
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 04 April 2022
Sec. Ecophysiology
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.848150
This article is part of the Research Topic Nutritional Ecology of Wild and Managed Bees View all 8 articles
Bees rely on their sensitive olfactory system to perform foraging activities in the surrounding environment. This ability is associated with the existence of olfactory receptors (ORs). In this study, we identified the AcerOr2 (ortholog to the Orco) protein in Apis cerana cerana, which contains a conserved, putative calmodulin (CaM)-binding site (CBS) indicating that CaM is involved in its function. We used immunofluorescence, Western blot, and Ca2 + imaging to monitor changes in the expression and activation of the signaling pathway associated with Ca2 + and Ca2 +/CaM-dependent protein kinase II (CaMKII) in Sf9 cells heterologously expressing AcerOr2 and a CaM-binding mutant. We used the synthetic Orco agonist VUAA1 to stimulate the cells or the antagonist W7 to inhibit CaM activity. The AcerOr2 CaM-binding mutant has a point mutation in the putative CBS (K331N). When heterologously expressed in Sf9 cells, the mutant should have less CaM activity. When the cells expressing AcerOr2 were treated with W7, the Ca2 + response of AceOr2 was similar to that of the mutant stimulated by VUAA1, and the expression of the CaM, CaMKII, and p-CaMKII has similar effects. Our results suggest that CaM activity affects the function of AceOr2 in vitro and can be used to further study the interaction between the AcerOr2 and calcium/CaM signaling pathway in the pollen collection behavior of bees.
Bees are among the most important economic insects and pollinators in nature. They can help pollinate plants and improve the yield and quality of crops. Bee pollination plays an indispensable role in protecting species diversity and ecological environment. Bees rely on their sensitive olfactory system to perform foraging activities in the surrounding environment. Olfactory receptors (ORs) are a primary requirement for odorant recognition and coding. In insects, volatile substances in the surrounding environment are recognized by ORs expressed in olfactory sensory neurons (OSNs) of the two specialized head olfactory appendages, namely, the maxillary palps and the antennae (Vosshall et al., 1999; de Bruyne et al., 2001). The matching of odors to their cognate ORs in the OSNs activates signal transduction cascades that control the quality, intensity, and temporal pattern of odorant-related action potentials. Studies in insects have demonstrated that ORs form ligand-gated, non-selective cation channels whose activation is the initial step in transduction, and lead to Ca2 + influx into the cell. In heterologous systems, this cascade promotes an increase in intracellular Ca2 + concentration (Sato et al., 2008; Wicher et al., 2008). In olfactory transduction, odorant-specific ORs and odorant receptor co-receptor (Orco) assemble into heterodimers or Orco homomers that contribute to activation of intracellular signaling (Nakagawa et al., 2012; Getahun et al., 2013). Orco can support OrX proteins located to the dendritic as a chaperone (Larsson et al., 2004). In total, 180 ORs in Apis florae (Snehal et al., 2016), 177 ORs in Apis mellifera (Robertson and Wanner, 2006; Wanner et al., 2007), and 119 ORs in Apis cerana cerana (Park et al., 2015) have been identified. However, there are only a few studies on their function. AmOr11 specifically binds to the queen pheromone 9-oxo-decenoic acid (9-ODA) (Wanner et al., 2007), and AmOr151 specifically binds to the floral odorant component linalool (Reinhard and Claudianos, 2012), indicating that the OR-binding odorant in honeybees is associated with scent detection.
Activation of Orco assembly plays an important role in forming the ion channels, which in turn modulate OR sensitivity to odorants but is not direct binding odorant and identified by a chemical compound VUAA1 (Chen and Luetje, 2012; Taylor et al., 2012). The sequences of amino acids are important for the gate control and cation selectivity of the Orco channel (Kumar et al., 2013). A short, conserved predicted amino acid sequence of the calmodulin (CaM) binding sequence was found in the conserved regions of Orco, indicating a potential CaM-dependent function (Yap et al., 2000). CaM is a highly conserved, 150 amino acid intracellular Ca2 + sensor that is ubiquitously expressed in all eukaryotic cells (Finn and Forsén, 1995). It consists of four EF-hand calcium-binding motifs that allow it to undergo conformational changes on binding Ca2 +. These conformational changes allow CaM to act as a Ca2 + sensor and affect the interactions with the protein it targets, thus modulating the mechanism of their biological functions (Hoeflich and Ikura, 2002). CaM-dependent protein kinase II (CaMKII) is an example of a downstream target protein of CaM.
The putative CaM binding site (CBS) in Orco proteins, combined with odorant-evoked olfactory neuronal activity, promotes Ca2 + entry into cells and increases levels of intracellular Ca2 + (Wang et al., 2003), which suggests that Ca2 + affects olfactory transduction via multiple pathways, some of which require the CaM/CaMKII signaling pathway. Therefore, odorant receptor response termination through CaM-mediated desensitization of the olfactory cyclic nucleotide-gated (CNG) channels and downregulation of intracellular free Ca2 + concentrations not only restores the ability of the olfactory neurons to perceive further stimuli but also protects the cell from Ca2 + overload that may harm it or even lead to cell death (Song et al., 2008). CaM is one of the key proteins connecting intercellular Ca2 + concentration to the control of Ca2 + influx or efflux (Song et al., 2008; Antolin et al., 2010; Faas et al., 2011). As insect OR activation is accompanied by Ca2 + influx into sensory cells, there is an obvious question as to whether CaM plays a role in regulating the function of insect ORs. In this study, to specify the role of CaM activity on the AcerOr2 (ortholog to the Orco) in vitro, we measured the changes in the Ca2 + levels and in the protein expression and activation of signaling pathways associated with Ca2 +/CaMKII in AcerOr2 and a CaM-binding mutant (on the putative CBS of AcerOr2) heterologously expressed in Sf9 cells that were treated with the synthetic Orco agonist VUAA1 in combination with the CaM antagonist W7.
The instandard strong honeybee colonies provided the 180 adult bees (Apis cerana cerana) for the experiment. The antennae of the bees were removed and frozen in liquid nitrogen and stored at −80°C. The total RNA was isolated from the antennae using the TRIzol reagent (Takara, Dalian, China). Notably, 10 μg of total RNA was used for the synthesis of first-strand cDNA using the PrimeScript RT Reagent Kit (TaKaRa). The amplification was carried out using a real-time PCR system (7500 Real-Time PCR System; ABI, Foster City, CA, United States) with the SYBR® Green Master Mix (ABI, Foster City, CA, United States).
The complete open reading frame (ORF) of AcerOr2 from honeybee antenna cDNA was PCR-amplified using gene-specific primers (F:5′-ATGAGCGTAAAAACAAGAAACATAA-3′, R:5′- TGGTGTTGGTGCAACTGAAGTGA-3′) with restriction sites for BamHI and EcoRI (NEB, Beverly, MA, United States) and cloned into a TA-cloning vector (Invitrogen, Carlsbad, CA, United States). The primer pair containing AcerOr2 cDNA of the mutant site was as follows: MUT F: 5′-CAA CGGTGGTGTTGGGCCAAATGGGTTAACCAAGAAACAGG AAATGC-3′ R:5′-ACGATATGCTTGTGCCTTCGTACCAGCAT TTCCTGTTTCT-3′. Each 20 μl reaction mixture consisted of 2 μl of cDNA, 10 μl of SYBR® Premix Ex Taq II TM (2 ×), 0.4 μl of ROX Reference Dye II (50 ×), 0.8 μl of each of the forward and reverse primers (10 μM), and 6.0 μl of ddH2O. The reaction conditions were as follows: pre-denaturation at 95°C for 10 min, followed by 1 cycle at 95°C for 30 s, and 40 cycles at 95°C for 5 s and 65°C for 34 s. The 2–ΔΔCt method was used to calculate the relative mRNA level of AcerOr2. All reactions were performed in triplicate.
The original cloning insert was subcloned into the pIB-V5-His expression vector to ensure a lack of nucleotide substitutions or other mutations that could have occurred in the secondary PCR amplifications. Spodoptera frugiperda Sf9 cells were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). The cells were grown in T-25 tissue culture flasks (Corning Inc., NY, United States) containing SF900 III-SFM insect culture medium (Gibco, Invitrogen, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum (Sijiqing, Hangzhou, China), 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator (Thermo Fisher Scientific, Cornelius, OR, United States) at 28°C in the absence of CO2. The AcerOr2 and Orco K331N CaM-binding mutant constructs were transfected at a 0.3–0.5 μg/well-concentration using the Cellfectin II® Reagent Kit (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions.
The AcerOr2 CBS K331N mutation was generated with site-directed mutagenesis. Two overlapping mutagenic oligonucleotides were designed to introduce a point mutation at K331 to change it to an asparagine (N) residue. The PCR products were then used to run a full-length PCR using AcerOr2 (full length) primers, and the final product was TA cloned into the pGEM®-T easy vector (Invitrogen Life Technologies). The mutated cDNA was then subcloned into the BamHI and EcoRI sites of pIB/V5-His expression vectors. We also subcloned AcerOr2 cDNA into the same vector as a control. The sequences were confirmed by double-strand DNA sequencing (Eurofins MWG operon). The reaction condition for cDNA synthesis and PCR is the same as in the section “Bee Preparation and AcerOr2 Expression in Cultured Sf9 Cells.”
The expression and function of AcerOr2 and AcerOr2 CaM mutant constructs in Sf9 cells were measured with Ca2 + imaging. Approximately 48 h after transfection with a plasmid containing the ORF of the OR gene, the medium was removed and the cells were washed three times with Hank’s Balanced Salt Solution (HBSS without Ca2 +). Fluo4-AM (2 μmol/L, Beyotime, Shanghai, China) was added to cells, which were then cultured at 37°C in the dark for 30 min before being stimulated by chemical odorants. Each test chemical ligand was applied at a final concentration of 1 μM to Fluo4-AM-loaded Sf9 cells expressing AcerOr2 or the AcerOr2 CaM mutant. Fluorescence was measured using an excitation wavelength of 494 nm and an emission wavelength of 516 nm. The results were recorded using a Synergy H1 microplate reader (BioTek, Winooski, VT, United States). To calculate the free intracellular Ca2 + concentration, we used the following equation: , where the minimal and maximal fluorescence ratio values (Fmax and Fmin) indicate the fluorescence under Ca2+-saturating conditions with 0.1% Triton X-100 and Ca2+-free conditions with 5 mM EGTA, respectively, and Kd is the fluo-4/Ca2+ binding at 37°C, which was 360 nM. The cells were stimulated using VUAA1 after incubation in the presence of W7 for 50 s. The experiment was repeated six times.
VUAA1 (N-(4-ethylphenyl)-2-((4-ethyl-5-(3-pyridinyl)-4H-1,2, 4-triazol-3-yl) thio) acetamide) and W7 (N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide, HCl) were purchased from Sigma-Aldrich (St. Louis, MO, United States). Stock solutions of W7 and VUAA1 were prepared at a final concentration of 10 mM in dimethylsulfoxide (DMSO) and were stored at −20°C.
For immunofluorescence measurements, the cells transfected with constructs expressing AcerOr2, and its variants were grown on poly-L-lysine-coated coverslips placed in six-well plates. The medium was removed from the wells containing transformed and untransformed cells, the cells were then washed with phosphate-buffered saline (PBS), and 1 ml of 4% paraformaldehyde (PFA) was added to the wells. The cells were incubated for 30 min at room temperature (27°C), after which PFA was removed and the cells were washed with PBS. The cells were then treated with 5% bovine serum albumin (BSA) for 1 h at room temperature to block non-specific binding. The primary antibody (1:2,000) prepared in 1% BSA was then added. The cells were incubated at 4°C overnight and then washed with PBS. Subsequently, a secondary antibody (goat anti-rabbit Alexa Fluor® 594; 1:10,000) prepared in 1% BSA was added to the cell preparations and incubated for 2 h at room temperature. The cells were again washed, and the coverslips with stained cells were removed for analysis by immunofluorescence microscopy. The images were analyzed using Image J software (National Institutes of Health, Bethesda, MD, United States).
The Sf9 cells expressing AcerOr2 and its variants were plated (6 × 105 cells/well) in a 6-well plate, grown for 24 h, and induced with non-permeable tetracycline (0.1 mg/ml for 24 h; proteins at the cell surface were labeled with EZLink-Sulfo-NHS-SS-Biotin (ThermoScience). Aliquots (100 μl) of cells were placed in pressure-equalization tubes and incubated at 28°C for 30 min with 100 μM VUAA1 and W7 in DMSO. Then, the reagent was removed, and the cells were lysed in RIPA buffer (25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EGTA, 1% Nonidet P-40, 12 mM deoxycholic acid, and 1 mM PMSF). The cells were then centrifuged at 5,000 × g for 15 min at 4°C. The supernatant was collected and incubated with avidin-conjugated agarose beads (Sigma) for 2 h at 4°C. The cell surface proteins captured by avidin-coupled beads were analyzed using Western blot.
For Western blotting, the protein was extracted from the harvested transfected cells using RIPA buffer. The total protein was quantified using the BCA Protein Assay Kit (Boster, Wuhan, China) according to the manufacturer’s instructions. The extracted proteins (100 μg per sample) were separated using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto a nitrocellulose filter membrane (Boster). The membranes were blocked for 1.5 h at room temperature in 5% skimmed milk (Boster) and then incubated overnight at 4°C with rabbit polyclonal anti-AcerOr2, rabbit anti-CaMKII, rabbit anti-p-CaMKII (1:1,000 v/v) (BioWorld, United States), and mouse anti-β-actin (1:500 v/v; Boster) antibodies. Subsequently, the membranes were incubated with donkey anti-rabbit IgG (1:5,000 v/v; Boster) and goat anti-mouse IgG (1:2,000 v/v; Boster) secondary antibodies for 2 h at room temperature. Bands were detected using the Super ECL Plus detection reagent (Boster) and analyzed using ImageJ version 1.49.
The transmembrane (TM) domains were predicted using TTHMM server version 2.0, and CaM motif prediction was performed using the CaM target database1. The Orco sequences from various insect species were aligned using the MUSCLE alignment tool (Genesis, Auckland, New Zealand). Statistical analysis was performed using Prism 6 software (GraphPad Software, Inc., La Jolla, CA, United States). Data were analyzed using one-way ANOVA and t-test using SPSS version 17.0 software.
A conserved CaM binding domain residue in TM7 of AcerOr2 is important for channel activity. Insect OR proteins have six to seven TM domains and an intracellular terminus (Lin et al., 2015). According to membrane topology predictions, AcerOr2 belongs to the TM7 class of OR proteins (Zhao et al., 2013, 2014). The role of CaM-regulated insect ORs depends on the regulation of Orco function; therefore, we first conducted a screen to identify a putative CaM binding domain in the primary amino acid sequence of the Apis cerana cerana Orco protein, AcerOr2. We identified a candidate amino acid motif 328SAIKYWVER336 within the second intracellular loop (ICL2) of the AcerOr2 protein (Figure 1A). This motif is highly conserved in Orco proteins from other insect species. In Drosophila melanogaster, the DmelOrco putative CaM binding domain is 336SAIKYWVER344 (Jain et al., 2021), and in Aedes albopictus, AalOrco is 329SAIKYWVER337 (Liu et al., 2016), indicating that CaM activity in ICL2 plays a crucial functional role in Orco channels (Mukunda et al., 2014). Orco CBS mutations including the single tryptophan deletion mutant (K341) can disrupt OR trafficking (Bahk and Jones, 2016), and the expression of an Orco mutant bearing a point mutation in the putative CaM site (K339N) results in much weaker responses to Ca2+ than those observed for wtOrco (Mukunda et al., 2014). To study CaM-dependent signaling in a heterologous system, we expressed AcerOr2 and modified the CaM-binding motif of the AcerOr2 mutant with a substitution at the K331N site in the putative CaM domain in Sf9 cells. The computationally designed mutation in the AcerOr2 CaM-binding domain stabilized the inactivated form of the Ca2+-binding domain in its “closed” Ca2+-free conformation (Figure 1B).
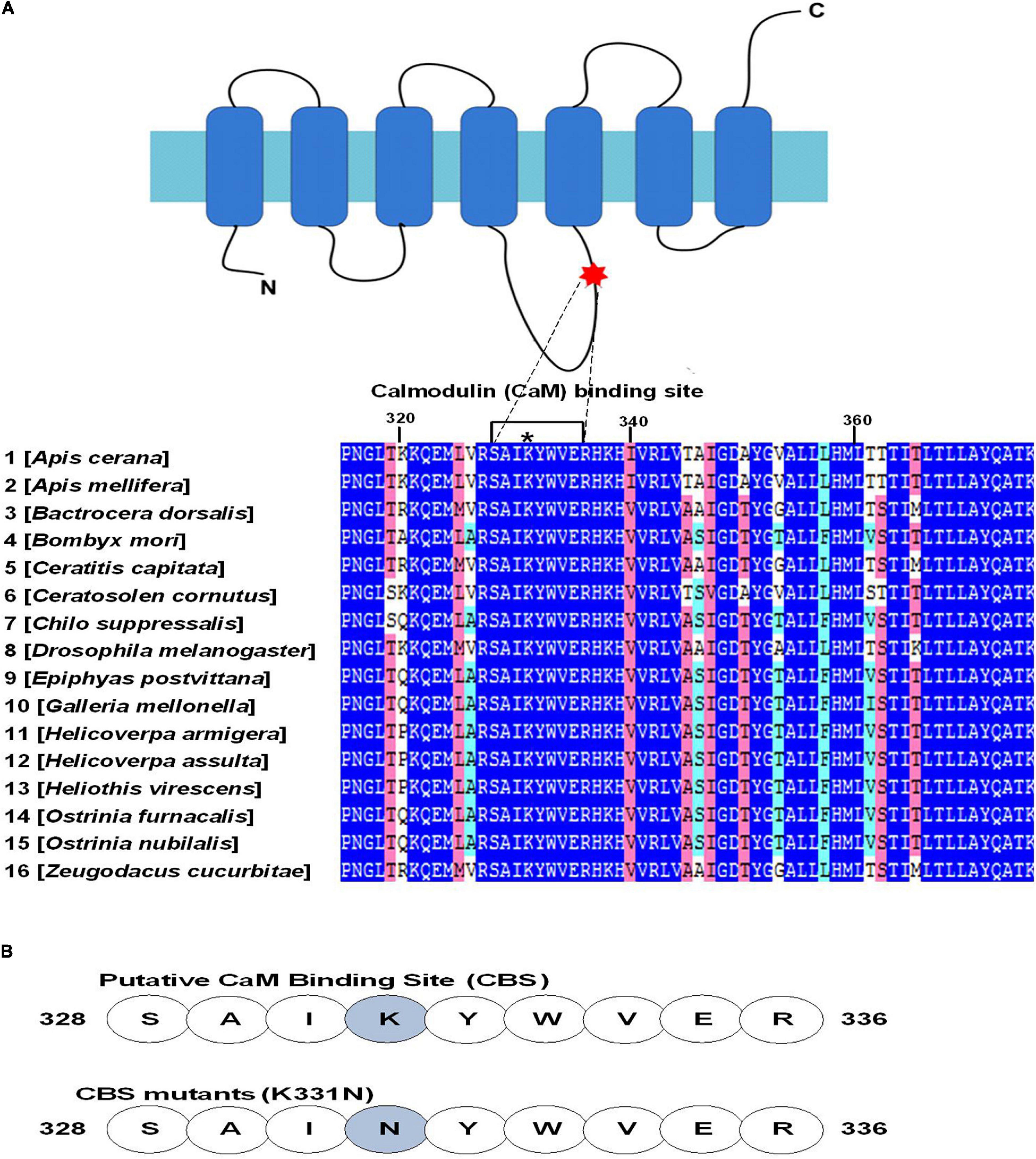
Figure 1. Conservation of putative calmodulin (CaM) binding sites (CBSs) in the insect odorant co-receptor Orco. (A) Alignment of amino acids at the indicated position in the second intracellular loop of AcerOr2. The amino acids 328SAIKYWVER336 have a maximum score for the CBS and are highly conserved among various insect species. The asterisk indicates the position of the point mutation (K331N) introduced into the AcerOr2 CaM protein. The NCBI accession number for the Orco protein sequences are as follows: Apis cerana AET85154.3; Bactrocera dorsalis ACC86853.1; Bombyx mori NP_001037060.1; Ceratitis capitata NP_001266301.1; Ceratosolen cornutus ACU31808.1; Chilo suppressalis AFQ94048.1; Drosophila melanogaster AAT71306.1; Epiphyas postvittana ACJ12928.1; Galleria mellonella ALM30348.1; Helicoverpa armigera ADQ13177.1; Helicoverpa assulta ABU45983.2; Heliothis virescens CAD31851.1; Ostrinia furnacalis BAR43445.1; Ostrinia nubilalis BAJ23263.1; and Zeugodacus cucurbitae ADK97803.1. (B) Sequences of the AcerOr2 CBS and the location of the point mutation (K331N) introduced in the AcerOr2 CaM-binding mutant.
To study the effect of CBS mutant (K331N) on the protein expression of AcerOr2, samples of total protein and the cell-surface protein were analyzed using Western blotting with anti-AcerOr2. AcerOr2 and its variant transfected cells presented a 56 kDa fragment band for both total protein and cell membrane protein. No obvious bands were observed in the pIB/V5-His transfection group. The level of the 56 kDa band was reduced in its K331N variant, compared with the AceOr2 in the total protein. The 56 kDa band was also decreased in the biotinylated cell surface protein of variant compared with both AcerOr2 (Figure 2A). These results indicate that a mutation in the CBS affects and leads to reduced expression of AcerOr2 protein at the surface membrane, which determines AcerOr2 function as a channel. Analysis of the subcellular localization of AcerOr2 has shown that the K331N mutant has a weaker localization at the cell surface compared with AcerOr2 expressed cells (Figure 2B). The lower expression of AcerOr2 in the CaM-binding mutant group may indicate that the mutation in the AcerOr2 CBS impaired the localization of AcerOr2 in the plasma membrane. This would be consistent with the results from in vivo experiments showing disruption of Orco localization to the outer ciliated dendrites of OSNs for knock-down CaM mutants of Orco CBS with mutations at the C-terminus (CaM C), N-terminus (CaM N), or both (CaM CN) of the EF-hand domains (Bahk and Jones, 2016).
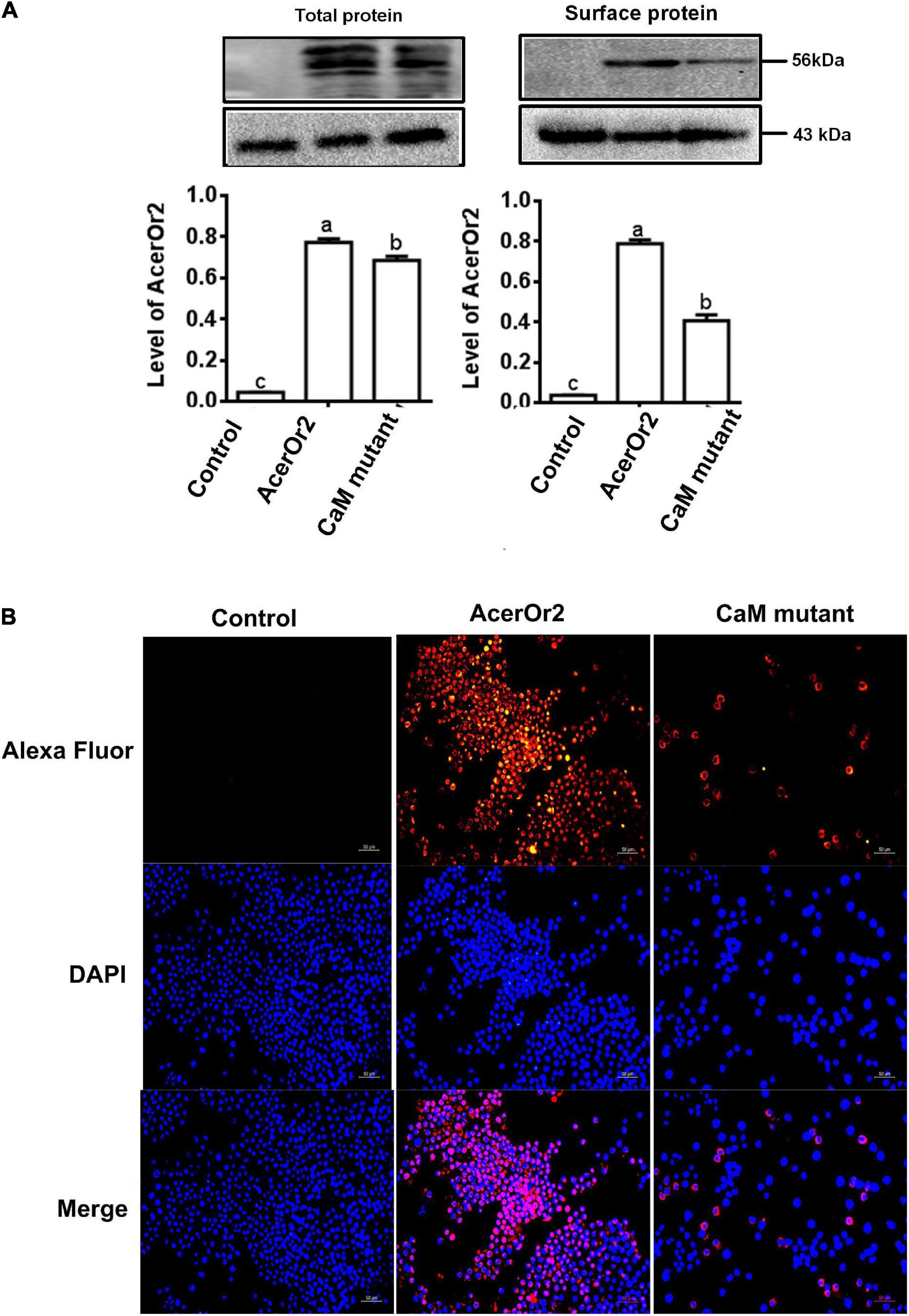
Figure 2. Effects of K331N mutant transporters in AcerOr2 protein expression, cellular distribution. (A) Comparison of the expression and cellular distribution of AcerOr2 and its mutants expressed in Sf9 cells. Samples of total protein and the cell-surface protein were analyzed by Western blotting by anti-AcerOr2, the molecular weights of AcerOr2 and β-actin are 56 and 43 kDa, and expression of AcerOr2 relative to that of β-actin. Bars represent the mean ± SEM from three independent experiments, and data were analyzed using one-way ANOVA. Different letters indicate the significant difference among groups (P < 0.05). (B) Immunofluorescence and membrane staining of AcerOr2 and AcerOr2 CaM (K331N) mutants expressed in Sf9 cells. Scale bar = 50 μm.
When intracellular Ca2+ levels are low, CaM is mainly in its inactive state, which is derived from its closed molecular structure. As Ca2+ levels increase, CaM binds Ca2+ to form a complex that causes the molecular structure to rearrange, thereby changing the state of CaM from inactive to active and allowing it to function as a switch that regulates cell activity. Ca2+ has long been known to be involved in the regulation of olfactory transduction, and CaM can bind AcerOr2. Since insect olfactory signal transduction was coupled with Ca2+, it had been reported that the influence of conserved CBS residues for channel function in AcerOr2 was researched by a single-site mutation and functional characterization using Ca2+ imaging assays. Substitution mutant of K331N was stably expressed in Sf9 cells and stimulated with VUAA1. The intracellular Ca2+ changes of AcerOr2 and its K331N substitution mutants following the addition of an agonist (100 μM VUAA1) in the absence/presence of 10 μM W7 are shown in Figure 3. Cells expressing the AcerOr2 CBS mutant are significantly less sensitive to agonist activation than AcerOr2 by 100 μM VUAA1 (Figure 3A). When the cells expressing AcerOr2 pretreated with the CaM inhibitor W7, the intracellular Ca2+ responses from cells expressing AcerOr2 became weaker than without treatment stimulated by VUAA1 (Figure 3B). While the cells expressing AcerOr2 CBS mutant (K331N) pretreated with the CaM inhibitor W7, there has been no difference in intracellular Ca2+ responses compared with cells expressing AcerOr2 CBS mutant (K331N) without pretreated with the CaM inhibitor W7 stimulated by VUAA1 (Figure 3C). Cells expressing the AcerOr2 and its mutant pretreated with the CaM inhibitor W7, there has been no difference stimulated by VUAA1 (Figure 3D). This may indicate that AcerOr2 agonist VUAA1 causes Ca2+ influx. Thus, the AcerOr2 protein acts as a Ca2+ permeable channel regulating the ligand response. The inhibitor W7 binds to CaM and expression CaM mutant in heterologously expressed Apis cerana cerana AcerOr2 show that inhibition of CaM activity can inhibit Ca2+ response of AcerOr2 ion channel. The results consist of previous studies in other insects (Mukunda et al., 2014, 2016; Bahk and Jones, 2016).
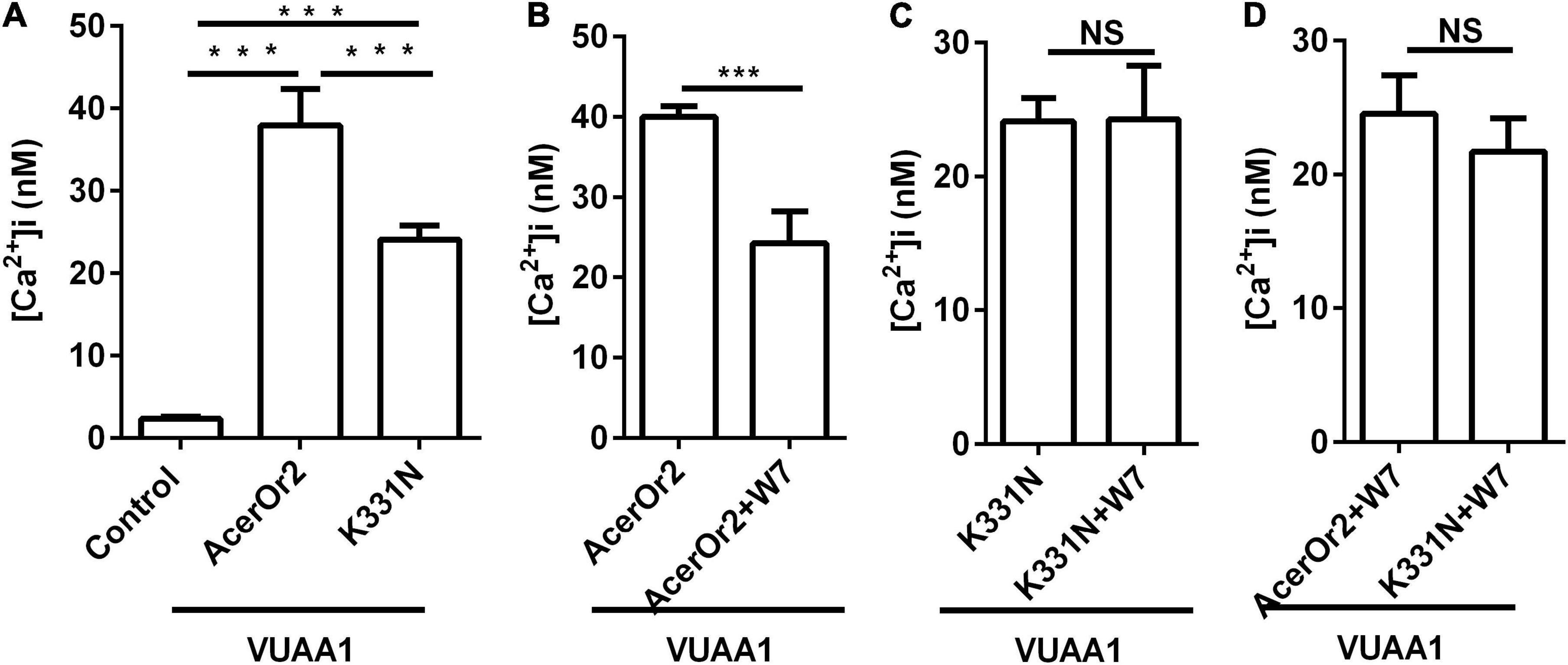
Figure 3. VUAA1-stimulated Ca2+ influx activity of AcerOr2 and its CaM-binding mutant (K331N) constructs expressed in Sf9 cells treated with the agonist VUAA1 in the absence/presence of 10 μM CaM inhibitor W7. (A) Average recording of [Ca2+]i in Sf9 cells expressing no AcerOr2 (empty vector), AcerOr2, or the CaM mutant stimulated with 100 μM VUAA1 in the absence of 10 μM W7. (B) Average recording of [Ca2 +]i in Sf9 cells expressing AcerOr2 stimulated with 100 μM VUAA1 in the absence or presence of 10 μM W7. (C) Average recording of [Ca2+]i in Sf9 cells expressing the AcerOr2 CaM mutant (K331N) stimulated with 100 μM VUAA1 in the absence or presence of 10 μM W7. (D) Average recording of [Ca2+]i in Sf9 cells expressing the AcerOr2 and its CaM mutant (K331N) stimulated with 100 μM VUAA1 in the presence of 10 μM W7. The data are expressed as the mean ± SEM (n = 15), and data were analyzed using one-way ANOVA (A) and t-test (B–D); ***P < 0.001. NS, not significant.
Ca2 +/CaM-dependent protein kinase II (CaMKII) is a downstream target of Ca2 +/CaM and is an important molecule that participates in insect learning and memory and is, therefore, referred to as a “memory switch” (Mizunami et al., 2014; Scholl et al., 2015). CaMKII is regulated by the Ca2 +/CaM complex, and when Ca2 + levels increase, CaMKII is activated by autophosphorylation. Therefore, intracellular CaMKII can be activated/deactivated through switching between phosphorylation and dephosphorylation states (Lisman et al., 2012; Coultrap and Bayer, 2012). Furthermore, in order to understand the details of the functional role of CaM AcerOr2 in regulating olfactory signaling, we then evaluated the expression of CaM, CaMKII, and p-CaKMII in Sf9 cells expressing AcerOr2 and the CaM-binding mutant by Western blot analysis in the absence or presence of 10 μM W7 stimulated by 100 μM VUAA1 (Figure 4). The expression of CaM, CaMKII, and p-CaMKI in cells expressing the AcerOr2 CBS mutant is significantly less than AcerOr2 stimulated by 100 μM VUAA1. When the cells expressing AcerOr2 pretreated with the CaM inhibitor W7, the expression of CaM, CaMKII, and p-CaMKI from cells expressing AcerOr2 became weaker than without treatment stimulated by VUAA1. Our results also provide indirect evidence for the role of the Ca2+/CaM/CaMKII pathway and the CaM functions in the regulation of AcerOr2 channel activity. Ca2 + channel change is a critical parameter that controls the function of Orco (Vardanush et al., 2011). Future studies of the characterized interaction between AcerOr2 CBS and CaM should be performed in studying the cellular and molecular mechanisms involved in honeybee olfactory signal transduction.
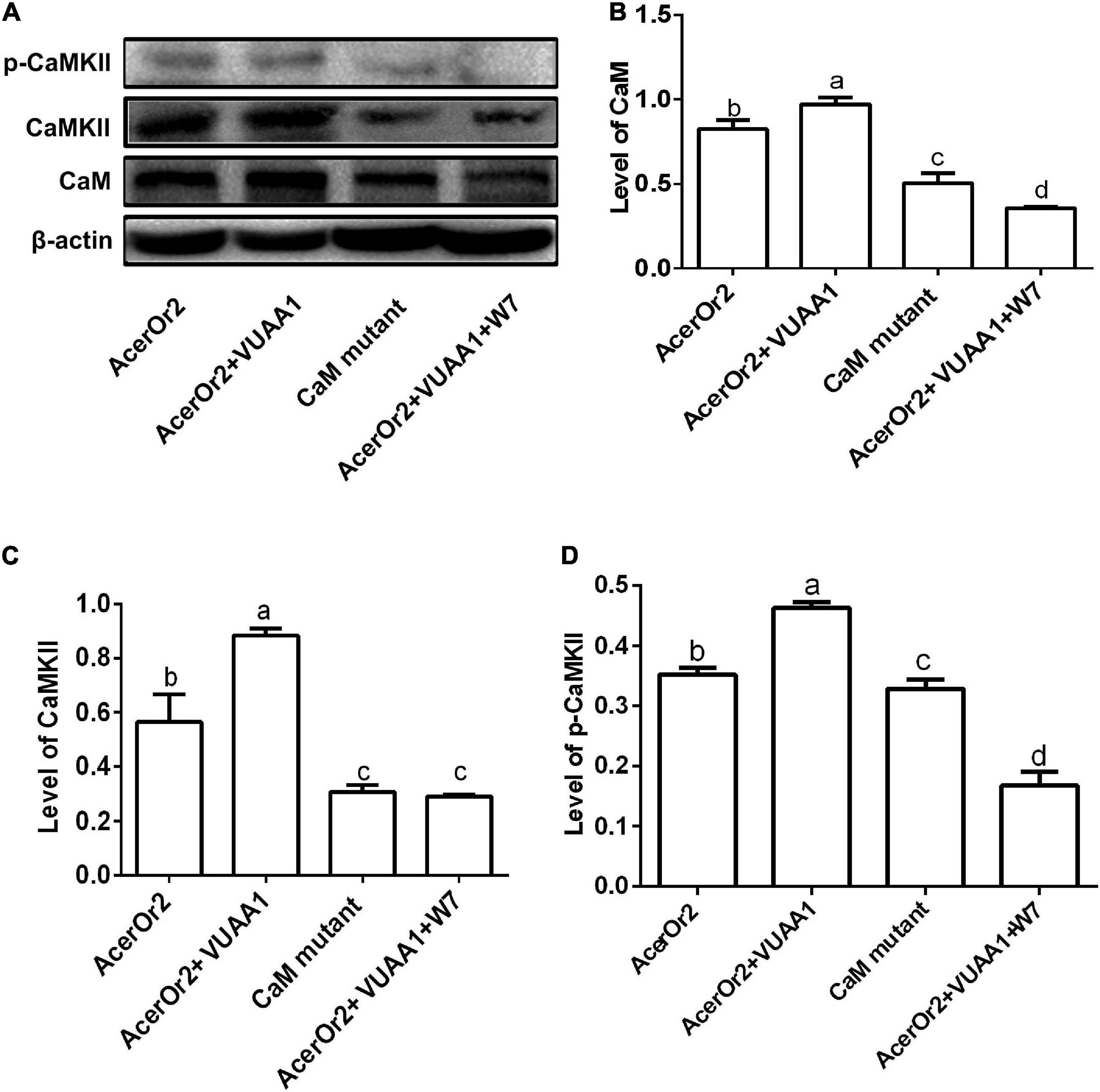
Figure 4. Effects of the VUAA1 agonist on CaM, CaMKII, and p-CaMKII protein expression in Sf9 cells transfected with the AcerOr2 and its CaM-binding mutant (K331N) constructs in the absence/presence of the CaM inhibitor W7, and the expression levels of CaM, CaMKII, and p-CaMKII in these cells were assessed by Western blot analysis at 12 h. (A) Western blot showing the expression of p-CaMKII, CaMKII, CaM, and β-actin. The molecular weight of p-CaMKII, CaMKII, CaM, and β-actin has molecular weights of 54, 72.7, or 56.3, 16.7, and 43 kDa, respectively. (B–D) Expression of CaM, CaMKII, and p-CaMKII relative to that of β-actin. Bars represent the mean ± SEM from 3 independent experiments, and data were analyzed using one-way ANOVA. Different letters indicate the significant difference among groups (P < 0.05).
This study provides insights into how CaM affects the function of AcerOr2. The results can be useful in the development of novel behavior modifiers for Apis cerana cerana, which are important for understanding the molecular mechanism of the pollen collection behavior of bees.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, AET85154.3; https://www.ncbi.nlm.nih.gov/, ACC86853.1; https://www.ncbi.nlm.nih.gov/, NP_001037060.1; https://www.ncbi.nlm.nih.gov/, NP_001266301.1; https://www.ncbi.nlm.nih.gov/, ACU31808.1; https://www.ncbi.nlm.nih.gov/, AFQ94048.1; https://www.ncbi.nlm.nih.gov/, AAT71306.1; https://www.ncbi.nlm.nih.gov/, ACJ12928.1; https://www.ncbi.nlm.nih.gov/, ALM30348.1; https://www.ncbi.nlm.nih.gov/, ADQ13177.1; https://www.ncbi.nlm.nih.gov/, ABU45983.2; https://www.ncbi.nlm.nih.gov/, CAD31851.1; https://www.ncbi.nlm.nih.gov/, NP_001037060.1; https://www.ncbi.nlm.nih.gov/, BAR43445.1; https://www.ncbi.nlm.nih.gov/, BAJ23263.1; and https://www.ncbi.nlm.nih.gov/, ADK97803.1.
LG: data curation, investigation, and writing original draft. HZ and BX: visualization and writing – review and editing. YG: contributed to reagents, materials, and analysis tools. YJ: supervision, visualization, conceptualization, and writing – review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by the Nature Science Institutes of Shanxi Province (Grant No. 20210302124360), Shanxi Province Science and Technology Innovation Project Foundation for Youths (Grant No. 2021L098), China’s Agricultural Research System of MOF and MARA (Grant No. CARS-44-KXJ2), and Award Project for Excellent Doctors (Grant No. SXYBKY2018043).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage [www.editage.cn] for English language editing.
Antolin, S., Reisert, J., and Matthews, H. R. (2010). Olfactory response termination involves Ca2 +-ATPase in vertebrate olfactory receptor neuron cilia. J. Gen. Physiol. 135, 367–378. doi: 10.1085/jgp.200910337
Bahk, S., and Jones, W. D. (2016). Insect odorant receptor trafficking requires calmodulin. BMC Biol. 14:83. doi: 10.1186/s12915-016-0306-x
Chen, S., and Luetje, C. W. (2012). Identification of new agonists and antagonists of the insect odorant receptor co-receptor subunit. PLoS One 7:e36784. doi: 10.1371/journal.pone.0036784
Coultrap, S. J., and Bayer, K. U. (2012). CaMKII regulation in information processing and storage. Trends Neurosci. 35, 607–618. doi: 10.1016/j.tins.2012.05.003
de Bruyne, M., Foster, K., and Carlson, J. R. (2001). Odor coding in the drosophila antenna. Neuron 30, 537–552. doi: 10.1016/S0896-6273(01)00289-6
Faas, G. C., Raghavachari, S., Lisman, J. E., and Mody, I. (2011). Calmodulin as a direct detector of Ca2 + signals. Nat. Neurosci. 14, 301–304. doi: 10.1038/nn.2746
Finn, B. E., and Forsén, S. (1995). The evolving model of calmodulin structure, function and activation. Structure 3, 7–11. doi: 10.1016/S0969-2126(01)00130-7
Getahun, M. N., Olsson, S. B., Lavista-Llanos, S., Hansson, B. S., Dieter, W., and David, N. R. (2013). Insect odorant response sensitivity is tuned by metabotropically autoregulated olfactory receptors. PLoS One 8:e58889. doi: 10.1371/journal.pone.0058889
Hoeflich, K. P., and Ikura, M. (2002). Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739–742. doi: 10.1016/S0092-8674(02)00682-7
Jain, K., Lavista-Llanos, S., Grabe, V., Hansson, B. S., and Wicher, D. (2021). Calmodulin regulates the olfactory performance in Drosophila melanogaster. Sci. Rep. 11:3747. doi: 10.1038/s41598-021-83296-9
Kumar, B. N., Taylor, R. W., Pask, G. M., Zwiebel, L. J., Newcomb, R. D., and Christie, D. L. (2013). A conserved aspartic acid is important for agonist (VUAA1) and odorant/tuning receptor-dependent activation of the insect odorant co-receptor (Orco). PLoS One 8:e70218. doi: 10.1371/journal.pone.0070218
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Lin, W., Yu, Y., Zhou, P., Zhang, J. H., and Zhu, S. F. (2015). Identification and knockdown of the olfactory receptor (OrCo) in gypsy moth. Lymantria dispar. Int. J. Biol. Sci. 11, 772–780. doi: 10.7150/ijbs.11898
Lisman, J., Yasuda, R., and Raghavachari, S. (2012). Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182. doi: 10.1038/nrn3192
Liu, H., Liu, T., Xie, L., Wang, X., Deng, Y., and Chen, C. H. (2016). Functional analysis of orco and odorant receptors in odor recognition in Aedes albopictus. Parasit. Vector. 9:363. doi: 10.1186/s13071-016-1644-9
Mizunami, M., Nemoto, Y., Terao, K., Hamanaka, Y., and Matsumoto, Y. (2014). Roles of calcium/calmodulin-dependent kinase II in long-term memory formation in crickets. PLoS One 9:e107442. doi: 10.1371/journal.pone.0107442
Mukunda, L., Miazzi, F., Kaltofen, S., Hansson, B. S., and Wicher, D. (2014). Calmodulin modulates insect odorant receptor function. Cell Calcium 55, 191–199. doi: 10.1016/j.ceca.2014.02.013
Mukunda, L., Miazzi, F., Sargsyan, V., Hansson, B. S., and Wicher, D. (2016). Calmodulin affects sensitization of Drosophila melanogaster odorant receptors. Front. Cell Neurosci. 10:28. doi: 10.3389/fncel.2016.00028
Nakagawa, T., Pellegrino, M., Sato, K., Vosshall, L. B., and Touhara, K. (2012). Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLoS One 7:e32372. doi: 10.1371/journal.pone.0032372
Park, D., Jung, J. W., Choi, B. S., Jayakodi, M., Lee, J., Lim, J., et al. (2015). Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genom. 16:1. doi: 10.1186/1471-2164-16-1
Reinhard, J., and Claudianos, C. (2012). Molecular Insights Into Honey Bee Brain Plasticity. Berlin: Springer Verlag 359–372.
Robertson, H. M., and Wanner, K. W. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403. doi: 10.1101/gr.5057506
Sato, K., Pellegrino, M., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Scholl, C., Kübert, N., Muenz, T. S., and Rössler, W. (2015). CaMKII knockdown affects both early and late phases of olfactory long-term memory in the honeybee. J. Exp. Biol. 218, 3788–3796. doi: 10.1242/jeb.124859
Snehal, D. K., Rikesh, J., Axel, B., and Ramanathan, S. (2016). Identification of complete repertoire of Apis florae odorant receptors reveals complex orthologous relationships with Apis mellifera. Genome Biol. Evol. 8, 2879–2895. doi: 10.1093/gbe/evw202
Song, Y. J., Cygnar, K. D., Sagdullaev, B., Valley, M., Hirsh, S., and Stephan, A. (2008). Olfactory CNG channel desensitization by Ca2 +/CaM via the B1b subunit affects response termination but not sensitivity to recurring stimulation. Neuron 58, 374–386. doi: 10.1016/j.neuron.2008.02.029
Taylor, R. W., Romaine, I. M., Liu, C., Murthi, P., Jones, P. L., Waterson, A. G., et al. (2012). Structure–activity relationship of a broad-spectrum insect odorant receptor agonist. ACS Chem. Biol. 7, 1647–1652. doi: 10.1021/cb300331z
Vardanush, S., Negash, G. M., Llanos, S. L., Olsson, S. B., Hansson, B. S., and Dieter, W. (2011). Phosphorylation via PKC regulates the function of the drosophila odorant co-receptor. Front. Cell Neurosci. 5:5. doi: 10.3389/fncel.2011.00005
Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A., and Axel, R. (1999). A spatial map of olfactory receptor expression in the drosophila antenna. Cell 96, 725–736. doi: 10.1016/S0092-8674(00)80582-6
Wang, J. W., Wong, A. M., Flores, J., Vosshall, L. B., and Axel, R. (2003). Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271–282. doi: 10.1016/S0092-8674(03)00004-7
Wanner, K. W., Nichols, A. S., Walden, K. K., Brockmann, A., Luetje, C. W., and Robertson, H. M. (2007). A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc. Natl. Acad. Sci. U S A 104, 14383–14388. doi: 10.1073/pnas.0705459104
Wicher, D., Schafer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Yap, K. L., Kim, J., Truong, K., Sherman, M., Tao, Y., and Ikura, M. (2000). Calmodulin target database. J. Struct. Funct. Genom. 1, 8–14. doi: 10.1023/A:1011320027914
Zhao, H. T., Gao, P. F., Du, H. Y., Ma, W. H., Tian, S. H., and Jiang, Y. S. (2014). Molecular characterization and differential expression of two duplicated odorant receptor genes, AcerOr1 and AcerOr3 in Apis cerana cerana. J. Genet. 93, 53–61. doi: 10.1007/s12041-014-0332-9
Keywords: Apis cerana cerana, AcerOr2, calmodulin, Ca2+/CaM-dependent protein kinase II, calcium/CaM signaling pathway
Citation: Guo L, Xu B, Zhao H, Guo Y and Jiang Y (2022) Calmodulin Activity Affects the Function of the Odorant Receptor AcerOr2 in Honeybees. Front. Ecol. Evol. 10:848150. doi: 10.3389/fevo.2022.848150
Received: 04 January 2022; Accepted: 23 February 2022;
Published: 04 April 2022.
Edited by:
Ying Wang, Shandong Agricultural University, ChinaReviewed by:
Zilong Wang, Southern University of Science and Technology, ChinaCopyright © 2022 Guo, Xu, Zhao, Guo and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Guo, eXlzZ3kzQDE2My5jb20=; Yusuo Jiang, amlhbmd5cy0wMDFAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.