- 1Centro de Ciências Naturais e Humanas, Universidade Federal do ABC, Santo André, SP, Brazil
- 2Laboratory of Chemical Ecology, Departamento de Química Ambiental, Facultad de Ciencias, Universidad Católica de la Santísima Concepción, Concepción, Chile
- 3Fort Lauderdale Research and Education Center, University of Florida, Institute of Food and Agricultural Sciences, Davie, FL, United States
- 4Laboratório de Termitologia, Departamento de Sistemática e Ecologia, Universidade Federal da Paraíba, João Pessoa, PB, Brazil
- 5Museu de Zoologia da Universidade de São Paulo, São Paulo, SP, Brazil
Little is known about the phylogeography of termites in the Neotropical region. Here, we explored the genetic patterns and phylogeographical processes in the evolutionary history of Nasutitermes kemneri, an endemic termite of the South American diagonal of open formations (DOF) formed by the Chaco, Cerrado, and Caatinga phytogeographic domains. We sampled 60 individuals across the three domains of the DOF, and using the mitochondrial genes 16S, COI, and COII, as well as the nuclear gene ITS, evaluated the genetic diversity and divergence time of the populations, along with their genetic structure. The results show a strong genetic and spatial structure within the samples, evidencing the existence of two well-differentiated genetic groups: the Northeastern and the Southwestern populations, which diverged about 2.5 Mya, during the Pliocene-Pleistocene boundary. The Northeastern population, which encompasses Caatinga and northern portions of Cerrado, has an intricate structure and seems to have suffered repetitive retraction-expansion events due to climactic fluctuations during the Quaternary. The Southwestern population, which ranges from central-south Cerrado to the northeast peripherical portions of the Chaco, displays a star-shaped haplotype structure, indicating that this region may have acted as a refugia during interglacial periods.
1. Introduction
There are more than 3,000 described species of termites (Blattaria: Isoptera) (Krishna et al., 2013), and they are one the most successful terrestrial organisms on Earth. This success is due to a sum of ecologically relevant characteristics. Termites may be poor land dispersers and weak flyers (< 2 km; Hu et al., 2007; Tonini et al., 2013; Mullins et al., 2015), however, transoceanic dispersal events mostly via flotsam (Bourguignon et al., 2017) facilitated their near world-wide distribution. Coupled with their ability to decompose cellulosic compounds (Jouquet et al., 2016), these eusocial insects thrived in new ecological opportunities and became keystone species and ecosystem engineers with an exceptional biomass in tropical ecosystems (Tuma et al., 2020).
Despite the extension of insects across the world and being the largest and most diverse group of organisms on Earth (Chapman, 2009), their phylogenetic relationships have only recently become evident due to advances in next generation technology (Trautwein et al., 2012; Yeates et al., 2016). Not much is known about the evolutionary history of species and their populations in South America, which is poorly studied in comparison to North America and Europe (see Tembrock et al., 2019), and what little is known mostly focuses on agricultural pests due to their economic relevance (i.e., Tembrock et al., 2019; Raszick et al., 2021; Vilardi et al., 2021). Phylogeographic studies would help to clarify the evolutionary history of species and their populations through the use of several resources such as population genetics and phylogenetics to understand the geological and climatic processes that determined the current geographic distribution of genealogical lineages (Avise et al., 1987; Avise, 2000).
Little is known about termites’ species evolutionary history, with most articles just focusing on species of the Rhinotermitidae family (e.g., Kutnik et al., 2004; Szalanski et al., 2004; Park et al., 2006; Tripodi et al., 2006; Jenkins et al., 2007; Luchetti et al., 2007; Austin et al., 2008; Lefebvre et al., 2008; Li et al., 2009; Scicchitano et al., 2018; Hyseni and Garrick, 2019). Although being the evolutionary most recent and most diverse family, the phylogeography of the Termitidae remains scarcely studied (Ozeki et al., 2007; de Faria Santos et al., 2017, 2022; Singham et al., 2017). Among these, there are only two phylogeographic studies for Nasutitermes species: N. corniger and N. ephratae (de Faria Santos et al., 2017, 2022), up to date. In both species, significant degrees of genetic structure were found across the different domains of the wide Neotropical region. However, another species of the genus, N. kemneri (Figure 1), seems to be restricted to a smaller area known as the South American “diagonal of open formations” (DOF) (Mathews, 1977).
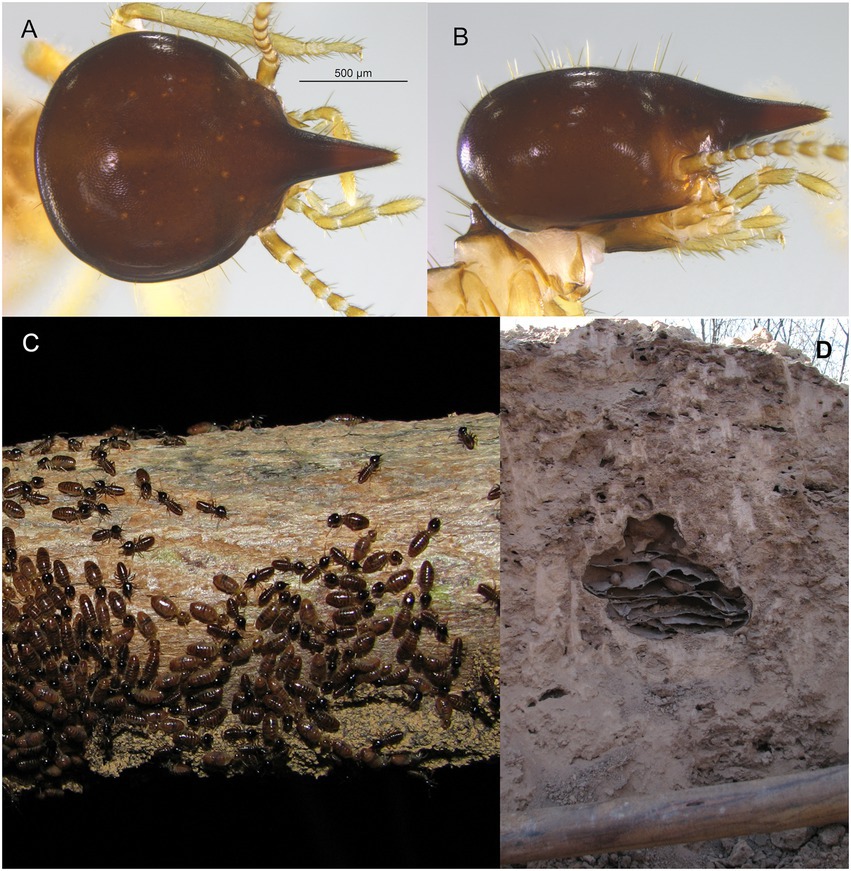
Figure 1. Nasutitermes kemneri. Soldier from Bolivia in dorsal (A) and lateral (B) views. Individuals foraging during twilight on a tree branch (C) and its nest inside what looks like an abandoned nest of Syntermes sp. (D) in the Caatinga phytogeographic domain, São Miguel do Tapuio, PI. As scale in (D), the diameter of the pickaxe handle is approximately 5 cm.
The DOF consists of three phytogeographic domains (or biomes) crossing the South America from southwest to northeast: Caatinga, seasonally dry tropical forests of northeastern Brazil; Cerrado, the central Brazilian savanna; and Chaco, seasonally dry subtropical forests in northern Argentina, Paraguay, and Bolivia (Vanzolini, 1963; Zanella, 2010). All these domains are characterized by marked seasonality with severe droughts, nevertheless, they are different physionomically and in vegetation, harboring unique biotas well adapted to such fluctuations (Werneck, 2011).
The current geographical arrangement of the DOF is the result of the Central Brazilian Plateau uplift since the Cenozoic (Martins-Ferreira et al., 2020), changing the extensions of the Caatinga, Cerrado, and Chaco domains, in addition to the periods of climatic fluctuations during the Quaternary, when the Earth experienced ice ages interspersed with warmer periods. During these fluctuations, dry vegetation would have expanded and contracted, respectively, during glacial maximums and minimums, while tropical forest regions would have followed the reverse pattern (Pinheiro and Monteiro, 2010; Zanella, 2010). Specifically, dry regions were connected during glacial periods, while when interglacial periods occurred, tropical forests expanded and disconnected dry regions (Pennington et al., 2004; Werneck et al., 2012). Consequently, these events not only shaped the current geomorphology of the region, but became important drivers of the diversification and distribution of biota across the DOF.
Nasutitermes kemneri Snyder and Emerson, 1949 is a termite species endemic to the DOF that feeds on wood in early stages of decomposition in a diversity of vegetation types (Mathews, 1977). Due to its well delimited area of distribution, this termite is a relevant study model to understand how species respond to the vegetational, elevational, and climatic shifts between the Caatinga–Cerrado–Chaco phytogeographic domains. Accordingly, this study aims to determine if there are divergences between the populations distributed across the DOF, and if these form a spatial structure in genetic differentiation. Secondarily, through demographic and time estimation analyses, we tested if populations of N. kemneri contracted in size and were disconnected during interglacial periods but survived in climatic refugia. For this purpose, we used both nuclear (ITS) and mitochondrial molecular markers (COI, COII, and16S rRNA).
2. Materials and methods
2.1. Sampling, DNA extraction, and sequencing
A total of 60 individuals of Nasutitermes kemneri (Figure 1) were sampled, from Bolivia to Rio Grande do Norte state in Brazil, covering most of the South American DOF, and the known distribution of the species (Supplementary Table S1). Samples studied were from the Museum of Zoology of the University of São Paulo (MZUSP), the Federal University of Paraíba (UFPB), and University of Florida Termite Collection (UFTC).
Genomic DNA was extracted from workers’ head and thorax using the DNeasy Blood & Tissue Kit—QIAGEN extraction kit following the manufacturer’s instructions. Amplification was performed with the Master Mix (Prodimol) for PCR, where primers used for PCR are detailed in Supplementary Table S2, and cycle settings are specified in references therein. We amplified both mitochondrial and nuclear markers for subsequent analyses: the mitochondrial genes 16S rRNA, and cytochrome oxidase I and II (COI and COII), and the entire ribosomal internal transcribed spacer region (ITS1 + 5.8S + ITS2; Supplementary Tables S1, S2). Samples with confirmed amplification through agarose gel electrophoresis underwent DNA purification using ExoSap (GE Technology Infrastructure). Fragments were sequenced forward and reverse using the BigDye reagent kit (Perkin-Elmer) in an automatic sequencer ABI 3730 XL DNA Analyzer (Applied Biosystems), according to manufacturer’s instructions.
We successfully amplified and sequenced 55 individuals, but not throughout all molecular markers, obtaining 55 sequences of 16S rRNA, 25 of COI, 53 of COII and 43 sequences of ITS. All sequences were deposited on GenBank database (access numbers in Supplementary Table S1).
2.2. Population genetics and phylogeography
To understand the genetic structure of N. kemneri, we performed various analyses with 16S rRNA, COII and ITS region sequences. We did not use the COI sequences to avoid incorporating many missing data to the matrix alignment.
Median-joining haplotype networks (Bandelt et al., 1999) were reconstructed in the PopART software (Leigh and Bryant, 2015), one for aligned and concatenated 16S rRNA and COII sequences through the MUSCLE algorithm in Geneious v9.1 (Biomatters Ltd., Auckland, New Zealand), and another for the ITS region sequences. Genetic diversity indexes (Hd, π) were estimated in DnaSP 5.0 (Librado and Rozas, 2009).
As ITS marker was highly conserved (only two parsimony informative sites), a Maximum Likelihood (ML) phylogenetic tree was inferred using a final alignment including only the 16S rRNA and COII molecular markers. Model optimization was performed with the R packages “ape” and “phangorn” (Schliep et al., 2018; Paradis and Schliep, 2019). The selected model for the entire alignment was GTR + I + G. The phylogeny was transformed into an ultrametric tree using the chronos function of the package “ape” (Paradis and Schliep, 2019) and plotted in the map using the package “phytools” (Revell, 2012).
To estimate the presence of spatial genetic structure within the sequences of N. kemneri we used Bayesian Analysis of Population Structure (BAPS) v6 (Corander et al., 2008) implementing the method of “spatial clustering of individuals,” testing up to 8 clusters (populations; K = 8). For comparison, we conducted analyses in GENELAND 4.9.2 (Guillot et al., 2008, 2012) using both the uncorrelated and correlated allele frequency models (1.000.000 iterations, 100 thinning), given that the correlated frequency model may be more powerful at detecting subtle differentiations, but is more sensitive to departures from model assumptions (i.e., isolation-by-distance) than the uncorrelated frequency model. Furthermore, an Analysis of Molecular Variance (AMOVA) and Tajima’s D neutrality test were performed with the package “pegas” (Paradis, 2010) to evaluate Fst magnitude and significance and detect signals of population expansion or retraction for the genetic groups of N. kemneri.
2.3. Dating analysis
To estimate the divergence time between the main populations of N. kemneri (Figure 2), we performed a phylogenetic analysis calibrated with fossils. For that, we used three mitochondrial markers (COI, COII, and 16SrRNA) from one individual of each main population, in addition to 32 sequences from GenBank (Supplementary Table S3). A GTR + G + I substitution model for the concatenated matrix was used.
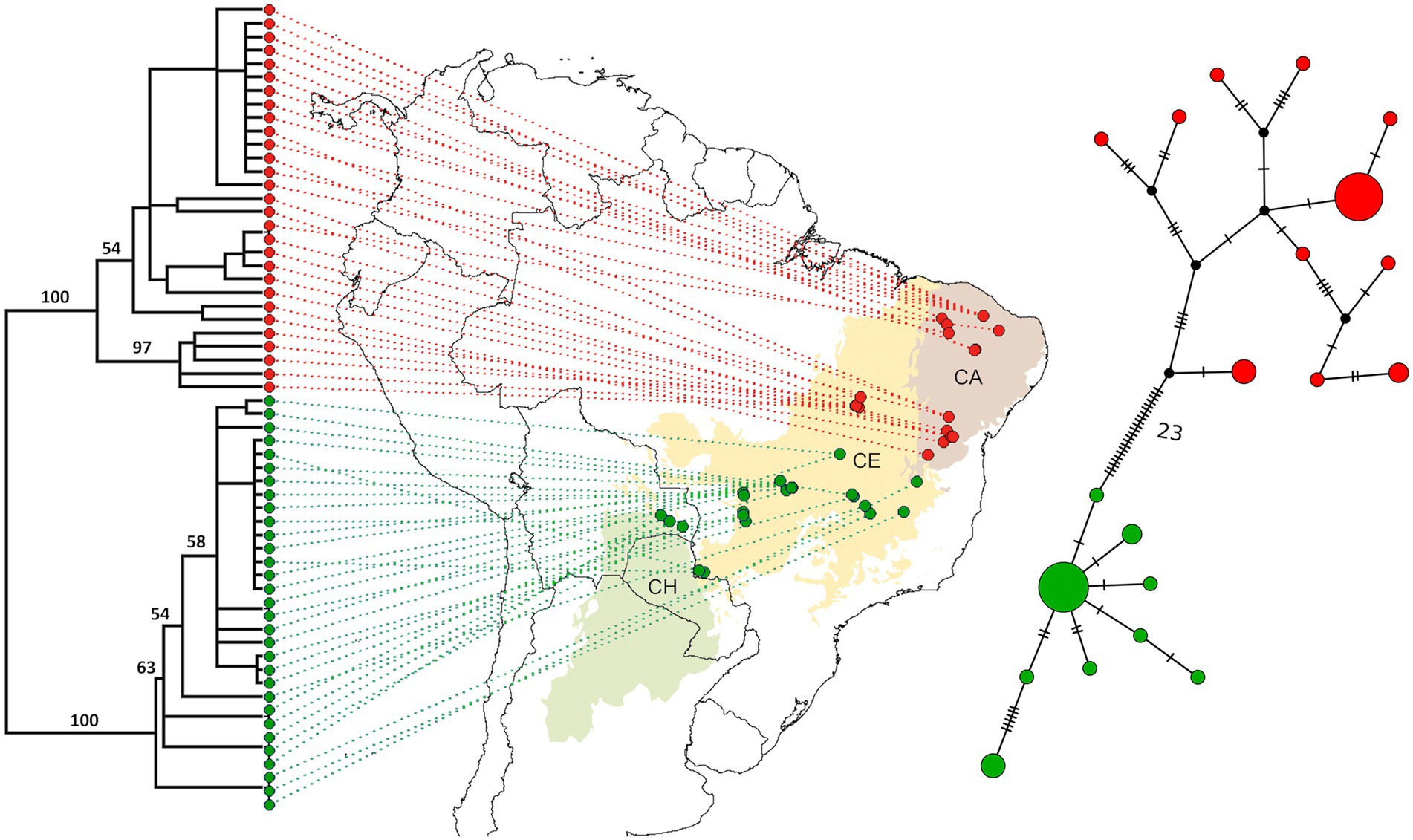
Figure 2. The two major groups of Nasutitermes kemneri: Northeastern (NE, in red) and Southwestern (SW, in green). In the left, a Maximum Likelihood (ML) phylogenetic tree with corresponding sampling sites, and in the right the haplotype network. Branch support of the phylogeny are bootstrap values; number of substitutions between the two major groups are included in the haplotype network. CH, Chaco; CE, Cerrado; and CA, Caatinga.
The phylogenetic analysis was carried out under Bayesian inference with BEAST 1.8.0 (Drummond et al., 2012). A Yule speciation process (Gernhard et al., 2008), with a random starting tree, and an uncorrelated lognormal relaxed clock (Drummond et al., 2006), were used as tree priors.
The molecular clock was calibrated using six calibration nodes that were set as monophyletic (Supplementary Table S4). The fossils were used as minimum age constrains and implemented as exponential priors of node time (Supplementary Table S4). Times for most recent common ancestor priors were selected to have an exponential distribution (Ho, 2007), with mean and offset or standard deviation are showed in Supplementary Table S4.
The analysis was performed with Monte Carlo Markov Chain searches conducted for 40,000,000 generations. Convergence and stationarity were assessed with Tracer 1.6 (Rambaut et al., 2018) and the first 1,000 trees (10%) were discarded as burn-in with TreeAnnotator 1.8.0 and visualized using FigTree 1.3.1.
3. Results
Sequencing efforts resulted in varying lengths: 321, 640, and 735 bp for 16 rRNA, COII, and ITS, respectively. ITS sequences were highly conserved in the 43 sampled individuals, with only 3 polymorphic sites. On the other hand, 14 polymorphic sites were present in the 55 16S rRNA sequences, 43 polymorphic sites were present in the 55 COII sequences, and the final alignment of 961 bp containing both 16S rRNA and COII genes revealed 53 polymorphic sites.
We used different approaches to estimate the degree of population differentiation. The genealogical relationships reconstructed by the ML phylogenetic tree evidenced high bootstrap support for the two mtDNA genetic groups of N. kemneri within the South American DOF, one at the Northeast (NE), and another one at the Southwest (SW; Figure 2).
The Median-Joining haplotype network of the two mitochondrial gene regions (16S rRNA + COII) also revealed the presence of these two groups, and that they are separated by 23 mutations, with no haplotypes in common (Figure 2). A total of 11 haplotypes were from the NE group, while 9 haplotypes were from the SW group. The ITS haplotype network illustrated one genetic group with only 4 closely related haplotypes (Supplementary Figure S1).
Furthermore, BAPS v6 and the correlated frequency model in GENELAND analyses used to infer population structure separated N. kemneri in the same two spatial clusters (Figure 3), further affirming the strong genetic differentiation found between these two genetic groups.
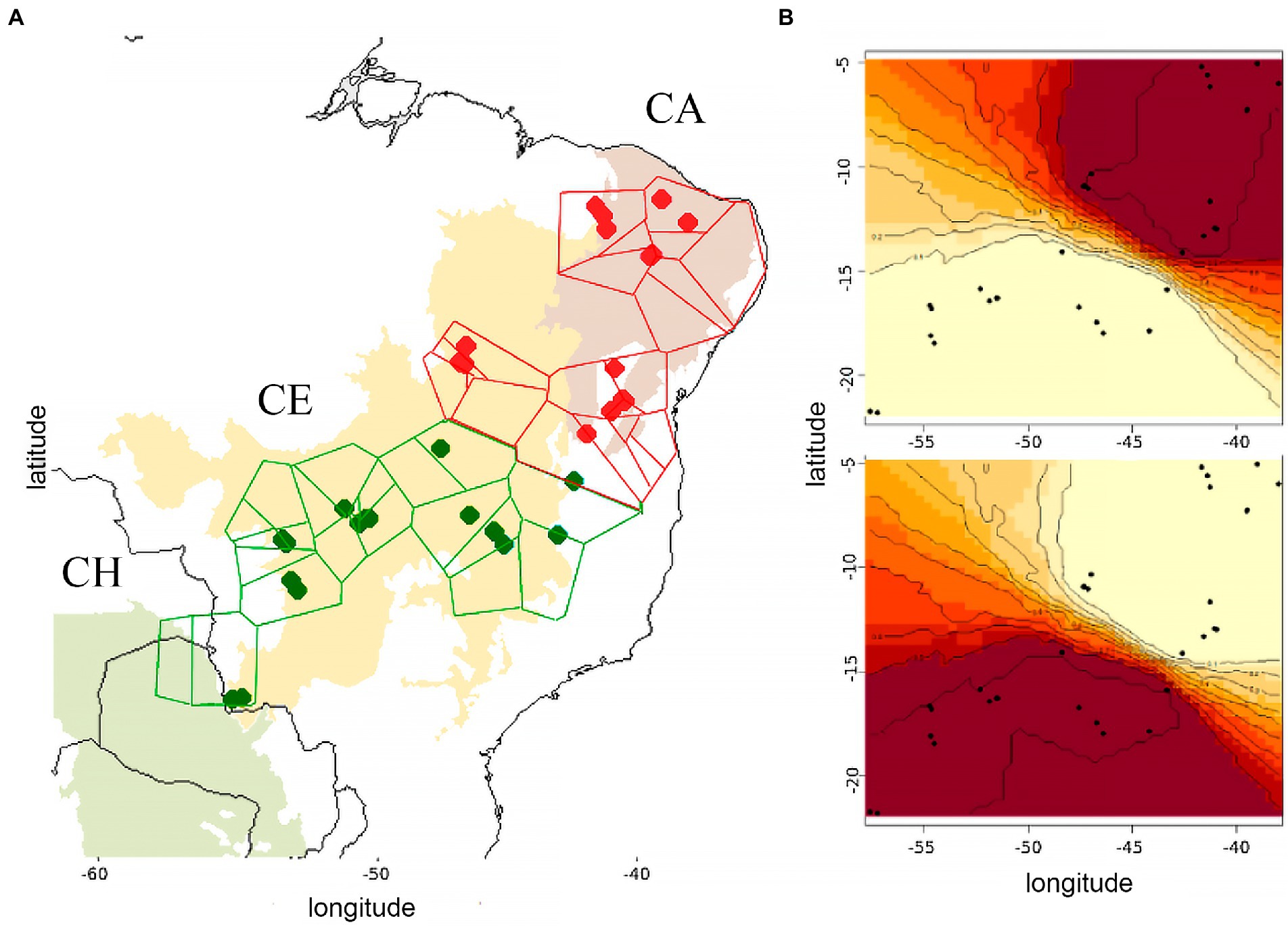
Figure 3. (A) Voronoi tessellation produced by the Bayesian spatial clustering analysis of 961 bp (16S rRNA + COII) sequences from Nasutitermes kemneri, as implemented in BAPS v6. resulting in K = 2, (ln(P) = −563.0329). (B) GENELAND map of posterior probabilities of cluster membership, correlated allele frequency model. CA, Caatinga; CE, Cerrado; and CH, Chaco.
Notably, some genetic substructure in N. kemneri was hinted by the uncorrelated frequency model from GENELAND and BAPS non-spatial clustering of individuals. These results illustrated 4 clusters, distinguishing the Caatinga from northern Cerrado within the NE group, and separating the Cerrado from samples near the Paraná biogeographic dominion (Morrone et al., 2022; Supplementary Figure S2). Moreover, the phylogenetic tree shows high bootstrap values(= 97) which support the subtle divergence of the Caatinga and Cerrado within the Northeastern group (NE), while the easternmost individuals are the most divergent samples within the SW group (Figure 2). A 2-level AMOVA distinguishing both NE–SW and these four clusters further evidences the presence of sub-structure (Supplementary Table S5). However, since Cluster B (N = 5) and Cluster C (N = 3) are comprised of few individuals, further sampling is needed to firmly ascertain the presence of genetic sub-structure within Nasutitermes kemneri.
Considering the highest level of differentiation broadly supported by previous analyses, the AMOVA partitioned in two groups (NE–SW) revealed a high variation among groups (82.74%) contrasting with a low variation within groups (17.26%; Table 1). The high Fst (0.827, p < 0.0001) confirms a strong genetic population structure as supported by the phylogenetic tree, haplotype network and Bayesian spatial clustering. Regarding the genetic diversity across the South American DOF, haplotype and nucleotide diversity was overall high (Hd = 0.867, π = 0.00107), and high but slightly lower within both genetic groups (NE Hd = 0.767, π = 0.00734, SW Hd = 0.723, π = 0.00922).
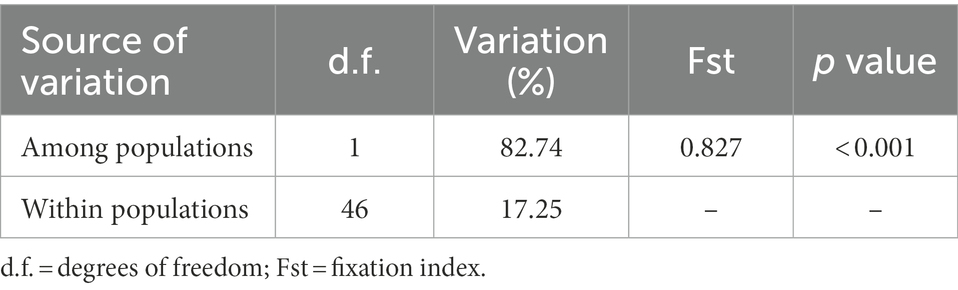
Table 1. Analysis of molecular variance results for Northeastern (NE) and Southwestern (SW) populations inferred by markers 16S rRNA and COII.
Demographic inferences through Tajima’s D neutrality test showed no signal of population expansion or retraction for the entire species range (D = 0.395, p = 0.69) nor for each separate group (NE, D = −1.496, p = 0.13/SW, D = −1.883, p = 0.059). Finally, the dated phylogenetic analyses inferred that the divergence between NE and SW lineages occurred around 2.41 Mya (3.59–1.47, 95% height posterior density), during the Pliocene–Pleistocene boundary.
4. Discussion
Results show a strong genetic and spatial structure within N. kemneri (Figure 2), evidenced by two highly differentiated groups, henceforth called populations. These populations are not restricted to the phytogeographic domain where they were sampled (Caatinga, Cerrado, and Chaco), instead, the NE population encompasses Caatinga and northern portions of Cerrado, while the Southwestern (SW) population ranges from central-south Cerrado to the northeast peripherical portions of the Chaco (Figure 2). This Northeast–Southwest pattern of spatial differentiation is shared with other taxonomic groups distributed along the DOF such as squamates (Werneck et al., 2012b; Recorder et al., 2014; Fonseca et al., 2018), amphibians (Oliveira et al., 2018), flies (Moraes et al., 2009), bees (Miranda et al., 2016), marsupials (Carvalho et al., 2011), trees (Resende-Moreira et al., 2017), and, to some extent, birds (Rocha et al., 2020).
Estimates suggest that these populations diverged ~ 2.5 Mya, during the Pliocene-Pleistocene boundary. During this period, the Cerrado plateaus were already uplifted to their present-day elevations while other areas within the Cerrado subsided, forming peripheral depressions (Del'Arco and Bezerra, 1989). The depressions inhibited gene flow between east and west of the plateau, and promoted diversification of subsequently isolated populations of flora and fauna. As discussed by Bonatelli et al. (2021), while several species across the DOF experienced range shifts, there is no single demographic response to Pleistocene climatic fluctuations. Some species experimented demographic expansion, others exhibited the opposite response, and some, like N. kemneri, underwent different processes since the separation of both populations (i.e., 59), where each experienced different evolutionary histories, illustrated by the different structures displayed by the haplotype network.
The NE population network has an intricate structure, with many “missing” haplotypes, which could be due to repetitive extinction-expansion events. Demographic inferences do not offer statistical support for this hypothesis, however, the geoclimatic history of northeastern Brazil does. During the Quaternary, major paleoenvironmental changes occurred during wet periods throughout the Pleistocene (Auler et al., 2004). Increased rainfalls facilitated the expansion of semi-deciduous forest to the Caatinga (Auler et al., 2004; Wang et al., 2004; Werneck, 2011), which possibly provided new ecological niches for N. kemneri to exploit besides xeric shrublands and an increase in biomass. On the contrary, arid periods led to regional extinction of taxa adapted to wet forests such as herpetofauna, as discussed by Gehara et al. (2017), and the cyclic nature of these humid-drier events left its mark on the NE population, accounting for the high genetic diversity and complex haplotype network. Moreover, recent studies have shown that termites are resistant to temperature shifts (Woon et al., 2019; Janowiecki et al., 2020; Woon et al., 2022), but precipitation would be the main factor in determining termite distribution, abundance, and survival rate (Woon et al., 2019; Pozo-Santiago et al., 2020). Although we did not measure this, it seems that N. kemneri is more abundant in the most humid phytogeographic domain, the Cerrado, than in the semiarid ones, Caatinga and Chaco. In the same way, elevation plays important roles in both restrictive and non-restrictive effects of temperature and humidity, as well as in the interaction between these factors, and while the Caatinga is composed mostly by flattened surfaces between 300 and 500 m above sea level (Silva et al., 2017), the Cerrado has higher plateaus in central and northeastern regions (Werneck et al., 2012).
The SW population displays a star-shaped haplotype group, which is characteristic of refugia with later population expansion, yet demographic inferences lack statistical support. Nevertheless, during the Quaternary climatic and vegetation fluctuations, the general area currently occupied by Cerrado appears to have been drier (Collevatti et al., 2020; Oliveira et al., 2020), paleoclimatic model reconstructions predicted areas of historical climatic stability (Werneck et al., 2012). These stable areas (the higher plateaus in central and north-eastern Cerrado) were important refugia for taxa such as trees (Ramos et al., 2007; Buzatti et al., 2017; Resende-Moreira et al., 2017; Camps et al., 2018), and may, consequently, also serve as refugia for wood feeding termites.
Finally, while the spatially explicit Bayesian clustering programs BAPS and GENELAND reached a consensus in dividing N. kemneri in two populations, the uncorrelated frequency model in GENELAND and the non-spatial BAPS clustering are also worth noting and briefly discussed. These less robust approaches hint at subtle population substructure within both the NE and SW populations, wholly distinguishing Caatinga (Cluster A) and northeastern Cerrado (Cluster B) in NE, and separating central western Cerrado (Cluster C) from the rest of Cerrado (Cluster D) in SW. Tree species with a similar distribution across the DOF (and some that extent to the Parana dominion), show fairly recent (~ 800.000 years BP) fine-scale genetic structure within the Chaco-Cerrado-Atlantic Forest, from 3 to up to 7 haplogroups/lineages (Collevatti et al., 2009; Novaes et al., 2010, 2013). These lineages present a similar pattern of distribution to Clusters A, B, C, and D of N. kemneri, and the recent divergence time between the main populations (NE/SW) at the Pliocene-Pleistocene transition (with confidence intervals spanning from Late Miocene to Middle Pleistocene) may explain the low resolution of these analyses, where subtle climatic or geographic differences within these phytogeographic domain may be driving this incipient or shallower population sub-structuring. In fact, biogeographic districts based on tree species (Françoso et al., 2020) partially match the distribution of these clusters, citing different temperatures, seasonal variation, radiation and the degree of cover transformation as responsible for the floristic differences. However, further sampling is needed to determine the extent and significance of these patterns in N. kemneri.
This study provides important inferences about the general panorama of the evolutionary history of N. kemneri in the DOF, its endemic region, offering important contributions to the understanding of biogeographic and phylogeographic issues in the Neotropics. Our results support an ancient divergence between two well-defined lineages (NE and SW) for N. kemneri, which occurred during the Plio-Pleistocene transition, reinforcing the role of climatic and vegetation variation events in the diversification of the DOF biota (Del'Arco and Bezerra, 1989; Wang et al., 2004). Further climatic fluctuations during the Quaternary had pronounced effects on the Caatinga biota (Bonatelli et al., 2021) and could have facilitated the expansion and diversification of N. kemneri across the NE region of the DOF, while refugia zones in the Southeastern region may account for the constricted diversity patterns within the Cerrado, and altogether explain the unique evolutionary history of these two separate populations. The findings presented in this work, however, came from a few mitochondrial markers, and the investigation using multi-locus nuclear data can reveal other relevant patterns in the population genetic structure in this species.
Most of the integrative phylogeographic and predictive studies in the DOF rely heavily on data from herpetofauna and plants, where insects are less than 15% of the species considered (Del'Arco and Bezerra, 1989). In this sense, data presented in this study not only aids the understanding of the evolutionary history of N. kemneri and add evidence to the response patterns described for this area (Del'Arco and Bezerra, 1989; Wang et al., 2004), but also partly accounts for our knowledge gap. Furthering our knowledge on biotic variables that influenced the demography and distribution of DOF endemic species is key, and especially insects (Wilson and Fox, 2021) for future decisions in conservation strategies of the DOF threatened phytogeographic domains and potential effects linked to climate warming.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
KK and RGS made the laboratory work. KK, VP-O, RGS, and TC did the analyses. AV, EC, and RHS provided the specimens. RHS provided photos of the N. kemneri soldier. KK, VP-O, and TC did the writing. RHS, AV, and EC edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Conselho Nacional de desenvolvimento Científico e Tecnológico (CNPq): productivity in research fellowship proc. #309820/2020-0 to AV and #308408/2019-5 to EC, and
São Paulo Research Foundation, Brazil (FAPESP): grants #13/20068-9 to EC and #13/03767-0 and #20/06041-4 to TC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1081114/full#supplementary-material
References
Auler, A. A., Wang, X., Edwards, R. L., Cheng, H., Cristalli, P. S., Smart, P. L., et al. (2004). Quaternary ecological and geomorphic changes associated with rainfall events in presently semi-arid northeastern Brazil. J. Quat. Sci. 19, 693–701. doi: 10.1002/jqs.876
Austin, J. W., Szalanski, A. L., McKern, J. A., and Gold, R. E. (2008). Molecular phylogeography of the subterranean termite Reticulitermes tibialis (Isoptera: Rhinotermitidae). J. Agric. Urban Entomol. 25, 63–79. doi: 10.3954/1523-5475-25.2.63
Avise, J. C. (2000). Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press, 447.
Avise, J. C., Arnold, J., Ball, J. R. M., Bermingham, E., Lamb, T., Neigel, J. E., et al. (1987). Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18, 489–522. doi: 10.1146/annurev.es.18.110187.002421
Bandelt, H., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bonatelli, I. A. S., Gehara, M., Carstens, B. C., Colli, G. R., and Moraes, E. M. (2021). Comparative and predictive phylogeography in the south American diagonal of open formations: unravelling the biological and environmental influences on multitaxon demography. Mol. Ecol. 31, 331–342. doi: 10.1111/mec.16210
Bourguignon, T., Lo, N., Šobotník, J., Ho, S. Y. W., Iqbal, N., Coissac, E., et al. (2017). Mitochondrial phylogenomics resolves the global spread of higher termites, ecosystem engineers of the tropics. Mol. Biol. Evol. 34, 589–597. doi: 10.1093/molbev/msw253
Buzatti, R. S. O., De Lemos-Filho, J. P., Bueno, M. L., and Lovato, M. B. (2017). Multiple Pleistocene refugia in the Brazilian cerrado: evidence from phylogeography and climatic nichemodelling of two Qualea species (Vochysiaceae). Bot. J. Linn. Soc. 185, 307–320. doi: 10.1093/botlinnean/box062
Camps, G. A., Martínez-Meyer, E., Verga, A. R., Sérsic, A. N., and Cosacov, A. (2018). Genetic and climatic approaches reveal effects of Pleistocene refugia and climatic stability in an old giant of the Neotropical dry Forest. Biol. J. Linn. Soc. 125, 401–420. doi: 10.1093/biolinnean/bly115
Carvalho, B. A., Oliveira, L. F. B., Langguth, A., Freygang, C. C., Ferraz, R. S., and Mattevi, M. S. (2011). Phylogenetic relationships and phylogeographic patterns in Monodelphis (Didelphimorphia: Didelphidae). J. Mammal. 92, 121–133. doi: 10.1644/10-MAMM-A-075.1
Chapman, A. D. (2009). Numbers of Living Species in Australia and the World. 2nd Edn. Toowoomba, Australia: Australian Biodiversity Information Services, 80.
Collevatti, R. G., Lima, N. E., and Vitorino, L. C. (2020). “The diversification of extant angiosperms in the South America dry diagonal,” in Neotropical Diversification: Patterns and Processes. Fascinating Life Sciences. eds. V. Rull and A. C. Carnaval (Cham, Switzerland: Springer International Publishing), 547–568.
Collevatti, R. G., Rabelo, S. G., and Vieira, R. F. (2009). Phylogeography and disjunct distribution in Lychnophora ericoides (Asteraceae), an endangered cerrado shrub species. Ann. Bot. 104, 655–664. doi: 10.1093/aob/mcp157
Corander, J., Marttinen, P., Sirén, J., and Tang, J. (2008). Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinform. 9:539. doi: 10.1186/1471-2105-9-539
de Faria Santos, A., Cancello, E. M., and Morales, A. C. (2022). Phylogeography of Nasutitermes ephratae (Termitidae: Nasutitermitinae) in Neotropical region. Sci. Rep. 12:11656. doi: 10.1038/s41598-022-15407-z
de Faria Santos, A., Fernandes Carrijo, T., Marques Cancello, E., Coletto Morales-Corrêa, E., and Castro, A. (2017). Phylogeography of Nasutitermes corniger (Isoptera: Termitidae) in the neotropical region. BMC Evol. Biol. 17, 1–12. doi: 10.1186/s12862-017-1079-8
Del'Arco, J. F., and Bezerra, P. E. L. (1989). “Geologia,” in Geografia do Brasil-Região Centro-Oeste. ed. A. C. Duarte (Río de Janeiro, Brazil: FIBGE-Directoria de Geociências), 35–51.
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., and Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. doi: 10.1371/journal.pbio.0040088
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Fonseca, E. M., Gehara, M., Werneck, F. P., Lanna, F. M., Colli, G. R., Sites, J. W. Jr., et al. (2018). Diversification with gene flow and niche divergence in a lizard species along the south American “diagonal of open formations”. J. Biogeogr. 45, 1688–1700. doi: 10.1111/jbi.13356
Françoso, R. D., Dexter, K. G., Machado, R. B., Pennington, R. T., Pinto, J. R. R., Brandão, R. A., et al. (2020). Delimiting floristic biogeographic districts in the Cerrado and assessing their conservation status. Biodivers. Conserv. 29, 1477–1500. doi: 10.1007/s10531-019-01819-3
Gehara, M., Garda, A. A., Werneck, F. P., Oliveira, E. F., da Fonseca, E. M., Camurugi, F., et al. (2017). Estimating synchronous demographic changes across populations using hABC and its application for a herpetological community from northeastern Brazil. Mol. Ecol. 26, 4756–4771. doi: 10.1111/mec.14239
Gernhard, T., Hartmann, K., and Steel, M. (2008). Stochastic properties of generalised yule models, with biodiversity applications. J. Math. Biol. 57, 713–735. doi: 10.1007/s00285-008-0186-y
Guillot, G., Renaud, S., Ledevin, R., Michaux, J., and Claude, J. (2012). A unifying model for the analysis of phenotypic, genetic, and geographic data. Syst. Biol. 61, 897–911. doi: 10.1093/sysbio/sys038
Guillot, G., Santos, F., and Estoup, A. (2008). Analysing georeferenced population genetics data with Geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 24, 1406–1407. doi: 10.1093/bioinformatics/btn136
Ho, S. Y. (2007). Calibrating molecular estimates of substitution rates and divergence times in birds. J. Avian Biol. 38, 409–414. doi: 10.1111/j.0908-8857.2007.04168.x
Hu, J., Zhong, J. H., and Guo, M. F. (2007). Alate dispersal distances of the black-winged subterranean termite Odontotermes formosanus (Isoptera: Termitidae) in southern China. Sociobiology 50, 1–8.
Hyseni, C., and Garrick, R. C. (2019). The role of glacial-interglacial climate change in shaping the genetic structure of eastern subterranean termites in the southern Appalachian Mountains, USA. Ecol. Evol. 9, 4621–4636. doi: 10.1002/ece3.5065
Janowiecki, M., Clifton, E., Avalos, A., and Vargo, E. L. (2020). Upper thermal tolerance of tropical and temperate termite species (Isoptera: Rhinotermitidae, Termitidae): a test of the climate variability hypothesis in termites. Insect. Soc. 67, 51–57. doi: 10.1007/s00040-019-00727-7
Jenkins, T. M., Jones, S. C., Lee, C. Y., Forschler, B. T., Chen, Z., Lopez-Martinez, G., et al. (2007). Phylogeography illuminates maternal origins of exotic Coptotermes gestroi (Isoptera: Rhinotermitidae). Mol. Phylogenet. Evol. 42, 612–621. doi: 10.1016/j.ympev.2006.11.024
Jouquet, P., Bottinelli, N., Shanbhag, R. R., Bourguignon, T., Traoré, S., and Abbasi, S. A. (2016). Termites: the neglected soil engineers of tropical soils. Soil Sci. 181, 157–165. doi: 10.1097/SS.0000000000000119
Krishna, K., Grimaldi, D. A., Krishna, V., and Engel, M. S. (2013). Treatise on the Isoptera of the world. Bull. Am. Mus. Nat. Hist. 377, 1–200. doi: 10.1206/377.1
Kutnik, M., Uva, P., Brinkworth, L., and Bagnères, A. G. (2004). Phylogeography of two European Reticulitermes (Isoptera) species: the Iberian refugium. Mol. Ecol. 13, 3099–3113. doi: 10.1111/j.1365-294X.2004.02294.x
Lefebvre, T., Châline, N., Limousin, D., Dupont, S., and Bagnères, A. G. (2008). From speciation to introgressive hybridization: the phylogeographic structure of an island subspecies of termite, Reticulitermes lucifugus corsicus. BMC Evol. Biol. 8, 1–13. doi: 10.1186/1471-2148-8-38
Leigh, J. W., and Bryant, D. (2015). PopART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Li, H. F., Ye, W., Su, N. Y., and Kanzaki, N. (2009). Phylogeography of Coptotermes gestroi and Coptotermes formosanus (Isoptera: Rhinotermitidae) in Taiwan. Ann. Entomol. Soc. Am. 102, 684–693. doi: 10.1603/008.102.0413
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Luchetti, A., Marini, M., and Mantovani, B. (2007). Filling the European gap: biosystematics of the eusocial system Reticulitermes (Isoptera, Rhinotermitidae) in the Balkanic peninsula and Aegean area. Mol. Phylogenet. Evol. 45, 377–383. doi: 10.1016/j.ympev.2007.07.015
Martins-Ferreira, M. A. C., Dias, A. N. C., Chemale, F. Jr., and Campos, J. E. G. (2020). Intracontinental uplift of the Brazilian central plateau linked to continental breakup, orogenies, and basin filling, supported by apatite and zircon fission-track data. Arab. J. Geosci. 13, 1–14. doi: 10.1007/s12517-020-05885-8
Mathews, A. (1977). Studies on Termites From Mato Grosso State, Brazil. Río de Janeiro: Academia Brasileira de Ciencias, 159–168.
Miranda, E. A., Batalha-Filho, H., Congrains, C., Carvalho, A. F., Ferreira, K. M., and Del Lama, M. A. (2016). Phylogeography of Partamona rustica (hymenoptera, Apidae), an endemic stingless bee from the neotropical dry forest diagonal. PLoS One 11:e0164441. doi: 10.1371/journal.pone.0164441
Moraes, E. M., Yotoko, K. S. C., Manfrin, M. H., Solferini, V. N., and Sene, F. M. (2009). Phylogeography of the cactophilic species Drosophila gouveai: demographic events and divergence timing in dry vegetation enclaves in eastern Brazil. J. Biogeogr. 36, 2136–2147. doi: 10.1111/j.1365-2699.2009.02145.x
Morrone, J. J., Escalante, T., Rodríguez-Tapia, G., Carmona, A., Arana, M., and Mercado-Gómez, J. D. (2022). Biogeographic regionalization of the Neotropical region: new map and shapefile. An. Acad. Bras. Cienc. 94:e20211167. doi: 10.1590/0001-3765202220211167
Mullins, A. J., Messenger, M. T., Hochmair, H. H., Tonini, F., Su, N. Y., and Riegel, C. (2015). Dispersal flights of the Formosan subterranean termite (Isoptera: Rhinotermitidae). J. Econ. Entomol. 108, 707–719. doi: 10.1093/jee/tov022
Novaes, R. M. L., De Lemos-Filho, J. P., Ribeiro, R. A., and Lovato, M. B. (2010). Phylogeography of Plathymenia reticulata (Leguminosae) reveals patterns of recent range expansion towards northeastern Brazil and southern Cerrados in eastern tropical South America. Mol. Ecol. 19, 985–998. doi: 10.1111/j.1365-294X.2010.04530.x
Novaes, R. M. L., Ribeiro, R. A., De Lemos-Filho, J. P., and Lovato, M. B. (2013). Concordance between Phylogeographical and biogeographical patterns in the Brazilian Cerrado: diversification of the endemic tree Dalbergia miscolobium (Fabaceae). PLoS One 8:e82198. doi: 10.1371/journal.pone.0082198
Oliveira, E. F., Gehara, M., São-Pedro, V. A., Costa, G. C., Burbrink, F. T., Colli, G. R., et al. (2018). Phylogeography of Muller's termite frog suggests the vicariant role of the central Brazilian plateau. J. Biogeogr. 45, 2508–2519. doi: 10.1111/jbi.13427
Oliveira, P. E. D., Raczka, M., McMichael, C. N. H., Pinaya, J. L. D., and Bush, M. B. (2020). Climate change and biogeographic connectivity across the Brazilian cerrado. J. Biogeogr. 47, 396–407. doi: 10.1111/jbi.13732
Ozeki, M., Isagi, Y., Tsubota, H., Jacklyn, P., and Bowman, D. M. (2007). Phylogeography of an Australian termite, Amitermes laurensis (Isoptera, Termitidae), with special reference to the variety of mound shapes. Mol. Phylogenet. Evol. 42, 236–247. doi: 10.1016/j.ympev.2006.06.024
Paradis, E. (2010). Pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Paradis, E., and Schliep, K. (2019). Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Park, Y. C., Kitade, O., Schwarz, M., Kim, J. P., and Kim, W. (2006). Intraspecific molecular phylogeny, genetic variation and phylogeography of Reticulitermes speratus (Isoptera: Rhinotermitidae). Mol. Cell 21, 89–103.
Pennington, R. T., Lavin, M., Prado, D. E., Pendry, C. A., Pell, S. K., and Butterworth, C. A. (2004). Historical climate change and speciation: neotropical seasonally dry forest plants show patterns of both tertiary and quaternary diversification. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 359, 515–538. doi: 10.1098/rstb.2003.1435
Pinheiro, M. H. O., and Monteiro, R. (2010). Contribution to the discussions on the origin of the cerrado biome: Brazilian savanna. Braz. J. Ecol. 70, 95–102. doi: 10.1590/s1519-69842010000100013
Pozo-Santiago, C. O., Velázquez-Msartínez, J. R., Torres-De la Cruz, M., De la Cruz-Perez, A., Capello-García, S., Sanchéz-Gutierrez, F., et al. (2020). El papel de la humedad relativa, temperatura y sustratos en la supervivencia de Nasutitermes corniger. Ecosistemas y Recursos Agropecuarios 7:e2742. doi: 10.19136/era.a7n3.2742
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ramos, A. C., Lemos-Filho, J. P., Ribeiro, R. A., Santos, F. R., and Lovato, M. B. (2007). Phylogeography of the tree Hymenaea stigonocarpa (Fabaceae: Caesalpinioideae) and the influence of quaternary climate changes in the Brazilian Cerrado. Ann. Bot. 100, 1219–1228. doi: 10.1093/aob/mcm221
Raszick, T. J., Dickens, C. M., Perkin, L. C., Tessnow, A. E., Suh, C. P.-C., Ruiz-Arce, R., et al. (2021). Population genomics and phylogeography of the boll weevil, Anthonomus grandis Boheman (Coleoptera: Curculionidae), in the United States, northern Mexico, and Argentina. Evol. Appl. 14, 1778–1793. doi: 10.1111/eva.13238
Recorder, R. S., Werneck, F. D. P., Teixeira, M. Jr., Colli, G. R., Sites, J. W. Jr., and Rodrigues, M. T. (2014). Geographic variation and systematic review of the lizard genus Vanzosaura (Squamata, Gymnophthalmidae), with the description of a new species. Zool. J. Linnean Soc. 171, 206–225. doi: 10.1111/zoj.12128
Resende-Moreira, L. C., Vasconcelos, P. N., Souto, A. P., Menezes, A. P. A., de Lemos-Filho, J. P., and Lovato, M. B. (2017). East-west divergence in central Brazilian Cerrado revealed by cpDNA sequences of a bird-dispersed tree species. Biochem. Syst. Ecol. 70, 247–253. doi: 10.1016/j.bse.2016.12.007
Revell, L. J. (2012). Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Rocha, A. V., Cabanne, G. S., Silveira, L. F., Tubaro, P., and Caparroz, R. (2020). Pleistocene climatic oscillations associated with landscape heterogeneity of the south American dry diagonal explains the phylogeographic structure of the narrow-billed woodcreeper (Lepidocolaptes angustirostris, Dendrocolaptidae). J. Avian Biol. 51:e02537. doi: 10.1111/jav.02537
Schliep, K., Emmanuel Paradis, E., de Oliveira Martins, L., Potts, A., White, T. W., Stachniss, C., et al. (2018). Phangorn: phylogenetic reconstruction and analysis. R package Version 2.0.
Scicchitano, V., Dedeine, F., Bagnères, A. G., Luchetti, A., and Mantovani, B. (2018). Genetic diversity and invasion history of the European subterranean termite Reticulitermes urbis (Blattodea, Termitoidae, Rhinotermitidae). Biol. Invasions 20, 33–44. doi: 10.1007/s10530-017-1510-5
Silva, J. M. C. D., Barbosa, L. C. F., Leal, I. R., and Tabarelli, M. (2017). “The Caatinga: understanding the challenges,” in Caatinga The largest tropical dry forest region in South America. eds. J. M. Cardoso da Silva, I. R. Leal, and M. Tabarelli (Cham: Springer), 3–19.
Singham, G., Othman, A. S., and Lee, C. Y. (2017). Phylogeography of the termite Macrotermes gilvus and insight into ancient dispersal corridors in Pleistocene Southeast Asia. PLoS One 12:e0186690. doi: 10.1371/journal.pone.0186690
Szalanski, A. L., Scheffrahn, R. H., Austin, J. W., Krecek, J., and Su, N. Y. (2004). Molecular phylogeny and biogeography of Heterotermes (Isoptera: Rhinotermitidae) in the West Indies. Ann. Entomol. Soc. Am. 97, 556–566. doi: 10.1603/0013-8746(2004)097[0556:MPABOH]2.0.CO;2
Tembrock, L. R., Timm, A. E., Zink, F. A., and Gilligan, T. M. (2019). Phylogeography of the recent expansion of Helicoverpa armigera (Lepidoptera: Noctuidae) in South America and the Caribbean Basin. Ann. Entomol. Soc. Am. 112, 388–401. doi: 10.1093/aesa/saz019
Tonini, F., Hochmair, H. H., Scheffrahn, R. H., and Deangelis, D. L. (2013). Simulating the spread of an invasive termite in an urban environment using a stochastic individual-based model. Environ. Entomol. 42, 412–423. doi: 10.1603/EN12325
Trautwein, M. D., Wiegmann, B. M., Beutel, R., Kjer, K. M., and Yeaters, D. K. (2012). Advances in insect phylogeny at the Dawn of the postgenomic era. Annu. Rev. Entomol. 57, 449–468. doi: 10.1146/annurev-ento-120710-100538
Tripodi, A. D., Austin, J. W., Szalanski, A. L., McKern, J., Carroll, M. K., Saran, R. K., et al. (2006). Phylogeography of Reticulitermes termites (Isoptera: Rhinotermitidae) in California inferred from mitochondrial DNA sequences. Ann. Entomol. Soc. Am. 99, 697–706. doi: 10.1603/0013-8746(2006)99[697:PORTIR]2.0.CO;2
Tuma, J., Eggleton, P., and Fayle, T. M. (2020). Ant-termite interactions: an important but under-explored ecological linkage. Biol. Rev. 95, 555–572. doi: 10.1111/brv.12577
Vanzolini, P. E. (1963). Problemas faunísticos do cerrado. 1963. Simposio sobre o Cerrado, São Paulo, EDUSP, Brazil.
Vilardi, J. C., Freilij, D., Ferreyra, L. I., and Gómez-Cendra, P. (2021). Ecological phylogeography and coalescent models suggest a linear population expansion of Anastrepha fraterculus (Diptera: Tephritidae) in southern South America. Biol. J. Linn. Soc. 133, 779–795. doi: 10.1093/biolinnean/blab029
Wang, X. F., Auler, A. S., Edwards, R. L., Cheng, H., Cristalli, P. S., Smart, P. L., et al. (2004). Wet periods in northeastern Brazil over the past 210 kyr linked to distant climate anomalies. Nature 432:740e743. doi: 10.1038/nature03067
Werneck, F. P. (2011). The diversification of eastern south American open vegetation biomes: historical biogeography and perspectives. Quat. Sci. Rev. 30, 1630–1648. doi: 10.1016/j.quascirev.2011.03.009
Werneck, F. P., Gamble, T., Colli, G. R., Rodrigues, M. T., and Sites, J. W. J. (2012b). Deep diversification and long-term persistence in the south American ‘dry diagonal’: integrating continent-wide phylogeography and distribution modeling of geckos. Evolution 66, 3014–3034. doi: 10.1111/j.1558-5646.2012.01682.x
Werneck, F. P., Nogueira, C., Colli, G. R., Sites, J. W. Jr., and Costa, G. C. (2012). Climatic stability in the Brazilian Cerrado: implications for biogeographical connections of south American savannas, species richness and conservation in a biodiversity hotspot. J. Biogeogr. 39, 1695–1706. doi: 10.1111/j.1365-2699.2012.02715.x
Wilson, R. J., and Fox, R. (2021). Insect responses to global change offer signposts for biodiversity and conservation. Ecol. Entomol. 46, 699–717. doi: 10.1111/een.12970
Woon, J. S., Atkinson, D., Adu-Bredu, S., Eggleton, P., and Parr, C. L. (2022). Termites have wider thermal limits to cope with environmental conditions in savannas. J. Anim. Ecol. 91, 766–779. doi: 10.1111/1365-2656.13673
Woon, J. S., Boyle, M. J. W., Ewers, R. M., Chung, A., and Eggleton, P. (2019). Termite environmental tolerances are more linked to desiccation than temperature in modified tropical forests. Insect. Soc. 66, 57–64. doi: 10.1007/s00040-018-0664-1
Yeates, D. K., Meusemann, K., Trautwein, M., Wiegmann, B., and Zwick, A. (2016). Power, resolution and bias: recent advances in insect phylogeny driven by the genomic revolution. Curr. Opin. Insect Sci. 13, 16–23. doi: 10.1016/j.cois.2015.10.007
Keywords: Isoptera, phylogeography, molecular phylogeny, biogeography, Chaco, Cerrado, Caatinga
Citation: Kohari KS, Palma-Onetto V, Scheffrahn RH, Vasconcellos A, Cancello EM, Santos RG and Carrijo TF (2022) Evolutionary history of Nasutitermes kemneri (Termitidae, Nasutitermitinae), a termite from the South American diagonal of open formations. Front. Ecol. Evol. 10:1081114. doi: 10.3389/fevo.2022.1081114
Edited by:
Daniel Aguilera-Olivares, Universidad de Concepción, ChileReviewed by:
Juan José Morrone, National Autonomous University of Mexico, MexicoEvandro Marsola Moraes, Federal University of Sao Carlos, Brazil
Copyright © 2022 Kohari, Palma-Onetto, Scheffrahn, Vasconcellos, Cancello, Santos and Carrijo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiago F. Carrijo, dGlhZ29jYXJyaWpvQGdtYWlsLmNvbQ==
†ORCID: Eliana M. Cancello https://orcid.org/0000-0003-3125-6335
 Karolina S. Kohari1
Karolina S. Kohari1 Valeria Palma-Onetto
Valeria Palma-Onetto Tiago F. Carrijo
Tiago F. Carrijo