
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 22 February 2022
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.829989
This article is part of the Research Topic Evolutionary Cytogenetics of Insects View all 7 articles
Recent phylogenetic and molecular data are changing our knowledge about the relations between species and evolutionary processes resulting in the chromosome variation observed in ants (Hymenoptera: Formicidae). Ants exhibit remarkable variations in morphology, behavior, karyotypes, and chromosome structure. By assembling genetic and chromosome information about the trap-jaw ants from the subfamily Ponerinae, we reconstructed the phylogenetic relationships that inferred the monophyletic condition between the Anochetus and Odontomachus genera and estimated their ancestral haploid chromosome number. According to our inferences, these clades have an ancestral haploid chromosome number n = 15. The most recent common ancestor of Anochetus and Odontomachus has arisen between the Early Paleocene and the Early Eocene periods (time of the most recent common ancestor). In the Anochetus genus, we observed maintenance of the ancestral chromosome number estimated here in most species. This also suggests that pericentric inversions were the primary chromosomal rearrangement modulating the karyotype evolution of this genus. However, a reduction from n = 15–14 is observed in Anochetus emarginatus and Anochetus cf. madaraszi, which likely occurred by centromeric fusion. In contrast, the increase from the ancestral karyotype number in Anochetus horridus suggested centromeric fissions. Odontomachus showed maintenance of the ancestral chromosome number in the “rixosus group” and several gains in all species from the “haematodus group.” Our findings suggest that centromeric fissions and pericentric rearrangements lead to chromosomal changes in trap-jaw ants. Considering the ancestral state estimated here, changes in chromosome morphology are likely due to pericentric inversions, and chromosome number increases are likely due to centric fissions. The higher number of acrocentric or telocentric chromosomes in the karyotypes with n < 15 haploid chromosomes supports such an idea.
Chromosomes are fundamental parts of genome organization that can be related to interspecific divergences. Morphological and numerical chromosomal traits are relevant to understanding the fundamental aspects of the genomic organization of eukaryotes, and are often key components in comprehending evolutionary pathways and karyotypic differentiation (Imai et al., 1986, 1988, 1994, 2001; Hirai et al., 1996; Goodisman et al., 2008; Lorite and Palomeque, 2010; Cardoso et al., 2014; Menezes et al., 2014; Velasco et al., 2014; de Aguiar et al., 2020; Micolino et al., 2020). For instance, chromosomes can differ in size, shape, and genetic information, such as in their number and redundancy. These variations can be observed in individual organisms of a population and between different species. Considering all such characteristics, they play a very important role in evolutionary studies (Schubert, 2007). Analyzing plants and vertebrates, Rieseberg (2001) assumed that chromosome rearrangements could reduce gene flow more effectively than other mechanisms. These thoughts are concordant with what has been postulated some decades before, when it was proposed that changes in chromosome patterns were very important in isolating lineages, promoting rapid speciation, and diversification (Bush et al., 1977).
Taking Formicidae into account, previous studies have been done to understand the evolution of chromosomal changes. Several hypotheses have been raised due to the diversity and variation in ant species and karyotypes (see Cardoso et al., 2018, reviewed in Lorite and Palomeque, 2010). The main expressive and discussed phenomena to chromosomal change in ants is based on the Minimum Interaction Theory proposed by Imai et al. (1988, 1994). According to this theory, karyotype will change in response to the potential of contact (interaction) inside the nucleus. It favors a mechanism that prevents deleterious bumps between long chromosomes, which promote fission rearrangements, chromosomal shortening, and chromosome number increase (see Imai et al., 1994 for details). The evidence of such a dynamic mechanism comes from studies of Myrmecia species that bear a single chromosome in haploid male and 84 chromosomes in the diploid female (Cardoso et al., 2018)1. It is important to mention that this theory can be considered too mechanistic and biased to speciose groups within eukaryotes.
Besides the karyotype description, studies at the species level are still poorly explored in ants, mainly when coupling cytogenetic and molecular data. Cardoso et al. (2014) reconstructed the ancestral chromosome number of the Mycetophylax genus. By matching these analyses with karyotype and chromosome banding, Cardoso et al. (2014) proposed that centromeric fusion and fission were responsible for the chromosomal variation in this genus. Their findings also suggested that the lineages of Mycetophylax morschi Emery, 1888, should be treated as two phylogenetically isolated species by the differences in their karyotype structure. In the subsequent work, cytogenetic and molecular techniques revealed differences between lineages of M. morschi including the localization of the 45S rDNA cluster, reinforcing the hypotheses that these lineages should be treated as a cryptic or even distinct species (Micolino et al., 2019). In addition, Cristiano et al. (2013) performed a study combining molecular and cytogenetic data, which suggested that Acromyrmex striatus Roger, 1863 should be allocated to a new genus. Thus, based on morphological characters, colony size, behavior, chromosome inferences, and phylogenetic analyses, Acromyrmex striatus and Acromyrmex silvestrii Emery, 1905, including the subspecies Acromyrmex silvestrii bruchi Forel, 1902, were allocated to the new genus Amoimyrmex (Cristiano et al., 2020). Studies considering cytogenetic and molecular data together are unavailable for most ant subfamilies, including Ponerinae, despite cytogenetic variation and phylogenetic position within Formicidae. For instance, Ponerinae includes the ant species with the highest chromosome number known (Dinoponera lucida Emery, 1901, 2n = 120; Mariano et al., 2008) and several ancient characteristics like solitary foraging, small colonies, small differentiation between workers and the queen, and monomorphic worker caste (Wilson and Holldobler, 2005; Schmidt, 2013). In their first phylogeny description by morphological analyses, the monophyletic condition of this subfamily was shown by Brown (1958) and corroborated by molecular phylogenetic studies (Brady et al., 2006; Moreau et al., 2006; Schmidt, 2013).
Ponerinae ants are diverse with karyotypes ranging from 3 to 60 chromosomes in the haploid set (see text footnote 1; Cardoso et al., 2018). There is a huge variation along genera and species, or even among populations (e.g., Mariano et al., 2008; Santos et al., 2010). Distinct cytotypes and chromosome morphology have been used to discuss about the polyphyly of certain Ponerinae clades, such as Pachycondyla (Mariano et al., 2012), contributing to systematics and taxonomy of this ant subfamily. Among the Ponerinae, the genera Odontomachus and Anochetus extend out due to slightly chromosomal number inertia in the former compared to the later, and long sword-like mandibles that can open 180° and close in a snapshot, being known as trap-jaw ants. Interestingly, these two genera are reciprocally monophyletic (Larabee et al., 2016; Fernandes et al., 2021). The most recent common ancestor (MRCA) of both genera probably emerged in the Neotropical or Afrotropical region at the late Cretaceous, about 65 Mya. It is supported that Odontomachus arose in the Neotropical or Afrotropical regions, dispersing about 37 Mya in the Eocene. The Anochetus species shared an MRCA that probably originated in the Neotropical region and radiated about 58 Mya in the Paleocene, earlier than the estimated radiation of Odontomachus (Fernandes et al., 2021).
Anochetus chromosome numbers vary from n = 14–23, while in Odontomachus, most species that have their karyotype described present n = 22, except for Odontomachus latidens and Odontomachus rixosus that exhibit n = 15, as can be seen in the Ant Chromosome database (ACdb) (see text footnote 1; Cardoso et al., 2018). Moreover, in Anochetus, we observed a high variability at the karyotype structure level, while in Odontomachus, the karyotype structure presented less variation in all cytogenetically analyzed species, notable by the absence of metacentric and the high presence of telocentric chromosomes, suggesting that fissions were likely the main chromosomal rearrangements involved in karyotypic changes in trap-jaw ants (Santos et al., 2010).
Despite the plethora of phylogenetic inference publications currently found in the literature, few studies utilize these inferences to reconstruct the possible ancestral chromosome states. In the present study, our framework combined phylogenetic and cytogenetic data to infer the ancestral chromosome number of Anochetus and Odontomachus trap-jaw ants. We have hypothesized that several lineages of both genera have undergone chromosomal rearrangements throughout their evolutionary times, being a low chromosome number below n = 22 the ancestral condition. From our analyses, we show a remarkable chromosomal diversification pattern in trap-jaw ants. Several events of changes in the number and morphology of chromosomes were identified and discussed in the light of the species evolution from the karyotype of the most recent common ancestor of trap-jaw ants.
Since we could not get the phylogenetic hypotheses (tree files) available in the literature, we searched for the gene sequences available on GenBank to reconstruct a phylogenetic hypothesis focusing on Odontomachus and Anochetus. In total, five molecular markers were identified to cover the majority of species: the mitochondrial gene cytochrome oxidase I (cox1), three nuclear protein-coding genes: wingless (wg), long-wave rhodopsin (LWRh), and rudimentary (CAD), and the nuclear large subunit ribosomal RNA gene (nRNA 28S) (Supplementary Table 1).
Sequences were assembled and edited in the MEGA X program (Kumar et al., 2018), aligned using the ClustalW algorithm for the 28S gene. WebPRANK (Löytynoja and Goldman, 2010) was used to align the other four protein-coding genes. The amino acid sequences were downloaded for these genes alongside their nucleotide sequence from GenBank. To ensure correct codon positions, protein alignments were constructed and mapped to nucleotide alignments using RevTrans (Wernersson and Pedersen, 2003). Ambiguously aligned regions of the genes and introns from LWRh and CAD genes were removed before further analyses. Cytogenetic information available for Anochetus and Odontomachus was downloaded from the Ant Chromosome database - ACdb (Cardoso et al., 2018) (see text footnote 1). Further information was also retrieved from the literature (Table 1).
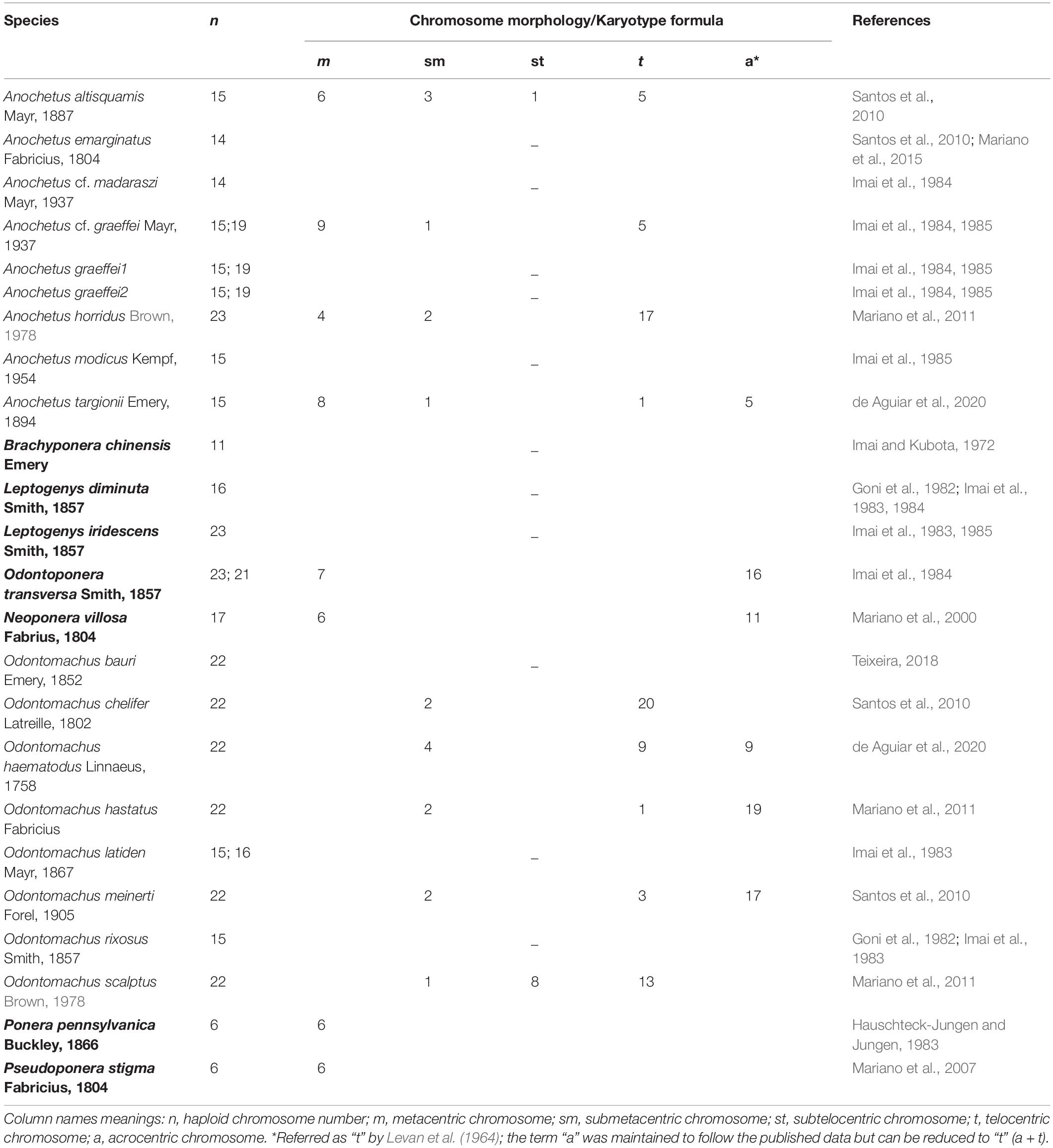
Table 1. Chromosome haploid numbers and karyotype formula for Anochetus and Odontomachus species found in the literature plus outgroups used in reconstruction analysis (in bold).
The following methods were applied for the phylogenetic inference. PartitionFinder 2 (Lanfear et al., 2017) was used to choose partitioning schemes and models of molecular evolution using the concatenated alignment of five genes. The concatenated alignment was divided into 13 subsets (one for 28S and three for each protein-coding gene), and the best model for each partition was determined (Supplementary Table 2). After that, the concatenated alignment was used as input in MrBayes 3.2.2 (Ronquist and Huelsenbeck, 2003) with a compound Dirichlet prior for more accurate branch lengths estimation [command line: “brlenspr = Unconstrained:GammaDir (1.0, 0.10, 1.0, 1.0)]”, and each partition set according to the results of the PartitionFinder 2 analyses. Base frequencies, substitution rates, and gamma-shaped parameters were unlinked across partitions. Each analysis consisted of two simultaneous Markov Chain Monte Carlo (MCMC) runs with four chains per run for 40 million generations. The first 25% of the sampled trees were discarded as burn-in. A Maximum Likelihood tree was recovered using the rapid bootstrapping algorithm (1,000 replicates) in the RAxML NG program (Kozlov et al., 2019). The same partition scheme used in MrBayes analyses was used for Maximum Likelihood inferences.
A user-friendly Python script was developed for pruning trees when only clades which chromosome numbers had been previously determined were needed from the full tree (see “Chromosome Ancestral Estimation” section below). This script was deployed in an iPython notebook (available on https://bit.ly/2W0vI3w and annexed in the Supporting Information), including detailed usage instructions for code reusability and transparency. The ETE 3 library (Huerta-Cepas et al., 2016) functions were used for the pruning method. A tree in the newick format was used as an input, and clade selections were done by a dropdown menu or text input widgets. The output was a pruned tree in the newick format, including only the selected clades and optionally summing branch lengths when required by the user for proper tree scaling.
Thus, 25 species from the Ponerinae subfamily were selected with chromosome numbers previously described in the literature, including 10 Anochetus and 10 Odontomachus species and five other species as an outgroup. This dataset was used to infer the ancestral chromosome condition of trap-jaw ants.
ChromEvol 2.0 (Glick and Mayrose, 2014) and ChromoSSE (Goldberg and Igić, 2012) were used to infer the best chromosome evolutionary model and the haploid ancestral number in the present study. Two independent runs were carried out: complete phylogeny, including missing cytogenetic data, and a pruned tree with complete cytogenetic data. Therefore, the Bayesian tree retrieved in our phylogenetic analysis was pruned using the iPython notebook to include only the taxa containing chromosome number descriptions. For the ancestral chromosome number estimation, the constant rate model presented itself as the best chromosome evolution hypothesis among the options displayed in ChromEvol 2.0 (see Glick and Mayrose, 2014) since other models included polyploid and demi-polyploid duplications, which are very rare in Metazoa. In the same way, the polyploid and demi-polyploid events were not set for ChromoSSE.
Analyses resulted in trees that displayed the same phylogenetic relationships, particularly within the genera Anochetus and Odontomachus. The topological structures of these trees were retained across Bayesian inference (Figure 1) and the Maximum Likelihood inference methods (Figure 1 and Supplementary Figure 1). The support values of Bayesian and Maximum Likelihood inferences for each clade are available in Table 2.
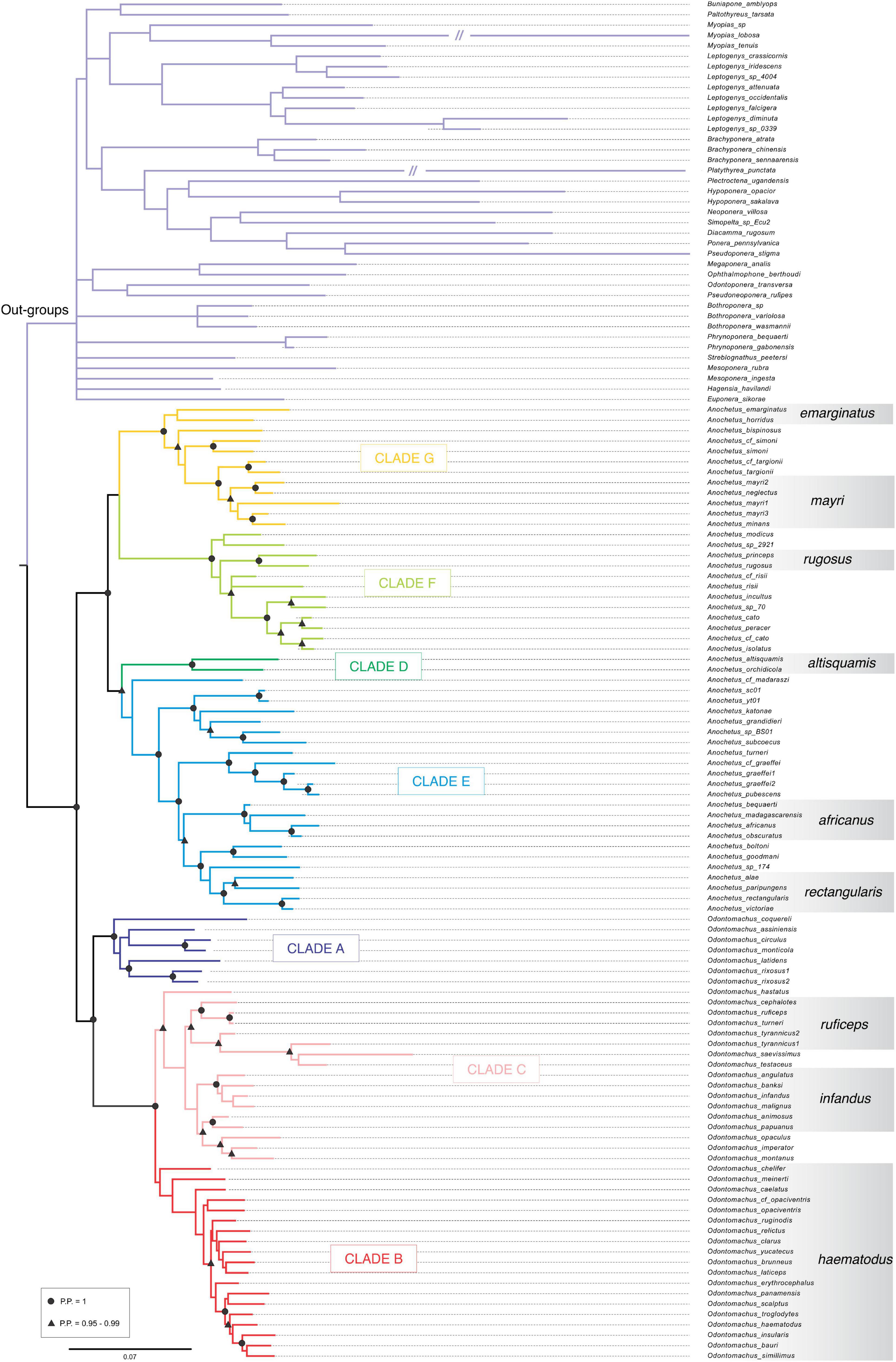
Figure 1. Global molecular phylogeny of trap-jaw ants reconstructed under Bayesian inference used to ancestral chromosome reconstruction. Clades A–G are highlighted in different colors. Morphological groups proposed by Brown (1978), are shown at the right side of the tree. The outgroup can be found at the top of the figure in light purple.
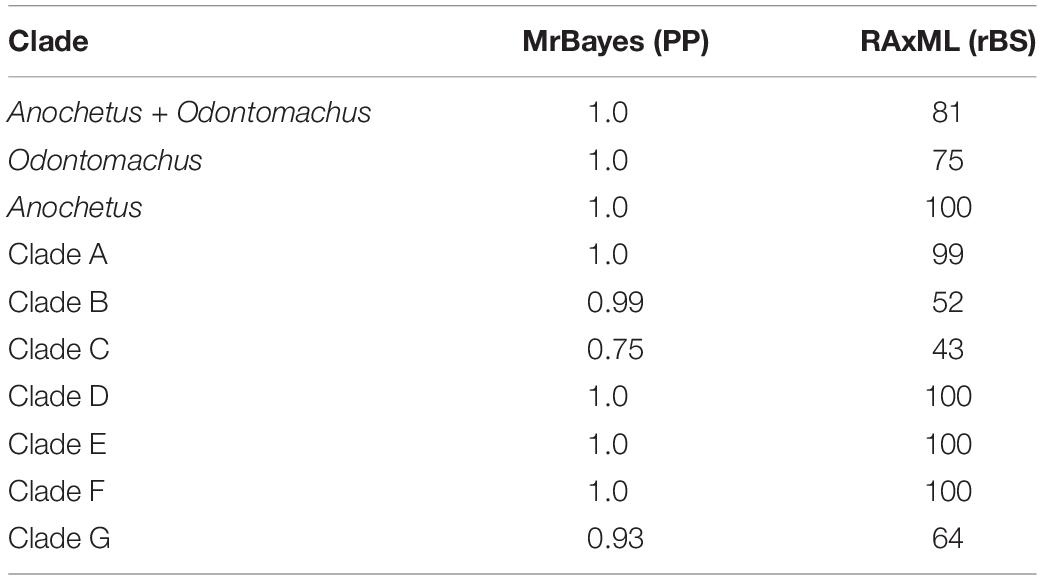
Table 2. Support values for Bayesian Inference Posterior Probability (PP) and Maximum Likelihood (BS) with the respective groups of the genus Anochetus and Odontomachus.
The clades A, B, and C are formed by species of the Odontomachus genus. Clade A displayed high support values in both trees with a PP: 1.00 and BS: 99. Clade B was represented by the hastatus group and showed PP: 0.99 and a low value of BS: 52. Clade C included the ruficeps and infandus groups with PP: 0.75 and BS: 43. The clades D, E, F, and G are composed of species of the Anochetus genus. Clade D was formed by the altisquamis group with PP: 1.0 and BS: 100. Clade E, to which the rectangularis group belongs, had BP: 1.0 and BS: 100. The clade F also contained high support values of BP: 1.0 and BS: 100. The clade G included the groups mayri and emarginatus with BP: 0.93 and rBS: 64.
Our results indicated that the chromosomal ancestral number of Odontomachus and Anochetus genera was n = 15 (PP = 0.98, Figure 2). In Odontomachus, n = 15 was retained as the ancestral chromosome number for the Odontomachus latidens and Odontomachus rixosus species. In the haematodus group, this number evolved to n = 22 (Figures 2, 3). Most ancestral chromosome numbers were inferred with high posterior probabilities (above 95%), with some notable exceptions. This was the case for two ancestors representing the transition between groups D and E in Anochetus. This can be explained by the existence of descriptions for both chromosome numbers n = 15 and n = 19 in Anochetus cf. graeffei and Anochetus graeffei.
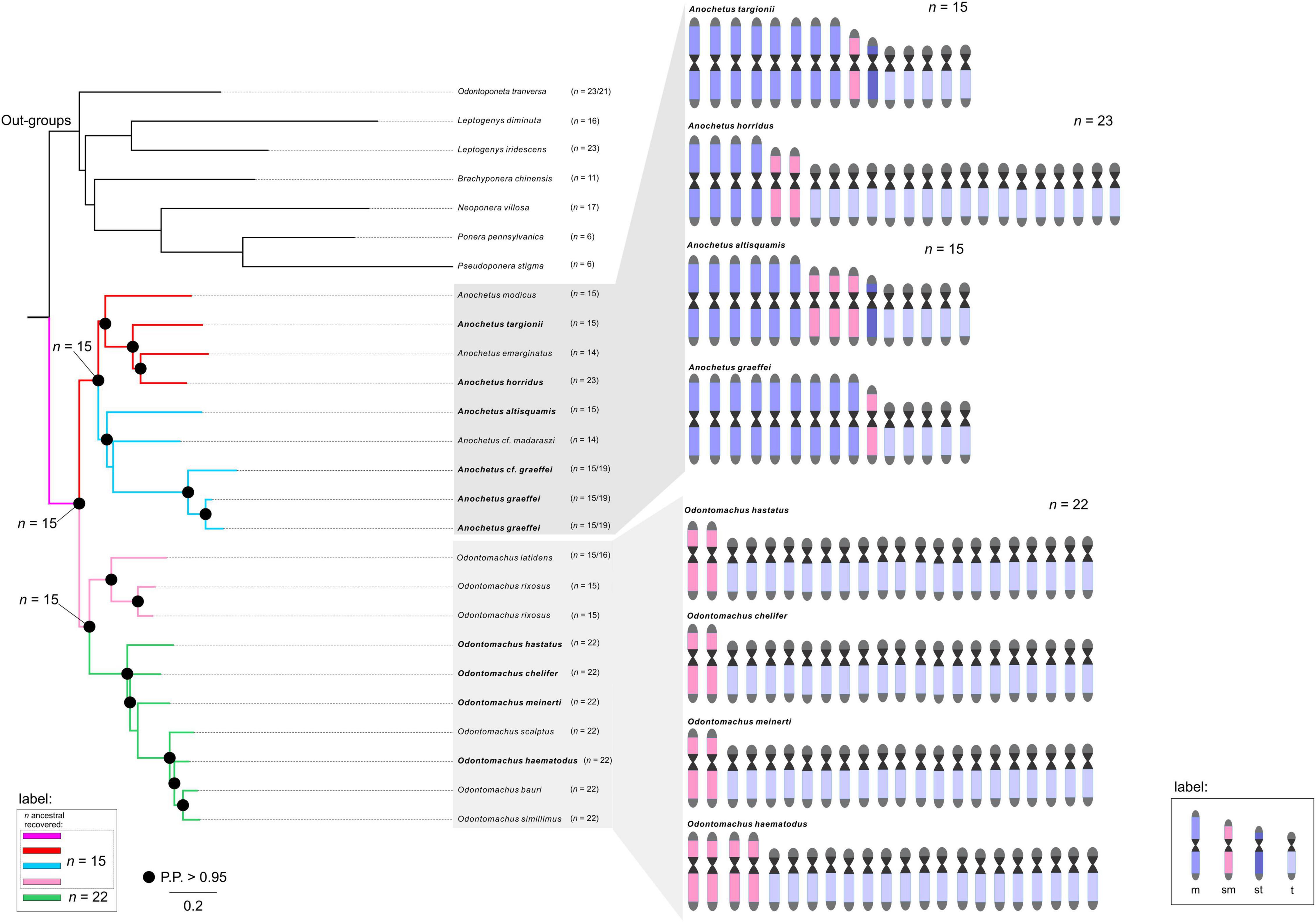
Figure 2. Ancestral haploid chromosome state reconstruction inferred under Bayesian Inference and Maximum Likelihood methods. The ancestral chromosome number with the highest probability or with the best likelihood are denoted at the main nodes. The colors of branches represent each given chromosome number according to the legend. The known karyotypes of species are given at the tip. An ideogram was estimated from the available karyotypic formulae (in bold), denoting the number of metacentric, submetacentric, subtelocentric, and telocentric chromosomes along the phylogeny.
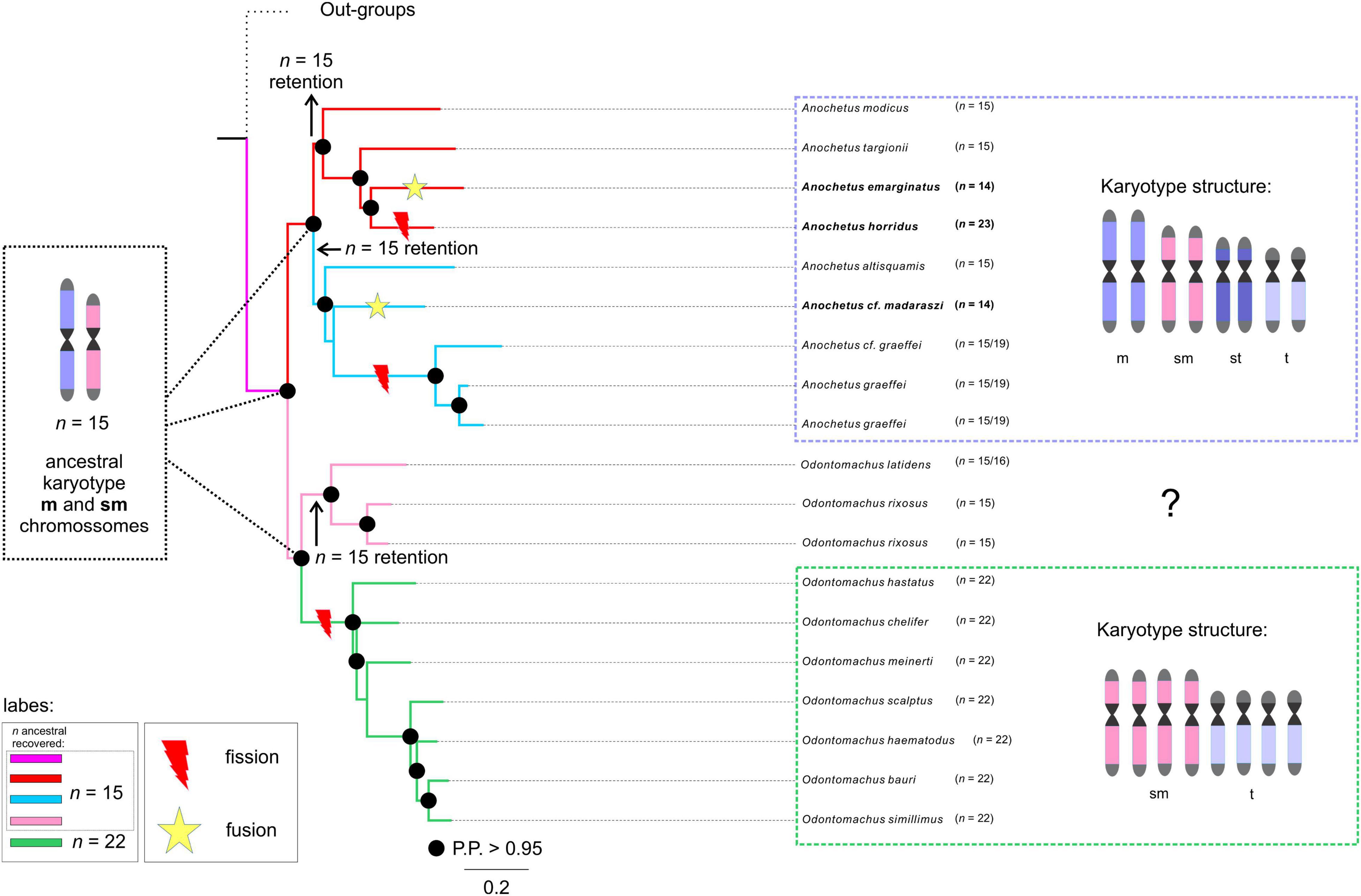
Figure 3. Chromosome number evolution and inferred ancestral chromosome state in the trap-jaw ants inferred under Bayesian and Maximum likelihood optimization. Number at the nodes and respective colors (see label) present the inferred ancestral haploid chromosome. Numbers at the tips are the known haploid chromosome numbers of species. At right of each clade of Anochetus and Odontomachus are depicted the general chromosome morphology comprising the karyotypes. At each branch are highlighted the likely rearrangements undergone during karyotype evolution (see “Discussion” for details).
From the ChromEvol best model, the final rate parameter values were 5.93, 1.87 × 10–5, and 39.37 for chromosomal duplication, gain, and loss events, respectively. This model was selected over models including missing data and all ChromoSSE models due to higher posterior probability values found. Both ChromoSSE and ChromEvol pruned ancestral chromosome number trees and the full data ChromEvol tree proposed similar ancestral haploid chromosome numbers and can be regarded as equivalent. We used Tracer 1.5 (Rambaut et al., 2018) to determine that our sampling of the posterior distribution had reached an effective sample size (ESS) and to ascertain that the number of generations required to reach stationarity of the posterior distribution had been reached. For all models, including the full phylogenies for the full data, see Supplementary Figures 2–4.
In this work, we identified the likely chromosome number of the MRCA of Anochetus and Odontomachus trap-jaw ants. We estimated the haploid number as n = 15 chromosomes with a strong posterior probability (Figure 2). We hypothesize that this ancestral karyotype would presumably have been characterized by metacentric/submetacentric chromosomes as this is the most likely way to elucidate the chromosome evolution of these sister genera and the respective karyotype structure. In this scenario, chromosomal rearrangements accompanied the diversification of both trap-jaw ant genera. Some indications for such an assumption must be mentioned. First, there may be a trend for an increase in the chromosome number, mediated by an unbalanced fission-fusion ratio, at least in some animal taxa, especially ants (Imai et al., 1994, 2001; de Vos et al., 2020). Second, the karyotyped Anochetus species, belonging to six morphological groups well distributed along its phylogenetic tree (see Fernandes et al., 2021), tend to have a conserved number of n = 15. Moreover, early splitting karyotyped species of Odontomachus in the tree also have n = 15 (Figure 2). Third, the species used here as outgroups, all within the informal Odontomachus genus group (see Schmidt and Shattuck, 2014), are recorded with a predominance of metacentric chromosomes (see Table 1). Considering these arguments, the ancestor of Anochetus and Odontomachus had n = 15 chromosomes, most of them putatively being metacentric/submetacentric (Figure 3). Apparently, the initial diversification of trap-jaw ants into two genera was not mediated by numerical chromosomal changes, and we do not have enough data to suppose any chromosomal speciation between Anochetus and Odontomachus. On the other hand, the karyotypes of current species can only be evolutionarily explained by chromosomal changes that numerically alter the karyotypes, such as fissions and fusions. Further, the massive number of subtelocentric and telocentric chromosomes within karyotypes with n > 15 (see Figure 2) agree with this assumption, as suggested by Santos et al. (2010). Here, based on our ancestral state reconstruction, we can corroborate the previous assumption, indicating that the alternative hypotheses of fusions from n = 22 to n < 22 in the observed karyotype today is unlikely. However, fusions are apparently involved in the karyotypes with n < 15 (see Figure 3).
Independent divergence-dating estimates show that the MRCA of Anochetus and Odontomachus would have arisen either in the Early Paleocene at about 64.8 Mya (Fernandes et al., 2021) or in the Early Eocene at about 52.5 Mya (Larabee et al., 2016). Biogeographical estimates, on the other hand, are still incongruous but leading toward the Neotropical region (Larabee et al., 2016; Fernandes et al., 2021). As increased taxon sampling leads to effective estimates of divergence time inference (reviewed by Bromham et al., 2018), Fernandes et al. (2021) have an advantage. According to these authors, Anochetus and Odontomachus appeared at around 53.9 and 51.4 Mya, respectively. Regardless of the variation of these divergence time estimates, a major global climatic event could be involved in the origin and diversification of both trap-jaw ants. We refer to the largest known climatic warming of the Cenozoic, the Paleocene–Eocene Thermal Maximum (PETM). The PETM was a transient hyperthermal event occurring around 55 Mya and characterized by a global temperature increase of 5–8°C (McInerney and Wing, 2011). Profound changes in biotic communities were identified during PETM, including the mass extinction of benthic foraminifera (Clyde and Gingerich, 1998; McInerney and Wing, 2011). Severe environmental disturbances, such as PETM, create unstable environments for natural populations. Some authors have shown that low recombination rates are favored in individuals living in unstable environments within rapidly developing populations (Escudero et al., 2012; Carta et al., 2018), in addition to the likely role of recombination suppression in speciation (Faria and Navarro, 2010). However, Anochetus and Odontomachus would have diversified without karyotype differentiation, while some descendant karyotypes must have evolved independently in specific clades/lineages from those having n = 15 chromosomes. The mechanisms of chromosomal change that provided such structural and numerical alterations were identified from the detailed analysis of the described karyotypes of some of these descendent lineages and our ancestral chromosome number estimates.
Anochetus has been morphologically and phylogenetically divided into 22 species groups. However, many of them were considered polyphyletic (Larabee et al., 2016; Fernandes et al., 2021). Thus, many of them (not all) should be disregarded from a phylogenetic point of view. Therefore, the correct way to analyze the karyotype differences is to place many of these lineages into major clades. Although it seems relatively scarce, our taxon sampling includes well-defined clades and species in the phylogenetic tree. The ancestral chromosome number of Anochetus was estimated as n = 15 (Figure 2). The maintenance of n = 15 in some Anochetus species, including A. altisquamis and A. modicus, which are shown as diverged earlier in the phylogenetic tree (see also Larabee et al., 2016; Fernandes et al., 2021), provides further evidence that this is a plesiomorphic condition. Such an idea has been suggested by de Aguiar et al. (2020) based on cytogenetic data. Our observations include not only Anochetus, but, as we have found out, Odontomachus as well. Interesting findings about two major clades of Anochetus must be considered. One clade comprises the emarginatus species group. In this clade, the ancestral n = 15 has undergone several chromosomal rearrangements, including putative pericentric inversions and centric fissions and fusions (Figure 3). As A. emarginatus and A. horridus are sister species, an ancestral karyotype with n = 15 had apparently undergone at least one fusion event in A. emarginatus and many fission events in A. horridus, increasing the chromosome number of the latter of n = 15 to n = 23 (Figure 3). The other clade of Anochetus refers to the so-called “Clade J” (see Fernandes et al., 2021), composed of various scattered species groups. Again, both fission and fusion seem to be characteristic of this clade. Previous cytogenetic studies have raised issues about chromosomal rearrangements having participated in the diversification of trap-jaw ants. Santos et al. (2010) conclude that chromosomal fission may have played an important role in the divergence between Anochetus and Odontomachus, while de Aguiar et al. (2020) suggest this rearrangement particularly in A. horridus. Here, we can suggest that in fact chromosomal fission seems to have played an important role in recent karyotype evolution, increasing the number of chromosomes in the karyotype of Odontomachus from an ancestral karyotype of n = 15.
Interestingly, A. graeffei had two distinct karyotypes described, one with n = 15 and the other with n = 19 (see text footnote 1; Cardoso et al., 2018). Unless these lineages have been misidentified, it is reasonable to think that these must be closely related distinct species, which indicate that at least fission rearrangements would have occurred throughout its evolutionary history. Descriptions of distinct number of chromosomes among populations of the same species are common in ants (see Lorite and Palomeque, 2010). As an example, three distinct karyotypes were reported for the same species of fungus-farming ants, Mycetophylax morschi. Integrative cytogenetic and molecular analyses propose that they must be independent lineages and that chromosomal rearrangements played a role in their diversification (Cardoso et al., 2014; Micolino et al., 2019). Similarly to Anochetus, the genus Odontomachus has been classified into 12 species groups (Larabee et al., 2016; Fernandes et al., 2021). Two major clades can be observed, each with mostly monophyletic species groups: on the one hand the rixosus group, for example, and on the other with the hastatus and the haematodus groups. Likewise, we presented the Odontomachus species with cytogenetic data that are well distributed along the phylogenetic tree. We also estimated the ancestral chromosome number of Odontomachus as n = 15 (Figure 2). Although there are no cytogenetic data on the chromosome morphology of Odontomachus species with n = 15 (those belonging to the rixosus species group), we predict that these must have metacentric and/or submetacentric chromosomes. In this scenario, we suggest that the ancestor of hastatus + haematodus species groups would have undergone at least six centric fission rearrangements, increasing from n = 15 to 22 chromosomes (Figure 3). The haematodus species group is the most diverse of Odontomachus, consisting of about 30 lineages, mainly distributed in the Neotropics (Bolton, 2021). Divergence dating reconstructions estimated its origin to have occurred at about 20 Mya in the early Miocene (Fernandes et al., 2021). The rapid radiation of this group may therefore be related to the severe climatic events that happened from this period onward. Previous studies with Neotropical ants raised hypotheses about a relationship between chromosomal changes and environmental disturbances, which could have somehow triggered and/or accompanied lineage diversification (Micolino et al., 2019). Species in the haematodus group with cytogenetic data available correspond to n = 22, most of which are telocentric chromosomes. Despite the conserved chromosome number, structural rearrangements, such as pericentric inversions, must have played a role in the diversification of this group. Inversions abound in natural populations, and it has been thought that inversions are central to the evolution of many species, with implications for adaptation and speciation (Wellenreuther and Bernatchez, 2018; Faria et al., 2019). In ants, inversion rearrangements have often been identified using cytogenetic and molecular techniques (Imai et al., 1994; Wang et al., 2013; Micolino et al., 2020).
We were able to indicate the major inversion rearrangements in the light of phylogenetic relationships among the species in the haematodus group. One way to determine whether there have been structural changes in karyotypes with the same chromosome number in closely related lineages is to look for disparities in the chromosome arms. The analysis of the number of chromosome arms, often known as the fundamental number (FN) (see Matthey, 1945), can be used for inferences about karyotype evolution, which lead to possibilities of both numerical and structural rearrangements. For example, cytogenetic analyses considering only the FN of Nothoscordum weed species indicate that the variation in the chromosome number (2n = 8–26) was provided by centric fusion-fission and polyploidy (Guerra, 2008). Likewise, cytogenetic data from Myrmecia ants exposed high variation in the number and structure of their karyotypes and suggested that many telocentric chromosomes were converted into bi-armed types (metacentric, submetacentric, and/or subtelocentric) by pericentric inversions (Imai et al., 1977). Furthermore, theoretically based cytogenetic inferences suggest that karyotypes changing only by pericentric inversion tend to evolve into the accumulation of bi-armed chromosomes (Imai and Maruyama, 1978). From our approach to the karyotype evolution and the well-established phylogenetic relationships within the haematodus species group, we identified the likely inversion rearrangements that occurred throughout cladogenesis events. The species O. chelifer diverge early in the phylogenetic tree, and its 22 haploid chromosomes are classified as 2 sm + 20 t. In just one cladogenesis event, three pericentric inversions were established, where three of the telocentric chromosomes changed to subtelocentric (KF = 2 sm + 3 st + 17 t in O. meinerti). At this point in evolutionary history, FN increased from 48 to 54, following a decrease in telocentric chromosomes. After a hiatus of four split events due to lack of available cytogenetic data, we again observed a decrease in telocentric chromosomes in O. scalptus and a consequent increase in FN (KF = 1 sm + 8 st + 13 t and FN = 62). The karyotype of this species must have undergone five pericentric inversions, four of them involving telocentrics and the remaining one involving a submetacentric chromosome. The most derived species from this clade, O. bauri and O. haematodus, also had karyotype changes shaped by pericentric inversions. While O. bauri had two inversions occurring only in the subtelocentric chromosomes (KF = 3 sm + 6 st + 13 t and FN = 62), O. haematodus appears to have undergone four inversions, three of them being from telocentric to subtelocentric and one from telocentric to submetacentric (KF = 4 sm + 9 st + 9 t and FN = 70). We emphasize that, at least in this ant clade, there was a tendency to increase in bi-armed chromosomes. In this way, we corroborate previous hypotheses demonstrating that chromosome evolution generally proceeds toward increasing the chromosome number by the combination of centric fissions and pericentric inversions, changing telocentric chromosomes to bi-armed ones (Imai et al., 2001).
The cytogenetic data available still cover a small number of trap-jaw species across the phylogeny, and this could be a limitation to our ancestral state reconstruction. However, we managed to discover valuable rearrangements in the course of the chromosome evolution of Anochetus and Odontomachus trap-jaw ants that likely contributed to their diversification. Further, species with available chromosome numbers within this group are still scarce, but they comprise a significant part of the major clades. Nevertheless, we noticed many problems that make comparative analyses difficult and these problems must be resolved during the future studies. Descriptions of many karyotypes lack essential details, which impedes the reliability of the data. Likewise, if only chromosome numbers are known for certain taxa, it substantially hampers evolutionary comparisons of these groups. Therefore, we strongly recommend a more complete description of the karyotypes, showing metaphases and karyotypes properly assembled by size and morphology. The karyomorphometric analyses (i.e., taking chromosome measurements) can be used present the karyotype structure more accurately (see Cristiano et al., 2017). Species identification is also very important, and it should be a critical step in publishing the cytogenetic data that could be included in future analysis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
PCA, MPC, and DCC performed, conceived, and designed the experiments and analyzed the data. MPC and DCC contributed materials and analysis tools. PCA, RM, MPC, and DCC wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
This research project was part of the M.Sc. Thesis of the PCA and was supported by the Brazilian Research Agency Coordenação de Pessoal de Nível Superior (CAPES – financial code: 001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We wish to thank Marcelo Querino, Tiago Degrandi, and the referees for their helpful comments and suggestions. We would like to thank the support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the fellowship of research productivity (PQ) conceived for DCC and MPC.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.829989/full#supplementary-material
Bolton, B. (2021). AntCat - An online catalog of the ants of the world. Available online at: https://www.antcat.org/about [Accessed December 25, 2020]
Brady, S. G., Schultz, T. R., Fisher, B. L., and Ward, P. S. (2006). Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc. Natl. Acad. Sci. U. S. A. 103, 18172–18177. doi: 10.1073/pnas.0605858103
Bromham, L., Duchêne, S., Hua, X., Ritchie, A. M., Duchêne, D. A., and Ho, S. Y. W. (2018). Bayesian molecular dating: opening up the black box. Biol. Rev. 93, 1165–1191. doi: 10.1111/brv.12390
Brown, W. L. (1958). Contributions toward a reclassification of the Formicidae. II. Tribe Ectatommini (Hymenoptera). Bull. Mus. Comp. Zool. 118, 175–362.
Brown, W. L. Jr. (1978). Contributions toward a reclassification of the Formicidae. Part VI. Ponerinae, tribe Ponerini, subtribe Odontomachiti. Section B. Genus Anochetus and bibliography. Stud. Entomol. 20, 549–638.
Bush, G. L., Case, S. M., Wilson, A. C., and Patton, J. L. (1977). Rapid speciation and chromosomal evolution in mammals. Proc. Natl. Acad. Sci. 74, 3942–3946. doi: 10.1073/pnas.74.9.3942
Cardoso, D. C., Das Graças Pompolo, S., Cristiano, M. P., and Tavares, M. G. (2014). The role of fusion in ant chromosome evolution: insights from cytogenetic analysis using a molecular phylogenetic approach in the genus Mycetophylax. PLoS One 9:e87473. doi: 10.1371/journal.pone.0087473
Cardoso, D. C., Santos, H. G., and Cristiano, M. P. (2018). The Ant Chromosome database – ACdb: an online resource for ant (Hymenoptera: formicidae) chromosome researchers. Myrmecol. News 27, 87–91. doi: 10.25849/myrmecol.news_027:087
Carta, A., Bedini, G., and Peruzzi, L. (2018). Unscrambling phylogenetic effects and ecological determinants of chromosome number in major angiosperm clades. Sci. Rep. 81, 1–14. doi: 10.1038/s41598-018-32515-x
Clyde, W. C., and Gingerich, P. D. (1998). Mammalian community response to the latest Paleocene thermal maximum: an isotaphonomic study in the northern Bighorn Basin, Wyoming. Geology 26:1011. doi: 10.1130/0091-76131998026<1011:MCRTTL<2.3.CO;2
Cristiano, M. P., Cardoso, D. C., and Fernandes-Salomão, T. M. (2013). Cytogenetic and Molecular Analyses Reveal a Divergence between Acromyrmex striatus (Roger, 1863) and Other Congeneric Species: taxonomic Implications. PLoS One 8:e59784. doi: 10.1371/journal.pone.0059784
Cristiano, M. P., Cardoso, D. C., Sandoval-Gómez, V. E., and Simões-Gomes, F. C. (2020). Amoimyrmex Cristiano, Cardoso & Sandoval, gen. nov. (Hymenoptera: formicidae): a new genus of leaf-cutting ants revealed by multilocus molecular phylogenetic and morphological analyses. Austral Entomol. 59, 643–676. doi: 10.1111/aen.12493
Cristiano, M. P., Pereira, T. T. P., Simões, L. P., Sandoval-Gómez, V. E., and Cardoso, D. C. (2017). Reassessing the chromosome number and morphology of the turtle ant Cephalotes pusillus (Klug, 1824) using karyomorphometrical analysis and observations of new nesting behavior. Insects 8:114. doi: 10.3390/insects8040114
de Aguiar, H. J. A. C., Barros, L. A. C., Silveira, L. I., Petitclerc, F., Etienne, S., and Orivel, J. (2020). Cytogenetic data for sixteen ant species from North-eastern Amazonia with phylogenetic insights into three subfamilies. Comp. Cytogenet. 14, 43–60. doi: 10.3897/CompCytogen.v14i1.46692
de Vos, J. M., Augustijnen, H., Bätscher, L., and Lucek, K. (2020). Speciation through chromosomal fusion and fission in Lepidoptera. Philos. Trans. R. Soc. B Biol. Sci. 375:20190539. doi: 10.1098/rstb.2019.0539
Escudero, M., Hipp, A. L., Hansen, T. F., Voje, K. L., and Luceño, M. (2012). Selection and inertia in the evolution of holocentric chromosomes in sedges (Carex. Cyperaceae). New Phytol. 195, 237–247. doi: 10.1111/J.1469-8137.2012.04137.X
Faria, R., Johannesson, K., Butlin, R. K., and Westram, A. M. (2019). Evolving inversions. Trends Ecol Evol. 34, 239–248. doi: 10.1016/j.tree.2018.12.005
Faria, R., and Navarro, A. (2010). Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 25, 660–669. doi: 10.1016/j.tree.2010.07.008
Fernandes, I. O., Larabee, F. J., Oliveira, M. L., Delabie, J. H. C., and Schultz, T. R. (2021). A global phylogenetic analysis of trap-jaw ants, Anochetus Mayr and Odontomachus Latreille (Hymenoptera: formicidae: ponerinae). Syst. Entomol. 46, 685–703. doi: 10.1111/SYEN.12483
Glick, L., and Mayrose, I. (2014) ChromEvol: assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Molecu. Biol. Evol. 31, 1914–1922. doi: 10.1093/molbev/msu122
Goldberg, E. E., and Igić, B. (2012). Tempo and mode in plant breeding system evolution. Evolution 66, 3701–3709. doi: 10.1111/j.1558-5646.2012.01730.x
Goni, B., Imai, H. T., Kubota, M., Rondo, M., Yong, H. S., and Tho, Y. D. (1982). Chromosome observations of tropical ants in Western Malaysia and Singapore. Ann. Rep. Natl. Inst. Genet. 32, 71–73.
Goodisman, M. A. D., Kovacs, J. L., and Hunt, B. G. (2008). Functional genetics and genomics in ants (Hymenoptera: formicidae): the interplay of and social life. Myrmecol. News 11, 107–117. doi: 10.1093/gbe/evz177
Guerra, M. (2008). Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenet. Genome Res. 120, 339–350. doi: 10.1159/000121083
Hauschteck-Jungen, E., and Jungen, H. (1983). Ant chromosomes. II. Karyotypes of Western Palearctic species. Insectes Sociaux 30, 149–164. doi: 10.1007/bf02223865
Hirai, H., Yamamoto, M. T., Taylor, R. W., and Imai, H. T. (1996). Genomic dispersion of 28S rDNA during karyotypic evolution in the ant genus Myrmecia (Formicidae). Chromosoma 105, 190–196. doi: 10.1007/BF02509500
Huerta-Cepas, J., Serra, F., and Bork, P. (2016). ETE 3: reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Imai, H. T., Brown, W. L. Jr., Kubota, M., Yong, H., and Tho, Y. (1983). Chromosome observations on tropical ants from western Malaysia. II. Annu. Rep. Natl. Inst. Genet. 34, 66–69.
Imai, H. T., Crozier, R. H., and Taylor, R. W. (1977). Karyotype evolution in Australian ants. Chromosoma 59, 341–393. doi: 10.1007/BF00327974
Imai, H. T., and Kubota, M. (1972). Karyological studies of Japanese ants (Hymenoptera, Formicidae) - III. Karyotypes of nine species in Ponerinae, Formicinae, and Myrmicinae. Chromosoma 37, 193–200. doi: 10.1007/BF00284938
Imai, H. T., Kubota, M., Brown, W. L. Jr., Ihara, M., Tohari, M., and Pranata, R. I. (1985). Chromosome observation on tropical ants from Indonesia. Ann. Rep. Natl. Inst. Genet 35, 46–48.
Imai, H. T., and Maruyama, T. (1978). Karyotype evolution by pericentric inversion as a stochastic process. J. Theor. Biol. 70, 253–261. doi: 10.1016/0022-5193(78)90375-2
Imai, H. T., Maruyama, T., Gojobori, T., Inoue, Y., and Crozier, R. H. (1986). Theoretical Bases for Karyotype Evolution. I. The Minimum-Interaction Hypothesis. Am. Nat. 128, 900–920. doi: 10.1086/284612
Imai, H. T., Satta, Y., and Takahata, N. (2001). Integrative study on chromosome evolution of mammals, ants and wasps based on the minimum interaction theory. J. Theor. Biol. 210, 475–497. doi: 10.1006/jtbi.2001.2327
Imai, H. T., Taylor, R. W., Crosland, M. W. J., and Crozier, R. H. (1988). Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to the minimum interaction hypothesis. Japanese J. Genet. 63, 159–185. doi: 10.1266/jjg.63.159
Imai, H. T., Taylor, R. W., and Crozier, R. H. (1994). Experimental bases for the minimum interaction theory. I. Chromosome evolution in ants of the Myrmecia pilosula species complex (Hymenoptera: formicidae: myrmeciinae). Japanese J. Genet. 69, 137–182. doi: 10.1266/jjg.69.137
Imai, H. T., Urbani, C. B., Kubota, M., Sharma, G. P., Narasimhanna, M. N., Das, B. C., et al. (1984). Karyological survey of Indian ants. Japanese J. Genet. 59, 1–32. doi: 10.1266/jjg.59.1
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., Stamatakis, A., and Wren, J. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). Partitionfinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Larabee, F. J., Fisher, B. K., Schmidt, C. A., Matos-Maraví, P., Janda, M., and Suarez, A. V. (2016). Molecular phylogenetics and diversification of trap-jaw ants in the genera Anochetus and Odontomachus (Hymenoptera: formicidae). Mol. Phylogenet. Evol. 103, 143–154. doi: 10.1016/j.ympev.2016.07.024
Levan, A., Fredga, K., and Sandberg, A. A. (1964). Nomeclature for centromeric position on chromosomes. Hereditas 52, 201–220. doi: 10.1111/j.1601-5223.1964.tb01953.x
Lorite, P., and Palomeque, T. (2010). Karyotype evolution in ants (Hymenoptera: formicidae), with a review of the known ant chromosome numbers. Myrmecol. News 13, 89–102.
Löytynoja, A., and Goldman, N. (2010). webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinform. 11:579. doi: 10.1186/1471-2105-11-579
Mariano, C., dos, S. F., Pompolo, S. D. G., Barros, L. A. C., Mariano-Neto, E., Campiolo, S., et al. (2008). A biogeographical study of the threatened ant Dinoponera lucida Emery (Hymenoptera: formicidae: ponerinae) using a cytogenetic approach. Insect Conserv. Divers. 1, 161–168. doi: 10.1111/j.1752-4598.2008.00022.x
Mariano, C., dos, S. F., Pompolo, S. D. G., and Delabie, J. H. C. (2000). Citogenética das espécies gêmeas e simpátricas Pachycondyla villosa e Pachycondyla sp ‘inversa’. (Ponerinae). Naturalia 24, 215–217.
Mariano, C. D. S. F., Pompolo, S. D. G., Silva, J. G., and Delabie, J. H. C. (2012). Contribution of cytogenetics to the debate on the paraphyly of Pachycondyla spp. (Hymenoptera. Formicidae, Ponerinae). Psyche. 973897, 1–9. doi: 10.1155/2012/973897
Mariano, C. S. F., Delabie, J. H. C., Santos, J. R. M., and Pompolo, S. G. (2007). Evolução cariotípica em Pachycondyla spp. (Ponerinae) Neotropicais. O Biológico 69, 409–412.
Mariano, C. S. F., Santos, I., da, S., Groc, S., Leroy, C., Malé, P. J., et al. (2011). The karyotypes of Gigantiops destructor (Fabricius) and other ants from French Guiana (Formicidae). Ann. Soc. Entomol. Fr. 47, 140–146. doi: 10.1080/00379271.2011.10697705
Mariano, C. S. F., Santos, I. S., Silva, J. D., Costa, M. A., Pompolo, S. G., and Delabie, J. H. C. (2015). As Formigas Poneromorfas do Brasil - Editus. Available online at: SciELO Books http://books.scielo.org (accessed November 13, 2021).
Matthey, R. (1945). L’évolution de la formule chromosomiale chez les Vertébrés. Experientia 1, 78–86. doi: 10.1007/BF02156807
McInerney, F. A., and Wing, S. L. (2011). The Paleocene-Eocene Thermal Maximum: a Perturbation of Carbon Cycle, Climate, and Biosphere with Implications for the Future. Annu. Rev. Earth Planet. Sci. 39, 489–516. doi: 10.1146/annurev-earth-040610-133431
Menezes, R. S. T., Carvalho, A. F., Correia, J. P. S. O., Silva, T. S., Somavilla, A., and Costa, M. A. (2014). Evolutionary trends in the chromosome numbers of swarm-founding social wasps. Insectes Soc. 61, 385–393. doi: 10.1007/s00040-014-0365-3
Micolino, R., Cristiano, M. P., and Cardoso, D. C. (2020). Karyotype and putative chromosomal inversion suggested by integration of cytogenetic and molecular data of the fungus-farming ant Mycetomoellerius iheringi Emery, 1888. Comp. Cytogenet. 14, 197–210. doi: 10.3897/compcytogen.v14i2.49846
Micolino, R., Cristiano, M. P., Travenzoli, N. M., Lopes, D. M., and Cardoso, D. C. (2019). Chromosomal dynamics in space and time: evolutionary history of Mycetophylax ants across past climatic changes in the Brazilian Atlantic coast. Sci. Rep. 9:18800. doi: 10.1038/s41598-019-55135-5
Moreau, C. S., Bell, C. D., Vila, R., Archibald, S. B., and Pierce, N. E. (2006). Phylogeny of the ants: diversification in the age of angiosperms. Science 312, 101–104. doi: 10.1126/science.1124891
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Rieseberg, L. H. (2001). Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358. doi: 10.1016/S0169-5347(01)02187-5
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Santos, I. S., Costa, M. A., Mariano, C. S. F., Delabie, J. H. C., Andrade-Souza, V., and Silva, J. G. (2010). A Cytogenetic Approach to the Study of Neotropical Odontomachus and Anochetus Ants (Hymenoptera: formicidae). Ann. Entomol. Soc. Am 103, 424–429. doi: 10.1603/an09101
Schmidt, C. (2013). Molecular phylogenetics of Ponerine ants (Hymenoptera: formicidae: ponerinae). Zootaxa 3647, 201–250. doi: 10.11646/zootaxa.3647.2.1
Schmidt, C. A., and Shattuck, S. O. (2014). The higher classification of the ant subfamily Ponerinae (Hymenoptera: formicidae), with a review of Ponerinae ecology and behavior. Zootaxa 3817, 1–242. doi: 10.11646/zootaxa.3817.1.1
Schubert, I. (2007). Chromosome evolution. Curr. Opin. Plant Biol. 10, 109–115. doi: 10.1016/j.pbi.2007.01.001
Teixeira, A. G. (2018). Citogenética Clássica e Molecular de Formigas Neotropicais. Brazil: Universidade Federal de Viçosa.
Velasco, Y. A. M., Delabie, J. H. C., Costa, M. A., Lacau, S., and Mariano, C. D. S. F. (2014). Studies on the Karyotype of the Ant Pachycondyla harpax (Formicidae: ponerinae: ponerini) in Southern Bahia, Brazil. Florida Entomol. 97, 1049–1055. doi: 10.1653/024.097.0307
Wang, J., Wurm, Y., Nipitwattanaphon, M., Riba-Grognuz, O., Huang, Y.-C., Shoemaker, D., et al. (2013). A Y-like social chromosome causes alternative colony organization in fire ants. Nature 4937434, 664–668. doi: 10.1038/nature11832
Wellenreuther, M., and Bernatchez, L. (2018). Eco-Evolutionary Genomics of Chromosomal Inversions. Trends Ecol. Evol. 33, 427–440. doi: 10.1016/j.tree.2018.04.002
Wernersson, R., and Pedersen, A. G. (2003). RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 31, 3537–3539. doi: 10.1093/nar/gkg609
Keywords: chromosome evolution, Formicidae, karyotype, ancestral reconstruction, Ponerinae ants, phylogenetic analysis
Citation: Afonso Neto PC, Micolino R, Cardoso DC and Cristiano MP (2022) Phylogenetic Reconstruction of the Ancestral Chromosome Number of the Genera Anochetus Mayr, 1861 and Odontomachus Latreille, 1804 (Hymenoptera: Formicidae: Ponerinae). Front. Ecol. Evol. 10:829989. doi: 10.3389/fevo.2022.829989
Received: 06 December 2021; Accepted: 20 January 2022;
Published: 22 February 2022.
Edited by:
Vladimir E. Gokhman, Lomonosov Moscow State University, RussiaReviewed by:
Eugenia E. Montiel, University of Jaén, SpainCopyright © 2022 Afonso Neto, Micolino, Cardoso and Cristiano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maykon Passos Cristiano, bWF5a29uQHVmb3AuZWR1LmJy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.