- 1Key Laboratory of Genetics and Germplasm Innovation of Tropical Special Forest Trees and Ornamental Plants, Ministry of Education, School of Forestry, Hainan University, Haikou, China
- 2Sanya Academy of Forestry, Sanya, China
More than half of the world's tropical lowland rainforests have been lost due to conversion to agricultural land (such as rubber plantations). Thus, ecological restoration in degraded tropical lowland rainforests is crucial. The first step to restoration is restoring soil functioning (i.e., soil fertility, carbon, and nitrogen cycling) to levels similar to those in the primary tropical lowland rainforest. This requires understanding soil nematode community assembly in primary tropical lowland rainforest, which has never been explored in this habitat. In this study, we measured species compositions of plant and soil nematode communities and soil characteristics (pH, total and available nitrogen, phosphorus, and soil water content) in a primary tropical lowland rainforest, which is located on Hainan Island, China. We performed two tests (the null-model test and distance-based Moran's eigenvector maps (MEM) and redundancy analysis-based variance partitioning) to quantify the relative contribution of the deterministic (abiotic filtering and biotic interactions) and stochastic processes (random processes and dispersal limitation) to the soil nematode community. We found that a deterministic process (habitat filtering) determined nematode community assembly in our tropical lowland rainforest. Moreover, soil properties, but not plant diversity, were the key determinants of nematode community assembly. We have, for the first time, managed to identify factors that contribute to the nematode community assembly in the tropical lowland rainforest. This quantified community assembly mechanism can guide future soil functioning recovery of the tropical lowland rainforest.
Introduction
Tropical lowland forests harbor the most biodiversity, but these forests are also easily destroyed due to their suitability for agricultural use (Corlett, 2011). As of now, over half of the tropical lowland rainforest has been lost due to its conversion into agricultural lands (i.e., rubber plantations and palm plantations). (Aleman et al., 2018). Thus, ecological restoration is crucial to restoring the percentage of tropical lowland rainforests worldwide (Brancalion et al., 2019). The first step in reforestation is restoring the soil functioning of degraded soil (Zhang et al., 2019). Soil nematodes are the most abundant soil-dwelling animal group on earth and are vital to soil function (i.e., processing soil organic nutrients and regulating soil microorganism populations) (Ferris et al., 2001; Ruess, 2003; Pausch et al., 2016; van den Hoogen et al., 2019). Thus, a deep understanding of soil nematode community assembly in the primary tropical lowland rainforest can aid the recovery of soil functioning (Wang et al., 2022). That is because soil nematode community assembly in the primary tropical lowland rainforest can easily establish a restoration standard (Wang et al., 2022). For example, soil nematode community assembly in the restored tropical lowland rainforest should be comparable to that in the primary tropical lowland rainforest.
Identification of different ecological processes that shape the community assembly of different communities is the primary focus of ecological research (Condit et al., 2002; Chave, 2013; Chase, 2014). For more than a century, community ecologists have debated whether the deterministic or stochastic processes play a primary role in structuring ecological communities (Hubbell, 2001; Chase and Myers, 2011; Trisos et al., 2014). This debate has led to conceptual ideas that invoke multiple processes ranging from niche-based habitat filtering and interspecific competition (niche differentiation) to random dispersal and demographic stochasticity (Weiher et al., 2011). The relative contribution of deterministic and stochastic processes to community assembly has been widely investigated for plant communities, soil bacterial, and fungal communities in different ecosystems (Zhang et al., 2015; Moroenyane et al., 2016; Tripathi et al., 2018; Jiao et al., 2021; Ni et al., 2021). However, till now, there has been no study investigating soil nematode community assembly in the tropical lowland rainforest.
A recent global meta-analysis indicated that a deterministic process may dominate soil nematode community assembly globally (Luan et al., 2020). Thus, we hypothesized that deterministic processes (habitat filtering) might also dominate nematode community assembly in the tropical lowland rainforest. To test this hypothesis, in the primary tropical lowland rainforest on Hainan Island, we evaluated the relative contributions of the deterministic and stochastic processes to soil nematode community assembly by examining the following: (i) species compositions of plant and soil nematode communities in 20 plots of 400 m2 each and (ii) soil characteristics (pH, total and available nitrogen, and phosphorus, and soil water content) in each plot. With this dataset, we first used a null model test to compare the observed beta diversity and the beta diversity of simulated random communities to determine if there is significant community convergence, divergence, or random distribution. Then, we used it to reveal the relative contributions of the deterministic process (soil variables) and stochastic processes (dispersal limitation and random process) to the beta diversity of the nematode community. Finally, by determining the beta diversity of the soil nematode community, we were able to quantify the relative importance of soil variables and plant diversity if a deterministic process dominated the nematode community assembly.
Materials and methods
Study site
The study site is the Xinglong National Forest Park (110.16°E, 18.77°N) in Hainan Province, southern China (Figure 1). It has a unique primary tropical lowland rainforest with high species diversity. Its altitude ranges from 5 to 300 m; the area has a mean annual temperature of 24.5°C, and the lowest and the highest mean monthly temperatures are 18.7°C (January) and 28.5°C (July), respectively (Guo et al., 2018). The mean annual precipitation is ~2,500 mm, and most of the precipitation is distributed between May and November.
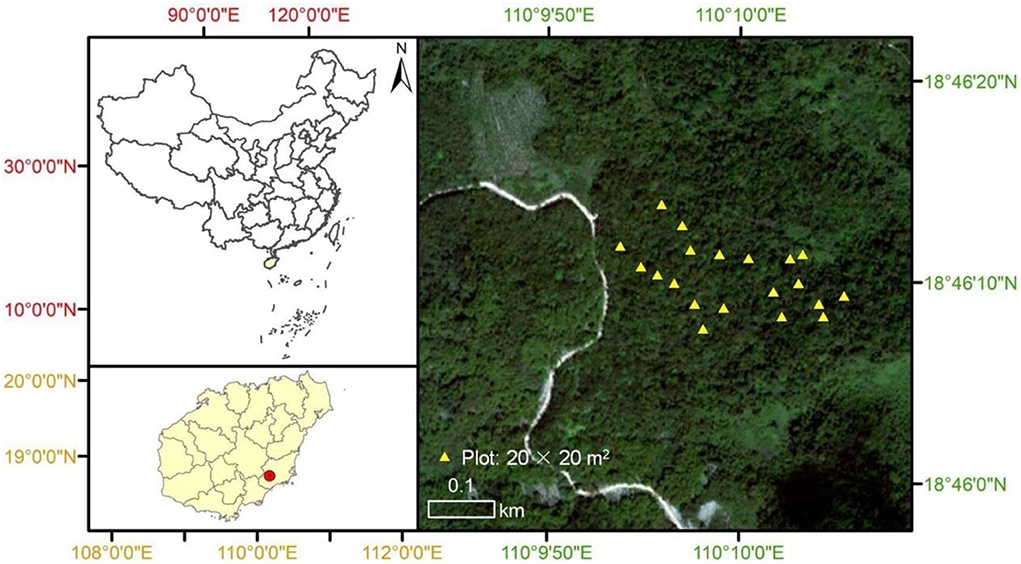
Figure 1. The map of our study site and detailed spatial location of our 20 × 20 m2 plots in the primary tropical lowland rainforest.
Sampling
On 10 August 2021 (at the peak of the growing season), we randomly sampled 20 plots in the adjacent undisturbed secondary tropical rainforest. Each of these plots was 20 × 20 m2 and at least 100–300 m apart (Figure 1). The elevation of all 20 plots ranged from 15 to 150 m. Within a plot, all freestanding trees with a DBH (diameter at breast height) of ≥1 cm were measured and identified as species. The spatial locations (x- and y- coordinates) for each plot were also recorded. We randomly collected five soil cores (depth diameter of soil cores, 0–20 cm) from each plot and mixed them into one soil sample. We then used 100 g of soil to measure the soil properties, and soil nematodes were extracted from another 100 g of soil using Baermann's funnel method (Barker et al., 1985). Then, we used a microscope (Leica DM500) to measure nematode abundance in each soil sample by identifying the first 100 nematodes to the genus level based on their morphological traits. All nematode images were identified to genus level for soils with fewer than 100 nematodes. We followed the method used by Yeates et al. (1993) to classify all the identified nematodes into five functional groups: phytophagous nematodes, bacterivores, fungivores, predators, and omnivores (Supplementary Table S1).
As described by Burt (2009), the soil N concentration was determined using the Kjeldahl digestion method and an OIFS 3000 autoanalyzer (OI Analytical, College Station, TX, USA). Both ammonium N and nitrate N were extracted from 2 g of soil using a 2.0 M KCl solution. The ammonium and nitrate N extracts were further analyzed colorimetrically using a KCl extract and an OIFS 3000 autoanalyzer (OI Analytical, College Station, TX, USA). To determine the total P in the soil, we took 2.5 g of fresh soil, added Mehlich III solution, and then evaluated the color change in the soil with a spectrophotometer after the soil was digested with perchloric acid. Elemental analysis of P was performed on the Mehlich III extracts using inductively coupled plasma atomic emission spectroscopy (ICP-AES). Soil pH was measured using a pH meter in a KCl suspension (UB-7, UltraBASIC, Denver, CO, USA). The water content of the soil was determined by measuring the amount of weight lost upon drying the soil at 100°C for 48 h in an oven.
Statistic methods
We first calculated Bray–Curtis dissimilarities to represent the beta diversity of the nematode community. To test whether any observed beta diversity pattern was a random distribution or showed species convergence or divergence, we first simulated null communities in which species abundance values were randomly assigned. Randomization procedures were applied to calculate “null” distributions for both species composition (i.e., on the species × quadrat matrix) and all species (Götzenberger et al., 2012). We reshuffled the species × quadrat matrices with three constraints simultaneously, following the method of Zhang et al. (2015, 2018); that is, keeping (1) the same number of species (species richness) per plot in the simulated and observed data; (2) the same number of total species occurrences per region (i.e., the number of plots where the species occur in each group of the five spatial scales); and (iii) the total abundance of species in a region constant (i.e., the sum of the number of quadrats occupied in all plots). We implemented this using the function “randomized matrix” in the “Picante” package in R software (Kembel et al., 2010). We then compared the observed beta diversity to the beta diversity simulated in 1,000 randomly assembled communities. Beta diversity was quantified using the standard effect size index (SES) developed by Gotelli and McCabe (2002):
where beta diversity observed and beta diversity random represent observed beta diversity and mean beta diversity values of the simulated null community, respectively. The beta diversitysd represents the standard deviation of beta diversity values generated from the 1,000 simulations. We used the Wilcoxon signed-rank tests to examine whether SES was significantly more than, less than, or ≈0, which indicated the prevalence of significant species divergence, species convergence, and random distribution, respectively.
Partitioning the spatial patterns in the beta diversity of nematodes
We first used the “diversity result” in the “biodiversity” package in r software to calculate plant diversity (richness and abundance). Then, we used Moran's eigenvector maps (MEM) and the spatial location of each plot to quantify spatial autocorrelations in nematode species composition in each plot. The MEM was based on the principal coordinates of neighbor matrix axes (Legendre and Legendre, 2012) and could be used to describe the spatial autocorrelations in beta diversity (Zhang et al., 2015, 2018). We used the function “cmdscale” in the “SpacemakerR” package in r software (Stephane, 2013) to calculate MEM. We then used the r function“poly” to quantify each variable's polynomial terms, including the linear and nonlinear relationships between soil variables and species composition. While performing redundancy analysis (RDA)-based variance partitioning, we used the method developed by Blanchet et al. (2008) to forward-select significant soil variables and purely spatial variables represented by MEM associated with beta diversity of the soil nematode community. Finally, we used variance partitioning to allocate variations in beta diversity of nematode community to eight complementary components: (a) “purely soil variables” (explained by soil factors only), (b)“purely spatial variables” (spatial autocorrelation in beta diversity independent of soil variables and induced by dispersal limitation and biotic interactions), (c) “purely plant diversity variables (explained by plant diversity (richness and abundance) only),” (d) “spatially structured abiotic variables” (spatial autocorrelation in beta diversity and merely induced by soil variables), (e) “spatially structured plant diversity” (spatial autocorrelation in beta diversity and merely induced by plant diversity), (f) variables explained by both soil variables and plant diversity, (g) variations in beta diversity explained by soil variables, spatial variables, and plant diversity, and “undetermined variables” (Legendre et al., 2009; Zhang et al., 2018). Variation in beta diversity of the soil nematode community explained by soil variables was represented by a + d + f + g, whereas b + d + e + g showed the variation in the soil nematode community explained by MEM. In addition, the variation of the soil nematode community explained by plant diversity was reflected by c+e+f+g. Variance partitioning was performed using the function “varpart” in the “vegan” package in R (Oksanen et al., 2016).
Results
There are a total of 29 genera of soil nematodes across all plots in our primary tropical lowland rainforest (Supplementary Table S1). Bacterivores had the most (12) genera, whereas predators had the fewest (2) genera (Supplementary Table S1). Both phytophagous nematodes and omnivores had six genera (Supplementary Table S1). In addition, fungivores had three genera (Supplementary Table S1).
The nematode community converged, as indicated by a significantly smaller SES than zero (Figure 2), indicating that the deterministic processes (habitat filtering or exclusion of weak competitors) dominated the nematode community assembly. The deterministic process (habitat filtering), represented by the soil variables, accounted for 63% of the variation in beta diversity, respectively (Figure 3). By contrast, the stochastic process (dispersal limitation), represented by spatial variables, explained 22% of the variation in beta diversity (Figure 3). In the tests of soil variables on beta diversity of soil nematode communities, the forward selection analysis revealed that the significant abiotic variables that predicted beta diversity of soil nematode communities included pH, soil total and available phosphorus, and soil water content (Supplementary Table S2). When comparing the relative contributions of all soil variables and plant diversity (richness and abundance) to the beta diversity of the soil nematode community, we found that soil variables explained 63% of the variation in beta diversity, whereas plant diversity merely accounted for 25% of the variations in beta diversity (Figure 3).
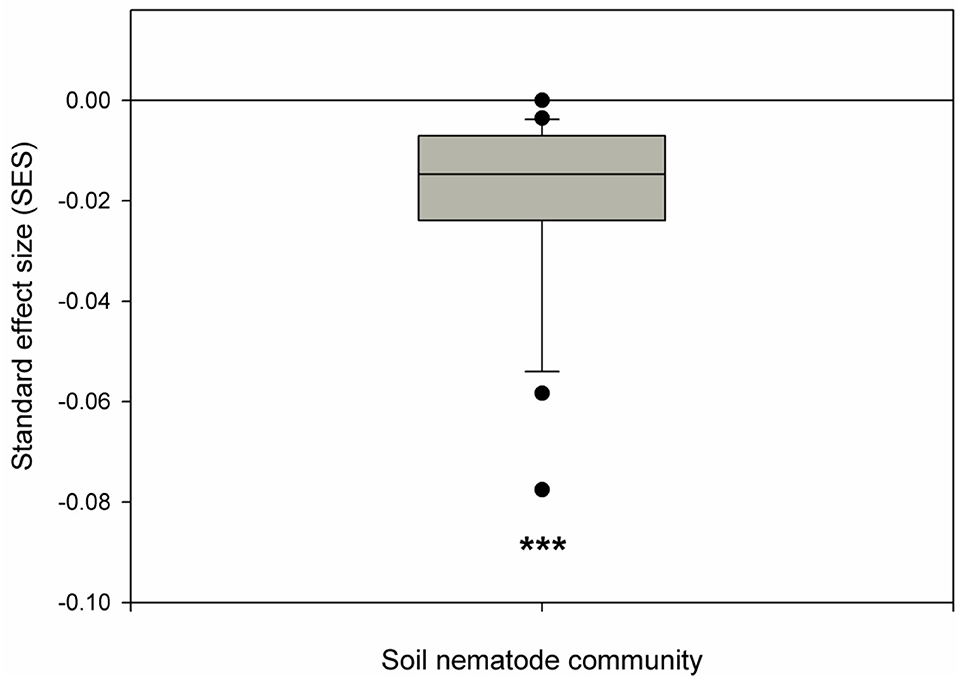
Figure 2. Distribution of standard effect size (SES) of beta diversity in our primary tropical lowland rainforest. SES that is significantly greater than, smaller than, or approaching zero indicates significant trait divergence, trait convergence, or random distribution, respectively. The box plot shows the median (line within the box), 25 and 75th percentiles (the boundaries of the box), and 90 and 10th percentiles (error bars) of SES at each spatial scale. ***Indicates a p-value < 0.001 based on the Wilcoxon signed-rank tests.
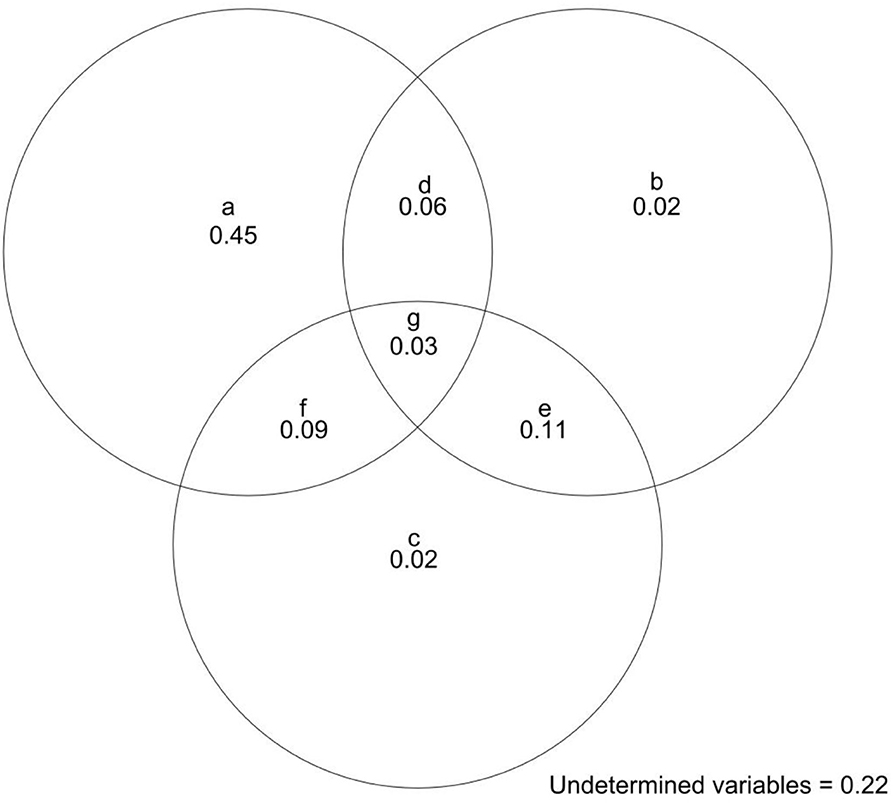
Figure 3. The variations of beta diversity are partitioned into eight complementary components if statistically significant (p < 0.05). Analysis was performed, including all quadrants. These eight components include (a) “purely soil variables” (explained by soil factors only), (b) “purely spatial variables” (spatial autocorrelation in beta diversity independent of soil variables and induced by dispersal limitation and biotic interactions), (c) “purely plant diversity variables (explained by plant diversity (richness and abundance) only),” (d) “spatially structured abiotic variables” (spatial autocorrelation in beta diversity and merely induced by soil variables), (e) “spatially structured plant diversity” (spatial autocorrelation in beta diversity and merely induced by plant diversity), (f) variables explained by both soil variables and plant diversity, (g) variations of beta diversity explained by soil variables, spatial variables, and plant diversity and “undetermined variables.” Variation in beta diversity of the soil nematode community explained by soil variables is represented by a + d + f + g, whereas b + d + e + g shows the variation of the soil nematode community explained by MEM. In addition, the variation of soil nematode community explained by plant diversity is reflected by c + e + f + g.
Discussion
We studied a primary tropical lowland rainforest to estimate the relative importance of the deterministic and stochastic processes in determining soil nematode community assembly in tropical lowland rainforests. As we hypothesized, habitat filtering was the major determinant of the soil nematode community assembly in tropical lowland rainforests. Moreover, soil properties determine soil nematode community assembly.
By comparing observed beta diversity to that simulated from null communities, we found significant convergence (SES < 0), indicating deterministic processes (habitat filtering or exclusion of weak competitors) dominated community assembly of soil nematodes (Kraft et al., 2008; De Bello et al., 2012; Zhang et al., 2015, 2018). We further showed that habitat filtering (indicated by soil variables) explained 63% of the variations in the beta diversity of soil nematodes. We noted that our attribution of the effect of dispersal limitation on the beta diversity of soil nematodes to purely spatial variables might be an overestimate (Legendre et al., 2009), which could be, despite our attempt to include the important soil nutrient variables, because some significant abiotic variables remain unaccounted for (Siefert et al., 2013). However, given the high variations in beta diversity explained by soil variables, we can easily judge habitat-filtering-dominated soil nematode community assembly; this is consistent with a recent meta-analysis, which showed that a deterministic process dominated soil nematode community assembly at the global scale (Luan et al., 2020). However, different results have also been found in other ecosystems. For instance, Zou et al. (2022) found that the stochastic processes are the primary determinants of soil nematode community assembly in three Asian mountains. In contrast, the deterministic process dominated the community assembly of nematodes in a tropical seasonal forest (Wang et al., 2022) and our tropical lowland rainforest. The different soil (i.e., N-limited or P-limited) and plant diversity (e.g., high or low) types in different ecosystems may result in inconsistent soil nematode community assembly patterns between our study and other studies. Thus, comparative research on the community assembly mechanisms of soil nematode communities across different ecosystems should be performed in the future to generate a general community assembly mechanism of soil nematodes at the global scale.
A null-model approach developed by Kraft et al. (2008) and distance-based Moran's eigenvector maps (MEM) and redundancy analysis-based variance partitioning have widely proven to be helpful in identifying community assembly mechanisms of plant communities (Kraft and Ackerly, 2010; De Bello et al., 2012; Myers et al., 2013; Zhang et al., 2015, 2018). However, no study used them to identify the relative contributions of deterministic and stochastic processes to community assembly of nematode communities. It remains unclear whether they can also be applied to dissolve soil nematode community assembly. In this study, the results clearly demonstrated that they could successfully determine the key determinants of community assembly of soil nematodes. We recommend that these two methods be used to reveal the community assembly of soil nematodes in future studies.
Soil properties and plant diversity are reported as the key influences on the soil nematode community (Neher, 2010; Viketoft et al., 2011; Liu et al., 2015; Wang et al., 2018). However, the relative contributions of soil characteristics and plant diversity to community assembly of the soil nematode community remain unknown. Our variance partitioning results clearly demonstrated that soil properties explained 63% of the variations in the beta diversity of soil nematodes, whereas plant diversity merely explained 25%. This indicated that soil properties, but not plant diversity, dominated the soil nematode community assembly in the tropical lowland rainforest. However, we only measured plant diversity, and it is highly possible that plant-derived resources (e.g., litter input) may be more important than plant diversity in determining the beta diversity of the soil nematode community. As a result, the plant-derived resources should be measured in the future to identify the relative contributions of soil properties and plant-derived resources in driving the variation in beta diversity of the soil nematode community.
Conclusion
Overall, we gained invaluable insights into soil nematode community assembly in tropical lowland forests. The first null-model test, distance-based Moran's eigenvector maps (MEM), and redundancy analysis-based variance partitioning revealed the community assembly of soil nematodes in tropical lowland rainforests. Second, the deterministic process (habitat filtering) dominated the community assembly of soil nematodes. Third, soil properties but not plant diversity were the primary determinants of soil nematode community assembly. This quantified community assembly mechanism can guide future soil functioning recovery of tropical lowland rainforests. For example, since soil properties are the key determinants of soil nematode communities, adding fertilization to improve soil fertility is one key step to recovering the soil nematode community and its induced key soil functioning in degraded tropical lowland rainforest.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
WH, TL, and JL designed the research. WH, MH, YQ, TL, and JL performed the research and wrote the manuscript. WH, MH, TL, and JL analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Hainan Province Science and Technology Special Fund (ZDYF2022SHFZ320), Hainan Provincial Natural Science Foundation of China (422CXTD508, 421MS013), the Education Department of Hainan Province (Hnjg2021ZD-9), the specific research fund of the Innovation Platform for Academicians of Hainan Province, the research project of the Hainan Academician Innovation Platform (YSPTZX202017), and the scientific research project for the ecological restoration of Baopoling Mountain in Sanya, China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1034829/full#supplementary-material
References
Aleman, J. C., Jarzyna, M. A., and Staver, A. C. (2018). Forest extent and deforestation in tropical Africa since 1900. Nat. Ecol. E2, 26–33. doi: 10.1038/s41559-017-0406-1
Barker, K. R., Carter, C. C., and Sasser, J. N. (1985). An Advanced Treatise on MELOIDOGYNE (vol.2): |Methodology. Raleigh: North Carolina State University Press.
Blanchet, F. G., Legendre, P., and Borcard, D. (2008). Forward selection of explanatory variables. Ecology 89, 2623–2632. doi: 10.1890/07-0986.1
Brancalion, P. H. S., Niamir, A., Broadbent, E., Crouzeilles, R., Barros, F. S. M., Almeyda Zambrano, A. M., et al. (2019). Global restoration opportunities in tropical rainforest landscapes. Sci. Adv. 5, eaav3223. doi: 10.1126/sciadv.aav3223
Burt, R. (2009). Soil Survey Field and Laboratory Methods Manual. Lincoln: National Soil Survey Center Press.
Chase, J. M. (2014). Spatial scale resolves the niche versus neutral theory debate. J. Veg. Sci. 25, 319–322. doi: 10.1111/jvs.12159
Chase, J. M., and Myers, J. A. (2011). Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B Biol. Sci. 366, 2351–2363. doi: 10.1098/rstb.2011.0063
Chave, J. (2013). The problem of pattern and scale in ecology: what have we learned in 20 years? Ecol. Lett. 16, 4–16. doi: 10.1111/ele.12048
Condit, R., Pitman, N., Leigh, E. G., Chave, J., Terborgh, J., Foster, R. B., et al. (2002). Beta-diversity in tropical forest trees. Science 295, 666–669. doi: 10.1126/science.1066854
Corlett, R. T. (2011). Impacts of warming on tropical lowland rainforests. Trends Ecol. E26, 606–613. doi: 10.1016/j.tree.2011.06.015
De Bello, F., Price, J. N., Münkemüller, T., Liira, J., Zobel, M., Thuiller, W., et al. (2012). Functional species pool framework to test for biotic effects on community assembly. Ecology 93, 2263–2273. doi: 10.1890/11-1394.1
Ferris, H., Bongers, T., and de Goede, R. G. M. (2001). A framework for soil food web diagnostics: extension of the nematode faunal analysis concept. Appl. Soil Ecol. 18, 13–29. doi: 10.1016/S0929-1393(01)00152-4
Gotelli, N. J., and McCabe, D. J. (2002). Species co-occurrence: a meta-analysis of JM Diamond's assembly rules model. Ecology 83, 2091–2096. doi: 10.1890/0012-9658(2002)083[2091:SCOAMA]2.0.CO;2
Götzenberger, L., de Bello, F., Bråthen, K. A., Davison, J., Dubuis, A., Guisan, A., et al. (2012). Ecological assembly rules in plant communities-approaches, patterns, and prospects. Biol. Rev. 87, 111–127. doi: 10.1111/j.1469-185X.2011.00187.x
Guo, J., Shang, S., and Zeng, J. (2018). Morphometric study confirms the presence of only Vatica mangachapoi on Hainan Island, China. J. For. Res. 29, 639–646. doi: 10.1007/s11676-017-0458-8
Hubbell, S. P. (2001). The Unified Neutral Theory of Biodiversity and Biogeography. Princeton: Princeton University Press.
Jiao, S., Zhang, B., Zhang, G., Chen, W., and Wei, G. (2021). Stochastic community assembly decreases soil fungal richness in arid ecosystems. Mol. Ecol. 30, 4338–4348. doi: 10.1111/mec.16047
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Kraft, N. J. B., and Ackerly, D. D. (2010). Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 80, 401–422. doi: 10.1890/09-1672.1
Kraft, N. J. B., Valencia, R., and Ackerly, D. D. (2008). Functional traits and niche-based tree community assembly in an amazonian forest. Science 322, 580–582. doi: 10.1126/science.1160662
Legendre, P., Mi, X., Ren, H., Ma, K., Yu, M., Sun, I-. F., et al. (2009). Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 90, 663–674. doi: 10.1890/07-1880.1
Liu, T., Guo, R., Ran, W., Whalen, J. K., and Li, H. (2015). Body size is a sensitive trait-based indicator of soil nematode community response to fertilization in rice and wheat agroecosystems. Soil Biol. Biochem. 88, 275–281. doi: 10.1016/j.soilbio.2015.05.027
Luan, L., Jiang, Y., Cheng, M., Dini-Andreote, F., Sui, Y., Xu, Q., et al. (2020). Organism body size structures the soil microbial and nematode community assembly at a continental and global scale. Nat. Commun. 11, 1–11. doi: 10.1038/s41467-020-20271-4
Moroenyane, I., Dong, K., Singh, D., Chimphango, S. B. M., and Adams, J. M. (2016). Deterministic processes dominate nematode community structure in the Fynbos Mediterranean heathland of South Africa. Evol. Ecol. 30, 685–701. doi: 10.1007/s10682-016-9837-4
Myers, J. A., Chase, J. M., Jiménez, I., Jørgensen, P. M., Araujo-Murakami, A., Paniagua-Zambrana, N., et al. (2013). Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 16, 151–157. doi: 10.1111/ele.12021
Neher, D. A. (2010). Ecology of Plant and Free-Living Nematodes in Natural and Agricultural Soil. Annu. Rev. Phytopathol. 48, 371–394. doi: 10.1146/annurev-phyto-073009-114439
Ni, Y., Yang, T., Ma, Y., Zhang, K., Soltis, P. S., Soltis, D. E., et al. (2021). Soil pH determines bacterial distribution and assembly processes in natural mountain forests of eastern China. Glob. Ecol. Biogeogr. 30, 2164–2177. doi: 10.1111/geb.13373
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2016). Vegan: Community Ecology Package. R Package Version 2.4-0. Available online at: https://CRAN.R-project.org/package=vegan (accessed August 15, 2022.).
Pausch, J., Hofmann, S., Scharroba, A., Kuzyakov, Y., and Ruess, L. (2016). Fluxes of root-derived carbon into the nematode micro-food web of an arable soil. Food Webs 9, 32–38. doi: 10.1016/j.fooweb.2016.05.001
Ruess, L. (2003). Nematode soil faunal analysis of decomposition pathways in different ecosystems. Nematology 5, 179–181. doi: 10.1163/156854103767139662
Siefert, A., Ravenscroft, C., Weiser, M. D., and Swenson, N. G. (2013). Functional beta diversity patterns reveal deterministic community assembly processes in eastern North American trees. Glob. Ecol. Biogeogr. 22, 682–691. doi: 10.1111/geb.12030
Stephane, D. (2013). SpacemakeR: Spatial Modelling. R package version 0.0-5/r113. Available online at: https://r-forge.r-project.org/projects/sedar/ (accessed August 10, 2022.).
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., Lee, Y. K., et al. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083. doi: 10.1038/s41396-018-0082-4
Trisos, C. H., Petchey, O. L., and Tobias, J. A. (2014). Unraveling the interplay of community assembly processes acting on multiple niche axes across spatial scales. Am. Nat. 184, 593–608. doi: 10.1086/678233
van den Hoogen, J., Geisen, S., Routh, D., Ferris, H., Traunspurger, W., Wardle, D. A., et al. (2019). Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198. doi: 10.1038/s41586-019-1418-6
Viketoft, M., Sohlenius, B., Boström, S., Palmborg, C., Bengtsson, J., Berg, M. P., et al. (2011). Temporal dynamics of soil nematode communities in a grassland plant diversity experiment. Soil Biol. Biochem. 43, 1063–1070. doi: 10.1016/j.soilbio.2011.01.027
Wang, W., Sun, Z., Mishra, S., Xia, S., Lin, L., Yang, X., et al. (2022). Body size determines multitrophic soil microbiota community assembly associated with soil and plant attributes in a tropical seasonal rainforest. Mol. Ecol. 1, 1–10. doi: 10.1111/mec.16585
Wang, X., Nielsen, U. N., Yang, X., Zhang, L., Zhou, X., Du, G., et al. (2018). Grazing induces direct and indirect shrub effects on soil nematode communities. Soil Biol. Biochem. 121, 193–201. doi: 10.1016/j.soilbio.2018.03.007
Weiher, E., Freund, D., Bunton, T., Stefanski, A., Lee, T., Bentivenga, S., et al. (2011). Advances, challenges and a developing synthesis of ecological community assembly theory. Philos. Trans. R. Soc. B Biol. Sci. 366, 2403–2413. doi: 10.1098/rstb.2011.0056
Yeates, G. W., Bongers, T., De Goede, R. G., Freckman, D. W., and Georgieva, S. S. (1993). Feeding habits in soil nematode families and genera-an outline for soil ecologists. J. Nematol. 25, 315–331.
Zhang, H., Chen, H. Y. H., Lian, J., John, R., Ronghua, L., Liu, H., et al. (2018). Using functional trait diversity patterns to disentangle the scale-dependent ecological processes in a subtropical forest. Funct. Ecol. 32, 1379–1389. doi: 10.1111/1365-2435.13079
Zhang, H., Qi, W., John, R., Wang, W., Song, F., Zhou, S., et al. (2015). Using functional trait diversity to evaluate the contribution of multiple ecological processes to community assembly during succession. Ecography 38, 1176–1186. doi: 10.1111/ecog.01123
Zhang, W., Zhang, H., Jian, S., and Liu, N. (2019). Tree plantations influence the abundance of ammonia-oxidizing bacteria in the soils of a coral island. Appl. Soil Ecol. 138, 220–222. doi: 10.1016/j.apsoil.2019.02.014
Keywords: biotic interaction, community convergence or divergence, dispersal limitation, habitat filtering, stochastic processes
Citation: Hou W, He M, Qi Y, Liu T and Luo J (2022) Soil nematode community assembly in a primary tropical lowland rainforest. Front. Ecol. Evol. 10:1034829. doi: 10.3389/fevo.2022.1034829
Received: 02 September 2022; Accepted: 22 September 2022;
Published: 07 November 2022.
Edited by:
Xiang Liu, Lanzhou University, ChinaReviewed by:
Youzheng Zhang, Second Institute of Oceanography, Ministry of Natural Resources, ChinaPei Zhang, Sichuan University, China
Copyright © 2022 Hou, He, Qi, Liu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiedong Liu, bGl1QGhhaW5hbnUuZWR1LmNu; Jinhuan Luo, NDQzNDY0MTk5QHFxLmNvbQ==
 Weichen Hou1
Weichen Hou1 Yanwen Qi
Yanwen Qi Tiedong Liu
Tiedong Liu