- 1Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
- 2Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 3National Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai, China
- 4Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
The spatial distribution of fish individuals is affected by habitat conditions and species ecological characteristics, and it also reflects the longtime adaptation to habitat at the phenotypic and genotypic level. As a typical river-lake migratory fish species, the silver carp habitat selection was determined by its migration preference and genetic features. In this study, 15 microsatellite fluorescent markers combined with capillary electrophoresis were used to analyze the genetic diversity, genetic differentiation, and structure of nature silver carp populations in the Sanzhou (SZ), Hukou (HK), Anqing (AQ), Zhenjiang (ZJ), and Rugao (RG) sections of the Yangtze River. The results showed that 15 microsatellite loci exhibited medium to high polymorphisms. The overall genetic diversity in the Yangtze River was high, with the average value of Shannon’s information index ranging from 1.559 to 1.668. The numbers of alleles (Ne) ranged from 1.630 to 10.100. The expected heterozygosity (He, 0.690–0.721) was higher than observed heterozygosity (Ho, 0.598–0.646), and the genetic variation mainly originated from within the population (94.69%). However, the entire population was in the state of heterozygous deletion, and HK, RG populations encountered the risk of inbreeding risk (F > 1). Interestingly, there was a distinct genetic structure for the population in the HK section, which indicated that local population has occurred to the silver carp in this river section, and they may also possess aggregation characteristics specific to the river-lake-connected (RLC) habitat. The results mostly support the conclusion that the RLC habitat is essential for geographic population formation. The potential impact of special habitats on natural populations should be considered, and continuous surveys on population dynamics should be performed.
Introduction
The silver carp (Hypophthalmichthys molitrix) belongs to the Cypriniformes order, Cyprinidae family, and Hypophthalmichthys genus, and it is one of four major Chinese carp (Ni and Wu, 2006). Paleontological analyses demonstrated that the silver carp originated in the Yangtze-Huanghe River basin (Li and Fang, 1990). Currently, silver carp is widely distributed and is found in the basin of the Red River, the Pearl River, and the Heilongjiang River in China (Lu et al., 2020).
Artificial cultivation technology of silver carp was founded in 1958, which got rid of the passive situation of relying on catching and promoted the development of aquaculture (Mao et al., 2010). Then silver carp were transplanted to rivers in Europe, the United States, and Africa (Pinter, 1980), and even became an invasive fish species (Kolar and Lodge, 2002; Conover et al., 2007). The overseas population of silver carp continued to expand, while the Chinese indigenous population exhibited a declining trend (Li, 1996). Because of the construction of Three Gorges Dam, water pollution, and overfishing, fishery output was 427,000 tons in 1954. From 1956 to 1960, the fishing volume decreased to 260,000 tons, 200,000 tons in the 1980s, and reduced to about 100,000 tons 2000s, less than 1/4 of the maximum annual output (Mai, 2003). The four major carps accounted for 46.15 percent of the catch weight, but that figure decreased to only 10 percent in 2001–2003. The proportion of silver carp and bighead Carp among the four major fish decreased significantly (Zeng, 1990; Liu et al., 2005; Li and Xu, 2008). With a 10-year ban on fishing in the Yangtze River initiated in January 2020 and the continuous implementation of stock enhancement, the populations of the four major Chinese carp are in the process of recovery. However, stock enhancement and unscientific artificial release may result in negative ecological impacts such as impaired growth, disease spread, and decrement of genetic diversity (Liu et al., 1997; Bell et al., 2006; Fang et al., 2021).
The Yangtze River is the longest and largest river in China, and is the main germplasm resource area for silver carp. Studies have reported that there are 11 spawning grounds for the four major Chinese carp in the middle and lower reaches of the Yangtze River (Xu et al., 2017), and silver carp spawning grounds are still being found (Tang et al., 2010; He et al., 2021). As a fish species that migrates between rivers and lakes, the silver carp spawns in the main stream of the Yangtze River every breeding season (from April to July). The postpartum parent fish and the young fish enter the lake connected to the Yangtze River for feeding (Xu et al., 2017).
Moving away from an unsuitable environment is one of the most important adaptive strategies for fish (Matter et al., 1989). During the process of migration, adult fish are able to be selective regarding their habitat, and they would choose to remain in habitats with appropriate environmental conditions, especially if there are abundant food sources and little interference (Bonte and Maelfait, 2004). For example, studies on the manini (Acanthurus triostegus and A. vicensis) fish habitat found that a covered shallow water area was its preferred habitat (Sale, 1968). Similarly, silver carp requires a great deal of energy input for the process of reproduction and fattening. When the environment lacks sufficient resources, this will lead to its continuous exploration to find a suitable habitat (McMahon and Matter, 2006). There is abundant plankton bait in river-lake-connected (RLC) habitats, which are ideal living places for silver carp (Liu et al., 2019). During the growth period for silver carp, adults will choose a habitat with rich bait for growth and fattening.
The Hukou section connects Poyang Lake with the Yangtze River, which is the largest lake connected to the Yangtze River in the lower reaches (He et al., 2021). The Anqing section connects the Wan River, which also forms a unique complex ecosystem with RLC characteristics. Because these bodies of water are rich in aquatic biological resources, they provide a key channel for fish migration behavior (Liu et al., 2019). These similar habitats provide perfect feeding and reproduction grounds for the silver carp in the Yangtze River. This habitat connectivity is vital for ecosystem function and the distribution of biota (Lindenmayer et al., 2008). However, studies on silver carp in the Yangtze River have mainly concentrated on germplasm genetic diversity, and it has been proven that there is genetic differentiation in fish from different river sections (Zhu et al., 2007; Chen et al., 2018; Sha et al., 2018). The value of connectivity in conservation has been poorly understood, and few genetic studies have linked population dynamics to habitat selection.
The simple sequence repeat (SSR) is a powerful genetic marker that has been widely used in fishery assessment. It has the advantages of large number of markers, wide distribution in gene sequence, neutrality, and co-dominance. It is one of the ideal molecular markers for genetic evaluation (Sun et al., 2008). It was previously reported that five microsatellite loci were used to investigate the population structure of Culterery thropterus from seven lakes in the middle and lower reaches of the Yangtze River (Wang et al., 2007). Additionally, sixteen SSR polymorphic loci were used to construct a partial genomic library of bighead carp (Aristichthys nobilis) (Cheng et al., 2008). In this study, fifteen microsatellite loci were used to analyze the genetic diversity and genetic structure of silver carp populations in the Sanzhou (SZ), Hukou (HK), Anqing (AQ), Zhenjiang (ZJ), and Rugao (RG) sections of the Yangtze River. The population’s genetic structure difference between sampled sections in the Yangtze River was clarified, so that we can deduce potential relation between the fish distribution pattern and habitat differences.
Materials and Methods
Sample Collection
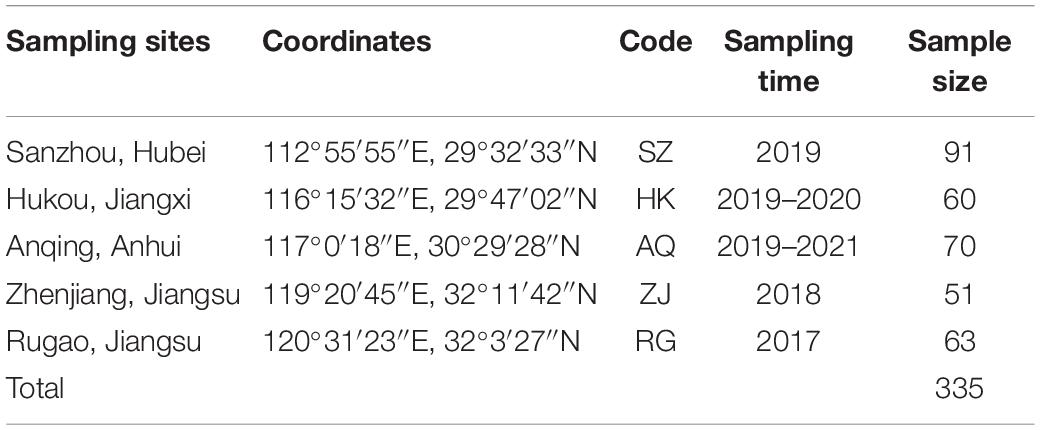
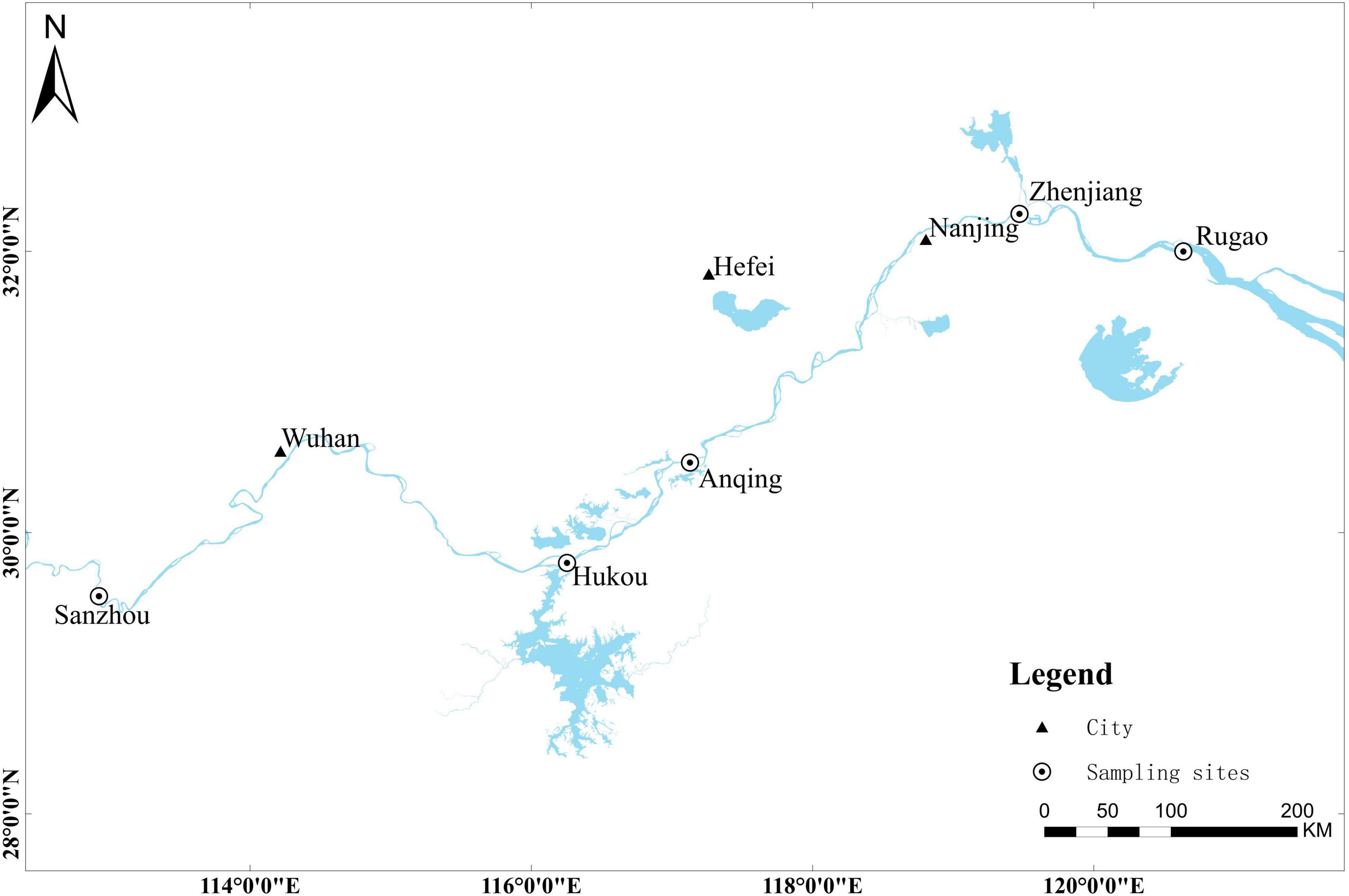
Five nature populations of adult silver carp in the Yangtze River were collected by a traditional net trap from 2017 to 2021 avoiding the stock enhancement period. Totally 335 tail fins of silver carp were non-destructively sampled and stored in absolute ethanol for further study. The sampling sites, coordinates, time and size were listed in Table 1, and the distribution of sampling sites was shown in Figure 1. SZ belongs to the middle reaches of the Yangtze River, HK, AQ, ZJ, RG belongs to the lower reaches of the Yangtze River.
DNA Extraction and Microsatellite Genotyping
Total genomic DNA was extracted using a TIANamp Marine Animals DNA Kit (TIANGEN Biotech Co., Ltd., Beijing, China) following the manufacturer’s instructions. The quality of the extracted DNA was assessed by 1.2% agarose gel electrophoresis to ensure that successful amplification could be accomplished for all DNA strands, and then, the DNA was stored at −20°C for further study.
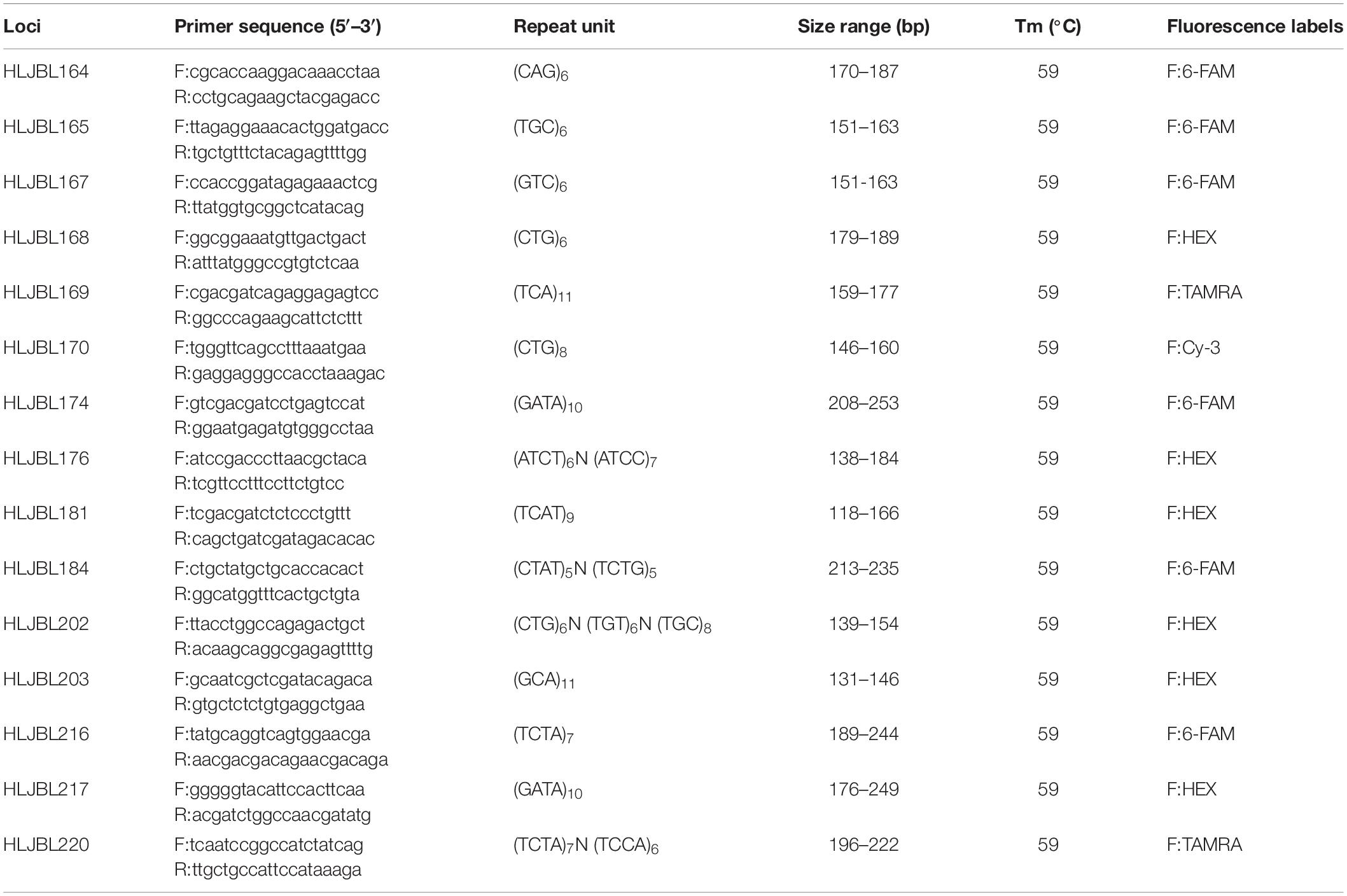
Fifteen polymorphic microsatellites were proactively identified and also used in another study (Tan et al., 2011). Each primer was synthesized by Sangon Biotech (Shanghai Co., Ltd., Shanghai, China) and fluorescently labeled at the 5′ terminus (Table 2). The PCR application of SSR markers was carried out in a total volume of 10 μL and consisted of 5 μL Taq Premix (TaKaRaTaq™ Version 2.0 plus dye, Takara, Dalian, China), 0.2 μL (10 μM/OD) primer pairs, 1 μL (200 ng/μL) genomic DNA, and 3.8 μL ddH2O. PCRs were performed in a 96-well thermal cycler (Gene Co., Ltd., Shanghai) using the following conditions: 94°C for 120 s; 30 cycles of 94°C for 20 s, 59°C for 20 s, 72°C for 20 s; extension at 72°C for 600 s, and finally, storage at 4°C. The PCR products were qualified by 2% agarose gel electrophoresis and the G:BOX automatic gel imaging system. Qualified PCR products were sent to Sangon Biotech (Shanghai Co., Ltd., Shanghai, China) for capillary electrophoresis sequencing and typing, which was performed using an ABI Prism 3730 XL automated sequencer (Rox-500 standard).
Genetic Analysis
After genotyping based on capillary electrophoresis technology, we manually confirmed the accuracy of genotypes. Pedigree analysis software Cervus 3.0.7 was used to analyze the polymorphic information content (PIC) (Kalinowski et al., 2007). GenAlEx 6.503 software was used to calculate sample sizes (n), the number of alleles (Na), effective numbers of alleles (Ne), numbers of private alleles (Ar), expected heterozygosity (He), observed heterozygosity (Ho), unbiased expected heterozygosity (uHe), Shannon’s information index (I), and inbreeding index (F) (Peakall and Smouse, 2006). Bootstrapping analysis (1,000 repeated samplings) was used to evaluate the paired F-statistics values (Fst) and gene flow (Nm) among populations.
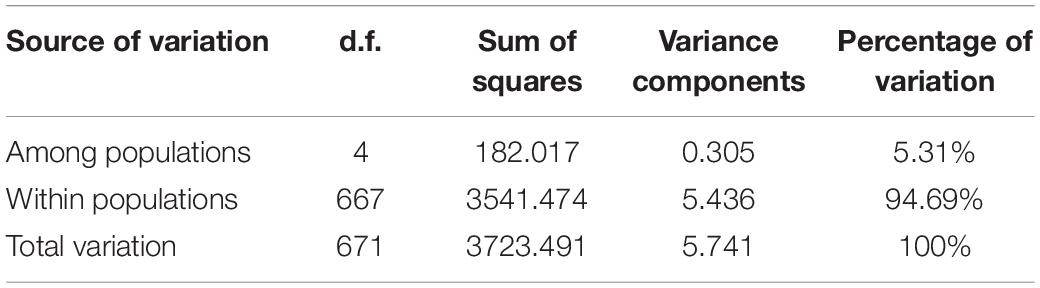
Analysis of molecular variance (AMOVA) was performed on the genetic variation of samples using the software Arlequin 3.1 to obtain the variation level difference between and within populations (Excoffier and Lischer, 2010), and the paired genetic distance (GD) was calculated at the same time. GenAlEx 6.503 was used to verify the principal coordinates analysis (PCoA) with GD as a parameter. Mega 5.0 software used the unweighted pair group method with the arithmetical mean (UPGMA) to construct a phylogenetic tree based on genetic distance (Tamura et al., 2011).
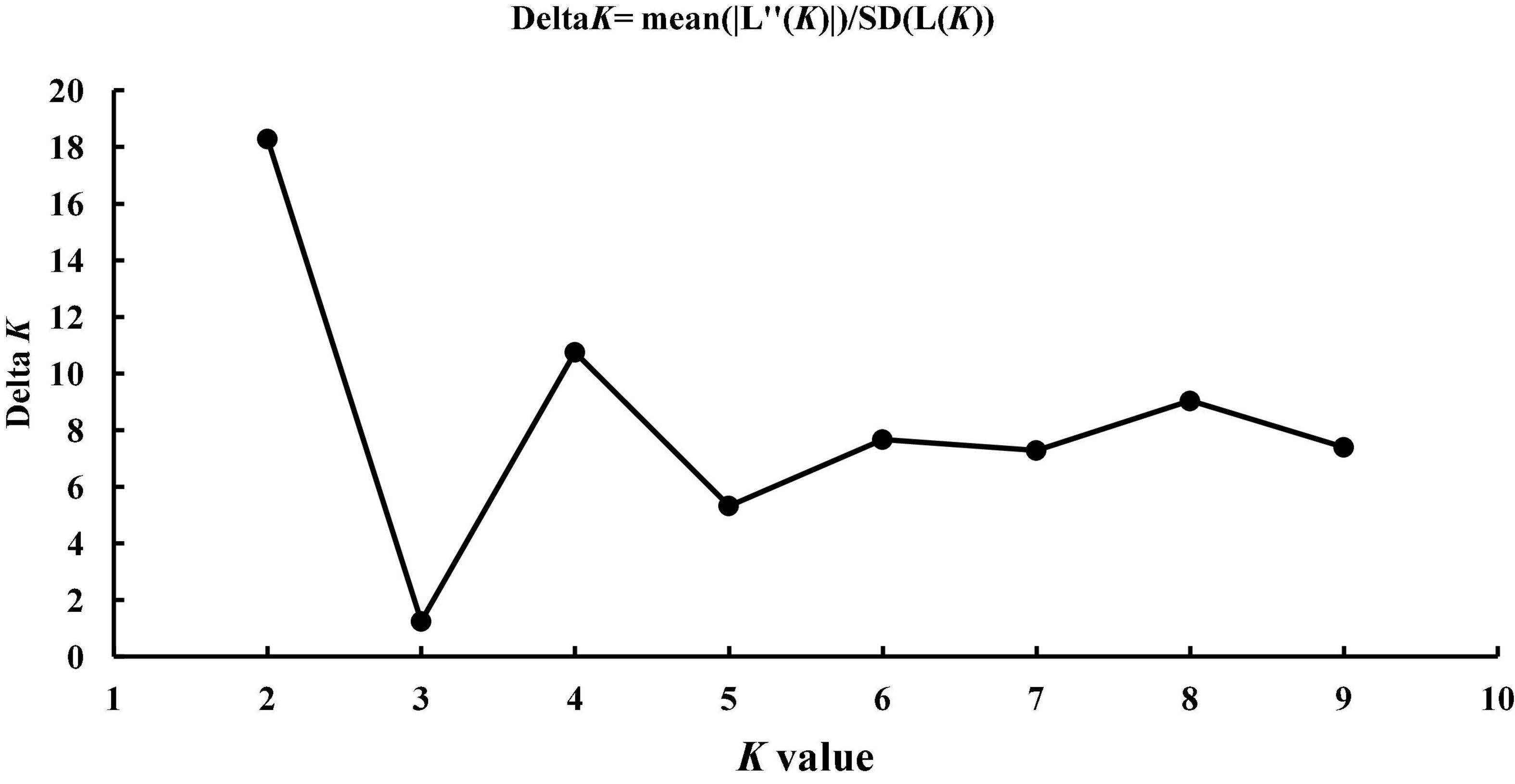
Structure 2.3.4 software was used to divide the population genetic structure (Smouse and Peakall, 1999). Based on the Bayesian model, the number of possible genetic cluster K values was set at 1–10, the length of the burn in-period for Markov chain Monte Carlo (MCMC) was set at 50,000 times, and each K value was repeated five times. The method of Evanno was used to calculate Delta K, and the most optimal K value was the number of clusters for the population (Evanno et al., 2005). Through the repeated sampling analysis of Clumpp 1.1.2 (Jakobsson and Rosenberg, 2007), a diagram was drawn by the software GraphPad Prism 8.0.2 to illustrate the population’s genetic structure (Swift, 1997).
Results
Genetic Diversity
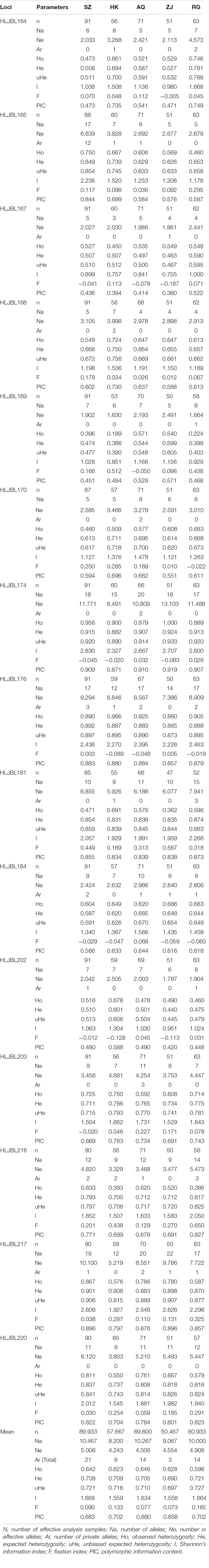
Fifteen microsatellites fluorescently labeled PCR products were genotyped by capillary electrophoresis. Cervus 3.0.7 analysis showed that the PIC (polymorphism information content) ranged from 0.380 (ZJ-HLJBL167) ∼ 0.919 (ZJ-HLJBL174). Every locus showed high polymorphism (PIC ≥ 0.5) in one or more silver carp populations, which indicated that the 15 SSR loci are suitable for the evaluation of genetic diversity of silver carp.
The genetic diversity results for different silver carp populations were based on the GenAlEx 6.503 analysis (Table 3). Na in every locus of each population ranged from 3 (HK, HLJBL167) to 22 (ZJ, HLJBL217). Ne ranged from 1.630 (HK, HLJBL169) to 10.100 (SZ, HLJBL217). Ar was the highest at the HLJBL165 loci in the SZ population (12). There were more private alleles in the SZ population (21), and fewer private alleles in the HK (8) and ZJ (3) populations, which indicated that SZ population has the highest gene abundance. The mean He of all populations was higher than the mean of Ho, which indicated that the proportion of homozygotes was larger than that of heterozygotes at the average level of these 15 loci. The mean value of uHe (i.e., gene diversity index) ranged from 0.697 (ZJ) to 0.727 (RG), which indicated that there was a similar level to the genetic diversity of all populations. The average value range of I was 1.559 (HK) to 1.668 (SZ). The mean of F was higher than zero and ranged from 0.073 (ZJ) to 0.169 (RG).
Genetic Differentiation
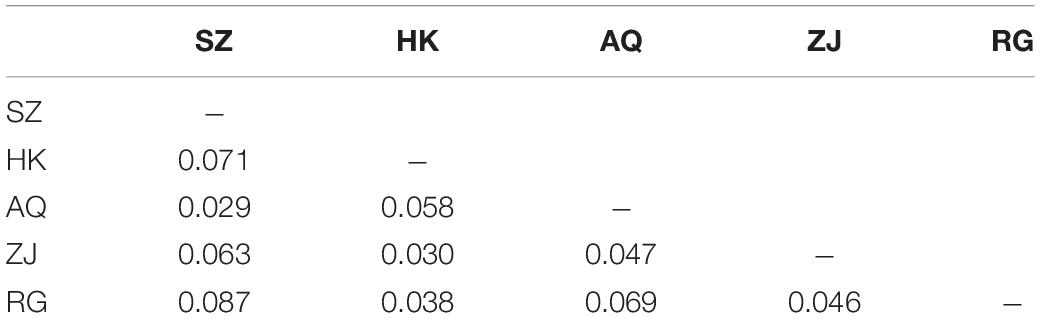
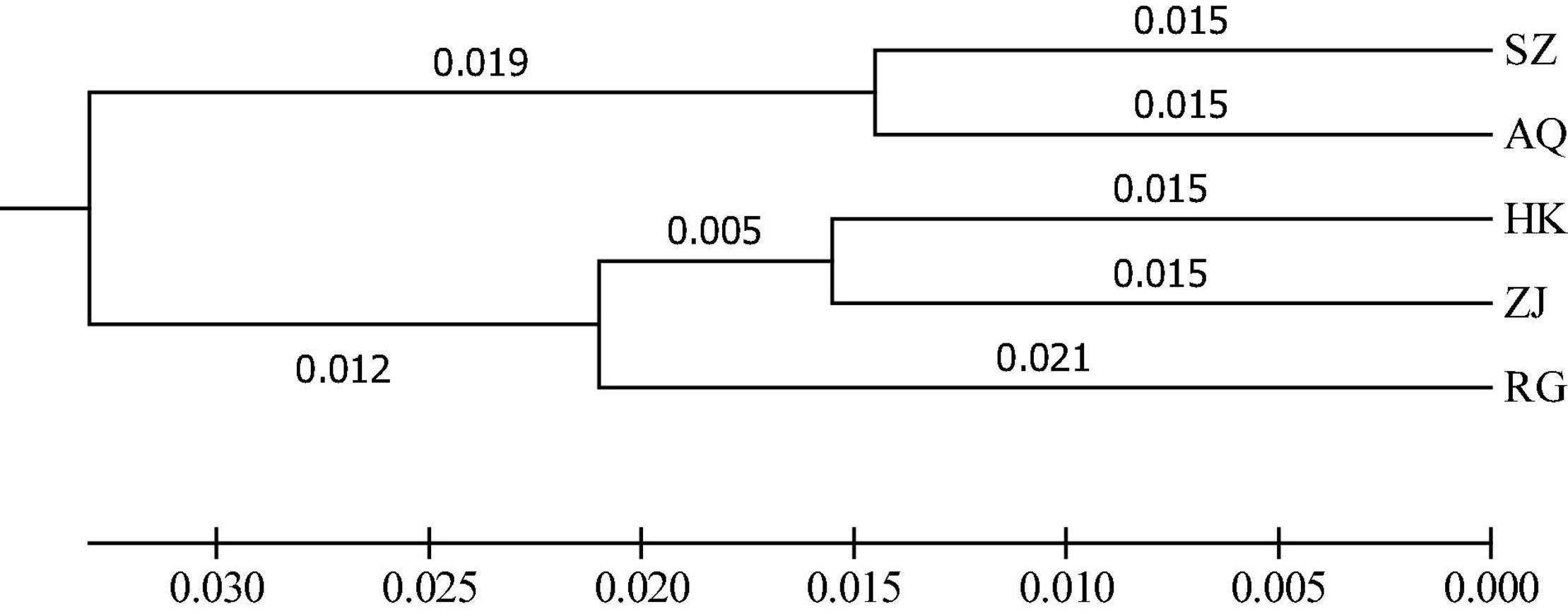
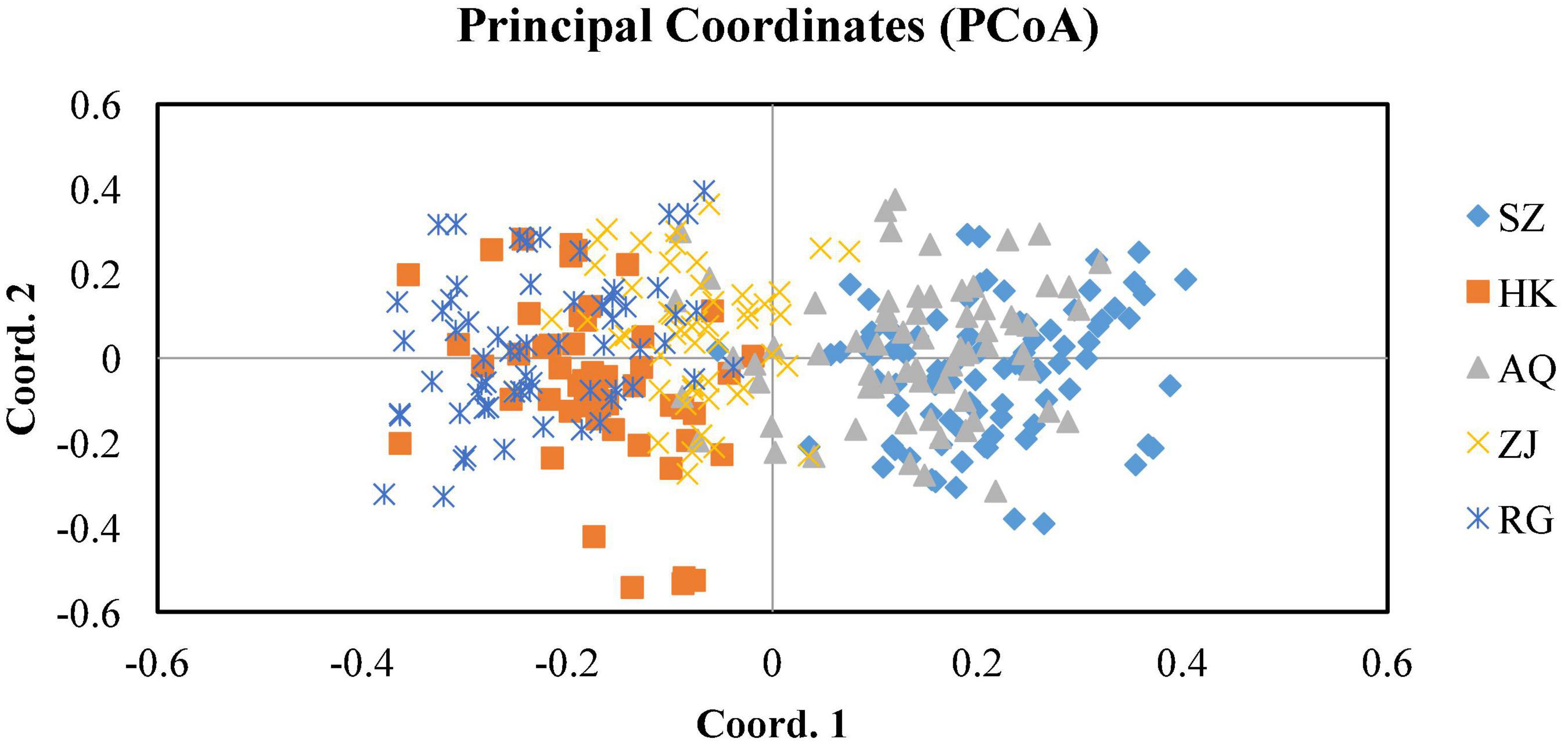
The results of AMOVA analysis showed that genetic variation mainly existed among different individuals within the population (94.69%), and a small part was contributed from different populations (5.31%) (Table 4). The pair-wise matrix of Fst and Nm showed that the Fst ranged from 0.016 (SZ and AQ) to 0.045 (SZ and RG), Nm was 6.365 (SZ and HK) to 15.850 (SZ and AQ) (Table 5). The genetic distance between populations ranged from 0.029 (SZ and AQ) to 0.087 (SZ and RG) (Table 6). Among them, the genetic distance between SZ and HK, SZ and ZJ, SZ and RG, HK and AQ, and AQ and RG was greater than 0.05. The genetic distance between the SZ and RG population was the largest (0.087), which was consistent with the geographical distance of the two populations. Based on the genetic distance, the population phylogenetic tree (Figure 2) showed that the SZ and AQ populations were first clustered, the HK and ZJ populations were also clustered, and then RG joined them together. The genetic distance between individuals was analyzed by PCoA (Figure 3). When the four sections were divided by the abscissa and ordinate as different habitats, HK, ZJ, and RG formed one group of habitats, and SZ and AQ formed another group of habitats. It was observed that some groups of AQ integrated into the first environment, and a small portion of ZJ individuals entered the second group of environments.
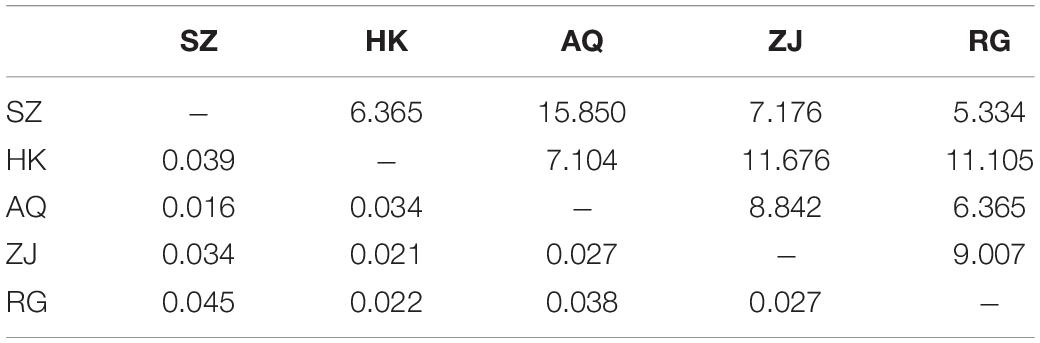
Table 5. F-statistics values (Fst) (below diagonal) and gene flow (Nm) (above diagonal) matrix of 5 silver carp populations.
Genetic Structure
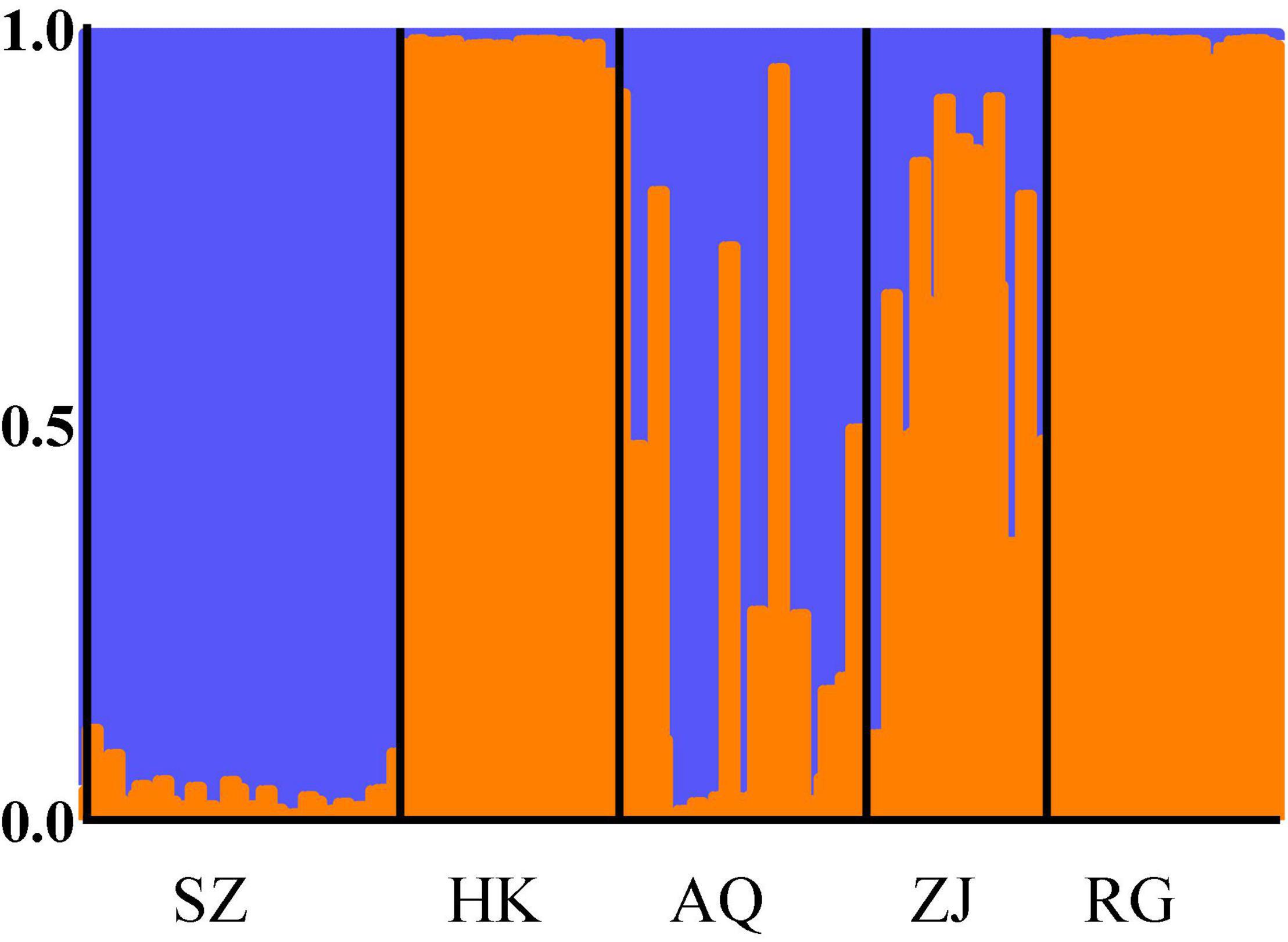
The most optimal K value was 2 when Delta K was the largest (Figure 4), indicating that 335 individuals can be divided into two potential groups (Figure 5). The results show that cluster 1 of the SZ population accounts for more than 90%. The HK and RG populations are nearly composed of cluster 2, which indicates that one potential group occupies most of the components in SZ and a part of AQ and ZJ. The other potential group occupies most of the components in HK and RG, and a part of AQ and ZJ.
Discussion
Genetic Diversity
Genetic diversity is the basis for long-term survival of a species as it adapts to the environment and maintains evolution. It provides an important basis for population resource assessment (Yuan et al., 2017). Heterozygosity of 0.500–0.800 indicates that the genetic diversity of this population was high (Takezaki and Nei, 1996). From the uHe of these five populations, the genetic diversity of all populations was at a middle to high level. From the comparison of Ar and I, the genetic diversity level of the SZ population was the highest, while the ZJ population was the lowest. Compared with the research of five silver carp populations (Shishou, Jianli, Jiujiang, Xiangjiang, and Anqing) in the middle and lower Yangtze River (Zhu et al., 2007) and two populations (Wanzhou and Jianli) in the middle and upper Yangtze River (Wang et al., 2008), it was found that the genetic diversity of silver carp in the middle and upper Yangtze River was generally higher than that in the lower reaches, which was consistent with the results of this study. This is due to the sharp decrease in silver carp biomass caused by overfishing. The decrease in biomass is closely related to the decline of genetic diversity (Xuan et al., 2021).
Na and Ne reflect the difference in population genetic variation. The more evenly alleles distributed in the population, the closer the Ne value to Na (Sun et al., 2014). In this study, alleles distributed in the population unevenly. The value of Ho and He reflect the excess or deletion of heterozygotes in the population (Li et al., 2006). From the average heterozygosity of the five populations, He was higher than Ho, which indicated that the entire population was in a state of heterozygous deletion. The F value of most loci (F > 0) in each population indicated that there was no bottleneck effect in the silver carp population of the Yangtze River. From the mean value of the population inbreeding coefficient, the HK and RG populations have encountered the risk of inbreeding inhibition (F > 0.1) (Moss et al., 2007).
Large numbers of young silver carp have been introduced into the Yangtze River every year to supplement the nature resources, which has a significant negative impact on the natural population genetics (Araki and Schmid, 2010). Genetic studies on the silver carp in four sections of the Yangtze River in Jiangsu also showed that large-scale stock enhancement would increase the gene flow among populations in the Yangtze River and increase the risk of inbreeding (Fang et al., 2021). It has been proved that stock enhancement in the lower Yangtze River can result in heterozygous deletion (Feng et al., 2020), which bring potential genetic risk and lead to a decrease in desirable traits.
Genetic Differentiation and Genetic Structure
In this study, Nm among the five populations was higher than 4 (6.365–15.850), which indicated that gene exchange can be carried out among the five populations. The higher the value, the greater the degree of gene exchange, which prevented the generation of genetic differentiation to a certain extent. Moreover, Fst among 5 populations (0.016–0.045) less than 0.15 indicates low genetic differentiation (0.05 < Fst < 0.15, low genetic differentiation, 0.15 < Fst < 0.25, moderate genetic differentiation) (Weight, 1978). The influence of genetic drift can be ignored, and each population can mate randomly (Hu et al., 2020), that is, the variation mainly came from within the population, which was consistent with the results of the AMOVA analysis. The high Nm was also obtained in a 2016 study among the three populations of the Ganjiang River, Poyang Lake, and the Yangtze River. This occurred because the three sampling points were close to each other, and the silver carp populations in the Ganjiang River and Yangtze River exchanged genes through Poyang Lake (Yu et al., 2016). However, the distance between the five populations in the current study was far, and therefore, it could be speculated that stock enhancement perhaps increased the gene exchange between populations in the Yangtze River. The genetic distance of fish at the three levels of genus, species, and population are 0.90, 0.30, and 0.05 respectively (Shaklee et al., 1982). There was population differentiation in SZ and HK, SZ and ZJ, SZ and RG, HK and AQ, and AQ and RG. The genetic distance between the SZ and RG populations was the largest, and it was positively correlated with the distance of the geographical location.
The results of the UPGMA phylogenetic tree and PCoA analysis divided the five silver carp populations into two groups. Among them, SZ and AQ were grouped together, HK and ZJ were grouped together, and RG was alone. It has been proven that the SZ and HK sections are the spawning grounds of silver carp (Xu et al., 2017; He et al., 2021). In the process of local fertilized eggs drifting downstream with the water, they develop into larvae and juveniles (Ren et al., 2016). When the juvenile fish have the ability to independently swim, they also have the ability to select adaptive habitats for feeding. Furthermore, the geographical distance between the SZ and AQ sections is similar to that from HK to ZJ (both approximately 600 km). Some supplementary populations in the AQ and ZJ sections were probably derived from the offspring of spawning populations in SZ and HK, respectively, which then locally formed corresponding populations.
HK is the dividing section between the middle and lower reaches. There was a single genetic cluster for the HK section in the genetic structure diagram. Poyang Lake is the largest lake connected to the Yangtze River. There were complex ecological conditions and abandoned bait resources (Wang et al., 2016). Silver carp likely entered Poyang Lake through the HK section for fattening after spawning. Some larvae and juveniles that developed from eggs may subsequently complete the life history of feeding and rearing in the lake until they return to the main stream of the Yangtze River for reproduction and spawning. In this case, the HK population possibly formed a distinct population composition that is different from the downstream populations. Another study that investigated the fish assemblage structure of Chinese carp in the Yangtze River indicated that the larvae and juveniles of the four major Chinese carp will passively enter the lake when the water flows backward and will grow for 3–4 years in the lake (Ru and Liu, 2013). Research on the habitat between coral reefs and mangroves indicated that the connectivity promoted fish abundance. The study also recommended that connected habitats should be considered as a high priority for conservation (Olds et al., 2012). Therefore, only by protecting the habitat and fishery resources of the HK section can we ensure the natural connectivity of the river and lake and thus successfully enable the silver carp to complete its reproduction and fattening process.
The AQ section was also one of the most important fishery waters and the key habitat for aquatic animals in the lower reaches of the Yangtze River (Tian et al., 2020). The silver carp migrate to the lower reaches of the Yangtze River for overwintering after finishing the reproductive migration in the middle reaches. Therefore, silver carp populations in AQ and ZJ contain more genetic clusters. The RG population showed the same single genetic structure in the structural diagram, while the UPGMA tree showed that it was clustered into one single class, which indicated that silver carp populations in this section had a further genetic relationship with other populations. The RG section is located at the estuary of the Yangtze River and far from the spawning ground in the middle and upper reaches. Besides, the silver carp in Rugao section experienced overfishing in the process during upward migration, so it is difficult to form a surplus group of multiple reproduction. Similar studies have been found in the biological investigation of Coilia nasus in the Yangtze River (Luo et al., 2021). Overfishing made it difficult for large-scale Coilia nasus to reach the Anqing section of the Yangtze River. Most of the silver carp in this section came from stock enhancement, and thus, the genetic structure was more singular than that of other river sections in the lower reaches.
Habitat Relevance and Conservation Strategies
Experiments on fish and other animals showed that local residents readily become immigrants when the resources required by a habitat are limited. Similarly, when the resources are sufficient, immigrant individuals become residents (Matter et al., 1989; Nelson et al., 2002). The HK section is located between the SZ and AQ section, but there is closer genetic distance between SZ and AQ with more frequent gene exchange, indicating that when migratory silver carp pass the HK section with rich bait resources after spawning and breeding in the middle reaches, some silver carp populations enter the lake for fattening and become residents of local Poyang Lake (Liu et al., 2019). Although seasonal migrations are easily confused with real migration, we have largely avoided this error through years of sampling (McMahon and Matter, 2006). Therefore, the populations preferred to select the RLC habitats until sexual maturity, and then formed a local geographical population. Research on Dongting Lake also showed that the larvae and juveniles of the four major Chinese carp will grow and mature in the lake for 3–4 years (Ru and Liu, 2013).
For the silver carp populations with distinct genetic structure and inbreeding inhibition such as HK and RG, excellent silver carp larvae and juveniles should be introduced from different sections of the Yangtze River to avoid sib or half-sib mating, and reduce inbreeding and avoid heterozygote loss. In addition, in order to increase the genetic diversity of each section and maintain the ecological balance of fish species reproduction, individuals from the RG section should be encouraged to enter the HK section. In this case, fish between these two populations can exchange their migratory habits. For instance, fish from the RG section may lead fish in the HK section to migrate downward, so as to increase the information exchange of the silver carp population in the Yangtze River and maintain the overall ecological balance. Populations with additional genetic structures can better adapt to a changing environment (Fang et al., 2021). For AQ, ZJ, and other river sections with diversified genetic structures, the genetic balance in the population is in a dynamic process of regulation. The parents of released fish should be derived from the natural population in the corresponding river basin to avoid potential genetic risk caused by gene mixing (Sha et al., 2021). Furthermore, maintaining distinctive habitats of the silver carp is vital importance to protect their species diversity radically.
Conclusion and Future Prospects
Based on 15 microsatellite loci, the genetic diversity, and genetic structure of five silver carp populations in the middle and lower Yangtze River were analyzed. The results showed that the genetic diversity level in the middle was higher than that of the lower reaches. There are potential genetic risks in distinct geographic populations. The RLC habitat characteristics perhaps have greatly contributed to forming local residents and the geographical population. Future stock enhancement of silver carp from different river sections should be considered genetic structure analysis. Furthermore, more valuable molecular markers should be developed to continuously evaluate the population dynamics of the silver carp in the Yangtze River as well as fishery resources assessment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Regulations for the Administration of Affairs Concerning Experimental Animals.
Author Contributions
Y-TL and Y-FZ conceived and designed the experiments. Y-TL and D-AF were responsible for data scoring and analyses, and writing the manuscript. D-PX, YY, Y-XP, and JX helped selecting the samples. C-CM, X-MT, and M-YZ helped DNA extraction and data analysis during manuscript preparation. All authors have read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (No. 2018YFD0900905), the National Infrastructure of Fishery Germplasm Resources (No. 2021DKA3047003), and Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD61).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.850183/full#supplementary-material
Supplementary Table S1 | Microsatellite loci data for the 335 silver carps.
References
Araki, H., and Schmid, C. (2010). Is hatchery stocking a help or harm? : evidence, limitations and future directions in ecological and genetic surveys. Aquaculture 308, S2–S11.
Bell, J. D., Bartley, D. M., Lorenzen, K., and Loneragan, N. R. (2006). Restocking and stock enhancement of coastal fisheries: potential, problems and progress. Fisheries Res. 80, 1–8. doi: 10.1016/j.fishres.2006.03.008
Bonte, D., and Maelfait, L. (2004). Lack of homeward orientation and increased mobility result in high emigration rates from low-quality fragments in a dune wolf spider. J. Animal Ecol. 73, 643–650. doi: 10.1111/j.0021-8790.2004.00838.x
Chen, H. J., Liu, M. D., Wang, D. Q., Dong, W. W., Lv, H., Chen, D. Q., et al. (2018). Analysis of genetic diversity for 4 Hypophthalmichthys molitrix populations from middle and upper Yangtze River. Freshwater Fish. 48, 20–25.
Cheng, L., Liu, L., Yu, X., and Tong, J. (2008). Sixteen polymorphic microsatellites in bighead carp (Aristichthys nobilis) and cross-amplification in silver carp (Hypophthalmichthys molitrix). Mol. Ecol. Resources 8, 656–658. doi: 10.1111/j.1471-8286.2007.02037.x
Conover, G., Simmons, R., and Whalen, N. (2007). Management and Control Plan for Bighead, Black, Grass, and Silver Carps in the United States. Washington, D.C: Aquatic Nuisance Species Task Force, 29–30.
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resources 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fang, D. A., Luo, Y. T., Xu, D. P., Yang, X. W., and Wang, X. H. (2021). Relationship between genetic risk and stock enhancement of the silver carp (Hypophthalmichthys molitrix) in the Yangtze River. Fisheries Res. 235, 1–9.
Feng, X. T., Zhang, G. N., Xue, X. P., Wang, X. Y., Zhou, Y. F., Fang, D. A., et al. (2020). Current germplasm situation of bighead carp (Aristichthys nobilis) candidate parent and parent from hatchery in the lower reaches of Changjiang River based on SSR markers. J. Fishery Sci. China 27, 589–597.
He, X. H., Tan, L. F., Peng, Y. X., Fang, D.-A., and Xu, D. P. (2021). Four major Chinese carps eggs resources and spawning grounds distribution at Hukou section of the Yangtze River. J. Fishery Sci. China 28, 420–430.
Hu, Y. T., Jiang, H., Duan, G. Q., Zhou, H. X., Ling, J., and Wang, H. (2020). Genetic diversity analysis of Pelteobagrusfulvidraco from two major drainage systems in Anhui Province based on microsatellite markers. South China Fisheries Sci. 16, 33–41.
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Kolar, C. S., and Lodge, D. M. (2002). Ecological predictions and risk assessment for alien fishes in North America. Science 298, 1233–1236. doi: 10.1126/science.1075753
Li, H. B., and Xu, D. P. (2008). Analysis on the change characteristics and causes of “four major Chinese carp” resources in Dongting Lake. J. Inland Fisheries 33, 34–36.
Li, P. F., Liu, P., Liu, X. Z., Gao, T., and Wang, Q. Y. (2006). Isozyme polymorphism in paralichthys, Paralichthys lethostigma. Fishery Sci. China 13, 13–19.
Li, S. F. (1996). Germplasm Resources and Protection of Freshwater Fish in China. Beijing: Freshwater fish germplasm resources and conservation in China.
Li, S. Z., and Fang, F. (1990). On the geographical distribution of the four kinds of pond-cultured carps in China. Acta ZoologicaSinica 36, 244–250.
Lindenmayer, D., Hobbs, R. J., Montague-Drake, R., and Alexandra, J. (2008). A checklist for ecological management of landscapes for conservation. Ecol. Lett. 11, 78–91. doi: 10.1111/j.1461-0248.2007.01114.x
Liu, S. P., Duan, X. B., Chen, D. Q., and Liao, M. J. (2005). Studies on status of fishery resources in the Yangtze River. Acta Hydro Biol. Sinica 29, 708–711.
Liu, S. P., Qiu, S. L., Chen, D. Q., and Huang, M. J. (1997). Protection andrational utilization of the germplasm resources of the four majorChinese carps in the Yangtze River system. Resources Environ. Yangtze 6, 127–131.
Liu, Y., Yang, X. W., Ren, P., Li, X. F., Fang, D. A., and Xu, D. P. (2019). Community characteristics of larvae and juvenile fish inHukou section of the Yangtze river in spring and summer. Acta Hydrobiol. Sinica 43, 142–154.
Lu, G. Q., Wang, C. H., Zhao, J. L., Liao, X. L., Wang, J., Luo, M. K., et al. (2020). Evolution and genetics of bighead and silver carps: native population conservation versus invasive species control. Evol. Appl. 13, 1351–1362. doi: 10.1111/eva.12982
Luo, Y. T., Dai, P., Liu, S. L., Ma, F. J., You, Y., and Liu, K. (2021). Age structure and growth characteristics of coilia nasus in anqing section of yangtze river during fishing season in 2018. J. Guangdong Ocean University 41, 1–8.
Mai, L. F. (2003). The fishery resources of the Yangtze River have declined seriously. South China Fisheries Sci. 5:4.
Mao, R. X., Zhang, Y. B., Zheng, W., Du, X. Y., and Sun, X. W. (2010). The progress of germplasm resources of four major chinese farmed carps. Chinese J. Fish. 23, 52–59.
Matter, W. J., Mannan, R. W., Bianchi, E. W., Mcmahon, T. E., Menke, J. H., and Tash, J. C. (1989). A laboratory approach for studying emigration. Ecology 70, 1543–1546. doi: 10.2307/1938212
McMahon, T. E., and Matter, W. J. (2006). Linking habitat selection, emigration and population dynamics of freshwater fishes: a synthesis of ideas and approaches. Ecol. Freshwater Fish 15, 200–210.
Moss, D. R., Arce, S. M., Otoshi, C. A., Doyle, R. W., and Moss, S. M. (2007). Effects of inbreeding on survival and growth of Pacific white shrimp Penaeus (Litopenaeus vannamei. Aquaculture 272, S30–S37.
Nelson, A. R., Johnson, C. L., Matter, W. J., and Mannan, R. W. (2002). Tests of emigration in small mammals under experimental conditions. Canadian J. Zool. 80, 2056–2060.
Olds, A. D., Connolly, R. M., Pitt, K. A., and Maxwell, P. S. (2012). Habitat connectivity improves reserve performance. Conservation Lett. 5, 56–63. doi: 10.1111/j.1755-263x.2011.00204.x
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1093/bioinformatics/bts460
Pinter, K. (1980). Exotic fishes in hungarian waters: their importance in fishery utilization of natural water bodies and fish farming. Aquaculture Res. 11, 163–167. doi: 10.1111/j.1365-2109.1980.tb00401.x
Ren, J., Peng, Q. D., Lin, J. Q., and Dong, G. D. (2016). An instantaneous point-source diffusion model for pelagic eggs for estimating early life stage fish resources. J. Hydrometeorol. 37, 82–87.
Ru, H. J., and Liu, X. Q. (2013). River-lake migration of fishes in the Dongting Lake area of the Yangtze floodplain. J. Appl. Ichthyol. 29, 594–601. doi: 10.1111/jai.12116
Sale, P. F. (1968). An experimental study of habitat selection in the juvenile manini, Acanthurus triostegus sandvicensis. Ecology 54, 616–623.
Sha, H., Luo, X. Z., Li, Z., Zhou, G. W., and Liang, H. W. (2018). Genetic diversity of six silver carp (Hypophthalmichthys molitrix) geographical populations based on mitochondrial CO? sequences. J. Fishery Sci. China 25, 783–792. doi: 10.3724/sp.j.1118.2018.18058
Sha, H., Luo, X. Z., Wang, D. Q., Li, X. H., Zou, G. W., and Liang, H. W. (2021). New insights to protection and utilization of silver carp (Hypophthalmichthys molitrix) in Yangtze River based on microsatellite analysis. Fish. Res. 241, 1–7.
Shaklee, J. B., Tamaru, C. S., and Waples, R. S. (1982). Speciation and evolution of marine fishes studied by the electrophoretic analysis of proteins. Pacific J. Optimization 36, 141–157.
Smouse, P. E., and Peakall, R. (1999). Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82, 561–573. doi: 10.1038/sj.hdy.6885180
Sun, L. Y., Guo, H. Y., Zhu, C. Y., Ma, Z. H., Jiang, S. G., and Zhang, D. C. (2014). Genetic polymorphism of breeding populations of goldenpompano (Trachinotusovatus). South China Fisheries Sci. 10, 67–71.
Sun, X. W., Zhang, X. F., Zhao, Y. Y., Zhang, Y., Jia, Z. Y., Chang, Y. M., et al. (2008). Development and application of microsatellite markers in aquatic species. J. Fishery Sci. China 15, 689–703.
Swift, M. L. (1997). GraphPad prism, data analysis, and scientific graphing. J. Chem. Inform. Model. 37, 411–412. doi: 10.1021/ci960402j
Takezaki, N., and Nei, M. (1996). Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144, 389–399. doi: 10.1093/genetics/144.1.389
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tan, Z. J., Zhang, T. Q., Lu, C. Y., Li, C., and Sun, X. W. (2011). Development and characteristics of tri- and tetra-nucleotide microsatellites for silver carp (Hypophthalmichthysmolitrix). J. Shanghai Ocean University 20, 328–335.
Tang, X. L., Chen, D. Q., Wang, K., Liu, S. P., Luo, H. W., and Duan, X. B. (2010). Spatial and temporal distribution of larval resources of fishes in the upper reaches of Yangtze River. Freshwater Aquaculture 40, 27–31.
Tian, J. L., Dai, P., Ren, P., Guo, W. J., Zhu, Z. B., Li, X. F., et al. (2020). Assemblage of larvae and juvenile fish in AnqingXinzhou section of the Yangtze River. J. Fishery Sci. China 27, 916–926.
Wang, C. Z., Liang, H. W., Zhou, G. W., Luo, X. Z., Li, Z., Tian, H., et al. (2008). Genetic variation analysis of two silver carp populations in the middle and upper Yangtze River by microsatellite. Hereditas(Beijing) 30, 1341–1348. doi: 10.3724/sp.j.1005.2008.01341
Wang, C., Yu, X., and Tong, J. G. (2007). Microsatellite diversity and population genetic structure of redfin culter (Culter erythropterus) in fragmented lakes of the Yangtze River. Hydrobiologia 586, 321–329. doi: 10.1007/s10750-007-0702-x
Wang, S., Duan, X. B., Chen, W. J., Zhang, H. X., Fu, P. F., He, G., et al. (2016). Status and changes of fish resources in the Hukou area of Poyang Lake. Freshwater Fisheries 46, 50–55.
Weight, S. (1978). Evolution and the Genetics of Population Variability Within and Among Natural Populations. Chicago, IL: University of Chicago Press.
Xu, C. S., Ai, Z. Q., and Xiao, M. (2017). A review of influencing factors on natural reproduction of four major Chinese carps in Yangtze River. J. China Three Gorges University (Natural Sciences) 39, 27–31.
Xuan, Z. Y., Jiang, T., Liu, H. B., Chen, X., and Yang, J. (2021). Mitochondrial DNA and microsatellite analyses reveal strong genetic differentiation between two types of estuarine tapertail anchovies (Coilia) in Yangtze River Basin, China. Hydrobiologia 848, 1409–1431. doi: 10.1007/s10750-021-04541-w
Yu, Y., Pang, M. X., Yu, X. M., Tong, J. G., and Shen, J. Z. (2016). Population genetic structure of silver carp from Yangtze River, Ganjiang River and Poyang Lake based on microsatellite markers. J. Huazhong Agricultural University 35, 104–110.
Yuan, J. G., Li, J. Q., Liu, L., Chen, Z. X., and Zhang, Y. P. (2017). Genetic diversity of GIFT strains of oreochromis niloticus based on 18S-ITS1-5.8S sequence. J. Guangdong Ocean University 37, 7–10.
Keywords: silver carp, the Yangtze River, habitat selection, SSR, stock enhancement
Citation: Luo YT, Fang DA, Zhou YF, Xu DP, Peng YX, Zhang MY, Mao CC, Tang XM, Xu J and You Y (2022) Genetic Diversity, Habitat Relevance and Conservation Strategies of the Silver Carp in the Yangtze River by Simple Sequence Repeat. Front. Ecol. Evol. 10:850183. doi: 10.3389/fevo.2022.850183
Received: 07 January 2022; Accepted: 22 March 2022;
Published: 26 May 2022.
Edited by:
Peng Xu, Xiamen University, ChinaReviewed by:
Hong Wei Liang, Chinese Academy of Fishery Sciences (CAFS), ChinaJian Xu, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2022 Luo, Fang, Zhou, Xu, Peng, Zhang, Mao, Tang, Xu and You. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Feng Zhou, emhvdXlmQGZmcmMuY24=
†These authors have contributed equally to this work
 Yu-Ting Luo1†
Yu-Ting Luo1† Di-An Fang
Di-An Fang Dong-Po Xu
Dong-Po Xu Jun Xu
Jun Xu