- 1Laboratory of Systematic Evolution and Biogeography of Woody Plants, School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, China
- 2Xianxialing Provincial Nature Reserve Management Center, Jiangshan, China
Bretschneidera sinensis is an endangered woody species found in East and South China. Comprehensive intraspecies chloroplast genome studies have demonstrated novel genetic resources to assess the genetic variation and diversity of this species. Using genome skimming method, we assembled the whole chloroplast genome of 12 genotypes of B. sinensis from different geographical locations, covering most wild populations. The B. sinensis chloroplast genome size ranged from 158,959 to 159,045 base pairs (bp) and displayed a typical circular quadripartite structure. Comparative analyses of 12 B. sinensis chloroplast genome revealed 33 polymorphic simple sequence repeats (SSRs), 105 polymorphic single nucleotide polymorphisms (SNPs), and 55 indels. Phylogenetic analysis showed that the 12 genotypes were grouped into 2 branches, which is consistent with the geographical distribution (Eastern clade and Western clade). Divergence time estimates showed that the two clades were divergent from 0.6 Ma in the late Pleistocene. Ex situ conservation is essential for this species. In this study, we identified SNPs, indels, and microsatellites of B. sinensis by comparative analyses of chloroplast genomes and determined genetic variation between populations using these genomic markers. Chloroplast genomic resources are also important for further domestication, population genetic, and phylogenetic analysis, possibly in combination with molecular markers of mitochondrial and/or nuclear genomes.
Introduction
Bretschneidera sinensis Hemsl. is an endangered species endemic to south China and the adjacent area in Vietnam, and is infrequent in evergreen broad-leaved or mixed evergreen and deciduous forests (Lu and Boufford, 2005). According to morphological evidence, this species was placed in Sapindaceae, as the only species of the monogeneric family, Bretschneideraceae (Tobe and Peng, 1990; Tobe and Raven, 1995). Phylogenetic studies have shown that Bretschneidera is a monotypic genus of Akaniaceae, which compose of only two species, B. sinensis and Akania bidwillii (Hend. ex R.Hogg) Mabb. (The Angiosperm Phylogeny, 2016).
Bretschneidera sinensis is a relict species, and its population has declined with habitat destruction and fragmentation, due to deforestation and destructive collection of seedlings (Wang et al., 2008; Qi et al., 2009). On the other hand, its slow growing speed, predominant outcrossing reproductive system and short-distant dispersal seeds are significant obstacles to its natural regeneration (Wang et al., 2011). This species has a low pollination rate and survivorship of seeds (Qi et al., 2009; Qiao et al., 2012), and global warming may shrink its distribution, further aggravating its extinction (Guo et al., 2020). B. sinensis is listed on the IUCN Red List of threatened species as “endangered,” and is included in the “Wild plants of national priority protection in China (Category-II).”
Delineation of genetic diversity is a critical step for endangered species in plant conservation, especially for determining management policies and protective units. Understanding genetic diversity and genetic divergence will also help avoid the potential risks of inbreeding depression and clarify the causes of declining population sizes (Escudero et al., 2003). Species with high genetic diversity have more alleles, and more polymorphisms may alter responses to environmental changes; thus, these plants may exhibit greater adaptability (Ellegren and Galtier, 2016). Plant species with lower genetic diversity may suffer bottlenecks or long periods with small population sizes and possible eventual extinction. Knowledge and evaluation of the genetic variation between and within populations provide important information for defining protection units and for the formulation of appropriate management strategies directed at species conservation.
Previously, several markers have been used to evaluate the genetic diversity of B. sinensis, including amplified fragment length polymorphism (AFLP) (Hu et al., 2017), random amplified polymorphic DNA (RAPD) (Wang et al., 2008), inter-simple sequence repeats (ISSRs) (Liang et al., 2012; Xu et al., 2013; Hu et al., 2014), microsatellite (SSR) (Guan et al., 2012; Li et al., 2016), and several chloroplast sequence markers (Wang et al., 2018). These results showed that the genetic diversity of B. sinensis was low within populations but relatively high at the species level (Hu et al., 2014, 2017; Wang et al., 2018). However, these markers exhibit low stability and reproducibility. Developing more effective genetic markers, such as the whole chloroplast genome, will be essential to assess genetic divergence and diversity in the genomic era.
Chloroplast genomes lack recombination and have a slower mutation rate compared with nuclear genomes. Chloroplast genome sequences in particular are a popular method in the evolutionary biology, such as the reconstruction of plant relationships at the systemic level (Dong et al., 2021a,2022a,b). Chloroplast genomes are typically inherited uniparentally, creating a complement for biparentally inherited markers, such as nuclear markers. Furthermore, uniparental inheritance shows a clear geographical structure and chloroplast genome markers are widely used in phylogeography studies. Whole chloroplast genomes contain numerous single nucleotide polymorphisms (SNPs), microsatellites (SSRs), and indels at the intraspecies level (Huang et al., 2014; Van Der Merwe et al., 2014; Perdereau et al., 2017; Cao et al., 2018; Cheng et al., 2019), which have been used to estimate population differentiation, genetic diversity, gene flow, and biogeographical structure. These chloroplast markers can also be used to investigate endangered plant’s conservation processes, such as assessing genetic structure and defining protection units.
It will be important to access whole chloroplast genome sequence variability, identify genomic resources, and determine the genetic divergence of endangered B. sinensis. This is the first study to use identified chloroplast genomic resources to examine the genetic variation and genetic diversity of B. sinensis. The results of this study provide valuable genomic resources that may be combined with ecological and evolutionary information to guide conservation of this species.
Materials and Methods
Plant Materials, DNA Extraction, and Sequencing
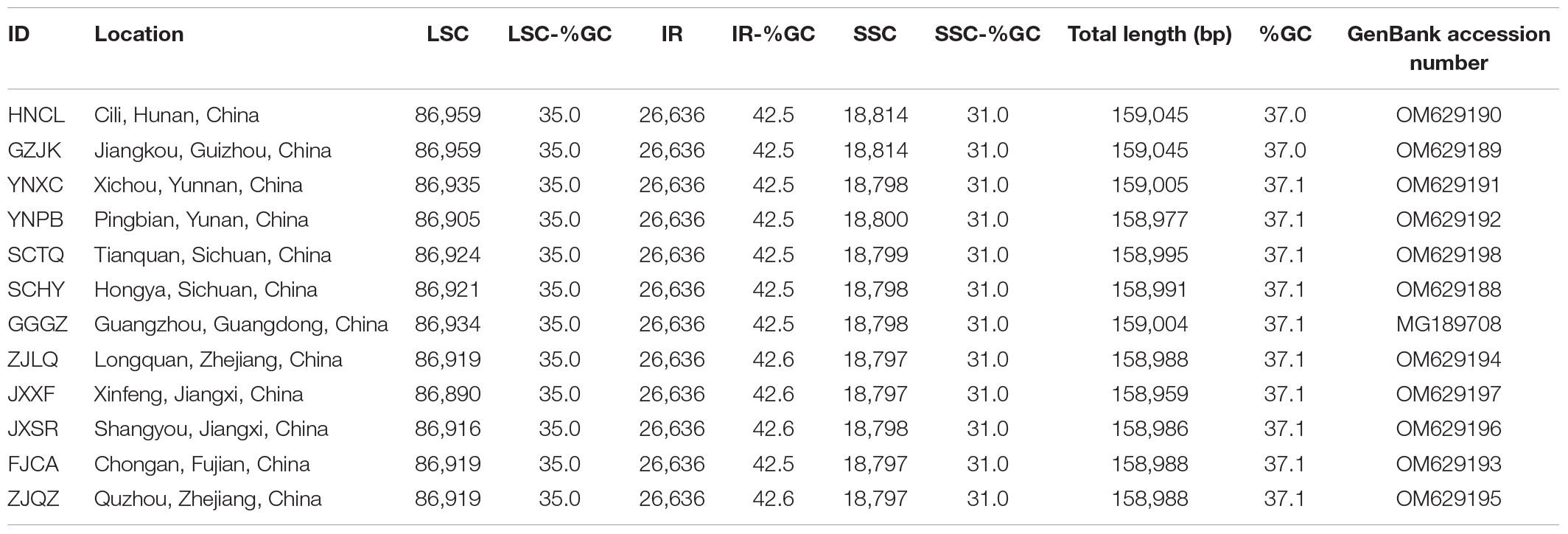
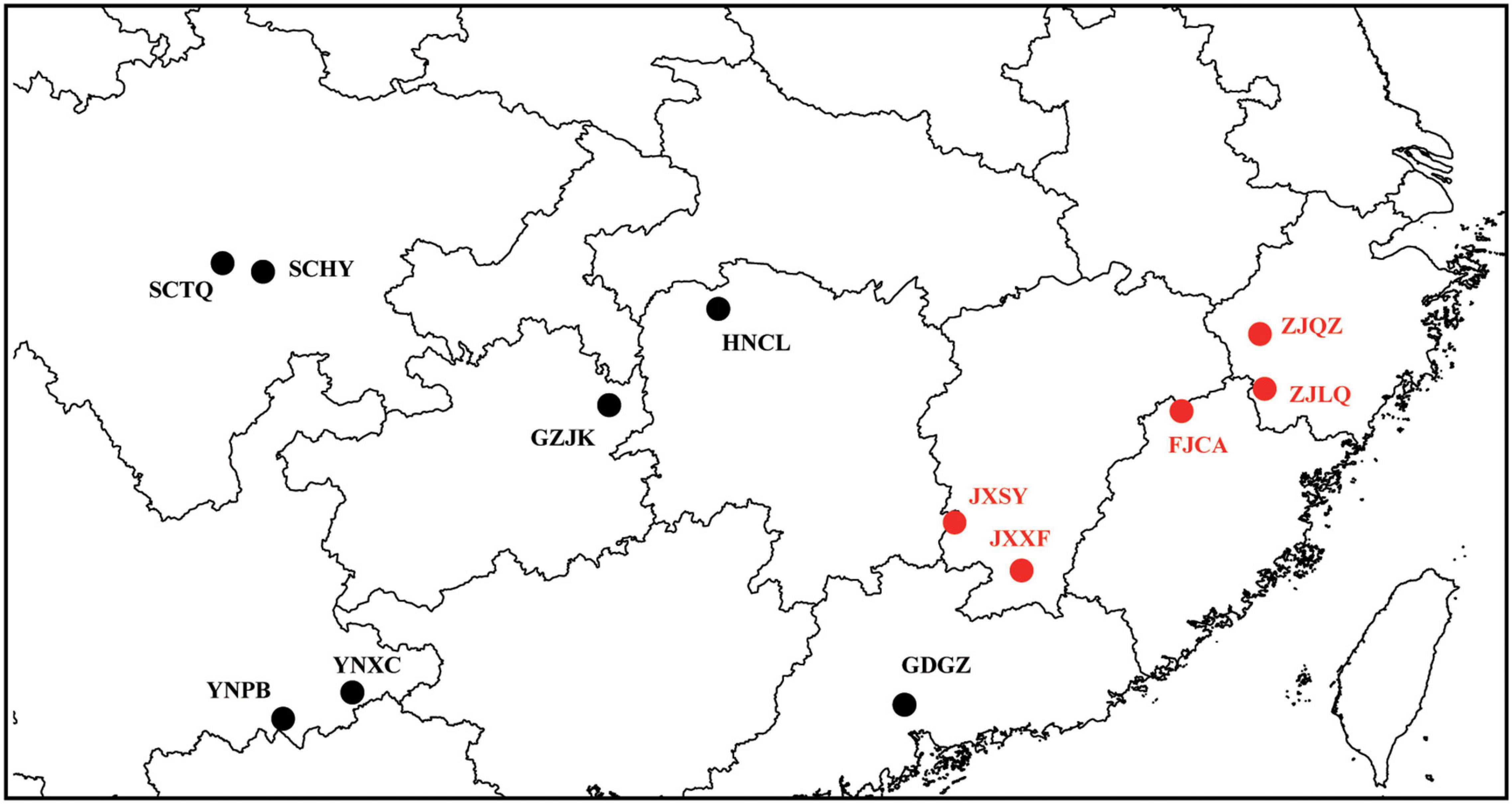
Twelve genotypes of B. sinensis, representing the geographical distribution of this species, were used in this study. Sample information is provided in Table 1 and Figure 1. Young leaves of B. sinensis were collected for silica gel conservation. Total DNA was extracted using a modified CTAB DNA extraction protocol (Li et al., 2013). DNA concentration was measured using the Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and DNA with a total concentration >1 μg were used for Illumina sequencing.
Genome skinning was used to sequence the chloroplast genomes of B. sinensis (Straub et al., 2012; Dong et al., 2022b). Total DNA was sheared to 350 bp fragments using an ultrasonicator, and 350 bp insert libraries were constructed for Illumina sequencing according to the manufacturer’s instructions (Illumina, San Diego, CA, United States). The library was paired-end sequenced (150 bp) on the Illumina HiSeq X-Ten at Novogene (Tianjin, China); each sample yielded approximately 5 Gb data.
Chloroplast Genome Assembly and Annotation
Raw data were cleaned and filtered using Trimmomatic 0.36 (Bolger et al., 2014), with the following parameters: LEADING, 20; TRAILING, 20; SLIDING WINDOW, 4:15; MIN LEN, 36; and AVG QUAL, 20. Two methods were used to assemble the chloroplast genomes. First, the clear data were directly inputted into GetOrganelle (Jin et al., 2020) with k-mer lengths of 85, 95, and 105. If the GetOrganelle was failed assembly the complete chloroplast genome, we selected the second method to finish it. For the second method, the clean data was assembled into contigs using the SPAdes 3.6.1 program (k-mer = 95) (Bankevich et al., 2012). Chloroplast genome contigs were selected using the Blast program (Altschul et al., 1990), with the complete B. sinensis chloroplast genome from GetOrganelle as the reference. The selected contigs were assembled using Sequencher 5.4.5 (Gene Codes Corporation, Ann Arbor, MI, United States1). The gaps between the contigs were filled using the clean reads that were mapped to the contigs using Geneious 8.1 (Kearse et al., 2012).
Whole chloroplast sequences were annotated using Plann (Huang and Cronk, 2015) and the published sequence of B. sinensis (GenBank accession number: MG189708) was used as the reference. Annotation was confirmed in Geneious. Complete chloroplast genomes were depicted using OGDRAW (Greiner et al., 2019) and annotated chloroplast genomes were deposited in GenBank under the accession numbers of OM629188 to OM629198.
Simple Sequence Repeats
Microsatellites were identified using Genome-wide Microsatellite Analyzing Tool Package (GMATA) software (Wang and Wang, 2016). The minimum number of repetitions was set to 10 for mononucleotides, 5 for dinucleotides, 4 for trinucleotides, and 3 repeat units for tetranucleotides, pentanucleotides, and hexanucleotides. The SCHY genotype was used to represent the species of B. sinensis to count the number of repeats and SSRs.
Intraspecific Variations of Bretschneidera sinensis
Single nucleotide polymorphisms and indels were identified to demonstrate intraspecific variations in the chloroplast genome sequences of B. sinensis. The chloroplast genome sequences were aligned with MAFFT 7 (Katoh and Standley, 2013) and adjusted manually using Se-Al 2.0 (Rambaut, 1996). SNPs were calculated using MEGA X (Kumar et al., 2018), and indels were identified using DnaSP 6 (Rozas et al., 2017). The number, location, and direction of SNPs and indels were determined using the SCHY genotype chloroplast genome as the standard reference.
Phylogenetic Inference
Maximum likelihood (ML) and Bayesian inference (BI) methods were used to infer phylogenetic relationships. The best-fit substitution mode was conducted using ModelFinder (Kalyaanamoorthy et al., 2017) under the Bayesian information criterion. ML tree was generated in RAxML-NG (Kozlov et al., 2019) and the node support values were determined using 500 rapid bootstrap replicates. Mrbayes v3.2 (Ronquist et al., 2012) was used to perform the BI tree. Markov Chain Monte Carlo (MCMC) analysis was run for 2 × 10,000,000 generations and sampled every 1,000 generations. Stationarity was considered to be reached when the average standard deviation of split frequencies remained below 0.001 after the 25% burn-in.
Divergence Time Analyses
We used BEAST v2.5.1 (Bouckaert et al., 2014) to estimate the divergence times of the two clades of B. sinensis (see section “Results”) based on the 79 coding genes in the chloroplast genomes. The dataset includes 41 samples of Brassicales (Supplementary Table 1). Coding genes were extracted using a custom Python script. Each gene was aligned using MAFFT independently and concatenated using PhyloSuite 1.2.2 (Zhang et al., 2020). The coding genes accD, ycf1, and ycf2 were not included because of the number of indels in the alignment. In total, this dataset included 79 genes and 45 samples.
Four priors were used to calibrate the analysis according the results of Edger et al. (2015): (1) the stem age of Brassicales (the root of the tree) was 112 Ma; (2) the crown age of Brassicales was 89.5 Ma; (3) the crown age of Brassicaceae was 31.8 Ma; and (4) the average age of the most recent common ancestor of Tropaeolaceae/Akaniaceae was 36.3 Ma. The prior distributions of the four calibration points were set as a normal distribution, with a standard deviation of 1.0.
GTR nucleotide and Yule speciation models were selected. MCMC analysis was run for 500 million generations with sampling every 1,000 generations. The stationary phase was examined using Tracer 1.6 (Rambaut et al., 2014) to evaluate convergence and to ensure a sufficient and effective sample size (ESS) for all parameters surpassing 200. The first 10% of the trees were discarded as burn-in, and the MCC tree was generated with mean heights in TreeAnnotator.
Results
Chloroplast Genome Features of Bretschneidera sinensis
The chloroplast genomes of B. sinensis ranged from 158,959 bp (JXXF) to 159,045 bp (GZJK) and displayed a typical circular quadripartite structure, consisting of a pair of inverted repeat (IR) regions (26,636 bp) separated by a large single copy (LSC) region (86,890–86,959 bp) and a small single copy (SSC) region (18,797–18,814 bp) (Figure 2). The overall GC content of the chloroplast genome was 37.0–37.1%, while the GC contents of the SSC, LSC, and IR regions were 31.0, 35, and 42.5–42.6%, respectively (Table 1). We mapped the reads of each sample to its own chloroplast genome (Supplementary Figures 1, 2). All reads of every sample were mostly consistent, suggesting that there was not a polymorphism structure.
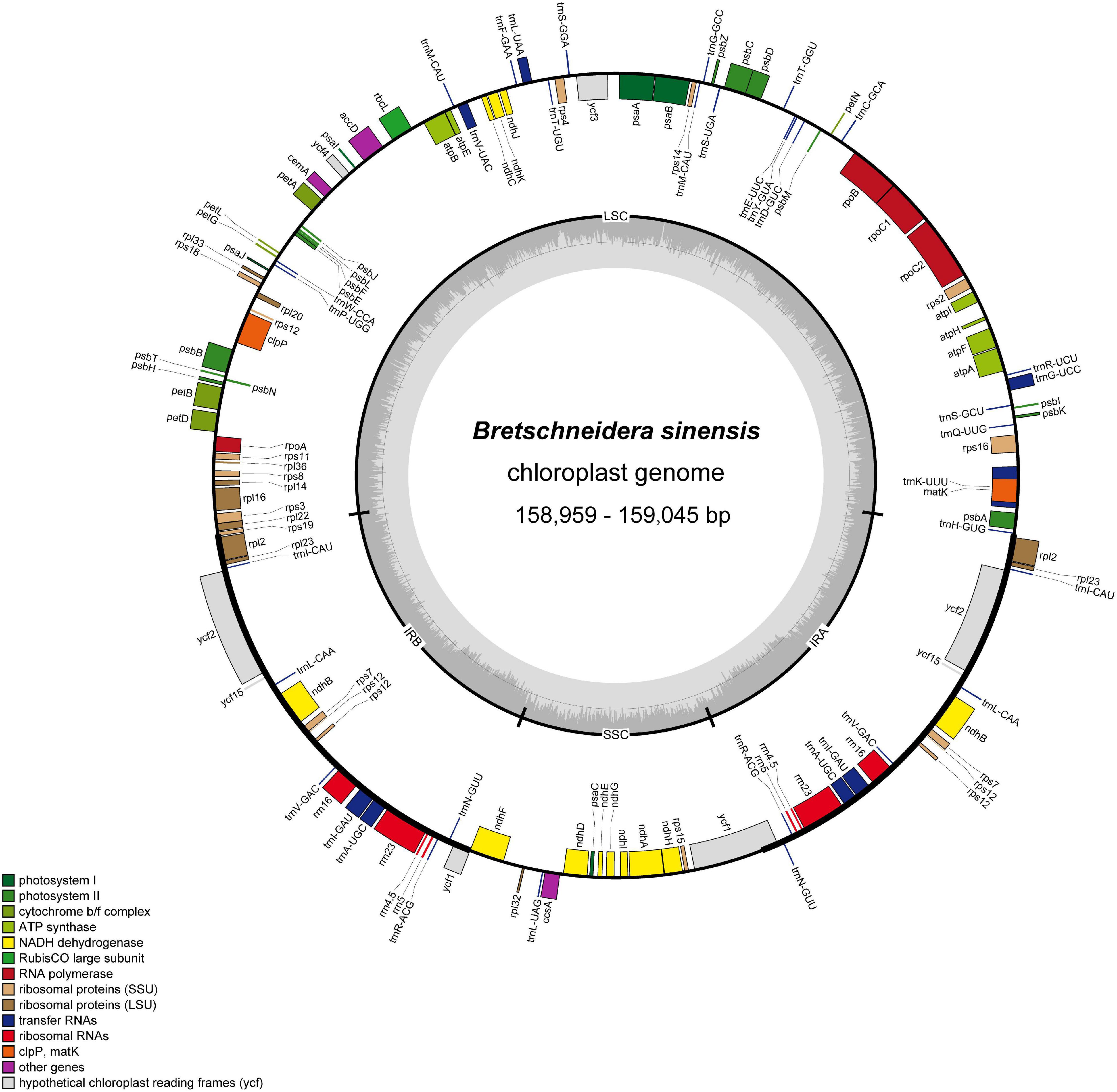
Figure 2. Gene maps of the B. sinensis chloroplast genomes. Genes on the inside of the large circle are transcribed clockwise, and those on the outside are transcribed counterclockwise. The genes are color-coded based on their functions. The dashed area represents the GC composition of the chloroplast genome.
We annotated 112 unique functional genes in the B. sinensis chloroplast genome, consisting of 78 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes. A total of 62 protein-coding and 22 tRNA genes were located in the LSC region, 12 protein-coding and 1 tRNA gene in the SSC region, and 6 protein-coding, 8 tRNA, and all 4 rRNA genes in the IR region. Among these genes, 14 had 1 intron and 2 (clpP and ycf3) had 2 introns. The matK gene was located in trnK-UUU, which is the largest intron in the chloroplast genome, and the rps12 gene was trans-spliced, with two copies of the 3′ end in the IR region with two copies, and the 5′ end in the LSC region.
We compared the IR boundary regions of the two species of Akaniaceae (Figure 3), and the results showed that the borders of the Akaniaceae chloroplast genome were very similar. In Akaniaceae, the boundary was in the rps19 gene on the LSC/IRb. The IRa/SSC border extended into ycf1, resulting in a pseudogene of 1,109 bp in B. sinensis and 1,097 bp in Akania lucens. The trnH-GUG gene was located downstream of the IRa/LSC border, a by 24 bp in B. sinensis and 23 bp in A. lucens.
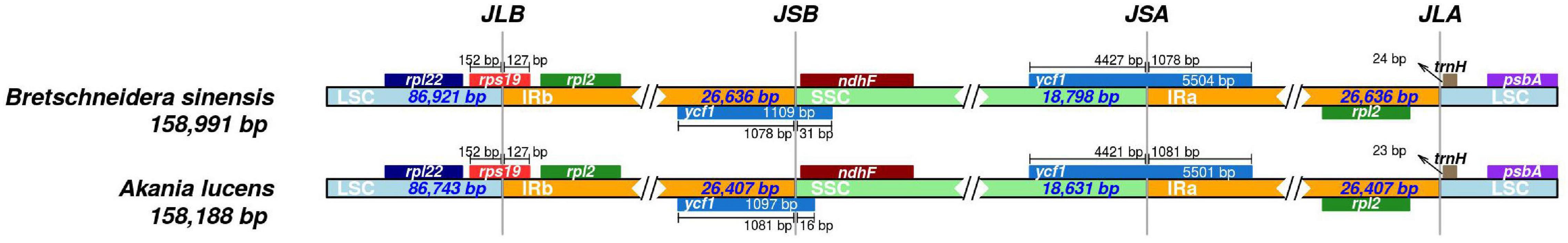
Figure 3. Comparison of the border positions of the LSC, SSC, and IR regions among the chloroplast genomes of two genera of Akaniaceae.
Simple Sequence Repeats in Bretschneidera sinensis Plastomes
A total of 98 SSRs were identified in the chloroplast genomes (Supplementary Table 2 and Figure 4). These SSRs included 65 mononucleotide, 14 dinucleotide, 12 tetranucleotide, and 7 trinucleotide SSRs. There were no pentanucleotide or hexanucleotide repeats in the B. sinensis plastomes. Most SSR types were composed of A/T with minimal G/C, e.g., all the mononucleotide SSRs were composed of A/T. The LSC region contained the most significant SSRs (78.26%), followed by the SSC region (13.04%). Non-coding regions (introns and spacers) contained the majority of SSRs (89.13%); only four coding genes (rpoC1, rpoB, cemA, and ycf1) included SSRs. After in silico comparative analysis, 33 SSRs were found to be polymorphic across the 12 genotypes of B. sinensis plastomes (Table 2). All polymorphic SSRs were located in the LSC region, except one each in the IR (trnV-rps7) and SSC (ndhD-psaC) regions. With the exception of one dinucleotide SSR (trnT-trnL), the other polymorphic SSRs were mononucleotide repeats. The space of ycf4-cemA contained the highest number of polymorphic SSRs (three), followed by trnE-trnT, trnT-trnL, psbE-petL, and rpl16 introns with two polymorphic SSRs. The remaining 18 spacers and 4 introns contained 1 polymorphic SSR each. The primers for the polymorphic SSRs are shown in Table 2.
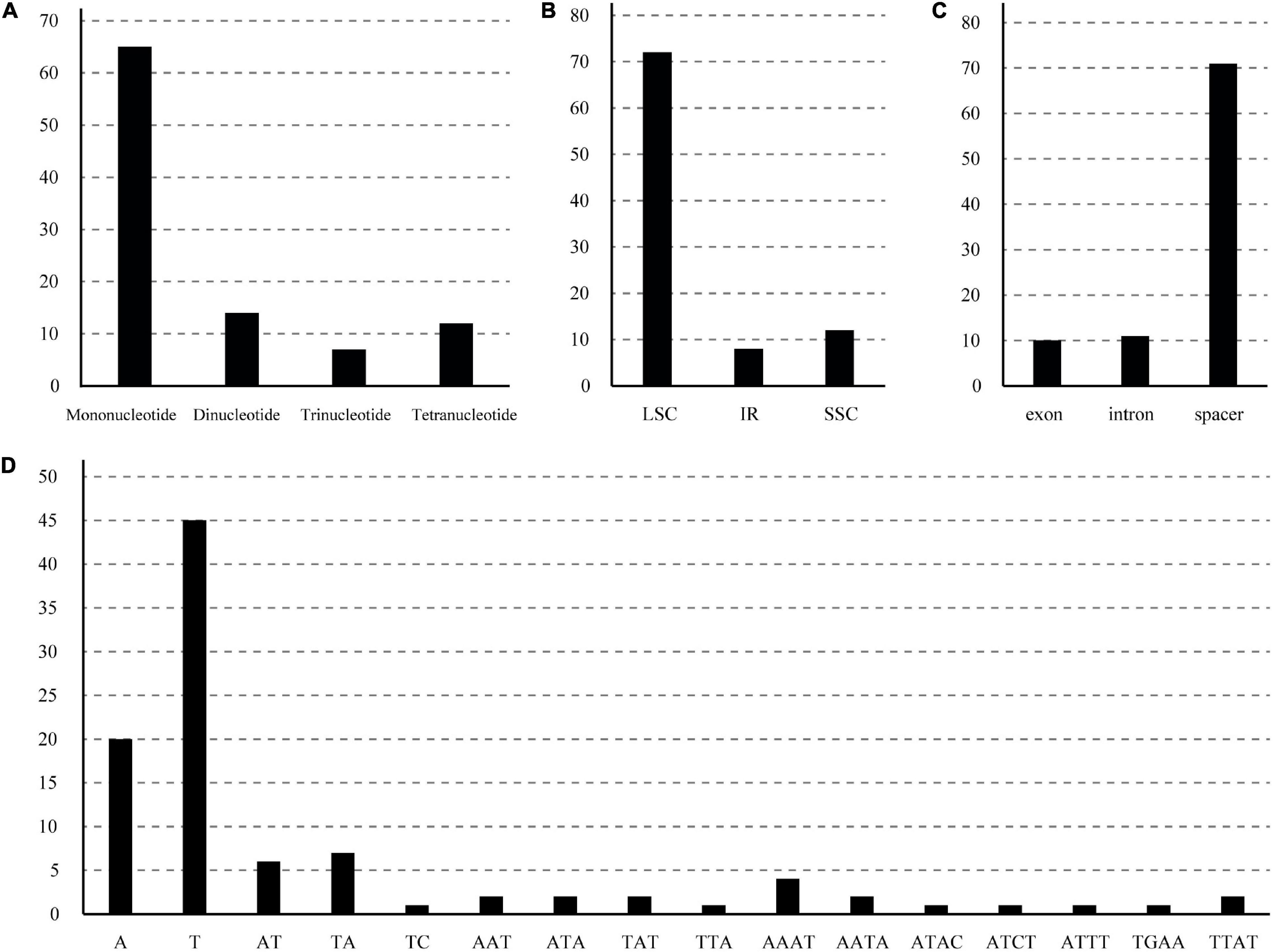
Figure 4. Type and distribution of simple sequence repeats (SSRs) in B. sinensis chloroplast genomes. (A) Number of different SSR types detected by MISA. (B) Number of SSRs in the LSC, SSC, and IR regions. (C) Number of SSRs in spacers, exons, and introns. (D) Frequencies of identified SSR motifs in the different repeat classes.
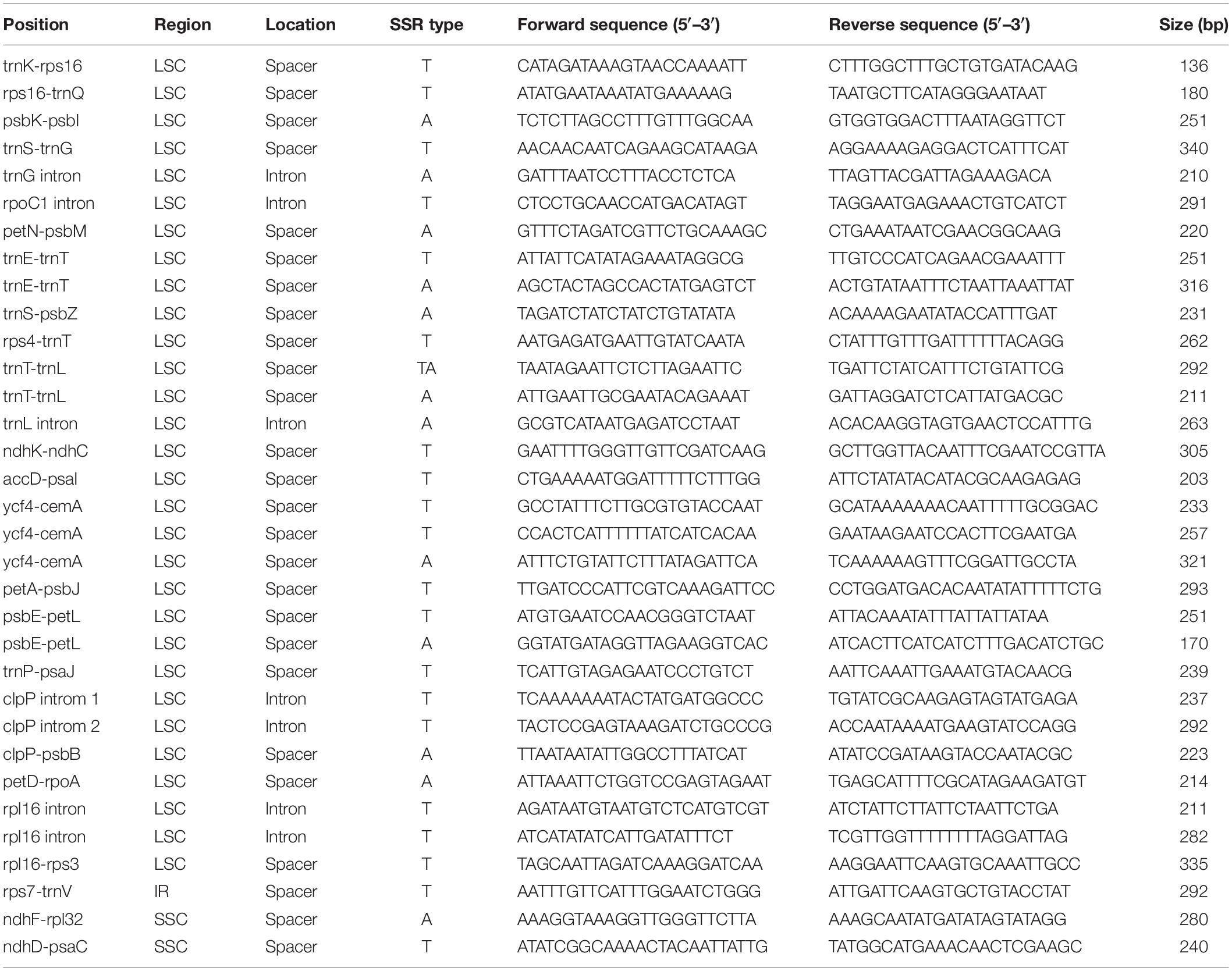
Table 2. Polymorphic SSRs identified from in silico comparative analysis of 12 genotypes of B. sinensis plastomes.
Numbers and Mutation Patterns of Single Nucleotide Polymorphisms
Alignments of the chloroplast genomes of the 12 B. sinensis samples showed 159,111 bp, including 105 polymorphic sites, 40 singleton variable sites, and 68 parsimony informative sites (Table 3 and Supplementary Table 3). The SNPs included 42 transition (Ts) and 63 transversion (Tv) sites, with a Tv to Ts ratio of 1: 1.5. The most frequent SNP mutation types were C to A and G to T; G to C or C to G mutations were less (Figure 5). There were 80 SNPs in LSC regions, 4 in IR regions, and 17 in SSC regions (Supplementary Table 2). Sixty-five sequence regions, comprising 34 spacer regions, 21 coding regions, and 10 intron regions, harbored SNPs. The ycf1 gene had the highest number of SNPs (6), followed by the psbM-trnD spacer (5).
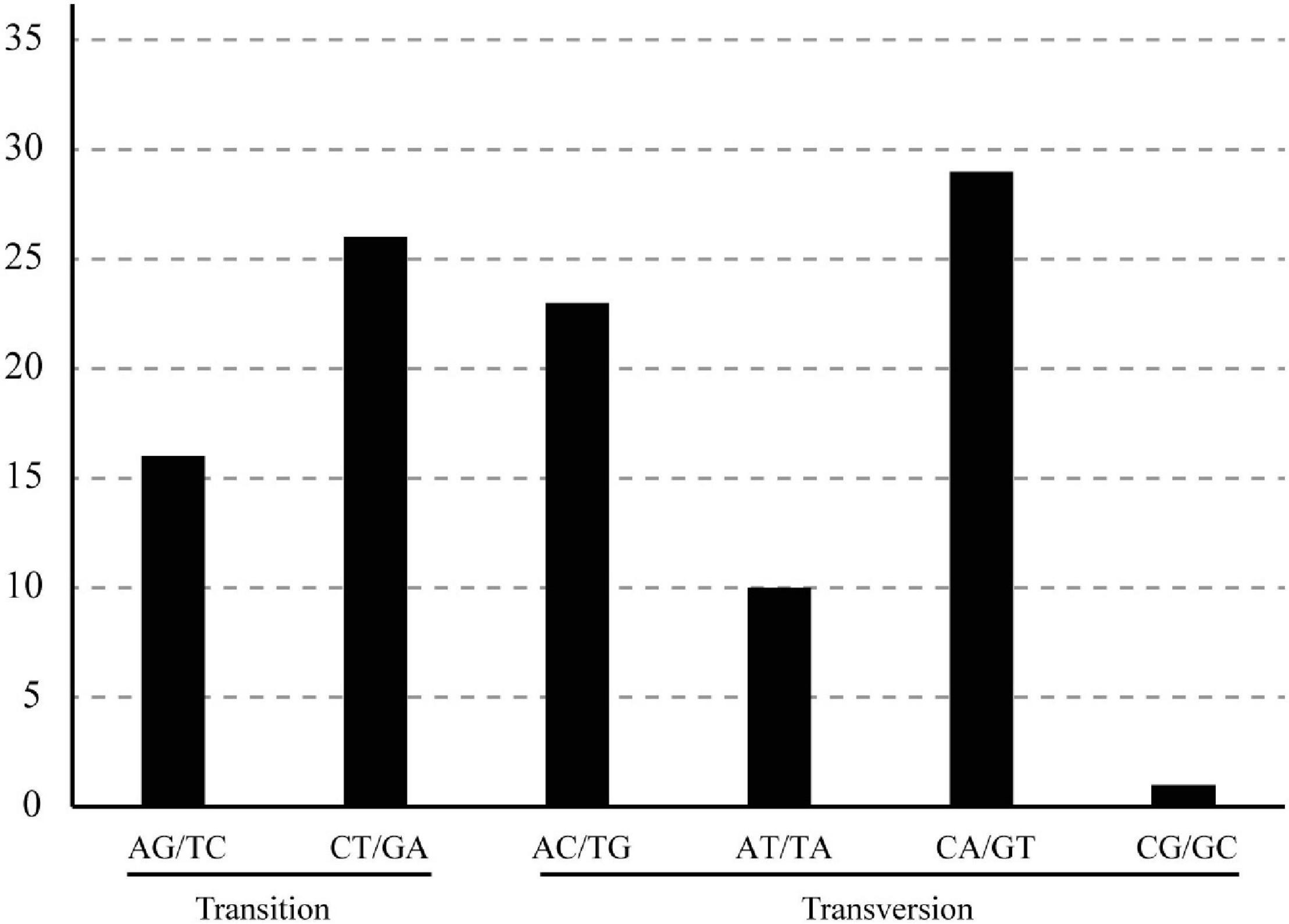
Figure 5. Patterns of SNPs among the B. sinensis samples. Nucleotide substitutions were divided into six types as indicated by the six non-strand-specific base-substitution types (i.e., numbers of G to A and C to T sites for each respective set of associated mutation types).
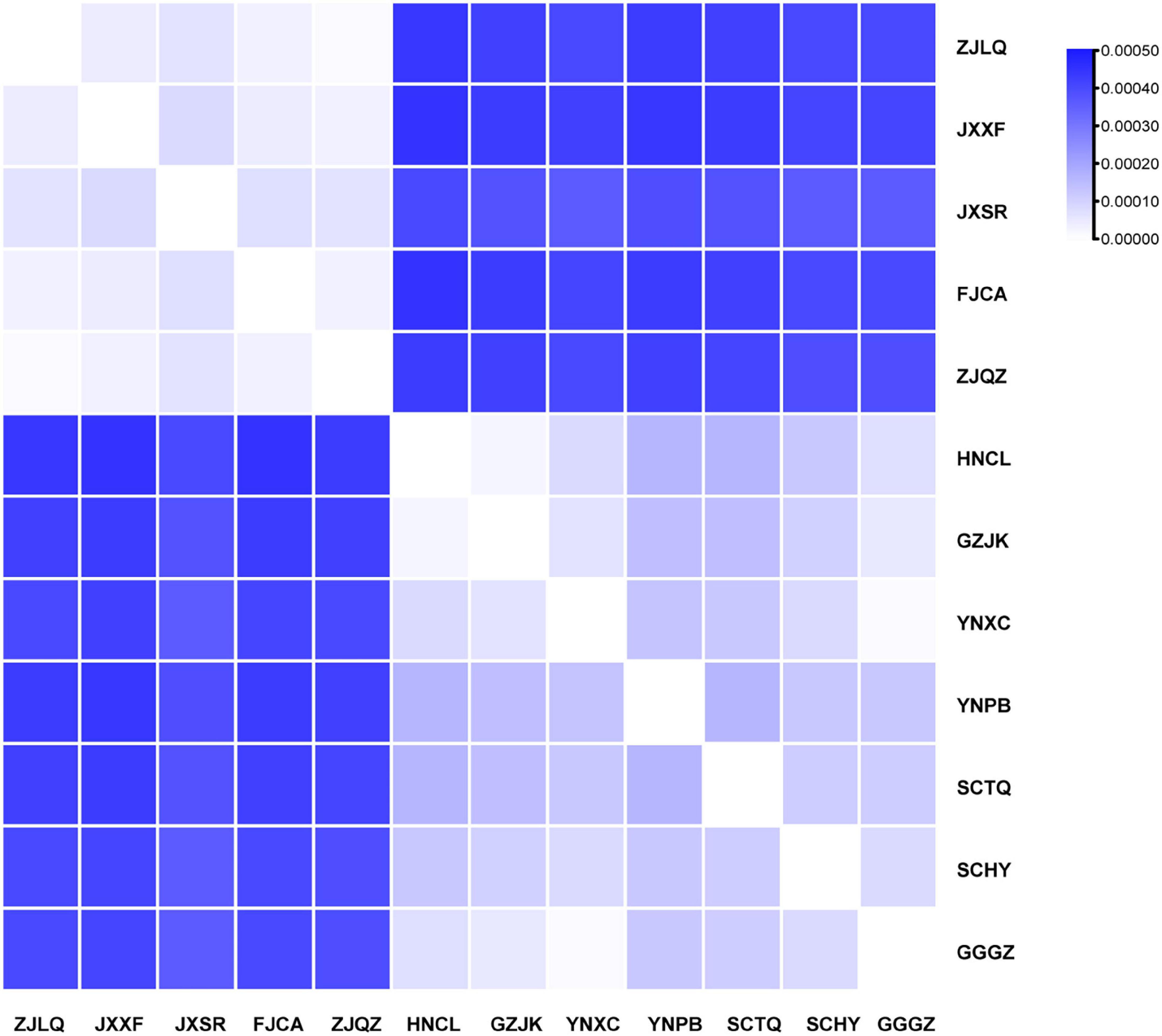
The IR region exhibited the lowest divergence according to nucleotide diversity and number of haplotypes; overall nucleotide diversity was 0.00026. Based on whole plastome sequences, each sample had a unique haplotype, and the IR region contained only five haplotypes. According to the genetic distance results, the largest divergence was observed between samples HNCL and JXXF and the lowest between ZJQZ and ZJLQ and between GGGZ and YNXC (Figure 6).
Numbers and Length of Indels
We identified 55 indels in the 12 B. sinensis chloroplast genomes (Figure 7 and Supplementary Table 4). All indels were located in non-coding regions. A total of 37 sequence regions harbored indels; psbE-petL and rpl32-trnL had the most indels (4), followed by trnT-trnL (3). The length of indels ranged from 1 to 30 bp, indels of 1, 2, and 6 bp were the most common. The largest indel (30 bp), found in the matK-trnK spacer, was an insertion in the GGGZ, YNXC, HNCL, and GZJK genotypes. The second largest indel (28 bp) was a deletion in the trnT-trnL spacer in the JXXF genotype.
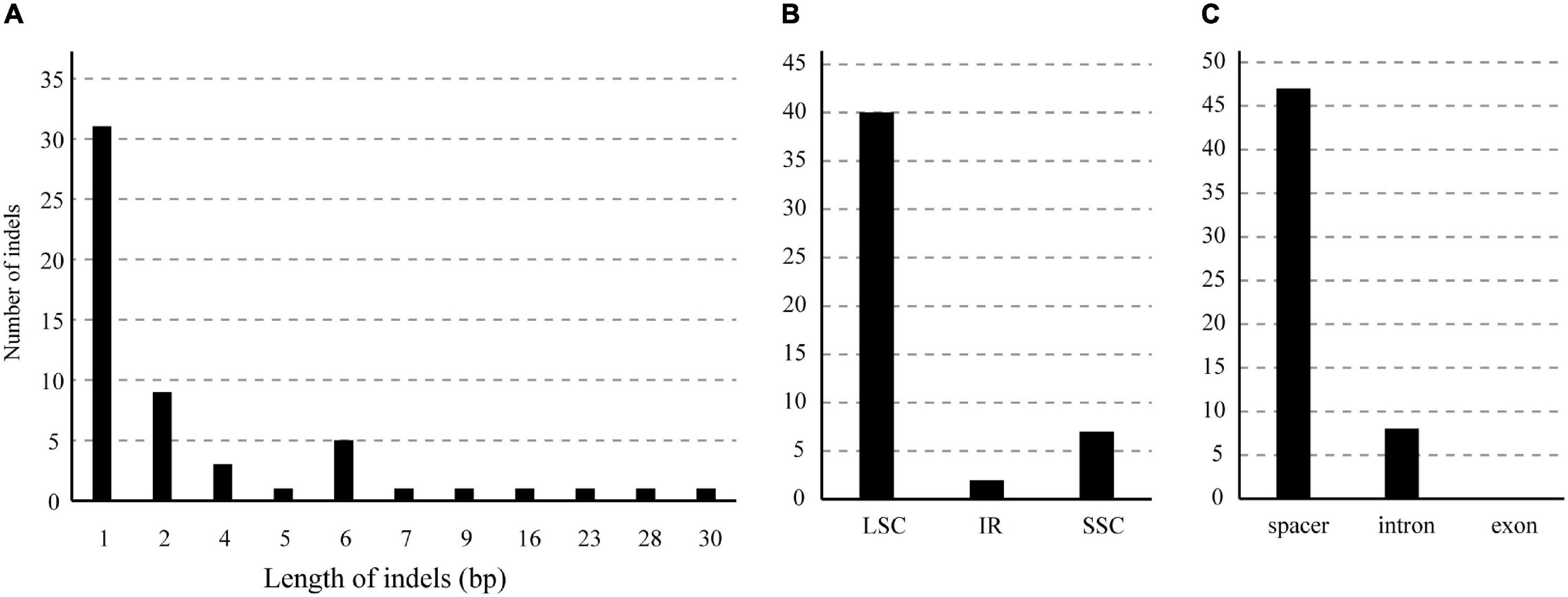
Figure 7. Numbers and patterns of indels among the B. sinensis chloroplast genomes. (A) Numbers and sizes of indels. (B) Numbers of indels in the LSC, SSC, and IR regions. (C) Numbers of SSRs in spacers, exons, and introns.
Phylogenetic Relationships
Phylogenetic relationships among B. sinensis genotypes were inferred using the chloroplast genomes; the phylogenetic tree is presented in Figure 8. The ML and BI trees were congruent, and both trees support grouping of the 12 genotypes into 2 branches with 100% bootstrap support and 1.0 posterior probabilities. The two branches were congruent with the locations of genotypes; these were defined as the Eastern and Western clades. The Eastern clade included five genotypes, and the JXSY genotype was the earliest diverging lineage. Two genotypes from Zhejiang (ZJLQ and ZJQZ) had closer relationships. The YNPB genotype was the earliest diverging lineage in the Western clade. The genotypes GGGZ and YNXC, and GZJK and HNCL, formed sister groups.
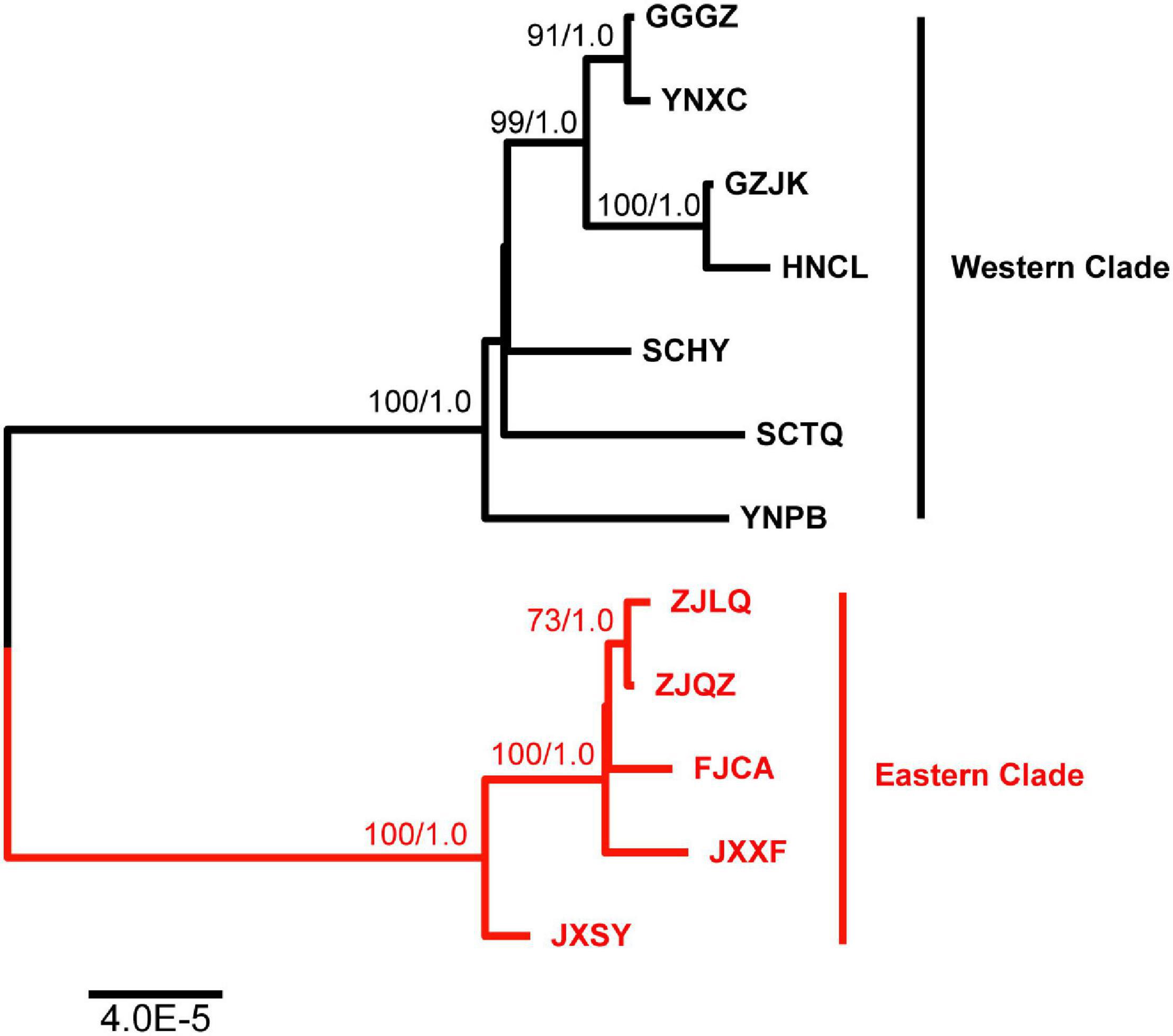
Figure 8. Phylogenetic relationships among B. sinensis samples. ML topology is shown with the ML bootstrap support value and Bayesian posterior probability at each node. ML values < 50 and BI values < 0.7 were not present.
Divergence Times
Divergence time estimates showed that the stem note of Bretschneideraceae was dated to 36.01 Ma during the Eocene (Figure 9). The crown age of Bretschneideraceae was 9.7 Ma in the late Miocene and the two clades of B. sinensis were divergent from 0.6 Ma in the late Pleistocene.
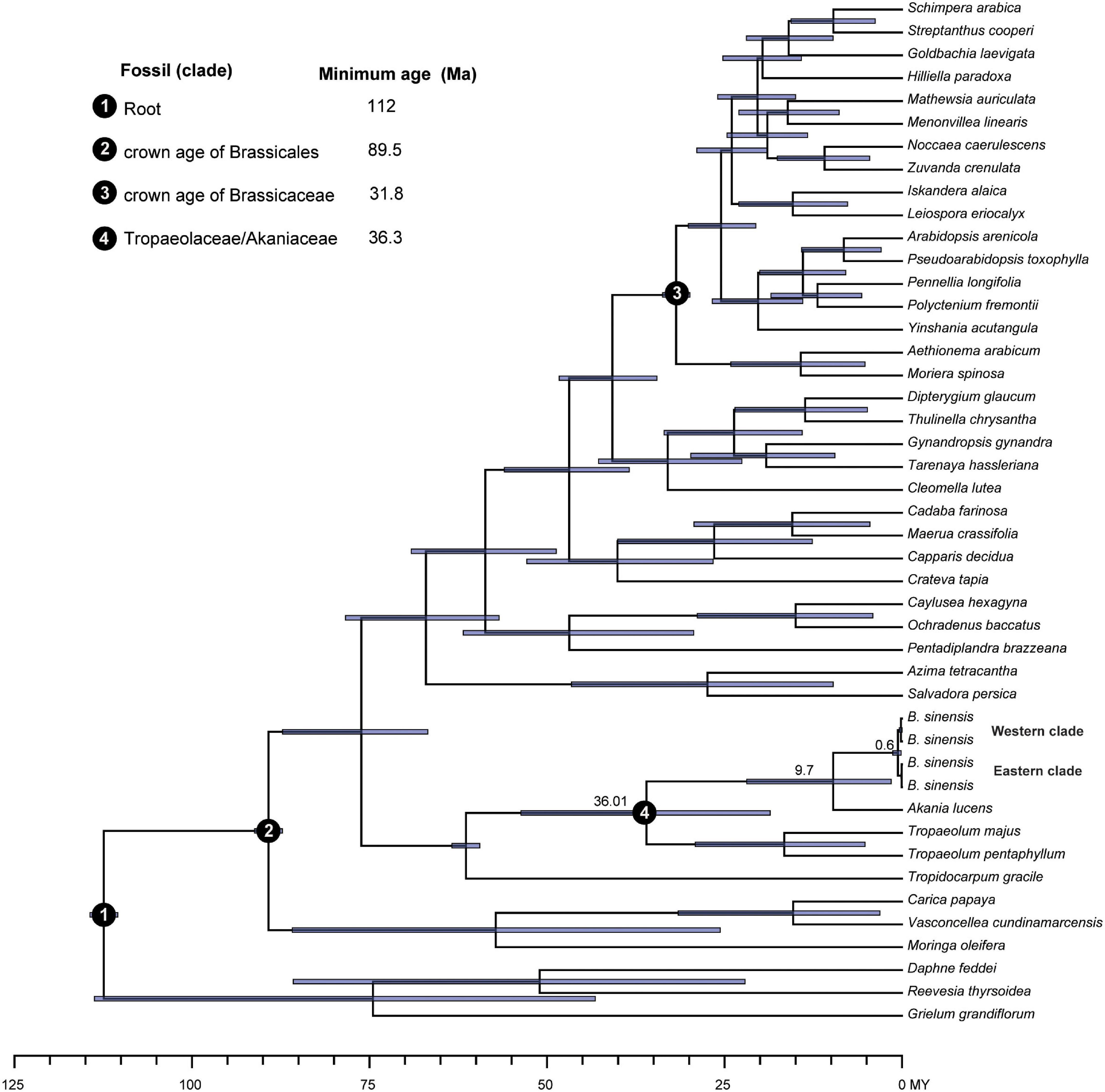
Figure 9. Divergence times of Brassicales obtained from BEAST analysis. Mean divergence time of the nodes is shown next to the nodes, while the blue bars correspond to the 95% highest posterior density.
Discussion
Intra-Species Chloroplast Genome Sequence Variation in Bretschneidera sinensis
Using genome skimming, we obtained complete chloroplast genome sequences for 12 B. sinensis genotypes, which will be important for the discovery of molecular markers and detection of genetic diversity. Using these sequences, we detected genetic variations, including SSRs, SNPs, and indels, which are important for population genetics studies.
Simple sequence repeats, which consist of tandemly repeated motifs of 6 bp or less, are widely used in population studies and are considered highly variable markers (Schlotterer, 2000). Chloroplast genome SSRs (cpSSRs) have become widely used chloroplast genome markers previously (Xu et al., 2002; Yang et al., 2011) for cultivated origins of crops and population studies of wild plants. As with other species (Dong et al., 2021a,b), mononucleotide repeats are the most common, ranging in size from 10 to 30 bp. Thirty-three polymorphic SSRs across the 12 genotypes of the B. sinensis chloroplast genome were identified after in silico comparative analysis, which investigated genetic divergence and gene flow among different populations and phylogeographical studies.
Only 105 SNPs were detected in the B. sinensis chloroplast genome and a mutation bias in the SNP patterns was observed. AT to TA and GC to CG occurred significantly less frequently (Figure 5). In general, because of less natural selection, non-coding regions tend to contain more SNPs. However, there was no significant difference in the distribution of SNPs in the Dioscorea polystachya chloroplast genome. The ycf1 and psbM-trnD markers included more SNPs and showed higher divergence at the species level (Dong et al., 2012, 2015).
Indels are common mutation events in the chloroplast genome (Dong et al., 2020), and studies have shown indels are derived from, and accelerate nucleotide substitutions mutations (Tian et al., 2008; Hollister et al., 2010; Mcdonald et al., 2011), revealing the importance of indels as a source of genetic variation. In the B. sinensis chloroplast genome, we identified 55 indel events, which is less than SNPs. Most indels occurred between the two clades indicating indels take a long time to be retained. The two regions containing a higher number of indels, psbE-petL and rpl32-trnL, were variable regions in the chloroplast genome, as indicated previously (Shaw et al., 2005, 2007). Adding indel information significantly increases resolution compared with simple substitution-based matrices of chloroplast DNA sequences (Dong et al., 2020).
Genetic Divergence of Bretschneidera sinensis
Analysis of the chloroplast genome sequences of B. sinensis revealed significant genetic divergences. There are two possible explanations for this divergence. First, B. sinensis is a long-lived species with insect-pollinated flowers and heterogeneous pollinators, which are suitable for maintaining rich genetic variation (Qiao et al., 2012). Second, the divergence time supported an origin of B. sinensis in the late Miocene (Figure 9), suggesting B. sinensis is a relict species of the Tertiary palaeotropical flora and was once widely distributed continuously in a subtropical region, which may be the basis of its genetic divergence. However, the genetic diversity within populations is quite low according to the ISSRs (Hu et al., 2014) and chloroplast markers (Wang et al., 2018). Seeds of B. sinensis with short-distance dispersal may contribute to the lower genetic divergence in certain populations (Wang et al., 2018).
The 12 genotypes were separated into two clades consistent with the geographical distribution (Eastern and Western clades) (Figure 1). The results were also supported by nuclear SNPs dataset (Liu et al., 2022). B. sinensis exhibited genetic divergence during the Quaternary glacial period, and in the meantime B. sinensis population size suffered a significant decline (Liu et al., 2022), suggesting at least two glacial refugia. The three genotypes of YNPB (Yunnan Province), SCTQ, and SCHY (Sichuan Province) were the early divergent lineages of the Western clade, and the tree nodes had short length, indicating rapid divergence over a short period. The Hengduan Mountains region may be the first glacial refugium of B. sinensis. The populations from Guizhou (QZJK), Hunan (HNCL), and Guangdong (GDGZ) Province were derived from the Hengduan Mountains, after the initial western colonization of B. sinensis. The second glacial refugium may be the Nanling Mountains and adjacent regions (Wang et al., 2018), and this region colonized the Eastern clade of B. sinensis. The populations of Fujian, Zhejiang, and even Taiwan Province were derived from this glacial refugium (Figures 1, 9; Wang et al., 2018). Besides Hengduan and Nanling Mountains, there may be other refugia preserving B. sinensis populations during the last glacial maximum. An alternative approach using Maxent model (Guo et al., 2020) provided evidence for putative glacial refugia of Jinfo Mountains and Dayao Mountains.
Conservation Implications for Bretschneidera sinensis
There are several ways a plant species may become endangered, such as lower intraspecific genetic variation, lower fecundity, poor adaptability, over-harvesting, and habitat destruction. Our results indicated that B. sinensis had higher genetic divergence among different natural populations and lower genetic diversity within populations according to SSRs, ISSRs, and several chloroplast markers (Xu et al., 2013; Li et al., 2016; Wang et al., 2018) suggesting that endangerment of this species is not due to the low evolutionary potential of the population. Several factors may be have contributed to the endangerment of B. sinensis. First, the Quaternary glaciation resulted in the fragmented distribution of B. sinensis. Second, the loss of habitat due to climate change and deforestation, especially the planting of fast-growing forests such as Cunninghamia lanceolata and Pinus massoniana over the past 50 years, has resulted in a serious loss of B. sinensis populations in ravines and streamside slopes. Third, biological characteristics such as low pollen transfer efficacy, weak fruit retention, and short flowering season are not suitable for the rehabilitation or reconstruction of natural populations (Qiao et al., 2012).
The ultimate goals of species conservation are to ensure sustainable survival of a species and to preserve populations with genetic divergence resulting in increased evolutionary potential (Moritz, 1999). According to a field survey, many populations were small with low genetic divergence, with limited renewal ability (Xu et al., 2013). Based on this and other genetic studies, ex situ conservation may be essential for endemic, rare, and relict species of B. sinensis. We can artificially increase gene flow among different populations with ex situ conservation, which substantially increases genetic diversity. All 12 populations exhibited genetic differences based on the chloroplast genome sequences (Figure 8) and possessed unique haplotypes. Protection of different populations could preserve the genetic variation of B. sinensis. On the other hand, it may be sufficient for each population to protect a few individuals owing to the lower genetic diversity within populations. Therefore, using genomic information to determine genetic variation within populations is important for establishing scientific conservation strategies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accessions: OM629188 to OM629198). Available at: https://www.ncbi.nlm.nih.gov/nuccore/OM629188.
Author Contributions
CS, ZY, and WD conceived the idea. ZY, ZC, LX, MW, and ZT collected the plant materials. EL, ML, and KL analyzed the data. CS and WD drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Technology Basic Resources Investigation Program of China (grant no. 2017FY100100) and the Fundamental Research Funds for the Central Universities (grant no. BLX201723).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the DNA Bank of China for providing some materials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.873100/full#supplementary-material
Supplementary Figure 1 | Schematic diagram of chloroplast genome sequence coverage from mapping reads to its own chloroplast genome.
Supplementary Figure 2 | Details of three sequence mapping results from Supplementary Figure 1. The results showed all the reads were consistent and there was not a polymorphism structure.
Supplementary Table 1 | A list of 41 samples of Brassicales from GenBank for phylogenetic and divergence time analysis.
Supplementary Table 2 | Types and numbers of SSRs in B. sinensis chloroplast genomes.
Supplementary Table 3 | Types and numbers of SNPs in B. sinensis chloroplast genomes.
Supplementary Table 4 | Sizes and numbers of indels in B. sinensis chloroplast genomes.
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mole. Biol. 215, 403–410.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comp. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouckaert, R., Heled, J., Kuhnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comp. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Cao, J., Jiang, D., Zhao, Z., Yuan, S., Zhang, Y., Zhang, T., et al. (2018). Development of Chloroplast Genomic Resources in Chinese Yam (Dioscorea polystachya). Biomed. Res. Int. 2018:6293847. doi: 10.1155/2018/6293847
Cheng, L., Nam, J., Chu, S.-H., Rungnapa, P., Min, M.-H., Cao, Y., et al. (2019). Signatures of differential selection in chloroplast genome between japonica and indica. Rice 12:65. doi: 10.1186/s12284-019-0322-x
Dong, W., Liu, J., Yu, J., Wang, L., and Zhou, S. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One 7:e35071. doi: 10.1371/journal.pone.0035071
Dong, W., Liu, Y., Xu, C., Gao, Y., Yuan, Q., Suo, Z., et al. (2021a). Chloroplast phylogenomic insights into the evolution of Distylium (Hamamelidaceae). BMC Gen. 22:293. doi: 10.1186/s12864-021-07590-6
Dong, W., Xu, C., Liu, Y., Shi, J., Li, W., and Suo, Z. (2021b). Chloroplast phylogenomics and divergence times of Lagerstroemia (Lythraceae). BMC Gen. 22:434. doi: 10.1186/s12864-021-07769-x
Dong, W., Liu, Y., Li, E., Xu, C., Sun, J., Li, W., et al. (2022a). Phylogenomics and biogeography of Catalpa (Bignoniaceae) reveal incomplete lineage sorting and three dispersal events. Mole. Phylogen. Evol. 166:107330. doi: 10.1016/j.ympev.2021.107330
Dong, W., Sun, J., Liu, Y., Xu, C., Wang, Y., Suo, Z., et al. (2022b). Phylogenomic relationships and species identification of the olive genus Olea (Oleaceae). J. Syst. Evol. 2022:12802. doi: 10.1111/jse.12802
Dong, W., Xu, C., Li, C., Sun, J., Zuo, Y., Shi, S., et al. (2015). ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 5:8348. doi: 10.1038/srep08348
Dong, W., Xu, C., Wen, J., and Zhou, S. (2020). Evolutionary directions of single nucleotide substitutions and structural mutations in the chloroplast genomes of the family Calycanthaceae. BMC Evol. Biol. 20:96. doi: 10.1186/s12862-020-01661-0
Edger, P. P., Heidel-Fischer, H. M., Bekaert, M., Rota, J., Glöckner, G., Platts, A. E., et al. (2015). The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl. Acad. Sci. 112:8362. doi: 10.1073/pnas.1503926112
Ellegren, H., and Galtier, N. (2016). Determinants of genetic diversity. Nat. Rev. Genet. 17, 422–433. doi: 10.1038/nrg.2016.58
Escudero, A., Iriondo, J. M., and Torres, M. E. (2003). Spatial analysis of genetic diversity as a tool for plant conservation. Biolog. Conserv. 113, 351–365. doi: 10.1016/s0006-3207(03)00122-8
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guan, B.-C., Song, G.-R., and Ge, G. (2012). Sixteen microsatellite markers developed from Bretschneidera sinensis (Bretschneideraceae). Conserv. Genet. Res. 4, 673–675. doi: 10.1007/s12686-012-9620-y
Guo, F., Xu, G., and Mu, H. (2020). Simulation of potential spatiotemporal population dynamics of Bretschneidera sinensis Hemsl. based on MaxEnt model. Plant Sci. J. 38, 185–194.
Hollister, J. D., Ross-Ibarra, J., and Gaut, B. S. (2010). Indel-Associated Mutation Rate Varies with Mating System in Flowering Plants. Mole. Biol. Evol. 27, 409–416. doi: 10.1093/molbev/msp249
Hu, P.-W., Duan, L., Wang, M.-N., Wang, Z.-F., and Chen, H.-F. (2017). Phylogeographic study on Bretschneidera sinensis inferred from AFLP data. Plant Sci. J. 35, 815–824.
Hu, Z.-Y., Lin, L., Deng, J.-F., and Wang, S.-H. (2014). Genetic diversity and differentiation among populations of Bretschneidera sinensis (Bretschneideraceae), a narrowly distributed and endemic species in China, detected by inter-simple sequence repeat (ISSR). Biochem. Syst. Ecol. 56, 104–110. doi: 10.1016/j.bse.2014.04.007
Huang, D. I., and Cronk, Q. C. B. (2015). Plann: a command-line application for annotating plastome sequences. Appl. Plant Sci. 3:1500026. doi: 10.3732/apps.1500026
Huang, D. I., Hefer, C. A., Kolosova, N., Douglas, C. J., and Cronk, Q. C. (2014). Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 204, 693–703. doi: 10.1111/nph.12956
Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., Depamphilis, C. W., Yi, T.-S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Gen. Biol. 21:241. doi: 10.1186/s13059-020-02154-5
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mole. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular Evolutionary Genetics Analysis across Computing Platforms. Mole. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, J., Wang, S., Jing, Y., Wang, L., and Zhou, S. (2013). A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 48, 72–78. doi: 10.3724/sp.j.1259.2013.00072
Li, M., Chen, H., Wang, Z., and Zhang, S. (2016). Isolation and characterization of polymorphic microsatellite markers in the endangered species Bretschneidera sinensis Hemsl. Genet. Mol. Res 2016:15. doi: 10.4238/gmr.15038234
Liang, Y., Xu, G., Zhang, H., Wu, X., Shen, X., and Wang, A. (2012). Genetic Diversity of Natural and Planted Populations of Bretschneidera sinensis from Nanling Region. Sci. Silvae Sinicae 48, 45–52.
Liu, H., Harris, A., Wang, Z., Chen, H., Li, Z., and Wei, X. (2022). The genome of the paleogene relic tree Bretschneidera sinensis : insights into trade-offs in gene family evolution, demographic history, and adaptive snps. DNA Res. 29:dsac003. doi: 10.1093/dnares/dsac003
Lu, L., and Boufford, D. (2005). “Bretschneideraceae,” in Flora of China, eds Z. Wu, P. Raven, and D. Hong (Beijing: Science Press Missouri Botanical Garden), 197.
Mcdonald, M. J., Wang, W. C., Huang, H. D., and Leu, J. Y. (2011). Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. 9:e1000622. doi: 10.1371/journal.pbio.1000622
Moritz, C. (1999). Conservation Units and Translocations: Strategies for Conserving Evolutionary Processes. Hereditas 130, 217–228. doi: 10.1111/j.1601-5223.1999.00217.x
Perdereau, A., Klaas, M., Barth, S., and Hodkinson, T. R. (2017). Plastid genome sequencing reveals biogeographical structure and extensive population genetic variation in wild populations of Phalaris arundinacea L. in north-western Europe. GCB Bioenergy 9, 46–56.
Qi, Q., Hong-Feng, C., Fu-Wu, X., Dong-Ming, L., and Xiao-Gai, H. (2009). Seed germination protocol for the threatened plant species, Bretschneidera sinensis Hemsl. Seed Sci. Technol. 37, 70–78. doi: 10.15258/sst.2009.37.1.09
Qiao, Q., Chen, H., Xing, F., Wang, F., Zhong, W., Wen, X., et al. (2012). Pollination ecology of Bretschneidera sinensis (Hemsley), a rare and endangered tree in China. Pak. J. Bot. 44, 1897–1903.
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sanchez-Delbarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mole. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Schlotterer, C. (2000). Evolutionary dynamics of microsatellite DNA. Chromosoma 109, 365–371. doi: 10.1007/s004120000089
Shaw, J., Lickey, E. B., Beck, J. T., Farmer, S. B., Liu, W. S., Miller, J., et al. (2005). The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 92, 142–166. doi: 10.3732/ajb.92.1.142
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Straub, S. C. K., Parks, M., Weitemier, K., Fishbein, M., Cronn, R. C., and Liston, A. (2012). Navigating the Tip of the Genomic Iceberg: next-Generation Sequencing for Plant Systematics. Am. J. Bot. 99, 349–364. doi: 10.3732/ajb.1100335
The Angiosperm Phylogeny, G. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20. doi: 10.1111/boj.12385
Tian, D. C., Wang, Q., Zhang, P. F., Araki, H., Yang, S. H., Kreitman, M., et al. (2008). Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature 455, 105–U170. doi: 10.1038/nature07175
Tobe, H., and Peng, C.-I. (1990). The embryology and taxonomic relationships of Bretschneidera (Bretschneideraceae). Botan. J. Linn. Soc. 103, 139–152.
Tobe, H., and Raven, P. H. (1995). Embryology and relationships of Akania (Akaniaceae). Bot. J. Linn. Soc. 118, 261–274. doi: 10.1111/j.1095-8339.1995.tb00471.x
Van Der Merwe, M., Mcpherson, H., Siow, J., and Rossetto, M. (2014). Next-Gen phylogeography of rainforest trees: exploring landscape-level cpDNA variation from whole-genome sequencing. Mole. Ecol. Resour. 14, 199–208. doi: 10.1111/1755-0998.12176
Wang, J., Liu, R.-L., and Liao, W.-M. (2008). A Study on Growth and Development Rhythm, Phenological Characteristics of Bretschneidera sinensis. Jiangxi Science 2008:4.
Wang, M., Qiao, Q., Zhang, R., Hu, P., Yang, G., and Chen, H. (2011). Studies on the community feature comparison and phylogeography of relic plant Bretschneidera sinensis in Nankun and Daling Mountain, Guangdong Province. Guangxi Zhiwu/Guihaia 31, 789–794.
Wang, M.-N., Duan, L., Qiao, Q., Wang, Z.-F., Zimmer, E. A., Li, Z.-C., et al. (2018). Phylogeography and conservation genetics of the rare and relict Bretschneidera sinensis (Akaniaceae). PLoS One 13:e0189034. doi: 10.1371/journal.pone.0189034
Wang, X. W., and Wang, L. (2016). GMATA: An Integrated Software Package for Genome-Scale SSR Mining, Marker Development and VIewing. Front. Plant Sci. 2016:7. doi: 10.3389/fpls.2016.01350
Xu, D., Abe, J., Gai, J., and Shimamoto, Y. (2002). Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theoret. Appl. Gen. 105, 645–653. doi: 10.1007/s00122-002-0972-7
Xu, G., Liang, Y., Jiang, Y., Liu, X., Hu, S., Xiao, Y., et al. (2013). Genetic diversity and population structure of Bretschneidera sinensis, an endangered species. Biodiv. Sci. 21:723. doi: 10.3724/sp.j.1003.2013.09117
Yang, A. H., Zhang, J. J., Yao, X. H., and Huang, H. W. (2011). Chloroplast microsatellite markers in Liriodendron Tulipifera (Magnoliaceae) and cross-species amplification in L. Chin. Am. J. Bot. 98, E123–E126. doi: 10.3732/ajb.1000532
Keywords: chloroplast genome, Bretschneidera sinensis, ex situ conservation, genetic resources, genetic variation
Citation: Shang C, Li E, Yu Z, Lian M, Chen Z, Liu K, Xu L, Tong Z, Wang M and Dong W (2022) Chloroplast Genomic Resources and Genetic Divergence of Endangered Species Bretschneidera sinensis (Bretschneideraceae). Front. Ecol. Evol. 10:873100. doi: 10.3389/fevo.2022.873100
Received: 10 February 2022; Accepted: 25 April 2022;
Published: 01 June 2022.
Edited by:
Yukio Nagano, Saga University, JapanReviewed by:
Yun Song, Chinese Academy of Inspection and Quarantine (CAIQ), ChinaPankaj Bhardwaj, Central University of Punjab, India
Copyright © 2022 Shang, Li, Yu, Lian, Chen, Liu, Xu, Tong, Wang and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ce Shang, Y2Vfc2hhbmdAYmpmdS5lZHUuY24=
 Ce Shang
Ce Shang Enze Li
Enze Li Zhucheng Yu2
Zhucheng Yu2 Wenpan Dong
Wenpan Dong