- Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, CA, United States
Many traits of industrial and basic biological interest arose long ago, and manifest now as fixed differences between a focal species and its reproductively isolated relatives. In these systems, extant individuals can hold clues to the mechanisms by which phenotypes evolved in their ancestors. We harnessed yeast thermotolerance as a test case for such molecular-genetic inferences. In viability experiments, we showed that extant Saccharomyces cerevisiae survived at temperatures where cultures of its sister species S. paradoxus died out. Then, focusing on loci that contribute to this difference, we found that the genetic mechanisms of high-temperature growth changed with temperature. We also uncovered an enrichment of low-frequency variants at thermotolerance loci in S. cerevisiae population sequences, suggestive of a history of non-neutral selective forces acting at these genes. We interpret these results in light of models of the evolutionary mechanisms by which the thermotolerance trait arose in the S. cerevisiae lineage. Together, our results and interpretation underscore the power of genetic approaches to explore how an ancient trait came to be.
Introduction
A central goal of evolutionary genetics is to understand how nature builds new phenotypes. Thanks to advances in statistical genetics and experimental evolution, mechanisms of trait evolution over relatively short timescales have come well within reach in the modern literature. By contrast, longer-term innovations have posed a more profound challenge for the field (Orr, 2001). In principle, for a phenotype that originated long ago and manifests now as a fixed difference between species, evolution could have refined the character along the entire divergence time of the respective taxa. In landmark cases, candidate-gene studies have shed light on suites of mutational changes of this kind between species, at a given model locus. This includes the order by which adaptive alleles were likely acquired, and/or the functional pressures that drove them (Bridgham et al., 2009; Baldwin et al., 2014; Dong et al., 2014; Sayou et al., 2014; Anderson et al., 2015; Daugherty et al., 2016; Sulak et al., 2016; Liu et al., 2018; Starr et al., 2018; Xie et al., 2018; Siddiq and Thornton, 2019; Pillai et al., 2020; Prieto-Godino et al., 2021). But, in most cases, any factor pursued by such a candidate-gene approach only represents part of the complex genetic architecture of an ancient trait. As a field, we still know relatively little about how evolution coordinates multiple adaptive loci over deep divergences.
In the search for evolutionary principles, genetically tractable model systems can be of great utility. Saccharomyces yeasts are well suited for this purpose, and environmental yeast isolates have been studied extensively for their innovations within and between species (Hittinger, 2013). Thermotolerance is of particular interest because it tracks with phylogeny across the 20 million years of the Saccharomyces radiation (Gonçalves et al., 2011; Salvadó et al., 2011). Even the two most recent branches of the phylogeny exhibit a robust difference in this phenotype: S. cerevisiae acquired the ability to grow at temperatures near 40°C in the five million years since it diverged from its sister species, S. paradoxus (Sweeney et al., 2004; Gonçalves et al., 2011; Salvadó et al., 2011; Williams et al., 2015). In previous work, a whole-genome mapping scheme identified housekeeping genes at which variation between S. cerevisiae and S. paradoxus impacts growth at the high end of the S. cerevisiae temperature range (Weiss et al., 2018). These loci exhibit striking sequence differences between the species, and conservation in S. cerevisiae, consistent with a history of positive selection on pro-thermotolerance alleles (Weiss et al., 2018; Abrams et al., 2021, 2022). However, we have as yet little insight into the ecological dynamics by which this model trait evolved along the S. cerevisiae lineage.
Cases of adaptation across a temperature gradient are a mainstay of the evolutionary genetics literature (Turner et al., 2008; Prasad et al., 2011; Mimura et al., 2013; Savolainen et al., 2013; Robin et al., 2017; Dudaniec et al., 2018; Key et al., 2018; Endler, 2020; Tepolt and Palumbi, 2020; Calfee et al., 2021; Machado et al., 2021). Here we sought to explore the relevance of such a mechanism to the events by which S. cerevisiae gained its maximal thermotolerance phenotype. Given that we have no access to genotypes representing ancient intermediates between this species and S. paradoxus, we designed a strategy to interrogate the genetics of extant strains, focusing on contributing genes of major effect. We surveyed gene-environment interactions by these thermotolerance loci across warm temperatures, complementing previous studies of interspecies variation at a single high temperature (Weiss et al., 2018; Abrams et al., 2022). And we investigated the frequency of variants at these loci with a population-genomic approach.
Methods
Viability Assay
For the survey of viability phenotypes at high temperatures across wild-type isolates in Supplementary Figure 1, each strain was streaked from a −80°C freezer stock onto a yeast peptone dextrose (YPD) agar plate and incubated at room temperature for 3 days. For each biological replicate, a single colony was inoculated into 5 mL liquid YPD and grown for 24 h at 28°C with shaking at 200 rpm to generate pre-cultures. Each pre-culture was back-diluted into 10 mL of YPD to reach an OD600/mL of 0.05 and then cultured for 24 h at the temperature of interest (28–39°C). The viability of both the precultures and the cultures after 24 h at the temperature of interest were measured with a spotting assay, where we diluted aliquots from the culture in a 1:10 series and spotting 3 μL of each dilution for growth on a solid YPD plate. After incubation at 28°C for 2 days, we used the dilution corresponding to the densest spot that was not a lawn for to determine viability: we counted the number of colonies in each of the two technical replicate spots, formulated the number of colony forming units per mL of undiluted culture (CFU/mL). We determined the change in viable cells by subtracting the number of cells in the culture at the initial time point from that at the final timespoints, based on the CFU/mL count and the culture volume. We evaluated the significance of the difference between S. cerevisiae and S. paradoxus at a given temperature using a one-sided Mann-Whitney U-test.
Dose Response Growth Assay
For growth measurements in Figure 1 we assayed S. paradoxus Z1; S. cerevisiae DBVPG1373; and S. cerevisiae DBVPG1373 with the S. paradoxus Z1 allele of ESP1, MYO1, AFG2, or CEP3 swapped in at the endogenous locus from Weiss et al. (2018) (Supplementary Table 1) as follows. Strains were streaked out and a colony of each was pre-cultured as for the viability assay above, except that the initial pre-culture lasted 24 h. Each pre-culture was back-diluted into YPD at an OD600/mL of 0.05 and grown for an additional 5.5–6 h at 28°C, shaking at 200 rpm, until reaching logarithmic phase. Each pre-culture was again back-diluted into 10 mL YPD in 1-inch diameter glass tubes with a target OD600/mL of 0.05; the actual OD600/mL of each was measured, after which it was grown at a temperature of interest (28°C or 35–38°C) with shaking at 200 rpm for 24 h, and OD600/mL was measured again. The growth efficiency for each replicate was calculated as the difference between these final and initial OD600/mL values. We used the growth efficiency from all days and all temperatures of a given pair of strains as input into a two-factor type 2 ANOVA test for a temperature-by-strain effect (Supplementary Table 2).
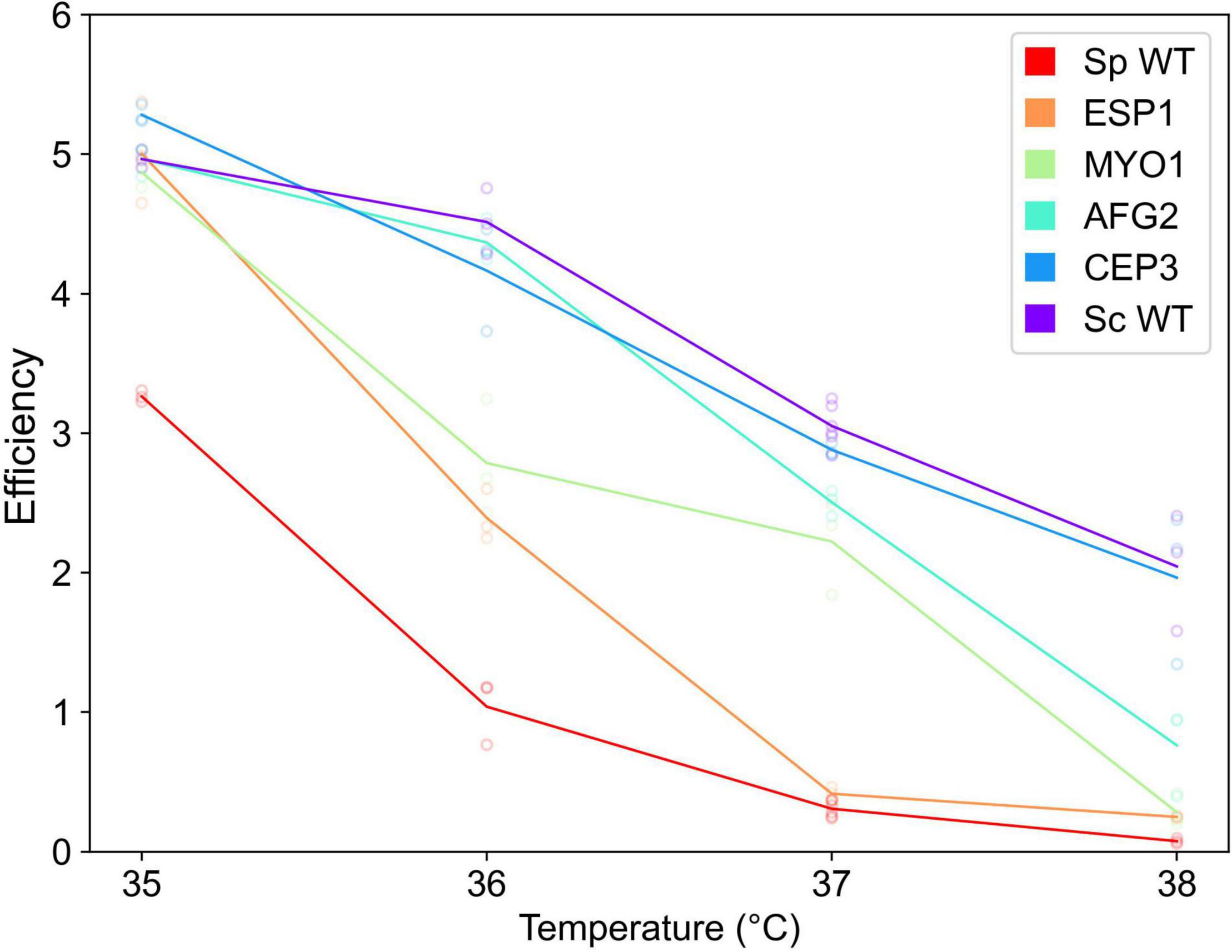
Figure 1. Saccharomyces paradoxus alleles of thermotolerance genes confer temperature-dependent defects in the S. cerevisiae background. Each line reports the results of growth experiments of a strain of S. cerevisiae DBVPG1373 harboring the indicated gene from S. paradoxus Z1, or the respective wild-type parent strains (WT), across a temperature gradient. The y-axis reports growth efficiency, the optical density reached by the culture after 24 h at the temperature indicated on the x-axis, as a difference from the analogous quantity at time zero. Each point reports results from one biological replicate (n = 3), and the line represents the average growth efficiency of the indicated strain across the temperature gradient. Statistical comparisons across strains are reported in Supplementary Table 2.
For growth measurements of ESP1 swap strains with different donors, as reported in Supplementary Figure 2, cultures were grown and measured as above, except that the only temperature was 36°C.
Tajima’s D in Wine/European Saccharomyces cerevisiae
Tajima’s D tabulates the difference between the average number of differences in pairs of sequences in a population sample and the number of variant sites in the sample. When the latter is of much bigger magnitude and D is negative, it indicates that variation in the sample is accounted for mostly by rare alleles. This pattern is expected some time after a selective sweep when de novo mutations arise on the swept haplotype; it can also reflect weak purifying selection or a history of population expansion (Suzuki, 2010). Taking as input the VCFs reporting inheritance in the strains of a given S. cerevisiae population from Peter et al. (2018), we used VCF-kit (Cook and Andersen, 2017) to calculate Tajima’s D for the region from coding start to coding stop for each gene. We then developed a resampling test to assess enrichment trends in Tajima’s D in genes of interest against a genomic null. This scheme normalizes out impacts on Tajima’s D values from events which affect the whole genome, such as a population expansion; that said, formally, any results from such an empirical outlier-based analysis serve as suggestive rather than conclusive evidence for non-neutral evolution (Teshima et al., 2006; Thornton and Jensen, 2007). For the test, we sampled 10,000 random cohorts of genes from the genome with the same number of essential and non-essential genes (Winzeler et al., 1999) as a given thermotolerance cohort, and we used as an empirical P-value the proportion of random cohorts where the median Tajima’s D was less than or equal to that of our thermotolerance cohort.
Statistical comparisons across strains are reported in Supplementary Table 2.
Results
For an initial study of the genetics of yeast species variation in temperature response, we chose to harness DBVPG1373, a homozygous diploid S. cerevisiae strain derived from an isolate from Dutch soil, and Z1, a homozygous diploid S. paradoxus strain derived from an isolate from an oak tree in England. We anticipated that detailed analyses using these strains, as representatives of their respective species, could accelerate the discovery of more general principles (Weiss et al., 2018; Abrams et al., 2021, 2022). We developed an assay quantifying cell viability in a given liquid culture before and after incubation at a temperature of interest, and we implemented this approach for each species in turn. The results revealed an advantage for S. cerevisiae over S. paradoxus at temperatures above 35°C (Supplementary Figure 1), consistent with previous growth-based surveys (Sweeney et al., 2004; Gonçalves et al., 2011; Salvadó et al., 2011; Williams et al., 2015). S. cerevisiae maintained viability at all temperatures tested, whereas by 37°C, S. paradoxus became inviable, with no evidence for spontaneous rescue even over long incubation times (Supplementary Figure 1). Using the latter as a window onto the phenotype of the common ancestor of S. cerevisiae and S. paradoxus, we would envision that the ancient population would have gone extinct rather than adapting, if exposed to temperatures at the high end of the range tolerated by modern S. cerevisiae.
We next aimed to investigate the genetics of temperature response as it differs between extant S. cerevisiae and S. paradoxus, to motivate inferences about the evolution of the maximal thermotolerance trait. We focused on four genes—the cell division factors ESP1, MYO1, and CEP3, and the ribosome maturation factor AFG2—where alleles from modern S. paradoxus compromise growth at 39°C (Weiss et al., 2018). We made use of homozygous diploid strains of the S. cerevisiae DBVPG1373 background in which the endogenous allele of each gene in turn was replaced with that of S. paradoxus Z1. In each, we measured the growth phenotype as a dose-response across temperatures, and observed a drop as temperature increased (Figure 1). In the S. cerevisiae background, S. paradoxus Z1 alleles eroded growth efficiency at temperatures well below the hard limit of viability for the Z1 wild-type (∼38°C), an effect that reached significance for three of our four focal genes (Figure 1). We conclude that many problems posed by S. paradoxus Z1 alleles at the high end of our temperature range also are relevant to a lesser extent at lower temperatures.
Inspecting the shape of the temperature dose-responses of allelic effects, we noted quantitative differences between our loci (Figure 1 and Supplementary Table 2). At the chromatid separase gene ESP1, the Z1 allele conferred an appreciable drop in growth efficiency at 36°C and supported almost no growth by 37°C. By contrast, at AFG2, the S. paradoxus Z1 allele was sufficient for growth approximating that of S. cerevisiae until 38°C. The dose-response of allelic effects at MYO1, encoding a class II myosin heavy chain, lay between these two extremes. Overall, all allele-replacement strains exhibited distinct dose-response behaviors across temperatures (Supplementary Table 2), likely reflecting distinguishing properties of their structure and function, and of the interspecies variants they harbor.
We reasoned that trends from our temperature dose-response approach would be most informative when they were conserved across S. paradoxus as a species. To pursue this, we earmarked ESP1, whose S. paradoxus Z1 allele had exhibited the sharpest falloff with temperature among the genes of our set (Figure 1). We repeated our growth efficiency experiments in strains of S. cerevisiae DBVPG1373 harboring ESP1 from other S. paradoxus donors beside the Z1 strain. These transgenics, which phenocopy wild-type DBVPG1373 at 28°C (Weiss et al., 2018), all dropped off in growth efficiency by 36°C (Supplementary Figure 2), as expected if the temperature preference of ESP1 were ancestral to, and shared across, extant S. paradoxus populations.
Together, these dose-response results establish that the functions of S. paradoxus alleles at our focal genes break down at distinct temperatures between 35 and 38°C—suggesting similar gene-by-environment effects in the ancestor of S. cerevisiae and S. paradoxus, if it sampled a range of temperature conditions over evolutionary history.
To gain insight into past and current selective pressures on thermotolerance loci, we turned to a molecular-evolution approach. Previous work on Saccharomyces thermotolerance genes has emphasized sequence divergence between species (Weiss et al., 2018; Abrams et al., 2021, 2022). For a complementary focus on population variation within S. cerevisiae, we analyzed the Tajima’s D statistic, which formulates properties of sequence variants in a population into a test of how well the sequence fits the expectations under a neutral evolutionary model (Tajima, 1989; Biswas and Akey, 2006). We first developed a resampling-based scheme that compared Tajima’s D between a gene cohort of interest and a genomic null. This enabled a non-parametric assessment of significance and normalized out potential effects of demography on the statistic (see section “Methods”). Expecting that our test would have maximal power in a data set of large sample size, we focused on the most deeply sampled S. cerevisiae population in current compendia, comprising isolates from vineyards and European soil (362 strains; Peter et al., 2018). Examining our four focal thermotolerance genes, we detected an enrichment for low, negative Tajima’s D at these loci across S. cerevisiae genomes (P = 0.0263; Supplementary Figure 3 and Supplementary Table 3A), reporting an excess of rare variants—as expected after a selective sweep, or under constraints from purifying selection (Biswas and Akey, 2006; Suzuki, 2010). We repeated this analysis using more comprehensive sets of hits from interspecies thermotolerance screens (Weiss et al., 2018; Abrams et al., 2021, 2022), and detected strong signal for low, negative Tajima’s D at these loci in vineyard/European S. cerevisiae in every case (Supplementary Tables 3B,C). Interestingly, ESP1 exhibited the most negative Tajima’s D value among all thermotolerance genes in these analyses (Supplementary Table 3), dovetailing with the strong effect of variation at this gene in phenotypic analyses (Weiss et al., 2018; Figure 1).
We also investigated patterns of Tajima’s D in thermotolerance genes beyond vineyard/European S. cerevisiae. For this purpose we harnessed the next best-sampled S. cerevisiae populations from Peter et al. (2018): Mosaic Region 3 (113 strains), Mixed Origin (72 strains), Sake (47 strains), and Brazilian Bioethanol (35 strains). Of these groups of isolates, tests for enrichment of low Tajima’s D among our four focal thermotolerance genes yielded significant signal only in the Brazilian Bioethanol population (Supplementary Table 3A). Thus, the trend for low, negative Tajima’s D at thermotolerance loci was detectable but not consistent across these populations, potentially reflecting limited power, or weaker or different selective forces, in the latter relative to vineyard/European strains.
Discussion
Most trait differences that define long-diverged species are likely the product of suites of unlinked variants that have come together over long timescales. Tracing the ancient evolutionary events at such loci remains a central challenge in the field. In this work, we have characterized thermotolerance genes in extant yeast strains and species, to inform models of the evolution of the trait in S. cerevisiae.
Our data have shown that, for the strains studied here, temperatures below the high end of the S. cerevisiae range are lethal for S. paradoxus. This complements previous surveys of the Saccharomyces clade using growth-based assays (Sweeney et al., 2004; Gonçalves et al., 2011; Salvadó et al., 2011) and reveals that, under high temperature conditions, a given culture of S. paradoxus will die off rather than adapting. We have also leveraged results from genetic mapping of S. cerevisiae thermotolerance to trace the temperature dependence of contributing genes. Our approach contrasts with the search for DNA sequence variants associated with environment or population variables (Rellstab et al., 2015; Hoban et al., 2016), in that we focus directly on bona fide causal determinants of the trait of interest. With this strategy, we discerned differences between thermotolerance loci in terms of the temperatures at which alleles from modern S. paradoxus fail in their growth functions. Our experiments for this purpose used transgenesis of one locus at a time in a single strain background, so the potential for epistasis involving S. paradoxus alleles remains untested. As they stand, our data do make clear that the genetic mechanisms of growth change with temperature in the S. cerevisiae–S. paradoxus system.
If we infer that these effects are representative of the genetics of a thermosensitive S. cerevisiae ancestor, long ago in history, we can formulate models of the mechanism by which thermotolerance evolved. In one scenario, an ancestral population of S. cerevisiae in temperate conditions could have acquired pro-thermotolerance alleles at many loci, which, once they had accumulated as standing variation, would have enabled a single jump to a hotter growth regime. Interestingly, however, experimental evolution studies suggest that events of the latter kind may be rare; by contrast, adaptation to a gradually increasing stress is evolutionarily easier and reduces the risk of extinction (Lindsey et al., 2013). Under a model of gradual adaptation, we would picture the ancestral S. cerevisiae population expanding its range stepwise along a temperature gradient, analogous to previous local-adaptation case studies in other systems (Mimura et al., 2013; Robin et al., 2017; Dudaniec et al., 2018; Key et al., 2018; Tepolt and Palumbi, 2020). As evolution proceeded along the S. cerevisiae lineage, ESP1, MYO1, and other underlying loci would each have come under selective pressure (and acquired variants de novo) at the temperatures where their defects manifested. This would represent an example of “whack-a-mole” dynamics (Shin and MacCarthy, 2015), whereby the weakest point in the genetic network targeted by evolution changes as conditions change. Given the genetic complexity of the thermotolerance trait (Weiss et al., 2018), its history may ultimately prove to involve both gradual adaptation and sudden jumps to new resistance states, each relevant for some subset of the underlying alleles.
What were the ecological forces driving the gain of thermotolerance in S. cerevisiae? The ancestral population could have undergone a change in the temperature of its growth conditions over space or time: that is, migrants from temperate physical locales could have advanced to warmer sites, or the environment could have warmed up around a static population. In either case, it is tempting to speculate that these ecological pressures were exerted in the hot East Asian niches to which the ancestor of modern S. cerevisiae has been traced (Peter et al., 2018). Features of human-associated environments could also have contributed later, in domesticated populations. Alternatively, the trait syndrome in S. cerevisiae—a unique ability to tolerate ethanol as well as heat, with both given off by fermentative metabolism—could have arisen as a specialization to kill off microbial competitors in nutrient-rich substrates, regardless of the endemic temperature of the location (Goddard, 2008; Salvadó et al., 2011). If so, variants would have been acquired, potentially over millions of generations, gradually to ratchet up fermentative activity and tolerance to its byproducts, with our genetic insights thus far largely restricted to the latter.
Our work leaves open the dating of any such events. In several S. cerevisiae populations, we have uncovered an enrichment of rare variants at thermotolerance genes. This signature of non-neutral evolution can be interpreted at a given locus as evidence for a relatively recent sweep of a positively selected haplotype, or for negative selection to maintain a fitness-relevant haplotype that arose long ago. We favor the latter hypothesis, given that prior work across populations has also shown S. cerevisiae alleles at our focal genes to be partly sufficient for maximal thermotolerance, conserved within the species, and divergent from S. paradoxus (Weiss et al., 2018; Abrams et al., 2022). We thus propose that many thermotolerance alleles were acquired in ancient selective sweeps, before the radiation of modern S. cerevisiae, and have been maintained since then by purifying selection. That said, later refinements in particular populations of S. cerevisiae may also have strengthened beneficial facets of the trait, added regulatory tuning, or eliminated antagonistic pleiotropic “side effects” that were niche-specific. A comprehensive genetic and ecological reconstruction of this history may be out of our current grasp, especially in light of the caveats of our approach using a laboratory setting and extant strain backgrounds. Nonetheless, our data add compelling detail to an emerging consensus view of how evolution built maximal thermotolerance in S. cerevisiae.
Data Availability Statement
Strains and plasmids are available upon request. Custom scripts for Tajima’s D analyses are available at https://github.com/melanieabrams-pub/.
Author Contributions
MA and RB designed the study and wrote the manuscript. MA conducted experimental work and performed data collection and analysis. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by NSF GRFP DGE 1752814 to MA and NIH R01 GM120430 to RB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Faisal AlZaben, Abel Duarte, and Jude Edwards for experimental support, and Adam Arkin for his generosity with computational resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.859904/full#supplementary-material
References
Abrams, M. B., Chuong, J. N., AlZaben, F., Dubin, C. A., Skerker, J. M., and Brem, R. B. (2022). Barcoded reciprocal hemizygosity analysis via sequencing illuminates the complex genetic basis of yeast thermotolerance. G3 Genes Genomes Genet. 12:jkab412. doi: 10.1101/2021.07.26.453780
Abrams, M. B., Dubin, C. A., AlZaben, F., Bravo, J., Joubert, P. M., Weiss, C. V., et al. (2021). Population and comparative genetics of thermotolerance divergence between yeast species. G3 Genes Genomes Genet. 11:jkab139. doi: 10.1093/g3journal/jkab139
Anderson, D. W., McKeown, A. N., and Thornton, J. W. (2015). Intermolecular epistasis shaped the function and evolution of an ancient transcription factor and its DNA binding sites. eLife 4:e07864. doi: 10.7554/eLife.07864
Baldwin, M. W., Toda, Y., Nakagita, T., O’Connell, M. J., Klasing, K. C., Misaka, T., et al. (2014). Sensory biology. Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science 345, 929–933. doi: 10.1126/science.1255097
Biswas, S., and Akey, J. M. (2006). Genomic insights into positive selection. Trends Genet. 22, 437–446. doi: 10.1016/j.tig.2006.06.005
Bridgham, J. T., Ortlund, E. A., and Thornton, J. W. (2009). An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461, 515–519. doi: 10.1038/nature08249
Calfee, E., Gates, D., Lorant, A., Perkins, M. T., Coop, G., and Ross-Ibarra, J. (2021). Selective sorting of ancestral introgression in maize and teosinte along an elevational cline. PLoS Genet. 17:e1009810. doi: 10.1371/journal.pgen.1009810
Cook, D. E., and Andersen, E. C. (2017). VCF-kit: assorted utilities for the variant call format. Bioinformatics 33, 1581–1582. doi: 10.1093/bioinformatics/btx011
Daugherty, M. D., Schaller, A. M., Geballe, A. P., and Malik, H. S. (2016). Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. eLife 5:e14228. doi: 10.7554/eLife.14228
Dong, S., Stam, R., Cano, L. M., Song, J., Sklenar, J., Yoshida, K., et al. (2014). Effector specialization in a lineage of the Irish potato famine pathogen. Science 343, 552–555. doi: 10.1126/science.1246300
Dudaniec, R. Y., Yong, C. J., Lancaster, L. T., Svensson, E. I., and Hansson, B. (2018). Signatures of local adaptation along environmental gradients in a range-expanding damselfly (Ischnura elegans). Mol. Ecol. 27, 2576–2593. doi: 10.1111/mec.14709
Endler, J. A. (2020). Geographic Variation, Speciation and Clines. (MPB-10), Volume 10. Princeton, NJ: Princeton University Press. doi: 10.2307/j.ctvx5wbdg
Goddard, M. R. (2008). Quantifying the complexities of Saccharomyces cerevisiae’s ecosystem engineering via fermentation. Ecology 89, 2077–2082. doi: 10.1890/07-2060.1
Gonçalves, P., Valério, E., Correia, C., de Almeida, J. M., and Sampaio, J. P. (2011). Evidence for divergent evolution of growth temperature preference in sympatric Saccharomyces species. PLoS One 6:e20739. doi: 10.1371/journal.pone.0020739
Hittinger, C. T. (2013). Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 29, 309–317. doi: 10.1016/j.tig.2013.01.002
Hoban, S., Kelley, J. L., Lotterhos, K. E., Antolin, M. F., Bradburd, G., Lowry, D. B., et al. (2016). Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am. Nat. 188, 379–397. doi: 10.1086/688018
Key, F. M., Abdul-Aziz, M. A., Mundry, R., Peter, B. M., Sekar, A., D’Amato, M., et al. (2018). Human local adaptation of the TRPM8 cold receptor along a latitudinal cline. PLoS Genet. 14:e1007298. doi: 10.1371/journal.pgen.1007298
Lindsey, H. A., Gallie, J., Taylor, S., and Kerr, B. (2013). Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature 494, 463–467. doi: 10.1038/nature11879
Liu, Q., Onal, P., Datta, R. R., Rogers, J. M., Schmidt-Ott, U., Bulyk, M. L., et al. (2018). Ancient mechanisms for the evolution of the bicoid homeodomain’s function in fly development. eLife 7:e34594. doi: 10.7554/eLife.34594
Machado, H. E., Bergland, A. O., Taylor, R., Tilk, S., Behrman, E., Dyer, K., et al. (2021). Broad geographic sampling reveals the shared basis and environmental correlates of seasonal adaptation in Drosophila. eLife 10:e67577. doi: 10.7554/eLife.67577
Mimura, M., Ono, K., Goka, K., and Hara, T. (2013). Standing variation boosted by multiple sources of introduction contributes to the success of the introduced species, Lotus corniculatus. Biol. Invasions 15, 2743–2754. doi: 10.1007/s10530-013-0488-x
Orr, H. A. (2001). The genetics of species differences. Trends Ecol. Evol. 16, 343–350. doi: 10.1016/s0169-5347(01)02167-x
Peter, J., Chiara, M. D., Friedrich, A., Yue, J.-X., Pflieger, D., Bergström, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Pillai, A. S., Chandler, S. A., Liu, Y., Signore, A. V., Cortez-Romero, C. R., Benesch, J. L. P., et al. (2020). Origin of complexity in haemoglobin evolution. Nature 581, 480–485. doi: 10.1038/s41586-020-2292-y
Prasad, A., Croydon-Sugarman, M. J. F., Murray, R. L., and Cutter, A. D. (2011). Temperature-dependent fecundity associates with latitude in Caenorhabditis briggsae. Evolution 65, 52–63. doi: 10.1111/j.1558-5646.2010.01110.x
Prieto-Godino, L. L., Schmidt, H. R., and Benton, R. (2021). Molecular reconstruction of recurrent evolutionary switching in olfactory receptor specificity. eLife 10:e69732. doi: 10.7554/eLife.69732
Rellstab, C., Gugerli, F., Eckert, A. J., Hancock, A. M., and Holderegger, R. (2015). A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 24, 4348–4370. doi: 10.1111/mec.13322
Robin, C., Andanson, A., Saint-Jean, G., Fabreguettes, O., and Dutech, C. (2017). What was old is new again: thermal adaptation within clonal lineages during range expansion in a fungal pathogen. Mol. Ecol. 26, 1952–1963. doi: 10.1111/mec.14039
Salvadó, Z., Arroyo-López, F. N., Guillamón, J. M., Salazar, G., Querol, A., and Barrio, E. (2011). Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl. Environ. Microbiol. 77, 2292–2302. doi: 10.1128/AEM.01861-10
Savolainen, O., Lascoux, M., and Merilä, J. (2013). Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820. doi: 10.1038/nrg3522
Sayou, C., Monniaux, M., Nanao, M. H., Moyroud, E., Brockington, S. F., Thévenon, E., et al. (2014). A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science 343, 645–648. doi: 10.1126/science.1248229
Shin, J., and MacCarthy, T. (2015). Antagonistic coevolution drives whack-a-mole sensitivity in gene regulatory networks. PLoS Comput. Biol. 11:e1004432. doi: 10.1371/journal.pcbi.1004432
Siddiq, M. A., and Thornton, J. W. (2019). Fitness effects but no temperature-mediated balancing selection at the polymorphic Adh gene of Drosophila melanogaster. PNAS 116, 21634–21640. doi: 10.1073/pnas.1909216116
Starr, T. N., Flynn, J. M., Mishra, P., Bolon, D. N. A., and Thornton, J. W. (2018). Pervasive contingency and entrenchment in a billion years of Hsp90 evolution. PNAS 115, 4453–4458. doi: 10.1073/pnas.1718133115
Sulak, M., Fong, L., Mika, K., Chigurupati, S., Yon, L., Mongan, N. P., et al. (2016). TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 5:e11994. doi: 10.7554/eLife.11994
Suzuki, Y. (2010). Statistical methods for detecting natural selection from genomic data. Genes Genet. Syst. 85, 359–376. doi: 10.1266/ggs.85.359
Sweeney, J. Y., Kuehne, H. A., and Sniegowski, P. D. (2004). Sympatric natural Saccharomyces cerevisiae and S. paradoxus populations have different thermal growth profiles. FEMS Yeast Res. 4, 521–525. doi: 10.1016/S1567-1356(03)00171-5
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Tepolt, C. K., and Palumbi, S. R. (2020). Rapid adaptation to temperature via a potential genomic island of divergence in the invasive green crab, Carcinus maenas. Front. Ecol. Evol. 8:411. doi: 10.3389/fevo.2020.580701
Teshima, K. M., Coop, G., and Przeworski, M. (2006). How reliable are empirical genomic scans for selective sweeps? Genome Res. 16, 702–712. doi: 10.1101/gr.5105206
Thornton, K. R., and Jensen, J. D. (2007). Controlling the false-positive rate in multilocus genome scans for selection. Genetics 175, 737–750. doi: 10.1534/genetics.106.064642
Turner, T. L., Levine, M. T., Eckert, M. L., and Begun, D. J. (2008). Genomic analysis of adaptive differentiation in drosophila melanogaster. Genetics 179, 455–473. doi: 10.1534/genetics.107.083659
Weiss, C. V., Roop, J. I., Hackley, R. K., Chuong, J. N., Grigoriev, I. V., Arkin, A. P., et al. (2018). Genetic dissection of interspecific differences in yeast thermotolerance. Nat. Genet. 50, 1501–1504. doi: 10.1038/s41588-018-0243-4
Williams, K. M., Liu, P., and Fay, J. C. (2015). Evolution of ecological dominance of yeast species in high-sugar environments. Evolution 69, 2079–2093. doi: 10.1111/evo.12707
Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., et al. (1999). Functional characterization of the s. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. doi: 10.1126/science.285.5429.901
Keywords: Saccharomyces, ancient, evolution, thermotolerance, adaptation
Citation: Abrams MB and Brem RB (2022) Temperature-Dependent Genetics of Thermotolerance Between Yeast Species. Front. Ecol. Evol. 10:859904. doi: 10.3389/fevo.2022.859904
Received: 21 January 2022; Accepted: 22 March 2022;
Published: 27 April 2022.
Edited by:
Jean-Baptiste Leducq, Université du Québec à Montréal, CanadaCopyright © 2022 Abrams and Brem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachel B. Brem, cmJyZW1AYmVya2VsZXkuZWR1
 Melanie B. Abrams
Melanie B. Abrams Rachel B. Brem
Rachel B. Brem