- 1Department of Biology, Faculty of Natural Sciences, Vytautas Magnus University, Kaunas, Lithuania
- 2Department of Mathematics and Statistics, Faculty of Informatics, Vytautas Magnus University, Kaunas, Lithuania
- 3Department of Environment and Ecology, Faculty of Forest Sciences and Ecology, Vytautas Magnus University, Kaunas, Lithuania
- 4Department of Mathematical Statistics, Faculty of Fundamental Sciences, Vilnius Gediminas Technical University, Vilnius, Lithuania
Data on alien species show that plant invasions are caused by a complex combination of characteristics of invasive species (invasiveness) and characteristics of invaded environment (invasibility). Impatiens parviflora is one of Europe’s top invasive species. The present study aimed to evaluate molecular diversity of populations of highly invasive in Lithuania I. parviflora by applying several DNA markers and relating genetic parameters to abiotic and biotic environment. For sampling, urban forests, riparian forests, and agrarian shrublands were selected. Three different DNA-based techniques, Simple Sequence Repeats (SSR), Inter Simple Sequence Repeat (ISSR) markers, and Random Amplified Polymorphic DNA (RAPD), were used for detecting genetic variation between 21 populations. All population individuals were monomorphic and homozygotic for four loci and heterozygotic for one locus by SSR analysis. Hierarchical analyses of molecular variance at ISSR and RAPD loci revealed significant differentiation of populations depending on geographic zones of the country. Bayesian Structure analyses of molecular data demonstrated existence of many genetic clusters and this finding is in support to multiple introduction of the species. The polymorphism extent at ISSR loci was positively correlated with the total coverage of herbaceous plant species. The coverage by I. parviflora was negatively correlated with the total number of herbaceous species and light in the sites. Our results indicate that Lithuanian sites with I. parviflora might be distinguished by high soil nutrient levels. According to the principal component analysis, the coverage by I. parviflora was a more important variable of populations compared to molecular data or parameters of abiotic environment. In conclusion, complexity of invasibility and invasiveness factors determine the variability of I. parviflora sites, including genetic traits, coverage of invasive species and conditions of environment that were significant and interrelated.
Introduction
The world is facing multiple environmental challenges simultaneously, including chemical and biological pollution, climate change, degradation, and loss of biological diversity. Tackling these challenges has become increasingly complicated due to the raise of temporal and spatial human-environment interactions (Wang, 2021). Phenomenon of species invasion is considered among the most harmful processes, creating threats to agriculture and forestry, health and the global economy (Schmidt et al., 2012). To date, an unusually large amount of time and money has been spent controlling invaders (Haubrock et al., 2021).
Forests provide valuable habitat for various living beings, supplying them with food and shelter, cleaning and enriching air, mitigating the influence of cold and droughts, ameliorating climate change and chemical pollution. However, forests are trampled, burned, felled, ignorantly replanted and otherwise modified by human, making them highly susceptible to plant invasion (Wagner et al., 2017, 2021). Forest alien species of herbaceous plants might be unfavorable for recreation, diminishing the number of local species, causing soil erosion and other changes of habitats. The impact of specific invaders on forest ecosystem services is not clear enough and requires wider insights. Exotic species interfering with tree communities are extremely difficult to eradicate.
European woodlands, covering a third of Europe’s terrestrial area, are prone to alien plant invasions because many sites show typical symptoms of hemeroby, such as disturbance, fragmentation, high concentration of soil nutrients and pressure of foreign propagules. The regulation of invasive alien species has become a top priority for conservation policy at the national and European levels (European Parliament, 2014; EU Regulation 1143/2014).
The first target of invasive alien species regulation is the most aggressive invaders. Small balsam (Impatiens parviflora from Balsaminaceae) could be classified in this category as alien species that is extremely penetrative into forest ecosystems (Coombe, 1956; Trepl, 1984; Eliáš, 1999; Tanner, 2008; Chmura, 2014). As summarized by Coombe (1956) in 1831 I. parviflora was cultivated in the Botanic Garden in Geneva and within the next few decades was recorded as an escape in many places in Europe, often near botanic gardens: Geneva, Vienna, Prague, Tartu, Königsberg, and Berlin. There is a lot of convincing data about the type of further random invasion of I. parviflora (Trepl, 1984). In Poland and Latvia naturalization of this species was recorded in the second half of 19th century (Tokarska-Guzik, 2005; Priede, 2009b) and the first records for this species occurrence in the current territory of Lithuania were published later (Wisnewski, 1934). From the second half of the former century I. parviflora has been listed as adventive species of Lithuania (Natkevicaite-Ivanauskiene, 1951) and nowadays it belongs to the most widely spread aliens in the country (The Ministry of Environment of the Republic of Lithuania, 2015). Different European countries have reported continued spread of this species within recent decades (Priede, 2009a; Reczyńska et al., 2015; Jarčuška et al., 2016; Wagner et al., 2017). Invasive strength of this plant is proven by both the size of occupied territory and the variety of habitats (Klimko and Piskorz, 2003; Chmura and Sierka, 2006). I. parviflora is an exceptionally successful invader of forest vegetation and is presently the most common alien plant in European woodlands (Wagner et al., 2017, 2021). This plant has been very frequently discussed when evaluating impact on forest regeneration (Langmaier and Lapin, 2020). Many studies have been carried out focusing on physiology and ecology of I. parviflora (Trepl, 1984; Chmura, 2014; Reczyńska et al., 2015; Jarčuška et al., 2016; Florianová and Münzbergová, 2017). Information about steps and vitality of populations of alien herbs in North-East parts of Europe still remains very fragmental.
To counteract expansion of harmful species to the new areas, comprehensive knowledge in both traditional and modern biology is required (Tokarska-Guzik, 2005; Cross et al., 2011). Neutral molecular genetic markers can be used as the tools to study ecological processes such as colonization, dispersal, or community interactions (Nybom et al., 2014). Data in molecular genetics may provide unique insights into the sources, routes and mechanisms of alien species spread. There is a growing understanding that integrating genetics and ecology is critical in the context of biological invasions, since the two are explicitly linked (Handley et al., 2011). Information on the genetic make-up of an invasive emerging population is important for answering one of the most fundamental questions in invasion biology: what determines the success of the spread of alien species? In addition, understanding these processes is seen as essential for implementing proper management policies. Molecular markers differ in terms of the number of fragments per primer, the extent of polymorphic fragments per primer. SSR loci are known as simple di- or tri-nucleotide repeats with higher rate of mutation compared to the average for the genome, ISSR cover genomic areas flanked with the inverted repeats of SSR loci, and RAPDs are related to anonymous sequences scattered all over the genome (Grover and Sharma, 2016; Bechtold and Field, 2018; Garrido-Cardenas et al., 2018; Younis et al., 2020). Studying closely related genotypes, the analysis of variability is more informative in case more than one DNA-based technique is applied (Costa et al., 2016; Yasui, 2020). Molecular genetics of populations has provided valuable information about Lupinus polyphyllus as alien forest species of Finland (Li et al., 2016), and Lithuania (Vyšniauskienė et al., 2011). When compared with other molecular techniques, ISSR markers are easy to use, specific enough, and are applicable for all plant groups. Overall, these markers showed great promise as a method of elucidating relationships among individuals within populations and between populations and is still much used nowadays (Nybom et al., 2014; Lazzaro et al., 2018). ISSRs have been employed for evaluation of genetic diversity of Alliaria petiolata which scope of invasion in North America is similar to Impatiens parviflora expansion in Europe (Meekins et al., 2001).
Information about steps and vitality of populations of alien herbs in North-East parts of Europe still remains very fragmental. Till now, I. parviflora remains among the least genetically examined alien plant species world-wide. Extensive investigations of I. parviflora has started long before the dawn of molecular methods for population studies (Coombe, 1956; Trepl, 1984). To the best of our knowledge, till the present study, genetic diversity of two Polish populations was evaluated by amplified fragment length polymorphism (AFLP) markers (Komosińska et al., 2006) and cpDNA from one United Kingdom plant was recently analyzed (Kurose et al., 2020). However, the relations between genetic data and assemblages of herbaceous species in sites with I. parviflora have never been traced for the species. This aspect might be important while searching for unique invasive plant abilities to extend into secondary areas. The present study is aimed at evaluation of genetic diversity of Lithuanian populations of I. parviflora using a set of molecular markers and relating genetic parameters to species abiotic and biotic environment.
Materials and Methods
Study Area and Species
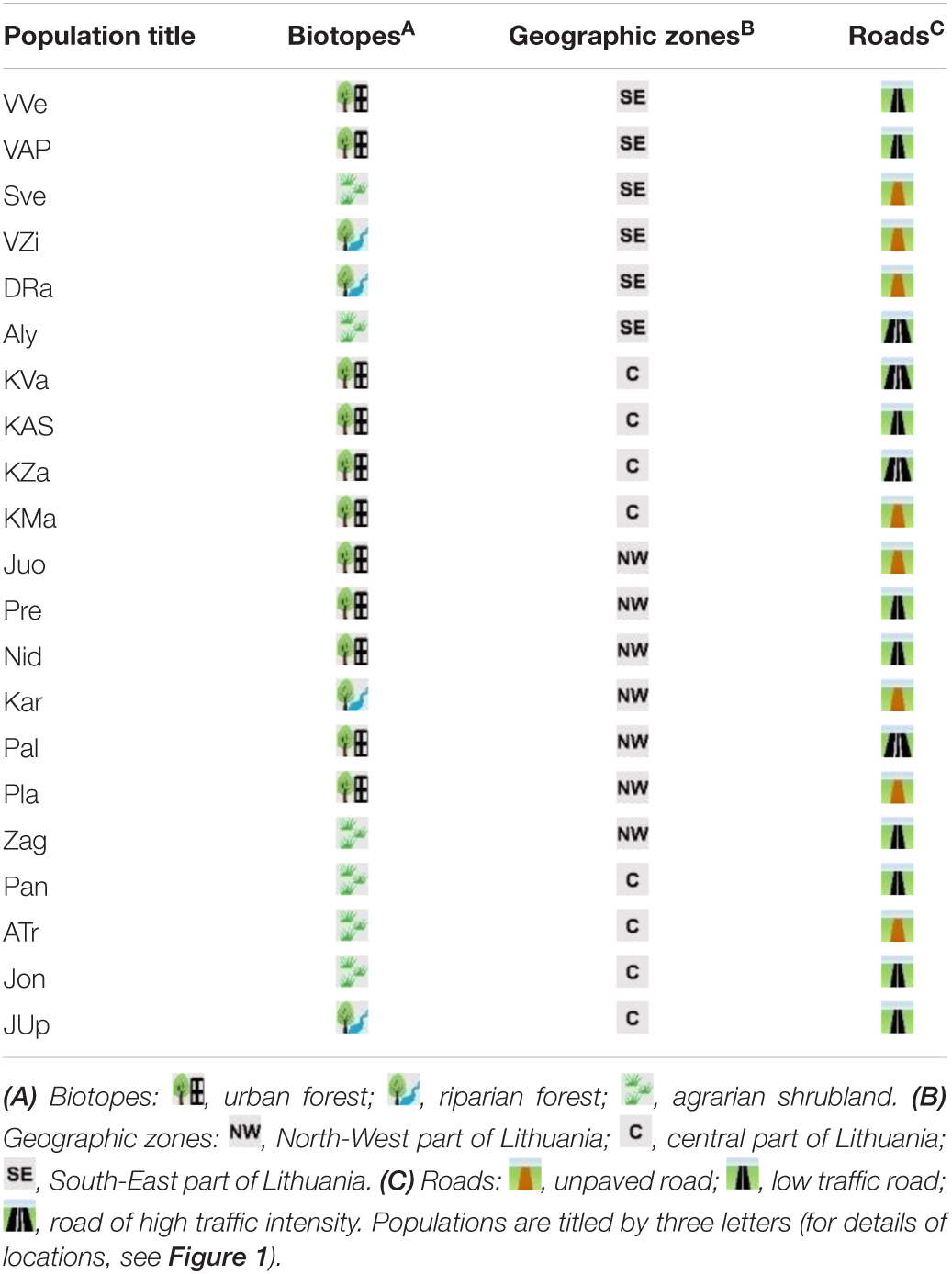
Twenty-one populations of small balsam (Impatiens parviflora DC.) were selected in the way to cover all the territory of Lithuania (Figure 1). Sites with I. parviflora were named by three letters, reflecting their geographical location. Geographical coordinates of the examination are ranged from 53°59′ to 56°21′ latitude (N), from 20°59′ to 26°09′ longitude (E), with annual rainfall of 722–936 mm, and mean annual temperature of 6.0–6.8°C (Kupcinskiene et al., 2013).
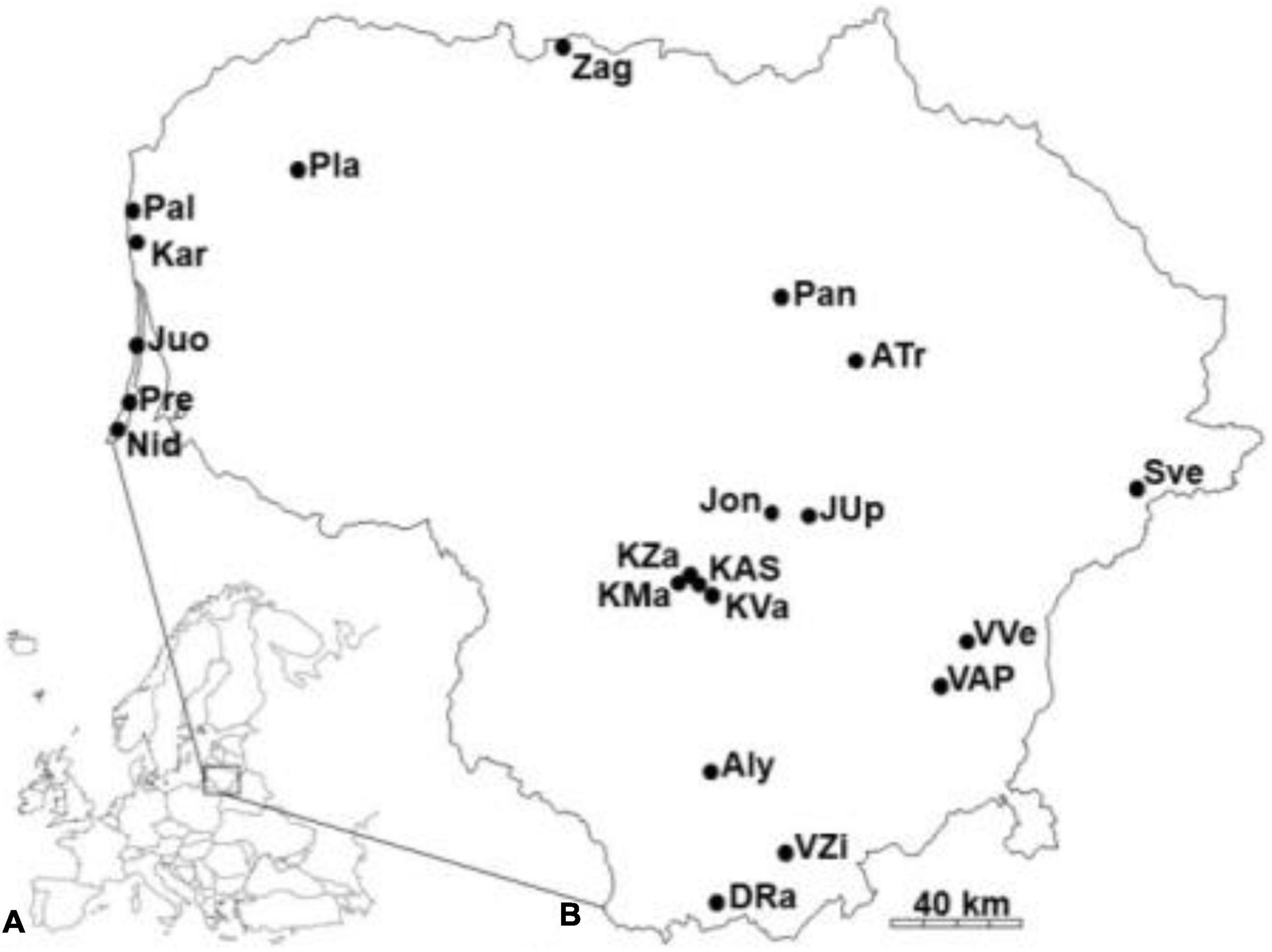
Figure 1. Location of Lithuania (A), and sites with Impatiens parviflora in the country (B). Populations are named by three letters.
Impatiens parviflora sites were described by a number of properties (Table 1). According to the biotope they were divided into urban forests, riparian forests and agrarian shrublands. Sites were divided into three groups based on the traffic intensity along the road close to the site: a road without blacktop in the forest, a blacktop road with low intensity traffic, and a blacktop road with intensive traffic. Herbaceous plant species growing along with populations of I. parviflora were recorded in 100 m2 plots at each site. The coverage of each species was calculated using Braun-Blanquet methodology (Braun-Blanquet, 1964) and transformed into mean-percentage values for each degree (0.1, 0.5, 3, 15, 38, 63, and 88%). For evaluation of environmental preferences of I. parviflora in each site, Ellenberg indicator values (EIV; Ellenberg et al., 1992), i.e. – light, temperature, continentality, soil moisture, soil reaction and soil nutrients, were quantified in proportion to the corresponding species coverage, employing weighted average (W-EIV) method, as it was described previously (Marozas, 2014; Marozas et al., 2015).
For molecular analyses 15 plants (315 individuals in total), were sampled from each site in the way described above (Kupcinskiene et al., 2013).
Molecular Analyses
Plant leaf DNA was extracted using DNA purification kit #K0512 (Thermo Fisher Scientific, Vilnius, Lithuania), following the instructions of the producer with some modifications (Zybartaite et al., 2011).
For SSR analyses the primer pairs IGNSSR101-EF025990, IGNSSR104-EF025992, IGNSSR106-EF025993, IGNSSR203-EF025994, IGNSSR210-EF025995, and IGNSSR240-EF025997 (Metabion International AG, Steinkirchen, Germany) were used following Provan et al. (2007). The polymerase chain reaction (PCR) was carried out using 6.25 μl PCR Master mix, 5 pmol of each primer in the pair and 25 ng of genomic DNA. Temperature conditions for PCR were as follows: initial denaturation at 94°C for 3 min, 35 cycles including denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 5 min.
Inter simple sequence repeat-polymerase chain reaction was carried out using primers former employed to develop microsatellites for Impatiens glandulifera (Provan et al., 2007): (1) GGCC(AG)8, (2) GGCC(AC)8, (3) CCGG(AG)8, (4) CCGG(AC)8, (5) GCGC(AG)8, and (6) GCGC(AC)8 (Metabion International AG, Steinkirchen, Germany). Of the six tested primers, four (2–3, 5–6) generated clear and reproducible fragments of DNA and were used for population studies. Eight RAPD markers (OPA-20, OPD-20, 222, 250, 269, 340, 474, and 516) applied for populations of I. glandulifera (Zybartaite et al., 2011) later were employed for Impatiens parviflora (Kupcinskiene et al., 2013).
For genetic analyses of 315 individuals of I. parviflora ISSR-PCR amplification was carried out in a final volume of 20 μL containing 100 ng of template DNA, 20 pmol primer, 1 × Taq buffer, 200 μM dNTPs, 2.5 mM MgCl2 and 1 U of Taq polymerase, following Hatcher et al. (2004) and Provan et al. (2007). DNA amplification was carried out in Eppendorf Mastercycler ® (“Eppendorf,” Germany) under such a program: initial denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C, annealing at 60°C for 1 min, extension at 72°C for 1 min and a final extension at 72°C for 5 min. The products of PCR were fractionated by electrophoresis on 1.5% TBE-agarose gels stained with ethidium bromide and imaged using UV transilluminator (“Herolab”, Germany).
Statistical Analysis
Both ISSR and RAPD data were evaluated by Mantel test, hierarchical analyses of molecular variance (AMOVA), principal coordinate analyses of individuals, and Bayesian clustering. In addition, genetic parameters of I. parviflora were related to biotic environment (herbaceous plant species data).
Standard parameters of genetic diversity were obtained using GENALEX version 6.5 (Peakall and Smouse, 2012). An unweighted pair group method with arithmetic mean (UPGMA) cluster analysis based on pairwise Nei’s unbiased genetic distances (Nei, 1978) was used to assess genetic relationships among populations. Mantel test (9999 permutations; Mantel, 1967) and principal coordinate analyses (PCoA) were done in GENALEX program, version 6.5. Hierarchical analyses AMOVA were done using GENALEX, which allowed partition the total genetic variation into three hierarchical levels: among regions, among sites within regions, and within sites. To explore patterns in genetic variation of I. parviflora, and to relate this parameter to the traits of abiotic and biotic environment, a hierarchical analysis of molecular variance (AMOVA at ISSR and RAPD loci) was conducted among populations depending on biotope (agrarian shrubland, urban forest, riparian forest), geographic zone (North-West, Central, and South-East of Lithuania), and road type.
For SSR data (21 population, 15 individuals in each) observed heterozygosity (Ho), expected heterozygosity (He) and inbreeding coefficient (FIS) were calculated using ARLEQUIN version 3.1 (Excoffier et al., 2005).
For identification of gene clusters of I. parviflora populations, Bayesian analysis was performed using STRUCTURE, version 2.3.1 (Pritchard et al., 2000; Falush et al., 2003, 2007). A priori estimation of clusters was set to K = 1–21, the highest probable number of the clusters corresponded to the number of populations. To test for significant patterns of clustering, the procedure described by Evanno et al. (2005) was implemented. Twenty runs were carried out for each K and the rate of change in the log probability of the data between successive likelihood values (ΔK) was estimated using a 104 steps burn-in period followed by 105 iterations of Markov chain Monte Carlo.
To assess the similarity of assemblages of herbaceous plant species in the sites, a cluster analysis was performed, applying program PC-ORD, version 6.0 (McCune and Mefford, 2011). For comparison of features of the sites, median values of W-EIV were used and differences among sites were calculated using STATISTICA, version 7.0 (StatSoft Inc, 2004).
To relate genetic data of I. parviflora and environment data, Spearman correlations were calculated, drawing correlogram by R package “corrplot,” version 0.90 (Wei and Simko, 2021). Principal component analysis was carried out using two genetic variables [percentage of polymorphic loci (PLP) at ISSR loci and RAPD loci], two biotic variables (coverage by I. parviflora, and the number of herbaceous species), six abiotic variables [W-EIV for light (L), temperature (T), continentality (K), soil moisture (F), soil reaction (R), and soil fertility (nutrients, hereinafter N) of herbaceous species] of sites employing statistical software R, version 4.1.1 (R Core Team, 2021).
Results
Profiles of Molecular Markers of Impatiens parviflora Populations
No alleles were obtained by IGNSSR104-EF025992, one allele was produced by each out of four primer pairs: IGNSSR101-EF025990, IGNSSR106-EF025993, IGNSSR203-EF025994, and IGNSSR240-EF025997, and two alleles were synthesized by IGNSSR210-EF025995. The size of DNA fragments ranged within 98–148 bp. The test of conformity of locus IGNSSR210-EF025995 heterozygosity to the Hardy-Weinberg equilibrium revealed, that for all populations inbreeding coefficient, (FIS) was −1, observed heterozygosity (HO) was 1 and expected heterozygosity (HE) was 0.517 (p < 0.001).
Among 21 populations examined, 108 alleles were generated at four ISSR loci and 218 alleles at eight RAPD loci. All ISSR and RAPD loci were polymorphic at the species level. In case of ISSR markers the range of polymorphic loci percentage (PLP–ISSR) per population was 10.2–25.9, the mean being 16.5 (Figure 2). Among populations Nei’s gene diversity (h–ISSR) range was 0.030–0.078 and Shannon’s index (I–ISSR) range was 0.046–0.121, the respective mean values per population were 0.047 and 0.073. For RAPD loci the range of PLP per population was 7.3–39.0, the mean being 21.0 (PLP–RAPD). Among populations Nei’s gene diversity (h–RAPD) range was 0.052–0.141 and Shannon’s index (I–RAPD) range was 0.079–0.225, the respective mean values per population were 0.095 and 0.146. There were 13 unique alleles at ISSR loci and seven alleles at RAPD loci. Of 20 unique (either for ISSR or RAPD loci) alleles, two alleles were identified in the I. parviflora populations in the South-East Lithuania, 10 alleles in the Central and eight alleles in the North-West Lithuania.
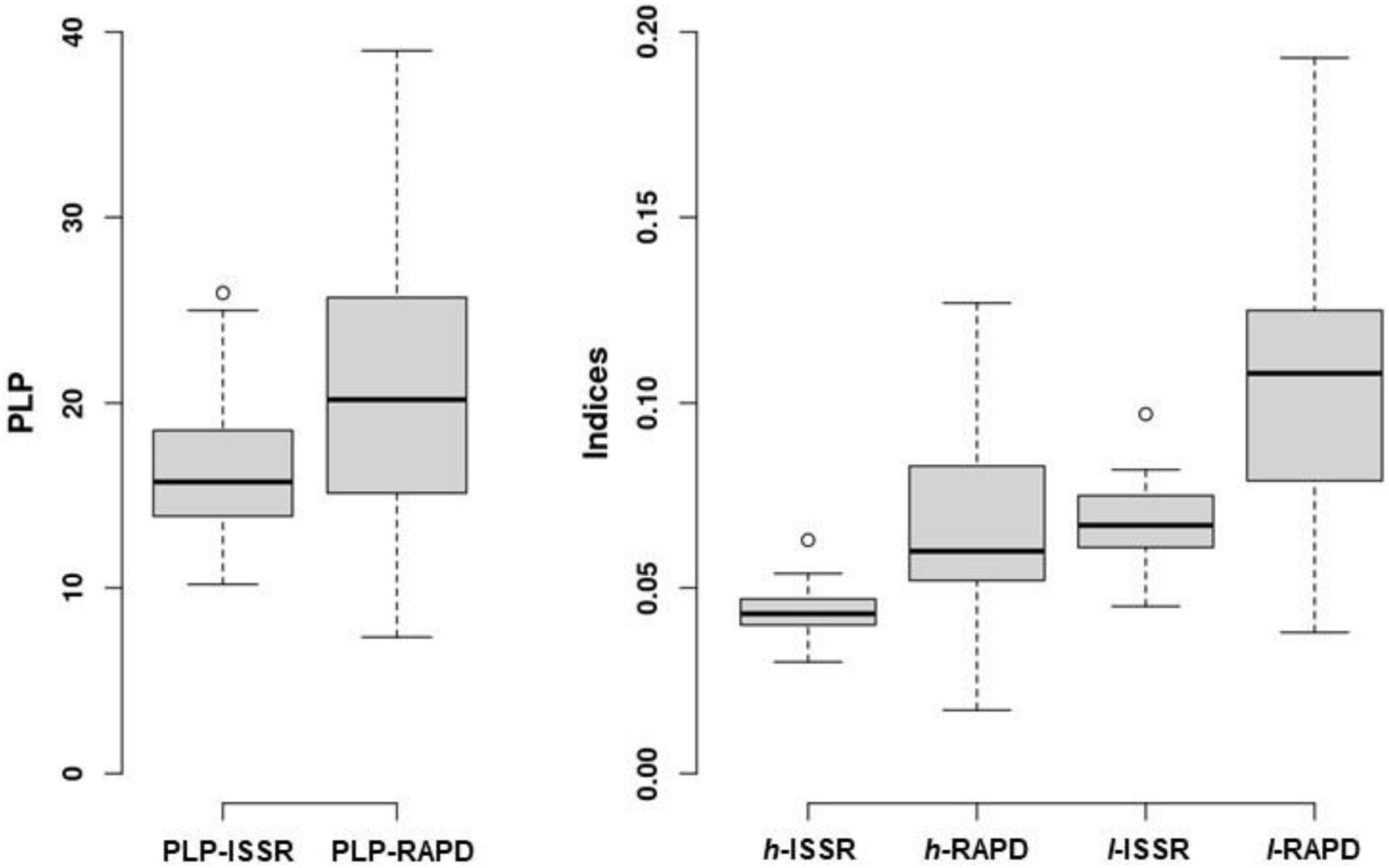
Figure 2. ISSR and RAPD loci based parameters of genetic diversity of 21 Impatiens parviflora populations: PLP, polymorphic loci percentage; indices: h, Nei’s gene diversity; I, Shannon’s information index. The central line of each box indicates the median value; the boxes, the lower (25%) and upper (75%) quartiles, and the whiskers, are from 10 to 90 percentiles (typical range), the points are outliers.
Estimated by ISSR markers, Nei’s (1978) pairwise genetic distances (GD) between the populations were in the range of 0.083–0.405. Mantel test for those markers did not show correlation between the genetic and geographic distances of populations (Figure 3). Interval of pairwise genetic distances at RAPD loci was 0.135–0.426. Mantel test showed significant correlation between the genetic and geographic distances of the populations. Dendrograms split populations into clades of 9th (at ISSR loci) or 12th order (at RAPD loci) (Figure 4). In both dendrograms of genetic relationships at ISSR and RAPD loci, one of the three most distinct populations were DRa and in opposite situation was VVe.
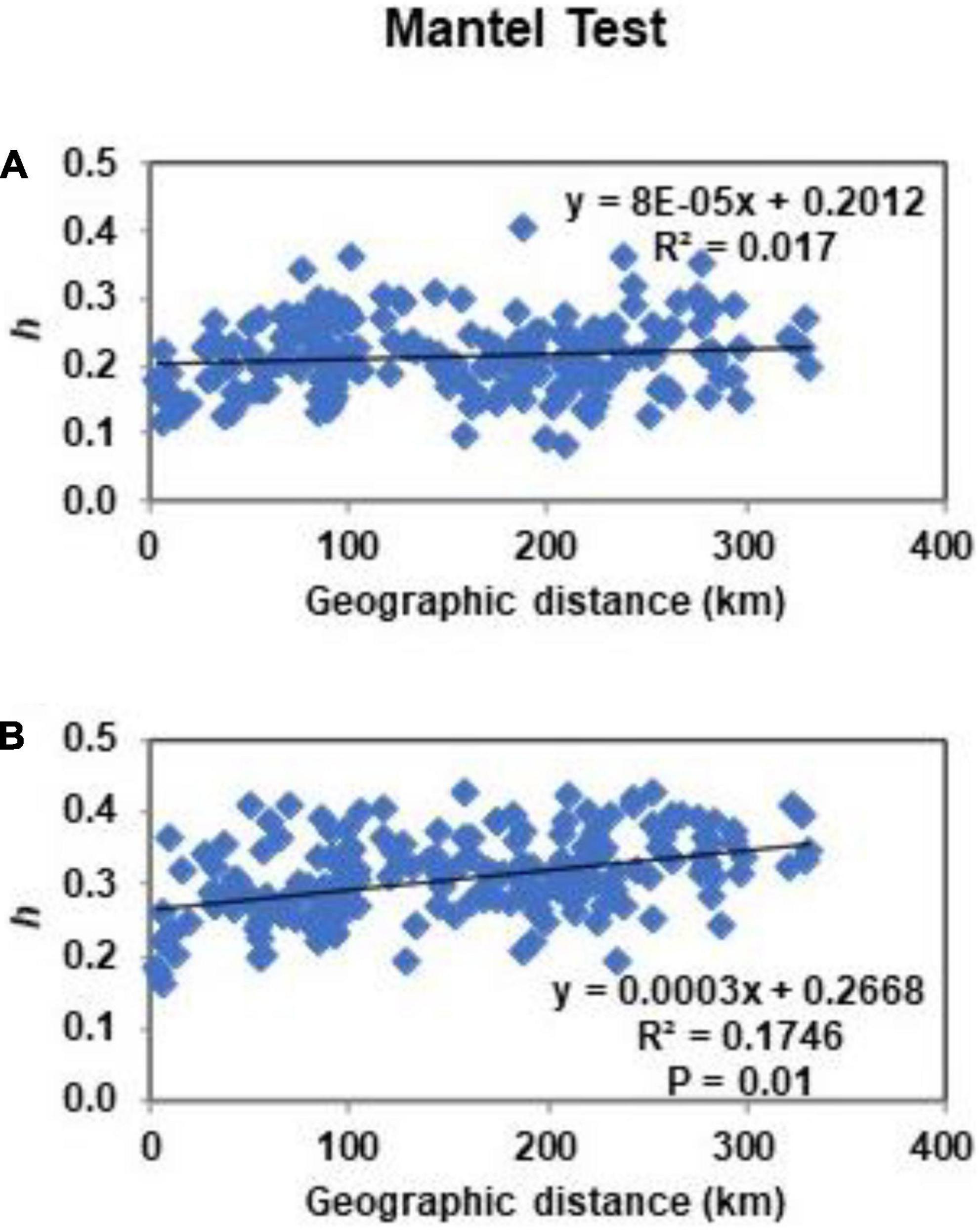
Figure 3. Mantel test of isolation by distance at ISSR loci (A) and RAPD loci (B) for populations of Impatiens parviflora. Each point represents Nei’s genetic distance (h) plotted against geographic distance.
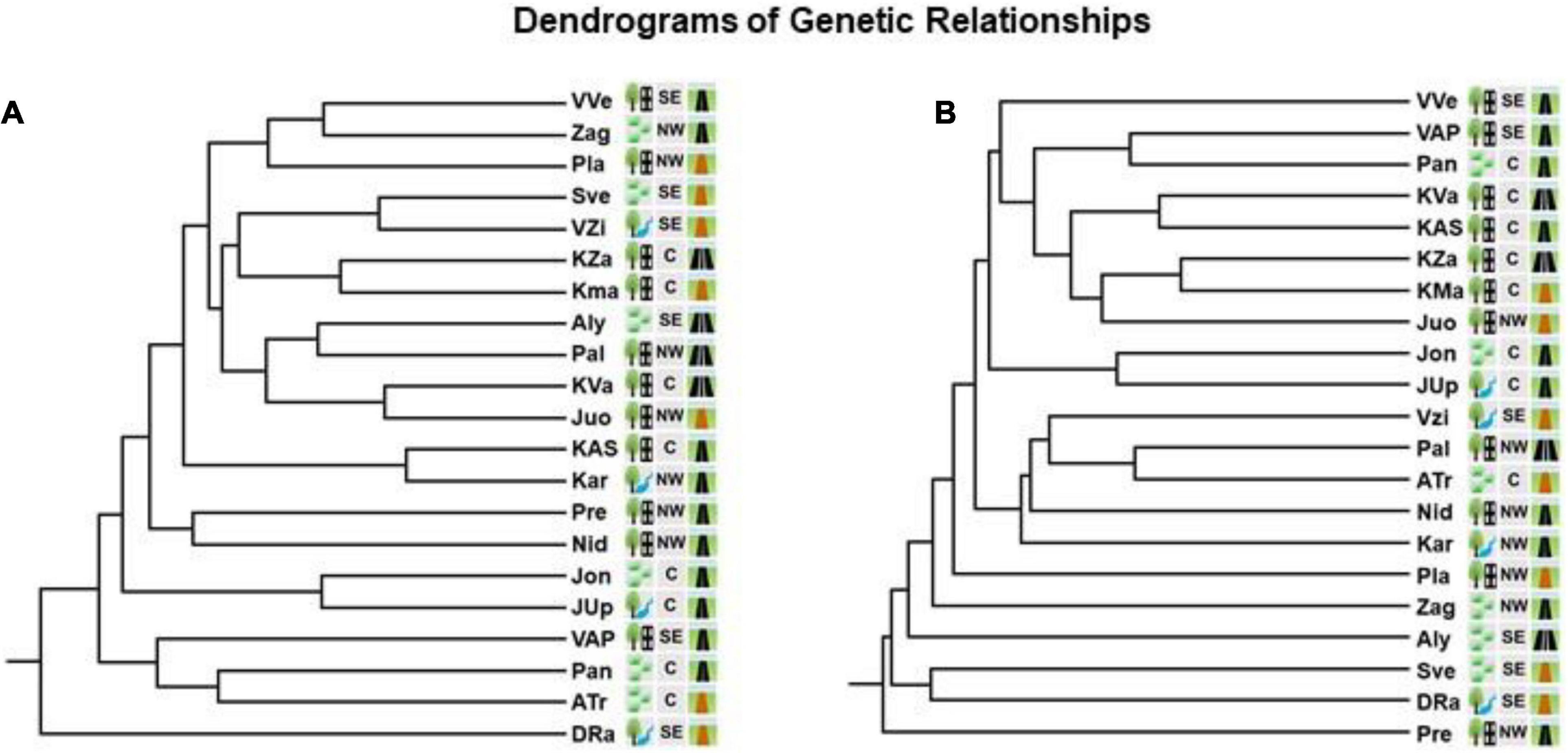
Figure 4. Dendrograms based on UPGMA cluster analysis of Nei’s genetic distances at ISSR loci (A) and RAPD loci (B) of genetic relationships between the populations of Impatiens parviflora. Three letters denote the populations (for details of their location, see Figure 1. Explanations of the pictogram are provided in the Table 1).
Hierarchical analyses of molecular variance (AMOVA) showed that I. parviflora populations, grouped according to the biotope (urban forest, riparian forest, and agrarian shrublands), differed significantly at RAPD loci accounting for 2% of the variation (Table 2).
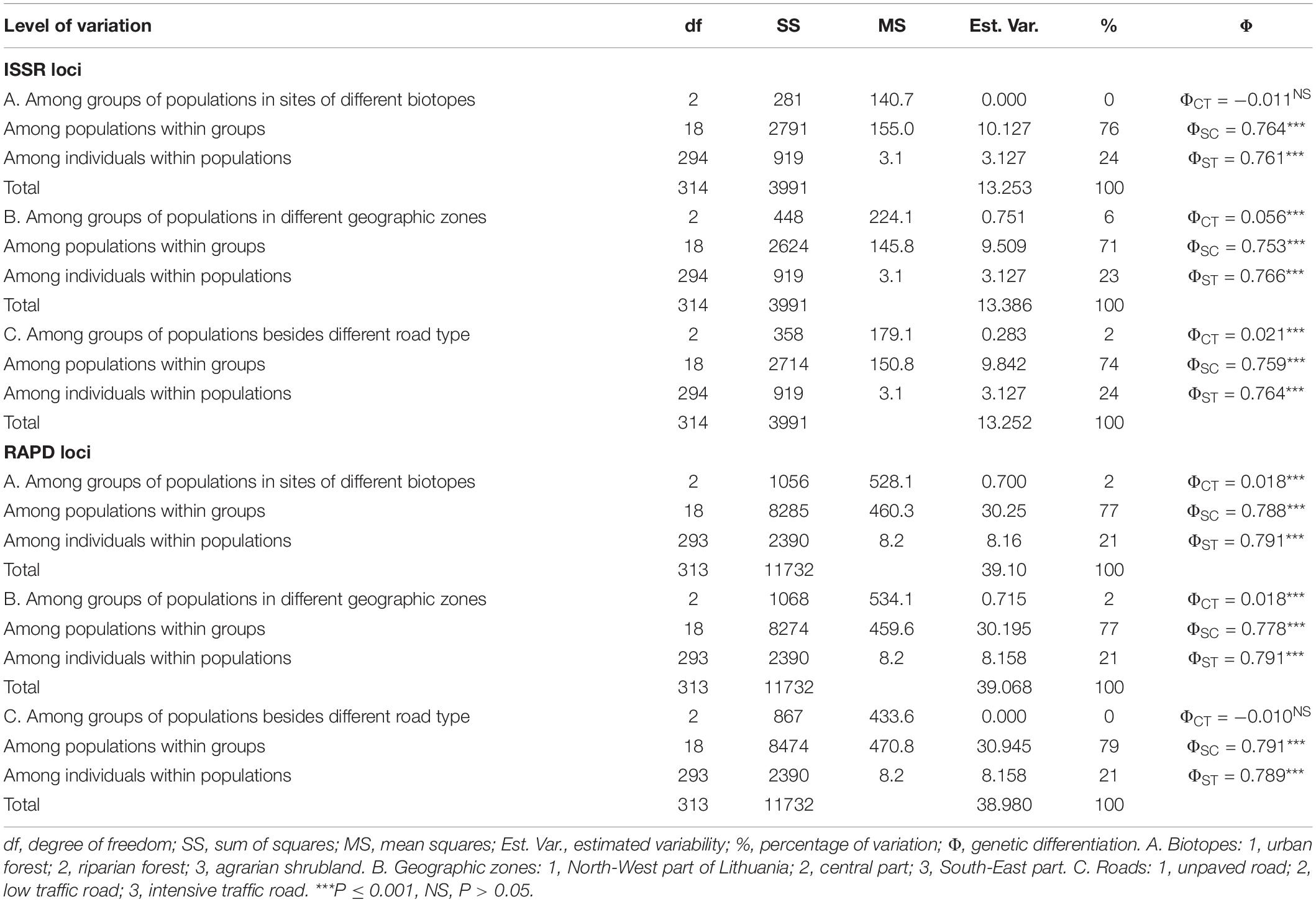
Table 2. ISSR and RAPD loci based hierarchical analyses of molecular variance (1) among groups of populations differing in biotope (A), geographic zone (B), type of neighboring road (C); (2) among populations within groups, and (3) within populations of Impatiens parviflora.
Hierarchical AMOVA for distinct geographic zone (North-West part, Central part, and South-East part of Lithuania) populations reflected significant genetic structure of the populations, accounting for 6% of the variation at ISSR loci and 2% of the variation at RAPD loci. Hierarchical AMOVA analysis of ISSR data showed significance of population grouping according to the neighboring road type: 2% of molecular variance was related to differences between the groups of populations. At RAPD loci, differentiation of population groups was not significant. Based on two types of molecular markers, the populations within groups (A–C cases of grouping) were significantly different (accounting for 71–79% of the genetic variation), while the within-population proportion of variance was smaller and very similar for all cases of grouping (21–24%, respectively).
According to the analysis PCoA of ISSR data, the first three coordinates accounted for about 28.2% of the genetic variation of individuals of I. parviflora populations (Figure 5), the first, the second, and the third coordinate explained, respectively, 10.6, 9.7, and 7.9% of the total variation. In all ISSR based PCoA plots (axis 1 vs. 2, 1 vs. 3, and 2 vs. 3) the most distinct individuals belonged to DRa, ATr, and Jon populations. Individuals of the North-West populations were mostly overlapping with individuals of the Central zone populations.
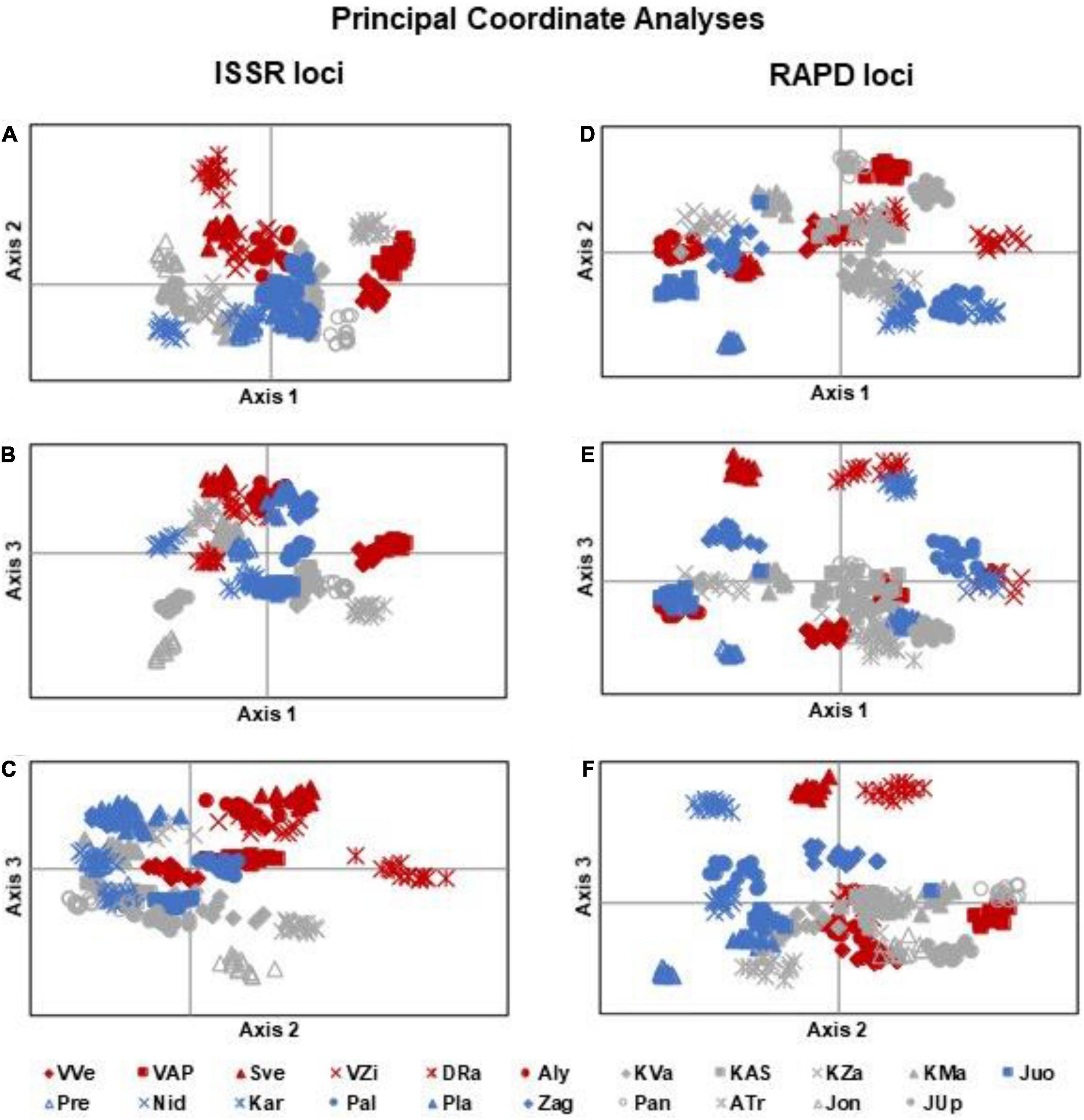
Figure 5. Principal coordinate analyses of Impatiens parviflora individuals of 21 populations according to: (A–C) ISSR loci, (A) – axis 1 vs. 2, (B) – axis 1 vs. 3, (C) axis 2 vs. 3; (D–F) RAPD loci, (D) – axis 1 vs. 2, (E) – axis 1 vs. 3, (F) – axis 2 vs. 3. Color of the labels denotes location of populations: blue – North-West, gray – Central, and red – South-East part of the country.
Based on RAPD data PCoA for the first three coordinates accounted for about 21.4% of the genetic variation of individuals of I. parviflora populations. The first, the second, and the third coordinate explained, respectively, 7.7, 7.1, and 6.7% of the total variation. Defined by PCoA at RAPD loci (axis 1 vs. 3, axis 2 vs. 3), the most distinct were individuals of the population DRa. For both systems of molecular markers (ISSR and RAPD), PCoA analyses did not differentiate populations into three distinct subgroups corresponding to the different geographic zones.
The Bayesian analysis of ISSR data of 315 individuals of I. parviflora revealed that a simulation equilibrium (likelihood of K’s) provided by the genomic structure was reached at K = 11 (Figure 6). The next likelihood of K was three genetic clusters (K = 3). The Bayesian analysis of RAPD data of the same populations suggested the most likely K value being 13.
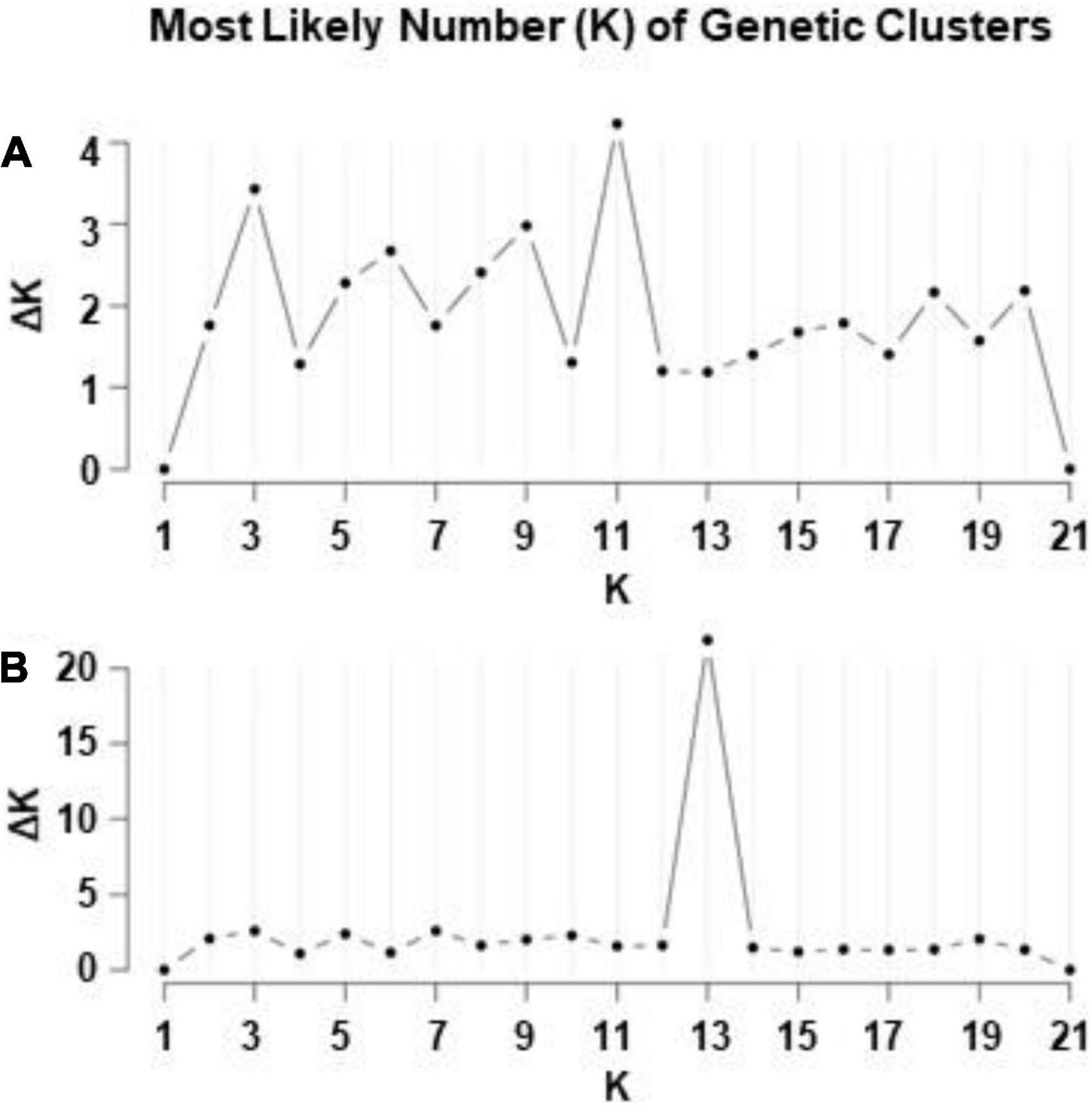
Figure 6. Evanno et al. (2005)ΔK statistics according to ISSR (A) and RAPD (B) markers for 21 populations of Impatiens parviflora.
For ISSR STRUCTURE barplot at K = 3, the 1st gene pool was predominant among five populations (24%), each of the remaining two gene pools was characteristic for groups of eight populations (Figure 7). Populations with the same prevalent gene pools were scattered along all the territory under investigation. For ISSR STRUCTURE barplot at K = 11, there were eight pairs of populations (VVe-VAP, SVe-VZi, KZa-KMa, Pre-Nid, Pla-Zag, Juo-Pal, Pan-ATr, and JUp-Jon) with the same prevailing gene cluster. For RAPD STRUCTURE barplot at K = 13, there were two the same pairs (like in case of ISSR, KZa-KMa and JUp-Jon) out of seven pairs of populations with the prevalence of identical gene cluster. The extent of admixture of gene pools was negligible for most of the populations, with the following exceptions: ATr at ISSR loci and Pal at RAPD loci.
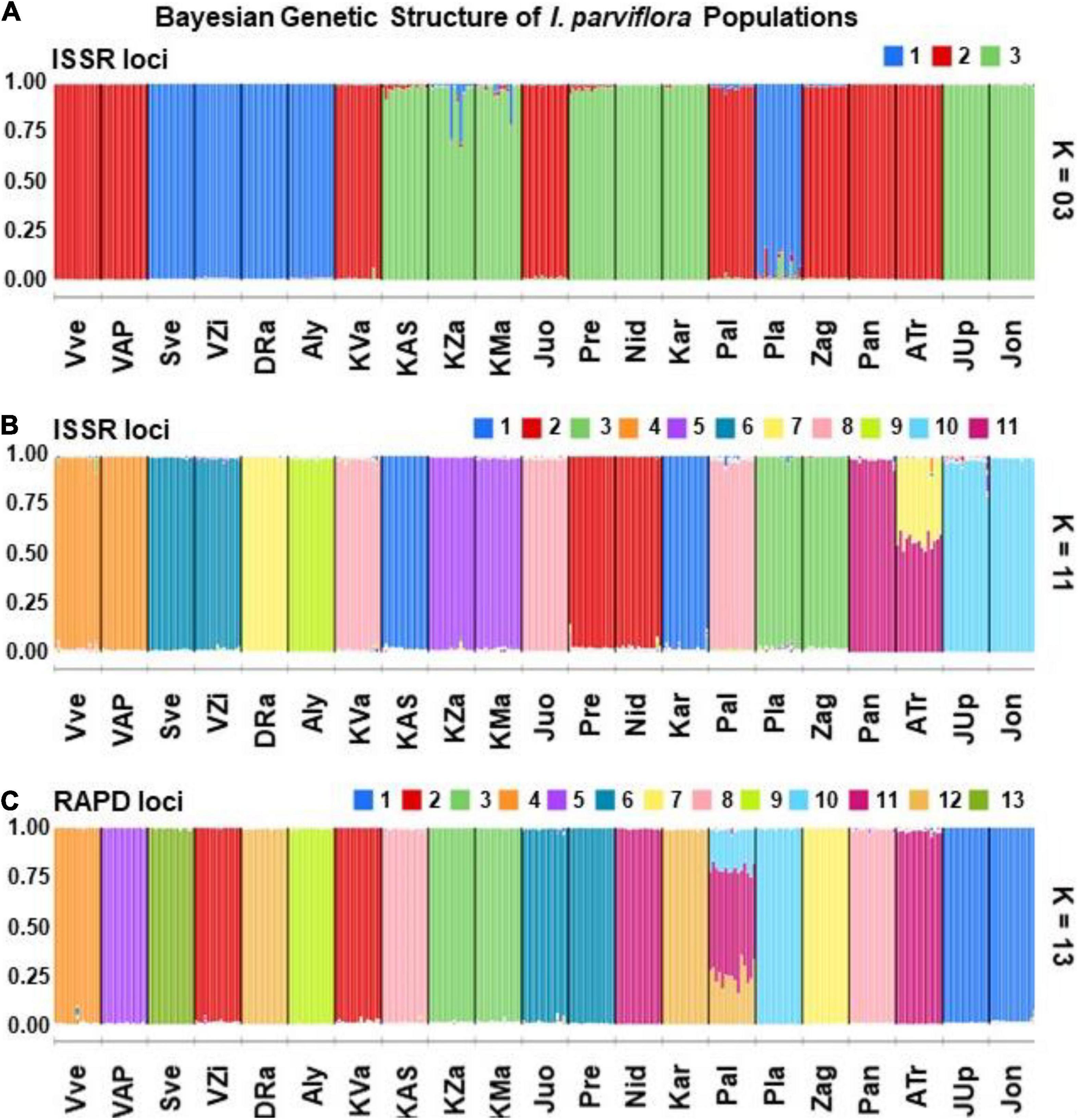
Figure 7. Barplots of the membership of Impatiens parviflora individuals/populations from the 21 sites in each of K genetic clusters identified from the ISSR or RAPD datasets using the Bayesian clustering in the STRUCTURE: (A) ISSR (K = 3, as next to the highest K); (B) ISSR (K = 11, the highest K); (C) RAPD (K = 13, the highest K). White lines separate bars of different individuals, black lines separate bars of different populations. Bars are partitioned into K colored segments that represent the individual’s/population’s estimated membership fractions (the amount of the genetic diversity) in K clusters. Three letters denote the populations (Pop) (for details of their locations, see Figure 1).
Analyses of Biotic Variables of Sites
In urban forests, the characteristic tree species were Pinus sylvestris, Acer pseudoplatanum, Tilia cordata, Quercus robur, Fraxinus excelsior, and Ulmus glabra. In riparian forests Salix alba, S. fragilis, and in some sites Alnus glutinosa were growing. Agrarian shrublands consisted of shrubs such as Corylus avellana, Ribes nigrum, Euonymus europaeus, E. verrucosus, and Lonicera xylosteum and/or single trees such as Fraxinus excelsior, Acer platanoides, Tilia cordata.
Evaluating the sites with I. parviflora according to the number of herbaceous plant species, 138 (62) species were recorded. The most frequent were species belonging to following families: Asteraceae (16), Fabaceae (14), Poaceae (12), Rosaceae (11), Lamiaceae (10), Apiaceae (8), Caryophyllaceae (7), Plantaginaceae (5), Polygonaceae (5), Ranunculaceae (5), Onagraceae (4), and Brassicaceae (4). The number of herbaceous plant species in the sites ranged from 13 to 32 (Figure 8). The most commonly species found near the I. parviflora individuals were hemicryptophytes, such as Urtica dioica (frequency of occurrence in the sites was 100%), Anthriscus sylvestris (76%), Chelidonium majus (67%), Geum urbanum (62%), Aegopodium podagraria (57%), and Alliaria petiolata (48%).
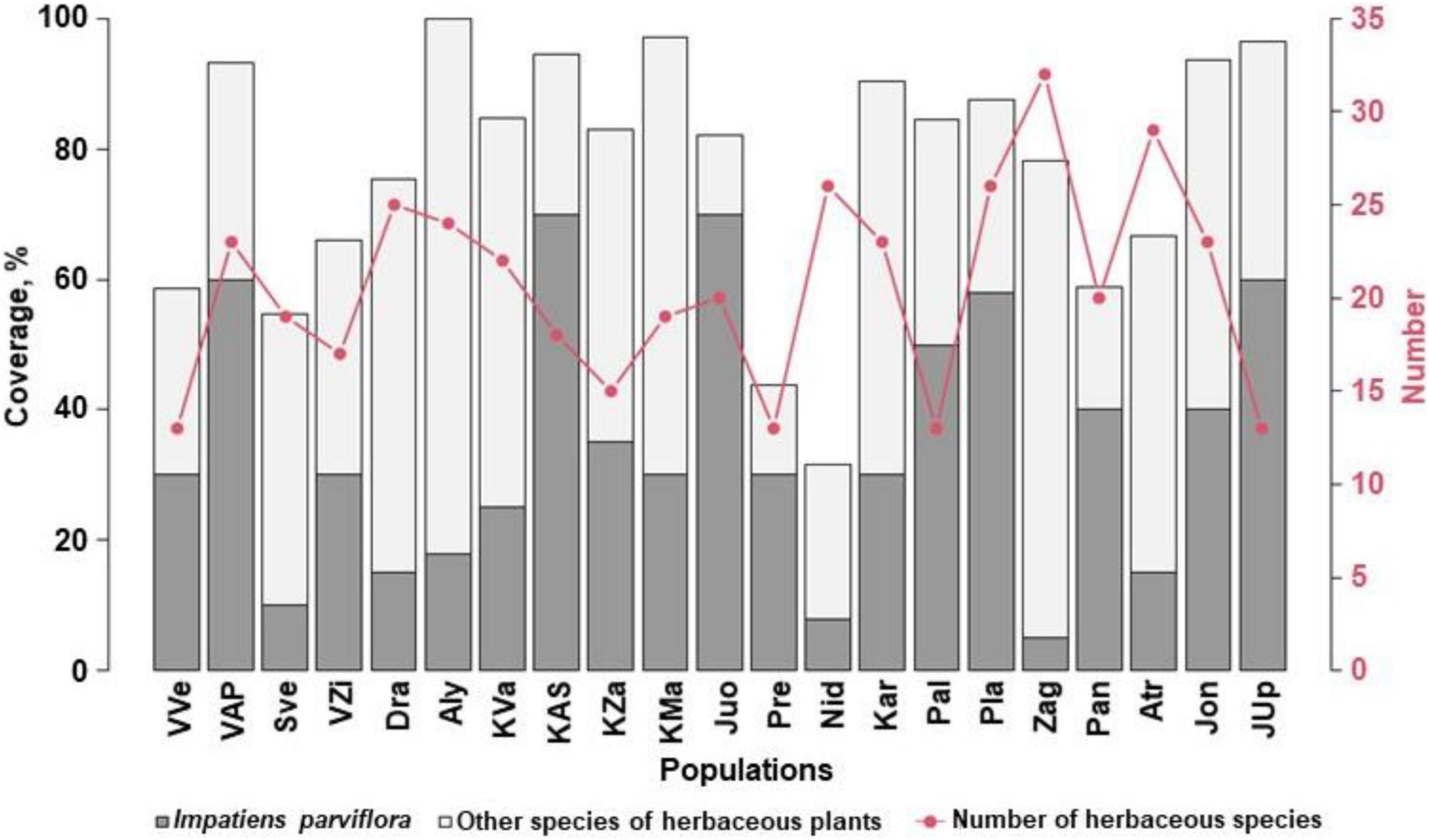
Figure 8. The number of herbaceous species and coverage by Impatiens parviflora and herbaceous plant species in the sites. Three letters denote populations (for details of their locations, see Figure 1).
The most diverse in species were the following sites: Zag with 32 species (23.2%), ATr – 29 (21.0%), Nid, Pla – 26 (18.8%). In the sites, the coverage by I. parviflora ranged from 5% (population Zag) to 70% (populations KAS and Juo), the mean for all populations was 35%. In the sites (KAS and Juo) with the most abundant I. parviflora (mean coverage 42.5%) the number of species was not large (13 species per site). Coverage by herbaceous plants other than I. parviflora ranged from 12.2 to 81.9%, the mean coverage being 42.5%. Total coverage by herbaceous plants (covT) ranged from 31.6 to 99.9% (populations Nid and Aly, respectively), the mean coverage being 76%.
In the cladogram based on the similarities of the assemblages of herbaceous plant species, the sites split into clusters of 14 orders (Figure 9). Clade of the 1st order involved Zag site only (it had the lowest coverage by I. parviflora – 5% and the highest number of herbaceous species – 32).
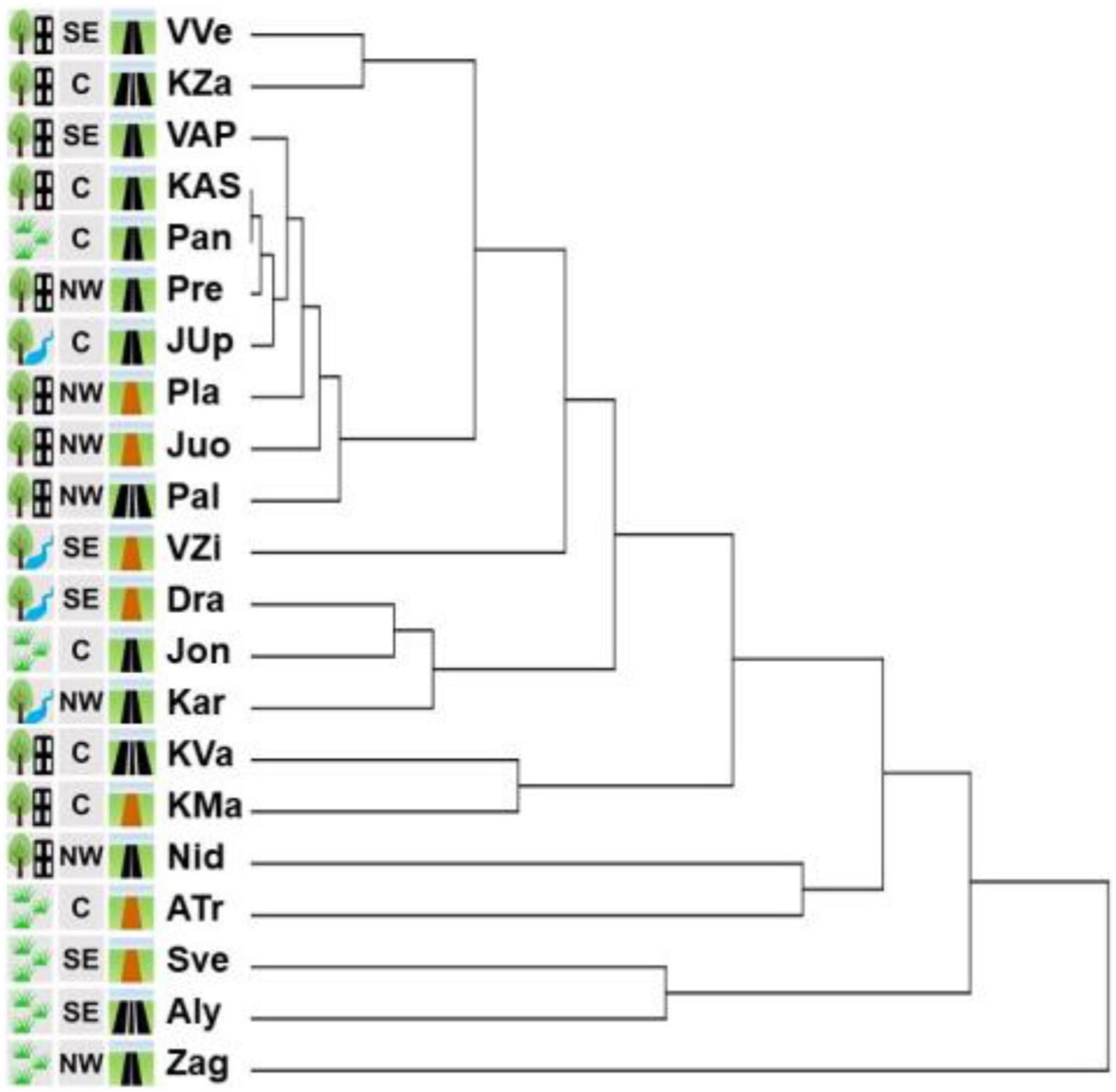
Figure 9. Cluster analysis of the similarity of the assemblages of herbaceous plant species in 21 sites with Impatiens parviflora. Three letters denote populations (location is provided in the Figure 1. Pictogram explanations are provided in the Table 1).
Clades of the 2nd–6th order (Aly, Sve, ATr, Nid, KMa, KVa, Kar, Jon, and DRa) encompassed the sites which had low I. parviflora coverage (mean 20%, range 8–30%), and the mean number of species in these sites was 23 (range 19–29). Clades of 7th–14th order included 10 sites of the most similar assemblages (Pal, Juo, Pla, JUp, Pre, Pan, KAS, VAP, KZa, and VVe) which had high I. parviflora coverage (mean 50%, range 30–70%) and the mean number of species was 17 (range 13–26). Neighboring clades encompassed the sites scattered geographically and located near the roads with different traffic intensity. Sites of the most distinct four clades (Zag, Aly, Sve, and ATr) belonged to the agrarian shrubland biotope, eight out of 10 sites of the 7th–14th order clades belonged to the urban forest biotope.
Assessing the abiotic environment of the sites with Ellenberg indicator values (W-EIV), the range of light (L) between the sites was 4.87–6.37 (Juo and Zag, respectively), temperature (T) – 5.20–6.00 (KZa and Juo, respectively), continentality (K) – 3.63–4.67 (Sve and JUp, respectively), soil moisture (F) – 4.81–6.30 (Nid and Kar, respectively), soil reaction (R) – 4.99–6.91 (KZa and VZi, respectively), and soil nutrients (N) – 5.78–7.22 (Pla and Aly, respectively) (Figure 10).
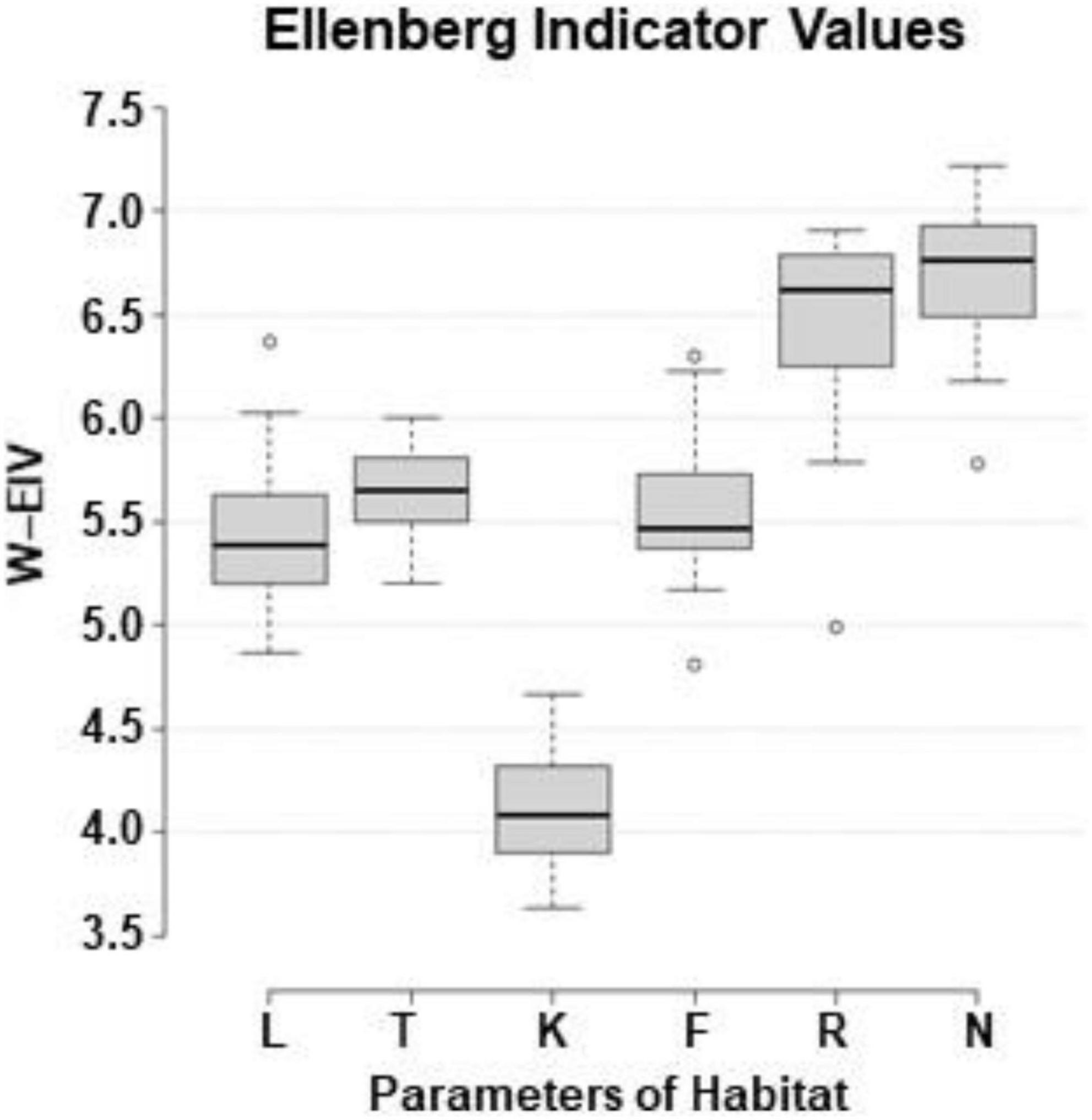
Figure 10. Box-and-whisker plots of weighted Ellenberg indicator values (W-EIV) of herbaceous species in the sites with Impatiens parviflora (W-EIV: L, light; T, temperature; K, continentality; F, soil moisture; R, soil reaction; N, soil nutrients). The central line of each box indicates the median value; the boxes, the lower (25%) and upper (75%) quartiles, and the whiskers, are from 10 to 90 percentiles (typical range), the points are outliers.
Interrelating parameters of the sites, PLP, coverage, and W-EIV data, Spearman correlations were calculated (Figure 11). The PLP-ISSR was positively correlated with the total coverage (covT) of herbaceous plant species (Rs = 0.44, P < 0.047). The covIP was positively correlated with covT (Rs = 0.45, P < 0.039), continentality (K) of the site (Rs = 0.58, P < 0.006), and was negatively correlated with the total number of herbaceous species (Rs = −0.23, P < 0.044), and light (L) in the site (Rs = −0.70, P < 0.001). Total number of herbaceous plant species showed positive correlation with light (Rs = 0.45, P < 0.042).
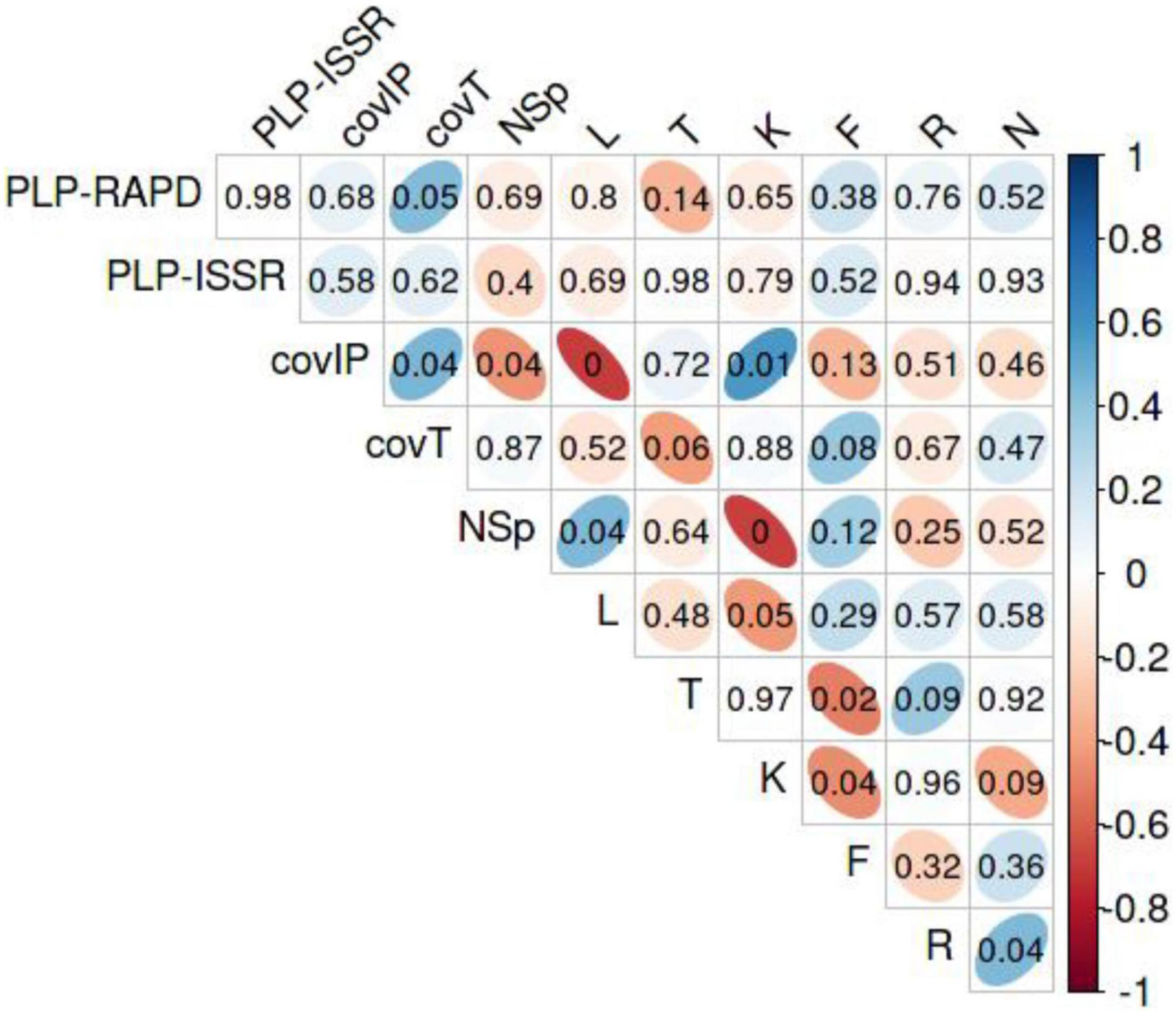
Figure 11. The correlogram represents the Spearman correlation coefficients (Rs) from –1 to 1 for all pairs of variables in the sites with Impatiens parviflora. Blue color represents positive and red color represents negative correlations. The intensity of the color indicates the strength of the correlation, so the stronger the correlation, the darker the figures. Correlations numbers inside colored figures indicate the level of significance, thin ellipsoid figures show significant (P < 0.05), figures toward spherical shape – insignificant correlations. PLP-RAPD, percentage of polymorphic loci at randomly amplified polymorphic DNA; PLP-ISSR, percentage of polymorphic loci at inter simple sequence repeats; covIP, coverage by I. parviflora; covT, total coverage by herbaceous plant species; NSp, number of herbaceous plant species; L, light; T, temperature; K, continentality; F, soil moisture; R, soil reaction; N, nutrients as weighted Ellenberg indicator values (W-EIV).
To explain the most important parameters for the variability of I. parviflora populations, principal component (PC) analysis was performed (Figure 12) on two genetic variables (PLP at ISSR loci and PLP at RAPD loci) and I. parviflora coverage, two biotic variables of I. parviflora environment (total coverage by herbaceous plant species, and total number of herbaceous species), and six abiotic variables of I. parviflora environment (W-EIV traits – light, temperature, continentality, moisture, acidity of the soil, and soil nutrients). The first four PCs were quite informative accounting for approximately 73.64% of the overall variance for the entire variables. For separate principal components PC1, PC2, PC3, and PC4 the variance (with eigenvalues) was: 30.08% (3.01), 18.96% (2.32), 14.35% (1.70), and 10.26% (1.06), respectively. PC1 variability was mainly caused by the number of herbaceous plant species (NSp), light (L), continentality (K), soil moisture (F) and I. parviflora coverage (covIP). The contribution of NSp, L, and F was positive while the contribution of K, and covIP was negative. For variability of PC2 PLP-RAPD, PLP-ISSR, the total coverage by herbaceous plant species (covT), soil moisture (F), temperature (T), and soil reaction (R) were the most important. The contribution of PLP-RAPD, PLP-ISSR, covT, and F was positive and the contribution of T, and R was negative. According to the variability of the parameters, displayed in the two principal component biplot by the vectors of different length, the importance of the variables (in descending order) in PC1 was as follows: covIP > L > K > NSp > F > T > N > PLP-ISSR > R > covT > PLP-RAPD. The order of importance of the variables in PC2 was slightly different: PC2: covT > T > F > R > PLP-RAPD > covIP > N > PLP-ISSR > L > NSp > K. Compared to the remaining sites, (1) the variables L, F, NSp, and covT had higher values and the variables T, R, K, and covIP had lower values for the sites Zag, ATr, DRa, Kar, Aly, and KVa; (2), the variables K, covT, and covIP had higher values, while the variables L, F, T, R, and NSp had lower values for the sites KZa, VAP, Pla, KAS, and Pal; (3) the variables T, R, K, and covIP had higher values and the variables L, F, NSp, and covT had lower values for the sites JUp, Juo, VVe, Pan, and Pre; (4) the variables L, F, T, R, and NSp had higher values, while the variables K, covT, and covIP had lower values for the sites Sve and Nid. The most extreme locations in the PC biplot were characteristic for the sites Kar, Pre, Nid, and Zag.
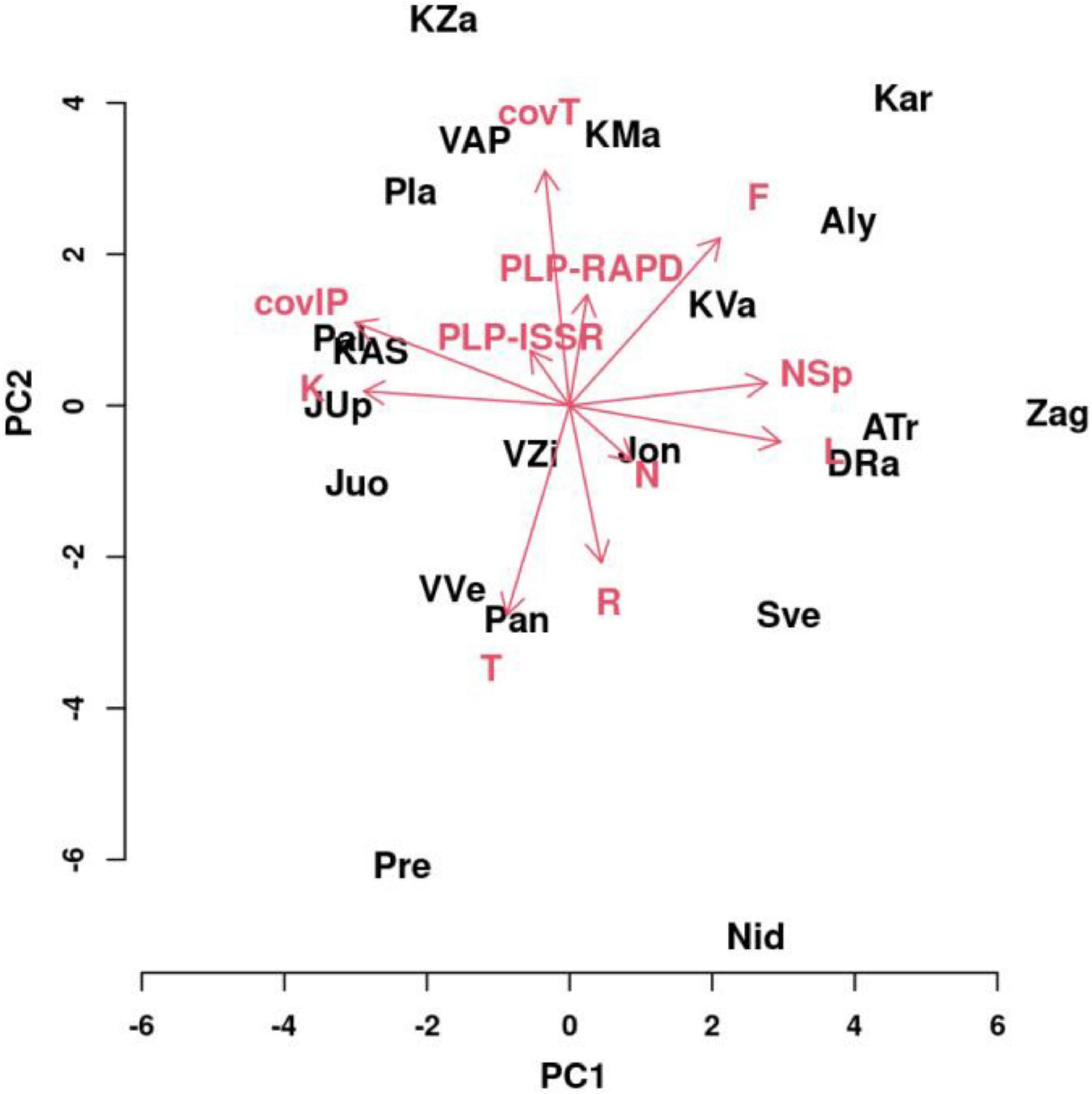
Figure 12. Biplot of the principal component analysis for the first two principal components of a model testing variation of populations depending on molecular markers (PLP at ISSR and RAPD loci), coverage of Impatiens parviflora, total coverage of herbaceous plant species, the number of herbaceous plant species, and abiotic environment features such as W-EIVs’ (L, light; T, temperature; K, continentality; F, moisture; R, pH; N, nutrients). Three black letters denote populations and red arrows with letters denote variables.
Discussion
Molecular Features of Impatiens parviflora Populations
Invasiveness of alien Impatiens parviflora was documented in various parts of Europe with 21% frequency of occurrence among alien plants (Wagner et al., 2017). The most comprehensive and numerous studies devoted exclusively for this species were performed in the countries of Central and Western Europe – Great Britain (Coombe, 1956), Germany (Trepl, 1984), Slovakia (Eliáš, 1999; Jarčuška et al., 2016), Poland (Obidziński and Symonides, 2000; Klimko and Piskorz, 2003; Chmura and Sierka, 2006; Komosińska et al., 2006; Chmura et al., 2007; Chmura, 2014; Reczyńska et al., 2015; Barabasz-Krasny et al., 2018; Grabowska et al., 2020), Belgium (Vervoort and Jacquemart, 2012), Hungary (Csiszar and Bartha, 2008; Csontos et al., 2012), Czechia (Hejda, 2012; Florianová and Münzbergová, 2017; Skálová et al., 2019), and summarized data for many aforementioned regions of the continent (Reczyńska et al., 2015). The first attempts to characterize populations of I. parviflora of the Baltic States were rather limited in terms of scope and quantification (Priede, 2009a; Dobravolskaitė, 2012). Congeneric for I. parviflora species, Impatiens glandulifera is also an annual plant existing in Europe already for the second century, known as top of widespread and worrying alien of the continent. Surprisingly, to date, molecular analyses of I. parviflora populations lag far behind I. glandulifera analyses performed in various countries by various types and sets of DNA markers (Walker, 2001; Provan et al., 2007; Walker et al., 2009; Zybartaite et al., 2011; Hagenblad et al., 2015; Jocienė, 2015; Nagy and Korpelainen, 2015; Cafa et al., 2020; Kurose et al., 2020; Korpelainen and Elshibli, 2021).
Based on successful application of SSR markers (Provan et al., 2007) for Lithuanian populations of I. glandulifera (Jocienė, 2015), analyses we employed the same simple sequence repeat markers for I. parviflora analyses. The decision was supported by the fact that SSR markers developed for the Impatiens lateristachys appeared to be applicable for the other four congeneric species I. oxyanthera, I. faberi, I. imbecilla, and I. rostellata (Li et al., 2008). In addition, such strategy was also based on the close position of Impatiens parviflora and Impatiens glandulifera in the Impatiens phylogenetic trees built on the basis of nDNA and cpDNA markers (Yuan et al., 2004; Janssens et al., 2006, 2007). In our study of I. parviflora no alleles were detected in case of one out of six SSR markers, four out of five remaining markers were monoallelic and one marker was diallelic (Kupcinskiene et al., 2013). According to the length of SSR locus (IGNSSR203-EF025994) I. parviflora populations in Lithuania were very similar by size of the amplified DNA fragments to those of I. glandulifera populations in Great Britain (Provan et al., 2007). The remaining loci were either shorter in several nucleotides (IGNSSR240-EF025997; IGNSSR101-EF025990) or longer (IGNSSR240-EF025997). Comparison of Impatiens parviflora and Impatiens glandulifera data of the same SSR loci revealed bigger numbers of alleles for populations of I. glandulifera examined in a similar scope study (Jocienė, 2015). Species specific structure of evaluated loci could explain the observed differences.
To compensate shortage of SSR markers for I. parviflora, inter simple sequence repeats (ISSRs) were chosen as SSR-related markers, located between microsatellite sites in the genome (Zietkiewicz et al., 1994). Possibly similar reasons for the selection of ISSR were in the numerous assessments of other Impatiens species (Hatcher et al., 2004; Gao et al., 2012; Li et al., 2013; Zhong et al., 2014), with the latest ISSR use approvement for analyses of genetic diversity on Impatiens (Tamboli et al., 2018).
Comparison of our I. parviflora molecular data (21 populations, 315 individuals) with I. glandulifera results obtained applying the same eight RAPD markers, examining similar numbers of individuals and populations of close geographical location (20 populations, 400 individuals) showed, that the percentage of polymorphisms at RAPD loci in I. parviflora populations (mean PLP 21.0; Figure 2) was twice lower than the I. glandulifera populations (mean PLP 46.1) (Zybartaite et al., 2011). The genetic diversity at ISSR loci in our populations of I. parviflora was even lower than that of RAPD loci (mean PLP – 16.5). Unique alleles were observed in I. parviflora populations located very close to Western (Pre, Nid, Kar, Pal), Northern (Zag), and Eastern borders (Sve) of the country. I. parviflora populations (KVa, KAS, KZa, and KMa) near the second largest city of Lithuania (Kaunas) were also characterized by unique alleles, whereas no unique alleles were found in the populations near the largest city (Vilnius) and the Southern border of the country. There are many factors that may determine the diversity level of populations. Absence of data about the native populations of I. parviflora does not allow to relate low genetic polymorphism at RAPD or ISSR loci either to species level features or population level fitness. Often, new populations are formed by a few individuals, which potentially lead to a decreased population-level genetic diversity as a result of genetic bottle necks and founder effects (Nei, 1978).
Although information on the diversity of I. parviflora populations at six SSR loci (Provan et al., 2007) was not obtained, application of other distinct marker systems (ISSR and RAPD) allowed us to reach the same conclusion about the possible multiple introductions of I. parviflora within the area of the investigation.
Data about I. parviflora performance in temperate climate forests of Europe (Chmura, 2014; Jarčuška et al., 2016) were complemented by our study on species behavior in hemiboreal forest zone (Figure 1). Levels of invasion of aliens have been found to increase with increasing proportion of urban land and proximity to roads (Wagner et al., 2021). In Latvia, I. parviflora was most commonly registered in the vicinity of roads in the semi-natural slightly disturbed forest (Priede, 2009b). Our study encompassed environmental characteristics of I. parviflora populations including several biotopes (urban forest, agricultural shrubland, and riparian forests, the main habitats in which this species usually grows in Lithuania) and traffic intensity differences along the road close to the site (Table 1). Species occurrence in Vilnius, 1934, was not supported by the botanical literature of the 1940s about species in the areas located west from Vilnius (toward the Baltic Sea). During the subsequent period of supposed intensive spread of I. parviflora, various socio-economic communications for nearly a half of century were limited to the Soviet Union. Due to this reason, geographical zonation within the country was also taken into account in our studies of invasion pathways. We determined genetic variation of I. parviflora in 21 sites across a longitudinal gradient (East-West) that might reflect the invasion history of the species within the country.
Presence of significant isolation of populations by distance (Mantel test) was documented only in case of RAPD loci (Figure 3). Based on ISSR and RAPD data, UPGMA clustering of populations did not reveal relations of populations neither to the geographical location nor to the traffic intensity along the road (Figure 4). In the case of RAPD loci based dendrogram, urban forest biotope was very rare among the most dissimilar populations and was prevailing among the most similar populations (three of 13 cases and seven of eight cases, respectively). With the exception of the Juo population, the populations (VVe, VAP) most similar at RAPD loci were sampled from the sites close to the biggest cities of Lithuania (Vilnius, Kaunas, and Panevėžys). The dendrograms of genetic relationships at ISSR and RAPD loci were similar according to the most distant populations: one of the three most distinct populations was located near the southern border of the country (DRa) while opposite results were detected for populations located near Vilnius (VVe).
An important factor influencing genetic diversity within the introduced range of invasive species is the number of introduction events (Bossdorf et al., 2005). The differences found among the population dendrograms built on RAPD and ISSR data could be partially explained by the distinct nature of RAPD and ISSR loci.
Unweighted pair group method with arithmetic mean cluster analyses at both ISSR and RAPD loci and the lack of correlation between genetic and geographic distances at ISSR loci (Figures 3, 4) suggest that the populations might have received genotypes from several donors. Hereby, our study supports the assumption that multiple introductions of invasive plants appear to be dominant pattern of species introduction (Hagenblad et al., 2015).
We used hierarchical AMOVA to group populations according to various environmental criteria to determine which of these factors would divide the populations into significant differentiation groups. Hierarchical analyses of molecular variance at both type markers’ loci revealed significant small extent differentiation (causing 6% of the total variation at ISSR loci and 2% of the total variation at RAPD loci) among population groups of distinct geographic zones (Figure 1 and Table 2). The least polymorphic at both ISSR and RAPD loci were populations located by the western border near the Baltic Sea (PLP 14.55 and 16.45, respectively). The geographical subdivision of our study populations into North-West, Central and South-East corresponded to the climatic zones based on several temperature parameters (average annual temperature, warmest and coldest months, absolute max and absolute min, annual precipitation, period with snow cover, and duration of sunshine) (Lithuanian Hydrometeorological Service under the Ministry of Environment, 2021). Hereby, significant differentiation among the populations of North-West, Central and South-East parts of the country might be related to climate effect. This observation was also supported by positive correlation of the covIP with continentality (K) of the site (Rs = 0.58, P < 0.006) (Figure 9).
Significant differentiation of population groups at one of the two markers loci was related to the road type (causing 2% of the total variation at ISSR loci), and biotope (causing 2% of the total variation at RAPD loci). Among population groups with different traffic intensity roads, the least polymorphic at ISSR loci (PLP 13.89) were I. parviflora populations located near the low intensity traffic road with a blacktop. The least polymorphic at RAPD loci (PLP 17.09) were I. parviflora population groups of riparian forest, when compared to population groups of urban forest or agrarian shrubland. The small among-region variance component implies that regions are only slightly differentiated from one another. Low differentiation among groups of populations belonging to distinct geographic zones was evident from PCoA plots (Figure 5). In agreement with AMOVA results were PCoA data, indicating the biggest differentiation among populations within groups, and much lower extent of differentiation among individuals within populations. For invasive species, low intrapopulation genetic differentiation may be due to a combination of genetic drift, founder effects, a short life span, autogamy or limited pollination or dispersal of seeds, small number of individuals in a population, and environmental variation (Barrett and Husband, 1990).
Based on the large number of genetic clusters identified by two different molecular markers (11 according to ISSR loci and 13 according to RAPD loci) (Figures 6, 7), Lithuanian I. parviflora populations highly differed from the number of gene clusters of the congeneric species Impatiens glandulifera populations sampled from three continents employing other molecular markers (three gene clusters at simple sequence repeats loci) (Hagenblad et al., 2015; Nagy and Korpelainen, 2015). Multiple introductions for European I. glandulifera were suggested based on numerous assessments (Hagenblad et al., 2015). We did not find relations between Bayesian genetic clusters and geography of the sites with I. parviflora, suggesting unrestricted geographically gene flow. Our Mantel test, PCoA and Bayesian clustering data (Figures 4–7) are in support for multiple introductions of I. parviflora. Since the naturalization of I. parviflora in all neighboring countries were registered earlier species might entered Lithuania from several places. We must point out three as the second most probable number of the gene clusters at ISSR loci (Figure 6) and this could be related to the probable external donors. In most cases the extreme locations in the biplot (Figure 12) of principal component analyses were characteristic for the populations close to the borders of the country, either western (Kar, Pre, Nid) or northern (Zag). Quite distant location in the biplot was also characteristic for the populations close to the eastern border (Sve) or the southern border (DRa). Therefore, our study of genetic diversity of I. parviflora populations indicates multiple introductions of this species in Lithuania. In addition, invaded areas might become re-colonized from the other sites due to high production of seeds (tens, hundreds up to 10,000 seeds might be produced annually by an individual) (Coombe, 1956; Trepl, 1984). Presence of two pairs of geographically close populations with the same prevailing gene cluster at two different loci (Jon and JUp, also KZa and KMa) (Figure 7) might be the evidence for occurrence of short distance unidirectional transport of seeds.
Indications Interrelating Invasiveness and Invasibility
Negative correlation (Figures 9, 11) between coverage of I. parviflora and herbaceous plant diversity of invaded forests supports the facts documented by other European forest studies (Obidziński and Symonides, 2000; Chmura and Sierka, 2006; Jarčuška et al., 2016). Coverage of I. parviflora was used as a measure of invader success (Jarčuška et al., 2016).
In the dendrogram of similarity of assemblages of herbaceous plant species in the sites with I. parviflora (Figure 9), with increasing similarity of the assemblages, I. parviflora coverage was also increased, whereas the number of herbaceous plant species in the sites (Figure 8) was decreased. Eight out of 10 the most similar sites according to herbaceous plant species belonged to urban forest. The dendrogram of similarity of assemblages of species was very similar to the genetic dendrogram (built for RAPD data) in terms of the most genetically similar populations (Figure 4B), among which seven of eight also belonged to urban forest. Based on our results, it might be concluded, that both the genetic diversity and herbaceous species diversity could be diminished in urban forest.
Biotope-related differences in association with environment conditions were noted among our study sites. Agricultural shrubs as the open areas with single trees and shrubs comprised numerous herbaceous plant species, especially those with the higher demand for light (Zag) and moisture. Riverside forests had higher numbers of species which require the soil of high humidity and fewer species with the higher demand for light than agrarian shrubs, but more than urban forests. Abundance of I. parviflora was highest in urban forests with the lowest number of herbaceous plant species with higher demand for light. In urban forests with I. parviflora signs of eutrophication were obvious, with the tree species such as Acer pseudoplatanum, Tilia cordata, Quercus robur, Fraxinus excelsior, and Ulmus glabra, which were typical for fairly fertile, non-acidic soils.
There are many studies which aimed to evaluate environment characteristics of European Impatiens parviflora based on EIV of neighboring herbaceous plant species (Chmura, 2014; Reczyńska et al., 2015; Jarčuška et al., 2016; Barabasz-Krasny et al., 2018). Despite difference in latitudes, defined by EIV (Figure 10) temperature conditions at our sites were similar to those in Poland (Reczyńska et al., 2015) or Slovakia (Jarčuška et al., 2016). Values of our I. parviflora sites for continentality, moisture and soil reaction were slightly higher compared to the aforementioned studies.
Impatiens parviflora is well-known as a species growing in semi-shaded, shaded habitats (Coombe, 1956; Trepl, 1984). Studies of I. parviflora with defined EIV were conducted in different biotopes with a wide variety of dominant tree species such as alders, poplars, willows, elms, oaks, beeches, fir, and fir-spruce forests (Chmura, 2014; Reczyńska et al., 2015; Jarčuška et al., 2016) which had different light conditions for understory herbaceous species. Our results coincide well with most these studies, although light demand was higher compared to those performed with underground cover of a tree layer ≥50% (Jarčuška et al., 2016). In support to many other studies, our data show that covIP was negatively correlated with light (L) in the site (Rs = −0.70, P < 0.001) (Figure 10).
Analyses of European woodlands disclosed the fact, that high soil nutrient levels are favorable factors for alien plant invasions (Wagner et al., 2017). Our results indicate that Lithuanian sites with I. parviflora might be distinguished by high levels of soil nutrients (Figure 8): the mean of Ellenberg indicator values (EIV) was very high (8 among possible top score 9) for the soil nutrients among 6 the most common herbaceous plant species growing near the I. parviflora (Ellenberg et al., 1992), while mean range for our populations was lower (6.49–6.93; Figure 10). Our results are in agreement with the positive association between nutrient (EIV) availability and I. parviflora coverage observed in understories in Western Carpathians (Jarčuška et al., 2016). In oak forests of Central Europe, I. parviflora was common in soils of medium or higher fertility and was absent or infrequent in less fertile areas (Reczyńska et al., 2015).
A wide range of information is available on genetic diversity and variability of invasive species at a geographic level (Meekins et al., 2001; Hagenblad et al., 2015). However, the interaction between genetic characteristics and neighboring species in sites has been poorly documented until now. Relationships between genetic differentiation and various environmental variables such as temperature, humidity, and nitrogen were documented for P. arundinacea within invasive distribution range (Kettenring et al., 2019). For a better interrelation of the genetic and community data, we conducted correlation and a principal component analyses (Figures 11, 12). In our study, the only case of significant relations was observed: the PLP-ISSR was positively correlated with total coverage (covT) of herbaceous plant species (Figure 11). We did not find significant relationships between I. parviflora molecular polymorphisms and site characteristics based on EIV of herbaceous plant species. Coverage by I. parviflora (covIP) was the most important variable of populations for the 1st principal component and total coverage by herbaceous plants (covT) was the most important variable for the 2nd principal component (PC2; Figure 12). Importance of molecular data for PC1 was less significant compared to weighed Ellenberg indicator values of herbaceous plant species for light, temperature, continentality, soil moisture, and nutrients. Importance of molecular data for PC2 was less compared to temperature, soil moisture, and soil reaction. According to the principal component analysis, the coverage by I. parviflora was more important variable of populations compared to molecular data or parameters of abiotic environment.
Conclusion
Use of non-indigenous species to study genetic variation is not only valuable in an evolutionary context, but might also be important for the management of invasion control and eradication. Complexities in human-nature interactions exist across multiple invasiveness and invasibility scales. The present study showed that the pervasion of I. parviflora in Lithuania was likely due to human-mediated activities, such as nature tourism, especially fishery, road building that are transporting seeds along road borders, which may also explain the lack of a relationship between genetic variation and neighboring species. Other molecular tools, including amplified fragment length polymorphism markers and chloroplast DNA, could also be used in conjunction with ISSR and RAPD data to present a more complete picture. Further studies of molecular, ecological, and geographic relationships of I. parviflora populations should examine sites across a broader range from Western Europe to Eastern part of Eurasia. This would help to evaluate the impact of multiple East-South Asian introductions of I. parviflora populations to Europe and provide a more extensive overview of I. parviflora genetic diversity in Europe.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
EKu conceived and designed the study. EKu, LJ, and RJ collected the material in the field. RJ, GR, and LJ obtained the laboratory data. TR, VM, EKr, and RJ did the statistical analyses and data visualization. RJ, LJ, EKu, EKr, VM, TR, and AP wrote the manuscript. All authors have read and agreed to the final version of the manuscript.
Funding
This study was funded by the Research Council of Lithuania, grant no. LEK-07/2012.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Barabasz-Krasny, B., Możdżeń, K., Sołtys-Lelek, A., and Stachurska-Swakoń, A. (2018). Biological Traits of Impatiens parviflora DC. under Different Habitat Conditions. Not. Bot. Horti Agrobot. Cluj Napoca 46, 277–285. doi: 10.15835/nbha46110970
Barrett, S. C., and Husband, B. C. (1990). Variation in outcrossing rates in Eichhornia paniculata: the role of demographic and reproductive factors. Plant Species Biol. 5, 41–55. doi: 10.1111/j.1442-1984.1990.tb00191.x
Bechtold, U., and Field, B. (2018). Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 69, 2753–2758. doi: 10.1093/jxb/ery157
Bossdorf, O., Auge, H., Lafuma, L., Rogers, W. E., Siemann, E., and Prati, D. (2005). Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144, 1–11. doi: 10.1007/s00442-005-0070-z
Braun-Blanquet, J. (1964). Pflanzensoziologie. Grundzüge der Vegetationskunde. 3. Aufl. New York: Springer.
Cafa, G., Baroncelli, R., Ellison, C. A., and Kurose, D. (2020). Impatiens glandulifera (Himalayan balsam) chloroplast genome sequence as a promising target for populations studies. PeerJ 8:e8739. doi: 10.7717/peerj.8739
Chmura, D. (2014). Biology and Ecology of an Invasion of Impatiens Parviflora DC. in Natural and Semi-Natural Habitats. Bialsko-Biela: Akademia Techniczno-Humanistyczna w Bielsku-Bialej.
Chmura, D., and Sierka, E. (2006). Relationships between invasive plant Impatiens parviflora and species richness of forest floor vegetation. Pol. J. Ecol. 54, 417–428.
Chmura, D., Sierka, E., and Orczewska, A. (2007). Autecology of Impatiens parviflora in natural forest stands. Bot. Stec. 11, 17–21.
Coombe, D. E. (1956). Biological flora of the British Isles, Impatiens parviflora DC. J. Ecol. 44, 701–713.
Costa, R., Pereira, G., Garrido, I., Tavares-de-Sousa, M. M., and Espinosa, F. (2016). Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify orchardgrass (Dactylis glomerata L.) germplasm variations. PLoS One 11:e0152972. doi: 10.1371/journal.pone.0152972
Cross, H. B., Lowe, A. J., and Gurgel, C. F. D. (2011). “DNA barcoding of invasive species,” in Fifty Years of Invasion Ecology: The Legacy of Charles Elton, ed. D. M. Richardson (Oxford: Wiley-Blackwell), 289–300.
Csiszar, A., and Bartha, D. (2008). “Small balsam (Impatiens parviflora DC.),” in The Most Important Invasive Plants in Hungary, eds Z. Botta-Dukat and L. Balogh (Vacratot: Institute of Ecology and Botany, Hungarian Academy of Sciences), 139–149.
Csontos, P., Chmura, D., and Sándor, A. (2012). Seed mass of biogeographically distinct populations of Impatiens parviflora DC. Tájökol. Lapok 10, 1–7.
Dobravolskaitė, R. (2012). Alien species Impatiens parviflora invasion into forest communities of Lithuania. Bot. Lith. 18, 3–12. doi: 10.2478/v10279-012-0001-y
Eliáš, P. (1999). Biological and ecological causes of invasion of Impatiens parviflora DC. into forest communities in Central Europe. Acta Hortic. 1, 1–3.
Ellenberg, H., Weber, H. E., Wirth, V., Werner, W., and Paulien, D. (1992). Zeigerwerte von Pflanzen in Mitteleuropa. Scripta Geobot. 18, 1–258.
European Parliament (2014). Regulation (EU) No 1143/2014 of the European Parliament and of the Council of 22 October 2014 on the prevention and management of the introduction and spread of invasive alien species. Available online at: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX:32014R1143 (Accessed October 18, 2021).
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. doi: 10.1177/117693430500100003
Falush, D., Stephens, M., and Pritchard, J. K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578. doi: 10.1111/j.1471-8286.2007.01758.x
Falush, D., Stephens, M., and Pritchard, K. J. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1093/genetics/164.4.1567
Florianová, A., and Münzbergová, Z. (2017). Invasive Impatiens parviflora has negative impact on native vegetation in oak-hornbeam forests. Flora 226, 10–16. doi: 10.1016/j.flora.2016.11.006
Gao, W. J., Su, J. X., Xie, L., Deng, C. L., Zhang, T., and Lu, L. D. (2012). The point mutation induced by the low-energy N+ ion implantation in Impatiens balsamine genome. Russ. J. Genet. 48, 1009–1014. doi: 10.1134/S1022795412070034
Garrido-Cardenas, J. A., Mesa-Valle, C., and Manzano-Agugliaro, F. (2018). Trends in plant research using molecular markers. Planta 247, 543–557. doi: 10.1007/s00425-017-2829-y
Grabowska, K. Wróbel, D. Żmudzki, P., and Podolak, I. (2020). Anti-inflammatory activity of saponins from roots of Impatiens parviflora DC. Nat. Prod. Res. 34, 1581–1585.
Grover, A., and Sharma, P. C. (2016). Development and use of molecular markers: past and present. Crit. Rev. Biotechnol. 36, 290–302. doi: 10.3109/07388551.2014.959891
Hagenblad, J., Hülskötter, J., Acharya, K. P., Brunet, J., Chabrerie, O., Cousins, S. A. O., et al. (2015). Low genetic diversity despite multiple introductions of the invasive plant species Impatiens glandulifera in Europe. BMC Genet. 16:103. doi: 10.1186/s12863-015-0242-8
Handley, L. J. L., Estoup, A., Evans, D. M., Thomas, C. E., Lombaert, E., Facon, B., et al. (2011). Ecological genetics of invasive alien species. BioControl 56, 409–428. doi: 10.1007/s10526-011-9386-2
Hatcher, P. E., Wilkinson, M. J., Albani, M. C., and Hebbern, C. A. (2004). Conserving marginal populations of the food plant (Impatiens noli-tangere) of an endangered moth (Eustroma reticulatum) in a changing climate. Biol. Conserv. 116, 305–317. doi: 10.1016/S0006-3207(03)00200-3
Haubrock, P. J., Turbelin, A. J., Cuthbert, R. N., Novoa, A., Taylor, N. G., Angulo, E., et al. (2021). Economic costs of invasive alien species across Europe. NeoBiota 67, 153–190. doi: 10.3897/neobiota.67.58196
Hejda, M. (2012). What is the impact of Impatiens parviflora on diversity and composition of herbal layer communities of temperate forests? PLoS One 7:e39571. doi: 10.1371/journal.pone.0039571
Janssens, S., Geuten, K., Viaene, T., Yuan, Y. M., Song, Y., and Smets, E. (2007). Phylogenetic utility of the AP3/DEF K-domain and its molecular evolution in Impatiens (Balsaminaceae). Mol. Phylogenet. Evol. 43, 225–239. doi: 10.1016/j.ympev.2006.11.016
Janssens, S., Geuten, K., Yuan, Y. M., Song, Y., Küpfer, P., and Smets, E. (2006). Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) using chloroplast atpB-rbcL spacer sequences. Syst. Bot. 31, 171–180. doi: 10.1600/036364406775971796
Jarčuška, B., Slezák, M., Hrivnák, R., and Senko, D. (2016). Invasibility of alien Impatiens parviflora in temperate forest understories. Flora 224, 14–23. doi: 10.1016/j.flora.2016.06.005
Jocienė, L. (2015). Genetic diversity of invasive populations of himalayan balsam (Impatiens glandulifera Royle). Ph.D. thesis. Kaunas: Vytautas Magnus University.
Kettenring, K. M., Menuz, D. R., and Mock, K. E. (2019). The nativity and distribution of the cryptic invader Phalaris arundinacea (reed canarygrass) in riparian areas of the Columbia and Missouri River Basins. Wetlands 39, 55–66. doi: 10.1007/s13157-018-1074-x
Klimko, M., and Piskorz, R. (2003). The usefulness of retrospective research along the road-forest border zone for reconstruction of invasion of oak-hombeam forest communities by Impatiens parviflora DC. Ecological Questions. Int. J. Controvers. Probl. Ecol. 3, 63–69.
Komosińska, E., Wòdkiewicz, M., Jarzyna, I., Jarochowska, E., Milanowski, R., Chwedorzewska, K., et al. (2006). Some attempts to detect genetic differences between populations of small balsam (Impatiens parviflora DC.). Biodivers. Res. Conserv. 3–4, 245–247.
Korpelainen, H., and Elshibli, S. (2021). Assessment of genetic relationships among native and introduced Himalayan balsam (Impatiens glandulifera) plants based on genome profiling. Ecol. Evol. 11, 13295–13304. doi: 10.1002/ece3.8051
Kupcinskiene, E., Zybartaite, L., Janulioniene, R., Zukauskiene, J., and Paulauskas, A. (2013). Molecular diversity of small balsam populations in relation to site characteristics. Cent. Eur. J. Biol. 8, 1048–1061. doi: 10.2478/s11535-013-0228-3
Kurose, D., Pollard, K. M., and Ellison, C. A. (2020). Chloroplast DNA analysis of the invasive weed, Himalayan balsam (Impatiens glandulifera), in the British Isles. Sci. Rep. 10:10966. doi: 10.1038/s41598-020-67871-0
Langmaier, M., and Lapin, K. (2020). A Systematic Review of the Impact of Invasive Alien Plants on Forest Regeneration in European Temperate Forests. Front. Plant Sci. 11:1349. doi: 10.3389/fpls.2020.524969
Lazzaro, L., Sarracco, E., Benesperi, R., and Coppi, A. (2018). A probable anthropic origin of Nerium oleander L. (Apocynaceae) population in Montecristo island (Italy, Tuscany): evidence from loci polymorphism and ISSR analysis. Caryologia 71, 50–57. doi: 10.1080/00087114.2017.1410634
Li, L. F., Chen, G. Q., Yuan, Y. M., and Ge, X. J. (2008). Development of thirteen microsatellite loci for Impatiens lateristachys (Balsaminaceae). Conserv. Genet. 9, 439–441. doi: 10.1007/s10592-007-9325-z
Li, S. L., Vasemägi, A., and Ramula, S. (2016). Genetic variation and population structure of the garden escaper Lupinus polyphyllus in Finland. Plant Syst. Evol. 302, 399–407. doi: 10.1007/s00606-015-1273-3
Li, X. J., Tang, W. X., and Huang, S. X. (2013). ISSR analysis for genetic diversity of Impatiens macrovexilla in Guilin. Guangdong Agric. Sci. 40, 142–144. doi: 10.16768/j.issn.1004-874x.2013.13.056
Lithuanian Hydrometeorological Service under the Ministry of Environment (2021). Lithuanian Hydrometeorological Service under the Ministry of Environment. Available online at: http://www.meteo.lt/en/web/guest/climate-regions-of-lithuania (Accessed December 28, 2021)
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220.
Marozas, V. (2014). Effect of the coniferous forest-grassland edge on ground vegetation in the mixed European forest zone, Lithuania. Dendrobiology 71, 15–22. doi: 10.12657/denbio.071.002
Marozas, V., Cekstere, G., Laivins, M., and Straigyte, L. (2015). Comparison of neophyte communities of Robinia pseudoacacia L. and Acer negundo L. in the eastern Baltic Sea region cities of Riga and Kaunas. Urban For. Urban Green. 14, 826–834. doi: 10.1016/J.UFUG.2015.08.003
McCune, B., and Mefford, M. J. (2011). PC-ORD. Multivariate analysis of Ecological Data, Version 6.0. Oregon: MJM Software.
Meekins, J. F., Ballard, H. E. Jr., and McCarthy, B. C. (2001). Genetic variation and molecular biogeography of a North American invasive plant species (Alliaria petiolata, Brassicaceae). Int. J. Plant Sci. 162, 161–169. doi: 10.1086/317903
Nagy, A. M., and Korpelainen, H. (2015). Population genetics of Himalayan balsam (Impatiens glandulifera): comparison of native and introduced populations. Plant Ecol. Divers. 8, 317–321. doi: 10.1080/17550874.2013.863407
Natkevicaite-Ivanauskiene, M. (1951). Adventive flora of Lithuanian Soviet Republic. Proc. Inst. Biol. 1, 77–124.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590. doi: 10.1093/genetics/89.3.583
Nybom, H., Weising, K., and Rotter, B. (2014). DNA fingerprinting in botany: past, present, future. Investig. Genet. 5, 1–35. doi: 10.1186/2041-2223-5-1
Obidziński, T., and Symonides, E. (2000). The influence of the groundlayer structure on the the invasion of small balsam (Impatiens parviflora DC.) to natural and degraded forests. Acta Soc. Bot. Pol. 69, 311–318. doi: 10.5586/asbp.2000.041
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Priede, A. (2009b). Invasive neophytes in the flora of Latvia: distribution and dynamics. Ph.D. thesis. Riga: University of Latvia.
Priede, A. (2009a). Distribution of some invasive alien plant species in riparian habitats in Latvia. Bot. Lith. 14, 137–150.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Provan, J. I. M., Love, H. M., and Maggs, C. A. (2007). Development of microsatellites for the invasive riparian plant Impatiens glandulifera (Himalayan balsam) using intersimple sequence repeat cloning. Mol. Ecol. Notes 7, 451–453. doi: 10.1111/j.1471-8286.2006.01614.x
R Core Team (2021). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reczyńska, K., Świerkosz, K., and Dajdok, Z. (2015). The spread of Impatiens parviflora DC. in Central European oak forests – another stage of invasion? Acta Soc. Bot. Pol. 84, 401–411. doi: 10.5586/asbp.2015.039
Schmidt, O., Padel, S., and Levidow, L. (2012). The bio-economy concept and knowledge base in a public goods and farmer perspective. Bio-based Appl. Econ. 1, 47–63. doi: 10.13128/BAE-10770
Skálová, H., Moravcová, L., Čuda, J., and Pyšek, P. (2019). Seed-bank dynamics of native and invasive Impatiens species during a five-year field experiment under various environmental conditions. NeoBiota 50, 75–95. doi: 10.3897/neobiota.50.34827
StatSoft Inc (2004). STATISTICA (Data Analysis Software System). Version, 7. Tulsa, OK: StatSoft Inc, 1984–2004.
Tamboli, A. S., Dalavi, J. V., Patil, S. M., Yadav, S. R., and Govindwar, S. P. (2018). Implication of ITS phylogeny for biogeographic analysis, and comparative study of morphological and molecular interspecies diversity in Indian Impatiens. Meta Gene 16, 108–116.
Tanner, R. (2008). Datasheets: Impatiens parviflora (small balsam). Available online at: http://www.cabi.org/isc/datasheet/28768 (Accessed October 18, 2021)
The Ministry of Environment of the Republic of Lithuania (2015). List of Invasive Species in Lithuania, Approved by the Minister of Environment of the Republic of Lithuania, Order No. D1-433 of 27 August 2004. Register Of Legal Acts, 2015-07-20, No. 11487. Available online at: https://www.e-tar.lt/portal/lt/legalAct/TAR.7B6390A69C91/asr (Accessed December 28, 2021)
Tokarska-Guzik, B. (2005). “Invasive ability of kenophytes occurrig in Poland: a tenative assesment,” in Biological Invasions – From Ecology to Control, eds W. Nentwig, S. Bacher, M. Cock, H. J. Dietz, A. Gigon, and R. Wittenberg (Berlin: NEOBIOTA), 47–65.
Trepl, L. (1984). Über Impatiens parviflora DC. als Agriophyt in Mitteleuropa. Diss. Bot. 73, 1–400.
Vervoort, A., and Jacquemart, A. L. (2012). Habitat overlap of the invasive Impatiens parviflora DC. with its native congener I. noli-tangere L. Phytocoenologia 42, 249–257. doi: 10.1127/0340-269X/2012/0042-0496
Vyšniauskienė, R., Rancelienė, V., Žvingila, D., and Patamsytė, J. (2011). Genetic diversity of invasive alien species Lupinus polyphyllus populations in Lithuania. Zemdirbyste Agric. 98, 383–390. doi: 10.3390/biology10111094
Wagner, V., Chytrý, M., Jiménez-Alfaro, B., Pergl, J., Hennekens, S., Biurrun, I., et al. (2017). Alien plant invasions in European woodlands. Divers. Distrib. 23, 969–981. doi: 10.1111/ddi.12592
Wagner, V., Večeřa, M., Jiménez-Alfaro, B., Pergl, J., Lenoir, J., Svenning, J. C., et al. (2021). Alien plant invasion hotspots and invasion debt in European woodlands. J. Veg. Sci. 32:e13014. doi: 10.1111/jvs.13014
Walker, N. F. (2001). Population Genetic Analysis of Two Species of Non-Indigenous Riparian Weeds in Northeast England in the Context of Their Spatial Ecology. Doctoral dissertation. Durham: Durham University.
Walker, N. F., Hulme, P. E., and Hoelzel, A. R. (2009). Population genetics of an invasive riparian species, Impatiens glandulifera. Plant Ecol. 203, 243–252.
Wang, C. (2021). Grand challenges in environmental systems engineering. Front. Environ. Sci. 9:809627. doi: 10.3389/fenvs.2021.809627
Wei, T., and Simko, V. (2021). R package ‘corrplot’: Visualization of a Correlation Matrix (Version 0.90). Available online at: https://github.com/taiyun/corrplot (Accessed December 20, 2021).
Wisnewski, P. (1934). Nienotowane stanowiska kilku rzadszych krajowych i zawleczonych roslin w Wilensczyznie. Acta Soc. Bot. Pol. 11, 207–211. doi: 10.5586/asbp.1934.051
Yasui, Y. (2020). History of the progressive development of genetic marker systems for common buckwheat. Breed. Sci. 70, 13–18. doi: 10.1270/jsbbs.19075
Younis, A., Ramzan, F., Ramzan, Y., Zulfiqar, F., Ahsan, M., and Lim, K. B. (2020). Molecular markers improve abiotic stress tolerance in crops: a review. Plants 9:1374. doi: 10.3390/plants9101374
Yuan, Y. M., Song, Y., Geuten, K., Rahelivololona, E., Wohlhauser, S., Fischer, E., et al. (2004). Phylogeny and biogeography of Balsaminaceae inferred from ITS sequence data. Taxon 53, 391–403. doi: 10.2307/4135617
Zhong, Y. F., Hu, X. Y., Song, X. Q., and Zhou, Z. D. (2014). ISSR analysis on population genetic diversity of Impatiens hainanensis Balsaminaceae, an endemic species to Hainan island. Chin. J. Trop. Crop. 35, 1041–1046.
Zietkiewicz, E., Rafalski, A., and Labuda, D. (1994). Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20, 176–183. doi: 10.1006/geno.1994.1151
Keywords: small balsam, genetic diversity, ISSR, Balsaminaceae, alien plants, herbaceous plants, phytosociology
Citation: Krokaitė E, Janulionienė R, Jocienė L, Rekašius T, Rajackaitė G, Paulauskas A, Marozas V and Kupčinskienė E (2022) Relating Invasibility and Invasiveness: Case Study of Impatiens parviflora. Front. Ecol. Evol. 10:845947. doi: 10.3389/fevo.2022.845947
Received: 30 December 2021; Accepted: 21 February 2022;
Published: 01 April 2022.
Edited by:
Damian Chmura, University of Bielsko-Biała, PolandReviewed by:
Ádám Lovas-Kiss, Institute of Ecology Research Center, Hungarian Academy of Sciences, HungaryEduardo Cires, University of Oviedo, Spain
Copyright © 2022 Krokaitė, Janulionienė, Jocienė, Rekašius, Rajackaitė, Paulauskas, Marozas and Kupčinskienė. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edvina Krokaitė, ZWR2aW5hLmtyb2thaXRlQGdtYWlsLmNvbQ==
 Edvina Krokaitė
Edvina Krokaitė Rasa Janulionienė
Rasa Janulionienė Lina Jocienė
Lina Jocienė Tomas Rekašius
Tomas Rekašius Giedrė Rajackaitė
Giedrė Rajackaitė Algimantas Paulauskas
Algimantas Paulauskas Vitas Marozas
Vitas Marozas Eugenija Kupčinskienė
Eugenija Kupčinskienė