- 1Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, China
- 2Qinghai Provincial Key Laboratory of Animal Ecological Genomics, Xining, China
- 3Ecological Research Center, Northwest Institute of Plateau Biology, University of Chinese Academy of Sciences, Beijing, China
The prevalence and transmission of antibiotic resistance genes (ARGs) and virulence factors (VFs) pose a great threat to public health. The importance of pollution in determining the occurrence of ARGs and VFs in wildlife is poorly understood. Using a metagenomic approach, this study investigates the composition and functional pathways of bacteria, ARGs, and VFs in the gut microbiome of Plateau pikas in regions of medical pollution (MPR), heavy tourist traffic (HTR), and no contamination (NCR). We found that the abundance of probiotic genera (Clostridium, Eubacterium, Faecalibacterium, and Roseburia) were significantly lower in the HTR. The metabolic pathways of replication and repair in the endocrine and nervous systems were significantly enriched in the MPR, whereas endocrine and metabolic diseases were significantly enriched in the NCR. The Shannon and Gini–Simpson α-diversity indices of ARGs were highest in the HTR, and there were significant differences in β-diversity among the three regions. The resistance of ARGs to glycopeptide antibiotics increased significantly in the MPR, whereas the ARGs for aminocoumarins increased significantly in the HTR. The diversity of mobile genetic elements (MGEs) was significantly higher in the MPR than in other regions. We observed a strong positive correlation between ARGs and pathogenic bacteria, and the network structure was the most complex in the MPR. There were significant differences in the β-diversity of VFs among the three regions. Medical pollution led to significant enrichment of fibronectin-binding protein and PhoP, whereas tourism-related pollution (in the HTR) led to significant enrichment of LPS and LplA1. Our study indicates that environmental pollution can affect the structure and function of gut microbes and disseminate ARGs and VFs via horizontal transmission, thereby posing a threat to the health of wild animals.
Introduction
With the rapid development of society and the economy, human activities are taking an increasing toll on the environment (Doyle et al., 2020). Antibiotic resistance genes (ARGs) are a novel anthropogenic pollutant that can directly affect physiology. Additionally, disposal of human drugs can have toxic effects on aquatic organisms; among these, antibiotics can severely alter immune function in humans and wildlife (Sanderson et al., 2004). ARGs can spread rapidly in the environment through drug-resistant strains and have become a global public health concern (Pruden et al., 2006). Exposure to ARGs is associated with human population density, with wildlife in areas of higher human density showing increased antibiotic resistance (Skurnik et al., 2006). A study found that the concentration of clarithromycin in the surface water of Wailingding Island (located in Zhuhai, China) increased with increasing tourist populations (Liu et al., 2020). Similarly, antibiotic-resistant bacteria in the guts of Galapagos wildlife also increased due to tourist arrivals (Wheeler et al., 2012). Hospitals are importance sources of ARGs, and wastewater discharged from hospitals is enriched with highly drug-resistant Pseudomonas, which increases the risk of resistance gene transmission in water bodies (Spindler et al., 2012). Consequently, multidrug resistant bacteria carrying virulence factors (VFs), such as Enterococcus faecium (CC17), Streptococcus pneumoniae (PMEN1), and Escherichia coli, have spread worldwide (Pan et al., 2020). Although pollution from hospitals and tourism (including transportation) play essential roles in the transmission of resistance genes and virulence factors, the associated effects on the gut microbes of wildlife are unclear.
The plateau pika (Ochotona curzoniae) is the most abundant and widely distributed small herbivorous mammal in Qinghai–Tibet. It is group-living and does not hibernate (Zhang et al., 1998; Smith and Foggin, 1999). Plateau pikas play a unique role in regulating competition among plant species, promoting mutualism between passerine birds and lizards, and maintaining the diversity of raptors and carnivorous mammals (Arthur et al., 2008; Qu et al., 2015; Smith et al., 2019; Zhao et al., 2020). As a key species in the local ecosystem, plateau pika coprophagous behavior and game behavior, in that they were selected as research object. Recent studies on gut microbes have shown that gut microbes provide physiological functions to the host, especially herbivores (Kohl et al., 2014, 2018). Most of the research on the plateau pika gut microbiome has focused on altitude, plant secondary compounds, and seasonal dynamics (Li et al., 2018, 2019; Fan et al., 2020, 2022; Ren et al., 2021; Yu et al., 2022). Few reports exist on the effects of resistance genes and virulence factors on intestinal flora composition and function, and whether these effects are exacerbated by pollution from anthropogenic activities.
In this study, we performed a metagenomic analysis of the gut of Plateau pikas occurring at three sites with varying anthropogenic activities. By profiling the bacteria, ARGs, and VFs of the gut microbiome, we aimed to determine the effects of different anthropogenic activities on ARG and VF occurrence. Our study findings emphasize the potential threats of medical and tourist-based pollution on wildlife health.
Materials and methods
Sample collection
The protocol was reviewed by the Ethics Committee of Northwest Institute of Plateau Biology. All animal procedures were conducted according to the Regulations for the Administration of Laboratory Animals established by the Ministry of Science and Technology of China (2017 Revision).
A total of 34 wild adult Plateau pikas were captured along the Qinghai–Tibet Highway in July 2019, including the Kunlun Mountain Pass (heavy-tourist-traffic region, HTR), Gouli Clinic (medical-pollution region, MPR), and Reshui Village (non-contaminated region, NCR; Figure 1). All habitats were typical alpine meadows. NCR is located in a grassland grazed by livestock which is 5–10 km away from Reshui Village. There is not tourist-based pollution, and no medical waste, so the sampling area of NCR can be ensured as an unpolluted area. MPR was the grassland near the clinic of Zhiyu village, Gouli town, where there was a large amount of medical waste such as cotton swab and medicine bottles. HTR located in Kunlun Mountain Pass and there is a highway across the sample sites, and nearly over 3,000 trucks and vehicles pass through the highway every day. At the same time, Kunlun Mountain Pass is a tourist attraction place, and there are a lot of waste food, plastic bottles and other household garbage in the area due to a large number of tourists. Twelve samples were collected in Reshui Village (NCR), 13 samples were collected in Gouli Clinic (MPR), and 9 samples were collected in Kunlun Mountain Pass (HTR).
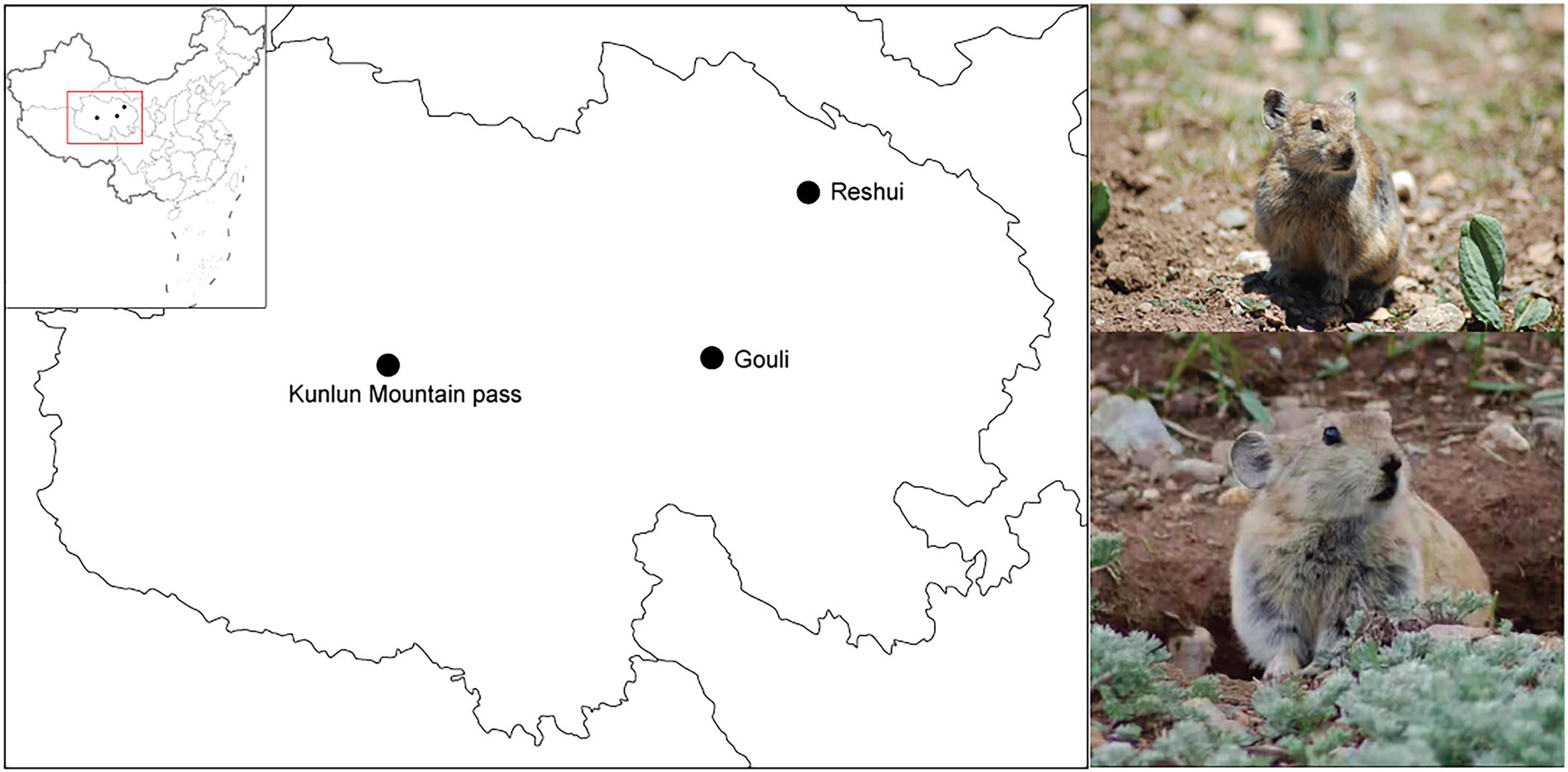
Figure 1. Sampling sites along the Qinghai–Tibet Highway. The three black dots represent the sampling site locations at the Kunlun Mountain Pass, Gouli Clinic, and Reshui Village. The photos to the right display the most obvious features of the Plateau Pikas.
After capture, each pika was euthanized and dissected on a sterile dissecting table. Fresh fecal contents were collected in 2 ml sterile tubes, immediately frozen in liquid nitrogen, and then transported to Northwest Plateau Institute of Biology, Chinese Academy of Sciences, Xining, China. Samples were stored at −80°C until DNA extraction. This project was approved by the Animal Care and Ethics Committee of the Northwest Institute of Plateau Biology, Chinese Academy of Sciences (China; nwipb2019110801).
DNA extraction, library construction, and metagenomic sequencing
Total genomic DNA was extracted from fecal content using a QIAamp DNA Stool Mini Kit (No. 51504; Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Then DNA was purified using QIAamp Mini Spin columns according to the standard protocol. Concentration and quality of DNA extract were determined by gel electrophoresis (1% agarose) and a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
DNA extract was fragmented to an average size of about 400 bp using Covaris M220 (Gene Company Limited, China) for paired-end library construction. Paired-end sequencing was performed on Illumina Hiseq platform (Illumina Inc., San Diego, CA, United States) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions.1 Finally, the sequencing produced an average of 540 million metagenome jointed reads (12 Gb) per sample.
Metagenomic data processing
The data were analyzed on the free online platform of Majorbio Cloud Platform.2 The paired-end Illumina reads were trimmed of adaptors, and low-quality reads (length < 50 bp or with a quality value <20 or having N bases) were removed by fastp[1] (version 0.20.0).3 Metagenomics data were assembled using MEGAHIT[3] (version 1.1.2),4 which makes use of succinct de Bruijn graphs. Contigs with the length being or over 300 bp were selected as the final assembling result, and then the contigs were used for further gene prediction and annotation.
Open reading frames (ORFs) from each assembled contig were predicted using MetaGene[4].5 The predicted ORFs with length being or over 100 bp were retrieved and translated into amino acid sequences using the NCBI translation table.6
A non-redundant gene catalog was constructed using CD-HIT[5] (version 4.6.1)7 with 90% sequence identity and 90% coverage. Reads after quality control were mapped to the non-redundant gene catalog with 95% identity using SOAPaligner[6] (version 2.21),8 and gene abundance in each sample were evaluated.
Representative sequences of non-redundant gene catalog were aligned to NCBI NR database with e-value cutoff of 1e−5 using Diamond[7] (version 0.8.35)9 for taxonomic annotations. The KEGG annotation was conducted using Diamond[7] (version 0.8.35)10 against the Kyoto Encyclopedia of Genes and Genomes database11 with an e-value cutoff of 1e−5.
Antibiotic resistance annotation was conducted using Diamond[7] (version 0.8.35)12 against ARDB database13 and CARD database14 with an e-value cutoff of 1e−5.
To identify MGEs, we used nucleotide BLAST (BLASTn; Altschul et al., 1990) with an E-value cut-off of 10-5-to align our sequences against databases of integrons (INTEGRALL; Moura et al., 2009),15 insertion sequences (ISfinder; Siguier et al., 2006),16 and plasmids (NCBI RefSeq; Kristiansson et al., 2011).17 A read was identified as an integron or insertion sequence if the BLAST hit had > 90% sequence identity and an alignment of at least 50 bp (Kristiansson et al., 2011). A read was identified as a plasmid if the BLAST hit had a > 95% sequence identity and an alignment of at least 90 bp.
Statistical analysis
Statistical analyses were performed using R v4.0.0 (The R Foundation, Vienna, Austria) and SPSS v20.0. (IBM SPSS, Chicago, IL, United States). α-Diversity of ARGs and VFs, including the observed species number, Shannon–Wiener, and Gini–Simpson index, was calculated using the “vegan” and “picante” packages in R. The Kruskal–Wallis test was used to assess the differences in the relative abundance of bacterial phyla and genera, level 2 KEGG pathways,ARGs, and VFs between the different regions, and level 2 KEGG pathways present with log2 relative abundance. The significant level in this study was determined according to the common standard (p > 0.05, no significance; *p < 0.05; **p < 0.01; ***p < 0.001). Correlation tests were performed using the “corrplot” package, and data were visualized using the “ggplot2” package in R and GraphPad Prism v7.0 (GraphPad Software, San Diego, CA, United States). Principal coordinate analysis (PCoA) was performed to compare the clustering of ARGs and VFs between sample sites (with an Adonis test for significance). PCoA based on Bray–Curtis and Jaccard distances was conducted to determine the effects of the different anthropogenic disturbances on the overall patterns of ARGs and VFs. The relative abundance of bacterial genera, level 2 KEGG pathways, ARGs, and VFs were visualized on a heatmap using the R package “pheatmap.” The linear discriminant analysis (LDA) effect size (LefSe) method was used to assess differences in microbe, ARG, and VF composition with an LDA threshold score of 2 (Segata et al., 2011). Network co-occurrence analysis of relative abundance of all samples was used to investigate the relationship between the microbial community and ARGs within each region. We performed a Spearman’s correlation between the top 30 bacterial genera and ARGs in each group “psych,” and presented the results as a network using Gephi v0.9.2 (Jacomy et al., 2014). Significance was set at p < 0.05.
Results
Gut microbial composition according to pollution type
We first characterized the gut microbiota composition at the phylum and genus levels (Figures 2A,B). The dominant phyla were Firmicutes, Bacteroidetes, and Proteobacteria. The dominant genera were unclassified (Firmicutes), Prevotella, and Bacteroides. We compared the gut microbiota compositions of the NCR, MPR, and HTR and determined the LDA score for the species (Figures 2C,D). At the phylum level, Candidatus_Wolfebacteria were enriched in the NCR; Candidatus-Yanofsky bacteria, and Tenericutes were enriched in the MPR; and Proteobacteria, Latescibacteria, Chlamydiae, Deferribacteres, Fibrobacteres, and Cyanobacteria were enriched in the HTR. At the genus level, Ruminococcus, Rikenella, Microcella, Enterococcus, Mycoplasma, Dermacoccus, Nosocomiicoccus, Saccharospirillum, and Pseudodonghicola were enriched in the MPR; Campylobacter, Neisseria, Chlamydia, Bacillus, Bilophila, Pseudoflavonifractor, Dysgonomonas, Listeria, Enterobacter, Clostridiales family XIII Incertae Sedis, Sphingorhabdus, Pectobacterium, Bacteroidetes (phylum), and Stenotrophomonas were enriched in the HTR; while Clostridiaceae (family) and Brachymonas were enriched in the NCR. Although 1,802 genera were shared between the three study sites, two, eight, and seven genera were unique to the NCR, MPR, and HTR, respectively (Figure 2E). A total of 1,814 genera were shared between the NCR and MPR, 1,805 between the NCR and HTR, and 1,809 between the MPR and HTR. For all 34 samples, the dominant genera were Firmicutes, Prevotella, Bacteroides, Clostridium, and Ruminococcus (p < 0.05; Figure 2F). At the phylum level (Figure 2G; Supplementary Table S1), the abundance of Cyanobacteria and Proteobacteria was significantly higher in the HTR than that in the MPR, whereas the abundance of Firmicutes and Tenericutes were significantly lower in the HTR than that in the MPR (p < 0.05). At the genus level (Figure 2H; Supplementary Table S2). We found that Blautia, Butyrivibrio, Clostridium, Eubacterium, Faecalibacterium, Roseburia, Ruminococcus, and Lachnospiraceae (family) were significantly more abundant in the NCR than in the HTR (p < 0.05).
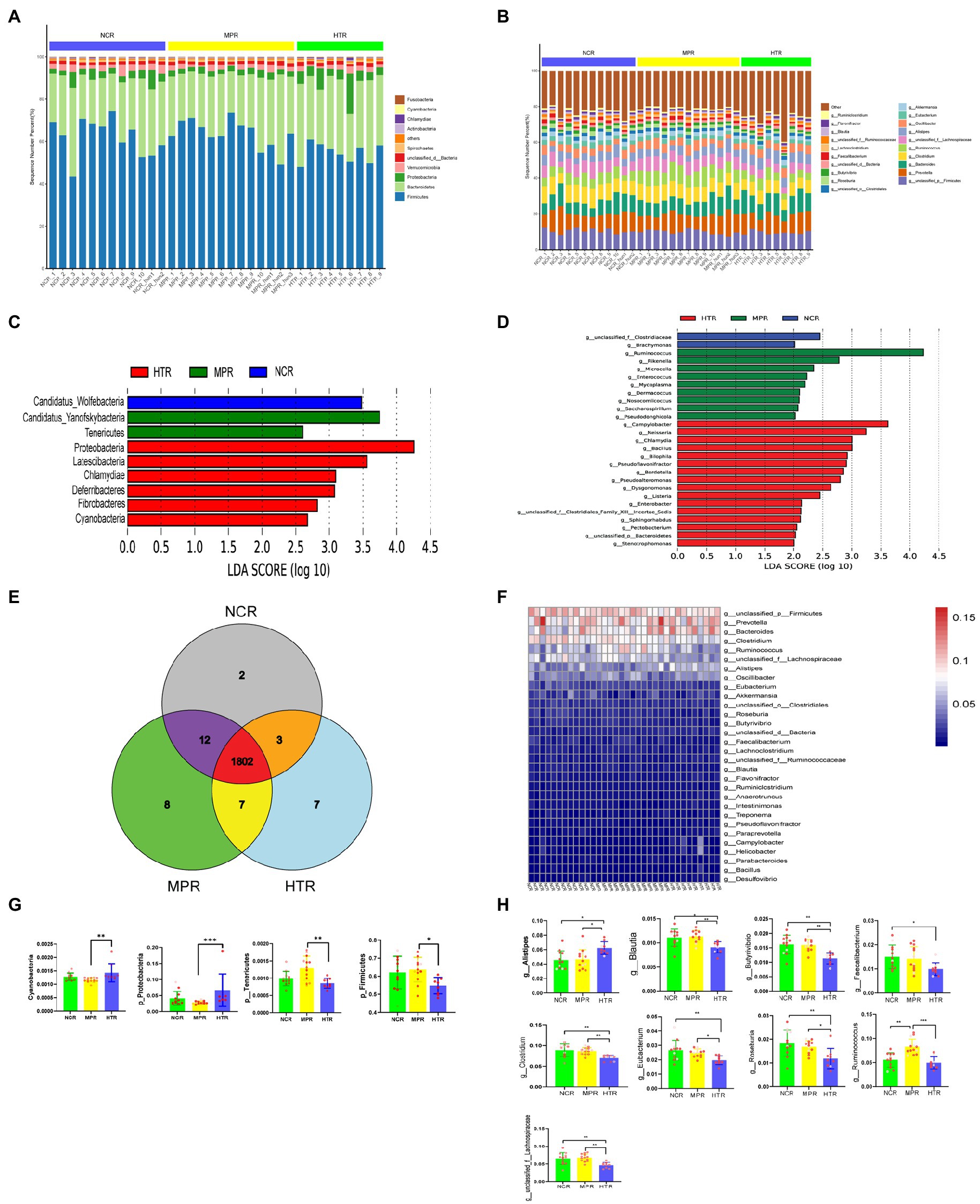
Figure 2. (A) Abundance of the top 10 gut bacteria phyla. (B) Abundance of the top 20 gut bacteria genera. Linear discriminant analysis effect size (LEfSe) showing the significantly different taxa between the study sites at the (C) phylum and (D) genus levels (LDA scores > 3.0, p < 0.05). NCR, non-contaminated region; MPR, medical pollution region; HTR, heavy-tourist traffic region. (E) Genus distribution between the three study sites. (F) Heatmap showing the relative bacterial abundance at the genus level. Comparison of the relative abundance of gut microbiota between the three study sites at the (G) phylum and (H) genus levels across top 20 bacteria. *p < 0.05; **p < 0.01; ***p < 0.001.
Functional comparison of the gut microbiome according to pollution type
To investigate the influence of different pollutants on gut microbiota function, we blasted unigenes to the relative abundance of level 2 KEGG pathways for the NCR, MPR, and HTR (Figure 3). The most dominant pathways were replication and repair, carbohydrate metabolism, and global overview maps (Figure 3A). The genes involved in most metabolic processes, such as Cellular community—eukaryotes, Sensory system, Metabolism of cofactors and vitamins, and Metabolism of other amino acids, were significantly more abundant in the HTR than in the MPR (p < 0.05). Most genes associated with pathways (cancer—overview, Endocrine system, replication and repair and Nervous system) were significantly more prevalent in the MPR than in the NCR (p < 0.05; Figure 3C; Supplementary Table S3). The heatmap based on the relative abundance of the KEGG pathways showed that carbohydrate metabolism, global overview maps, and amino acid metabolism were the most important pathways in the gut microbiome (Figure 3D).
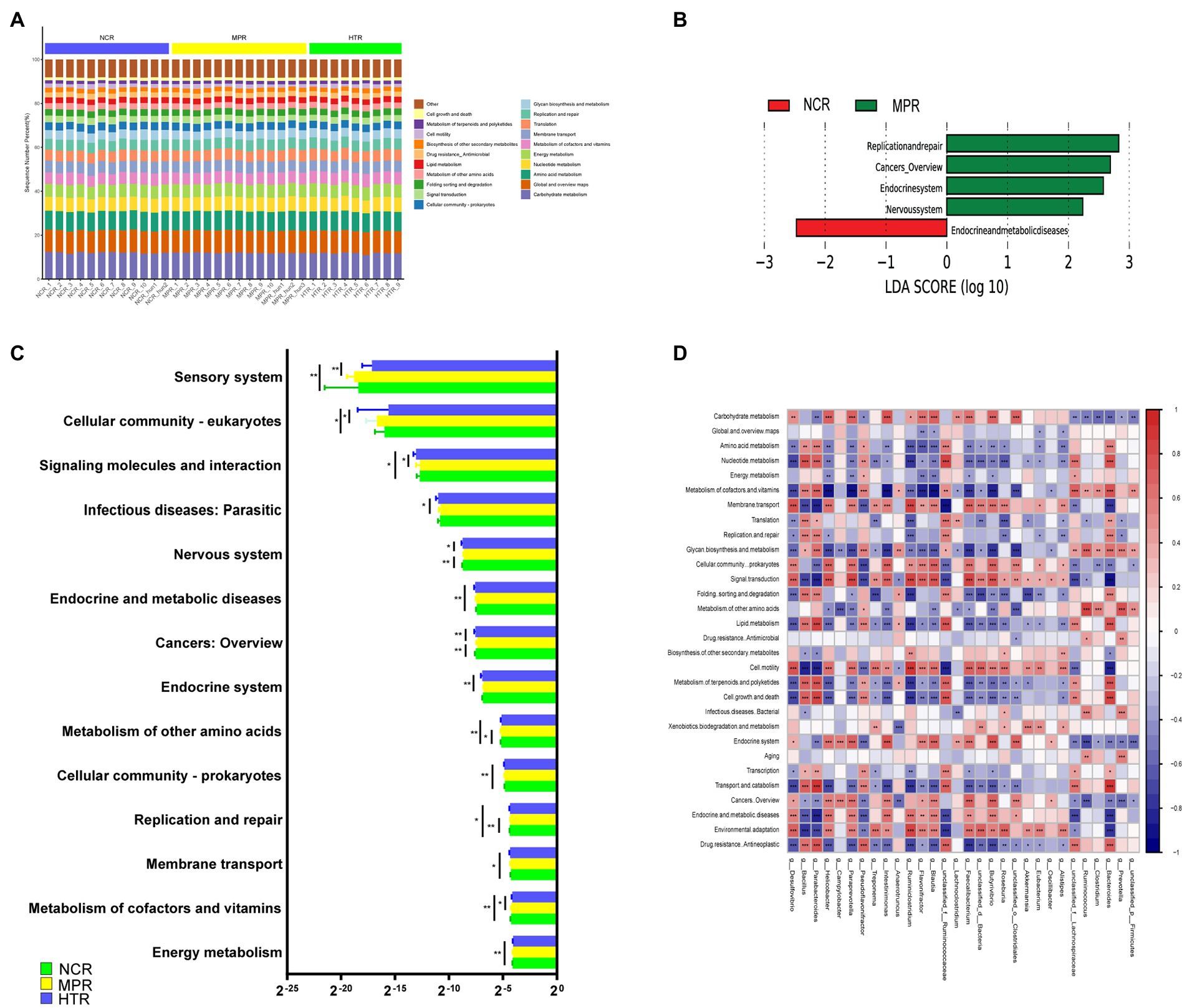
Figure 3. (A) Main metabolic pathways in the gut of Plateau Pikas. (B) The difference in metabolic pathway abundance by LEfSe (p < 0.05 and LDA > 2). (C) Comparison of the main level-2 KEGG pathways. (D) Distribution of the main metabolic pathways.
Antibiotic resistance gene composition according to pollution type
In total, 368 ARGs were detected in the gut microbiome, and each study site corresponded with a different number of ARGs (Figure 4A). The Simpson and Shannon indices were significantly higher in the HTR than those in the NCR and MPR. Differences in ARG composition were determined using PcoA (Figure 4B). The 34 samples were separated into three clusters, indicating that the composition of ARGs differed significantly between the study sites. An ANOSIM revealed similar results, indicating that ARG composition was affected differently by medical and tourist traffic pollution. We identified 20 differentially expressed ARGs between the three regions, of which the abundance for 5 (efrA, lmrC, Saur_gyrA_FLO, vanTG, and desR) was highest in the NCR, 5 (evgS, arlR, mtrA, macB, and bcrA) was highest in the MPR, 7 (mfd, Efac_EFTu_GE2A, PmrE, Saur_rpoC_DAP, Ecol_rpoB_RIF, and aminocoumarin-resistant alaS) was highest in the HTR, and 3 (sav1866, ImrD and msbA was lowest in the HTR (p < 0.05; Figure 4D; Supplementary Table S4). Notably, ARG density was the lowest in the NCR. According to the LefSe, the ARGs in the NCR were enriched in Saur_gyrA_FLO, hmrM, Ecol_folP_SLF, etc. ARGs in the MPR were enriched in bcrA, macB, cea, etc. ARGs in the HTR were enriched in Saur_rpoC_DAP, aminocoumarin-resistance alaS, and Saur_rpoB_RIF (Figure 4E). Nine unique plasmids were recovered from the 34 samples, with higher plasmid α-diversity (Shannon and observed indices) found in the MPR than in the NCR and HTR (Figure 4F).
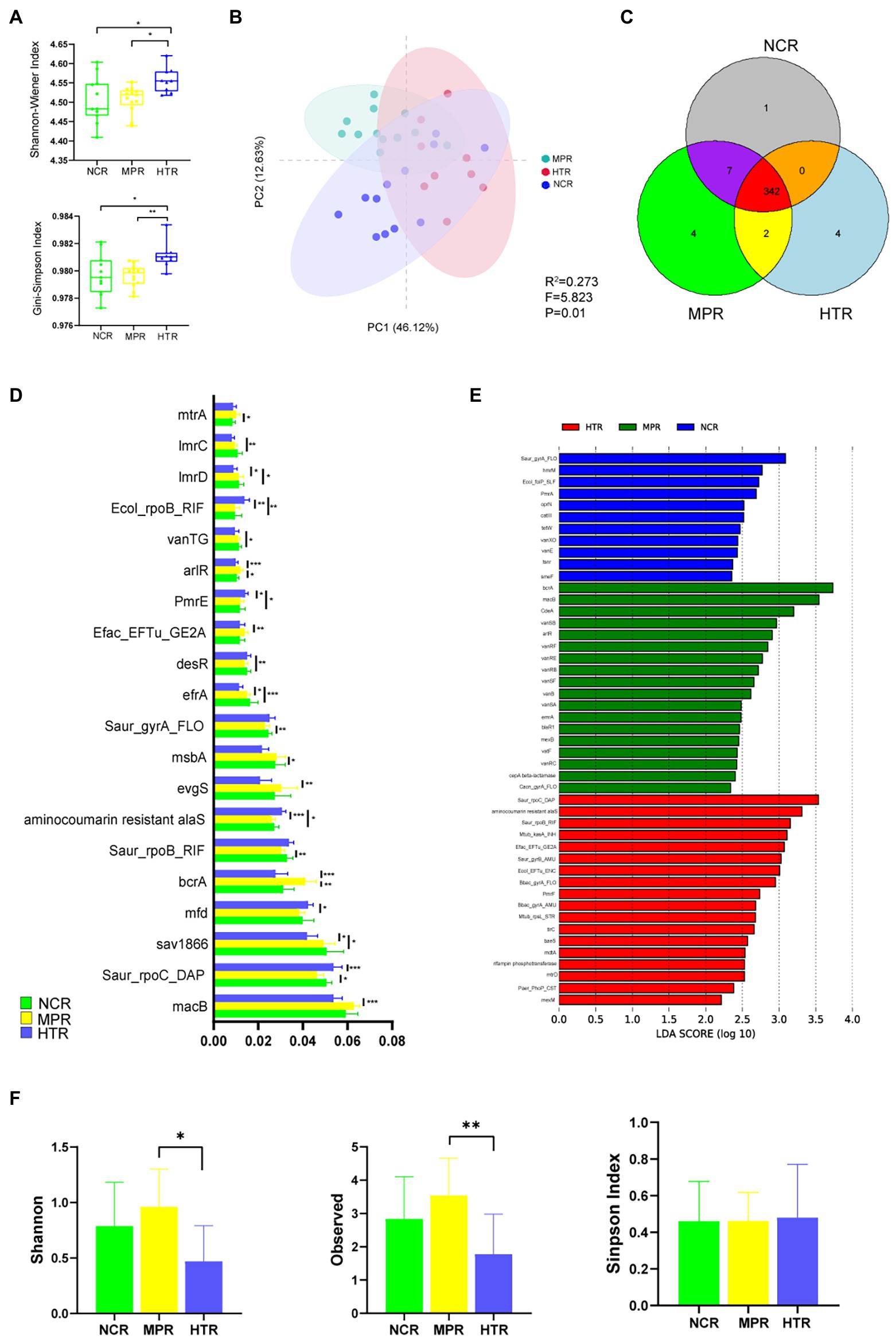
Figure 4. The alpha diversity of ARGs in the three study sites. (A) Shannon–Wiener and Gini–Simpson indices. (B) Bray–Curtis dissimilarity of gut microbial community composition between the three study sites; ellipses with 95% confidence intervals are displayed for the PCoA. (C) Shared ARGs between the NCR, MPR, and HTR. (D) Comparison of the main ARGs. (E) Difference in ARG abundance by LEfSe (p < 0.05 and LDA > 2). (F) Alpha diversity of plasmids in the three study sites according to observed species richness, Shannon–Wiener, and Gini–Simpson indices. *p < 0.05; **p < 0.01; ***p < 0.001.
Potential correlations between ARGs and the gut microbiome
Co-occurrence networks were constructed based on the abundance of ARGs and bacterial genera to determine the potential associations between drug resistance and the gut microbiome (Figure 5). The most complex network structures had 34 nodes and 39 links in the NCR, 48 nodes and 74 links in the MPR, and 40 nodes and 48 links in the HTR (Supplementary Figure 1). The results suggested that the ARGs for multidrug, fluoroquinolones, aminocoumarin, MLS, rifampicin, and tetracycline resistance showed strong relationships with seven, four, one, two, two, and eight bacterial genera, respectively, in the NCR. It is worth noting that in the NCR, there was a strong association between Helicobacter and multidrug resistance, while Helicobacter had little correlation with ARGs in the MPR and HTR. Pathogenic bacteria, such as Campylobacter and Ruminiclostridium, showed strong positive correlations with ARGs in the MPR. Bacteroides, Parabacteroides, Prevotella, and Faecalibacterium showed strong positive correlations with multidrug resistance genes (msbA, sav1866, and efrA) in the HTR. Self-resistance to antibiotics (desR and oleR) was unique to the MPR, and desR showed strong relationships with 20 genera. Multidrug (arlR, mtrA, efrA, efrB, lmrC, and sav1866), fluoroquinolones (PatB and Saur_gyrB_AMU), nitroimidazole (msbA), and rifampicin resistance was mainly observed in the HTR, and the occurrence of msbA was unique to the HTR.
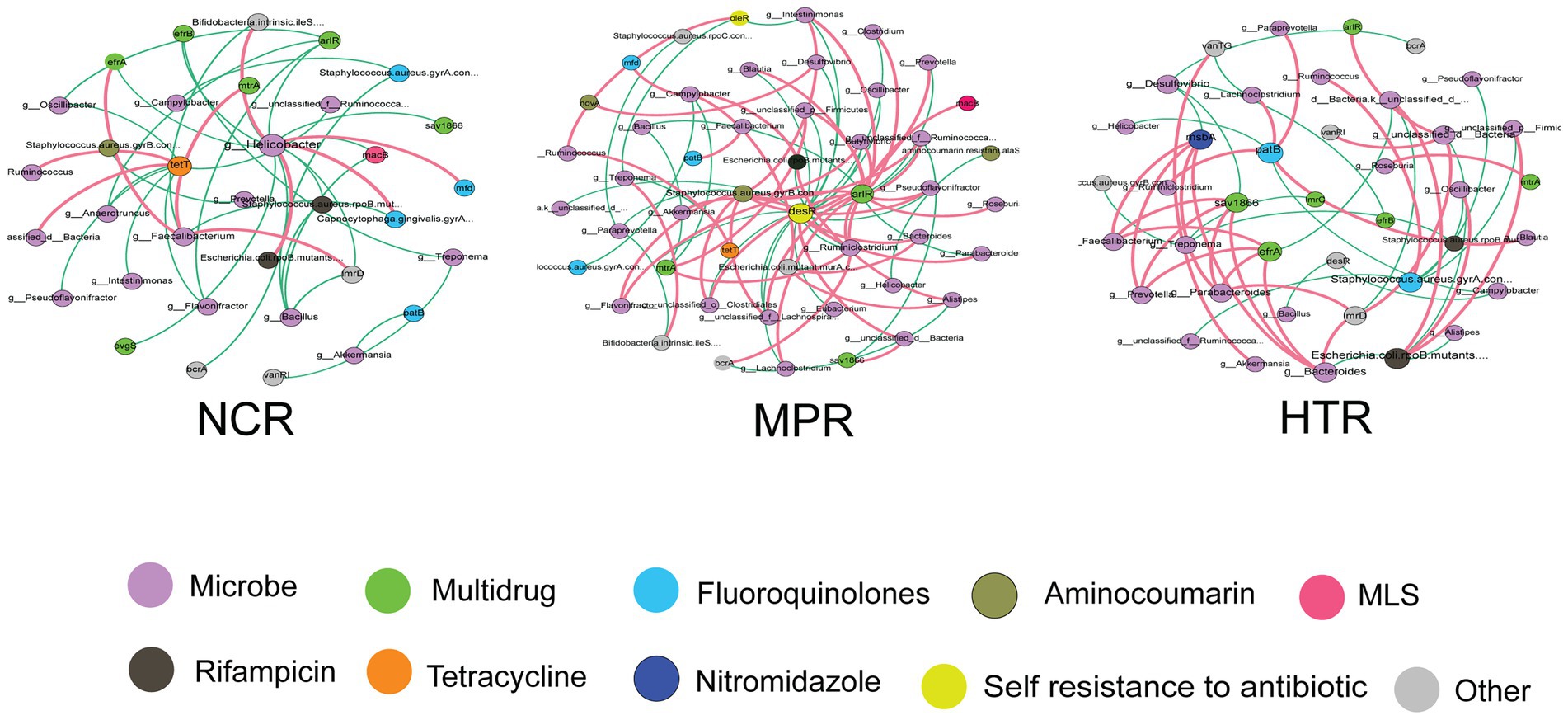
Figure 5. Network analysis depicting the patterns of co-occurrence among ARG subtypes and bacterial genera. A link represents a strong correlation (Spearman’s correlation coefficient ρ ≥ 0.6 and p < 0.01). The size of each node and the edge thickness is proportional to the number of connections and the value of ρ, respectively. Line color reflects direction (green: negative; red: positive).
Virulence factor gene (VFG) composition according to pollution type
The pairwise Adonis test revealed significant differences in VFG composition between the three study sites. There were notable differences between the NCR and MPR (R2 = 0.121, F = 3.166, p = 0.006), NCR and HTR (R2 = 0.115, F = 2.458, p = 0.033), and MPR and HTR (R2 = 0.197, F = 4.897, p = 0.001; Figure 6A). VFG abundance and density were the highest in the MPR, followed by the HTR, and then the NCR (Figure 6B). The abundance of VFGs in MPR samples was significantly higher than that in the HTR, and included alginate, fibronectin-binding protein, GacS/GacA two-component system, hemolysin, PhoP, and Type IV pili. The abundance of the GacS/GacA two-component system, Type IV pili, and VirR/VirS two-component system was significantly higher in the NCR than in the HTR. VFGs for fibronectin-binding protein, PhoP, and polysaccharide capsule were markedly higher in the MPR than in the NCR. Regardless, the abundance of MOMP in the NCR was significantly higher than that in the MPR (p < 0.05; Figure 6D; Supplementary Table S5).
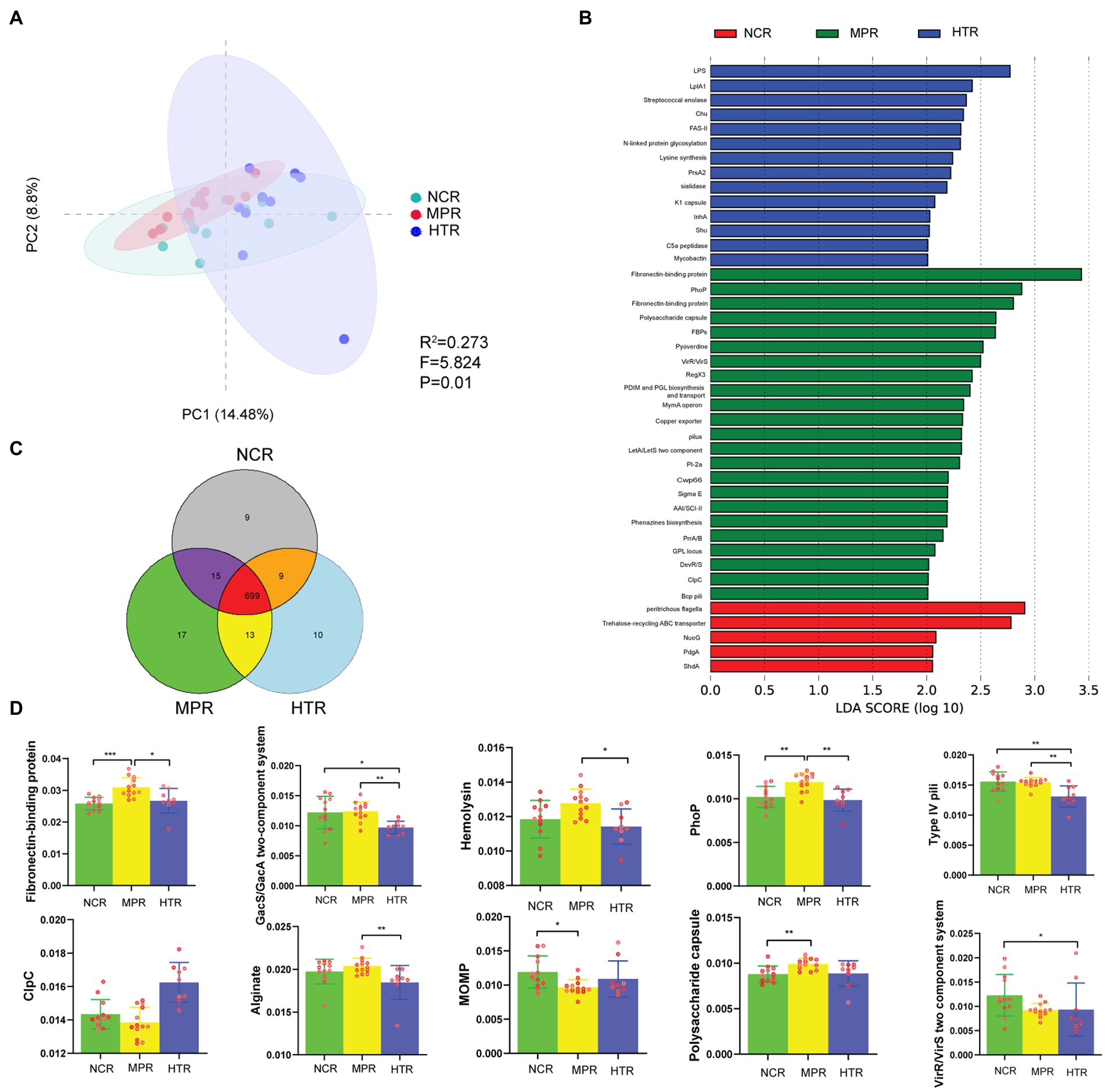
Figure 6. (A) PCoA plot based on the Bray–Curtis distance of VF relative abundance between the NCR, MPR, and HTR (Adonis, R2 = 0.273, F = 5.824, p = 0.01). (B) The difference in VF abundance by LEfSe (p < 0.05 and LDA > 2). (C) Distribution of VFs for the three study sites. (D) Relative abundance of VFs indicating significant discrepancies among the three study sites and top 25 samples, *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
In this study, we observed significant differences in the dominant phyla and genera between the three study sites. The abundance of probiotic genera, such as Butyrivibrio, Eubacterium, Faecalibacterium, Roseburia, Ruminococcus, and Blautia, was lowest in the gut microbiome of Plateau pikas at the HTR (Figure 2). More than 3,000 trucks and vehicles pass through the HTR each day. Transportation and tourism can significantly increase domestic waste and exhaust emissions. Recent studies have demonstrated that waste plastics lead to a lower abundance of probiotics in the gut of Procambarus clarkia (Han et al., 2022). Similarly, exposure to lead (Pb) and cadmium (Cd) significantly reduced the abundance of probiotics in carp gut (Chang et al., 2019; Zhang et al., 2021). These studies emphasize the potential of domestic waste and exhaust emissions caused by transportation and tourism to alter the composition of plateau pika intestinal flora, consistent with the reduction in the abundance of probiotics observed in this study.
The functions of the flora also varied between the study sites (Figure 3). Metabolic pathways, such as genetic information processing and human diseases, were significantly enriched in the MPR. Studies have shown that antibiotics can substantially increase the expression of the genetic information processing genes of gut microbes in humans (Francino, 2016) and piglets (Lourenco et al., 2021). These findings are consistent with those of our study, indicating that antibiotic residues can affect the function of plateau pika intestinal flora through horizontal transmission.
We found that the α-diversity of ARGs were significantly higher in the HTR than in the NCR; the β-diversity in the HTR and MPR was also significantly different from that in the NCR (Figure 4). The most important sources of pollution in the HTR are also important reservoirs of ARGs, such as food waste, plastic bottles, and carton packaging (Yang et al., 2019; Kanger et al., 2020). High-protein foods, such as meat, can increase the diversity of ARGs during the decomposition process, thereby increasing the risk of their transmission and diffusion in the environment (Liang et al., 2020). Marine researchers have found that plastics support a significantly higher diversity and abundance of ARGs than the surrounding environment (Yang et al., 2019), and their ingestion by several aquatic and terrestrial animals leads to a considerable accumulation of ARGs (Jeong and Choi, 2019; Ma et al., 2020; Sarker et al., 2020; Cheng et al., 2021). These results confirm that microplastics can promote the occurrence of resistance genes (Wang et al., 2020a), thereby increasing the probability of transmission to wildlife, such as Plateau pikas. There were significant differences in β-diversity between the MPR, HTR, and NCR, indicating that the type of environmental pollution was related to the occurrence/distribution of resistance genes, which is consistent with the effects of different anthropogenic disturbances on ARGs in the Yarlung Tsangpo River on the Tibetan Plateau (Liu et al., 2021).
We found that a total of 47 ARG subtypes were enriched in the three study sites, and 18 different ARG subtypes were found in the MPR and HTR. Similarly, a study on the impact of municipal drainage and agricultural runoff on river basins also found that the enrichment of ARG subtypes varied with pollution source (Liang et al., 2020). Glycopeptide resistance genes are the most abundant in clinics (~ 44% of all resistance genes), followed by efflux pump resistance genes (33%). Glycopeptides—the most important markers of medical pollution—are the most frequently used antibiotics in hospitals, and the levels of their resistance genes are significantly higher in hospital wastewater than in other sources of pollution (Szekeres et al., 2017; Zhang et al., 2020). Szekeres et al. (2017) surveyed antibiotic-resistant bacteria in 17 Romanian hospitals and observed a high incidence of glycopeptide-resistant E. faecium. These studies support our finding that medical pollution can lead to a high abundance of glycopeptide resistance genes in the gut microbiome of Plateau pikas. The most abundant resistance genes in the HTR were associated with aminocoumarins, which may be related to domestic pollution. Plateau pikas exhibit typical coprophagous behavior, in that they forage for conspecific and heterogeneous feces (Speakman et al., 2021). Recent studies have shown that the primary pollution source in the Yamuna River is municipal waste, and aminocoumarin resistance genes are the most important resistance genes in the microbial community of the water body (Mittal et al., 2019). Shotgun sequencing and metagenomic assembly analyses have shown that human fecal contamination leads to substantial enrichment of aminocoumarin resistance genes in rivers around cities (Chen et al., 2019). This indicates that domestic pollution caused by traffic and tourism could promote the accumulation of aminocoumarin resistance genes in the gut microbiomes of Plateau pikas.
Analysis of the network structure between ARGs and bacterial genera showed that pathogenic bacteria were positively correlated with ARGs in the MPR, consistent with previous investigations showing that the zoonotic pathogen Campylobacter is widespread in the gut of wildlife (Altekruse and Tollefson, 2003; Humphrey et al., 2007). Fluoroquinolones are the most frequently used antimicrobials for the treatment of Campylobacter infections. However, Campylobacter is becoming increasingly resistant to clinically important antibiotics, such as rifampicin (Guo et al., 2010), thus requiring increased dosage (Luangtongkum et al., 2009). Kabir et al. (2015) found that 25% of Campylobacter strains isolated from Indian poultry were resistant to fosfomycin. These results confirm the importance of medical pollution to the enrichment of Campylobacter resistance genes in the gut of Plateau pikas. Interestingly for the HTR, Bacteroides, Parabacteroides, Prevotella, and Faecalibacterium were positively correlated with multidrug resistance genes (msbA, sav1866, and efrA), while these bacteria had little correlation with ARGs in the NCR. This is due to tourism-related pollution, which leads to increased resistance of bacteria and promotes the spread of ARGs (Abe et al., 2020; Ratajczak et al., 2021). Bacteroides strains have been reported to cause multidrug-resistant infections (Katsandri et al., 2006; Kalapila et al., 2013); Parabacteroides strains have been isolated in many laboratories, and researchers have confirmed their multidrug resistance capabilities (Molina et al., 2014; Rong et al., 2021); and approximately 10% of Prevotella spp. exhibit multidrug resistance (Toprak et al., 2018). Foditsch et al. (2014) isolated a multidrug-resistant strain of Faecalibacterium from healthy calves and piglets. There was a strong association between Helicobacter and multidrug resistance (Bifidobacteria intrinsic ileS conferring resistance to mupirocin, Cgin_gyrA_FLO, Ecol_rpoB_RIF and mfd) in NCR, while Helicobacter had little correlation with ARGs in the MPR and HTR. Helicobacter colonized the gastrointestinal tracts of humans and animals(Duarte et al., 2021), and it have been isolated from the feces of dolphins as well as detected in the feces of seals(Harper et al., 2002; Goldman et al., 2009). NCR is a typical grassland grazed by livestock, and there is amount of cattle and sheep feces. As plateau pikas engage in typical coprophagy which shows as eating conspecific and other species’ feces, so it may promote the spread of ARGs. There was positive correlation between Campylobacter and Ecol_rpoB_RIF, Ecol_murA_FOF, and mfd in MPR, while Campylobacter had little correlation with ARGs in the NCR and HTR. The reason for this phenomenon is due to the rise of drug-resistant bacteria caused by medical contamination, which promotes the spread of ARGs (Wang et al., 2020b). The environmental bacterial resistome as an important reservoir for the dissemination of antibiotic resistance. Previous studies have shown that an increase of the resistance gene pool in environmental bacteria due to pollution is likely to increase the probability of the transmission of these genes into pathogenic bacteria (Buelow et al., 2021) Medical and domestic pollution have different impacts on the environment, resulting in differences in pathogenic bacteria and opportunistic pathogens in the gut microbiome of Plateau pikas, and leading to the occurrence of different resistance genes.
The β-diversity of VFs varied significantly between the MPR and HTR, thus suggesting a strong association between pollution source and VF distribution patterns. We identified 23 and 14 VFGs that were significantly enriched in the MPR and HTR, respectively (Figure 6). This showed that medical pollution was more important than domestic pollution in determining the accumulation of VFGs in the gut microbiomes of Plateau pikas. A study comparing the VFs of captive and wild pangolins found that captivity resulted in a greater accumulation of VFGs in the host’s gut microbiome, owing to chronic antibiotic use (Liu et al., 2021). Liang et al. (2020) found that the abundance and density of VFs were the highest in agricultural areas, followed by wastewater from cities, and lowest in mountainous areas, which was mainly related to the use of antibiotics (Liang et al., 2020). VirR/VirS two-component system have been thought to be very important for its pathogenicity by controlling the production of various known virulence factors in Clostridium perfringens (Ohtani et al., 2010). In our result, Clostridiaceae (family) was enriched in the NCR, which explained the results that abundance of VirR/VirS two-component system in NCR were higher than that of HTR. The abundance of VFGs including alginate, fibronectin-binding protein, GacS/GacA two component system, hemolysin, PhoP, and Type IV pili in MPR was significantly higher than that in the HTR. We speculated that medical pollution led to an increase in the abundance of drug-resistant bacteria, so this result achieved. Researcher investigated the microbial gene catalogue in samples of phosphinothricin (PPT)-utilized soils from South China, many common virulence factors with high abundance were found in two samples, including Alginate (Lou et al., 2019); VFs were detected in ARG pollution covered two constructed wetlands and their receiving river, and found that Type IV pili was main VFs (Ohtani et al., 2010); we found Enterococcus was enriched in samples from MPR, this result correlated well with previous study that hemolysin plays an important role in Enterococcal virulence (Franz et al., 2001). These researches support our supposition. In this study, the quantities of antibiotics used in the MPR were higher than that in the HTR and NCR, which was associated with increased levels of VFs in the gut microbiome of Plateau pikas.
In summary, we revealed the structure, function, ARGs, and VFs of the gut microbial community in Plateau pikas under different anthropogenic disturbances on the Qinghai–Tibet Plateau. The results showed that the abundance of probiotics significantly decreased in the HTR. The enrichment of metabolic pathways differed between medical and domestic sources of contamination. The metabolic pathways of replication and repair, endocrine system, and nervous system were significantly enriched in the MPR, while endocrine and metabolic diseases were significantly enriched in the NCR. The α-diversity of ARGs was highest in the HTR, and there were significant differences in β-diversity between the study sites. Resistance to glycopeptide antibiotics increased significantly in the MPR, while the ARGs for aminocoumarins increased significantly in the HTR. Furthermore, the α-diversity of MGEs was the highest in the MPR. We observed strong positive correlations between ARGs and pathogenic bacteria, and the network structure was the most complex in the MPR. The β-diversity of VFs varied significantly between the three sites. The MPR, which was affected by a greater accumulation of antibiotics, contributed considerably to the enrichment of VFs. Our study highlights the dissemination of ARGs and VFs in wildlife through horizontal transmission and provides an important theoretical basis for the conservation management of wild animals in national parks based on relevant sources of pollution.
The disturbance of human activities on the Qinghai-Tibet Plateau is diverse, including returning farmland to forest, artificial grassland, overgrazing, etc. We only selected typical interference of medical and traffic-based pollution, and we need to pay more attention to the impact of more human activities on wild animals. Plateau pika plays a pivotal role in the food web structure of the alpine grassland ecosystem. In the next step, we will target the predators of the plateau pika, such as Tibetan fox and giant buzzard, to explore the transmission and diffusion of ARGs and VFs along the food chain.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA903767.
Ethics statement
The animal study was reviewed and approved by Northwest Institute of Plateau Biology, Chinese Academy of Sciences.
Author contributions
YZ designed the study. YZ-Q, LZ, XT, and SR collected samples. LZ was responsible for metagenomic assays and quality control. YZ, LZ, YZ-Q, XT, and SR analyzed the data. YZ and YZ-Q revised the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Second Tibetan Plateau Scientific Expedition and Research Program (No. 2019QZKK0501). the National Natural Science Foundation of China (No. 31670394), the project of western light for interdisciplinary teams, and the Science and Technology Department of Qinghai Province Major Project “Sanjiangyuan National Park Animal Genome Program.”
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1027941/full#supplementary-material
Footnotes
3. ^https://github.com/OpenGene/fastp
4. ^https://github.com/voutcn/megahit
5. ^http://metagene.cb.k.u-tokyo.ac.jp/
6. ^http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#SG1
7. ^http://www.bioinformatics.org/cd-hit/
8. ^http://soap.genomics.org.cn/
9. ^http://www.diamondsearch.org/index.phphttp://www.diamondsearch.org/index.php
10. ^http://www.diamondsearch.org/index.php
11. ^http://www.genome.jp/keeg/
12. ^http://www.diamondsearch.org/index.php
13. ^http://ardb.cbcb.umd.edu/
14. ^https://card.mcmaster.ca/home
15. ^http://integrall.bio.ua.pt/
References
Abe, K., Nomura, N., and Suzuki, S. (2020). Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 96:fiaa031. doi: 10.1093/femsec/fiaa031
Altekruse, S. F., and Tollefson, L. K. (2003). Human campylobacteriosis: a challenge for the veterinary profession. J. Am. Vet. Med. Assoc. 223, 445–452. doi: 10.2460/javma.2003.223.445
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arthur, A. D., Pech, R. P., Davey, C., Yanming, Z., and Hui, L. (2008). Livestock grazing, plateau pikas and the conservation of avian biodiversity on the Tibetan plateau. Biol. Conserv. 141, 1972–1981. doi: 10.1016/j.biocon.2008.05.010
Buelow, E., Ploy, M.-C., and Dagot, C. (2021). Role of pollution on the selection of antibiotic resistance and bacterial pathogens in the environment. Curr. Opin. Microbiol. 64, 117–124. doi: 10.1016/j.mib.2021.10.005
Chang, X., Li, H., Feng, J., Chen, Y., Nie, G., and Zhang, J. (2019). Effects of cadmium exposure on the composition and diversity of the intestinal microbial community of common carp (Cyprinus carpio L.). Ecotoxicol. Environ. Saf. 171, 92–98. doi: 10.1016/j.ecoenv.2018.12.066
Chen, H., Bai, X., Li, Y., Jing, L., Chen, R., and Teng, Y. (2019). Source identification of antibiotic resistance genes in a peri-urban river using novel crAssphage marker genes and metagenomic signatures. Water Res. 167:115098. doi: 10.1016/j.watres.2019.115098
Cheng, Y., Song, W., Tian, H., Zhang, K., Li, B., Du, Z., et al. (2021). The effects of high-density polyethylene and polypropylene microplastics on the soil and earthworm Metaphire guillelmi gut microbiota. Chemosphere 267:129219. doi: 10.1016/j.chemosphere.2020.129219
Doyle, S., Meade, E., Fowley, C., and Garvey, M. (2020). A comprehensive review of current environmental pollutants of pharmaceutical, agricultural and industrial origin. Eur. J. Exp. Biol 10, 1–13. doi: 10.36648/2248-9215.10.2.102
Duarte, N., Salazar, V., Casanova, G., Suárez, P., and Fernández-Delgado, M. (2021). Occurrence of helicobacter spp. and fecal bacterial contamination in high altitude aquatic environments from the Andes. Bull. Environ. Contam. Toxicol. 107, 433–440. doi: 10.1007/s00128-021-03347-9
Fan, C., Zhang, L., Fu, H., Liu, C., Li, W., Cheng, Q., et al. (2020). Enterotypes of the gut microbial community and their response to plant secondary compounds in plateau pikas. Microorganisms 8:1311. doi: 10.3390/microorganisms8091311
Fan, C., Zhang, L., Jia, S., Tang, X., Fu, H., Li, W., et al. (2022). Seasonal variations in the composition and functional profiles of gut microbiota reflect dietary changes in plateau pikas. Integr. Zool. 17, 379–395. doi: 10.3390/microorganisms8091311
Foditsch, C., Santos, T. M., Teixeira, A. G., Pereira, R. V., Dias, J. M., Gaeta, N., et al. (2014). Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets? PLoS one 9:e116465. doi: 10.1371/journal.pone.0116465
Francino, M. (2016). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6:1543. doi: 10.3389/fmicb.2015.01543
Franz, C. M., Muscholl-Silberhorn, A. B., Yousif, N. M., Vancanneyt, M., Swings, J., and Holzapfel, W. H. (2001). Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67, 4385–4389. doi: 10.1128/AEM.67.9.4385-4389.2001
Goldman, C., Matteo, M., Loureiro, J., Degrossi, J., Teves, S., Heredia, S. R., et al. (2009). Detection of helicobacter and campylobacter spp. from the aquatic environment of marine mammals. Vet. Microbiol. 133, 287–291. doi: 10.1016/j.vetmic.2008.06.023
Guo, B., Lin, J., Reynolds, D. L., and Zhang, Q. (2010). Contribution of the multidrug efflux transporter CmeABC to antibiotic resistance in different campylobacter species. Foodborne Pathog. Dis. 7, 77–83. doi: 10.1089/fpd.2009.0354
Han, M., Gao, T., Liu, G., Zhu, C., Zhang, T., Sun, M., et al. (2022). The effect of a polystyrene nanoplastic on the intestinal microbes and oxidative stress defense of the freshwater crayfish, Procambarus clarkii. Sci. Total Environ. 833:155722. doi: 10.1016/j.scitotenv.2022.155722
Harper, C., Feng, Y., Xu, S., Taylor, N., Kinsel, M., Dewhirst, F., et al. (2002). Helicobacter cetorum sp. nov., a urease-positive helicobacter species isolated from dolphins and whales. J. Clin. Microbiol. 40, 4536–4543. doi: 10.1128/JCM.40.12.4536-4543.2002
Humphrey, T., O'Brien, S., and Madsen, M. (2007). Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117, 237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006
Jacomy, M., Venturini, T., Heymann, S., and Bastian, M. (2014). ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One 9:e98679. doi: 10.1016/j.ijfoodmicro.2007.01.006
Jeong, J., and Choi, J. (2019). Adverse outcome pathways potentially related to hazard identification of microplastics based on toxicity mechanisms. Chemosphere 231, 249–255. doi: 10.1016/j.chemosphere.2019.05.003
Kabir, S. L., Asakura, M., Shiramaru, S., Pal, A., Hinenoya, A., and Yamasaki, S. (2015). Molecular identification and antimicrobial resistance profiles of Campylobacter strains of poultry origin in India with special emphasis on fluoroquinolone resistance? Asian Journal of Medical and Biological Research 1, 1–8. doi: 10.3329/ajmbr.v1i1.25491
Kalapila, A., Pergam, S., Pottinger, P., Butler-Wu, S., Whimbey, E., and Duchin, J. (2013). Multidrug-resistant Bacteroides fragilis—Seattle, Washington, 2013. Morb. Mortal. Wkly Rep. 62:694.
Kanger, K., Guilford, N. G., Lee, H., Nesbø, C. L., Truu, J., and Edwards, E. A. (2020). Antibiotic resistome and microbial community structure during anaerobic co-digestion of food waste, paper and cardboard. FEMS Microbiol. Ecol. 96:fiaa006. doi: 10.1093/femsec/fiaa006
Katsandri, A., Papaparaskevas, J., Pantazatou, A., Petrikkos, G. L., Thomopoulos, G., Houhoula, D. P., et al. (2006). Two cases of infections due to multidrug-resistant Bacteroides fragilis group strains. J. Clin. Microbiol. 44, 3465–3467. doi: 10.1128/JCM.00316-06
Kohl, K. D., Varner, J., Wilkening, J. L., and Dearing, M. D. (2018). Gut microbial communities of American pikas (O chotona princeps): evidence for phylosymbiosis and adaptations to novel diets. J. Anim. Ecol. 87, 323–330. doi: 10.1111/1365-2656.12692
Kohl, K. D., Weiss, R. B., Cox, J., Dale, C., and Denise Dearing, M. (2014). Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 17, 1238–1246. doi: 10.1111/ele.12329
Kristiansson, E., Fick, J., Janzon, A., Grabic, R., Rutgersson, C., Weijdegård, B., et al. (2011). Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One 6:e17038. doi: 10.1371/journal.pone.0017038
Li, H., Qu, J., Li, T., Wirth, S., Zhang, Y., Zhao, X., et al. (2018). Diet simplification selects for high gut microbial diversity and strong fermenting ability in high-altitude pikas. Appl. Microbiol. Biotechnol. 102, 6739–6751. doi: 10.1007/s00253-018-9097-z
Li, H., Zhou, R., Zhu, J., Huang, X., and Qu, J. (2019). Environmental filtering increases with elevation for the assembly of gut microbiota in wild pikas. Microb. Biotechnol. 12, 976–992. doi: 10.1111/1751-7915.13450
Liang, J., Mao, G., Yin, X., Ma, L., Liu, L., Bai, Y., et al. (2020). Identification and quantification of bacterial genomes carrying antibiotic resistance genes and virulence factor genes for aquatic microbiological risk assessment. Water Res. 168:115160. doi: 10.1016/j.watres.2019.115160
Liang, Z., Zhang, Y., He, T., Yu, Y., Liao, W., Li, G., et al. (2020). The formation mechanism of antibiotic-resistance genes associated with bacterial communities during biological decomposition of household garbage. J. Hazard. Mater. 398:122973. doi: 10.1016/j.jhazmat.2020.122973
Liu, C., Hu, J., Wu, Y., Irwin, D. M., Chen, W., Zhang, Z., et al. (2021). Comparative study of gut microbiota from captive and confiscated-rescued wild pangolins. J. Genet. Genomics 48, 825–835. doi: 10.1016/j.jgg.2021.07.009
Liu, S., Su, H., Pan, Y.-F., and Xu, X.-R. (2020). Spatial and seasonal variations of antibiotics and antibiotic resistance genes and ecological risks in the coral reef regions adjacent to two typical islands in South China Sea. Mar. Pollut. Bull. 158:111424. doi: 10.1016/j.marpolbul.2020.111424
Liu, S., Wang, P., Wang, C., Wang, X., and Chen, J. (2021). Anthropogenic disturbances on antibiotic resistome along the Yarlung Tsangpo River on the Tibetan plateau: ecological dissemination mechanisms of antibiotic resistance genes to bacterial pathogens. Water Res. 202:117447. doi: 10.1016/j.watres.2021.117447
Lourenco, J. M., Hampton, R. S., Johnson, H. M., Callaway, T. R., Rothrock, M. J. Jr., and Azain, M. J. (2021). The effects of feeding antibiotic on the intestinal microbiota of weanling pigs. Front. Vet. Sci. 8:2021. doi: 10.3389/fvets.2021.601394.eCollection
Lou, J., Liu, M., Gu, J., Liu, Q., Zhao, L., Ma, Y., et al. (2019). Metagenomic sequencing reveals microbial gene catalogue of phosphinothricin-utilized soils in South China. Gene 711:143942. doi: 10.1016/j.gene.2019.143942
Luangtongkum, T., Jeon, B., Han, J., Plummer, P., Logue, C. M., and Zhang, Q. (2009). Antibiotic resistance in campylobacter: Emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189
Ma, H., Pu, S., Liu, S., Bai, Y., Mandal, S., and Xing, B. (2020). Microplastics in aquatic environments: toxicity to trigger ecological consequences. Environ. Pollut. 261:114089. doi: 10.1016/j.envpol.2020.114089
Mittal, P., Prasoodanan, P. K. V., Dhakan, D. B., Kumar, S., and Sharma, V. K. (2019). Metagenome of a polluted river reveals a reservoir of metabolic and antibiotic resistance genes. Environ. Microbiome 14, 5–12. doi: 10.1186/s40793-019-0345-3
Molina, J., Barrantes, G., Quesada-Gómez, C., Rodríguez, C., and Rodríguez-Cavallini, E. (2014). Phenotypic and genotypic characterization of multidrug-resistant Bacteroides, Parabacteroides spp., and Pseudoflavonifractor from a Costa Rican hospital. Microb. Drug Resist. 20, 478–484. doi: 10.1089/mdr.2013.0180
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Ohtani, K., Hirakawa, H., Tashiro, K., Yoshizawa, S., Kuhara, S., and Shimizu, T. (2010). Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16, 258–264. doi: 10.1016/j.anaerobe.2009.10.003
Pan, Y., Zeng, J., Li, L., Yang, J., Tang, Z., Xiong, W., et al. (2020). Coexistence of antibiotic resistance genes and virulence factors deciphered by large-scale complete genome analysis. Msystems 5, e00821–e00819. doi: 10.1128/mSystems.00821-19
Pruden, A., Pei, R., Storteboom, H., and Carlson, K. H. (2006). Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40, 7445–7450. doi: 10.1021/es060413l
Qu, J., Liu, M., Yang, M., Zhang, Z., and Zhang, Y. (2015). Effects of fertility control in plateau pikas (Ochotona curzoniae) on diversity of native birds on Tibetan plateau. Acta Theriologica Sinica 35:164. doi: 10.16829/j.slxb.2015.02.006
Ratajczak, M., Kaminska, D., Dlugaszewska, J., and Gajecka, M. (2021). Antibiotic resistance, biofilm formation, and presence of genes encoding virulence factors in strains isolated from the pharmaceutical production environment. Pathogens 10:130. doi: 10.3390/pathogens10020130
Ren, S., Fan, C., Zhang, L., Tang, X., Fu, H., Liu, C., et al. (2021). The plant secondary compound swainsonine reshapes gut microbiota in plateau pikas (Ochotona curzoniae). Appl. Microbiol. Biotechnol. 105, 6419–6433. doi: 10.1007/s00253-021-11478-6
Rong, S. M. M., Rodloff, A. C., and Stingu, C.-S. (2021). Diversity of antimicrobial resistance genes in Bacteroides and Parabacteroides strains isolated in Germany. J. Glob. Antimicrob. Resist. 24, 328–334. doi: 10.1016/j.jgar.2021.01.007
Sanderson, H., Brain, R. A., Johnson, D. J., Wilson, C. J., and Solomon, K. R. (2004). Toxicity classification and evaluation of four pharmaceuticals classes: antibiotics, antineoplastics, cardiovascular, and sex hormones. Toxicology 203, 27–40. doi: 10.1016/j.tox.2004.05.015
Sarker, A., Deepo, D. M., Nandi, R., Rana, J., Islam, S., Rahman, S., et al. (2020). A review of microplastics pollution in the soil and terrestrial ecosystems: a global and Bangladesh perspective. Sci. Total Environ. 733:139296. doi: 10.1016/j.scitotenv.2020.139296
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60–R18. doi: 10.1186/gb-2011-12-6-r60
Siguier, P., Pérochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Skurnik, D., Ruimy, R., Andremont, A., Amorin, C., Rouquet, P., Picard, B., et al. (2006). Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57, 1215–1219. doi: 10.1093/jac/dkl122
Smith, A. T., and Foggin, J. M. (1999). The plateau pika (Ochotona curzoniae) is a keystone species for biodiversity on the Tibetan plateau. Anim. Conserv. 2, 235–240. doi: 10.1111/j.1469-1795.1999.tb00069.x
Smith, A. T., Wilson, M. C., and Hogan, B. W. (2019). Functional-trait ecology of the plateau pika Ochotona curzoniae in the Qinghai–Tibetan plateau ecosystem. Integr. Zool. 14, 87–103. doi: 10.1111/1749-4877.12300
Speakman, J. R., Chi, Q., Ołdakowski, Ł., Fu, H., Fletcher, Q. E., Hambly, C., et al. (2021). Surviving winter on the Qinghai-Tibetan plateau: Pikas suppress energy demands and exploit yak feces to survive winter. Proc. Natl. Acad. Sci. 118:e2100707118. doi: 10.1073/pnas.2100707118
Spindler, A., Otton, L. M., Fuentefria, D. B., and Corção, G. (2012). Beta-lactams resistance and presence of class 1 integron in pseudomonas spp. isolated from untreated hospital effluents in Brazil. Antonie Van Leeuwenhoek 102, 73–81. doi: 10.1007/s10482-012-9714-2
Szekeres, E., Baricz, A., Chiriac, C. M., Farkas, A., Opris, O., Soran, M.-L., et al. (2017). Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. 225, 304–315. doi: 10.1016/j.envpol.2017.01.054
Toprak, N. U., Veloo, A. C., Urban, E., Wybo, I., Justesen, U. S., Jean-Pierre, H., et al. (2018). A multicenter survey of antimicrobial susceptibility of Prevotella species as determined by Etest methodology. Anaerobe 52, 9–15. doi: 10.1016/j.anaerobe.2018.05.005
Wang, Y., Lu, J., Engelstädter, J., Zhang, S., Ding, P., Mao, L., et al. (2020b). Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation. ISME J. 14, 2179–2196. doi: 10.1038/s41396-020-0679-2
Wang, S., Xue, N., Li, W., Zhang, D., Pan, X., and Luo, Y. (2020a). Selectively enrichment of antibiotics and ARGs by microplastics in river, estuary and marine waters. Sci. Total Environ. 708:134594. doi: 10.1016/j.scitotenv.2019.134594
Wheeler, E., Hong, P.-Y., Bedon, L. C., and Mackie, R. I. (2012). Carriage of antibiotic-resistant enteric bacteria varies among sites in Galapagos reptiles. J. Wildl. Dis. 48, 56–67. doi: 10.7589/0090-3558-48.1.56
Yang, Y., Liu, G., Song, W., Ye, C., Lin, H., Li, Z., et al. (2019). Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ. Int. 123, 79–86. doi: 10.1016/j.envint.2018.11.061
Yu, Q., Li, G., and Li, H. (2022). Two community types occur in gut microbiota of large-sample wild plateau pikas (Ochotona curzoniae). Integr. Zool. 17, 366–378. doi: 10.1111/1749-4877.12575
Zhang, Y., Fan, N., Wang, Q., and Jing, Z. (1998). The changing ecological process of rodent communities during rodent pest managements in alpine meadow. Acta Theriologica Sinica 18:137. doi: 10.16829/j.slxb.1998.02.010
Zhang, S., Huang, J., Zhao, Z., Cao, Y., and Li, B. (2020). Hospital wastewater as a reservoir for antibiotic resistance genes: a meta-analysis. Front. Public Health 8:574968. doi: 10.3389/fpubh.2020.574968
Zhang, Y., Zhang, P., Shang, X., Lu, Y., and Li, Y. (2021). Exposure of lead on intestinal structural integrity and the diversity of gut microbiota of common carp. Comp. Biochem. Physiol. C 239:108877. doi: 10.1016/j.cbpc.2020.108877
Keywords: plateau pika, environmental pollution, antibiotic resistance gene, virulence factor, horizontal transmission
Citation: Zhao Y, Zhang L, Tang X, Ren S and Zhang Y (2022) Anthropogenic disturbance promotes the diversification of antibiotic resistance genes and virulence factors in the gut of plateau pikas (Ochotona curzoniae). Front. Ecol. Evol. 10:1027941. doi: 10.3389/fevo.2022.1027941
Edited by:
Lixing Huang, Jimei University, ChinaReviewed by:
Tongtong Li, Zhejiang University of Technology, ChinaYoulu Su, Zhongkai University of Agriculture and Engineering, China
Copyright © 2022 Zhao, Zhang, Tang, Ren and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanming Zhang, emhhbmd5bUBud2lwYi5jYXMuY24=
†These authors have contributed equally to this work
 Yaqi Zhao
Yaqi Zhao Liangzhi Zhang
Liangzhi Zhang Xianjiang Tang
Xianjiang Tang Shien Ren
Shien Ren Yanming Zhang
Yanming Zhang