
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 14 February 2022
Sec. Behavioral and Evolutionary Ecology
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.834422
This article is part of the Research TopicSensory Ecology of Phenotypic Plasticity: from Receptors via Modulators to EffectorsView all 8 articles
Cryptochromes are evolutionary ancient blue-light photoreceptors that are part of the circadian clock in the nervous system of many organisms. Cryptochromes transfer information of the predominant light regime to the clock which results in the fast adjustment to photoperiod. Therefore, the clock is sensitive to light changes and can be affected by anthropogenic Artificial Light At Night (ALAN). This in turn has consequences for clock associated behavioral processes, e.g., diel vertical migration (DVM) of zooplankton. In freshwater ecosystems, the zooplankton genus Daphnia performs DVM in order to escape optically hunting predators and to avoid UV light. Concomitantly, Daphnia experience circadian changes in food-supply during DVM. Daphnia play the keystone role in the carbon-transfer to the next trophic level. Therefore, the whole ecosystem is affected during the occurrence of cyanobacteria blooms as cyanobacteria reduce food quality due to their production of digestive inhibitors (e.g., protease inhibitors). In other organisms, digestion is linked to the circadian clock. If this is also the case for Daphnia, the expression of protease genes should show a rhythmic expression following circadian expression of clock genes (e.g., cryptochrome 2). We tested this hypothesis and demonstrated that gene expression of the clock and of proteases was affected by ALAN. Contrary to our expectations, the activity of one type of proteases (chymotrypsins) was increased by ALAN. This indicates that higher protease activity might improve the diet utilization. Therefore, we treated D. magna with a chymotrypsin-inhibitor producing cyanobacterium and found that ALAN actually led to an increase in Daphnia’s growth rate in comparison to growth on the same cyanobacterium in control light conditions. We conclude that this increased tolerance to protease inhibitors putatively enables Daphnia populations to better control cyanobacterial blooms that produce chymotrypsin inhibitors in the Anthropocene, which is defined by light pollution and by an increase of cyanobacterial blooms due to eutrophication.
Cryptochromes were detected in a range of organisms from plants to arthropods to mammals (Sancar, 2004; Yoshii et al., 2008; Tilden et al., 2011). In animals, those cryptochromes can be found in the retina and the brain of mammals (Sancar, 2004) and in pacemaker neurons and in photoreceptor cells of the compound eyes in Drosophila (Yoshii et al., 2008). These blue-light sensitive molecules are associated and probably co-evolved with the circadian clock (Gehring and Rosbash, 2003). In Drosophila, cryptochromes act both as photoreceptors and as part of the core clockwork (Yoshii et al., 2008). Drosophila only possesses type 1 cryptochromes, whereas in Daphnia, a keystone crustacean genus in freshwater ecosystems, cryptochromes were detected that belong either to type 2 (which putatively are part of the core clock) or type 1 that code for photoreceptors molecules that participate in the input of the clock (Tilden et al., 2011). Daphnia possess four different cryptochrome genes of both types that showed different subcellular localization when expressed in the Drosophila clock neuron (Nitta et al., 2019). The cryptochromes of different Daphnia species show a rhythmic gene expression pattern (D. pulex: homologs of type 1 and 2; Bernatowicz et al., 2016, D. pulex and D. magna: homolog of type 2; Schwarzenberger and Wacker, 2015; Schwarzenberger et al., 2021a). Also rhodopsins might play a role in the synchronization of the clock to the predominant photoperiod since a rhodopsin was detected to play a role in the induction of resting stage induction in a QTL study (Roulin et al., 2016).
Beside cryptochrome 2 (Tomioka and Matsumoto, 2010), the main genes that constitute the clock’s major feedback loop (the core clock) in most organisms and also in Daphnia are period, timeless, cycle and clock (Tilden et al., 2011). Those genes are expressed in a circadian manner in Daphnia (Schwarzenberger and Wacker, 2015; Bernatowicz et al., 2016; Coldsnow et al., 2017) and are involved in different physiological and behavioral rhythms (Sainath et al., 2013).
The clock plays a role in termination of diapause in Daphnia (Schwarzenberger et al., 2020a) and the molt cycle (Lipcius and Herrnkind, 1982), reproduction, development and metabolism of different crustaceans (Strauss and Dircksen, 2010). The information on photoperiod—in order to regulate those processes—is mediated by the hormone melatonin (Sainath et al., 2013), which is an output of the clock. Melatonin is cyclically expressed in Daphnia following the circadian gene expression of clock and melatonin-synthesis genes (Schwarzenberger and Wacker, 2015). In vertebrates, this indeloamine hormone is produced in the brain’s pineal gland (Foulkes et al., 1997). Similarly, the highest concentration of melatonin in Daphnia has been detected in the head region (Markowska et al., 2009). In crustaceans, many rhythmically expressed genes have been detected that are probably controlled by the clock, e.g., immune genes in Daphnia (Rund et al., 2016).
Behavior can also be governed by the crustacean clock. Many Daphnia clones are migrating to deeper water layers during the day in order to escape UV light and fish predation (Loose and Dawidowicz, 1994; Rhode et al., 2001; Ekvall et al., 2015); thus, they are confronted with diurnal changes in food quality and quantity. In Daphnia, the involvement of the clock in diel vertical migration (DVM) is likely (Cellier-Michel et al., 2003), especially because DVM is inhibited by the application of exogenous melatonin (Bentkowski et al., 2010). Since food quantity often is high at the surface of freshwater bodies (where Daphnia are situated during night) and low in deeper water levels (to which Daphnia sink to during day) a circadian expression and activity of digestive enzymes is expected. Actually, clock-mediated rhythmic expression and activity of different metabolic enzymes are known for krill (Teschke et al., 2011), whereas this was not the case for a clone of D. pulex (Rund et al., 2016). However, it is not known whether the clone from a study of Rund et al. (2016) shows DVM or whether it is a non-migrating clone. We propose that digestive enzymes are rhythmically expressed in other Daphnia clones and species, e.g., D. magna, and that gene expression and activity peaks at night (when food quantity is high during DVM).
The circadian clock is not triggered but adjusted by different photoperiods and therefore responds to light changes by elongation or shortening of its cycle and by increasing or decreasing the clock’s gene expression (e.g., Yasuo et al., 2003; Muguruma et al., 2010; Schwarzenberger et al., 2021a). Following expectations, the clock has been shown to be sensitive to human light pollution (Dominoni, 2015; Walker et al., 2020). Artificial Light At Night (ALAN) has a different spectral composition than moonlight or starlight with LEDs showing a spectrum that is similar to daylight or shifted to blue-light which activates blue-light receptors like cryptochromes. Therefore, we hypothesize that Daphnia’s gene expression of clock and especially of cryptochrome genes are disrupted due to ALAN with a high proportion of blue light.
ALAN has been shown to cause diapause inhibition in a terrestrial arthropod (i.e., moths of the species Mamestra brassicae, van Geffen et al., 2014). Furthermore, also movement, behavior and development of many different insects are affected by ALAN which impacts their ecology, population dynamics and species-interactions (reviewed in Desouhant et al., 2019). Similarly, birds have been shown to sing earlier in light polluted cities (Miller, 2006; da Silva et al., 2015). Since half of all humans, responsible for the light pollution, live within 3 km of surface water bodies, ALAN should also affect freshwater organisms. However, only few studies have investigated the effect of ALAN in freshwater: Frogs are attracted by light sources at night and show a higher activity when ALAN is switched on (Baker and Richardson, 2006). The crustacean genus Gammarus shows a reduced activity in the presence of ALAN (Perkin et al., 2014). Also the biomass and the composition of periphyton changes when illuminated (Hölker et al., 2015). ALAN has also been shown to differentially affect the behavior of Daphnia and their fish predators (Talanda et al., 2018). This might result in a mismatch of Daphnia-fish interaction. Furthermore, Daphnia migrate to deeper water levels due to ALAN (Moore et al., 2000).
In fish, ALAN leads to higher costs due to increased nest-hatching behavior (Foster et al., 2016), desynchronized hatching of eggs and lower production of the sleeping hormone melatonin (Brüning et al., 2015). ALAN and a reduction of melatonin concentrations might exacerbate the response to other stressors. In Daphnia, melatonin leads to attenuation of stress responses (Schwarzenberger et al., 2014). If melatonin production is disrupted by ALAN, a stressor might have a stronger effect on Daphnia than in dark nights. We hypothesize that when Daphnia are stressed by low food-quality, e.g., by the ingestion of cyanobacteria that produce digestive inhibitors, ALAN should lead to even lower growth rates and to a changed expression of digestive enzymes (cf. Schwarzenberger et al., 2010; Schwarzenberger and Fink, 2018).
ALAN is often not present as a constant light source at night. Street lamps illuminating small water bodies in the evening might be switched off at night in order to save energy. Illuminated ships, ferries and boats cross lakes more often in summer than in winter and illuminate water bodies only for a short time interval, Cars pass by water bodies mostly at dusk and dawn illuminating the water in short light pulses. We were especially interested in theses short light pulses, and hypothesized that exposure to ALAN affects Daphnia’s clock.
In our experiments we grew clones of D. magna in a 16:8 h day-night cycle as a control light treatment and switched on ALAN for 5 min every hour of the night. We measured gene expression of clock genes (e.g., cryptochrome 2) and digestive enzymes, and activity of gut proteases over a whole 24 h day-night cycle under control and ALAN conditions. Furthermore, we grew two clones of D. magna on a diet consisting of either 100% high-quality green alga or on a mixture containing 20% cyanobacteria that produce protease inhibitors in order to investigate the effect of ALAN on digestion of low-quality food.
The Daphnia magna clones “CH-H-5” (Switzerland) and “FR-LR 6-1” (France) were kept in a day-night cycle of 16:8 h at 20°C for several generations. Fifteen animals per liter were kept in filtrated (< 0.2 μm) and aerated Lake Constance water and fed with the green alga Acutodesmus obliquus ad libitum. The clones were transferred to new medium every other day. Only neonates from the second or third clutch were used for the experiments.
The green alga A. obliquus was used as a high-quality food source and was cultivated semi-continuously in cyanophycean medium (Von Elert and Jüttner, 1997) at 20°C in constant light. Two cyanobacterial strains, i.e., Microcystis sp. BM25 (originating from Lake Bysjön in Sweden), which produces chymotrypsin inhibitors (Schwarzenberger et al., 2013), and a microcystin knock-out mutant of Microcystis aeruginosa PCC7806 (referred to as PCC7806 Mut) producing trypsin inhibitors (Dittmann et al., 2001), were cultivated in cyanophycean medium. Both cyanobacterial strains do not produce microcystins; furthermore, BM25 does not produce trypsin inhibitors (Schwarzenberger et al., 2013). PCC7806 Mut also inhibits chymotrypsins but to a much lower extent than trypsins (Agrawal et al., 2005). We did not detect any other toxins (i.e., anatoxin-α or carboxypeptidase inhibitors) in our cultures.
Several days before the start of the experiments, aliquots of A. obliquus and of the cyanobacteria were transferred to the climate chamber of the D. magna clones in control light conditions (16:8 h day-night cycle) in order to adjust the phytoplankton’s clock systems to the control day-night cycle of the experiments.
The 24 h experiment with clone “CH-H-5” was similar to a previously published setup with another D. magna clone [“Binnensee” (Germany); Schwarzenberger et al., 2021a]. The animals that were used in the experiments were placed in two separate Styrofoam compartments in a climate chamber (at 20°C) and were isolated from each other and from outer light. At the top of the compartments we placed aquarium lamps (sera LED X-Change Tube cool daylight 965 that contain a high proportion of blue light) that were programmed with a day–night cycle of 16:8 h (corresponding to a middle-European summer photoperiod) and a light intensity of 3,000–4,000 lux during day and 0 lux in the night. The compartment that was used for the ALAN experiment was programmed with the same dark-light cycle but with additional light pulses (∼3,000 lux ≈ 4.4 W m–2) for 5 min every hour of the night.
Fifteen to twenty 3rd clutch neonates of “CH-H-5” were transferred to 12 open 1 L glasses (two glasses per each of three replicates) with 1 L filtrated and aerated Lake Constance water and 2 mg C L–1 A. obliquus. The animals were transferred to new medium every other day. Starting at the first day of the experiment, half of the glasses were kept in the control compartment, the other half in the ALAN compartment during the following consecutive days. On the sixth day of the experiment the animals were sampled (always one or two animals per replicate) and frozen at −80°C previous to RNA extraction (collected at ZT1, 4, 5.5, 8.5, 11, 13, 15, 16, 17, 18, 19.5, 21, and 23) and protease activity measurements (collected at ZT 15, 16, 17, and 20). The night samples were collected in dim red light.
Before the start of the experiment, 15 8 day old mothers of clone “CH-H-5” and 15 8 day old mothers of clone “FR-LR 6-1” were kept in 1 L aerated and filtrated Lake Constance water in a 16:8 h day-night cycle and were fed with A. obliquus ad libitum. Thus, we made sure that the experimental animals were born in the same start conditions. At the start of the experiment, per replicate five 3rd clutch neonates of those mothers were transferred to 200 ml beakers with filtrated and aerated Lake Constance water and 2 mg C L–1 of one of three food mixtures. The animals were fed with either 100% A. obliquus, 80% A. obliquus and 20% of BM25 or 80% A. obliquus and 20% PCC 7806 Mut. Three replicates of each of the food treatment groups were kept in a 16:8 day-night cycle (control) or in the same light cycle with 5 min ALAN each hour of the night for 6 days. The medium was exchanged every other day. As our results indicated an ALAN-induced higher tolerance of Daphnia against cyanobacterial chymotrypsin inhibitors but not trypsin inhibitors, for a further clone (“FR-LR 6-1”) we tested the chymotrypsin containing diets only. At the start and at day 6 of the experiment animals were collected for determination of dry weight. The juvenile somatic growth rate (g) was calculated as according to Marzetz et al. (2017) by using the following formula (with mend = individual dry weight at the end of the experiment; mstart = individual dry weight at the start of the experiment; Δt = total duration of the experiment):
RNA was extracted with the NucleoSpin® RNA Kit (Macherey-Nagel) as according to the manufacturer’s instructions. The extracted RNA of the samples was stored at −80°C. The RNA concentration of each sample was measured either with a Thermo Fisher Scientific™ NanoDrop™ 2000 Spectrometer (Thermo Fisher Scientific) or the Qubit™ 4 Fluorometer (Thermo Fisher Scientific) with the respective dye from the Qubit RNA HS Assay Kit from Thermo Fischer Scientific following the manufacturer’s instructions. The High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems)™ was used for the transcription of the RNA in cDNA. For qPCR measurements each reaction in a 96 well plate contained 10 μL PowerSYBR Green PCR Master Mix (Applied Biosystems) 5 ng cDNA, 0, 5 μL of a forward primer (10 μM) and 0, 5 μL of a reverse primer (10 μM). RNase free H2O was added to the reaction mix to gain a final volume of 20 μL; ubc served as endogenous control (Heckmann et al., 2006). The target genes were an enzyme of the melatonin synthesis pathway (aanat5), the clock genes period, cycle, cry2, clock and timeless (Schwarzenberger et al., 2021a) and the protease genes CT448 (Schwarzenberger et al., 2010) and trypsin (Schwarzenberger and Fink, 2018). Gene expression was measured with the QuantStudio 5 qPCR-System (Applied Biosystems). The qPCR reaction was run in three steps: A hold stage at 95°C for 5 min, a PCR stage and a melt curve stage to ensure that only cDNA had been amplified and the sample was free of primer dimers and genomic DNA. The PCR stage consisted of 40 cycles with 30 s at 95°C for melting and amplification and 1 min at 60°C for primer binding. The melt curve stage ran as follows: 15 s at 95°C, 1 min at 60°C and again 15 s at 95°C. The temperature for the melting curve increased by 0.1°C/s (cf. Schwarzenberger et al., 2021a).
One animal of “CH-H-5” per replicate of each point in time at day 6 of the 24 h experiment was transferred to 50 μL of 0.1 M potassium-phosphate buffer (pH7.5), homogenized with a pestle, and centrifuged for 3 min at 14,000 × g. The protein concentration of the supernatant of the homogenates was analyzed using a Qubit fluorometer and the appropriate Quant-iT™Protein Assay Kit (Invitrogen) as according to the manufacturer’s instructions. Enzyme activities were measured photometrically using the artificial substrates N-Benzoyl-Arginine-para-Nitroanilide (BApNA, trypsin substrate; Sigma-Aldrich) or N-Succinyl-alanine-alanine-proline-phenylalanine-para-Nitroanilide (SucpNA, chymotrypsin substrate; Sigma Aldrich), as described in Schwarzenberger et al. (2010). Briefly, 1 μL Daphnia homogenate was mixed with 99 μL 0.1 M potassium phosphate buffer (pH 7.5). The buffer contained 1.88 mM of BApNA or 125 μM of SucpNA and 3% DMSO. The change in absorption was measured continuously over 6 min at 390 nm at 30°C.
After verifying homogenous variances (Levene’s test) the juvenile somatic growth rates were analyzed via two-way ANOVA and Tukey’s HSD post hoc tests and the protease activities across different points in time were analyzed via two-way ANOVA. ANOVAs were calculated with the program STATISTICA (StatSoft, Inc. 2011, version 10.0, Tulsa, OK, United States). The level of significance was p < 0.05.
“CH-H-5” presented three cryptochrome 2 expression peaks over 24 h in the control light treatment (Figure 1; see also five other clock or melatonin synthesis genes in Supplementary Figure 1). One at the switch from day to night (ZT 16), one in the middle of the night (ZT 19.5) and the highest one in the first third of the day (ZT 4). In the ALAN treatment, the first of the three gene expression peaks was missing (Figure 1).
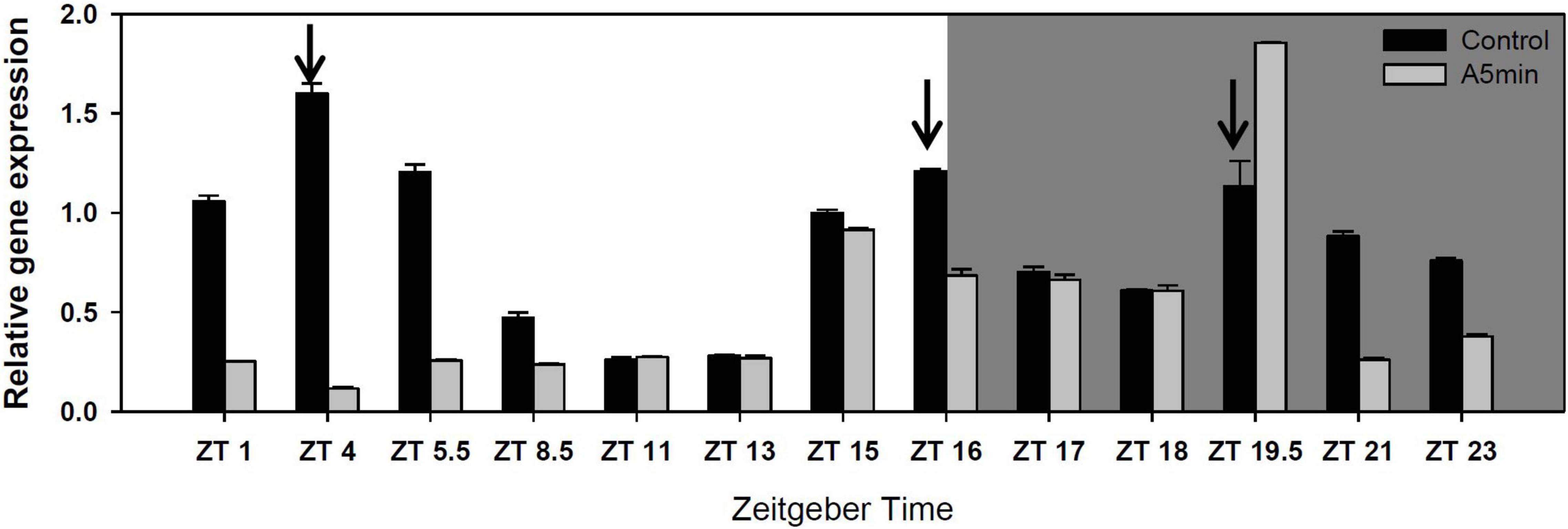
Figure 1. Gene expression of cryptochrome 2 over 24 h in a day-night cycle of 16:8 h (control) and in ALAN (A5 min: 5 min light every hour of the night; N = 3, mean + SD). Shaded area: night, white area: day. Arrows indicate the highest peaks of gene expression in the control treatment.
Both trypsin and the chymotrypsin gene CT448 showed a cyclical gene expression pattern with two peaks of gene expression in the control light treatment (Figure 2); the highest peak in the middle of the night (trypsin: ZT 18; CT448: ZT 17), the second peak during day (trypsin: ZT 5.5–8.5; CT448: ZT 1). In the ALAN treatment, trypsin gene expression was strongly decreased and was on low levels throughout the whole night, whereas CT448 still showed a cyclical expression pattern with the night peak shifted to ZT 19.5. At each point in time, gene expression was mostly lower or slightly higher (ZT 18, 19.5, 23) in the ALAN treatment.
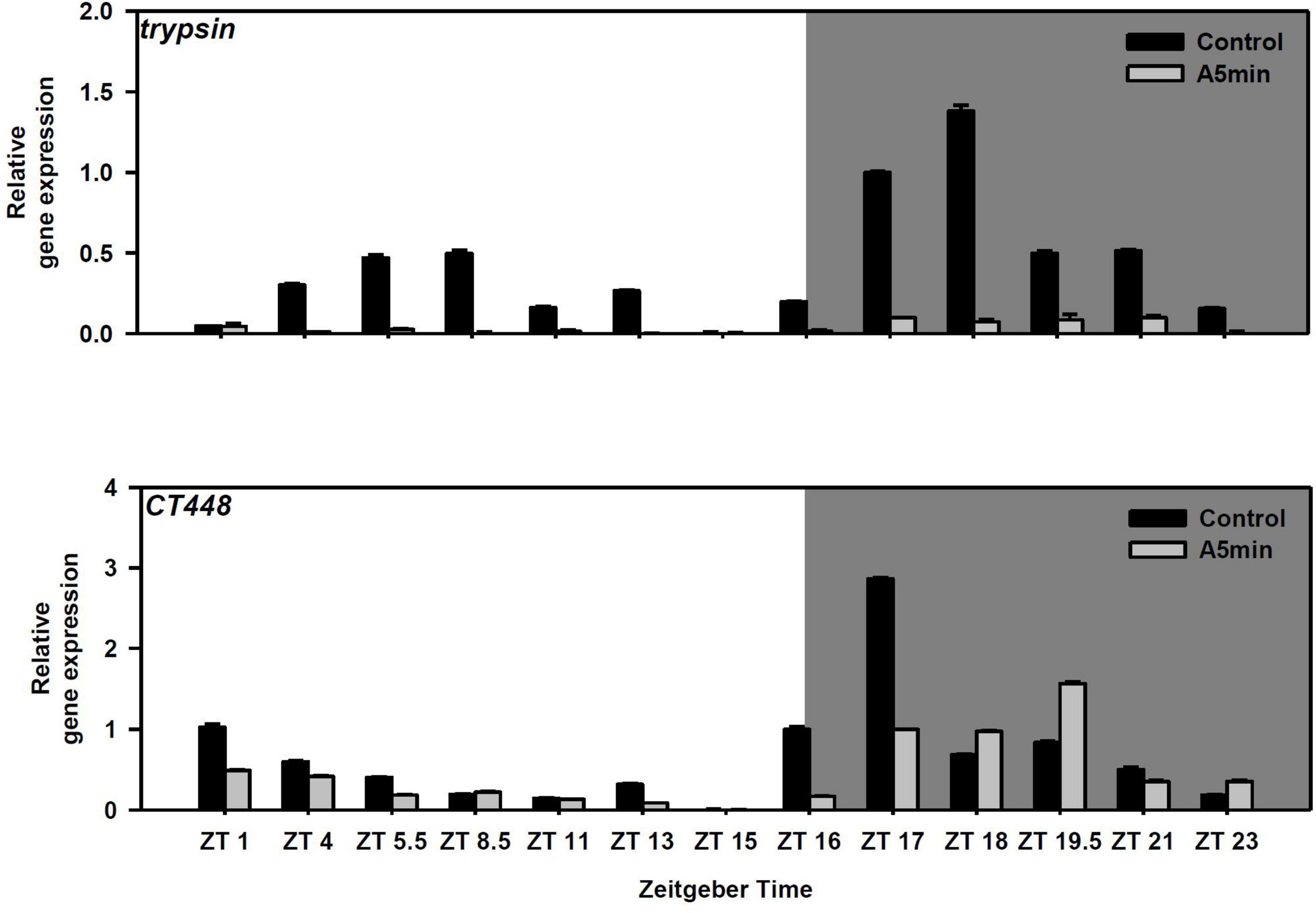
Figure 2. Gene expression of trypsin and a chymotrypsin gene (CT448) over 24 h in a day-night cycle of 16:8 h (control) and in ALAN (A5 min: 5 min light every hour of the night; N = 3, mean + SD). Shaded area: night, white area: day.
Trypsin activity did not differ between points in time and among light treatments (Figure 3; two-way ANOVA, Table 1; cf. clone “Binnensee” Supplementary Figure 2). Similarly, chymotrypsin activity did not differ between most of the points in time but the ALAN treatment showed a slightly higher activity than the control light treatment (Figure 3, two-way ANOVA, Table 1). As there was no significant interaction between the factors “light treatment” and “point in time” in the two-way ANOVA (p = 0.766, Table 1), this light effect did not change along the time axis, clearly indicating the effect of ALAN could be detected throughout.
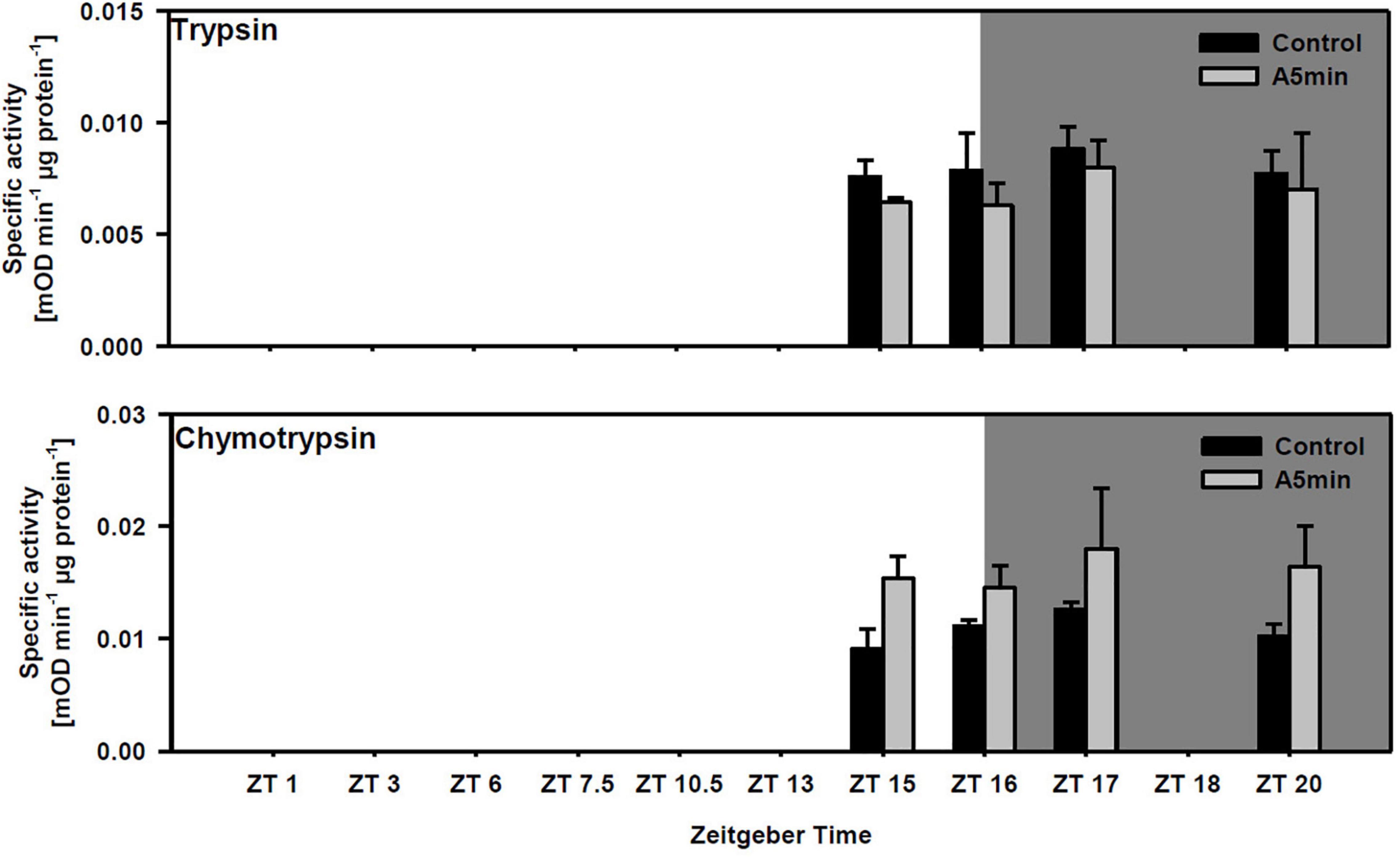
Figure 3. Specific trypsin and chymotrypsin activity at four points in time over 24 h in a day-night cycle of 16:8 h (control) and in ALAN (A5 min: 5 min light every hour of the night; N = 3, mean + SD). Shaded area: night, white area: day. Control and A5 min treatment were significantly different (see Table 1).
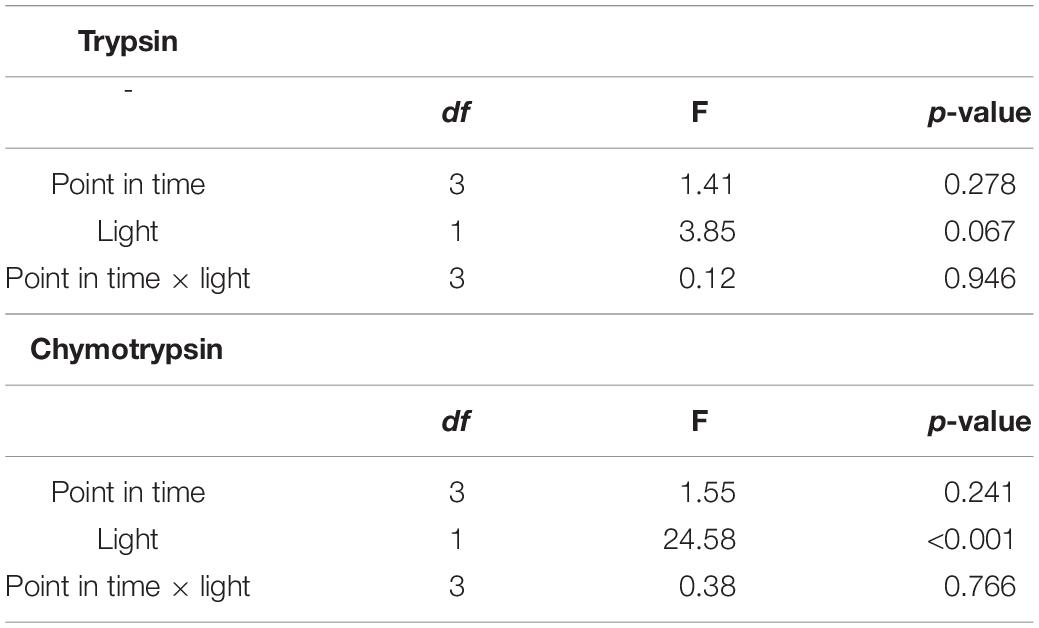
Table 1. Results of the two-way ANOVA for trypsin and chymotrypsin activities between different points in time in control or ALAN light conditions (df = degrees of freedom).
Juvenile somatic growth rates of clone “CH-H-5” did not differ between food and light treatments except for the food treatment with the chymotrypsin inhibitor-producing cyanobacterium BM25 in control light conditions (Figure 4 upper panel; Tukey’ HSD after two-way ANOVA, Table 2). In this treatment, the rate of juvenile somatic growth was lower in the control light treatment than in all other treatments including the one on the same food in the ALAN treatment. Also in clone “FR-LR-6-1” growth on BM25 was higher in ALAN than in the control light treatment (Figure 4 lower panel; Tukey’ HSD after two-way ANOVA, Table 2), but did not differ in comparison to growth on A. obliquus in both light treatments.
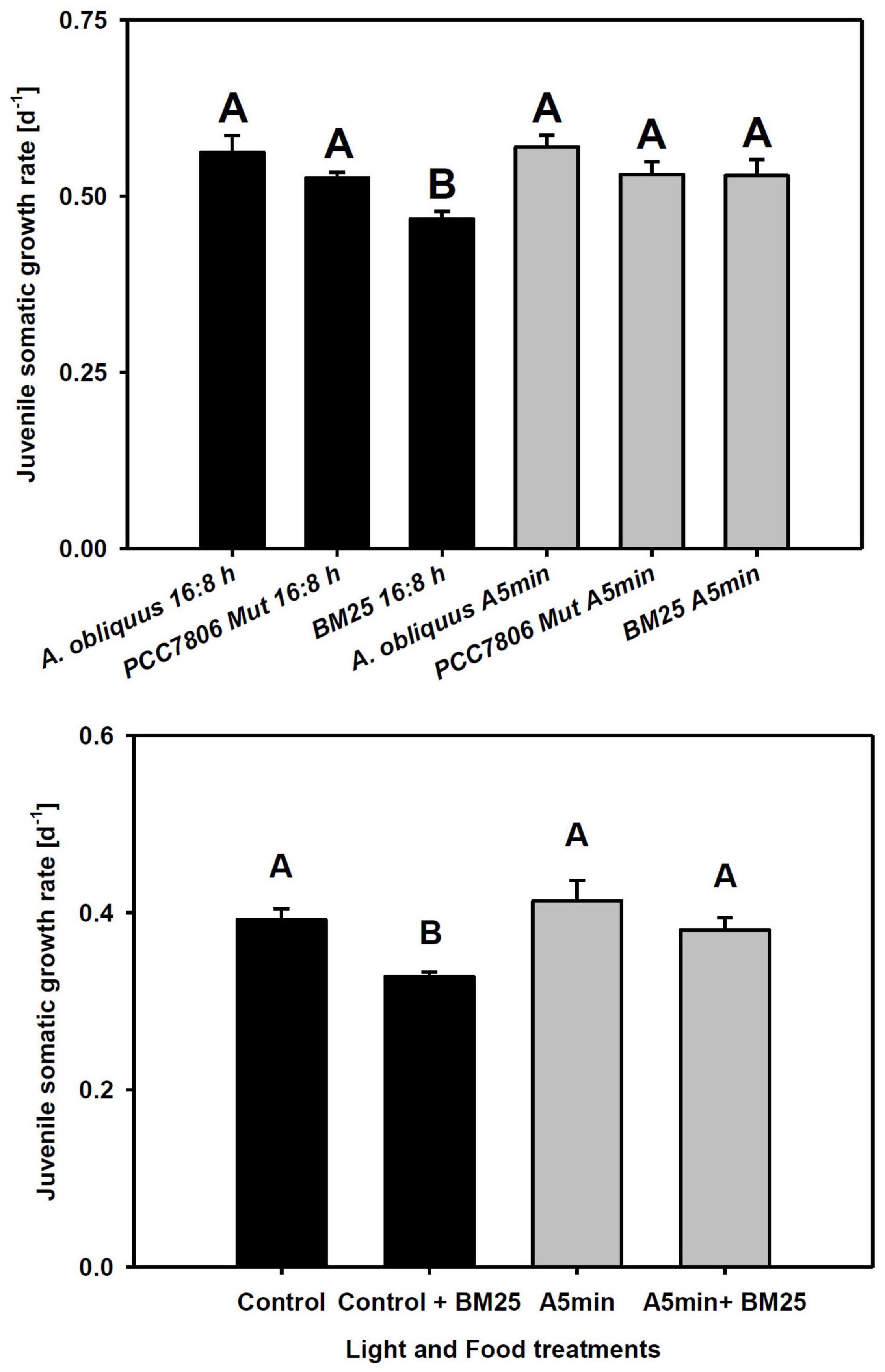
Figure 4. Upper panel: Juvenile somatic growth rates of clone “CH-H-5” fed with 100% A. obliquus, or 80% A. obliquus with 20% of either M. aeruginosa PCC7806 Mut (producing trypsin inhibitors) or Microcystis sp. BM25 (producing chymotrypsin inhibitors). Lower panel: Juvenile somatic growth rates of clone “FR-LR 6-1” fed with 100% A. obliquus, or 80% A. obliquus with 20% of Microcystis sp. BM25 (producing chymotrypsin inhibitors) over 6 days in a day-night cycle of 16:8 h (black columns) and in ALAN (gray columns; A5 min: 5 min light every hour of the night; N = 3, mean + SD). Different letters indicate statistically significant differences between light and food treatments (Tukey’s HSD after two-way ANOVA). Shaded area: night, white area: day.
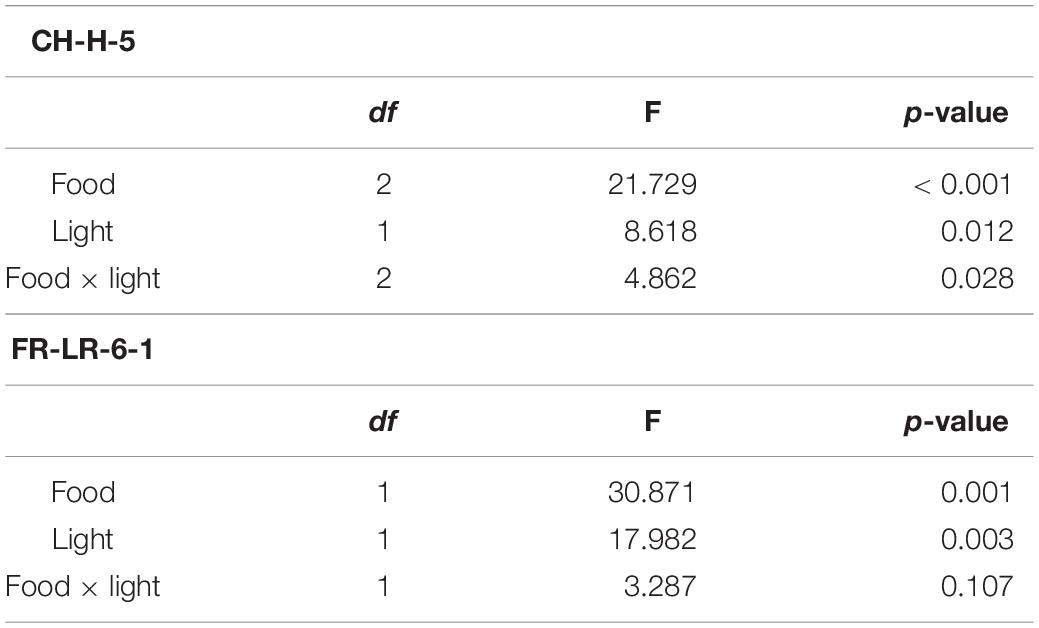
Table 2. Results of the two-way ANOVA for the somatic growth rates of clones “CH-H-5” and “FR-LR-6-1” on different food treatments and in control or ALAN light conditions (df = degrees of freedom).
Both in a D. pulex and a D. magna clone we had demonstrated earlier that clock genes show the highest peak of gene expression 1 h after the beginning of the night (Schwarzenberger and Wacker, 2015; Schwarzenberger et al., 2021a). In D. pulex most genes expressed an additional smaller peak during day (and period and timeless a third peak in the middle of the night), whereas in the D. magna clone (“Binnensee”) only one peak had been observed. In the present study we used a different D. magna clone (“CH-H-5”) and found that this clone differed in its gene expression pattern in a 16:8 h day-night cycle in comparison to “Binnensee,” and possessed more similarities with the D. pulex clone: (i) The genes either expressed two or three peaks over 24 h (except for timeless with only one peak). (ii) All genes had a peak in the first third of the day at ZT 4. (iii) The highest peak of gene expression appeared at the switch from day to night (clock, cycle, timeless), or 1 h before the onset of night (period), except for cryptochrome 2 with the highest peak at ZT 4. Therefore, gene expression of the clock is not only dependent on the height and the latitudinal origin of clones (cf. Schwarzenberger et al., 2021a) but also differs in its 24 h expression pattern in clones of the same species.
The circadian clock is adjusted by the predominant light regime and might therefore be affected by light pollution. In order to investigate the effect of light pollution on Daphnia’s clock, we measured gene expression in control light conditions and in ALAN. The gene expression pattern of the clock genes (and the melatonin synthesis gene aanat5; c.f. Supplementary Figure 1) was clearly influenced by the applied 5 min of ALAN-pulses. The peaks around the switch from day to night and in the middle of the night were shifted to a later point in time. Except for period, the highest peak of gene expression was clearly higher in the ALAN treatment than in the control. In cryptochrome 2 the peak during day vanished (although it had been the highest peak of gene expression in the control). It seems that this blue-light receptor was more significantly influenced by ALAN than the other genes. It might be that cryptochrome 2 was phase-shifted due to the repetitive perception of the light pulses at night. The phase-shift of cryptochrome 2 gene expression might have been translated to the other core clock genes and might have led to the clock’s misinterpretation of the light pulses at night as a different photoperiod, i.e., a shift to longer days and shorter nights. This can explain why the gene expression peaks of both the clock and aanat 5 were higher in ALAN: In shorter nights Daphnia need to increase clock gene expression in order to produce the same amount of signal molecules, i.e., melatonin, in shorter time than in long nights.
A clonal or species difference could also be found for the rhythmic expression of diverse genes that are under clock control. Rund et al. (2016) did not find any rhythms in the expression of metabolic processes genes in D. pulex, whereas we measured a bimodal gene expression of both types of digestive proteases, i.e., trypsin and chymotrypsin, in D. magna. A bimodal expression of trypsin is also known in Atlantic krill (Teschke et al., 2011). In our Daphnia experiment, the highest peak was expressed during night (trypsin at ZT 18, chymotrypsin at ZT 17), which followed our expectations. This is also in accordance with findings from Pfenning-Butterworth et al. (2021) who described that also in the absence of fish or kairomones D. dentifera feeding rates—and thus digestion—shows a rhythmic pattern following the day-night cycle with a peak at night. This is because migrating Daphnia should have a high supply of food particles during night (when they are in the euphotic zone of lakes) and should then be able to produce a high concentration of digestive protease molecules. During day, Daphnia sink to deeper water layers in order to escape optically hunting predators like fish and to avoid deleterious UV light. In this deeper water layers temperature and phytoplankton growth is low. Therefore, less digestive enzymes are needed.
Surprisingly, we did not find any differences in protease activity between points in time (neither in “CH-H-5” nor in “Binnensee”) although in krill the peak in trypsin gene expression translated into a peak of trypsin activity some hours later (Teschke et al., 2011). One explanation might be that we missed the peaks of activity because of few data points; however, this is not likely since we measured activity both several hours before the point in time of the highest gene expression (ZT 15–17, at which we would expect low activity) and 2 h after the peak (ZT 20, at which a time-shifted increase of activity would most probably already be observed). Another explanation might be that the D. magna clone did not actually migrate but was kept in 1 l glasses and at high light intensity where no escape to darker water layers was possible. From literature it is hypothesized that D. magna that are threatened by fish (by application of kairomones) and that would normally migrate, switch from one predator avoidance strategy (DVM) to another (changes in life-history; Effertz and Von Elert, 2014). Therefore, if Daphnia cannot migrate to areas with low food supply, it is also not necessary to produce digestive enzymes in a rhythmic manner following the circadian clock. Still, the gene expression is probably under clock control and cannot change its rhythmic pattern. Probably, the translation of protease mRNA to protease enzymes [or the activation of enzymatic activity by turning (chymo) trysinogen into (chymo) trypsin] is dependent on actual food supply (which in our experiments was constant over 24 h). Furthermore, also feeding rates showed a rhythmic pattern in Daphnia although no kairomones or fish were present (Pfenning-Butterworth et al., 2021).
ALAN also influenced protease gene expression and chymotrypsin activity. Although trypsin gene expression is strongly reduced in ALAN, no change in activity was observed. Eventually, trypsin is stored in the midgut gland in high amounts and the rhythmic expression of trypsin genes is only necessary to fill up the daily amount; after only 6 d of ALAN, the storage might still be full, so that activity was similar to the control. However, it might be that the activity will decrease in comparison to the control after more than 6 d of ALAN (actually trypsin activity showed a tendency to be lower in the ALAN treatment). Another explanation might be that in ALAN a different trypsin gene that produces a different isozyme was switched-on and led to the observed stable trypsin activity over time and among light treatments.
Chymotrypsin gene expression showed a shift of the peak at night similar to the clock genes. In most cases, CT448 expression was lower than in the control except at ZT 18, 19.5, and 23. Still activity was significantly increased in the ALAN treatment. It might be that also the activation of the inactive precursor of chymotrypsins (i.e., chymotrypsinogen) is influenced by ALAN which results into increased chymotrypsin activity. To our knowledge no positive effect of ALAN had been observed before. Therefore, we tested whether the observed increase of chymotrypsin activity (but not of trypsin) was only an insignificant artifact or whether it actually increased Daphnia’s tolerance.
Cyanobacteria are known to produce a diverse array of toxic secondary metabolites (Carmichael, 1994; Gademann and Portmann, 2008; Schwarzenberger et al., 2020c). One of those toxins is the group of protease inhibitors (ahp-cyclodepsipeptides; Köcher et al., 2019). It has been shown that Daphnia are negatively affected by protease inhibitors which can be measured as decreased growth (Schwarzenberger et al., 2021b). Daphnia are able to respond to protease inhibitors by increasing gene expression and protease activity and by switching on protease isoforms (Von Elert et al., 2012). Furthermore, tolerant Daphnia populations had been selected by protease inhibitors (Schwarzenberger et al., 2020b); this positive selection had led to more resistant proteases and decreased copy numbers (Schwarzenberger et al., 2017).
Here, we grew “CH-H-5” on diets with cyanobacteria that produced either trypsin or chymotrypsin inhibitors (and probably no other toxin types). It has often been shown that the presence of protease inhibitors decreases Daphnia growth rates (Von Elert et al., 2012; Schwarzenberger et al., 2021b), which we could also observe in our results when Daphnia were grown on BM25 in comparison to control food without ALAN. Actually, we found that the combination of ALAN conditions with cyanobacterial food containing chymotrypsin inhibitors led to increased Daphnia growth. This was not the case for growth on the trypsin-inhibitor producing cyanobacterium. We were unsure whether this effect on Daphnia growth-rate was an artifact of our study with a single clone grown in a day-night cycle with frequent and high intensity light pulses. Therefore, we investigated, whether this response could also be found in another clone (“FR-LR-6-1”) for which we only tested the chymotrypsin-producing cyanobacterium. This clone does not only originate from an ecosystem that is several hundred kilometers apart from the Swiss lake but also shows physiological differences in comparison to “CH-H-5,” e.g., a different growth rate in control conditions (cf. Figure 4). Again, we found increased growth in ALAN. This suggests an increased chymotrypsin activity as a general response of Daphnia to ALAN which leads to higher tolerance to cyanobacterial chymotrypsin inhibitors.
The amount of light pollution will further increase in the proceeding Anthropocene. Currently, the use of artificial light increases by about 6% and spreads into new areas by 2.2% per year world-wide (reviewed by Grubisic, 2018). The total irradiance increases by 2% per year globally and already 88% of Europe is light polluted. Here, we found that Daphnia’s circadian clock and melatonin-synthesis genes are influenced by ALAN, which is also the case for genes that are under clock-control (i.e., the proteases trypsin and chymotrypsin). Furthermore, we demonstrated that chymotrypsin activity was increased in ALAN, which led to higher tolerance to cyanobacterial chymotrypsin-inhibitors. Therefore, light-polluted Daphnia populations might be able to better control a cyanobacterial bloom if chymotrypsin-inhibitor producing cyanobacteria established in a lake.
However, although we found that ALAN is advantageous for Daphnia‘s digestion when grown on cyanobacterial protease inhibitors, we cannot rule out that the increase in protease activity is associated with a yet unknown fitness cost. In future studies, we are going to investigate trade-offs caused by ALAN. Furthermore, we are going to elucidate the mechanism underlying increased protease activity due to ALAN. Such a mechanism could be a changed microbiome in the transparent Daphnia gut (which plays a role in tolerance to the cyanobacterial toxin microcystin; Macke et al., 2017), or a general accumulation of proteases in the midgut gland in light.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
AS and AW designed the experiments that were conducted by RC and AS. All authors read and contributed to the manuscript that was written by AS and RC. This study was financed by a grant to AS by the German Research Foundation (DFG; SCHW 1830/4-1) and by a grant to AS by the University of Konstanz’s Young Scholar Fund for Excellent Postdocs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Patrick Bartolin for help in conducting the experiments. We thank Dieter Ebert for provision of the clones, and Bettina Zeis for stimulating discussions. We would also like to thank the reviewers for their very helpfull comments on our manuscript. We thank to Lutz Becks, Laura Epp, Dominik Martin-Creuzburg, Bernard Lepetit and Peter Kroth for access to their laboratory infrastructure.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.834422/full#supplementary-material
Agrawal, M. K., Zitt, A., Bagchi, D., Weckesser, J., Bagchi, S. N., and Von Elert, E. (2005). Characterization of proteases in guts of Daphnia magna and their inhibition by Microcystis aeruginosa PCC 7806. Environ. Toxicol. 20, 314–322. doi: 10.1002/tox.20123
Baker, B. J., and Richardson, J. M. L. (2006). The effect of artificial light on male breeding-season behaviour in green frogs Rana clamitans melanota. Can. J. Zool. 84, 1528–1532. doi: 10.1139/z06-142
Bentkowski, P., Markowska, M., and Pijanowska, J. (2010). Role of melatonin in the control of depth distribution of Daphnia magna. Hydrobiologia 643, 43–50. doi: 10.1007/s10750-010-0134-x
Bernatowicz, P. P., Kotwica-Rolinska, J., Joachimiak, E., Sikora, A., Polanska, M. A., Pijanowska, J., et al. (2016). Temporal expression of the clock genes in the water flea Daphnia pulex (Crustacea: Cladocera). J. Exp. Zool. Ecol. Genet. Physiol. 325, 233–254. doi: 10.1002/jez.2015
Brüning, A., Hölker, F., Franke, S., Preuer, T., and Kloas, W. (2015). Spotlight on fish: light pollution affects circadian rhythms of European perch but does not cause stress. Sci. Tot. Environ. 511, 516–522. doi: 10.1016/j.scitotenv.2014.12.094
Cellier-Michel, S., Rehailia, M., and Berthon, J. L. (2003). Is the rhythm of vertical migration of Daphnia longispina circadian or simply nycthemeral? Ann. Limnol. Int. J. Limnol. 39, 265–272. doi: 10.1051/limn/2003021
Coldsnow, K. D., Relyea, R. A., and Hurley, J. M. (2017). Evolution to environmental contamination ablates the circadian clock of an aquatic sentinel species. Ecol. Evol. 7, 10339–10349. doi: 10.1002/ece3.3490
da Silva, A., Valcu, M., and Kempenaers, B. (2015). Light pollution alters the phenology of dawn and dusk singing in common European songbirds. Philos. Trans. R. Soc. B 370:20140126. doi: 10.1098/rstb.2014.0126
Desouhant, E., Gomes, E., Mondy, N., and Amat, I. (2019). Mechanistic, ecological, and evolutionary consequences of artificial light at night for insects: review and prospective. Entomol. Exp. Appl. 167, 37–58.
Dittmann, E., Erhard, M., Kaebernick, M., Scheler, C., Neilan, B. A., von Dohren, H., et al. (2001). Altered expression of two light-dependent genes in a microcystin-lacking mutant of Microcystis aeruginosa PCC 7806. Microbiol. Sgm 147, 3113–3119. doi: 10.1099/00221287-147-11-3113
Dominoni, D. M. (2015). The effects of light pollution on biological rhythms of birds: an integrated, mechanistic perspective. J. Ornithol. 156, 409–418. doi: 10.1007/s10336-015-1196-3
Effertz, C., and Von Elert, E. (2014). Light intensity controls anti-predator defences in Daphnia: the suppression of life-history changes. Proc. Biol. Sci. Roy. Soc. 281:20133250. doi: 10.1098/rspb.2013.3250
Ekvall, M. T., Hylander, S., Walles, T., Yang, X., and Hansson, L. A. (2015). Diel vertical migration, size distribution and photoprotection in zooplankton as response to UV-A radiation. Limnol. Oceanogr. 60, 2048–2058. doi: 10.1002/lno.10151
Foster, J. G., Algera, D. A., Brownscombe, J. W., Zolderdo, A. J., and Cooke, S. J. (2016). Consequences of different types of littoral zone light pollution on the parental care behaviour of a freshwater teleost fish. Water Air Soil Pollut. 227:404.
Foulkes, N. S., Borjigin, J., Snyder, S. H., and SassoneCorsi, P. (1997). Rhythmic transcription: the molecular basis of circadian melatonin synthesis. Trends Neurosci. 20, 487–492.
Gademann, K., and Portmann, C. (2008). Secondary metabolites from cyanobacteria: complex structures and powerful bioactivities. Curr. Org. Chem. 12, 326–341.
Gehring, W., and Rosbash, M. (2003). The coevolution of blue-light photoreception and circadian rhythms. J. Mol. Evol. 57, S286–S289. doi: 10.1007/s00239-003-0038-8
Grubisic, M. (2018). Waters under artificial lights: does light pollution matter for aquatic primary producers? Limnol. Oceanogr. Bull. 27, 76–81.
Heckmann, L. H., Connon, R., Hutchinson, T. H., Maund, S. J., Sibly, R. M., and Callaghan, A. (2006). Expression of target and reference genes in Daphnia magna exposed to ibuprofen. BMC Genomics 7:175. doi: 10.1186/1471-2164-7-175
Hölker, F., Wurzbacher, C., Weißenborn, C., Monaghan, M. T., Holzhauer, S. I., and Premke, K. (2015). Microbial diversity and community respiration in freshwater sediments influenced by artificial light at night. Philos. Trans. Roy. Soc. Lond. Ser. B 370:20140130. doi: 10.1098/rstb.2014.0130
Köcher, S., Resch, S., Kessenbrock, T., Schrapp, L., Ehrmann, M., and Kaiser, M. (2019). From dolastatin 13 to cyanopeptolins, micropeptins and lyngbyastatins: the chemical biology of Ahp-cyclodepsipeptides. Nat. Prod. Rep. 37, 163–174. doi: 10.1039/c9np00033j
Lipcius, R. N., and Herrnkind, W. F. (1982). Molt cycle alterations in behavior, feeding and diel rhythms of a decapod crustacean, the spiny lobster Panulirus argus. Mar. Biol. 68, 241–252. doi: 10.1007/bf00409591
Loose, C. J., and Dawidowicz, P. (1994). Trade-offs in diel vertical migration by zooplankton: the costs of predator avoidance. Ecology 75, 2255–2263. doi: 10.1111/j.1365-2656.2005.01033.x
Macke, E., Callens, M., De Meester, L., and Decaestecker, E. (2017). Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 8:1608. doi: 10.1038/s41467-017-01714-x
Markowska, M., Bentkowski, P., Kloc, M., and Pijanowska, J. (2009). Presence of melatonin in Daphnia magna. J. Pineal Res. 46, 242–244.
Marzetz, V., Koussoroplis, A.-M., Martin-Creuzburg, D., Striebel, M., and Wacker, A. (2017). Linking primary producer diversity and food quality effects on herbivores: a biochemical perspective. Sci. Rep. 7:11035. doi: 10.1038/s41598-017-11183-3
Miller, M. W. (2006). Apparent effects of light pollution on singing behavior of American robins. Condor 108, 130–139. doi: 10.1650/0010-5422(2006)108[0130:aeolpo]2.0.co;2
Moore, M. V., Pierce, S. M., Walsh, H. M., Kvalvik, S. K., and Lim, J. D. (2000). Urban light pollution alters the diel vertical migration of Daphnia. Int. Ver. Theor. Angew. Limnol. Verh. 27, 779–782.
Muguruma, F., Goto, S. G., Numata, H., and Shiga, S. (2010). Effect of photoperiod on clock gene expression and subcellular distribution of PERIOD in the circadian clock neurons of the blow fly Protophormia terraenovae. Cell Tissue Res. 340, 497–507. doi: 10.1007/s00441-010-0966-8
Nitta, Y., Matsui, S., Kato, Y., Kaga, Y., Sugimoto, K., and Sugie, A. (2019). Analysing the evolutional and functional differentiation of four types of Daphnia magna cryptochrome in Drosophila circadian clock. Sci. Rep. 9:8857. doi: 10.1038/s41598-019-45410-w
Perkin, E. K., Hölker, F., Heller, S., and Berghahn, R. (2014). Artificial light and nocturnal activity in gammarids. PeerJ 2:e279. doi: 10.7717/peerj.279
Pfenning-Butterworth, A. C., Amato, K., and Cressler, C. E. (2021). Circadian rhythm in feeding behavior of Daphnia dentifera. J. Biol. Rhythms 36, 589–594. doi: 10.1177/07487304211054404
Rhode, S. C., Pawlowski, M., and Tollrian, R. (2001). The impact of ultraviolet radiation on the vertical distribution of zooplankton of the genus Daphnia. Nature 412, 69–72. doi: 10.1038/35083567
Roulin, A. C., Bourgeois, Y., Stiefel, U., Walser, J. C., and Ebert, D. (2016). A photoreceptor contributes to the natural variation of diapause induction in Daphnia Magna. Mol. Biol. Evol. 33, 3194–3204. doi: 10.1093/molbev/msw200
Rund, S. S. C., Yoo, B., Alam, C., Green, T., Stephens, M. T., Zeng, E., et al. (2016). Genome-wide profiling of 24 hr diel rhythmicity in the water flea. BMC Genom. 17:653. doi: 10.1186/s12864-016-2998-2
Sainath, S., Swetha, C., and Reddy, P. S. (2013). What do we (need to) know about the melatonin in crustaceans? J. Exp. Zool. A Ecol. Genet. Physiol. 319, 365–377. doi: 10.1002/jez.1800
Sancar, A. (2004). Regulation of the mammalian circadian clock by cryptochrome. J. Biol. Chem. 279, 34079–34082. doi: 10.1074/jbc.r400016200
Schwarzenberger, A., and Fink, P. (2018). Gene expression and activity of digestive enzymes of Daphnia pulex in response to food quality differences. Comp. Biochem. Physiol. B 218, 23–29. doi: 10.1016/j.cbpb.2018.01.009
Schwarzenberger, A., and Wacker, A. (2015). Melatonin synthesis follows a daily cycle in Daphnia. J. Plankton Res. 37, 636–644.
Schwarzenberger, A., Christjani, M., and Wacker, A. (2014). Longevity of Daphnia and the attenuation of stress responses by melatonin. BMC Physiol. 14:8. doi: 10.1186/s12899-014-0008-y
Schwarzenberger, A., Sadler, T., and Von Elert, E. (2013). Effect of nutrient limitation of cyanobacteria on protease inhibitor production and fitness of Daphnia magna. J. Exp. Biol. 216, 3649–3655.
Schwarzenberger, A., Handke, N. H., Romer, T., and Wacker, A. (2021a). Geographic clines in Daphnia magna’s circadian clock gene expression: local adaptation to photoperiod. Zoology 114:125856. doi: 10.1016/j.zool.2020.125856
Schwarzenberger, A., Chen, L., and Weiss, L. C. (2020a). The expression of circadian clock genes in Daphnia magna diapause. Sci. Rep. 10:19928. doi: 10.1038/s41598-020-77065-3
Schwarzenberger, A., Hasselmann, M., and Von Elert, E. (2020b). Positive selection of digestive proteases in Daphnia: a mechanism for local adaptation to cyanobacterial protease inhibitors. Mol. Ecol. 29, 912–919. doi: 10.1111/mec.15375
Schwarzenberger, A., Ilić, M., and Von Elert, E. (2021b). Daphnia populations are similar but not identical in tolerance to different protease inhibitors. Harmful Algae 106:102062. doi: 10.1016/j.hal.2021.102062
Schwarzenberger, A., Keith, N. R., Jackson, C. E., and Von Elert, E. (2017). Copy number variation of a protease gene of Daphnia: its role in population tolerance. J. Exp. Zool. A 327, 119–126. doi: 10.1002/jez.2077
Schwarzenberger, A., Kurmayer, R., and Martin-Creuzburg, D. (2020c). Toward disentangling the multiple nutritional constraints imposed by Planktothrix: the significance of harmful secondary metabolites and sterol limitation. Front. Microbiol. 11:586120. doi: 10.3389/fmicb.2020.586120
Schwarzenberger, A., Zitt, A., Kroth, P., Mueller, S., and Von Elert, E. (2010). Gene expression and activity of digestive proteases in Daphnia: effects of cyanobacterial protease inhibitors. BMC Physiol. 10:6. doi: 10.1186/1472-6793-10-6
Strauss, J., and Dircksen, H. (2010). Circadian clocks in crustaceans: identified neuronal and cellular systems. Front. Biosci. 15:1040–1074. doi: 10.2741/3661
Talanda, L., Maszczyk, P., and Babkiewicz, E. (2018). The reaction distance of a planktivorous fish (Scardinius erythrophthalmus) and the evasiveness of its prey (Daphnia pulex x pulicaria) under different artificial light spectra. Limnology 19, 311–319.
Teschke, M., Wendt, S., Kawaguchi, S., Kramer, A., and Meyer, B. (2011). A circadian clock in Antarctic krill: an endogenous timing system governs metabolic output rhythms in the euphausid species Euphausia superba. PLoS One 6:e26090. doi: 10.1371/journal.pone.0026090
Tilden, A. R., McCoole, M. D., Harmon, S. M., Baer, K. N., and Christie, A. E. (2011). Genomic identification of a putative circadian system in the cladoceran crustacean Daphnia pulex. Comp. Biochem. Physiol. D Genomics Proteomics 6, 282–309. doi: 10.1016/j.cbd.2011.06.002
Tomioka, K., and Matsumoto, A. (2010). A comparative view of insect circadian clock systems. Cell. Mol. Life Sci. 67, 1397–1406. doi: 10.1007/s00018-009-0232-y
van Geffen, K. G., Van Grunsven, R. H. A., Van Ruijven, J., Van Berendse, F., and Veenendaal, E. M. (2014). Artificial light at night causes diapause inhibition and sex-specific life history changes in a moth. Ecol. Evol. 4, 2082–2089. doi: 10.1002/ece3.1090
Von Elert, E., and Jüttner, F. (1997). Phosphorus limitation not light controls the exudation of allelopathic compounds by Trichormus doliolum. Limnol. Oceanogr. 42, 1796–1802.
Von Elert, E., Zitt, A., and Schwarzenberger, A. (2012). Inducible tolerance in Daphnia magna to dietary protease inhibitors. J. Exp. Biol. 215, 2051–2059. doi: 10.1242/jeb.068742
Walker, W. H., Bumgarner, J. R., Walton, J. C., Liu, J. A., Meléndez-Fernández, O. H., Nelson, R. J., et al. (2020). Light pollution and cancer. Int. J. Mol. Sci. 21:9360.
Yasuo, S., Watanabe, M., Okabayashi, N., Ebihara, S., and Yoshimura, T. (2003). Circadian clock genes and photoperiodism: comprehensive analysis of clock gene expression in the mediobasal hypothalamus, the suprachiasmatic nucleus, and the pineal gland of Japanese quail under various light schedules. Endocrinology 144, 3742–3748. doi: 10.1210/en.2003-0435
Keywords: circadian clock, light pollution, cyanobacterial protease inhibitors, cryptochrome, protease activity, melatonin
Citation: Cremer R, Wacker A and Schwarzenberger A (2022) More Light Please: Daphnia Benefit From Light Pollution by Increased Tolerance Toward Cyanobacterial Chymotrypsin Inhibitors. Front. Ecol. Evol. 10:834422. doi: 10.3389/fevo.2022.834422
Received: 13 December 2021; Accepted: 20 January 2022;
Published: 14 February 2022.
Edited by:
Mingbo Yin, Fudan University, ChinaReviewed by:
Piotr Dawidowicz, University of Warsaw, PolandCopyright © 2022 Cremer, Wacker and Schwarzenberger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anke Schwarzenberger, YW5rZS5zY2h3YXJ6ZW5iZXJnZXJAdW5pLWtvbnN0YW56LmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.