- 1The Co-Innovation Center for Sustainable Forestry in Southern China, College of Biology and the Environment, Nanjing Forestry University, Nanjing, China
- 2College of Animal Science and Technology, Zhejiang A&F University, Linan, China
- 3Institute of Qinghai Tibetan Plateau, Southwest Minzu University, Chengdu, China
- 4Department of Agriculture, Zhejiang Open University, Hangzhou, China
The red-crowned crane (Grus japonensis) has been demoted to “vulnerable species” because its populations have apparently stabilized in Japan and Korea. Low variation and genetic drift may cause damage to the nascent recovery of the G. japonensis population. The major histocompatibility complex (MHC) is one of the most polymorphic gene families in the vertebrate genome and can reflect information on the adaptive evolution of endangered species. In this study, variations in MHC I exon 3 of captive G. japonensis in China were assessed and compared with those in cranes from Japan. Forty MHC alleles of 274 base pairs were isolated from 32 individuals from two captive populations in China. There was high variability in the nucleotide and amino acid composition, showing the proportion of polymorphic sites of 18.98 and 32.97%, respectively. Comparative analyses of the Chinese and Japanese populations based on 222 base pair sequences revealed more alleles and higher variation in the Chinese population. The lack of significant geographical differentiation of G. japonensis was supported by the genetic differentiation coefficient (0.04506) between the Chinese and Japanese populations. Positive selection of antigen-binding sites was observed, which contributed to maintaining the diversity of MHC class I genes. Phylogenetic analysis suggested the persistence of trans-species polymorphisms among MHC class I genes in Gruidae species. Our results may contribute to optimizing the management of G. japonensis populations and population recovery of this threatened species.
Introduction
The red-crowned crane (Grus japonensis) is a large wading bird distributed in East Asia and far east Russia (BirdLife International, 2021). This bird has been a long-term and auspicious symbol of East Asian culture (Lee et al., 2020). G. japonensis can be divided into two main populations based on differences in their geographical distribution and life history, namely the migratory population on the Asian continent and the non-migratory population in Japan. Continental populations can be further divided into a subpopulation that winters in coastal intertidal zones and lakes along the middle and lower reaches of the Yangtze River in China and a subpopulation that winters in the demilitarized zone of the central Korean Peninsula (BirdLife International, 2021). With the rapid decline of the G. japonensis population because of the loss and degradation of suitable wetlands through conversion of land for agriculture and industrial development purposes, this crane was listed as an endangered species by the International Union for Conservation of Nature from 2000 to 2020 (Ye et al., 2021; Zhou et al., 2021). To protect birds from anthropogenic threats, governmental and non-governmental actions have been taken, such as enacting protection laws and setting up nature reserves and supplementary food in winter (Gong et al., 2020). Population trends in Japan and subpopulation wintering in Korea have rapidly increased, whereas subpopulation wintering in China continues to decline. With the apparent stabilization of populations in Japan and Korea, G. japonensis was downgraded to the vulnerable level by the International Union for Conservation of Nature in 2021 (BirdLife International, 2021).
Monitoring changes in the genetic diversity of endangered species over time contributes to understanding potential risks associated with genetic drift and reduced genetic variation (Zhang et al., 2015; Williams et al., 2020). Compared with neutral markers, the genetic variation of major histocompatibility complexes (MHC) under selection pressure can provide more accurate information on the adaptive evolution of endangered species (Sutton et al., 2015; He et al., 2017; Manlik et al., 2019; Minias et al., 2021). MHC form one of the most polymorphic gene families in the vertebrate genome, and their protein are involved in triggering immune response. MHC molecules can be divided into class I (MHC I) and class II (MHC II) molecules according to the antigen they can recognize. Exons 2–3 of MHC IA genes and exon 2 of MHC IIB genes encode the peptide-binding regions of MHC protein molecules, respectively (Minias et al., 2019; Williams et al., 2020). The level of variation of these MHC exons is often used in conservation genetic studies of endangered species, such as population differentiation, mate choice, and disease association (Luo et al., 2012; Sutton et al., 2015, 2016; Zhang B. Y. et al., 2020; Zhu et al., 2020).
The genetic diversity of G. japonensis has been widely examined, which is beneficial for population management and conservation (Miura et al., 2013a,b; Sugimoto et al., 2015; Akiyama et al., 2017a,b; Sun et al., 2021). However, most studies have been performed on island populations from Japan, whereas the genetic information between continent and island populations has not been extensively compared. Moreover, the ethology (vocalization patterns) and morphology (coloration and egg size) differ between the continental and island populations (BirdLife International, 2021). Studies focusing on the genetic diversity of the continental population are needed because of the rapid decline of the wintering subpopulation in China. In this study, we assessed the genetic diversity of captive G. japonensis in China by cloning and sequencing of MHC I exon 3. We then compared the genetic information between the Chinese (CHN population) and Japanese (JPN population) populations. This study was conducted to understand the genetic diversity and differentiation of MHC class I genes in G. japonensis populations and contribute to the conservation efforts and recovery of this threatened bird.
Materials and Methods
Sample Collection and DNA Extraction
The Jiangsu-Yancheng Coastal Wetlands Rare Birds National Nature Reserve (JYNNR) is one of the world’s largest winter habitats for red-crowned cranes. In addition to the wild population of red-crowned cranes, there are large captive populations in Jiangsu province, which are important complementary populations of west-flying red-crowned cranes. The captive populations of JYNNR in Yancheng and Nanjing Hong Shan Forest Zoo (NHSFZ) in Nanjing are the two largest populations in Jiangsu Province. A certain number of chicks hatch and are raised in these two locations each year. In this study, 16 pairs of captive G. japonensis (11 pairs from JYNNR and five pairs from NHSFZ) involved in reproduction were sampled. These cranes contributed to the gene pool of captive and even wild populations of the west-flying red-crowned crane. Blood samples were collected during periodic physical examination, labeled, and stored at −80°C. Total genomic DNA was extracted using DNAiso reagent (Takara, Beijing, China) and used for subsequent PCR amplification.
PCR Amplification and Cloning Sequencing
A pair of universal primers (MHC I-E3-F: 5′-TCA GCC CCR TCT CCC TGG TC-3′, MHC I-E3-R: 5′-GTA GAA GCC GTA AGC GCG GCA-3′) to amplify the complete exon 3 of Gruidae MHC class I genes was designed according to five MHC sequences from Grus grus (Sequence ID: Grgr-UA*01, Grgr-UA*02; GenBank accession: MK034099.1, MK034100.1) and Bugeranus carunculatus (Sequence ID: Buca-UA*01, Buca-UA*02, Buca-UA*03; GenBank accession: MK034101.1, MK034102.1, MK034103.1). PCR amplification was performed in a 50 μL volume containing 25 μL Premix Taq™ (Takara, Beijing, China), 5 μL DNA template (∼100 ng), 1.5 μL MHC I-E3-F (1 μmol/L), 1.5 μL MHC I-E3-R (1 μmol/L), and 17 μL sterile water. The PCR program consisted of initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. After 0.5% agarose gel electrophoresis, the target products were excised from the agarose gel and purified using a MiniBEST agarose gel DNA extraction kit (Takara, Beijing, China). The purified PCR products were inserted into the pMD 18-T vector (Takara, Beijing, China) and introduced into Escherichia coli DH 5α competent cells. Positive clones were identified by PCR using the original and universal M13 primers. Ten positive clones per individual were sequenced unidirectionally by Tsingke Biotechnology (Nanjing, China) using the M13 primer.
Statistical Analysis
The MHC sequences of G. japonensis were identified using BLAST for the MHC sequences of G. japonensis and other cranes in GenBank.1 The generated MHC sequences were longer than those (partial exon 3) isolated in previous studies of the genetic diversity of G. japonensis in Japan (GenBank accessions: LC132723.1–LC132738.1) (Akiyama et al., 2017a). Comparative analyses were performed based on 274-bp sequences (complete exon 3) for two populations in China and analyses based on 222-bp sequences for six populations between China and Japan. LaserGene software (version 7.0; DNASTAR, Inc., Madison, WI, United States) was used to align and edit the obtained MHC sequences (Burland, 2000). The variable sites in the nucleotide and amino acid sequences were counted using MEGA v10.0 software (Kumar et al., 2018). The nucleotide diversity (Pi) and overall mean distance of nucleotide sequences were calculated using DnaSP 6.0, and Mega v10.0, respectively (Rozas et al., 2017; Kumar et al., 2018). Antigen-binding sites (ABSs) were predicted from MHC class I molecules from Nipponia nippon (Chen et al., 2015). Non-synonymous substitution (dN) and synonymous substitution (dS) were calculated using MEGA v10.0 (Kumar et al., 2018). Evidence for positive, purifying or neutral selection was assessed using two methods. First, dN/dS with z-test in Mega v10.0 was employed to assess selection of the ABSs, non-ABSs, and entire codons, respectively (Kumar et al., 2018). Second, the selection coefficient of each codon was estimated using the mixed effects model of evolution (MEME), fixed effects likelihood, and fast unconstrained Bayesian approximation on Datamonkey online2 (Weaver et al., 2018). The population differentiation of G. japonensis was evaluated based on the genetic differentiation coefficient (Fst) using Arlequin v3.5 (Excoffier and Lischer, 2010). A phylogenetic tree of the MHC alleles of G. japonensis was reconstructed based on Bayesian inference using the PhyloSuite software package (Zhang D. et al., 2020). The allele sequences from B. carunculatus, G. grus, and Grus canadensis (Sequence ID: Grca-UA*01; GenBank accession: AF033106.1) as outgroups were also used for phylogenetic analysis. The phylogenetic tree was visualized and edited using iTOL v5.0 (Letunic and Bork, 2021).
Results
Allelic Diversity of MHC Class I Genes in Red-Crowned Crane
MHC class I fragments of 450 bp in length were obtained through cloning and sequencing of the PCR products. After comparing the nucleotide sequences of Grgr-UA and Buca-UA, we removed the redundancies and confirmed 40 complete exon 3 sequences of MHC class I that were 274 bp in length. There were 2–7 unique sequences per individual, with an average of four sequences per individual (Supplementary Table 1). This result indicates the existence of four MHC class I genes in red-crowned cranes. As 16 partial exon 3 sequences of Grja-UA from four sampling locations of the JPN population were previously isolated, these sequences were labeled as Grja-UA*17–56 (GenBank accessions: OM772895–OM772934). Fourteen alleles were shared between two sampling locations of the CHN population, whereas 7 and 19 alleles were private to Nanjing and Yancheng, respectively. The exon 3 sequences exhibited high variability in nucleotide composition, as 52 of 274 bases were variable among the 40 alleles from the CHN population (Table 1). No pseudogene sequences were detected. All Grja-UA alleles could be translated into amino acid sequences. The amino acid sequences also showed high variability, with 30 variable amino acids among the 91 residues (Figure 1). To synthetically analyze the allelic diversity of MHC class I genes in red-crowned cranes, nucleotide and amino acid sequences from the CHN population were shortened to sequence length from the JPN population. As a result, some sequences were regarded as identical alleles, and 43 alleles were used for analysis (Supplementary Table 2). Nucleotide and amino acid diversity in the CHN population was higher than that in the JPN population (Table 2). Overall, the allelic diversity of MHC class I genes was high in red-crowned cranes, with an overall proportion of polymorphic nucleotide and amino acid of 18.55 and 31.51%, respectively (Table 2).
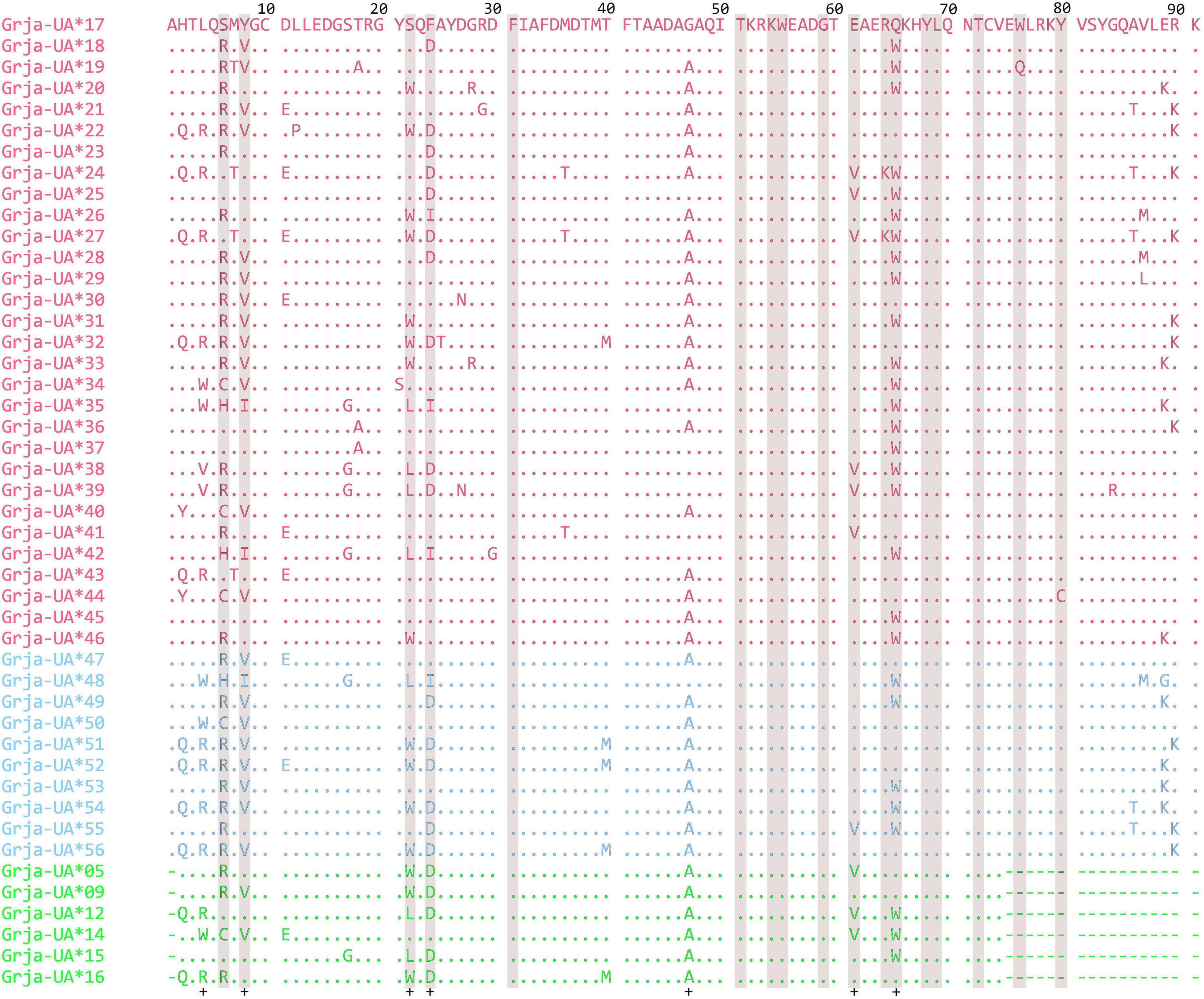
Figure 1. Amino acid sequences of MHC class I exon 3 molecules from Grus japonensis. Dots indicate identity to reference sequence. Dashes indicate missing amino acid residues. Shaded areas present antigen-binding sites (ABSs) predicted from MHC class I molecules of N. nippon (Chen et al., 2015). Crosses present sites deduced to be under positive selection estimated by mixed effects model of evolution (MEME) analysis. Private alleles of the Chinese (CHN) population, private alleles of Japanese (JPN) population, and common alleles are marked as red, green, and blue, respectively.
Genetic Structure of Red-Crowned Crane Populations
Ten alleles were common to the CHN and JPN populations (Supplementary Table 2). Furthermore, Grja-UA*47, Grja-UA*49, Grja-UA*50, and Grja-UA*51 were shared by all six G. japonensis populations (Figure 2). However, the frequencies of the common alleles greatly differed between the CHN and JPN populations. The frequency of Grja-UA*47 ranged from 20% (in the Hongshan population) to 100% (in the Nemuro and Abashiri populations), whereas the frequencies of Grja-UA*49, Grja-UA*50, and Grja-UA*51 ranged from 10% (in the Hongshan population) to 100% (in the Abashiri population) (Figure 2B). Fst analysis revealed slight differentiation between the CHN and JPN populations in MHC class I genes (Fst = 0.04506, P = 0.00000). The minimum and maximum pairwise Fst values were 0.02490 (between the Yancheng and Abashiri populations) and 0.07170 (between the Nanjing and Tokachi populations), respectively. In contrast, the pairwise Fst values were all less than zero within the four sampling locations of the JPN population and 0.01038 between the two sampling locations of the CHN population (Table 3). These results suggest that gene flow occurred internally in the JPN and CHN populations.
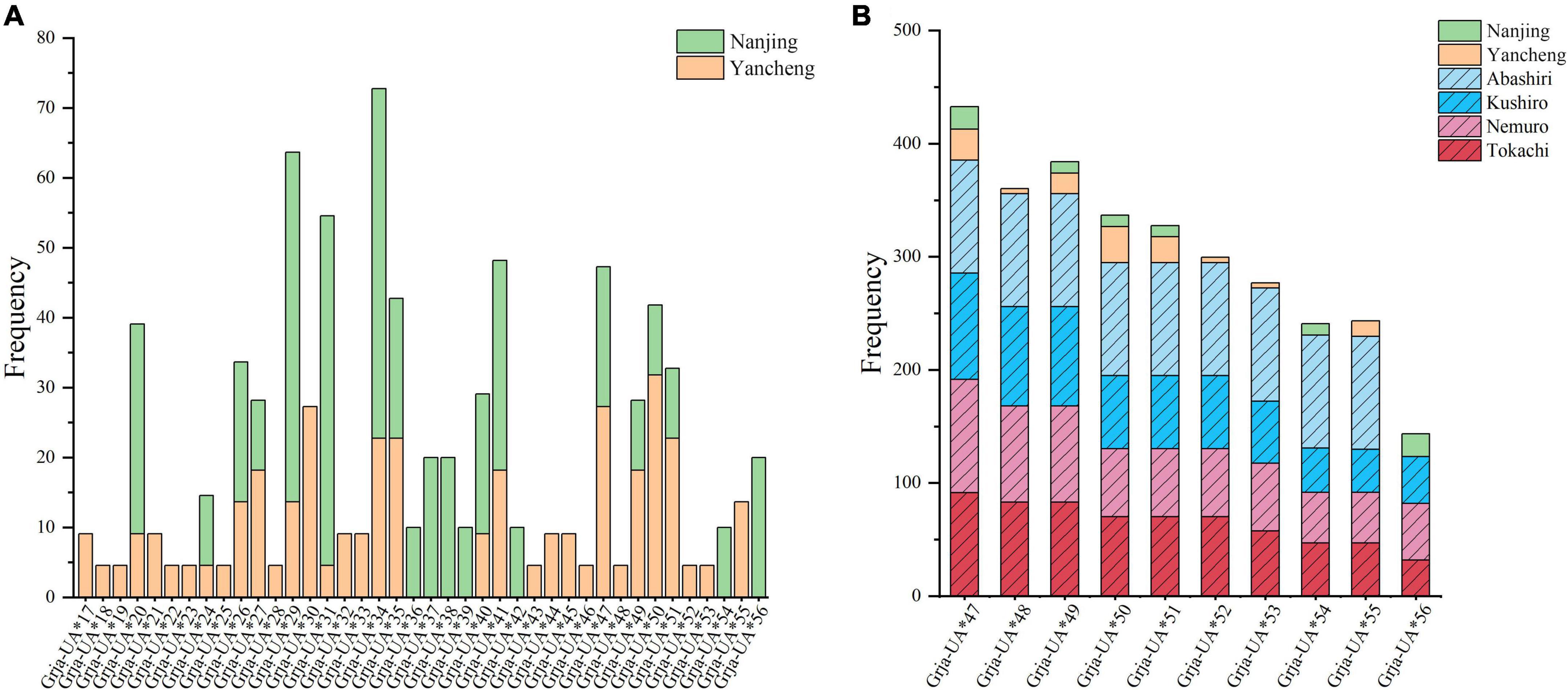
Figure 2. Distributions and frequencies of MHC class I alleles in Grus japonensis populations. (A) Distributions and frequencies of alleles in the Chinese (CHN) population. (B) Distributions and frequencies of common alleles in the CHN and Japanese (JPN) populations. Data on the JPN population (including: Abashiri, Kushiro, Nemuro, Tokachi) was cited from Akiyama et al. (2017a).
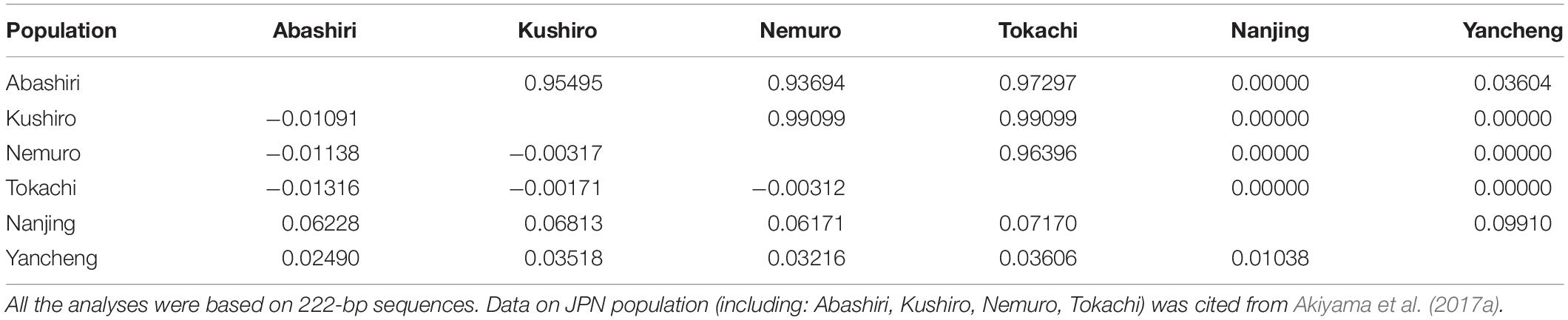
Table 3. Population pairwise Fst values (below the diagonal) and P-values (above the diagonal) between six populations of Grus japonensis.
Selection on MHC Class I Exon 3
Seventeen codon positions were inferred as ABSs compared to the N. nippon MHC sequences (Figure 1). The dN/dS values of the putative ABSs were 1.280, 1.176, and 1.271 for the MHC in the CHN, JPN, and entire populations, respectively, suggesting that these sites are under positive selection (Table 2). Positive selection also occurred at two non-ABS sites (3rd and 46th within the 73 codons), supporting that positive selection has affected Grja-UA. The dN/dS values of non-ABS and all sites were less than 1.0 supporting purifying selection (Table 2). In addition, MEME analysis indicated that three codons (3rd, 27th, and 56th among the 91 codons) were under strong purifying selection (Figure 3).
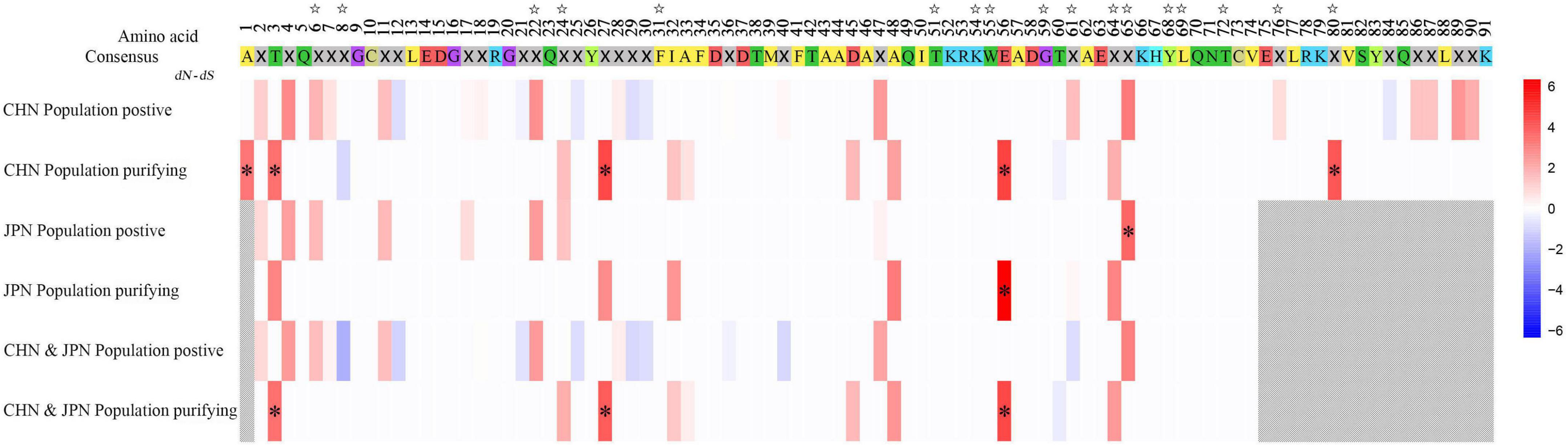
Figure 3. Selection on amino acid residues of MHC class I exon 3 in different populations of Grus japonensis. Stars above the consensus amino acid sequence indicate antigen-binding sites. Crosses in the consensus amino acid sequence indicate variable sites. Asterisks indicate strong selection (| dN–dS| > 10). Gray slashes indicate missing data.
Phylogenetic Relationships Among MHC Class I Alleles
A Bayesian phylogenetic tree was constructed to illustrate the phylogenetic relationships among MHC class I alleles within Gruidae species (Figure 4). Grja-UA*41 and Grja-UA*50 first clustered with Grca-UA*01 and Grgr-UA*01, respectively, indicating that the alleles did not cluster by species. MHC alleles from some species were closer to alleles from other species than to alleles from the same species, indicating an evolutionary pattern of trans-species polymorphism within Gruidae. Additionally, the relationships among the Grja-UA alleles were not congruent with the sampling locations of the CHN and JPN populations. Private alleles did not group into a clade but were mixed with common alleles. Grja-UA*16 clustered with Grja-UA*51, and Grja-UA*35 clustered with Grja-UA*42. However, no clear and overall phylogenetic relationships were identified because of the many parallel clades and low posterior probabilities in the Bayesian tree.
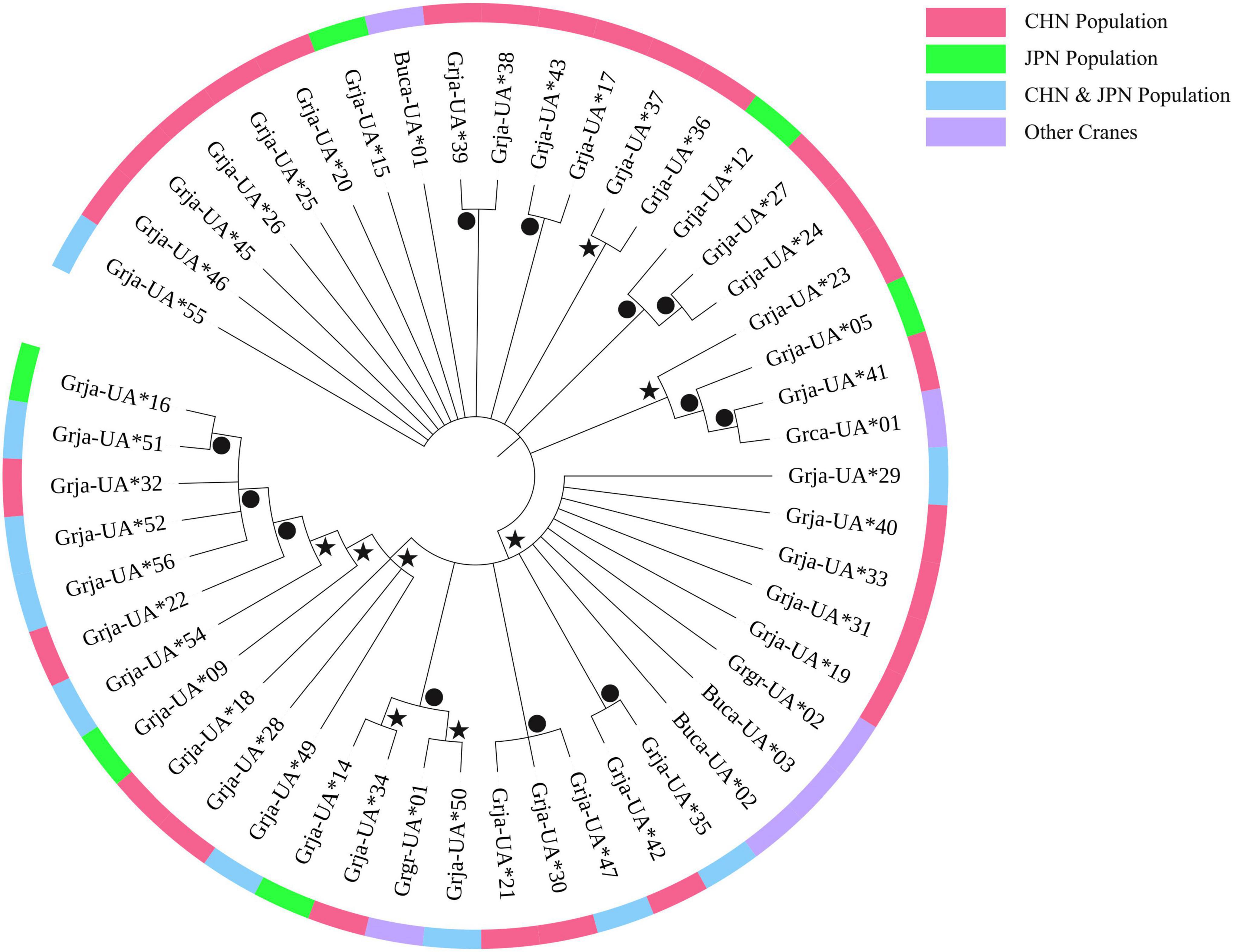
Figure 4. Bayesian phylogenetic tree for MHC class I alleles (222-bp sequences) from Grus japonensis and other three species in Gruidae. Dots indicate posterior probabilities >0.8. A star indicates 0.5 < posterior probabilities <0.8.
Discussion
Genetic Diversity and Differentiation of Red-Crowned Crane
In this study, we examined the MHC class I allelic variation in the threatened red-crowned crane G. japonensis. We identified 40 alleles (complete exon 3), including 52 nucleotide variable sites, from 32 individuals in two CHN subpopulations (Table 1). These results highlight the high diversity of MHC class I genes in the CHN population of red-crowned cranes. However, previous data indicated low genetic variability in MHC class I genes in the JPN population, as only 16 alleles were isolated from 152 individuals (Akiyama et al., 2017a). The difference in genetic variability between the CHN and JPN populations were congruent with previous results based on neutral markers (Hasegawa et al., 1999; Miura et al., 2013b). In addition, shorter amplified fragments in the JPN populations may also prevent the identification of certain alleles. The effects of bottlenecks on genetic variation differ widely among different species. Low genetic variation was generally observed in most bottlenecked species (Babik et al., 2012; Kubota and Watanabe, 2013; Sutton et al., 2013, 2015; Li et al., 2017; Saka et al., 2018; Lan et al., 2019; Arauco-Shapiro et al., 2020), whereas high variation was also detected (Galaverni et al., 2013; Newhouse and Balakrishnan, 2015; Fulton et al., 2017; MacDougall-Shackleton, 2020; Cruz-Lopez et al., 2021). Multiple MHC copies may be a major cause of this high variation (Wan et al., 2011; Stervander et al., 2020; He et al., 2021). However, the copy number of MHC class I genes in G. japonensis has not been precisely defined because the number of exon 3 sequences varies greatly among individuals. It remains unclear whether multiple gene copies or multiple sequences at a certain locus lead to high diversity.
Evolutionary significant units and management units have been developed to better protect endangered species populations on separate evolutionary trajectories (Moritz, 1994; Zhu et al., 2013; Carlson et al., 2016). The definition of evolutionarily significant units highlights the high distinctiveness of separate populations, specifically the monophyly for mitochondrial alleles as well as the significant divergence of frequency for nuclear alleles. In practical situations, management units have often been defined and used and only revealed the significant divergence of allele frequencies at mitochondrial or nuclear loci but neglected the phylogenetic distinctiveness of the alleles (Hoglund et al., 2011). Although there were differences in the ethology and morphology of red-crowned cranes, MHC allele frequencies (Fst = 0.04506, P = 0.00000) and evolutionary relationship of haplotypes did not support the separate evolution of the continental and island populations (Hasegawa et al., 1999; Miura et al., 2013b). Nevertheless, a larger gene pool in the continental population can be inferred because of the higher diversity of MHC and other neutral markers (Hasegawa et al., 1999; Miura et al., 2013b; Akiyama et al., 2017a). The low genetic variation in the JPN population can be increased via translocations by introducing continental individuals. According to recent reports, continental individuals flew to Japan and may have enriched the low gene pool of the JPN population (Akiyama et al., 2017a; Kawasaki et al., 2022). Recovering G. japonensis may be more difficult, as the CHN population with high genetic variation is still decreasing.
Historical Balancing Selection and Trans-Species Polymorphism
Nucleotide sites under positive selection can accumulate more non-synonymous sites, which can alter the encoded amino acid sequence, structure, and function of the protein. The ABS region, subjected to positive selection, is a well-known feature of MHC alleles (Luo et al., 2012; Zeng et al., 2016). We found significant evidence for positive selection of the ABS region of MHC class I alleles in G. japonensis, as five of seven positively selected sites were ABSs and the dN/dS value for the ABS region was more than 1 (Figure 1 and Table 2). This indicates that balancing selection for immunological responses to pathogens is involved in generating and maintaining MHC diversity in G. japonensis undergoing a bottleneck. However, multiple MHC gene copies were included in the analysis because of the difficulty of locus assignment, which may artificially inflate MHC variation and bias our conclusion (Knafler et al., 2014).
Trans-species polymorphisms are another well-known feature of MHC alleles (Kohyama et al., 2015; Qin et al., 2021). The phylogenetic tree showed that the allelic lineage of cranes did not form monophyletic groups among species, suggesting that Grja-UA alleles are involved in several instances of trans-species polymorphism (Figure 4). This may be because allelic lineages present in a common ancestor were retained by balancing selection, even after species-level diversification (Akiyama et al., 2017a; Nishita et al., 2018). But sharing of polymorphism between unrelated or distantly related species may be owing to convergent evolution rather than maintenance of ancestral sequence (Yeager and Hughes, 1999). The analyzed G. japonensis samples were restricted to populations in Japan and captive populations in China. However, wild populations of this species are distributed over a much wider area from Far East Russia to Korea and northeast and east China. Only the three species were used in phylogenetic analysis. In further studies, novel MHC class I sequences could be detected in other G. japonensis populations and in Gruidae species, which will more precisely reveal the relationships between species divergence and evolution of MHC class I genes among Gruidae species.
Conclusion
In this study, 40 MHC class I exon 3 alleles with 274 bp were isolated from 32 individuals from two captive populations in China. High polymorphism was found at both the nucleotide and amino acid levels. Comparative analyses between the CHN and JPN populations based on 222 bp sequences showed more alleles and higher variation in the former. The low Fst value (Fst = 0.04506) suggests no significant geographical differentiation of G. japonensis between the CHN and JPN populations. Low genetic variation in the JPN population may be increased via translocations by introducing individuals from the CHN population. Well-known features of MHC class I genes are found in G. japonensis, such as positive selection on ABSs and trans-species polymorphism within Gruidae species. Our study suggests that genuine rejuvenescence of G. japonensis requires conservation of the CHN population.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/genbank/, OM772895–OM772934.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of Nanjing Forestry University.
Author Contributions
HyL conceived the study. HyL and NX acquired the funds. NX and CS conducted the experiments. WY and KH carried out the bioinformatics analysis. NX drafted the manuscript. YZ, HL, and CL reviewed and revised the manuscript. All authors approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31800453), the Natural Science Foundation of Jiangsu Province (BK20160927), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX21_0916).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dawei Liu (JYNNR), Yongqiang Zhao (JYNNR) and Rong Chen (NHSFZ) for collecting samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.898581/full#supplementary-material
Footnotes
References
Akiyama, T., Kohyama, T. I., Nishida, C., Onuma, M., Momose, K., and Masuda, R. (2017a). Genetic variation of major histocompatibility complex genes in the endangered red-crowned crane. Immunogenetics 69, 451–462. doi: 10.1007/s00251-017-0994-6
Akiyama, T., Momose, K., Onuma, M., Matsumoto, F., and Masuda, R. (2017b). Low genetic variation of red-crowned cranes on Hokkaido Island, Japan, over the hundred years. Zool. Sci. 34, 211–216. doi: 10.2108/zs160194
Arauco-Shapiro, G., Schumacher, K. I., Boersma, D., and Bouzat, J. L. (2020). The role of demographic history and selection in shaping genetic diversity of the Galapagos penguin (Spheniscus mendiculus). PLoS One 15:e0226439. doi: 10.1371/journal.pone.0226439
Babik, W., Kawalko, A., Wojcik, J. M., and Radwan, J. (2012). Low major histocompatibility complex class I (MHC I) variation in the European bison (Bison bonasus). J. Heredity 103, 349–359. doi: 10.1093/jhered/ess005
BirdLife International (2021). Grus Japonensis. The IUCN Red List of Threatened Species 2021: e.T22692167A175614850. Available online at: https://dx.doi.org/10.2305/IUCN.UK.2021-3.RLTS.T22692167A175614850.en. Accessed [March 17, 2022].
Burland, T. G. (2000). DNASTAR’s lasergene sequence analysis software. Methods Mol. Biol. 132, 71–91. doi: 10.1385/1-59259-192-2:71
Carlson, E., MacDonald, A. J., Adamack, A., McGrath, T., Doucette, L. I., Osborne, W. S., et al. (2016). How many conservation units are there for the endangered grassland earless dragons? Conserv. Genet. 17, 761–774. doi: 10.1007/s10592-016-0819-4
Chen, L. C., Lan, H., Sun, L., Deng, Y. L., Tang, K. Y., and Wan, Q. H. (2015). Genomic organization of the crested ibis MHC provides new insight into ancestral avian MHC structure. Sci. Rep. 5, 7963. doi: 10.1038/srep07963
Cruz-Lopez, M., Fernandez, G., Hipperson, H., Palacios, E., Cavitt, J., Galindo-Espinosa, D., et al. (2021). Allelic diversity and patterns of selection at the major histocompatibility complex class I and II loci in a threatened shorebird, the Snowy Plover (Charadrius nivosus). BMC Evol. Biol. 20:114. doi: 10.1186/s12862-020-01676-7
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fulton, J. E., Berres, M. E., Kantanen, J., and Honkatukia, M. (2017). MHC-B variability within the Finnish Landrace chicken conservation program. Poultry Sci. 96, 3026–3030. doi: 10.3382/ps/pex102
Galaverni, M., Caniglia, R., Fabbri, E., Lapalombella, S., and Randi, E. (2013). MHC variability in an isolated wolf population in Italy. J. Heredity 104, 601–612. doi: 10.1093/jhered/est045
Gong, Y. Z., Bi, X., and Wu, J. (2020). Willingness to pay for the conservation of the endangered red-crowned crane in China: roles of conservation attitudes and income. For. Policy Econ. 120:102296. doi: 10.1016/j.forpol.2020.102296
Hasegawa, O., Takada, S., Yoshida, M., and Abe, S. (1999). Variation of mitochondrial control region sequences in three cranes species, the red-crowned crane Grus japonensis, the common crane Grus grus and the hooded crane Grus monacha. Zool. Sci. 16, 685–692. doi: 10.2108/zsj.16.685
He, K., Liu, H. Y., Ge, Y. F., Wu, S. Y., and Wan, Q. H. (2017). Historical gene flow and profound spatial genetic structure among golden pheasant populations suggested by multi-locus analysis. Mol. Phylogenet. Evol. 110, 93–103. doi: 10.1016/j.ympev.2017.03.009
He, K., Minias, P., and Dunn, P. O. (2021). Long-read genome assemblies reveal extraordinary variation in the number and structure of MHC loci in birds. Genome Biol. Evol. 13:evaa270. doi: 10.1093/gbe/evaa270
Hoglund, J., Larsson, J. K., Corrales, C., Santafe, G., Baines, D., and Segelbacher, G. (2011). Genetic structure among black grouse in Britain: implications for designing conservation units. Anim. Conserv. 14, 400–408.
Kawasaki, E., Hasebe, M., Hwang, J. H., Kim, E. Y., Lee, K., Momose, K., et al. (2022). Origin of a pair of red-crowned cranes (Grus japonensis) found in Sarobetsu Wetland, northwestern Hokkaido, Japan: a possible crossbreeding between the island and the mainland population. J. Vet. Med. Sci. 84, 233–237. doi: 10.1292/jvms.21-0500
Knafler, G. J., Fidler, A., Jamieson, I. G., and Robertson, B. C. (2014). Evidence for multiple MHC class II β loci in New Zealand’s critically endangered kakapo, Strigops habroptilus. Immunogenetics 66, 115–121. doi: 10.1007/s00251-013-0750-5
Kohyama, T. I., Akiyama, T., Nishida, C., Takami, K., Onuma, M., Momose, K., et al. (2015). Isolation and characterization of major histocompatibility complex class II B genes in cranes. Immunogenetics 67, 705–710. doi: 10.1007/s00251-015-0874-x
Kubota, H., and Watanabe, K. (2013). Loss of genetic diversity at an MHC locus in the endangered Tokyo bitterling Tanakia tanago (Teleostei: Cyprinidae). Zool. Sci. 30, 1092–1101. doi: 10.2108/zsj.30.1092
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lan, H., Zhou, T., Wan, Q. H., and Fang, S. G. (2019). Genetic diversity and differentiation at structurally varying MHC haplotypes and microsatellites in bottlenecked populations of endangered crested ibis. Cells 8:377. doi: 10.3390/cells8040377
Lee, H., Kim, J., Weber, J. A., Chung, O., Cho, Y. S., Jho, S., et al. (2020). Whole genome analysis of the red-crowned crane provides insight into avian longevity. Mol. Cells 43, 86–95. doi: 10.14348/molcells.2019.0190
Letunic, I., and Bork, P. (2021). Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, D., Sun, K. P., Zhao, Y. J., Lin, A. Q., Li, S., Jiang, Y. L., et al. (2017). Polymorphism in the major histocompatibility complex (MHC class II B) genes of the Rufous-backed Bunting (Emberiza jankowskii). PeerJ 5:e2917. doi: 10.7717/peerj.2917
Luo, M. F., Pan, H. J., Liu, Z. J., and Li, M. (2012). Balancing selection and genetic drift at major histocompatibility complex class II genes in isolated populations of golden snub-nosed monkey (Rhinopithecus roxellana). BMC Evol. Biol. 12:207. doi: 10.1186/1471-2148-12-207
MacDougall-Shackleton, E. A. (2020). Many loci make light work: high individual diversity despite low population diversity and random mating at class I MHC in a critically endangered island songbird. Mol. Ecol. 29, 3575–3577. doi: 10.1111/mec.15578
Manlik, O., Krutzen, M., Kopps, A. M., Mann, J., Bejder, L., Allen, S. J., et al. (2019). Is MHC diversity a better marker for conservation than neutral genetic diversity? A case study of two contrasting dolphin populations. Ecol. Evol. 9, 6986–6998. doi: 10.1002/ece3.5265
Minias, P., Janiszewska, A., Pikus, E., Zadworny, T., and Anderwald, D. (2021). MHC reflects fine-scale habitat structure in white-tailed eagles, Haliaeetus albicilla. Journal of Heredity. 112, 335–345. doi: 10.1093/jhered/esab026
Minias, P., Pikus, E., and Anderwald, D. (2019). Allelic diversity and selection at the MHC class I and class II in a bottlenecked bird of prey. BMC Evol. Biol. 19:2. doi: 10.1186/s12862-018-1338-3
Miura, Y., Shiomi, A., Shiraishi, J., Makita, K., Asakawa, M., Kitazawa, T., et al. (2013a). Large-scale survey of mitochondrial D-Loop of the red-crowned crane Grus japonensis in Hokkaido, Japan by convenient genotyping method. J. Vet. Med. Sci. 75, 43–47. doi: 10.1292/jvms.12-0193
Miura, Y., Shiraishi, J., Shiomi, A., Kitazawa, T., Hiraga, T., Matsumoto, F., et al. (2013b). Origin of three red-crowned cranes Grus japonensis found in northeast Honshu and west Hokkaido, Japan, from 2008 to 2012. J. Vet. Med. Sci. 75, 1241–1244. doi: 10.1292/jvms.13-0090
Moritz, C. (1994). Defining ‘evolutionarily significant units’ for conservation. Trends Ecol. Evol. 9, 373–375. doi: 10.1016/0169-5347(94)90057-4
Newhouse, D. J., and Balakrishnan, C. N. (2015). High major histocompatibility complex class I polymorphism despite bottlenecks in wild and domesticated populations of the zebra finch (Taeniopygia guttata). BMC Evol. Biol. 15:265. doi: 10.1186/s12862-015-0546-3
Nishita, Y., Spassov, N., Peeva, S., Raichev, E. G., Kaneko, Y., and Masuda, R. (2018). Genetic diversity of MHC class II DRB alleles in the marbled polecat, Vormela peregusna, in Bulgaria. Ethol. Ecol. Evol. 31, 59–72. doi: 10.1080/03949370.2018.1486887
Qin, S. D., Dunn, P. O., Yang, Y., Liu, H. Y., and He, K. (2021). Polymorphism and varying selection within the MHC class I of four Anas species. Immunogenetics 73, 395–404. doi: 10.1007/s00251-021-01222-9
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Saka, T., Nishita, Y., and Masuda, R. (2018). Low genetic variation in the MHC class II DRB gene and MHC-linked microsatellites in endangered island populations of the leopard cat (Prionailurus bengalensis) in Japan. Immunogenetics 70, 115–124. doi: 10.1007/s00251-017-1020-8
Stervander, M., Dierickx, E. G., Thorley, J., Brooke, M. D., and Westerdahl, H. (2020). High MHC gene copy number maintains diversity despite homozygosity in a critically endangered single-island endemic bird, but no evidence of MHC-based mate choice. Mol. Ecol. 29, 3578–3592. doi: 10.1111/mec.15471
Sugimoto, T., Hasegawa, O., Azuma, N., Masatomi, H., Sato, F., Matsumoto, F., et al. (2015). Genetic structure of the endangered red-crowned cranes in Hokkaido, Japan and conservation implications. Conserv. Genet. 16, 1395–1401. doi: 10.1007/s10592-015-0748-7
Sun, C. H., Liu, H. Y., Xu, P., and Lu, C. H. (2021). Genetic diversity of wild wintering red-crowned crane (Grus japonensis) by microsatellite markers and mitochondrial Cyt B gene sequence in the Yancheng reserve. Anim. Biotechnol. 32, 531–536. doi: 10.1080/10495398.2020.1725538
Sutton, J. T., Castro, I., Robertson, B. C., Tompkins, D. M., Stanton, J. A. L., and Jamieson, I. G. (2016). MHC genetic diversity and avian malaria prevalence in Mokoia Island saddlebacks. New Zealand J. Ecol. 40, 351–360. doi: 10.20417/nzjecol.40.34
Sutton, J. T., Robertson, B. C., and Jamieson, I. G. (2015). MHC variation reflects the bottleneck histories of New Zealand passerines. Mol. Ecol. 24, 362–373. doi: 10.1111/mec.13039
Sutton, J. T., Robertson, B. C., Grueber, C. E., Stanton, J. A. L., and Jamieson, I. G. (2013). Characterization of MHC class II B polymorphism in bottlenecked New Zealand saddlebacks reveals low levels of genetic diversity. Immunogenetics 65, 619–633. doi: 10.1007/s00251-013-0708-7
Wan, Q. H., Zhang, P., Ni, X. W., Wu, H. L., Chen, Y. Y., Kuang, Y. Y., et al. (2011). A novel HURRAH protocol reveals high numbers of monomorphic MHC class II loci and two asymmetric multi-locus haplotypes in the Pere David’s deer. PLoS One 6:e14518. doi: 10.1371/journal.pone.0014518
Weaver, S., Shank, S. D., Spielman, S. J., Li, M., Muse, S. V., and Pond, S. L. K. (2018). Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 35, 773–777. doi: 10.1093/molbev/msx335
Williams, S. T., Haas, C. A., and Taylor, S. S. (2020). Depauperate major histocompatibility complex variation in the endangered reticulated flatwoods salamander (Ambystoma bishopi). Immunogenetics 72, 263–274. doi: 10.1007/s00251-020-01160-y
Ye, W. T., Xu, W., Xu, N., Chen, R., Lu, C. H., and Liu, H. Y. (2021). Comprehensive transcriptome characterization of Grus japonensis using PacBio SMRT and Illumina sequencing. Sci. Rep. 11:23927. doi: 10.1038/s41598-021-03474-7
Yeager, M., and Hughes, A. L. (1999). Evolution of the mammalian MHC: natural selection, recombination, and convergent evolution. Immunol. Rev. 167, 45–58. doi: 10.1111/j.1600-065x.1999.tb01381.x
Zeng, Q. Q., He, K., Sun, D. D., Ma, M. Y., Ge, Y. F., Fang, S. G., et al. (2016). Balancing selection and recombination as evolutionary forces caused population genetic variations in golden pheasant MHC class I genes. BMC Evol. Biol. 16:42. doi: 10.1186/s12862-016-0609-0
Zhang, B. Y., Hu, H. Y., Song, C. M., Huang, K., Dunn, D. W., Yang, X., et al. (2020a). MHC-based mate choice in wild golden snub-nosed monkeys. Front. Genet. 11:609414. doi: 10.3389/fgene.2020.609414
Zhang, D., Gao, F. L., Jakovlic, I., Zou, H., Zhang, J., Li, W. X., et al. (2020b). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, W., Luo, Z. H., Zhao, M., and Wu, H. (2015). High genetic diversity in the endangered and narrowly distributed amphibian species Leptobrachium leishanense. Integr. Zool. 10, 465–481. doi: 10.1111/1749-4877.12142
Zhou, D. Q., Zhang, H. N., Zhang, X. S., Zhang, W. W., Zhang, T. T., and Lu, C. H. (2021). Habitat changes in the most important stopover sites for the endangered red-crowned crane in China: a large-scale study. Environ. Sci. Pollut. Res. 28, 54719–54727. doi: 10.1007/s11356-021-14488-z
Zhu, Y., Grueber, C., Li, Y. D., He, M., Hu, L., He, K., et al. (2020). MHC-associated Baylisascaris schroederi load informs the giant panda reintroduction program. Int. J. Parasitol. Parasites Wildlife 12, 113–120. doi: 10.1016/j.ijppaw.2020.05.010
Keywords: Grus japonensis, major histocompatibility complex, genetic structure, balancing selection, trans-species polymorphism
Citation: Xu N, Ye W, Sun C, He K, Zhu Y, Lan H, Lu C and Liu H (2022) Genetic Diversity and Differentiation of MHC Class I Genes in Red-Crowned Crane Populations. Front. Ecol. Evol. 10:898581. doi: 10.3389/fevo.2022.898581
Received: 17 March 2022; Accepted: 25 April 2022;
Published: 25 May 2022.
Edited by:
Thales Renato Ochotorena De Freitas, Federal University of Rio Grande do Sul, BrazilReviewed by:
Nicolas Dussex, Swedish Museum of Natural History, SwedenElena A. Mudrik, Vavilov Institute of General Genetics (RAS), Russia
Copyright © 2022 Xu, Ye, Sun, He, Zhu, Lan, Lu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyi Liu, aG9uZ3lpX2xpdUBuamZ1LmVkdS5jbg==
†These authors have contributed equally to this work
 Nan Xu1†
Nan Xu1† Chenghe Sun
Chenghe Sun Ke He
Ke He Changhu Lu
Changhu Lu Hongyi Liu
Hongyi Liu