- 1College of Life Sciences, Qufu Normal University, Qufu, China
- 2College of Animal Science and Technology, Shandong Agricultural University, Taian, China
As key pollinators, bees are frequently exposed to multiple environmental stresses and have developed crucial mechanisms by which they adapt to these stressors. However, the molecular bases mediated at the gene level remain to be discovered. Here, we found four heat shock protein DnaJB subfamily genes, DnaJB6, DnaJshv, DnaJB13, and DnaJB14, from Apis cerana cerana, that all have J domains in their protein sequences. The expression levels of DnaJB6 and DnaJshv were upregulated by different degrees of heat stress, and the transcript level of DnaJB14 was gradually upregulated as the degree of heat stress increased, while the mRNA level of DnaJB13 was downregulated at multiple time points during heat stress treatment. The mRNA levels of all four DnaJBs were upregulated by cold and UV stress. In addition, the expression levels of DnaJB6, DnaJshv and DnaJB13 were reduced under abamectin, imidacloprid, cypermethrin, bifenthrin, spirodiclofen, and methomyl stresses. The transcript level of DnaJB14 was decreased by imidacloprid, cypermethrin, spirodiclofen, and methomyl exposure but increased by abamectin and bifenthrin exposure. These results indicate that the demand of A. cerana cerana for these four DnaJBs differs under various stress conditions. To further explore the role of DnaJBs in the stress response, we successfully silenced DnaJshv and DnaJB14. The content of protein carbonyl was increased, while the content of VC, the enzymatic activities of CAT, GST, and SOD, the mRNA levels of many antioxidant-related genes, and the total antioxidant capacity were reduced after knockdown of DnaJshv and DnaJB14 in A. cerana cerana. These results indicate that silencing DnaJshv and DnaJB14 increases oxidative damage and decreases the antioxidant ability of A. cerana cerana. Taken together, our results demonstrate that DnaJB6, DnaJshv, DnaJB13, and DnaJB14 are differentially expressed under stress conditions and play crucial roles in response to various stressors, possibly through the antioxidant signalling pathway. These findings will be conducive to understanding the molecular basis of bee responses to environmental stresses and are beneficial for improving bee protection.
Introduction
Previous reports on regional bee declines are alarming, especially because they are accompanied by the increasing pollination service requirements of conventional intensive agriculture. Increasing evidence has indicated that different environmental stresses, such as parasites, pathogens, heat, cold, and agrochemicals are key drivers of bee declines (Potts et al., 2010; Goulson et al., 2015). Therefore, it is vital to understand how bees cope with various stressors, and breed bees with enhanced resistance to different stressors.
Many studies have revealed the adverse effects of various environmental stresses on bees, which will help to understand the reason or the contributions of different environmental stresses to bee declines. For example, longer durations of cold stress during the capped brood stage result in shorter worker longevity, more misorientation inside cells and increased mortality rates for bees (Wang et al., 2016). The immunocompetence of drones, queens and workers of bees is affected by heat stress during development, and heat-stressed workers have a significantly shorter lifespan after infecting Metarhizium anisopliae than drones and queens (Medina et al., 2020). The synergistic interaction of agrochemicals increases bee mortality (Siviter et al., 2021). Clothianidin can decrease the immune response of bees and is implicated in increased Varroa destructor fertility (Annoscia et al., 2020). Despite these adverse effects of various stressors on bee fitness, the molecular defence systems by which bees defend against stressors still require further study.
Sophisticated genes and protein-dependent regulatory mechanisms play crucial roles in the defence responses of animals to multiple stressors. Heat shock proteins (Hsps), also known as stress proteins and molecular chaperones, are known to be important in stress response (Feder and Hofmann, 1999; King and MacRae, 2015). Based on differences in their molecular weights, Hsps are divided into six major groups, including small Hsps (sHsps), Hsp40, Hsp60, Hsp70, Hsp90, and Hsp110 (Sørensen et al., 2003). Based on its name, Hsp40 is often thought to have a molecular weight of 40 kDa, but the molecular weights of most DnaJ members differ substantially from 40 kDa. Due to the existence of the J domain (a helical hairpin structure of approximately 70 amino acid residues), Hsp40 is often known as the DnaJ protein (Kampinga and Craig, 2010). It is important to note that the J domain is crucial for the function of DnaJ protein, especially for those functioning as cochaperones of Hsp104, Hsp90, and Hsp70 (Kampinga and Craig, 2010; Craig and Marszalek, 2017).
DnaJs include DnaJA, DnaJB, and DnaJC, which are three classes, also known as class I, class II, and class III, based on differences in domains and motifs, and each class contains several members. It is worth mentioning that the DnaJ subfamily is the most diverse among all Hsp subfamilies. A total of 45, 36, 34, 27, 22, 17, and 7 DnaJ members (or putative members) have been found in Homo sapiens, Drosophila melanogaster, Apis cerana cerana, Apis mellifera, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Escherichia coli, respectively (Craig and Marszalek, 2017; McKinstry et al., 2017; Li et al., 2020a). Fully exploring the functions of DnaJs will help to understand the variation in DnaJ gene numbers across various organisms.
Many studies have revealed that DnaJs play a variety of important roles in organisms. For example, DnaJB6 plays a crucial role in neuronal sensitivity to amyloidogenesis. Knockdown of DnaJB6 causes spontaneous polyglutamine aggregation in progenitors, and increased expression of DnaJB6 antagonises glutamate-dependent aggregation in neurons (Thiruvalluvan et al., 2020). Gastric cancer patients with high DnaJC12 transcript levels were shown to have obviously reduced overall survival compared to those with low levels, and DnaJC12 knockdown significantly reduced the invasion and proliferation of gastric cancer cells (Uno et al., 2019). Mutation of DnaJB13 triggers primary ciliary dyskinesia and male infertility, and male fertility in asthenozoospermia is involved in missense mutation of DnaJB13 (El Khouri et al., 2016; Li et al., 2020b). Autosomal-dominant polycystic kidney disease is triggered by DnaJB11 monoallelic mutations (Cornec-Le Gall et al., 2018). The expression of DnaJB14 and DnaJB12 results in coordinated nuclear invasion through membranes connected to a novel nuclear pore structure (Goodwin et al., 2014). The interaction between DnaJA1 and mutant p53 promotes tumour metastasis, and the expression levels of DnaJA1 in head and neck squamous cell carcinoma tissues are higher than those in normal tissues (Kaida et al., 2021). However, these studies mainly focused on the function of DnaJs in diseases. It has been shown that the Hsps family plays key roles in tuning animal stress responses (Feder and Hofmann, 1999; King and MacRae, 2015; Li et al., 2020a; Chand et al., 2021). Nevertheless, the precise functions of DnaJ subfamily members in response to stress have been poorly investigated.
Apis cerana cerana is a vital indigenous bee species in China, and Chinese beekeepers began raising it at least 1,700 years ago. Although A. cerana cerana is immensely economically beneficial for Chinese agriculture, obvious A. cerana cerana colony loss has been observed (Theisen-Jones and Bienefeld, 2016; Chen et al., 2017). The drivers of A. cerana cerana colony loss also include environmental stresses, but the molecular mechanisms by which A. cerana cerana defends against stressors need to be clarified. Our previous studies identified five DnaJBs (DnaJB6, DnaJshv, DnaJB12, DnaJB13, and DnaJ14) via the annotation of transcriptome data and found DnaJB12 to be important for A. cerana cerana to cope with stressors (Li et al., 2018, 2020a). However, the functions of DnaJB6, DnaJBshv, DnaJB13, and DnaJ14 under stress conditions still need to be investigated. Herein, we aimed to further explore the expression patterns and functions of these four DnaJBs under different stresses in A. cerana cerana. We hope that our results will contribute to further understanding the defence mechanism of A. cerana cerana against environmental stresses.
Materials and Methods
Isolation of the Coding Sequences of DnaJB6, DnaJshv, DnaJB13, and DnaJB14
To clone the coding sequences (CDSs) of A. cerana cerana DnaJB6, dnaJ homolog shv (DnaJshv), DnaJB13, and DnaJB14, PCR was performed using specific primers (Supplementary Table 1) designed based on the CDSs of A. cerana DnaJB6 (Gene ID: 108002258), DnaJshv (Gene ID: 108000047), DnaJB13 (Gene ID: 107995408), and DnaJB14 (Gene ID: 107994739) deposited in NCBI. TransFast®® Taq DNA Polymerase (TransGen Biotech, China) was used to carry out the PCR amplifications according to the manufacturer’s instructions.
Bioinformatics Analysis of DnaJB6, DnaJshv, DnaJB13, and DnaJB14
Phylogenetic analyses of A. cerana cerana DnaJs were conducted using MEGA. The J domains of DnaJBs were determined based on SMART, NCBI Conserved Domain and described previously (Qiu et al., 2006). SWISS-MODEL1 and SPDBV were used to build and analyse the tertiary structures of the J domains of DnaJB6, DnaJshv, DnaJB13, and DnaJB14. DNAMAN was used for multiple alignments of amino acid and nucleotide sequences. The transmembrane helices of DnaJshv and DnaJB14 were analysed using TMHMM and SMART. The protein name, protein ID and species of DnaJs used for bioinformatics analysis are listed in Supplementary Table 2.
Apis cerana cerana Treatments
Adults (15-day-old A. cerana cerana) were collected from a healthy colony located at Shandong Agricultural University (Shandong, China) with no agrochemical use. To obtain 15-day-old A. cerana cerana, we used paint (B&Q, China) to mark newly emerged workers, which were collected 15 days later. The collected bees were randomly divided into 14 groups, and each group contained 50 bees. Our previous studies revealed that the mRNA levels of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 were differentially regulated under 47°C treatment for 2 h (Li et al., 2020a). To further investigate the expression levels of these four DnaJBs under heat stress conditions, we subjected A. cerana cerana from groups 1–3 to different degrees of heat stress, including 40, 43, and 46°C, for 5 h. Groups 4–5 were exposed to ultraviolet light (UV, 30 mJ/cm2) and cold (4°C) stress. Each bee from groups 6–11 was administered 1 μL of field-realistic doses of abamectin, imidacloprid, cypermethrin, bifenthrin, spirodiclofen, and methomyl. The field-realistic doses of agrochemicals were prepared in accordance with the manufacturers’ protocol. Group 12 served as the control for groups 1–11 and was untreated. The bees from groups 1–12 were sampled at 0, 1, 2, 3, 4, and 5 h after treatment. Bees from group 13 were subjected to 40°C treatment for 48 h. Bees from group 14 served as the control for group 13, and were maintained at 33°C. Groups 13–14 were sampled at 0, 12, 24, 36, and 48 h after treatment. The culture temperature of all the treatment groups was 33°C except for the 4, 40, 43, and 46°C treatment groups. In addition, bees in all groups were kept in constant darkness at 70% relative humidity and fed 30% sucrose syrup. The collected bees were immediately frozen in liquid nitrogen and then stored at −80°C.
RNA Extraction, Reverse Transcription, and Quantitative Real Time PCR
Total RNA from four pooled A. cerana cerana was prepared using TRIzol (TaKaRa, China) as described by the manufacturer. HiScript® II Q RT SuperMix for qPCR (+ gDNA Wiper) (TaKaRa, China) was used to reverse transcribe RNA according to the manufacturer’s instructions. The SYBR® Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology Co. Ltd., China) was used for quantitative real-time PCR (qRT–PCR) analysis. The 20 μL reaction mixtures were composed of 1 μL of cDNA, 0.5 μL of each primer, 8 μL of RNase free water and 10 μL of 2X SYBR® Green Pro Taq HS Premix. The qRT–PCR protocol was as follows: 95°C for 30 s; 40 cycles of 95°C for 5 s and 60°C for 30 s; and an increase from 65 to 95°C at a 5°C interval every 5 s. A final melting curve was generated, and the relative levels of gene expression were calculated using the 2–ΔΔCT method. β-actin, which has a stable transcript level in bees, was used as an endogenous control. Three biological replicates were performed for this part of the experiments.
RNA Interference
To knock down DnaJshv and DnaJB14 in A. cerana cerana, double-stranded RNAs (dsRNAs) targeting DnaJshv and DnaJB14 were generated by the T7 RiboMAX™ Express RNAi System (Promega, United States) according to the manufacturer’s instructions. DNA templates for generating dsRNAs targeting DnaJshv (6–507 nt) and DnaJB14 (43–542 nt) were amplified from A. cerana cerana cDNA by PCR using specific primers, which included the T7 RNA polymerase promoter sequence at their 5′ ends (Supplementary Table 1). The dsRNA targeting the synthetic construct with a modified green fluorescent protein (GFP) coding sequence (151–650 nt, GenBank: U87974.1) was used as a control. The 15-day-old adults were collected and then randomly divided into three groups (n = 30/group). A. cerana cerana from the three groups were fed 5 μg dsRNA-DnaJshv, dsRNA-DnaJB14, and dsRNA-GFP. One day after being fed the dsRNAs, bees from these three groups were collected, and the RNAi efficiencies, expression levels of antioxidant-related genes, total antioxidant capacity (T-AOC), enzymatic activities, and levels of oxidant and antioxidant-related substances were determined.
Expression Levels of Some Antioxidant Genes After Knockdown of DnaJshv and DnaJB14
Quantitative real-time PCR was used to detect the transcription levels of some antioxidant genes when silenced DnaJshv and DnaJB14. These antioxidant genes include thioredoxin peroxidase 3 (Tpx3), thioredoxin peroxidase 4 (Tpx4), thioredoxin peroxidase 5 (Tpx5), thioredoxin reductase 1 (TrxR1), thioredoxin 1 (Trx1), thioredoxin 2 (Trx2), thioredoxin-like 1 (Trx-like1), omega class glutathione S-transferase 2 (GSTO2), delta class glutathione S-transferase (GSTD), theta class glutathione S-transferase 1 (GSTT1), sigma class glutathione S-transferase 4 (GSTS4), zeta class glutathione S-transferase 1 (GSTZ1), cyclin-dependent kinase 5 (CDK5), mitogen-activated protein kinase kinase 4 (MKK4), superoxide dismutase 1 (SOD1), cyclin-dependent kinase 5 regulatory subunit 1 (CDK5r), cytochrome P450 4G11 (CYP4G11), p38 MAPK isoform (p38b), small heat shock proteins 22.6 (Hsp22.6), and methionine sulfoxide reductase B (MsrB).
Determination of the Contents of Oxidant- and Antioxidant-Related Substances, Enzymatic Activity, and Total Antioxidant Capacity
Whole A. cerana cerana tissue homogenates were extracted using 0.9% normal saline. The quantitative total protein in tissue homogenates, substance contents, enzymatic activities, and total antioxidant capacities of the samples were then determined. The contents of malondialdehyde (MDA), vitamin C (VC) and protein carbonyl; the enzymatic activities of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and glutathione S-transferase (GST); and the T-AOC were detected using kits from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
Statistical Analysis
All experiments were performed with at least three biological duplicates and three technical repeats. Standard Student’s t-tests were used for statistical analyses of two groups, and P < 0.05 (one asterisk) and P < 0.01 (two asterisks) were considered to indicate significant differences. Statistical analyses of multiple groups were performed based on one-way ANOVA followed by Tukey’s correction. Significant differences are indicated by different letters, the same letter indicates a lack of significance, and overlapping letters indicate differences among multiple groups that are not significant.
Results
Molecular Characterisation of DnaJB6, DnaJshv, DnaJB13, and DnaJB14
The CDSs of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 are 984, 1,101, 113, and 636 bp and encode 327, 366, 370, and 211 amino acids, respectively. Phylogenetic analysis showed that DnaJB6 and DnaJB13 of A. cerana cerana had high homology with A. mellifera DnaJB6 and Apis dorsata DnaJB13, respectively (Figure 1). In our recent studies, DnaJshv was named DnaJB11 based on the NR annotation of genes identified in the transcriptome data of A. cerana cerana (Li et al., 2020a). In the NCBI database, we found that the homologous genes of DnaJshv in A. mellifera, Apis florea, A. dorsata, and A. cerana were all named DnaJshv. However, upon subjecting the protein sequences of DnaJshv from A. cerana cerana to a BLAST search, we identified DnaJB11 as a homologous protein in many species, such as Acromyrmex echinatior, Habropoda laboriosa, H. sapiens, and Danio rerio. Phylogenetic analysis revealed that DnaJshv from bees has a high homology with DnaJB proteins, indicating that DnaJshv belongs to the DnaJB subfamily (Figure 1). A BLAST search of the DnaJB13 protein sequence from A. cerana cerana revealed homologous proteins other than DnaJB13 in various species as follows: DnaJB9 in species such as H. sapiens and A. florea; DnaJB14 in species such as A. dorsata; and DnaJC18 in species such as A. mellifera. Phylogenetic analysis showed that DnaJB13 of A. cerana cerana had a high homology with DnaJB proteins, suggesting that DnaJB13 belongs to the DnaJB subfamily (Figure 1). However, these results also indicate that the name of some DnaJ proteins may need further refinement to better display their subfamily classification.
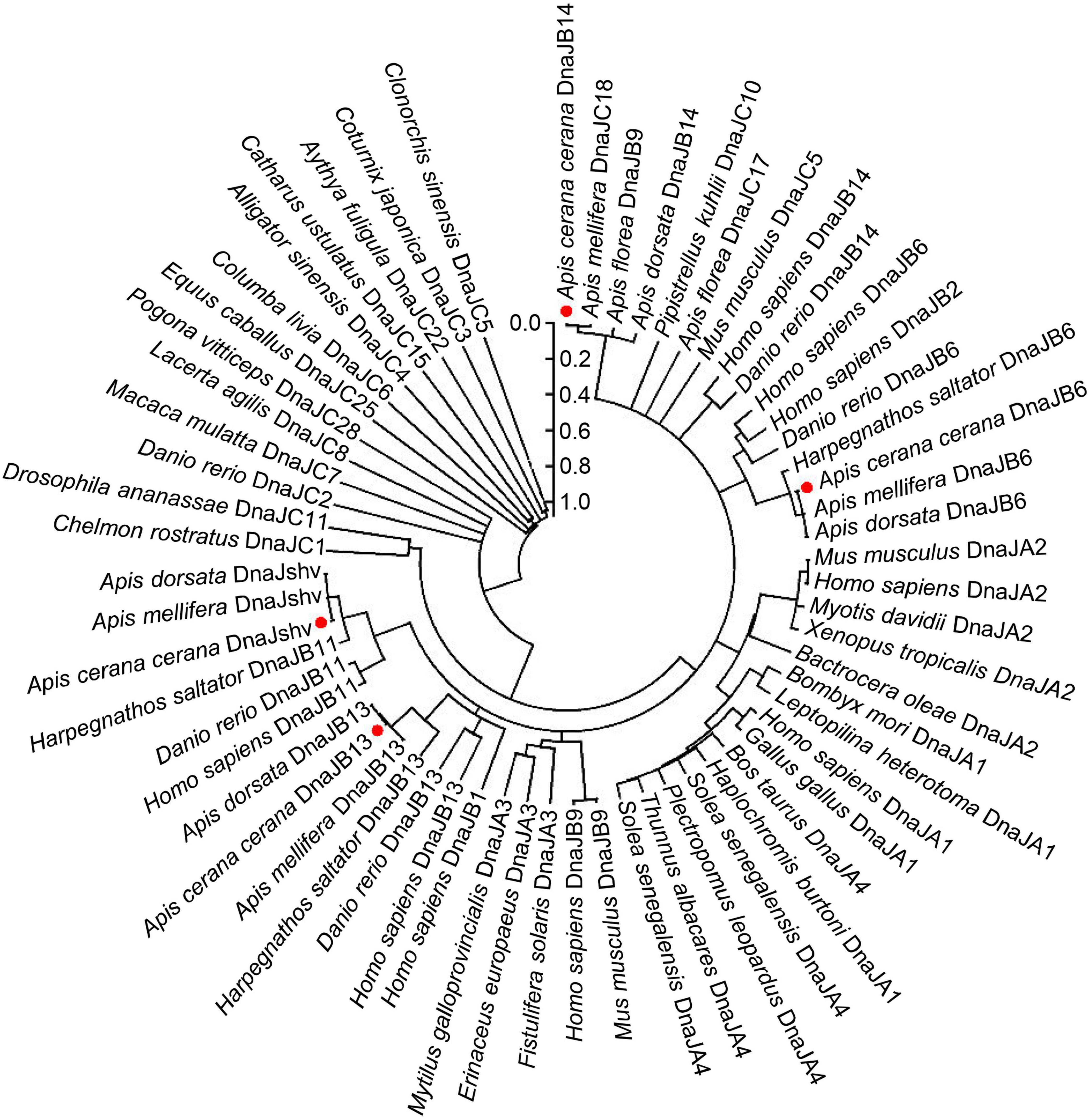
Figure 1. Phylogenetic relationships among DnaJs from different animal species. The positions of A. cerana cerana DnaJB6, DnaJshv, DnaJB13, and DnaJB14 are indicated by red dots. The protein sequences of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 in A. cerana cerana were acquired in this study. The protein sequences of other DnaJs were acquired from NCBI, and their protein IDs are shown in Supplementary Table 2.
The protein sequences of DnaJshv and DnaJB14 of A. cerana cerana were shown to have one transmembrane domain, and the transmembrane regions of DnaJshv and DnaJB14 are located in aa 12–31 and aa 172–194, respectively (Supplementary Figure 1). These results indicate that DnaJshv and DnaJB14 are transmembrane proteins. In addition, the J domain was found in the amino acid sequences of the A. cerana cerana proteins DnaJB6, DnaJshv, DnaJB13, and DnaJB14, and four α-helices (helix I, helix II, helix III, and helix IV) were shown to be located in the J domain (Figures 2, 3 and Supplementary Figures 2–5). A conserved HPD (His, Pro, and Asp) tripeptide was located between helix II and helix III in the J domain of DnaJB6, DnaJshv, and DnaJB14, while the J domain of DnaJB13 lacked the HPD tripeptide (Figures 2, 3). Multiple sequence alignments indicated that the J domains of the A. cerana cerana proteins DnaJB6, DnaJshv, DnaJB13, and DnaJB14 were more homologous to those of other animals than other protein regions (Supplementary Figures 2–5). Tertiary structure analysis of the proteins further revealed the structural characteristics of the J domains of DnaJB6, DnaJBshv, DnaJB13, and DnaJB14 (Figure 3).
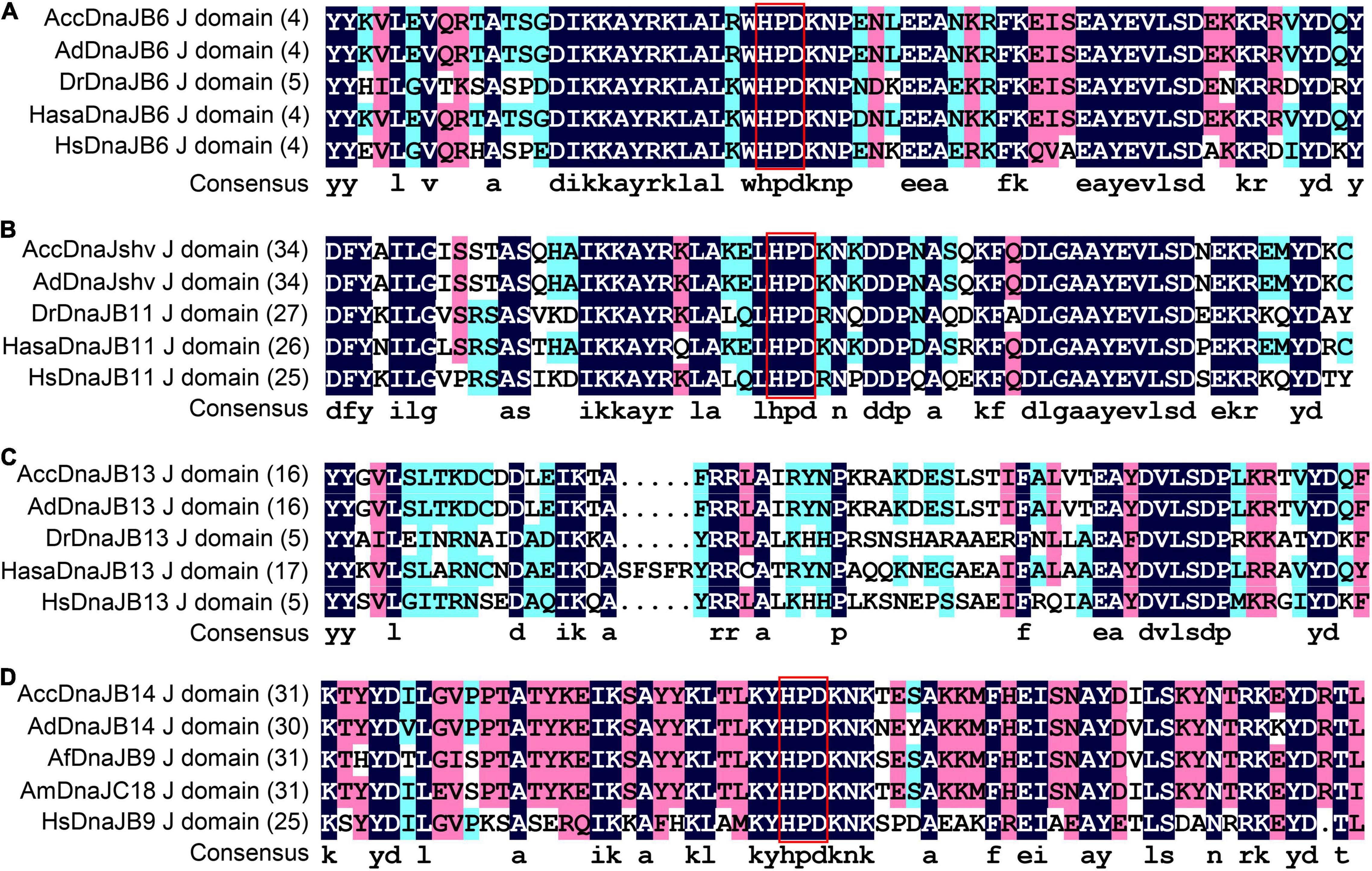
Figure 2. Multiple alignment of the J domain amino acid sequences of DnaJs. Multiple alignment of the J domains of DnaJ6 (A), DnaJshv (B), DnaJB13 (C), and DnaJB14 (D) from A. cerana cerana with those of other animal DnaJ proteins. The HPD tripeptide in DnaJs is shown in the red box. Acc, A. cerana cerana; Ad, Apis dorsata; Dr, Danio rerio; Hasa, Harpegnathos saltator; Hs, Homo sapiens; Af, Apis florea; Am, Apis mellifera.
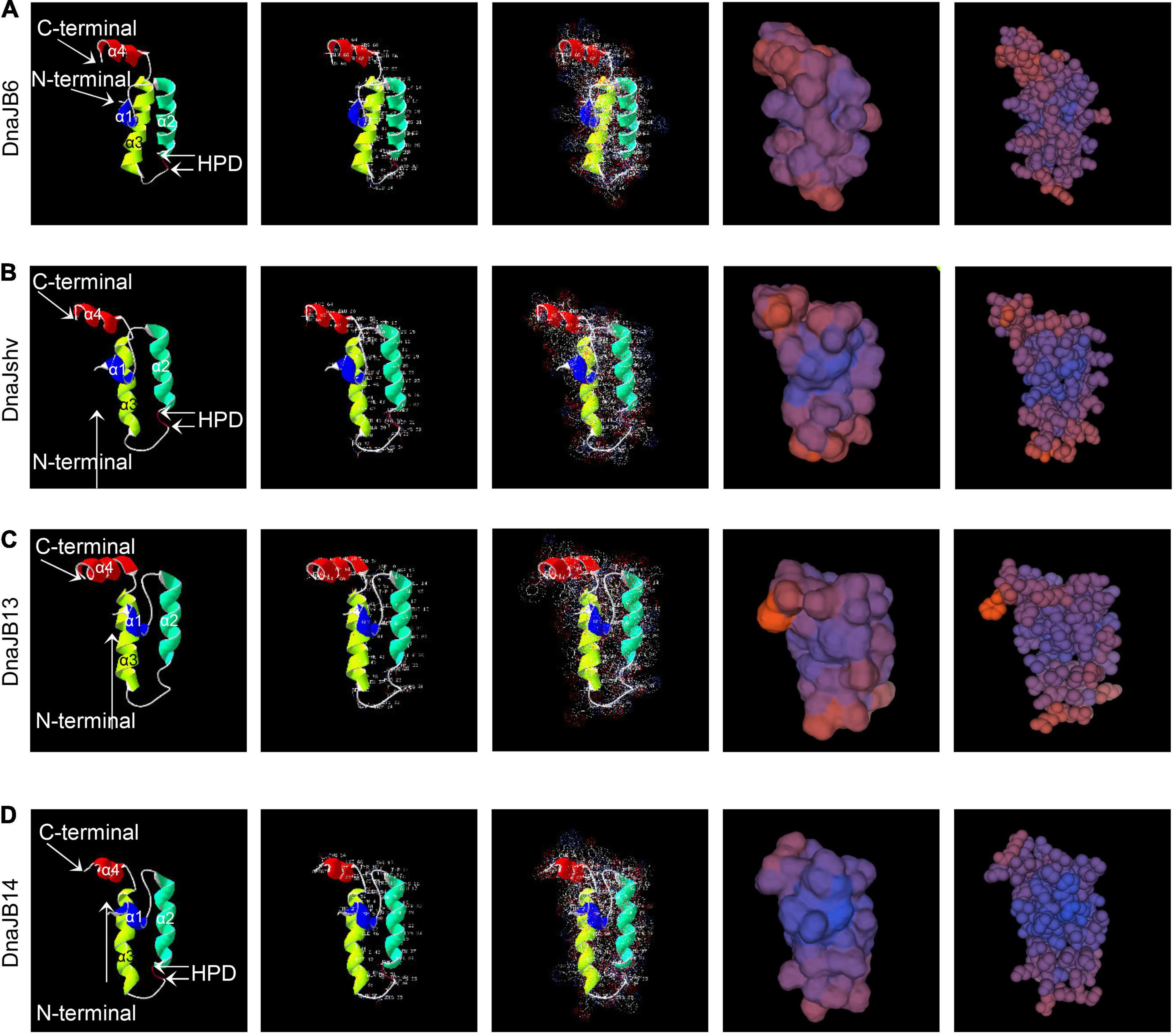
Figure 3. Tertiary structures of the J domains of A. cerana cerana DnaJB6 (A), DnaJshv (B), DnaJB13 (C), and DnaJB14 (D). The amino acid residues, surface electrostatic distributions, surfaces, and spacefill of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 are shown in the second, third, fourth, and fifth columns, respectively.
Expression Profiles of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 in Response to Heat Stress
The mRNA level of DnaJB6 was not significantly changed in the 33°C control group but was upregulated under 40, 43, and 46°C treatment within 5 h, and the degree of induction of DnaJB6 increased with increasing temperature from 4 to 5 h (Figure 4A). The expression level of DnaJshv was induced when A. cerana cerana was exposed to 40, 43, and 46°C compared to 33°C, and the degree of induction of DnaJshv was higher under the 43°C treatment than under the 40 and 46°C treatments from 2 to 5 h (Figure 4B). Although the expression level of DnaJB13 was downregulated at 1 and 2 h, its transcript level was upregulated at 4 and 5 h after 40, 43, and 46°C treatment (Figure 4C). The transcript level of DnaJB14 was increased under 43 and 46°C treatment but was not significantly changed at 33 and 40°C (Figure 4D). These findings reveal differential expression patterns of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 under different degrees of heat stress within 5 h.
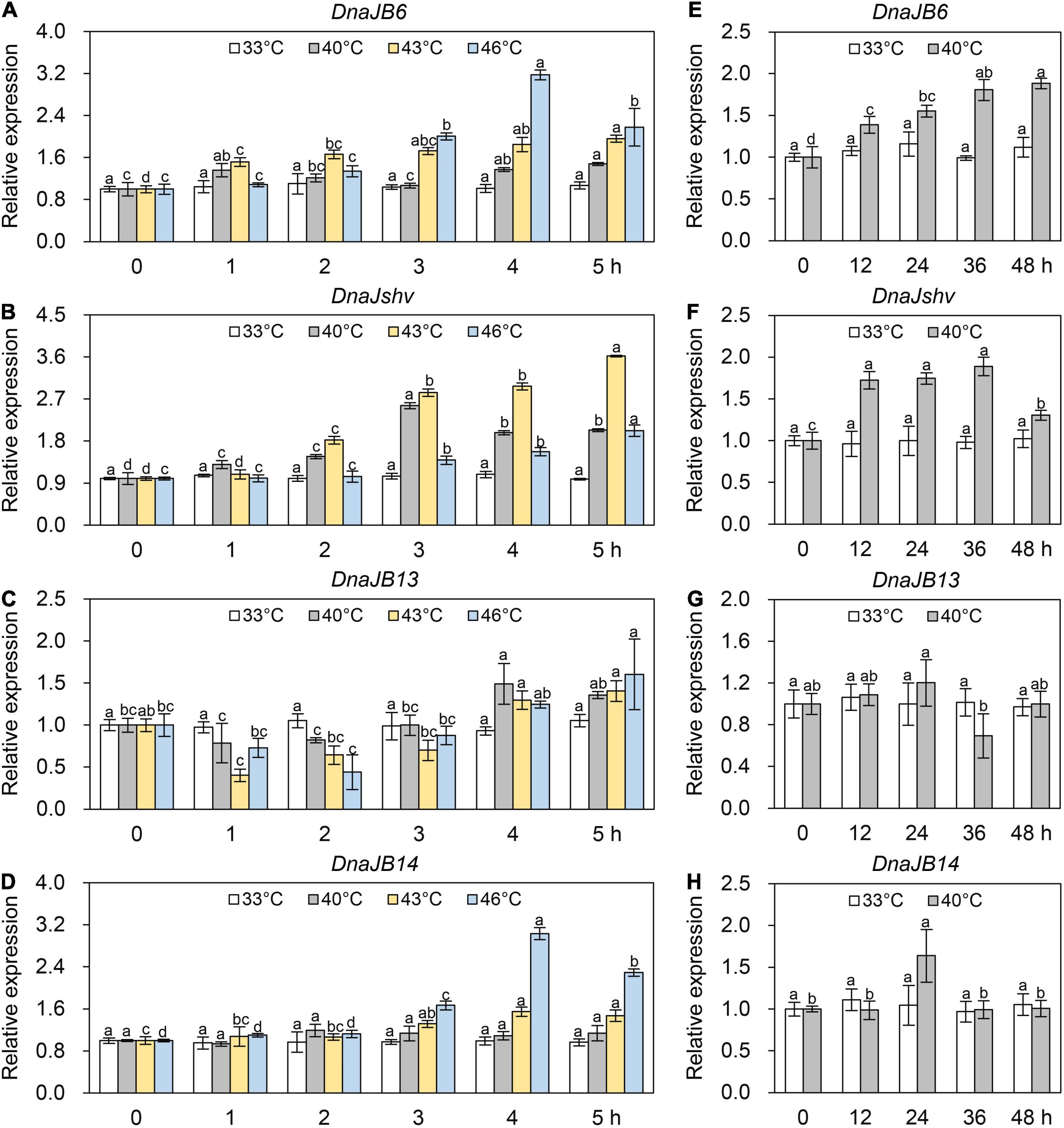
Figure 4. The expression levels of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 under heat stress. The mRNA levels of DnaJB6 (A), DnaJshv (B), DnaJB13 (C), and DnaJB14 (D) in A. cerana cerana exposed to 33, 40, 43, and 46°C treatment for 5 h. The expression profiles of DnaJB6 (E), DnaJshv (F), DnaJB13 (G), and DnaJB14 (H) in A. cerana cerana subjected to 33 and 40°C treatment for 48 h. β-actin served as the internal control. Different letters above the columns indicate significant differences between groups.
We prolonged the heat treatment time to further investigate the roles of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 under heat stress. As presented in Figure 4E, the mRNA level of DnaJB6 was not significantly altered by up to 48 h at 33°C but was induced at 40°C and reached its maximum levels after 48 h of treatment. The expression level of DnaJshv was increased under 40°C treatment for 48 h compared to 33°C (Figure 4F). The mRNA level of DnaJB13 was not significantly altered at 33°C but was inhibited after 36 h of 40°C treatment (Figure 4G). The transcript level of DnaJB14 was not significantly changed when A. cerana cerana was maintained at 33°C for 48 h and was upregulated at only 24 h after 40°C treatment (Figure 4H). These results further indicate that DnaJB6, DnaJshv, DnaJB13, and DnaJB14 respond to heat stress differently.
Expression Patterns of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 Under UV, Cold, and Various Agrochemical Stress Conditions
To investigate the response of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 to stresses other than heat, qRT–PCR was used. The expression levels of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 were induced at different levels under cold and UV stress (Figure 5). The mRNA level of DnaJB6 was inhibited under abamectin, imidacloprid, cypermethrin, bifenthrin, spirodiclofen, and methomyl stress, and reached its minimum level at 3, 1, 4, 5, 4, and 5 h, respectively (Figure 6A). The expression level of DnaJshv was decreased following abamectin, imidacloprid, cypermethrin, bifenthrin, spirodiclofen, and methomyl treatment, and reached its minimum level at 4, 4, 5, 5, 5, and 5 h, respectively (Figure 6B). Treatment with abamectin, imidacloprid, cypermethrin, bifenthrin, spirodiclofen, and methomyl for 5 h downregulated the transcription level of DnaJB13 (Figure 6C). The mRNA level of DnaJB14 was induced under abamectin and bifenthrin stress but suppressed under imidacloprid, cypermethrin, spirodiclofen, and methomyl stress (Figure 6D). These results indicate that DnaJB6, DnaJshv, DnaJB13, and DnaJB14 are crucial in the responses of A. cerana cerana to different stressors, and A. cerana cerana differentially requires four DnaJBs under different stress conditions.
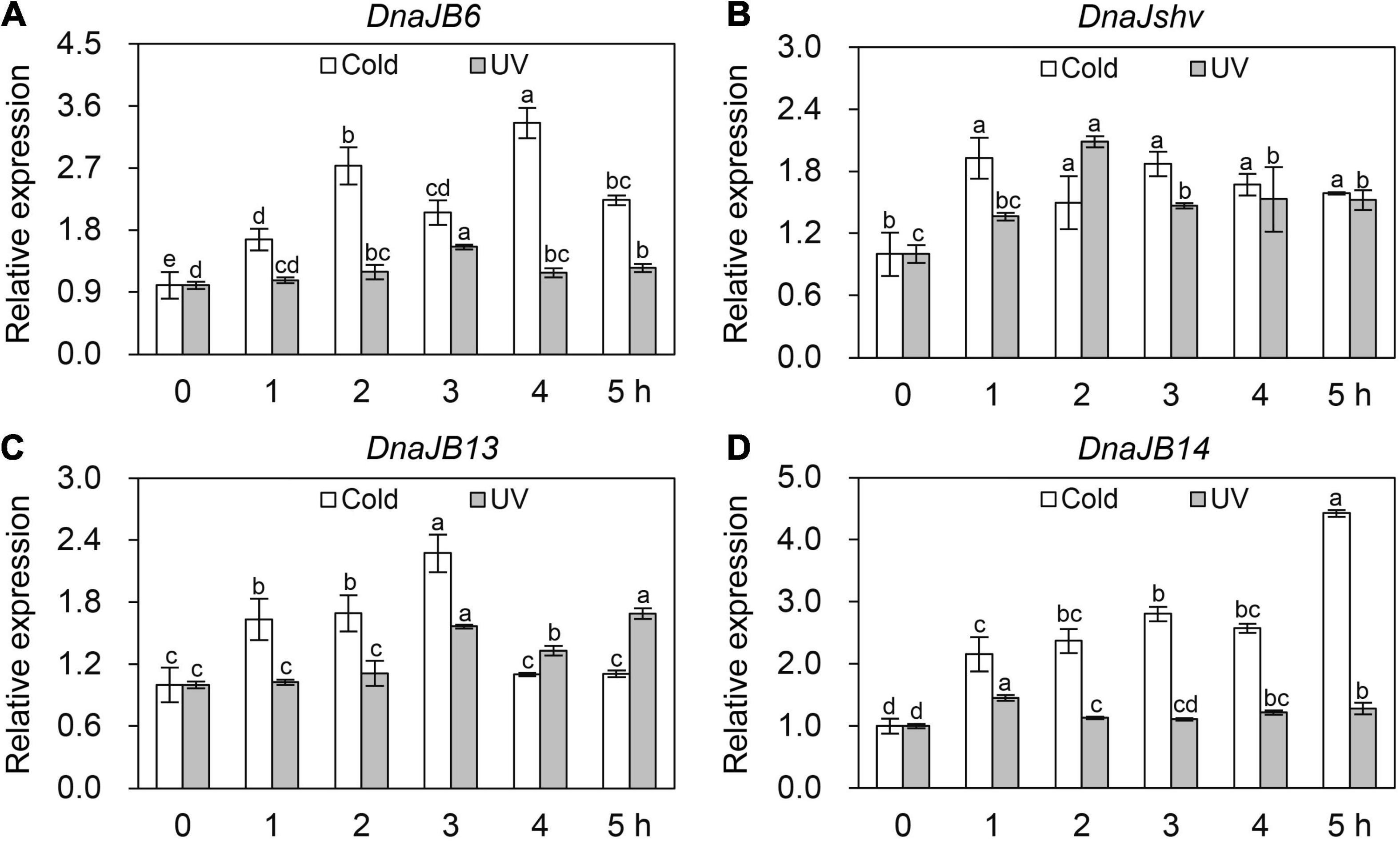
Figure 5. The transcript levels of DnaJB6 (A), DnaJshv (B), DnaJB13 (C) and DnaJB14 (D) under cold and UV stresses. Different letters above the columns indicate significant differences among the groups. β-actin served as the internal control.
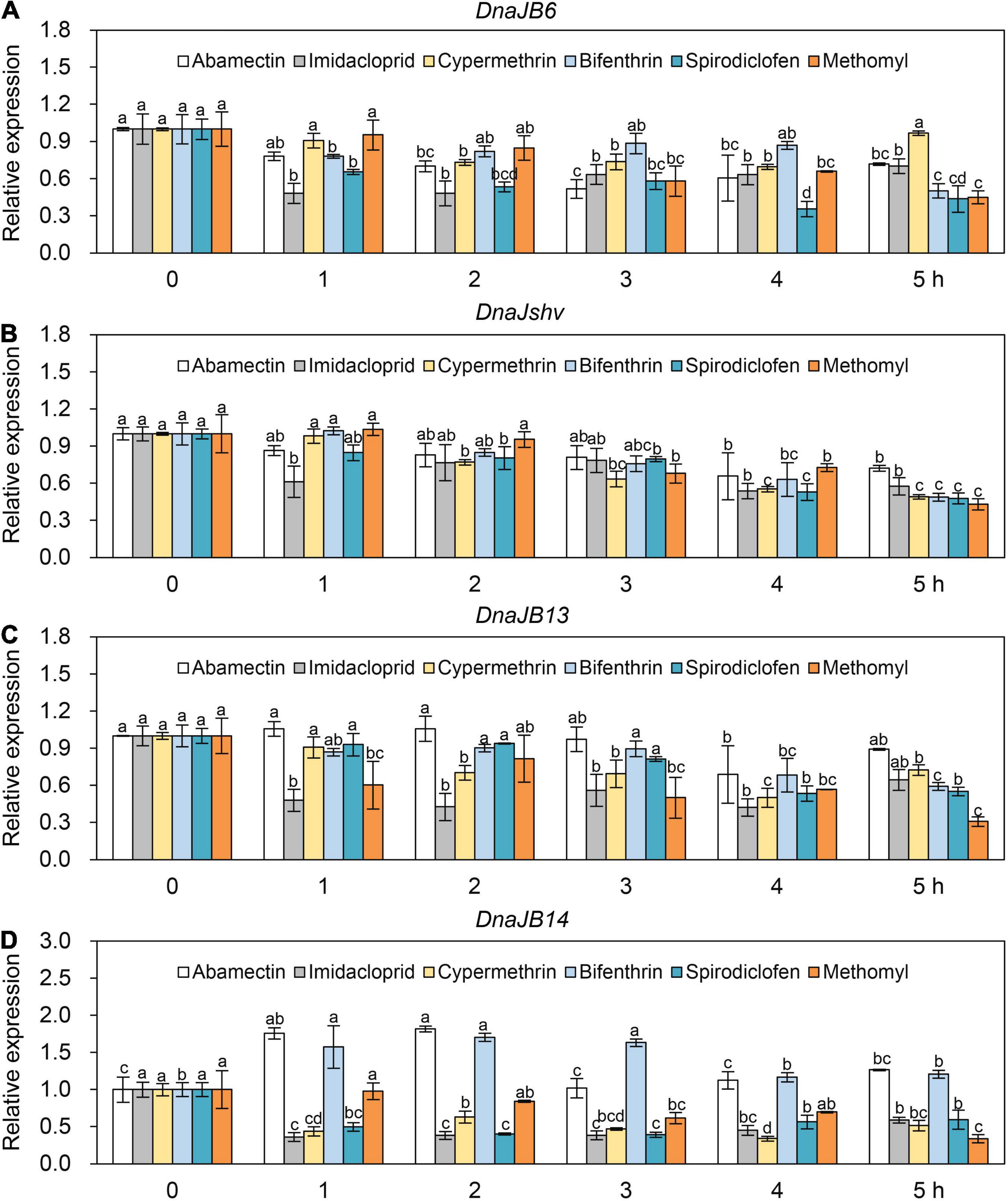
Figure 6. The mRNA levels of DnaJB6 (A), DnaJshv (B), DnaJB13 (C), and DnaJB14 (D) under various agrochemical stresses. The agrochemicals used here included abamectin, imidacloprid, cypermethrin, bifenthrin, spirodiclofen, and methomyl. qRT–PCR was performed to detect the expression levels of DnaJB6, DnaJshv, DnaJB13, and DnaJB14, with β-actin serving as the internal control. Significant differences among various groups are indicated by different letters above the columns.
Knockdown of DnaJshv and DnaJB14
To further explore the role of DnaJBs in A. cerana cerana, dsRNA-mediated RNAi experiments were performed. A. cerana cerana was fed dsRNAs targeting DnaJshv or DnaJB14, and the control was fed dsRNA targeting GFP. We then used qRT–PCR to determine the RNAi knockdown efficiencies of DnaJshv and DnaJB14 in A. cerana cerana. The mRNA levels of both DnaJshv and DnaJB14 were downregulated compared to the dsRNA-GFP control group after feeding with dsRNA-DnaJshv and dsRNA-DnaJB14, respectively (Supplementary Figure 6). These results indicate that DnaJshv and DnaJB14 can be successfully silenced by using dsRNA-DnaJshv and dsRNA-DnaJB14, respectively.
Influence of DnaJshv and DnaJB14 Knockdown on the mRNA Levels of Antioxidant Genes
To explore the influences of silencing DnaJshv and DnaJB14 on the mRNA levels of other antioxidant genes, we performed qRT–PCR assays. As shown in Figure 7A, knockdown of DnaJshv resulted in the downregulation of many antioxidant genes, including Tpx3, Trx1, Trx2, Trx-like1, GSTO2, GSTD, GSTS4, SOD1, CYP4G11, p38b, Hsp22.6, and MsrB. In contrast to DnaJshv, DnaJB14 silencing triggered the upregulation of many antioxidant genes, such as Tpx4, Tpx5, Trx2, GSTD, GSTT1, CDK5, SOD1, CDK5r, CYP4G11, and MsrB, and the downregulation of some antioxidant genes, including Tpx3, TrxR1, Trx-like1, GSTO2, GSTZ1, MKK4, and p38b (Figure 7B). These results indicate that the roles of DnaJshv and DnaJB14 in the stress response may involve the antioxidant signalling pathway by influencing the expression of antioxidant genes.
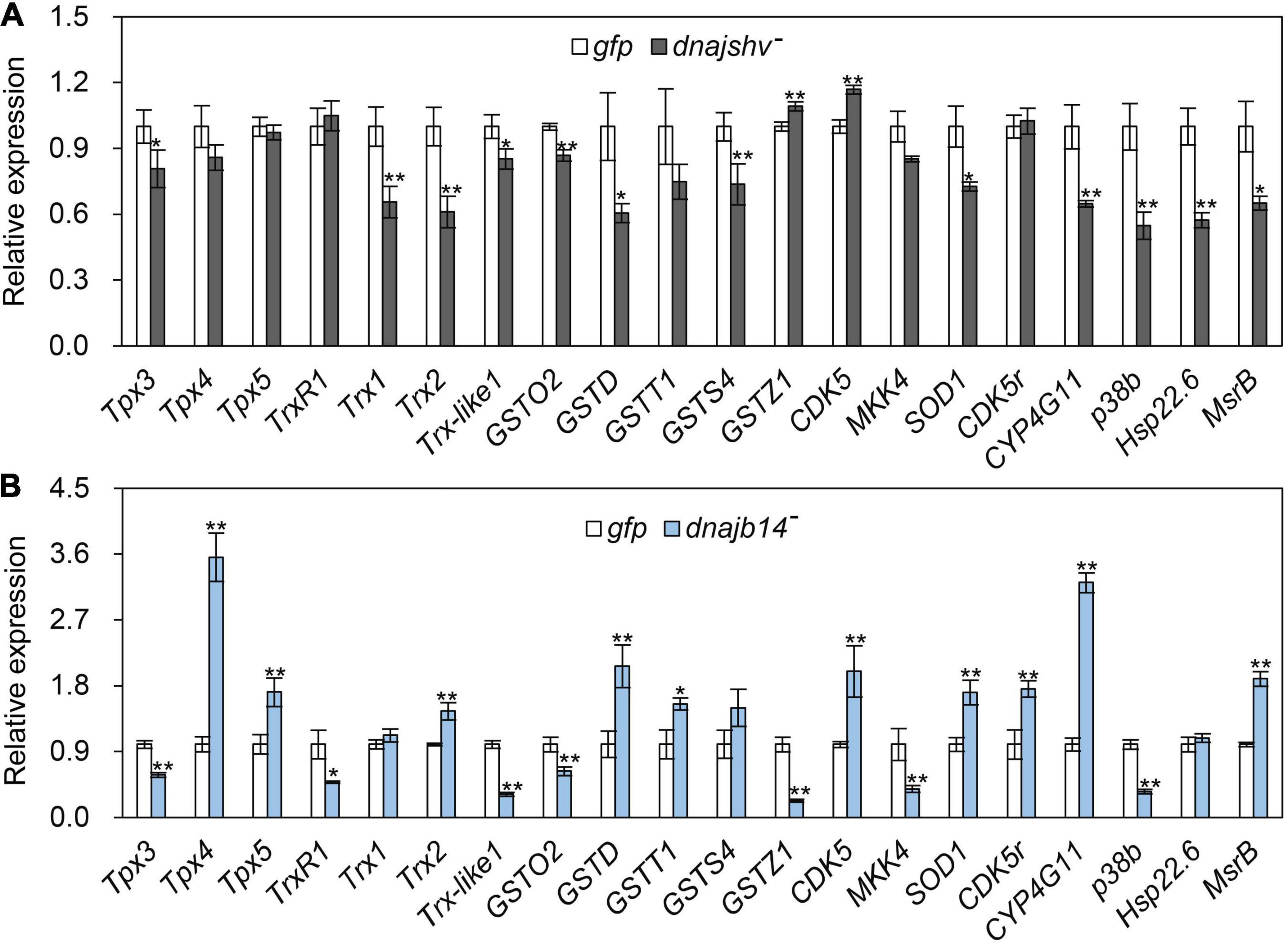
Figure 7. The influences of DnaJshv (A) and DnaJB14 (B) knockdown on antioxidant genes. The detected antioxidant genes included Tpx3, Tpx4, Tpx5, TrxR1, Trx1, Trx2, Trx-like1, GSTO2, GSTD, GSTT1, GSTS4, GSTZ1, CDK5, MKK4, SOD1, CDK5r, CYP4G11, p38b, Hsp22.6, and MsrB. β-actin was used as an internal control. **P < 0.01 and *P < 0.05 were considered statistically significant as determined by Student’s t-test. The abbreviations dnajshv–, dnajb14–, and gfp indicate DnaJshv knockdown, DnaJB14 knockdown, and the GFP control, respectively.
Effects of Silencing DnaJshv and DnaJB14 on the Enzymatic Activities of Antioxidant Enzymes and the Contents of Antioxidant- and Oxidant-Related Substances
To further investigate the mechanism underlying DnaJBs in response to stress conditions, we detected the enzymatic activities of antioxidant enzymes after the knockdown of DnaJshv and DnaJB14. As shown in Figure 8, DnaJshv knockdown decreased the enzymatic activities of POD, CAT, GST, and SOD. Silencing DnaJB14 reduced the enzymatic activities of CAT, GST, and SOD but had no significant effect on the enzymatic activity of POD (Figure 8). In addition, DnaJshv knockdown had no obvious impact on the MDA content but increased the protein carbonyl content and decreased the VC content (Figures 9A–C). Silencing DnaJB14 increased the contents of MDA and protein carbonyl and reduced the content of VC (Figures 9A–C). These results indicate that silencing DnaJshv and DnaJB14 may affect the antioxidant ability of A. cerana cerana by influencing antioxidant enzymes and oxidant- or antioxidant-related substances.
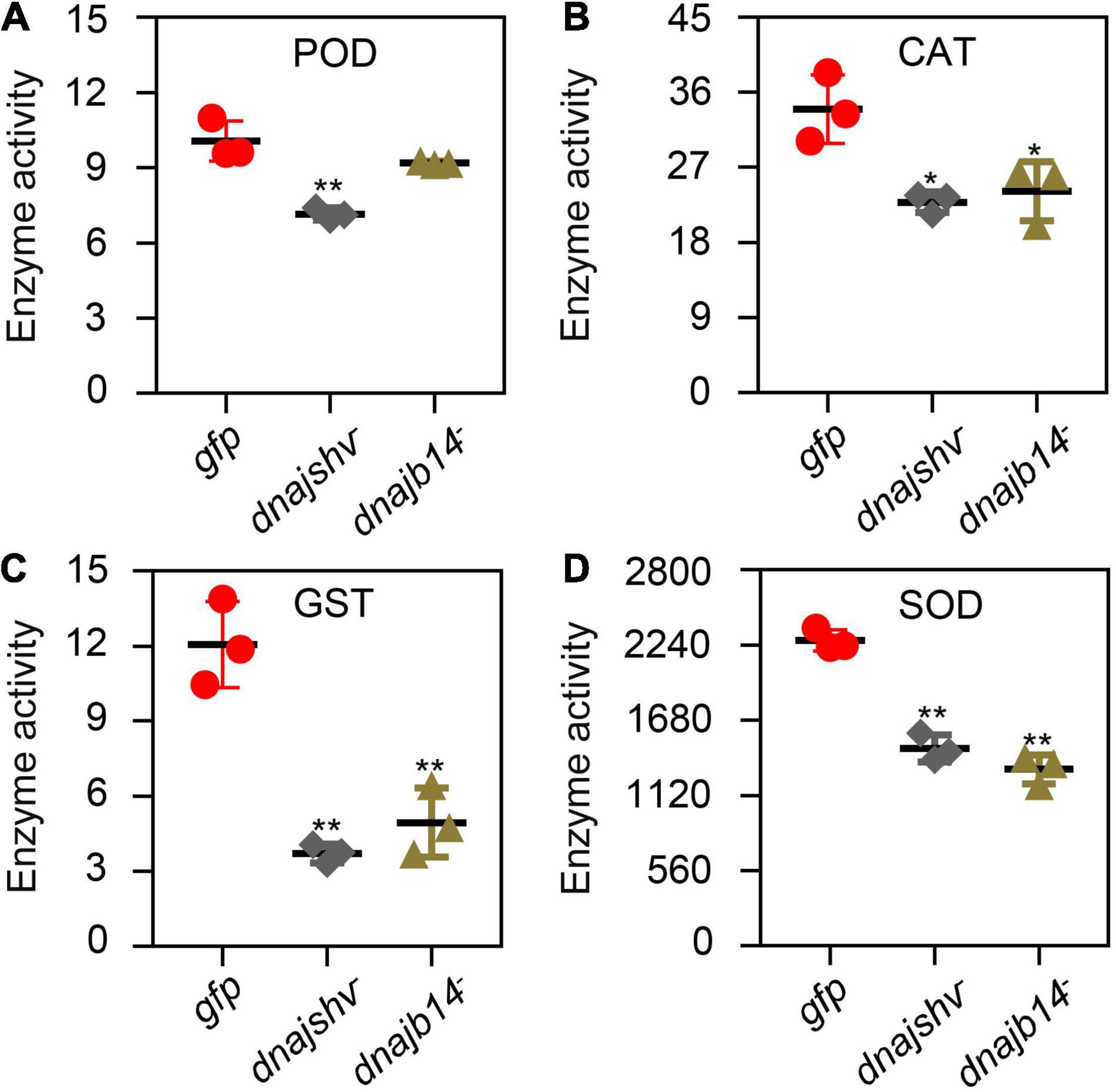
Figure 8. The enzymatic activities of antioxidant enzymes after DnaJshv and DnaJB14 knockdown. The enzymatic activities of POD (A), CAT (B), GST (C), and SOD (D) were detected after silencing DnaJshv and DnaJB14. **P < 0.01 and *P < 0.05 indicate significant differences as determined by Student’s t-test. dnajshv–, DnaJshv knockdown; dnajb14–, DnaJB14 knockdown; gfp, GFP control.
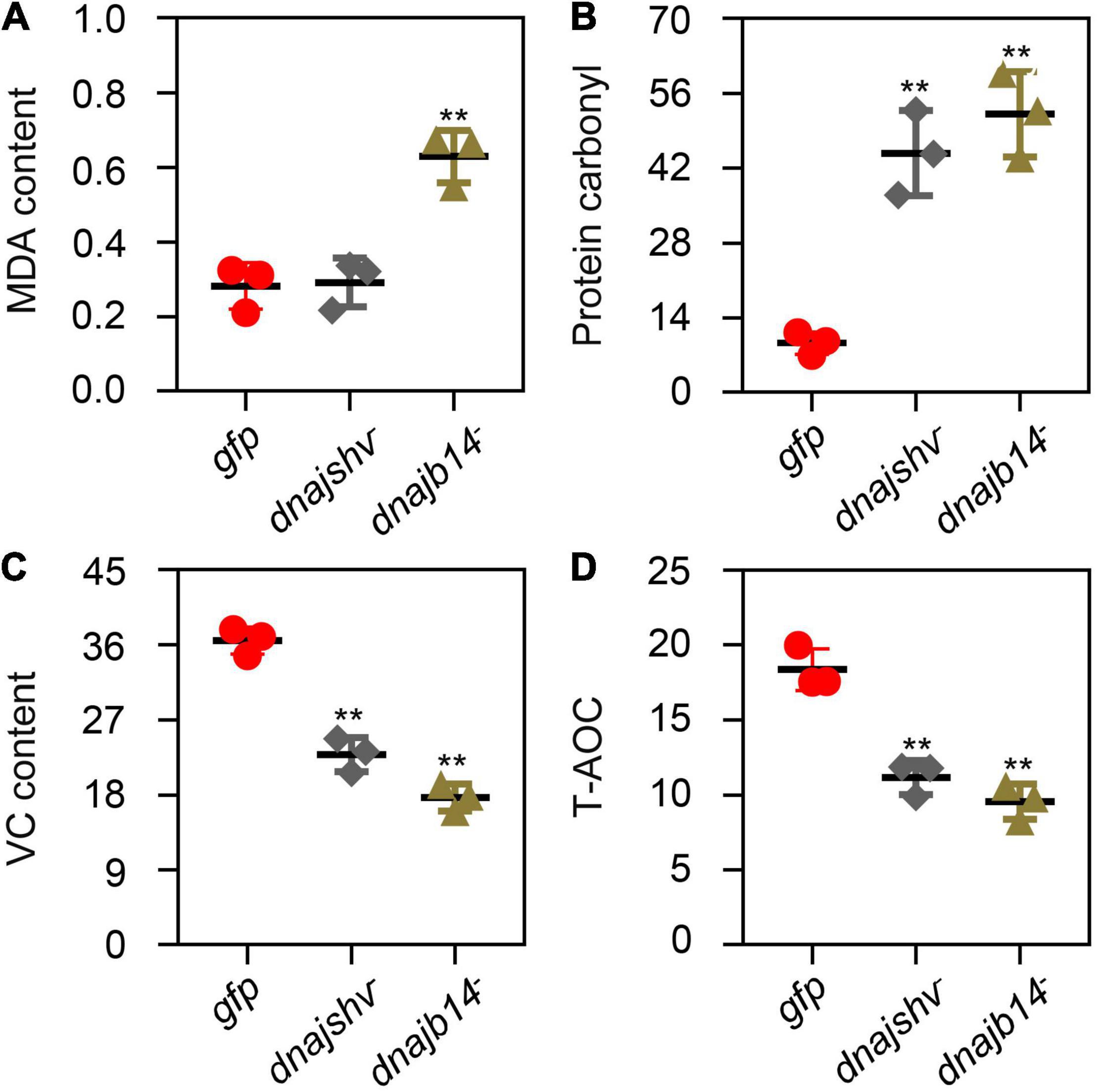
Figure 9. The content of oxidant- and antioxidant-related substances and the T-AOC of A. cerana cerana after DnaJshv and DnaJB14 knockdown. The contents of MDA (A), protein carbonyl (B), and VC (C) after silencing DnaJshv and DnaJB14. (D) The T-AOC of A. cerana cerana after DnaJshv and DnaJB14 knockdown. Significant differences are indicated by **P < 0.01 and *P < 0.05 as determined by Student’s t-test. DnaJshv knockdown, DnaJB14 knockdown and the GFP control are shown by dnajshv–, dnajb14–, and gfp, respectively.
Impacts of DnaJshv and DnaJB14 Knockdown on Bee Antioxidant Ability
To determine whether DnaJshv and DnaJB14 affect the antioxidant ability of bees, we detected the T-AOC of A. cerana cerana after silencing DnaJshv and DnaJB14. As presented in Figure 9D, knockdown of DnaJshv and DnaJB14 reduced the T-AOC of A. cerana cerana, indicating that DnaJshv and DnaJB14 play vital roles in the stress response by affecting the antioxidant ability of A. cerana cerana.
Discussion
Different stresses, such as heat, cold, UV, and agrochemicals, are the primary threat to the population, health, and fitness of bees (Potts et al., 2010; Even et al., 2012). Understanding the molecular defence systems used by bees is vital for decreasing the negative impacts of various stresses. Nonetheless, data on the mechanisms by which bees protect themselves from stresses at the molecular level remain limited, especially those mediated by key genes. Here, we showed that the expression profiles of DnaJB6, DnaJshv, DnaJB13, and DnaJ14 differed under different stress conditions, and these four DnaJBs played crucial roles in the A. cerana cerana stress response.
The typical feature of DnaJ protein is that it contains a J domain (Craig and Marszalek, 2017). We found that all four DnaJBs have a J domain with four α-helices (Figure 3). An HPD tripeptide is located between helix II and helix III of the J domain of DnaJB6, DnaJshv, and DnaJB14, while the J domain of DnaJB13 lacks this tripeptide (Figures 2, 3). The HPD tripeptide is crucial for the J domain, which can stimulate the ATPase activity of HSP70 (Kampinga and Craig, 2010). The orthologs of DnaJB13 from D. rerio, Harpegnathos saltator and H. sapiens also lack the HPD tripeptide (Figure 2C), while those of DnaJB13 from Chlamydomonas reinhardtii have the HPD tripeptide (Yang et al., 2005). The lack of HPD tripeptide in the J domain may increase the functional significance of protein portions other than the J domain.
The functions of some DnaJB subfamily members have been revealed by previous studies. For example, it has been shown that DnaJBs inhibit protein aggregation by supporting Hsp70-dependent degradation of the double mutant isoform of firefly luciferase (McMahon et al., 2021). DnaJB8 and DnaJB6 decrease polyglutamine aggregation by binding the polyQ tract (Gillis et al., 2013). Primary ciliary dyskinesia and male infertility are caused by mutation of DnaJB13 (El Khouri et al., 2016). Nevertheless, these studies are mainly focused on the role of DnaJB in diseases. Our previous studies annotated five DnaJBs, DnaJB6, DnaJshv, DnaJB12, DnaJB13, and DnaJB14, in A. cerana cerana, revealed that DnaJB12 plays a vital role in A. cerana cerana under different stress conditions, and silencing DnaJB12 increases the mortality of A. cerana cerana under heat stress and lambda-cyhalothrin stress (Li et al., 2018, 2020a). However, the specific functions and regulatory mechanisms of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 in bees have not been clearly elucidated. To investigate the functions of DnaJBs, we first assessed their expression profiles under heat stress. DnaJB6, DnaJshv, DnaJB13, and DnaJB14 respond differently to various degrees of heat stress, and the transcript levels of DnaJB6, DnaJshv, and DnaJB14 were significantly increased by heat stress (Figure 4). It has been shown that 50% or more of the sperm stored in live queen spermathecae are killed when the temperature exceeds 40°C (Pettis et al., 2016). Recent studies have demonstrated that heat stress during development influences honey bee immunocompetence (Medina et al., 2020), and thermal stress causes a regenerative signalling pathway shift and tissue damage in bees (Bach et al., 2021). Therefore, the upregulation of DnaJBs may play crucial roles in helping bees reduce the adverse effects of heat stress.
Although Hsps are important for heat stress responses, some evidence suggests that they also respond to agrochemical stresses (King and MacRae, 2015; Perez-Rodriguez et al., 2020). For example, the mRNA level of Hsp70 was significantly induced by toxaphene treatment in D. rerio early-stage (Perez-Rodriguez et al., 2020). The female dung beetle Euoniticellus intermedius exhibited increased expression of Hsp70 upon exposure to ivermectin (Villada-Bedoya et al., 2021). The mRNA levels of Hsp60, Hsp70, and Hsp90 in Eriocheir sinensis were shown to be upregulated by deltamethrin treatment (Hong et al., 2019). The transcription of DnaJA1, DnaJB12, and DnaJC8 was induced by the subjection of A. cerana cerana to paraquat, emamectin benzoate, lambda-cyhalothrin, avermectin, and spirodiclofen treatment (Li et al., 2018). We found that the expression levels of DnaJB6, DnaJshv, and DnaJB13 were inhibited under various agrochemical stresses, while the transcript level of DnaJB14 was induced under abamectin and bifenthrin stress (Figure 6). These results indicate that DnaJB6, DnaJshv, DnaJB13, and DnaJB14 respond to agrochemical stresses, and their regulatory methods differ under different agrochemical conditions.
Except for heat and agrochemical stresses, we found that the expression levels of DnaJB6, DnaJshv, DnaJB13, and DnaJB14 were increased to different degrees under cold and UV stress (Figure 5), indicating that these four DnaJBs function in the defence of A. cerana cerana against cold and UV stress by increasing their expression. Previous studies have demonstrated that many environmental stresses, including heat, agrochemical, cold, and UV stress, can generate high levels of reactive oxygen species (ROS), which mainly include H2O2, superoxide anions and hydroxyl radicals. The physiological levels of ROS have beneficial impacts on health, such as acting as second messengers and promoting host defence mechanisms. However, high levels of ROS have adverse health effects, including triggering oxidative damage to cellular proteins, DNA, and lipids (Finkel and Holbrook, 2000; Venditti et al., 2004; Paravani et al., 2019; Atli Sekeroglu et al., 2020). Changes in DnaJB6, DnaJshv, DnaJB13, and DnaJB14 at the transcript level may help A. cerana cerana cope with oxidative damage triggered by environmental stresses. In addition, MDA and protein carbonyl have been used as biomarkers of oxidative damage (Nabeshi et al., 2006; Lykkesfeldt, 2007). We found that knockdown of DnaJshv increased the content of protein carbonyl and that silencing DnaJB14 increased the contents of both protein carbonyl and MDA (Figures 9A–C), indicating that knockdown of DnaJshv and DnaJB14 increased the oxidative damage in A. cerana cerana.
The degree of oxidative damage was mainly determined by assessing the balance between antioxidant defence and the production of ROS. A sophisticated antioxidant defence system of non-enzymes, such as VC, and enzymes, including POD, CAT, GST, and SOD, plays a crucial role in maintaining this balance (Finkel and Holbrook, 2000; Khan et al., 2014). Previous research has suggested that VC can upregulate the expression levels of Hsp70, Hsp60, and Hsp27 and increase the antioxidant ability of H9C2 cells to relieve heat stress (Yin et al., 2018). POD is related to oxyradical oxidation, and plays an important role in the antioxidant defence systems of living organisms (Finkel and Holbrook, 2000). CAT can prevent cellular oxidative damage by catalysing the conversion of H2O2 into oxygen and water, and CAT deficiency triggers ROS-mediated cell death and pexophagy during prolonged fasting in the mouse liver (Glorieux and Calderon, 2017; Dutta et al., 2021). Previous research has suggested that GST, as an antioxidant defence agent, promotes pyrethroid resistance in Nilaparvata lugens (Vontas et al., 2001). SOD can convert superoxide anions into oxygen and H2O2, and H2O2 is then reduced to water by peroxiredoxins, CAT and glutathione peroxidases, thereby playing important roles in the antioxidant defence system (Fukai and Ushio-Fukai, 2011). Knockdown of mitochondrial manganese SOD increases oxidative stress in Litopenaeus vannamei under hypoxic, reoxygenation, and high-temperature conditions (Gonzalez-Ruiz et al., 2021). We found that the mRNA levels of many antioxidant-related genes were altered, the enzymatic activities of SOD, GST, and CAT, the content of VC and T-AOC were reduced after DnaJshv and DnaJB14 knockdown (Figures 7–9). In addition, silencing DnaJshv reduced the enzymatic activity of POD (Figure 8A). Therefore, we conclude that DnaJB11 and DnaJB14 play vital roles in reducing the oxidative damage triggered by various stresses through the antioxidant defence system.
Conclusion
In conclusion, our findings suggest that DnaJB6, DnaJshv, DnaJB13, and DnaJB14 play vital roles in the stress response, and A. cerana cerana needs these four genes to different extents under various environmental stress conditions. Knockdown of DnaJshv and DnaJB14 decreased the T-AOC of A. cerana cerana, and DnaJBs may have alleviated the oxidative damage triggered by environmental stresses through the antioxidant defence system. Our results will contribute to understanding the underlying regulatory mechanism mediated by DnaJBs upon the exposure of A. cerana cerana to diverse environmental stresses.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HZ and BX designed the experiments. GL, HZ, and SZ performed the experimental work. GL, HZ, BX, HW, and LL analysed the data. GL and HZ wrote the manuscript. All authors approved the final version of the manuscript.
Funding
This work was financially supported by the earmarked fund for the China Agriculture Research System (No. CARS-44) and the Special Funds for Taishan Industry Leading Talent Project (No. LJNY202003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.873791/full#supplementary-material
Footnotes
References
Annoscia, D., Di Prisco, G., Becchimanzi, A., Caprio, E., Frizzera, D., Linguadoca, A., et al. (2020). Neonicotinoid Clothianidin reduces honey bee immune response and contributes to Varroa mite proliferation. Nat. Commun. 11:5887.
Atli Sekeroglu, Z., Sekeroglu, V., Aydin, B., Kontas Yedier, S., and Ilkun, E. (2020). Clothianidin induces DNA damage and oxidative stress in bronchial epithelial cells. Environ. Mol. Mutagen. 61, 647–655.
Bach, D. M., Holzman, M. A., Wague, F., Miranda, J. L., Lopatkin, A. J., Mansfield, J. H., et al. (2021). Thermal stress induces tissue damage and a broad shift in regenerative signaling pathways in the honey bee digestive tract. J. Exper. Biol. 224:jeb242262.
Chand, K., Iyer, K., and Mitra, D. (2021). Comparative analysis of differential gene expression of HSP40 and HSP70 family isoforms during heat stress and HIV-1 infection in T-cells. Cell Stress Chaperones 26, 403–416.
Chen, C., Liu, Z., Luo, Y., Xu, Z., Wang, S., Zhang, X., et al. (2017). Managed honeybee colony losses of the Eastern honeybee (Apis cerana) in China (2011–2014). Apidologie 48, 692–702.
Cornec-Le Gall, E., Olson, R. J., Besse, W., Heyer, C. M., Gainullin, V. G., Smith, J. M., et al. (2018). Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am. J. Hum. Genet. 102, 832–844.
Craig, E. A., and Marszalek, J. (2017). How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 42, 355–368.
Dutta, R. K., Maharjan, Y., Lee, J. N., Park, C., Ho, Y. S., and Park, R. (2021). Catalase deficiency induces reactive oxygen species mediated pexophagy and cell death in the liver during prolonged fasting. BioFactors 47, 112–125.
El Khouri, E., Thomas, L., Jeanson, L., Bequignon, E., Vallette, B., Duquesnoy, P., et al. (2016). Mutations in DNAJB13, encoding an HSP40 family member, cause primary ciliary dyskinesia and male infertility. Am. J. Hum. Gen. 99, 489–500.
Even, N., Devaud, J. M., and Barron, A. B. (2012). General stress responses in the honey bee. Insects 3, 1271–1298.
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Ann. Rev. Physiol. 61, 243–282.
Finkel, T., and Holbrook, N. J. (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247.
Fukai, T., and Ushio-Fukai, M. (2011). Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 15, 1583–1606.
Gillis, J., Schipper-Krom, S., Juenemann, K., Gruber, A., Coolen, S., van den Nieuwendijk, R., et al. (2013). The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J. Biol. Chem. 288, 17225–17237.
Glorieux, C., and Calderon, P. B. (2017). Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 398, 1095–1108.
Gonzalez-Ruiz, R., Peregrino-Uriarte, A. B., Valenzuela-Soto, E. M., Cinco-Moroyoqui, F. J., Martinez-Tellez, M. A., and Yepiz-Plascencia, G. (2021). Mitochondrial manganese superoxide dismutase knock-down increases oxidative stress and caspase-3 activity in the white shrimp Litopenaeus vannamei exposed to high temperature, hypoxia, and reoxygenation. Comp. Biochem. Physiol. Part A 252:110826.
Goodwin, E. C., Motamedi, N., Lipovsky, A., Fernandez-Busnadiego, R., and DiMaio, D. (2014). Expression of DNAJB12 or DNAJB14 causes coordinate invasion of the nucleus by membranes associated with a novel nuclear pore structure. PloS One 9:e94322. doi: 10.1371/journal.pone.0094322
Goulson, D., Nicholls, E., Botias, C., and Rotheray, E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347:1255957.
Hong, Y., Huang, Y., Yan, G., and Huang, Z. (2019). Effects of deltamethrin on the antioxidant defense and heat shock protein expression in Chinese mitten crab. Eriocheir sinensis. Environ. Toxicol. Pharmacol. 66, 1–6.
Kaida, A., Yamamoto, S., Parrales, A., Young, E. D., Ranjan, A., Alalem, M. A., et al. (2021). DNAJA1 promotes cancer metastasis through interaction with mutant p53. Oncogene 40, 5013–5025.
Kampinga, H. H., and Craig, E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev Mol. Cell Biol. 11, 579–592.
Khan, A. A., Rahmani, A. H., Aldebasi, Y. H., and Aly, S. M. (2014). Biochemical and pathological studies on peroxidases -an updated review. Glob. J. Health Sci. 6, 87–98.
King, A. M., and MacRae, T. H. (2015). Insect heat shock proteins during stress and diapause. Ann. Rev. Entomol. 60, 59–75.
Li, G., Zhao, H., Guo, H., Wang, Y., Cui, X., Li, H., et al. (2020a). Analyses of the function of DnaJ family proteins reveal an underlying regulatory mechanism of heat tolerance in honeybee. Sci. Total Environ. 716:137036.
Li, G., Zhao, H., Zhang, X., Zhang, Y., Zhao, H., Yang, X., et al. (2018). Environmental stress responses of DnaJA1. DnaJB12 and DnaJC8 in Apis cerana cerana. Front. Genet. 9:445. doi: 10.3389/fgene.2018.00445
Li, W. N., Zhu, L., Jia, M. M., Yin, S. L., Lu, G. X., and Liu, G. (2020b). Missense mutation in DNAJB13 gene correlated with male fertility in asthenozoospermia. Andrology 8, 299–306.
Lykkesfeldt, J. (2007). Malondialdehyde as biomarker of oxidative damage to lipids caused by smoking. Clin. Chim. Acta 380, 50–58.
McKinstry, M., Chung, C., Truong, H., Johnston, B. A., and Snow, J. W. (2017). The heat shock response and humoral immune response are mutually antagonistic in honey bees. Scie. Rep. 7:8850.
McMahon, S., Bergink, S., Kampinga, H. H., and Ecroyd, H. (2021). DNAJB chaperones suppress destabilised protein aggregation via a region distinct from that used to inhibit amyloidogenesis. J. Cell Sci. 134:jcs255596
Medina, R. G., Paxton, R. J., Hernandez-Sotomayor, S. M. T., Pech-Jimenez, C., Medina-Medina, L. A., and Quezada-Euan, J. J. G. (2020). Heat stress during development affects immunocompetence in workers, queens and drones of Africanized honey bees (Apis mellifera L.) (Hymenoptera: Apidae). J. Therm. Biol. 89:102541.
Nabeshi, H., Oikawa, S., Inoue, S., Nishino, K., and Kawanishi, S. (2006). Proteomic analysis for protein carbonyl as an indicator of oxidative damage in senescence-accelerated mice. Free Radic. Res. 40, 1173–1181.
Paravani, E. V., Simoniello, M. F., Poletta, G. L., and Casco, V. H. (2019). Cypermethrin induction of DNA damage and oxidative stress in zebrafish gill cells. Ecotoxicol. Environ. Safety 173, 1–7.
Perez-Rodriguez, V., Wu, N., de la Cova, A., Schmidt, J., Denslow, N. D., and Martyniuk, C. J. (2020). The organochlorine pesticide toxaphene reduces non-mitochondrial respiration and induces heat shock protein 70 expression in early-staged zebrafish (Danio rerio). Comp. Biochem. Physiol. Toxicol. Pharmacol. 228:108669.
Pettis, J. S., Rice, N., Joselow, K., vanEngelsdorp, D., and Chaimanee, V. (2016). Colony failure linked to low sperm viability in honey bee (Apis mellifera) queens and an exploration of potential causative factors. PloS One 11:e0147220. doi: 10.1371/journal.pone.0147220
Potts, S. G., Biesmeijer, J. C., Kremen, C., Neumann, P., Schweiger, O., and Kunin, W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353.
Qiu, X. B., Shao, Y. M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570.
Siviter, H., Bailes, E. J., Martin, C. D., Oliver, T. R., Koricheva, J., Leadbeater, E., et al. (2021). Agrochemicals interact synergistically to increase bee mortality. Nature 596, 389–392.
Sørensen, J. G., Kristensen, T. N., and Loeschcke, V. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6, 1025–1037.
Theisen-Jones, H., and Bienefeld, K. (2016). The Asian honey bee (Apis cerana) is significantly in decline. Bee World 93, 90–97.
Thiruvalluvan, A., de Mattos, E. P., Brunsting, J. F., Bakels, R., Serlidaki, D., Barazzuol, L., et al. (2020). DNAJB6, a key factor in neuronal sensitivity to amyloidogenesis. Mol. Cell 78, 346–358.e9.
Uno, Y., Kanda, M., Miwa, T., Umeda, S., Tanaka, H., Tanaka, C., et al. (2019). Increased expression of DNAJC12 is associated with aggressive phenotype of gastric cancer. Ann. Surg. Oncol. 26, 836–844.
Venditti, P., De Rosa, R., Portero-Otin, M., Pamplona, R., and Di Meo, S. (2004). Cold-induced hyperthyroidism produces oxidative damage in rat tissues and increases susceptibility to oxidants. Int. J. Biochem. Cell Biol. 36, 1319–1331.
Villada-Bedoya, S., Chavez-Rios, J. R., Montoya, B., Castelan, F., Cordoba-Aguilar, A., Escobar, F., et al. (2021). Heat shock proteins and antioxidants as mechanisms of response to ivermectin in the dung beetle Euoniticellus intermedius. Chemosphere 269:128707.
Vontas, J. G., Small, G. J., and Hemingway, J. (2001). Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 357, 65–72.
Wang, Q., Xu, X., Zhu, X., Chen, L., Zhou, S., Huang, Z. Y., et al. (2016). Low-temperature stress during capped brood stage increases pupal mortality, misorientation and adult mortality in honey bees. PloS One 11:e0154547. doi: 10.1371/journal.pone.0154547
Yang, C., Compton, M. M., and Yang, P. (2005). Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol. Biol. Cell 16, 637–648.
Keywords: bees, environmental stresses, DnaJBs, expression profile, antioxidant
Citation: Li G, Zhang S, Wang H, Liang L, Liu Z, Wang Y, Xu B and Zhao H (2022) Differential Expression Characterisation of the Heat Shock Proteins DnaJB6, DnaJshv, DnaJB13, and DnaJB14 in Apis cerana cerana Under Various Stress Conditions. Front. Ecol. Evol. 10:873791. doi: 10.3389/fevo.2022.873791
Received: 11 February 2022; Accepted: 29 March 2022;
Published: 28 April 2022.
Edited by:
Vera Maria Fonseca Almeida-Val, National Institute of Amazonian Research (INPA), BrazilCopyright © 2022 Li, Zhang, Wang, Liang, Liu, Wang, Xu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Zhao, aGFuZ3poYW9sZ2xAMTYzLmNvbQ==; Baohua Xu, Ymh4dUBzZGF1LmVkdS5jbg==
 Guilin Li
Guilin Li Shuxin Zhang
Shuxin Zhang Hongfang Wang
Hongfang Wang Li Liang
Li Liang Zhenguo Liu
Zhenguo Liu Ying Wang
Ying Wang Baohua Xu
Baohua Xu Hang Zhao
Hang Zhao