Abstract
Microorganisms play a crucial role in regulating the turnover and transformation of soil organic carbon (SOC), whereas microbial contribution to SOC formation and storage is still unclear in coastal wetlands. In this study, we collected topsoil (0–20 cm) with 7 salinity concentrations and explored the shifts in microbial residues [represented by amino sugar (AS)] and their contribution to the SOC pool of coastal wetlands in the Yellow River delta. The gradually increasing soil salinity reduced soil water content (SWC), SOC, and soil nitrogen (N), especially in high salinity soils of coastal wetlands. Total ASs and their ratio to SOC, respectively, decreased by 90.56 and 66.35% from low salinity to high salinity soils, indicating that coastal wetlands with high salinity restrained microbial residue accumulation and microbial residue-C retention in the SOC pool. Together with redundancy analysis and path analysis, we found that SWC, pH, SOC, soil N, and glucosamine/muramic arid were positively associated with the ratio of ASs to SOC. The higher available soil resource (i.e., water, C substrate, and nutrient) increased microbial residue accumulation, promoting microbial derived-C contribution to SOC in low salinity coastal wetlands. The greatly decreased microbial residue contribution to SOC might be ascribed to microbial stress strategy and low available C substrate in coastal wetlands with high salinity concentration. Additionally, the gradually increasing salinity reduced fungal residue contribution to SOC but did not change bacterial residue contribution to SOC. These findings indicated that changed fungal residues would substantially influence SOC storage. Our study elucidates microbial contribution to SOC pool through residue reservoir in coastal wetlands and pushes microbial metabolites to a new application in global wetland SOC cycling.
Introduction
Coastal wetlands act as the important carbon (C) sink, which plays a crucial role in contributing to soil organic carbon (SOC) storage and regulating global C cycling (Chmura et al., 2003; Macreadie et al., 2019). However, the disturbance would greatly influence the structure and function of coastal wetlands. Naturally environmental disturbances (e.g., climate change and sea-level rise) or anthropogenic activities (e.g., agricultural reclamation and dam construction) result in the degradation of coastal wetlands, aggravating soil salinization (Xu et al., 2019; Haywood et al., 2020). Increased salinity stress results in the decline in biodiversity, loss of soil nutrients, and SOC and alters plant and microbial mediation in SOC transformation and accumulation (Crooks et al., 2018; Lewis et al., 2019; Chen et al., 2021).
Coastal wetlands are the transitional zone between terrestrial and aquatic ecosystems. The drastically changing habitats alter water and salt conditions, soil nutrition, and plant community, influencing soil microbial community structure and functions in coastal wetlands (Hu et al., 2014; Spivak et al., 2019). To date, the studies on microorganisms in coastal wetlands mainly focus on active soil microbial community structure and functions using the technologies and methods of phospholipid fatty acid, enzyme, high-throughput sequencing, and metagenome (Bossio et al., 2006; Wang et al., 2020). Soil microorganisms are key participants and drivers in SOC transformation, formation, and storage through microbial metabolic processes (Trivedi et al., 2013; Spivak et al., 2019). Microbial community composition (e.g., fungi and bacteria) and extracellular enzymes govern litter decomposition, soil nutrient release, and recycling, which further influence SOC turnover in coastal wetlands (Hill et al., 2018; Zhao et al., 2020). However, it is not clear how microbes directly contribute to SOC accumulation through microbial metabolites (e.g., microbial necromass) in coastal wetlands.
Microorganisms utilize plant and soil C substrates to synthesize microbial cells, and the dead microbial cell that persists in the soil as relative stable soil organic matter (SOM) contributes to the SOC pool via microbial residues (Angst et al., 2021). Microbial residues can account for 30–60% of SOC in the forest, grassland, and agricultural ecosystems (Liang et al., 2019). Coastal wetlands have special soil conditions, such as low oxygen, high salinity, or dramatically fluctuant hydrology, which are different from terrestrial ecosystems (Lyu et al., 2018; Steinmuller et al., 2019). Therefore, it is necessary to explore the contribution of microbial residues to the SOC pool in coastal wetlands.
Amino sugars (ASs), as the main components of microbial cell walls, are usually used to represent microbial residues (Ding et al., 2019). As the biomarker of microbial residues, AS, such as glucosamine (GluN), galactosamine (GalN), and muramic acid (MurA), are derived from different soil microbial communities. GluN is mainly detected in fungi; MurA is solely extracted in bacteria; whereas GalN can be synthesized by fungi and bacteria (Glaser et al., 2004; Joergensen, 2018). Based on the origin of AS monomer, the ratio of AS monomer not only represents the ratio of fungal residues to bacterial residues but also reflects the changes in fungi and bacteria (Joergensen, 2018).
Salinity is the main limiting factor, affecting soil microbial community structure and functions in coastal wetlands (Ma et al., 2017; Zhang et al., 2021). Phragmites australis is widely distributed in coastal wetlands due to their salt-rejection and salt-adaption, which is a suitable plant community to study the responses of microbial residues to salinity. Using the biomarker of AS, we explored the dynamic of microbial residues and their contribution to SOC under coastal wetlands along a natural salinity gradient in the Yellow River delta, China. The goals of this study were (1) to detect the impacts of salinity on microbial residues and their contributions to SOC; (2) to examine the variation in fungal and bacterial residues and their contributions to microbial residues; and (3) to clarify how abiotic and biotic parameters drive microbial residue-C retention in wetland soils.
Materials and Methods
Site Description and Sampling
Our study sites were located in the Yellow River Delta National Nature Reserve (118°33′–119°20′E, 37°35′–38°12′N), near Bohai and Laizhou Bay, China (Figure 1). Mean annual temperature and precipitation are approximately 12.4°C and 550 mm, respectively. Soil types are classified as fluvial soil formed by alluvial sediment of the Yellow River and saline soil formed by long-term seawater intrusion. In the water-salt interaction regions, we selected seven salinity concentrations (represented by electrical conductivity), including S1 (1.61 dS m–1), S2 (2.47 dS m–1), S3 (3.67 dS m–1), S4 (5.21 dS m–1), S5 (9.24 dS m–1), S6 (14.53 dS m–1), and S7 (29.90 dS m–1) (Table 1 and Figure 1). The sites of S1 and S2 are the interaction regions of the Yellow River and Bohai bay; S3 and S4 are affected by the seasonal tide; and S5, S6, and S7 are the degraded wetlands due to dam construction. Plant communities are dominated by Phragmites australis, following Suaeda glauca, Tamarix chinensis, Cynanchum, etc. We established study plots in August 2019 using a randomized within-wetland nested design. In each of the seven salinity concentrations, four plots (each approximately 10 m × 10 m) were randomly selected to collect soil samples (0–20 cm), with at least a 100-m distance between each plot. To alleviate spatial heterogeneity, we randomly took five soil cores using a soil auger (diameter of 5 cm) within each plot and pooled them into one soil sample (n = 4 samples per salinity concentration). After sieving soils to 2 mm to remove fine root, plant, and animal debris, soils were homogenized, and analyzed as below.
FIGURE 1
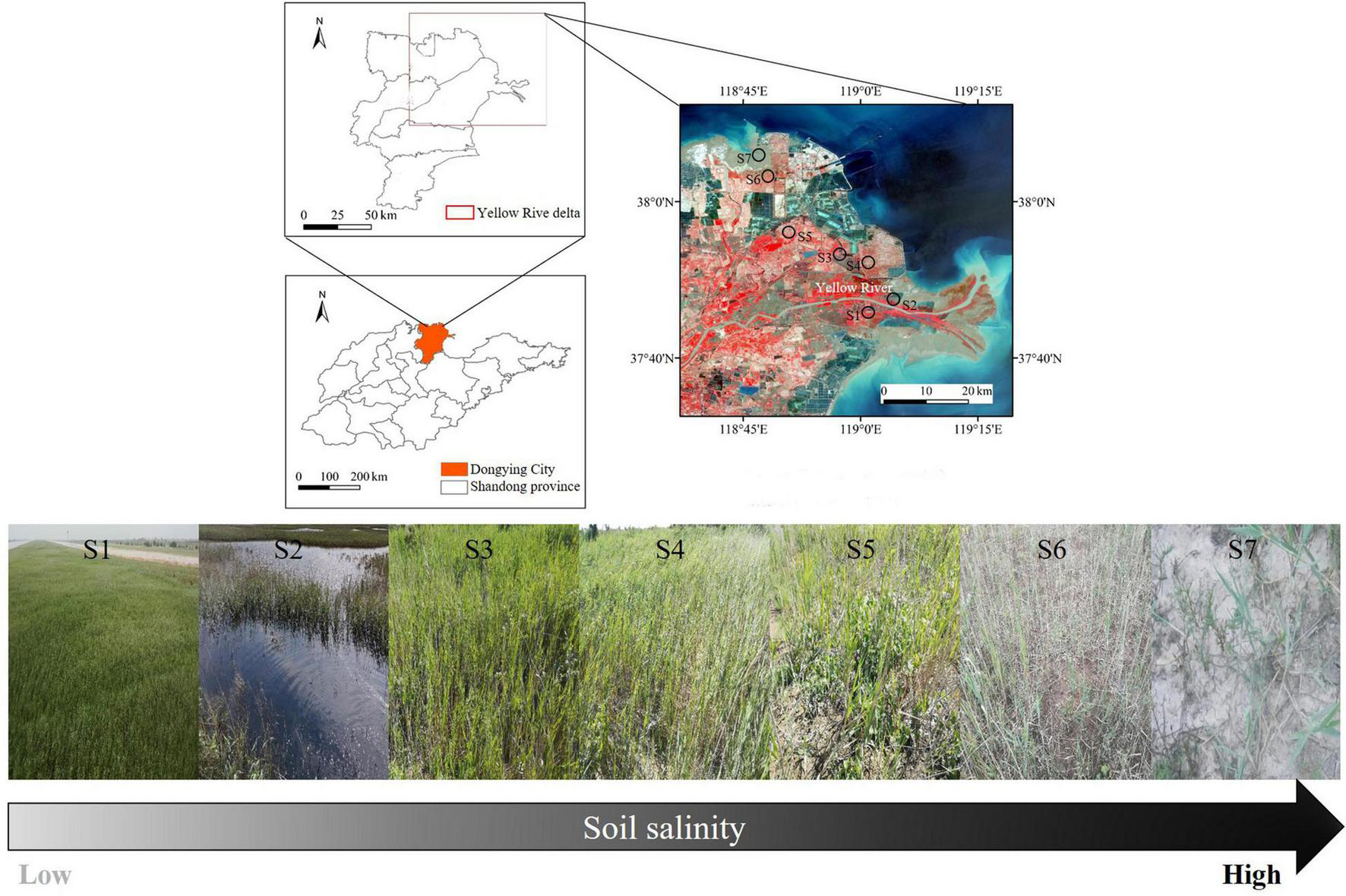
Sampling site of coastal wetlands in the Yellow River delta and photos of Phragmites australis along a soil salinity gradient.
TABLE 1
| Salinity gradient | Statistical value | ||||||||
|
|
|
||||||||
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | F | P | |
| EC (dS m–1) | 1.61 ± 0.13d | 2.47 ± 0.19d | 3.67 ± 0.11d | 5.21 ± 0.47d | 9.24 ± 0.31c | 14.53 ± 1.63b | 29.90 ± 2.71a | 68.06 | <0.001 |
| pH | 7.83 ± 0.06a | 7.57 ± 0.06ab | 7.64 ± 0.10ab | 7.69 ± 0.09ab | 7.36 ± 0.13b | 7.38 ± 0.156b | 7.40 ± 0.12b | 2.84 | 0.04 |
| SWC (%) | 29.41 ± 0.40ab | 32.03 ± 3.19a | 30.94 ± 3.63a | 30.22 ± 1.28a | 28.93 ± 4.21ab | 25.03 ± 1.34ab | 21.73 ± 1.58b | 2.2 | 0.08 |
| SOC (g kg–1) | 10.13 ± 1.64a | 8.12 ± 0.50a | 5.13 ± 0.28b | 4.98 ± 0.60b | 3.83 ± 0.32b | 3.05 ± 0.22b | 2.93 ± 0.40b | 13.89 | <0.001 |
| TN (g kg–1) | 1.07 ± 0.12a | 0.92 ± 0.06ab | 0.65 ± 0.06bc | 0.70 ± 0.10bc | 0.54 ± 0.05cd | 0.41 ± 0.02d | 0.47 ± 0.04d | 11.24 | <0.001 |
| SOC/TN | 9.62 ± 1.60a | 8.83 ± 0.31a | 7.99 ± 0.21a | 7.47 ± 1.22a | 8.11 ± 1.50a | 7.41 ± 0.38a | 6.34 ± 1.00a | 1.08 | 0.41 |
| TP (g kg–1) | 0.59 ± 0.07a | 0.55 ± 0.07a | 0.47 ± 0.03a | 0.50 ± 0.06a | 0.53 ± 0.08a | 0.56 ± 0.09a | 0.51 ± 0.06a | 0.29 | 0.94 |
Soil physicochemical properties with 7 salinity concentrations under coastal wetlands in the Yellow River delta.
EC, electrical conductivity; SWC, soil water content; SOC, soil organic carbon; TN, total nitrogen; SOC/TN ratio of SOC to TN; TP, total phosphorus. The superscript letters represent statistical significance.
Soil Physicochemical Analysis
Air-dried soils (sieved to 2 mm) were used to prepare soil slurries (1:5 w/v), oscillate for 30 min, and subsequently measure electrical conductivity (EC) and pH on an electrode meter (Jenco 6173, Jenco, United States). Fresh soil samples (sieved to 2 mm) were oven-dried (105°C for 48 h) to determine soil water content (SWC). Air-dried soil samples (sieved to 0.15 mm) were prepared to analyze SOC using the potassium dichromate oxidation method (Nelson and Sommers, 1982). Total nitrogen (TN) was measured on a Vario MACRO Cube (Elementar vario EL III, Germany). Total phosphorus (TP) was detected using a UV-VIS spectrophotometer (PERSEE, WA, United States) after soil digestion using H2SO4 and HClO4 (Cheng et al., 2016).
Soil Amino Sugar Analysis
Soil ASs, including GluN, GalN, and MurA, were determined according to a modified procedure referring to Zhang and Amelung (1996). In brief, air-dried soil samples (sieved to 0.15 mm) containing 0.3 mg N at least were hydrolyzed using 15 ml 6 M HCl at 105°C for 8 h. Each hydrolysate was filtered and dried at 52°C on a rotary vacuum evaporator after adding 100 μl of myoinositol (internal standard 1 with response factors, Rf of 1). The dried residue was dissolved using deionized water and purified with KOH neutralization. The solution was fully dried and dissolved with absolute methanol. The dissolved residue was dried by N2 gas at 45°C, subsequently redissolved with 1 ml deionized water and 100 μl N-methylglucamine (internal standard 2), and then lyophilized.
Each lyophilized residue was derivatized with a 300-μl derivatization reagent. The reactive solution was completely mixed and heated at 75–80°C for 35 min. The derivative was heated (75–80°C, 25 min) for acetylation after cooling and adding 1 ml acetic anhydride. After cooling to room temperature, we orderly added 1.5 ml dichloromethane and 1 ml 1 M HCl and fully mixed to isolate the organic phase. The organic phase was washed with deionized water three times to absolutely remove residual anhydride. The remaining organic phase was dried with N2 gas at 45°C and then dissolved with 100 μl ethyl acetate-hexane (1:1 v/v). Finally, the extracted ASs were detected and analyzed on an Agilent 7890B GC (Agilent Technologies, Santa Clara, CA, United States). In terms of the response factors of standards (i.e., GluN, GalN, MurA, and mannosamine) relative to the internal standard 1 (Rf = 1), we can calculate the concentration of soil ASs; the internal standard 2 is used to check the derivatization process and calculate the recovery of soil ASs (Zhang and Amelung, 1996; Liang et al., 2012).
Statistical Analysis
One-way ANOVA was conducted to assess the effects of salinity on soil physicochemical properties and AS parameters using R software (R version 3.6.0). We performed redundancy analysis to detect the relationships of GluN/MurA, GluN/SOC, GalN/SOC, and MurA/SOC (log-transformed) with soil physicochemical properties on Canoco (version 4.5 for Windows; Ithaca, NY, United States). Path analysis was used to evaluate the causality among soil physicochemical properties and ASs along a salinity gradient, using AMOS software (AMOS 17.0.2 student version; Amos Development, Crawfordville, FL, United States). The maximum likelihood method was used to fit measured data to the model. The adequate model goodness of fit was tested using root-mean-square error of approximation (RMSEA).
Results
Variation of Soil Physicochemical Properties Along a Salinity Gradient
Salinity resulted in the significant changes in soil pH (F = 2.84, P = 0.04), SOC (F = 13.89, P < 0.001), and TN (F = 11.24, P < 0.001), whereas there was no change in SWC, SOC/TN, and TP (Table 1). High salinity soils (S5, S6, and S7) had lower pH than low salinity soils (S1), decreasing by 5.82%. SOC and TN gradually decreased along with increased salinity, from 10.13 to 2.93 g kg–1 and from 1.07 to 0.41 g kg–1, respectively. Although salinity did not change SWC, high salinity coastal wetlands had low SWC (Table 1).
Effects of Salinity on Soil Amino Sugar Content and Their Contributions to Soil Organic Carbon
Salinity greatly influenced the contents of soil AS (F = 29.37, P < 0.001), GluN (F = 25.84, P < 0.001), GalN (F = 35.65, P < 0.001), and MurA (F = 10.77, P < 0.001) (Figure 2). The contents of AS, GluN, and GalN gradually declined across a salinity gradient, which decreased by 90.56% (Figure 2A), 91.11% (Figure 2B), and 90.62% (Figure 2C), respectively. The contents of MurA were higher in low salinity soils (S1 and S2) than that of other soil salinity concentrations. Additionally, MurA showed no change from S3 to S7 (Figure 2D).
FIGURE 2
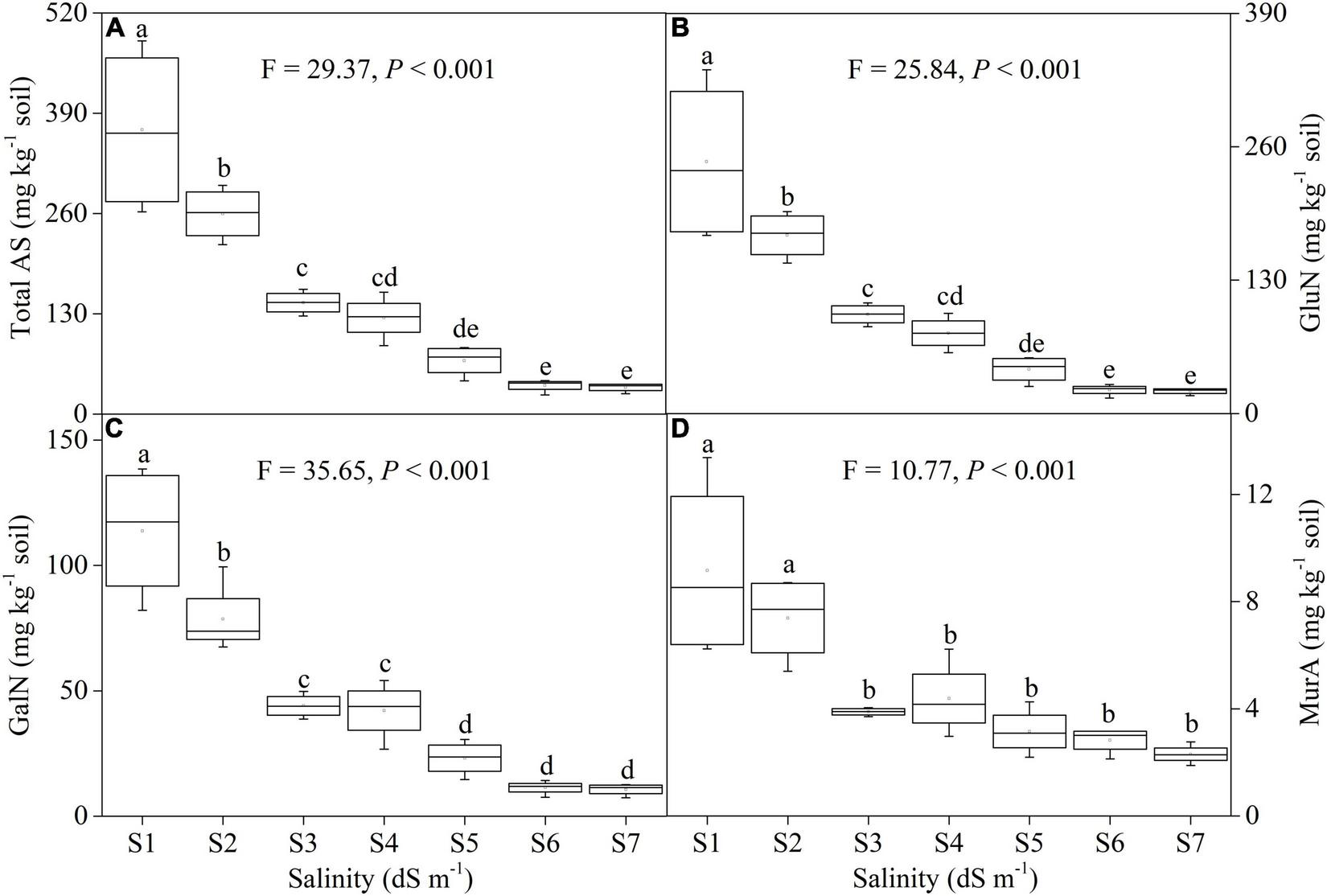
Changes in the content of soil total amino sugar (AS, A), glucosamine (GluN, B), galactosamine (GalN, C), and muramic acid (MurA, D) along a soil salinity gradient.
The ratios of AS, GluN, and GalN to SOC showed significant responses to salinity (P < 0.001), whereas there was no change in the ratio of MurA to SOC (Figure 3). The ratios of AS, GluN, and GalN to SOC showed the similar trend with AS content across a salinity gradient. The AS/SOC decreased from 36.62 to 12.32 g kg–1 (Figure 3A; F = 39.20, P < 0.001); GluN/SOC showed a decline from 24.22 to 7.76 g kg–1 (Figure 3B; F = 43.27, P < 0.001); and GalN/SOC reduced from 11.50 to 3.75 g kg–1 (Figure 3C; F = 24.00, P < 0.001) from S1 to S7.
FIGURE 3
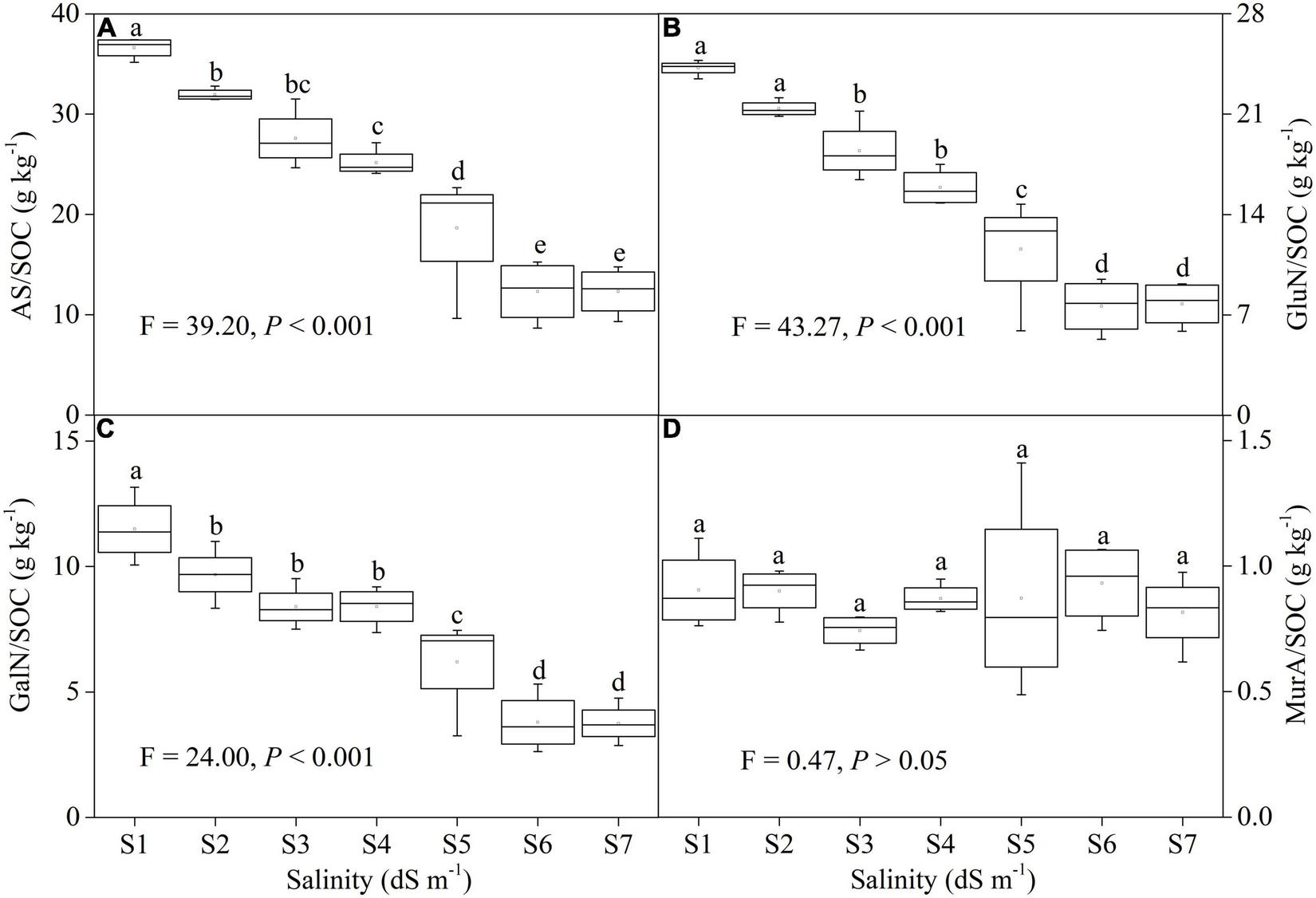
The ratio of total AS (A), GluN (B), GalN (C), and MurA (D) to SOC across a soil salinity gradient.
Shifts in GluN/GalN and GluN/MurA Under Different Salinity Concentrations
Salinity resulted in a significant change in GluN/MurA (Figure 4B; F = 31.32, P < 0.001), but there was no change in GluN/MurA (Figure 4A). Low salinity soils had high GluN/MurA, and high salinity lowered GluN/MurA in soils (Figure 4B).
FIGURE 4
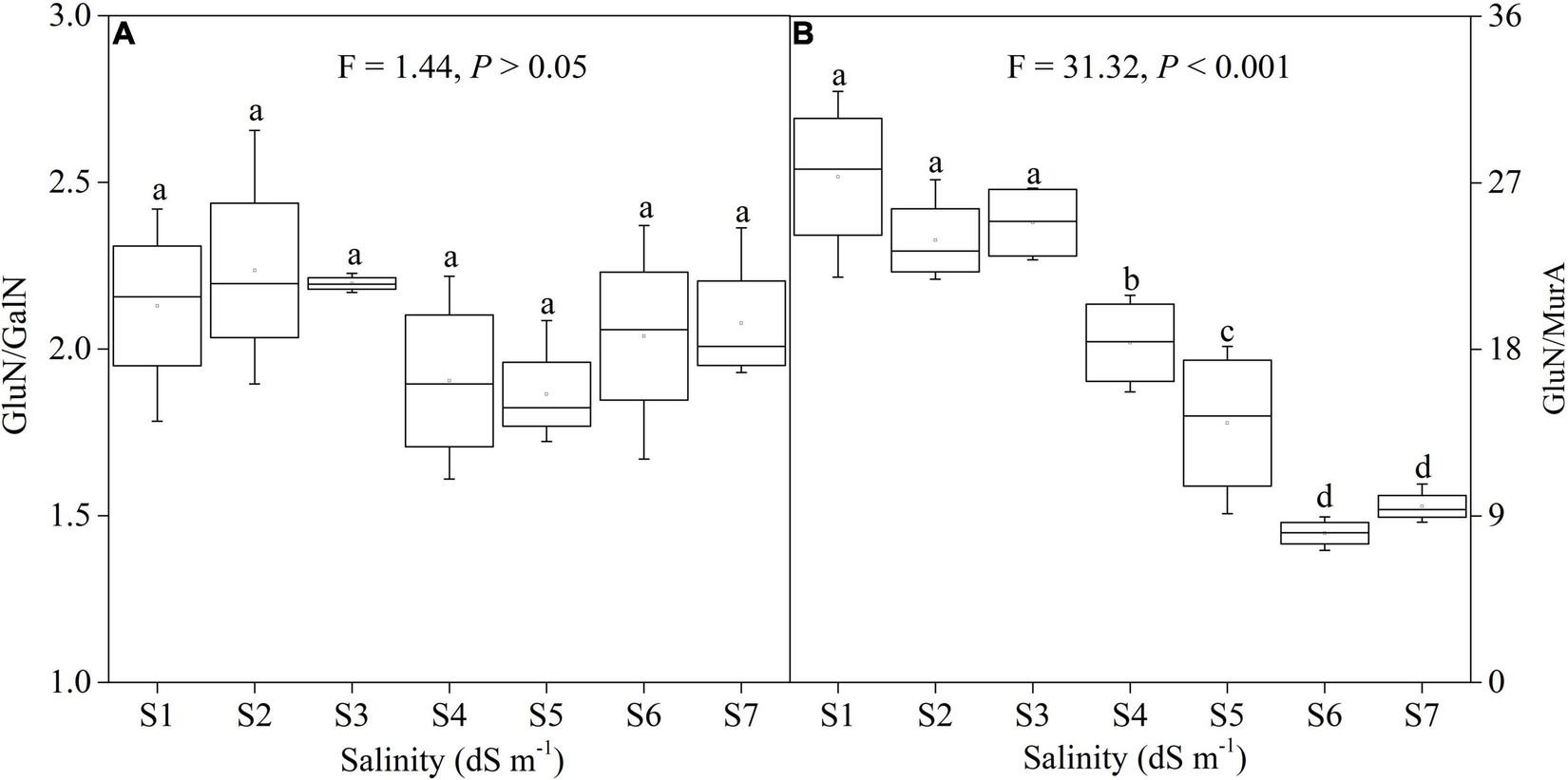
Variation of GluN/GalN (A) and GluN/MurA (B) under different salinity concentrations.
Relationships Between Soil Physicochemical Properties and Amino Sugar
Redundancy analysis demonstrated that soil physicochemical properties explained 82.19% (axis 1 of 80.1% and axis 2 of 2.1%) variation of the GluN/MurA, GluN/SOC, GalN/SOC, and MurA/SOC (Figure 5). The ratios of GluN/SOC, GalN/SOC, and GluN/MurA were negatively related to soil salinity (represented by EC) and positively associated with SWC, pH, SOC, and TN, whereas not related to SOC/TN and TP. Soil physicochemical properties showed no impact on the MurA/SOC (Figure 5).
FIGURE 5
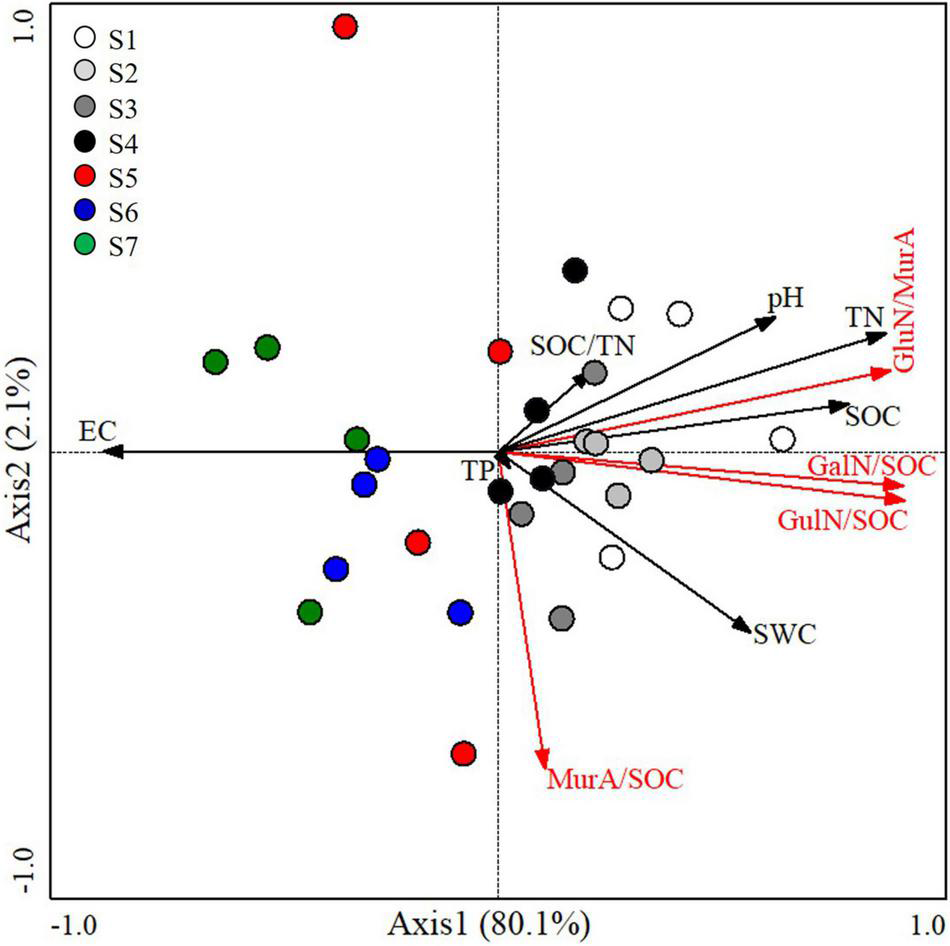
Redundancy analysis detecting the relationships of AS parameters with soil physicochemical properties along a salinity gradient (white circle, S1; light gray circle, S2; gray circle, S3; black circle, S4; red circle, S5; blue circle, S6; green circle, S7). Black vectors indicate trajectories of soil chemical parameters as independent variables; red vectors indicate AS parameters, including the ratio of GluN, GalN, and MurA to SOC and GluN/MurA. The acute and obtuse angles among variables represent positive and negative relationships, respectively. The numbers in brackets indicate the percentage of the total variance explained by the first and second axes. Soil physicochemical properties with significant effects on AS parameters are evaluated at the 0.05 level (Monte Carlo tests with 499 permutations).
Path analysis further evaluated the causal relationships among soil physicochemical properties, GluN/MurA, and AS/SOC across a salinity gradient in coastal wetlands and elucidated the relative contributions of these soil variables to explain changed AS/SOC (Figure 6). Salinity, pH, SOC, TN, and GluN/MurA were the main abiotic and biotic factors influencing AS/SOC, accounting for 84% variation of the AS/SOC. Soil salinity showed a negative effect on soil chemical properties and AS/SOC; soil pH and TN indirectly and positively influenced AS/SOC via changing the GluN/MurA; and SOC showed a positive direct impact on AS/SOC (Figure 6).
FIGURE 6
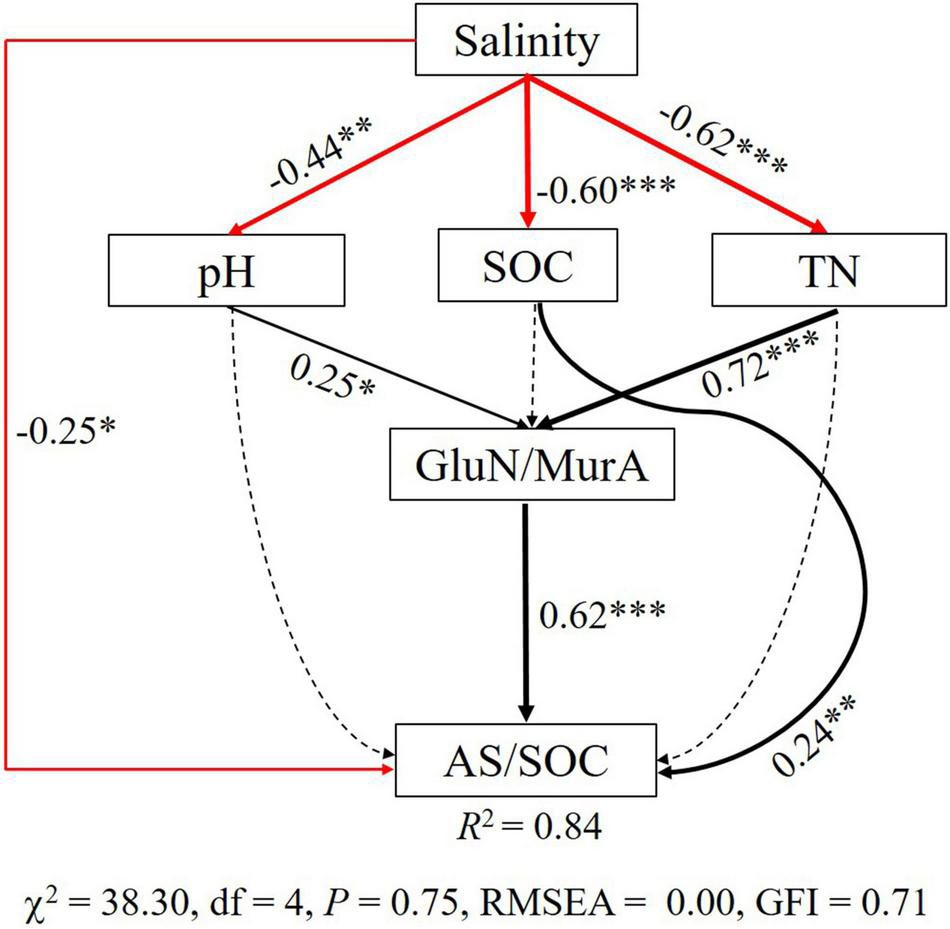
Path analysis evaluating the underlying causality between soil physicochemical properties and AS/SOC along a salinity gradient under coastal wetlands in the Yellow River delta. Solid and dashed lines indicate significant (P < 0.05) and non-significant (P > 0.05) effects (P-values inside brackets), respectively. Standardized path coefficients are listed beside each path (black and red lines represent positive and negative paths, respectively; line width indicates the proportion of factorial contribution). R2 values indicate the strength of explanation by independent variables. The symbols of ***, **, *represent statistical signifiance with p < 0.001, p < 0.01, and p < 0.05, respectively.
Discussion
Responses of Soil Microbial Residues to Salinity in Coastal Wetlands
In this study, we found that the AS content was 34.82–368.6 mg kg–1 along a salinity gradient under coastal wetlands in the Yellow River delta, which was several or even tens of times lower than AS content in terrestrial ecosystems (i.e., forest, grassland, and agriculture) (Lauer et al., 2011; Xia et al., 2019; Chen et al., 2020). Coastal wetlands are predominately controlled by water and salinity due to tides and underground seawater (Ma et al., 2017; Steinmuller et al., 2019). The restrained plant productivity leads to low SOM (Zhao et al., 2020), decreasing the accumulation of microbial AS. Also, plant phenology shows a quick response to changed soil salinity (Sun et al., 2021). High salinity negatively influences plant phenology and alters the seasonality of plant photosynthesis or productivity. Especially in dry seasons, the exacerbated soil salinity stress restrains aboveground plant growth, root germination, and reproduction (related to plant reproduction the next year) (Baldwin et al., 2014; Sun et al., 2021), reducing C input to soils, consequently resulting in the decrease in microbial AS. In addition, the SOM was not easily utilized by soil microorganisms with a low decomposition rate (Steinmuller and Chambers, 2019; Ward, 2020), which is likely to result in low AS synthesis.
As the main component of the microbial cell wall, soil AS has certain specificity and stability, which cannot only represent microbial residues but also reflect the dynamic changes of ecosystems (Lauer et al., 2011; Shao et al., 2019). Increased soil salinity decreased AS content, indicating that salinity stress would suppress microbial residue accumulation in coastal wetland soils. Microbial residues are mainly originated from the fast iteration of microbial cell production and death, thus high microbial biomass could promote microbial residue retention in soils (Liang et al., 2017). A recent study has reported that increased salinity results in a decline in microbial biomass, associated with the decreased microbial residues in the saline-alkali soils (Chen et al., 2021). In addition, microorganisms utilize more SOM to synthesize several stress-resistant substances (e.g., extracellular polysaccharides) to maintain microbial survival and integrity and less SOM investment in microbial cell growth and metabolism (Malik et al., 2020), thus resulting in low microbial residues in high salinity soils.
The GluN and MurA can be used to represent fungal and bacterial residues, respectively (Joergensen, 2018). The GluN accounted for 65.67% of total AS; MurA accounted for 3.19% of total AS; and the content of GluN was approximately 20 times of MurA in coastal wetland soils. These findings are similar to the previous studies that the content of soil GluN was approximately 15–20 times of MurA in terrestrial ecosystems (Shao et al., 2019; Wang et al., 2021). Although increased salinity stress greatly decreased the contents of GluN and MurA, the proportion of GluN and MurA in total AS showed an increment and decline, respectively. The inconsistent changes in fungal and bacterial residues are probably ascribed to the different responses of fungal and bacterial communities to salinity. The 80% of fungi in soil systems are symbiotic with plants; and changed plant productivity and traits would influence fungal activity and biomass (Zheng et al., 2021). In our studied regions, increased soil salinity inhibits the growth and photosynthate synthesis of Phragmites australis; and few Phragmites australis survive in high or extreme salinity soils (Guan et al., 2017). The decreased plant litterfall and root exudates lead to less available C input to soil fungi, particularly plant symbiotic fungi, to assimilate less C into fungal biomass, thus reducing fungal residue retention in soils. MurA is uniquely derived from bacteria, which can reflect the change in the bacterial community (Glaser et al., 2004). Low salinity soils had higher bacterial residues, and there was no change in bacterial residues from medium salinity to high salinity. The dynamic of bacterial residues along with soil salinity gradient may be attributed to the salinity tolerance of the bacterial community. Some specific bacterial taxa (e.g., Halomonas and Chloroflexi) can survive in high salinity soil (Xu et al., 2021), which maintains the balance between production and degradation of bacterial residues, resulting in no change in bacterial residues.
Microbial Residue Contributions to Soil Organic Carbon Pool Along a Salinity Gradient in Coastal Wetlands
With the developments of microbial biomarkers (i.e., AS) and microbial C pump theory (Zhang and Amelung, 1996; Liang et al., 2017), more studies concentrate on microbial residue contribution to SOC pool (Kallenbach et al., 2016; Ma et al., 2018; Deng and Liang, 2022). The AS extracted from soils accounts for a major percentage of dead microbial biomass (Joergensen, 2018). Therefore, AS levels are used to evaluate the extent to which microbial residue contributes to the SOC pool, providing significant information on the “chronic” responses of microbial residues to changing ecosystems (Shao et al., 2019). The AS/SOC can represent microbial residue contributions to SOC. The AS/SOC was gradually declined with increasing salinity, approximately decreasing by 70% from low salinity to high salinity, indicating that high salinity stress greatly inhibits microbial residue contributions to the SOC pool. We found that salinity negatively affected fungal residue contributions to SOC, and there was no change in bacterial residue contributions to SOC. These findings imply that fungal residues predominately drive SOC storage, which is consistent with previous views (Wang et al., 2021). The different effects of salinity on fungal and bacterial residue contributions to SOC are likely ascribed to change soil physicochemical properties. Our study shows the positive relationship of GluN/SOC with SWC, pH, SOC, and TN, suggesting that changed soil water and nutrient conditions regulate fungal residue accumulation in the SOC pool. Additionally, the chemical stability of bacterial residues is lower than fungal residues, which are easily decomposed and assimilated by soil microorganisms (Joergensen, 2018; Ma et al., 2022). The fast turnover of bacterial residues is not conducive to the retention of microbial residue-C belowground in coastal wetlands.
Current studies pay more attention to the plant community effect on SOC cycling, and plant-derived C mainly drives wetland soil C storage (Duarte et al., 2013; Xia et al., 2021). Microbial C pump theory has proposed that microorganisms utilize the decomposed plant detritus and micromolecular SOM to synthesize microbial cells, contributing to the SOC pool via microbial necromass (Kästner and Miltner, 2018). Low salinity held high AS/SOC, which indicates more soil microbial residue-C accumulation in low salinity coastal wetlands. Abundant soil water, SOC, and N promote the synthesis of microbial products, enhancing the contribution of microbial residues to existing SOC stocks. High soil water and nutrient stimulate plant growth and photosynthetic C input into soils (Chu et al., 2019); and microbes assimilate more plant-derived C to synthesize microbial biomass, ultimately increasing microbial residue-C storage in coastal wetland soils. In addition, we also found that AS/SOC was positively related to pH. Some studies have clarified that pH is tightly associated with soil cation, such as calcium and aluminum ions (Gruba and Mulder, 2015). Microbial residues can persist in soils for decades or even centuries in the form of stable SOM through mineral association (Sokol et al., 2019; Buckeridge et al., 2020), substantially enhancing soil C storage. Overall, the abundant soil resource (i.e., water, C substrate, and N nutrient) can promote SOC storage by increasing microbial residue accumulation and stability in low salinity coastal wetlands.
In high salinity coastal wetlands, salinity stress greatly restrained the contribution of microbial residues to SOC. This might be explained by the two mechanisms, namely, microbial stress tolerance strategy and low available C. In extreme environmental stress (e.g., high salinity or drought), microorganisms select a stress tolerance strategy responding to environmental changes (Krause et al., 2014; Malik et al., 2020). Microorganisms invest more C to synthesize osmolytes (e.g., mycose) or extracellular polymeric substances (e.g., polysaccharides) to protect microbial cell integrity and to enhance microbial stress-resistance in high salinity soils. Meanwhile, less C was used for microbial cell growth and proliferation, thus reducing the source of microbial residues. The low microbial cell death and microbial residue production both lower the retention of microbial residues in the SOC pool of coastal wetlands. In addition, high salinity soils held few SOC and low SOC/TN, which decrease microbial C availability. Low soil available C inhibits the production of microbial biomass and residues, resulting in the low contribution of microbial residues to SOC storage in high salinity coastal wetlands.
Path analysis was used to evaluate the causal relationships among microbial residue-C contribution, soil physicochemical properties, and fungal/bacterial residues, clarifying the main factors driving microbial residue contribution to SOC. Changed soil salinity, pH, SOC, TN, and GluN/MurA explained 84% variation of microbial residue-C contribution to SOC. Soil nutrient regulates microbial residue contribution to SOC through varied fungal and bacterial residues associated with their stability along a salinity gradient in coastal wetlands.
Although soil physicochemical properties (e.g., salinity, water, SOC, and N) were substantially different among coastal wetlands with different salinity concentrations, the changes in microbial residues and their contributions to SOC were influenced. However, we cannot neglect the impacts of system experience conditions (e.g., hydrology, microclimate, and topography) on microbial residues in different geographical locations. Together, our study presents supporting evidence on how salinity affects the fate and magnitude of microbial residues in coastal wetlands through changed soil properties. Further research is needed to accurately assess microbial residues in changing wetlands considering system experience conditions and other fluctuant environmental variants.
Responses of Fungal/Bacterial Residues to Salinity in Coastal Wetlands
The ratio of GluN to MurA cannot only indicate the responses of fungal and bacterial communities to environmental changes but also represent the relative contributions of fungal and bacterial residues to the SOC pool (Liang et al., 2015; Joergensen, 2018). We found a gradual reduction in GluN/MurA across increasing salinity, implying that fungi are more competitive in low salinity soils, and bacteria are more tolerant in high salinity soils of coastal wetlands. The positive relationships of GluN/MurA to SWC, SOC, and TN indicate that suitable soil water, salinity, and nutrients favor fungi and stimulate fungal residue accumulation, reserving more microbial residue-C in the SOC pool.
Conclusion
Microbial residues (represented by AS) play a crucial role in coastal wetland SOC accumulation. In our study, we found that low-salinity soils held high microbial residues and microbial residue contribution to SOC, while high-salinity stress greatly inhibited microbial residue accumulation and microbial residue retention in the SOC pool in coastal wetlands. Changed soil resource (i.e., water, C substrate, and nitrogen) availability and microbial strategy to salinity stress explained the accumulation of microbial residues along a salinity gradient in coastal wetlands. Additionally, the different responses of fungal and bacterial residues to salinity and the positive relationship between fungal/bacterial residues and microbial residue C contribution indicate that fungal residues predominately contributed to the SOC pool. We suggest that clarifying microbial necromass contribution to SOC pool will strengthen the C persistence in soils and a more accurate understanding of global wetland soil C cycling.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
PS wrote the manuscript. HH, JS, HY, and HX revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (32101387, 32001134, and 42171059) and the Natural Science Foundation of Shandong Province (ZR2020QD004, ZR2020MD005, and ZR2020QC040).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Angst G. Mueller K. E. Nierop K. G. Simpson M. J. (2021). Plant-or microbial-derived? A review on the molecular composition of stabilized soil organic matter.Soil Biol. Biochem.156:108189. 10.1016/j.soilbio.2021.108189
2
Baldwin A. H. Jensen K. Schönfeldt M. (2014). Warming increases plant biomass and reduces diversity across continents, latitudes, and species migration scenarios in experimental wetland communities.Glob. Chang. Biol.20835–850. 10.1111/gcb.12378
3
Bossio D. A. Fleck J. A. Scow K. M. Fujii R. (2006). Alteration of soil microbial communities and water quality in restored wetlands.Soil Biol. Biochem.381223–1233. 10.1016/j.soilbio.2005.09.027
4
Buckeridge K. M. La Rosa A. F. Mason K. E. Whitaker J. McNamara N. P. Grant H. K. et al (2020). Sticky dead microbes: rapid abiotic retention of microbial necromass in soil.Soil Biol. Biochem.149:107929. 10.1016/j.soilbio.2020.107929
5
Chen G. Ma S. Tian D. Xiao W. Jiang L. Xing A. et al (2020). Patterns and determinants of soil microbial residues from tropical to boreal forests.Soil Biol. Biochem.151:108059. 10.1016/j.soilbio.2020.108059
6
Chen J. Wang H. Hu G. Li X. Dong Y. Zhuge Y. et al (2021). Distinct accumulation of bacterial and fungal residues along a salinity gradient in coastal salt-affected soils.Soil Biol. Biochem.158:108266. 10.1016/j.soilbio.2021.108266
7
Cheng Y. Li P. Xu G. Gao H. (2016). Spatial distribution of soil total phosphorus in Yingwugou watershed of the Dan River, China.Catena136175–181. 10.1016/j.catena.2015.02.015
8
Chmura G. L. Anisfeld S. C. Cahoon D. R. Lynch J. C. (2003). Global carbon sequestration in tidal, saline wetland soils.Glob. Biogeochem. Cycles17:1111.
9
Chu X. Han G. Xing Q. Xia J. Sun B. Li X. et al (2019). Changes in plant biomass induced by soil moisture variability drive interannual variation in the net ecosystem CO2 exchange over a reclaimed coastal wetland.Agric. For. Meteorol.264138–148. 10.1016/j.agrformet.2018.09.013
10
Crooks S. Sutton-Grier A. E. Troxler T. G. Herold N. Bernal B. Schile-Beers L. et al (2018). Coastal wetland management as a contribution to the US National Greenhouse Gas Inventory.Nat. Clim. Change81109–1112. 10.1038/s41558-018-0345-0
11
Deng F. Liang C. (2022). Revisiting the quantitative contribution of microbial necromass to soil carbon pool: stoichiometric control by microbes and soil.Soil Biol. Biochem.165:108486. 10.1016/j.soilbio.2021.108486
12
Ding X. Zhang B. Filley T. R. Tian C. Zhang X. He H. (2019). Changes of microbial residues after wetland cultivation and restoration.Biol. Fertil. Soils55405–409. 10.1007/s00374-019-01341-2
13
Duarte C. M. Losada I. J. Hendriks I. E. Mazarrasa I. Marbà N. (2013). The role of coastal plant communities for climate change mitigation and adaptation.Nat. Clim. Change3961–968. 10.1038/nclimate1970
14
Glaser B. Turrión M.-B. Alef K. (2004). Amino sugars and muramic acid—biomarkers for soil microbial community structure analysis.Soil Biol. Biochem.36399–407. 10.1016/j.soilbio.2003.10.013
15
Gruba P. Mulder J. (2015). Tree species affect cation exchange capacity (CEC) and cation binding properties of organic matter in acid forest soils.Sci. Total Environ.511655–662. 10.1016/j.scitotenv.2015.01.013
16
Guan B. Yu J. Hou A. Han G. Wang G. Qu F. et al (2017). The ecological adaptability of Phragmites australis to interactive effects of water level and salt stress in the Yellow River Delta.Aquat. Ecol.51107–116. 10.1007/s10452-016-9602-3
17
Haywood B. J. Hayes M. P. White J. R. Cook R. L. (2020). Potential fate of wetland soil carbon in a deltaic coastal wetland subjected to high relative sea level rise.Sci. Total Environ.711:135185. 10.1016/j.scitotenv.2019.135185
18
Hill B. H. Elonen C. M. Herlihy A. T. Jicha T. M. Serenbetz G. (2018). Microbial ecoenzyme stoichiometry, nutrient limitation, and organic matter decomposition in wetlands of the conterminous United States.Wetl. Ecol. Manage.26425–439. 10.1007/s11273-017-9584-5
19
Hu Y. Wang L. Tang Y. Li Y. Chen J. Xi X. et al (2014). Variability in soil microbial community and activity between coastal and riparian wetlands in the Yangtze River estuary–Potential impacts on carbon sequestration.Soil Biol. Biochem.70221–228. 10.1016/j.soilbio.2013.12.025
20
Joergensen R. G. (2018). Amino sugars as specific indices for fungal and bacterial residues in soil.Biol. Fertil. Soils54559–568. 10.1007/s00374-018-1288-3
21
Kallenbach C. M. Frey S. D. Grandy A. S. (2016). Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls.Nat. Commun.7:13630.
22
Kästner M. Miltner A. (2018). “SOM and Microbes-What is Left from Microbial Life,” in The Future of Soil Carbon, edsGarciaC.NannipieriP.HernandezT. (Cambridge, MA: Academic Press), 125–163. 10.1016/b978-0-12-811687-6.00005-5
23
Krause S. Le Roux X. Niklaus P. A. Van Bodegom P. M. Lennon J. T. Bertilsson S. et al (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning.Front. Microbiol.5:251. 10.3389/fmicb.2014.00251
24
Lauer F. Kösters R. Du Preez C. C. Amelung W. (2011). Microbial residues as indicators of soil restoration in South African secondary pastures.Soil Biol. Biochem.43787–794. 10.1016/j.soilbio.2010.12.012
25
Lewis C. J. E. Baldock J. A. Hawke B. Gadd P. S. Zawadzki A. Heijnis H. et al (2019). Impacts of land reclamation on tidal marsh ‘blue carbon’stocks.Sci. Total Environ.672427–437. 10.1016/j.scitotenv.2019.03.345
26
Liang C. Amelung W. Lehmann J. Kästner M. (2019). Quantitative assessment of microbial necromass contribution to soil organic matter.Glob. Chang. Biol.253578–3590. 10.1111/gcb.14781
27
Liang C. Gutknecht J. Balser T. (2015). Microbial lipid and amino sugar responses to long-term simulated global environmental changes in a California annual grassland.Front. Microbiol.6:385. 10.3389/fmicb.2015.00385
28
Liang C. Read H. W. Balser T. C. (2012). GC-based detection of aldononitrile acetate derivatized glucosamine and muramic acid for microbial residue determination in soil.J. Vis. Exp.63:e3767. 10.3791/3767
29
Liang C. Schimel J. P. Jastrow J. D. (2017). The importance of anabolism in microbial control over soil carbon storage.Nat. Microbiol.2:17105. 10.1038/nmicrobiol.2017.105
30
Lyu Z. Genet H. He Y. Zhuang Q. McGuire A. D. Bennett A. et al (2018). The role of environmental driving factors in historical and projected carbon dynamics of wetland ecosystems in Alaska.Ecol. Appl.281377–1395. 10.1002/eap.1755
31
Ma T. Zhu S. Wang Z. Chen D. Dai G. Feng B. et al (2018). Divergent accumulation of microbial necromass and plant lignin components in grassland soils.Nat. Commun.9:3480. 10.1038/s41467-018-05891-1
32
Ma X. Zhang W. Zhang X. Bao X. Xie H. Li J. et al (2022). Dynamics of microbial necromass in response to reduced fertilizer application mediated by crop residue return.Soil Biol. Biochem.165:108512. 10.1016/j.soilbio.2021.108512
33
Ma Z. Zhang M. Xiao R. Cui Y. Yu F. (2017). Changes in soil microbial biomass and community composition in coastal wetlands affected by restoration projects in a Chinese delta.Geoderma289124–134. 10.1016/j.geoderma.2016.11.037
34
Macreadie P. I. Anton A. Raven J. A. Beaumont N. Connolly R. M. Friess D. A. et al (2019). The future of Blue Carbon science.Nat. Commun.10:3998.
35
Malik A. A. Martiny J. B. Brodie E. L. Martiny A. C. Treseder K. K. Allison S. D. (2020). Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change.ISME J.141–9. 10.1038/s41396-019-0510-0
36
Nelson D. W. Sommers L. E. (1982). “Total carbon, organic carbon and organic matter. In Methods of Soil Analysis Part 3-Chemical Methods,” in Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties,ed.PageA. L. (Madison: SSSA)
37
Shao P. Liang C. Lynch L. Xie H. Bao X. (2019). Reforestation accelerates soil organic carbon accumulation: evidence from microbial biomarkers.Soil Biol. Biochem.131182–190. 10.1016/j.soilbio.2019.01.012
38
Sokol N. W. Sanderman J. Bradford M. A. (2019). Pathways of mineral-associated soil organic matter formation: integrating the role of plant carbon source, chemistry, and point of entry.Glob. Chang. Biol.2512–24. 10.1111/gcb.14482
39
Spivak A. C. Sanderman J. Bowen J. L. Canuel E. A. Hopkinson C. S. (2019). Global-change controls on soil-carbon accumulation and loss in coastal vegetated ecosystems.Nat. Geosci.12685–692. 10.1038/s41561-019-0435-2
40
Steinmuller H. E. Chambers L. G. (2019). Characterization of coastal wetland soil organic matter: implications for wetland submergence.Sci. Total Environ.677648–659. 10.1016/j.scitotenv.2019.04.405
41
Steinmuller H. E. Dittmer K. M. White J. R. Chambers L. G. (2019). Understanding the fate of soil organic matter in submerging coastal wetland soils: a microcosm approach.Geoderma3371267–1277. 10.1016/j.geoderma.2018.08.020
42
Sun B. Yan L. Jiang M. Li X. Han G. Xia J. (2021). Reduced magnitude and shifted seasonality of CO2 sink by experimental warming in a coastal wetland.Ecology102:e03236. 10.1002/ecy.3236
43
Trivedi P. Anderson I. C. Singh B. K. (2013). Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction.Trends Microbiol.21641–651. 10.1016/j.tim.2013.09.005
44
Wang B. An S. Liang C. Liu Y. Kuzyakov Y. (2021). Microbial necromass as the source of soil organic carbon in global ecosystems.Soil Biol. Biochem.162:108422. 10.1111/gcb.14070
45
Wang J. Wang J. Zhang Z. Li Z. Zhang Z. Zhao D. et al (2020). Shifts in the bacterial population and ecosystem functions in response to vegetation in the Yellow River Delta wetlands.mSystems5:e00412-20. 10.1128/mSystems.00412-20
46
Ward R. D. (2020). Carbon sequestration and storage in Norwegian Arctic coastal wetlands: impacts of climate change.Sci. Total Environ.748:141343. 10.1016/j.scitotenv.2020.141343
47
Xia S. Song Z. Li Q. Guo L. Yu C. Singh B. P. et al (2021). Distribution, sources, and decomposition of soil organic matter along a salinity gradient in estuarine wetlands characterized by C: N ratio, d13C-d15N, and lignin biomarker.Glob. Chang. Biol.27417–434. 10.1111/gcb.15403
48
Xia Y. Chen X. Hu Y. Zheng S. Ning Z. Guggenberger G. et al (2019). Contrasting contribution of fungal and bacterial residues to organic carbon accumulation in paddy soils across eastern China.Biol. Fertil. Soils55767–776. 10.1007/s00374-019-01390-7
49
Xu J. Gao W. Zhao B. Chen M. Ma L. Jia Z. et al (2021). Bacterial community composition and assembly along a natural sodicity/salinity gradient in surface and subsurface soils.Appl. Soil Ecol.157:103731. 10.1016/j.apsoil.2020.103731
50
Xu W. Fan X. Ma J. Pimm S. L. Kong L. Zeng Y. et al (2019). Hidden loss of wetlands in China.Curr. Biol.293065–3071. 10.1016/j.cub.2019.07.053
51
Zhang G. Bai J. Tebbe C. C. Zhao Q. Jia J. Wang W. et al (2021). Salinity controls soil microbial community structure and function in coastal estuarine wetlands.Environ. Microbiol.231020–1037. 10.1111/1462-2920.15281
52
Zhang X. Amelung W. (1996). Gas chromatographic determination of muramic acid, glucosamine, mannosamine, and galactosamine in soils.Soil Biol. Biochem.281201–1206.
53
Zhao Q. Zhao H. Gao Y. Zheng L. Wang J. Bai J. (2020). Alterations of bacterial and archaeal communities by freshwater input in coastal wetlands of the Yellow River Delta, China.Appl. Soil Ecol.153:103581. 10.1016/j.apsoil.2020.103581
54
Zheng Y. Chen L. Ji N. N. Wang Y. L. Gao C. Jin S. S. et al (2021). Assembly processes lead to divergent soil fungal communities within and among twelve forest ecosystems along a latitudinal gradient.New Phytol.2311183–1194. 10.1111/nph.17457
Summary
Keywords
amino sugar, soil salinity, soil organic carbon, microbial necromass, coastal wetland
Citation
Shao P, Han H, Sun J, Yang H and Xie H (2022) Salinity Effects on Microbial Derived-C of Coastal Wetland Soils in the Yellow River Delta. Front. Ecol. Evol. 10:872816. doi: 10.3389/fevo.2022.872816
Received
10 February 2022
Accepted
28 March 2022
Published
27 April 2022
Volume
10 - 2022
Edited by
Yijian Zeng, University of Twente, Netherlands
Reviewed by
Yuntao Hu, Lawrence Berkeley National Laboratory, United States; Ying Huang, East China Normal University, China
Updates
Copyright
© 2022 Shao, Han, Sun, Yang and Xie.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongtu Xie, xieht@iae.ac.cn
This article was submitted to Conservation and Restoration Ecology, a section of the journal Frontiers in Ecology and Evolution
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.