- 1Guangzhou Key Laboratory of Forensic Multi-Omics for Precision Identification, School of Forensic Medicine, Southern Medical University, Guangzhou, China
- 2Clinical Research Center of Shaanxi Province for Dental and Maxillofacial Diseases, College of Stomatology, Xi’an Jiaotong University, Xi’an, China
The utilization of the ancestry informative markers to disclose the ancestral composition of a certain population and explore the genetic affinities between diverse populations is beneficial to inferring the biogeographic ancestry of unknown individuals and assisting in case detection, as well as avoiding the impacts of population stratification during genome-wide association analysis studies. In the present study, we applied an in-house ancestry informative deletion/insertion polymorphic multiplex amplification system to investigate the ancestral compositions of the Beijing Han population and analyze the genetic relationships between the Beijing Han population and 31 global reference populations. The results demonstrated that 32 loci of this self-developed panel containing 39 loci significantly contributed to the inference of genetic information for the Beijing Han population. The results of multiple population genetics statistical analyses indicated that the ancestral component and genetic architecture of the Beijing Han population were analogous to the reference East Asian populations, and that the Beijing Han population was genetically close to the reference East Asian populations.
Introduction
In recent years, ancestry informative markers (AIMs) have caught the interest of scholars in population genetics, forensic genetics, and other fields. The analysis of AIMs enables the inference of possible biogeographic ancestral origin in unknown individual and the assessment of percentage composition of ancestral information component in an admixed population or individual (Enoch et al., 2006; Kersbergen et al., 2009; Wang H. Y. et al., 2021). Correctly classifying unknown individuals into appropriate populations by identifying the population structure is a very effective means of inferring biogeographic ancestry and characterizing the phenotype, which can provide directional clue for case detection and narrow the scope of criminal investigation, as well as rectify the impact of population stratification on genome-wide association studies (GWAS) (Galanter et al., 2012; Freire-Aradas et al., 2014; Qin et al., 2014; Chen et al., 2021).
The allele frequencies of single nucleotide polymorphisms (SNPs), deletion/insertion polymorphisms (DIPs), and other genetic markers have significant population differences and show different distribution characteristics in different ethnic groups as well as geographic regions, which means that they can be used as AIMs (Guo et al., 2021; Jin et al., 2022). Presently, AIM-SNPs are widely used in the field of ancestry inference, but there are still limitations (Yahya et al., 2020). Compared with SNPs, the DIPs exhibit fragment length polymorphisms, and the genotyping data are easy to detect and analyze, which are suitable for promotion in primary laboratories. DIPs are valuable genetic markers for forensic science and have an important role in forensic individual identification, parentage testing, and biogeographic ancestry inference (Pereira et al., 2012; Romanini et al., 2015; Xie et al., 2020; Fan et al., 2021).
The Han Chinese is the main nationality of China and the most populous ethnic group in the world. The distributions of the Han nationality in China were characterized by dense regions in the east and sparse regions in the west. Beijing is situated in the northern part of the North China Plain, adjoining Tianjin city in the east and Hebei province in the rest. It is the capital of China, a world-famous ancient metropolis and a modern international city. Beijing has a long history and is one of the four ancient capitals of China, with many famous heritage sites. The discovery of the Peking Man site in the Zhoukoudian area of Beijing is crucial to the investigation of the history of ancient human evolution and provides reliable evidence for the exploration of human origins1.
The exploration of the genetic background of Han Chinese in Beijing based on ancestry informative deletion/insertion polymorphisms (AIDIPs) is of great significance. However, there were few studies to explore the genetic background of the Beijing Han population by a set of AIDIPs. Our laboratory pre-screened 39 highly polymorphic AIDIPs suitable for the Chinese populations and successfully constructed a multiplex amplification system based on the platform of capillary electrophoresis, which could predict ancestry components for individuals from three diverse biogeographic zones (East Asia, Europe, and Africa) (Lan et al., 2019; Zhang X. et al., 2021). In this study, a sample of 299 individuals of Beijing Han population was gathered to further evaluate the efficacy of the panel for forensic applications, and the genetic relationships between the Beijing Han and 31 worldwide reference populations were revealed using a variety of statistical analyses to probe the genetic structures of the Beijing Han population.
Materials and Methods
Sample Information and Reference Populations
Bloodstain samples were gathered from 299 unrelated healthy Han Chinese individuals living in Beijing, China, besides, all volunteers signed the informed consent documents before sample collections and declared that they were unrelated within three generations. The study was authorized by the ethics committee of Xi’an Jiaotong University (No. XJTULAC201), and the sample collection procedure was in accordance with the requirements of medical research. The 26 intercontinental populations from the International 1,000 Genomes Project Phase III and five previously reported populations were used as reference populations to investigate the genetic relationships between the Beijing Han population and these 31 reference populations (Auton et al., 2015; Jin et al., 2020; Xie et al., 2020; Zhang X. et al., 2021). Among these 39 AIDIP loci, genetic data for locus rs3034941 were not obtainable from the 1,000 Genomes Project Phase III, so this locus was excluded from the subsequent interpopulation genetic comparison study. The detailed information including the abbreviations and geographical positions of these continental populations was given in Supplementary Table 1.
PCR Amplification and AIDIP Genotyping
Without extraction of genomic DNA, bloodstain samples of 299 individuals from the Beijing Han population were directly amplified through the multiplex amplification system constructed in our lab (Lan et al., 2019). The multiplex PCR amplification of 39 AIDIPs was conducted in the Beijing Han population on the GeneAmp PCR System 9700 Thermal Cycler (Applied Biosystems, Foster City, CA, United States) with final reaction volume and thermal cycling condition in line with former research (Lan et al., 2019). The ABI 3500xL Genetic Analyzer (Applied Biosystem, Foster City, CA, United States) was utilized to isolate and analyze the above PCR products by capillary electrophoresis. Subsequently, the 39 AIDIPs were genotyped by the GeneMapper ID-X v1.5 program (Applied Biosystem, Foster City, CA, United States). Throughout the whole experiment, the DNA F312 and nuclear-free water were utilized as positive control and negative control, respectively.
Statistical Analysis
STRAF v1.0.5 tool was employed to carry out the Hardy-Weinberg equilibrium (HWE) tests for 39 AIDIPs, linkage disequilibrium (LD) analyses for pairwise AIDIPs, allele frequencies of 39 AIDIPs, and forensic parameters including the polymorphism information content (PIC), match probability (MP), power of discrimination (PD), probability of exclusion (PE), typical paternity index (TPI), expected heterozygosity (He), observed heterozygosity (Ho) of 39 AIDIPs in the Beijing Han population (Gouy and Zieger, 2017). Corrections for HWE and LD tests were performed according to Bonferroni statistical tests, and the obtained results of allele frequencies and forensic parameters of 39 AIDIPs were visualized in the circle diagram through R v4.1.2 software (Goeman and Solari, 2014). Based on the 38 overlapped AIDIPs, the locus-by-locus analyses of molecular variance (AMOVA) between the Beijing Han population and 31 reference populations, and pairwise fixation index (FST) values of the 32 worldwide populations were worked out through the Arlequin software v3.5 (Excoffier and Lischer, 2010). The Nei’s genetic distances (DA distances) of pairwise populations derived from the 38 overlapped AIDIPs were assessed by the DISPAN program and the phylogenetic tree was constructed by MEGA software v10.2.5 based on the obtained DA distances (Tateno et al., 1982; Nei et al., 1983; Kumar et al., 2018). The heat maps of insertion allele frequencies, pairwise FST values and DA distances of 38 AIDIPs in the 32 populations were constructed by R v4.1.2 software and TBtools software, respectively (Chen et al., 2020). Furthermore, principal component analysis (PCA), the cos2 values of 38 overlapped AIDIPs, and multidimensional scaling (MDS) analysis derived from pairwise FST values between the Beijing Han population and 31 reference populations were also carried out by R v4.1.2 software. STRUCTURE program v2.3.4 was applied to conduct population genetic structure analysis (Evanno et al., 2005). The number of iterations and the hypothetical ancestry cluster (K) values were set at 15, and 2 to 7, respectively. The results obtained from STRUCTURE software v2.3.4 were uploaded to the online tool Structure Harvester to measure the optimal K value (Earl and Vonholdt, 2012). The CLUMPP software v1.1.2 was used to assess the average Q-matrices for the above 15 iterations (Jakobsson and Rosenberg, 2007). And the evaluated ancestry compositions of 4,391 individuals and 32 populations were visualized by the Distruct software v1.1, respectively (Rosenberg, 2004). The cross-validation estimation success ratios and informativeness for assignment (In) values were carried out by the online website, which was named snipper 2.5 app suite2. Specifically, the population-specific divergence (PSD) value obtained from the snipper 2.5 app suite multiplied by 0.693 is the In value (Rosenberg et al., 2003, 2005).
Results
HWE Tests and LD Analyses of 39 AIDIPs in the Beijing Han Population
The P values of HWE tests of 39 AIDIPs in the Beijing Han population were presented in Supplementary Table 2. After the Bonferroni correction (P = 0.05/39 = 0.0013), no significant deviation from HWE was obtained in all the 39 AIDIPs in the Beijing Han population. The results of pairwise LD analyses of 39 AIDIPs in the Beijing Han population were manifested in Supplementary Table 3. After applying the multiple corrections (P = 0.05/741 = 0.00006748), all pairwise loci apart from one pair (rs3033760 and rs36038238) were observed to conform to linkage equilibrium in the Beijing Han population.
Allele Frequencies and Forensic Parameters of 39 AIDIPs in the Beijing Han Population
The allele frequencies and forensic parameters of 39 AIDIPs in the Beijing Han population were exhibited in Supplementary Table 2 and visualized by the circle diagram in Figure 1. As shown in Figure 1, the outermost circle represented the chromosomes on which these 39 AIDIPs were distributed, and the inner circles from the outside to the inside represented heat maps of insertion allele frequencies, deletion allele frequencies, scatter plots of PIC values (red points for values higher than 0.25 and black points for values lower than 0.25), histograms of MP, PD, PE, and TPI values, and network plots of He and Ho values, respectively. In the Beijing Han population, the insertion allele frequencies of 39 AIDIPs varied from 0.0017 (rs5896844) to 0.9799 (rs146391383), and the deletion allele frequencies of 39 AIDIPs ranged from 0.0201 (rs146391383) to 0.9983 (rs5896844). The values of PIC, PM, PD, PE, TPI, He and Ho of 39 AIDIPs in the Beijing Han population spanned from 0.0033 (rs5896844) to 0.3747 (rs3839348); 0.3599 (rs3839348) to 0.9933 (rs5896844); 0.0067 (rs5896844) to 0.6401 (rs3839348); 0.00001 (rs5896844) to 0.2041 (rs5788207); 0.5017 (rs5896844) to 1.0382 (rs5788207); 0.0033 (rs5896844) to 0.5003 (rs3839348); and 0.0033 (rs5896844) to 0.5184 (rs5788207), respectively. Since a pair of loci, rs3033760 and rs36038238, couldn’t be considered independent in the Beijing Han population based on the outcome of the LD analyses, 38 AIDIPs (except for the rs3033760 loci) were utilized to calculate the cumulative power of discrimination (CPD) and the combined probability of exclusion (CPE) values according to relevant formulae. The obtained CPD and CPE values of 38 AIDIPs in the Beijing Han population were 0.999999999961369 and 0.9747, respectively.
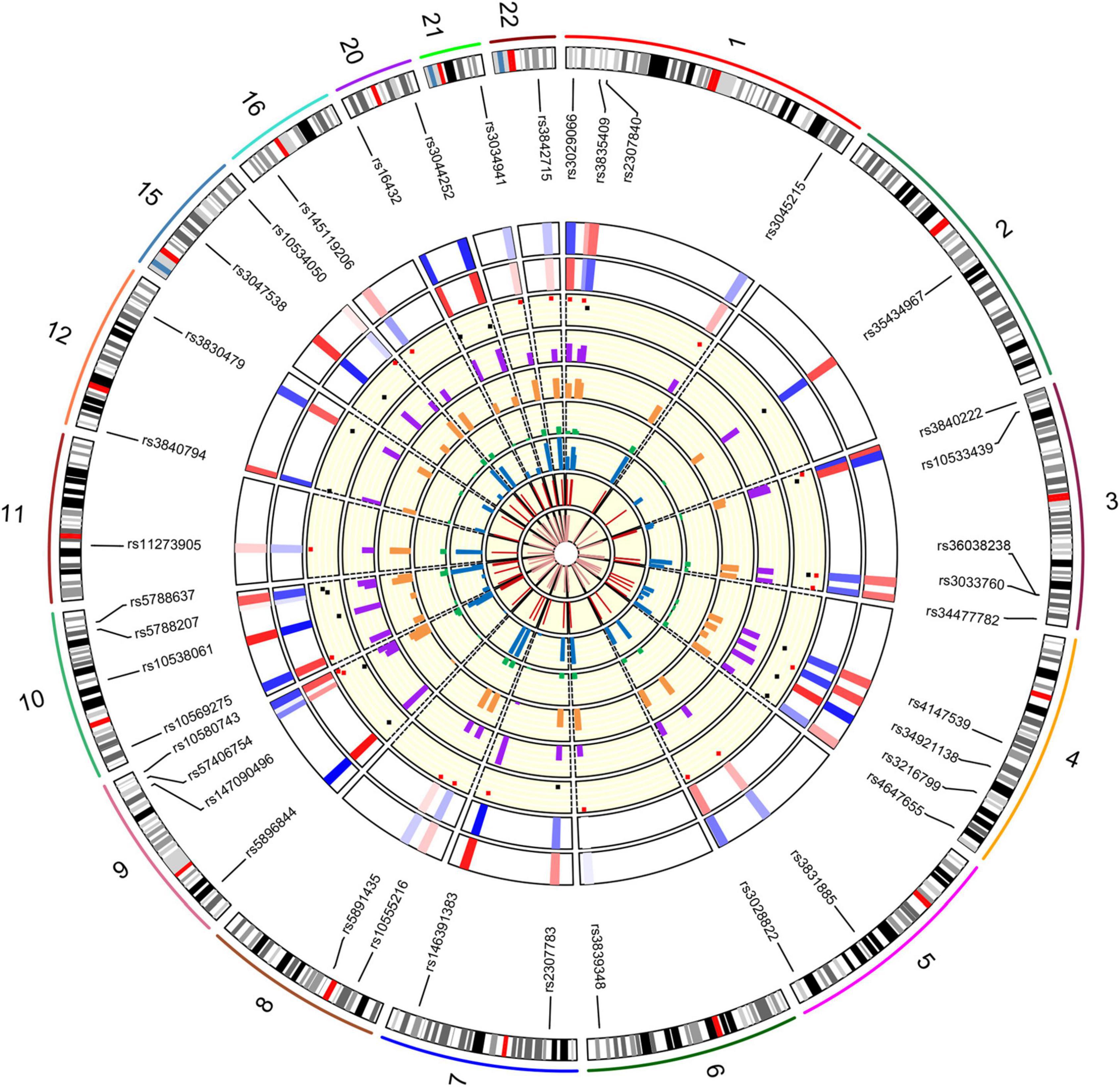
Figure 1. The circle diagram of the chromosome distribution positions, allele frequencies and forensic parameters of the 39 AIDIPs in the Beijing Han population. The inner circles from the outside to the inside represented insertion allele frequencies, deletion allele frequencies, PIC, MP, PD, PE, and TPI, He and Ho values, respectively.
Interpopulation Genetic Differentiations Among the Beijing Han Population and 31 Reference Populations
The insertion allele frequencies based on the 38 AIDIPs among the Beijing Han population and 31 reference populations were displayed in Supplementary Table 4, and to better visualize the allele frequency distributions of the 38 AIDIPs in the 32 populations, the figure of heat map and cluster analysis derived from the above insertion allele frequencies of 38 AIDIPs was drawn. And as shown in Figure 2, the 38 AIDIPs were divided into four branches and demoed different allele frequency distributions among the Beijing Han population and five intercontinental populations. The rs3029066, rs5891435, rs10569275, rs3830479, rs3216799, and rs5896844 loci exhibited relatively high insertion allele frequencies but the rs2307783, rs3840222, and rs34921138 loci manifested relatively low insertion allele frequencies in the African populations. The rs3831885 locus showed relatively small insertion allele frequencies while the rs4647655, rs10555216, and rs36038238 loci presented large insertion allele frequencies in the Beijing Han population and other East Asian populations. The rs34477782 and rs35434967 loci appeared relatively low insertion allele frequencies but the rs3028822 locus conveyed relatively high insertion allele frequencies in the European populations. Furthermore, the 32 populations were classified into four branches, where the Beijing Han population and nine East Asian populations gathered together; five South Asian populations and one East Asian population (CHU) and two American populations (MXL and PEL) grouped together; five European populations and other two American populations (CLM and PUR) clustered together; and seven African populations converged together.
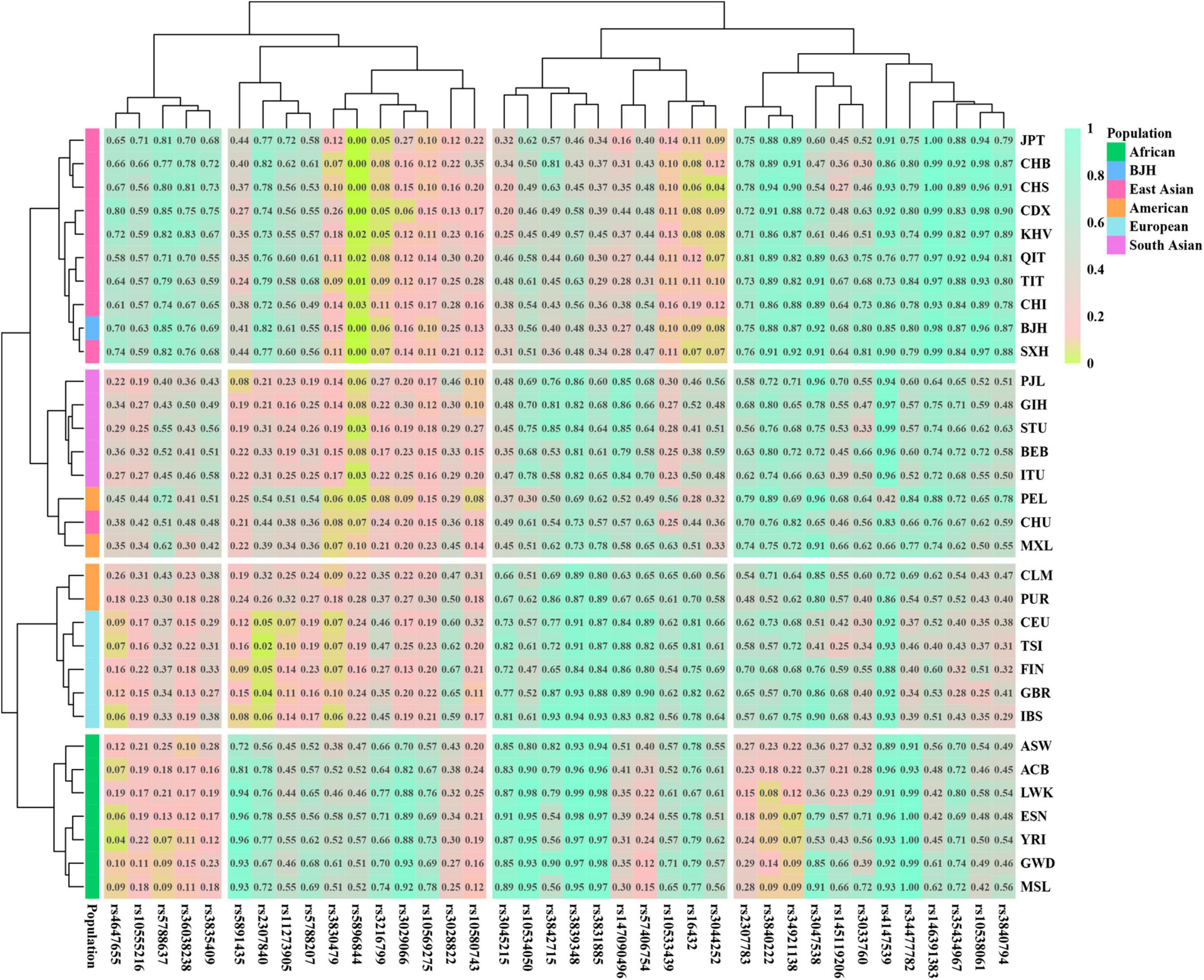
Figure 2. The heat map of insertion allele frequencies of 38 AIDIPs among the Beijing Han population and 31 reference populations.
The locus-by-locus P values computed through the AMOVA method were presented in Supplementary Table 5, and the significance level was modified to 0.0013 (P = 0.05/39 = 0.0013) after Bonferroni correction. The Beijing Han population manifested the numbers of significant difference loci with East Asian populations at 0–31. Specifically, there was no statistically significant difference between the Beijing Han population and the Shaanxi Han population. Additionally, regarding the pairwise population comparisons between the Beijing Han population and the remaining populations, the significant difference loci were 30–33 in five populations from Europe, 22–33 in four populations from America, 27–33 in five populations from South Asia, and 29–36 in seven populations from Africa.
Comparisons of Population Genetic Differences and Phylogenetic Tree Among the Beijing Han Population and 31 Reference Populations
The pairwise FST values and DA distances among the Beijing Han population and 31 global reference populations were calculated and the outcomes were exhibited in Supplementary Tables 6, 7, respectively. Moreover, two heat maps on the basis of the pairwise FST values and DA distances were intuitively presented by different colors in Figures 3A,B, respectively. The Beijing Han population showed the smallest FST value with the Shaanxi Han population (0.0006), followed by the Qinghai Tibetan group (0.0096) and Tibet Tibetan group (0.0169). And the largest FST value was found between the Beijing Han population and LWK (0.4248), then came with the GWD (0.4219), YRI (0.4195), and ESN (0.4154). In the meanwhile, the pairwise DA distances among the Beijing Han population and 31 reference populations demonstrated similar outcomes with the pairwise FST values. The closest DA distance was between the Beijing Han population and Shaanxi Han population (0.0007), next came the Qinghai Tibetan group (0.0028) and Tibet Tibetan group (0.0042); while the farthest DA distance was between the Beijing Han population and YRI (0.1512), followed by LWK (0.1508), ESN (0.1484), and GWD (0.1474).
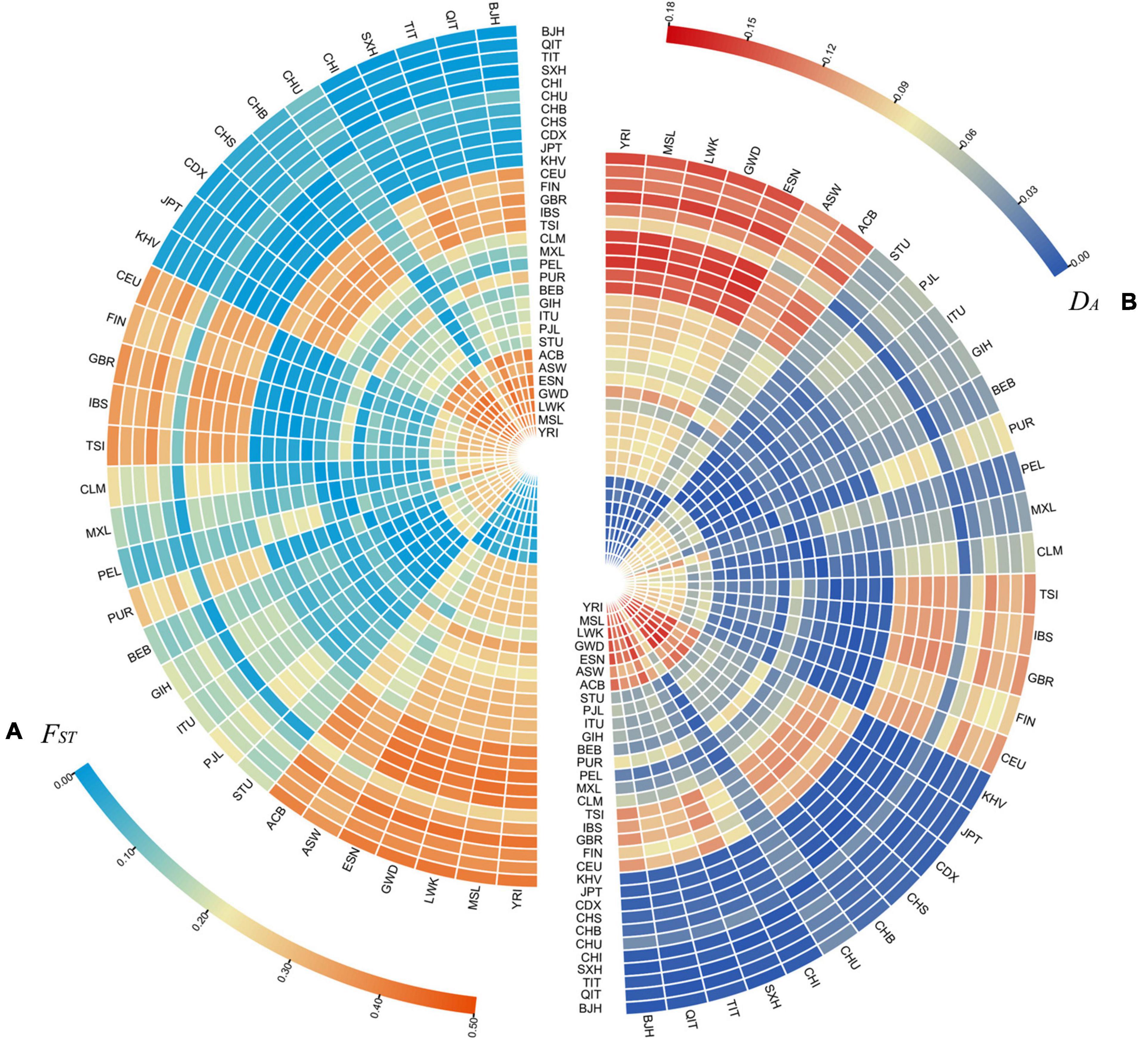
Figure 3. The heat maps of the pairwise FST values (A) and DA distances (B) among the Beijing Han populations and 31 reference populations.
To better visualize the genetic distances among the Beijing Han population and 31 reference populations, one phylogenetic tree on the basis of pairwise DA distances was constructed and exhibited in Figure 4. The phylogenetic tree was divided into two branches, and the first branch comprised two subbranches, of which one subbranch included an American population (CLM) and another subbranch included five South Asian populations, two American populations (MXL and PEL), Beijing Han population, and ten East Asian populations. The second branch also incorporated two subbranches, one of which contained five European populations, and the other subbranch contained one American population (PUR) and seven African populations. Particularly, the Beijing Han population was first clustered with the Shaanxi Han population, then gathered with the Qinghai Tibetan group, Tibet Tibetan group, and other East Asian populations (except the CHU group), which further indicated that the Beijing Han population exhibited close genetic relationships with East Asian populations.

Figure 4. The phylogenetic tree conducted on the basis of the pairwise DA distances among the Beijing Han population and 31 reference populations.
PCA and MDS Analyses Among the Beijing Han Population and 31 Reference Populations
The results of PCA analysis based on insertion allele frequencies and allele genotyping at 38 AIDIPs in the Beijing Han population and 31 worldwide reference populations were shown in Figures 5A,B, respectively. In the PCA plot of 32 populations at the population level, in where each point represented a population, PC1 and PC2 could explain 54.11% and 27.30% of the total variation, respectively. PC1 could distinguish Beijing Han, reference East Asian (except the CHU group), and African populations from other intercontinental populations, PC2 could separate European, Beijing Han, reference East Asian (except the CHU group), and African populations from other intercontinental populations, while the Beijing Han population grouped with other East Asian populations. In the PCA plot of 4,391 individuals at the individual level, PC1 and PC2 accounted for 21.77% and 9.49% of the total variation. Four clusters could be observed in general, namely the African cluster, the American and South Asian cluster, the European cluster, and the East Asian cluster. Moreover, individuals from the Beijing Han almost overlapped with other individuals from East Asia. The cos2 values of 38 AIDIPs were exhibited in Figure 5C, with rs10580743, rs4147539, rs145119206, rs3047538, rs3033760, and rs3842715 loci showed relatively low cos2 values and located in the innermost part of the circle. The results of MDS analysis based on the pairwise FST values were presented in Figure 5D, and a population distribution pattern similar to that of PCA analysis was obtained. As shown in the bi-dimensional scatter plot, the Beijing Han population had close genetic relationships with other East Asian populations. Besides, to further explore the distinguishing ability of this multiplex amplification system between the Beijing Han population and other 10 East Asian populations, PCA analysis on the population level and cos2 values of these 38 AIDIPs were conducted, and the results were presented in Supplementary Figure 1. As illustrated in Supplementary Figure 1A, the Beijing Han population had relatively close genetic relationships with most East Asian populations, and except for the rs10580743, rs3830479, rs3029066, rs2307783, and rs384275 loci, the remaining 33 loci exhibited comparatively high cos2 values in Supplementary Figure 1B.

Figure 5. The principal component analysis on the population level (A) and individual level (B) among the Beijing Han population and 31 reference populations; the cos2 values of 38 AIDIPs (C); the multidimensional scaling analysis based on the pairwise FST values among the Beijing Han population and 31 reference populations (D).
Genetic Structure Analyses Among the Beijing Han Population and 31 Reference Populations
Population genetic structure analyses were developed to further assess the genetic structure of the Beijing Han population, and the findings were manifested in Figure 6, Supplementary Figure 2, and Figure 7, respectively. The genetic structure results in the individual level and the population level were listed in Figure 6, Supplementary Table 8, and Supplementary Figure 2A, respectively. The assumed numbers of subgroups (K) were set to 2–7, and the optimum K value presented in Supplementary Figure 3 was estimated at K = 3. The ancestry component compositions of the Beijing Han population were alike to the East Asian populations at K = 2–7. When K = 3, East Asian, European, and African ancestry components were further identified, and the Beijing Han population showed the dominant ancestry component of East Asia. The specific ancestral compositions (K = 3) of the 32 populations were exhibited in the column chart of Supplementary Figure 2B, and the European, African, and East Asian ancestral components of the Beijing Han population were 0.0233, 0.0183, 0.9585, respectively. The results of clustering analysis (K = 3) for individual ancestry estimation were exhibited in Figures 7A,B. Figure 7A incorporated the Beijing Han population and three continental populations, while Figure 7B contained the Beijing Han population and five continental populations, and the spots representing the Beijing Han population were mostly overlapped with those of East Asian populations.
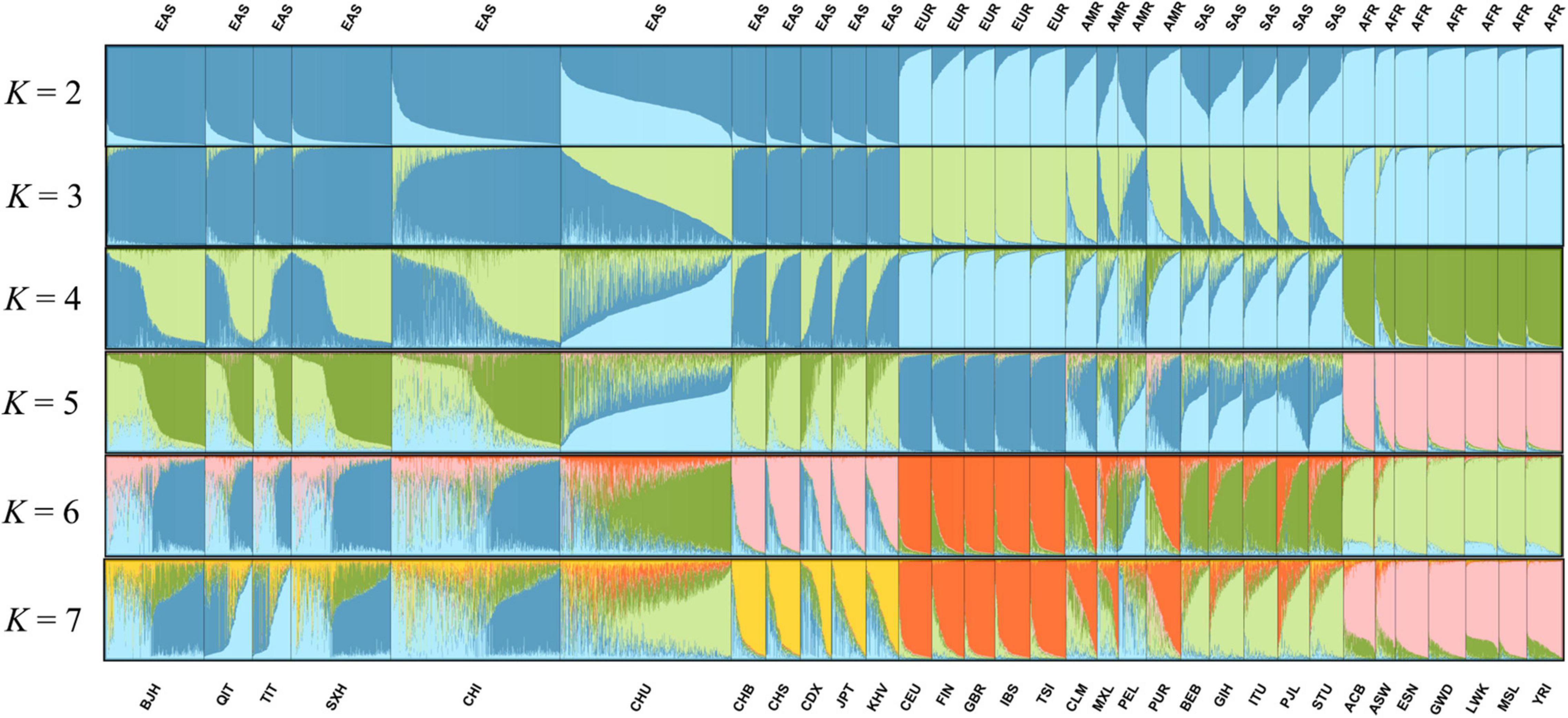
Figure 6. The result of population genetic structure analysis on the individual level among the Beijing Han population and 31 reference populations.
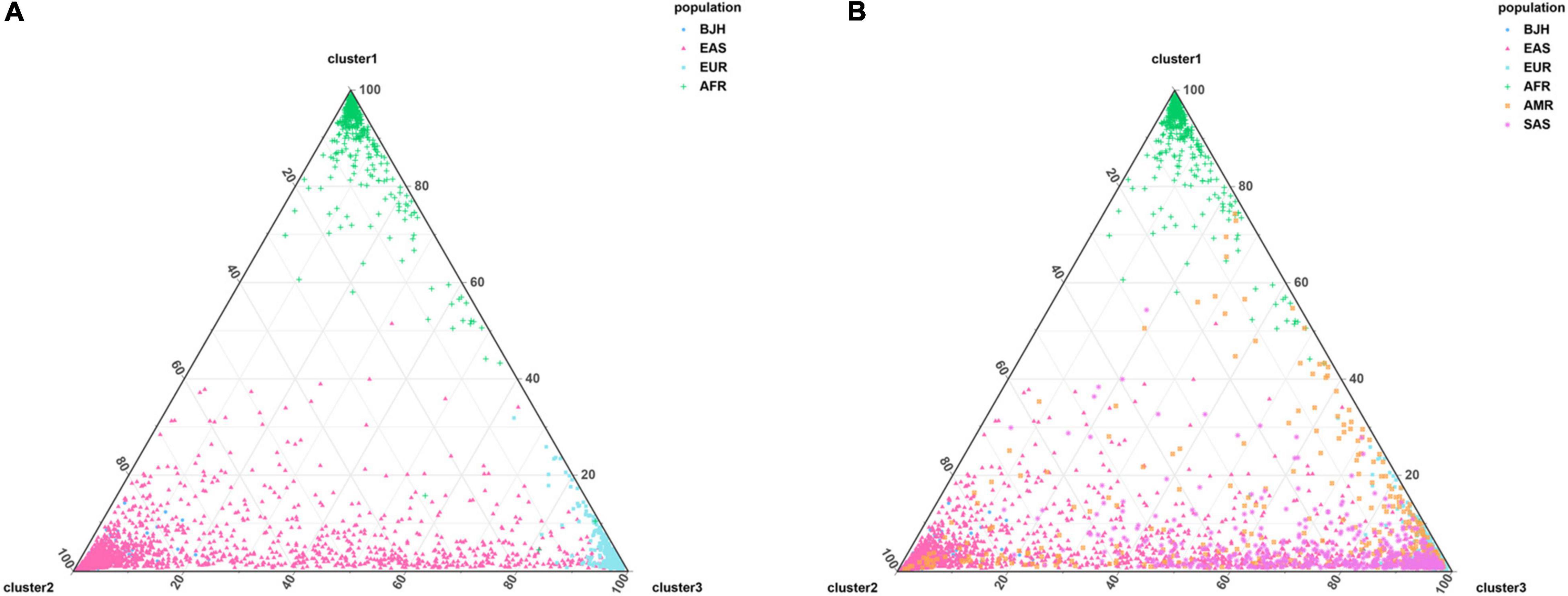
Figure 7. Clustering analysis (K = 3) on the individual level among the Beijing Han population and three continental populations (A); clustering analysis (K = 3) on the individual level among the Beijing Han population and five continental populations (B).
The In values of these 38 AIDIPs in five intercontinental populations were presented in Supplementary Table 9 and shown in a boxplot of Figure 8A. Two AIDIPs (rs16432 and rs3831885), one AIDIP (rs2307840) and 11 AIDIPs (rs10569275, rs2307783, rs3029066, rs3216799, rs34921138, rs3830479, rs3831885, rs3840222, rs5788637, rs5891435 and rs5896844) exhibited relatively higher In values (>0.1) in East Asian, European and African populations, respectively. The cumulative In values derived from the data of 38 AIDIPs in 31 reference populations were 2.4896 in East Asian populations, 1.5115 in European populations, 2.8650 in African populations, 0.2800 in American populations, and 0.4118 in South Asian populations. Cross-validations were conducted to measure success ratios of the system in estimating the continental population classifications, and the results were shown in Figures 8B,C. When only three continental populations were included, the estimation of success ratios for the East Asians, Europeans, and Africans were 94.74%, 100%, and 99.09%, respectively. When five continental populations were contained, the estimation of success ratios of the East Asians, Europeans, and Africans dropped to 81.17%, 89.86%, and 98.64%, respectively. In addition, the classification success ratios were 82.21% for South Asians and 53.60% for Americans. Therefore, a classification model based on 38 AIDIPs data from the reference three intercontinental populations (Africa, East Asia, and Europe) was constructed, and as the results listed in Supplementary Table 10, 299 (100%) Beijing Han individuals were categorized into East Asians.
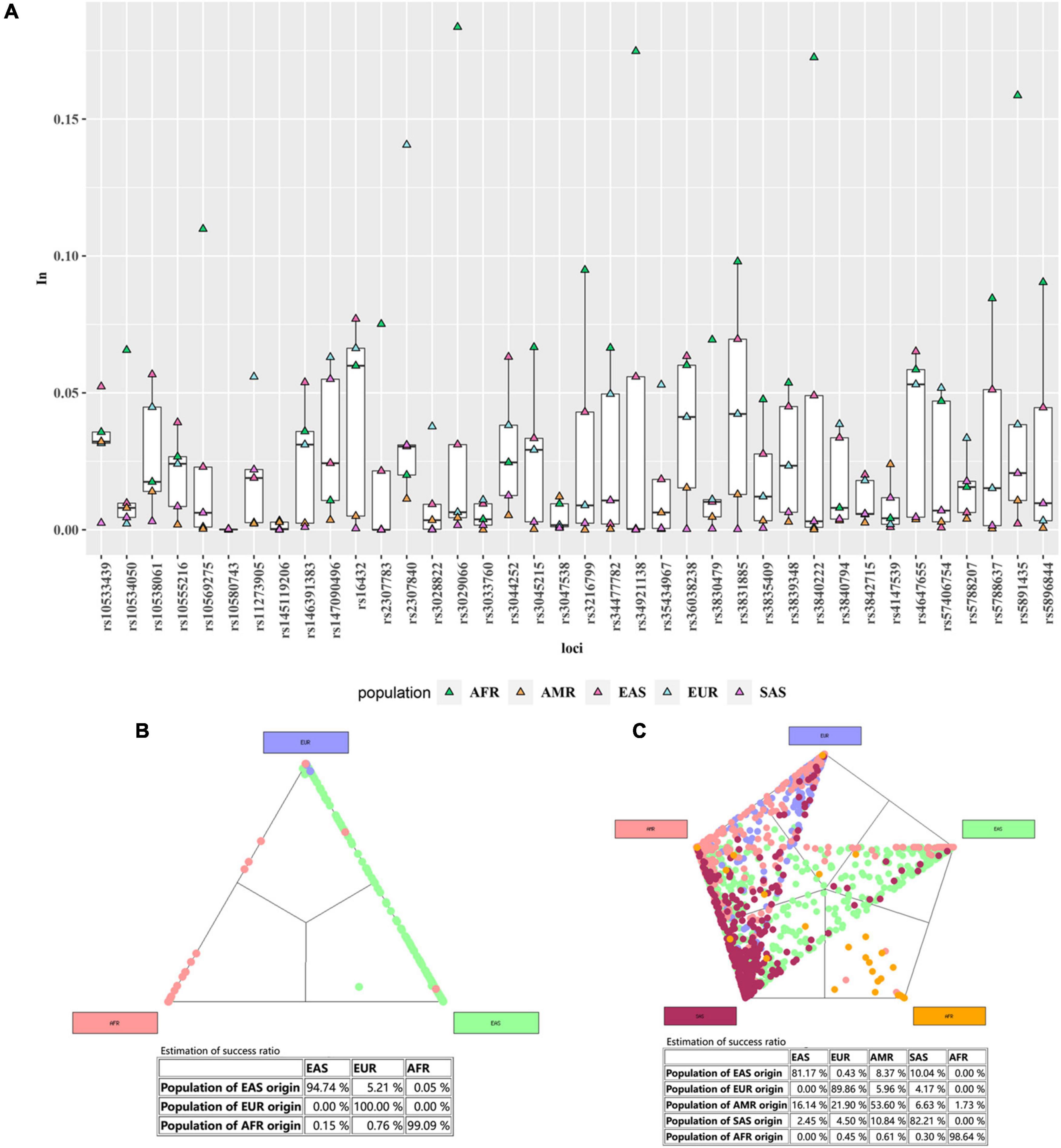
Figure 8. The In values of 38 AIDIPs in five intercontinental populations (A); the cross-validation success ratios of three continental populations (B); the cross-validation success ratios of five continental populations (C).
Discussion
China is a unified multi-ethnic country, and during the long historical development process of five thousand years, ethnic groups have been continuously integrated and multiplied (Black et al., 2006). The distributions of the various ethnic groups are generally dominated by the Han nationality, forming a situation in which the diverse ethnic groups live in mixed communities. The Han nationality is the largest population with a very complicated origin, and the exploration of the origin for the Han Chinese has been the focus of attention by anthropologists and geneticists (Wang M. et al., 2017; Chen et al., 2019; Li et al., 2020). The Han Chinese is a typical agricultural nationality, and the Chinese language and Chinese characters have played an active role in the formation of the Han nationality. It is commonly believed that the Han nationality originated from the Huaxia people, who were active in the Central Plains, and that the Huaxia people continued to interact and integrate with other ethnic groups, eventually forming the modern Han nationality (Wang et al., 2000; Chen et al., 2009; Zhao et al., 2015). However, the exact process and degree of integration of the Huaxia peoples with other neighboring ethnic groups is still not completely clear. The utilization of AIDIPs to exploit the genetic characteristics and genetic makeups of contemporary Han nationality is crucial for understanding its origin and evolution.
In the current study, we employed the novel in-house 39 AIDIPs panel to explore the genetic polymorphism and forensic efficiency of this system in the Beijing Han population. Results showed that 39 AIDIPs were no significant HWE deviations, and all pairwise AIDIPs were conformed to linkage equilibrium except for one pair (rs36038238 and rs3033760) in the Beijing Han population. In general, loci located on different chromosomes or the same chromosome but far apart are not linkage. The two loci, rs36038238 and rs3033760, are separated by within 1 Mb of each other and are both located on the third chromosome. Therefore, the presence of LD at these two loci might be due to genetic linkage, which was also in agreement with our earlier studies (Jin et al., 2020; Zhang W. et al., 2021; Zhang X. et al., 2021). Since these pairwise AIDIPs could not be considered as mutually independent loci in the Beijing Han population, only rs36038238 loci was selected to calculate the subsequent CPD and CPE values. The mean values of PIC, MP, PD, PE, TPI, He, and Ho for the 39 AIDIPs in the Beijing Han population were 0.2526, 0.5547, 0.4453, 0.0898, 0.7669, 0.3122, and 0.3173, respectively. Though these 39 AIDIPs were primarily utilized for ancestry inference in diverse intercontinental populations, the CPD value of these 38 AIDIPs was 0.99999999999961369, revealing that this panel could also be used for individual identification in the Beijing Han population. The relatively low CPE value which was 0.9747 demonstrated that this multiplex system could be adopted as an auxiliary approach for paternity testing in the Han Chinese population in Beijing.
For the purpose of exploring the genetic architecture of the Beijing Han population in greater depth, we employed 31 global populations as reference populations and adopted multiple population genetic analysis approaches based on 38 overlapped AIDIPs to explore the genetic relationships between the Beijing Han population and the 31 reference populations. Apart from the rs10580743, rs4147539, rs145119206, rs3047538, rs3033760, and rs3842715 loci, the insertion allele frequencies at the remaining AIDIPs loci were relatively significant distinct between different continents. The findings of AMOVA indicated that the significant discrepancy loci between the Beijing Han population and East Asian populations were the fewest, especially the Shaanxi Han population which had no significant discrepancy loci with the Beijing Han population. The outcomes of both pairwise FST values and DA distances among the Beijing Han population and 31 reference populations indicated that the Beijing Han population had the least genetic variation with the Shaanxi Han population, then followed by the Qinghai Tibetan and Tibet Tibetan groups. Our findings were in agreement with the previous results which showed that northern Han populations were close to one another and then other East Asian populations (Yao et al., 2004, 2021; Wang H. et al., 2017; Song et al., 2020). Tibetans in China are mainly distributed in the Tibet Autonomous Region, Qinghai, and Gansu provinces, and their settlements are situated on the Tibetan Plateau, the highest elevation on average in the world. Genetic studies of Han nationality and Tibetan group confirmed that modern Han Chinese and contemporary Tibetans shared the similar origins, which would also explained the comparatively close genetic affinities between the Beijing Han population and the two Tibetan groups in our research (Lu et al., 2016; Wang et al., 2020; Wang C. C. et al., 2021; Yu and Li, 2021).
And in accordance with the findings of population genetics, that was, the genetic makeups of populations residing in different geographic regions generally differs greatly due to their remote geographical distances and relatively little genetic exchanges among populations, while populations living in the same or neighboring areas tend to have more genetic exchanges, and their genetic compositions were more similar (Ramachandran et al., 2005; Jay et al., 2013). The results of phylogenetic tree reconstruction, PCA, and MDS all revealed that the Beijing Han population grouped with the reference East Asian populations except the CHU group. Besides, the larger the cos2 value, that was, the closer the AIDIP locus to the side of the ring in the PCA plot, the greater the capability of this AIDIP locus to discriminate between different populations and the higher their contributions. Furthermore, the low cos2 values of rs10580743, rs4147539, rs145119206, rs3047538, rs3033760, and rs3842715 loci were due to the small differences in allele frequency distributions among five intercontinental populations, which indicated that these six AIDIPs of the 38 AIDIPs were less effective in the ancestry inference. The outcomes of the population genetic structure analyses also corroborated with the findings of other cluster analyses, demonstrating that the ancestral composition proportions of the Beijing Han population were comparable to those of the reference East Asian populations (except the CHU group). Besides, the K values in structure analysis refer to the subgroups into which the hypothetical total population can be divided. When at the optimal K value (K = 3), the proportion of East Asian ancestral components of the Beijing Han population was 96.99%. And the cluster analysis results illustrated the Beijing Han individuals were overlapped with majority of individuals from East Asia.
Moreover, In value can be used to assess the validity of locus in differentiating across populations (Rosenberg et al., 2003, 2005). The relatively low In values of rs10580743, rs4147539, rs145119206, rs3047538, rs3033760, and rs3842715 loci were in line with the results of cos2 values. And the relatively low performance loci of rs10580743 and rs4147539 were consistent with a previous study on the Chinese Kyrgyz group (Zhang W. et al., 2021). In the future studies of constructing new multiplex amplification systems, we will remove AIDIPs with low potency of ancestral inference and screen more AIDIPs with high efficacy in East Asian populations to further exploit the genetic structure and background of different populations in China. Besides, this amplification system containing 38 AIDIPs was identified to vary in distinguishing populations from five continents by assessing the In value. Specifically, the cumulative In values were greater than 1.5 in the populations of Africa, East Asia, and Europe, while less than 0.5 in the populations of America and South Asia, indicating that this panel was more effective in discriminating African, European, and East Asian populations. The cross-validation findings of the continental population classifications also revealed that the African populations had the largest estimation of success ratio. In addition, the follow-up taxonomy model based on three continents (East Asian, European, and African) categorized 100% of all Beijing Han individuals as being of East Asian origin, which further indicated the close genetic relationships between the Beijing Han population and the reference East Asian populations.
Conclusion
In conclusion, we employed the self-developed multiplex amplification system containing 39 AIDIPs to further validate the forensic efficacy of the panel in the Beijing Han population. The ancestral components of the Beijing Han population and genetic relationships between the Beijing Han population and 31 worldwide reference populations were also analyzed. The acquired results showed that the Beijing Han population had the close genetic affinities and similar ancestral components with reference East Asian populations. Furthermore, this in-house AIDIP panel can be used for personal identification in the Beijing Han population and can effectively discriminate continental populations containing African, European, and East Asian populations.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Xi’an Jiaotong University (No. XJTULAC201). The participants provided their written informed consent to participate in this study.
Author Contributions
BZ conducted the study design. HX performed the genotyping, statistical analysis, and wrote the manuscript. YF, MZ, and QL helped with the statistical analysis. SM, LL, and XB helped with the sample collections and genotyping. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81930055).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the volunteers in this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.890153/full#supplementary-material
Footnotes
References
Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., Korbel, J. O., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi: 10.1038/nature15393
Black, M. L., Wise, C. A., Wang, W., and Bittles, A. H. (2006). Combining genetics and population history in the study of ethnic diversity in the People’s Republic of China. Hum. Biol. 78, 277–293. doi: 10.1353/hub.2006.0041
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Jin, X., Zhang, X., Zhang, W., Guo, Y., Tao, R., et al. (2021). Comprehensive insights into forensic features and genetic background of chinese northwest hui group using six distinct categories of 231 molecular markers. Front. Genet. 12:705753. doi: 10.3389/fgene.2021.705753
Chen, J., Zheng, H., Bei, J. X., Sun, L., Jia, W. H., Li, T., et al. (2009). Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am. J. Hum. Genet. 85, 775–785. doi: 10.1016/j.ajhg.2009.10.016
Chen, P., Wu, J., Luo, L., Gao, H., Wang, M., Zou, X., et al. (2019). Population genetic analysis of modern and ancient DNA variations yields new insights into the formation, genetic structure, and phylogenetic relationship of Northern Han Chinese. Front. Genet. 10:1045. doi: 10.3389/fgene.2019.01045
Earl, D. A., and Vonholdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Enoch, M. A., Shen, P. H., Xu, K., Hodgkinson, C., and Goldman, D. (2006). Using ancestry-informative markers to define populations and detect population stratification. J. Psychopharmacol. 20(Suppl.), 19–26. doi: 10.1177/1359786806066041
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fan, H., He, Y., Li, S., Xie, Q., Wang, F., Du, Z., et al. (2021). Systematic evaluation of a novel 6-dye direct and multiplex PCR-CE-based indel typing system for forensic purposes. Front. Genet. 12:744645. doi: 10.3389/fgene.2021.744645
Freire-Aradas, A., Ruiz, Y., Phillips, C., Maroñas, O., Söchtig, J., Tato, A. G., et al. (2014). Exploring iris colour prediction and ancestry inference in admixed populations of South America. Forensic Sci. Int. Genet. 13, 3–9. doi: 10.1016/j.fsigen.2014.06.007
Galanter, J. M., Fernandez-Lopez, J. C., Gignoux, C. R., Barnholtz-Sloan, J., Fernandez-Rozadilla, C., Via, M., et al. (2012). Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet. 8:e1002554. doi: 10.1371/journal.pgen.1002554
Goeman, J. J., and Solari, A. (2014). Multiple hypothesis testing in genomics. Stat. Med. 33, 1946–1978. doi: 10.1002/sim.6082
Gouy, A., and Zieger, M. (2017). STRAF-A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci. Int. Genet. 30, 148–151. doi: 10.1016/j.fsigen.2017.07.007
Guo, X. Y., Sun, C. C., Xue, S. Y., Zhao, H., Jiang, L., and Li, C. X. (2021). 49AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia. Yi Chuan 43, 880–889. doi: 10.16288/j.yczz.21-073
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Jay, F., Sjödin, P., Jakobsson, M., and Blum, M. G. (2013). Anisotropic isolation by distance: the main orientations of human genetic differentiation. Mol. Biol. Evol. 30, 513–525. doi: 10.1093/molbev/mss259
Jin, X. Y., Liu, Y. F., Cui, W., Chen, C., Zhang, X. R., Huang, J., et al. (2022). Development a multiplex panel of AISNPs, multi-allelic InDels, microhaplotypes, and Y-SNP/InDel loci for multiple forensic purposes via the NGS. Electrophoresis 43, 632–644. doi: 10.1002/elps.202100253
Jin, X. Y., Shen, C. M., Chen, C., Guo, Y. X., Cui, W., Wang, Y. J., et al. (2020). Ancestry informative DIP loci for dissecting genetic structure and ancestry proportions of Qinghai Tibetan and Tibet Tibetan groups. Mol. Biol. Rep. 47, 1079–1087. doi: 10.1007/s11033-019-05202-x
Kersbergen, P., van Duijn, K., Kloosterman, A. D., den Dunnen, J. T., Kayser, M., and de Knijff, P. (2009). Developing a set of ancestry-sensitive DNA markers reflecting continental origins of humans. BMC Genet. 10:69. doi: 10.1186/1471-2156-10-69
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lan, Q., Shen, C., Jin, X., Guo, Y., Xie, T., Chen, C., et al. (2019). Distinguishing three distinct biogeographic regions with an in-house developed 39-AIM-InDel panel and further admixture proportion estimation for Uyghurs. Electrophoresis 40, 1525–1534. doi: 10.1002/elps.201800448
Li, L., Zou, X., Zhang, G., Wang, H., Su, Y., Wang, M., et al. (2020). Population genetic analysis of Shaanxi male Han Chinese population reveals genetic differentiation and homogenization of East Asians. Mol. Genet. Genomic Med. 8:e1209. doi: 10.1002/mgg3.1209
Lu, D., Lou, H., Yuan, K., Wang, X., Wang, Y., Zhang, C., et al. (2016). Ancestral origins and genetic history of tibetan highlanders. Am. J. Hum. Genet. 99, 580–594. doi: 10.1016/j.ajhg.2016.07.002
Nei, M., Tajima, F., and Tateno, Y. (1983). Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol. 19, 153–170. doi: 10.1007/bf02300753
Pereira, R., Phillips, C., Pinto, N., Santos, C., dos Santos, S. E., Amorim, A., et al. (2012). Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS One 7:e29684. doi: 10.1371/journal.pone.0029684
Qin, P., Li, Z., Jin, W., Lu, D., Lou, H., Shen, J., et al. (2014). A panel of ancestry informative markers to estimate and correct potential effects of population stratification in Han Chinese. Eur. J. Hum. Genet. 22, 248–253. doi: 10.1038/ejhg.2013.111
Ramachandran, S., Deshpande, O., Roseman, C. C., Rosenberg, N. A., Feldman, M. W., and Cavalli-Sforza, L. L. (2005). Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. U. S. A. 102, 15942–15947. doi: 10.1073/pnas.0507611102
Romanini, C., Romero, M., Salado Puerto, M., Catelli, L., Phillips, C., Pereira, R., et al. (2015). Ancestry informative markers: inference of ancestry in aged bone samples using an autosomal AIM-Indel multiplex. Forensic Sci. Int. Genet. 16, 58–63. doi: 10.1016/j.fsigen.2014.11.025
Rosenberg, N. A. (2004). DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Rosenberg, N. A., Li, L. M., Ward, R., and Pritchard, J. K. (2003). Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 73, 1402–1422. doi: 10.1086/380416
Rosenberg, N. A., Mahajan, S., Ramachandran, S., Zhao, C., Pritchard, J. K., and Feldman, M. W. (2005). Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 1:e70. doi: 10.1371/journal.pgen.0010070
Song, F., Lang, M., Li, L., Luo, H., and Hou, Y. (2020). Forensic features and genetic background exploration of a new 47-autosomal InDel panel in five representative Han populations residing in Northern China. Mol. Genet. Genomic Med. 8:e1224. doi: 10.1002/mgg3.1224
Tateno, Y., Nei, M., and Tajima, F. (1982). Accuracy of estimated phylogenetic trees from molecular data. I. Distantly related species. J. Mol. Evol. 18, 387–404. doi: 10.1007/bf01840887
Wang, C. C., Yeh, H. Y., Popov, A. N., Zhang, H. Q., Matsumura, H., Sirak, K., et al. (2021). Genomic insights into the formation of human populations in East Asia. Nature 591, 413–419. doi: 10.1038/s41586-021-03336-2
Wang, H. Y., Hu, Y. H., Cao, Y. Y., Zhu, Q., Huang, Y. G., Li, X., et al. (2021). AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations. Yi Chuan 43, 938–948. doi: 10.16288/j.yczz.21-185
Wang, H., Ba, H., Yang, C., Zhang, J., and Tai, Y. (2017). Inner and inter population structure construction of Chinese Jiangsu Han population based on Y23 STR system. PLoS One 12:e0180921. doi: 10.1371/journal.pone.0180921
Wang, M., Wang, Z., Zhang, Y., He, G., Liu, J., and Hou, Y. (2017). Forensic characteristics and phylogenetic analysis of two Han populations from the southern coastal regions of China using 27 Y-STR loci. Forensic Sci. Int. Genet. 31, e17–e23. doi: 10.1016/j.fsigen.2017.10.009
Wang, L., Oota, H., Saitou, N., Jin, F., Matsushita, T., and Ueda, S. (2000). Genetic structure of a 2,500-year-old human population in China and its spatiotemporal changes. Mol. Biol. Evol. 17, 1396–1400. doi: 10.1093/oxfordjournals.molbev.a026422
Wang, M., Du, W., He, G., Wang, S., Zou, X., Liu, J., et al. (2020). Revisiting the genetic background and phylogenetic structure of five Sino-Tibetan-speaking populations: insights from autosomal InDels. Mol. Genet. Genomics 295, 969–979. doi: 10.1007/s00438-020-01673-x
Xie, T., Shen, C., Jin, X., Lan, Q., Fang, Y., and Zhu, B. (2020). Genetic structural differentiation analyses of intercontinental populations and ancestry inference of the chinese hui group based on a novel developed autosomal AIM-InDel genotyping system. Biomed. Res. Int. 2020:2124370. doi: 10.1155/2020/2124370
Yahya, P., Sulong, S., Harun, A., Wangkumhang, P., Wilantho, A., Ngamphiw, C., et al. (2020). Ancestry-informative marker (AIM) SNP panel for the Malay population. Int. J. Legal Med. 134, 123–134. doi: 10.1007/s00414-019-02184-0
Yao, H., Wang, M., Zou, X., Li, Y., Yang, X., Li, A., et al. (2021). New insights into the fine-scale history of western-eastern admixture of the northwestern Chinese population in the Hexi Corridor via genome-wide genetic legacy. Mol. Genet. Genomics 296, 631–651. doi: 10.1007/s00438-021-01767-0
Yao, Y. G., Kong, Q. P., Wang, C. Y., Zhu, C. L., and Zhang, Y. P. (2004). Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in china. Mol. Biol. Evol. 21, 2265–2280. doi: 10.1093/molbev/msh238
Yu, X., and Li, H. (2021). Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome. Mol. Genet. Genomics 296, 783–797. doi: 10.1007/s00438-021-01794-x
Zhang, W., Jin, X., Wang, Y., Chen, C., and Zhu, B. (2021). Genetic structure analyses and ancestral information inference of Chinese Kyrgyz group via a panel of 39 AIM-DIPs. Genomics 113, 2056–2064. doi: 10.1016/j.ygeno.2021.03.008
Zhang, X., Shen, C., Jin, X., Guo, Y., Xie, T., and Zhu, B. (2021). Developmental validations of a self-developed 39 AIM-InDel panel and its forensic efficiency evaluations in the Shaanxi Han population. Int. J. Legal Med. 135, 1359–1367. doi: 10.1007/s00414-021-02600-4
Keywords: deletion/insertion polymorphism, genetic characteristic, population structure, genetic relationship, Beijing Han nationality
Citation: Xu H, Fang Y, Zhao M, Lan Q, Mei S, Liu L, Bai X and Zhu B (2022) Forensic Features and Genetic Structure Analyses of the Beijing Han Nationality Disclosed by a Self-Developed Panel Containing a Series of Ancestry Informative Deletion/Insertion Polymorphism Loci. Front. Ecol. Evol. 10:890153. doi: 10.3389/fevo.2022.890153
Received: 08 March 2022; Accepted: 13 April 2022;
Published: 31 May 2022.
Edited by:
Dennis McNevin, University of Technology Sydney, AustraliaReviewed by:
Jiang Huang, Guizhou Medical University, ChinaHector Rangel-Villalobos, University of Guadalajara, Mexico
Copyright © 2022 Xu, Fang, Zhao, Lan, Mei, Liu, Bai and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bofeng Zhu, emh1Ym9mZW5nNzM3MkAxMjYuY29t
 Hui Xu1
Hui Xu1 Bofeng Zhu
Bofeng Zhu