- 1College of Landscape Architecture and Art, Henan Agricultural University, Zhengzhou, China
- 2College of Life Sciences, Henan Agricultural University, Zhengzhou, China
Although soil microbes play an important role in the functioning of the forest ecosystem, our understanding of the spatial distribution characteristics of soil microbes among different vegetation types in urban forest ecosystems is poor. In this study, with the help of high-throughput sequencing, we examined the vegetation type preferences of soil microbes (fungi and bacteria) and then analyzed the microbe–environment (plant community, light availability, soil properties) relations in a temperate urban forest in China. Our results showed that the soil microbial (bacterial and fungal) richness of deciduous forest was higher than that of evergreen, and mixed forests. The spatial distribution of fungi was more specialized than that of bacteria among different vegetation types. The driving forces of environmental factors on soil bacteria and fungi were different. Our findings suggest that different vegetation types favor the occurrence of different microbes, and the relationships between soil microbes and environmental factors depend on different vegetation types in this temperate urban forest. These findings shed new light on the biodiversity conservation of microbes in temperate urban forests and point to the potential importance of vegetation types for microbe formation.
Introduction
Forests in the urban landscape provide many ecosystem services that directly (e.g., reduce noise, improve air quality) and indirectly (e.g., promote soil nutrient cycling, absorb greenhouse gases) benefit human beings (Sergio et al., 2021). Evergreen and deciduous forests are the main vegetation types in urban forest ecosystems (Jing, 2017). Different vegetation types contain varying canopy structure, litter, and understory environment (Song et al., 2017; Fu et al., 2021). For instance, the soil organic carbon and soil nutrients are significantly lower in deciduous forests than in evergreen forests (Sheng et al., 2019). The litter is different between evergreen and deciduous forests and is correlated with microbial diversity (Lin et al., 2018; Simon et al., 2018). Previous studies have focused on the aboveground plant communities in urban forest ecosystems, such as plant diversity (Bardgett and van der Putten, 2014), space shaping of the plant community (Wagg et al., 2014), and ecological service functions (Wardle et al., 2004). The aboveground plant community and underground microbes are interrelated and influence each other (Wardle et al., 2004). Although soil microbes play an important role in urban forest ecosystem stability and ecosystem function (Wardle et al., 2004), our understanding of the association between plants and soil microbes is poor. Moreover, the spatial distribution characteristics of soil microbes in the urban ecosystem among different vegetation types are still unclear.
Soil microbes are the richest component of terrestrial biodiversity and play major roles in ecosystem processes, such as carbon and nitrogen cycling and soil formation (Kang et al., 2021). They play a role in the growth of plants and ecosystem functioning (Singh et al., 2004). They can directly affect plant roots through the underground grazing food chain and affect soil physical and chemical properties (Kreuzer et al., 2004). Plants influence microbial communities through microhabitats (Hooper et al., 2000; Wardle, 2006; Dickie, 2007; Wang et al., 2020). Conversely, according to the plant–soil feedback theory, soil microbes may influence plant communities by altering soil trophic and physicochemical properties or regulating plant coexistence (Bever et al., 2015; Bennett and Cahill, 2016). Exploring the relationships between aboveground plants and subsurface soil microbial diversity can help us develop strategies to improve stress resistance and maintain the stability of urban forest ecosystems. Previous studies have shown that bacteria and fungi have substantially different growth habitats and dispersal capability (Powell et al., 2015; Wang et al., 2021). Fungi generally tolerate drought better than many bacteria (Schimel et al., 2007; Yuste et al., 2011; Rivest et al., 2015). However, whether the spatial distribution characteristics of soil fungal and bacterial communities in the urban ecosystem is consistent under different vegetation types remains to be elucidated.
According to the habitat partitioning hypothesis, the species composition of different habitats depends on the difference in environmental requirements and spatial distribution of environmental conditions (Svenning, 1999; Chen et al., 2020). Different vegetation types provide different habitats for understory organisms (Sheng et al., 2019). Therefore, the spatial distribution characteristics of soil microbes under different vegetation types may be different. Moreover, whether the distribution of soil microbes in urban ecosystems among different vegetation types is random or habitat-special remains unknown. With the rapid development of urban construction, more and more attention has been paid to the health and management of urban green space. However, the relationship between aboveground vegetation and underground microorganisms in the urban ecosystem remains unknown. Zhengzhou, the capital city of Henan Province and one of the largest transportation hubs in China, is an ideal area to study the relationship between urban green space and soil microorganisms. In this study, we hypothesized that (H1) the main factors affecting microbial diversities and community compositions in different vegetation habitats would differ and that (H2) the relationships between microbial and woody plant assemblages would vary among different vegetation habitat types in the urban forest. To test these hypotheses, we examined soil microbial communities in plots representing the three vegetation habitat types in the temperate urban forest using Illumina-based high-throughput sequencing (Wang et al., 2018). We delineated vegetation habitat types with similarities in vegetation conditions and then analyzed the relationships among microbial, plant, and abiotic variables in the delineated microbial habitats in the urban forest. The purpose of this study was (1) to compare the differences of spatial distribution characteristics between soil bacteria and fungi across vegetation type changes in a temperate urban forest and (2) explore the relationship between soil microbes and environmental factors in a temperate urban forest ecosystem.
Materials and Methods
Study Site and Sample Collection
The study site was located in Zhengzhou, China (112°42′–114°13′ E, 34°16′–34°58′ N, Figures 1A,B). The city has a continental monsoon climate in the north temperate zone. This region has a mean annual temperature of 15.6°C and a mean annual rainfall of 690 mm. Vegetation belongs to warm temperate deciduous broad-leaved forest. Deciduous vegetation includes Styphnolobium japonicum, Platanus acerifolia, Ginkgo biloba, etc. Evergreen vegetation includes Cedrus deodara, Ligustrum lucidum, Eriobotrya japonica, etc.
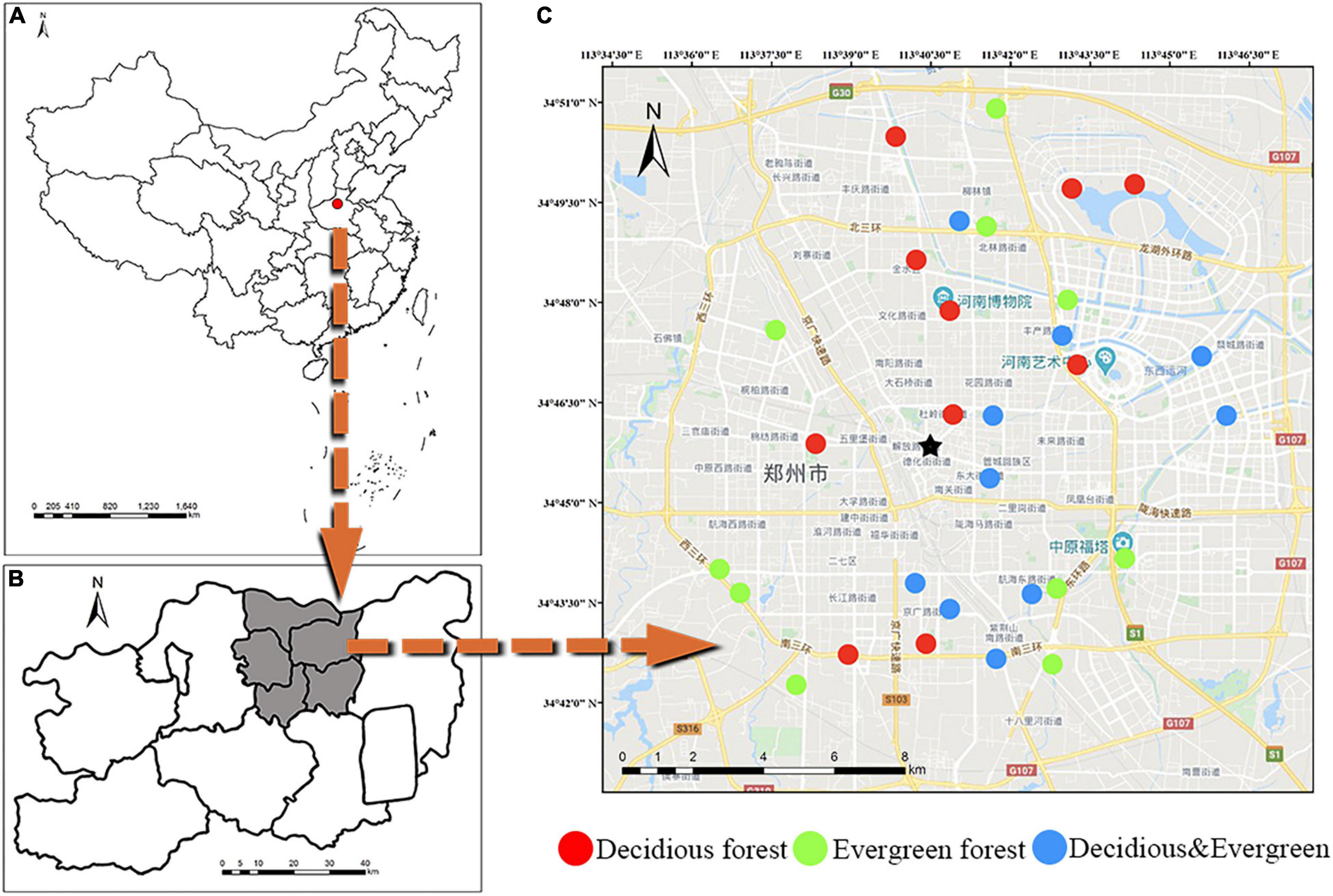
Figure 1. Study area and location of sample plots. General location (A) of the study area within Zhengzhou, China; location of all plots (B) and detailed view of an example of plots and stands (C).
A total of 30 plots (10 m × 10 m) were established in Zhengzhou City (Figure 1C), including 10 deciduous forest plots, 10 evergreen forest plots, and 10 mixed forest plots (deciduous and evergreen). Detailed information of each plot is shown in Supplementary Table 1. These plots are mainly distributed in Zhengzhou City. To avoid the effects of human activities on soil microbial composition, the following conditions should be met to establish a plot: First, the green area should be greater than 1 ha. Second, vegetation should be planted over 3 years. Finally, the distance from the road to the plot should be more than 3 m, and the distance between quadrats must be at least 500 m. For each plot, plant species, number of trees, plant height, and DBH (diameter at the breast) were recorded for woody plants with DBH greater than 1 cm. The quantitative characteristics of plants are described by abundance (PA: total number of individual plants in the plot) and density (PD: the number of individual plants per unit area; Chen et al., 2018).
Each 10 m × 10 m plot was divided into four small plots (5 m × 5 m), and soil samples were taken at the central point of each small plot. Four soil cores were collected (10–20 cm depth) and mixed in each plot. Plant roots and stones were removed using a mesh (2 mm) sieve. The mixed soil samples were divided into two parts. One part was transported in an icebox and stored at –80°C until DNA was extracted. The other was stored at 4°C for the measurement of soil physicochemical parameters. All soil samples for analyses were collected within July 2020.
Environmental Factors
Light environment data were collected using an SLM9-UM-1.2 canopy analyzer in accordance with the method of Chen et al. (2018). The leaf area index (LAI), canopy coverage (CC), total transmission ratio (VS), mean leaf angle (MLA), direct radiation (DR), and scatter radiation (SR) were measured by using HemiView digital canopy analysis software.
The air-dried soil samples were sieved through an 80-mesh sieve (0.18 mm hole) before soil physicochemical analysis. Soil pH (pH) was measured (soil-to-water ratio of 1:2.5) using a pH meter (PHSJ-6L, REX, China; Cong et al., 2015). The soil moisture content (SMC) was measured by weighing after drying in an oven at 105°C for 15 h (Cong et al., 2015). The content of soil total nitrogen (N) was measured by a modified Kjeldahl procedure (Ning et al., 2020). Soil organic matter (SOM) content was determined by the potassium dichromate oxidation method (Wang et al., 2021). The sodium bicarbonate extraction method (Shan et al., 2018) was used to determine the content of soil available phosphorus (P).
DNA Extraction, Polymerase Chain Reaction, and Bioinformatic Analysis
Microbial community genomic DNA was extracted using the Soil DNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s instructions. The DNA extract was checked on 1% agarose gel, and DNA concentration and purity were determined with a NanoDrop 2000 UV-Vis spectrophotometer. Primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTA CHVGGGTWTCTAAT-3′), specific to the V4–V5 region of the archaeal 16S rRNA gene, were used to create the archaeal libraries (Caporaso et al., 2012). Primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′), specific to the ITS I region of the ITS gene, were used to create fungal libraries (Gardes and Bruns, 1993).
PCR amplification of the gene (16S rRNA and ITS rRNA) was performed according to the methods of Xiao et al. (2021) and Yang et al. (2019), respectively. PCRs were performed in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions and quantified using the Quantus™ Fluorometer (Promega, United States).
Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, United States) according to the standard protocols by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The 16S rRNA and ITS gene sequencing reads were demultiplexed, quality-filtered by Trimmomatic, and merged by FLASH with the following criteria: (i) The 300 bp reads were truncated at any site receiving an average quality score of <20 over a 50 bp sliding window, and truncated reads shorter than 50 bp and reads containing ambiguous characters were discarded. (ii) Only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of the overlap region is 0.2. Reads that could not be assembled were discarded. (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted (exact barcode matching, two nucleotide mismatches in primer matching). Operational taxonomic units were clustered with 97% similarity (Franciska et al., 2020), and chimeric sequences were identified and removed based on a prediction by UPARSE (Benjamin et al., 2016; Xiao et al., 2021). The taxonomy of each OTU representative sequence was analyzed by RDP Classifier (Wang et al., 2007) against the 16S rRNA database (Silva SSU138; Christian et al., 2013) and ITS rRNA database (Unite7.2; Nilsson et al., 2019) using confidence threshold of 0.7.
Statistical Analyses
Before the analysis, the number of OTUs in each sample was normalized according to the minimum sample sequence number. To understand the species information of different groups, three microbial groups were constructed: all species, core species (abundance above 10% of total abundance), and dominant species (abundance above 0.5% of total abundance; Jiao et al., 2020; Mo et al., 2021). Different groups may have different functions in soil microbial community. Dominant species were more abundant in soil microbial community than core species. Moreover, core species may play a particular role in the decomposition of matter or the growth of aboveground plants, whereas dominant species do not.
Venn diagrams were constructed to count the number of common and unique OTUs in different vegetation types using the R “VennDiagram” package (Chen and Boutros, 2011). The differences in species richness among the vegetation types were determined by the Tukey test and phylogenetic tree at the OTU level. Rarefaction analysis by the “vegan” package (Oksanen et al., 2007) was used to determine whether the sequencing output for each sample is sufficient.
The ecological interactions between functionally different partners can be assessed by network analysis. Network analyses inherently account for the fact that all components of an ecosystem are interconnected (Hacquard, 2016). The program Gephi (0.9.2) was used to visualize the architecture of the plant–microbe network. We used the H2’ metric of specialization to evaluate the relationship between microbes and different vegetation types (Blüthgen et al., 2007). The H2’ metric of specialization (Svenning, 1999) was calculated by the “bipartite” package (Heegaard, 2017).
The torus-translation test is a method for determining the association between species and habitat (Harms et al., 2001; Gunatilleke et al., 2006; Comita et al., 2007). The species distribution preference in the three vegetation types was analyzed by the torus-translation test. The concept of the torus-translation test is to randomly calculate the real distribution probability of a species in each habitat (Chen et al., 2018). Given that the method considers spatial autocorrelation to a large extent, it is more reasonable to compare the goodness-of-fit test or random habitat method. Associations between species and habitat were tested for all species in the plot. A total of 9305 bacterial OTUs and 3931 fungal OTUs were used for torus-translation test analysis. The torus-translation test was conducted according to the methods of Harms et al. (2001) and Comita et al. (2007).
Nonmetric multidimensional scaling (NMDS) plots based on UniFrac dissimilarity and displacement multivariate analysis were used to compare the differences of soil microbial communities among different vegetation types, and the habitat types were fitted as centroids onto the NMDS graph using the envfit function (Chen et al., 2018). Permutational multivariate analysis of variance was applied to explore the significant differences based on 999 permutations.
Linear mixed-effect models were performed to estimate whether microbial species richness depended on environmental factors. In the model, vegetation types (deciduous, evergreen, mixed forest) were used as random variables. The environmental factors included physicochemical characters of soil, light environment, and plant abundance (PA). The linear mixed-effect model was established with the “glmm.hp” package (Lai et al., 2022).
All statistical analyses and plotting were performed with R statistical software version 4.0.3 (R Core Team, 2020).
Results
Biodiversity of Soil Microbes
A total of 1,872,421 effective bacterial sequences, with an average length of 417 bp, were obtained from 30 soil samples. The complete data set was divided into 40 phyla, 143 classes, 344 orders, 545 families, 1032 genera, and 9305 bacterial OTUs. The dominant phyla were Actinobacteriota, Proteobacteria, and Acidobacteriota. Venn diagrams showed that the number of detected OTUs varied across the three vegetation types, and these vegetation habitats shared 5396 OTUs, accounting for 57.99% of total bacterial OTUs (Figure 2A). The bacterial (all species) richness in the evergreen forest was significantly different from that in the deciduous forest and mixed forest (p < 0.05; Figure 2B). The bacterial diversity indices of each plot are shown in Supplementary Table 2.
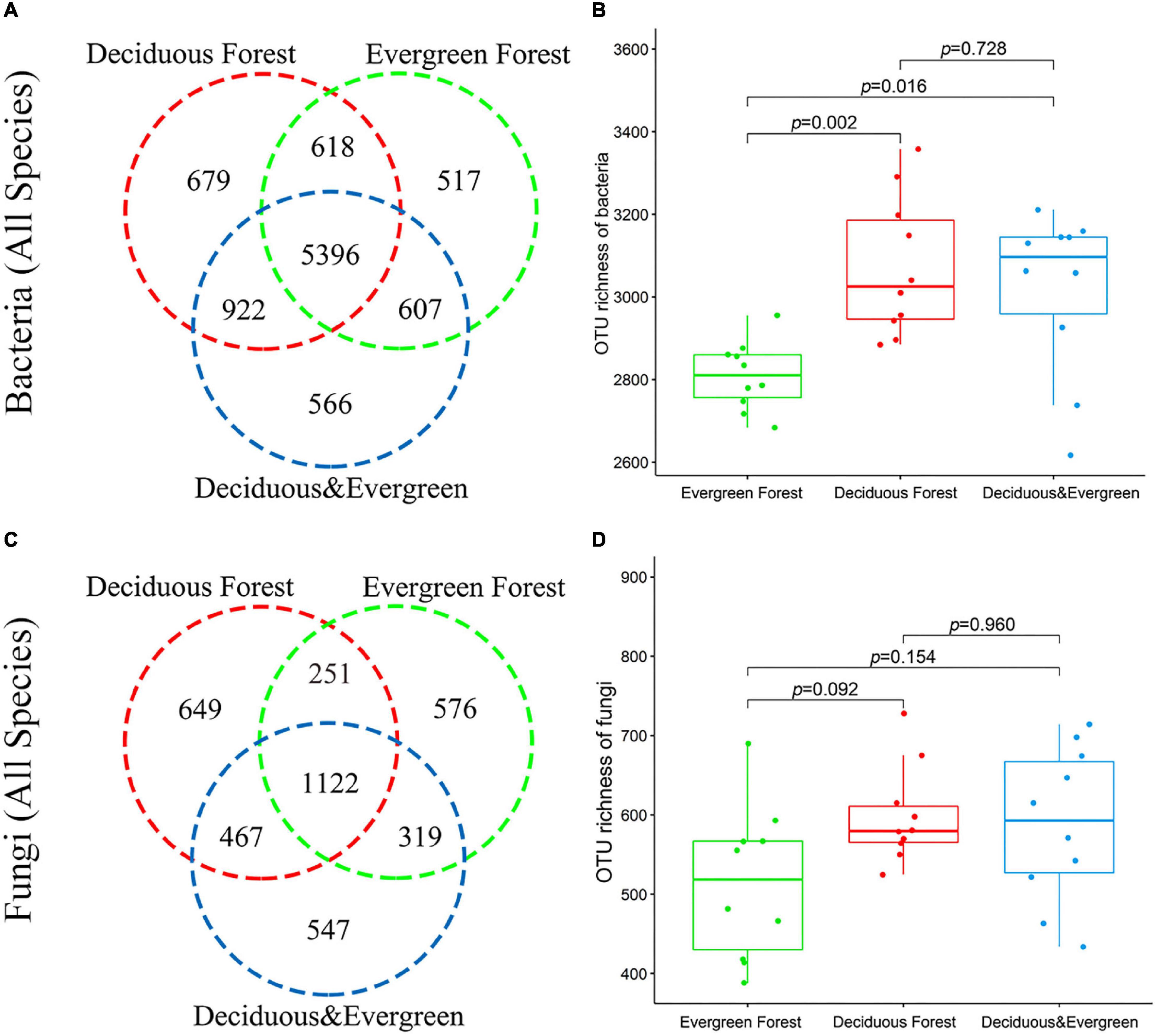
Figure 2. Biodiversity of soil microbes in the temperate urban forest in China. Number of bacterial (A) and fungal (C) OTUs in different vegetation types. OTU richness of bacteria (B) and fungi (D) in different vegetation types.
A total of 2,561,818 effective fungal sequences, with an average length of 236 bp, were obtained from 30 soil samples. Only 14 phyla, 41 classes, 106 orders, 260 families, 538 genera, and 3931 fungal OTUs were detected. Ascomycota and Basidiomycota were the dominant phyla. Moreover, 1122 OTUs were recorded in three vegetation types, accounting for 28.52% of the total fungal OTUs (Figure 2C). There was no significant difference in fungal richness among the three vegetation types. (p > 0.05, Figure 2D). The fungal diversity indices of each plot are shown in Supplementary Table 3. The sparse curve results showed that the sequencing output for each sample is sufficient (Supplementary Figure 2).
Relationships Between Microbe Species and Vegetation Types
The results of network analysis showed that the specialization index (all species) was 0.091 (bacteria) and 0.486 (fungi). For core species, the specialization index was 0.062 (bacteria) and 0.467 (fungi). The specialization index of dominant species was 0.028 (bacteria) and 0.399 (fungi). Obviously, the specialization index of soil fungi was higher than that of soil bacteria at the community level (Figure 3).
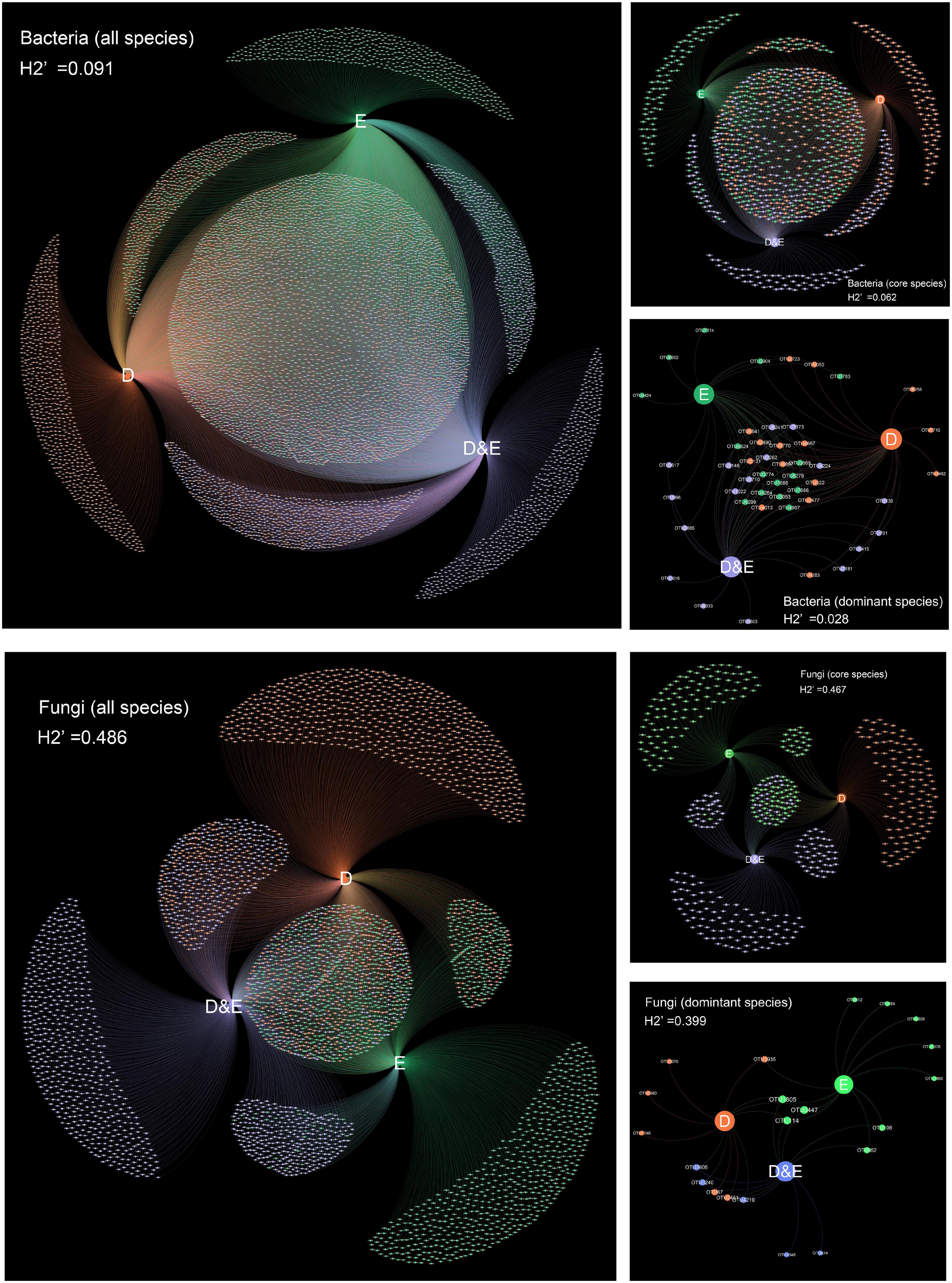
Figure 3. Microbial community network relationships among different vegetation types in the temperate urban forest in China. D is the deciduous forest, E is the evergreen forest, D&E is the deciduous and evergreen mixed forest. Each circle represents an OTU, different colors represent different vegetation types, and the circle size represents OTU richness. H2’ is the specialization index.
On the basis of the torus-translation test, 58.63% (5669/9305) OTUs of bacteria showed significant correlations, among which 5280 OTUs were positively correlated and 389 OTUs were negatively correlated (Figures 4A,B). The torus-translation test showed that 76.19% (2992/3927) OTUs of fungi had significant correlations, among which 2894 OTUs were positively correlated and 98 OTUs were negatively correlated (Figures 4D,E). The proportion of positively correlated OTUs of bacteria (28.1%) in the deciduous forest was higher than that in the evergreen forest (25.2%) and mixed forest (24.1%). Moreover, the proportion of negatively correlated OTUs of bacteria (0.9%) in the deciduous forest was lower than that in the evergreen forest (3.3%) and mixed forest (1.2%; Figure 4C). Meanwhile, the proportion of positively correlated OTUs of soil fungi was the highest in the deciduous forest (42.3%), followed by evergreen forest (41.1%) and mixed forest (39.5%). The proportion of negatively correlated OTUs of fungi in the deciduous forest was 0.9%, which was lower than that in the evergreen forest (1.6%) and mixed forest (1.6%; Figure 4F).
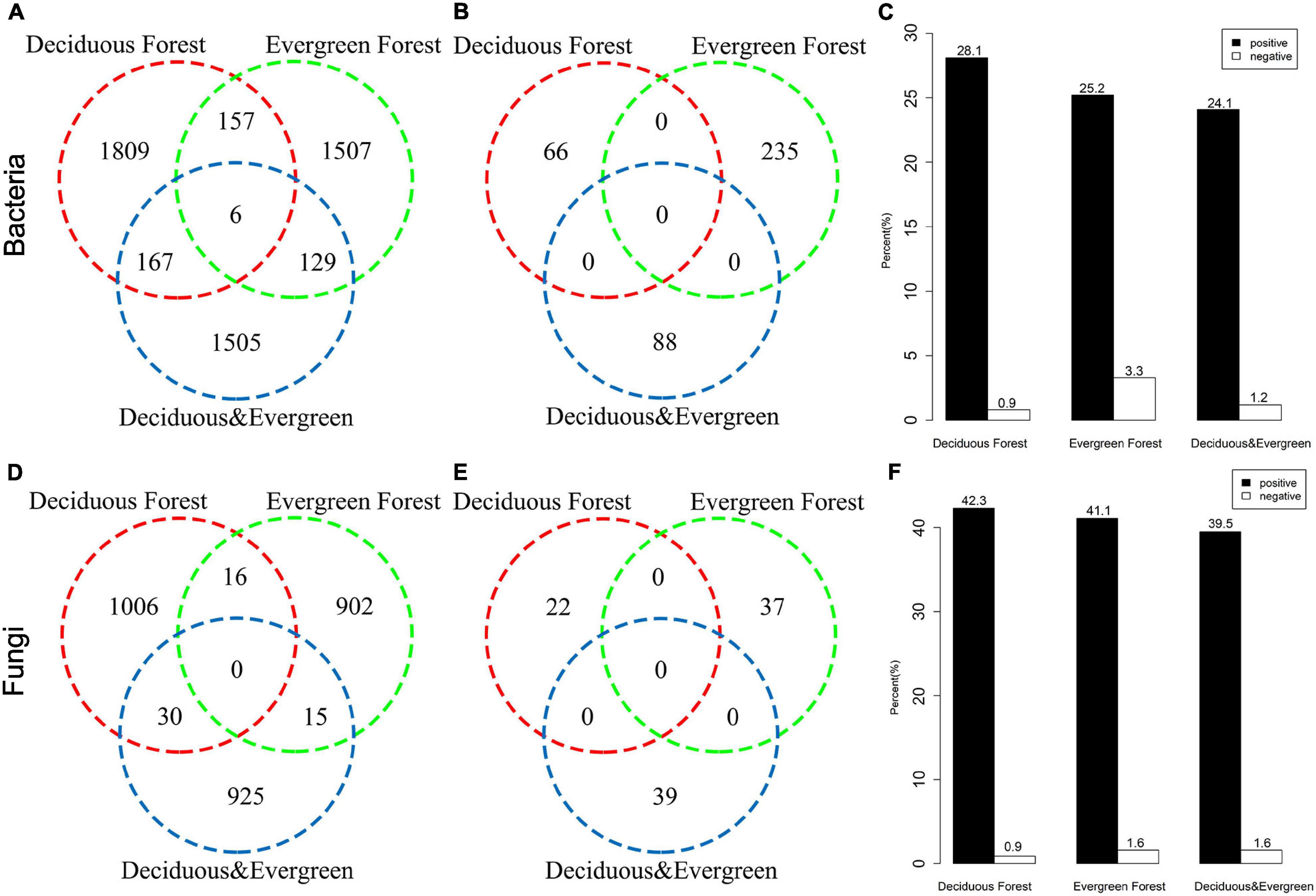
Figure 4. Associations between bacteria (A,B,C) and fungi (D,E,F) with vegetation types in the temperate urban forest in China based on the torus-translation test. (A,D) are the number of positively correlated OTUs, (B,E) are the number of negatively correlated OTUs, (C,F) are the proportion of positively and negatively correlated OTUs.
Associations Between Environmental Factors and Microbe Species
The environmental factors of each plot are shown in Supplementary Table 4. NMDS analysis showed that there were significant differences in microbial community composition among different vegetation types (p < 0.05). The phylogenetic tree at the OTU level is shown in Supplementary Figure 3. The detailed p-values of each factor are shown in Supplementary Table 5. The canopy structure (CC, p = 0.011; MLA, p = 0.028; VS, p = 0.013; SR, p = 0.023; DR, p = 0.025) had a significant influence on bacteria (Figure 5A). However, fungi were significantly influenced by SOM (p = 0.037) and CC (p = 0.041; Figure 5B). The results showed that environmental factors had different effects on the soil microbial community.
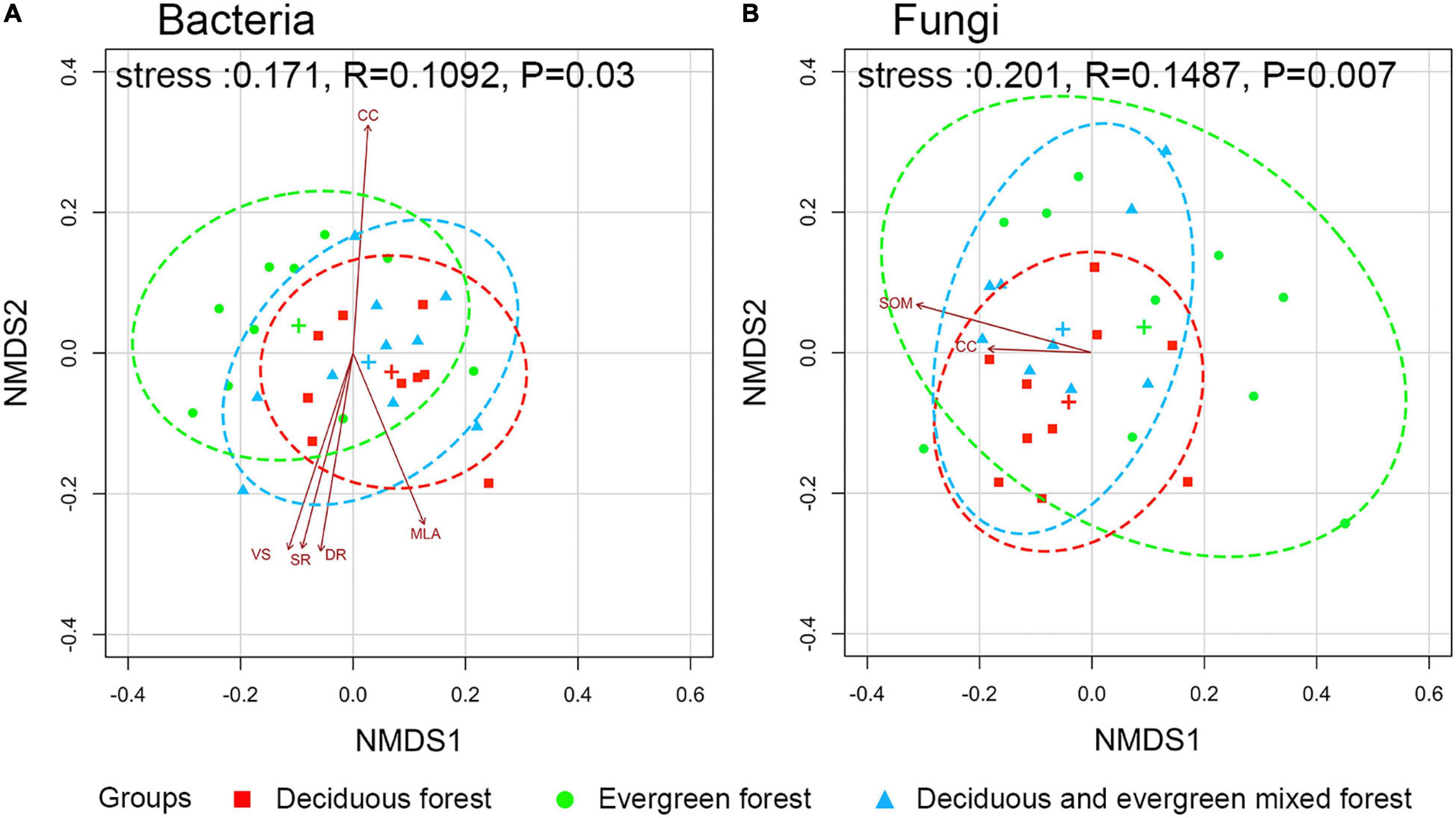
Figure 5. Ordination plot of bacterial (A) and fungal (B) communities from non-metric multidimensional scaling analysis based on unifrac dissimilarities among different vegetation types. (The ellipses are the 95% confidence interval, and the significance between communities was evaluated by permutational multivariate analyses of variance. The relationships between the community and environmental factors only show strongly correlated (p ≤ 0.05) variables. SOM, soil organic matter; CC, canopy cover degrees; VS, total transmission ratio; SR, scatter radiation under the canopy; DR, direct radiation under the canopy; MLA, mean leaf angle.)
Fixed variables (environmental factors) explained 0.33 (R2m) of the variation in soil bacterial communities while vegetation types explained a large proportion of variation (R2c - R2m = 0.52; Supplementary Figure 4). Bacterial richness was significantly influenced by P content (β = 2.16, p < 0.001), SOM (β = –3.51, p = 0.005), VS (β = 2.27, p = 0.002) and LAI (β = 4.55, p = 0.002; Table 1).
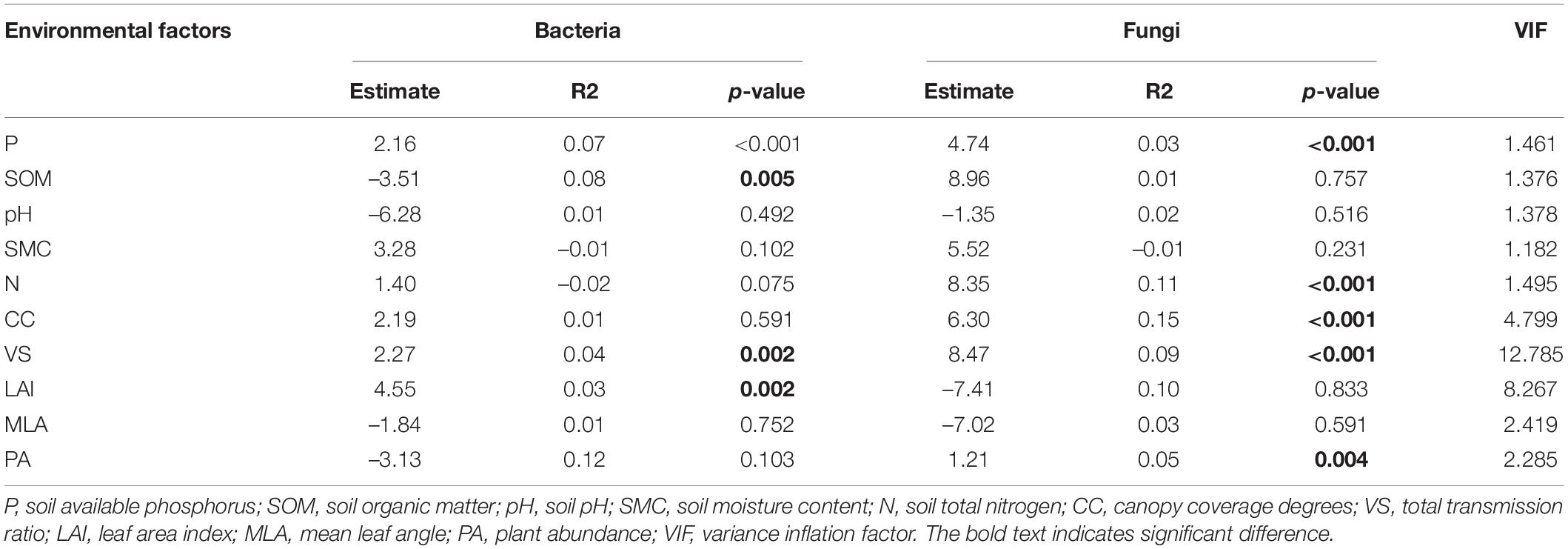
Table 1. Linear mixed-effects model for the relationships between multiple factors (physicochemical characters of soil, light environment, and plant abundance) with considering vegetation types as random terms.
Fixed variables (environmental factors) explained 0.57 (R2m) of the variation in soil fungal communities while vegetation types explained a large proportion of variation (R2c - R2m = 0.32; Supplementary Figure 4B). Fungal richness was significantly influenced by P content (β = 4.74, p < 0.001), N (β = 8.35, p < 0.001), CC (β = 6.30, p < 0.001), VS (β = 8.47, p < 0.001) and PA (β = 1.21, p = 0.004; Table 1).
Discussion
Our results showed that the spatial distribution characteristics of soil microbes differed among different vegetation types in the urban forest. We also found that the driving forces of environmental factors on soil bacteria and fungi were different. Together, these findings suggest the importance of different vegetation types in maintaining local diversity in a soil microbial community in the urban forest.
Fungi Are More Specialized Than Bacteria
The Venn diagram showed that the number of fungal and bacterial OTUs in deciduous forest was higher than that in other vegetation types. On the basis of the results of network analysis, the soil fungal community specialization index (H2’) of different vegetation types was 0.486, which was higher than that of the plant root–fungus network (0.265; Toju et al., 2014). The specialization index (H2’) of the bacterial community among vegetation types was only 0.091. The findings suggest that fungi are more specialized than bacteria. Tree species are known to strongly affect ecosystem conditions including soil properties and microclimate (Menyailo et al., 2002; Reich et al., 2005; Russell et al., 2007). Bacteria usually grow single-celled, whereas fungi usually grow filamentous (Powell et al., 2015). Due to the size and number of propagules, bacteria have a greater ability to spread than fungi (Powell et al., 2015). Bacteria are also generally more resilient than fungi in the face of disturbance (Wardle, 2002). This phenomenon may be an important reason why fungal communities are more sensitive than bacterial communities in the face of vegetation type changes in the temperate urban forest. Different vegetation types form different soil microhabitats and affect the distribution characteristics of soil microbes. Hence, the spatial distribution of fungi is more specialized than that of bacteria among different vegetation types in this temperate urban forest.
Preferences of Different Vegetation Types
Based on the results of the torus-translation test, the composition characteristics of soil fungi and bacteria were different among different vegetation types. About 56.74% (5280/9305) species of bacteria and 73.69% (2894/3927) species of fungi were positively associated with vegetation types. Therefore, different soil microbes show different ecological preferences of different vegetation types in the temperate urban forest.
The number of positively correlated OTUs of fungi (42.3%) and bacteria (28.1%) was higher in the deciduous forest than in the evergreen and mixed forests. Moreover, the number of negatively correlated OTUs of fungi (0.9%) and bacteria (0.9%) was lower in the deciduous forest than in other vegetation types. Relevant studies have proved that forest gap can be related to the growth of soil pathogenic oomycetes and the occurrence of diseases (Reinhart et al., 2010). In this study, light intensity under deciduous forest is higher than that in the other two types, which may promote the growth and development of microbes. This phenomenon may be an important reason why microbial species exhibited ecological preferences for different vegetation types. At the same time, the release of various compounds by plant roots into the surrounding soil creates a unique environment in the rhizosphere for microbes that are symbiotic with plant roots (Garbeva et al., 2004), which may also lead to different habitat preferences of soil microbes.
Driving Forces of Environmental Factors on Soil Bacteria and Fungi Were Different
In this study, we found that the environmental drivers of fungi and bacteria are different in the temperate urban forest. Previous studies have shown that the survival and reproduction of soil microbes were affected by soil physical and chemical properties (Wang et al., 2013; Wang et al., 2018; Huang et al., 2019; Wang et al., 2021). Soil nutrients greatly differ in different vegetation types (Sheng et al., 2019), so the different physical and chemical properties of soil have different effects on fungi or bacteria. According to our study results, N was negatively correlated with bacteria in urban forests. However, it was positively correlated with fungi. Cheng et al. (2017) found that the relationships between soil fungi and woody plants are different in different habitats. In this study, environmental factors have different driving effects on bacteria and fungi, which may be related to the habitat. Our results also indicated that SR and CC influenced fungal richness. This result is consistent with the findings of Chen et al. (2018), who found that light is the major driver for soil fungi. Many studies have shown that N, P, and SOM have important effects on microbial community composition (Yoshimura et al., 2013; Van Geel et al., 2016). Overall, the driving forces of environmental factors on soil bacteria and fungi were different in temperate urban forest.
Conclusion
The results of this study indicate that habitat-specific vegetation types are important for microbes and potentially important for the maintenance of microbial species diversity in the urban ecosystem. This study provides a scientific basis for the formulation of soil biodiversity conservation strategies in urban ecosystems. At the same time, it also provides a certain ecological basis for garden designers in the planting and configuration of landscape plants. We should pay attention not only to the construction of aboveground landscape but also to the influence of soil microorganisms on plants in urban ecosystems.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJNA783475].
Author Contributions
YY, ZY, YC, YS, FL, and GT planned and designed the research. QF and SW performed experiments, conducted fieldwork, and analyzed data, etc. QF, YS, and SW wrote the manuscript. YC and ZY provide this idea. QF and YS contributed equally. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Project funded by China Postdoctoral Science Foundation (2021M693400); Young Talents project funded by Henan Agricultural University (111/30500744); Youth Foundation of Natural Science Foundation of Henan Province (212300410153).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.858254/full#supplementary-material
Supplementary Figure 1 | Bar plot of microbial community composition at the phylum and family level.
Supplementary Figure 2 | Sparse curves of bacteria and fungi.
Supplementary Figure 3 | Phylogenetic tree at the OTU level.
Supplementary Figure 4 | Bar plot of the contribution of different environmental factors to fixed effects. pH, soil pH; SMC, soil moisture content; P, soil available phosphorus; SOM, soil organic matter; N, soil total nitrogen; CC, canopy cover degrees; VS, total transmission ratio; LAI, leaf area index; MLA, mean leaf angle; PA, plant abundance; R2m, R squared of fixed effect; R2c, total R squared for fixed effects and random effects.
References
Bardgett, R. D., and van der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511.
Benjamin, J. C., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Bennett, J. A., and Cahill, J. F. (2016). Fungal effects on plant-plant interactions contribute to grassland plant abundances: evidence from the field. J. Ecol. 104, 755–764.
Bever, J. D., Mangan, S., and Alexander, H. (2015). Maintenance of plant species diversity by pathogens. Ann. Rev. Ecol. Evol. Syst. 46, 305–325. doi: 10.1146/annurev-ecolsys-112414-054306
Blüthgen, N., Menzel, F., Hovestadt, T., Fiala, B., and Blüthgen, N. (2007). Specialization, constraints, and conflflicting interests in mutualistic networks. Curr. Biol 17, 341–346. doi: 10.1016/j.cub.2006.12.039
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Chen, H. B., and Boutros, P. C. (2011). Venn diagram: a package for the generation of highly-customizable venn and euler diagrams in R. BMC Bioinform. 12:35. doi: 10.1186/1471-2105-12-35
Chen, Y., Yizhen, S., Jingjing, X., Zhiliang, Y., Yongzhong, Y., and Ting, W. (2020). Community Preferences of Woody Plant Species in a Heterogeneous Temperate Forest, China. Frontiers in Ecology and Evolution 8:165.
Chen, Y., Zhiliang, Y., Shuai, B., Xueying, W., Yongzhong, Y., and Jens-Christian, S. (2018). Macrofungal species distributions depend on habitat partitioning of topography, light, and vegetation in a temperate mountain forest. Scientific Reports 8, 13589. doi: 10.1038/s41598-018-31795-7
Cheng, G., Shi, N., Chen, L., Ji, N., Wu, B., Wang, Y., et al. (2017). Relationships between soil fungal and woody plant assemblages differ between ridge and valley habitats in a subtropical mountain forest. New Phytol. 213, 1874–1885. doi: 10.1111/nph.14287
Christian, Q., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Comita, L. S., Condit, R., and Hubbell, S. P. (2007). Developmental changes in habitat associations of tropical trees. J. Ecol. 95, 482–492. doi: 10.1111/j.1365-2745.2007.01229.x
Cong, J., Yunfeng, Y., Xueduan, L., Hui, L., Xiao, L., Jizhong, Z., et al. (2015). Analyses of soil microbial community compositions and functional genes reveal potential consequences of natural forest succession. Sci. Rep. 5:10007. doi: 10.1038/srep10007
Dickie, I. A. (2007). Host preference, niches and fungal diversity. New Phytol. 174, 230–233. doi: 10.1111/j.1469-8137.2007.02055.x
Franciska, M., Benedettia, Y., and Callaghan, C. T. (2020). Ecological specialization and population trends in European breeding birds. Global Ecol. Conserv. 22:e00996. doi: 10.1016/j.scitotenv.2017.12.150
Fu, Q., Nan, W., Man, X., Jingjing, X., Yizhen, S., Hongru, J., et al. (2021). Canopy structure and illumination characteristics of different man-made interference communities in baiyun mountain national forest park. Acta Ecol. Sin. 41, 7830–7837.
Garbeva, P., van Veen, J. A., and van Elsas, J. D. (2004). Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Ann. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Gardes, M., and Bruns, T. D. (1993). Its primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x
Gunatilleke, C. V. S., Gunatilleke, I. A. U. N., Esufali, S., Harms, K. E., Ashton, P. M. S., Burslem, D. F. R. P., et al. (2006). Species-habitat associations in a Sri Lankan dipterocarp forest. J. Trop. Ecol. 22, 371–384. doi: 10.1017/s0266467406003282
Hacquard, S. (2016). Disentangling the factors shaping microbiota composition across the plant holobiont. New Phytol. 209, 454–457. doi: 10.1111/nph.13760
Harms, K. E., Condit, R., Hubbell, S. P., and Foster, R. B. (2001). Habitat associations of trees and shrubs in a 50-ha neotropical forest plot. J. Ecol. 89, 947–959. doi: 10.1111/j.1365-2745.2001.00615.x
Heegaard, E. (2017). Fine-scale spatiotemporal dynamics of fungal fruiting: prevalence, amplitude, range and continuity. Ecography 40, 947–959. doi: 10.1111/ecog.02256
Hooper, D., Bignell, D., Brown, V., Brussard, L., Coleman, D., Giller, K., et al. (2000). Interactions between aboveground and belowground biodiversity in terrestrial ecosystems: patterns, mechanisms, and feedbacks. BioScience 50, 1049–1061. doi: 10.1641/0006-3568(2000)050[1049:ibaabb]2.0.co;2
Huang, Z., Zhao, F., Wang, M., Qi, K., Wu, J., and Zhang, S. (2019). Soil chemical properties and geographical distance exerted effects on arbuscular mycorrhizal fungal community composition in pear orchards in jiangsu province. China. Appl. Soil Ecol. 142, 18–24. doi: 10.1016/j.apsoil.2019.05.017
Jiao, S., Yunfeng, Y., Yiqin, X., Jie, Z., and Yahai, L. (2020). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Jing, J. (2017). Study on Urban Garden Plant Community In Southern Warm Zone Based On Economization And Beauty Evaluation. China: Zhejiang Sci-Tech University.
Kang, H., Wenjuan, Y., Somak, D., and Huanhuan, G. (2021). Soil microbial community composition and function are closely associated with soil organic matter chemistry along a latitudinal gradient. Geoderma 383:114744. doi: 10.1016/j.geoderma.2020.114744
Kreuzer, K., Bonkowski, M., and Langel, R. (2004). Decomposer animals (lumbricidae, collembola) and organic matter distribution affect the performance of Lolium perenne (poaceae) and Trifolium repens (fabaceae). Soil Biol. Biochem. 36, 2005–2011. doi: 10.1016/j.soilbio.2004.05.019
Lai, J. S., Zou, Y., Zhang, J. L., and Peres-Neto, P. R. (2022) Generalizinghierarchical and variation partitioning in multiple regression and canonicalanalyses using the rdacca.hp R package. Methods Ecol. Evol. 13, 782–788. doi: 10.1111/2041-210X.13800
Lin, D., Mei, P., Fanin, N., Hongjuan, W., Shenhua, Q., Liang, Z., et al. (2018). Fungi participate in driving home-field advantage of litter decomposition in a subtropical forest. Plant Soil 434, 467–480. doi: 10.1007/s11104-018-3865-5
Menyailo, O. V., Hungate, B. A., and Zech, W. (2002). Tree species mediated soil chemical changes in a siberian artificial afforestation experiment: tree species and soil chemistry. Plant Soil 242, 171–182.
Mo, Y., Feng, P., Xiaofei, G., Xiao, P., Logares, R., Jeppesen, E., et al. (2021). Low shifts in salinity determined assembly processes and network stability of microeukaryotic plankton communities in a subtropical urban reservoir. Microbiome 9:1079. doi: 10.1186/s40168-021-01079-w
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Ning, R., Wang, Y., Ye, Y., Zhao, Y., Huang, Y., Fu, W., et al. (2020). Effects of continuous nitrogen fertilizer application on the diversity and composition of rhizosphere soil bacteria. Front. Microbiol. 11:1948. doi: 10.3389/fmicb.2020.01948
Oksanen, J., Kindt, R., Legendre, P., and O’Hara, B. (2007). Vegan: The Community Ecology Package. 1, 8–4.
Powell, J. R., Karunaratne, S., Campbell, C. D., Yao, H., Robinson, L., and Singh, B. K. (2015). Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat. Commun. 6, 1–10.
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Reich, P. B., Oleksyn, J., Modrzynski, J., Mrozinski, P., Hobbie, S. E., Eissenstat, D. M., et al. (2005). Linking litter calcium, earthworms and soil properties: acommon garden test with 14 tree species. Ecol. Lett. 8, 811–818. doi: 10.1111/j.1461-0248.2005.00779.x
Reinhart, K. O., Royo, A. A., Kageyama, S. A., and Clay, K. (2010). Canopy gaps decrease microbial densities and disease risk for a shade-intolerant tree species. Acta Oecol. 36, 530–536. doi: 10.1016/j.actao.2010.07.006
Rivest, D., Paquette, A., Shipley, B., Reich, P. B., and Messier, C. (2015). Tree communities rapidly alter soil microbial resistance and resilience to drought. Function. Ecol. 29, 570–578. doi: 10.1111/1365-2435.12364
Russell, A. E., Raich, J. W., Valverde-Barrantes, O. J., and Fisher, R. F. (2007). Tree species effects on soil properties in experimental plantations in tropical moist forest. Soil Sci. Soc. Am. J. 71, 1389–1397. doi: 10.2136/sssaj2006.0069
Schimel, J., Balser, T. C., and Wallenstein, M. (2007). Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. doi: 10.1890/06-0219
Sergio, A., Sotob, J. R., Escobedoc, F. J., Laid, J., Kibriab, A. S. M. G., and Adamse, D. C. (2021). Heterogeneous preferences and economic values for urban forest structural and functional attributes. Land. Urban Plan. 215:104234. doi: 10.1016/j.landurbplan.2021.104234
Shan, L., Shakoor, A., Wubet, T., Zhang, N., Liang, Y., and Ma, K. (2018). Fine-scale variations of fungal community in a heterogeneous grassland in inner mongolia: effects of the plant community and edaphic parameters. Soil Biol. Biochem. 122, 104–110. doi: 10.1016/j.soilbio.2018.04.007
Sheng, Y., Jing, C., Hui, L., Linsen, Y., Qiang, L., Li, D., et al. (2019). Broad-leaved forest types affect soil fungal community structure and soil organic carbon contents. MicrobiologyOpen 2019:e874. doi: 10.1002/mbo3.874
Simon, T., Frey, D., Renée-ClaireLeBayon Zanetta, A., Rasche, F., Fliessbach, A., et al. (2018). Litter decomposition driven by soil fauna,plant diversity and soil management in urban gardens. Sci. Environ. 12:235. doi: 10.1016/j.scitotenv.2018.12.235
Singh, B. K., Millard, P., Whiteley, A. S., and Murrell, J. C. (2004). Unravelling rhizosphere-microbial interactions: opportunities and limitations. Trends Microbiol. 12, 386–393. doi: 10.1016/j.tim.2004.06.008
Song, X., Kimberley, M. O., Guomo, Z., and Hailong, W. (2017). Soil carbon dynamics in successional and plantation forests in subtropical China. J. Soils Sedim. 17:9.
Svenning, J. C. (1999). Microhabitat specialization in a species-rich palm community in amazonian ecuador. J. Ecol. 87, 55–65. doi: 10.1046/j.1365-2745.1999.00329.x
Toju, H., Guimaraes, P. R., Olesen, J. M., and Thompson, J. N. (2014). Assembly of complex plant–fungus networks. Nat. Commun. 5:5273. doi: 10.1038/ncomms6273
Van Geel, M., De Beenhouwer, M., Ceulemans, T., Caes, K., Ceustermans, A., Bylemans, D., et al. (2016). Application of slow-release phosphorus fertilizers increases arbuscular mycorrhizal fungal diversity in the roots of apple trees. Plant Soil 402, 291–301. doi: 10.1007/s11104-015-2777-x
Wagg, C., Bender, S. F., Widmer, F., and van der Heijden, M. G. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. 111, 5266–5270. doi: 10.1073/pnas.1320054111
Wang, B., Adachi, Y., and Sugiyama, S. (2018). Soil productivity and structure of bacterial and fungal communities in unfertilized arable soil. PLoS One 13:e0204085. doi: 10.1371/journal.pone.0204085
Wang, J., Yin, W., Nianpeng, H., Ziqi, Y., Chen, C., Runguo, Z., et al. (2020). Plant functional traits regulate soil bacterial diversity across temperate deserts. Sci. Total Environ. 715:136976. doi: 10.1016/j.scitotenv.2020.136976
Wang, N., Qiang, F., Ziyu, Z., Yizhen, S., Jing, W., Wang, L., et al. (2021). Humus microhabitat affects distributions of soil fungi and bacteria in a temperate mountain forest. Ecol. Evol. 11, 9148–9158.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, Q., He, T., Wang, S., and Liu, L. (2013). Carbon input manipulation affects soil respiration and microbial community composition in a subtropical coniferous forest. Agric. Forest Meteorol. 178-179, 152–160. doi: 10.1016/j.agrformet.2013.04.021
Wardle, D. A. (2002). Communities and Ecosystems: Linking the Aboveground and Belowground Components. Princeton, NJ: Princeton University Press.
Wardle, D. A. (2006). The influence of biotic interactions on soil biodiversity. Ecol. Lett. 9, 870–886. doi: 10.1111/j.1461-0248.2006.00931.x
Wardle, D. A., Bardgett, R. D., Klironomos, J. N., Set€al€a, H., van der Putten, W. H., and Wall, D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633. doi: 10.1126/science.1094875
Xiao, X., Na, Z., Haowei, N., Yunfeng, Y., Jizhong, Z., Bo, S., et al. (2021). A latitudinal gradient of microbial β-diversity in continental paddy soils. Global Ecol. Biogeogr. 4:13267. doi: 10.1111/GEB.13267
Yang, J., Yanfen, W., Xiaoyong, C., Kai, X., Yiming, Z., and Zhisheng, Y. (2019). Habitat filtering shapes the differential structure of microbial communities in the Xilingol grassland. Sci. Rep. 1:1598. doi: 10.1038/S41598-019-55940-Y
Yoshimura, Y., Ido, A., Iwase, K., Matsumoto, T., and Yamato, M. (2013). Communities of arbuscular mycorrhizal fungi in the roots of Pyrus pyrifolia var. culta (Japanese pear) in orchards with variable amounts of soil available phosphorus. Microbes Environ. 28, 105–111. doi: 10.1264/jsme2.me12118
Keywords: bacteria, fungi, urban green space, species diversity, soil ecosystem
Citation: Fu Q, Shao Y, Wang S, Liu F, Tian G, Chen Y, Yuan Z and Ye Y (2022) Soil Microbial Distribution Depends on Different Types of Landscape Vegetation in Temperate Urban Forest Ecosystems. Front. Ecol. Evol. 10:858254. doi: 10.3389/fevo.2022.858254
Received: 19 January 2022; Accepted: 25 April 2022;
Published: 20 May 2022.
Edited by:
Ke Dong, Seoul National University, South KoreaReviewed by:
Seung-Yoon Oh, Changwon National University, South KoreaAnvar Sanaei, University of Tehran, Iran
Copyright © 2022 Fu, Shao, Wang, Liu, Tian, Chen, Yuan and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Chen, Y3llY29sb2d5QDE2My5jb20=; Zhiliang Yuan, emx5dWFuMTYzQDE2My5jb20=
†These authors have contributed equally to this work
 Qiang Fu
Qiang Fu Yizhen Shao2†
Yizhen Shao2† Senlin Wang
Senlin Wang Guohang Tian
Guohang Tian Yun Chen
Yun Chen