- Department of Natural and Life Sciences, The Open University of Israel, Ra’anana, Israel
Nuclear pseudogenes of mitochondrial origin (numts) are common in all eukaryotes. Our previous scan of numts in sequenced nuclear genomes suggested that the highest numt content currently known in animals is that in the gray short-tailed opossum. The present work sought to determine numt content in marsupials and to compare it to those in placental and monothematic mammals as well as in non-mammalian vertebrates. To achieve this, 70 vertebrate species with available nuclear and mitochondrial genomes were scanned for numt content. An extreme numt content was found in the Dasyuridae, with 3,450 in Sarcophilus harrisii (1,955 kb) and 2,813 in Antechinus flavipes (847 kb). The evolutionarily closest species analyzed, the extinct Thylacinus cynocephalus belonging to the Thylacindae family, had only 435 numts (238 kb). These two Dasyuridae genomes featured the highest numt content identified in animals to date. A phylogenetic analysis of numts longer than 300 bp, using a Diprotodonita mitochondrial tree, indicated a burst of numt insertion that began before the divergence of the Dasyurini and Phascogalini, reaching a peak in the early evolution of the two tribes. No comparable increase was found in the early divergent species T. cynocephalus. Divergence of the Dasyuridae tribes has been previously dated to shortly after the Miocene climate transition, characterized by a rapid temperature decline. Interestingly, deviation from optimal growth temperature is one of the environmental factors reported to increase numt insertions in a laboratory setting.
Introduction
Mitochondria are descended from free-living proteobacteria. Mitochondria encode only a small portion of the genes (3–60) needed for mitochondrial function (Timmis et al., 2004), while the majority of mitochondrial proteins are encoded on genes in the nuclear genome and their products are then targeted into the organelles (Meisinger et al., 2008; Kleine et al., 2009). This reduction in gene number encoded within the mitochondria is explained by the occurrence of endosymbiotic gene transfer (EGT): the transfer of genes from the organelle ancestor to the nuclear genome during evolution (Allen, 2015). EGT also explains the transfer of genes from a plastid ancestor, a cyanobacterium, to the nuclear genome in plants.
The transfer of DNA from the organelles to the nuclear genome is an ongoing process and occurs in almost all eukaryote genomes (reviewed in Hazkani-Covo et al., 2010). These nuclear sequences of mitochondrial origin (numts), ranging in size from a few bases to almost a full organelle, are now recognized as typical components of eukaryotic genomes (Mourier et al., 2001; Timmis et al., 2004; Kleine et al., 2009; Hazkani-Covo et al., 2010). A recent analysis of next generation sequencing demonstrated that numts constitute an important component of human and primate genomic variation, both through normal evolution (Dayama et al., 2014, 2020) and through cancer dynamics (Ju et al., 2014, 2015).
Although numts can be identical to the current mitochondrion or resemble an older one, due to the differences in the genetic code between the nuclear and mitochondrial genomes, metazoan numts are considered “dead on arrival” pseudogenes (Bensasson et al., 2001). The time of their insertion can be dated using either genome alignment (Hazkani-Covo and Graur, 2007; Hazkani-Covo, 2009) when dealing with very close genomes or, for more distant genomes, by dating numt insertion based on a mitochondrial phylogenetic tree (Hazkani-Covo et al., 2003, 2010). The latter analysis is only possible where the mitochondrial rate of evolution is lower than the nuclear rate of evolution.
The rate of evolution varies significantly between the nuclear and mitochondrial genomes in animals, with a mitochondrial rate about 10-fold higher than the nuclear one (Brown et al., 1979, 1982; Haag-Liautard et al., 2008). Although numts are not functional elements, their rate decreases when they are integrated in the nuclear genome and they are therefore considered “mitochondrial fossils” that can resemble ancient mitochondria (Perna and Kocher, 1996; Zhang and Hewitt, 1996). The similarity of numts to mitochondria challenges mitochondrial research with numts occasionally misidentified as bone fide mitochondrial DNA (Hazkani-Covo et al., 2010) in mitochondrial (Wallace et al., 1997), ancient DNA (Collura and Stewart, 1995) and taxonomic studies (Song et al., 2008). It is therefore important to identify and map numts in genomes in order to distinguish them from mitochondrial variations.
Our current knowledge of the mechanism of numt insertion is only partial. It has been shown that they are integrated into DNA double-strand breaks in the nuclear genome via the non-homologous end joining mechanism (Blanchard and Schmidt, 1996; Ricchetti et al., 1999, 2004; Hazkani-Covo and Covo, 2008). Several conditions have been shown to increase numt insertion rate, of which aging is the best established (Wright and Cummings, 1983; Richter, 1988; Caro et al., 2010; Cheng and Ivessa, 2010, 2012). Environmental conditions are also associated with increased mitochondrial transfer into the nuclear genome, with a change of temperature as a contributing factor. Thorsness and Fox (1990) established a selection system in yeast to examine DNA escape from the mitochondria to the nucleus. Cells that were grown at non-optimal temperature showed an increased rate of escape. Similarly, in tobacco plants, the numt (as well as nupts—nuclear sequences of plastid origin) rate of transfer from the organelles increased following only a mild heat stress (Wang et al., 2012a,b).
The number of numts varies significantly among genomes, ranging from only a few bases to hundreds of thousands of bases (Hazkani-Covo et al., 2010). The reason for this variation is poorly understand, as although larger genomes tend to have more numts, species with similar genome size can differ greatly in numt content. Several animals have been identified to date as possessing a high content, including insects (e.g., grasshoppers—Song et al., 2014; and honeybees—Behura, 2007; Pamilo et al., 2007). Our previous survey of numt content in 85 genomes suggested that the gray short-tailed opossum is the animal with the highest numt content known at present (Hazkani-Covo et al., 2010). Since the latter survey, many genomes, including those of several marsupials, have been sequenced. In the present work, the numt content in 70 vertebrate genomes was studied to identify cases with extreme content, with specific emphasis on marsupials. Specifically, while the previous analysis related only to the short-tailed opossum (belonging to the order Didelphimorphia), the present analyses incorporated eight marsupials from four of the seven marsupial orders (Microbiotheria, Didelphimorphia, Diprotodontia and Diprotodontia) for which both nuclear and mitochondrial genomes are available.
Methods
Data
The genomes of 70 vertebrates with available mitochondria from the National Center for Biotechnology Information organelle dataset and corresponding nuclear genomes were downloaded. The list comprised a lamprey, 20 bony fish, 3 amphibians, 3 turtles, 3 lizards, 14 birds, 8 marsupial mammals, 2 monothematic mammals, and 16 placental mammals. Accession numbers are provided in Supplementary Table 1.
Identification of Nuclear Pseudogenes of Mitochondrial Origins
The mitochondrial genome was BLASTed against the nuclear genome using BLASTN with an E-value of 0.0001 (Camacho et al., 2009). Numts were inferred using a permissive concatenation (Hazkani-Covo and Martin, 2017) with a nuclear distance of up to 500 bp. To achieve this, BLAST hits were concatenated regardless of the position and orientation of hits on the mitochondria, with only distances between nuclear coordinates being scored. Other concatenation distances and stringency are provided in Supplementary Table 1. Numts are considered simple if they are composed of one BLAST hit and complex if composed of two or more BLAST hits.
Phylogenetic Analysis and Identification of Accumulation Rate
Twenty-nine fully sequenced mitochondrial sequences were selected for phylogenetic analysis and aligned using MAFFT (Katoh et al., 2009). They comprised 10 Dasyuromorphia species, 12 Diprotodonita species, and seven Didelphimorphia species. The taxa were chosen on the basis of complete-mitochondrial sequence availability and the possibility of constructing a taxonomically valid phylogenetic tree. First, to date the numt accumulation, MAFFT was used to align each numt above 300 bp to the mitochondria alignment (using the—addfragment flag).
Each numt was then added to each of the branches on the lineage leading to either Sarcophilus harrisii, Antechinus flavipes or Thylacinus cynocephalus genomes, and RAxML with GTR gamma was used to compute the likelihood of appearance of each of the resulting trees (Tavaré, 1986; Stamatakis, 2014). Two scores were calculated: (1) the best score was the number of times that each of the trees emerged as the most likely tree; and (2) the weighted score was calculated as follows: if the likelihood of the best tree significantly differed from the other trees by a log likelihood test (S-H test, Shimodaira and Hasegawa, 1999) with p < 0.05, the tree was given a score of 1. If two trees could not be shown to differ from one another, each of the two trees was given a score of 0.5. Similarly, if three trees could not be shown to differ from each other, each of them was given a score of 0.33, and so on.
For each of the trees, scores for all tested numts were summed. The analysis was performed separately for S. harrisii with seven user trees; A. flavipes with eight trees; and T. cynocephalus with three possible trees. For complex numts, each fragment longer than 300 bp was mapped separately and best assignment was determined only if all fragments agreed. Similarly, for weighted scores, only phylogenetic placements shared by all fragments of the numt were considered for score calculation.
Numt accumulation rate was calculated by dividing the number of numts mapped to a specific branch by the mitochondrial tree branch length. An outlier was identified using MATLAB “isoutlier” function, as a value that constituted more than three scaled median absolute deviations from the median.
Identification of Duplicated Numts
To identify numts within a genome that are the result of post insertion duplications rather than mitochondrial insertions a duplicated flanking region analysis was preformed (Liang et al., 2018; Wang et al., 2020). All-against-all BLAST search of numts longer than 300 bp within a genome with a flanking region of 200 bp from each side was performed. Numts identified as matching with another numt, with at least 50 bp from each side of the two, were marked as possible duplication. These numts were subtracted from the overall pool and insertion rate was calculated.
Results
Nuclear Pseudogenes of Mitochondrial Origin Content Varied Dramatically, With the Highest Number Found in Dasyuromorphia
Simple and complex numts were inferred in 70 vertebrates. First, the full mitochondrial genome was BLASTed against the nuclear genomes. BLAST hits were then concatenated to the inferred full numts using a concatenation distance of 500 bp and a permissive strategy (Hazkani-Covo and Martin, 2017). BLAST hits were thus concatenated depending on nuclear distance, allowing non-linear organelle hits and hits on both nuclear strands (Table 1). Inferred numt number together with other concatenation distances and stringencies are provided in Supplementary Table 1. The number of numts identified in a genome and the coverage of identified numts varied from none to thousands of events and up to 1,955 kb. The majority of identified numts were simple and comprised only one BLAST fragment per numt. Complex numts composed of several BLAST hits that are not continuous with organelle DNA were, however, very common (Hazkani-Covo and Martin, 2017). For example, in seven species of placental mammals that featured at least 100 numts, 20% or more of these were complex: Felis catus genome (31%), Papio anubis (22%), Physeter catodon (38%), and Sus scrofa (27%); in marsupials: Tursiops truncatus (39%), and the extinct Thylacinus cynocephalus (51%); and one lizard, Lacerta agilis (20%). When considering the smaller numt number of an additional nine birds, lizards, and turtles with at least 20 numts, 20% or more of these were complex.
Figure 1 presents the number of numts as a function of genome size on a log-log scale. The species are color coded: placental mammals, marsupial mammals, monotremata mammals, birds, turtles, lizards, amphibians, bony fish, and a lamprey. The data reveal a strong correlation between genome size and total number of inferred numts (Spearman non-parametric rho = 0.7838, P = 1.0347e-15), with the highest numt number found in marsupial and placental mammals. The highest overall number of numts was identified in the marsupial order Dasyuromorphia, with extreme numbers belonging to the Tasmanian devil, Sarcophilus harrisii (3,450) and the yellow-footed antechinus, Antechinus flavipes (2,813). The number of inferred numts in S. harrisii was fourfold higher than the species with the next highest number (except A. flavipes). S. harrisii also showed an extreme numt base count in the nuclear genome, with 1,955 kb, about 2.3-fold that of A. flavipes (847 kb).
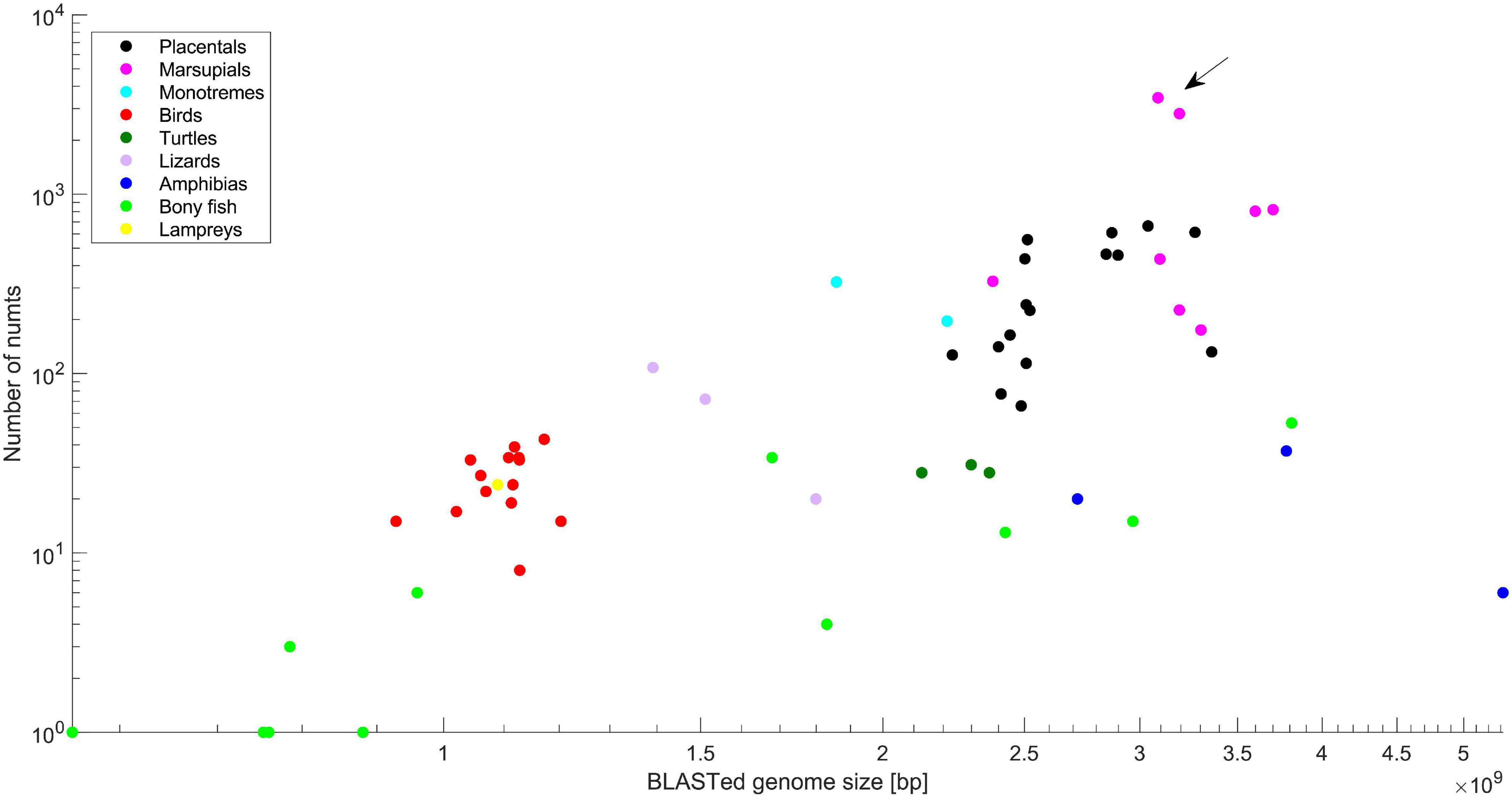
Figure 1. A log–log scale graph showing the dependency between inferred number of numts and BLASTed genome size. Species are color-coded. The extreme numt content in Dasyuridae is indicated by an arrow.
The gray short-tailed opossum (Monodelphis domestica) genome previously identified as possessing 2,093 kb of numts is reassessed here to possess 804 numts, 104 of which are complex, with a genome coverage of 752 kb. While this coverage is among the highest identified, it is smaller than that identified in 2010. This was predicted, as most numts identified previously were not mapped to chromosomes (Hazkani-Covo et al., 2010). In the current analysis, based on a recent genome version, only a minority of M. domestica numts (5%) were not mapped to chromosomes.
Nuclear Pseudogenes of Mitochondrial Origin Insertion Rate Increased Before the Dasyurini and Phascogalini Split, Reaching a Peak During Early Dasyurini and Phascogalini Evolution
The extreme numt numbers identified in the Dasyuridae were further studied to identify the dynamics of their insertion rate. The order Dasyuromorphia is second to Diprotodonita in the number of Australian and New Guinean marsupial species, with about 80 known species (Westerman et al., 2016; Kealy and Beck, 2017). The order comprises three families: Myrmecobiidae and Thylacinidae (Miller et al., 2009), each with one species; and Dasyuridae, with four tribes: Dasyurini, Phascogalini, Sminthopsini, and Planigalini. The phylogeny of Dasyuromorphia has been resolved following many studies, based on both molecular and morphological data (Mitchell et al., 2014; Woolley et al., 2015; Westerman et al., 2016; Kealy and Beck, 2017; Duchêne et al., 2018). When considering high numt content, two of the Dasyuridae tribes are represented in the present analysis: Dasyurini (S. harrisii with 3,452) and Phascogalini (A. flavipes with 2,815). The numt content in the extinct thylacine, T. cynocephalus of the Thylacinidae family, featuring a much smaller numt number (435), was also analyzed. While S. harrisii and A. flavipes possess only a small fraction of complex numts (6–8%), T. cynocephalus features over 50% of complex numts.
The nuclear genomes of S. harrisii and A. flavipes include assembled chromosomes. The distribution of numts in chromosomes was tested and found not to deviate significantly from a random distribution (S. harrisii χ2 = 7.0326, df = 7, p = 0.425; A. flavipes, χ2 = 9.5894, df = 6 the Y chromosome was unavailable, p = 0.143). This finding is in agreement with chromosome distribution in humans (Mourier et al., 2001; Hazkani-Covo et al., 2003). In contrast to chromosomal distribution, the distribution of numts on the mitochondrial genomes is uneven with some regions either over or underrepresented (Figure 2).
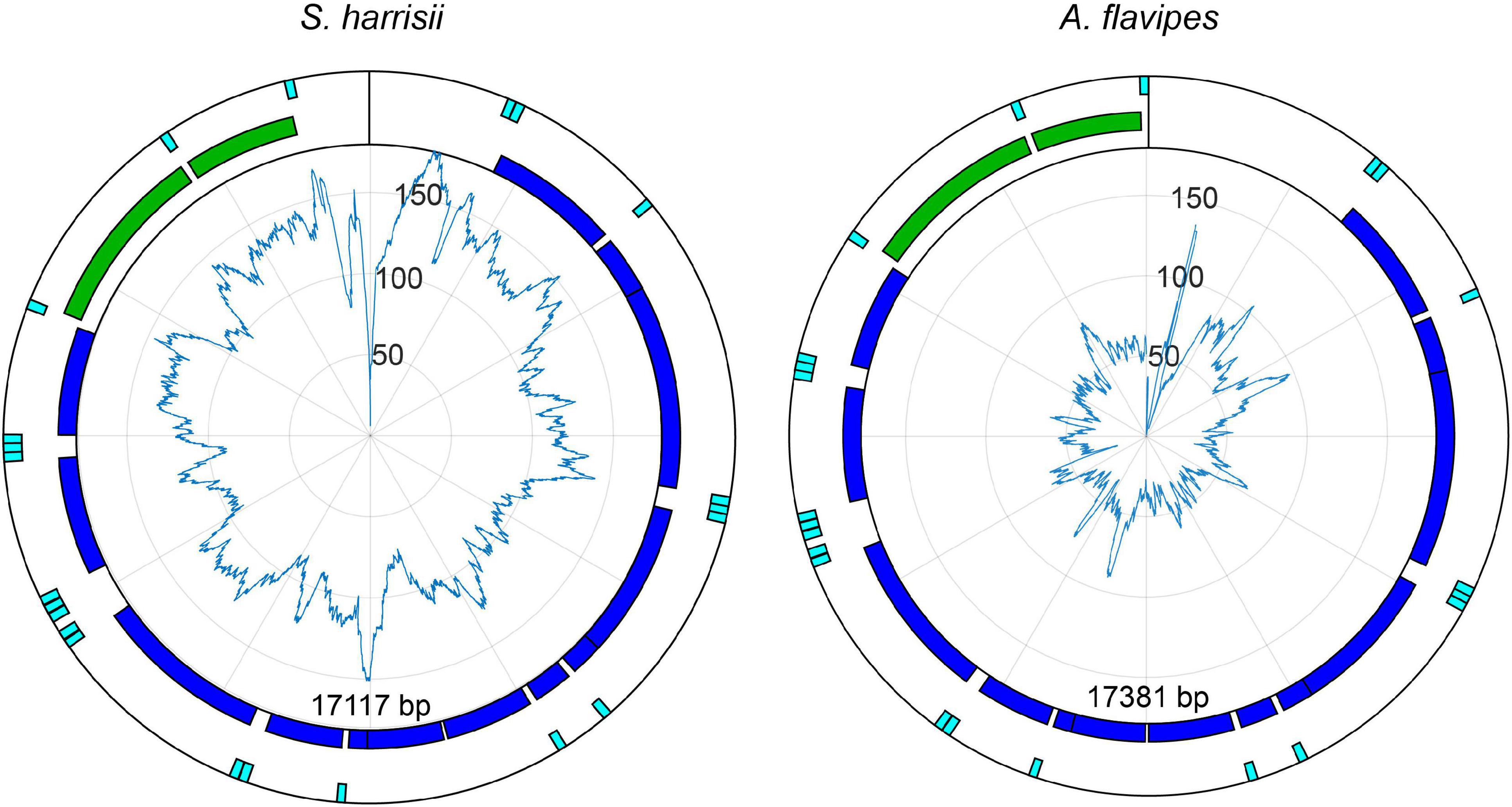
Figure 2. Mitochondria of S. harrisii and A. flavipes drawn in a counter-clockwise orientatiom. Coding genes are shown in blue, rRNA in green, and tRNA in teal. Polar plots indicate the coverage of each mitochondrial positions within numts.
In order to identify the dynamics of numt accumulation rate during Dasyuromorphia evolution, a methodology based on mitochondrial phylogenetic was applied (Hazkani-Covo et al., 2003, 2010). A maximum likelihood mitochondrial tree was first reconstructed based on 29 fully sequenced mitochondrial genomes: 10 Dasyuromorphia from the three families—Myrmecobiidae, Thylacinidae and Dasyuridae; and three of the four Dasyuridae tribes (no mitochondrial genome was available for Planigalini). The tree also included 12 Diprotodonita and seven Didelphimorphia species (Figure 3A). The taxa were chosen on the basis of complete mitochondrial sequence availability and the possibility of constructing a taxonomically valid phylogenetic tree.

Figure 3. (A) Maximum likelihood tree based on mitochondria of Dasyuromorphia with Diphodnita and Didelpmorphia outgroups. Branch lengths are measured in units of nucleotide substitution per site. Numbers of numts that originated during various periods of evolution are shown on branches, with red for S. harrisii (branches a-b; f-h; j-k), blue for A. flavipes (branches c-h; j-k), and green for Thylacinus cynocephalus (branches i-k). (B) Dynamics of numt accumulation in S. harrisii (red for all, orange without duplication) A. flavipes (blue), and T. cynocephalus (green). Values achieved by dividing the number of numts mapped to the branch by the branch length. Highest values appear on branches b for S. harrisii and e for A. flavipes.
The tree was then used to position the accumulation times of a total of 1,736 numts that are 300 bp or longer: S. harrisii (1,059), A. flavipes (518) and T. cynocephalus (158). Using maximum likelihood methodology, it was possible to position each numt in its temporal evolutionary context (Figure 3 and Supplementary Table 2) in one of the lineages leading to A. flavipes, S. harrisii, or T. cynocephalus. A. flavipes numts were assigned to eight possible trees, S. harrisii numts to seven possible trees, and T. cynocephalus numts to three possible trees. Numts on the linage leading to S. harrisii could be mapped to branches a-b, f-h, j-k; those on the linage leading to A. flavipes to branches c-h, j-k; and those on the linage leading to T. cynocephalus to branches i-k (Figure 3A). After each numt was assigned to the best scoring maximum likelihood tree, the total number of numts supporting each tree was counted to yield the number of numts accumulated on each branch, (shown on tree branches: Figure 3A and Supplementary Table 2). Since each fragment in a complex numt was mapped separately, and best assignment was determined only if all fragments agreed on the best tree, 34 of the 1,736 numts are not shown on the tree as their fragments disagree. Additionally, weighted numt number was calculated to account for cases when positioning on adjacent branches of the phylogenetic tree could not be differentiated from one another with sufficient statistical confidence. In this case, equal probability of numt origin on each of the possible indistinguishable branches was assigned. Results for weighted numts are given in Supplementary Figure 1.
The analysis indicates that most numts in S. harrisii and A. flavipes are recent and dated to after the split of the two tribes. S. harrisii was estimated to possess 735–858 (69–81%) numts after the divergence of the Dasyurini and Phascogalini (branches a-b), while A. flavipes was estimated to possess 242–354 (47–68%) numts after the divergence of the Dasyurini and the Phascogalini (branches c-e). While it is possible that numts that were too short for phylogenetic analysis dominated among the older ones, there was no correlation between length and percentage identity for the simple numts of A. flavipes and S. harrisii.
The rate of numt accumulation was calculated next, based on the number of numts assigned to the branch as best tree divided by the branch length of the mitochondrial tree (Figure 3B): i.e., the number of numts per nucleotide substitution. Their accumulation rate started to climb before the divergence of Dasyurini and Phascogalini and reached a peak in early Dasyurini and Phascogalini evolution, where the two rates on branches b and e are identified as significant outliers. S. harrisii reveals the highest rate on branch b while A. flavipes reveals a high but more moderate rate on branch e. These two branches represent the first split after the divergence of Dasyurini and Phascogalini and before the divergence of species within the Pascogalini and Dasyurini. The rates on branches b and e are 114- and 98-fold higher than the rate on branch j, indicating the emergence of Dasyuromorphia. Weighted analysis revealed a similar dynamic but indicated a rate that is only 25-fold higher in branches b and e than in branch j (Supplementary Figure 1). Following the peak of numt accumulation, the rate then slowed down to close to its base level. In contrast to the Dasyuridae clade, which revealed a substantial variation in the rate of numt accumulation, the lineage leading to T. cynocephalus revealed no such changes.
The high rate on numt accumulation could be due to an increase in the rate of independent transfers from the mitochondria to the nucleus or due to post-insertion duplications within the nucleus (Hazkani-Covo et al., 2003). While numts have no intrinsic ability to transpose, they can duplicate as part of genomic segmental duplication, or in events mediated by transposable elements. Numts were shown to be enriched in the regions of transposable elements (Mishmar et al., 2004; Tsuji et al., 2012; Michalovova et al., 2013; Hazkani-Covo and Martin, 2017). Moreover, transposable elements were shown to be a driving force for bursts of post-insertion numt duplication (Wang et al., 2020). To determine whether the observed enrichment of numt accumulation was the result of increased insertions from the mitochondria or of numt duplications within the nucleus, numts followed by similar flanking regions of at least 50 bp were identified. Only, 22 S. harrisii numts were identified as potentially involved in such duplications, and all of them are mapped to recent branches a and b. No comparable A. flavipes duplicated numts were identified. Because the number of duplicated numts is shown to have a minimal effect on their accumulation (Figure 3), it appears that numt accumulation in Dasyuridae is a result of increased insertion rate rather than that of post-insertion duplications.
Discussion
The current analysis sought to identify recent sequenced genomes featuring an extreme numt content, and specifically to determine whether the high numt content previously identified in the gray short-tailed opossum is also evident in other marsupials. Numt content in vertebrates is correlated with genome size (Figure 1), and placental and marsupial mammals feature the highest such content. Two genomes revealed a very high numt number, belonging to two marsupials from the family Dasyuridae in the order Dasyuromorphia: S. harrisii belonging to the Dasyurini tribe (with 3,452 numts, 1,956 kb); and A. flavipes (with 2,815 numts, 848 kb), belonging to the Phascogalini tribe. It is also notable that the numt content identified in genomes changes with the level of genome completion and assembly curation over the years. In our previous analysis, the majority of numts in the gray short-tailed opossum did not map to chromosomes, whereas almost all those in two Dasyuridae genomes mapped to chromosomes, suggesting that the current high numt content in Dasyuridae is stable.
Kealy and Beck (2017) have shown that an increased rate of species diversification in the Dasyuromorphias occurred around the late to middle Miocene, driven by the radiation of the three Dasyurine tribes: Dasyurini, Phascogalini, and Sminthopsini. Their morphological and molecular data indicate that the three tribes began to radiate simultaneously 11.5–13.1 MYA (Mitchell et al., 2014; Kealy and Beck, 2017). The latter study also indicated that this species’ radiation occurred shortly after a rapid decline in global temperature, known as the Miocene climate transition, dated to 15–17 MYA (Zachos et al., 2001; Hansen et al., 2013). The findings from the current study indicate that Dasyurini and Phascogalini radiation also co-occurred, with a significant expansion in numt content (Figure 3, Supplementary Figure 1, and Supplementary Table 2).
Numt expansion can be either the result of increased insertion rate or of post-insertion amplification. Numts are enriched near retrotransposons (Tsuji et al., 2012; Hazkani-Covo and Martin, 2017) and post-insertion amplification of numts can be mediated by transposable elements (Wang et al., 2020). Moreover, transposable element amplification was specifically shown to coexist with both species diversification and environmental stress (Belyayev, 2014). In the case of Dasyuromorphias, the findings indicate that recent insertions rather than numt duplications are the source of their amplification. When nuclear genomic data will become available, it will be interesting to determine whether an increased rate of numt integration also took place in the Sminthopsini.
Numt integration is known to increase with a change in environmental factors. Interestingly, their integration rate was shown to increase with a change in growth temperature in a laboratory setting. Saccharomyces cerevisiae colonies that grew under non-optimal conditions of either higher or lower temperatures demonstrated an increased escape of DNA from the mitochondria to the nuclear genome (Thorsness and Fox, 1990). A similar phenomenon was reported in a different laboratory system in regard to the transfer of mitochondria and plastid DNA into the nuclear genome of Arabidopsis thaliana. Organelle integration was shown to increase up to 10-fold following mild heat stress (Wang et al., 2012a,b; Wang and Timmis, 2013). While the high numt content observed in the Dasyuridae correlates with a past temperature change, it does not indicate causation. However, this offers a unique example of environmental conditions agreeing with our limited data on mitochondrial escape and integration. To establish the connection between numt influx and environmental aspects it is important to continue to improve our understanding of the factors that contribute to increasing numt integration.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
EH-C conceived, designed the study, collected, analyzed the data, and wrote the manuscript.
Funding
EH-C was supported by the German-Israeli Foundation grant I-1321-203.13/2015 and the Open University of Israel Research Fund.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.844443/full#supplementary-material
Supplementary Figure 1 | (A) Maximum likelihood tree based on mitochondria of Dasyuromorphia with Diphodnita and Didelpmorphia outgroups. Branch lengths are measured in units of nucleotide substitution per site. Numbers of weighted numts are shown on branches, with red for S. harrisii (branches a-b; f-h; j-k), blue for A. flavipes (branches c-h; j-k), and green for Thylacinus cynocephalus (branches i-k). (B) Dynamics of numt accumulation in S. harrisii (red for all, orange without duplication) A. flavipes (blue), and T. cynocephalus (green). Values achieved by dividing the number of weighted numts mapped to the branch by the branch length.
References
Allen, J. F. (2015). Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. U.S.A. 112, 10231–10238. doi: 10.1073/pnas.1500012112
Behura, S. K. (2007). Analysis of nuclear copies of mitochondrial sequences in honeybee (Apis mellifera) genome. Mol. Biol. Evol. 24, 1492–1505. doi: 10.1093/molbev/msm068
Belyayev, A. (2014). Bursts of transposable elements as an evolutionary driving force. J. Evol. Biol. 27, 2573–2584. doi: 10.1111/jeb.12513
Bensasson, D., Zhang, D., Hartl, D. L., and Hewitt, G. M. (2001). Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol. Evol. 16, 314–321. doi: 10.1016/s0169-5347(01)02151-6
Blanchard, J. L., and Schmidt, G. W. (1996). Mitochondrial DNA migration events in yeast and humans: integration by a common end-joining mechanism and alternative perspectives on nucleotide substitution patterns. Mol. Biol. Evol. 13:893.
Brown, W. M., George, M. Jr., and Wilson, A. C. (1979). Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 76, 1967–1971.
Brown, W. M., Prager, E. M., Wang, A., and Wilson, A. C. (1982). Mitochondrial DNA sequences of primates: tempo and mode of evolution. J. Mol. Evol. 18, 225–239. doi: 10.1007/BF01734101
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Caro, P., Gómez, J., Arduini, A., González-Sánchez, M., González-García, M., Borrás, C., et al. (2010). Mitochondrial DNA sequences are present inside nuclear DNA in rat tissues and increase with age. Mitochondrion 10, 479–486. doi: 10.1016/j.mito.2010.05.004
Cheng, X., and Ivessa, A. S. (2010). The migration of mitochondrial DNA fragments to the nucleus affects the chronological aging process of Saccharomyces cerevisiae. Aging Cell 9, 919–923. doi: 10.1111/j.1474-9726.2010.00607.x
Cheng, X., and Ivessa, A. S. (2012). Accumulation of linear mitochondrial DNA fragments in the nucleus shortens the chronological life span of yeast. Eur. J. Cell Biol. 91, 782–788. doi: 10.1016/j.ejcb.2012.06.005
Collura, R. V., and Stewart, C. B. (1995). Insertions and duplications of mtDNA in the nuclear genomes of Old World monkeys and hominoids. Nature 378, 485–489. doi: 10.1038/378485a0
Dayama, G., Emery, S. B., Kidd, J. M., and Mills, R. E. (2014). The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 42, 12640–12649. doi: 10.1093/nar/gku1038
Dayama, G., Zhou, W., Prado-Martinez, J., Marques-Bonet, T., and Mills, R. E. (2020). Characterization of nuclear mitochondrial insertions in the whole genomes of primates. NAR Genom. Bioinform. 2:lqaa089. doi: 10.1093/nargab/lqaa089
Duchêne, D. A., Bragg, J. G., Duchêne, S., Neaves, L. E., Potter, S., Moritz, C., et al. (2018). Analysis of Phylogenomic Tree Space Resolves Relationships Among Marsupial Families. Syst. Biol. 67, 400–412. doi: 10.1093/sysbio/syx076
Haag-Liautard, C., Coffey, N., Houle, D., Lynch, M., Charlesworth, B., and Keightley, P. D. (2008). Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 6:e204. doi: 10.1371/journal.pbio.0060204
Hansen, J., Sato, M., Russell, G., and Kharecha, P. (2013). Climate sensitivity, sea level and atmospheric carbon dioxide. Philos. Trans. Math. Phys. Eng. Sci. 371:20120294. doi: 10.1098/rsta.2012.0294
Hazkani-Covo, E. (2009). Mitochondrial insertions into primate nuclear genomes suggest the use of numts as a tool for phylogeny. Mol. Biol. Evol. 26, 2175–2179. doi: 10.1093/molbev/msp131
Hazkani-Covo, E., and Covo, S. (2008). Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 4:e1000237. doi: 10.1371/journal.pgen.1000237
Hazkani-Covo, E., and Graur, D. (2007). A comparative analysis of numt evolution in human and chimpanzee. Mol. Biol. Evol. 24, 13–18. doi: 10.1093/molbev/msl149
Hazkani-Covo, E., and Martin, W. F. (2017). Quantifying the Number of Independent Organelle DNA Insertions in Genome Evolution and Human Health. Genome. Biol. Evol. 9, 1190–1203. doi: 10.1093/gbe/evx078
Hazkani-Covo, E., Sorek, R., and Graur, D. (2003). Evolutionary dynamics of large numts in the human genome: rarity of independent insertions and abundance of post-insertion duplications. J. Mol. Evol. 56, 169–174. doi: 10.1007/s00239-002-2390-5
Hazkani-Covo, E., Zeller, R. M., and Martin, W. (2010). Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 6:e1000834. doi: 10.1371/journal.pgen.1000834
Ju, Y. S., Alexandrov, L. B., Gerstung, M., Martincorena, I., Nik-Zainal, S., Ramakrishna, M., et al. (2014). Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife 3:e02935. doi: 10.7554/eLife.02935
Ju, Y. S., Tubio, J. M., Mifsud, W., Fu, B., Davies, H. R., Ramakrishna, M., et al. (2015). Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome. Res. 25, 814–824.
Katoh, K., Asimenos, G., and Toh, H. (2009). Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537, 39–64. doi: 10.1007/978-1-59745-251-9_3
Kealy, S., and Beck, R. (2017). Total evidence phylogeny and evolutionary timescale for Australian faunivorous marsupials (Dasyuromorphia). BMC Evol. Biol. 17:240. doi: 10.1186/s12862-017-1090-0
Kleine, T., Maier, U. G., and Leister, D. (2009). DNA Transfer from Organelles to the Nucleus: the Idiosyncratic Genetics of Endosymbiosis. Ann. Rev. Plant Biol. 60, 115–138. doi: 10.1146/annurev.arplant.043008.092119
Liang, B., Wang, N., Li, N., Kimball, R. T., and Braun, E. L. (2018). Comparative Genomics Reveals a Burst of Homoplasy-Free Numt Insertions. Mol. Biol. Evol. 35, 2060–2064. doi: 10.1093/molbev/msy112
Meisinger, C., Sickmann, A., and Pfanner, N. (2008). The mitochondrial proteome: from inventory to function. Cell 134, 22–24. doi: 10.1016/j.cell.2008.06.043
Michalovova, M., Vyskot, B., and Kejnovsky, E. (2013). Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: size, relative age and chromosomal localization. Heredity 111, 314–320. doi: 10.1038/hdy.2013.51
Miller, W., Drautz, D. I., Janecka, J. E., Lesk, A. M., Ratan, A., Tomsho, L. P., et al. (2009). The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus). Genome. Res. 19, 213–220. doi: 10.1101/gr.082628.108
Mishmar, D., Ruiz-Pesini, E., Brandon, M., and Wallace, D. C. (2004). Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum. Mutat. 23, 125–133. doi: 10.1002/humu.10304
Mitchell, K. J., Pratt, R. C., Watson, L. N., Gibb, G. C., Llamas, B., Kasper, M., et al. (2014). Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Mol. Biol. Evol. 31, 2322–2330. doi: 10.1093/molbev/msu176
Mourier, T., Hansen, A. J., Willerslev, E., and Arctander, P. (2001). The Human Genome Project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol. Biol. Evol. 18, 1833–1837. doi: 10.1093/oxfordjournals.molbev.a003971
Pamilo, P., Viljakainen, L., and Vihavainen, A. (2007). Exceptionally high density of NUMTs in the honeybee genome. Mol. Biol. Evol. 24, 1340–1346. doi: 10.1093/molbev/msm055
Perna, N. T., and Kocher, T. D. (1996). Mitochondrial DNA: molecular fossils in the nucleus. Curr. Biol. 6, 128–129. doi: 10.1016/s0960-9822(02)00441-4
Ricchetti, M., Fairhead, C., and Dujon, B. (1999). Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402, 96–100. doi: 10.1038/47076
Ricchetti, M., Tekaia, F., and Dujon, B. (2004). Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2:E273. doi: 10.1371/journal.pbio.0020273
Richter, C. (1988). Do mitochondrial DNA fragments promote cancer and aging? FEBS Lett. 241, 1–5. doi: 10.1016/0014-5793(88)81018-4
Shimodaira, H., and Hasegawa, M. (1999). Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. In. Molecular. Biol. Evol. 16, 1114–1116.
Song, H., Buhay, J. E., Whiting, M. F., and Crandall, K. A. (2008). Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. U.S.A. 105, 13486–13491. doi: 10.1073/pnas.0803076105
Song, H., Moulton, M. J., and Whiting, M. F. (2014). Rampant nuclear insertion of mtDNA across diverse lineages within Orthoptera (Insecta). PLoS One 9:e110508. doi: 10.1371/journal.pone.0110508.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tavaré, S. (1986). Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Mathe. Life Sci. 17, 57–86.
Thorsness, P. E., and Fox, T. D. (1990). Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature 346, 376–379. doi: 10.1038/346376a0
Timmis, J. N., Ayliffe, M. A., Huang, C. Y., and Martin, W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. doi: 10.1038/nrg1271
Tsuji, J., Frith, M. C., Tomii, K., and Horton, P. (2012). Mammalian NUMT insertion is non-random. Nucleic Acids Res. 40, 9073–9088. doi: 10.1093/nar/gks424
Wallace, D. C., Stugard, C., Murdock, D., Schurr, T., and Brown, M. D. (1997). Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc. Natl. Acad. Sci. U.S.A. 94, 14900–14905. doi: 10.1073/pnas.94.26.14900
Wang, D., Lloyd, A. H., and Timmis, J. N. (2012a). Environmental stress increases the entry of cytoplasmic organellar DNA into the nucleus in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 2444–2448.
Wang, D., Lloyd, A. H., and Timmis, J. N. (2012b). Nuclear genome diversity in somatic cells is accelerated by environmental stress. Plant Sign. Behav. 7, 595–597. doi: 10.4161/psb.19871
Wang, D., and Timmis, J. N. (2013). Cytoplasmic organelle DNA preferentially inserts into open chromatin. Genome. Evol. 5, 1060–1064. doi: 10.1093/gbe/evt070
Wang, J. X., Liu, J., Miao, Y. H., Huang, D. W., and Xiao, J. H. (2020). Tracking the Distribution and Burst of Nuclear Mitochondrial DNA Sequences (NUMTs) in Fig Wasp Genomes. Insects 11:680. doi: 10.3390/insects11100680
Westerman, M., Blacket, M., Hintz, A., Armstrong, K., Woolley, P., and Krajewski, C. (2016). A plethora of planigales: genetic variability and cryptic species in a genus of dasyurid marsupials from northern Australia. Aus. J. Zool. 64, 303–311.
Woolley, P. A., Krajewski, C., and Westerman, M. (2015). Phylogenetic relationships within Dasyurus (Dasyuromorphia: Dasyuridae):quoll systematics based on molecular evidence and male characteristics. J. Mammal. 96, 37–46.
Wright, R. M., and Cummings, D. J. (1983). Integration of mitochondrial gene sequences within the nuclear genome during senescence in a fungus. Nature 302, 86–88. doi: 10.1038/302086a0
Zachos, J., Pagani, M., Sloan, L., Thomas, E., and Billups, K. (2001). Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292, 686–693. doi: 10.1126/science.1059412
Keywords: numts, marsupials, mitochondria, organelle insertions, promiscuous DNA, Dasyuridae
Citation: Hazkani-Covo E (2022) A Burst of Numt Insertion in the Dasyuridae Family During Marsupial Evolution. Front. Ecol. Evol. 10:844443. doi: 10.3389/fevo.2022.844443
Received: 28 December 2021; Accepted: 20 January 2022;
Published: 11 February 2022.
Edited by:
Stephane Boissinot, New York University Abu Dhabi, United Arab EmiratesReviewed by:
Jeremy Timmis, University of Adelaide, AustraliaJustin Wilcox, New York University Abu Dhabi, United Arab Emirates
Copyright © 2022 Hazkani-Covo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Einat Hazkani-Covo, ZWluYXRjb0BvcGVudS5hYy5pbA==, orcid.org/0000-0002-4878-6001
 Einat Hazkani-Covo
Einat Hazkani-Covo