
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 08 February 2022
Sec. Chemical Ecology
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.823119
This article is part of the Research Topic The Morphology and Physiology of Insect Chemosensory Systems – Its Origin and Evolution View all 17 articles
Aldehyde oxidases (AOXs) are common detoxifying enzymes in several organisms. In insects, AOXs act in xenobiotic metabolism and as odorant-degrading enzymes (ODEs). These last appear as crucial enzymes in the life cycle of insects, helping to reset their olfactory system, particularly in lepidopterans, which fulfill important ecological roles (e.g., pollination or destructive life cycles). A comprehensive understanding of their olfactory system has provided opportunities to study key chemosensory proteins. However, no significant advance has been made around lepidopteran AOXs research, and even less around butterflies, a recently evolved lineage. In this study we have identified novel AOX gene families in moths and butterflies in order to understand their role as ODEs. Eighteen genomes from both moths and butterflies were used for phylogenetics, molecular evolution and sequence analyses. We identified 164 AOXs, from which 91 are new. Their phylogeny showed two main clades that are potentially related to odorant-degrading function, where both moths and butterflies have AOXs. A first ODE-related clade seems to have a non-ditrysian origin, likely related to plant volatiles. A second ODE-related clade could be more pheromone-biased. Molecular evolution analysis suggests a slight purifying selection process, though a number of sites appeared under positive selection. ODE-related AOXs have changed a phenylalanine residue by proline in the active site. Finally, this study could serve as a reference for further evolutionary and functional studies around Lepidopteran AOXs.
The study of gene evolution in insects has provided outstanding advances in the understanding of evolutionary processes, such as expansion or contraction of gene families (Li et al., 2019). Particularly, lepidopterans represent an extraordinary target due to a clear diversification into moth and butterflies lineages (Kawahara et al., 2019). Thus, the impact of gene evolution can be seen even within moths. For instance, regulation of desaturase genes in two sibling Helicoverpa species (i.e., H. armigera and H. assulta) results in reproductive isolation (Li et al., 2015). Nowadays, the enormous amount of genomic and transcriptomic datasets for insects has provided an opportunity to elucidate novel genes and their evolutionary relationships (Oppenheim et al., 2015), something that can support our understanding of ecological aspects of insects, such as behavior. For many insect species, behavior is mainly driven by olfaction. Olfaction is primarily processed by insect antennae and their small hair-like structures called sensilla, in which a set of proteins work synergistically to maintain an extremely sensitive and dynamic system (Hansson and Stensmyr, 2011; Leal, 2013; He et al., 2019). For instance, odorant-binding proteins (OBPs) and chemosensory proteins (CSPs) function as transporters that carry odorants across the sensillar lymph (Zhou, 2010; Leal, 2013). These odorants reach an heteromeric complex of receptors, such as odorant receptors (ORs), an odorant receptor co-receptor (Orco) and a sensory neuron membrane protein (SNMP), as recently reported (Zhang et al., 2020), to unleash depolarization in olfactory neuron membranes that triggers a behavioral response (Kaissling, 2013). Along with these olfactory proteins, odorant-degrading enzymes (ODEs), such as carboxylesterases (CXEs), glutathione-S-transferases (GSTs) and aldehyde oxidases (AOXs), are responsible for resetting the insect olfactory system through the degradation of odorant molecules (Chertemps and Meïbèche, 2021; Godoy et al., 2021).
Among ODEs, CXEs and GSTs have received particular attention due to their role in sex pheromone degradation in moths. For example, CXEs have been reported to degrade ester-type molecules (e.g., sex pheromones and plant volatiles) in moths Plodia interpunctella, Spodoptera exigua, Grapholita molesta, Plutella xylostella, and Athetis lepigone (He et al., 2014a,b, c, 2015; Zhang et al., 2017a; Liu et al., 2019; Wei et al., 2020; Wang et al., 2021a). Likewise, GSTs have been well characterized in terms of function, being the delta class likely related to odorant degrading functions (Durand et al., 2018). This is supported by the reported degrading function of G. molesta GST (GmolGSTd1) (Li et al., 2018) and Cydia pomonella GST (CpomGSTd2) (Huang et al., 2017). The AOXs, on the other hand, have received less attention so far. However, some species that use aldehydes as semiochemicals (i.e., chemicals that mediate communication between two organisms), have been studied, such as Manduca sexta, Bombyx mori, Antheraea polyphemus, Amyelois transitella, and H. armigera, among others (Riddiford, 1967; Kasang et al., 1978; Coffelt et al., 1979; Zhang et al., 2012). Particularly, AOXs catalyze the oxidation of aldehydes to carboxylic acids (Garattini et al., 2009; Garattini and Terao, 2012). In that sense, a few studies have functionally evaluated that process against aldehyde-based semiochemicals: an early study in M. sexta AOX reports that it catalyzes (E,Z)-10,12-hexadecadienal (bombykal) (Rybczynski et al., 1989), and more recently, A. transitella AOX2 (AtraAOX2) was reported to hydrolyze plant volatiles (e.g., propanal, hexanal, and heptanal) as well as a sex pheromone component (Z,Z)-11,13-hexadecadienal (Choo et al., 2013). Further evidence in terms of enzymatic activity of AOXs is still lacking. Nevertheless, important aspects of their function and structural features are underpinned by xanthine dehydrogenases (XDHs), an enzyme that catalyzes the oxidation of purines, pterin and aldehydes (Wang et al., 2016).
Among insects, lepidopterans have attracted special attention due to their establishment as crop pests, some with worldwide distribution. It is known that moths rely heavily on the sense of smell (Weiss, 2001), developing long distance attraction based on volatile chemicals (e.g., sex pheromones) (Chemnitz et al., 2015). In fact, hundreds of these volatiles have been identified since the first one reported for B. mori, the sex pheromone (E,Z)-10,12-hexadecadien-1-ol (bombykol) (Butenandt et al., 1959). On the contrary, butterflies rely heavily on visual cues and short-range chemical communication, understood as a multi-sensory integration (Costanzo and Monteiro, 2007), and hence have received less attention in terms of olfaction. Moreover, butterflies represent an interesting group for comparative studies considering their transition from moths approximately 98 Mya (million years ago) (Kawahara et al., 2019). Thus, it is believed that comparing AOXs between moths and butterflies might deepen our understanding of their odorant-degrading function.
Considering the difference in olfactory integration during the life cycle of moths and butterflies, we hypothesize that there is a specific clade of AOXs for both moths and butterflies that could be related to odorant-degrading function as well as both moth- and butterfly-specific gene expansions. Therefore, the objective of this study was to identify novel AOX genes from moths and butterflies using genomic and transcriptomic data and analyze them in terms of gene location, phylogeny, evolutionary processes, and structure.
Publicly available genomic data were retrieved from NCBI Genome database1 and InsectBase2 for major lepidopteran families, such as Bombycidae, Sphingidae, Noctuidae, Pyralidae, Crambidae, and Plutellidae for moths, whereas Nymphalidae, Pieridae and Papilionidae were used for butterflies (Table 1). Each fully represented genome assembly with Reference Sequence (RefSeq) was downloaded from NCBI Assembly database at either contig, scaffold or chromosome level (Supplementary Table 1).
Bioinformatics pipeline BITACORA (Vizueta et al., 2020) was used to identify already annotated AOX genes and potentially novel related genes from both moth and butterfly genomes. A database for AOX gene family was built using reported protein sequences for lepidopterans (Rybczynski et al., 1989; Merlin et al., 2005; Pelletier et al., 2007; Choo et al., 2013; Ou et al., 2014; Zhang et al., 2014, 2017b; Yang et al., 2015; Huang et al., 2016; He et al., 2017; Xu and Liao, 2017; Wang et al., 2021b). To identify family and structural domains for AOXs, InterPro server was used3. The identified profile was used to search for HMM profile in PFAM database4 (PF01315; ID Ald_Xan_dh_C). This process increased the likelihood of identifying sequences encoding members of the AOX gene family. Further processing included the trimming of isoforms (98% cutoff) using a provided script in BITACORA pipeline. Subsequently, BLAST searches were run with the identified proteins for manual annotation. Protein domain finder CDvist5 (Adebali et al., 2015) was used to identify conserved domains of AOXs, namely two (2Fe-2S), one flavin-containing region (FAD-binding domain) and one molybdenum cofactor/substrate-binding domain. All proteins identified in this study are provided in Supplementary Table 1.
The genomic organization of identified AOX genes from both moth and butterfly species that use aldehyde-based semiochemicals was analyzed based on Vogt et al. (2015) and Xu and Liao (2017) including some modifications. Moths Bombyx mandarina, P. xylostella, and A. transitella, and butterflies Heliconius melpomene and Bicyclus anynana, were selected for this task. Annotated gene features (in GFF3 format) were retrieved from the gene identification protocol based on the BITACORA pipeline and analyzed manually. Thus, species name as well as source, start, end, strand and attributes were used for each AOX gene. Finally, gene localization was prepared in image editor Inkscape 0.48 software.
To both take advantage of transcriptomic data and include Tortricidae and Eriocraniidae families, we retrieved antennal RNA-seq data for moth Lobesia botrana (data from our laboratory) and non-ditrysian moth Eriocrania semipurpurella (SRR5328787). FASTQ files for both moths, one containing left-pair reads and other the right-pair reads, were used for assembly. Ribosomal RNA reads were removed by mapping the libraries using Bowtie2 v.2.3.3.1 (Langmead et al., 2009) against a custom rRNA database created from insect ribosomal sequences downloaded from NCBI6, and keeping non-mapped reads using SAMtools v.1.6 (Li et al., 2009). Low-quality reads were removed based on their q-score composition using NGSQC Toolkit v.2.3 (Patel and Jain, 2012), and high-quality reads were concatenated to build de novo transcriptomes using Trinity v.2.6.5 (Grabherr et al., 2011) with a P-value of 0.05 and fold-change value of 2.
A phylogeny for the identified AOX genes in moths and butterflies, including XDHs and AOXs from mosquitoes, beetles and bees as outgroups, was built. Full-length amino acid sequences that include conserved domains were aligned using MAFFT server7 (Katoh et al., 2019). GUIDANCE2 server8 was used to check consistency of the multiple sequence alignment (Sela et al., 2015). Briefly, the consistency of the alignment was measured with a score less than 0.5, in which sequences were deleted. It is worth noting that confidence scores near 1 and 0, suggest a highly and poorly consistent alignment, respectively. Finally, phylogenetic analysis was performed using maximum-likelihood method with FastTree software (Price et al., 2010). To highlight clades, specific taxa and functional evidence, the phylogenetic tree was edited using FigTree software9 and image editor Inkscape 0.48 software.
In order to identify putative selective pressures on AOXs, a molecular evolution analysis was performed based on the methodologies reported by Engsontia et al. (2014) and Soffan et al. (2018). Two models were used through EasyCodeML software (Gao et al., 2019) to elucidate selective pressures acting on the evolutionary process of 93 lepidopteran AOX genes, 9 XDHs and 11 AOXs from other insect orders. First, site model was applied to detect positive selection for a set of 113 sequences (Yang et al., 2000). Additionally, a branch-site model was applied to test the presence of amino acids that evolved under positive selection in a specific clade represented by 8 AOX sequences (most of them functionally studied). All the amino acid sequences were aligned by ClustalW10, and converted to DNA alignment with PAL2NAL server11. A maximum likelihood tree was prepared using the DNA alignment by FastTree software under default parameters. Briefly, the software estimated the ratio of normalized non-synonymous (dN) to synonymous (dS) (e.g., dN/dS or ω) substitution rate via the maximum likelihood method. The ω value indicates the mode of evolution, where ω > 1 suggests evidence of positive selection with amino acid replacement, whereas ω < 1 refers to purifying selection, and ω = 0 indicates neutral selection. The specific models (M0, M3, M1a, M2a, M7, M8, and M8a) used under the “site model” method are described in detail in previous reports (Yang et al., 2000; Yang and Nielsen, 2002; Swanson et al., 2003). For the branch-site model, the 8 AOX sequences were labeled in the phylogenetic tree as foreground branch with the remaining clades as background branches. The change in ω was evaluated for a set of sites in each foreground branch through an alternative model, whereas neutral evolution was evaluated through a null model. Likelihood ratio tests (LRTs) were used to compare both models and significant results were determined using χ2-tests. Finally, Bayes Empirical Bayes (BEB) analysis was used when LRT was significant to identify positive selected sites (PSSs) within each amino acid sequence (Yang et al., 2005).
First, a multiple sequence alignment (MSA) was built with 7 AOX sequences also used in molecular evolution analyses, belonging to A. transitella, B. mori, P. xylostella, H. armigera, Papilio xuthus, Papilio machaon, and B. anynana, in which PSSs were identified and indicated (Supplementary Table 1). Sequences from B. mandarina AOX5 (BmanAOX5), Drosophila melanogaster AOX2 (DmelAOX2) and mammal AOXs, such as Mus musculus AOX2 (MmAOX2) and AOX3 (MmAOX3), and Homo sapiens AOX1 (HsAOX1), were also included. MSA was built in Multalin server12 and ESPript 3.013 (Corpet, 1988). The amino acid sequence of AtraAOX2 and BanyAOX2 were submitted to BLASTp available on the NCBI website14 for template selection. To optimize the structural information available for AOXs, a multiple template-based homology modeling approach was considered as it was reported to increase accuracy in predicted protein models (Sokkar et al., 2011). First, multiple structure alignments were generated by SALIGN command, which is implemented in Modeler 10.1. Five hundred models of each AOX were obtained using Modeler 10.115. The best models were selected according to the lowest discrete optimized protein energy (DOPE) score provided by the software. The coordinates were analyzed via ProCheck16 to check stereochemical quality. Lepidopteran AOXs were visualized through PyMOL software17. The 3D structure of mammal AOXs were retrieved from Protein Data Bank18 and DmelAOX2 was downloaded from AlphaFold database19.
Eighteen genome assemblies were retrieved from NCBI assembly database and InsectBase server. From those, 6 were assembled at chromosome level, whereas 11 at scaffold and 1 at contig level (Supplementary Table 1). The use of BITACORA pipeline resulted in a raw amount of 163 putative AOX genes for moths and 100 for butterflies. After homology searches through BLAST followed by conserved domain analyses, 99 AOX genes were left for moths and 65 for butterflies. The average amino acid length of AOXs is 1272 and 1407 for moths and butterflies, respectively. On the other hand, the moth species that showed a higher number of AOX were Spodoptera frugiperda with 20 sequences, M. sexta with 19 sequences and Trichoplusia ni with 14. In butterflies, B. anynana and Danaus plexippus showed 11 and 10 AOX sequences, respectively. The specific number of AOX genes for each species is summarized in Table 1. Overall, 58 novel AOX genes were identified for moths whereas 33 AOX genes were identified for butterflies. BLAST hits for most of the novel AOX genes were either AOXs from other lepidopteran species or XDHs from the same species. In that sense, 6 new AOXs were identified for B. mandarina and B. mori, 16 for M. sexta, 17 for S. frugiperda and 10 for D. plexippus, as the greater numbers found. No novel AOX genes were found for butterflies Papilio polytes and P. machaon. Although most of the lepidopteran species studied here have a few annotated AOX genes, several of them are not fully annotated nor studied in terms of function. It is worth noting that the amount and length of AOX genes might be dependent on genome sequencing and annotation quality, therefore, previous estimates should be taken into account with caution.
Phylogenetic analysis (Figure 1) suggests the presence of 5 main clades, 2 being related to either antennae specificity or odorant degradation (clades A and B in Figure 1). A clear diversification from insect XDHs and non-lepidopteran AOXs is observable. A clade with putative odorant-degrading function (labeled as A in Figure 1) appears to be a group of AOX genes evolved in ditrysian species, having the non-ditrysian moth E. semipurpurella EsemAOX1 at the base of the clade. Here, 9 moth AOX genes are reported to be enriched in antennae (Figure 1, species indicated with black circles next to their name), from which only M. sexta AOX1 (MsexAOX1) has been related to aldehyde-degrading function (Rybczynski et al., 1989). A secondary odorant degrading-related clade (labeled as B in Figure 1) seems to have evolved by gene duplication. This clade includes 5 moth AOX genes enriched in antennae (Figure 1, species indicated with black circles next to their name), and includes A. transitella (AtraAOX2), P. xylostella (PxylAOX3), and B. mori (BmorAOX5) which were functionally studied (Choo et al., 2013; Zhang et al., 2020; Wang et al., 2021b). Furthermore, butterfly- and moth-specific AOX lineages were identified (highlighted in red and gray, respectively, in Figure 1), but no reported odorant-degrading function was found for these.
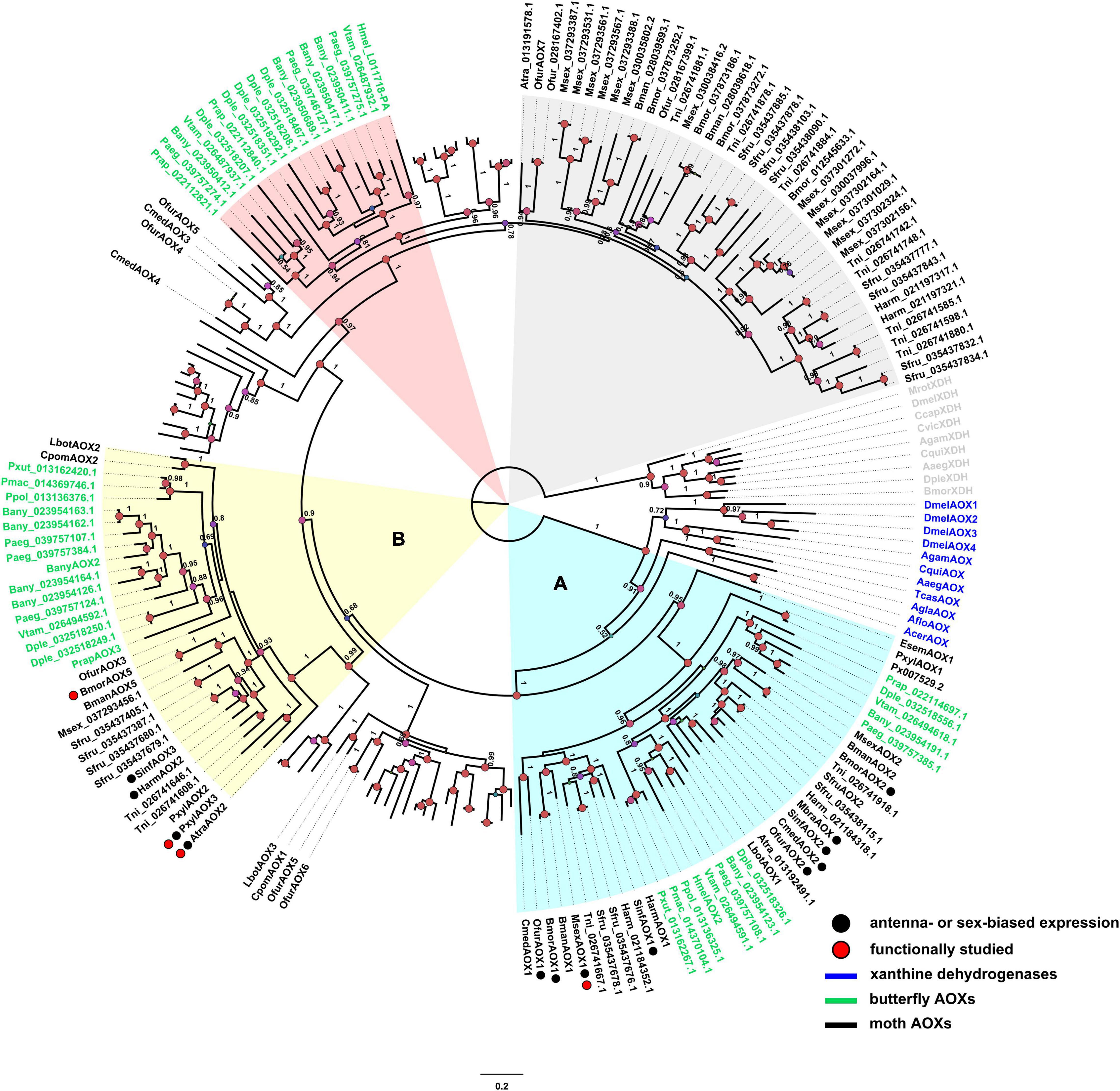
Figure 1. Phylogenetic tree of AOXs identified from moth and butterfly genomes as well as some AOXs and XDHs from other insect species. Clades A (light blue) and B (yellow) are highlighted as lineage with putative ODE function and lineage with ODE function, respectively. Species in green correspond to butterflies, species in black are moths, and species in blue are AOX outgroups. The clade shaded in red color corresponds to butterfly specific AOXs with no ODE described function, and the clade shaded in gray correspond to moth specific AOXs with no ODE described function. Red circles next to the sequence name represent AOXs that have been functionally studied [MsexAOX1, (E,Z)-10,12-hexadecadienal; AtraAOX2, (Z,Z)-11,13-hexadecadienal; PxylAOX3, (Z)-11-hexadecenal; BmorAOX5, benzaldehyde, salicylaldehyde, vanillic aldehyde, propanal, and heptanal]. Black circles next to the sequence name indicate antennae- or sex-biased expression. Confidence scores are indicated as circles (> 70%) in nodes. All annotated genes and their amino acid sequences are in Supplementary Table 1.
Interestingly, butterflies that have aldehyde-related pheromones have AOXs present in at least one of the odorant-related clades (A or B). There are AOXs of some butterflies that are in these clades, but no aldehyde-related semiochemical has been reported yet, such as those present in D. plexippus, Pararge aegeria, Pieris rapae, P. polytes, P. xuthus, P. machaon, and Vanessa tameamea. On the contrary, H. melpomene, that uses aldehyde-based semiochemicals, showed only 1 AOX (HmelAOX2) in clade A. Likewise, B. anynana (with hexadecanal as a pheromone component), has 2 AOXs in clade A, while 5 in clade B.
In terms of gene location, B. mandarina, A. transitella and P. xylostella, that use aldehydes as semiochemicals and have AOX genes related to ODE function, are far from other AOX genes, with the exception of PxylAOX3 (Figure 2). H. melpomene has 2 grouped AOX genes that suggest the same origin for both. Likewise, B. anynana has two big clusters of 4 and 7 AOX genes. Interestingly, from the bigger cluster of B. anynana, the 7 AOX genes were distributed in odorant-degrading clades A and B. Similarly, AOX1, AOX2 and AOX5 of B. mandarina that are grouped in a single cluster, are distributed in clades A and B. Besides HmelAOX2 present in clade A, gene HMEL011718-PA clustered with HmelAOX2, which appeared in the previously mentioned butterfly-specific clade (red clade in Figure 1).
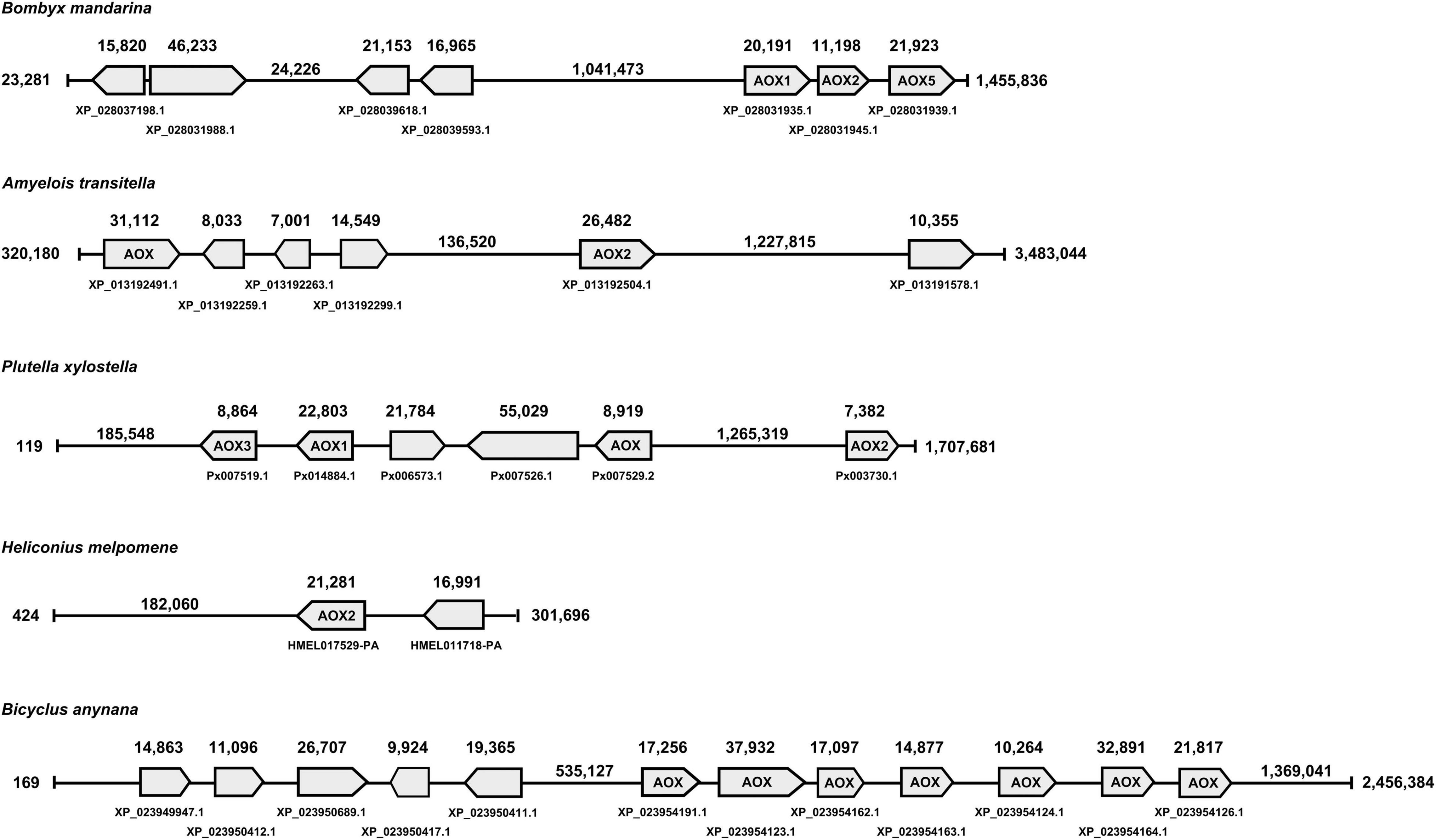
Figure 2. Gene location analysis for AOXs identified from lepidopterans that use aldehydes as semiochemicals, such as moths B. mandarina, A. transitella, P. xylostella and butterflies H. melpomene and B. anynana.
Positive selection was first evaluated for a set of 113 sequences that included XDHs and AOXs of not only butterflies and moths, but also beetles, mosquitoes, and flies (Table 2). The four models implemented (e.g., M3 vs. M0, M1a vs. M2a, M7 vs. M8 and M8a vs. M8) showed significant differences according to LRT analysis. Interestingly, a purifying selection was suggested as site model (M0) resulted in ω = 0.89.
Additionally, a branch-site model was used to test selective pressures on specific sites (i.e., codons) among 8 closely related AOX sequences, including moths A. transitella AtraAOX2, B. mori BmorAOX5, P. xylostella PxylAOX3, H. armigera HarmAOX2 and Sesamia inferens SinfAOX3, and butterflies B. anynana BanyAOX2, P. xuthus PxutAOX2 and P. machaon PmacAOX2 (Table 3). As expected, most of the enzymes were found to be under positive selection at many sites. For instance, AtraAOX2 resulted in 23.5% of their amino acids as PSSs, from which 105 sites showed either P < 0.01 or P < 0.001. Similarly, 22.2% of residues in BmorAOX5, 11.9% in HarmAOX, and 11.2% in SinfAOX3 were PSSs, with more than 20 sites identified with P < 0.001. In terms of butterflies, BanyAOX2 resulted in PSSs distributed in 40% of the entire sequence. However, less PSSs resulted for PxutAOX2 and PmacAOX2, representing only a 2–3% of the amino acid sequence length.
To complement our previous methods that included annotation, phylogeny and molecular evolution analyses, a MSA was built followed by AOX structure prediction. The MSA was based on the same 8 lepidopteran AOX sequences detailed above, as well as B. mandarina BmanAOX5, D. melanogaster DmelAOX2, and mammal AOXs M. musculus MmAOX2 and MmAOX3, and H. sapiens HsAOX1, which have been well characterized in terms of structural features and active sites.
As reference, the active site of MmAOX3 comprise Gln772, Glu1266, Lys889, Phe919 and Phe1014 (Terao et al., 2020). From those, Gln772 and Glu1266 at positions equivalent to 739 and 1209 in Figure 3, were found to be conserved between all AOXs (red triangles in Figure 3). Interestingly, Phe919 (in vertebrates) at position 884 (in Lepidoptera) in Figure 3 was conserved among mammal AOXs, but replaced by Pro in all lepidopteran AOXs as well as DmelAOX2 (blue triangle in Figure 3). Lys889 (at position 855 in Figure 3) was also not conserved, with SinfAOX3, HarmAOX2, AtraAOX2, PmacAOX2, PxutAOX2 and PxylAOX3 having Gly, and BmorAOX5, BmanAOX5 and BanyAOX5 having Ser instead (purple triangle in Figure 3).
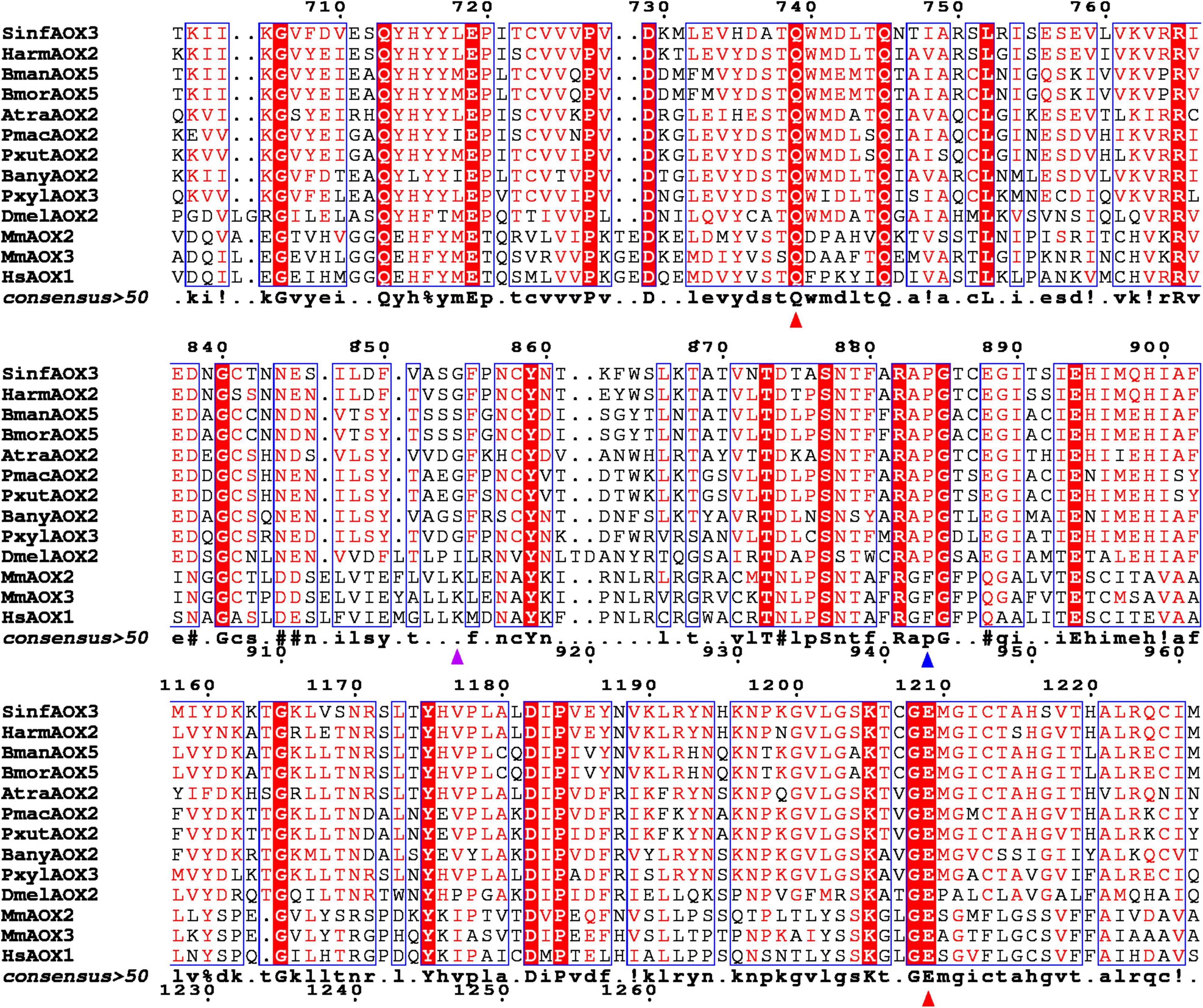
Figure 3. Fragment of a multiple sequence alignment between vertebrate and invertebrate AOXs. Identical residues are highlighted in white letters with red background. Similar residues are highlighted in red and framed in blue. Triangles indicate conserved sites according to Terao et al. (2020). Red triangles show conserved residues across all analyzed species. Blue triangle shows residue conserved only in insect species. Purple triangle shows an active site with different residues within Lepidoptera species. Full alignment can be found in Supplementary Figure 1.
In terms of structure, we could predict the 3D arrangements for AtraAOX2 and BanyAOX2, which were used to corroborate the identified conserved residues at the active site (Figure 4). The active sites equivalent to MmAOX3 Gln772 and Glu1266 were identified in both Lepidoptera species as well as in DmelAOX2 and vertebrate HsAOX1. Differences in conformation were observed, which are found in large structures that have not been relaxed through molecular dynamics. Nevertheless, our results are consistent with residue locations, supporting, for instance, the role that the insect specific Pro884 plays in the active site.
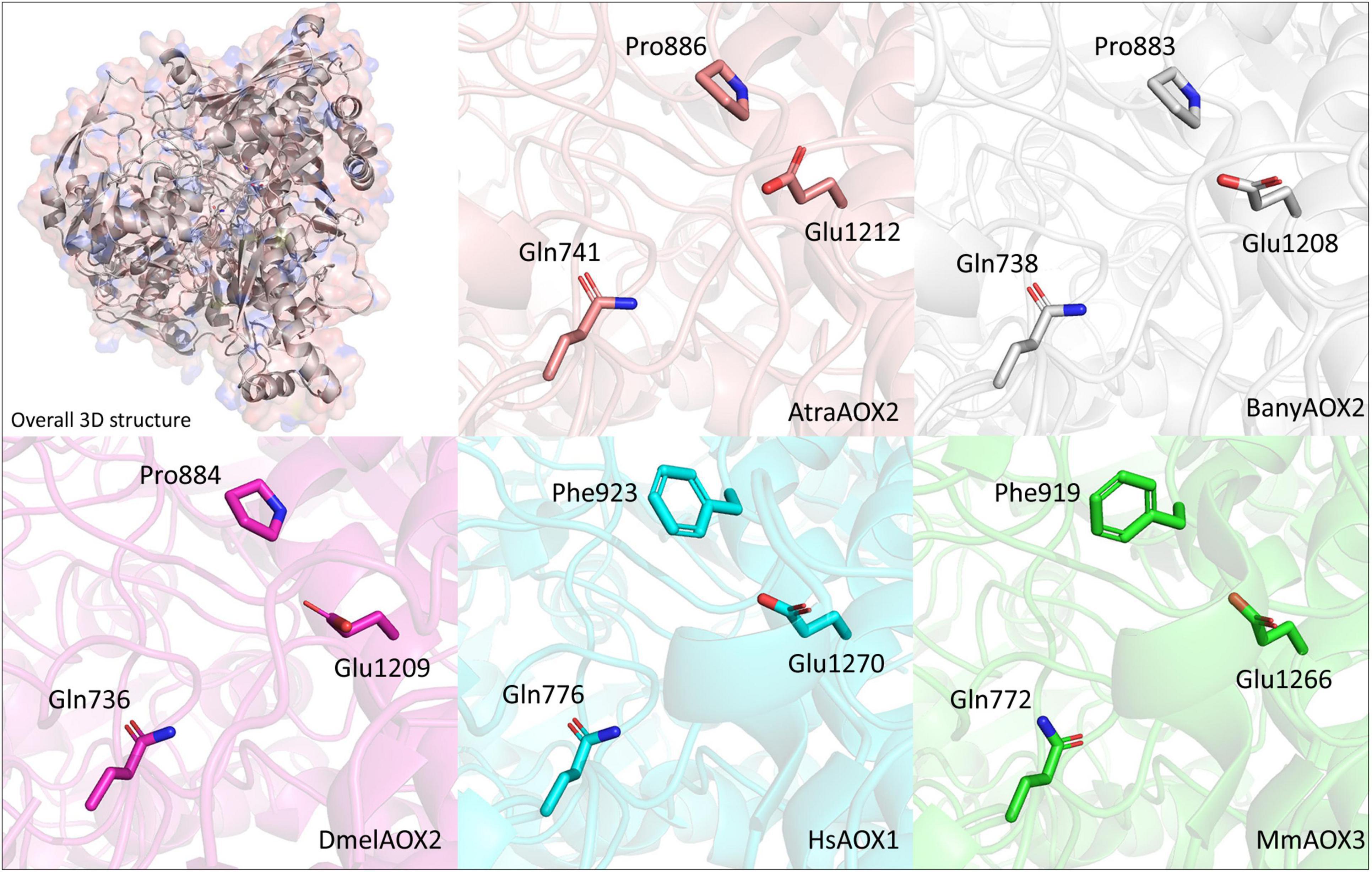
Figure 4. Visualization of the location of conserved residues in 3D representations of the active site of A. transitella AOX2 (AtraAOX2—orange), B. anynana AOX2 (BanyAOX2—gray), D. melanogaster AOX2 (DmelAOX2—pink), H. sapiens AOX1 (HsAOX1—cyan) and M. musculus AOX3 (MmAOX3—green). Residues and their positions are indicated in each AOX. Mammal AOXs were retrieved from Protein Data Bank (https://www.rcsb.org/) and DmelAOX2 was downloaded from AlphaFold database (https://alphafold.ebi.ac.uk/).
In this study we identified a total of 164 AOX sequences from both moths and butterflies. In the context of an increasing amount of data from genomic studies, we have taken advantage of publicly available genome assemblies to identify and analyze AOX gene families in Lepidoptera. Particularly, AOXs are metal-containing enzymes that metabolize aldehydes into their corresponding carboxylic acids and other sub products (Krenitsky et al., 1972). Their role in insect chemosensation has been studied since 1989 when M. sexta AOX (MsexAOX1) was reported to catalyze (E,Z)-10,12-hexadecadienal (bombykal), the sex pheromone of this species (Rybczynski et al., 1989). However, reports about insect AOXs and their function toward aldehydes took more than 20 years to be published again, when A. transitella AOX2 (AtraAOX2) was comprehensively studied (Choo et al., 2013).
Although AOX genes have been related to metabolism of xenobiotics in mammals as well as in Culex mosquitoes (Hemingway et al., 2000; Coleman et al., 2002; Terao et al., 2020), recent efforts have been focused on insect AOXs that can act as ODEs in olfactory organs, such as antennae and maxillary palps. Here, we report a profile of sequences related to AOX gene family that provides new data sets for several lepidopterans (Table 1 and Supplementary Table 1). For instance, our analyses revealed 5 full-length and 1 partial AOX sequences for P. xylostella, including the only identified AOX so far (PxylAOX3) (Wang et al., 2021b). Similarly, 9 full-length sequences were identified for B. mori, including BmorAOX1, BmorAOX2, and BmorAOX5, the only AOXs reported so far (Pelletier et al., 2007; Zhang et al., 2020). Likewise, we report 2 new AOXs for H. armigera, in which 6 AOXs had previously been reported, including HarmAOX2, suggested to be a candidate pheromone-degrading enzyme (Xu and Liao, 2017). For butterflies, no AOX-related studies have been published to our knowledge. Hence, this study would be the first to report such enzymes in this group.
In terms of number of AOXs identified in both moths and butterflies, it is interesting to notice that generalist moth species, such as S. frugiperda, M. sexta and T. ni resulted in the highest number of AOXs. However, we could not establish a direct relationship between number of AOXs and the condition of generalist vs. specialist species, something that has been proposed for other chemosensory proteins, such as ORs (Venthur and Zhou, 2018). Thus, we can highlight that, overall, moths resulted in a similar number of AOXs compared with butterflies, excluding S. frugiperda, M. sexta, and T. ni. On the one hand, the amount of identified AOXs could have been determined by the unavailability of well-assembled genomes in both moths and butterflies. On the other hand, this can also be explained because moths are largely dependent on chemosensation (at short and long range) whereas butterflies use pheromones for short range communication and visual cues (Costanzo and Monteiro, 2007). In that sense, it can be suggested that butterflies have some AOXs related to odorant degradation and to a lesser extent for the metabolism of xenobiotics. This assumption is supported by our phylogenetic analysis, where two clades related to odorant-degrading function showed the presence of both moth and butterfly AOXs.
To date, two studies have reported a phylogeny for insect AOXs with focus on moths P. xylostella and H. armigera (He et al., 2017; Xu and Liao, 2017). Both analyses confirm XDHs as common ancestors followed by Dipteran AOXs, and support lepidopteran AOXs as more recently evolved enzymes. In that sense, our phylogenetic analysis is consistent with both. Furthermore, this analysis showed the two ODE-related clades already mentioned as well as a common ancestor in E. semipurpurella AOX1 (EsemAOX1), the only AOX identified from its antennal transcriptome. Interestingly, as an old lineage of moths (i.e., non-ditrysia), E. semipurpurella represents a model for evolutionary studies. Yuvaraj et al. (2017) showed that moth pheromone receptors could have evolved from plant volatile-related ORs, since two E. semipurpurella ORs (EsemOR3 and EsemOR5) that are phylogenetically close to plant volatile-responding ORs, respond to its sex pheromone (2S,6Z-6-nonen-2-ol), which resemble plant volatiles. The lack of more AOXs in E. semipurpurella could indicate that gene duplication events in other moths, and likely butterflies, happened in response to the use of more specialized aldehyde-related volatiles, such as sex pheromones. Furthermore, those that are close to EsemAOX1, in ODE-related clade (clade A in Figure 1), could likely be more plant volatile-biased.
It can be argued that moths with functionally studied AOXs, namely BmorAOX5, PxylAOX3 and AtraAOX2, are not strictly related to sex pheromone degradation. In fact, AtraAOX2 has not showed specificity for A. transitella sex pheromone [(Z,Z)-11,13-hexadecadienal], being also able to catalyze aldehyde-related plant volatiles (Choo et al., 2013). Similarly, PxylAOX3 was reported able to degrade sex pheromone (Z)-11-hexadecenal as well as plant-derived aldehydes, such as phenylacetaldehyde and non-anal (Wang et al., 2021b). Butterflies, H. melpomene with (Z)-9-octadecenal, octadecanal, (Z)-11-icosenal, icosanal and (Z)-13-docosenal as sex pheromone components (Darragh et al., 2017), and B. anynana with hexadecanal (Nieberding et al., 2008), represent the only butterflies that use aldehydes as semiochemicals in our data sets. Interestingly, one AOX (i.e., HmelAOX2) was present in an ODE-related clade according to our phylogenetic analysis, while B. anynana had seven. It is worth noting that H. melpomene as pollinator (Andersson and Dobson, 2003) and B. anynana as a fruit-feeding butterfly (Lewis and Wedell, 2007), are both exposed to more aldehydes emitted by plants and fruits. For B. anynana, it is noticeable that the seven AOXs (from a total of 12) appear to have emerged independently from the rest. Something similar to what is found in the gene location of moth species, such as B. mandarina, A. transitella and P. xylostella, where their AOX genes potentially related to ODE function, are far from other AOX genes, with the exception of PxylAOX3.
From the two ODE-related clades in our phylogenetic analysis, clade B resulted highly supported by both functional studies and antennal-enriched expression (Figure 1). The fact that AOXs from butterflies were also present in this clade, further suggests that these could use aldehyde-based volatiles as semiochemicals. Although fewer studies have exploited the semiochemistry of butterflies compared with moths, increasing evidence suggests that several species of butterflies, including H. melpomene and B. anynana, use volatiles as semiochemicals. For example, an early study reported strong antennal responses of H. melpomene to several tropical plant-derived volatiles, such as linalool, linalool oxide I and II, oxoisophoroneoxide and phenylacetaldehyde (Andersson and Dobson, 2003). Recently, 55 compounds exclusive of androconia (specialized units where secretory glands are found) in sympatric Pieridae butterflies that would play a role in mating orientation, were reported (Nobre et al., 2021). On the other hand, some moths and butterflies can share pheromone biosynthetic pathways. It has been reported that in B. anynana the synthesis of hexadecanal and (Z)-9-tetradecenol is mediated by conserved fatty acyl Δ11-desaturases (Liénard et al., 2014). In that sense, it is expected that other enzymes, such as AOXs, could be conserved between moths and butterflies. Therefore, it appears that AOXs in ODE-related clades could function for aldehyde-related semiochemicals whether derived from plants or conspecific species. Thus, more functional studies focused on both moths and butterflies would be necessary to support a monophyletic pheromone-degrading clade.
In general, the function of AOXs toward aldehydes might resemble the function of XDHs, which are their evolutionary ancestors (Kurosaki et al., 2013; Wang et al., 2016). High levels of similarity between vertebrate XDHs and AOXs have been reported (Terao et al., 2020). For instance, sequence identity between mammal AOXs, namely HsAOX1 and MmAOX1, reaches 83%. On the other hand, sequence identity between lepidopteran and mammal AOXs is ∼30%. More specifically, among lepidopteran AOXs, sequence identity starts decreasing at 67%. This divergence within lepidopterans is evidenced in an important amount of PSSs among those phylogenetically close AOXs that were selected for our molecular evolution analyses. Nevertheless, and as expected, residues that are conserved were not PSSs, such as those from the active site.
Our MSA analysis revealed that highly conserved residues in vertebrate AOXs, namely Glu1266, Phe919, Lys889 and Gln772, may or may not be conserved in lepidopteran and D. melanogaster AOXs. Thus, Glu1266 (at position 1,209 in our MSA, Figure 3) that is reported to be crucial for catalytic activity resulted highly conserved (Coelho et al., 2012), while Phe919 from vertebrates changes to Pro in insect AOXs (at position 884 in our MSA, Figures 3, 4). It is difficult to predict the effect of Pro instead of Phe in insect AOXs. The change from an aromatic side chain toward an aliphatic portion like the one present in Pro, might have some effects on selectivity and stability of aldehyde substrates. In our structural analyses, Pro884 was found in the active site of AtraAOX2, BanyAOX2 and DmelAOX2, keeping the region hydrophobic. In other studies, enzymes such as HCV NS5b polymerase, Pro197 along with Arg200, Cys366, Met414 and Tyr448, were reported to be crucial for ligand selectivity (Li et al., 2010). On the contrary, Pro substitutions in human carbonic anhydrase II led to an increased rigidity of the enzyme and subsequent decreased catalytic activity (Boone et al., 2015). In fact, it is well accepted that Pro restricts protein backbones with the lack of a hydrogen bond donor, disrupting α-helices (Woolfson and Williams, 1990; Van Arnam et al., 2011).
Overall, we believe this study represents the first to group a comprehensive set of AOX genes for several lepidopteran species. We have validated AOX sequences previously described and added 58 more in moths and 33 more in butterflies. We have also uncovered the potential importance of aldehydes as semiochemicals in butterflies, as reflected by the number of AOX present in this group. The information presented herein is a helpful reference for further evolutionary and functional studies in this highly biodiverse order.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
RG, AM, and HV conceived the idea. RG and HV performed genome and structural analysis. LCP and AM analyzed phylogenetics and evolution data. HV and LCP wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Agencia Nacional de Investigación y Desarrollo (ANID) project N° 11200349 and ANID scholarship N° 21190666. LCP was supported by Wellcome Trust Investigator Award 110117/Z/15/Z.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank reviewers for the valuable comments that helped to improve this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.823119/full#supplementary-material
Adebali, O., Ortega, D., and Zhulin, I. (2015). CDvist: a webserver for identification and visualization of conserved domains in protein sequences. Bioinformatics 31, 1475–1477. doi: 10.1093/bioinformatics/btu836
Andersson, S., and Dobson, H. (2003). Antennal responses to floral scents in the butterfly. J. Chem. Ecol. 29, 2319–2330. doi: 10.1023/A:1026278531806
Boone, C., Rasi, V., Tu, C., and McKenna, R. (2015). Structural and catalytic effects of proline substitution and surface loop deletion in the extended active site of human carbonic anhydrase II. FEBS J. 282, 1445–1457. doi: 10.1111/febs.13232
Butenandt, A., Beckman, R., Stamm, D., and Hecker, E. (1959). Über den sexuallockstoff des seidenspinners Bombyx mori. Reindarstellung und konstitution. Z Naturforsch B. 14, 283–284.
Chemnitz, J., Jentschke, P. C., Ayasse, M., and Steiger, S. (2015). Beyond species recognition: somatic state affects long-distance sex pheromone communication. Proc. R. Soc. B 282:20150832. doi: 10.1098/rspb.2015.0832
Chertemps, T., and Meïbèche, M. (2021). “19 - Odor degrading enzymes and signal termination,” in Insect Pheromone Biochemistry and Molecular Biology, 2nd Edn, eds J. Gary, Blomquist and G. Richard, and Vogt (Cambridge, MA: Academic Press), 619–644.
Choo, Y. M., Pelletier, J., Atungulu, E., and Leal, W. S. (2013). Identification and characterization of an antennae-specific aldehyde oxidase from the navel orangeworm. PLoS One 8:e67794. doi: 10.1371/journal.pone.0067794
Coelho, C., Mahro, M., Trincão, J., Carvalho, A., Ramos, M., Terao, M., et al. (2012). The first mammalian aldehyde oxidase crystal structure: insights into substrate specificity. J. Biol. Chem. 287, 40690–40702. doi: 10.1074/jbc.M112.390419
Coffelt, J. A., Vick, K. W., Sonnet, P. E., and Doolittle, R. E. (1979). Isolation, identification, and synthesis of a female sex pheromone of the navel orangeworm, Amyelois transitella (Lepidoptera: Pyralidae). J. Chem. Ecol. 5, 955–966.
Coleman, M., Vontas, J. G., and Hemingway, J. (2002). Molecular characterization of the amplified aldehyde oxidase from insecticide resistant Culex quinquefasciatus. Eur. J. Biochem. 269, 768–779. doi: 10.1046/j.0014-2956.2001.02682.x
Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. doi: 10.1093/nar/16.22.10881
Costanzo, K., and Monteiro, A. (2007). The use of chemical and visual cues in female choice in the butterfly Bicyclus anynana. Proc. R. Soc. B 274, 845–851. doi: 10.1098/rspb.2006.3729
Darragh, K., Vanjari, S., Mann, F., Gonzalez-Rojas, M. F., Morrison, C. R., Salazar, C., et al. (2017). Male sex pheromone components in Heliconius butterflies released by the androconia affect female choice. PeerJ 5:e3953. doi: 10.7717/peerj.3953
Durand, N., Pottier, M. A., Siaussat, D., Bozzolan, F., Maïbèche, M., and Chertemps, T. (2018). Glutathione-S-transferases in the olfactory organ of the noctuid moth Spodoptera littoralis, diversity and conservation of chemosensory clades. Front. Physiol. 9:1283. doi: 10.3389/fphys.2018.01283
Engsontia, P., Sangket, U., Chotigeat, W., and Satasook, C. (2014). Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: implications for their adaptation and speciation. J. Mol. Evol. 79, 21–39. doi: 10.1007/s00239-014-9633-0
Gao, F., Chen, C., Arab, D. A., Du, Z., He, Y., and Ho, S. (2019). EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
Garattini, E., and Terao, M. (2012). The role of aldehyde oxidase in drug metabolism. Expert Opin. 8, 487–503. doi: 10.1517/17425255.2012.663352
Garattini, E., Fratelli, M., and Terao, M. (2009). The mammalian aldehyde oxidase gene family. Hum. Genom. 4, 119–130. doi: 10.1186/1479-7364-4-2-119
Godoy, R., Machuca, J., Venthur, H., Quiroz, A., and Mutis, A. (2021). An overview of antennal esterases in lepidoptera. Front. Physiol. 12:643281. doi: 10.3389/fphys.2021.643281
Grabherr, M., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hansson, B., and Stensmyr, M. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
He, P., Zhang, Y. N., Li, Z. Q., Yang, K., Zhu, J. Y., Liu, S. J., et al. (2014a). An antennae-enriched carboxylesterase from Spodoptera exigua displays degradation activity in both plant volatiles and female sex pheromones. Insect Mol. Biol. 23, 475–486. doi: 10.1111/imb.12095
He, P., Zhang, J., Li, Z. Q., Zhang, Y. N., Yang, K., and Dong, S. L. (2014b). Functional characterization of an antennal esterase from the noctuid moth, Spodoptera exigua. Arch. Insect Biochem. Physiol. 86, 85–99. doi: 10.1002/arch.21164
He, P., Li, Z. Q., Liu, C. C., Liu, S. J., and Dong, S. L. (2014c). Two esterases from the genus Spodoptera degrade sex pheromones and plant volatiles. Genome 57, 201–208. doi: 10.1139/gen-2014-0041
He, P., Zhang, Y. N., Yang, K., Li, Z. Q., and Dong, S. L. (2015). An antenna-biased carboxylesterase is specifically active to plant volatiles in Spodoptera exigua. Pestic. Biochem. Physiol. 123, 93–100.
He, P., Zhang, Y. F., Hong, D. Y., Wang, J., Wang, X. L., Zuo, L. H., et al. (2017). A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genomics 18:219. doi: 10.1186/s12864-017-3592-y
He, P., Mang, D. Z., Wang, H., Wang, M. M., Ma, Y. F., Wang, J., et al. (2019). Molecular characterization and functional analysis of a novel candidate of cuticle carboxylesterase in Spodoptera exigua degrading sex pheromones and plant volatiles esters. Pestic. Biochem. Physiol. 163, 227–234. doi: 10.1016/j.pestbp.2019.11.022
Hemingway, J., Coleman, M., Paton, M., McCarroll, L., Vaughan, A., and DeSilva, D. (2000). Aldehyde oxidase is coamplified with the World’s most common Culex mosquito insecticide resistance-associated esterases. Insect Mol. Biol. 9, 93–99. doi: 10.1046/j.1365-2583.2000.00160.x
Huang, X., Fan, D., Liu, L., and Feng, J. (2017). Identification and characterization of glutathione S-Transferase genes in the antennae of codling moth (Lepidoptera: Tortricidae). Ann. Entomol. Soc. Am. 110, 409–416. doi: 10.1093/aesa/sax041
Huang, X., Liu, L., Su, X., and Feng, J. (2016). Identification f biotransformation enzymes in the antennae of codling moth Cydia pomonella. Gene 580, 73–79. doi: 10.1016/j.gene.2016.01.008
Kaissling, K. E. (2013). Kinetics of olfactory responses might largely depend on the odorant-receptor interaction and the odorant deactivation postulated for flux detectors. J. Comp. Physiol. A Neuroethol. Sensory Neural Behav. Physiol. 199, 879–896. doi: 10.1007/s00359-013-0812-z
Kasang, G., Kaissling, K. E., Vostrowsky, O., and Bestmann, H. J. (1978). Bombykal, a second pheromone component of the silkworm moth Bombyx mori L. Z. Angew. Chem. Int. Ed. Engl. 17:60. doi: 10.1002/anie.197800601
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kawahara, A. Y., Plotkin, D., Espeland, M., Meusemann, K., Toussaint, E., Donath, A., et al. (2019). Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. PNAS 116, 22657–22663. doi: 10.1073/pnas.1907847116
Krenitsky, T. A., Neil, S. M., Elion, G. B., and Hitchings, G. H. (1972). A comparison of the specificities of xanthine oxidase and aldehyde oxidase. Arch. Biochem. Biophys. 150, 585–599. doi: 10.1016/0003-9861(72)90078-1
Kurosaki, M., Bolis, M., Fratelli, M., Barzago, M. M., Pattini, L., Perretta, G., et al. (2013). Structure and evolution of vertebrate aldehyde oxidases: from gene duplication to gene suppression. Cell. Mol. Life Sci. 70, 1807–1830. doi: 10.1007/s00018-012-1229-5
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Leal, W. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Lewis, Z., and Wedell, N. (2007). Effect of adult feeding on male mating behaviour in the butterfly, Bicyclus anynana (Lepidoptera: Nymphalidae). J. Insect Behav. 20, 201–213. doi: 10.1007/s10905-007-9075-2
Li, F., Zhao, X., Li, M., He, K., Huang, C., Zhou, Y., et al. (2019). Insect genomes: progress and challenges. Insect Mol. Biol. 28, 739–758. doi: 10.1111/imb.12599
Li, G. W., Chen, X. L., Xu, X. L., and Wu, J. X. (2018). Degradation of sex pheromone and plant volatile components by an antennal glutathione S-transferase in the oriental fruit moth, Grapholita molesta Busck (Lepidoptera: Tortricidae). Arch. Insect Biochem. Physiol. 99:e21512. doi: 10.1002/arch.21512
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homar, N., et al. (2009). The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, T., Froeyen, M., and Herdewijn, P. (2010). Insight into ligand selectivity in HCV NS5B polymerase: molecular dynamics simulations, free energy decomposition and docking. J. Mol. Model. 16, 49–59. doi: 10.1007/s00894-009-0519-9
Li, Z. Q., Zhang, S. Z., Luo, J. Y., Wang, C. Y., Lv, L. M., Dong, S. L., et al. (2015). Transcriptome comparison of the sex pheromone glands from two sibling Helicoverpa species with opposite sex pheromone components. Sci. Rep. 5:9324. doi: 10.1038/srep09324
Liénard, M., Wang, H. L., Lassance, J. M., and Löfstedt, C. (2014). Sex pheromone biosynthetic pathways are conserved between moths and the butterfly Bicyclus anynana. Nat. Commun. 5:3957. doi: 10.1038/ncomms4957
Liu, H., Lei, X., Du, L., Yin, J., Shi, H., Zhang, T., et al. (2019). Antennae-specific carboxylesterase genes from Indian meal moth: Identification, tissue distribution and the response to semiochemicals. J. Stored Prod. Res. 84:101528. doi: 10.1016/j.jspr.2019.101528
Merlin, C., François, M. C., Bozzolan, F., Pelletier, J., Jacquin-Joly, E., and Maïbèche-Coisne, M. (2005). A new aldehyde oxidase selectivity expressed in chemosensory organs of insects. Biochem. Biophys. Res. Commun. 332, 4–10. doi: 10.1016/j.bbrc.2005.04.084
Nieberding, C. M., de Vos, H., Schneider, M. V., Lassance, J. M., Estramil, N., Andersson, J., et al. (2008). The male sex pheromone of the butterfly Bicyclus anynana: towards an evolutionary analysis. PLoS One 3:e2751. doi: 10.1371/journal.pone.0002751
Nobre, C. E. B., Lucas, L. A. D., Padilha, R. J. R., Navarro, D. M., Alves, L. C., and Maia, A. C. D. (2021). Specialized androconial scales conceal species-specific semiochemicals of sympatric Sulphur butterflies (Lepidoptera: Pieridae: Coliadinae). Org. Divers Evol. 21, 93–105. doi: 10.1007/s13127-020-00475-8
Oppenheim, S., Baker, R., Simon, S., and DeSalle, R. (2015). We can’t all be supermodels: the value of comparative transcriptome to the study of non-model insects. Insect Mol. Biol. 24, 139–154. doi: 10.1111/imb.12154
Ou, J., Deng, H. M., Zheng, S. C., Huang, L. H., Feng, Q. L., and Liu, L. (2014). Transcriptomic analysis of developmental features of Bombyx mori wing disc during metamorphosis. BMC Genomics 15:820. doi: 10.1186/1471-2164-15-820
Patel, R., and Jain, M. (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Pelletier, J., Bozzolan, F., Solvar, M., Françoise, M. C., Jacquin-Joly, E., and Maïbèche-Coisne, M. (2007). Identification of candidate aldehyde oxidases from the silkworm Bombyx mori potentially involved in antennal pheromone degradation. Gene 404, 31–40. doi: 10.1016/j.gene.2007.08.022
Price, M., Dehal, P., and Arkin, A. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Riddiford, L. M. (1967). Trans-2-hexenal: mating stimulant for Polyphemus moths. Science 158, 139–141. doi: 10.1126/science.158.3797.139
Rybczynski, R., Reagan, J., and Lerner, M. (1989). A pheromone-degrading aldehyde oxidase in the antennae of the moth Manduca sexta. J. Neurosci. 9, 1341–1353.
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Soffan, A., Subandiyah, S., Makino, H., Watanabe, T., and Horiike, T. (2018). Evolutionary analysis of the highly conserved insect odorant coreceptor (Orco) revealed a positive selection mode, implying functional flexibility. J. Insect Sci. 18, 1–8. doi: 10.1093/jisesa/iey120
Sokkar, P., Mohandass, S., and Ramachandran, M. (2011). Multiple templates-based homology modeling enhances structures quality of AT1 receptor: validation by molecular dynamics and antagonist docking. J. Mol. Model. 17, 1565–1577. doi: 10.1007/s00894-010-0860-z
Swanson, W. J., Nielsen, R., and Yang, Q. (2003). Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 20, 18–20. doi: 10.1093/oxfordjournals.molbev.a004233
Terao, M., Garattini, E., Romão, M. J., and Leimkühler, S. (2020). Evolution, expression, and substrate specificities of aldehyde oxidase enzymes in eukaryotes. J. Biol. Chem. 295, 5377–5389. doi: 10.1074/jbc.REV119.007741
Van Arnam, E. B., Lester, H. A., and Dougherty, D. A. (2011). Dissecting the functions of conserved prolines within transmembrane helices of the D2 dopamine receptor. ACS Chem. Biol. 6, 1063–1068. doi: 10.1021/cb200153g
Venthur, H., and Zhou, J. J. (2018). Odorant receptors and odorant-binding proteins as insect pest control targets: a comparative analysis. Front. Physiol. 9:1163. doi: 10.3389/fphys.2018.01163
Vizueta, J., Sánchez-Gracia, A., and Rozas, J. (2020). BITACORA: a comprehensive tool for the identification and annotation of gene families in genome assemblies. Mol. Ecol. Resour. 20, 1445–1452. doi: 10.1111/1755-0998.13202
Vogt, R., Grosse-Wilde, E., and Zhou, J. J. (2015). The lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 62, 142–153. doi: 10.1016/j.ibmb.2015.03.003
Wang, C. H., Zhang, C., and Hing, X. H. (2016). Xanthine dehydrogenase: an old enzyme with new knowledge and prospects. Bioengineered 7, 395–405. doi: 10.1080/21655979.2016.1206168
Wang, M. M., Long, G. J., Guo, H., Liu, X. Z., Wang, H., Dewer, Y., et al. (2021a). Two carboxylesterase genes in Plutella xylostella associated with sex pheromones and plant volatiles degradation. Pest Manag. Sci. 77, 2737–2746. doi: 10.1002/ps.6302
Wang, M. M., He, M., Wang, H., Ma, Y. F., Dewer, Y., Zhang, F., et al. (2021b). A candidate aldehyde oxidase in the antennae of the diamondback moth, Plutella xylostella (L.), is potentially involved volatiles and the detoxification of xenobiotics. Pestic. Biochem. Phys. 171:104726. doi: 10.1016/j.pestbp.2020.104726
Wei, H., Tan, S., Li, Z., Li, J., Moural, T. W., Zhu, F., et al. (2020). Odorant degrading carboxylesterases modulate foraging and mating behaviors of Grapholita molesta. Chemosphere 270:128647. doi: 10.1016/j.chemosphere.2020.128647
Weiss, M. R. (2001). “Vision and learning in some neglected pollinators: beetles, flies, moths and butterflies,” in Cognitive Ecology of Pollination. Animal Behavior and Floral Evolution, eds L. Chittka and J. D. Thomson (Cambridge: Cambridge University Press).
Woolfson, D. N., and Williams, D. H. (1990). The influence of proline residues on α-helical structure. FEBS Lett. 277, 185–188. doi: 10.1016/0014-5793(90)80839-b
Xu, W., and Liao, Y. (2017). Identification and characterization of aldehyde oxidases (AOXs) in the cotton bollworm. Sci. Nat. 104:94. doi: 10.1007/s00114-017-1515-z
Yang, B., Ozaki, K., Ishikawa, Y., and Matsuo, T. (2015). Identification of candidate odorant receptors in asian corn borer ostrinia furnacalis. PLoS One 10:e0121261. doi: 10.1371/journal.pone.0121261
Yang, Z., and Nielsen, R. (2002). Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917. doi: 10.1093/oxfordjournals.molbev.a004148
Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A. M. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449. doi: 10.1093/genetics/155.1.431
Yang, Z., Wong, W., and Nielsen, R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yuvaraj, J. K., Corcoran, J. A., Andersson, M. N., Newcomb, R. D., Anderbrant, O., and Löfstedt, C. (2017). Characterization of odorant receptors from a non-ditrysian moth, Eriocrania semipurpurella sheds light on the origin of sex pheromone receptors in lepidoptera. Mol. Biol. Evol. 34, 2733–2746. doi: 10.1093/molbev/msx215
Zhang, J. P., Salcedo, C., Fang, Y. L., Zhang, R. J., and Zhang, Z. N. (2012). An overlooked component: (Z)-9-tetradecenal as a sex pheromone in Helicoverpa armigera. J. Insect Physiol. 58, 1209–1216.
Zhang, Y. N., Xia, Y. H., Zhu, J. Y., Li, S. Y., and Dong, S. L. (2014). Putative pathway of sex pheromone biosynthesis and degradation by expression patterns of genes identified from female pheromone gland and adult antenna of Sesamia inferens (Walker). J. Chem. Ecol. 40, 439–451. doi: 10.1007/s10886-014-0433-1
Zhang, Y. N., Li, Z. Q., Zhu, X. Y., Qian, J. L., Dong, Z. P., and Xu, L. (2017a). Identification and tissue distribution of carboxylesterase (CXE) genes in Athetis lepigone (Lepidoptera: Noctuidae) by RNA-seq. J. Asia-Pacif. Entomol. 20, 1150–1155.
Zhang, Y. X., Wang, W. L., Li, M. Y., Li, S. H., and Liu, S. (2017b). Identification of putative carboxylesterase and aldehyde oxidase genes from the antennae of the rice leaffolder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). J. Asia Pac. Entomol. 20, 907–913. doi: 10.1016/j.aspen.2017.06.001
Zhang, Y., Yang, Y., Shen, G., Mao, X., Jiao, M., and Lin, Y. (2020). Identification and characterization of aldehyde oxidase 5 in the pheromone gland of the silkworm (Lepidoptera: Bombycidae). J. Insect Sci. 20, 1–10. doi: 10.1093/jisesa/ieaa132
Keywords: Lepidoptera, insect olfaction, aldehyde oxidase, genome, phylogenetics
Citation: Godoy R, Mutis A, Carabajal Paladino L and Venthur H (2022) Genome-Wide Identification of Aldehyde Oxidase Genes in Moths and Butterflies Suggests New Insights Into Their Function as Odorant-Degrading Enzymes. Front. Ecol. Evol. 10:823119. doi: 10.3389/fevo.2022.823119
Received: 26 November 2021; Accepted: 14 January 2022;
Published: 08 February 2022.
Edited by:
Xin-Cheng Zhao, Henan Agricultural University, ChinaReviewed by:
Peng He, Guizhou University, ChinaCopyright © 2022 Godoy, Mutis, Carabajal Paladino and Venthur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Herbert Venthur, aGVyYmVydC52ZW50aHVyQHVmcm9udGVyYS5jbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.