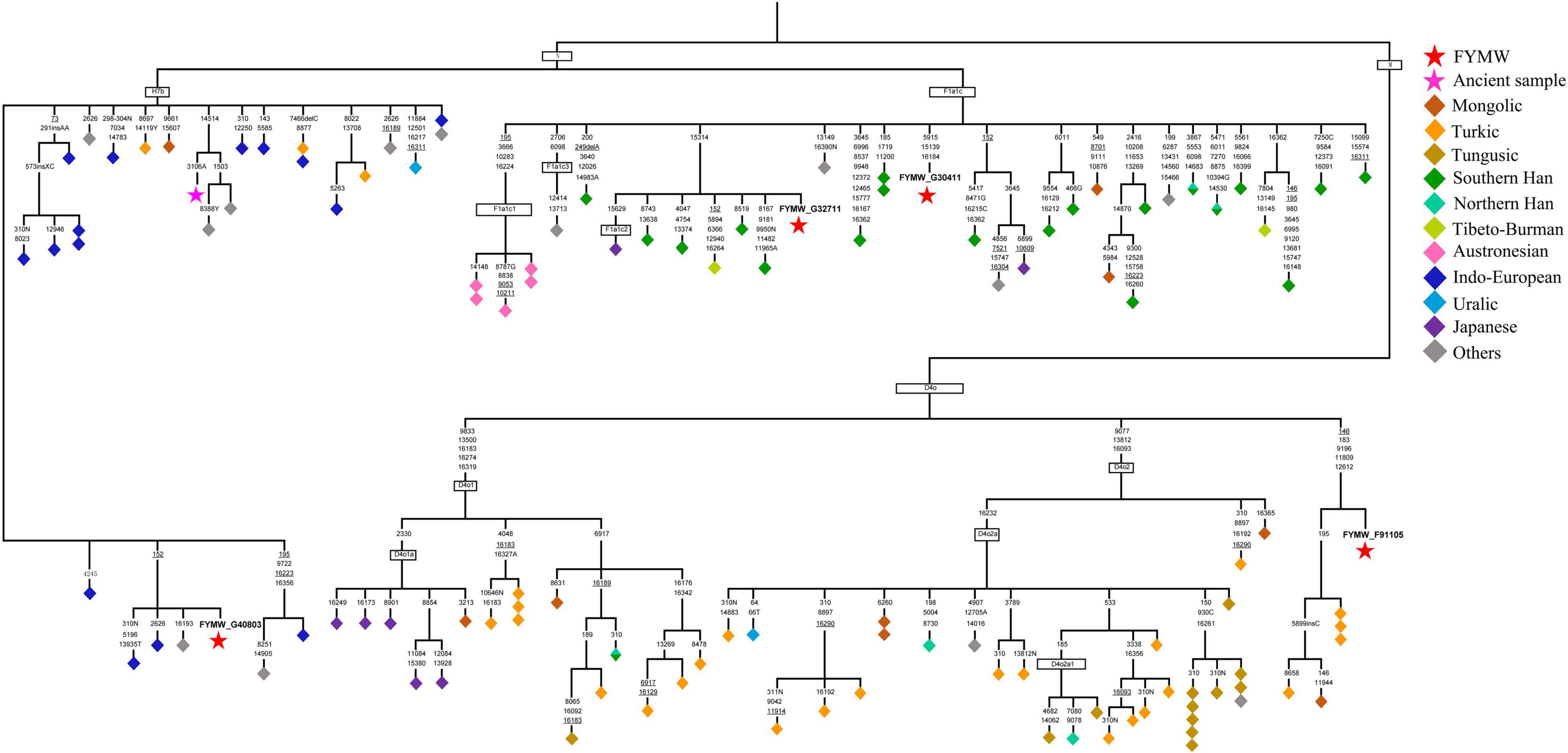
Figure 6. Maximum-parsimony phylogenetic trees of complete mtDNA sequences belonging to haplogroup Haplogroups D4o, F1a1c, and H7b*. Transversions are further specified; ins and del denote insertions and deletions of nucleotides, respectively, back mutations are underlined.
Our study supports the hypothesis that Foyemiaowan population have multiple potential ancestral sources. In our study, we analyzed the Y-chromosome and mtDNA of 34 Cao-Wei to Sui-Tang period individuals from the Foyemiaowan site, located in the west of the Hexi Corridor. After interpopulation comparison using PCA plots, we observed that the Foyemiaowan population was closely clustered with Tibeto-Burman populations on the paternal side, but intimately associated with Mongolic-speaking populations from the maternal aspect. Furthermore, we also observed the fine structure of Foyemiaowan population via lineage analysis. Y chromosome profiles of Foyemiaowan population revealed mainly Yellow River Valley origins related to Tibeto-Burman and Han Chinese populations, partially North Eurasian origins associated with Altaic speaking population and a small degree of southern East Asian origins. Similar to paternal structure, the maternal gene pool consisted of ancestries from northern East Asian (including haplogroups A, CZ, D, G, and M9), southern East Asian (including haplogroups B and F) and western Eurasian (including haplogroups H and J) groups. We can conclude that the genetic diversity of Foyemiaowan population was relatively high, and that western Eurasia lineages indicate the eastward migration of Hu people from ancient western Regions along the Silk Road.
Previous archeological and genetic studies argue that the central Hexi Corridor was densely populated by immigrants from Middle and Yellow River during the Han Dynasty (Chen et al., 2019; Xiong et al., 2022). This Han cultural element continued to play a predominant role in the Hexi Corridor even after the collapse of the dynasty. We see use of daily-use potteries, tomb guardian vases (镇墓瓶) and bronze mirrors at Foyemiaowan, items exhibiting a marked Han Chinese style (Chen et al., 2022). This study, contrary to what might be expected, found that population composition gradually trended toward diversification in the west of Hexi Corridor. An elevated value of δ15N and greater use of also C3 foods suggest that the dietary structure of Foyemiaowan population differed strikingly from farming populations consisting of migrants from the cultural core of China, such as the Heishuiguo population (Li, 2021), and veered closer to paleodietary patterns observed for nomadic populations (Wang et al., 2016; Zhang et al., 2016). Archeological findings such as painted murals unearthed in the Hexi Corridor and dating to the Cao-Wei and Sixteen Kingdoms periods support our results by showing the prevalence of non-Han customs (Gansu Province Cultural Relics Team, and Gansu Province Museum, 1985; Ma, 2000; E et al., 2009a,b). Other recorded customs also reflected the influence of a non-Han (Hu) population (Supplementary Figure 18). Hair style, attire, customs and facial features suggest that this population belonged to different ancient ethnic groups, such as Di peoples, Qiang peoples, Xianbei peoples, and other non-Han groups (Li, 2010).
Historical records further illustrate the multiethnic nature of the Hexi corridor during the Cao-Wei. At this time, the area included Han, Di, Qiang, Hexi Xianbei (i.e., Tufa Xianbei [秃发鲜卑], Yifu Xianbei [乙弗鲜卑], Yiyun Xianbei [意云鲜卑] etc.), Lushuihu (卢水胡), Western Region Hu (西域胡) and Tuge Hu (屠各胡) (Gao et al., 2018). The Han, Qiang and Di had settled in Hexi Corridor during the Han Dynasty (Zhao et al., 2011; Gao et al., 2018; Xiong et al., 2022). The Tufa Xianbei would later emerge as one of most powerful forces among the Hexi Xianbei. Migrating to the Hexi Corridor from the Yin Mountains in 219–256 CE (Zhou, 1987), they established the Southern Liang under Tufawugu (秃发乌孤) in 397CE. At its peak, the Tufa Xianbei commanded most of northwestern China. The Lushuihu settled along the Hexi Corridor during the Han and Wei Dynasties, founding the Northern Liang in 397 CE. This ethnic group was very complex and may have Xiongnu, Xiao Yuezhi (小月氏), Yiqu (义渠), Zahu (杂胡), Western Rong (西戎) or Zilu (赀虏) connections (Gao et al., 2018). Hu peoples from Western Region also can be found along Hexi corridor during Three Kingdoms periods (220–280 CE). The Dunhuang manuscripts (敦煌文书) record intermarriage between Han Chinese and Hu peoples (Lu, 1996). Sogdian correspondence (311CE) unearthed from Dunhuang indicates that a substantial population of Sogdian merchants migrated from Samarkand in this period (Bai, 2011). The discovery of western Eurasian haplogroups in the Foyemiaowan maternal gene pool may provide corroboration of these documents. These ethnic groups lived and intermarried at Dunhuang, strongly affecting the population history of the Hexi Corridor.
In conclusion, we have found that despite the similarity between archeological cultures at Foyemiaowan and Central Plains sites, the genetic structure and dietary structure of Foyemiaowan population differed strikingly from those of Han Chinese. This suggests that archeological culture is not consistent with ethnicity at Foyemiaowan. Yet although in this period dynastic governments had gradually relinquished control of the Hexi Corridor, and regional political forces established by various non-Han peoples became the dominant factors of Hexi history, Han cultural factors still strongly affected the Foyemiaowan peoples.
The FASTA files of mtDNA reported in this article have been deposited in the Genome Warehouse in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession number: GWHBHOV01000000-GWHBHPS01000000 that is publicly accessible at https://ngdc.cncb.ac.cn/gwh.
The studies involving human participants were reviewed and approved by the Ethics Committee of Fudan University of Life Sciences. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
GC, HL, and SW designed the study. HL and SW supervised the study. GC, YiY, and HW provided the materials and resources. HM collected the samples. GC and YiY performed the archeological data analysis. JX and YX performed the genetic laboratory work. YT, MB, YaY, and BZ performed the genetic data analysis. JX, HB, YT, and PD integrated the genetic data. JX, PD, SW, YT, and EA wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Key R&D Program of China (2020YFE0201600 and 2020YFC1521607), the National Social Science Fund of China (19VJX074), the National Natural Science Foundation of China (32070576), B&R Joint Laboratory of Eurasian Anthropology (18490750300), the Major Research Program of National Natural Science Foundation of China (91731303), the Major Project of National Social Science Foundation of China granted to Shaoqing Wen (20&ZD212), and the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), the 111 Project (B13016).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.901295/full#supplementary-material
Supplementary Figure 1 | Pattern of DNA degradation.
Supplementary Figure 2 | Phylogenetic relationship of Y-chromosome haplogroups in this study and their frequencies in Cao-Wei and Western Jin, Sixteen Kingdoms, Sui and Tang Dynasties, Undetermined.
Supplementary Figure 3 | Phylogenetic relationship of mtDNA haplogroups in this study and their frequencies in Cao-Wei and Western Jin, Sixteen Kingdoms, Sui and Tang Dynasties, Undetermined.
Supplementary Figure 4 | PCA plot based on mtDNA haplogroups frequency of Foyemiaowan populations and ancient reference populations.
Supplementary Figure 5 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup A6.
Supplementary Figure 6 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup D4.
Supplementary Figure 7 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup D4b2b.
Supplementary Figure 8 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup D4h1c.
Supplementary Figure 9 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup D4o.
Supplementary Figure 10 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup F1a1c.
Supplementary Figure 11 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup F2a.
Supplementary Figure 12 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup G1a2’3.
Supplementary Figure 13 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup H7b.
Supplementary Figure 14 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup J1b1a1 + 146.
Supplementary Figure 15 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup M9a1a.
Supplementary Figure 16 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup Z3.
Supplementary Figure 17 | Bayesian maximum clade credibility (MCC) tree of complete mtDNA sequences belonging to haplogroup Z3 + 709.
Supplementary Figure 18 | (A) Xincheng cemetery (Wei to Jin Dynasties), Burial 6, showing two people picking mulberry (E et al., 2009a). (B) Xigou cemetery (Wei to Jin Dynasties), containing two individuals, a male was riding a horse on the left side, a woman with long hair standing on his right side (Ma, 2000). (C) Xincheng mural tomb (Wei to Jin Dynasties), Burial 3, depicts two people living inside the yurt, one sleeping, the other cooking (Gansu Province Cultural Relics Team, and Gansu Province Museum, 1985). (D) Dingjiazha cemetery (Sixteen Kingdoms), Burial5, showing a man plowing (E et al., 2009b).
Allentoft, M. E., Sikora, M., Sjögren, K. G., Rasmussen, S., Rasmussen, M., Stenderup, J., et al. (2015). Population genomics of bronze age Eurasia. Nature 522, 167–172. doi: 10.1038/nature14507
Anthony, D. (2007). The Horse, the Wheel and Language. How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. Princeton, NJ: Princeton University Press.
Bai, X. (2011). The study of ethnic structure in the Hexi Corridor during Wei and Jin Dynasties. Soc. Sci. 169, 31–34.
Buikstra, J. E. (1994). Standards for Data Collection from Human Skeletal Remains, Research Series no. 44. Fayetteville: Arkansas Archeological Survey.
Caramelli, D., Posth, C., and Rickards, O. (2021). Reconstruction of the human peopling of Europe: a genetic insight. Ann. Hum. Biol. 48, 175–178. doi: 10.1080/03014460.2021.1955472
Chen, G. K., Ma, H. L., and Wang, Y. A. (2022). Dunhuang: The Excavation Report of Foyemiaowan-Xindiantai Cemetery in 2015. Lanzhou: Gansu Education Publishing House.
Chen, G. K., Yang, Y., and Liu, B. (2019). Ganzhou, Zhangye: The Excavation Report of Han Dynasty Cemetery in Heishuiguo Site (Volume II). Lanzhou: Gansu Education Publishing House.
Damgaard, P. B., Marchi, N., Rasmussen, S., Peyrot, M., Renaud, G., Korneliussen, T., et al. (2018). 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374. doi: 10.1038/s41586-018-0094-2
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
De Angelis, F., Veltre, V., Romboni, M., Di Corcia, T., Scano, G., Martínez-Labarga, C., et al. (2021). Ancient genomes from a rural site in Imperial Rome (1st-3rd cent. CE): a genetic junction in the Roman Empire. Ann. Hum. Biol. 48, 234–246. doi: 10.1080/03014460.2021.1944313
Derenko, M., Malyarchuk, B., Grzybowski, T., Denisova, G., Rogalla, U., Perkova, M., et al. (2010). Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in Northern Asia. PLoS One 5:e15214. doi: 10.1371/journal.pone.0015214
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
E, J., Zheng, B. L., and Gao, G. X. (2009a). Murals Unearthed from Wei, Jin and Tang Tombs in Gansu Province (Volume I). Lanzhou: Gansu University Press.
E, J., Zheng, B. L., and Gao, G. X. (2009b). Murals Unearthed from Wei, Jin and Tang Tombs in Gansu Province (Volume III). Lanzhou: Gansu University Press.
Fei, X. T. (1982). On the question of deepening the development of ethnographic surveys. J. South Cent. Univ. Natl. 3, 2–6. doi: 10.19898/j.cnki.42-1704/c.1982.03.001
Fu, Q., Mittnik, A., Johnson, P., Bos, K., Lari, M., Bollongino, R., et al. (2013). A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 23, 553–559. doi: 10.1016/j.cub.2013.02.044
Furholt, M. (2019). Re-integrating archaeology: a contribution to aDNA studies and the migration discourse on the 3rd millennium BC in Europe. Proc. Prehistor. Soc. 85, 115–129. doi: 10.1017/ppr.2019.4
Gansu Province Cultural Relics Team, and Gansu Province Museum (1985). Jiayuguan Mural Cemetery Excavation Report. Beijing: Cultural Relics Publishing House.
Gao, R., Jia, X., and Pu, Z. (2018). Sinification and Minoritization: The National Fusion of Hexi Population in the Han and Tang Dynasties. Beijing: China Social Sciences Press.
Gayden, T., Cadenas, A. M., Regueiro, M., Singh, N. B., Zhivotovsky, L. A., Underhill, P. A., et al. (2007). The Himalayas as a directional barrier to gene flow. Am. J. Hum. Genet. 80, 884–894. doi: 10.1086/516757
Helga, T., James, T. R., and Jill, P. M. (2013). Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. doi: 10.1093/bib/bbs017
Hu, K., Yan, S., Liu, K., Ning, C., Wei, L., Li, S., et al. (2015). The dichotomy structure of Y chromosome Haplogroup N. arXiv [Preprint] doi: 10.48550/arXiv.1504.06463
Ilumäe, A. M., Reidla, M., Chukhryaeva, M., Järve, M., Post, H., Karmin, M., et al. (2016). Human Y chromosome Haplogroup N: a non-trivial time-resolved phylogeography that cuts across language families. Am. J. Hum. Genet. 99, 163–173. doi: 10.1016/j.ajhg.2016.05.025
Ingman, M., and Gyllensten, U. (2007). A recent genetic link between Sami and the Volga-Ural region of Russia. Eur. J. Hum. Genet. 15, 115–120. doi: 10.1038/sj.ejhg.5201712
Jones, S. (1997). The Archaeology of Ethnicity: Constructing Identities in the Past and Present, 1st Edn. Abingdon: Routledge, 108.
Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L., and Orlando, L. (2013). mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684. doi: 10.1093/bioinformatics/btt193
Kang, L., Lu, Y., Wang, C., Hu, K., Chen, F., Liu, K., et al. (2012). Y-chromosome O3 Haplogroup diversity in Sino-Tibetan populations reveals two migration routes into the eastern Himalayas. Ann. Hum. Genet. 76, 92–99. doi: 10.1111/j.1469-1809.2011.00690.x
Katoh, K., Rozewicki, J., and Yamada, K. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Klales, A. R., Ousley, S. D., and Vollner, J. M. (2012). A revised method of sexing the human innominate using Phenice’s nonmetric traits and statistical methods. Am. J. Phys. Anthropol. 149, 104–114. doi: 10.1002/ajpa.22102
Ko, A. M. S., Chen, C. Y., Fu, Q., Delfin, F., Li, M., Chiu, H. L., et al. (2014). Early Austronesians: into and out of Taiwan. Am. J. Hum. Genet. 94, 426–436. doi: 10.1016/j.ajhg.2014.02.003
Kutanan, W., Kampuansai, J., Srikummool, M., Kangwanpong, D., Ghirotto, S., Brunelli, A., et al. (2017). Complete mitochondrial genomes of Thai and Lao populations indicate an ancient origin of Austroasiatic groups and demic diffusion in the spread of Tai–Kadai languages. Hum. Genet. 136, 85–98. doi: 10.1007/s00439-016-1742-y
Lang, M., Liu, H., Song, F., Qiao, X., Ye, Y., Ren, H., et al. (2019). Forensic characteristics and genetic analysis of both 27 Y-STRs and 143 Y-SNPs in Eastern Han Chinese population. Forensic Sci. Int. Genet. 42, e13–e20. doi: 10.1016/j.fsigen.2019.07.011
Li, S. H. (2010). Discussing the image of minority ethnic-groups in mural cemetery at Wei and Jin dynasties. Huaxia Archaeol. 4, 122–125. doi: 10.16143/j.cnki.1001-9928.2010.04.015
Li, X. (2021). Human Diets and its Influencing Factors During Han and Jin Periods in the Hexi Corridor and its Adjacent Areas. Doctoral Dissertation. Lanzhou: Lanzhou University.
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with burrows-wheeler transform. Bioinform. 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, Y. C., Ye, W. J., Jiang, C. G., Zeng, Z., Tian, J. Y., Yang, L. Q., et al. (2019). River valleys shaped the maternal genetic landscape of Han Chinese. Mol. Biol. Evol. 36, 1643–1652. doi: 10.1093/molbev/msz072
Liu, Y. J. (2012). Research on Migrants in Hexi During the Han Dynasty. Doctoral Dissertation. Lznhou: Northwest Normal University.
Lu, Q. F. (1996). Sinification of sogdians at dunhuang from tang to song dynasties. Hist. Res. 6, 25–34.
Lv, X. D. (2017). The history of Hexi immigrant during the Wuliang period from the view of a minority culture corridor. J. Hexi Univ. 33, 61–65.
Ma, J. H. (2000). Painted Bricks of Wei and Jin Tombs in Xigou, Jiuquan, Gansu Province. Chongqing: Chongqing Publishing Group.
Ma, P., Yang, X., Yan, S., Li, C., Gao, S., Han, B., et al. (2021). Ancient Y-DNA with reconstructed phylogeny provides insights into the demographic history of paternal Haplogroup N1a2-F1360. J. Genet. Genomics 48, 1130–1133. doi: 10.1016/j.jgg.2021.07.018
Margaryan, A., Derenko, M., Hovhannisyan, H., Malyarchuk, B., Heller, R., Khachatryan, Z., et al. (2017). Eight millennia of matrilineal genetic continuity in the South Caucasus. Curr. Biol. 27, 2023–2028. doi: 10.1016/j.cub.2017.05.087
Nei, M., and Tajima, F. (1981). DNA polymorphism detectable by restriction endonucleases. Genetics 97, 145–163. doi: 10.1093/genetics/97.1.145
Pamjav, H., Fóthi, Á., Fehér, T., and Fóthi, E. (2017). A study of the Bodrogköz population in north-eastern Hungary by Y chromosomal haplotypes and haplogroups. Mol. Genet. Genomics 292, 883–894. doi: 10.1007/s00438-017-1319-z
Peltzer, A., Jäger, G., Herbig, A., Seitz, A., Kniep, C., Krause, J., et al. (2016). EAGER: efficient ancient genome reconstruction. Genome Biol. 17:60. doi: 10.1186/s13059-016-0918-z
Pereira, L., Richards, M., Goios, A., Alonso, A., Albarrán, C., Garcia, O., et al. (2005). High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 15, 19–24. doi: 10.1101/gr.3182305
Pliss, L., Tambets, K., Loogväli, E. L., Pronina, N., Lazdins, M., Krumina, A., et al. (2006). Mitochondrial DNA portrait of Latvians: towards the understanding of the genetic structure of Baltic-speaking populations. Ann. Hum. Genet. 70(Pt 4), 439–458. doi: 10.1111/j.1469-1809.2005.00238.x
Qi, X., Cui, C., Peng, Y., Zhang, X., Yang, Z., Zhong, H., et al. (2013). Genetic evidence of paleolithic colonization and neolithic expansion of modern humans on the Tibetan Plateau. Mol. Biol. Evol. 30, 1761–1778. doi: 10.1093/molbev/mst093
Ralf, A., Montiel González, D., Zhong, K., and Kayser, M. (2018). Yleaf: software for human Y-chromosomal haplogroup inference from next-generation sequencing data. Mol. Biol. Evol. 35, 1291–1294. doi: 10.1093/molbev/msy032
Renaud, G., Slon, V., Duggan, A. T., and Kelso, J. (2015). Schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 16:224. doi: 10.1186/s13059-015-0776-0
Renfrew, C., and Bahn, P. G. (2007). Archaeology Essentials: Theories, Methods, and Practice. London: Thames & Hudson College.
Rieux, A., Eriksson, A., Li, M., Sobkowiak, B., Weinert, L. A., Warmuth, V., et al. (2014). Improved calibration of the human mitochondrial clock using ancient genomes. Mol. Biol. Evol. 31, 2780–2792. doi: 10.1093/molbev/msu222
Schurr, T. G., Sukernik, R. I., Starikovskaya, Y. B., and Wallace, D. C. (1999). Mitochondrial DNA variation in Koryaks and Itel’men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am. J. Phys. Anthropol. 108, 1–39. doi: 10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1
Si, M. Q. Han Dynasty. (2002b). Shiji. Biography of Chulizi and Gan Mao. Changsha: Yuelu Publishing House, 436.
Sun, X. F., Wen, S. Q., Lu, C. Q., Zhou, B. Y., Curnoe, D., Lu, H. Y., et al. (2021). Ancient DNA and multimethod dating confirm the late arrival of anatomically modern humans in southern China. Proc. Natl. Acad. Sci. U.S.A. 118:e2019158118. doi: 10.1073/pnas.2019158118
Tambets, K., Rootsi, S., Kivisild, T., Help, H., Serk, P., Loogväli, E. L., et al. (2004). The western and eastern roots of the Saami–the story of genetic “outliers” told by mitochondrial DNA and Y chromosomes. Am. J. Hum. Genet. 74, 661–682. doi: 10.1086/383203
Unterländer, M., Palstra, F., Lazaridis, I., Pilipenko, A., Hofmanová, Z., Groß, M., et al. (2017). Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nat. Commun. 8:14615. doi: 10.1038/ncomms14615
Upton, D. (1996). Ethnicity, authenticity, and invented traditions. Hist. Archaeol. 30, 1–7. doi: 10.1007/bf03373584
Vidrová, V., Tesarová, M., Trefilova, E., Honzík, T., Magner, M., and Zeman, J. (2008). Mitochondrial DNA haplogroups in the Czech population compared to other European countries. Hum. Biol. 80, 669–674. doi: 10.3378/1534-6617-80.6.669
Wang, C. C., and Li, H. (2013). Inferring human history in East Asia from Y chromosomes. Investig. Genet. 4:11. doi: 10.1186/2041-2223-4-11
Wang, C. C., Wang, L. X., Shrestha, R., Zhang, M., Huang, X. Y., Hu, K., et al. (2014). Genetic structure of qiangic populations residing in the Western Sichuan corridor. PLoS One 9:e103772. doi: 10.1371/journal.pone.0103772
Wang, T. T., Fuller, B. T., Wei, D., Chang, X. E., and Hu, Y. W. (2016). Investigating dietary patterns with stable isotope ratios of collagen and starch grain analysis of dental calculus at the Iron Age cemetery site of Heigouliang, Xinjiang, China. Int. J. Osteoarchaeol. 26, 693–704. doi: 10.1002/oa.2467
Weissensteiner, H., Pacher, D., Kloss-Brandstätter, A., Forer, L., Specht, G., Bandelt, H. J., et al. (2016). HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63. doi: 10.1093/nar/gkw233
Wen, B., Li, H., Gao, S., Mao, X., Gao, Y., Li, F., et al. (2005). Genetic structure of Hmong-Mien speaking populations in East Asia as revealed by mtDNA lineages. Mol. Biol. Evol. 22, 725–734. doi: 10.1093/molbev/msi055
Wen, S. Q., Bao, R. Y., Zhou, B. Y., Du, P. X., Sun, C., Chen, L., et al. (2019). China National DNA Martyry: a beacon of hope for the martyrs’ coming home. J. Hum. Genet. 64, 1045–1047. doi: 10.1038/s10038-019-0649-6
Xia, Z., Yan, S., Wang, C., Zheng, H., Zhang, F., Liu, Y., et al. (2019). Inland-coastal bifurcation of southern East Asians revealed by Hmong-Mien genomic history. bioRxiv [Preprint] doi: 10.1101/730903
Xiong, J. X., Du, P. X., Chen, G. K., Tao, Y. C., Zhou, B. Y., and Yang, Y. S. (2022). Sex-biased population admixture mediated subsistence strategy transition of Heishuiguo people in Han dynasty Hexi corridor. Front. Genet. 13:827277. doi: 10.3389/fgene.2022.827277
Xue, Y., Zerjal, T., Bao, W., Zhu, S., Shu, Q., Xu, J., et al. (2006). Male demography in East Asia: a north-south contrast in human population expansion times. Genetics 172, 2431–2439. doi: 10.1534/genetics.105.054270
Yan, S., Wang, C. C., Wang, C. C., Li, H., Li, S. L., and Jin, L. (2011). An updated tree of Y-chromosome haplogroup O and revised phylogenetic positions of mutations P164 and PK4. Eur. J. Hum. Genet. 19, 1013–1015. doi: 10.1038/ejhg.2011.64
Zhang, X., Wei, D., Wu, Y., Nie, Y., and Hu, Y. (2016). Carbon and nitrogen stable isotope ratio analysis of Bronze Age humans from the Xiabandi cemetery, Xinjiang, China: implications for cultural interactions between the East and West. Chin. Sci. Bull. 61, 3509–3519. doi: 10.1360/N972016-00514
Zhao, Y. B., Li, H. J., Li, S. N., Yu, C. C., Gao, S. Z., Xu, Z., et al. (2011). Ancient DNA evidence supports the contribution of Di-Qiang people to the Han Chinese gene pool. Am. J. Phys. Anthropol. 144, 258–268. doi: 10.1002/ajpa.21399
Keywords: Dunhuang, dynastic transitions, archeological culture, genetic diversity, multi-ethnicity
Citation: Xiong J, Tao Y, Ben M, Yang Y, Du P, Allen E, Wang H, Xu Y, Yu Y, Meng H, Bao H, Zhou B, Chen G, Li H and Wen S (2022) Uniparental Genetic Analyses Reveal Multi-Ethnic Background of Dunhuang Foyemiaowan Population (220–907 CE) With Typical Han Chinese Archaological Culture. Front. Ecol. Evol. 10:901295. doi: 10.3389/fevo.2022.901295
Received: 21 March 2022; Accepted: 14 April 2022;
Published: 27 June 2022.
Edited by:
Chuan-Chao Wang, Xiamen University, ChinaCopyright © 2022 Xiong, Tao, Ben, Yang, Du, Allen, Wang, Xu, Yu, Meng, Bao, Zhou, Chen, Li and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoke Chen, Y2hlbmd1b2tlMTk4MEBzaW5hLmNvbQ==; Hui Li, bGhjYUBmdWRhbi5lZHUuY24=; Shaoqing Wen, d2Vuc2hhb3FpbmcxOTgyQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.