
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Ecol. Evol. , 14 April 2022
Sec. Chemical Ecology
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.900818
This article is part of the Research Topic The Morphology and Physiology of Insect Chemosensory Systems – Its Origin and Evolution View all 17 articles
This article is a correction to:
Sex Pheromone Receptors of Lepidopteran Insects
A Corrigendum on
Sex Pheromone Receptors of Lepidopteran Insects
by Yang, C., Cheng, J., Lin, J., Zheng, Y., Yu, X., and Sun, J. (2022). Front. Ecol. Evol. 10:797287. doi: 10.3389/fevo.2022.797287
In the original article, the sentence “The identified SPRs and their specific sex pheromone ligands were summarized (Table 1).” should have been removed.
A correction has been made to Introduction, paragraph 3:
“In this review, we summarized SPR and Orco information from 10 families (i.e., Bombycidae, Plutellidae, Sphingidae, Saturniidae, Geometridae, Nymphalidae, Noctuidae, Tortricidae, Pyralidae, and Crambidae) of Lepidopteran insects (Krieger et al., 2004, 2005; Sakurai et al., 2004; Miura et al., 2005; Nakagawa et al., 2005; Grosse-Wilde et al., 2007, 2010, 2011; Mitsuno et al., 2008; Jordan et al., 2009; Patch et al., 2009; Zhang et al., 2009, 2010, 2013, 2014, 2015; Wanner et al., 2010; Legeai et al., 2011; Wang et al., 2011; Yasukochi et al., 2011; Zhan et al., 2011; Bengtsson et al., 2012; Carraher et al., 2012; Leary et al., 2012; Liu et al., 2012, 2014; Montagne et al., 2012; Xu et al., 2012, 2015; Liu C. et al., 2013; Liu Y. et al., 2013; Sun et al., 2013; Wu et al., 2013; Zhang and Lofstedt, 2013; Jiang et al., 2014; Corcoran et al., 2015; De Fouchier et al., 2015; Feng et al., 2015; Garczynski and Leal, 2015; Lin et al., 2015; Steinwender et al., 2015; Chang et al., 2016; Ge et al., 2016; Jia et al., 2016; Walker et al., 2016; Zhang D.D. et al., 2016; Zhang Y.N. et al., 2016; Gonzalez et al., 2017; Li et al., 2017; Wicher et al., 2017; Yang et al., 2017; Du et al., 2018; Grapputo et al., 2018; Rojas et al., 2018; Table 1). Among all the Lepidopteran SPRs, several of them have been characterized to be sex pheromone sensing receptors. First of all, we reviewed the phylogenetic analyses of Lepidopteran SPRs, and the evolution of the summarized Lepidopteran SPRs was analyzed through MEGA X (Whelan and Goldman, 2001; Kumar et al., 2018). Second, we reviewed the transmembrane predictions of Lepidopteran SPRs, and the protein structure of Lepidopteran SPRs was predicted by online software Consensus Constrained TOPology Prediction (CCTOP) (Dobson et al., 2015) and SwissModel (Bertoni et al., 2017; Bienert et al., 2017; Waterhouse et al., 2018; Guex et al., 2019; Studer et al., 2020). Third, the interaction of Lepidopteran SPM and SPR was reviewed. Finally, the research status of downstream signaling responses and ligand-gated ion channels by the coupling of SPR and Orco was depicted.”
In the original article, Figures 3 and 4 did not match their captions because the figures were erroneously interchanged. The corrected Figures 3 and 4 with their captions appear below.
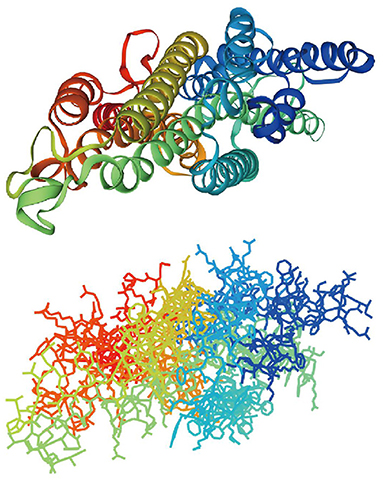
Figure 3. Predicted structure of Lepidopteran SPRs BmOR3 and Orcos through SwissModel. In total, 7 transmembrane helixes were colored rainbow in the form of silk ribbon (Cartoon) and lines.
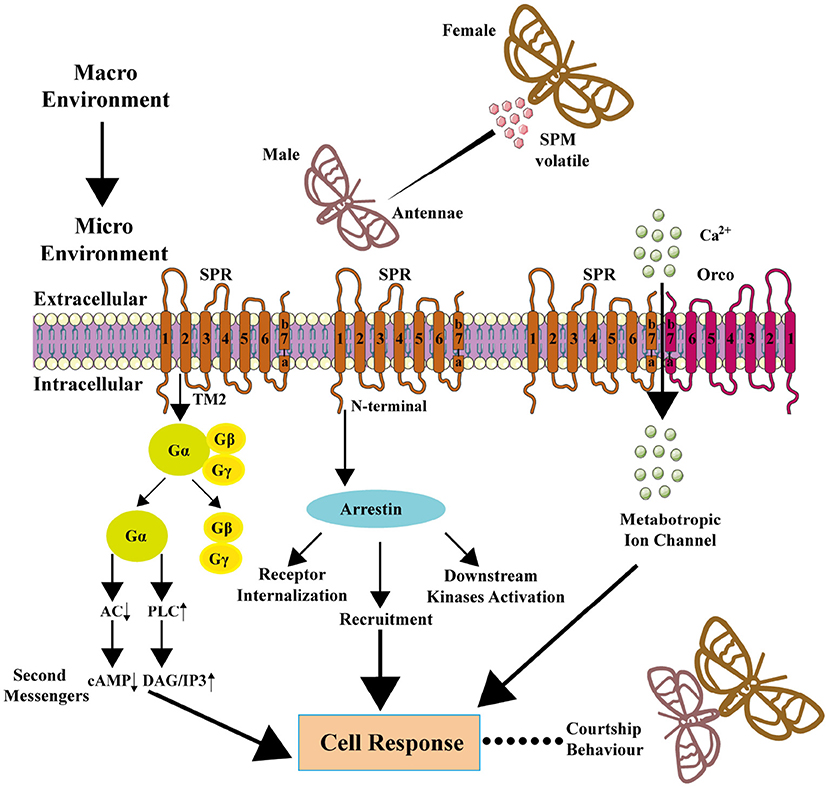
Figure 4. Downstream signaling pathway of SPR and metabotropic channel of SPR-Orco. In macro environment, male insects sense the SPM released by female insects through an olfactory sensory neuron in antennae, and SPM works as an agonist to SPR, and signal transduction leads to ultimate mating behavior.
In the original article, the citation “[18]” should have been replaced with “Wicher et al., 2017”. The citation has now been inserted in “Downstream Signaling Pathways of Sex Pheromone Receptors”, paragraph 1 and should read:
“Early research revealed the presence of G-protein, belonging to the Gαq family, in antennal preparations (especially the pheromone-sensitive sensilla trichodea) of B. mori and Antheraea pernyi, implied a participation of G-protein of the Gαq family in the signal transduction of OR cells in moths (Laue et al., 1997; Nakagawa et al., 2005). The bombykol stimulation of Xenopus laevis oocytes expressing BmOR-1 and BmGαq elicited robust dose-dependent inward Ca2+-dependent Cl− currents on two-electrode voltage-clamp recordings, demonstrating that the binding of bombykol to BmOR-1 leads to the activation of a BmGαq-mediated signaling cascade (Sakurai et al., 2004). MsexOR1 and MsexOrco coexpressed in HEK293 and CHO cells caused bombykal-dependent increases in the intracellular free Ca2+ concentration, and inhibitor evidence showed that phospholipase C (PLC) and protein kinase C (PKC) activities are involved in the bombykal-receptor-mediated Ca2+ signals of hawk moths. It could be hypothesized that MsexOrs couple to Gαq proteins, requiring the activation of PLC for pheromone transduction (Wicher et al., 2017). Immunocytochemistry research showed that anti-Gαq and anti-Gαs antisera stained the inner and outer dendritic segments of the putative OR neuron in male and female antennae, which suggested that each subunit mediates a subset of the odorant response (Miura et al., 2005). In addition, a computational model of the insect pheromone transduction cascade had been used to calculate the presence of the G-protein pathway in pheromone detection (Gu et al., 2009). Furthermore, recent research showed that in HEK293A cells expressing BmOR3 and human Gαi, the dose-dependent coupling of BmOR3 and Gαi on bombykal stimulation was detected through BRET (Lin et al., 2021). From the biophysical perspective, a conservation residue W103 in transmembrane 2 of BmOR3 is the key that determines receptor-Gi coupling (Lin et al., 2021). Pretreatment with specific Gi inhibitor PTX had no significant effects on bombykal-induced BmOR3-BmOrco complex formation or complex-regulated calcium influx, suggesting that Gi coupling and BmOrco coupling are the two independent processes in the case of BmOR3 (Lin et al., 2021).”
Due to a production error, bombykol was erroneously described as the agonist of MsexOR1 and BmOR3; the correct agonist is bombykal.
A correction has been made to “Downstream Signaling Pathways of Sex Pheromone Receptors”, paragraphs 1 and 2:
“Early research revealed the presence of G-protein, belonging to the Gαq family, in antennal preparations (especially the pheromone-sensitive sensilla trichodea) of B. mori and Antheraea pernyi, implied a participation of G-protein of the Gαq family in the signal transduction of OR cells in moths (Laue et al., 1997; Nakagawa et al., 2005). The bombykol stimulation of Xenopus laevis oocytes expressing BmOR-1 and BmGαq elicited robust dose-dependent inward Ca2+-dependent Cl− currents on two-electrode voltage-clamp recordings, demonstrating that the binding of bombykol to BmOR-1 leads to the activation of a BmGαq-mediated signaling cascade (Sakurai et al., 2004). MsexOR1 and MsexOrco coexpressed in HEK293 and CHO cells caused bombykal-dependent increases in the intracellular free Ca2+ concentration, and inhibitor evidence showed that phospholipase C (PLC) and protein kinase C (PKC) activities are involved in the bombykal-receptor-mediated Ca2+ signals of hawk moths. It could be hypothesized that MsexORs couple to Gαq proteins, requiring the activation of PLC for pheromone transduction (Wicher et al., 2017). Immunocytochemistry research showed that anti-Gαq and anti-Gαs antisera stained the inner and outer dendritic segments of the putative OR neuron in male and female antennae, which suggested that each subunit mediates a subset of the odorant response (Miura et al., 2005). In addition, a computational model of the insect pheromone transduction cascade had been used to calculate the presence of the G-protein pathway in pheromone detection (Gu et al., 2009). Furthermore, recent research showed that in HEK293A cells expressing BmOR3 and human Gαi, the dose-dependent coupling of BmOR3 and Gαi on bombykal stimulation was detected through BRET (Lin et al., 2021). From the biophysical perspective, a conservation residue W103 in transmembrane 2 of BmOR3 is the key that determines receptor-Gi coupling (Lin et al., 2021). Pretreatment with specific Gi inhibitor PTX had no significant effects on bombykal-induced BmOR3-BmOrco complex formation or complex-regulated calcium influx, suggesting that Gi coupling and BmOrco coupling are the two independent processes in the case of BmOR3 (Lin et al., 2021).
The GPCRs usually direct the recruitment, activation, and scaffolding of the cytoplasmic signaling complexes via two multifunctional adaptor and transducer molecules, β-arrestins 1 and 2, and arrestins also function to activate signaling cascades independently of G-protein activation or mediate receptor desensitization (Lefkowitz and Shenoy, 2005; DeWire et al., 2007). Individual arrestins had been reported to function in both olfactory and visual pathways in Dipteran insects (Merrill et al., 2001) but not in Lepidopteran insects. Recent research reported that bombykal robustly stimulated the recruitment of human β-arrestin-1/2 and B. mori intrinsic arrestin to BmOR3 in HEK293A cells in a concentration-dependent manner, and the arrestin, in turn, regulated BmOR3 internalization (Lin et al., 2021). Bombykal also induced downstream kinase (i.e., ERK, SRC, AKT, and JNK) activation (phosphorylation) through arrestin (Lin et al., 2021). These results confirmed the arrestin-mediated signaling downstream of BmOR3. The knockdown of β-arrestins significantly reduced bombykal-induced calcium influx through BmOR3-BmOR2, which was accompanied by the collapse of the receptor complex, suggesting that the β-arrestins mediate Ca2+ response mainly by regulating the structural and functional integrity of the BmOR3-BmOR2 complex (Lin et al., 2021). The summarized researches show that insect pheromone receptors may both have G-protein and arrestin downstream pathways (Figure 4).”
The same correction has also been made to “Metabotropic Ion Channel by the Coupling of Lepidopteran Sex Pheromone Receptors and Orcos”, paragraphs 1 and 2:
“BmOR1 and BmOR3 of B. mori are mutually exclusively expressed in a pair of adjacent pheromone-sensitive neurons of male antennae, and both of which are coexpressed with the highly conserved insect Orco. Heterologous cells coexpressing BmOR2 can greatly enhance the sensitivity of BmOR1 to bombykol, and the current-voltage analysis showed that bombykol activated a non-selective cation channel in oocytes expressing BmOR1 and BmOR2, which is different from Ca2+-activated Cl− channel through BmGαq, and the non-selective cation channel activity in response to bombykol was also observed when BmOR1 was coexpressed with HvirOR2 or Or83b (Whelan and Goldman, 2001; Nakagawa et al., 2005; Figure 4). As reported in MsexOR1, the PLC/PKC activity is a prerequisite to bombykal-receptor-mediated Ca2+ signals in HEK293 and CHO cells, and it could be hypothesized that MsexOR1 and MsexOrco need to be phosphorylated before they can be gated by bombykal as an ionotropic odor receptor-ion channel complex (Wicher et al., 2017).
In a recent study, the BmOR3-BmOR2 combination elicited a response to bombykal and showed similar channel properties, and the coupling of BmOR3 and BmOR2 forms a cation channel with the detection of calcium influx (Lin et al., 2021). From the view of biophysics, there was also physical interaction between BmOR3 and its Orco BmOR2. On bombykal stimulation, the cytoplasmic parts intracellular loop 1 (ICL1), ICL2, and ICL3 moved away from the N-terminus, while the C-terminal helical kink moved close to the N-terminus of BmOR3. On the contrary, the lower part of loop7a-7b moved away from the N-terminus in BmOR2. ICL1, ICL2, and ICL3 also moved away from the N-terminus of BmOR2 (Lin et al., 2021). The replacement of transmembrane 7 in both receptors confirmed its indispensable role in BmOR3-BmOrco coupling for ionotropic functions (Lin et al., 2021). Several key motifs determine the BmOR3-BmOR2 coupling, the charged residue pair of BmOR3-E403 and BmOrco-K437 represents an important “ionic lock” in regard to mediating BmOR3-BmOrco coupling, and the hydrophobic patches F428/F433 of BmOR3 and zipper Y464/V467/L468/L471 of BmOrco are spatially close to each other, suggesting that they might form hydrophobic interactions (Lin et al., 2021). These reports suggest that the coupling of both SPR and Orco plays a vital role in sex pheromone signal detection and transduction.”
The authors and publisher apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bengtsson, J. M., Trona, F., Montagne, N., Anfora, G., Ignell, R., Witzgall, P., et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS One 7:e31620. doi: 10.1371/journal.pone.0031620
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., and Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7:10480. doi: 10.1038/s41598-017-09654-8
Bienert, S., Waterhouse, A., de Beer, T. A. P., Tauriello, G., Studer, G., Bordoli, L., et al. (2017). The SWISS-MODEL Repository - new features and functionality. Nucleic Acids Res. 45, D313–D319. doi: 10.1093/nar/gkw1132
Carraher, C., Authier, A., Steinwender, B., and Newcomb, R. D. (2012). Sequence comparisons of odorant receptors among tortricid moths reveal different rates of molecular evolution among family members. PLoS One 7:e38391. doi: 10.1371/journal.pone.0038391
Chang, H., Guo, M., Wang, B., Liu, Y., Dong, S., and Wang, G. (2016). Sensillar expression and responses of olfactory receptors reveal different peripheral coding in two Helicoverpa species using the same pheromone components. Sci. Rep. 6:18742. doi: 10.1038/srep18742
Corcoran, J. A., Jordan, M. D., Thrimawithana, A. H., Crowhurst, R. N., and Newcomb, R. D. (2015). The peripheral olfactory repertoire of the lightbrown apple moth, Epiphyas postvittana. PLoS One 10:e0128596. doi: 10.1371/journal.pone.0128596
De Fouchier, A., Sun, X., Monsempes, C., Mirabeau, O., Jacquin-Joly, E., and Montagné, N. (2015). Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus Spodoptera. Front. Ecol. Evol. 3:95. doi: 10.3389/fevo.2015.00095
DeWire, S. M., Ahn, S., Lefkowitz, R. J., and Shenoy, S. K. (2007). Beta-arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510.
Dobson, L., Reményi, I., and Tusnády, G. E. (2015). CCTOP: a consensus constrained TOPology prediction web server. Nucleic Acids Res. 43, W408–W412. doi: 10.1093/nar/gkv451
Du, L. X., Liu, Y., Zhang, J., Gao, X. W., Wang, B., and Wang, G. R. (2018). Identification and characterization of chemosensory genes in the antennal transcriptome of Spodoptera exigua. Comp. Biochem. Physiol. Part D Genom. Proteom. 27, 54–65. doi: 10.1016/j.cbd.2018.05.001
Feng, B., Lin, X., Zheng, K., Qian, K., Chang, Y., and Du, Y. (2015). Transcriptome and expression profiling analysis link patterns of gene expression to antennal responses in Spodoptera litura. BMC Genomics 16:269. doi: 10.1186/s12864-015-1375-x
Garczynski, S. F., and Leal, W. S. (2015). Alternative splicing produces two transcripts encoding female-biased pheromone subfamily receptors in the navel orangeworm, Amyelois transitella. Front. Ecol. Evol. 3:115. doi: 10.3389/fevo.2015.00115
Ge, X., Zhang, T. T., Wang, Z. Y., He, K. L., and Bai, S. X. (2016). Identification of putative chemosensory receptor genes from yellow peach moth Conogethes punctiferalis (Guenée) antennae transcriptome. Sci. Rep. 6, 32636–32648. doi: 10.1038/srep32636
Gonzalez, F., Witzgall, P., and Walker, W. B. (2017). Antennal transcriptomes of three tortricid moths reveal putative conserved chemosensory receptors for social and habitat olfactory cues. Sci. Rep. 7, 41829–41840. doi: 10.1038/srep41829
Grapputo, A., Thrimawithana, A. H., Steinwender, B., and Newcomb, R. D. (2018). Differential gene expression in the evolution of sex pheromone communication in New Zealand's endemic leafroller moths of the genera Ctenopseustis and Planotortrix. BMC Genomics 19:94. doi: 10.1186/s12864-018-4451-1
Grosse-Wilde, E., Gohl, T., Bouche, E., Breer, H., and Krieger, J. (2007). Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 25, 2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x
Grosse-Wilde, E., Kuebler, L. S., Bucks, S., Vogel, H., Wicher, D., and Hansson, B. S. (2011). Antennal transcriptome of Manduca sexta. PNAS 108, 7449–7454.
Grosse-Wilde, E., Stieber, R., Forstner, M., Krieger, J., Wicher, D., and Hansson, B. S. (2010). Sex-specific odorant receptors of the tobacco hornworm manduca sexta. Front. Cell Neurosci. 4:22. doi: 10.3389/fncel.2010.00022
Gu, Y., Lucas, P., and Rospars, J. (2009). Computational model of the insect pheromone transduction cascade. PLoS Comput. Biol. 5:e1000321. doi: 10.1371/journal.pcbi.1000321
Guex, N., Peitsch, M. C., and Schwede, T. (2019). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30, S162–S173. doi: 10.1002/elps.200900140
Jia, X. J., Wang, H. X., Yan, Z. G., Zhang, M. Z., Wei, C. H., Qin, X. C., et al. (2016). Antennal transcriptome and differential expression of olfactory genes in the yellow peach moth, Conogethes punctiferalis (Lepidoptera: Crambidae). Sci. Rep. 6, 29067–29082. doi: 10.1038/srep29067
Jiang, X. J., Guo, H., Di, C., Yu, S., Zhu, L., Huang, L. Q., et al. (2014). Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem. Mol. Biol. 48, 63–74. doi: 10.1016/j.ibmb.2014.02.010
Jordan, M. D., Anderson, A., Begum, D., Carraher, C., Authier, A., Marshall, S. D., et al. (2009). Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem. Senses 34, 383–394. doi: 10.1093/chemse/bjp010
Krieger, J., Grosse-Wilde, E., Gohl, T., and Breer, H. (2005). Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurosci. 21, 2167–2176.
Krieger, J., Grosse-Wilde, E., Gohl, T., Dewer, Y. M. E., Raming, K., and Breer, H. (2004). Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). PNAS 101, 11845–11850. doi: 10.1073/pnas.0403052101
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Laue, M., Maida, R., and Redkozubov, A. (1997). G-protein activation, identification and immunolocalization in pheromone-sensitive sensilla trichodea of moths. Cell Tissue Res. 288, 149–158. doi: 10.1007/s004410050802
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. PNAS 109, 14081–14086. doi: 10.1073/pnas.1204661109
Lefkowitz, R. J., and Shenoy, S. K. (2005). Transduction of receptor signals by β-Arrestins. Science 308, 512–517. doi: 10.1126/science.1109237
Legeai, F., Malpel, S., Montagné, N., Monsempes, C., Cousserans, F., Merlin, C., et al. (2011). An expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics 12:86. doi: 10.1186/1471-2164-12-86
Li, Z. Q., Luo, Z. X., Cai, X. M., Bian, L., Xin, Z. J., Liu, Y., et al. (2017). Chemosensory gene families in ectropis grisescens and candidates for detection of Type-II sex pheromones. Front. Physiol. 8:953. doi: 10.3389/fphys.2017.00953
Lin, J. Y., Yang, Z., Yang, C., Du, J. X., Yang, F., Cheng, J., et al. (2021). An ionic lock and a hydrophobic zipper mediate the coupling between an insect pheromone receptor BmOR3 and downstream effectors. J. Biol. Chem. 297:101160. doi: 10.1016/j.jbc.2021.101160
Lin, X., Zhang, Q., Wu, Z., and Du, Y. (2015). Identification and differential expression of a candidate sex pheromone receptor in natural populations of Spodoptera litura. PLoS One 10:e0131407. doi: 10.1371/journal.pone.0131407
Liu, C., Liu, Y., Walker, W. B., Dong, S., and Wang, G. (2013). Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hubner). Insect Biochem. Mol. Biol. 43, 747–754. doi: 10.1016/j.ibmb.2013.05.009
Liu, N. Y., Xu, W., Papanicolaou, A., Dong, S. L., and Anderson, A. (2014). Identification and characterization of three chemosensory receptor families in the cotton bollworm Helicoverpa armigera. BMC Genomics 15:597. doi: 10.1186/1471-2164-15-597
Liu, Y., Gu, S., Zhang, Y., Guo, Y., and Wang, G. (2012). Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS One 7:e48260. doi: 10.1371/journal.pone.0048260
Liu, Y., Liu, C., Lin, K., and Wang, G. (2013). Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS One 8:e62094. doi: 10.1371/journal.pone.0062094
Merrill, C. E., Riesgo-Escovar, J., Pitts, R. J., Kafatos, F. C., Carlson, J. R., and Zwiebel, L. J. (2001). Visual arrestins in olfactory pathways of Drosophila and the malaria vector mosquito Anopheles gambiae. PNAS 99, 1633–1638. doi: 10.1073/pnas.022505499
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Ozawa, R., et al. (2008). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Miura, N., Atsumi, S., Tabunoki, H., and Sato, R. (2005). Expression and localization of three G protein alpha subunits, Go, Gq, and Gs, in adult antennae of the silkmoth (Bombyx mori). J. Comp. Neurol. 485, 143–152. doi: 10.1002/cne.20488
Montagne, N., Chertemps, T., Brigaud, I., Francois, A., Francois, M. C., Fouchier, A., et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596. doi: 10.1111/j.1460-9568.2012.08183.x
Nakagawa, T., Sakurai, T., Nishioka, T., and Touhara, K. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1641. doi: 10.1126/science.1106267
Patch, H. M., Velarde, R. A., Walden, K. K., and Robertson, H. M. (2009). A candidate pheromone receptor and two odorant receptors of the hawkmoth Manduca sexta. Chem. Senses 34, 305–316. doi: 10.1093/chemse/bjp002
Rojas, V., Jimenez, H., Palma-Millanao, R., Gonzalez-Gonzalez, A., Machuca, J., Godoy, R., et al. (2018). Analysis of the grapevine moth Lobesia botrana antennal transcriptome and expression of odorant-binding and chemosensory proteins. Comp. Biochem. Physiol. Part D 27, 1–12. doi: 10.1016/j.cbd.2018.04.003
Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. PNAS 101, 16653–16658. doi: 10.1073/pnas.0407596101
Steinwender, B., Crowhurst, R. N., Thrimawithana, A. H., and Newcomb, R. D. (2015). Pheromone receptor evolution in the cryptic leafroller species, Ctenopseustis obliquana and C. herana. J. Mol. Evol. 80, 42–56. doi: 10.1007/s00239-014-9650-z
Studer, G., Rempfer, C., Waterhouse, A. M., Gumienny, G., Haas, J., and Schwede, T. (2020). QMEANDisCo - distance constraints applied on model quality estimation. Bioinformatics 36, 1765–1771.
Sun, M., Liu, Y., Walker, W. B., Liu, C., Lin, K., Gu, S., et al. (2013). Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PLoS One 8:e62098. doi: 10.1371/journal.pone.0062098
Walker, W. B., Gonzalez, F., Garczynski, S. F., and Witzgall, P. (2016). The chemosensory receptors of codling moth Cydia pomonella-expression in larvae and adults. Sci. Rep. 6, 23518–23534. doi: 10.1038/srep23518
Wang, G., Vasquez, G. M., Schal, C., Zwiebel, L. J., and Gould, F. (2011). Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol. Biol. 20, 125–133. doi: 10.1111/j.1365-2583.2010.01045.x
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One 5:e8685. doi: 10.1371/journal.pone.0008685
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Wicher, D., Morinaga, S., Halty-deLeon, L., Funk, N., Hansson, B., Touhara, K., et al. (2017). Identification and characterization of the bombykal receptor in the hawkmoth Manduca sexta. J. Exp. Biol. 220, 1781–1786. doi: 10.1242/jeb.154260
Wu, Z. N., Chen, X., Du, Y. J., Zhou, J. J., and ZhuGe, Q. C. (2013). Molecular identification and characterization of the Orco orthologue of Spodoptera litura. Insect Sci. 20, 175–182. doi: 10.1111/j.1744-7917.2011.01483.x
Xu, P., Garczynski, S. F., Atungulu, E., Syed, Z., Choo, Y. M., Vidal, D. M., et al. (2012). Moth sex pheromone receptors and deceitful parapheromones. PLoS One 7:e41653. doi: 10.1371/journal.pone.0041653
Xu, W., Papanicolaou, A., Liu, N. Y., Dong, S. L., and Anderson, A. (2015). Chemosensory receptor genes in the Oriental tobacco budworm Helicoverpa assulta. Insect Mol. Biol. 24, 253–263. doi: 10.1111/imb.12153
Yang, S., Cao, D., Wang, G., and Liu, Y. (2017). Identification of genes involved in chemoreception in Plutella xyllostella by antennal transcriptome analysis. Sci. Rep. 7:11941. doi: 10.1038/s41598-017-11646-7
Yasukochi, Y., Miura, N., Nakano, R., Sahara, K., and Ishikawa, Y. (2011). Sex-linked pheromone receptor genes of the European corn borer, Ostrinia nubilalis, are in tandem arrays. PLoS One 6:e18843. doi: 10.1371/journal.pone.0018843
Zhan, S., Merlin, C., Boore, J. L., and Reppert, S. M. (2011). The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185. doi: 10.1016/j.cell.2011.09.052
Zhang, D.D., Wang, H. L., Schultze, A., Fross, H., Francke, W., Krieger, J., et al. (2016). Receptor for detection of a Type II sex pheromone in the winter moth Operophtera brumata. Sci. Rep. 6:18576. doi: 10.1038/srep18576
Zhang, D. D., and Lofstedt, C. (2013). Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PLoS One 8:e77345. doi: 10.1371/journal.pone.0077345
Zhang, D. D., Zhu, K. Y., and Wang, C. Z. (2010). Sequencing and characterization of six cDNAs putatively encoding three pairs of pheromone receptors in two sibling species, Helicoverpa armigera and Helicoverpa assulta. J. Insect Physiol. 56, 586–593. doi: 10.1016/j.jinsphys.2009.12.002
Zhang, J., Yan, S., Liu, Y., Jacquin-Joly, E., Dong, S., and Wang, G. (2015). Identification and functional characterization of sex pheromone receptors in the common cutworm (Spodoptera litura). Chem. Senses 40, 7–16. doi: 10.1093/chemse/bju052
Zhang, S. X., Xu, S. Q., Kong, L. F., Sima, Y. H., and Cui, W. Z. (2009). Cloning of pbp1, or1 and or3 from wild silkworm Bombyx mandarina and evolutional analysis with the orthologous genes of domesticated silkworm Bombyx mori. Acta Entomol. Sinica 52, 917–922.
Zhang, Y.N., Ma, J. F., Sun, L., Dong, Z. P., Li, Z. Q., Zhu, X. Y., et al. (2016). Molecular identification and sex distribution of two chemosensory receptor families in Athetis lepigone by antennal transcriptome analysis. J. Asia-Pac Entomol. 19, 571–580. doi: 10.1016/j.aspen.2016.05.009
Zhang, Y. N., Jin, J. Y., Jin, R., Xia, Y. H., Zhou, J. J., Deng, J. Y., et al. (2013). Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS One 8:e69715. doi: 10.1371/journal.pone.0069715
Keywords: sex pheromone receptor, structure, signal transduction, Lepidopteran insects, evolution, function
Citation: Yang C, Cheng J, Lin J, Zheng Y, Yu X and Sun J (2022) Corrigendum: Sex Pheromone Receptors of Lepidopteran Insects. Front. Ecol. Evol. 10:900818. doi: 10.3389/fevo.2022.900818
Received: 21 March 2022; Accepted: 23 March 2022;
Published: 14 April 2022.
Edited and reviewed by: Rui Tang, Guangdong Academy of Science (CAS), China
Copyright © 2022 Yang, Cheng, Lin, Zheng, Yu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Yu, eXV4aWFvQHNkdS5lZHUuY24=; Jinpeng Sun, c3VuamlucGVuZ0BzZHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.