- 1Divison of Vegetable Crops, Bengaluru, India
- 2Divison of Crop Protection, ICAR-Indian Institute of Horticultural Research, Bengaluru, India
Interactions of a virus with its vector and host plant have challenged entomologists, pathologists and biologists alike. Phytophagous insects depend on specific host volatile cues to locate suitable host plants for feeding and oviposition. Several studies have revealed that plant viruses modify their insect vector’s orientation toward specific host plants to facilitate their spread and survival. The ecological and molecular basis of this vector behavior modification remains largely unknown and was therefore explored in this study. Interestingly, host volatile preference for non-viruliferous female whiteflies [Bemisia tabaci (Genn.)] was found to be preferentially oriented toward infected chili plant [with chili leaf curl (ChLCV)] volatiles, while viruliferous whiteflies preferred healthy chili plant (Capsicum annum L.) volatiles in olfactometer. The electrophysiological studies involving electroantennogram (EAG) assays exhibited similar trend in EAG response amplitudes. Gas Chromatography linked electroantennodetection (GC EAD) revealed specific plant volatile cues responsible for altered host orientation behavior of the vector. Transcriptome profiling of the viruliferous and non-viruliferous whiteflies and Realtime qPCR validation showed differential expression of certain odorant binding proteins (OBPs) in viruliferous whiteflies. Our results suggest that there is a plant virus mediated altered chemoecological behavior in the vector with respect to orientation toward its host plant. Based on the findings we speculate that the virus mediates such change in the vector for a continued transmission success to the host.
Introduction
The majority of plant viruses thrive and spread from one host to another by exploiting insects as vectors. Interactions between a virus and its insect vector are not only intricate but also multimodal in nature as the virus can significantly alter the diverse traits of its vector as well as host plant (Moreno-Delafuente et al., 2013). More importantly, performance of a vector with respect to fecundity (Jiu et al., 2007), survival, host choice (Stafford et al., 2011; Ingwell et al., 2012; Moreno-Delafuente et al., 2013) and behavior (Mauck et al., 2012) can be influenced by the virus. Whitefly, Bemicia tabaci (Genn.) has been found to feed more efficiently after the acquisition of Tomato yellow leaf curl virus (TYLCV) compared to their non-viruliferous counterparts (Moreno-Delafuente et al., 2013). Studies reported in the past show that virus acquisition can change host plant preference of insect vectors in a way that in turn benefits the virus (Stafford et al., 2011; Ingwell et al., 2012; Moreno-Delafuente et al., 2013; Stafford-Banks et al., 2014). The aphid, Rhopalosiphum padi (L.) exhibits feeding preference toward healthy wheat plants after acquiring Barley Yellow Dwarf Virus (BYDV) whereas non-viruliferous aphids prefer to feed on infected plants (Ingwell et al., 2012). Further, several independent studies on vectors like whiteflies and aphids have shown virus mediated host plant manipulation by bringing about physiological and biochemical changes in the host plant that are known to cause variations in its volatile emissions favoring vector attraction to infected or healthy plants (Fereres and Moreno, 2009; Ingwell et al., 2012; Mauck et al., 2012).
Phytophagous insects (= herbivore) developed the ability to perceive specific volatile cues from host plants over millions of years (Jermy, 1984). This has substantially helped them in locating their ideal hosts and any change in these host volatile patterns could easily disorient them from locating their hosts. This selective discrimination of host plants by herbivores is typically attributed to their specialized sensory system involving various olfactory proteins (such as odorant binding proteins, OBPs and odorant receptors, ORs) (Kamala Jayanthi et al., 2014). These olfactory proteins, process volatile cues received by herbivores which consequently help them identify a specific host plant (Bruce et al., 2005).
The studies mentioned above indicate a pattern suggesting vector and host plant manipulation by the virus to increase its transmission efficiency. To achieve this, the vector will have to preferentially be attracted to the host plant of virus interest. If the above-mentioned dynamics between the virus-vector and the host plant were believed to be true, are the interactions effective enough to result in such manipulative changes? Considering the fact that there is enough evidence to show that viruses manipulate vector and host plant to suit their transmission patterns, how virus-induced changes in plants impact herbivore orientation remains largely unexplored.
Using the whitefly, Bemisia tabaci: Chili Leaf Curl Virus (ChLCV) as the vector—pathogen model system for its host plant (Chili, Capsicum annum L.), the interaction of ChLCV with its host plant and the consequent effect on host location behavior of its vector, B. tabaci was investigated in this study. Chili Leaf Curl Virus is a single stranded DNA begomovirus transmitted in a persistent manner by B. tabaci which is a complex of 39 globally distributed cryptic species. The highly invasive biotypes (B and Q) of B. tabaci display adaptability to extreme climatic conditions and are major contributors to its widespread distribution (De Barro et al., 2011). Introduction of the highly polyphagous B biotype of B. tabaci has therefore facilitated global transmission of begomoviruses, the largest species in the family Geminiviridae (Lazarowitz and Shepherd, 1992) into potential new hosts (Navas-Castillo et al., 2011).
In this study, chemical ecology and behavioral response of viruliferous and non-viruliferous B. tabaci (ChLCV vector) was explored using virus-infected and healthy host plant volatiles in olfactometer bioassays and electrophysiological techniques like Coupled Gas Chromatography-Electroantennographic Detection (GC-EAD). Since the chemo-ecological response in an insect is guided by the olfactory system, the differential expression of olfactory related genes in viruliferous and non-viruliferous B. tabaci was explored using antennal transcriptomics.
Materials and Methods
The study was conducted at ICAR-Indian Institute of Horticultural Research (ICAR-IIHR), Bengaluru (13.1348°N, 77.4960°E; 900 MSL), Karnataka, India during 2015–17.
Host Plant Maintenance
A high yielding chili variety IIHR 3909 was used for this study. The seeds were collected from the Division of Vegetable Crops, ICAR-IIHR and were sown in coco-peat filled protrays (50 cavity). After 1 week (when they germinated), the protrays were shifted to an insect-proof polyhouse.
Virus Inoculum Maintenance
Chili Leaf Curl Virus (ChLCV)-infected plants were collected from Capsicum annum experimental plots at ICAR-IIHR Bengaluru. Infected plants were transplanted in pots and maintained separately in a screen house.
Insect Cultures
The ChLCV vector, B. tabaci, Biotype B that transmits the virus in a semi-persistent manner were collected from the Division of Entomology and Nematology, ICAR-IIHR, Bengaluru. These insects were further cultured as explained below to obtain non-viruliferous and viruliferous whiteflies. Whiteflies that attained the age of 20-25 days (both males and females) were used for all the experiments.
Non-viruliferous Vectors
Whiteflies were maintained on healthy host plants inside an insect-proof net house.
Viruliferous Vectors
Mass-inoculation method was used to obtain viruliferous B. tabaci. Non-viruliferous whiteflies were released on to ChLCV infected plants placed in a cage (30 × 30 × 30 CM L, B, H) and allowed to feed for a minimum of 7 days.
Healthy Host Plants
Healthy seedlings were transplanted into pots and maintained inside an insect-proof net house to protect them from viruliferous whiteflies.
Infected Host Plants
Four-week-old seedlings (2–4 true leaf stage) were exposed to ∼10 viruliferous whiteflies per seedling. After 3–5 days of Inoculation Access Period (IAP), the seedlings were shifted to a virus-free net house (40 threads/cm2). The plants were allowed 10–15 days to express disease symptoms. The time period was increased to 20 days depending upon the climatic conditions. Such infected seedlings were used for further studies.
Chemicals
Authentic chemical standards (>95%) n-Undecane, 1 octanol 3,7 dimethyl, n-Dodecane, Methyl eugenol, 2,3, butanediol 2,3, dimethyl, cis-3-hexenyl isobutyrate, 2-hexyl-1-decanol, n-Nonadecane, Limonene, ethyl benzaldehyde, cis-geranyl acetate, and Phytane were procured from Sigma Aldrich, United States. Porapak extracts (10 μL) were directly used as test samples after elution without further processing. Compounds for which no standards were available (Supplementary Table 1) were tentatively identified using the NIST-database 14.
Volatile Sampling of Healthy and Virus Infected Plants
Headspace volatiles from both ChLCV infected chili and healthy chili plants (n = 3, 65 days old age) were collected for 24 h using a customized air entrainment system (Shivaramu et al., 2017). Headspace volatiles trapped in Porapak Q, (50 mg, 60/80 mesh; Supelco, Sigma Aldrich St Louis, United States) placed in a glass tube (5 mm dia) were eluted with 750 μL of redistilled diethyl ether (99.7% pure, Merck) and stored in a freezer (−20°C) until further use.
Olfactometer Bioassays With Infected and Healthy Chili Plant Headspace Volatiles
The behavioral responses of viruliferous and non-viruliferous B. tabaci to headspace volatiles of both ChLCV-infected and healthy chili plants were studied through, dual-choice behavioral assays using a circular Perspex four-arm olfactometer (Kamala Jayanthi et al., 2015). Each set consisted of two treated arms (healthy chili headspace sample and infected chili headspace sample) and two control arms (diethyl ether, 99.7% pure, Merck). The test samples (10 μL) were directly applied to a filter paper strip (4 × 2 cm, L × B) and allowed to evaporate before placing in the treatment arms. Filter paper strips with just the solvent (diethyl ether, 10 μL) served as control. Ten (n = 10) replicates were carried out. Observations on the time spent and number of entries into each arm were recorded using Olfa software (F. Nazzi, Udine, Italy). The apparatus was rotated 90° for every 2 min to eliminate any directional bias in the bioassay cage.
Electrophysiological Studies
Preparation of Insect Antenna
Viruliferous and non-viruliferous female whiteflies were collected from the respective virus infected and healthy chili plants in plastic vials (Eppendorf, United States) and allowed to starve for 2 h prior to conducting Electroantennogram (EAG) study. For antennal preparation, an individual female whitefly was placed into a plastic micropipette (10 μL) tip. Slight air pressure was provided to wedge the whitefly into the tip, with the head turned forward. Insect was forced to protrude the head out of the plastic tip. The head was excised using a pair of micro-scissors and placed on the EAG (Electroantennogram)/EAD (Electroantennodetection) probe holder (Syntech, Germany), with a small amount of electrode gel (Sigma Gel, United States) ensuring that the base of the antenna was placed on the indifferent electrode, while the antennae tip touched the recording electrode. The prepared EAG probe containing live antenna was then inserted into the pre-amplifier with a constant stream of humidified air over the antenna at 200 mL/min.
Electroantennogram Bioassay
Electroantennogram recordings (n = 10) were made as described in Kamala Jayanthi et al. (2014). In this bioassay humidified air and honey were used as negative and positive controls, respectively. Odor stimulation data was recorded using AutoSpike software included with the Syntech EAG Model IDAC-4 (Intelligent Data Acquisition Controller). Antennal responses for headspace plant volatiles (healthy and infected) were recorded based on the downward deflection signal (in mV) using SYNTECH Electroantennogram software. Signal means (in millivolts, mV) of whitefly antenna for headspace plant volatiles were obtained to compare the response between healthy and infected plant volatiles.
Coupled Gas Chromatography-Electroantennographic Detection
Electroantennodetection recordings (n = 10) (Kamala Jayanthi et al., 2012; Shivaramu et al., 2017). The antennal preparations were same as described above. A customized software package (Syntech, Germany) was used to analyze the signal passing through a high impedance amplifier. The coupled GC-EAD system, in which the effluent from the GC column is simultaneously directed to the antennal preparation and the GC detector, has been described previously (Wadhams, 1990). Separation of the host plant volatiles was achieved on an Agilent, GC-7890 gas chromatography system equipped with a flame ionization detector (FID) and an Agilent HP-5 (5%-phenyl)-methylpolysiloxane non-polar fused silica capillary tubing column (30 m length, 0.25 mm Diameter, and 0.25 μm film thickness). The thermal program was set at an initial step of 60°C for 1 min with inlet temperature maintained at 250°C through splitless mode (40 mL/min ratio) and later ramped at 15°C/min up to 240°C, held for 2 min with Nitrogen as the carrier gas at a flow rate of 1 mL/min.
Gas Chromatography-Mass Spectrometry Analysis
The Porapak Q elutes of headspace plant volatiles (both healthy and infected) collected in the solvent (Diethyl ether, Merck, 99.97%) were analyzed to identify specific compounds that elicited antennal response in B. tabaci using GC-MS, Agilent 7890B GC system apparatus equipped with Mass Spectrometry, MS (Agilent 5977 MSD). An Agilent (HP-5 MS UI) capillary column (30 m length, 0.25 mm Diameter, and 0.25 μm film thickness) was used to examine the samples. The thermal setting was similar to the one described above. Helium was used as a carrier gas at a flow rate of 1 mL/min. MS was set to full scan mode (70 eV) with AMU range of 40–450. One microliter of the sample was injected at a temperature of 250°C in a splitless mode ratio (40 mL/min). Individual volatile compounds were identified by comparing the GC retention time, Kovats index using homologous series of n-alkanes (C7–C30 procured from Sigma-Aldrich) as standard (Kovats, 1965) and comparing the MS spectra with spectral library NIST 14. Total volatile production was estimated by a sum of all GC-FID peak areas in the chromatogram and individual compounds were quantified as relative percent area. Identified EAD compounds were authenticated by comparing the retention time and mass spectra of the obtained data with commercially available standards and the remaining VOCs for which standards were unavailable were identified tentatively based on NIST spectral library 14.
Comparison of Viruliferous and Non-viruliferous Whitefly Odorant Binding Proteins and Olfactory Receptor Related Gene Expression
A whole-body transcriptomics study was done for both viruliferous and non-viruliferous whiteflies to understand the differential expression of OBPs and Olfactory Receptors (ORs). The insects for the study were collected as previously described.
RNA Isolation
Synchronous mixed populations comprising both sexes were used for Transcriptome studies. RNA extraction was carried out for both viruliferous (N = 200) and non-viruliferous (N = 200) whiteflies using RNeasy Mini Kit (Qiagen) according to manufacturer’s protocol with slight modifications. Concentration and quality of RNA were checked using NanoDrop (DeNovix DS-11 spectrophotometer) and also visualized on 1.5% gel-electrophoresis.
cDNA Library Preparation
The mRNA sequencing libraries were prepared using TruSeq RNA Library Prep Kit v2 (Illumina ®) following the manufacturer’s instructions. Briefly, 800 ng of the total RNA was used as input for poly (A) mRNA enrichment followed by fragmentation and reverse transcription to generate cDNA. Adapter index primers with unique barcodes for each sample were ligated to fragmented double-strand cDNA. Ampure beads were used to purify adapter-ligated fragments and the purified product was amplified using Illumina primers to generate sequencing library. The libraries were quantified using Qubit DNA HS kit and library quality was checked on Agilent Bioanalyzer 2100 using DNA 1000 kit.
Transcriptome Sequencing
After quality inspections, cDNA libraries were denatured and diluted to Illumina recommended 1.8 pM concentration using 0.2 nM NaOH and HT1 reagent, respectively. The denatured and diluted libraries were loaded onto NextSeq reagent cartridge. Cluster generation and sequencing were performed on Illumina NextSeq 500 to generate 2 × 150 paired-end reads. Soon after the completion of a run, demultiplexed sequence data were collected in FastQ format and checked for the data quality. The sequencing data were quality checked using Fast QC (Krueger, 2015 Krueger F: Trim Galore)1 and MultiQC (Ewels et al., 2016) software. The data were checked for various parameters such as base call quality distribution, % bases above Q20, Q30, %GC, sequencing adapter contamination etc. Raw sequence reads were processed to remove adapter sequences and low-quality reads.
Transcriptome Assembly and Annotation
The raw reads were checked for regions with low quality, adapter contamination, polynucleotide repeats and were trimmed accordingly using after a quality check (QC) (Chen et al., 2017) and Trim galore (Babraham Bioinformatics). Quality passed reads with minimum length of 75 were taken for all downstream analysis. Going forward the QC passed reads were then checked for reads originating from rRNA using sortmeRNA (Kopylova et al., 2012) tool by blasting the reads against SILVA and Rfam eukaryotic rRNA database and were excluded from further analysis. After filtering the reads for low quality, polynucleotide regions and rRNA, the sequence reads from both the samples were combined and assembled into 1,05, 818 transcripts belonging to 61,176 unigenes using Trinity v.2.5.1 (Haas et al., 2013) with default parameters (Kmer = 25, Min contig length 300). Coding regions (ORFs) within the transcripts were identified and putative protein sequences were predicted using TransDecoder.2 The protein sequences were annotated against NCBI nr, Uniprot, Swissprot and Uniref100 protein database using BLASTp (Altschup et al., 1990) module of Diamond (Buchfink et al., 2014). Any transcript that mapped onto the database with E-value < 1e-04 with confident annotations were considered for downstream analysis.
Differential Expression Analysis
To extract raw expression counts of the unigenes, the reads were mapped onto the unigenes using hisat2 aligner (Kim et al., 2015) and using feature counts (Liao et al., 2014) the raw expression count, i.e., the number of counts mapping onto each transcript were estimated. Approximately greater than 80% of reads from all the samples aligned onto the unigenes. Differential expression analysis was performed using DESeq3 and R package for non-viruliferous and viruliferous samples to identify differentially expressed genes (DEG). Unigenes were considered as significantly differentially expressed if the absolute log2 Fold Change was > 2 and p-value < 0.05. The objective here was to explore the expression of the odorant binding proteins (OBPs) and other olfaction related protein-encoding genes based on the fact that the insects have altered behavior to the infected and healthy plants.
Validation of Differential Expression Analysis Through Quantitative Realtime PCR
Eight differentially expressed olfactory related transcript sequences (Supplementary Table 1) were selected for validation in viruliferous and non-viruliferous whiteflies. Two reference genes namely actin (accession no: KJ913697.1) and 60S ribosomal protein L13a (accession no: XM_019052223.1) were used for the study and were obtained from NCBI database.4 All the primers were designed using Real Time Design qPCR Assay Design Software5 and tested for PCR efficiency. Single strand cDNA was prepared with 1 μg total RNA using the Qiagen Inverse strand kit (Qiagen catalog no. 205310: United States) following the manufacturer’s protocol. Quantitative Realtime PCR (qRT-PCR) analysis was done on the Applied Biosystems Step One Plus machine. Two biological replicates with three technical replicates each were used for the expression analysis. Ct values obtained were exported to Excel and the ddCt method was followed to compare the expression profile. 2^-ddCt value was represented as a histogram against the target gene for the viruliferous and non-viruliferous samples. A t-test was performed to estimate the p-values and Standard Deviation (SD) was plotted as error bars on the histograms.
Statistical Data Analysis
Dual-choice olfactometer bioassays were subjected to repeated measures (mixed model) three-way ANOVA with Tukey’s multiple comparisons, while EAG results analyzed through repeated measures one-way ANOVA. The boxplots and heatmaps constructed here were carried out using GraphPad Prism software (Ver. 9.0) for Windows 10.
Results
A plant virus induces several changes in its host as well as its vector during transmission. In this study the behavioral responses of non-viruliferous as well as viruliferous (ChLCV infected) whiteflies toward healthy and virus infected host plant volatiles was explored and the specific host volatile cues that elicited these behavior variations in B. tabaci were identified. The differential expression of olfactory genes in both viruliferous and non-viruliferous whiteflies was also investigated.
Behavioral Response of Bemisia tabaci to Plant Volatiles
The magnitude of dependency of herbivores on volatile cues to locate their hosts could potentially be exploited by plant viruses to their advantage, resulting in the vectors’ altered host preference that favors virus transmission. To test the orientation of non-viruliferous and viruliferous B. tabaci to ChLCV-infected and healthy plant volatiles, four-arm olfactometer (dual-choice) assays were carried out.
Analyses of data on time spent by test insects using three-way ANOVA revealed significant differences with respect to whitefly status [viruliferous/non-viruliferous; P = 0.05, F(1, 56) = 3.76], whitefly sex [female/male: P = 0.04, F(1, 56) = 4.20] and plant status (healthy/infected) × whitefly status [P < 0.0001, F(1, 56) = 28.38]. Similarly, the data analysis on number of entries made by test insects revealed significant differences with respect to whitefly status [viruliferous/non-viruliferous; P = 0.05, F(1, 56) = 3.84] and plant status (healthy/infected) x whitefly status [P < 0.0001, F(1, 56) = 25.63].
Repeated measures three-way ANOVA revealed statistically significant behavioral interactions among the non-viruliferous and viruliferous B. tabaci. The time spent and the number of entries made by non-viruliferous female B. tabaci was significantly higher in the virus infected plant volatile treated region compared to healthy plant volatile treated region (Figure 1A; P = 0.05 and Figure 1B; P = 0.03). In contrast, viruliferous female B. tabaci spent more time in the healthy plant volatile treated region (Figure 1C; P = 0.02) compared to the infected plant volatile region, however, no significant difference was observed for the number of entries made between both the treated regions (Figure 1D; P = 0.54). Non-viruliferous and viruliferous male whiteflies showed no significant difference in response to infected and healthy plant volatiles with respect to time spent and also the number of entries made (Figures 2A–D, Time, non-viruliferous: P = 0.28; Entries, non-viruliferous: P = 0.06; Time, viruliferous: P = 0.26; Entries, viruliferous: P = 0.64). The interactions revealed that non-viruliferous females were more attracted to infected plant volatiles as they spent significantly more time in treated region compared to their counterparts that were tested against healthy plant volatiles (P = 0.05). Further, these non-viruliferous females exhibited significantly higher attraction to infected plant volatiles as they spent more time (P = 0.001) and made a greater number of entries (P = 0.02) in treated region compared to their viruliferous female counterparts. The non-viruliferous females were significantly attracted toward infected plant volatiles compared to viruliferous males [P = 0.0002 (time spent); P = 0.001 (number of entries made)]. In contrast, viruliferous females were found to be more attractive to healthy plant volatiles when compared to viruliferous males [P = 0.03 (time spent)] when tested against infected plant volatiles. Further, non-viruliferous females were found to be more attractive to infected plant volatiles compared to non-viruliferous males when tested against healthy plant volatiles [P = 0.001 (time spent), P = 0.008 (entries)]. In case of males, non-viruliferous whiteflies made a greater number of entries in to the infected plant volatile region compared to their viruliferous counterparts (P = 0.01).
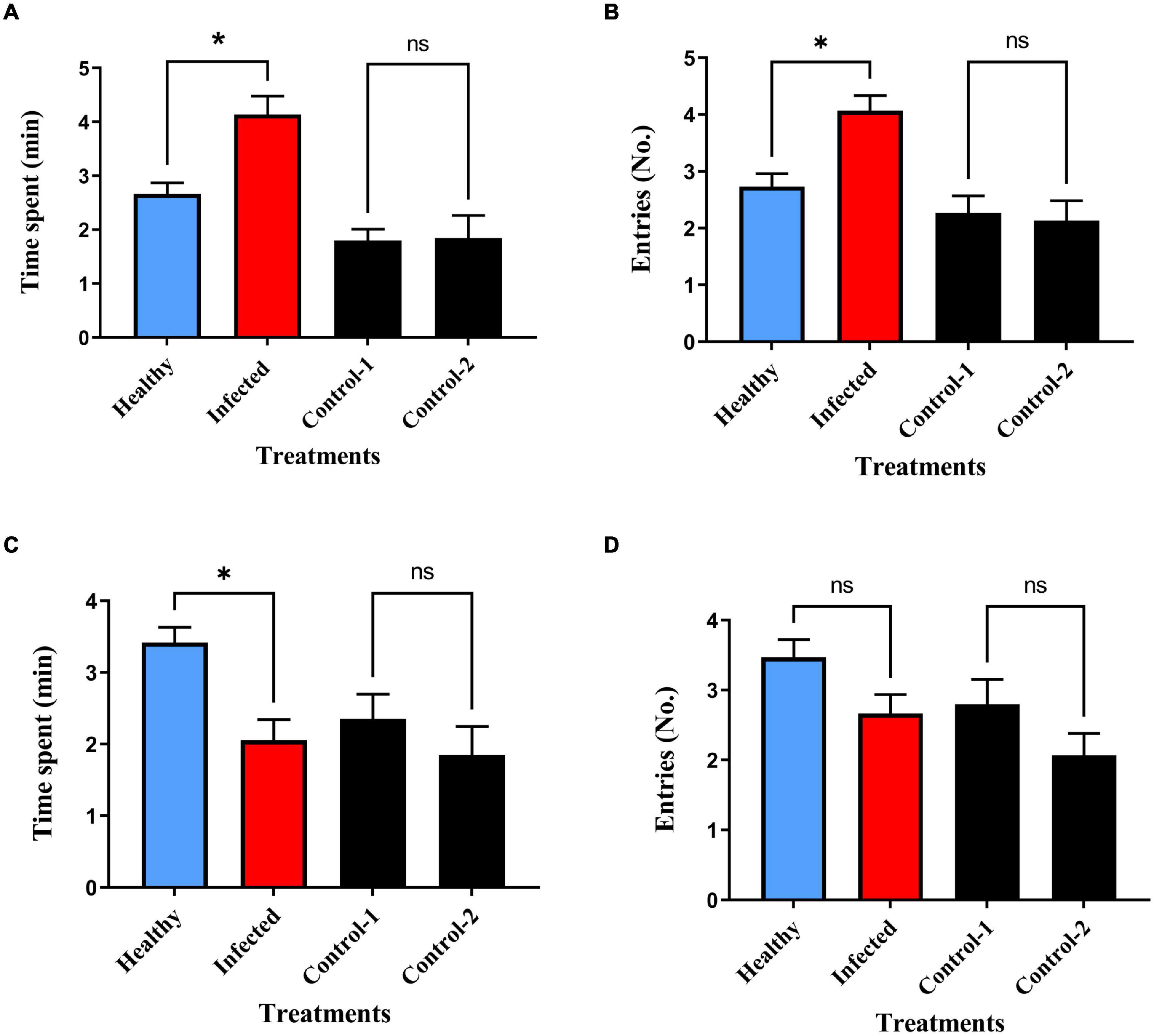
Figure 1. Behavioral response of B. tabaci to chili plant volatiles in olfactometer bioassays (A,B) non-viruliferous females to healthy (blue bars) and ChLCV infected plant (red bars) volatiles, * in (A,B) indicates P = 0.05, 0.03, respectively. (C,D) Viruliferous females to healthy and ChLCV infected plant volatiles, * in (C) indicates P = 0.02. *denotes P ≤ 0.05; ns = non-significant.
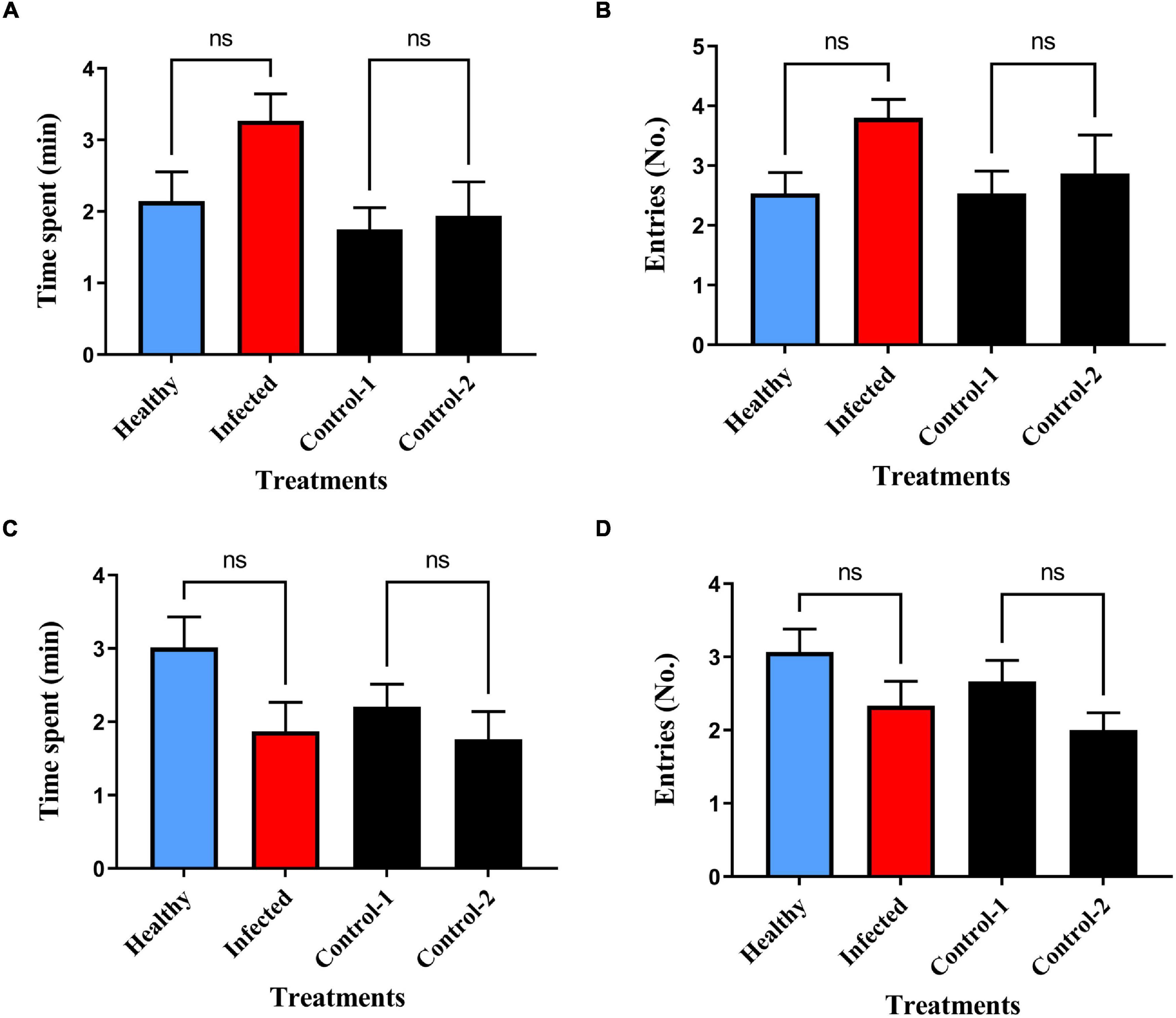
Figure 2. Behavioral response of B. tabaci to chili plant volatiles in olfactometer bioassays (A,B) non-viruliferous males to healthy (blue bars) and ChLCV infected plant (red bars) volatiles. (C,D) Viruliferous males to healthy and ChLCV infected plant volatiles. ns = non-significant.
Antennal Response of Female Non-viruliferous/Viruliferous Whiteflies to Plant Volatiles
While the olfactometer assays measure the overall behavioral response of the insect to host volatile cues, it is important to understand the specific antennal response to healthy as well as ChLCV infected plant volatiles. Electroantennogram (EAG) studies carried out in this regard revealed significant differences in the antennal response of non-viruliferous and viruliferous B. tabaci females to healthy and ChLCV infected plant volatiles (Figure 3). Viruliferous females significantly responded to healthy plant volatiles (Figure 3A; P < 0.0001, healthy plant volatiles, Mean ± SE: 0.74 ± 0.05 mV; infected plant volatiles, Mean ± SE: 0.28 ± 0.01 mV; control, Diethyl ether, Mean ± SE: 0.15 ± 0.02 mV) while, non-viruliferous females showed significant response to infected plant volatiles (Figure 3B; P < 0.0001, healthy plant volatiles, Mean ± SE: 0.35 ± 0.04 mV; infected plant volatiles, Mean ± SE: 0.68 ± 0.03 mV; control, Diethyl ether, Mean ± SE: 0.16 ± 0.02 mV). This study is in congruent with the olfactometer bioassay studies presented earlier.
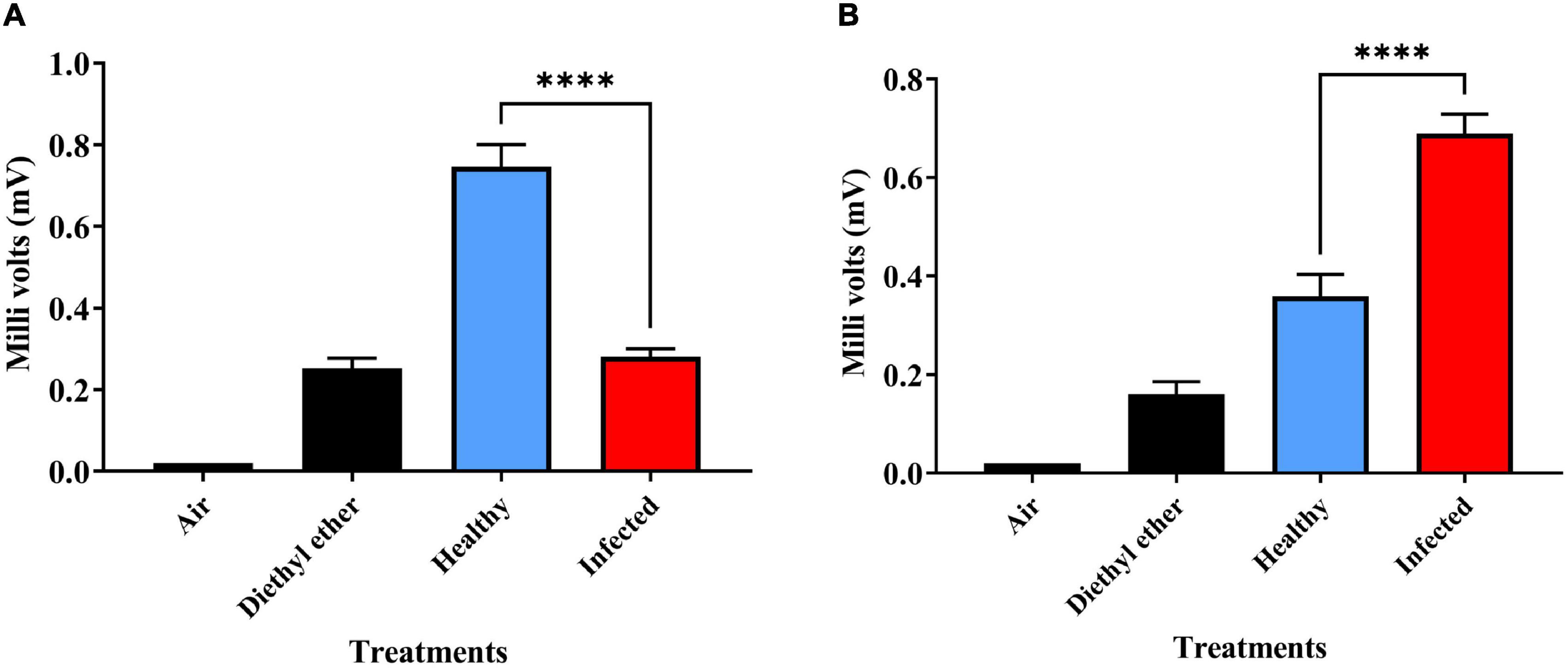
Figure 3. Electroanntenogram response of B. tabaci to healthy and ChLCV infected chili plant volatiles, (A) viruliferous females to healthy (blue bars) and ChLCV infected volatiles (red bars) (B) non-viruliferous females to healthy and ChLCV infected plant volatiles. ****denotes P < 0.0001.
Electrophysiological Response of Non-viruliferous/Viruliferous Whiteflies to Plant Volatiles
Butanoic acid, 2-ethyl-2- methyl-, methyl ester; 2,3,4-trimethylhexane; undecane; 1-octanol,3,7-dimethyl; 2,6,11-trimethyldodecane; methyl eugenol; phytane and heptadecane, 2,6,10,15-tetramethyl were the major EAD active compounds that elicited antennal response in non-viruliferous whiteflies when exposed to healthy plant volatiles. The compounds, dodecane; methyl eugenol; 3, 4, 5-trimethoxy benzaldehyde; isoelemicin and heptadecane, 2,6,10,15-tetramethyl elicited an antennal response in non-viruliferous whiteflies when exposed to infected plant volatiles (Figure 4).
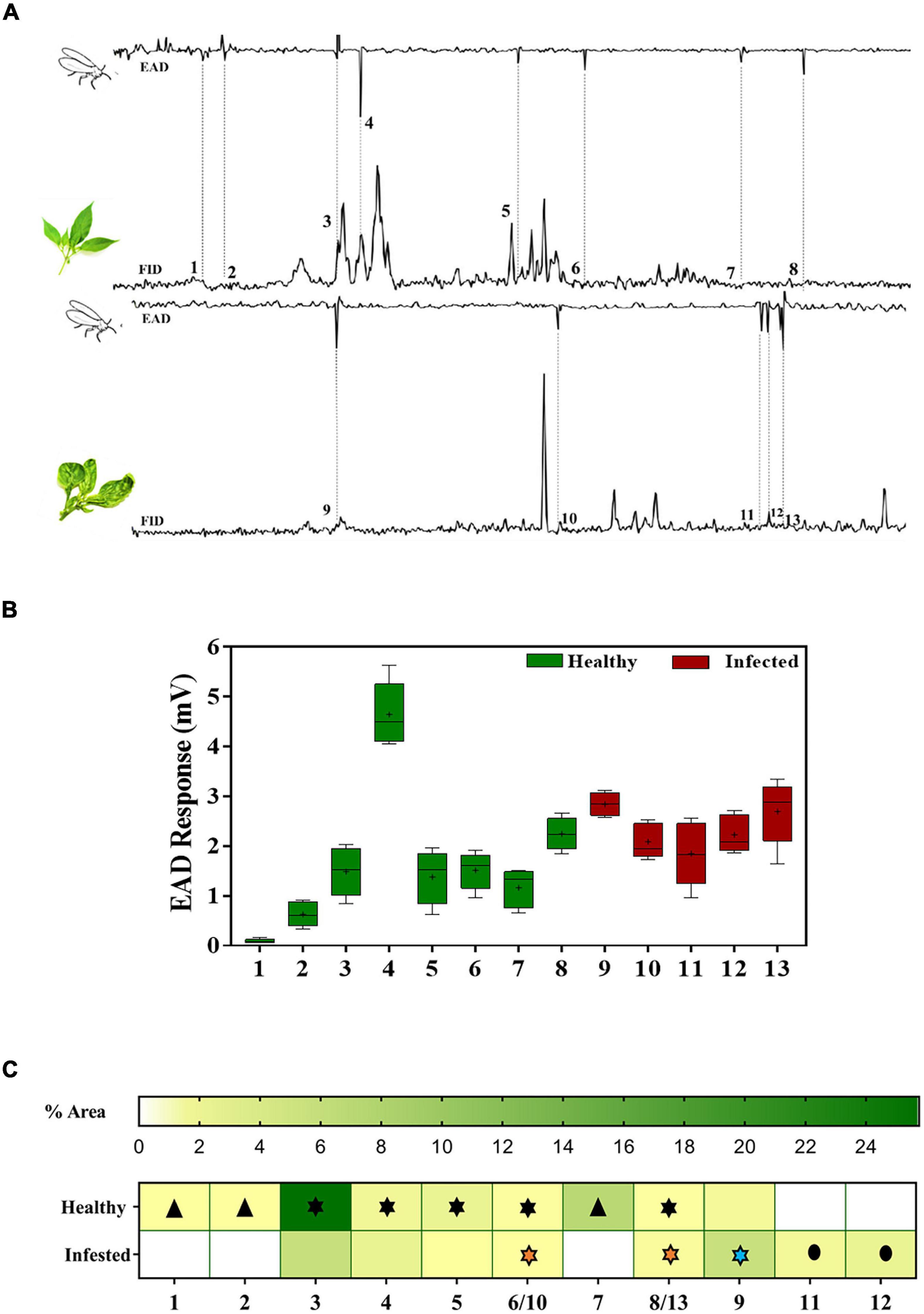
Figure 4. Antennal response of non-viruliferous B. tabaci to healthy and infected chili volatiles. (A) Representative GC-EAD trace showing EAD active peaks. (B) Box and Whisker plot showing the compounds that elicited antennal response. (C) Heat map showing the differences in the volatile emissions of chili plants under different treatments. “ ” – indicates antennal response to compounds exclusively present in healthy plant volatiles; “
” – indicates antennal response to compounds exclusively present in healthy plant volatiles; “ ” – indicates antennal response to compounds (with varied concentrations) from “healthy,” “healthy/infected” and “infected” plant volatiles, respectively; “
” – indicates antennal response to compounds (with varied concentrations) from “healthy,” “healthy/infected” and “infected” plant volatiles, respectively; “ ” – indicates the antennal response to compounds exclusively present in infected plant volatiles. Serial numbers in the x-axis of (A–C) correspond to EAD active chemical compounds and given here [(1) Butanoic acid, 2-ethyl-2- methyl-, methyl ester; (2) 2,3,4 Trimethyl hexane; (3) n-Undecane; (4) 1-octanol-3, 7 dimethyl; (5) 2,6,11 Trimethyl dodecane; (6,10) Methyl eugenol; (7) Phytane; (8,13) Heptadecane 2,6,10,15, tetramethyl; (9) Dodecane; (11) 3,4,5 Trimethoxy benzaldehyde; (12) Isoelemicin].
” – indicates the antennal response to compounds exclusively present in infected plant volatiles. Serial numbers in the x-axis of (A–C) correspond to EAD active chemical compounds and given here [(1) Butanoic acid, 2-ethyl-2- methyl-, methyl ester; (2) 2,3,4 Trimethyl hexane; (3) n-Undecane; (4) 1-octanol-3, 7 dimethyl; (5) 2,6,11 Trimethyl dodecane; (6,10) Methyl eugenol; (7) Phytane; (8,13) Heptadecane 2,6,10,15, tetramethyl; (9) Dodecane; (11) 3,4,5 Trimethoxy benzaldehyde; (12) Isoelemicin].
The chemical cues, 3,4,5-trimethoxy benzaldehyde (1.21%) and isoelemicin (2.03%), present only in ChLCV infected plants elicited an antennal response in non-viruliferous B. tabaci. Compounds like butanoic acid 2-ethyl-2-methyl methyl ester, 2,3,4-trimethylhexane, phytane which elicited a response in non-viruliferous whiteflies (Supplementary Figure 1) were absent in ChLCV infected plant volatiles but present in healthy plant volatiles (0.22, 0.06, and 6.99%, respectively). Undecane, which elicited an antennal response in non-viruliferous whiteflies, was found in higher concentration in healthy plant volatiles (25.75%) but its concentration was reduced in ChLCV infected plants (5.66%). Similar concentration-dependent response was noticed for methyl eugenol (0.34 and 0.13% in healthy and ChLCV infected plant volatiles, respectively). Non-viruliferous whiteflies responded to heptadecane 2, 6, 10, 15-tetramethyl- in both infected (0.64%) and healthy (0.18%) plant volatiles.
The chemical cues, 2,3-butanediol; 2,3-dimethyl, butanoic acid; 2-ethyl-2-methyl- methyl ester; cis-3-hexenyl iso-butyrate; 2-hexyl-1-decanol; 2-methylhexadecane; phytane; heptadecane 2,6,10,15-tetramethyl- and n-Nonadecane were the major EAD active compounds that elicited antennal response in viruliferous whiteflies when exposed to healthy plant volatiles. Volatile compounds such as limonene; 1-octanol 3, 7-dimethyl; ethyl-benzaldehyde; cis-geranyl acetate and 2-Octen-1-ol, 3,7- dimethyl-, isobutyrate, (Z) were the EAD active compounds that elicited antennal response in viruliferous whiteflies when exposed to infected plant volatiles (Figure 5). The chemical cues, 2, 3-butanediol 2, 3-dimethyl (1.19%); butanoic acid 2-ethyl-2- methyl-, methyl ester (0.22%); cis-3-hexenyl iso-butyrate (2.35%); 2-methylhexadecane (0.02%); phytane (6.99%) and n-Nonadecane (0.26%) elicited an antennal response in viruliferous whiteflies and were present only in healthy plants. The compounds, cis-geranyl acetate; 2-Octen-1-ol, 3, 7- dimethyl-, isobutyrate (Z) and limonene, present only in ChLCV infected plant volatiles (1.38, 5.81, and 0.34%, respectively) elicited a response in viruliferous B. tabaci. The isoprenoid alkane, phytane, which elicited an antennal response in viruliferous whiteflies was found in higher concentration in healthy plant volatiles (6.99%) but absent in ChLCV infected plants. Viruliferous whiteflies exhibited concentration-dependent response to 1-octanol 3, 7-dimethyl (1.40% in healthy; 2.18% in infected); 2-hexyl-1-decanol (0.23% in healthy; 1.67% in infected); heptadecane 2, 6, 10, 15-tetramethyl- (0.18 in healthy; 0.64% in infected) and ethyl-benzaldehyde (0.58% in healthy; 0.62% in infected).
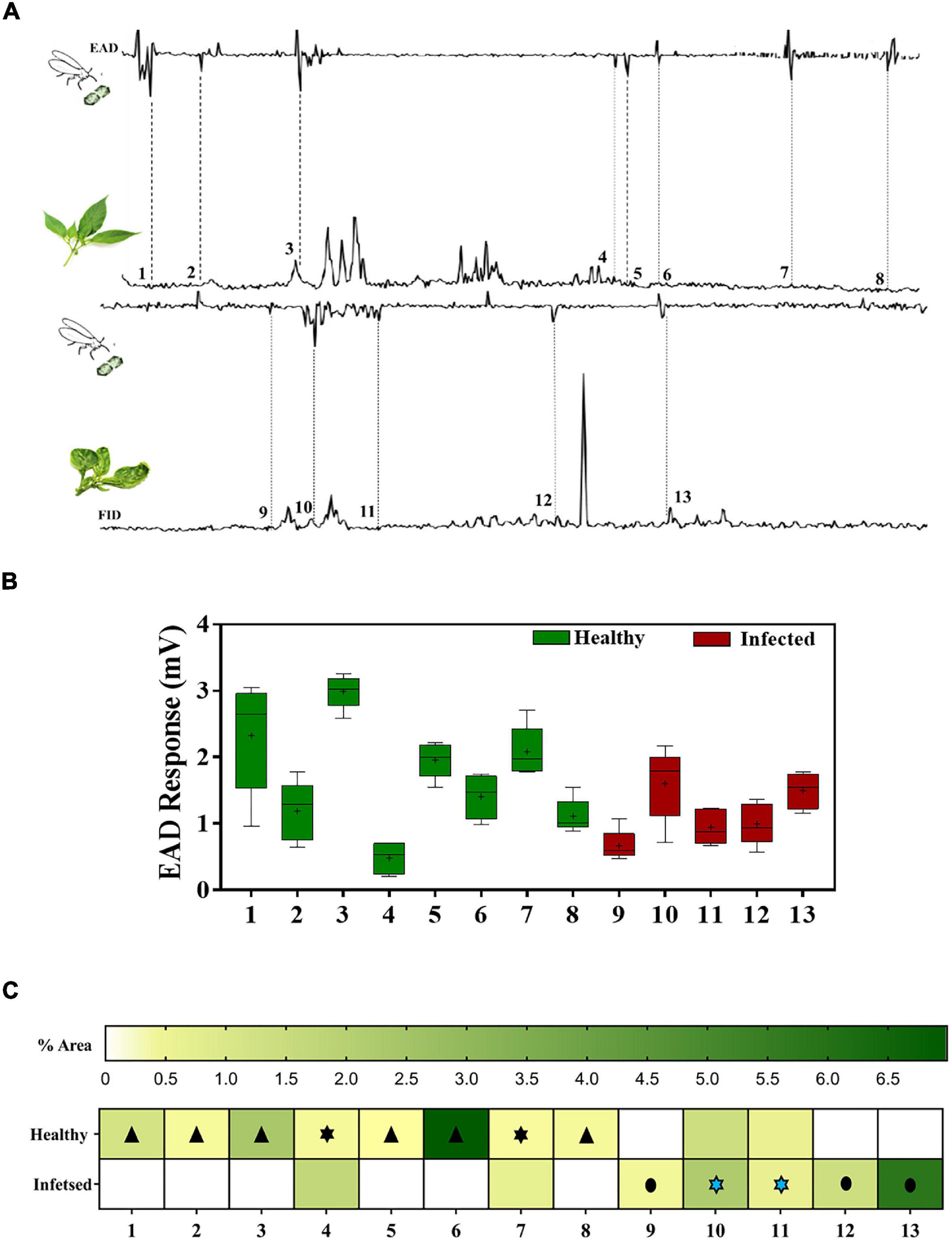
Figure 5. Antennal response of viruliferous B. tabaci to healthy and infected chili volatiles. (A) Representative GC-EAD trace showing EAD active peaks. (B) Box and Whisker plot showing the compounds that elicited antennal response. (C) Heat map showing the differences in the volatile emissions of chili plants under different treatments. “ ” – indicates antennal response to compounds exclusively present in healthy plant volatiles; “
” – indicates antennal response to compounds exclusively present in healthy plant volatiles; “ ” – indicates antennal response to compounds (with varied concentrations) from “healthy” and “infected” plant volatiles, respectively; “
” – indicates antennal response to compounds (with varied concentrations) from “healthy” and “infected” plant volatiles, respectively; “ ” – indicates the antennal response to compounds exclusively present in infected plant volatiles. Serial numbers in the x-axis of (A–C) correspond to EAD active chemical compounds and given here [1 2,3, Butanediol 2,3,dimethyl; 2 Butanoic acid, 2-ethyl-2- methyl-, methyl ester; 3. cis-3-Hexenyl isobutyrate; 4. 2-Hexyl-1-decanol; 5. 2-Methyl hexadecane; 6. Phytane; 7. Heptadecane 2, 6,10,15, tetramethyl; 8. n-Nonadecane; 9. Limonene; 10. 1-octanol- 3,7 dimethyl; 11. Ethyl benzaldehyde; 12. cis-Geranyl acetate; 13. 2 octenol-1-ol 3,7 dimethyl isobutyrate (Z)].
” – indicates the antennal response to compounds exclusively present in infected plant volatiles. Serial numbers in the x-axis of (A–C) correspond to EAD active chemical compounds and given here [1 2,3, Butanediol 2,3,dimethyl; 2 Butanoic acid, 2-ethyl-2- methyl-, methyl ester; 3. cis-3-Hexenyl isobutyrate; 4. 2-Hexyl-1-decanol; 5. 2-Methyl hexadecane; 6. Phytane; 7. Heptadecane 2, 6,10,15, tetramethyl; 8. n-Nonadecane; 9. Limonene; 10. 1-octanol- 3,7 dimethyl; 11. Ethyl benzaldehyde; 12. cis-Geranyl acetate; 13. 2 octenol-1-ol 3,7 dimethyl isobutyrate (Z)].
GC-MS Analysis of Headspace Volatiles From Healthy and Infected Plants
A detailed analysis of the volatiles could potentially help identify specific compounds and hence GC-MS analysis was carried out. The volatile compounds varied significantly between healthy and ChLCV infected plants (Supplementary Figure 1 and Supplementary Table 2). The chemical functional group, acid esters (50.54%) dominated the ChLCV infected plant volatiles; whereas alkanes (88.91%) dominated the healthy plant volatiles. The functional groups namely, acids; esters; alcohols; aromatic hydrocarbons; aldehydes; phenols; phenyl propenes; and sesquiterpenes were overrepresented in ChLCV infected plants compared to healthy plants. Terpenes, terpenoids and other miscellaneous groups could be identified only in the infected plants. Alkanes were reduced in infected plants (25.5%) compared to healthy plants (88.91%). A cyclic monoterpene, limonene was found significantly in infected plants (0.34%), but could not be detected in healthy plant volatiles.
Whitefly Transcriptomics to Identify Olfactory Related Genes That Show Altered Expression
The chemical ecology results presented above indicated that virus induced changes in host plant affect vector behavior. Since attraction to specific host cues is linked to olfaction or chemo-sensation related genes, RNA sequencing data of viruliferous and non-viruliferous whiteflies was explored to study the expression profile of such genes. Assembly and annotation of the transcriptomes led to identification of one hundred and thirty OBPs, ORs, and related proteins. They were further classified into receptors and binding proteins. Some of these were shown to be differentially expressed in the viruliferous and the non-viruliferous whiteflies. A few of the differentially expressed transcripts were odorant receptor co-receptor (B. tabaci), protein D3-like isoform X2 (B. tabaci), protein D1-like (B. tabaci), protein D3-like isoform X3 (B. tabaci), putative odorant-binding protein A5 isoform X1 (B. tabaci), odorant binding protein 5, partial (B. tabaci), putative odorant-binding protein A5 (B. tabaci), putative odorant-binding protein A10 (B. tabaci), ejaculatory bulb-specific protein 3-like (B. tabaci), protein D2-like (B. tabaci), protein D3-like isoform X1 (B. tabaci), chemosensory protein 12, partial (B. tabaci), OBP1 (B. tabaci), odorant binding protein 2, partial (B. tabaci), neuropeptide Y receptor type 6-like (B. tabaci) and odorant-binding protein 8 (B. tabaci).
Realtime Expression Analysis
As observed in the Transcriptome profile, some of the olfactory related genes that were differentially expressed were chosen for validation based on two criteria. First, the genes should be involved in the olfaction related chemosensory functions and second, the extent to which these genes were differentially expressed. Realtime analysis of eight putative candidates that were mis-expressed based on the Transcriptome data provided interesting cues. While expression of Fragile X (FragX), neuropeptide Y receptor type 6-like (NeuroB), Odorant binding protein 1 (OBP1) and Odorant binding protein 8 (OBP8) transcripts was not altered (Figure 6A), mRNA levels of Chemosensory protein1 (CSP1), Odorant binding protein 5 (OBP5), phosphatidylethanolamine-binding protein 1-like (PHOSB1) and Protein D1 like (Prod1), were upregulated in viruliferous whitefly (Figure 6B). The above results provide interesting information on how the acquisition of chili leaf curl virus could affect olfactory gene expression suggesting transcriptional modulation in viruliferous whiteflies.
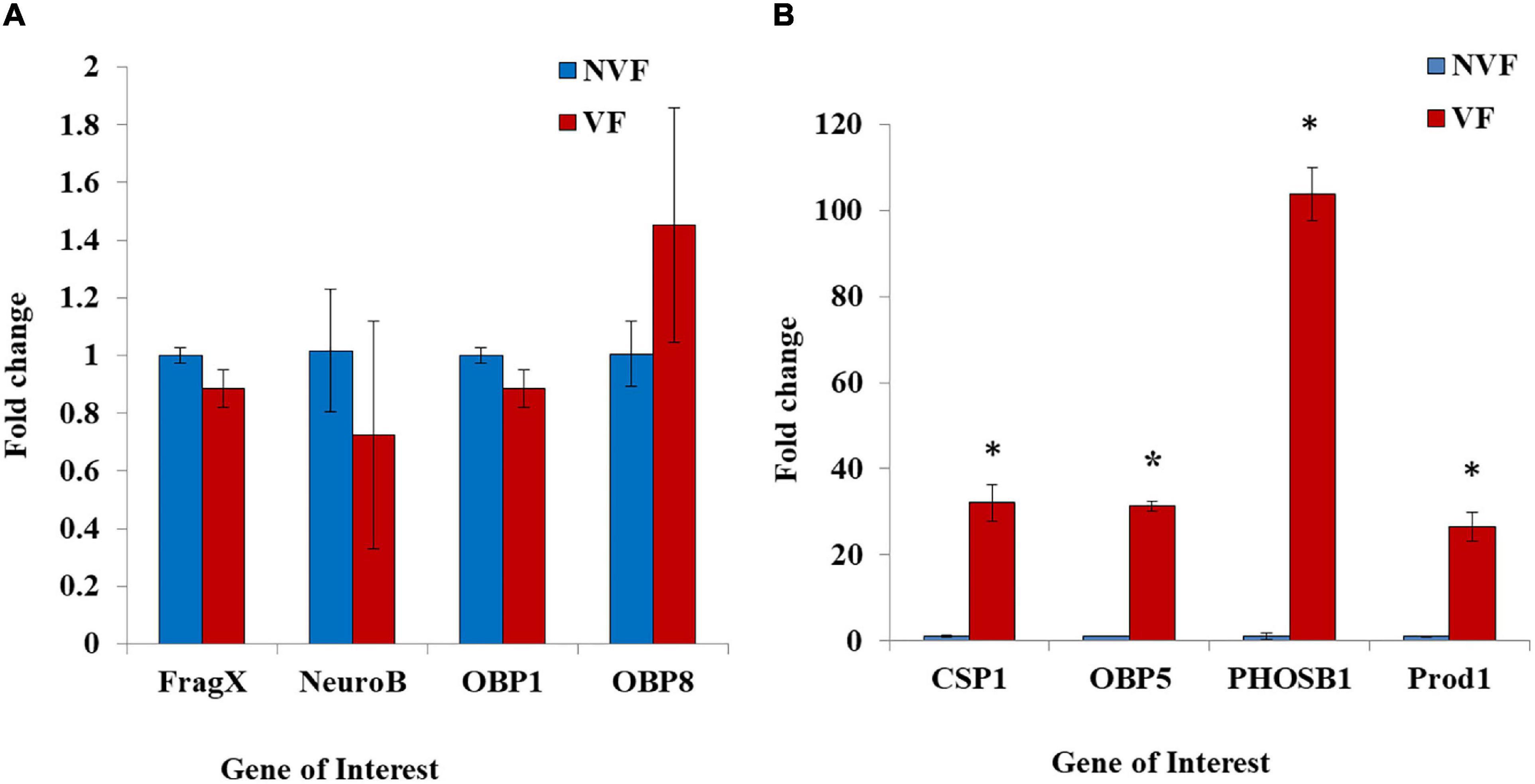
Figure 6. Expression analysis of olfactory related proteins: Realtime expression analysis of OBPs was done for eight OBP related genes. Each experiment had two biological replicates with three technical replicates each. Actin and ribosomal protein 13L were used as reference genes for normalization of transcripts from viruliferous (VF- represented as red bars) and non-viruliferous (NVF- represented as blue bars) whiteflies. The expression was validated for FragileX mental retardation related protein 1 (FragX), neuropeptide Y receptor type B (NeuroB), Odorant binding protein1 (OBP1), Odorant binding protein 8 (OBP8) in (A) and Chemosensory protein 1 (CSP1), Odorant binding protein5 (OBP5), Phosphotydylethalonamine- binding protein 1 like (PhosB1), and protein D1 like (prod1) in (B). The genes in (B) were found to be upregulated in the viruliferous whiteflies. Significance of the data was estimated by an unpaired t-test. The standard deviations (SD) were used to plot the error bars. *denotes P < 0.05.
Discussion
Previous studies have indicated that viruses induce direct and indirect effects on their vectors after acquisition leading to enhanced transmission efficiency and their spread (Moreno-Delafuente et al., 2013). The results presented in this study highlight virus (ChLCV) induced behavioral changes in the vector, B. tabaci where the viruliferous and non-viruliferous female B. tabaci clearly made their choice by orienting toward- healthy and ChLCV infected chili plant volatiles, respectively. Antennal response data from the EAG studies further corroborated this. Virus induced behavioral changes have been reported in aphids (Ingwell et al., 2012), thrips (Stafford et al., 2011), and whiteflies (Moreno-Delafuente et al., 2013). However, these studies were mainly exploratory and do not provide an explanation for virus induced changes in the vector. The Tomato yellow leaf curl virus (TYLCV) infected B. tabaci differ from their non-infected counterparts in settling, probing and feeding behaviors (Moreno-Delafuente et al., 2013). A study conducted by Fereres et al. (2016) reported various contrast changes induced in host plant volatile profiles and vector behavior when tomato plants were infected by circulative and non-circulative whitefly-transmitted viruses.
Behavioral and electrophysiological studies conducted, revealed that varied compounds elicited antennal response in viruliferous and non-viruliferous B. tabaci when exposed to healthy and ChLCV infected host plant volatiles. The volatile composition of these plants also differed significantly. This emphasizes that the presence/absence of certain chemical cues could possibly play a role in attracting viruliferous and non-viruliferous whiteflies toward healthy or ChLCV infected plants. The variations observed in the abundance of certain chemical cues like n-undecane, methyl eugenol and heptadecane 2, 6, 10, 15-tetramethyl in healthy and ChLCV infected plants might have potentially caused the altered behavioral response in B. tabaci. Our results are in concurrence with earlier reports which suggested that high undecane concentrations in tomato cultivars attract B. tabaci adults resulting in higher oviposition and nymphal density (Fernandes et al., 2011). Presence of limonene, a reported putative attractant (Sadeh et al., 2017) for B. tabaci in ChLCV infected plants but its absence in healthy plant volatiles indicates a systematic manipulation of host plant volatiles. Changes in plant defense chemistry have been reported to affect subsequent virus transmission and disease severity (Carmo-Sousa et al., 2016; Mauck, 2016; Mauck et al., 2019). Some Gemini viruses affect the host sumoylation and ubiquitination pathways such that, the host signaling machinery is modulated leading to changes in protein modifications. Gemini viruses also use the host machinery to their benefit and may starve the host of essential proteins that can bring about biochemical changes in the plants (Ascencio-Ibáñez et al., 2008; Hanley-Bowdoin et al., 2013; Pierce and Rey, 2013). It is worth noting that the plant virus induced behavior manipulations were consistently exhibited only by female B. tabaci suggesting a possible sex-biased preference of host cues. Several factors come into play in the herbivore decision-making process with respect to host plant preference and acceptability. However, the progeny performance and host plant defense response decide the subsequent oviposition attempts by herbivores (Singer, 1986). A greater fitness cost associated with females in terms of enhanced fecundity, survival and population increase (Jiu et al., 2007; Ingwell et al., 2012; Moreno-Delafuente et al., 2013) could provide an explanation to the above results. Studies by Fernandes et al. (2011) established that visual cues are more important than olfactory stimuli for host location in B. tabaci. Our study cannot corroborate this finding as the dual-choice bioassays were done using volatiles of the ChLCV infected and healthy chili plants in an experimental arena avoiding the interference of any external visual stimuli and B. tabaci would still make a choice.
Volatile cue recognition depends on an array of olfactory related genes like OBPs, receptors and neuropeptides which might be involved in processing of signals. Transcriptome and Real time data revealed differential expression of some such genes as observed previously in behavioral studies of other insects. Interestingly, Chemosensory protein1 (CSP1), Odorant binding protein 5 (OBP5), phosphatidylethanolamine-binding protein 1-like (PHOSB1), and Protein D1 like (Prod1), were upregulated in ChLCV infected whiteflies suggesting their possible role in B. tabaci’s altered volatile detection. Olfactory related gene changes possibly affecting vector behavior have been reported in a few other insect-systems. OBP2 has been reported to be responsible for the preference of Sogatella furcifera to southern rice black-streaked dwarf virus infected rice plants (Hu et al., 2019). Therefore OBP5, which showed altered expression in viruliferous whiteflies might have a similar role. Similar results for certain odor binding proteins like pherokine 2 and 3 have been reported in Drosophila infected with virus and bacteria (Sabatier et al., 2003). CSP1 is a known olfactory related protein with diverse functions (Pelosi et al., 2018) and hence it is not surprising that its expression is changed under viruliferous conditions. Phosphatidylethanolamine-binding protein 1-like protein which had altered expression in viruliferous whiteflies is isolated from antennal proteins of other insects (Justice et al., 2003). These proteins have an immune response role (Reumer et al., 2009) and have been speculated to bind hydrophobic ligands (Justice et al., 2003) which could potentially be receptors for volatiles. The pathways which trigger these changes in gene expression is not clear. A virus might mediate transcription regulation by affecting the regulatory mechanisms like DNA methylation. Decrease or increase in DNA methylation could switch genes on or off. Some reports have suggested increase in methylation of DNA-by-DNA 5 cytosine methyltransferase (Mikovits et al., 1998). Others have observed down-regulation of 5 cytosine DNA methyltransferase in virus infected whitefly (Li et al., 2011). Changes in regulation of mRNA degradation might result in accumulation of mRNA levels for certain genes (Steckelberg et al., 2018).
Most plant viruses show fulfillment of the two criteria needed to be classified as manipulators of vector behavior, (a) increased host palatability for persistent transmission and decreased palatability and vector performance for non-persistent transmission, (b) Molecular interaction between virus and host or vector proteins indicating a genetic aspect to this manipulation (Mauck et al., 2019). Our results clearly show that as a whitefly vectored, persistent virus, ChLCV is able to indirectly modulate the non-viruliferous insects to feed on infected plants and make viruliferous insects feed on healthy plants, therefore, increasing transmission efficiency. The gene expression data also provides evidence for a possible molecular interaction. While the current findings suggest that volatiles are important to herbivores for host recognition, it does not negate the possibility of their dependence on visual and chemosensory cues after landing on the host, for acceptance (Alyokhin et al., 2000). This report opens up huge knowledge gaps in the field of virus-vector-host plant interaction for further research and at the same time also indicates that virus resistant cultivar breeding needs to be focused on variations in plant volatile emissions as they can critically influence herbivore choice (Fernandes et al., 2011). While a deeper understanding of the molecular effectors and their functional aspects is need of the hour, based on the results in this report, we propose a model for the virus mediated vector and host modulation (Figure 7). The semiochemical perception of host volatiles guides an insect pest to it, when a foreign or pathogenic entity like a virus (ChLCV) enters a host, it potentially has the ability to alter the host plant volatile profile thereby affecting the non-viruliferous vector host orientation response resulting in an event promoting virus transmission by the host to a healthy vector. This modulation could be mediated by several molecular effectors that come into effect when a host is infected. Vice versa, when a viruliferous vector carries the virus, its olfactory senses are altered by the virus possibly mediated by interactions with vector proteins that result is altered expression of odorant binding and receptor proteins. This molecular interaction potentially results in a favorable event for virus transmission from the viruliferous vector to a healthy host plant. The two aspects together could potentially ensure a continuous cycle for successful virus transmission. Keeping this model as a base trophic level interactions among the virus, host plant and vector need to be explored further to reckon sustainable vector and disease management strategies.
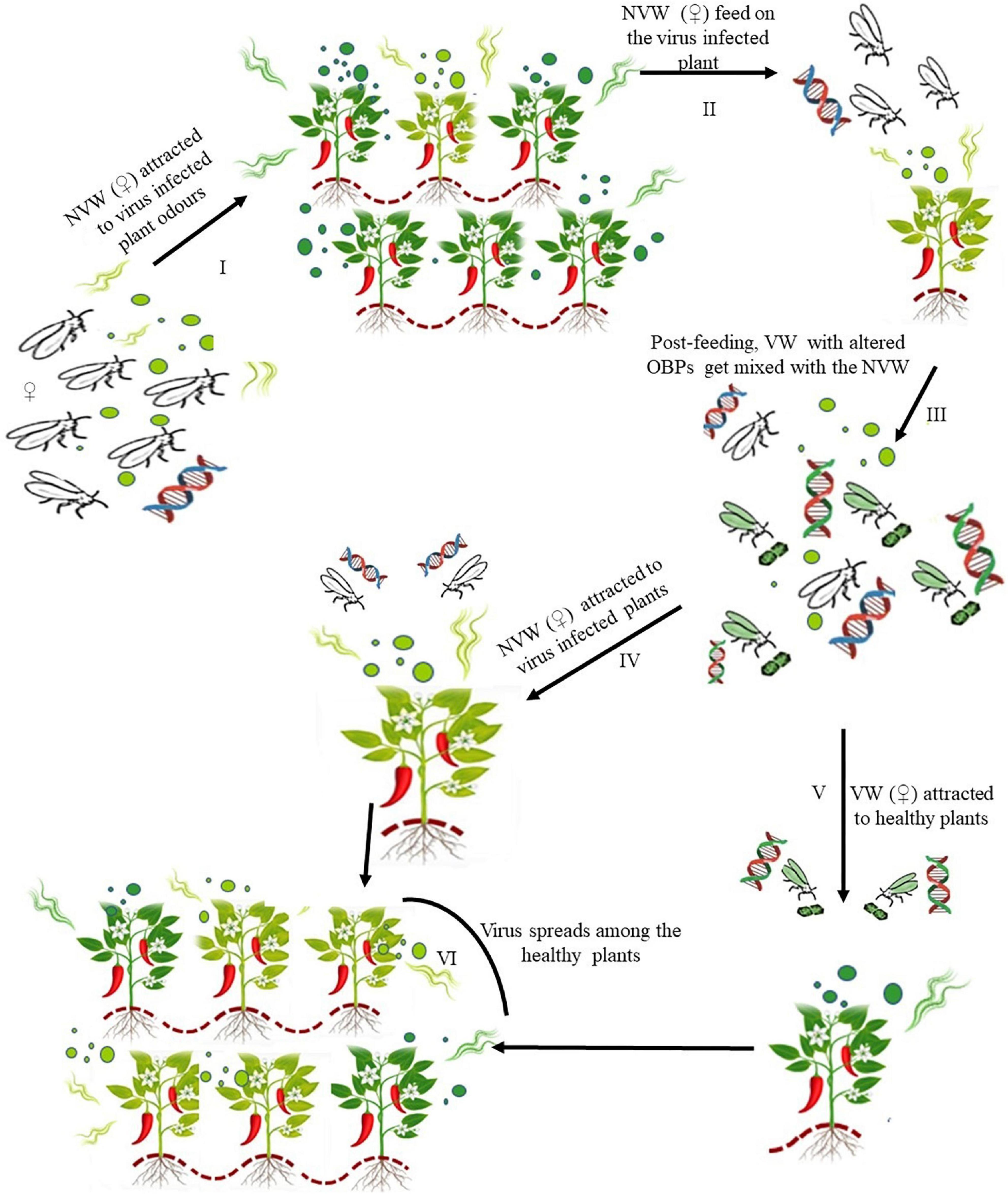
Figure 7. Hypothetical representation showing how ChLCV spreads among the plants by manipulating virus infected plant odors and influencing the host location behavior of viruliferous (VW) and non-viruliferous (NVW) whiteflies.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: www.ncbi.nlm.nih. gov/sra/, PRJNA795317.
Author Contributions
RY performed the experiments, analyzed the data, and drafted the manuscript. PDKJ designed the experiments, provided resources and funding, interpretation of data, drafted, and edited the manuscript. SP performed the experiments, analysis, and interpretation of data and edited the manuscript. MV designed the experiments, performed the experiments, analysis, and interpretation of data, drafted, and edited manuscript. MK and KM provided resources and funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Fellow Project—Studies on Phyto-Semiochemicals involved in insect-plant interactions of major horticulture pests: Deciphering the chemical cues (Award no. 27(3)/2010-HRD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Director, Indian Institute of Horticultural Research, Bangalore for providing research facilities. RY acknowledges the University Grant Commission (India), New Delhi for providing financial support. Financial support from National Fellow Project, Indian Council of Agricultural Research, New Delhi is duly acknowledged. We thank immensely the reviewers whose constructive suggestions helped us to improve the manuscript tremendously.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.819023/full#supplementary-material
Footnotes
- ^ http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- ^ http://transdecoder.sf.net
- ^ https://genomebiology.biomedcentral.com/articles/10.1186/s13059-014-0550-8
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.biosearchtech.com/support/tools/design-software/realtimedesign-software
References
Altschup, S. F., Gish, W., Pennsylvania, T., and Park, U. (1990). Basic local alignment search tool. Dep. Comput. Sci. 2, 403–410.
Alyokhin, A. V., Messing, R. H., and Duan, J. J. (2000). Visual and olfactory stimuli and fruit maturity affect trap captures of oriental fruit flies (Diptera: Tephritidae). J. Econ. Entomol. 93, 644–649. doi: 10.1603/0022-0493-93.3.644
Ascencio-Ibáñez, J. T., Sozzani, R., Lee, T. J., Chu, T. M., Wolfinger, R. D., Cella, R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. doi: 10.1104/pp.108.121038
Bruce, T. J. A., Wadhams, L. J., and Woodcock, C. M. (2005). Insect host location: a volatile situation. Trends Plant Sci. 10, 269–274. doi: 10.1016/j.tplants.2005.04.003
Buchfink, B., Xie, C., and Huson, D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Carmo-Sousa, M., Moreno, A., Plaza, M., Garzo, E., and Fereres, A. (2016). Cucurbit aphid-borne yellows virus (CABYV) modifies the alighting, settling and probing behaviour of its vector Aphis gossypii favouring its own spread. Ann. Appl. Biol. 169, 284–297. doi: 10.1111/aab.12300
Chen, S., Huang, T., Zhou, Y., Han, Y., Xu, M., and Gu, J. (2017). AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics 18:80. doi: 10.1186/s12859-017-1469-3
De Barro, P. J., Liu, S. S., Boykin, L. M., and Dinsdale, A. B. (2011). Bemisia tabaci: a statement of species status. Annu. Rev. Entomol. 56, 1–19. doi: 10.1146/annurev-ento-112408-085504
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fereres, A., and Moreno, A. (2009). Behavioural aspects influencing plant virus transmission by homopteran insects. Virus research, 141, 158–168.
Fereres, A., Peñaflor, M. F. G. V., Favaro, C. F., Azevedo, K. E. X., Landi, C. H., Maluta, N. K. P., et al. (2016). Tomato infection by whitefly-transmitted circulative and non-circulative viruses induce contrasting changes in plant volatiles and vector behaviour. Viruses 8:225. doi: 10.3390/v8080225
Fernandes, M. E., de, S., da Silva, D. J. H., Picanço, M. C., Fernandes, F. L., Jham, G. N., et al. (2011). Resistance of tomato subsamples to Bemisia tabaci biotype B (Genn.) (Hemiptera: Aleyrodidae). Agron. J. 103, 1849–1861. doi: 10.2134/agronj2011.0049
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hanley-Bowdoin, L., Bejarano, E. R., Robertson, D., and Mansoor, S. (2013). Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. doi: 10.1038/nrmicro3117
Hu, K., Yang, H., Liu, S., He, H., Ding, W., Qiu, L., et al. (2019). Odorant-binding protein 2 is involved in the preference of Sogatella furcifera (Hemiptera: Delphacidae) for rice plants infected with the southern rice black-streaked dwarf virus. Fla. Entomol. 102, 353–358. doi: 10.1653/024.102.0210
Ingwell, L. L., Eigenbrode, S. D., and Bosque-Pérez, N. A. (2012). Plant viruses alter insect behavior to enhance their spread. Sci. Rep. 2:578. doi: 10.1038/srep00578
Jiu, M., Zhou, X. P., Tong, L., Xu, J., Yang, X., Wan, F. H., et al. (2007). Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS One 2:e182. doi: 10.1371/journal.pone.0000182
Justice, R. W., Dimitratos, S., Walter, M. F., Woods, D. F., and Biessmann, H. (2003). Sexual dimorphic expression of putative antennal carrier protein genes in the malaria vector Anopheles gambiae. Insect Mol. Biol. 12, 581–594. doi: 10.1046/j.1365-2583.2003.00443.x
Kamala Jayanthi, P. D., Aurade, R. M., Kempraj, V., Roy, T. K., Shivashankara, K. S., and Verghese, A. (2015). Salicylic acid induces changes in mango fruit that affect oviposition behavior and development of the oriental fruit fly, Bactrocera dorsalis. PLoS One 10:e0139124. doi: 10.1371/journal.pone.0139124
Kamala Jayanthi, P. D., Kempraj, V., Aurade, R. M., Venkataramanappa, R. K., Nandagopal, B., Verghese, A., et al. (2014). Oviposition site-selection by Bactrocera dorsalis is mediated through an innate recognition template tuned to γ-octalactone. PLoS One 9:e85764. doi: 10.1371/journal.pone.0085764
Kamala Jayanthi, P. D., Woodcock, C. M., Caulfield, J., Birkett, M. A., and Bruce, T. J. A. (2012). Isolation and identification of host cues from mango, Mangifera indica, that attract gravid female oriental fruit fly, Bactrocera dorsalis. J. Chem. Ecol. 38, 361–369. doi: 10.1007/s10886-012-0093-y
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kopylova, E., Noé, L., and Touzet, H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Kovats, E. (1965). Gas chromatographic characterization of organic substances in the retention index system. Adv. Chromatogr. 1, 229–247.
Krueger, F. (2015). Trim Galore! Available Online at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed May 08, 2019).
Lazarowitz, S. G. and Shepherd, R. J. (1992). Gemini viruses: genome structure and gene function. Crit. Rev. Plant Sci. 11, 327–349. doi: 10.1080/07352689209382350
Li, J. M., Ruan, Y. M., Li, F. F., Liu, S. S., and Wang, X. W. (2011). Gene expression profiling of the whitefly (Bemisia tabaci) Middle East - Asia Minor 1 feeding on healthy and Tomato yellow leaf curl China virus-infected tobacco. Insect Sci. 18, 11–22. doi: 10.1111/j.1744-7917.2010.01386.x
Liao, Y., Smyth, G. K., and Shi, W. (2014). Feature Counts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Mauck, K., Bosque-Pérez, N. A., Eigenbrode, S. D., De Moraes, C. M., and Mescher, M. C. (2012). Transmission mechanisms shape pathogen effects on host-vector interactions: evidence from plant viruses. Funct. Ecol. 26, 1162–1175. doi: 10.1111/j.1365-2435.2012.02026.x
Mauck, K. E. (2016). Variation in virus effects on host plant phenotypes and insect vector behavior: what can it teach us about virus evolution? Curr. Opin. Virol. 21, 114–123. doi: 10.1016/j.coviro.2016.09.002
Mauck, K. E., Kenney, J., and Chesnais, Q. (2019). Progress and challenges in identifying molecular mechanisms underlying host and vector manipulation by plant viruses. Curr. Opin. Insect Sci. 33, 7–18. doi: 10.1016/j.cois.2019.01.001
Mikovits, J. A., Young, H. A., Vertino, P., Issa, J.-P. J., Pitha, P. M., Turcoski-Corrales, S., et al. (1998). Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in De Novo methylation of the gamma interferon (IFN-γ) promoter and subsequent downregulation of IFN-γ production. Mol. Cell. Biol. 18, 5166–5177. doi: 10.1128/mcb.18.9.5166
Moreno-Delafuente, A., Garzo, E., Moreno, A., and Fereres, A. (2013). A plant virus manipulates the behavior of its whitefly vector to enhance its transmission efficiency and spread. PLoS One 8:e61543. doi: 10.1371/journal.pone.0061543
Navas-Castillo, J., Fiallo-Olivé, E., and Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Pelosi, P., Iovinella, I., Zhu, J., Wang, G., and Dani, F. R. (2018). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 93, 184–200. doi: 10.1111/brv.12339
Pierce, E. J., and Rey, M. E. C. (2013). Assessing global transcriptome changes in response to South African cassava mosaic virus [ZA-99] infection in susceptible Arabidopsis thaliana. PLoS One 8:e67534. doi: 10.1371/journal.pone.0067534
Reumer, A., Bogaerts, A., Van Loy, T., Husson, S. J., Temmerman, L., Choi, C., et al. (2009). Unraveling the protective effect of a Drosophila phosphatidylethanolamine-binding protein upon bacterial infection by means of proteomics. Dev. Comp. Immunol. 33, 1186–1195. doi: 10.1016/j.dci.2009.06.010
Sabatier, L., Jouanguy, E., Dostert, C., Zachary, D., Dimarcq, J. L., Bulet, P., et al. (2003). Pherokine-2 and -3: two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biochem. 270, 3398–3407. doi: 10.1046/j.1432-1033.2003.03725.x
Sadeh, D., Nitzan, N., Shachter, A., Chaimovitsh, D., Dudai, N., and Ghanim, M. (2017). Whitefly attraction to rosemary (Rosmarinus officinialis L.) is associated with volatile composition and quantity. PLoS One 12:e0177483. doi: 10.1371/journal.pone.0177483
Shivaramu, S., Jayanthi, P. D. K., Kempraj, V., Anjinappa, R., Nandagopal, B., and Chakravarty, A. K. (2017). What signals do herbivore-induced plant volatiles provide conspecific herbivores? Arthropod Plant Interact. 11, 815–823. doi: 10.1007/s11829-017-9536-2
Singer, M. C. (1986). “The definition and measurement of oviposition preference in plant-feeding insects,” in Insect-Plant Interactions. Springer Series in Experimental Entomology, eds J. R. Miller and T. A. Miller (New York, NY: Springer), 65–94. doi: 10.1007/978-1-4612-4910-8_3
Stafford, C. A., Walker, G. P., and Ullman, D. E. (2011). Infection with a plant virus modifies vector feeding behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 9350–9355. doi: 10.1073/pnas.1100773108
Stafford-Banks, C. A., Yang, L. H., Mcmunn, M. S., and Ullman, D. E. (2014). Virus infection alters the predatory behavior of an omnivorous vector. Oikos 123, 1384–1390. doi: 10.1111/oik.01148
Steckelberg, A. L., Akiyama, B. M., Costantino, D. A., Sit, T. L., Nix, J. C., and Kieft, J. S. (2018). A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure. Proc. Natl. Acad. Sci. U.S.A. 115, 6404–6409. doi: 10.1073/pnas.1802429115
Keywords: Chili, ChLCV, Bemisia tabaci, behavioral bioassays, host cues, OBPs, GC-EAD, virus-vector
Citation: Yadav RK, Kambham MR, Parepally SK, Vyas M, Manem KR and Kamala Jayanthi PD (2022) Encounter With a Selfish Virus Sabotages Its Vector to Orient Toward Requisite Host Plant: A Case Study With Chili Leaf Curl Virus-Whitefly. Front. Ecol. Evol. 10:819023. doi: 10.3389/fevo.2022.819023
Received: 20 November 2021; Accepted: 10 March 2022;
Published: 06 April 2022.
Edited by:
Michael Wink, Heidelberg University, GermanyReviewed by:
Lukasz Lech Stelinski, University of Florida, United StatesHong Lei, Arizona State University, United States
Copyright © 2022 Yadav, Kambham, Parepally, Vyas, Manem and Kamala Jayanthi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Madhavi Reddy Kambham, TWFkaGF2aVJlZGR5LktAaWNhci5nb3YuaW4=, a21yZWRkeTE0QGdtYWlsLmNvbQ==; Krishna Reddy Manem, bWtyZWRkeTYwQGdtYWlsLmNvbQ==, S3Jpc2hhbmFSZWRkeS5NQGljYXIuZ292Lmlu; Pagadala Damodaram Kamala Jayanthi, amFpaW5zZWN0QGdtYWlsLmNvbQ==, S2FtYWxhSmF5YW50aGkuUERAaWNhci5nb3YuaW4=
 Rajeev Kumar Yadav
Rajeev Kumar Yadav Madhavi Reddy Kambham
Madhavi Reddy Kambham Saravan Kumar Parepally
Saravan Kumar Parepally Meenal Vyas
Meenal Vyas Krishna Reddy Manem
Krishna Reddy Manem Pagadala Damodaram Kamala Jayanthi
Pagadala Damodaram Kamala Jayanthi