- 1Environmental Microbial Genomics Group, École Centrale de Lyon, Université de Lyon, Écully, France
- 2Université Grenoble Alpes, CNRS, IRD, Grenoble INP, IGE UMR 5001, HyDRIMZ Group, Grenoble, France
Introduction: Micro-scale analysis of microbes in soil is essential to the overall understanding of microbial organization, interactions, and ecosystem functioning. Soil fractionation according to its aggregated structure has been used to access microbial habitats. While bacterial communities have been extensively described, little is known about the fungal communities at scales relevant to microbial interactions.
Methods: We applied a gentle soil fractionation method to preserve stable aggregated structures within the range of micro-aggregates and studied fungal and bacterial communities as well as nitrogen cycling potentials in the pristine Rothamsted Park Grass soil (bulk soil) as well as in its particle size fractions (PSFs; >250 μm, 250–63 μm, 63–20 μm, 20–2 μm, <2 μm, and supernatant).
Results: Overall bacterial and fungal community structures changed in PSFs below 20 μm. The relative abundance of Basidiomycota decreased with decreasing particle size over the entire measure range, while Ascomycota showed an increase and Mucoromycota became more prominent in particles below 20 μm. Bacterial diversity was found highest in the < 2 μm fraction, but only a few taxa were washed-off during the procedure and found in supernatant samples. These taxa have been associated with exopolysaccharide production and biofilm formation (e.g., Pseudomonas, Massilia, Mucilaginibacter, Edaphobaculum, Duganella, Janthinobacterium, and Variovorax). The potential for nitrogen reduction was found elevated in bigger aggregates.
Discussion: The observed changes below 20 μm particle are in line with scales where microbes operate and interact, highlighting the potential to focus on little researched sub-fractions of micro-aggregates. The applied method shows potential for use in studies focusing on the role of microbial biofilms in soil and might also be adapted to research various other soil microbial functions. Technical advances in combination with micro-sampling methods in soil promise valuable output in soil studies when particles below 20 μm are included.
1 Introduction
Soil harbors the highest microbial densities and diversity relative to other habitats and thus represents an important environment with a global impact on biochemical mass balances (Bardgett and Van Der Putten, 2014). The complexity and compartmentalization of soil at the microscopic scale makes it one of the most challenging microbial habitats to study (Young and Crawford, 2004). This compartmentalization, or soil structure, itself defined by single connected particles or so called aggregates, affects basic physical and mechanical properties such as water retention and movement, erodibility, and aeration and thus ultimately soil fertility and productivity (Rabot et al., 2018). The aggregated micro-structure of soil, in combination with the fluctuating water saturation of the available soil pore space, creates a mosaic of microenvironments varying in chemical reactivity (Spadini et al., 2018) or in respiration processes (different electron acceptors). This, in turn, can generate a range of microhabitats in which specific reactions within the N-cycle might be favored. While reductive processes (denitrification) might be linked to inner parts of bigger aggregates where oxygen becomes limited, oxidative processes (ammonia oxidation) could preferentially occur on the surface regions of these aggregates (Sexstone et al., 1985; Ebrahimi and Or, 2016). Reactive forms of inorganic nitrogen would therefore be continuously shuttled between oxidation states in close proximity and eventually emitted as N2O, NO, or N2 gas.
The formation, turnover and stability of aggregates depend on factors such as vegetation, soil fauna, microorganisms, cations, the interaction of clay particles and OM (Bronick and Lal, 2005; Rillig and Mummey, 2006; Costa et al., 2018; Totsche et al., 2018). Recent work highlights the importance of microbial activity in soil structuring processes (Lehndorff et al., 2021) and a hierarchical model where smaller building blocks are linked together to form larger aggregated structures has been widely accepted in the field (Edwards and Bremner, 1967; Totsche et al., 2018). Physical fractionation methods generating particle size fractions (PSFs) of soil have shown that different physical-chemical and microbiological signatures exist among the different aggregates (Jocteur Monrozier et al., 1991; Bailey et al., 2013a,b; Bach and Hofmockel, 2014; Hemkemeyer et al., 2015; Zhang et al., 2015). However, most of these studies addressed differences between macro- (>2000 and 2000 - 250 μm) and microaggregates (<250 μm), but few have focused on the different size classes found within microaggregates. In studies on macroaggregates, it has been shown that stabilization capacity vary among fungal taxa and are linked to specific physiological traits such as mycelial/hyphal density and enzymes production (Lehmann et al., 2020). In the microaggregate range less is known, although these size classes (250—2 μm) are (i) more relevant to the individual organisms themselves given the scales at which they operate (Raynaud and Nunan, 2014) and (ii) high microbial abundances were linked to their habitats (Wilpiszeski et al., 2019).
An important factor in soil stabilization and aggregation is the ability to excrete extracellular polymeric substances (EPS) mainly containing polysaccharides, proteins and nucleic acids (Donot et al., 2012; Pandit et al., 2020). In addition to their physical property of acting as a glue to bind soil particles, thus favoring cell attachment and biofilm formation, these polymers provide crucial functions for microbes such as protection against drought, antibiotics and heavy metals, entrapment of nutrients and carbon storage, signaling, genetic exchange, adhesion and aggregation (Costa et al., 2018). Unlike bacteria, fungi are not only capable of excreting EPS and thus influencing their immediate environment, but their hyphal growth enmeshes microaggregates and stabilizes soil structures at scales up to 2 mm (Chenu and Cosentino, 2011). Furthermore, a correlation between hyphal length and aggregate stability has often been observed (Beare et al., 1997; Caesar-TonThat and Cochran, 2000; Rillig and Mummey, 2006; Tisdall et al., 2012). Therefore, communities which excrete biofilms and thus contribute to soil micro-structure can be expected to be located in both macro and micro aggregates due to their hierarchical nature, whereas fungi involved in enmeshment and stabilization of macroaggregates are preferentially found in larger PSFs.
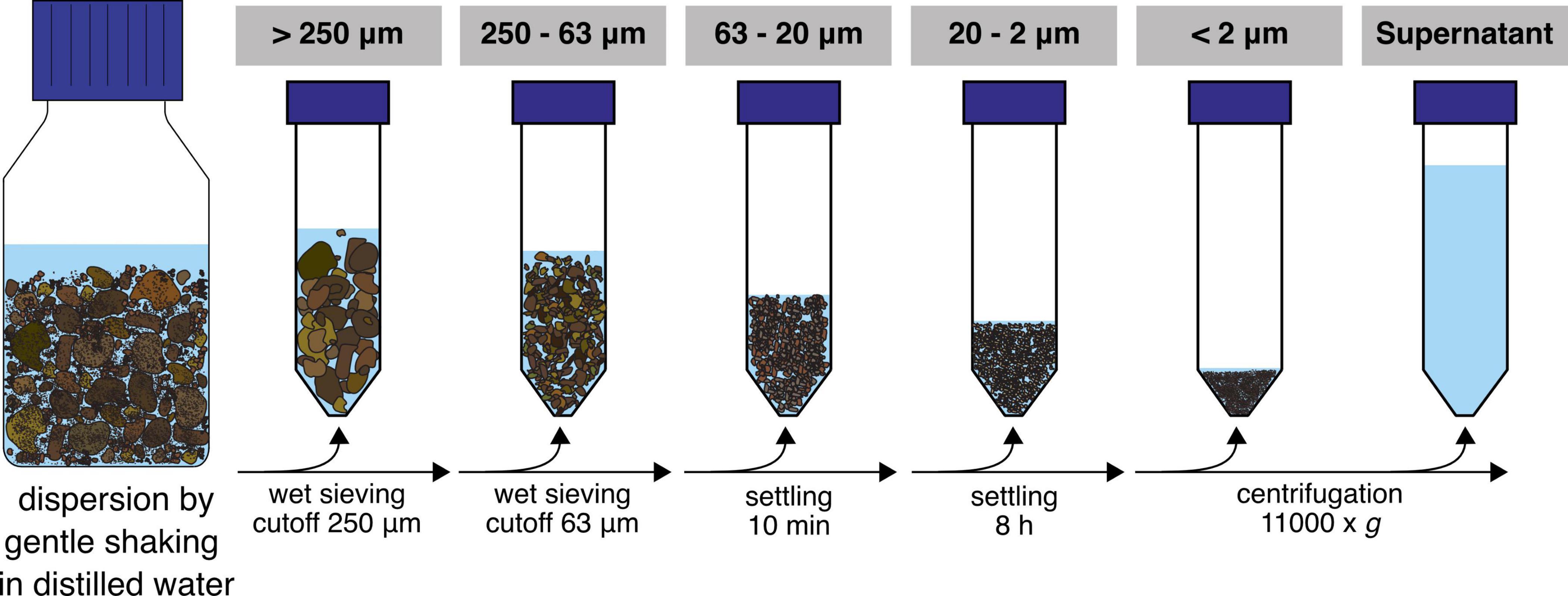
In this study, we applied a gentle soil washing and separation method (Figure 1) aiming to disperse the soil structure and retrieve stable aggregated structures of different sizes to investigate their microbial communities. We hypothesized that the resulting microaggregate fractions consist of different microhabitats which is reflected by a change in community structure and nitrogen cycling potentials.
2 Materials and methods
2.1 Soil sampling and physical fractionation
Soil samples were collected from the top 21 cm at multiple locations in the untreated control plot 3D of the Park Grass Experiment, Rothamsted Research, Hertfordshire, UK (Silvertown et al., 2006) in October 2013 by using sterile manual corers and put in a sterile plastic bag. This soil has not been manipulated (i.e., tillage, fertilization and liming) for over 150 years at the time of sampling, therefore representing natural soil structuring processes. After sampling, the soil was dried at ambient temperature and subsequently sieved (cutoff = 2 mm) to exclude plant roots and homogenized manually through physical mixing. Thirty grams of this sample were fractionated in PSFs (n = 8) and analyzed in parallel with non-fractionated samples (referred to as “bulk soil” throughout the manuscript). Six replicates were used for biological and two for physical and chemical analysis. A fractionation procedure from Jocteur Monrozier et al. (1991) was adapted to optimize preservation of soil microaggregation (beads were not used in the macroaggregate disruption step) and yield PSFs. Briefly, 200 ml of ultra-pure water was added to 30 g of bulk soil in 500 ml Duran® bottles and subsequently placed on an orbital shaker for 1 h at 250 rpm. The water addition was repeated two more times and subsequent fractionation steps were carried out as previously described and consisted of a sequence of (i) wet sieving of the dispersed soils to separate fractions of particle sizes >250 μm and 250–63 μm, (ii) settling of 63–20 μm diameter particles at 1 g (approx. 10 min), (iii) sedimentation of 20–2 μm diameter particles at 1 g (5–6 h) and (iv) centrifugation at 11,000 g to collect particles of <2 μm in diameter (Jocteur Monrozier et al., 1991; Navel and Martins, 2013). The supernatant from the centrifugation step was included in the further biological analysis and represented all organisms and DNA washed from the soil matrix and still in solution after centrifugation. Sterile material was used throughout the entire procedure. All fractions were stored at −80°C until further analysis. Soil physical analysis was carried out directly on fractions after the completed fractionation procedure.
2.2 Soil physical and chemical analysis
The particle size distributions of bulk and fractionated soil samples were determined by laser granulometry with a Mastersizer 2,000 laser granulometer (Malvern Panalytical Ltd., Malvern, UK) with and without ultrasonication treatment to determine the presence of stable aggregates. A refractive index of 1.55 was used as a model parameter according to the procedure described by Navel and Martins (2013). Total organic carbon and nitrogen concentrations were determined in duplicate on 15 mg of soil with a FlashEA1112/FLASH 2,000 Elemental Analyzer equipped with a thermal conductivity detector. For ammonium (NH4+) and nitrate (NO3–) measurements, 1 g dry weight of soil sample was extracted with 5 ml of 1 M KCl solution in a 15 ml Greiner tube on a spinning wheel for 30 min, subsequently centrifuged (5 min at maximum speed) and the supernatant used for quantification. Colorimetric measurements were performed as described previously (Hood-Nowotny et al., 2010). Major and trace elements were quantified in duplicate with 0.2 g soil after microwave-assisted acid digestion. Samples were suspended in a mixed solution of HF:HNO3 (6:2, v:v) in a pressurized closed-vessel microwave system (NovaWAVE, SCP SCIENCE). The digestion program consisted of a 10 min gradual increase to 180°C, a 10 min step at 180°C followed by a cooling step. Quantification was performed by ICP-AES (Perkin Elmer; Optima 3000 DV, PerkinElmer, Waltham, MA, USA).
2.3 DNA extraction and sequencing
DNA was extracted from 100 to 300 mg of homogenized bulk and fractionated soil samples (n = 6; DNeasy Power Soil extraction kit, QIAGEN GmbH, Hilden, Germany). Soil water content of each sample was determined gravimetrically by drying 1 g of sample at 60°C for 24 h. The supernatant samples were filtered on sterile cellulose membranes (MF-Millipore™ membrane filters; pore size 0.45 μm, Merck KGaA, Darmstadt, Germany) with a sterile filtration unit and DNA was subsequently extracted from the membranes (n = 6; DNeasy PowerWater Kit, QIAGEN GmbH, Hilden, Germany). DNA was quantified with the Qubit® dsDNA HS Assay Kit on a Qubit® Fluorometer (Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA) and stored at −20°C for further analysis.
2.4 Ribosomal intergenic spacer analysis analysis
Internal transcribed spacer regions (ITS) of bacterial and fungal communities were amplified with primer pairs FGPL 132-38/FGPS 1490-72 and fITS7/ITS4, respectively (Ranjard et al., 2000; Bokulich and Mills, 2013). Each 25 μl reaction contained 12.5 pmol of the primers and 2 ng of template DNA. Cycling conditions for the bacterial assay were: 94°C for 10 min; 29 cycles of 94°C for 20 s, 55°C for 30 s and 72°C for 1.5 min; final elongation step at 72°C for 10 min. Cycling conditions for the fungal assay were: 94°C for 10 min, 35 cycles at 94°C for 20 s, 57°C for 30 s and 72°C for 1.5 min; final elongation step at 72°C for 10 min. PCR products were run on DNA 1,000 chips on an Agilent 2,100 Bioanalyzer system (Agilent Technologies, Inc., Santa Clara, CA, USA). Tables of band profile traces were imported in R and NDMS on Bray-Curtis dissimilarity matrices were performed with the metaMDS function from the “vegan” package and plots generated with the ggplot2 package (R Core Team, 2019). PERMANOVA analysis was done on Bray-Curtis dissimilarity matrices with the adonis2 function with 999 permutations. Test on multivariate homogeneity of group dispersions was done by the betadisper and permutest function of the vegan package.
2.5 Amplicon sequence generation and analysis
Ribosomal intergenic spacer analysis (RISA) band profiles suggested distinct community profiles in the fractions. DNA samples were pooled according to fractions before library preparation for amplicon sequencing as follows: Fungal V7-V8 region of the 18S rRNA gene was amplified using FR1/FF390 primers (5′-AICCATTCAATCGGTAIT-3′—5′-CGATAACGAACGAGACCT-3′; 348 bp amplicon). Bacterial V4-V6 region of the 16S rRNA gene was amplified using 16S-0515F/16S-1061R primers (5′-TGYCAGCMGCCGCGGTA-3′—5′-TCACGRCACGAGCTGACG-3′; 540 bp amplicon). Primers were tailed with Roche multiplex identifiers (MIDs). Each reaction mix (50 μl) contained 10 pmol of each primer, 40 nmol of dNTPs, 1 x reaction buffer, 10 ng of template DNA and 1 x Titanium Taq DNA Polymerase (Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA). Cycling conditions were: 95°C for 1 min; 35 cycles of 95°C for 30 s and 68°C for 1 min; final elongation step at 68°C for 3 min. PCR reactions were loaded on an agarose gel and bands cut out and purified (Gfx—DNA Purification Kit, GE-Healthcare, Chicago, USA). Concentration of amplified DNA was quantified with the Qubit® dsDNA HS Assay Kit on a Qubit® Fluorometer (Invitrogen™) and an equimolar pool (60 ng DNA per sample) was sent for 454-pyrosequencing (Beckman Coulter GmbH, Krefeld, Germany).
All steps conducted from raw sequence files to phyloseq objects used to create plots are documented in the Supplementary material. Briefly, initial quality filtering and sequence primer trimming was performed with PRINSEQ and pyrosequencing pipeline of RDP (Schmieder and Edwards, 2011; Cole et al., 2014). Further steps followed standard analysis steps in the dada2 pipeline using recommended parameters for reads generated with 454-pyrosequencing. For bacterial amplicons, 187,465 raw reads gave 116,090 filtered, 103,405 denoised and 99,814 chimera filtered reads which resulted in 717 amplicon sequence variants (ASVs). For fungal 18S rRNA gene amplicons, 135,025 raw reads gave 130,136 filtered, 126,333 denoised and 91,080 chimera filtered reads which resulted in 1,216 ASVs. The training set for taxonomic annotation of prokaryotic reads was downloaded from the dada2 repository. For fungal 18S annotation, a training set was generated from the SILVA small subunit (SSU) reference database clustered to 99% similarity including only fungal reads and taxonomic strings in the fasta-header format were changed to match requirements of dada2. Detailed steps involved in database generation are documented in the Supplementary material. Phylogenetic trees used in the analysis were generated with FastTree from multiple sequence alignments of ASV sequences generated with MAFFT (Katoh et al., 2002; Price et al., 2009). Plots were generated within the R package phyloseq (McMurdie and Holmes, 2013).
2.6 Quantitative PCR
Quantitative PCR was performed on the 6 biological replicates using a Corbett Rotor-Gene 6,000 real-time PCR cycler and QuantiTect SYBR® Green PCR Kit (QIAGEN GmbH, Hilden, Germany). Each reaction (25 μl) contained 12.5 μ, 2x QuantiTect SYBR® Green Mix; 0.3–1.8 μl of each primer (10 μM); 100 ng of T4 gene protein 32 (Thermo Fisher Scientific, Waltham, MA, USA); 2–5 ng of template DNA and PCR grade water. Primer sequences are summarized in Supplementary Table 1. In each run three non-template control reactions were included to test for contamination of the master mix. Standard curves for all reactions were derived from serial dilutions of linearized pGEM-T plasmids (Promega, Madison, AL, USA) with the target inserted sequences (Bru et al., 2011). All standard curves were linear and had comparable R-square (0.96–0.99) and efficiency (0.85–1.05) values. Melting curve analysis was used as quality control of qPCR runs. Results from runs showing no amplification in the non-template controls and only one melting peak overlapping with the peaks from standards were considered for further analysis. Plotting and statistical analysis were performed in R using ggplot2 (R Core Team, 2019).
3 Results
3.1 Quality of soil fractionation
The particle size distributions of the obtained fractions (Figure 1) and bulk soil samples were analyzed by laser granulometry (Dynamic Light Scattering, DLS) to ensure that the targeted size ranges were captured (Table 1 and Supplementary Figure 1). The distribution curves showed a shift to smaller particles with ultrasonication (US) applied in the fractions >250 μm and 250–63 μm, but not below (Supplementary Figure 1). The main physical and chemical parameters obtained from the separate fractions and the bulk soil are shown in Table 1. As a quality estimate for the fractionation procedure, values from the single PSFs were multiplied by their mass fraction and the sum was compared to the values from bulk soil samples. While extracted DNA was overestimated with this approach, we found similar results for total carbon and nitrogen as well as NH4+ measurements. This was also the case for most major cations and trace elements measured (Supplementary Table 2). Nitrate was found to be lower in the summed-up fractions than in the bulk soil samples. The observed median diameters of the PSFs were within the expected size range, except in the <2 μm fraction where d50 was 3.1.
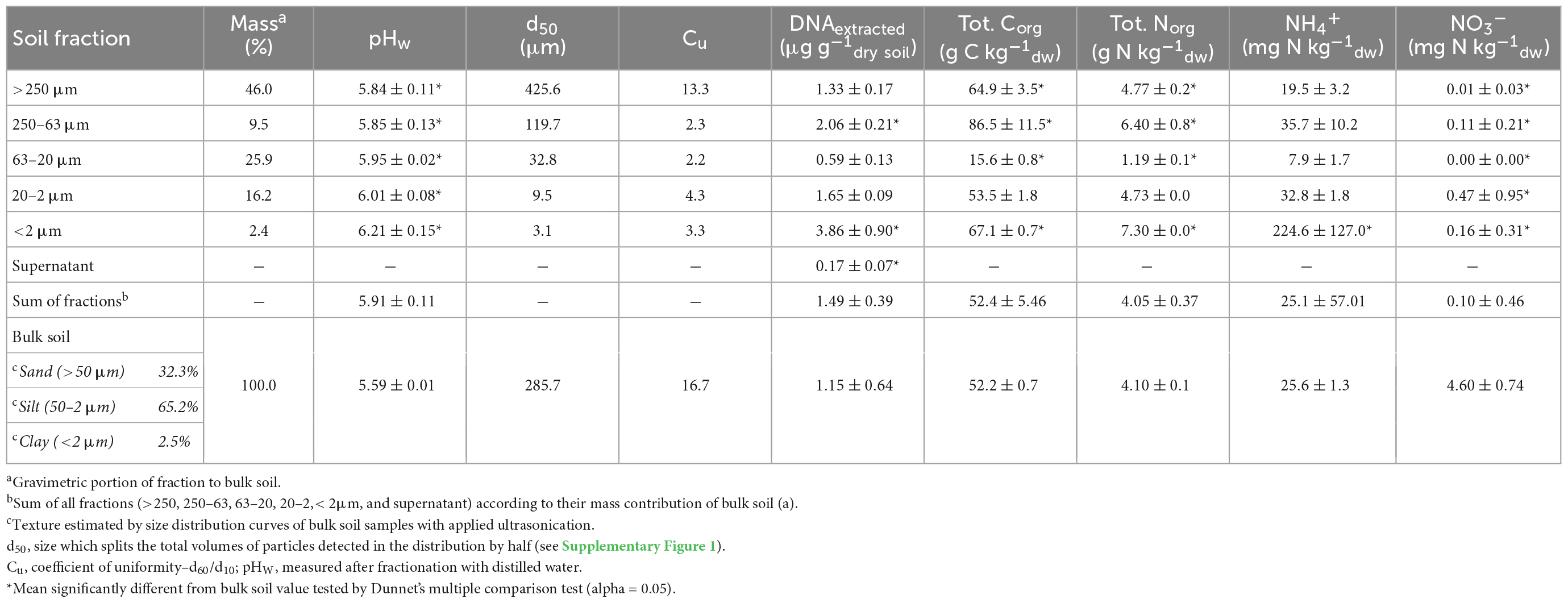
Table 1. Summary of main soil physical and chemical parameters as well as extracted DNA of particle size fractions (PSFs) and bulk samples of Rothamsted Park Grass soil.
3.2 Microbial community structures in PSFs
Non-metric multidimensional scaling plots of RISA band profiles from PSFs and bulk soil samples (Figure 2) showed that bacterial as well as fungal community structures differed between the sample groups. PERMANOVA and subsequent multivariate homogeneity of group dispersions analysis on Bray-Curtis dissimilarity matrices (Supplementary Table 3) showed that the group means were significantly different in the data set. However, it could not be excluded that these differences were due to a heterogeneity of variance among the different groups.
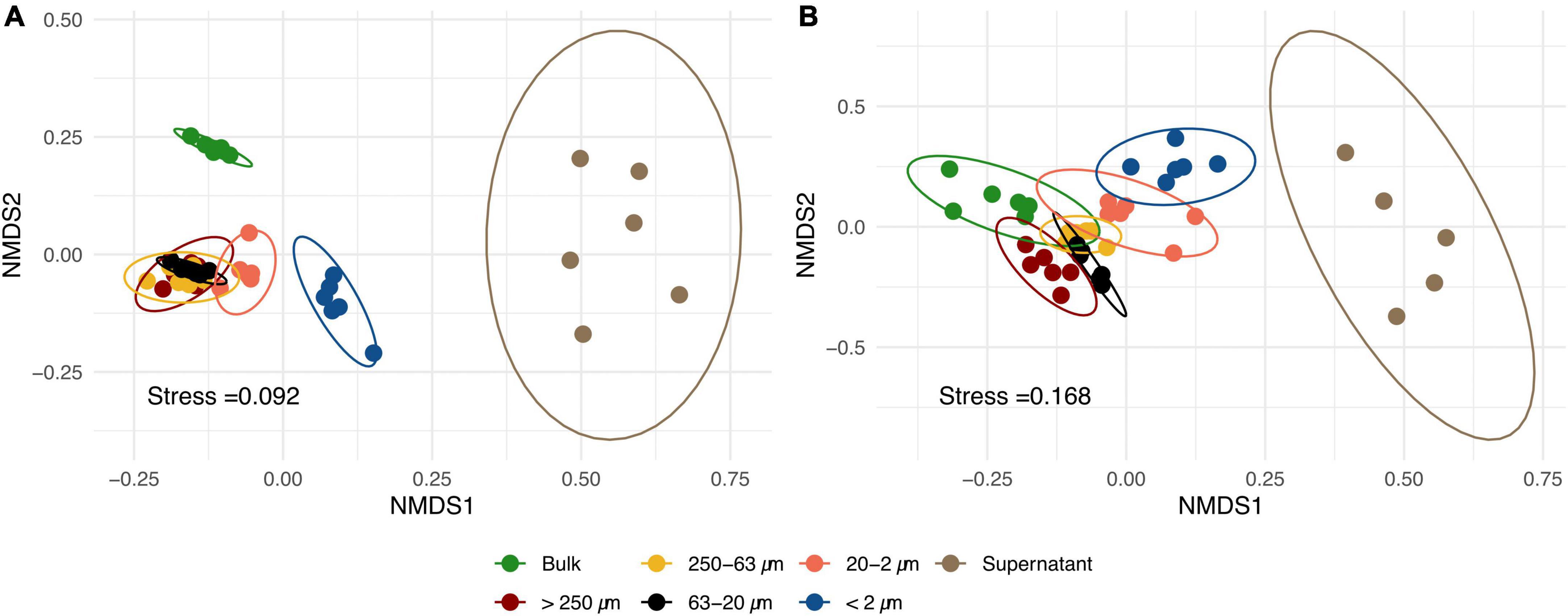
Figure 2. Non-metric multidimensional scaling (NMDS) of bacterial (A) and fungal (B) ribosomal intergenic spacer analysis (RISA) band profiles of different particle size fractions (PSFs) and bulk soil samples based on Bray-Curtis dissimilarity matrices. Each point represents one replicate (n = 6) and confidence ellipses are plotted for each sample group.
Results from annotated bacterial and fungal SSU rRNA amplicons are shown in Figures 3, 4, respectively. The order of taxa on the y-axis as well as the PSFs and bulk soil samples on the x-axis of the heatmap plots are based on a PCoA ordination of unweighted unifrac distances. Therefore, the left and right columns of the heatmap plots are adjacent in a continuous ordering and more discriminating groups are placed in the middle of the heatmap. Taking all bacterial ASVs at genus level into account (Figure 3A), PSFs with bigger aggregated structures (>250 and 250–63 μm) showed communities closer to bulk soil samples based on relative abundance. The smaller PSFs (<63 μm) showed a distinct community composition which placed them central of the heatmap plot. The supernatant clearly separated from all other PSFs with Bacteroidota (mainly Mucilaginibacter and Edaphobaculum) and Proteobacteria (mainly Pseudomonas, Massilia, Duganella, Janthinobacterium, and Variovorax) dominating the community (Figure 3D).
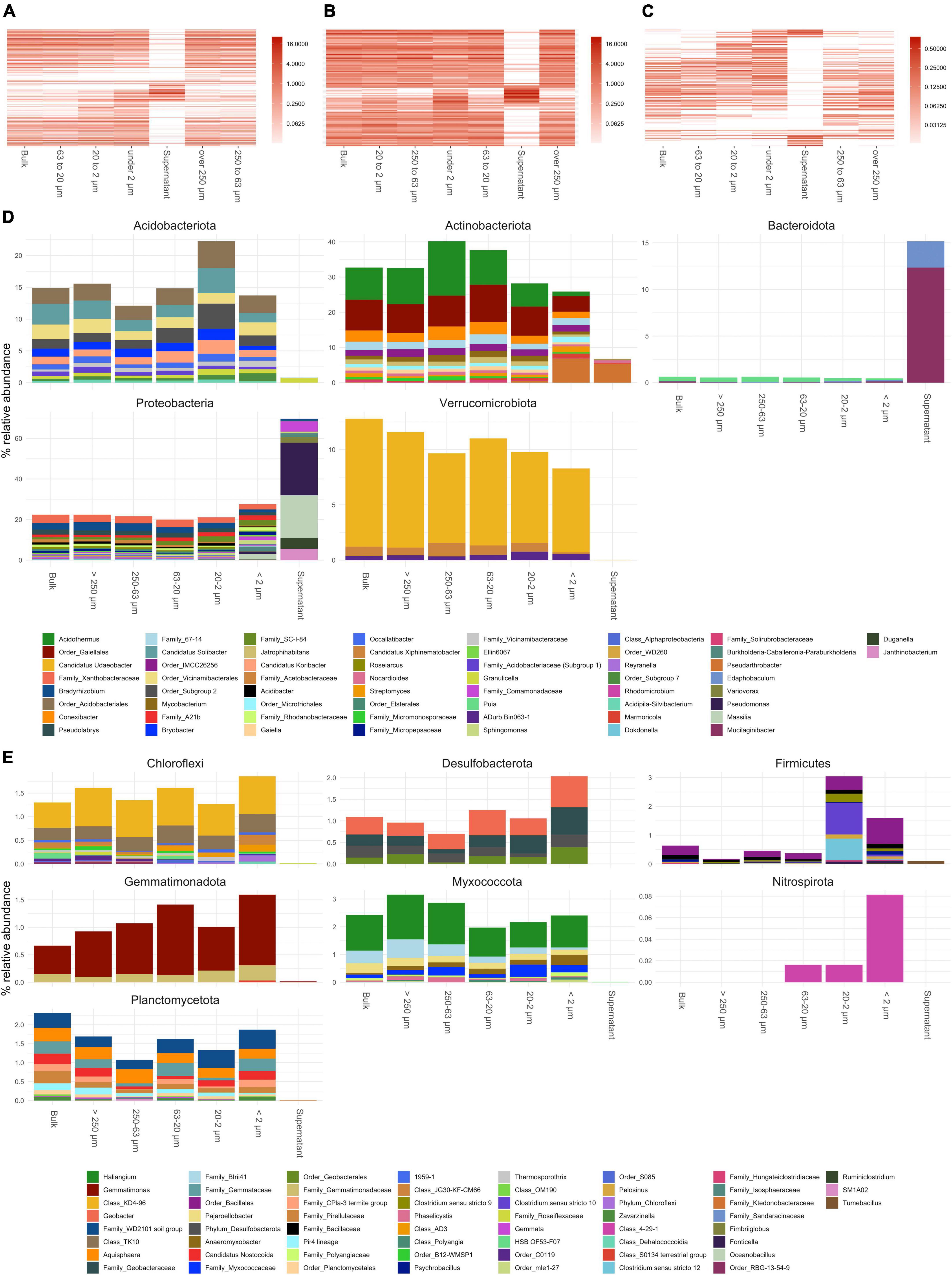
Figure 3. Annotated bacterial amplicon sequence variants (ASVs) of small subunit (SSU) rRNA gene amplicons collapsed to genus level. (A–C) Show heatmaps of all genera detected, genera present over and under 1% in relative abundance, respectively. Order of x and y-axis of heatmaps are based on a PCoA ordination of unweighted unifrac distances. (D) Bar plots of higher abundant phyla showing genera over 0.3% in relative abundance. (E) Bar plots of lower abundant phyla (unfiltered). If taxa unit could not be annotated at the genus level the name of the last higher rank where an annotation at a bootstrap value of 0.8 was possible is shown.
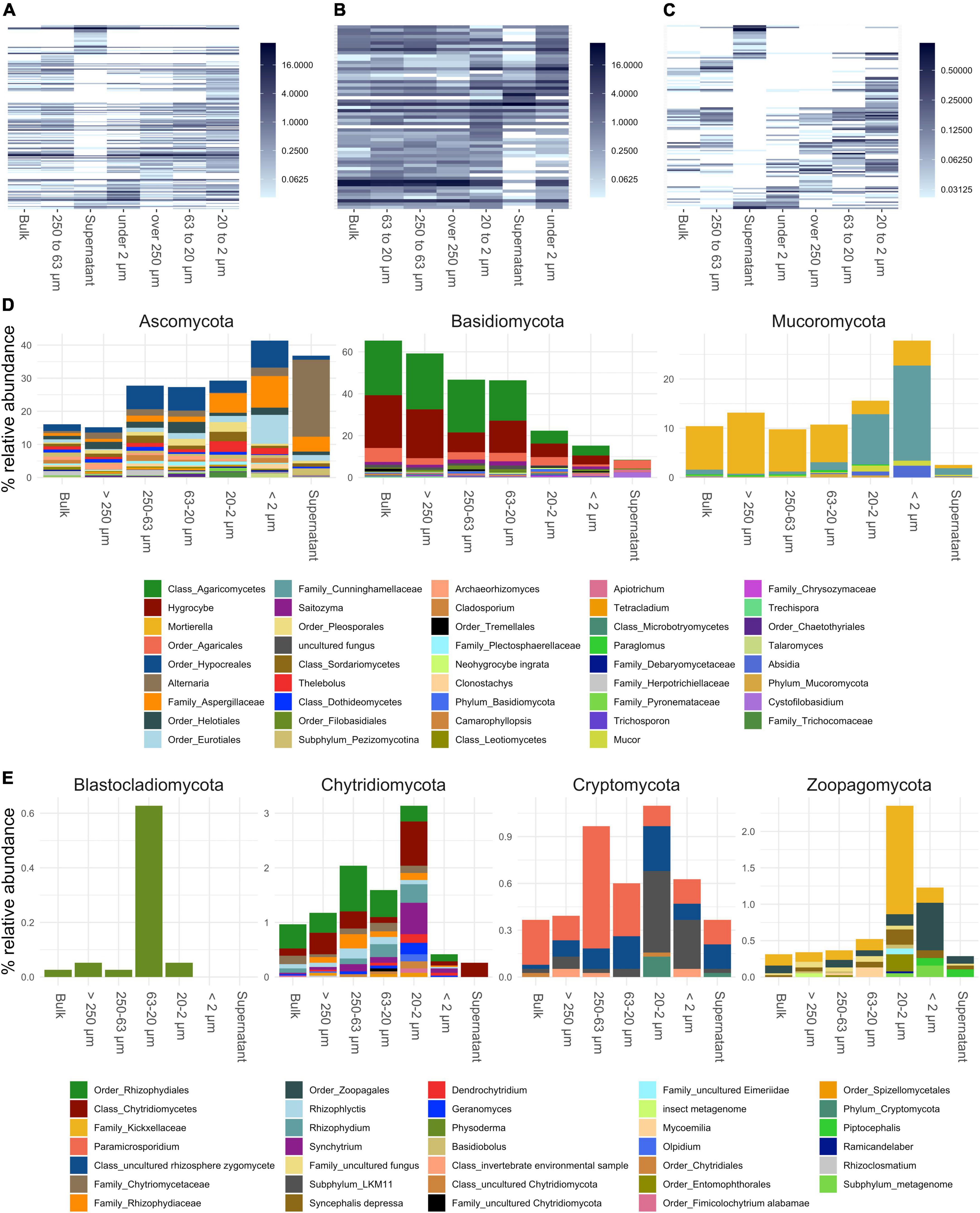
Figure 4. Annotated fungal amplicon sequence variants (ASVs) of small subunit (SSU) rRNA gene amplicons collapsed to genus level. (A–C) Show heatmaps of all genera detected, genera present over and under 1% in relative abundance, respectively. Order of x and y-axis of heatmaps are based on a PCoA ordination of unweighted unifrac distances. (D) Bar plots of higher abundant phyla showing genera over 0.2% in relative abundance. (E) Bar plots of lower abundant phyla (unfiltered). If taxa unit could not be annotated at the genus level the name of the last higher rank where an annotation at a bootstrap value of 0.8 was possible is shown.
Fungal communities showed a shift in community structure from larger to smaller PSFs at the phylum level (Figure 3). The relative abundance of Basidiomycota (mainly Hygrocybe and other Agaricomycetes) decreased by about 45% (from >250 to <2 μm), whereas that of Ascomycota (mainly Hypocreales, Eurotiales, Aspergillaceae, and Alternaria) and Mucoromycota (mainly Mortierella and Cunninghamellaceae) increased within these size classes by about 25 and 15%, respectively. Within the Mucoromycota annotated reads, a shift from Mortierella to taxa in the Mucorales order (Cunninghamellaceae) was observed in smaller PSFs. All fungal phyla showed a decrease in relative abundance in the supernatant samples with the exception of the Ascomycota, which were dominated by Alternaria and Aspergillaceae reads.
3.3 Abundance of bacterial, fungal, and N-cycling genes
The highest abundances of bacterial, fungal and archaeal SSU rRNA gene copies per gram of dry soil were found in the <2 μm fraction (Figures 5A–C, respectively). The lowest were found in the 63–20 μm and in the supernatant. The other fractions showed comparable abundances to the bulk soil samples. The proportion of nitrogen cycle marker genes in reductive pathways [nitrite reductases (nirK and nirS) and nitrous oxide reductases (nosZ clade 1 and 2; Figures 2C, D, respectively] relative to the total estimated abundance of bacteria was higher in the >250 μm and 250–63 μm PSFs compared to the bulk soil samples. Overall, the proportion of marker genes for reductive nitrogen cycling decreased with particle size (Figure 5J; R2 = 0.46, Supplementary Table 4). Marker genes for oxidative pathways in the nitrogen cycle (bacterial and archaeal ammonia monooxygenase (amoA); Figures 5H, I) did not change across the distinct sample groups and were generally low (∼104 copies g–1dry soil). However, we were not able to detect ammonia oxidizing bacterial genes in the <2 μm and supernatant fractions, whereas this was not the case for the ammonia-oxidizing archaeal genes.
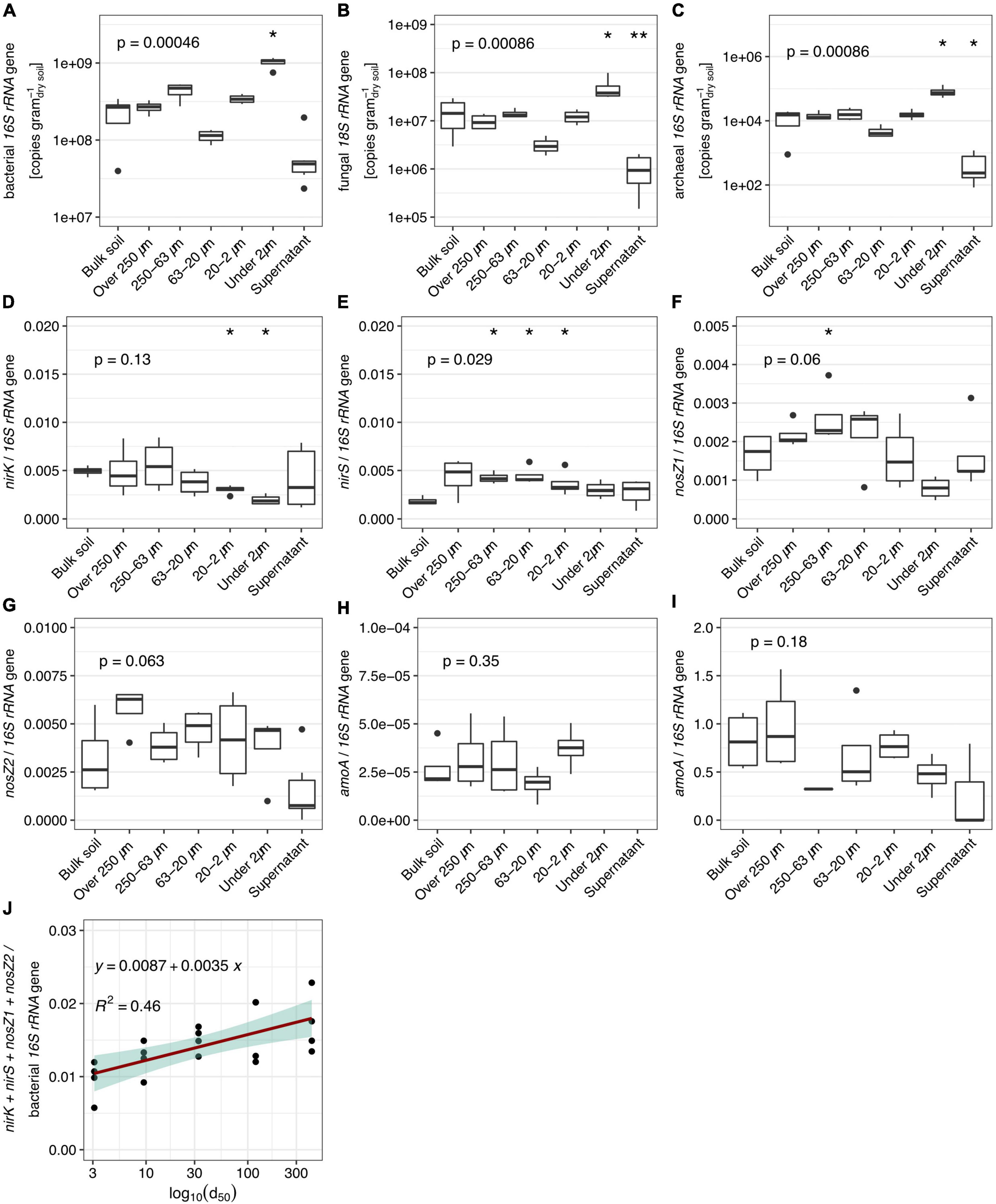
Figure 5. Abundance estimated by quantitative PCR (n = 6) of bacterial, fungal, and archaeal SSU rRNA genes [(A–C), respectively] and ratios of various nitrogen cycling maker gene abundances in obtained fractions and bulk soil. (D,E) Nitrite reductases to bacterial SSU rRNA. (F,G) Nitrous oxide reductases to bacterial SSU rRNA. (H) Ammonia oxidizing bacteria to bacterial SSU rRNA. (I) Ammonia oxidizing archaea to bacterial SSU rRNA. (J) Linear regression (p-value = 0.026) of ratio of summed up reductive N-cycle marker to bacterial SSU rRNA. Values presented in the supernatant group were obtained by calculating the amount washed-off DNA from 1 g of soil during the fractionation procedure to allow comparison. P-values correspond to non-parametric ANOVA Kruskal–Wallis tests. Asterisks indicate significance from Wilcoxon tests between the sample group and bulk soil samples (* and ** stand for alpha = 0.05 and 0.01, respectively).
4 Discussion
4.1 DNA retrieved from bulk and fractionated soil samples
The perfect fractionation of a soil sample would result in equal values of a parameter measured in both the bulk soil sample and in each PSF multiplied with its mass fraction and totaled. Calculations in Table 1 and Supplementary Figure 2 show that this was the case for almost all measured parameters. However, if the totaled value was found higher or lower than the one in the bulk soil, a cross-contamination between fractions could have occurred. For example, if the measured analyte would be transferred to a fraction with a high (i.e., >250 or 63–20 μm) or a low (i.e., <2 μm) mass fraction, the totaled value from the fractions would deviate from the one found in the bulk soil samples.
For extracted DNA, the totaled value was found higher but not significantly different from the bulk soil sample. Highest amounts of extracted DNA and abundance of bacterial and fungal communities were found in the <2 μm fraction. Due to its low mass fraction (∼2.5%) one would expect lower totaled values if a significant transfer of DNA from other fractions would have occurred. This suggests that DNA based parameters are connected to the respective fractions in this study and are not critically biased by cross-contamination effects. However, we cannot exclude that multiple cross-contaminations cancel each other out or that DNA originating from other fractions binds to clay during the dispersion step and is thus collected in the <2 μm fraction. Although these scenarios are difficult to trace back, we believe that the approach to compare totaled vs. bulk soil values adds a level of confidence in the microbiological interpretation of our dataset.
4.2 Microbial particle size preferences
Microbial community structures found in the fractionated samples differed from those of bulk soil (Figure 2). These differences increased with deceasing particle size class and was highest in the supernatant samples. Therefore, the investigation of size classes within the microaggregates (<250 μm) shows potential to gain knowledge on the spatial organization of microbial communities in soil. Results are further discussed based on the different PSFs:
Over 250 and 250–63 μm: The shift to smaller particle sizes induced by ultrasonic (US) treatment showed that we were able to collect still intact aggregates after soil dispersion (Supplementary Figure 1). It is therefore likely that the microbial community found in these aggregates contributes to their stability. Macroaggregate stability has been linked to hyphal abundance of saprophytic and lignin degrading basidiomycetes previously (Caesar-TonThat and Cochran, 2000; Tisdall et al., 2012). Our data confirms this as Basidiomycota (Hygrocybe and other taxa within the Agaricomycetes) dominated the fractions larger than 250, and between 250 and 63 μm but decreased in smaller PSFs (Figure 4D). Hygrocybe was previously identified in nutrient poor, unmanaged grasslands (Griffith et al., 2004; Silvertown et al., 2006; Griffith and Roderick, 2008) and members of the Agaricales can establish large, long lived mycelial networks, which likely have a stabilizing effect in these aggregates obtained in these PSFs through hyphal meshing of smaller microaggregates and production of binding agents (Six et al., 2000; Christensen, 2001; Kleber et al., 2007).
Ascomycota increased in relative abundance with decreasing particle size (Figure 4D). This phylum has been shown to be efficient at forming aggregates at a macroscopic scale, due to its high hyphal density (Lehmann et al., 2020). However, our data suggests that small microaggregates constitute preferred habitats for Ascomycota were specific taxa increased in relative abundance like Aspergillaceae and Eurotiales.
63–20 μm: This fraction was characterized by silt-organomineral microaggregates that had the lowest concentrations of organic carbon, measured ions, extracted DNA and microbial abundances but constituted 25% of the total soil mass. This suggests the presence of inert particles with a low negative surface charge that would limit adhesion of organics, ions and microbes (Chenu and Stotzky, 2002; Mills, 2003). Blastocladiomycota, although found only in low relative abundance (∼ 0.02%) in bulk soil communities, were preferentially detected. These organisms, along with Chytridiomycota, belong to the zoosporic true fungi group (Gleason et al., 2012). Their ability to produce zoospores with a flagellum (Sparrow, 1960; Barr, 2001), their survival in nutrient depleted environments for long periods of time (Gleason et al., 2010) and their attachment to mineral surfaces and insoluble minerals may explain why they are mainly detected in the silt-organomineral microaggregates.
20–2 μm: Bacterial communities were found comparable in higher PSFs but discriminated in this fraction and below (Figure 2A). This might indicate that we can access microbial niches more specifically looking at particles below 20 μm, which is supported by previous findings stating that the majority of bacterial interactions occur at that scale (Raynaud and Nunan, 2014). For example, an increase in sequences annotated to Clostridium was observed, which could indicate oxygen-limited conditions, as Clostridium was previously linked to the fermentation of organic matter in the soil (Boutard et al., 2014). However, as Clostridium is spore forming, its elevated abundance could also be due to an accumulation of spores in that fraction.
For fungi a shift within the Mucoromycota was observed from Mortiella dominating bigger PSF to Mucorales related taxa (Cunninghamellaceae, Mucor, and Absidia) below 20 μm (Figure 4D). Mucoraceae have previously been reported to dominate non-Dikarya in similar PSFs (Hemkemeyer et al., 2019; Bonfante and Venice, 2020). However, the increase of these fungi in small microaggregates remains speculative. Following our observation of a general decrease of Basidiomycota involved in enmeshment of larger aggregate classes, different adhesive forces might take over in order to connect single particles to microaggregates, such as excretion of exopolysaccharides (EPS), for attachment (Flemming and Wingender, 2010; Tisdall et al., 2012; Costa et al., 2018; Sevinç Şengör, 2019; Pandit et al., 2020). A possible mechanism involving the binding of chitosan present on cell walls of Mucorales to clay particles is described in the <2 μm section below. Glomeromycota, an important sub-group of Mucoromycota expected to be present in an unmanaged soil, was not found in our study. The detection of this group with the universal fungal primers used is not without difficulties (detection limit when Glomeromycota are present at low abundance) as explained in the primer design study (Chemidlin Prévost-Bouré et al., 2011), which might explain the unexpected outcome.
Below 2 μm: This fraction showed the highest abundances of bacteria and fungi and an accumulation of organic C and N and cations such as ammonium, iron, calcium, copper and chromium. Several studies have also found that clay fractions contained the highest gene copy numbers or biomass (Jocteur Monrozier et al., 1991; Neumann et al., 2013; Hemkemeyer et al., 2018). Clay particles have varying degrees of negative charges on their surface depending on their chemical composition and are known to interact with microbes in multiple ways (Mills, 2003; Mueller, 2015). This allows microbial cells to adhere through electrostatic forces where enhanced adhesion aided by EPS-like structures could occur (Chenu and Stotzky, 2002; Flemming and Wingender, 2010). EPS exhibit and enhance key criteria for microbial life such as nutrients and water binding, physical and chemical protection and signaling (Costa et al., 2018; Watteau and Villemin, 2018). One explanation for the high microbial abundance and nutrient levels found could therefore be that we collected EPS bound communities by centrifugation in this fraction and that binding of cells and free DNA on clay particles might be even enhanced during the fractionation procedure. Nevertheless, the microbial community we find here were the most discriminating compared to bulk soil samples and preferentially selected a part of the total community which is detected at lower relative abundance in the bulk samples. Therefore, we think that studying microbial communities in this fraction harbors potential in studies focusing on soil microbiome functions.
Another explanation for the higher abundance could be that the total available surface for microbes to attach increases when particle size decreases. This is supported by increasing estimates of abundances for bacteria, fungi and archaea when going from 63–20 to <2 μm fractions (Figures 5A–C). While the high numbers of fungi could also be explained by an accumulation of spores in this fraction, as suggested by Hemkemeyer et al., 2019, we were surprised to find that the relative abundance of Mucoromycota increased to ∼27%. The cell wall of fungi withing the Mucorales is considered unique as it is the only natural source of the biopolymer chitosan known to date (in contrast to chitin, which is abundant) (De Oliveira Franco et al., 2004; Sahariah and Másson, 2017; Elsoud and Kady, 2019; Kaczmarek et al., 2019). Chitosan has been intensively studied in engineering applications due to its binding properties to clay, resulting in nanocomposites with unique properties such as strong absorption (Wan Ngah et al., 2011). It seems plausible that a similar mechanism is responsible for the increased binding of Mucorales to clay particles in soil, which would explain the observed accumulation in this size class.
Supernatant: Almost all bacterial taxa detected in the this fraction were previously reported to be involved in EPS production and biofilm formation (Pankratov et al., 2007; Pantanella et al., 2007; Urai et al., 2008; Andrews et al., 2010; Fish and Boxall, 2018; Sevinç Şengör, 2019; Fredendall et al., 2020; Pandit et al., 2020) and dominated by the phyla Proteobacteria (Pseudomonas, Massilia, Variovorax, Burkholderia, Duganella, and Janthinobacterium) and Bacteroidota (Mucilaginibacter and Edaphobaculum) which were present in the PSFs only at low abundances (Figure 3 and Supplementary Figure 2). Adding centrifugation steps to soil fractionation protocols could therefore be of use in studies focusing on biofilm forming microorganisms.
The settlement of particles in a solution depends in part on density (Li and Yuan, 2002; Yamada et al., 2013). Thus, one explanation for the observed occurrence of specific taxa could be the presence of gas vesicles, which can provide buoyancy (Walsby, 1972; Pfeifer, 2012). Extracellular DNA (eDNA) could also remain in solution rather than settling during centrifugation. In a recent study, it was shown that Pseudomonas forming biofilm on fungal hyphae used eDNA filaments to create stability (Guennoc et al., 2018). This could explain the higher abundance of Pseudomonas sequences in the supernatant, although it would imply that the excreted DNA contains the targeted SSU rRNA gene used in our study. However, the amount of 16S rRNA gene copies per ng DNA were found to be similar in the supernatant and bulk soil samples, suggesting a comparable ratio of rRNA genes to total DNA.
Filamentous fungi were not expected to be detected in the supernatant, or if so, only in low abundances. Indeed, the abundance of fungi in this fraction was found to be significantly lower (Figure 5B) compared to the PSFs. The dominance of Alternaria related sequences might be due to spores from this genus which do not settle during the final centrifugation step. Yeasts or fungi which can reproduce in yeast form might also end up in this fraction depending on buoyancy as discussed for bacteria before. ASVs annotated to yeasts included Cystobasidium, Malassezia, Sporidiobulus, Schefferomyces, Pichia, and Filobasidium (Supplementary Figure 3B). Saitozyma, another ubiquitous yeast often found in soil (Yurkov, 2018), was linked to bigger PSFs (Supplementary Figure 3A) which could point to differences in preferred habitats.
4.3 Nitrogen cycling potentials in obtained particle size fractions
Microbes driving oxidative and reductive reactions with the nitrogen cycle depend on the availability of their preferred electron donors and acceptors to thrive, which in turn describes micro-niches for these organisms. Therefore, we measured marker genes which served as a proxy to access spatial niches within the nitrogen cycle. Overall, the proportion of nitrite reductase (nirK and nirS) and nitrous oxide reductase (nosZ clade 1 and 2) copies to the overall bacterial community (Figures 5D–G) was consistent with previous studies on unmanaged soils (Henry et al., 2006; Philippot et al., 2009; Cuhel et al., 2010; Bru et al., 2011; Di et al., 2014). The observed relationship of a potential N-oxide reducing community decreasing in relative abundance with particle size (Figure 5J) suggests that denitrifying organisms find optimal conditions in larger stable aggregate structures. This is in agreement with a previous study, which showed that denitrification activity was higher in larger PSFs while copy number of marker genes was comparable (Miller et al., 2009). In contrast, nitrogen cycle oxidative pathways are uniformly distributed in PSFs, albeit at low levels. The low abundance of ammonia oxidation has been previously demonstrated in pristine grasslands compared to managed soils (Stienstra et al., 1994; Leininger et al., 2006; Adair and Schwartz, 2008). In unfertilized soils, nitrogen mineralization from decomposing organic matter and biological N-fixation are critical for N-supply (De Vries et al., 2011; Fowler et al., 2013). The low abundance of amoA genes detected (Figures 5H, I) suggests that this NH4+ pool was not accessible to ammonia oxidizers and thus could be primarily used in an assimilatory manner by plants and other microbes (Norton and Ouyang, 2019).
Altogether, the distribution of nitrogen cycling communities in PSFs did not show a clear pattern other than a preference for reductive pathways in larger aggregated structures where oxygen might periodically become limited and the switch to other electron acceptors constitutes an advantage. In an unmanaged natural grassland, nitrogen cycling microbes find limiting resources which results in an ecological equilibrium. The observed patterns might therefore change when the same fractionation method is applied to fertilized soils.
5 Conclusion
This study confirms that distinct habitat preferences of microbial communities are reflected by the size of soil particle aggregates and extend this by showing that a further separation of microaggregates (<250 μm) leads to bigger observed discrimination of community structures. Advances in data generation from smaller samples in combination with sampling techniques at micrometer scale will therefore be key to knowledge gain in a complex environment like soil. The observed shift in fungal taxa is in line with previous findings where hyphal meshes of Basidiomycota stabilized larger aggregates. We showed that fungal communities change in particles below 20 μm. This points to a different role of fungi at those scales. Particularly, similar fractionation approaches might be relevant in studies focusing on bacterial-fungal interactions, as close proximity is a key feature (Kohlmeier et al., 2005; Deveau et al., 2018; Sharma et al., 2020).
Only a few taxa from two bacterial and one fungal phyla (Proteobacteria, Bacteroidota, and Ascomycota, respectively) were recovered in the supernatant fraction. By rinsing soil through sieves during the applied wet fractionation, these microbes are likely washed-off the soil matrix and thus can be seen as loosely attached. This shows that the presence of microbial communities found in a specific fraction does not necessarily mean that we find them with the particles we isolate. The probability of cross-contamination should therefore be evaluated differently for each obtained fraction. For example, loosely attached communities in the supernatant likely stem from particles of random size, whereas those in the bigger PSFs can be linked to stable cores of bigger aggregated structures. The biggest risk of cross-contamination effects might be in the very fine <2 μm fraction, where binding of cells and eDNA to clay particles could lead to misinterpretation (Mueller, 2015; Fomina and Skorochod, 2020). Such a scenario is supported by the high microbial densities and diversities usually found in that fraction using similar approaches (Hemkemeyer et al., 2018, 2019). In our study, taxa found in the supernatant were often also detected in elevated relative abundances in the <2 μm PSF. It seems therefore plausible that of i.e., Pseudomonas, Massilia, or Pseudoathrobacter were detached from the soil matrix and then partly bound to clay particles until saturation and settled in the <2 μm fraction during centrifugation. Remaining cells and eDNA can then be found in the supernatant. However, data from Bacteroidota and fungi do not support this theory. It has previously been shown that the nature of the fractionation method has an influence on the distribution of extracellular enzyme activity, which is likely also the case in studies focusing on microbes in soil (Bach and Hofmockel, 2014). As dry fractionation approaches usually obtain no clay fractions, a comparison of data to estimate these cross-contamination effects proves difficult.
Overall, the gentle fractionation of soil in fractions below the size of microaggregates shows potential to answer relevant research questions in soil microbial ecology by accessing communities at scales where they operate. Specifically, the attempt to wash-off and analyze microbes with varying degrees of detachment forces from soil might generate insight in how communities are organized in biofilm-like structures in soil.
Data availability statement
The datasets presented in this study can be found in online repositories. The link to the data can be found below: https://www.mmnt.net/db/0/0/ftp.ec-lyon.fr/pub/ADN/Keuschnig_2021_AEM_microaggregates_data.
Author contributions
CK, CL, PS, and JM: conceptualization. CK and AN: analysis. CK: writing—original draft. CK, JM, AN, PS, and CL: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by NORA (Nitrogen Oxide Research Alliance) Marie Skłodowska-Curie ITN and research project under the EU’s Seventh Framework Program (FP7), 316472.
Acknowledgments
The authors acknowledge the help of Frédéric Lehembre in experimental design and soil chemical analysis. The authors thank Laurent Philippot and David Bru from the Department of Agroecology in Dijon for fruitful discussion of the results and provision of qPCR standards used in this study, respectively. The authors also thank Sandrine Demaneche and Lorna Dawson for access to samples from the Rothamsted Park Grass experiment. Chemical analytical aspects were supported at IGE by the Air-O-Sol and MOME technical platforms, within the Labex OSUG@2020 (ANR10 LABX56).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1091773/full#supplementary-material
References
Adair, K. L., and Schwartz, E. (2008). Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb. Ecol. 56, 420–426. doi: 10.1007/s00248-007-9360-9
Andrews, J. S., Rolfe, S. A., Huang, W. E., Scholes, J. D., and Banwart, S. A. (2010). Biofilm formation in environmental bacteria is influenced by different macromolecules depending on genus and species. Environ. Microbiol. 12, 2496–2507. doi: 10.1111/j.1462-2920.2010.02223.x
Bach, E. M., and Hofmockel, K. S. (2014). Soil aggregate isolation method affects measures of intra-aggregate extracellular enzyme activity. Soil Biol. Biochem. 69, 54–62. doi: 10.1016/j.soilbio.2013.10.033
Bailey, V. L., Fansler, S. J., Stegen, J. C., and McCue, L. A. (2013a). Linking microbial community structure to β-glucosidic function in soil aggregates. ISME J. 7, 2044–2053. doi: 10.1038/ismej.2013.87
Bailey, V. L., McCue, L. A., Fansler, S., Boyanov, M., DeCarlo, F., and Kemner, K. (2013b). Micrometer-scale physical structure and microbial composition of soil macroaggregates. Soil Biol. Biochem. 65, 60–68. doi: 10.1016/j.soilbio.2013.02.005
Bardgett, R. D., and Van Der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Barr, D. J. S. (2001). “Chytridiomycota BT,” in Systematics and evolution: Part A, eds D. J. McLaughlin, E. G. McLaughlin, and P. A. Lemke (Berlin, Heidelberg: Springer Berlin Heidelberg), 93–112. doi: 10.1007/978-3-662-10376-0_5
Beare, M. H., Hu, S., Coleman, D., and Hendrix, P. (1997). Influences of mycelial fungi on soil aggregation and organic matter storage in conventional and no-tillage soils. Appl. Soil Ecol. 5, 211–219. doi: 10.1016/S0929-1393(96)00142-4
Bokulich, N. A., and Mills, D. A. (2013). Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 79, 2519–2526. doi: 10.1128/AEM.03870-12
Bonfante, P., and Venice, F. (2020). Mucoromycota: Going to the roots of plant-interacting fungi. Fungal Biol. Rev. 34, 100–113. doi: 10.1016/j.fbr.2019.12.003
Boutard, M., Cerisy, T., Nogue, P., Alberti, A., Weissenbach, J., Salanoubat, M., et al. (2014). Functional diversity of carbohydrate-active enzymes enabling a bacterium to ferment plant biomass. PLoS Genet. 10:e1004773. doi: 10.1371/journal.pgen.1004773
Bronick, C. J., and Lal, R. (2005). Soil structure and management: A review. Geoderma 124, 3–22. doi: 10.1016/j.geoderma.2004.03.005
Bru, D., Ramette, A., Saby, N. P., Dequiedt, S., Ranjard, L., Jolivet, C., et al. (2011). Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 5, 532–542. doi: 10.1038/ismej.2010.130
Caesar-TonThat, T. C., and Cochran, V. L. (2000). Soil aggregate stabilization by a saprophytic lignin-decomposing basidiomycete fungus I. Microbiological aspects. Biol. Fertil. Soils 32, 374–380. doi: 10.1007/s003740000263
Chemidlin Prévost-Bouré, N., Christen, R., Dequiedt, S., Mougel, C., Lelièvre, M., Jolivet, C., et al. (2011). Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS One 6:e24166. doi: 10.1371/journal.pone.0024166
Chenu, C., and Cosentino, D. (2011). “Microbial regulation of soil structural dynamics,” in The architecture and biology of soils: Life in inner space, eds K. Ritz and I. Young (Wallingford: CABI), 37–70. doi: 10.1079/9781845935320.0037
Chenu, C., and Stotzky, G. (2002). “Interactions between microorganisms and soil particles: An overview,” in Interactions between soil particles and microorganisms: Impact on the terrestrial ecosystem, eds P. M. Huang, J.-M. Bollag, and N. Senesi (Hoboken, NJ: John Wiley & Sons).
Christensen, B. T. (2001). Physical fractionation of soil and structural and functional complexity in organic matter turnover. Eur. J. Soil Sci. 52, 345–353.
Cole, J. R., Wang, Q., Fish, J., Chai, B., McGarrell, D., Sun, Y., et al. (2014). Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, 633–642. doi: 10.1093/nar/gkt1244
Costa, O. Y. A., Raaijmakers, J. M., and Kuramae, E. E. (2018). Microbial extracellular polymeric substances: Ecological function and impact on soil aggregation. Front. Microbiol. 9:1636. doi: 10.3389/fmicb.2018.01636
Cuhel, J., Simek, M., Laughlin, R. J., Bru, D., Chèneby, D., Watson, C. J., et al. (2010). Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl. Environ. Microbiol. 76, 1870–1878. doi: 10.1128/AEM.02484-09
Deveau, A., Bonito, G., Uehling, J., Paoletti, M., Becker, M., Bindschedler, S., et al. (2018). Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 42, 335–352. doi: 10.1093/femsre/fuy008
Di, H. J., Cameron, K. C., Podolyan, A., and Robinson, A. (2014). Effect of soil moisture status and a nitrification inhibitor, dicyandiamide, on ammonia oxidizer and denitrifier growth and nitrous oxide emissions in a grassland soil. Soil Biol. Biochem. 73, 59–68. doi: 10.1016/J.SOILBIO.2014.02.011
Donot, F., Fontana, A., Baccou, J. C., and Schorr-Galindo, S. (2012). Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohydr. Polym. 87, 951–962. doi: 10.1016/j.carbpol.2011.08.083
Ebrahimi, A., and Or, D. (2016). Microbial community dynamics in soil aggregates shape biogeochemical gas fluxes from soil profiles - upscaling an aggregate biophysical model. Glob. Change Biol. 22, 3141–3156. doi: 10.1111/gcb.13345
Edwards, A. P., and Bremner, J. M. (1967). Microaggregates in soils. J. Soil Sci. 18, 64–73. doi: 10.1111/j.1365-2389.1967.tb01488.x
Elsoud, M. M. A., and Kady, E. M. (2019). Current trends in fungal biosynthesis of chitin and chitosan. Bull. Natl. Res. Centre 43, 1–12.
Fish, K. E., and Boxall, J. B. (2018). Biofilm microbiome (re)growth dynamics in drinking water distribution systems are impacted by chlorine concentration. Front. Microbiol. 9:2519. doi: 10.3389/fmicb.2018.02519
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Fomina, M., and Skorochod, I. (2020). Microbial interaction with clay minerals and its environmental and biotechnological implications. Minerals 10, 1–54. doi: 10.3390/min10100861
Fowler, D., Coyle, M., Skiba, U., Sutton, M. A., Cape, J. N., Reis, S., et al. (2013). The global nitrogen cycle in the twentyfirst century. Philos. Trans. R Soc. B Biol. Sci. 368:20130164. doi: 10.1098/rstb.2013.0164
Franco, L. O., Maia, R. C., Porto, A., Messias, A., Fukushima, K., and Campos-Takaki, G. M. (2004). Heavy metal biosorption by chitin and chitosan isolated from Cunninghamella elegans (IFM 46109). Brazil. J. Microbiol. 35, 243–247. doi: 10.1590/s1517-83822004000200013
Fredendall, R. J., Stone, J. L., Pehl, M. J., and Orwin, P. M. (2020). Transcriptome profiling of variovorax paradoxus EPS under different growth conditions reveals regulatory and structural novelty in biofilm formation. Access Microbiol. 2, 1–11. doi: 10.1099/acmi.0.000121
Gleason, F. H., Crawford, J. W., Neuhauser, S., Henderson, L. E., and Lilje, O. (2012). Resource seeking strategies of zoosporic true fungi in heterogeneous soil habitats at the microscale level. Soil Biol. Biochem. 45, 79–88. doi: 10.1016/j.soilbio.2011.10.011
Gleason, F. H., Schmidt, S. K., and Marano, A. V. (2010). Can zoosporic true fungi grow or survive in extreme or stressful environments ? Extremophiles 14, 417–425. doi: 10.1007/s00792-010-0323-6
Griffith, G. W., and Roderick, K. (2008). “Chapter 15 Saprotrophic basidiomycetes in grasslands: Distribution and function,” in British mycological society symposia series, eds L. Boddy, J. Frankland, and P. V. West (Amsterdam: Elsevier Ltd), 277–299. doi: 10.1016/S0275-0287(08)80017-3
Griffith, G. W., Bratton, J. H., and Easton, G. (2004). Charismatic megafungi - The conservation of waxcap grasslands. Br. Wildlife 16, 31–43.
Guennoc, C. M., Rose, C., Labb, J., and Deveau, A. (2018). Bacterial biofilm formation on the hyphae of ectomycorrhizal fungi: A widespread ability under controls? FEMS Microbiol. Ecol. 94, 1–14. doi: 10.1093/femsec/fiy093
Hemkemeyer, M., Christensen, B. T., Martens, R., and Tebbe, C. C. (2015). Soil particle size fractions harbour distinct microbial communities and differ in potential for microbial mineralisation of organic pollutants. Soil Biol. Biochem. 90, 255–265. doi: 10.1016/j.soilbio.2015.08.018
Hemkemeyer, M., Christensen, B., and Tebbe, C. (2019). Taxon-specific fungal preference for distinct soil particle size fractions. Eur. J. Soil Biol. 94:103103. doi: 10.1016/j.ejsobi.2019.103103
Hemkemeyer, M., Dohrmann, A. B., Christensen, B. T., and Tebbe, C. C. (2018). Bacterial preferences for specific soil particle size fractions revealed by community analyses. Front. Microbiol. 9:149. doi: 10.3389/fmicb.2018.00149
Henry, S., Bru, D., Stres, B., Hallet, S., and Philippot, L. (2006). Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72, 5181–5189. doi: 10.1128/AEM.00231-06
Hood-Nowotny, R., Umana, N. H. N., Inselbacher, E., Lachouani, P., and Wanek, W. (2010). Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci. Soc. Am. J. 74, 1018–1027. doi: 10.2136/sssaj2009.0389
Jocteur Monrozier, L., Ladd, J., Fitzpatrick, R., Foster, R. C., and Rapauch, M. (1991). Components and microbial biomass content of size fractions in soils of contrasting aggregation. Geoderma 50, 37–62. doi: 10.1016/0016-7061(91)90025-O
Kaczmarek, M. B., Struszczyk-Swita, K., Li, X., Szczêsna-Antczak, M., and Daroch, M. (2019). Enzymatic modifications of chitin, chitosan, and chitooligosaccharides. Front. Bioeng. Biotechnol. 7:243. doi: 10.3389/fbioe.2019.00243
Katoh, K., Misawa, K., Kuma, K.-I., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kleber, M., Sollins, P., and Sutton, R. (2007). A conceptual model of organo-mineral interactions in soils: Self-assembly of organic molecular fragments into zonal structures on mineral surfaces. Biogeochemistry 85, 9–24. doi: 10.1007/s10533-007-9103-5
Kohlmeier, S., Smits, T. H., Ford, R. M., Keel, C., Harms, H., and Wick, L. Y. (2005). Taking the fungal highway: Mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 39, 4640–4646. doi: 10.1021/es047979z
Lehmann, A., Zheng, W., Ryo, M., Soutschek, K., Roy, J., Rongstock, R., et al. (2020). Fungal traits important for soil aggregation. Front. Microbiol. 10:2904. doi: 10.3389/fmicb.2019.02904
Lehndorff, E., Rodionov, A., Plümer, L., Rottmann, P., Spiering, B., Dultz, S., et al. (2021). Spatial organization of soil microaggregates. Geoderma 386:114915. doi: 10.1016/j.geoderma.2020.114915
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G. W., et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809. doi: 10.1038/nature04983
Li, X., and Yuan, Y. (2002). Settling velocities and permeabilities of microbial aggregates. Water Res. 36, 3110–3120. doi: 10.1016/S0043-1354(01)00541-3
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Miller, M. N., Zebarth, B. J., Dandie, C. E., Burton, D. L., Goyer, and Trevors, J. T. (2009). Denitrifier community dynamics in soil aggregates under permanent grassland and arable cropping systems. Soil Sci. Soc. Am. J. 73, 1843–1851. doi: 10.2136/sssaj2008.0357
Mills, A. L. (2003). Keeping in touch: Microbial life on soil particle surfaces. Adv. Agron. 78, 1–43. doi: 10.1016/S0065-2113(02)78001-2
Mueller, B. (2015). Experimental interactions between clay minerals and bacteria: A review. Pedosphere 25, 799–810. doi: 10.1016/S1002-0160(15)30061-8
Navel, A., and Martins, J. M. F. (2013). Effect of long term organic amendments and vegetation of vineyard soils on the microscale distribution and biogeochemistry of copper. Sci. Total Environ. 46, 681–689. doi: 10.1016/j.scitotenv.2013.07.064
Neumann, D., Heuer, A., Hemkemeyer, M., Martens, R., and Tebbe, C. C. (2013). Response of microbial communities to long-term fertilization depends on their microhabitat. FEMS Microbiol. Ecol. 86, 71–84. doi: 10.1111/1574-6941.12092
Norton, J., and Ouyang, Y. (2019). Controls and adaptive management of nitrification in agricultural soils. Front. Microbiol. 10:1931. doi: 10.3389/fmicb.2019.01931
Pandit, A., Adholeya, A., Cahill, D., Brau, L., and Kochar, M. (2020). Microbial biofilms in nature: Unlocking their potential for agricultural applications. J. Appl. Microbiol. 129, 199–211. doi: 10.1111/jam.14609
Pankratov, T. A., Tindall, B. J., Liesack, W., and Dedysh, S. N. (2007). Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 57, 2349–2354. doi: 10.1099/ijs.0.65100-0
Pantanella, F., Berlutti, F., Passariello, C., Sarli, S., Morea, C., and Schippa, S. (2007). Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 102, 992–999. doi: 10.1111/j.1365-2672.2006.03155.x
Pfeifer, F. (2012). Distribution, formation and regulation of gas vesicles. Nat. Rev. Microbiol. 10, 705–715. doi: 10.1038/nrmicro2834
Philippot, L., Bru, D., Saby, N. P., Cuhel, J., Arrouays, D., Simek, M., et al. (2009). Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ. Microbiol. 11, 3096–3104. doi: 10.1111/j.1462-2920.2009.02014.x
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
R Core Team. (2019). A language and environment for statistical computing, R foundation for statistical computing. Vienna: R Core Team.
Rabot, E., Wiesmeier, M., Schlüter, S., and Vogel, H. (2018). Soil structure as an indicator of soil functions: A review. Geoderma 314, 122–137. doi: 10.1016/j.geoderma.2017.11.009
Ranjard, L., Brothier, E., and Nazaret, S. (2000). Sequencing bands of ribosomal intergenic spacer analysis fingerprints for characterization and microscale distribution of soil bacterium populations responding to mercury spiking. Appl. Environ. Microbiol. 66, 5334–5339. doi: 10.1128/AEM.66.12.5334-5339.2000
Raynaud, X., and Nunan, N. (2014). Spatial ecology of bacteria at the microscale in soil. PLoS One 9:e87217. doi: 10.1371/journal.pone.0087217
Rillig, M. C., and Mummey, D. L. (2006). Mycorrhizas and soil structure. New Phytol. 171, 41–53. doi: 10.1111/j.1469-8137.2006.01750.x
Sahariah, P., and Másson, M. (2017). Antimicrobial chitosan and chitosan derivatives: A Review of the structure–activity relationship. Biomacromolecules 18, 3846–3868. doi: 10.1021/acs.biomac.7b01058
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Sevinç Şengör, S. (2019). “Review of current applications of microbial biopolymers in soil and future perspectives,” in ACS Symposium series, eds R. N. Rathinam and R. K. Sani (Washington, DC: American Chemical Society), 275–299. doi: 10.1021/bk-2019-1323.ch013
Sexstone, A. J., Revsbech, N. P., Parkin, T. B., and Tiedje, J. M. (1985). Direct measurement of oxygen profiles and denitrification rates in soil aggregates. Soil Sci. Soc. Am. J. 49, 645–651. doi: 10.2136/sssaj1985.03615995004900030024x
Sharma, K., Palatinszky, M., Nikolov, G., Berry, D., and Shank, E. (2020). Transparent soil microcosms for live-cell imaging and non-destructive stable isotope probing of soil microorganisms. Elife 9, 1–28. doi: 10.7554/eLife.56275
Silvertown, J., Poulton, P., Johnston, E., Edwards, G., Heard, M., and Biss, P. (2006). The park grass experiment 1856-2006: Its contribution to ecology. J. Ecol. 94, 801–814. doi: 10.1111/j.1365-2745.2006.01145.x
Six, J., Elliott, E. T., and Paustian, K. (2000). Soil macroaggregate turnover and microaggregate formation: A mechanism for C sequestration under no-tillage agriculture. Soil Biol. Biochem. 32, 2099–2103. doi: 10.1016/S0038-0717(00)00179-6
Spadini, L., Navel, A., Martins, J. M. F., Vince, E., and Lamy, I. (2018). Soil aggregates: A scale to investigate the densities of metal and proton reactive sites of organic matter and clay phases in soil. Eur. J. Soil Sci. 69, 953–961. doi: 10.1111/ejss.12695
Sparrow, F. K. (1960). Aguatic phycomycetes. Second revised and enlarged edition. Ann Arbor, MI: Univ Michigan Press.
Stienstra, A. W., Klein Gunnewiek, P., and Laanbroek, H. J. (1994). Repression of nitrification in soils under a climax grassland vegetation. FEMS Microbiol. Ecol. 14, 45–52. doi: 10.1111/j.1574-6941.1994.tb00089.x
Tisdall, J. M., Nelson, S., Wilkinson, K., and Smith, S. (2012). Stabilisation of soil against wind erosion by six saprotrophic fungi. Soil Biol. Biochem. 50, 134–141. doi: 10.1016/j.soilbio.2012.02.035
Totsche, K. U., Amelung, W., Gerzabek, M., Guggenberger, G., Klumpp, E., Knief, C., et al. (2018). Microaggregates in soils. J. Plant Nutr. Soil Sci. 181, 104–136. doi: 10.1002/jpln.201600451
Urai, M., Aizawa, T., Nakagawa, Y., Nakajima, M., and Sunairi, M. (2008). Mucilaginibacter kameinonensis sp., nov., isolated from garden soil. Int. J. Syst. Evol. Microbiol. 58, 2046–2050. doi: 10.1099/ijs.0.65777-0
Vries, W d, Leip, A., Reinds, G. J., Kros, J., Lesschen, J. P., and Bouwman, A. F. (2011). Comparison of land nitrogen budgets for European agriculture by various modeling approaches. Environ. Pollut. 159, 3254–3268. doi: 10.1016/J.ENVPOL.2011.03.038
Walsby, A. E. (1972). Structure and function of gas vacuoles. Bacteriol. Rev. 36, 1–32. doi: 10.1128/mmbr.36.1.1-32.1972
Wan Ngah, W. S., Teong, L. C., and Hanafiah, M. A. K. M. (2011). Adsorption of dyes and heavy metal ions by chitosan composites: A review. Carbohydr. Polym. 83, 1446–1456. doi: 10.1016/j.carbpol.2010.11.004
Watteau, F., and Villemin, G. (2018). Soil microstructures examined through transmission electron microscopy reveal soil-microorganisms interactions. Front. Environ. Sci. 6:106. doi: 10.3389/fenvs.2018.00106
Wilpiszeski, R. L., Aufrecht, J. A., Retterer, S. T., Sullivan, M. B., Graham, D. E., Pierce, E. M., et al. (2019). Soil aggregate microbial communities: Towards understanding microbiome interactions at biologically relevant scales. Appl. Environ. Microbiol. 85, 1–18. doi: 10.1128/AEM.00324-19
Yamada, Y., Fukuda, H., Inoue, K., Kogure, K., and Nagata, T. (2013). Effects of attached bacteria on organic aggregate settling velocity in seawater. Aquat. Microb. Ecol. 70, 261–272.
Young, I. M., and Crawford, J. W. (2004). Interactions and self-organization in the soil-microbe complex. Science 304, 1634–1637.
Yurkov, A. M. (2018). Yeasts of the soil – obscure but precious. Yeast 35, 369–378. doi: 10.1002/yea.3310
Keywords: soil fractionation, micro-scale, fungi, nitrogen cycling, aggregates, biofilm, soil
Citation: Keuschnig C, Martins JMF, Navel A, Simonet P and Larose C (2022) Micro-fractionation shows microbial community changes in soil particles below 20 μm. Front. Ecol. Evol. 10:1091773. doi: 10.3389/fevo.2022.1091773
Received: 07 November 2022; Accepted: 07 December 2022;
Published: 21 December 2022.
Edited by:
Miles T. Wetherington, Cornell University, United StatesReviewed by:
Michael Kertesz, The University of Sydney, AustraliaArtur Banach, The John Paul II Catholic University of Lublin, Poland
Copyright © 2022 Keuschnig, Martins, Navel, Simonet and Larose. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christoph Keuschnig, ✉ Y2hyaXN0b3BoLmtldXNjaG5pZ0BnZnotcG90c2RhbS5kZQ==
†Present address: Christoph Keuschnig, Interface Geochemistry, GFZ German Research Centre for Geosciences, Potsdam, Germany
 Christoph Keuschnig
Christoph Keuschnig Jean M. F. Martins
Jean M. F. Martins Aline Navel2
Aline Navel2 Pascal Simonet
Pascal Simonet Catherine Larose
Catherine Larose