- 1Graduate School of Science and Engineering, Chiba University, Chiba, Japan
- 2Graduate School of Science, Chiba University, Chiba, Japan
Organisms have endogenous timekeeping system(s) to coordinate their biological processes with environmental cycles, allowing adaptation to external rhythmic changes in their environment. The change in endogenous rhythms could contribute to range expansion in a novel rhythmic environment. We hypothesized that populations of the freshwater snail near estuaries show a circatidal rhythm to synchronize with the tidal cycle. We compared the behavioral and gene expression rhythms between non-tidal and tidal populations of the freshwater snail, Semisulcospira reiniana. Individuals inhabiting tidal areas exhibited a rhythmic activity pattern coordinated with the tidal cycle under both field and laboratory conditions, but individuals inhabiting upstream non-tidal areas showed a circadian activity pattern. The proportion of circadian oscillating genes was greater in non-tidal than in tidal individuals, while that of circatidal oscillating genes was greater in tidal than in non-tidal individuals. Additionally, transcriptome-wide population genetic analyses revealed that these two adjacent populations can be clearly distinguished genetically, though the genetic distance was very small. Our results provide evidence of the shift in an endogenous rhythm via range expansion to a novel rhythmic environment. The changes in a small number of genes and/or phenotypic plasticity may contribute to the difference in the endogenous rhythms between non-tidal and tidal populations.
Introduction
Life on Earth is exposed to a variety of periodic environmental changes. The Earth’s rotation provides a daily cycle and seasonal variation, resulting in periodic changes in environmental factors, such as light and temperature. In marine environments, the rise and fall of sea levels every 12.4 h and the neap-spring tidal cycle are caused by the combination of the inertial force of the Earth and the gravitational forces of the sun and moon. Organisms have endogenous timekeeping system(s) that coordinates their biological processes in rhythmic environments. Circadian clocks are the best characterized among biological clocks and are ubiquitous in organisms (Pittendrigh, 1960; Sharma, 2003). Even in the absence of environmental time cues, circadian clocks sustain a period of approximately 24 h. Circadian clocks can be entrained to the environmental cues and are temperature-compensated (Dunlap, 1999; Bell-Pedersen et al., 2005). To adapt to complicated environmental cycles, some species have endogenous rhythms other than circadian rhythms. Some marine species show rhythmic behaviors with a period of approximately 12.4 h synchronized to the tidal cycle (i.e., circatidal rhythm) (Barnwell, 1966; Chabot et al., 2004; Satoh et al., 2008; Zhang et al., 2013). Several species exhibit semi-lunar or lunar rhythmicity (Takemura et al., 2010; Zantke et al., 2014) and annual rhythmicity (Heideman and Bronson, 1994; Guyomarc’h and Guyomarc’h, 1995).
Biological clocks could contribute to adaptation to rhythmic environments by allowing organisms to anticipate environmental changes and prepare accordingly (Yerushalmi and Green, 2009). For instance, Apteronemobius asahinai, which inhabits mangrove forest floors, can anticipate high tides by the endogenous clock and escape from submergence (Satoh et al., 2008). Biological clocks also play important roles in the regulation of genes known as clock-controlled genes (CCGs). They are associated with the temporal regulation of many physiological processes synchronized with periodic environmental variation (Miller et al., 2007; Bozek et al., 2009; Gamble et al., 2014). Given that phase and period of biological clocks are suggested to affect fitness in the natural condition (Yerushalmi et al., 2011; Rubin et al., 2017, 2018), modification of these parameters could coincide with the range expansion to a novel rhythmic environment. The mutation of genes governing biological clocks causes the adaptive evolution of phenological traits and could lead to range expansion to novel habitats with different rhythmic environments (Urbanski et al., 2012; Zakhrabekova et al., 2012). In addition, the genetic and/or non-genetic modification of endogenous rhythm should contribute to local adaptation and subsequent range expansion as well (Kaiser, 2014; Kaiser et al., 2021).
Individuals may have endogenous rhythms specific to dominant environmental cycles in their habitats. Because a river is adjacent to the sea, there is a steep gradient of the strength of periodic variation caused by tidal cycles. In addition to the light-dark (LD) cycle of 24 h period and daily changes in water temperature, animals near the estuary are exposed to an environmental cycle of a 12.4 h period in salinity and water level. Because the tidal cycle causes drastic environmental changes in both biotic and abiotic factors, it is advantageous for a given species to anticipate tidal changes by the circatidal clock (Tessmar-Raible et al., 2011). Thus, we hypothesized that populations near the estuary showed a circatidal rhythm to synchronize with the tidal cycle. The relationship between the modification of endogenous rhythms and range expansion can be revealed by focusing on the adaptation of freshwater species to tidal areas in a river.
To investigate the accurate endogenous rhythm, the analysis of gene expression rhythms should be informative for us because the activity rhythm is sometimes irregular owing to the effect of environmental variables other than zeitgeber, a process termed masking (Mrosovsky, 2009; Helm et al., 2017; Kronfeld-Schor et al., 2017). Unfortunately, little is known about the molecular basis of circatidal rhythm despite recent advances in molecular techniques (Zhang et al., 2013). However, examining transcriptome dynamics helps in understanding the rhythmicity of gene expression and detecting candidate circatidal clock genes or associated genes. The identification of oscillators that generate circatidal rhythmicity can be a key step in understanding the molecular mechanism by which the circatidal clock regulates the rhythmic process of physiology and activity.
The freshwater snail Semisulcospira reiniana mainly inhabits freshwater areas in Japan, but it can also inhabit tidal areas in several rivers. To evaluate whether populations established near the estuary have a circatidal rhythm, we compared the endogenous rhythms of snails between non-tidal and tidal populations. First, we investigated the activity rhythms of each population in the field and the laboratory. We then performed a transcriptome analysis to reveal the gene expression rhythm of each population under constant conditions. Finally, we investigated the genetic differentiation between the two populations using single nucleotide polymorphism (SNP) data obtained from the transcriptome analysis.
Materials and methods
Species and sampling sites
Semisulcospira reiniana is a common freshwater snail that inhabits freshwater areas in Japan. Sampling sites were set in non-tidal (3–5 m above sea level) and tidal areas (<1 m above sea level) of the Kiso River in Japan. In the present study, we refer to river areas where the water level fluctuates due to the tidal cycle as tidal areas and individuals inhabiting there as tidal individuals. These populations were approximately 20 km apart along the river. Fluctuations in the water level and salt concentration caused by the tidal cycle occur only in tidal areas. While snails are submerged at high tide, they go under the mud at low tide because the riverbed is exposed to air. The salt concentration is nearly 0% but slightly increases at high tide. Therefore, snails in tidal areas are exposed to both daily and tidal environmental cycles. In May 2018, June 2019, and June 2021, we collected snails from two areas of the Kiso River for behavioral assays and transcriptome analysis in the laboratory. In August 2020, we collected snails and conducted behavioral observations in the field. The animal study was reviewed and approved by the ethics committee of Chiba University.
Activity rhythm in the field
To investigate the activity patterns of adult S. reiniana exposed to field conditions, the activity of individuals collected from non-tidal and tidal areas was examined in August 2020. To collect non-tidal and tidal individuals whose habitats were located 20 km apart from each other just before the onset of the series of observations, the non-tidal individuals were kept in tanks in the tidal area near our research base. The tanks were filled with river water collected from a non-tidal area and kept in the shade on the riverbank. The tidal individuals collected were enclosed in mesh bags, which were placed in the river to expose the stimulus of the tidal cycle. Ten individuals of non-tidal and tidal populations, respectively, were collected simultaneously for behavioral observations at 13:30 (low tide), 19:30 (high tide), 1:30 (low tide), and 7:30 (high tide). Snails between 23 and 33 mm in shell height were used for subsequent analysis. All observations were conducted at room temperature (approximately 25°C) without tidal stimulation. The first and fourth observations were conducted under light conditions, and the second and third observations were conducted under dark conditions corresponding to the natural photoperiod. Individuals were separately placed into individual bowls (φ13 cm × 3.5 cm) filled with freshwater from the river and monitored using a model 400-CAM061 camera (Sanwa Direct, Japan) for 60 min. Images were taken every 30 s and were used to create time-lapse movies. We tracked the position of each individual using UMATracker tracking software (Yamanaka and Takeuchi, 2018). Using the snail trajectories, the total locomotion distance traveled in the hour was calculated for each individual.
Activity rhythm under constant conditions
To investigate the endogenous activity rhythm of S. reiniana, individuals collected from non-tidal and tidal populations were examined under constant laboratory conditions. Eight snails per population were individually placed in containers (13 cm × 11.7 cm, 4.7 cm in height) filled with freshwater. Snails were observed under the constant dark (DD) condition at a temperature of 23°C in the laboratory. No food was supplied during the observation. Water was circulated using a pump to keep the water clean. Individuals were monitored for 120 h (10 tidal cycles) and images were taken every 30 s. After averaging the coordinates of each individual every three frames, the locomotion distance of each individual for every hour was calculated using the same procedure as the activity rhythm in the field.
RNA sampling
Adult individuals were collected from non-tidal and tidal areas around the time of low tide. They were preserved in containers without water under the DD condition at a constant temperature (23°C) for tissue collection. Starting at subjective high tide in the evening on the same day, three individuals per population were dissected every 3.1 h for 49.6 h (i.e., 17 sampling time points). The epidermis of each individual was preserved in 750 μL of RNAlater Stabilization Solution (Invitrogen, USA). The samples were stored at 4°C for approximately 3 h and then stored at −80°C until total RNA was extracted.
RNA extraction and sequencing
Total RNA was extracted using the Maxwell 16 LEV Plant RNA Kit with the Maxwell 16 Research Instrument (Promega, USA), according to the manufacturer’s instructions. Electrophoresis was performed on 1% agarose gel to evaluate RNA degradation. RNA concentrations were estimated using a Qubit 2.0 fluorometer (Invitrogen, USA). RNA purity was estimated using BioSpec-nano (Shimadzu, Japan). Since our experiment was not designed to compare the expression level of genes between groups, biological replicates for each time point were not sampled. However, to reduce variations between individuals, three individuals were pooled per sample. Total RNA from three individuals in the same population at one sampling time point was pooled in equal amounts prior to RNA sequencing (pooled RNA-seq). RNA integrity was checked by Bioanalyser. A coding DNA (cDNA) library was constructed using the (TruSeq RNA Sample Prep Kit, Illumina). Paired-end (150 bp) pooled RNA-seq was performed using the Illumina NovaSeq6000 platform. After the removal of adaptor sequences and low-quality reads using Trimmomatic (version 0.38) (Bolger et al., 2014), quality control was performed using FastQC (version 0.11.8).1 The remaining high-quality reads of all samples were used for de novo assembly using Trinity software (version 2.6.5) (Grabherr et al., 2011). To estimate gene expression levels, all reads from each sample were mapped to the reference transcripts using RSEM software (version 1.3.0) (Li and Dewey, 2014), providing the FPKM of each gene. Putative coding regions and coding DNA sequences (CDS) were extracted from the reference transcripts using TransDecoder (version 5.5.0),2 providing reference transcripts containing only CDS. Using all CDS transcripts, we searched for homologs of every S. reiniana gene using BLAST for all protein sequences of Crassostrea gigas (oyster_v9, Wang et al., 2012). Genes with the best hit and an e-value < 0.0001 were used to analyze gene expression rhythmicity.
Population genetics
Reference transcripts containing only CDS were clustered based on sequence identities of 90% to remove redundancy using the CD-HIT program (Li and Godzik, 2006). All reads from each sample were mapped to the clustered CDS reference using the STAR program. We then used the GATK4 (version 4.2.0) (McKenna et al., 2010) for SNPs calling. Because three individuals were pooled per sample, the sample ploidy option of GATK4 was set to six. Homologs of C. gigas detected in the same manner as described in the previous section were used to estimate the genetic structure of the two populations. We performed discriminant analysis of principal components (DAPC) and principal component analysis (PCA) using adegenet (Jombart, 2008), and calculated FST between the two populations using PopGenome (Pfeifer et al., 2014).
Statistical analyses
We evaluated whether the locomotor distances of the snails in the field differed between the times corresponding to high and low tides using the general linear model. Because the activity level tended to increase with time after the onset of the experiment (i.e., the time after the enclosure in a tank or mesh bags), irrespective of the population, probably because of acclimation, the residuals of the regression of the activity level at the time after the onset of the experiment were used to detect the daily rhythm of activity. The LD condition and tidal levels were included as explanatory variables. The activity rhythm of each population in the laboratory was determined via Lomb–Scargle periodogram using ActogramJ, a software package based on ImageJ for the analysis and visualization of chronobiological data (Schmid et al., 2011), using the mean locomotor distance of all individuals for each population. Gene expression rhythmicity was analyzed using two programs, JTK_CYCLE (Hughes et al., 2010) and RAIN (Thaben and Westermark, 2014) to identify rhythmic components and estimate their phase. FPKM data of genes with average value > 0 and oscillation amplitude (peak/trough) ≥ 1.3 were used. Genes with significant rhythmic expression in the 12.4 and 24.8 h period (P < 0.01) detected by at least one of two programs were identified as circatidal oscillating genes and circadian oscillating genes, respectively. To investigate whether pathways in salinity response are regulated by circatidal rhythm, we listed circatidal oscillating genes involved in salinity response using transcriptome data from our previous study (Yokomizo and Takahashi, 2020), referring to data in the Dryad Data Archive at https://doi.org/10.5061/dryad.jdfn2z37w. All statistical analyses were performed using R 4.0.4.
Results
Activity rhythm in the field
The activity of individuals from the tidal area was relatively higher at 19:30 and 7:30, and lower at 13:30 and 1:30 (Supplementary Figure 1). The level of activity corresponded to the tidal cycle rather than the LD cycle, and it was higher during high tide (tide: F = 4.27, P = 0.05; LD: F = 4.81, P = 0.91; tide × LD: F = 0.01, P = 0.93; Figure 1A). In contrast, non-tidal individuals were more active at 19:30, 1:30, and 7:30 than at 13:30 (Supplementary Figure 1). The level of activity changed with the LD cycle rather than the tidal cycle and was higher under dark conditions (tide: F = 2.56, P = 0.12; LD: F = 0.01, P = 0.04; tide × LD: F = 0.14, P = 0.71; Figure 1A), indicating nocturnal activity patterns.
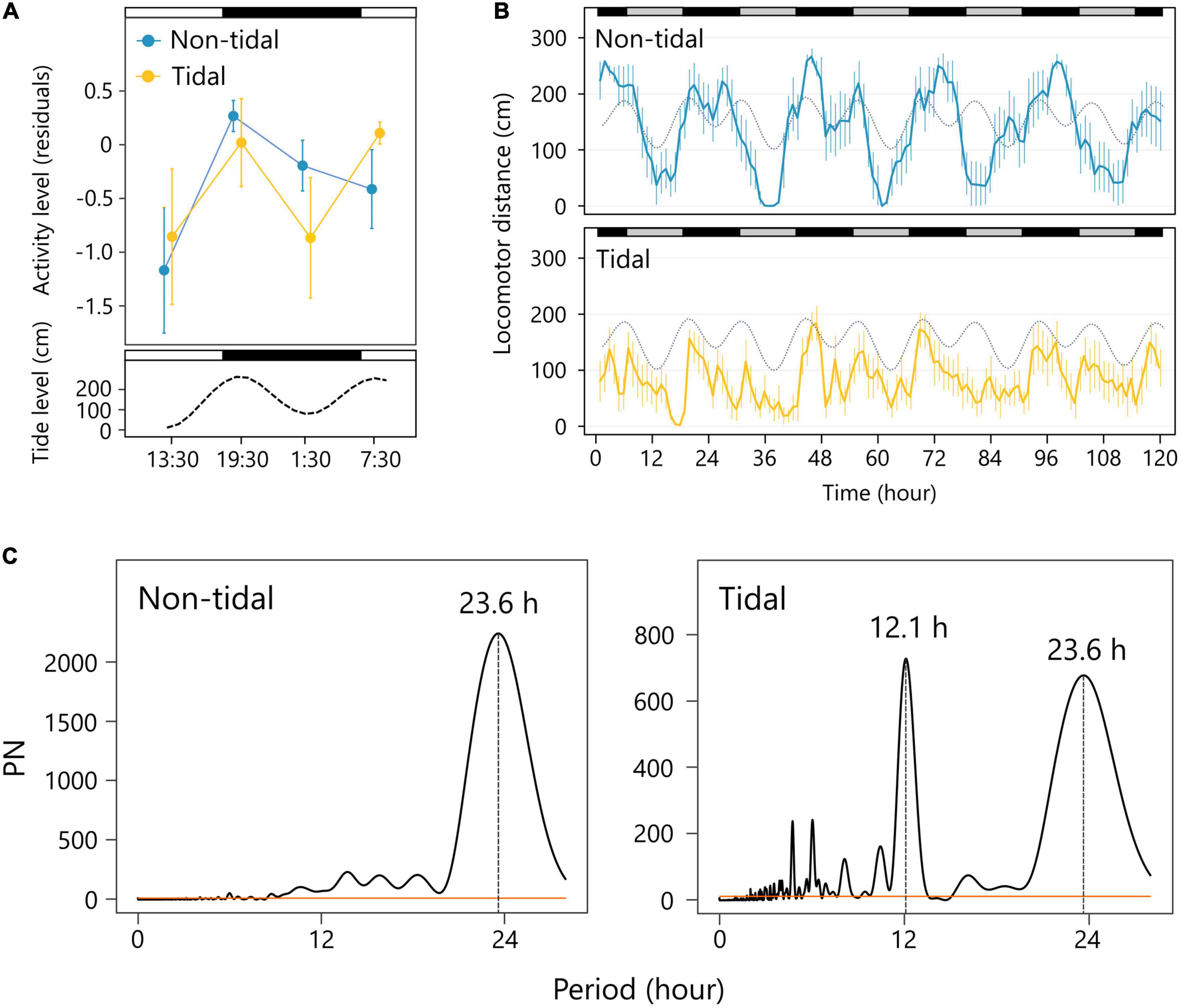
Figure 1. Activity patterns of non-tidal and tidal individuals. (A) Daily changes in the activity level (residuals of the regression of locomotion distance on the time after the onset of the experiment) in the field. Error bars depict the standard error of the mean (SEM). The non-tidal and tidal populations in each observation time are shown in blue and yellow, respectively. The white and black bars represent light and dark conditions, respectively. The lower panel represents a tidal level. (B) Locomotor distance of the non-tidal and tidal population for 5 days under the constant dark (DD) condition in the laboratory. Error bars are SEM. Subjective day and night are shown as gray and black bars above. The tide level of the natural environment is shown as a gray dotted line. (C) Lomb–Scargle periodogram analysis. Peaks in the circatidal and circadian range are noted.
Activity rhythm in the laboratory
Snails exhibited rhythmic activity patterns in the laboratory under DD conditions. Non-tidal individuals exhibited circadian activity rhythm with a period of 23.6 h (Figures 1B,C), indicating that their activity patterns were synchronized with the daily cycle, but not with the tidal cycle. In contrast, the activity of individuals from the tidal population increased with the time of high tide in the laboratory under DD conditions, despite no tidal stimulation (Figure 1B). The significant periodicities of 12.1 and 23.6 h were detected in the mean locomotor distance, indicating that endogenous activity rhythms were synchronized with both the daily and tidal cycles (Figure 1C).
Gene expression rhythm under constant conditions
We obtained 1,686,925 contigs, with a mean length of 556 bp. Among the 978 metazoan core gene orthologs, 975 (99.1%) were identified. Using all the contigs, 225,139 putative CDS were estimated using TransDecoder. The CDS contained 99,784 contigs annotated against all protein sequences of C. gigas. These annotated sequences comprised 40,506 genes, of which 11,967 and 13,044 in non-tidal and tidal populations, respectively, met the requirements mentioned in the “Materials and methods” section.
We detected 546 (4.0%) and 333 (2.3%) circadian oscillating genes in non-tidal and tidal samples, respectively (Figures 2A,B and Supplementary Dataset 1). The proportion of genes oscillating in the circadian period (i.e., 24.8 h) was greater in non-tidal individuals than in tidal individuals (Fisher’s exact test, P < 0.001). Eight circadian oscillating genes kept the same period in the two populations (Supplementary Figure 2). The distribution of peak phase estimated by the JTK_CYCLE and RAIN programs revealed that genes whose expression peaked in the evening were most frequent among the circadian oscillating genes (Figure 2C). In the non-tidal population, peak expression was coordinated with their activity patterns. We detected 303 (2.2%) and 364 (2.6%) circatidal oscillating genes in non-tidal and tidal samples, respectively (Figures 2A,B and Supplementary Dataset 1). The proportion of genes oscillating in a circatidal period (i.e., 12.4 h) was greater in tidal individuals than in non-tidal individuals (Fisher’s exact test, P = 0.03). Eight circatidal oscillating genes kept the same period in the two populations (Supplementary Figure 2). The expression peaks of most circatidal oscillating genes in the tidal population coincided with subjective high tide (Figure 2C), which was synchronized with the activity pattern shown in Figure 1. In non-tidal individuals, the majority of circatidal oscillating genes showed the expression peak of time of low or rising tide. These timings correspond to midnight and early afternoon.
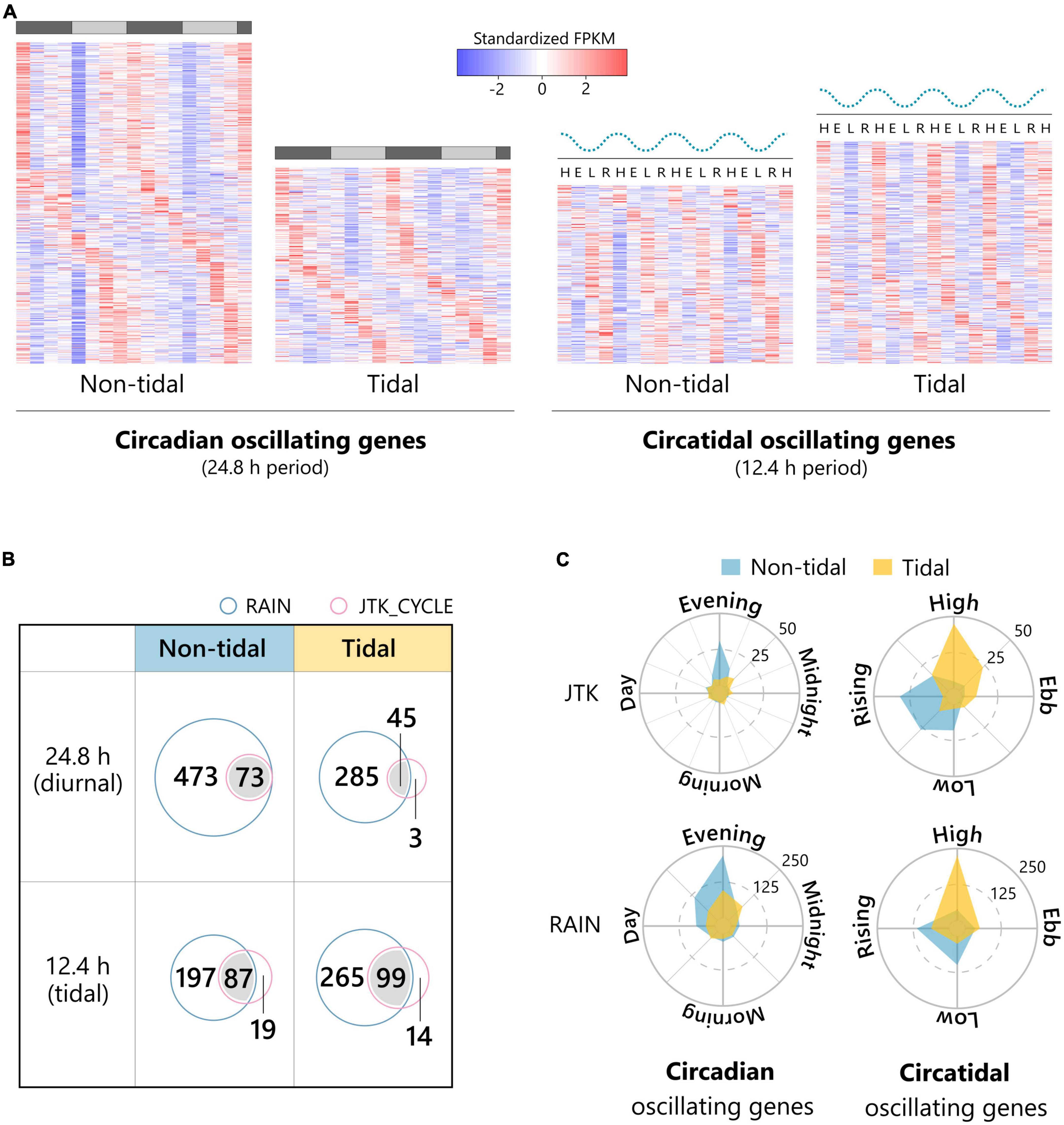
Figure 2. Rhythmic transcripts with circadian and circatidal period in the non-tidal and tidal individuals. (A) Heatmaps of standardized expression patterns of rhythmic transcripts with 24.8 h (diurnal) or 12.4 h (tidal) period in the non-tidal and tidal individuals under DD conditions detected by JTK_CYCLE or RAIN. The dotted wavy lines above the heatmaps of tidal transcripts represent the tide level of the natural environment. H: subjective high tide, E: subjective ebb tide, L: subjective low tide, R: subjective rising tide. The gray and black bars above the heatmaps of diurnal transcripts represent the subjective day and night, respectively. (B) Venn diagrams detailing the number of rhythmic transcripts with 24.8 h (diurnal), or 12.4 h (tidal) period detected by JTK_CYCLE (pink circle) and/or RAIN (light blue circle). (C) The distribution of the phase of circadian and circatidal oscillating genes in the non-tidal and tidal individuals. Oscillating genes detected by both JTK_CYCLE and RAIN were included in both upper and bottom radar charts.
We identified some core clock genes. None showed circadian rhythmicity in either population (Figure 3 and Supplementary Figure 3). Among significantly oscillating transcripts, 16 genes exhibited circadian rhythmicity in their expression in the non-tidal population, but circatidal rhythmicity in the tidal population (Figure 3 and Supplementary Figure 4). Three genes with clearly different rhythmicity between populations are shown in Figure 3 (Wnt glycoprotein, chromatin remodeling ATPase INO80, and VPRBP protein). The Wnt glycoprotein functions as a ligand for the Wnt pathway. This glycoprotein is involved in a variety of cellular responses (Arend et al., 2013). Chromatin remodeling ATPase INO80 is associated with chromatin remodeling by either sliding nucleosomes along with DNA or exchanging histones within nucleosomes (Bao and Shen, 2011). VPRBP is a component of the DDB1-CUL4AVPRBP E3 ubiquitin ligase complex and is involved in the induction of G2 arrest (Belzile et al., 2007). As these genes do not function as clock genes, they are likely to be CCGs. The presence of these genes indicates the existence of different biological clocks in non-tidal and tidal populations or indicates that they reflect the dominant cycle that snails usually face in their environment.
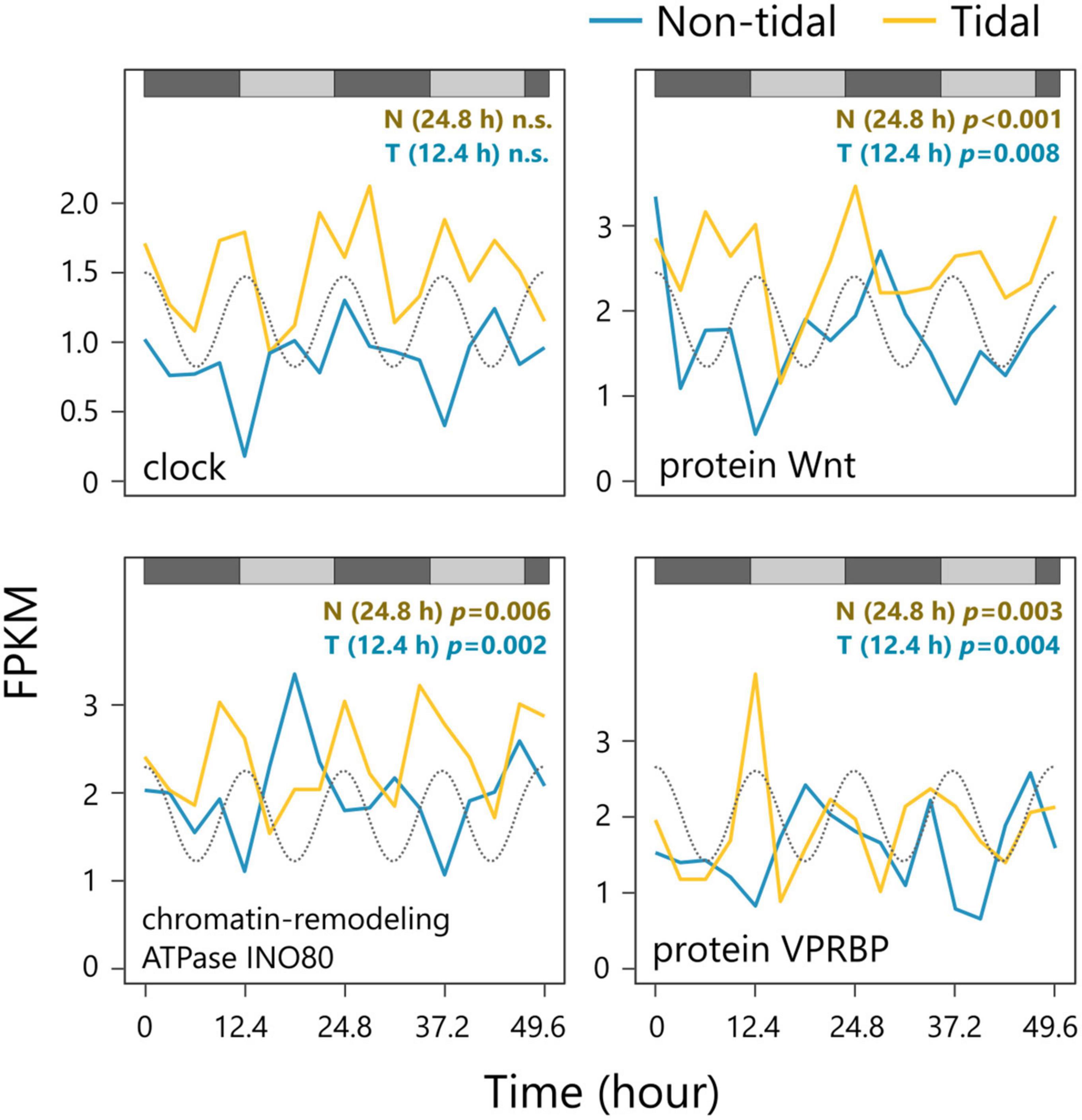
Figure 3. Examples of the expression patterns of the circadian clock gene (upper left) and oscillating genes with a different period between non-tidal and tidal populations (upper right, bottom right, and bottom left). The tide level of the natural environment is shown as a gray dotted line. The gray and black bars represent the subjective day and night, respectively. Rhythmicity of a period of 12.4 and 24.8 h were tested using RAIN for tidal (T) and non-tidal (N) populations, respectively.
Since salt concentration in the sampling site of the tidal population slightly fluctuates by the tidal cycle, salinity response pathways may be regulated by the circatidal rhythm. Among the circatidal oscillating genes, 41 were found in the list of genes involved in the salinity response (Supplementary Table 1). Most of these genes were highly expressed under high salinity conditions. Although we found rhythmic genes with various expression phases, there were few genes with expression peaks during rising tide.
Population differentiation
We identified 35,864 SNPs on the contigs annotated against all protein sequences of C. gigas using a BLAST search. Composition plot by DAPC using these SNPs distinguished non-tidal and tidal populations (Supplementary Figure 5). The two populations separated along PC1 and overlapped along PC2 (Figure 4). Although PCA indicated that non-tidal and tidal populations could be distinguished, the proportion of variance of PC1 was small (4.8%). The bayesian information criterion (BIC) value was lowest when one cluster was estimated, indicating that the most likely number of groups in the sample was one (Supplementary Figure 6). FST value between the non-tidal and tidal populations was estimated at 0.018, indicating quite low genetic differentiation.
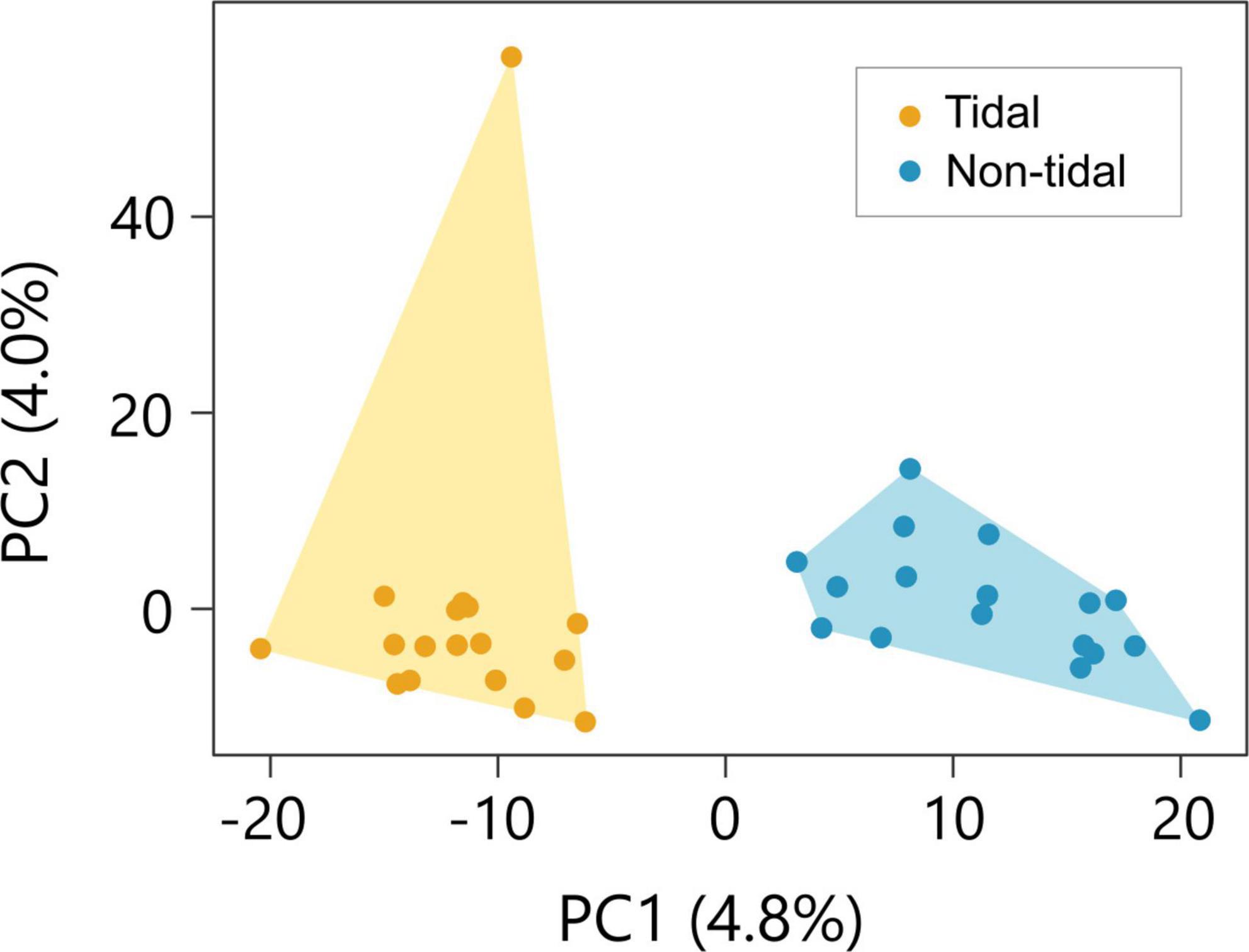
Figure 4. Genetic structure of the non-tidal and tidal population. Samples collected in the tidal area and non-tidal area are shown in yellow and blue, respectively. Principal component analysis (PCA) on the non-tidal and tidal individuals. The two populations can be distinguished along the PC1.
Discussion
Biological clocks could contribute to the improvement of fitness via the entrainment of biological processes and the environmental cycle (Yerushalmi and Green, 2009). Circadian clocks are considered adaptive in an environment with a diurnal cycle. An additional or modified timekeeping system synchronized with the tidal cycle can be helpful in the tidal environment. Although much evidence suggests that the circatidal rhythm is adaptive in intertidal organisms (Tessmar-Raible et al., 2011; Goto and Takekata, 2015; Häfker and Tessmar-Raible, 2020), the mechanism of the evolution of the circatidal clock during tidal adaptation remains unclear. In the present study, we focused on freshwater organisms, which have expanded their distribution to tidal reaches, and investigated their endogenous rhythms. Our results concerning the difference in activity and gene expression rhythm between populations could be evidence of the expression of habitat-specific endogenous rhythms in a single species. The small genetic distance between the two populations suggests that they have a similar genetic basis of clocks and plastically show different endogenous rhythms depending on the environmental cycle. Although we cannot deny that the evolution of a small number of genes contributing to distinguishing two populations results in differentiated clocks, the further experiment would be needed to prove the hypothesis.
First, we clarified the activity patterns of S. reiniana under natural and DD conditions. Snails in a non-tidal population exhibited nocturnal activity patterns in the field, consistent with previous research (Urabe, 1998). Given that this observation was conducted in summer, the low daytime activity could be attributed to high temperatures. To assess the endogenous activity rhythms of non-tidal individuals, we conducted behavioral observations under DD conditions. Periodogram analysis detected unimodal rhythmicity of the circadian period, suggesting that non-tidal individuals have an endogenous rhythm corresponding to the natural LD cycle. Snails in tidal populations tended to be active at high tide, suggesting that they responded to the rising water level caused by the tidal cycle. This finding also suggests that a slight increase in salt concentration at high tide does not reduce snails’ activity. This activity pattern was also observed under DD conditions in the laboratory, with no tidal stimulation. We detected a free-running period close to the natural tidal cycle in addition to the circadian period, supporting our hypothesis that snails in tidal populations have a circatidal rhythm. Additionally, tidal individuals exhibited an activity rhythm with a period of 24 h. Although we cannot rule out the possibility that this periodicity appears computationally because 24 is twice as large as 12, this is more likely to be a circadian rhythm since activity at night was higher than during the day. Taken together, the data indicate that S. reiniana expresses habitat-specific endogenous rhythm.
In addition to the activity rhythm, the present study focused on gene expression rhythms under DD conditions with no tidal stimulation. The proportion of circadian oscillating genes was greater in non-tidal individuals than in tidal individuals, whereas the opposite was true for circatidal oscillating genes. We identified some genes whose expression showed circadian rhythmicity in the non-tidal population but showed circatidal rhythmicity in the tidal population. These findings suggest that endogenous rhythms could differ in their periods between non-tidal and tidal populations, and some of the CCGs are regulated by putative different clocks. In the non-tidal population, however, we also found hundreds of rhythmic transcripts with a 12.4-h period. This might be a conservative rhythm among species derived from the circadian clock, as reported in mammals (Hughes et al., 2009; Zhu et al., 2017; Zhu, 2020). Rhythmicity with a period of 12 h in mammalian clocks is suggested to be harmonics of the circadian clock (Hughes et al., 2009). In our experiments, the beginning of the dark phase and the main high tide coincide, and thus, we could not distinguish between genes with 12 h periodicity driven by harmonics and those synchronized with tides.
We did not find any significant rhythmicity in the identified core clock genes in either population. This finding is not surprising in tidal populations, as the lack of circadian rhythmicity in core clock genes has also been reported in other intertidal animals (Zhang et al., 2013; Schnytzer et al., 2018; Satoh and Terai, 2019). Alternatively, the absence of circadian rhythmicity in core clock genes was due to the weak peripheral clocks in the epidermis or samples including more than one tissue with not synchronized peripheral clocks leading to a masking effect. Few common circadian or circatidal oscillating genes in the two populations may also result from this limitation. Although homologs of the core clock genes identified here might lack the function associated with the circadian clock, the circadian activity rhythm under constant conditions suggests the presence of a circadian timekeeping system in S. reiniana. It is unlikely that they have a novel timekeeping system without circadian clock genes. We cannot deny that the arrhythmic expression pattern in core circadian clock genes is caused by the lack of information on variability between biological replicates due to the pooled RNA-seq. The mechanism that governs the activity rhythm with a period of 23.6 h in the non-tidal population is unclear. Transcriptome analysis revealed that some genes exhibited different expression rhythms between populations. We found 16 genes, including those encoding Wnt, which showed circadian and circatidal oscillations in non-tidal and tidal populations, respectively. Previous studies have reported that the Wnt signaling pathway is regulated by circadian clock genes (Guo et al., 2012; Fuwei et al., 2013). The fact that genes directly regulated by the circadian clock exhibit circatidal rhythmicity in their expression supports the hypothesis of a shift in the endogenous rhythm between populations. Considering the results of the behavioral assay, S. reiniana in the tidal area likely has a circatidal rhythm.
In addition to the water level, the salt concentration at the tidal site fluctuated from 0 to 0.5%, according to the tidal cycle. We detected 41 genes that were related to the salinity response and oscillated in circatidal rhythmicity. Given that S. reiniana is active at high tide and low salinity conditions (Yokomizo and Takahashi, 2020), the one gene that was highly expressed under such conditions should be associated with locomotor function. In contrast, the 10 genes highly expressed under low tide and high salinity conditions should be expressed when snails are inactive. We found eight genes that were highly expressed under high tide and high salinity conditions, indicating that they are related to a direct response to salinity. These findings suggest that some of the pathways involved in the salinity response are controlled by the circatidal rhythm. Therefore, we hypothesized that S. reiniana in a tidal area could anticipate the variation in salt concentration due to the tidal cycle and regulate gene expression via the circatidal rhythm. Further investigation is needed on the relationship between circatidal rhythm and salinity response in freshwater snails.
Although our study indicates that S. reiniana in a tidal area has a circatidal rhythm, the nature of the circatidal clock remains controversial in any organism. Based on previous studies of tidal animals, three major hypotheses have been proposed to explain tidally synchronized biological rhythms. The first hypothesis is that there is a biological clock with a 12.4-h period other than the circadian clock (Naylor, 2010). In the second hypothesis, Palmer (1995) argued for the existence of two unimodal clocks with 24.8-h coupled in antiphase, termed circadian clocks. Finally, the third hypothesis suggests that a single oscillator governs both circadian and circatidal patterns (Enright, 1976; Mat et al., 2016). We could not evaluate whether the circadian and circatidal rhythms of snails are regulated by a single or multiple biological clock(s). Previous research in mangrove crickets showed that silencing the circadian core clock genes does not disrupt the circatidal rhythm (Takekata et al., 2012, 2014), and molecular mechanisms different from the circadian clock might exist in intertidal animals. In contrast, C. gigas under subtidal field conditions exhibits tidal rhythmicity of circadian clock genes, suggesting that a single clock could entrain behavioral patterns at tidal and daily rhythms (Tran et al., 2020). Revealing the molecular mechanisms of the circatidal clock is an ongoing challenge in chronobiology. Given that the first proto-clocks evolved in an aquatic environment, developing a comprehensive understanding of the circatidal clocks should contribute to a more holistic understanding of the evolution of the biological clock (Rock et al., 2022).
Our results revealed a circatidal rhythm only in snails in tidal areas. Although genetic differentiation estimated by FST was quite small, DAPC and PCA indicated that non-tidal and tidal populations were apparently isolated, suggesting differentiation of a small number of genes. Therefore, our results from the population genetic analyses suggest recent isolation and subsequent slight divergence between the two populations. We provide two hypotheses for the fact that different endogenous rhythms were observed between tidal and non-tidal populations. The first hypothesis is that only the tidal population has the circatidal clock resulted from the adaptive evolution of a small number of genes. Such genetic adaptation may lead to the clear but slight genetic differentiation between tidal and non-tidal populations. The second hypothesis is that differences in environmental cycles in habitats plastically determine the endogenous rhythm of S. reiniana. Even non-tidal individuals have the potential to show the circatidal rhythm when exposed to a tidal environment. If it is true, however, we cannot deny that the ability of plasticity of biological clocks evolved only in the river studied or only in the lower reaches of the river. Further investigations on the ability of plasticity on endogenous rhythm in the individuals derived from other populations of Kiso River and populations in the independent rivers can provide a deeper understanding of the evolution of biological clocks.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving animals were reviewed and approved by the Ethics Committee of Chiba University.
Author contributions
TY performed the experiments, drafted the manuscript, and conducted the data analysis. Both authors designed the experiments, edited the manuscript, and approved the submitted version.
Funding
This work was supported by the research grants JSPS and KAKENHI (grant numbers JP17H03729 and JP20K06822), Fujiwara Natural History Public Interest Incorporated Foundation and Research Institute of Marine Invertebrates to YT, and Grant-in-aid for JSPS fellows (grant number JP21J20682) to TY.
Acknowledgments
Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1078234/full#supplementary-material
Footnotes
- ^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ https://github.com/TransDecoder/TransDecoder/
References
Arend, R. C., Londoño-Joshi, A. I., Straughn, J. M., and Buchsbaum, D. J. (2013). The Wnt/β-catenin pathway in ovarian cancer: A review. Gynecol. Oncol. 131, 772–779. doi: 10.1016/j.ygyno.2013.09.034
Bao, Y., and Shen, X. (2011). SnapShot: Chromatin remodeling: INO80 and SWR1. Cell 144, 158–158.e2. doi: 10.1016/j.cell.2010.12.024
Barnwell, F. H. (1966). Daily and tidal patterns of activity in individual fiddler crab (Genus Uca) from the Woods Hole region. Biol. Bull. 130, 1–17. doi: 10.2307/1539948
Bell-Pedersen, D., Cassone, V. M., Earnest, D. J., Golden, S. S., Hardin, P. E., Thomas, T. L., et al. (2005). Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556. doi: 10.1038/nrg1633
Belzile, J. P., Duisit, G., Rougeau, N., Mercier, J., Finzi, A., and Cohen, É. A. (2007). HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 3:e85. doi: 10.1371/journal.ppat.0030085
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bozek, K., Relógio, A., Kielbasa, S. M., Heine, M., Dame, C., Kramer, A., et al. (2009). Regulation of clock-controlled genes in mammals. PLoS One 4:e4882. doi: 10.1371/journal.pone.0004882
Chabot, C. C., Kent, J., and Watson, W. H. I. (2004). Circatidal and circadian rhythms of locomotion in Limulus polyphemus. Biol. Bull. 207, 72–75. doi: 10.2307/1543630
Dunlap, J. C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. doi: 10.1016/S0092-8674(00)80566-8
Enright, J. T. (1976). Plasticity in an isopod’s clockworks: Shaking shapes form and affects phase and frequency. J. Comp. Physiol. A 107, 13–37. doi: 10.1007/BF00663916
Fuwei, L., Yijia, C., Xiaolong, L., Qing, Z., and Zhen, T. (2013). Over-expression of circadian clock gene Bmal1 affects proliferation and the canonical Wnt pathway in NIH-3T3 cells. Cell Biochem. Funct. 31, 166–172. doi: 10.1002/cbf.2871
Gamble, K. L., Berry, R., Frank, S. J., and Young, M. E. (2014). Circadian clock control of endocrine factors. Nat. Rev. Endocrinol. 10, 466–475.
Goto, S. G., and Takekata, H. (2015). Circatidal rhythm and the veiled clockwork. Curr. Opin. Insect Sci. 7, 92–97. doi: 10.1016/j.cois.2014.12.004
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, B., Chatterjee, S., Li, L., Kim, J. M., Lee, J., Yechoor, V. K., et al. (2012). The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 26, 3453–3463. doi: 10.1096/fj.12-205781
Guyomarc’h, C., and Guyomarc’h, J.-C. (1995). Moulting cycles in European quail (Coturnix coturnix coturnix) under constant photoperiodic conditions. Biol. Rhythm Res. 26, 292–305.
Häfker, N. S., and Tessmar-Raible, K. (2020). Rhythms of behavior: Are the times changin’? Curr. Opin. Neurobiol. 60, 55–66. doi: 10.1016/j.conb.2019.10.005
Heideman, P. D., and Bronson, F. H. (1994). An endogenous circannual rhythm of reproduction in a tropical bat, Anoura Geoffroyi, is not entrained by photoperiod. Biol. Reprod. 50, 607–614. doi: 10.1095/biolreprod50.3.607
Helm, B., Visser, M. E., Schwartz, W., Kronfeld-Schor, N., Gerkema, M., Piersma, T., et al. (2017). Two sides of a coin: Ecological and chronobiological perspectives of timing in the wild. Philos. Trans. R. Soc. B Biol. Sci. 372:20160246. doi: 10.1098/rstb.2016.0246
Hughes, M. E., DiTacchio, L., Hayes, K. R., Vollmers, C., Pulivarthy, S., Baggs, J. E., et al. (2009). Harmonics of circadian gene transcription in mammals. PLoS Genet. 5:e1000442. doi: 10.1371/journal.pgen.1000442
Hughes, M. E., Hogenesch, J. B., and Kornacker, K. (2010). JTK-CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380. doi: 10.1177/0748730410379711
Jombart, T. (2008). adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/BIOINFORMATICS/BTN129
Kaiser, T. S. (2014). “Local adaptations of circalunar and circadian clocks: The case of Clunio marinus,” in Annual, lunar, and tidal clocks, eds H. Numata and B. Helm (Tokyo: Springer), 121–141. doi: 10.1007/978-4-431-55261-1_7
Kaiser, T. S., von Haeseler, A., Tessmar-Raible, K., and Heckel, D. G. (2021). Timing strains of the marine insect Clunio marinus diverged and persist with gene flow. Mol. Ecol. 30, 1264–1280. doi: 10.1111/MEC.15791
Kronfeld-Schor, N., Visser, M. E., Salis, L., and van Gils, J. A. (2017). Chronobiology of interspecific interactions in a changing world. Philos. Trans. R. Soc. B Biol. Sci. 372:20160248. doi: 10.1098/rstb.2016.0248
Li, B., and Dewey, C. N. (2014). RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, W., and Godzik, A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Mat, A. M., Perrigault, M., Massabuau, J. C., and Tran, D. (2016). Role and expression of cry1 in the adductor muscle of the oyster Crassostrea gigas during daily and tidal valve activity rhythms. Chronobiol. Int. 33, 949–963. doi: 10.1080/07420528.2016.1181645
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110.20
Miller, B. H., McDearmon, E. L., Panda, S., Hayes, K. R., Zhang, J., Andrews, J. L., et al. (2007). Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 104, 3342–3347. doi: 10.1073/pnas.0611724104
Mrosovsky, N. (2009). Masking: History, definitions, and measurement. Chronobiol. Int. 16, 415–429. doi: 10.3109/07420529908998717
Naylor, E. (2010). Chronobiology of marine organisms. Cambridge: Cambridge University Press. doi: 10.1017/CBO9780511803567
Palmer, J. D. (1995). Review of the dual-clock control of tidal rhythms and the hypothesis that the same clock governs both circatidal and circadian rhythms. Chronobiol. Int. 12, 299–310. doi: 10.3109/07420529509057279
Pfeifer, B., Wittelsbürger, U., Ramos-Onsins, S. E., and Lercher, M. J. (2014). PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936. doi: 10.1093/MOLBEV/MSU136
Pittendrigh, C. S. (1960). Circadian rhythms and the circadian organization of living systems. Cold Spring Harb. Symp. Quant. Biol. 25, 159–184.
Rock, A., Wilcockson, D., and Last, K. S. (2022). Towards an understanding of circatidal clocks. Front. Physiol. 13:830107. doi: 10.3389/fphys.2022.830107
Rubin, M. J., Brock, M. T., Baker, R. L., Wilcox, S., Anderson, K., Davis, S. J., et al. (2018). Circadian rhythms are associated with shoot architecture in natural settings. New Phytol. 219, 246–258. doi: 10.1111/nph.15162
Rubin, M. J., Brock, M. T., Davis, A. M., German, Z. M., Knapp, M., Welch, S. M., et al. (2017). Circadian rhythms vary over the growing season and correlate with fitness components. Mol. Ecol. 26, 5528–5540. doi: 10.1111/mec.14287
Satoh, A., and Terai, Y. (2019). Circatidal gene expression in the mangrove cricket Apteronemobius asahinai. Sci. Rep. 9:3719. doi: 10.1038/s41598-019-40197-2
Satoh, A., Yoshioka, E., and Numata, H. (2008). Circatidal activity rhythm in the mangrove cricket Apteronemobius asahinai. Biol. Lett. 4, 233–236. doi: 10.1098/rsbl.2008.0036
Schmid, B., Helfrich-Förster, C., and Yoshii, T. (2011). A new ImageJ plug-in “actogramJ” for chronobiological analyses. J. Biol. Rhythms 26, 464–467. doi: 10.1177/0748730411414264
Schnytzer, Y., Simon-Blecher, N., Li, J., Ben-Asher, H. W., Salmon-Divon, M., Achituv, Y., et al. (2018). Tidal and diel orchestration of behaviour and gene expression in an intertidal mollusc. Sci. Rep. 8, 4917. doi: 10.1038/s41598-018-23167-y
Sharma, V. K. (2003). Adaptive significance of circadian clocks. Chronobiol. Int. 20, 901–919. doi: 10.1081/CBI-120026099
Takekata, H., Matsuura, Y., Goto, S. G., Satoh, A., and Numata, H. (2012). RNAi of the circadian clock gene period disrupts the circadian rhythm but not the circatidal rhythm in the mangrove cricket. Biol. Lett. 8, 488–491. doi: 10.1098/rsbl.2012.0079
Takekata, H., Numata, H., Shiga, S., and Goto, S. G. (2014). Silencing the circadian clock gene Clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J. Insect Physiol. 68, 16–22. doi: 10.1016/j.jinsphys.2014.06.012
Takemura, A., Rahman, M. S., and Park, Y. J. (2010). External and internal controls of lunar-related reproductive rhythms in fishes. J. Fish Biol. 76, 7–26. doi: 10.1111/j.1095-8649.2009.02481.x
Tessmar-Raible, K., Raible, F., and Arboleda, E. (2011). Another place, another timer: Marine species and the rhythms of life. BioEssays 33, 165–172. doi: 10.1002/bies.201000096
Thaben, P. F., and Westermark, P. O. (2014). Detecting rhythms in time series with rain. J. Biol. Rhythms 29, 391–400. doi: 10.1177/0748730414553029
Tran, D., Perrigault, M., Ciret, P., and Payton, L. (2020). Bivalve mollusc circadian clock genes can run at tidal frequency. Proc. R. Soc. B Biol. Sci. 287:20192440. doi: 10.1098/rspb.2019.2440
Urabe, M. (1998). Diel change of activity and movement on natural river beds in Semisulcospira reiniana. Jpn. J. Malacol. 57, 17–27. doi: 10.18941/venusjjm.57.1_17
Urbanski, J., Mogi, M., Donnell, D. O., Decotiis, M., and Armbruster, P. (2012). Rapid adaptive evolution of photoperiodic response during invasion and range expansion across a climatic gradient. Am. Nat. 179, 490–500. doi: 10.1086/664709
Wang, J., Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Yamanaka, O., and Takeuchi, R. (2018). UMATracker: An intuitive image-based tracking platform. J. Exp. Biol. 221:jeb182469. doi: 10.1242/jeb.182469
Yerushalmi, S., and Green, R. M. (2009). Evidence for the adaptive significance of circadian rhythms. Ecol. Lett. 12, 970–981. doi: 10.1111/j.1461-0248.2009.01343.x
Yerushalmi, S., Yakir, E., and Green, R. M. (2011). Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 20, 1155–1165. doi: 10.1111/j.1365-294X.2010.04962.x
Yokomizo, T., and Takahashi, Y. (2020). Changes in transcriptomic response to salinity stress induce the brackish water adaptation in a freshwater snail. Sci. Rep. 10:16049. doi: 10.1038/s41598-020-73000-8
Zakhrabekova, S., Gough, S. P., Braumann, I., Mul̈ler, A. H., Lundqvist, J., Ahmann, K., et al. (2012). Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. U.S.A. 109, 4326–4331. doi: 10.1073/pnas.1113009109
Zantke, J., Oberlerchner, H., and Tessmar-Raible, K. (2014). “Circadian and circalunar clock interactions and the impact of light in Platynereis dumerilii,” in Annual, lunar, and tidal clocks, eds H. Numata and B. Helm (Tokyo: Springer), 143–162. doi: 10.1007/978-4-431-55261-1_8
Zhang, L., Hastings, M. H., Green, E. W., Tauber, E., Sladek, M., Webster, S. G., et al. (2013). Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr. Biol. 23, 1863–1873. doi: 10.1016/j.cub.2013.08.038
Zhu, B. (2020). Decoding the function and regulation of the mammalian 12-h clock. J. Mol. Cell Biol. 12, 752–758. doi: 10.1093/jmcb/mjaa021
Keywords: circatidal rhythm, circadian rhythm, transcriptome, tidal environments, biological clock, activity pattern
Citation: Yokomizo T and Takahashi Y (2022) Endogenous rhythm variation and adaptation to the tidal environment in the freshwater snail, Semisulcospira reiniana. Front. Ecol. Evol. 10:1078234. doi: 10.3389/fevo.2022.1078234
Received: 24 October 2022; Accepted: 25 November 2022;
Published: 08 December 2022.
Edited by:
Naoki Osada, Hokkaido University, JapanReviewed by:
Alberto Biscontin, University of Padua, ItalyElena Frigato, University of Ferrara, Italy
Copyright © 2022 Yokomizo and Takahashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuma Takahashi, dGFrYWhhc2hpLnl1bUBnbWFpbC5jb20=
 Takumi Yokomizo1
Takumi Yokomizo1 Yuma Takahashi
Yuma Takahashi